Figure 1
Florian Wuennemann
Last updated: 2024-03-21
Checks: 7 0
Knit directory: mi_spatialomics/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20230612) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version e6213a5. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rproj.user/
Ignored: analysis/.DS_Store
Ignored: analysis/deprecated/.DS_Store
Ignored: analysis/molecular_cartography_python/.DS_Store
Ignored: analysis/seqIF_python/.DS_Store
Ignored: analysis/seqIF_python/pixie/.DS_Store
Ignored: analysis/seqIF_python/pixie/cell_clustering/
Ignored: annotations/.DS_Store
Ignored: annotations/SeqIF/.DS_Store
Ignored: annotations/molkart/.DS_Store
Ignored: annotations/molkart/Figure1_regions/.DS_Store
Ignored: annotations/molkart/Supplementary_Figure4_regions/.DS_Store
Ignored: data/.DS_Store
Ignored: data/140623.calcagno_et_al.seurat_object.rds
Ignored: data/Calcagno2022_int_logNorm_annot.h5Seurat
Ignored: data/IC_03_IF_CCR2_CD68 cell numbers.xlsx
Ignored: data/Traditional_IF_absolute_cell_counts.csv
Ignored: data/Traditional_IF_relative_cell_counts.csv
Ignored: data/pixie.cell_table_size_normalized_cell_labels.csv
Ignored: data/results_cts_100.sqm
Ignored: data/seqIF_regions_annotations/
Ignored: data/seurat/
Ignored: output/.DS_Store
Ignored: output/mol_cart.harmony_object.h5Seurat
Ignored: output/molkart/
Ignored: output/proteomics/
Ignored: output/results_cts.lowres.125.sqm
Ignored: output/seqIF/
Ignored: pipeline_configs/.DS_Store
Ignored: plots/
Ignored: references/.DS_Store
Ignored: renv/.DS_Store
Ignored: renv/library/
Ignored: renv/staging/
Untracked files:
Untracked: analysis/deprecated/figures.supplementary_figureX.Rmd
Untracked: analysis/deprecated/figures.supplementary_figure_X.MistyR.Rmd
Unstaged changes:
Deleted: analysis/figures.supplementary_figureX.Rmd
Deleted: analysis/figures.supplementary_figure_X.MistyR.Rmd
Deleted: analysis/figures.supplementary_figure_X.proteomics_qc.Rmd
Deleted: figures/Figure_5.eps
Deleted: figures/Figure_5.pdf
Deleted: figures/Figure_5.png
Deleted: figures/Figure_5.svg
Deleted: figures/Supplementary_Figure_1_Molecular_Cartography_ROIs.png
Deleted: figures/Supplementary_figure_5.segmentation_metrics.poster.eps
Modified: figures/Supplementary_figure_X.proteomics.eps
Modified: figures/Supplementary_figure_X.proteomics.png
Deleted: results_cts.lowres.125.sqm
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/figures.Figure1.Rmd) and
HTML (docs/figures.Figure1.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 56559c7 | FloWuenne | 2024-03-21 | Cleaned up repository. |
| Rmd | a49803c | FloWuenne | 2024-02-29 | Updated a number of figures. |
| Rmd | b06dcd3 | FloWuenne | 2024-02-25 | Updated Figure 1,4 and S4 and 5 code. |
| Rmd | af64c40 | FloWuenne | 2024-01-30 | Updated analysis for Figure 1 and 2. |
| Rmd | 86e53f0 | FloWuenne | 2024-01-22 | Finalized new Figure 1 plots. |
| Rmd | 82f107f | FloWuenne | 2024-01-21 | Updates to Molkart analysis. |
| html | b267494 | FloWuenne | 2023-12-06 | Build site. |
| Rmd | 2dcd178 | FloWuenne | 2023-12-06 | wflow_publish("*") |
| html | 2dcd178 | FloWuenne | 2023-12-06 | wflow_publish("*") |
| Rmd | 5dee03d | FloWuenne | 2023-09-04 | Latest code update. |
Introduction
library(tidyverse)── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ ggplot2 3.4.4 ✔ tibble 3.2.1
✔ lubridate 1.9.3 ✔ tidyr 1.3.0
✔ purrr 1.0.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorslibrary(cowplot)
Attaching package: 'cowplot'
The following object is masked from 'package:lubridate':
stamplibrary(Seurat)Loading required package: SeuratObject
Loading required package: sp
'SeuratObject' was built with package 'Matrix' 1.6.3 but the current
version is 1.6.5; it is recomended that you reinstall 'SeuratObject' as
the ABI for 'Matrix' may have changed
Attaching package: 'SeuratObject'
The following object is masked from 'package:base':
intersectlibrary(SCpubr)
── SCpubr 2.0.0.9000 ───────────────────────────────────────────────────────────
ℹ Have a look at extensive tutorials in SCpubr's book.
✔ If you use SCpubr in your research, please cite it accordingly.
★ If the package is useful to you, consider leaving a Star in the GitHub repository.
! Keep track of the package updates on Twitter (@Enblacar) or in the Official NEWS website.
♥ Happy plotting!
── Package version ──
CRAN: 2.0.2
Installed: 2.0.0.9000
⚠ There is a new version available onCRAN!
── Required packages ──
✔ AnnotationDbi 1.64.1 | 1.58.0 ✔ assertthat 0.2.1 | 0.2.1 ✖ AUCell
✔ circlize 0.4.15 | 0.4.16 ✔ cluster 2.1.6 | 2.1.6 ✖ clusterProfiler
✔ colorspace 2.1.0 | 2.1-0 ✔ decoupleR 2.8.0 | 2.2.2 ✔ dplyr 1.1.4 | 1.1.4
✖ enrichplot ✔ forcats 1.0.0 | 1.0.0 ✖ ggalluvial
✔ ggbeeswarm 0.7.2 | 0.7.2 ✖ ggdist ✖ ggExtra
✖ ggnewscale ✔ ggplot2 3.4.4 | 3.5.0 ✔ ggplotify 0.1.2 | 0.1.2
✔ ggrastr 1.0.2 | 1.0.2 ✔ ggrepel 0.9.5 | 0.9.5 ✔ ggridges 0.5.5 | 0.5.6
✔ ggsignif 0.6.4 | 0.6.4 ✔ labeling 0.4.3 | 0.4.3 ✖ liana
✔ magrittr 2.0.3 | 2.0.3 ✔ MASS 7.3.60.0.1 | 7.3-60.0.1 ✔ Matrix 1.6.5 | 1.6-5
✔ Nebulosa 1.12.0 | 1.6.0 ✔ patchwork 1.2.0 | 1.2.0 ✔ pbapply 1.7.2 | 1.7-2
✔ plyr 1.8.9 | 1.8.9 ✔ RColorBrewer 1.1.3 | 1.1-3 ✔ rlang 1.1.3 | 1.1.3
✔ scales 1.3.0 | 1.3.0 ✔ scattermore 1.2 | 1.2 ✔ Seurat 5.0.1 | 5.0.3
✔ SeuratObject 5.0.1 | 5.0.1 ✔ stringr 1.5.1 | 1.5.1 ✔ svglite 2.1.3 | 2.1.3
✔ tibble 3.2.1 | 3.2.1 ✔ tidyr 1.3.0 | 1.3.1 ✖ UCell
✔ viridis 0.6.4 | 0.6.5 ✔ withr 2.5.2 | 3.0.0
ℹ Installed packages are denoted by a tick (✔) and missing packages by a cross (✖).
ℹ Installed packages that still require an update to correctly run SCpubr have an exclamation mark (!).
ℹ Packages version are displayed as: Installed | Available.
── Available functions ──
✔ do_AffinityAnalysisPlot | DEV ✖ do_AlluvialPlot ✔ do_BarPlot
✔ do_BeeSwarmPlot ✔ do_BoxPlot ✖ do_CellularStatesPlot
✔ do_ChordDiagramPlot ✔ do_ColorPalette ✖ do_CopyNumberVariantPlot
✔ do_CorrelationPlot ✔ do_DiffusionMapPlot | DEV ✔ do_DimPlot
✔ do_DotPlot ✖ do_EnrichmentHeatmap ✔ do_ExpressionHeatmap
✔ do_FeaturePlot ✖ do_FunctionalAnnotationPlot ✖ do_GeyserPlot
✖ do_GroupedGOTermPlot ✔ do_GroupwiseDEPlot ✖ do_LigandReceptorPlot | DEV
✔ do_LoadingsPlot ✔ do_MetadataPlot | DEV ✔ do_NebulosaPlot
✔ do_PathwayActivityPlot ✔ do_RidgePlot ✖ do_SCEnrichmentHeatmap | DEV
✔ do_SCExpressionHeatmap | DEV ✔ do_TermEnrichmentPlot ✔ do_TFActivityPlot
✔ do_ViolinPlot ✔ do_VolcanoPlot ✔ save_Plot | DEV
ℹ Functions tied to development builds of SCpubr are marked by the (| DEV) tag.
ℹ You can install development builds of SCpubr by following the instructions in the Releases page.
ℹ Check the package requirements function-wise with: SCpubr::check_dependencies()
── Tips! ──
ℹ To adjust package messages to dark mode themes, use: options("SCpubr.darkmode" = TRUE)
ℹ To remove the white and black end from continuous palettes, use: options("SCpubr.ColorPaletteEnds" = FALSE)
✖ To suppress this startup message, use: suppressPackageStartupMessages(library(SCpubr))
✖ Alternatively, you can also set the following option: options("SCpubr.verbose" = FALSE)
And then load the package normally (and faster) as: library(SCpubr)
────────────────────────────────────────────────────────────────────────────────library(pals)
library(patchwork)
Attaching package: 'patchwork'
The following object is masked from 'package:cowplot':
align_plotslibrary(ggbeeswarm)
library(viridis)Loading required package: viridisLite
Attaching package: 'viridisLite'
The following objects are masked from 'package:pals':
cividis, inferno, magma, plasma, turbo, viridis
Attaching package: 'viridis'
The following objects are masked from 'package:pals':
cividis, inferno, magma, plasma, turbo, viridislibrary(ggdark)
source("./code/functions.R")here() starts at /Users/florian_wuennemann/1_Projects/MI_project/mi_spatialomics## If the object has already been computed
seurat_object <- readRDS(file = "./output/molkart/molkart.seurat_object.rds")## How many cells did we recover per sample?
cells_per_sample <- seurat_object@meta.data %>%
group_by(sample_ID) %>%
tally() %>%
ungroup()
mean(cells_per_sample$n)[1] 8628.5Umap plot
seurat_object@meta.data$anno_cell_type_lvl2 <- gsub("_"," ",seurat_object@meta.data$anno_cell_type_lvl2)
#
# pal.bands(alphabet, alphabet2, cols25, glasbey, kelly, polychrome,
# stepped, tol, watlington, tableau20,
# show.names=FALSE)
cell_types <- unique(seurat_object@meta.data$anno_cell_type_lvl2)
colors <- kelly(n=22)
## Replace white ish color as we can't see it well in plots
# Create a named vector for cell_types and colors to use in plots later
# Code from scanpy
# https://github.com/scverse/scanpy/commit/58fae77cc15893503c0c34ce0295dd6f67af2bd7
vega10_scanpy <- c("#1f77b4","#ff7f0e","#2ca02c","#d62728","#9467bd","#8c564b","#e377c2","#7f7f7f","#bcbd22","#17becf")
# Use vega10 color palette for clusters
# named_colors <- c("Cardiac fibroblasts" = "#1f77b4",
# "Cardiomyocytes" = "#d62728",
# "Cardiomyocytes Nppa+" = "#ff7f0e",
# "Endocardial cells" = "#17becf",
# "Endothelial cells" = "#8c564b",
# "Lymphatic endothelial cells" = "lightgrey",
# "Lymphoid cells" = "#bcbd22",
# "Myeloid cells" = "#2ca02c",
# "Pericytes" = "#e377c2",
# "Smooth muscle cells" = "#9467bd"
# )
named_colors <- c("Cardiac fibroblasts" = "#1f77b4",
"Cardiomyocytes" = "#ff7697",
"Cardiomyocytes Nppa+" = "#ff9966",
"Endocardial cells" = "#17becf",
"Endothelial cells" = "#8c564b",
"Lymphoid cells" = "#bcbd22",
"Myeloid cells" = "#2ca02c",
"Pericytes" = "#9467bd",
"Smooth muscle cells" = "#e377c2")Figure 1C : UMAP plot of cell types
## Single total UMAP plot
library(scCustomize)scCustomize v2.0.1
If you find the scCustomize useful please cite.
See 'samuel-marsh.github.io/scCustomize/articles/FAQ.html' for citation info.## Set color palette
Idents(object = seurat_object) <- "anno_cell_type_lvl2"
umap_plot <- SCpubr::do_DimPlot(sample = seurat_object,
label = FALSE, label.box = TRUE,
group.by = "anno_cell_type_lvl2",
repel = TRUE, legend.position = "none",
colors.use = named_colors,
plot_cell_borders = TRUE,
plot_density_contour = FALSE, plot.axes = FALSE, raster.dpi = 300, shuffle = FALSE, pt.size = 0.4,
legend.icon.size = 5, legend.byrow = TRUE) +
theme(legend.text = element_text(size = 18))
umap_plot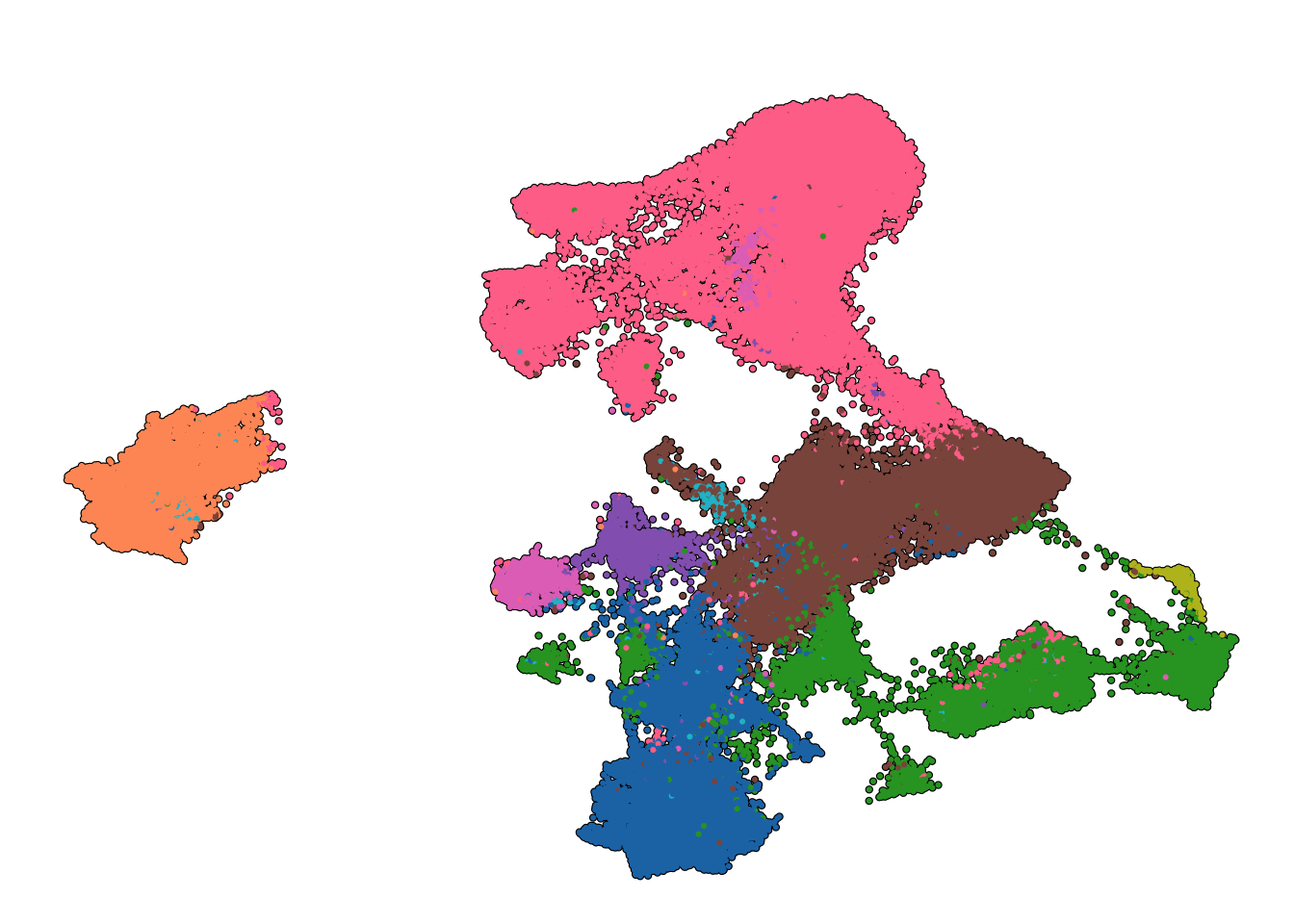
| Version | Author | Date |
|---|---|---|
| 2dcd178 | FloWuenne | 2023-12-06 |
# save_plot(umap_plot,
# file = "./plots/Figure1.umap_plot.pdf",
# base_height = 6)
#
#
# save_plot(umap_plot,
# file = "./plots/Figure1.umap_plot.png",
# base_height = 6)## UMAP split by time
library(scCustomize)
## Set color palette
Idents(object = seurat_object) <- "anno_cell_type_lvl2"
seurat_object_ctrl <- subset(seurat_object, timepoint == "control")
seurat_object_4h <- subset(seurat_object, timepoint == "4h")
seurat_object_2d <- subset(seurat_object, timepoint == "2d")
seurat_object_4d <- subset(seurat_object, timepoint == "4d")
umap_plot_control <- SCpubr::do_DimPlot(sample = seurat_object_ctrl,
label = FALSE, label.box = FALSE,
group.by = "anno_cell_type_lvl2",
repel = TRUE, legend.position = "none",
colors.use = named_colors,
plot_cell_borders = TRUE,
plot_density_contour = FALSE, plot.axes = FALSE,
raster.dpi = 300, shuffle = FALSE, pt.size = 0.4,
legend.icon.size = 5)
umap_plot_4h <- SCpubr::do_DimPlot(sample = seurat_object_4h,
label = FALSE, label.box = FALSE,
group.by = "anno_cell_type_lvl2",
repel = TRUE, legend.position = "none",
colors.use = named_colors,
plot_cell_borders = TRUE,
plot_density_contour = FALSE, plot.axes = FALSE,
raster.dpi = 300, shuffle = FALSE, pt.size = 0.4,
legend.icon.size = 5)
umap_plot_2d <- SCpubr::do_DimPlot(sample = seurat_object_2d,
label = FALSE, label.box = FALSE,
group.by = "anno_cell_type_lvl2",
repel = TRUE, legend.position = "none",
colors.use = named_colors,
plot_cell_borders = TRUE,
plot_density_contour = FALSE, plot.axes = FALSE,
raster.dpi = 300, shuffle = FALSE, pt.size = 0.4,
legend.icon.size = 5)
umap_plot_4d <- SCpubr::do_DimPlot(sample = seurat_object_4d,
label = FALSE, label.box = FALSE,
group.by = "anno_cell_type_lvl2",
repel = TRUE, legend.position = "none",
colors.use = named_colors,
plot_cell_borders = TRUE,
plot_density_contour = FALSE, plot.axes = FALSE,
raster.dpi = 300, shuffle = FALSE, pt.size = 0.4,
legend.icon.size = 5)full_plot <- umap_plot_control | umap_plot_4h | umap_plot_2d | umap_plot_4d
save_plot(full_plot,
file = "./plots/Figure1.umap_plot.png",
base_height = 4,
base_asp = 4,
dpi = 300)
save_plot(full_plot,
file = "./plots/Figure1.umap_plot.pdf",
base_height = 4,
base_asp = 4,
dpi = 300)Figure 1F : Overview of spatial distribution of cell types within 1 sample
sample_highlight <- "sample_2d_r2_s1"
#sample_highlight <- "sample_control_r2_s1"
meta_sample <- subset(seurat_object@meta.data,sample_ID == sample_highlight)
full_overview <- ggplot(meta_sample,aes(X_centroid,Y_centroid)) +
geom_point(aes(color = anno_cell_type_lvl2),size = 0.6) +
theme_classic() +
dark_theme_void() +
labs(x = "Spatial 1",
y = "Spatial 2") +
theme(axis.title = element_blank(),
axis.ticks = element_blank(),
axis.text = element_blank(),
axis.line = element_blank(),
legend.position = "none") +
scale_color_manual(values = named_colors)Inverted geom defaults of fill and color/colour.
To change them back, use invert_geom_defaults().# Custom function to return an empty string for each label
blank_labels <- function(value) {
return(rep("", length(value)))
}
cell_type_views <- ggplot(meta_sample,aes(X_centroid,Y_centroid)) +
dark_theme_void() +
geom_point(aes(color = anno_cell_type_lvl2),size = 0.4) +
scale_color_manual(values = named_colors) +
facet_wrap(~ anno_cell_type_lvl2,labeller = as_labeller(blank_labels)) +
theme(strip.text = element_text(size = 18, color = "white"),
legend.position = "none"
)
full_plot <- (full_overview | cell_type_views) + plot_layout(ncol = 2,widths = c(0.6, 1))
full_plot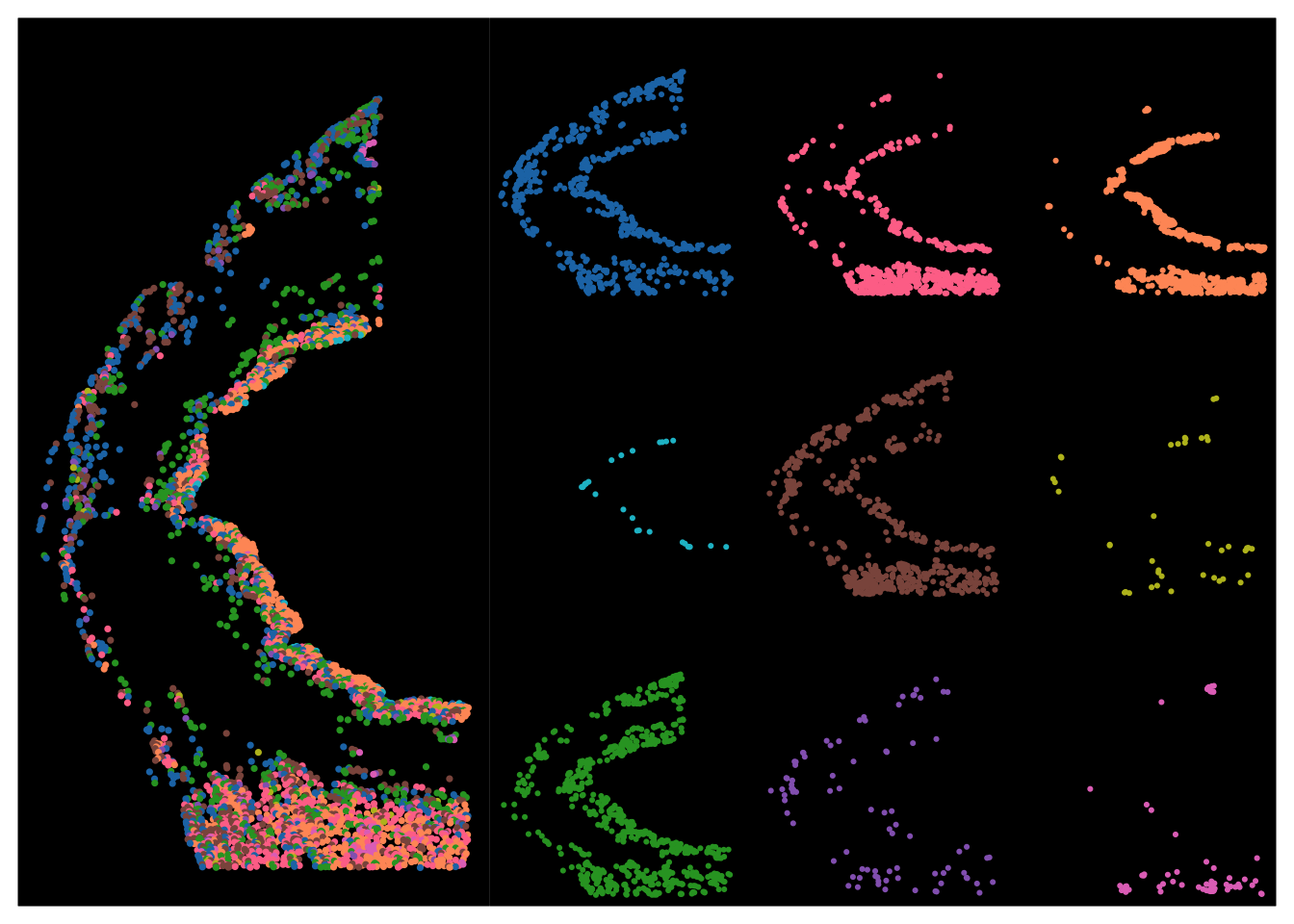
| Version | Author | Date |
|---|---|---|
| 2dcd178 | FloWuenne | 2023-12-06 |
save_plot(full_plot,filename = here("./plots/molkart.Figure_1.spatial_distribution.pdf"),
base_height = 4,
base_asp = 2)library(ggplot2)
library(dplyr)
# Assuming meta_sample is your data frame and named_colors is your color vector
# Get unique cell types
unique_cell_types <- sort(unique(meta_sample$anno_cell_type_lvl2))
max_x <- max(meta_sample$X_centroid)
max_y <- max(meta_sample$Y_centroid)
# Loop through each cell type and generate a plot
plots <- lapply(unique_cell_types, function(cell_type) {
# Generate the plot
## Add a line of blank space on top of the plot
p <- ggplot(meta_sample, aes(x = X_centroid, y =
Y_centroid)) +
dark_theme_void() +
# Set x and y limits
xlim(0,max_x) +
ylim(0,max_y) +
# geom_point(data = subset(meta_sample,anno_cell_type_lvl2 != cell_type),
# size = 0.6, color = "white") +
geom_point(data = subset(meta_sample,anno_cell_type_lvl2 == cell_type),
size = 0.6,
aes(color = anno_cell_type_lvl2)) +
scale_color_manual(values = named_colors) +
theme(legend.position = "none") # Hide legend or adjust as needed
# Add a line of blank space on top of the plot
p <- p + theme(plot.margin = margin(0.5,0,0,0, "cm"))
# Return the plot
return(p)
})
# Optionally, print plots or save them to files
# for (p in plots) print(p)
cell_type_views <- wrap_plots(plots, nrow = 3, ncol = 3)
save_plot(full_overview,
filename = "./plots/molkart.Figure_1.spatial_distribution.full.pdf",
base_height = 5,
base_asp = 0.9)
save_plot(cell_type_views,
filename = "./plots/molkart.Figure_1.spatial_distribution.cts.pdf",
base_height = 5,
base_asp = 1.3)
sessionInfo()R version 4.3.1 (2023-06-16)
Platform: aarch64-apple-darwin20 (64-bit)
Running under: macOS Sonoma 14.1.2
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.11.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: Europe/Berlin
tzcode source: internal
attached base packages:
[1] stats graphics grDevices datasets utils methods base
other attached packages:
[1] scCustomize_2.0.1 RColorBrewer_1.1-3 here_1.0.1 ggsci_3.0.0
[5] ggdark_0.2.1 viridis_0.6.4 viridisLite_0.4.2 ggbeeswarm_0.7.2
[9] patchwork_1.2.0 pals_1.8 SCpubr_2.0.0.9000 Seurat_5.0.1
[13] SeuratObject_5.0.1 sp_2.1-2 cowplot_1.1.2 lubridate_1.9.3
[17] forcats_1.0.0 stringr_1.5.1 dplyr_1.1.4 purrr_1.0.2
[21] readr_2.1.5 tidyr_1.3.0 tibble_3.2.1 ggplot2_3.4.4
[25] tidyverse_2.0.0 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] fs_1.6.3 matrixStats_1.2.0
[3] spatstat.sparse_3.0-3 bitops_1.0-7
[5] httr_1.4.7 tools_4.3.1
[7] sctransform_0.4.1 utf8_1.2.4
[9] R6_2.5.1 lazyeval_0.2.2
[11] uwot_0.1.16 withr_2.5.2
[13] gridExtra_2.3 progressr_0.14.0
[15] textshaping_0.3.7 cli_3.6.2
[17] Biobase_2.62.0 spatstat.explore_3.2-5
[19] fastDummies_1.7.3 labeling_0.4.3
[21] sass_0.4.8 mvtnorm_1.2-4
[23] spatstat.data_3.0-3 ggridges_0.5.5
[25] pbapply_1.7-2 systemfonts_1.0.5
[27] yulab.utils_0.1.3 svglite_2.1.3
[29] dichromat_2.0-0.1 parallelly_1.36.0
[31] maps_3.4.2 rstudioapi_0.15.0
[33] RSQLite_2.3.4 generics_0.1.3
[35] gridGraphics_0.5-1 shape_1.4.6
[37] ica_1.0-3 spatstat.random_3.2-2
[39] Matrix_1.6-5 fansi_1.0.6
[41] S4Vectors_0.40.2 abind_1.4-5
[43] lifecycle_1.0.4 whisker_0.4.1
[45] yaml_2.3.8 snakecase_0.11.1
[47] SummarizedExperiment_1.32.0 SparseArray_1.2.3
[49] Rtsne_0.17 paletteer_1.5.0
[51] grid_4.3.1 blob_1.2.4
[53] promises_1.2.1 crayon_1.5.2
[55] miniUI_0.1.1.1 lattice_0.22-5
[57] KEGGREST_1.42.0 mapproj_1.2.11
[59] pillar_1.9.0 knitr_1.45
[61] GenomicRanges_1.54.1 future.apply_1.11.1
[63] codetools_0.2-19 leiden_0.4.3.1
[65] glue_1.7.0 getPass_0.2-4
[67] data.table_1.14.10 vctrs_0.6.5
[69] png_0.1-8 spam_2.10-0
[71] gtable_0.3.4 rematch2_2.1.2
[73] assertthat_0.2.1 cachem_1.0.8
[75] ks_1.14.2 xfun_0.41
[77] S4Arrays_1.2.0 mime_0.12
[79] pracma_2.4.4 survival_3.5-7
[81] SingleCellExperiment_1.24.0 ellipsis_0.3.2
[83] fitdistrplus_1.1-11 ROCR_1.0-11
[85] nlme_3.1-164 bit64_4.0.5
[87] RcppAnnoy_0.0.21 GenomeInfoDb_1.38.5
[89] rprojroot_2.0.4 bslib_0.6.1
[91] irlba_2.3.5.1 vipor_0.4.7
[93] KernSmooth_2.23-22 colorspace_2.1-0
[95] BiocGenerics_0.48.1 DBI_1.2.0
[97] ggrastr_1.0.2 tidyselect_1.2.0
[99] processx_3.8.3 bit_4.0.5
[101] compiler_4.3.1 git2r_0.33.0
[103] DelayedArray_0.28.0 plotly_4.10.4
[105] scales_1.3.0 lmtest_0.9-40
[107] callr_3.7.3 digest_0.6.34
[109] goftest_1.2-3 spatstat.utils_3.0-4
[111] rmarkdown_2.25 XVector_0.42.0
[113] decoupleR_2.8.0 htmltools_0.5.7
[115] pkgconfig_2.0.3 MatrixGenerics_1.14.0
[117] highr_0.10 fastmap_1.1.1
[119] rlang_1.1.3 GlobalOptions_0.1.2
[121] htmlwidgets_1.6.4 shiny_1.8.0
[123] farver_2.1.1 jquerylib_0.1.4
[125] zoo_1.8-12 jsonlite_1.8.8
[127] mclust_6.0.1 RCurl_1.98-1.14
[129] magrittr_2.0.3 GenomeInfoDbData_1.2.11
[131] ggplotify_0.1.2 dotCall64_1.1-1
[133] munsell_0.5.0 Rcpp_1.0.12
[135] reticulate_1.34.0 stringi_1.8.3
[137] zlibbioc_1.48.0 MASS_7.3-60.0.1
[139] plyr_1.8.9 parallel_4.3.1
[141] listenv_0.9.0 ggrepel_0.9.5
[143] deldir_2.0-2 Biostrings_2.70.1
[145] splines_4.3.1 tensor_1.5
[147] hms_1.1.3 circlize_0.4.15
[149] ps_1.7.6 igraph_1.6.0
[151] spatstat.geom_3.2-7 ggsignif_0.6.4
[153] RcppHNSW_0.5.0 reshape2_1.4.4
[155] stats4_4.3.1 evaluate_0.23
[157] Nebulosa_1.12.0 renv_1.0.3
[159] BiocManager_1.30.22 ggprism_1.0.4
[161] tzdb_0.4.0 httpuv_1.6.14
[163] RANN_2.6.1 polyclip_1.10-6
[165] future_1.33.1 scattermore_1.2
[167] janitor_2.2.0 xtable_1.8-4
[169] RSpectra_0.16-1 later_1.3.2
[171] ragg_1.2.7 memoise_2.0.1
[173] beeswarm_0.4.0 AnnotationDbi_1.64.1
[175] IRanges_2.36.0 cluster_2.1.6
[177] timechange_0.2.0 globals_0.16.2