Last updated: 2024-06-09
Checks: 6 1
Knit directory: Cinquina_2024/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R
Markdown file created these results, you’ll want to first commit it to
the Git repo. If you’re still working on the analysis, you can ignore
this warning. When you’re finished, you can run
wflow_publish to commit the R Markdown file and build the
HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20240320) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version cd64ad0. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rproj.user/
Untracked files:
Untracked: analysis/cortex.Rmd
Untracked: analysis/cortex_visualisation.Rmd
Untracked: analysis/figure/
Untracked: data/SCP1290/
Untracked: data/azimuth_integrated.rds
Untracked: data/cfg.txt
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with
wflow_publish() to start tracking its development.
library(here)here() starts at /data/Cinquina_2024library(RColorBrewer)
library(Seurat)Loading required package: SeuratObjectLoading required package: sp
Attaching package: 'SeuratObject'The following object is masked from 'package:base':
intersectlibrary(scCustomize)scCustomize v2.0.1
If you find the scCustomize useful please cite.
See 'samuel-marsh.github.io/scCustomize/articles/FAQ.html' for citation info.library(SeuratData)Using cached data manifest, last updated at 2024-05-07 11:22:16.949269── Installed datasets ──────────────────────────────── SeuratData v0.2.2.9001 ──✔ mousecortexref 1.0.0 ────────────────────────────────────── Key ─────────────────────────────────────✔ Dataset loaded successfully
❯ Dataset built with a newer version of Seurat than installed
❓ Unknown version of Seurat installedlibrary(SeuratWrappers)
library(Azimuth)Registered S3 method overwritten by 'SeuratDisk':
method from
as.sparse.H5Group SeuratAttaching shinyBSlibrary(BPCells)
library(dplyr)
Attaching package: 'dplyr'The following objects are masked from 'package:stats':
filter, lagThe following objects are masked from 'package:base':
intersect, setdiff, setequal, unionlibrary(magrittr)
library(stringr)
library(readr)
library(ggplot2)
library(cowplot)
library(patchwork)
Attaching package: 'patchwork'The following object is masked from 'package:cowplot':
align_plotsoptions(future.globals.maxSize = 1e9)
options(Seurat.object.assay.version = "v5")# Create a vector with the stage of development for each object
stage_info <- c("E11.5", "E12.5", "E13.5", "E14.5", "E15.5", "E16", "E18.5", "E18", "P1", "P1", "E10", "E17.5", "P4")merged_cortex_2 <- SeuratObject::LoadSeuratRds(here::here("data/azimuth_integrated.rds"))
merged_cortex_2$cell_name <- Cells(merged_cortex_2)
merged_cortex_2An object of class Seurat
28186 features across 82415 samples within 5 assays
Active assay: RNA (27998 features, 2000 variable features)
25 layers present: data.E11.5, data.E12.5, data.E13.5, data.E14.5, data.E15.5, data.E16, data.E18.5, data.E18, data.P1, data.E10, data.E17.5, data.P4, scale.data, counts.E11.5, counts.E12.5, counts.E13.5, counts.E14.5, counts.E15.5, counts.E16, counts.E18.5, counts.E18, counts.P1, counts.E10, counts.E17.5, counts.P4
4 other assays present: prediction.score.class, prediction.score.cluster, prediction.score.subclass, prediction.score.cross_species_cluster
7 dimensional reductions calculated: pca, integrated_dr, ref.umap, integrated.cca, umap.cca, harmony, umap.harmonyorig_umap <- readr::read_tsv(
here("data/SCP1290/cluster/cluster_scDevSC.merged.umap.txt"),
skip = 2,
col_names = c("cell_name", "UMAP_1", "UMAP_2"),
col_types = list(col_character(), col_double(), col_double())
)
glimpse(orig_umap)Rows: 98,047
Columns: 3
$ cell_name <chr> "E10_v1_AAACCTGAGGGTCTCC-1", "E10_v1_AAACCTGCACAACGCC-1", "E…
$ UMAP_1 <dbl> -3.0025911, -3.6729214, -3.8859395, -3.9020242, -2.9312939, …
$ UMAP_2 <dbl> -10.453364, -6.552985, -10.773631, -10.869657, -10.769403, -…orig_umap %<>% tibble::column_to_rownames("cell_name")
orig_umap %<>% as.matrix()
orig_tsne <- readr::read_tsv(
here("data/SCP1290/cluster/cluster_scDevSC.merged.tsne.txt"),
skip = 2,
col_names = c("cell_name", "tSNE_1", "tSNE_2"),
col_types = list(col_character(), col_double(), col_double())
)
glimpse(orig_tsne)Rows: 98,047
Columns: 3
$ cell_name <chr> "E10_v1_AAACCTGAGGGTCTCC-1", "E10_v1_AAACCTGCACAACGCC-1", "E…
$ tSNE_1 <dbl> 15.442958, 10.373660, 14.828413, 16.307658, 18.062250, 13.72…
$ tSNE_2 <dbl> -19.603245, -17.062466, -20.102599, -20.003542, -18.636268, …orig_tsne %<>% tibble::column_to_rownames("cell_name")
orig_tsne %<>% as.matrix()
orig_metadata <- readr::read_tsv(here(
"data/SCP1290/metadata/metaData_scDevSC.txt"))Rows: 98048 Columns: 28
── Column specification ────────────────────────────────────────────────────────
Delimiter: "\t"
chr (28): NAME, orig_ident, nCount_RNA, nFeature_RNA, percent_mito, n_hkgene...
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.orig_metadata %<>% rename("cell_name" = "NAME")
orig_metadata_types <- orig_metadata[1,] |> purrr::simplify()
orig_metadata %<>% filter(!cell_name == "TYPE")
glimpse(orig_metadata)Rows: 98,047
Columns: 28
$ cell_name <chr> "E10_v1_AAACCTGAGGGTCTCC-…
$ orig_ident <chr> "E10", "E10", "E10", "E10…
$ nCount_RNA <chr> "1544", "1157", "2081", "…
$ nFeature_RNA <chr> "1022", "783", "1200", "1…
$ percent_mito <chr> "0.02007772", "0.01469317…
$ n_hkgene <chr> "51", "39", "67", "71", "…
$ S_Score <chr> "0.356987282", "0.4538538…
$ G2M_Score <chr> "0.330795055", "0.2605599…
$ Phase <chr> "S", "S", "S", "G2M", "S"…
$ CC_Difference <chr> "0.026192226", "0.1932938…
$ seurat_clusters <chr> "34", "34", "34", "37", "…
$ RNA_snn_res_1 <chr> "20", "20", "20", "20", "…
$ scrublet_doublet <chr> "FALSE", "FALSE", "FALSE"…
$ RNA_snn_res_2 <chr> "34", "34", "34", "37", "…
$ Doublet_intersect <chr> NA, NA, NA, NA, NA, NA, N…
$ Gral_cellType <chr> NA, NA, NA, NA, NA, NA, N…
$ New_cellType <chr> "Apical progenitors", "In…
$ biosample_id <chr> "E10", "E10", "E10", "E10…
$ donor_id <chr> "mouse_E10", "mouse_E10",…
$ species <chr> "NCBITaxon_10090", "NCBIT…
$ disease <chr> "PATO_0000461", "PATO_000…
$ disease__ontology_label <chr> "normal", "normal", "norm…
$ organ <chr> "UBERON_0008930", "UBERON…
$ organ__ontology_label <chr> "somatosensory cortex", "…
$ library_preparation_protocol <chr> "EFO_0009899", "EFO_00098…
$ library_preparation_protocol__ontology_label <chr> "10X 3' v2 sequencing", "…
$ sex <chr> "mixed", "mixed", "mixed"…
$ species__ontology_label <chr> "Mus musculus", "Mus musc…change_column_types <- function(df, types) {
for (col_name in names(types)) {
col_type <- types[col_name]
if (col_type == "character") {
df[[col_name]] <- as.character(df[[col_name]])
} else if (col_type == "numeric") {
df[[col_name]] <- as.numeric(df[[col_name]])
} else if (col_type == "integer") {
df[[col_name]] <- as.integer(df[[col_name]])
} else if (col_type == "logical") {
df[[col_name]] <- as.logical(df[[col_name]])
} else if (col_type == "factor") {
df[[col_name]] <- as.factor(df[[col_name]])
} else if (col_type == "group") {
df[[col_name]] <- as.factor(df[[col_name]])
} else {
warning(paste("Unknown type:", col_type, "for column", col_name))
}
}
return(df)
}
# Apply the function to the metadata
orig_metadata <- change_column_types(orig_metadata, orig_metadata_types)
# Print the modified metadata
glimpse(orig_metadata)Rows: 98,047
Columns: 28
$ cell_name <chr> "E10_v1_AAACCTGAGGGTCTCC-…
$ orig_ident <fct> E10, E10, E10, E10, E10, …
$ nCount_RNA <dbl> 1544, 1157, 2081, 2490, 2…
$ nFeature_RNA <dbl> 1022, 783, 1200, 1430, 14…
$ percent_mito <dbl> 0.020077720, 0.014693172,…
$ n_hkgene <dbl> 51, 39, 67, 71, 70, 50, 4…
$ S_Score <dbl> 0.35698728, 0.45385381, 0…
$ G2M_Score <dbl> 0.33079506, 0.26055995, 0…
$ Phase <fct> S, S, S, G2M, S, S, S, S,…
$ CC_Difference <dbl> 0.026192226, 0.193293862,…
$ seurat_clusters <fct> 34, 34, 34, 37, 37, 34, 4…
$ RNA_snn_res_1 <fct> 20, 20, 20, 20, 20, 20, 3…
$ scrublet_doublet <fct> FALSE, FALSE, FALSE, FALS…
$ RNA_snn_res_2 <fct> 34, 34, 34, 37, 37, 34, 4…
$ Doublet_intersect <fct> NA, NA, NA, NA, NA, NA, N…
$ Gral_cellType <fct> NA, NA, NA, NA, NA, NA, N…
$ New_cellType <fct> Apical progenitors, Inter…
$ biosample_id <fct> E10, E10, E10, E10, E10, …
$ donor_id <fct> mouse_E10, mouse_E10, mou…
$ species <fct> NCBITaxon_10090, NCBITaxo…
$ disease <fct> PATO_0000461, PATO_000046…
$ disease__ontology_label <fct> normal, normal, normal, n…
$ organ <fct> UBERON_0008930, UBERON_00…
$ organ__ontology_label <fct> somatosensory cortex, som…
$ library_preparation_protocol <fct> EFO_0009899, EFO_0009899,…
$ library_preparation_protocol__ontology_label <fct> 10X 3' v2 sequencing, 10X…
$ sex <fct> mixed, mixed, mixed, mixe…
$ species__ontology_label <fct> Mus musculus, Mus musculu…orig_srt <- Read10X(data.dir = here("data/SCP1290/expression/601ae2f4771a5b0d72588bfb"))
# Convert the log1p normalized matrix to a standard matrix if it's not already
normalized_matrix <- as.matrix(orig_srt)
# Reverse the log1p transformation to get the count matrix
count_matrix <- expm1(normalized_matrix)
# Convert the count matrix to a sparse matrix format (dgCMatrix) if needed
count_matrix_sparse <- as(count_matrix, "dgCMatrix")
# Create a Seurat object using the recovered count matrix
merged_cortex <- CreateSeuratObject(counts = count_matrix_sparse, meta.data = orig_metadata)
merged_cortex[["umap"]] <- CreateDimReducObject(embeddings = orig_umap, key = "UMAP_", assay = DefaultAssay(merged_cortex))
merged_cortex[["tsne"]] <- CreateDimReducObject(embeddings = orig_tsne, key = "tSNE_", assay = DefaultAssay(merged_cortex))
merged_cortex$stage <- merged_cortex$orig.ident
table(merged_cortex$New_cellType)
Apical progenitors Astrocytes Cajal Retzius cells
18491 2976 532
CThPN Cycling glial cells DL CPN
4607 1004 3106
DL_CPN_1 DL_CPN_2 Doublet
422 146 1854
Endothelial cells Ependymocytes Immature neurons
291 35 3092
Intermediate progenitors Interneurons Layer 4
8490 10469 5317
Layer 6b Low quality cells Microglia
194 4545 263
Migrating neurons NP Oligodendrocytes
12332 424 1098
Pericytes Red blood cells SCPN
236 330 2987
UL CPN VLMC
14041 765 Idents(merged_cortex) <- "New_cellType"
merged_cortex <- subset(merged_cortex, idents = c("Doublet", "Low quality cells", "Red blood cells"), invert = TRUE)
merged_cortex <-
Store_Palette_Seurat(
seurat_object = merged_cortex,
palette = rev(brewer.pal(n = 11, name = "Spectral")),
palette_name = "expr_Colour_Pal"
)Seurat Object now contains the following items in @misc slot:
ℹ 'expr_Colour_Pal'# Get the list of S100 family genes
s100_genes <- grep("^S100", rownames(merged_cortex), value = TRUE)
genes.embed <- c(
"Abcd1",
"Abcd2",
"Abcd3",
"Acaa1",
"Acaa2",
"Acox1",
"Agrn",
"Agt",
"Alcam",
"Aldh1a1",
"Aldh1l1",
"Aldoc",
"Angpt1",
"Apoe",
"App",
"Aqp4",
"Arf1",
"Bmp7",
"Bsg",
"Cacybp",
"Caf4",
"Ccl25",
"Ckb",
"Cnr1",
"Cnr2",
"Col4a5",
"Cst3",
"Dagla",
"Daglb",
"Decr2",
"Dcc",
"Dnm1",
"Drp1",
"Ech1",
"Efna5",
"Egfr",
"Enho",
"Eno1",
"Faah",
"Fgf1",
"Fgfr3",
"Fis1",
"Fos",
"Fth1",
"Ftl1",
"Gfap",
"Gja1",
"Gli1",
"Glul",
"Gnai2",
"Gnas",
"H2-K1",
"Hacd2",
"Hadhb",
"Hbegf",
"Hepacam",
"Hif1",
"Htra1",
"Igsf1",
"Il18",
"Il1rapl1",
"Itgav",
"Jam2",
"Lama2",
"Lamb2",
"Lcat",
"Lgi1",
"Lgi4",
"Lpcat3",
"Lrpap1",
"Lrrc4b",
"Lxn",
"Mdk",
"Mdv1",
"Mfn1",
"Mfn2",
"Mgll",
"Mief1",
"Napepld",
"Ncam1",
"Ncan",
"Ndrg2",
"Nfasc",
"Nfia",
"Nlgn3",
"Nrxn1",
"Nrxn2",
"Ntn1",
"Ntrk3",
"Opa1",
"Otp",
"Pex1",
"Pex10",
"Pex12",
"Pex13",
"Pex14",
"Pex16",
"Pex2",
"Pex26",
"Pex3",
"Pex6",
"Pkm",
"Pla2g7",
"Plcb1",
"Psap",
"Ptn",
"Pygb",
"Ralyl",
"Rgma",
"Rtn4",
"S100a1",
"S100a6",
"S100b",
"Siah1a",
"Siah1b",
"Scd2",
"Sdc2",
"Sema6a",
"Sema6d",
"Sgcd",
"Sirpa",
"Slc1a2",
"Slc1a3",
"Slc38a1",
"Slc4a4",
"Slc6a11",
"Slc7a10",
"Slit1",
"Slit2",
"Slitrk2",
"Sorbs1",
"Sox9",
"Sparc",
"Spon1",
"Tafa1",
"Timp3",
"Tkt",
"Trpv1",
"Vcam1",
"Vegfa"
) %>% .[. %in% rownames(merged_cortex)]
merged_cortex <- FindVariableFeatures(merged_cortex, nfeatures = 5000, verbose = FALSE)
merged_cortex <- NormalizeData(
merged_cortex,
features = c(
VariableFeatures(merged_cortex),
s100_genes,
genes.embed),
verbose = FALSE)
# Scale data
merged_cortex <- ScaleData(
merged_cortex,
features = c(
VariableFeatures(merged_cortex),
s100_genes,
genes.embed),
verbose = FALSE)# Create DimPlot
p1 <- DimPlot(
merged_cortex,
reduction = "umap",
group.by = c("stage", "New_cellType"),
combine = FALSE, label.size = 2
)
p2 <- DimPlot(
merged_cortex,
reduction = "tsne",
group.by = c("stage", "New_cellType"),
combine = FALSE, label.size = 2
)wrap_plots(c(p1, p2), ncol = 2, byrow = F)
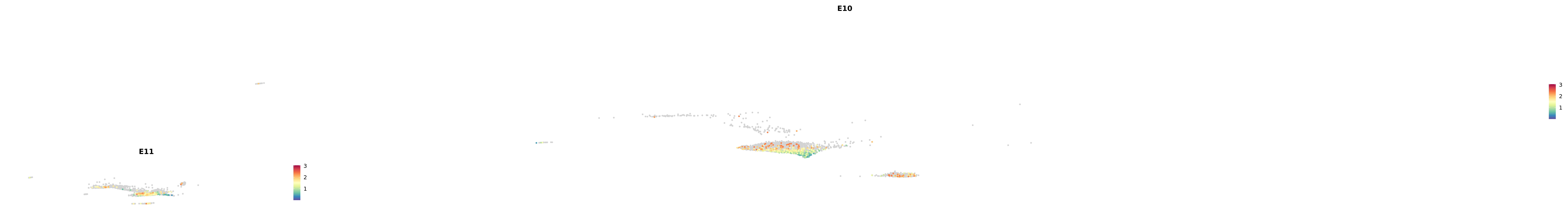
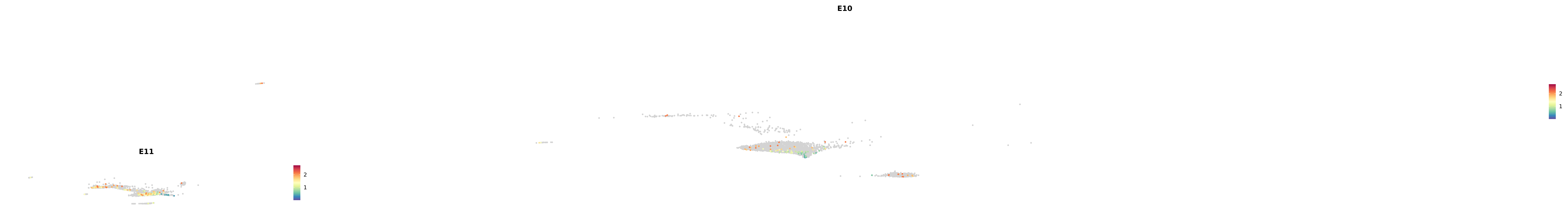
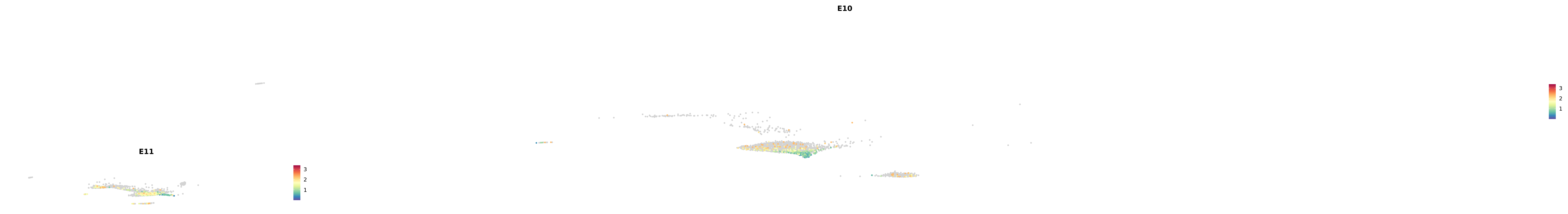
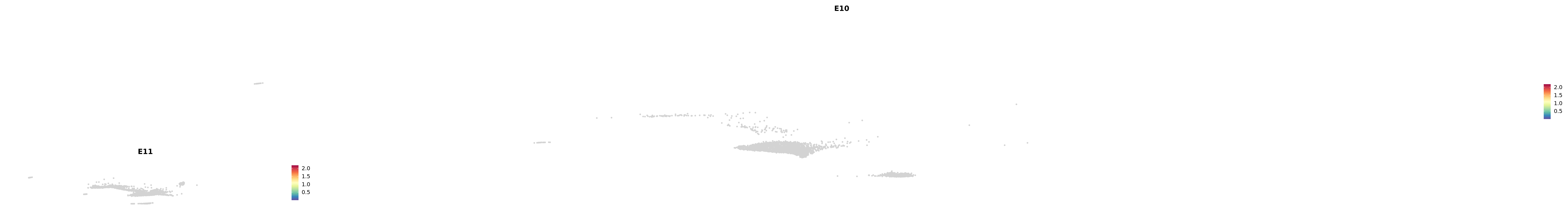
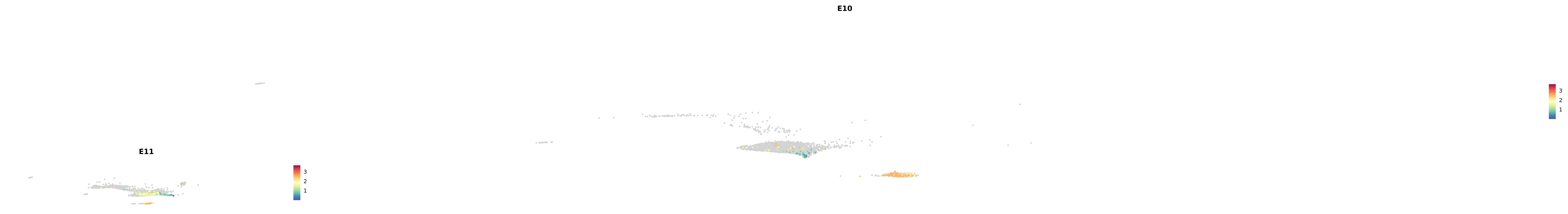
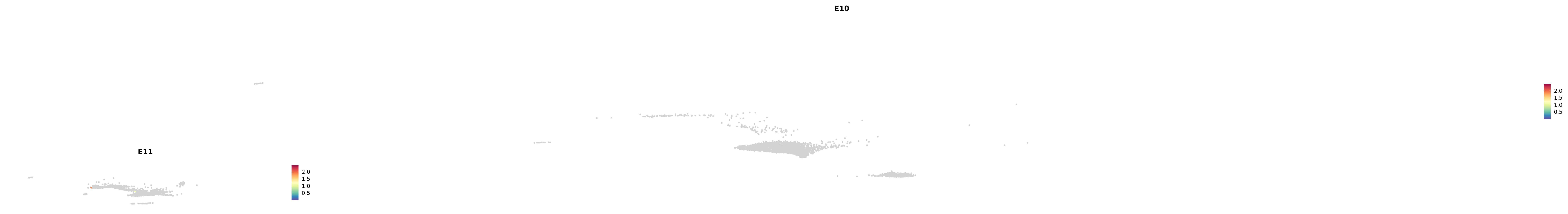
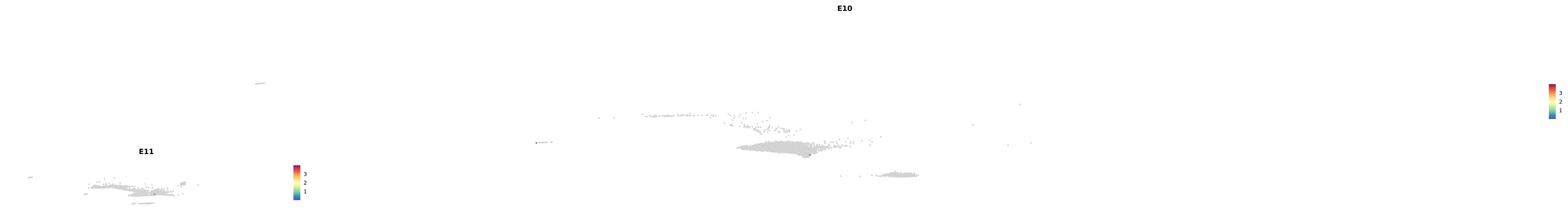
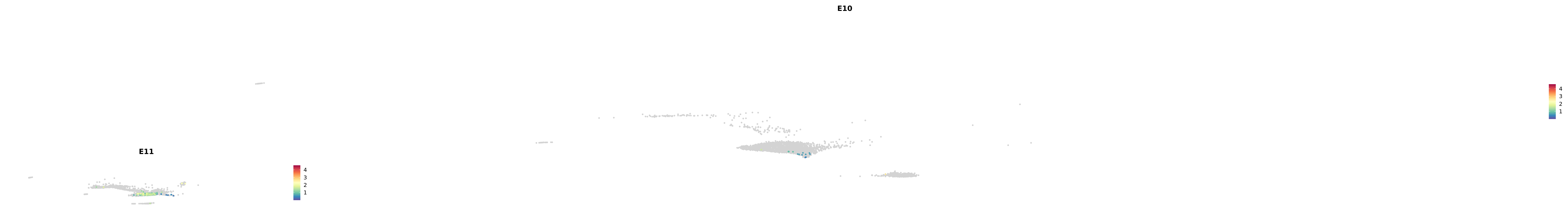
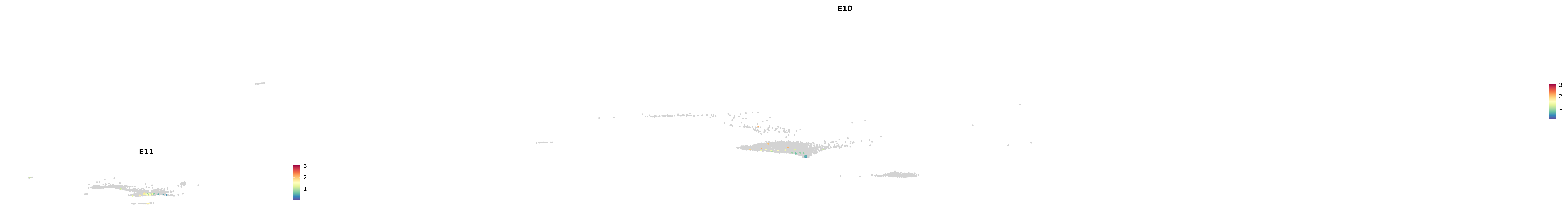
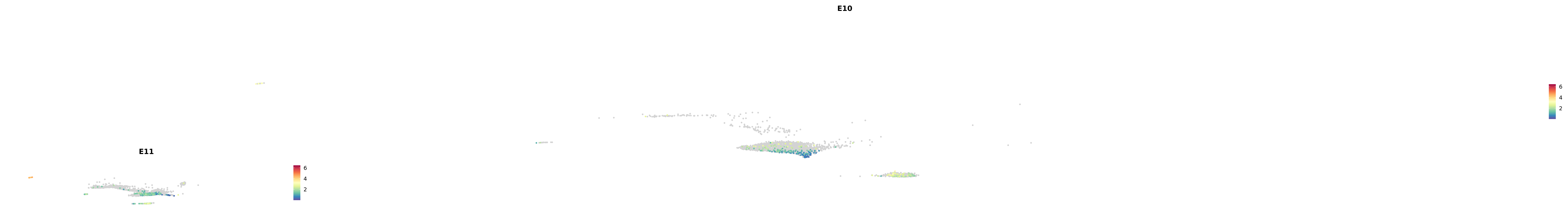
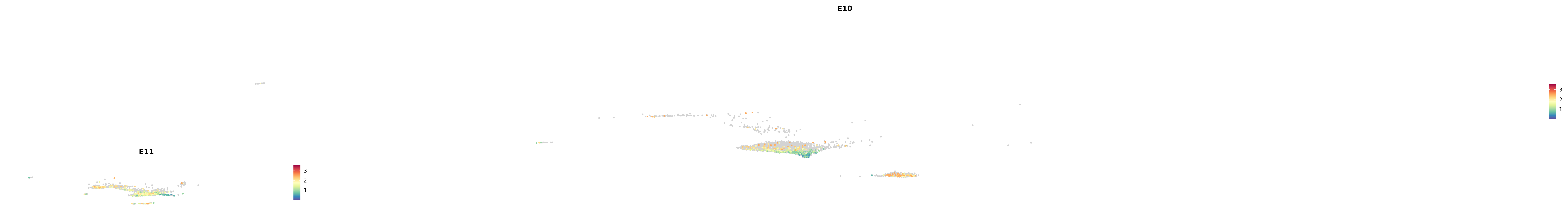
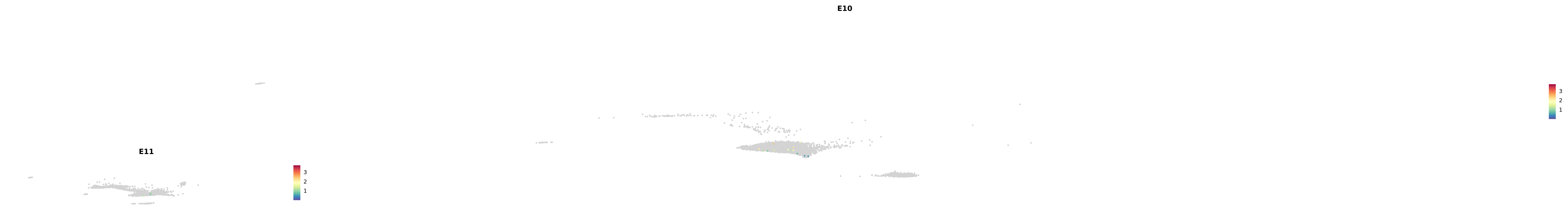
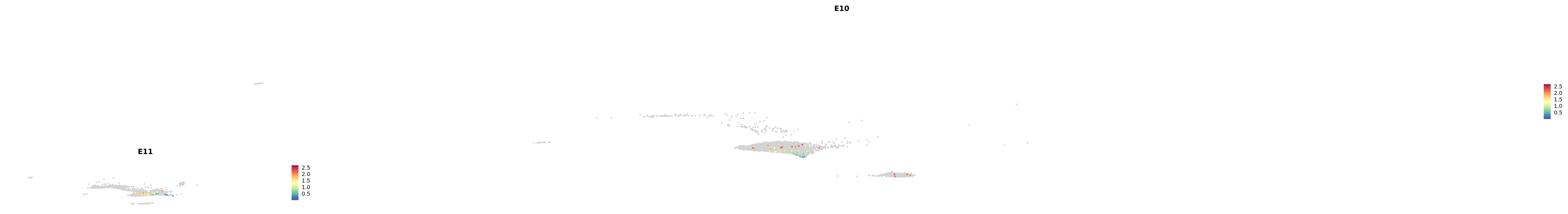
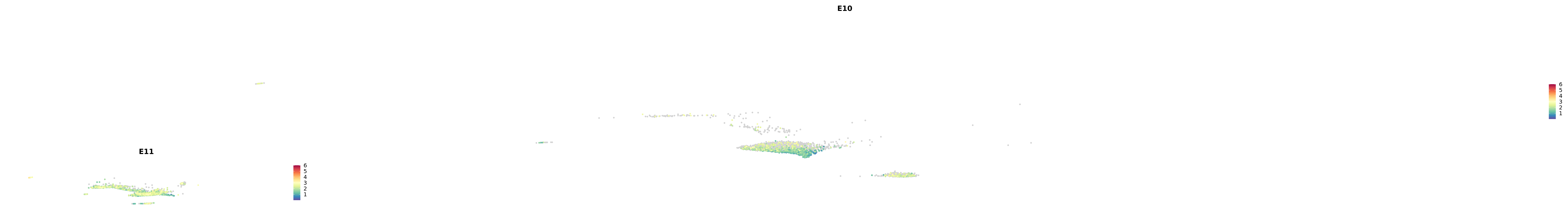
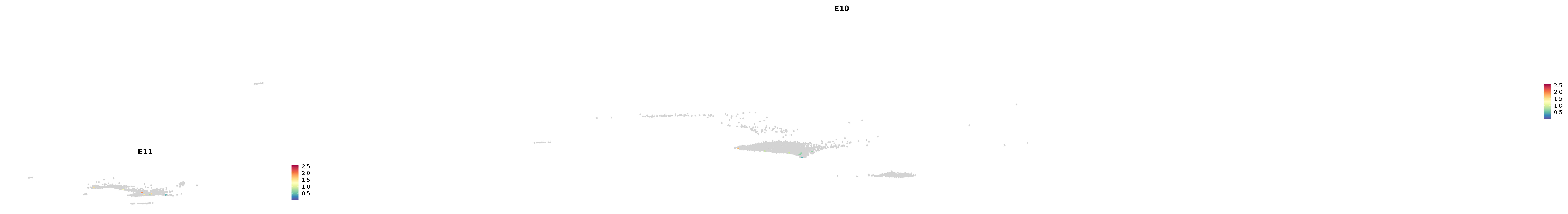
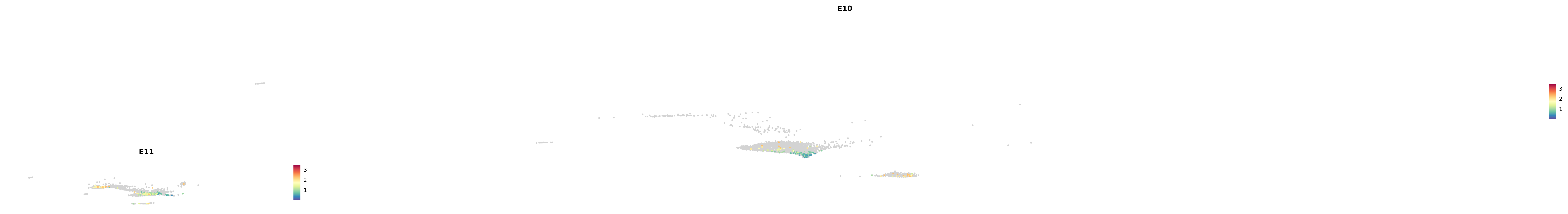
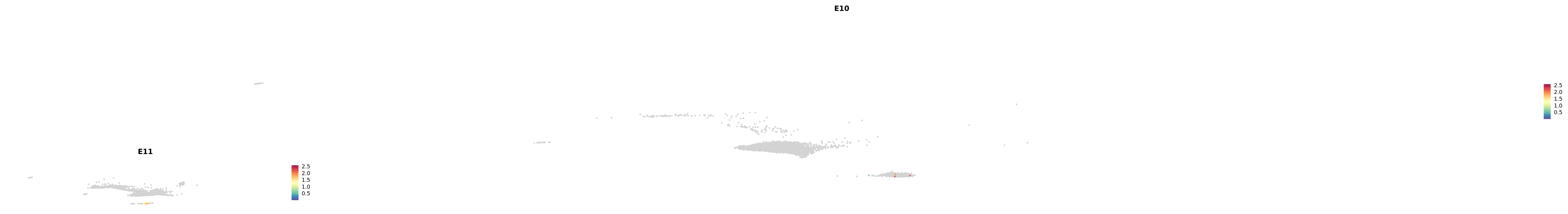
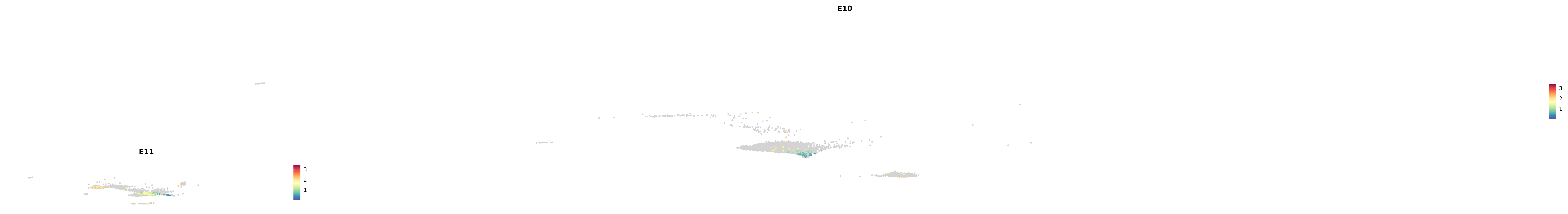
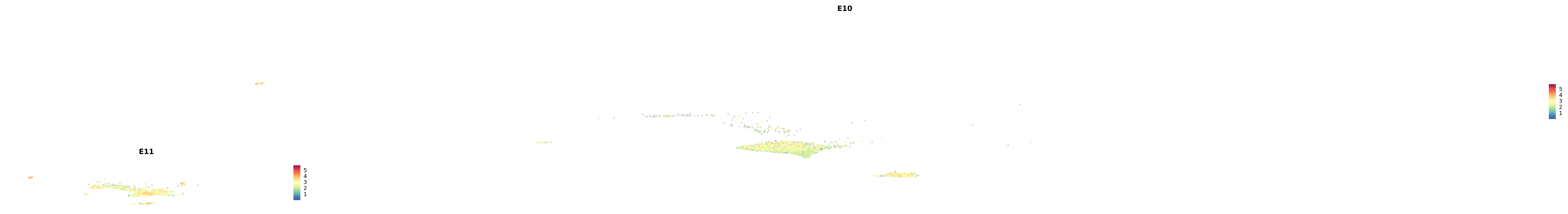
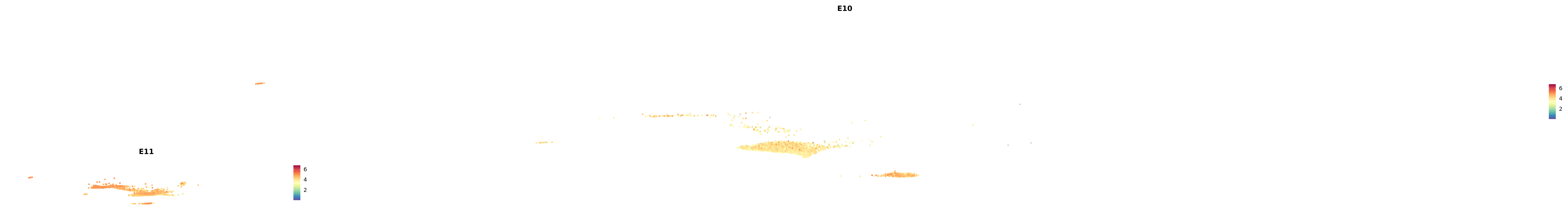
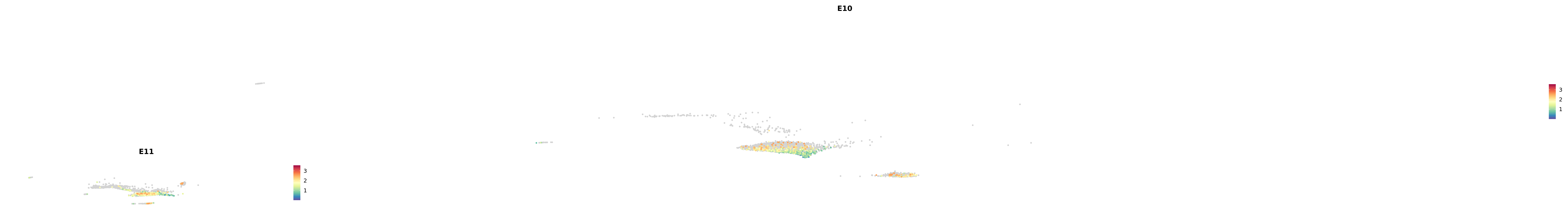
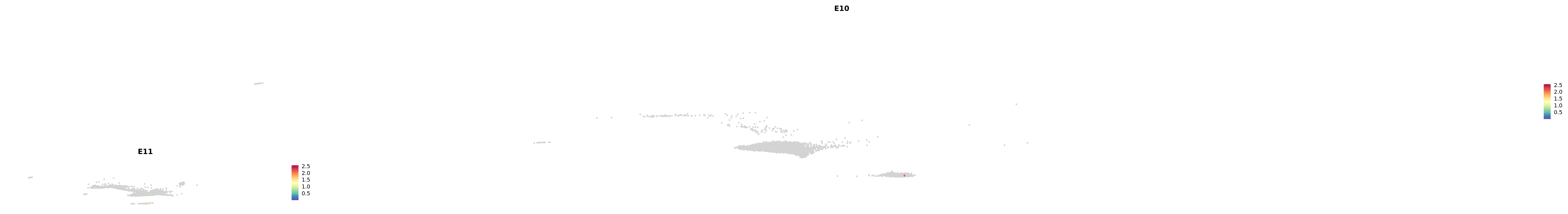
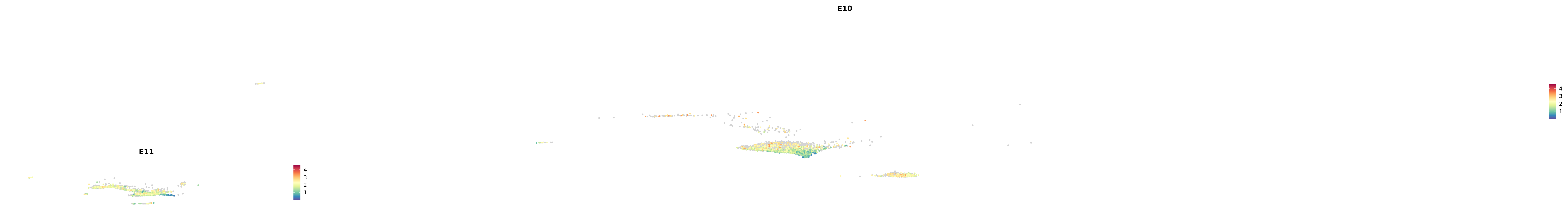
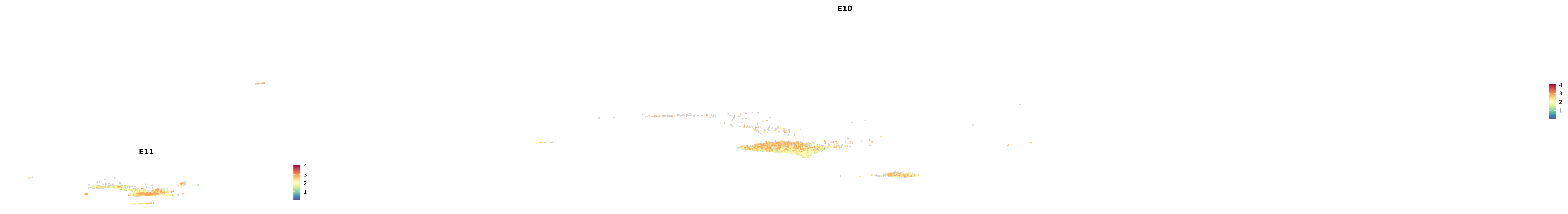
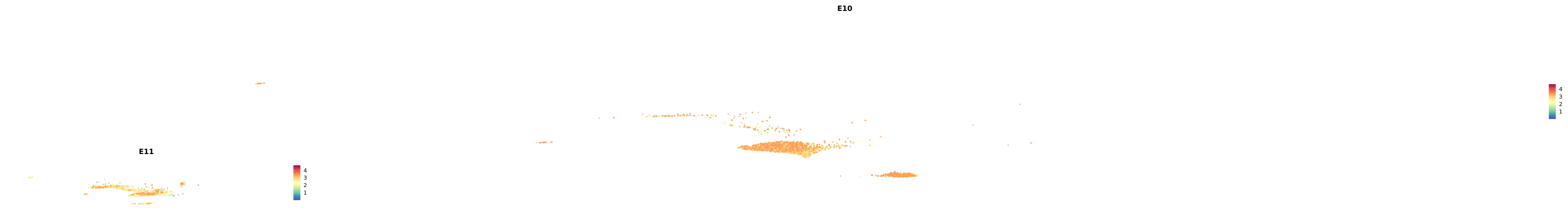
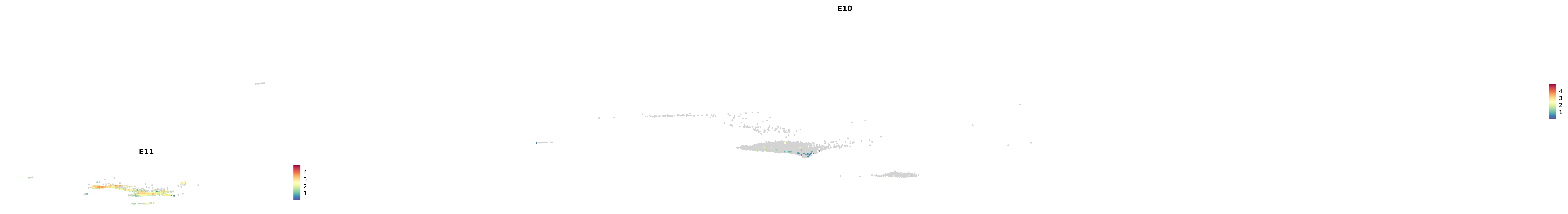
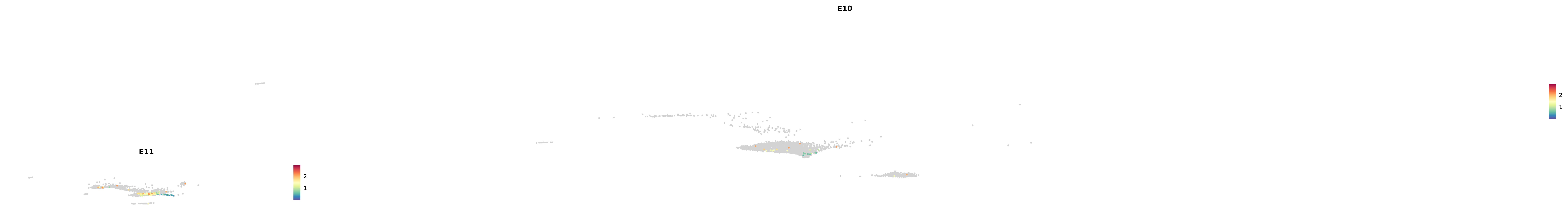
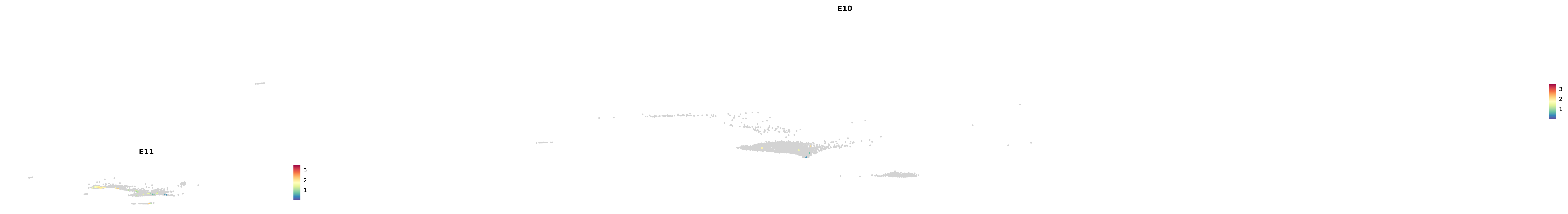
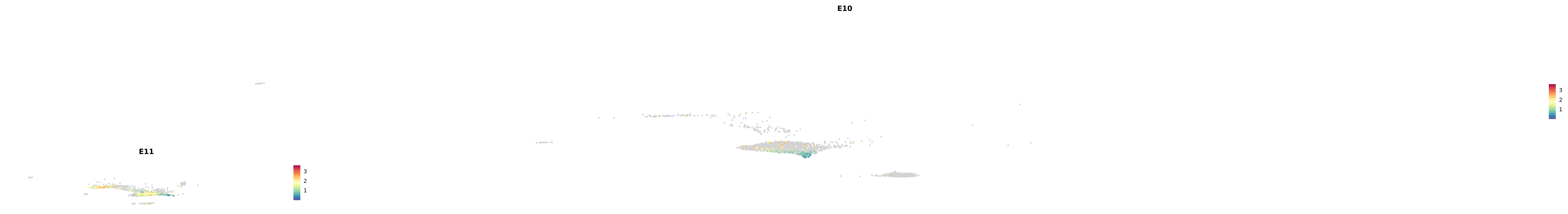
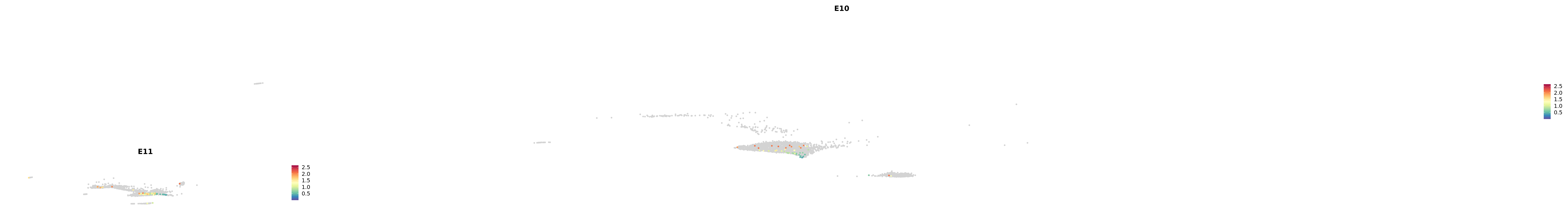
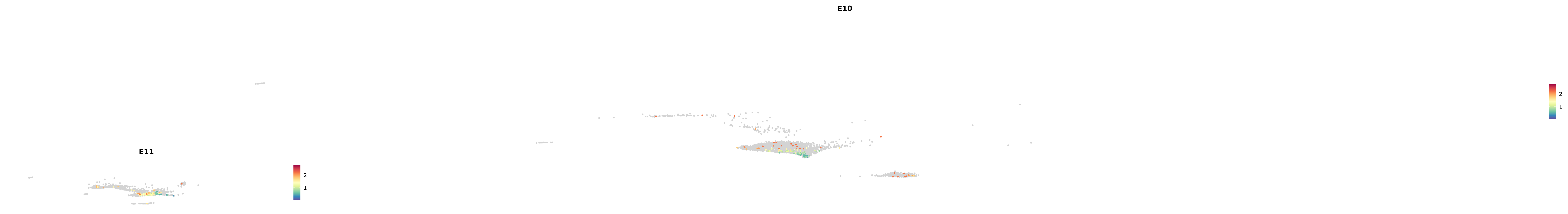
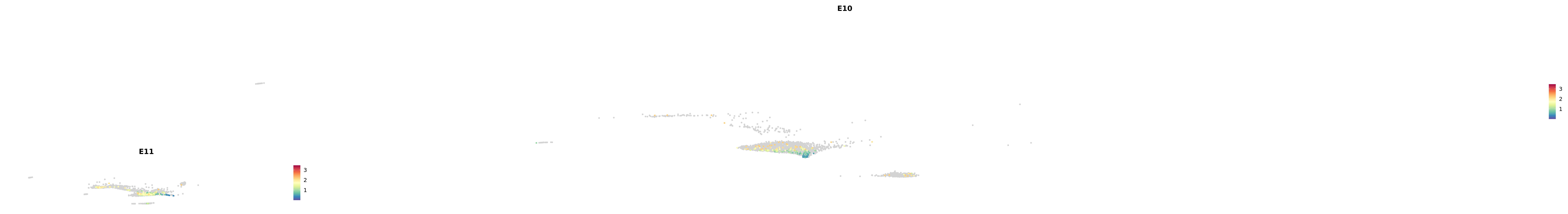
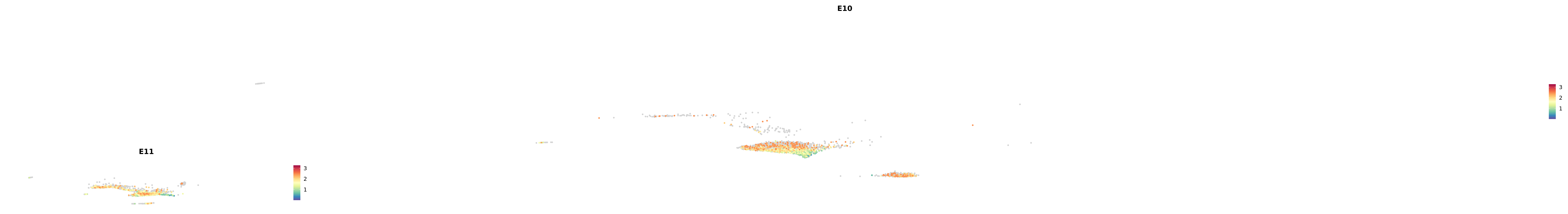
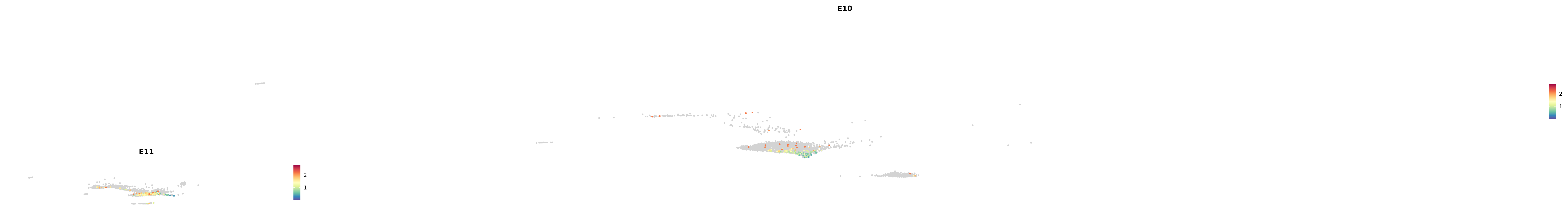
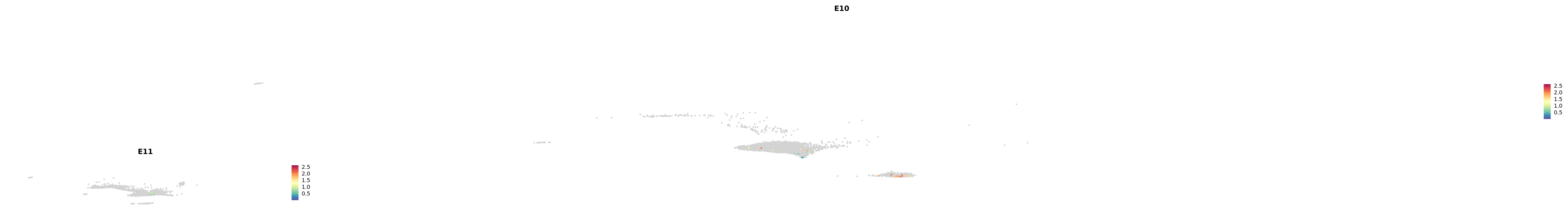
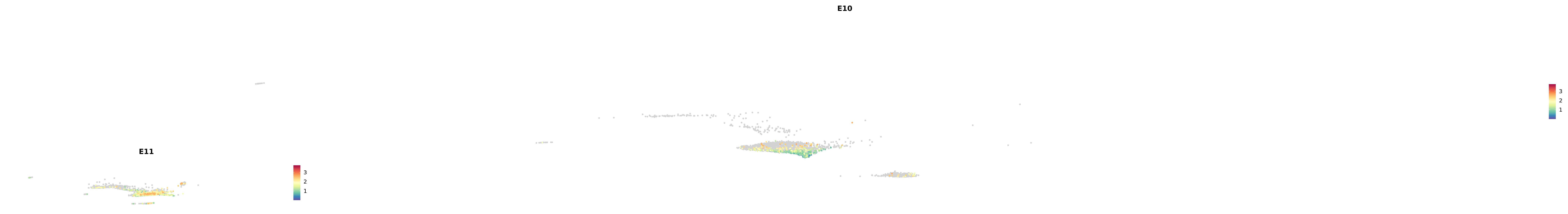
# Create a custom FeaturePlot for each S100 gene
plot_list <-
lapply(
c(s100_genes, "Cacybp", "Siah1a", "Siah1b"),
function(gene) {
FeaturePlot_scCustom(
seurat_object = merged_cortex,
features = gene,
colors_use = merged_cortex@misc$expr_Colour_Pal,
na_color = "lightgray",
layer = "data",
order = TRUE,
pt.size = 1,
reduction = "umap",
split.by = "stage",
split_collect = FALSE,
label = F,
label_feature_yaxis = TRUE,
combine = FALSE
)
})
NOTE: FeaturePlot_scCustom uses a specified `na_cutoff` when plotting to
color cells with no expression as background color separate from color scale.
Please ensure `na_cutoff` value is appropriate for feature being plotted.
Default setting is appropriate for use when plotting from 'RNA' assay.
When `na_cutoff` not appropriate (e.g., module scores) set to NULL to
plot all cells in gradient color palette.
-----This message will be shown once per session.-----# Combine the plots into a single grid
combined_plot <- patchwork::wrap_plots(plot_list, ncol = 1)
# Display the combined plot
print(combined_plot)
# Create a compact DotPlot
compact_plot <- DotPlot(
object = merged_cortex,
features = c(s100_genes,
"Cacybp",
"Siah1a",
"Siah1b"),
group.by = "stage",
cluster.idents = FALSE,
scale = TRUE,
dot.scale = 12
) + RotatedAxis()
# Display the compact plot
print(compact_plot)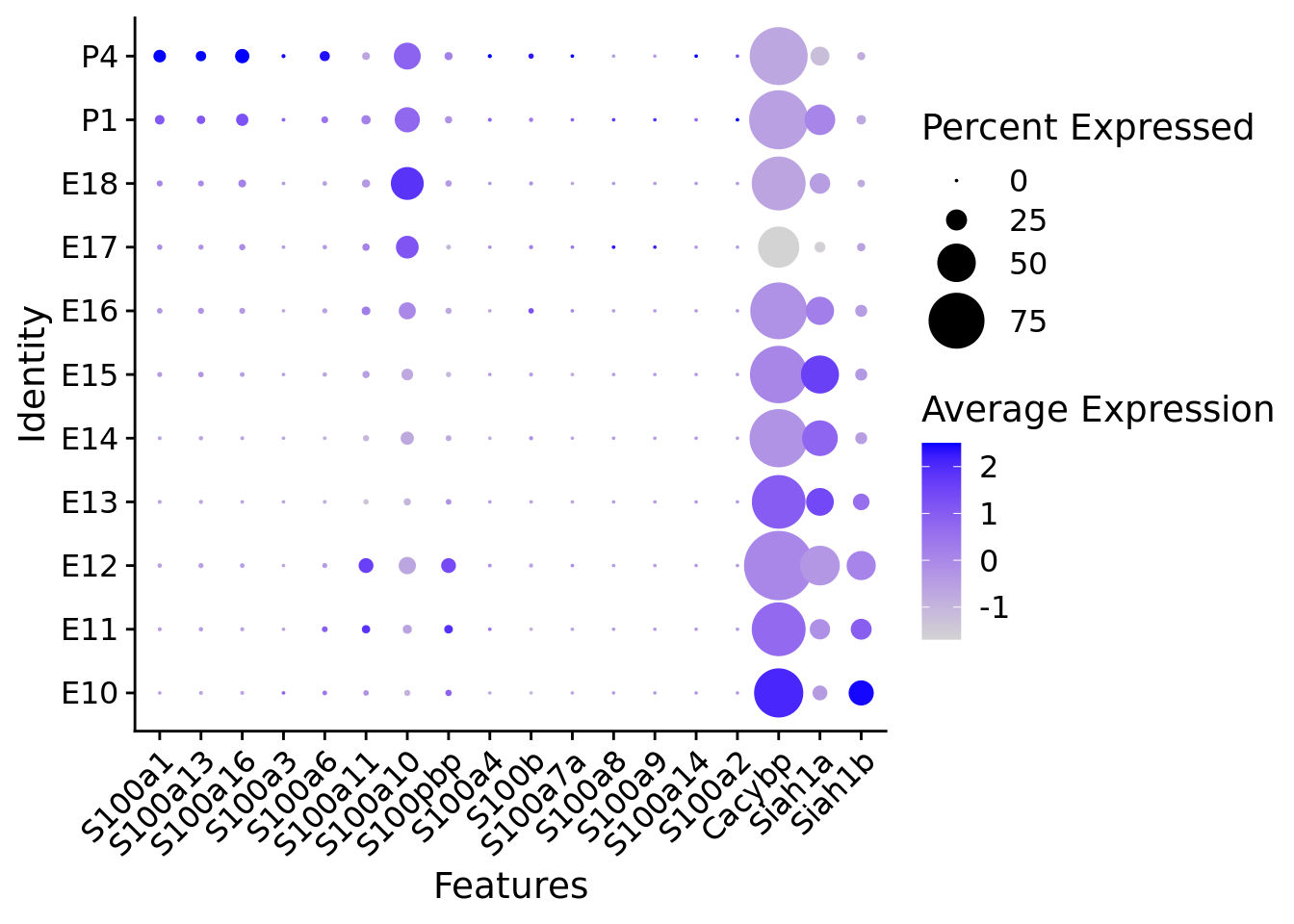
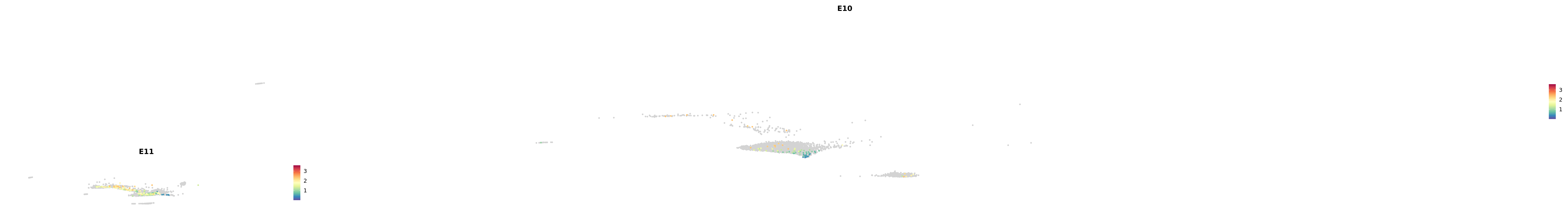
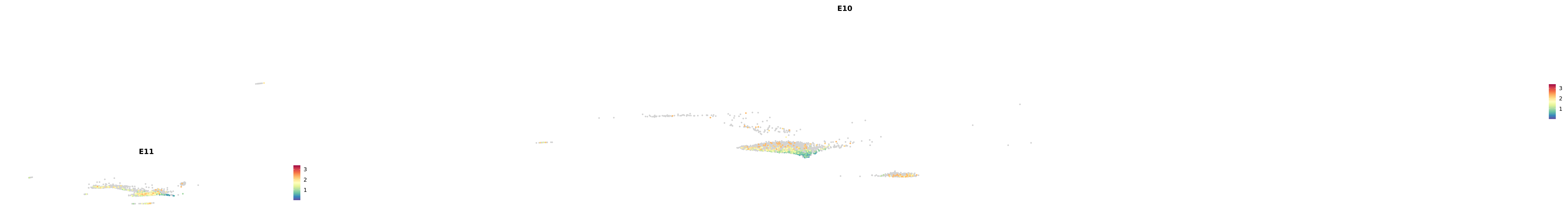
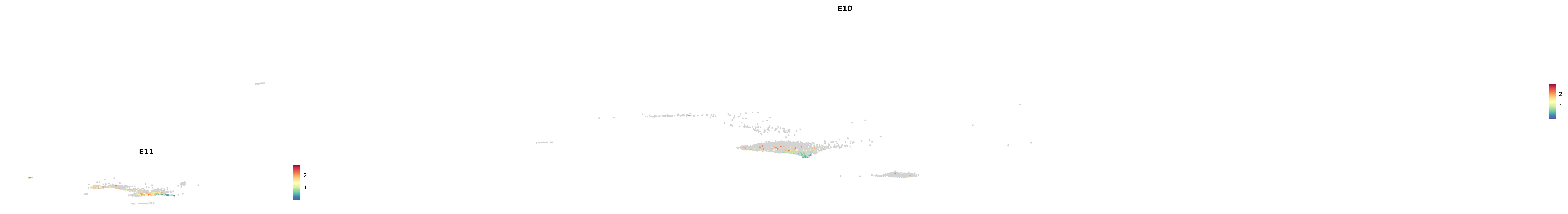
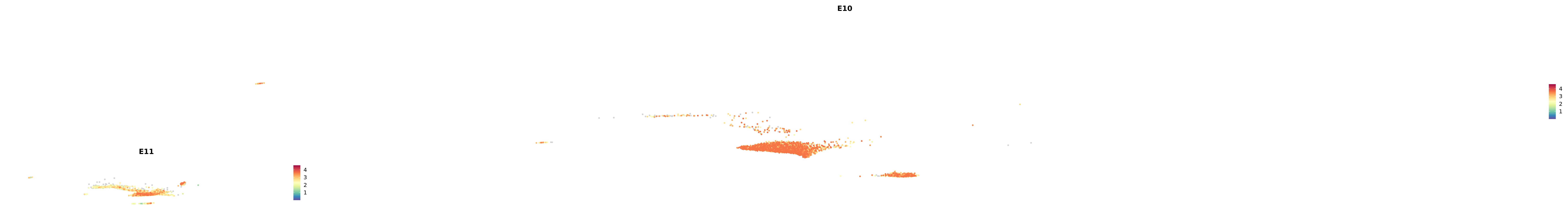
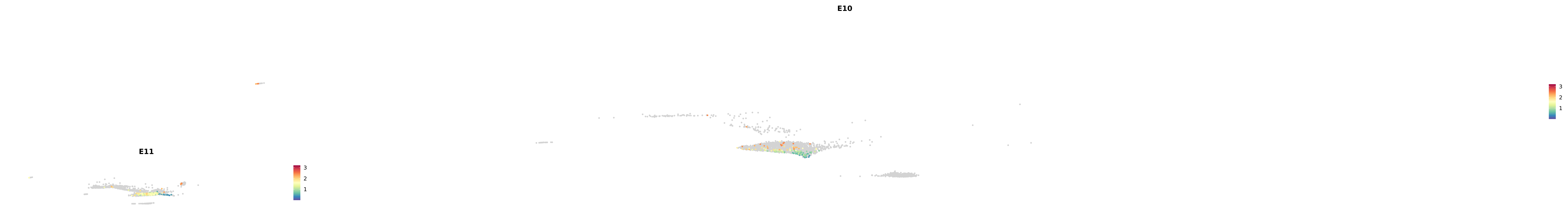
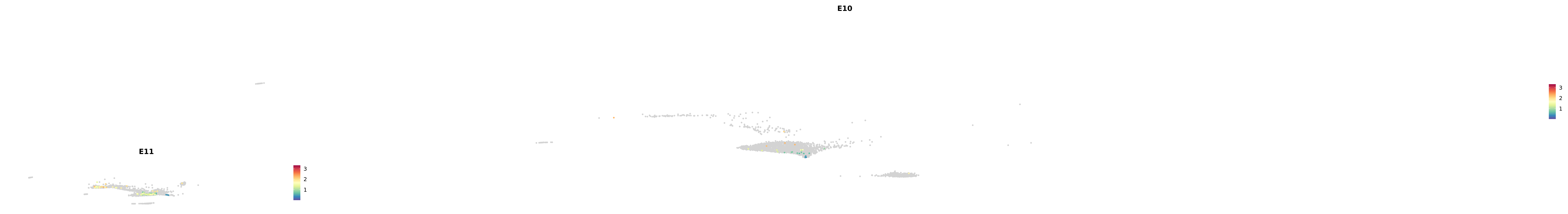
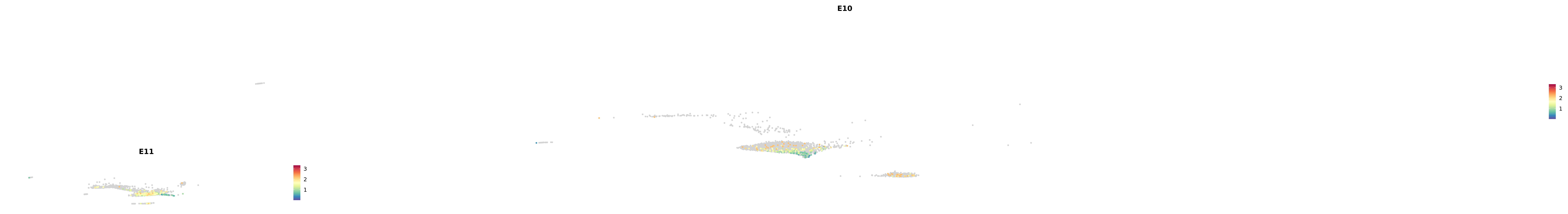
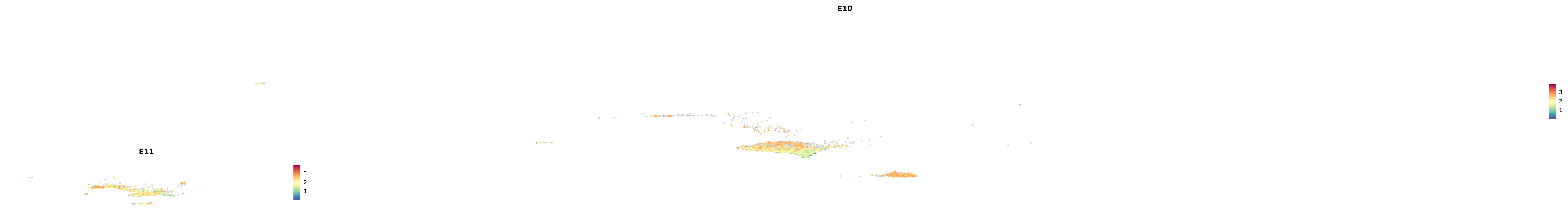
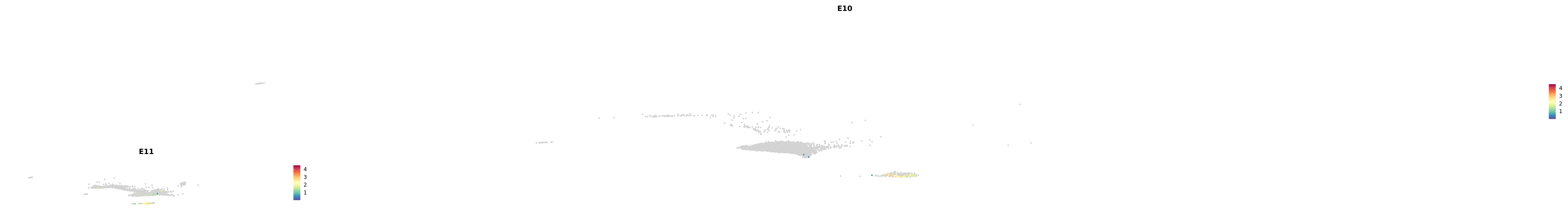
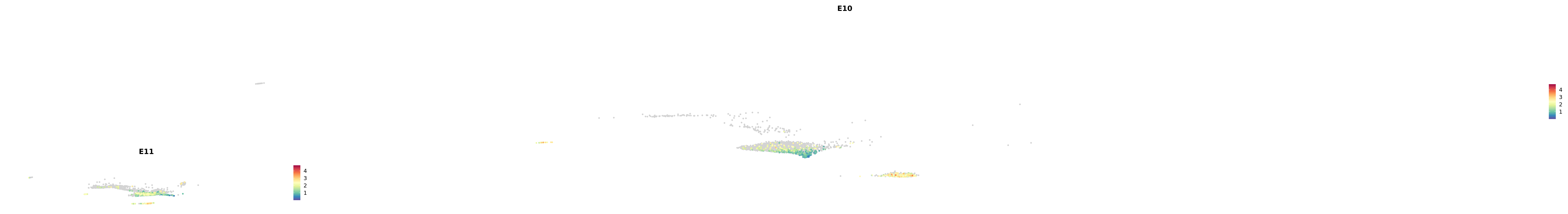
plot_gene_by_dev <- function(x) {
f_plot <- FeaturePlot_scCustom(
merged_cortex,
colors_use = merged_cortex@misc$expr_Colour_Pal,
features = x,
layer = "data",
max.cutoff = "q99",
na_color = "lightgray",
figure_plot = T,
pt.size = 1,
reduction = "umap",
split.by = "stage",
split_collect = FALSE,
label = F,
label_feature_yaxis = TRUE,
combine = FALSE
)
print(f_plot)
}
genes.embed |> purrr::walk(plot_gene_by_dev)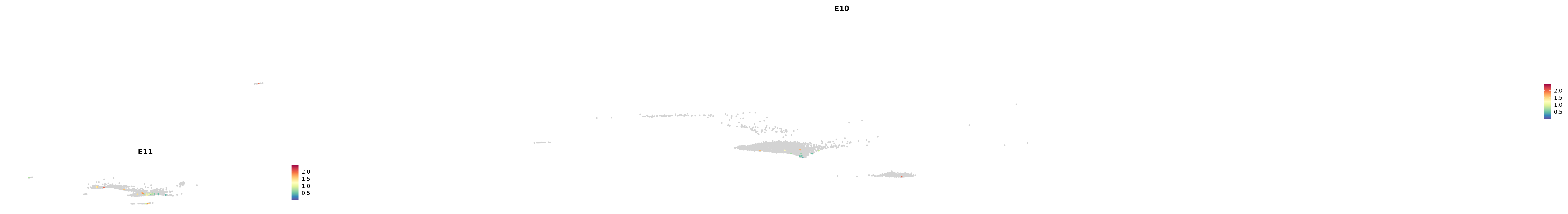
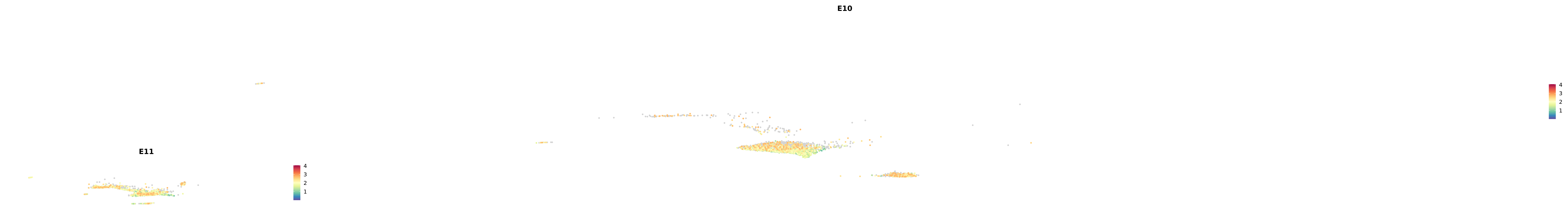
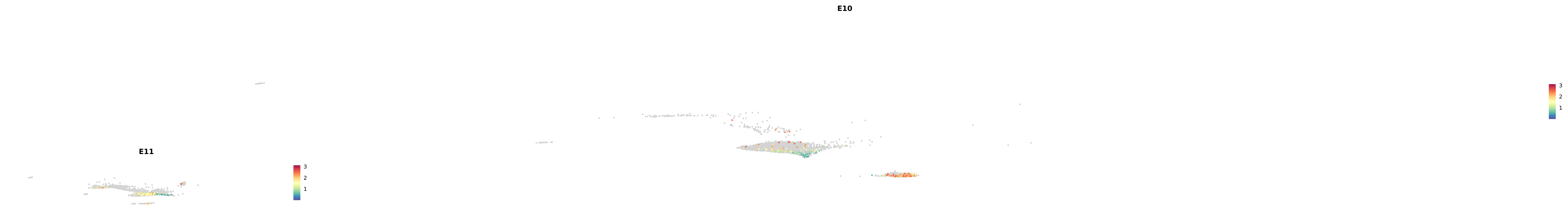
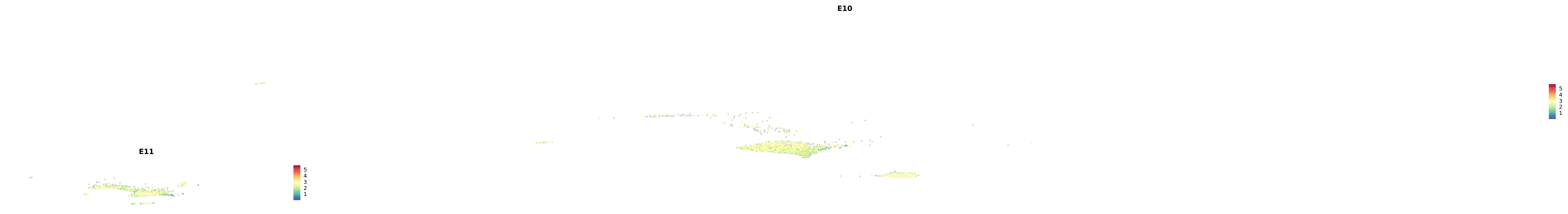
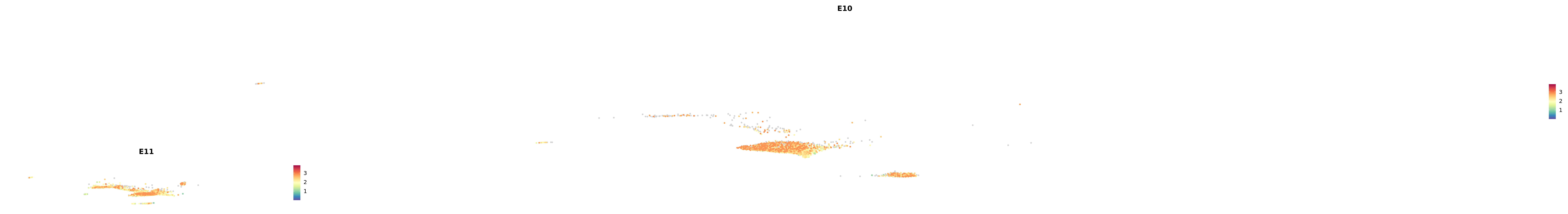
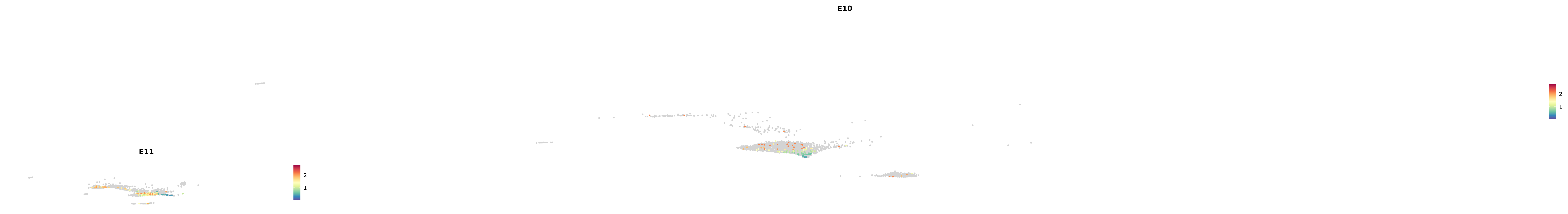
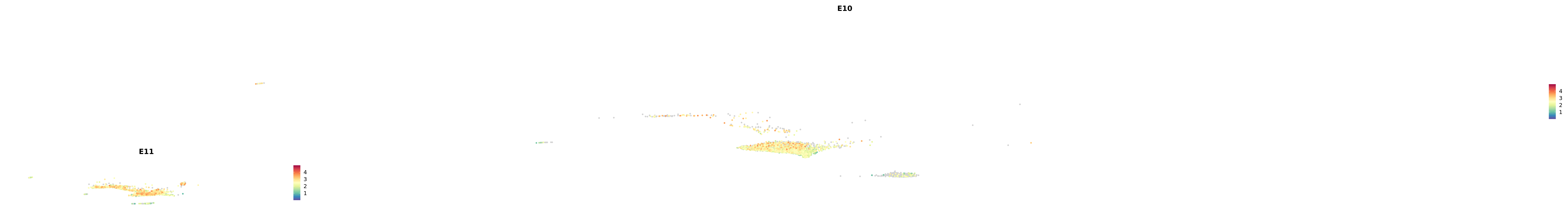
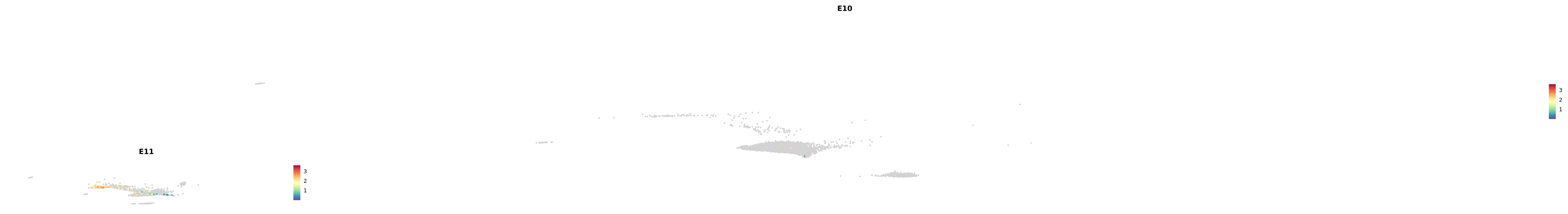
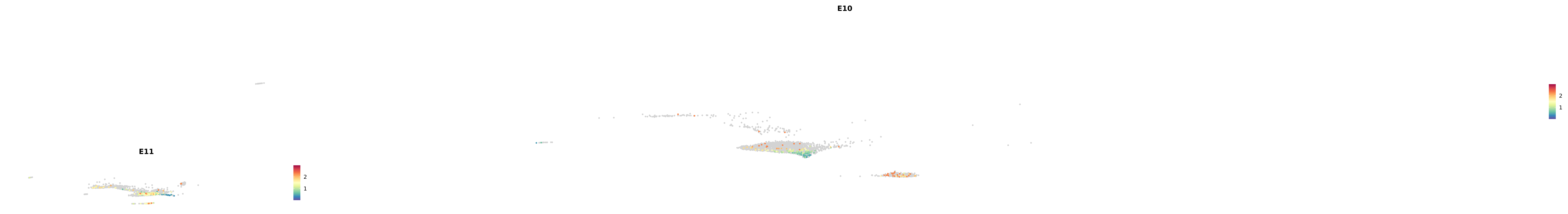
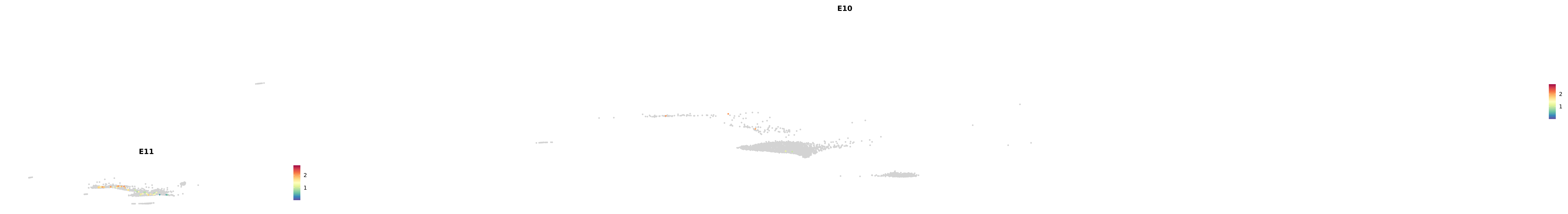
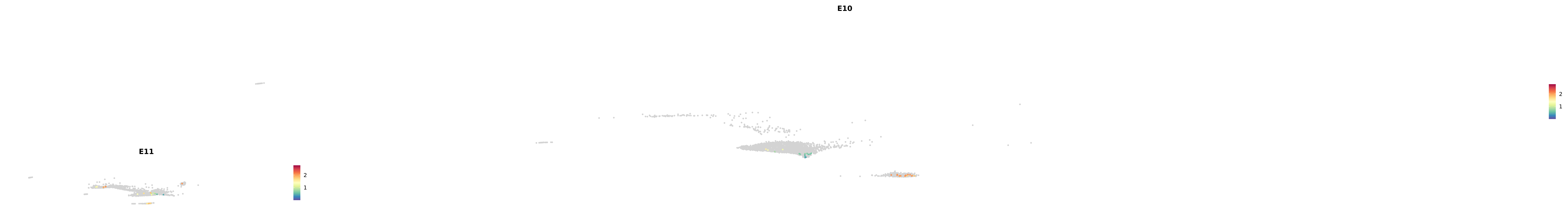
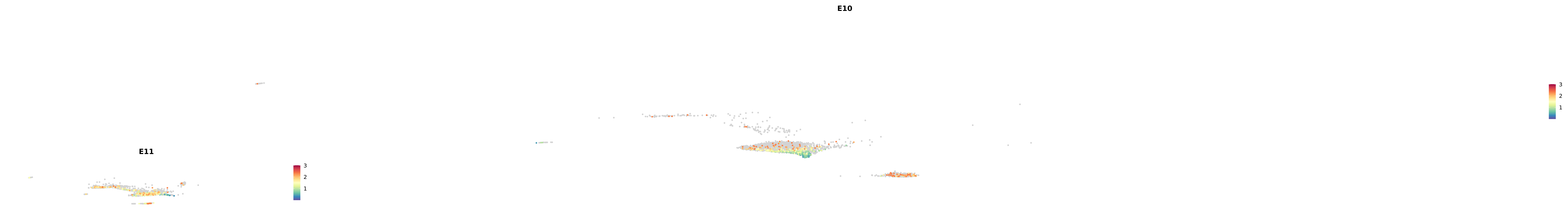

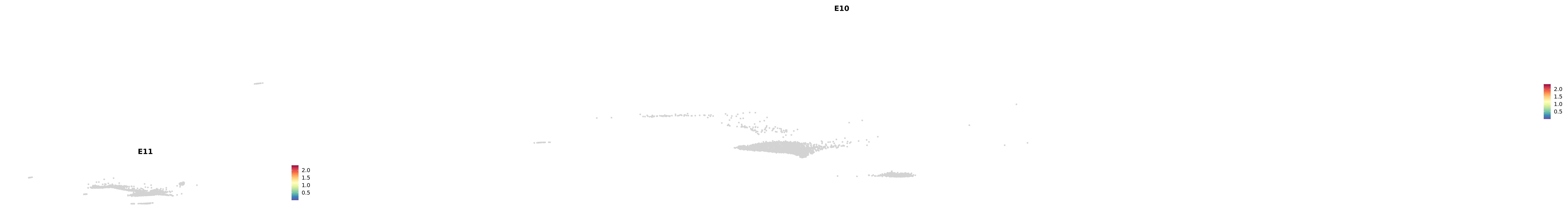
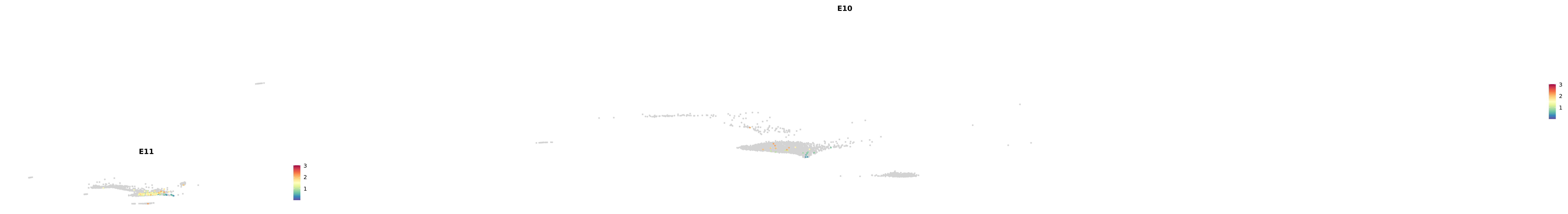
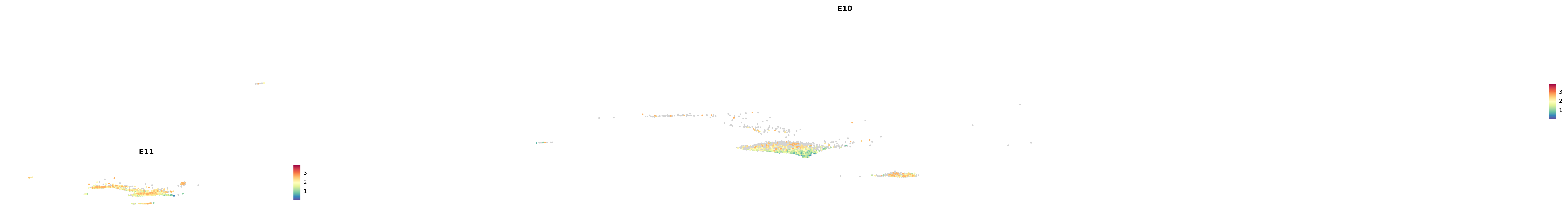
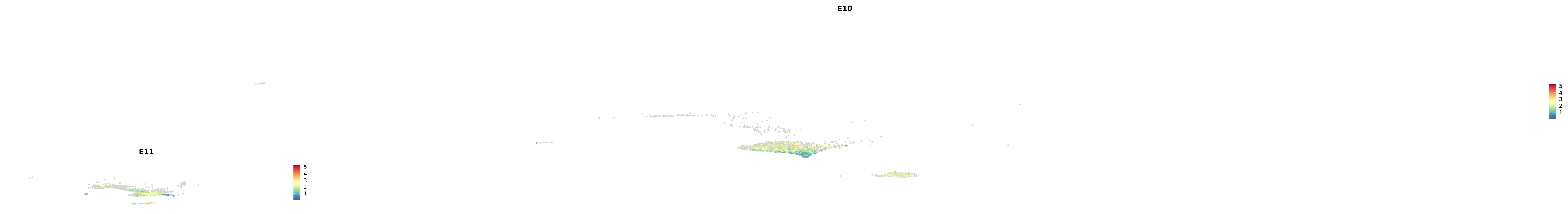

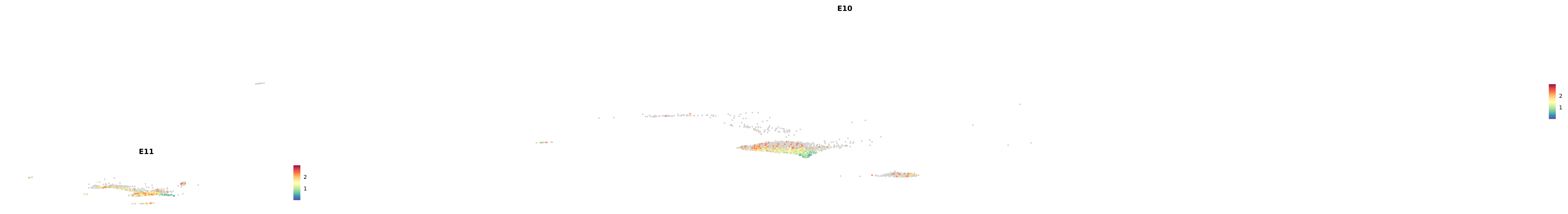
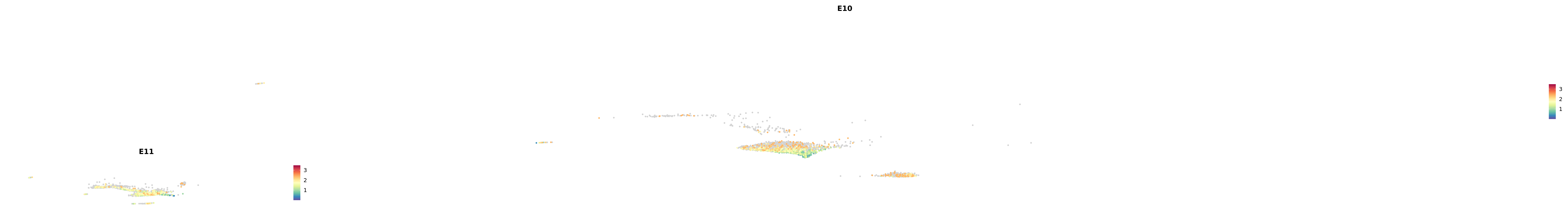
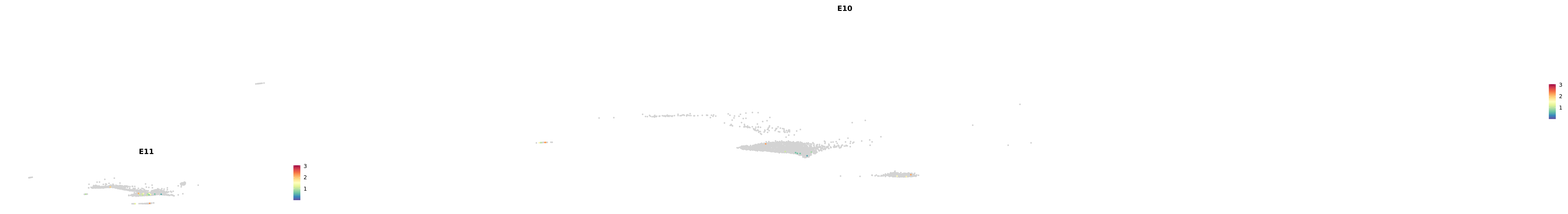


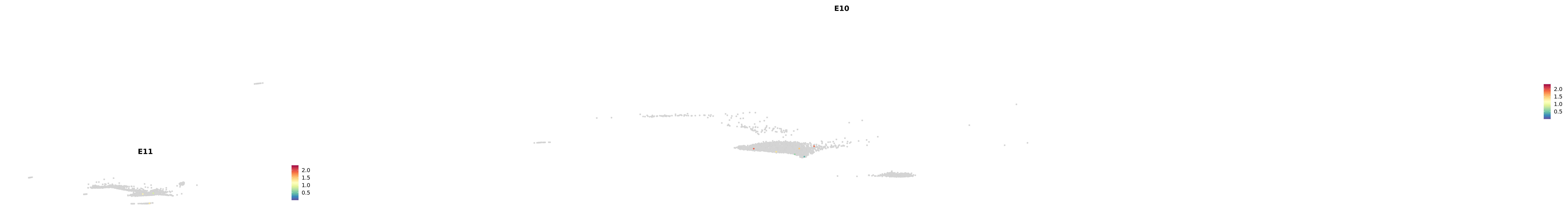
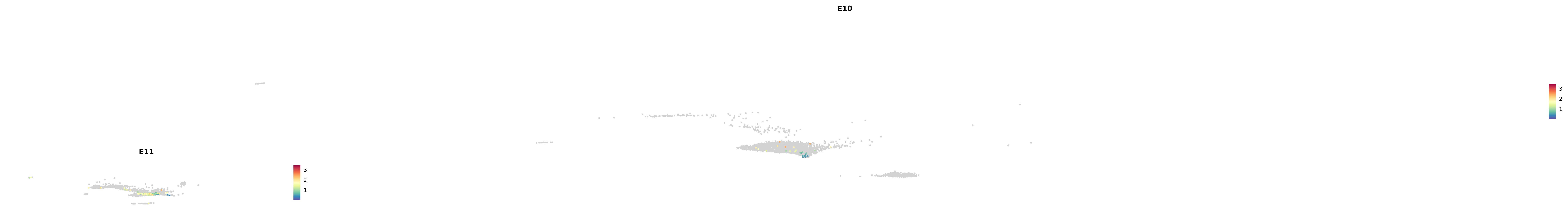
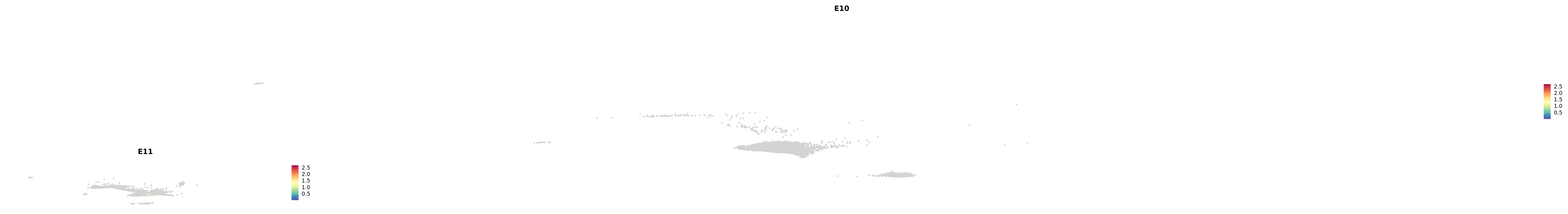
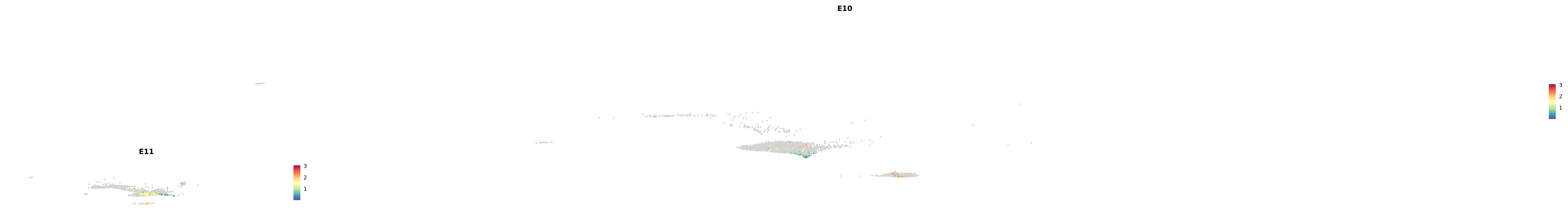
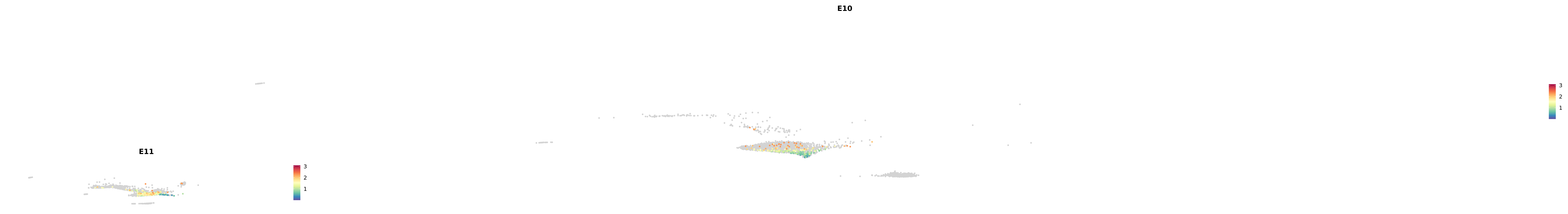
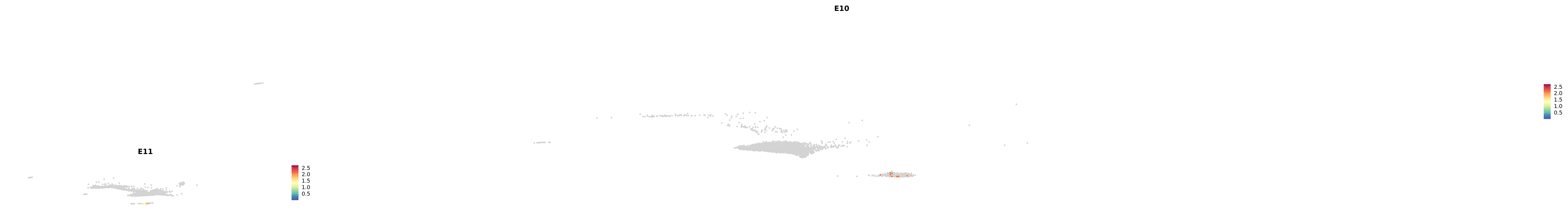
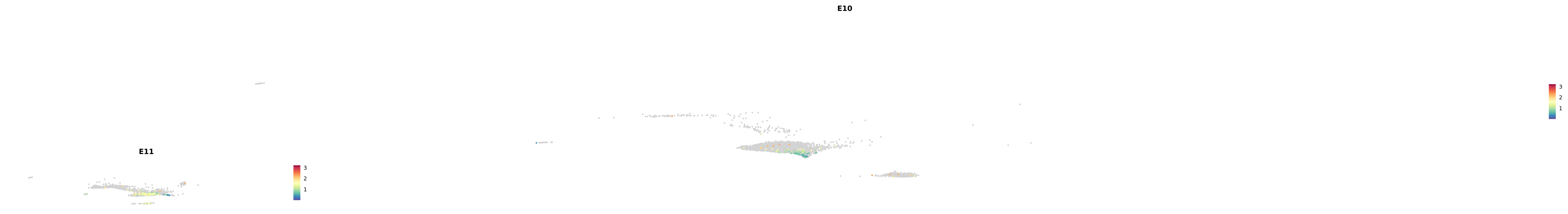
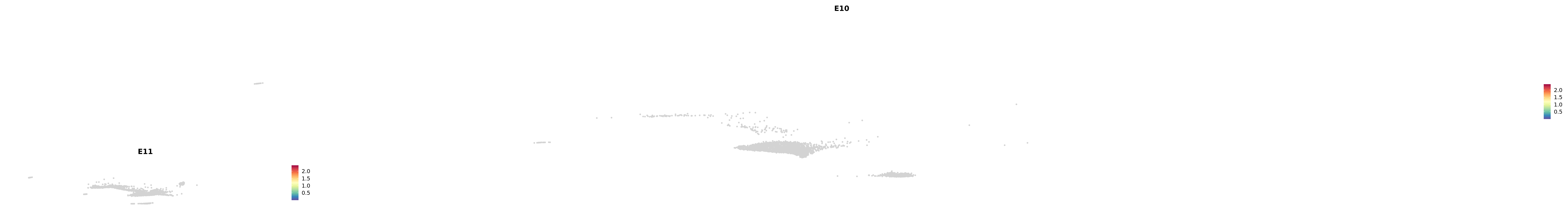


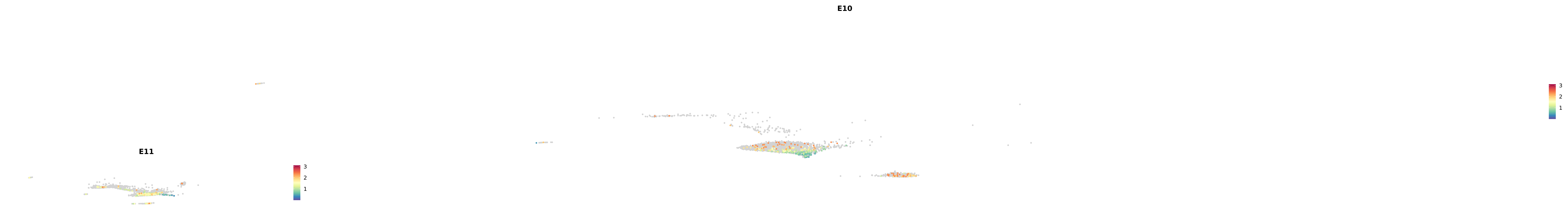
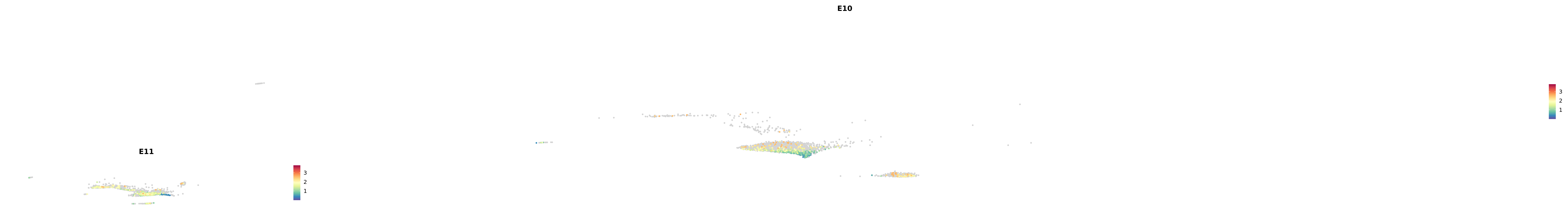
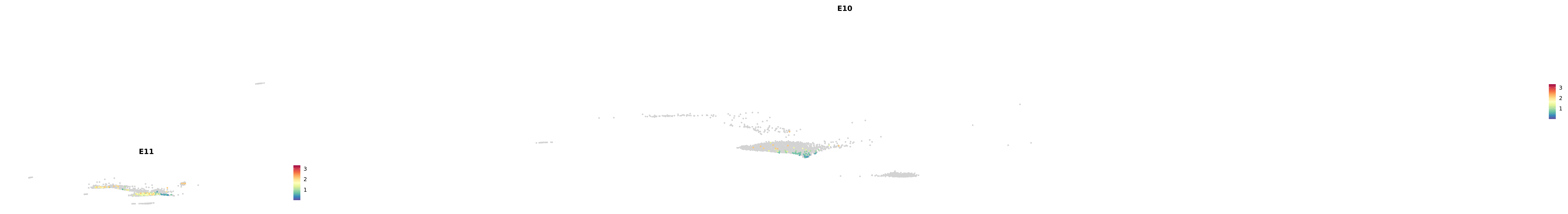
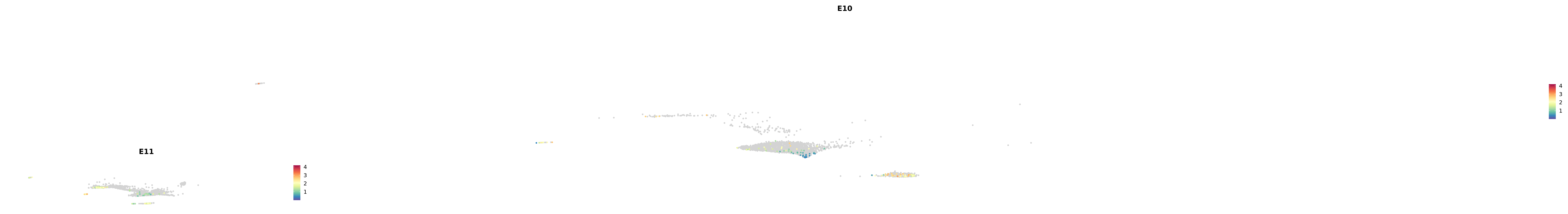
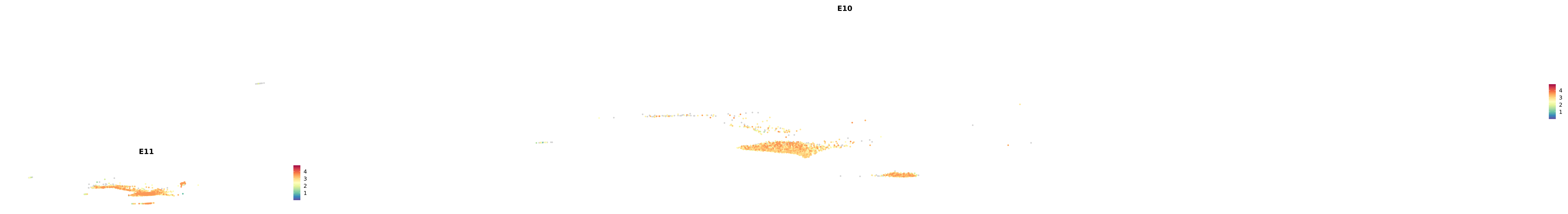
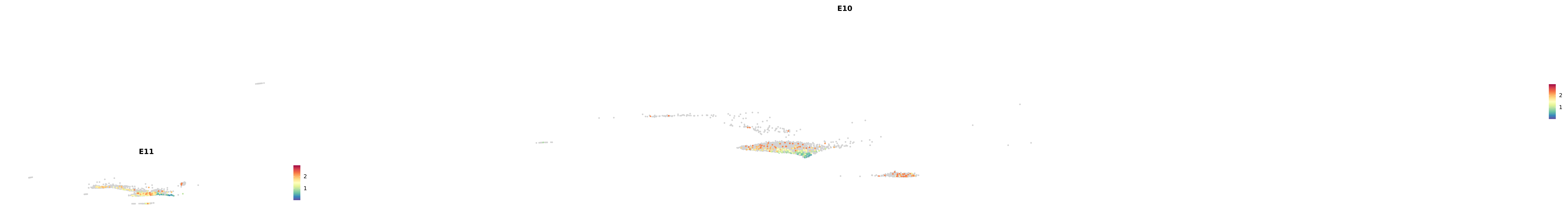
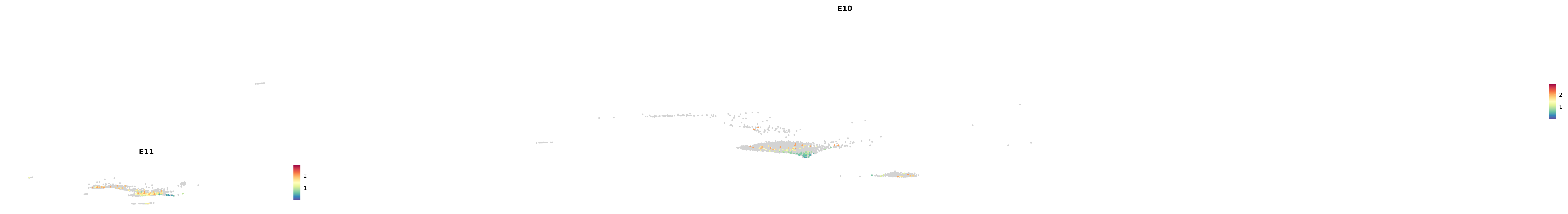
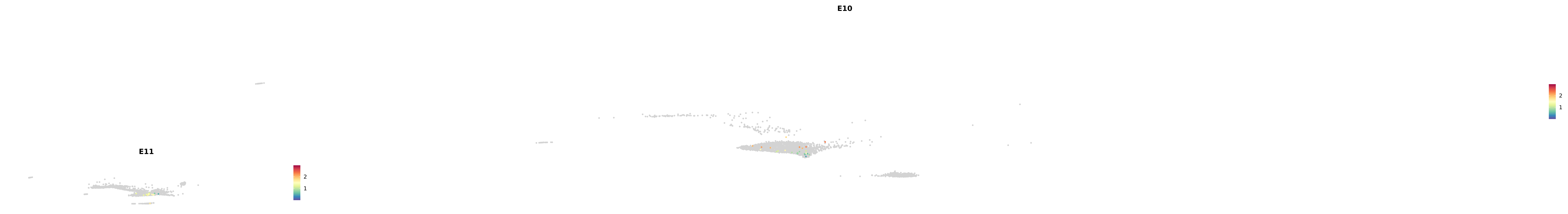
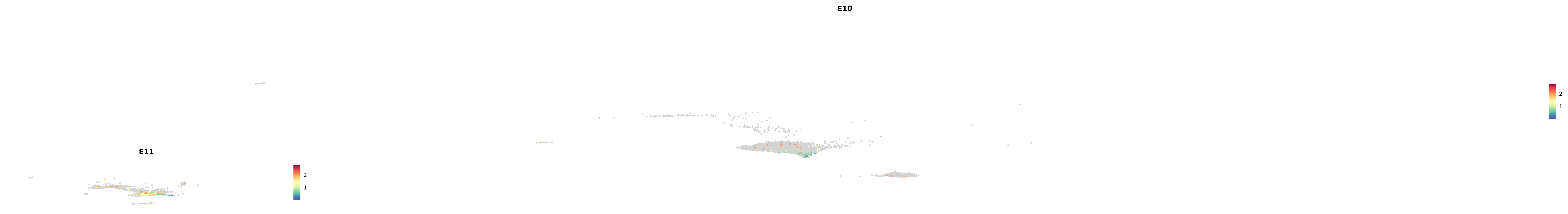
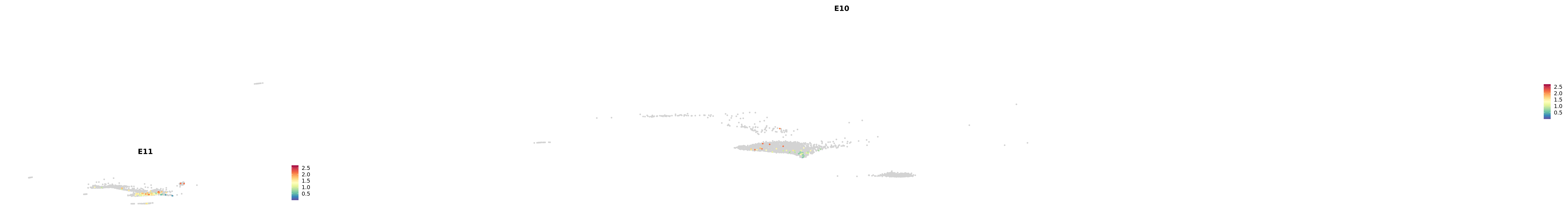
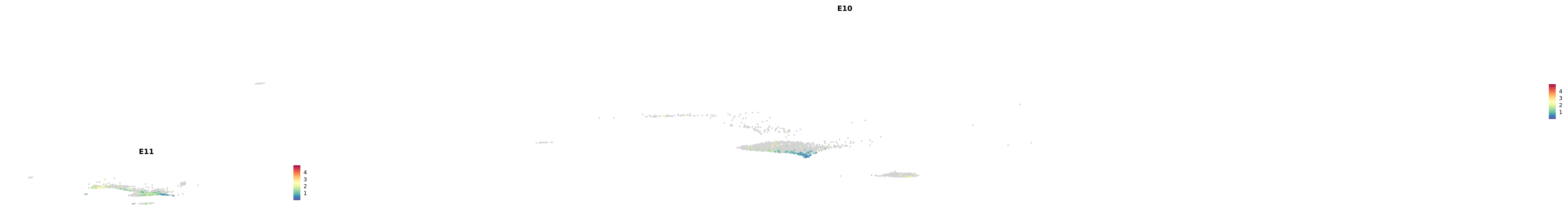

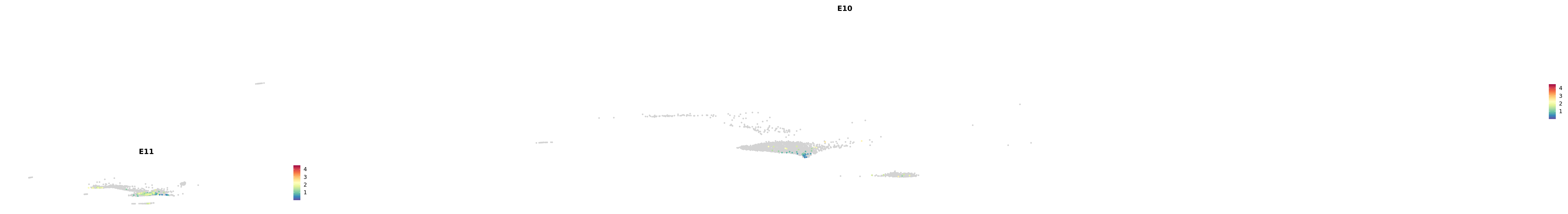
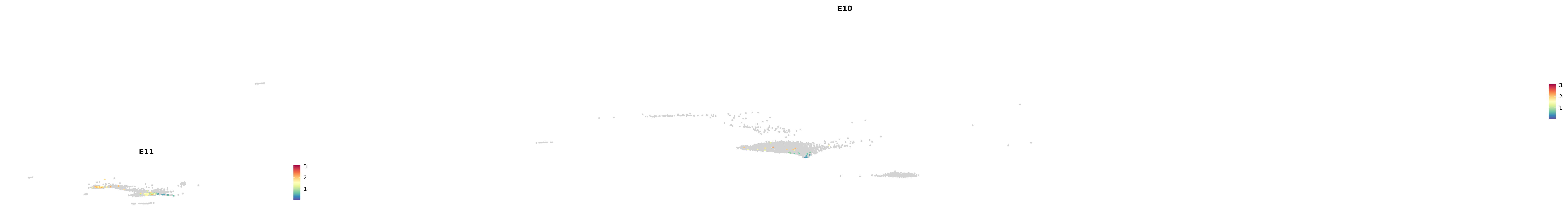
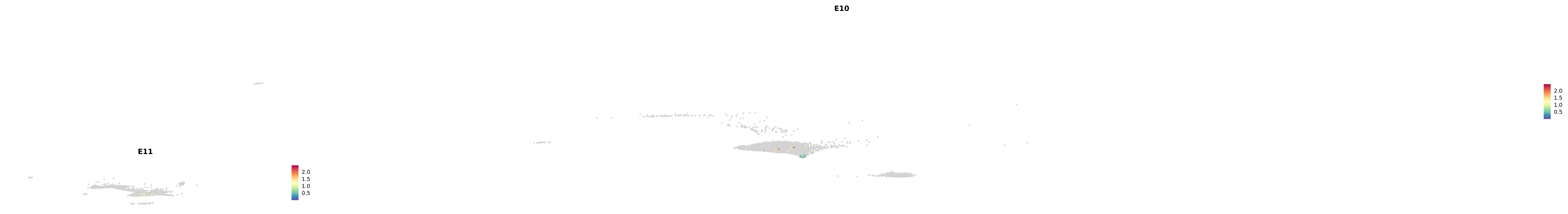
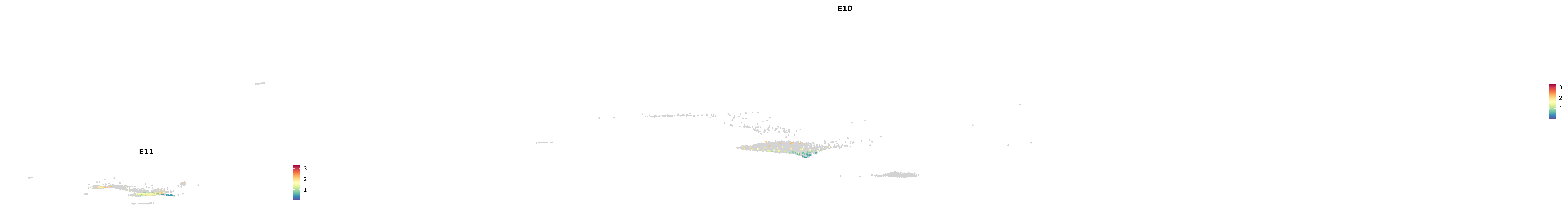
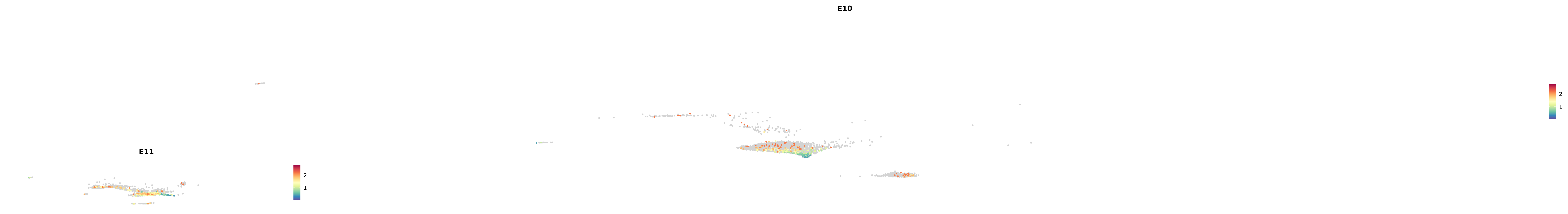
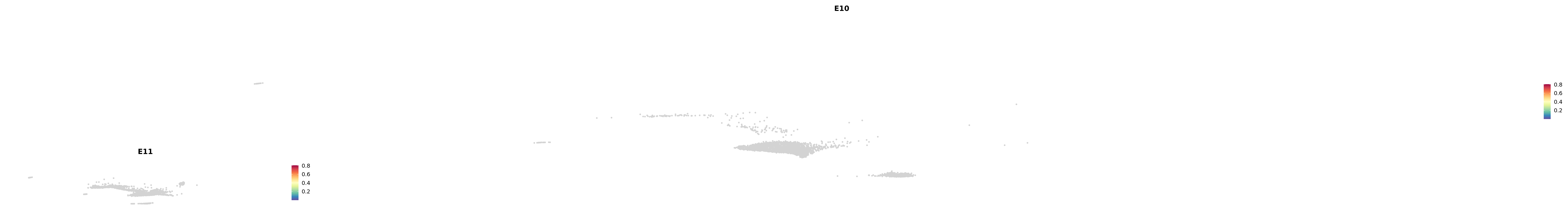
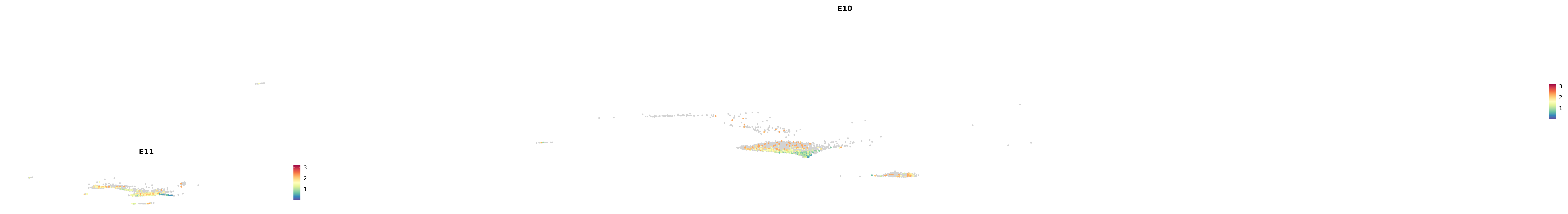
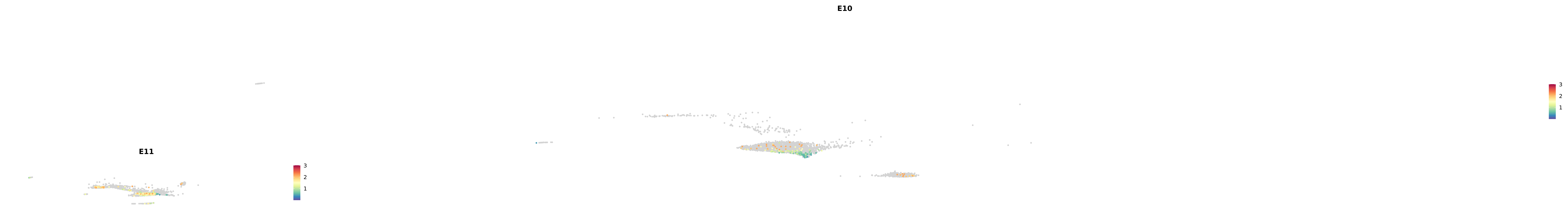
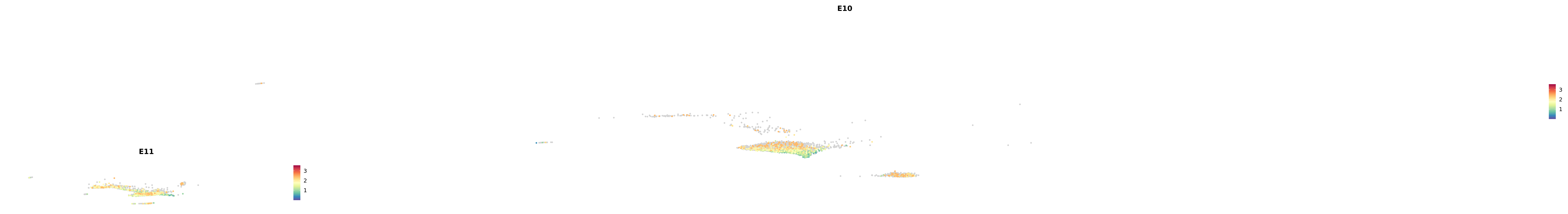
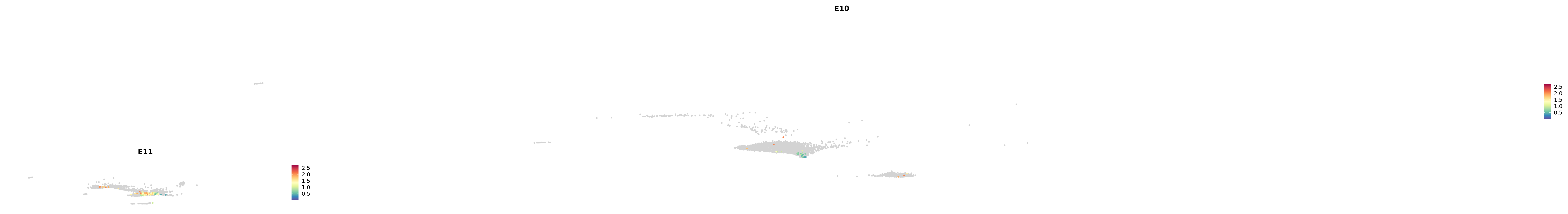
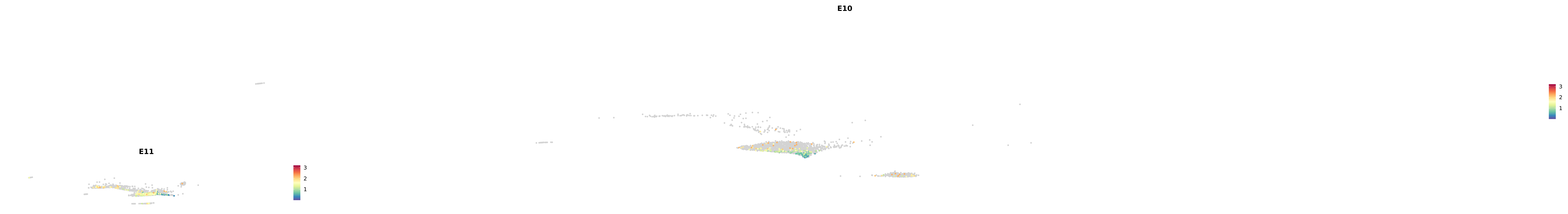
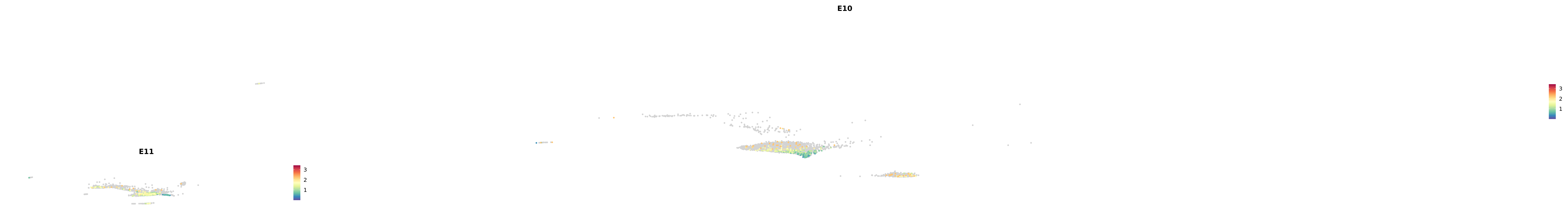
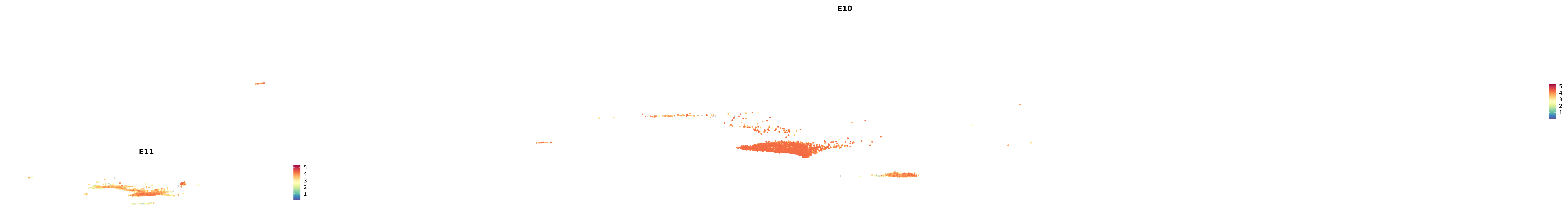
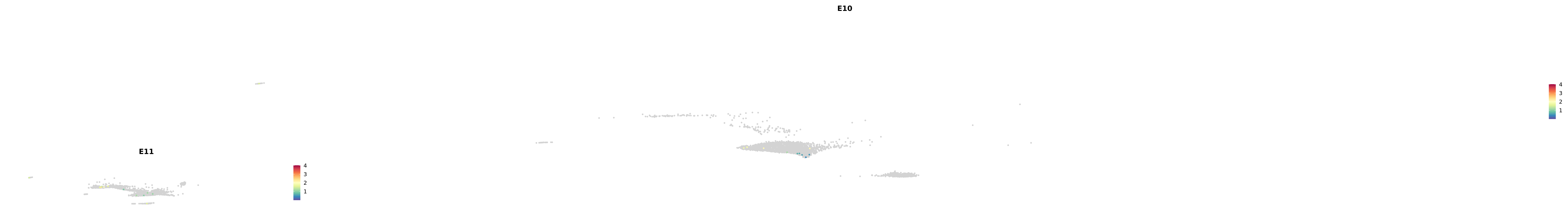
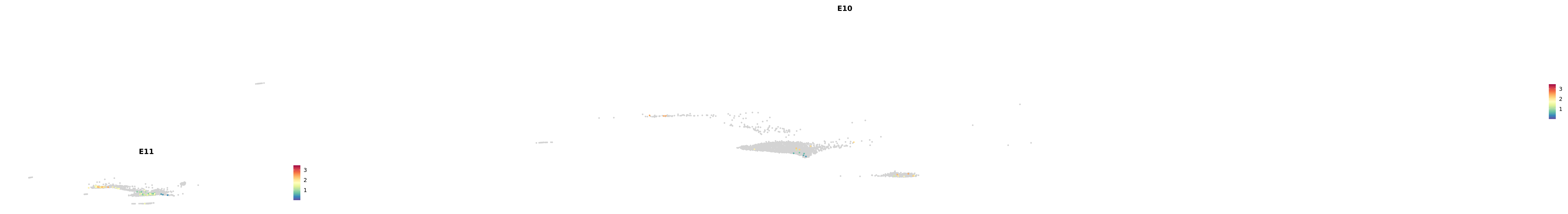
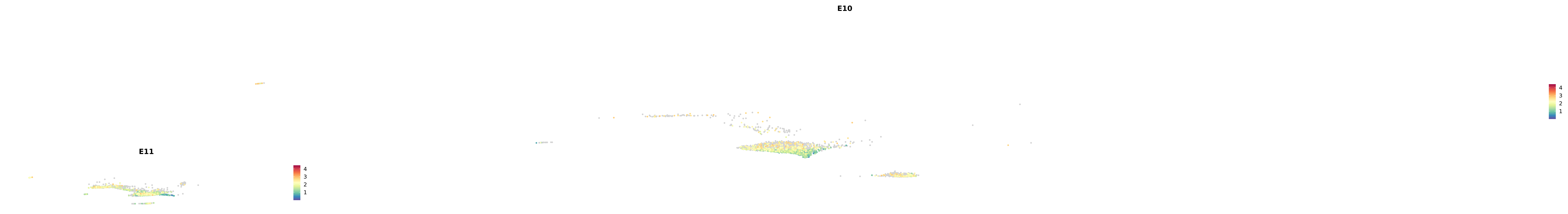
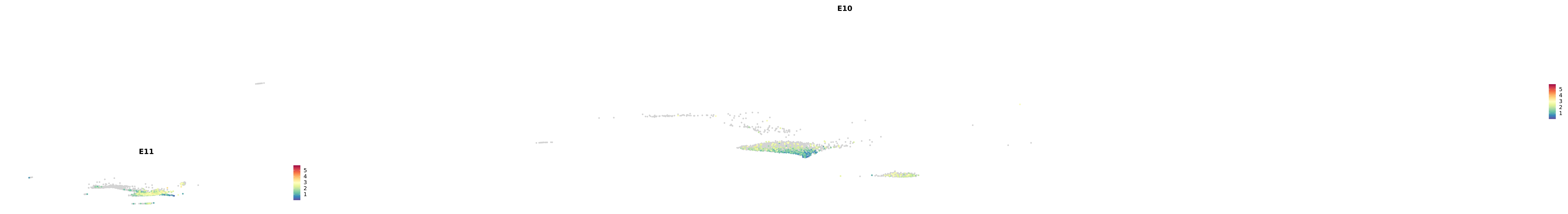
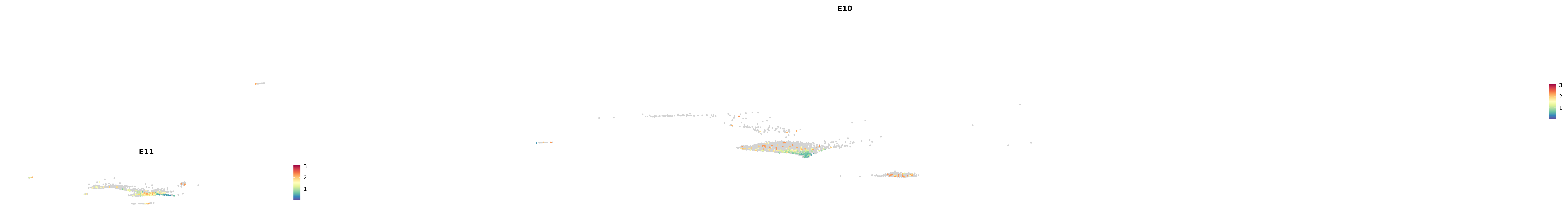


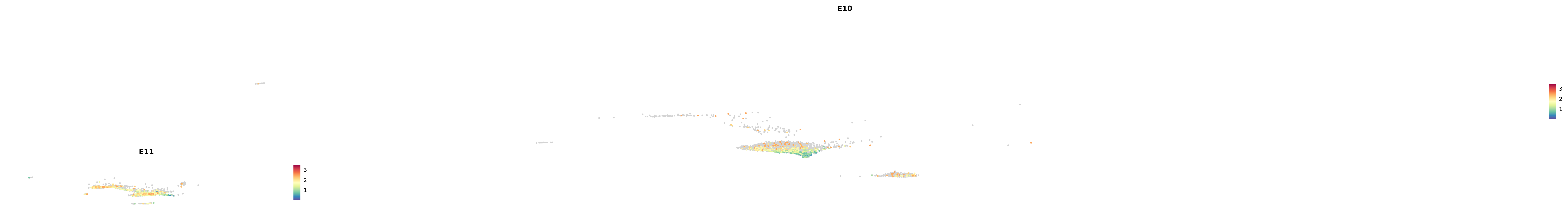
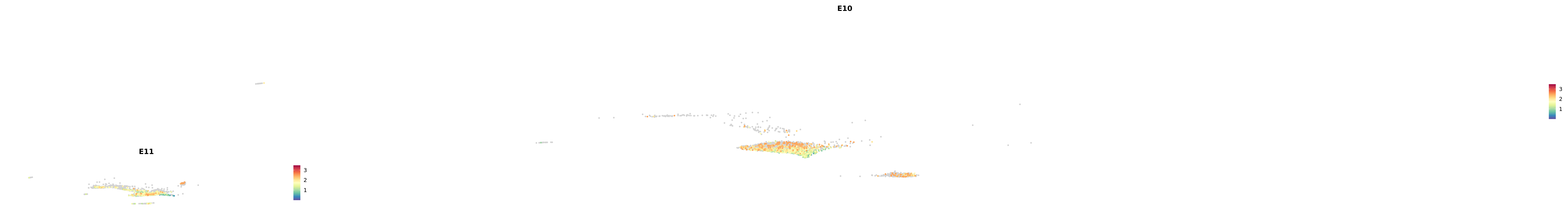
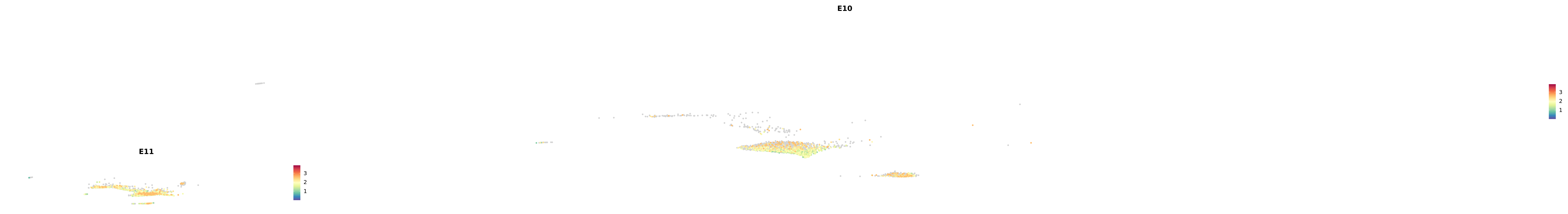
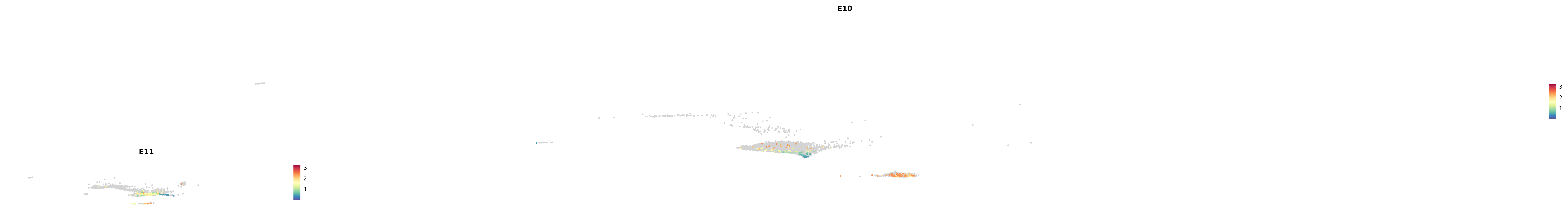
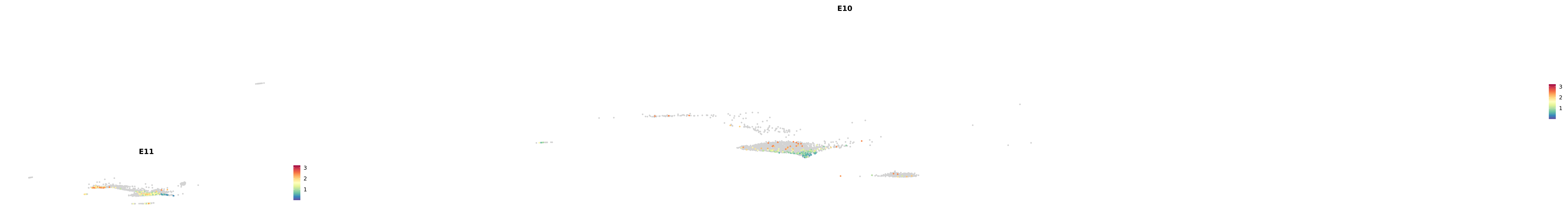
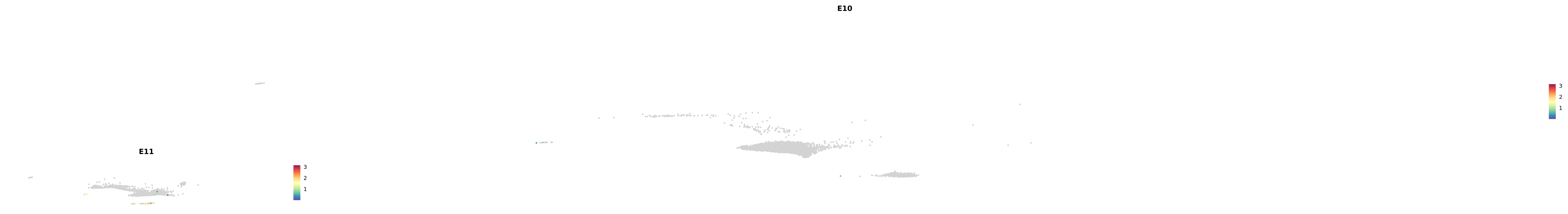

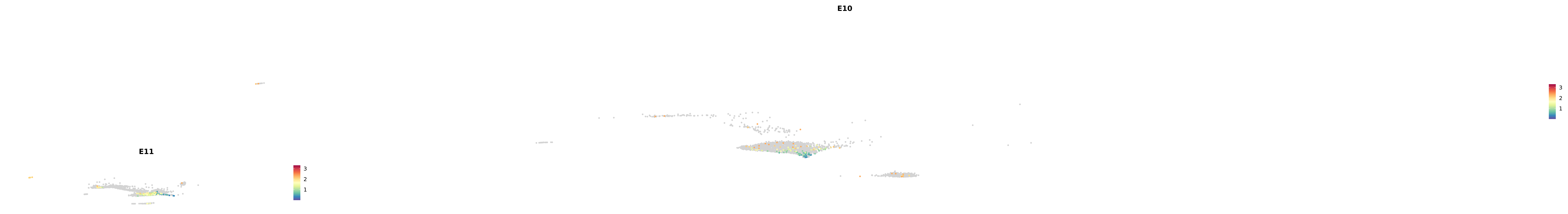
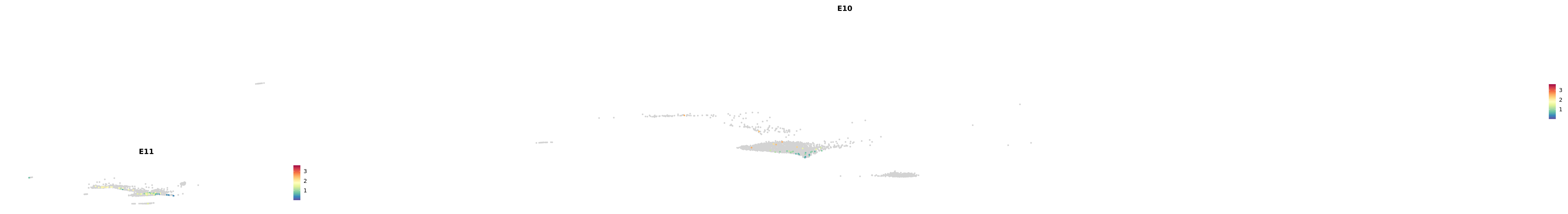
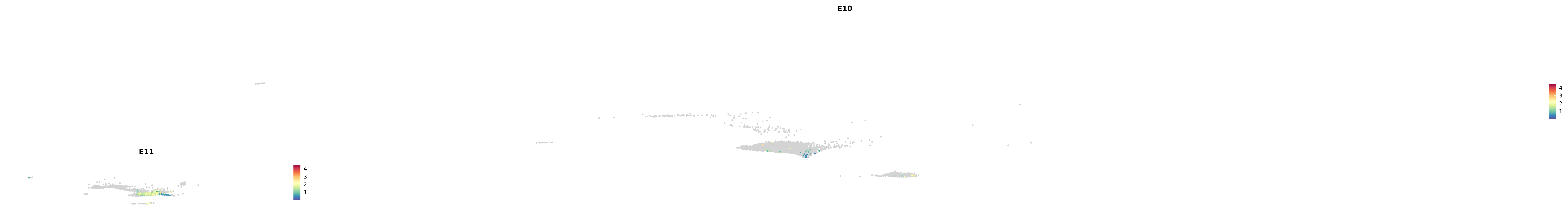
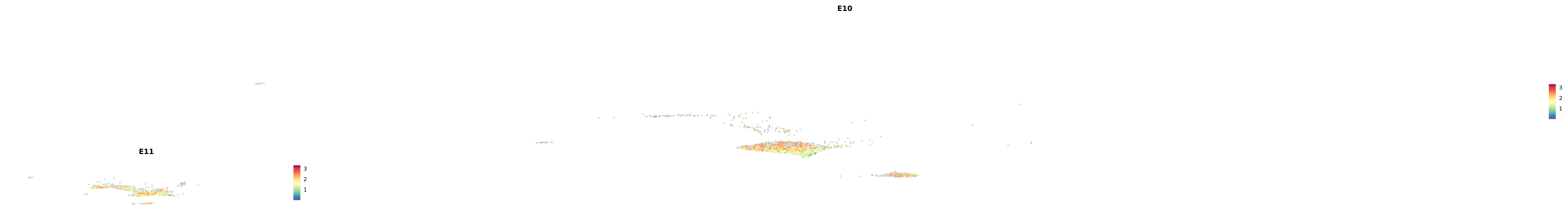
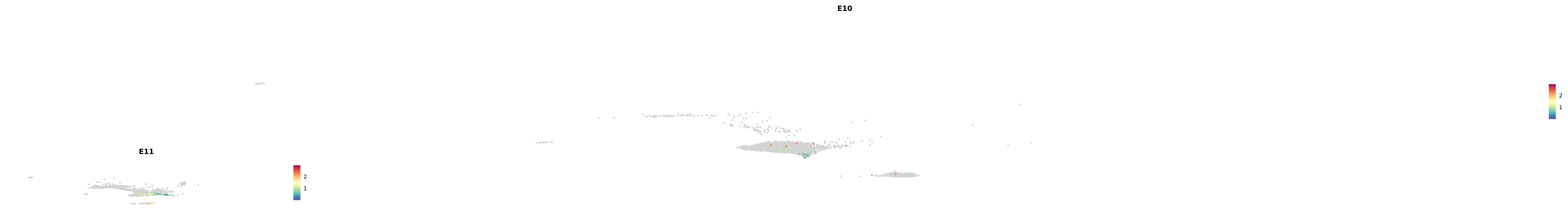




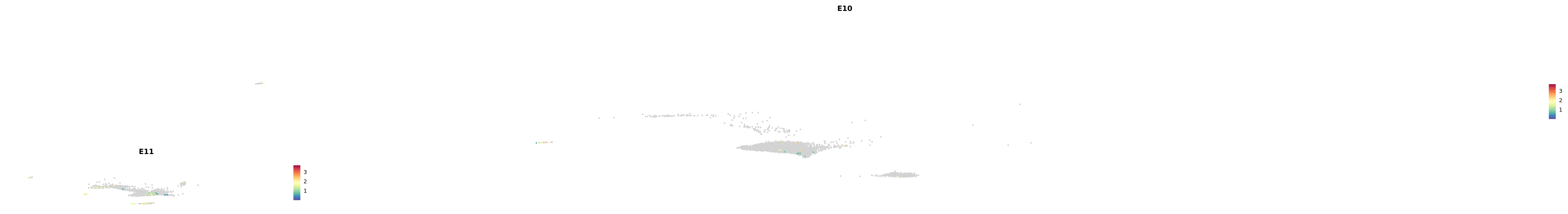
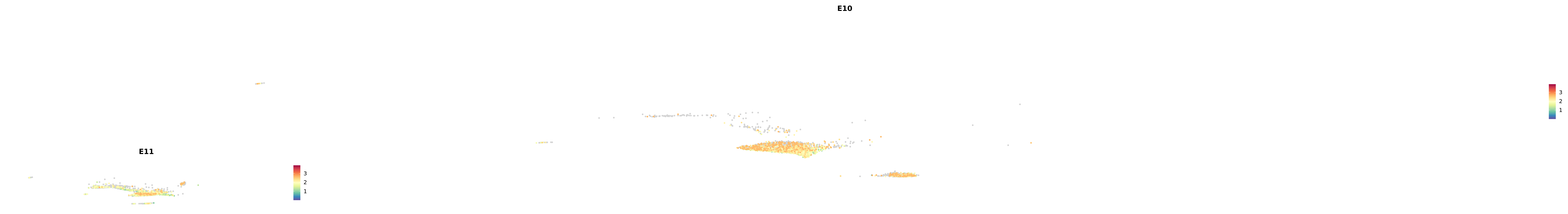
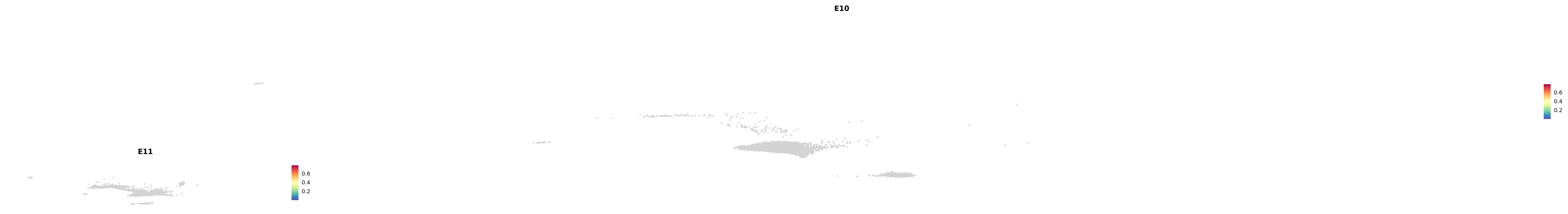
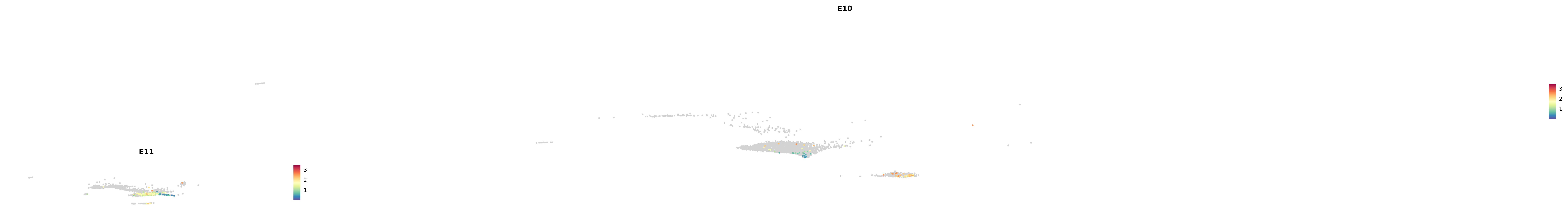
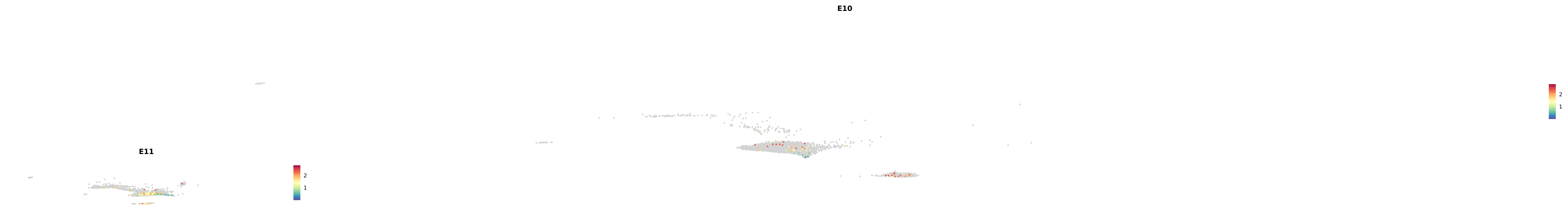
astro <- subset(
x = merged_cortex,
subset = New_cellType == c("Apical progenitors",
"Cycling glial cells",
"Astrocytes"))
astro <- FindVariableFeatures(astro, nfeatures = 5000, verbose = FALSE)
# Scale data
astro <- ScaleData(
astro,
features = c(
VariableFeatures(astro),
s100_genes,
genes.embed),
verbose = FALSE)
# Run PCA
astro <- RunPCA(astro, verbose = FALSE)
# Find neighbors
astro <- FindNeighbors(astro, reduction = "pca", dims = 1:30)Computing nearest neighbor graphComputing SNN# Find clusters
astro <- FindClusters(astro, resolution = 0.7, cluster.name = "astro_clusters", algorithm = 4, random.seed = 42)# Create DimPlot
p1 <- DimPlot(
astro,
reduction = "umap",
group.by = c("stage", "New_cellType"),
combine = FALSE, label.size = 2
)
p2 <- DimPlot(
astro,
reduction = "tsne",
group.by = c("stage", "New_cellType"),
combine = FALSE, label.size = 2
)wrap_plots(c(p1, p2), ncol = 2, byrow = F)
DimPlot(
astro,
reduction = "umap",
group.by = c("astro_clusters"),
combine = FALSE, label.size = 2,
label = T
)[[1]]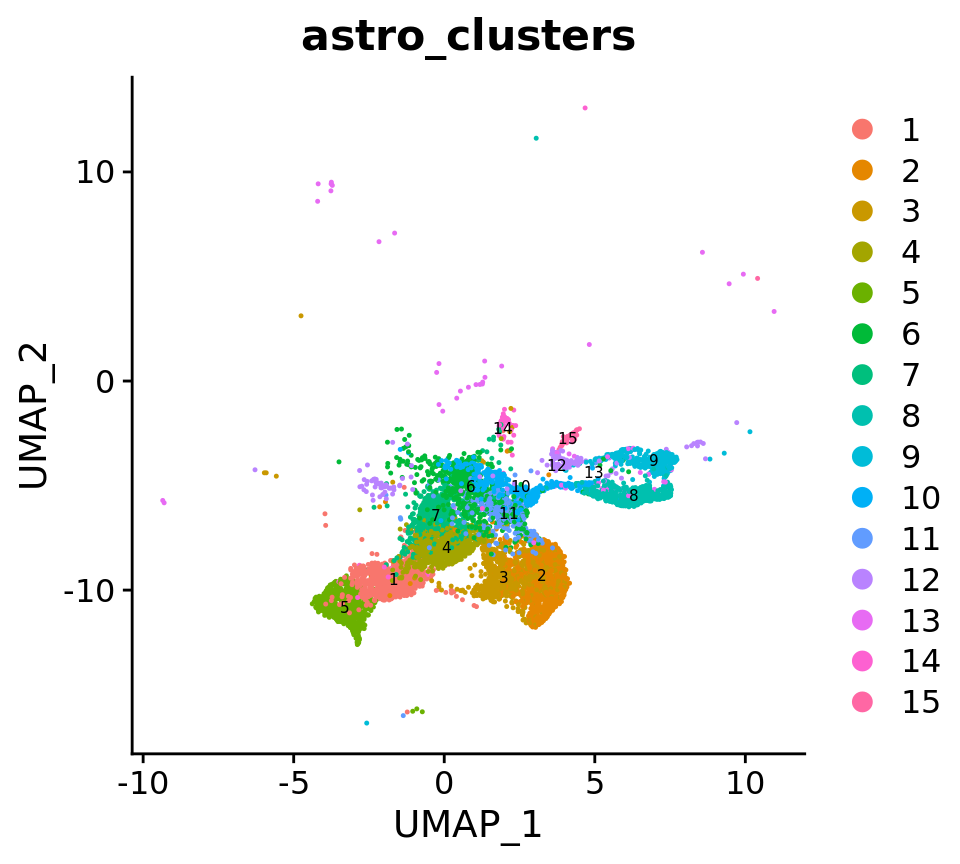
# Create a custom FeaturePlot for each S100 gene
# plot_list <-
# lapply(
# c(s100_genes, "Cacybp", "Siah1a", "Siah1b"),
# function(gene) {
# FeaturePlot_scCustom(
# seurat_object = astro,
# features = gene,
# colors_use = merged_cortex@misc$expr_Colour_Pal,
# na_color = "lightgray",
# layer = "data",
# order = TRUE,
# pt.size = 1,
# reduction = "umap",
# split.by = "stage",
# split_collect = FALSE,
# label = F,
# label_feature_yaxis = TRUE,
# combine = FALSE
# )
# })
#
#
# # Combine the plots into a single grid
# combined_plot <- patchwork::wrap_plots(plot_list, ncol = 1)
#
# # Display the combined plot
# print(combined_plot)# Create a compact DotPlot
compact_plot <- DotPlot(
object = astro,
features = c(s100_genes,
"Cacybp",
"Siah1a",
"Siah1b"),
group.by = "stage",
cluster.idents = F,
scale = TRUE,
dot.scale = 12
) + RotatedAxis()
# Display the compact plot
print(compact_plot)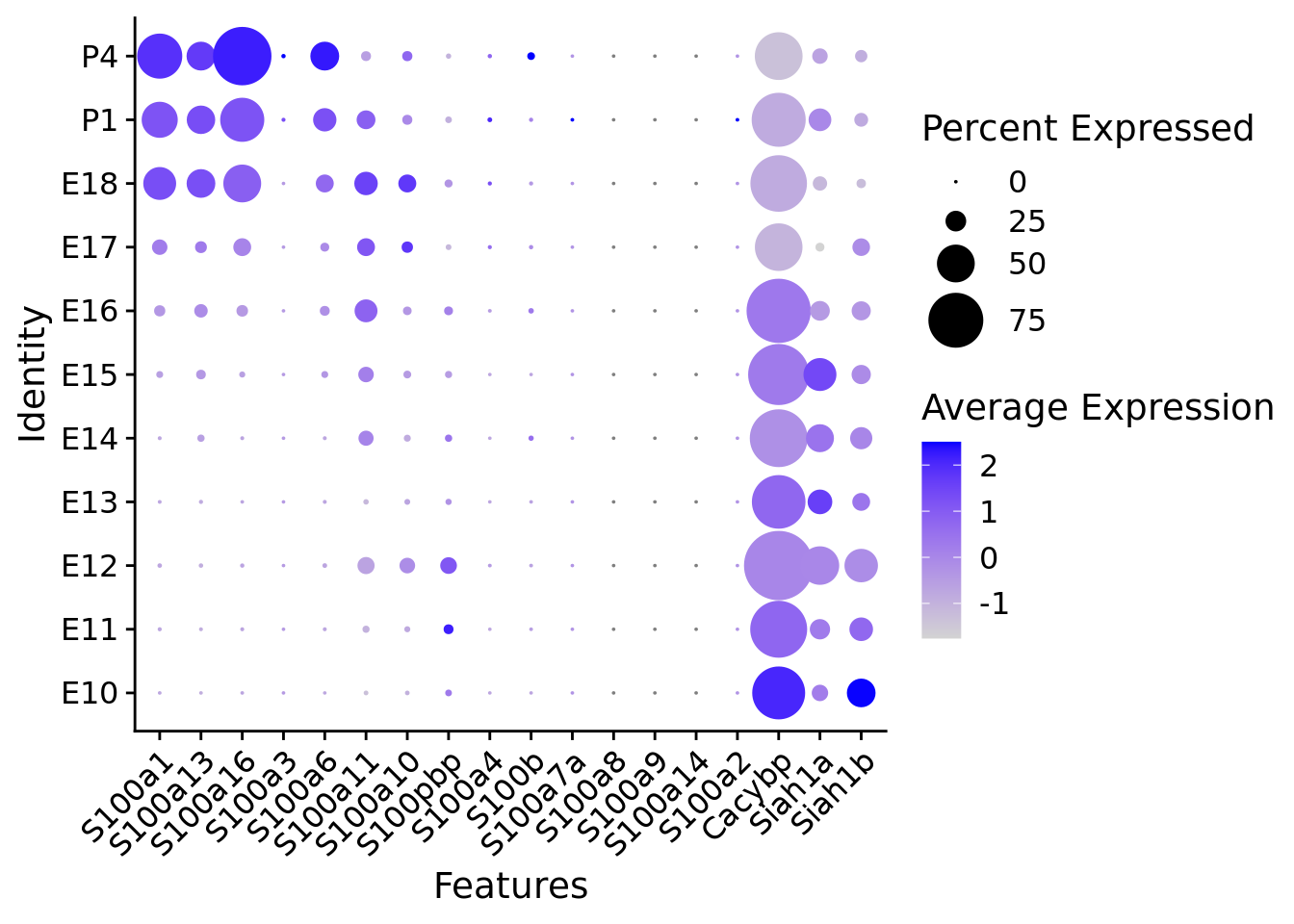
# Create a compact DotPlot
compact_plot <- DotPlot(
object = astro,
features = c(s100_genes,
"Cacybp",
"Siah1a",
"Siah1b"),
group.by = "stage",
cluster.idents = F,
cols = c("yellow", "cyan", "magenta"),
scale = TRUE,
split.by = "New_cellType",
dot.scale = 12
) + RotatedAxis()
# Display the compact plot
print(compact_plot)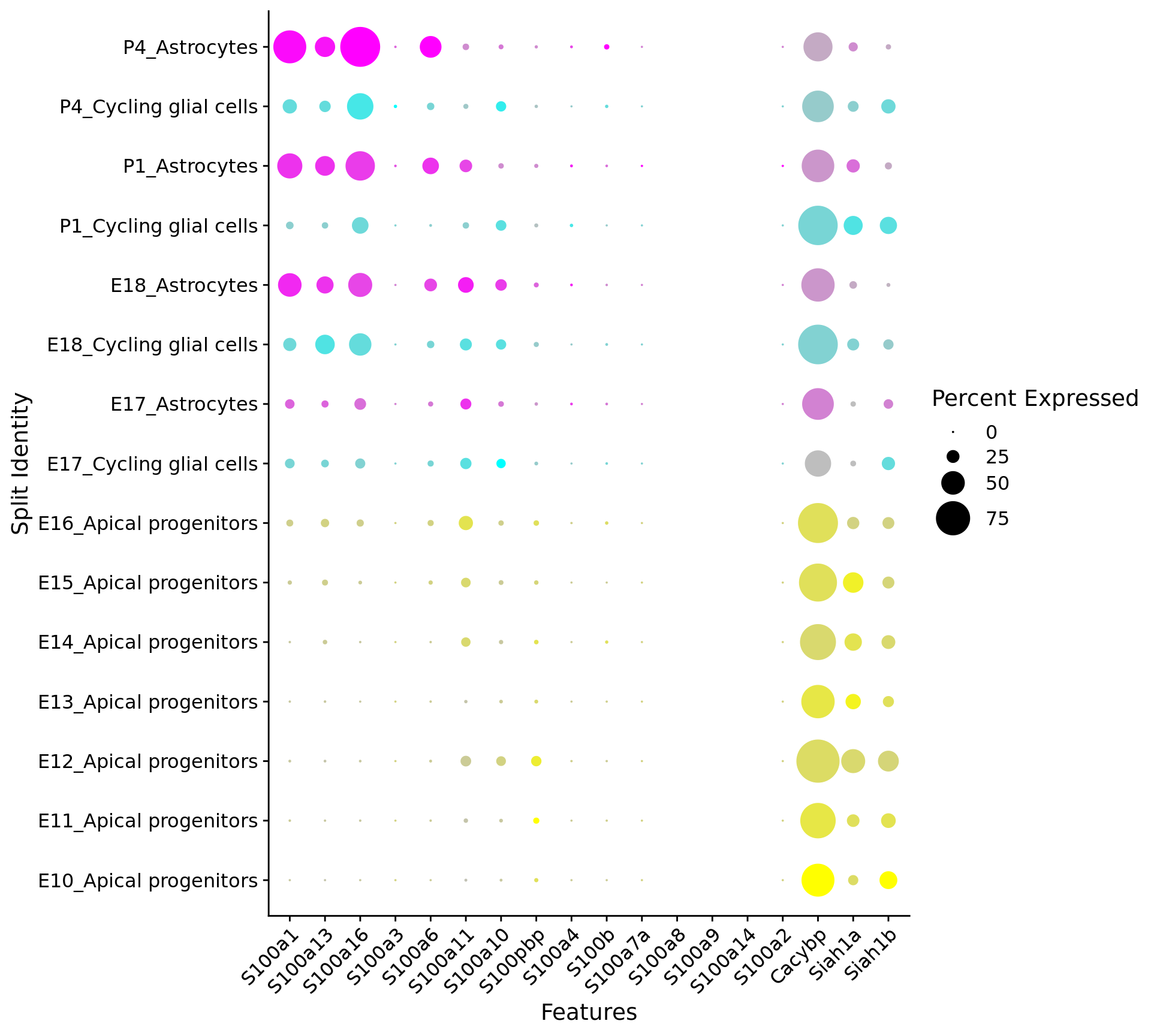
FeaturePlot(astro,
blend = TRUE,
features = c("S100a6", "Gja1"))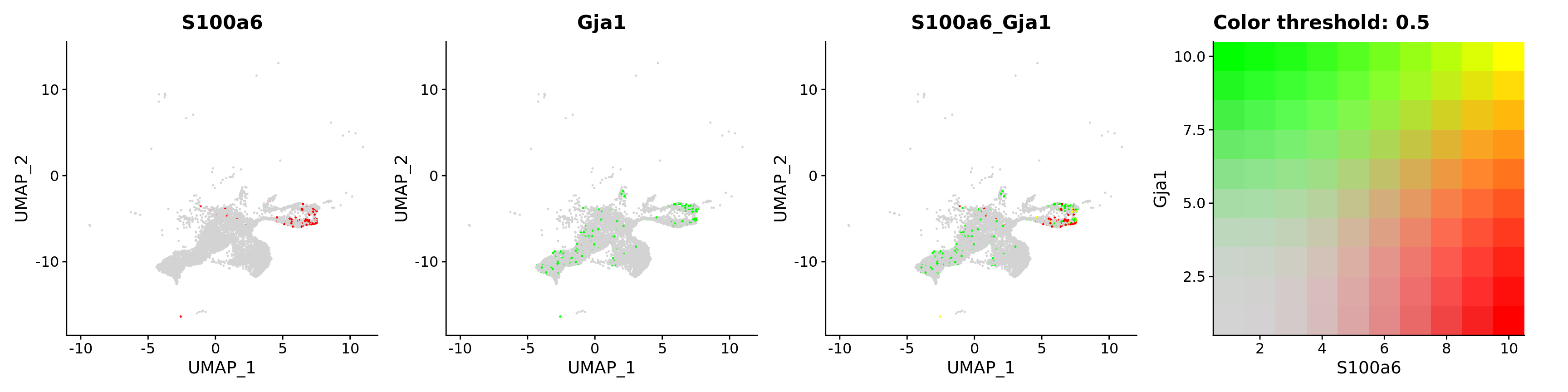
FeaturePlot(astro,
blend = TRUE,
features = c("Cacybp", "Gja1"))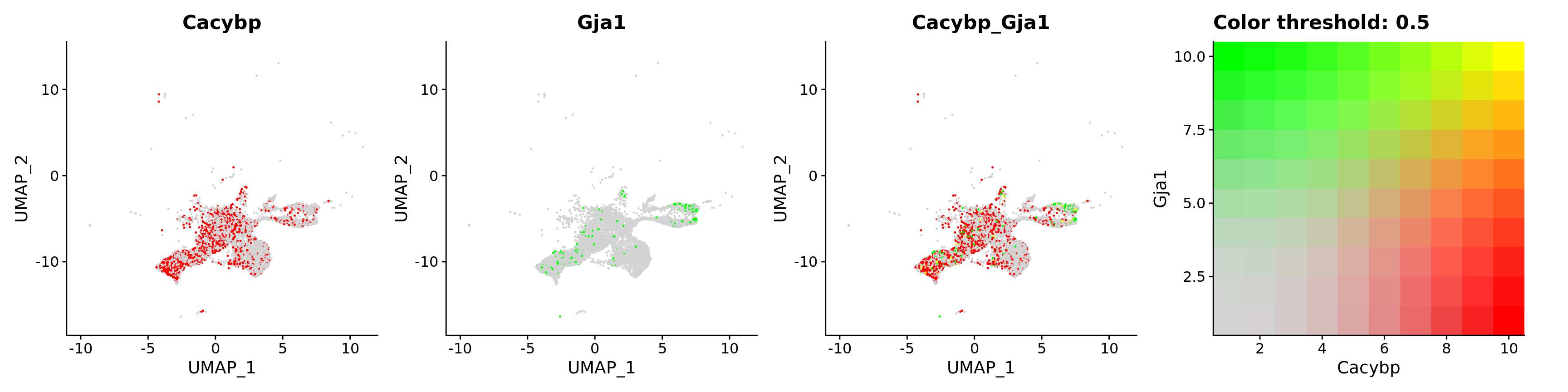
FeaturePlot(astro,
blend = TRUE,
features = c("S100a6", "Glul"))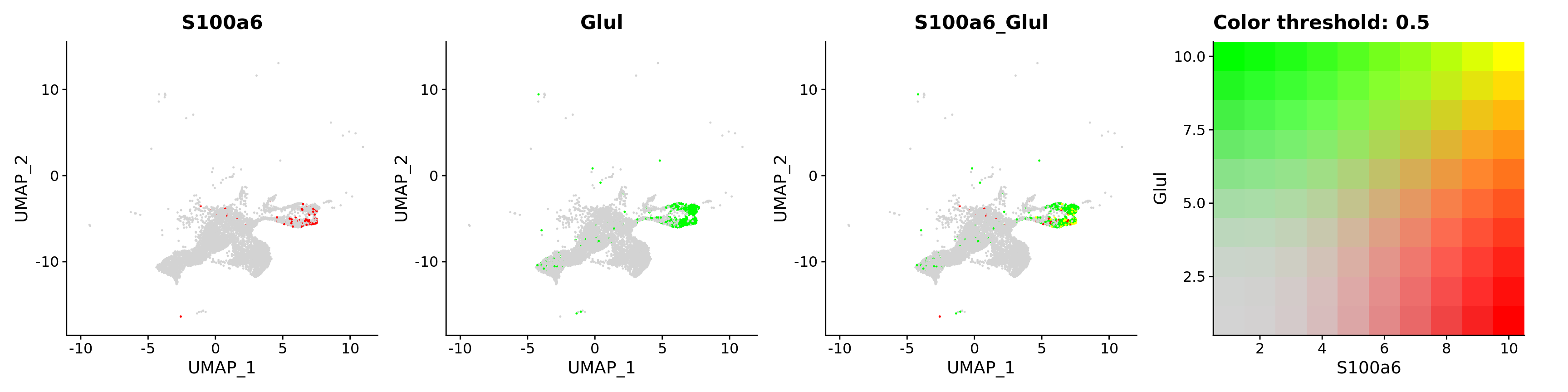
FeaturePlot(astro,
blend = TRUE,
features = c("Cacybp", "Glul"))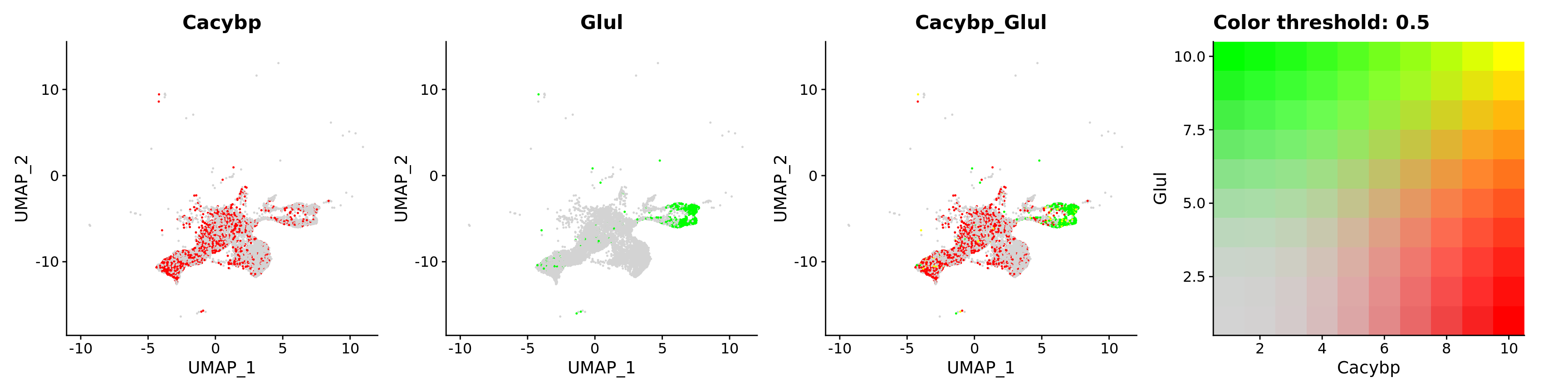
FeaturePlot(astro,
blend = TRUE,
features = c("S100a6", "Apoe"))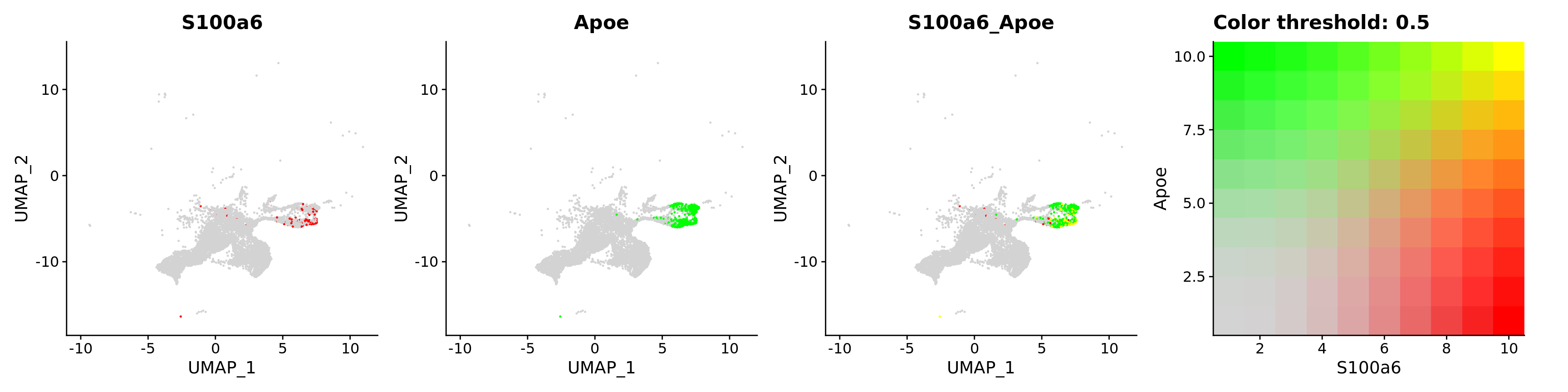
FeaturePlot(astro,
blend = TRUE,
features = c("Cacybp", "Apoe"))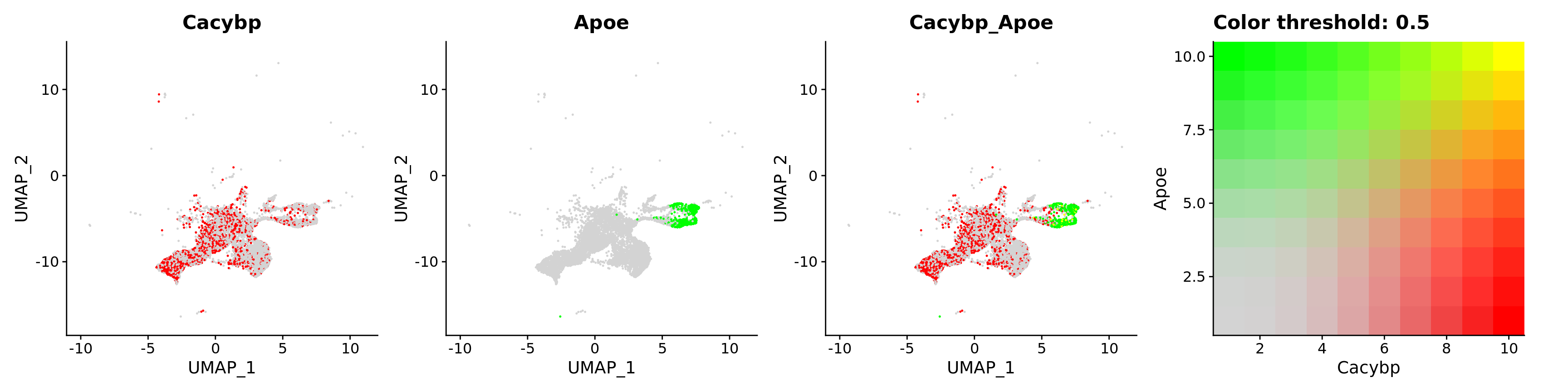
FeaturePlot(astro,
blend = TRUE,
features = c("S100a6", "Ntrk2"))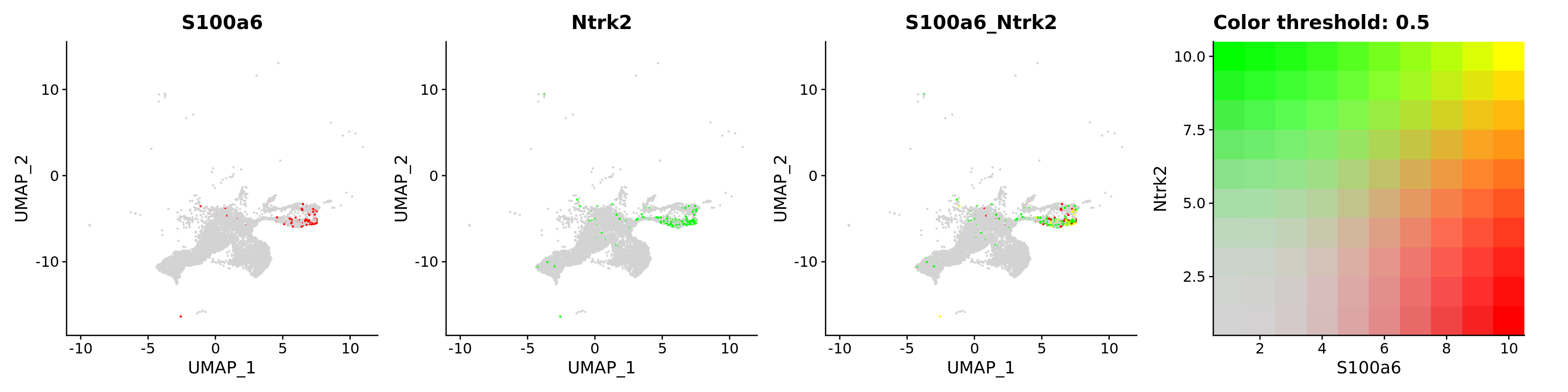
FeaturePlot(astro,
blend = TRUE,
features = c("Cacybp", "Ntrk2"))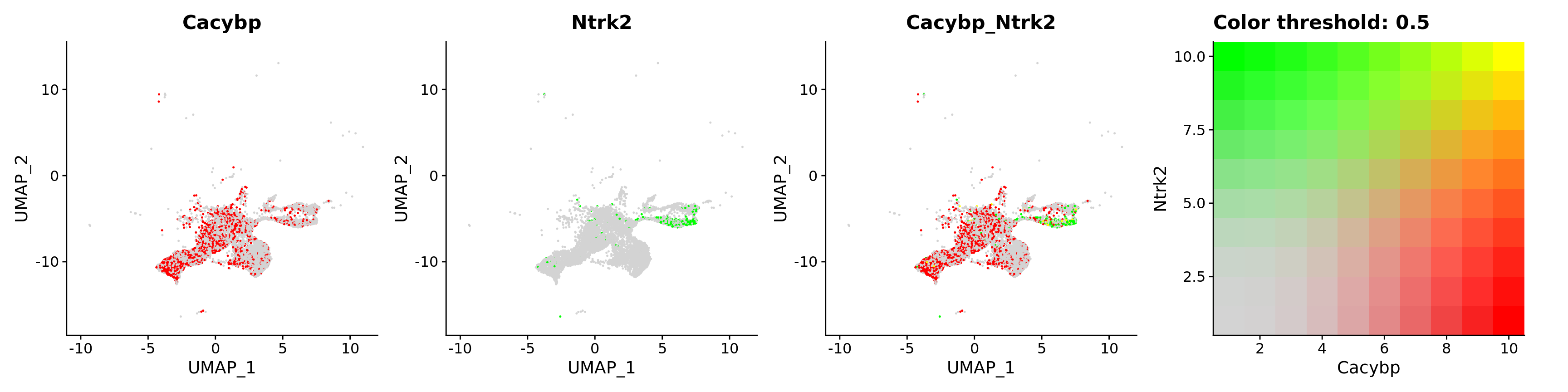
FeaturePlot(astro,
blend = TRUE,
features = c("S100a6", "Ntsr2"))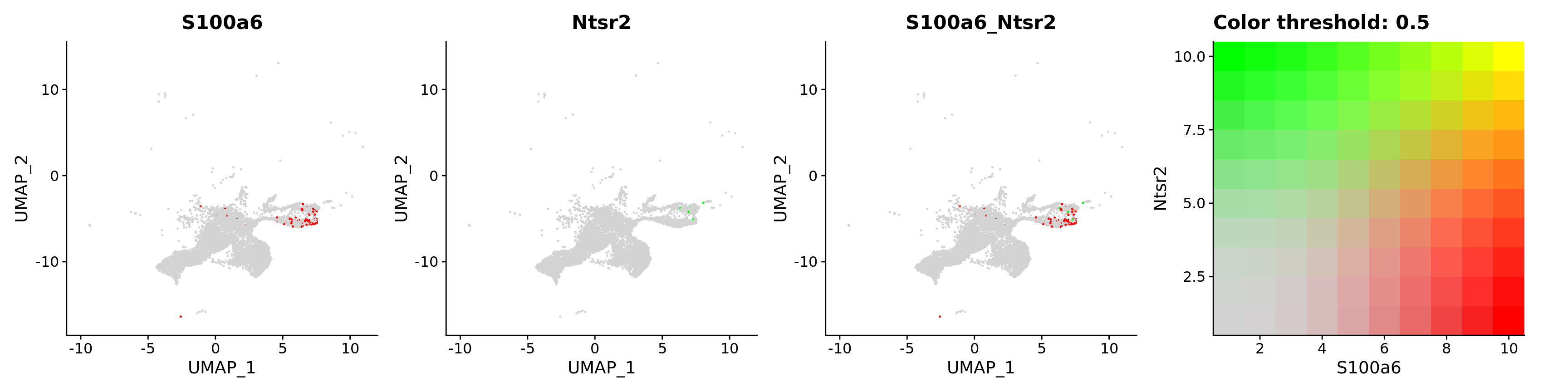
FeaturePlot(astro,
blend = TRUE,
features = c("Cacybp", "Ntsr2"))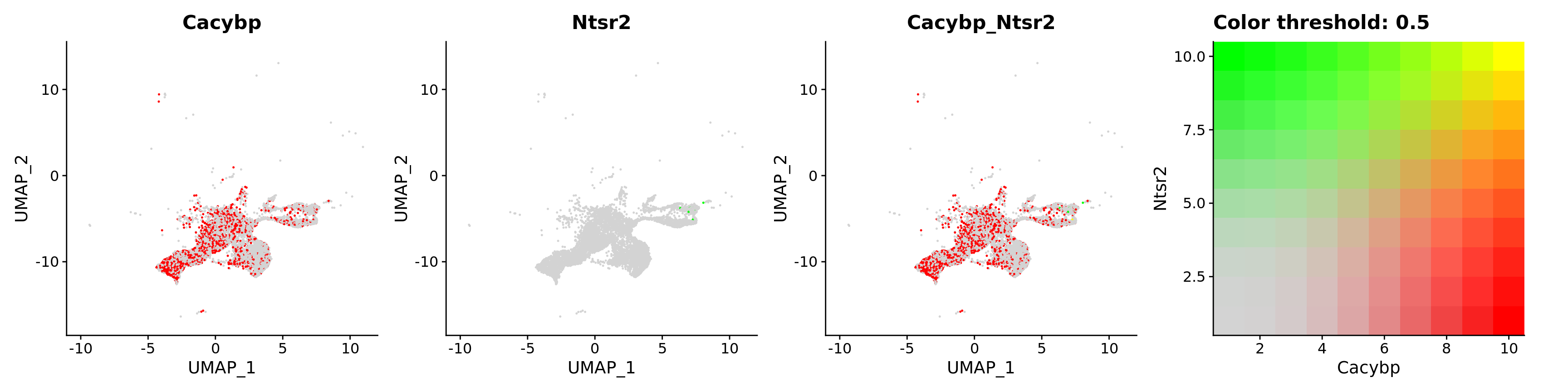
FeaturePlot(astro,
blend = TRUE,
features = c("S100a6", "Ndrg2"))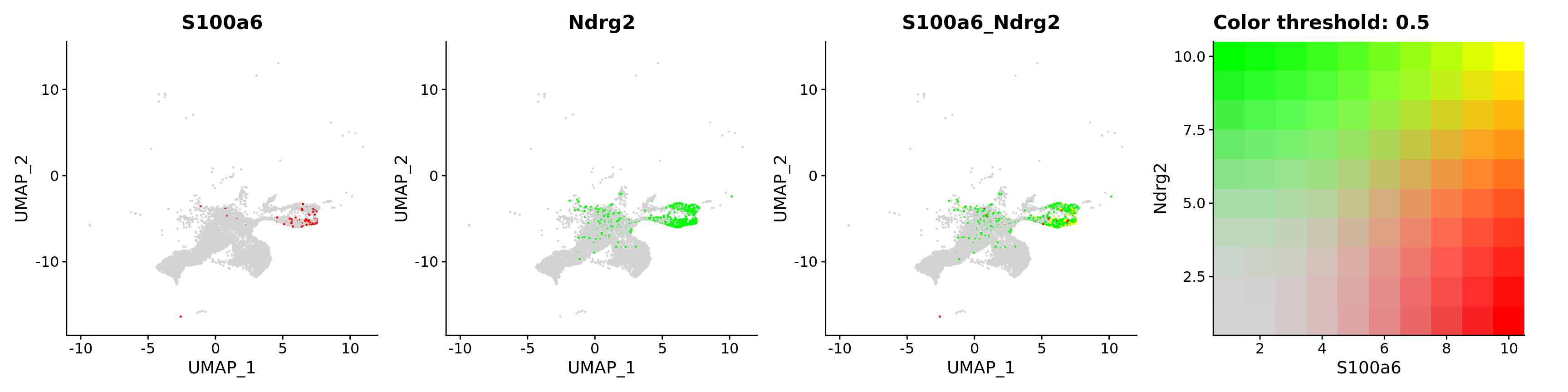
FeaturePlot(astro,
blend = TRUE,
features = c("Cacybp", "Ndrg2"))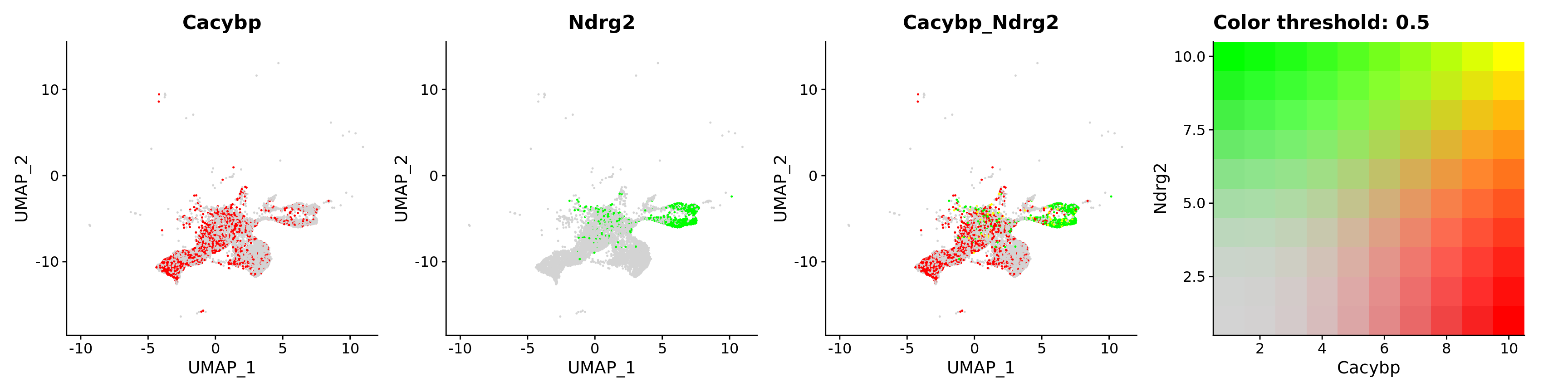
FeaturePlot(astro,
blend = TRUE,
features = c("S100a6", "Aldoc"))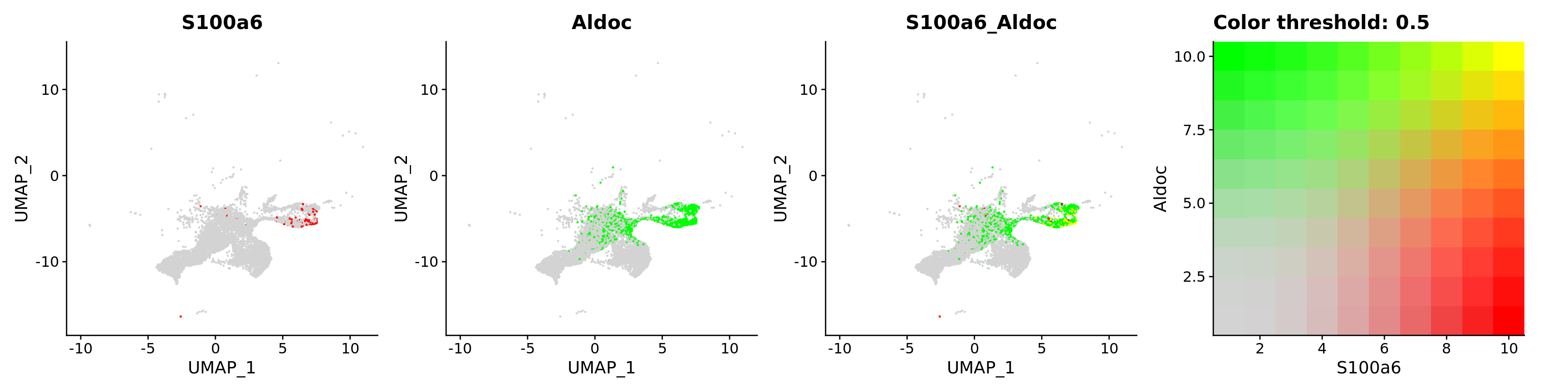
FeaturePlot(astro,
blend = TRUE,
features = c("Cacybp", "Aldoc"))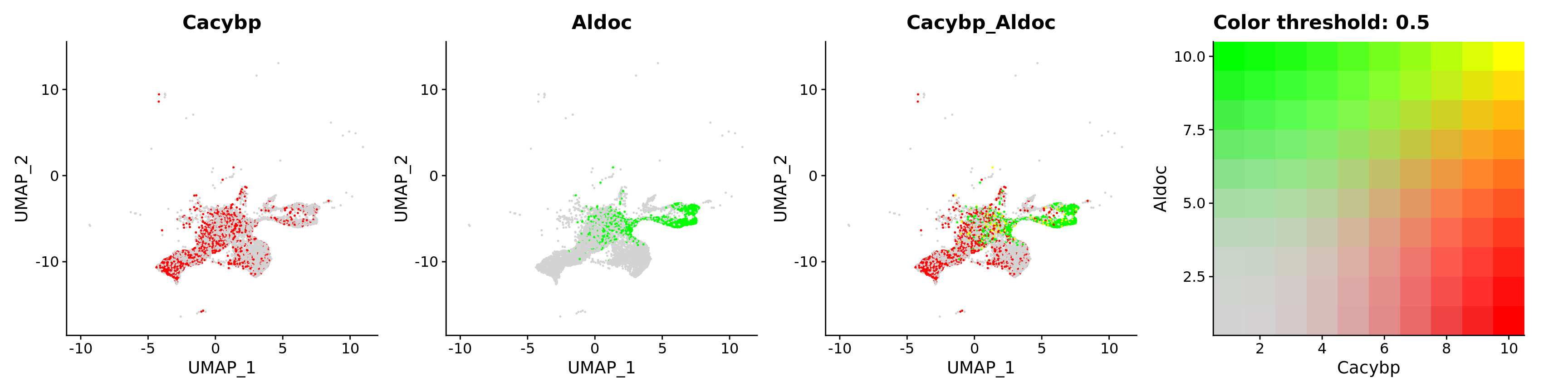
FeaturePlot(astro,
blend = TRUE,
features = c("S100a6", "Slc1a3"))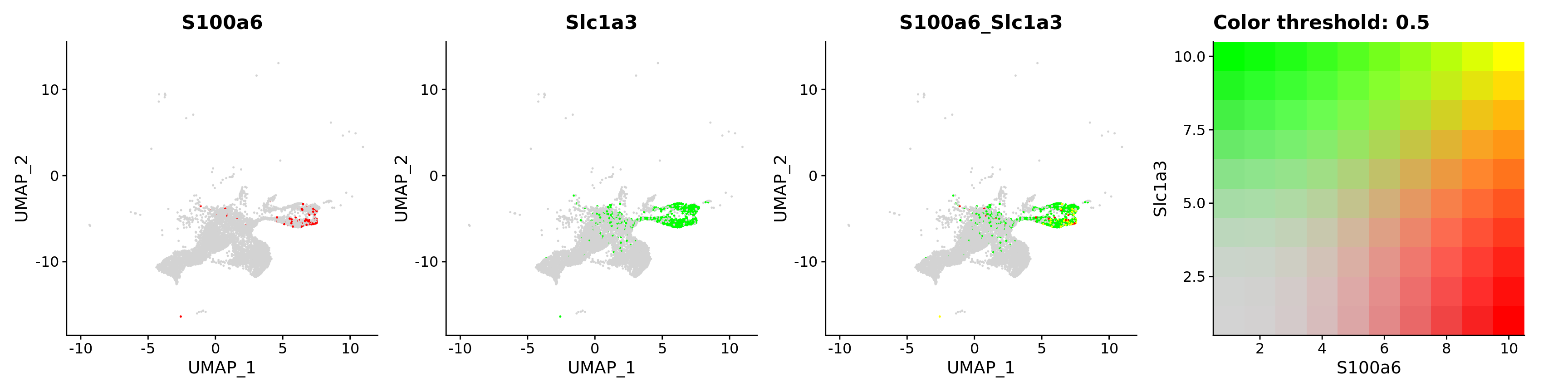
FeaturePlot(astro,
blend = TRUE,
features = c("Cacybp", "Slc1a3"))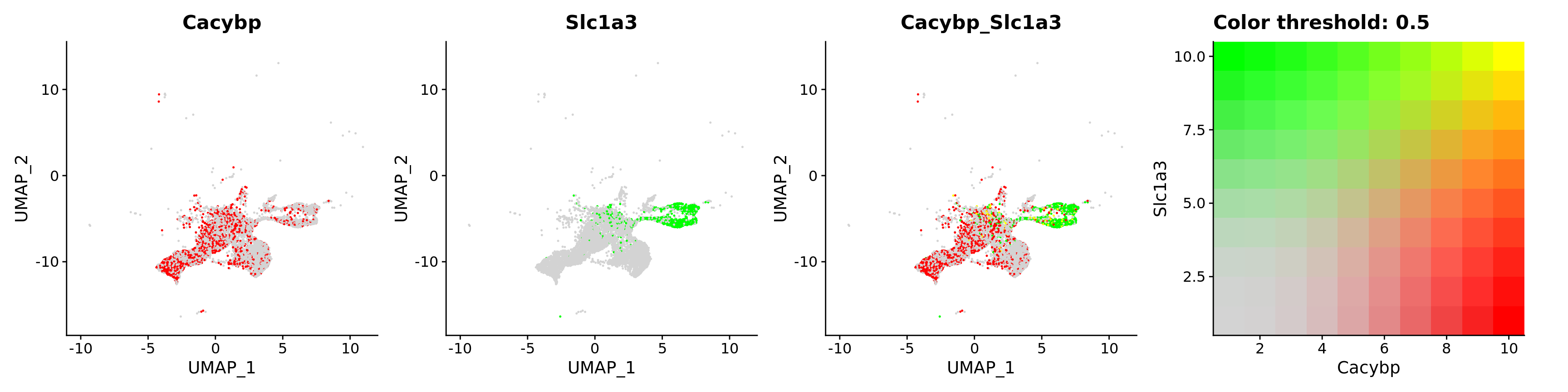
FeaturePlot(astro,
blend = TRUE,
features = c("S100a6", "Gfap"))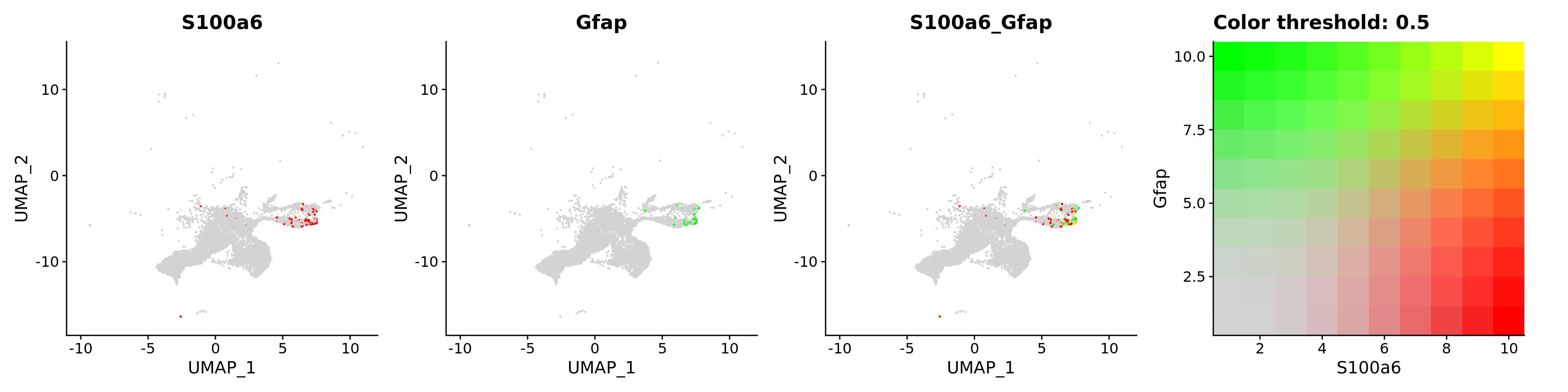
FeaturePlot(astro,
blend = TRUE,
features = c("Cacybp", "Gfap"))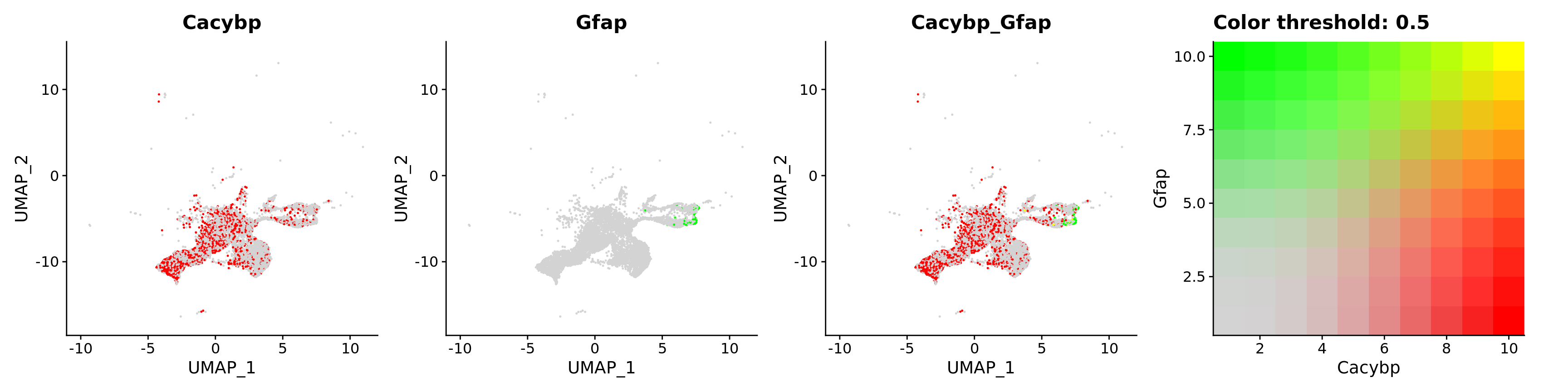
FeaturePlot(astro,
blend = TRUE,
features = c("S100a6", "Htra1"))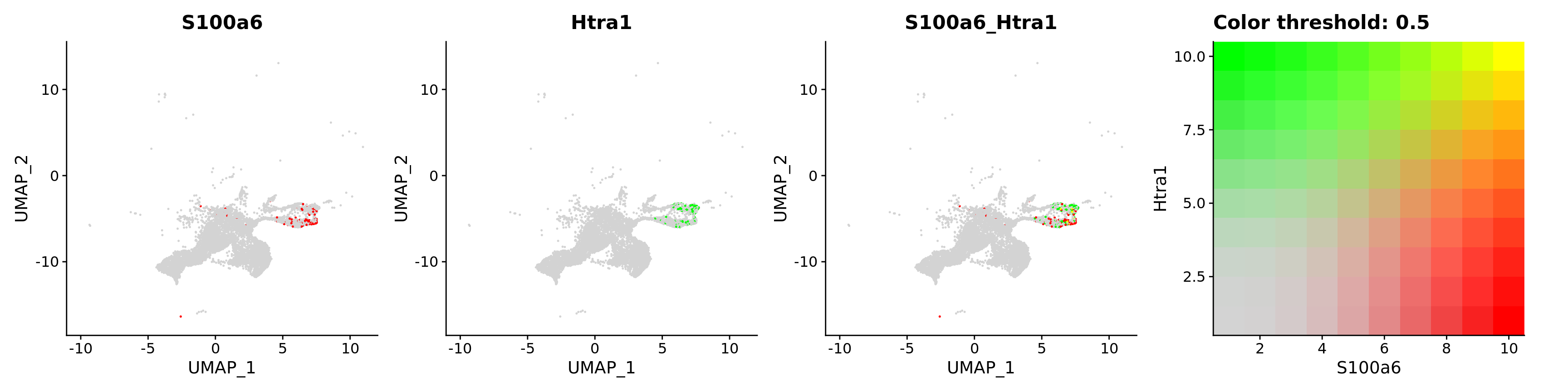
FeaturePlot(astro,
blend = TRUE,
features = c("Cacybp", "Htra1"))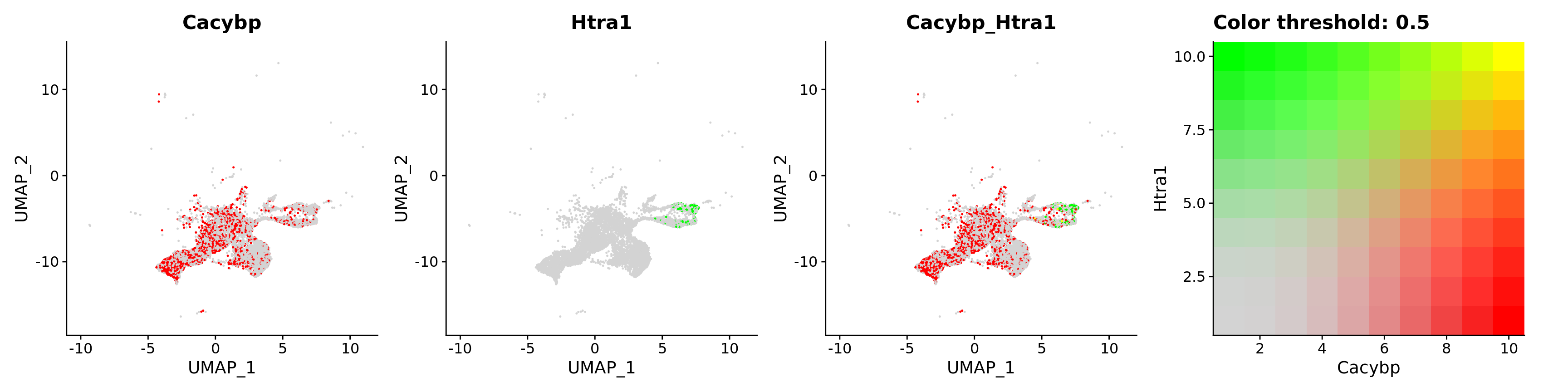
FeaturePlot(astro,
blend = TRUE,
features = c("S100a6", "Aqp4"))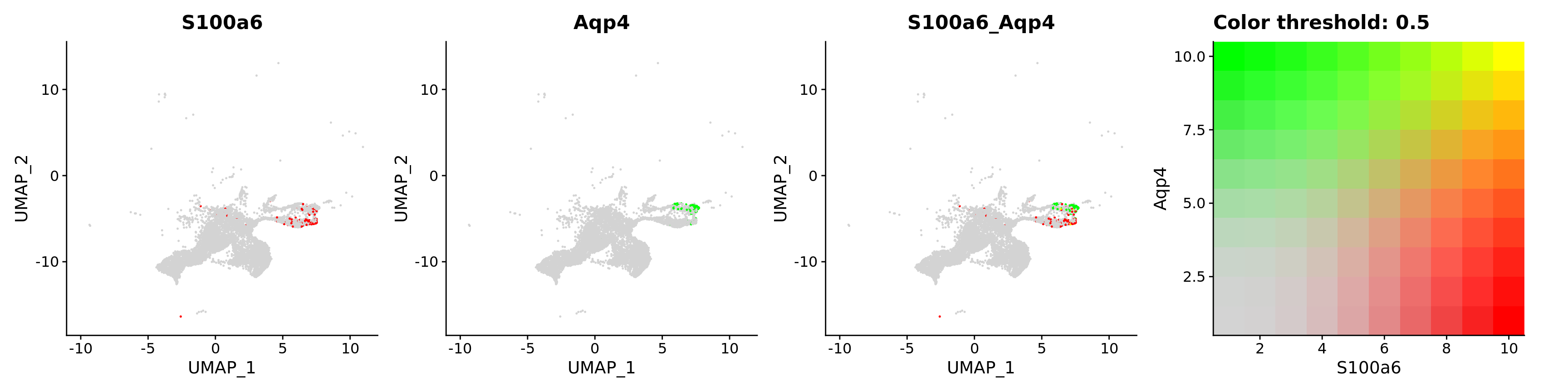
FeaturePlot(astro,
blend = TRUE,
features = c("Cacybp", "Aqp4"))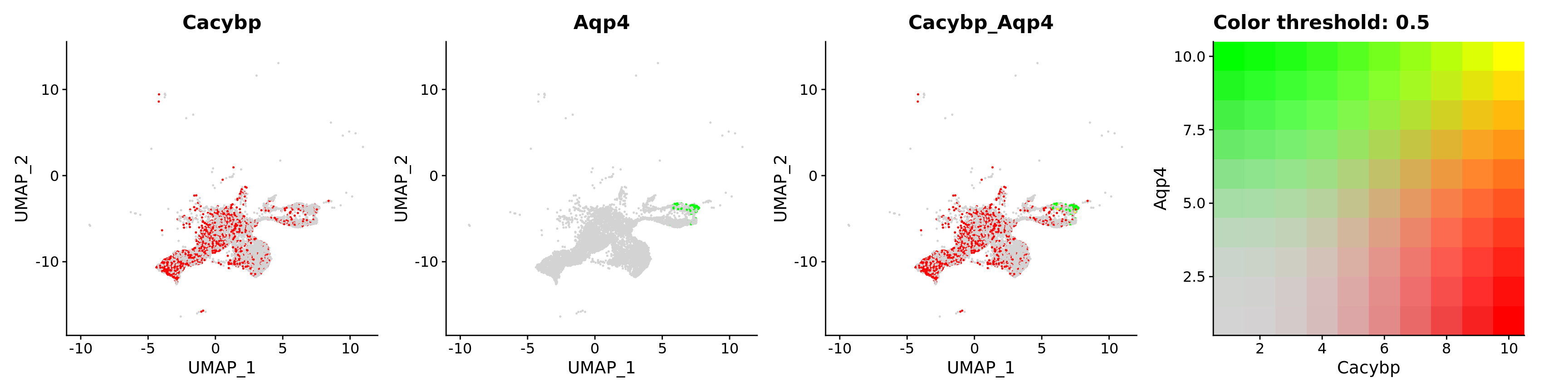
sessioninfo::session_info()─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.3.2 (2023-10-31)
os Ubuntu 22.04.3 LTS
system x86_64, linux-gnu
ui X11
language en_US:en
collate en_US.UTF-8
ctype en_US.UTF-8
tz Etc/UTC
date 2024-06-09
pandoc 3.1.3 @ /opt/python/3.8.8/bin/ (via rmarkdown)
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
abind 1.4-5 2016-07-21 [2] RSPM (R 4.3.0)
annotate 1.80.0 2023-10-24 [2] RSPM (R 4.3.2)
AnnotationDbi 1.64.1 2023-11-03 [2] RSPM (R 4.3.2)
AnnotationFilter 1.26.0 2023-10-24 [2] RSPM (R 4.3.2)
Azimuth * 0.5.0 2024-01-27 [2] Github (satijalab/azimuth@243ee5d)
beeswarm 0.4.0 2021-06-01 [2] RSPM (R 4.3.0)
Biobase 2.62.0 2023-10-24 [2] RSPM (R 4.3.2)
BiocFileCache 2.10.1 2023-10-26 [2] RSPM (R 4.3.2)
BiocGenerics 0.48.1 2023-11-01 [2] RSPM (R 4.3.2)
BiocIO 1.12.0 2023-10-24 [2] RSPM (R 4.3.2)
BiocManager 1.30.22 2023-08-08 [2] RSPM (R 4.3.0)
BiocParallel 1.36.0 2023-10-24 [2] RSPM (R 4.3.2)
biomaRt 2.58.0 2023-10-24 [2] RSPM (R 4.3.2)
Biostrings 2.70.1 2023-10-25 [2] RSPM (R 4.3.2)
bit 4.0.5 2022-11-15 [2] RSPM (R 4.3.0)
bit64 4.0.5 2020-08-30 [2] RSPM (R 4.3.0)
bitops 1.0-7 2021-04-24 [2] RSPM (R 4.3.0)
blob 1.2.4 2023-03-17 [2] RSPM (R 4.3.0)
BPCells * 0.1.0 2024-01-27 [2] Github (bnprks/BPCells@0d56524)
BSgenome 1.70.1 2023-11-01 [2] RSPM (R 4.3.2)
BSgenome.Hsapiens.UCSC.hg38 1.4.5 2024-01-26 [2] RSPM (R 4.3.2)
bslib 0.6.1 2023-11-28 [2] RSPM (R 4.3.0)
cachem 1.0.8 2023-05-01 [2] RSPM (R 4.3.0)
callr 3.7.3 2022-11-02 [2] RSPM (R 4.3.0)
caTools 1.18.2 2021-03-28 [2] RSPM (R 4.3.0)
cellranger 1.1.0 2016-07-27 [2] RSPM (R 4.3.0)
circlize 0.4.16 2024-01-26 [2] Github (jokergoo/circlize@9b21578)
cli 3.6.2 2023-12-11 [2] RSPM (R 4.3.0)
cluster 2.1.6 2023-12-01 [2] RSPM (R 4.3.0)
CNEr 1.38.0 2023-10-24 [2] RSPM (R 4.3.2)
codetools 0.2-19 2023-02-01 [2] RSPM (R 4.3.0)
colorspace 2.1-0 2023-01-23 [2] RSPM (R 4.3.0)
cowplot * 1.1.3 2024-01-22 [2] RSPM (R 4.3.0)
crayon 1.5.2 2022-09-29 [2] RSPM (R 4.3.0)
curl 5.2.0 2023-12-08 [2] RSPM (R 4.3.0)
data.table 1.14.10 2023-12-08 [2] RSPM (R 4.3.0)
DBI 1.2.1 2024-01-12 [2] RSPM (R 4.3.0)
dbplyr 2.4.0 2023-10-26 [2] RSPM (R 4.3.0)
DelayedArray 0.28.0 2023-10-24 [2] RSPM (R 4.3.2)
deldir 2.0-2 2023-11-23 [2] RSPM (R 4.3.0)
digest 0.6.34 2024-01-11 [2] RSPM (R 4.3.0)
DirichletMultinomial 1.44.0 2023-10-24 [2] RSPM (R 4.3.2)
dotCall64 1.1-1 2023-11-28 [2] RSPM (R 4.3.0)
dplyr * 1.1.4 2023-11-17 [2] RSPM (R 4.3.0)
DT 0.31 2023-12-09 [2] RSPM (R 4.3.0)
ellipsis 0.3.2 2021-04-29 [2] RSPM (R 4.3.0)
EnsDb.Hsapiens.v86 2.99.0 2024-01-26 [2] RSPM (R 4.3.2)
ensembldb 2.26.0 2023-10-24 [2] RSPM (R 4.3.2)
evaluate 0.23 2023-11-01 [2] RSPM (R 4.3.0)
fansi 1.0.6 2023-12-08 [2] RSPM (R 4.3.0)
farver 2.1.1 2022-07-06 [2] RSPM (R 4.3.0)
fastDummies 1.7.3 2023-07-06 [2] RSPM (R 4.3.0)
fastmap 1.1.1 2023-02-24 [2] RSPM (R 4.3.0)
fastmatch 1.1-4 2023-08-18 [2] RSPM (R 4.3.0)
filelock 1.0.3 2023-12-11 [2] RSPM (R 4.3.0)
fitdistrplus 1.1-11 2023-04-25 [2] RSPM (R 4.3.0)
forcats 1.0.0 2023-01-29 [2] RSPM (R 4.3.0)
fs 1.6.3 2023-07-20 [2] RSPM (R 4.3.0)
future 1.33.1 2023-12-22 [2] RSPM (R 4.3.0)
future.apply 1.11.1 2023-12-21 [2] RSPM (R 4.3.0)
gargle 1.5.2 2023-07-20 [2] RSPM (R 4.3.0)
generics 0.1.3 2022-07-05 [2] RSPM (R 4.3.0)
GenomeInfoDb 1.38.5 2023-12-28 [2] RSPM (R 4.3.2)
GenomeInfoDbData 1.2.11 2024-01-26 [2] RSPM (R 4.3.2)
GenomicAlignments 1.38.2 2024-01-16 [2] RSPM (R 4.3.2)
GenomicFeatures 1.54.1 2023-10-29 [2] RSPM (R 4.3.2)
GenomicRanges 1.54.1 2023-10-29 [2] RSPM (R 4.3.2)
getPass 0.2-4 2023-12-10 [2] RSPM (R 4.3.0)
ggbeeswarm 0.7.2 2024-01-26 [2] Github (eclarke/ggbeeswarm@3cf58a9)
ggplot2 * 3.4.4.9000 2024-01-26 [2] Github (tidyverse/ggplot2@a4be39d)
ggprism 1.0.4 2024-01-26 [2] Github (csdaw/ggprism@0e411f4)
ggrastr 1.0.2 2024-01-26 [2] Github (VPetukhov/ggrastr@50ca3e0)
ggrepel 0.9.5.9999 2024-01-26 [2] Github (slowkow/ggrepel@1144585)
ggridges 0.5.6 2024-01-23 [2] RSPM (R 4.3.0)
git2r 0.33.0 2023-11-26 [2] RSPM (R 4.3.0)
GlobalOptions 0.1.2 2020-06-10 [2] RSPM (R 4.3.0)
globals 0.16.2 2022-11-21 [2] RSPM (R 4.3.0)
glue 1.7.0 2024-01-09 [2] RSPM (R 4.3.0)
GO.db 3.18.0 2024-01-26 [2] RSPM (R 4.3.2)
goftest 1.2-3 2021-10-07 [2] RSPM (R 4.3.0)
googledrive 2.1.1 2023-06-11 [2] RSPM (R 4.3.0)
googlesheets4 1.1.1 2023-06-11 [2] RSPM (R 4.3.0)
gridExtra 2.3 2017-09-09 [2] RSPM (R 4.3.0)
gtable 0.3.4 2023-08-21 [2] RSPM (R 4.3.0)
gtools 3.9.5 2023-11-20 [2] RSPM (R 4.3.0)
hdf5r 1.3.9 2024-01-14 [2] RSPM (R 4.3.2)
here * 1.0.1 2020-12-13 [2] RSPM (R 4.3.0)
highr 0.10 2022-12-22 [2] RSPM (R 4.3.0)
hms 1.1.3 2023-03-21 [2] RSPM (R 4.3.0)
htmltools 0.5.7 2023-11-03 [2] RSPM (R 4.3.0)
htmlwidgets 1.6.4 2023-12-06 [2] RSPM (R 4.3.0)
httpuv 1.6.13 2023-12-06 [2] RSPM (R 4.3.0)
httr 1.4.7 2023-08-15 [2] RSPM (R 4.3.0)
ica 1.0-3 2022-07-08 [2] RSPM (R 4.3.0)
igraph 1.6.0 2023-12-11 [2] RSPM (R 4.3.0)
IRanges 2.36.0 2023-10-24 [2] RSPM (R 4.3.2)
irlba 2.3.5.1 2022-10-03 [2] RSPM (R 4.3.0)
janitor 2.2.0.9000 2024-01-26 [2] Github (sfirke/janitor@ad52765)
JASPAR2020 0.99.10 2024-01-26 [2] RSPM (R 4.3.2)
jquerylib 0.1.4 2021-04-26 [2] RSPM (R 4.3.0)
jsonlite 1.8.8 2023-12-04 [2] RSPM (R 4.3.0)
KEGGREST 1.42.0 2023-10-24 [2] RSPM (R 4.3.2)
KernSmooth 2.23-22 2023-07-10 [2] RSPM (R 4.3.0)
knitr 1.45 2023-10-30 [2] RSPM (R 4.3.0)
labeling 0.4.3 2023-08-29 [2] RSPM (R 4.3.0)
later 1.3.2 2023-12-06 [2] RSPM (R 4.3.0)
lattice 0.22-5 2023-10-24 [2] RSPM (R 4.3.0)
lazyeval 0.2.2 2019-03-15 [2] RSPM (R 4.3.0)
leiden 0.4.3.1 2023-11-17 [2] RSPM (R 4.3.0)
lifecycle 1.0.4 2023-11-07 [2] RSPM (R 4.3.0)
listenv 0.9.0 2022-12-16 [2] RSPM (R 4.3.0)
lmtest 0.9-40 2022-03-21 [2] RSPM (R 4.3.0)
lubridate 1.9.3 2023-09-27 [2] RSPM (R 4.3.0)
magrittr * 2.0.3 2022-03-30 [2] RSPM (R 4.3.0)
MASS 7.3-60.0.1 2024-01-13 [2] RSPM (R 4.3.0)
Matrix 1.6-5 2024-01-11 [2] RSPM (R 4.3.0)
MatrixGenerics 1.14.0 2023-10-24 [2] RSPM (R 4.3.2)
matrixStats 1.2.0 2023-12-11 [2] RSPM (R 4.3.0)
memoise 2.0.1 2021-11-26 [2] RSPM (R 4.3.0)
mime 0.12 2021-09-28 [2] RSPM (R 4.3.0)
miniUI 0.1.1.1 2018-05-18 [2] RSPM (R 4.3.0)
mousecortexref.SeuratData * 1.0.0 2023-10-20 [1] local
munsell 0.5.0 2018-06-12 [2] RSPM (R 4.3.0)
nlme 3.1-164 2023-11-27 [2] RSPM (R 4.3.0)
paletteer 1.6.0 2024-01-21 [2] RSPM (R 4.3.0)
parallelly 1.36.0 2023-05-26 [2] RSPM (R 4.3.0)
patchwork * 1.2.0.9000 2024-01-26 [2] Github (thomasp85/patchwork@d943757)
pbapply 1.7-2 2023-06-27 [2] RSPM (R 4.3.0)
pillar 1.9.0 2023-03-22 [2] RSPM (R 4.3.0)
pkgconfig 2.0.3 2019-09-22 [2] RSPM (R 4.3.0)
plotly 4.10.4 2024-01-13 [2] RSPM (R 4.3.0)
plyr 1.8.9 2023-10-02 [2] RSPM (R 4.3.0)
png 0.1-8 2022-11-29 [2] RSPM (R 4.3.0)
polyclip 1.10-6 2023-09-27 [2] RSPM (R 4.3.0)
poweRlaw 0.80.0 2024-01-25 [2] RSPM (R 4.3.2)
pracma 2.4.4 2023-11-10 [2] RSPM (R 4.3.0)
presto 1.0.0 2024-01-26 [2] Github (immunogenomics/presto@31dc97f)
prettyunits 1.2.0 2023-09-24 [2] RSPM (R 4.3.0)
processx 3.8.3 2023-12-10 [2] RSPM (R 4.3.0)
progress 1.2.3 2023-12-06 [2] RSPM (R 4.3.0)
progressr 0.14.0 2023-08-10 [2] RSPM (R 4.3.0)
promises 1.2.1 2023-08-10 [2] RSPM (R 4.3.0)
ProtGenerics 1.34.0 2023-10-24 [2] RSPM (R 4.3.2)
ps 1.7.6 2024-01-18 [2] RSPM (R 4.3.0)
purrr 1.0.2 2023-08-10 [2] RSPM (R 4.3.0)
R.methodsS3 1.8.2 2022-06-13 [2] RSPM (R 4.3.0)
R.oo 1.26.0 2024-01-24 [2] RSPM (R 4.3.0)
R.utils 2.12.3 2023-11-18 [2] RSPM (R 4.3.0)
R6 2.5.1 2021-08-19 [2] RSPM (R 4.3.0)
RANN 2.6.1 2019-01-08 [2] RSPM (R 4.3.0)
rappdirs 0.3.3 2021-01-31 [2] RSPM (R 4.3.0)
RColorBrewer * 1.1-3 2022-04-03 [2] RSPM (R 4.3.0)
Rcpp 1.0.12 2024-01-09 [2] RSPM (R 4.3.0)
RcppAnnoy 0.0.22 2024-01-23 [2] RSPM (R 4.3.0)
RcppHNSW 0.5.0 2023-09-19 [2] RSPM (R 4.3.0)
RcppRoll 0.3.0 2018-06-05 [2] RSPM (R 4.3.0)
RCurl 1.98-1.14 2024-01-09 [2] RSPM (R 4.3.0)
readr * 2.1.5 2024-01-10 [2] RSPM (R 4.3.0)
rematch2 2.1.2 2020-05-01 [2] RSPM (R 4.3.0)
remotes 2.4.2.1 2023-07-18 [2] RSPM (R 4.3.0)
reshape2 1.4.4 2020-04-09 [2] RSPM (R 4.3.0)
restfulr 0.0.15 2022-06-16 [2] RSPM (R 4.3.2)
reticulate 1.34.0 2023-10-12 [2] RSPM (R 4.3.0)
rhdf5 2.46.1 2023-11-29 [2] RSPM (R 4.3.2)
rhdf5filters 1.14.1 2023-11-06 [2] RSPM (R 4.3.2)
Rhdf5lib 1.24.1 2023-12-11 [2] RSPM (R 4.3.2)
rjson 0.2.21 2022-01-09 [2] RSPM (R 4.3.0)
rlang 1.1.3 2024-01-10 [2] RSPM (R 4.3.0)
rmarkdown 2.25 2023-09-18 [2] RSPM (R 4.3.0)
ROCR 1.0-11 2020-05-02 [2] RSPM (R 4.3.0)
rprojroot 2.0.4 2023-11-05 [2] RSPM (R 4.3.0)
Rsamtools 2.18.0 2023-10-24 [2] RSPM (R 4.3.2)
RSpectra 0.16-1 2022-04-24 [2] RSPM (R 4.3.0)
RSQLite 2.3.5 2024-01-21 [2] RSPM (R 4.3.0)
rstudioapi 0.15.0 2023-07-07 [2] RSPM (R 4.3.0)
rsvd 1.0.5 2021-04-16 [2] RSPM (R 4.3.0)
rtracklayer 1.62.0 2024-01-26 [2] bioc_git2r (@58efbf9)
Rtsne 0.17 2023-12-07 [2] RSPM (R 4.3.0)
S4Arrays 1.2.0 2023-10-24 [2] RSPM (R 4.3.2)
S4Vectors 0.40.2 2023-11-23 [2] RSPM (R 4.3.2)
sass 0.4.8 2023-12-06 [2] RSPM (R 4.3.0)
scales 1.3.0 2023-11-28 [2] RSPM (R 4.3.0)
scattermore 1.2 2023-06-12 [2] RSPM (R 4.3.0)
scCustomize * 2.0.1 2024-01-26 [2] Github (samuel-marsh/scCustomize@0aefbe9)
sctransform 0.4.1 2023-10-19 [2] RSPM (R 4.3.0)
seqLogo 1.68.0 2023-10-24 [2] RSPM (R 4.3.2)
sessioninfo 1.2.2 2021-12-06 [2] RSPM (R 4.3.0)
Seurat * 5.0.1.9003 2024-01-27 [2] Github (satijalab/seurat@938698c)
SeuratData * 0.2.2.9001 2024-01-26 [2] Github (satijalab/seurat-data@4dc08e0)
SeuratDisk 0.0.0.9021 2024-01-26 [2] Github (mojaveazure/seurat-disk@877d4e1)
SeuratObject * 5.0.1 2024-01-27 [2] Github (satijalab/seurat-object@4d3739b)
SeuratWrappers * 0.3.3 2024-01-26 [2] Github (satijalab/seurat-wrappers@17b8d5a)
shape 1.4.6 2021-05-19 [2] RSPM (R 4.3.0)
shiny 1.8.0 2023-11-17 [2] RSPM (R 4.3.0)
shinyBS * 0.61.1 2022-04-17 [2] RSPM (R 4.3.0)
shinydashboard 0.7.2 2021-09-30 [2] RSPM (R 4.3.0)
shinyjs 2.1.0 2021-12-23 [2] RSPM (R 4.3.0)
Signac 1.12.9004 2024-01-27 [2] Github (stuart-lab/signac@0c43d88)
snakecase 0.11.1 2023-08-27 [2] RSPM (R 4.3.0)
sp * 2.1-2 2023-11-26 [2] RSPM (R 4.3.0)
spam 2.10-0 2023-10-23 [2] RSPM (R 4.3.0)
SparseArray 1.2.3 2023-12-25 [2] RSPM (R 4.3.2)
spatstat.data 3.0-4 2024-01-15 [2] RSPM (R 4.3.0)
spatstat.explore 3.2-5 2023-10-22 [2] RSPM (R 4.3.0)
spatstat.geom 3.2-7 2023-10-20 [2] RSPM (R 4.3.0)
spatstat.random 3.2-2 2023-11-29 [2] RSPM (R 4.3.0)
spatstat.sparse 3.0-3 2023-10-24 [2] RSPM (R 4.3.0)
spatstat.utils 3.0-4 2023-10-24 [2] RSPM (R 4.3.0)
stringi 1.8.3 2023-12-11 [2] RSPM (R 4.3.0)
stringr * 1.5.1 2023-11-14 [2] RSPM (R 4.3.0)
SummarizedExperiment 1.32.0 2023-10-24 [2] RSPM (R 4.3.2)
survival 3.5-7 2023-08-14 [2] RSPM (R 4.3.0)
tensor 1.5 2012-05-05 [2] RSPM (R 4.3.0)
TFBSTools 1.40.0 2023-10-24 [2] RSPM (R 4.3.2)
TFMPvalue 0.0.9 2022-10-21 [2] RSPM (R 4.3.0)
tibble 3.2.1 2023-03-20 [2] RSPM (R 4.3.0)
tidyr 1.3.1 2024-01-24 [2] RSPM (R 4.3.0)
tidyselect 1.2.0 2022-10-10 [2] RSPM (R 4.3.0)
timechange 0.3.0 2024-01-18 [2] RSPM (R 4.3.0)
tzdb 0.4.0 2023-05-12 [2] RSPM (R 4.3.0)
utf8 1.2.4 2023-10-22 [2] RSPM (R 4.3.0)
uwot 0.1.16 2023-06-29 [2] RSPM (R 4.3.0)
vctrs 0.6.5 2023-12-01 [2] RSPM (R 4.3.0)
vipor 0.4.7 2023-12-18 [2] RSPM (R 4.3.0)
viridisLite 0.4.2 2023-05-02 [2] RSPM (R 4.3.0)
vroom 1.6.5 2023-12-05 [2] RSPM (R 4.3.0)
whisker 0.4.1 2022-12-05 [2] RSPM (R 4.3.0)
withr 3.0.0 2024-01-16 [2] RSPM (R 4.3.0)
workflowr * 1.7.1 2023-08-23 [2] RSPM (R 4.3.0)
xfun 0.41 2023-11-01 [2] RSPM (R 4.3.0)
XML 3.99-0.16.1 2024-01-22 [2] RSPM (R 4.3.0)
xml2 1.3.6 2023-12-04 [2] RSPM (R 4.3.0)
xtable 1.8-4 2019-04-21 [2] RSPM (R 4.3.0)
XVector 0.42.0 2023-10-24 [2] RSPM (R 4.3.2)
yaml 2.3.8 2023-12-11 [2] RSPM (R 4.3.0)
zlibbioc 1.48.0 2023-10-24 [2] RSPM (R 4.3.2)
zoo 1.8-12 2023-04-13 [2] RSPM (R 4.3.0)
[1] /home/etretiakov/R/x86_64-pc-linux-gnu-library/4.3
[2] /opt/R/4.3.2/lib/R/library
─ Python configuration ───────────────────────────────────────────────────────
python: /opt/python/3.8.8/bin/python
libpython: /opt/python/3.8.8/lib/libpython3.8.so
pythonhome: /opt/python/3.8.8:/opt/python/3.8.8
version: 3.8.8 | packaged by conda-forge | (default, Feb 20 2021, 16:22:27) [GCC 9.3.0]
numpy: /opt/python/3.8.8/lib/python3.8/site-packages/numpy
numpy_version: 1.23.5
leidenalg: /opt/python/3.8.8/lib/python3.8/site-packages/leidenalg
NOTE: Python version was forced by RETICULATE_PYTHON
──────────────────────────────────────────────────────────────────────────────
sessionInfo()R version 4.3.2 (2023-10-31)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 22.04.3 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
time zone: Etc/UTC
tzcode source: system (glibc)
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] patchwork_1.2.0.9000 cowplot_1.1.3
[3] ggplot2_3.4.4.9000 readr_2.1.5
[5] stringr_1.5.1 magrittr_2.0.3
[7] dplyr_1.1.4 BPCells_0.1.0
[9] Azimuth_0.5.0 shinyBS_0.61.1
[11] SeuratWrappers_0.3.3 mousecortexref.SeuratData_1.0.0
[13] SeuratData_0.2.2.9001 scCustomize_2.0.1
[15] Seurat_5.0.1.9003 SeuratObject_5.0.1
[17] sp_2.1-2 RColorBrewer_1.1-3
[19] here_1.0.1 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] IRanges_2.36.0 R.methodsS3_1.8.2
[3] vroom_1.6.5 progress_1.2.3
[5] poweRlaw_0.80.0 goftest_1.2-3
[7] DT_0.31 Biostrings_2.70.1
[9] vctrs_0.6.5 spatstat.random_3.2-2
[11] digest_0.6.34 png_0.1-8
[13] shape_1.4.6 git2r_0.33.0
[15] ggrepel_0.9.5.9999 deldir_2.0-2
[17] parallelly_1.36.0 MASS_7.3-60.0.1
[19] Signac_1.12.9004 reshape2_1.4.4
[21] httpuv_1.6.13 BiocGenerics_0.48.1
[23] withr_3.0.0 ggrastr_1.0.2
[25] xfun_0.41 ellipsis_0.3.2
[27] survival_3.5-7 EnsDb.Hsapiens.v86_2.99.0
[29] memoise_2.0.1 ggbeeswarm_0.7.2
[31] janitor_2.2.0.9000 zoo_1.8-12
[33] GlobalOptions_0.1.2 gtools_3.9.5
[35] pbapply_1.7-2 R.oo_1.26.0
[37] prettyunits_1.2.0 rematch2_2.1.2
[39] KEGGREST_1.42.0 promises_1.2.1
[41] httr_1.4.7 restfulr_0.0.15
[43] rhdf5filters_1.14.1 globals_0.16.2
[45] fitdistrplus_1.1-11 rhdf5_2.46.1
[47] ps_1.7.6 rstudioapi_0.15.0
[49] miniUI_0.1.1.1 generics_0.1.3
[51] processx_3.8.3 curl_5.2.0
[53] S4Vectors_0.40.2 zlibbioc_1.48.0
[55] polyclip_1.10-6 GenomeInfoDbData_1.2.11
[57] SparseArray_1.2.3 xtable_1.8-4
[59] pracma_2.4.4 evaluate_0.23
[61] S4Arrays_1.2.0 BiocFileCache_2.10.1
[63] hms_1.1.3 GenomicRanges_1.54.1
[65] irlba_2.3.5.1 colorspace_2.1-0
[67] filelock_1.0.3 hdf5r_1.3.9
[69] ROCR_1.0-11 reticulate_1.34.0
[71] spatstat.data_3.0-4 lmtest_0.9-40
[73] snakecase_0.11.1 later_1.3.2
[75] lattice_0.22-5 spatstat.geom_3.2-7
[77] future.apply_1.11.1 getPass_0.2-4
[79] scattermore_1.2 XML_3.99-0.16.1
[81] matrixStats_1.2.0 RcppAnnoy_0.0.22
[83] pillar_1.9.0 nlme_3.1-164
[85] caTools_1.18.2 compiler_4.3.2
[87] RSpectra_0.16-1 stringi_1.8.3
[89] tensor_1.5 SummarizedExperiment_1.32.0
[91] lubridate_1.9.3 GenomicAlignments_1.38.2
[93] plyr_1.8.9 crayon_1.5.2
[95] abind_1.4-5 BiocIO_1.12.0
[97] googledrive_2.1.1 bit_4.0.5
[99] fastmatch_1.1-4 whisker_0.4.1
[101] codetools_0.2-19 bslib_0.6.1
[103] paletteer_1.6.0 plotly_4.10.4
[105] mime_0.12 splines_4.3.2
[107] circlize_0.4.16 Rcpp_1.0.12
[109] fastDummies_1.7.3 dbplyr_2.4.0
[111] cellranger_1.1.0 knitr_1.45
[113] blob_1.2.4 utf8_1.2.4
[115] seqLogo_1.68.0 AnnotationFilter_1.26.0
[117] fs_1.6.3 listenv_0.9.0
[119] tibble_3.2.1 Matrix_1.6-5
[121] callr_3.7.3 tzdb_0.4.0
[123] pkgconfig_2.0.3 tools_4.3.2
[125] cachem_1.0.8 RSQLite_2.3.5
[127] viridisLite_0.4.2 DBI_1.2.1
[129] fastmap_1.1.1 rmarkdown_2.25
[131] scales_1.3.0 grid_4.3.2
[133] ica_1.0-3 shinydashboard_0.7.2
[135] Rsamtools_2.18.0 sass_0.4.8
[137] ggprism_1.0.4 BiocManager_1.30.22
[139] dotCall64_1.1-1 RANN_2.6.1
[141] farver_2.1.1 yaml_2.3.8
[143] MatrixGenerics_1.14.0 rtracklayer_1.62.0
[145] cli_3.6.2 purrr_1.0.2
[147] stats4_4.3.2 leiden_0.4.3.1
[149] lifecycle_1.0.4 uwot_0.1.16
[151] Biobase_2.62.0 sessioninfo_1.2.2
[153] presto_1.0.0 BSgenome.Hsapiens.UCSC.hg38_1.4.5
[155] BiocParallel_1.36.0 annotate_1.80.0
[157] timechange_0.3.0 gtable_0.3.4
[159] rjson_0.2.21 ggridges_0.5.6
[161] progressr_0.14.0 parallel_4.3.2
[163] jsonlite_1.8.8 RcppHNSW_0.5.0
[165] TFBSTools_1.40.0 bitops_1.0-7
[167] bit64_4.0.5 Rtsne_0.17
[169] spatstat.utils_3.0-4 CNEr_1.38.0
[171] highr_0.10 jquerylib_0.1.4
[173] shinyjs_2.1.0 SeuratDisk_0.0.0.9021
[175] R.utils_2.12.3 lazyeval_0.2.2
[177] shiny_1.8.0 htmltools_0.5.7
[179] GO.db_3.18.0 sctransform_0.4.1
[181] rappdirs_0.3.3 ensembldb_2.26.0
[183] glue_1.7.0 TFMPvalue_0.0.9
[185] spam_2.10-0 googlesheets4_1.1.1
[187] XVector_0.42.0 RCurl_1.98-1.14
[189] rprojroot_2.0.4 BSgenome_1.70.1
[191] gridExtra_2.3 JASPAR2020_0.99.10
[193] igraph_1.6.0 R6_2.5.1
[195] tidyr_1.3.1 labeling_0.4.3
[197] forcats_1.0.0 RcppRoll_0.3.0
[199] GenomicFeatures_1.54.1 cluster_2.1.6
[201] Rhdf5lib_1.24.1 gargle_1.5.2
[203] GenomeInfoDb_1.38.5 DirichletMultinomial_1.44.0
[205] DelayedArray_0.28.0 tidyselect_1.2.0
[207] vipor_0.4.7 ProtGenerics_1.34.0
[209] xml2_1.3.6 AnnotationDbi_1.64.1
[211] future_1.33.1 rsvd_1.0.5
[213] munsell_0.5.0 KernSmooth_2.23-22
[215] data.table_1.14.10 htmlwidgets_1.6.4
[217] biomaRt_2.58.0 rlang_1.1.3
[219] spatstat.sparse_3.0-3 spatstat.explore_3.2-5
[221] remotes_2.4.2.1 fansi_1.0.6
[223] beeswarm_0.4.0