Modeling Changes in Microbiome
Last updated: 2020-05-07
Checks: 6 1
Knit directory: Fiber_Intervention_Study/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20191210) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 2d97c53. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: reference-papers/Dietary_Variables.xlsx
Ignored: reference-papers/Johnson_2019.pdf
Untracked files:
Untracked: analysis/microbiome_diet_trends.Rmd
Untracked: fig/figure2.pdf
Untracked: fig/figure2_legend.pdf
Untracked: fig/figure3.pdf
Untracked: fig/figure3_legend.pdf
Untracked: fig/figure4.pdf
Untracked: fig/figure4_legend.pdf
Unstaged changes:
Modified: analysis/analysis_beta_diversity.Rmd
Modified: analysis/change_in_alpha_diversity.Rmd
Deleted: analysis/dietary_intake_figure.Rmd
Modified: analysis/lme_microbiome.Rmd
Modified: code/load_packages.R
Modified: data/analysis-data/Dietary_Variables.xlsx
Modified: fig/figure1.pdf
Modified: fig/figure1_legend.pdf
Modified: fig/predicted_Actinobacteria_model5.png
Modified: fig/predicted_Actinobacteria_model7.png
Modified: fig/predicted_Bacteroidetes_model5.png
Modified: fig/predicted_Bacteroidetes_model7.png
Modified: fig/predicted_Cyanobacteria_model5.png
Modified: fig/predicted_Cyanobacteria_model7.png
Modified: fig/predicted_Euryarchaeota_model5.png
Modified: fig/predicted_Euryarchaeota_model7.png
Modified: fig/predicted_Firmicutes_model5.png
Modified: fig/predicted_Firmicutes_model7.png
Modified: fig/predicted_Fusobacteria_model5.png
Modified: fig/predicted_Fusobacteria_model7.png
Modified: fig/predicted_Lentisphaerae_model5.png
Modified: fig/predicted_Lentisphaerae_model7.png
Modified: fig/predicted_Proteobacteria_model5.png
Modified: fig/predicted_Proteobacteria_model7.png
Modified: fig/predicted_Tenericutes_model5.png
Modified: fig/predicted_Tenericutes_model7.png
Modified: fig/predicted_Verrucomicrobia_model5.png
Modified: fig/predicted_Verrucomicrobia_model7.png
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/lme_microbiome.Rmd) and HTML (docs/lme_microbiome.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | 51b6275 | noah-padgett | 2020-02-20 | Build site. |
| Rmd | 1c719ff | noah-padgett | 2020-02-20 | updated output window size |
| html | 1c719ff | noah-padgett | 2020-02-20 | updated output window size |
| html | a6488a7 | noah-padgett | 2020-02-06 | Build site. |
| Rmd | 0674e1b | noah-padgett | 2020-02-06 | fixed index |
| html | 6de5819 | noah-padgett | 2020-02-06 | Build site. |
| Rmd | 77daa75 | noah-padgett | 2020-02-06 | fixed index |
| Rmd | dd420c9 | noah-padgett | 2020-02-06 | updated glmm results and index |
| html | dd420c9 | noah-padgett | 2020-02-06 | updated glmm results and index |
This page contains the investigation of the changes over time.
Data prep
This is only to recode some variables to ease interpretation.
microbiome_data$meta.dat <- microbiome_data$meta.dat %>%
mutate(intB = ifelse(Intervention=="B", 1,0),
time = as.numeric(Week) - 1,
female = ifelse(Gender == "F", 1, 0),
hispanic = ifelse(Ethnicity %in% c("White", "Asian", "Native America"), 1, 0))Phylum Level Microbiome and Intervention
We will build up to the effect of the intervention by investigating covariates to see if we can exclude them from model with intervention. Due to only having 11 people, we will only estimate 1 random effect per model (random intercepts). All other effects are fixed, but we will only estimate up to 2 effects. Meaning we can only include two covariates in each model at a time.
In all models, \(h(.)\) is an arbitrary link function. We selected the Poisson model for the outcomes so the link function is the log link.
For all significance test, we will use \(p < 0.05\) as the level to declare significance, but we will largely focus on magnitude of effects.
Unconditional Model
This is a random intercepts model (worst case). No covariates are included.
\[\begin{align*} h\left(Y_{ij}\right)&=\beta_{0j} + r_{ij}\\ \beta_{0j}&= \gamma_{00} + u_{0j}\\ \end{align*}\]
phylum.fit0 <- glmm_microbiome(mydata=microbiome_data,model.number=0,
taxa.level="Phylum", link="poisson",
model="1 + (1|SubjectID)")
#########################################
#########################################
Model: Actinobacteria ~ 1 + (1 | SubjectID)
<environment: 0x0000000041f2dbb0>
Link: poisson
Intraclass Correlation (ICC): 0.5588572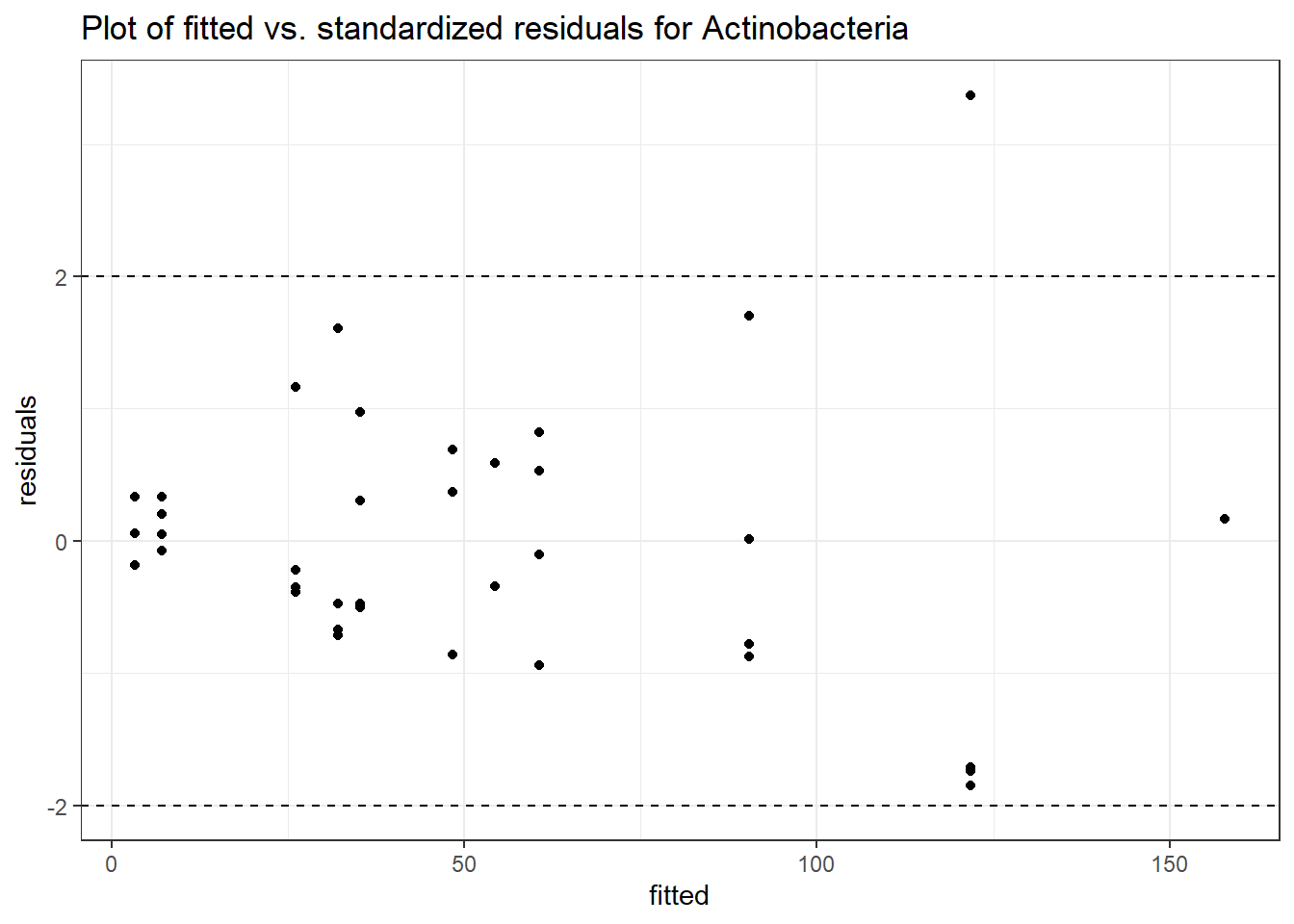
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Actinobacteria 3.60892 0.34173 10.56075 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.1255
Significance test not available
#########################################
#########################################
Model: Bacteroidetes ~ 1 + (1 | SubjectID)
<environment: 0x0000000041f2dbb0>
Link: poisson
Intraclass Correlation (ICC): 0.1012137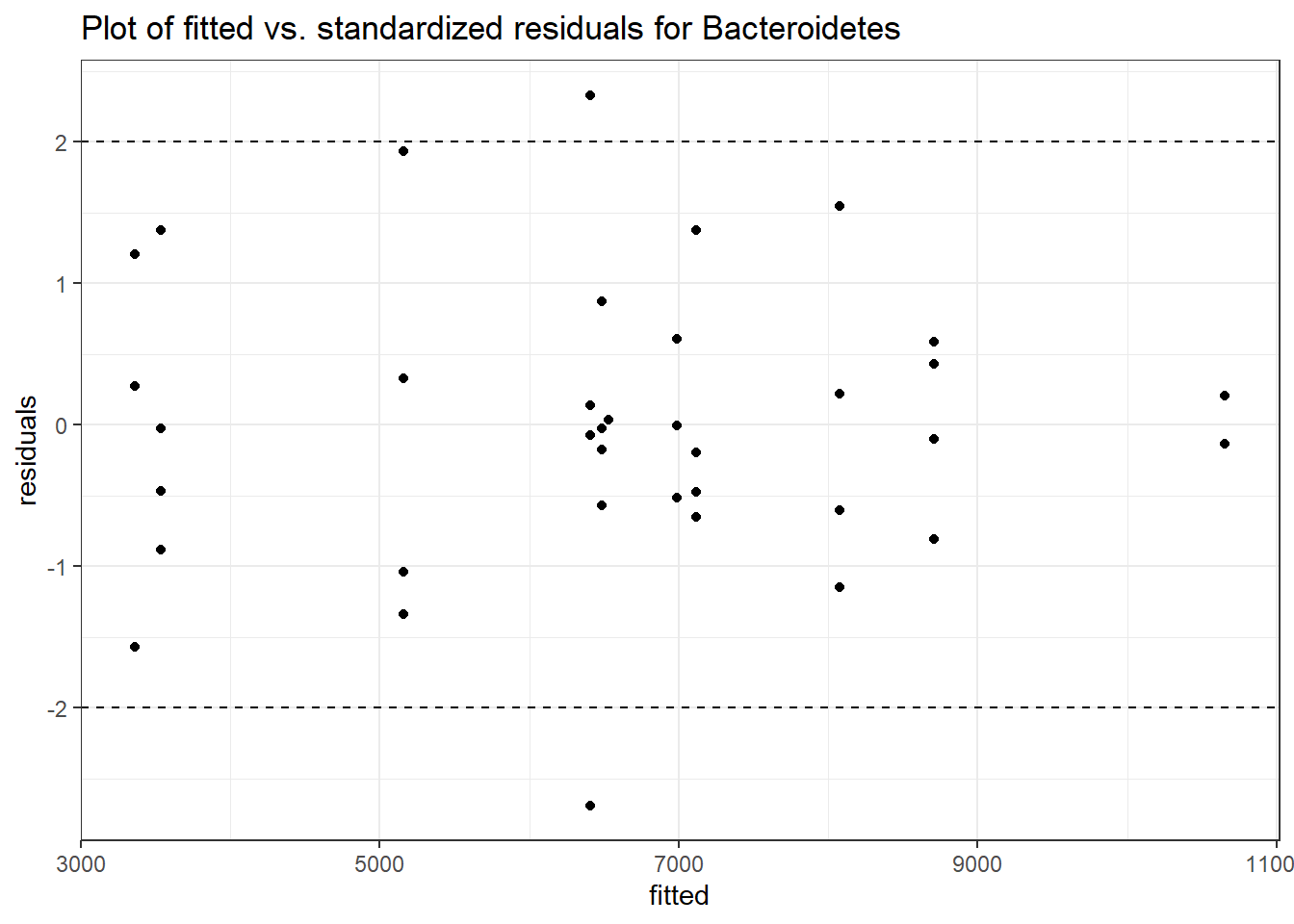
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroidetes 8.74822 0.10121 86.43962 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.33558
Significance test not available
#########################################
#########################################
Model: Cyanobacteria ~ 1 + (1 | SubjectID)
<environment: 0x0000000041f2dbb0>
Link: poisson
Intraclass Correlation (ICC): 0.8456119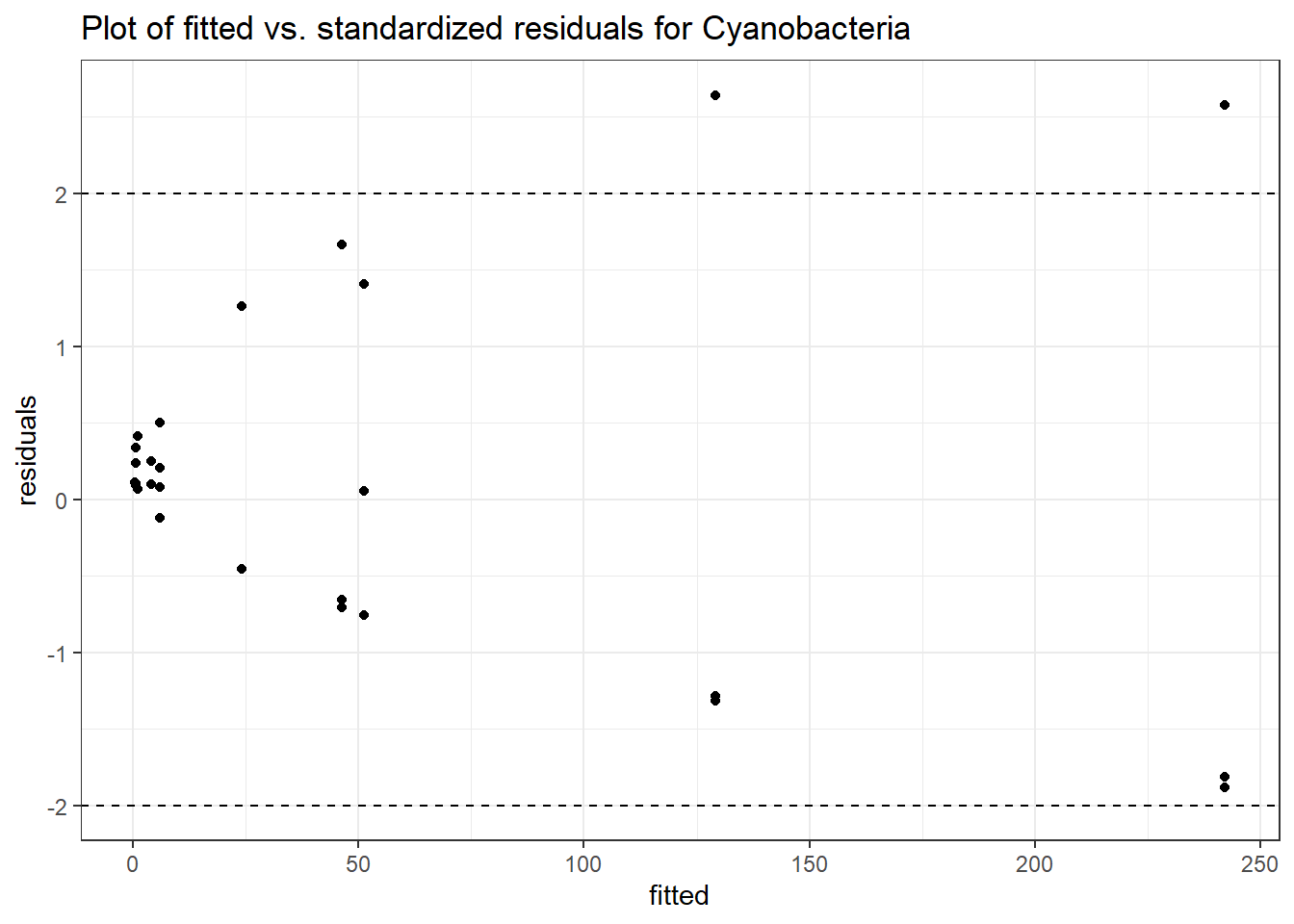
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Cyanobacteria 1.99 0.72467 2.74608 0.00603
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.3403
Significance test not available
#########################################
#########################################
Model: Euryarchaeota ~ 1 + (1 | SubjectID)
<environment: 0x0000000041f2dbb0>
Link: poisson
Intraclass Correlation (ICC): 0.7888934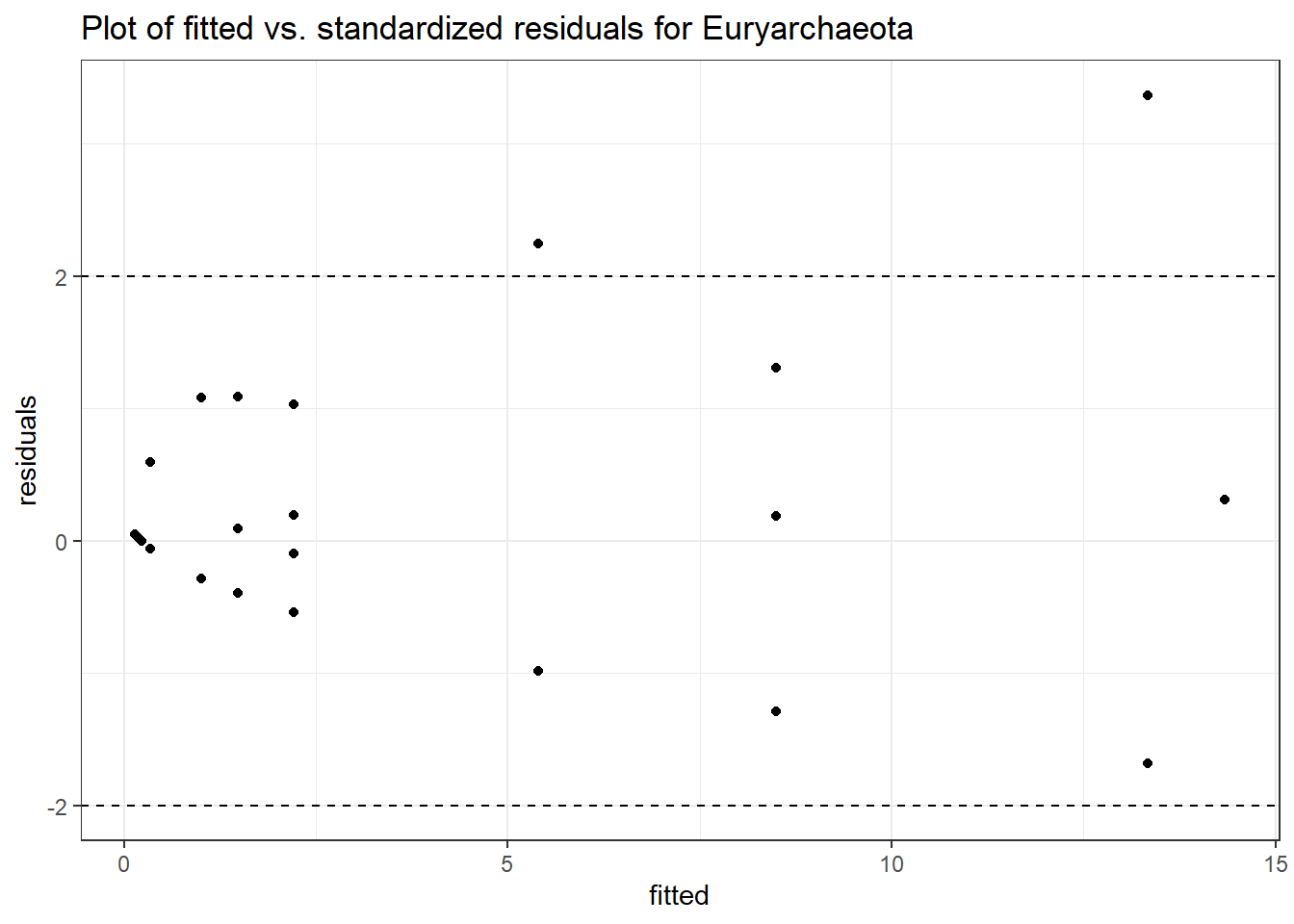
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Euryarchaeota 0.20274 0.63788 0.31784 0.75061
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.9331
Significance test not available
#########################################
#########################################
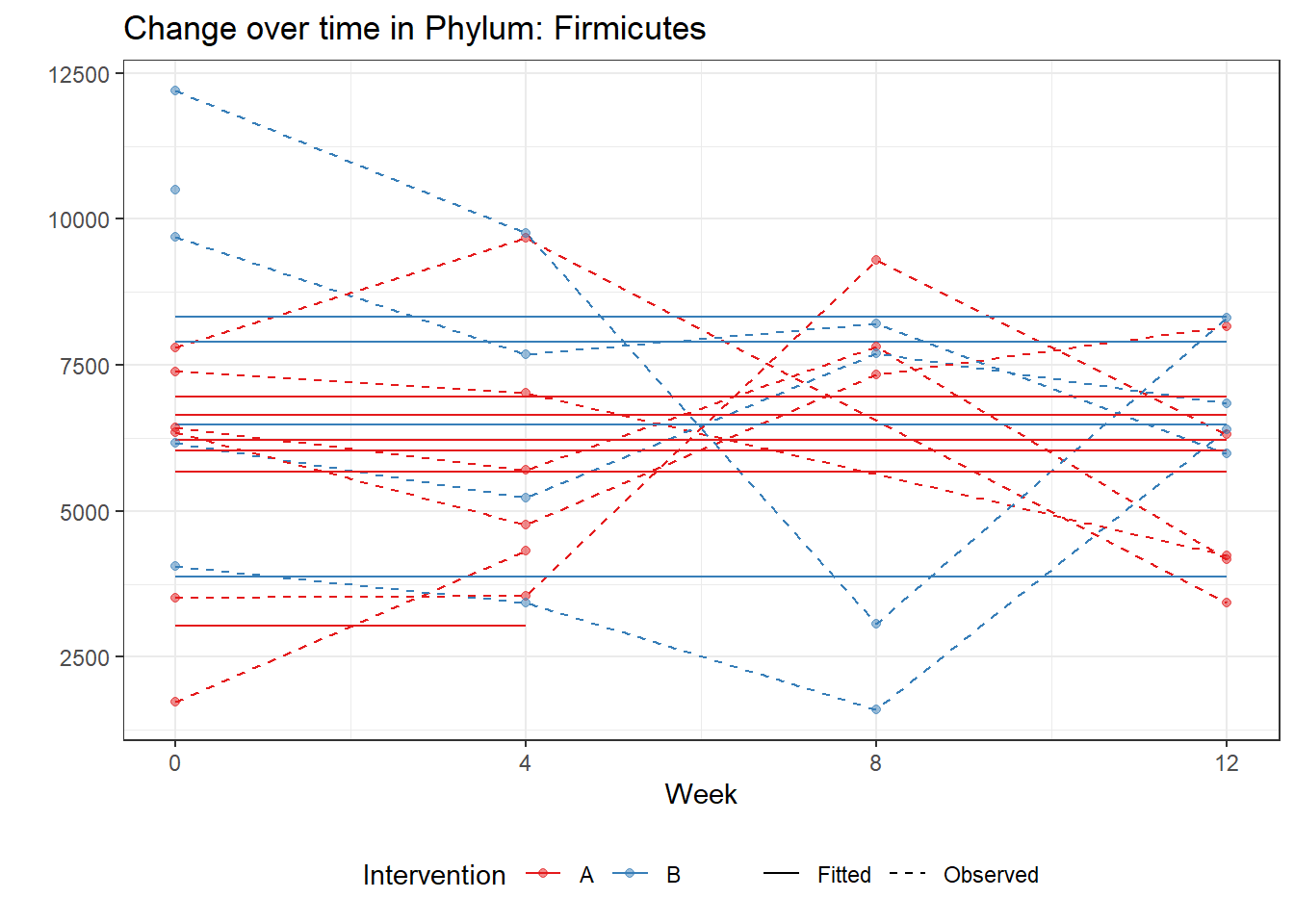
Model: Firmicutes ~ 1 + (1 | SubjectID)
<environment: 0x0000000041f2dbb0>
Link: poisson
Intraclass Correlation (ICC): 0.09724719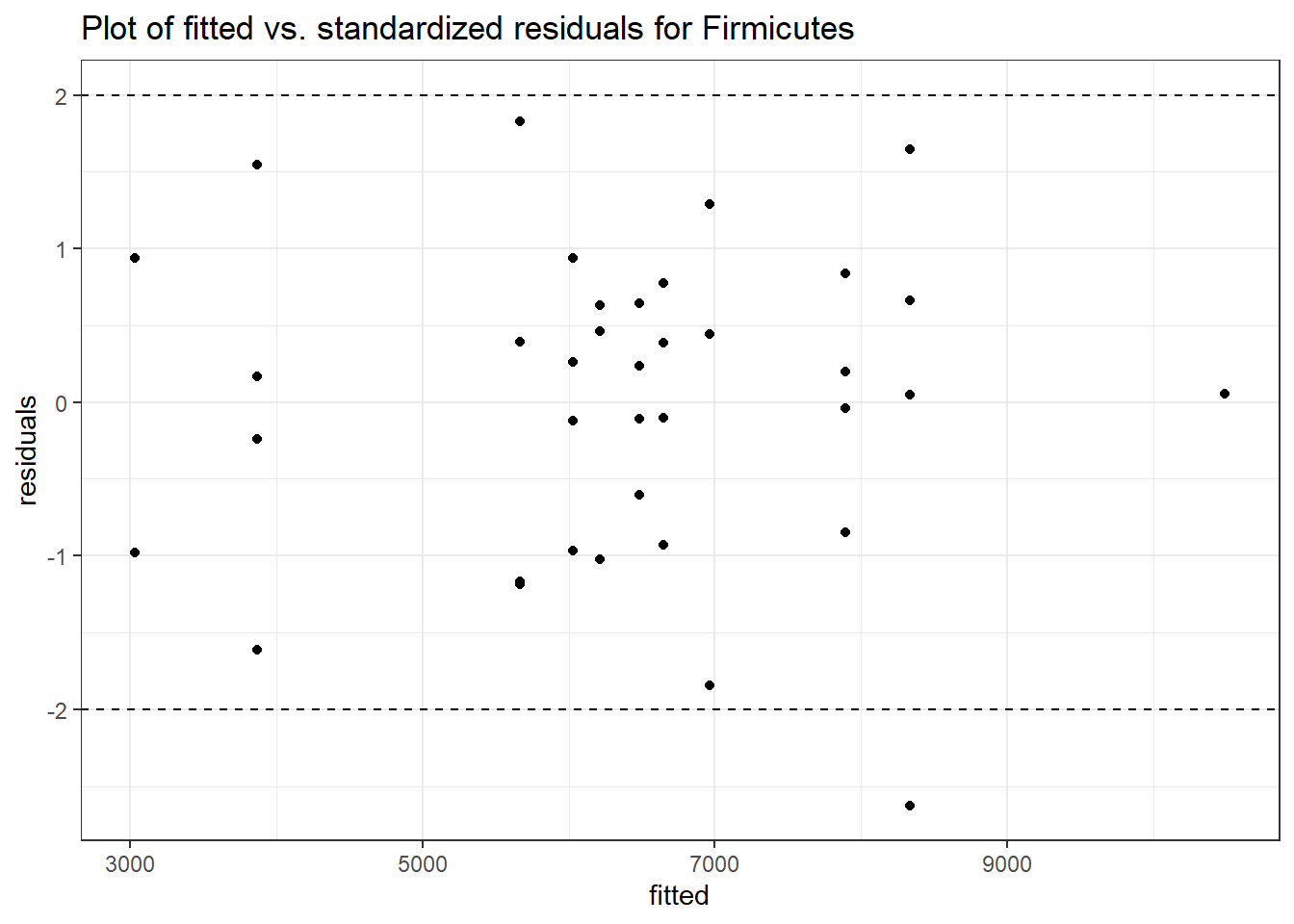
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Firmicutes 8.73145 0.09899 88.20897 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.32821
Significance test not available
#########################################
#########################################
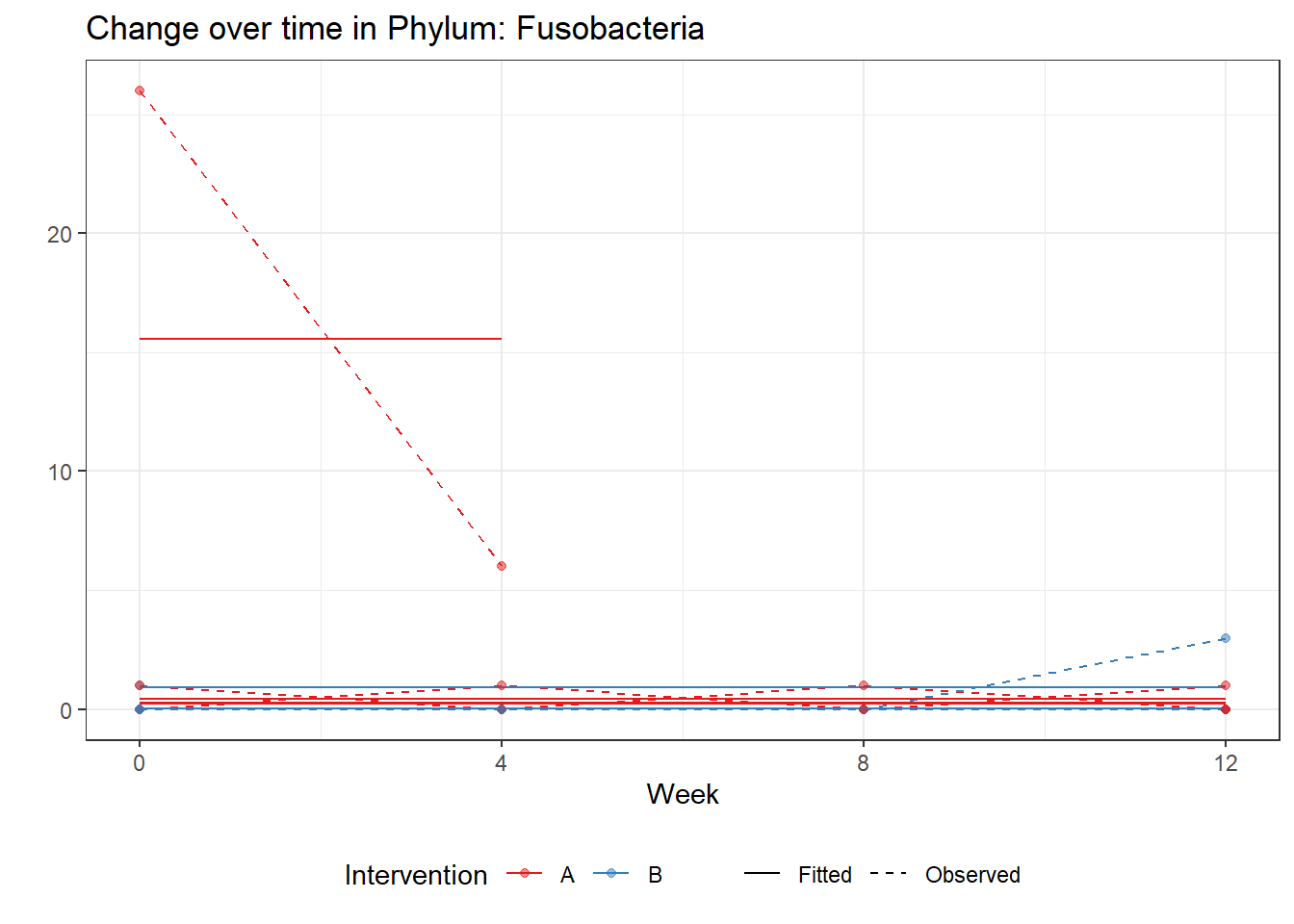
Model: Fusobacteria ~ 1 + (1 | SubjectID)
<environment: 0x0000000041f2dbb0>
Link: poisson
Intraclass Correlation (ICC): 0.854714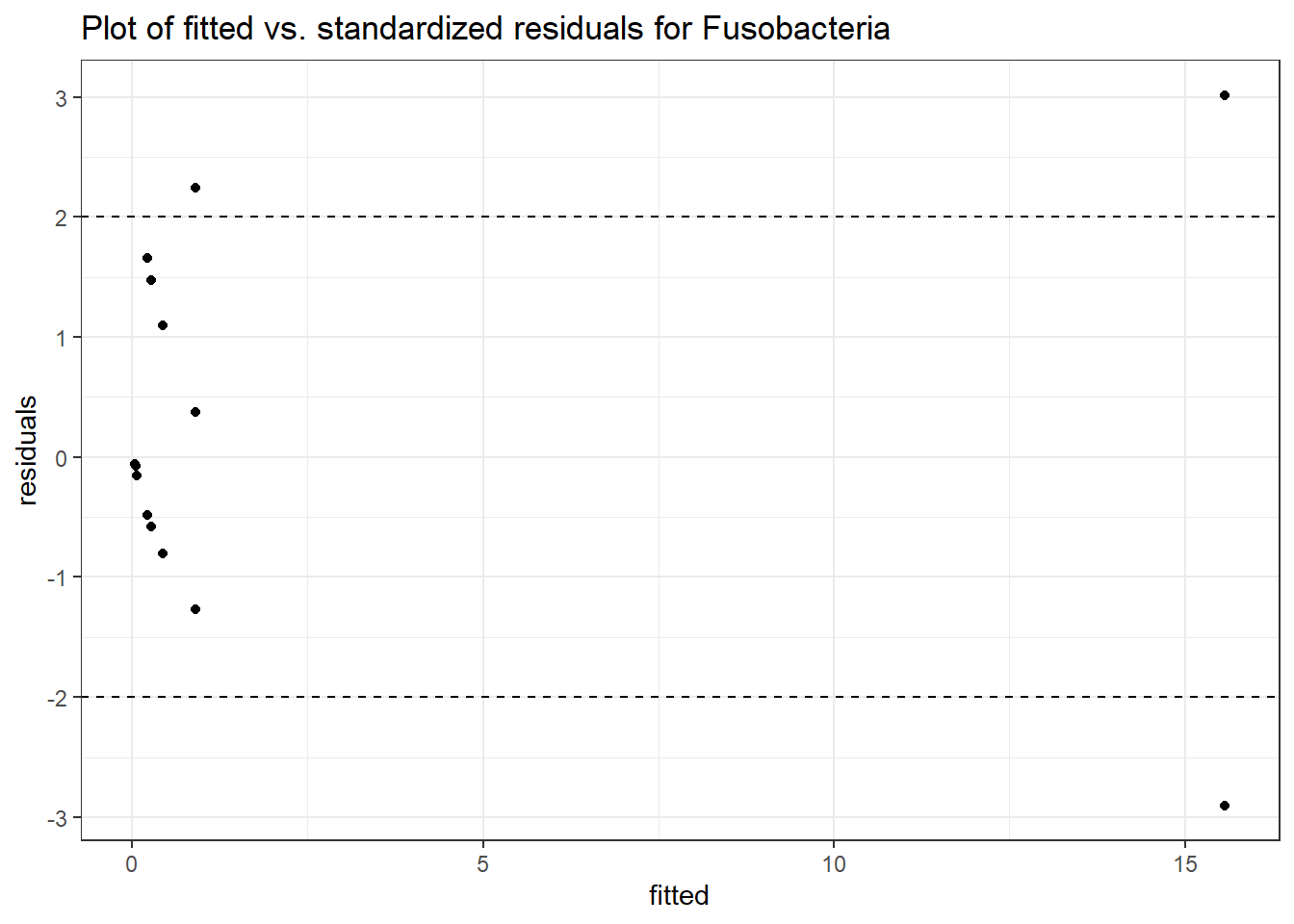
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Fusobacteria -2.28008 1.12843 -2.02057 0.04332
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.4255
Significance test not available
#########################################
#########################################
Model: Lentisphaerae ~ 1 + (1 | SubjectID)
<environment: 0x0000000041f2dbb0>
Link: poisson
Intraclass Correlation (ICC): 0.6843902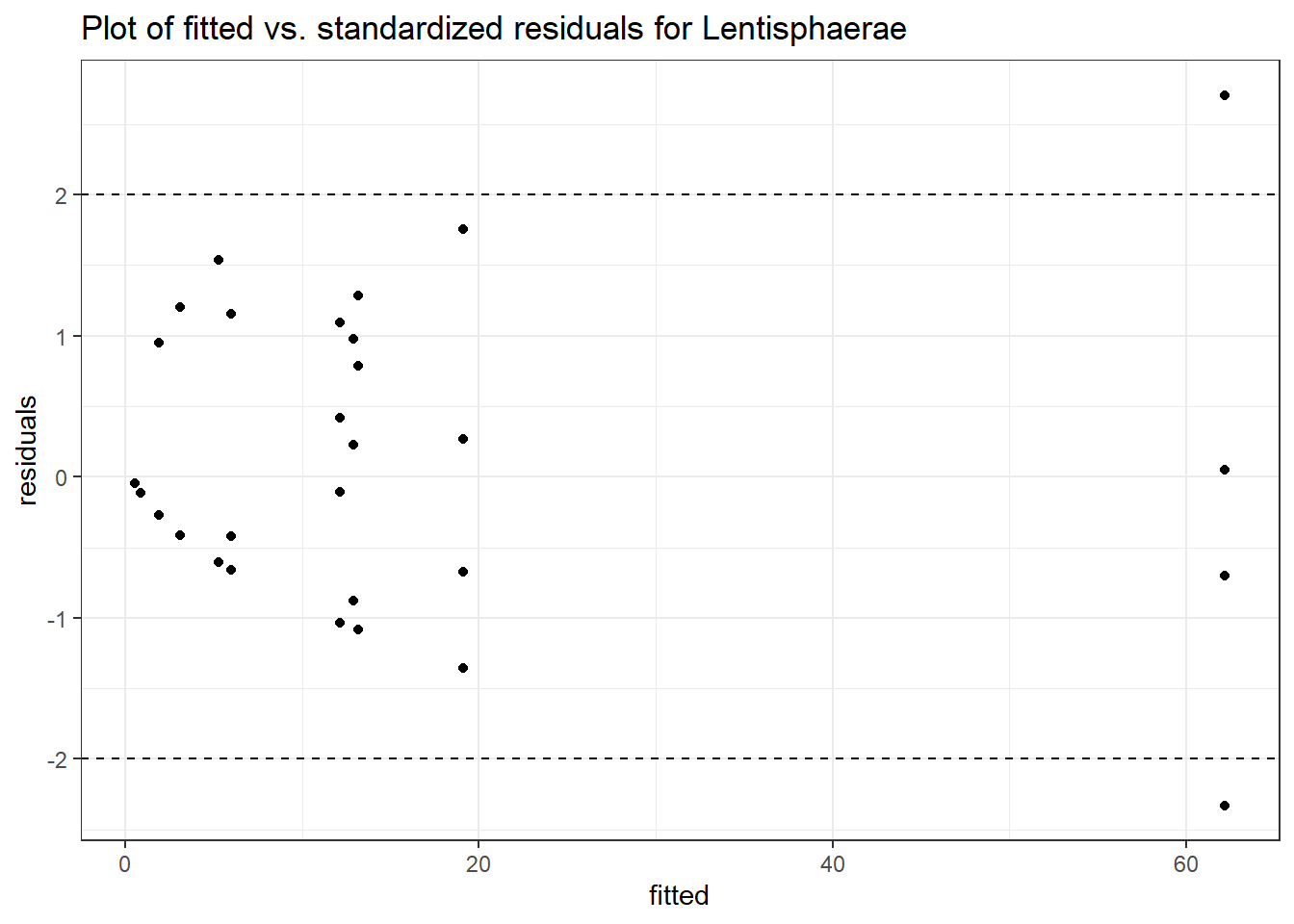
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lentisphaerae 1.67757 0.46839 3.58158 0.00034
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.4726
Significance test not available
#########################################
#########################################
Model: Proteobacteria ~ 1 + (1 | SubjectID)
<environment: 0x0000000041f2dbb0>
Link: poisson
Intraclass Correlation (ICC): 0.09326096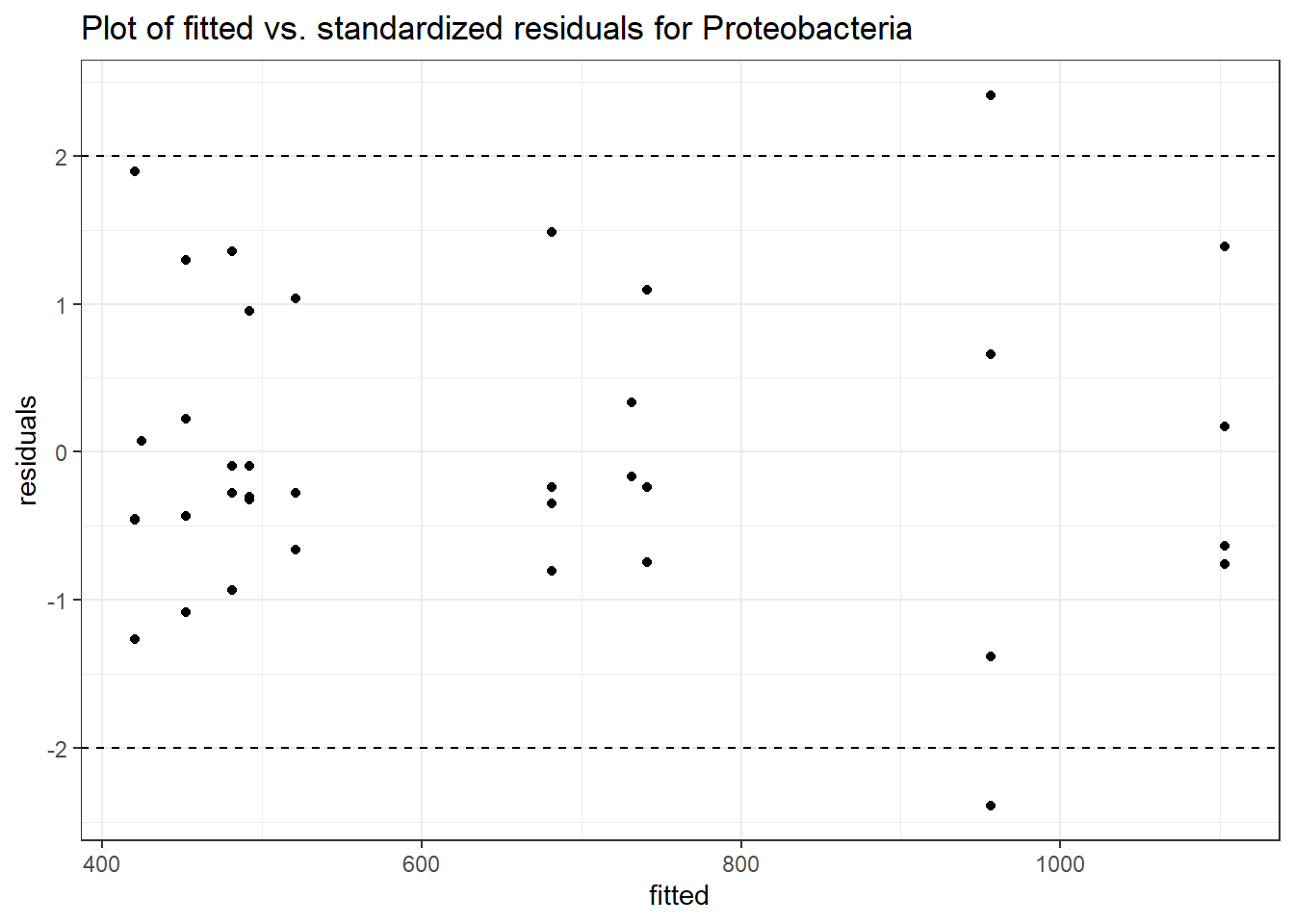
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Proteobacteria 6.40274 0.097 66.0065 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.32071
Significance test not available
#########################################
#########################################
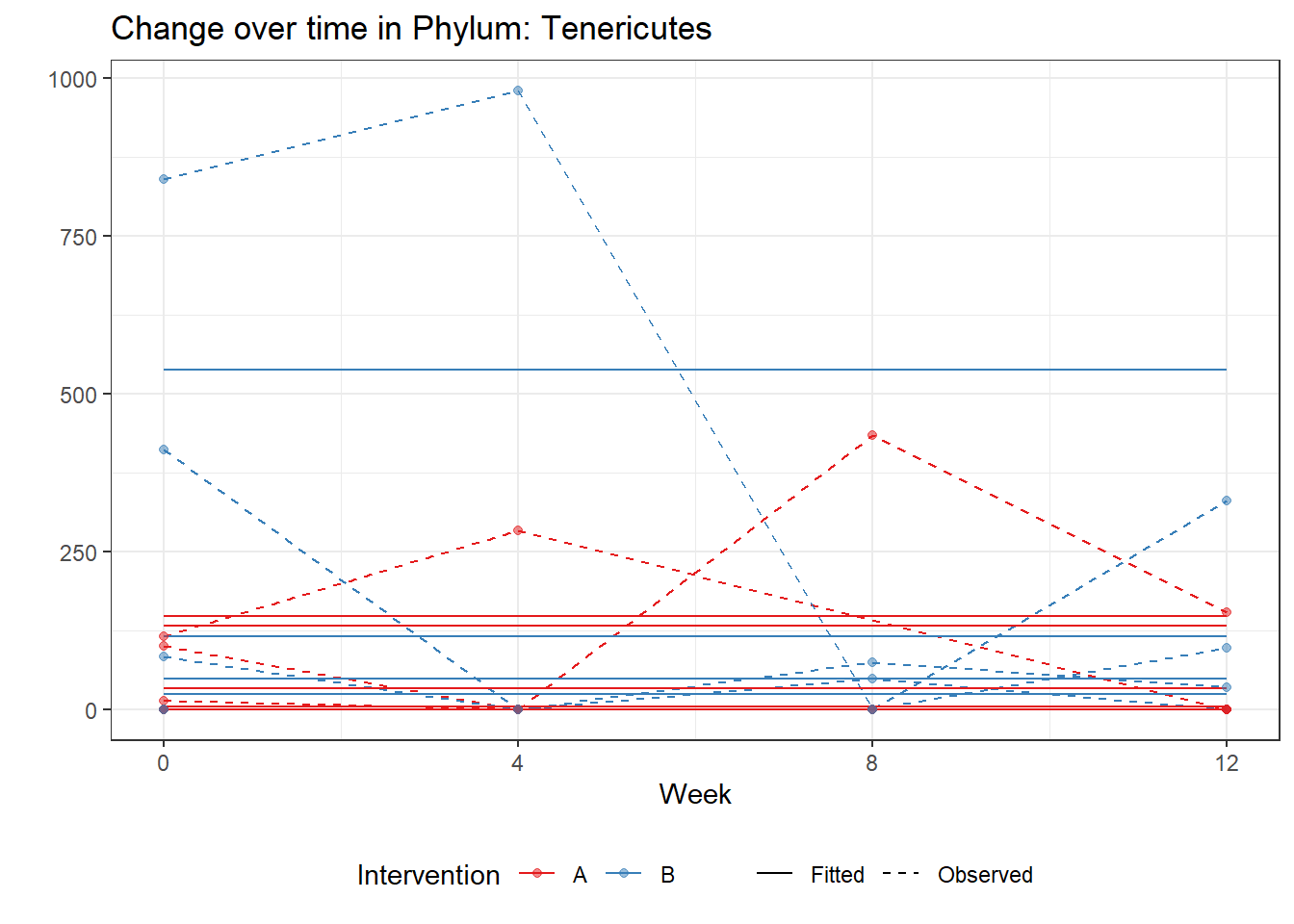
Model: Tenericutes ~ 1 + (1 | SubjectID)
<environment: 0x0000000041f2dbb0>
Link: poisson
Intraclass Correlation (ICC): 0.9129633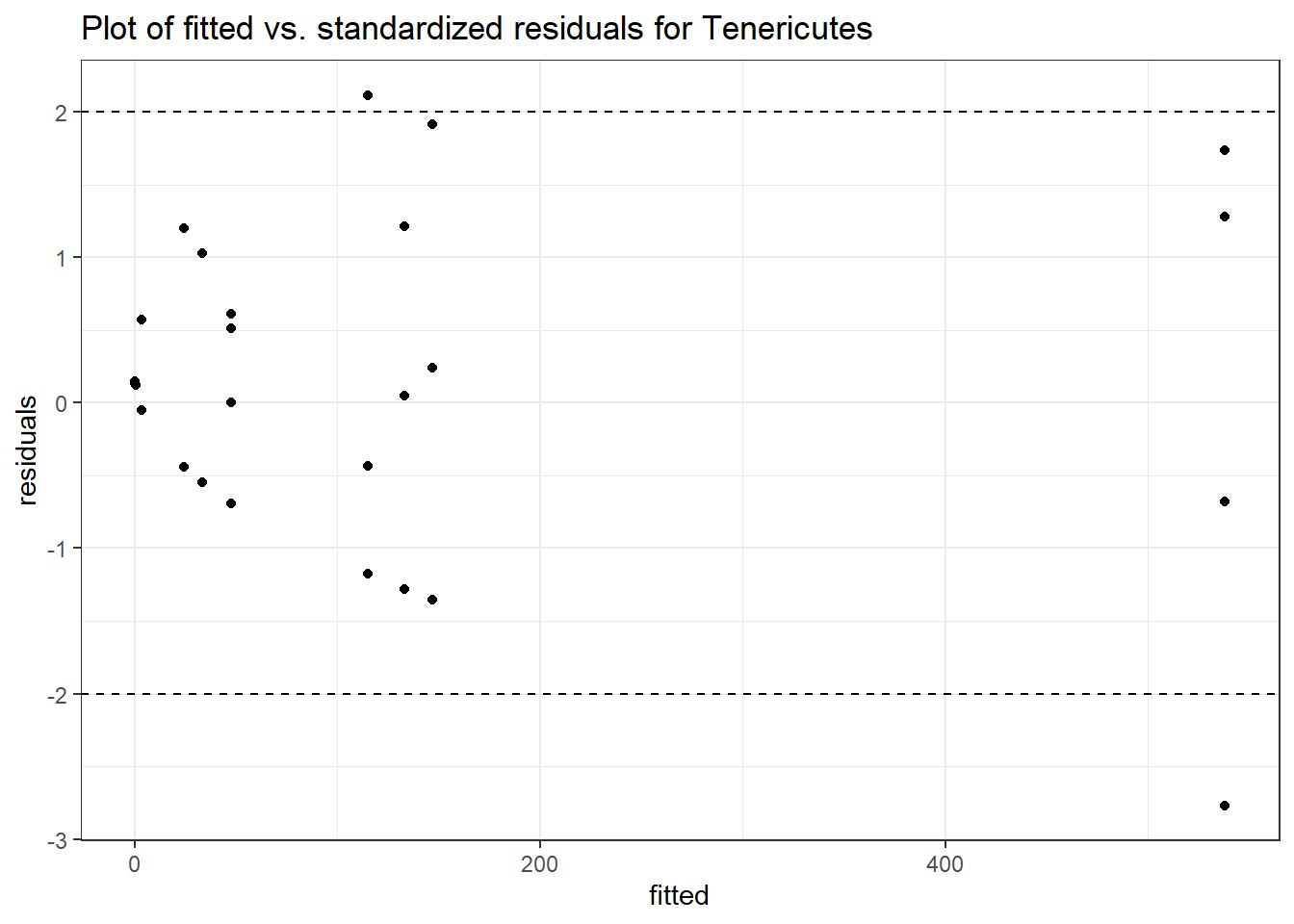
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Tenericutes 2.29296 1.01091 2.2682 0.02332
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 3.2387
Significance test not available
#########################################
#########################################
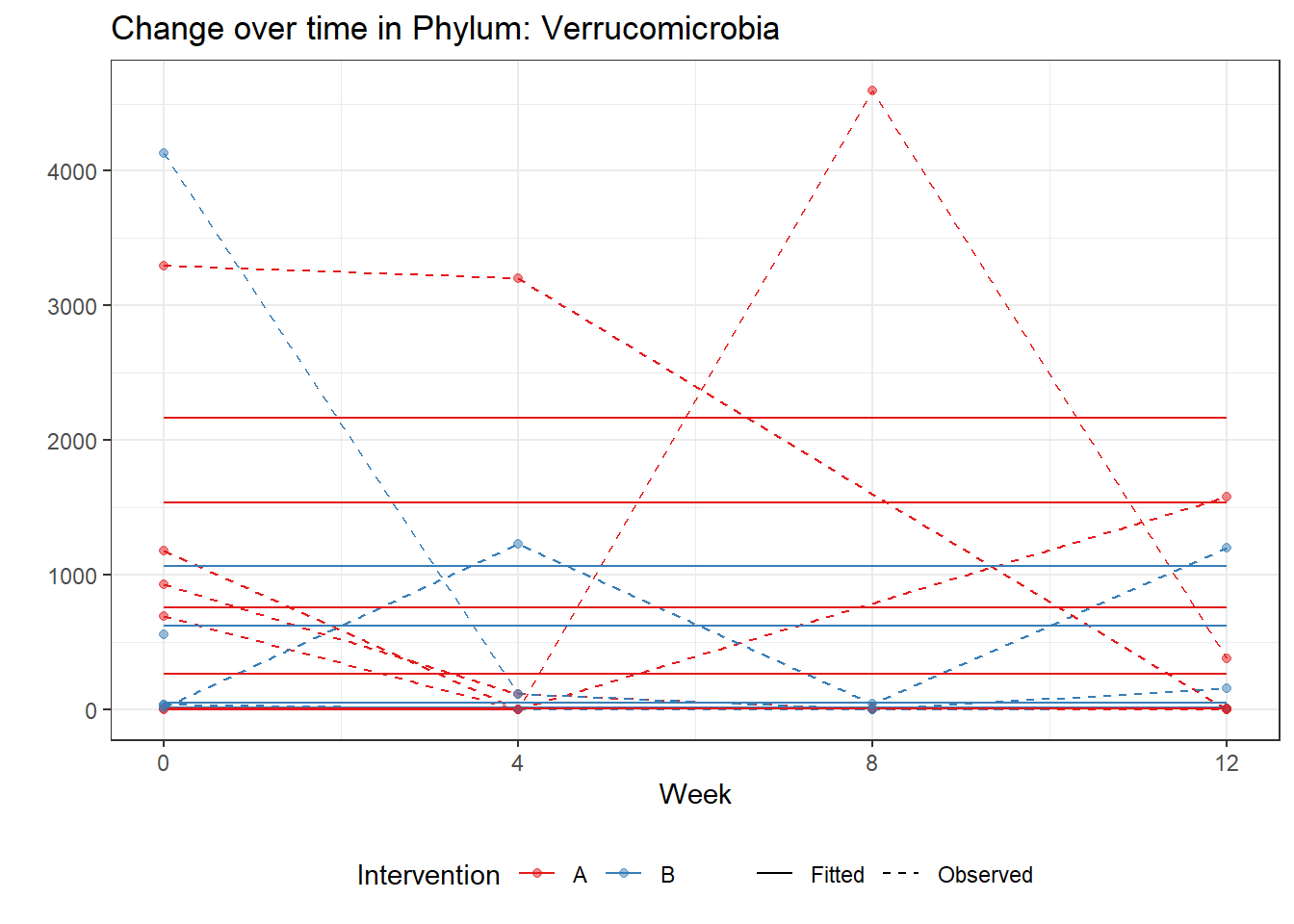
Model: Verrucomicrobia ~ 1 + (1 | SubjectID)
<environment: 0x0000000041f2dbb0>
Link: poisson
Intraclass Correlation (ICC): 0.8745356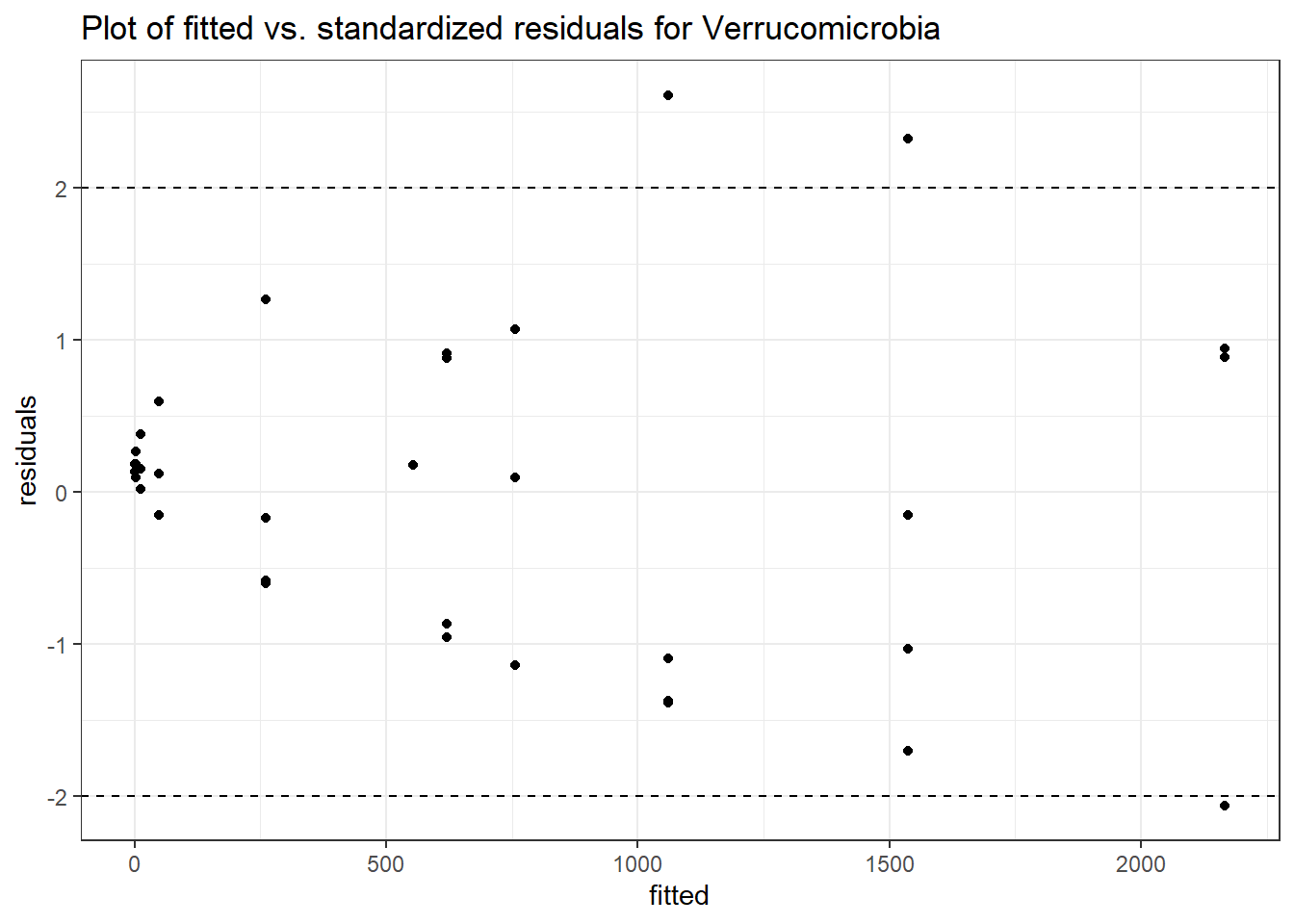
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Verrucomicrobia 4.89307 0.79979 6.11792 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.6401
Significance test not availableFixed Effect of Time
\[\begin{align*} h\left(Y_{ij}\right)&=\beta_{0j} + \beta_{1j}*(Week)_{ij}+ r_{ij}\\ \beta_{0j}&= \gamma_{00} + u_{0j}\\ \beta_{1j}&= \gamma_{10}\\ \end{align*}\]
phylum.fit1 <- glmm_microbiome(mydata=microbiome_data,model.number=1,
taxa.level="Phylum", link="poisson",
model="1 + time + (1|SubjectID)")
#########################################
#########################################
Model: Actinobacteria ~ 1 + time + (1 | SubjectID)
<environment: 0x00000000405d6288>
Link: poisson
Intraclass Correlation (ICC): 0.5426055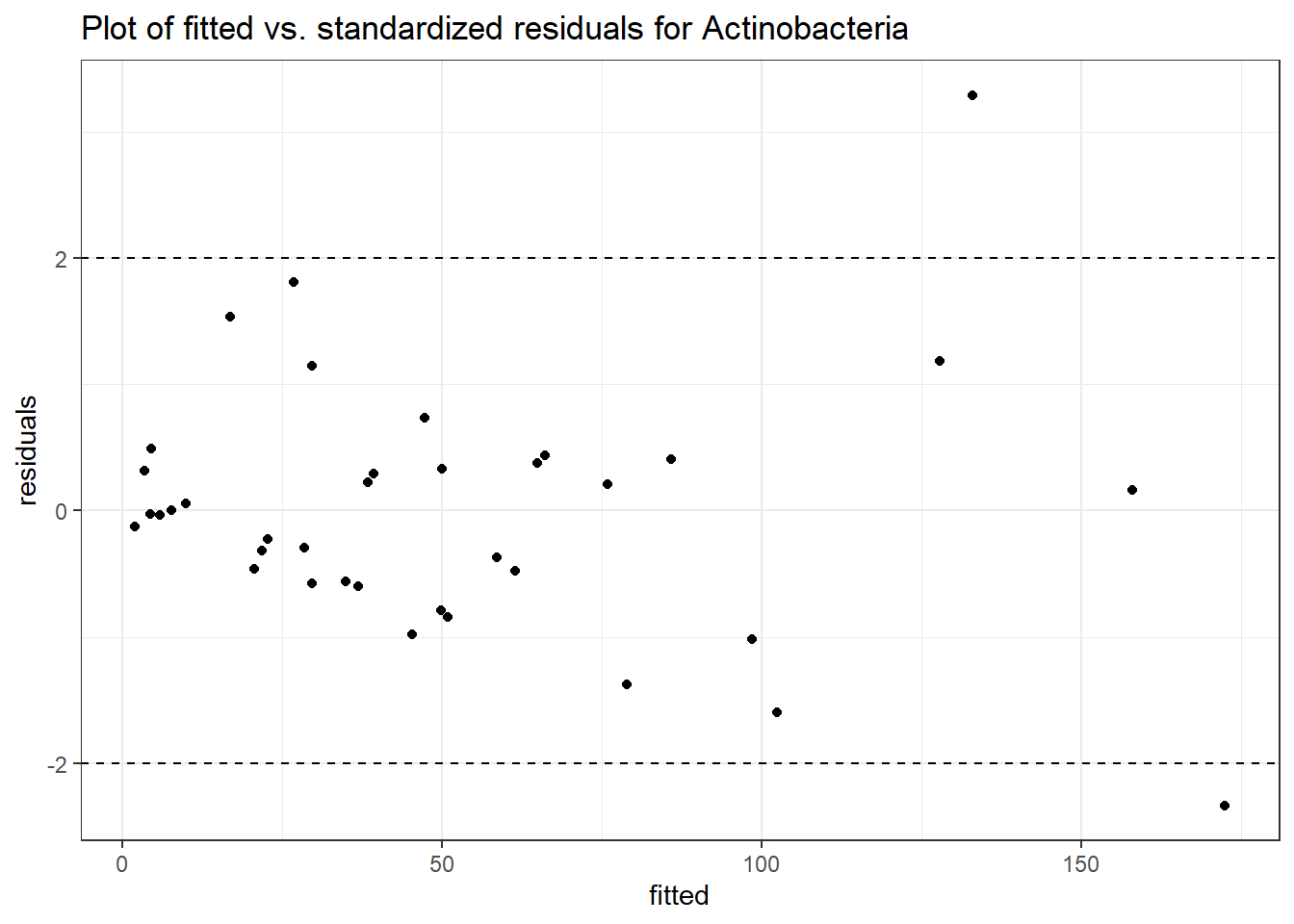
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Actinobacteria 3.89720 0.33155 11.75456 0
time Actinobacteria -0.26049 0.02241 -11.62621 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.0892
Significance test not available
#########################################
#########################################
Model: Bacteroidetes ~ 1 + time + (1 | SubjectID)
<environment: 0x00000000405d6288>
Link: poisson
Intraclass Correlation (ICC): 0.09952787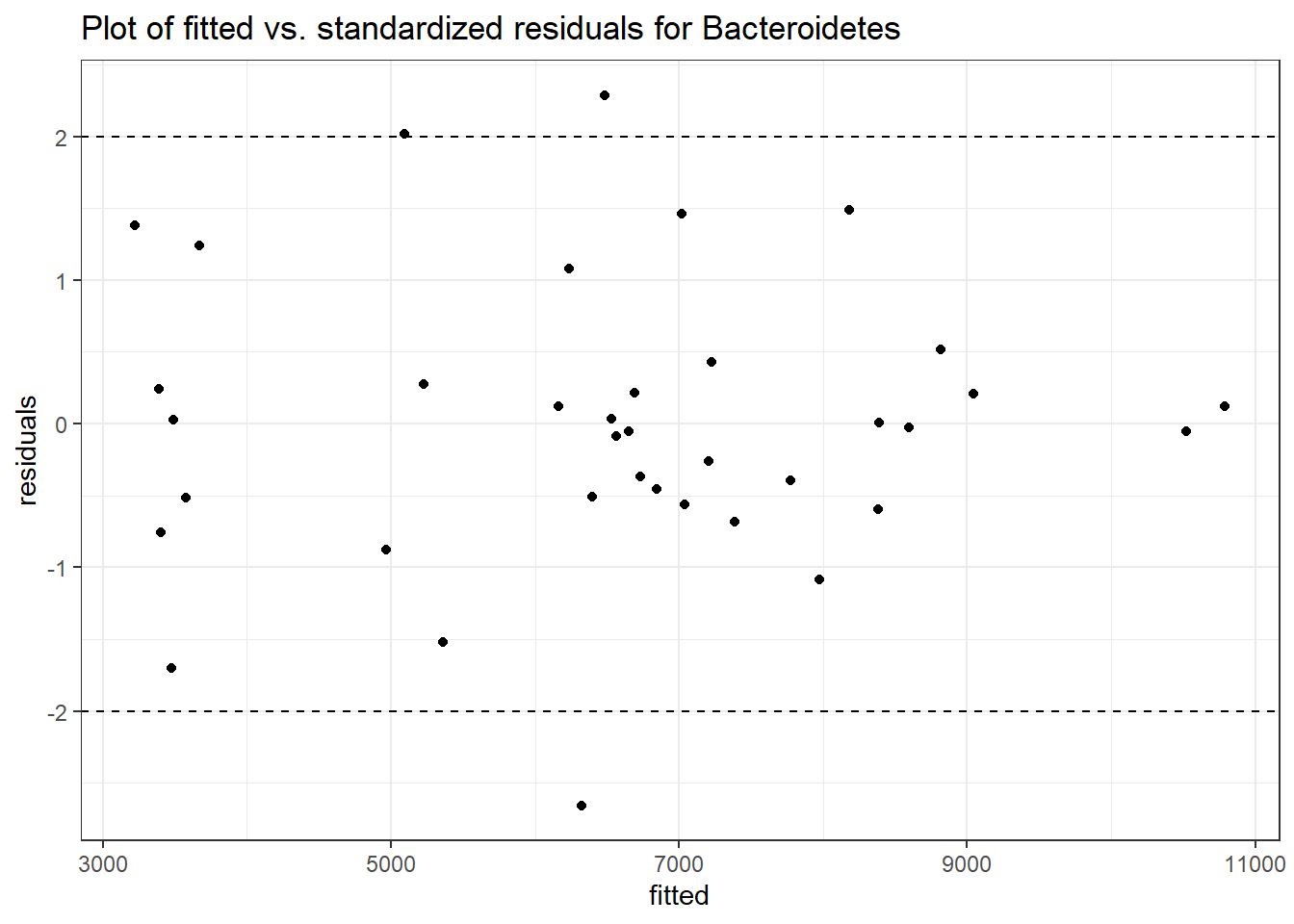
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroidetes 8.77944 0.10029 87.53848 0
time Bacteroidetes -0.02541 0.00189 -13.45403 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.33246
Significance test not available
#########################################
#########################################
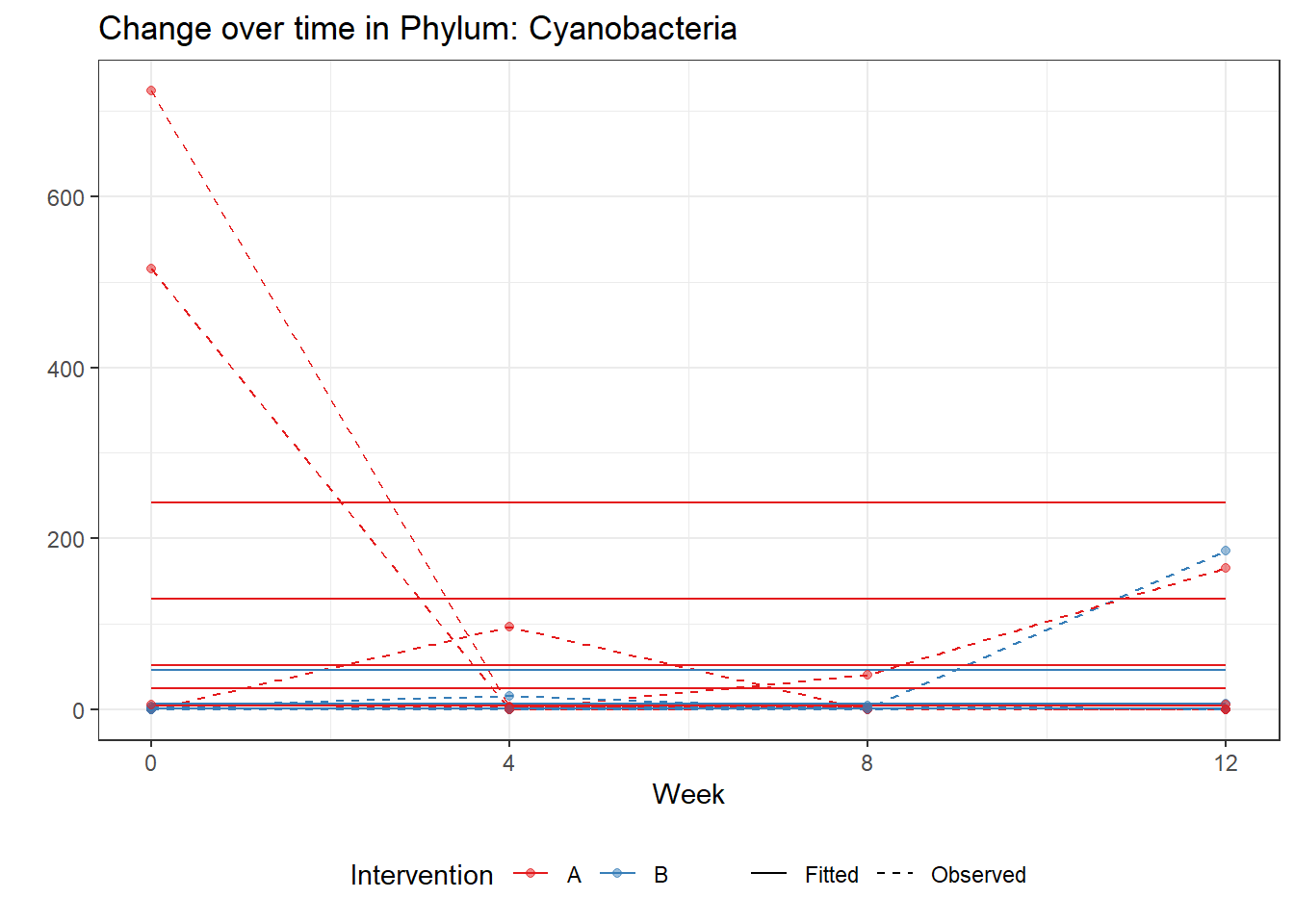
Model: Cyanobacteria ~ 1 + time + (1 | SubjectID)
<environment: 0x00000000405d6288>
Link: poisson
Intraclass Correlation (ICC): 0.8578909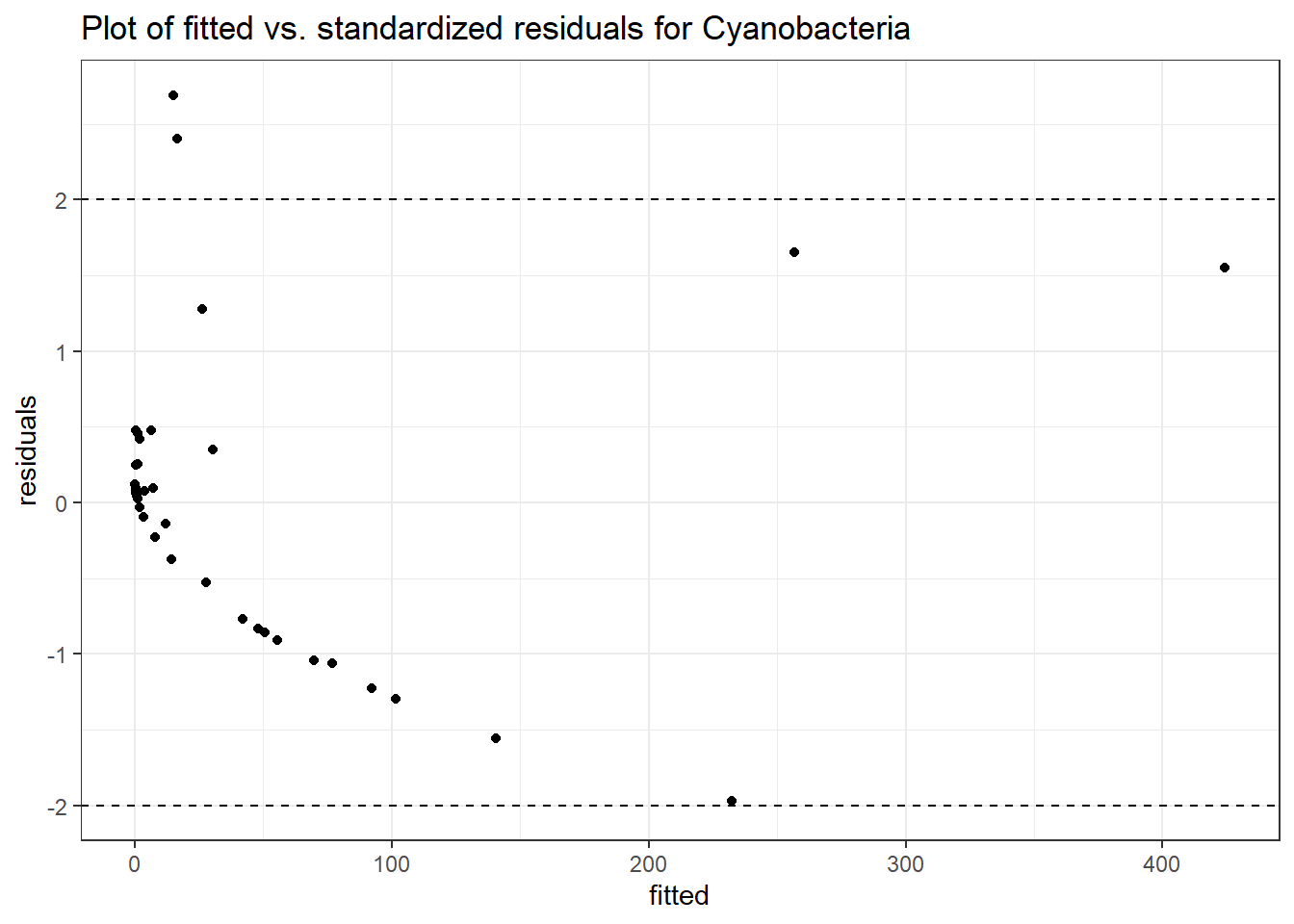
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Cyanobacteria 2.55435 0.75897 3.36553 0.00076
time Cyanobacteria -0.60192 0.02518 -23.90231 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.457
Significance test not available
#########################################
#########################################
Model: Euryarchaeota ~ 1 + time + (1 | SubjectID)
<environment: 0x00000000405d6288>
Link: poisson
Intraclass Correlation (ICC): 0.7860171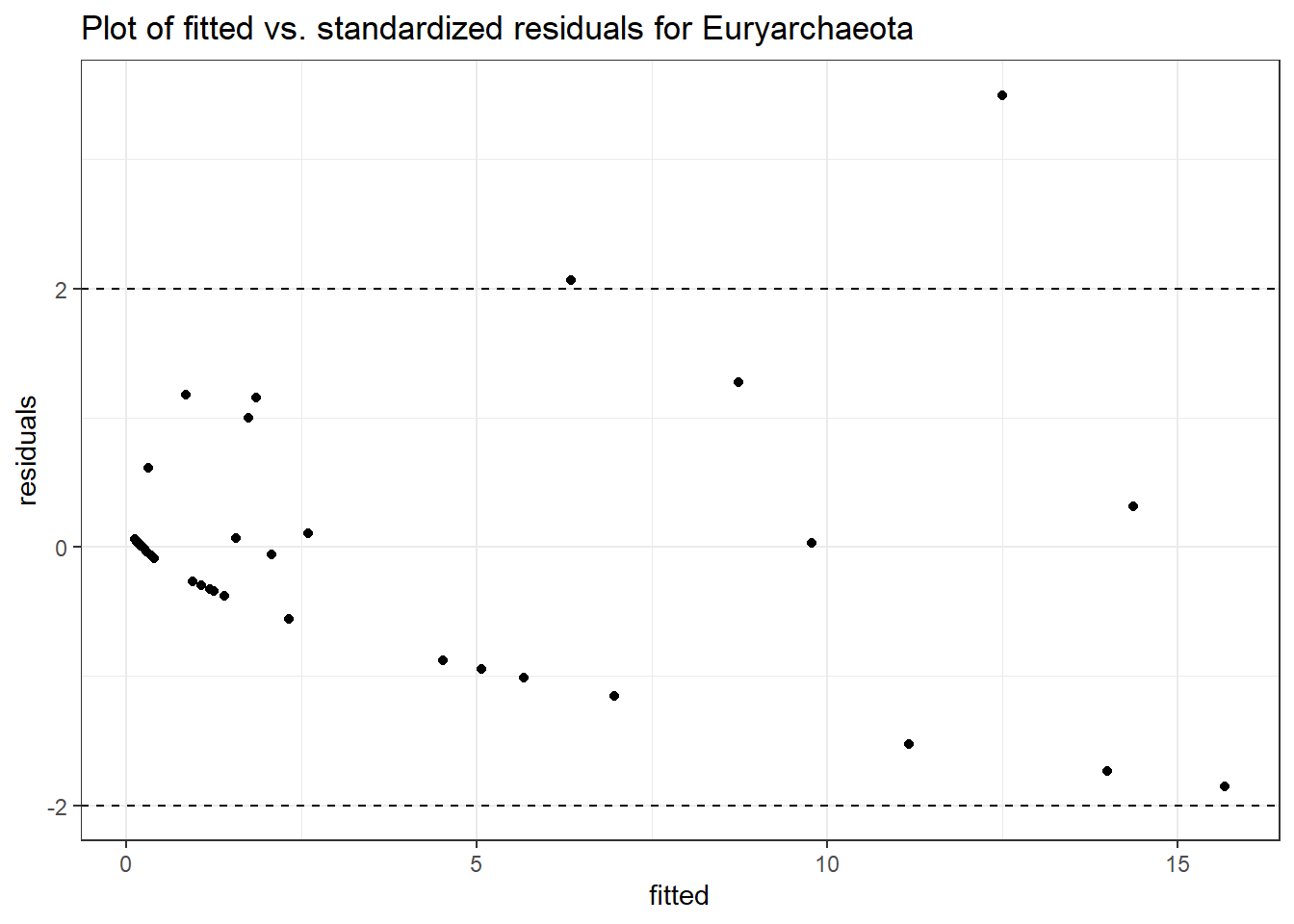
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Euryarchaeota 0.34352 0.63946 0.53719 0.59113
time Euryarchaeota -0.11323 0.07861 -1.44043 0.14975
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.9166
Significance test not available
#########################################
#########################################
Model: Firmicutes ~ 1 + time + (1 | SubjectID)
<environment: 0x00000000405d6288>
Link: poisson
Intraclass Correlation (ICC): 0.096963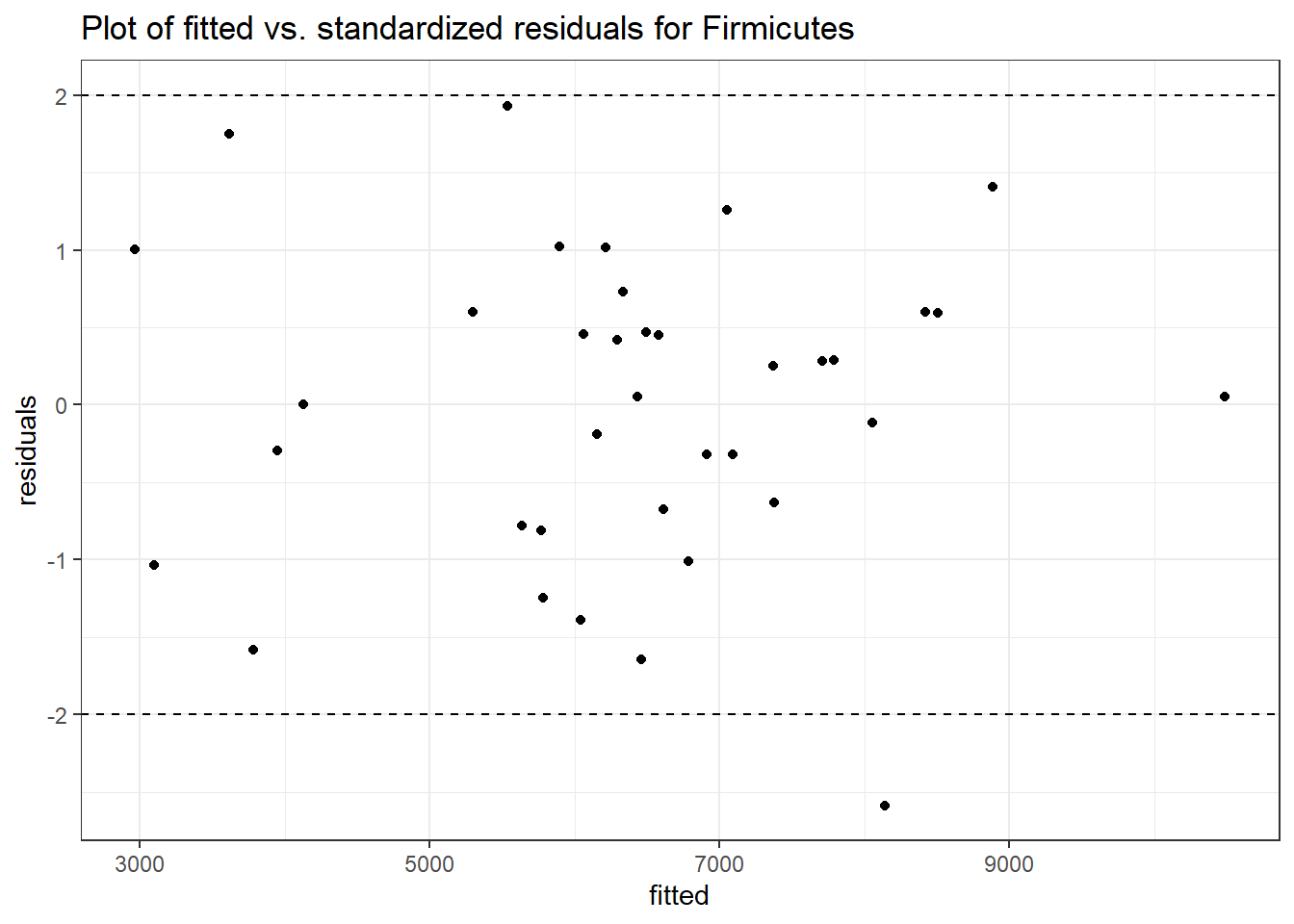
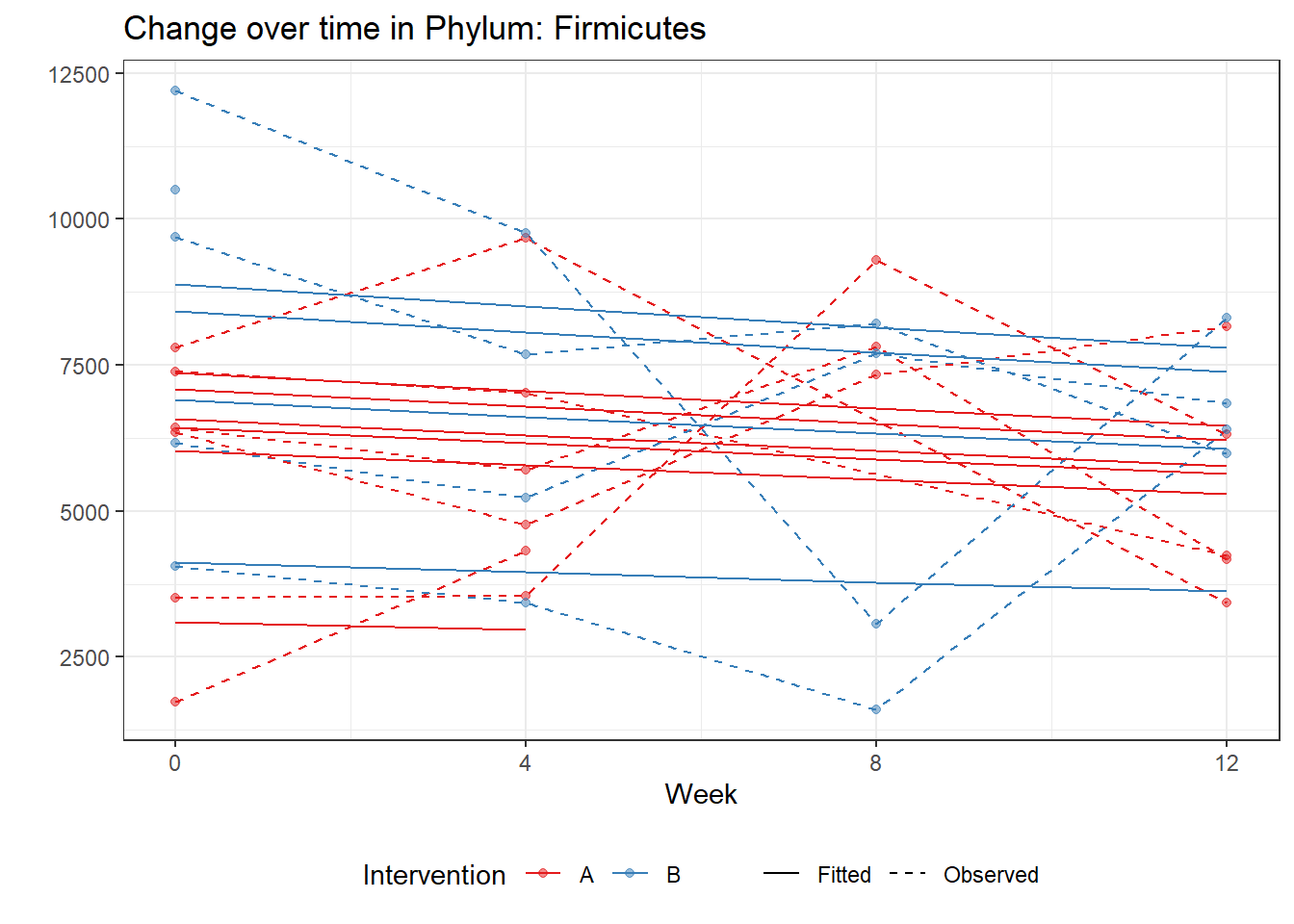
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Firmicutes 8.78496 0.09885 88.86992 0
time Firmicutes -0.04391 0.00187 -23.47528 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.32768
Significance test not available
#########################################
#########################################
Model: Fusobacteria ~ 1 + time + (1 | SubjectID)
<environment: 0x00000000405d6288>
Link: poisson
Intraclass Correlation (ICC): 0.8416402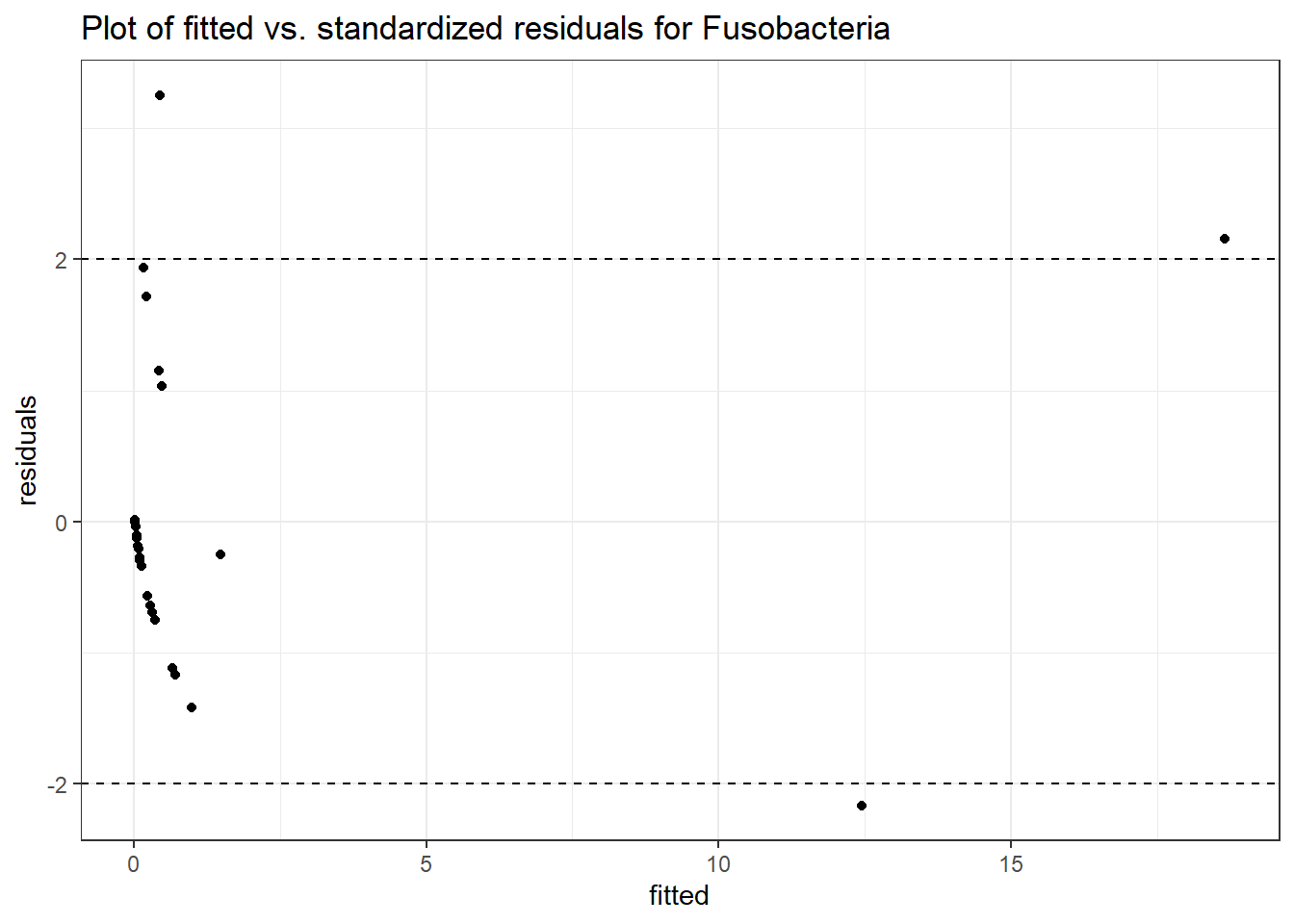
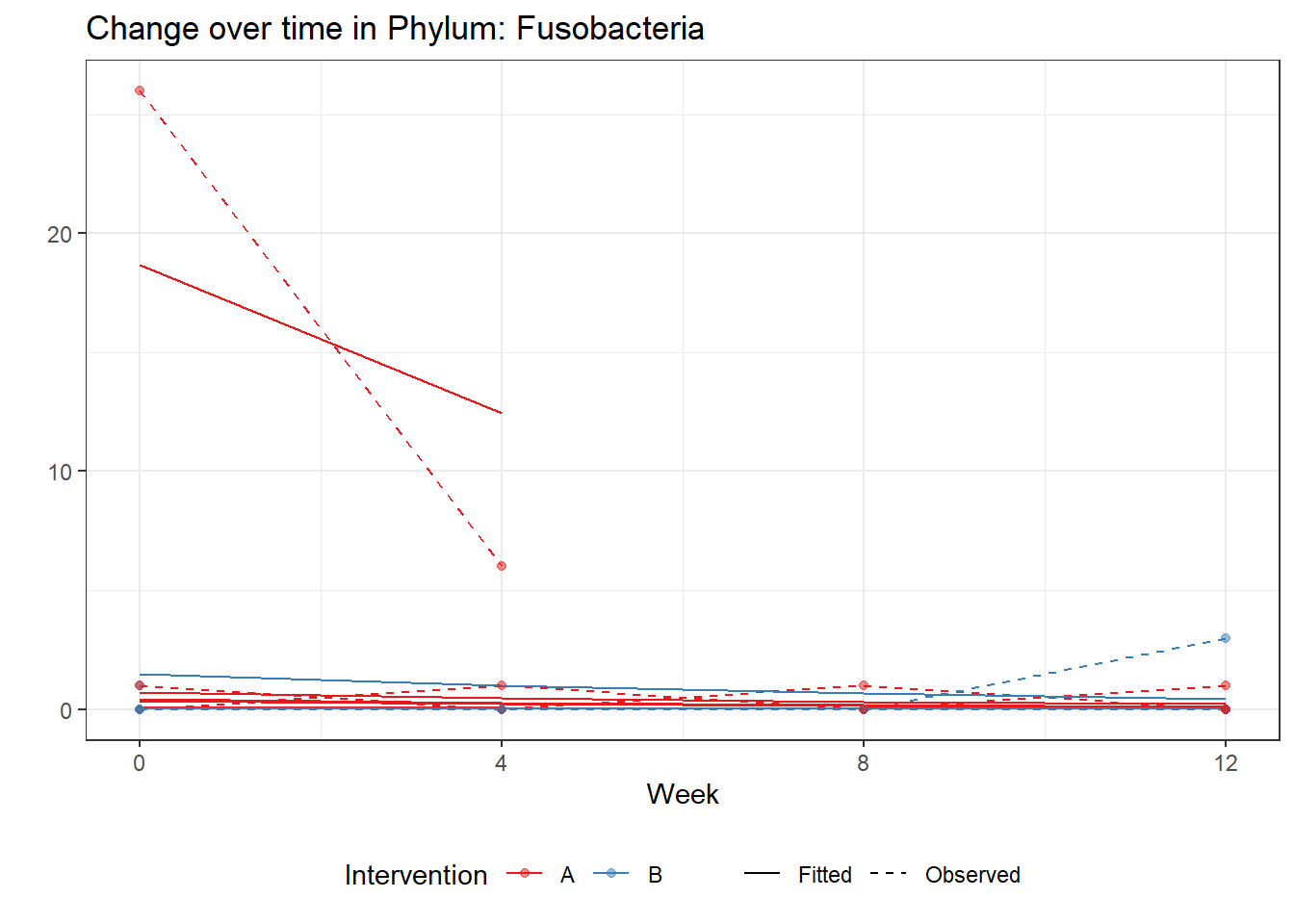
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Fusobacteria -1.77193 1.10786 -1.59941 0.10973
time Fusobacteria -0.40467 0.24249 -1.66879 0.09516
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.3054
Significance test not available
#########################################
#########################################
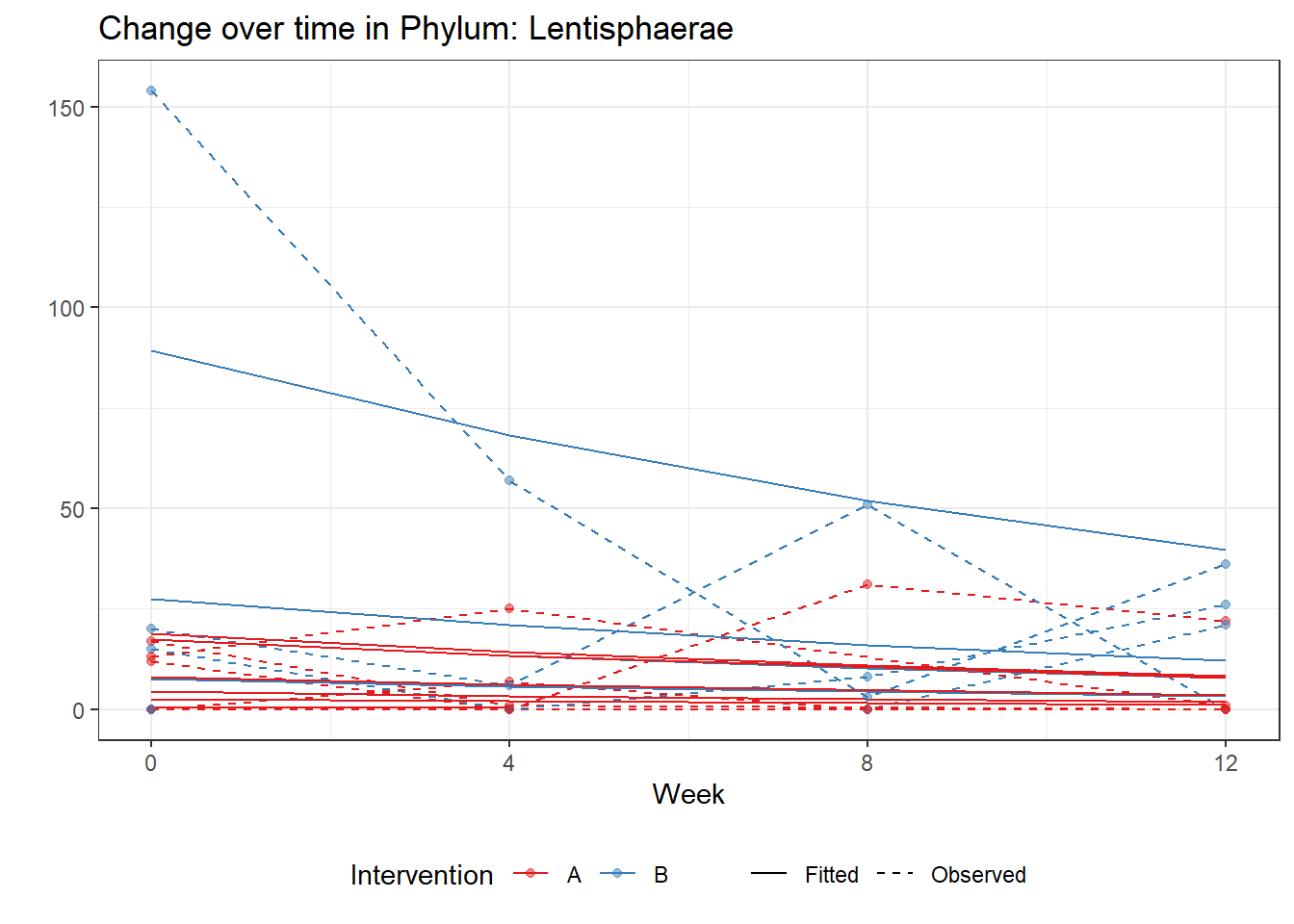
Model: Lentisphaerae ~ 1 + time + (1 | SubjectID)
<environment: 0x00000000405d6288>
Link: poisson
Intraclass Correlation (ICC): 0.7110489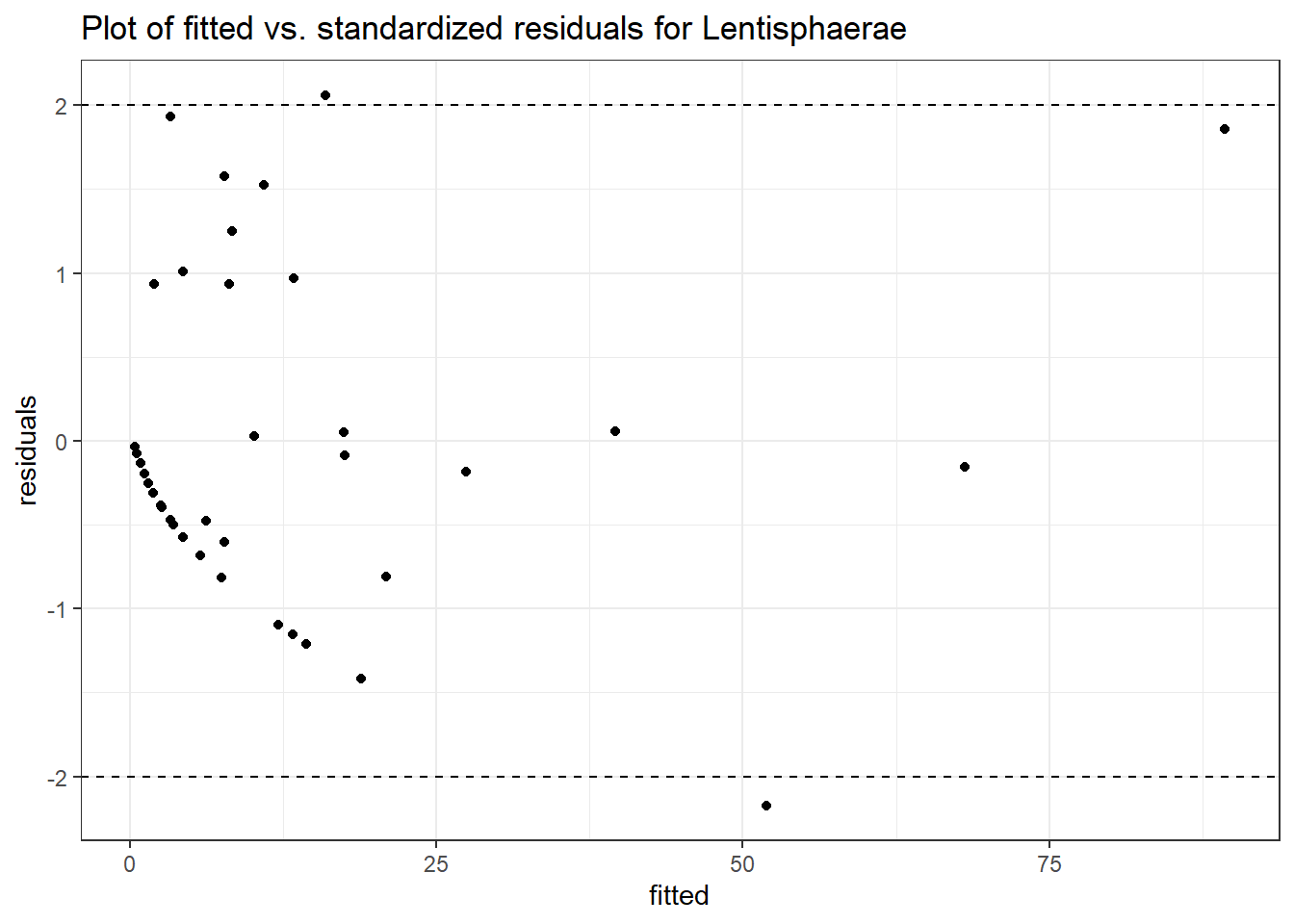
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lentisphaerae 1.96678 0.49800 3.94935 8e-05
time Lentisphaerae -0.27109 0.03998 -6.78139 0e+00
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.5687
Significance test not available
#########################################
#########################################
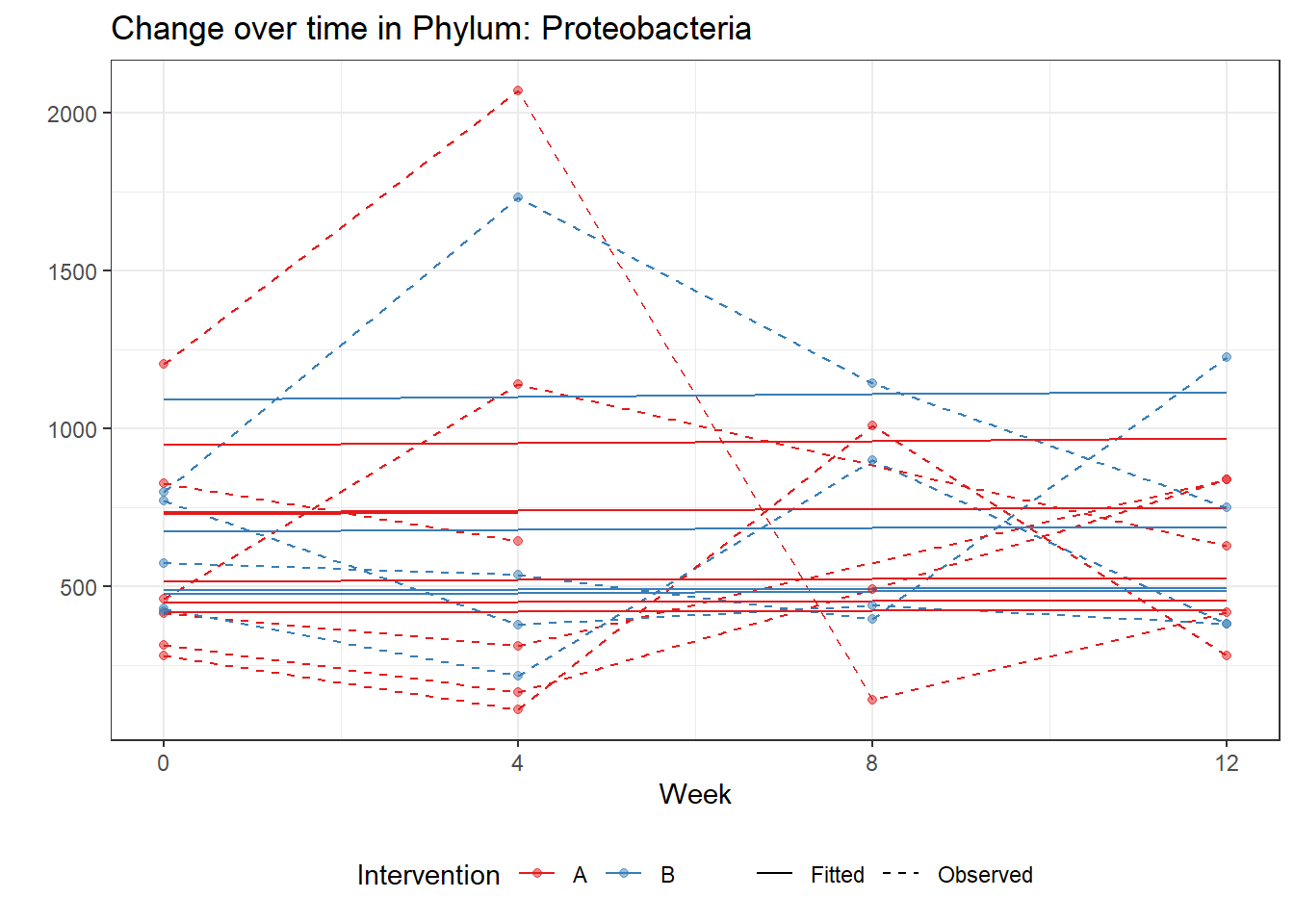
Model: Proteobacteria ~ 1 + time + (1 | SubjectID)
<environment: 0x00000000405d6288>
Link: poisson
Intraclass Correlation (ICC): 0.09290682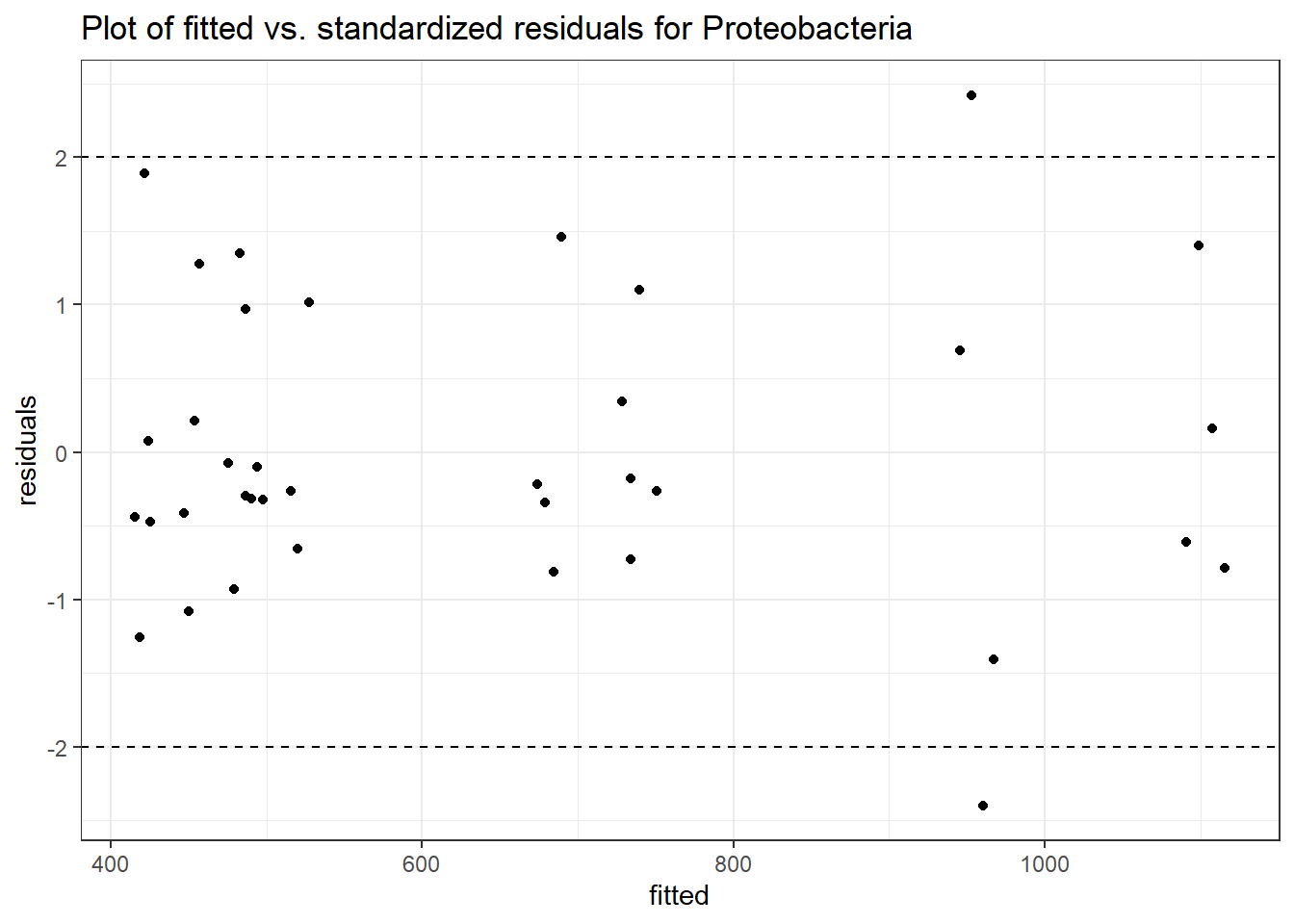
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Proteobacteria 6.39332 0.09708 65.85861 0.00000
time Proteobacteria 0.00755 0.00585 1.29033 0.19694
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.32004
Significance test not available
#########################################
#########################################
Model: Tenericutes ~ 1 + time + (1 | SubjectID)
<environment: 0x00000000405d6288>
Link: poisson
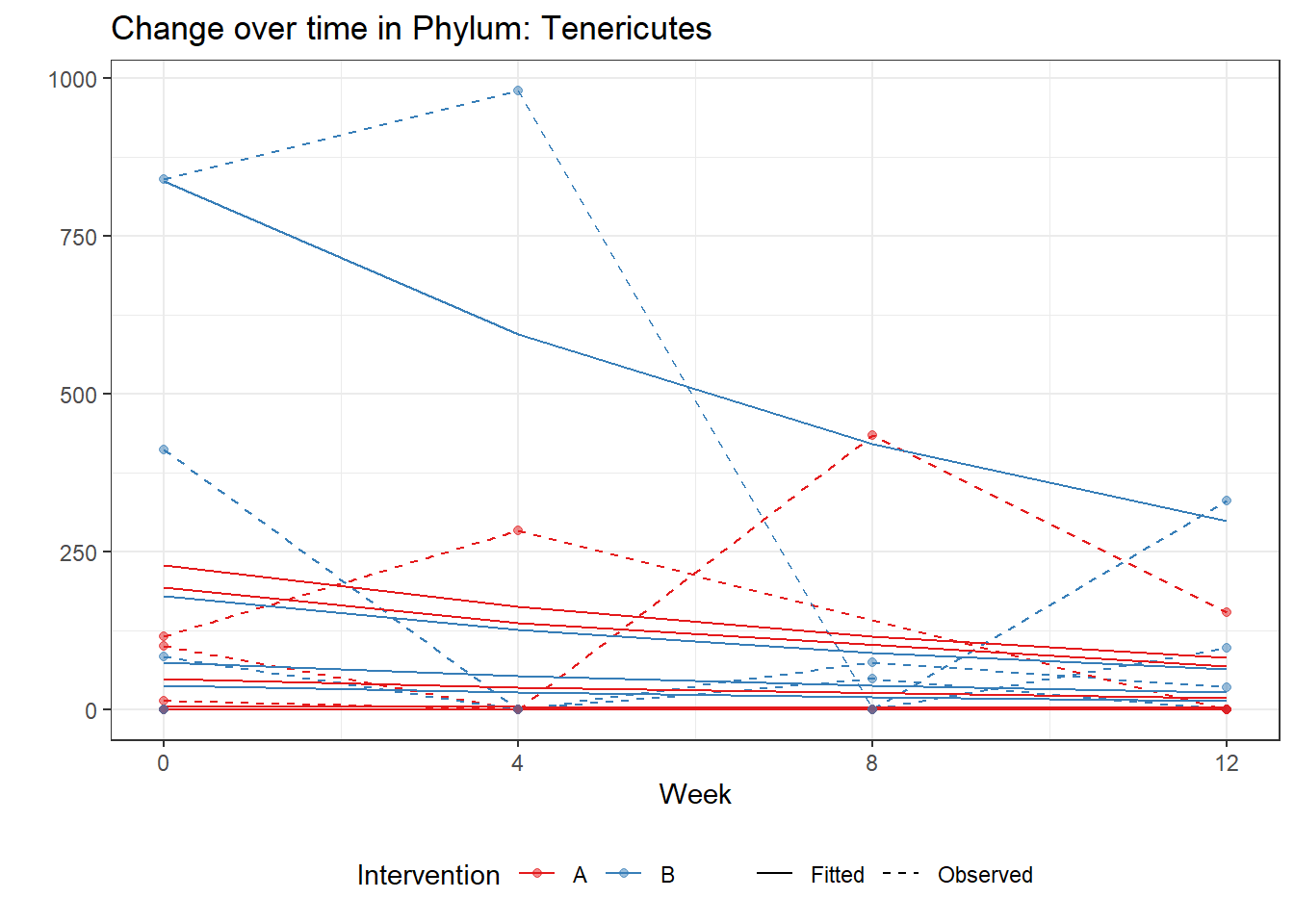
Intraclass Correlation (ICC): 0.9180696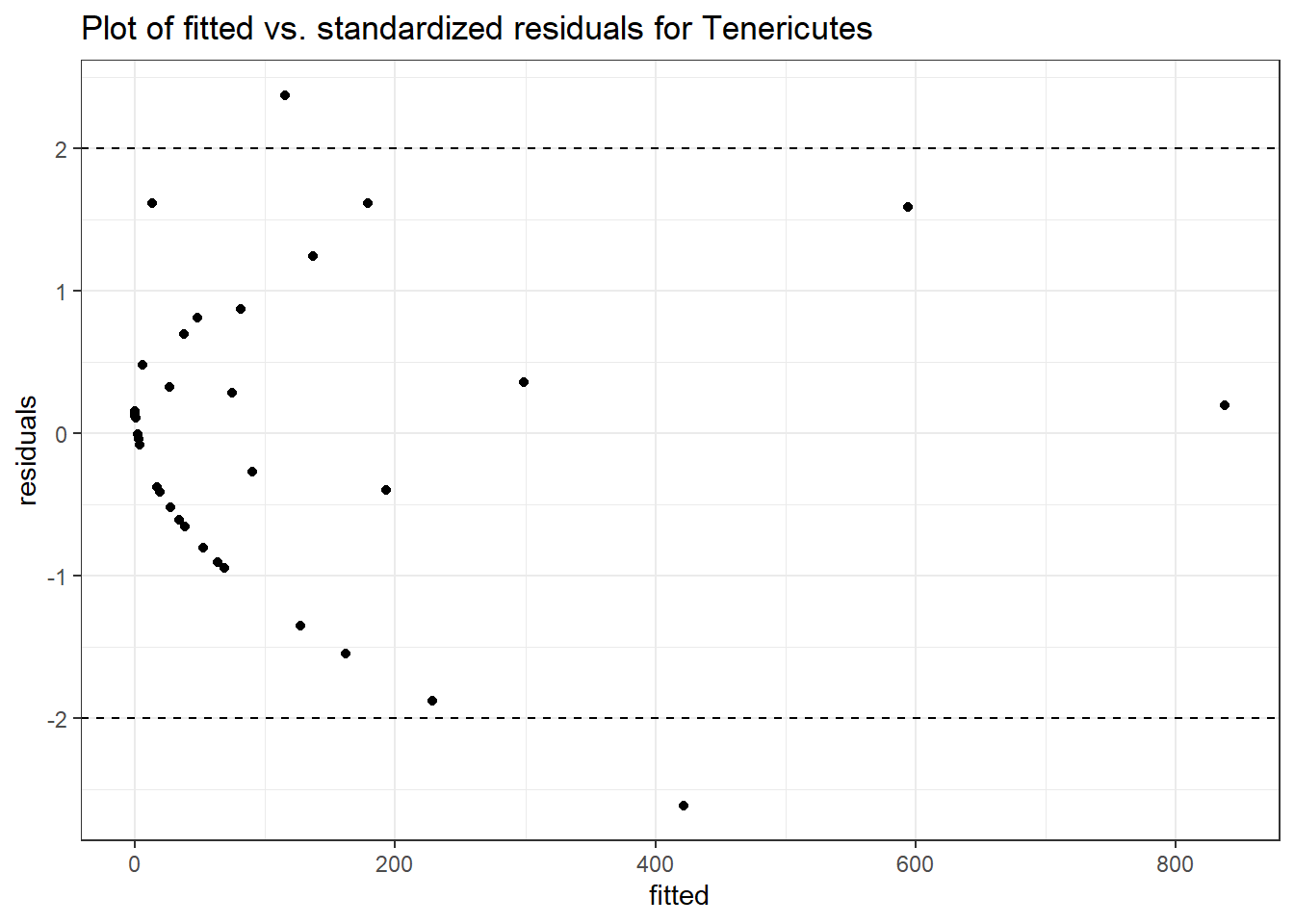
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Tenericutes 2.64267 1.04333 2.53291 0.01131
time Tenericutes -0.34365 0.01482 -23.18291 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 3.3475
Significance test not available
#########################################
#########################################
Model: Verrucomicrobia ~ 1 + time + (1 | SubjectID)
<environment: 0x00000000405d6288>
Link: poisson
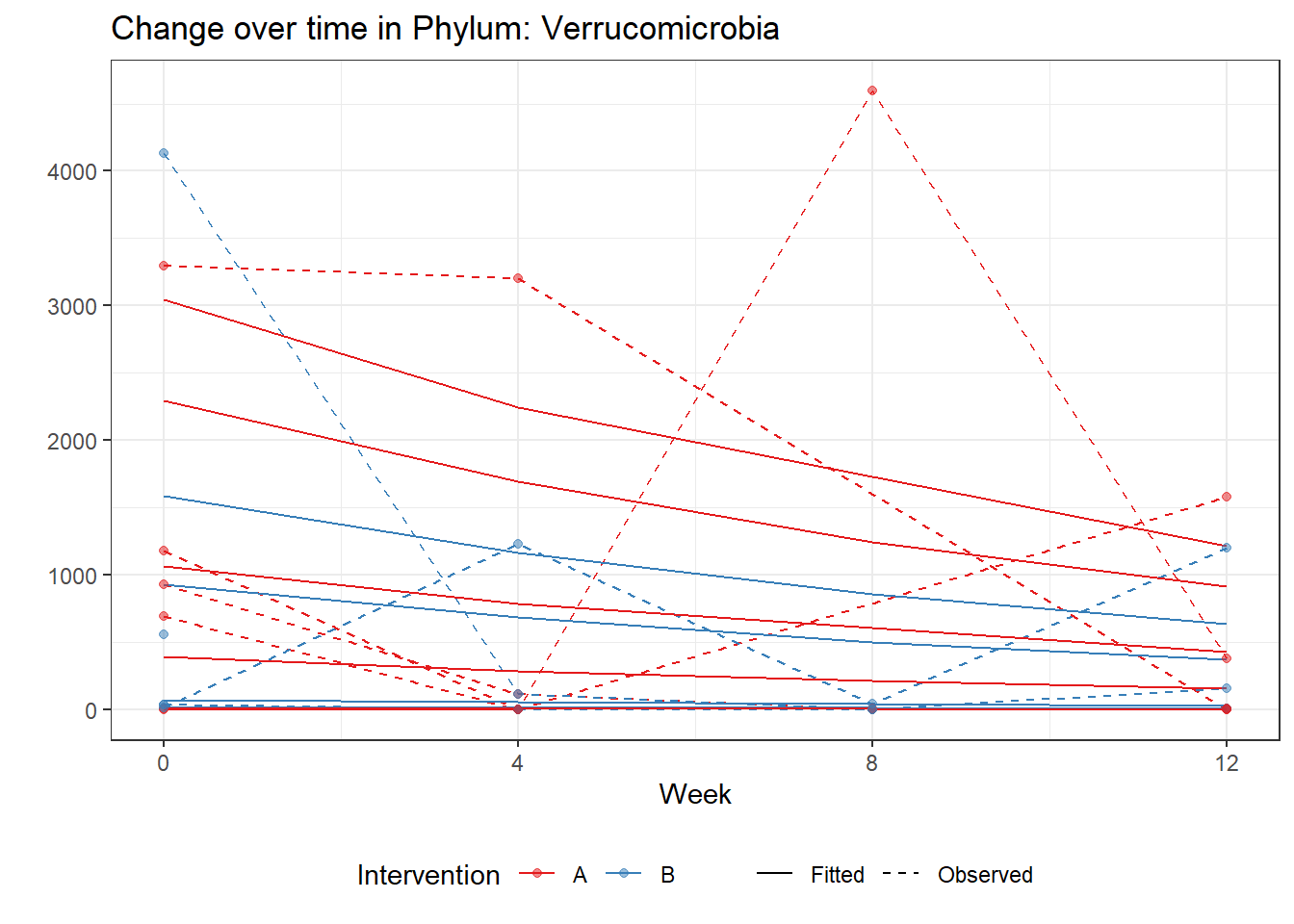
Intraclass Correlation (ICC): 0.8761372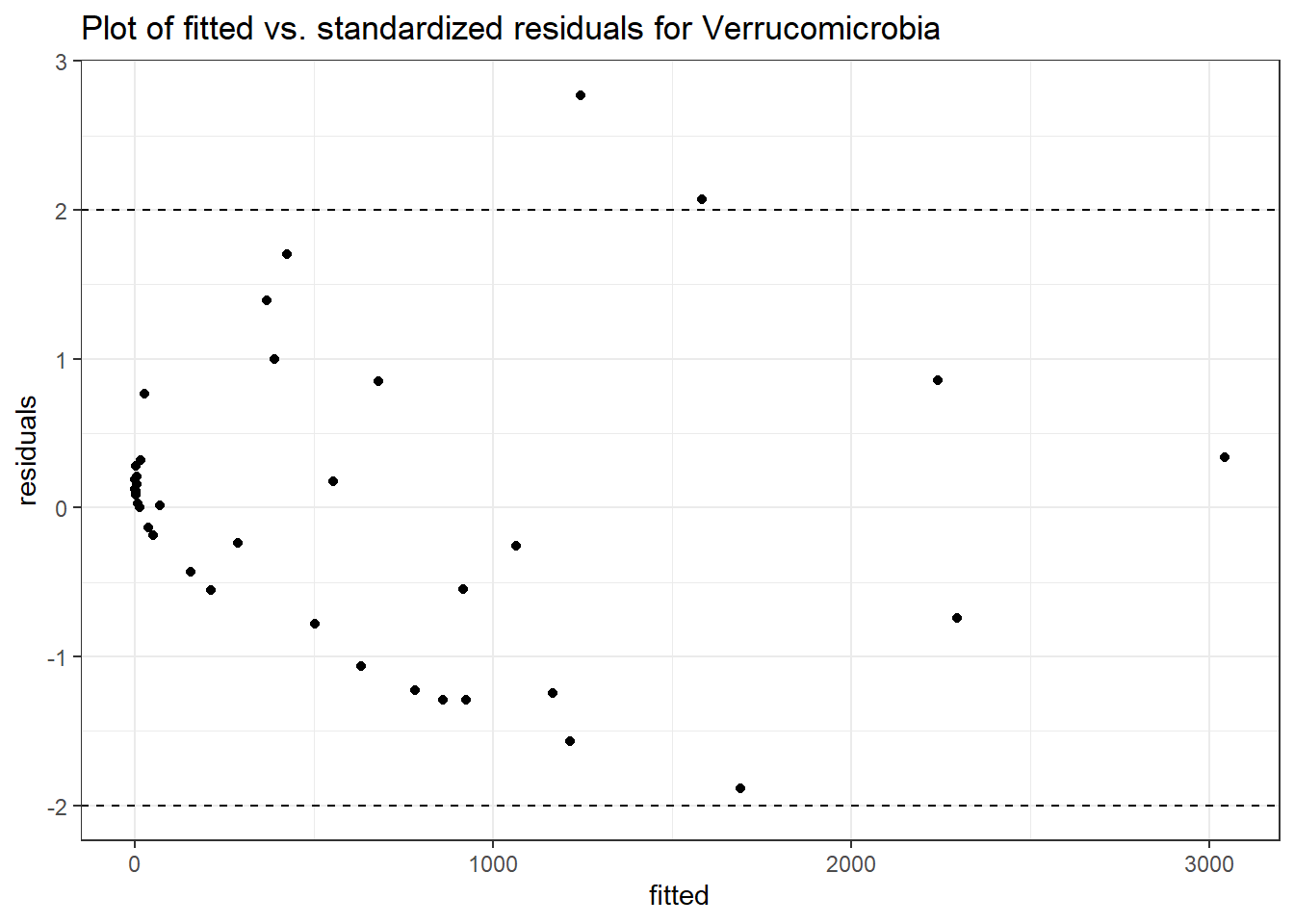
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Verrucomicrobia 5.22355 0.80562 6.48390 0
time Verrucomicrobia -0.30578 0.00609 -50.22346 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.6596
Significance test not availableSo, we found that time does significantly influence at least some of the bacteria over time. This is contrary to the results when we used the normal theory model.
Covariates: Time + Gender
\[\begin{align*} h\left(Y_{ij}\right)&=\beta_{0j} + \beta_{1j}*(Week)_{ij} + r_{ij}\\ \beta_{0j}&= \gamma_{00} + \gamma_{01}(Gender)_{j} + u_{0j}\\ \beta_{1j}&= \gamma_{10}\\ \end{align*}\]
Gender is coded as female (effect of being female compared to males).
phylum.fit2 <- glmm_microbiome(mydata=microbiome_data,model.number=2,
taxa.level="Phylum", link="poisson",
model="1 + time + female + (1|SubjectID)")
#########################################
#########################################
Model: Actinobacteria ~ 1 + time + female + (1 | SubjectID)
<environment: 0x000000002f679a68>
Link: poisson
Intraclass Correlation (ICC): 0.4883953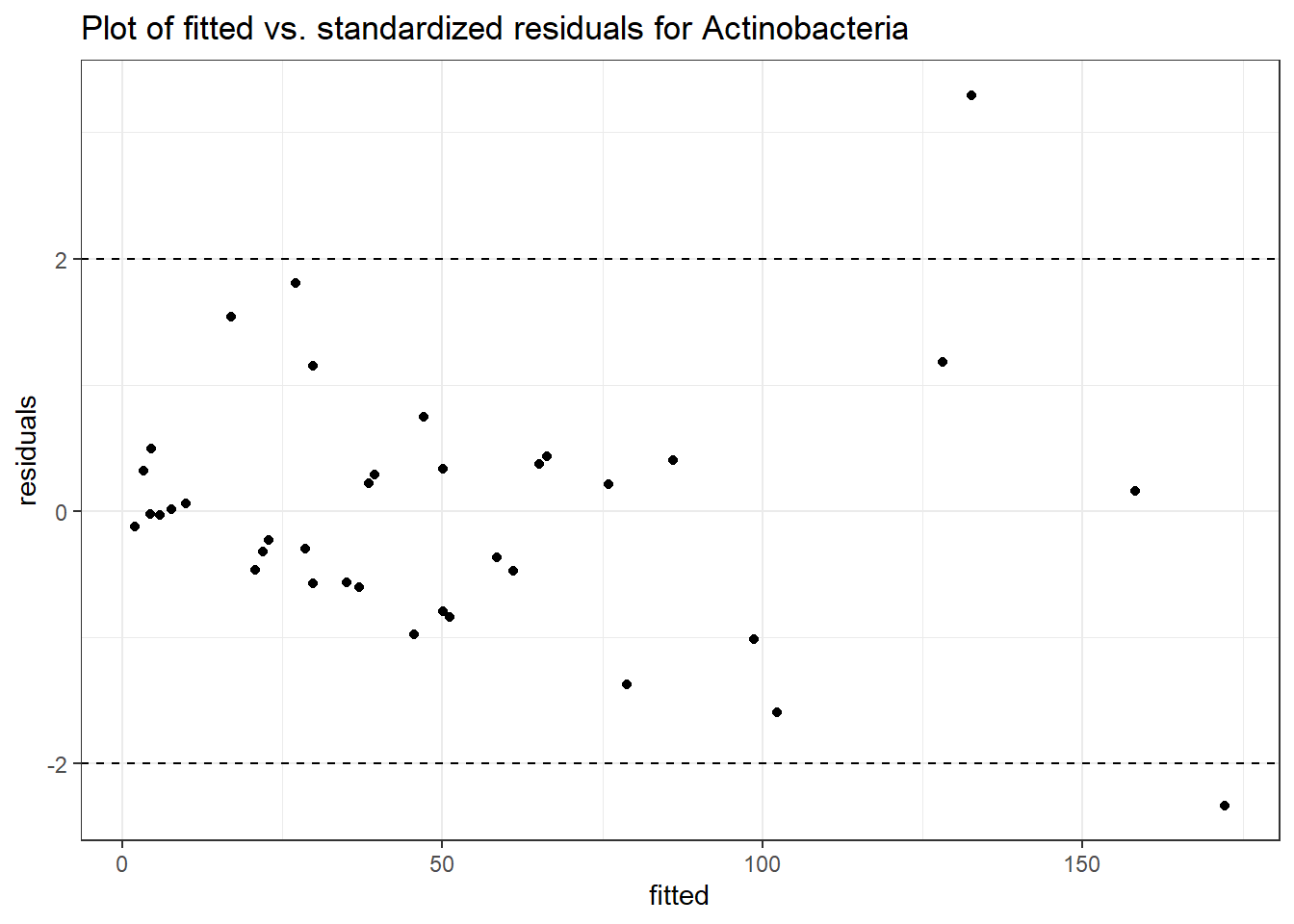
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Actinobacteria 3.24428 0.49851 6.50796 0.00000
time Actinobacteria -0.26066 0.02240 -11.63501 0.00000
female Actinobacteria 1.02306 0.62074 1.64812 0.09933
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.97705
Significance test not available
#########################################
#########################################
Model: Bacteroidetes ~ 1 + time + female + (1 | SubjectID)
<environment: 0x000000002f679a68>
Link: poisson
Intraclass Correlation (ICC): 0.07091696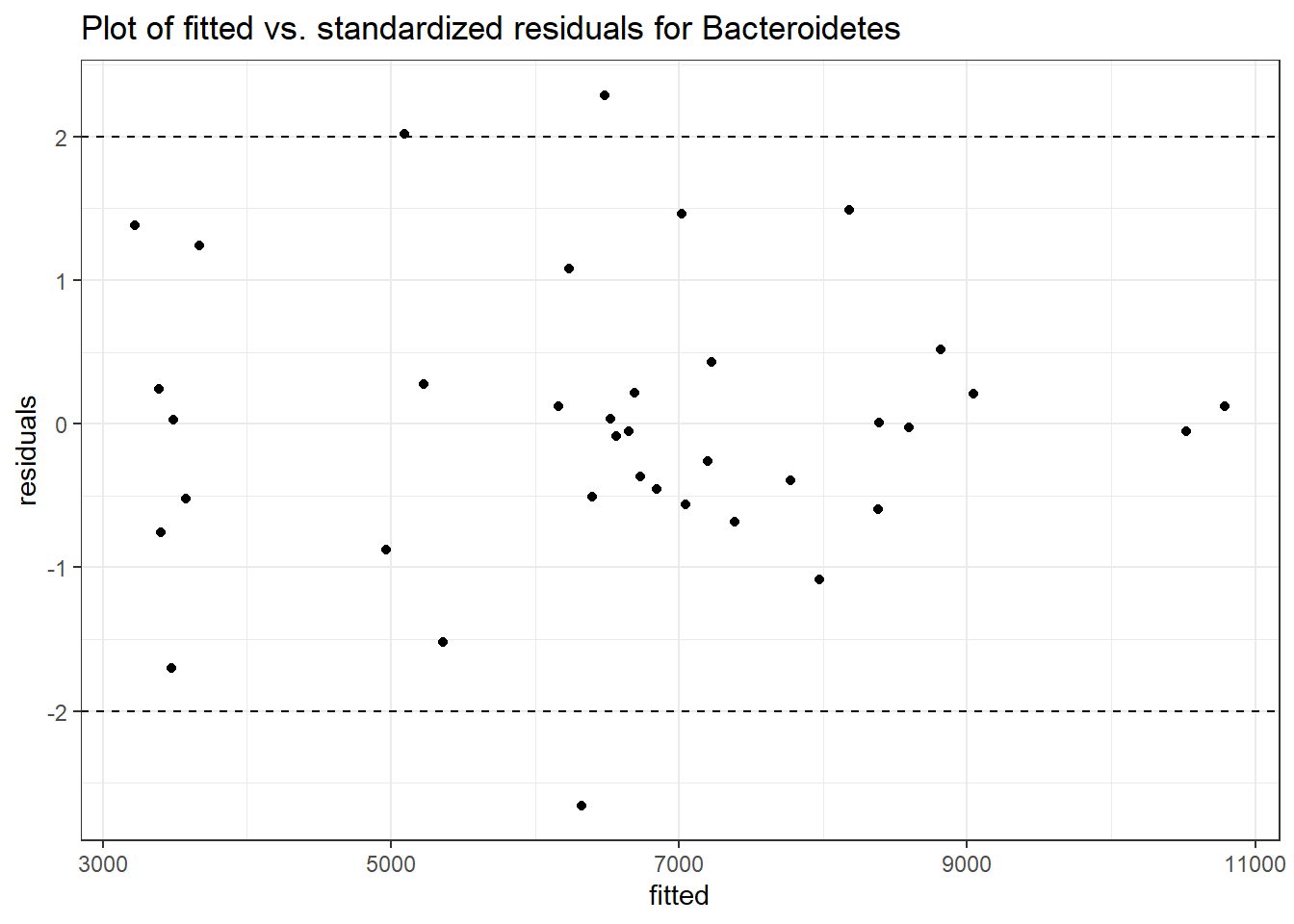
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroidetes 9.02407 0.13819 65.29994 0.00000
time Bacteroidetes -0.02541 0.00189 -13.45618 0.00000
female Bacteroidetes -0.38442 0.17322 -2.21917 0.02647
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.27628
Significance test not available
#########################################
#########################################
Model: Cyanobacteria ~ 1 + time + female + (1 | SubjectID)
<environment: 0x000000002f679a68>
Link: poisson
Intraclass Correlation (ICC): 0.8578945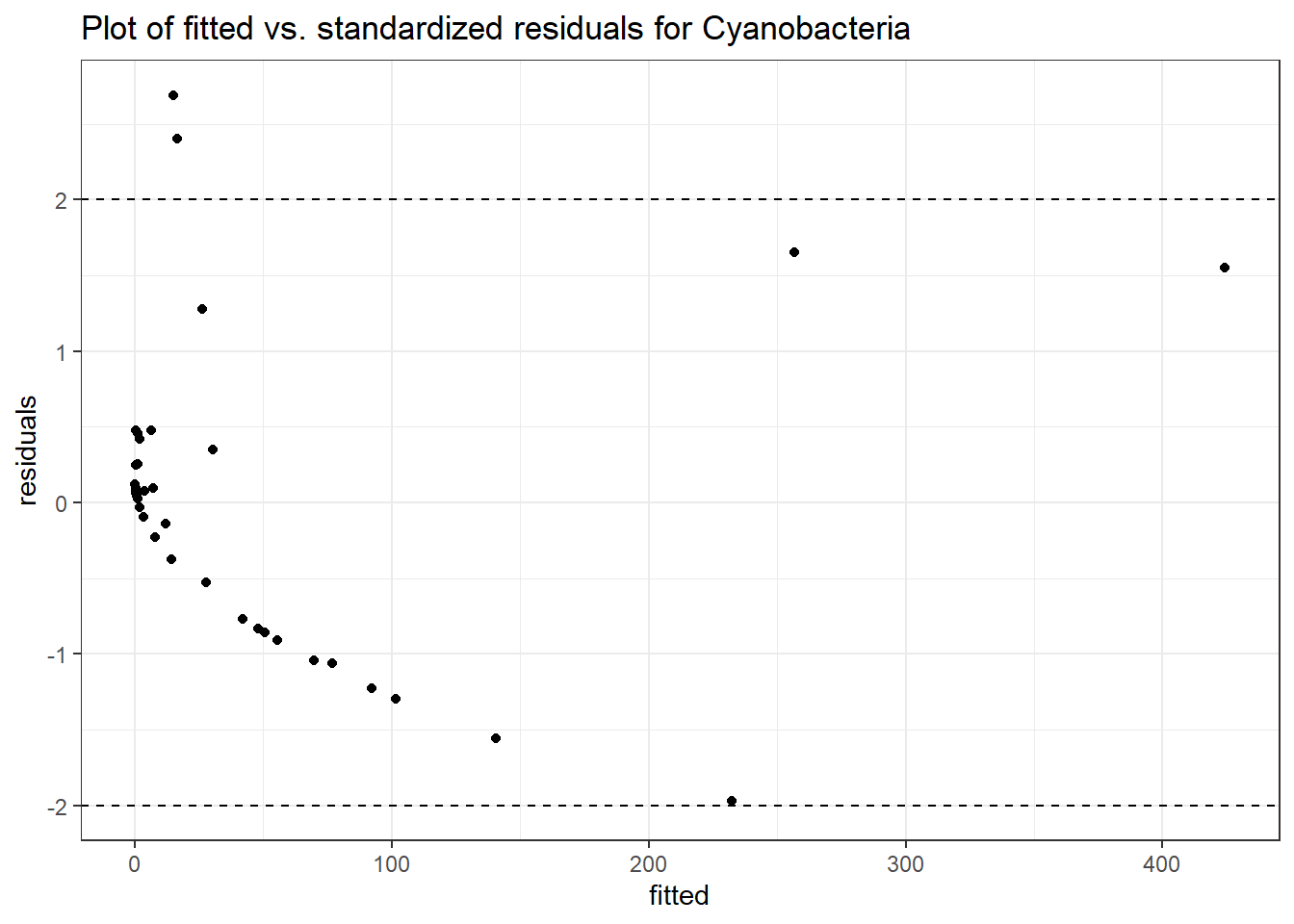
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Cyanobacteria 2.55302 1.25648 2.03188 0.04217
time Cyanobacteria -0.60192 0.02518 -23.90230 0.00000
female Cyanobacteria 0.00203 1.57224 0.00129 0.99897
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.457
Significance test not available
#########################################
#########################################
Model: Euryarchaeota ~ 1 + time + female + (1 | SubjectID)
<environment: 0x000000002f679a68>
Link: poisson
Intraclass Correlation (ICC): 0.7363739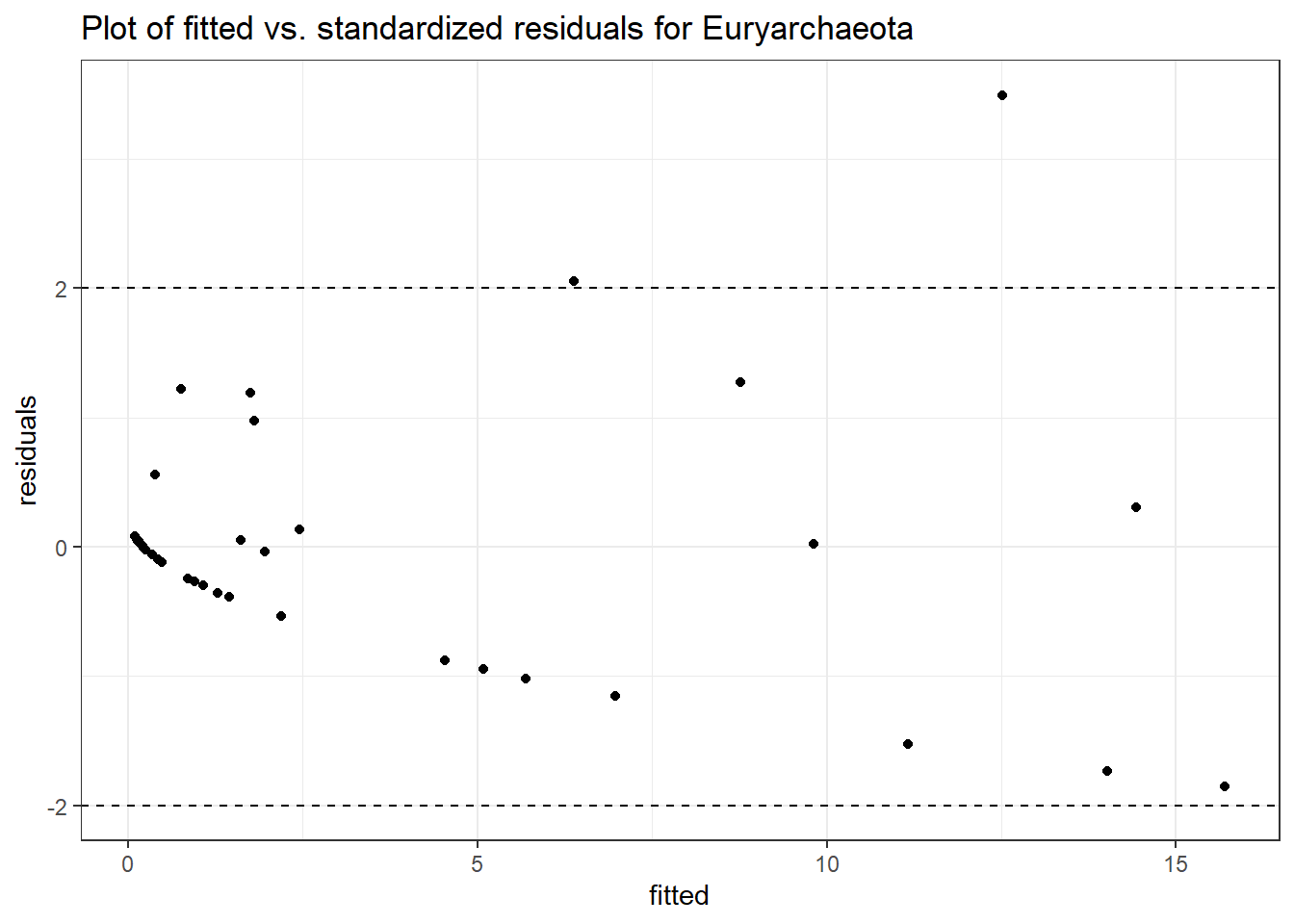
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Euryarchaeota -0.90825 1.05155 -0.86373 0.38774
time Euryarchaeota -0.11360 0.07975 -1.42442 0.15432
female Euryarchaeota 1.96695 1.20950 1.62625 0.10390
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.6713
Significance test not available
#########################################
#########################################
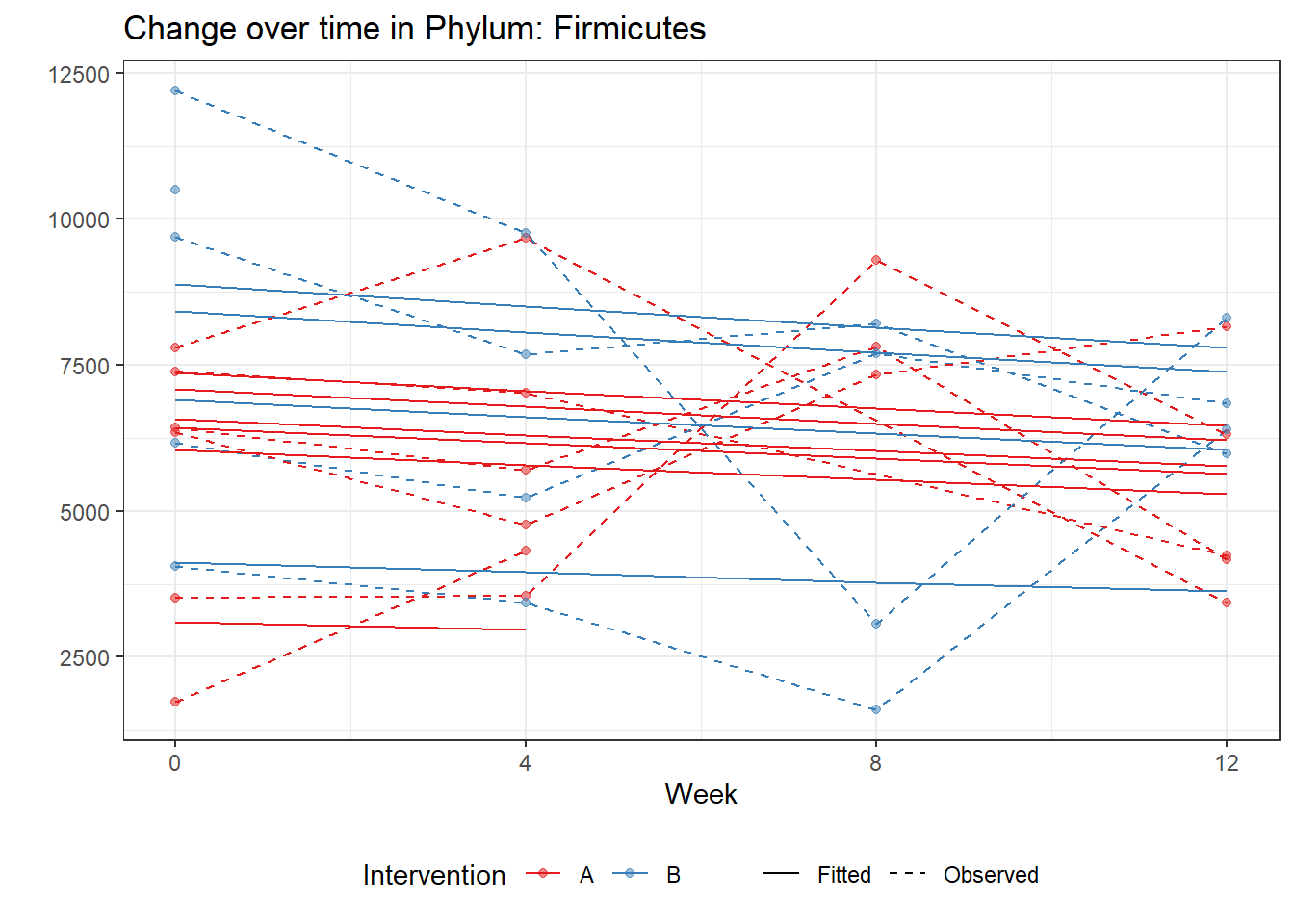
Model: Firmicutes ~ 1 + time + female + (1 | SubjectID)
<environment: 0x000000002f679a68>
Link: poisson
Intraclass Correlation (ICC): 0.05726281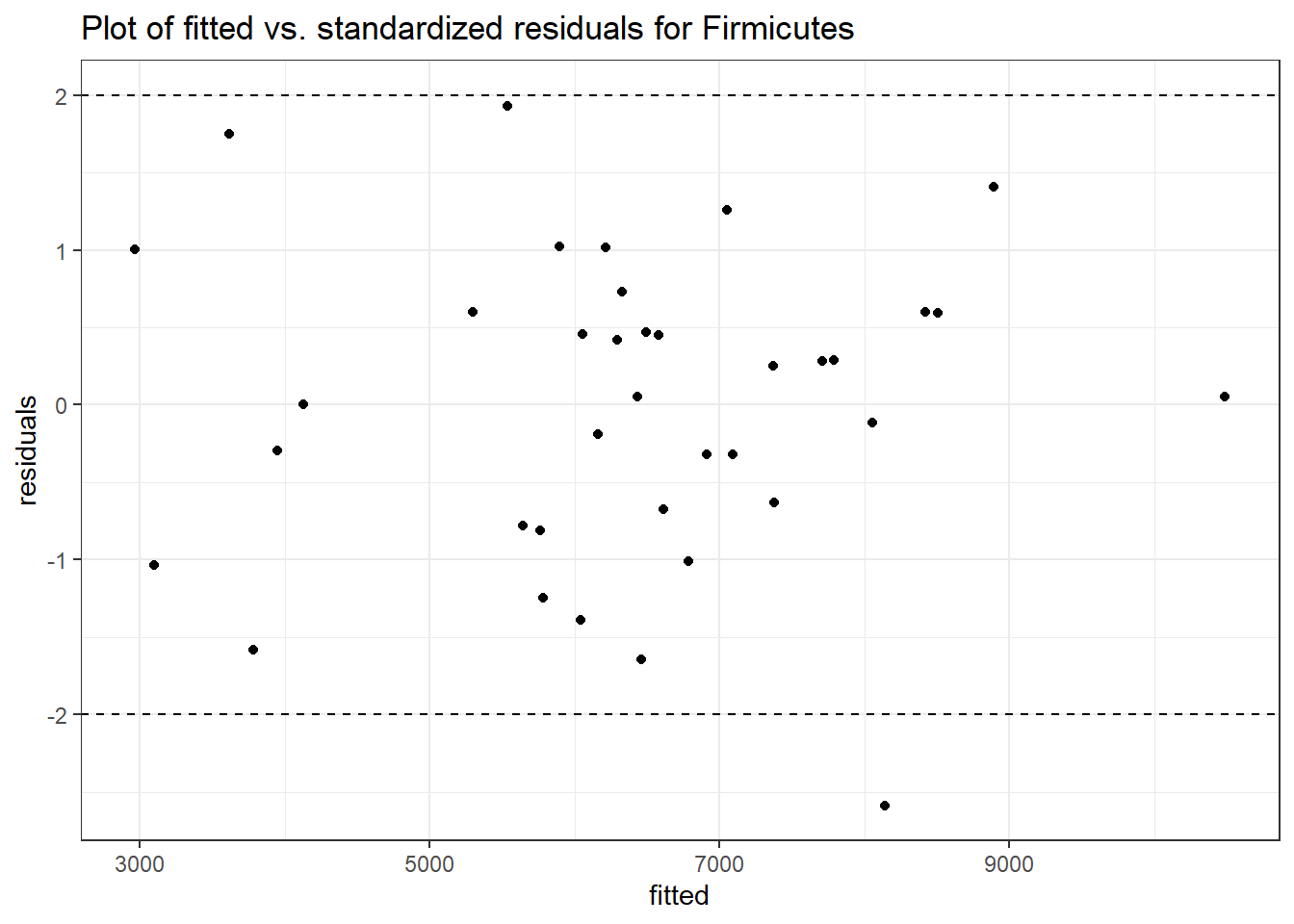
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Firmicutes 8.49933 0.12333 68.91608 0.00000
time Firmicutes -0.04391 0.00187 -23.47765 0.00000
female Firmicutes 0.44885 0.15456 2.90402 0.00368
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.24646
Significance test not available
#########################################
#########################################
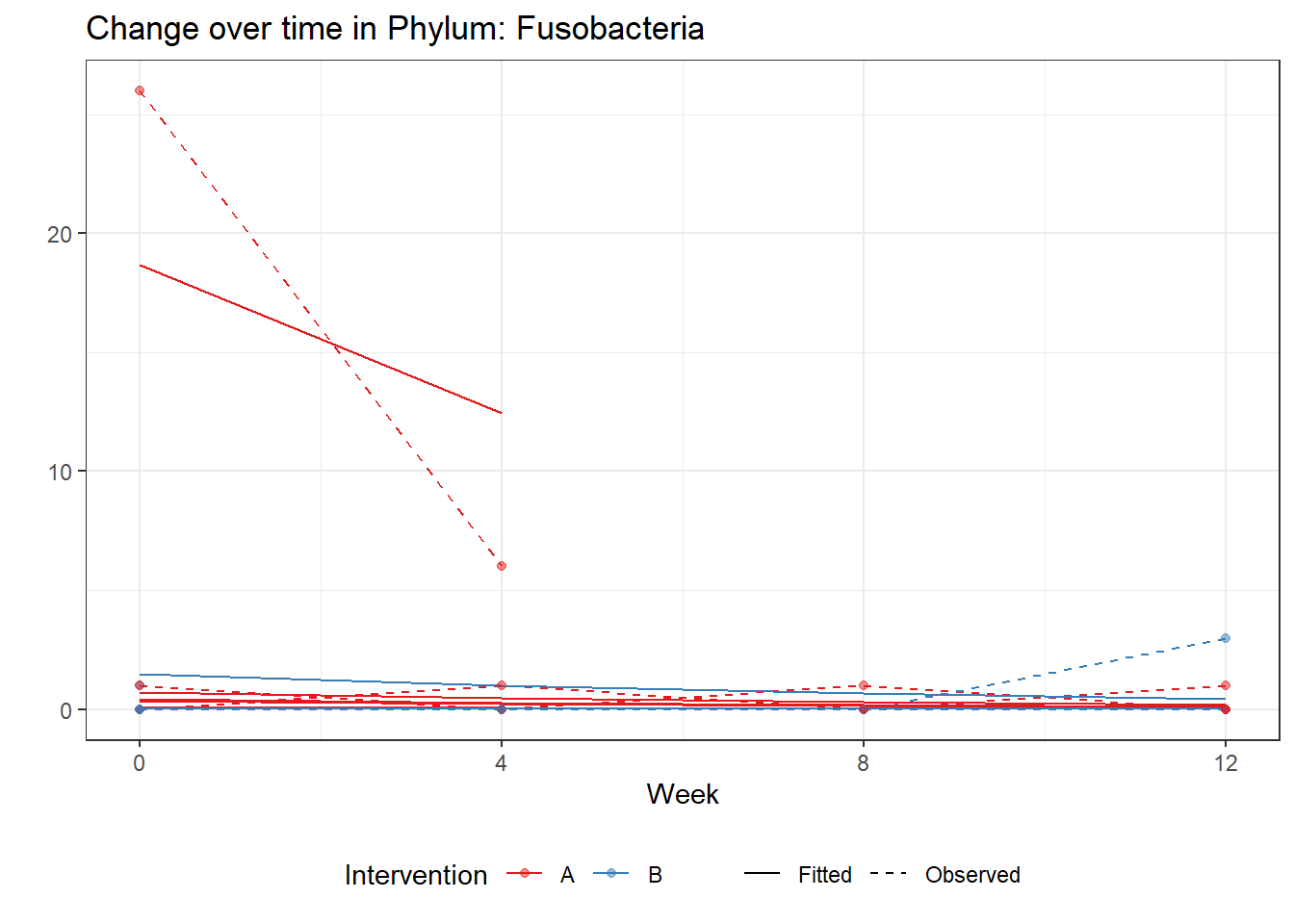
Model: Fusobacteria ~ 1 + time + female + (1 | SubjectID)
<environment: 0x000000002f679a68>
Link: poisson
Intraclass Correlation (ICC): 0.8398348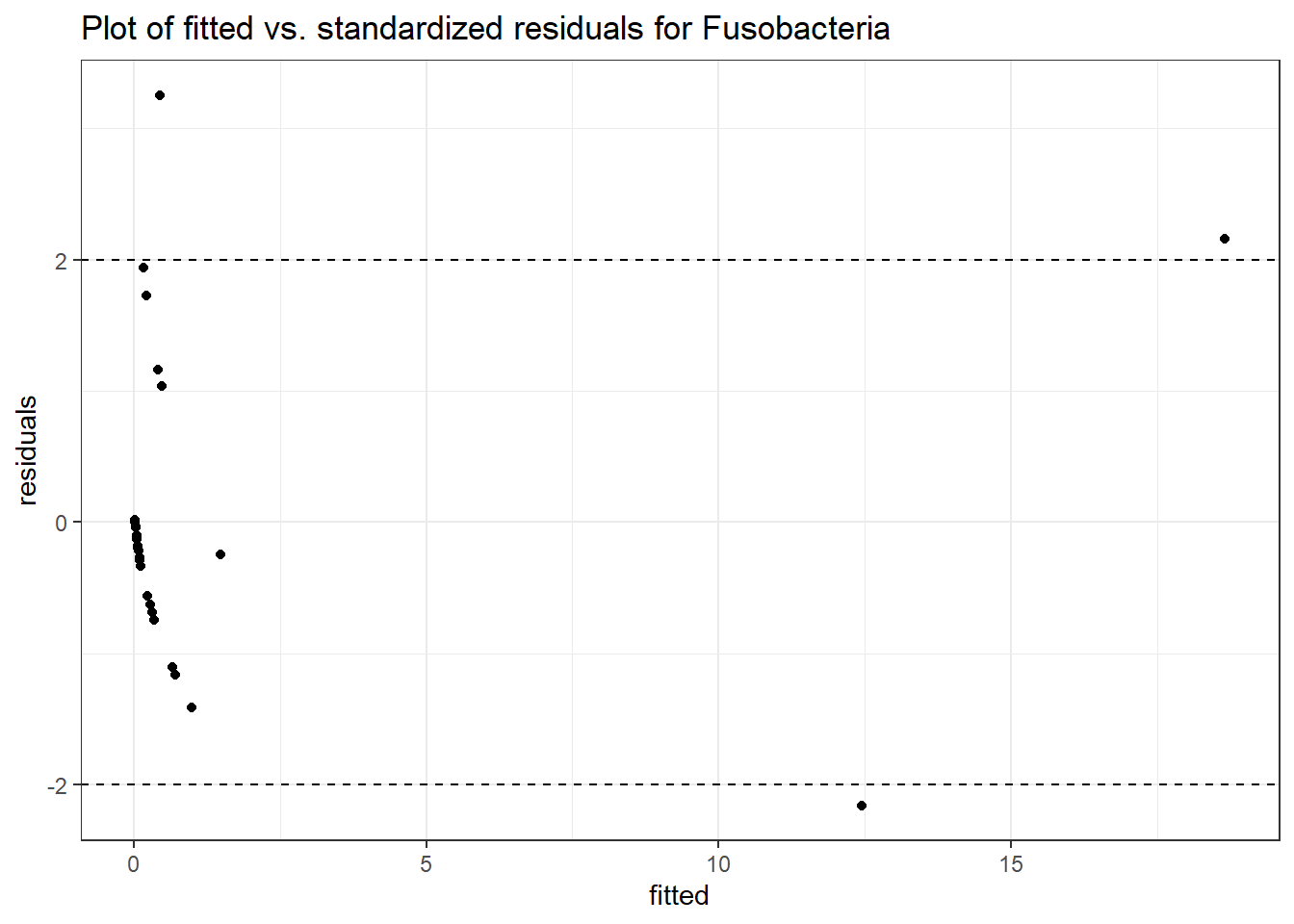
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Fusobacteria -1.70091 1.82355 -0.93274 0.35095
time Fusobacteria -0.40465 0.24247 -1.66883 0.09515
female Fusobacteria -0.08897 1.87964 -0.04733 0.96225
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.2899
Significance test not available
#########################################
#########################################
Model: Lentisphaerae ~ 1 + time + female + (1 | SubjectID)
<environment: 0x000000002f679a68>
Link: poisson
Intraclass Correlation (ICC): 0.697862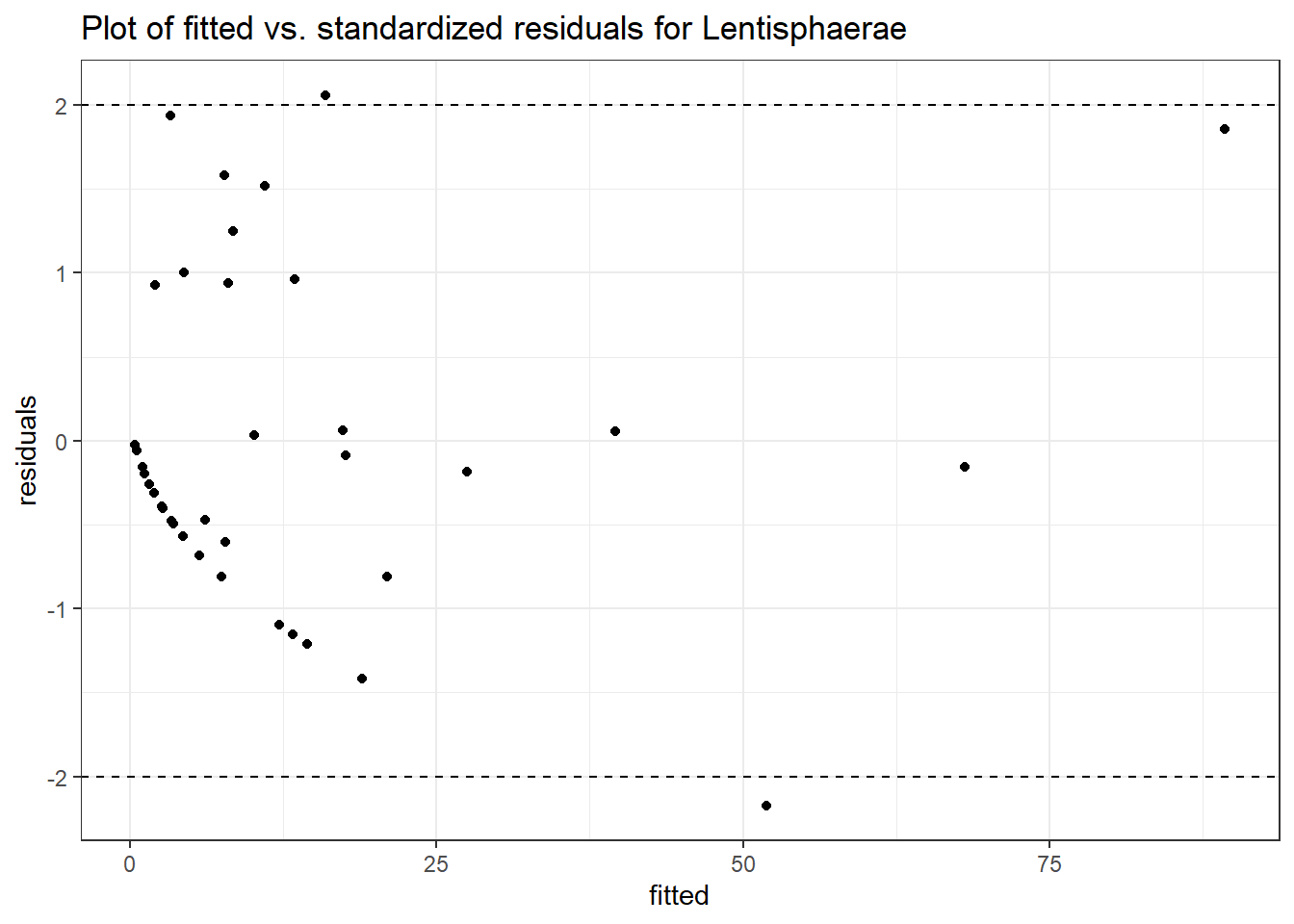
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lentisphaerae 1.50001 0.80365 1.86649 0.06197
time Lentisphaerae -0.27101 0.03998 -6.77909 0.00000
female Lentisphaerae 0.74300 0.99183 0.74912 0.45378
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.5198
Significance test not available
#########################################
#########################################
Model: Proteobacteria ~ 1 + time + female + (1 | SubjectID)
<environment: 0x000000002f679a68>
Link: poisson
Intraclass Correlation (ICC): 0.09278889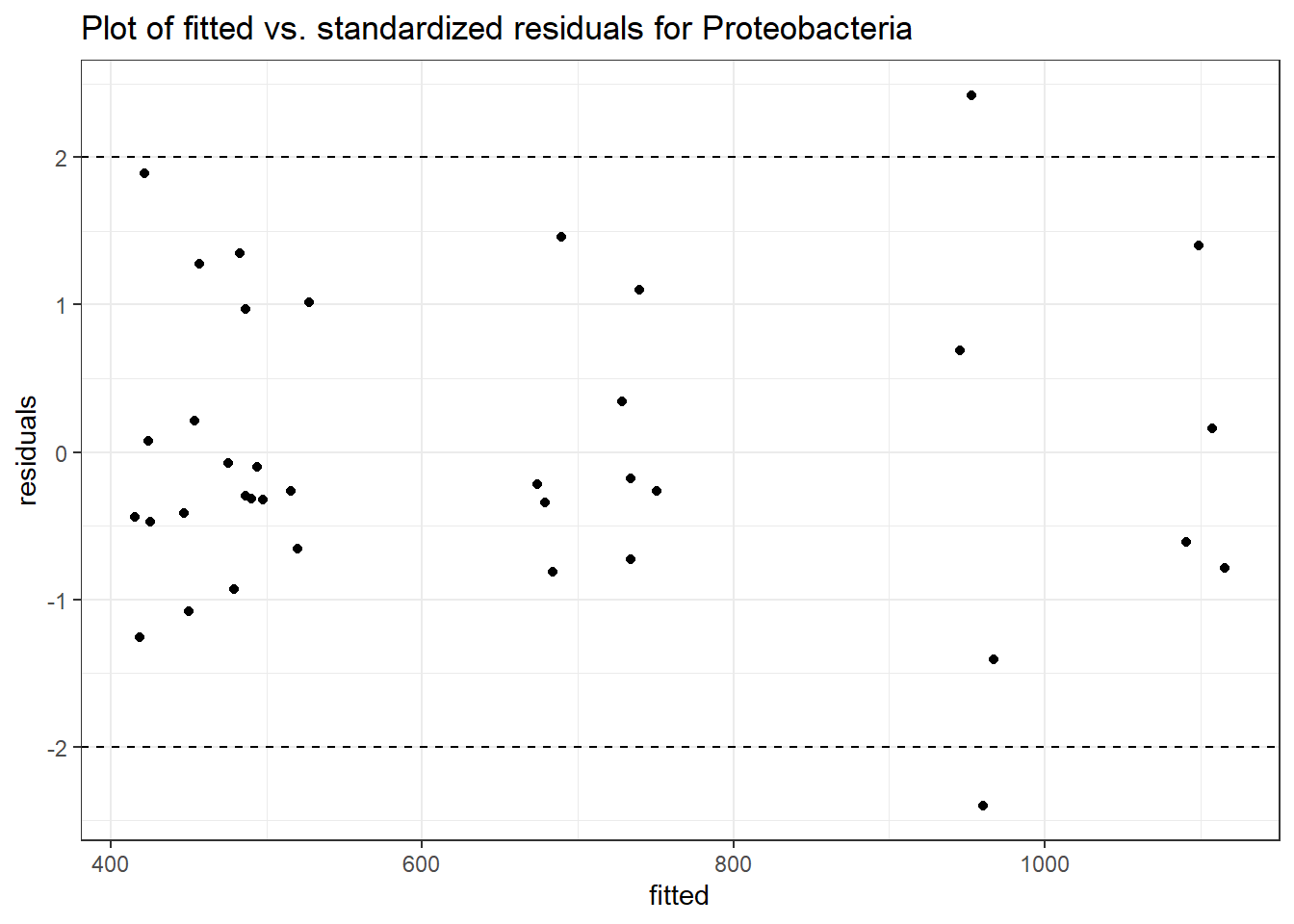
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Proteobacteria 6.37843 0.16050 39.74222 0.00000
time Proteobacteria 0.00754 0.00585 1.29010 0.19702
female Proteobacteria 0.02341 0.20105 0.11642 0.90732
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.31981
Significance test not available
#########################################
#########################################
Model: Tenericutes ~ 1 + time + female + (1 | SubjectID)
<environment: 0x000000002f679a68>
Link: poisson
Intraclass Correlation (ICC): 0.9173448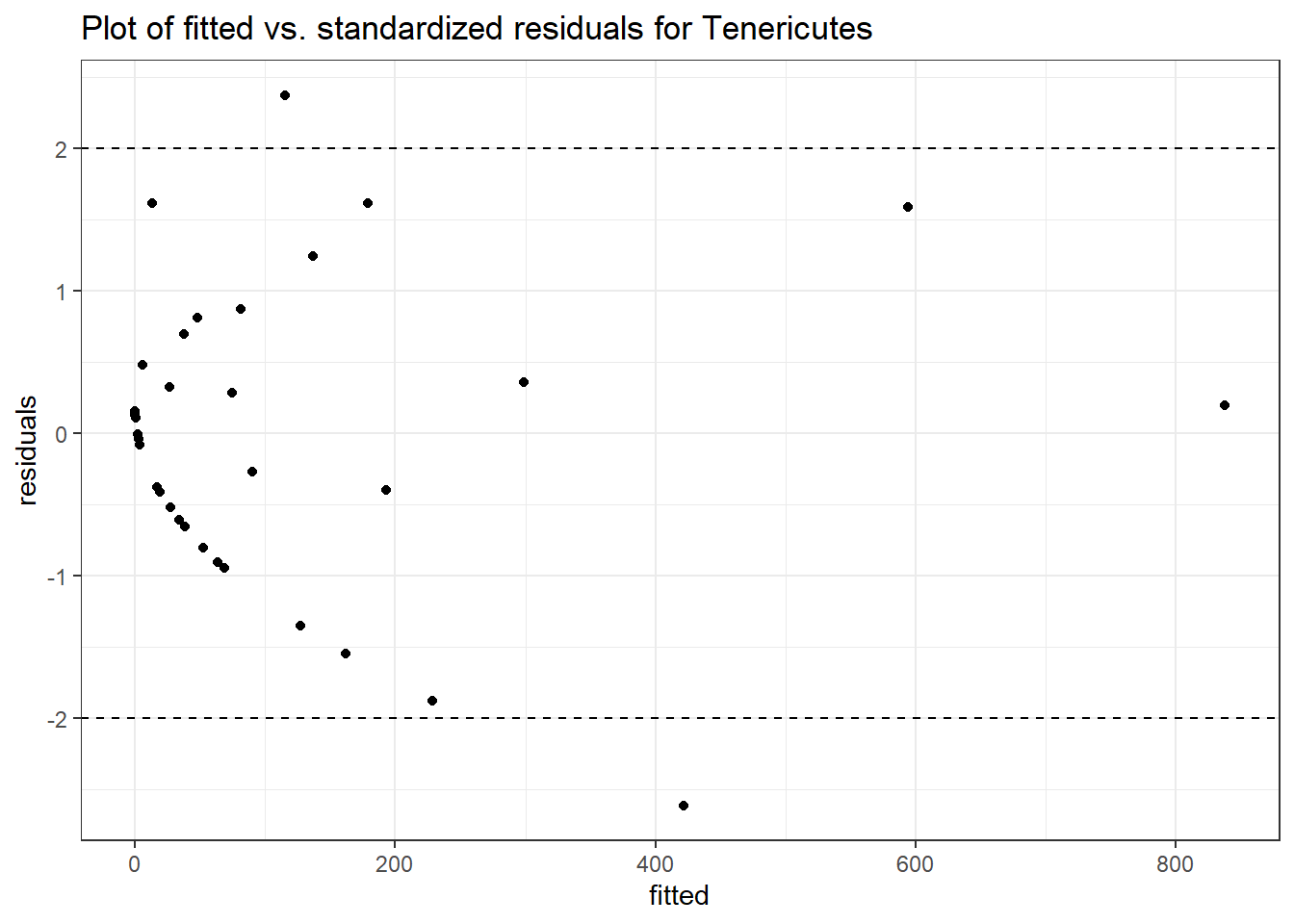
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Tenericutes 2.34245 1.70805 1.37141 0.17025
time Tenericutes -0.34365 0.01482 -23.18289 0.00000
female Tenericutes 0.48030 2.13654 0.22480 0.82213
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 3.3314
Significance test not available
#########################################
#########################################
Model: Verrucomicrobia ~ 1 + time + female + (1 | SubjectID)
<environment: 0x000000002f679a68>
Link: poisson
Intraclass Correlation (ICC): 0.8645285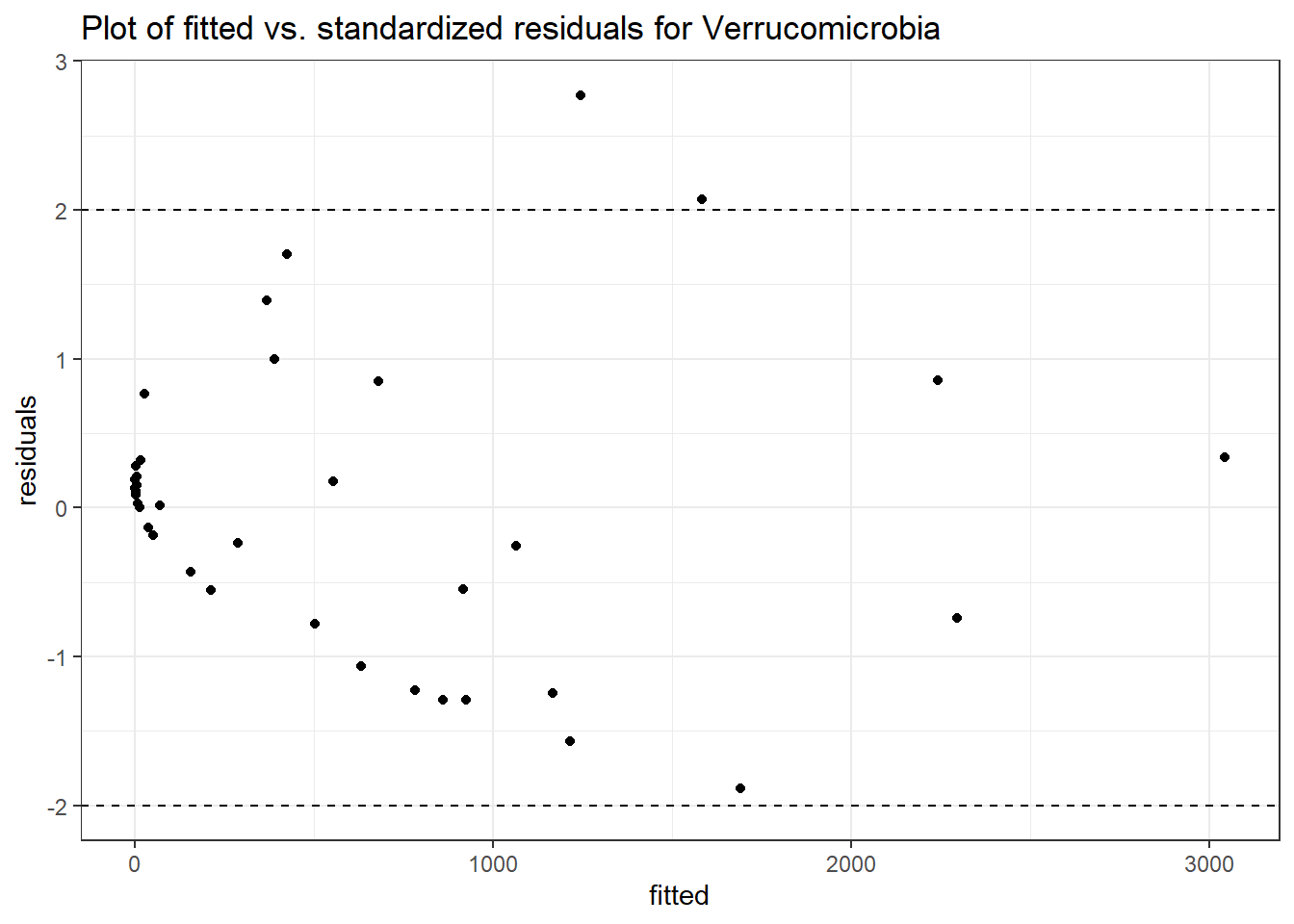
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Verrucomicrobia 4.07726 1.27761 3.19130 0.00142
time Verrucomicrobia -0.30578 0.00609 -50.22344 0.00000
female Verrucomicrobia 1.79441 1.59433 1.12550 0.26038
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.5262
Significance test not availableControlling for the effect of time, Gender was only signficant for Bacteroidetes and Firmicutes. We will take out gender out of model to test other effects, but will come back when testing intervention.
Covariates: Time + Ethnicity
\[\begin{align*} h\left(Y_{ij}\right)&=\beta_{0j} + \beta_{1j}*(Week)_{ij}+ r_{ij}\\ \beta_{0j}&= \gamma_{00} + \gamma_{01}(Ethnicity)_{j} + u_{0j}\\ \beta_{1j}&= \gamma_{10}\\ \end{align*}\]
Ethnicity is coded as Hispanic (effect of being Hispanic compared to White, Asian or Native American). Groups were coded this way due to sample size.
phylum.fit3 <- glmm_microbiome(mydata=microbiome_data,model.number=3,
taxa.level="Phylum", link="poisson",
model="1 + time + hispanic + (1|SubjectID)")
#########################################
#########################################
Model: Actinobacteria ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x00000000065308b0>
Link: poisson
Intraclass Correlation (ICC): 0.5267952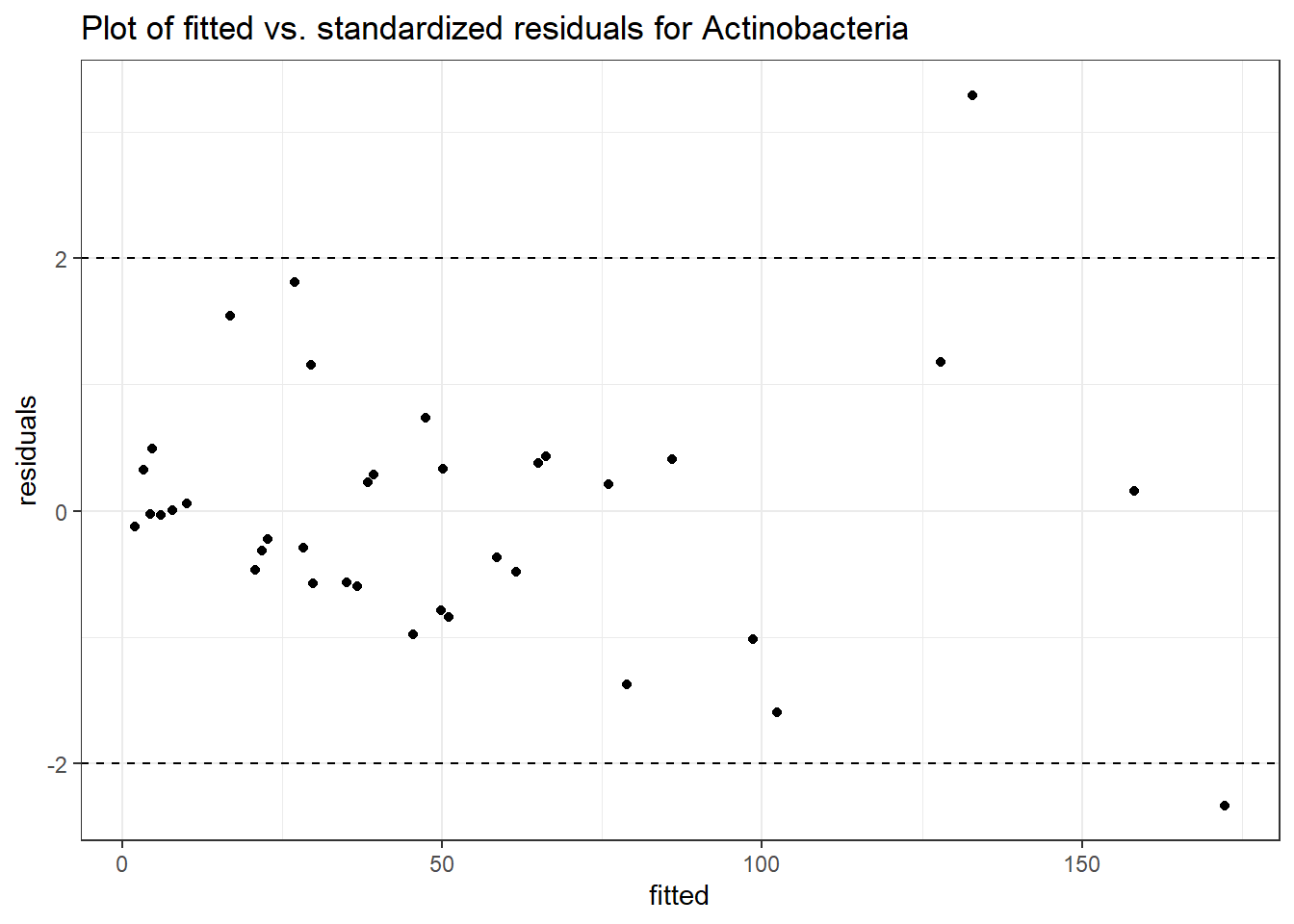
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Actinobacteria 3.52412 0.53525 6.58404 0.00000
time Actinobacteria -0.26035 0.02241 -11.61946 0.00000
hispanic Actinobacteria 0.58475 0.66803 0.87534 0.38139
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.0551
Significance test not available
#########################################
#########################################
Model: Bacteroidetes ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x00000000065308b0>
Link: poisson
Intraclass Correlation (ICC): 0.07964047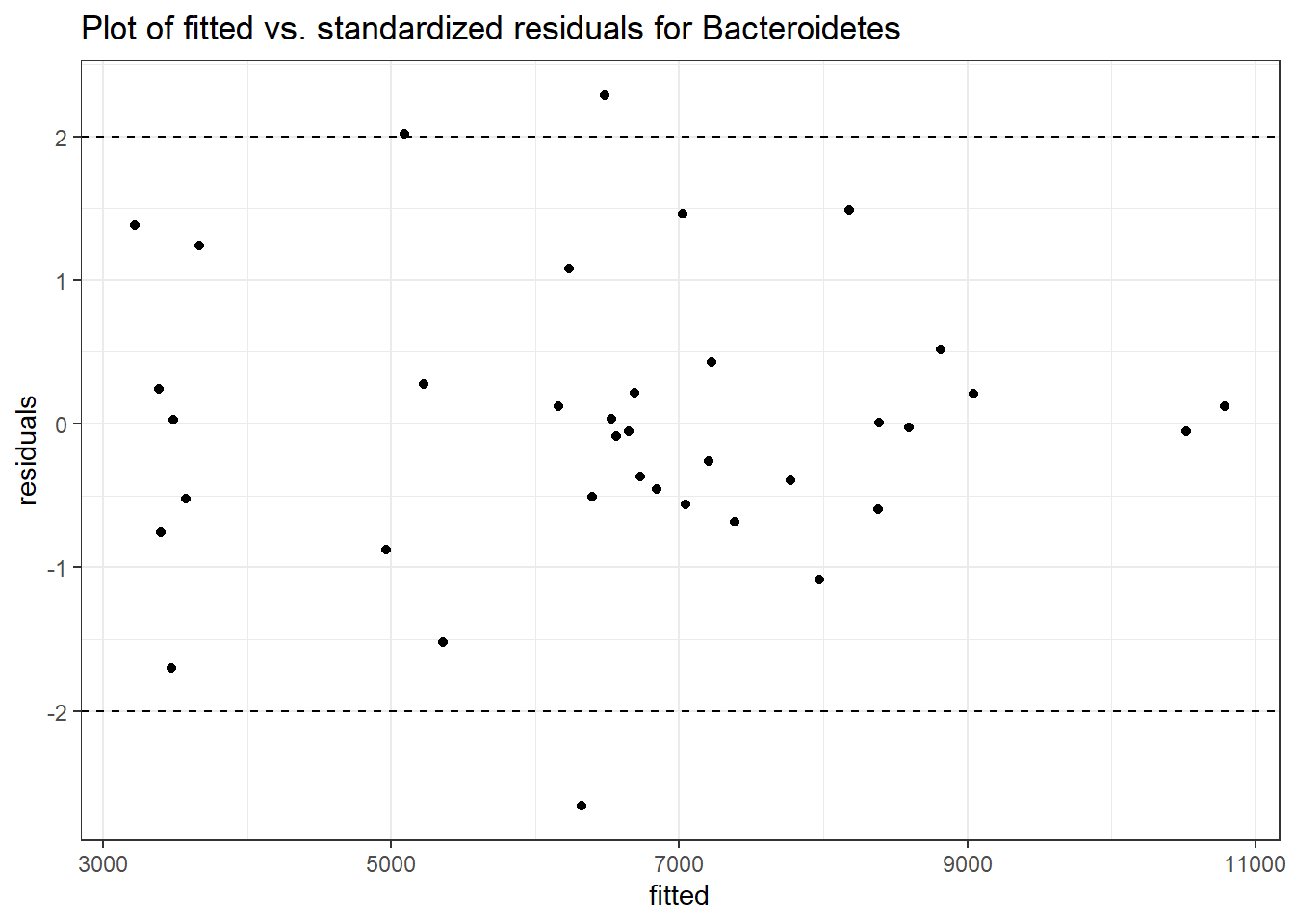
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroidetes 8.98436 0.14713 61.06205 0.00000
time Bacteroidetes -0.02542 0.00189 -13.46185 0.00000
hispanic Bacteroidetes -0.32200 0.18443 -1.74594 0.08082
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.29416
Significance test not available
#########################################
#########################################
Model: Cyanobacteria ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x00000000065308b0>
Link: poisson
Intraclass Correlation (ICC): 0.8325719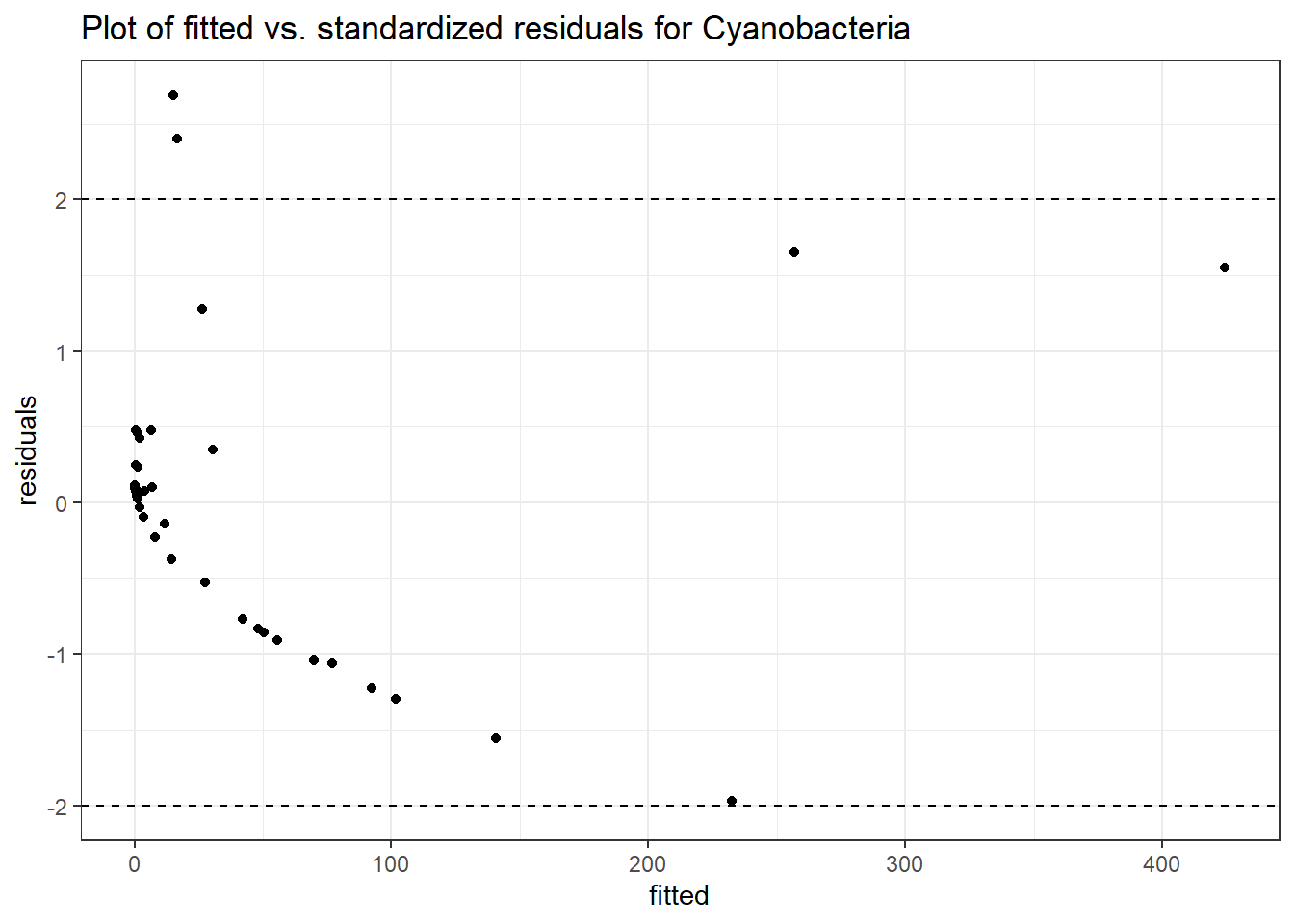
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Cyanobacteria 3.90294 1.12565 3.46729 0.00053
time Cyanobacteria -0.60194 0.02518 -23.90315 0.00000
hispanic Cyanobacteria -2.10908 1.42385 -1.48125 0.13854
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.23
Significance test not available
#########################################
#########################################
Model: Euryarchaeota ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x00000000065308b0>
Link: poisson
Intraclass Correlation (ICC): 0.7021741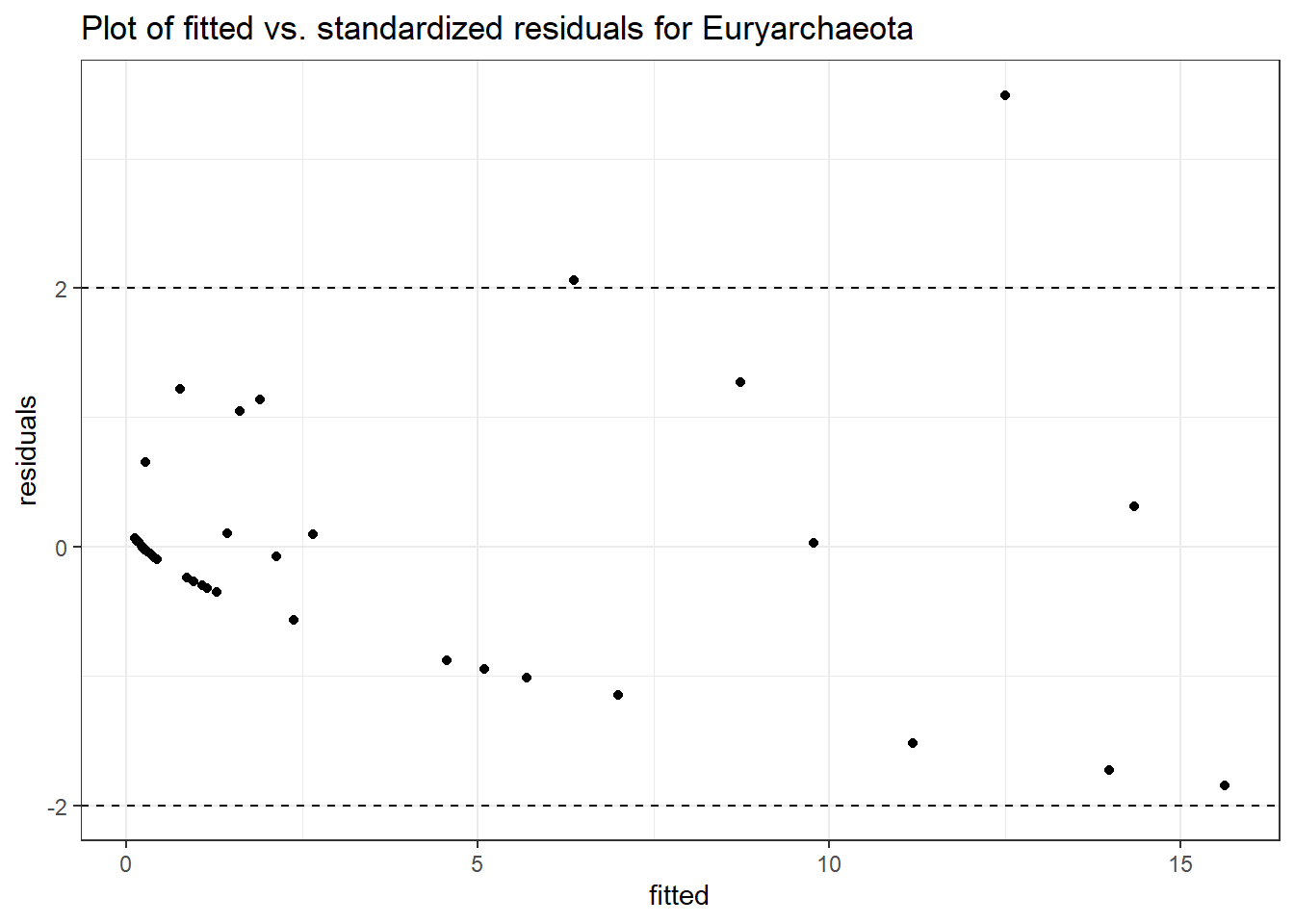
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Euryarchaeota -0.75656 0.88533 -0.85455 0.39280
time Euryarchaeota -0.11145 0.07869 -1.41636 0.15667
hispanic Euryarchaeota 1.87423 1.06086 1.76671 0.07728
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.5355
Significance test not available
#########################################
#########################################
Model: Firmicutes ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x00000000065308b0>
Link: poisson
Intraclass Correlation (ICC): 0.09257982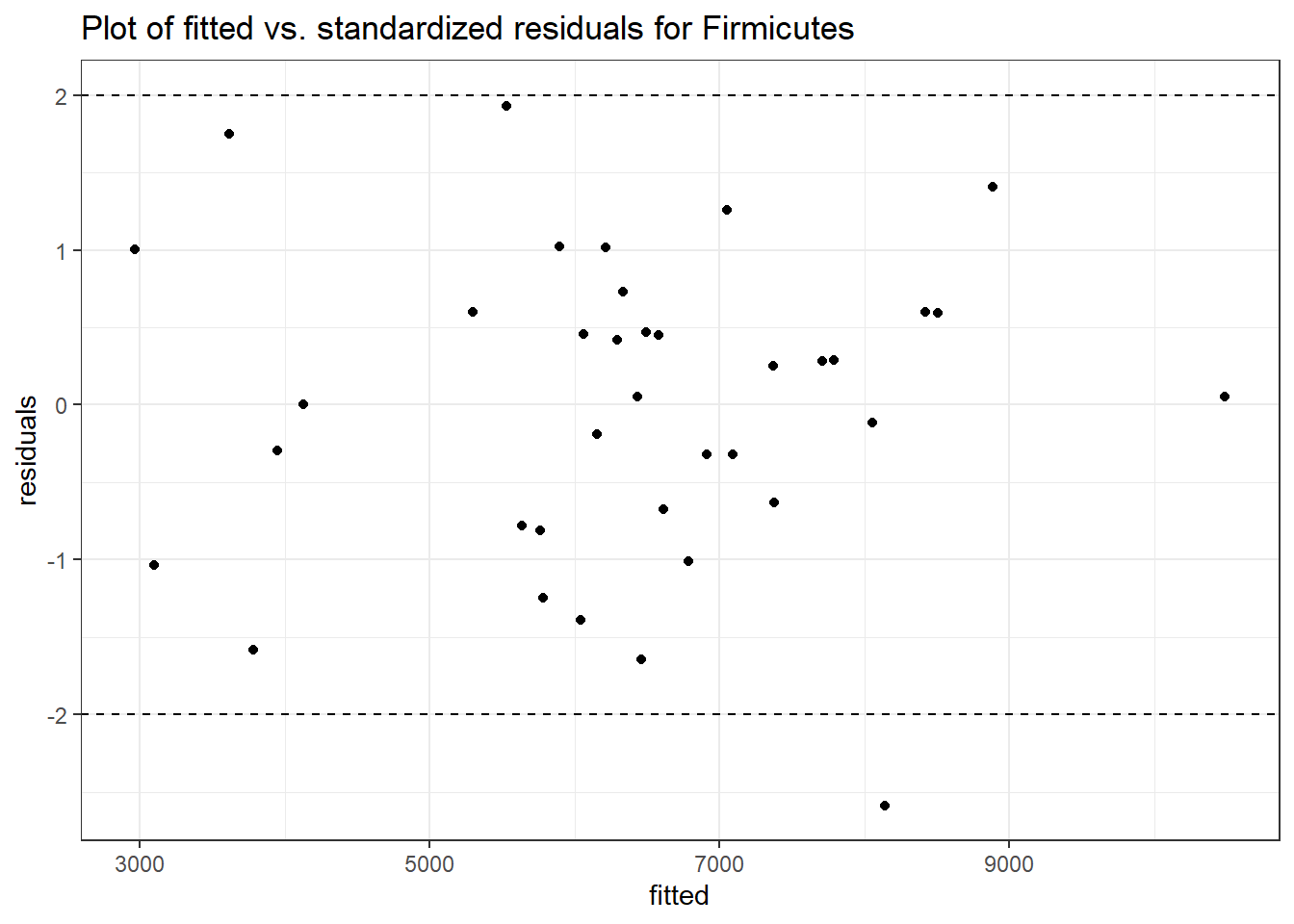
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Firmicutes 8.68835 0.15977 54.38205 0.00000
time Firmicutes -0.04390 0.00187 -23.47284 0.00000
hispanic Firmicutes 0.15182 0.20026 0.75814 0.44837
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.31941
Significance test not available
#########################################
#########################################
Model: Fusobacteria ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x00000000065308b0>
Link: poisson
Intraclass Correlation (ICC): 0.8273398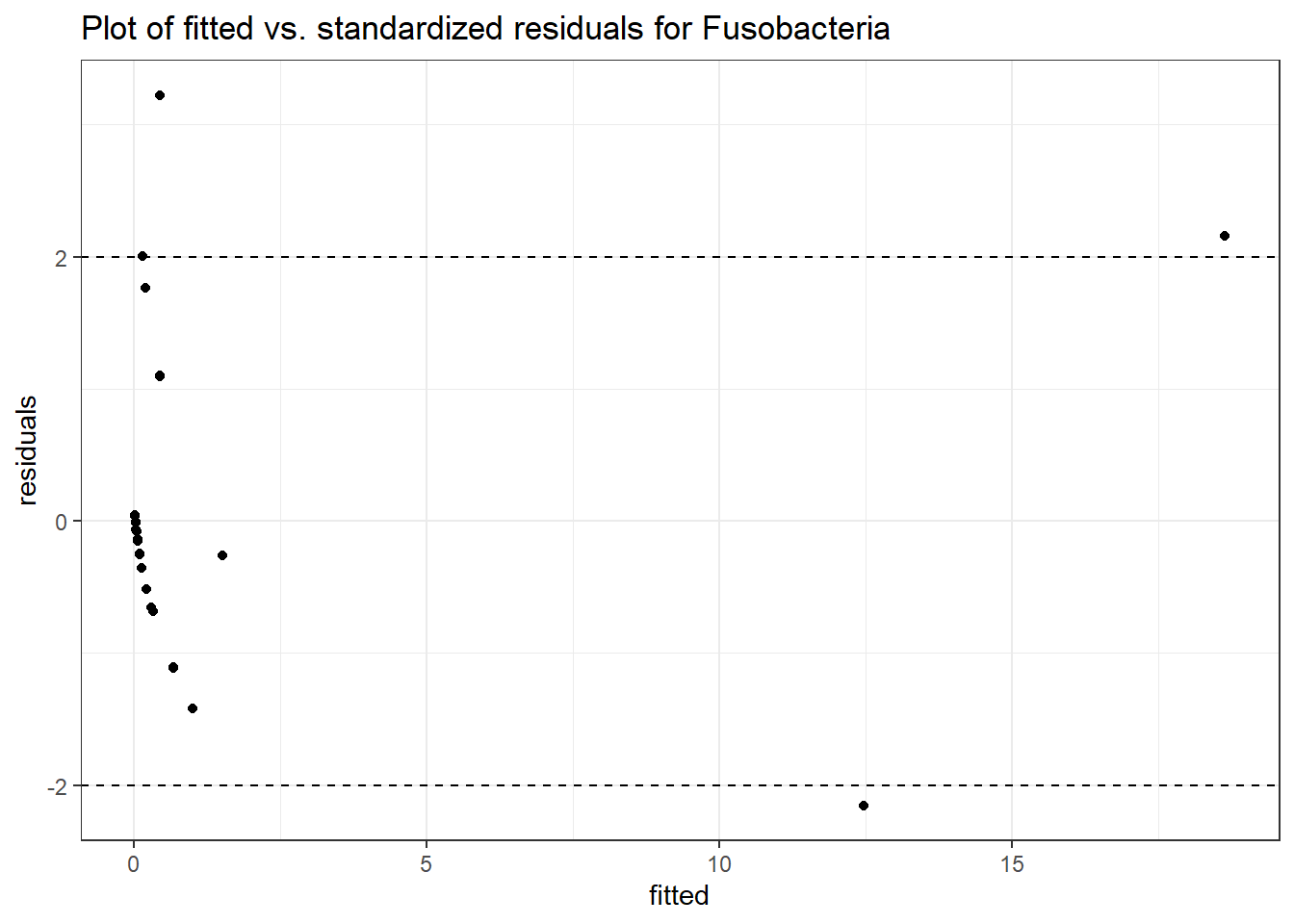
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Fusobacteria -2.25724 1.49545 -1.50941 0.13119
time Fusobacteria -0.40251 0.24257 -1.65936 0.09704
hispanic Fusobacteria 0.88922 1.71475 0.51857 0.60406
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.189
Significance test not available
#########################################
#########################################
Model: Lentisphaerae ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x00000000065308b0>
Link: poisson
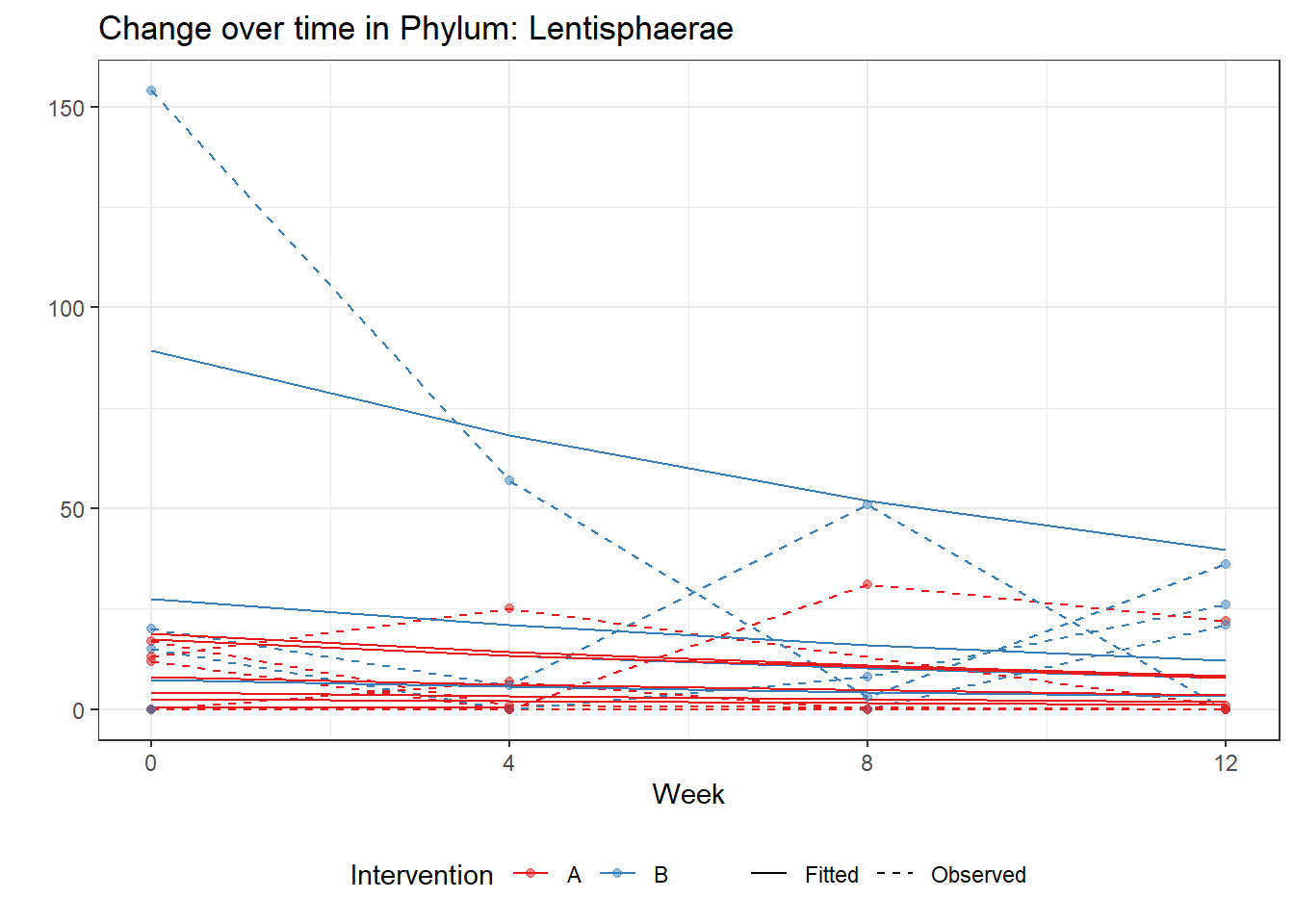
Intraclass Correlation (ICC): 0.6850713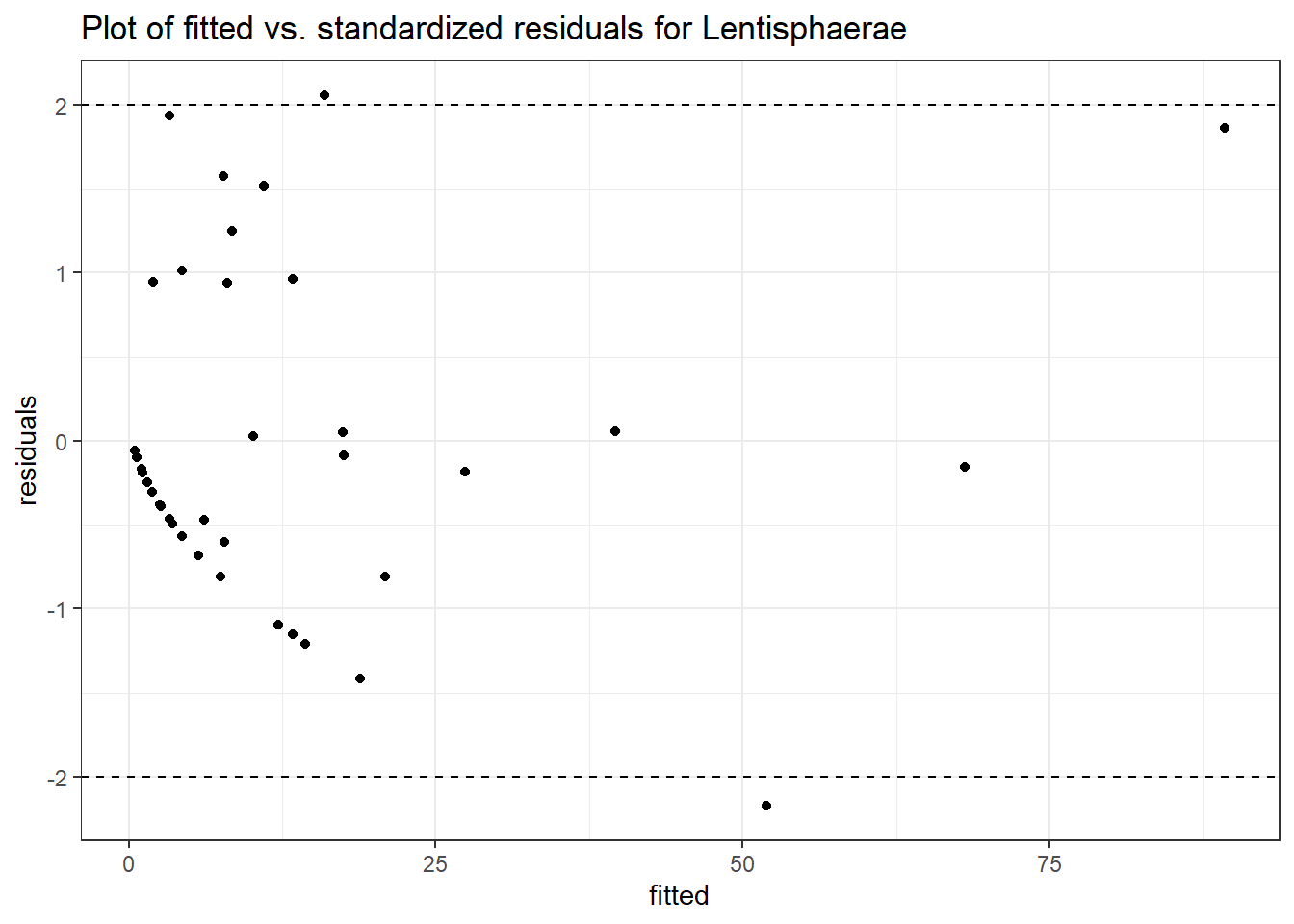
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lentisphaerae 1.59306 0.75228 2.11762 0.03421
time Lentisphaerae -0.27062 0.03999 -6.76680 0.00000
hispanic Lentisphaerae 0.63780 0.95946 0.66476 0.50621
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.4749
Significance test not available
#########################################
#########################################
Model: Proteobacteria ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x00000000065308b0>
Link: poisson
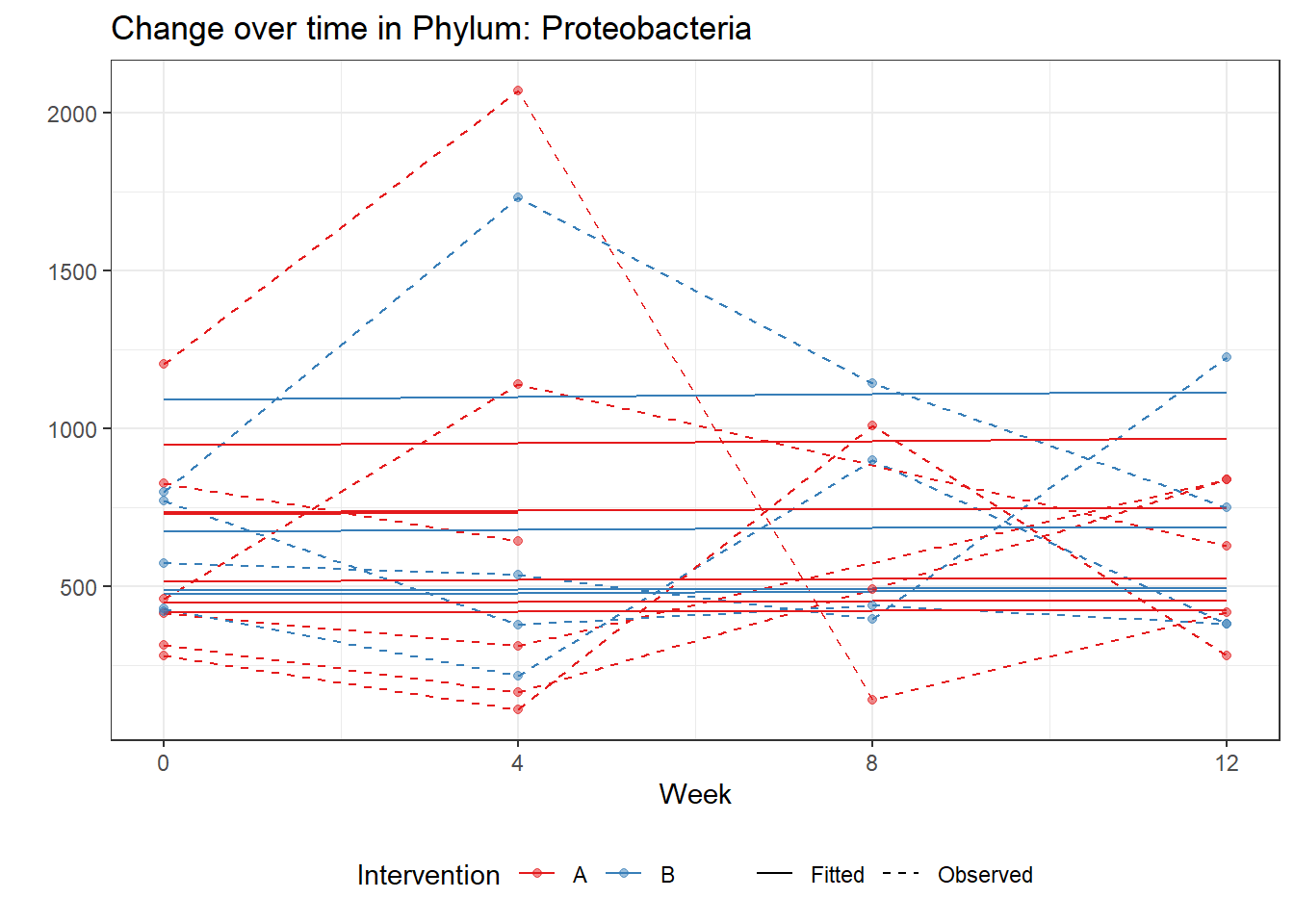
Intraclass Correlation (ICC): 0.09234374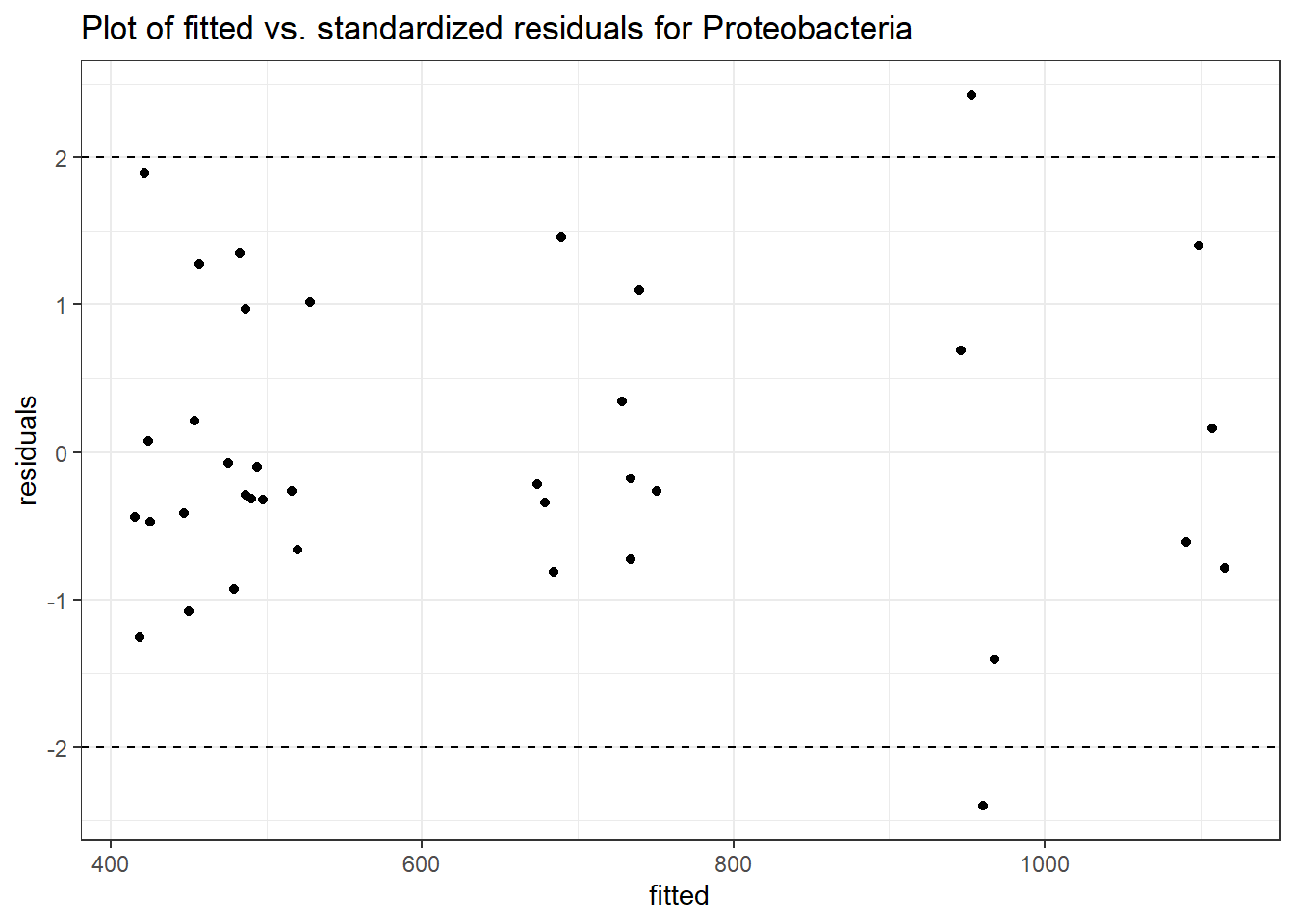
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Proteobacteria 6.42834 0.16007 40.15939 0.00000
time Proteobacteria 0.00753 0.00585 1.28766 0.19786
hispanic Proteobacteria -0.05503 0.20049 -0.27450 0.78370
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.31896
Significance test not available
#########################################
#########################################
Model: Tenericutes ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x00000000065308b0>
Link: poisson
Intraclass Correlation (ICC): 0.9091899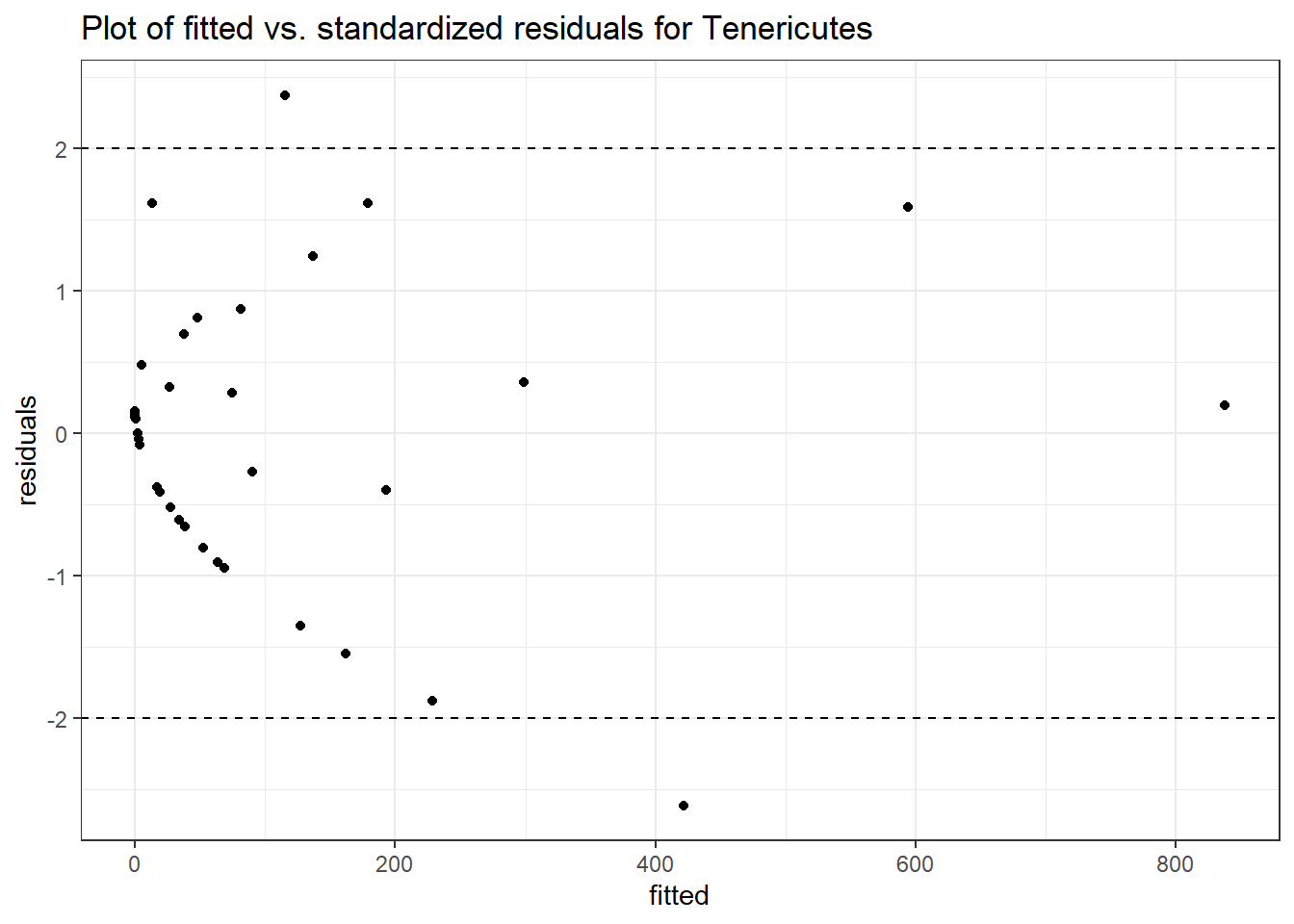
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Tenericutes 1.59152 1.62705 0.97816 0.32799
time Tenericutes -0.34362 0.01482 -23.18060 0.00000
hispanic Tenericutes 1.73375 2.03217 0.85315 0.39358
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 3.1642
Significance test not available
#########################################
#########################################
Model: Verrucomicrobia ~ 1 + time + hispanic + (1 | SubjectID)
<environment: 0x00000000065308b0>
Link: poisson
Intraclass Correlation (ICC): 0.8662343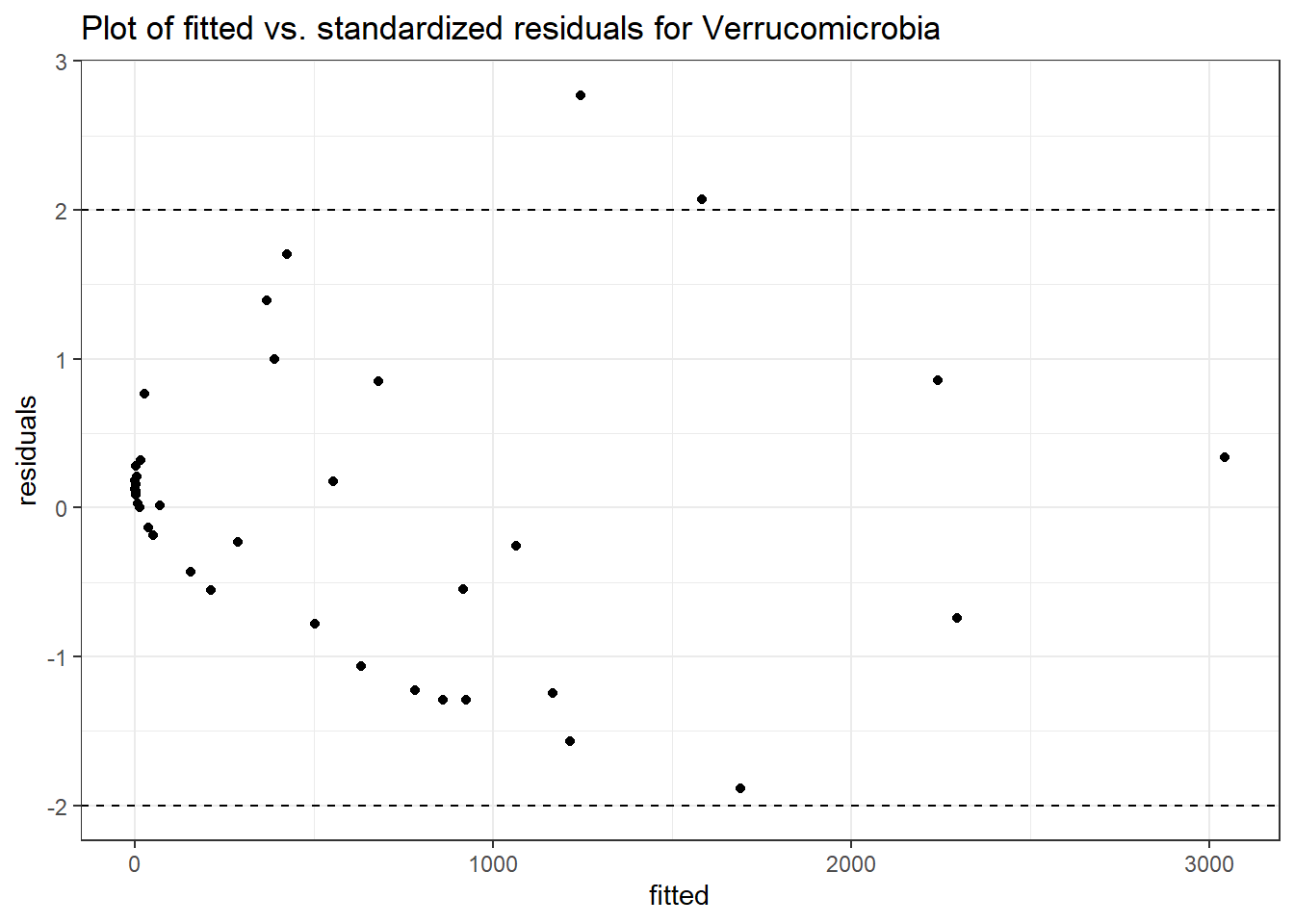
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Verrucomicrobia 4.28983 1.27709 3.35906 0.00078
time Verrucomicrobia -0.30577 0.00609 -50.22242 0.00000
hispanic Verrucomicrobia 1.47957 1.60311 0.92294 0.35604
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.5448
Significance test not availableControlling for the effect of time, Ethnicity was not significant for all Phylum.
Effect of Intervention
Next, we invested the effect of intervention, however, the results are parsed between which covariates are included in the model.
Intervention was coded as (group A = 0, and group B = 1). So, the results are interpretted as the effect of being in intervention group B.
Covariate(s): Time
\[\begin{align*} h\left(Y_{ij}\right)&=\beta_{0j} + \beta_{1j}*(Week)_{ij} + r_{ij}\\ \beta_{0j}&= \gamma_{00} + \gamma_{01}(Intervention)_{j} + u_{0j}\\ \beta_{1j}&= \gamma_{10}\\ \end{align*}\]
The effect of intervention is the main outcome of this study.
phylum.fit4 <- glmm_microbiome(mydata=microbiome_data,model.number=4,
taxa.level="Phylum", link="poisson",
model="1 + intB + time + (1|SubjectID)")
#########################################
#########################################
Model: Actinobacteria ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x000000003ca59558>
Link: poisson
Intraclass Correlation (ICC): 0.4992711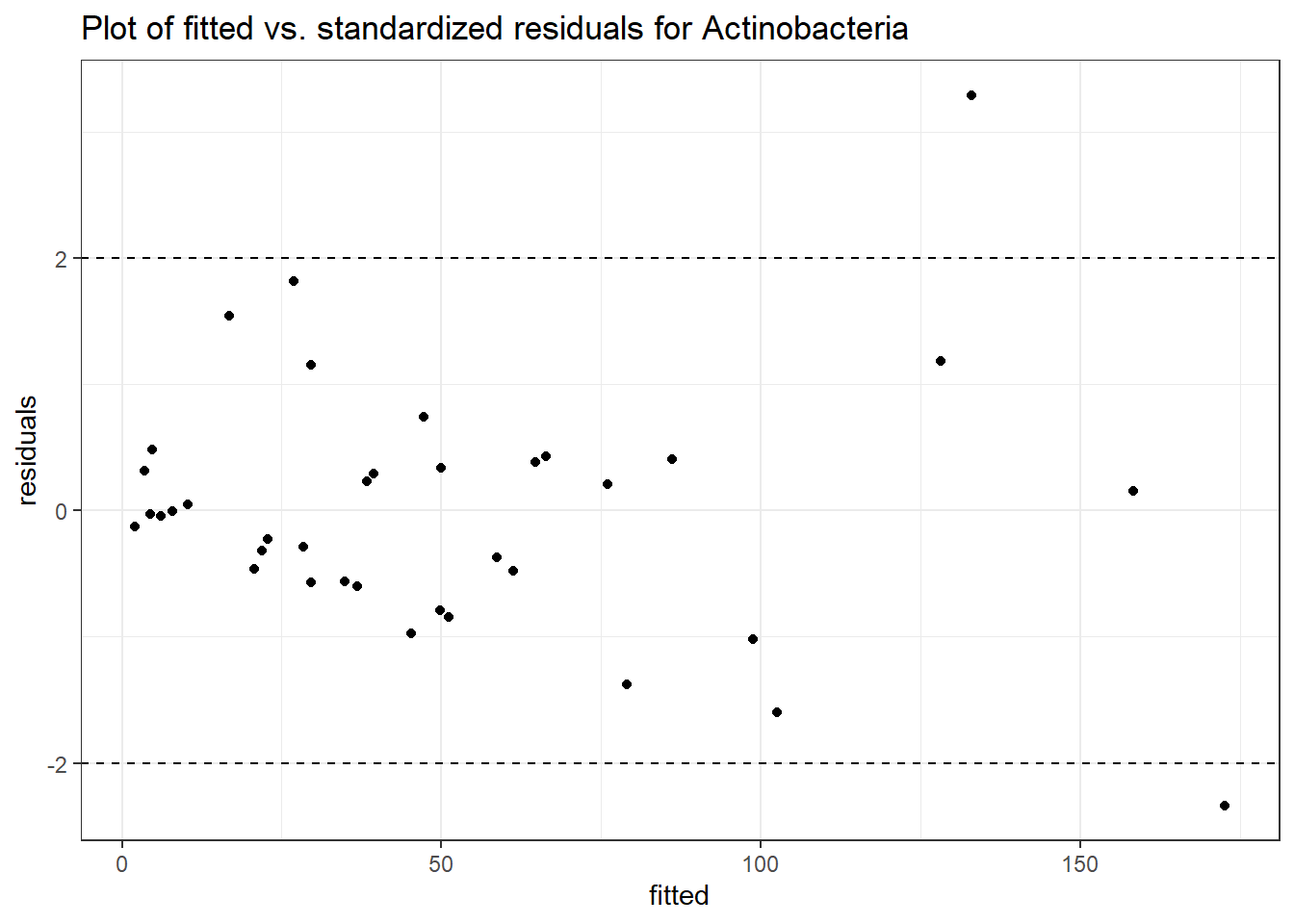
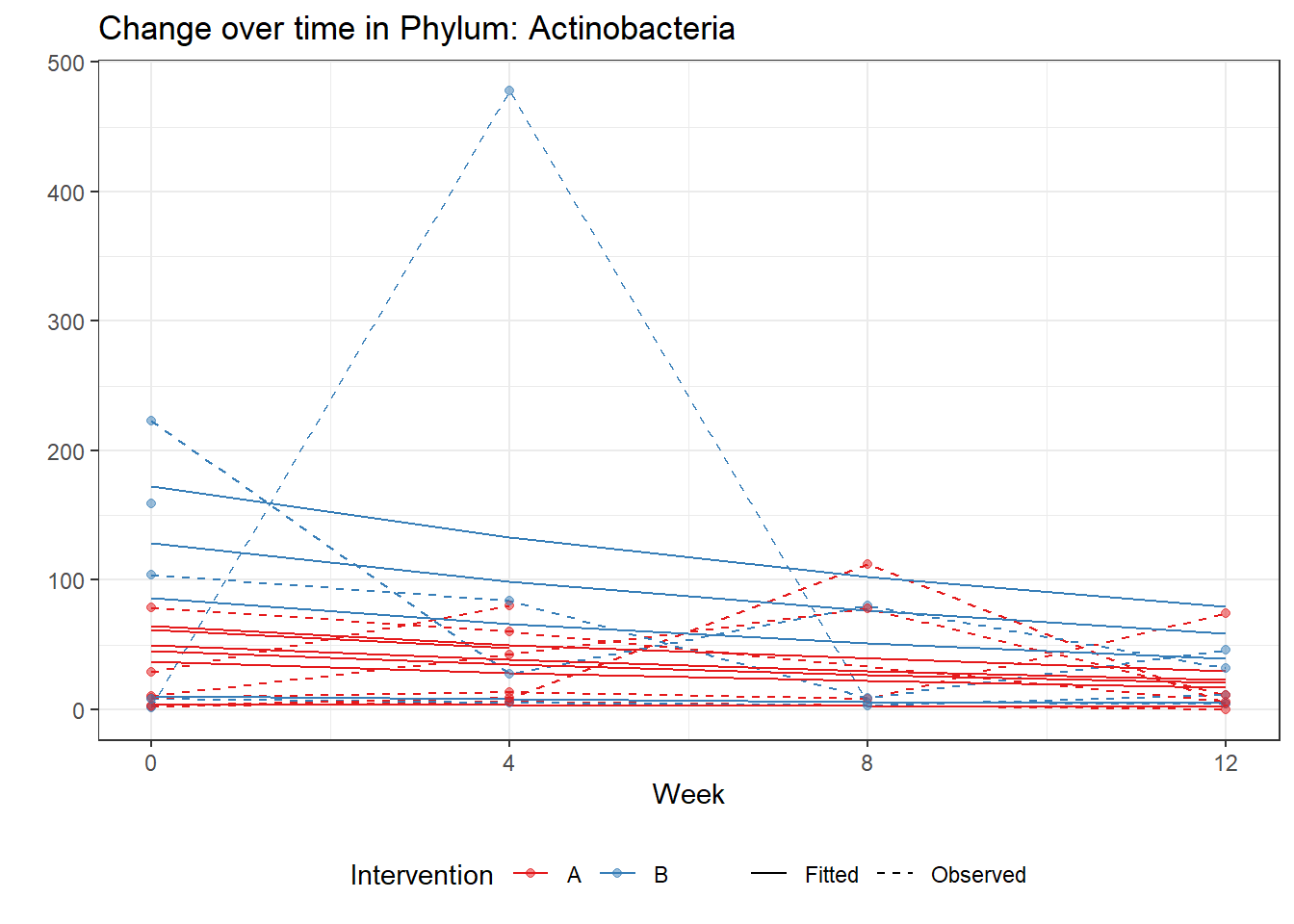
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Actinobacteria 3.50884 0.41300 8.49601 0.00000
intB Actinobacteria 0.85738 0.60937 1.40699 0.15943
time Actinobacteria -0.26053 0.02240 -11.62836 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.99854
Significance test not available
#########################################
#########################################
Model: Bacteroidetes ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x000000003ca59558>
Link: poisson
Intraclass Correlation (ICC): 0.09531271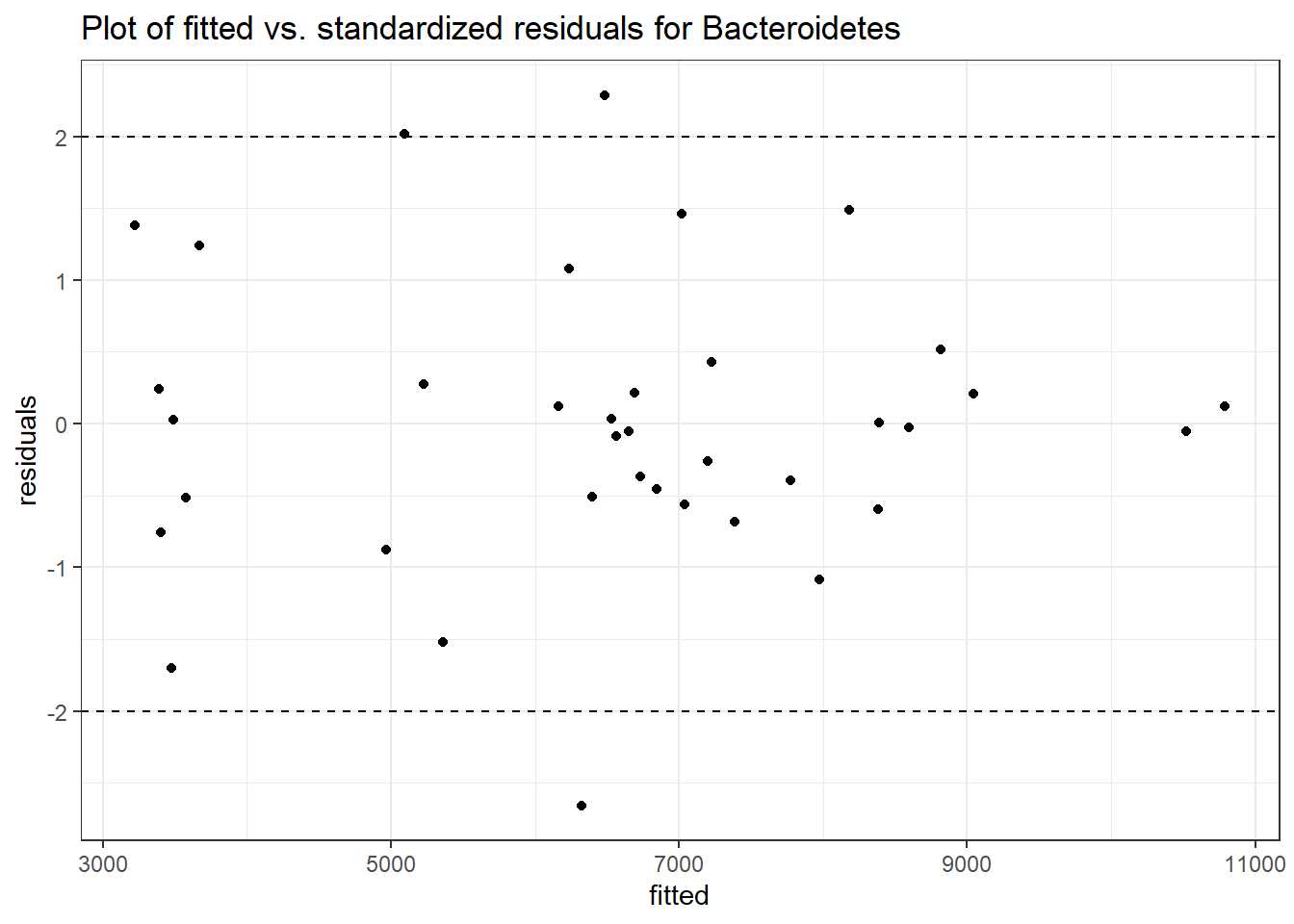
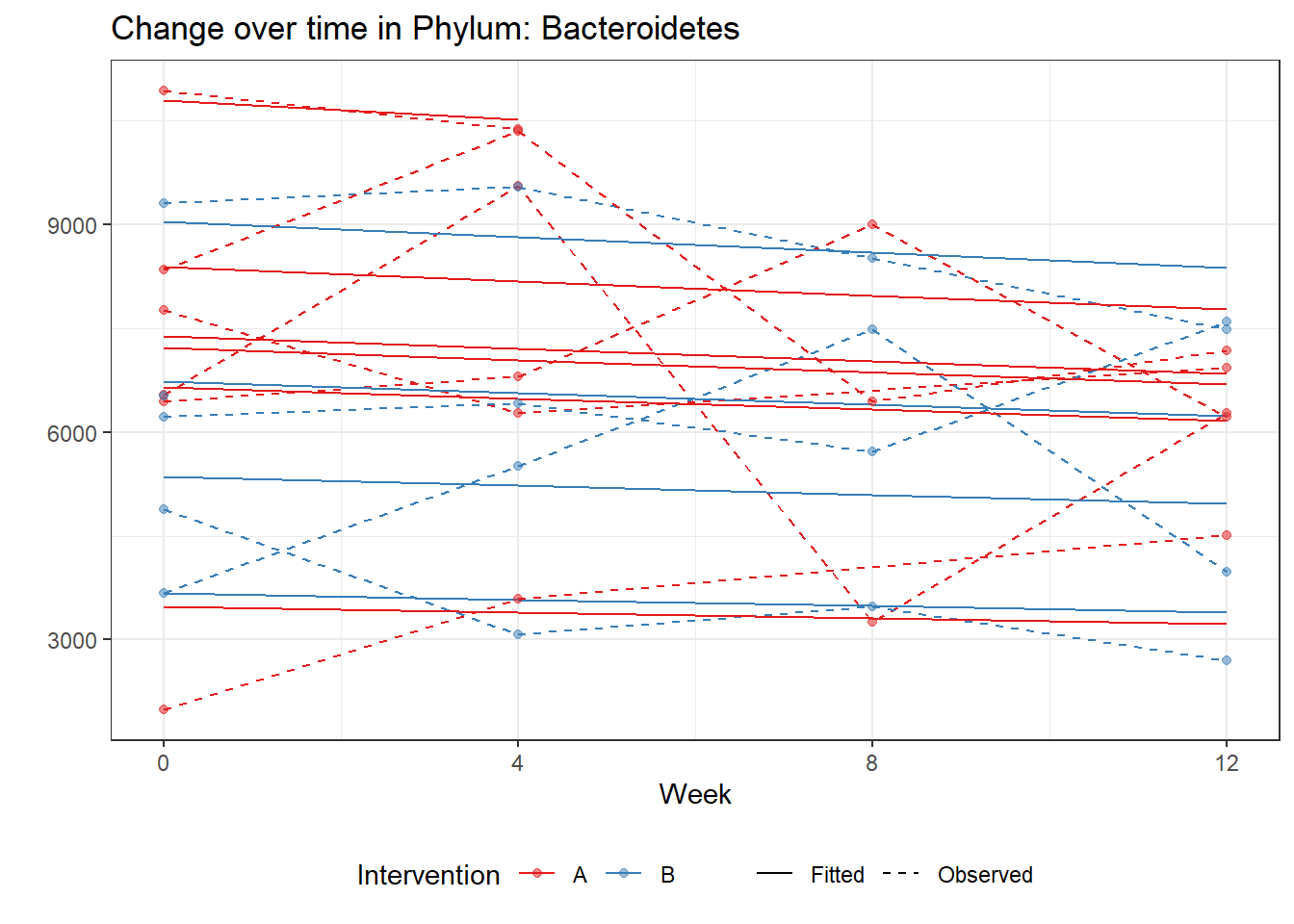
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroidetes 8.84509 0.13256 66.72348 0.00000
intB Bacteroidetes -0.14444 0.19660 -0.73466 0.46254
time Bacteroidetes -0.02541 0.00189 -13.45500 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.32458
Significance test not available
#########################################
#########################################
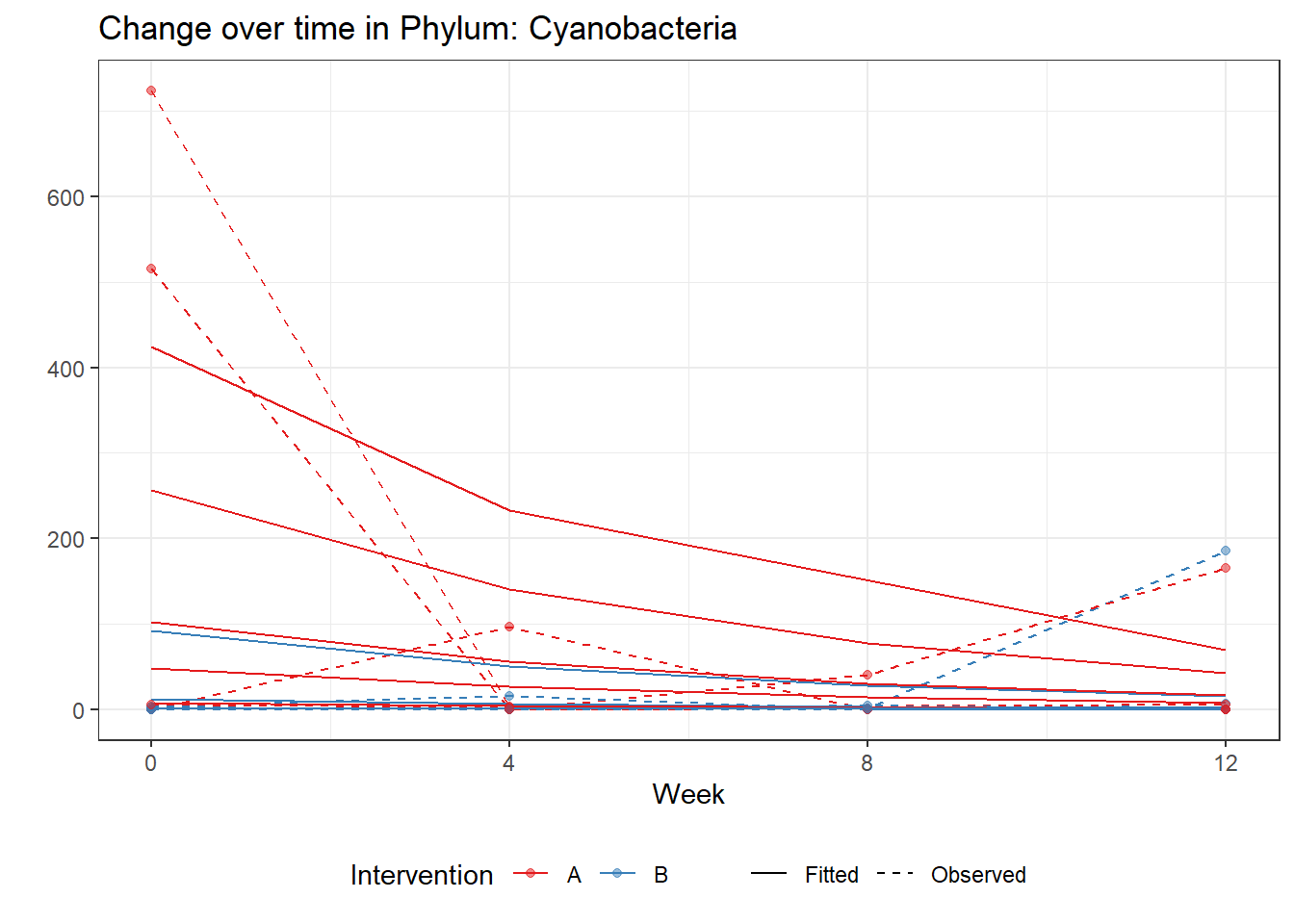
Model: Cyanobacteria ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x000000003ca59558>
Link: poisson
Intraclass Correlation (ICC): 0.8210815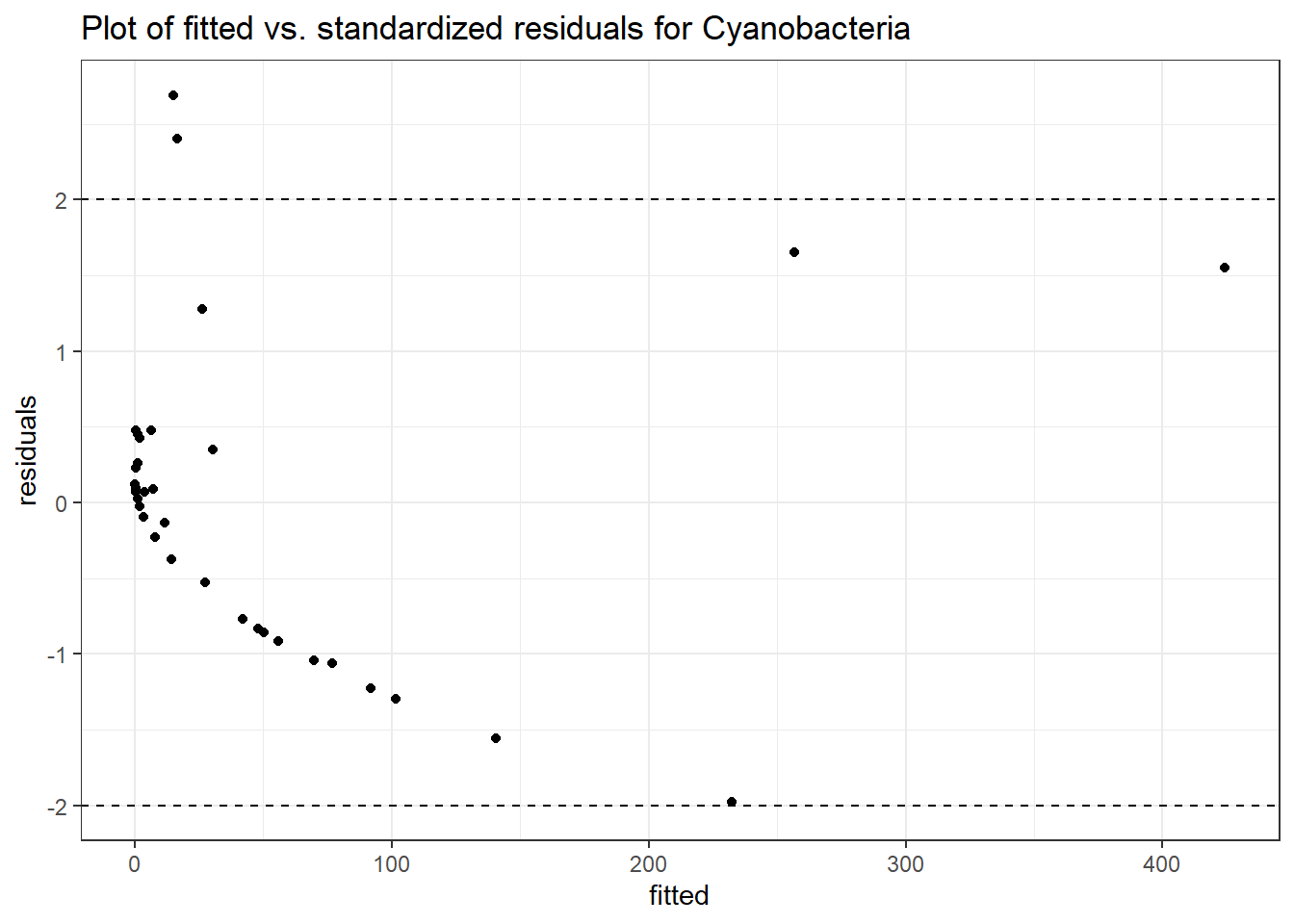
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Cyanobacteria 3.65573 0.88488 4.13131 0.00004
intB Cyanobacteria -2.41428 1.33763 -1.80489 0.07109
time Cyanobacteria -0.60182 0.02518 -23.89744 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.1422
Significance test not available
#########################################
#########################################
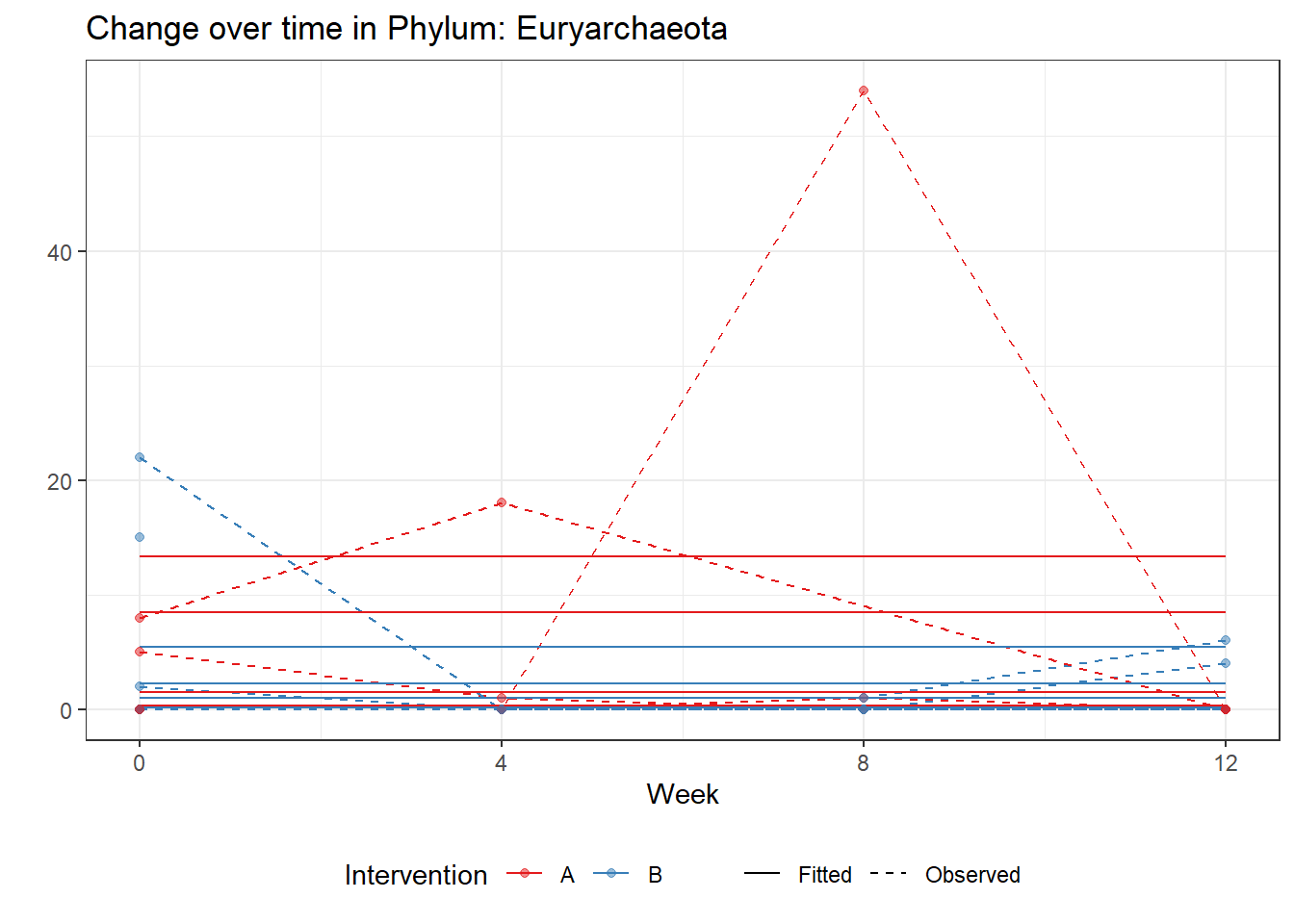
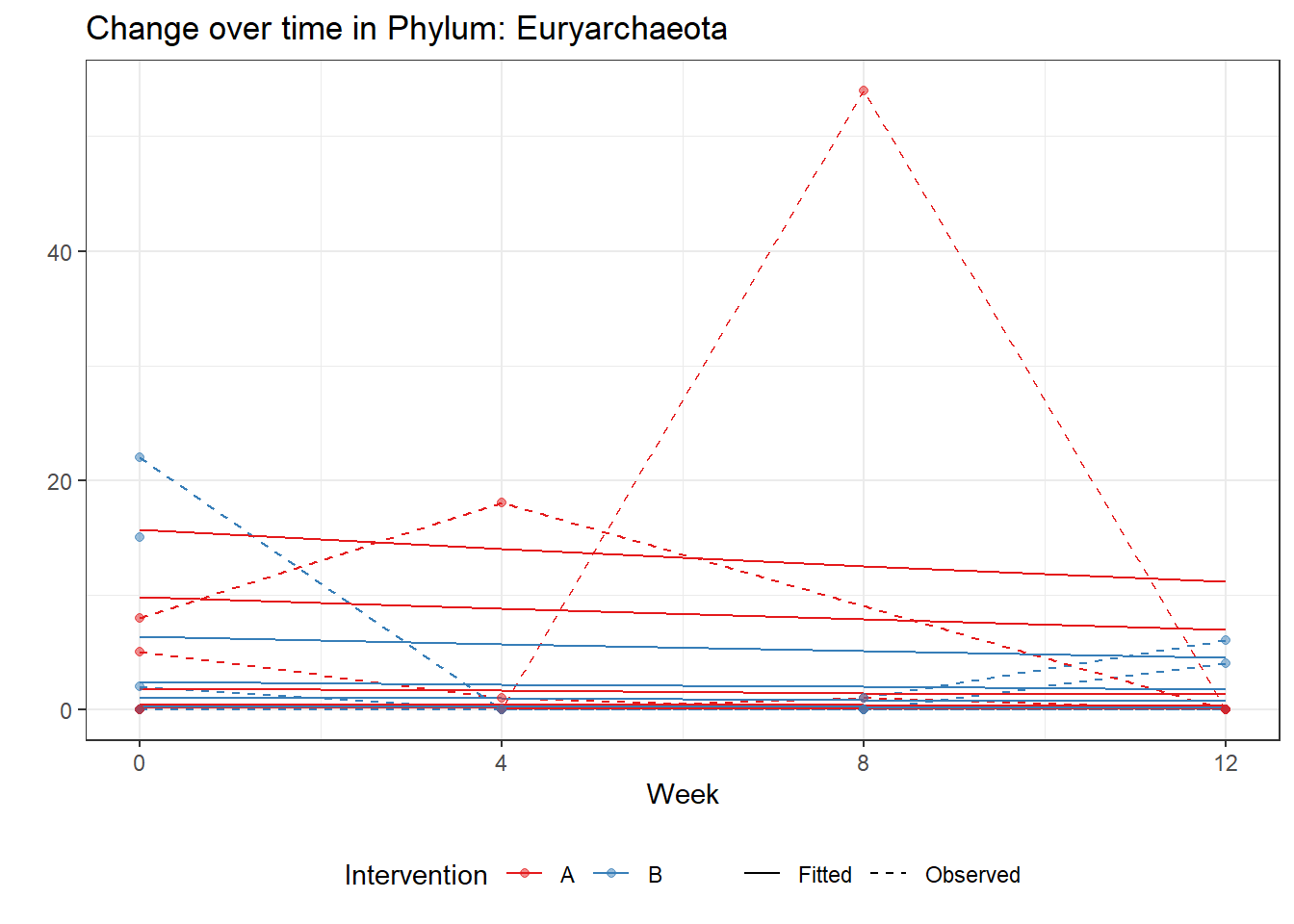
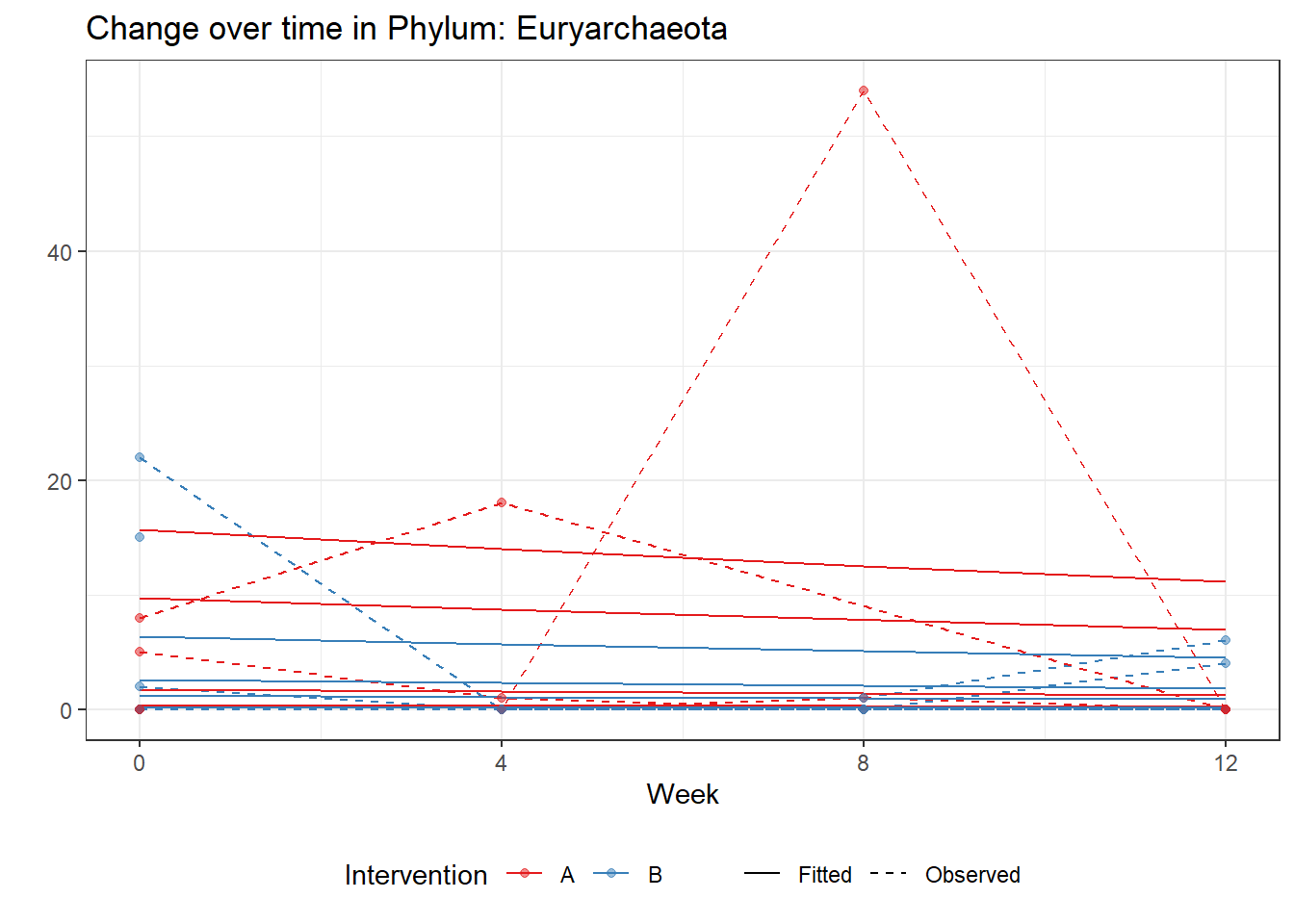
Model: Euryarchaeota ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x000000003ca59558>
Link: poissonWarning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model failed to converge with max|grad| = 0.0419304 (tol = 0.002, component 1)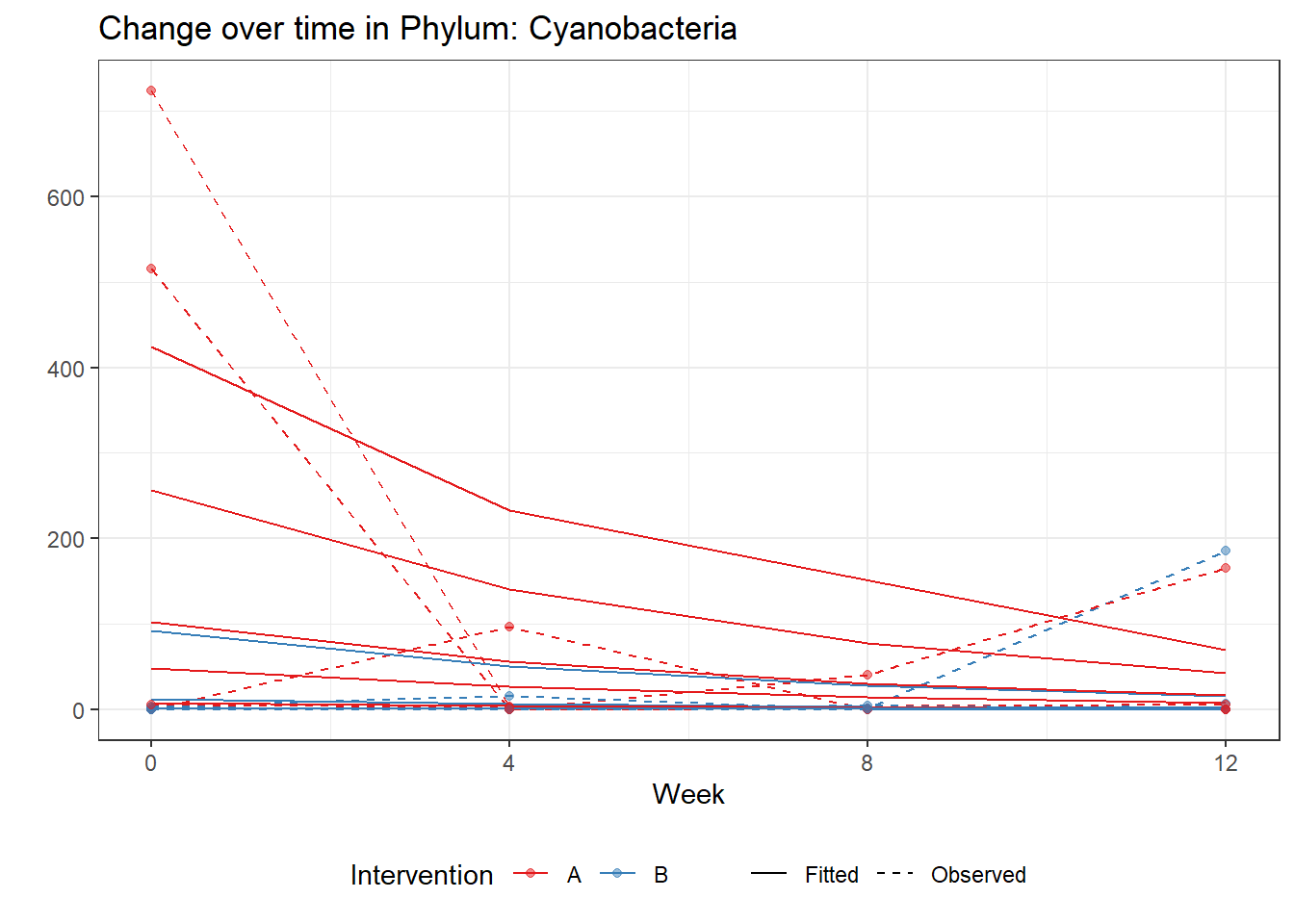
Intraclass Correlation (ICC): 0.7885852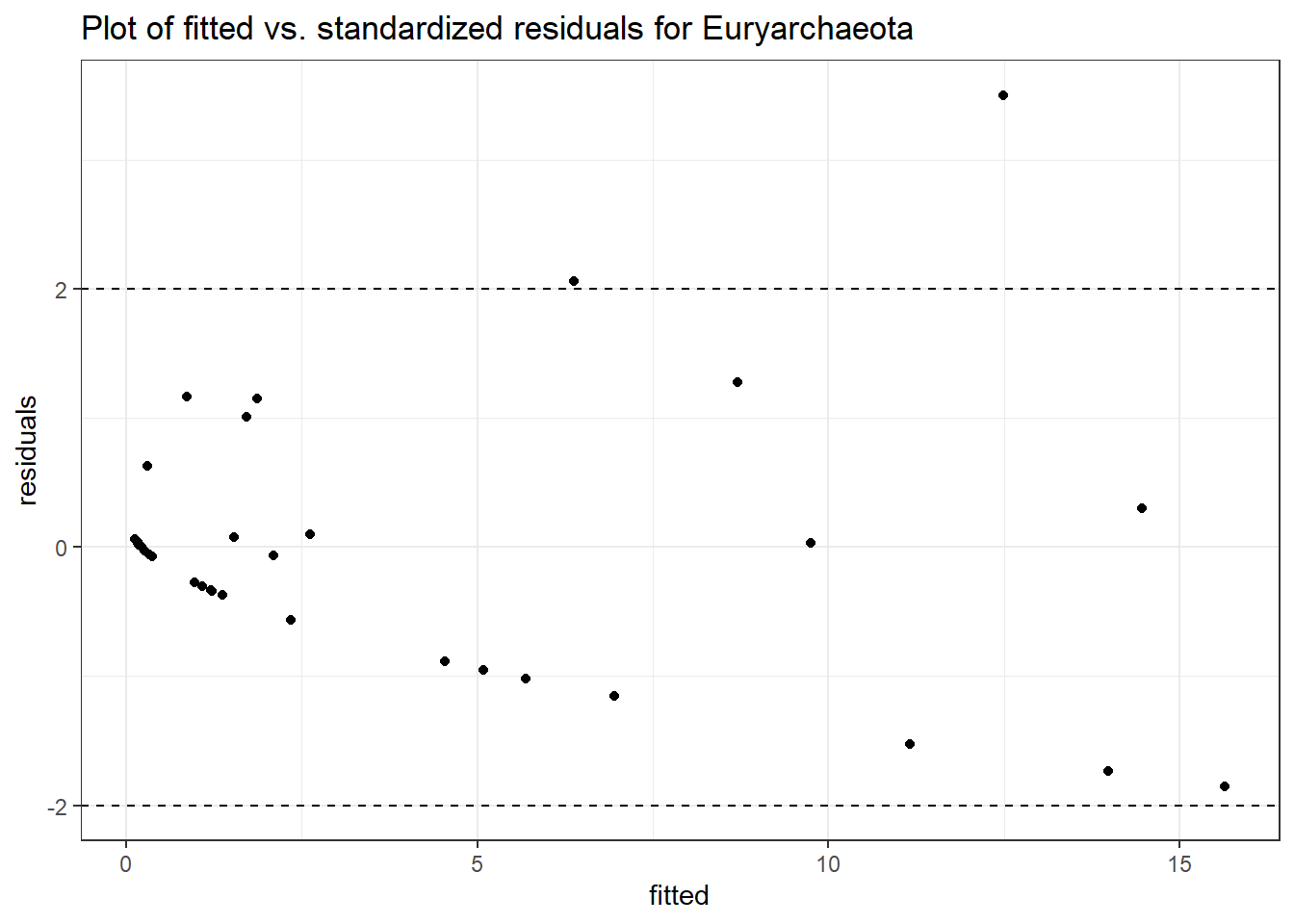
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Euryarchaeota -0.00149 0.00169 -0.88305 0.37721
intB Euryarchaeota 0.67878 0.00169 402.52163 0.00000
time Euryarchaeota -0.11280 0.00169 -66.88817 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.9313
Significance test not available
#########################################
#########################################
Model: Firmicutes ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x000000003ca59558>
Link: poisson
Intraclass Correlation (ICC): 0.0859897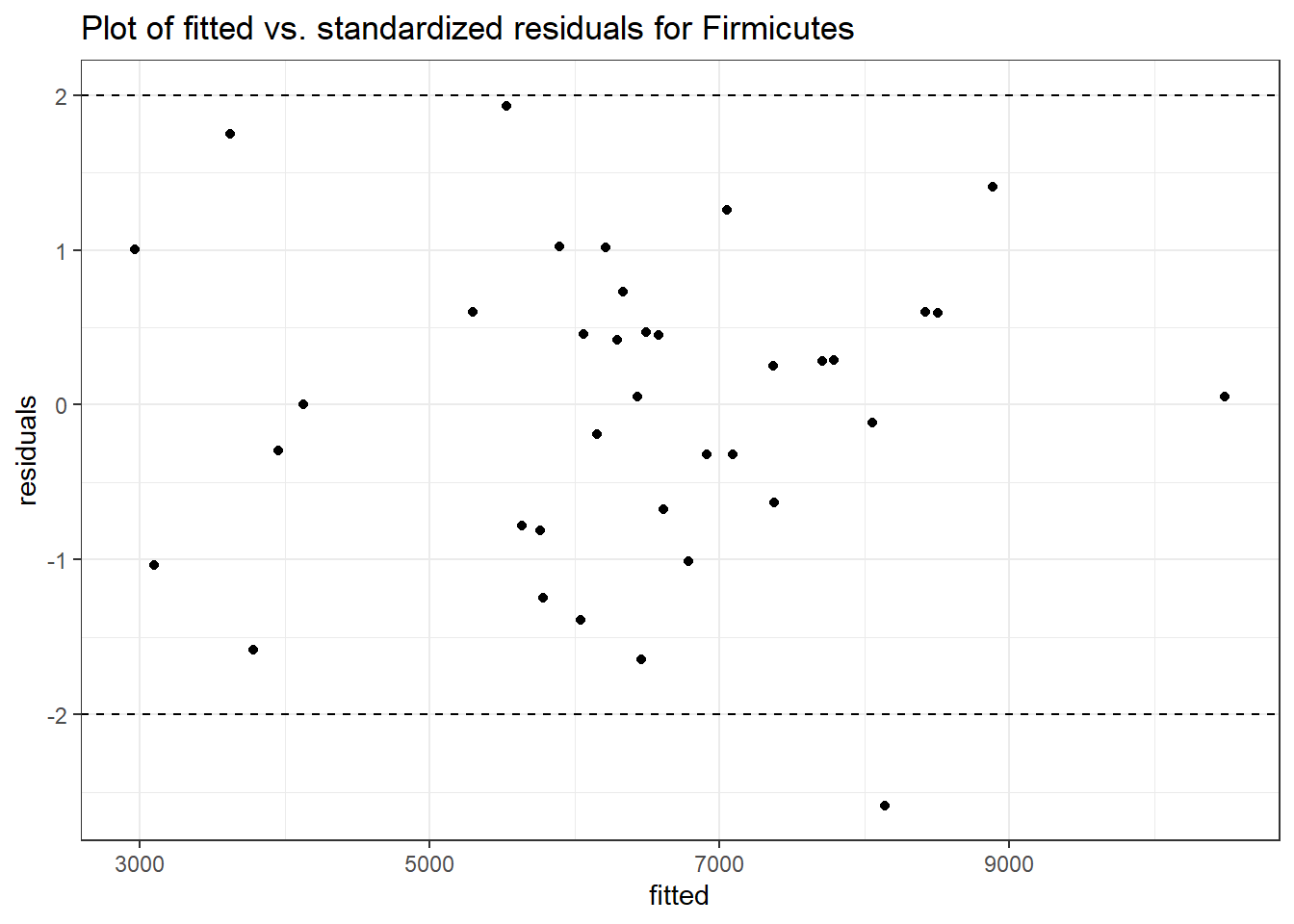
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Firmicutes 8.67971 0.12528 69.28039 0.00000
intB Firmicutes 0.23156 0.18579 1.24636 0.21263
time Firmicutes -0.04391 0.00187 -23.47439 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.30672
Significance test not available
#########################################
#########################################
Model: Fusobacteria ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x000000003ca59558>
Link: poisson
Intraclass Correlation (ICC): 0.8326759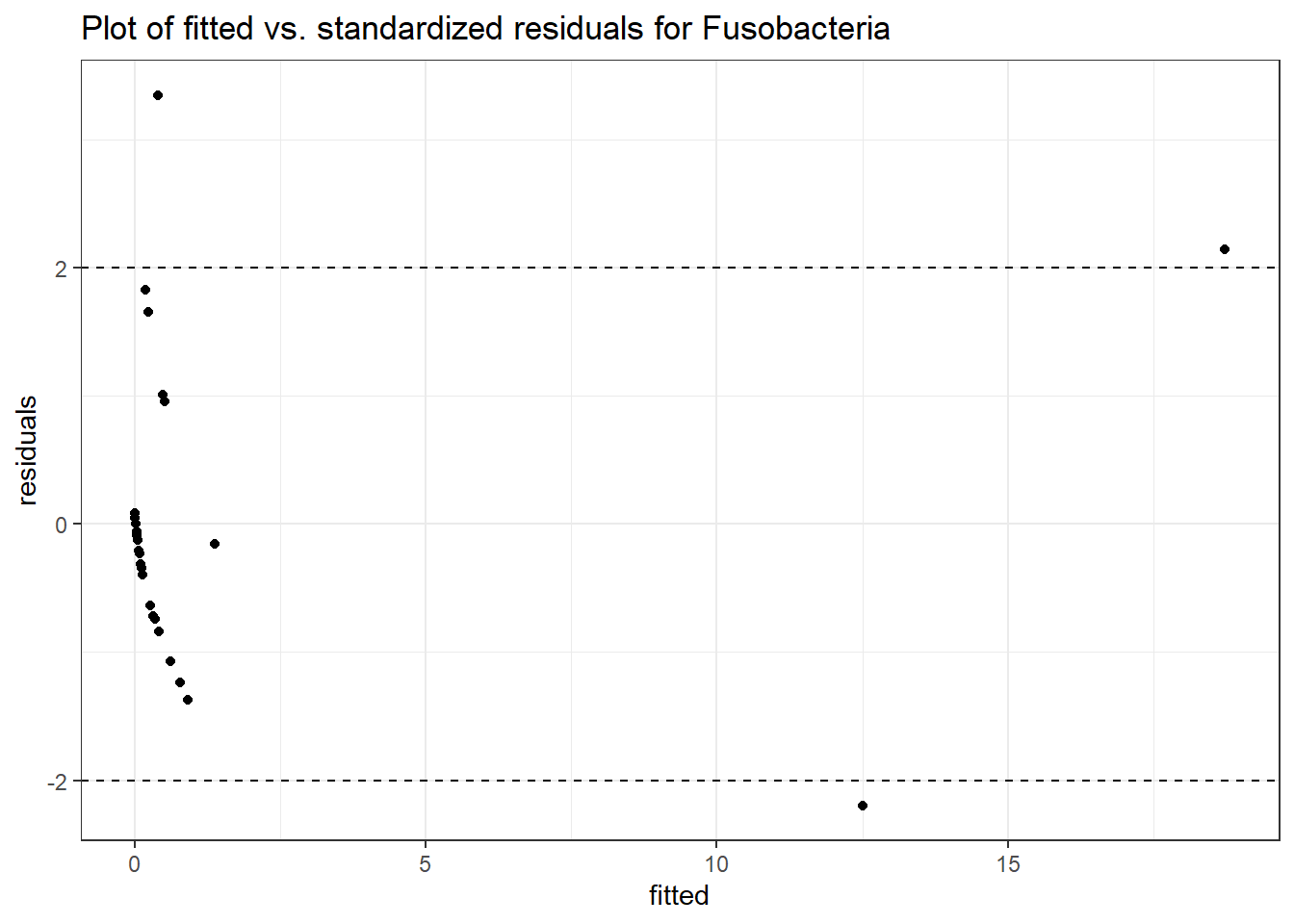
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Fusobacteria -0.89901 1.14984 -0.78185 0.43430
intB Fusobacteria -2.13150 1.82425 -1.16843 0.24263
time Fusobacteria -0.40385 0.24243 -1.66586 0.09574
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.2308
Significance test not available
#########################################
#########################################
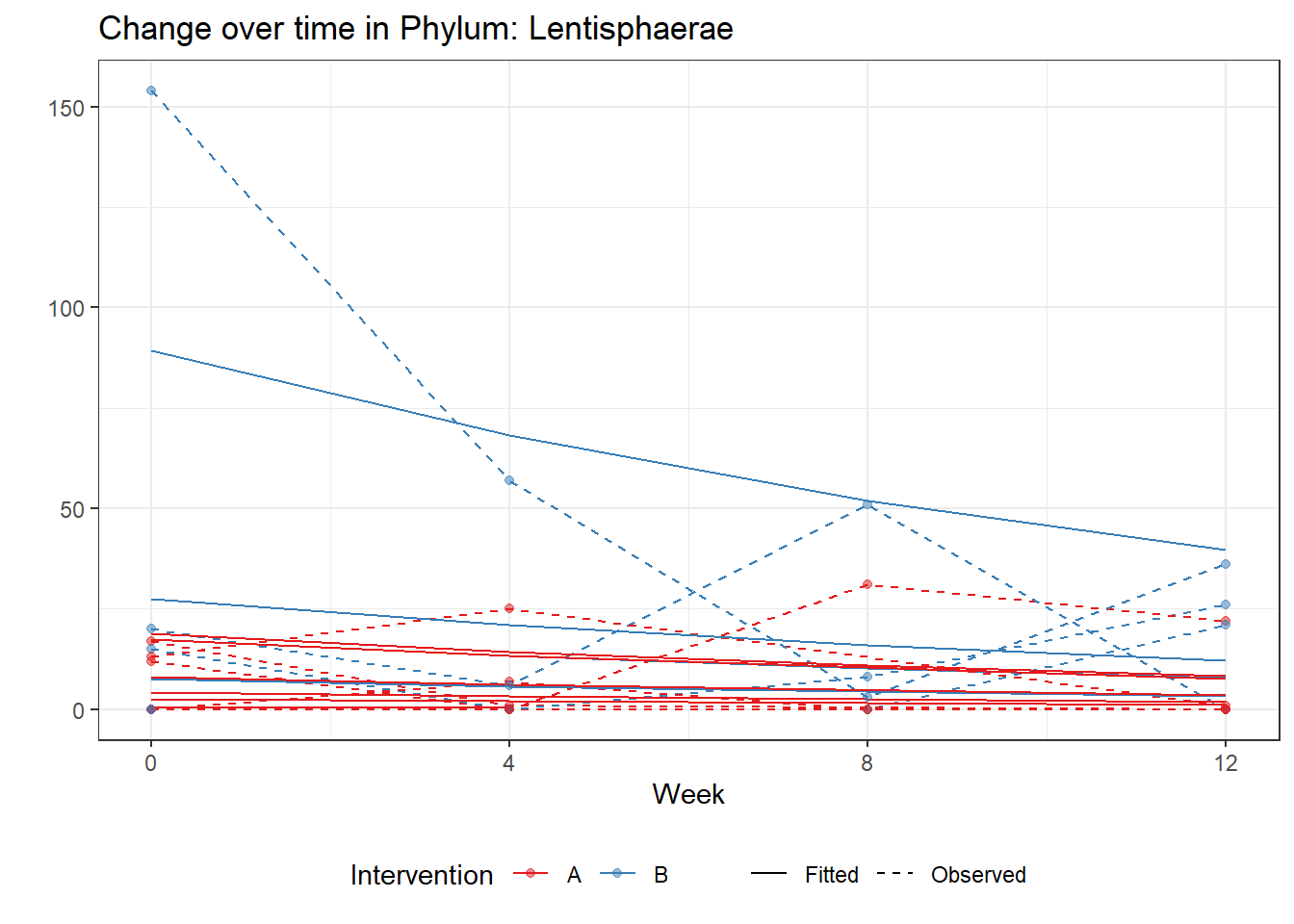
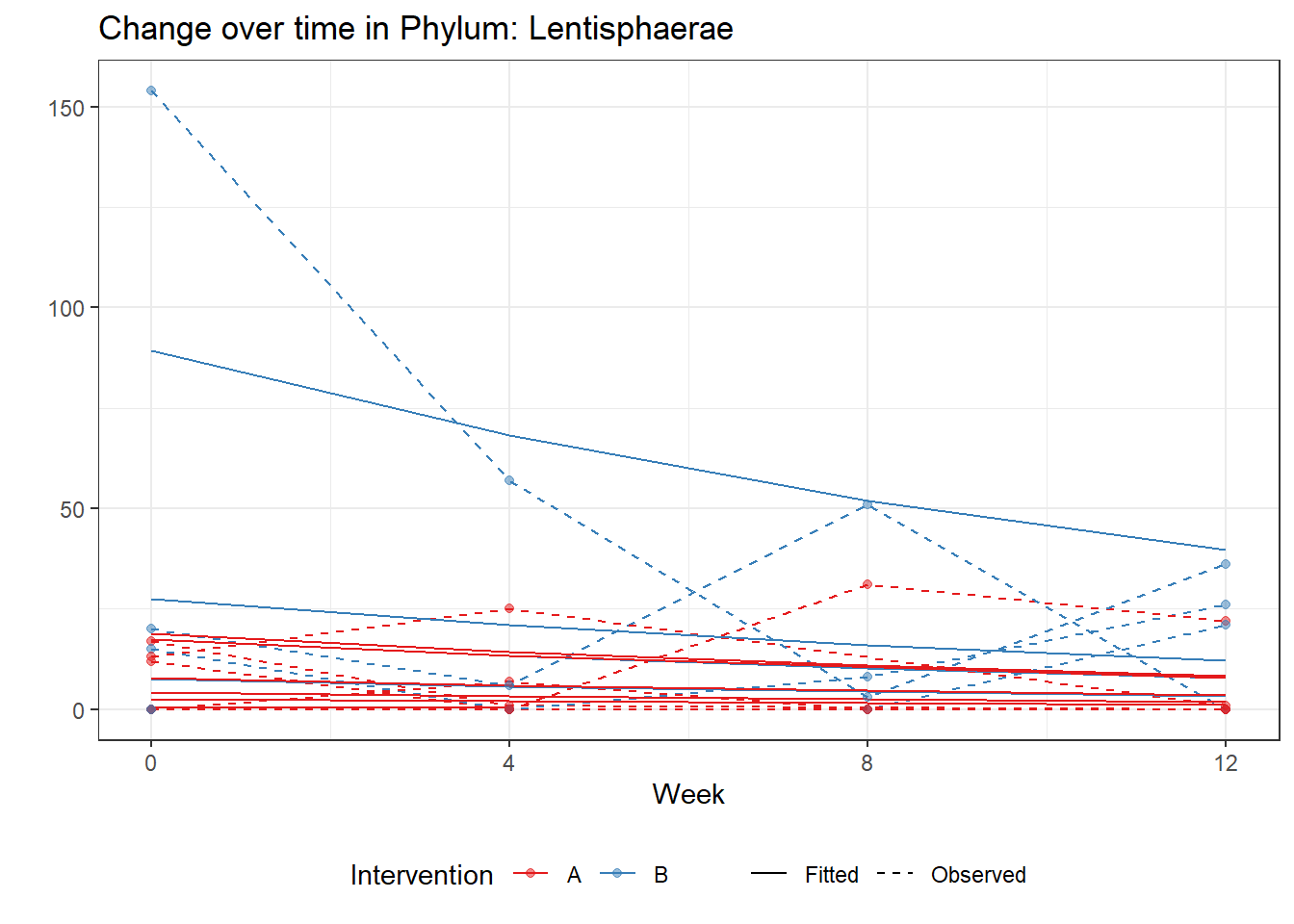
Model: Lentisphaerae ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x000000003ca59558>
Link: poisson
Intraclass Correlation (ICC): 0.6748463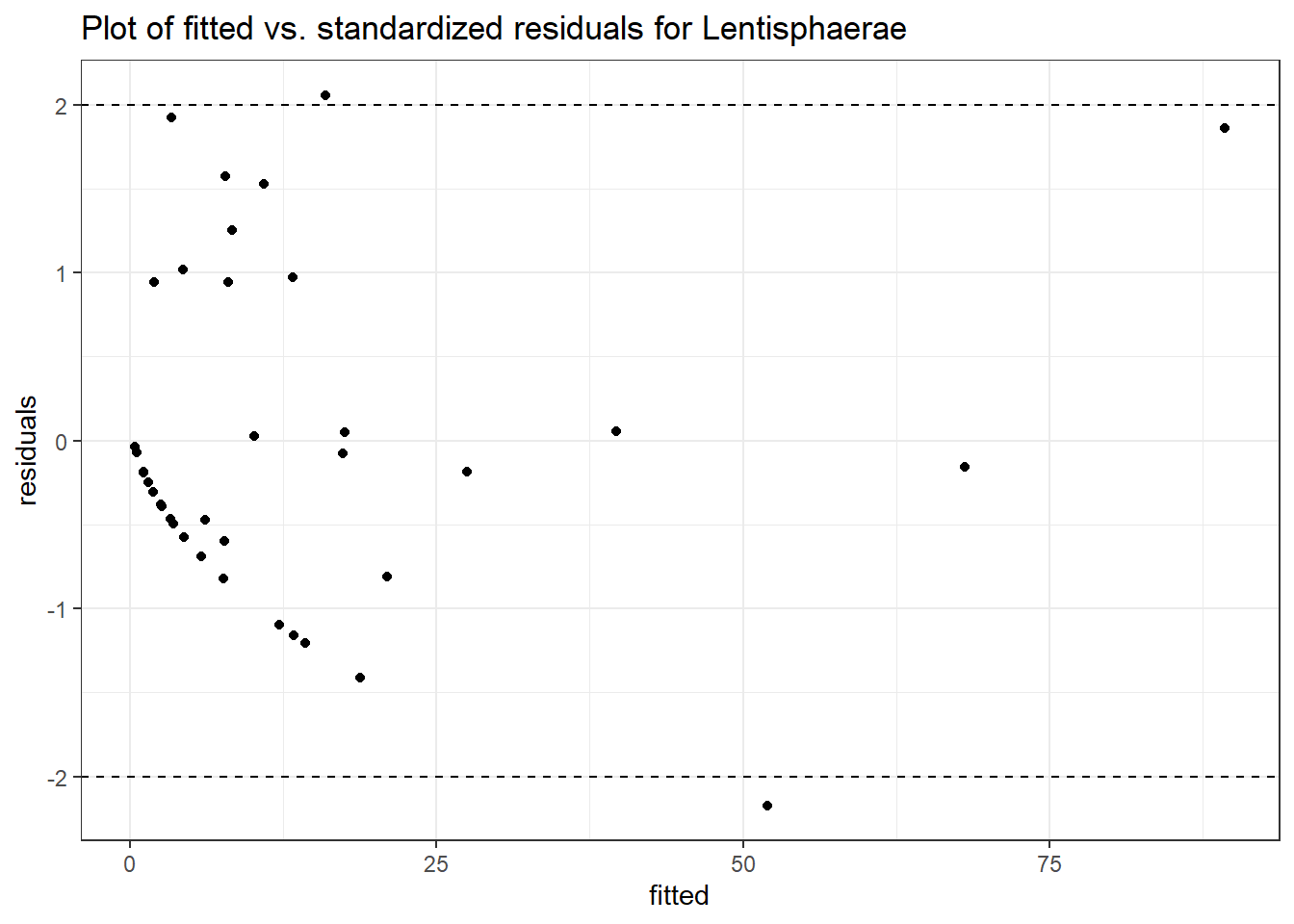
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lentisphaerae 1.56301 0.61744 2.53142 0.01136
intB Lentisphaerae 0.95309 0.90400 1.05431 0.29174
time Lentisphaerae -0.27072 0.03999 -6.77006 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.4406
Significance test not available
#########################################
#########################################
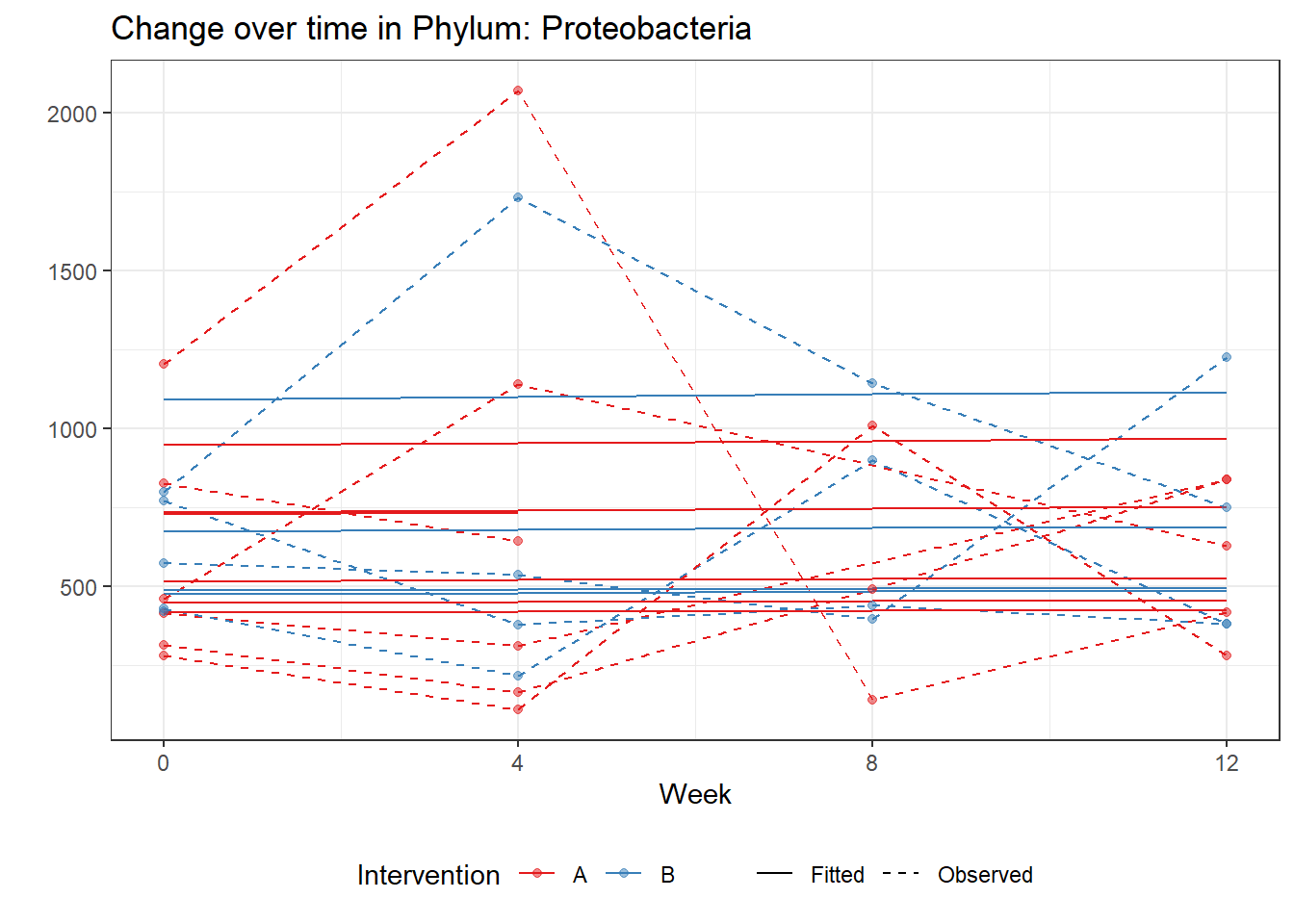
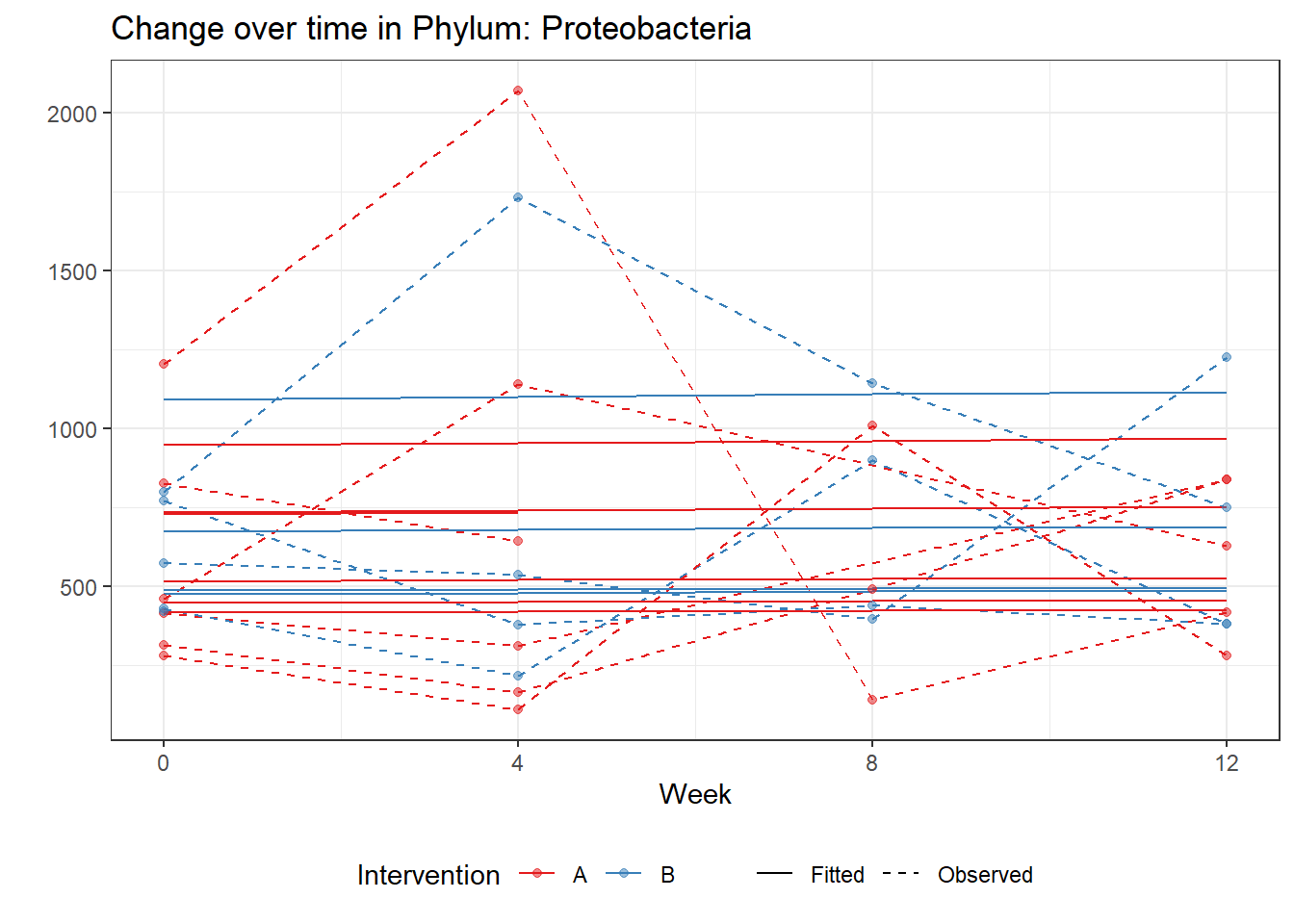
Model: Proteobacteria ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x000000003ca59558>
Link: poisson
Intraclass Correlation (ICC): 0.09281958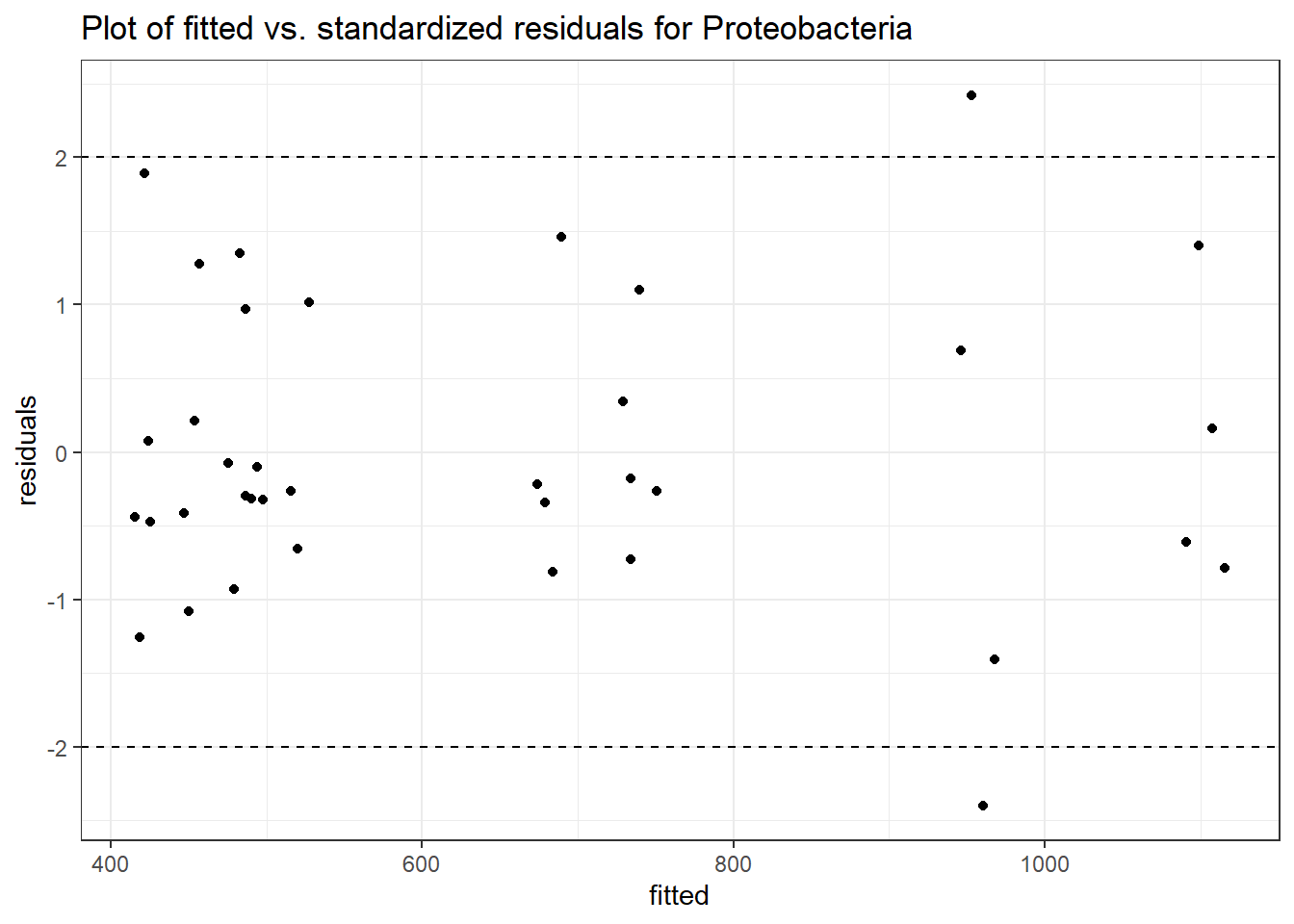
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Proteobacteria 6.40308 0.13114 48.82631 0.00000
intB Proteobacteria -0.02151 0.19433 -0.11068 0.91187
time Proteobacteria 0.00754 0.00585 1.29008 0.19702
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.31987
Significance test not available
#########################################
#########################################
Model: Tenericutes ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x000000003ca59558>
Link: poisson
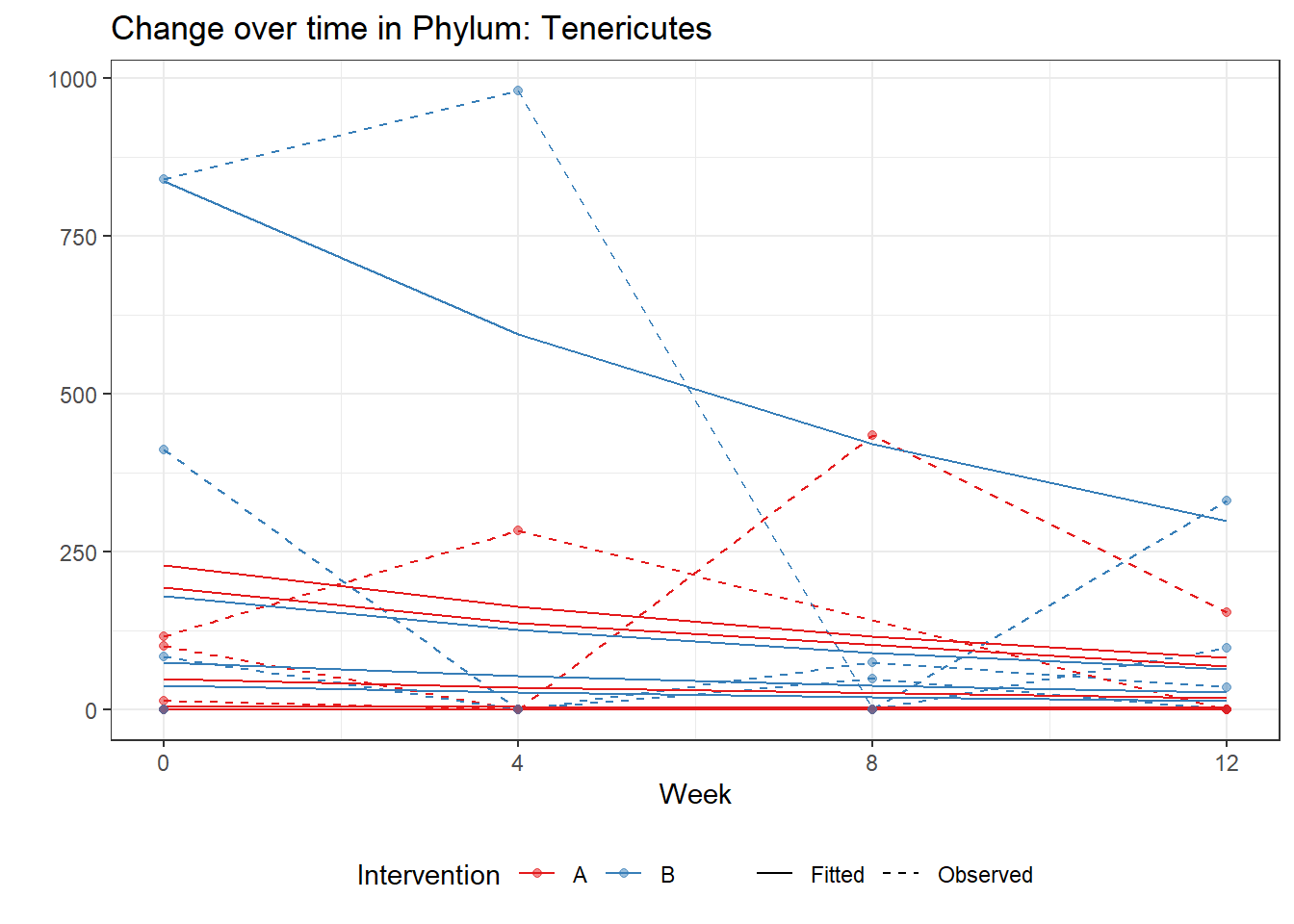
Intraclass Correlation (ICC): 0.9107768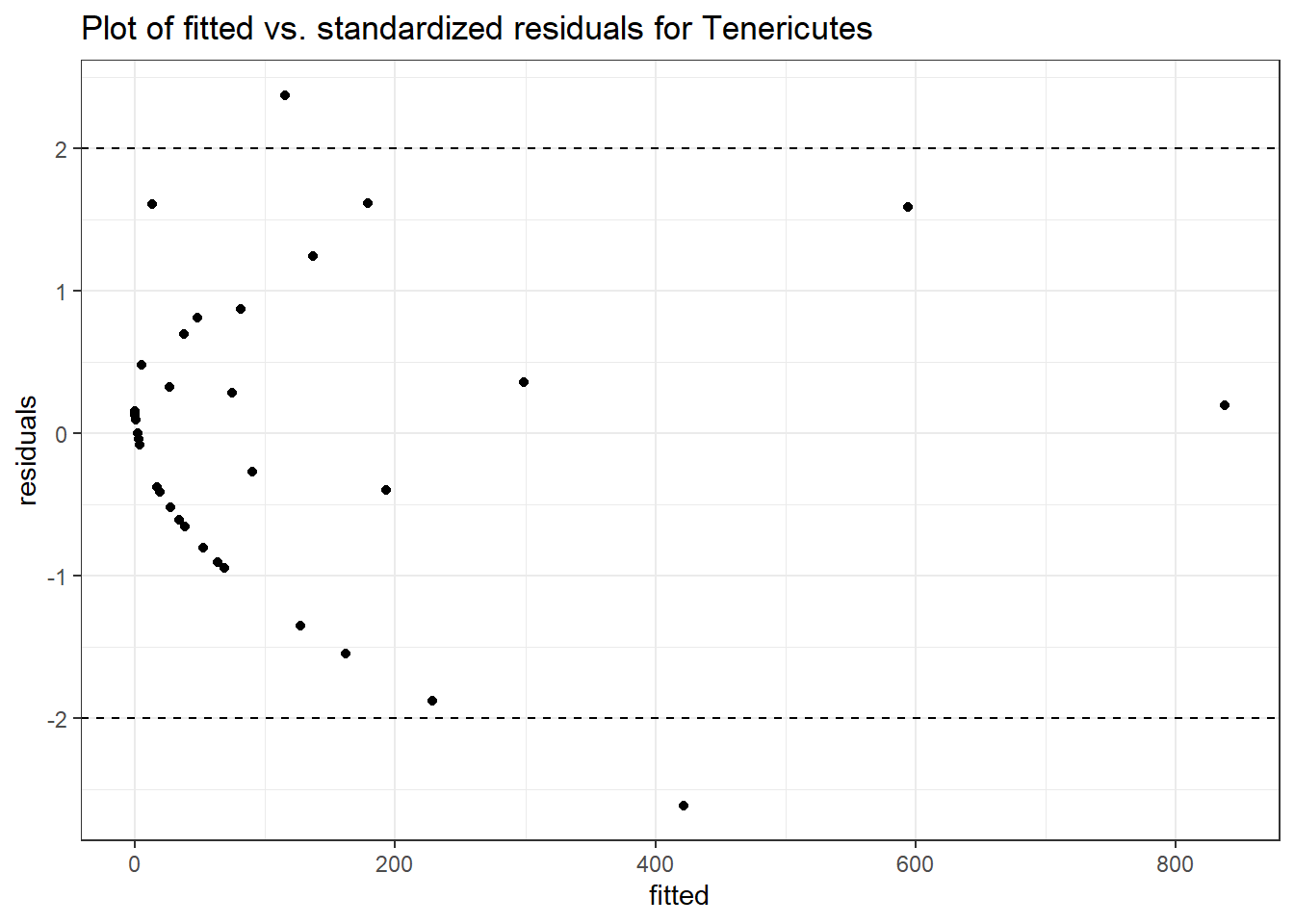
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Tenericutes 1.83646 1.35864 1.35169 0.17648
intB Tenericutes 1.82166 1.98222 0.91900 0.35810
time Tenericutes -0.34364 0.01482 -23.18197 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 3.195
Significance test not available
#########################################
#########################################
Model: Verrucomicrobia ~ 1 + intB + time + (1 | SubjectID)
<environment: 0x000000003ca59558>
Link: poisson
Intraclass Correlation (ICC): 0.8753226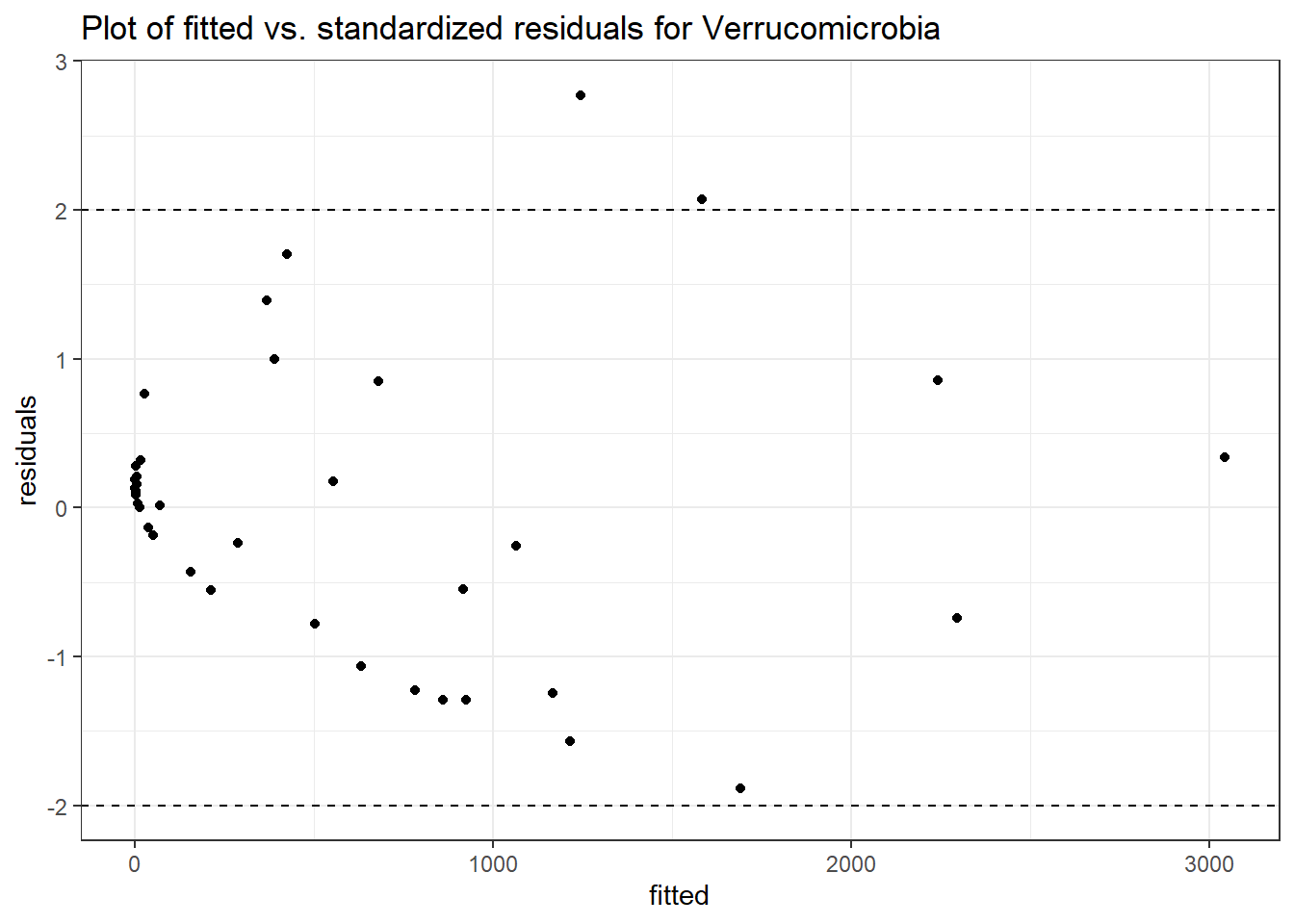
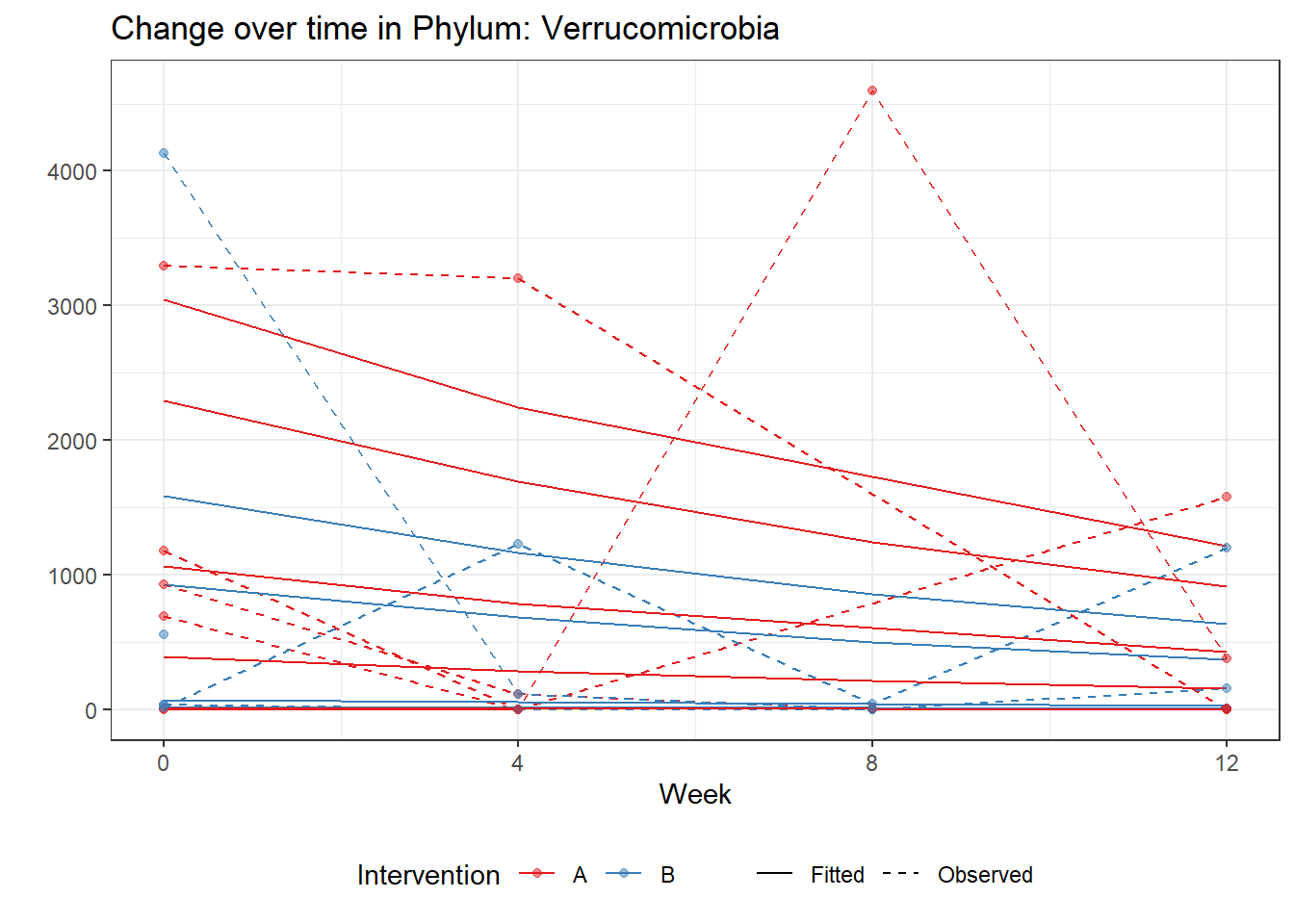
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Verrucomicrobia 4.97431 1.09055 4.56127 0.00001
intB Verrucomicrobia 0.54433 1.61144 0.33779 0.73552
time Verrucomicrobia -0.30578 0.00609 -50.22337 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.6497
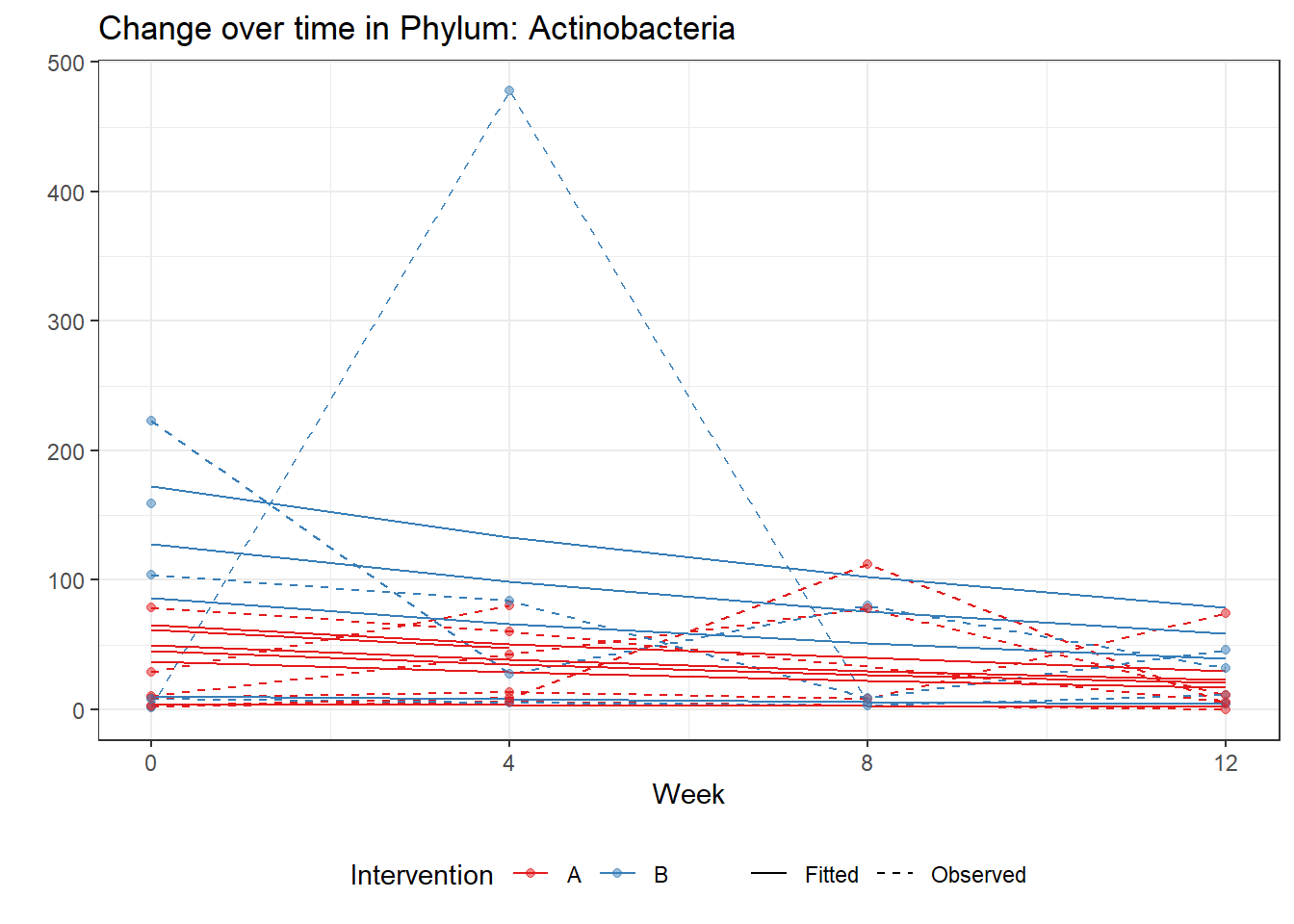
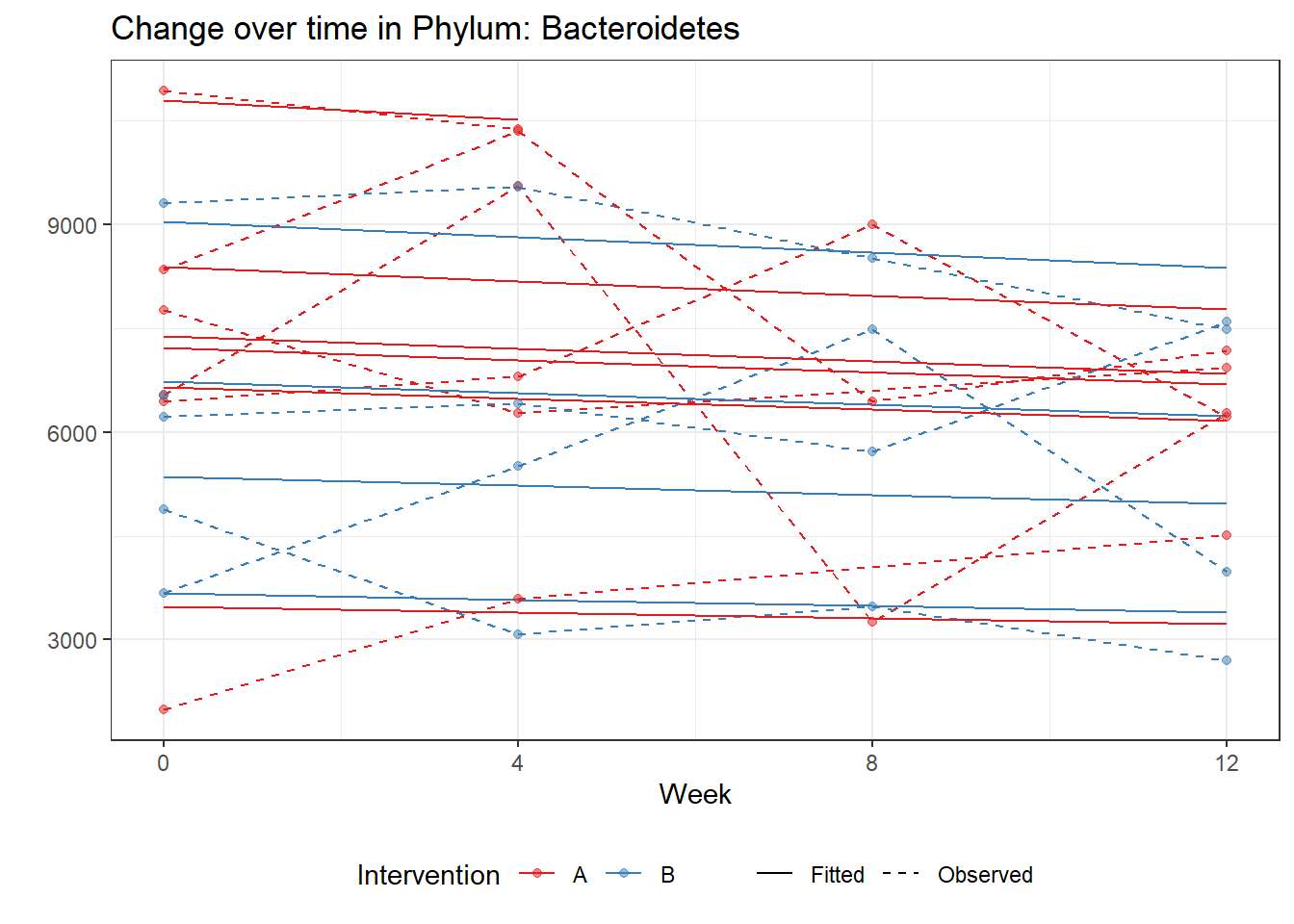
Significance test not availableControlling for the effect of time, the intervention resulted in a significant increase in abundance of Phylums: Euryarchaeota. This is in contrast to the effect of time, which was negatively associated with abundance. So, abundance of Euryarchaeota increased in intervention group B but not in group A.
Other interesting but not significant results. The effect of the intervention was in the opposite direction (positive) compared to the effect of time (negative) is Phylums: Actinobacteria, Firmicutes, Lentisphaerae, and Tenericutes.
Covariate(s): Time by intervention interaction
Here, the model tests whether the effect of the intervention is constant across time or does the effect of the intervention get stronger (or weaker) at time progress. And remember, time is coded 0, 1, 2, 3, so a 1 unit increase in time corresponds to 1 month or 4 weeks of the intervention.
\[\begin{align*} h\left(Y_{ij}\right)&=\beta_{0j} + \beta_{1j}(Week)_{ij} + r_{ij}\\ \beta_{0j}&= \gamma_{00} + \gamma_{01}(Intervention)_{j} + u_{0j}\\ \beta_{1j}&= \gamma_{10} + \gamma_{11}(Intervention)_{j}\\ \end{align*}\]
It could be that the intervention is directly related to time so included a average effect of time through the main effect effect is just adding more parameters and is not informative. If the effect of the intervention is still essentially zero, we will re-estimate the model without the main effect.
phylum.fit5 <- glmm_microbiome(mydata=microbiome_data,model.number=5,
taxa.level="Phylum", link="poisson",
model="1 + intB + time + intB:time + (1|SubjectID)")
#########################################
#########################################
Model: Actinobacteria ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x00000000071c9878>
Link: poisson
Intraclass Correlation (ICC): 0.5000466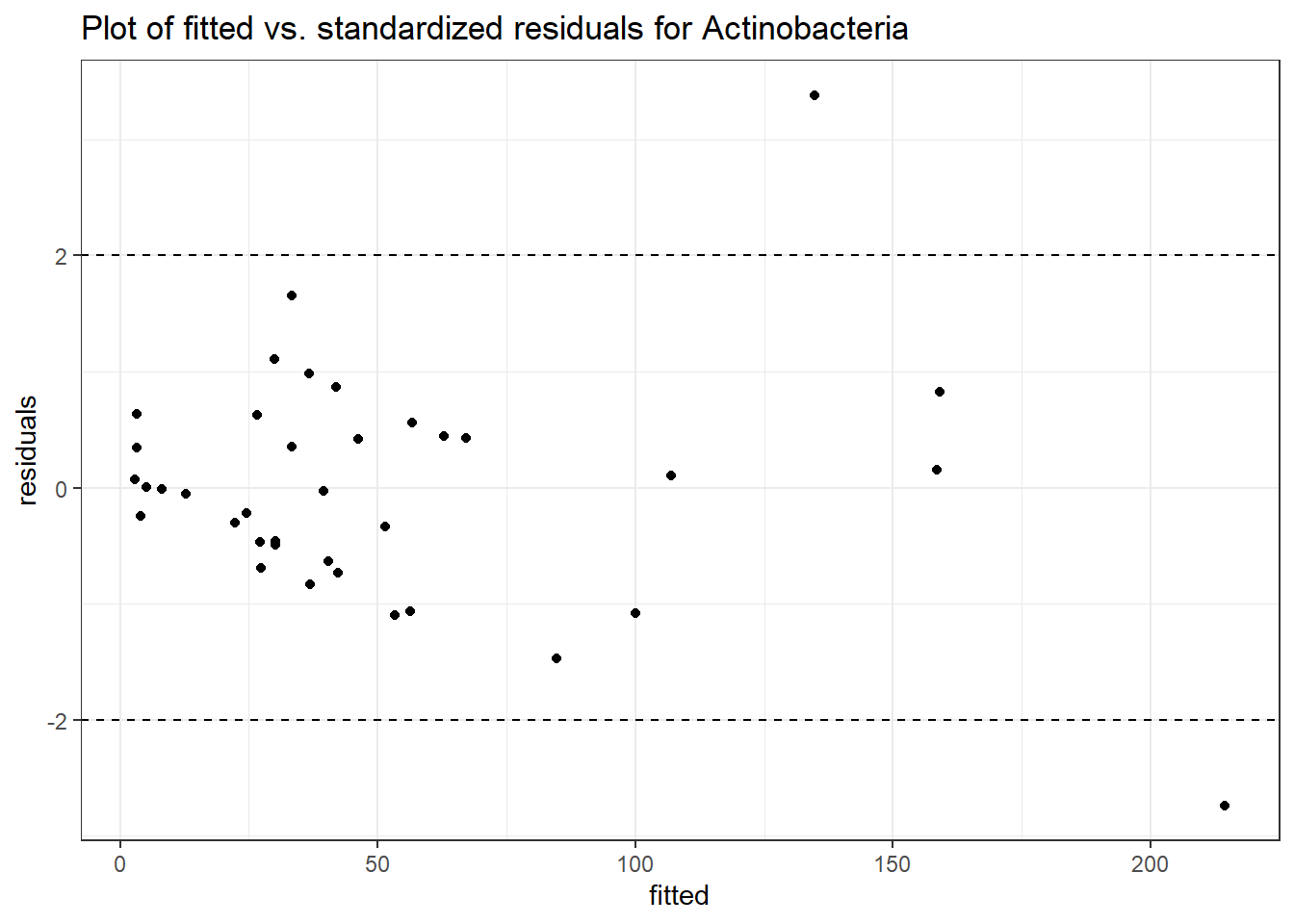
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Actinobacteria 3.08368 0.41620 7.40904 0.00000
intB Actinobacteria 1.45591 0.61285 2.37563 0.01752
time Actinobacteria 0.09832 0.03672 2.67733 0.00742
intB:time Actinobacteria -0.56276 0.04688 -12.00542 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.0001
Significance test not available
#########################################
#########################################
Model: Bacteroidetes ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x00000000071c9878>
Link: poisson
Intraclass Correlation (ICC): 0.09532667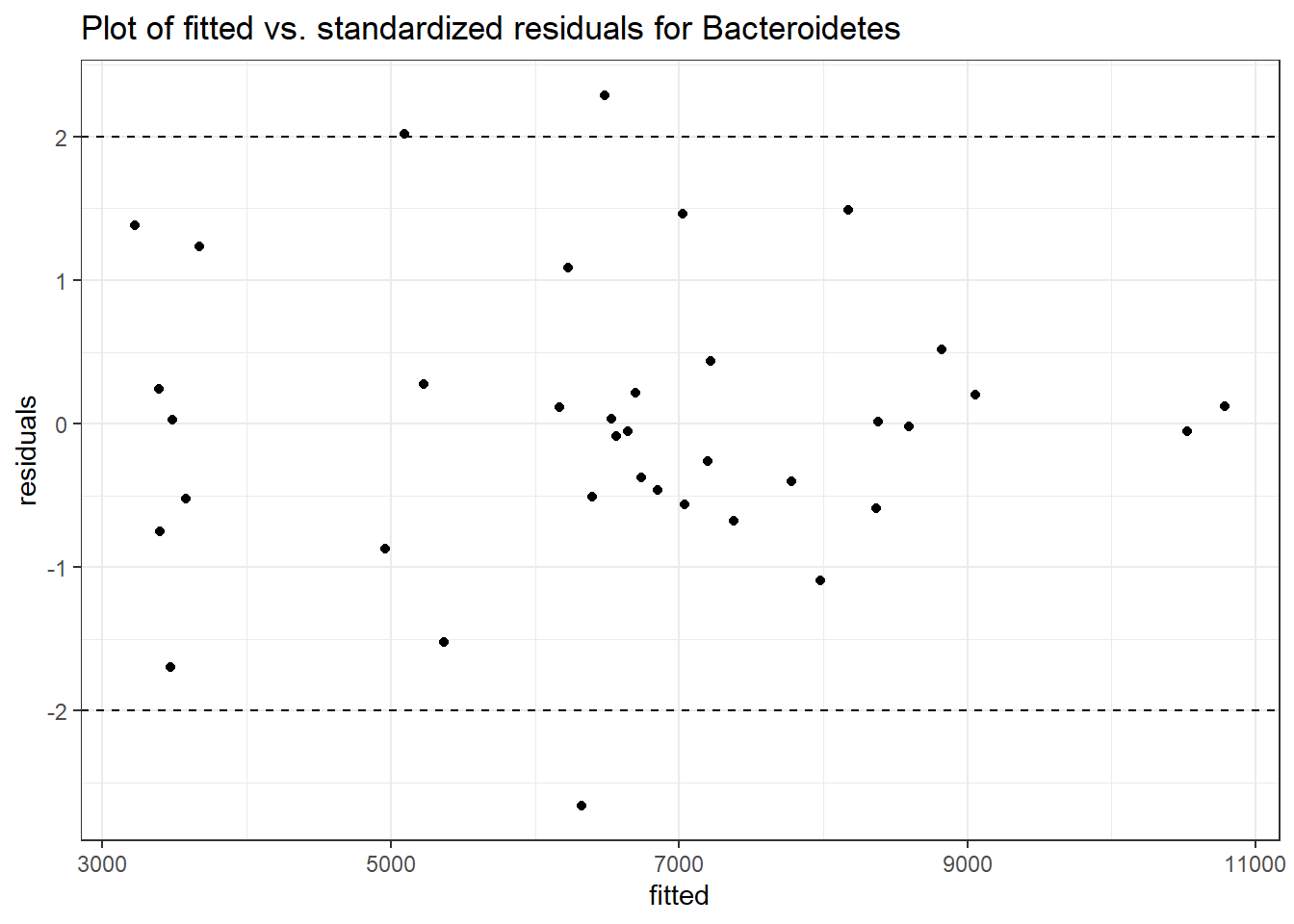
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroidetes 8.84429 0.13259 66.70503 0.00000
intB Bacteroidetes -0.14262 0.19667 -0.72516 0.46835
time Bacteroidetes -0.02477 0.00249 -9.93904 0.00000
intB:time Bacteroidetes -0.00150 0.00382 -0.39278 0.69448
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.32461
Significance test not available
#########################################
#########################################
Model: Cyanobacteria ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x00000000071c9878>
Link: poisson
Intraclass Correlation (ICC): 0.8022182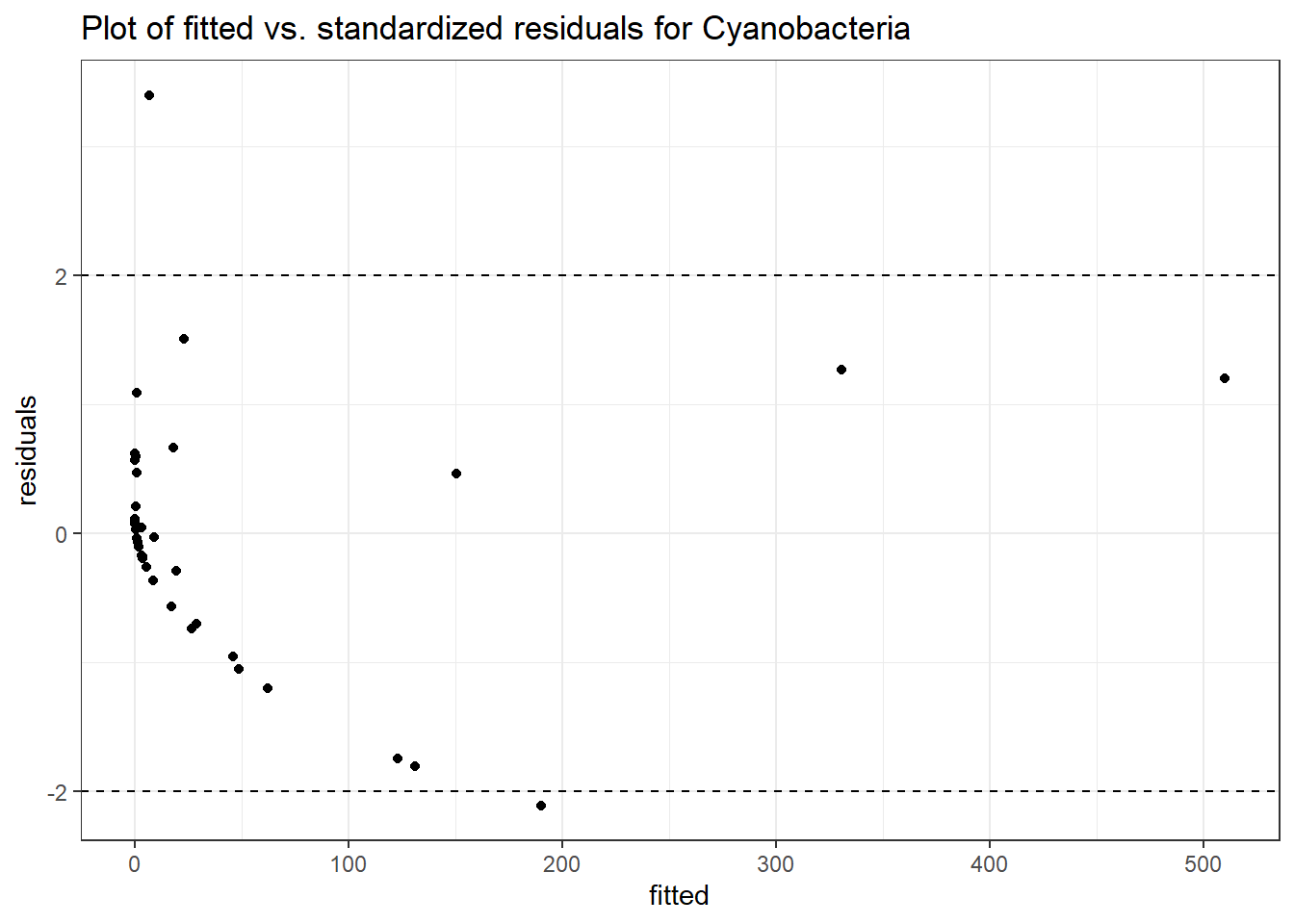
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Cyanobacteria 3.87746 0.83283 4.65578 0
intB Cyanobacteria -6.58632 1.32883 -4.95647 0
time Cyanobacteria -0.98832 0.03438 -28.74885 0
intB:time Cyanobacteria 2.64708 0.13435 19.70301 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.014
Significance test not available
#########################################
#########################################
Model: Euryarchaeota ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x00000000071c9878>
Link: poissonWarning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model failed to converge with max|grad| = 0.0409519 (tol = 0.002, component 1)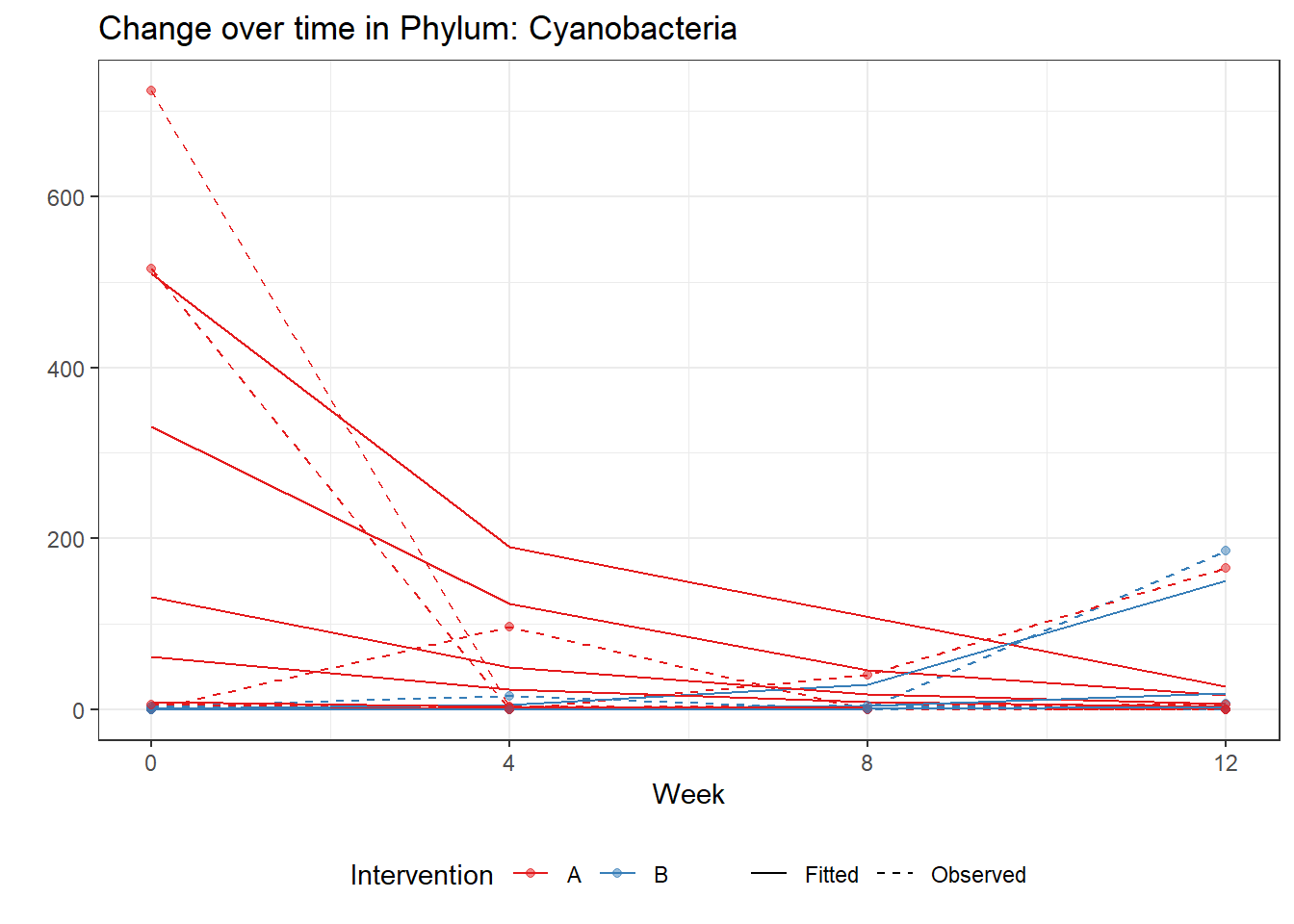
Intraclass Correlation (ICC): 0.7741486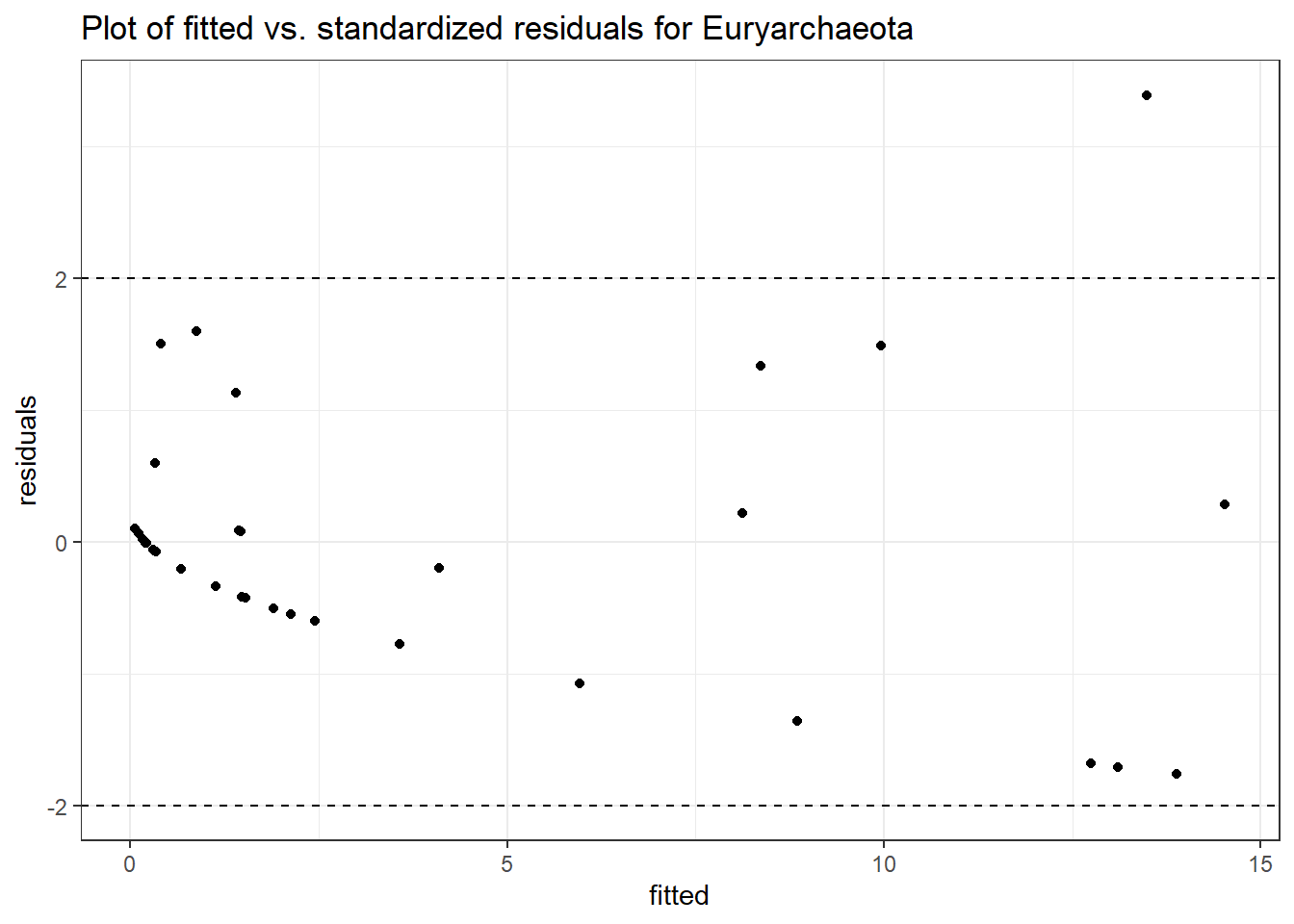
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
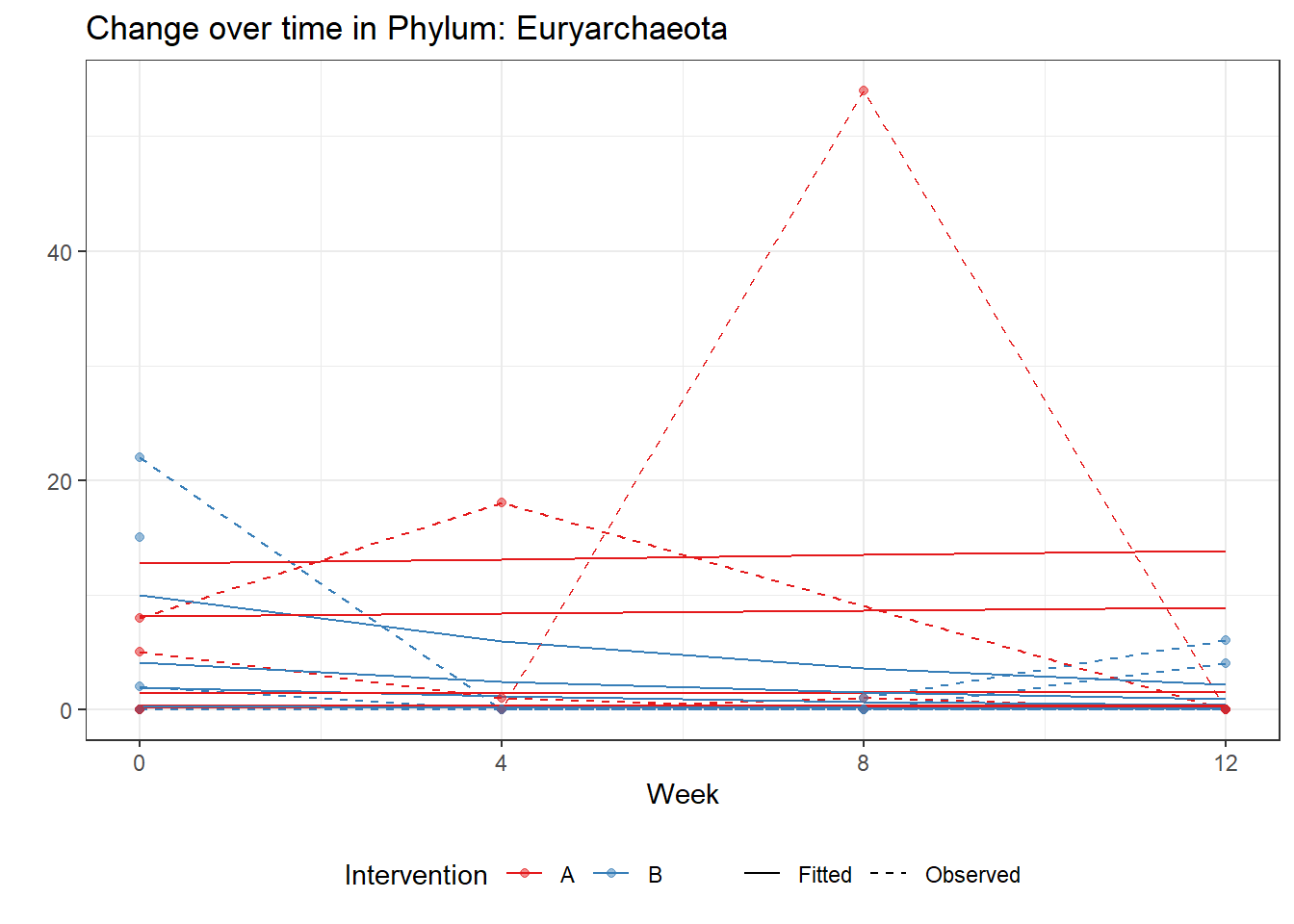
(Intercept) Euryarchaeota -0.14217 0.00173 -82.31957 0
intB Euryarchaeota 1.18925 0.00173 688.79114 0
time Euryarchaeota 0.02855 0.00173 16.53565 0
intB:time Euryarchaeota -0.54150 0.00173 -313.64265 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.8514
Significance test not available
#########################################
#########################################
Model: Firmicutes ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x00000000071c9878>
Link: poisson
Intraclass Correlation (ICC): 0.08185485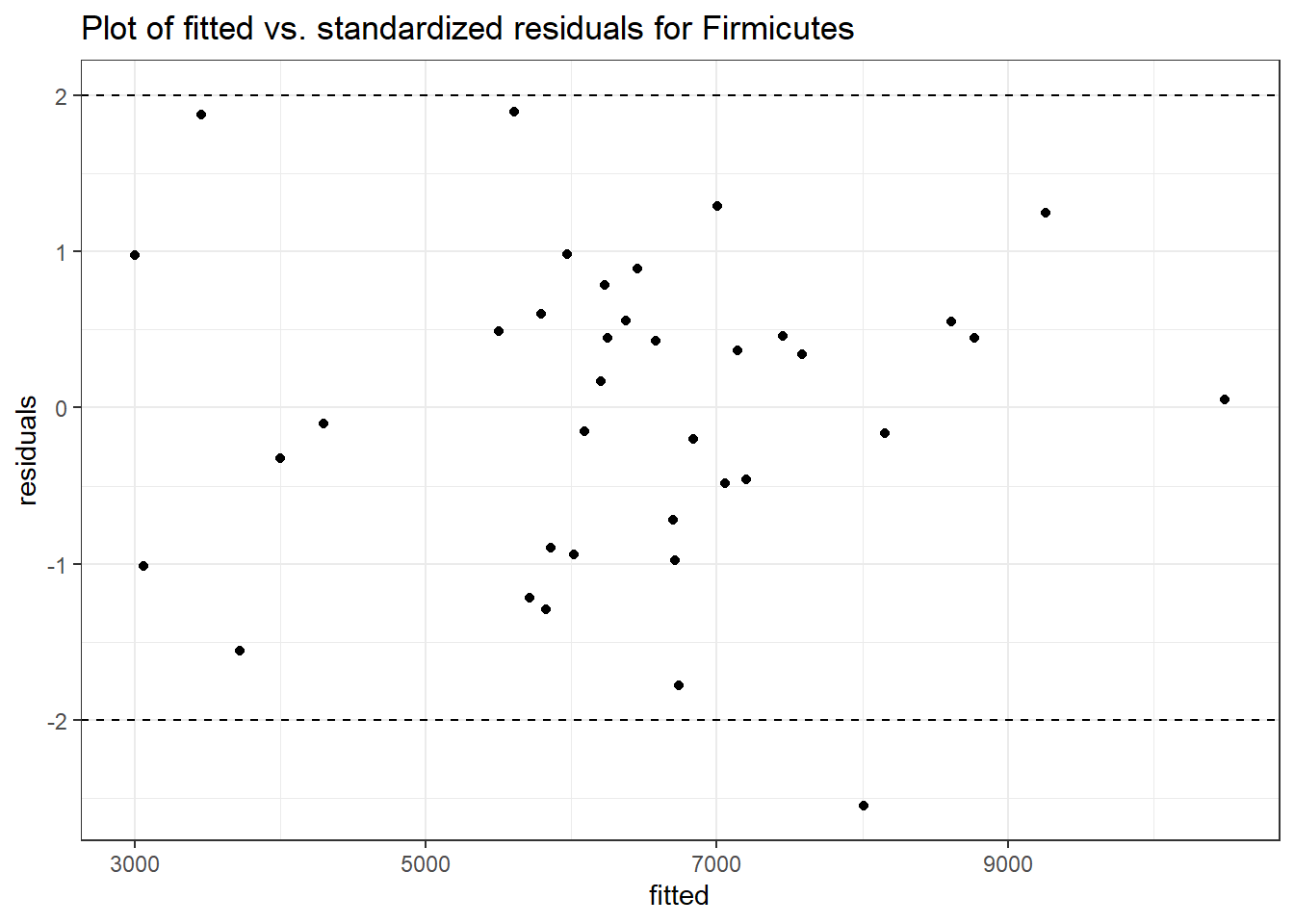
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Firmicutes 8.64916 0.12198 70.90592 0.00000
intB Firmicutes 0.29471 0.18091 1.62903 0.10331
time Firmicutes -0.01928 0.00255 -7.56656 0.00000
intB:time Firmicutes -0.05319 0.00375 -14.19120 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.29858
Significance test not available
#########################################
#########################################
Model: Fusobacteria ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x00000000071c9878>
Link: poisson
Intraclass Correlation (ICC): 0.8177454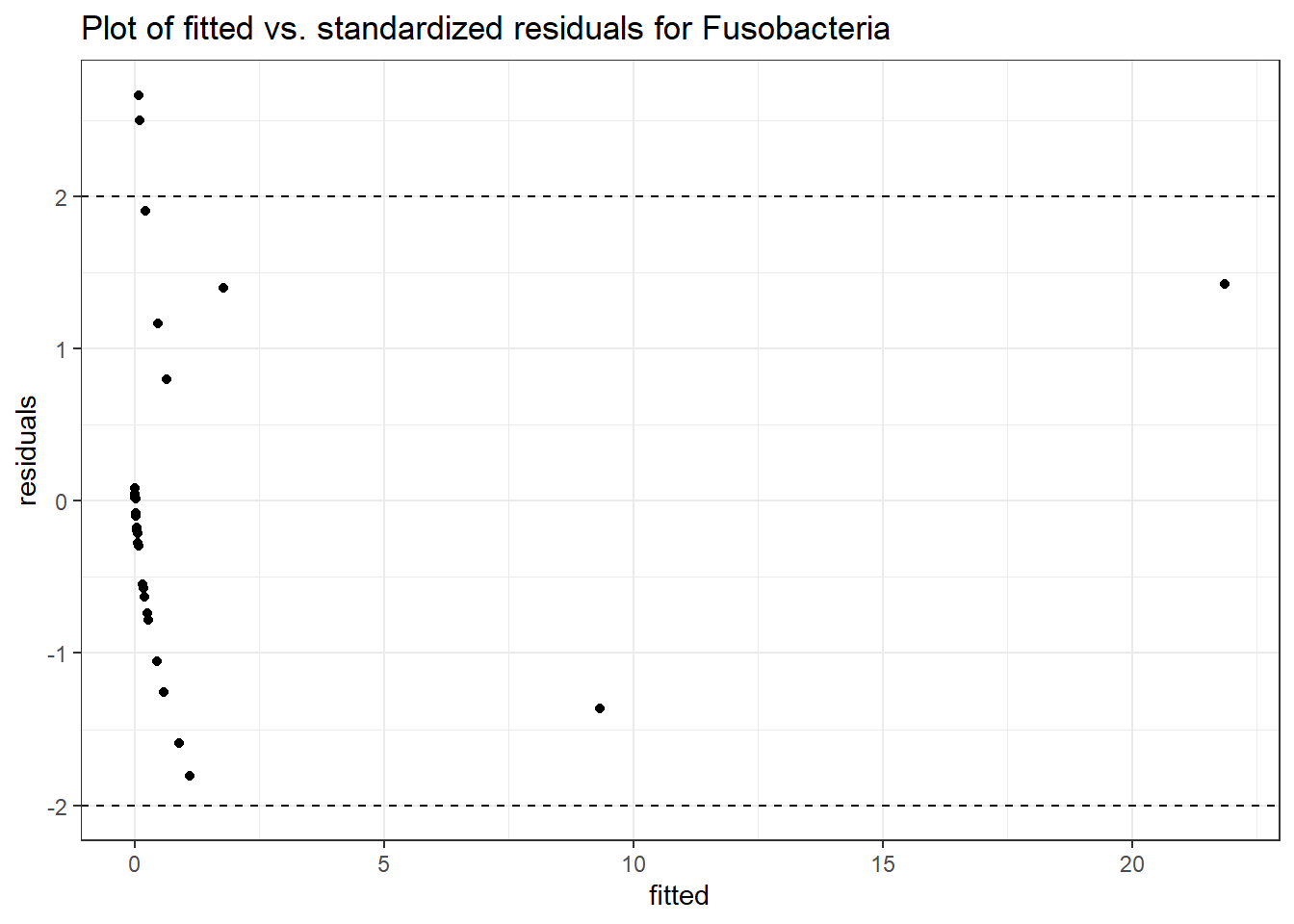
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Fusobacteria -0.55743 1.09231 -0.51033 0.60982
intB Fusobacteria -4.00449 2.10974 -1.89810 0.05768
time Fusobacteria -0.85229 0.31941 -2.66830 0.00762
intB:time Fusobacteria 1.54615 0.61913 2.49729 0.01251
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.1182
Significance test not available
#########################################
#########################################
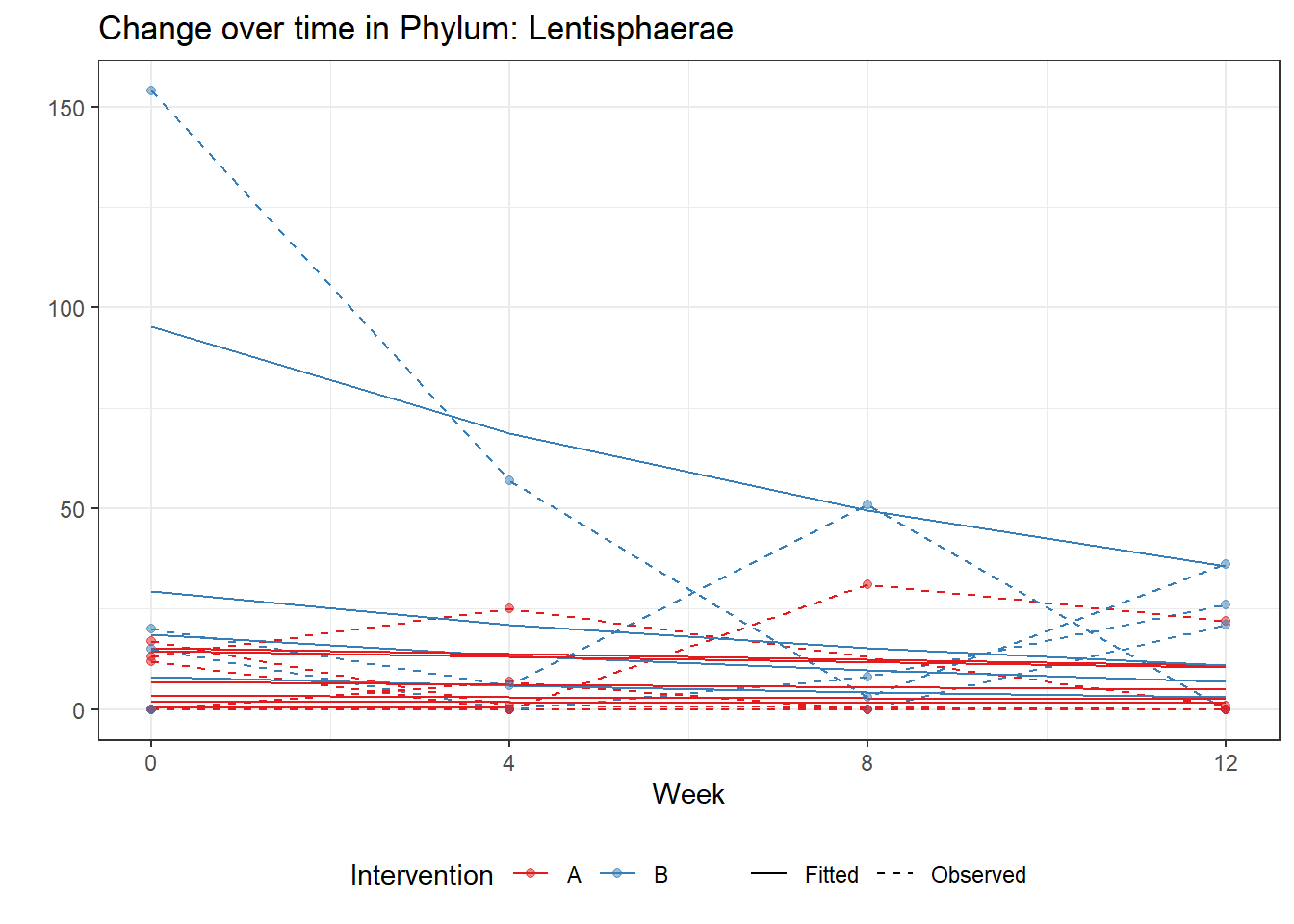
Model: Lentisphaerae ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x00000000071c9878>
Link: poisson
Intraclass Correlation (ICC): 0.6737277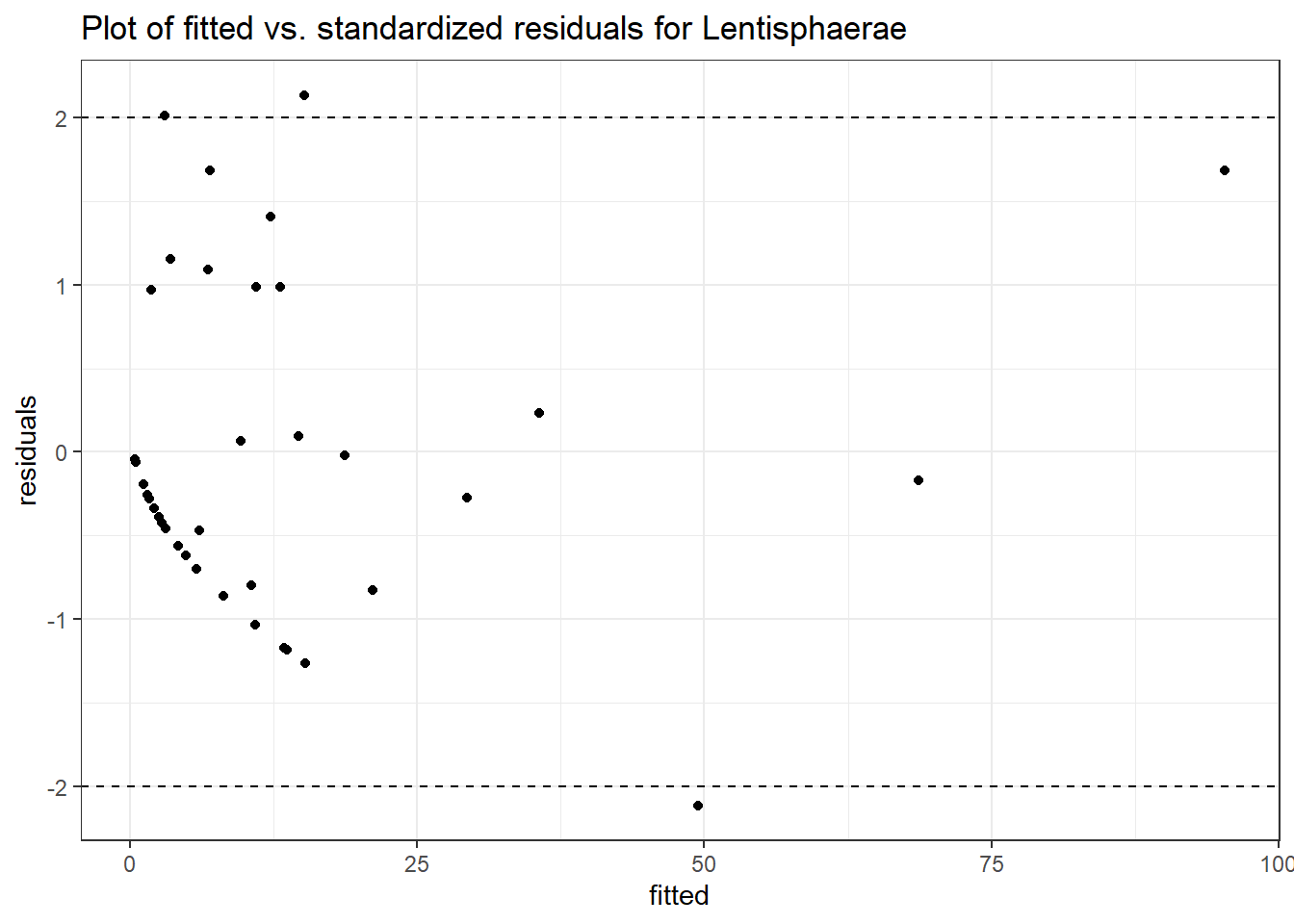
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lentisphaerae 1.38710 0.62140 2.23221 0.02560
intB Lentisphaerae 1.18566 0.90736 1.30672 0.19131
time Lentisphaerae -0.10926 0.07580 -1.44144 0.14946
intB:time Lentisphaerae -0.21895 0.08911 -2.45711 0.01401
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.437
Significance test not available
#########################################
#########################################
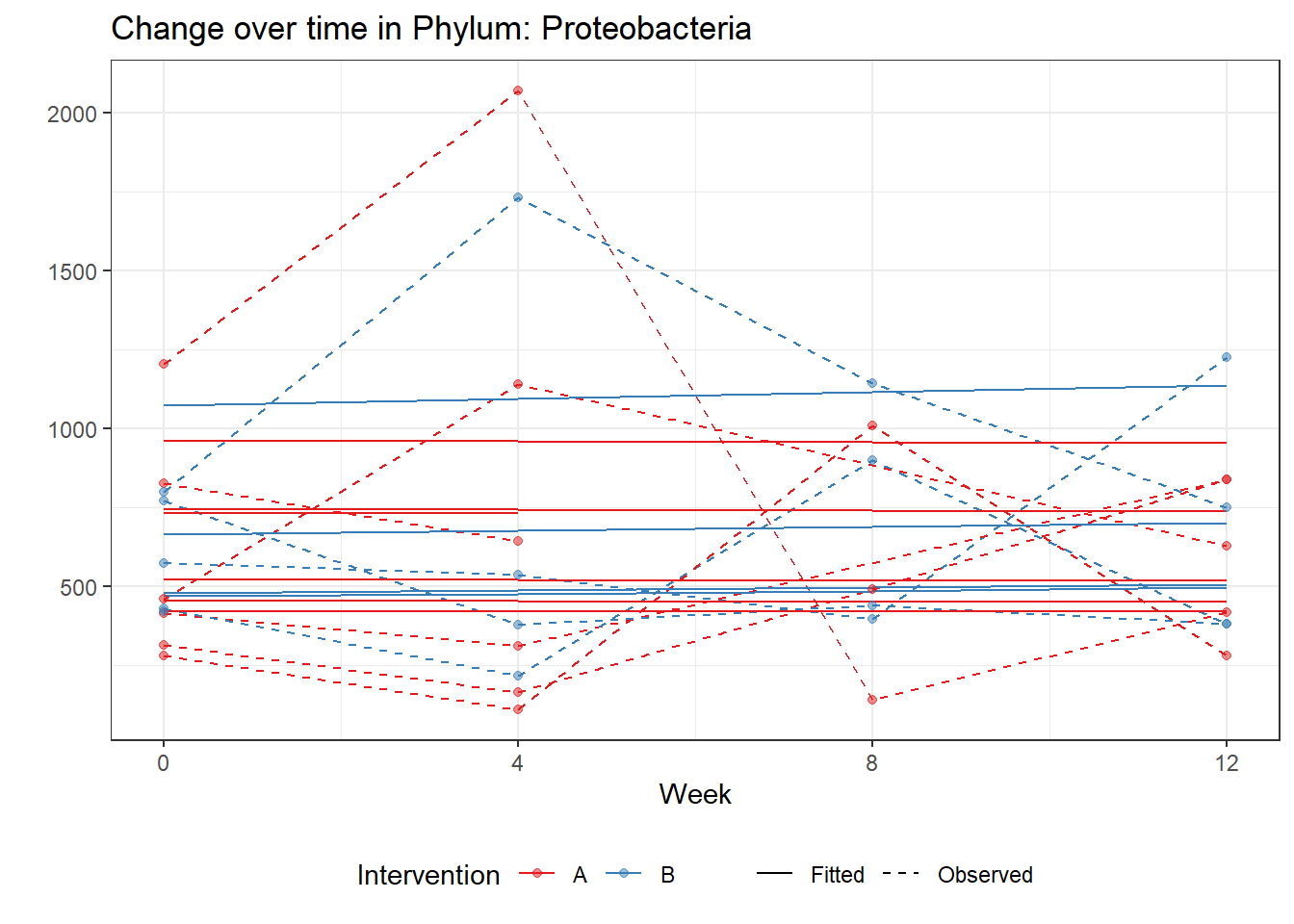
Model: Proteobacteria ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x00000000071c9878>
Link: poisson
Intraclass Correlation (ICC): 0.09168786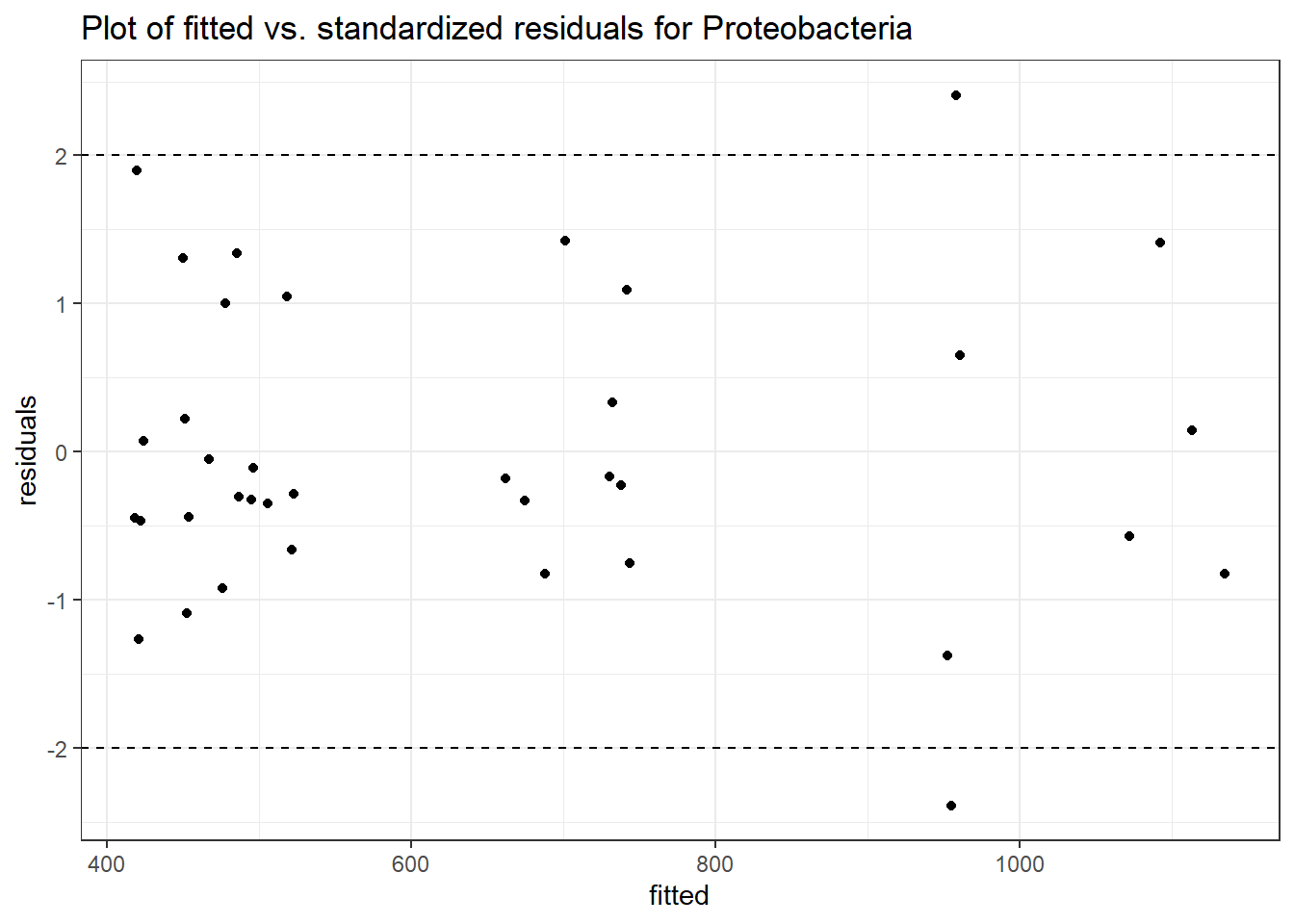
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Proteobacteria 6.41625 0.13045 49.18486 0.00000
intB Proteobacteria -0.04862 0.19358 -0.25118 0.80168
time Proteobacteria -0.00273 0.00806 -0.33866 0.73486
intB:time Proteobacteria 0.02175 0.01172 1.85541 0.06354
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.31772
Significance test not available
#########################################
#########################################
Model: Tenericutes ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x00000000071c9878>
Link: poisson
Intraclass Correlation (ICC): 0.9108551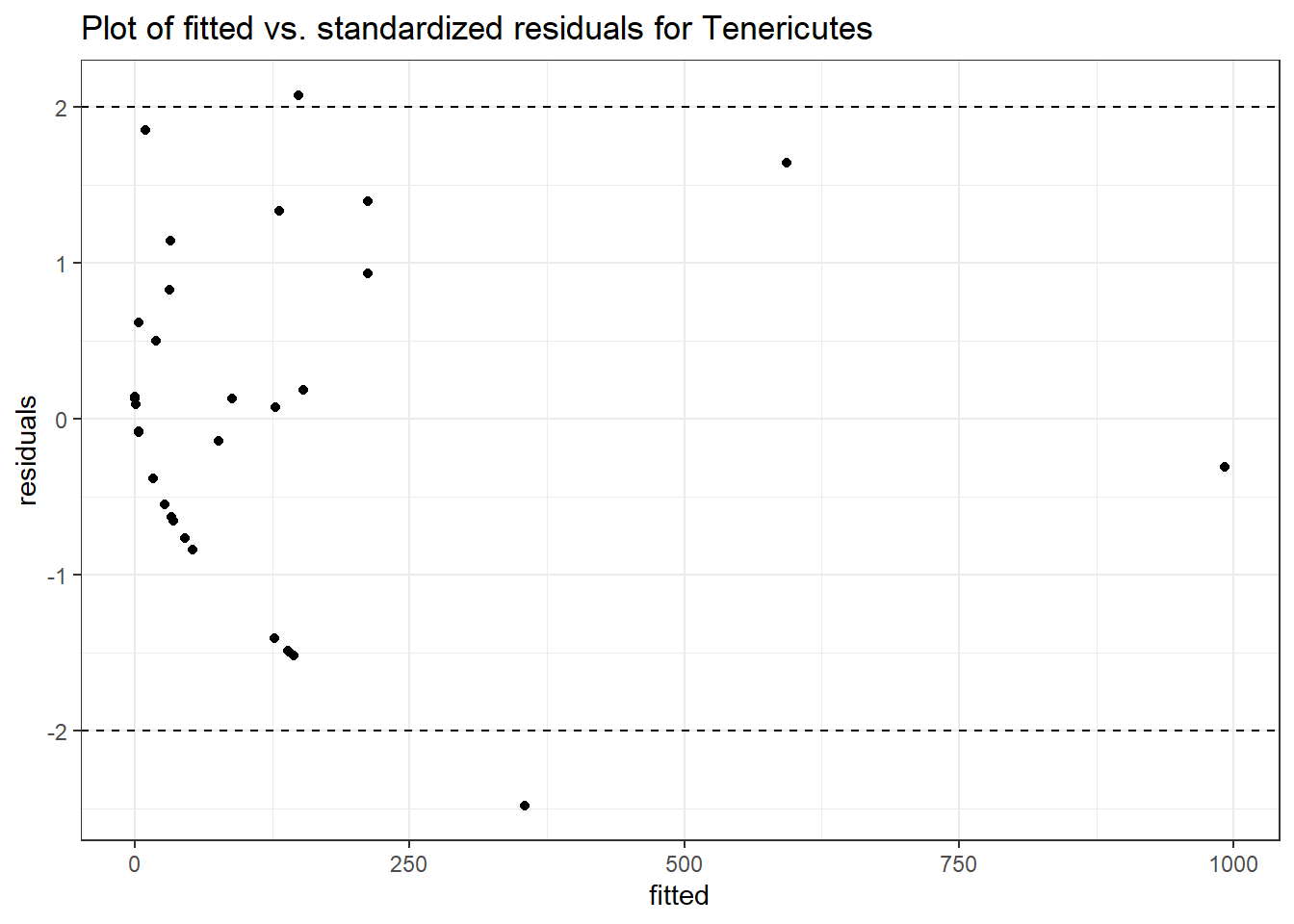
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Tenericutes 1.41504 1.35995 1.04051 0.29810
intB Tenericutes 2.38553 1.98298 1.20300 0.22898
time Tenericutes 0.02999 0.02547 1.17734 0.23906
intB:time Tenericutes -0.54430 0.03146 -17.29975 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 3.1965
Significance test not available
#########################################
#########################################
Model: Verrucomicrobia ~ 1 + intB + time + intB:time + (1 | SubjectID)
<environment: 0x00000000071c9878>
Link: poisson
Intraclass Correlation (ICC): 0.8737435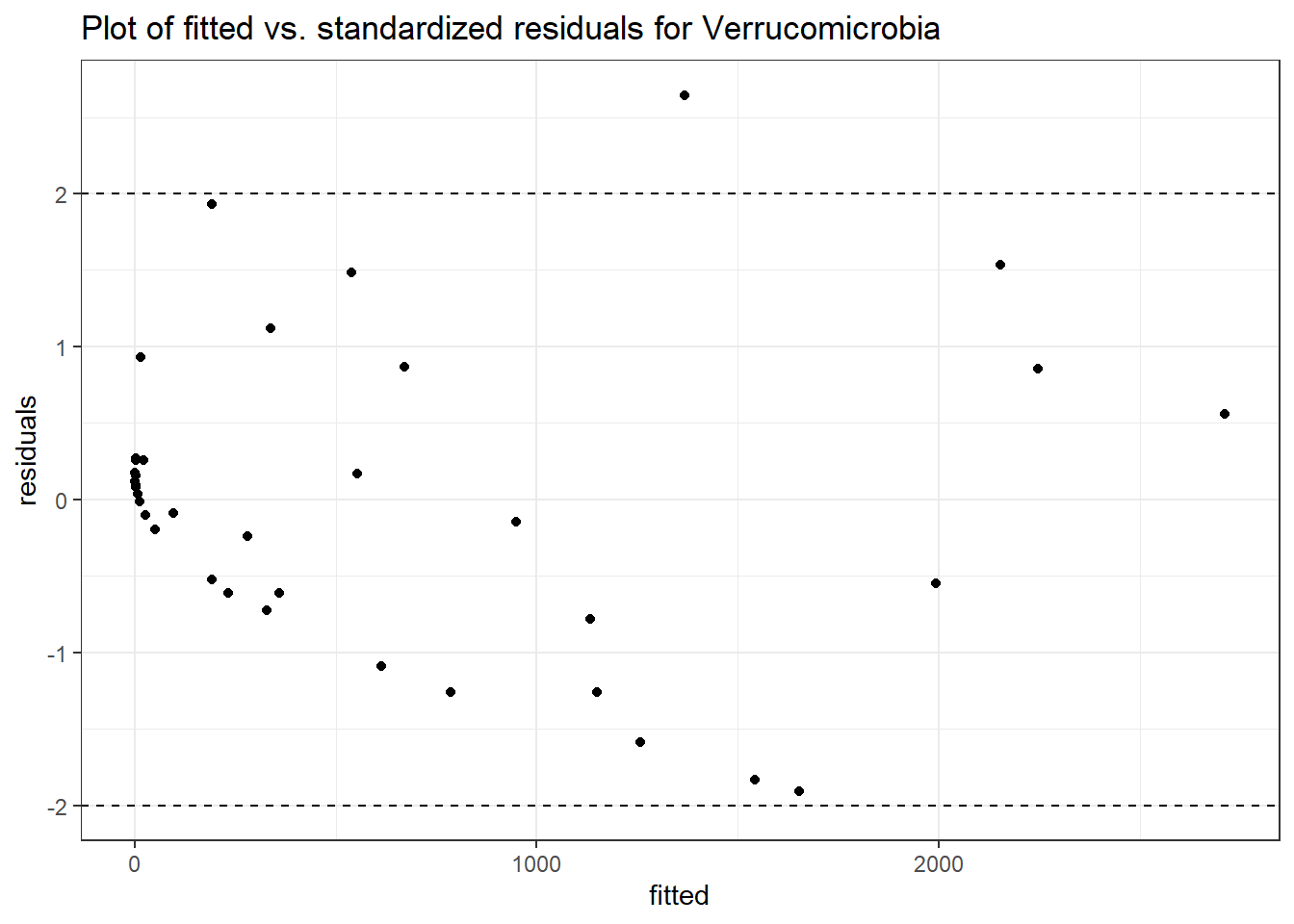
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Verrucomicrobia 4.85709 1.08191 4.48938 0.00001
intB Verrucomicrobia 0.90672 1.59709 0.56773 0.57022
time Verrucomicrobia -0.18803 0.00695 -27.05413 0.00000
intB:time Verrucomicrobia -0.43929 0.01431 -30.69182 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.6307
Significance test not availableCovariate(s): Time by intervention interaction (only)
Only the interaction term.
\[\begin{align*} h\left(Y_{ij}\right)&=\beta_{0j} + \beta_{1j}(Week)_{ij}+ r_{ij}\\ \beta_{0j}&= \gamma_{00} + u_{0j}\\ \beta_{1j}&= \gamma_{10} + \gamma_{11}(Intervention)_{j}\\ \end{align*}\]
phylum.fit6 <- glmm_microbiome(mydata=microbiome_data,model.number=6,
taxa.level="Phylum", link="poisson",
model="1 + time + intB:time + (1|SubjectID)")
#########################################
#########################################
Model: Actinobacteria ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x0000000038c463e8>
Link: poisson
Intraclass Correlation (ICC): 0.606858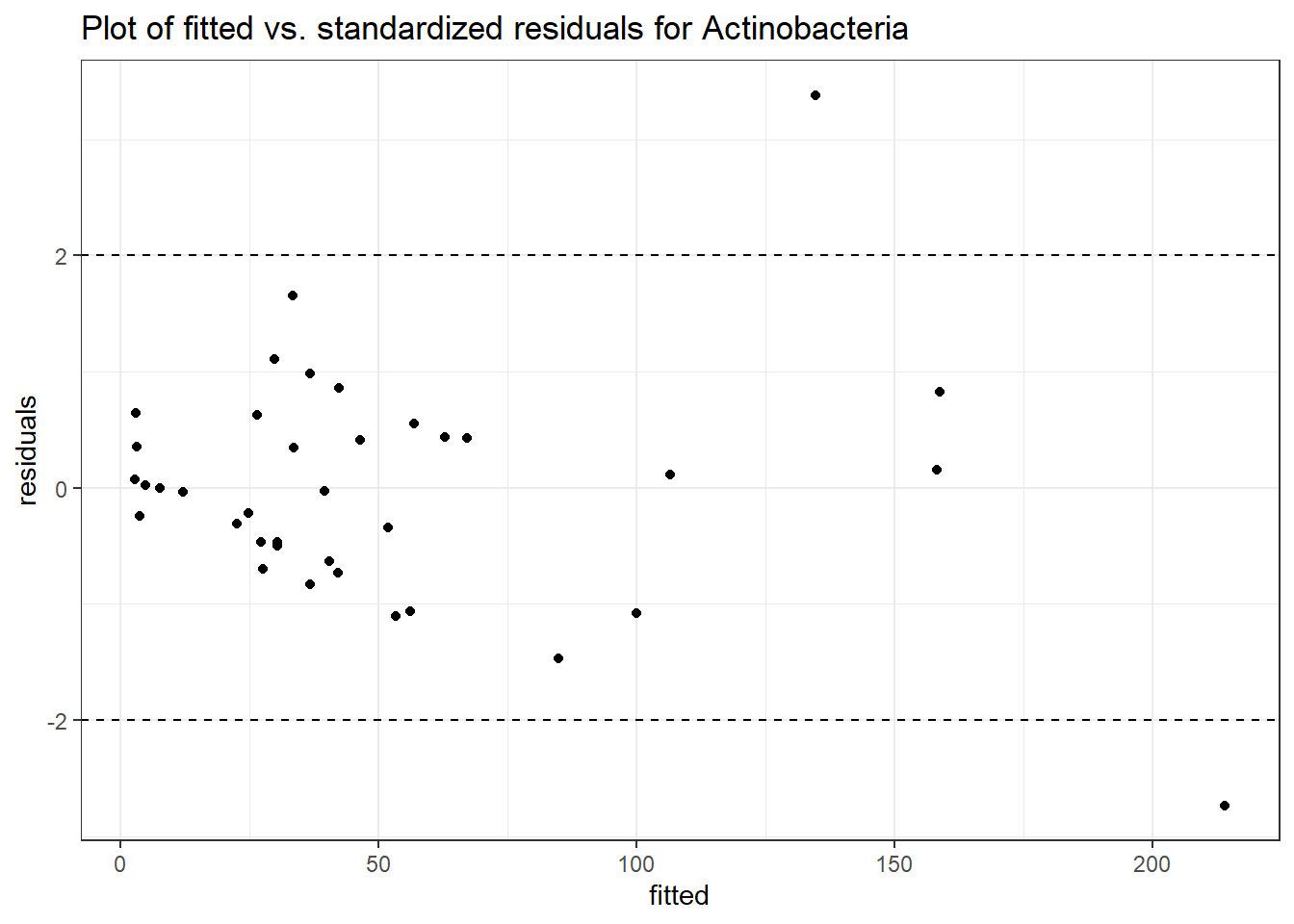
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Actinobacteria 3.74307 0.37795 9.90364 0.00000
time Actinobacteria 0.09388 0.03665 2.56140 0.01043
time:intB Actinobacteria -0.55653 0.04681 -11.88910 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.2424
Significance test not available
#########################################
#########################################
Model: Bacteroidetes ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x0000000038c463e8>
Link: poisson
Intraclass Correlation (ICC): 0.09943526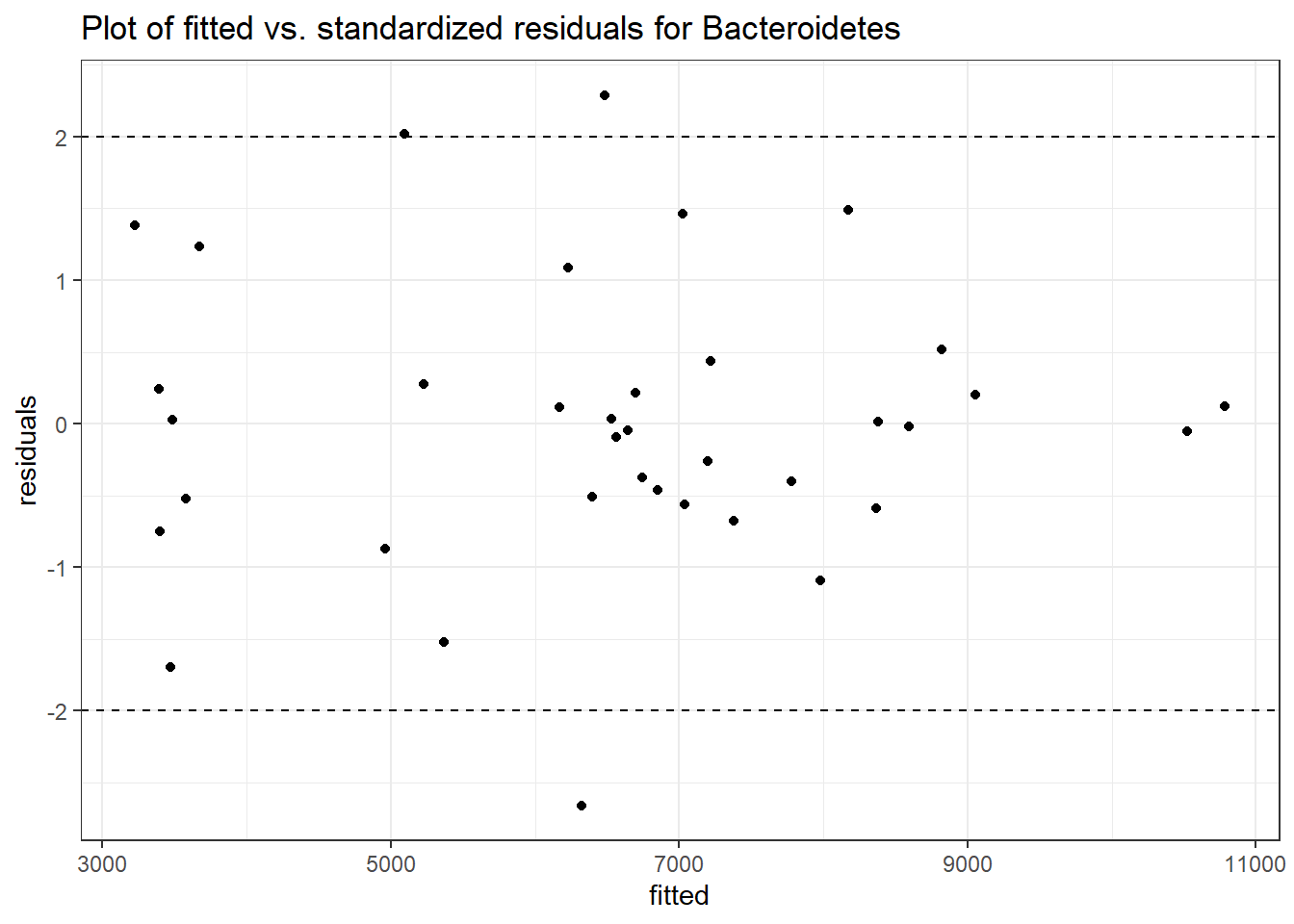
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroidetes 8.77946 0.10024 87.58375 0.00000
time Bacteroidetes -0.02474 0.00249 -9.92886 0.00000
time:intB Bacteroidetes -0.00156 0.00382 -0.40921 0.68238
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.33229
Significance test not available
#########################################
#########################################
Model: Cyanobacteria ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x0000000038c463e8>
Link: poisson
Intraclass Correlation (ICC): 0.9355782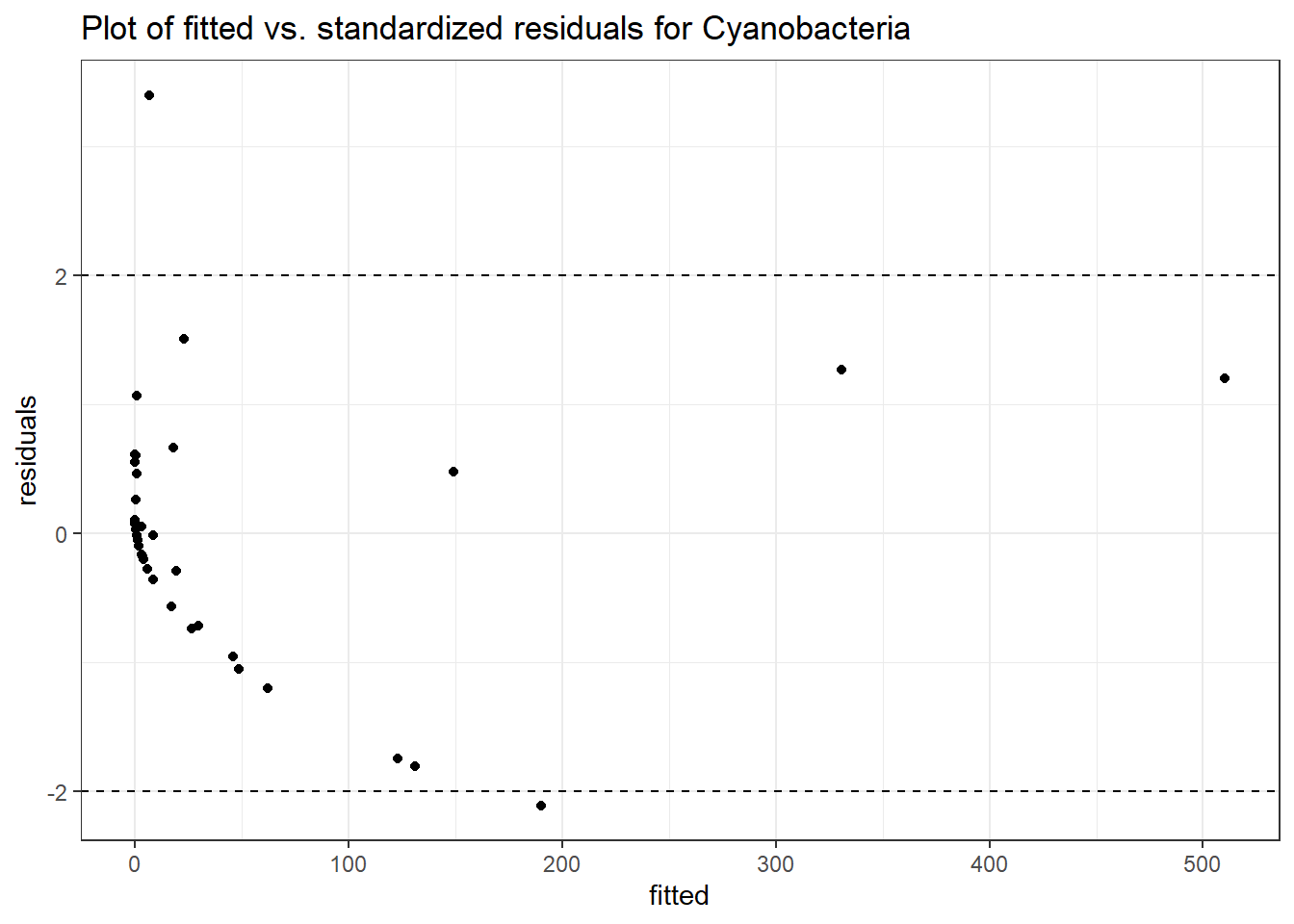
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Cyanobacteria 0.86458 1.17438 0.73620 0.46161
time Cyanobacteria -0.98789 0.03438 -28.73852 0.00000
time:intB Cyanobacteria 2.59883 0.13098 19.84202 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 3.8109
Significance test not available
#########################################
#########################################
Model: Euryarchaeota ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x0000000038c463e8>
Link: poisson
Intraclass Correlation (ICC): 0.7854791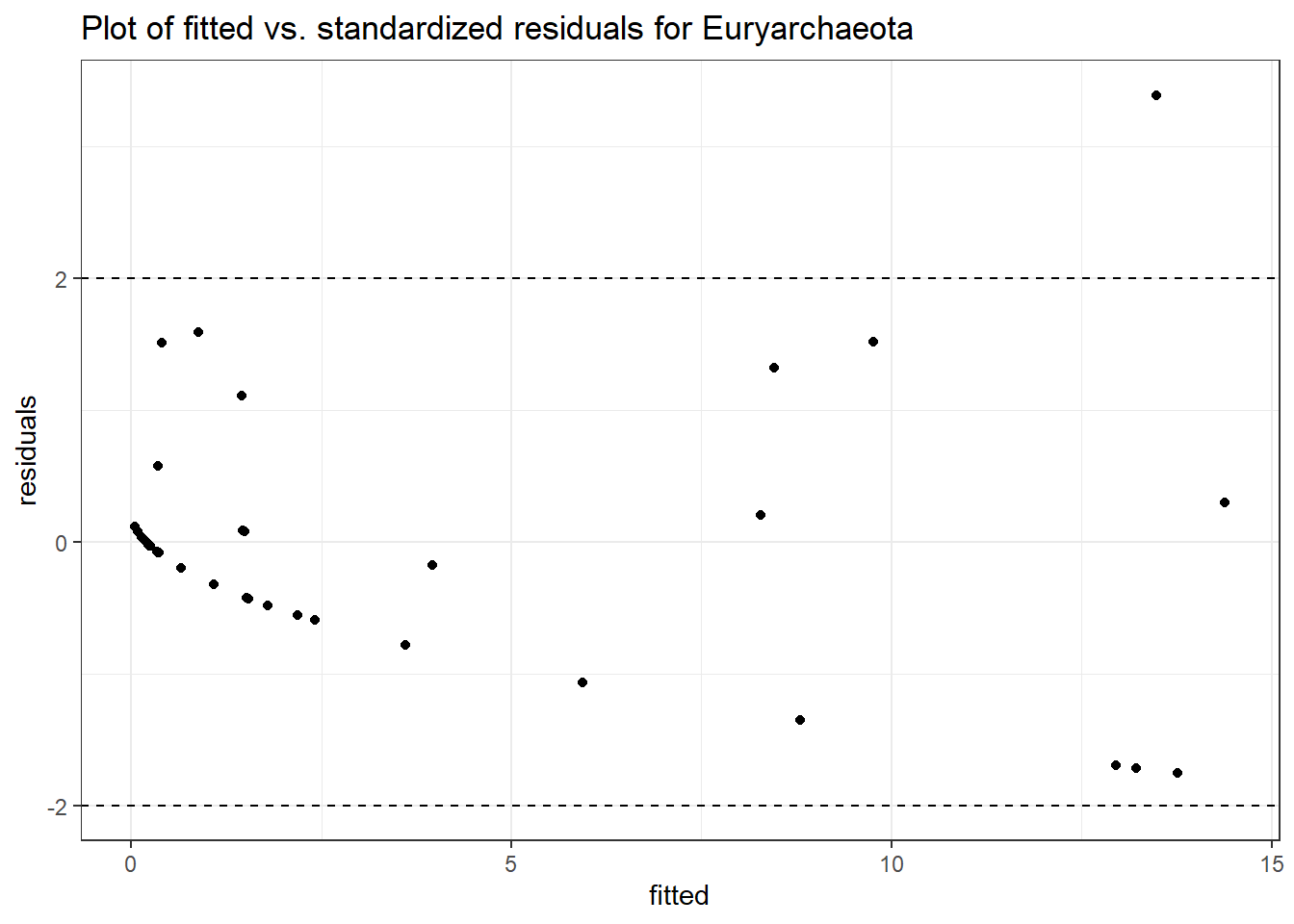
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Euryarchaeota 0.41699 0.63809 0.65350 0.51344
time Euryarchaeota 0.02006 0.09071 0.22113 0.82499
time:intB Euryarchaeota -0.51748 0.18542 -2.79082 0.00526
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.9135
Significance test not available
#########################################
#########################################
Model: Firmicutes ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x0000000038c463e8>
Link: poisson
Intraclass Correlation (ICC): 0.09965168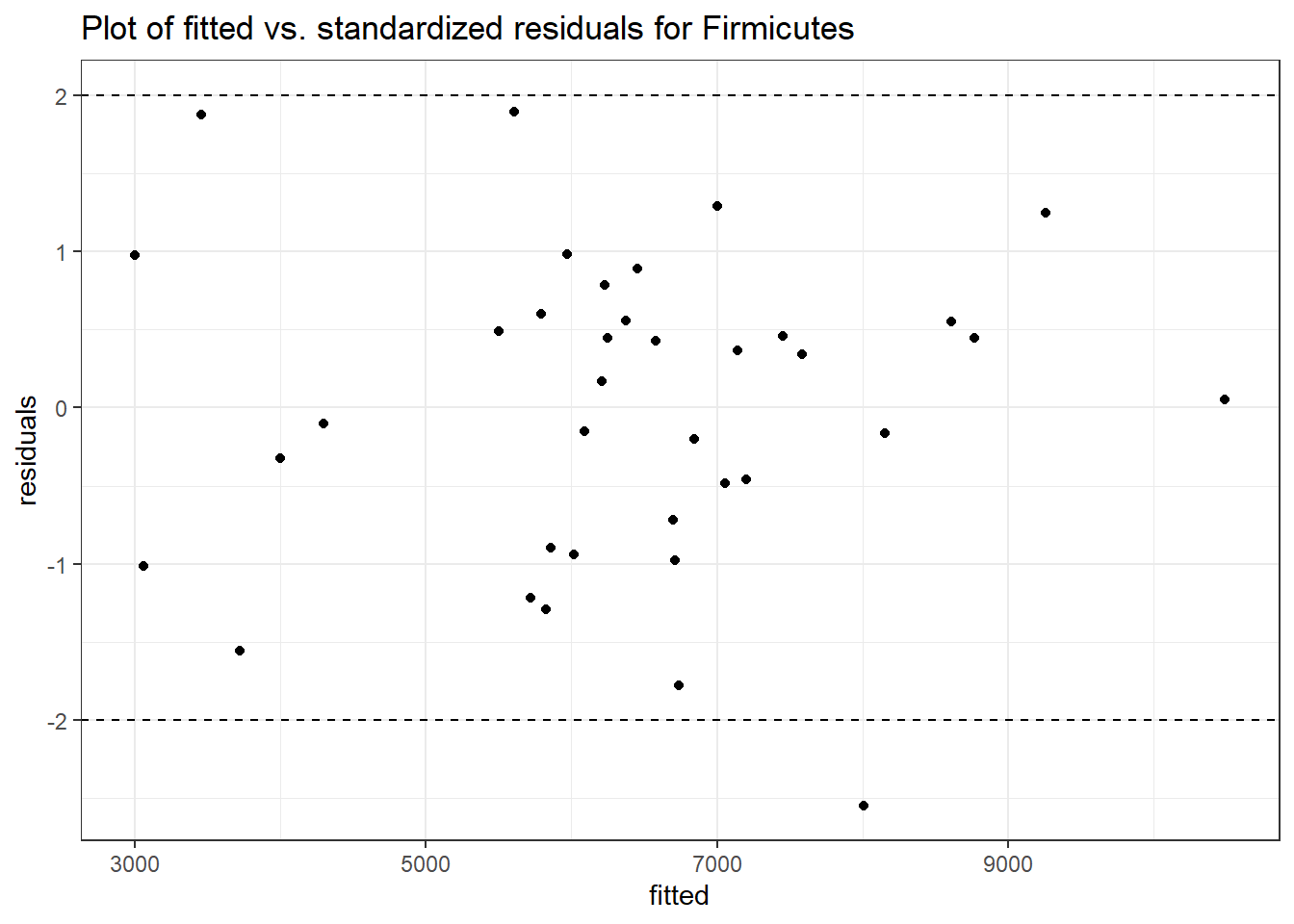
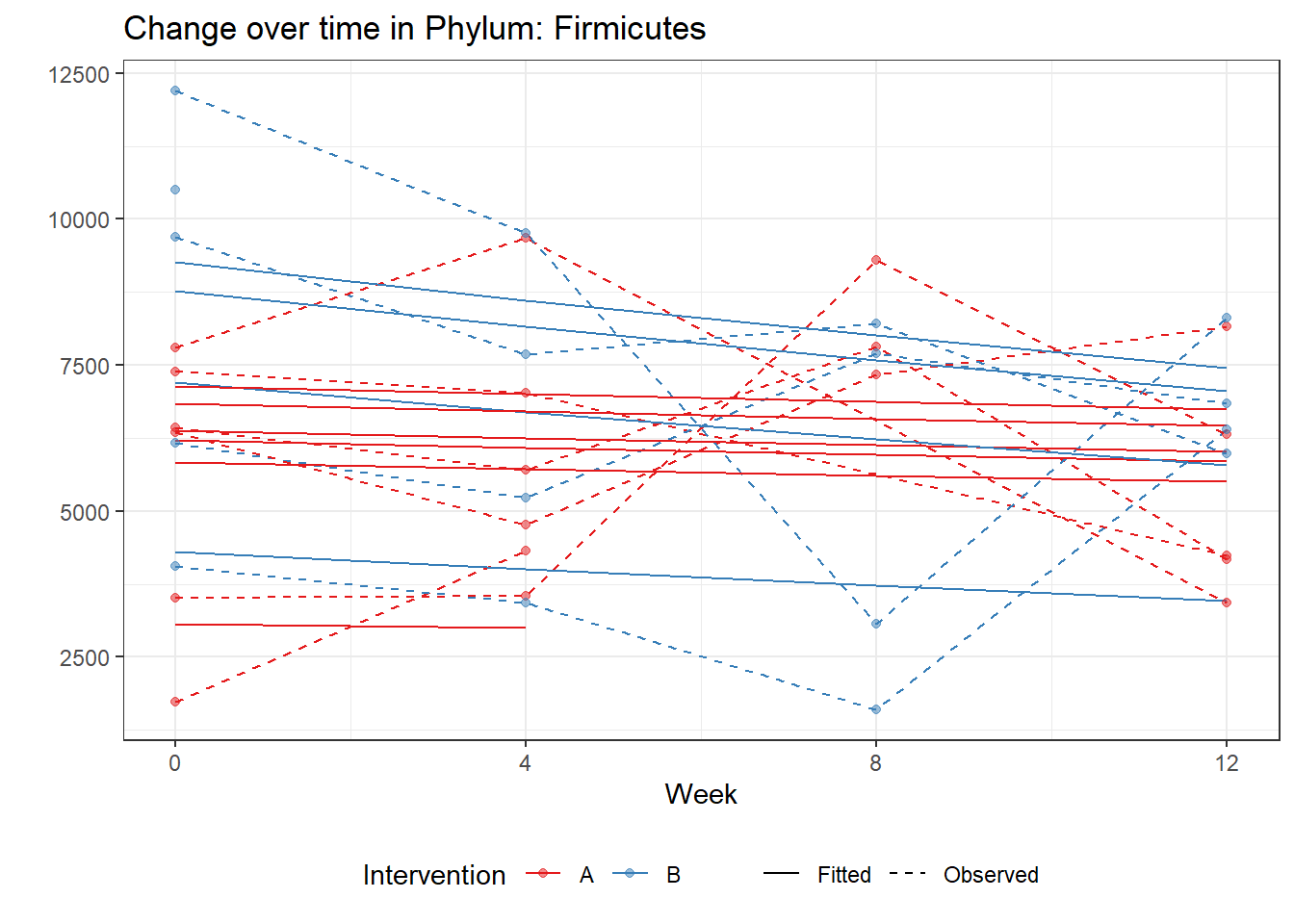
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Firmicutes 8.78313 0.10036 87.51536 0
time Firmicutes -0.01934 0.00255 -7.59396 0
time:intB Firmicutes -0.05305 0.00375 -14.15847 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.33269
Significance test not available
#########################################
#########################################
Model: Fusobacteria ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x0000000038c463e8>
Link: poisson
Intraclass Correlation (ICC): 0.8757236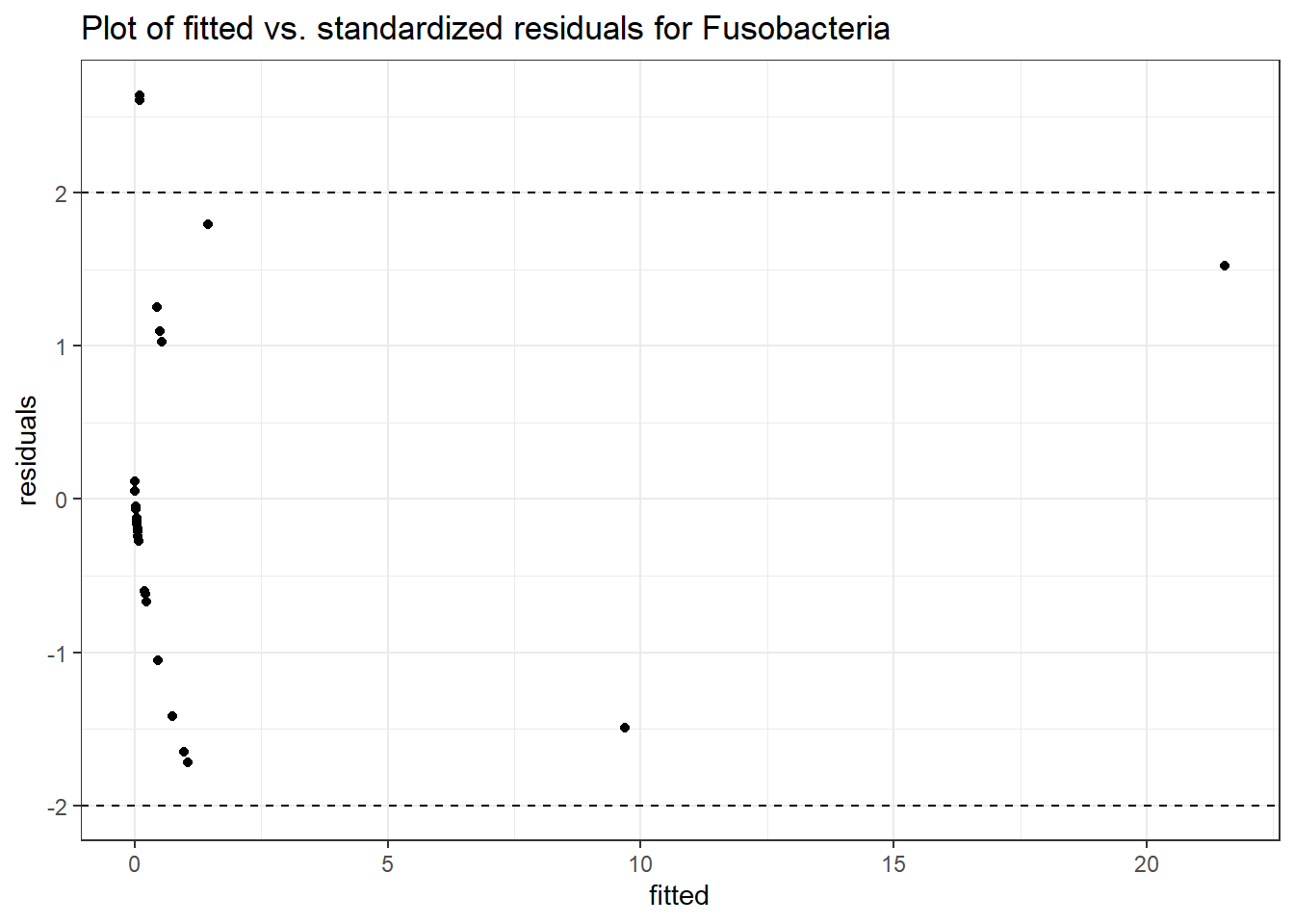
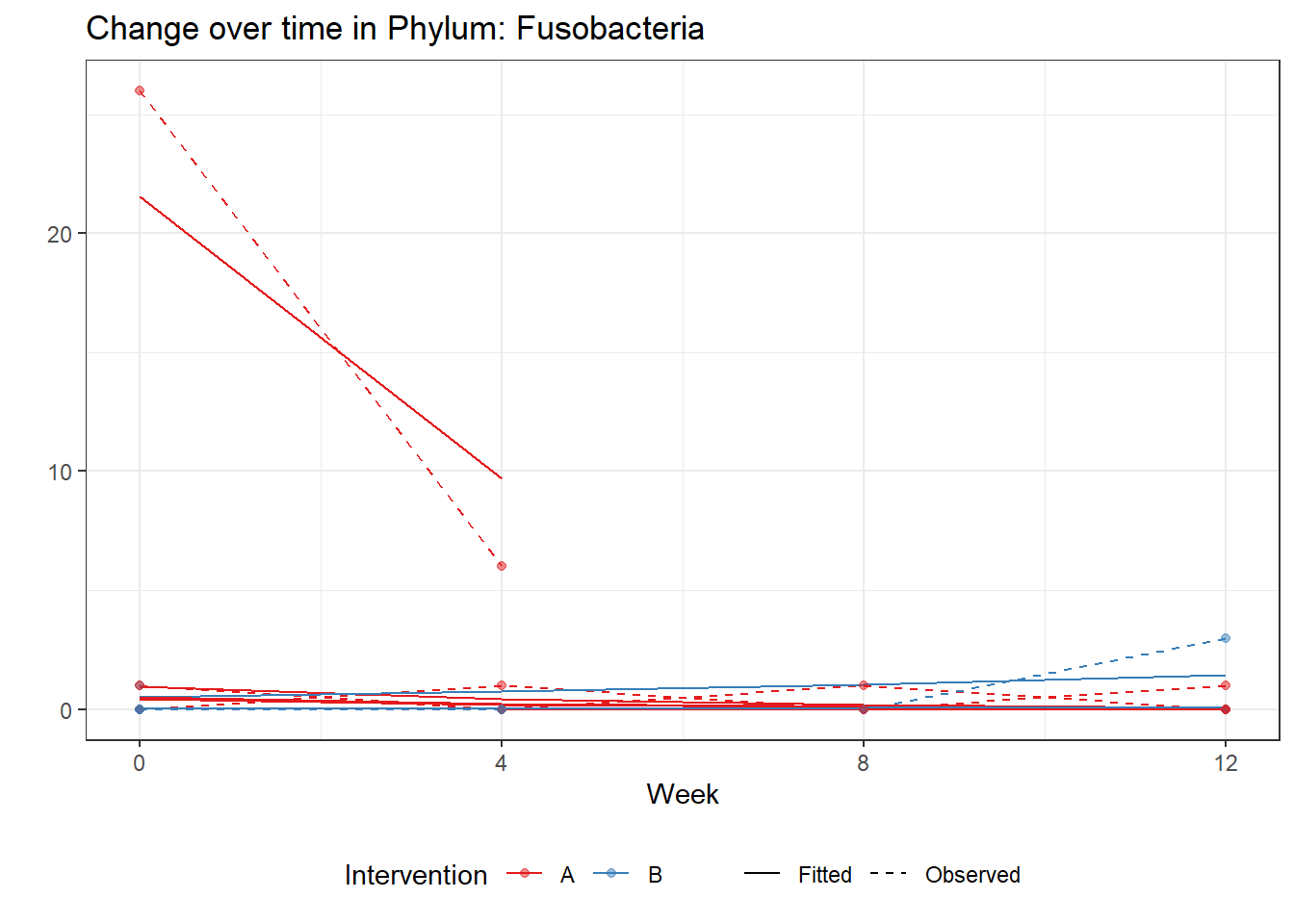
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Fusobacteria -2.23959 1.31770 -1.69962 0.08920
time Fusobacteria -0.79848 0.32354 -2.46798 0.01359
time:intB Fusobacteria 1.13030 0.52474 2.15404 0.03124
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.6545
Significance test not available
#########################################
#########################################
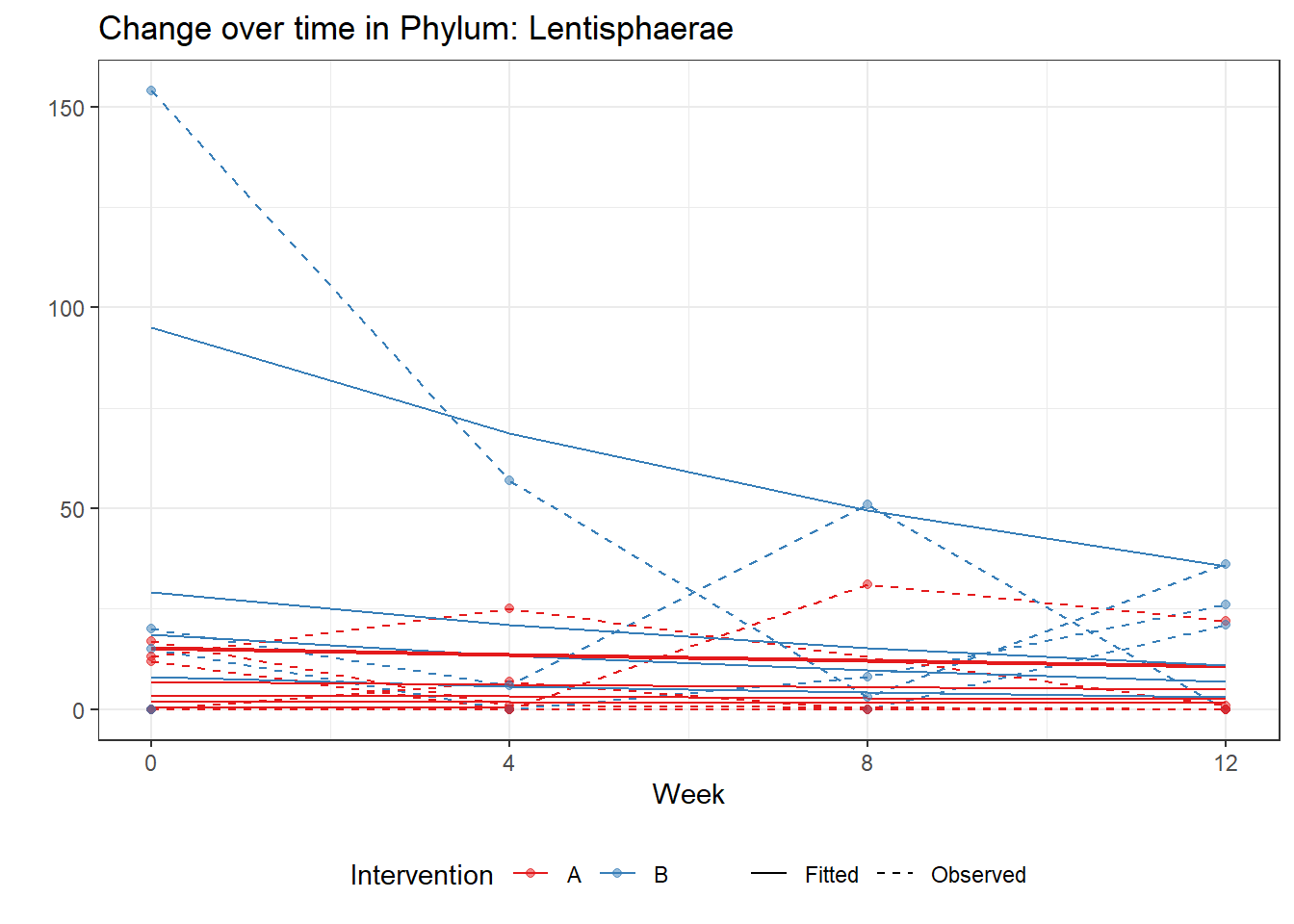
Model: Lentisphaerae ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x0000000038c463e8>
Link: poisson
Intraclass Correlation (ICC): 0.7249303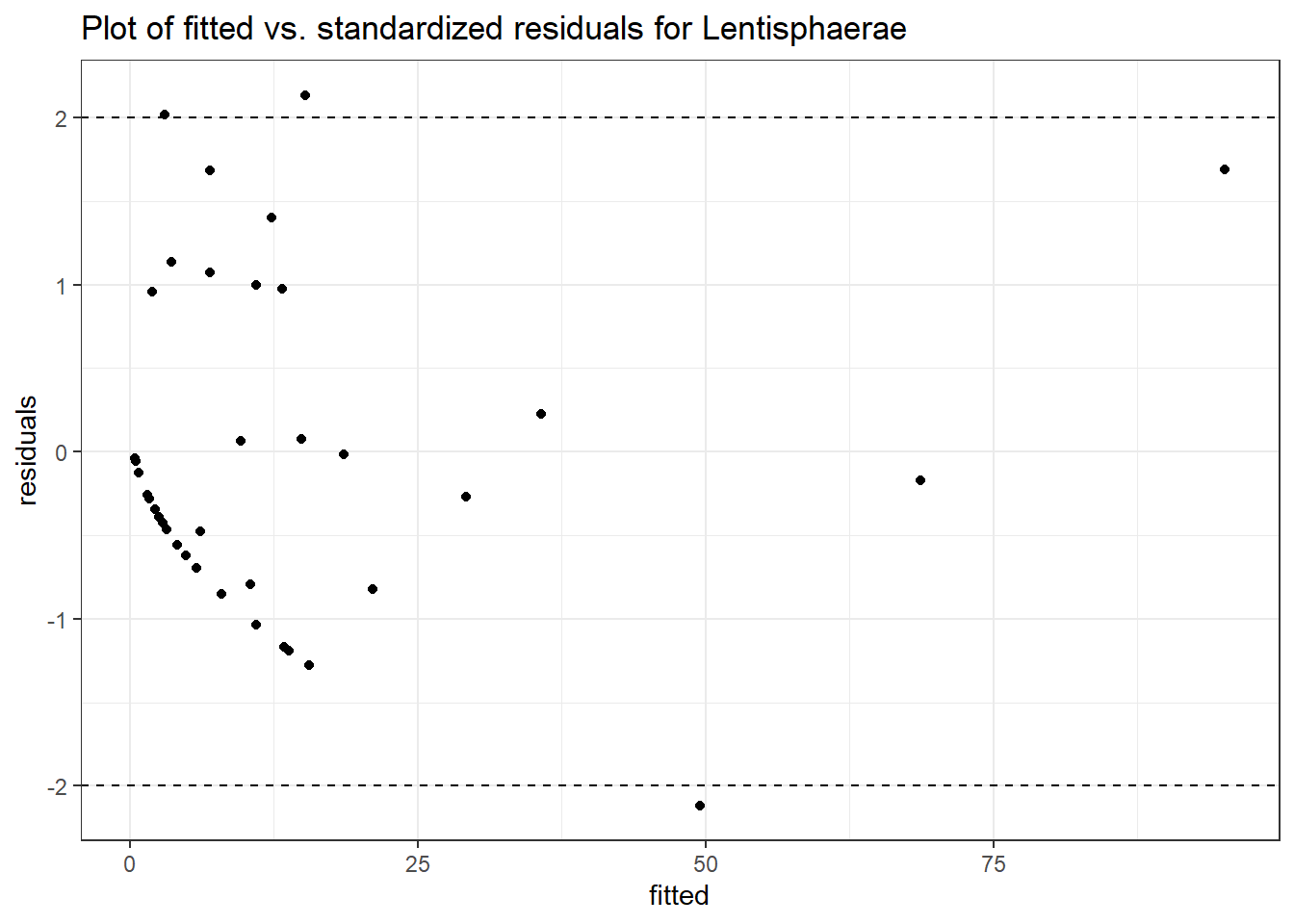
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lentisphaerae 1.88736 0.51580 3.65906 0.00025
time Lentisphaerae -0.11763 0.07535 -1.56114 0.11849
time:intB Lentisphaerae -0.20874 0.08868 -2.35381 0.01858
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.6234
Significance test not available
#########################################
#########################################
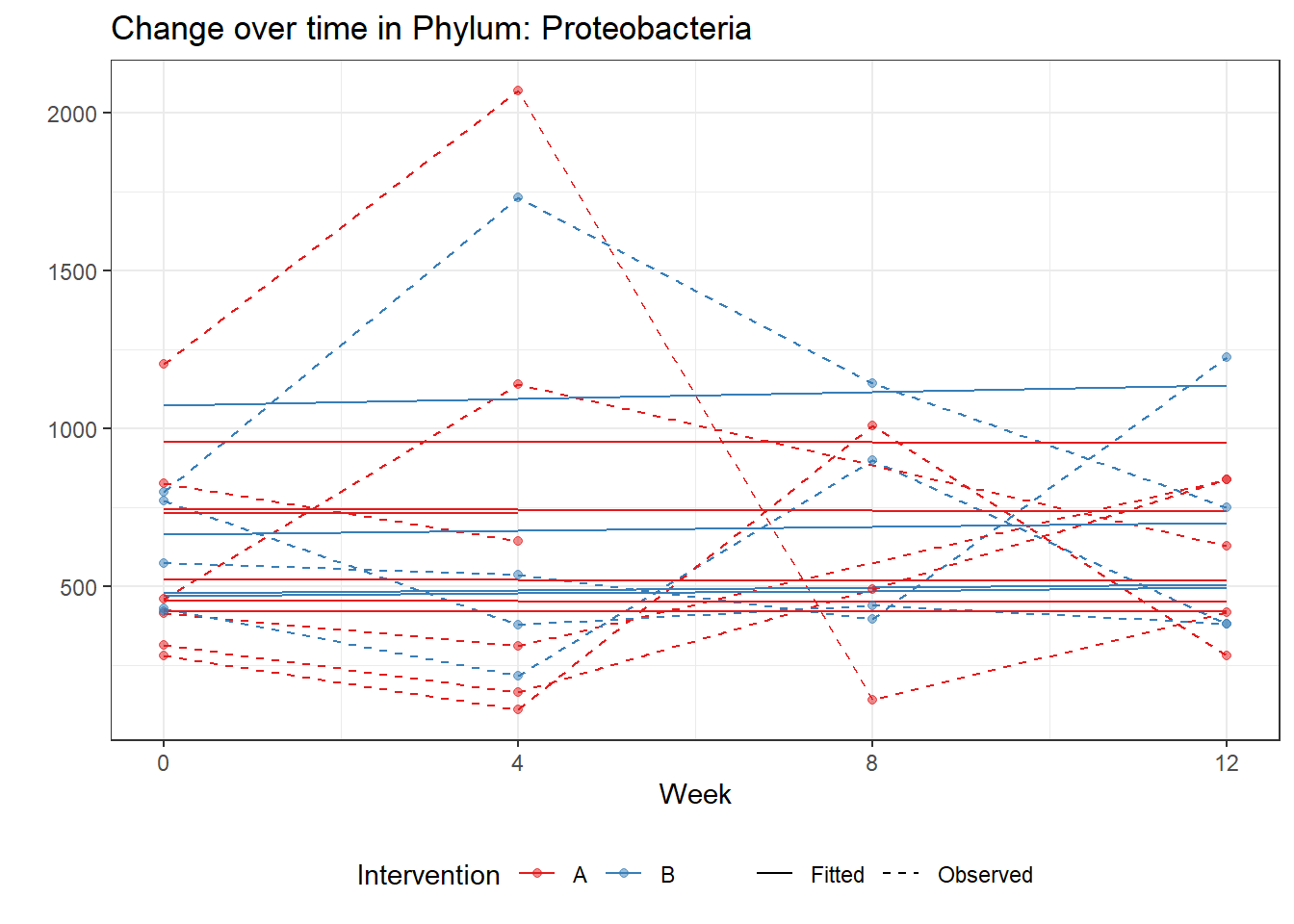
Model: Proteobacteria ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x0000000038c463e8>
Link: poisson
Intraclass Correlation (ICC): 0.09214969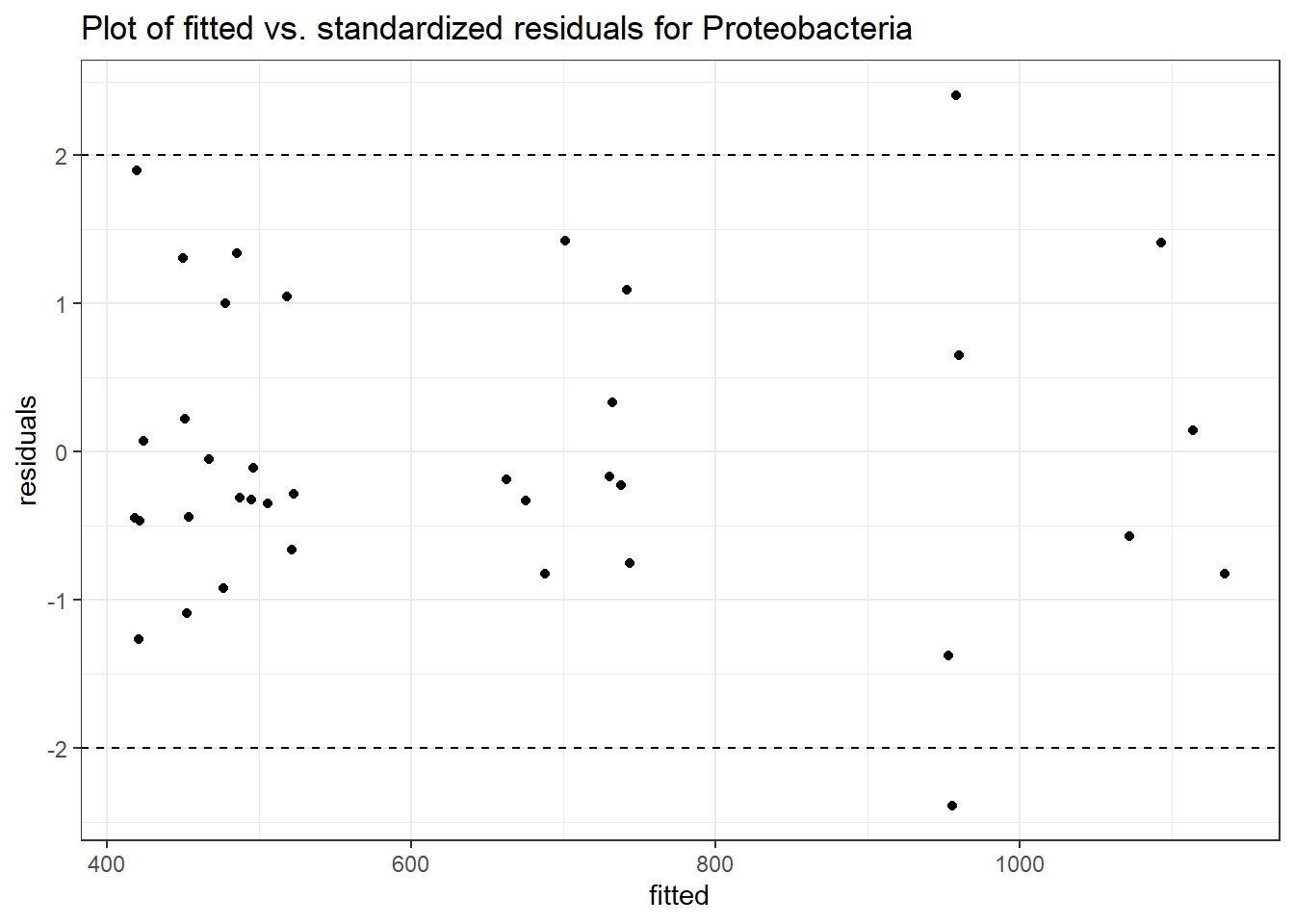
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Proteobacteria 6.39416 0.09665 66.16092 0.00000
time Proteobacteria -0.00262 0.00805 -0.32575 0.74462
time:intB Proteobacteria 0.02152 0.01169 1.84176 0.06551
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.3186
Significance test not available
#########################################
#########################################
Model: Tenericutes ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x0000000038c463e8>
Link: poisson
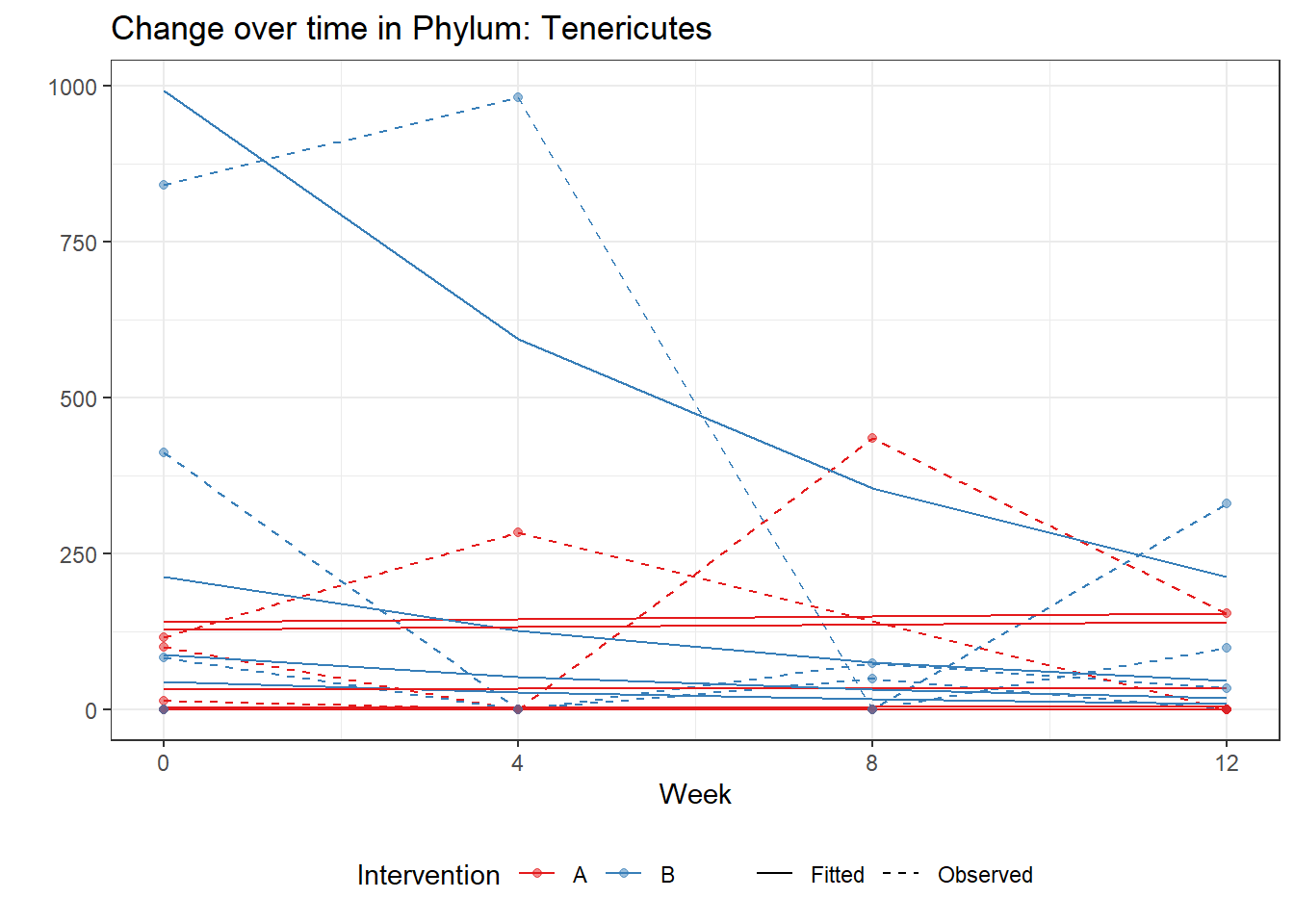
Intraclass Correlation (ICC): 0.9233551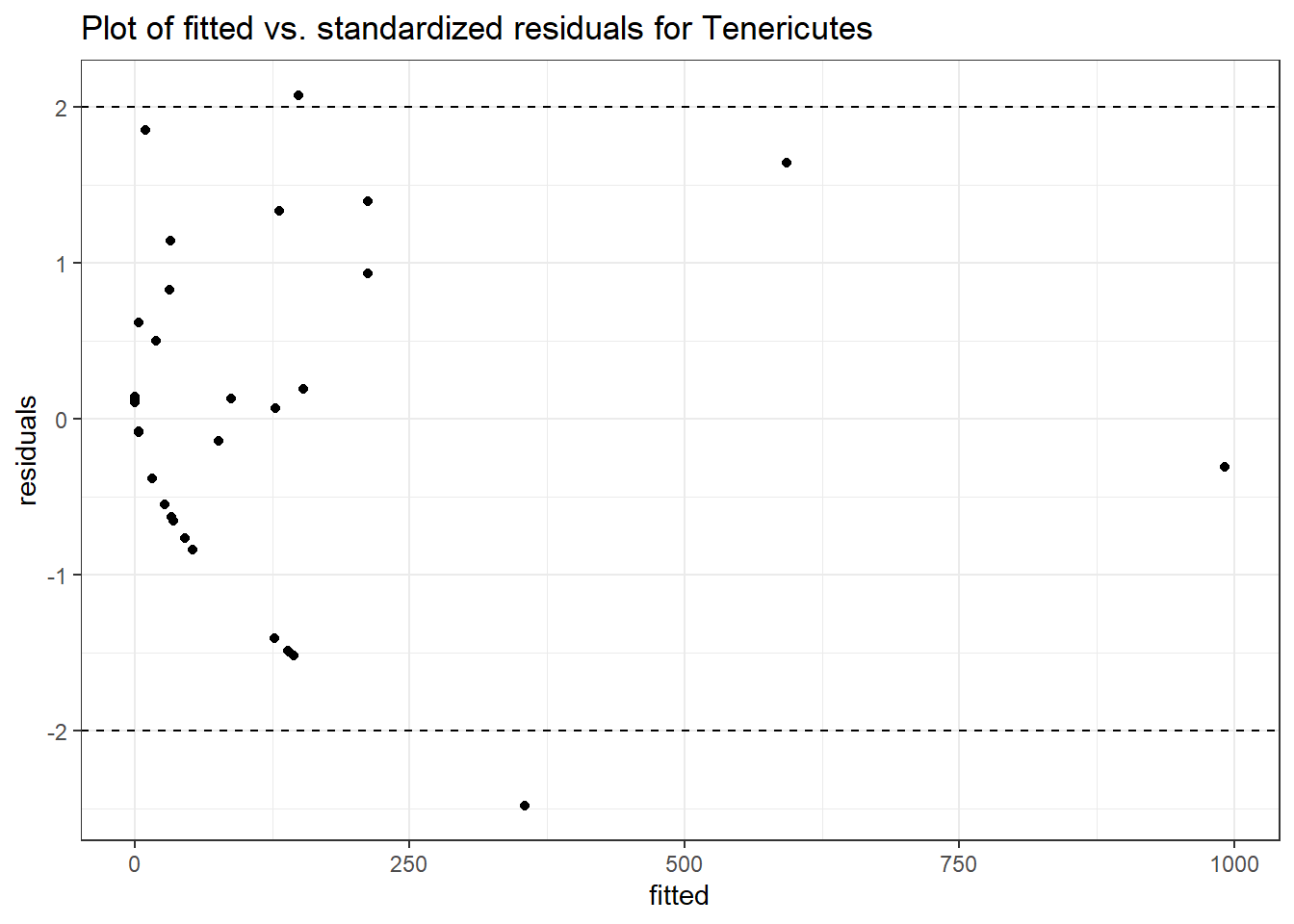
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Tenericutes 2.45099 1.08069 2.26799 0.02333
time Tenericutes 0.02952 0.02546 1.15933 0.24632
time:intB Tenericutes -0.54372 0.03146 -17.28516 0.00000
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 3.4709
Significance test not available
#########################################
#########################################
Model: Verrucomicrobia ~ 1 + time + intB:time + (1 | SubjectID)
<environment: 0x0000000038c463e8>
Link: poisson
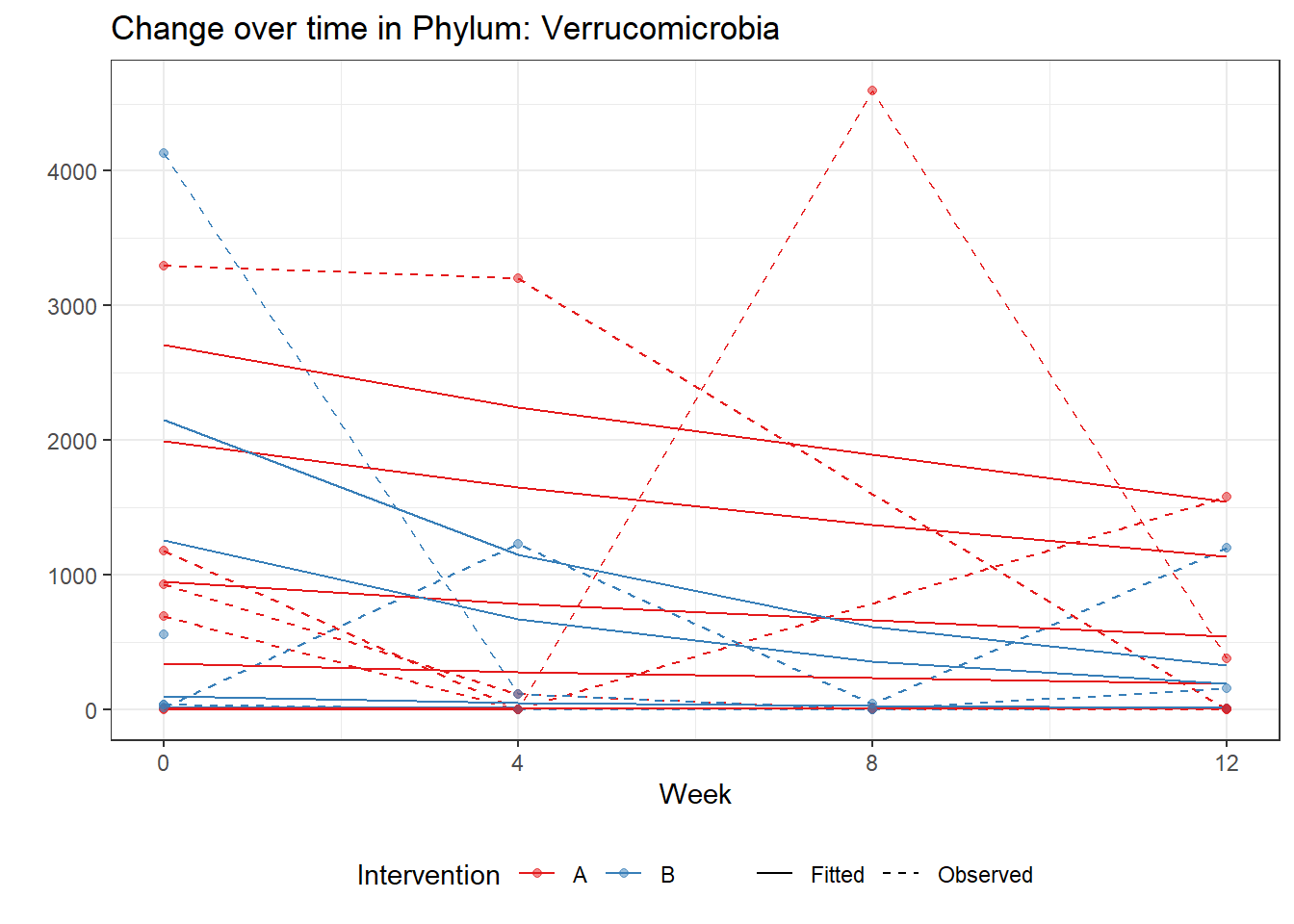
Intraclass Correlation (ICC): 0.8764792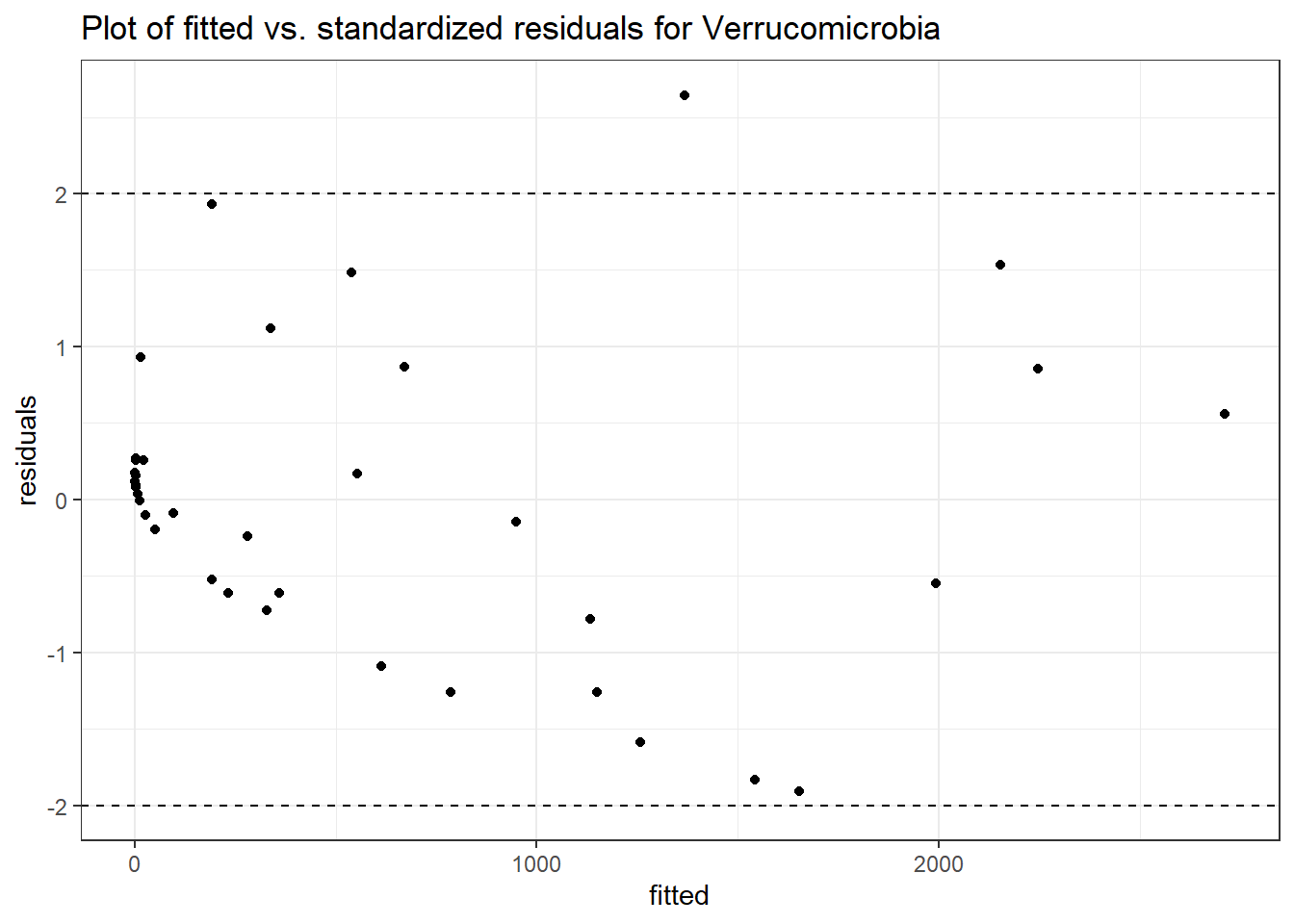
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Verrucomicrobia 5.27169 0.80683 6.53380 0
time Verrucomicrobia -0.18805 0.00695 -27.05719 0
time:intB Verrucomicrobia -0.43924 0.01431 -30.68870 0
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.6638
Significance test not availableCovariate(s): Time and Gender
\[\begin{align*} h\left(Y_{ij}\right)&=\beta_{0j} + \beta_{1j}(Week)_{ij} + r_{ij}\\ \beta_{0j}&= \gamma_{00} + \gamma_{01}(Intervention)_{j} + \gamma_{02}(Gender)_{j} + u_{0j}\\ \beta_{1j}&= \gamma_{10}\\ \beta_{2j}&= \gamma_{20}\\ \end{align*}\]
phylum.fit7 <- glmm_microbiome(mydata=microbiome_data, model.number=7,
taxa.level="Phylum", link="poisson",
model="1 + intB + time + female + (1|SubjectID)")
#########################################
#########################################
Model: Actinobacteria ~ 1 + intB + time + female + (1 | SubjectID)
<environment: 0x000000000b5e7848>
Link: poisson
Intraclass Correlation (ICC): 0.4239553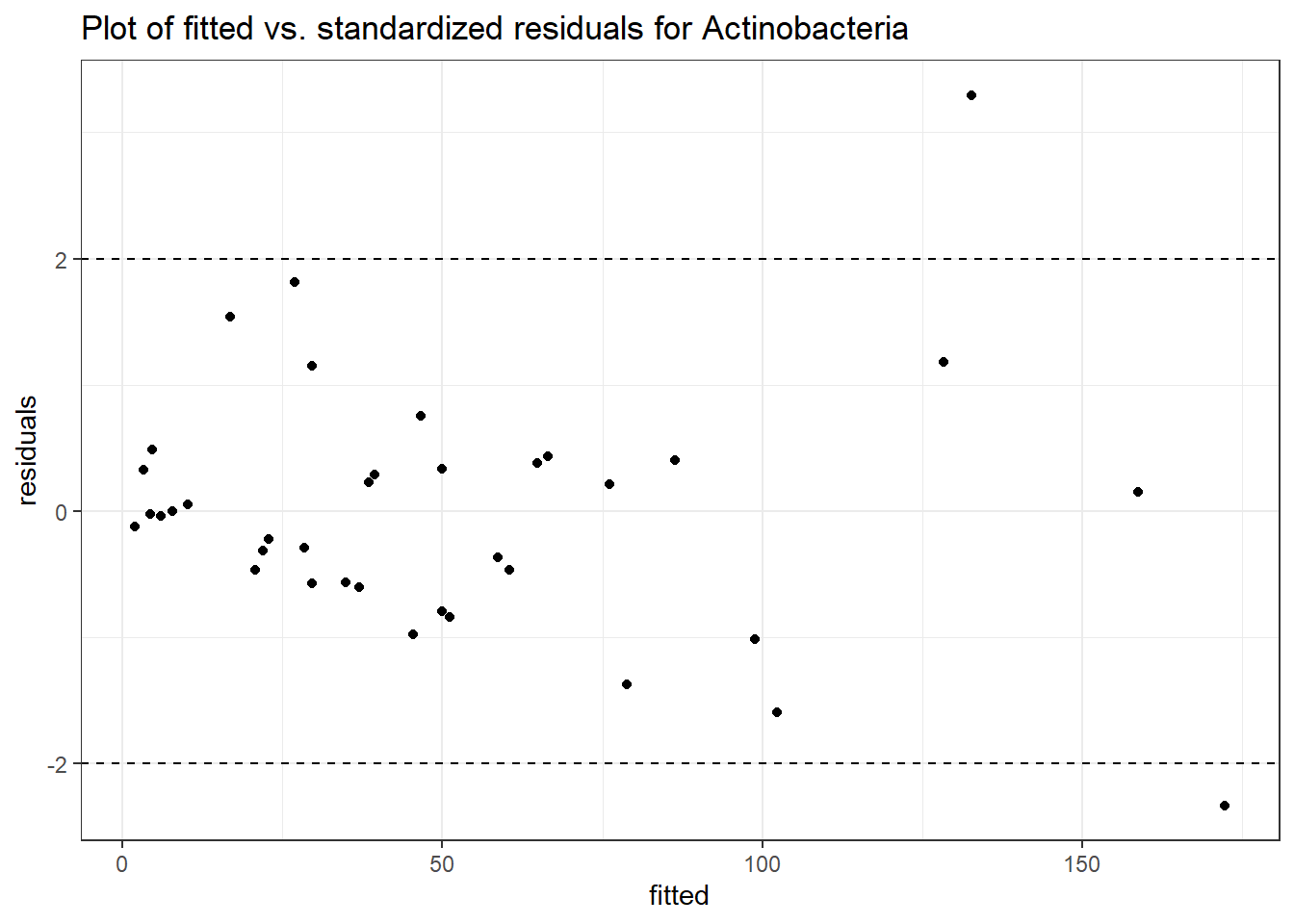
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Actinobacteria 2.78265 0.51627 5.38988 0.00000
intB Actinobacteria 0.93295 0.52659 1.77168 0.07645
time Actinobacteria -0.26077 0.02240 -11.64121 0.00000
female Actinobacteria 1.08413 0.54889 1.97512 0.04825
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.85789
Significance test not available
#########################################
#########################################
Model: Bacteroidetes ~ 1 + intB + time + female + (1 | SubjectID)
<environment: 0x000000000b5e7848>
Link: poisson
Intraclass Correlation (ICC): 0.06463683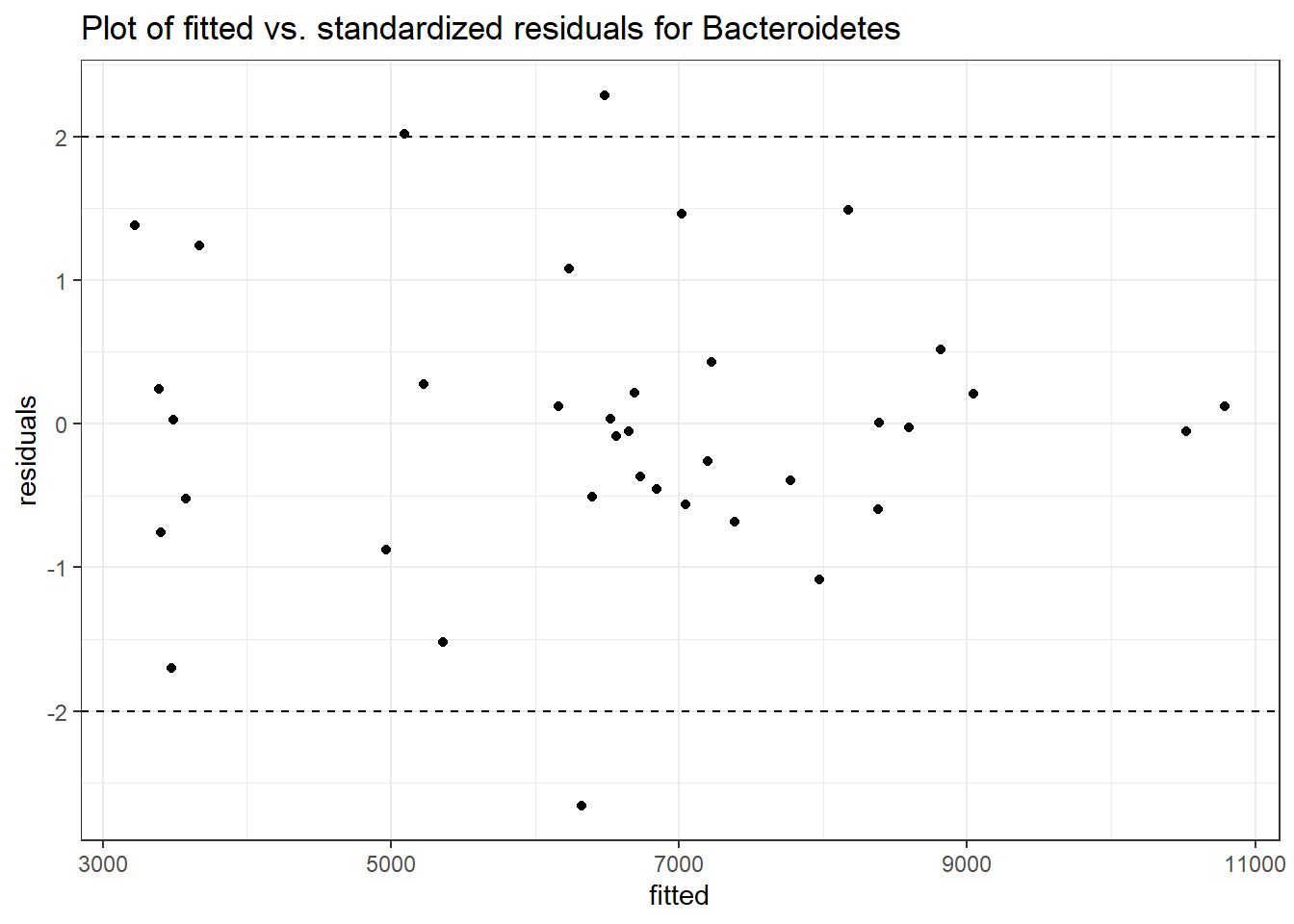
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Bacteroidetes 9.10955 0.15383 59.21996 0.00000
intB Bacteroidetes -0.17097 0.15963 -1.07102 0.28416
time Bacteroidetes -0.02542 0.00189 -13.45806 0.00000
female Bacteroidetes -0.39664 0.16522 -2.40071 0.01636
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.26288
Significance test not available
#########################################
#########################################
Model: Cyanobacteria ~ 1 + intB + time + female + (1 | SubjectID)
<environment: 0x000000000b5e7848>
Link: poisson
Intraclass Correlation (ICC): 0.8205922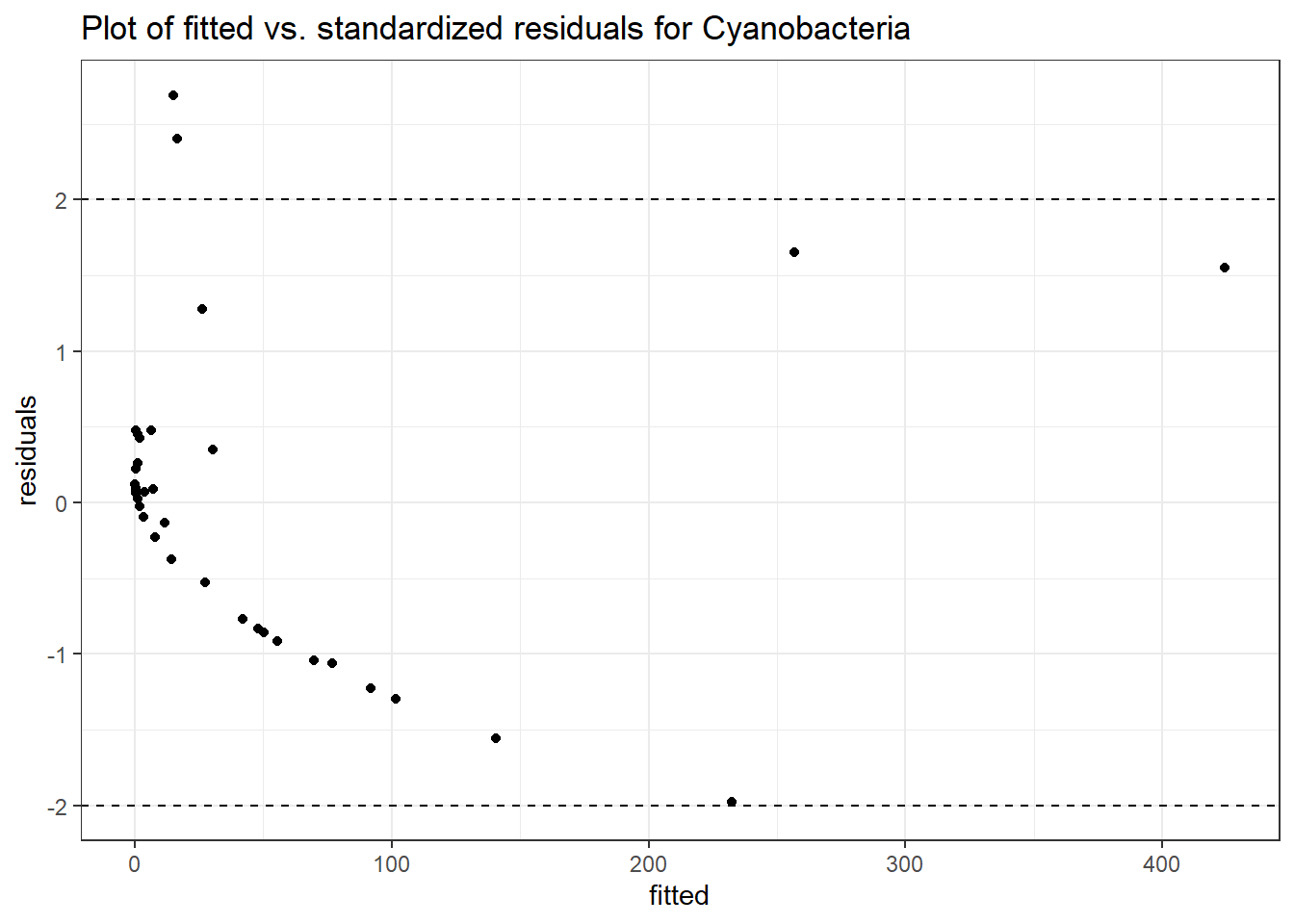
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Cyanobacteria 3.83226 1.28717 2.97728 0.00291
intB Cyanobacteria -2.44114 1.34246 -1.81841 0.06900
time Cyanobacteria -0.60181 0.02518 -23.89714 0.00000
female Cyanobacteria -0.25919 1.38186 -0.18757 0.85122
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.1387
Significance test not available
#########################################
#########################################
Model: Euryarchaeota ~ 1 + intB + time + female + (1 | SubjectID)
<environment: 0x000000000b5e7848>
Link: poisson
Intraclass Correlation (ICC): 0.7253927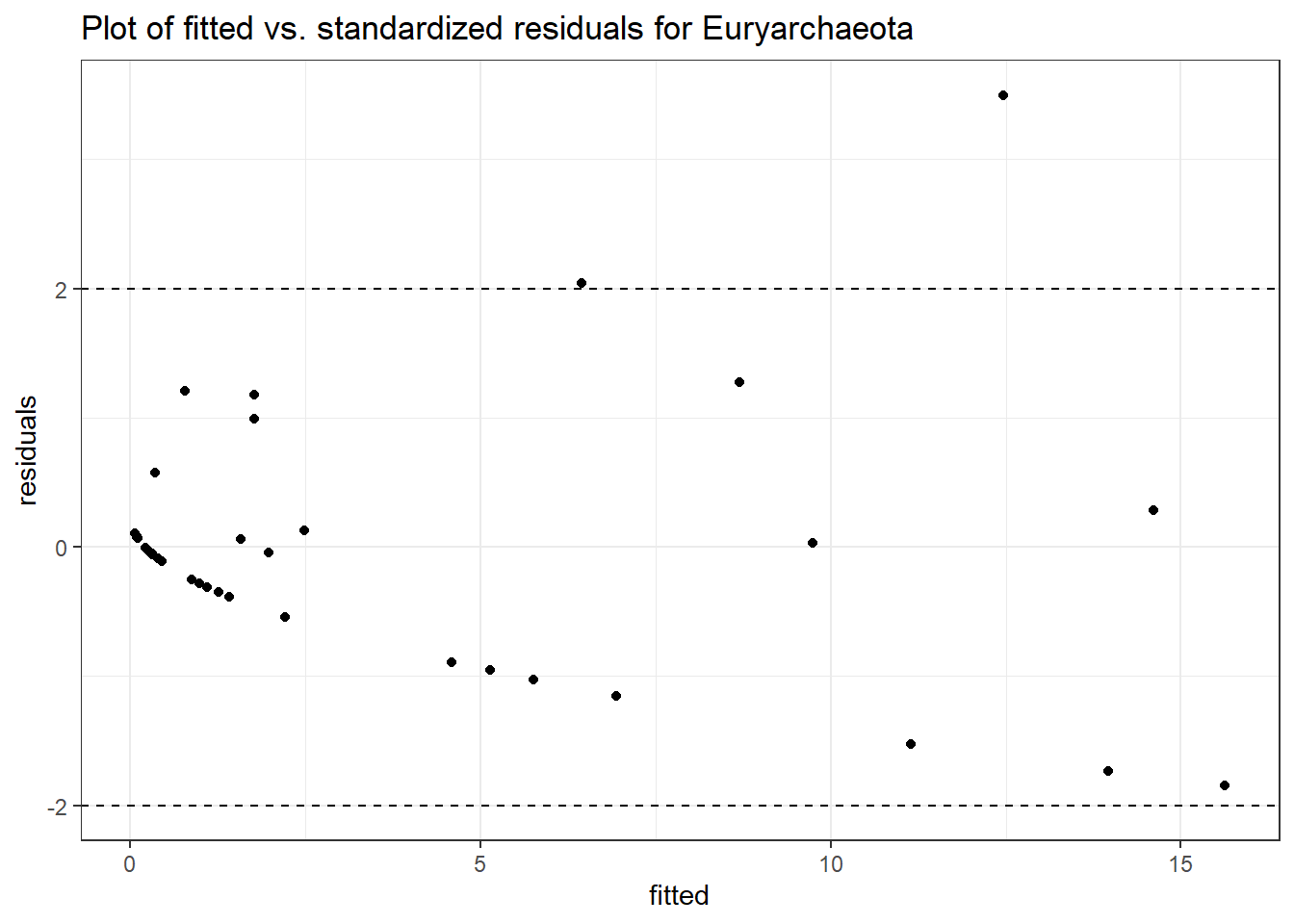
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Euryarchaeota -1.56218 1.32619 -1.17795 0.23882
intB Euryarchaeota 1.02337 1.12826 0.90704 0.36439
time Euryarchaeota -0.11287 0.07978 -1.41467 0.15717
female Euryarchaeota 2.20529 1.23630 1.78378 0.07446
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.6253
Significance test not available
#########################################
#########################################
Model: Firmicutes ~ 1 + intB + time + female + (1 | SubjectID)
<environment: 0x000000000b5e7848>
Link: poisson
Intraclass Correlation (ICC): 0.04187562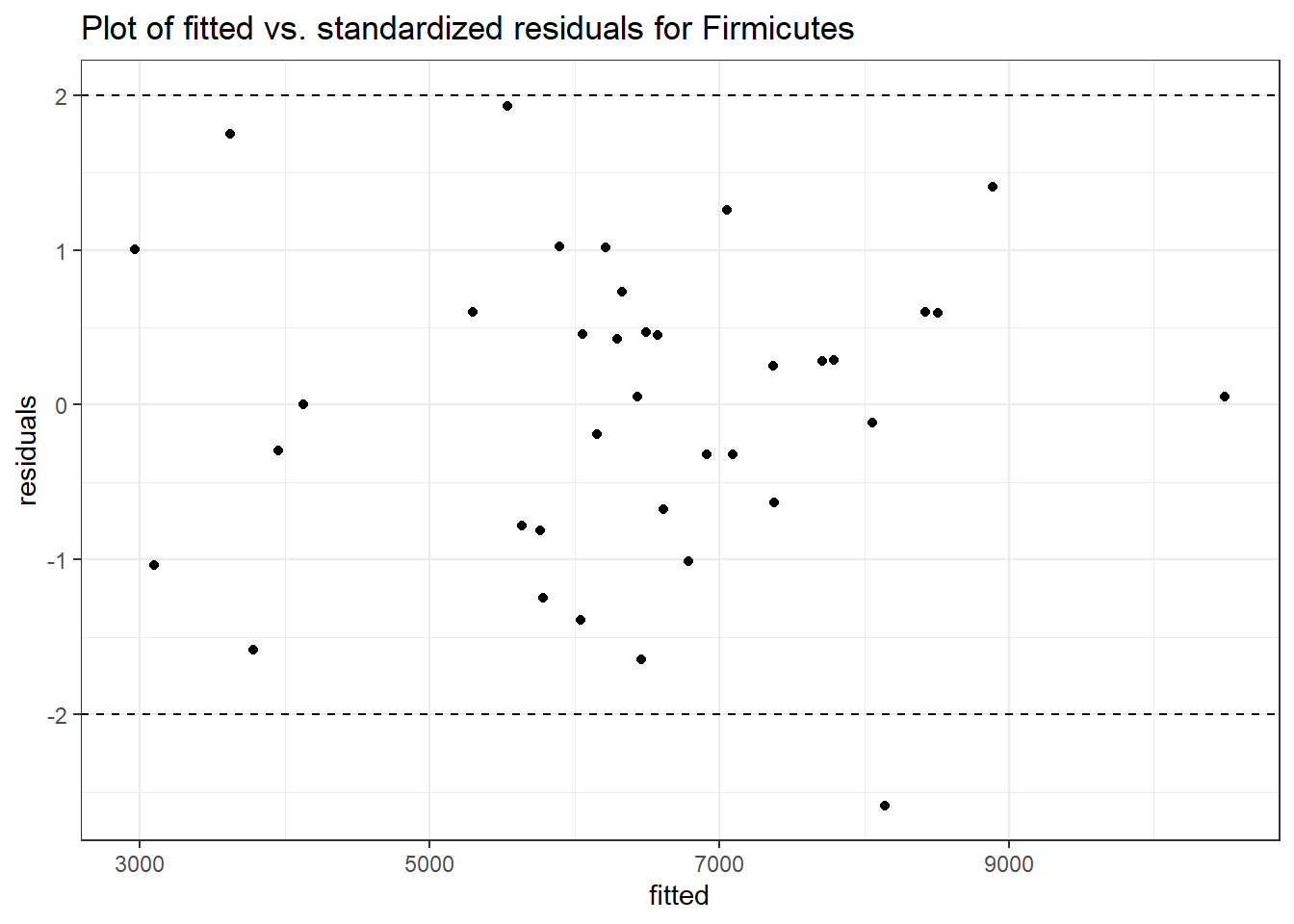
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Firmicutes 8.36797 0.12242 68.35276 0.00000
intB Firmicutes 0.26273 0.12698 2.06907 0.03854
time Firmicutes -0.04391 0.00187 -23.47675 0.00000
female Firmicutes 0.46761 0.13145 3.55723 0.00037
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.20906
Significance test not available
#########################################
#########################################
Model: Fusobacteria ~ 1 + intB + time + female + (1 | SubjectID)
<environment: 0x000000000b5e7848>
Link: poisson
Intraclass Correlation (ICC): 0.829431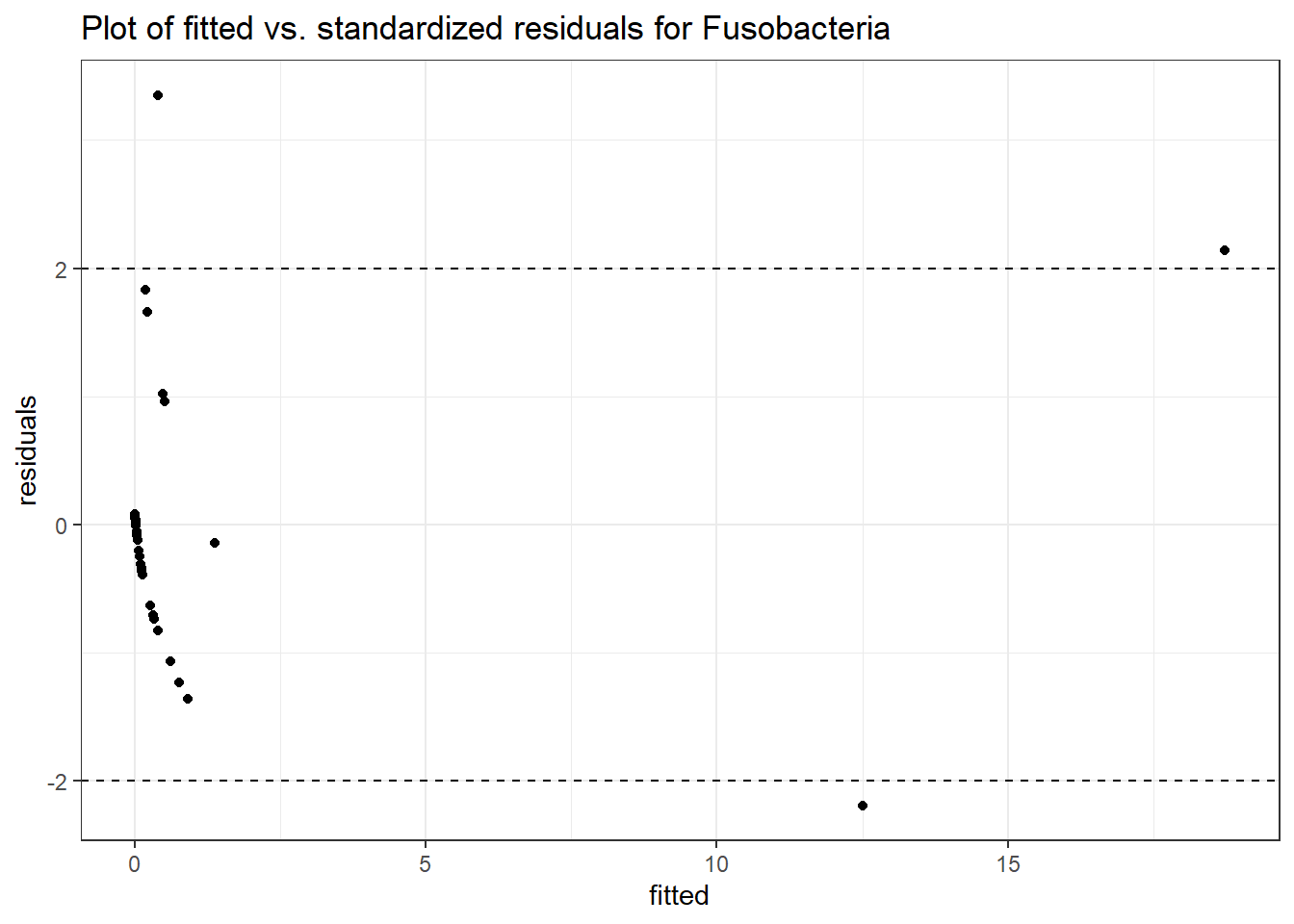
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Fusobacteria -0.78714 1.75543 -0.44840 0.65386
intB Fusobacteria -2.11435 1.81618 -1.16417 0.24435
time Fusobacteria -0.40368 0.24242 -1.66517 0.09588
female Fusobacteria -0.15009 1.82866 -0.08208 0.93459
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.2052
Significance test not available
#########################################
#########################################
Model: Lentisphaerae ~ 1 + intB + time + female + (1 | SubjectID)
<environment: 0x000000000b5e7848>
Link: poisson
Intraclass Correlation (ICC): 0.647492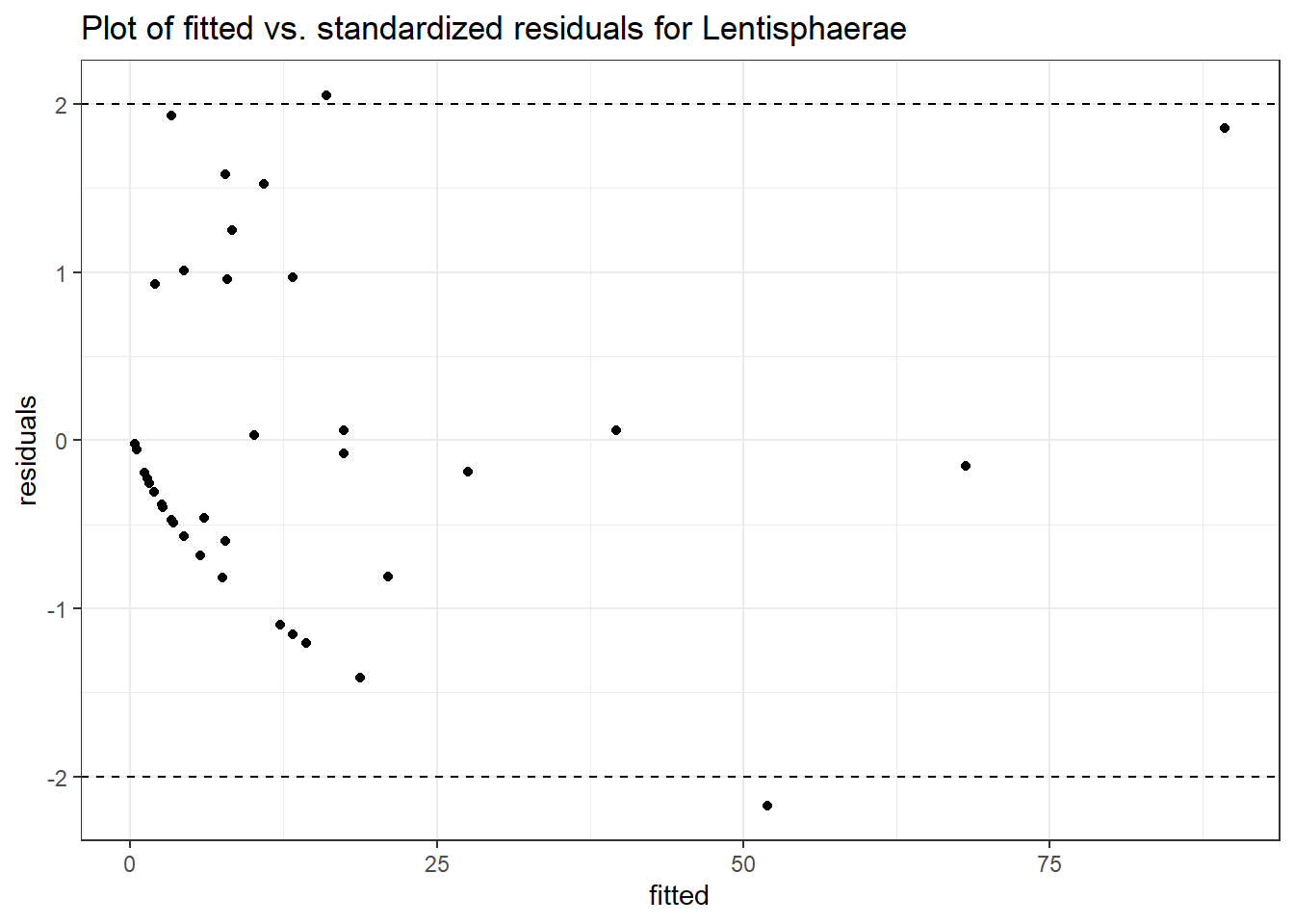
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Lentisphaerae 0.96215 0.86377 1.11390 0.26532
intB Lentisphaerae 1.07069 0.85960 1.24557 0.21292
time Lentisphaerae -0.27048 0.03999 -6.76290 0.00000
female Lentisphaerae 0.87684 0.89764 0.97682 0.32866
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 1.3553
Significance test not available
#########################################
#########################################
Model: Proteobacteria ~ 1 + intB + time + female + (1 | SubjectID)
<environment: 0x000000000b5e7848>
Link: poisson
Intraclass Correlation (ICC): 0.09272157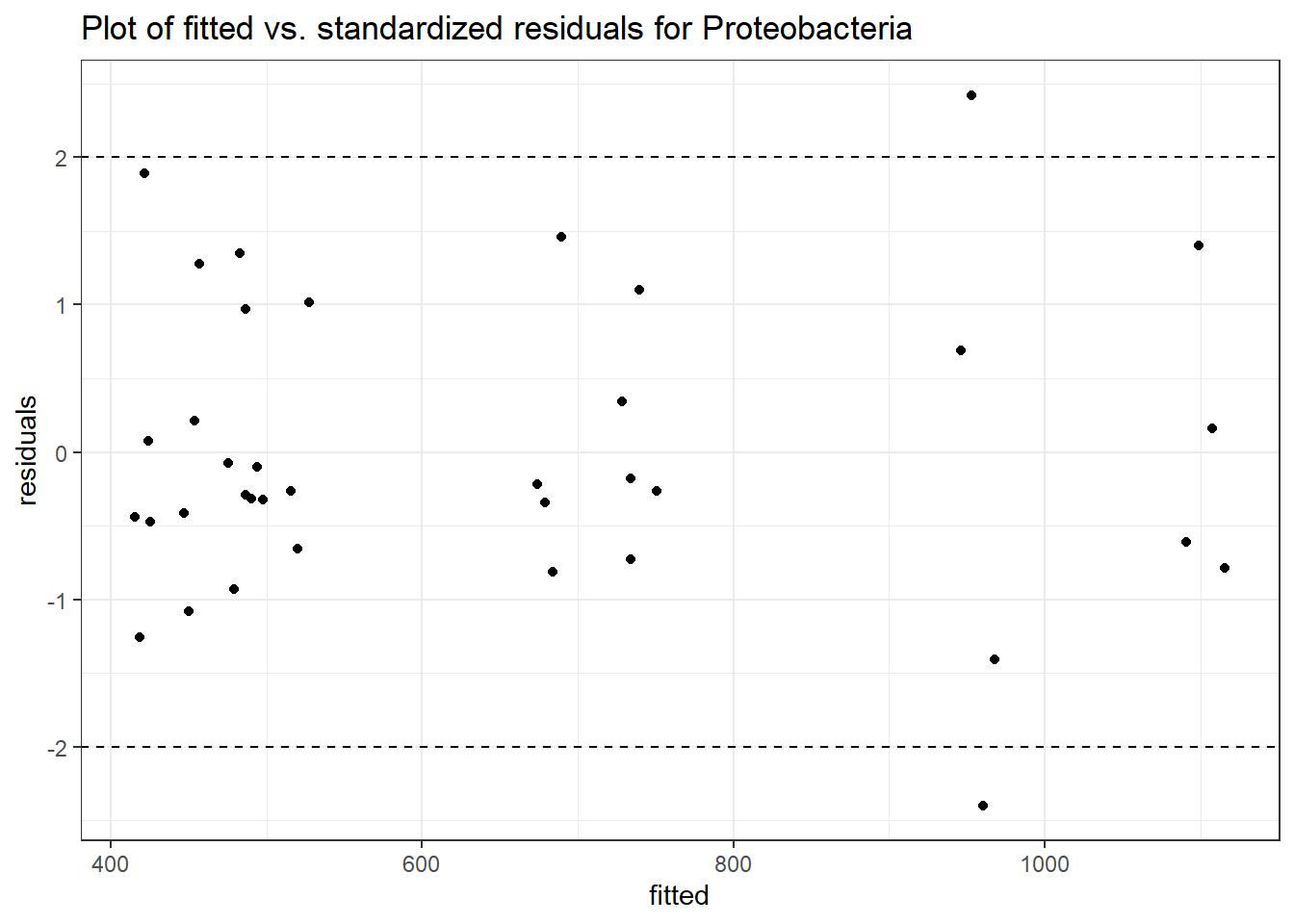
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Proteobacteria 6.38847 0.18772 34.03204 0.00000
intB Proteobacteria -0.02001 0.19470 -0.10279 0.91813
time Proteobacteria 0.00754 0.00585 1.28995 0.19707
female Proteobacteria 0.02193 0.20147 0.10884 0.91333
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 0.31968
Significance test not available
#########################################
#########################################
Model: Tenericutes ~ 1 + intB + time + female + (1 | SubjectID)
<environment: 0x000000000b5e7848>
Link: poisson
Intraclass Correlation (ICC): 0.9092078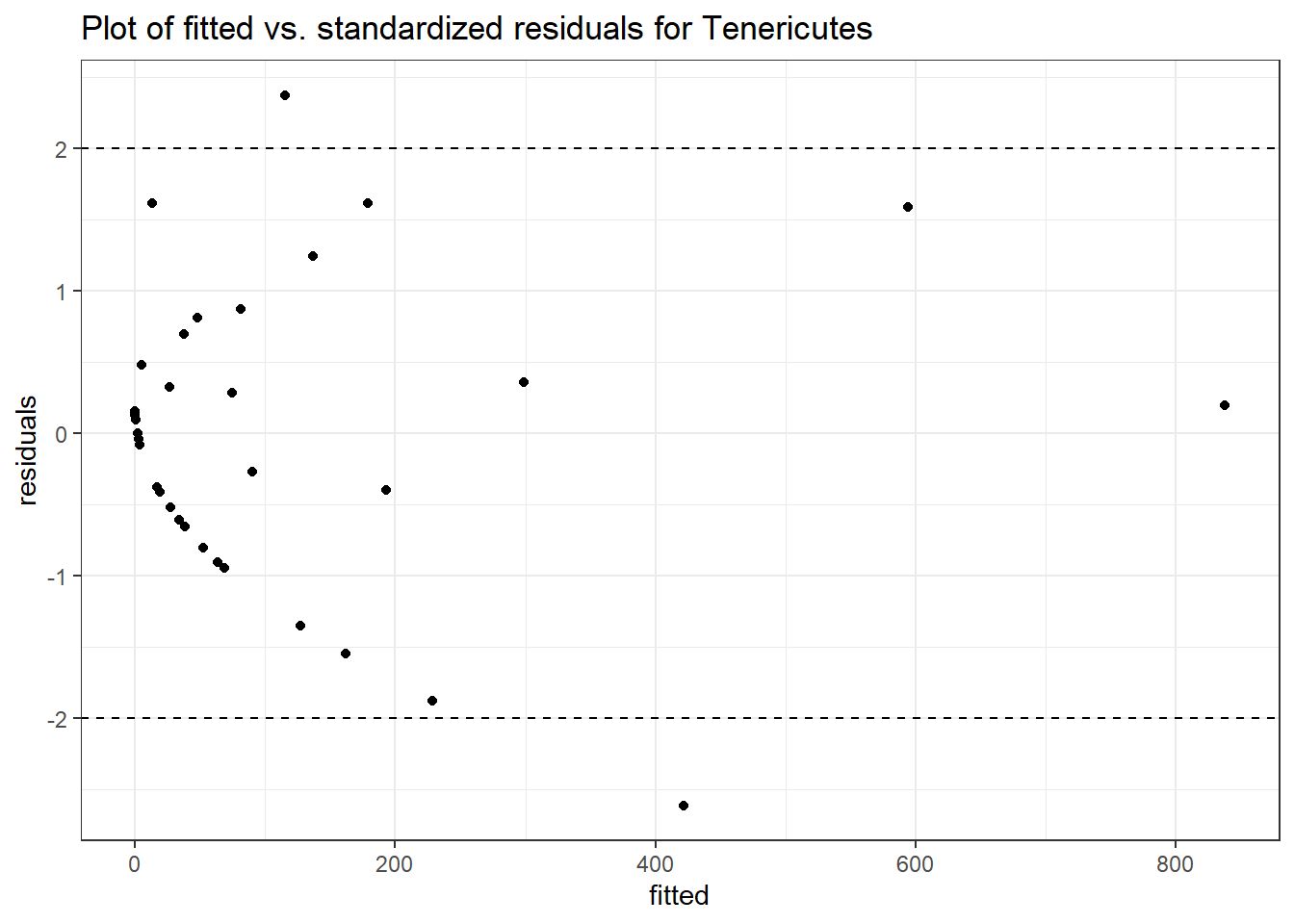
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Tenericutes 1.35734 1.93308 0.70216 0.48258
intB Tenericutes 1.89536 1.97158 0.96134 0.33638
time Tenericutes -0.34364 0.01482 -23.18186 0.00000
female Tenericutes 0.71077 2.04269 0.34796 0.72787
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 3.1645
Significance test not available
#########################################
#########################################
Model: Verrucomicrobia ~ 1 + intB + time + female + (1 | SubjectID)
<environment: 0x000000000b5e7848>
Link: poisson
Intraclass Correlation (ICC): 0.8629325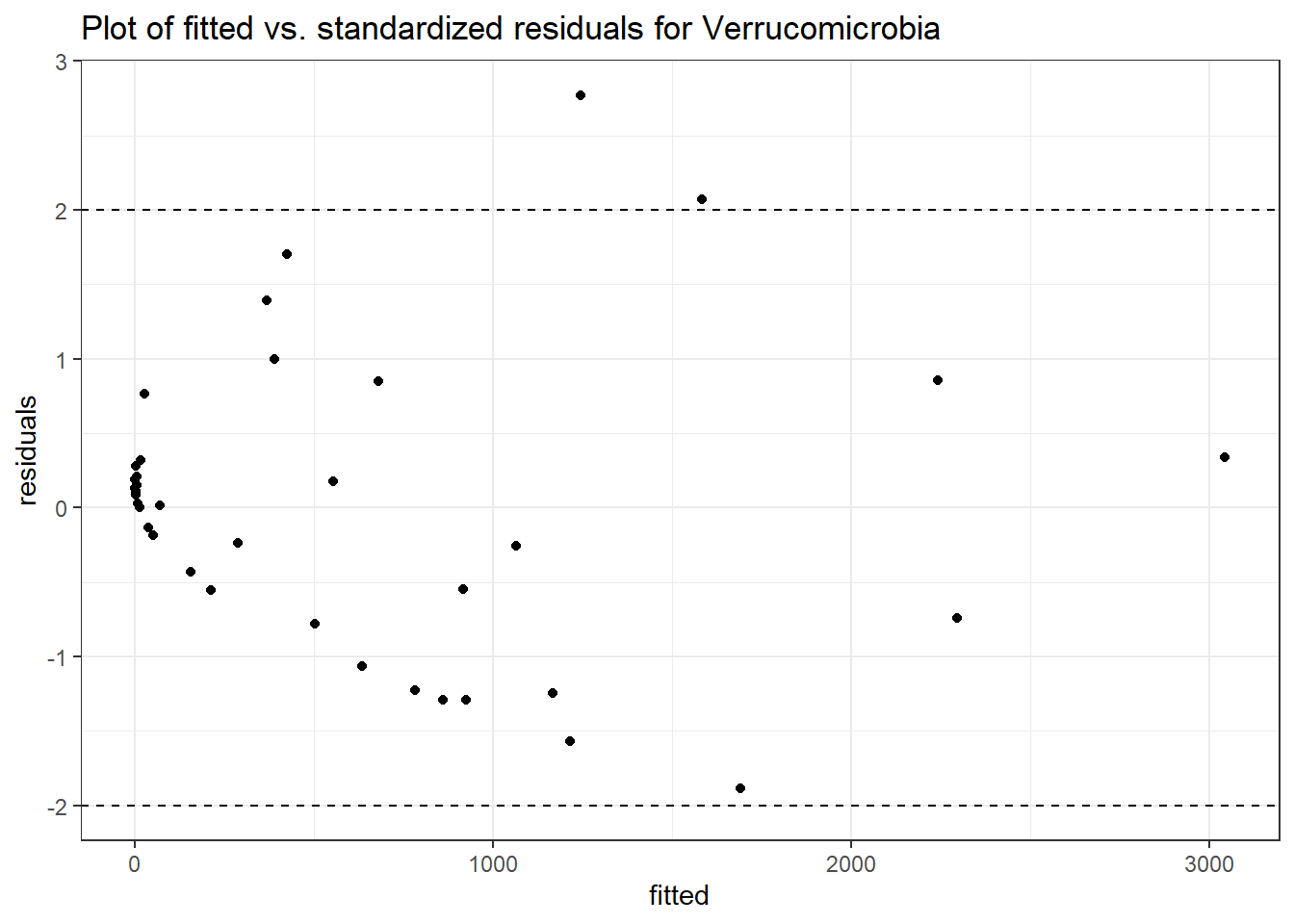
Fixed Effects (change in log)
Outcome Estimate Std. Error z value Pr(>|z|)
(Intercept) Verrucomicrobia 3.73169 1.49123 2.50242 0.01233
intB Verrucomicrobia 0.67678 1.53031 0.44225 0.65831
time Verrucomicrobia -0.30578 0.00609 -50.22331 0.00000
female Verrucomicrobia 1.84951 1.58984 1.16333 0.24470
Random Effects (SD on log scale)
Groups Name Std.Dev.
SubjectID (Intercept) 2.5091
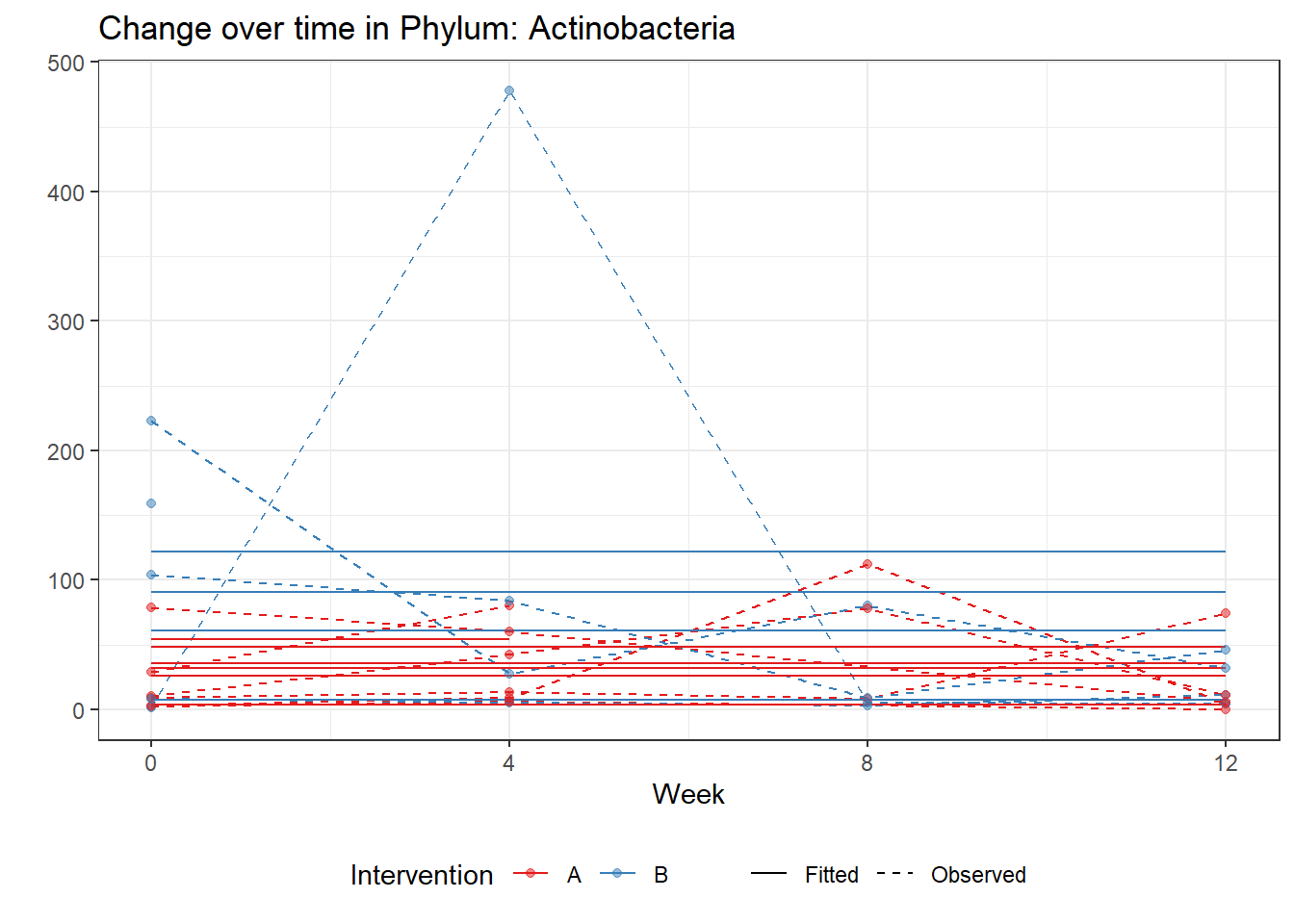
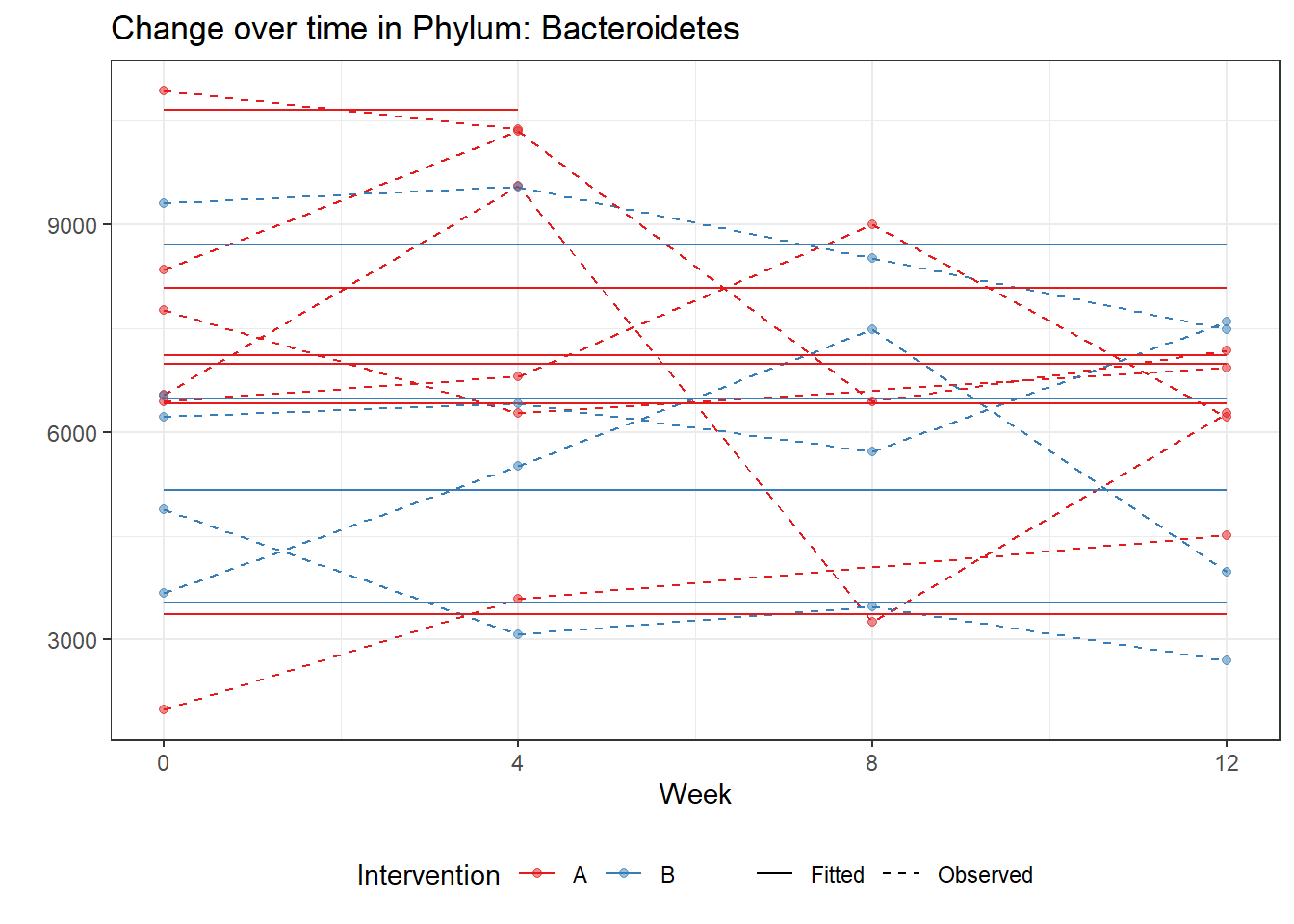
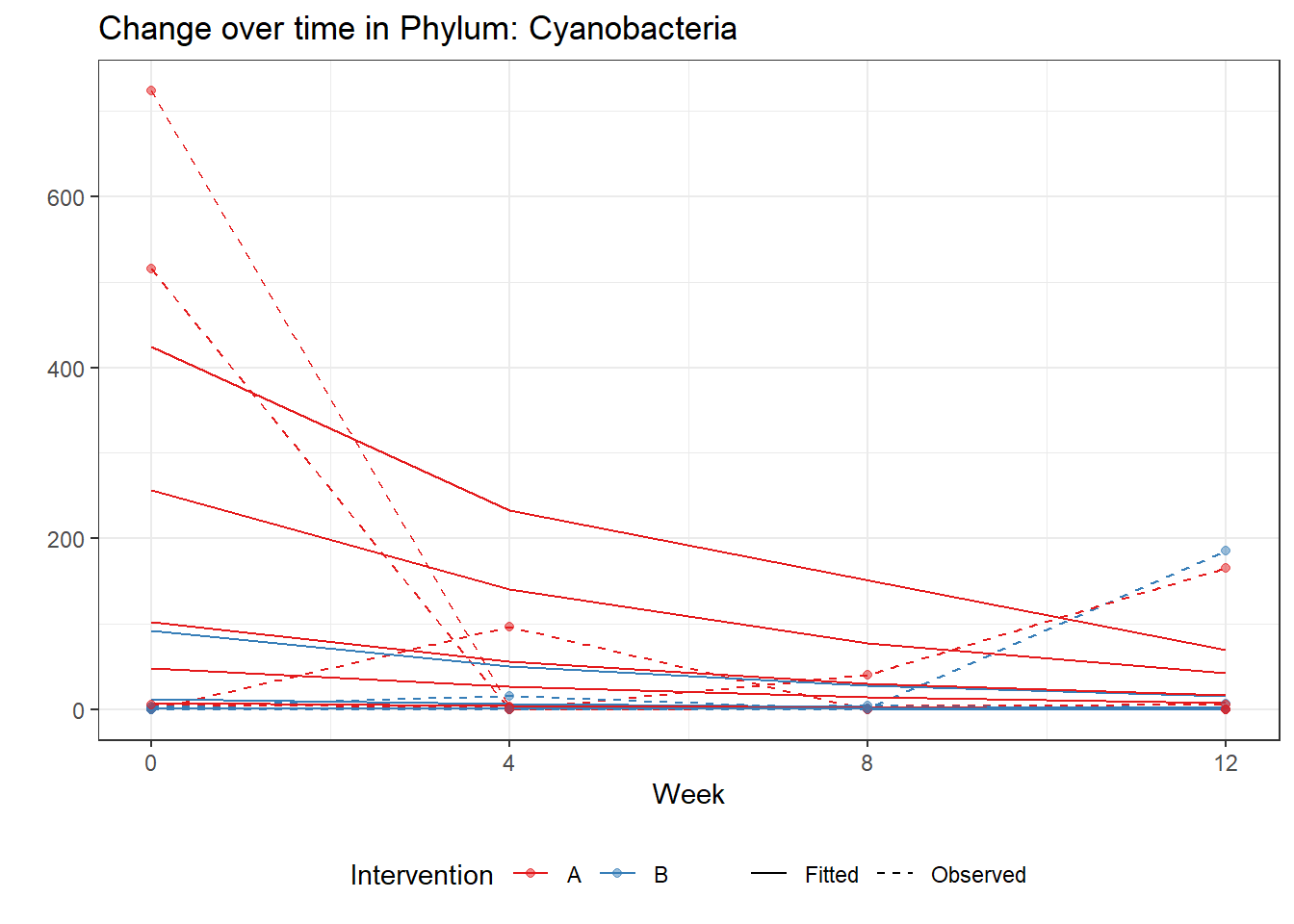
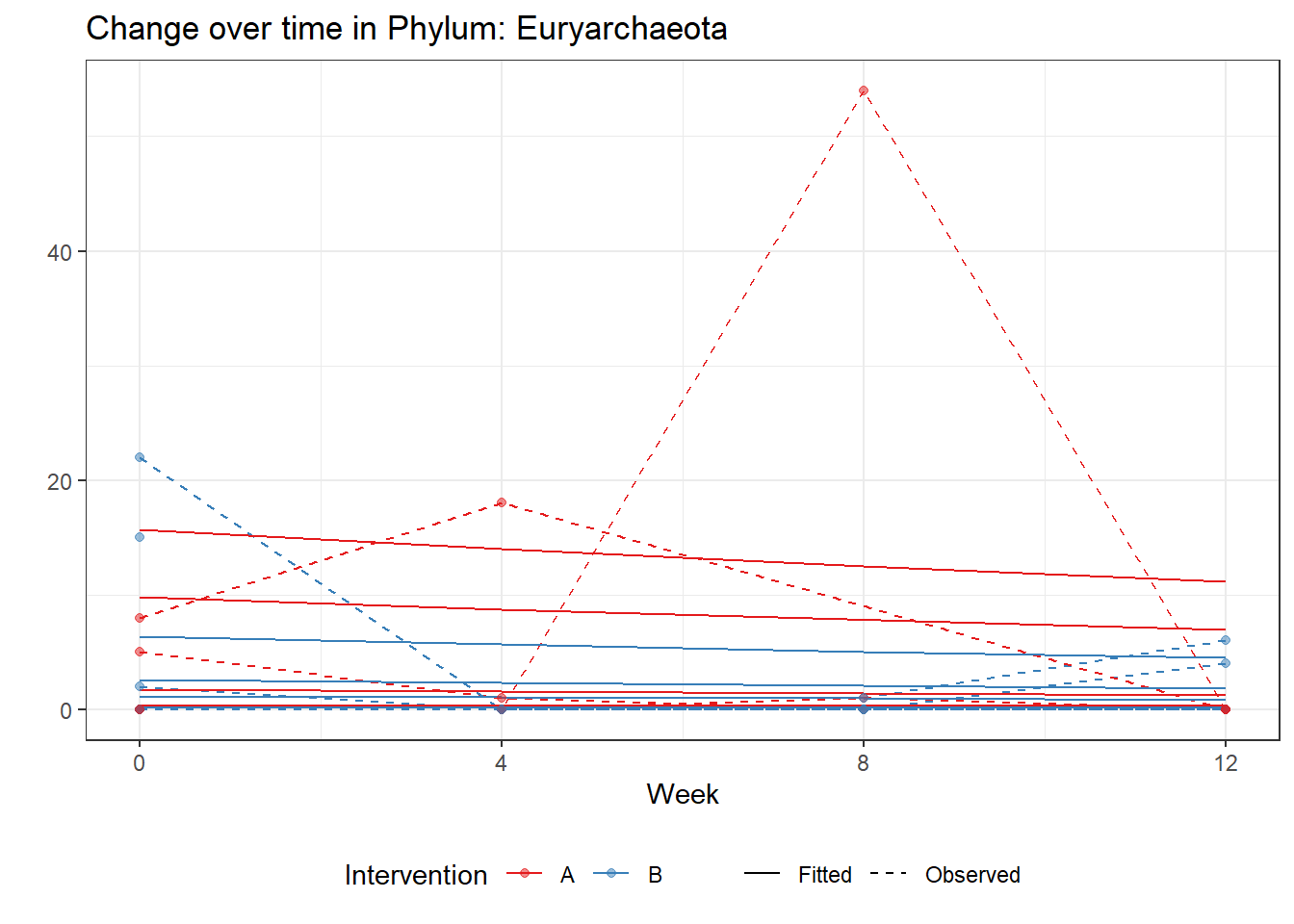
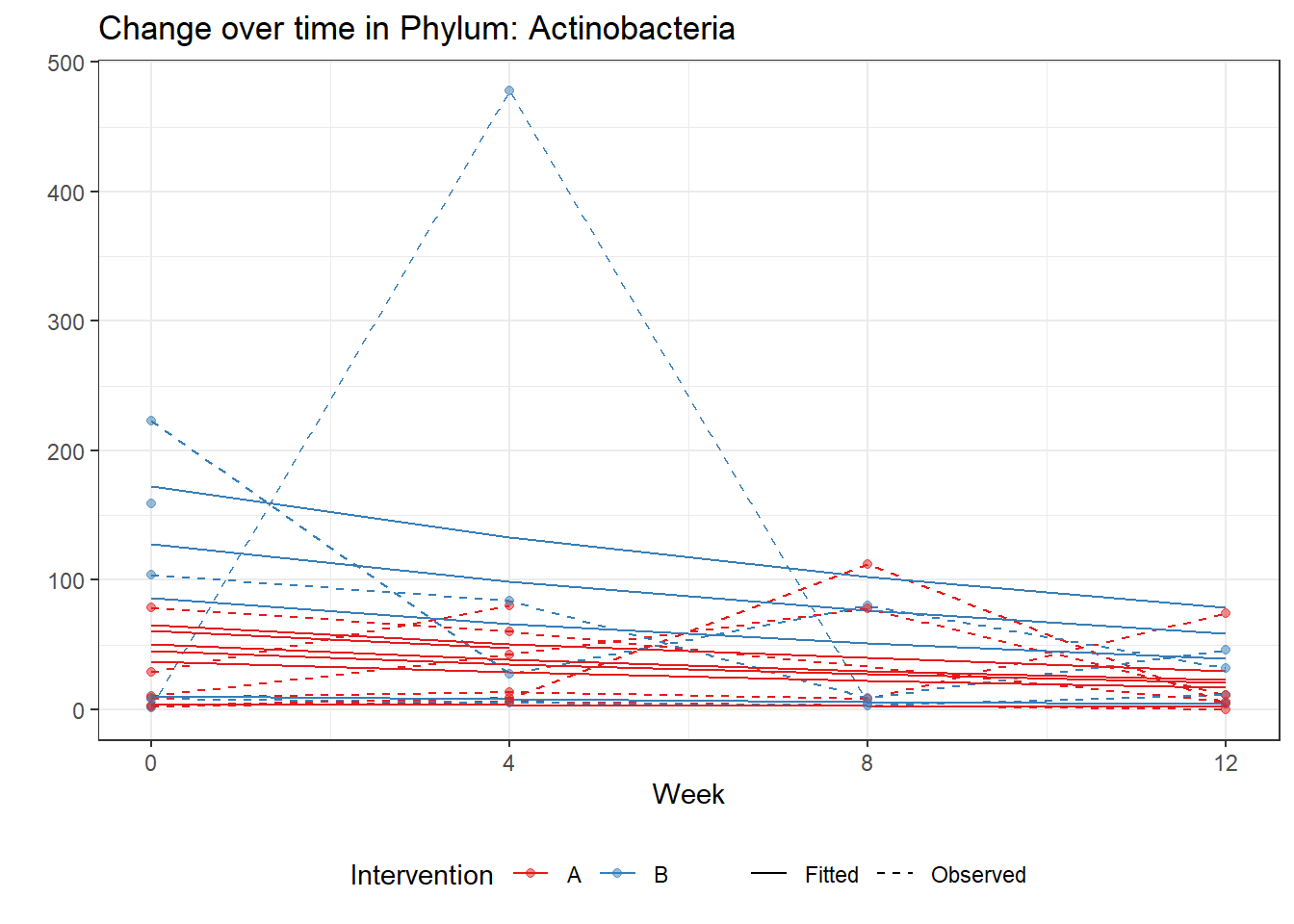
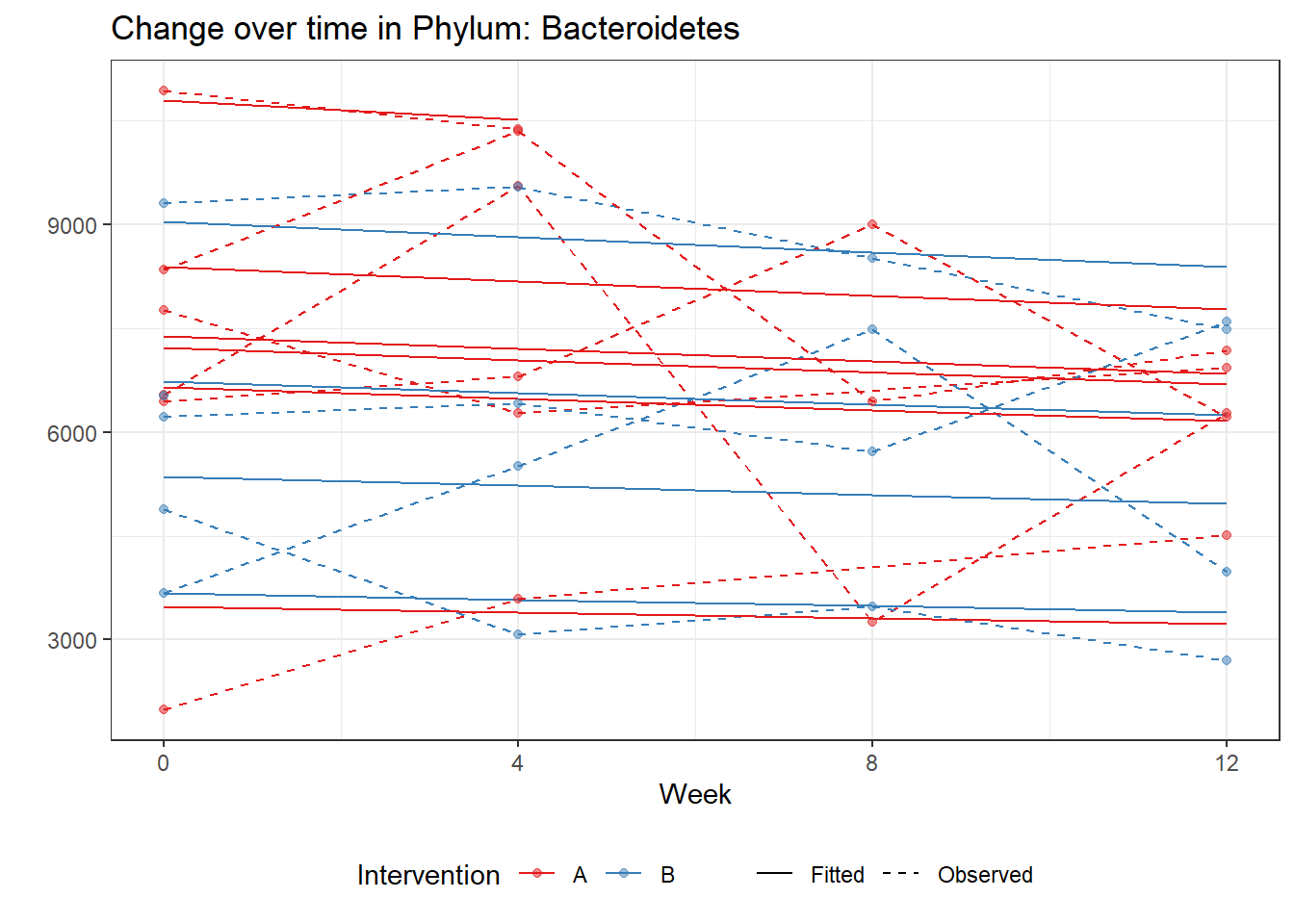
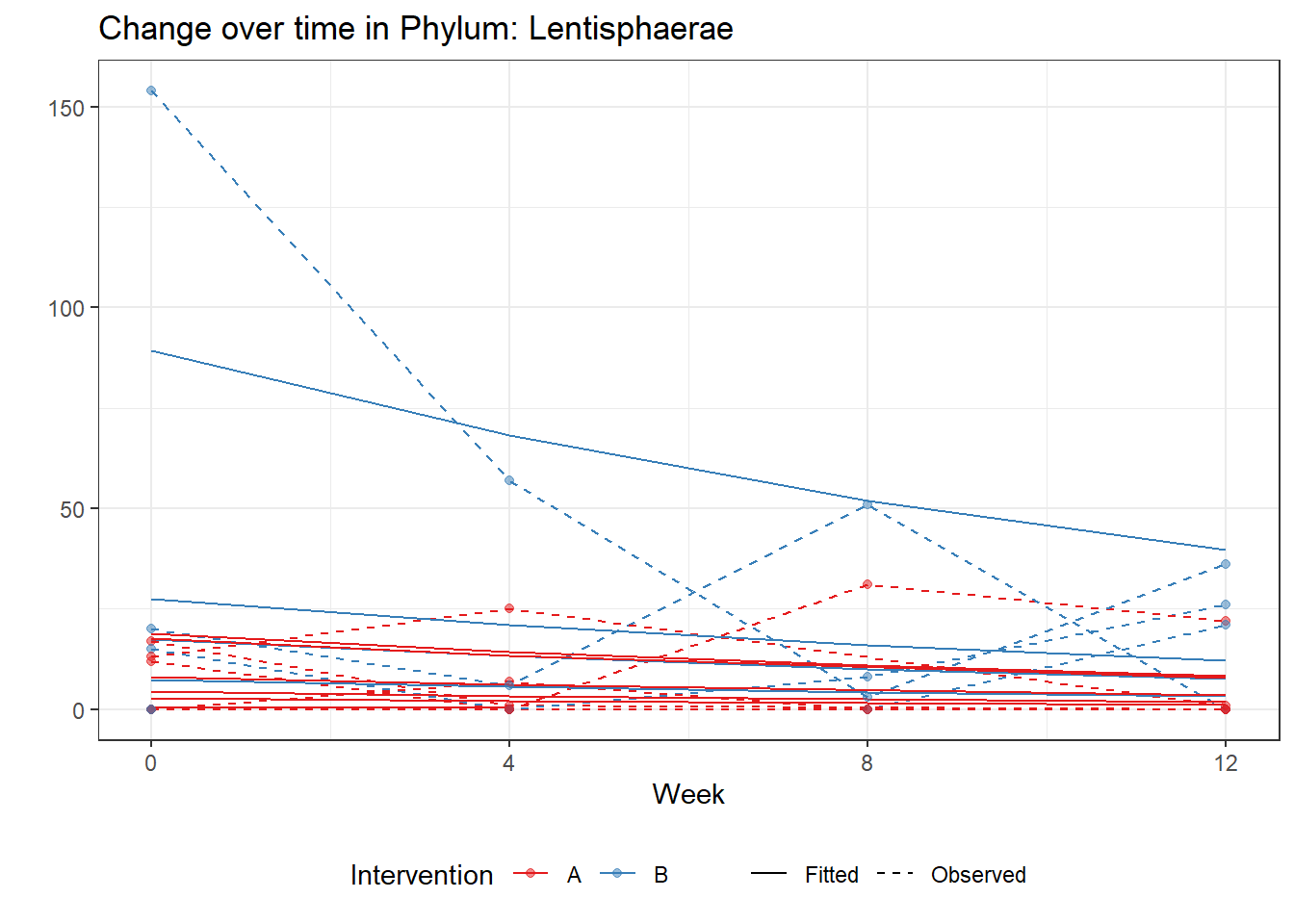
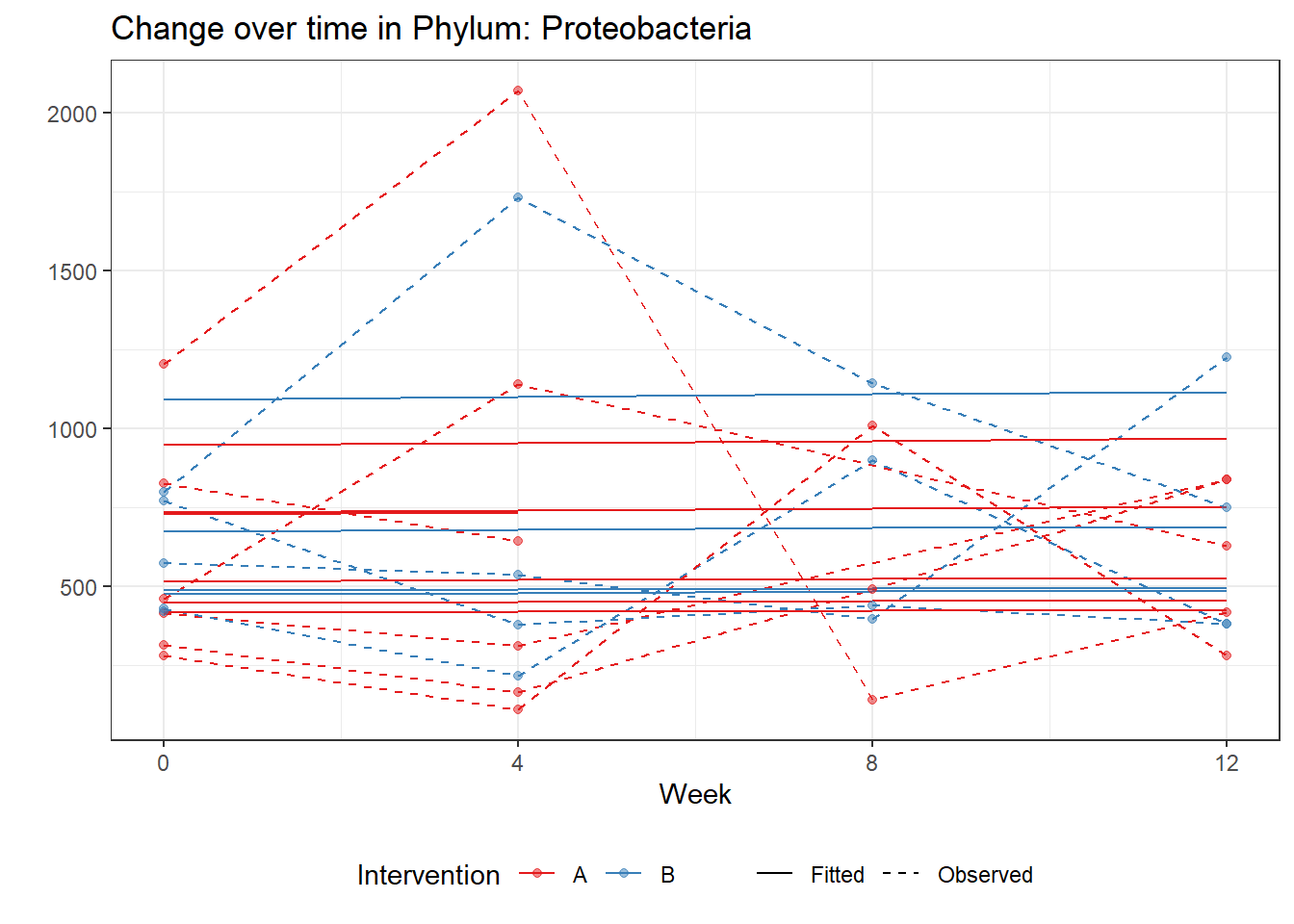
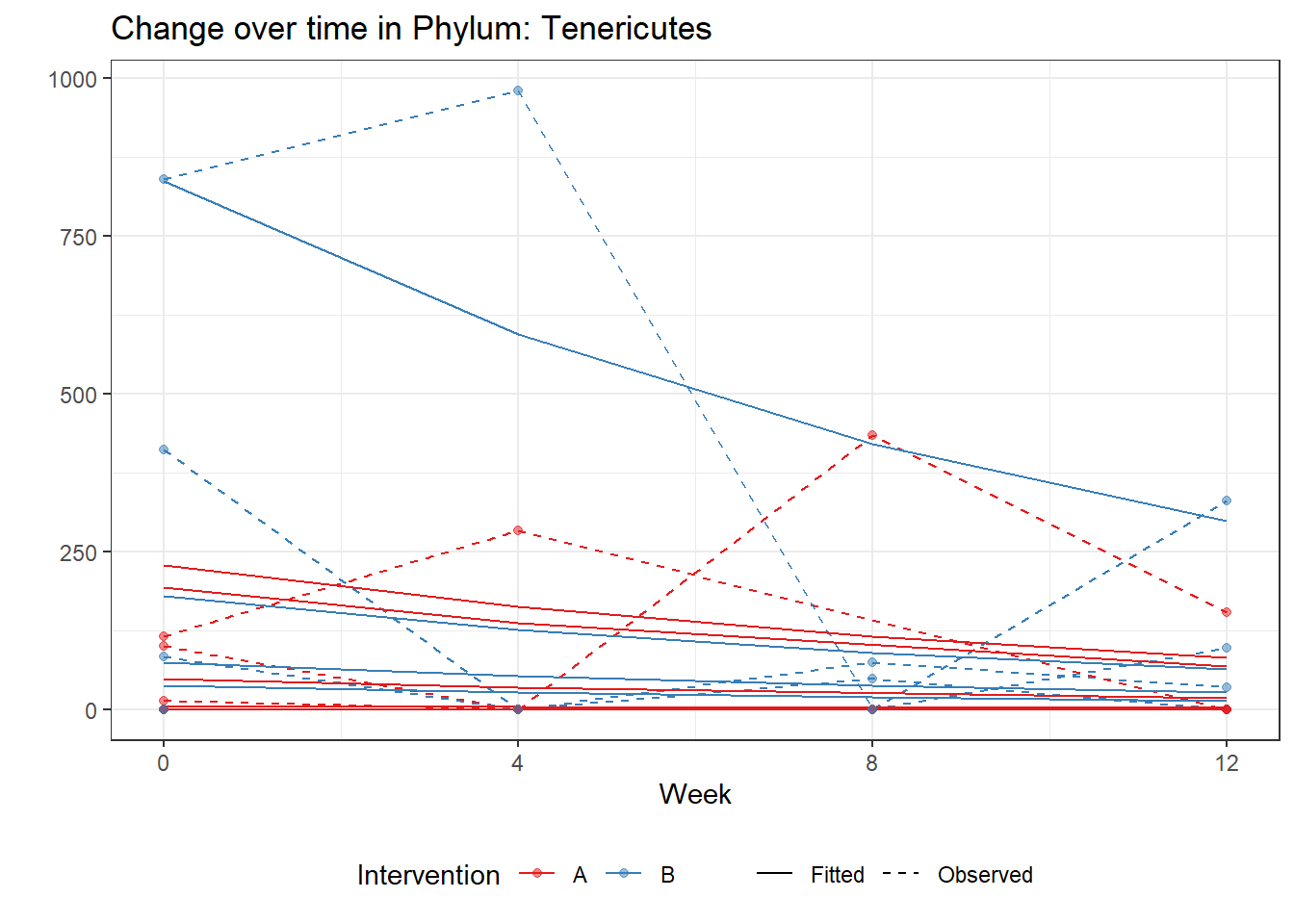
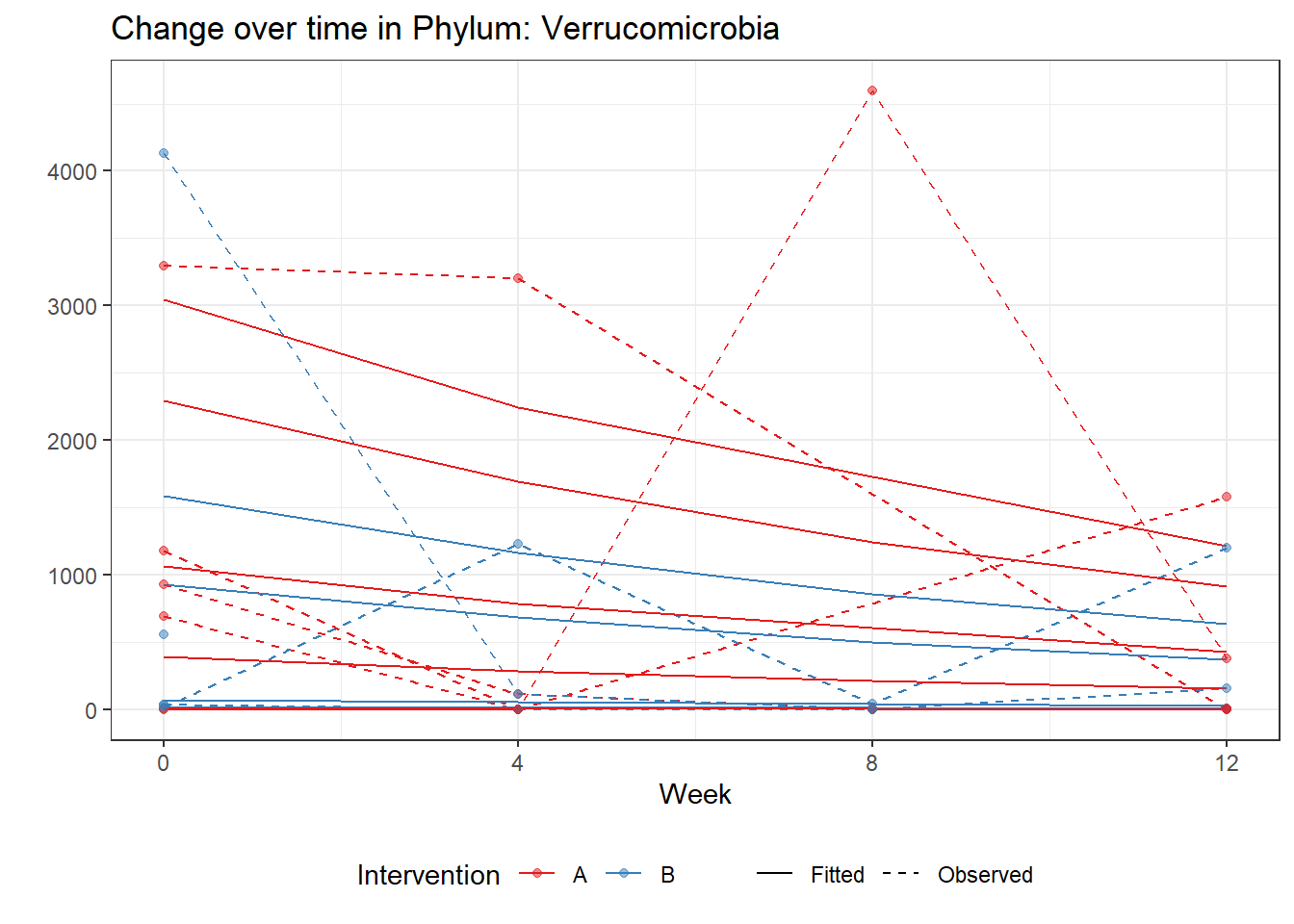
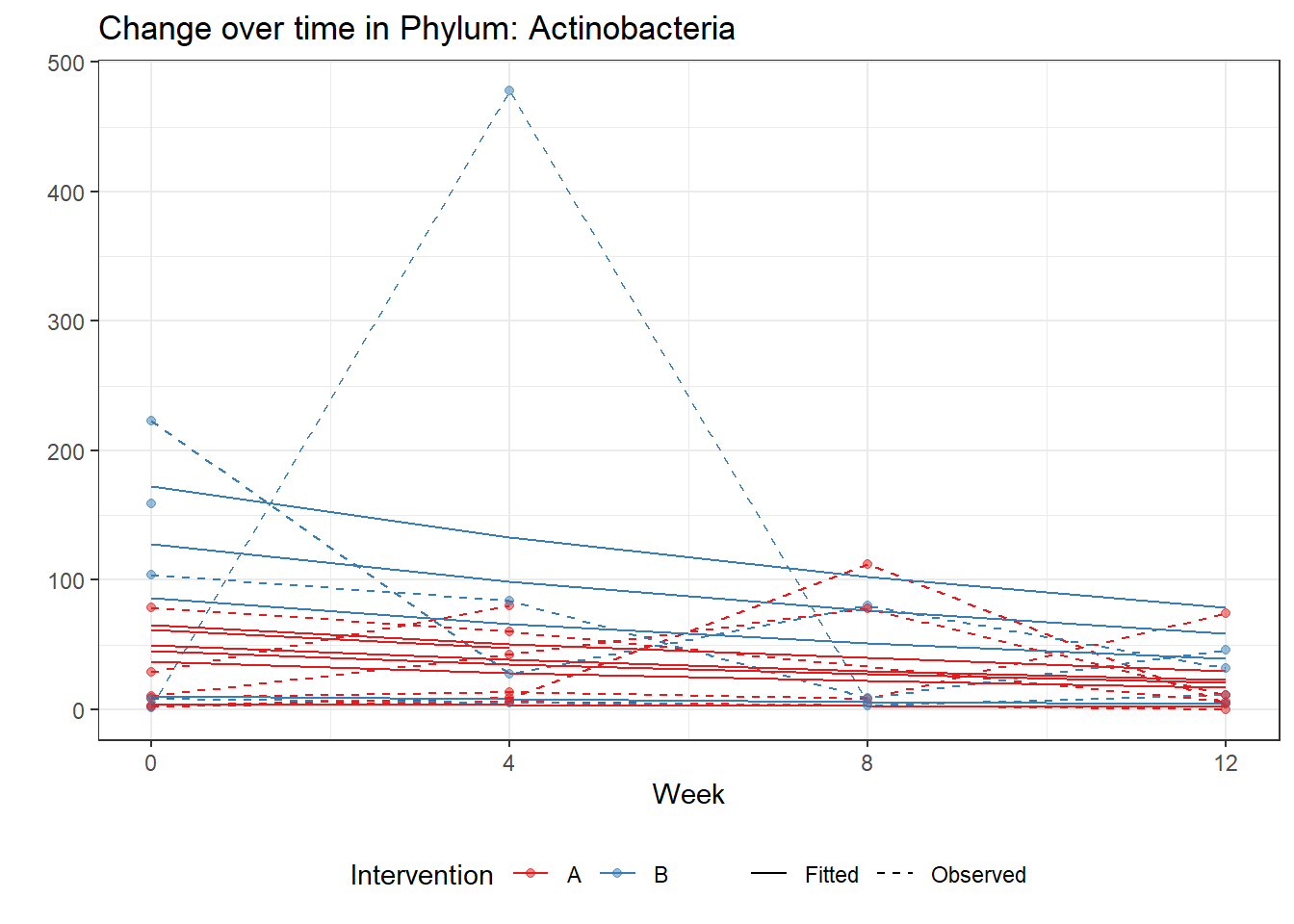
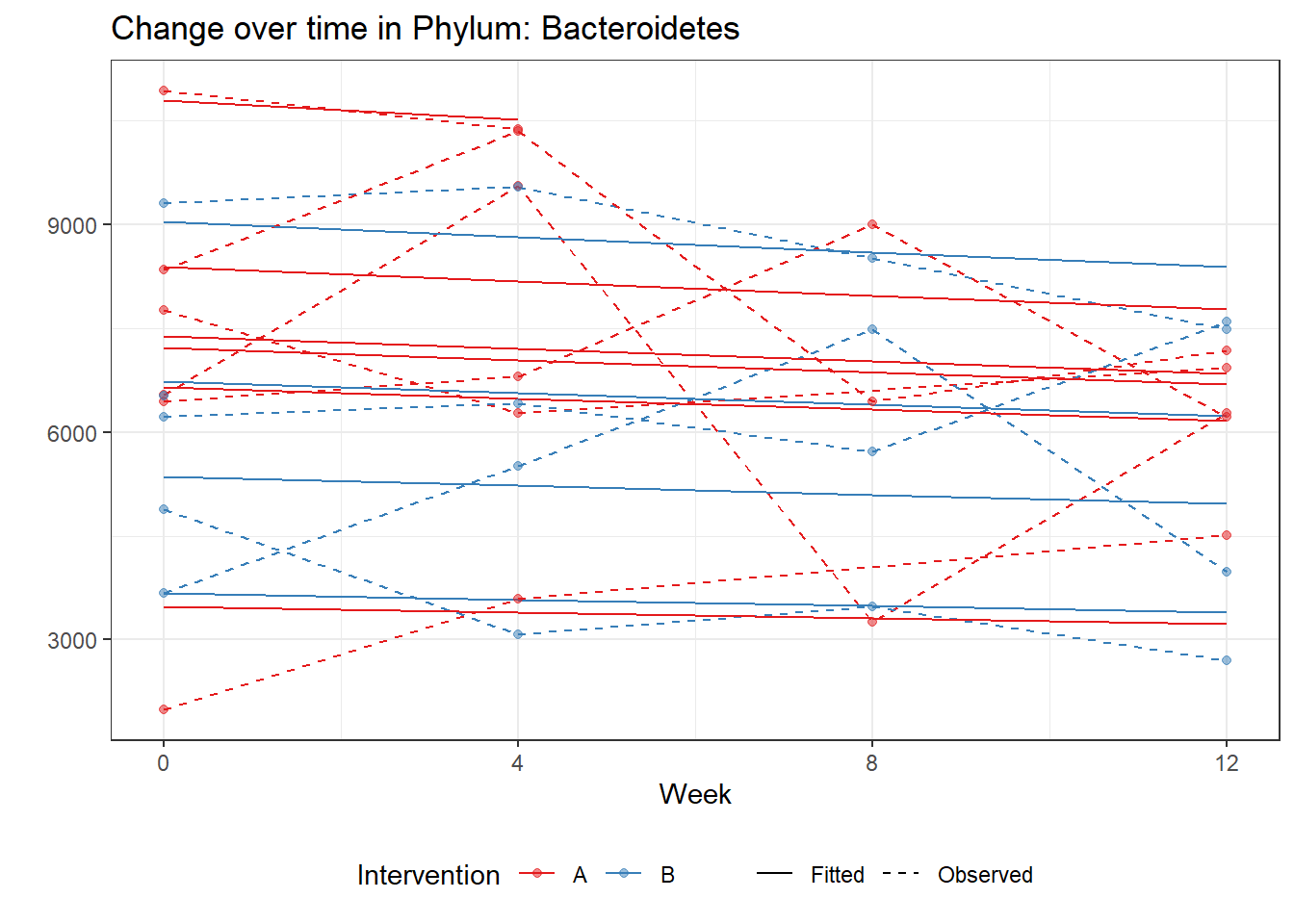
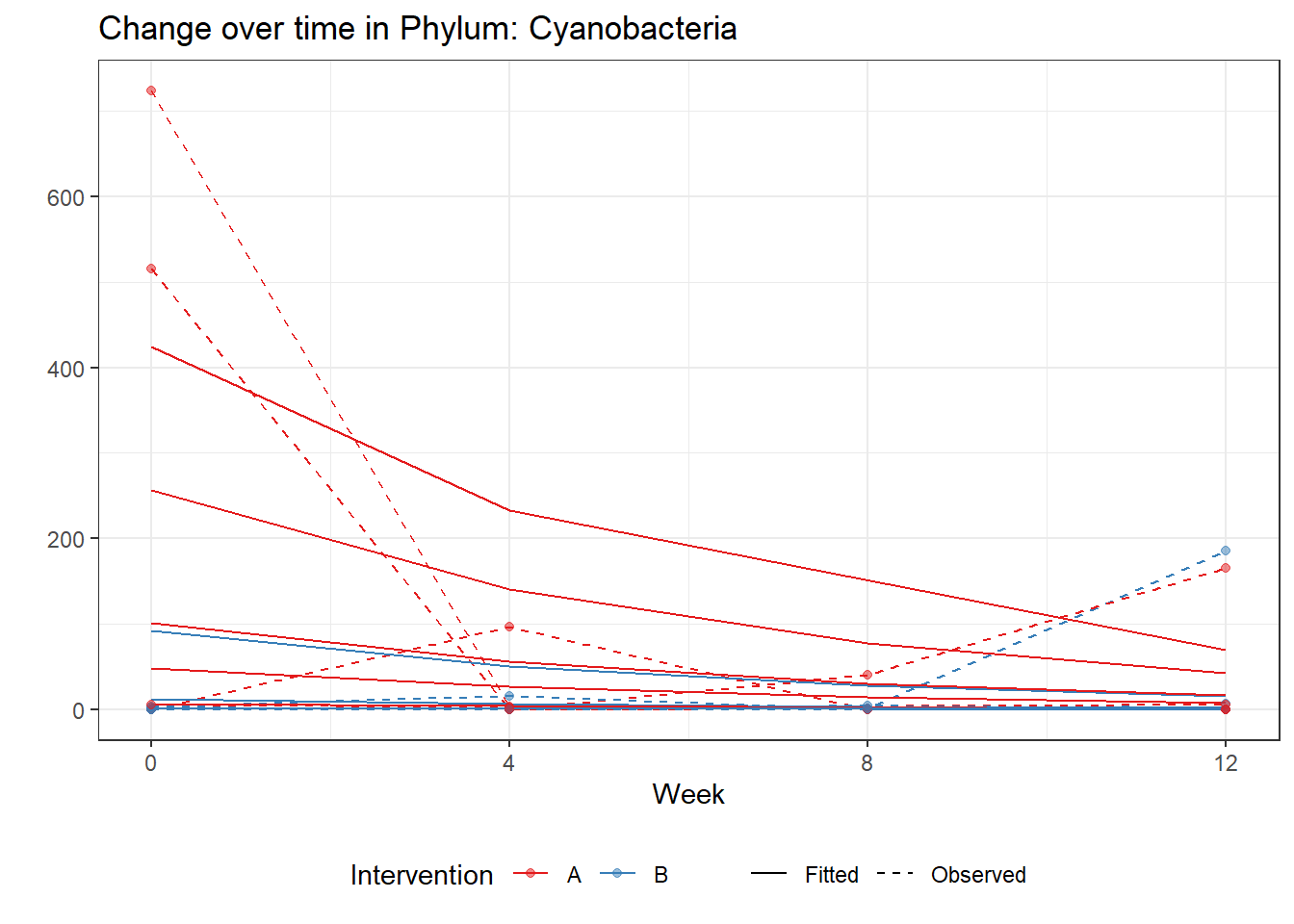
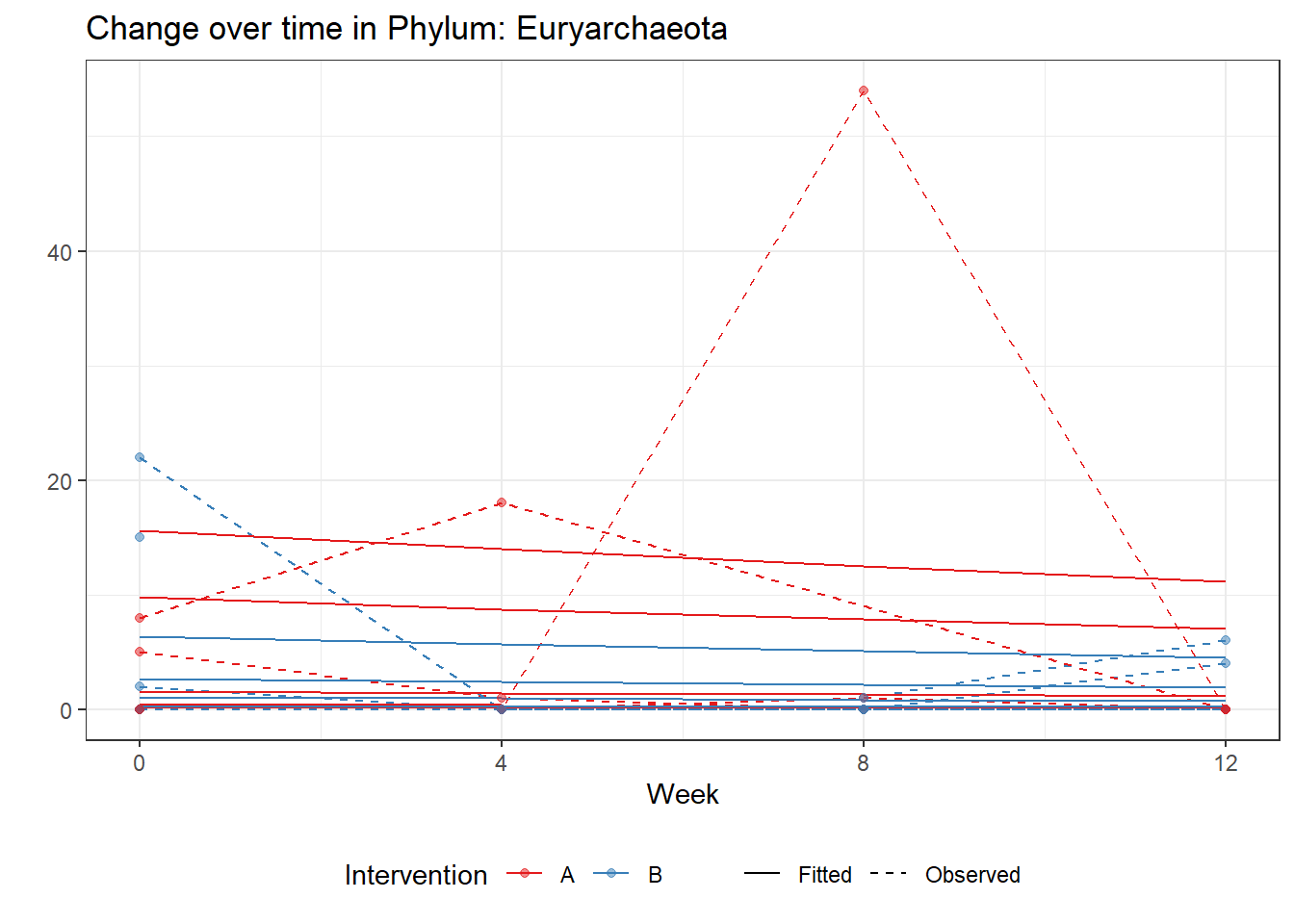
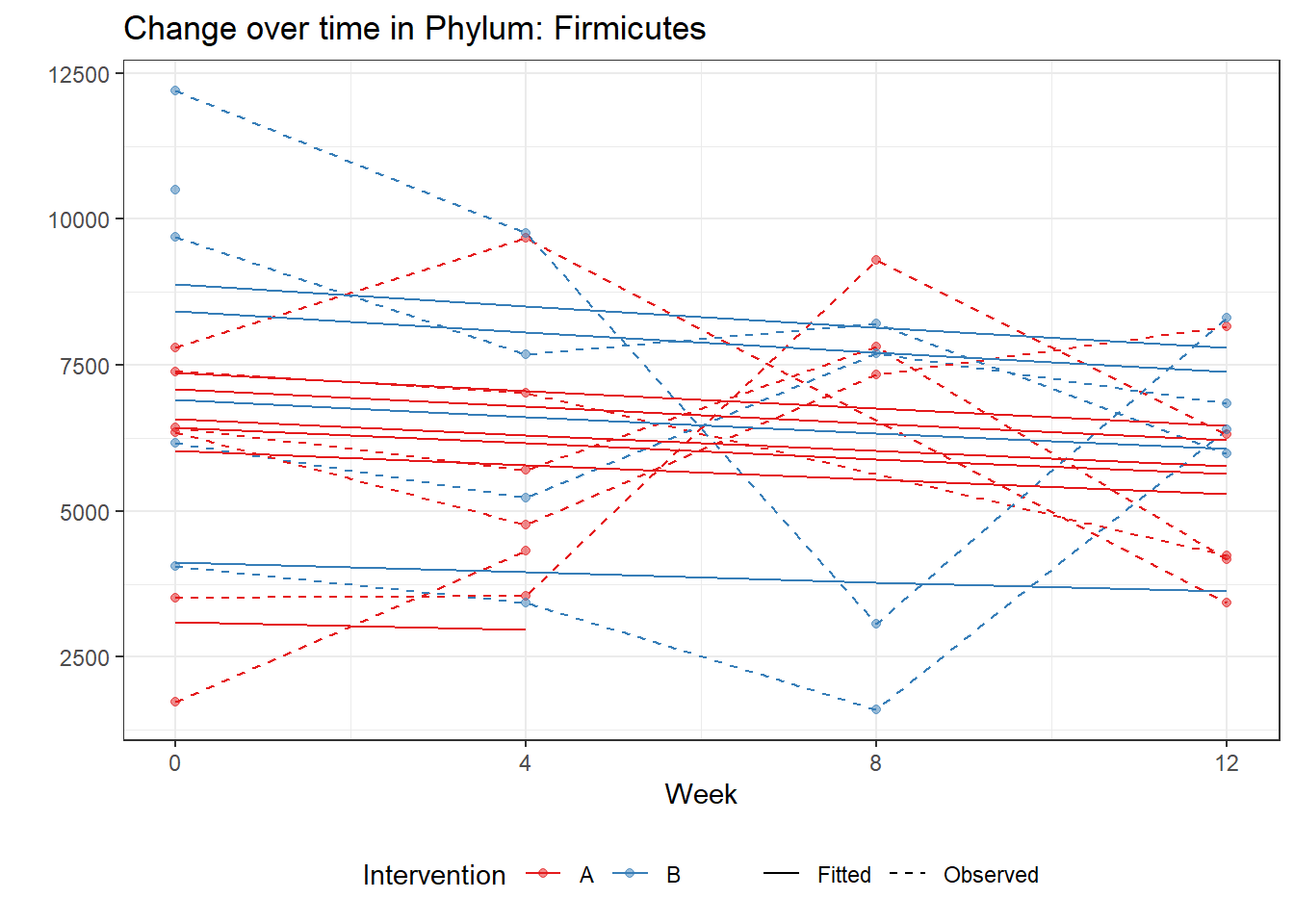
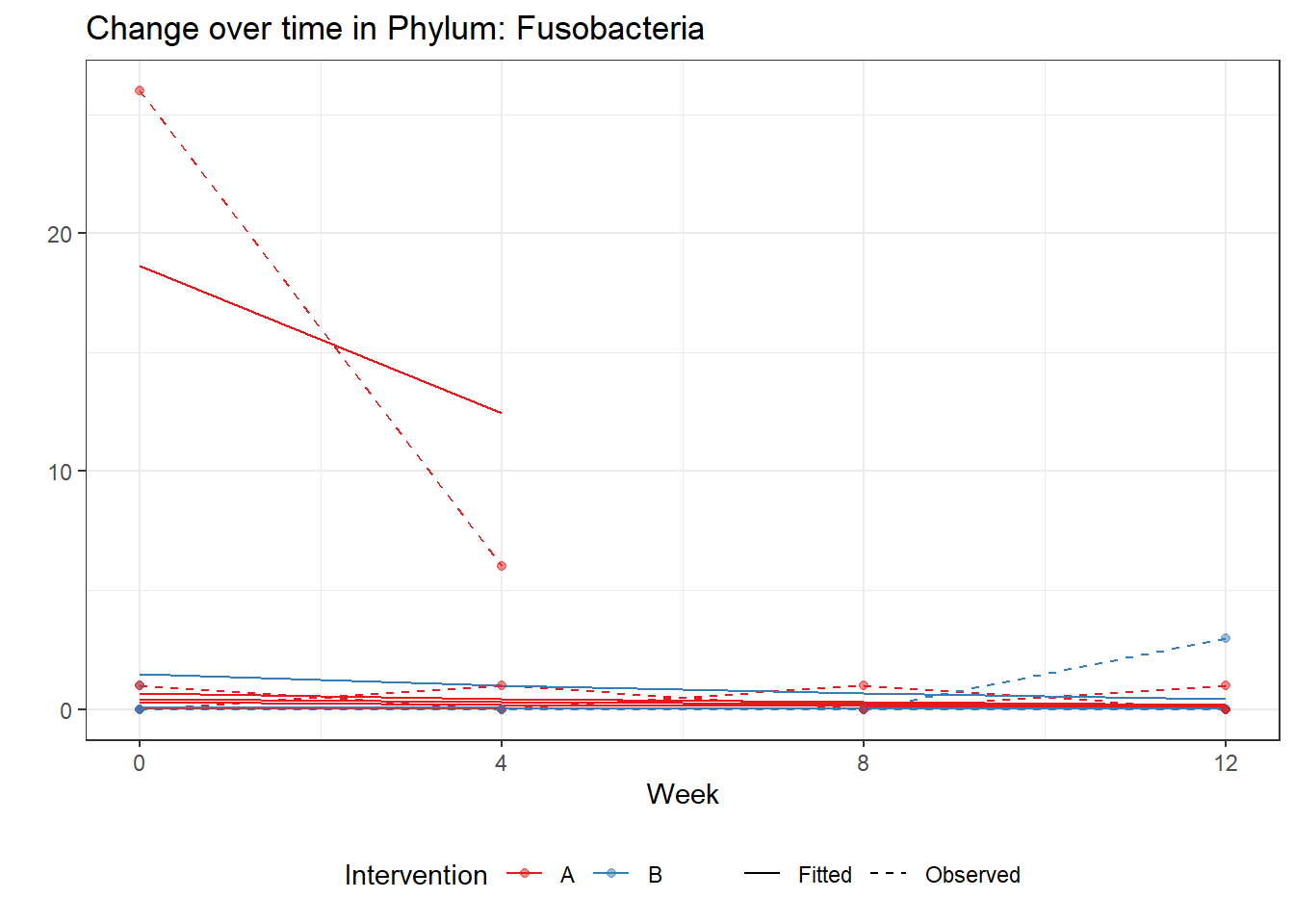
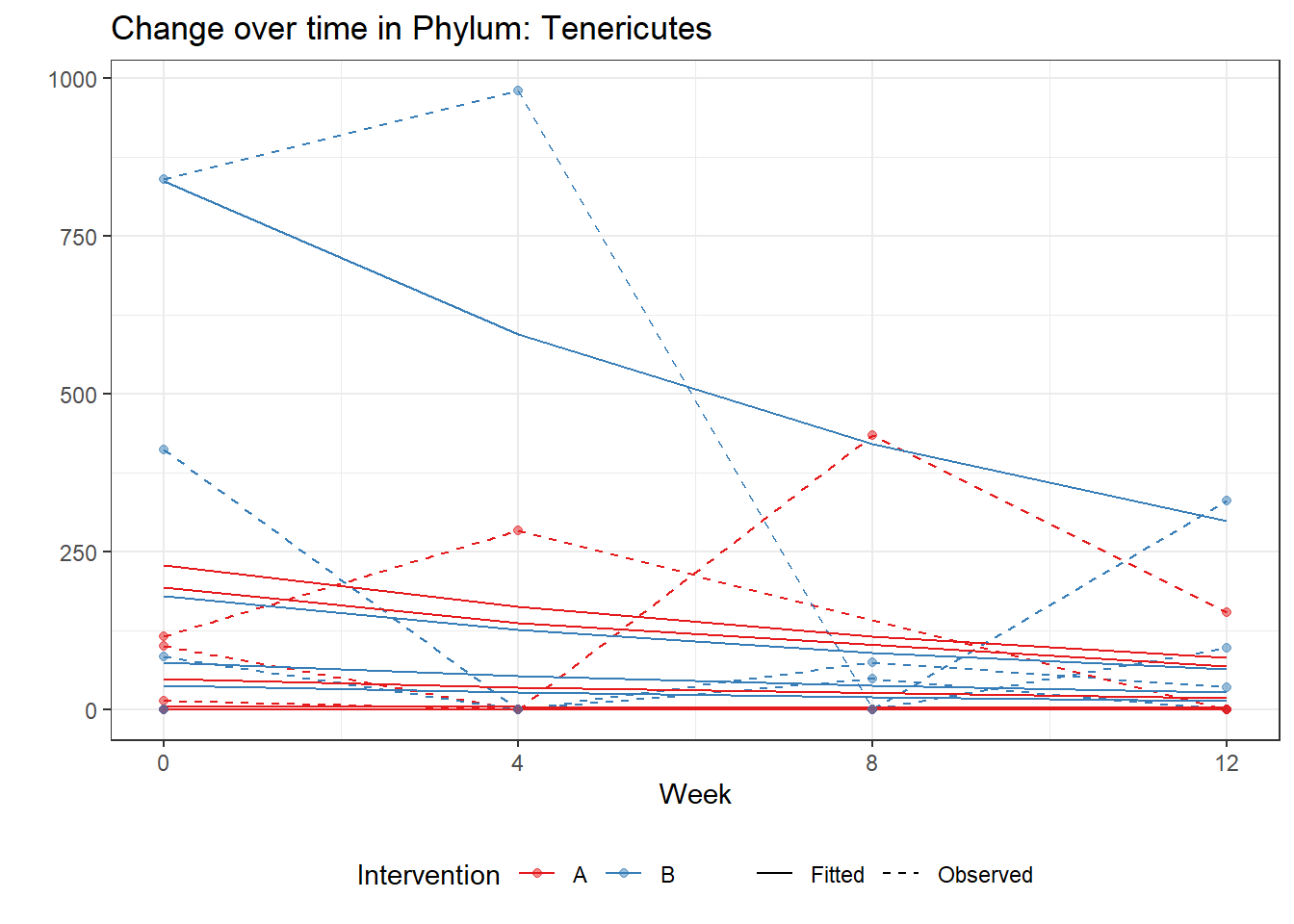
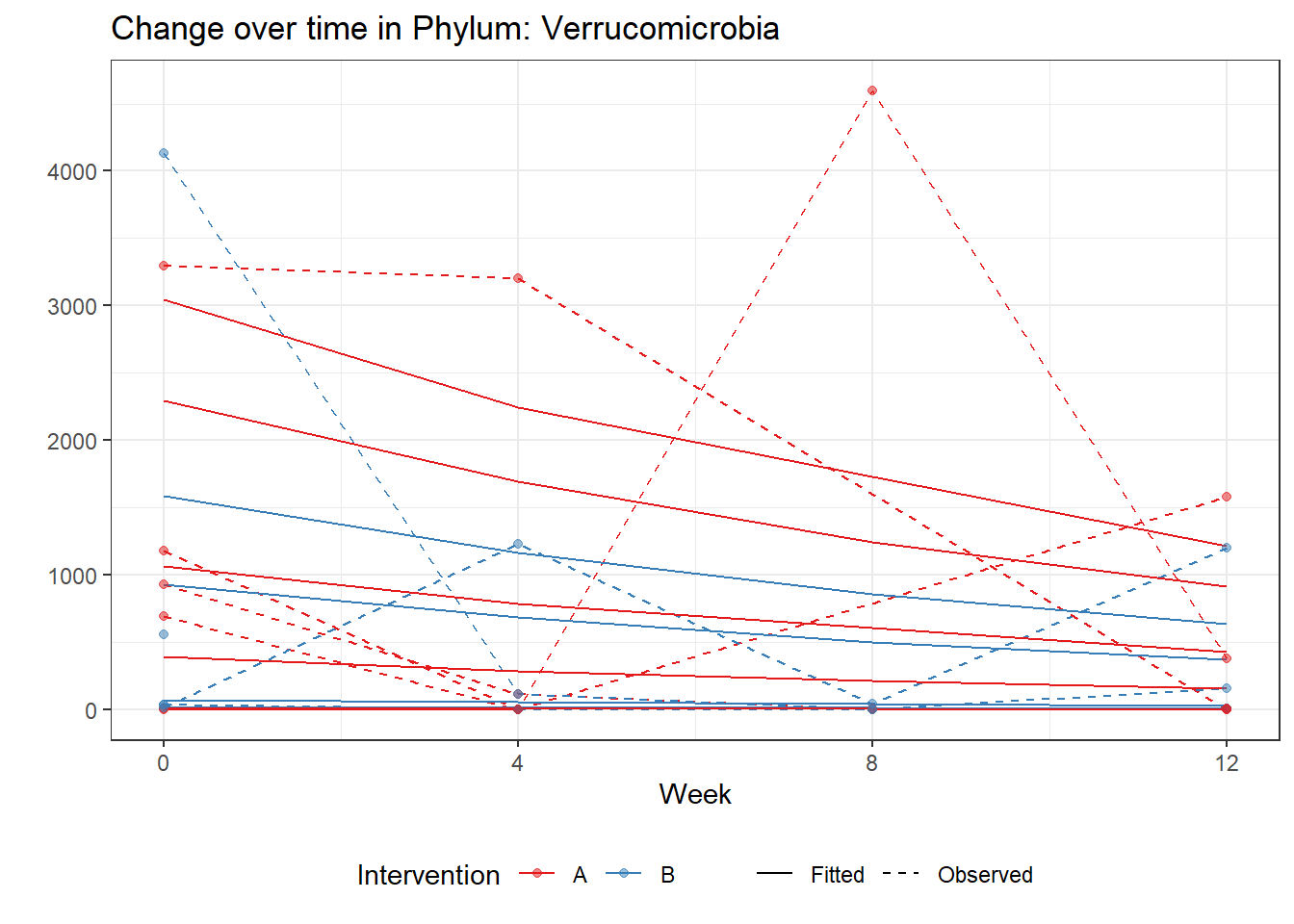
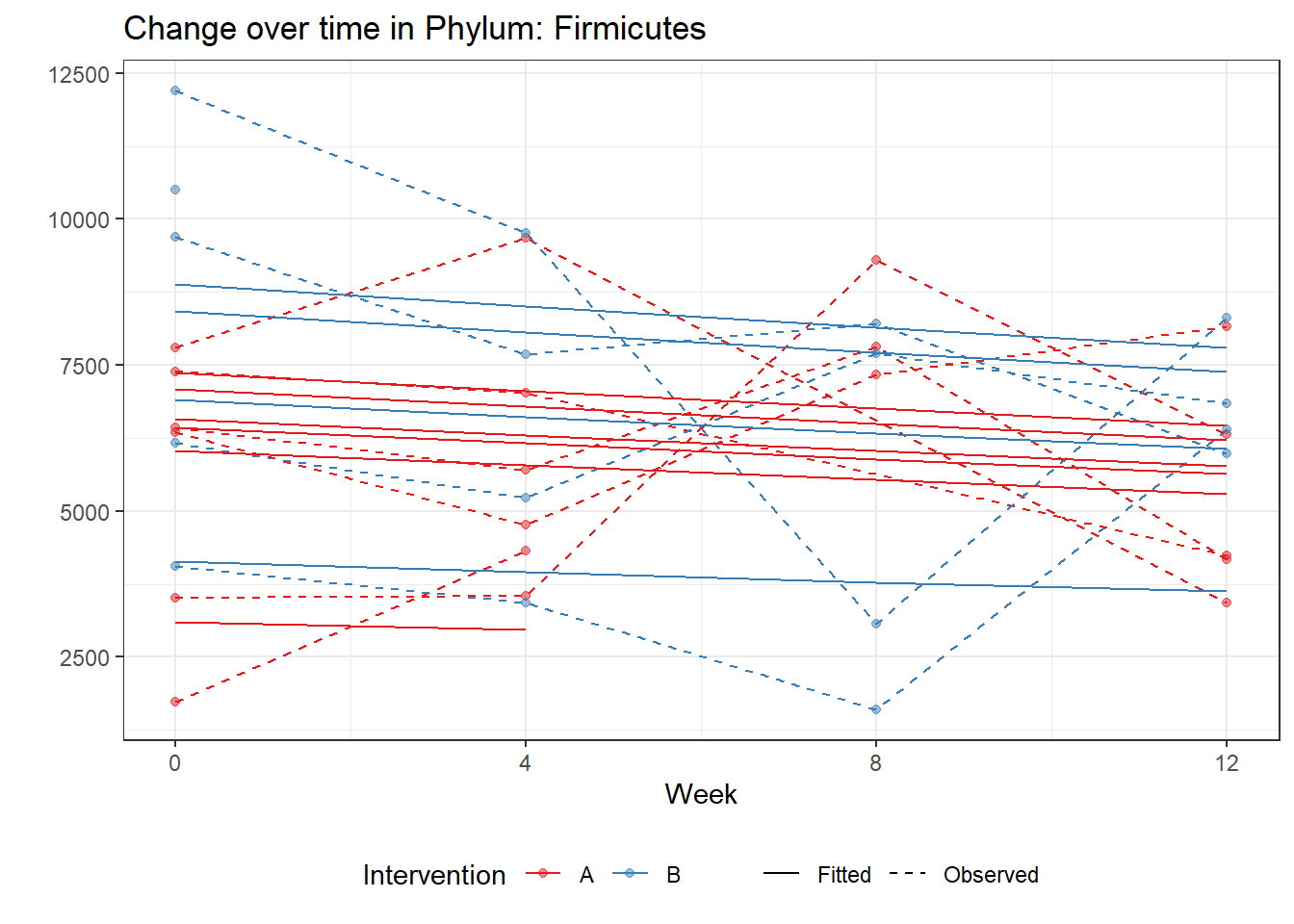
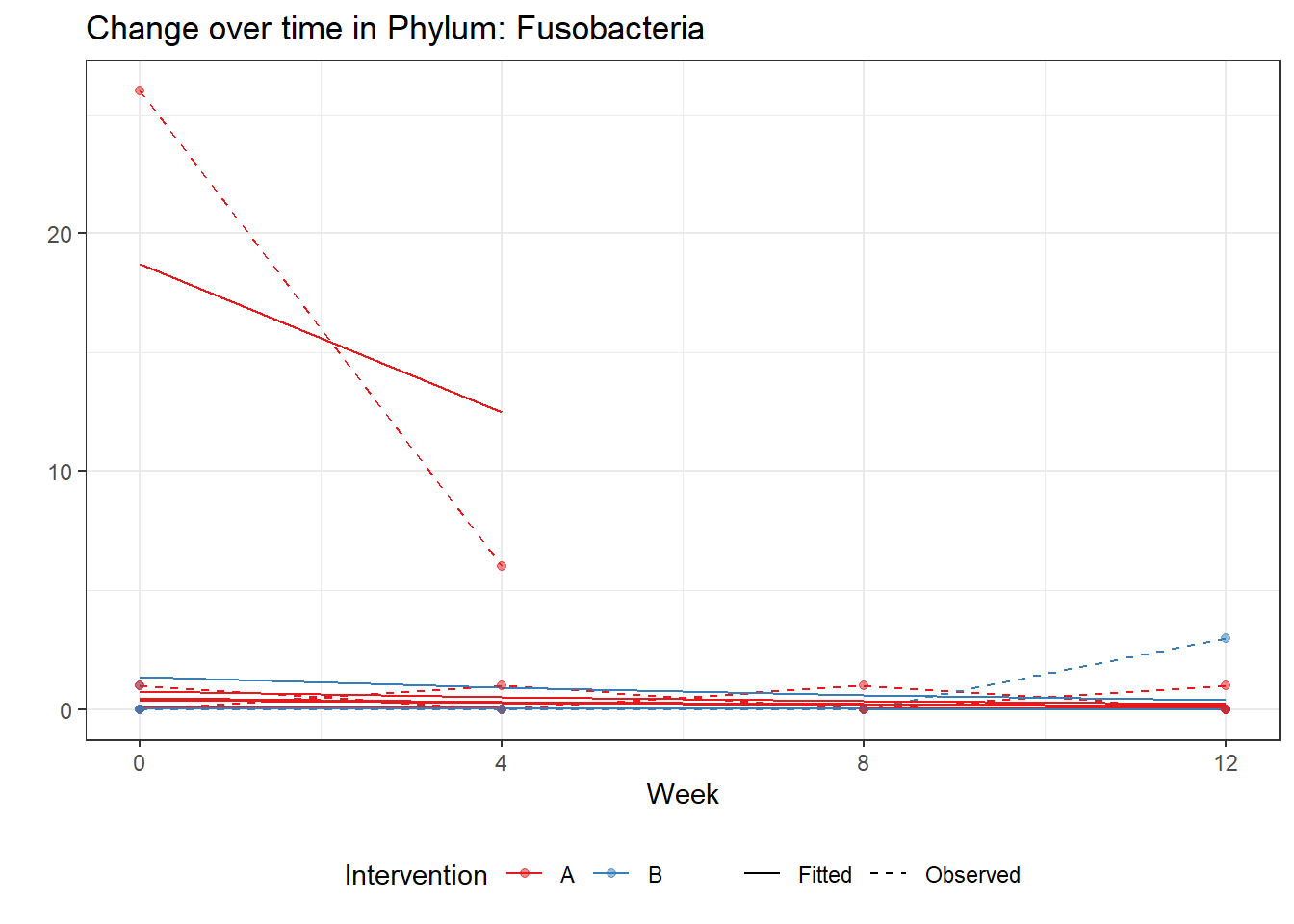
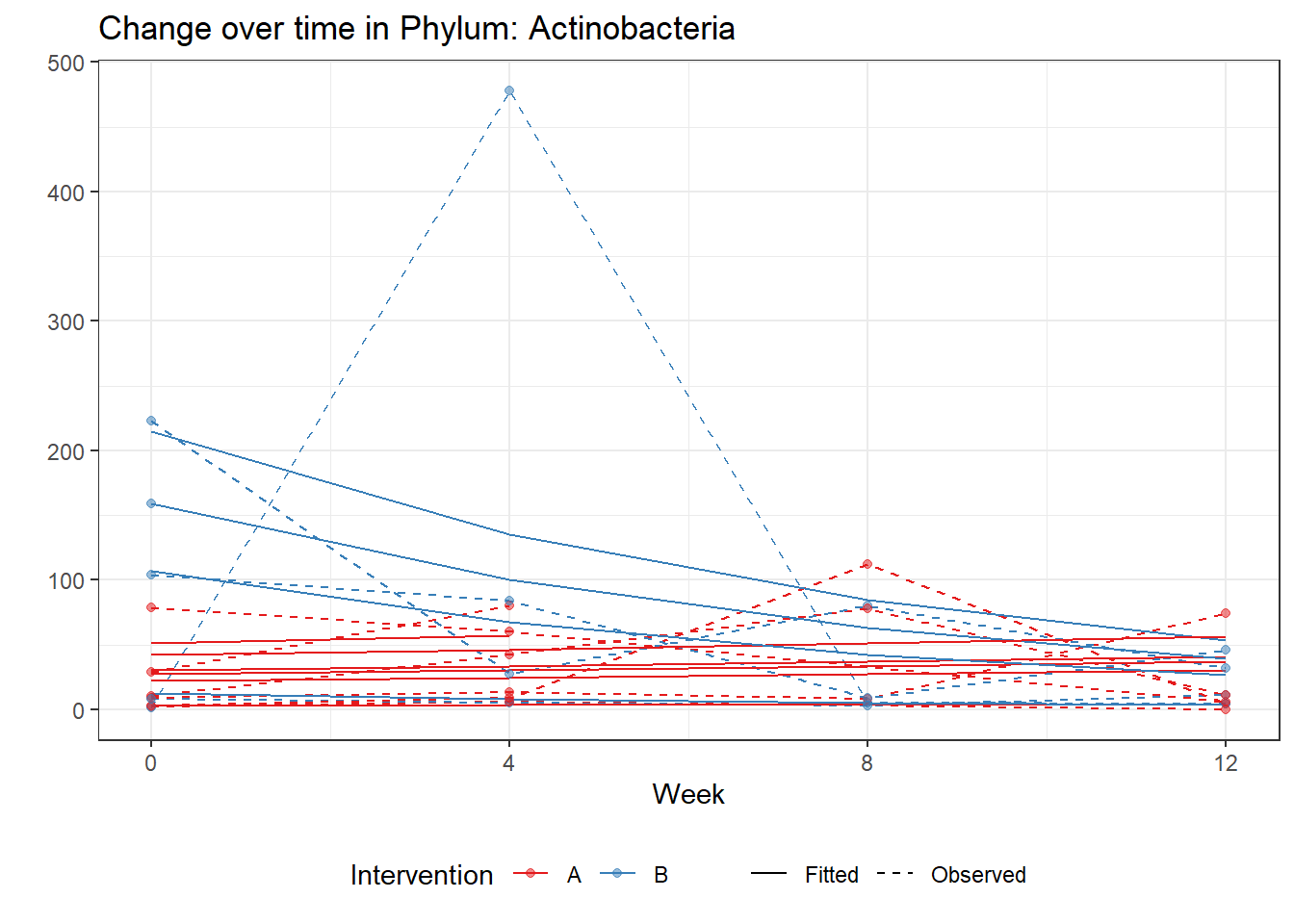
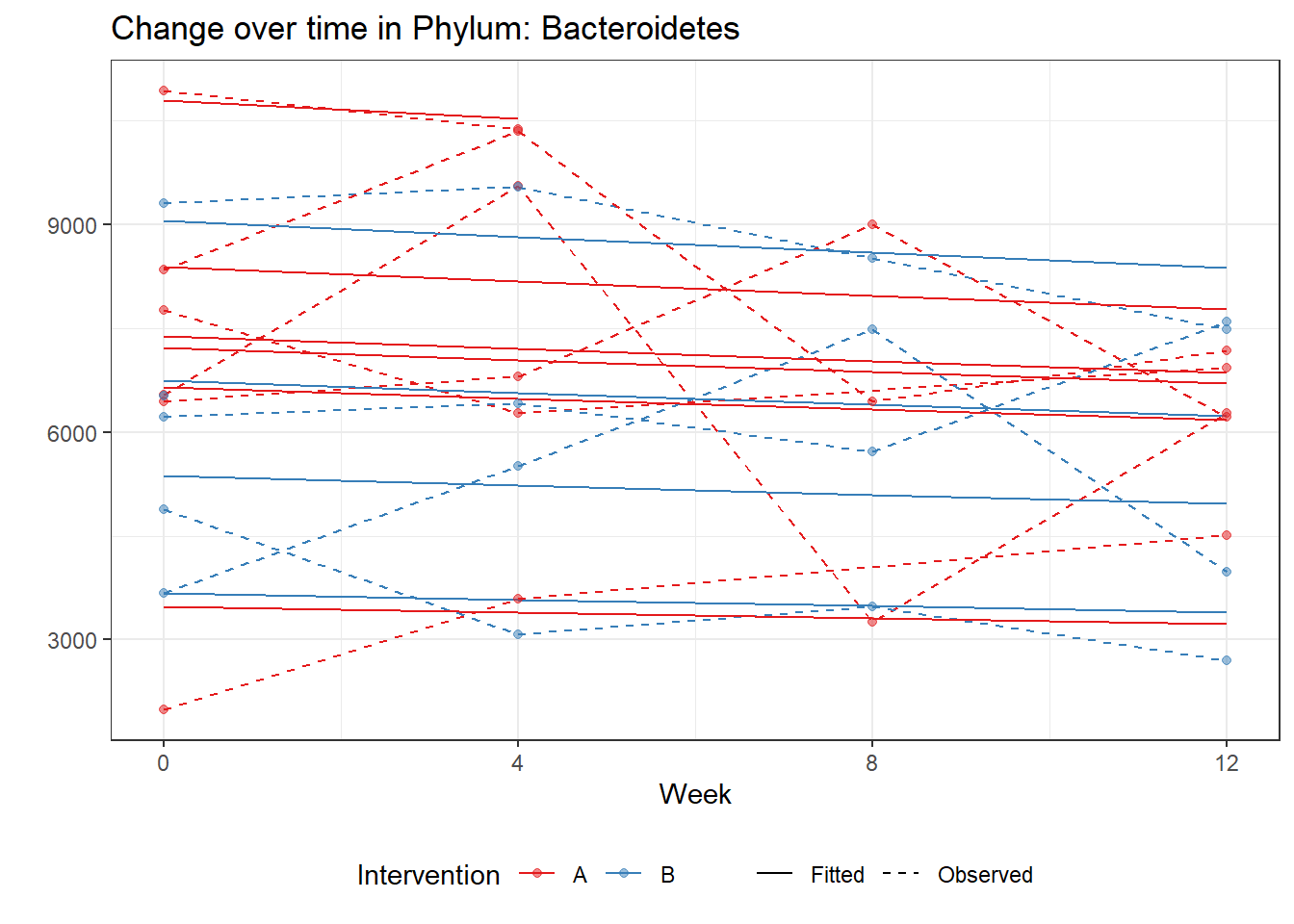
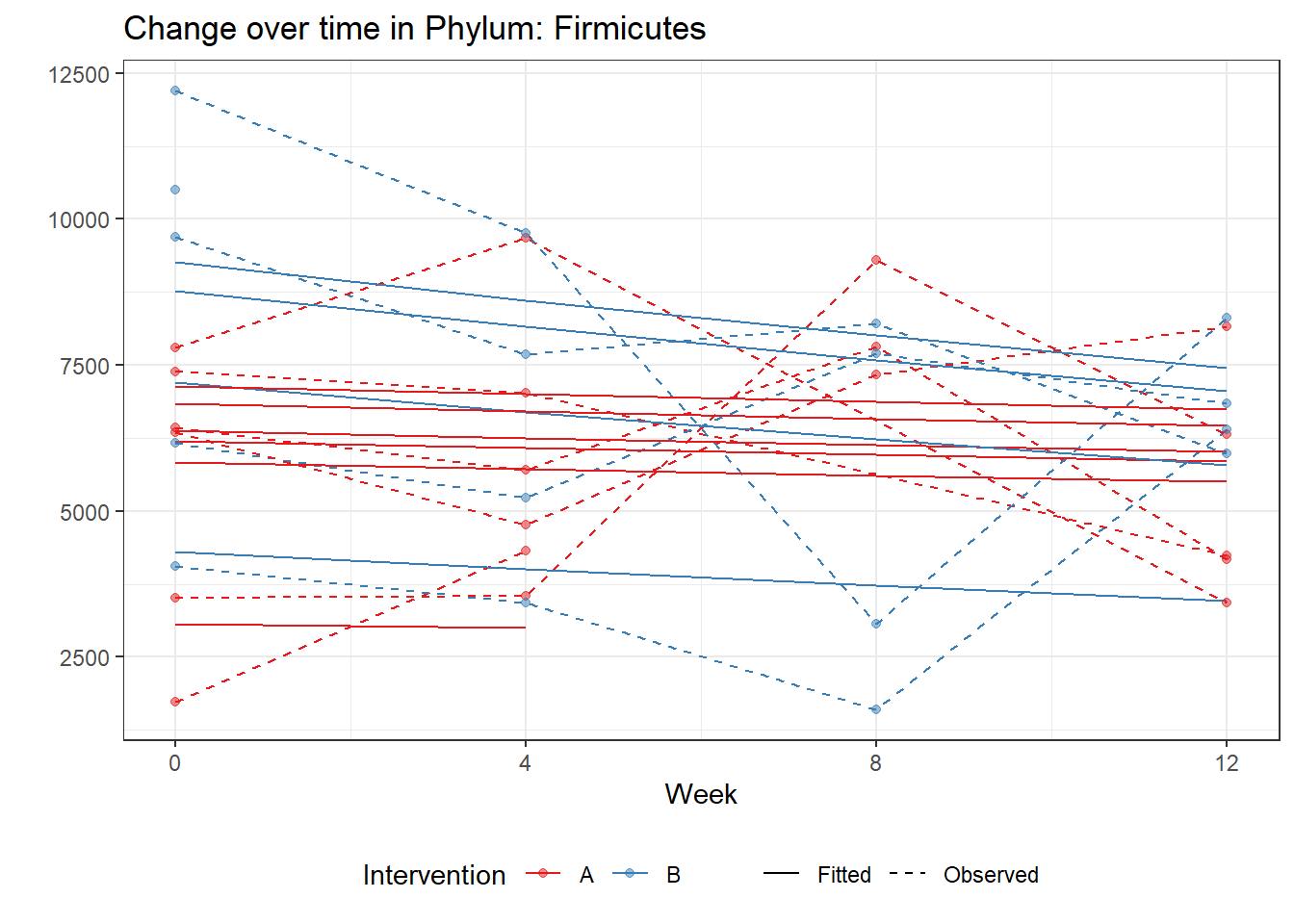
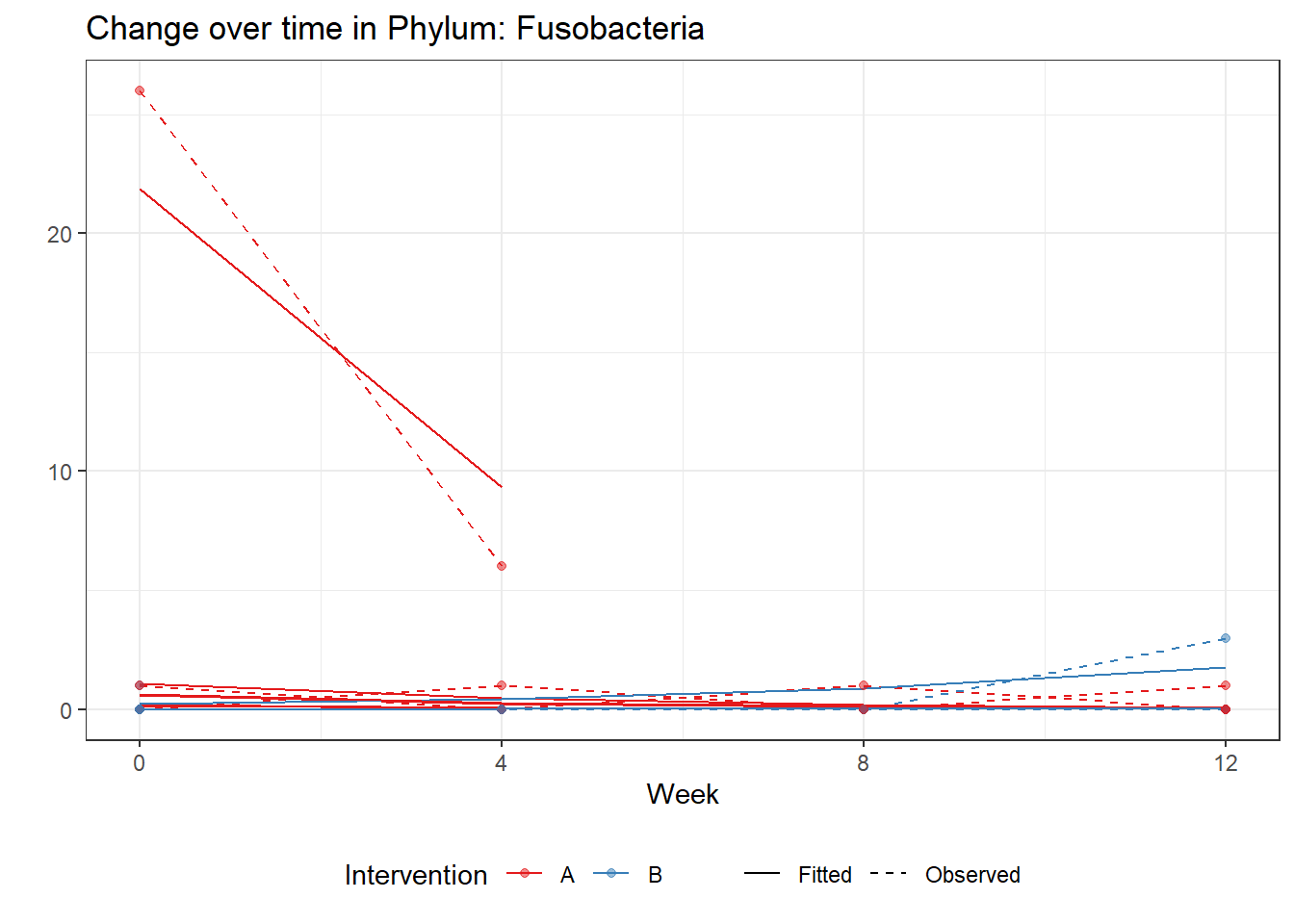
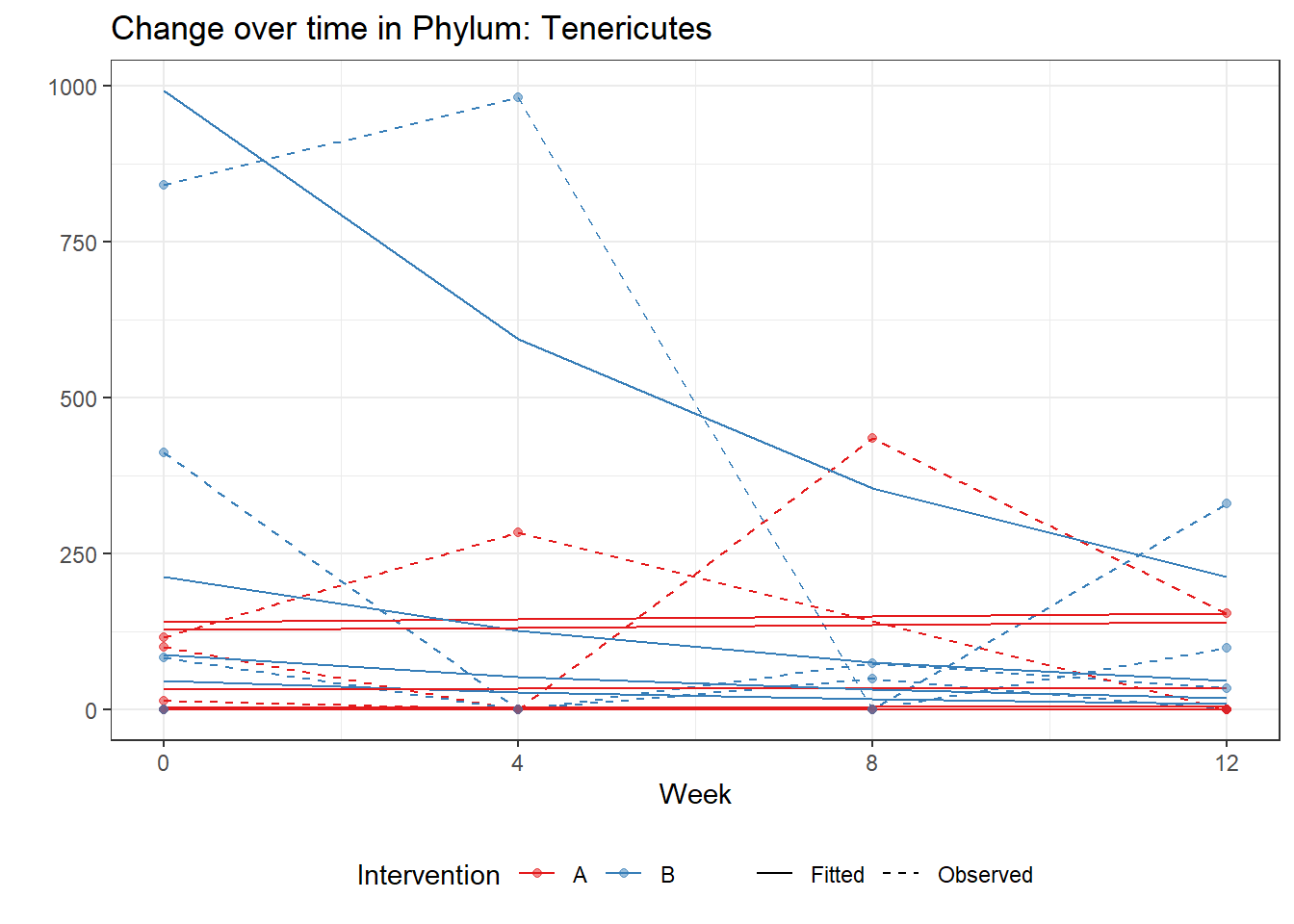
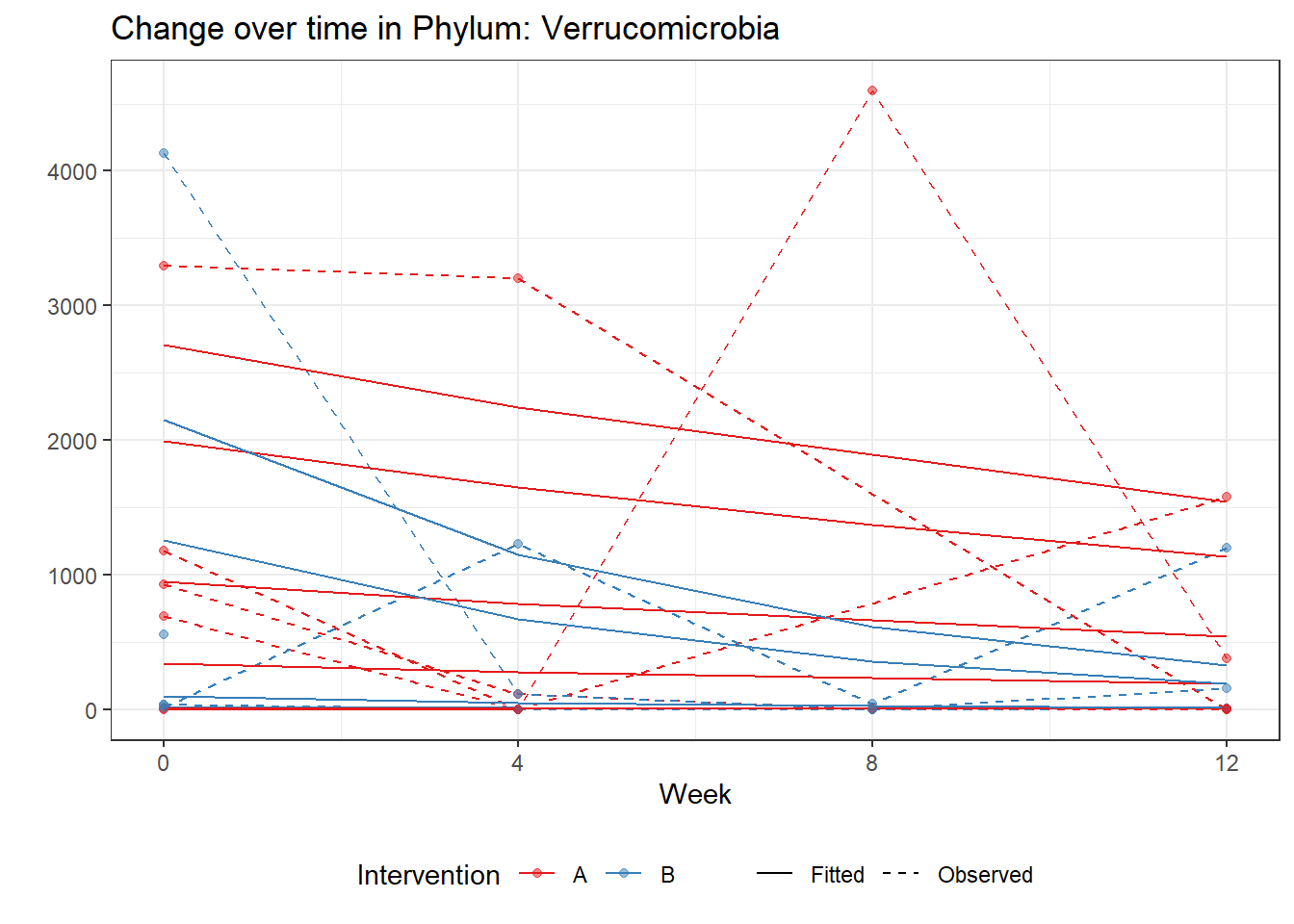
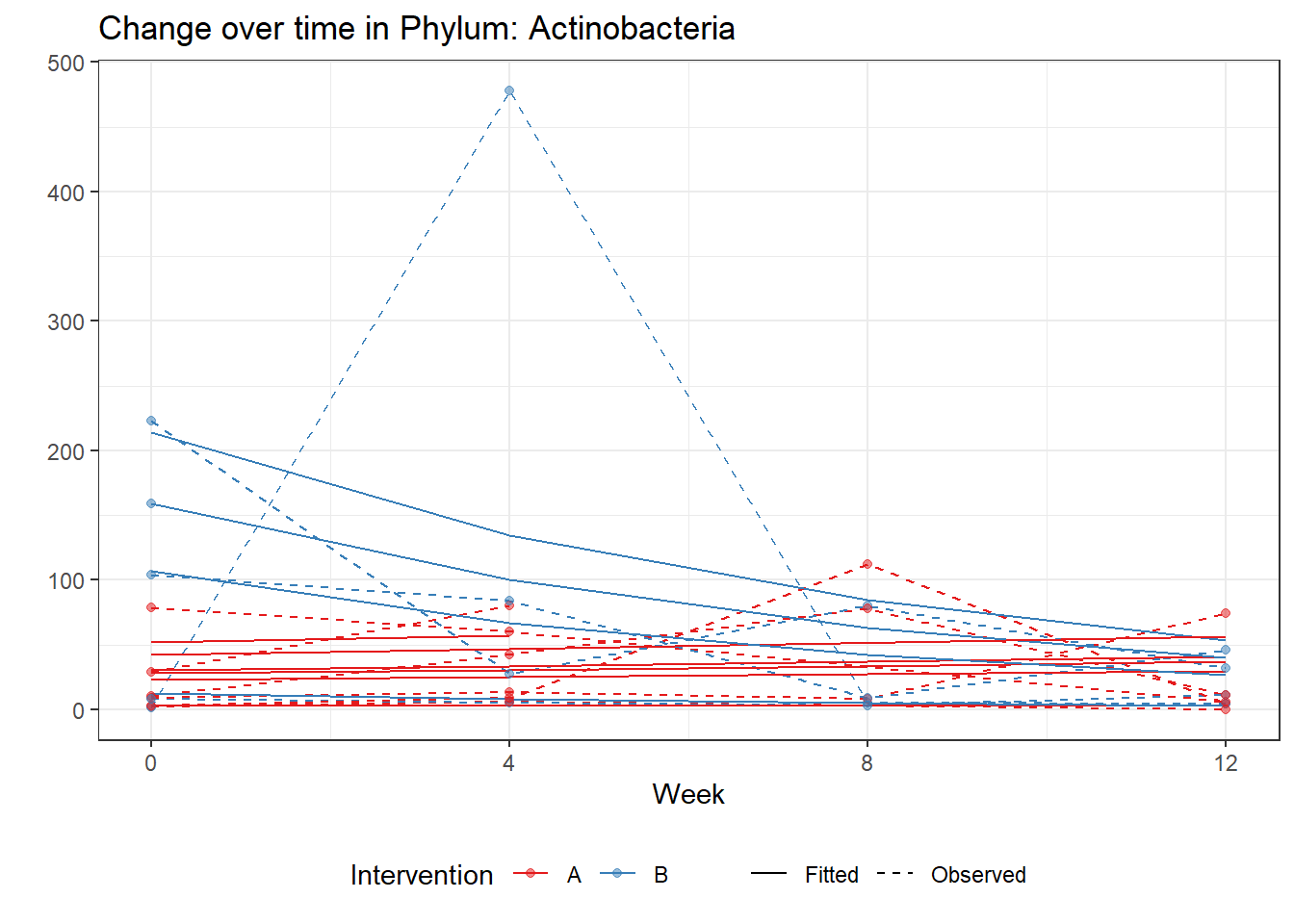
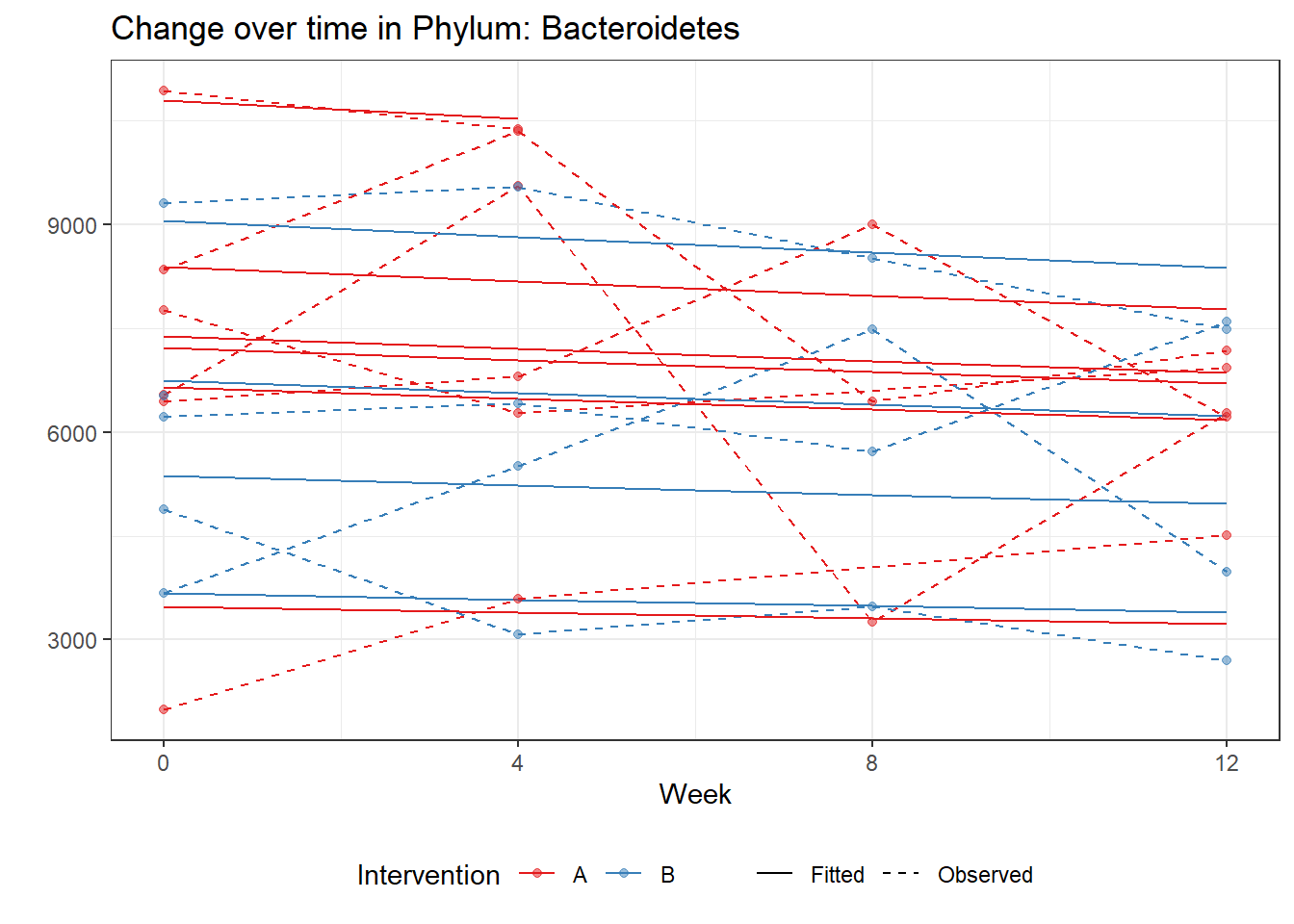
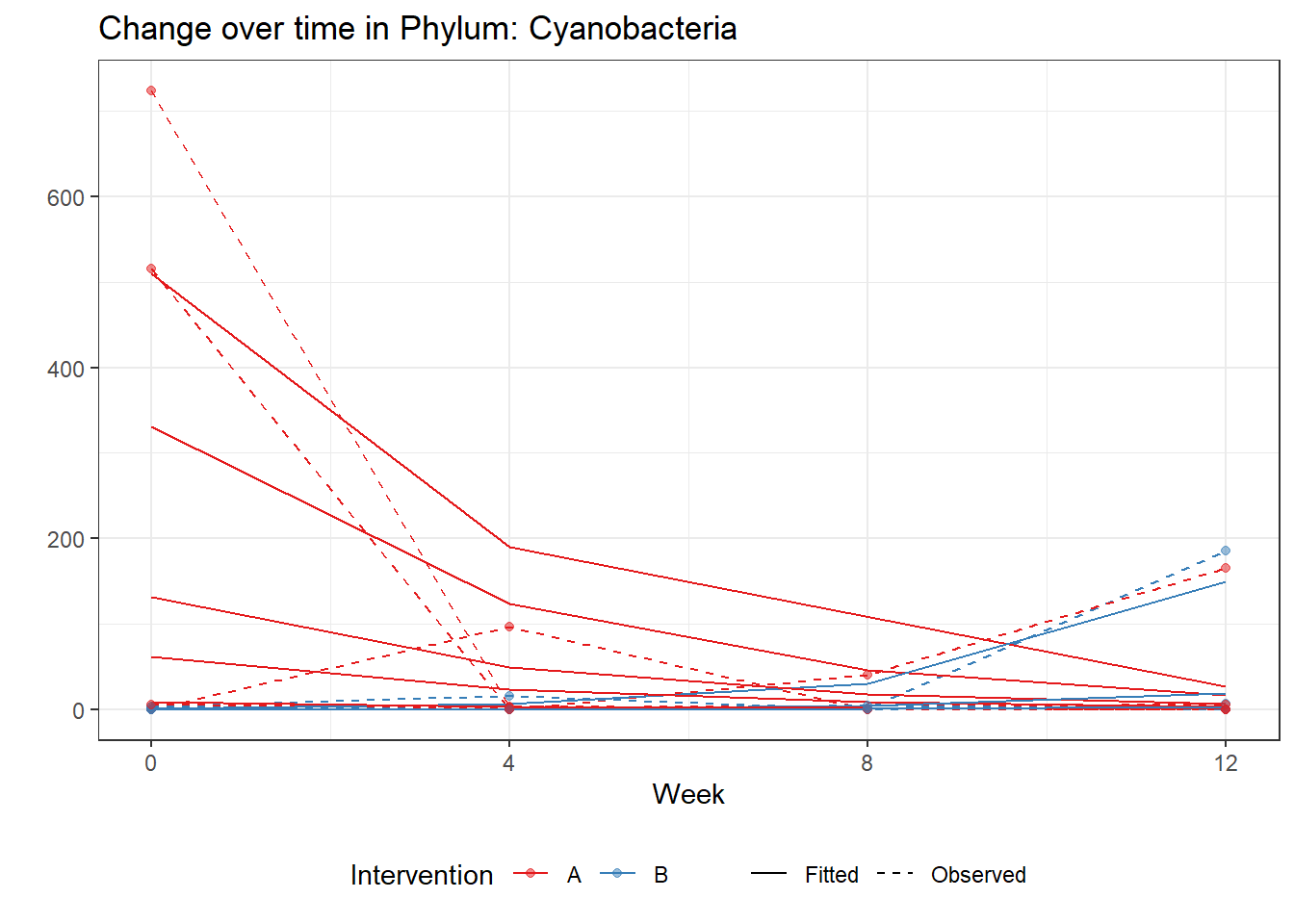
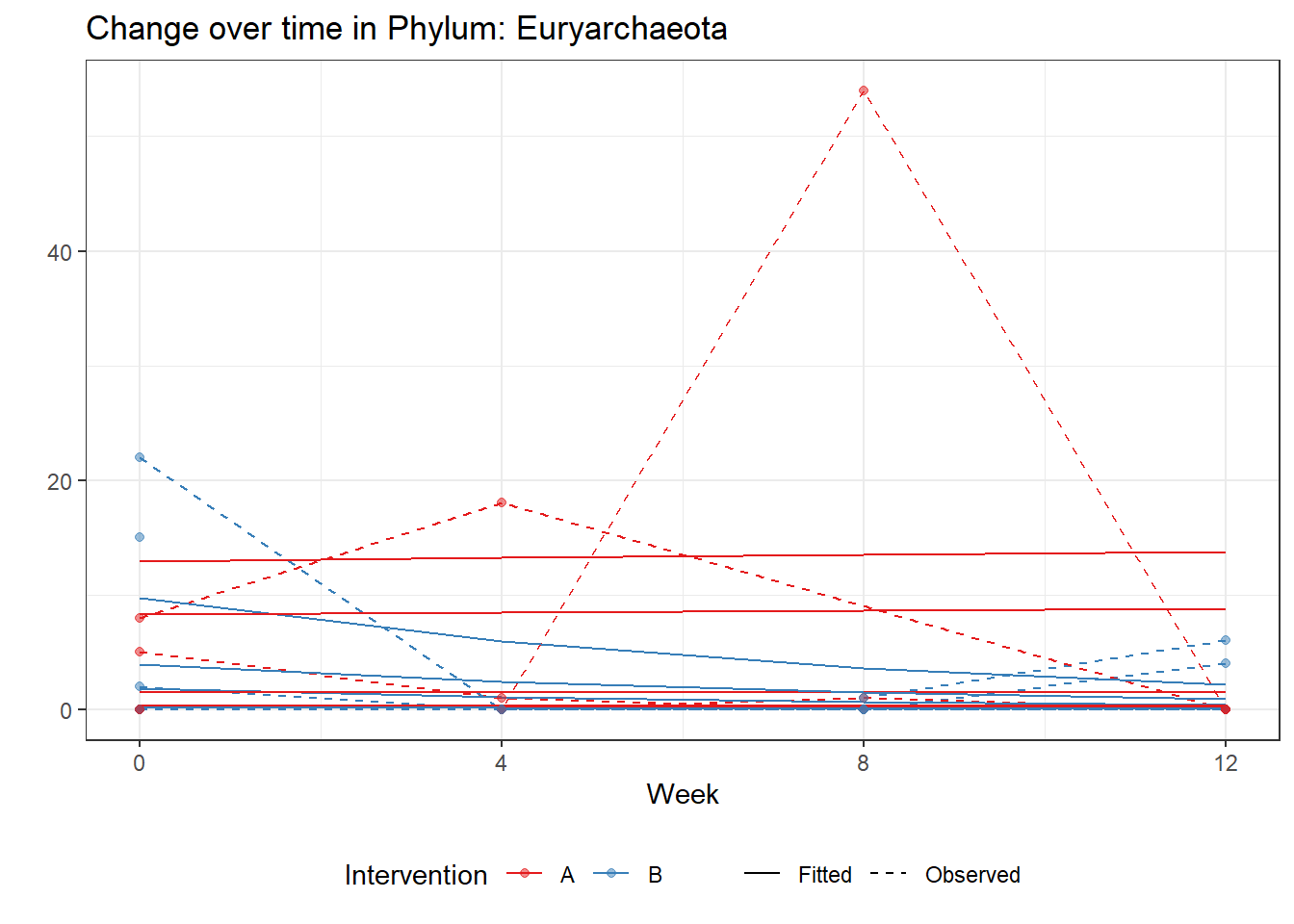
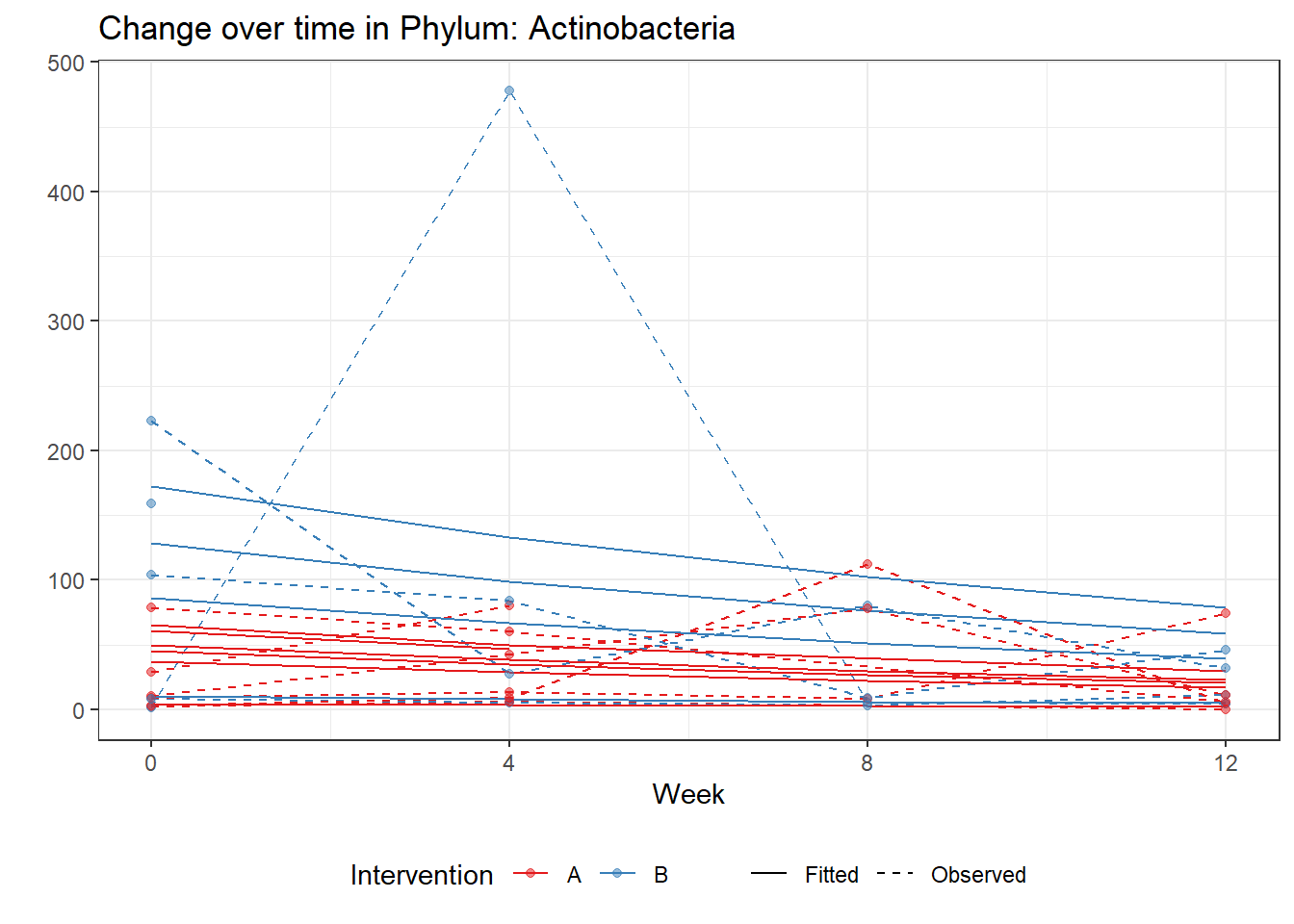
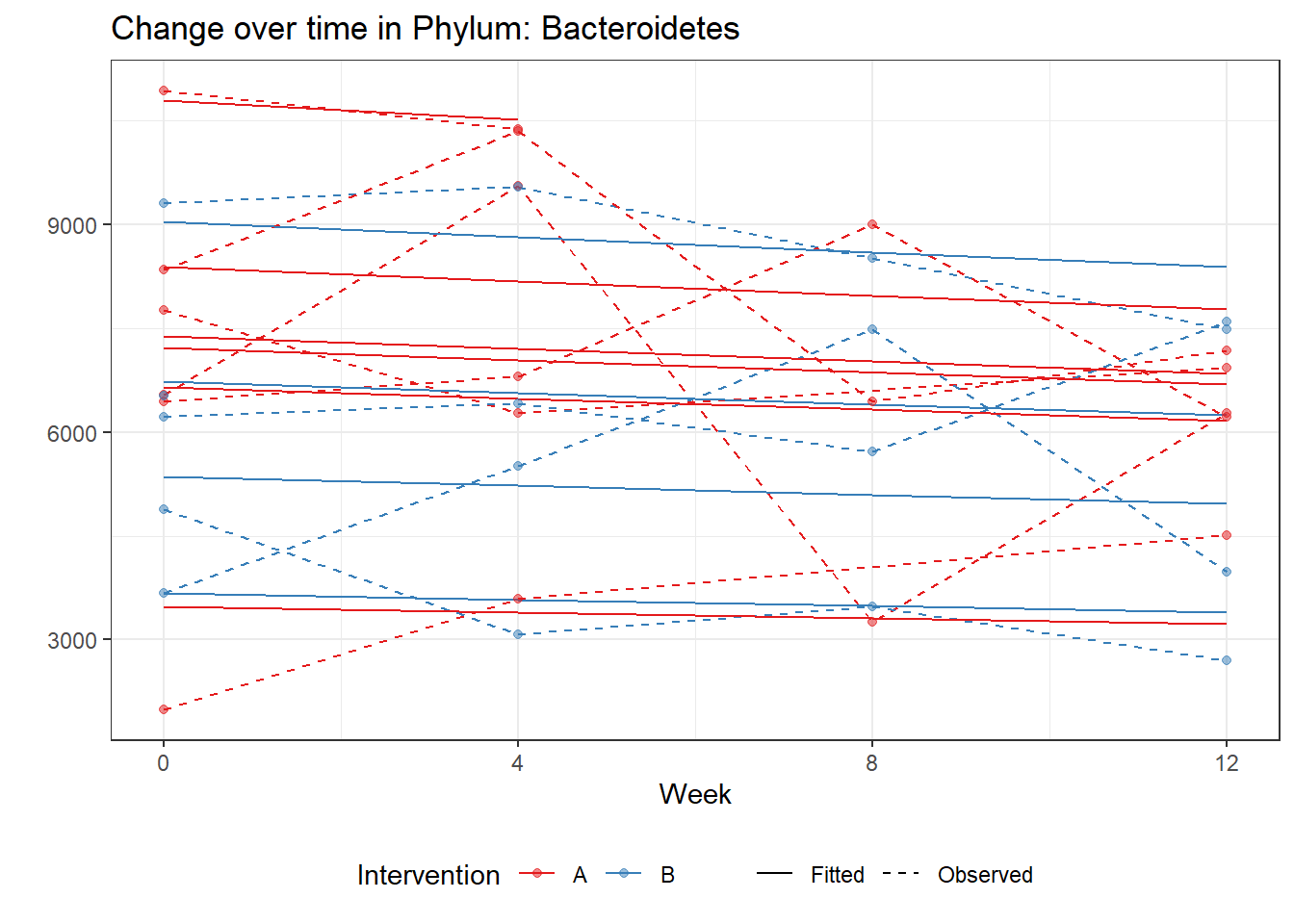
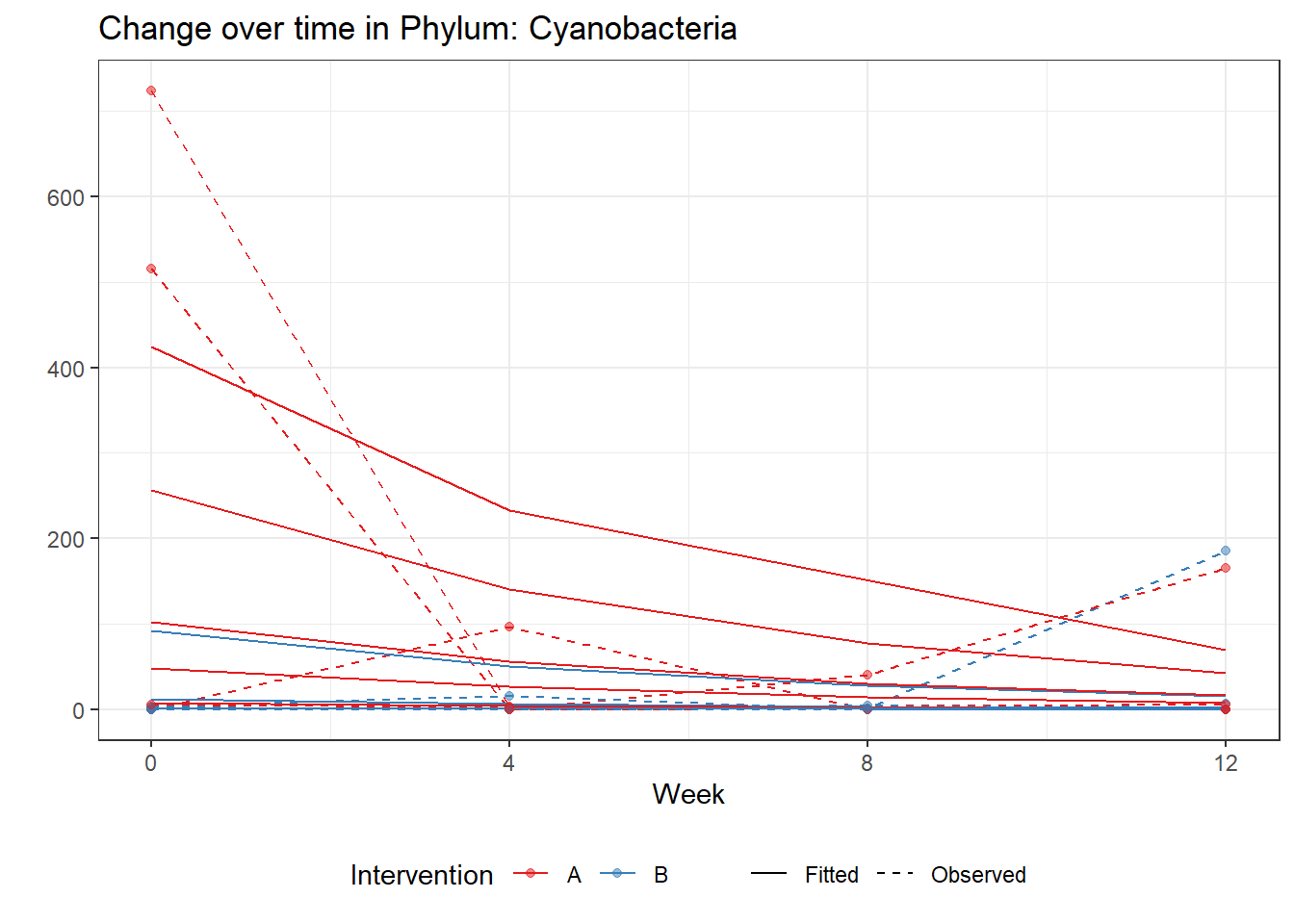
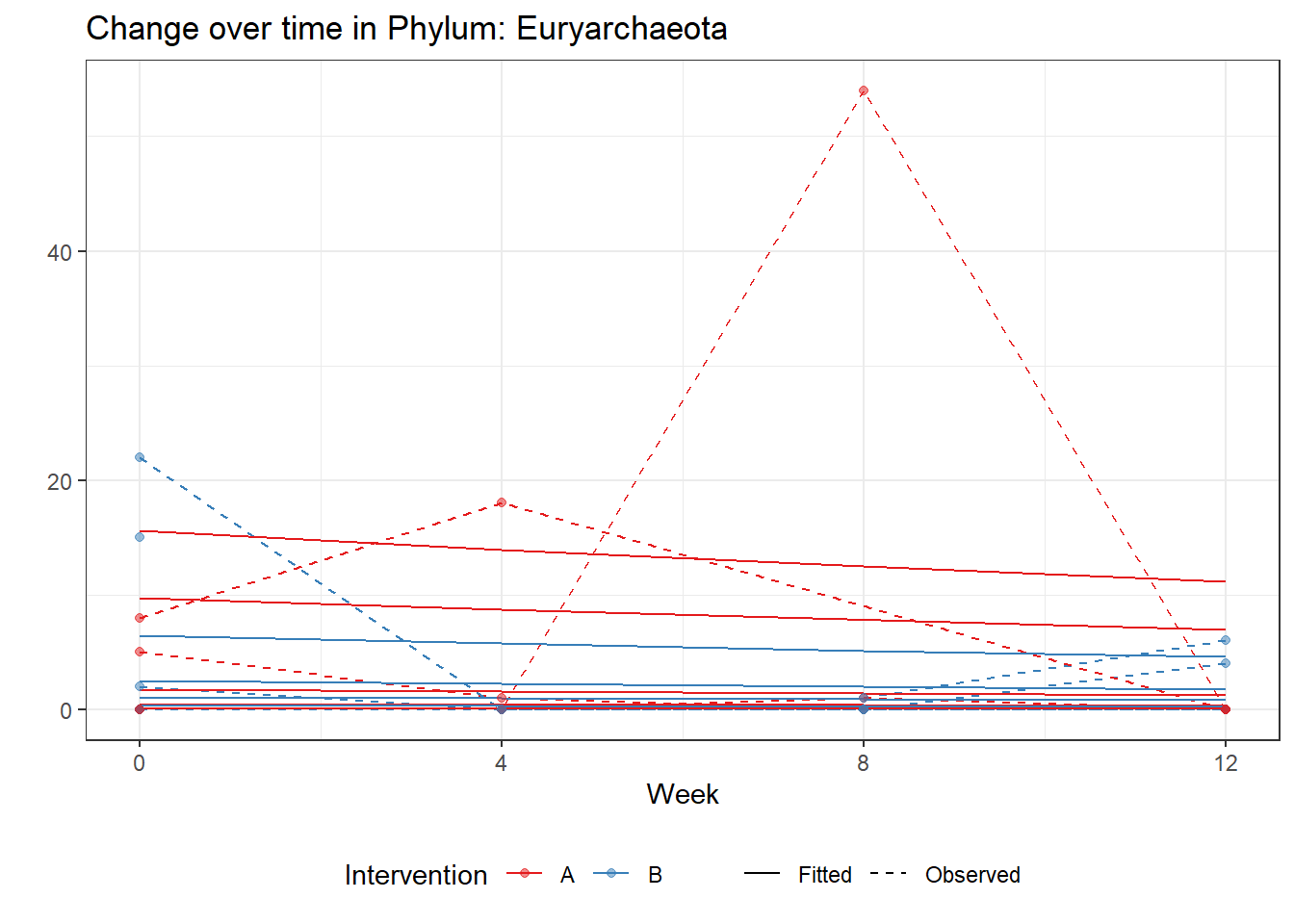
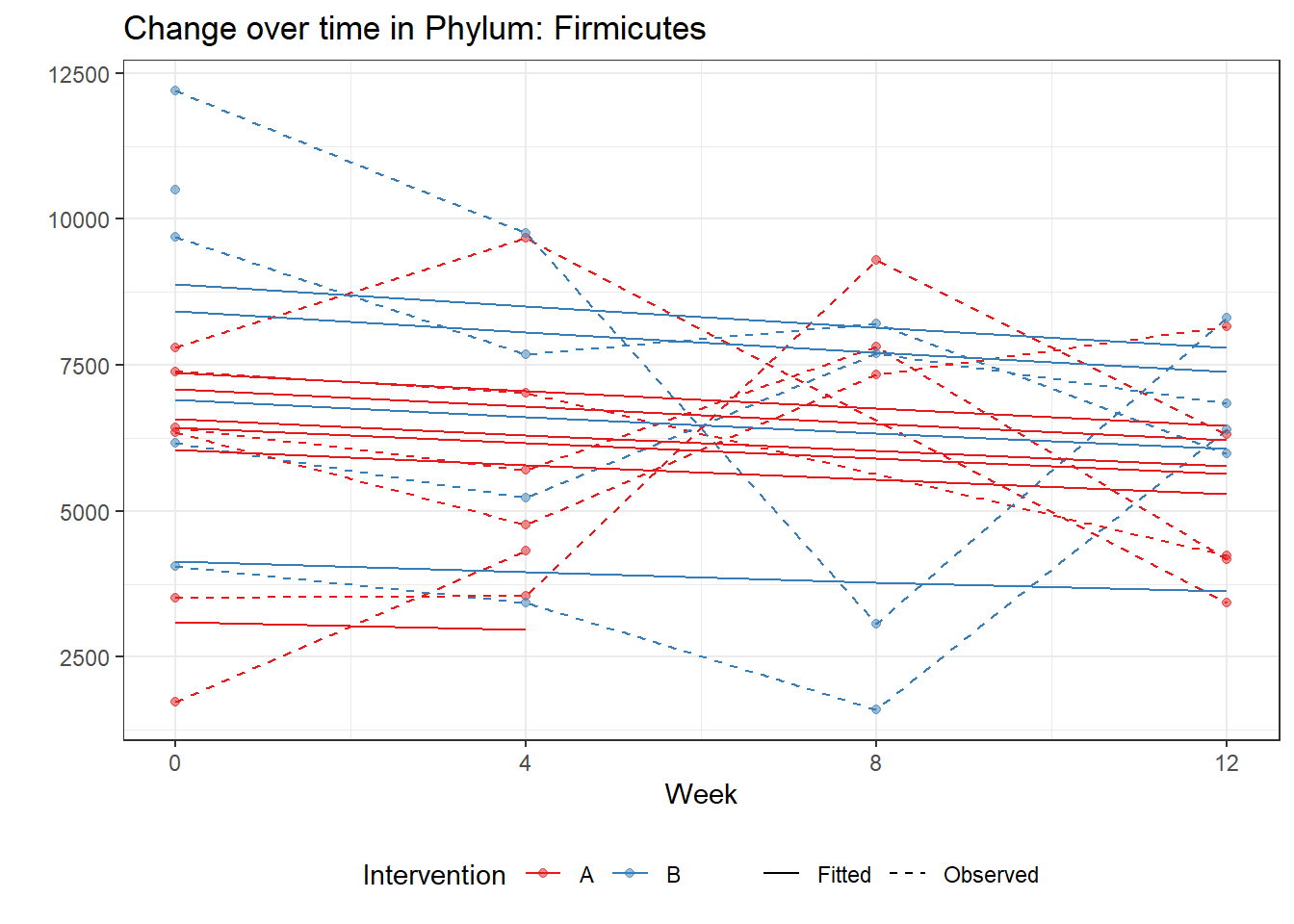
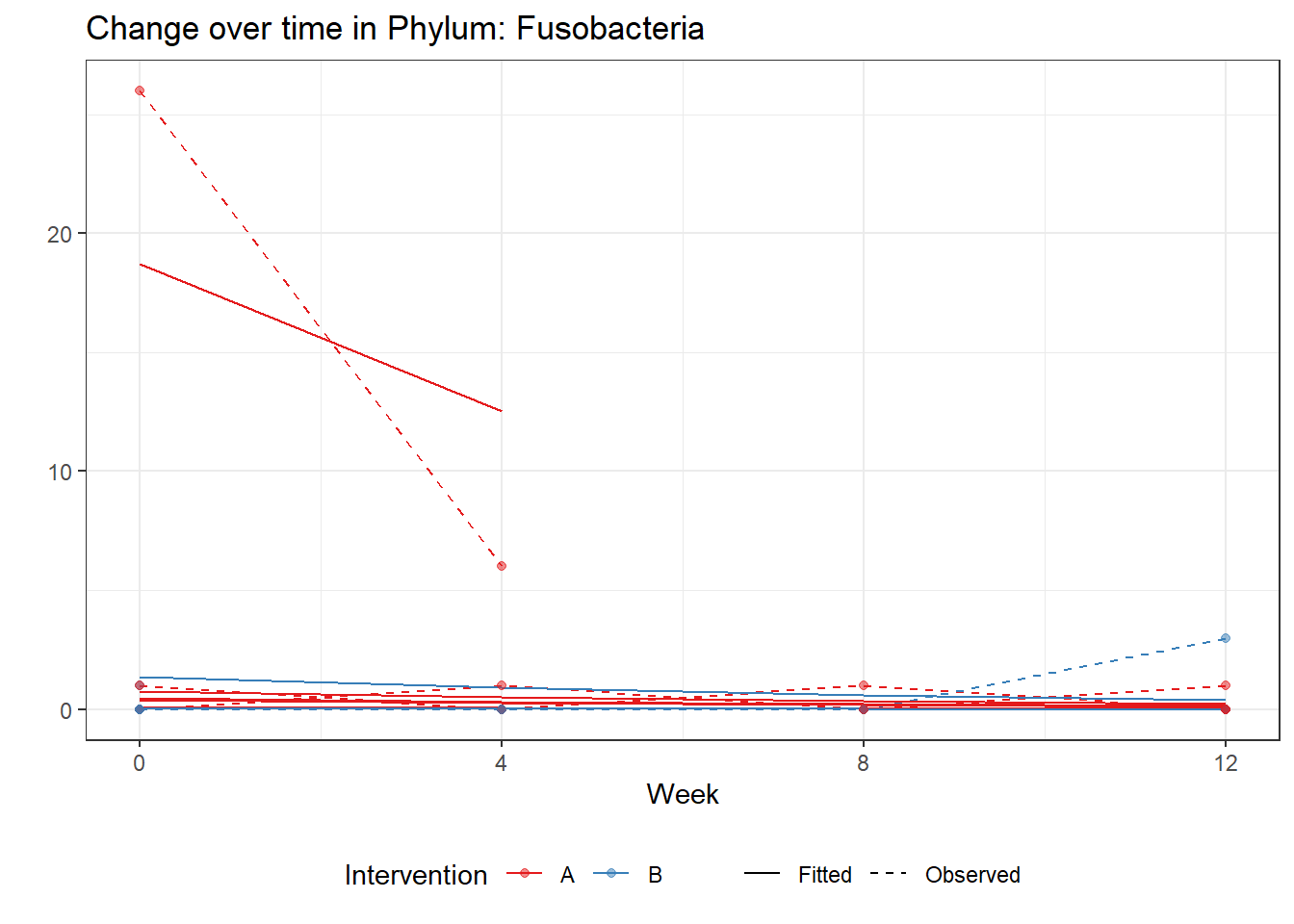
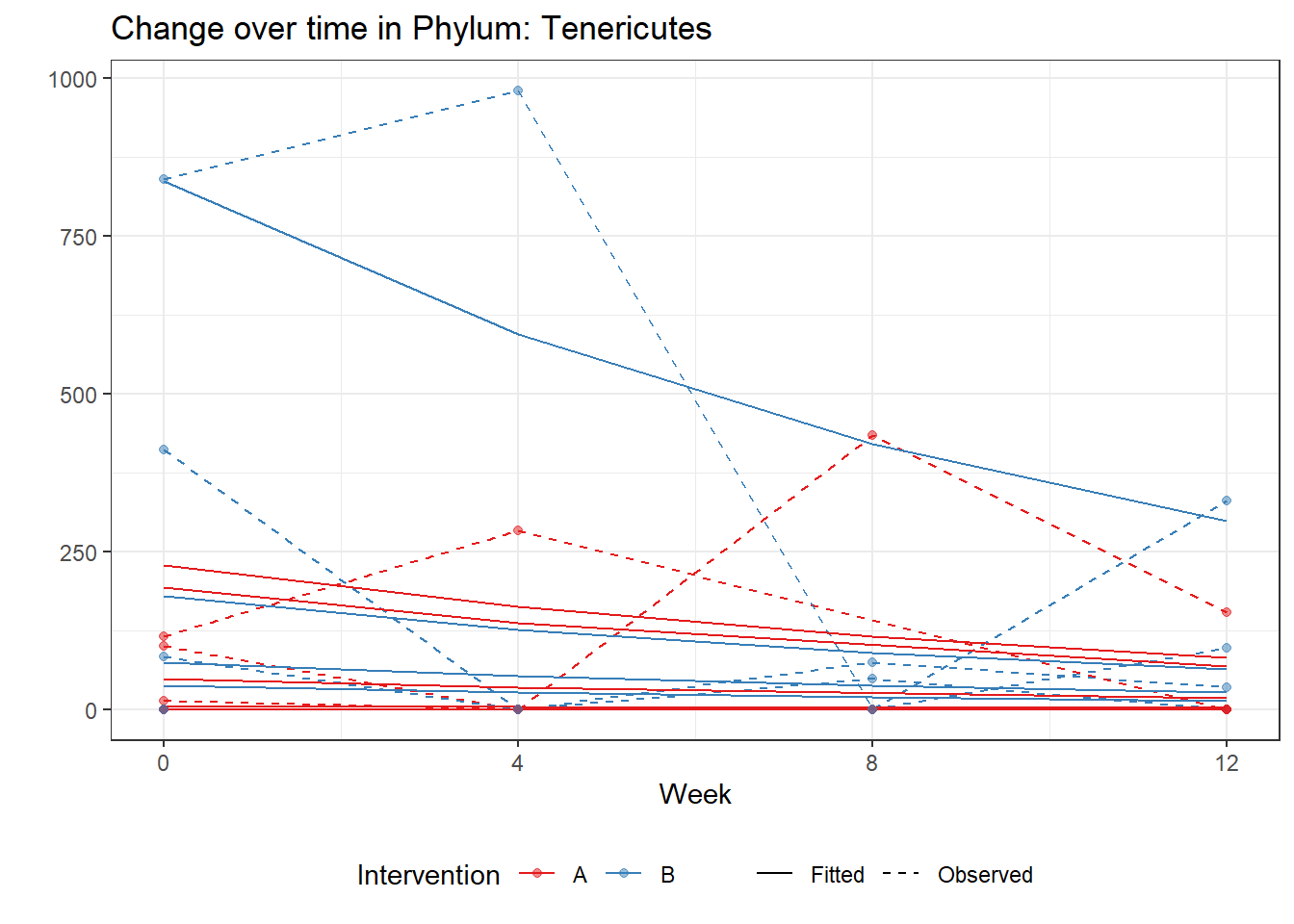
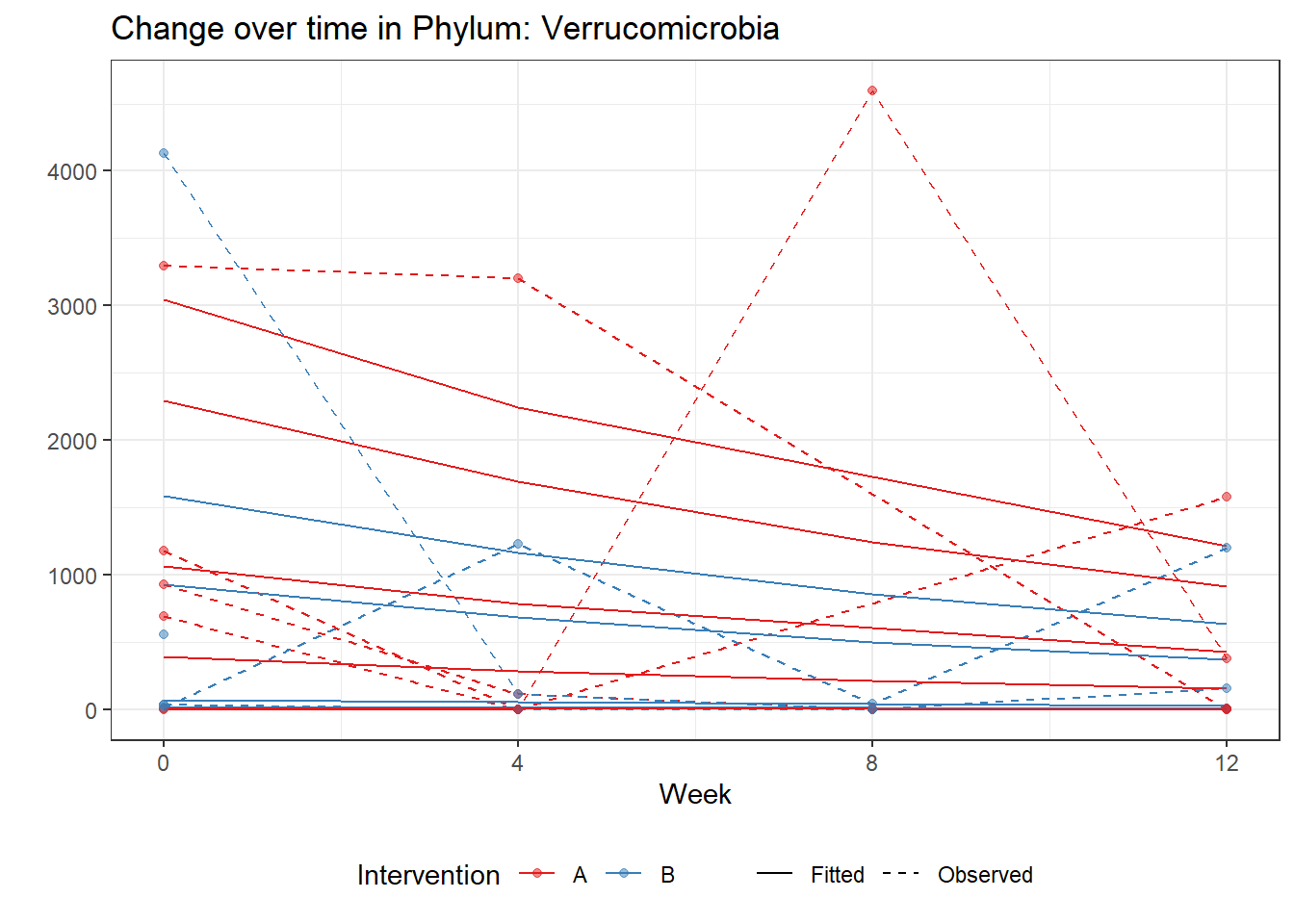
Significance test not availableMicrobiome Figure
mphyseq = psmelt(phylo_data)
mphyseq2 <- mphyseq %>%
dplyr::group_by(SubjectID, Week) %>%
dplyr::mutate(Total = sum(Abundance)) %>%
dplyr::ungroup()%>%
dplyr::group_by(SubjectID, Week, Phylum) %>%
dplyr::mutate(PhylumAbund = sum(Abundance),
RelAbund = PhylumAbund/Total)
mphyseq2 <- mphyseq2 %>% distinct(SubjectID, Week, Phylum, .keep_all = T)
keepVar <- c("SubjectID", "Week", "Intervention", "Phylum", "PhylumAbund", "RelAbund")
mphyseq2 <- mphyseq2[, keepVar]
# take out "__" at start of names
mphyseq2$Phylum <- substring(mphyseq2$Phylum, 3)
# get right number of colors for plotting
no_cols <- length(unique(mphyseq2$Phylum))
## Some Colors
colors_micro <- rev(c("grey90", rev(c("#00a2f2", "#c91acb", "#7f5940", "#cc5200", "#00d957", "#40202d", "#e60099", "#006fa6", "#f29d3d", "#300059"))))
# Now get the intervention by time model
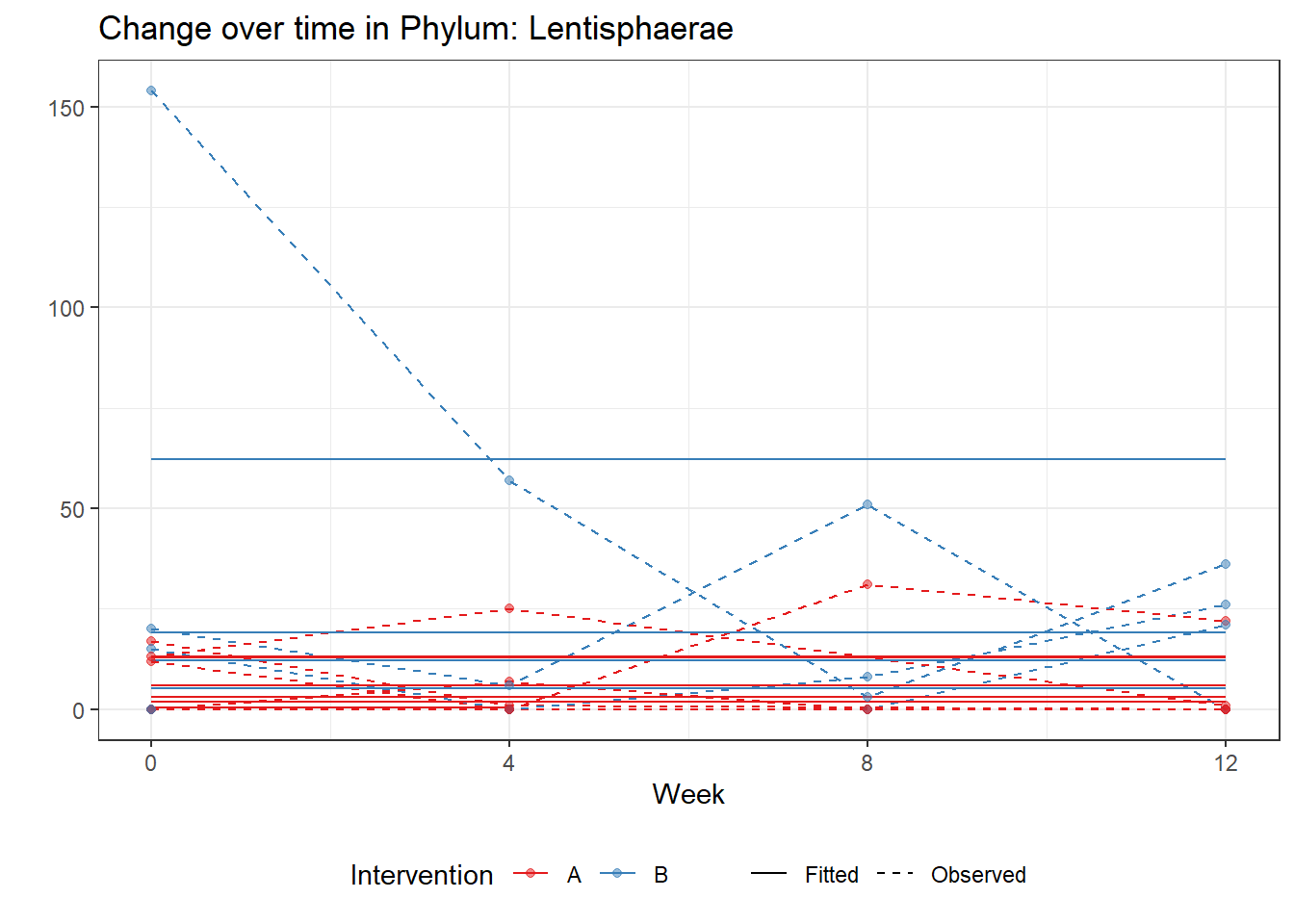
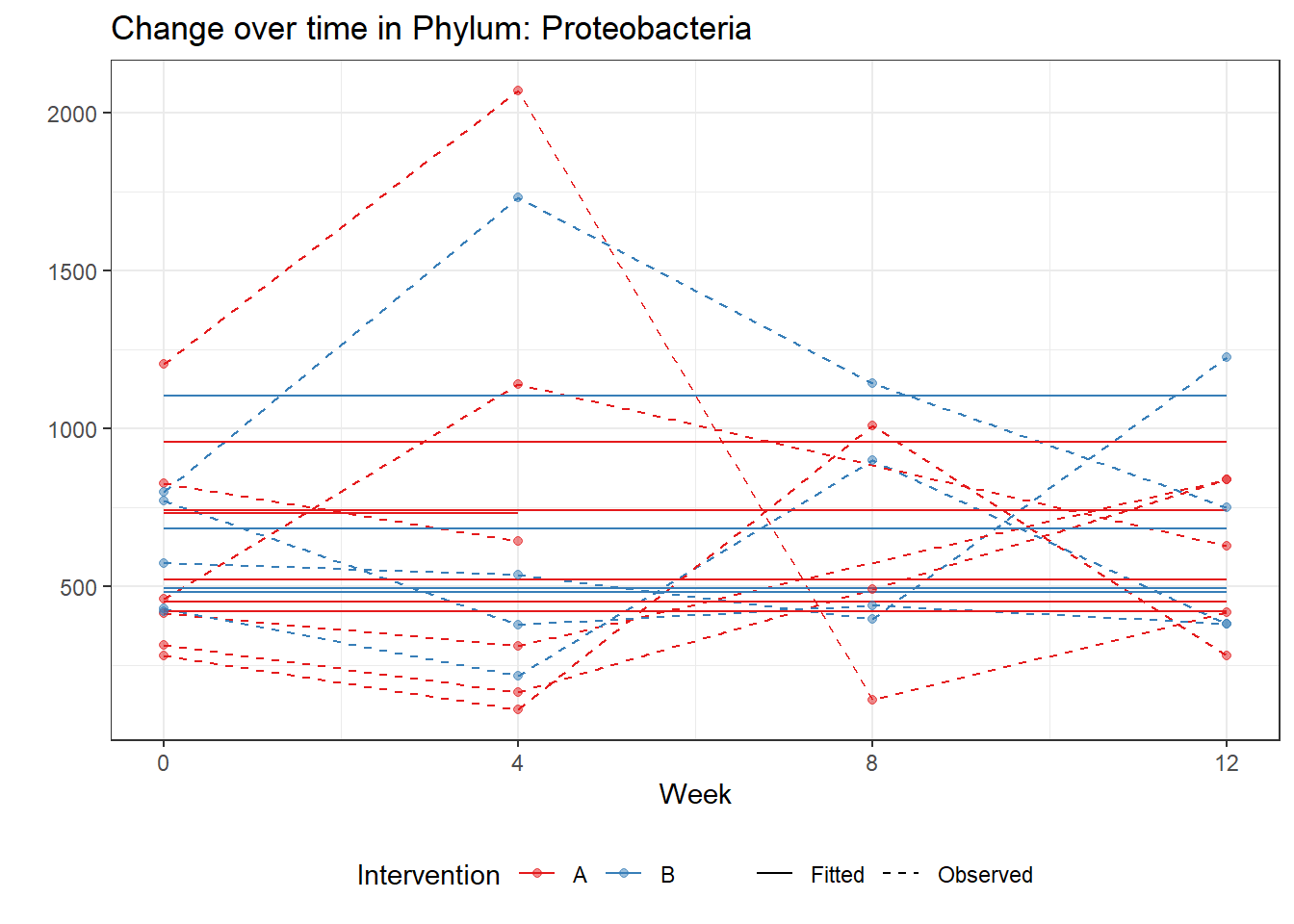
fe <- phylum.fit5$`Fixed Effects`$Actinobacteria
fe[,2][1] 3.08368 1.45591 0.09832 -0.56276get.data.with.trend <- function(data, fit){
data$Y <- 0
ph <- names(fit[[1]])
i <- 1
for(i in 1:length(ph)){
fe <- phylum.fit5[[1]][[ph[i]]][,2]
dat <- filter(data, Phylum == ph[i])
Y <- exp(fe[1] + fe[2]*as.numeric(as.factor(dat$Intervention)) + fe[3]*(as.numeric(dat$Week) - 1) + fe[4]*as.numeric(as.factor(dat$Intervention))*(as.numeric(dat$Week) - 1))
data$Y[data$Phylum == ph[i]] <- Y
}
return(data)
}
micro_data <- get.data.with.trend(mphyseq2, phylum.fit5)
micro_data$Week <- (as.numeric(micro_data$Week)-1)*4
p <- ggplot(micro_data, aes(Week, PhylumAbund))+
geom_jitter(height=0, width=0.25) +
#geom_line(data=micro_data, aes(x=Week, y=Y, group=Intervention, color=Intervention)) +
facet_wrap(.~Phylum, ncol = 5, scales = "free") +
labs(y="Phylum Abundance") +
theme(panel.grid = element_blank(),
strip.background =element_blank(),
legend.position = "bottom")
p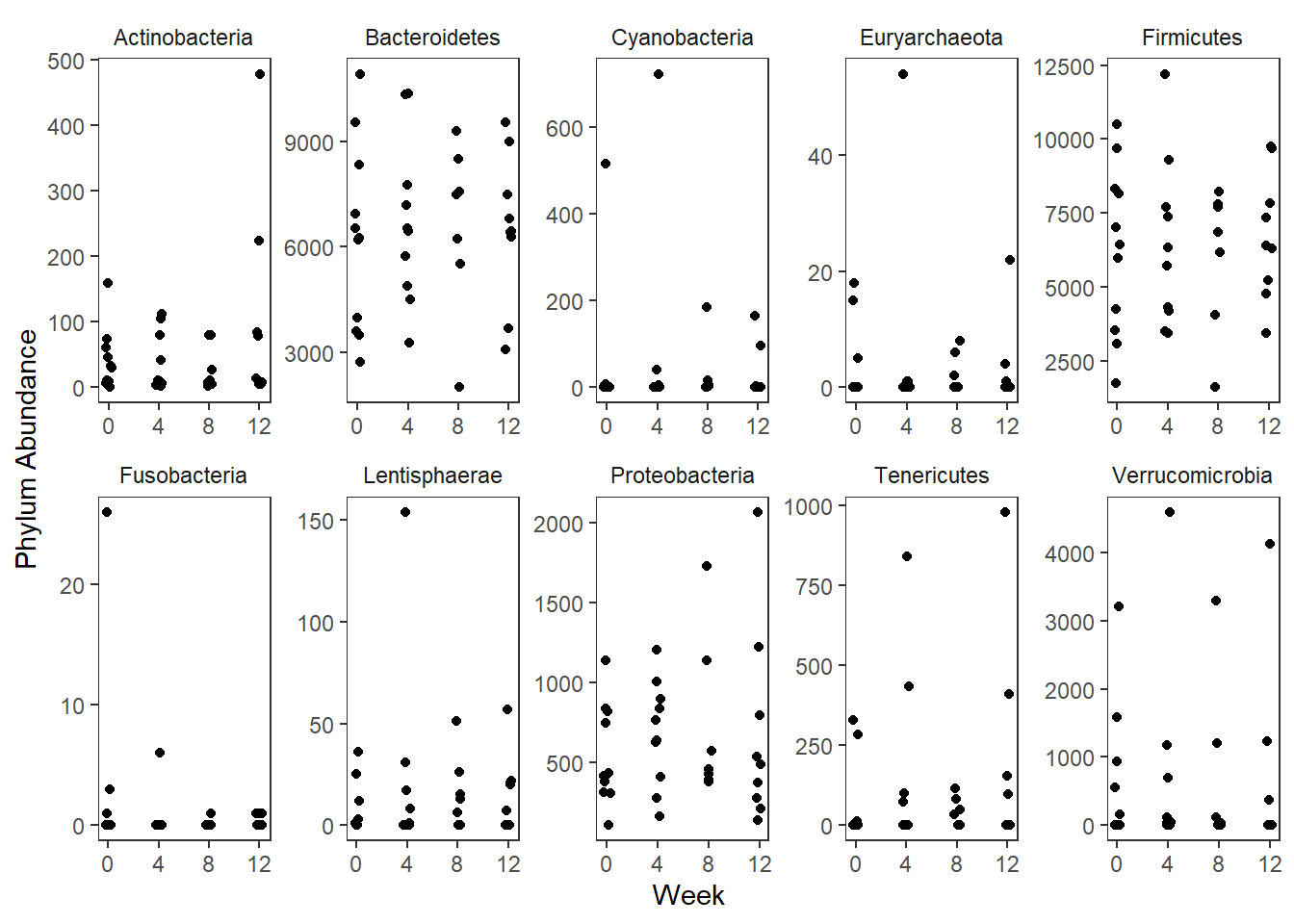
sessionInfo()R version 3.6.3 (2020-02-29)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 18362)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.1252
[2] LC_CTYPE=English_United States.1252
[3] LC_MONETARY=English_United States.1252
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.1252
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] car_3.0-7 carData_3.0-3 gvlma_1.0.0.3 patchwork_1.0.0
[5] viridis_0.5.1 viridisLite_0.3.0 gridExtra_2.3 xtable_1.8-4
[9] kableExtra_1.1.0 plyr_1.8.6 data.table_1.12.8 readxl_1.3.1
[13] forcats_0.5.0 stringr_1.4.0 dplyr_0.8.5 purrr_0.3.4
[17] readr_1.3.1 tidyr_1.0.2 tibble_3.0.1 ggplot2_3.3.0
[21] tidyverse_1.3.0 lmerTest_3.1-2 lme4_1.1-23 Matrix_1.2-18
[25] vegan_2.5-6 lattice_0.20-38 permute_0.9-5 phyloseq_1.30.0
[29] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] minqa_1.2.4 colorspace_1.4-1 rio_0.5.16
[4] ellipsis_0.3.0 rprojroot_1.3-2 XVector_0.26.0
[7] fs_1.4.1 rstudioapi_0.11 farver_2.0.3
[10] fansi_0.4.1 lubridate_1.7.8 xml2_1.3.2
[13] codetools_0.2-16 splines_3.6.3 knitr_1.28
[16] ade4_1.7-15 jsonlite_1.6.1 nloptr_1.2.2.1
[19] broom_0.5.6 cluster_2.1.0 dbplyr_1.4.3
[22] compiler_3.6.3 httr_1.4.1 backports_1.1.6
[25] assertthat_0.2.1 cli_2.0.2 later_1.0.0
[28] htmltools_0.4.0 tools_3.6.3 igraph_1.2.5
[31] gtable_0.3.0 glue_1.4.0 reshape2_1.4.4
[34] Rcpp_1.0.4.6 Biobase_2.46.0 cellranger_1.1.0
[37] vctrs_0.2.4 Biostrings_2.54.0 multtest_2.42.0
[40] ape_5.3 nlme_3.1-144 iterators_1.0.12
[43] xfun_0.13 openxlsx_4.1.4 rvest_0.3.5
[46] lifecycle_0.2.0 statmod_1.4.34 zlibbioc_1.32.0
[49] MASS_7.3-51.5 scales_1.1.0 hms_0.5.3
[52] promises_1.1.0 parallel_3.6.3 biomformat_1.14.0
[55] rhdf5_2.30.1 RColorBrewer_1.1-2 curl_4.3
[58] yaml_2.2.1 stringi_1.4.6 S4Vectors_0.24.4
[61] foreach_1.5.0 BiocGenerics_0.32.0 zip_2.0.4
[64] boot_1.3-24 rlang_0.4.5 pkgconfig_2.0.3
[67] evaluate_0.14 Rhdf5lib_1.8.0 labeling_0.3
[70] tidyselect_1.0.0 magrittr_1.5 R6_2.4.1
[73] IRanges_2.20.2 generics_0.0.2 DBI_1.1.0
[76] foreign_0.8-75 pillar_1.4.4 haven_2.2.0
[79] whisker_0.4 withr_2.2.0 mgcv_1.8-31
[82] abind_1.4-5 survival_3.1-8 modelr_0.1.7
[85] crayon_1.3.4 rmarkdown_2.1 grid_3.6.3
[88] git2r_0.27.1 reprex_0.3.0 digest_0.6.25
[91] webshot_0.5.2 httpuv_1.5.2 numDeriv_2016.8-1.1
[94] stats4_3.6.3 munsell_0.5.0