PICRUSt Stratified Analyses - Prevotella spp. (genus level only)
Last updated: 2021-04-15
Checks: 6 1
Knit directory: esoph-micro-cancer-workflow/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200916) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version b11a4b5. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/
Untracked files:
Untracked: analysis/picrust-campy.Rmd
Untracked: analysis/picrust-fuso.Rmd
Untracked: analysis/picrust-prevo.Rmd
Untracked: analysis/picrust-strepto.Rmd
Untracked: code/results-question-1.Rmd
Untracked: code/results-question-2.Rmd
Untracked: code/results-question-3.Rmd
Untracked: output/picrust_ec_stratefied_campy_data_results.csv
Untracked: output/picrust_ec_stratefied_fuso_data_results.csv
Untracked: output/picrust_ko_stratefied_campy_data_results.csv
Untracked: output/picrust_ko_stratefied_fuso_data_results.csv
Unstaged changes:
Modified: analysis/index.Rmd
Modified: analysis/picrust-analyses.Rmd
Deleted: analysis/results-question-1.Rmd
Deleted: analysis/results-question-2.Rmd
Deleted: analysis/results-question-3.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with wflow_publish() to start tracking its development.
i. Mean relative abundance in Tumors
ii. Standard error of mean in Tumors
iii. Mean relative abundance in Non-tumors
iv. Standard error of mean in Non-tumors
v. GLM differential abundance analysis between Tumor vs Non-tumor (Patient=Fixed effect)
1. P-value
2. FDR corrected q-value
vi. GLMM differential abundance analysis between Tumor vs Non-tumor (Patient=Random Effect)
1.P-value
2. FDR corrected q-valuePart 1 EC Data
# note: must rename function to func (or something else) because "function" is a reserved name in R and can't be used.
descriptions <- readr::read_tsv("data/PICRUST/ec_pred_metagenome_unstrat_descrip.tsv")[,1:2] %>%
distinct()
-- Column specification --------------------------------------------------------
cols(
.default = col_double(),
`function` = col_character(),
description = col_character()
)
i Use `spec()` for the full column specifications.colnames(descriptions) <- c("func", "description")
pi.dat <- readxl::read_xlsx("data/PICRUST-Stratified/EC_prevo.xlsx")
colnames(pi.dat) <- c("func", colnames(pi.dat)[-c(1)])
# add descriptions to dataframe
pi.dat <- left_join(pi.dat, descriptions, by="func")
pi.dat <- pi.dat[,c(1,161,3:160)]
# aggregate descriptions to get 1 description per row - unique;y defined.
pi.dat <- pi.dat %>%
group_by(description) %>%
summarise(across(`1.S37.Jun172016`:`99.D01.S37.Jun232016`,.fns = mean))
# make long format
pi.dat <- pi.dat %>%
pivot_longer(
cols=`1.S37.Jun172016`:`99.D01.S37.Jun232016`,
names_to = "ID",
values_to = "Abundance"
)
d <- as.data.frame(dat.16s)
mydata <- full_join(pi.dat, d)Joining, by = "ID"Warning in class(x) <- c(setdiff(subclass, tibble_class), tibble_class): Setting
class(x) to multiple strings ("tbl_df", "tbl", ...); result will no longer be an
S4 objectmydata <- mydata %>%
mutate(
ID.n = as.numeric(as.factor(ID)),
description.n = as.numeric(as.factor(description))
) %>%
group_by(description)%>%
mutate(avgA = mean(Abundance))Abundance
I needed to change from relative abundance to abundance due to so many individuals having 0 counts for all descriptions.
d <- mydata %>%
filter(description=="DNA-directed DNA polymerase")
t.test(d$Abundance ~ d$tumor.cat)
Welch Two Sample t-test
data: d$Abundance by d$tumor.cat
t = 0.808, df = 156, p-value = 0.42
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-2.575 6.142
sample estimates:
mean in group Non-Tumor mean in group Tumor
8.7621 6.9787 # Run on all descriptions
tb.ra1 <- mydata %>%
group_by(description) %>%
summarise(ng = n(),
Overall.M = mean(Abundance),
Overall.SE = sd(Abundance)/sqrt(ng))
tb.ra2m <- mydata %>%
group_by(description, tumor.cat) %>%
summarise(M = mean(Abundance)) %>%
pivot_wider(id_cols = description,
names_from = tumor.cat,
values_from = M)`summarise()` has grouped output by 'description'. You can override using the `.groups` argument.tb.ra2se <- mydata %>%
group_by(description, tumor.cat) %>%
summarise(ng=n(),SE = sd(Abundance)/sqrt(ng)) %>%
pivot_wider(id_cols = description,
names_from = tumor.cat,
values_from = SE)`summarise()` has grouped output by 'description'. You can override using the `.groups` argument.tb.ra2var <- mydata %>%
group_by(description, tumor.cat) %>%
summarise(ng=n(), VAR = var(Abundance)) %>%
pivot_wider(id_cols = description,
names_from = tumor.cat,
values_from = VAR)`summarise()` has grouped output by 'description'. You can override using the `.groups` argument.tb.ra2ng <- mydata %>%
group_by(description, tumor.cat) %>%
summarise(ng=n()) %>%
pivot_wider(id_cols = description,
names_from = tumor.cat,
values_from = ng)`summarise()` has grouped output by 'description'. You can override using the `.groups` argument.tb.ra <- left_join(tb.ra1, tb.ra2m)Joining, by = "description"tb.ra <- cbind(tb.ra, tb.ra2se[,-1])
tb.ra <- cbind(tb.ra, tb.ra2var[,-1])
tb.ra <- cbind(tb.ra, tb.ra2ng[,-1])
colnames(tb.ra) <- c("description", "ng", "Overall Mean", "Overall SE", "Non-Tumor Mean", "Tumor Mean", "Non-Tumor SE", "Tumor SE","Non-Tumor Var", "Tumor Var", "Non-Tumor Ng", "Tumor Ng")
tb.ra <- tb.ra %>%
arrange(desc(`Overall Mean`))
tb.ra <- tb.ra[, c("description", "Overall Mean", "Overall SE","Tumor Mean","Tumor Var", "Tumor SE","Tumor Ng", "Non-Tumor Mean","Non-Tumor Var", "Non-Tumor SE", "Non-Tumor Ng")]
# compute t-test
tb.ra <- tb.ra %>%
mutate(
SEpooled = sqrt(`Tumor Var`/`Tumor Ng` + `Non-Tumor Var`/`Non-Tumor Ng`),
t = (`Tumor Mean` - `Non-Tumor Mean`)/(SEpooled),
df = ((`Tumor Var`/`Tumor Ng` + `Non-Tumor Var`/`Non-Tumor Ng`)**2)/(((`Tumor Var`/`Tumor Ng`)**2)/(`Tumor Ng`-1) + ((`Non-Tumor Var`/`Non-Tumor Ng`)**2)/(`Non-Tumor Ng`-1)),
p = pt(q = abs(t), df=df, lower.tail = F)*2,
fdr_p = p.adjust(p, method="fdr")
)
kable(tb.ra, format="html", digits=5, caption="Stratefied EC Data - Prevotella spp. (genus level only): Average Abundance of Each Description (sorted in descending order)") %>%
kable_styling(full_width = T) %>%
scroll_box(width = "100%", height="600px")| description | Overall Mean | Overall SE | Tumor Mean | Tumor Var | Tumor SE | Tumor Ng | Non-Tumor Mean | Non-Tumor Var | Non-Tumor SE | Non-Tumor Ng | SEpooled | t | df | p | fdr_p |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NADH:ubiquinone reductase (H(+)-translocating) | 8.87320 | 1.27123 | 7.69867 | 168.38125 | 1.60950 | 65 | 9.69410 | 316.93644 | 1.84605 | 93 | 2.44916 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| DNA helicase | 8.70286 | 1.26020 | 7.51459 | 163.32620 | 1.58515 | 65 | 9.53337 | 312.88664 | 1.83422 | 93 | 2.42427 | -0.83274 | 155.808 | 0.40627 | 0.57933 |
| Peptidylprolyl isomerase | 8.38237 | 1.19184 | 7.34589 | 152.31948 | 1.53081 | 65 | 9.10679 | 275.75335 | 1.72194 | 93 | 2.30401 | -0.76427 | 155.375 | 0.44586 | 0.57933 |
| 23S rRNA (adenine(2085)-N(6))-dimethyltransferase | 8.24684 | 1.49040 | 8.93846 | 312.33990 | 2.19208 | 65 | 7.76344 | 381.07387 | 2.02425 | 93 | 2.98376 | 0.39381 | 145.890 | 0.69430 | 0.75992 |
| DNA-directed DNA polymerase | 8.02844 | 1.14496 | 6.97867 | 136.88722 | 1.45119 | 65 | 8.76214 | 256.92136 | 1.66211 | 93 | 2.20648 | -0.80829 | 155.680 | 0.42016 | 0.57933 |
| 2-oxoglutarate synthase | 5.19332 | 0.75914 | 4.48637 | 59.69762 | 0.95834 | 65 | 5.68743 | 113.25740 | 1.10355 | 93 | 1.46159 | -0.82175 | 155.751 | 0.41247 | 0.57933 |
| Glycerol-3-phosphate dehydrogenase | 4.02373 | 0.65081 | 3.49615 | 31.16307 | 0.69241 | 65 | 4.39247 | 92.19076 | 0.99564 | 93 | 1.21274 | -0.73909 | 151.551 | 0.46100 | 0.57933 |
| DNA topoisomerase | 3.73034 | 0.59433 | 3.10525 | 34.46261 | 0.72814 | 65 | 4.16722 | 70.79682 | 0.87250 | 93 | 1.13642 | -0.93449 | 156.000 | 0.35150 | 0.57933 |
| 2-oxoacid oxidoreductase (ferredoxin) | 3.41868 | 0.50521 | 2.94663 | 26.36358 | 0.63686 | 65 | 3.74861 | 50.21162 | 0.73479 | 93 | 0.97237 | -0.82477 | 155.774 | 0.41077 | 0.57933 |
| Methylaspartate mutase | 3.33755 | 0.92571 | 2.91282 | 77.93846 | 1.09501 | 65 | 3.63441 | 176.62334 | 1.37811 | 93 | 1.76018 | -0.40995 | 155.652 | 0.68241 | 0.75116 |
| Site-specific DNA-methyltransferase (adenine-specific) | 3.29688 | 0.48730 | 2.89114 | 26.44815 | 0.63788 | 65 | 3.58046 | 45.42989 | 0.69892 | 93 | 0.94625 | -0.72848 | 154.752 | 0.46742 | 0.58202 |
| Type I site-specific deoxyribonuclease | 3.17729 | 0.46624 | 2.74544 | 20.66831 | 0.56389 | 65 | 3.47911 | 44.00984 | 0.68791 | 93 | 0.88949 | -0.82482 | 155.955 | 0.41073 | 0.57933 |
| Beta-galactosidase | 3.01906 | 0.43442 | 2.59337 | 16.24369 | 0.49990 | 65 | 3.31658 | 39.36714 | 0.65062 | 93 | 0.82049 | -0.88144 | 155.023 | 0.37945 | 0.57933 |
| 5’ to 3’ exodeoxyribonuclease (nucleoside 3’-phosphate-forming) | 2.78481 | 1.14192 | 1.63077 | 36.67404 | 0.75114 | 65 | 3.59140 | 324.48340 | 1.86790 | 93 | 2.01328 | -0.97385 | 119.662 | 0.33210 | 0.57933 |
| Formyltetrahydrofolate deformylase | 2.78481 | 1.14192 | 1.63077 | 36.67404 | 0.75114 | 65 | 3.59140 | 324.48340 | 1.86790 | 93 | 2.01328 | -0.97385 | 119.662 | 0.33210 | 0.57933 |
| Alpha-L-fucosidase | 2.74904 | 0.39463 | 2.36477 | 14.25551 | 0.46831 | 65 | 3.01761 | 31.89585 | 0.58563 | 93 | 0.74985 | -0.87062 | 155.737 | 0.38530 | 0.57933 |
| DNA-directed RNA polymerase | 2.70495 | 0.39129 | 2.35257 | 16.76654 | 0.50788 | 65 | 2.95124 | 29.46928 | 0.56292 | 93 | 0.75817 | -0.78962 | 155.051 | 0.43095 | 0.57933 |
| 23S rRNA pseudouridine(1911/1915/1917) synthase | 2.66196 | 0.38137 | 2.30960 | 15.15431 | 0.48285 | 65 | 2.90823 | 28.52428 | 0.55382 | 93 | 0.73475 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| C-terminal processing peptidase | 2.66196 | 0.38137 | 2.30960 | 15.15431 | 0.48285 | 65 | 2.90823 | 28.52428 | 0.55382 | 93 | 0.73475 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Beta-N-acetylhexosaminidase | 2.54759 | 0.33420 | 2.23600 | 10.03300 | 0.39288 | 65 | 2.76538 | 23.01861 | 0.49751 | 93 | 0.63393 | -0.83507 | 155.560 | 0.40496 | 0.57933 |
| Long-chain-fatty-acid–CoA ligase | 2.48114 | 0.36259 | 2.12976 | 12.59593 | 0.44021 | 65 | 2.72673 | 26.53775 | 0.53418 | 93 | 0.69220 | -0.86243 | 155.978 | 0.38977 | 0.57933 |
| Galactoside O-acetyltransferase | 2.44304 | 0.43807 | 2.35385 | 23.69096 | 0.60372 | 65 | 2.50538 | 35.25356 | 0.61569 | 93 | 0.86229 | -0.17573 | 151.987 | 0.86074 | 0.91444 |
| [Formate-C-acetyltransferase]-activating enzyme | 2.36772 | 0.34270 | 2.08987 | 12.52266 | 0.43893 | 65 | 2.56192 | 22.86272 | 0.49582 | 93 | 0.66219 | -0.71287 | 155.456 | 0.47700 | 0.58389 |
| Phosphomethylpyrimidine synthase | 2.21751 | 0.41729 | 2.23641 | 21.65211 | 0.57716 | 65 | 2.20430 | 31.88716 | 0.58555 | 93 | 0.82218 | 0.03905 | 151.729 | 0.96890 | 0.97226 |
| Acid phosphatase | 2.12922 | 0.31788 | 2.03846 | 10.04537 | 0.39312 | 65 | 2.19265 | 20.24728 | 0.46660 | 93 | 0.61013 | -0.25272 | 155.985 | 0.80082 | 0.86239 |
| Glutathione peroxidase | 2.08575 | 0.30445 | 1.84212 | 9.82200 | 0.38873 | 65 | 2.25603 | 18.08841 | 0.44102 | 93 | 0.58788 | -0.70406 | 155.532 | 0.48245 | 0.58808 |
| All-trans-retinol 13,14-reductase | 2.06962 | 0.51497 | 1.30000 | 7.26781 | 0.33438 | 65 | 2.60753 | 65.73679 | 0.84074 | 93 | 0.90480 | -1.44510 | 119.123 | 0.15105 | 0.57933 |
| Ribonucleoside-diphosphate reductase | 2.05690 | 0.29130 | 1.86589 | 10.08857 | 0.39397 | 65 | 2.19040 | 15.81714 | 0.41240 | 93 | 0.57034 | -0.56898 | 153.168 | 0.57021 | 0.66830 |
| Undecaprenyl-diphosphate phosphatase | 2.00941 | 0.29161 | 1.69501 | 8.38834 | 0.35924 | 65 | 2.22914 | 16.97370 | 0.42722 | 93 | 0.55818 | -0.95691 | 155.990 | 0.34009 | 0.57933 |
| Cyclic pyranopterin phosphate synthase | 1.95570 | 0.59054 | 1.80000 | 15.53750 | 0.48892 | 65 | 2.06452 | 83.19144 | 0.94580 | 93 | 1.06469 | -0.24844 | 133.985 | 0.80417 | 0.86239 |
| Protein-L-isoaspartate(D-aspartate) O-methyltransferase | 1.95570 | 0.59054 | 1.80000 | 15.53750 | 0.48892 | 65 | 2.06452 | 83.19144 | 0.94580 | 93 | 1.06469 | -0.24844 | 133.985 | 0.80417 | 0.86239 |
| UDP-glucuronate decarboxylase | 1.95570 | 0.59054 | 1.80000 | 15.53750 | 0.48892 | 65 | 2.06452 | 83.19144 | 0.94580 | 93 | 1.06469 | -0.24844 | 133.985 | 0.80417 | 0.86239 |
| Succinate–CoA ligase (ADP-forming) | 1.93639 | 0.30827 | 1.74308 | 11.18452 | 0.41481 | 65 | 2.07151 | 17.79801 | 0.43747 | 93 | 0.60286 | -0.54478 | 153.468 | 0.58670 | 0.67955 |
| Alpha-amylase | 1.92010 | 0.27559 | 1.64690 | 7.13167 | 0.33124 | 65 | 2.11105 | 15.42784 | 0.40730 | 93 | 0.52498 | -0.88413 | 155.905 | 0.37799 | 0.57933 |
| Chondroitin-sulfate-ABC endolyase | 1.89557 | 0.58745 | 1.20769 | 10.04603 | 0.39313 | 65 | 2.37634 | 85.49270 | 0.95879 | 93 | 1.03626 | -1.12776 | 120.634 | 0.26166 | 0.57933 |
| Chondroitin-sulfate-ABC exolyase | 1.89557 | 0.58745 | 1.20769 | 10.04603 | 0.39313 | 65 | 2.37634 | 85.49270 | 0.95879 | 93 | 1.03626 | -1.12776 | 120.634 | 0.26166 | 0.57933 |
| Fructuronate reductase | 1.89557 | 0.58745 | 1.20769 | 10.04603 | 0.39313 | 65 | 2.37634 | 85.49270 | 0.95879 | 93 | 1.03626 | -1.12776 | 120.634 | 0.26166 | 0.57933 |
| Phosphoglycerate mutase (2,3-diphosphoglycerate-independent) | 1.86338 | 0.28190 | 1.55473 | 7.36391 | 0.33659 | 65 | 2.07911 | 16.19044 | 0.41724 | 93 | 0.53608 | -0.97818 | 155.834 | 0.32950 | 0.57933 |
| Thiamine-phosphate diphosphorylase | 1.84301 | 0.30387 | 1.77550 | 11.57318 | 0.42196 | 65 | 1.89020 | 16.84047 | 0.42554 | 93 | 0.59927 | -0.19139 | 151.422 | 0.84848 | 0.90307 |
| DNA (cytosine-5-)-methyltransferase | 1.83761 | 0.27317 | 1.58082 | 6.34957 | 0.31255 | 65 | 2.01709 | 15.62402 | 0.40988 | 93 | 0.51545 | -0.84638 | 154.840 | 0.39865 | 0.57933 |
| Beta-glucosidase | 1.83754 | 0.37510 | 1.42643 | 10.79776 | 0.40758 | 65 | 2.12488 | 30.22281 | 0.57007 | 93 | 0.70078 | -0.99666 | 152.728 | 0.32050 | 0.57933 |
| S-adenosylmethionine:tRNA ribosyltransferase-isomerase | 1.81763 | 0.26441 | 1.58271 | 7.84342 | 0.34737 | 65 | 1.98183 | 13.32825 | 0.37857 | 93 | 0.51379 | -0.77682 | 154.598 | 0.43845 | 0.57933 |
| Peroxiredoxin | 1.79577 | 0.25581 | 1.55236 | 6.81598 | 0.32382 | 65 | 1.96589 | 12.83220 | 0.37146 | 93 | 0.49279 | -0.83916 | 155.701 | 0.40267 | 0.57933 |
| Carbamoyl-phosphate synthase (glutamine-hydrolyzing) | 1.79498 | 0.25900 | 1.55629 | 7.00384 | 0.32825 | 65 | 1.96181 | 13.14606 | 0.37597 | 93 | 0.49911 | -0.81250 | 155.680 | 0.41775 | 0.57933 |
| GTP diphosphokinase | 1.79472 | 0.25899 | 1.55618 | 7.00416 | 0.32826 | 65 | 1.96144 | 13.14542 | 0.37596 | 93 | 0.49910 | -0.81197 | 155.680 | 0.41805 | 0.57933 |
| Pyridoxal 5’-phosphate synthase (glutamine hydrolyzing) | 1.79417 | 0.26143 | 1.56835 | 7.09991 | 0.33050 | 65 | 1.95200 | 13.42770 | 0.37998 | 93 | 0.50360 | -0.76181 | 155.731 | 0.44733 | 0.57933 |
| H(+)-transporting two-sector ATPase | 1.78402 | 0.25443 | 1.54133 | 6.73809 | 0.32197 | 65 | 1.95365 | 12.69662 | 0.36949 | 93 | 0.49009 | -0.84133 | 155.707 | 0.40145 | 0.57933 |
| Mannose-1-phosphate guanylyltransferase | 1.77468 | 0.25424 | 1.53984 | 6.73493 | 0.32189 | 65 | 1.93882 | 12.67746 | 0.36921 | 93 | 0.48983 | -0.81453 | 155.700 | 0.41658 | 0.57933 |
| Acyl-[acyl-carrier-protein]–UDP-N-acetylglucosamine O-acyltransferase | 1.77464 | 0.25425 | 1.53973 | 6.73525 | 0.32190 | 65 | 1.93882 | 12.67746 | 0.36921 | 93 | 0.48983 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Dipeptidyl-peptidase IV | 1.77464 | 0.25425 | 1.53973 | 6.73525 | 0.32190 | 65 | 1.93882 | 12.67746 | 0.36921 | 93 | 0.48983 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| DNA topoisomerase (ATP-hydrolyzing) | 1.77464 | 0.25425 | 1.53973 | 6.73525 | 0.32190 | 65 | 1.93882 | 12.67746 | 0.36921 | 93 | 0.48983 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Exodeoxyribonuclease VII | 1.77464 | 0.25425 | 1.53973 | 6.73525 | 0.32190 | 65 | 1.93882 | 12.67746 | 0.36921 | 93 | 0.48983 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Fumarate reductase (quinol) | 1.77464 | 0.25425 | 1.53973 | 6.73525 | 0.32190 | 65 | 1.93882 | 12.67746 | 0.36921 | 93 | 0.48983 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| IMP cyclohydrolase | 1.77464 | 0.25425 | 1.53973 | 6.73525 | 0.32190 | 65 | 1.93882 | 12.67746 | 0.36921 | 93 | 0.48983 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Phenylalanine–tRNA ligase | 1.77464 | 0.25425 | 1.53973 | 6.73525 | 0.32190 | 65 | 1.93882 | 12.67746 | 0.36921 | 93 | 0.48983 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Phosphoribosylaminoimidazolecarboxamide formyltransferase | 1.77464 | 0.25425 | 1.53973 | 6.73525 | 0.32190 | 65 | 1.93882 | 12.67746 | 0.36921 | 93 | 0.48983 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Ribonuclease H | 1.77464 | 0.25425 | 1.53973 | 6.73525 | 0.32190 | 65 | 1.93882 | 12.67746 | 0.36921 | 93 | 0.48983 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Succinate dehydrogenase (quinone) | 1.77464 | 0.25425 | 1.53973 | 6.73525 | 0.32190 | 65 | 1.93882 | 12.67746 | 0.36921 | 93 | 0.48983 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| tRNA dimethylallyltransferase | 1.77464 | 0.25425 | 1.53973 | 6.73525 | 0.32190 | 65 | 1.93882 | 12.67746 | 0.36921 | 93 | 0.48983 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Uridine kinase | 1.77464 | 0.25425 | 1.53973 | 6.73525 | 0.32190 | 65 | 1.93882 | 12.67746 | 0.36921 | 93 | 0.48983 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| XTP/dITP diphosphatase | 1.77464 | 0.25425 | 1.53973 | 6.73525 | 0.32190 | 65 | 1.93882 | 12.67746 | 0.36921 | 93 | 0.48983 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Xaa-Pro aminopeptidase | 1.77289 | 0.25428 | 1.53602 | 6.74360 | 0.32210 | 65 | 1.93845 | 12.67577 | 0.36919 | 93 | 0.48995 | -0.82138 | 155.690 | 0.41269 | 0.57933 |
| Peptidyl-dipeptidase Dcp | 1.76264 | 0.26389 | 1.52647 | 7.76356 | 0.34560 | 65 | 1.92770 | 13.30841 | 0.37829 | 93 | 0.51239 | -0.78305 | 154.723 | 0.43479 | 0.57933 |
| 5’-nucleotidase | 1.75347 | 0.25311 | 1.52700 | 6.66960 | 0.32033 | 65 | 1.91175 | 12.57260 | 0.36768 | 93 | 0.48765 | -0.78900 | 155.710 | 0.43131 | 0.57933 |
| Sialate O-acetylesterase | 1.74466 | 0.26559 | 1.46302 | 5.38439 | 0.28781 | 65 | 1.94151 | 15.17863 | 0.40399 | 93 | 0.49603 | -0.96464 | 152.585 | 0.33625 | 0.57933 |
| Glycine dehydrogenase (aminomethyl-transferring) | 1.74106 | 0.27045 | 1.55050 | 8.64640 | 0.36472 | 65 | 1.87424 | 13.66341 | 0.38330 | 93 | 0.52909 | -0.61187 | 153.329 | 0.54153 | 0.63989 |
| Bleomycin hydrolase | 1.73339 | 0.24859 | 1.51056 | 6.42249 | 0.31434 | 65 | 1.88914 | 12.13470 | 0.36122 | 93 | 0.47884 | -0.79062 | 155.725 | 0.43037 | 0.57933 |
| Aldose 1-epimerase | 1.70764 | 0.25264 | 1.46971 | 6.59844 | 0.31861 | 65 | 1.87393 | 12.55117 | 0.36737 | 93 | 0.48629 | -0.83125 | 155.767 | 0.40710 | 0.57933 |
| 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase | 1.69935 | 0.25200 | 1.47342 | 6.59059 | 0.31842 | 65 | 1.85725 | 12.47603 | 0.36627 | 93 | 0.48533 | -0.79086 | 155.737 | 0.43023 | 0.57933 |
| 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine diphosphokinase | 1.68931 | 0.24805 | 1.45698 | 6.34169 | 0.31235 | 65 | 1.85169 | 12.11350 | 0.36091 | 93 | 0.47730 | -0.82696 | 155.791 | 0.40952 | 0.57933 |
| Dephospho-CoA kinase | 1.68603 | 0.25086 | 1.46493 | 6.48323 | 0.31582 | 65 | 1.84056 | 12.39950 | 0.36514 | 93 | 0.48277 | -0.77807 | 155.798 | 0.43771 | 0.57933 |
| Choline-phosphate cytidylyltransferase | 1.66878 | 0.46286 | 1.45641 | 19.48462 | 0.54751 | 65 | 1.81720 | 44.15584 | 0.68905 | 93 | 0.88009 | -0.40995 | 155.652 | 0.68241 | 0.75116 |
| Methylaspartate ammonia-lyase | 1.66878 | 0.46286 | 1.45641 | 19.48462 | 0.54751 | 65 | 1.81720 | 44.15584 | 0.68905 | 93 | 0.88009 | -0.40995 | 155.652 | 0.68241 | 0.75116 |
| Tryptophanase | 1.66878 | 0.46286 | 1.45641 | 19.48462 | 0.54751 | 65 | 1.81720 | 44.15584 | 0.68905 | 93 | 0.88009 | -0.40995 | 155.652 | 0.68241 | 0.75116 |
| Coproporphyrinogen dehydrogenase | 1.66377 | 0.23703 | 1.44637 | 5.81089 | 0.29900 | 65 | 1.81572 | 11.04920 | 0.34469 | 93 | 0.45630 | -0.80946 | 155.765 | 0.41948 | 0.57933 |
| Maltose alpha-D-glucosyltransferase | 1.65506 | 0.61136 | 1.09231 | 14.98353 | 0.48012 | 65 | 2.04839 | 89.97318 | 0.98359 | 93 | 1.09452 | -0.87352 | 130.421 | 0.38399 | 0.57933 |
| Glycerol kinase | 1.64376 | 0.27456 | 1.57143 | 7.20663 | 0.33297 | 65 | 1.69432 | 15.30550 | 0.40568 | 93 | 0.52483 | -0.23415 | 155.961 | 0.81518 | 0.87083 |
| Phosphoglycolate phosphatase | 1.63605 | 0.22237 | 1.42621 | 4.85721 | 0.27336 | 65 | 1.78272 | 9.90094 | 0.32628 | 93 | 0.42566 | -0.83755 | 155.997 | 0.40356 | 0.57933 |
| 2-dehydropantoate 2-reductase | 1.61850 | 0.25853 | 1.41368 | 5.91609 | 0.30169 | 65 | 1.76165 | 13.85538 | 0.38598 | 93 | 0.48990 | -0.71030 | 155.382 | 0.47858 | 0.58460 |
| Fructokinase | 1.59684 | 0.23344 | 1.37824 | 5.35937 | 0.28714 | 65 | 1.74962 | 10.90726 | 0.34247 | 93 | 0.44692 | -0.83097 | 155.996 | 0.40726 | 0.57933 |
| Non-specific serine/threonine protein kinase | 1.59396 | 0.24475 | 1.55308 | 8.62888 | 0.36435 | 65 | 1.62254 | 10.14689 | 0.33031 | 93 | 0.49179 | -0.14124 | 144.521 | 0.88788 | 0.94070 |
| Aspartate transaminase | 1.59236 | 0.22338 | 1.37716 | 4.39429 | 0.26001 | 65 | 1.74276 | 10.34194 | 0.33347 | 93 | 0.42286 | -0.86460 | 155.335 | 0.38859 | 0.57933 |
| Methionyl aminopeptidase | 1.59022 | 0.22308 | 1.38536 | 4.92936 | 0.27538 | 65 | 1.73341 | 9.93905 | 0.32691 | 93 | 0.42744 | -0.81426 | 155.985 | 0.41674 | 0.57933 |
| Glucosamine-6-phosphate deaminase | 1.57069 | 0.22173 | 1.36710 | 4.29694 | 0.25711 | 65 | 1.71299 | 10.21754 | 0.33146 | 93 | 0.41949 | -0.82454 | 155.232 | 0.41090 | 0.57933 |
| Alpha-N-acetylglucosaminidase | 1.56804 | 0.57661 | 0.87308 | 9.23266 | 0.37688 | 65 | 2.05376 | 82.64518 | 0.94269 | 93 | 1.01523 | -1.16297 | 119.376 | 0.24716 | 0.57933 |
| Methylmalonyl-CoA mutase | 1.56044 | 0.23473 | 1.34000 | 4.60666 | 0.26622 | 65 | 1.71452 | 11.59286 | 0.35306 | 93 | 0.44218 | -0.84697 | 154.541 | 0.39832 | 0.57933 |
| Cobaltochelatase | 1.56013 | 0.31434 | 1.25897 | 7.86721 | 0.34790 | 65 | 1.77061 | 21.05973 | 0.47587 | 93 | 0.58948 | -0.86795 | 153.565 | 0.38678 | 0.57933 |
| Aspartate kinase | 1.55792 | 0.22318 | 1.33549 | 4.84715 | 0.27308 | 65 | 1.71339 | 9.99837 | 0.32789 | 93 | 0.42671 | -0.88560 | 156.000 | 0.37720 | 0.57933 |
| Mannan endo-1,4-beta-mannosidase | 1.49324 | 0.25655 | 1.21474 | 4.20653 | 0.25439 | 65 | 1.68789 | 14.72714 | 0.39794 | 93 | 0.47231 | -1.00179 | 147.217 | 0.31809 | 0.57933 |
| 8-oxo-dGTP diphosphatase | 1.48703 | 0.23488 | 1.37615 | 6.41118 | 0.31406 | 65 | 1.56452 | 10.40014 | 0.33441 | 93 | 0.45876 | -0.41059 | 153.832 | 0.68195 | 0.75116 |
| Leucine–tRNA ligase | 1.44723 | 0.20945 | 1.26218 | 4.66661 | 0.26794 | 65 | 1.57657 | 8.54070 | 0.30304 | 93 | 0.40451 | -0.77721 | 155.478 | 0.43821 | 0.57933 |
| Uracil-DNA glycosylase | 1.42115 | 0.21151 | 1.25422 | 4.39841 | 0.26013 | 65 | 1.53782 | 8.96938 | 0.31056 | 93 | 0.40511 | -0.70007 | 155.997 | 0.48493 | 0.58986 |
| Glucose-1-phosphatase | 1.41667 | 0.21186 | 1.35726 | 4.45679 | 0.26185 | 65 | 1.45818 | 8.99823 | 0.31105 | 93 | 0.40660 | -0.24820 | 155.987 | 0.80430 | 0.86239 |
| Hydroxymethylpyrimidine kinase | 1.40152 | 0.21274 | 1.29254 | 5.40085 | 0.28825 | 65 | 1.47770 | 8.43109 | 0.30109 | 93 | 0.41683 | -0.44422 | 153.077 | 0.65751 | 0.73071 |
| Phosphomethylpyrimidine kinase | 1.40152 | 0.21274 | 1.29254 | 5.40085 | 0.28825 | 65 | 1.47770 | 8.43109 | 0.30109 | 93 | 0.41683 | -0.44422 | 153.077 | 0.65751 | 0.73071 |
| Alpha-glucuronidase | 1.39304 | 0.57095 | 0.81692 | 9.16612 | 0.37552 | 65 | 1.79570 | 81.12085 | 0.93395 | 93 | 1.00662 | -0.97234 | 119.656 | 0.33284 | 0.57933 |
| Beta-ketoacyl-[acyl-carrier-protein] synthase I | 1.38633 | 0.32063 | 0.84215 | 2.57519 | 0.19904 | 65 | 1.76667 | 25.57225 | 0.52438 | 93 | 0.56088 | -1.64832 | 116.932 | 0.10197 | 0.57933 |
| Glutamate–ammonia ligase | 1.38595 | 0.19733 | 1.22302 | 4.19249 | 0.25397 | 65 | 1.49981 | 7.55094 | 0.28494 | 93 | 0.38170 | -0.72516 | 155.324 | 0.46945 | 0.58303 |
| 2-dehydro-3-deoxygluconokinase | 1.38101 | 0.38355 | 0.94593 | 8.18439 | 0.35484 | 65 | 1.68510 | 33.74515 | 0.60237 | 93 | 0.69912 | -1.05728 | 142.297 | 0.29217 | 0.57933 |
| Acetolactate synthase | 1.37107 | 0.37877 | 0.95912 | 8.19332 | 0.35504 | 65 | 1.65899 | 32.77952 | 0.59369 | 93 | 0.69175 | -1.01173 | 143.235 | 0.31337 | 0.57933 |
| Serine O-acetyltransferase | 1.36414 | 0.19280 | 1.24018 | 3.82836 | 0.24269 | 65 | 1.45079 | 7.34149 | 0.28096 | 93 | 0.37127 | -0.56727 | 155.813 | 0.57135 | 0.66830 |
| Alpha-galactosidase | 1.35490 | 0.18980 | 1.22355 | 3.46868 | 0.23101 | 65 | 1.44670 | 7.27929 | 0.27977 | 93 | 0.36282 | -0.61507 | 155.985 | 0.53940 | 0.63989 |
| Glutamate dehydrogenase (NADP(+)) | 1.35029 | 0.19046 | 1.22951 | 4.15986 | 0.25298 | 65 | 1.43472 | 6.86956 | 0.27178 | 93 | 0.37130 | -0.55268 | 154.145 | 0.58128 | 0.67719 |
| UDP-glucuronate 4-epimerase | 1.34494 | 0.37843 | 1.30000 | 13.55312 | 0.45663 | 65 | 1.37634 | 29.18291 | 0.56017 | 93 | 0.72271 | -0.10564 | 155.921 | 0.91601 | 0.94070 |
| Arylsulfatase | 1.33931 | 0.22182 | 1.23761 | 4.97468 | 0.27665 | 65 | 1.41039 | 9.79395 | 0.32452 | 93 | 0.42643 | -0.40519 | 155.927 | 0.68589 | 0.75292 |
| Alpha-glucosidase | 1.32077 | 0.19689 | 1.23077 | 3.89749 | 0.24487 | 65 | 1.38368 | 7.73111 | 0.28832 | 93 | 0.37827 | -0.40422 | 155.950 | 0.68660 | 0.75292 |
| Glycerol-3-phosphate cytidylyltransferase | 1.27848 | 0.34367 | 1.55385 | 17.46971 | 0.51843 | 65 | 1.08602 | 19.60122 | 0.45909 | 93 | 0.69248 | 0.67558 | 142.692 | 0.50040 | 0.59952 |
| Pectinesterase | 1.26456 | 0.39162 | 0.80718 | 4.46182 | 0.26200 | 65 | 1.58423 | 37.99675 | 0.63919 | 93 | 0.69080 | -1.12485 | 120.617 | 0.26289 | 0.57933 |
| 5-dehydro-4-deoxy-D-glucuronate isomerase | 1.26414 | 0.39163 | 0.80615 | 4.46329 | 0.26204 | 65 | 1.58423 | 37.99675 | 0.63919 | 93 | 0.69082 | -1.12631 | 120.625 | 0.26227 | 0.57933 |
| Altronate dehydratase | 1.26414 | 0.39163 | 0.80615 | 4.46329 | 0.26204 | 65 | 1.58423 | 37.99675 | 0.63919 | 93 | 0.69082 | -1.12631 | 120.625 | 0.26227 | 0.57933 |
| Tagaturonate reductase | 1.26414 | 0.39163 | 0.80615 | 4.46329 | 0.26204 | 65 | 1.58423 | 37.99675 | 0.63919 | 93 | 0.69082 | -1.12631 | 120.625 | 0.26227 | 0.57933 |
| N-acetylneuraminate lyase | 1.24747 | 0.17966 | 1.09949 | 2.55363 | 0.19821 | 65 | 1.35090 | 6.90057 | 0.27240 | 93 | 0.33688 | -0.74629 | 153.397 | 0.45663 | 0.57933 |
| Gingipain R | 1.24684 | 0.64160 | 1.24615 | 84.15721 | 1.13786 | 65 | 1.24731 | 52.44904 | 0.75098 | 93 | 1.36334 | -0.00085 | 116.519 | 0.99932 | 0.99932 |
| Fumarate hydratase | 1.23258 | 0.20541 | 1.07119 | 4.53949 | 0.26427 | 65 | 1.34538 | 8.18734 | 0.29671 | 93 | 0.39733 | -0.69007 | 155.338 | 0.49118 | 0.59125 |
| O-acetylhomoserine aminocarboxypropyltransferase | 1.22934 | 0.18238 | 1.08240 | 3.28065 | 0.22466 | 65 | 1.33204 | 6.66094 | 0.26762 | 93 | 0.34942 | -0.71445 | 155.994 | 0.47602 | 0.58389 |
| Sulfate adenylyltransferase | 1.22785 | 0.61341 | 0.73846 | 24.82115 | 0.61795 | 65 | 1.56989 | 83.89995 | 0.94982 | 93 | 1.13314 | -0.73374 | 148.198 | 0.46427 | 0.57933 |
| Methionine synthase | 1.21907 | 0.18450 | 1.07449 | 3.26570 | 0.22415 | 65 | 1.32012 | 6.88104 | 0.27201 | 93 | 0.35246 | -0.69689 | 155.978 | 0.48691 | 0.59000 |
| Hyaluronoglucosaminidase | 1.20863 | 0.18683 | 1.03736 | 2.53613 | 0.19753 | 65 | 1.32834 | 7.61253 | 0.28610 | 93 | 0.34767 | -0.83694 | 151.219 | 0.40394 | 0.57933 |
| Asparaginase | 1.20334 | 0.19490 | 1.02451 | 3.97096 | 0.24717 | 65 | 1.32833 | 7.44163 | 0.28287 | 93 | 0.37565 | -0.80879 | 155.669 | 0.41987 | 0.57933 |
| 4-O-beta-D-mannosyl-D-glucose phosphorylase | 1.19937 | 0.33040 | 0.81538 | 6.28179 | 0.31087 | 65 | 1.46774 | 24.88754 | 0.51731 | 93 | 0.60353 | -1.08090 | 143.538 | 0.28155 | 0.57933 |
| D-lactate dehydrogenase | 1.19488 | 0.16852 | 1.13333 | 2.77166 | 0.20650 | 65 | 1.23790 | 5.72496 | 0.24811 | 93 | 0.32280 | -0.32395 | 155.999 | 0.74641 | 0.80931 |
| 3-oxoacyl-[acyl-carrier-protein] reductase | 1.17501 | 0.17336 | 0.94743 | 2.41854 | 0.19289 | 65 | 1.33407 | 6.35848 | 0.26148 | 93 | 0.32493 | -1.18994 | 153.873 | 0.23590 | 0.57933 |
| Glycerate 3-kinase | 1.17495 | 0.18401 | 1.03538 | 3.52709 | 0.23294 | 65 | 1.27250 | 6.65223 | 0.26745 | 93 | 0.35467 | -0.66854 | 155.713 | 0.50478 | 0.60261 |
| DNA-3-methyladenine glycosylase I | 1.17245 | 0.18913 | 1.01953 | 3.52469 | 0.23286 | 65 | 1.27934 | 7.16494 | 0.27757 | 93 | 0.36231 | -0.71708 | 155.995 | 0.47440 | 0.58318 |
| DNA-(apurinic or apyrimidinic site) lyase | 1.17169 | 0.16825 | 1.07809 | 3.36481 | 0.22752 | 65 | 1.23712 | 5.28178 | 0.23831 | 93 | 0.32948 | -0.48265 | 153.192 | 0.63003 | 0.70833 |
| Exodeoxyribonuclease III | 1.17169 | 0.16825 | 1.07809 | 3.36481 | 0.22752 | 65 | 1.23712 | 5.28178 | 0.23831 | 93 | 0.32948 | -0.48265 | 153.192 | 0.63003 | 0.70833 |
| Signal peptidase I | 1.15362 | 0.16620 | 1.00605 | 2.79378 | 0.20732 | 65 | 1.25677 | 5.47819 | 0.24270 | 93 | 0.31920 | -0.78547 | 155.913 | 0.43337 | 0.57933 |
| Alkaline phosphatase | 1.14684 | 0.25842 | 0.88923 | 3.79879 | 0.24175 | 65 | 1.32688 | 15.28416 | 0.40540 | 93 | 0.47200 | -0.92722 | 143.060 | 0.35538 | 0.57933 |
| Alanine dehydrogenase | 1.14557 | 0.30192 | 0.74615 | 3.00877 | 0.21515 | 65 | 1.42473 | 22.29454 | 0.48962 | 93 | 0.53480 | -1.26883 | 124.297 | 0.20687 | 0.57933 |
| Threonine ammonia-lyase | 1.13063 | 0.25296 | 0.88985 | 3.75645 | 0.24040 | 65 | 1.29892 | 14.57011 | 0.39581 | 93 | 0.46310 | -0.88335 | 144.189 | 0.37852 | 0.57933 |
| 3-hydroxyacyl-[acyl-carrier-protein] dehydratase | 1.12634 | 0.16778 | 0.91507 | 2.21757 | 0.18471 | 65 | 1.27401 | 5.99412 | 0.25388 | 93 | 0.31396 | -1.14328 | 153.392 | 0.25470 | 0.57933 |
| Carbonate dehydratase | 1.12238 | 0.18217 | 1.01287 | 3.32961 | 0.22633 | 65 | 1.19892 | 6.61767 | 0.26675 | 93 | 0.34983 | -0.53185 | 155.956 | 0.59559 | 0.68286 |
| Prolyl oligopeptidase | 1.09605 | 0.17348 | 0.96352 | 3.09898 | 0.21835 | 65 | 1.18868 | 5.93799 | 0.25268 | 93 | 0.33396 | -0.67425 | 155.809 | 0.50115 | 0.59952 |
| Thymidylate synthase (FAD) | 1.09367 | 0.29995 | 0.72385 | 3.00696 | 0.21508 | 65 | 1.35215 | 22.00304 | 0.48641 | 93 | 0.53184 | -1.18138 | 124.645 | 0.23970 | 0.57933 |
| Cob(I)yrinic acid a,c-diamide adenosyltransferase | 1.09107 | 0.17600 | 0.93105 | 3.31354 | 0.22578 | 65 | 1.20291 | 6.01597 | 0.25434 | 93 | 0.34010 | -0.79934 | 155.403 | 0.42531 | 0.57933 |
| Glycine C-acetyltransferase | 1.08936 | 0.19753 | 0.88974 | 2.30356 | 0.18825 | 65 | 1.22888 | 8.86981 | 0.30883 | 93 | 0.36168 | -0.93766 | 144.411 | 0.34998 | 0.57933 |
| Indole-3-glycerol-phosphate synthase | 1.08875 | 0.22804 | 0.85846 | 5.14544 | 0.28135 | 65 | 1.24970 | 10.37860 | 0.33406 | 93 | 0.43676 | -0.89578 | 155.986 | 0.37175 | 0.57933 |
| Serine-type D-Ala-D-Ala carboxypeptidase | 1.08466 | 0.16430 | 0.90631 | 2.54642 | 0.19793 | 65 | 1.20931 | 5.46909 | 0.24250 | 93 | 0.31302 | -0.96796 | 155.930 | 0.33456 | 0.57933 |
| Glucokinase | 1.08069 | 0.15300 | 0.91523 | 2.08960 | 0.17930 | 65 | 1.19633 | 4.82502 | 0.22778 | 93 | 0.28988 | -0.96973 | 155.508 | 0.33369 | 0.57933 |
| Superoxide reductase | 1.07949 | 0.16837 | 0.97557 | 3.03351 | 0.21603 | 65 | 1.15212 | 5.52058 | 0.24364 | 93 | 0.32562 | -0.54220 | 155.426 | 0.58846 | 0.67955 |
| Inositol-3-phosphate synthase | 1.06705 | 0.17302 | 0.97795 | 3.53452 | 0.23319 | 65 | 1.12933 | 5.60300 | 0.24545 | 93 | 0.33856 | -0.44713 | 153.392 | 0.65541 | 0.73071 |
| Phosphoserine phosphatase | 1.06604 | 0.16400 | 0.92092 | 3.01917 | 0.21552 | 65 | 1.16747 | 5.12606 | 0.23477 | 93 | 0.31870 | -0.77362 | 154.585 | 0.44034 | 0.57933 |
| Formimidoyltetrahydrofolate cyclodeaminase | 1.05802 | 0.17069 | 0.95855 | 3.47823 | 0.23132 | 65 | 1.12754 | 5.42424 | 0.24151 | 93 | 0.33442 | -0.50533 | 153.056 | 0.61405 | 0.69713 |
| Glutamate formimidoyltransferase | 1.05802 | 0.17069 | 0.95855 | 3.47823 | 0.23132 | 65 | 1.12754 | 5.42424 | 0.24151 | 93 | 0.33442 | -0.50533 | 153.056 | 0.61405 | 0.69713 |
| Histidine ammonia-lyase | 1.05802 | 0.17069 | 0.95855 | 3.47823 | 0.23132 | 65 | 1.12754 | 5.42424 | 0.24151 | 93 | 0.33442 | -0.50533 | 153.056 | 0.61405 | 0.69713 |
| Imidazolonepropionase | 1.05802 | 0.17069 | 0.95855 | 3.47823 | 0.23132 | 65 | 1.12754 | 5.42424 | 0.24151 | 93 | 0.33442 | -0.50533 | 153.056 | 0.61405 | 0.69713 |
| Urocanate hydratase | 1.05802 | 0.17069 | 0.95855 | 3.47823 | 0.23132 | 65 | 1.12754 | 5.42424 | 0.24151 | 93 | 0.33442 | -0.50533 | 153.056 | 0.61405 | 0.69713 |
| Nicotinamidase | 1.05680 | 0.16226 | 0.99676 | 3.08655 | 0.21791 | 65 | 1.09875 | 4.94772 | 0.23065 | 93 | 0.31731 | -0.32143 | 153.610 | 0.74832 | 0.80986 |
| 2’,3’-cyclic-nucleotide 2’-phosphodiesterase | 1.03813 | 0.15560 | 0.86960 | 2.12907 | 0.18098 | 65 | 1.15591 | 5.01277 | 0.23217 | 93 | 0.29437 | -0.97261 | 155.332 | 0.33226 | 0.57933 |
| 3’-nucleotidase | 1.03813 | 0.15560 | 0.86960 | 2.12907 | 0.18098 | 65 | 1.15591 | 5.01277 | 0.23217 | 93 | 0.29437 | -0.97261 | 155.332 | 0.33226 | 0.57933 |
| Beta-ketoacyl-[acyl-carrier-protein] synthase III | 1.03420 | 0.16752 | 0.90143 | 3.04246 | 0.21635 | 65 | 1.12699 | 5.42925 | 0.24162 | 93 | 0.32432 | -0.69548 | 155.226 | 0.48780 | 0.59000 |
| Glutamate synthase (NADH) | 1.03284 | 0.15847 | 0.84330 | 2.11659 | 0.18045 | 65 | 1.16532 | 5.25552 | 0.23772 | 93 | 0.29845 | -1.07898 | 154.723 | 0.28227 | 0.57933 |
| Glutamate synthase (NADPH) | 1.03284 | 0.15847 | 0.84330 | 2.11659 | 0.18045 | 65 | 1.16532 | 5.25552 | 0.23772 | 93 | 0.29845 | -1.07898 | 154.723 | 0.28227 | 0.57933 |
| Aspartate ammonia-lyase | 1.02337 | 0.15977 | 0.87191 | 2.22413 | 0.18498 | 65 | 1.12923 | 5.30824 | 0.23891 | 93 | 0.30215 | -0.85164 | 155.194 | 0.39573 | 0.57933 |
| Butyrate kinase | 1.02182 | 0.16349 | 0.93441 | 3.15985 | 0.22048 | 65 | 1.08291 | 4.99974 | 0.23186 | 93 | 0.31996 | -0.46411 | 153.355 | 0.64323 | 0.71897 |
| Phosphate butyryltransferase | 1.02182 | 0.16349 | 0.93441 | 3.15985 | 0.22048 | 65 | 1.08291 | 4.99974 | 0.23186 | 93 | 0.31996 | -0.46411 | 153.355 | 0.64323 | 0.71897 |
| 23S rRNA pseudouridine(746) synthase | 1.02173 | 0.16584 | 0.89071 | 2.97830 | 0.21406 | 65 | 1.11330 | 5.32313 | 0.23924 | 93 | 0.32103 | -0.69337 | 155.243 | 0.48912 | 0.59000 |
| tRNA pseudouridine(32) synthase | 1.02173 | 0.16584 | 0.89071 | 2.97830 | 0.21406 | 65 | 1.11330 | 5.32313 | 0.23924 | 93 | 0.32103 | -0.69337 | 155.243 | 0.48912 | 0.59000 |
| Pyridoxal kinase | 1.02128 | 0.15197 | 0.91167 | 2.41098 | 0.19259 | 65 | 1.09789 | 4.53521 | 0.22083 | 93 | 0.29301 | -0.63556 | 155.695 | 0.52600 | 0.62665 |
| UDP-N-acetylglucosamine 2-epimerase (non-hydrolyzing) | 1.01984 | 0.14519 | 0.85273 | 1.82545 | 0.16758 | 65 | 1.13663 | 4.38041 | 0.21703 | 93 | 0.27420 | -1.03538 | 155.135 | 0.30210 | 0.57933 |
| Histidinol-phosphate transaminase | 1.01905 | 0.14116 | 0.92515 | 1.83094 | 0.16783 | 65 | 1.08468 | 4.08823 | 0.20967 | 93 | 0.26857 | -0.59398 | 155.750 | 0.55339 | 0.65257 |
| 1-deoxy-D-xylulose-5-phosphate synthase | 1.00849 | 0.14842 | 0.88589 | 2.52045 | 0.19692 | 65 | 1.09418 | 4.16802 | 0.21170 | 93 | 0.28913 | -0.72041 | 154.169 | 0.47236 | 0.58318 |
| L-rhamnose isomerase | 0.99831 | 0.38400 | 0.57538 | 4.10133 | 0.25119 | 65 | 1.29391 | 36.69046 | 0.62811 | 93 | 0.67647 | -1.06216 | 119.391 | 0.29031 | 0.57933 |
| Rhamnulokinase | 0.99831 | 0.38400 | 0.57538 | 4.10133 | 0.25119 | 65 | 1.29391 | 36.69046 | 0.62811 | 93 | 0.67647 | -1.06216 | 119.391 | 0.29031 | 0.57933 |
| Rhamnulose-1-phosphate aldolase | 0.99831 | 0.38400 | 0.57538 | 4.10133 | 0.25119 | 65 | 1.29391 | 36.69046 | 0.62811 | 93 | 0.67647 | -1.06216 | 119.391 | 0.29031 | 0.57933 |
| N-acetylmuramoyl-L-alanine amidase | 0.99535 | 0.15085 | 0.83353 | 2.30607 | 0.18836 | 65 | 1.10845 | 4.50038 | 0.21998 | 93 | 0.28960 | -0.94932 | 155.895 | 0.34392 | 0.57933 |
| Protoporphyrinogen oxidase | 0.99084 | 0.14020 | 0.85900 | 2.02211 | 0.17638 | 65 | 1.08298 | 3.87268 | 0.20406 | 93 | 0.26972 | -0.83043 | 155.806 | 0.40757 | 0.57933 |
| D-Ala-D-Ala dipeptidase | 0.99079 | 0.13819 | 0.87704 | 2.03436 | 0.17691 | 65 | 1.07029 | 3.71838 | 0.19996 | 93 | 0.26698 | -0.72384 | 155.466 | 0.47025 | 0.58303 |
| Acetate–CoA ligase | 0.98474 | 0.14425 | 0.86939 | 1.81708 | 0.16720 | 65 | 1.06537 | 4.33031 | 0.21578 | 93 | 0.27298 | -0.71790 | 155.209 | 0.47390 | 0.58318 |
| 4-carboxymuconolactone decarboxylase | 0.97848 | 0.29526 | 0.90154 | 3.88172 | 0.24437 | 65 | 1.03226 | 20.79786 | 0.47290 | 93 | 0.53231 | -0.24557 | 133.963 | 0.80639 | 0.86303 |
| Nitrite reductase (cytochrome; ammonia-forming) | 0.97651 | 0.14137 | 0.85444 | 2.10075 | 0.17978 | 65 | 1.06183 | 3.90918 | 0.20502 | 93 | 0.27268 | -0.76057 | 155.617 | 0.44807 | 0.57933 |
| N-acylglucosamine 2-epimerase | 0.96313 | 0.13569 | 0.83077 | 1.43388 | 0.14853 | 65 | 1.05565 | 3.94579 | 0.20598 | 93 | 0.25394 | -0.88553 | 153.059 | 0.37726 | 0.57933 |
| L-arabinose isomerase | 0.96234 | 0.29423 | 0.61615 | 2.50509 | 0.19632 | 65 | 1.20430 | 21.45645 | 0.48033 | 93 | 0.51890 | -1.13346 | 120.471 | 0.25927 | 0.57933 |
| L-ribulose-5-phosphate 4-epimerase | 0.96234 | 0.29423 | 0.61615 | 2.50509 | 0.19632 | 65 | 1.20430 | 21.45645 | 0.48033 | 93 | 0.51890 | -1.13346 | 120.471 | 0.25927 | 0.57933 |
| Levanase | 0.95660 | 0.14949 | 0.83417 | 1.91419 | 0.17161 | 65 | 1.04216 | 4.67583 | 0.22423 | 93 | 0.28236 | -0.73662 | 154.930 | 0.46247 | 0.57933 |
| Type II site-specific deoxyribonuclease | 0.95640 | 0.21683 | 0.77094 | 1.69015 | 0.16125 | 65 | 1.08602 | 11.46005 | 0.35104 | 93 | 0.38630 | -0.81564 | 126.806 | 0.41624 | 0.57933 |
| Triose-phosphate isomerase | 0.95567 | 0.14137 | 0.82408 | 2.22079 | 0.18484 | 65 | 1.04765 | 3.82301 | 0.20275 | 93 | 0.27436 | -0.81484 | 154.782 | 0.41641 | 0.57933 |
| Cu(2+)-exporting ATPase | 0.95476 | 0.13169 | 0.83194 | 1.80421 | 0.16660 | 65 | 1.04060 | 3.40276 | 0.19128 | 93 | 0.25366 | -0.82260 | 155.713 | 0.41199 | 0.57933 |
| Formate C-acetyltransferase | 0.95416 | 0.13648 | 0.83621 | 1.95387 | 0.17338 | 65 | 1.03660 | 3.64631 | 0.19801 | 93 | 0.26319 | -0.76139 | 155.638 | 0.44758 | 0.57933 |
| Methylenetetrahydrofolate reductase (NAD(P)H) | 0.95074 | 0.14143 | 0.80742 | 1.69554 | 0.16151 | 65 | 1.05092 | 4.18928 | 0.21224 | 93 | 0.26670 | -0.91298 | 154.787 | 0.36267 | 0.57933 |
| Maltose O-acetyltransferase | 0.95002 | 0.15103 | 0.82438 | 2.14875 | 0.18182 | 65 | 1.03784 | 4.63628 | 0.22328 | 93 | 0.28794 | -0.74134 | 155.914 | 0.45960 | 0.57933 |
| Thymidylate synthase | 0.94975 | 0.14176 | 0.85538 | 2.65241 | 0.20201 | 65 | 1.01570 | 3.56296 | 0.19573 | 93 | 0.28128 | -0.56995 | 149.138 | 0.56957 | 0.66830 |
| Thiamine diphosphokinase | 0.94759 | 0.14099 | 0.83917 | 2.06207 | 0.17811 | 65 | 1.02337 | 3.91100 | 0.20507 | 93 | 0.27162 | -0.67813 | 155.749 | 0.49869 | 0.59905 |
| Cystathionine beta-lyase | 0.93790 | 0.14370 | 0.79458 | 2.27348 | 0.18702 | 65 | 1.03806 | 3.96174 | 0.20640 | 93 | 0.27852 | -0.87418 | 154.944 | 0.38337 | 0.57933 |
| Hydroxylamine reductase | 0.93453 | 0.13627 | 0.82348 | 1.94746 | 0.17309 | 65 | 1.01215 | 3.63721 | 0.19776 | 93 | 0.26281 | -0.71789 | 155.644 | 0.47390 | 0.58318 |
| Cadmium-exporting ATPase | 0.93141 | 0.12843 | 0.82345 | 1.75378 | 0.16426 | 65 | 1.00686 | 3.21315 | 0.18588 | 93 | 0.24805 | -0.73940 | 155.487 | 0.46078 | 0.57933 |
| Dihydrofolate reductase | 0.93141 | 0.12843 | 0.82345 | 1.75378 | 0.16426 | 65 | 1.00686 | 3.21315 | 0.18588 | 93 | 0.24805 | -0.73940 | 155.487 | 0.46078 | 0.57933 |
| GTP cyclohydrolase I | 0.93141 | 0.12843 | 0.82345 | 1.75378 | 0.16426 | 65 | 1.00686 | 3.21315 | 0.18588 | 93 | 0.24805 | -0.73940 | 155.487 | 0.46078 | 0.57933 |
| Zinc-exporting ATPase | 0.93141 | 0.12843 | 0.82345 | 1.75378 | 0.16426 | 65 | 1.00686 | 3.21315 | 0.18588 | 93 | 0.24805 | -0.73940 | 155.487 | 0.46078 | 0.57933 |
| Dihydroorotase | 0.93031 | 0.13799 | 0.81284 | 2.28793 | 0.18761 | 65 | 1.01242 | 3.52594 | 0.19471 | 93 | 0.27039 | -0.73812 | 152.800 | 0.46157 | 0.57933 |
| Endopeptidase La | 0.93031 | 0.13799 | 0.81284 | 2.28793 | 0.18761 | 65 | 1.01242 | 3.52594 | 0.19471 | 93 | 0.27039 | -0.73812 | 152.800 | 0.46157 | 0.57933 |
| Isoleucine–tRNA ligase | 0.93031 | 0.13799 | 0.81284 | 2.28793 | 0.18761 | 65 | 1.01242 | 3.52594 | 0.19471 | 93 | 0.27039 | -0.73812 | 152.800 | 0.46157 | 0.57933 |
| Phosphoribosylformylglycinamidine synthase | 0.93031 | 0.13799 | 0.81284 | 2.28793 | 0.18761 | 65 | 1.01242 | 3.52594 | 0.19471 | 93 | 0.27039 | -0.73812 | 152.800 | 0.46157 | 0.57933 |
| Transketolase | 0.92748 | 0.13732 | 0.80276 | 1.98606 | 0.17480 | 65 | 1.01465 | 3.68428 | 0.19904 | 93 | 0.26490 | -0.79988 | 155.592 | 0.42500 | 0.57933 |
| Aspartate–ammonia ligase | 0.92260 | 0.13172 | 0.80930 | 1.52721 | 0.15328 | 65 | 1.00179 | 3.60065 | 0.19677 | 93 | 0.24942 | -0.77173 | 155.318 | 0.44145 | 0.57933 |
| Inorganic diphosphatase | 0.92032 | 0.14160 | 0.79362 | 1.73841 | 0.16354 | 65 | 1.00887 | 4.17766 | 0.21195 | 93 | 0.26770 | -0.80408 | 155.119 | 0.42258 | 0.57933 |
| 1,4-beta-mannosyl-N-acetylglucosamine phosphorylase | 0.91968 | 0.12914 | 0.79634 | 1.29454 | 0.14112 | 65 | 1.00589 | 3.57808 | 0.19615 | 93 | 0.24164 | -0.86720 | 152.975 | 0.38719 | 0.57933 |
| Beta-1,4-mannooligosaccharide phosphorylase | 0.91968 | 0.12914 | 0.79634 | 1.29454 | 0.14112 | 65 | 1.00589 | 3.57808 | 0.19615 | 93 | 0.24164 | -0.86720 | 152.975 | 0.38719 | 0.57933 |
| Exo-alpha-sialidase | 0.91968 | 0.12914 | 0.79634 | 1.29454 | 0.14112 | 65 | 1.00589 | 3.57808 | 0.19615 | 93 | 0.24164 | -0.86720 | 152.975 | 0.38719 | 0.57933 |
| L-aspartate oxidase | 0.91959 | 0.14626 | 0.82835 | 2.54680 | 0.19794 | 65 | 0.98336 | 3.98623 | 0.20703 | 93 | 0.28643 | -0.54116 | 153.133 | 0.58918 | 0.67955 |
| Nicotinate-nucleotide diphosphorylase (carboxylating) | 0.91959 | 0.14626 | 0.82835 | 2.54680 | 0.19794 | 65 | 0.98336 | 3.98623 | 0.20703 | 93 | 0.28643 | -0.54116 | 153.133 | 0.58918 | 0.67955 |
| Quinolinate synthase | 0.91959 | 0.14626 | 0.82835 | 2.54680 | 0.19794 | 65 | 0.98336 | 3.98623 | 0.20703 | 93 | 0.28643 | -0.54116 | 153.133 | 0.58918 | 0.67955 |
| Nicotinate phosphoribosyltransferase | 0.91896 | 0.13166 | 0.79725 | 1.80641 | 0.16671 | 65 | 1.00403 | 3.39979 | 0.19120 | 93 | 0.25367 | -0.81516 | 155.699 | 0.41623 | 0.57933 |
| 3-deoxy-manno-octulosonate cytidylyltransferase | 0.91384 | 0.12900 | 0.78658 | 1.71297 | 0.16234 | 65 | 1.00278 | 3.27573 | 0.18768 | 93 | 0.24815 | -0.87127 | 155.798 | 0.38495 | 0.57933 |
| Methionine adenosyltransferase | 0.91384 | 0.12900 | 0.78658 | 1.71297 | 0.16234 | 65 | 1.00278 | 3.27573 | 0.18768 | 93 | 0.24815 | -0.87127 | 155.798 | 0.38495 | 0.57933 |
| Amidophosphoribosyltransferase | 0.91268 | 0.13560 | 0.79756 | 2.08608 | 0.17915 | 65 | 0.99314 | 3.49048 | 0.19373 | 93 | 0.26387 | -0.74121 | 154.361 | 0.45969 | 0.57933 |
| UDP-glucose 6-dehydrogenase | 0.91268 | 0.13560 | 0.79756 | 2.08608 | 0.17915 | 65 | 0.99314 | 3.49048 | 0.19373 | 93 | 0.26387 | -0.74121 | 154.361 | 0.45969 | 0.57933 |
| Phosphoribosylglycinamide formyltransferase | 0.91264 | 0.13560 | 0.79745 | 2.08624 | 0.17915 | 65 | 0.99314 | 3.49048 | 0.19373 | 93 | 0.26387 | -0.74160 | 154.360 | 0.45946 | 0.57933 |
| Ribonuclease III | 0.90929 | 0.13167 | 0.79571 | 1.80479 | 0.16663 | 65 | 0.98867 | 3.40350 | 0.19130 | 93 | 0.25370 | -0.76058 | 155.712 | 0.44806 | 0.57933 |
| 3-methyl-2-oxobutanoate hydroxymethyltransferase | 0.90861 | 0.13122 | 0.79736 | 1.80623 | 0.16670 | 65 | 0.98637 | 3.37159 | 0.19040 | 93 | 0.25306 | -0.74686 | 155.640 | 0.45627 | 0.57933 |
| Aspartate 1-decarboxylase | 0.90861 | 0.13122 | 0.79736 | 1.80623 | 0.16670 | 65 | 0.98637 | 3.37159 | 0.19040 | 93 | 0.25306 | -0.74686 | 155.640 | 0.45627 | 0.57933 |
| Pantoate–beta-alanine ligase (AMP-forming) | 0.90861 | 0.13122 | 0.79736 | 1.80623 | 0.16670 | 65 | 0.98637 | 3.37159 | 0.19040 | 93 | 0.25306 | -0.74686 | 155.640 | 0.45627 | 0.57933 |
| Phosphopantothenate–cysteine ligase | 0.90861 | 0.13122 | 0.79736 | 1.80623 | 0.16670 | 65 | 0.98637 | 3.37159 | 0.19040 | 93 | 0.25306 | -0.74686 | 155.640 | 0.45627 | 0.57933 |
| Phosphopantothenoylcysteine decarboxylase | 0.90861 | 0.13122 | 0.79736 | 1.80623 | 0.16670 | 65 | 0.98637 | 3.37159 | 0.19040 | 93 | 0.25306 | -0.74686 | 155.640 | 0.45627 | 0.57933 |
| UDP-glucose 4-epimerase | 0.90853 | 0.12890 | 0.78271 | 1.72754 | 0.16303 | 65 | 0.99648 | 3.25921 | 0.18720 | 93 | 0.24824 | -0.86115 | 155.715 | 0.39048 | 0.57933 |
| 16S rRNA (cytidine(1402)-2’-O)-methyltransferase | 0.90849 | 0.12890 | 0.78260 | 1.72771 | 0.16303 | 65 | 0.99648 | 3.25921 | 0.18720 | 93 | 0.24824 | -0.86156 | 155.715 | 0.39025 | 0.57933 |
| 3’(2’),5’-bisphosphate nucleotidase | 0.90849 | 0.12890 | 0.78260 | 1.72771 | 0.16303 | 65 | 0.99648 | 3.25921 | 0.18720 | 93 | 0.24824 | -0.86156 | 155.715 | 0.39025 | 0.57933 |
| 3-deoxy-manno-octulosonate-8-phosphatase | 0.90740 | 0.13199 | 0.78631 | 1.82382 | 0.16751 | 65 | 0.99203 | 3.41117 | 0.19152 | 93 | 0.25444 | -0.80851 | 155.655 | 0.42003 | 0.57933 |
| Glutamine–fructose-6-phosphate transaminase (isomerizing) | 0.90699 | 0.13614 | 0.78071 | 1.91050 | 0.17144 | 65 | 0.99524 | 3.64942 | 0.19809 | 93 | 0.26198 | -0.81890 | 155.792 | 0.41410 | 0.57933 |
| Fructan beta-fructosidase | 0.90382 | 0.13636 | 0.76536 | 1.91458 | 0.17162 | 65 | 1.00060 | 3.65856 | 0.19834 | 93 | 0.26229 | -0.89689 | 155.794 | 0.37116 | 0.57933 |
| Protein-tyrosine-phosphatase | 0.90025 | 0.13104 | 0.77758 | 1.79285 | 0.16608 | 65 | 0.98598 | 3.36442 | 0.19020 | 93 | 0.25251 | -0.82533 | 155.679 | 0.41045 | 0.57933 |
| Diaminopimelate dehydrogenase | 0.89821 | 0.12715 | 0.78033 | 1.67989 | 0.16076 | 65 | 0.98061 | 3.17398 | 0.18474 | 93 | 0.24489 | -0.81781 | 155.725 | 0.41472 | 0.57933 |
| Phosphoglycerate dehydrogenase | 0.89821 | 0.12715 | 0.78033 | 1.67989 | 0.16076 | 65 | 0.98061 | 3.17398 | 0.18474 | 93 | 0.24489 | -0.81781 | 155.725 | 0.41472 | 0.57933 |
| Phosphoserine transaminase | 0.89821 | 0.12715 | 0.78033 | 1.67989 | 0.16076 | 65 | 0.98061 | 3.17398 | 0.18474 | 93 | 0.24489 | -0.81781 | 155.725 | 0.41472 | 0.57933 |
| Uroporphyrinogen-III synthase | 0.89736 | 0.12823 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.98647 | 3.24299 | 0.18674 | 93 | 0.24653 | -0.87860 | 155.836 | 0.38097 | 0.57933 |
| Bacterial non-heme ferritin | 0.89708 | 0.13071 | 0.78418 | 1.77498 | 0.16525 | 65 | 0.97600 | 3.35692 | 0.18999 | 93 | 0.25180 | -0.76181 | 155.731 | 0.44733 | 0.57933 |
| Enoyl-[acyl-carrier-protein] reductase (NADH) | 0.89708 | 0.13071 | 0.78418 | 1.77498 | 0.16525 | 65 | 0.97600 | 3.35692 | 0.18999 | 93 | 0.25180 | -0.76181 | 155.731 | 0.44733 | 0.57933 |
| Enoyl-[acyl-carrier-protein] reductase (NADPH, Si-specific) | 0.89708 | 0.13071 | 0.78418 | 1.77498 | 0.16525 | 65 | 0.97600 | 3.35692 | 0.18999 | 93 | 0.25180 | -0.76181 | 155.731 | 0.44733 | 0.57933 |
| o-succinylbenzoate–CoA ligase | 0.89708 | 0.13071 | 0.78418 | 1.77498 | 0.16525 | 65 | 0.97600 | 3.35692 | 0.18999 | 93 | 0.25180 | -0.76181 | 155.731 | 0.44733 | 0.57933 |
| Phosphoribosylanthranilate isomerase | 0.89708 | 0.13071 | 0.78418 | 1.77498 | 0.16525 | 65 | 0.97600 | 3.35692 | 0.18999 | 93 | 0.25180 | -0.76181 | 155.731 | 0.44733 | 0.57933 |
| Lipoyl synthase | 0.89695 | 0.13680 | 0.77860 | 2.16725 | 0.18260 | 65 | 0.97966 | 3.52170 | 0.19460 | 93 | 0.26685 | -0.75348 | 153.863 | 0.45231 | 0.57933 |
| Aminodeoxychorismate lyase | 0.88973 | 0.14169 | 0.80853 | 2.36590 | 0.19078 | 65 | 0.94648 | 3.75928 | 0.20105 | 93 | 0.27717 | -0.49771 | 153.439 | 0.61940 | 0.70008 |
| Aminodeoxychorismate synthase | 0.88973 | 0.14169 | 0.80853 | 2.36590 | 0.19078 | 65 | 0.94648 | 3.75928 | 0.20105 | 93 | 0.27717 | -0.49771 | 153.439 | 0.61940 | 0.70008 |
| 3,4-dihydroxy-2-butanone-4-phosphate synthase | 0.88928 | 0.12710 | 0.77358 | 1.68098 | 0.16081 | 65 | 0.97015 | 3.17056 | 0.18464 | 93 | 0.24485 | -0.80281 | 155.713 | 0.42331 | 0.57933 |
| Histidine kinase | 0.88907 | 0.12710 | 0.77358 | 1.68098 | 0.16081 | 65 | 0.96978 | 3.17023 | 0.18463 | 93 | 0.24485 | -0.80132 | 155.713 | 0.42417 | 0.57933 |
| GTP cyclohydrolase II | 0.88754 | 0.12713 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96978 | 3.16970 | 0.18462 | 93 | 0.24492 | -0.81623 | 155.700 | 0.41562 | 0.57933 |
| (2E,6E)-farnesyl diphosphate synthase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase (ferredoxin) | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase (flavodoxin) | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| (Kdo)-lipid IV(A) 3-deoxy-D-manno-octulosonic acid transferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| (Kdo)(2)-lipid IV(A) (2-8) 3-deoxy-D-manno-octulosonic acid transferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| (Kdo)(3)-lipid IV(A) (2-4) 3-deoxy-D-manno-octulosonic acid transferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| [Acyl-carrier-protein] S-malonyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| [Ribosomal protein S12] (aspartate(89)-C(3))-methylthiotransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 1-acylglycerol-3-phosphate O-acyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 1-deoxy-D-xylulose-5-phosphate reductoisomerase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 1,4-alpha-glucan branching enzyme | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 1,4-dihydroxy-2-naphthoate polyprenyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 1,4-dihydroxy-2-naphthoyl-CoA synthase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 16S rRNA (adenine(1518)-N(6)/adenine(1519)-N(6))-dimethyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 16S rRNA (cytosine(1402)-N(4))-methyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 16S rRNA (guanine(527)-N(7))-methyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 16S rRNA (uracil(1498)-N(3))-methyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 2-iminobutanoate/2-iminopropanoate deaminase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 2-methoxy-6-polyprenyl-1,4-benzoquinol methylase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 23S rRNA (adenine(2503)-C(2))-methyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 23S rRNA (cytosine(1962)-C(5))-methyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 23S rRNA (pseudouridine(1915)-N(3))-methyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 23S rRNA (uracil(1939)-C(5))-methyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 23S rRNA pseudouridine(2605) synthase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 3-dehydroquinate dehydratase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 3-dehydroquinate synthase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 3-deoxy-8-phosphooctulonate synthase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 3-phosphoshikimate 1-carboxyvinyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 4-(cytidine 5’-diphospho)-2-C-methyl-D-erythritol kinase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 4-alpha-glucanotransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 4-hydroxy-3-methylbut-2-enyl diphosphate reductase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 4-hydroxy-tetrahydrodipicolinate reductase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 4-hydroxy-tetrahydrodipicolinate synthase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 4-hydroxythreonine-4-phosphate dehydrogenase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 5-(carboxyamino)imidazole ribonucleotide mutase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 5-amino-6-(5-phosphoribosylamino)uracil reductase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 5-formyltetrahydrofolate cyclo-ligase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 6-phosphofructokinase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 6,7-dimethyl-8-ribityllumazine synthase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| 7-cyano-7-deazaguanine synthase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Acetate kinase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| acetylglucosaminyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Adenine phosphoribosyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Adenosylhomocysteine nucleosidase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Adenylate kinase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Adenylosuccinate lyase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Adenylosuccinate synthase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Alanine–tRNA ligase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Alanine racemase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| All-trans-octaprenyl-diphosphate synthase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Aminoacyl-tRNA hydrolase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Arabinose-5-phosphate isomerase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Arginine–tRNA ligase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Arginine decarboxylase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Asparagine–tRNA ligase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Aspartate–tRNA ligase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Aspartate-semialdehyde dehydrogenase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Aspartate carbamoyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Beta-ketoacyl-[acyl-carrier-protein] synthase II | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Biotin–[acetyl-CoA-carboxylase] ligase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Branched-chain-amino-acid transaminase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Carboxynorspermidine decarboxylase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| CDP-diacylglycerol–serine O-phosphatidyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Chorismate mutase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Chorismate synthase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Crossover junction endodeoxyribonuclease | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| CTP synthase (glutamine hydrolyzing) | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Cysteine–tRNA ligase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Cysteine desulfurase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Cytidine deaminase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| D-alanine–D-alanine ligase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| dCMP deaminase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Demethylmenaquinone methyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Deoxyribose-phosphate aldolase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| dGTPase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Diamine N-acetyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Diaminohydroxyphosphoribosylaminopyrimidine deaminase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Dihydrofolate synthase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Dihydroneopterin aldolase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Dihydroorotate dehydrogenase (NAD(+)) | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Dihydropteroate synthase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Dimethylallyltranstransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Dipeptidyl-peptidase III | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Diphosphate–fructose-6-phosphate 1-phosphotransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| diphosphate specific) | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| DNA ligase (NAD(+)) | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Dolichyl-phosphate beta-D-mannosyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| dTDP-4-dehydrorhamnose 3,5-epimerase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| dTDP-4-dehydrorhamnose reductase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| dTDP-glucose 4,6-dehydratase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| dUTP diphosphatase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Endopeptidase Clp | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Epoxyqueuosine reductase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| FAD synthetase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| FMN reductase (NAD(P)H) | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Fructose-bisphosphatase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Fructose-bisphosphate aldolase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| GDP-L-fucose synthase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| GDP-mannose 4,6-dehydratase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Geranylgeranyl diphosphate synthase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Glucose-1-phosphate thymidylyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Glucose-6-phosphate isomerase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Glutamate–tRNA ligase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Glutamate racemase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Glutamine–tRNA ligase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Glycerate dehydrogenase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Glycerol-3-phosphate dehydrogenase (NAD(P)(+)) | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Glycine–tRNA ligase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Glycine hydroxymethyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Glycogen phosphorylase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| GMP synthase (glutamine-hydrolyzing) | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Guanylate kinase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Histidine–tRNA ligase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Hypoxanthine phosphoribosyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| IMP dehydrogenase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Iron-chelate-transporting ATPase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Isochorismate synthase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Kdo(2)-lipid IV(A) lauroyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| L-serine ammonia-lyase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| L-threonylcarbamoyladenylate synthase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Lipid-A-disaccharide synthase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Lipid IV(A) 3-deoxy-D-manno-octulosonic acid transferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Lysine–tRNA ligase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Malate dehydrogenase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Malate dehydrogenase (oxaloacetate-decarboxylating) (NADP(+)) | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Mannose-6-phosphate isomerase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Methenyltetrahydrofolate cyclohydrolase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Methionine–tRNA ligase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Methionyl-tRNA formyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Methylated-DNA–[protein]-cysteine S-methyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Methylenetetrahydrofolate dehydrogenase (NADP(+)) | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| N-carbamoylputrescine amidase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| N(6)-L-threonylcarbamoyladenine synthase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| NAD(+) diphosphatase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| NAD(+) kinase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| NAD(+) synthase (glutamine-hydrolyzing) | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| NAD(P)H-hydrate epimerase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Nicotinamide-nucleotide amidase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Nicotinate-nucleotide adenylyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Oligonucleotidase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Orotate phosphoribosyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Orotidine-5’-phosphate decarboxylase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Pantetheine-phosphate adenylyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Pantothenate kinase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Peptide-methionine (R)-S-oxide reductase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Peptide-methionine (S)-S-oxide reductase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Peptide chain release factor N(5)-glutamine methyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Peptide deformylase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Phosphate acetyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Phosphatidate cytidylyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Phosphatidylserine decarboxylase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Phospho-N-acetylmuramoyl-pentapeptide-transferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Phosphoenolpyruvate carboxykinase (ATP) | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Phosphoglucomutase (alpha-D-glucose-1,6-bisphosphate-dependent) | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Phosphoglycerate kinase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Phosphomannomutase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Phosphopyruvate hydratase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Phosphoribosyl 1,2-cyclic phosphate phosphodiesterase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Phosphoribosylamine–glycine ligase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Phosphoribosylaminoimidazolesuccinocarboxamide synthase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Phosphoribosylformylglycinamidine cyclo-ligase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Polyribonucleotide nucleotidyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| PreQ(1) synthase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Proline–tRNA ligase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Protein-disulfide reductase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Purine-nucleoside phosphorylase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Pyruvate carboxylase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Pyruvate kinase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Pyruvate synthase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Pyruvate, phosphate dikinase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Riboflavin kinase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Riboflavin synthase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Ribonuclease P | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Ribonuclease Z | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Ribonucleoside-triphosphate reductase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Ribose-5-phosphate isomerase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Ribose-phosphate diphosphokinase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Ribulose-phosphate 3-epimerase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| RNA helicase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Saccharopine dehydrogenase (NAD(+), L-lysine-forming) | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Selenocysteine lyase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Serine–tRNA ligase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Shikimate dehydrogenase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Shikimate kinase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Signal peptidase II | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Starch synthase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| synthase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Tetraacyldisaccharide 4’-kinase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Tetrahydrofolate synthase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Thiamine-phosphate kinase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Thioredoxin-disulfide reductase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Threonine–tRNA ligase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Thymidine kinase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Tripeptide aminopeptidase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| tRNA-2-methylthio-N(6)-dimethylallyladenosine synthase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| tRNA-guanine(34) transglycosylase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| tRNA (guanine(37)-N(1))-methyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| tRNA (guanine(46)-N(7))-methyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| tRNA (N(6)-L-threonylcarbamoyladenosine(37)-C(2))-methylthiotransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| tRNA pseudouridine(38-40) synthase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| tRNA pseudouridine(55) synthase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| tRNA(1)(Val) (adenine(37)-N(6))-methyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| tRNA(adenine(34)) deaminase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| tRNA(Ile)-lysidine synthetase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Tryptophan–tRNA ligase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Tyrosine–tRNA ligase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| UDP-2,3-diacylglucosamine diphosphatase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| UDP-3-O-(3-hydroxymyristoyl)glucosamine N-acyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| UDP-3-O-acyl-N-acetylglucosamine deacetylase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| UDP-N-acetylglucosamine 1-carboxyvinyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| UDP-N-acetylmuramate–L-alanine ligase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| UDP-N-acetylmuramate dehydrogenase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| UDP-N-acetylmuramoyl-L-alanine–D-glutamate ligase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| UDP-N-acetylmuramoyl-L-alanyl-D-glutamate–2,6-diaminopimelate ligase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| UDP-N-acetylmuramoyl-tripeptide–D-alanyl-D-alanine ligase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| UMP kinase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| UMP/CMP kinase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Uracil phosphoribosyltransferase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Uridine phosphorylase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Valine–tRNA ligase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.57933 |
| Pullulanase | 0.88296 | 0.13528 | 0.75567 | 1.87450 | 0.16982 | 65 | 0.97192 | 3.61109 | 0.19705 | 93 | 0.26013 | -0.83133 | 155.837 | 0.40706 | 0.57933 |
| Tryptophan synthase | 0.87661 | 0.12769 | 0.76128 | 1.60201 | 0.15699 | 65 | 0.95721 | 3.26565 | 0.18739 | 93 | 0.24446 | -0.80148 | 155.997 | 0.42407 | 0.57933 |
| Formate–tetrahydrofolate ligase | 0.87335 | 0.13145 | 0.74187 | 1.77574 | 0.16528 | 65 | 0.96525 | 3.40283 | 0.19128 | 93 | 0.25280 | -0.88361 | 155.809 | 0.37827 | 0.57933 |
| S-ribosylhomocysteine lyase | 0.87335 | 0.13145 | 0.74187 | 1.77574 | 0.16528 | 65 | 0.96525 | 3.40283 | 0.19128 | 93 | 0.25280 | -0.88361 | 155.809 | 0.37827 | 0.57933 |
| Aminomethyltransferase | 0.87053 | 0.13523 | 0.77525 | 2.16160 | 0.18236 | 65 | 0.93712 | 3.41585 | 0.19165 | 93 | 0.26455 | -0.61187 | 153.329 | 0.54153 | 0.63989 |
| Dihydrolipoyl dehydrogenase | 0.87053 | 0.13523 | 0.77525 | 2.16160 | 0.18236 | 65 | 0.93712 | 3.41585 | 0.19165 | 93 | 0.26455 | -0.61187 | 153.329 | 0.54153 | 0.63989 |
| Anthranilate synthase | 0.86390 | 0.14139 | 0.78231 | 2.54045 | 0.19770 | 65 | 0.92092 | 3.61461 | 0.19715 | 93 | 0.27920 | -0.49648 | 150.822 | 0.62028 | 0.70008 |
| Galactokinase | 0.84006 | 0.10980 | 0.73174 | 1.12392 | 0.13150 | 65 | 0.91577 | 2.45463 | 0.16246 | 93 | 0.20901 | -0.88050 | 155.866 | 0.37994 | 0.57933 |
| Calcium-transporting ATPase | 0.82149 | 0.11978 | 0.74746 | 1.68326 | 0.16092 | 65 | 0.87324 | 2.69117 | 0.17011 | 93 | 0.23417 | -0.53717 | 153.559 | 0.59193 | 0.68001 |
| Xanthine phosphoribosyltransferase | 0.82149 | 0.11978 | 0.74746 | 1.68326 | 0.16092 | 65 | 0.87324 | 2.69117 | 0.17011 | 93 | 0.23417 | -0.53717 | 153.559 | 0.59193 | 0.68001 |
| NADH dehydrogenase | 0.81739 | 0.11537 | 0.71828 | 1.18617 | 0.13509 | 65 | 0.88667 | 2.75203 | 0.17202 | 93 | 0.21872 | -0.76987 | 155.467 | 0.44254 | 0.57933 |
| Non-reducing end alpha-L-arabinofuranosidase | 0.81190 | 0.23710 | 0.51200 | 1.60636 | 0.15720 | 65 | 1.02151 | 13.93258 | 0.38706 | 93 | 0.41776 | -1.21960 | 120.156 | 0.22500 | 0.57933 |
| Mannonate dehydratase | 0.81165 | 0.23711 | 0.51138 | 1.60692 | 0.15723 | 65 | 1.02151 | 13.93258 | 0.38706 | 93 | 0.41777 | -1.22105 | 120.165 | 0.22446 | 0.57933 |
| Xylulokinase | 0.79873 | 0.22090 | 0.55949 | 2.78800 | 0.20710 | 65 | 0.96595 | 11.14889 | 0.34624 | 93 | 0.40345 | -1.00746 | 143.250 | 0.31541 | 0.57933 |
| Cysteine synthase | 0.78473 | 0.11908 | 0.66808 | 1.20008 | 0.13588 | 65 | 0.86626 | 2.97209 | 0.17877 | 93 | 0.22455 | -0.88261 | 154.757 | 0.37882 | 0.57933 |
| Diaminopimelate decarboxylase | 0.77790 | 0.11572 | 0.65612 | 1.15146 | 0.13310 | 65 | 0.86301 | 2.79168 | 0.17326 | 93 | 0.21848 | -0.94695 | 155.019 | 0.34514 | 0.57933 |
| Homoserine dehydrogenase | 0.77790 | 0.11572 | 0.65612 | 1.15146 | 0.13310 | 65 | 0.86301 | 2.79168 | 0.17326 | 93 | 0.21848 | -0.94695 | 155.019 | 0.34514 | 0.57933 |
| Homoserine O-succinyltransferase | 0.77790 | 0.11572 | 0.65612 | 1.15146 | 0.13310 | 65 | 0.86301 | 2.79168 | 0.17326 | 93 | 0.21848 | -0.94695 | 155.019 | 0.34514 | 0.57933 |
| Nitric-oxide reductase (cytochrome c) | 0.77532 | 0.18790 | 0.80000 | 4.34219 | 0.25846 | 65 | 0.75806 | 6.49790 | 0.26433 | 93 | 0.36969 | 0.11343 | 152.123 | 0.90984 | 0.94070 |
| Beta-lactamase | 0.76582 | 0.21622 | 0.63590 | 1.76640 | 0.16485 | 65 | 0.85663 | 11.35603 | 0.34944 | 93 | 0.38637 | -0.57130 | 128.367 | 0.56880 | 0.66830 |
| Glucosylceramidase | 0.73418 | 0.49184 | 0.80000 | 41.60000 | 0.80000 | 65 | 0.68817 | 36.28214 | 0.62460 | 93 | 1.01495 | 0.11018 | 131.751 | 0.91243 | 0.94070 |
| (4S)-4-hydroxy-2-oxoglutarate aldolase | 0.72622 | 0.19784 | 0.47956 | 2.04833 | 0.17752 | 65 | 0.89862 | 9.05571 | 0.31205 | 93 | 0.35901 | -1.16727 | 140.092 | 0.24509 | 0.57933 |
| 2-dehydro-3-deoxy-phosphogluconate aldolase | 0.72622 | 0.19784 | 0.47956 | 2.04833 | 0.17752 | 65 | 0.89862 | 9.05571 | 0.31205 | 93 | 0.35901 | -1.16727 | 140.092 | 0.24509 | 0.57933 |
| Alpha-D-xyloside xylohydrolase | 0.72369 | 0.19004 | 0.48659 | 2.04561 | 0.17740 | 65 | 0.88940 | 8.24762 | 0.29780 | 93 | 0.34663 | -1.16205 | 142.995 | 0.24715 | 0.57933 |
| Dihydroxy-acid dehydratase | 0.68553 | 0.18938 | 0.47956 | 2.04833 | 0.17752 | 65 | 0.82949 | 8.19488 | 0.29685 | 93 | 0.34588 | -1.01173 | 143.235 | 0.31337 | 0.57933 |
| Gluconate 5-dehydrogenase | 0.68553 | 0.18951 | 0.48176 | 2.07329 | 0.17860 | 65 | 0.82796 | 8.19102 | 0.29678 | 93 | 0.34637 | -0.99951 | 143.624 | 0.31923 | 0.57933 |
| Ketol-acid reductoisomerase (NADP(+)) | 0.68553 | 0.18938 | 0.47956 | 2.04833 | 0.17752 | 65 | 0.82949 | 8.19488 | 0.29685 | 93 | 0.34588 | -1.01173 | 143.235 | 0.31337 | 0.57933 |
| Glycerophosphodiester phosphodiesterase | 0.68291 | 0.16114 | 0.44462 | 0.89958 | 0.11764 | 65 | 0.84946 | 6.30692 | 0.26042 | 93 | 0.28576 | -1.41676 | 125.847 | 0.15902 | 0.57933 |
| 6-carboxytetrahydropterin synthase | 0.68241 | 0.14779 | 0.53108 | 1.53653 | 0.15375 | 65 | 0.78817 | 4.79279 | 0.22701 | 93 | 0.27418 | -0.93769 | 150.299 | 0.34991 | 0.57933 |
| 6-pyruvoyltetrahydropterin synthase | 0.68241 | 0.14779 | 0.53108 | 1.53653 | 0.15375 | 65 | 0.78817 | 4.79279 | 0.22701 | 93 | 0.27418 | -0.93769 | 150.299 | 0.34991 | 0.57933 |
| UDP-galactopyranose mutase | 0.66237 | 0.13315 | 0.53594 | 1.44317 | 0.14901 | 65 | 0.75073 | 3.75708 | 0.20099 | 93 | 0.25020 | -0.85846 | 154.033 | 0.39197 | 0.57933 |
| Arabinogalactan endo-beta-1,4-galactanase | 0.63648 | 0.16168 | 0.46601 | 1.35653 | 0.14446 | 65 | 0.75562 | 6.06953 | 0.25547 | 93 | 0.29349 | -0.98679 | 139.710 | 0.32545 | 0.57933 |
| Threonine synthase | 0.63386 | 0.16633 | 0.42538 | 1.56647 | 0.15524 | 65 | 0.77957 | 6.31739 | 0.26063 | 93 | 0.30336 | -1.16753 | 142.987 | 0.24494 | 0.57933 |
| Arabinan endo-1,5-alpha-L-arabinosidase | 0.62841 | 0.14992 | 0.43179 | 1.32576 | 0.14282 | 65 | 0.76583 | 5.09164 | 0.23398 | 93 | 0.27413 | -1.21855 | 144.489 | 0.22500 | 0.57933 |
| 4-phosphoerythronate dehydrogenase | 0.61392 | 0.30670 | 0.36923 | 6.20529 | 0.30898 | 65 | 0.78495 | 20.97499 | 0.47491 | 93 | 0.56657 | -0.73374 | 148.198 | 0.46427 | 0.57933 |
| 8-amino-7-oxononanoate synthase | 0.61392 | 0.30670 | 0.36923 | 6.20529 | 0.30898 | 65 | 0.78495 | 20.97499 | 0.47491 | 93 | 0.56657 | -0.73374 | 148.198 | 0.46427 | 0.57933 |
| Adenylyl-sulfate kinase | 0.61392 | 0.30670 | 0.36923 | 6.20529 | 0.30898 | 65 | 0.78495 | 20.97499 | 0.47491 | 93 | 0.56657 | -0.73374 | 148.198 | 0.46427 | 0.57933 |
| AMP nucleosidase | 0.61392 | 0.30670 | 0.36923 | 6.20529 | 0.30898 | 65 | 0.78495 | 20.97499 | 0.47491 | 93 | 0.56657 | -0.73374 | 148.198 | 0.46427 | 0.57933 |
| Exodeoxyribonuclease V | 0.61392 | 0.30670 | 0.36923 | 6.20529 | 0.30898 | 65 | 0.78495 | 20.97499 | 0.47491 | 93 | 0.56657 | -0.73374 | 148.198 | 0.46427 | 0.57933 |
| Nucleoside-diphosphate kinase | 0.61392 | 0.30670 | 0.36923 | 6.20529 | 0.30898 | 65 | 0.78495 | 20.97499 | 0.47491 | 93 | 0.56657 | -0.73374 | 148.198 | 0.46427 | 0.57933 |
| Polynucleotide adenylyltransferase | 0.61392 | 0.30670 | 0.36923 | 6.20529 | 0.30898 | 65 | 0.78495 | 20.97499 | 0.47491 | 93 | 0.56657 | -0.73374 | 148.198 | 0.46427 | 0.57933 |
| Pyridoxine 5’-phosphate synthase | 0.61392 | 0.30670 | 0.36923 | 6.20529 | 0.30898 | 65 | 0.78495 | 20.97499 | 0.47491 | 93 | 0.56657 | -0.73374 | 148.198 | 0.46427 | 0.57933 |
| Adenosylcobalamin/alpha-ribazole phosphatase | 0.59351 | 0.15114 | 0.37731 | 0.75533 | 0.10780 | 65 | 0.74462 | 5.57742 | 0.24489 | 93 | 0.26757 | -1.37279 | 124.393 | 0.17229 | 0.57933 |
| Adenosylcobinamide-GDP ribazoletransferase | 0.59351 | 0.15114 | 0.37731 | 0.75533 | 0.10780 | 65 | 0.74462 | 5.57742 | 0.24489 | 93 | 0.26757 | -1.37279 | 124.393 | 0.17229 | 0.57933 |
| Adenosylcobinamide-phosphate guanylyltransferase | 0.59351 | 0.15114 | 0.37731 | 0.75533 | 0.10780 | 65 | 0.74462 | 5.57742 | 0.24489 | 93 | 0.26757 | -1.37279 | 124.393 | 0.17229 | 0.57933 |
| Adenosylcobinamide kinase | 0.59351 | 0.15114 | 0.37731 | 0.75533 | 0.10780 | 65 | 0.74462 | 5.57742 | 0.24489 | 93 | 0.26757 | -1.37279 | 124.393 | 0.17229 | 0.57933 |
| Nicotinate-nucleotide–dimethylbenzimidazole phosphoribosyltransferase | 0.59351 | 0.15114 | 0.37731 | 0.75533 | 0.10780 | 65 | 0.74462 | 5.57742 | 0.24489 | 93 | 0.26757 | -1.37279 | 124.393 | 0.17229 | 0.57933 |
| Superoxide dismutase | 0.58763 | 0.11111 | 0.45680 | 0.92527 | 0.11931 | 65 | 0.67907 | 2.66422 | 0.16926 | 93 | 0.20708 | -1.07334 | 152.144 | 0.28482 | 0.57933 |
| Alpha-L-rhamnosidase | 0.58228 | 0.27015 | 0.47692 | 9.34712 | 0.37921 | 65 | 0.65591 | 13.16293 | 0.37621 | 93 | 0.53417 | -0.33508 | 150.536 | 0.73803 | 0.80172 |
| Nitrous-oxide reductase | 0.58228 | 0.27015 | 0.47692 | 9.34712 | 0.37921 | 65 | 0.65591 | 13.16293 | 0.37621 | 93 | 0.53417 | -0.33508 | 150.536 | 0.73803 | 0.80172 |
| Prephenate dehydratase | 0.54437 | 0.11659 | 0.35530 | 0.54727 | 0.09176 | 65 | 0.67652 | 3.24171 | 0.18670 | 93 | 0.20803 | -1.54412 | 130.839 | 0.12497 | 0.57933 |
| CDP-glycerol glycerophosphotransferase | 0.52532 | 0.36271 | 0.55385 | 16.84471 | 0.50907 | 65 | 0.50538 | 23.75269 | 0.50538 | 93 | 0.71732 | 0.06757 | 150.573 | 0.94622 | 0.95115 |
| Phosphatidylglycerophosphatase | 0.52363 | 0.14626 | 0.51897 | 1.94007 | 0.17276 | 65 | 0.52688 | 4.41865 | 0.21797 | 93 | 0.27814 | -0.02843 | 155.616 | 0.97736 | 0.97905 |
| Lysozyme | 0.51899 | 0.11395 | 0.53077 | 1.59279 | 0.15654 | 65 | 0.51075 | 2.39255 | 0.16039 | 93 | 0.22412 | 0.08931 | 152.213 | 0.92895 | 0.94070 |
| Leucyltransferase | 0.51730 | 0.12525 | 0.53436 | 1.92882 | 0.17226 | 65 | 0.50538 | 2.88795 | 0.17622 | 93 | 0.24643 | 0.11761 | 152.136 | 0.90653 | 0.94070 |
| N-acetylmuramic acid 6-phosphate etherase | 0.47710 | 0.11628 | 0.28839 | 0.40133 | 0.07858 | 65 | 0.60899 | 3.32371 | 0.18905 | 93 | 0.20473 | -1.56599 | 121.329 | 0.11995 | 0.57933 |
| (R)-2-methylmalate dehydratase | 0.47532 | 0.26003 | 0.44769 | 10.49972 | 0.40191 | 65 | 0.49462 | 10.92660 | 0.34277 | 93 | 0.52823 | -0.08885 | 139.587 | 0.92933 | 0.94070 |
| 3-isopropylmalate dehydratase | 0.47532 | 0.26003 | 0.44769 | 10.49972 | 0.40191 | 65 | 0.49462 | 10.92660 | 0.34277 | 93 | 0.52823 | -0.08885 | 139.587 | 0.92933 | 0.94070 |
| Xylose isomerase | 0.47152 | 0.26006 | 0.44615 | 10.50096 | 0.40194 | 65 | 0.48925 | 10.92923 | 0.34281 | 93 | 0.52827 | -0.08157 | 139.592 | 0.93510 | 0.94325 |
| Glucuronate isomerase | 0.46722 | 0.12602 | 0.26338 | 0.40408 | 0.07885 | 65 | 0.60968 | 3.95088 | 0.20611 | 93 | 0.22068 | -1.56921 | 117.285 | 0.11929 | 0.57933 |
| CDP-diacylglycerol–glycerol-3-phosphate 3-phosphatidyltransferase | 0.46076 | 0.11582 | 0.40077 | 1.08496 | 0.12920 | 65 | 0.50269 | 2.85801 | 0.17530 | 93 | 0.21777 | -0.46802 | 153.840 | 0.64044 | 0.71863 |
| Dipeptidase E | 0.41845 | 0.10157 | 0.27811 | 0.46453 | 0.08454 | 65 | 0.51654 | 2.43470 | 0.16180 | 93 | 0.18255 | -1.30611 | 134.660 | 0.19374 | 0.57933 |
| L-rhamnose mutarotase | 0.41139 | 0.17535 | 0.23077 | 1.67248 | 0.16041 | 65 | 0.53763 | 7.08824 | 0.27608 | 93 | 0.31929 | -0.96108 | 141.431 | 0.33815 | 0.57933 |
| Phosphoglycerate mutase (2,3-diphosphoglycerate-dependent) | 0.39873 | 0.09404 | 0.40769 | 1.08314 | 0.12909 | 65 | 0.39247 | 1.63098 | 0.13243 | 93 | 0.18494 | 0.08229 | 152.271 | 0.93452 | 0.94325 |
| Cellobiose phosphorylase | 0.36772 | 0.24592 | 0.40154 | 10.39890 | 0.39998 | 65 | 0.34409 | 9.07054 | 0.31230 | 93 | 0.50746 | 0.11322 | 131.756 | 0.91003 | 0.94070 |
| Diaminopimelate epimerase | 0.35994 | 0.06710 | 0.28031 | 0.45305 | 0.08349 | 65 | 0.41559 | 0.89130 | 0.09790 | 93 | 0.12866 | -1.05147 | 155.925 | 0.29467 | 0.57933 |
| LL-diaminopimelate aminotransferase | 0.35994 | 0.06710 | 0.28031 | 0.45305 | 0.08349 | 65 | 0.41559 | 0.89130 | 0.09790 | 93 | 0.12866 | -1.05147 | 155.925 | 0.29467 | 0.57933 |
| Anthranilate phosphoribosyltransferase | 0.31477 | 0.17337 | 0.29846 | 4.66654 | 0.26794 | 65 | 0.32616 | 4.85744 | 0.22854 | 93 | 0.35217 | -0.07866 | 139.597 | 0.93741 | 0.94393 |
| Lipoate–protein ligase | 0.30922 | 0.09671 | 0.19121 | 0.96829 | 0.12205 | 65 | 0.39171 | 1.83137 | 0.14033 | 93 | 0.18598 | -1.07805 | 155.732 | 0.28268 | 0.57933 |
| Creatininase | 0.27215 | 0.16573 | 0.04615 | 0.04471 | 0.02623 | 65 | 0.43011 | 7.31300 | 0.28042 | 93 | 0.28164 | -1.36327 | 93.606 | 0.17607 | 0.57933 |
| Exo-1,4-beta-D-glucosaminidase | 0.27215 | 0.16573 | 0.04615 | 0.04471 | 0.02623 | 65 | 0.43011 | 7.31300 | 0.28042 | 93 | 0.28164 | -1.36327 | 93.606 | 0.17607 | 0.57933 |
| N-acetylneuraminate synthase | 0.27215 | 0.16573 | 0.04615 | 0.04471 | 0.02623 | 65 | 0.43011 | 7.31300 | 0.28042 | 93 | 0.28164 | -1.36327 | 93.606 | 0.17607 | 0.57933 |
| N-acylneuraminate cytidylyltransferase | 0.27215 | 0.16573 | 0.04615 | 0.04471 | 0.02623 | 65 | 0.43011 | 7.31300 | 0.28042 | 93 | 0.28164 | -1.36327 | 93.606 | 0.17607 | 0.57933 |
| RNA ligase (ATP) | 0.27215 | 0.16573 | 0.04615 | 0.04471 | 0.02623 | 65 | 0.43011 | 7.31300 | 0.28042 | 93 | 0.28164 | -1.36327 | 93.606 | 0.17607 | 0.57933 |
| Beta-fructofuranosidase | 0.26846 | 0.05911 | 0.19487 | 0.46751 | 0.08481 | 65 | 0.31989 | 0.61040 | 0.08102 | 93 | 0.11729 | -1.06595 | 148.231 | 0.28818 | 0.57933 |
| Dextranase | 0.26772 | 0.06391 | 0.22462 | 0.67001 | 0.10153 | 65 | 0.29785 | 0.63304 | 0.08250 | 93 | 0.13082 | -0.55979 | 135.370 | 0.57654 | 0.67302 |
| L-threonine aldolase | 0.26498 | 0.09697 | 0.23897 | 1.40165 | 0.14685 | 65 | 0.28315 | 1.55965 | 0.12950 | 93 | 0.19579 | -0.22565 | 142.358 | 0.82180 | 0.87628 |
| (R)-citramalate synthase | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94070 |
| 2-isopropylmalate synthase | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94070 |
| 3-isopropylmalate dehydrogenase | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94070 |
| Acetylglutamate kinase | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94070 |
| Acetylornithine deacetylase | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94070 |
| Acetylornithine transaminase | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94070 |
| Argininosuccinate lyase | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94070 |
| Argininosuccinate synthase | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94070 |
| ATP phosphoribosyltransferase | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94070 |
| Histidinol-phosphatase | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94070 |
| Histidinol dehydrogenase | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94070 |
| Imidazoleglycerol-phosphate dehydratase | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94070 |
| isomerase | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94070 |
| N-acetyl-gamma-glutamyl-phosphate reductase | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94070 |
| N-succinylornithine carbamoyltransferase | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94070 |
| Phosphoribosyl-AMP cyclohydrolase | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94070 |
| Phosphoribosyl-ATP diphosphatase | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94070 |
| Lactoylglutathione lyase | 0.23259 | 0.08954 | 0.11154 | 0.39069 | 0.07753 | 65 | 0.31720 | 1.87247 | 0.14189 | 93 | 0.16169 | -1.27195 | 137.511 | 0.20554 | 0.57933 |
| Pyrroline-5-carboxylate reductase | 0.19968 | 0.12364 | 0.21615 | 2.60298 | 0.20011 | 65 | 0.18817 | 2.31067 | 0.15763 | 93 | 0.25474 | 0.10985 | 132.556 | 0.91270 | 0.94070 |
| Hippurate hydrolase | 0.17089 | 0.13785 | 0.05385 | 0.18846 | 0.05385 | 65 | 0.25269 | 4.97622 | 0.23132 | 93 | 0.23750 | -0.83722 | 101.811 | 0.40443 | 0.57933 |
| Potassium-transporting ATPase | 0.15190 | 0.13414 | 0.32308 | 6.78462 | 0.32308 | 65 | 0.03226 | 0.09677 | 0.03226 | 93 | 0.32468 | 0.89570 | 65.278 | 0.37370 | 0.57933 |
| Type III site-specific deoxyribonuclease | 0.14620 | 0.13620 | 0.00154 | 0.00015 | 0.00154 | 65 | 0.24731 | 4.97622 | 0.23132 | 93 | 0.23132 | -1.06247 | 92.008 | 0.29080 | 0.57933 |
| Polar-amino-acid-transporting ATPase | 0.13671 | 0.08286 | 0.02462 | 0.01126 | 0.01316 | 65 | 0.21505 | 1.82825 | 0.14021 | 93 | 0.14083 | -1.35230 | 93.618 | 0.17954 | 0.57933 |
| Aconitate hydratase | 0.06677 | 0.04503 | 0.05077 | 0.08043 | 0.03518 | 65 | 0.07796 | 0.49046 | 0.07262 | 93 | 0.08069 | -0.33693 | 129.952 | 0.73671 | 0.80172 |
| Isocitrate dehydrogenase (NADP(+)) | 0.06677 | 0.04503 | 0.05077 | 0.08043 | 0.03518 | 65 | 0.07796 | 0.49046 | 0.07262 | 93 | 0.08069 | -0.33693 | 129.952 | 0.73671 | 0.80172 |
| Citrate CoA-transferase | 0.05063 | 0.04471 | 0.10769 | 0.75385 | 0.10769 | 65 | 0.01075 | 0.01075 | 0.01075 | 93 | 0.10823 | 0.89570 | 65.278 | 0.37370 | 0.57933 |
| Citrate lyase holo-[acyl-carrier protein] synthase | 0.05063 | 0.04471 | 0.10769 | 0.75385 | 0.10769 | 65 | 0.01075 | 0.01075 | 0.01075 | 93 | 0.10823 | 0.89570 | 65.278 | 0.37370 | 0.57933 |
| Citryl-CoA lyase | 0.05063 | 0.04471 | 0.10769 | 0.75385 | 0.10769 | 65 | 0.01075 | 0.01075 | 0.01075 | 93 | 0.10823 | 0.89570 | 65.278 | 0.37370 | 0.57933 |
| RNA-directed DNA polymerase | 0.05063 | 0.04471 | 0.10769 | 0.75385 | 0.10769 | 65 | 0.01075 | 0.01075 | 0.01075 | 93 | 0.10823 | 0.89570 | 65.278 | 0.37370 | 0.57933 |
| 2-iminoacetate synthase | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.57933 |
| Asparagine synthase (glutamine-hydrolyzing) | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.57933 |
| Aspartyl aminopeptidase | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.57933 |
| Cellulase | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.57933 |
| Citrate (Si)-synthase | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.57933 |
| D-glycero-alpha-D-manno-heptose-1,7-bisphosphate 7-phosphatase | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.57933 |
| D-glycero-alpha-D-manno-heptose-7-phosphate kinase | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.57933 |
| D-glycero-beta-D-manno-heptose 1,7-bisphosphate 7-phosphatase | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.57933 |
| D-sedoheptulose 7-phosphate isomerase | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.57933 |
| Endo-1,4-beta-xylanase | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.57933 |
| Glutamate-5-semialdehyde dehydrogenase | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.57933 |
| Glutamate 5-kinase | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.57933 |
| Phosphoadenylyl-sulfate reductase (thioredoxin) | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.57933 |
| Prolyl aminopeptidase | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.57933 |
| Thiazole synthase | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.57933 |
plot.dat <- tb.ra %>%
arrange(p) %>%
slice_head(n=50)%>%
mutate(
description = fct_reorder(description, `Overall Mean`),
description = factor(description, levels = levels(description), ordered=T)
) %>%
arrange(description) %>%
mutate(
id = 1:n(),
step = ifelse(id%%2 == 0, 1, 0),
Mean_diff = `Tumor Mean` - `Non-Tumor Mean`,
diff_se = SEpooled,
Mean_diff_ll = Mean_diff - qt(0.975, df)*diff_se,
Mean_diff_ul = Mean_diff + qt(0.975, df)*diff_se
) %>%
pivot_longer(
cols=contains("Mean"),
names_to = "group",
values_to = "mean"
)
p1.d <- plot.dat %>%
filter(group %in% c("Tumor Mean","Non-Tumor Mean")) %>%
mutate(
group = ifelse(group == "Tumor Mean", "Tumor", "Non-Tumor"),
col = ifelse(step == 1, "grey90", "white"),
h=1, w=Inf
)
p1 <- ggplot()+
geom_tile(data = p1.d,
aes(y = description, x=0,
height=h, width=w),
fill = p1.d$col, color=p1.d$col)+
geom_bar(data=p1.d,
aes(x=mean, y=description,
group=group, color=group,
fill=group),
stat="identity",position = "dodge",
alpha = 1)+
labs(x="Mean Abundance")+
theme_classic()+
theme(
legend.position = "bottom",
plot.margin = unit(c(1,0,1,1), "lines")
)
p2.d <- plot.dat %>%
filter(group %in% c("Mean_diff", "Mean_diff_ll", "Mean_diff_ul")) %>%
pivot_wider(
names_from = group,
values_from = mean
) %>%
mutate(
group = ifelse(Mean_diff > 0, "Tumor", "Non-Tumor"),
p = sprintf("%.3f", round(p,3)),
ll = min(Mean_diff_ll)-0.01,
ul = max(Mean_diff_ul)+0.01
)
p2<-ggplot(p2.d, aes(x=Mean_diff, y=description))+
geom_tile(data = p1.d,
aes(y = description, x=0,
height=h, width=w),
fill = p1.d$col, color=p1.d$col)+
geom_vline(xintercept = 0, linetype="dashed", alpha=0.5)+
geom_segment(aes(x=Mean_diff_ll, y=description, xend=Mean_diff_ul, yend=description))+
geom_point(aes(fill=group, color=group))+
geom_text(aes(label=p, x=unique(ul)+0.1))+
coord_cartesian(xlim = c(unique(p2.d$ll), unique(p2.d$ul)),
clip = 'off') +
annotate("text", x=unique(p2.d$ul)+0.2,y = 25,
angle=90,
label="p-value (uncorrected)")+
labs(x="Mean Difference in Abundance")+
theme_classic()+
theme(
legend.position = "bottom",
axis.title.y = element_blank(),
axis.text.y = element_blank(),
axis.line.y = element_blank(),
axis.ticks.y = element_blank(),
plot.margin = unit(c(1,4,1,0), "lines")
)
# plot
p1 + p2 +
plot_annotation(title="Stratefied EC Data - Prevotella spp. (genus level only): First 50 descriptions with lowest p-value (uncorrected)")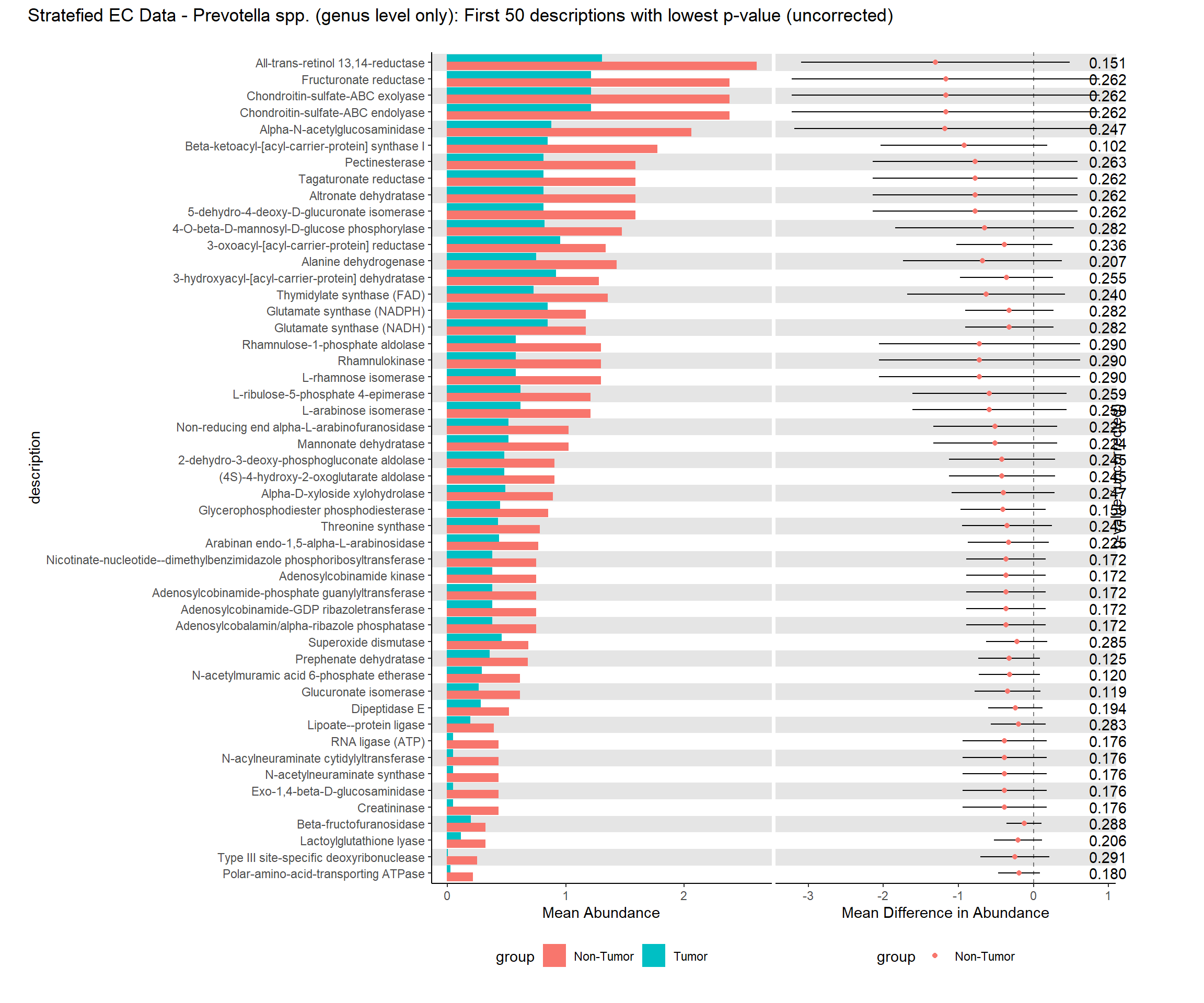
Modeling Difference between Tumor and Non-Tumor
For the modeling, we used a generalized linear mixed model (GLMM).
First, we looked at the biserial correlation between the abundance of each description and the tumor status.
tb <- mydata %>%
group_by(description)%>%
summarise(
r = cor(tumor, Abundance)
) %>%
mutate(
M=mean(r)
)
ggplot(tb, aes(x=r))+
geom_density()+
geom_vline(aes(xintercept = M))+
labs(x="Biserial Correlation",title="Relationship between description abundance and tumor (tumor vs. non-tumor)")+
theme(panel.grid = element_blank())
Next, we need the data to be on an interpretable scale. First, let’s use the raw abundance like scale.
p <- ggplot(mydata, aes(x=Abundance))+
geom_density()
p
mydata <- mydata %>%
mutate(Abundance.dich=ifelse(Abundance==0, 0, 1))
table(mydata$Abundance.dich)
0 1
38726 52756 Let’s first run models by description so that we can avoid the nesting issue initially. We will come back to this to conduct the final model (it will be more powerful).
DESCRIPTIONS <- unique(mydata$description)
i<-1
dat0 <- mydata %>% filter(description==DESCRIPTIONS[i])
# quasipossion (approximately negative binom) give an approx. answer.
fit0 <- glm(
Abundance ~ 1 + tumor,
data= dat0,
family=quasipoisson(link = "log")
)
summary(fit0)
Call:
glm(formula = Abundance ~ 1 + tumor, family = quasipoisson(link = "log"),
data = dat0)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.392 -1.241 -0.906 0.232 5.513
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.0311 0.1770 -0.18 0.86
tumor -0.2305 0.2963 -0.78 0.44
(Dispersion parameter for quasipoisson family taken to be 2.8254)
Null deviance: 293.35 on 157 degrees of freedom
Residual deviance: 291.61 on 156 degrees of freedom
AIC: NA
Number of Fisher Scoring iterations: 6Run over all descriptors
results.out <- as.data.frame(matrix(ncol=4, nrow=length(DESCRIPTIONS)))
colnames(results.out) <- c("description", "Est", "SE", "p")
#results.out$description <-DESCRIPTIONS
i <- 1
for(i in 1:length(DESCRIPTIONS)){
#for(i in 1:5){
dat0 <- mydata %>%
filter(description == DESCRIPTIONS[i])
fit0 <- glm(
Abundance ~ 1 + tumor,
data= dat0,
family=quasipoisson(link = "log")
)
fit.sum <- summary(fit0)
results.out[i, 1] <- DESCRIPTIONS[i]
results.out[i, 2:4] <- fit.sum$coefficients[2, c(1,2,4)]
}
results.out$fdr_p <- p.adjust(results.out$p, method="fdr")
kable(results.out, format="html", digits=3) %>%
kable_styling(full_width = T)%>%
scroll_box(width="100%", height="600px")| description | Est | SE | p | fdr_p |
|---|---|---|---|---|
| (2E,6E)-farnesyl diphosphate synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| (4S)-4-hydroxy-2-oxoglutarate aldolase | -0.628 | 0.582 | 0.282 | 0.635 |
| (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase (ferredoxin) | -0.230 | 0.296 | 0.438 | 0.635 |
| (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase (flavodoxin) | -0.230 | 0.296 | 0.438 | 0.635 |
| (Kdo)-lipid IV(A) 3-deoxy-D-manno-octulosonic acid transferase | -0.230 | 0.296 | 0.438 | 0.635 |
| (Kdo)(2)-lipid IV(A) (2-8) 3-deoxy-D-manno-octulosonic acid transferase | -0.230 | 0.296 | 0.438 | 0.635 |
| (Kdo)(3)-lipid IV(A) (2-4) 3-deoxy-D-manno-octulosonic acid transferase | -0.230 | 0.296 | 0.438 | 0.635 |
| (R)-2-methylmalate dehydratase | -0.100 | 1.127 | 0.930 | 0.968 |
| (R)-citramalate synthase | -0.100 | 1.127 | 0.930 | 0.968 |
| [Acyl-carrier-protein] S-malonyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| [Formate-C-acetyltransferase]-activating enzyme | -0.204 | 0.299 | 0.496 | 0.635 |
| [Ribosomal protein S12] (aspartate(89)-C(3))-methylthiotransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| 1-acylglycerol-3-phosphate O-acyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| 1-deoxy-D-xylulose-5-phosphate reductoisomerase | -0.230 | 0.296 | 0.438 | 0.635 |
| 1-deoxy-D-xylulose-5-phosphate synthase | -0.211 | 0.305 | 0.489 | 0.635 |
| 1,4-alpha-glucan branching enzyme | -0.230 | 0.296 | 0.438 | 0.635 |
| 1,4-beta-mannosyl-N-acetylglucosamine phosphorylase | -0.234 | 0.288 | 0.419 | 0.635 |
| 1,4-dihydroxy-2-naphthoate polyprenyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| 1,4-dihydroxy-2-naphthoyl-CoA synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| 16S rRNA (adenine(1518)-N(6)/adenine(1519)-N(6))-dimethyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| 16S rRNA (cytidine(1402)-2’-O)-methyltransferase | -0.242 | 0.294 | 0.412 | 0.635 |
| 16S rRNA (cytosine(1402)-N(4))-methyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| 16S rRNA (guanine(527)-N(7))-methyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| 16S rRNA (uracil(1498)-N(3))-methyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| 2’,3’-cyclic-nucleotide 2’-phosphodiesterase | -0.285 | 0.310 | 0.360 | 0.635 |
| 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine diphosphokinase | -0.240 | 0.304 | 0.431 | 0.635 |
| 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase | -0.232 | 0.307 | 0.452 | 0.635 |
| 2-dehydro-3-deoxy-phosphogluconate aldolase | -0.628 | 0.582 | 0.282 | 0.635 |
| 2-dehydro-3-deoxygluconokinase | -0.577 | 0.590 | 0.329 | 0.635 |
| 2-dehydropantoate 2-reductase | -0.220 | 0.329 | 0.504 | 0.635 |
| 2-iminoacetate synthase | 18.519 | 3411.091 | 0.996 | 0.997 |
| 2-iminobutanoate/2-iminopropanoate deaminase | -0.230 | 0.296 | 0.438 | 0.635 |
| 2-isopropylmalate synthase | -0.100 | 1.127 | 0.930 | 0.968 |
| 2-methoxy-6-polyprenyl-1,4-benzoquinol methylase | -0.230 | 0.296 | 0.438 | 0.635 |
| 2-oxoacid oxidoreductase (ferredoxin) | -0.241 | 0.306 | 0.433 | 0.635 |
| 2-oxoglutarate synthase | -0.237 | 0.303 | 0.434 | 0.635 |
| 23S rRNA (adenine(2085)-N(6))-dimethyltransferase | 0.141 | 0.367 | 0.701 | 0.790 |
| 23S rRNA (adenine(2503)-C(2))-methyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| 23S rRNA (cytosine(1962)-C(5))-methyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| 23S rRNA (pseudouridine(1915)-N(3))-methyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| 23S rRNA (uracil(1939)-C(5))-methyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| 23S rRNA pseudouridine(1911/1915/1917) synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| 23S rRNA pseudouridine(2605) synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| 23S rRNA pseudouridine(746) synthase | -0.223 | 0.336 | 0.508 | 0.635 |
| 3’-nucleotidase | -0.285 | 0.310 | 0.360 | 0.635 |
| 3’(2’),5’-bisphosphate nucleotidase | -0.242 | 0.294 | 0.412 | 0.635 |
| 3-dehydroquinate dehydratase | -0.230 | 0.296 | 0.438 | 0.635 |
| 3-dehydroquinate synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| 3-deoxy-8-phosphooctulonate synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| 3-deoxy-manno-octulosonate-8-phosphatase | -0.232 | 0.301 | 0.441 | 0.635 |
| 3-deoxy-manno-octulosonate cytidylyltransferase | -0.243 | 0.292 | 0.407 | 0.635 |
| 3-hydroxyacyl-[acyl-carrier-protein] dehydratase | -0.331 | 0.309 | 0.285 | 0.635 |
| 3-isopropylmalate dehydratase | -0.100 | 1.127 | 0.930 | 0.968 |
| 3-isopropylmalate dehydrogenase | -0.100 | 1.127 | 0.930 | 0.968 |
| 3-methyl-2-oxobutanoate hydroxymethyltransferase | -0.213 | 0.298 | 0.477 | 0.635 |
| 3-oxoacyl-[acyl-carrier-protein] reductase | -0.342 | 0.306 | 0.265 | 0.635 |
| 3-phosphoshikimate 1-carboxyvinyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| 3,4-dihydroxy-2-butanone-4-phosphate synthase | -0.226 | 0.295 | 0.445 | 0.635 |
| 4-(cytidine 5’-diphospho)-2-C-methyl-D-erythritol kinase | -0.230 | 0.296 | 0.438 | 0.635 |
| 4-alpha-glucanotransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| 4-carboxymuconolactone decarboxylase | -0.135 | 0.612 | 0.825 | 0.905 |
| 4-hydroxy-3-methylbut-2-enyl diphosphate reductase | -0.230 | 0.296 | 0.438 | 0.635 |
| 4-hydroxy-tetrahydrodipicolinate reductase | -0.230 | 0.296 | 0.438 | 0.635 |
| 4-hydroxy-tetrahydrodipicolinate synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| 4-hydroxythreonine-4-phosphate dehydrogenase | -0.230 | 0.296 | 0.438 | 0.635 |
| 4-O-beta-D-mannosyl-D-glucose phosphorylase | -0.588 | 0.587 | 0.318 | 0.635 |
| 4-phosphoerythronate dehydrogenase | -0.754 | 1.120 | 0.502 | 0.635 |
| 5’-nucleotidase | -0.225 | 0.298 | 0.452 | 0.635 |
| 5’ to 3’ exodeoxyribonuclease (nucleoside 3’-phosphate-forming) | -0.789 | 0.881 | 0.372 | 0.635 |
| 5-(carboxyamino)imidazole ribonucleotide mutase | -0.230 | 0.296 | 0.438 | 0.635 |
| 5-amino-6-(5-phosphoribosylamino)uracil reductase | -0.230 | 0.296 | 0.438 | 0.635 |
| 5-dehydro-4-deoxy-D-glucuronate isomerase | -0.676 | 0.652 | 0.302 | 0.635 |
| 5-formyltetrahydrofolate cyclo-ligase | -0.230 | 0.296 | 0.438 | 0.635 |
| 6-carboxytetrahydropterin synthase | -0.395 | 0.451 | 0.383 | 0.635 |
| 6-phosphofructokinase | -0.230 | 0.296 | 0.438 | 0.635 |
| 6-pyruvoyltetrahydropterin synthase | -0.395 | 0.451 | 0.383 | 0.635 |
| 6,7-dimethyl-8-ribityllumazine synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| 7-cyano-7-deazaguanine synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| 8-amino-7-oxononanoate synthase | -0.754 | 1.120 | 0.502 | 0.635 |
| 8-oxo-dGTP diphosphatase | -0.128 | 0.324 | 0.693 | 0.790 |
| Acetate–CoA ligase | -0.203 | 0.301 | 0.500 | 0.635 |
| Acetate kinase | -0.230 | 0.296 | 0.438 | 0.635 |
| Acetolactate synthase | -0.548 | 0.584 | 0.350 | 0.635 |
| acetylglucosaminyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| Acetylglutamate kinase | -0.100 | 1.127 | 0.930 | 0.968 |
| Acetylornithine deacetylase | -0.100 | 1.127 | 0.930 | 0.968 |
| Acetylornithine transaminase | -0.100 | 1.127 | 0.930 | 0.968 |
| Acid phosphatase | -0.073 | 0.305 | 0.811 | 0.902 |
| Aconitate hydratase | -0.429 | 1.387 | 0.758 | 0.845 |
| Acyl-[acyl-carrier-protein]–UDP-N-acetylglucosamine O-acyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| Adenine phosphoribosyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| Adenosylcobalamin/alpha-ribazole phosphatase | -0.680 | 0.538 | 0.208 | 0.635 |
| Adenosylcobinamide-GDP ribazoletransferase | -0.680 | 0.538 | 0.208 | 0.635 |
| Adenosylcobinamide-phosphate guanylyltransferase | -0.680 | 0.538 | 0.208 | 0.635 |
| Adenosylcobinamide kinase | -0.680 | 0.538 | 0.208 | 0.635 |
| Adenosylhomocysteine nucleosidase | -0.230 | 0.296 | 0.438 | 0.635 |
| Adenylate kinase | -0.230 | 0.296 | 0.438 | 0.635 |
| Adenylosuccinate lyase | -0.230 | 0.296 | 0.438 | 0.635 |
| Adenylosuccinate synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| Adenylyl-sulfate kinase | -0.754 | 1.120 | 0.502 | 0.635 |
| Alanine–tRNA ligase | -0.230 | 0.296 | 0.438 | 0.635 |
| Alanine dehydrogenase | -0.647 | 0.554 | 0.244 | 0.635 |
| Alanine racemase | -0.230 | 0.296 | 0.438 | 0.635 |
| Aldose 1-epimerase | -0.243 | 0.306 | 0.429 | 0.635 |
| Alkaline phosphatase | -0.400 | 0.466 | 0.392 | 0.635 |
| All-trans-octaprenyl-diphosphate synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| All-trans-retinol 13,14-reductase | -0.696 | 0.523 | 0.185 | 0.635 |
| Alpha-amylase | -0.248 | 0.296 | 0.404 | 0.635 |
| Alpha-D-xyloside xylohydrolase | -0.603 | 0.561 | 0.284 | 0.635 |
| Alpha-galactosidase | -0.168 | 0.287 | 0.561 | 0.685 |
| Alpha-glucosidase | -0.117 | 0.305 | 0.702 | 0.790 |
| Alpha-glucuronidase | -0.788 | 0.881 | 0.373 | 0.635 |
| Alpha-L-fucosidase | -0.244 | 0.296 | 0.412 | 0.635 |
| Alpha-L-rhamnosidase | -0.319 | 0.983 | 0.746 | 0.837 |
| Alpha-N-acetylglucosaminidase | -0.855 | 0.801 | 0.287 | 0.635 |
| Altronate dehydratase | -0.676 | 0.652 | 0.302 | 0.635 |
| Amidophosphoribosyltransferase | -0.219 | 0.308 | 0.477 | 0.635 |
| Aminoacyl-tRNA hydrolase | -0.230 | 0.296 | 0.438 | 0.635 |
| Aminodeoxychorismate lyase | -0.158 | 0.328 | 0.632 | 0.736 |
| Aminodeoxychorismate synthase | -0.158 | 0.328 | 0.632 | 0.736 |
| Aminomethyltransferase | -0.190 | 0.321 | 0.556 | 0.680 |
| AMP nucleosidase | -0.754 | 1.120 | 0.502 | 0.635 |
| Anthranilate phosphoribosyltransferase | -0.089 | 1.133 | 0.938 | 0.969 |
| Anthranilate synthase | -0.163 | 0.338 | 0.630 | 0.736 |
| Arabinan endo-1,5-alpha-L-arabinosidase | -0.573 | 0.507 | 0.260 | 0.635 |
| Arabinogalactan endo-beta-1,4-galactanase | -0.483 | 0.529 | 0.363 | 0.635 |
| Arabinose-5-phosphate isomerase | -0.230 | 0.296 | 0.438 | 0.635 |
| Arginine–tRNA ligase | -0.230 | 0.296 | 0.438 | 0.635 |
| Arginine decarboxylase | -0.230 | 0.296 | 0.438 | 0.635 |
| Argininosuccinate lyase | -0.100 | 1.127 | 0.930 | 0.968 |
| Argininosuccinate synthase | -0.100 | 1.127 | 0.930 | 0.968 |
| Arylsulfatase | -0.131 | 0.339 | 0.701 | 0.790 |
| Asparaginase | -0.260 | 0.336 | 0.441 | 0.635 |
| Asparagine–tRNA ligase | -0.230 | 0.296 | 0.438 | 0.635 |
| Asparagine synthase (glutamine-hydrolyzing) | 18.519 | 3411.091 | 0.996 | 0.997 |
| Aspartate–ammonia ligase | -0.213 | 0.293 | 0.468 | 0.635 |
| Aspartate–tRNA ligase | -0.230 | 0.296 | 0.438 | 0.635 |
| Aspartate-semialdehyde dehydrogenase | -0.230 | 0.296 | 0.438 | 0.635 |
| Aspartate 1-decarboxylase | -0.213 | 0.298 | 0.477 | 0.635 |
| Aspartate ammonia-lyase | -0.259 | 0.322 | 0.423 | 0.635 |
| Aspartate carbamoyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| Aspartate kinase | -0.249 | 0.296 | 0.402 | 0.635 |
| Aspartate transaminase | -0.235 | 0.289 | 0.416 | 0.635 |
| Aspartyl aminopeptidase | 18.519 | 3411.091 | 0.996 | 0.997 |
| ATP phosphoribosyltransferase | -0.100 | 1.127 | 0.930 | 0.968 |
| Bacterial non-heme ferritin | -0.219 | 0.301 | 0.468 | 0.635 |
| Beta-1,4-mannooligosaccharide phosphorylase | -0.234 | 0.288 | 0.419 | 0.635 |
| Beta-fructofuranosidase | -0.496 | 0.487 | 0.311 | 0.635 |
| Beta-galactosidase | -0.246 | 0.296 | 0.408 | 0.635 |
| Beta-glucosidase | -0.399 | 0.427 | 0.352 | 0.635 |
| Beta-ketoacyl-[acyl-carrier-protein] synthase I | -0.741 | 0.488 | 0.131 | 0.635 |
| Beta-ketoacyl-[acyl-carrier-protein] synthase II | -0.230 | 0.296 | 0.438 | 0.635 |
| Beta-ketoacyl-[acyl-carrier-protein] synthase III | -0.223 | 0.335 | 0.506 | 0.635 |
| Beta-lactamase | -0.298 | 0.574 | 0.604 | 0.718 |
| Beta-N-acetylhexosaminidase | -0.212 | 0.270 | 0.432 | 0.635 |
| Biotin–[acetyl-CoA-carboxylase] ligase | -0.230 | 0.296 | 0.438 | 0.635 |
| Bleomycin hydrolase | -0.224 | 0.296 | 0.452 | 0.635 |
| Branched-chain-amino-acid transaminase | -0.230 | 0.296 | 0.438 | 0.635 |
| Butyrate kinase | -0.147 | 0.329 | 0.655 | 0.755 |
| C-terminal processing peptidase | -0.230 | 0.296 | 0.438 | 0.635 |
| Cadmium-exporting ATPase | -0.201 | 0.284 | 0.481 | 0.635 |
| Calcium-transporting ATPase | -0.156 | 0.300 | 0.605 | 0.718 |
| Carbamoyl-phosphate synthase (glutamine-hydrolyzing) | -0.232 | 0.298 | 0.439 | 0.635 |
| Carbonate dehydratase | -0.169 | 0.333 | 0.614 | 0.727 |
| Carboxynorspermidine decarboxylase | -0.230 | 0.296 | 0.438 | 0.635 |
| CDP-diacylglycerol–glycerol-3-phosphate 3-phosphatidyltransferase | -0.227 | 0.517 | 0.662 | 0.761 |
| CDP-diacylglycerol–serine O-phosphatidyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| CDP-glycerol glycerophosphotransferase | 0.092 | 1.404 | 0.948 | 0.979 |
| Cellobiose phosphorylase | 0.154 | 1.349 | 0.909 | 0.968 |
| Cellulase | 18.519 | 3411.091 | 0.996 | 0.997 |
| Choline-phosphate cytidylyltransferase | -0.221 | 0.572 | 0.699 | 0.790 |
| Chondroitin-sulfate-ABC endolyase | -0.677 | 0.652 | 0.301 | 0.635 |
| Chondroitin-sulfate-ABC exolyase | -0.677 | 0.652 | 0.301 | 0.635 |
| Chorismate mutase | -0.230 | 0.296 | 0.438 | 0.635 |
| Chorismate synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| Citrate (Si)-synthase | 18.519 | 3411.091 | 0.996 | 0.997 |
| Citrate CoA-transferase | 2.304 | 1.989 | 0.248 | 0.635 |
| Citrate lyase holo-[acyl-carrier protein] synthase | 2.304 | 1.989 | 0.248 | 0.635 |
| Citryl-CoA lyase | 2.304 | 1.989 | 0.248 | 0.635 |
| Cob(I)yrinic acid a,c-diamide adenosyltransferase | -0.256 | 0.335 | 0.446 | 0.635 |
| Cobaltochelatase | -0.341 | 0.419 | 0.416 | 0.635 |
| Coproporphyrinogen dehydrogenase | -0.227 | 0.294 | 0.441 | 0.635 |
| Creatininase | -2.232 | 1.933 | 0.250 | 0.635 |
| Crossover junction endodeoxyribonuclease | -0.230 | 0.296 | 0.438 | 0.635 |
| CTP synthase (glutamine hydrolyzing) | -0.230 | 0.296 | 0.438 | 0.635 |
| Cu(2+)-exporting ATPase | -0.224 | 0.285 | 0.434 | 0.635 |
| Cyclic pyranopterin phosphate synthase | -0.137 | 0.613 | 0.823 | 0.905 |
| Cystathionine beta-lyase | -0.267 | 0.319 | 0.403 | 0.635 |
| Cysteine–tRNA ligase | -0.230 | 0.296 | 0.438 | 0.635 |
| Cysteine desulfurase | -0.230 | 0.296 | 0.438 | 0.635 |
| Cysteine synthase | -0.260 | 0.313 | 0.407 | 0.635 |
| Cytidine deaminase | -0.230 | 0.296 | 0.438 | 0.635 |
| D-Ala-D-Ala dipeptidase | -0.199 | 0.288 | 0.490 | 0.635 |
| D-alanine–D-alanine ligase | -0.230 | 0.296 | 0.438 | 0.635 |
| D-glycero-alpha-D-manno-heptose-1,7-bisphosphate 7-phosphatase | 18.519 | 3411.091 | 0.996 | 0.997 |
| D-glycero-alpha-D-manno-heptose-7-phosphate kinase | 18.519 | 3411.091 | 0.996 | 0.997 |
| D-glycero-beta-D-manno-heptose 1,7-bisphosphate 7-phosphatase | 18.519 | 3411.091 | 0.996 | 0.997 |
| D-lactate dehydrogenase | -0.088 | 0.288 | 0.760 | 0.846 |
| D-sedoheptulose 7-phosphate isomerase | 18.519 | 3411.091 | 0.996 | 0.997 |
| dCMP deaminase | -0.230 | 0.296 | 0.438 | 0.635 |
| Demethylmenaquinone methyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| Deoxyribose-phosphate aldolase | -0.230 | 0.296 | 0.438 | 0.635 |
| Dephospho-CoA kinase | -0.228 | 0.308 | 0.459 | 0.635 |
| Dextranase | -0.282 | 0.509 | 0.580 | 0.704 |
| dGTPase | -0.230 | 0.296 | 0.438 | 0.635 |
| Diamine N-acetyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| Diaminohydroxyphosphoribosylaminopyrimidine deaminase | -0.230 | 0.296 | 0.438 | 0.635 |
| Diaminopimelate decarboxylase | -0.274 | 0.307 | 0.374 | 0.635 |
| Diaminopimelate dehydrogenase | -0.228 | 0.293 | 0.436 | 0.635 |
| Diaminopimelate epimerase | -0.394 | 0.395 | 0.320 | 0.635 |
| Dihydrofolate reductase | -0.201 | 0.284 | 0.481 | 0.635 |
| Dihydrofolate synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| Dihydrolipoyl dehydrogenase | -0.190 | 0.321 | 0.556 | 0.680 |
| Dihydroneopterin aldolase | -0.230 | 0.296 | 0.438 | 0.635 |
| Dihydroorotase | -0.220 | 0.308 | 0.477 | 0.635 |
| Dihydroorotate dehydrogenase (NAD(+)) | -0.230 | 0.296 | 0.438 | 0.635 |
| Dihydropteroate synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| Dihydroxy-acid dehydratase | -0.548 | 0.584 | 0.350 | 0.635 |
| Dimethylallyltranstransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| Dipeptidase E | -0.619 | 0.514 | 0.230 | 0.635 |
| Dipeptidyl-peptidase III | -0.230 | 0.296 | 0.438 | 0.635 |
| Dipeptidyl-peptidase IV | -0.230 | 0.296 | 0.438 | 0.635 |
| Diphosphate–fructose-6-phosphate 1-phosphotransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| diphosphate specific) | -0.230 | 0.296 | 0.438 | 0.635 |
| DNA-(apurinic or apyrimidinic site) lyase | -0.138 | 0.295 | 0.642 | 0.744 |
| DNA-3-methyladenine glycosylase I | -0.227 | 0.333 | 0.497 | 0.635 |
| DNA-directed DNA polymerase | -0.228 | 0.295 | 0.441 | 0.635 |
| DNA-directed RNA polymerase | -0.227 | 0.300 | 0.450 | 0.635 |
| DNA (cytosine-5-)-methyltransferase | -0.244 | 0.306 | 0.427 | 0.635 |
| DNA helicase | -0.238 | 0.300 | 0.428 | 0.635 |
| DNA ligase (NAD(+)) | -0.230 | 0.296 | 0.438 | 0.635 |
| DNA topoisomerase | -0.294 | 0.331 | 0.376 | 0.635 |
| DNA topoisomerase (ATP-hydrolyzing) | -0.230 | 0.296 | 0.438 | 0.635 |
| Dolichyl-phosphate beta-D-mannosyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| dTDP-4-dehydrorhamnose 3,5-epimerase | -0.230 | 0.296 | 0.438 | 0.635 |
| dTDP-4-dehydrorhamnose reductase | -0.230 | 0.296 | 0.438 | 0.635 |
| dTDP-glucose 4,6-dehydratase | -0.230 | 0.296 | 0.438 | 0.635 |
| dUTP diphosphatase | -0.230 | 0.296 | 0.438 | 0.635 |
| Endo-1,4-beta-xylanase | 18.519 | 3411.091 | 0.996 | 0.997 |
| Endopeptidase Clp | -0.230 | 0.296 | 0.438 | 0.635 |
| Endopeptidase La | -0.220 | 0.308 | 0.477 | 0.635 |
| Enoyl-[acyl-carrier-protein] reductase (NADH) | -0.219 | 0.301 | 0.468 | 0.635 |
| Enoyl-[acyl-carrier-protein] reductase (NADPH, Si-specific) | -0.219 | 0.301 | 0.468 | 0.635 |
| Epoxyqueuosine reductase | -0.230 | 0.296 | 0.438 | 0.635 |
| Exo-1,4-beta-D-glucosaminidase | -2.232 | 1.933 | 0.250 | 0.635 |
| Exo-alpha-sialidase | -0.234 | 0.288 | 0.419 | 0.635 |
| Exodeoxyribonuclease III | -0.138 | 0.295 | 0.642 | 0.744 |
| Exodeoxyribonuclease V | -0.754 | 1.120 | 0.502 | 0.635 |
| Exodeoxyribonuclease VII | -0.230 | 0.296 | 0.438 | 0.635 |
| FAD synthetase | -0.230 | 0.296 | 0.438 | 0.635 |
| FMN reductase (NAD(P)H) | -0.230 | 0.296 | 0.438 | 0.635 |
| Formate–tetrahydrofolate ligase | -0.263 | 0.312 | 0.401 | 0.635 |
| Formate C-acetyltransferase | -0.215 | 0.295 | 0.468 | 0.635 |
| Formimidoyltetrahydrofolate cyclodeaminase | -0.162 | 0.333 | 0.626 | 0.734 |
| Formyltetrahydrofolate deformylase | -0.789 | 0.881 | 0.372 | 0.635 |
| Fructan beta-fructosidase | -0.268 | 0.313 | 0.394 | 0.635 |
| Fructokinase | -0.239 | 0.302 | 0.431 | 0.635 |
| Fructose-bisphosphatase | -0.230 | 0.296 | 0.438 | 0.635 |
| Fructose-bisphosphate aldolase | -0.230 | 0.296 | 0.438 | 0.635 |
| Fructuronate reductase | -0.677 | 0.652 | 0.301 | 0.635 |
| Fumarate hydratase | -0.228 | 0.345 | 0.510 | 0.635 |
| Fumarate reductase (quinol) | -0.230 | 0.296 | 0.438 | 0.635 |
| Galactokinase | -0.224 | 0.269 | 0.406 | 0.635 |
| Galactoside O-acetyltransferase | -0.062 | 0.367 | 0.865 | 0.945 |
| GDP-L-fucose synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| GDP-mannose 4,6-dehydratase | -0.230 | 0.296 | 0.438 | 0.635 |
| Geranylgeranyl diphosphate synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| Gingipain R | -0.001 | 1.049 | 0.999 | 0.999 |
| Glucokinase | -0.268 | 0.292 | 0.361 | 0.635 |
| Gluconate 5-dehydrogenase | -0.542 | 0.584 | 0.355 | 0.635 |
| Glucosamine-6-phosphate deaminase | -0.226 | 0.290 | 0.438 | 0.635 |
| Glucose-1-phosphatase | -0.072 | 0.305 | 0.815 | 0.904 |
| Glucose-1-phosphate thymidylyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| Glucose-6-phosphate isomerase | -0.230 | 0.296 | 0.438 | 0.635 |
| Glucosylceramidase | 0.151 | 1.352 | 0.911 | 0.968 |
| Glucuronate isomerase | -0.839 | 0.582 | 0.151 | 0.635 |
| Glutamate–ammonia ligase | -0.204 | 0.294 | 0.489 | 0.635 |
| Glutamate–tRNA ligase | -0.230 | 0.296 | 0.438 | 0.635 |
| Glutamate-5-semialdehyde dehydrogenase | 18.519 | 3411.091 | 0.996 | 0.997 |
| Glutamate 5-kinase | 18.519 | 3411.091 | 0.996 | 0.997 |
| Glutamate dehydrogenase (NADP(+)) | -0.154 | 0.290 | 0.596 | 0.718 |
| Glutamate formimidoyltransferase | -0.162 | 0.333 | 0.626 | 0.734 |
| Glutamate racemase | -0.230 | 0.296 | 0.438 | 0.635 |
| Glutamate synthase (NADH) | -0.323 | 0.318 | 0.311 | 0.635 |
| Glutamate synthase (NADPH) | -0.323 | 0.318 | 0.311 | 0.635 |
| Glutamine–fructose-6-phosphate transaminase (isomerizing) | -0.243 | 0.311 | 0.436 | 0.635 |
| Glutamine–tRNA ligase | -0.230 | 0.296 | 0.438 | 0.635 |
| Glutathione peroxidase | -0.203 | 0.301 | 0.502 | 0.635 |
| Glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) | -0.230 | 0.296 | 0.438 | 0.635 |
| Glycerate 3-kinase | -0.206 | 0.323 | 0.524 | 0.647 |
| Glycerate dehydrogenase | -0.230 | 0.296 | 0.438 | 0.635 |
| Glycerol-3-phosphate cytidylyltransferase | 0.358 | 0.550 | 0.516 | 0.641 |
| Glycerol-3-phosphate dehydrogenase | -0.228 | 0.331 | 0.492 | 0.635 |
| Glycerol-3-phosphate dehydrogenase (NAD(P)(+)) | -0.230 | 0.296 | 0.438 | 0.635 |
| Glycerol kinase | -0.075 | 0.341 | 0.826 | 0.905 |
| Glycerophosphodiester phosphodiesterase | -0.647 | 0.496 | 0.194 | 0.635 |
| Glycine–tRNA ligase | -0.230 | 0.296 | 0.438 | 0.635 |
| Glycine C-acetyltransferase | -0.323 | 0.372 | 0.387 | 0.635 |
| Glycine dehydrogenase (aminomethyl-transferring) | -0.190 | 0.321 | 0.556 | 0.680 |
| Glycine hydroxymethyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| Glycogen phosphorylase | -0.230 | 0.296 | 0.438 | 0.635 |
| GMP synthase (glutamine-hydrolyzing) | -0.230 | 0.296 | 0.438 | 0.635 |
| GTP cyclohydrolase I | -0.201 | 0.284 | 0.481 | 0.635 |
| GTP cyclohydrolase II | -0.231 | 0.296 | 0.437 | 0.635 |
| GTP diphosphokinase | -0.231 | 0.299 | 0.439 | 0.635 |
| Guanylate kinase | -0.230 | 0.296 | 0.438 | 0.635 |
| H(+)-transporting two-sector ATPase | -0.237 | 0.295 | 0.423 | 0.635 |
| Hippurate hydrolase | -1.546 | 2.070 | 0.456 | 0.635 |
| Histidine–tRNA ligase | -0.230 | 0.296 | 0.438 | 0.635 |
| Histidine ammonia-lyase | -0.162 | 0.333 | 0.626 | 0.734 |
| Histidine kinase | -0.226 | 0.296 | 0.445 | 0.635 |
| Histidinol-phosphatase | -0.100 | 1.127 | 0.930 | 0.968 |
| Histidinol-phosphate transaminase | -0.159 | 0.284 | 0.576 | 0.702 |
| Histidinol dehydrogenase | -0.100 | 1.127 | 0.930 | 0.968 |
| Homoserine dehydrogenase | -0.274 | 0.307 | 0.374 | 0.635 |
| Homoserine O-succinyltransferase | -0.274 | 0.307 | 0.374 | 0.635 |
| Hyaluronoglucosaminidase | -0.247 | 0.317 | 0.437 | 0.635 |
| Hydroxylamine reductase | -0.206 | 0.301 | 0.494 | 0.635 |
| Hydroxymethylpyrimidine kinase | -0.134 | 0.312 | 0.669 | 0.765 |
| Hypoxanthine phosphoribosyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| Imidazoleglycerol-phosphate dehydratase | -0.100 | 1.127 | 0.930 | 0.968 |
| Imidazolonepropionase | -0.162 | 0.333 | 0.626 | 0.734 |
| IMP cyclohydrolase | -0.230 | 0.296 | 0.438 | 0.635 |
| IMP dehydrogenase | -0.230 | 0.296 | 0.438 | 0.635 |
| Indole-3-glycerol-phosphate synthase | -0.376 | 0.442 | 0.397 | 0.635 |
| Inorganic diphosphatase | -0.240 | 0.317 | 0.450 | 0.635 |
| Inositol-3-phosphate synthase | -0.144 | 0.334 | 0.667 | 0.765 |
| Iron-chelate-transporting ATPase | -0.230 | 0.296 | 0.438 | 0.635 |
| Isochorismate synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| Isocitrate dehydrogenase (NADP(+)) | -0.429 | 1.387 | 0.758 | 0.845 |
| Isoleucine–tRNA ligase | -0.220 | 0.308 | 0.477 | 0.635 |
| isomerase | -0.100 | 1.127 | 0.930 | 0.968 |
| Kdo(2)-lipid IV(A) lauroyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| Ketol-acid reductoisomerase (NADP(+)) | -0.548 | 0.584 | 0.350 | 0.635 |
| L-arabinose isomerase | -0.670 | 0.642 | 0.298 | 0.635 |
| L-aspartate oxidase | -0.172 | 0.328 | 0.602 | 0.718 |
| L-rhamnose isomerase | -0.810 | 0.830 | 0.330 | 0.635 |
| L-rhamnose mutarotase | -0.846 | 0.965 | 0.382 | 0.635 |
| L-ribulose-5-phosphate 4-epimerase | -0.670 | 0.642 | 0.298 | 0.635 |
| L-serine ammonia-lyase | -0.230 | 0.296 | 0.438 | 0.635 |
| L-threonine aldolase | -0.170 | 0.761 | 0.824 | 0.905 |
| L-threonylcarbamoyladenylate synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| Lactoylglutathione lyase | -1.045 | 0.919 | 0.257 | 0.635 |
| Leucine–tRNA ligase | -0.222 | 0.299 | 0.458 | 0.635 |
| Leucyltransferase | 0.056 | 0.493 | 0.910 | 0.968 |
| Levanase | -0.223 | 0.321 | 0.489 | 0.635 |
| Lipid-A-disaccharide synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| Lipid IV(A) 3-deoxy-D-manno-octulosonic acid transferase | -0.230 | 0.296 | 0.438 | 0.635 |
| Lipoate–protein ligase | -0.717 | 0.722 | 0.322 | 0.635 |
| Lipoyl synthase | -0.230 | 0.317 | 0.469 | 0.635 |
| LL-diaminopimelate aminotransferase | -0.394 | 0.395 | 0.320 | 0.635 |
| Long-chain-fatty-acid–CoA ligase | -0.247 | 0.302 | 0.414 | 0.635 |
| Lysine–tRNA ligase | -0.230 | 0.296 | 0.438 | 0.635 |
| Lysozyme | 0.038 | 0.447 | 0.932 | 0.968 |
| Malate dehydrogenase | -0.230 | 0.296 | 0.438 | 0.635 |
| Malate dehydrogenase (oxaloacetate-decarboxylating) (NADP(+)) | -0.230 | 0.296 | 0.438 | 0.635 |
| Maltose alpha-D-glucosyltransferase | -0.629 | 0.781 | 0.422 | 0.635 |
| Maltose O-acetyltransferase | -0.230 | 0.328 | 0.484 | 0.635 |
| Mannan endo-1,4-beta-mannosidase | -0.329 | 0.354 | 0.354 | 0.635 |
| Mannonate dehydratase | -0.692 | 0.616 | 0.263 | 0.635 |
| Mannose-1-phosphate guanylyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| Mannose-6-phosphate isomerase | -0.230 | 0.296 | 0.438 | 0.635 |
| Methenyltetrahydrofolate cyclohydrolase | -0.230 | 0.296 | 0.438 | 0.635 |
| Methionine–tRNA ligase | -0.230 | 0.296 | 0.438 | 0.635 |
| Methionine adenosyltransferase | -0.243 | 0.292 | 0.407 | 0.635 |
| Methionine synthase | -0.206 | 0.312 | 0.510 | 0.635 |
| Methionyl-tRNA formyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| Methionyl aminopeptidase | -0.224 | 0.289 | 0.440 | 0.635 |
| Methylaspartate ammonia-lyase | -0.221 | 0.572 | 0.699 | 0.790 |
| Methylaspartate mutase | -0.221 | 0.572 | 0.699 | 0.790 |
| Methylated-DNA–[protein]-cysteine S-methyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| Methylenetetrahydrofolate dehydrogenase (NADP(+)) | -0.230 | 0.296 | 0.438 | 0.635 |
| Methylenetetrahydrofolate reductase (NAD(P)H) | -0.264 | 0.307 | 0.391 | 0.635 |
| Methylmalonyl-CoA mutase | -0.246 | 0.310 | 0.427 | 0.635 |
| N-acetyl-gamma-glutamyl-phosphate reductase | -0.100 | 1.127 | 0.930 | 0.968 |
| N-acetylmuramic acid 6-phosphate etherase | -0.747 | 0.519 | 0.152 | 0.635 |
| N-acetylmuramoyl-L-alanine amidase | -0.285 | 0.315 | 0.367 | 0.635 |
| N-acetylneuraminate lyase | -0.206 | 0.295 | 0.486 | 0.635 |
| N-acetylneuraminate synthase | -2.232 | 1.933 | 0.250 | 0.635 |
| N-acylglucosamine 2-epimerase | -0.240 | 0.289 | 0.409 | 0.635 |
| N-acylneuraminate cytidylyltransferase | -2.232 | 1.933 | 0.250 | 0.635 |
| N-carbamoylputrescine amidase | -0.230 | 0.296 | 0.438 | 0.635 |
| N-succinylornithine carbamoyltransferase | -0.100 | 1.127 | 0.930 | 0.968 |
| N(6)-L-threonylcarbamoyladenine synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| NAD(+) diphosphatase | -0.230 | 0.296 | 0.438 | 0.635 |
| NAD(+) kinase | -0.230 | 0.296 | 0.438 | 0.635 |
| NAD(+) synthase (glutamine-hydrolyzing) | -0.230 | 0.296 | 0.438 | 0.635 |
| NAD(P)H-hydrate epimerase | -0.230 | 0.296 | 0.438 | 0.635 |
| NADH dehydrogenase | -0.211 | 0.290 | 0.469 | 0.635 |
| NADH:ubiquinone reductase (H(+)-translocating) | -0.230 | 0.296 | 0.438 | 0.635 |
| Nicotinamidase | -0.097 | 0.315 | 0.757 | 0.845 |
| Nicotinamide-nucleotide amidase | -0.230 | 0.296 | 0.438 | 0.635 |
| Nicotinate-nucleotide–dimethylbenzimidazole phosphoribosyltransferase | -0.680 | 0.538 | 0.208 | 0.635 |
| Nicotinate-nucleotide adenylyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| Nicotinate-nucleotide diphosphorylase (carboxylating) | -0.172 | 0.328 | 0.602 | 0.718 |
| Nicotinate phosphoribosyltransferase | -0.231 | 0.296 | 0.438 | 0.635 |
| Nitric-oxide reductase (cytochrome c) | 0.054 | 0.493 | 0.913 | 0.968 |
| Nitrite reductase (cytochrome; ammonia-forming) | -0.217 | 0.299 | 0.469 | 0.635 |
| Nitrous-oxide reductase | -0.319 | 0.983 | 0.746 | 0.837 |
| Non-reducing end alpha-L-arabinofuranosidase | -0.691 | 0.615 | 0.263 | 0.635 |
| Non-specific serine/threonine protein kinase | -0.044 | 0.314 | 0.889 | 0.968 |
| Nucleoside-diphosphate kinase | -0.754 | 1.120 | 0.502 | 0.635 |
| O-acetylhomoserine aminocarboxypropyltransferase | -0.208 | 0.306 | 0.498 | 0.635 |
| o-succinylbenzoate–CoA ligase | -0.219 | 0.301 | 0.468 | 0.635 |
| Oligonucleotidase | -0.230 | 0.296 | 0.438 | 0.635 |
| Orotate phosphoribosyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| Orotidine-5’-phosphate decarboxylase | -0.230 | 0.296 | 0.438 | 0.635 |
| Pantetheine-phosphate adenylyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| Pantoate–beta-alanine ligase (AMP-forming) | -0.213 | 0.298 | 0.477 | 0.635 |
| Pantothenate kinase | -0.230 | 0.296 | 0.438 | 0.635 |
| Pectinesterase | -0.674 | 0.651 | 0.302 | 0.635 |
| Peptide-methionine (R)-S-oxide reductase | -0.230 | 0.296 | 0.438 | 0.635 |
| Peptide-methionine (S)-S-oxide reductase | -0.230 | 0.296 | 0.438 | 0.635 |
| Peptide chain release factor N(5)-glutamine methyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| Peptide deformylase | -0.230 | 0.296 | 0.438 | 0.635 |
| Peptidyl-dipeptidase Dcp | -0.233 | 0.311 | 0.453 | 0.635 |
| Peptidylprolyl isomerase | -0.215 | 0.294 | 0.466 | 0.635 |
| Peroxiredoxin | -0.236 | 0.295 | 0.424 | 0.635 |
| Phenylalanine–tRNA ligase | -0.230 | 0.296 | 0.438 | 0.635 |
| Phosphate acetyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| Phosphate butyryltransferase | -0.147 | 0.329 | 0.655 | 0.755 |
| Phosphatidate cytidylyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| Phosphatidylglycerophosphatase | -0.015 | 0.569 | 0.979 | 0.997 |
| Phosphatidylserine decarboxylase | -0.230 | 0.296 | 0.438 | 0.635 |
| Phospho-N-acetylmuramoyl-pentapeptide-transferase | -0.230 | 0.296 | 0.438 | 0.635 |
| Phosphoadenylyl-sulfate reductase (thioredoxin) | 18.519 | 3411.091 | 0.996 | 0.997 |
| Phosphoenolpyruvate carboxykinase (ATP) | -0.230 | 0.296 | 0.438 | 0.635 |
| Phosphoglucomutase (alpha-D-glucose-1,6-bisphosphate-dependent) | -0.230 | 0.296 | 0.438 | 0.635 |
| Phosphoglycerate dehydrogenase | -0.228 | 0.293 | 0.436 | 0.635 |
| Phosphoglycerate kinase | -0.230 | 0.296 | 0.438 | 0.635 |
| Phosphoglycerate mutase (2,3-diphosphoglycerate-dependent) | 0.038 | 0.480 | 0.937 | 0.969 |
| Phosphoglycerate mutase (2,3-diphosphoglycerate-independent) | -0.291 | 0.314 | 0.356 | 0.635 |
| Phosphoglycolate phosphatase | -0.223 | 0.280 | 0.427 | 0.635 |
| Phosphomannomutase | -0.230 | 0.296 | 0.438 | 0.635 |
| Phosphomethylpyrimidine kinase | -0.134 | 0.312 | 0.669 | 0.765 |
| Phosphomethylpyrimidine synthase | 0.014 | 0.383 | 0.970 | 0.997 |
| Phosphopantothenate–cysteine ligase | -0.213 | 0.298 | 0.477 | 0.635 |
| Phosphopantothenoylcysteine decarboxylase | -0.213 | 0.298 | 0.477 | 0.635 |
| Phosphopyruvate hydratase | -0.230 | 0.296 | 0.438 | 0.635 |
| Phosphoribosyl-AMP cyclohydrolase | -0.100 | 1.127 | 0.930 | 0.968 |
| Phosphoribosyl-ATP diphosphatase | -0.100 | 1.127 | 0.930 | 0.968 |
| Phosphoribosyl 1,2-cyclic phosphate phosphodiesterase | -0.230 | 0.296 | 0.438 | 0.635 |
| Phosphoribosylamine–glycine ligase | -0.230 | 0.296 | 0.438 | 0.635 |
| Phosphoribosylaminoimidazolecarboxamide formyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| Phosphoribosylaminoimidazolesuccinocarboxamide synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| Phosphoribosylanthranilate isomerase | -0.219 | 0.301 | 0.468 | 0.635 |
| Phosphoribosylformylglycinamidine cyclo-ligase | -0.230 | 0.296 | 0.438 | 0.635 |
| Phosphoribosylformylglycinamidine synthase | -0.220 | 0.308 | 0.477 | 0.635 |
| Phosphoribosylglycinamide formyltransferase | -0.219 | 0.308 | 0.477 | 0.635 |
| Phosphoserine phosphatase | -0.237 | 0.319 | 0.459 | 0.635 |
| Phosphoserine transaminase | -0.228 | 0.293 | 0.436 | 0.635 |
| Polar-amino-acid-transporting ATPase | -2.168 | 1.874 | 0.249 | 0.635 |
| Polynucleotide adenylyltransferase | -0.754 | 1.120 | 0.502 | 0.635 |
| Polyribonucleotide nucleotidyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| Potassium-transporting ATPase | 2.304 | 1.989 | 0.248 | 0.635 |
| Prephenate dehydratase | -0.644 | 0.452 | 0.157 | 0.635 |
| PreQ(1) synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| Proline–tRNA ligase | -0.230 | 0.296 | 0.438 | 0.635 |
| Prolyl aminopeptidase | 18.519 | 3411.091 | 0.996 | 0.997 |
| Prolyl oligopeptidase | -0.210 | 0.327 | 0.521 | 0.645 |
| Protein-disulfide reductase | -0.230 | 0.296 | 0.438 | 0.635 |
| Protein-L-isoaspartate(D-aspartate) O-methyltransferase | -0.137 | 0.613 | 0.823 | 0.905 |
| Protein-tyrosine-phosphatase | -0.237 | 0.301 | 0.432 | 0.635 |
| Protoporphyrinogen oxidase | -0.232 | 0.293 | 0.430 | 0.635 |
| Pullulanase | -0.252 | 0.318 | 0.429 | 0.635 |
| Purine-nucleoside phosphorylase | -0.230 | 0.296 | 0.438 | 0.635 |
| Pyridoxal 5’-phosphate synthase (glutamine hydrolyzing) | -0.219 | 0.301 | 0.468 | 0.635 |
| Pyridoxal kinase | -0.186 | 0.306 | 0.545 | 0.671 |
| Pyridoxine 5’-phosphate synthase | -0.754 | 1.120 | 0.502 | 0.635 |
| Pyrroline-5-carboxylate reductase | 0.139 | 1.250 | 0.912 | 0.968 |
| Pyruvate carboxylase | -0.230 | 0.296 | 0.438 | 0.635 |
| Pyruvate kinase | -0.230 | 0.296 | 0.438 | 0.635 |
| Pyruvate synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| Pyruvate, phosphate dikinase | -0.230 | 0.296 | 0.438 | 0.635 |
| Quinolinate synthase | -0.172 | 0.328 | 0.602 | 0.718 |
| Rhamnulokinase | -0.810 | 0.830 | 0.330 | 0.635 |
| Rhamnulose-1-phosphate aldolase | -0.810 | 0.830 | 0.330 | 0.635 |
| Riboflavin kinase | -0.230 | 0.296 | 0.438 | 0.635 |
| Riboflavin synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| Ribonuclease H | -0.230 | 0.296 | 0.438 | 0.635 |
| Ribonuclease III | -0.217 | 0.299 | 0.469 | 0.635 |
| Ribonuclease P | -0.230 | 0.296 | 0.438 | 0.635 |
| Ribonuclease Z | -0.230 | 0.296 | 0.438 | 0.635 |
| Ribonucleoside-diphosphate reductase | -0.160 | 0.292 | 0.584 | 0.707 |
| Ribonucleoside-triphosphate reductase | -0.230 | 0.296 | 0.438 | 0.635 |
| Ribose-5-phosphate isomerase | -0.230 | 0.296 | 0.438 | 0.635 |
| Ribose-phosphate diphosphokinase | -0.230 | 0.296 | 0.438 | 0.635 |
| Ribulose-phosphate 3-epimerase | -0.230 | 0.296 | 0.438 | 0.635 |
| RNA-directed DNA polymerase | 2.304 | 1.989 | 0.248 | 0.635 |
| RNA helicase | -0.230 | 0.296 | 0.438 | 0.635 |
| RNA ligase (ATP) | -2.232 | 1.933 | 0.250 | 0.635 |
| S-adenosylmethionine:tRNA ribosyltransferase-isomerase | -0.225 | 0.301 | 0.457 | 0.635 |
| S-ribosylhomocysteine lyase | -0.263 | 0.312 | 0.401 | 0.635 |
| Saccharopine dehydrogenase (NAD(+), L-lysine-forming) | -0.230 | 0.296 | 0.438 | 0.635 |
| Selenocysteine lyase | -0.230 | 0.296 | 0.438 | 0.635 |
| Serine–tRNA ligase | -0.230 | 0.296 | 0.438 | 0.635 |
| Serine-type D-Ala-D-Ala carboxypeptidase | -0.288 | 0.314 | 0.360 | 0.635 |
| Serine O-acetyltransferase | -0.157 | 0.290 | 0.590 | 0.713 |
| Shikimate dehydrogenase | -0.230 | 0.296 | 0.438 | 0.635 |
| Shikimate kinase | -0.230 | 0.296 | 0.438 | 0.635 |
| Sialate O-acetylesterase | -0.283 | 0.313 | 0.368 | 0.635 |
| Signal peptidase I | -0.223 | 0.297 | 0.456 | 0.635 |
| Signal peptidase II | -0.230 | 0.296 | 0.438 | 0.635 |
| Site-specific DNA-methyltransferase (adenine-specific) | -0.214 | 0.306 | 0.485 | 0.635 |
| Starch synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| Succinate–CoA ligase (ADP-forming) | -0.173 | 0.329 | 0.600 | 0.718 |
| Succinate dehydrogenase (quinone) | -0.230 | 0.296 | 0.438 | 0.635 |
| Sulfate adenylyltransferase | -0.754 | 1.120 | 0.502 | 0.635 |
| Superoxide dismutase | -0.396 | 0.395 | 0.317 | 0.635 |
| Superoxide reductase | -0.166 | 0.321 | 0.605 | 0.718 |
| synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| Tagaturonate reductase | -0.676 | 0.652 | 0.302 | 0.635 |
| Tetraacyldisaccharide 4’-kinase | -0.230 | 0.296 | 0.438 | 0.635 |
| Tetrahydrofolate synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| Thiamine-phosphate diphosphorylase | -0.063 | 0.337 | 0.853 | 0.934 |
| Thiamine-phosphate kinase | -0.230 | 0.296 | 0.438 | 0.635 |
| Thiamine diphosphokinase | -0.198 | 0.307 | 0.519 | 0.643 |
| Thiazole synthase | 18.519 | 3411.091 | 0.996 | 0.997 |
| Thioredoxin-disulfide reductase | -0.230 | 0.296 | 0.438 | 0.635 |
| Threonine–tRNA ligase | -0.230 | 0.296 | 0.438 | 0.635 |
| Threonine ammonia-lyase | -0.378 | 0.462 | 0.414 | 0.635 |
| Threonine synthase | -0.606 | 0.561 | 0.282 | 0.635 |
| Thymidine kinase | -0.230 | 0.296 | 0.438 | 0.635 |
| Thymidylate synthase | -0.172 | 0.309 | 0.579 | 0.704 |
| Thymidylate synthase (FAD) | -0.625 | 0.575 | 0.278 | 0.635 |
| Transketolase | -0.234 | 0.306 | 0.446 | 0.635 |
| Triose-phosphate isomerase | -0.240 | 0.307 | 0.435 | 0.635 |
| Tripeptide aminopeptidase | -0.230 | 0.296 | 0.438 | 0.635 |
| tRNA-2-methylthio-N(6)-dimethylallyladenosine synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| tRNA-guanine(34) transglycosylase | -0.230 | 0.296 | 0.438 | 0.635 |
| tRNA (guanine(37)-N(1))-methyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| tRNA (guanine(46)-N(7))-methyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| tRNA (N(6)-L-threonylcarbamoyladenosine(37)-C(2))-methylthiotransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| tRNA dimethylallyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| tRNA pseudouridine(32) synthase | -0.223 | 0.336 | 0.508 | 0.635 |
| tRNA pseudouridine(38-40) synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| tRNA pseudouridine(55) synthase | -0.230 | 0.296 | 0.438 | 0.635 |
| tRNA(1)(Val) (adenine(37)-N(6))-methyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| tRNA(adenine(34)) deaminase | -0.230 | 0.296 | 0.438 | 0.635 |
| tRNA(Ile)-lysidine synthetase | -0.230 | 0.296 | 0.438 | 0.635 |
| Tryptophan–tRNA ligase | -0.230 | 0.296 | 0.438 | 0.635 |
| Tryptophan synthase | -0.229 | 0.301 | 0.447 | 0.635 |
| Tryptophanase | -0.221 | 0.572 | 0.699 | 0.790 |
| Type I site-specific deoxyribonuclease | -0.237 | 0.303 | 0.435 | 0.635 |
| Type II site-specific deoxyribonuclease | -0.343 | 0.461 | 0.459 | 0.635 |
| Type III site-specific deoxyribonuclease | -5.080 | 10.935 | 0.643 | 0.744 |
| Tyrosine–tRNA ligase | -0.230 | 0.296 | 0.438 | 0.635 |
| UDP-2,3-diacylglucosamine diphosphatase | -0.230 | 0.296 | 0.438 | 0.635 |
| UDP-3-O-(3-hydroxymyristoyl)glucosamine N-acyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| UDP-3-O-acyl-N-acetylglucosamine deacetylase | -0.230 | 0.296 | 0.438 | 0.635 |
| UDP-galactopyranose mutase | -0.337 | 0.418 | 0.421 | 0.635 |
| UDP-glucose 4-epimerase | -0.241 | 0.294 | 0.412 | 0.635 |
| UDP-glucose 6-dehydrogenase | -0.219 | 0.308 | 0.477 | 0.635 |
| UDP-glucuronate 4-epimerase | -0.057 | 0.574 | 0.921 | 0.968 |
| UDP-glucuronate decarboxylase | -0.137 | 0.613 | 0.823 | 0.905 |
| UDP-N-acetylglucosamine 1-carboxyvinyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| UDP-N-acetylglucosamine 2-epimerase (non-hydrolyzing) | -0.287 | 0.294 | 0.330 | 0.635 |
| UDP-N-acetylmuramate–L-alanine ligase | -0.230 | 0.296 | 0.438 | 0.635 |
| UDP-N-acetylmuramate dehydrogenase | -0.230 | 0.296 | 0.438 | 0.635 |
| UDP-N-acetylmuramoyl-L-alanine–D-glutamate ligase | -0.230 | 0.296 | 0.438 | 0.635 |
| UDP-N-acetylmuramoyl-L-alanyl-D-glutamate–2,6-diaminopimelate ligase | -0.230 | 0.296 | 0.438 | 0.635 |
| UDP-N-acetylmuramoyl-tripeptide–D-alanyl-D-alanine ligase | -0.230 | 0.296 | 0.438 | 0.635 |
| UMP kinase | -0.230 | 0.296 | 0.438 | 0.635 |
| UMP/CMP kinase | -0.230 | 0.296 | 0.438 | 0.635 |
| Undecaprenyl-diphosphate phosphatase | -0.274 | 0.301 | 0.364 | 0.635 |
| Uracil-DNA glycosylase | -0.204 | 0.307 | 0.507 | 0.635 |
| Uracil phosphoribosyltransferase | -0.230 | 0.296 | 0.438 | 0.635 |
| Uridine kinase | -0.230 | 0.296 | 0.438 | 0.635 |
| Uridine phosphorylase | -0.230 | 0.296 | 0.438 | 0.635 |
| Urocanate hydratase | -0.162 | 0.333 | 0.626 | 0.734 |
| Uroporphyrinogen-III synthase | -0.248 | 0.296 | 0.403 | 0.635 |
| Valine–tRNA ligase | -0.230 | 0.296 | 0.438 | 0.635 |
| Xaa-Pro aminopeptidase | -0.233 | 0.297 | 0.434 | 0.635 |
| Xanthine phosphoribosyltransferase | -0.156 | 0.300 | 0.605 | 0.718 |
| XTP/dITP diphosphatase | -0.230 | 0.296 | 0.438 | 0.635 |
| Xylose isomerase | -0.092 | 1.135 | 0.935 | 0.969 |
| Xylulokinase | -0.546 | 0.585 | 0.352 | 0.635 |
| Zinc-exporting ATPase | -0.201 | 0.284 | 0.481 | 0.635 |
# merge with tb.ra for full results table
full.res <- left_join(tb.ra, results.out, by = "description")
write.csv(full.res, "output/picrust_ec_stratefied_prevo_data_results.csv", row.names = F)Part 2 KO Data
descriptions <- readr::read_tsv("data/PICRUST/ko_pred_metagenome_unstrat_descrip.tsv")[,1:2] %>%
distinct()
-- Column specification --------------------------------------------------------
cols(
.default = col_double(),
`function` = col_character(),
description = col_character()
)
i Use `spec()` for the full column specifications.colnames(descriptions) <- c("func", "description")
pi.dat <- readxl::read_xlsx("data/PICRUST-Stratified/KO_prevo.xlsx")
colnames(pi.dat) <- c("func", colnames(pi.dat)[-c(1)])
# add descriptions to dataframe
pi.dat <- left_join(pi.dat, descriptions, by="func")
pi.dat <- pi.dat[,c(1,161,3:160)]
# aggregate descriptions to get 1 description per row - unique;y defined.
pi.dat <- pi.dat %>%
group_by(description) %>%
summarise(across(`X1.S37.Jun172016`:`X99.D01.S37.Jun232016`,.fns = mean))
# make long format
pi.dat <- pi.dat %>%
pivot_longer(
cols=`X1.S37.Jun172016`:`X99.D01.S37.Jun232016`,
names_to = "ID",
values_to = "Abundance"
) %>%
mutate(
ID = substr(ID, 2, 99)
)
d <- as.data.frame(dat.16s)
mydata <- full_join(pi.dat, d)Joining, by = "ID"Warning in class(x) <- c(setdiff(subclass, tibble_class), tibble_class): Setting
class(x) to multiple strings ("tbl_df", "tbl", ...); result will no longer be an
S4 objectmydata <- mydata %>%
mutate(
ID.n = as.numeric(as.factor(ID)),
description.n = as.numeric(as.factor(description))
) %>%
group_by(description)%>%
mutate(avgA = mean(Abundance))Abundance
# Run on all descriptions
tb.ra1 <- mydata %>%
group_by(description) %>%
summarise(ng = n(),
Overall.M = mean(Abundance),
Overall.SE = sd(Abundance)/sqrt(ng))
tb.ra2m <- mydata %>%
group_by(description, tumor.cat) %>%
summarise(M = mean(Abundance)) %>%
pivot_wider(id_cols = description,
names_from = tumor.cat,
values_from = M)`summarise()` has grouped output by 'description'. You can override using the `.groups` argument.tb.ra2se <- mydata %>%
group_by(description, tumor.cat) %>%
summarise(ng=n(),SE = sd(Abundance)/sqrt(ng)) %>%
pivot_wider(id_cols = description,
names_from = tumor.cat,
values_from = SE)`summarise()` has grouped output by 'description'. You can override using the `.groups` argument.tb.ra2var <- mydata %>%
group_by(description, tumor.cat) %>%
summarise(ng=n(), VAR = var(Abundance)) %>%
pivot_wider(id_cols = description,
names_from = tumor.cat,
values_from = VAR)`summarise()` has grouped output by 'description'. You can override using the `.groups` argument.tb.ra2ng <- mydata %>%
group_by(description, tumor.cat) %>%
summarise(ng=n()) %>%
pivot_wider(id_cols = description,
names_from = tumor.cat,
values_from = ng)`summarise()` has grouped output by 'description'. You can override using the `.groups` argument.tb.ra <- left_join(tb.ra1, tb.ra2m)Joining, by = "description"tb.ra <- cbind(tb.ra, tb.ra2se[,-1])
tb.ra <- cbind(tb.ra, tb.ra2var[,-1])
tb.ra <- cbind(tb.ra, tb.ra2ng[,-1])
colnames(tb.ra) <- c("description", "ng", "Overall Mean", "Overall SE", "Non-Tumor Mean", "Tumor Mean", "Non-Tumor SE", "Tumor SE","Non-Tumor Var", "Tumor Var", "Non-Tumor Ng", "Tumor Ng")
tb.ra <- tb.ra %>%
arrange(desc(`Overall Mean`))
tb.ra <- tb.ra[, c("description", "Overall Mean", "Overall SE","Tumor Mean","Tumor Var", "Tumor SE","Tumor Ng", "Non-Tumor Mean","Non-Tumor Var", "Non-Tumor SE", "Non-Tumor Ng")]
# compute t-test
tb.ra <- tb.ra %>%
mutate(
SEpooled = sqrt(`Tumor Var`/`Tumor Ng` + `Non-Tumor Var`/`Non-Tumor Ng`),
t = (`Tumor Mean` - `Non-Tumor Mean`)/(SEpooled),
df = ((`Tumor Var`/`Tumor Ng` + `Non-Tumor Var`/`Non-Tumor Ng`)**2)/(((`Tumor Var`/`Tumor Ng`)**2)/(`Tumor Ng`-1) + ((`Non-Tumor Var`/`Non-Tumor Ng`)**2)/(`Non-Tumor Ng`-1)),
p = pt(q = abs(t), df=df, lower.tail = F)*2,
fdr_p = p.adjust(p, method="fdr")
)
kable(tb.ra, format="html", digits=5, caption="Stratefied KO Data - Prevotella spp. (genus level only): Average Abundance of Each Description (sorted in descending order)") %>%
kable_styling(full_width = T) %>%
scroll_box(width = "100%", height="600px")| description | Overall Mean | Overall SE | Tumor Mean | Tumor Var | Tumor SE | Tumor Ng | Non-Tumor Mean | Non-Tumor Var | Non-Tumor SE | Non-Tumor Ng | SEpooled | t | df | p | fdr_p |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| APEX1; AP endonuclease 1 [EC:4.2.99.18] | 8.24684 | 1.49040 | 8.93846 | 312.33990 | 2.19208 | 65 | 7.76344 | 381.07387 | 2.02425 | 93 | 2.98376 | 0.39381 | 145.890 | 0.69430 | 0.75849 |
| ermC, ermA; 23S rRNA (adenine-N6)-dimethyltransferase [EC:2.1.1.184] | 8.24684 | 1.49040 | 8.93846 | 312.33990 | 2.19208 | 65 | 7.76344 | 381.07387 | 2.02425 | 93 | 2.98376 | 0.39381 | 145.890 | 0.69430 | 0.75849 |
| mvpA, vapC; tRNA(fMet)-specific endonuclease VapC [EC:3.1.-.-] | 8.24684 | 1.49040 | 8.93846 | 312.33990 | 2.19208 | 65 | 7.76344 | 381.07387 | 2.02425 | 93 | 2.98376 | 0.39381 | 145.890 | 0.69430 | 0.75849 |
| rpoE; RNA polymerase sigma-70 factor, ECF subfamily | 7.26942 | 1.07943 | 6.13316 | 109.99455 | 1.30086 | 65 | 8.06359 | 236.09686 | 1.59332 | 93 | 2.05692 | -0.93851 | 155.932 | 0.34943 | 0.59014 |
| TC.FEV.OM; iron complex outermembrane recepter protein | 6.70618 | 0.96552 | 5.84387 | 96.82517 | 1.22050 | 65 | 7.30886 | 183.11004 | 1.40318 | 93 | 1.85972 | -0.78775 | 155.731 | 0.43204 | 0.59014 |
| aqpZ; aquaporin Z | 4.40823 | 0.75967 | 4.78846 | 85.01998 | 1.14368 | 65 | 4.14247 | 96.28587 | 1.01751 | 93 | 1.53079 | 0.42200 | 143.062 | 0.67366 | 0.75298 |
| TC.AGCS; alanine or glycine:cation symporter, AGCS family | 4.27772 | 0.73490 | 4.16431 | 56.82331 | 0.93499 | 65 | 4.35699 | 106.07639 | 1.06799 | 93 | 1.41944 | -0.13574 | 155.641 | 0.89220 | 0.93415 |
| ABCB-BAC; ATP-binding cassette, subfamily B, bacterial | 3.96416 | 0.59975 | 3.31289 | 34.02772 | 0.72354 | 65 | 4.41935 | 72.80635 | 0.88480 | 93 | 1.14296 | -0.96806 | 155.942 | 0.33451 | 0.59014 |
| wbpO; UDP-N-acetyl-D-galactosamine dehydrogenase [EC:1.1.1.-] | 3.93882 | 0.63902 | 3.76410 | 43.01987 | 0.81354 | 65 | 4.06093 | 80.13997 | 0.92829 | 93 | 1.23433 | -0.24048 | 155.625 | 0.81028 | 0.85611 |
| fklB; FKBP-type peptidyl-prolyl cis-trans isomerase FklB [EC:5.2.1.8] | 3.59337 | 0.50939 | 3.13305 | 27.15857 | 0.64639 | 65 | 3.91509 | 50.81504 | 0.73919 | 93 | 0.98195 | -0.79642 | 155.658 | 0.42700 | 0.59014 |
| hlpA, ompH; outer membrane protein | 3.54928 | 0.50849 | 3.07947 | 26.94100 | 0.64380 | 65 | 3.87764 | 50.70983 | 0.73842 | 93 | 0.97967 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ABC-2.P; ABC-2 type transport system permease protein | 3.53269 | 0.53036 | 3.00202 | 29.05643 | 0.66860 | 65 | 3.90360 | 55.29069 | 0.77105 | 93 | 1.02056 | -0.88342 | 155.769 | 0.37837 | 0.59014 |
| K07054; uncharacterized protein | 3.47679 | 0.60965 | 3.37692 | 39.28930 | 0.77746 | 65 | 3.54659 | 72.87039 | 0.88519 | 93 | 1.17814 | -0.14402 | 155.591 | 0.88567 | 0.92815 |
| acm; lysozyme | 3.41403 | 0.48975 | 3.00721 | 25.05267 | 0.62083 | 65 | 3.69837 | 47.04435 | 0.71123 | 93 | 0.94408 | -0.73210 | 155.683 | 0.46521 | 0.59070 |
| K07729; putative transcriptional regulator | 3.30380 | 0.59987 | 3.20000 | 35.79792 | 0.74212 | 65 | 3.37634 | 72.10924 | 0.88055 | 93 | 1.15157 | -0.15313 | 155.984 | 0.87849 | 0.92146 |
| tonB; periplasmic protein TonB | 2.99358 | 0.40215 | 2.67459 | 16.16316 | 0.49866 | 65 | 3.21654 | 32.23904 | 0.58878 | 93 | 0.77157 | -0.70240 | 155.964 | 0.48348 | 0.60131 |
| K08987; putative membrane protein | 2.90633 | 0.50347 | 2.97231 | 31.53985 | 0.69658 | 65 | 2.86022 | 46.40068 | 0.70635 | 93 | 0.99205 | 0.11299 | 151.703 | 0.91019 | 0.94278 |
| topB; DNA topoisomerase III [EC:5.99.1.2] | 2.84302 | 0.47030 | 2.33538 | 21.14334 | 0.57034 | 65 | 3.19781 | 44.61939 | 0.69266 | 93 | 0.89725 | -0.96119 | 155.975 | 0.33795 | 0.59014 |
| cas3; CRISPR-associated endonuclease/helicase Cas3 [EC:3.1.-.- 3.6.4.-] | 2.78481 | 1.14192 | 1.63077 | 36.67404 | 0.75114 | 65 | 3.59140 | 324.48340 | 1.86790 | 93 | 2.01328 | -0.97385 | 119.662 | 0.33210 | 0.59014 |
| cas4; CRISPR-associated exonuclease Cas4 [EC:3.1.12.1] | 2.78481 | 1.14192 | 1.63077 | 36.67404 | 0.75114 | 65 | 3.59140 | 324.48340 | 1.86790 | 93 | 2.01328 | -0.97385 | 119.662 | 0.33210 | 0.59014 |
| purU; formyltetrahydrofolate deformylase [EC:3.5.1.10] | 2.78481 | 1.14192 | 1.63077 | 36.67404 | 0.75114 | 65 | 3.59140 | 324.48340 | 1.86790 | 93 | 2.01328 | -0.97385 | 119.662 | 0.33210 | 0.59014 |
| rhaT; L-rhamnose-H+ transport protein | 2.78481 | 1.14192 | 1.63077 | 36.67404 | 0.75114 | 65 | 3.59140 | 324.48340 | 1.86790 | 93 | 2.01328 | -0.97385 | 119.662 | 0.33210 | 0.59014 |
| hupB; DNA-binding protein HU-beta | 2.74369 | 0.39029 | 2.35098 | 15.08710 | 0.48178 | 65 | 3.01817 | 30.39124 | 0.57165 | 93 | 0.74759 | -0.89245 | 155.984 | 0.37353 | 0.59014 |
| ABC-2.A; ABC-2 type transport system ATP-binding protein | 2.72351 | 0.40871 | 2.27724 | 15.33992 | 0.48580 | 65 | 3.03541 | 34.12899 | 0.60579 | 93 | 0.77652 | -0.97637 | 155.771 | 0.33039 | 0.59014 |
| lacZ; beta-galactosidase [EC:3.2.1.23] | 2.71339 | 0.38345 | 2.36663 | 13.31689 | 0.45263 | 65 | 2.95575 | 30.23717 | 0.57020 | 93 | 0.72802 | -0.80922 | 155.639 | 0.41963 | 0.59014 |
| yjdF; putative membrane protein | 2.67563 | 0.45828 | 2.54423 | 22.15793 | 0.58386 | 65 | 2.76747 | 41.19178 | 0.66552 | 93 | 0.88533 | -0.25216 | 155.609 | 0.80125 | 0.85280 |
| E3.4.21.102, prc, ctpA; carboxyl-terminal processing protease [EC:3.4.21.102] | 2.66196 | 0.38137 | 2.30960 | 15.15431 | 0.48285 | 65 | 2.90823 | 28.52428 | 0.55382 | 93 | 0.73475 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rluD; 23S rRNA pseudouridine1911/1915/1917 synthase [EC:5.4.99.23] | 2.66196 | 0.38137 | 2.30960 | 15.15431 | 0.48285 | 65 | 2.90823 | 28.52428 | 0.55382 | 93 | 0.73475 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| THI4, THI1; cysteine-dependent adenosine diphosphate thiazole synthase [EC:2.4.2.60] | 2.66076 | 0.50075 | 2.68308 | 31.18237 | 0.69262 | 65 | 2.64516 | 45.91750 | 0.70266 | 93 | 0.98664 | 0.03843 | 151.727 | 0.96940 | 0.97191 |
| K07133; uncharacterized protein | 2.60452 | 0.39305 | 2.23125 | 15.38840 | 0.48656 | 65 | 2.86541 | 30.78182 | 0.57531 | 93 | 0.75348 | -0.84164 | 155.971 | 0.40128 | 0.59014 |
| rpoD; RNA polymerase primary sigma factor | 2.57584 | 0.37879 | 2.24085 | 14.91242 | 0.47898 | 65 | 2.80997 | 28.17801 | 0.55044 | 93 | 0.72967 | -0.77998 | 155.725 | 0.43659 | 0.59014 |
| pqqL; zinc protease [EC:3.4.24.-] | 2.55596 | 0.37836 | 2.25989 | 15.85698 | 0.49392 | 65 | 2.76288 | 27.46255 | 0.54341 | 93 | 0.73434 | -0.68496 | 154.863 | 0.49439 | 0.60550 |
| HEXA_B; hexosaminidase [EC:3.2.1.52] | 2.54759 | 0.33420 | 2.23600 | 10.03300 | 0.39288 | 65 | 2.76538 | 23.01861 | 0.49751 | 93 | 0.63393 | -0.83507 | 155.560 | 0.40496 | 0.59014 |
| ACSL, fadD; long-chain acyl-CoA synthetase [EC:6.2.1.3] | 2.48114 | 0.36259 | 2.12976 | 12.59593 | 0.44021 | 65 | 2.72673 | 26.53775 | 0.53418 | 93 | 0.69220 | -0.86243 | 155.978 | 0.38977 | 0.59014 |
| lacA; galactoside O-acetyltransferase [EC:2.3.1.18] | 2.44304 | 0.43807 | 2.35385 | 23.69096 | 0.60372 | 65 | 2.50538 | 35.25356 | 0.61569 | 93 | 0.86229 | -0.17573 | 151.987 | 0.86074 | 0.90366 |
| slo; thiol-activated cytolysin | 2.43987 | 0.45834 | 2.22212 | 20.01331 | 0.55488 | 65 | 2.59207 | 42.66462 | 0.67732 | 93 | 0.87559 | -0.42252 | 155.952 | 0.67323 | 0.75298 |
| recG; ATP-dependent DNA helicase RecG [EC:3.6.4.12] | 2.32095 | 0.34957 | 2.03586 | 13.80326 | 0.46082 | 65 | 2.52021 | 23.24809 | 0.49998 | 93 | 0.67995 | -0.71232 | 154.464 | 0.47734 | 0.59693 |
| K07025; putative hydrolase of the HAD superfamily | 2.29437 | 0.34449 | 1.98928 | 11.80667 | 0.42619 | 65 | 2.50760 | 23.67258 | 0.50452 | 93 | 0.66044 | -0.78480 | 155.975 | 0.43376 | 0.59014 |
| ampG; MFS transporter, PAT family, beta-lactamase induction signal transducer AmpG | 2.28627 | 0.32814 | 2.00912 | 11.75174 | 0.42520 | 65 | 2.47998 | 20.76603 | 0.47254 | 93 | 0.63568 | -0.74071 | 155.115 | 0.45999 | 0.59014 |
| higA; HTH-type transcriptional regulator / antitoxin HigA | 2.26076 | 0.38985 | 2.12154 | 15.89797 | 0.49455 | 65 | 2.35806 | 29.89637 | 0.56698 | 93 | 0.75236 | -0.31438 | 155.693 | 0.75366 | 0.80734 |
| yfbK; Ca-activated chloride channel homolog | 2.22776 | 0.32567 | 1.91146 | 11.15516 | 0.41427 | 65 | 2.44883 | 20.71792 | 0.47199 | 93 | 0.62801 | -0.85568 | 155.602 | 0.39349 | 0.59014 |
| thiC; phosphomethylpyrimidine synthase [EC:4.1.99.17] | 2.21751 | 0.41729 | 2.23641 | 21.65211 | 0.57716 | 65 | 2.20430 | 31.88716 | 0.58555 | 93 | 0.82218 | 0.03905 | 151.729 | 0.96890 | 0.97191 |
| gmuG; mannan endo-1,4-beta-mannosidase [EC:3.2.1.78] | 2.21519 | 0.61468 | 1.43077 | 14.98341 | 0.48012 | 65 | 2.76344 | 90.71381 | 0.98763 | 93 | 1.09815 | -1.21356 | 130.171 | 0.22711 | 0.59014 |
| lrp; Lrp/AsnC family transcriptional regulator, leucine-responsive regulatory protein | 2.19139 | 0.37266 | 2.13754 | 14.60691 | 0.47405 | 65 | 2.22903 | 27.27991 | 0.54160 | 93 | 0.71976 | -0.12712 | 155.644 | 0.89901 | 0.94043 |
| phoN; acid phosphatase (class A) [EC:3.1.3.2] | 2.12922 | 0.31788 | 2.03846 | 10.04537 | 0.39312 | 65 | 2.19265 | 20.24728 | 0.46660 | 93 | 0.61013 | -0.25272 | 155.985 | 0.80082 | 0.85280 |
| ompX; outer membrane protein X | 2.12579 | 0.34008 | 2.00673 | 13.00038 | 0.44722 | 65 | 2.20901 | 22.12241 | 0.48772 | 93 | 0.66173 | -0.30568 | 154.618 | 0.76026 | 0.81366 |
| chrA; chromate transporter | 2.11580 | 0.30578 | 1.84870 | 9.84472 | 0.38918 | 65 | 2.30248 | 18.27595 | 0.44330 | 93 | 0.58989 | -0.76926 | 155.598 | 0.44290 | 0.59014 |
| FUCA; alpha-L-fucosidase [EC:3.2.1.51] | 2.10873 | 0.30611 | 1.81154 | 8.68035 | 0.36544 | 65 | 2.31644 | 19.12088 | 0.45343 | 93 | 0.58236 | -0.86698 | 155.825 | 0.38728 | 0.59014 |
| gpx; glutathione peroxidase [EC:1.11.1.9] | 2.08575 | 0.30445 | 1.84212 | 9.82200 | 0.38873 | 65 | 2.25603 | 18.08841 | 0.44102 | 93 | 0.58788 | -0.70406 | 155.532 | 0.48245 | 0.60073 |
| RETSAT; all-trans-retinol 13,14-reductase [EC:1.3.99.23] | 2.06962 | 0.51497 | 1.30000 | 7.26781 | 0.33438 | 65 | 2.60753 | 65.73679 | 0.84074 | 93 | 0.90480 | -1.44510 | 119.123 | 0.15105 | 0.59014 |
| lacI, galR; LacI family transcriptional regulator | 2.06038 | 0.29688 | 1.77812 | 8.80637 | 0.36808 | 65 | 2.25767 | 17.54288 | 0.43432 | 93 | 0.56931 | -0.84233 | 155.961 | 0.40089 | 0.59014 |
| TC.FEV.OM2, cirA, cfrA, hmuR; outer membrane receptor for ferrienterochelin and colicins | 2.02785 | 0.30127 | 1.78005 | 10.52108 | 0.40232 | 65 | 2.20104 | 17.07975 | 0.42855 | 93 | 0.58781 | -0.71619 | 153.845 | 0.47496 | 0.59524 |
| ENDOG; endonuclease G, mitochondrial | 1.99900 | 0.29592 | 1.76944 | 9.19099 | 0.37603 | 65 | 2.15944 | 17.15463 | 0.42949 | 93 | 0.57084 | -0.68319 | 155.639 | 0.49550 | 0.60550 |
| K07481; transposase, IS5 family | 1.95886 | 0.41971 | 1.67231 | 17.61860 | 0.52063 | 65 | 2.15914 | 35.14184 | 0.61471 | 93 | 0.80556 | -0.60434 | 155.964 | 0.54650 | 0.64313 |
| E2.1.1.77, pcm; protein-L-isoaspartate(D-aspartate) O-methyltransferase [EC:2.1.1.77] | 1.95570 | 0.59054 | 1.80000 | 15.53750 | 0.48892 | 65 | 2.06452 | 83.19144 | 0.94580 | 93 | 1.06469 | -0.24844 | 133.985 | 0.80417 | 0.85291 |
| moaA, CNX2; GTP 3’,8-cyclase [EC:4.1.99.22] | 1.95570 | 0.59054 | 1.80000 | 15.53750 | 0.48892 | 65 | 2.06452 | 83.19144 | 0.94580 | 93 | 1.06469 | -0.24844 | 133.985 | 0.80417 | 0.85291 |
| UXS1, uxs; UDP-glucuronate decarboxylase [EC:4.1.1.35] | 1.95570 | 0.59054 | 1.80000 | 15.53750 | 0.48892 | 65 | 2.06452 | 83.19144 | 0.94580 | 93 | 1.06469 | -0.24844 | 133.985 | 0.80417 | 0.85291 |
| capA, pgsA; gamma-polyglutamate biosynthesis protein CapA | 1.95359 | 0.32110 | 1.82051 | 9.89523 | 0.39017 | 65 | 2.04659 | 20.89454 | 0.47400 | 93 | 0.61393 | -0.36826 | 155.974 | 0.71318 | 0.77765 |
| parA, soj; chromosome partitioning protein | 1.95118 | 0.29226 | 1.62854 | 7.58564 | 0.34162 | 65 | 2.17668 | 17.62959 | 0.43539 | 93 | 0.55341 | -0.99046 | 155.452 | 0.32349 | 0.59014 |
| hsdS; type I restriction enzyme, S subunit [EC:3.1.21.3] | 1.93693 | 0.28025 | 1.66355 | 7.26802 | 0.33439 | 65 | 2.12800 | 16.03147 | 0.41519 | 93 | 0.53310 | -0.87122 | 155.818 | 0.38497 | 0.59014 |
| cas6; CRISPR-associated endoribonuclease Cas6 [EC:3.1.-.-] | 1.89557 | 0.58745 | 1.20769 | 10.04603 | 0.39313 | 65 | 2.37634 | 85.49270 | 0.95879 | 93 | 1.03626 | -1.12776 | 120.634 | 0.26166 | 0.59014 |
| K08961; chondroitin-sulfate-ABC endolyase/exolyase [EC:4.2.2.20 4.2.2.21] | 1.89557 | 0.58745 | 1.20769 | 10.04603 | 0.39313 | 65 | 2.37634 | 85.49270 | 0.95879 | 93 | 1.03626 | -1.12776 | 120.634 | 0.26166 | 0.59014 |
| uxuB; fructuronate reductase [EC:1.1.1.57] | 1.89557 | 0.58745 | 1.20769 | 10.04603 | 0.39313 | 65 | 2.37634 | 85.49270 | 0.95879 | 93 | 1.03626 | -1.12776 | 120.634 | 0.26166 | 0.59014 |
| ccrM; modification methylase [EC:2.1.1.72] | 1.87975 | 0.62564 | 1.56154 | 24.60553 | 0.61526 | 65 | 2.10215 | 88.30195 | 0.97441 | 93 | 1.15240 | -0.46912 | 146.506 | 0.63968 | 0.72339 |
| K07318; adenine-specific DNA-methyltransferase [EC:2.1.1.72] | 1.87975 | 0.62564 | 1.56154 | 24.60553 | 0.61526 | 65 | 2.10215 | 88.30195 | 0.97441 | 93 | 1.15240 | -0.46912 | 146.506 | 0.63968 | 0.72339 |
| uvrD, pcrA; DNA helicase II / ATP-dependent DNA helicase PcrA [EC:3.6.4.12] | 1.86146 | 0.27463 | 1.59056 | 7.81293 | 0.34670 | 65 | 2.05080 | 14.81346 | 0.39910 | 93 | 0.52866 | -0.87058 | 155.747 | 0.38533 | 0.59014 |
| thiE; thiamine-phosphate pyrophosphorylase [EC:2.5.1.3] | 1.84301 | 0.30387 | 1.77550 | 11.57318 | 0.42196 | 65 | 1.89020 | 16.84047 | 0.42554 | 93 | 0.59927 | -0.19139 | 151.422 | 0.84848 | 0.89240 |
| DNMT1, dcm; DNA (cytosine-5)-methyltransferase 1 [EC:2.1.1.37] | 1.83761 | 0.27317 | 1.58082 | 6.34957 | 0.31255 | 65 | 2.01709 | 15.62402 | 0.40988 | 93 | 0.51545 | -0.84638 | 154.840 | 0.39865 | 0.59014 |
| bglX; beta-glucosidase [EC:3.2.1.21] | 1.83754 | 0.37510 | 1.42643 | 10.79776 | 0.40758 | 65 | 2.12488 | 30.22281 | 0.57007 | 93 | 0.70078 | -0.99666 | 152.728 | 0.32050 | 0.59014 |
| uvrA; excinuclease ABC subunit A | 1.81873 | 0.25528 | 1.59332 | 6.85442 | 0.32473 | 65 | 1.97627 | 12.74171 | 0.37015 | 93 | 0.49240 | -0.77773 | 155.609 | 0.43791 | 0.59014 |
| K08303; putative protease [EC:3.4.-.-] | 1.81763 | 0.26441 | 1.58271 | 7.84342 | 0.34737 | 65 | 1.98183 | 13.32825 | 0.37857 | 93 | 0.51379 | -0.77682 | 154.598 | 0.43845 | 0.59014 |
| queA; S-adenosylmethionine:tRNA ribosyltransferase-isomerase [EC:2.4.99.17] | 1.81763 | 0.26441 | 1.58271 | 7.84342 | 0.34737 | 65 | 1.98183 | 13.32825 | 0.37857 | 93 | 0.51379 | -0.77682 | 154.598 | 0.43845 | 0.59014 |
| dnaQ; DNA polymerase III subunit epsilon [EC:2.7.7.7] | 1.81698 | 0.25532 | 1.58960 | 6.86317 | 0.32494 | 65 | 1.97590 | 12.74006 | 0.37012 | 93 | 0.49252 | -0.78433 | 155.598 | 0.43404 | 0.59014 |
| recQ; ATP-dependent DNA helicase RecQ [EC:3.6.4.12] | 1.80677 | 0.25605 | 1.55523 | 6.82229 | 0.32397 | 65 | 1.98257 | 12.85513 | 0.37179 | 93 | 0.49314 | -0.86659 | 155.707 | 0.38750 | 0.59014 |
| relA; GTP pyrophosphokinase [EC:2.7.6.5] | 1.79472 | 0.25899 | 1.55618 | 7.00416 | 0.32826 | 65 | 1.96144 | 13.14542 | 0.37596 | 93 | 0.49910 | -0.81197 | 155.680 | 0.41805 | 0.59014 |
| K07483; transposase | 1.79114 | 0.42529 | 1.39077 | 15.30304 | 0.48521 | 65 | 2.07097 | 37.93100 | 0.63864 | 93 | 0.80205 | -0.84807 | 154.746 | 0.39771 | 0.59014 |
| manC, cpsB; mannose-1-phosphate guanylyltransferase [EC:2.7.7.13] | 1.77468 | 0.25424 | 1.53984 | 6.73493 | 0.32189 | 65 | 1.93882 | 12.67746 | 0.36921 | 93 | 0.48983 | -0.81453 | 155.700 | 0.41658 | 0.59014 |
| clsA_B; cardiolipin synthase A/B [EC:2.7.8.-] | 1.77464 | 0.25425 | 1.53973 | 6.73525 | 0.32190 | 65 | 1.93882 | 12.67746 | 0.36921 | 93 | 0.48983 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| DPP4, CD26; dipeptidyl-peptidase 4 [EC:3.4.14.5] | 1.77464 | 0.25425 | 1.53973 | 6.73525 | 0.32190 | 65 | 1.93882 | 12.67746 | 0.36921 | 93 | 0.48983 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| fusA, GFM, EFG; elongation factor G | 1.77464 | 0.25425 | 1.53973 | 6.73525 | 0.32190 | 65 | 1.93882 | 12.67746 | 0.36921 | 93 | 0.48983 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| lolC_E; lipoprotein-releasing system permease protein | 1.77464 | 0.25425 | 1.53973 | 6.73525 | 0.32190 | 65 | 1.93882 | 12.67746 | 0.36921 | 93 | 0.48983 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| lpxA; UDP-N-acetylglucosamine acyltransferase [EC:2.3.1.129] | 1.77464 | 0.25425 | 1.53973 | 6.73525 | 0.32190 | 65 | 1.93882 | 12.67746 | 0.36921 | 93 | 0.48983 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| miaA, TRIT1; tRNA dimethylallyltransferase [EC:2.5.1.75] | 1.77464 | 0.25425 | 1.53973 | 6.73525 | 0.32190 | 65 | 1.93882 | 12.67746 | 0.36921 | 93 | 0.48983 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| mrcA; penicillin-binding protein 1A [EC:2.4.1.129 3.4.16.4] | 1.77464 | 0.25425 | 1.53973 | 6.73525 | 0.32190 | 65 | 1.93882 | 12.67746 | 0.36921 | 93 | 0.48983 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| pepD; dipeptidase D [EC:3.4.13.-] | 1.77464 | 0.25425 | 1.53973 | 6.73525 | 0.32190 | 65 | 1.93882 | 12.67746 | 0.36921 | 93 | 0.48983 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| purH; phosphoribosylaminoimidazolecarboxamide formyltransferase / IMP cyclohydrolase [EC:2.1.2.3 3.5.4.10] | 1.77464 | 0.25425 | 1.53973 | 6.73525 | 0.32190 | 65 | 1.93882 | 12.67746 | 0.36921 | 93 | 0.48983 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rdgB; XTP/dITP diphosphohydrolase [EC:3.6.1.66] | 1.77464 | 0.25425 | 1.53973 | 6.73525 | 0.32190 | 65 | 1.93882 | 12.67746 | 0.36921 | 93 | 0.48983 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| surA; peptidyl-prolyl cis-trans isomerase SurA [EC:5.2.1.8] | 1.77464 | 0.25425 | 1.53973 | 6.73525 | 0.32190 | 65 | 1.93882 | 12.67746 | 0.36921 | 93 | 0.48983 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| udk, UCK; uridine kinase [EC:2.7.1.48] | 1.77464 | 0.25425 | 1.53973 | 6.73525 | 0.32190 | 65 | 1.93882 | 12.67746 | 0.36921 | 93 | 0.48983 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| pepP; Xaa-Pro aminopeptidase [EC:3.4.11.9] | 1.77289 | 0.25428 | 1.53602 | 6.74360 | 0.32210 | 65 | 1.93845 | 12.67577 | 0.36919 | 93 | 0.48995 | -0.82138 | 155.690 | 0.41269 | 0.59014 |
| dcp; peptidyl-dipeptidase Dcp [EC:3.4.15.5] | 1.76264 | 0.26389 | 1.52647 | 7.76356 | 0.34560 | 65 | 1.92770 | 13.30841 | 0.37829 | 93 | 0.51239 | -0.78305 | 154.723 | 0.43479 | 0.59014 |
| exbB; biopolymer transport protein ExbB | 1.75347 | 0.25311 | 1.52700 | 6.66960 | 0.32033 | 65 | 1.91175 | 12.57260 | 0.36768 | 93 | 0.48765 | -0.78900 | 155.710 | 0.43131 | 0.59014 |
| K06950; uncharacterized protein | 1.75347 | 0.25311 | 1.52700 | 6.66960 | 0.32033 | 65 | 1.91175 | 12.57260 | 0.36768 | 93 | 0.48765 | -0.78900 | 155.710 | 0.43131 | 0.59014 |
| SIAE; sialate O-acetylesterase [EC:3.1.1.53] | 1.74466 | 0.26559 | 1.46302 | 5.38439 | 0.28781 | 65 | 1.94151 | 15.17863 | 0.40399 | 93 | 0.49603 | -0.96464 | 152.585 | 0.33625 | 0.59014 |
| BLMH, pepC; bleomycin hydrolase [EC:3.4.22.40] | 1.73339 | 0.24859 | 1.51056 | 6.42249 | 0.31434 | 65 | 1.88914 | 12.13470 | 0.36122 | 93 | 0.47884 | -0.79062 | 155.725 | 0.43037 | 0.59014 |
| K07095; uncharacterized protein | 1.73291 | 0.26228 | 1.47879 | 7.10645 | 0.33065 | 65 | 1.91052 | 13.52636 | 0.38137 | 93 | 0.50475 | -0.85533 | 155.771 | 0.39368 | 0.59014 |
| ABC-2.TX; HlyD family secretion protein | 1.72881 | 0.25380 | 1.48244 | 6.66557 | 0.32023 | 65 | 1.90100 | 12.65810 | 0.36893 | 93 | 0.48852 | -0.85679 | 155.757 | 0.39288 | 0.59014 |
| K09975; uncharacterized protein | 1.71941 | 0.29950 | 1.54872 | 9.80744 | 0.38844 | 65 | 1.83871 | 17.32864 | 0.43166 | 93 | 0.58070 | -0.49938 | 155.113 | 0.61822 | 0.70276 |
| K07098; uncharacterized protein | 1.71571 | 0.25360 | 1.48881 | 6.68619 | 0.32072 | 65 | 1.87430 | 12.62784 | 0.36849 | 93 | 0.48852 | -0.78912 | 155.722 | 0.43124 | 0.59014 |
| fur, zur, furB; Fur family transcriptional regulator, ferric uptake regulator | 1.71370 | 0.25715 | 1.45516 | 6.76393 | 0.32258 | 65 | 1.89439 | 13.04342 | 0.37450 | 93 | 0.49428 | -0.88862 | 155.842 | 0.37558 | 0.59014 |
| asnC; Lrp/AsnC family transcriptional regulator, regulator for asnA, asnC and gidA | 1.70938 | 0.25260 | 1.47342 | 6.59059 | 0.31842 | 65 | 1.87430 | 12.55290 | 0.36739 | 93 | 0.48618 | -0.82456 | 155.775 | 0.41088 | 0.59014 |
| korA, oorA, oforA; 2-oxoglutarate/2-oxoacid ferredoxin oxidoreductase subunit alpha [EC:1.2.7.3 1.2.7.11] | 1.70934 | 0.25260 | 1.47332 | 6.59090 | 0.31843 | 65 | 1.87430 | 12.55290 | 0.36739 | 93 | 0.48619 | -0.82477 | 155.774 | 0.41077 | 0.59014 |
| korB, oorB, oforB; 2-oxoglutarate/2-oxoacid ferredoxin oxidoreductase subunit beta [EC:1.2.7.3 1.2.7.11] | 1.70934 | 0.25260 | 1.47332 | 6.59090 | 0.31843 | 65 | 1.87430 | 12.55290 | 0.36739 | 93 | 0.48619 | -0.82477 | 155.774 | 0.41077 | 0.59014 |
| galM, GALM; aldose 1-epimerase [EC:5.1.3.3] | 1.70764 | 0.25264 | 1.46971 | 6.59844 | 0.31861 | 65 | 1.87393 | 12.55117 | 0.36737 | 93 | 0.48629 | -0.83125 | 155.767 | 0.40710 | 0.59014 |
| TC.MATE, SLC47A, norM, mdtK, dinF; multidrug resistance protein, MATE family | 1.70218 | 0.25314 | 1.48032 | 6.65813 | 0.32005 | 65 | 1.85725 | 12.58751 | 0.36790 | 93 | 0.48763 | -0.77299 | 155.729 | 0.44070 | 0.59014 |
| ispD; 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [EC:2.7.7.60] | 1.69935 | 0.25200 | 1.47342 | 6.59059 | 0.31842 | 65 | 1.85725 | 12.47603 | 0.36627 | 93 | 0.48533 | -0.79086 | 155.737 | 0.43023 | 0.59014 |
| folK; 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine diphosphokinase [EC:2.7.6.3] | 1.68931 | 0.24805 | 1.45698 | 6.34169 | 0.31235 | 65 | 1.85169 | 12.11350 | 0.36091 | 93 | 0.47730 | -0.82696 | 155.791 | 0.40952 | 0.59014 |
| coaE; dephospho-CoA kinase [EC:2.7.1.24] | 1.68603 | 0.25086 | 1.46493 | 6.48323 | 0.31582 | 65 | 1.84056 | 12.39950 | 0.36514 | 93 | 0.48277 | -0.77807 | 155.798 | 0.43771 | 0.59014 |
| algI; alginate O-acetyltransferase complex protein AlgI | 1.66878 | 0.46286 | 1.45641 | 19.48462 | 0.54751 | 65 | 1.81720 | 44.15584 | 0.68905 | 93 | 0.88009 | -0.40995 | 155.652 | 0.68241 | 0.75298 |
| cobA, btuR; cob(I)alamin adenosyltransferase [EC:2.5.1.17] | 1.66878 | 0.46286 | 1.45641 | 19.48462 | 0.54751 | 65 | 1.81720 | 44.15584 | 0.68905 | 93 | 0.88009 | -0.40995 | 155.652 | 0.68241 | 0.75298 |
| dcuB; anaerobic C4-dicarboxylate transporter DcuB | 1.66878 | 0.46286 | 1.45641 | 19.48462 | 0.54751 | 65 | 1.81720 | 44.15584 | 0.68905 | 93 | 0.88009 | -0.40995 | 155.652 | 0.68241 | 0.75298 |
| E4.2.1.2AA, fumA; fumarate hydratase subunit alpha [EC:4.2.1.2] | 1.66878 | 0.46286 | 1.45641 | 19.48462 | 0.54751 | 65 | 1.81720 | 44.15584 | 0.68905 | 93 | 0.88009 | -0.40995 | 155.652 | 0.68241 | 0.75298 |
| E4.2.1.2AB, fumB; fumarate hydratase subunit beta [EC:4.2.1.2] | 1.66878 | 0.46286 | 1.45641 | 19.48462 | 0.54751 | 65 | 1.81720 | 44.15584 | 0.68905 | 93 | 0.88009 | -0.40995 | 155.652 | 0.68241 | 0.75298 |
| glmE, mutE, mamB; methylaspartate mutase epsilon subunit [EC:5.4.99.1] | 1.66878 | 0.46286 | 1.45641 | 19.48462 | 0.54751 | 65 | 1.81720 | 44.15584 | 0.68905 | 93 | 0.88009 | -0.40995 | 155.652 | 0.68241 | 0.75298 |
| glmS, mutS, mamA; methylaspartate mutase sigma subunit [EC:5.4.99.1] | 1.66878 | 0.46286 | 1.45641 | 19.48462 | 0.54751 | 65 | 1.81720 | 44.15584 | 0.68905 | 93 | 0.88009 | -0.40995 | 155.652 | 0.68241 | 0.75298 |
| inlA; internalin A | 1.66878 | 0.46286 | 1.45641 | 19.48462 | 0.54751 | 65 | 1.81720 | 44.15584 | 0.68905 | 93 | 0.88009 | -0.40995 | 155.652 | 0.68241 | 0.75298 |
| mal; methylaspartate ammonia-lyase [EC:4.3.1.2] | 1.66878 | 0.46286 | 1.45641 | 19.48462 | 0.54751 | 65 | 1.81720 | 44.15584 | 0.68905 | 93 | 0.88009 | -0.40995 | 155.652 | 0.68241 | 0.75298 |
| PCYT1; choline-phosphate cytidylyltransferase [EC:2.7.7.15] | 1.66878 | 0.46286 | 1.45641 | 19.48462 | 0.54751 | 65 | 1.81720 | 44.15584 | 0.68905 | 93 | 0.88009 | -0.40995 | 155.652 | 0.68241 | 0.75298 |
| tnaA; tryptophanase [EC:4.1.99.1] | 1.66878 | 0.46286 | 1.45641 | 19.48462 | 0.54751 | 65 | 1.81720 | 44.15584 | 0.68905 | 93 | 0.88009 | -0.40995 | 155.652 | 0.68241 | 0.75298 |
| icmF; isobutyryl-CoA mutase [EC:5.4.99.13] | 1.66561 | 0.25721 | 1.44786 | 5.53394 | 0.29178 | 65 | 1.81780 | 13.93145 | 0.38704 | 93 | 0.48470 | -0.76323 | 154.536 | 0.44649 | 0.59014 |
| hemN, hemZ; oxygen-independent coproporphyrinogen III oxidase [EC:1.3.98.3] | 1.66377 | 0.23703 | 1.44637 | 5.81089 | 0.29900 | 65 | 1.81572 | 11.04920 | 0.34469 | 93 | 0.45630 | -0.80946 | 155.765 | 0.41948 | 0.59014 |
| treS; maltose alpha-D-glucosyltransferase / alpha-amylase [EC:5.4.99.16 3.2.1.1] | 1.65506 | 0.61136 | 1.09231 | 14.98353 | 0.48012 | 65 | 2.04839 | 89.97318 | 0.98359 | 93 | 1.09452 | -0.87352 | 130.421 | 0.38399 | 0.59014 |
| GLPF; glycerol uptake facilitator protein | 1.64376 | 0.27456 | 1.57143 | 7.20663 | 0.33297 | 65 | 1.69432 | 15.30550 | 0.40568 | 93 | 0.52483 | -0.23415 | 155.961 | 0.81518 | 0.85972 |
| glpK, GK; glycerol kinase [EC:2.7.1.30] | 1.64376 | 0.27456 | 1.57143 | 7.20663 | 0.33297 | 65 | 1.69432 | 15.30550 | 0.40568 | 93 | 0.52483 | -0.23415 | 155.961 | 0.81518 | 0.85972 |
| gph; phosphoglycolate phosphatase [EC:3.1.3.18] | 1.63605 | 0.22237 | 1.42621 | 4.85721 | 0.27336 | 65 | 1.78272 | 9.90094 | 0.32628 | 93 | 0.42566 | -0.83755 | 155.997 | 0.40356 | 0.59014 |
| panE, apbA; 2-dehydropantoate 2-reductase [EC:1.1.1.169] | 1.61850 | 0.25853 | 1.41368 | 5.91609 | 0.30169 | 65 | 1.76165 | 13.85538 | 0.38598 | 93 | 0.48990 | -0.71030 | 155.382 | 0.47858 | 0.59720 |
| csm1, cas10; CRISPR-associated protein Csm1 | 1.61392 | 1.05415 | 0.55385 | 3.68846 | 0.23821 | 65 | 2.35484 | 295.70968 | 1.78316 | 93 | 1.79901 | -1.00110 | 95.269 | 0.31931 | 0.59014 |
| csm2; CRISPR-associated protein Csm2 | 1.61392 | 1.05415 | 0.55385 | 3.68846 | 0.23821 | 65 | 2.35484 | 295.70968 | 1.78316 | 93 | 1.79901 | -1.00110 | 95.269 | 0.31931 | 0.59014 |
| csm3; CRISPR-associated protein Csm3 | 1.61392 | 1.05415 | 0.55385 | 3.68846 | 0.23821 | 65 | 2.35484 | 295.70968 | 1.78316 | 93 | 1.79901 | -1.00110 | 95.269 | 0.31931 | 0.59014 |
| K07088; uncharacterized protein | 1.59991 | 0.26952 | 1.51648 | 7.31741 | 0.33552 | 65 | 1.65822 | 14.48800 | 0.39470 | 93 | 0.51804 | -0.27360 | 155.945 | 0.78475 | 0.83755 |
| sanA; SanA protein | 1.59991 | 0.26952 | 1.51648 | 7.31741 | 0.33552 | 65 | 1.65822 | 14.48800 | 0.39470 | 93 | 0.51804 | -0.27360 | 155.945 | 0.78475 | 0.83755 |
| E2.7.1.4, scrK; fructokinase [EC:2.7.1.4] | 1.59684 | 0.23344 | 1.37824 | 5.35937 | 0.28714 | 65 | 1.74962 | 10.90726 | 0.34247 | 93 | 0.44692 | -0.83097 | 155.996 | 0.40726 | 0.59014 |
| aspB; aspartate aminotransferase [EC:2.6.1.1] | 1.59236 | 0.22338 | 1.37716 | 4.39429 | 0.26001 | 65 | 1.74276 | 10.34194 | 0.33347 | 93 | 0.42286 | -0.86460 | 155.335 | 0.38859 | 0.59014 |
| map; methionyl aminopeptidase [EC:3.4.11.18] | 1.59022 | 0.22308 | 1.38536 | 4.92936 | 0.27538 | 65 | 1.73341 | 9.93905 | 0.32691 | 93 | 0.42744 | -0.81426 | 155.985 | 0.41674 | 0.59014 |
| pflA, pflC, pflE; pyruvate formate lyase activating enzyme [EC:1.97.1.4] | 1.59006 | 0.23298 | 1.41778 | 5.82845 | 0.29945 | 65 | 1.71047 | 10.54574 | 0.33674 | 93 | 0.45063 | -0.64953 | 155.370 | 0.51695 | 0.62047 |
| nagB, GNPDA; glucosamine-6-phosphate deaminase [EC:3.5.99.6] | 1.57069 | 0.22173 | 1.36710 | 4.29694 | 0.25711 | 65 | 1.71299 | 10.21754 | 0.33146 | 93 | 0.41949 | -0.82454 | 155.232 | 0.41090 | 0.59014 |
| NAGLU; alpha-N-acetylglucosaminidase [EC:3.2.1.50] | 1.56804 | 0.57661 | 0.87308 | 9.23266 | 0.37688 | 65 | 2.05376 | 82.64518 | 0.94269 | 93 | 1.01523 | -1.16297 | 119.376 | 0.24716 | 0.59014 |
| cobN; cobaltochelatase CobN [EC:6.6.1.2] | 1.56013 | 0.31434 | 1.25897 | 7.86721 | 0.34790 | 65 | 1.77061 | 21.05973 | 0.47587 | 93 | 0.58948 | -0.86795 | 153.565 | 0.38678 | 0.59014 |
| acrA, mexA, adeI, smeD, mtrC, cmeA; membrane fusion protein, multidrug efflux system | 1.54522 | 0.20843 | 1.35289 | 4.34873 | 0.25866 | 65 | 1.67964 | 8.64356 | 0.30486 | 93 | 0.39981 | -0.81728 | 155.956 | 0.41502 | 0.59014 |
| TC.HAE1; hydrophobic/amphiphilic exporter-1 (mainly G- bacteria), HAE1 family | 1.54522 | 0.20843 | 1.35289 | 4.34873 | 0.25866 | 65 | 1.67964 | 8.64356 | 0.30486 | 93 | 0.39981 | -0.81728 | 155.956 | 0.41502 | 0.59014 |
| mrdA; penicillin-binding protein 2 [EC:3.4.16.4] | 1.51008 | 0.22165 | 1.34122 | 5.26436 | 0.28459 | 65 | 1.62811 | 9.54999 | 0.32045 | 93 | 0.42858 | -0.66939 | 155.395 | 0.50424 | 0.60887 |
| SPP; sucrose-6-phosphatase [EC:3.1.3.24] | 1.50707 | 0.21096 | 1.32647 | 4.14615 | 0.25256 | 65 | 1.63330 | 9.07591 | 0.31239 | 93 | 0.40172 | -0.76378 | 155.855 | 0.44615 | 0.59014 |
| pepO; putative endopeptidase [EC:3.4.24.-] | 1.50314 | 0.22186 | 1.33655 | 5.27597 | 0.28490 | 65 | 1.61958 | 9.56843 | 0.32076 | 93 | 0.42902 | -0.65971 | 155.393 | 0.51042 | 0.61441 |
| ytfE, scdA; regulator of cell morphogenesis and NO signaling | 1.49790 | 0.20673 | 1.31162 | 4.19327 | 0.25399 | 65 | 1.62811 | 8.56459 | 0.30347 | 93 | 0.39573 | -0.79975 | 155.998 | 0.42507 | 0.59014 |
| mutT, NUDT15, MTH2; 8-oxo-dGTP diphosphatase [EC:3.6.1.55] | 1.48703 | 0.23488 | 1.37615 | 6.41118 | 0.31406 | 65 | 1.56452 | 10.40014 | 0.33441 | 93 | 0.45876 | -0.41059 | 153.832 | 0.68195 | 0.75298 |
| E1.17.4.1A, nrdA, nrdE; ribonucleoside-diphosphate reductase alpha chain [EC:1.17.4.1] | 1.48586 | 0.21233 | 1.33273 | 5.31387 | 0.28592 | 65 | 1.59288 | 8.43176 | 0.30110 | 93 | 0.41523 | -0.62652 | 153.411 | 0.53191 | 0.63304 |
| fsr; MFS transporter, FSR family, fosmidomycin resistance protein | 1.46559 | 0.24027 | 1.39615 | 5.79984 | 0.29871 | 65 | 1.51411 | 11.52566 | 0.35204 | 93 | 0.46169 | -0.25549 | 155.955 | 0.79868 | 0.85163 |
| ada; AraC family transcriptional regulator, regulatory protein of adaptative response / methylated-DNA-[protein]-cysteine methyltransferase [EC:2.1.1.63] | 1.45781 | 0.39995 | 0.96410 | 5.34765 | 0.28683 | 65 | 1.80287 | 39.11651 | 0.64854 | 93 | 0.70914 | -1.18279 | 124.655 | 0.23914 | 0.59014 |
| LARS, leuS; leucyl-tRNA synthetase [EC:6.1.1.4] | 1.44723 | 0.20945 | 1.26218 | 4.66661 | 0.26794 | 65 | 1.57657 | 8.54070 | 0.30304 | 93 | 0.40451 | -0.77721 | 155.478 | 0.43821 | 0.59014 |
| dam; DNA adenine methylase [EC:2.1.1.72] | 1.44051 | 0.24781 | 1.29538 | 5.87607 | 0.30067 | 65 | 1.54194 | 12.44550 | 0.36582 | 93 | 0.47352 | -0.52067 | 155.968 | 0.60333 | 0.69037 |
| SERPINB; serpin B | 1.43631 | 0.23694 | 1.32692 | 5.60239 | 0.29358 | 65 | 1.51277 | 11.22542 | 0.34742 | 93 | 0.45486 | -0.40858 | 155.974 | 0.68341 | 0.75298 |
| UNG, UDG; uracil-DNA glycosylase [EC:3.2.2.27] | 1.42115 | 0.21151 | 1.25422 | 4.39841 | 0.26013 | 65 | 1.53782 | 8.96938 | 0.31056 | 93 | 0.40511 | -0.70007 | 155.997 | 0.48493 | 0.60187 |
| acpP; acyl carrier protein | 1.42010 | 0.21826 | 1.12647 | 3.31796 | 0.22593 | 65 | 1.62532 | 10.43263 | 0.33493 | 93 | 0.40401 | -1.23475 | 150.100 | 0.21885 | 0.59014 |
| agp; glucose-1-phosphatase [EC:3.1.3.10] | 1.41667 | 0.21186 | 1.35726 | 4.45679 | 0.26185 | 65 | 1.45818 | 8.99823 | 0.31105 | 93 | 0.40660 | -0.24820 | 155.987 | 0.80430 | 0.85291 |
| E5.2.1.8; peptidylprolyl isomerase [EC:5.2.1.8] | 1.40625 | 0.23569 | 1.34038 | 5.58130 | 0.29303 | 65 | 1.45228 | 11.08962 | 0.34532 | 93 | 0.45289 | -0.24708 | 155.955 | 0.80517 | 0.85305 |
| K07126; uncharacterized protein | 1.40506 | 0.35581 | 1.18462 | 11.17632 | 0.41466 | 65 | 1.55914 | 26.30217 | 0.53181 | 93 | 0.67436 | -0.55538 | 155.336 | 0.57944 | 0.67436 |
| thiD; hydroxymethylpyrimidine/phosphomethylpyrimidine kinase [EC:2.7.1.49 2.7.4.7] | 1.40152 | 0.21274 | 1.29254 | 5.40085 | 0.28825 | 65 | 1.47770 | 8.43109 | 0.30109 | 93 | 0.41683 | -0.44422 | 153.077 | 0.65751 | 0.73851 |
| aguA; alpha-glucuronidase [EC:3.2.1.139] | 1.39304 | 0.57095 | 0.81692 | 9.16612 | 0.37552 | 65 | 1.79570 | 81.12085 | 0.93395 | 93 | 1.00662 | -0.97234 | 119.656 | 0.33284 | 0.59014 |
| fabB; 3-oxoacyl-[acyl-carrier-protein] synthase I [EC:2.3.1.41] | 1.38633 | 0.32063 | 0.84215 | 2.57519 | 0.19904 | 65 | 1.76667 | 25.57225 | 0.52438 | 93 | 0.56088 | -1.64832 | 116.932 | 0.10197 | 0.59014 |
| glnA, GLUL; glutamine synthetase [EC:6.3.1.2] | 1.38595 | 0.19733 | 1.22302 | 4.19249 | 0.25397 | 65 | 1.49981 | 7.55094 | 0.28494 | 93 | 0.38170 | -0.72516 | 155.324 | 0.46945 | 0.59347 |
| kdgK; 2-dehydro-3-deoxygluconokinase [EC:2.7.1.45] | 1.38101 | 0.38355 | 0.94593 | 8.18439 | 0.35484 | 65 | 1.68510 | 33.74515 | 0.60237 | 93 | 0.69912 | -1.05728 | 142.297 | 0.29217 | 0.59014 |
| kup; KUP system potassium uptake protein | 1.37880 | 0.20989 | 1.30000 | 4.69031 | 0.26862 | 65 | 1.43387 | 8.60800 | 0.30424 | 93 | 0.40585 | -0.32985 | 155.503 | 0.74196 | 0.79850 |
| moxR; MoxR-like ATPase [EC:3.6.3.-] | 1.37241 | 0.18981 | 1.23735 | 3.74018 | 0.23988 | 65 | 1.46681 | 7.09041 | 0.27612 | 93 | 0.36576 | -0.62737 | 155.746 | 0.53134 | 0.63301 |
| cysE; serine O-acetyltransferase [EC:2.3.1.30] | 1.36414 | 0.19280 | 1.24018 | 3.82836 | 0.24269 | 65 | 1.45079 | 7.34149 | 0.28096 | 93 | 0.37127 | -0.56727 | 155.813 | 0.57135 | 0.66696 |
| mviM; virulence factor | 1.35979 | 0.21656 | 1.15621 | 3.95654 | 0.24672 | 65 | 1.50207 | 9.84250 | 0.32532 | 93 | 0.40829 | -0.84708 | 154.698 | 0.39826 | 0.59014 |
| K15977; putative oxidoreductase | 1.35970 | 0.18726 | 1.31987 | 4.04504 | 0.24946 | 65 | 1.38754 | 6.63884 | 0.26718 | 93 | 0.36554 | -0.18513 | 154.040 | 0.85337 | 0.89673 |
| hipA; serine/threonine-protein kinase HipA [EC:2.7.11.1] | 1.35886 | 0.21962 | 1.34752 | 7.16264 | 0.33196 | 65 | 1.36679 | 8.02275 | 0.29371 | 93 | 0.44324 | -0.04346 | 142.623 | 0.96539 | 0.96958 |
| E3.2.1.22B, galA, rafA; alpha-galactosidase [EC:3.2.1.22] | 1.35490 | 0.18980 | 1.22355 | 3.46868 | 0.23101 | 65 | 1.44670 | 7.27929 | 0.27977 | 93 | 0.36282 | -0.61507 | 155.985 | 0.53940 | 0.63858 |
| E1.4.1.4, gdhA; glutamate dehydrogenase (NADP+) [EC:1.4.1.4] | 1.35029 | 0.19046 | 1.22951 | 4.15986 | 0.25298 | 65 | 1.43472 | 6.86956 | 0.27178 | 93 | 0.37130 | -0.55268 | 154.145 | 0.58128 | 0.67515 |
| PPIB, ppiB; peptidyl-prolyl cis-trans isomerase B (cyclophilin B) [EC:5.2.1.8] | 1.34567 | 0.20077 | 1.23163 | 4.43243 | 0.26113 | 65 | 1.42538 | 7.76911 | 0.28903 | 93 | 0.38953 | -0.49739 | 155.017 | 0.61962 | 0.70276 |
| E5.1.3.6; UDP-glucuronate 4-epimerase [EC:5.1.3.6] | 1.34494 | 0.37843 | 1.30000 | 13.55312 | 0.45663 | 65 | 1.37634 | 29.18291 | 0.56017 | 93 | 0.72271 | -0.10564 | 155.921 | 0.91601 | 0.94278 |
| glpA, glpD; glycerol-3-phosphate dehydrogenase [EC:1.1.5.3] | 1.34124 | 0.21694 | 1.16538 | 3.46256 | 0.23080 | 65 | 1.46416 | 10.24342 | 0.33188 | 93 | 0.40425 | -0.73909 | 151.551 | 0.46100 | 0.59014 |
| glpB; glycerol-3-phosphate dehydrogenase subunit B [EC:1.1.5.3] | 1.34124 | 0.21694 | 1.16538 | 3.46256 | 0.23080 | 65 | 1.46416 | 10.24342 | 0.33188 | 93 | 0.40425 | -0.73909 | 151.551 | 0.46100 | 0.59014 |
| glpC; glycerol-3-phosphate dehydrogenase subunit C [EC:1.1.5.3] | 1.34124 | 0.21694 | 1.16538 | 3.46256 | 0.23080 | 65 | 1.46416 | 10.24342 | 0.33188 | 93 | 0.40425 | -0.73909 | 151.551 | 0.46100 | 0.59014 |
| E3.1.6.1, aslA; arylsulfatase [EC:3.1.6.1] | 1.33931 | 0.22182 | 1.23761 | 4.97468 | 0.27665 | 65 | 1.41039 | 9.79395 | 0.32452 | 93 | 0.42643 | -0.40519 | 155.927 | 0.68589 | 0.75500 |
| malZ; alpha-glucosidase [EC:3.2.1.20] | 1.32077 | 0.19689 | 1.23077 | 3.89749 | 0.24487 | 65 | 1.38368 | 7.73111 | 0.28832 | 93 | 0.37827 | -0.40422 | 155.950 | 0.68660 | 0.75507 |
| hsdM; type I restriction enzyme M protein [EC:2.1.1.72] | 1.30624 | 0.19202 | 1.13584 | 3.60830 | 0.23561 | 65 | 1.42533 | 7.39726 | 0.28203 | 93 | 0.36750 | -0.78773 | 156.000 | 0.43205 | 0.59014 |
| ybgC; acyl-CoA thioester hydrolase [EC:3.1.2.-] | 1.29832 | 0.20019 | 1.02753 | 2.82302 | 0.20840 | 65 | 1.48758 | 8.75422 | 0.30681 | 93 | 0.37089 | -1.24037 | 150.443 | 0.21677 | 0.59014 |
| K07387; putative metalloprotease [EC:3.4.24.-] | 1.29625 | 0.19954 | 1.11243 | 2.96081 | 0.21343 | 65 | 1.42473 | 8.63519 | 0.30472 | 93 | 0.37202 | -0.83947 | 151.868 | 0.40252 | 0.59014 |
| dpo; DNA polymerase bacteriophage-type [EC:2.7.7.7] | 1.27848 | 0.34367 | 1.55385 | 17.46971 | 0.51843 | 65 | 1.08602 | 19.60122 | 0.45909 | 93 | 0.69248 | 0.67558 | 142.692 | 0.50040 | 0.60550 |
| epsD; glycosyltransferase EpsD [EC:2.4.-.-] | 1.27848 | 0.34367 | 1.55385 | 17.46971 | 0.51843 | 65 | 1.08602 | 19.60122 | 0.45909 | 93 | 0.69248 | 0.67558 | 142.692 | 0.50040 | 0.60550 |
| K06904; uncharacterized protein | 1.27848 | 0.34367 | 1.55385 | 17.46971 | 0.51843 | 65 | 1.08602 | 19.60122 | 0.45909 | 93 | 0.69248 | 0.67558 | 142.692 | 0.50040 | 0.60550 |
| K07270; glycosyl transferase, family 25 | 1.27848 | 0.34367 | 1.55385 | 17.46971 | 0.51843 | 65 | 1.08602 | 19.60122 | 0.45909 | 93 | 0.69248 | 0.67558 | 142.692 | 0.50040 | 0.60550 |
| K09137; uncharacterized protein | 1.27848 | 0.34367 | 1.55385 | 17.46971 | 0.51843 | 65 | 1.08602 | 19.60122 | 0.45909 | 93 | 0.69248 | 0.67558 | 142.692 | 0.50040 | 0.60550 |
| kch, trkA, mthK, pch; voltage-gated potassium channel | 1.27848 | 0.34367 | 1.55385 | 17.46971 | 0.51843 | 65 | 1.08602 | 19.60122 | 0.45909 | 93 | 0.69248 | 0.67558 | 142.692 | 0.50040 | 0.60550 |
| mcrA; 5-methylcytosine-specific restriction enzyme A [EC:3.1.21.-] | 1.27848 | 0.34367 | 1.55385 | 17.46971 | 0.51843 | 65 | 1.08602 | 19.60122 | 0.45909 | 93 | 0.69248 | 0.67558 | 142.692 | 0.50040 | 0.60550 |
| mcrC; 5-methylcytosine-specific restriction enzyme subunit McrC | 1.27848 | 0.34367 | 1.55385 | 17.46971 | 0.51843 | 65 | 1.08602 | 19.60122 | 0.45909 | 93 | 0.69248 | 0.67558 | 142.692 | 0.50040 | 0.60550 |
| PNC1; nicotinamidase [EC:3.5.1.19] | 1.27848 | 0.34367 | 1.55385 | 17.46971 | 0.51843 | 65 | 1.08602 | 19.60122 | 0.45909 | 93 | 0.69248 | 0.67558 | 142.692 | 0.50040 | 0.60550 |
| rgpB; rhamnosyltransferase [EC:2.4.1.-] | 1.27848 | 0.34367 | 1.55385 | 17.46971 | 0.51843 | 65 | 1.08602 | 19.60122 | 0.45909 | 93 | 0.69248 | 0.67558 | 142.692 | 0.50040 | 0.60550 |
| tagD; glycerol-3-phosphate cytidylyltransferase [EC:2.7.7.39] | 1.27848 | 0.34367 | 1.55385 | 17.46971 | 0.51843 | 65 | 1.08602 | 19.60122 | 0.45909 | 93 | 0.69248 | 0.67558 | 142.692 | 0.50040 | 0.60550 |
| MAN; mannan endo-1,4-beta-mannosidase [EC:3.2.1.78] | 1.27563 | 0.18545 | 1.13648 | 3.40006 | 0.22871 | 65 | 1.37289 | 6.88497 | 0.27209 | 93 | 0.35544 | -0.66510 | 155.991 | 0.50697 | 0.61090 |
| K06871; uncharacterized protein | 1.27416 | 0.20132 | 1.04891 | 3.24064 | 0.22328 | 65 | 1.43159 | 8.61287 | 0.30432 | 93 | 0.37745 | -1.01385 | 153.689 | 0.31225 | 0.59014 |
| E3.1.1.11; pectinesterase [EC:3.1.1.11] | 1.26456 | 0.39162 | 0.80718 | 4.46182 | 0.26200 | 65 | 1.58423 | 37.99675 | 0.63919 | 93 | 0.69080 | -1.12485 | 120.617 | 0.26289 | 0.59014 |
| kduI; 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase [EC:5.3.1.17] | 1.26414 | 0.39163 | 0.80615 | 4.46329 | 0.26204 | 65 | 1.58423 | 37.99675 | 0.63919 | 93 | 0.69082 | -1.12631 | 120.625 | 0.26227 | 0.59014 |
| PTS-Gat-EIIC, gatC, sgcC; PTS system, galactitol-specific IIC component | 1.26414 | 0.39163 | 0.80615 | 4.46329 | 0.26204 | 65 | 1.58423 | 37.99675 | 0.63919 | 93 | 0.69082 | -1.12631 | 120.625 | 0.26227 | 0.59014 |
| uxaA; altronate hydrolase [EC:4.2.1.7] | 1.26414 | 0.39163 | 0.80615 | 4.46329 | 0.26204 | 65 | 1.58423 | 37.99675 | 0.63919 | 93 | 0.69082 | -1.12631 | 120.625 | 0.26227 | 0.59014 |
| uxaB; tagaturonate reductase [EC:1.1.1.58] | 1.26414 | 0.39163 | 0.80615 | 4.46329 | 0.26204 | 65 | 1.58423 | 37.99675 | 0.63919 | 93 | 0.69082 | -1.12631 | 120.625 | 0.26227 | 0.59014 |
| K02477; two-component system, LytTR family, response regulator | 1.25190 | 0.34713 | 1.09308 | 10.95843 | 0.41060 | 65 | 1.36290 | 24.83766 | 0.51679 | 93 | 0.66005 | -0.40880 | 155.651 | 0.68325 | 0.75298 |
| E4.1.3.3, nanA, NPL; N-acetylneuraminate lyase [EC:4.1.3.3] | 1.24747 | 0.17966 | 1.09949 | 2.55363 | 0.19821 | 65 | 1.35090 | 6.90057 | 0.27240 | 93 | 0.33688 | -0.74629 | 153.397 | 0.45663 | 0.59014 |
| K07454; putative restriction endonuclease | 1.24684 | 0.64160 | 1.24615 | 84.15721 | 1.13786 | 65 | 1.24731 | 52.44904 | 0.75098 | 93 | 1.36334 | -0.00085 | 116.519 | 0.99932 | 0.99932 |
| rgpA_B; gingipain R [EC:3.4.22.37] | 1.24684 | 0.64160 | 1.24615 | 84.15721 | 1.13786 | 65 | 1.24731 | 52.44904 | 0.75098 | 93 | 1.36334 | -0.00085 | 116.519 | 0.99932 | 0.99932 |
| metY; O-acetylhomoserine (thiol)-lyase [EC:2.5.1.49] | 1.22934 | 0.18238 | 1.08240 | 3.28065 | 0.22466 | 65 | 1.33204 | 6.66094 | 0.26762 | 93 | 0.34942 | -0.71445 | 155.994 | 0.47602 | 0.59592 |
| licD; lipopolysaccharide cholinephosphotransferase [EC:2.7.8.-] | 1.22844 | 0.18331 | 1.10355 | 3.79335 | 0.24158 | 65 | 1.31572 | 6.40294 | 0.26239 | 93 | 0.35666 | -0.59487 | 154.497 | 0.55280 | 0.64989 |
| cusB, silB; membrane fusion protein, Cu(I)/Ag(I) efflux system | 1.22152 | 0.31319 | 1.25385 | 12.96190 | 0.44656 | 65 | 1.19892 | 17.43011 | 0.43292 | 93 | 0.62196 | 0.08830 | 149.171 | 0.92975 | 0.94278 |
| metH, MTR; 5-methyltetrahydrofolate–homocysteine methyltransferase [EC:2.1.1.13] | 1.21907 | 0.18450 | 1.07449 | 3.26570 | 0.22415 | 65 | 1.32012 | 6.88104 | 0.27201 | 93 | 0.35246 | -0.69689 | 155.978 | 0.48691 | 0.60286 |
| hya; hyaluronoglucosaminidase [EC:3.2.1.35] | 1.20863 | 0.18683 | 1.03736 | 2.53613 | 0.19753 | 65 | 1.32834 | 7.61253 | 0.28610 | 93 | 0.34767 | -0.83694 | 151.219 | 0.40394 | 0.59014 |
| E3.5.1.1, ansA, ansB; L-asparaginase [EC:3.5.1.1] | 1.20334 | 0.19490 | 1.02451 | 3.97096 | 0.24717 | 65 | 1.32833 | 7.44163 | 0.28287 | 93 | 0.37565 | -0.80879 | 155.669 | 0.41987 | 0.59014 |
| cbe, mbe; cellobiose epimerase [EC:5.1.3.11] | 1.19937 | 0.33040 | 0.81538 | 6.28179 | 0.31087 | 65 | 1.46774 | 24.88754 | 0.51731 | 93 | 0.60353 | -1.08090 | 143.538 | 0.28155 | 0.59014 |
| mgp; 4-O-beta-D-mannosyl-D-glucose phosphorylase [EC:2.4.1.281] | 1.19937 | 0.33040 | 0.81538 | 6.28179 | 0.31087 | 65 | 1.46774 | 24.88754 | 0.51731 | 93 | 0.60353 | -1.08090 | 143.538 | 0.28155 | 0.59014 |
| nhaA; Na+:H+ antiporter, NhaA family | 1.19753 | 0.17902 | 1.05628 | 3.67865 | 0.23790 | 65 | 1.29626 | 6.05811 | 0.25523 | 93 | 0.34891 | -0.68783 | 154.098 | 0.49259 | 0.60550 |
| ABC.CD.P; putative ABC transport system permease protein | 1.19611 | 0.28547 | 0.78201 | 6.08754 | 0.30603 | 65 | 1.48553 | 17.53267 | 0.43419 | 93 | 0.53120 | -1.32437 | 152.139 | 0.18736 | 0.59014 |
| ldhA; D-lactate dehydrogenase [EC:1.1.1.28] | 1.19488 | 0.16852 | 1.13333 | 2.77166 | 0.20650 | 65 | 1.23790 | 5.72496 | 0.24811 | 93 | 0.32280 | -0.32395 | 155.999 | 0.74641 | 0.80237 |
| fabG; 3-oxoacyl-[acyl-carrier protein] reductase [EC:1.1.1.100] | 1.17501 | 0.17336 | 0.94743 | 2.41854 | 0.19289 | 65 | 1.33407 | 6.35848 | 0.26148 | 93 | 0.32493 | -1.18994 | 153.873 | 0.23590 | 0.59014 |
| glxK, garK; glycerate 2-kinase [EC:2.7.1.165] | 1.17495 | 0.18401 | 1.03538 | 3.52709 | 0.23294 | 65 | 1.27250 | 6.65223 | 0.26745 | 93 | 0.35467 | -0.66854 | 155.713 | 0.50478 | 0.60889 |
| tag; DNA-3-methyladenine glycosylase I [EC:3.2.2.20] | 1.17245 | 0.18913 | 1.01953 | 3.52469 | 0.23286 | 65 | 1.27934 | 7.16494 | 0.27757 | 93 | 0.36231 | -0.71708 | 155.995 | 0.47440 | 0.59518 |
| E3.1.11.2, xthA; exodeoxyribonuclease III [EC:3.1.11.2] | 1.17169 | 0.16825 | 1.07809 | 3.36481 | 0.22752 | 65 | 1.23712 | 5.28178 | 0.23831 | 93 | 0.32948 | -0.48265 | 153.192 | 0.63003 | 0.71387 |
| fucP; MFS transporter, FHS family, L-fucose permease | 1.16825 | 0.16725 | 0.97793 | 2.68789 | 0.20335 | 65 | 1.30126 | 5.62901 | 0.24602 | 93 | 0.31919 | -1.01298 | 155.988 | 0.31264 | 0.59014 |
| hlyIII; hemolysin III | 1.16781 | 0.19075 | 1.03154 | 3.10997 | 0.21874 | 65 | 1.26306 | 7.62508 | 0.28634 | 93 | 0.36033 | -0.64252 | 154.884 | 0.52148 | 0.62384 |
| K08884; serine/threonine protein kinase, bacterial [EC:2.7.11.1] | 1.16715 | 0.18260 | 1.10070 | 3.29820 | 0.22526 | 65 | 1.21359 | 6.69079 | 0.26822 | 93 | 0.35026 | -0.32229 | 155.993 | 0.74766 | 0.80237 |
| nosL; copper chaperone NosL | 1.16456 | 0.54031 | 0.95385 | 37.38846 | 0.75842 | 65 | 1.31183 | 52.65171 | 0.75243 | 93 | 1.06834 | -0.33508 | 150.536 | 0.73803 | 0.79501 |
| oprO_P; phosphate-selective porin OprO and OprP | 1.15605 | 0.16847 | 1.03558 | 2.23565 | 0.18546 | 65 | 1.24026 | 6.08012 | 0.25569 | 93 | 0.31587 | -0.64799 | 153.280 | 0.51796 | 0.62091 |
| lepB; signal peptidase I [EC:3.4.21.89] | 1.15362 | 0.16620 | 1.00605 | 2.79378 | 0.20732 | 65 | 1.25677 | 5.47819 | 0.24270 | 93 | 0.31920 | -0.78547 | 155.913 | 0.43337 | 0.59014 |
| mntH; manganese transport protein | 1.14789 | 0.17859 | 1.00000 | 2.42437 | 0.19313 | 65 | 1.25125 | 6.88687 | 0.27213 | 93 | 0.33369 | -0.75295 | 152.428 | 0.45264 | 0.59014 |
| E3.1.3.1, phoA, phoB; alkaline phosphatase [EC:3.1.3.1] | 1.14684 | 0.25842 | 0.88923 | 3.79879 | 0.24175 | 65 | 1.32688 | 15.28416 | 0.40540 | 93 | 0.47200 | -0.92722 | 143.060 | 0.35538 | 0.59014 |
| ald; alanine dehydrogenase [EC:1.4.1.1] | 1.14557 | 0.30192 | 0.74615 | 3.00877 | 0.21515 | 65 | 1.42473 | 22.29454 | 0.48962 | 93 | 0.53480 | -1.26883 | 124.297 | 0.20687 | 0.59014 |
| K07058; membrane protein | 1.13994 | 0.17017 | 0.99015 | 3.23971 | 0.22325 | 65 | 1.24462 | 5.52682 | 0.24378 | 93 | 0.33056 | -0.76981 | 154.655 | 0.44258 | 0.59014 |
| K06885; uncharacterized protein | 1.13515 | 0.16799 | 1.05928 | 3.06616 | 0.21719 | 65 | 1.18817 | 5.46965 | 0.24251 | 93 | 0.32555 | -0.39593 | 155.222 | 0.69270 | 0.75849 |
| E4.3.1.19, ilvA, tdcB; threonine dehydratase [EC:4.3.1.19] | 1.13063 | 0.25296 | 0.88985 | 3.75645 | 0.24040 | 65 | 1.29892 | 14.57011 | 0.39581 | 93 | 0.46310 | -0.88335 | 144.189 | 0.37852 | 0.59014 |
| K07727; putative transcriptional regulator | 1.12785 | 0.31577 | 0.99077 | 9.50929 | 0.38249 | 65 | 1.22366 | 20.24770 | 0.46660 | 93 | 0.60334 | -0.38600 | 155.955 | 0.70002 | 0.76402 |
| ybaZ; methylated-DNA-protein-cysteine methyltransferase related protein | 1.12243 | 0.17770 | 0.98462 | 2.70905 | 0.20415 | 65 | 1.21875 | 6.60687 | 0.26654 | 93 | 0.33574 | -0.69738 | 154.949 | 0.48661 | 0.60286 |
| cynT, can; carbonic anhydrase [EC:4.2.1.1] | 1.12238 | 0.18217 | 1.01287 | 3.32961 | 0.22633 | 65 | 1.19892 | 6.61767 | 0.26675 | 93 | 0.34983 | -0.53185 | 155.956 | 0.59559 | 0.68219 |
| bcrC; undecaprenyl-diphosphatase [EC:3.6.1.27] | 1.12209 | 0.16612 | 0.92515 | 2.57597 | 0.19907 | 65 | 1.25973 | 5.60195 | 0.24543 | 93 | 0.31602 | -1.05876 | 155.884 | 0.29134 | 0.59014 |
| mrr; restriction system protein | 1.11392 | 0.31049 | 0.92308 | 3.93149 | 0.24594 | 65 | 1.24731 | 23.21535 | 0.49963 | 93 | 0.55688 | -0.58224 | 130.934 | 0.56141 | 0.65734 |
| abgT; aminobenzoyl-glutamate transport protein | 1.10814 | 0.16890 | 1.02879 | 3.19546 | 0.22172 | 65 | 1.16359 | 5.46169 | 0.24234 | 93 | 0.32846 | -0.41040 | 154.682 | 0.68208 | 0.75298 |
| glpT; MFS transporter, OPA family, glycerol-3-phosphate transporter | 1.10601 | 0.29987 | 0.75000 | 3.01562 | 0.21539 | 65 | 1.35484 | 21.99636 | 0.48633 | 93 | 0.53190 | -1.13714 | 124.733 | 0.25766 | 0.59014 |
| mod; adenine-specific DNA-methyltransferase [EC:2.1.1.72] | 1.09620 | 0.24077 | 0.98769 | 5.41679 | 0.28868 | 65 | 1.17204 | 11.84870 | 0.35694 | 93 | 0.45906 | -0.40158 | 155.859 | 0.68854 | 0.75648 |
| PREP; prolyl oligopeptidase [EC:3.4.21.26] | 1.09605 | 0.17348 | 0.96352 | 3.09898 | 0.21835 | 65 | 1.18868 | 5.93799 | 0.25268 | 93 | 0.33396 | -0.67425 | 155.809 | 0.50115 | 0.60578 |
| terC; tellurite resistance protein TerC | 1.09367 | 0.29995 | 0.72385 | 3.00696 | 0.21508 | 65 | 1.35215 | 22.00304 | 0.48641 | 93 | 0.53184 | -1.18138 | 124.645 | 0.23970 | 0.59014 |
| thyX, thy1; thymidylate synthase (FAD) [EC:2.1.1.148] | 1.09367 | 0.29995 | 0.72385 | 3.00696 | 0.21508 | 65 | 1.35215 | 22.00304 | 0.48641 | 93 | 0.53184 | -1.18138 | 124.645 | 0.23970 | 0.59014 |
| AMY, amyA, malS; alpha-amylase [EC:3.2.1.1] | 1.08983 | 0.16030 | 0.92148 | 2.14612 | 0.18171 | 65 | 1.20749 | 5.40175 | 0.24100 | 93 | 0.30183 | -0.94758 | 154.539 | 0.34482 | 0.59014 |
| kbl, GCAT; glycine C-acetyltransferase [EC:2.3.1.29] | 1.08936 | 0.19753 | 0.88974 | 2.30356 | 0.18825 | 65 | 1.22888 | 8.86981 | 0.30883 | 93 | 0.36168 | -0.93766 | 144.411 | 0.34998 | 0.59014 |
| trpC; indole-3-glycerol phosphate synthase [EC:4.1.1.48] | 1.08875 | 0.22804 | 0.85846 | 5.14544 | 0.28135 | 65 | 1.24970 | 10.37860 | 0.33406 | 93 | 0.43676 | -0.89578 | 155.986 | 0.37175 | 0.59014 |
| dacB; serine-type D-Ala-D-Ala carboxypeptidase/endopeptidase (penicillin-binding protein 4) [EC:3.4.16.4 3.4.21.-] | 1.08466 | 0.16430 | 0.90631 | 2.54642 | 0.19793 | 65 | 1.20931 | 5.46909 | 0.24250 | 93 | 0.31302 | -0.96796 | 155.930 | 0.33456 | 0.59014 |
| dps; starvation-inducible DNA-binding protein | 1.08310 | 0.15848 | 0.94482 | 2.65002 | 0.20191 | 65 | 1.17976 | 4.90594 | 0.22968 | 93 | 0.30581 | -0.76825 | 155.576 | 0.44350 | 0.59014 |
| glk; glucokinase [EC:2.7.1.2] | 1.08069 | 0.15300 | 0.91523 | 2.08960 | 0.17930 | 65 | 1.19633 | 4.82502 | 0.22778 | 93 | 0.28988 | -0.96973 | 155.508 | 0.33369 | 0.59014 |
| dfx; superoxide reductase [EC:1.15.1.2] | 1.07949 | 0.16837 | 0.97557 | 3.03351 | 0.21603 | 65 | 1.15212 | 5.52058 | 0.24364 | 93 | 0.32562 | -0.54220 | 155.426 | 0.58846 | 0.67888 |
| dcuA; anaerobic C4-dicarboxylate transporter DcuA | 1.07665 | 0.24521 | 0.88205 | 6.09795 | 0.30629 | 65 | 1.21266 | 11.92422 | 0.35807 | 93 | 0.47120 | -0.70164 | 155.903 | 0.48395 | 0.60131 |
| INO1, ISYNA1; myo-inositol-1-phosphate synthase [EC:5.5.1.4] | 1.06705 | 0.17302 | 0.97795 | 3.53452 | 0.23319 | 65 | 1.12933 | 5.60300 | 0.24545 | 93 | 0.33856 | -0.44713 | 153.392 | 0.65541 | 0.73686 |
| serB, PSPH; phosphoserine phosphatase [EC:3.1.3.3] | 1.06604 | 0.16400 | 0.92092 | 3.01917 | 0.21552 | 65 | 1.16747 | 5.12606 | 0.23477 | 93 | 0.31870 | -0.77362 | 154.585 | 0.44034 | 0.59014 |
| K07005; uncharacterized protein | 1.06111 | 0.17224 | 0.92148 | 3.22300 | 0.22268 | 65 | 1.15870 | 5.73368 | 0.24830 | 93 | 0.33352 | -0.71126 | 155.192 | 0.47799 | 0.59710 |
| ftsI; cell division protein FtsI (penicillin-binding protein 3) [EC:3.4.16.4] | 1.05995 | 0.16396 | 0.92053 | 2.90314 | 0.21134 | 65 | 1.15740 | 5.20582 | 0.23659 | 93 | 0.31724 | -0.74665 | 155.278 | 0.45640 | 0.59014 |
| FTCD; glutamate formiminotransferase / formiminotetrahydrofolate cyclodeaminase [EC:2.1.2.5 4.3.1.4] | 1.05802 | 0.17069 | 0.95855 | 3.47823 | 0.23132 | 65 | 1.12754 | 5.42424 | 0.24151 | 93 | 0.33442 | -0.50533 | 153.056 | 0.61405 | 0.69919 |
| hutH, HAL; histidine ammonia-lyase [EC:4.3.1.3] | 1.05802 | 0.17069 | 0.95855 | 3.47823 | 0.23132 | 65 | 1.12754 | 5.42424 | 0.24151 | 93 | 0.33442 | -0.50533 | 153.056 | 0.61405 | 0.69919 |
| hutI, AMDHD1; imidazolonepropionase [EC:3.5.2.7] | 1.05802 | 0.17069 | 0.95855 | 3.47823 | 0.23132 | 65 | 1.12754 | 5.42424 | 0.24151 | 93 | 0.33442 | -0.50533 | 153.056 | 0.61405 | 0.69919 |
| hutU, UROC1; urocanate hydratase [EC:4.2.1.49] | 1.05802 | 0.17069 | 0.95855 | 3.47823 | 0.23132 | 65 | 1.12754 | 5.42424 | 0.24151 | 93 | 0.33442 | -0.50533 | 153.056 | 0.61405 | 0.69919 |
| pncA; nicotinamidase/pyrazinamidase [EC:3.5.1.19 3.5.1.-] | 1.05680 | 0.16226 | 0.99676 | 3.08655 | 0.21791 | 65 | 1.09875 | 4.94772 | 0.23065 | 93 | 0.31731 | -0.32143 | 153.610 | 0.74832 | 0.80237 |
| K07003; uncharacterized protein | 1.05654 | 0.17686 | 0.90667 | 2.63456 | 0.20132 | 65 | 1.16129 | 6.57433 | 0.26588 | 93 | 0.33350 | -0.76349 | 154.657 | 0.44634 | 0.59014 |
| rnfG; electron transport complex protein RnfG | 1.05371 | 0.16546 | 0.91902 | 2.82409 | 0.20844 | 65 | 1.14785 | 5.39544 | 0.24086 | 93 | 0.31853 | -0.71838 | 155.792 | 0.47360 | 0.59518 |
| ahpF; alkyl hydroperoxide reductase subunit F [EC:1.6.4.-] | 1.04830 | 0.16066 | 0.97530 | 3.06455 | 0.21713 | 65 | 1.09932 | 4.82177 | 0.22770 | 93 | 0.31463 | -0.39416 | 153.241 | 0.69401 | 0.75849 |
| tex; protein Tex | 1.04739 | 0.15224 | 0.92890 | 2.45406 | 0.19431 | 65 | 1.13020 | 4.52556 | 0.22059 | 93 | 0.29397 | -0.68479 | 155.544 | 0.49450 | 0.60550 |
| ABC-2.OM, wza; polysaccharide biosynthesis/export protein | 1.03813 | 0.15560 | 0.86960 | 2.12907 | 0.18098 | 65 | 1.15591 | 5.01277 | 0.23217 | 93 | 0.29437 | -0.97261 | 155.332 | 0.33226 | 0.59014 |
| cpdB; 2’,3’-cyclic-nucleotide 2’-phosphodiesterase / 3’-nucleotidase [EC:3.1.4.16 3.1.3.6] | 1.03813 | 0.15560 | 0.86960 | 2.12907 | 0.18098 | 65 | 1.15591 | 5.01277 | 0.23217 | 93 | 0.29437 | -0.97261 | 155.332 | 0.33226 | 0.59014 |
| fabH; 3-oxoacyl-[acyl-carrier-protein] synthase III [EC:2.3.1.180] | 1.03420 | 0.16752 | 0.90143 | 3.04246 | 0.21635 | 65 | 1.12699 | 5.42925 | 0.24162 | 93 | 0.32432 | -0.69548 | 155.226 | 0.48780 | 0.60286 |
| gltD; glutamate synthase (NADPH/NADH) small chain [EC:1.4.1.13 1.4.1.14] | 1.03280 | 0.15847 | 0.84319 | 2.11677 | 0.18046 | 65 | 1.16532 | 5.25552 | 0.23772 | 93 | 0.29846 | -1.07934 | 154.724 | 0.28212 | 0.59014 |
| aspA; aspartate ammonia-lyase [EC:4.3.1.1] | 1.02337 | 0.15977 | 0.87191 | 2.22413 | 0.18498 | 65 | 1.12923 | 5.30824 | 0.23891 | 93 | 0.30215 | -0.85164 | 155.194 | 0.39573 | 0.59014 |
| buk; butyrate kinase [EC:2.7.2.7] | 1.02182 | 0.16349 | 0.93441 | 3.15985 | 0.22048 | 65 | 1.08291 | 4.99974 | 0.23186 | 93 | 0.31996 | -0.46411 | 153.355 | 0.64323 | 0.72528 |
| ptb; phosphate butyryltransferase [EC:2.3.1.19] | 1.02182 | 0.16349 | 0.93441 | 3.15985 | 0.22048 | 65 | 1.08291 | 4.99974 | 0.23186 | 93 | 0.31996 | -0.46411 | 153.355 | 0.64323 | 0.72528 |
| rluA; tRNA pseudouridine32 synthase / 23S rRNA pseudouridine746 synthase [EC:5.4.99.28 5.4.99.29] | 1.02173 | 0.16584 | 0.89071 | 2.97830 | 0.21406 | 65 | 1.11330 | 5.32313 | 0.23924 | 93 | 0.32103 | -0.69337 | 155.243 | 0.48912 | 0.60319 |
| pdxK, pdxY; pyridoxine kinase [EC:2.7.1.35] | 1.02128 | 0.15197 | 0.91167 | 2.41098 | 0.19259 | 65 | 1.09789 | 4.53521 | 0.22083 | 93 | 0.29301 | -0.63556 | 155.695 | 0.52600 | 0.62749 |
| ATOX1, ATX1, copZ, golB; copper chaperone | 1.02056 | 0.15883 | 0.87906 | 2.58386 | 0.19938 | 65 | 1.11945 | 4.98020 | 0.23141 | 93 | 0.30545 | -0.78698 | 155.839 | 0.43249 | 0.59014 |
| wecB; UDP-N-acetylglucosamine 2-epimerase (non-hydrolysing) [EC:5.1.3.14] | 1.01984 | 0.14519 | 0.85273 | 1.82545 | 0.16758 | 65 | 1.13663 | 4.38041 | 0.21703 | 93 | 0.27420 | -1.03538 | 155.135 | 0.30210 | 0.59014 |
| K07052; uncharacterized protein | 1.01969 | 0.13517 | 0.87385 | 1.91481 | 0.17164 | 65 | 1.12162 | 3.56846 | 0.19588 | 93 | 0.26044 | -0.95135 | 155.628 | 0.34290 | 0.59014 |
| hisC; histidinol-phosphate aminotransferase [EC:2.6.1.9] | 1.01905 | 0.14116 | 0.92515 | 1.83094 | 0.16783 | 65 | 1.08468 | 4.08823 | 0.20967 | 93 | 0.26857 | -0.59398 | 155.750 | 0.55339 | 0.64992 |
| radA, sms; DNA repair protein RadA/Sms | 1.01805 | 0.15154 | 0.85316 | 1.99073 | 0.17500 | 65 | 1.13330 | 4.77463 | 0.22658 | 93 | 0.28630 | -0.97849 | 155.141 | 0.32936 | 0.59014 |
| crcB, FEX; fluoride exporter | 1.01696 | 0.15512 | 0.87756 | 2.30760 | 0.18842 | 65 | 1.11439 | 4.85906 | 0.22858 | 93 | 0.29623 | -0.79948 | 155.979 | 0.42523 | 0.59014 |
| fes; enterochelin esterase and related enzymes | 1.00904 | 0.15526 | 0.88949 | 2.15418 | 0.18205 | 65 | 1.09259 | 4.98431 | 0.23151 | 93 | 0.29451 | -0.68964 | 155.490 | 0.49145 | 0.60478 |
| dxs; 1-deoxy-D-xylulose-5-phosphate synthase [EC:2.2.1.7] | 1.00849 | 0.14842 | 0.88589 | 2.52045 | 0.19692 | 65 | 1.09418 | 4.16802 | 0.21170 | 93 | 0.28913 | -0.72041 | 154.169 | 0.47236 | 0.59518 |
| eptC; heptose-I-phosphate ethanolaminephosphotransferase [EC:2.7.8.-] | 1.00264 | 0.13762 | 0.86687 | 1.96035 | 0.17366 | 65 | 1.09754 | 3.72060 | 0.20002 | 93 | 0.26489 | -0.87084 | 155.753 | 0.38518 | 0.59014 |
| rhaA; L-rhamnose isomerase [EC:5.3.1.14] | 0.99831 | 0.38400 | 0.57538 | 4.10133 | 0.25119 | 65 | 1.29391 | 36.69046 | 0.62811 | 93 | 0.67647 | -1.06216 | 119.391 | 0.29031 | 0.59014 |
| rhaB; rhamnulokinase [EC:2.7.1.5] | 0.99831 | 0.38400 | 0.57538 | 4.10133 | 0.25119 | 65 | 1.29391 | 36.69046 | 0.62811 | 93 | 0.67647 | -1.06216 | 119.391 | 0.29031 | 0.59014 |
| rhaD; rhamnulose-1-phosphate aldolase [EC:4.1.2.19] | 0.99831 | 0.38400 | 0.57538 | 4.10133 | 0.25119 | 65 | 1.29391 | 36.69046 | 0.62811 | 93 | 0.67647 | -1.06216 | 119.391 | 0.29031 | 0.59014 |
| TC.KEF; monovalent cation:H+ antiporter-2, CPA2 family | 0.99360 | 0.14904 | 0.88651 | 2.70026 | 0.20382 | 65 | 1.06844 | 4.09676 | 0.20988 | 93 | 0.29256 | -0.62187 | 152.447 | 0.53496 | 0.63602 |
| PPOX, hemY; protoporphyrinogen/coproporphyrinogen III oxidase [EC:1.3.3.4 1.3.3.15] | 0.99084 | 0.14020 | 0.85900 | 2.02211 | 0.17638 | 65 | 1.08298 | 3.87268 | 0.20406 | 93 | 0.26972 | -0.83043 | 155.806 | 0.40757 | 0.59014 |
| vanX; zinc D-Ala-D-Ala dipeptidase [EC:3.4.13.22] | 0.99079 | 0.13819 | 0.87704 | 2.03436 | 0.17691 | 65 | 1.07029 | 3.71838 | 0.19996 | 93 | 0.26698 | -0.72384 | 155.466 | 0.47025 | 0.59353 |
| cutC; copper homeostasis protein | 0.98563 | 0.14506 | 0.82949 | 1.97063 | 0.17412 | 65 | 1.09476 | 4.27329 | 0.21436 | 93 | 0.27616 | -0.96055 | 155.895 | 0.33826 | 0.59014 |
| ACSS, acs; acetyl-CoA synthetase [EC:6.2.1.1] | 0.98474 | 0.14425 | 0.86939 | 1.81708 | 0.16720 | 65 | 1.06537 | 4.33031 | 0.21578 | 93 | 0.27298 | -0.71790 | 155.209 | 0.47390 | 0.59518 |
| pcaC; 4-carboxymuconolactone decarboxylase [EC:4.1.1.44] | 0.97848 | 0.29526 | 0.90154 | 3.88172 | 0.24437 | 65 | 1.03226 | 20.79786 | 0.47290 | 93 | 0.53231 | -0.24557 | 133.963 | 0.80639 | 0.85356 |
| nrfA; nitrite reductase (cytochrome c-552) [EC:1.7.2.2] | 0.97651 | 0.14137 | 0.85444 | 2.10075 | 0.17978 | 65 | 1.06183 | 3.90918 | 0.20502 | 93 | 0.27268 | -0.76057 | 155.617 | 0.44807 | 0.59014 |
| nrfH; cytochrome c nitrite reductase small subunit | 0.97651 | 0.14137 | 0.85444 | 2.10075 | 0.17978 | 65 | 1.06183 | 3.90918 | 0.20502 | 93 | 0.27268 | -0.76057 | 155.617 | 0.44807 | 0.59014 |
| tatD; TatD DNase family protein [EC:3.1.21.-] | 0.97593 | 0.13336 | 0.84467 | 1.84709 | 0.16857 | 65 | 1.06767 | 3.48953 | 0.19371 | 93 | 0.25679 | -0.86843 | 155.724 | 0.38650 | 0.59014 |
| lemA; LemA protein | 0.97009 | 0.13585 | 0.84187 | 1.92458 | 0.17207 | 65 | 1.05972 | 3.61748 | 0.19722 | 93 | 0.26174 | -0.83231 | 155.690 | 0.40651 | 0.59014 |
| sucC; succinyl-CoA synthetase beta subunit [EC:6.2.1.5] | 0.96820 | 0.15414 | 0.87154 | 2.79613 | 0.20741 | 65 | 1.03575 | 4.44950 | 0.21873 | 93 | 0.30143 | -0.54478 | 153.468 | 0.58670 | 0.67888 |
| sucD; succinyl-CoA synthetase alpha subunit [EC:6.2.1.5] | 0.96820 | 0.15414 | 0.87154 | 2.79613 | 0.20741 | 65 | 1.03575 | 4.44950 | 0.21873 | 93 | 0.30143 | -0.54478 | 153.468 | 0.58670 | 0.67888 |
| E2.1.1.72; site-specific DNA-methyltransferase (adenine-specific) [EC:2.1.1.72] | 0.96632 | 0.15067 | 0.77026 | 1.49461 | 0.15164 | 65 | 1.10335 | 5.03540 | 0.23269 | 93 | 0.27774 | -1.19929 | 148.289 | 0.23233 | 0.59014 |
| gpmI; 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [EC:5.4.2.12] | 0.96549 | 0.14717 | 0.82782 | 2.22262 | 0.18492 | 65 | 1.06172 | 4.27066 | 0.21429 | 93 | 0.28305 | -0.82638 | 155.823 | 0.40985 | 0.59014 |
| RENBP; N-acylglucosamine 2-epimerase [EC:5.1.3.8] | 0.96313 | 0.13569 | 0.83077 | 1.43388 | 0.14853 | 65 | 1.05565 | 3.94579 | 0.20598 | 93 | 0.25394 | -0.88553 | 153.059 | 0.37726 | 0.59014 |
| araA; L-arabinose isomerase [EC:5.3.1.4] | 0.96234 | 0.29423 | 0.61615 | 2.50509 | 0.19632 | 65 | 1.20430 | 21.45645 | 0.48033 | 93 | 0.51890 | -1.13346 | 120.471 | 0.25927 | 0.59014 |
| araD, ulaF, sgaE, sgbE; L-ribulose-5-phosphate 4-epimerase [EC:5.1.3.4] | 0.96234 | 0.29423 | 0.61615 | 2.50509 | 0.19632 | 65 | 1.20430 | 21.45645 | 0.48033 | 93 | 0.51890 | -1.13346 | 120.471 | 0.25927 | 0.59014 |
| sacC, levB; levanase [EC:3.2.1.65] | 0.95660 | 0.14949 | 0.83417 | 1.91419 | 0.17161 | 65 | 1.04216 | 4.67583 | 0.22423 | 93 | 0.28236 | -0.73662 | 154.930 | 0.46247 | 0.59014 |
| E3.1.21.4; type II restriction enzyme [EC:3.1.21.4] | 0.95640 | 0.21683 | 0.77094 | 1.69015 | 0.16125 | 65 | 1.08602 | 11.46005 | 0.35104 | 93 | 0.38630 | -0.81564 | 126.806 | 0.41624 | 0.59014 |
| TPI, tpiA; triosephosphate isomerase (TIM) [EC:5.3.1.1] | 0.95567 | 0.14137 | 0.82408 | 2.22079 | 0.18484 | 65 | 1.04765 | 3.82301 | 0.20275 | 93 | 0.27436 | -0.81484 | 154.782 | 0.41641 | 0.59014 |
| copB; Cu2+-exporting ATPase [EC:3.6.3.4] | 0.95476 | 0.13169 | 0.83194 | 1.80421 | 0.16660 | 65 | 1.04060 | 3.40276 | 0.19128 | 93 | 0.25366 | -0.82260 | 155.713 | 0.41199 | 0.59014 |
| E2.3.1.54, pflD; formate C-acetyltransferase [EC:2.3.1.54] | 0.95416 | 0.13648 | 0.83621 | 1.95387 | 0.17338 | 65 | 1.03660 | 3.64631 | 0.19801 | 93 | 0.26319 | -0.76139 | 155.638 | 0.44758 | 0.59014 |
| K09704; uncharacterized protein | 0.95269 | 0.13225 | 0.83077 | 1.38056 | 0.14574 | 65 | 1.03790 | 3.73760 | 0.20047 | 93 | 0.24785 | -0.83573 | 153.363 | 0.40461 | 0.59014 |
| metF, MTHFR; methylenetetrahydrofolate reductase (NADPH) [EC:1.5.1.20] | 0.95074 | 0.14143 | 0.80742 | 1.69554 | 0.16151 | 65 | 1.05092 | 4.18928 | 0.21224 | 93 | 0.26670 | -0.91298 | 154.787 | 0.36267 | 0.59014 |
| maa; maltose O-acetyltransferase [EC:2.3.1.79] | 0.95002 | 0.15103 | 0.82438 | 2.14875 | 0.18182 | 65 | 1.03784 | 4.63628 | 0.22328 | 93 | 0.28794 | -0.74134 | 155.914 | 0.45960 | 0.59014 |
| thyA, TYMS; thymidylate synthase [EC:2.1.1.45] | 0.94975 | 0.14176 | 0.85538 | 2.65241 | 0.20201 | 65 | 1.01570 | 3.56296 | 0.19573 | 93 | 0.28128 | -0.56995 | 149.138 | 0.56957 | 0.66555 |
| fic; cell filamentation protein | 0.94843 | 0.13963 | 0.83194 | 2.33880 | 0.18969 | 65 | 1.02985 | 3.61389 | 0.19713 | 93 | 0.27357 | -0.72344 | 152.858 | 0.47051 | 0.59353 |
| thiN, TPK1, THI80; thiamine pyrophosphokinase [EC:2.7.6.2] | 0.94759 | 0.14099 | 0.83917 | 2.06207 | 0.17811 | 65 | 1.02337 | 3.91100 | 0.20507 | 93 | 0.27162 | -0.67813 | 155.749 | 0.49869 | 0.60550 |
| xapB; MFS transporter, NHS family, xanthosine permease | 0.94620 | 0.22482 | 0.96154 | 5.62740 | 0.29424 | 65 | 0.93548 | 9.71318 | 0.32318 | 93 | 0.43706 | 0.05961 | 154.818 | 0.95254 | 0.95750 |
| K06872; uncharacterized protein | 0.94536 | 0.23737 | 0.80821 | 4.95858 | 0.27620 | 65 | 1.04122 | 11.72081 | 0.35501 | 93 | 0.44980 | -0.51804 | 155.293 | 0.60517 | 0.69179 |
| HSP20; HSP20 family protein | 0.94443 | 0.14656 | 0.80722 | 2.42157 | 0.19302 | 65 | 1.04032 | 4.08414 | 0.20956 | 93 | 0.28490 | -0.81818 | 154.485 | 0.41451 | 0.59014 |
| pyrP, uraA; uracil permease | 0.94365 | 0.13585 | 0.82604 | 1.91169 | 0.17150 | 65 | 1.02585 | 3.62981 | 0.19756 | 93 | 0.26161 | -0.76377 | 155.756 | 0.44616 | 0.59014 |
| xlyAB; N-acetylmuramoyl-L-alanine amidase [EC:3.5.1.28] | 0.94304 | 0.53233 | 0.30000 | 0.94375 | 0.12050 | 65 | 1.39247 | 75.25462 | 0.89955 | 93 | 0.90758 | -1.20372 | 95.287 | 0.23168 | 0.59014 |
| rlpA; rare lipoprotein A | 0.94292 | 0.13655 | 0.82507 | 1.94114 | 0.17281 | 65 | 1.02529 | 3.66028 | 0.19839 | 93 | 0.26310 | -0.76099 | 155.712 | 0.44781 | 0.59014 |
| feoB; ferrous iron transport protein B | 0.94232 | 0.12869 | 0.82610 | 1.75707 | 0.16441 | 65 | 1.02354 | 3.22699 | 0.18628 | 93 | 0.24846 | -0.79468 | 155.509 | 0.42801 | 0.59014 |
| oxyR; LysR family transcriptional regulator, hydrogen peroxide-inducible genes activator | 0.94222 | 0.13609 | 0.82678 | 1.94269 | 0.17288 | 65 | 1.02290 | 3.62596 | 0.19746 | 93 | 0.26244 | -0.74728 | 155.639 | 0.45602 | 0.59014 |
| K06889; uncharacterized protein | 0.94048 | 0.14384 | 0.81151 | 2.59305 | 0.19973 | 65 | 1.03062 | 3.75495 | 0.20094 | 93 | 0.28332 | -0.77340 | 151.296 | 0.44049 | 0.59014 |
| patB, malY; cystathione beta-lyase [EC:4.4.1.8] | 0.93790 | 0.14370 | 0.79458 | 2.27348 | 0.18702 | 65 | 1.03806 | 3.96174 | 0.20640 | 93 | 0.27852 | -0.87418 | 154.944 | 0.38337 | 0.59014 |
| K01163; uncharacterized protein | 0.93770 | 0.12889 | 0.87596 | 1.66601 | 0.16010 | 65 | 0.98085 | 3.31579 | 0.18882 | 93 | 0.24756 | -0.42368 | 155.959 | 0.67238 | 0.75298 |
| hcp; hydroxylamine reductase [EC:1.7.99.1] | 0.93453 | 0.13627 | 0.82348 | 1.94746 | 0.17309 | 65 | 1.01215 | 3.63721 | 0.19776 | 93 | 0.26281 | -0.71789 | 155.644 | 0.47390 | 0.59518 |
| DHFR, folA; dihydrofolate reductase [EC:1.5.1.3] | 0.93141 | 0.12843 | 0.82345 | 1.75378 | 0.16426 | 65 | 1.00686 | 3.21315 | 0.18588 | 93 | 0.24805 | -0.73940 | 155.487 | 0.46078 | 0.59014 |
| fldA, nifF, isiB; flavodoxin I | 0.93141 | 0.12843 | 0.82345 | 1.75378 | 0.16426 | 65 | 1.00686 | 3.21315 | 0.18588 | 93 | 0.24805 | -0.73940 | 155.487 | 0.46078 | 0.59014 |
| GCH1, folE; GTP cyclohydrolase IA [EC:3.5.4.16] | 0.93141 | 0.12843 | 0.82345 | 1.75378 | 0.16426 | 65 | 1.00686 | 3.21315 | 0.18588 | 93 | 0.24805 | -0.73940 | 155.487 | 0.46078 | 0.59014 |
| RP-L17, MRPL17, rplQ; large subunit ribosomal protein L17 | 0.93141 | 0.12843 | 0.82345 | 1.75378 | 0.16426 | 65 | 1.00686 | 3.21315 | 0.18588 | 93 | 0.24805 | -0.73940 | 155.487 | 0.46078 | 0.59014 |
| zntA; Cd2+/Zn2+-exporting ATPase [EC:3.6.3.3 3.6.3.5] | 0.93141 | 0.12843 | 0.82345 | 1.75378 | 0.16426 | 65 | 1.00686 | 3.21315 | 0.18588 | 93 | 0.24805 | -0.73940 | 155.487 | 0.46078 | 0.59014 |
| xanP; xanthine permease XanP | 0.93081 | 0.14043 | 0.86014 | 2.31371 | 0.18867 | 65 | 0.98021 | 3.70162 | 0.19951 | 93 | 0.27459 | -0.43726 | 153.572 | 0.66254 | 0.74343 |
| cusA, silA; Cu(I)/Ag(I) efflux system membrane protein CusA/SilA | 0.93038 | 0.21345 | 1.01538 | 6.32788 | 0.31201 | 65 | 0.87097 | 7.87447 | 0.29098 | 93 | 0.42664 | 0.33850 | 146.597 | 0.73547 | 0.79501 |
| IARS, ileS; isoleucyl-tRNA synthetase [EC:6.1.1.5] | 0.93031 | 0.13799 | 0.81284 | 2.28793 | 0.18761 | 65 | 1.01242 | 3.52594 | 0.19471 | 93 | 0.27039 | -0.73812 | 152.800 | 0.46157 | 0.59014 |
| lon; ATP-dependent Lon protease [EC:3.4.21.53] | 0.93031 | 0.13799 | 0.81284 | 2.28793 | 0.18761 | 65 | 1.01242 | 3.52594 | 0.19471 | 93 | 0.27039 | -0.73812 | 152.800 | 0.46157 | 0.59014 |
| purL, PFAS; phosphoribosylformylglycinamidine synthase [EC:6.3.5.3] | 0.93031 | 0.13799 | 0.81284 | 2.28793 | 0.18761 | 65 | 1.01242 | 3.52594 | 0.19471 | 93 | 0.27039 | -0.73812 | 152.800 | 0.46157 | 0.59014 |
| rpoA; DNA-directed RNA polymerase subunit alpha [EC:2.7.7.6] | 0.93031 | 0.13799 | 0.81284 | 2.28793 | 0.18761 | 65 | 1.01242 | 3.52594 | 0.19471 | 93 | 0.27039 | -0.73812 | 152.800 | 0.46157 | 0.59014 |
| URA4, pyrC; dihydroorotase [EC:3.5.2.3] | 0.93031 | 0.13799 | 0.81284 | 2.28793 | 0.18761 | 65 | 1.01242 | 3.52594 | 0.19471 | 93 | 0.27039 | -0.73812 | 152.800 | 0.46157 | 0.59014 |
| uup; ABC transport system ATP-binding/permease protein | 0.93031 | 0.13799 | 0.81284 | 2.28793 | 0.18761 | 65 | 1.01242 | 3.52594 | 0.19471 | 93 | 0.27039 | -0.73812 | 152.800 | 0.46157 | 0.59014 |
| pbuG; putative MFS transporter, AGZA family, xanthine/uracil permease | 0.93026 | 0.13556 | 0.81311 | 1.90908 | 0.17138 | 65 | 1.01215 | 3.61019 | 0.19703 | 93 | 0.26113 | -0.76223 | 155.731 | 0.44708 | 0.59014 |
| ybdG, mscM; miniconductance mechanosensitive channel | 0.92843 | 0.13560 | 0.80923 | 1.91412 | 0.17160 | 65 | 1.01175 | 3.60920 | 0.19700 | 93 | 0.26126 | -0.77516 | 155.711 | 0.43942 | 0.59014 |
| E2.2.1.1, tktA, tktB; transketolase [EC:2.2.1.1] | 0.92748 | 0.13732 | 0.80276 | 1.98606 | 0.17480 | 65 | 1.01465 | 3.68428 | 0.19904 | 93 | 0.26490 | -0.79988 | 155.592 | 0.42500 | 0.59014 |
| SLC13A2_3_5; solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2/3/5 | 0.92330 | 0.13956 | 0.81275 | 2.20743 | 0.18428 | 65 | 1.00058 | 3.70162 | 0.19951 | 93 | 0.27159 | -0.69158 | 154.395 | 0.49024 | 0.60393 |
| asnA; aspartate–ammonia ligase [EC:6.3.1.1] | 0.92260 | 0.13172 | 0.80930 | 1.52721 | 0.15328 | 65 | 1.00179 | 3.60065 | 0.19677 | 93 | 0.24942 | -0.77173 | 155.318 | 0.44145 | 0.59014 |
| lolA; outer membrane lipoprotein carrier protein | 0.92194 | 0.37171 | 0.55641 | 2.97507 | 0.21394 | 65 | 1.17742 | 35.02464 | 0.61368 | 93 | 0.64991 | -0.95554 | 113.315 | 0.34134 | 0.59014 |
| K09155; uncharacterized protein | 0.92077 | 0.13164 | 0.80110 | 1.80337 | 0.16657 | 65 | 1.00442 | 3.40072 | 0.19122 | 93 | 0.25360 | -0.80174 | 155.712 | 0.42393 | 0.59014 |
| hppA; K(+)-stimulated pyrophosphate-energized sodium pump [EC:3.6.1.1] | 0.92032 | 0.14160 | 0.79362 | 1.73841 | 0.16354 | 65 | 1.00887 | 4.17766 | 0.21195 | 93 | 0.26770 | -0.80408 | 155.119 | 0.42258 | 0.59014 |
| mp2; beta-1,4-mannooligosaccharide/beta-1,4-mannosyl-N-acetylglucosamine phosphorylase [EC:2.4.1.319 2.4.1.320] | 0.91968 | 0.12914 | 0.79634 | 1.29454 | 0.14112 | 65 | 1.00589 | 3.57808 | 0.19615 | 93 | 0.24164 | -0.86720 | 152.975 | 0.38719 | 0.59014 |
| NEU1; sialidase-1 [EC:3.2.1.18] | 0.91968 | 0.12914 | 0.79634 | 1.29454 | 0.14112 | 65 | 1.00589 | 3.57808 | 0.19615 | 93 | 0.24164 | -0.86720 | 152.975 | 0.38719 | 0.59014 |
| nadA; quinolinate synthase [EC:2.5.1.72] | 0.91959 | 0.14626 | 0.82835 | 2.54680 | 0.19794 | 65 | 0.98336 | 3.98623 | 0.20703 | 93 | 0.28643 | -0.54116 | 153.133 | 0.58918 | 0.67888 |
| nadB; L-aspartate oxidase [EC:1.4.3.16] | 0.91959 | 0.14626 | 0.82835 | 2.54680 | 0.19794 | 65 | 0.98336 | 3.98623 | 0.20703 | 93 | 0.28643 | -0.54116 | 153.133 | 0.58918 | 0.67888 |
| nadC, QPRT; nicotinate-nucleotide pyrophosphorylase (carboxylating) [EC:2.4.2.19] | 0.91959 | 0.14626 | 0.82835 | 2.54680 | 0.19794 | 65 | 0.98336 | 3.98623 | 0.20703 | 93 | 0.28643 | -0.54116 | 153.133 | 0.58918 | 0.67888 |
| pncB, NAPRT1; nicotinate phosphoribosyltransferase [EC:6.3.4.21] | 0.91896 | 0.13166 | 0.79725 | 1.80641 | 0.16671 | 65 | 1.00403 | 3.39979 | 0.19120 | 93 | 0.25367 | -0.81516 | 155.699 | 0.41623 | 0.59014 |
| PRDX2_4, ahpC; peroxiredoxin (alkyl hydroperoxide reductase subunit C) [EC:1.11.1.15] | 0.91896 | 0.13166 | 0.79725 | 1.80641 | 0.16671 | 65 | 1.00403 | 3.39979 | 0.19120 | 93 | 0.25367 | -0.81516 | 155.699 | 0.41623 | 0.59014 |
| cbpA; curved DNA-binding protein | 0.91887 | 0.14107 | 0.79716 | 2.05755 | 0.17792 | 65 | 1.00393 | 3.91646 | 0.20521 | 93 | 0.27160 | -0.76130 | 155.771 | 0.44763 | 0.59014 |
| K07011; uncharacterized protein | 0.91831 | 0.13226 | 0.78897 | 1.82753 | 0.16768 | 65 | 1.00871 | 3.42549 | 0.19192 | 93 | 0.25485 | -0.86226 | 155.670 | 0.38987 | 0.59014 |
| mltD, dniR; membrane-bound lytic murein transglycosylase D [EC:4.2.2.-] | 0.91720 | 0.13171 | 0.79352 | 1.81119 | 0.16693 | 65 | 1.00365 | 3.39889 | 0.19117 | 93 | 0.25379 | -0.82796 | 155.679 | 0.40896 | 0.59014 |
| cidA; holin-like protein | 0.91642 | 0.13156 | 0.85788 | 1.62602 | 0.15816 | 65 | 0.95733 | 3.53175 | 0.19487 | 93 | 0.25098 | -0.39621 | 155.889 | 0.69249 | 0.75849 |
| K07085; putative transport protein | 0.91574 | 0.12900 | 0.78589 | 1.72448 | 0.16288 | 65 | 1.00649 | 3.26720 | 0.18743 | 93 | 0.24832 | -0.88838 | 155.742 | 0.37571 | 0.59014 |
| ina; immune inhibitor A [EC:3.4.24.-] | 0.91497 | 0.14053 | 0.78059 | 2.02659 | 0.17657 | 65 | 1.00889 | 3.89311 | 0.20460 | 93 | 0.27026 | -0.84475 | 155.822 | 0.39955 | 0.59014 |
| purT; phosphoribosylglycinamide formyltransferase 2 [EC:2.1.2.2] | 0.91447 | 0.13111 | 0.81273 | 1.52673 | 0.15326 | 65 | 0.98558 | 3.56072 | 0.19567 | 93 | 0.24855 | -0.69546 | 155.420 | 0.48781 | 0.60286 |
| kdsB; 3-deoxy-manno-octulosonate cytidylyltransferase (CMP-KDO synthetase) [EC:2.7.7.38] | 0.91384 | 0.12900 | 0.78658 | 1.71297 | 0.16234 | 65 | 1.00278 | 3.27573 | 0.18768 | 93 | 0.24815 | -0.87127 | 155.798 | 0.38495 | 0.59014 |
| metK; S-adenosylmethionine synthetase [EC:2.5.1.6] | 0.91384 | 0.12900 | 0.78658 | 1.71297 | 0.16234 | 65 | 1.00278 | 3.27573 | 0.18768 | 93 | 0.24815 | -0.87127 | 155.798 | 0.38495 | 0.59014 |
| purF, PPAT; amidophosphoribosyltransferase [EC:2.4.2.14] | 0.91268 | 0.13560 | 0.79756 | 2.08608 | 0.17915 | 65 | 0.99314 | 3.49048 | 0.19373 | 93 | 0.26387 | -0.74121 | 154.361 | 0.45969 | 0.59014 |
| UGDH, ugd; UDPglucose 6-dehydrogenase [EC:1.1.1.22] | 0.91268 | 0.13560 | 0.79756 | 2.08608 | 0.17915 | 65 | 0.99314 | 3.49048 | 0.19373 | 93 | 0.26387 | -0.74121 | 154.361 | 0.45969 | 0.59014 |
| RP-L36, MRPL36, rpmJ; large subunit ribosomal protein L36 | 0.91160 | 0.13501 | 0.84630 | 1.81642 | 0.16717 | 65 | 0.95725 | 3.64575 | 0.19799 | 93 | 0.25913 | -0.42815 | 155.977 | 0.66913 | 0.75010 |
| K07007; uncharacterized protein | 0.91085 | 0.13488 | 0.79271 | 1.89393 | 0.17070 | 65 | 0.99343 | 3.57105 | 0.19596 | 93 | 0.25988 | -0.77238 | 155.711 | 0.44106 | 0.59014 |
| rnc, DROSHA, RNT1; ribonuclease III [EC:3.1.26.3] | 0.90929 | 0.13167 | 0.79571 | 1.80479 | 0.16663 | 65 | 0.98867 | 3.40350 | 0.19130 | 93 | 0.25370 | -0.76058 | 155.712 | 0.44806 | 0.59014 |
| K08999; uncharacterized protein | 0.90874 | 0.13082 | 0.79556 | 1.77252 | 0.16514 | 65 | 0.98785 | 3.36592 | 0.19024 | 93 | 0.25192 | -0.76334 | 155.757 | 0.44642 | 0.59014 |
| coaBC, dfp; phosphopantothenoylcysteine decarboxylase / phosphopantothenate—cysteine ligase [EC:4.1.1.36 6.3.2.5] | 0.90861 | 0.13122 | 0.79736 | 1.80623 | 0.16670 | 65 | 0.98637 | 3.37159 | 0.19040 | 93 | 0.25306 | -0.74686 | 155.640 | 0.45627 | 0.59014 |
| panB; 3-methyl-2-oxobutanoate hydroxymethyltransferase [EC:2.1.2.11] | 0.90861 | 0.13122 | 0.79736 | 1.80623 | 0.16670 | 65 | 0.98637 | 3.37159 | 0.19040 | 93 | 0.25306 | -0.74686 | 155.640 | 0.45627 | 0.59014 |
| panC; pantoate–beta-alanine ligase [EC:6.3.2.1] | 0.90861 | 0.13122 | 0.79736 | 1.80623 | 0.16670 | 65 | 0.98637 | 3.37159 | 0.19040 | 93 | 0.25306 | -0.74686 | 155.640 | 0.45627 | 0.59014 |
| panD; aspartate 1-decarboxylase [EC:4.1.1.11] | 0.90861 | 0.13122 | 0.79736 | 1.80623 | 0.16670 | 65 | 0.98637 | 3.37159 | 0.19040 | 93 | 0.25306 | -0.74686 | 155.640 | 0.45627 | 0.59014 |
| ftsZ; cell division protein FtsZ | 0.90853 | 0.12890 | 0.78271 | 1.72754 | 0.16303 | 65 | 0.99648 | 3.25921 | 0.18720 | 93 | 0.24824 | -0.86115 | 155.715 | 0.39048 | 0.59014 |
| rsmI; 16S rRNA (cytidine1402-2’-O)-methyltransferase [EC:2.1.1.198] | 0.90849 | 0.12890 | 0.78260 | 1.72771 | 0.16303 | 65 | 0.99648 | 3.25921 | 0.18720 | 93 | 0.24824 | -0.86156 | 155.715 | 0.39025 | 0.59014 |
| carB, CPA2; carbamoyl-phosphate synthase large subunit [EC:6.3.5.5] | 0.90744 | 0.13199 | 0.78642 | 1.82366 | 0.16750 | 65 | 0.99203 | 3.41117 | 0.19152 | 93 | 0.25443 | -0.80811 | 155.655 | 0.42026 | 0.59014 |
| kdsC; 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase (KDO 8-P phosphatase) [EC:3.1.3.45] | 0.90740 | 0.13199 | 0.78631 | 1.82382 | 0.16751 | 65 | 0.99203 | 3.41117 | 0.19152 | 93 | 0.25444 | -0.80851 | 155.655 | 0.42003 | 0.59014 |
| ssb; single-strand DNA-binding protein | 0.90740 | 0.13199 | 0.78631 | 1.82382 | 0.16751 | 65 | 0.99203 | 3.41117 | 0.19152 | 93 | 0.25444 | -0.80851 | 155.655 | 0.42003 | 0.59014 |
| glmS, GFPT; glucosamine—fructose-6-phosphate aminotransferase (isomerizing) [EC:2.6.1.16] | 0.90699 | 0.13614 | 0.78071 | 1.91050 | 0.17144 | 65 | 0.99524 | 3.64942 | 0.19809 | 93 | 0.26198 | -0.81890 | 155.792 | 0.41410 | 0.59014 |
| lldE; L-lactate dehydrogenase complex protein LldE | 0.90572 | 0.14000 | 0.78473 | 2.02147 | 0.17635 | 65 | 0.99028 | 3.86073 | 0.20375 | 93 | 0.26947 | -0.76279 | 155.791 | 0.44674 | 0.59014 |
| lldF; L-lactate dehydrogenase complex protein LldF | 0.90572 | 0.14000 | 0.78473 | 2.02147 | 0.17635 | 65 | 0.99028 | 3.86073 | 0.20375 | 93 | 0.26947 | -0.76279 | 155.791 | 0.44674 | 0.59014 |
| lldG; L-lactate dehydrogenase complex protein LldG | 0.90572 | 0.14000 | 0.78473 | 2.02147 | 0.17635 | 65 | 0.99028 | 3.86073 | 0.20375 | 93 | 0.26947 | -0.76279 | 155.791 | 0.44674 | 0.59014 |
| MMAB, pduO; cob(I)alamin adenosyltransferase [EC:2.5.1.17] | 0.90565 | 0.13632 | 0.76923 | 1.90988 | 0.17141 | 65 | 1.00100 | 3.65956 | 0.19837 | 93 | 0.26217 | -0.88403 | 155.809 | 0.37804 | 0.59014 |
| fruA; fructan beta-fructosidase [EC:3.2.1.80] | 0.90382 | 0.13636 | 0.76536 | 1.91458 | 0.17162 | 65 | 1.00060 | 3.65856 | 0.19834 | 93 | 0.26229 | -0.89689 | 155.794 | 0.37116 | 0.59014 |
| umuC; DNA polymerase V | 0.90138 | 0.13197 | 0.80596 | 1.61397 | 0.15758 | 65 | 0.96808 | 3.56233 | 0.19572 | 93 | 0.25127 | -0.64520 | 155.815 | 0.51975 | 0.62241 |
| E3.1.3.48; protein-tyrosine phosphatase [EC:3.1.3.48] | 0.90025 | 0.13104 | 0.77758 | 1.79285 | 0.16608 | 65 | 0.98598 | 3.36442 | 0.19020 | 93 | 0.25251 | -0.82533 | 155.679 | 0.41045 | 0.59014 |
| TC.BASS; bile acid:Na+ symporter, BASS family | 0.89830 | 0.12715 | 0.78055 | 1.67955 | 0.16075 | 65 | 0.98061 | 3.17398 | 0.18474 | 93 | 0.24488 | -0.81695 | 155.726 | 0.41521 | 0.59014 |
| dapdh; diaminopimelate dehydrogenase [EC:1.4.1.16] | 0.89821 | 0.12715 | 0.78033 | 1.67989 | 0.16076 | 65 | 0.98061 | 3.17398 | 0.18474 | 93 | 0.24489 | -0.81781 | 155.725 | 0.41472 | 0.59014 |
| K07139; uncharacterized protein | 0.89821 | 0.12715 | 0.78033 | 1.67989 | 0.16076 | 65 | 0.98061 | 3.17398 | 0.18474 | 93 | 0.24489 | -0.81781 | 155.725 | 0.41472 | 0.59014 |
| malY, malT; maltose/moltooligosaccharide transporter | 0.89821 | 0.12715 | 0.78033 | 1.67989 | 0.16076 | 65 | 0.98061 | 3.17398 | 0.18474 | 93 | 0.24489 | -0.81781 | 155.725 | 0.41472 | 0.59014 |
| RP-L34, MRPL34, rpmH; large subunit ribosomal protein L34 | 0.89821 | 0.12715 | 0.78033 | 1.67989 | 0.16076 | 65 | 0.98061 | 3.17398 | 0.18474 | 93 | 0.24489 | -0.81781 | 155.725 | 0.41472 | 0.59014 |
| serA, PHGDH; D-3-phosphoglycerate dehydrogenase / 2-oxoglutarate reductase [EC:1.1.1.95 1.1.1.399] | 0.89821 | 0.12715 | 0.78033 | 1.67989 | 0.16076 | 65 | 0.98061 | 3.17398 | 0.18474 | 93 | 0.24489 | -0.81781 | 155.725 | 0.41472 | 0.59014 |
| serC, PSAT1; phosphoserine aminotransferase [EC:2.6.1.52] | 0.89821 | 0.12715 | 0.78033 | 1.67989 | 0.16076 | 65 | 0.98061 | 3.17398 | 0.18474 | 93 | 0.24489 | -0.81781 | 155.725 | 0.41472 | 0.59014 |
| greA; transcription elongation factor GreA | 0.89740 | 0.12823 | 0.76997 | 1.68365 | 0.16094 | 65 | 0.98647 | 3.24299 | 0.18674 | 93 | 0.24652 | -0.87819 | 155.836 | 0.38119 | 0.59014 |
| hemD, UROS; uroporphyrinogen-III synthase [EC:4.2.1.75] | 0.89736 | 0.12823 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.98647 | 3.24299 | 0.18674 | 93 | 0.24653 | -0.87860 | 155.836 | 0.38097 | 0.59014 |
| K07000; uncharacterized protein | 0.89713 | 0.13071 | 0.78429 | 1.77481 | 0.16524 | 65 | 0.97600 | 3.35692 | 0.18999 | 93 | 0.25180 | -0.76138 | 155.732 | 0.44758 | 0.59014 |
| coaW; type II pantothenate kinase [EC:2.7.1.33] | 0.89708 | 0.13071 | 0.78418 | 1.77498 | 0.16525 | 65 | 0.97600 | 3.35692 | 0.18999 | 93 | 0.25180 | -0.76181 | 155.731 | 0.44733 | 0.59014 |
| E3.1.3.5; 5’-nucleotidase [EC:3.1.3.5] | 0.89708 | 0.13071 | 0.78418 | 1.77498 | 0.16525 | 65 | 0.97600 | 3.35692 | 0.18999 | 93 | 0.25180 | -0.76181 | 155.731 | 0.44733 | 0.59014 |
| fabI; enoyl-[acyl-carrier protein] reductase I [EC:1.3.1.9 1.3.1.10] | 0.89708 | 0.13071 | 0.78418 | 1.77498 | 0.16525 | 65 | 0.97600 | 3.35692 | 0.18999 | 93 | 0.25180 | -0.76181 | 155.731 | 0.44733 | 0.59014 |
| ftnA, ftn; ferritin [EC:1.16.3.2] | 0.89708 | 0.13071 | 0.78418 | 1.77498 | 0.16525 | 65 | 0.97600 | 3.35692 | 0.18999 | 93 | 0.25180 | -0.76181 | 155.731 | 0.44733 | 0.59014 |
| K09125; uncharacterized protein | 0.89708 | 0.13071 | 0.78418 | 1.77498 | 0.16525 | 65 | 0.97600 | 3.35692 | 0.18999 | 93 | 0.25180 | -0.76181 | 155.731 | 0.44733 | 0.59014 |
| menE; O-succinylbenzoic acid—CoA ligase [EC:6.2.1.26] | 0.89708 | 0.13071 | 0.78418 | 1.77498 | 0.16525 | 65 | 0.97600 | 3.35692 | 0.18999 | 93 | 0.25180 | -0.76181 | 155.731 | 0.44733 | 0.59014 |
| pdxS, pdx1; pyridoxal 5’-phosphate synthase pdxS subunit [EC:4.3.3.6] | 0.89708 | 0.13071 | 0.78418 | 1.77498 | 0.16525 | 65 | 0.97600 | 3.35692 | 0.18999 | 93 | 0.25180 | -0.76181 | 155.731 | 0.44733 | 0.59014 |
| pdxT, pdx2; 5’-phosphate synthase pdxT subunit [EC:4.3.3.6] | 0.89708 | 0.13071 | 0.78418 | 1.77498 | 0.16525 | 65 | 0.97600 | 3.35692 | 0.18999 | 93 | 0.25180 | -0.76181 | 155.731 | 0.44733 | 0.59014 |
| trpF; phosphoribosylanthranilate isomerase [EC:5.3.1.24] | 0.89708 | 0.13071 | 0.78418 | 1.77498 | 0.16525 | 65 | 0.97600 | 3.35692 | 0.18999 | 93 | 0.25180 | -0.76181 | 155.731 | 0.44733 | 0.59014 |
| zapA; cell division protein ZapA | 0.89708 | 0.13071 | 0.78418 | 1.77498 | 0.16525 | 65 | 0.97600 | 3.35692 | 0.18999 | 93 | 0.25180 | -0.76181 | 155.731 | 0.44733 | 0.59014 |
| lipA; lipoyl synthase [EC:2.8.1.8] | 0.89695 | 0.13680 | 0.77860 | 2.16725 | 0.18260 | 65 | 0.97966 | 3.52170 | 0.19460 | 93 | 0.26685 | -0.75348 | 153.863 | 0.45231 | 0.59014 |
| comM; magnesium chelatase family protein | 0.89675 | 0.12737 | 0.77156 | 1.68510 | 0.16101 | 65 | 0.98424 | 3.18329 | 0.18501 | 93 | 0.24526 | -0.86714 | 155.724 | 0.38720 | 0.59014 |
| ATPF1A, atpA; F-type H+-transporting ATPase subunit alpha [EC:3.6.3.14] | 0.89670 | 0.12737 | 0.77146 | 1.68526 | 0.16102 | 65 | 0.98424 | 3.18329 | 0.18501 | 93 | 0.24527 | -0.86755 | 155.723 | 0.38697 | 0.59014 |
| ppnN; pyrimidine/purine-5’-nucleotide nucleosidase [EC:3.2.2.10 3.2.2.-] | 0.89528 | 0.13334 | 0.75505 | 1.82431 | 0.16753 | 65 | 0.99328 | 3.50140 | 0.19403 | 93 | 0.25635 | -0.92929 | 155.818 | 0.35418 | 0.59014 |
| pabB; para-aminobenzoate synthetase component I [EC:2.6.1.85] | 0.88973 | 0.14169 | 0.80853 | 2.36590 | 0.19078 | 65 | 0.94648 | 3.75928 | 0.20105 | 93 | 0.27717 | -0.49771 | 153.439 | 0.61940 | 0.70276 |
| pabC; 4-amino-4-deoxychorismate lyase [EC:4.1.3.38] | 0.88973 | 0.14169 | 0.80853 | 2.36590 | 0.19078 | 65 | 0.94648 | 3.75928 | 0.20105 | 93 | 0.27717 | -0.49771 | 153.439 | 0.61940 | 0.70276 |
| ribBA; 3,4-dihydroxy 2-butanone 4-phosphate synthase / GTP cyclohydrolase II [EC:4.1.99.12 3.5.4.25] | 0.88754 | 0.12713 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96978 | 3.16970 | 0.18462 | 93 | 0.24492 | -0.81623 | 155.700 | 0.41562 | 0.59014 |
| ogt, MGMT; methylated-DNA-[protein]-cysteine S-methyltransferase [EC:2.1.1.63] | 0.88753 | 0.12961 | 0.80769 | 1.97763 | 0.17443 | 65 | 0.94332 | 3.14608 | 0.18393 | 93 | 0.25348 | -0.53507 | 153.462 | 0.59337 | 0.68100 |
| yjbB; phosphate:Na+ symporter | 0.88736 | 0.12712 | 0.76997 | 1.68365 | 0.16094 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24491 | -0.81432 | 155.700 | 0.41670 | 0.59014 |
| AARS, alaS; alanyl-tRNA synthetase [EC:6.1.1.7] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ABC.FEV.A; iron complex transport system ATP-binding protein [EC:3.6.3.34] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ABC.FEV.P; iron complex transport system permease protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ABC.FEV.S; iron complex transport system substrate-binding protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ABC.SN.S; NitT/TauT family transport system substrate-binding protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ABCF3; ATP-binding cassette, subfamily F, member 3 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ackA; acetate kinase [EC:2.7.2.1] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| adk, AK; adenylate kinase [EC:2.7.4.3] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| aguB; N-carbamoylputrescine amidase [EC:3.5.1.53] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| AIBP, nnrE; NAD(P)H-hydrate epimerase [EC:5.1.99.6] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| alr; alanine racemase [EC:5.1.1.1] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| amiABC; N-acetylmuramoyl-L-alanine amidase [EC:3.5.1.28] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| apbE; FAD:protein FMN transferase [EC:2.7.1.180] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| APRT, apt; adenine phosphoribosyltransferase [EC:2.4.2.7] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| argR, ahrC; transcriptional regulator of arginine metabolism | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| aroA; 3-phosphoshikimate 1-carboxyvinyltransferase [EC:2.5.1.19] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| AROA1, aroA; chorismate mutase [EC:5.4.99.5] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| aroB; 3-dehydroquinate synthase [EC:4.2.3.4] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| aroC; chorismate synthase [EC:4.2.3.5] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| aroE; shikimate dehydrogenase [EC:1.1.1.25] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| aroQ, qutE; 3-dehydroquinate dehydratase II [EC:4.2.1.10] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| asd; aspartate-semialdehyde dehydrogenase [EC:1.2.1.11] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| aspS; aspartyl-tRNA synthetase [EC:6.1.1.12] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ATPF0A, atpB; F-type H+-transporting ATPase subunit a | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ATPF0B, atpF; F-type H+-transporting ATPase subunit b | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ATPF0C, atpE; F-type H+-transporting ATPase subunit c | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ATPF1B, atpD; F-type H+-transporting ATPase subunit beta [EC:3.6.3.14] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ATPF1D, atpH; F-type H+-transporting ATPase subunit delta | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ATPF1E, atpC; F-type H+-transporting ATPase subunit epsilon | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ATPF1G, atpG; F-type H+-transporting ATPase subunit gamma | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| bacA; undecaprenyl-diphosphatase [EC:3.6.1.27] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| bamD; outer membrane protein assembly factor BamD | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| BCP, PRXQ, DOT5; peroxiredoxin Q/BCP [EC:1.11.1.15] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| birA; BirA family transcriptional regulator, biotin operon repressor / biotin—[acetyl-CoA-carboxylase] ligase [EC:6.3.4.15] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| carA, CPA1; carbamoyl-phosphate synthase small subunit [EC:6.3.5.5] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| CARS, cysS; cysteinyl-tRNA synthetase [EC:6.1.1.16] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| cdd, CDA; cytidine deaminase [EC:3.5.4.5] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| CHO1, pssA; CDP-diacylglycerol—serine O-phosphatidyltransferase [EC:2.7.8.8] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| clpB; ATP-dependent Clp protease ATP-binding subunit ClpB | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| clpC; ATP-dependent Clp protease ATP-binding subunit ClpC | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| clpP, CLPP; ATP-dependent Clp protease, protease subunit [EC:3.4.21.92] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| clpX, CLPX; ATP-dependent Clp protease ATP-binding subunit ClpX | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| cmk; CMP/dCMP kinase [EC:2.7.4.25] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| comEB; dCMP deaminase [EC:3.5.4.12] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| comEC; competence protein ComEC | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| corA; magnesium transporter | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| cydA; cytochrome bd ubiquinol oxidase subunit I [EC:1.10.3.14] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| cydB; cytochrome bd ubiquinol oxidase subunit II [EC:1.10.3.14] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| dapA; 4-hydroxy-tetrahydrodipicolinate synthase [EC:4.3.3.7] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| dapB; 4-hydroxy-tetrahydrodipicolinate reductase [EC:1.17.1.8] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ddl; D-alanine-D-alanine ligase [EC:6.3.2.4] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| deaD, cshA; ATP-dependent RNA helicase DeaD [EC:3.6.4.13] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| deoC, DERA; deoxyribose-phosphate aldolase [EC:4.1.2.4] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| dgt; dGTPase [EC:3.1.5.1] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| dinB; DNA polymerase IV [EC:2.7.7.7] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| dnaA; chromosomal replication initiator protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| dnaB; replicative DNA helicase [EC:3.6.4.12] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| dnaE; DNA polymerase III subunit alpha [EC:2.7.7.7] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| dnaG; DNA primase [EC:2.7.7.-] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| dnaJ; molecular chaperone DnaJ | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| dnaK, HSPA9; molecular chaperone DnaK | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| dnaN; DNA polymerase III subunit beta [EC:2.7.7.7] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| dnaX; DNA polymerase III subunit gamma/tau [EC:2.7.7.7] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| DPM1; dolichol-phosphate mannosyltransferase [EC:2.4.1.83] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| DPP3; dipeptidyl-peptidase III [EC:3.4.14.4] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| dsbD; thiol:disulfide interchange protein DsbD [EC:1.8.1.8] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| dtd, DTD1; D-tyrosyl-tRNA(Tyr) deacylase [EC:3.1.-.-] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| dut, DUT; dUTP pyrophosphatase [EC:3.6.1.23] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| dxr; 1-deoxy-D-xylulose-5-phosphate reductoisomerase [EC:1.1.1.267] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| E1.1.1.40, maeB; malate dehydrogenase (oxaloacetate-decarboxylating)(NADP+) [EC:1.1.1.40] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| E2.3.1.8, pta; phosphate acetyltransferase [EC:2.3.1.8] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| E2.6.1.42, ilvE; branched-chain amino acid aminotransferase [EC:2.6.1.42] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| E2.7.1.71, aroK, aroL; shikimate kinase [EC:2.7.1.71] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| E2.7.4.8, gmk; guanylate kinase [EC:2.7.4.8] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| E2.7.7.24, rfbA, rffH; glucose-1-phosphate thymidylyltransferase [EC:2.7.7.24] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| E2.7.7.3A, coaD, kdtB; pantetheine-phosphate adenylyltransferase [EC:2.7.7.3] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| E2.7.7.41, CDS1, CDS2, cdsA; phosphatidate cytidylyltransferase [EC:2.7.7.41] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| E3.2.1.1A; alpha-amylase [EC:3.2.1.1] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| E3.6.1.22, NUDT12, nudC; NAD+ diphosphatase [EC:3.6.1.22] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| E4.1.1.49, pckA; phosphoenolpyruvate carboxykinase (ATP) [EC:4.1.1.49] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| E4.2.1.2A, fumA, fumB; fumarate hydratase, class I [EC:4.2.1.2] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| E4.2.1.46, rfbB, rffG; dTDP-glucose 4,6-dehydratase [EC:4.2.1.46] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| E4.3.1.17, sdaA, sdaB, tdcG; L-serine dehydratase [EC:4.3.1.17] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| E6.3.5.1, NADSYN1, QNS1, nadE; NAD+ synthase (glutamine-hydrolysing) [EC:6.3.5.1] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| E6.5.1.2, ligA, ligB; DNA ligase (NAD+) [EC:6.5.1.2] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| EARS, gltX; glutamyl-tRNA synthetase [EC:6.1.1.17] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| efp; elongation factor P | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| engA, der; GTPase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| engB; GTP-binding protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ENO, eno; enolase [EC:4.2.1.11] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| entC; isochorismate synthase [EC:5.4.4.2] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| era, ERAL1; GTPase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| fabD; [acyl-carrier-protein] S-malonyltransferase [EC:2.3.1.39] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| fabF; 3-oxoacyl-[acyl-carrier-protein] synthase II [EC:2.3.1.179] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| FARSA, pheS; phenylalanyl-tRNA synthetase alpha chain [EC:6.1.1.20] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| FARSB, pheT; phenylalanyl-tRNA synthetase beta chain [EC:6.1.1.20] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| FBA, fbaA; fructose-bisphosphate aldolase, class II [EC:4.1.2.13] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| fbp3; fructose-1,6-bisphosphatase III [EC:3.1.3.11] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| fixA, etfB; electron transfer flavoprotein beta subunit | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| fixB, etfA; electron transfer flavoprotein alpha subunit | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| folB; 7,8-dihydroneopterin aldolase/epimerase/oxygenase [EC:4.1.2.25 5.1.99.8 1.13.11.81] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| folC; dihydrofolate synthase / folylpolyglutamate synthase [EC:6.3.2.12 6.3.2.17] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| folD; methylenetetrahydrofolate dehydrogenase (NADP+) / methenyltetrahydrofolate cyclohydrolase [EC:1.5.1.5 3.5.4.9] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| folP; dihydropteroate synthase [EC:2.5.1.15] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| frr, MRRF, RRF; ribosome recycling factor | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ftsA; cell division protein FtsA | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ftsE; cell division transport system ATP-binding protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ftsH, hflB; cell division protease FtsH [EC:3.4.24.-] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ftsK, spoIIIE; DNA segregation ATPase FtsK/SpoIIIE, S-DNA-T family | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ftsQ; cell division protein FtsQ | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ftsW, spoVE; cell division protein FtsW | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ftsX; cell division transport system permease protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ftsY; fused signal recognition particle receptor | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| galE, GALE; UDP-glucose 4-epimerase [EC:5.1.3.2] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| GAPDH, gapA; glyceraldehyde 3-phosphate dehydrogenase [EC:1.2.1.12] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| GARS, glyS1; glycyl-tRNA synthetase [EC:6.1.1.14] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| GBE1, glgB; 1,4-alpha-glucan branching enzyme [EC:2.4.1.18] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| gcpE, ispG; (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase [EC:1.17.7.1 1.17.7.3] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| GGPS; geranylgeranyl diphosphate synthase, type II [EC:2.5.1.1 2.5.1.10 2.5.1.29] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| gidA, mnmG, MTO1; tRNA uridine 5-carboxymethylaminomethyl modification enzyme | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| gidB, rsmG; 16S rRNA (guanine527-N7)-methyltransferase [EC:2.1.1.170] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| glgA; starch synthase [EC:2.4.1.21] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| glyA, SHMT; glycine hydroxymethyltransferase [EC:2.1.2.1] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| gmd, GMDS; GDPmannose 4,6-dehydratase [EC:4.2.1.47] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| GPI, pgi; glucose-6-phosphate isomerase [EC:5.3.1.9] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| gpsA; glycerol-3-phosphate dehydrogenase (NAD(P)+) [EC:1.1.1.94] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| groEL, HSPD1; chaperonin GroEL | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| groES, HSPE1; chaperonin GroES | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| GRPE; molecular chaperone GrpE | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| guaA, GMPS; GMP synthase (glutamine-hydrolysing) [EC:6.3.5.2] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| gyrA; DNA gyrase subunit A [EC:5.99.1.3] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| gyrB; DNA gyrase subunit B [EC:5.99.1.3] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| HARS, hisS; histidyl-tRNA synthetase [EC:6.1.1.21] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| hemK, prmC, HEMK; release factor glutamine methyltransferase [EC:2.1.1.297] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| hflX; GTPase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| HINT1, hinT, hit; histidine triad (HIT) family protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| holA; DNA polymerase III subunit delta [EC:2.7.7.7] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| holB; DNA polymerase III subunit delta’ [EC:2.7.7.7] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| hprA; glycerate dehydrogenase [EC:1.1.1.29] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| hprT, hpt, HPRT1; hypoxanthine phosphoribosyltransferase [EC:2.4.2.8] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| hslR; ribosome-associated heat shock protein Hsp15 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| HSP90A, htpG; molecular chaperone HtpG | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| IMPDH, guaB; IMP dehydrogenase [EC:1.1.1.205] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| infA; translation initiation factor IF-1 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| infB, MTIF2; translation initiation factor IF-2 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| infC, MTIF3; translation initiation factor IF-3 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ispB; octaprenyl-diphosphate synthase [EC:2.5.1.90] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ispE; 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase [EC:2.7.1.148] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ispF; 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [EC:4.6.1.12] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ispH, lytB; 4-hydroxy-3-methylbut-2-en-1-yl diphosphate reductase [EC:1.17.7.4] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| K00243; uncharacterized protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| K06890; uncharacterized protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| K07027; glycosyltransferase 2 family protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| K07037; uncharacterized protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| K07082; UPF0755 protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| K07137; uncharacterized protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| K07164; uncharacterized protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| K09861; uncharacterized protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| KAE1, tsaD, QRI7; N6-L-threonylcarbamoyladenine synthase [EC:2.3.1.234] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| KARS, lysS; lysyl-tRNA synthetase, class II [EC:6.1.1.6] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| kdsA; 2-dehydro-3-deoxyphosphooctonate aldolase (KDO 8-P synthase) [EC:2.5.1.55] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| kdsD, kpsF; arabinose-5-phosphate isomerase [EC:5.3.1.13] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| kdtA, waaA; 3-deoxy-D-manno-octulosonic-acid transferase [EC:2.4.99.12 2.4.99.13 2.4.99.14 2.4.99.15] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| korC, oorC; 2-oxoglutarate ferredoxin oxidoreductase subunit gamma [EC:1.2.7.3] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| korD, oorD; 2-oxoglutarate ferredoxin oxidoreductase subunit delta [EC:1.2.7.3] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ksgA; 16S rRNA (adenine1518-N6/adenine1519-N6)-dimethyltransferase [EC:2.1.1.182] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| lepA; GTP-binding protein LepA | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| lolD; lipoprotein-releasing system ATP-binding protein [EC:3.6.3.-] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| lptB; lipopolysaccharide export system ATP-binding protein [EC:3.6.3.-] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| lptF; lipopolysaccharide export system permease protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| lptG; lipopolysaccharide export system permease protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| lpxB; lipid-A-disaccharide synthase [EC:2.4.1.182] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| lpxC-fabZ; UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase / 3-hydroxyacyl-[acyl-carrier-protein] dehydratase [EC:3.5.1.108 4.2.1.59] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| lpxD; UDP-3-O-[3-hydroxymyristoyl] glucosamine N-acyltransferase [EC:2.3.1.191] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| lpxH; UDP-2,3-diacylglucosamine hydrolase [EC:3.6.1.54] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| lpxK; tetraacyldisaccharide 4’-kinase [EC:2.7.1.130] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| lpxL, htrB; Kdo2-lipid IVA lauroyltransferase [EC:2.3.1.241] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| lspA; signal peptidase II [EC:3.4.23.36] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| LYS1; saccharopine dehydrogenase (NAD+, L-lysine forming) [EC:1.5.1.7] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| lysC; aspartate kinase [EC:2.7.2.4] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| maf; septum formation protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| malQ; 4-alpha-glucanotransferase [EC:2.4.1.25] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| manA, MPI; mannose-6-phosphate isomerase [EC:5.3.1.8] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| manB; phosphomannomutase [EC:5.4.2.8] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| MARS, metG; methionyl-tRNA synthetase [EC:6.1.1.10] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| mdh; malate dehydrogenase [EC:1.1.1.37] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| menA; 1,4-dihydroxy-2-naphthoate octaprenyltransferase [EC:2.5.1.74 2.5.1.-] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| menB; naphthoate synthase [EC:4.1.3.36] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| menD; 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase [EC:2.2.1.9] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| mfd; transcription-repair coupling factor (superfamily II helicase) [EC:3.6.4.-] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| miaB; tRNA-2-methylthio-N6-dimethylallyladenosine synthase [EC:2.8.4.3] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| mlaD, linM; phospholipid/cholesterol/gamma-HCH transport system substrate-binding protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| mlaE, linK; phospholipid/cholesterol/gamma-HCH transport system permease protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| mlaF, linL, mkl; phospholipid/cholesterol/gamma-HCH transport system ATP-binding protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| mnmA, trmU; tRNA-uridine 2-sulfurtransferase [EC:2.8.1.13] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| mnmE, trmE, MSS1; tRNA modification GTPase [EC:3.6.-.-] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| motB; chemotaxis protein MotB | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| mraW, rsmH; 16S rRNA (cytosine1402-N4)-methyltransferase [EC:2.1.1.199] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| mraY; phospho-N-acetylmuramoyl-pentapeptide-transferase [EC:2.7.8.13] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| mreB; rod shape-determining protein MreB and related proteins | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| mreC; rod shape-determining protein MreC | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| mrp, NUBPL; ATP-binding protein involved in chromosome partitioning | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| msbA; ATP-binding cassette, subfamily B, bacterial MsbA [EC:3.6.3.-] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| mscL; large conductance mechanosensitive channel | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| msrAB; peptide methionine sulfoxide reductase msrA/msrB [EC:1.8.4.11 1.8.4.12] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| mtaB; threonylcarbamoyladenosine tRNA methylthiotransferase MtaB [EC:2.8.4.5] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| MTFMT, fmt; methionyl-tRNA formyltransferase [EC:2.1.2.9] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| mtgA; monofunctional glycosyltransferase [EC:2.4.1.129] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| MTHFS; 5-formyltetrahydrofolate cyclo-ligase [EC:6.3.3.2] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| mtnN, mtn, pfs; adenosylhomocysteine nucleosidase [EC:3.2.2.9] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| murA; UDP-N-acetylglucosamine 1-carboxyvinyltransferase [EC:2.5.1.7] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| murB; UDP-N-acetylmuramate dehydrogenase [EC:1.3.1.98] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| murC; UDP-N-acetylmuramate–alanine ligase [EC:6.3.2.8] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| murD; UDP-N-acetylmuramoylalanine–D-glutamate ligase [EC:6.3.2.9] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| murE; UDP-N-acetylmuramoyl-L-alanyl-D-glutamate–2,6-diaminopimelate ligase [EC:6.3.2.13] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| murF; UDP-N-acetylmuramoyl-tripeptide–D-alanyl-D-alanine ligase [EC:6.3.2.10] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| murG; UDP-N-acetylglucosamine–N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase [EC:2.4.1.227] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| murI; glutamate racemase [EC:5.1.1.3] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| mutL; DNA mismatch repair protein MutL | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| mutS; DNA mismatch repair protein MutS | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| mutS2; DNA mismatch repair protein MutS2 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| mutY; A/G-specific adenine glycosylase [EC:3.2.2.31] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| nadD; nicotinate-nucleotide adenylyltransferase [EC:2.7.7.18] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| NARS, asnS; asparaginyl-tRNA synthetase [EC:6.1.1.22] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| nfrA2; FMN reductase [NAD(P)H] [EC:1.5.1.39] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| nqrA; Na+-transporting NADH:ubiquinone oxidoreductase subunit A [EC:1.6.5.8] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| nqrB; Na+-transporting NADH:ubiquinone oxidoreductase subunit B [EC:1.6.5.8] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| nqrC; Na+-transporting NADH:ubiquinone oxidoreductase subunit C [EC:1.6.5.8] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| nqrD; Na+-transporting NADH:ubiquinone oxidoreductase subunit D [EC:1.6.5.8] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| nqrE; Na+-transporting NADH:ubiquinone oxidoreductase subunit E [EC:1.6.5.8] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| nqrF; Na+-transporting NADH:ubiquinone oxidoreductase subunit F [EC:1.6.5.8] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| nrdG; anaerobic ribonucleoside-triphosphate reductase activating protein [EC:1.97.1.4] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| nrnA; bifunctional oligoribonuclease and PAP phosphatase NrnA [EC:3.1.3.7 3.1.13.3] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| nspC; carboxynorspermidine decarboxylase [EC:4.1.1.96] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| NTH; endonuclease III [EC:4.2.99.18] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| nuoA; NADH-quinone oxidoreductase subunit A [EC:1.6.5.3] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| nuoB; NADH-quinone oxidoreductase subunit B [EC:1.6.5.3] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| nuoCD; NADH-quinone oxidoreductase subunit C/D [EC:1.6.5.3] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| nuoH; NADH-quinone oxidoreductase subunit H [EC:1.6.5.3] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| nuoI; NADH-quinone oxidoreductase subunit I [EC:1.6.5.3] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| nuoJ; NADH-quinone oxidoreductase subunit J [EC:1.6.5.3] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| nuoK; NADH-quinone oxidoreductase subunit K [EC:1.6.5.3] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| nuoL; NADH-quinone oxidoreductase subunit L [EC:1.6.5.3] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| nuoM; NADH-quinone oxidoreductase subunit M [EC:1.6.5.3] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| nuoN; NADH-quinone oxidoreductase subunit N [EC:1.6.5.3] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| nupG; MFS transporter, NHS family, nucleoside permease | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| nusA; N utilization substance protein A | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| nusB; N utilization substance protein B | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| nusG; transcriptional antiterminator NusG | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| obgE, cgtA; GTPase [EC:3.6.5.-] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| parB, spo0J; chromosome partitioning protein, ParB family | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| parC; topoisomerase IV subunit A [EC:5.99.1.-] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| parE; topoisomerase IV subunit B [EC:5.99.1.-] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| PARS, proS; prolyl-tRNA synthetase [EC:6.1.1.15] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| PDF, def; peptide deformylase [EC:3.5.1.88] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| pdxA; 4-hydroxythreonine-4-phosphate dehydrogenase [EC:1.1.1.262] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| pepT; tripeptide aminopeptidase [EC:3.4.11.4] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| pfkA, PFK; 6-phosphofructokinase 1 [EC:2.7.1.11] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| pfp, PFP; diphosphate-dependent phosphofructokinase [EC:2.7.1.90] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| PGK, pgk; phosphoglycerate kinase [EC:2.7.2.3] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| pgm; phosphoglucomutase [EC:5.4.2.2] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| phnP; phosphoribosyl 1,2-cyclic phosphate phosphodiesterase [EC:3.1.4.55] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| phoH, phoL; phosphate starvation-inducible protein PhoH and related proteins | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| phoR; two-component system, OmpR family, phosphate regulon sensor histidine kinase PhoR [EC:2.7.13.3] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| PK, pyk; pyruvate kinase [EC:2.7.1.40] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| plsC; 1-acyl-sn-glycerol-3-phosphate acyltransferase [EC:2.3.1.51] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| pncC; nicotinamide-nucleotide amidase [EC:3.5.1.42] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| pnp, PNPT1; polyribonucleotide nucleotidyltransferase [EC:2.7.7.8] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| pnuC; nicotinamide mononucleotide transporter | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| polA; DNA polymerase I [EC:2.7.7.7] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| por, nifJ; pyruvate-ferredoxin/flavodoxin oxidoreductase [EC:1.2.7.1 1.2.7.-] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ppdK; pyruvate, orthophosphate dikinase [EC:2.7.9.1] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ppiD; peptidyl-prolyl cis-trans isomerase D [EC:5.2.1.8] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ppnK, NADK; NAD+ kinase [EC:2.7.1.23] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| prfA, MTRF1, MRF1; peptide chain release factor 1 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| prfB; peptide chain release factor 2 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| prfC; peptide chain release factor 3 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| priA; primosomal protein N’ (replication factor Y) (superfamily II helicase) [EC:3.6.4.-] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| prmA; ribosomal protein L11 methyltransferase [EC:2.1.1.-] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| PRPS, prsA; ribose-phosphate pyrophosphokinase [EC:2.7.6.1] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| psd, PISD; phosphatidylserine decarboxylase [EC:4.1.1.65] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| pstS; phosphate transport system substrate-binding protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| PTH1, pth, spoVC; peptidyl-tRNA hydrolase, PTH1 family [EC:3.1.1.29] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| punA, PNP; purine-nucleoside phosphorylase [EC:2.4.2.1] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| purA, ADSS; adenylosuccinate synthase [EC:6.3.4.4] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| purB, ADSL; adenylosuccinate lyase [EC:4.3.2.2] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| purC; phosphoribosylaminoimidazole-succinocarboxamide synthase [EC:6.3.2.6] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| purD; phosphoribosylamine—glycine ligase [EC:6.3.4.13] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| purE; 5-(carboxyamino)imidazole ribonucleotide mutase [EC:5.4.99.18] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| purM; phosphoribosylformylglycinamidine cyclo-ligase [EC:6.3.3.1] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| pycB; pyruvate carboxylase subunit B [EC:6.4.1.1] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| PYG, glgP; glycogen phosphorylase [EC:2.4.1.1] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| pyrB, PYR2; aspartate carbamoyltransferase catalytic subunit [EC:2.1.3.2] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| pyrDI; dihydroorotate dehydrogenase (NAD+) catalytic subunit [EC:1.3.1.14] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| pyrDII; dihydroorotate dehydrogenase electron transfer subunit | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| pyrE; orotate phosphoribosyltransferase [EC:2.4.2.10] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| pyrF; orotidine-5’-phosphate decarboxylase [EC:4.1.1.23] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| pyrG, CTPS; CTP synthase [EC:6.3.4.2] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| pyrH; uridylate kinase [EC:2.7.4.22] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| pyrI; aspartate carbamoyltransferase regulatory subunit | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| QARS, glnS; glutaminyl-tRNA synthetase [EC:6.1.1.18] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| queC; 7-cyano-7-deazaguanine synthase [EC:6.3.4.20] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| queF; 7-cyano-7-deazaguanine reductase [EC:1.7.1.13] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| queG; epoxyqueuosine reductase [EC:1.17.99.6] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| radC; DNA repair protein RadC | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RARS, argS; arginyl-tRNA synthetase [EC:6.1.1.19] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rbfA; ribosome-binding factor A | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| recA; recombination protein RecA | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| recF; DNA replication and repair protein RecF | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| recJ; single-stranded-DNA-specific exonuclease [EC:3.1.-.-] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| recN; DNA repair protein RecN (Recombination protein N) | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| recO; DNA repair protein RecO (recombination protein O) | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| recR; recombination protein RecR | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| recX; regulatory protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rfbC, rmlC; dTDP-4-dehydrorhamnose 3,5-epimerase [EC:5.1.3.13] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rfbD, rmlD; dTDP-4-dehydrorhamnose reductase [EC:1.1.1.133] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rho; transcription termination factor Rho | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ribD; diaminohydroxyphosphoribosylaminopyrimidine deaminase / 5-amino-6-(5-phosphoribosylamino)uracil reductase [EC:3.5.4.26 1.1.1.193] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ribE, RIB5; riboflavin synthase [EC:2.5.1.9] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ribF; riboflavin kinase / FMN adenylyltransferase [EC:2.7.1.26 2.7.7.2] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ribH, RIB4; 6,7-dimethyl-8-ribityllumazine synthase [EC:2.5.1.78] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ridA, tdcF, RIDA; 2-iminobutanoate/2-iminopropanoate deaminase [EC:3.5.99.10] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rimM; 16S rRNA processing protein RimM | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rimO; ribosomal protein S12 methylthiotransferase [EC:2.8.4.4] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rimP; ribosome maturation factor RimP | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rlmH; 23S rRNA (pseudouridine1915-N3)-methyltransferase [EC:2.1.1.177] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rlmI; 23S rRNA (cytosine1962-C5)-methyltransferase [EC:2.1.1.191] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rlmN; 23S rRNA (adenine2503-C2)-methyltransferase [EC:2.1.1.192] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rluB; 23S rRNA pseudouridine2605 synthase [EC:5.4.99.22] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rmuC; DNA recombination protein RmuC | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rng, cafA; ribonuclease G [EC:3.1.26.-] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rnhA, RNASEH1; ribonuclease HI [EC:3.1.26.4] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rnhB; ribonuclease HII [EC:3.1.26.4] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rnpA; ribonuclease P protein component [EC:3.1.26.5] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rnr, vacB; ribonuclease R [EC:3.1.-.-] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rny; ribonucrease Y [EC:3.1.-.-] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rnz; ribonuclease Z [EC:3.1.26.11] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rodA, mrdB; rod shape determining protein RodA | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-L1, MRPL1, rplA; large subunit ribosomal protein L1 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-L10, MRPL10, rplJ; large subunit ribosomal protein L10 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-L11, MRPL11, rplK; large subunit ribosomal protein L11 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-L13, MRPL13, rplM; large subunit ribosomal protein L13 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-L14, MRPL14, rplN; large subunit ribosomal protein L14 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-L15, MRPL15, rplO; large subunit ribosomal protein L15 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-L16, MRPL16, rplP; large subunit ribosomal protein L16 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-L18, MRPL18, rplR; large subunit ribosomal protein L18 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-L19, MRPL19, rplS; large subunit ribosomal protein L19 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-L2, MRPL2, rplB; large subunit ribosomal protein L2 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-L20, MRPL20, rplT; large subunit ribosomal protein L20 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-L21, MRPL21, rplU; large subunit ribosomal protein L21 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-L22, MRPL22, rplV; large subunit ribosomal protein L22 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-L23, MRPL23, rplW; large subunit ribosomal protein L23 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-L24, MRPL24, rplX; large subunit ribosomal protein L24 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-L25, rplY; large subunit ribosomal protein L25 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-L27, MRPL27, rpmA; large subunit ribosomal protein L27 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-L28, MRPL28, rpmB; large subunit ribosomal protein L28 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-L29, rpmC; large subunit ribosomal protein L29 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-L3, MRPL3, rplC; large subunit ribosomal protein L3 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-L30, MRPL30, rpmD; large subunit ribosomal protein L30 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-L31, rpmE; large subunit ribosomal protein L31 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-L32, MRPL32, rpmF; large subunit ribosomal protein L32 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-L33, MRPL33, rpmG; large subunit ribosomal protein L33 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-L35, MRPL35, rpmI; large subunit ribosomal protein L35 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-L4, MRPL4, rplD; large subunit ribosomal protein L4 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-L5, MRPL5, rplE; large subunit ribosomal protein L5 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-L6, MRPL6, rplF; large subunit ribosomal protein L6 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-L7, MRPL12, rplL; large subunit ribosomal protein L7/L12 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-L9, MRPL9, rplI; large subunit ribosomal protein L9 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-S1, rpsA; small subunit ribosomal protein S1 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-S10, MRPS10, rpsJ; small subunit ribosomal protein S10 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-S11, MRPS11, rpsK; small subunit ribosomal protein S11 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-S12, MRPS12, rpsL; small subunit ribosomal protein S12 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-S13, rpsM; small subunit ribosomal protein S13 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-S14, MRPS14, rpsN; small subunit ribosomal protein S14 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-S15, MRPS15, rpsO; small subunit ribosomal protein S15 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-S16, MRPS16, rpsP; small subunit ribosomal protein S16 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-S17, MRPS17, rpsQ; small subunit ribosomal protein S17 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-S18, MRPS18, rpsR; small subunit ribosomal protein S18 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-S19, rpsS; small subunit ribosomal protein S19 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-S2, MRPS2, rpsB; small subunit ribosomal protein S2 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-S20, rpsT; small subunit ribosomal protein S20 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-S21, MRPS21, rpsU; small subunit ribosomal protein S21 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-S3, rpsC; small subunit ribosomal protein S3 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-S4, rpsD; small subunit ribosomal protein S4 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-S5, MRPS5, rpsE; small subunit ribosomal protein S5 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-S6, MRPS6, rpsF; small subunit ribosomal protein S6 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-S7, MRPS7, rpsG; small subunit ribosomal protein S7 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-S8, rpsH; small subunit ribosomal protein S8 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| RP-S9, MRPS9, rpsI; small subunit ribosomal protein S9 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rpe, RPE; ribulose-phosphate 3-epimerase [EC:5.1.3.1] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rpiB; ribose 5-phosphate isomerase B [EC:5.3.1.6] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rpoB; DNA-directed RNA polymerase subunit beta [EC:2.7.7.6] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rpoC; DNA-directed RNA polymerase subunit beta’ [EC:2.7.7.6] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rpoN; RNA polymerase sigma-54 factor | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rseP; regulator of sigma E protease [EC:3.4.24.-] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rsgA, engC; ribosome biogenesis GTPase / thiamine phosphate phosphatase [EC:3.6.1.- 3.1.3.100] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rsmE; 16S rRNA (uracil1498-N3)-methyltransferase [EC:2.1.1.193] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rtpR; ribonucleoside-triphosphate reductase (thioredoxin) [EC:1.17.4.2] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| rumA; 23S rRNA (uracil1939-C5)-methyltransferase [EC:2.1.1.190] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ruvA; holliday junction DNA helicase RuvA [EC:3.6.4.12] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ruvB; holliday junction DNA helicase RuvB [EC:3.6.4.12] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ruvC; crossover junction endodeoxyribonuclease RuvC [EC:3.1.22.4] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ruvX; putative holliday junction resolvase [EC:3.1.-.-] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| SAM50, TOB55, bamA; outer membrane protein insertion porin family | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| SARS, serS; seryl-tRNA synthetase [EC:6.1.1.11] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| sdhA, frdA; succinate dehydrogenase / fumarate reductase, flavoprotein subunit [EC:1.3.5.1 1.3.5.4] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| sdhB, frdB; succinate dehydrogenase / fumarate reductase, iron-sulfur subunit [EC:1.3.5.1 1.3.5.4] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| sdhC, frdC; succinate dehydrogenase / fumarate reductase, cytochrome b subunit | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| secA; preprotein translocase subunit SecA | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| secDF; SecD/SecF fusion protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| secG; preprotein translocase subunit SecG | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| secY; preprotein translocase subunit SecY | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| slyD; FKBP-type peptidyl-prolyl cis-trans isomerase SlyD [EC:5.2.1.8] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| smf; DNA processing protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| smpB; SsrA-binding protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| speA; arginine decarboxylase [EC:4.1.1.19] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| speG, SAT; diamine N-acetyltransferase [EC:2.3.1.57] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| spoU; RNA methyltransferase, TrmH family | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| sppA; protease IV [EC:3.4.21.-] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| SRP54, ffh; signal recognition particle subunit SRP54 [EC:3.6.5.4] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| sufB; Fe-S cluster assembly protein SufB | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| sufC; Fe-S cluster assembly ATP-binding protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| sufD; Fe-S cluster assembly protein SufD | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| sufE; cysteine desulfuration protein SufE | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| sufS; cysteine desulfurase / selenocysteine lyase [EC:2.8.1.7 4.4.1.16] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| surE; 5’-nucleotidase [EC:3.1.3.5] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| tadA; tRNA(adenine34) deaminase [EC:3.5.4.33] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| TARS, thrS; threonyl-tRNA synthetase [EC:6.1.1.3] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| TC.CIC; chloride channel protein, CIC family | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| TC.POT; proton-dependent oligopeptide transporter, POT family | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| tdk, TK; thymidine kinase [EC:2.7.1.21] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| tgt, QTRT1; queuine tRNA-ribosyltransferase [EC:2.4.2.29] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| thiJ; protein deglycase [EC:3.5.1.124] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| thiL; thiamine-monophosphate kinase [EC:2.7.4.16] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| tig; trigger factor | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| tilS, mesJ; tRNA(Ile)-lysidine synthase [EC:6.3.4.19] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| topA; DNA topoisomerase I [EC:5.99.1.2] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| trkA, ktrA; trk system potassium uptake protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| trkH, trkG, ktrB; trk system potassium uptake protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| trmB, METTL1; tRNA (guanine-N7-)-methyltransferase [EC:2.1.1.33] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| trmD; tRNA (guanine37-N1)-methyltransferase [EC:2.1.1.228] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| truA, PUS1; tRNA pseudouridine38-40 synthase [EC:5.4.99.12] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| truB, PUS4, TRUB1; tRNA pseudouridine55 synthase [EC:5.4.99.25] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| trxA; thioredoxin 1 | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| trxB, TRR; thioredoxin reductase (NADPH) [EC:1.8.1.9] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| tsaB; tRNA threonylcarbamoyladenosine biosynthesis protein TsaB | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| tsaC, rimN, SUA5; L-threonylcarbamoyladenylate synthase [EC:2.7.7.87] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| tsaE; tRNA threonylcarbamoyladenosine biosynthesis protein TsaE | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| tsf, TSFM; elongation factor Ts | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| TSTA3, fcl; GDP-L-fucose synthase [EC:1.1.1.271] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| tuf, TUFM; elongation factor Tu | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| typA, bipA; GTP-binding protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ubiE; demethylmenaquinone methyltransferase / 2-methoxy-6-polyprenyl-1,4-benzoquinol methylase [EC:2.1.1.163 2.1.1.201] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| udp, UPP; uridine phosphorylase [EC:2.4.2.3] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| upp, UPRT; uracil phosphoribosyltransferase [EC:2.4.2.9] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| uppS; undecaprenyl diphosphate synthase [EC:2.5.1.31] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| uvrB; excinuclease ABC subunit B | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| uvrC; excinuclease ABC subunit C | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| VARS, valS; valyl-tRNA synthetase [EC:6.1.1.9] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| WARS, trpS; tryptophanyl-tRNA synthetase [EC:6.1.1.2] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| xerC; integrase/recombinase XerC | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| xerD; integrase/recombinase XerD | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| xseA; exodeoxyribonuclease VII large subunit [EC:3.1.11.6] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| xseB; exodeoxyribonuclease VII small subunit [EC:3.1.11.6] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| yajC; preprotein translocase subunit YajC | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| YARS, tyrS; tyrosyl-tRNA synthetase [EC:6.1.1.1] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ybeB; ribosome-associated protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ycaJ; putative ATPase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ychF; ribosome-binding ATPase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| yfiC, trmX; tRNA1Val (adenine37-N6)-methyltransferase [EC:2.1.1.223] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| yfiH; polyphenol oxidase [EC:1.10.3.-] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| yggS, PROSC; PLP dependent protein | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| yidC, spoIIIJ, OXA1, ccfA; YidC/Oxa1 family membrane protein insertase | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| ypsC; putative N6-adenine-specific DNA methylase [EC:2.1.1.-] | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| yraN; putative endonuclease | 0.88732 | 0.12712 | 0.76987 | 1.68381 | 0.16095 | 65 | 0.96941 | 3.16936 | 0.18461 | 93 | 0.24492 | -0.81474 | 155.700 | 0.41647 | 0.59014 |
| lctP; lactate permease | 0.88713 | 0.23359 | 0.74103 | 4.88458 | 0.27413 | 65 | 0.98925 | 11.28913 | 0.34841 | 93 | 0.44332 | -0.55991 | 155.500 | 0.57635 | 0.67167 |
| gpmB; probable phosphoglycerate mutase [EC:5.4.2.12] | 0.88413 | 0.13159 | 0.75168 | 1.77177 | 0.16510 | 65 | 0.97670 | 3.41497 | 0.19163 | 93 | 0.25294 | -0.88963 | 155.839 | 0.37504 | 0.59014 |
| mraZ; MraZ protein | 0.88413 | 0.13159 | 0.75168 | 1.77177 | 0.16510 | 65 | 0.97670 | 3.41497 | 0.19163 | 93 | 0.25294 | -0.88963 | 155.839 | 0.37504 | 0.59014 |
| pulA; pullulanase [EC:3.2.1.41] | 0.88296 | 0.13528 | 0.75567 | 1.87450 | 0.16982 | 65 | 0.97192 | 3.61109 | 0.19705 | 93 | 0.26013 | -0.83133 | 155.837 | 0.40706 | 0.59014 |
| fhs; formate–tetrahydrofolate ligase [EC:6.3.4.3] | 0.87335 | 0.13145 | 0.74187 | 1.77574 | 0.16528 | 65 | 0.96525 | 3.40283 | 0.19128 | 93 | 0.25280 | -0.88361 | 155.809 | 0.37827 | 0.59014 |
| luxS; S-ribosylhomocysteine lyase [EC:4.4.1.21] | 0.87335 | 0.13145 | 0.74187 | 1.77574 | 0.16528 | 65 | 0.96525 | 3.40283 | 0.19128 | 93 | 0.25280 | -0.88361 | 155.809 | 0.37827 | 0.59014 |
| npdA; NAD-dependent deacetylase [EC:3.5.1.-] | 0.87335 | 0.13145 | 0.74187 | 1.77574 | 0.16528 | 65 | 0.96525 | 3.40283 | 0.19128 | 93 | 0.25280 | -0.88361 | 155.809 | 0.37827 | 0.59014 |
| yhbH; putative sigma-54 modulation protein | 0.87335 | 0.13145 | 0.74187 | 1.77574 | 0.16528 | 65 | 0.96525 | 3.40283 | 0.19128 | 93 | 0.25280 | -0.88361 | 155.809 | 0.37827 | 0.59014 |
| TC.ZIP, zupT, ZRT3, ZIP2; zinc transporter, ZIP family | 0.87071 | 0.23720 | 0.74725 | 6.72689 | 0.32170 | 65 | 0.95699 | 10.47284 | 0.33558 | 93 | 0.46487 | -0.45117 | 153.020 | 0.65250 | 0.73430 |
| DLD, lpd, pdhD; dihydrolipoamide dehydrogenase [EC:1.8.1.4] | 0.87053 | 0.13523 | 0.77525 | 2.16160 | 0.18236 | 65 | 0.93712 | 3.41585 | 0.19165 | 93 | 0.26455 | -0.61187 | 153.329 | 0.54153 | 0.63858 |
| gcvH, GCSH; glycine cleavage system H protein | 0.87053 | 0.13523 | 0.77525 | 2.16160 | 0.18236 | 65 | 0.93712 | 3.41585 | 0.19165 | 93 | 0.26455 | -0.61187 | 153.329 | 0.54153 | 0.63858 |
| gcvPA; glycine dehydrogenase subunit 1 [EC:1.4.4.2] | 0.87053 | 0.13523 | 0.77525 | 2.16160 | 0.18236 | 65 | 0.93712 | 3.41585 | 0.19165 | 93 | 0.26455 | -0.61187 | 153.329 | 0.54153 | 0.63858 |
| gcvPB; glycine dehydrogenase subunit 2 [EC:1.4.4.2] | 0.87053 | 0.13523 | 0.77525 | 2.16160 | 0.18236 | 65 | 0.93712 | 3.41585 | 0.19165 | 93 | 0.26455 | -0.61187 | 153.329 | 0.54153 | 0.63858 |
| gcvT, AMT; aminomethyltransferase [EC:2.1.2.10] | 0.87053 | 0.13523 | 0.77525 | 2.16160 | 0.18236 | 65 | 0.93712 | 3.41585 | 0.19165 | 93 | 0.26455 | -0.61187 | 153.329 | 0.54153 | 0.63858 |
| yrbG; cation:H+ antiporter | 0.85430 | 0.12458 | 0.77723 | 1.82080 | 0.16737 | 65 | 0.90817 | 2.91077 | 0.17691 | 93 | 0.24354 | -0.53766 | 153.557 | 0.59159 | 0.68001 |
| hsdR; type I restriction enzyme, R subunit [EC:3.1.21.3] | 0.85248 | 0.13447 | 0.71933 | 1.76907 | 0.16497 | 65 | 0.94554 | 3.62356 | 0.19739 | 93 | 0.25725 | -0.87930 | 155.999 | 0.38059 | 0.59014 |
| cynR; LysR family transcriptional regulator, cyn operon transcriptional activator | 0.85190 | 0.20611 | 0.69275 | 3.64304 | 0.23674 | 65 | 0.96313 | 8.88993 | 0.30918 | 93 | 0.38941 | -0.69436 | 154.942 | 0.48850 | 0.60307 |
| E1.17.4.1B, nrdB, nrdF; ribonucleoside-diphosphate reductase beta chain [EC:1.17.4.1] | 0.85074 | 0.11871 | 0.78974 | 1.37919 | 0.14567 | 65 | 0.89337 | 2.83584 | 0.17462 | 93 | 0.22740 | -0.45570 | 156.000 | 0.64924 | 0.73135 |
| TC.NSS; neurotransmitter:Na+ symporter, NSS family | 0.84059 | 0.12909 | 0.72406 | 1.43319 | 0.14849 | 65 | 0.92204 | 3.47976 | 0.19343 | 93 | 0.24386 | -0.81190 | 155.002 | 0.41809 | 0.59014 |
| galK; galactokinase [EC:2.7.1.6] | 0.84006 | 0.10980 | 0.73174 | 1.12392 | 0.13150 | 65 | 0.91577 | 2.45463 | 0.16246 | 93 | 0.20901 | -0.88050 | 155.866 | 0.37994 | 0.59014 |
| K08998; uncharacterized protein | 0.84006 | 0.10980 | 0.73174 | 1.12392 | 0.13150 | 65 | 0.91577 | 2.45463 | 0.16246 | 93 | 0.20901 | -0.88050 | 155.866 | 0.37994 | 0.59014 |
| K09924; uncharacterized protein | 0.82505 | 0.19910 | 0.69341 | 3.57425 | 0.23450 | 65 | 0.91705 | 8.18149 | 0.29660 | 93 | 0.37810 | -0.59149 | 155.578 | 0.55505 | 0.65055 |
| trpG; anthranilate synthase component II [EC:4.1.3.27] | 0.82455 | 0.12974 | 0.74500 | 1.98318 | 0.17467 | 65 | 0.88015 | 3.15142 | 0.18408 | 93 | 0.25377 | -0.53259 | 153.440 | 0.59509 | 0.68219 |
| secE; preprotein translocase subunit SecE | 0.82271 | 0.11954 | 0.72024 | 1.54410 | 0.15413 | 65 | 0.89433 | 2.76620 | 0.17246 | 93 | 0.23130 | -0.75269 | 155.268 | 0.45277 | 0.59014 |
| E3.6.3.8; Ca2+-transporting ATPase [EC:3.6.3.8] | 0.82149 | 0.11978 | 0.74746 | 1.68326 | 0.16092 | 65 | 0.87324 | 2.69117 | 0.17011 | 93 | 0.23417 | -0.53717 | 153.559 | 0.59193 | 0.68001 |
| xpt; xanthine phosphoribosyltransferase [EC:2.4.2.22] | 0.82149 | 0.11978 | 0.74746 | 1.68326 | 0.16092 | 65 | 0.87324 | 2.69117 | 0.17011 | 93 | 0.23417 | -0.53717 | 153.559 | 0.59193 | 0.68001 |
| K07001; NTE family protein | 0.82033 | 0.12053 | 0.68282 | 1.23652 | 0.13793 | 65 | 0.91644 | 3.03441 | 0.18063 | 93 | 0.22727 | -1.02795 | 154.873 | 0.30558 | 0.59014 |
| ndh; NADH dehydrogenase [EC:1.6.99.3] | 0.81739 | 0.11537 | 0.71828 | 1.18617 | 0.13509 | 65 | 0.88667 | 2.75203 | 0.17202 | 93 | 0.21872 | -0.76987 | 155.467 | 0.44254 | 0.59014 |
| abfA; alpha-N-arabinofuranosidase [EC:3.2.1.55] | 0.81190 | 0.23710 | 0.51200 | 1.60636 | 0.15720 | 65 | 1.02151 | 13.93258 | 0.38706 | 93 | 0.41776 | -1.21960 | 120.156 | 0.22500 | 0.59014 |
| K09955; uncharacterized protein | 0.81190 | 0.23710 | 0.51200 | 1.60636 | 0.15720 | 65 | 1.02151 | 13.93258 | 0.38706 | 93 | 0.41776 | -1.21960 | 120.156 | 0.22500 | 0.59014 |
| exuT; MFS transporter, ACS family, hexuronate transporter | 0.81165 | 0.23711 | 0.51138 | 1.60692 | 0.15723 | 65 | 1.02151 | 13.93258 | 0.38706 | 93 | 0.41777 | -1.22105 | 120.165 | 0.22446 | 0.59014 |
| TC.SSS; solute:Na+ symporter, SSS family | 0.81165 | 0.23711 | 0.51138 | 1.60692 | 0.15723 | 65 | 1.02151 | 13.93258 | 0.38706 | 93 | 0.41777 | -1.22105 | 120.165 | 0.22446 | 0.59014 |
| uxuA; mannonate dehydratase [EC:4.2.1.8] | 0.81165 | 0.23711 | 0.51138 | 1.60692 | 0.15723 | 65 | 1.02151 | 13.93258 | 0.38706 | 93 | 0.41777 | -1.22105 | 120.165 | 0.22446 | 0.59014 |
| djlA; DnaJ like chaperone protein | 0.81110 | 0.12860 | 0.72962 | 1.94256 | 0.17287 | 65 | 0.86806 | 3.09963 | 0.18256 | 93 | 0.25143 | -0.55062 | 153.521 | 0.58269 | 0.67611 |
| mgtC; putative Mg2+ transporter-C (MgtC) family protein | 0.80820 | 0.12061 | 0.67897 | 1.25391 | 0.13889 | 65 | 0.89852 | 3.02985 | 0.18050 | 93 | 0.22775 | -0.96398 | 155.058 | 0.33655 | 0.59014 |
| perR; Fur family transcriptional regulator, peroxide stress response regulator | 0.80322 | 0.12456 | 0.71520 | 1.82770 | 0.16769 | 65 | 0.86473 | 2.90274 | 0.17667 | 93 | 0.24358 | -0.61389 | 153.429 | 0.54020 | 0.63858 |
| xylB, XYLB; xylulokinase [EC:2.7.1.17] | 0.79873 | 0.22090 | 0.55949 | 2.78800 | 0.20710 | 65 | 0.96595 | 11.14889 | 0.34624 | 93 | 0.40345 | -1.00746 | 143.250 | 0.31541 | 0.59014 |
| cysK; cysteine synthase A [EC:2.5.1.47] | 0.78473 | 0.11908 | 0.66808 | 1.20008 | 0.13588 | 65 | 0.86626 | 2.97209 | 0.17877 | 93 | 0.22455 | -0.88261 | 154.757 | 0.37882 | 0.59014 |
| arnE; undecaprenyl phosphate-alpha-L-ara4N flippase subunit ArnE | 0.78165 | 0.20123 | 0.49487 | 1.34720 | 0.14397 | 65 | 0.98208 | 9.88253 | 0.32598 | 93 | 0.35636 | -1.36719 | 124.575 | 0.17403 | 0.59014 |
| trpB; tryptophan synthase beta chain [EC:4.2.1.20] | 0.77916 | 0.11350 | 0.67658 | 1.26594 | 0.13956 | 65 | 0.85086 | 2.58026 | 0.16657 | 93 | 0.21730 | -0.80199 | 155.997 | 0.42378 | 0.59014 |
| lysA; diaminopimelate decarboxylase [EC:4.1.1.20] | 0.77790 | 0.11572 | 0.65612 | 1.15146 | 0.13310 | 65 | 0.86301 | 2.79168 | 0.17326 | 93 | 0.21848 | -0.94695 | 155.019 | 0.34514 | 0.59014 |
| metA; homoserine O-succinyltransferase/O-acetyltransferase [EC:2.3.1.46 2.3.1.31] | 0.77790 | 0.11572 | 0.65612 | 1.15146 | 0.13310 | 65 | 0.86301 | 2.79168 | 0.17326 | 93 | 0.21848 | -0.94695 | 155.019 | 0.34514 | 0.59014 |
| thrA; bifunctional aspartokinase / homoserine dehydrogenase 1 [EC:2.7.2.4 1.1.1.3] | 0.77790 | 0.11572 | 0.65612 | 1.15146 | 0.13310 | 65 | 0.86301 | 2.79168 | 0.17326 | 93 | 0.21848 | -0.94695 | 155.019 | 0.34514 | 0.59014 |
| K07504; predicted type IV restriction endonuclease | 0.77532 | 0.18790 | 0.80000 | 4.34219 | 0.25846 | 65 | 0.75806 | 6.49790 | 0.26433 | 93 | 0.36969 | 0.11343 | 152.123 | 0.90984 | 0.94278 |
| norB; nitric oxide reductase subunit B [EC:1.7.2.5] | 0.77532 | 0.18790 | 0.80000 | 4.34219 | 0.25846 | 65 | 0.75806 | 6.49790 | 0.26433 | 93 | 0.36969 | 0.11343 | 152.123 | 0.90984 | 0.94278 |
| TC.FEV.OM3, tbpA, hemR, lbpA, hpuB, bhuR, hugA, hmbR; hemoglobin/transferrin/lactoferrin receptor protein | 0.77532 | 0.18790 | 0.80000 | 4.34219 | 0.25846 | 65 | 0.75806 | 6.49790 | 0.26433 | 93 | 0.36969 | 0.11343 | 152.123 | 0.90984 | 0.94278 |
| fadL; long-chain fatty acid transport protein | 0.77342 | 0.20157 | 0.50564 | 1.33280 | 0.14319 | 65 | 0.96057 | 9.94227 | 0.32697 | 93 | 0.35695 | -1.27451 | 124.112 | 0.20486 | 0.59014 |
| penP; beta-lactamase class A [EC:3.5.2.6] | 0.76582 | 0.21622 | 0.63590 | 1.76640 | 0.16485 | 65 | 0.85663 | 11.35603 | 0.34944 | 93 | 0.38637 | -0.57130 | 128.367 | 0.56880 | 0.66532 |
| MuB; ATP-dependent target DNA activator [EC:3.6.1.3] | 0.75949 | 0.33255 | 0.64615 | 21.03690 | 0.56890 | 65 | 0.83871 | 15.16935 | 0.40387 | 93 | 0.69768 | -0.27599 | 123.027 | 0.78302 | 0.83724 |
| pspC; phage shock protein C | 0.75105 | 0.19464 | 0.57094 | 1.88653 | 0.17036 | 65 | 0.87694 | 8.86405 | 0.30873 | 93 | 0.35261 | -0.86781 | 138.146 | 0.38700 | 0.59014 |
| AXY8, FUC95A, afcA; alpha-L-fucosidase 2 [EC:3.2.1.51] | 0.74549 | 0.10071 | 0.67590 | 1.06265 | 0.12786 | 65 | 0.79413 | 1.98973 | 0.14627 | 93 | 0.19428 | -0.60858 | 155.663 | 0.54369 | 0.64048 |
| gpr; L-glyceraldehyde 3-phosphate reductase [EC:1.1.1.-] | 0.74367 | 0.19057 | 0.82308 | 4.43305 | 0.26115 | 65 | 0.68817 | 6.70062 | 0.26842 | 93 | 0.37450 | 0.36023 | 152.361 | 0.71918 | 0.78345 |
| cytR; LacI family transcriptional regulator, repressor for deo operon, udp, cdd, tsx, nupC, and nupG | 0.73418 | 0.49184 | 0.80000 | 41.60000 | 0.80000 | 65 | 0.68817 | 36.28214 | 0.62460 | 93 | 1.01495 | 0.11018 | 131.751 | 0.91243 | 0.94278 |
| GBA, srfJ; glucosylceramidase [EC:3.2.1.45] | 0.73418 | 0.49184 | 0.80000 | 41.60000 | 0.80000 | 65 | 0.68817 | 36.28214 | 0.62460 | 93 | 1.01495 | 0.11018 | 131.751 | 0.91243 | 0.94278 |
| K09973; uncharacterized protein | 0.73418 | 0.49184 | 0.80000 | 41.60000 | 0.80000 | 65 | 0.68817 | 36.28214 | 0.62460 | 93 | 1.01495 | 0.11018 | 131.751 | 0.91243 | 0.94278 |
| tetM, tetO; ribosomal protection tetracycline resistance protein | 0.72785 | 0.17690 | 0.56923 | 1.32326 | 0.14268 | 65 | 0.83871 | 7.48729 | 0.28374 | 93 | 0.31759 | -0.84850 | 132.254 | 0.39769 | 0.59014 |
| eda; 2-dehydro-3-deoxyphosphogluconate aldolase / (4S)-4-hydroxy-2-oxoglutarate aldolase [EC:4.1.2.14 4.1.3.42] | 0.72622 | 0.19784 | 0.47956 | 2.04833 | 0.17752 | 65 | 0.89862 | 9.05571 | 0.31205 | 93 | 0.35901 | -1.16727 | 140.092 | 0.24509 | 0.59014 |
| xylS, yicI; alpha-D-xyloside xylohydrolase [EC:3.2.1.177] | 0.72369 | 0.19004 | 0.48659 | 2.04561 | 0.17740 | 65 | 0.88940 | 8.24762 | 0.29780 | 93 | 0.34663 | -1.16205 | 142.995 | 0.24715 | 0.59014 |
| K06975; uncharacterized protein | 0.72038 | 0.14227 | 0.54800 | 1.50188 | 0.15201 | 65 | 0.84086 | 4.37679 | 0.21694 | 93 | 0.26489 | -1.10558 | 151.885 | 0.27066 | 0.59014 |
| fabZ; 3-hydroxyacyl-[acyl-carrier-protein] dehydratase [EC:4.2.1.59] | 0.69316 | 0.16032 | 0.42108 | 0.64380 | 0.09952 | 65 | 0.88333 | 6.39306 | 0.26219 | 93 | 0.28044 | -1.64832 | 116.932 | 0.10197 | 0.59014 |
| ihfA, himA; integration host factor subunit alpha | 0.68861 | 0.17835 | 0.54735 | 2.94573 | 0.21288 | 65 | 0.78734 | 6.50313 | 0.26444 | 93 | 0.33948 | -0.70693 | 155.814 | 0.48067 | 0.59915 |
| K06926; uncharacterized protein | 0.68662 | 0.11928 | 0.49315 | 0.87604 | 0.11609 | 65 | 0.82185 | 3.18210 | 0.18498 | 93 | 0.21839 | -1.50512 | 146.154 | 0.13445 | 0.59014 |
| E2.2.1.6L, ilvB, ilvG, ilvI; acetolactate synthase I/II/III large subunit [EC:2.2.1.6] | 0.68553 | 0.18938 | 0.47956 | 2.04833 | 0.17752 | 65 | 0.82949 | 8.19488 | 0.29685 | 93 | 0.34588 | -1.01173 | 143.235 | 0.31337 | 0.59014 |
| E2.2.1.6S, ilvH, ilvN; acetolactate synthase I/III small subunit [EC:2.2.1.6] | 0.68553 | 0.18938 | 0.47956 | 2.04833 | 0.17752 | 65 | 0.82949 | 8.19488 | 0.29685 | 93 | 0.34588 | -1.01173 | 143.235 | 0.31337 | 0.59014 |
| idnO; gluconate 5-dehydrogenase [EC:1.1.1.69] | 0.68553 | 0.18951 | 0.48176 | 2.07329 | 0.17860 | 65 | 0.82796 | 8.19102 | 0.29678 | 93 | 0.34637 | -0.99951 | 143.624 | 0.31923 | 0.59014 |
| ilvC; ketol-acid reductoisomerase [EC:1.1.1.86] | 0.68553 | 0.18938 | 0.47956 | 2.04833 | 0.17752 | 65 | 0.82949 | 8.19488 | 0.29685 | 93 | 0.34588 | -1.01173 | 143.235 | 0.31337 | 0.59014 |
| ilvD; dihydroxy-acid dehydratase [EC:4.2.1.9] | 0.68553 | 0.18938 | 0.47956 | 2.04833 | 0.17752 | 65 | 0.82949 | 8.19488 | 0.29685 | 93 | 0.34588 | -1.01173 | 143.235 | 0.31337 | 0.59014 |
| tolA; colicin import membrane protein | 0.68375 | 0.10467 | 0.61902 | 1.18828 | 0.13521 | 65 | 0.72898 | 2.12244 | 0.15107 | 93 | 0.20274 | -0.54238 | 155.236 | 0.58833 | 0.67888 |
| E3.1.4.46, glpQ, ugpQ; glycerophosphoryl diester phosphodiesterase [EC:3.1.4.46] | 0.68291 | 0.16114 | 0.44462 | 0.89958 | 0.11764 | 65 | 0.84946 | 6.30692 | 0.26042 | 93 | 0.28576 | -1.41676 | 125.847 | 0.15902 | 0.59014 |
| queD, ptpS, PTS; 6-pyruvoyltetrahydropterin/6-carboxytetrahydropterin synthase [EC:4.2.3.12 4.1.2.50] | 0.68241 | 0.14779 | 0.53108 | 1.53653 | 0.15375 | 65 | 0.78817 | 4.79279 | 0.22701 | 93 | 0.27418 | -0.93769 | 150.299 | 0.34991 | 0.59014 |
| wcaJ; putative colanic acid biosysnthesis UDP-glucose lipid carrier transferase | 0.67608 | 0.17518 | 0.51415 | 1.52777 | 0.15331 | 65 | 0.78925 | 7.17988 | 0.27785 | 93 | 0.31734 | -0.86686 | 138.140 | 0.38752 | 0.59014 |
| glf; UDP-galactopyranose mutase [EC:5.4.99.9] | 0.66237 | 0.13315 | 0.53594 | 1.44317 | 0.14901 | 65 | 0.75073 | 3.75708 | 0.20099 | 93 | 0.25020 | -0.85846 | 154.033 | 0.39197 | 0.59014 |
| vsr; DNA mismatch endonuclease, patch repair protein [EC:3.1.-.-] | 0.65759 | 0.15451 | 0.54615 | 1.78909 | 0.16590 | 65 | 0.73548 | 5.17710 | 0.23594 | 93 | 0.28843 | -0.65641 | 152.038 | 0.51255 | 0.61634 |
| htpX; heat shock protein HtpX [EC:3.4.24.-] | 0.65535 | 0.13230 | 0.49231 | 1.24481 | 0.13839 | 65 | 0.76931 | 3.82152 | 0.20271 | 93 | 0.24544 | -1.12856 | 150.687 | 0.26088 | 0.59014 |
| ABC.CD.TX; HlyD family secretion protein | 0.65479 | 0.16521 | 0.45209 | 3.06882 | 0.21728 | 65 | 0.79647 | 5.17557 | 0.23591 | 93 | 0.32072 | -1.07375 | 154.484 | 0.28461 | 0.59014 |
| K09797; uncharacterized protein | 0.65190 | 0.13030 | 0.50549 | 1.93388 | 0.17249 | 65 | 0.75422 | 3.20657 | 0.18569 | 93 | 0.25344 | -0.98142 | 154.213 | 0.32792 | 0.59014 |
| cas1; CRISP-associated protein Cas1 | 0.64887 | 0.13600 | 0.44103 | 1.01234 | 0.12480 | 65 | 0.79413 | 4.23077 | 0.21329 | 93 | 0.24712 | -1.42890 | 141.873 | 0.15523 | 0.59014 |
| cas2; CRISPR-associated protein Cas2 | 0.64887 | 0.13600 | 0.44103 | 1.01234 | 0.12480 | 65 | 0.79413 | 4.23077 | 0.21329 | 93 | 0.24712 | -1.42890 | 141.873 | 0.15523 | 0.59014 |
| ABC.X2.P; putative ABC transport system permease protein | 0.64747 | 0.13376 | 0.46154 | 0.93772 | 0.12011 | 65 | 0.77742 | 4.13068 | 0.21075 | 93 | 0.24257 | -1.30220 | 140.207 | 0.19498 | 0.59014 |
| E3.2.1.89; arabinogalactan endo-1,4-beta-galactosidase [EC:3.2.1.89] | 0.63648 | 0.16168 | 0.46601 | 1.35653 | 0.14446 | 65 | 0.75562 | 6.06953 | 0.25547 | 93 | 0.29349 | -0.98679 | 139.710 | 0.32545 | 0.59014 |
| apgM; 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [EC:5.4.2.12] | 0.63386 | 0.16633 | 0.42538 | 1.56647 | 0.15524 | 65 | 0.77957 | 6.31739 | 0.26063 | 93 | 0.30336 | -1.16753 | 142.987 | 0.24494 | 0.59014 |
| TC.GPH; glycoside/pentoside/hexuronide:cation symporter, GPH family | 0.63386 | 0.16633 | 0.42538 | 1.56647 | 0.15524 | 65 | 0.77957 | 6.31739 | 0.26063 | 93 | 0.30336 | -1.16753 | 142.987 | 0.24494 | 0.59014 |
| thrC; threonine synthase [EC:4.2.3.1] | 0.63386 | 0.16633 | 0.42538 | 1.56647 | 0.15524 | 65 | 0.77957 | 6.31739 | 0.26063 | 93 | 0.30336 | -1.16753 | 142.987 | 0.24494 | 0.59014 |
| abnA; arabinan endo-1,5-alpha-L-arabinosidase [EC:3.2.1.99] | 0.62841 | 0.14992 | 0.43179 | 1.32576 | 0.14282 | 65 | 0.76583 | 5.09164 | 0.23398 | 93 | 0.27413 | -1.21855 | 144.489 | 0.22500 | 0.59014 |
| amn; AMP nucleosidase [EC:3.2.2.4] | 0.61392 | 0.30670 | 0.36923 | 6.20529 | 0.30898 | 65 | 0.78495 | 20.97499 | 0.47491 | 93 | 0.56657 | -0.73374 | 148.198 | 0.46427 | 0.59014 |
| bioF; 8-amino-7-oxononanoate synthase [EC:2.3.1.47] | 0.61392 | 0.30670 | 0.36923 | 6.20529 | 0.30898 | 65 | 0.78495 | 20.97499 | 0.47491 | 93 | 0.56657 | -0.73374 | 148.198 | 0.46427 | 0.59014 |
| coaX; type III pantothenate kinase [EC:2.7.1.33] | 0.61392 | 0.30670 | 0.36923 | 6.20529 | 0.30898 | 65 | 0.78495 | 20.97499 | 0.47491 | 93 | 0.56657 | -0.73374 | 148.198 | 0.46427 | 0.59014 |
| cysC; adenylylsulfate kinase [EC:2.7.1.25] | 0.61392 | 0.30670 | 0.36923 | 6.20529 | 0.30898 | 65 | 0.78495 | 20.97499 | 0.47491 | 93 | 0.56657 | -0.73374 | 148.198 | 0.46427 | 0.59014 |
| cysD; sulfate adenylyltransferase subunit 2 [EC:2.7.7.4] | 0.61392 | 0.30670 | 0.36923 | 6.20529 | 0.30898 | 65 | 0.78495 | 20.97499 | 0.47491 | 93 | 0.56657 | -0.73374 | 148.198 | 0.46427 | 0.59014 |
| cysN; sulfate adenylyltransferase subunit 1 [EC:2.7.7.4] | 0.61392 | 0.30670 | 0.36923 | 6.20529 | 0.30898 | 65 | 0.78495 | 20.97499 | 0.47491 | 93 | 0.56657 | -0.73374 | 148.198 | 0.46427 | 0.59014 |
| cysQ, MET22, BPNT1; 3’(2’), 5’-bisphosphate nucleotidase [EC:3.1.3.7] | 0.61392 | 0.30670 | 0.36923 | 6.20529 | 0.30898 | 65 | 0.78495 | 20.97499 | 0.47491 | 93 | 0.56657 | -0.73374 | 148.198 | 0.46427 | 0.59014 |
| E3.1.11.5; exodeoxyribonuclease V [EC:3.1.11.5] | 0.61392 | 0.30670 | 0.36923 | 6.20529 | 0.30898 | 65 | 0.78495 | 20.97499 | 0.47491 | 93 | 0.56657 | -0.73374 | 148.198 | 0.46427 | 0.59014 |
| EIF1, SUI1; translation initiation factor 1 | 0.61392 | 0.30670 | 0.36923 | 6.20529 | 0.30898 | 65 | 0.78495 | 20.97499 | 0.47491 | 93 | 0.56657 | -0.73374 | 148.198 | 0.46427 | 0.59014 |
| K06973; uncharacterized protein | 0.61392 | 0.30670 | 0.36923 | 6.20529 | 0.30898 | 65 | 0.78495 | 20.97499 | 0.47491 | 93 | 0.56657 | -0.73374 | 148.198 | 0.46427 | 0.59014 |
| K07043; uncharacterized protein | 0.61392 | 0.30670 | 0.36923 | 6.20529 | 0.30898 | 65 | 0.78495 | 20.97499 | 0.47491 | 93 | 0.56657 | -0.73374 | 148.198 | 0.46427 | 0.59014 |
| K07150; uncharacterized protein | 0.61392 | 0.30670 | 0.36923 | 6.20529 | 0.30898 | 65 | 0.78495 | 20.97499 | 0.47491 | 93 | 0.56657 | -0.73374 | 148.198 | 0.46427 | 0.59014 |
| marC; multiple antibiotic resistance protein | 0.61392 | 0.30670 | 0.36923 | 6.20529 | 0.30898 | 65 | 0.78495 | 20.97499 | 0.47491 | 93 | 0.56657 | -0.73374 | 148.198 | 0.46427 | 0.59014 |
| mbtN, fadE14; acyl-ACP dehydrogenase [EC:1.3.99.-] | 0.61392 | 0.30670 | 0.36923 | 6.20529 | 0.30898 | 65 | 0.78495 | 20.97499 | 0.47491 | 93 | 0.56657 | -0.73374 | 148.198 | 0.46427 | 0.59014 |
| mepS, spr; murein DD-endopeptidase / murein LD-carboxypeptidase [EC:3.4.-.- 3.4.17.13] | 0.61392 | 0.30670 | 0.36923 | 6.20529 | 0.30898 | 65 | 0.78495 | 20.97499 | 0.47491 | 93 | 0.56657 | -0.73374 | 148.198 | 0.46427 | 0.59014 |
| mscS; small conductance mechanosensitive channel | 0.61392 | 0.30670 | 0.36923 | 6.20529 | 0.30898 | 65 | 0.78495 | 20.97499 | 0.47491 | 93 | 0.56657 | -0.73374 | 148.198 | 0.46427 | 0.59014 |
| ndk, NME; nucleoside-diphosphate kinase [EC:2.7.4.6] | 0.61392 | 0.30670 | 0.36923 | 6.20529 | 0.30898 | 65 | 0.78495 | 20.97499 | 0.47491 | 93 | 0.56657 | -0.73374 | 148.198 | 0.46427 | 0.59014 |
| pcnB; poly(A) polymerase [EC:2.7.7.19] | 0.61392 | 0.30670 | 0.36923 | 6.20529 | 0.30898 | 65 | 0.78495 | 20.97499 | 0.47491 | 93 | 0.56657 | -0.73374 | 148.198 | 0.46427 | 0.59014 |
| pdxB; erythronate-4-phosphate dehydrogenase [EC:1.1.1.290] | 0.61392 | 0.30670 | 0.36923 | 6.20529 | 0.30898 | 65 | 0.78495 | 20.97499 | 0.47491 | 93 | 0.56657 | -0.73374 | 148.198 | 0.46427 | 0.59014 |
| pdxJ; pyridoxine 5-phosphate synthase [EC:2.6.99.2] | 0.61392 | 0.30670 | 0.36923 | 6.20529 | 0.30898 | 65 | 0.78495 | 20.97499 | 0.47491 | 93 | 0.56657 | -0.73374 | 148.198 | 0.46427 | 0.59014 |
| tpx; thiol peroxidase, atypical 2-Cys peroxiredoxin [EC:1.11.1.15] | 0.61392 | 0.30670 | 0.36923 | 6.20529 | 0.30898 | 65 | 0.78495 | 20.97499 | 0.47491 | 93 | 0.56657 | -0.73374 | 148.198 | 0.46427 | 0.59014 |
| ybaK, ebsC; Cys-tRNA(Pro)/Cys-tRNA(Cys) deacylase [EC:3.1.1.-] | 0.61392 | 0.30670 | 0.36923 | 6.20529 | 0.30898 | 65 | 0.78495 | 20.97499 | 0.47491 | 93 | 0.56657 | -0.73374 | 148.198 | 0.46427 | 0.59014 |
| purN; phosphoribosylglycinamide formyltransferase 1 [EC:2.1.2.2] | 0.59863 | 0.14194 | 0.47692 | 2.30979 | 0.18851 | 65 | 0.68369 | 3.80745 | 0.20234 | 93 | 0.27654 | -0.74769 | 154.115 | 0.45578 | 0.59014 |
| cobC, phpB; alpha-ribazole phosphatase [EC:3.1.3.73] | 0.59351 | 0.15114 | 0.37731 | 0.75533 | 0.10780 | 65 | 0.74462 | 5.57742 | 0.24489 | 93 | 0.26757 | -1.37279 | 124.393 | 0.17229 | 0.59014 |
| cobP, cobU; adenosylcobinamide kinase / adenosylcobinamide-phosphate guanylyltransferase [EC:2.7.1.156 2.7.7.62] | 0.59351 | 0.15114 | 0.37731 | 0.75533 | 0.10780 | 65 | 0.74462 | 5.57742 | 0.24489 | 93 | 0.26757 | -1.37279 | 124.393 | 0.17229 | 0.59014 |
| E2.4.2.21, cobU, cobT; nicotinate-nucleotide–dimethylbenzimidazole phosphoribosyltransferase [EC:2.4.2.21] | 0.59351 | 0.15114 | 0.37731 | 0.75533 | 0.10780 | 65 | 0.74462 | 5.57742 | 0.24489 | 93 | 0.26757 | -1.37279 | 124.393 | 0.17229 | 0.59014 |
| E2.7.8.26, cobS, cobV; adenosylcobinamide-GDP ribazoletransferase [EC:2.7.8.26] | 0.59351 | 0.15114 | 0.37731 | 0.75533 | 0.10780 | 65 | 0.74462 | 5.57742 | 0.24489 | 93 | 0.26757 | -1.37279 | 124.393 | 0.17229 | 0.59014 |
| znuB; zinc transport system permease protein | 0.59017 | 0.11499 | 0.44166 | 0.93124 | 0.11969 | 65 | 0.69396 | 2.89104 | 0.17631 | 93 | 0.21310 | -1.18395 | 150.415 | 0.23830 | 0.59014 |
| znuC; zinc transport system ATP-binding protein [EC:3.6.3.-] | 0.59017 | 0.11499 | 0.44166 | 0.93124 | 0.11969 | 65 | 0.69396 | 2.89104 | 0.17631 | 93 | 0.21310 | -1.18395 | 150.415 | 0.23830 | 0.59014 |
| SOD2; superoxide dismutase, Fe-Mn family [EC:1.15.1.1] | 0.58763 | 0.11111 | 0.45680 | 0.92527 | 0.11931 | 65 | 0.67907 | 2.66422 | 0.16926 | 93 | 0.20708 | -1.07334 | 152.144 | 0.28482 | 0.59014 |
| ABC.CD.A; putative ABC transport system ATP-binding protein | 0.58685 | 0.13839 | 0.39101 | 1.52189 | 0.15302 | 65 | 0.72374 | 4.05946 | 0.20893 | 93 | 0.25897 | -1.28485 | 153.627 | 0.20078 | 0.59014 |
| nosD; nitrous oxidase accessory protein | 0.58228 | 0.27015 | 0.47692 | 9.34712 | 0.37921 | 65 | 0.65591 | 13.16293 | 0.37621 | 93 | 0.53417 | -0.33508 | 150.536 | 0.73803 | 0.79501 |
| nosF; Cu-processing system ATP-binding protein | 0.58228 | 0.27015 | 0.47692 | 9.34712 | 0.37921 | 65 | 0.65591 | 13.16293 | 0.37621 | 93 | 0.53417 | -0.33508 | 150.536 | 0.73803 | 0.79501 |
| nosY; Cu-processing system permease protein | 0.58228 | 0.27015 | 0.47692 | 9.34712 | 0.37921 | 65 | 0.65591 | 13.16293 | 0.37621 | 93 | 0.53417 | -0.33508 | 150.536 | 0.73803 | 0.79501 |
| nosZ; nitrous-oxide reductase [EC:1.7.2.4] | 0.58228 | 0.27015 | 0.47692 | 9.34712 | 0.37921 | 65 | 0.65591 | 13.16293 | 0.37621 | 93 | 0.53417 | -0.33508 | 150.536 | 0.73803 | 0.79501 |
| ramA; alpha-L-rhamnosidase [EC:3.2.1.40] | 0.58228 | 0.27015 | 0.47692 | 9.34712 | 0.37921 | 65 | 0.65591 | 13.16293 | 0.37621 | 93 | 0.53417 | -0.33508 | 150.536 | 0.73803 | 0.79501 |
| rnj; ribonuclease J [EC:3.1.-.-] | 0.58228 | 0.27015 | 0.47692 | 9.34712 | 0.37921 | 65 | 0.65591 | 13.16293 | 0.37621 | 93 | 0.53417 | -0.33508 | 150.536 | 0.73803 | 0.79501 |
| spsF; spore coat polysaccharide biosynthesis protein SpsF | 0.58228 | 0.27015 | 0.47692 | 9.34712 | 0.37921 | 65 | 0.65591 | 13.16293 | 0.37621 | 93 | 0.53417 | -0.33508 | 150.536 | 0.73803 | 0.79501 |
| tolC; outer membrane protein | 0.58146 | 0.13394 | 0.39473 | 1.74187 | 0.16370 | 65 | 0.71198 | 3.58390 | 0.19631 | 93 | 0.25561 | -1.24119 | 156.000 | 0.21640 | 0.59014 |
| mltF; membrane-bound lytic murein transglycosylase F [EC:4.2.2.-] | 0.56293 | 0.10702 | 0.41670 | 0.80029 | 0.11096 | 65 | 0.66513 | 2.50577 | 0.16415 | 93 | 0.19813 | -1.25385 | 150.205 | 0.21184 | 0.59014 |
| K09117; uncharacterized protein | 0.56144 | 0.11437 | 0.41207 | 0.89689 | 0.11747 | 65 | 0.66584 | 2.87606 | 0.17586 | 93 | 0.21148 | -1.19997 | 149.601 | 0.23205 | 0.59014 |
| pheA2; prephenate dehydratase [EC:4.2.1.51] | 0.54437 | 0.11659 | 0.35530 | 0.54727 | 0.09176 | 65 | 0.67652 | 3.24171 | 0.18670 | 93 | 0.20803 | -1.54412 | 130.839 | 0.12497 | 0.59014 |
| yoeB; toxin YoeB [EC:3.1.-.-] | 0.53376 | 0.12571 | 0.56923 | 1.97473 | 0.17430 | 65 | 0.50896 | 2.88549 | 0.17614 | 93 | 0.24780 | 0.24322 | 151.530 | 0.80817 | 0.85466 |
| eamB; cysteine/O-acetylserine efflux protein | 0.52532 | 0.36271 | 0.55385 | 16.84471 | 0.50907 | 65 | 0.50538 | 23.75269 | 0.50538 | 93 | 0.71732 | 0.06757 | 150.573 | 0.94622 | 0.95197 |
| K09124; uncharacterized protein | 0.52532 | 0.36271 | 0.55385 | 16.84471 | 0.50907 | 65 | 0.50538 | 23.75269 | 0.50538 | 93 | 0.71732 | 0.06757 | 150.573 | 0.94622 | 0.95197 |
| tagF; CDP-glycerol glycerophosphotransferase [EC:2.7.8.12] | 0.52532 | 0.36271 | 0.55385 | 16.84471 | 0.50907 | 65 | 0.50538 | 23.75269 | 0.50538 | 93 | 0.71732 | 0.06757 | 150.573 | 0.94622 | 0.95197 |
| pgpA; phosphatidylglycerophosphatase A [EC:3.1.3.27] | 0.52363 | 0.14626 | 0.51897 | 1.94007 | 0.17276 | 65 | 0.52688 | 4.41865 | 0.21797 | 93 | 0.27814 | -0.02843 | 155.616 | 0.97736 | 0.97905 |
| E3.2.1.17; lysozyme [EC:3.2.1.17] | 0.51899 | 0.11395 | 0.53077 | 1.59279 | 0.15654 | 65 | 0.51075 | 2.39255 | 0.16039 | 93 | 0.22412 | 0.08931 | 152.213 | 0.92895 | 0.94278 |
| aat; leucyl/phenylalanyl-tRNA—protein transferase [EC:2.3.2.6] | 0.51730 | 0.12525 | 0.53436 | 1.92882 | 0.17226 | 65 | 0.50538 | 2.88795 | 0.17622 | 93 | 0.24643 | 0.11761 | 152.136 | 0.90653 | 0.94278 |
| clpA; ATP-dependent Clp protease ATP-binding subunit ClpA | 0.51730 | 0.12525 | 0.53436 | 1.92882 | 0.17226 | 65 | 0.50538 | 2.88795 | 0.17622 | 93 | 0.24643 | 0.11761 | 152.136 | 0.90653 | 0.94278 |
| clpS; ATP-dependent Clp protease adaptor protein ClpS | 0.51730 | 0.12525 | 0.53436 | 1.92882 | 0.17226 | 65 | 0.50538 | 2.88795 | 0.17622 | 93 | 0.24643 | 0.11761 | 152.136 | 0.90653 | 0.94278 |
| yafQ; mRNA interferase YafQ [EC:3.1.-.-] | 0.50497 | 0.16244 | 0.30769 | 0.63567 | 0.09889 | 65 | 0.64286 | 6.62555 | 0.26691 | 93 | 0.28464 | -1.17749 | 115.853 | 0.24141 | 0.59014 |
| K09805; uncharacterized protein | 0.48133 | 0.12772 | 0.31077 | 0.64144 | 0.09934 | 65 | 0.60054 | 3.91698 | 0.20523 | 93 | 0.22801 | -1.27088 | 129.910 | 0.20604 | 0.59014 |
| murQ; N-acetylmuramic acid 6-phosphate etherase [EC:4.2.1.126] | 0.47710 | 0.11628 | 0.28839 | 0.40133 | 0.07858 | 65 | 0.60899 | 3.32371 | 0.18905 | 93 | 0.20473 | -1.56599 | 121.329 | 0.11995 | 0.59014 |
| xylA; xylose isomerase [EC:5.3.1.5] | 0.47152 | 0.26006 | 0.44615 | 10.50096 | 0.40194 | 65 | 0.48925 | 10.92923 | 0.34281 | 93 | 0.52827 | -0.08157 | 139.592 | 0.93510 | 0.94558 |
| uxaC; glucuronate isomerase [EC:5.3.1.12] | 0.46722 | 0.12602 | 0.26338 | 0.40408 | 0.07885 | 65 | 0.60968 | 3.95088 | 0.20611 | 93 | 0.22068 | -1.56921 | 117.285 | 0.11929 | 0.59014 |
| pgsA, PGS1; CDP-diacylglycerol—glycerol-3-phosphate 3-phosphatidyltransferase [EC:2.7.8.5] | 0.46076 | 0.11582 | 0.40077 | 1.08496 | 0.12920 | 65 | 0.50269 | 2.85801 | 0.17530 | 93 | 0.21777 | -0.46802 | 153.840 | 0.64044 | 0.72354 |
| czcA; cobalt-zinc-cadmium resistance protein CzcA | 0.46034 | 0.11758 | 0.59077 | 2.19943 | 0.18395 | 65 | 0.36918 | 2.17745 | 0.15301 | 93 | 0.23927 | 0.92612 | 137.435 | 0.35601 | 0.59014 |
| padR; PadR family transcriptional regulator, regulatory protein PadR | 0.45253 | 0.20371 | 0.18462 | 1.55132 | 0.15449 | 65 | 0.63978 | 10.02373 | 0.32830 | 93 | 0.36283 | -1.25448 | 128.217 | 0.21195 | 0.59014 |
| csn1, cas9; CRISPR-associated endonuclease Csn1 [EC:3.1.-.-] | 0.44321 | 0.09509 | 0.32991 | 0.94998 | 0.12089 | 65 | 0.52240 | 1.76175 | 0.13764 | 93 | 0.18319 | -1.05075 | 155.590 | 0.29500 | 0.59014 |
| bgaB, lacA; beta-galactosidase [EC:3.2.1.23] | 0.44143 | 0.09679 | 0.28249 | 0.30667 | 0.06869 | 65 | 0.55252 | 2.28238 | 0.15666 | 93 | 0.17105 | -1.57866 | 124.176 | 0.11696 | 0.59014 |
| agaR; DeoR family transcriptional regulator, aga operon transcriptional repressor | 0.42722 | 0.15725 | 0.26154 | 2.38365 | 0.19150 | 65 | 0.54301 | 4.97639 | 0.23132 | 93 | 0.30030 | -0.93730 | 155.992 | 0.35005 | 0.59014 |
| pepE; dipeptidase E [EC:3.4.13.21] | 0.41845 | 0.10157 | 0.27811 | 0.46453 | 0.08454 | 65 | 0.51654 | 2.43470 | 0.16180 | 93 | 0.18255 | -1.30611 | 134.660 | 0.19374 | 0.59014 |
| rhaM; L-rhamnose mutarotase [EC:5.1.3.32] | 0.41139 | 0.17535 | 0.23077 | 1.67248 | 0.16041 | 65 | 0.53763 | 7.08824 | 0.27608 | 93 | 0.31929 | -0.96108 | 141.431 | 0.33815 | 0.59014 |
| PGAM, gpmA; 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase [EC:5.4.2.11] | 0.39873 | 0.09404 | 0.40769 | 1.08314 | 0.12909 | 65 | 0.39247 | 1.63098 | 0.13243 | 93 | 0.18494 | 0.08229 | 152.271 | 0.93452 | 0.94558 |
| troR; DtxR family transcriptional regulator, Mn-dependent transcriptional regulator | 0.39873 | 0.09404 | 0.40769 | 1.08314 | 0.12909 | 65 | 0.39247 | 1.63098 | 0.13243 | 93 | 0.18494 | 0.08229 | 152.271 | 0.93452 | 0.94558 |
| pnbA; para-nitrobenzyl esterase [EC:3.1.1.-] | 0.39557 | 0.15968 | 0.28462 | 2.45288 | 0.19426 | 65 | 0.47312 | 5.15416 | 0.23542 | 93 | 0.30522 | -0.61760 | 155.983 | 0.53774 | 0.63858 |
| dinD; DNA-damage-inducible protein D | 0.37046 | 0.11444 | 0.32872 | 1.31388 | 0.14217 | 65 | 0.39964 | 2.61542 | 0.16770 | 93 | 0.21986 | -0.32259 | 155.959 | 0.74744 | 0.80237 |
| E2.4.1.20; cellobiose phosphorylase [EC:2.4.1.20] | 0.36772 | 0.24592 | 0.40154 | 10.39890 | 0.39998 | 65 | 0.34409 | 9.07054 | 0.31230 | 93 | 0.50746 | 0.11322 | 131.756 | 0.91003 | 0.94278 |
| xylE; MFS transporter, SP family, xylose:H+ symportor | 0.36772 | 0.24592 | 0.40154 | 10.39890 | 0.39998 | 65 | 0.34409 | 9.07054 | 0.31230 | 93 | 0.50746 | 0.11322 | 131.756 | 0.91003 | 0.94278 |
| dapF; diaminopimelate epimerase [EC:5.1.1.7] | 0.35994 | 0.06710 | 0.28031 | 0.45305 | 0.08349 | 65 | 0.41559 | 0.89130 | 0.09790 | 93 | 0.12866 | -1.05147 | 155.925 | 0.29467 | 0.59014 |
| E2.6.1.83; LL-diaminopimelate aminotransferase [EC:2.6.1.83] | 0.35994 | 0.06710 | 0.28031 | 0.45305 | 0.08349 | 65 | 0.41559 | 0.89130 | 0.09790 | 93 | 0.12866 | -1.05147 | 155.925 | 0.29467 | 0.59014 |
| znuA; zinc transport system substrate-binding protein | 0.35732 | 0.07854 | 0.28615 | 0.45511 | 0.08368 | 65 | 0.40707 | 1.34070 | 0.12007 | 93 | 0.14635 | -0.82619 | 151.646 | 0.40999 | 0.59014 |
| zraR, hydG; two-component system, NtrC family, response regulator HydG | 0.33228 | 0.15465 | 0.23846 | 1.71959 | 0.16265 | 65 | 0.39785 | 5.24217 | 0.23742 | 93 | 0.28779 | -0.55384 | 150.855 | 0.58051 | 0.67493 |
| K06921; uncharacterized protein | 0.32743 | 0.10964 | 0.22154 | 1.32262 | 0.14265 | 65 | 0.40143 | 2.30751 | 0.15752 | 93 | 0.21251 | -0.84653 | 154.959 | 0.39856 | 0.59014 |
| trpA; tryptophan synthase alpha chain [EC:4.2.1.20] | 0.31477 | 0.17337 | 0.29846 | 4.66654 | 0.26794 | 65 | 0.32616 | 4.85744 | 0.22854 | 93 | 0.35217 | -0.07866 | 139.597 | 0.93741 | 0.94558 |
| trpD; anthranilate phosphoribosyltransferase [EC:2.4.2.18] | 0.31477 | 0.17337 | 0.29846 | 4.66654 | 0.26794 | 65 | 0.32616 | 4.85744 | 0.22854 | 93 | 0.35217 | -0.07866 | 139.597 | 0.93741 | 0.94558 |
| trpE; anthranilate synthase component I [EC:4.1.3.27] | 0.31477 | 0.17337 | 0.29846 | 4.66654 | 0.26794 | 65 | 0.32616 | 4.85744 | 0.22854 | 93 | 0.35217 | -0.07866 | 139.597 | 0.93741 | 0.94558 |
| lplA, lplJ; lipoate—protein ligase [EC:6.3.1.20] | 0.30922 | 0.09671 | 0.19121 | 0.96829 | 0.12205 | 65 | 0.39171 | 1.83137 | 0.14033 | 93 | 0.18598 | -1.07805 | 155.732 | 0.28268 | 0.59014 |
| bdhAB; butanol dehydrogenase [EC:1.1.1.-] | 0.30759 | 0.15335 | 0.18615 | 1.55090 | 0.15447 | 65 | 0.39247 | 5.24375 | 0.23745 | 93 | 0.28327 | -0.72834 | 148.191 | 0.46756 | 0.59173 |
| sbcD, mre11; DNA repair protein SbcD/Mre11 | 0.30759 | 0.15335 | 0.18615 | 1.55090 | 0.15447 | 65 | 0.39247 | 5.24375 | 0.23745 | 93 | 0.28327 | -0.72834 | 148.191 | 0.46756 | 0.59173 |
| TC.SULP; sulfate permease, SulP family | 0.30759 | 0.15335 | 0.18615 | 1.55090 | 0.15447 | 65 | 0.39247 | 5.24375 | 0.23745 | 93 | 0.28327 | -0.72834 | 148.191 | 0.46756 | 0.59173 |
| K07014; uncharacterized protein | 0.29430 | 0.13507 | 0.23846 | 2.33678 | 0.18961 | 65 | 0.33333 | 3.28986 | 0.18808 | 93 | 0.26707 | -0.35523 | 150.528 | 0.72291 | 0.78678 |
| dnaC; DNA replication protein DnaC | 0.29114 | 0.27240 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.49462 | 19.90486 | 0.46263 | 93 | 0.46263 | -1.06915 | 92.000 | 0.28780 | 0.59014 |
| K06915; uncharacterized protein | 0.29114 | 0.27240 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.49462 | 19.90486 | 0.46263 | 93 | 0.46263 | -1.06915 | 92.000 | 0.28780 | 0.59014 |
| K07078; uncharacterized protein | 0.29114 | 0.27240 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.49462 | 19.90486 | 0.46263 | 93 | 0.46263 | -1.06915 | 92.000 | 0.28780 | 0.59014 |
| utp; urea transporter | 0.29114 | 0.27240 | 0.00000 | 0.00000 | 0.00000 | 65 | 0.49462 | 19.90486 | 0.46263 | 93 | 0.46263 | -1.06915 | 92.000 | 0.28780 | 0.59014 |
| K07317; adenine-specific DNA-methyltransferase [EC:2.1.1.72] | 0.28797 | 0.18249 | 0.33077 | 4.36935 | 0.25927 | 65 | 0.25806 | 5.93811 | 0.25269 | 93 | 0.36204 | 0.20082 | 149.496 | 0.84111 | 0.88546 |
| fecA; Fe(3+) dicitrate transport protein | 0.28727 | 0.05021 | 0.25385 | 0.24190 | 0.06100 | 65 | 0.31063 | 0.51015 | 0.07406 | 93 | 0.09595 | -0.59182 | 155.976 | 0.55483 | 0.65055 |
| K07090; uncharacterized protein | 0.27255 | 0.05523 | 0.22500 | 0.40415 | 0.07885 | 65 | 0.30578 | 0.53851 | 0.07609 | 93 | 0.10958 | -0.73716 | 148.884 | 0.46218 | 0.59014 |
| bshA; L-malate glycosyltransferase [EC:2.4.1.-] | 0.27215 | 0.16573 | 0.04615 | 0.04471 | 0.02623 | 65 | 0.43011 | 7.31300 | 0.28042 | 93 | 0.28164 | -1.36327 | 93.606 | 0.17607 | 0.59014 |
| csxA; exo-1,4-beta-D-glucosaminidase [EC:3.2.1.165] | 0.27215 | 0.16573 | 0.04615 | 0.04471 | 0.02623 | 65 | 0.43011 | 7.31300 | 0.28042 | 93 | 0.28164 | -1.36327 | 93.606 | 0.17607 | 0.59014 |
| E2.5.1.56, neuB; N-acetylneuraminate synthase [EC:2.5.1.56] | 0.27215 | 0.16573 | 0.04615 | 0.04471 | 0.02623 | 65 | 0.43011 | 7.31300 | 0.28042 | 93 | 0.28164 | -1.36327 | 93.606 | 0.17607 | 0.59014 |
| E3.5.2.10; creatinine amidohydrolase [EC:3.5.2.10] | 0.27215 | 0.16573 | 0.04615 | 0.04471 | 0.02623 | 65 | 0.43011 | 7.31300 | 0.28042 | 93 | 0.28164 | -1.36327 | 93.606 | 0.17607 | 0.59014 |
| FLOT; flotillin | 0.27215 | 0.16573 | 0.04615 | 0.04471 | 0.02623 | 65 | 0.43011 | 7.31300 | 0.28042 | 93 | 0.28164 | -1.36327 | 93.606 | 0.17607 | 0.59014 |
| higB-1; toxin HigB-1 | 0.27215 | 0.16573 | 0.04615 | 0.04471 | 0.02623 | 65 | 0.43011 | 7.31300 | 0.28042 | 93 | 0.28164 | -1.36327 | 93.606 | 0.17607 | 0.59014 |
| K13652; AraC family transcriptional regulator | 0.27215 | 0.16573 | 0.04615 | 0.04471 | 0.02623 | 65 | 0.43011 | 7.31300 | 0.28042 | 93 | 0.28164 | -1.36327 | 93.606 | 0.17607 | 0.59014 |
| neuA; N-acylneuraminate cytidylyltransferase [EC:2.7.7.43] | 0.27215 | 0.16573 | 0.04615 | 0.04471 | 0.02623 | 65 | 0.43011 | 7.31300 | 0.28042 | 93 | 0.28164 | -1.36327 | 93.606 | 0.17607 | 0.59014 |
| RTCB, rtcB; tRNA-splicing ligase RtcB (3’-phosphate/5’-hydroxy nucleic acid ligase) [EC:6.5.1.8] | 0.27215 | 0.16573 | 0.04615 | 0.04471 | 0.02623 | 65 | 0.43011 | 7.31300 | 0.28042 | 93 | 0.28164 | -1.36327 | 93.606 | 0.17607 | 0.59014 |
| TC.CPA1; monovalent cation:H+ antiporter, CPA1 family | 0.27215 | 0.16573 | 0.04615 | 0.04471 | 0.02623 | 65 | 0.43011 | 7.31300 | 0.28042 | 93 | 0.28164 | -1.36327 | 93.606 | 0.17607 | 0.59014 |
| INV, sacA; beta-fructofuranosidase [EC:3.2.1.26] | 0.26846 | 0.05911 | 0.19487 | 0.46751 | 0.08481 | 65 | 0.31989 | 0.61040 | 0.08102 | 93 | 0.11729 | -1.06595 | 148.231 | 0.28818 | 0.59014 |
| dexA; dextranase [EC:3.2.1.11] | 0.26772 | 0.06391 | 0.22462 | 0.67001 | 0.10153 | 65 | 0.29785 | 0.63304 | 0.08250 | 93 | 0.13082 | -0.55979 | 135.370 | 0.57654 | 0.67167 |
| tatA; sec-independent protein translocase protein TatA | 0.26582 | 0.10644 | 0.18974 | 1.09017 | 0.12951 | 65 | 0.31900 | 2.28965 | 0.15691 | 93 | 0.20345 | -0.63531 | 155.984 | 0.52616 | 0.62749 |
| tatC; sec-independent protein translocase protein TatC | 0.26582 | 0.10644 | 0.18974 | 1.09017 | 0.12951 | 65 | 0.31900 | 2.28965 | 0.15691 | 93 | 0.20345 | -0.63531 | 155.984 | 0.52616 | 0.62749 |
| ltaE; threonine aldolase [EC:4.1.2.48] | 0.26498 | 0.09697 | 0.23897 | 1.40165 | 0.14685 | 65 | 0.28315 | 1.55965 | 0.12950 | 93 | 0.19579 | -0.22565 | 142.358 | 0.82180 | 0.86592 |
| ABC.X2.A; putative ABC transport system ATP-binding protein | 0.24051 | 0.11969 | 0.06923 | 0.14748 | 0.04763 | 65 | 0.36022 | 3.72481 | 0.20013 | 93 | 0.20572 | -1.41447 | 102.247 | 0.16026 | 0.59014 |
| argB; acetylglutamate kinase [EC:2.7.2.8] | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94278 |
| argC; N-acetyl-gamma-glutamyl-phosphate reductase [EC:1.2.1.38] | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94278 |
| argE; acetylornithine deacetylase [EC:3.5.1.16] | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94278 |
| argF; N-succinyl-L-ornithine transcarbamylase [EC:2.1.3.11] | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94278 |
| argG, ASS1; argininosuccinate synthase [EC:6.3.4.5] | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94278 |
| argH, ASL; argininosuccinate lyase [EC:4.3.2.1] | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94278 |
| cimA; (R)-citramalate synthase [EC:2.3.1.182] | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94278 |
| E2.6.1.11, argD; acetylornithine aminotransferase [EC:2.6.1.11] | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94278 |
| hisA; phosphoribosylformimino-5-aminoimidazole carboxamide ribotide isomerase [EC:5.3.1.16] | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94278 |
| hisB; imidazoleglycerol-phosphate dehydratase / histidinol-phosphatase [EC:4.2.1.19 3.1.3.15] | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94278 |
| hisD; histidinol dehydrogenase [EC:1.1.1.23] | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94278 |
| hisF; cyclase [EC:4.1.3.-] | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94278 |
| hisG; ATP phosphoribosyltransferase [EC:2.4.2.17] | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94278 |
| hisH; glutamine amidotransferase [EC:2.4.2.-] | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94278 |
| hisIE; phosphoribosyl-ATP pyrophosphohydrolase / phosphoribosyl-AMP cyclohydrolase [EC:3.6.1.31 3.5.4.19] | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94278 |
| leuA, IMS; 2-isopropylmalate synthase [EC:2.3.3.13] | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94278 |
| leuB, IMDH; 3-isopropylmalate dehydrogenase [EC:1.1.1.85] | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94278 |
| leuC, IPMI-L; 3-isopropylmalate/(R)-2-methylmalate dehydratase large subunit [EC:4.2.1.33 4.2.1.35] | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94278 |
| leuD, IPMI-S; 3-isopropylmalate/(R)-2-methylmalate dehydratase small subunit [EC:4.2.1.33 4.2.1.35] | 0.23766 | 0.13002 | 0.22385 | 2.62493 | 0.20096 | 65 | 0.24731 | 2.73165 | 0.17138 | 93 | 0.26411 | -0.08885 | 139.587 | 0.92933 | 0.94278 |
| GLO1, gloA; lactoylglutathione lyase [EC:4.4.1.5] | 0.23259 | 0.08954 | 0.11154 | 0.39069 | 0.07753 | 65 | 0.31720 | 1.87247 | 0.14189 | 93 | 0.16169 | -1.27195 | 137.511 | 0.20554 | 0.59014 |
| cvrA, nhaP2; cell volume regulation protein A | 0.20886 | 0.17482 | 0.09231 | 0.55385 | 0.09231 | 65 | 0.29032 | 7.83871 | 0.29032 | 93 | 0.30464 | -0.64999 | 109.926 | 0.51706 | 0.62047 |
| proC; pyrroline-5-carboxylate reductase [EC:1.5.1.2] | 0.19968 | 0.12364 | 0.21615 | 2.60298 | 0.20011 | 65 | 0.18817 | 2.31067 | 0.15763 | 93 | 0.25474 | 0.10985 | 132.556 | 0.91270 | 0.94278 |
| umuD; DNA polymerase V [EC:3.4.21.-] | 0.18819 | 0.10575 | 0.01641 | 0.00500 | 0.00877 | 65 | 0.30824 | 2.97642 | 0.17890 | 93 | 0.17911 | -1.62933 | 92.442 | 0.10665 | 0.59014 |
| hipO; hippurate hydrolase [EC:3.5.1.32] | 0.17089 | 0.13785 | 0.05385 | 0.18846 | 0.05385 | 65 | 0.25269 | 4.97622 | 0.23132 | 93 | 0.23750 | -0.83722 | 101.811 | 0.40443 | 0.59014 |
| K07496; putative transposase | 0.15190 | 0.13414 | 0.32308 | 6.78462 | 0.32308 | 65 | 0.03226 | 0.09677 | 0.03226 | 93 | 0.32468 | 0.89570 | 65.278 | 0.37370 | 0.59014 |
| res; type III restriction enzyme [EC:3.1.21.5] | 0.14620 | 0.13620 | 0.00154 | 0.00015 | 0.00154 | 65 | 0.24731 | 4.97622 | 0.23132 | 93 | 0.23132 | -1.06247 | 92.008 | 0.29080 | 0.59014 |
| ABC.PA.A; polar amino acid transport system ATP-binding protein [EC:3.6.3.21] | 0.13671 | 0.08286 | 0.02462 | 0.01126 | 0.01316 | 65 | 0.21505 | 1.82825 | 0.14021 | 93 | 0.14083 | -1.35230 | 93.618 | 0.17954 | 0.59014 |
| ACO, acnA; aconitate hydratase [EC:4.2.1.3] | 0.06677 | 0.04503 | 0.05077 | 0.08043 | 0.03518 | 65 | 0.07796 | 0.49046 | 0.07262 | 93 | 0.08069 | -0.33693 | 129.952 | 0.73671 | 0.79501 |
| IDH1, IDH2, icd; isocitrate dehydrogenase [EC:1.1.1.42] | 0.06677 | 0.04503 | 0.05077 | 0.08043 | 0.03518 | 65 | 0.07796 | 0.49046 | 0.07262 | 93 | 0.08069 | -0.33693 | 129.952 | 0.73671 | 0.79501 |
| citD; citrate lyase subunit gamma (acyl carrier protein) | 0.05063 | 0.04471 | 0.10769 | 0.75385 | 0.10769 | 65 | 0.01075 | 0.01075 | 0.01075 | 93 | 0.10823 | 0.89570 | 65.278 | 0.37370 | 0.59014 |
| citE; citrate lyase subunit beta / citryl-CoA lyase [EC:4.1.3.34] | 0.05063 | 0.04471 | 0.10769 | 0.75385 | 0.10769 | 65 | 0.01075 | 0.01075 | 0.01075 | 93 | 0.10823 | 0.89570 | 65.278 | 0.37370 | 0.59014 |
| citF; citrate lyase subunit alpha / citrate CoA-transferase [EC:2.8.3.10] | 0.05063 | 0.04471 | 0.10769 | 0.75385 | 0.10769 | 65 | 0.01075 | 0.01075 | 0.01075 | 93 | 0.10823 | 0.89570 | 65.278 | 0.37370 | 0.59014 |
| citX; holo-ACP synthase [EC:2.7.7.61] | 0.05063 | 0.04471 | 0.10769 | 0.75385 | 0.10769 | 65 | 0.01075 | 0.01075 | 0.01075 | 93 | 0.10823 | 0.89570 | 65.278 | 0.37370 | 0.59014 |
| dndC; DNA sulfur modification protein DndC | 0.05063 | 0.04471 | 0.10769 | 0.75385 | 0.10769 | 65 | 0.01075 | 0.01075 | 0.01075 | 93 | 0.10823 | 0.89570 | 65.278 | 0.37370 | 0.59014 |
| dndE; DNA sulfur modification protein DndE | 0.05063 | 0.04471 | 0.10769 | 0.75385 | 0.10769 | 65 | 0.01075 | 0.01075 | 0.01075 | 93 | 0.10823 | 0.89570 | 65.278 | 0.37370 | 0.59014 |
| elaA; ElaA protein | 0.05063 | 0.04471 | 0.10769 | 0.75385 | 0.10769 | 65 | 0.01075 | 0.01075 | 0.01075 | 93 | 0.10823 | 0.89570 | 65.278 | 0.37370 | 0.59014 |
| K02481; two-component system, NtrC family, response regulator | 0.05063 | 0.04471 | 0.10769 | 0.75385 | 0.10769 | 65 | 0.01075 | 0.01075 | 0.01075 | 93 | 0.10823 | 0.89570 | 65.278 | 0.37370 | 0.59014 |
| K07220; uncharacterized protein | 0.05063 | 0.04471 | 0.10769 | 0.75385 | 0.10769 | 65 | 0.01075 | 0.01075 | 0.01075 | 93 | 0.10823 | 0.89570 | 65.278 | 0.37370 | 0.59014 |
| K07491; putative transposase | 0.05063 | 0.04471 | 0.10769 | 0.75385 | 0.10769 | 65 | 0.01075 | 0.01075 | 0.01075 | 93 | 0.10823 | 0.89570 | 65.278 | 0.37370 | 0.59014 |
| kdpA; K+-transporting ATPase ATPase A chain [EC:3.6.3.12] | 0.05063 | 0.04471 | 0.10769 | 0.75385 | 0.10769 | 65 | 0.01075 | 0.01075 | 0.01075 | 93 | 0.10823 | 0.89570 | 65.278 | 0.37370 | 0.59014 |
| kdpB; K+-transporting ATPase ATPase B chain [EC:3.6.3.12] | 0.05063 | 0.04471 | 0.10769 | 0.75385 | 0.10769 | 65 | 0.01075 | 0.01075 | 0.01075 | 93 | 0.10823 | 0.89570 | 65.278 | 0.37370 | 0.59014 |
| kdpC; K+-transporting ATPase ATPase C chain [EC:3.6.3.12] | 0.05063 | 0.04471 | 0.10769 | 0.75385 | 0.10769 | 65 | 0.01075 | 0.01075 | 0.01075 | 93 | 0.10823 | 0.89570 | 65.278 | 0.37370 | 0.59014 |
| kdpD; two-component system, OmpR family, sensor histidine kinase KdpD [EC:2.7.13.3] | 0.05063 | 0.04471 | 0.10769 | 0.75385 | 0.10769 | 65 | 0.01075 | 0.01075 | 0.01075 | 93 | 0.10823 | 0.89570 | 65.278 | 0.37370 | 0.59014 |
| ltrA; RNA-directed DNA polymerase [EC:2.7.7.49] | 0.05063 | 0.04471 | 0.10769 | 0.75385 | 0.10769 | 65 | 0.01075 | 0.01075 | 0.01075 | 93 | 0.10823 | 0.89570 | 65.278 | 0.37370 | 0.59014 |
| norB, norC; MFS transporter, DHA2 family, multidrug resistance protein | 0.05063 | 0.04471 | 0.10769 | 0.75385 | 0.10769 | 65 | 0.01075 | 0.01075 | 0.01075 | 93 | 0.10823 | 0.89570 | 65.278 | 0.37370 | 0.59014 |
| raxB, cvaB; ATP-binding cassette, subfamily B, bacterial RaxB | 0.05063 | 0.04471 | 0.10769 | 0.75385 | 0.10769 | 65 | 0.01075 | 0.01075 | 0.01075 | 93 | 0.10823 | 0.89570 | 65.278 | 0.37370 | 0.59014 |
| ribB, RIB3; 3,4-dihydroxy 2-butanone 4-phosphate synthase [EC:4.1.99.12] | 0.05063 | 0.04471 | 0.10769 | 0.75385 | 0.10769 | 65 | 0.01075 | 0.01075 | 0.01075 | 93 | 0.10823 | 0.89570 | 65.278 | 0.37370 | 0.59014 |
| TC.PIT; inorganic phosphate transporter, PiT family | 0.05063 | 0.04471 | 0.10769 | 0.75385 | 0.10769 | 65 | 0.01075 | 0.01075 | 0.01075 | 93 | 0.10823 | 0.89570 | 65.278 | 0.37370 | 0.59014 |
| wcaK, amsJ; colanic acid/amylovoran biosynthesis protein | 0.05063 | 0.04471 | 0.10769 | 0.75385 | 0.10769 | 65 | 0.01075 | 0.01075 | 0.01075 | 93 | 0.10823 | 0.89570 | 65.278 | 0.37370 | 0.59014 |
| dndD; DNA sulfur modification protein DndD | 0.02595 | 0.02236 | 0.05538 | 0.18845 | 0.05384 | 65 | 0.00538 | 0.00269 | 0.00538 | 93 | 0.05411 | 0.92417 | 65.278 | 0.35880 | 0.59014 |
| amt, AMT, MEP; ammonium transporter, Amt family | 0.00253 | 0.00253 | 0.00615 | 0.00246 | 0.00615 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00615 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| ABC.PA.S; polar amino acid transport system substrate-binding protein | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| asnB, ASNS; asparagine synthase (glutamine-hydrolysing) [EC:6.3.5.4] | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| CS, gltA; citrate synthase [EC:2.3.3.1] | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| cysH; phosphoadenosine phosphosulfate reductase [EC:1.8.4.8 1.8.4.10] | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| czcB; membrane fusion protein, cobalt-zinc-cadmium efflux system | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| DNPEP; aspartyl aminopeptidase [EC:3.4.11.21] | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| E3.2.1.4; endoglucanase [EC:3.2.1.4] | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| E3.2.1.8, xynA; endo-1,4-beta-xylanase [EC:3.2.1.8] | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| emrA; membrane fusion protein, multidrug efflux system | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| glnB; nitrogen regulatory protein P-II 1 | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| gltB; glutamate synthase (NADPH/NADH) large chain [EC:1.4.1.13 1.4.1.14] | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| gmhA, lpcA; D-sedoheptulose 7-phosphate isomerase [EC:5.3.1.28] | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| gmhB; D-glycero-D-manno-heptose 1,7-bisphosphate phosphatase [EC:3.1.3.82 3.1.3.83] | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| hddA; D-glycero-alpha-D-manno-heptose-7-phosphate kinase [EC:2.7.1.168] | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| K07166; ACT domain-containing protein | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| K07484; transposase | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| K09157; uncharacterized protein | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| K09922; uncharacterized protein | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| modF; molybdate transport system ATP-binding protein | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| nhaC; Na+:H+ antiporter, NhaC family | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| pip; proline iminopeptidase [EC:3.4.11.5] | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| proA; glutamate-5-semialdehyde dehydrogenase [EC:1.2.1.41] | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| proB; glutamate 5-kinase [EC:2.7.2.11] | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| rnfA; electron transport complex protein RnfA | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| rnfC; electron transport complex protein RnfC | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| rnfD; electron transport complex protein RnfD | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| rnfE; electron transport complex protein RnfE | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| rseC; sigma-E factor negative regulatory protein RseC | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| thiG; thiazole synthase [EC:2.8.1.10] | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| thiH; 2-iminoacetate synthase [EC:4.1.99.19] | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| thiS; sulfur carrier protein | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
| yaeR; glyoxylase I family protein | 0.00127 | 0.00127 | 0.00308 | 0.00062 | 0.00308 | 65 | 0.00000 | 0.00000 | 0.00000 | 93 | 0.00308 | 1.00000 | 64.000 | 0.32108 | 0.59014 |
plot.dat <- tb.ra %>%
arrange(p) %>%
slice_head(n=50)%>%
mutate(
description = fct_reorder(description, `Overall Mean`),
description = factor(description, levels = levels(description), ordered=T)
) %>%
arrange(description) %>%
mutate(
id = 1:n(),
step = ifelse(id%%2 == 0, 1, 0),
Mean_diff = `Tumor Mean` - `Non-Tumor Mean`,
diff_se = SEpooled,
Mean_diff_ll = Mean_diff - qt(0.975, df)*diff_se,
Mean_diff_ul = Mean_diff + qt(0.975, df)*diff_se
) %>%
pivot_longer(
cols=contains("Mean"),
names_to = "group",
values_to = "mean"
)
p1.d <- plot.dat %>%
filter(group %in% c("Tumor Mean","Non-Tumor Mean")) %>%
mutate(
group = ifelse(group == "Tumor Mean", "Tumor", "Non-Tumor"),
col = ifelse(step == 1, "grey90", "white"),
h=1, w=Inf
)
p1 <- ggplot()+
geom_tile(data = p1.d,
aes(y = description, x=0,
height=h, width=w),
fill = p1.d$col, color=p1.d$col)+
geom_bar(data=p1.d,
aes(x=mean, y=description,
group=group, color=group,
fill=group),
stat="identity",position = "dodge",
alpha = 1)+
labs(x="Mean Abundance")+
theme_classic()+
theme(
legend.position = "bottom",
plot.margin = unit(c(1,0,1,1), "lines")
)
p2.d <- plot.dat %>%
filter(group %in% c("Mean_diff", "Mean_diff_ll", "Mean_diff_ul")) %>%
pivot_wider(
names_from = group,
values_from = mean
) %>%
mutate(
group = ifelse(Mean_diff > 0, "Tumor", "Non-Tumor"),
p = sprintf("%.3f", round(p,3)),
ll = min(Mean_diff_ll)-0.01,
ul = max(Mean_diff_ul)+0.01
)
p2<-ggplot(p2.d, aes(x=Mean_diff, y=description))+
geom_tile(data = p1.d,
aes(y = description, x=0,
height=h, width=w),
fill = p1.d$col, color=p1.d$col)+
geom_vline(xintercept = 0, linetype="dashed", alpha=0.5)+
geom_segment(aes(x=Mean_diff_ll, y=description, xend=Mean_diff_ul, yend=description))+
geom_point(aes(fill=group, color=group))+
geom_text(aes(label=p, x=unique(ul)+0.1))+
coord_cartesian(xlim = c(unique(p2.d$ll), unique(p2.d$ul)),
clip = 'off') +
annotate("text", x=unique(p2.d$ul)+0.2,y = 25,
angle=90,
label="p-value (uncorrected)")+
labs(x="Mean Difference in Abundance")+
theme_classic()+
theme(
legend.position = "bottom",
axis.title.y = element_blank(),
axis.text.y = element_blank(),
axis.line.y = element_blank(),
axis.ticks.y = element_blank(),
plot.margin = unit(c(1,4,1,0), "lines")
)
# plot
p1 + p2 +
plot_annotation(title="Stratefied KO Data - Prevotella spp. (genus level only): First 50 descriptions with lowest p-value (uncorrected)")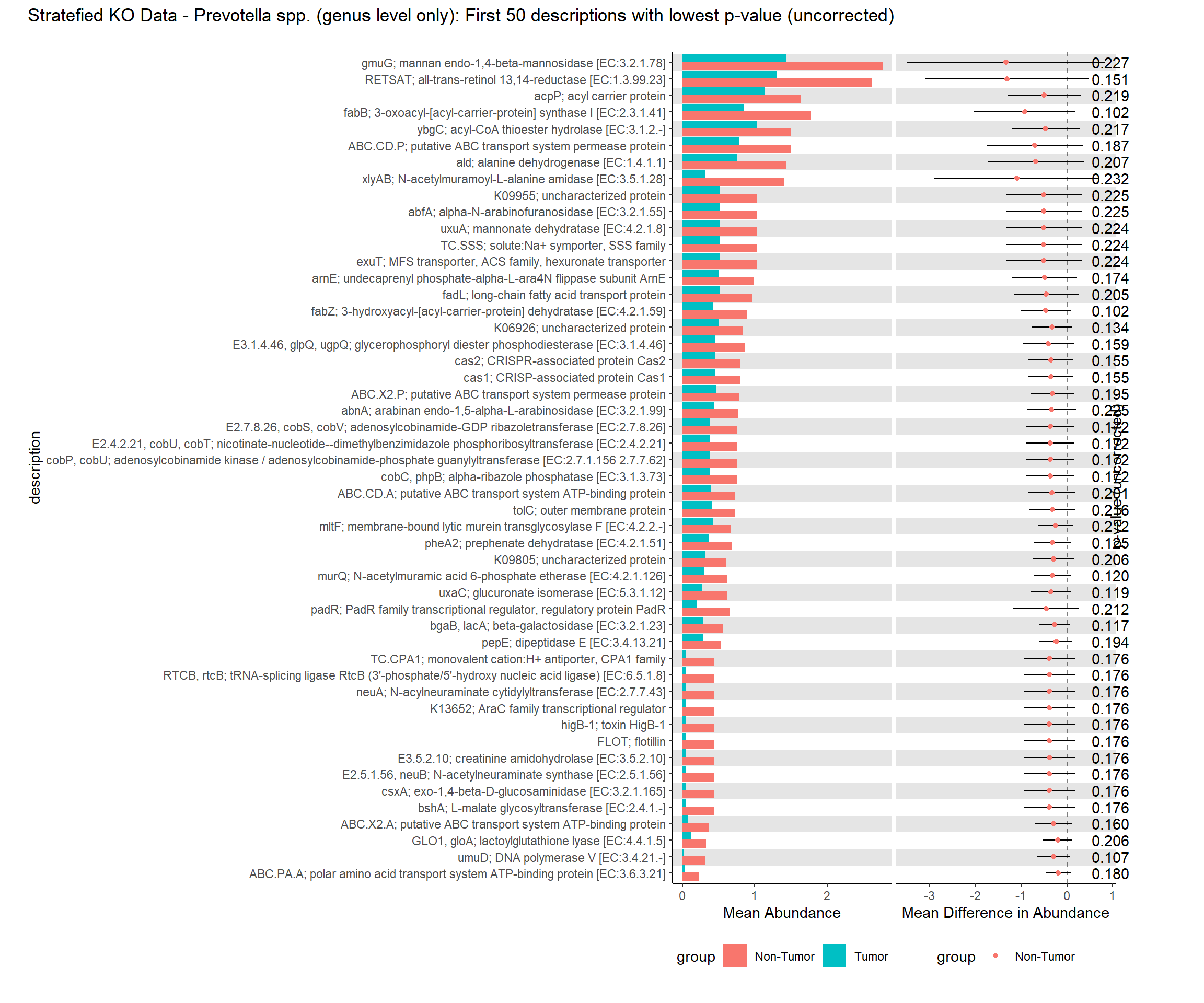
Modeling Difference between Tumor and Non-Tumor
For the modeling, we used a generalized linear mixed model (GLMM).
First, we looked at the biserial correlation between the abundance of each description and the tumor status.
tb <- mydata %>%
group_by(description)%>%
summarise(
r = cor(tumor, Abundance)
) %>%
mutate(
M=mean(r)
)
ggplot(tb, aes(x=r))+
geom_density()+
geom_vline(aes(xintercept = M))+
labs(x="Biserial Correlation",title="Relationship between description abundance and tumor (tumor vs. non-tumor)")+
theme(panel.grid = element_blank())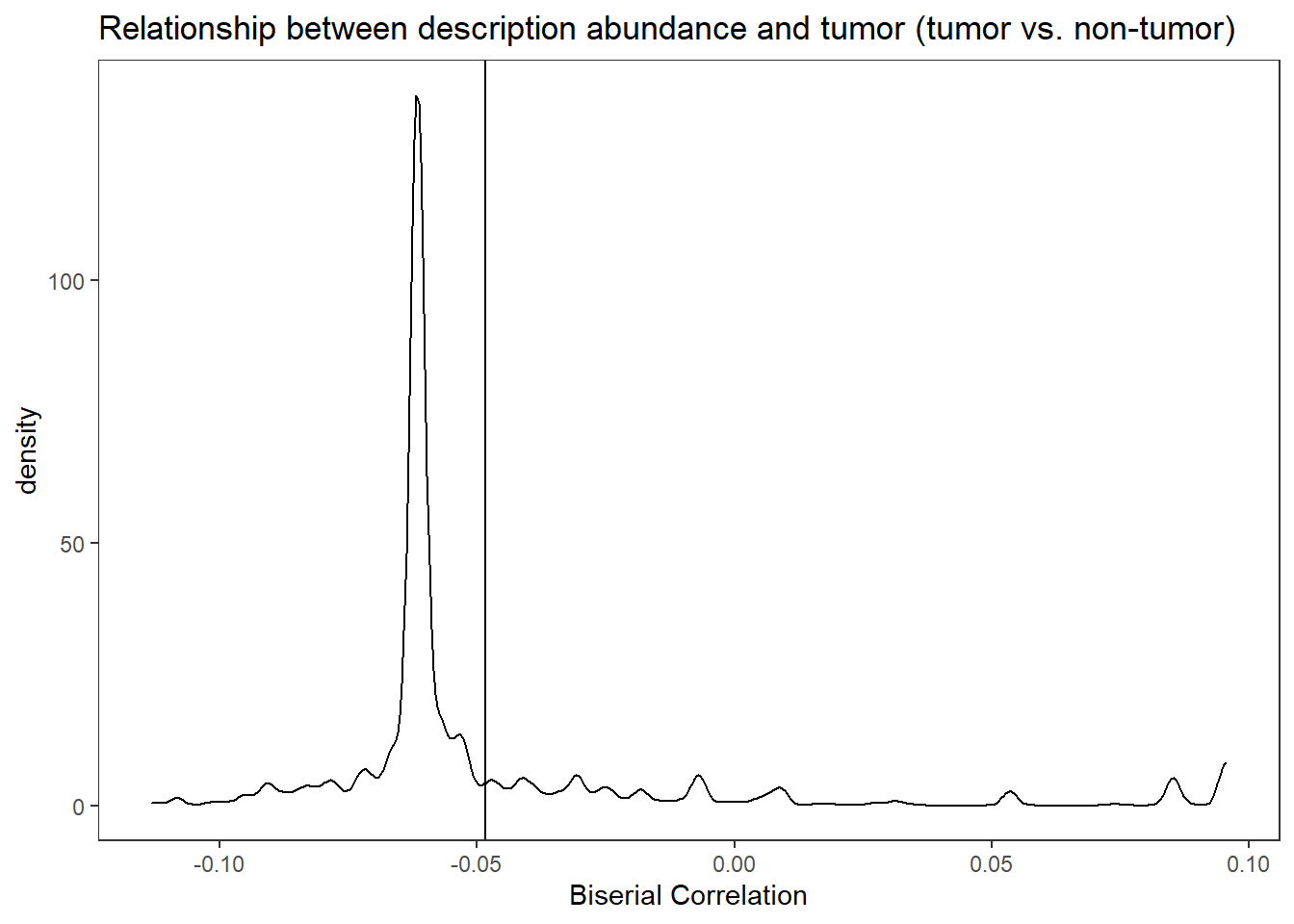
Next, we need the data to be on an interpretable scale. First, let’s use the raw abundance like scale.
p <- ggplot(mydata, aes(x=Abundance))+
geom_density()
p
mydata <- mydata %>%
mutate(Abundance.dich=ifelse(Abundance==0, 0, 1))
table(mydata$Abundance.dich)
0 1
80642 102322 Let’s first run models by description so that we can avoid the nesting issue initially. We will come back to this to conduct the final model (it will be more powerful).
DESCRIPTIONS <- unique(mydata$description)
i<-1
dat0 <- mydata %>% filter(description==DESCRIPTIONS[i])
# quasipossion (approximately negative binom) give an approx. answer.
fit0 <- glm(
Abundance ~ 1 + tumor,
data= dat0,
family=quasipoisson(link = "log")
)
summary(fit0)
Call:
glm(formula = Abundance ~ 1 + tumor, family = quasipoisson(link = "log"),
data = dat0)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.392 -1.241 -0.906 0.232 5.513
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.0311 0.1770 -0.18 0.86
tumor -0.2305 0.2963 -0.78 0.44
(Dispersion parameter for quasipoisson family taken to be 2.8254)
Null deviance: 293.35 on 157 degrees of freedom
Residual deviance: 291.61 on 156 degrees of freedom
AIC: NA
Number of Fisher Scoring iterations: 6NExt, let’s model the percent relative abundance. This will allow us to make inference about the difference average relative abundance. Which is a simpler interpretation than trying to model differences in the log relative abundance which will need to be interpreted as the multiplicative relative change.
results.out <- as.data.frame(matrix(ncol=4, nrow=length(DESCRIPTIONS)))
colnames(results.out) <- c("description", "Est", "SE", "p")
#results.out$description <-DESCRIPTIONS
i <- 1
for(i in 1:length(DESCRIPTIONS)){
#for(i in 1:5){
dat0 <- mydata %>%
filter(description == DESCRIPTIONS[i])
fit0 <- glm(
Abundance ~ 1 + tumor,
data= dat0,
family=quasipoisson(link = "log")
)
fit.sum <- summary(fit0)
results.out[i, 1] <- DESCRIPTIONS[i]
results.out[i, 2:4] <- fit.sum$coefficients[2, c(1,2,4)]
}
results.out$fdr_p <- p.adjust(results.out$p, method="fdr")
kable(results.out, format="html", digits=3) %>%
kable_styling(full_width = T)%>%
scroll_box(width="100%", height="600px")| description | Est | SE | p | fdr_p |
|---|---|---|---|---|
| AARS, alaS; alanyl-tRNA synthetase [EC:6.1.1.7] | -0.230 | 0.296 | 0.438 | 0.651 |
| aat; leucyl/phenylalanyl-tRNA—protein transferase [EC:2.3.2.6] | 0.056 | 0.493 | 0.910 | 0.976 |
| ABC-2.A; ABC-2 type transport system ATP-binding protein | -0.287 | 0.311 | 0.357 | 0.651 |
| ABC-2.OM, wza; polysaccharide biosynthesis/export protein | -0.285 | 0.310 | 0.360 | 0.651 |
| ABC-2.P; ABC-2 type transport system permease protein | -0.263 | 0.312 | 0.401 | 0.651 |
| ABC-2.TX; HlyD family secretion protein | -0.249 | 0.304 | 0.415 | 0.651 |
| ABC.CD.A; putative ABC transport system ATP-binding protein | -0.616 | 0.516 | 0.234 | 0.651 |
| ABC.CD.P; putative ABC transport system permease protein | -0.642 | 0.523 | 0.221 | 0.651 |
| ABC.CD.TX; HlyD family secretion protein | -0.566 | 0.561 | 0.314 | 0.651 |
| ABC.FEV.A; iron complex transport system ATP-binding protein [EC:3.6.3.34] | -0.230 | 0.296 | 0.438 | 0.651 |
| ABC.FEV.P; iron complex transport system permease protein | -0.230 | 0.296 | 0.438 | 0.651 |
| ABC.FEV.S; iron complex transport system substrate-binding protein | -0.230 | 0.296 | 0.438 | 0.651 |
| ABC.PA.A; polar amino acid transport system ATP-binding protein [EC:3.6.3.21] | -2.168 | 1.874 | 0.249 | 0.651 |
| ABC.PA.S; polar amino acid transport system substrate-binding protein | 18.519 | 3411.091 | 0.996 | 0.998 |
| ABC.SN.S; NitT/TauT family transport system substrate-binding protein | -0.230 | 0.296 | 0.438 | 0.651 |
| ABC.X2.A; putative ABC transport system ATP-binding protein | -1.649 | 1.326 | 0.215 | 0.651 |
| ABC.X2.P; putative ABC transport system permease protein | -0.521 | 0.433 | 0.230 | 0.651 |
| ABCB-BAC; ATP-binding cassette, subfamily B, bacterial | -0.288 | 0.314 | 0.360 | 0.651 |
| ABCF3; ATP-binding cassette, subfamily F, member 3 | -0.230 | 0.296 | 0.438 | 0.651 |
| abfA; alpha-N-arabinofuranosidase [EC:3.2.1.55] | -0.691 | 0.615 | 0.263 | 0.651 |
| abgT; aminobenzoyl-glutamate transport protein | -0.123 | 0.313 | 0.694 | 0.797 |
| abnA; arabinan endo-1,5-alpha-L-arabinosidase [EC:3.2.1.99] | -0.573 | 0.507 | 0.260 | 0.651 |
| ackA; acetate kinase [EC:2.7.2.1] | -0.230 | 0.296 | 0.438 | 0.651 |
| acm; lysozyme | -0.207 | 0.296 | 0.486 | 0.651 |
| ACO, acnA; aconitate hydratase [EC:4.2.1.3] | -0.429 | 1.387 | 0.758 | 0.843 |
| acpP; acyl carrier protein | -0.367 | 0.318 | 0.251 | 0.651 |
| acrA, mexA, adeI, smeD, mtrC, cmeA; membrane fusion protein, multidrug efflux system | -0.216 | 0.278 | 0.438 | 0.651 |
| ACSL, fadD; long-chain acyl-CoA synthetase [EC:6.2.1.3] | -0.247 | 0.302 | 0.414 | 0.651 |
| ACSS, acs; acetyl-CoA synthetase [EC:6.2.1.1] | -0.203 | 0.301 | 0.500 | 0.651 |
| ada; AraC family transcriptional regulator, regulatory protein of adaptative response / methylated-DNA-[protein]-cysteine methyltransferase [EC:2.1.1.63] | -0.626 | 0.575 | 0.278 | 0.651 |
| adk, AK; adenylate kinase [EC:2.7.4.3] | -0.230 | 0.296 | 0.438 | 0.651 |
| agaR; DeoR family transcriptional regulator, aga operon transcriptional repressor | -0.731 | 0.848 | 0.390 | 0.651 |
| agp; glucose-1-phosphatase [EC:3.1.3.10] | -0.072 | 0.305 | 0.815 | 0.896 |
| aguA; alpha-glucuronidase [EC:3.2.1.139] | -0.788 | 0.881 | 0.373 | 0.651 |
| aguB; N-carbamoylputrescine amidase [EC:3.5.1.53] | -0.230 | 0.296 | 0.438 | 0.651 |
| ahpF; alkyl hydroperoxide reductase subunit F [EC:1.6.4.-] | -0.120 | 0.315 | 0.704 | 0.799 |
| AIBP, nnrE; NAD(P)H-hydrate epimerase [EC:5.1.99.6] | -0.230 | 0.296 | 0.438 | 0.651 |
| ald; alanine dehydrogenase [EC:1.4.1.1] | -0.647 | 0.554 | 0.244 | 0.651 |
| algI; alginate O-acetyltransferase complex protein AlgI | -0.221 | 0.572 | 0.699 | 0.797 |
| alr; alanine racemase [EC:5.1.1.1] | -0.230 | 0.296 | 0.438 | 0.651 |
| amiABC; N-acetylmuramoyl-L-alanine amidase [EC:3.5.1.28] | -0.230 | 0.296 | 0.438 | 0.651 |
| amn; AMP nucleosidase [EC:3.2.2.4] | -0.754 | 1.120 | 0.502 | 0.651 |
| ampG; MFS transporter, PAT family, beta-lactamase induction signal transducer AmpG | -0.211 | 0.297 | 0.479 | 0.651 |
| amt, AMT, MEP; ammonium transporter, Amt family | 18.212 | 2925.911 | 0.995 | 0.998 |
| AMY, amyA, malS; alpha-amylase [EC:3.2.1.1] | -0.270 | 0.303 | 0.374 | 0.651 |
| apbE; FAD:protein FMN transferase [EC:2.7.1.180] | -0.230 | 0.296 | 0.438 | 0.651 |
| APEX1; AP endonuclease 1 [EC:4.2.99.18] | 0.141 | 0.367 | 0.701 | 0.797 |
| apgM; 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [EC:5.4.2.12] | -0.606 | 0.561 | 0.282 | 0.651 |
| APRT, apt; adenine phosphoribosyltransferase [EC:2.4.2.7] | -0.230 | 0.296 | 0.438 | 0.651 |
| aqpZ; aquaporin Z | 0.145 | 0.349 | 0.679 | 0.789 |
| araA; L-arabinose isomerase [EC:5.3.1.4] | -0.670 | 0.642 | 0.298 | 0.651 |
| araD, ulaF, sgaE, sgbE; L-ribulose-5-phosphate 4-epimerase [EC:5.1.3.4] | -0.670 | 0.642 | 0.298 | 0.651 |
| argB; acetylglutamate kinase [EC:2.7.2.8] | -0.100 | 1.127 | 0.930 | 0.976 |
| argC; N-acetyl-gamma-glutamyl-phosphate reductase [EC:1.2.1.38] | -0.100 | 1.127 | 0.930 | 0.976 |
| argE; acetylornithine deacetylase [EC:3.5.1.16] | -0.100 | 1.127 | 0.930 | 0.976 |
| argF; N-succinyl-L-ornithine transcarbamylase [EC:2.1.3.11] | -0.100 | 1.127 | 0.930 | 0.976 |
| argG, ASS1; argininosuccinate synthase [EC:6.3.4.5] | -0.100 | 1.127 | 0.930 | 0.976 |
| argH, ASL; argininosuccinate lyase [EC:4.3.2.1] | -0.100 | 1.127 | 0.930 | 0.976 |
| argR, ahrC; transcriptional regulator of arginine metabolism | -0.230 | 0.296 | 0.438 | 0.651 |
| arnE; undecaprenyl phosphate-alpha-L-ara4N flippase subunit ArnE | -0.685 | 0.544 | 0.210 | 0.651 |
| aroA; 3-phosphoshikimate 1-carboxyvinyltransferase [EC:2.5.1.19] | -0.230 | 0.296 | 0.438 | 0.651 |
| AROA1, aroA; chorismate mutase [EC:5.4.99.5] | -0.230 | 0.296 | 0.438 | 0.651 |
| aroB; 3-dehydroquinate synthase [EC:4.2.3.4] | -0.230 | 0.296 | 0.438 | 0.651 |
| aroC; chorismate synthase [EC:4.2.3.5] | -0.230 | 0.296 | 0.438 | 0.651 |
| aroE; shikimate dehydrogenase [EC:1.1.1.25] | -0.230 | 0.296 | 0.438 | 0.651 |
| aroQ, qutE; 3-dehydroquinate dehydratase II [EC:4.2.1.10] | -0.230 | 0.296 | 0.438 | 0.651 |
| asd; aspartate-semialdehyde dehydrogenase [EC:1.2.1.11] | -0.230 | 0.296 | 0.438 | 0.651 |
| asnA; aspartate–ammonia ligase [EC:6.3.1.1] | -0.213 | 0.293 | 0.468 | 0.651 |
| asnB, ASNS; asparagine synthase (glutamine-hydrolysing) [EC:6.3.5.4] | 18.519 | 3411.091 | 0.996 | 0.998 |
| asnC; Lrp/AsnC family transcriptional regulator, regulator for asnA, asnC and gidA | -0.241 | 0.306 | 0.433 | 0.651 |
| aspA; aspartate ammonia-lyase [EC:4.3.1.1] | -0.259 | 0.322 | 0.423 | 0.651 |
| aspB; aspartate aminotransferase [EC:2.6.1.1] | -0.235 | 0.289 | 0.416 | 0.651 |
| aspS; aspartyl-tRNA synthetase [EC:6.1.1.12] | -0.230 | 0.296 | 0.438 | 0.651 |
| ATOX1, ATX1, copZ, golB; copper chaperone | -0.242 | 0.322 | 0.454 | 0.651 |
| ATPF0A, atpB; F-type H+-transporting ATPase subunit a | -0.230 | 0.296 | 0.438 | 0.651 |
| ATPF0B, atpF; F-type H+-transporting ATPase subunit b | -0.230 | 0.296 | 0.438 | 0.651 |
| ATPF0C, atpE; F-type H+-transporting ATPase subunit c | -0.230 | 0.296 | 0.438 | 0.651 |
| ATPF1A, atpA; F-type H+-transporting ATPase subunit alpha [EC:3.6.3.14] | -0.244 | 0.294 | 0.409 | 0.651 |
| ATPF1B, atpD; F-type H+-transporting ATPase subunit beta [EC:3.6.3.14] | -0.230 | 0.296 | 0.438 | 0.651 |
| ATPF1D, atpH; F-type H+-transporting ATPase subunit delta | -0.230 | 0.296 | 0.438 | 0.651 |
| ATPF1E, atpC; F-type H+-transporting ATPase subunit epsilon | -0.230 | 0.296 | 0.438 | 0.651 |
| ATPF1G, atpG; F-type H+-transporting ATPase subunit gamma | -0.230 | 0.296 | 0.438 | 0.651 |
| AXY8, FUC95A, afcA; alpha-L-fucosidase 2 [EC:3.2.1.51] | -0.161 | 0.278 | 0.562 | 0.689 |
| bacA; undecaprenyl-diphosphatase [EC:3.6.1.27] | -0.230 | 0.296 | 0.438 | 0.651 |
| bamD; outer membrane protein assembly factor BamD | -0.230 | 0.296 | 0.438 | 0.651 |
| BCP, PRXQ, DOT5; peroxiredoxin Q/BCP [EC:1.11.1.15] | -0.230 | 0.296 | 0.438 | 0.651 |
| bcrC; undecaprenyl-diphosphatase [EC:3.6.1.27] | -0.309 | 0.308 | 0.317 | 0.651 |
| bdhAB; butanol dehydrogenase [EC:1.1.1.-] | -0.746 | 1.115 | 0.505 | 0.651 |
| bgaB, lacA; beta-galactosidase [EC:3.2.1.23] | -0.671 | 0.462 | 0.148 | 0.651 |
| bglX; beta-glucosidase [EC:3.2.1.21] | -0.399 | 0.427 | 0.352 | 0.651 |
| bioF; 8-amino-7-oxononanoate synthase [EC:2.3.1.47] | -0.754 | 1.120 | 0.502 | 0.651 |
| birA; BirA family transcriptional regulator, biotin operon repressor / biotin—[acetyl-CoA-carboxylase] ligase [EC:6.3.4.15] | -0.230 | 0.296 | 0.438 | 0.651 |
| BLMH, pepC; bleomycin hydrolase [EC:3.4.22.40] | -0.224 | 0.296 | 0.452 | 0.651 |
| bshA; L-malate glycosyltransferase [EC:2.4.1.-] | -2.232 | 1.933 | 0.250 | 0.651 |
| buk; butyrate kinase [EC:2.7.2.7] | -0.147 | 0.329 | 0.655 | 0.768 |
| capA, pgsA; gamma-polyglutamate biosynthesis protein CapA | -0.117 | 0.336 | 0.728 | 0.823 |
| carA, CPA1; carbamoyl-phosphate synthase small subunit [EC:6.3.5.5] | -0.230 | 0.296 | 0.438 | 0.651 |
| carB, CPA2; carbamoyl-phosphate synthase large subunit [EC:6.3.5.5] | -0.232 | 0.301 | 0.441 | 0.651 |
| CARS, cysS; cysteinyl-tRNA synthetase [EC:6.1.1.16] | -0.230 | 0.296 | 0.438 | 0.651 |
| cas1; CRISP-associated protein Cas1 | -0.588 | 0.445 | 0.188 | 0.651 |
| cas2; CRISPR-associated protein Cas2 | -0.588 | 0.445 | 0.188 | 0.651 |
| cas3; CRISPR-associated endonuclease/helicase Cas3 [EC:3.1.-.- 3.6.4.-] | -0.789 | 0.881 | 0.372 | 0.651 |
| cas4; CRISPR-associated exonuclease Cas4 [EC:3.1.12.1] | -0.789 | 0.881 | 0.372 | 0.651 |
| cas6; CRISPR-associated endoribonuclease Cas6 [EC:3.1.-.-] | -0.677 | 0.652 | 0.301 | 0.651 |
| cbe, mbe; cellobiose epimerase [EC:5.1.3.11] | -0.588 | 0.587 | 0.318 | 0.651 |
| cbpA; curved DNA-binding protein | -0.231 | 0.318 | 0.469 | 0.651 |
| ccrM; modification methylase [EC:2.1.1.72] | -0.297 | 0.684 | 0.664 | 0.776 |
| cdd, CDA; cytidine deaminase [EC:3.5.4.5] | -0.230 | 0.296 | 0.438 | 0.651 |
| CHO1, pssA; CDP-diacylglycerol—serine O-phosphatidyltransferase [EC:2.7.8.8] | -0.230 | 0.296 | 0.438 | 0.651 |
| chrA; chromate transporter | -0.220 | 0.299 | 0.463 | 0.651 |
| cidA; holin-like protein | -0.110 | 0.293 | 0.709 | 0.803 |
| cimA; (R)-citramalate synthase [EC:2.3.1.182] | -0.100 | 1.127 | 0.930 | 0.976 |
| citD; citrate lyase subunit gamma (acyl carrier protein) | 2.304 | 1.989 | 0.248 | 0.651 |
| citE; citrate lyase subunit beta / citryl-CoA lyase [EC:4.1.3.34] | 2.304 | 1.989 | 0.248 | 0.651 |
| citF; citrate lyase subunit alpha / citrate CoA-transferase [EC:2.8.3.10] | 2.304 | 1.989 | 0.248 | 0.651 |
| citX; holo-ACP synthase [EC:2.7.7.61] | 2.304 | 1.989 | 0.248 | 0.651 |
| clpA; ATP-dependent Clp protease ATP-binding subunit ClpA | 0.056 | 0.493 | 0.910 | 0.976 |
| clpB; ATP-dependent Clp protease ATP-binding subunit ClpB | -0.230 | 0.296 | 0.438 | 0.651 |
| clpC; ATP-dependent Clp protease ATP-binding subunit ClpC | -0.230 | 0.296 | 0.438 | 0.651 |
| clpP, CLPP; ATP-dependent Clp protease, protease subunit [EC:3.4.21.92] | -0.230 | 0.296 | 0.438 | 0.651 |
| clpS; ATP-dependent Clp protease adaptor protein ClpS | 0.056 | 0.493 | 0.910 | 0.976 |
| clpX, CLPX; ATP-dependent Clp protease ATP-binding subunit ClpX | -0.230 | 0.296 | 0.438 | 0.651 |
| clsA_B; cardiolipin synthase A/B [EC:2.7.8.-] | -0.230 | 0.296 | 0.438 | 0.651 |
| cmk; CMP/dCMP kinase [EC:2.7.4.25] | -0.230 | 0.296 | 0.438 | 0.651 |
| coaBC, dfp; phosphopantothenoylcysteine decarboxylase / phosphopantothenate—cysteine ligase [EC:4.1.1.36 6.3.2.5] | -0.213 | 0.298 | 0.477 | 0.651 |
| coaE; dephospho-CoA kinase [EC:2.7.1.24] | -0.228 | 0.308 | 0.459 | 0.651 |
| coaW; type II pantothenate kinase [EC:2.7.1.33] | -0.219 | 0.301 | 0.468 | 0.651 |
| coaX; type III pantothenate kinase [EC:2.7.1.33] | -0.754 | 1.120 | 0.502 | 0.651 |
| cobA, btuR; cob(I)alamin adenosyltransferase [EC:2.5.1.17] | -0.221 | 0.572 | 0.699 | 0.797 |
| cobC, phpB; alpha-ribazole phosphatase [EC:3.1.3.73] | -0.680 | 0.538 | 0.208 | 0.651 |
| cobN; cobaltochelatase CobN [EC:6.6.1.2] | -0.341 | 0.419 | 0.416 | 0.651 |
| cobP, cobU; adenosylcobinamide kinase / adenosylcobinamide-phosphate guanylyltransferase [EC:2.7.1.156 2.7.7.62] | -0.680 | 0.538 | 0.208 | 0.651 |
| comEB; dCMP deaminase [EC:3.5.4.12] | -0.230 | 0.296 | 0.438 | 0.651 |
| comEC; competence protein ComEC | -0.230 | 0.296 | 0.438 | 0.651 |
| comM; magnesium chelatase family protein | -0.243 | 0.294 | 0.409 | 0.651 |
| copB; Cu2+-exporting ATPase [EC:3.6.3.4] | -0.224 | 0.285 | 0.434 | 0.651 |
| corA; magnesium transporter | -0.230 | 0.296 | 0.438 | 0.651 |
| cpdB; 2’,3’-cyclic-nucleotide 2’-phosphodiesterase / 3’-nucleotidase [EC:3.1.4.16 3.1.3.6] | -0.285 | 0.310 | 0.360 | 0.651 |
| crcB, FEX; fluoride exporter | -0.239 | 0.315 | 0.449 | 0.651 |
| CS, gltA; citrate synthase [EC:2.3.3.1] | 18.519 | 3411.091 | 0.996 | 0.998 |
| csm1, cas10; CRISPR-associated protein Csm1 | -1.447 | 1.576 | 0.360 | 0.651 |
| csm2; CRISPR-associated protein Csm2 | -1.447 | 1.576 | 0.360 | 0.651 |
| csm3; CRISPR-associated protein Csm3 | -1.447 | 1.576 | 0.360 | 0.651 |
| csn1, cas9; CRISPR-associated endonuclease Csn1 [EC:3.1.-.-] | -0.460 | 0.462 | 0.321 | 0.651 |
| csxA; exo-1,4-beta-D-glucosaminidase [EC:3.2.1.165] | -2.232 | 1.933 | 0.250 | 0.651 |
| cusA, silA; Cu(I)/Ag(I) efflux system membrane protein CusA/SilA | 0.153 | 0.466 | 0.742 | 0.834 |
| cusB, silB; membrane fusion protein, Cu(I)/Ag(I) efflux system | 0.045 | 0.522 | 0.932 | 0.976 |
| cutC; copper homeostasis protein | -0.277 | 0.305 | 0.364 | 0.651 |
| cvrA, nhaP2; cell volume regulation protein A | -1.146 | 1.935 | 0.555 | 0.683 |
| cydA; cytochrome bd ubiquinol oxidase subunit I [EC:1.10.3.14] | -0.230 | 0.296 | 0.438 | 0.651 |
| cydB; cytochrome bd ubiquinol oxidase subunit II [EC:1.10.3.14] | -0.230 | 0.296 | 0.438 | 0.651 |
| cynR; LysR family transcriptional regulator, cyn operon transcriptional activator | -0.330 | 0.504 | 0.514 | 0.651 |
| cynT, can; carbonic anhydrase [EC:4.2.1.1] | -0.169 | 0.333 | 0.614 | 0.731 |
| cysC; adenylylsulfate kinase [EC:2.7.1.25] | -0.754 | 1.120 | 0.502 | 0.651 |
| cysD; sulfate adenylyltransferase subunit 2 [EC:2.7.7.4] | -0.754 | 1.120 | 0.502 | 0.651 |
| cysE; serine O-acetyltransferase [EC:2.3.1.30] | -0.157 | 0.290 | 0.590 | 0.717 |
| cysH; phosphoadenosine phosphosulfate reductase [EC:1.8.4.8 1.8.4.10] | 18.519 | 3411.091 | 0.996 | 0.998 |
| cysK; cysteine synthase A [EC:2.5.1.47] | -0.260 | 0.313 | 0.407 | 0.651 |
| cysN; sulfate adenylyltransferase subunit 1 [EC:2.7.7.4] | -0.754 | 1.120 | 0.502 | 0.651 |
| cysQ, MET22, BPNT1; 3’(2’), 5’-bisphosphate nucleotidase [EC:3.1.3.7] | -0.754 | 1.120 | 0.502 | 0.651 |
| cytR; LacI family transcriptional regulator, repressor for deo operon, udp, cdd, tsx, nupC, and nupG | 0.151 | 1.352 | 0.911 | 0.976 |
| czcA; cobalt-zinc-cadmium resistance protein CzcA | 0.470 | 0.526 | 0.372 | 0.651 |
| czcB; membrane fusion protein, cobalt-zinc-cadmium efflux system | 18.519 | 3411.091 | 0.996 | 0.998 |
| dacB; serine-type D-Ala-D-Ala carboxypeptidase/endopeptidase (penicillin-binding protein 4) [EC:3.4.16.4 3.4.21.-] | -0.288 | 0.314 | 0.360 | 0.651 |
| dam; DNA adenine methylase [EC:2.1.1.72] | -0.174 | 0.353 | 0.623 | 0.740 |
| dapA; 4-hydroxy-tetrahydrodipicolinate synthase [EC:4.3.3.7] | -0.230 | 0.296 | 0.438 | 0.651 |
| dapB; 4-hydroxy-tetrahydrodipicolinate reductase [EC:1.17.1.8] | -0.230 | 0.296 | 0.438 | 0.651 |
| dapdh; diaminopimelate dehydrogenase [EC:1.4.1.16] | -0.228 | 0.293 | 0.436 | 0.651 |
| dapF; diaminopimelate epimerase [EC:5.1.1.7] | -0.394 | 0.395 | 0.320 | 0.651 |
| dcp; peptidyl-dipeptidase Dcp [EC:3.4.15.5] | -0.233 | 0.311 | 0.453 | 0.651 |
| dcuA; anaerobic C4-dicarboxylate transporter DcuA | -0.318 | 0.477 | 0.505 | 0.651 |
| dcuB; anaerobic C4-dicarboxylate transporter DcuB | -0.221 | 0.572 | 0.699 | 0.797 |
| ddl; D-alanine-D-alanine ligase [EC:6.3.2.4] | -0.230 | 0.296 | 0.438 | 0.651 |
| deaD, cshA; ATP-dependent RNA helicase DeaD [EC:3.6.4.13] | -0.230 | 0.296 | 0.438 | 0.651 |
| deoC, DERA; deoxyribose-phosphate aldolase [EC:4.1.2.4] | -0.230 | 0.296 | 0.438 | 0.651 |
| dexA; dextranase [EC:3.2.1.11] | -0.282 | 0.509 | 0.580 | 0.706 |
| dfx; superoxide reductase [EC:1.15.1.2] | -0.166 | 0.321 | 0.605 | 0.723 |
| dgt; dGTPase [EC:3.1.5.1] | -0.230 | 0.296 | 0.438 | 0.651 |
| DHFR, folA; dihydrofolate reductase [EC:1.5.1.3] | -0.201 | 0.284 | 0.481 | 0.651 |
| dinB; DNA polymerase IV [EC:2.7.7.7] | -0.230 | 0.296 | 0.438 | 0.651 |
| dinD; DNA-damage-inducible protein D | -0.195 | 0.637 | 0.759 | 0.844 |
| djlA; DnaJ like chaperone protein | -0.174 | 0.327 | 0.596 | 0.722 |
| DLD, lpd, pdhD; dihydrolipoamide dehydrogenase [EC:1.8.1.4] | -0.190 | 0.321 | 0.556 | 0.683 |
| dnaA; chromosomal replication initiator protein | -0.230 | 0.296 | 0.438 | 0.651 |
| dnaB; replicative DNA helicase [EC:3.6.4.12] | -0.230 | 0.296 | 0.438 | 0.651 |
| dnaC; DNA replication protein DnaC | -17.599 | 3454.822 | 0.996 | 0.998 |
| dnaE; DNA polymerase III subunit alpha [EC:2.7.7.7] | -0.230 | 0.296 | 0.438 | 0.651 |
| dnaG; DNA primase [EC:2.7.7.-] | -0.230 | 0.296 | 0.438 | 0.651 |
| dnaJ; molecular chaperone DnaJ | -0.230 | 0.296 | 0.438 | 0.651 |
| dnaK, HSPA9; molecular chaperone DnaK | -0.230 | 0.296 | 0.438 | 0.651 |
| dnaN; DNA polymerase III subunit beta [EC:2.7.7.7] | -0.230 | 0.296 | 0.438 | 0.651 |
| dnaQ; DNA polymerase III subunit epsilon [EC:2.7.7.7] | -0.218 | 0.290 | 0.455 | 0.651 |
| dnaX; DNA polymerase III subunit gamma/tau [EC:2.7.7.7] | -0.230 | 0.296 | 0.438 | 0.651 |
| dndC; DNA sulfur modification protein DndC | 2.304 | 1.989 | 0.248 | 0.651 |
| dndD; DNA sulfur modification protein DndD | 2.332 | 1.962 | 0.236 | 0.651 |
| dndE; DNA sulfur modification protein DndE | 2.304 | 1.989 | 0.248 | 0.651 |
| DNMT1, dcm; DNA (cytosine-5)-methyltransferase 1 [EC:2.1.1.37] | -0.244 | 0.306 | 0.427 | 0.651 |
| DNPEP; aspartyl aminopeptidase [EC:3.4.11.21] | 18.519 | 3411.091 | 0.996 | 0.998 |
| DPM1; dolichol-phosphate mannosyltransferase [EC:2.4.1.83] | -0.230 | 0.296 | 0.438 | 0.651 |
| dpo; DNA polymerase bacteriophage-type [EC:2.7.7.7] | 0.358 | 0.550 | 0.516 | 0.651 |
| DPP3; dipeptidyl-peptidase III [EC:3.4.14.4] | -0.230 | 0.296 | 0.438 | 0.651 |
| DPP4, CD26; dipeptidyl-peptidase 4 [EC:3.4.14.5] | -0.230 | 0.296 | 0.438 | 0.651 |
| dps; starvation-inducible DNA-binding protein | -0.222 | 0.303 | 0.464 | 0.651 |
| dsbD; thiol:disulfide interchange protein DsbD [EC:1.8.1.8] | -0.230 | 0.296 | 0.438 | 0.651 |
| dtd, DTD1; D-tyrosyl-tRNA(Tyr) deacylase [EC:3.1.-.-] | -0.230 | 0.296 | 0.438 | 0.651 |
| dut, DUT; dUTP pyrophosphatase [EC:3.6.1.23] | -0.230 | 0.296 | 0.438 | 0.651 |
| dxr; 1-deoxy-D-xylulose-5-phosphate reductoisomerase [EC:1.1.1.267] | -0.230 | 0.296 | 0.438 | 0.651 |
| dxs; 1-deoxy-D-xylulose-5-phosphate synthase [EC:2.2.1.7] | -0.211 | 0.305 | 0.489 | 0.651 |
| E1.1.1.40, maeB; malate dehydrogenase (oxaloacetate-decarboxylating)(NADP+) [EC:1.1.1.40] | -0.230 | 0.296 | 0.438 | 0.651 |
| E1.17.4.1A, nrdA, nrdE; ribonucleoside-diphosphate reductase alpha chain [EC:1.17.4.1] | -0.178 | 0.295 | 0.546 | 0.680 |
| E1.17.4.1B, nrdB, nrdF; ribonucleoside-diphosphate reductase beta chain [EC:1.17.4.1] | -0.123 | 0.286 | 0.667 | 0.778 |
| E1.4.1.4, gdhA; glutamate dehydrogenase (NADP+) [EC:1.4.1.4] | -0.154 | 0.290 | 0.596 | 0.722 |
| E2.1.1.72; site-specific DNA-methyltransferase (adenine-specific) [EC:2.1.1.72] | -0.359 | 0.322 | 0.266 | 0.651 |
| E2.1.1.77, pcm; protein-L-isoaspartate(D-aspartate) O-methyltransferase [EC:2.1.1.77] | -0.137 | 0.613 | 0.823 | 0.900 |
| E2.2.1.1, tktA, tktB; transketolase [EC:2.2.1.1] | -0.234 | 0.306 | 0.446 | 0.651 |
| E2.2.1.6L, ilvB, ilvG, ilvI; acetolactate synthase I/II/III large subunit [EC:2.2.1.6] | -0.548 | 0.584 | 0.350 | 0.651 |
| E2.2.1.6S, ilvH, ilvN; acetolactate synthase I/III small subunit [EC:2.2.1.6] | -0.548 | 0.584 | 0.350 | 0.651 |
| E2.3.1.54, pflD; formate C-acetyltransferase [EC:2.3.1.54] | -0.215 | 0.295 | 0.468 | 0.651 |
| E2.3.1.8, pta; phosphate acetyltransferase [EC:2.3.1.8] | -0.230 | 0.296 | 0.438 | 0.651 |
| E2.4.1.20; cellobiose phosphorylase [EC:2.4.1.20] | 0.154 | 1.349 | 0.909 | 0.976 |
| E2.4.2.21, cobU, cobT; nicotinate-nucleotide–dimethylbenzimidazole phosphoribosyltransferase [EC:2.4.2.21] | -0.680 | 0.538 | 0.208 | 0.651 |
| E2.5.1.56, neuB; N-acetylneuraminate synthase [EC:2.5.1.56] | -2.232 | 1.933 | 0.250 | 0.651 |
| E2.6.1.11, argD; acetylornithine aminotransferase [EC:2.6.1.11] | -0.100 | 1.127 | 0.930 | 0.976 |
| E2.6.1.42, ilvE; branched-chain amino acid aminotransferase [EC:2.6.1.42] | -0.230 | 0.296 | 0.438 | 0.651 |
| E2.6.1.83; LL-diaminopimelate aminotransferase [EC:2.6.1.83] | -0.394 | 0.395 | 0.320 | 0.651 |
| E2.7.1.4, scrK; fructokinase [EC:2.7.1.4] | -0.239 | 0.302 | 0.431 | 0.651 |
| E2.7.1.71, aroK, aroL; shikimate kinase [EC:2.7.1.71] | -0.230 | 0.296 | 0.438 | 0.651 |
| E2.7.4.8, gmk; guanylate kinase [EC:2.7.4.8] | -0.230 | 0.296 | 0.438 | 0.651 |
| E2.7.7.24, rfbA, rffH; glucose-1-phosphate thymidylyltransferase [EC:2.7.7.24] | -0.230 | 0.296 | 0.438 | 0.651 |
| E2.7.7.3A, coaD, kdtB; pantetheine-phosphate adenylyltransferase [EC:2.7.7.3] | -0.230 | 0.296 | 0.438 | 0.651 |
| E2.7.7.41, CDS1, CDS2, cdsA; phosphatidate cytidylyltransferase [EC:2.7.7.41] | -0.230 | 0.296 | 0.438 | 0.651 |
| E2.7.8.26, cobS, cobV; adenosylcobinamide-GDP ribazoletransferase [EC:2.7.8.26] | -0.680 | 0.538 | 0.208 | 0.651 |
| E3.1.1.11; pectinesterase [EC:3.1.1.11] | -0.674 | 0.651 | 0.302 | 0.651 |
| E3.1.11.2, xthA; exodeoxyribonuclease III [EC:3.1.11.2] | -0.138 | 0.295 | 0.642 | 0.755 |
| E3.1.11.5; exodeoxyribonuclease V [EC:3.1.11.5] | -0.754 | 1.120 | 0.502 | 0.651 |
| E3.1.21.4; type II restriction enzyme [EC:3.1.21.4] | -0.343 | 0.461 | 0.459 | 0.651 |
| E3.1.3.1, phoA, phoB; alkaline phosphatase [EC:3.1.3.1] | -0.400 | 0.466 | 0.392 | 0.651 |
| E3.1.3.48; protein-tyrosine phosphatase [EC:3.1.3.48] | -0.237 | 0.301 | 0.432 | 0.651 |
| E3.1.3.5; 5’-nucleotidase [EC:3.1.3.5] | -0.219 | 0.301 | 0.468 | 0.651 |
| E3.1.4.46, glpQ, ugpQ; glycerophosphoryl diester phosphodiesterase [EC:3.1.4.46] | -0.647 | 0.496 | 0.194 | 0.651 |
| E3.1.6.1, aslA; arylsulfatase [EC:3.1.6.1] | -0.131 | 0.339 | 0.701 | 0.797 |
| E3.2.1.17; lysozyme [EC:3.2.1.17] | 0.038 | 0.447 | 0.932 | 0.976 |
| E3.2.1.1A; alpha-amylase [EC:3.2.1.1] | -0.230 | 0.296 | 0.438 | 0.651 |
| E3.2.1.22B, galA, rafA; alpha-galactosidase [EC:3.2.1.22] | -0.168 | 0.287 | 0.561 | 0.689 |
| E3.2.1.4; endoglucanase [EC:3.2.1.4] | 18.519 | 3411.091 | 0.996 | 0.998 |
| E3.2.1.8, xynA; endo-1,4-beta-xylanase [EC:3.2.1.8] | 18.519 | 3411.091 | 0.996 | 0.998 |
| E3.2.1.89; arabinogalactan endo-1,4-beta-galactosidase [EC:3.2.1.89] | -0.483 | 0.529 | 0.363 | 0.651 |
| E3.4.21.102, prc, ctpA; carboxyl-terminal processing protease [EC:3.4.21.102] | -0.230 | 0.296 | 0.438 | 0.651 |
| E3.5.1.1, ansA, ansB; L-asparaginase [EC:3.5.1.1] | -0.260 | 0.336 | 0.441 | 0.651 |
| E3.5.2.10; creatinine amidohydrolase [EC:3.5.2.10] | -2.232 | 1.933 | 0.250 | 0.651 |
| E3.6.1.22, NUDT12, nudC; NAD+ diphosphatase [EC:3.6.1.22] | -0.230 | 0.296 | 0.438 | 0.651 |
| E3.6.3.8; Ca2+-transporting ATPase [EC:3.6.3.8] | -0.156 | 0.300 | 0.605 | 0.723 |
| E4.1.1.49, pckA; phosphoenolpyruvate carboxykinase (ATP) [EC:4.1.1.49] | -0.230 | 0.296 | 0.438 | 0.651 |
| E4.1.3.3, nanA, NPL; N-acetylneuraminate lyase [EC:4.1.3.3] | -0.206 | 0.295 | 0.486 | 0.651 |
| E4.2.1.2A, fumA, fumB; fumarate hydratase, class I [EC:4.2.1.2] | -0.230 | 0.296 | 0.438 | 0.651 |
| E4.2.1.2AA, fumA; fumarate hydratase subunit alpha [EC:4.2.1.2] | -0.221 | 0.572 | 0.699 | 0.797 |
| E4.2.1.2AB, fumB; fumarate hydratase subunit beta [EC:4.2.1.2] | -0.221 | 0.572 | 0.699 | 0.797 |
| E4.2.1.46, rfbB, rffG; dTDP-glucose 4,6-dehydratase [EC:4.2.1.46] | -0.230 | 0.296 | 0.438 | 0.651 |
| E4.3.1.17, sdaA, sdaB, tdcG; L-serine dehydratase [EC:4.3.1.17] | -0.230 | 0.296 | 0.438 | 0.651 |
| E4.3.1.19, ilvA, tdcB; threonine dehydratase [EC:4.3.1.19] | -0.378 | 0.462 | 0.414 | 0.651 |
| E5.1.3.6; UDP-glucuronate 4-epimerase [EC:5.1.3.6] | -0.057 | 0.574 | 0.921 | 0.976 |
| E5.2.1.8; peptidylprolyl isomerase [EC:5.2.1.8] | -0.080 | 0.342 | 0.815 | 0.896 |
| E6.3.5.1, NADSYN1, QNS1, nadE; NAD+ synthase (glutamine-hydrolysing) [EC:6.3.5.1] | -0.230 | 0.296 | 0.438 | 0.651 |
| E6.5.1.2, ligA, ligB; DNA ligase (NAD+) [EC:6.5.1.2] | -0.230 | 0.296 | 0.438 | 0.651 |
| eamB; cysteine/O-acetylserine efflux protein | 0.092 | 1.404 | 0.948 | 0.986 |
| EARS, gltX; glutamyl-tRNA synthetase [EC:6.1.1.17] | -0.230 | 0.296 | 0.438 | 0.651 |
| eda; 2-dehydro-3-deoxyphosphogluconate aldolase / (4S)-4-hydroxy-2-oxoglutarate aldolase [EC:4.1.2.14 4.1.3.42] | -0.628 | 0.582 | 0.282 | 0.651 |
| efp; elongation factor P | -0.230 | 0.296 | 0.438 | 0.651 |
| EIF1, SUI1; translation initiation factor 1 | -0.754 | 1.120 | 0.502 | 0.651 |
| elaA; ElaA protein | 2.304 | 1.989 | 0.248 | 0.651 |
| emrA; membrane fusion protein, multidrug efflux system | 18.519 | 3411.091 | 0.996 | 0.998 |
| ENDOG; endonuclease G, mitochondrial | -0.199 | 0.305 | 0.515 | 0.651 |
| engA, der; GTPase | -0.230 | 0.296 | 0.438 | 0.651 |
| engB; GTP-binding protein | -0.230 | 0.296 | 0.438 | 0.651 |
| ENO, eno; enolase [EC:4.2.1.11] | -0.230 | 0.296 | 0.438 | 0.651 |
| entC; isochorismate synthase [EC:5.4.4.2] | -0.230 | 0.296 | 0.438 | 0.651 |
| epsD; glycosyltransferase EpsD [EC:2.4.-.-] | 0.358 | 0.550 | 0.516 | 0.651 |
| eptC; heptose-I-phosphate ethanolaminephosphotransferase [EC:2.7.8.-] | -0.236 | 0.284 | 0.407 | 0.651 |
| era, ERAL1; GTPase | -0.230 | 0.296 | 0.438 | 0.651 |
| ermC, ermA; 23S rRNA (adenine-N6)-dimethyltransferase [EC:2.1.1.184] | 0.141 | 0.367 | 0.701 | 0.797 |
| exbB; biopolymer transport protein ExbB | -0.225 | 0.298 | 0.452 | 0.651 |
| exuT; MFS transporter, ACS family, hexuronate transporter | -0.692 | 0.616 | 0.263 | 0.651 |
| fabB; 3-oxoacyl-[acyl-carrier-protein] synthase I [EC:2.3.1.41] | -0.741 | 0.488 | 0.131 | 0.651 |
| fabD; [acyl-carrier-protein] S-malonyltransferase [EC:2.3.1.39] | -0.230 | 0.296 | 0.438 | 0.651 |
| fabF; 3-oxoacyl-[acyl-carrier-protein] synthase II [EC:2.3.1.179] | -0.230 | 0.296 | 0.438 | 0.651 |
| fabG; 3-oxoacyl-[acyl-carrier protein] reductase [EC:1.1.1.100] | -0.342 | 0.306 | 0.265 | 0.651 |
| fabH; 3-oxoacyl-[acyl-carrier-protein] synthase III [EC:2.3.1.180] | -0.223 | 0.335 | 0.506 | 0.651 |
| fabI; enoyl-[acyl-carrier protein] reductase I [EC:1.3.1.9 1.3.1.10] | -0.219 | 0.301 | 0.468 | 0.651 |
| fabZ; 3-hydroxyacyl-[acyl-carrier-protein] dehydratase [EC:4.2.1.59] | -0.741 | 0.488 | 0.131 | 0.651 |
| fadL; long-chain fatty acid transport protein | -0.642 | 0.547 | 0.242 | 0.651 |
| FARSA, pheS; phenylalanyl-tRNA synthetase alpha chain [EC:6.1.1.20] | -0.230 | 0.296 | 0.438 | 0.651 |
| FARSB, pheT; phenylalanyl-tRNA synthetase beta chain [EC:6.1.1.20] | -0.230 | 0.296 | 0.438 | 0.651 |
| FBA, fbaA; fructose-bisphosphate aldolase, class II [EC:4.1.2.13] | -0.230 | 0.296 | 0.438 | 0.651 |
| fbp3; fructose-1,6-bisphosphatase III [EC:3.1.3.11] | -0.230 | 0.296 | 0.438 | 0.651 |
| fecA; Fe(3+) dicitrate transport protein | -0.202 | 0.360 | 0.576 | 0.703 |
| feoB; ferrous iron transport protein B | -0.214 | 0.282 | 0.449 | 0.651 |
| fes; enterochelin esterase and related enzymes | -0.206 | 0.316 | 0.516 | 0.651 |
| fhs; formate–tetrahydrofolate ligase [EC:6.3.4.3] | -0.263 | 0.312 | 0.401 | 0.651 |
| fic; cell filamentation protein | -0.213 | 0.305 | 0.486 | 0.651 |
| fixA, etfB; electron transfer flavoprotein beta subunit | -0.230 | 0.296 | 0.438 | 0.651 |
| fixB, etfA; electron transfer flavoprotein alpha subunit | -0.230 | 0.296 | 0.438 | 0.651 |
| fklB; FKBP-type peptidyl-prolyl cis-trans isomerase FklB [EC:5.2.1.8] | -0.223 | 0.293 | 0.448 | 0.651 |
| fldA, nifF, isiB; flavodoxin I | -0.201 | 0.284 | 0.481 | 0.651 |
| FLOT; flotillin | -2.232 | 1.933 | 0.250 | 0.651 |
| folB; 7,8-dihydroneopterin aldolase/epimerase/oxygenase [EC:4.1.2.25 5.1.99.8 1.13.11.81] | -0.230 | 0.296 | 0.438 | 0.651 |
| folC; dihydrofolate synthase / folylpolyglutamate synthase [EC:6.3.2.12 6.3.2.17] | -0.230 | 0.296 | 0.438 | 0.651 |
| folD; methylenetetrahydrofolate dehydrogenase (NADP+) / methenyltetrahydrofolate cyclohydrolase [EC:1.5.1.5 3.5.4.9] | -0.230 | 0.296 | 0.438 | 0.651 |
| folK; 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine diphosphokinase [EC:2.7.6.3] | -0.240 | 0.304 | 0.431 | 0.651 |
| folP; dihydropteroate synthase [EC:2.5.1.15] | -0.230 | 0.296 | 0.438 | 0.651 |
| frr, MRRF, RRF; ribosome recycling factor | -0.230 | 0.296 | 0.438 | 0.651 |
| fruA; fructan beta-fructosidase [EC:3.2.1.80] | -0.268 | 0.313 | 0.394 | 0.651 |
| fsr; MFS transporter, FSR family, fosmidomycin resistance protein | -0.081 | 0.335 | 0.809 | 0.893 |
| FTCD; glutamate formiminotransferase / formiminotetrahydrofolate cyclodeaminase [EC:2.1.2.5 4.3.1.4] | -0.162 | 0.333 | 0.626 | 0.741 |
| ftnA, ftn; ferritin [EC:1.16.3.2] | -0.219 | 0.301 | 0.468 | 0.651 |
| ftsA; cell division protein FtsA | -0.230 | 0.296 | 0.438 | 0.651 |
| ftsE; cell division transport system ATP-binding protein | -0.230 | 0.296 | 0.438 | 0.651 |
| ftsH, hflB; cell division protease FtsH [EC:3.4.24.-] | -0.230 | 0.296 | 0.438 | 0.651 |
| ftsI; cell division protein FtsI (penicillin-binding protein 3) [EC:3.4.16.4] | -0.229 | 0.320 | 0.476 | 0.651 |
| ftsK, spoIIIE; DNA segregation ATPase FtsK/SpoIIIE, S-DNA-T family | -0.230 | 0.296 | 0.438 | 0.651 |
| ftsQ; cell division protein FtsQ | -0.230 | 0.296 | 0.438 | 0.651 |
| ftsW, spoVE; cell division protein FtsW | -0.230 | 0.296 | 0.438 | 0.651 |
| ftsX; cell division transport system permease protein | -0.230 | 0.296 | 0.438 | 0.651 |
| ftsY; fused signal recognition particle receptor | -0.230 | 0.296 | 0.438 | 0.651 |
| ftsZ; cell division protein FtsZ | -0.241 | 0.294 | 0.412 | 0.651 |
| FUCA; alpha-L-fucosidase [EC:3.2.1.51] | -0.246 | 0.300 | 0.413 | 0.651 |
| fucP; MFS transporter, FHS family, L-fucose permease | -0.286 | 0.297 | 0.338 | 0.651 |
| fur, zur, furB; Fur family transcriptional regulator, ferric uptake regulator | -0.264 | 0.311 | 0.398 | 0.651 |
| fusA, GFM, EFG; elongation factor G | -0.230 | 0.296 | 0.438 | 0.651 |
| galE, GALE; UDP-glucose 4-epimerase [EC:5.1.3.2] | -0.230 | 0.296 | 0.438 | 0.651 |
| galK; galactokinase [EC:2.7.1.6] | -0.224 | 0.269 | 0.406 | 0.651 |
| galM, GALM; aldose 1-epimerase [EC:5.1.3.3] | -0.243 | 0.306 | 0.429 | 0.651 |
| GAPDH, gapA; glyceraldehyde 3-phosphate dehydrogenase [EC:1.2.1.12] | -0.230 | 0.296 | 0.438 | 0.651 |
| GARS, glyS1; glycyl-tRNA synthetase [EC:6.1.1.14] | -0.230 | 0.296 | 0.438 | 0.651 |
| GBA, srfJ; glucosylceramidase [EC:3.2.1.45] | 0.151 | 1.352 | 0.911 | 0.976 |
| GBE1, glgB; 1,4-alpha-glucan branching enzyme [EC:2.4.1.18] | -0.230 | 0.296 | 0.438 | 0.651 |
| GCH1, folE; GTP cyclohydrolase IA [EC:3.5.4.16] | -0.201 | 0.284 | 0.481 | 0.651 |
| gcpE, ispG; (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase [EC:1.17.7.1 1.17.7.3] | -0.230 | 0.296 | 0.438 | 0.651 |
| gcvH, GCSH; glycine cleavage system H protein | -0.190 | 0.321 | 0.556 | 0.683 |
| gcvPA; glycine dehydrogenase subunit 1 [EC:1.4.4.2] | -0.190 | 0.321 | 0.556 | 0.683 |
| gcvPB; glycine dehydrogenase subunit 2 [EC:1.4.4.2] | -0.190 | 0.321 | 0.556 | 0.683 |
| gcvT, AMT; aminomethyltransferase [EC:2.1.2.10] | -0.190 | 0.321 | 0.556 | 0.683 |
| GGPS; geranylgeranyl diphosphate synthase, type II [EC:2.5.1.1 2.5.1.10 2.5.1.29] | -0.230 | 0.296 | 0.438 | 0.651 |
| gidA, mnmG, MTO1; tRNA uridine 5-carboxymethylaminomethyl modification enzyme | -0.230 | 0.296 | 0.438 | 0.651 |
| gidB, rsmG; 16S rRNA (guanine527-N7)-methyltransferase [EC:2.1.1.170] | -0.230 | 0.296 | 0.438 | 0.651 |
| glf; UDP-galactopyranose mutase [EC:5.4.99.9] | -0.337 | 0.418 | 0.421 | 0.651 |
| glgA; starch synthase [EC:2.4.1.21] | -0.230 | 0.296 | 0.438 | 0.651 |
| glk; glucokinase [EC:2.7.1.2] | -0.268 | 0.292 | 0.361 | 0.651 |
| glmE, mutE, mamB; methylaspartate mutase epsilon subunit [EC:5.4.99.1] | -0.221 | 0.572 | 0.699 | 0.797 |
| glmS, GFPT; glucosamine—fructose-6-phosphate aminotransferase (isomerizing) [EC:2.6.1.16] | -0.243 | 0.311 | 0.436 | 0.651 |
| glmS, mutS, mamA; methylaspartate mutase sigma subunit [EC:5.4.99.1] | -0.221 | 0.572 | 0.699 | 0.797 |
| glnA, GLUL; glutamine synthetase [EC:6.3.1.2] | -0.204 | 0.294 | 0.489 | 0.651 |
| glnB; nitrogen regulatory protein P-II 1 | 18.519 | 3411.091 | 0.996 | 0.998 |
| GLO1, gloA; lactoylglutathione lyase [EC:4.4.1.5] | -1.045 | 0.919 | 0.257 | 0.651 |
| glpA, glpD; glycerol-3-phosphate dehydrogenase [EC:1.1.5.3] | -0.228 | 0.331 | 0.492 | 0.651 |
| glpB; glycerol-3-phosphate dehydrogenase subunit B [EC:1.1.5.3] | -0.228 | 0.331 | 0.492 | 0.651 |
| glpC; glycerol-3-phosphate dehydrogenase subunit C [EC:1.1.5.3] | -0.228 | 0.331 | 0.492 | 0.651 |
| GLPF; glycerol uptake facilitator protein | -0.075 | 0.341 | 0.826 | 0.900 |
| glpK, GK; glycerol kinase [EC:2.7.1.30] | -0.075 | 0.341 | 0.826 | 0.900 |
| glpT; MFS transporter, OPA family, glycerol-3-phosphate transporter | -0.591 | 0.565 | 0.297 | 0.651 |
| gltB; glutamate synthase (NADPH/NADH) large chain [EC:1.4.1.13 1.4.1.14] | 18.519 | 3411.091 | 0.996 | 0.998 |
| gltD; glutamate synthase (NADPH/NADH) small chain [EC:1.4.1.13 1.4.1.14] | -0.324 | 0.318 | 0.311 | 0.651 |
| glxK, garK; glycerate 2-kinase [EC:2.7.1.165] | -0.206 | 0.323 | 0.524 | 0.659 |
| glyA, SHMT; glycine hydroxymethyltransferase [EC:2.1.2.1] | -0.230 | 0.296 | 0.438 | 0.651 |
| gmd, GMDS; GDPmannose 4,6-dehydratase [EC:4.2.1.47] | -0.230 | 0.296 | 0.438 | 0.651 |
| gmhA, lpcA; D-sedoheptulose 7-phosphate isomerase [EC:5.3.1.28] | 18.519 | 3411.091 | 0.996 | 0.998 |
| gmhB; D-glycero-D-manno-heptose 1,7-bisphosphate phosphatase [EC:3.1.3.82 3.1.3.83] | 18.519 | 3411.091 | 0.996 | 0.998 |
| gmuG; mannan endo-1,4-beta-mannosidase [EC:3.2.1.78] | -0.658 | 0.589 | 0.265 | 0.651 |
| gph; phosphoglycolate phosphatase [EC:3.1.3.18] | -0.223 | 0.280 | 0.427 | 0.651 |
| GPI, pgi; glucose-6-phosphate isomerase [EC:5.3.1.9] | -0.230 | 0.296 | 0.438 | 0.651 |
| gpmB; probable phosphoglycerate mutase [EC:5.4.2.12] | -0.262 | 0.309 | 0.398 | 0.651 |
| gpmI; 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [EC:5.4.2.12] | -0.249 | 0.316 | 0.432 | 0.651 |
| gpr; L-glyceraldehyde 3-phosphate reductase [EC:1.1.1.-] | 0.179 | 0.522 | 0.732 | 0.826 |
| gpsA; glycerol-3-phosphate dehydrogenase (NAD(P)+) [EC:1.1.1.94] | -0.230 | 0.296 | 0.438 | 0.651 |
| gpx; glutathione peroxidase [EC:1.11.1.9] | -0.203 | 0.301 | 0.502 | 0.651 |
| greA; transcription elongation factor GreA | -0.248 | 0.296 | 0.404 | 0.651 |
| groEL, HSPD1; chaperonin GroEL | -0.230 | 0.296 | 0.438 | 0.651 |
| groES, HSPE1; chaperonin GroES | -0.230 | 0.296 | 0.438 | 0.651 |
| GRPE; molecular chaperone GrpE | -0.230 | 0.296 | 0.438 | 0.651 |
| guaA, GMPS; GMP synthase (glutamine-hydrolysing) [EC:6.3.5.2] | -0.230 | 0.296 | 0.438 | 0.651 |
| gyrA; DNA gyrase subunit A [EC:5.99.1.3] | -0.230 | 0.296 | 0.438 | 0.651 |
| gyrB; DNA gyrase subunit B [EC:5.99.1.3] | -0.230 | 0.296 | 0.438 | 0.651 |
| HARS, hisS; histidyl-tRNA synthetase [EC:6.1.1.21] | -0.230 | 0.296 | 0.438 | 0.651 |
| hcp; hydroxylamine reductase [EC:1.7.99.1] | -0.206 | 0.301 | 0.494 | 0.651 |
| hddA; D-glycero-alpha-D-manno-heptose-7-phosphate kinase [EC:2.7.1.168] | 18.519 | 3411.091 | 0.996 | 0.998 |
| hemD, UROS; uroporphyrinogen-III synthase [EC:4.2.1.75] | -0.248 | 0.296 | 0.403 | 0.651 |
| hemK, prmC, HEMK; release factor glutamine methyltransferase [EC:2.1.1.297] | -0.230 | 0.296 | 0.438 | 0.651 |
| hemN, hemZ; oxygen-independent coproporphyrinogen III oxidase [EC:1.3.98.3] | -0.227 | 0.294 | 0.441 | 0.651 |
| HEXA_B; hexosaminidase [EC:3.2.1.52] | -0.212 | 0.270 | 0.432 | 0.651 |
| hflX; GTPase | -0.230 | 0.296 | 0.438 | 0.651 |
| higA; HTH-type transcriptional regulator / antitoxin HigA | -0.106 | 0.353 | 0.765 | 0.849 |
| higB-1; toxin HigB-1 | -2.232 | 1.933 | 0.250 | 0.651 |
| HINT1, hinT, hit; histidine triad (HIT) family protein | -0.230 | 0.296 | 0.438 | 0.651 |
| hipA; serine/threonine-protein kinase HipA [EC:2.7.11.1] | -0.014 | 0.330 | 0.966 | 0.998 |
| hipO; hippurate hydrolase [EC:3.5.1.32] | -1.546 | 2.070 | 0.456 | 0.651 |
| hisA; phosphoribosylformimino-5-aminoimidazole carboxamide ribotide isomerase [EC:5.3.1.16] | -0.100 | 1.127 | 0.930 | 0.976 |
| hisB; imidazoleglycerol-phosphate dehydratase / histidinol-phosphatase [EC:4.2.1.19 3.1.3.15] | -0.100 | 1.127 | 0.930 | 0.976 |
| hisC; histidinol-phosphate aminotransferase [EC:2.6.1.9] | -0.159 | 0.284 | 0.576 | 0.703 |
| hisD; histidinol dehydrogenase [EC:1.1.1.23] | -0.100 | 1.127 | 0.930 | 0.976 |
| hisF; cyclase [EC:4.1.3.-] | -0.100 | 1.127 | 0.930 | 0.976 |
| hisG; ATP phosphoribosyltransferase [EC:2.4.2.17] | -0.100 | 1.127 | 0.930 | 0.976 |
| hisH; glutamine amidotransferase [EC:2.4.2.-] | -0.100 | 1.127 | 0.930 | 0.976 |
| hisIE; phosphoribosyl-ATP pyrophosphohydrolase / phosphoribosyl-AMP cyclohydrolase [EC:3.6.1.31 3.5.4.19] | -0.100 | 1.127 | 0.930 | 0.976 |
| hlpA, ompH; outer membrane protein | -0.230 | 0.296 | 0.438 | 0.651 |
| hlyIII; hemolysin III | -0.202 | 0.335 | 0.547 | 0.680 |
| holA; DNA polymerase III subunit delta [EC:2.7.7.7] | -0.230 | 0.296 | 0.438 | 0.651 |
| holB; DNA polymerase III subunit delta’ [EC:2.7.7.7] | -0.230 | 0.296 | 0.438 | 0.651 |
| hppA; K(+)-stimulated pyrophosphate-energized sodium pump [EC:3.6.1.1] | -0.240 | 0.317 | 0.450 | 0.651 |
| hprA; glycerate dehydrogenase [EC:1.1.1.29] | -0.230 | 0.296 | 0.438 | 0.651 |
| hprT, hpt, HPRT1; hypoxanthine phosphoribosyltransferase [EC:2.4.2.8] | -0.230 | 0.296 | 0.438 | 0.651 |
| hsdM; type I restriction enzyme M protein [EC:2.1.1.72] | -0.227 | 0.303 | 0.455 | 0.651 |
| hsdR; type I restriction enzyme, R subunit [EC:3.1.21.3] | -0.273 | 0.327 | 0.405 | 0.651 |
| hsdS; type I restriction enzyme, S subunit [EC:3.1.21.3] | -0.246 | 0.299 | 0.411 | 0.651 |
| hslR; ribosome-associated heat shock protein Hsp15 | -0.230 | 0.296 | 0.438 | 0.651 |
| HSP20; HSP20 family protein | -0.254 | 0.323 | 0.433 | 0.651 |
| HSP90A, htpG; molecular chaperone HtpG | -0.230 | 0.296 | 0.438 | 0.651 |
| htpX; heat shock protein HtpX [EC:3.4.24.-] | -0.446 | 0.424 | 0.294 | 0.651 |
| hupB; DNA-binding protein HU-beta | -0.250 | 0.294 | 0.397 | 0.651 |
| hutH, HAL; histidine ammonia-lyase [EC:4.3.1.3] | -0.162 | 0.333 | 0.626 | 0.741 |
| hutI, AMDHD1; imidazolonepropionase [EC:3.5.2.7] | -0.162 | 0.333 | 0.626 | 0.741 |
| hutU, UROC1; urocanate hydratase [EC:4.2.1.49] | -0.162 | 0.333 | 0.626 | 0.741 |
| hya; hyaluronoglucosaminidase [EC:3.2.1.35] | -0.247 | 0.317 | 0.437 | 0.651 |
| IARS, ileS; isoleucyl-tRNA synthetase [EC:6.1.1.5] | -0.220 | 0.308 | 0.477 | 0.651 |
| icmF; isobutyryl-CoA mutase [EC:5.4.99.13] | -0.228 | 0.317 | 0.474 | 0.651 |
| IDH1, IDH2, icd; isocitrate dehydrogenase [EC:1.1.1.42] | -0.429 | 1.387 | 0.758 | 0.843 |
| idnO; gluconate 5-dehydrogenase [EC:1.1.1.69] | -0.542 | 0.584 | 0.355 | 0.651 |
| ihfA, himA; integration host factor subunit alpha | -0.364 | 0.544 | 0.505 | 0.651 |
| ilvC; ketol-acid reductoisomerase [EC:1.1.1.86] | -0.548 | 0.584 | 0.350 | 0.651 |
| ilvD; dihydroxy-acid dehydratase [EC:4.2.1.9] | -0.548 | 0.584 | 0.350 | 0.651 |
| IMPDH, guaB; IMP dehydrogenase [EC:1.1.1.205] | -0.230 | 0.296 | 0.438 | 0.651 |
| ina; immune inhibitor A [EC:3.4.24.-] | -0.257 | 0.319 | 0.422 | 0.651 |
| infA; translation initiation factor IF-1 | -0.230 | 0.296 | 0.438 | 0.651 |
| infB, MTIF2; translation initiation factor IF-2 | -0.230 | 0.296 | 0.438 | 0.651 |
| infC, MTIF3; translation initiation factor IF-3 | -0.230 | 0.296 | 0.438 | 0.651 |
| inlA; internalin A | -0.221 | 0.572 | 0.699 | 0.797 |
| INO1, ISYNA1; myo-inositol-1-phosphate synthase [EC:5.5.1.4] | -0.144 | 0.334 | 0.667 | 0.778 |
| INV, sacA; beta-fructofuranosidase [EC:3.2.1.26] | -0.496 | 0.487 | 0.311 | 0.651 |
| ispB; octaprenyl-diphosphate synthase [EC:2.5.1.90] | -0.230 | 0.296 | 0.438 | 0.651 |
| ispD; 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [EC:2.7.7.60] | -0.232 | 0.307 | 0.452 | 0.651 |
| ispE; 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase [EC:2.7.1.148] | -0.230 | 0.296 | 0.438 | 0.651 |
| ispF; 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [EC:4.6.1.12] | -0.230 | 0.296 | 0.438 | 0.651 |
| ispH, lytB; 4-hydroxy-3-methylbut-2-en-1-yl diphosphate reductase [EC:1.17.7.4] | -0.230 | 0.296 | 0.438 | 0.651 |
| K00243; uncharacterized protein | -0.230 | 0.296 | 0.438 | 0.651 |
| K01163; uncharacterized protein | -0.113 | 0.281 | 0.688 | 0.797 |
| K02477; two-component system, LytTR family, response regulator | -0.221 | 0.571 | 0.700 | 0.797 |
| K02481; two-component system, NtrC family, response regulator | 2.304 | 1.989 | 0.248 | 0.651 |
| K06871; uncharacterized protein | -0.311 | 0.327 | 0.343 | 0.651 |
| K06872; uncharacterized protein | -0.253 | 0.518 | 0.626 | 0.741 |
| K06885; uncharacterized protein | -0.115 | 0.303 | 0.705 | 0.799 |
| K06889; uncharacterized protein | -0.239 | 0.319 | 0.455 | 0.651 |
| K06890; uncharacterized protein | -0.230 | 0.296 | 0.438 | 0.651 |
| K06904; uncharacterized protein | 0.358 | 0.550 | 0.516 | 0.651 |
| K06915; uncharacterized protein | -17.599 | 3454.822 | 0.996 | 0.998 |
| K06921; uncharacterized protein | -0.594 | 0.750 | 0.429 | 0.651 |
| K06926; uncharacterized protein | -0.511 | 0.365 | 0.164 | 0.651 |
| K06950; uncharacterized protein | -0.225 | 0.298 | 0.452 | 0.651 |
| K06973; uncharacterized protein | -0.754 | 1.120 | 0.502 | 0.651 |
| K06975; uncharacterized protein | -0.428 | 0.414 | 0.303 | 0.651 |
| K07000; uncharacterized protein | -0.219 | 0.301 | 0.469 | 0.651 |
| K07001; NTE family protein | -0.294 | 0.304 | 0.334 | 0.651 |
| K07003; uncharacterized protein | -0.248 | 0.345 | 0.474 | 0.651 |
| K07005; uncharacterized protein | -0.229 | 0.336 | 0.497 | 0.651 |
| K07007; uncharacterized protein | -0.226 | 0.306 | 0.462 | 0.651 |
| K07011; uncharacterized protein | -0.246 | 0.298 | 0.412 | 0.651 |
| K07014; uncharacterized protein | -0.335 | 0.976 | 0.732 | 0.826 |
| K07025; putative hydrolase of the HAD superfamily | -0.232 | 0.310 | 0.456 | 0.651 |
| K07027; glycosyltransferase 2 family protein | -0.230 | 0.296 | 0.438 | 0.651 |
| K07037; uncharacterized protein | -0.230 | 0.296 | 0.438 | 0.651 |
| K07043; uncharacterized protein | -0.754 | 1.120 | 0.502 | 0.651 |
| K07052; uncharacterized protein | -0.250 | 0.275 | 0.365 | 0.651 |
| K07054; uncharacterized protein | -0.049 | 0.358 | 0.891 | 0.966 |
| K07058; membrane protein | -0.229 | 0.309 | 0.461 | 0.651 |
| K07078; uncharacterized protein | -17.599 | 3454.822 | 0.996 | 0.998 |
| K07082; UPF0755 protein | -0.230 | 0.296 | 0.438 | 0.651 |
| K07085; putative transport protein | -0.247 | 0.292 | 0.398 | 0.651 |
| K07088; uncharacterized protein | -0.089 | 0.344 | 0.796 | 0.879 |
| K07090; uncharacterized protein | -0.307 | 0.429 | 0.475 | 0.651 |
| K07095; uncharacterized protein | -0.256 | 0.314 | 0.416 | 0.651 |
| K07098; uncharacterized protein | -0.230 | 0.306 | 0.452 | 0.651 |
| K07126; uncharacterized protein | -0.275 | 0.524 | 0.601 | 0.723 |
| K07133; uncharacterized protein | -0.250 | 0.312 | 0.424 | 0.651 |
| K07137; uncharacterized protein | -0.230 | 0.296 | 0.438 | 0.651 |
| K07139; uncharacterized protein | -0.228 | 0.293 | 0.436 | 0.651 |
| K07150; uncharacterized protein | -0.754 | 1.120 | 0.502 | 0.651 |
| K07164; uncharacterized protein | -0.230 | 0.296 | 0.438 | 0.651 |
| K07166; ACT domain-containing protein | 18.519 | 3411.091 | 0.996 | 0.998 |
| K07220; uncharacterized protein | 2.304 | 1.989 | 0.248 | 0.651 |
| K07270; glycosyl transferase, family 25 | 0.358 | 0.550 | 0.516 | 0.651 |
| K07317; adenine-specific DNA-methyltransferase [EC:2.1.1.72] | 0.248 | 1.294 | 0.848 | 0.924 |
| K07318; adenine-specific DNA-methyltransferase [EC:2.1.1.72] | -0.297 | 0.684 | 0.664 | 0.776 |
| K07387; putative metalloprotease [EC:3.4.24.-] | -0.247 | 0.316 | 0.435 | 0.651 |
| K07454; putative restriction endonuclease | -0.001 | 1.049 | 0.999 | 0.999 |
| K07481; transposase, IS5 family | -0.256 | 0.444 | 0.566 | 0.693 |
| K07483; transposase | -0.398 | 0.499 | 0.426 | 0.651 |
| K07484; transposase | 18.519 | 3411.091 | 0.996 | 0.998 |
| K07491; putative transposase | 2.304 | 1.989 | 0.248 | 0.651 |
| K07496; putative transposase | 2.304 | 1.989 | 0.248 | 0.651 |
| K07504; predicted type IV restriction endonuclease | 0.054 | 0.493 | 0.913 | 0.976 |
| K07727; putative transcriptional regulator | -0.211 | 0.577 | 0.715 | 0.809 |
| K07729; putative transcriptional regulator | -0.054 | 0.371 | 0.885 | 0.961 |
| K08303; putative protease [EC:3.4.-.-] | -0.225 | 0.301 | 0.457 | 0.651 |
| K08884; serine/threonine protein kinase, bacterial [EC:2.7.11.1] | -0.098 | 0.320 | 0.761 | 0.844 |
| K08961; chondroitin-sulfate-ABC endolyase/exolyase [EC:4.2.2.20 4.2.2.21] | -0.677 | 0.652 | 0.301 | 0.651 |
| K08987; putative membrane protein | 0.038 | 0.353 | 0.913 | 0.976 |
| K08998; uncharacterized protein | -0.224 | 0.269 | 0.406 | 0.651 |
| K08999; uncharacterized protein | -0.216 | 0.297 | 0.468 | 0.651 |
| K09117; uncharacterized protein | -0.480 | 0.429 | 0.265 | 0.651 |
| K09124; uncharacterized protein | 0.092 | 1.404 | 0.948 | 0.986 |
| K09125; uncharacterized protein | -0.219 | 0.301 | 0.468 | 0.651 |
| K09137; uncharacterized protein | 0.358 | 0.550 | 0.516 | 0.651 |
| K09155; uncharacterized protein | -0.226 | 0.296 | 0.445 | 0.651 |
| K09157; uncharacterized protein | 18.519 | 3411.091 | 0.996 | 0.998 |
| K09704; uncharacterized protein | -0.223 | 0.285 | 0.435 | 0.651 |
| K09797; uncharacterized protein | -0.400 | 0.427 | 0.350 | 0.651 |
| K09805; uncharacterized protein | -0.659 | 0.562 | 0.243 | 0.651 |
| K09861; uncharacterized protein | -0.230 | 0.296 | 0.438 | 0.651 |
| K09922; uncharacterized protein | 18.519 | 3411.091 | 0.996 | 0.998 |
| K09924; uncharacterized protein | -0.280 | 0.500 | 0.577 | 0.703 |
| K09955; uncharacterized protein | -0.691 | 0.615 | 0.263 | 0.651 |
| K09973; uncharacterized protein | 0.151 | 1.352 | 0.911 | 0.976 |
| K09975; uncharacterized protein | -0.172 | 0.359 | 0.633 | 0.746 |
| K13652; AraC family transcriptional regulator | -2.232 | 1.933 | 0.250 | 0.651 |
| K15977; putative oxidoreductase | -0.050 | 0.281 | 0.859 | 0.934 |
| KAE1, tsaD, QRI7; N6-L-threonylcarbamoyladenine synthase [EC:2.3.1.234] | -0.230 | 0.296 | 0.438 | 0.651 |
| KARS, lysS; lysyl-tRNA synthetase, class II [EC:6.1.1.6] | -0.230 | 0.296 | 0.438 | 0.651 |
| kbl, GCAT; glycine C-acetyltransferase [EC:2.3.1.29] | -0.323 | 0.372 | 0.387 | 0.651 |
| kch, trkA, mthK, pch; voltage-gated potassium channel | 0.358 | 0.550 | 0.516 | 0.651 |
| kdgK; 2-dehydro-3-deoxygluconokinase [EC:2.7.1.45] | -0.577 | 0.590 | 0.329 | 0.651 |
| kdpA; K+-transporting ATPase ATPase A chain [EC:3.6.3.12] | 2.304 | 1.989 | 0.248 | 0.651 |
| kdpB; K+-transporting ATPase ATPase B chain [EC:3.6.3.12] | 2.304 | 1.989 | 0.248 | 0.651 |
| kdpC; K+-transporting ATPase ATPase C chain [EC:3.6.3.12] | 2.304 | 1.989 | 0.248 | 0.651 |
| kdpD; two-component system, OmpR family, sensor histidine kinase KdpD [EC:2.7.13.3] | 2.304 | 1.989 | 0.248 | 0.651 |
| kdsA; 2-dehydro-3-deoxyphosphooctonate aldolase (KDO 8-P synthase) [EC:2.5.1.55] | -0.230 | 0.296 | 0.438 | 0.651 |
| kdsB; 3-deoxy-manno-octulosonate cytidylyltransferase (CMP-KDO synthetase) [EC:2.7.7.38] | -0.243 | 0.292 | 0.407 | 0.651 |
| kdsC; 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase (KDO 8-P phosphatase) [EC:3.1.3.45] | -0.232 | 0.301 | 0.441 | 0.651 |
| kdsD, kpsF; arabinose-5-phosphate isomerase [EC:5.3.1.13] | -0.230 | 0.296 | 0.438 | 0.651 |
| kdtA, waaA; 3-deoxy-D-manno-octulosonic-acid transferase [EC:2.4.99.12 2.4.99.13 2.4.99.14 2.4.99.15] | -0.230 | 0.296 | 0.438 | 0.651 |
| kduI; 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase [EC:5.3.1.17] | -0.676 | 0.652 | 0.302 | 0.651 |
| korA, oorA, oforA; 2-oxoglutarate/2-oxoacid ferredoxin oxidoreductase subunit alpha [EC:1.2.7.3 1.2.7.11] | -0.241 | 0.306 | 0.433 | 0.651 |
| korB, oorB, oforB; 2-oxoglutarate/2-oxoacid ferredoxin oxidoreductase subunit beta [EC:1.2.7.3 1.2.7.11] | -0.241 | 0.306 | 0.433 | 0.651 |
| korC, oorC; 2-oxoglutarate ferredoxin oxidoreductase subunit gamma [EC:1.2.7.3] | -0.230 | 0.296 | 0.438 | 0.651 |
| korD, oorD; 2-oxoglutarate ferredoxin oxidoreductase subunit delta [EC:1.2.7.3] | -0.230 | 0.296 | 0.438 | 0.651 |
| ksgA; 16S rRNA (adenine1518-N6/adenine1519-N6)-dimethyltransferase [EC:2.1.1.182] | -0.230 | 0.296 | 0.438 | 0.651 |
| kup; KUP system potassium uptake protein | -0.098 | 0.312 | 0.753 | 0.841 |
| lacA; galactoside O-acetyltransferase [EC:2.3.1.18] | -0.062 | 0.367 | 0.865 | 0.940 |
| lacI, galR; LacI family transcriptional regulator | -0.239 | 0.298 | 0.424 | 0.651 |
| lacZ; beta-galactosidase [EC:3.2.1.23] | -0.222 | 0.291 | 0.446 | 0.651 |
| LARS, leuS; leucyl-tRNA synthetase [EC:6.1.1.4] | -0.222 | 0.299 | 0.458 | 0.651 |
| lctP; lactate permease | -0.289 | 0.546 | 0.598 | 0.723 |
| ldhA; D-lactate dehydrogenase [EC:1.1.1.28] | -0.088 | 0.288 | 0.760 | 0.844 |
| lemA; LemA protein | -0.230 | 0.290 | 0.428 | 0.651 |
| lepA; GTP-binding protein LepA | -0.230 | 0.296 | 0.438 | 0.651 |
| lepB; signal peptidase I [EC:3.4.21.89] | -0.223 | 0.297 | 0.456 | 0.651 |
| leuA, IMS; 2-isopropylmalate synthase [EC:2.3.3.13] | -0.100 | 1.127 | 0.930 | 0.976 |
| leuB, IMDH; 3-isopropylmalate dehydrogenase [EC:1.1.1.85] | -0.100 | 1.127 | 0.930 | 0.976 |
| leuC, IPMI-L; 3-isopropylmalate/(R)-2-methylmalate dehydratase large subunit [EC:4.2.1.33 4.2.1.35] | -0.100 | 1.127 | 0.930 | 0.976 |
| leuD, IPMI-S; 3-isopropylmalate/(R)-2-methylmalate dehydratase small subunit [EC:4.2.1.33 4.2.1.35] | -0.100 | 1.127 | 0.930 | 0.976 |
| licD; lipopolysaccharide cholinephosphotransferase [EC:2.7.8.-] | -0.176 | 0.308 | 0.568 | 0.695 |
| lipA; lipoyl synthase [EC:2.8.1.8] | -0.230 | 0.317 | 0.469 | 0.651 |
| lldE; L-lactate dehydrogenase complex protein LldE | -0.233 | 0.320 | 0.468 | 0.651 |
| lldF; L-lactate dehydrogenase complex protein LldF | -0.233 | 0.320 | 0.468 | 0.651 |
| lldG; L-lactate dehydrogenase complex protein LldG | -0.233 | 0.320 | 0.468 | 0.651 |
| lolA; outer membrane lipoprotein carrier protein | -0.750 | 0.852 | 0.380 | 0.651 |
| lolC_E; lipoprotein-releasing system permease protein | -0.230 | 0.296 | 0.438 | 0.651 |
| lolD; lipoprotein-releasing system ATP-binding protein [EC:3.6.3.-] | -0.230 | 0.296 | 0.438 | 0.651 |
| lon; ATP-dependent Lon protease [EC:3.4.21.53] | -0.220 | 0.308 | 0.477 | 0.651 |
| lplA, lplJ; lipoate—protein ligase [EC:6.3.1.20] | -0.717 | 0.722 | 0.322 | 0.651 |
| lptB; lipopolysaccharide export system ATP-binding protein [EC:3.6.3.-] | -0.230 | 0.296 | 0.438 | 0.651 |
| lptF; lipopolysaccharide export system permease protein | -0.230 | 0.296 | 0.438 | 0.651 |
| lptG; lipopolysaccharide export system permease protein | -0.230 | 0.296 | 0.438 | 0.651 |
| lpxA; UDP-N-acetylglucosamine acyltransferase [EC:2.3.1.129] | -0.230 | 0.296 | 0.438 | 0.651 |
| lpxB; lipid-A-disaccharide synthase [EC:2.4.1.182] | -0.230 | 0.296 | 0.438 | 0.651 |
| lpxC-fabZ; UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase / 3-hydroxyacyl-[acyl-carrier-protein] dehydratase [EC:3.5.1.108 4.2.1.59] | -0.230 | 0.296 | 0.438 | 0.651 |
| lpxD; UDP-3-O-[3-hydroxymyristoyl] glucosamine N-acyltransferase [EC:2.3.1.191] | -0.230 | 0.296 | 0.438 | 0.651 |
| lpxH; UDP-2,3-diacylglucosamine hydrolase [EC:3.6.1.54] | -0.230 | 0.296 | 0.438 | 0.651 |
| lpxK; tetraacyldisaccharide 4’-kinase [EC:2.7.1.130] | -0.230 | 0.296 | 0.438 | 0.651 |
| lpxL, htrB; Kdo2-lipid IVA lauroyltransferase [EC:2.3.1.241] | -0.230 | 0.296 | 0.438 | 0.651 |
| lrp; Lrp/AsnC family transcriptional regulator, leucine-responsive regulatory protein | -0.042 | 0.347 | 0.904 | 0.976 |
| lspA; signal peptidase II [EC:3.4.23.36] | -0.230 | 0.296 | 0.438 | 0.651 |
| ltaE; threonine aldolase [EC:4.1.2.48] | -0.170 | 0.761 | 0.824 | 0.900 |
| ltrA; RNA-directed DNA polymerase [EC:2.7.7.49] | 2.304 | 1.989 | 0.248 | 0.651 |
| luxS; S-ribosylhomocysteine lyase [EC:4.4.1.21] | -0.263 | 0.312 | 0.401 | 0.651 |
| LYS1; saccharopine dehydrogenase (NAD+, L-lysine forming) [EC:1.5.1.7] | -0.230 | 0.296 | 0.438 | 0.651 |
| lysA; diaminopimelate decarboxylase [EC:4.1.1.20] | -0.274 | 0.307 | 0.374 | 0.651 |
| lysC; aspartate kinase [EC:2.7.2.4] | -0.230 | 0.296 | 0.438 | 0.651 |
| maa; maltose O-acetyltransferase [EC:2.3.1.79] | -0.230 | 0.328 | 0.484 | 0.651 |
| maf; septum formation protein | -0.230 | 0.296 | 0.438 | 0.651 |
| mal; methylaspartate ammonia-lyase [EC:4.3.1.2] | -0.221 | 0.572 | 0.699 | 0.797 |
| malQ; 4-alpha-glucanotransferase [EC:2.4.1.25] | -0.230 | 0.296 | 0.438 | 0.651 |
| malY, malT; maltose/moltooligosaccharide transporter | -0.228 | 0.293 | 0.436 | 0.651 |
| malZ; alpha-glucosidase [EC:3.2.1.20] | -0.117 | 0.305 | 0.702 | 0.797 |
| MAN; mannan endo-1,4-beta-mannosidase [EC:3.2.1.78] | -0.189 | 0.299 | 0.528 | 0.663 |
| manA, MPI; mannose-6-phosphate isomerase [EC:5.3.1.8] | -0.230 | 0.296 | 0.438 | 0.651 |
| manB; phosphomannomutase [EC:5.4.2.8] | -0.230 | 0.296 | 0.438 | 0.651 |
| manC, cpsB; mannose-1-phosphate guanylyltransferase [EC:2.7.7.13] | -0.230 | 0.296 | 0.438 | 0.651 |
| map; methionyl aminopeptidase [EC:3.4.11.18] | -0.224 | 0.289 | 0.440 | 0.651 |
| marC; multiple antibiotic resistance protein | -0.754 | 1.120 | 0.502 | 0.651 |
| MARS, metG; methionyl-tRNA synthetase [EC:6.1.1.10] | -0.230 | 0.296 | 0.438 | 0.651 |
| mbtN, fadE14; acyl-ACP dehydrogenase [EC:1.3.99.-] | -0.754 | 1.120 | 0.502 | 0.651 |
| mcrA; 5-methylcytosine-specific restriction enzyme A [EC:3.1.21.-] | 0.358 | 0.550 | 0.516 | 0.651 |
| mcrC; 5-methylcytosine-specific restriction enzyme subunit McrC | 0.358 | 0.550 | 0.516 | 0.651 |
| mdh; malate dehydrogenase [EC:1.1.1.37] | -0.230 | 0.296 | 0.438 | 0.651 |
| menA; 1,4-dihydroxy-2-naphthoate octaprenyltransferase [EC:2.5.1.74 2.5.1.-] | -0.230 | 0.296 | 0.438 | 0.651 |
| menB; naphthoate synthase [EC:4.1.3.36] | -0.230 | 0.296 | 0.438 | 0.651 |
| menD; 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase [EC:2.2.1.9] | -0.230 | 0.296 | 0.438 | 0.651 |
| menE; O-succinylbenzoic acid—CoA ligase [EC:6.2.1.26] | -0.219 | 0.301 | 0.468 | 0.651 |
| mepS, spr; murein DD-endopeptidase / murein LD-carboxypeptidase [EC:3.4.-.- 3.4.17.13] | -0.754 | 1.120 | 0.502 | 0.651 |
| metA; homoserine O-succinyltransferase/O-acetyltransferase [EC:2.3.1.46 2.3.1.31] | -0.274 | 0.307 | 0.374 | 0.651 |
| metF, MTHFR; methylenetetrahydrofolate reductase (NADPH) [EC:1.5.1.20] | -0.264 | 0.307 | 0.391 | 0.651 |
| metH, MTR; 5-methyltetrahydrofolate–homocysteine methyltransferase [EC:2.1.1.13] | -0.206 | 0.312 | 0.510 | 0.651 |
| metK; S-adenosylmethionine synthetase [EC:2.5.1.6] | -0.243 | 0.292 | 0.407 | 0.651 |
| metY; O-acetylhomoserine (thiol)-lyase [EC:2.5.1.49] | -0.208 | 0.306 | 0.498 | 0.651 |
| mfd; transcription-repair coupling factor (superfamily II helicase) [EC:3.6.4.-] | -0.230 | 0.296 | 0.438 | 0.651 |
| mgp; 4-O-beta-D-mannosyl-D-glucose phosphorylase [EC:2.4.1.281] | -0.588 | 0.587 | 0.318 | 0.651 |
| mgtC; putative Mg2+ transporter-C (MgtC) family protein | -0.280 | 0.308 | 0.365 | 0.651 |
| miaA, TRIT1; tRNA dimethylallyltransferase [EC:2.5.1.75] | -0.230 | 0.296 | 0.438 | 0.651 |
| miaB; tRNA-2-methylthio-N6-dimethylallyladenosine synthase [EC:2.8.4.3] | -0.230 | 0.296 | 0.438 | 0.651 |
| mlaD, linM; phospholipid/cholesterol/gamma-HCH transport system substrate-binding protein | -0.230 | 0.296 | 0.438 | 0.651 |
| mlaE, linK; phospholipid/cholesterol/gamma-HCH transport system permease protein | -0.230 | 0.296 | 0.438 | 0.651 |
| mlaF, linL, mkl; phospholipid/cholesterol/gamma-HCH transport system ATP-binding protein | -0.230 | 0.296 | 0.438 | 0.651 |
| mltD, dniR; membrane-bound lytic murein transglycosylase D [EC:4.2.2.-] | -0.235 | 0.297 | 0.430 | 0.651 |
| mltF; membrane-bound lytic murein transglycosylase F [EC:4.2.2.-] | -0.468 | 0.400 | 0.244 | 0.651 |
| MMAB, pduO; cob(I)alamin adenosyltransferase [EC:2.5.1.17] | -0.263 | 0.312 | 0.401 | 0.651 |
| mnmA, trmU; tRNA-uridine 2-sulfurtransferase [EC:2.8.1.13] | -0.230 | 0.296 | 0.438 | 0.651 |
| mnmE, trmE, MSS1; tRNA modification GTPase [EC:3.6.-.-] | -0.230 | 0.296 | 0.438 | 0.651 |
| mntH; manganese transport protein | -0.224 | 0.319 | 0.483 | 0.651 |
| moaA, CNX2; GTP 3’,8-cyclase [EC:4.1.99.22] | -0.137 | 0.613 | 0.823 | 0.900 |
| mod; adenine-specific DNA-methyltransferase [EC:2.1.1.72] | -0.171 | 0.451 | 0.705 | 0.799 |
| modF; molybdate transport system ATP-binding protein | 18.519 | 3411.091 | 0.996 | 0.998 |
| motB; chemotaxis protein MotB | -0.230 | 0.296 | 0.438 | 0.651 |
| moxR; MoxR-like ATPase [EC:3.6.3.-] | -0.170 | 0.284 | 0.551 | 0.683 |
| mp2; beta-1,4-mannooligosaccharide/beta-1,4-mannosyl-N-acetylglucosamine phosphorylase [EC:2.4.1.319 2.4.1.320] | -0.234 | 0.288 | 0.419 | 0.651 |
| mraW, rsmH; 16S rRNA (cytosine1402-N4)-methyltransferase [EC:2.1.1.199] | -0.230 | 0.296 | 0.438 | 0.651 |
| mraY; phospho-N-acetylmuramoyl-pentapeptide-transferase [EC:2.7.8.13] | -0.230 | 0.296 | 0.438 | 0.651 |
| mraZ; MraZ protein | -0.262 | 0.309 | 0.398 | 0.651 |
| mrcA; penicillin-binding protein 1A [EC:2.4.1.129 3.4.16.4] | -0.230 | 0.296 | 0.438 | 0.651 |
| mrdA; penicillin-binding protein 2 [EC:3.4.16.4] | -0.194 | 0.303 | 0.523 | 0.657 |
| mreB; rod shape-determining protein MreB and related proteins | -0.230 | 0.296 | 0.438 | 0.651 |
| mreC; rod shape-determining protein MreC | -0.230 | 0.296 | 0.438 | 0.651 |
| mrp, NUBPL; ATP-binding protein involved in chromosome partitioning | -0.230 | 0.296 | 0.438 | 0.651 |
| mrr; restriction system protein | -0.301 | 0.567 | 0.596 | 0.722 |
| msbA; ATP-binding cassette, subfamily B, bacterial MsbA [EC:3.6.3.-] | -0.230 | 0.296 | 0.438 | 0.651 |
| mscL; large conductance mechanosensitive channel | -0.230 | 0.296 | 0.438 | 0.651 |
| mscS; small conductance mechanosensitive channel | -0.754 | 1.120 | 0.502 | 0.651 |
| msrAB; peptide methionine sulfoxide reductase msrA/msrB [EC:1.8.4.11 1.8.4.12] | -0.230 | 0.296 | 0.438 | 0.651 |
| mtaB; threonylcarbamoyladenosine tRNA methylthiotransferase MtaB [EC:2.8.4.5] | -0.230 | 0.296 | 0.438 | 0.651 |
| MTFMT, fmt; methionyl-tRNA formyltransferase [EC:2.1.2.9] | -0.230 | 0.296 | 0.438 | 0.651 |
| mtgA; monofunctional glycosyltransferase [EC:2.4.1.129] | -0.230 | 0.296 | 0.438 | 0.651 |
| MTHFS; 5-formyltetrahydrofolate cyclo-ligase [EC:6.3.3.2] | -0.230 | 0.296 | 0.438 | 0.651 |
| mtnN, mtn, pfs; adenosylhomocysteine nucleosidase [EC:3.2.2.9] | -0.230 | 0.296 | 0.438 | 0.651 |
| MuB; ATP-dependent target DNA activator [EC:3.6.1.3] | -0.261 | 0.938 | 0.781 | 0.865 |
| murA; UDP-N-acetylglucosamine 1-carboxyvinyltransferase [EC:2.5.1.7] | -0.230 | 0.296 | 0.438 | 0.651 |
| murB; UDP-N-acetylmuramate dehydrogenase [EC:1.3.1.98] | -0.230 | 0.296 | 0.438 | 0.651 |
| murC; UDP-N-acetylmuramate–alanine ligase [EC:6.3.2.8] | -0.230 | 0.296 | 0.438 | 0.651 |
| murD; UDP-N-acetylmuramoylalanine–D-glutamate ligase [EC:6.3.2.9] | -0.230 | 0.296 | 0.438 | 0.651 |
| murE; UDP-N-acetylmuramoyl-L-alanyl-D-glutamate–2,6-diaminopimelate ligase [EC:6.3.2.13] | -0.230 | 0.296 | 0.438 | 0.651 |
| murF; UDP-N-acetylmuramoyl-tripeptide–D-alanyl-D-alanine ligase [EC:6.3.2.10] | -0.230 | 0.296 | 0.438 | 0.651 |
| murG; UDP-N-acetylglucosamine–N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase [EC:2.4.1.227] | -0.230 | 0.296 | 0.438 | 0.651 |
| murI; glutamate racemase [EC:5.1.1.3] | -0.230 | 0.296 | 0.438 | 0.651 |
| murQ; N-acetylmuramic acid 6-phosphate etherase [EC:4.2.1.126] | -0.747 | 0.519 | 0.152 | 0.651 |
| mutL; DNA mismatch repair protein MutL | -0.230 | 0.296 | 0.438 | 0.651 |
| mutS; DNA mismatch repair protein MutS | -0.230 | 0.296 | 0.438 | 0.651 |
| mutS2; DNA mismatch repair protein MutS2 | -0.230 | 0.296 | 0.438 | 0.651 |
| mutT, NUDT15, MTH2; 8-oxo-dGTP diphosphatase [EC:3.6.1.55] | -0.128 | 0.324 | 0.693 | 0.797 |
| mutY; A/G-specific adenine glycosylase [EC:3.2.2.31] | -0.230 | 0.296 | 0.438 | 0.651 |
| mviM; virulence factor | -0.262 | 0.328 | 0.427 | 0.651 |
| mvpA, vapC; tRNA(fMet)-specific endonuclease VapC [EC:3.1.-.-] | 0.141 | 0.367 | 0.701 | 0.797 |
| nadA; quinolinate synthase [EC:2.5.1.72] | -0.172 | 0.328 | 0.602 | 0.723 |
| nadB; L-aspartate oxidase [EC:1.4.3.16] | -0.172 | 0.328 | 0.602 | 0.723 |
| nadC, QPRT; nicotinate-nucleotide pyrophosphorylase (carboxylating) [EC:2.4.2.19] | -0.172 | 0.328 | 0.602 | 0.723 |
| nadD; nicotinate-nucleotide adenylyltransferase [EC:2.7.7.18] | -0.230 | 0.296 | 0.438 | 0.651 |
| nagB, GNPDA; glucosamine-6-phosphate deaminase [EC:3.5.99.6] | -0.226 | 0.290 | 0.438 | 0.651 |
| NAGLU; alpha-N-acetylglucosaminidase [EC:3.2.1.50] | -0.855 | 0.801 | 0.287 | 0.651 |
| NARS, asnS; asparaginyl-tRNA synthetase [EC:6.1.1.22] | -0.230 | 0.296 | 0.438 | 0.651 |
| ndh; NADH dehydrogenase [EC:1.6.99.3] | -0.211 | 0.290 | 0.469 | 0.651 |
| ndk, NME; nucleoside-diphosphate kinase [EC:2.7.4.6] | -0.754 | 1.120 | 0.502 | 0.651 |
| NEU1; sialidase-1 [EC:3.2.1.18] | -0.234 | 0.288 | 0.419 | 0.651 |
| neuA; N-acylneuraminate cytidylyltransferase [EC:2.7.7.43] | -2.232 | 1.933 | 0.250 | 0.651 |
| nfrA2; FMN reductase [NAD(P)H] [EC:1.5.1.39] | -0.230 | 0.296 | 0.438 | 0.651 |
| nhaA; Na+:H+ antiporter, NhaA family | -0.205 | 0.309 | 0.509 | 0.651 |
| nhaC; Na+:H+ antiporter, NhaC family | 18.519 | 3411.091 | 0.996 | 0.998 |
| norB, norC; MFS transporter, DHA2 family, multidrug resistance protein | 2.304 | 1.989 | 0.248 | 0.651 |
| norB; nitric oxide reductase subunit B [EC:1.7.2.5] | 0.054 | 0.493 | 0.913 | 0.976 |
| nosD; nitrous oxidase accessory protein | -0.319 | 0.983 | 0.746 | 0.834 |
| nosF; Cu-processing system ATP-binding protein | -0.319 | 0.983 | 0.746 | 0.834 |
| nosL; copper chaperone NosL | -0.319 | 0.983 | 0.746 | 0.834 |
| nosY; Cu-processing system permease protein | -0.319 | 0.983 | 0.746 | 0.834 |
| nosZ; nitrous-oxide reductase [EC:1.7.2.4] | -0.319 | 0.983 | 0.746 | 0.834 |
| npdA; NAD-dependent deacetylase [EC:3.5.1.-] | -0.263 | 0.312 | 0.401 | 0.651 |
| nqrA; Na+-transporting NADH:ubiquinone oxidoreductase subunit A [EC:1.6.5.8] | -0.230 | 0.296 | 0.438 | 0.651 |
| nqrB; Na+-transporting NADH:ubiquinone oxidoreductase subunit B [EC:1.6.5.8] | -0.230 | 0.296 | 0.438 | 0.651 |
| nqrC; Na+-transporting NADH:ubiquinone oxidoreductase subunit C [EC:1.6.5.8] | -0.230 | 0.296 | 0.438 | 0.651 |
| nqrD; Na+-transporting NADH:ubiquinone oxidoreductase subunit D [EC:1.6.5.8] | -0.230 | 0.296 | 0.438 | 0.651 |
| nqrE; Na+-transporting NADH:ubiquinone oxidoreductase subunit E [EC:1.6.5.8] | -0.230 | 0.296 | 0.438 | 0.651 |
| nqrF; Na+-transporting NADH:ubiquinone oxidoreductase subunit F [EC:1.6.5.8] | -0.230 | 0.296 | 0.438 | 0.651 |
| nrdG; anaerobic ribonucleoside-triphosphate reductase activating protein [EC:1.97.1.4] | -0.230 | 0.296 | 0.438 | 0.651 |
| nrfA; nitrite reductase (cytochrome c-552) [EC:1.7.2.2] | -0.217 | 0.299 | 0.469 | 0.651 |
| nrfH; cytochrome c nitrite reductase small subunit | -0.217 | 0.299 | 0.469 | 0.651 |
| nrnA; bifunctional oligoribonuclease and PAP phosphatase NrnA [EC:3.1.3.7 3.1.13.3] | -0.230 | 0.296 | 0.438 | 0.651 |
| nspC; carboxynorspermidine decarboxylase [EC:4.1.1.96] | -0.230 | 0.296 | 0.438 | 0.651 |
| NTH; endonuclease III [EC:4.2.99.18] | -0.230 | 0.296 | 0.438 | 0.651 |
| nuoA; NADH-quinone oxidoreductase subunit A [EC:1.6.5.3] | -0.230 | 0.296 | 0.438 | 0.651 |
| nuoB; NADH-quinone oxidoreductase subunit B [EC:1.6.5.3] | -0.230 | 0.296 | 0.438 | 0.651 |
| nuoCD; NADH-quinone oxidoreductase subunit C/D [EC:1.6.5.3] | -0.230 | 0.296 | 0.438 | 0.651 |
| nuoH; NADH-quinone oxidoreductase subunit H [EC:1.6.5.3] | -0.230 | 0.296 | 0.438 | 0.651 |
| nuoI; NADH-quinone oxidoreductase subunit I [EC:1.6.5.3] | -0.230 | 0.296 | 0.438 | 0.651 |
| nuoJ; NADH-quinone oxidoreductase subunit J [EC:1.6.5.3] | -0.230 | 0.296 | 0.438 | 0.651 |
| nuoK; NADH-quinone oxidoreductase subunit K [EC:1.6.5.3] | -0.230 | 0.296 | 0.438 | 0.651 |
| nuoL; NADH-quinone oxidoreductase subunit L [EC:1.6.5.3] | -0.230 | 0.296 | 0.438 | 0.651 |
| nuoM; NADH-quinone oxidoreductase subunit M [EC:1.6.5.3] | -0.230 | 0.296 | 0.438 | 0.651 |
| nuoN; NADH-quinone oxidoreductase subunit N [EC:1.6.5.3] | -0.230 | 0.296 | 0.438 | 0.651 |
| nupG; MFS transporter, NHS family, nucleoside permease | -0.230 | 0.296 | 0.438 | 0.651 |
| nusA; N utilization substance protein A | -0.230 | 0.296 | 0.438 | 0.651 |
| nusB; N utilization substance protein B | -0.230 | 0.296 | 0.438 | 0.651 |
| nusG; transcriptional antiterminator NusG | -0.230 | 0.296 | 0.438 | 0.651 |
| obgE, cgtA; GTPase [EC:3.6.5.-] | -0.230 | 0.296 | 0.438 | 0.651 |
| ogt, MGMT; methylated-DNA-[protein]-cysteine S-methyltransferase [EC:2.1.1.63] | -0.155 | 0.301 | 0.607 | 0.723 |
| ompX; outer membrane protein X | -0.096 | 0.328 | 0.770 | 0.853 |
| oprO_P; phosphate-selective porin OprO and OprP | -0.180 | 0.298 | 0.546 | 0.680 |
| oxyR; LysR family transcriptional regulator, hydrogen peroxide-inducible genes activator | -0.213 | 0.298 | 0.476 | 0.651 |
| pabB; para-aminobenzoate synthetase component I [EC:2.6.1.85] | -0.158 | 0.328 | 0.632 | 0.746 |
| pabC; 4-amino-4-deoxychorismate lyase [EC:4.1.3.38] | -0.158 | 0.328 | 0.632 | 0.746 |
| padR; PadR family transcriptional regulator, regulatory protein PadR | -1.243 | 1.127 | 0.272 | 0.651 |
| panB; 3-methyl-2-oxobutanoate hydroxymethyltransferase [EC:2.1.2.11] | -0.213 | 0.298 | 0.477 | 0.651 |
| panC; pantoate–beta-alanine ligase [EC:6.3.2.1] | -0.213 | 0.298 | 0.477 | 0.651 |
| panD; aspartate 1-decarboxylase [EC:4.1.1.11] | -0.213 | 0.298 | 0.477 | 0.651 |
| panE, apbA; 2-dehydropantoate 2-reductase [EC:1.1.1.169] | -0.220 | 0.329 | 0.504 | 0.651 |
| parA, soj; chromosome partitioning protein | -0.290 | 0.310 | 0.351 | 0.651 |
| parB, spo0J; chromosome partitioning protein, ParB family | -0.230 | 0.296 | 0.438 | 0.651 |
| parC; topoisomerase IV subunit A [EC:5.99.1.-] | -0.230 | 0.296 | 0.438 | 0.651 |
| parE; topoisomerase IV subunit B [EC:5.99.1.-] | -0.230 | 0.296 | 0.438 | 0.651 |
| PARS, proS; prolyl-tRNA synthetase [EC:6.1.1.15] | -0.230 | 0.296 | 0.438 | 0.651 |
| patB, malY; cystathione beta-lyase [EC:4.4.1.8] | -0.267 | 0.319 | 0.403 | 0.651 |
| pbuG; putative MFS transporter, AGZA family, xanthine/uracil permease | -0.219 | 0.301 | 0.468 | 0.651 |
| pcaC; 4-carboxymuconolactone decarboxylase [EC:4.1.1.44] | -0.135 | 0.612 | 0.825 | 0.900 |
| pcnB; poly(A) polymerase [EC:2.7.7.19] | -0.754 | 1.120 | 0.502 | 0.651 |
| PCYT1; choline-phosphate cytidylyltransferase [EC:2.7.7.15] | -0.221 | 0.572 | 0.699 | 0.797 |
| PDF, def; peptide deformylase [EC:3.5.1.88] | -0.230 | 0.296 | 0.438 | 0.651 |
| pdxA; 4-hydroxythreonine-4-phosphate dehydrogenase [EC:1.1.1.262] | -0.230 | 0.296 | 0.438 | 0.651 |
| pdxB; erythronate-4-phosphate dehydrogenase [EC:1.1.1.290] | -0.754 | 1.120 | 0.502 | 0.651 |
| pdxJ; pyridoxine 5-phosphate synthase [EC:2.6.99.2] | -0.754 | 1.120 | 0.502 | 0.651 |
| pdxK, pdxY; pyridoxine kinase [EC:2.7.1.35] | -0.186 | 0.306 | 0.545 | 0.680 |
| pdxS, pdx1; pyridoxal 5’-phosphate synthase pdxS subunit [EC:4.3.3.6] | -0.219 | 0.301 | 0.468 | 0.651 |
| pdxT, pdx2; 5’-phosphate synthase pdxT subunit [EC:4.3.3.6] | -0.219 | 0.301 | 0.468 | 0.651 |
| penP; beta-lactamase class A [EC:3.5.2.6] | -0.298 | 0.574 | 0.604 | 0.723 |
| pepD; dipeptidase D [EC:3.4.13.-] | -0.230 | 0.296 | 0.438 | 0.651 |
| pepE; dipeptidase E [EC:3.4.13.21] | -0.619 | 0.514 | 0.230 | 0.651 |
| pepO; putative endopeptidase [EC:3.4.24.-] | -0.192 | 0.304 | 0.529 | 0.663 |
| pepP; Xaa-Pro aminopeptidase [EC:3.4.11.9] | -0.233 | 0.297 | 0.434 | 0.651 |
| pepT; tripeptide aminopeptidase [EC:3.4.11.4] | -0.230 | 0.296 | 0.438 | 0.651 |
| perR; Fur family transcriptional regulator, peroxide stress response regulator | -0.190 | 0.321 | 0.555 | 0.683 |
| pfkA, PFK; 6-phosphofructokinase 1 [EC:2.7.1.11] | -0.230 | 0.296 | 0.438 | 0.651 |
| pflA, pflC, pflE; pyruvate formate lyase activating enzyme [EC:1.97.1.4] | -0.188 | 0.302 | 0.535 | 0.670 |
| pfp, PFP; diphosphate-dependent phosphofructokinase [EC:2.7.1.90] | -0.230 | 0.296 | 0.438 | 0.651 |
| PGAM, gpmA; 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase [EC:5.4.2.11] | 0.038 | 0.480 | 0.937 | 0.977 |
| PGK, pgk; phosphoglycerate kinase [EC:2.7.2.3] | -0.230 | 0.296 | 0.438 | 0.651 |
| pgm; phosphoglucomutase [EC:5.4.2.2] | -0.230 | 0.296 | 0.438 | 0.651 |
| pgpA; phosphatidylglycerophosphatase A [EC:3.1.3.27] | -0.015 | 0.569 | 0.979 | 0.998 |
| pgsA, PGS1; CDP-diacylglycerol—glycerol-3-phosphate 3-phosphatidyltransferase [EC:2.7.8.5] | -0.227 | 0.517 | 0.662 | 0.775 |
| pheA2; prephenate dehydratase [EC:4.2.1.51] | -0.644 | 0.452 | 0.157 | 0.651 |
| phnP; phosphoribosyl 1,2-cyclic phosphate phosphodiesterase [EC:3.1.4.55] | -0.230 | 0.296 | 0.438 | 0.651 |
| phoH, phoL; phosphate starvation-inducible protein PhoH and related proteins | -0.230 | 0.296 | 0.438 | 0.651 |
| phoN; acid phosphatase (class A) [EC:3.1.3.2] | -0.073 | 0.305 | 0.811 | 0.894 |
| phoR; two-component system, OmpR family, phosphate regulon sensor histidine kinase PhoR [EC:2.7.13.3] | -0.230 | 0.296 | 0.438 | 0.651 |
| pip; proline iminopeptidase [EC:3.4.11.5] | 18.519 | 3411.091 | 0.996 | 0.998 |
| PK, pyk; pyruvate kinase [EC:2.7.1.40] | -0.230 | 0.296 | 0.438 | 0.651 |
| plsC; 1-acyl-sn-glycerol-3-phosphate acyltransferase [EC:2.3.1.51] | -0.230 | 0.296 | 0.438 | 0.651 |
| pnbA; para-nitrobenzyl esterase [EC:3.1.1.-] | -0.508 | 0.875 | 0.562 | 0.689 |
| PNC1; nicotinamidase [EC:3.5.1.19] | 0.358 | 0.550 | 0.516 | 0.651 |
| pncA; nicotinamidase/pyrazinamidase [EC:3.5.1.19 3.5.1.-] | -0.097 | 0.315 | 0.757 | 0.843 |
| pncB, NAPRT1; nicotinate phosphoribosyltransferase [EC:6.3.4.21] | -0.231 | 0.296 | 0.438 | 0.651 |
| pncC; nicotinamide-nucleotide amidase [EC:3.5.1.42] | -0.230 | 0.296 | 0.438 | 0.651 |
| pnp, PNPT1; polyribonucleotide nucleotidyltransferase [EC:2.7.7.8] | -0.230 | 0.296 | 0.438 | 0.651 |
| pnuC; nicotinamide mononucleotide transporter | -0.230 | 0.296 | 0.438 | 0.651 |
| polA; DNA polymerase I [EC:2.7.7.7] | -0.230 | 0.296 | 0.438 | 0.651 |
| por, nifJ; pyruvate-ferredoxin/flavodoxin oxidoreductase [EC:1.2.7.1 1.2.7.-] | -0.230 | 0.296 | 0.438 | 0.651 |
| ppdK; pyruvate, orthophosphate dikinase [EC:2.7.9.1] | -0.230 | 0.296 | 0.438 | 0.651 |
| PPIB, ppiB; peptidyl-prolyl cis-trans isomerase B (cyclophilin B) [EC:5.2.1.8] | -0.146 | 0.307 | 0.634 | 0.747 |
| ppiD; peptidyl-prolyl cis-trans isomerase D [EC:5.2.1.8] | -0.230 | 0.296 | 0.438 | 0.651 |
| ppnK, NADK; NAD+ kinase [EC:2.7.1.23] | -0.230 | 0.296 | 0.438 | 0.651 |
| ppnN; pyrimidine/purine-5’-nucleotide nucleosidase [EC:3.2.2.10 3.2.2.-] | -0.274 | 0.310 | 0.377 | 0.651 |
| PPOX, hemY; protoporphyrinogen/coproporphyrinogen III oxidase [EC:1.3.3.4 1.3.3.15] | -0.232 | 0.293 | 0.430 | 0.651 |
| pqqL; zinc protease [EC:3.4.24.-] | -0.201 | 0.306 | 0.512 | 0.651 |
| PRDX2_4, ahpC; peroxiredoxin (alkyl hydroperoxide reductase subunit C) [EC:1.11.1.15] | -0.231 | 0.296 | 0.438 | 0.651 |
| PREP; prolyl oligopeptidase [EC:3.4.21.26] | -0.210 | 0.327 | 0.521 | 0.656 |
| prfA, MTRF1, MRF1; peptide chain release factor 1 | -0.230 | 0.296 | 0.438 | 0.651 |
| prfB; peptide chain release factor 2 | -0.230 | 0.296 | 0.438 | 0.651 |
| prfC; peptide chain release factor 3 | -0.230 | 0.296 | 0.438 | 0.651 |
| priA; primosomal protein N’ (replication factor Y) (superfamily II helicase) [EC:3.6.4.-] | -0.230 | 0.296 | 0.438 | 0.651 |
| prmA; ribosomal protein L11 methyltransferase [EC:2.1.1.-] | -0.230 | 0.296 | 0.438 | 0.651 |
| proA; glutamate-5-semialdehyde dehydrogenase [EC:1.2.1.41] | 18.519 | 3411.091 | 0.996 | 0.998 |
| proB; glutamate 5-kinase [EC:2.7.2.11] | 18.519 | 3411.091 | 0.996 | 0.998 |
| proC; pyrroline-5-carboxylate reductase [EC:1.5.1.2] | 0.139 | 1.250 | 0.912 | 0.976 |
| PRPS, prsA; ribose-phosphate pyrophosphokinase [EC:2.7.6.1] | -0.230 | 0.296 | 0.438 | 0.651 |
| psd, PISD; phosphatidylserine decarboxylase [EC:4.1.1.65] | -0.230 | 0.296 | 0.438 | 0.651 |
| pspC; phage shock protein C | -0.429 | 0.536 | 0.424 | 0.651 |
| pstS; phosphate transport system substrate-binding protein | -0.230 | 0.296 | 0.438 | 0.651 |
| ptb; phosphate butyryltransferase [EC:2.3.1.19] | -0.147 | 0.329 | 0.655 | 0.768 |
| PTH1, pth, spoVC; peptidyl-tRNA hydrolase, PTH1 family [EC:3.1.1.29] | -0.230 | 0.296 | 0.438 | 0.651 |
| PTS-Gat-EIIC, gatC, sgcC; PTS system, galactitol-specific IIC component | -0.676 | 0.652 | 0.302 | 0.651 |
| pulA; pullulanase [EC:3.2.1.41] | -0.252 | 0.318 | 0.429 | 0.651 |
| punA, PNP; purine-nucleoside phosphorylase [EC:2.4.2.1] | -0.230 | 0.296 | 0.438 | 0.651 |
| purA, ADSS; adenylosuccinate synthase [EC:6.3.4.4] | -0.230 | 0.296 | 0.438 | 0.651 |
| purB, ADSL; adenylosuccinate lyase [EC:4.3.2.2] | -0.230 | 0.296 | 0.438 | 0.651 |
| purC; phosphoribosylaminoimidazole-succinocarboxamide synthase [EC:6.3.2.6] | -0.230 | 0.296 | 0.438 | 0.651 |
| purD; phosphoribosylamine—glycine ligase [EC:6.3.4.13] | -0.230 | 0.296 | 0.438 | 0.651 |
| purE; 5-(carboxyamino)imidazole ribonucleotide mutase [EC:5.4.99.18] | -0.230 | 0.296 | 0.438 | 0.651 |
| purF, PPAT; amidophosphoribosyltransferase [EC:2.4.2.14] | -0.219 | 0.308 | 0.477 | 0.651 |
| purH; phosphoribosylaminoimidazolecarboxamide formyltransferase / IMP cyclohydrolase [EC:2.1.2.3 3.5.4.10] | -0.230 | 0.296 | 0.438 | 0.651 |
| purL, PFAS; phosphoribosylformylglycinamidine synthase [EC:6.3.5.3] | -0.220 | 0.308 | 0.477 | 0.651 |
| purM; phosphoribosylformylglycinamidine cyclo-ligase [EC:6.3.3.1] | -0.230 | 0.296 | 0.438 | 0.651 |
| purN; phosphoribosylglycinamide formyltransferase 1 [EC:2.1.2.2] | -0.360 | 0.503 | 0.475 | 0.651 |
| purT; phosphoribosylglycinamide formyltransferase 2 [EC:2.1.2.2] | -0.193 | 0.294 | 0.513 | 0.651 |
| purU; formyltetrahydrofolate deformylase [EC:3.5.1.10] | -0.789 | 0.881 | 0.372 | 0.651 |
| pycB; pyruvate carboxylase subunit B [EC:6.4.1.1] | -0.230 | 0.296 | 0.438 | 0.651 |
| PYG, glgP; glycogen phosphorylase [EC:2.4.1.1] | -0.230 | 0.296 | 0.438 | 0.651 |
| pyrB, PYR2; aspartate carbamoyltransferase catalytic subunit [EC:2.1.3.2] | -0.230 | 0.296 | 0.438 | 0.651 |
| pyrDI; dihydroorotate dehydrogenase (NAD+) catalytic subunit [EC:1.3.1.14] | -0.230 | 0.296 | 0.438 | 0.651 |
| pyrDII; dihydroorotate dehydrogenase electron transfer subunit | -0.230 | 0.296 | 0.438 | 0.651 |
| pyrE; orotate phosphoribosyltransferase [EC:2.4.2.10] | -0.230 | 0.296 | 0.438 | 0.651 |
| pyrF; orotidine-5’-phosphate decarboxylase [EC:4.1.1.23] | -0.230 | 0.296 | 0.438 | 0.651 |
| pyrG, CTPS; CTP synthase [EC:6.3.4.2] | -0.230 | 0.296 | 0.438 | 0.651 |
| pyrH; uridylate kinase [EC:2.7.4.22] | -0.230 | 0.296 | 0.438 | 0.651 |
| pyrI; aspartate carbamoyltransferase regulatory subunit | -0.230 | 0.296 | 0.438 | 0.651 |
| pyrP, uraA; uracil permease | -0.217 | 0.297 | 0.467 | 0.651 |
| QARS, glnS; glutaminyl-tRNA synthetase [EC:6.1.1.18] | -0.230 | 0.296 | 0.438 | 0.651 |
| queA; S-adenosylmethionine:tRNA ribosyltransferase-isomerase [EC:2.4.99.17] | -0.225 | 0.301 | 0.457 | 0.651 |
| queC; 7-cyano-7-deazaguanine synthase [EC:6.3.4.20] | -0.230 | 0.296 | 0.438 | 0.651 |
| queD, ptpS, PTS; 6-pyruvoyltetrahydropterin/6-carboxytetrahydropterin synthase [EC:4.2.3.12 4.1.2.50] | -0.395 | 0.451 | 0.383 | 0.651 |
| queF; 7-cyano-7-deazaguanine reductase [EC:1.7.1.13] | -0.230 | 0.296 | 0.438 | 0.651 |
| queG; epoxyqueuosine reductase [EC:1.17.99.6] | -0.230 | 0.296 | 0.438 | 0.651 |
| radA, sms; DNA repair protein RadA/Sms | -0.284 | 0.308 | 0.358 | 0.651 |
| radC; DNA repair protein RadC | -0.230 | 0.296 | 0.438 | 0.651 |
| ramA; alpha-L-rhamnosidase [EC:3.2.1.40] | -0.319 | 0.983 | 0.746 | 0.834 |
| RARS, argS; arginyl-tRNA synthetase [EC:6.1.1.19] | -0.230 | 0.296 | 0.438 | 0.651 |
| raxB, cvaB; ATP-binding cassette, subfamily B, bacterial RaxB | 2.304 | 1.989 | 0.248 | 0.651 |
| rbfA; ribosome-binding factor A | -0.230 | 0.296 | 0.438 | 0.651 |
| rdgB; XTP/dITP diphosphohydrolase [EC:3.6.1.66] | -0.230 | 0.296 | 0.438 | 0.651 |
| recA; recombination protein RecA | -0.230 | 0.296 | 0.438 | 0.651 |
| recF; DNA replication and repair protein RecF | -0.230 | 0.296 | 0.438 | 0.651 |
| recG; ATP-dependent DNA helicase RecG [EC:3.6.4.12] | -0.213 | 0.312 | 0.495 | 0.651 |
| recJ; single-stranded-DNA-specific exonuclease [EC:3.1.-.-] | -0.230 | 0.296 | 0.438 | 0.651 |
| recN; DNA repair protein RecN (Recombination protein N) | -0.230 | 0.296 | 0.438 | 0.651 |
| recO; DNA repair protein RecO (recombination protein O) | -0.230 | 0.296 | 0.438 | 0.651 |
| recQ; ATP-dependent DNA helicase RecQ [EC:3.6.4.12] | -0.243 | 0.293 | 0.409 | 0.651 |
| recR; recombination protein RecR | -0.230 | 0.296 | 0.438 | 0.651 |
| recX; regulatory protein | -0.230 | 0.296 | 0.438 | 0.651 |
| relA; GTP pyrophosphokinase [EC:2.7.6.5] | -0.231 | 0.299 | 0.439 | 0.651 |
| RENBP; N-acylglucosamine 2-epimerase [EC:5.1.3.8] | -0.240 | 0.289 | 0.409 | 0.651 |
| res; type III restriction enzyme [EC:3.1.21.5] | -5.080 | 10.935 | 0.643 | 0.756 |
| RETSAT; all-trans-retinol 13,14-reductase [EC:1.3.99.23] | -0.696 | 0.523 | 0.185 | 0.651 |
| rfbC, rmlC; dTDP-4-dehydrorhamnose 3,5-epimerase [EC:5.1.3.13] | -0.230 | 0.296 | 0.438 | 0.651 |
| rfbD, rmlD; dTDP-4-dehydrorhamnose reductase [EC:1.1.1.133] | -0.230 | 0.296 | 0.438 | 0.651 |
| rgpA_B; gingipain R [EC:3.4.22.37] | -0.001 | 1.049 | 0.999 | 0.999 |
| rgpB; rhamnosyltransferase [EC:2.4.1.-] | 0.358 | 0.550 | 0.516 | 0.651 |
| rhaA; L-rhamnose isomerase [EC:5.3.1.14] | -0.810 | 0.830 | 0.330 | 0.651 |
| rhaB; rhamnulokinase [EC:2.7.1.5] | -0.810 | 0.830 | 0.330 | 0.651 |
| rhaD; rhamnulose-1-phosphate aldolase [EC:4.1.2.19] | -0.810 | 0.830 | 0.330 | 0.651 |
| rhaM; L-rhamnose mutarotase [EC:5.1.3.32] | -0.846 | 0.965 | 0.382 | 0.651 |
| rhaT; L-rhamnose-H+ transport protein | -0.789 | 0.881 | 0.372 | 0.651 |
| rho; transcription termination factor Rho | -0.230 | 0.296 | 0.438 | 0.651 |
| ribB, RIB3; 3,4-dihydroxy 2-butanone 4-phosphate synthase [EC:4.1.99.12] | 2.304 | 1.989 | 0.248 | 0.651 |
| ribBA; 3,4-dihydroxy 2-butanone 4-phosphate synthase / GTP cyclohydrolase II [EC:4.1.99.12 3.5.4.25] | -0.231 | 0.296 | 0.437 | 0.651 |
| ribD; diaminohydroxyphosphoribosylaminopyrimidine deaminase / 5-amino-6-(5-phosphoribosylamino)uracil reductase [EC:3.5.4.26 1.1.1.193] | -0.230 | 0.296 | 0.438 | 0.651 |
| ribE, RIB5; riboflavin synthase [EC:2.5.1.9] | -0.230 | 0.296 | 0.438 | 0.651 |
| ribF; riboflavin kinase / FMN adenylyltransferase [EC:2.7.1.26 2.7.7.2] | -0.230 | 0.296 | 0.438 | 0.651 |
| ribH, RIB4; 6,7-dimethyl-8-ribityllumazine synthase [EC:2.5.1.78] | -0.230 | 0.296 | 0.438 | 0.651 |
| ridA, tdcF, RIDA; 2-iminobutanoate/2-iminopropanoate deaminase [EC:3.5.99.10] | -0.230 | 0.296 | 0.438 | 0.651 |
| rimM; 16S rRNA processing protein RimM | -0.230 | 0.296 | 0.438 | 0.651 |
| rimO; ribosomal protein S12 methylthiotransferase [EC:2.8.4.4] | -0.230 | 0.296 | 0.438 | 0.651 |
| rimP; ribosome maturation factor RimP | -0.230 | 0.296 | 0.438 | 0.651 |
| rlmH; 23S rRNA (pseudouridine1915-N3)-methyltransferase [EC:2.1.1.177] | -0.230 | 0.296 | 0.438 | 0.651 |
| rlmI; 23S rRNA (cytosine1962-C5)-methyltransferase [EC:2.1.1.191] | -0.230 | 0.296 | 0.438 | 0.651 |
| rlmN; 23S rRNA (adenine2503-C2)-methyltransferase [EC:2.1.1.192] | -0.230 | 0.296 | 0.438 | 0.651 |
| rlpA; rare lipoprotein A | -0.217 | 0.299 | 0.469 | 0.651 |
| rluA; tRNA pseudouridine32 synthase / 23S rRNA pseudouridine746 synthase [EC:5.4.99.28 5.4.99.29] | -0.223 | 0.336 | 0.508 | 0.651 |
| rluB; 23S rRNA pseudouridine2605 synthase [EC:5.4.99.22] | -0.230 | 0.296 | 0.438 | 0.651 |
| rluD; 23S rRNA pseudouridine1911/1915/1917 synthase [EC:5.4.99.23] | -0.230 | 0.296 | 0.438 | 0.651 |
| rmuC; DNA recombination protein RmuC | -0.230 | 0.296 | 0.438 | 0.651 |
| rnc, DROSHA, RNT1; ribonuclease III [EC:3.1.26.3] | -0.217 | 0.299 | 0.469 | 0.651 |
| rnfA; electron transport complex protein RnfA | 18.519 | 3411.091 | 0.996 | 0.998 |
| rnfC; electron transport complex protein RnfC | 18.519 | 3411.091 | 0.996 | 0.998 |
| rnfD; electron transport complex protein RnfD | 18.519 | 3411.091 | 0.996 | 0.998 |
| rnfE; electron transport complex protein RnfE | 18.519 | 3411.091 | 0.996 | 0.998 |
| rnfG; electron transport complex protein RnfG | -0.222 | 0.324 | 0.494 | 0.651 |
| rng, cafA; ribonuclease G [EC:3.1.26.-] | -0.230 | 0.296 | 0.438 | 0.651 |
| rnhA, RNASEH1; ribonuclease HI [EC:3.1.26.4] | -0.230 | 0.296 | 0.438 | 0.651 |
| rnhB; ribonuclease HII [EC:3.1.26.4] | -0.230 | 0.296 | 0.438 | 0.651 |
| rnj; ribonuclease J [EC:3.1.-.-] | -0.319 | 0.983 | 0.746 | 0.834 |
| rnpA; ribonuclease P protein component [EC:3.1.26.5] | -0.230 | 0.296 | 0.438 | 0.651 |
| rnr, vacB; ribonuclease R [EC:3.1.-.-] | -0.230 | 0.296 | 0.438 | 0.651 |
| rny; ribonucrease Y [EC:3.1.-.-] | -0.230 | 0.296 | 0.438 | 0.651 |
| rnz; ribonuclease Z [EC:3.1.26.11] | -0.230 | 0.296 | 0.438 | 0.651 |
| rodA, mrdB; rod shape determining protein RodA | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-L1, MRPL1, rplA; large subunit ribosomal protein L1 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-L10, MRPL10, rplJ; large subunit ribosomal protein L10 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-L11, MRPL11, rplK; large subunit ribosomal protein L11 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-L13, MRPL13, rplM; large subunit ribosomal protein L13 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-L14, MRPL14, rplN; large subunit ribosomal protein L14 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-L15, MRPL15, rplO; large subunit ribosomal protein L15 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-L16, MRPL16, rplP; large subunit ribosomal protein L16 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-L17, MRPL17, rplQ; large subunit ribosomal protein L17 | -0.201 | 0.284 | 0.481 | 0.651 |
| RP-L18, MRPL18, rplR; large subunit ribosomal protein L18 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-L19, MRPL19, rplS; large subunit ribosomal protein L19 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-L2, MRPL2, rplB; large subunit ribosomal protein L2 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-L20, MRPL20, rplT; large subunit ribosomal protein L20 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-L21, MRPL21, rplU; large subunit ribosomal protein L21 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-L22, MRPL22, rplV; large subunit ribosomal protein L22 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-L23, MRPL23, rplW; large subunit ribosomal protein L23 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-L24, MRPL24, rplX; large subunit ribosomal protein L24 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-L25, rplY; large subunit ribosomal protein L25 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-L27, MRPL27, rpmA; large subunit ribosomal protein L27 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-L28, MRPL28, rpmB; large subunit ribosomal protein L28 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-L29, rpmC; large subunit ribosomal protein L29 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-L3, MRPL3, rplC; large subunit ribosomal protein L3 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-L30, MRPL30, rpmD; large subunit ribosomal protein L30 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-L31, rpmE; large subunit ribosomal protein L31 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-L32, MRPL32, rpmF; large subunit ribosomal protein L32 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-L33, MRPL33, rpmG; large subunit ribosomal protein L33 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-L34, MRPL34, rpmH; large subunit ribosomal protein L34 | -0.228 | 0.293 | 0.436 | 0.651 |
| RP-L35, MRPL35, rpmI; large subunit ribosomal protein L35 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-L36, MRPL36, rpmJ; large subunit ribosomal protein L36 | -0.123 | 0.303 | 0.685 | 0.796 |
| RP-L4, MRPL4, rplD; large subunit ribosomal protein L4 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-L5, MRPL5, rplE; large subunit ribosomal protein L5 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-L6, MRPL6, rplF; large subunit ribosomal protein L6 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-L7, MRPL12, rplL; large subunit ribosomal protein L7/L12 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-L9, MRPL9, rplI; large subunit ribosomal protein L9 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-S1, rpsA; small subunit ribosomal protein S1 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-S10, MRPS10, rpsJ; small subunit ribosomal protein S10 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-S11, MRPS11, rpsK; small subunit ribosomal protein S11 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-S12, MRPS12, rpsL; small subunit ribosomal protein S12 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-S13, rpsM; small subunit ribosomal protein S13 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-S14, MRPS14, rpsN; small subunit ribosomal protein S14 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-S15, MRPS15, rpsO; small subunit ribosomal protein S15 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-S16, MRPS16, rpsP; small subunit ribosomal protein S16 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-S17, MRPS17, rpsQ; small subunit ribosomal protein S17 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-S18, MRPS18, rpsR; small subunit ribosomal protein S18 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-S19, rpsS; small subunit ribosomal protein S19 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-S2, MRPS2, rpsB; small subunit ribosomal protein S2 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-S20, rpsT; small subunit ribosomal protein S20 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-S21, MRPS21, rpsU; small subunit ribosomal protein S21 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-S3, rpsC; small subunit ribosomal protein S3 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-S4, rpsD; small subunit ribosomal protein S4 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-S5, MRPS5, rpsE; small subunit ribosomal protein S5 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-S6, MRPS6, rpsF; small subunit ribosomal protein S6 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-S7, MRPS7, rpsG; small subunit ribosomal protein S7 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-S8, rpsH; small subunit ribosomal protein S8 | -0.230 | 0.296 | 0.438 | 0.651 |
| RP-S9, MRPS9, rpsI; small subunit ribosomal protein S9 | -0.230 | 0.296 | 0.438 | 0.651 |
| rpe, RPE; ribulose-phosphate 3-epimerase [EC:5.1.3.1] | -0.230 | 0.296 | 0.438 | 0.651 |
| rpiB; ribose 5-phosphate isomerase B [EC:5.3.1.6] | -0.230 | 0.296 | 0.438 | 0.651 |
| rpoA; DNA-directed RNA polymerase subunit alpha [EC:2.7.7.6] | -0.220 | 0.308 | 0.477 | 0.651 |
| rpoB; DNA-directed RNA polymerase subunit beta [EC:2.7.7.6] | -0.230 | 0.296 | 0.438 | 0.651 |
| rpoC; DNA-directed RNA polymerase subunit beta’ [EC:2.7.7.6] | -0.230 | 0.296 | 0.438 | 0.651 |
| rpoD; RNA polymerase primary sigma factor | -0.226 | 0.304 | 0.458 | 0.651 |
| rpoE; RNA polymerase sigma-70 factor, ECF subfamily | -0.274 | 0.308 | 0.375 | 0.651 |
| rpoN; RNA polymerase sigma-54 factor | -0.230 | 0.296 | 0.438 | 0.651 |
| rseC; sigma-E factor negative regulatory protein RseC | 18.519 | 3411.091 | 0.996 | 0.998 |
| rseP; regulator of sigma E protease [EC:3.4.24.-] | -0.230 | 0.296 | 0.438 | 0.651 |
| rsgA, engC; ribosome biogenesis GTPase / thiamine phosphate phosphatase [EC:3.6.1.- 3.1.3.100] | -0.230 | 0.296 | 0.438 | 0.651 |
| rsmE; 16S rRNA (uracil1498-N3)-methyltransferase [EC:2.1.1.193] | -0.230 | 0.296 | 0.438 | 0.651 |
| rsmI; 16S rRNA (cytidine1402-2’-O)-methyltransferase [EC:2.1.1.198] | -0.242 | 0.294 | 0.412 | 0.651 |
| RTCB, rtcB; tRNA-splicing ligase RtcB (3’-phosphate/5’-hydroxy nucleic acid ligase) [EC:6.5.1.8] | -2.232 | 1.933 | 0.250 | 0.651 |
| rtpR; ribonucleoside-triphosphate reductase (thioredoxin) [EC:1.17.4.2] | -0.230 | 0.296 | 0.438 | 0.651 |
| rumA; 23S rRNA (uracil1939-C5)-methyltransferase [EC:2.1.1.190] | -0.230 | 0.296 | 0.438 | 0.651 |
| ruvA; holliday junction DNA helicase RuvA [EC:3.6.4.12] | -0.230 | 0.296 | 0.438 | 0.651 |
| ruvB; holliday junction DNA helicase RuvB [EC:3.6.4.12] | -0.230 | 0.296 | 0.438 | 0.651 |
| ruvC; crossover junction endodeoxyribonuclease RuvC [EC:3.1.22.4] | -0.230 | 0.296 | 0.438 | 0.651 |
| ruvX; putative holliday junction resolvase [EC:3.1.-.-] | -0.230 | 0.296 | 0.438 | 0.651 |
| sacC, levB; levanase [EC:3.2.1.65] | -0.223 | 0.321 | 0.489 | 0.651 |
| SAM50, TOB55, bamA; outer membrane protein insertion porin family | -0.230 | 0.296 | 0.438 | 0.651 |
| sanA; SanA protein | -0.089 | 0.344 | 0.796 | 0.879 |
| SARS, serS; seryl-tRNA synthetase [EC:6.1.1.11] | -0.230 | 0.296 | 0.438 | 0.651 |
| sbcD, mre11; DNA repair protein SbcD/Mre11 | -0.746 | 1.115 | 0.505 | 0.651 |
| sdhA, frdA; succinate dehydrogenase / fumarate reductase, flavoprotein subunit [EC:1.3.5.1 1.3.5.4] | -0.230 | 0.296 | 0.438 | 0.651 |
| sdhB, frdB; succinate dehydrogenase / fumarate reductase, iron-sulfur subunit [EC:1.3.5.1 1.3.5.4] | -0.230 | 0.296 | 0.438 | 0.651 |
| sdhC, frdC; succinate dehydrogenase / fumarate reductase, cytochrome b subunit | -0.230 | 0.296 | 0.438 | 0.651 |
| secA; preprotein translocase subunit SecA | -0.230 | 0.296 | 0.438 | 0.651 |
| secDF; SecD/SecF fusion protein | -0.230 | 0.296 | 0.438 | 0.651 |
| secE; preprotein translocase subunit SecE | -0.216 | 0.300 | 0.472 | 0.651 |
| secG; preprotein translocase subunit SecG | -0.230 | 0.296 | 0.438 | 0.651 |
| secY; preprotein translocase subunit SecY | -0.230 | 0.296 | 0.438 | 0.651 |
| serA, PHGDH; D-3-phosphoglycerate dehydrogenase / 2-oxoglutarate reductase [EC:1.1.1.95 1.1.1.399] | -0.228 | 0.293 | 0.436 | 0.651 |
| serB, PSPH; phosphoserine phosphatase [EC:3.1.3.3] | -0.237 | 0.319 | 0.459 | 0.651 |
| serC, PSAT1; phosphoserine aminotransferase [EC:2.6.1.52] | -0.228 | 0.293 | 0.436 | 0.651 |
| SERPINB; serpin B | -0.131 | 0.338 | 0.699 | 0.797 |
| SIAE; sialate O-acetylesterase [EC:3.1.1.53] | -0.283 | 0.313 | 0.368 | 0.651 |
| SLC13A2_3_5; solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2/3/5 | -0.208 | 0.313 | 0.507 | 0.651 |
| slo; thiol-activated cytolysin | -0.154 | 0.385 | 0.690 | 0.797 |
| slyD; FKBP-type peptidyl-prolyl cis-trans isomerase SlyD [EC:5.2.1.8] | -0.230 | 0.296 | 0.438 | 0.651 |
| smf; DNA processing protein | -0.230 | 0.296 | 0.438 | 0.651 |
| smpB; SsrA-binding protein | -0.230 | 0.296 | 0.438 | 0.651 |
| SOD2; superoxide dismutase, Fe-Mn family [EC:1.15.1.1] | -0.396 | 0.395 | 0.317 | 0.651 |
| speA; arginine decarboxylase [EC:4.1.1.19] | -0.230 | 0.296 | 0.438 | 0.651 |
| speG, SAT; diamine N-acetyltransferase [EC:2.3.1.57] | -0.230 | 0.296 | 0.438 | 0.651 |
| spoU; RNA methyltransferase, TrmH family | -0.230 | 0.296 | 0.438 | 0.651 |
| SPP; sucrose-6-phosphatase [EC:3.1.3.24] | -0.208 | 0.288 | 0.471 | 0.651 |
| sppA; protease IV [EC:3.4.21.-] | -0.230 | 0.296 | 0.438 | 0.651 |
| spsF; spore coat polysaccharide biosynthesis protein SpsF | -0.319 | 0.983 | 0.746 | 0.834 |
| SRP54, ffh; signal recognition particle subunit SRP54 [EC:3.6.5.4] | -0.230 | 0.296 | 0.438 | 0.651 |
| ssb; single-strand DNA-binding protein | -0.232 | 0.301 | 0.441 | 0.651 |
| sucC; succinyl-CoA synthetase beta subunit [EC:6.2.1.5] | -0.173 | 0.329 | 0.600 | 0.723 |
| sucD; succinyl-CoA synthetase alpha subunit [EC:6.2.1.5] | -0.173 | 0.329 | 0.600 | 0.723 |
| sufB; Fe-S cluster assembly protein SufB | -0.230 | 0.296 | 0.438 | 0.651 |
| sufC; Fe-S cluster assembly ATP-binding protein | -0.230 | 0.296 | 0.438 | 0.651 |
| sufD; Fe-S cluster assembly protein SufD | -0.230 | 0.296 | 0.438 | 0.651 |
| sufE; cysteine desulfuration protein SufE | -0.230 | 0.296 | 0.438 | 0.651 |
| sufS; cysteine desulfurase / selenocysteine lyase [EC:2.8.1.7 4.4.1.16] | -0.230 | 0.296 | 0.438 | 0.651 |
| surA; peptidyl-prolyl cis-trans isomerase SurA [EC:5.2.1.8] | -0.230 | 0.296 | 0.438 | 0.651 |
| surE; 5’-nucleotidase [EC:3.1.3.5] | -0.230 | 0.296 | 0.438 | 0.651 |
| tadA; tRNA(adenine34) deaminase [EC:3.5.4.33] | -0.230 | 0.296 | 0.438 | 0.651 |
| tag; DNA-3-methyladenine glycosylase I [EC:3.2.2.20] | -0.227 | 0.333 | 0.497 | 0.651 |
| tagD; glycerol-3-phosphate cytidylyltransferase [EC:2.7.7.39] | 0.358 | 0.550 | 0.516 | 0.651 |
| tagF; CDP-glycerol glycerophosphotransferase [EC:2.7.8.12] | 0.092 | 1.404 | 0.948 | 0.986 |
| TARS, thrS; threonyl-tRNA synthetase [EC:6.1.1.3] | -0.230 | 0.296 | 0.438 | 0.651 |
| tatA; sec-independent protein translocase protein TatA | -0.520 | 0.870 | 0.551 | 0.683 |
| tatC; sec-independent protein translocase protein TatC | -0.520 | 0.870 | 0.551 | 0.683 |
| tatD; TatD DNase family protein [EC:3.1.21.-] | -0.234 | 0.283 | 0.408 | 0.651 |
| TC.AGCS; alanine or glycine:cation symporter, AGCS family | -0.045 | 0.351 | 0.898 | 0.972 |
| TC.BASS; bile acid:Na+ symporter, BASS family | -0.228 | 0.293 | 0.437 | 0.651 |
| TC.CIC; chloride channel protein, CIC family | -0.230 | 0.296 | 0.438 | 0.651 |
| TC.CPA1; monovalent cation:H+ antiporter, CPA1 family | -2.232 | 1.933 | 0.250 | 0.651 |
| TC.FEV.OM; iron complex outermembrane recepter protein | -0.224 | 0.298 | 0.453 | 0.651 |
| TC.FEV.OM2, cirA, cfrA, hmuR; outer membrane receptor for ferrienterochelin and colicins | -0.212 | 0.308 | 0.491 | 0.651 |
| TC.FEV.OM3, tbpA, hemR, lbpA, hpuB, bhuR, hugA, hmbR; hemoglobin/transferrin/lactoferrin receptor protein | 0.054 | 0.493 | 0.913 | 0.976 |
| TC.GPH; glycoside/pentoside/hexuronide:cation symporter, GPH family | -0.606 | 0.561 | 0.282 | 0.651 |
| TC.HAE1; hydrophobic/amphiphilic exporter-1 (mainly G- bacteria), HAE1 family | -0.216 | 0.278 | 0.438 | 0.651 |
| TC.KEF; monovalent cation:H+ antiporter-2, CPA2 family | -0.187 | 0.310 | 0.548 | 0.681 |
| TC.MATE, SLC47A, norM, mdtK, dinF; multidrug resistance protein, MATE family | -0.227 | 0.307 | 0.462 | 0.651 |
| TC.NSS; neurotransmitter:Na+ symporter, NSS family | -0.242 | 0.316 | 0.446 | 0.651 |
| TC.PIT; inorganic phosphate transporter, PiT family | 2.304 | 1.989 | 0.248 | 0.651 |
| TC.POT; proton-dependent oligopeptide transporter, POT family | -0.230 | 0.296 | 0.438 | 0.651 |
| TC.SSS; solute:Na+ symporter, SSS family | -0.692 | 0.616 | 0.263 | 0.651 |
| TC.SULP; sulfate permease, SulP family | -0.746 | 1.115 | 0.505 | 0.651 |
| TC.ZIP, zupT, ZRT3, ZIP2; zinc transporter, ZIP family | -0.247 | 0.568 | 0.664 | 0.776 |
| tdk, TK; thymidine kinase [EC:2.7.1.21] | -0.230 | 0.296 | 0.438 | 0.651 |
| terC; tellurite resistance protein TerC | -0.625 | 0.575 | 0.278 | 0.651 |
| tetM, tetO; ribosomal protection tetracycline resistance protein | -0.388 | 0.498 | 0.437 | 0.651 |
| tex; protein Tex | -0.196 | 0.300 | 0.514 | 0.651 |
| tgt, QTRT1; queuine tRNA-ribosyltransferase [EC:2.4.2.29] | -0.230 | 0.296 | 0.438 | 0.651 |
| THI4, THI1; cysteine-dependent adenosine diphosphate thiazole synthase [EC:2.4.2.60] | 0.014 | 0.383 | 0.970 | 0.998 |
| thiC; phosphomethylpyrimidine synthase [EC:4.1.99.17] | 0.014 | 0.383 | 0.970 | 0.998 |
| thiD; hydroxymethylpyrimidine/phosphomethylpyrimidine kinase [EC:2.7.1.49 2.7.4.7] | -0.134 | 0.312 | 0.669 | 0.779 |
| thiE; thiamine-phosphate pyrophosphorylase [EC:2.5.1.3] | -0.063 | 0.337 | 0.853 | 0.928 |
| thiG; thiazole synthase [EC:2.8.1.10] | 18.519 | 3411.091 | 0.996 | 0.998 |
| thiH; 2-iminoacetate synthase [EC:4.1.99.19] | 18.519 | 3411.091 | 0.996 | 0.998 |
| thiJ; protein deglycase [EC:3.5.1.124] | -0.230 | 0.296 | 0.438 | 0.651 |
| thiL; thiamine-monophosphate kinase [EC:2.7.4.16] | -0.230 | 0.296 | 0.438 | 0.651 |
| thiN, TPK1, THI80; thiamine pyrophosphokinase [EC:2.7.6.2] | -0.198 | 0.307 | 0.519 | 0.653 |
| thiS; sulfur carrier protein | 18.519 | 3411.091 | 0.996 | 0.998 |
| thrA; bifunctional aspartokinase / homoserine dehydrogenase 1 [EC:2.7.2.4 1.1.1.3] | -0.274 | 0.307 | 0.374 | 0.651 |
| thrC; threonine synthase [EC:4.2.3.1] | -0.606 | 0.561 | 0.282 | 0.651 |
| thyA, TYMS; thymidylate synthase [EC:2.1.1.45] | -0.172 | 0.309 | 0.579 | 0.705 |
| thyX, thy1; thymidylate synthase (FAD) [EC:2.1.1.148] | -0.625 | 0.575 | 0.278 | 0.651 |
| tig; trigger factor | -0.230 | 0.296 | 0.438 | 0.651 |
| tilS, mesJ; tRNA(Ile)-lysidine synthase [EC:6.3.4.19] | -0.230 | 0.296 | 0.438 | 0.651 |
| tnaA; tryptophanase [EC:4.1.99.1] | -0.221 | 0.572 | 0.699 | 0.797 |
| tolA; colicin import membrane protein | -0.164 | 0.315 | 0.604 | 0.723 |
| tolC; outer membrane protein | -0.590 | 0.508 | 0.248 | 0.651 |
| tonB; periplasmic protein TonB | -0.185 | 0.276 | 0.505 | 0.651 |
| topA; DNA topoisomerase I [EC:5.99.1.2] | -0.230 | 0.296 | 0.438 | 0.651 |
| topB; DNA topoisomerase III [EC:5.99.1.2] | -0.314 | 0.345 | 0.363 | 0.651 |
| TPI, tpiA; triosephosphate isomerase (TIM) [EC:5.3.1.1] | -0.240 | 0.307 | 0.435 | 0.651 |
| tpx; thiol peroxidase, atypical 2-Cys peroxiredoxin [EC:1.11.1.15] | -0.754 | 1.120 | 0.502 | 0.651 |
| treS; maltose alpha-D-glucosyltransferase / alpha-amylase [EC:5.4.99.16 3.2.1.1] | -0.629 | 0.781 | 0.422 | 0.651 |
| trkA, ktrA; trk system potassium uptake protein | -0.230 | 0.296 | 0.438 | 0.651 |
| trkH, trkG, ktrB; trk system potassium uptake protein | -0.230 | 0.296 | 0.438 | 0.651 |
| trmB, METTL1; tRNA (guanine-N7-)-methyltransferase [EC:2.1.1.33] | -0.230 | 0.296 | 0.438 | 0.651 |
| trmD; tRNA (guanine37-N1)-methyltransferase [EC:2.1.1.228] | -0.230 | 0.296 | 0.438 | 0.651 |
| troR; DtxR family transcriptional regulator, Mn-dependent transcriptional regulator | 0.038 | 0.480 | 0.937 | 0.977 |
| trpA; tryptophan synthase alpha chain [EC:4.2.1.20] | -0.089 | 1.133 | 0.938 | 0.977 |
| trpB; tryptophan synthase beta chain [EC:4.2.1.20] | -0.229 | 0.301 | 0.447 | 0.651 |
| trpC; indole-3-glycerol phosphate synthase [EC:4.1.1.48] | -0.376 | 0.442 | 0.397 | 0.651 |
| trpD; anthranilate phosphoribosyltransferase [EC:2.4.2.18] | -0.089 | 1.133 | 0.938 | 0.977 |
| trpE; anthranilate synthase component I [EC:4.1.3.27] | -0.089 | 1.133 | 0.938 | 0.977 |
| trpF; phosphoribosylanthranilate isomerase [EC:5.3.1.24] | -0.219 | 0.301 | 0.468 | 0.651 |
| trpG; anthranilate synthase component II [EC:4.1.3.27] | -0.167 | 0.324 | 0.608 | 0.725 |
| truA, PUS1; tRNA pseudouridine38-40 synthase [EC:5.4.99.12] | -0.230 | 0.296 | 0.438 | 0.651 |
| truB, PUS4, TRUB1; tRNA pseudouridine55 synthase [EC:5.4.99.25] | -0.230 | 0.296 | 0.438 | 0.651 |
| trxA; thioredoxin 1 | -0.230 | 0.296 | 0.438 | 0.651 |
| trxB, TRR; thioredoxin reductase (NADPH) [EC:1.8.1.9] | -0.230 | 0.296 | 0.438 | 0.651 |
| tsaB; tRNA threonylcarbamoyladenosine biosynthesis protein TsaB | -0.230 | 0.296 | 0.438 | 0.651 |
| tsaC, rimN, SUA5; L-threonylcarbamoyladenylate synthase [EC:2.7.7.87] | -0.230 | 0.296 | 0.438 | 0.651 |
| tsaE; tRNA threonylcarbamoyladenosine biosynthesis protein TsaE | -0.230 | 0.296 | 0.438 | 0.651 |
| tsf, TSFM; elongation factor Ts | -0.230 | 0.296 | 0.438 | 0.651 |
| TSTA3, fcl; GDP-L-fucose synthase [EC:1.1.1.271] | -0.230 | 0.296 | 0.438 | 0.651 |
| tuf, TUFM; elongation factor Tu | -0.230 | 0.296 | 0.438 | 0.651 |
| typA, bipA; GTP-binding protein | -0.230 | 0.296 | 0.438 | 0.651 |
| ubiE; demethylmenaquinone methyltransferase / 2-methoxy-6-polyprenyl-1,4-benzoquinol methylase [EC:2.1.1.163 2.1.1.201] | -0.230 | 0.296 | 0.438 | 0.651 |
| udk, UCK; uridine kinase [EC:2.7.1.48] | -0.230 | 0.296 | 0.438 | 0.651 |
| udp, UPP; uridine phosphorylase [EC:2.4.2.3] | -0.230 | 0.296 | 0.438 | 0.651 |
| UGDH, ugd; UDPglucose 6-dehydrogenase [EC:1.1.1.22] | -0.219 | 0.308 | 0.477 | 0.651 |
| umuC; DNA polymerase V | -0.183 | 0.301 | 0.543 | 0.678 |
| umuD; DNA polymerase V [EC:3.4.21.-] | -2.933 | 2.379 | 0.219 | 0.651 |
| UNG, UDG; uracil-DNA glycosylase [EC:3.2.2.27] | -0.204 | 0.307 | 0.507 | 0.651 |
| upp, UPRT; uracil phosphoribosyltransferase [EC:2.4.2.9] | -0.230 | 0.296 | 0.438 | 0.651 |
| uppS; undecaprenyl diphosphate synthase [EC:2.5.1.31] | -0.230 | 0.296 | 0.438 | 0.651 |
| URA4, pyrC; dihydroorotase [EC:3.5.2.3] | -0.220 | 0.308 | 0.477 | 0.651 |
| utp; urea transporter | -17.599 | 3454.822 | 0.996 | 0.998 |
| uup; ABC transport system ATP-binding/permease protein | -0.220 | 0.308 | 0.477 | 0.651 |
| uvrA; excinuclease ABC subunit A | -0.215 | 0.290 | 0.459 | 0.651 |
| uvrB; excinuclease ABC subunit B | -0.230 | 0.296 | 0.438 | 0.651 |
| uvrC; excinuclease ABC subunit C | -0.230 | 0.296 | 0.438 | 0.651 |
| uvrD, pcrA; DNA helicase II / ATP-dependent DNA helicase PcrA [EC:3.6.4.12] | -0.254 | 0.306 | 0.407 | 0.651 |
| uxaA; altronate hydrolase [EC:4.2.1.7] | -0.676 | 0.652 | 0.302 | 0.651 |
| uxaB; tagaturonate reductase [EC:1.1.1.58] | -0.676 | 0.652 | 0.302 | 0.651 |
| uxaC; glucuronate isomerase [EC:5.3.1.12] | -0.839 | 0.582 | 0.151 | 0.651 |
| UXS1, uxs; UDP-glucuronate decarboxylase [EC:4.1.1.35] | -0.137 | 0.613 | 0.823 | 0.900 |
| uxuA; mannonate dehydratase [EC:4.2.1.8] | -0.692 | 0.616 | 0.263 | 0.651 |
| uxuB; fructuronate reductase [EC:1.1.1.57] | -0.677 | 0.652 | 0.301 | 0.651 |
| vanX; zinc D-Ala-D-Ala dipeptidase [EC:3.4.13.22] | -0.199 | 0.288 | 0.490 | 0.651 |
| VARS, valS; valyl-tRNA synthetase [EC:6.1.1.9] | -0.230 | 0.296 | 0.438 | 0.651 |
| vsr; DNA mismatch endonuclease, patch repair protein [EC:3.1.-.-] | -0.298 | 0.485 | 0.540 | 0.676 |
| WARS, trpS; tryptophanyl-tRNA synthetase [EC:6.1.1.2] | -0.230 | 0.296 | 0.438 | 0.651 |
| wbpO; UDP-N-acetyl-D-galactosamine dehydrogenase [EC:1.1.1.-] | -0.076 | 0.332 | 0.819 | 0.899 |
| wcaJ; putative colanic acid biosysnthesis UDP-glucose lipid carrier transferase | -0.429 | 0.535 | 0.425 | 0.651 |
| wcaK, amsJ; colanic acid/amylovoran biosynthesis protein | 2.304 | 1.989 | 0.248 | 0.651 |
| wecB; UDP-N-acetylglucosamine 2-epimerase (non-hydrolysing) [EC:5.1.3.14] | -0.287 | 0.294 | 0.330 | 0.651 |
| xanP; xanthine permease XanP | -0.131 | 0.310 | 0.674 | 0.784 |
| xapB; MFS transporter, NHS family, xanthosine permease | 0.027 | 0.484 | 0.955 | 0.992 |
| xerC; integrase/recombinase XerC | -0.230 | 0.296 | 0.438 | 0.651 |
| xerD; integrase/recombinase XerD | -0.230 | 0.296 | 0.438 | 0.651 |
| xlyAB; N-acetylmuramoyl-L-alanine amidase [EC:3.5.1.28] | -1.535 | 1.399 | 0.274 | 0.651 |
| xpt; xanthine phosphoribosyltransferase [EC:2.4.2.22] | -0.156 | 0.300 | 0.605 | 0.723 |
| xseA; exodeoxyribonuclease VII large subunit [EC:3.1.11.6] | -0.230 | 0.296 | 0.438 | 0.651 |
| xseB; exodeoxyribonuclease VII small subunit [EC:3.1.11.6] | -0.230 | 0.296 | 0.438 | 0.651 |
| xylA; xylose isomerase [EC:5.3.1.5] | -0.092 | 1.135 | 0.935 | 0.977 |
| xylB, XYLB; xylulokinase [EC:2.7.1.17] | -0.546 | 0.585 | 0.352 | 0.651 |
| xylE; MFS transporter, SP family, xylose:H+ symportor | 0.154 | 1.349 | 0.909 | 0.976 |
| xylS, yicI; alpha-D-xyloside xylohydrolase [EC:3.2.1.177] | -0.603 | 0.561 | 0.284 | 0.651 |
| yaeR; glyoxylase I family protein | 18.519 | 3411.091 | 0.996 | 0.998 |
| yafQ; mRNA interferase YafQ [EC:3.1.-.-] | -0.737 | 0.680 | 0.280 | 0.651 |
| yajC; preprotein translocase subunit YajC | -0.230 | 0.296 | 0.438 | 0.651 |
| YARS, tyrS; tyrosyl-tRNA synthetase [EC:6.1.1.1] | -0.230 | 0.296 | 0.438 | 0.651 |
| ybaK, ebsC; Cys-tRNA(Pro)/Cys-tRNA(Cys) deacylase [EC:3.1.1.-] | -0.754 | 1.120 | 0.502 | 0.651 |
| ybaZ; methylated-DNA-protein-cysteine methyltransferase related protein | -0.213 | 0.325 | 0.513 | 0.651 |
| ybdG, mscM; miniconductance mechanosensitive channel | -0.223 | 0.302 | 0.460 | 0.651 |
| ybeB; ribosome-associated protein | -0.230 | 0.296 | 0.438 | 0.651 |
| ybgC; acyl-CoA thioester hydrolase [EC:3.1.2.-] | -0.370 | 0.319 | 0.249 | 0.651 |
| ycaJ; putative ATPase | -0.230 | 0.296 | 0.438 | 0.651 |
| ychF; ribosome-binding ATPase | -0.230 | 0.296 | 0.438 | 0.651 |
| yfbK; Ca-activated chloride channel homolog | -0.248 | 0.303 | 0.415 | 0.651 |
| yfiC, trmX; tRNA1Val (adenine37-N6)-methyltransferase [EC:2.1.1.223] | -0.230 | 0.296 | 0.438 | 0.651 |
| yfiH; polyphenol oxidase [EC:1.10.3.-] | -0.230 | 0.296 | 0.438 | 0.651 |
| yggS, PROSC; PLP dependent protein | -0.230 | 0.296 | 0.438 | 0.651 |
| yhbH; putative sigma-54 modulation protein | -0.263 | 0.312 | 0.401 | 0.651 |
| yidC, spoIIIJ, OXA1, ccfA; YidC/Oxa1 family membrane protein insertase | -0.230 | 0.296 | 0.438 | 0.651 |
| yjbB; phosphate:Na+ symporter | -0.230 | 0.296 | 0.438 | 0.651 |
| yjdF; putative membrane protein | -0.084 | 0.350 | 0.811 | 0.894 |
| yoeB; toxin YoeB [EC:3.1.-.-] | 0.112 | 0.479 | 0.816 | 0.896 |
| ypsC; putative N6-adenine-specific DNA methylase [EC:2.1.1.-] | -0.230 | 0.296 | 0.438 | 0.651 |
| yraN; putative endonuclease | -0.230 | 0.296 | 0.438 | 0.651 |
| yrbG; cation:H+ antiporter | -0.156 | 0.300 | 0.605 | 0.723 |
| ytfE, scdA; regulator of cell morphogenesis and NO signaling | -0.216 | 0.284 | 0.448 | 0.651 |
| zapA; cell division protein ZapA | -0.219 | 0.301 | 0.468 | 0.651 |
| zntA; Cd2+/Zn2+-exporting ATPase [EC:3.6.3.3 3.6.3.5] | -0.201 | 0.284 | 0.481 | 0.651 |
| znuA; zinc transport system substrate-binding protein | -0.352 | 0.456 | 0.441 | 0.651 |
| znuB; zinc transport system permease protein | -0.452 | 0.409 | 0.271 | 0.651 |
| znuC; zinc transport system ATP-binding protein [EC:3.6.3.-] | -0.452 | 0.409 | 0.271 | 0.651 |
| zraR, hydG; two-component system, NtrC family, response regulator HydG | -0.512 | 0.991 | 0.606 | 0.723 |
# merge with tb.ra for full results table
full.res <- left_join(tb.ra, results.out, by = "description")
write.csv(full.res, "output/picrust_ko_stratefied_prevo_data_results.csv", row.names = F)
sessionInfo()R version 4.0.5 (2021-03-31)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19042)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.1252
[2] LC_CTYPE=English_United States.1252
[3] LC_MONETARY=English_United States.1252
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.1252
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] cowplot_1.1.1 dendextend_1.14.0 ggdendro_0.1.22 reshape2_1.4.4
[5] car_3.0-10 carData_3.0-4 gvlma_1.0.0.3 patchwork_1.1.1
[9] viridis_0.5.1 viridisLite_0.3.0 gridExtra_2.3 xtable_1.8-4
[13] kableExtra_1.3.4 MASS_7.3-53.1 data.table_1.14.0 readxl_1.3.1
[17] forcats_0.5.1 stringr_1.4.0 dplyr_1.0.5 purrr_0.3.4
[21] readr_1.4.0 tidyr_1.1.3 tibble_3.1.0 ggplot2_3.3.3
[25] tidyverse_1.3.0 lmerTest_3.1-3 lme4_1.1-26 Matrix_1.3-2
[29] vegan_2.5-7 lattice_0.20-41 permute_0.9-5 phyloseq_1.34.0
[33] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] minqa_1.2.4 colorspace_2.0-0 rio_0.5.26
[4] ellipsis_0.3.1 rprojroot_2.0.2 XVector_0.30.0
[7] fs_1.5.0 rstudioapi_0.13 farver_2.1.0
[10] fansi_0.4.2 lubridate_1.7.10 xml2_1.3.2
[13] codetools_0.2-18 splines_4.0.5 knitr_1.31
[16] ade4_1.7-16 jsonlite_1.7.2 nloptr_1.2.2.2
[19] broom_0.7.5 cluster_2.1.1 dbplyr_2.1.0
[22] BiocManager_1.30.10 compiler_4.0.5 httr_1.4.2
[25] backports_1.2.1 assertthat_0.2.1 cli_2.3.1
[28] later_1.1.0.1 htmltools_0.5.1.1 prettyunits_1.1.1
[31] tools_4.0.5 igraph_1.2.6 gtable_0.3.0
[34] glue_1.4.2 Rcpp_1.0.6 Biobase_2.50.0
[37] cellranger_1.1.0 jquerylib_0.1.3 vctrs_0.3.6
[40] Biostrings_2.58.0 rhdf5filters_1.2.0 multtest_2.46.0
[43] svglite_2.0.0 ape_5.4-1 nlme_3.1-152
[46] iterators_1.0.13 xfun_0.21 ps_1.6.0
[49] openxlsx_4.2.3 rvest_1.0.0 lifecycle_1.0.0
[52] statmod_1.4.35 zlibbioc_1.36.0 scales_1.1.1
[55] hms_1.0.0 promises_1.2.0.1 parallel_4.0.5
[58] biomformat_1.18.0 rhdf5_2.34.0 curl_4.3
[61] yaml_2.2.1 sass_0.3.1 stringi_1.5.3
[64] highr_0.8 S4Vectors_0.28.1 foreach_1.5.1
[67] BiocGenerics_0.36.0 zip_2.1.1 boot_1.3-27
[70] systemfonts_1.0.1 rlang_0.4.10 pkgconfig_2.0.3
[73] evaluate_0.14 Rhdf5lib_1.12.1 labeling_0.4.2
[76] tidyselect_1.1.0 plyr_1.8.6 magrittr_2.0.1
[79] R6_2.5.0 IRanges_2.24.1 generics_0.1.0
[82] DBI_1.1.1 foreign_0.8-81 pillar_1.5.1
[85] haven_2.3.1 withr_2.4.1 mgcv_1.8-34
[88] abind_1.4-5 survival_3.2-10 modelr_0.1.8
[91] crayon_1.4.1 utf8_1.1.4 rmarkdown_2.7
[94] progress_1.2.2 grid_4.0.5 git2r_0.28.0
[97] webshot_0.5.2 reprex_1.0.0 digest_0.6.27
[100] httpuv_1.5.5 numDeriv_2016.8-1.1 stats4_4.0.5
[103] munsell_0.5.0 bslib_0.2.4