Exploratory analysis of principal sensory trigeminal nucleus and ventrobasal thalamus
Evgenii O. Tretiakov
2023-01-24
Last updated: 2023-01-24
Checks: 7 0
Knit directory: Hevesi_2023/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20230121) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 2d7093f. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: .cache/
Ignored: .config/
Ignored: .nv/
Ignored: .snakemake/
Ignored: analysis/figure/
Ignored: cellbender/
Ignored: cellbender_latest.sif
Ignored: cellranger/
Ignored: fastq/
Ignored: mm10_optimized/
Ignored: souporcell/
Ignored: souporcell_latest.sif
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/eda.Rmd) and HTML
(docs/eda.html) files. If you’ve configured a remote Git
repository (see ?wflow_git_remote), click on the hyperlinks
in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 2d7093f | Evgenii O. Tretiakov | 2023-01-24 | workflowr::wflow_publish("analysis/eda.Rmd", all = TRUE, verbose = TRUE) |
| Rmd | 45efe1a | Evgenii O. Tretiakov | 2023-01-24 | workflowr::wflow_publish("analysis/eda.Rmd", all = TRUE, verbose = TRUE) |
| Rmd | 52ce403 | Evgenii O. Tretiakov | 2023-01-24 | workflowr::wflow_publish("analysis/eda.Rmd", all = TRUE, verbose = TRUE) |
Combined analysis of scRNA-seq datasets derived from the same experiment
Since it was derived from the same two pooled mice we might try to merge them together.
souporcell_THP7 <-
read_tsv(here("souporcell/THP7", "clusters.tsv")) %>%
mutate(origin = "THP7",
cell_name = str_c("THP7_", barcode))
souporcell_Pr5P7 <-
read_tsv(here("souporcell/Pr5P7", "clusters.tsv")) %>%
mutate(origin = "Pr5P7",
cell_name = str_c("Pr5P7_", barcode))
souporcell <-
bind_rows(
souporcell_THP7,
souporcell_Pr5P7)
souporcell %>%
janitor::tabyl(status, assignment, origin)$Pr5P7
status 0 0/1 1 1/0
singlet 294 0 321 0
unassigned 37 9 65 5
$THP7
status 0 0/1 1 1/0
singlet 492 0 318 0
unassigned 70 5 29 9cell_bender_merged <-
Read_CellBender_h5_Multi_Directory(
base_path = here("cellbender"),
custom_name = "_output_filtered.h5",
sample_names = c("THP7", "Pr5P7"),
merge = TRUE)
cell_ranger_merged <-
Read10X_h5_Multi_Directory(
base_path = here("cellranger"),
default_10X_path = TRUE,
h5_filename = "filtered_feature_bc_matrix.h5",
merge = TRUE,
sample_names = c("THP7", "Pr5P7"),
parallel = TRUE,
num_cores = 16)
combined.srt <-
Create_CellBender_Merged_Seurat(
raw_cell_bender_matrix = cell_bender_merged,
raw_counts_matrix = cell_ranger_merged,
raw_assay_name = "RAW")After running troublet (genotyping-based detection of doublets), it seems that we don’t have much of doublets.
Elimination of ambient RNA
| orig.ident | nCount_RNA | nFeature_RNA | nCount_RAW | nFeature_RAW | nFeature_Diff | nCount_Diff | |
|---|---|---|---|---|---|---|---|
| THP7_AAACCCAAGCTGACAG-1 | THP7 | 5526 | 2614 | 5542 | 2615 | 1 | 16 |
| THP7_AAACCCAGTCTTGTCC-1 | THP7 | 6876 | 2954 | 6898 | 2954 | 0 | 22 |
| THP7_AAACGAAGTGTTCCTC-1 | THP7 | 12076 | 4603 | 12110 | 4604 | 1 | 34 |
| THP7_AAACGCTTCCCTCGAT-1 | THP7 | 10009 | 3885 | 10046 | 3890 | 5 | 37 |
| THP7_AAAGGATGTTGCATGT-1 | THP7 | 3277 | 1840 | 3283 | 1840 | 0 | 6 |
| orig.ident | Median_nCount_RNA | Median_nFeature_RNA | Median_nCount_Diff | Median_nFeature_Diff |
|---|---|---|---|---|
| Pr5P7 | 8434 | 3476 | 89 | 14 |
| THP7 | 5142 | 2481 | 27 | 3 |
| Totals (All Cells) | 6489 | 2902 | 50 | 9 |
| Raw_Counts | CellBender_Counts | Count_Diff | Pct_Diff | |
|---|---|---|---|---|
| 1700054A03Rik | 357 | 134 | 223 | 62.46499 |
| Mt2 | 148 | 73 | 75 | 50.67568 |
| Gm50306 | 28 | 14 | 14 | 50.00000 |
| Hist2h2bb | 9 | 5 | 4 | 44.44444 |
| Ly6a | 12 | 7 | 5 | 41.66667 |
Plot feature differences
In addition to returning the data.frame it can be useful to visually examine the changes/trends after running CellBender.
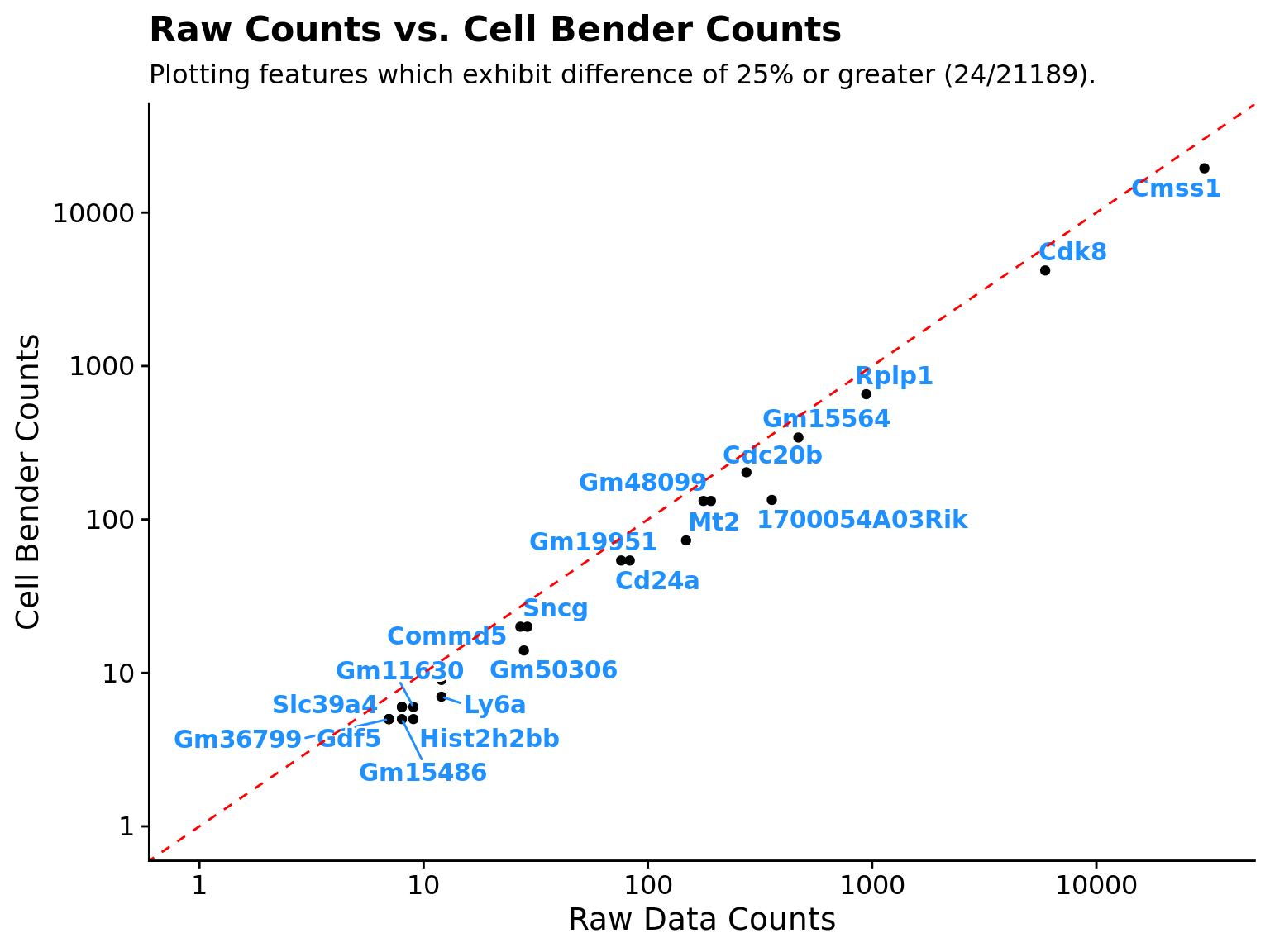
sex_genes <-
str_to_title(c('EHD2', 'ESPL1', 'JARID1D', 'PNPLA4',
'RPS4Y1', 'XIST','tsix', 'Eif2s3y',
'Ddx3y', 'Uty', 'Kdm5d')) %>% .[. %in% rownames(combined.srt)]
stress_genes <-
str_to_title(c('Rpl26','Gstp1','Rpl35a','Erh',
'Slc25a5','Pgk1','Eno1',
'Tubb2a','Emc4','Scg5')) %>% .[. %in% rownames(combined.srt)]
combined.srt[["percent_mt"]] <- PercentageFeatureSet(combined.srt, pattern = "^mt-")
combined.srt[["percent_rb"]] <- PercentageFeatureSet(combined.srt, pattern = "^Rp[sl]")
combined.srt[["percent_hb"]] <- PercentageFeatureSet(combined.srt, pattern = "^Hb[^(p)]")
combined.srt$log10GenesPerUMI <-
log10(combined.srt$nFeature_RNA) / log10(combined.srt$nCount_RNA)
# Visualize QC metrics as a violin plot
VlnPlot(combined.srt,
features = c("log10GenesPerUMI", "nFeature_RNA", "nCount_RNA",
"percent_mt", "percent_rb", "percent_hb"),
ncol = 3) &
theme(plot.title = element_text(size=16))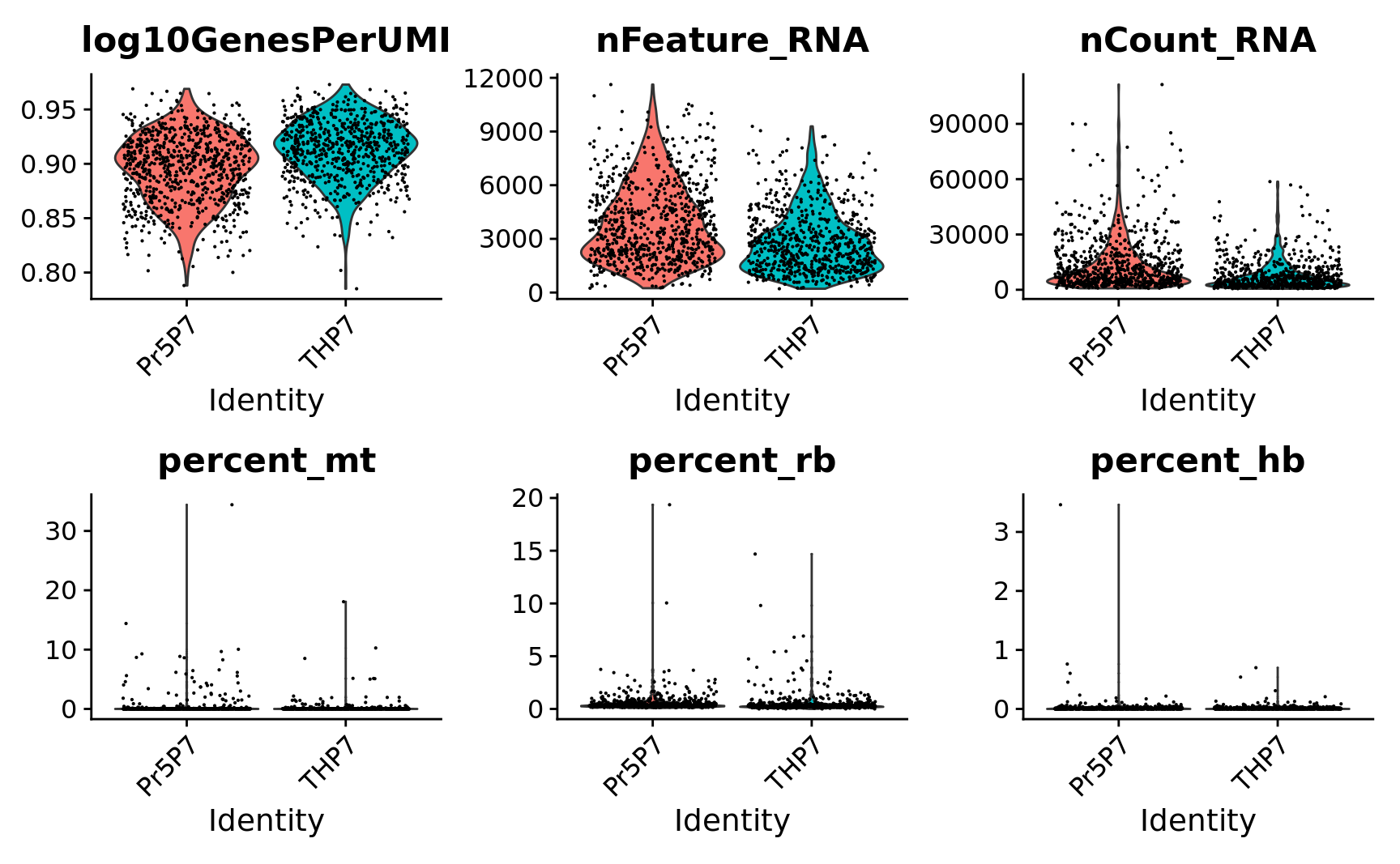
plot1 <- FeatureScatter(combined.srt, feature1 = "nCount_RNA", feature2 = "percent_mt") + NoLegend()
plot2 <- FeatureScatter(combined.srt, feature1 = "nCount_RNA", feature2 = "nFeature_RNA") + NoLegend()
plot3 <- FeatureScatter(combined.srt, feature1 = "nCount_RNA", feature2 = "percent_rb") + NoLegend()
plot4 <- FeatureScatter(combined.srt, feature1 = "percent_rb", feature2 = "percent_mt")
(plot1 + plot2) / (plot3 + plot4)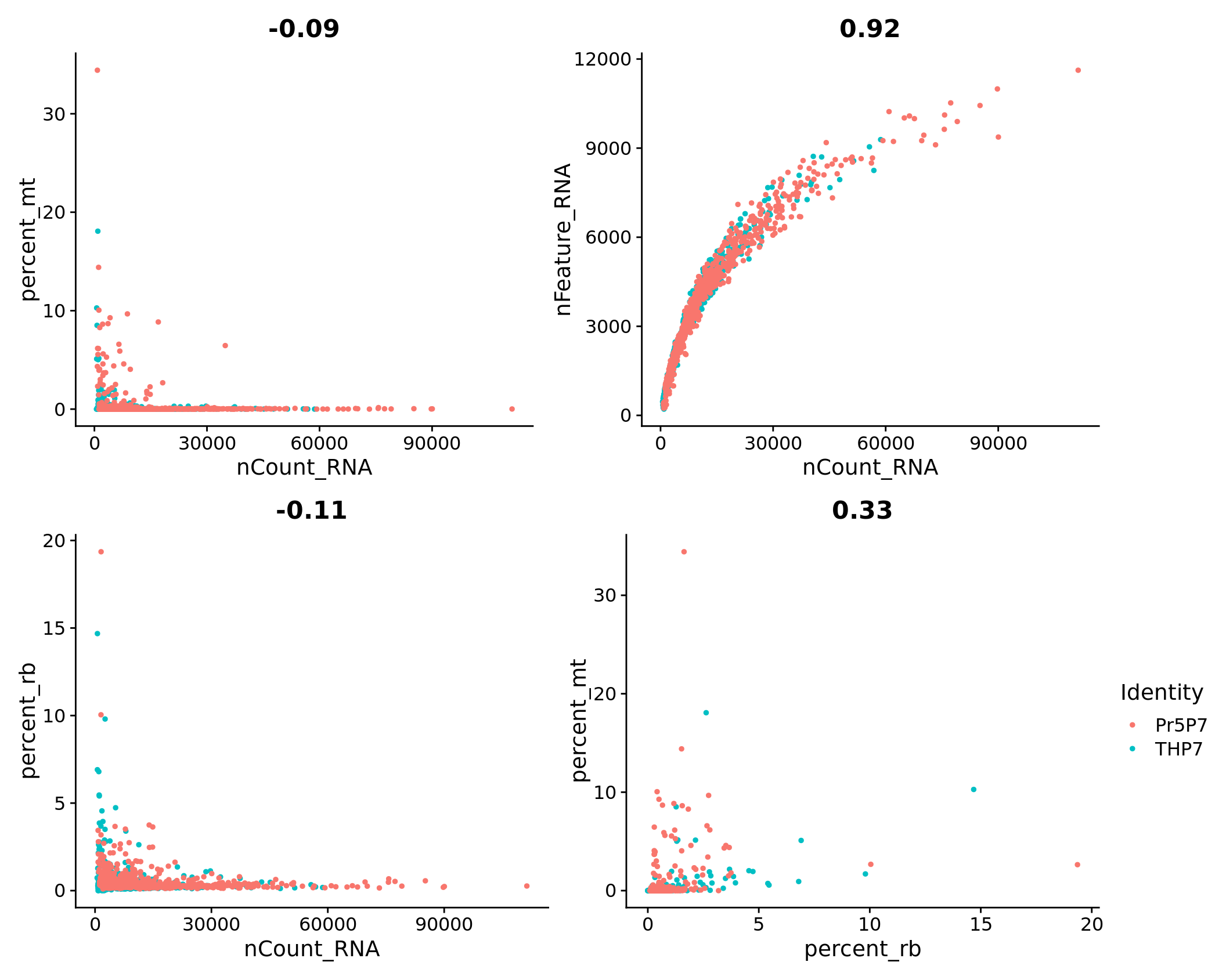
combined.srt[['QC']] <-
ifelse(
combined.srt@meta.data$nFeature_RNA < 500 &
combined.srt@meta.data$QC == 'Pass',
'Low_nFeature',
combined.srt@meta.data$QC
)
combined.srt[['QC']] <-
ifelse(
combined.srt@meta.data$nFeature_RNA < 500 &
combined.srt@meta.data$QC != 'Pass' &
combined.srt@meta.data$QC != 'Low_nFeature',
paste('Low_nFeature', combined.srt@meta.data$QC, sep = ','),
combined.srt@meta.data$QC
)
combined.srt[['QC']] <-
ifelse(
combined.srt@meta.data$percent_mt > 5 &
combined.srt@meta.data$QC == 'Pass',
'High_MT',
combined.srt@meta.data$QC
)
combined.srt[['QC']] <-
ifelse(
combined.srt@meta.data$percent_mt > 5 &
combined.srt@meta.data$QC != 'Pass' &
combined.srt@meta.data$QC != 'High_MT',
paste('High_MT', combined.srt@meta.data$QC, sep = ','),
combined.srt@meta.data$QC
)
combined.srt[['QC']] <-
ifelse(
combined.srt@meta.data$nCount_RNA > 35000 &
combined.srt@meta.data$QC == 'Pass',
'High_UMIs',
combined.srt@meta.data$QC
)
combined.srt[['QC']] <-
ifelse(
combined.srt@meta.data$nCount_RNA > 35000 &
combined.srt@meta.data$QC != 'Pass' &
combined.srt@meta.data$QC != 'High_UMIs',
paste('High_UMIs', combined.srt@meta.data$QC, sep = ','),
combined.srt@meta.data$QC
)
table(combined.srt[['QC']])QC
High_MT High_MT,Low_nFeature High_UMIs
11 13 72
Low_nFeature Pass
9 1382 VlnPlot(subset(combined.srt, subset = QC == 'Pass'),
features = c("log10GenesPerUMI", "nFeature_RNA", "nCount_RNA",
"percent_mt", "percent_rb", "percent_hb"),
ncol = 3) &
theme(plot.title = element_text(size=16))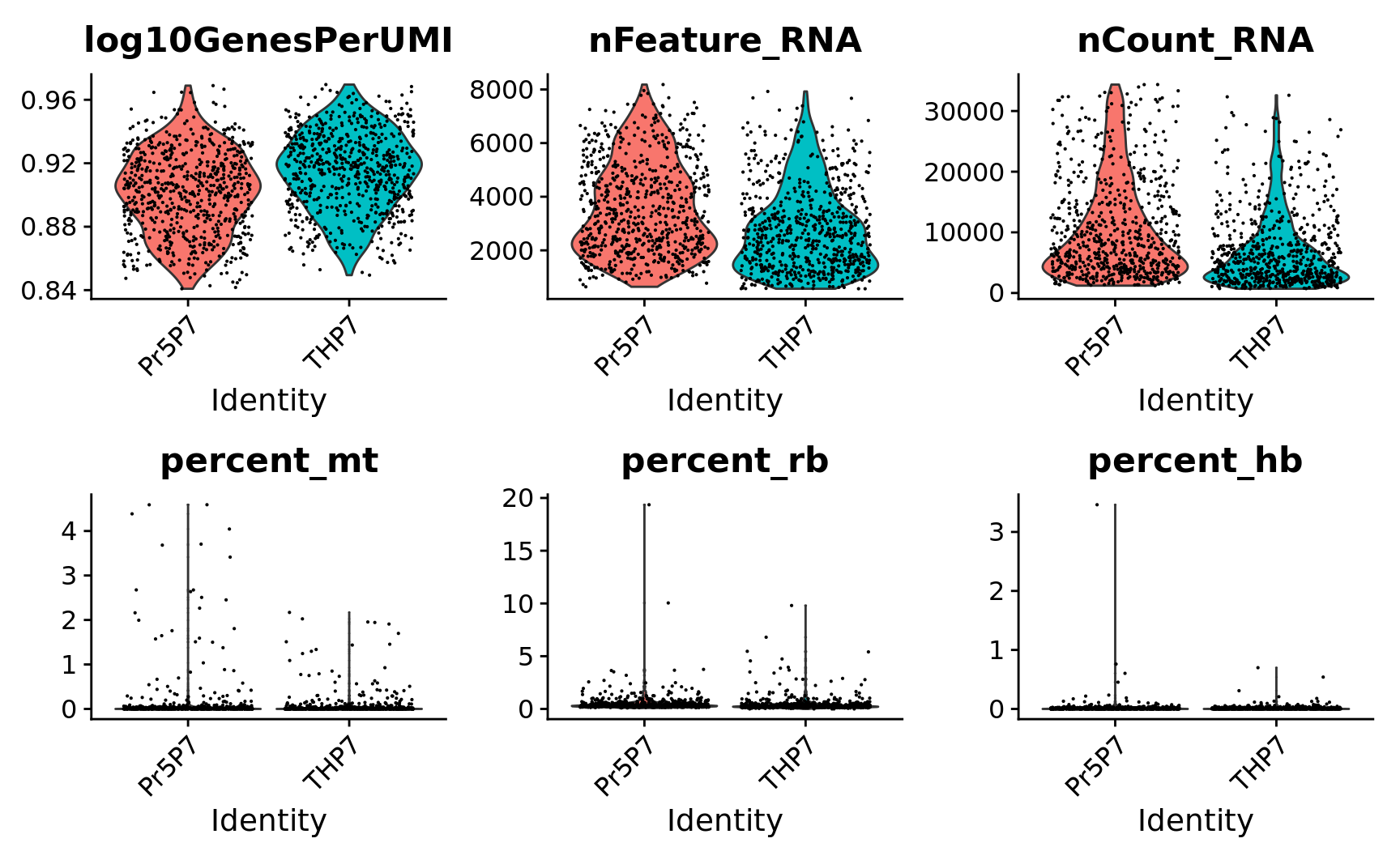
combined.srt <- NormalizeData(combined.srt)
combined.srt <-
FindVariableFeatures(
combined.srt,
selection.method = "vst",
nfeatures = 3000)
top10 <- head(VariableFeatures(combined.srt), 10)
top10 [1] "Ttr" "Htr2c" "Ptgds" "Bnc2" "Mbp" "Npsr1" "St18"
[8] "Dnah12" "Flt1" "Cfap299"plot5 <- VariableFeaturePlot(combined.srt)
LabelPoints(plot = plot5, points = top10, repel = TRUE, xnudge = 0, ynudge = 0)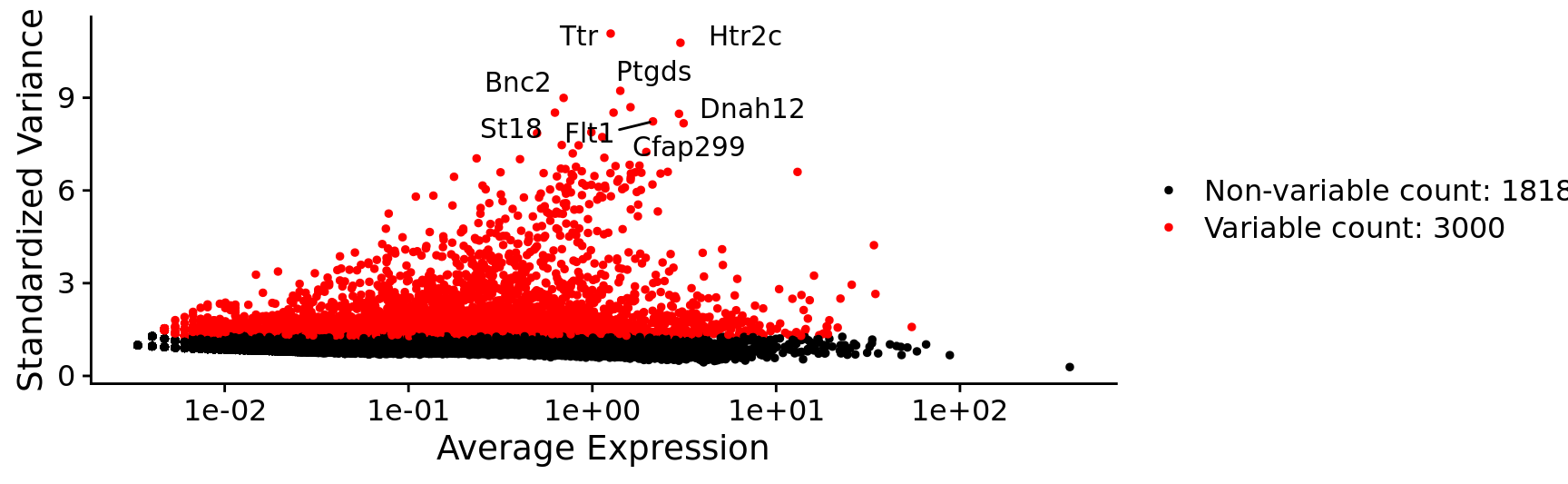
all.genes <- rownames(combined.srt)
hvg <- VariableFeatures(combined.srt)
var_regex <- '^Hla-|^Ig[hjkl]|^Rna|^mt-|^Rp[sl]|^Hb[^(p)]|^Gm'
hvg <- hvg[str_detect(pattern = var_regex, string = hvg, negate = T)]
combined.srt[["var_regex"]] <-
PercentageFeatureSet(combined.srt, pattern = var_regex)
combined.srt <- ScaleData(combined.srt,
features = all.genes,
vars.to.regress = c("var_regex"))
npcs <- 30
combined.srt <- RunPCA(combined.srt,
features = hvg,
npcs = npcs,
seed.use = reseed,
verbose = TRUE)
VizDimLoadings(combined.srt, dims = 1:9, reduction = "pca") &
theme(axis.text = element_text(size = 5),
axis.title = element_text(size = 8, face = "bold"))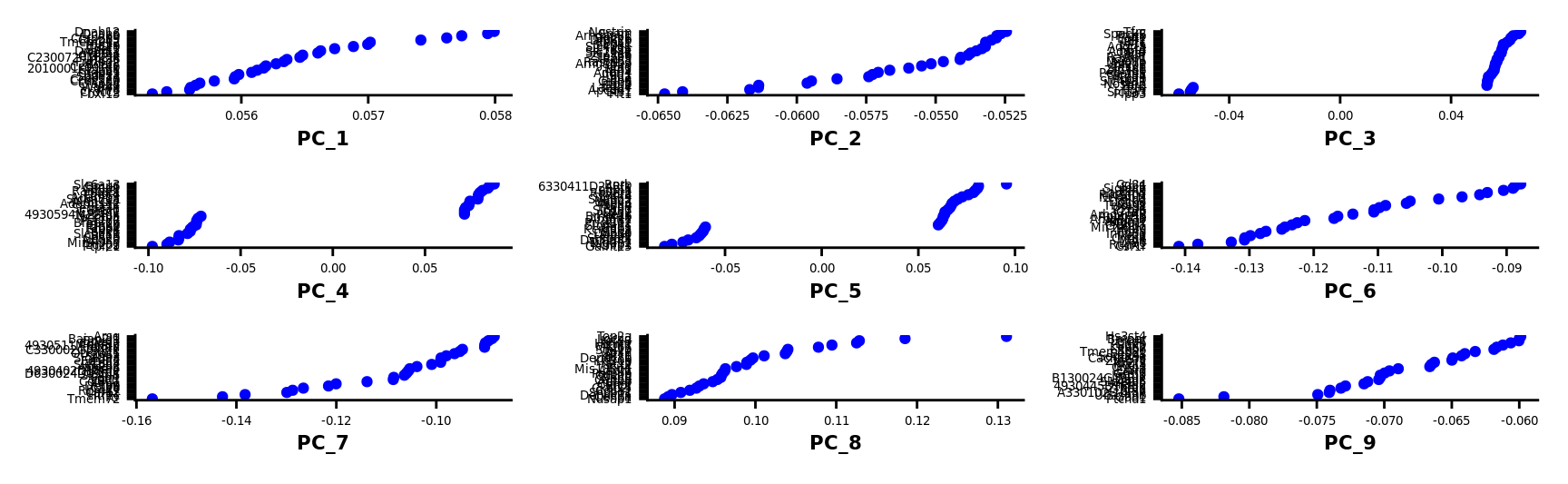
DimHeatmap(combined.srt, dims = 1:6, nfeatures = 20, cells = 500, balanced = T)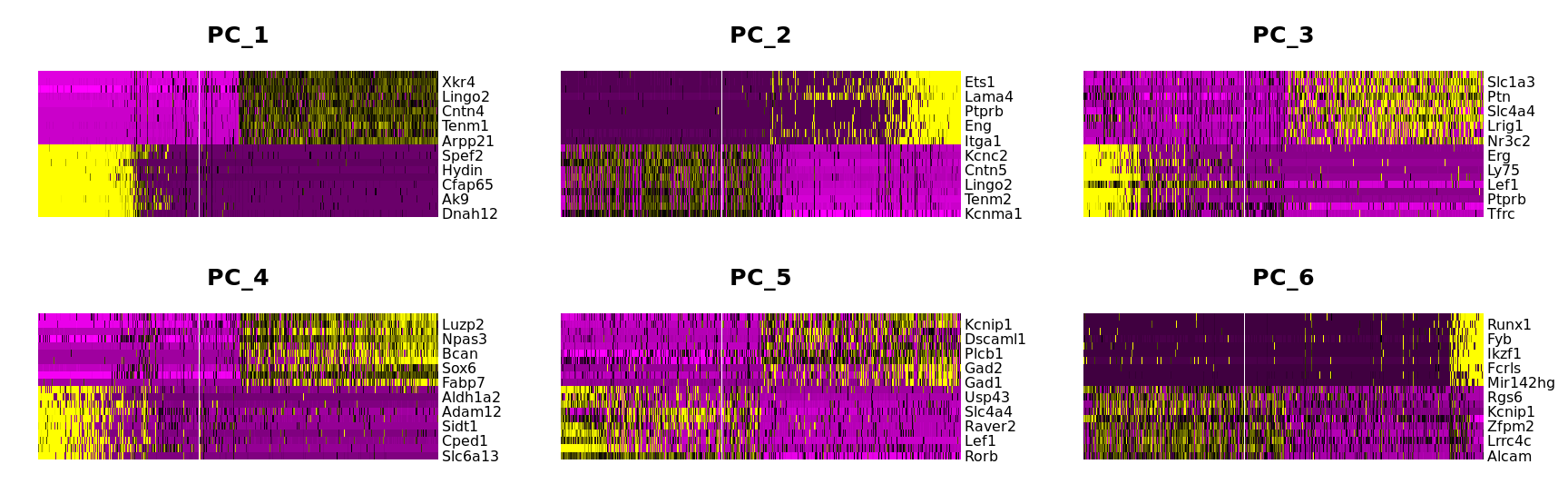
DimPlot(combined.srt, reduction = "pca")
ElbowPlot(combined.srt)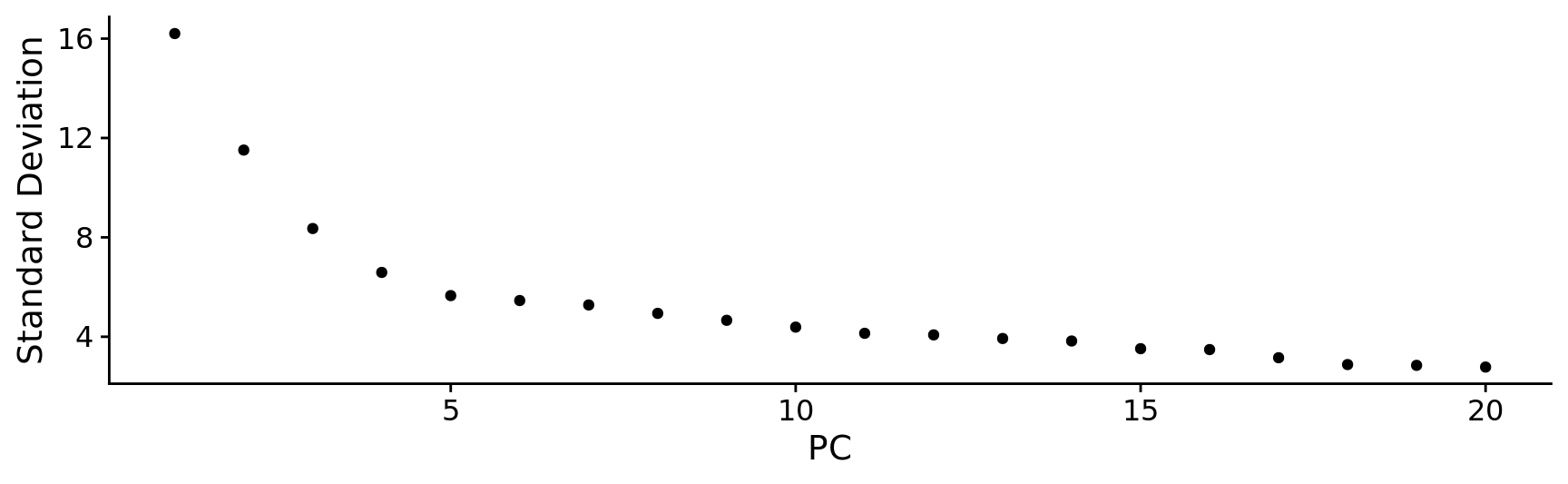
combined.srt <-
JackStraw(
object = combined.srt,
assay = "RNA",
reduction = "pca",
dims = npcs,
num.replicate = 100,
prop.freq = 0.02,
maxit = 1000)
combined.srt <-
ScoreJackStraw(combined.srt,
dims = seq_along(combined.srt[["pca"]]@stdev))
JackStrawPlot(combined.srt, dims = seq_along(combined.srt[["pca"]]@stdev))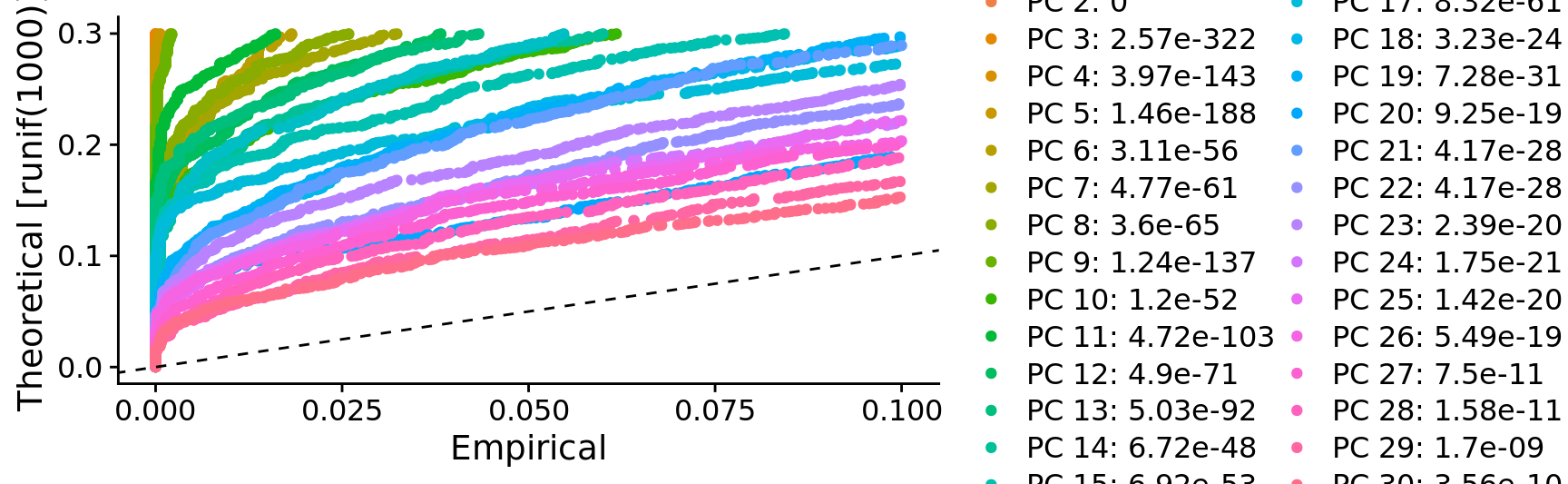
test_pc <-
PCScore(object = combined.srt,
PCs = seq_along(combined.srt[["pca"]]@stdev),
score.thresh = 1e-05)
selected_pcs <-
seq_along(
combined.srt[["pca"]]@stdev
)[test_pc$Score <= 1e-03 &
combined.srt[["pca"]]@stdev > quantile(combined.srt[["pca"]]@stdev, .25)]
selected_pcs [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22combined.srt <-
combined.srt |>
FindNeighbors(
dims = selected_pcs,
k.param = 15,
annoy.metric = "euclidean",
n.trees = 100,
verbose = FALSE) |>
RunUMAP(
dims = selected_pcs,
reduction.name = "umap",
reduction.key = "UMAP_",
return.model = FALSE,
umap.method = "umap-learn",
densmap = TRUE,
dens.lambda = 1L,
dens.frac = 0.3,
n.epochs = 1000L,
n.neighbors = 15L,
min.dist = 0.01,
spread = 2L,
metric = "correlation",
init = "pca",
seed.use = reseed,
verbose = FALSE)
metadata <- combined.srt@meta.data
rownames(metadata) <- colnames(combined.srt)
ref.labels <- metadata$orig.ident
resolutions <-
modularity_event_sampling(
A = combined.srt@graphs$RNA_snn,
n.res = 20,
gamma.min = 0.1,
gamma.max = 3.000001
) # sample based on the similarity matrix
# clustering using Suerat
combined.srt <- combined.srt |>
FindClusters(algorithm = 4, method = "igraph",
resolution = resolutions, random.seed = reseed,
verbose = FALSE)
# initial cluster tree from Seurat flat clustering
plot_clustree(
labelmat = combined.srt@meta.data,
prefix = 'RNA_snn_res.',
ref.labels = ref.labels,
plot.ref = FALSE
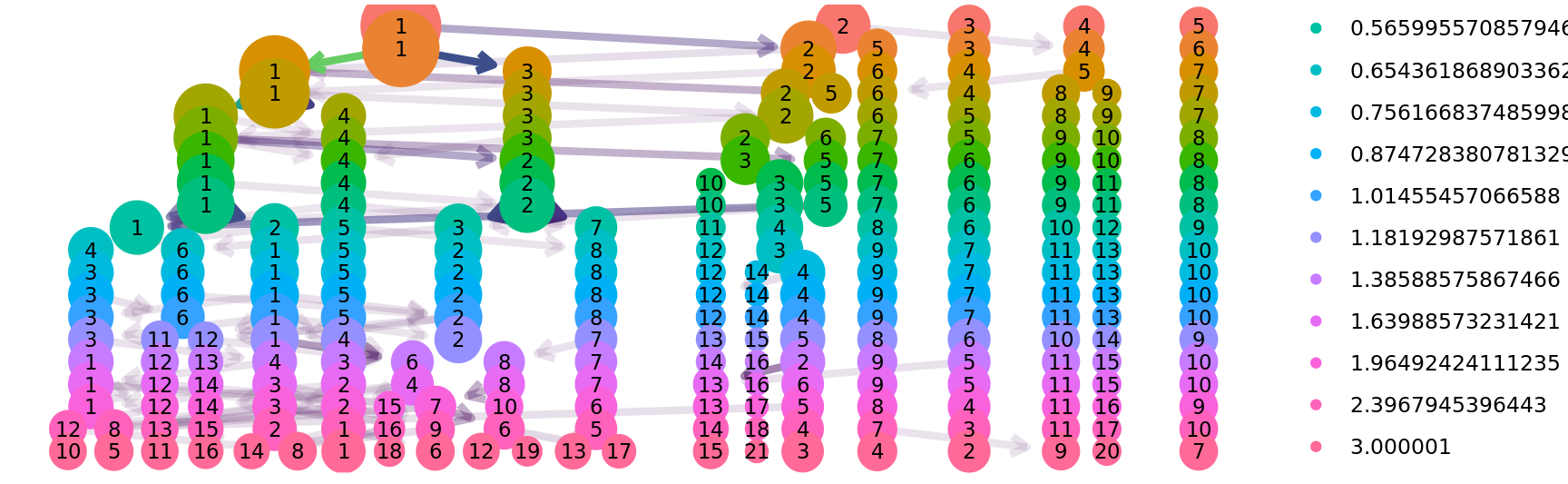
)
out <- mrtree(
combined.srt,
prefix = 'RNA_snn_res.',
n.cores = n_cores,
consensus = FALSE,
augment.path = FALSE
)
|
|======================================================================| 100%
|
|= | 2%
|
|=== | 4%
|
|==== | 5%
|
|===== | 7%
|
|====== | 9%
|
|======== | 11%
|
|========= | 13%
|
|========== | 15%
|
|=========== | 16%
|
|============== | 20%
|
|=============== | 22%
|
|================= | 24%
|
|================== | 25%
|
|=================== | 27%
|
|==================== | 29%
|
|====================== | 31%
|
|======================= | 33%
|
|======================== | 35%
|
|========================= | 36%
|
|============================ | 40%
|
|============================= | 42%
|
|=============================== | 44%
|
|================================ | 45%
|
|================================= | 47%
|
|================================== | 49%
|
|====================================== | 55%
|
|========================================= | 58%
|
|========================================== | 60%
|
|================================================== | 71%
|
|=================================================== | 73%
|
|==================================================== | 75%
|
|===================================================== | 76%
|
|======================================================= | 78%
|
|=========================================================== | 84%
|
|============================================================ | 85%
|
|============================================================== | 89%
|
|================================================================ | 91%
|
|================================================================= | 93%
|
|================================================================== | 95%
|
|=================================================================== | 96%
|
|===================================================================== | 98%
|
|======================================================================| 100%# if there are few partitions per k, within resolution consensus step can speed up the algorithm
# weight per sample is encoraged if the classes are imbalanced
plot_tree(
labelmat = out$labelmat.mrtree,
ref.labels = ref.labels,
plot.piechart = TRUE,
node.size = 0.2,
tip.label.dist = 10,
bottom.margin = 30
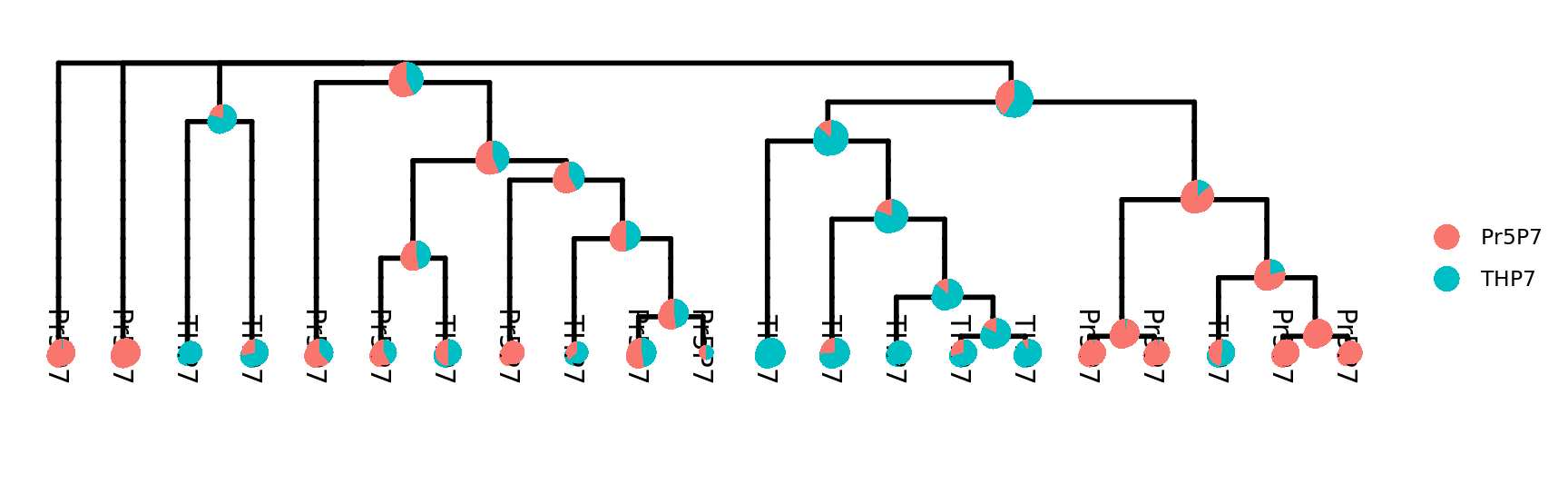
)
# Adjusted Multiresolution Rand Index (AMRI)
ks.flat <- apply(
out$labelmat.flat,
2,
FUN = function(x)
length(unique(x))
)
ks.mrtree <- apply(
out$labelmat.mrtree,
2,
FUN = function(x)
length(unique(x))
)
amri.flat <- sapply(1:ncol(out$labelmat.flat), function(i)
AMRI(out$labelmat.flat[, i], ref.labels)$amri)
amri.flat <- aggregate(amri.flat, by = list(k = ks.flat), FUN = mean)
amri.recon <- sapply(1:ncol(out$labelmat.mrtree), function(i)
AMRI(out$labelmat.mrtree[, i], ref.labels)$amri)
df <- rbind(
data.frame(
k = amri.flat$k,
amri = amri.flat$x,
method = 'Seurat flat'
),
data.frame(k = ks.mrtree, amri = amri.recon, method = 'MRtree')
)
ggplot2::ggplot(data = df, aes(x = k, y = amri, color = method)) + geom_line() + theme_bw()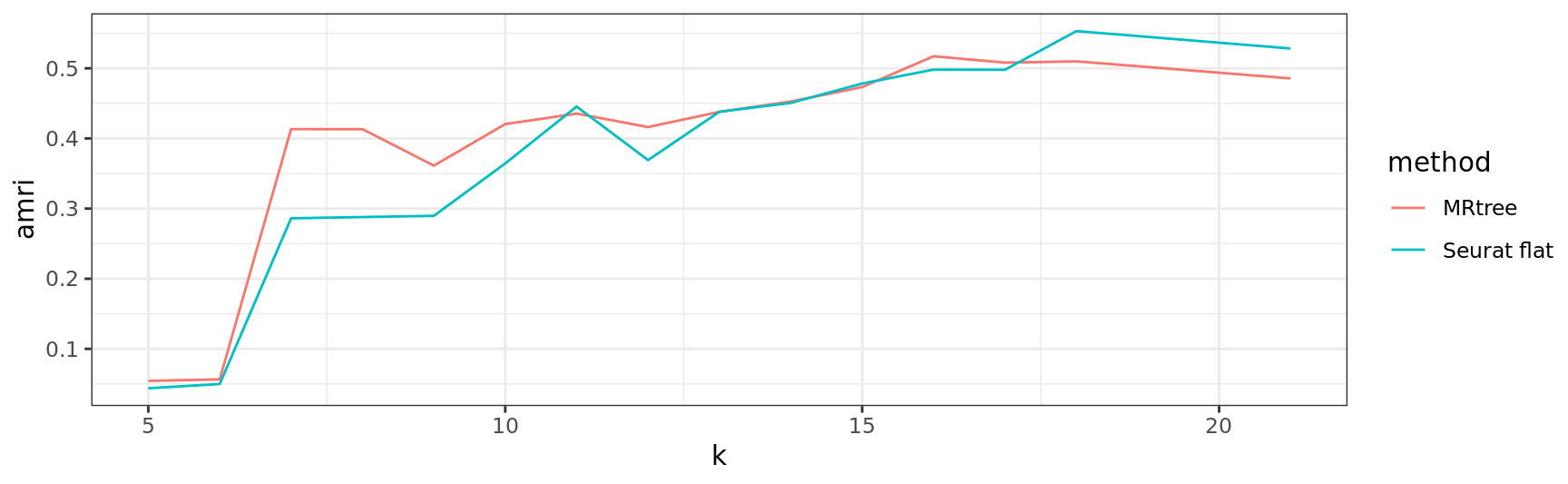
stab.out <- stability_plot(out)
stab.out$plot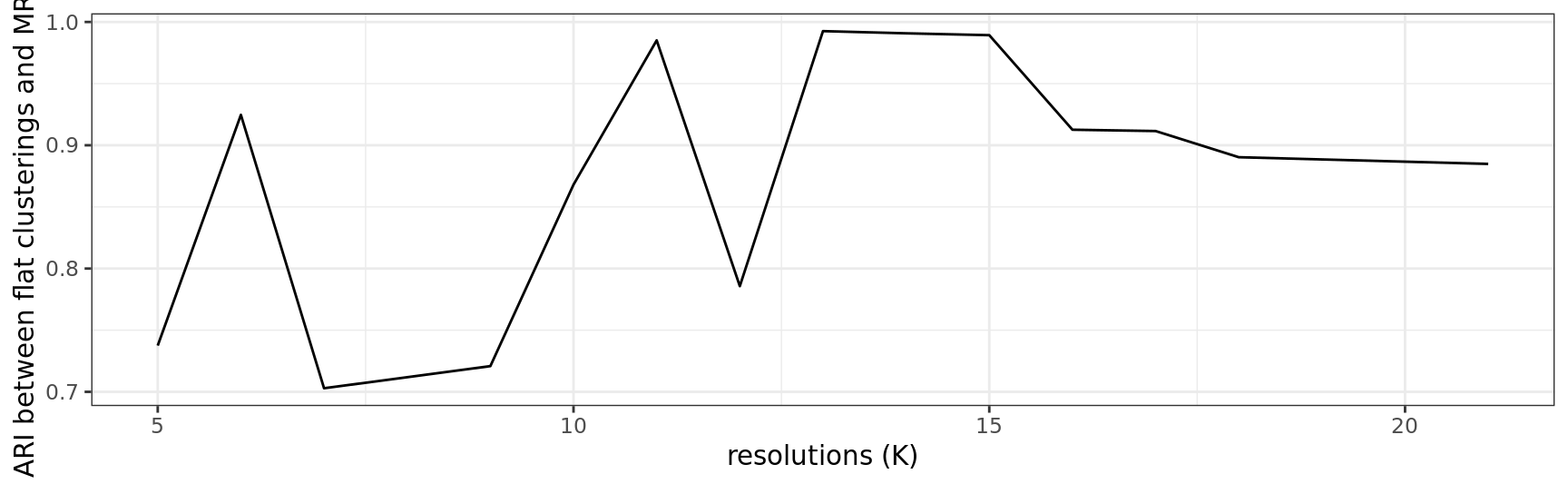
kable_material(
kable(
stab.out$df,
"html"),
bootstrap_options = c("bordered",
"condensed",
"responsive",
"striped"),
position = "left",
font_size = 14
)| resolution | ari |
|---|---|
| 5 | 0.7375092 |
| 6 | 0.9246521 |
| 7 | 0.7029071 |
| 9 | 0.7208427 |
| 10 | 0.8680305 |
| 11 | 0.9850789 |
| 12 | 0.7857161 |
| 13 | 0.9925812 |
| 14 | 0.9908067 |
| 15 | 0.9892876 |
| 16 | 0.9126044 |
| 17 | 0.9115270 |
| 18 | 0.8903003 |
| 21 | 0.8848603 |
resK <- SelectResolution(stab.out$df)
resK[1] 14kable_material(
kable(
table(
out$labelmat.mrtree[, which.min(
abs(as.integer(
str_remove(dimnames(
out$labelmat.mrtree)[[2]], "K"
)
) - resK)
)]
),
"html"),
bootstrap_options = c("bordered",
"condensed",
"responsive",
"striped"),
position = "left",
font_size = 14
)| Var1 | Freq |
|---|---|
| 1 | 185 |
| 2 | 175 |
| 3 | 147 |
| 4 | 143 |
| 5 | 143 |
| 6 | 130 |
| 7 | 118 |
| 8 | 115 |
| 9 | 92 |
| 10 | 80 |
| 11 | 76 |
| 12 | 32 |
| 13 | 29 |
| 14 | 22 |
combined.srt$k_tree <- out$labelmat.mrtree[, which.min(
abs(as.integer(
str_remove(dimnames(
out$labelmat.mrtree)[[2]], "K"
)
) - resK)
)]
p1 <- DimPlot(combined.srt, label = T, repel = T, pt.size = 2) + ggtitle("Unsupervised clustering") + NoLegend()
p2 <- DimPlot(combined.srt, label = T, repel = T, group.by = "k_tree", pt.size = 2) + ggtitle("MRTree") + NoLegend()
p1 | p2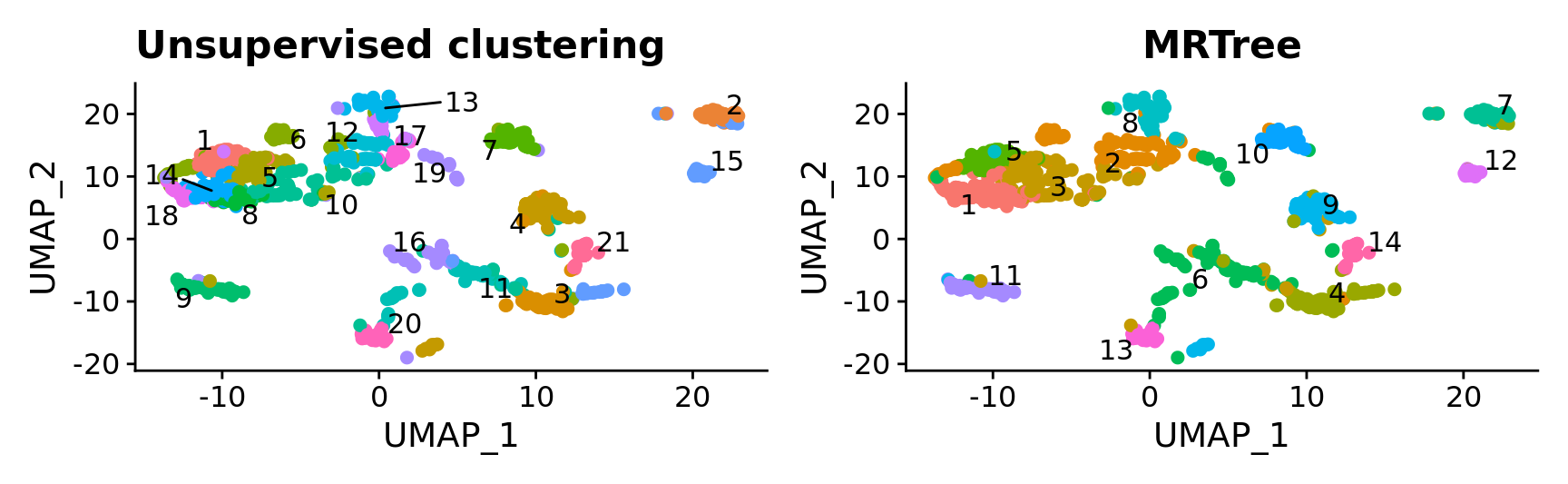
Idents(combined.srt) <- "k_tree"FeaturePlot(combined.srt, features = "percent_mt") &
theme(plot.title = element_text(size = 16)) &
scale_colour_gradientn(colours = rev(brewer.pal(n = 11, name = "RdYlGn")))
FeaturePlot(combined.srt, features = "nFeature_RNA") &
theme(plot.title = element_text(size = 16)) &
scale_colour_gradientn(colours = rev(brewer.pal(n = 11, name = "RdYlGn")))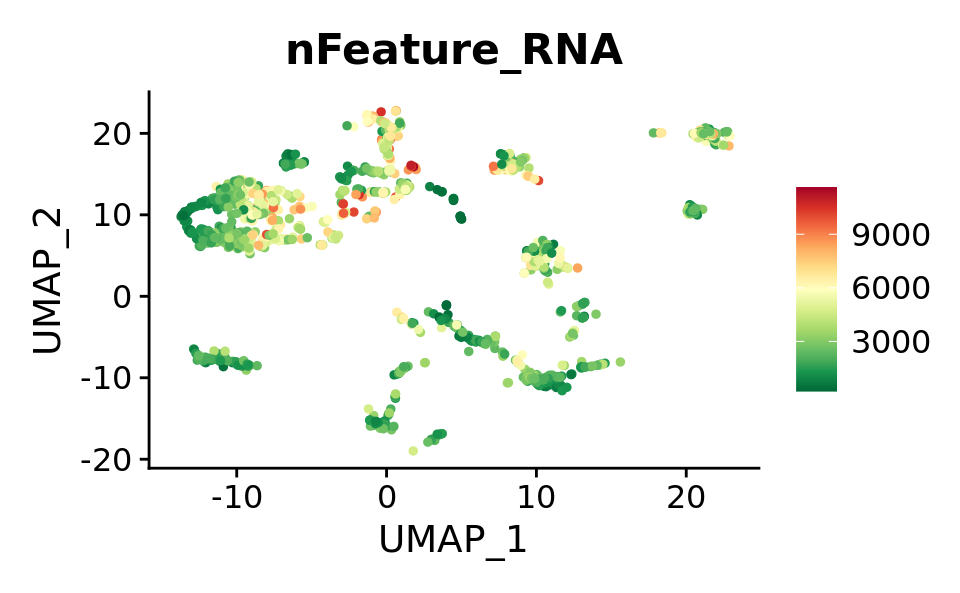
Dual Assay Plotting
For Cell Bender especially, but also potentially for other assays as well, it can be helpful during analysis to plot the corrected and uncorrected counts for given feature.
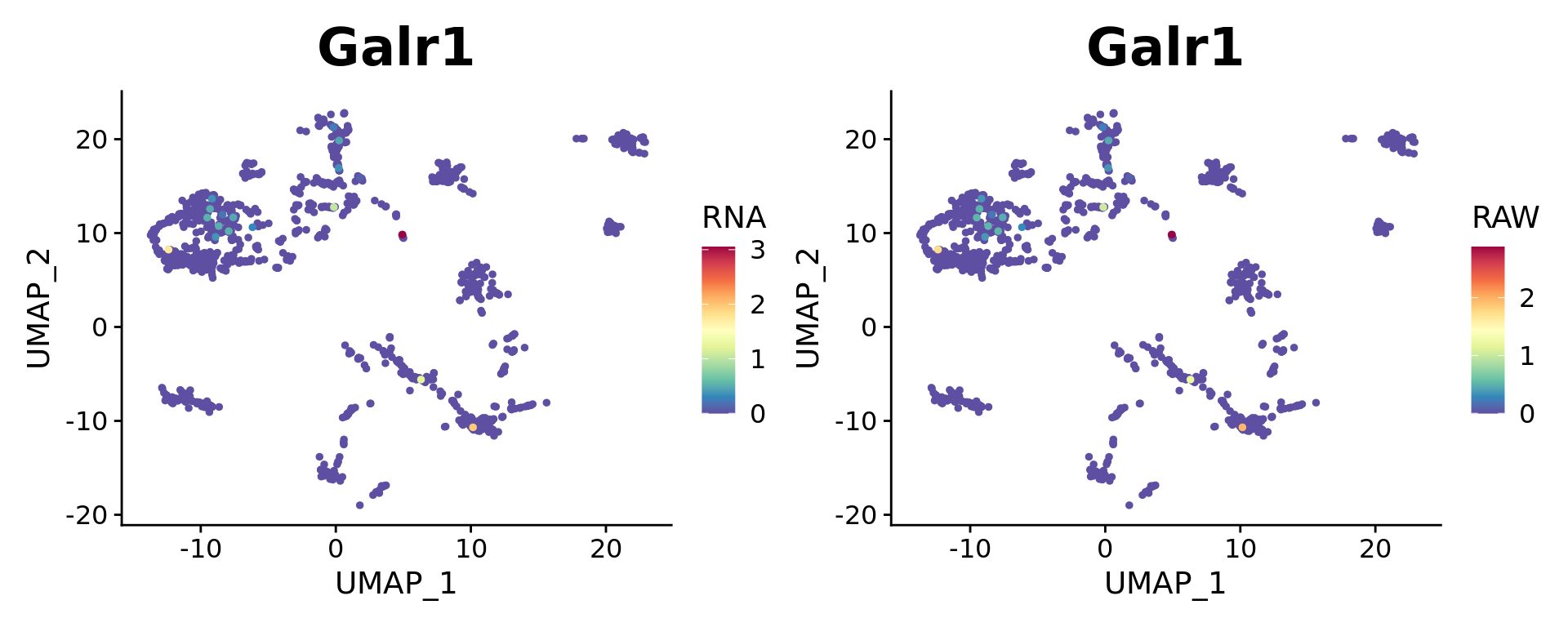
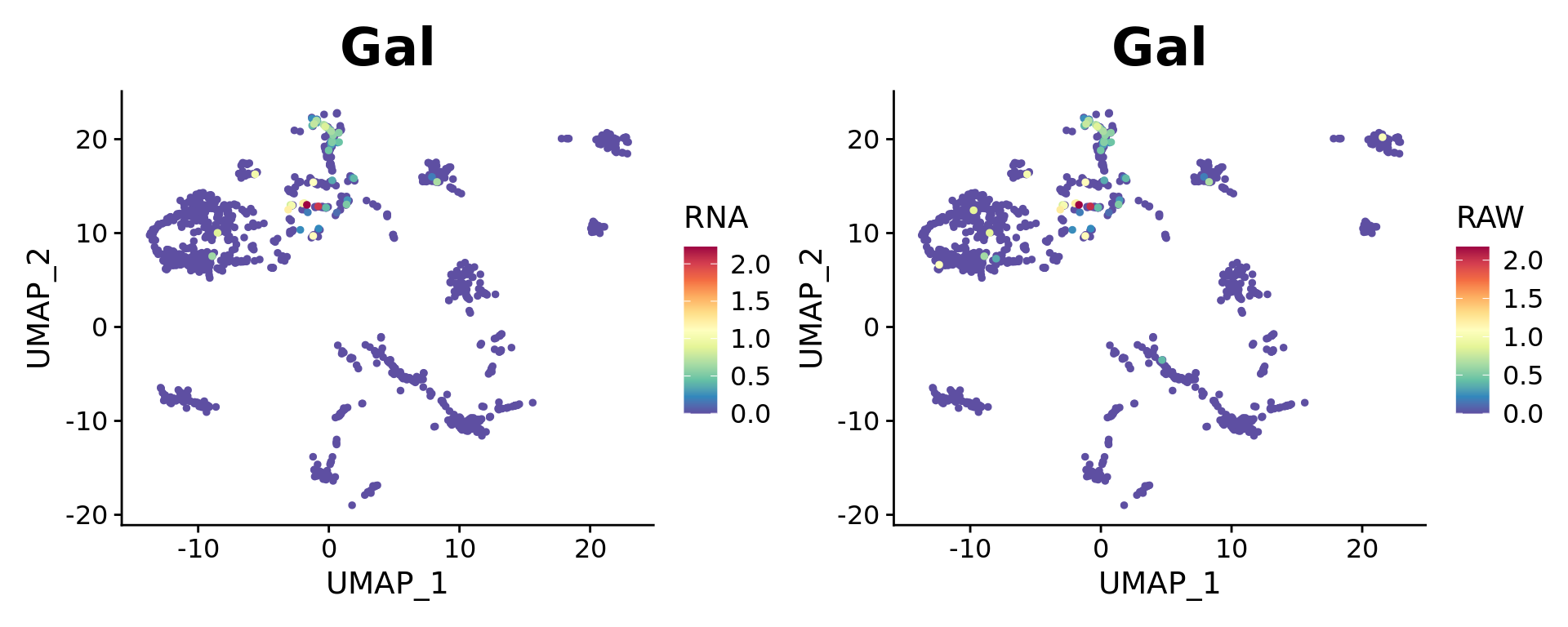 Both targets look fine.
Both targets look fine.
DimPlot(combined.srt, label.size = 4, repel = T, pt.size = 3, label = T)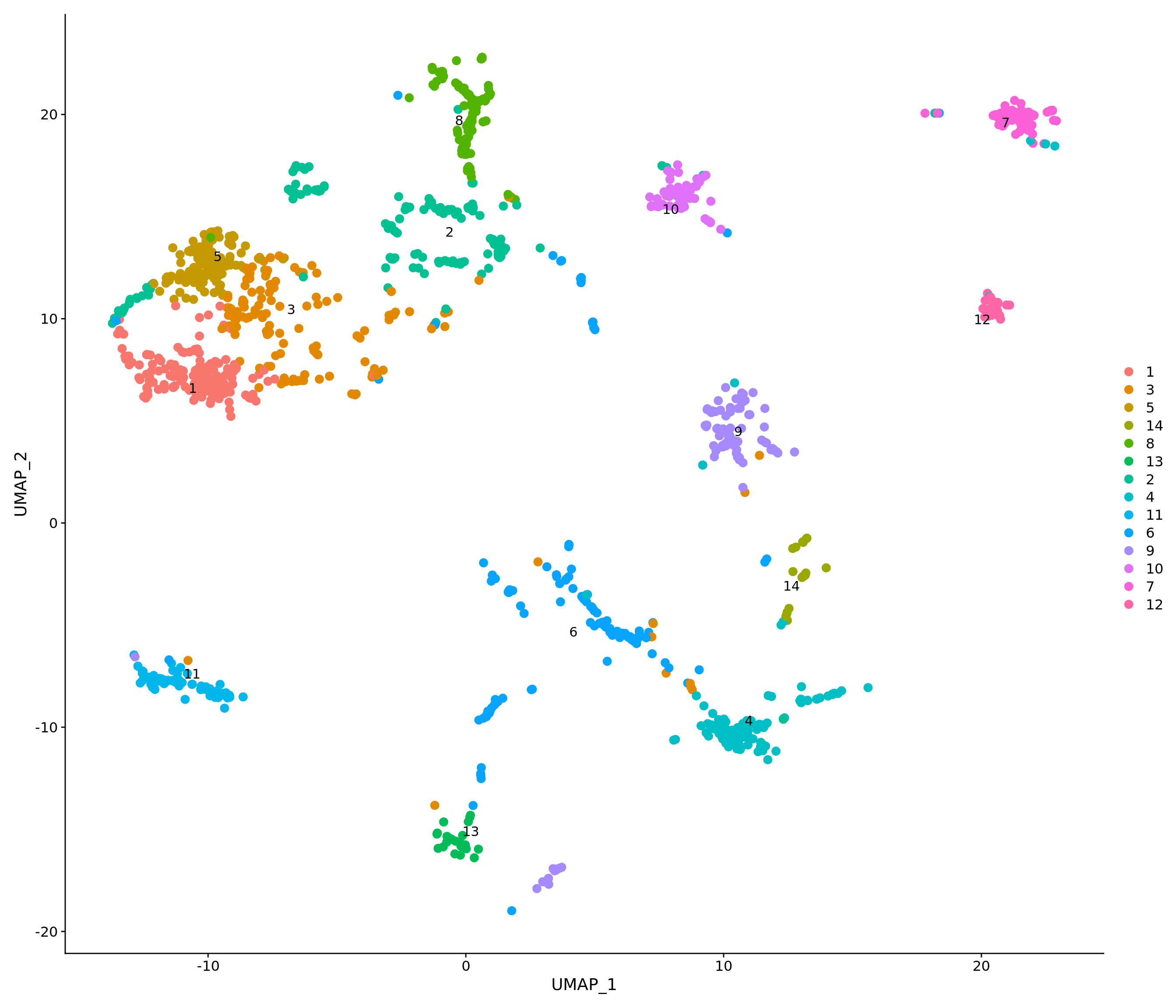
combined.srt <- subset(combined.srt, subset = QC == 'Pass')
DimPlot(combined.srt, label.size = 4, repel = T, pt.size = 3, label = T)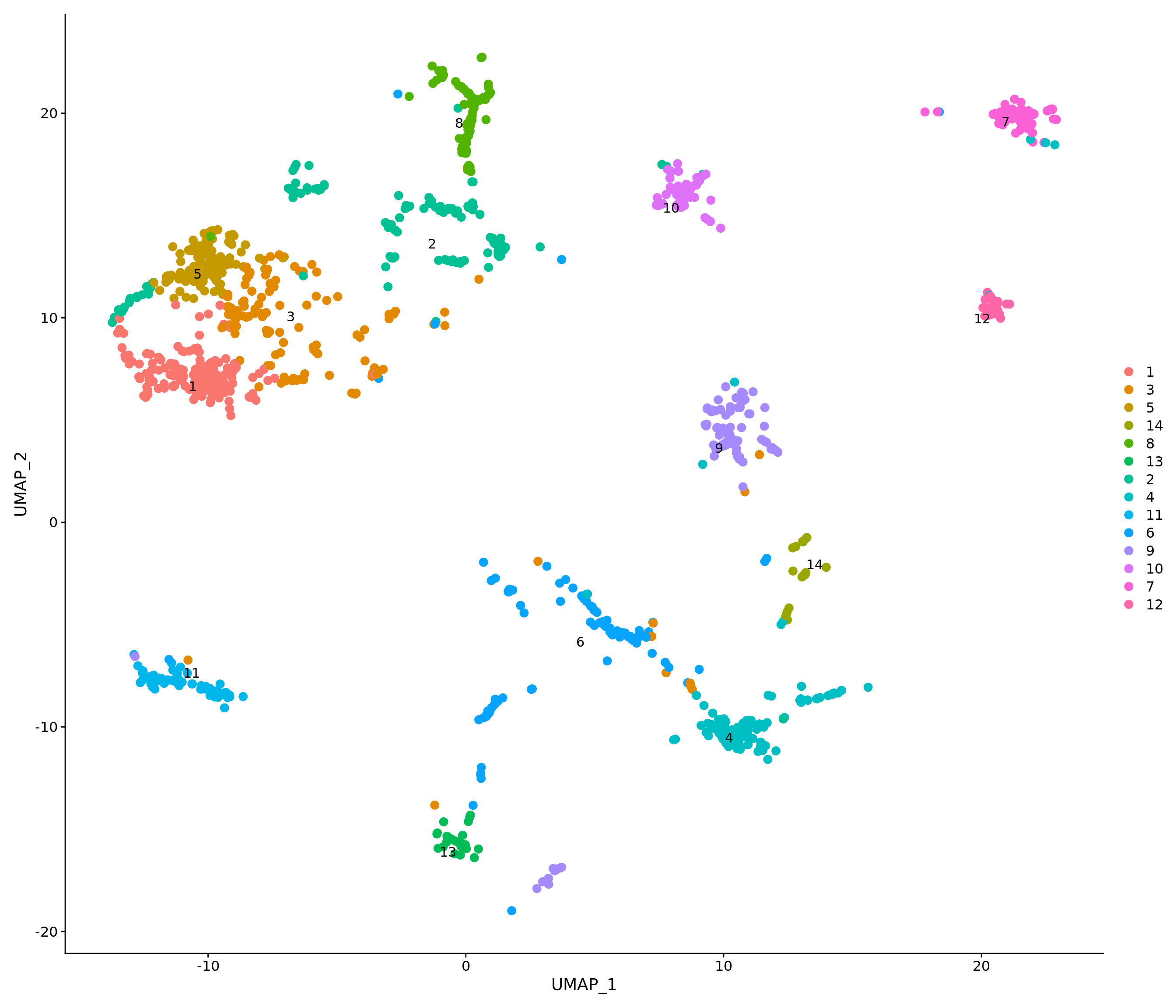 ### Reevaluate after subsetting low-quality cells
### Reevaluate after subsetting low-quality cells
s.genes = gorth(cc.genes.updated.2019$s.genes, source_organism = "hsapiens", target_organism = "mmusculus")$ortholog_name
g2m.genes = gorth(cc.genes.updated.2019$g2m.genes, source_organism = "hsapiens", target_organism = "mmusculus")$ortholog_name
combined.srt <-
CellCycleScoring(combined.srt,
s.features = s.genes,
g2m.features = g2m.genes)
table(combined.srt[[]]$Phase)
G1 G2M S
640 227 515 FeaturePlot(combined.srt,features = "percent_mt", label.size = 4, repel = T, pt.size = 3, label = T) &
theme(plot.title = element_text(size = 16)) &
scale_colour_gradientn(colours = rev(brewer.pal(n = 11, name = "RdYlGn")))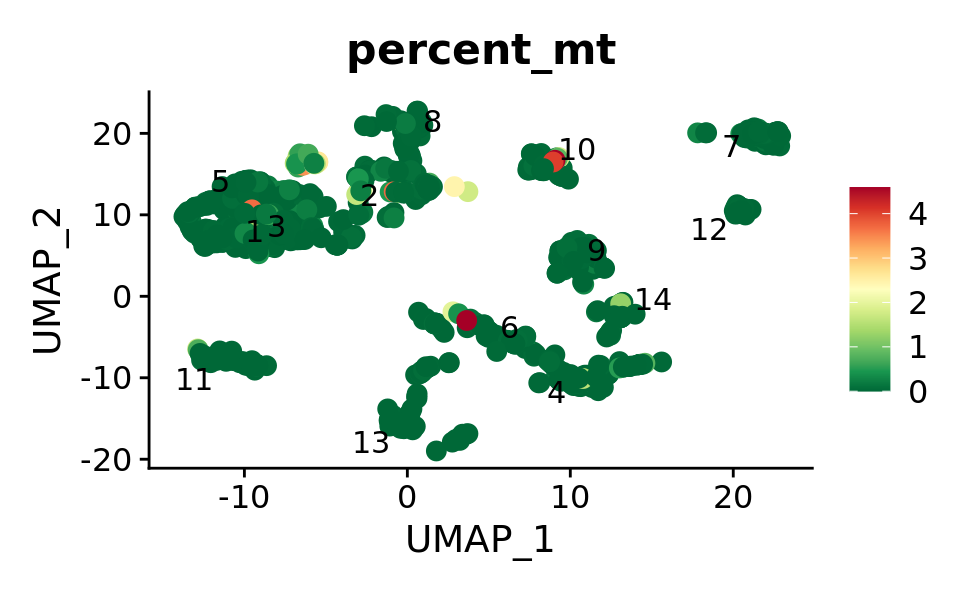
VlnPlot(combined.srt, features = "percent_mt") & theme(plot.title = element_text(size = 16))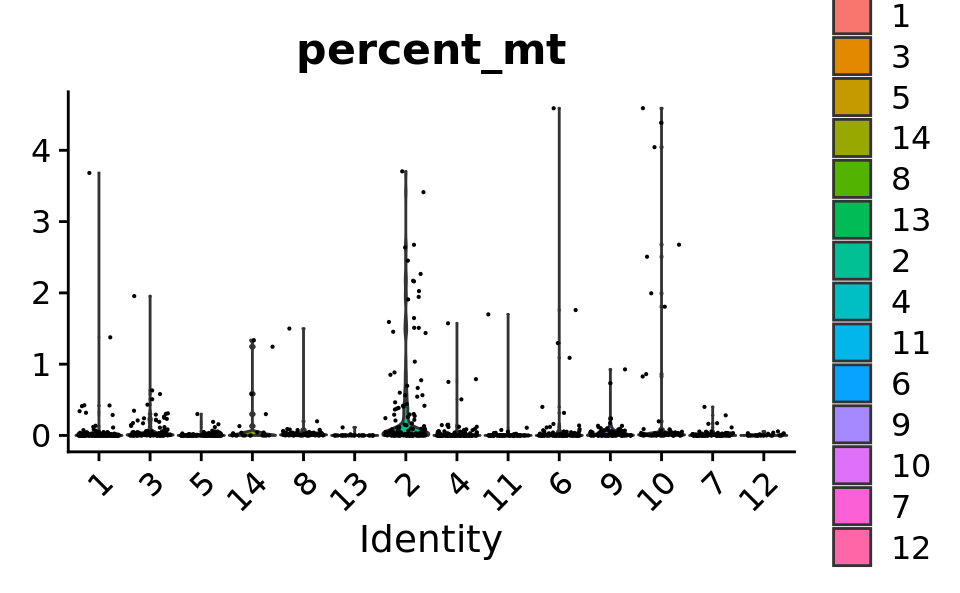
FeaturePlot(combined.srt, features = "percent_rb",
label.size = 4,repel = T,pt.size = 3,label = T) &
theme(plot.title = element_text(size = 16)) &
scale_colour_gradientn(colours = rev(brewer.pal(n = 11, name = "RdYlGn")))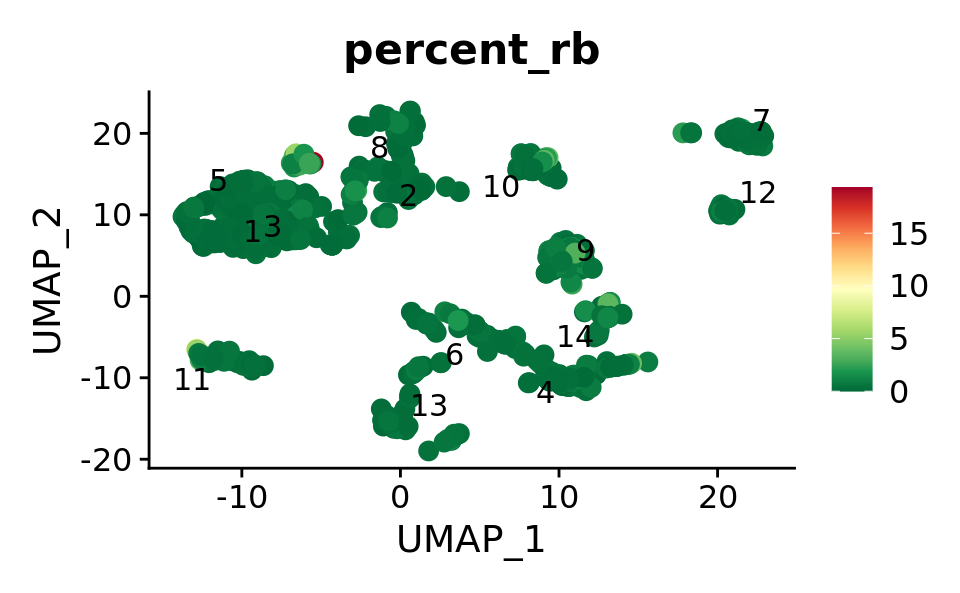
VlnPlot(combined.srt, features = "percent_rb") & theme(plot.title = element_text(size=16))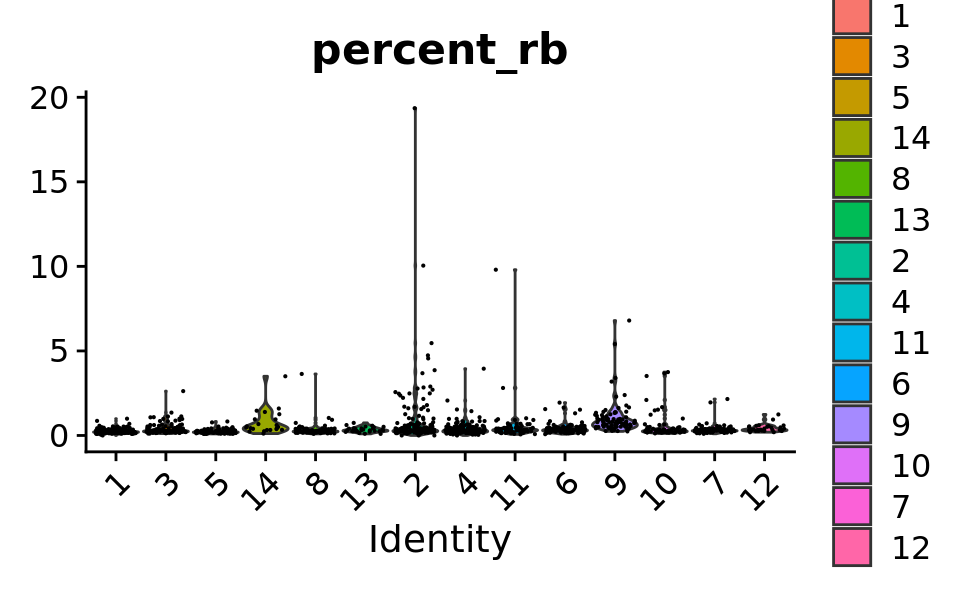
VlnPlot(combined.srt, features = c("nCount_RNA", "nFeature_RNA")) & theme(plot.title = element_text(size = 16))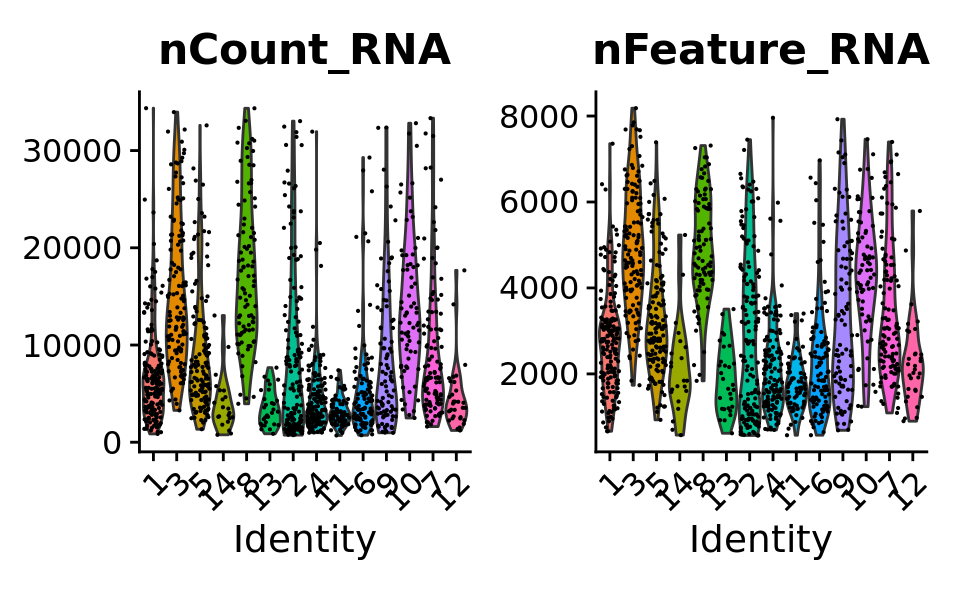
FeaturePlot(combined.srt,
features = c("S.Score", "G2M.Score"),
label.size = 4,
repel = T,
pt.size = 3,
label = T) &
theme(plot.title = element_text(size = 16))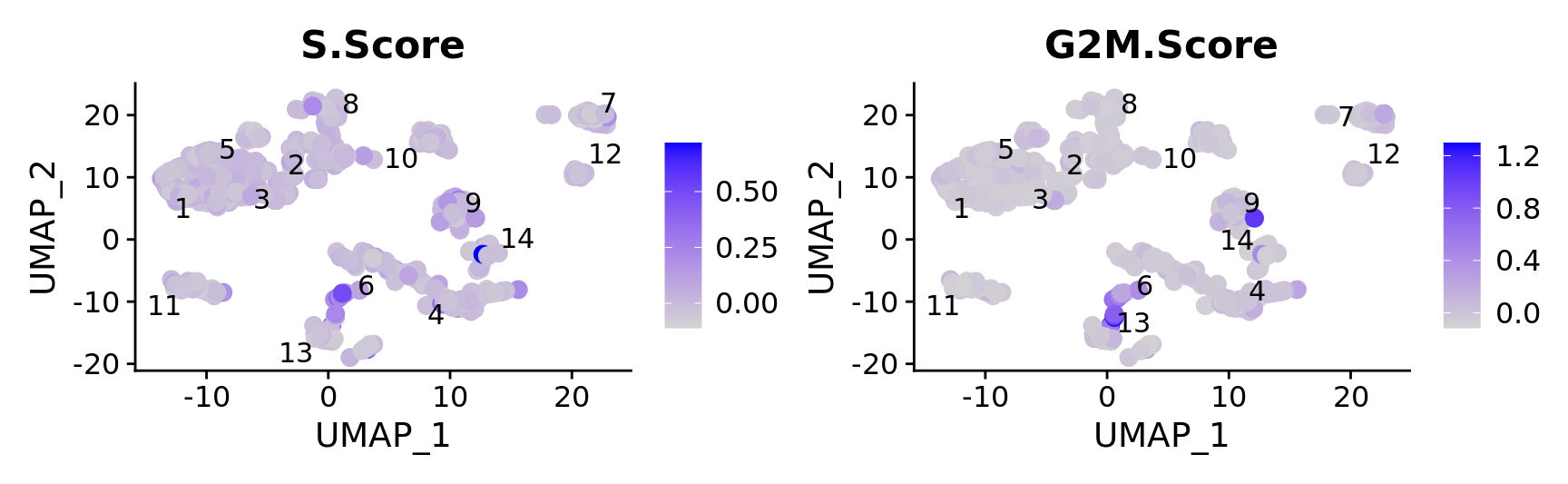
VlnPlot(combined.srt,
features = c("S.Score", "G2M.Score")) &
theme(plot.title = element_text(size=16))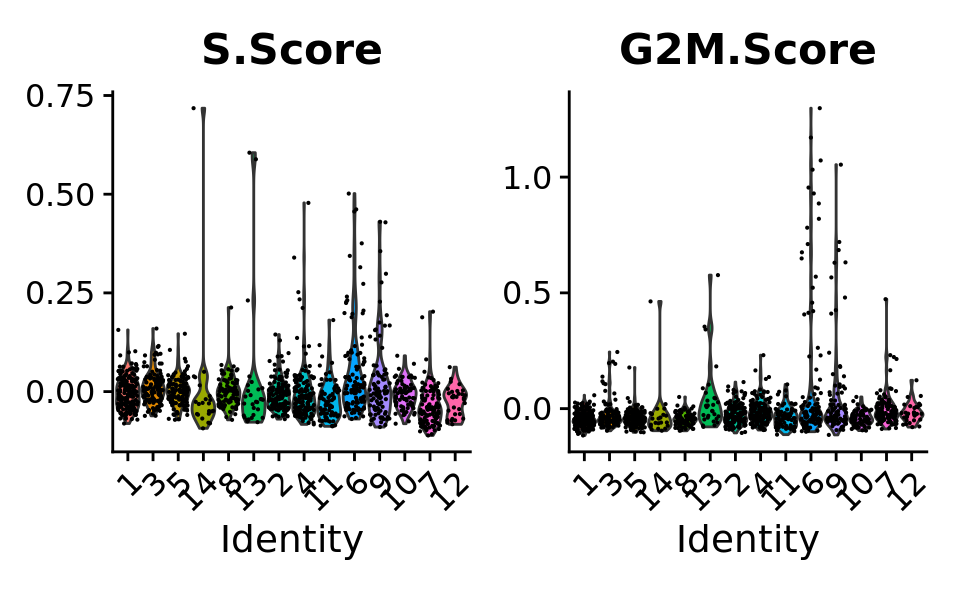
Apply SCTransform pipeline
# normalize and run dimensionality reduction on control dataset
npcs <- 30
metadata = combined.srt@meta.data
rownames(metadata) = colnames(combined.srt)
combined.srt <-
SCTransform(
combined.srt,
vst.flavor = "v2",
ncells = ncol(combined.srt),
variable.features.n = 3500,
vars.to.regress = c("var_regex", "log10GenesPerUMI",
"S.Score", "G2M.Score"),
return.only.var.genes = FALSE,
seed.use = reseed,
verbose = FALSE
)
hvg <- VariableFeatures(combined.srt)
var_regex <- '^Hla-|^Ig[hjkl]|^Rna|^mt-|^Rp[sl]|^Hb[^(p)]|^Gm'
hvg <- hvg[str_detect(pattern = var_regex, string = hvg, negate = T)]
combined.srt <- combined.srt %>%
RunPCA(features = hvg, npcs = npcs, seed.use = reseed, verbose = FALSE)print(combined.srt[["pca"]], dims = 1:5, nfeatures = 5)PC_ 1
Positive: Ntng1, Tenm2, Cntnap2, Meg3, Cntn5
Negative: Dnah6, Dnah12, Cfap299, Cfap54, Ak9
PC_ 2
Positive: Flt1, Lama4, Ets1, Dlc1, Apcdd1
Negative: Cntnap2, Meg3, Tenm2, Ntng1, Snhg11
PC_ 3
Positive: Ebf1, Ntng1, Mecom, Flt1, Hmcn1
Negative: Slc4a4, Npas3, Ptprz1, Luzp2, Wdr17
PC_ 4
Positive: Trpm3, Adam12, Ranbp3l, Slc6a13, Cped1
Negative: Npas3, Ptprz1, Qk, Luzp2, Myo10
PC_ 5
Positive: Lrrc4c, Dpp10, Erbb4, Galntl6, Galnt13
Negative: Ntng1, 6330411D24Rik, Arpp21, Rorb, Lef1 VizDimLoadings(combined.srt, dims = 1:4, reduction = "pca")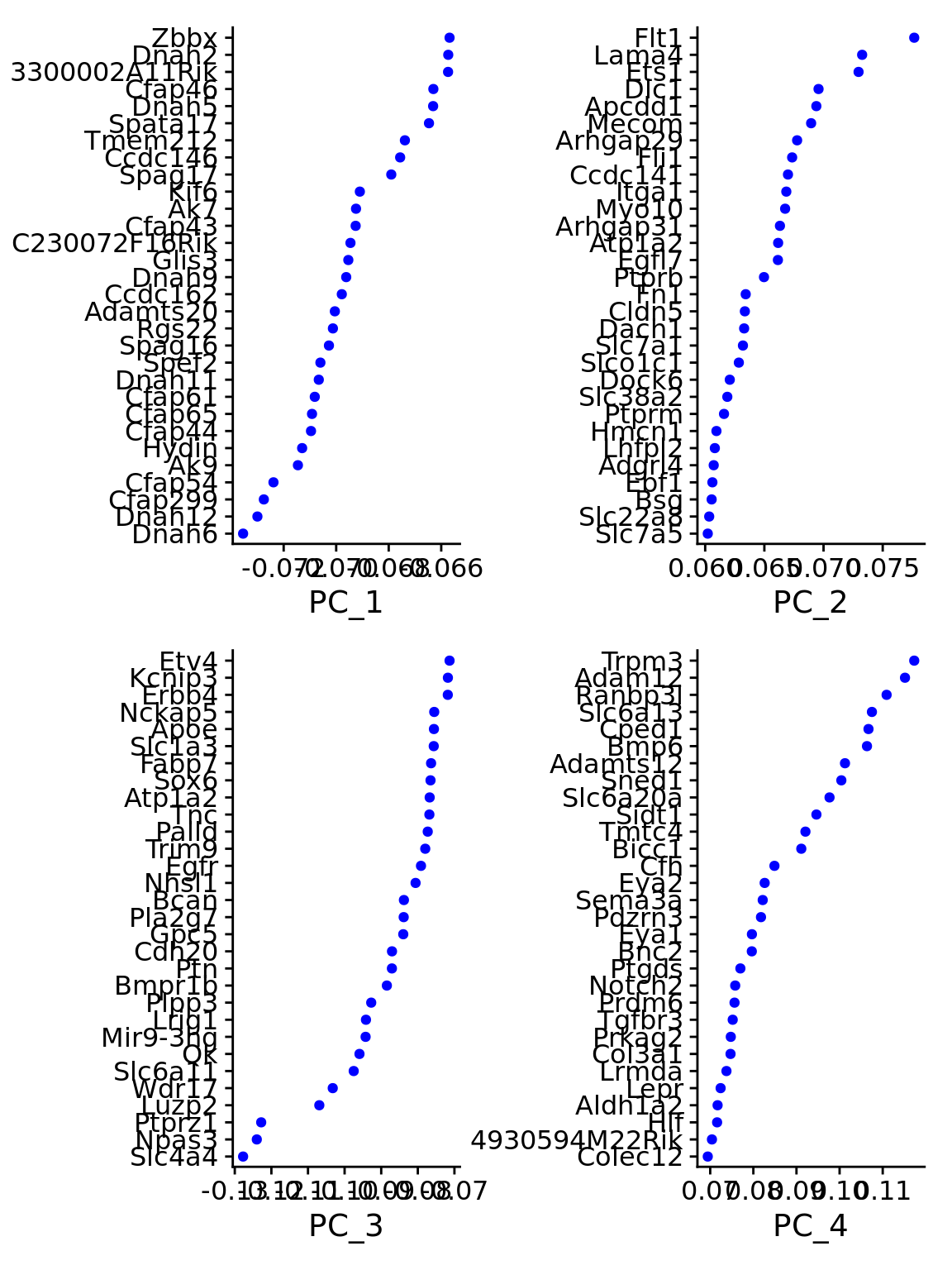
DimHeatmap(combined.srt, dims = 1:15, cells = 500, balanced = TRUE)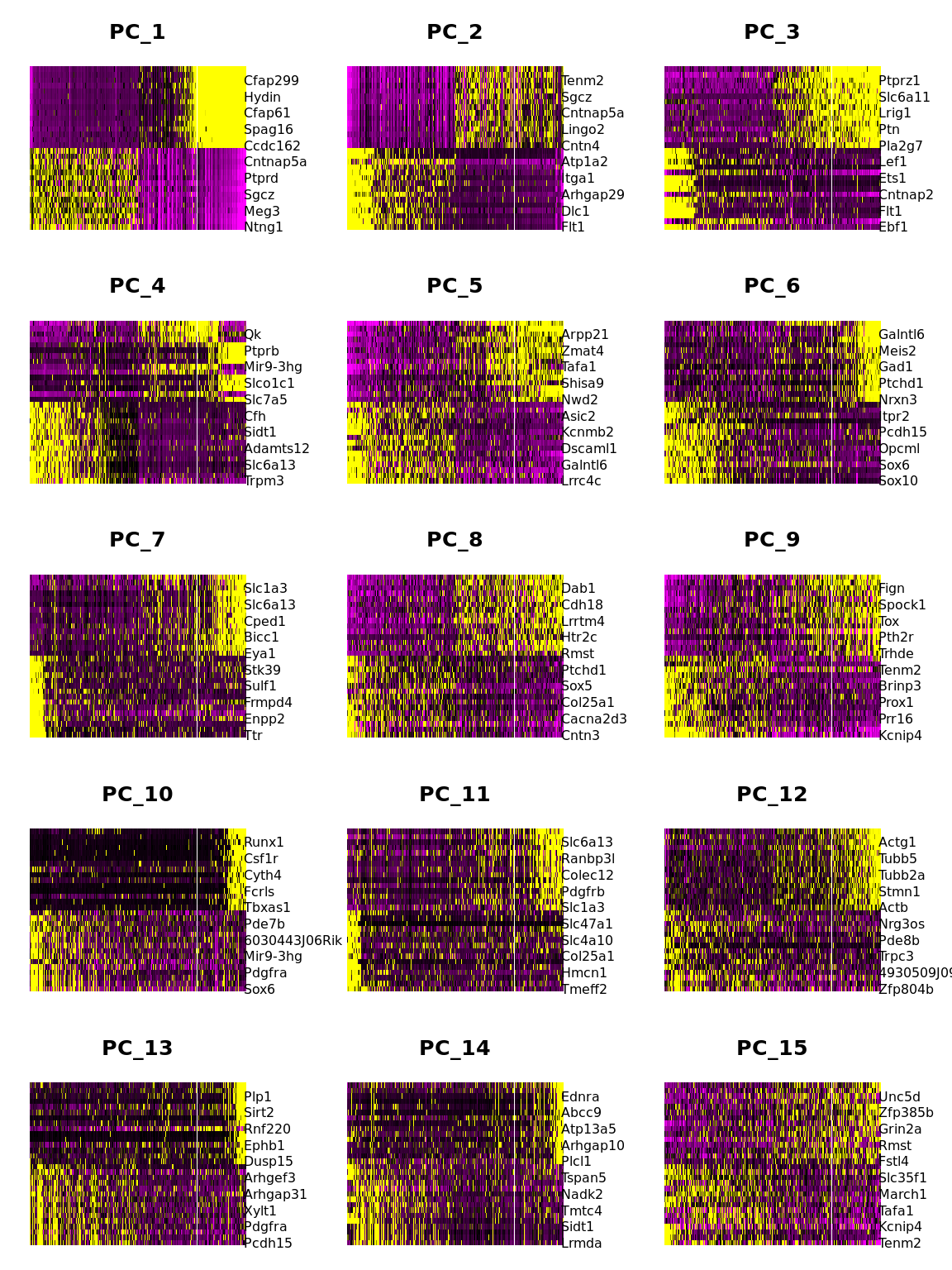
ElbowPlot(combined.srt, ndims = npcs)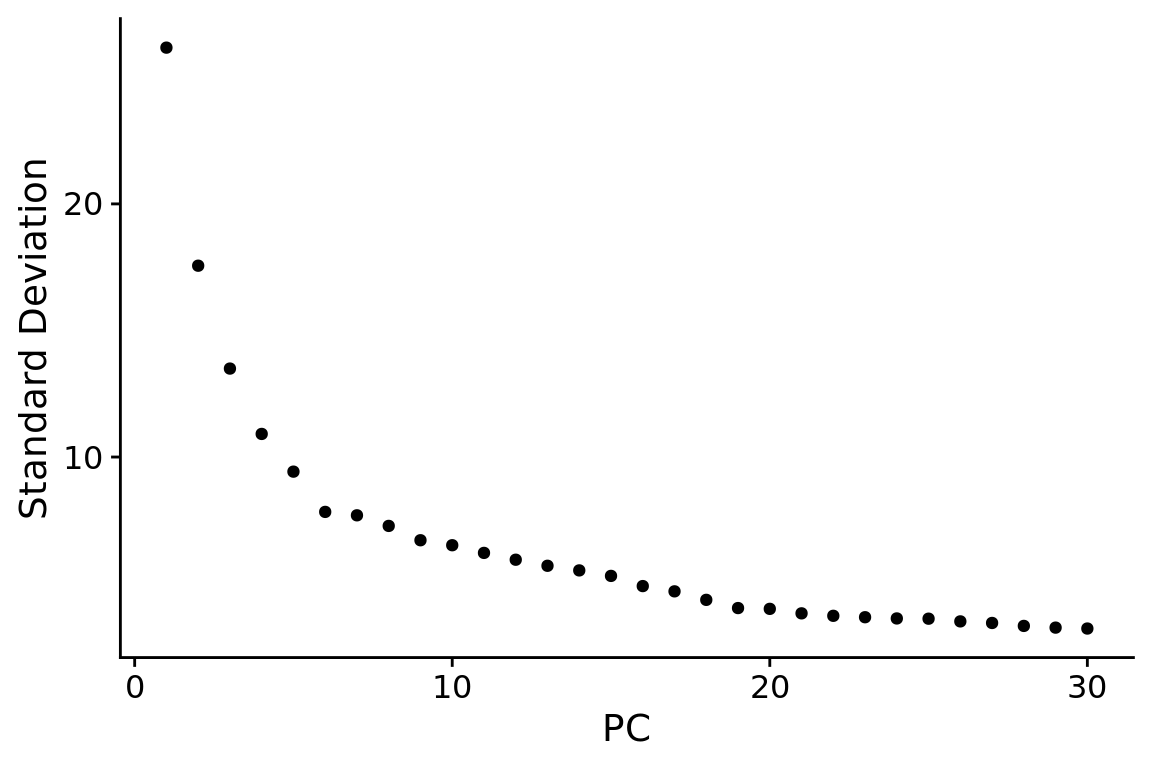
combined.srt <-
combined.srt |>
FindNeighbors(
dims = seq_along(combined.srt[["pca"]]@stdev),
k.param = 20,
annoy.metric = "euclidean",
n.trees = 100,
verbose = FALSE) |>
RunUMAP(
dims = seq_along(combined.srt[["pca"]]@stdev),
reduction.name = "umap",
reduction.key = "UMAP_",
return.model = TRUE,
umap.method = "umap-learn",
densmap = TRUE,
dens.lambda = 1L,
dens.frac = 0.1,
n.epochs = 1000L,
n.neighbors = 20L,
min.dist = 0.01,
spread = 4L,
metric = "correlation",
init = "pca",
seed.use = reseed,
verbose = FALSE) |>
FindNeighbors(
reduction = "umap",
dims = 1:2,
force.recalc = TRUE,
k.param = 20,
annoy.metric = "euclidean",
n.trees = 100,
verbose = FALSE)Plot by source after clean up
plEmbCombBatch <- DimPlot(combined.srt, reduction = "umap",
group.by = "orig.ident", pt.size = 3,
label = TRUE, repel = TRUE) + NoLegend()
plEmbCombBatch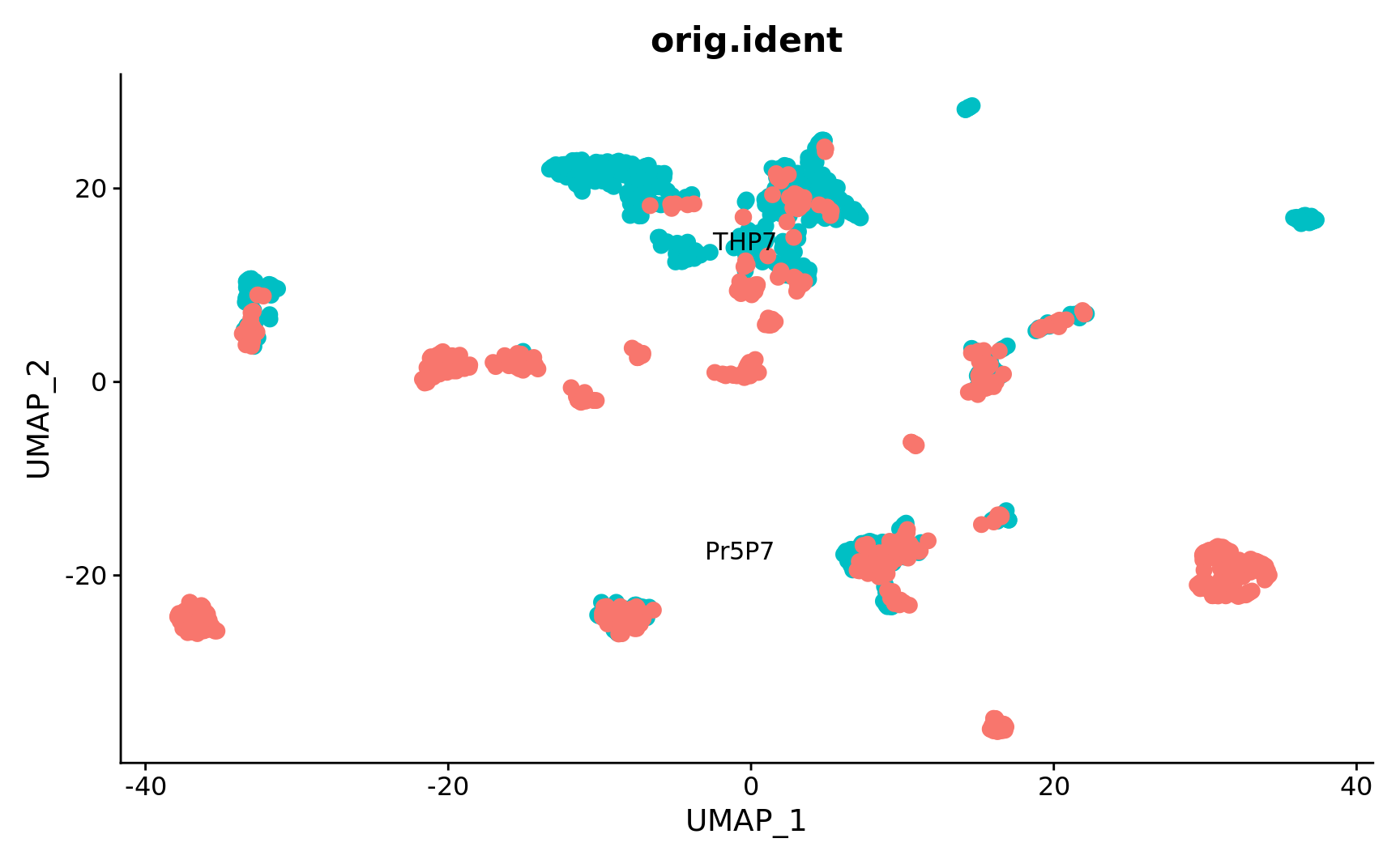
metadata <- combined.srt@meta.data
rownames(metadata) <- colnames(combined.srt)
ref.labels <- metadata$k_tree
resolutions <-
modularity_event_sampling(
A = combined.srt@graphs$SCT_snn,
n.res = 70,
gamma.min = 0.05,
gamma.max = 4.000001
) # sample based on the similarity matrix
# clustering using Suerat
combined.srt <- combined.srt |>
FindClusters(algorithm = 4, method = "igraph",
resolution = resolutions, random.seed = reseed,
verbose = FALSE)
# initial cluster tree from Seurat flat clustering
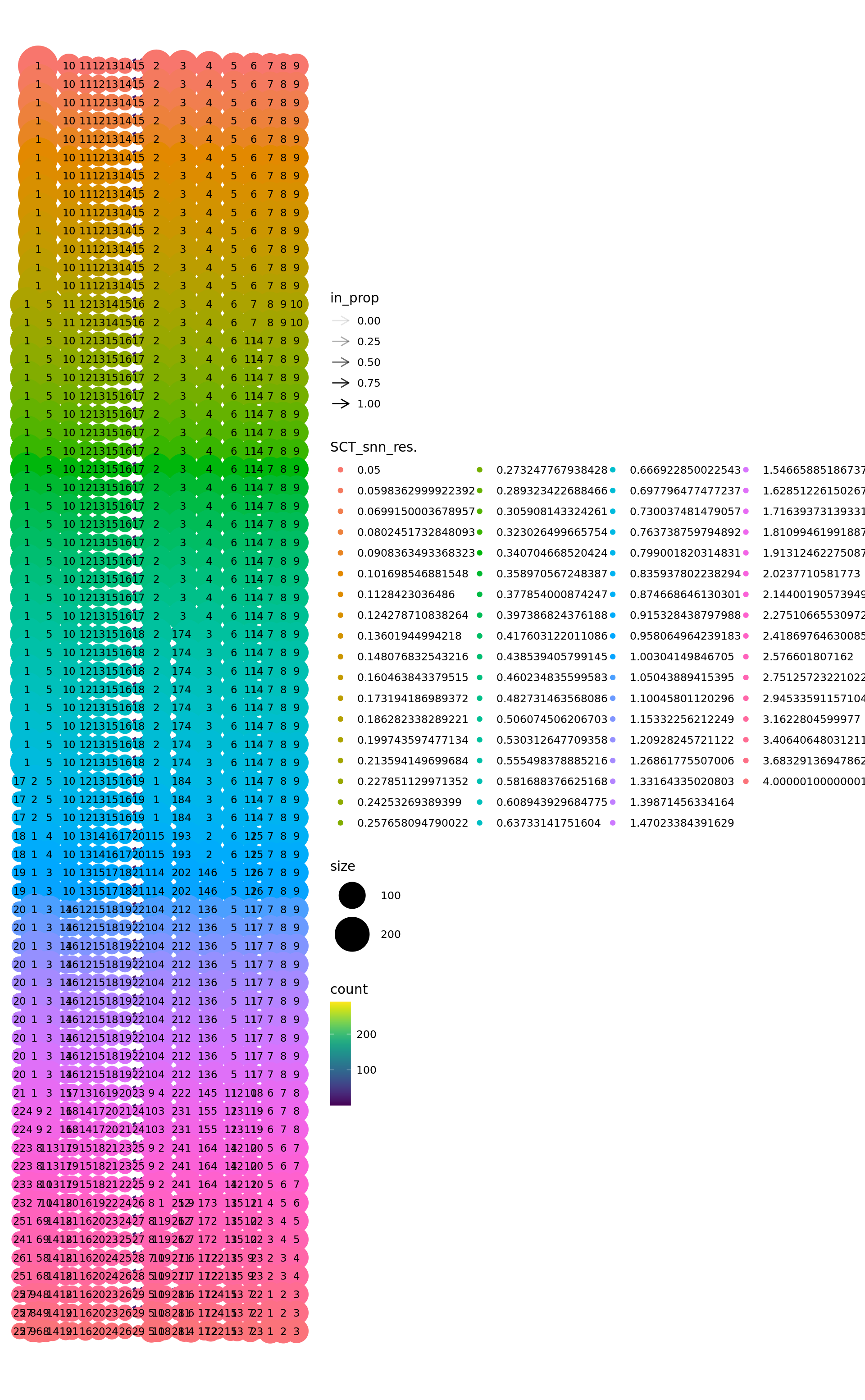
plot_clustree(
labelmat = combined.srt@meta.data,
prefix = 'SCT_snn_res.',
ref.labels = ref.labels,
plot.ref = FALSE
)
# Adjusted Multiresolution Rand Index (AMRI)
ks.flat <- apply(
out$labelmat.flat,
2,
FUN = function(x)
length(unique(x))
)
ks.mrtree <- apply(
out$labelmat.mrtree,
2,
FUN = function(x)
length(unique(x))
)
amri.flat <- sapply(1:ncol(out$labelmat.flat), function(i)
AMRI(out$labelmat.flat[, i], ref.labels)$amri)
amri.flat <- aggregate(amri.flat, by = list(k = ks.flat), FUN = mean)
amri.recon <- sapply(1:ncol(out$labelmat.mrtree), function(i)
AMRI(out$labelmat.mrtree[, i], ref.labels)$amri)
df <- rbind(
data.frame(
k = amri.flat$k,
amri = amri.flat$x,
method = 'Seurat flat'
),
data.frame(k = ks.mrtree, amri = amri.recon, method = 'MRtree')
)
ggplot2::ggplot(data = df, aes(x = k, y = amri, color = method)) + geom_line() + theme_bw()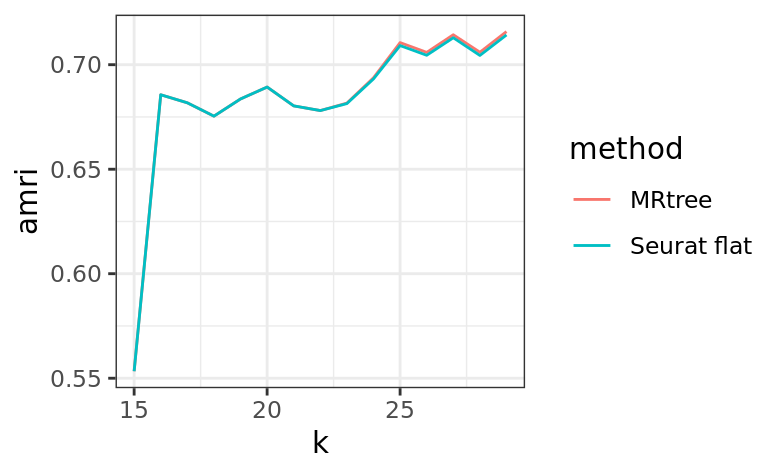
stab.out <- stability_plot(out)
stab.out$plot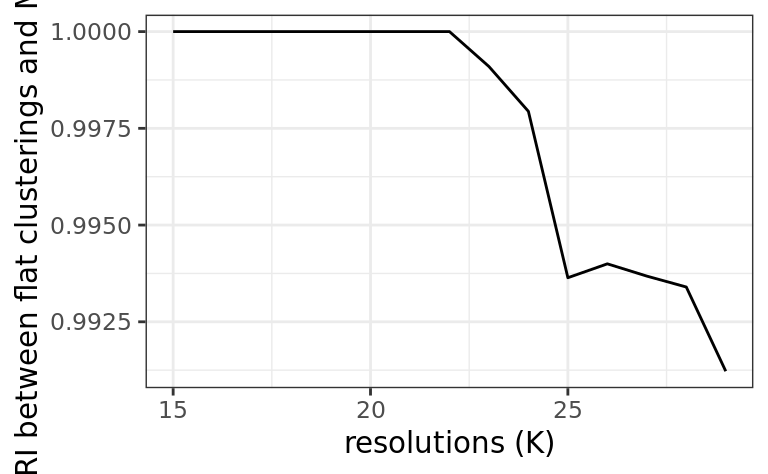
kable_material(
kable(
stab.out$df,
"html"),
bootstrap_options = c("bordered",
"condensed",
"responsive",
"striped"),
position = "left",
font_size = 14
)| resolution | ari |
|---|---|
| 15 | 1.0000000 |
| 16 | 1.0000000 |
| 17 | 1.0000000 |
| 18 | 1.0000000 |
| 19 | 1.0000000 |
| 20 | 1.0000000 |
| 21 | 1.0000000 |
| 22 | 1.0000000 |
| 23 | 0.9990972 |
| 24 | 0.9979382 |
| 25 | 0.9936390 |
| 26 | 0.9939979 |
| 27 | 0.9936809 |
| 28 | 0.9933988 |
| 29 | 0.9912227 |
resK <- SelectResolution(stab.out$df)
resK[1] 22 21 20 19 18 17 16 15kable_material(
kable(
table(
out$labelmat.mrtree[, which.min(
abs(as.integer(
str_remove(dimnames(
out$labelmat.mrtree)[[2]], "K"
)
) - resK)
)]
),
"html"),
bootstrap_options = c("bordered",
"condensed",
"responsive",
"striped"),
position = "left",
font_size = 14
)| Var1 | Freq |
|---|---|
| 1 | 185 |
| 2 | 162 |
| 3 | 124 |
| 4 | 121 |
| 5 | 105 |
| 6 | 93 |
| 7 | 81 |
| 8 | 79 |
| 9 | 74 |
| 10 | 73 |
| 11 | 60 |
| 12 | 42 |
| 13 | 37 |
| 14 | 32 |
| 15 | 32 |
| 16 | 30 |
| 17 | 28 |
| 18 | 24 |
combined.srt$k_tree <- out$labelmat.mrtree[, which.min(
abs(as.integer(
str_remove(dimnames(
out$labelmat.mrtree)[[2]], "K"
)
) - resK)
)]
p1 <- DimPlot(combined.srt, label = T, repel = T, pt.size = 2) + ggtitle("Unsupervised clustering") + NoLegend()
p2 <- DimPlot(combined.srt, label = T, repel = T, group.by = "k_tree", pt.size = 2) + ggtitle("MRTree") + NoLegend()
p1 | p2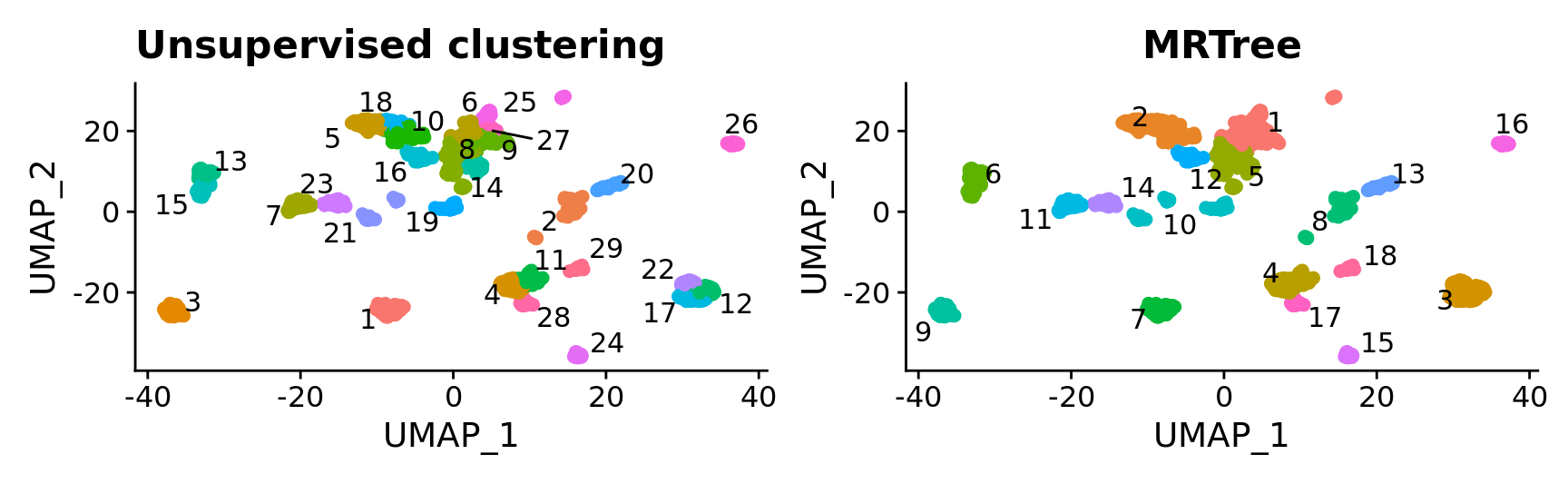
FeaturePlot(combined.srt, "Galr1", pt.size = 2, order = T) +
scale_colour_gradientn(colours = rev(brewer.pal(n = 11, name = "Spectral"))) + ggtitle("Galr1: ") + theme(plot.title = element_text(size = 24))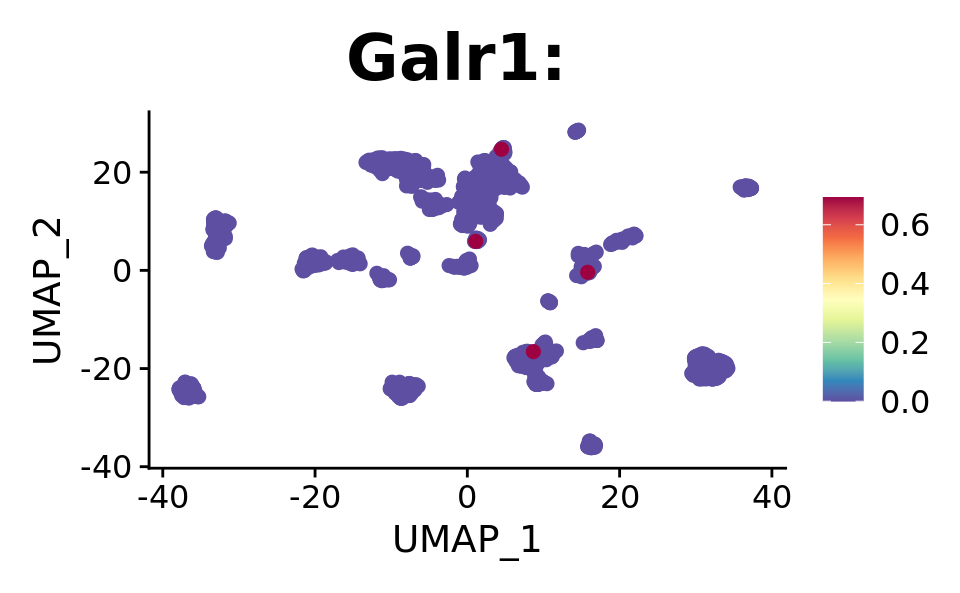
FeaturePlot(combined.srt, "Gal", pt.size = 2, order = T) +
scale_colour_gradientn(colours = rev(brewer.pal(n = 11, name = "Spectral"))) + ggtitle("Gal: ") + theme(plot.title = element_text(size = 24))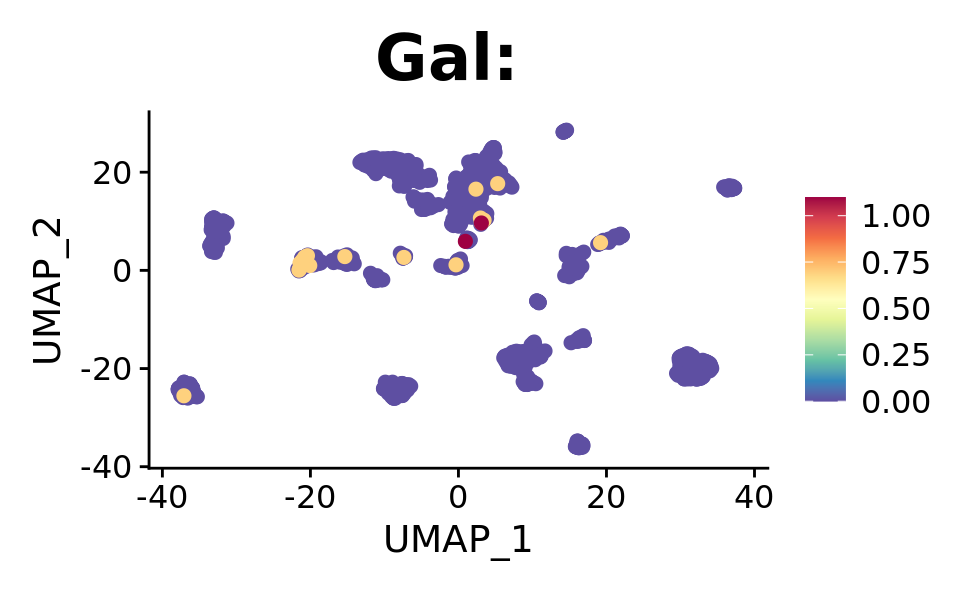
DotPlot(combined.srt, assay = "RNA", features = c("Galr1", "Gal"), split.by = "orig.ident")
We see the spread of our targets across derived clusters, which isn’t optimal. Lets see if we will see some significant hits with proper statistical testing.
Idents(combined.srt) <- "k_tree"
combined.srt <-
PrepSCTFindMarkers(combined.srt, assay = "SCT")
markers.logreg <-
FindAllMarkers(
combined.srt,
assay = "SCT",
verbose = FALSE,
random.seed = reseed,
only.pos = TRUE,
min.pct = 0.2,
base = 10,
logfc.threshold = 0.2,
densify = TRUE,
test.use = "LR")
write_csv(markers.logreg,
here(tables_dir,
'hevesi2023-all-mrk_logreg-sct_combined.csv'))
markers.logreg %>%
group_by(cluster) %>%
slice_max(n = 20, order_by = avg_log10FC) %>%
kable("html") %>%
kable_material(
bootstrap_options = c("bordered",
"condensed",
"responsive",
"striped"),
position = "left",
font_size = 14
)| p_val | avg_log10FC | pct.1 | pct.2 | p_val_adj | cluster | gene |
|---|---|---|---|---|---|---|
| 0.0000000 | 0.5461860 | 0.789 | 0.255 | 0.0000000 | 1 | Gm15398 |
| 0.0000000 | 0.5379832 | 0.849 | 0.472 | 0.0000000 | 1 | Cdh18 |
| 0.0000000 | 0.5250328 | 0.789 | 0.309 | 0.0000000 | 1 | Unc5d |
| 0.0000000 | 0.4923348 | 0.984 | 0.708 | 0.0000000 | 1 | Lrrtm4 |
| 0.0000000 | 0.4297117 | 0.838 | 0.306 | 0.0000000 | 1 | Tafa2 |
| 0.0000000 | 0.4267031 | 1.000 | 0.741 | 0.0000000 | 1 | Dlg2 |
| 0.0000000 | 0.4199060 | 0.800 | 0.337 | 0.0000000 | 1 | Gm26871 |
| 0.0000000 | 0.4185715 | 0.962 | 0.769 | 0.0000000 | 1 | Ralyl |
| 0.0000000 | 0.4176032 | 0.795 | 0.211 | 0.0000000 | 1 | Dlgap2 |
| 0.0000000 | 0.4117576 | 0.892 | 0.655 | 0.0000000 | 1 | Galntl6 |
| 0.0000000 | 0.4024267 | 0.730 | 0.231 | 0.0000000 | 1 | 4930555F03Rik |
| 0.0000000 | 0.4002103 | 0.805 | 0.398 | 0.0000000 | 1 | Grm8 |
| 0.0000000 | 0.3966198 | 0.946 | 0.758 | 0.0000000 | 1 | Dab1 |
| 0.0000000 | 0.3965816 | 0.978 | 0.744 | 0.0000000 | 1 | Nrg1 |
| 0.0000000 | 0.3867786 | 0.827 | 0.452 | 0.0000000 | 1 | Gria1 |
| 0.0000000 | 0.3867692 | 0.789 | 0.282 | 0.0000000 | 1 | Grin2a |
| 0.0000000 | 0.3856435 | 0.519 | 0.385 | 0.0003061 | 1 | Adarb2 |
| 0.0000000 | 0.3846172 | 1.000 | 0.804 | 0.0000000 | 1 | Ahi1 |
| 0.0000000 | 0.3658777 | 0.611 | 0.199 | 0.0000000 | 1 | Gm45321 |
| 0.0000000 | 0.3657352 | 0.946 | 0.679 | 0.0000000 | 1 | Sgcz |
| 0.0000000 | 0.6659039 | 1.000 | 0.680 | 0.0000000 | 2 | Tenm1 |
| 0.0000000 | 0.6523820 | 0.969 | 0.556 | 0.0000000 | 2 | Tafa1 |
| 0.0000000 | 0.6200753 | 1.000 | 0.657 | 0.0000000 | 2 | Ntng1 |
| 0.0000000 | 0.6149381 | 1.000 | 0.678 | 0.0000000 | 2 | Rnf220 |
| 0.0000000 | 0.5863378 | 0.969 | 0.635 | 0.0000000 | 2 | Shisa9 |
| 0.0000000 | 0.5635203 | 0.901 | 0.393 | 0.0000000 | 2 | Thsd7b |
| 0.0000000 | 0.5548733 | 0.883 | 0.260 | 0.0000000 | 2 | Gm48749 |
| 0.0000000 | 0.5513788 | 0.975 | 0.465 | 0.0000000 | 2 | Samd5 |
| 0.0000000 | 0.5407102 | 0.988 | 0.580 | 0.0000000 | 2 | Cntnap5a |
| 0.0000000 | 0.5233261 | 0.864 | 0.251 | 0.0000000 | 2 | Gm32647 |
| 0.0000000 | 0.4882065 | 0.981 | 0.584 | 0.0000000 | 2 | Tox |
| 0.0000000 | 0.4625518 | 0.988 | 0.670 | 0.0000000 | 2 | Egfem1 |
| 0.0000000 | 0.4594009 | 0.827 | 0.259 | 0.0000000 | 2 | Trhde |
| 0.0000000 | 0.4589436 | 1.000 | 0.737 | 0.0000000 | 2 | Cntnap2 |
| 0.0000000 | 0.4544066 | 1.000 | 0.875 | 0.0000000 | 2 | Ptprd |
| 0.0000000 | 0.4516664 | 0.951 | 0.372 | 0.0000000 | 2 | Foxp2 |
| 0.0000000 | 0.4458370 | 1.000 | 0.758 | 0.0000000 | 2 | Grm7 |
| 0.0000000 | 0.4429434 | 1.000 | 0.634 | 0.0000000 | 2 | Arpp21 |
| 0.0000000 | 0.4278149 | 0.994 | 0.845 | 0.0000000 | 2 | Grik2 |
| 0.0000000 | 0.4269539 | 0.988 | 0.675 | 0.0000000 | 2 | Cntn4 |
| 0.0000000 | 0.7270600 | 0.747 | 0.153 | 0.0000000 | 8 | 4930588A03Rik |
| 0.0000000 | 0.7259301 | 0.823 | 0.436 | 0.0000000 | 8 | Pcdh15 |
| 0.0000000 | 0.7098871 | 0.772 | 0.114 | 0.0000000 | 8 | Gm38505 |
| 0.0000000 | 0.6501237 | 0.873 | 0.498 | 0.0000000 | 8 | 6030443J06Rik |
| 0.0000000 | 0.6287367 | 0.873 | 0.412 | 0.0000000 | 8 | Sox6 |
| 0.0000000 | 0.6071638 | 0.696 | 0.121 | 0.0000000 | 8 | Pdgfra |
| 0.0000000 | 0.5930285 | 0.823 | 0.514 | 0.0000000 | 8 | Sox2ot |
| 0.0000000 | 0.5898584 | 0.835 | 0.439 | 0.0000000 | 8 | Lhfpl3 |
| 0.0001452 | 0.5890911 | 0.430 | 0.382 | 1.0000000 | 8 | Dlc1 |
| 0.0000000 | 0.5524413 | 0.747 | 0.236 | 0.0000000 | 8 | Dscaml1 |
| 0.0000000 | 0.5146779 | 0.861 | 0.503 | 0.0000000 | 8 | Epn2 |
| 0.0000000 | 0.5044968 | 0.835 | 0.477 | 0.0000000 | 8 | Xylt1 |
| 0.0000000 | 0.4766786 | 0.684 | 0.075 | 0.0000000 | 8 | Sox10 |
| 0.0000000 | 0.4746780 | 0.772 | 0.299 | 0.0000000 | 8 | Mir9-3hg |
| 0.0000000 | 0.4493794 | 0.911 | 0.675 | 0.0000000 | 8 | Sgcd |
| 0.0000000 | 0.4420664 | 0.810 | 0.531 | 0.0000000 | 8 | Pcdh11x |
| 0.0000000 | 0.4347889 | 0.899 | 0.642 | 0.0000000 | 8 | Dcc |
| 0.0000000 | 0.4181992 | 0.696 | 0.226 | 0.0000000 | 8 | Megf11 |
| 0.0000000 | 0.4159086 | 0.861 | 0.523 | 0.0000000 | 8 | Nxph1 |
| 0.0000000 | 0.4044997 | 0.759 | 0.239 | 0.0000000 | 8 | Arhgap31 |
| 0.0000000 | 0.8626318 | 0.667 | 0.258 | 0.0000000 | 18 | Rbpj |
| 0.0000000 | 0.8141688 | 1.000 | 0.378 | 0.0000000 | 18 | Mef2c |
| 0.0000000 | 0.7664959 | 0.292 | 0.043 | 0.0000001 | 18 | F13a1 |
| 0.0000000 | 0.7558552 | 0.792 | 0.351 | 0.0000000 | 18 | Slc9a9 |
| 0.0000000 | 0.7488395 | 1.000 | 0.415 | 0.0000000 | 18 | Zeb2 |
| 0.0000000 | 0.7469816 | 0.500 | 0.175 | 0.0000201 | 18 | Dab2 |
| 0.0000000 | 0.7250744 | 0.458 | 0.015 | 0.0000000 | 18 | Mrc1 |
| 0.0000000 | 0.7167972 | 0.792 | 0.019 | 0.0000000 | 18 | Runx1 |
| 0.0000000 | 0.7165791 | 0.958 | 0.356 | 0.0000000 | 18 | Mbnl1 |
| 0.0000000 | 0.6897250 | 0.833 | 0.309 | 0.0000040 | 18 | Apoe |
| 0.0000000 | 0.6816252 | 0.958 | 0.152 | 0.0000000 | 18 | Dock2 |
| 0.0000000 | 0.6738016 | 0.958 | 0.046 | 0.0000000 | 18 | Inpp5d |
| 0.0000000 | 0.6581980 | 0.875 | 0.686 | 0.0000000 | 18 | Plxdc2 |
| 0.0000000 | 0.6533039 | 0.917 | 0.312 | 0.0000000 | 18 | Rreb1 |
| 0.0000000 | 0.6422074 | 0.917 | 0.099 | 0.0000000 | 18 | Fli1 |
| 0.0000000 | 0.6255837 | 0.708 | 0.024 | 0.0000000 | 18 | Apbb1ip |
| 0.0000000 | 0.6238467 | 0.875 | 0.039 | 0.0000000 | 18 | Fyb |
| 0.0000000 | 0.6165447 | 0.833 | 0.141 | 0.0000000 | 18 | Maf |
| 0.0000000 | 0.6133034 | 0.792 | 0.015 | 0.0000000 | 18 | Ly86 |
| 0.0000000 | 0.6102902 | 0.875 | 0.273 | 0.0000000 | 18 | Pip4k2a |
| 0.0000000 | 1.3713943 | 1.000 | 0.078 | 0.0000000 | 16 | Bnc2 |
| 0.0000000 | 0.9347756 | 1.000 | 0.452 | 0.0000000 | 16 | Fbxl7 |
| 0.0000000 | 0.9258599 | 0.867 | 0.052 | 0.0000000 | 16 | Adamtsl3 |
| 0.0000000 | 0.9149946 | 0.933 | 0.148 | 0.0000000 | 16 | Thsd4 |
| 0.0000000 | 0.9028647 | 1.000 | 0.700 | 0.0000000 | 16 | Foxp1 |
| 0.0000000 | 0.9013542 | 0.900 | 0.513 | 0.0000000 | 16 | Slc4a10 |
| 0.0000000 | 0.9007700 | 0.933 | 0.310 | 0.0000000 | 16 | Nr3c2 |
| 0.0000000 | 0.8648775 | 1.000 | 0.376 | 0.0000000 | 16 | Slc38a2 |
| 0.0000000 | 0.8521231 | 0.833 | 0.290 | 0.0000000 | 16 | Col25a1 |
| 0.0000000 | 0.8468100 | 0.933 | 0.061 | 0.0000000 | 16 | Trabd2b |
| 0.0000000 | 0.8042045 | 0.833 | 0.173 | 0.0000000 | 16 | Itgbl1 |
| 0.0000000 | 0.7824174 | 0.933 | 0.346 | 0.0000000 | 16 | Gulp1 |
| 0.0000000 | 0.7498879 | 0.967 | 0.424 | 0.0000000 | 16 | Tmtc1 |
| 0.0000000 | 0.7471613 | 0.933 | 0.111 | 0.0000000 | 16 | Eya2 |
| 0.0000000 | 0.7423676 | 1.000 | 0.613 | 0.0000000 | 16 | Tmeff2 |
| 0.0000000 | 0.7290641 | 0.867 | 0.250 | 0.0000000 | 16 | Sh3pxd2a |
| 0.0000000 | 0.7133837 | 0.800 | 0.091 | 0.0000000 | 16 | Dock5 |
| 0.0000000 | 0.6949514 | 0.967 | 0.314 | 0.0000000 | 16 | Nxn |
| 0.0000000 | 0.6757103 | 0.967 | 0.209 | 0.0000000 | 16 | Hmcn1 |
| 0.0000000 | 0.6698614 | 0.567 | 0.067 | 0.0000000 | 16 | Crispld1 |
| 0.0000000 | 0.5858833 | 0.543 | 0.135 | 0.0000000 | 5 | mt-Co3 |
| 0.0000000 | 0.5707633 | 0.857 | 0.498 | 0.0000000 | 5 | Tuba1a |
| 0.0000000 | 0.5623345 | 0.886 | 0.570 | 0.0000000 | 5 | Cmss1 |
| 0.0000000 | 0.5326416 | 0.505 | 0.099 | 0.0000000 | 5 | mt-Atp6 |
| 0.0000000 | 0.5034257 | 0.971 | 0.892 | 0.0000000 | 5 | Gm42418 |
| 0.0000000 | 0.4866638 | 0.562 | 0.213 | 0.0000000 | 5 | mt-Co1 |
| 0.0000000 | 0.4762082 | 0.505 | 0.121 | 0.0000000 | 5 | mt-Co2 |
| 0.0000000 | 0.3906273 | 0.657 | 0.307 | 0.0000000 | 5 | Tubb5 |
| 0.0000000 | 0.3896130 | 0.610 | 0.325 | 0.0000000 | 5 | Tubb2a |
| 0.0000000 | 0.3794212 | 0.400 | 0.077 | 0.0000000 | 5 | mt-Cytb |
| 0.0000000 | 0.3791696 | 0.857 | 0.549 | 0.0000000 | 5 | Calm1 |
| 0.0000000 | 0.3721275 | 0.600 | 0.303 | 0.0000000 | 5 | Actg1 |
| 0.0000000 | 0.3716602 | 0.629 | 0.204 | 0.0000000 | 5 | Atp6v0c |
| 0.0000000 | 0.3669284 | 0.790 | 0.557 | 0.0000000 | 5 | Cdk8 |
| 0.0000000 | 0.3621710 | 0.886 | 0.641 | 0.0000000 | 5 | Basp1 |
| 0.0000000 | 0.3596345 | 0.371 | 0.077 | 0.0000000 | 5 | mt-Nd4 |
| 0.0000000 | 0.3590900 | 0.438 | 0.070 | 0.0000000 | 5 | mt-Nd1 |
| 0.0000000 | 0.3583674 | 0.438 | 0.084 | 0.0000000 | 5 | mt-Nd2 |
| 0.0000000 | 0.3576392 | 0.552 | 0.208 | 0.0000000 | 5 | Tmsb4x |
| 0.0000000 | 0.3566092 | 0.867 | 0.511 | 0.0000000 | 5 | Gap43 |
| 0.0000000 | 0.6689559 | 1.000 | 0.822 | 0.0000000 | 12 | Gm26917 |
| 0.0000000 | 0.5710436 | 0.952 | 0.463 | 0.0000000 | 12 | Rbfox3 |
| 0.0000000 | 0.4941579 | 0.976 | 0.852 | 0.0000000 | 12 | Camta1 |
| 0.0000000 | 0.4925956 | 0.690 | 0.437 | 0.0000032 | 12 | Cdh4 |
| 0.0000000 | 0.4665196 | 0.929 | 0.541 | 0.0000000 | 12 | Gpi1 |
| 0.0000000 | 0.4356404 | 0.238 | 0.043 | 0.0002533 | 12 | Ebf3 |
| 0.0000000 | 0.4338614 | 0.857 | 0.419 | 0.0000000 | 12 | Pacsin1 |
| 0.0000000 | 0.4276850 | 0.929 | 0.388 | 0.0000000 | 12 | Camk2b |
| 0.0000000 | 0.4197312 | 0.786 | 0.338 | 0.0000000 | 12 | Gse1 |
| 0.0000000 | 0.4127750 | 0.690 | 0.276 | 0.0000000 | 12 | Dlgap2 |
| 0.0000000 | 0.4107957 | 0.857 | 0.486 | 0.0000000 | 12 | Kif5a |
| 0.0000000 | 0.4102879 | 0.881 | 0.547 | 0.0000000 | 12 | Ksr2 |
| 0.0000000 | 0.4058210 | 0.762 | 0.485 | 0.0000000 | 12 | Xkr6 |
| 0.0000000 | 0.4048061 | 0.738 | 0.322 | 0.0000000 | 12 | Ank1 |
| 0.0000000 | 0.3964670 | 0.786 | 0.419 | 0.0000000 | 12 | Chd5 |
| 0.0000000 | 0.3959419 | 0.952 | 0.466 | 0.0000000 | 12 | Shank1 |
| 0.0000000 | 0.3942990 | 0.690 | 0.457 | 0.0000000 | 12 | Cux2 |
| 0.0000000 | 0.3934189 | 0.786 | 0.522 | 0.0000000 | 12 | Atp2b2 |
| 0.0000000 | 0.3919585 | 0.833 | 0.418 | 0.0000000 | 12 | Slc4a3 |
| 0.0000000 | 0.3910830 | 0.667 | 0.370 | 0.0000063 | 12 | Kif26b |
| 0.0000000 | 0.9391914 | 0.992 | 0.304 | 0.0000000 | 4 | Gm3764 |
| 0.0000000 | 0.8585705 | 1.000 | 0.609 | 0.0000000 | 4 | Ptprz1 |
| 0.0000000 | 0.8547763 | 1.000 | 0.415 | 0.0000000 | 4 | Slc4a4 |
| 0.0000000 | 0.7667826 | 1.000 | 0.712 | 0.0000000 | 4 | Npas3 |
| 0.0000000 | 0.7346128 | 0.934 | 0.642 | 0.0000000 | 4 | Luzp2 |
| 0.0000000 | 0.7284913 | 0.992 | 0.373 | 0.0000000 | 4 | Gm48747 |
| 0.0000000 | 0.6781296 | 0.868 | 0.171 | 0.0000000 | 4 | Slc6a11 |
| 0.0000000 | 0.6575202 | 0.959 | 0.611 | 0.0000000 | 4 | Gpc5 |
| 0.0000000 | 0.6438764 | 0.983 | 0.514 | 0.0000000 | 4 | Trim9 |
| 0.0000000 | 0.6425576 | 0.926 | 0.364 | 0.0000000 | 4 | Nhsl1 |
| 0.0000000 | 0.6229149 | 0.967 | 0.480 | 0.0000000 | 4 | Wdr17 |
| 0.0000000 | 0.6162126 | 0.992 | 0.623 | 0.0000000 | 4 | Grm3 |
| 0.0000000 | 0.5919610 | 0.917 | 0.186 | 0.0000000 | 4 | Lrig1 |
| 0.0000000 | 0.5791123 | 0.959 | 0.466 | 0.0000000 | 4 | Slc1a2 |
| 0.0000000 | 0.5751882 | 1.000 | 0.733 | 0.0000000 | 4 | Qk |
| 0.0000000 | 0.5560484 | 0.901 | 0.258 | 0.0000000 | 4 | Plpp3 |
| 0.0000000 | 0.5550735 | 1.000 | 0.580 | 0.0000000 | 4 | Ptn |
| 0.0000000 | 0.5515565 | 0.860 | 0.163 | 0.0000000 | 4 | Bmpr1b |
| 0.0000000 | 0.5466761 | 0.934 | 0.268 | 0.0000000 | 4 | Mir9-3hg |
| 0.0000000 | 0.5424140 | 0.777 | 0.141 | 0.0000000 | 4 | Pla2g7 |
| 0.0000000 | 1.3003979 | 0.645 | 0.041 | 0.0000000 | 6 | Ptgds |
| 0.0000000 | 1.0801231 | 0.892 | 0.047 | 0.0000000 | 6 | Ranbp3l |
| 0.0000000 | 1.0305402 | 0.935 | 0.860 | 0.0000000 | 6 | Trpm3 |
| 0.0000000 | 0.9166646 | 0.914 | 0.209 | 0.0000000 | 6 | Adam12 |
| 0.0000000 | 0.8684877 | 0.849 | 0.058 | 0.0000000 | 6 | Slc6a20a |
| 0.0000000 | 0.8575784 | 0.828 | 0.104 | 0.0000000 | 6 | Adamts12 |
| 0.0000000 | 0.8390771 | 0.860 | 0.147 | 0.0000000 | 6 | Sidt1 |
| 0.0000000 | 0.7826376 | 0.753 | 0.133 | 0.0000000 | 6 | Bmp6 |
| 0.0000000 | 0.7707784 | 0.935 | 0.318 | 0.0000000 | 6 | Atp1a2 |
| 0.0000000 | 0.7576284 | 0.882 | 0.016 | 0.0000000 | 6 | Slc6a13 |
| 0.0000000 | 0.7518776 | 0.903 | 0.274 | 0.0000000 | 6 | Bicc1 |
| 0.0000000 | 0.7271961 | 0.742 | 0.178 | 0.0000000 | 6 | Slc7a11 |
| 0.0000000 | 0.6886652 | 0.839 | 0.689 | 0.0000000 | 6 | Nnat |
| 0.0000000 | 0.6813297 | 0.828 | 0.099 | 0.0000000 | 6 | Cped1 |
| 0.0000000 | 0.6799306 | 0.763 | 0.140 | 0.0000000 | 6 | Lrmda |
| 0.0000000 | 0.6614480 | 0.828 | 0.257 | 0.0000000 | 6 | Sned1 |
| 0.0000000 | 0.6610377 | 0.849 | 0.147 | 0.0000000 | 6 | Tmtc4 |
| 0.0000000 | 0.6212833 | 0.871 | 0.220 | 0.0000000 | 6 | Pdzrn3 |
| 0.0000000 | 0.6208102 | 0.903 | 0.115 | 0.0000000 | 6 | Arhgap29 |
| 0.0000000 | 0.5748568 | 0.796 | 0.141 | 0.0000000 | 6 | Colec12 |
| 0.0000000 | 1.3273780 | 0.973 | 0.168 | 0.0000000 | 13 | Mbp |
| 0.0000000 | 1.1083179 | 0.946 | 0.083 | 0.0000000 | 13 | Plp1 |
| 0.0000000 | 1.0948631 | 0.865 | 0.138 | 0.0000000 | 13 | 9630013A20Rik |
| 0.0000000 | 1.0547744 | 0.865 | 0.041 | 0.0000000 | 13 | St18 |
| 0.0000000 | 0.9724562 | 0.892 | 0.131 | 0.0000000 | 13 | Bcas1 |
| 0.0000000 | 0.9608507 | 0.865 | 0.419 | 0.0000000 | 13 | Tnr |
| 0.0000000 | 0.8971909 | 0.919 | 0.592 | 0.0000000 | 13 | Nfasc |
| 0.0000000 | 0.8740391 | 0.892 | 0.200 | 0.0000000 | 13 | Tns3 |
| 0.0000000 | 0.8032452 | 0.541 | 0.117 | 0.0000000 | 13 | Cnksr3 |
| 0.0000000 | 0.7645961 | 0.946 | 0.290 | 0.0000000 | 13 | Itpr2 |
| 0.0000000 | 0.7406776 | 0.784 | 0.045 | 0.0000000 | 13 | Bcas1os2 |
| 0.0000000 | 0.7232176 | 0.973 | 0.541 | 0.0000000 | 13 | 9530059O14Rik |
| 0.0000000 | 0.7120959 | 0.946 | 0.613 | 0.0000000 | 13 | Tmeff2 |
| 0.0000000 | 0.6873725 | 0.324 | 0.010 | 0.0000000 | 13 | Mobp |
| 0.0000000 | 0.6853668 | 0.703 | 0.372 | 0.0000000 | 13 | Pik3r3 |
| 0.0000000 | 0.6850883 | 0.973 | 0.725 | 0.0000000 | 13 | Fyn |
| 0.0000000 | 0.6749825 | 0.946 | 0.266 | 0.0000000 | 13 | Sirt2 |
| 0.0000000 | 0.6671552 | 0.595 | 0.014 | 0.0000000 | 13 | Mag |
| 0.0000000 | 0.6400321 | 1.000 | 0.797 | 0.0000000 | 13 | Opcml |
| 0.0000000 | 0.6309468 | 0.811 | 0.024 | 0.0000000 | 13 | Enpp6 |
| 0.0000000 | 1.3145263 | 0.975 | 0.055 | 0.0000000 | 7 | Flt1 |
| 0.0000000 | 1.0884493 | 0.963 | 0.101 | 0.0000000 | 7 | Mecom |
| 0.0000000 | 0.9486995 | 0.926 | 0.145 | 0.0000000 | 7 | Slc7a1 |
| 0.0000000 | 0.9292332 | 0.926 | 0.203 | 0.0000000 | 7 | Dach1 |
| 0.0000000 | 0.8974278 | 0.901 | 0.115 | 0.0000000 | 7 | Slco1c1 |
| 0.0000000 | 0.8812812 | 0.914 | 0.020 | 0.0000000 | 7 | Egfl7 |
| 0.0000000 | 0.8764783 | 0.889 | 0.058 | 0.0000000 | 7 | Ets1 |
| 0.0000000 | 0.8709040 | 0.852 | 0.075 | 0.0000000 | 7 | Slc7a5 |
| 0.0000000 | 0.8412937 | 0.889 | 0.033 | 0.0000000 | 7 | Ptprb |
| 0.0000000 | 0.8406755 | 0.988 | 0.352 | 0.0000000 | 7 | Slc38a2 |
| 0.0000000 | 0.8162503 | 0.840 | 0.188 | 0.0000000 | 7 | Hmcn1 |
| 0.0000000 | 0.8072117 | 0.877 | 0.030 | 0.0000000 | 7 | Adgrl4 |
| 0.0000000 | 0.8001894 | 0.938 | 0.227 | 0.0000000 | 7 | Ccdc141 |
| 0.0000000 | 0.7961759 | 0.975 | 0.298 | 0.0000000 | 7 | Myo10 |
| 0.0000000 | 0.7894780 | 0.889 | 0.065 | 0.0000000 | 7 | Fli1 |
| 0.0000000 | 0.7816369 | 0.852 | 0.214 | 0.0000000 | 7 | Tfrc |
| 0.0000000 | 0.7684155 | 0.864 | 0.056 | 0.0000000 | 7 | Apcdd1 |
| 0.0000000 | 0.7588962 | 0.975 | 0.220 | 0.0000000 | 7 | Ebf1 |
| 0.0000000 | 0.7524957 | 0.901 | 0.142 | 0.0000000 | 7 | Bsg |
| 0.0000000 | 0.7500444 | 0.951 | 0.027 | 0.0000000 | 7 | Cldn5 |
| 0.0000000 | 0.7893187 | 0.857 | 0.058 | 0.0000000 | 17 | Gfap |
| 0.0000000 | 0.7228016 | 0.857 | 0.306 | 0.0000000 | 17 | Apoe |
| 0.0000000 | 0.6978836 | 0.857 | 0.154 | 0.0000000 | 17 | Aqp4 |
| 0.0000000 | 0.6241780 | 0.786 | 0.213 | 0.0000000 | 17 | Adamts9 |
| 0.0000000 | 0.6175138 | 0.821 | 0.153 | 0.0000000 | 17 | Tnc |
| 0.0000000 | 0.6100070 | 0.893 | 0.515 | 0.0000008 | 17 | Wdr17 |
| 0.0000000 | 0.5967415 | 0.929 | 0.624 | 0.0000000 | 17 | Gpm6b |
| 0.0000000 | 0.5821490 | 0.857 | 0.256 | 0.0000000 | 17 | Arhgef4 |
| 0.0000000 | 0.5364629 | 1.000 | 0.230 | 0.0000000 | 17 | Rfx4 |
| 0.0000000 | 0.5263381 | 0.857 | 0.203 | 0.0000000 | 17 | Slc7a11 |
| 0.0000000 | 0.5246198 | 0.964 | 0.456 | 0.0000000 | 17 | Slc4a4 |
| 0.0000000 | 0.5039781 | 0.857 | 0.294 | 0.0000000 | 17 | Sparcl1 |
| 0.0000000 | 0.5017582 | 0.786 | 0.151 | 0.0000000 | 17 | Ednrb |
| 0.0000000 | 0.4928392 | 0.893 | 0.219 | 0.0000000 | 17 | Igsf11 |
| 0.0000000 | 0.4915714 | 0.893 | 0.210 | 0.0000000 | 17 | Bmpr1b |
| 0.0000000 | 0.4883654 | 0.571 | 0.083 | 0.0000000 | 17 | Mamdc2 |
| 0.0000000 | 0.4864835 | 0.821 | 0.419 | 0.0000001 | 17 | Slc38a1 |
| 0.0000000 | 0.4645177 | 1.000 | 0.751 | 0.0000000 | 17 | Qk |
| 0.0000000 | 0.4630787 | 1.000 | 0.508 | 0.0000000 | 17 | Nfib |
| 0.0000000 | 0.4576987 | 0.714 | 0.411 | 0.0004846 | 17 | Rgs20 |
| 0.0000000 | 0.9198733 | 0.959 | 0.403 | 0.0000000 | 9 | Hs3st4 |
| 0.0000000 | 0.8866135 | 1.000 | 0.783 | 0.0000000 | 9 | Lrrc4c |
| 0.0000000 | 0.8366687 | 0.730 | 0.183 | 0.0000000 | 9 | 4930445B16Rik |
| 0.0000000 | 0.8309037 | 1.000 | 0.669 | 0.0000000 | 9 | Galntl6 |
| 0.0000000 | 0.8045590 | 1.000 | 0.673 | 0.0000000 | 9 | Dpp10 |
| 0.0000000 | 0.7311357 | 0.919 | 0.270 | 0.0000000 | 9 | Kcnmb2 |
| 0.0000000 | 0.6734903 | 0.878 | 0.188 | 0.0000000 | 9 | Meis2 |
| 0.0000000 | 0.6623663 | 0.946 | 0.281 | 0.0000000 | 9 | Ubash3b |
| 0.0000000 | 0.6594364 | 0.905 | 0.135 | 0.0000000 | 9 | Gm15155 |
| 0.0000000 | 0.6562089 | 0.973 | 0.764 | 0.0000000 | 9 | Nrg1 |
| 0.0000000 | 0.6413744 | 0.959 | 0.787 | 0.0000000 | 9 | Asic2 |
| 0.0000000 | 0.5928127 | 0.851 | 0.070 | 0.0000000 | 9 | Gad1 |
| 0.0000000 | 0.5917066 | 1.000 | 0.868 | 0.0000000 | 9 | Nrxn3 |
| 0.0000000 | 0.5894474 | 0.946 | 0.095 | 0.0000000 | 9 | Gad2 |
| 0.0000000 | 0.5869991 | 0.892 | 0.122 | 0.0000000 | 9 | Ptchd1 |
| 0.0000000 | 0.5862208 | 0.959 | 0.404 | 0.0000000 | 9 | Fign |
| 0.0000000 | 0.5701178 | 0.986 | 0.575 | 0.0000000 | 9 | Zfp804a |
| 0.0000000 | 0.5684483 | 0.959 | 0.513 | 0.0000000 | 9 | Kctd16 |
| 0.0000000 | 0.5644685 | 0.689 | 0.135 | 0.0000000 | 9 | Sema3e |
| 0.0000000 | 0.5629603 | 0.878 | 0.319 | 0.0000000 | 9 | Ak5 |
| 0.0000000 | 0.5987902 | 0.969 | 0.404 | 0.0000000 | 14 | Pcsk2 |
| 0.0000000 | 0.5748534 | 0.906 | 0.231 | 0.0000000 | 14 | Stxbp6 |
| 0.0000000 | 0.5567000 | 1.000 | 0.363 | 0.0000000 | 14 | Lef1 |
| 0.0000000 | 0.5496401 | 1.000 | 0.690 | 0.0000000 | 14 | Ntng1 |
| 0.0000000 | 0.5329311 | 1.000 | 0.280 | 0.0000000 | 14 | Cpne7 |
| 0.0000000 | 0.5155800 | 1.000 | 0.491 | 0.0000000 | 14 | Ryr3 |
| 0.0000000 | 0.4949808 | 0.906 | 0.395 | 0.0000000 | 14 | Cntn3 |
| 0.0000000 | 0.4879512 | 0.969 | 0.746 | 0.0000000 | 14 | Mgat4c |
| 0.0000000 | 0.4749312 | 0.969 | 0.720 | 0.0000000 | 14 | Lingo2 |
| 0.0000000 | 0.4726185 | 0.969 | 0.558 | 0.0000000 | 14 | Efna5 |
| 0.0000000 | 0.4724820 | 1.000 | 0.492 | 0.0000000 | 14 | Zmat4 |
| 0.0000000 | 0.4715426 | 1.000 | 0.733 | 0.0000000 | 14 | Syt1 |
| 0.0000000 | 0.4680961 | 1.000 | 0.762 | 0.0000000 | 14 | Cntnap2 |
| 0.0000000 | 0.4644152 | 0.906 | 0.280 | 0.0000002 | 14 | 6330411D24Rik |
| 0.0000000 | 0.4558477 | 0.844 | 0.393 | 0.0000442 | 14 | Adarb2 |
| 0.0000000 | 0.4508117 | 0.938 | 0.190 | 0.0000000 | 14 | Prox1 |
| 0.0000000 | 0.4376333 | 0.969 | 0.699 | 0.0000000 | 14 | Sox5 |
| 0.0000000 | 0.4347482 | 1.000 | 0.819 | 0.0000000 | 14 | Kcnip4 |
| 0.0000000 | 0.4342838 | 1.000 | 0.895 | 0.0000000 | 14 | Cadm2 |
| 0.0000000 | 0.4232970 | 1.000 | 0.768 | 0.0000000 | 14 | Lrrc7 |
| 0.0000000 | 1.3569120 | 0.960 | 0.040 | 0.0000000 | 3 | Cfap299 |
| 0.0000000 | 1.3376229 | 0.968 | 0.027 | 0.0000000 | 3 | Dnah12 |
| 0.0000000 | 1.2419726 | 0.952 | 0.102 | 0.0000000 | 3 | Adamts20 |
| 0.0000000 | 1.1665717 | 0.960 | 0.040 | 0.0000000 | 3 | Dnah6 |
| 0.0000000 | 1.1267706 | 0.968 | 0.098 | 0.0000000 | 3 | Cfap54 |
| 0.0000000 | 1.1096854 | 0.968 | 0.117 | 0.0000000 | 3 | Spag16 |
| 0.0000000 | 1.0928477 | 0.960 | 0.037 | 0.0000000 | 3 | Hydin |
| 0.0000000 | 1.0834740 | 0.960 | 0.076 | 0.0000000 | 3 | Ccdc162 |
| 0.0000000 | 1.0808259 | 0.960 | 0.059 | 0.0000000 | 3 | Rgs22 |
| 0.0000000 | 1.0784167 | 0.968 | 0.056 | 0.0000000 | 3 | Cfap61 |
| 0.0000000 | 1.0729439 | 0.935 | 0.074 | 0.0000000 | 3 | Cfap44 |
| 0.0000000 | 1.0546277 | 0.992 | 0.865 | 0.0000000 | 3 | Syne1 |
| 0.0000000 | 1.0507064 | 0.952 | 0.114 | 0.0000000 | 3 | Gm973 |
| 0.0000000 | 1.0437030 | 0.968 | 0.052 | 0.0000000 | 3 | Spef2 |
| 0.0000000 | 1.0400440 | 0.968 | 0.051 | 0.0000000 | 3 | Ak7 |
| 0.0000000 | 1.0318951 | 0.976 | 0.131 | 0.0000000 | 3 | Dnah9 |
| 0.0000000 | 1.0267513 | 0.976 | 0.060 | 0.0000000 | 3 | Kif6 |
| 0.0000000 | 1.0247695 | 0.952 | 0.032 | 0.0000000 | 3 | Ak9 |
| 0.0000000 | 1.0199062 | 0.952 | 0.043 | 0.0000000 | 3 | Dnah11 |
| 0.0000000 | 0.9844545 | 0.968 | 0.027 | 0.0000000 | 3 | Cfap65 |
| 0.0000000 | 1.6589135 | 0.875 | 0.167 | 0.0000000 | 15 | Htr2c |
| 0.0000000 | 1.4673531 | 1.000 | 0.087 | 0.0000000 | 15 | Ttr |
| 0.0000000 | 0.9553699 | 1.000 | 0.511 | 0.0000000 | 15 | Wdr17 |
| 0.0000000 | 0.9178587 | 0.781 | 0.024 | 0.0000000 | 15 | Gmnc |
| 0.0000000 | 0.8962658 | 0.875 | 0.181 | 0.0000000 | 15 | Enpp2 |
| 0.0000000 | 0.8825383 | 0.969 | 0.064 | 0.0000000 | 15 | Rbm47 |
| 0.0000000 | 0.8818117 | 0.875 | 0.233 | 0.0000000 | 15 | Vat1l |
| 0.0000000 | 0.8731853 | 0.938 | 0.179 | 0.0000000 | 15 | Otx2os1 |
| 0.0000000 | 0.8621759 | 0.906 | 0.158 | 0.0000000 | 15 | Sulf1 |
| 0.0000000 | 0.8325165 | 0.938 | 0.637 | 0.0000000 | 15 | Stk39 |
| 0.0000000 | 0.8169684 | 1.000 | 0.861 | 0.0000000 | 15 | Trpm3 |
| 0.0000000 | 0.7779916 | 0.969 | 0.219 | 0.0000000 | 15 | C330002G04Rik |
| 0.0000000 | 0.7569651 | 0.875 | 0.316 | 0.0000000 | 15 | Nhsl2 |
| 0.0000000 | 0.7382019 | 1.000 | 0.673 | 0.0000000 | 15 | Frmpd4 |
| 0.0000000 | 0.7310722 | 0.938 | 0.367 | 0.0000000 | 15 | Itpr1 |
| 0.0000000 | 0.6973879 | 0.781 | 0.207 | 0.0000000 | 15 | Atp2b3 |
| 0.0000000 | 0.6920626 | 0.875 | 0.169 | 0.0000000 | 15 | Prdm16 |
| 0.0000000 | 0.6890443 | 0.812 | 0.245 | 0.0000000 | 15 | Cab39l |
| 0.0000000 | 0.6844702 | 0.969 | 0.567 | 0.0000000 | 15 | Rfx3 |
| 0.0000000 | 0.6690855 | 0.938 | 0.627 | 0.0000000 | 15 | Tmem108 |
| 0.0000000 | 0.8211787 | 0.973 | 0.591 | 0.0000000 | 10 | Zfp804b |
| 0.0000000 | 0.8144616 | 1.000 | 0.813 | 0.0000000 | 10 | Kcnip4 |
| 0.0000000 | 0.8054150 | 0.644 | 0.074 | 0.0000000 | 10 | Npsr1 |
| 0.0000000 | 0.6461324 | 1.000 | 0.707 | 0.0000000 | 10 | Cntn5 |
| 0.0000000 | 0.6316225 | 1.000 | 0.759 | 0.0000000 | 10 | Tenm2 |
| 0.0000000 | 0.6087797 | 1.000 | 0.578 | 0.0000000 | 10 | Gm20754 |
| 0.0000000 | 0.6024773 | 0.973 | 0.566 | 0.0000000 | 10 | Kcnq5 |
| 0.0000000 | 0.5777072 | 0.973 | 0.458 | 0.0000000 | 10 | St6galnac5 |
| 0.0000000 | 0.5522407 | 0.575 | 0.207 | 0.0000000 | 10 | Pcsk5 |
| 0.0000000 | 0.5460744 | 0.945 | 0.740 | 0.0000000 | 10 | Mgat4c |
| 0.0000000 | 0.5258432 | 0.986 | 0.690 | 0.0000000 | 10 | Sox5 |
| 0.0000000 | 0.5065918 | 0.986 | 0.474 | 0.0000000 | 10 | Cdh8 |
| 0.0000000 | 0.4842790 | 0.699 | 0.319 | 0.0000000 | 10 | Cntnap5c |
| 0.0000000 | 0.4796889 | 0.658 | 0.182 | 0.0000000 | 10 | Prox1 |
| 0.0000000 | 0.4747579 | 0.836 | 0.661 | 0.0000000 | 10 | Brinp3 |
| 0.0000000 | 0.4657125 | 1.000 | 0.867 | 0.0000000 | 10 | Kcnma1 |
| 0.0000000 | 0.4578456 | 0.699 | 0.380 | 0.0000000 | 10 | 4930509J09Rik |
| 0.0000000 | 0.4474035 | 0.808 | 0.326 | 0.0000000 | 10 | Prr16 |
| 0.0000000 | 0.4269801 | 0.973 | 0.649 | 0.0000000 | 10 | Rasgef1b |
| 0.0000000 | 0.4182427 | 0.781 | 0.354 | 0.0000000 | 10 | Tafa2 |
| 0.0000000 | 0.9804342 | 0.950 | 0.265 | 0.0000000 | 11 | 6330411D24Rik |
| 0.0000000 | 0.8601788 | 0.967 | 0.304 | 0.0000000 | 11 | Pex5l |
| 0.0000000 | 0.6614258 | 1.000 | 0.488 | 0.0000000 | 11 | Spock1 |
| 0.0000000 | 0.6572678 | 0.983 | 0.481 | 0.0000000 | 11 | Ryr3 |
| 0.0000000 | 0.6184067 | 1.000 | 0.756 | 0.0000000 | 11 | Hs6st3 |
| 0.0000000 | 0.6148524 | 0.917 | 0.381 | 0.0000000 | 11 | Adarb1 |
| 0.0000000 | 0.6014458 | 0.867 | 0.151 | 0.0000000 | 11 | 4930419G24Rik |
| 0.0000000 | 0.5629957 | 0.983 | 0.477 | 0.0000000 | 11 | Edil3 |
| 0.0000000 | 0.5581578 | 1.000 | 0.666 | 0.0000000 | 11 | Frmpd4 |
| 0.0000000 | 0.5554781 | 0.933 | 0.383 | 0.0000000 | 11 | Cntn3 |
| 0.0000000 | 0.5453907 | 0.933 | 0.225 | 0.0000000 | 11 | Ptpn3 |
| 0.0000000 | 0.5336730 | 1.000 | 0.481 | 0.0000000 | 11 | Zmat4 |
| 0.0000000 | 0.5239501 | 1.000 | 0.856 | 0.0000000 | 11 | Prickle2 |
| 0.0000000 | 0.5223944 | 0.983 | 0.634 | 0.0000000 | 11 | Cacnb4 |
| 0.0000000 | 0.5198736 | 1.000 | 0.593 | 0.0000000 | 11 | Zfp804b |
| 0.0000000 | 0.5068030 | 1.000 | 0.620 | 0.0000000 | 11 | Unc13c |
| 0.0000000 | 0.5063109 | 0.967 | 0.390 | 0.0000000 | 11 | Epb41l4b |
| 0.0000000 | 0.5059856 | 0.933 | 0.370 | 0.0000000 | 11 | Zfp385b |
| 0.0000000 | 0.5027454 | 0.967 | 0.463 | 0.0000000 | 11 | Camk4 |
| 0.0000000 | 0.5009991 | 0.950 | 0.547 | 0.0000000 | 11 | Pak7 |
markers.wilcox <-
FindAllMarkers(
combined.srt,
assay = "SCT",
verbose = FALSE,
random.seed = reseed,
only.pos = TRUE,
min.pct = 0.2,
base = 10,
logfc.threshold = 0.2,
densify = TRUE,
test.use = "wilcox")
write_csv(markers.wilcox,
here(tables_dir,
'hevesi2023-all-mrk_wilcox-sct_combined.csv'))
markers.wilcox %>%
group_by(cluster) %>%
slice_max(n = 20, order_by = avg_log10FC) %>%
kable("html") %>%
kable_material(
bootstrap_options = c("bordered",
"condensed",
"responsive",
"striped"),
position = "left",
font_size = 14
)| p_val | avg_log10FC | pct.1 | pct.2 | p_val_adj | cluster | gene |
|---|---|---|---|---|---|---|
| 0.0e+00 | 0.5461860 | 0.789 | 0.255 | 0.0000000 | 1 | Gm15398 |
| 0.0e+00 | 0.5379832 | 0.849 | 0.472 | 0.0000000 | 1 | Cdh18 |
| 0.0e+00 | 0.5250328 | 0.789 | 0.309 | 0.0000000 | 1 | Unc5d |
| 0.0e+00 | 0.4923348 | 0.984 | 0.708 | 0.0000000 | 1 | Lrrtm4 |
| 0.0e+00 | 0.4297117 | 0.838 | 0.306 | 0.0000000 | 1 | Tafa2 |
| 0.0e+00 | 0.4267031 | 1.000 | 0.741 | 0.0000000 | 1 | Dlg2 |
| 0.0e+00 | 0.4199060 | 0.800 | 0.337 | 0.0000000 | 1 | Gm26871 |
| 0.0e+00 | 0.4185715 | 0.962 | 0.769 | 0.0000000 | 1 | Ralyl |
| 0.0e+00 | 0.4176032 | 0.795 | 0.211 | 0.0000000 | 1 | Dlgap2 |
| 0.0e+00 | 0.4117576 | 0.892 | 0.655 | 0.0000000 | 1 | Galntl6 |
| 0.0e+00 | 0.4024267 | 0.730 | 0.231 | 0.0000000 | 1 | 4930555F03Rik |
| 0.0e+00 | 0.4002103 | 0.805 | 0.398 | 0.0000000 | 1 | Grm8 |
| 0.0e+00 | 0.3966198 | 0.946 | 0.758 | 0.0000000 | 1 | Dab1 |
| 0.0e+00 | 0.3965816 | 0.978 | 0.744 | 0.0000000 | 1 | Nrg1 |
| 0.0e+00 | 0.3867786 | 0.827 | 0.452 | 0.0000000 | 1 | Gria1 |
| 0.0e+00 | 0.3867692 | 0.789 | 0.282 | 0.0000000 | 1 | Grin2a |
| 6.2e-06 | 0.3856435 | 0.519 | 0.385 | 0.1276437 | 1 | Adarb2 |
| 0.0e+00 | 0.3846172 | 1.000 | 0.804 | 0.0000000 | 1 | Ahi1 |
| 0.0e+00 | 0.3658777 | 0.611 | 0.199 | 0.0000000 | 1 | Gm45321 |
| 0.0e+00 | 0.3657352 | 0.946 | 0.679 | 0.0000000 | 1 | Sgcz |
| 0.0e+00 | 0.6659039 | 1.000 | 0.680 | 0.0000000 | 2 | Tenm1 |
| 0.0e+00 | 0.6523820 | 0.969 | 0.556 | 0.0000000 | 2 | Tafa1 |
| 0.0e+00 | 0.6200753 | 1.000 | 0.657 | 0.0000000 | 2 | Ntng1 |
| 0.0e+00 | 0.6149381 | 1.000 | 0.678 | 0.0000000 | 2 | Rnf220 |
| 0.0e+00 | 0.5863378 | 0.969 | 0.635 | 0.0000000 | 2 | Shisa9 |
| 0.0e+00 | 0.5635203 | 0.901 | 0.393 | 0.0000000 | 2 | Thsd7b |
| 0.0e+00 | 0.5548733 | 0.883 | 0.260 | 0.0000000 | 2 | Gm48749 |
| 0.0e+00 | 0.5513788 | 0.975 | 0.465 | 0.0000000 | 2 | Samd5 |
| 0.0e+00 | 0.5407102 | 0.988 | 0.580 | 0.0000000 | 2 | Cntnap5a |
| 0.0e+00 | 0.5233261 | 0.864 | 0.251 | 0.0000000 | 2 | Gm32647 |
| 0.0e+00 | 0.4882065 | 0.981 | 0.584 | 0.0000000 | 2 | Tox |
| 0.0e+00 | 0.4625518 | 0.988 | 0.670 | 0.0000000 | 2 | Egfem1 |
| 0.0e+00 | 0.4594009 | 0.827 | 0.259 | 0.0000000 | 2 | Trhde |
| 0.0e+00 | 0.4589436 | 1.000 | 0.737 | 0.0000000 | 2 | Cntnap2 |
| 0.0e+00 | 0.4544066 | 1.000 | 0.875 | 0.0000000 | 2 | Ptprd |
| 0.0e+00 | 0.4516664 | 0.951 | 0.372 | 0.0000000 | 2 | Foxp2 |
| 0.0e+00 | 0.4458370 | 1.000 | 0.758 | 0.0000000 | 2 | Grm7 |
| 0.0e+00 | 0.4429434 | 1.000 | 0.634 | 0.0000000 | 2 | Arpp21 |
| 0.0e+00 | 0.4278149 | 0.994 | 0.845 | 0.0000000 | 2 | Grik2 |
| 0.0e+00 | 0.4269539 | 0.988 | 0.675 | 0.0000000 | 2 | Cntn4 |
| 0.0e+00 | 0.7270600 | 0.747 | 0.153 | 0.0000000 | 8 | 4930588A03Rik |
| 0.0e+00 | 0.7259301 | 0.823 | 0.436 | 0.0000000 | 8 | Pcdh15 |
| 0.0e+00 | 0.7098871 | 0.772 | 0.114 | 0.0000000 | 8 | Gm38505 |
| 0.0e+00 | 0.6501237 | 0.873 | 0.498 | 0.0000000 | 8 | 6030443J06Rik |
| 0.0e+00 | 0.6287367 | 0.873 | 0.412 | 0.0000000 | 8 | Sox6 |
| 0.0e+00 | 0.6071638 | 0.696 | 0.121 | 0.0000000 | 8 | Pdgfra |
| 0.0e+00 | 0.5930285 | 0.823 | 0.514 | 0.0000000 | 8 | Sox2ot |
| 0.0e+00 | 0.5898584 | 0.835 | 0.439 | 0.0000000 | 8 | Lhfpl3 |
| 0.0e+00 | 0.5524413 | 0.747 | 0.236 | 0.0000000 | 8 | Dscaml1 |
| 0.0e+00 | 0.5146779 | 0.861 | 0.503 | 0.0000000 | 8 | Epn2 |
| 0.0e+00 | 0.5044968 | 0.835 | 0.477 | 0.0000000 | 8 | Xylt1 |
| 0.0e+00 | 0.4766786 | 0.684 | 0.075 | 0.0000000 | 8 | Sox10 |
| 0.0e+00 | 0.4746780 | 0.772 | 0.299 | 0.0000000 | 8 | Mir9-3hg |
| 0.0e+00 | 0.4493794 | 0.911 | 0.675 | 0.0000000 | 8 | Sgcd |
| 0.0e+00 | 0.4420664 | 0.810 | 0.531 | 0.0000000 | 8 | Pcdh11x |
| 0.0e+00 | 0.4347889 | 0.899 | 0.642 | 0.0000000 | 8 | Dcc |
| 0.0e+00 | 0.4181992 | 0.696 | 0.226 | 0.0000000 | 8 | Megf11 |
| 0.0e+00 | 0.4159086 | 0.861 | 0.523 | 0.0000000 | 8 | Nxph1 |
| 0.0e+00 | 0.4044997 | 0.759 | 0.239 | 0.0000000 | 8 | Arhgap31 |
| 0.0e+00 | 0.4038512 | 0.785 | 0.410 | 0.0000000 | 8 | Tnr |
| 1.0e-07 | 0.8626318 | 0.667 | 0.258 | 0.0014715 | 18 | Rbpj |
| 0.0e+00 | 0.8141688 | 1.000 | 0.378 | 0.0000000 | 18 | Mef2c |
| 0.0e+00 | 0.7664959 | 0.292 | 0.043 | 0.0000426 | 18 | F13a1 |
| 0.0e+00 | 0.7558552 | 0.792 | 0.351 | 0.0000013 | 18 | Slc9a9 |
| 0.0e+00 | 0.7488395 | 1.000 | 0.415 | 0.0000000 | 18 | Zeb2 |
| 3.7e-06 | 0.7469816 | 0.500 | 0.175 | 0.0766912 | 18 | Dab2 |
| 0.0e+00 | 0.7250744 | 0.458 | 0.015 | 0.0000000 | 18 | Mrc1 |
| 0.0e+00 | 0.7167972 | 0.792 | 0.019 | 0.0000000 | 18 | Runx1 |
| 0.0e+00 | 0.7165791 | 0.958 | 0.356 | 0.0000000 | 18 | Mbnl1 |
| 0.0e+00 | 0.6897250 | 0.833 | 0.309 | 0.0000111 | 18 | Apoe |
| 0.0e+00 | 0.6816252 | 0.958 | 0.152 | 0.0000000 | 18 | Dock2 |
| 0.0e+00 | 0.6738016 | 0.958 | 0.046 | 0.0000000 | 18 | Inpp5d |
| 2.0e-07 | 0.6581980 | 0.875 | 0.686 | 0.0038059 | 18 | Plxdc2 |
| 0.0e+00 | 0.6533039 | 0.917 | 0.312 | 0.0000000 | 18 | Rreb1 |
| 0.0e+00 | 0.6422074 | 0.917 | 0.099 | 0.0000000 | 18 | Fli1 |
| 0.0e+00 | 0.6255837 | 0.708 | 0.024 | 0.0000000 | 18 | Apbb1ip |
| 0.0e+00 | 0.6238467 | 0.875 | 0.039 | 0.0000000 | 18 | Fyb |
| 0.0e+00 | 0.6165447 | 0.833 | 0.141 | 0.0000000 | 18 | Maf |
| 0.0e+00 | 0.6133034 | 0.792 | 0.015 | 0.0000000 | 18 | Ly86 |
| 0.0e+00 | 0.6102902 | 0.875 | 0.273 | 0.0000000 | 18 | Pip4k2a |
| 0.0e+00 | 1.3713943 | 1.000 | 0.078 | 0.0000000 | 16 | Bnc2 |
| 0.0e+00 | 0.9347756 | 1.000 | 0.452 | 0.0000000 | 16 | Fbxl7 |
| 0.0e+00 | 0.9258599 | 0.867 | 0.052 | 0.0000000 | 16 | Adamtsl3 |
| 0.0e+00 | 0.9149946 | 0.933 | 0.148 | 0.0000000 | 16 | Thsd4 |
| 0.0e+00 | 0.9028647 | 1.000 | 0.700 | 0.0000000 | 16 | Foxp1 |
| 0.0e+00 | 0.9013542 | 0.900 | 0.513 | 0.0000000 | 16 | Slc4a10 |
| 0.0e+00 | 0.9007700 | 0.933 | 0.310 | 0.0000000 | 16 | Nr3c2 |
| 0.0e+00 | 0.8648775 | 1.000 | 0.376 | 0.0000000 | 16 | Slc38a2 |
| 0.0e+00 | 0.8521231 | 0.833 | 0.290 | 0.0000000 | 16 | Col25a1 |
| 0.0e+00 | 0.8468100 | 0.933 | 0.061 | 0.0000000 | 16 | Trabd2b |
| 0.0e+00 | 0.8042045 | 0.833 | 0.173 | 0.0000000 | 16 | Itgbl1 |
| 0.0e+00 | 0.7824174 | 0.933 | 0.346 | 0.0000000 | 16 | Gulp1 |
| 0.0e+00 | 0.7498879 | 0.967 | 0.424 | 0.0000000 | 16 | Tmtc1 |
| 0.0e+00 | 0.7471613 | 0.933 | 0.111 | 0.0000000 | 16 | Eya2 |
| 0.0e+00 | 0.7423676 | 1.000 | 0.613 | 0.0000000 | 16 | Tmeff2 |
| 0.0e+00 | 0.7290641 | 0.867 | 0.250 | 0.0000000 | 16 | Sh3pxd2a |
| 0.0e+00 | 0.7133837 | 0.800 | 0.091 | 0.0000000 | 16 | Dock5 |
| 0.0e+00 | 0.6949514 | 0.967 | 0.314 | 0.0000000 | 16 | Nxn |
| 0.0e+00 | 0.6757103 | 0.967 | 0.209 | 0.0000000 | 16 | Hmcn1 |
| 0.0e+00 | 0.6698614 | 0.567 | 0.067 | 0.0000000 | 16 | Crispld1 |
| 0.0e+00 | 0.5858833 | 0.543 | 0.135 | 0.0000000 | 5 | mt-Co3 |
| 0.0e+00 | 0.5707633 | 0.857 | 0.498 | 0.0000000 | 5 | Tuba1a |
| 0.0e+00 | 0.5623345 | 0.886 | 0.570 | 0.0000000 | 5 | Cmss1 |
| 0.0e+00 | 0.5326416 | 0.505 | 0.099 | 0.0000000 | 5 | mt-Atp6 |
| 0.0e+00 | 0.5034257 | 0.971 | 0.892 | 0.0000000 | 5 | Gm42418 |
| 0.0e+00 | 0.4866638 | 0.562 | 0.213 | 0.0000000 | 5 | mt-Co1 |
| 0.0e+00 | 0.4762082 | 0.505 | 0.121 | 0.0000000 | 5 | mt-Co2 |
| 0.0e+00 | 0.3906273 | 0.657 | 0.307 | 0.0000000 | 5 | Tubb5 |
| 0.0e+00 | 0.3896130 | 0.610 | 0.325 | 0.0000000 | 5 | Tubb2a |
| 0.0e+00 | 0.3794212 | 0.400 | 0.077 | 0.0000000 | 5 | mt-Cytb |
| 0.0e+00 | 0.3791696 | 0.857 | 0.549 | 0.0000000 | 5 | Calm1 |
| 0.0e+00 | 0.3721275 | 0.600 | 0.303 | 0.0000000 | 5 | Actg1 |
| 0.0e+00 | 0.3716602 | 0.629 | 0.204 | 0.0000000 | 5 | Atp6v0c |
| 0.0e+00 | 0.3669284 | 0.790 | 0.557 | 0.0000000 | 5 | Cdk8 |
| 0.0e+00 | 0.3621710 | 0.886 | 0.641 | 0.0000000 | 5 | Basp1 |
| 0.0e+00 | 0.3596345 | 0.371 | 0.077 | 0.0000000 | 5 | mt-Nd4 |
| 0.0e+00 | 0.3590900 | 0.438 | 0.070 | 0.0000000 | 5 | mt-Nd1 |
| 0.0e+00 | 0.3583674 | 0.438 | 0.084 | 0.0000000 | 5 | mt-Nd2 |
| 0.0e+00 | 0.3576392 | 0.552 | 0.208 | 0.0000000 | 5 | Tmsb4x |
| 0.0e+00 | 0.3566092 | 0.867 | 0.511 | 0.0000000 | 5 | Gap43 |
| 0.0e+00 | 0.6689559 | 1.000 | 0.822 | 0.0000000 | 12 | Gm26917 |
| 0.0e+00 | 0.5710436 | 0.952 | 0.463 | 0.0000000 | 12 | Rbfox3 |
| 0.0e+00 | 0.4941579 | 0.976 | 0.852 | 0.0000000 | 12 | Camta1 |
| 3.0e-07 | 0.4925956 | 0.690 | 0.437 | 0.0060376 | 12 | Cdh4 |
| 0.0e+00 | 0.4665196 | 0.929 | 0.541 | 0.0000000 | 12 | Gpi1 |
| 0.0e+00 | 0.4356404 | 0.238 | 0.043 | 0.0000586 | 12 | Ebf3 |
| 0.0e+00 | 0.4338614 | 0.857 | 0.419 | 0.0000000 | 12 | Pacsin1 |
| 0.0e+00 | 0.4276850 | 0.929 | 0.388 | 0.0000000 | 12 | Camk2b |
| 0.0e+00 | 0.4197312 | 0.786 | 0.338 | 0.0000000 | 12 | Gse1 |
| 0.0e+00 | 0.4127750 | 0.690 | 0.276 | 0.0000000 | 12 | Dlgap2 |
| 0.0e+00 | 0.4107957 | 0.857 | 0.486 | 0.0000000 | 12 | Kif5a |
| 0.0e+00 | 0.4102879 | 0.881 | 0.547 | 0.0000000 | 12 | Ksr2 |
| 0.0e+00 | 0.4058210 | 0.762 | 0.485 | 0.0000040 | 12 | Xkr6 |
| 0.0e+00 | 0.4048061 | 0.738 | 0.322 | 0.0000000 | 12 | Ank1 |
| 0.0e+00 | 0.3964670 | 0.786 | 0.419 | 0.0000000 | 12 | Chd5 |
| 0.0e+00 | 0.3959419 | 0.952 | 0.466 | 0.0000000 | 12 | Shank1 |
| 0.0e+00 | 0.3942990 | 0.690 | 0.457 | 0.0003113 | 12 | Cux2 |
| 0.0e+00 | 0.3934189 | 0.786 | 0.522 | 0.0000026 | 12 | Atp2b2 |
| 0.0e+00 | 0.3919585 | 0.833 | 0.418 | 0.0000000 | 12 | Slc4a3 |
| 0.0e+00 | 0.3910830 | 0.667 | 0.370 | 0.0008577 | 12 | Kif26b |
| 0.0e+00 | 0.9391914 | 0.992 | 0.304 | 0.0000000 | 4 | Gm3764 |
| 0.0e+00 | 0.8585705 | 1.000 | 0.609 | 0.0000000 | 4 | Ptprz1 |
| 0.0e+00 | 0.8547763 | 1.000 | 0.415 | 0.0000000 | 4 | Slc4a4 |
| 0.0e+00 | 0.7667826 | 1.000 | 0.712 | 0.0000000 | 4 | Npas3 |
| 0.0e+00 | 0.7346128 | 0.934 | 0.642 | 0.0000000 | 4 | Luzp2 |
| 0.0e+00 | 0.7284913 | 0.992 | 0.373 | 0.0000000 | 4 | Gm48747 |
| 0.0e+00 | 0.6781296 | 0.868 | 0.171 | 0.0000000 | 4 | Slc6a11 |
| 0.0e+00 | 0.6575202 | 0.959 | 0.611 | 0.0000000 | 4 | Gpc5 |
| 0.0e+00 | 0.6438764 | 0.983 | 0.514 | 0.0000000 | 4 | Trim9 |
| 0.0e+00 | 0.6425576 | 0.926 | 0.364 | 0.0000000 | 4 | Nhsl1 |
| 0.0e+00 | 0.6229149 | 0.967 | 0.480 | 0.0000000 | 4 | Wdr17 |
| 0.0e+00 | 0.6162126 | 0.992 | 0.623 | 0.0000000 | 4 | Grm3 |
| 0.0e+00 | 0.5919610 | 0.917 | 0.186 | 0.0000000 | 4 | Lrig1 |
| 0.0e+00 | 0.5791123 | 0.959 | 0.466 | 0.0000000 | 4 | Slc1a2 |
| 0.0e+00 | 0.5751882 | 1.000 | 0.733 | 0.0000000 | 4 | Qk |
| 0.0e+00 | 0.5560484 | 0.901 | 0.258 | 0.0000000 | 4 | Plpp3 |
| 0.0e+00 | 0.5550735 | 1.000 | 0.580 | 0.0000000 | 4 | Ptn |
| 0.0e+00 | 0.5515565 | 0.860 | 0.163 | 0.0000000 | 4 | Bmpr1b |
| 0.0e+00 | 0.5466761 | 0.934 | 0.268 | 0.0000000 | 4 | Mir9-3hg |
| 0.0e+00 | 0.5424140 | 0.777 | 0.141 | 0.0000000 | 4 | Pla2g7 |
| 0.0e+00 | 1.3003979 | 0.645 | 0.041 | 0.0000000 | 6 | Ptgds |
| 0.0e+00 | 1.0801231 | 0.892 | 0.047 | 0.0000000 | 6 | Ranbp3l |
| 0.0e+00 | 1.0305402 | 0.935 | 0.860 | 0.0000000 | 6 | Trpm3 |
| 0.0e+00 | 0.9166646 | 0.914 | 0.209 | 0.0000000 | 6 | Adam12 |
| 0.0e+00 | 0.8684877 | 0.849 | 0.058 | 0.0000000 | 6 | Slc6a20a |
| 0.0e+00 | 0.8575784 | 0.828 | 0.104 | 0.0000000 | 6 | Adamts12 |
| 0.0e+00 | 0.8390771 | 0.860 | 0.147 | 0.0000000 | 6 | Sidt1 |
| 0.0e+00 | 0.7826376 | 0.753 | 0.133 | 0.0000000 | 6 | Bmp6 |
| 0.0e+00 | 0.7707784 | 0.935 | 0.318 | 0.0000000 | 6 | Atp1a2 |
| 0.0e+00 | 0.7576284 | 0.882 | 0.016 | 0.0000000 | 6 | Slc6a13 |
| 0.0e+00 | 0.7518776 | 0.903 | 0.274 | 0.0000000 | 6 | Bicc1 |
| 0.0e+00 | 0.7271961 | 0.742 | 0.178 | 0.0000000 | 6 | Slc7a11 |
| 0.0e+00 | 0.6886652 | 0.839 | 0.689 | 0.0000000 | 6 | Nnat |
| 0.0e+00 | 0.6813297 | 0.828 | 0.099 | 0.0000000 | 6 | Cped1 |
| 0.0e+00 | 0.6799306 | 0.763 | 0.140 | 0.0000000 | 6 | Lrmda |
| 0.0e+00 | 0.6614480 | 0.828 | 0.257 | 0.0000000 | 6 | Sned1 |
| 0.0e+00 | 0.6610377 | 0.849 | 0.147 | 0.0000000 | 6 | Tmtc4 |
| 0.0e+00 | 0.6212833 | 0.871 | 0.220 | 0.0000000 | 6 | Pdzrn3 |
| 0.0e+00 | 0.6208102 | 0.903 | 0.115 | 0.0000000 | 6 | Arhgap29 |
| 0.0e+00 | 0.5748568 | 0.796 | 0.141 | 0.0000000 | 6 | Colec12 |
| 0.0e+00 | 1.3273780 | 0.973 | 0.168 | 0.0000000 | 13 | Mbp |
| 0.0e+00 | 1.1083179 | 0.946 | 0.083 | 0.0000000 | 13 | Plp1 |
| 0.0e+00 | 1.0948631 | 0.865 | 0.138 | 0.0000000 | 13 | 9630013A20Rik |
| 0.0e+00 | 1.0547744 | 0.865 | 0.041 | 0.0000000 | 13 | St18 |
| 0.0e+00 | 0.9724562 | 0.892 | 0.131 | 0.0000000 | 13 | Bcas1 |
| 0.0e+00 | 0.9608507 | 0.865 | 0.419 | 0.0000000 | 13 | Tnr |
| 0.0e+00 | 0.8971909 | 0.919 | 0.592 | 0.0000000 | 13 | Nfasc |
| 0.0e+00 | 0.8740391 | 0.892 | 0.200 | 0.0000000 | 13 | Tns3 |
| 0.0e+00 | 0.8032452 | 0.541 | 0.117 | 0.0000000 | 13 | Cnksr3 |
| 0.0e+00 | 0.7645961 | 0.946 | 0.290 | 0.0000000 | 13 | Itpr2 |
| 0.0e+00 | 0.7406776 | 0.784 | 0.045 | 0.0000000 | 13 | Bcas1os2 |
| 0.0e+00 | 0.7232176 | 0.973 | 0.541 | 0.0000000 | 13 | 9530059O14Rik |
| 0.0e+00 | 0.7120959 | 0.946 | 0.613 | 0.0000000 | 13 | Tmeff2 |
| 0.0e+00 | 0.6873725 | 0.324 | 0.010 | 0.0000000 | 13 | Mobp |
| 0.0e+00 | 0.6853668 | 0.703 | 0.372 | 0.0000543 | 13 | Pik3r3 |
| 0.0e+00 | 0.6850883 | 0.973 | 0.725 | 0.0000000 | 13 | Fyn |
| 0.0e+00 | 0.6749825 | 0.946 | 0.266 | 0.0000000 | 13 | Sirt2 |
| 0.0e+00 | 0.6671552 | 0.595 | 0.014 | 0.0000000 | 13 | Mag |
| 0.0e+00 | 0.6400321 | 1.000 | 0.797 | 0.0000000 | 13 | Opcml |
| 0.0e+00 | 0.6309468 | 0.811 | 0.024 | 0.0000000 | 13 | Enpp6 |
| 0.0e+00 | 1.3145263 | 0.975 | 0.055 | 0.0000000 | 7 | Flt1 |
| 0.0e+00 | 1.0884493 | 0.963 | 0.101 | 0.0000000 | 7 | Mecom |
| 0.0e+00 | 0.9486995 | 0.926 | 0.145 | 0.0000000 | 7 | Slc7a1 |
| 0.0e+00 | 0.9292332 | 0.926 | 0.203 | 0.0000000 | 7 | Dach1 |
| 0.0e+00 | 0.8974278 | 0.901 | 0.115 | 0.0000000 | 7 | Slco1c1 |
| 0.0e+00 | 0.8812812 | 0.914 | 0.020 | 0.0000000 | 7 | Egfl7 |
| 0.0e+00 | 0.8764783 | 0.889 | 0.058 | 0.0000000 | 7 | Ets1 |
| 0.0e+00 | 0.8709040 | 0.852 | 0.075 | 0.0000000 | 7 | Slc7a5 |
| 0.0e+00 | 0.8412937 | 0.889 | 0.033 | 0.0000000 | 7 | Ptprb |
| 0.0e+00 | 0.8406755 | 0.988 | 0.352 | 0.0000000 | 7 | Slc38a2 |
| 0.0e+00 | 0.8162503 | 0.840 | 0.188 | 0.0000000 | 7 | Hmcn1 |
| 0.0e+00 | 0.8072117 | 0.877 | 0.030 | 0.0000000 | 7 | Adgrl4 |
| 0.0e+00 | 0.8001894 | 0.938 | 0.227 | 0.0000000 | 7 | Ccdc141 |
| 0.0e+00 | 0.7961759 | 0.975 | 0.298 | 0.0000000 | 7 | Myo10 |
| 0.0e+00 | 0.7894780 | 0.889 | 0.065 | 0.0000000 | 7 | Fli1 |
| 0.0e+00 | 0.7816369 | 0.852 | 0.214 | 0.0000000 | 7 | Tfrc |
| 0.0e+00 | 0.7684155 | 0.864 | 0.056 | 0.0000000 | 7 | Apcdd1 |
| 0.0e+00 | 0.7588962 | 0.975 | 0.220 | 0.0000000 | 7 | Ebf1 |
| 0.0e+00 | 0.7524957 | 0.901 | 0.142 | 0.0000000 | 7 | Bsg |
| 0.0e+00 | 0.7500444 | 0.951 | 0.027 | 0.0000000 | 7 | Cldn5 |
| 0.0e+00 | 0.7893187 | 0.857 | 0.058 | 0.0000000 | 17 | Gfap |
| 0.0e+00 | 0.7228016 | 0.857 | 0.306 | 0.0000000 | 17 | Apoe |
| 0.0e+00 | 0.6978836 | 0.857 | 0.154 | 0.0000000 | 17 | Aqp4 |
| 0.0e+00 | 0.6241780 | 0.786 | 0.213 | 0.0000000 | 17 | Adamts9 |
| 0.0e+00 | 0.6175138 | 0.821 | 0.153 | 0.0000000 | 17 | Tnc |
| 0.0e+00 | 0.6100070 | 0.893 | 0.515 | 0.0000046 | 17 | Wdr17 |
| 0.0e+00 | 0.5967415 | 0.929 | 0.624 | 0.0000000 | 17 | Gpm6b |
| 0.0e+00 | 0.5821490 | 0.857 | 0.256 | 0.0000000 | 17 | Arhgef4 |
| 0.0e+00 | 0.5364629 | 1.000 | 0.230 | 0.0000000 | 17 | Rfx4 |
| 0.0e+00 | 0.5263381 | 0.857 | 0.203 | 0.0000000 | 17 | Slc7a11 |
| 0.0e+00 | 0.5246198 | 0.964 | 0.456 | 0.0000000 | 17 | Slc4a4 |
| 0.0e+00 | 0.5039781 | 0.857 | 0.294 | 0.0000000 | 17 | Sparcl1 |
| 0.0e+00 | 0.5017582 | 0.786 | 0.151 | 0.0000000 | 17 | Ednrb |
| 0.0e+00 | 0.4928392 | 0.893 | 0.219 | 0.0000000 | 17 | Igsf11 |
| 0.0e+00 | 0.4915714 | 0.893 | 0.210 | 0.0000000 | 17 | Bmpr1b |
| 0.0e+00 | 0.4883654 | 0.571 | 0.083 | 0.0000000 | 17 | Mamdc2 |
| 0.0e+00 | 0.4864835 | 0.821 | 0.419 | 0.0000036 | 17 | Slc38a1 |
| 0.0e+00 | 0.4645177 | 1.000 | 0.751 | 0.0000000 | 17 | Qk |
| 0.0e+00 | 0.4630787 | 1.000 | 0.508 | 0.0000000 | 17 | Nfib |
| 2.9e-06 | 0.4576987 | 0.714 | 0.411 | 0.0602560 | 17 | Rgs20 |
| 0.0e+00 | 0.9198733 | 0.959 | 0.403 | 0.0000000 | 9 | Hs3st4 |
| 0.0e+00 | 0.8866135 | 1.000 | 0.783 | 0.0000000 | 9 | Lrrc4c |
| 0.0e+00 | 0.8366687 | 0.730 | 0.183 | 0.0000000 | 9 | 4930445B16Rik |
| 0.0e+00 | 0.8309037 | 1.000 | 0.669 | 0.0000000 | 9 | Galntl6 |
| 0.0e+00 | 0.8045590 | 1.000 | 0.673 | 0.0000000 | 9 | Dpp10 |
| 0.0e+00 | 0.7311357 | 0.919 | 0.270 | 0.0000000 | 9 | Kcnmb2 |
| 0.0e+00 | 0.6734903 | 0.878 | 0.188 | 0.0000000 | 9 | Meis2 |
| 0.0e+00 | 0.6623663 | 0.946 | 0.281 | 0.0000000 | 9 | Ubash3b |
| 0.0e+00 | 0.6594364 | 0.905 | 0.135 | 0.0000000 | 9 | Gm15155 |
| 0.0e+00 | 0.6562089 | 0.973 | 0.764 | 0.0000000 | 9 | Nrg1 |
| 0.0e+00 | 0.6413744 | 0.959 | 0.787 | 0.0000000 | 9 | Asic2 |
| 0.0e+00 | 0.5928127 | 0.851 | 0.070 | 0.0000000 | 9 | Gad1 |
| 0.0e+00 | 0.5917066 | 1.000 | 0.868 | 0.0000000 | 9 | Nrxn3 |
| 0.0e+00 | 0.5894474 | 0.946 | 0.095 | 0.0000000 | 9 | Gad2 |
| 0.0e+00 | 0.5869991 | 0.892 | 0.122 | 0.0000000 | 9 | Ptchd1 |
| 0.0e+00 | 0.5862208 | 0.959 | 0.404 | 0.0000000 | 9 | Fign |
| 0.0e+00 | 0.5701178 | 0.986 | 0.575 | 0.0000000 | 9 | Zfp804a |
| 0.0e+00 | 0.5684483 | 0.959 | 0.513 | 0.0000000 | 9 | Kctd16 |
| 0.0e+00 | 0.5644685 | 0.689 | 0.135 | 0.0000000 | 9 | Sema3e |
| 0.0e+00 | 0.5629603 | 0.878 | 0.319 | 0.0000000 | 9 | Ak5 |
| 0.0e+00 | 0.5987902 | 0.969 | 0.404 | 0.0000000 | 14 | Pcsk2 |
| 0.0e+00 | 0.5748534 | 0.906 | 0.231 | 0.0000000 | 14 | Stxbp6 |
| 0.0e+00 | 0.5567000 | 1.000 | 0.363 | 0.0000000 | 14 | Lef1 |
| 0.0e+00 | 0.5496401 | 1.000 | 0.690 | 0.0000000 | 14 | Ntng1 |
| 0.0e+00 | 0.5329311 | 1.000 | 0.280 | 0.0000000 | 14 | Cpne7 |
| 0.0e+00 | 0.5155800 | 1.000 | 0.491 | 0.0000000 | 14 | Ryr3 |
| 0.0e+00 | 0.4949808 | 0.906 | 0.395 | 0.0000000 | 14 | Cntn3 |
| 0.0e+00 | 0.4879512 | 0.969 | 0.746 | 0.0000007 | 14 | Mgat4c |
| 0.0e+00 | 0.4749312 | 0.969 | 0.720 | 0.0000001 | 14 | Lingo2 |
| 0.0e+00 | 0.4726185 | 0.969 | 0.558 | 0.0000000 | 14 | Efna5 |
| 0.0e+00 | 0.4724820 | 1.000 | 0.492 | 0.0000000 | 14 | Zmat4 |
| 0.0e+00 | 0.4715426 | 1.000 | 0.733 | 0.0000000 | 14 | Syt1 |
| 0.0e+00 | 0.4680961 | 1.000 | 0.762 | 0.0000000 | 14 | Cntnap2 |
| 0.0e+00 | 0.4644152 | 0.906 | 0.280 | 0.0000000 | 14 | 6330411D24Rik |
| 0.0e+00 | 0.4558477 | 0.844 | 0.393 | 0.0000061 | 14 | Adarb2 |
| 0.0e+00 | 0.4508117 | 0.938 | 0.190 | 0.0000000 | 14 | Prox1 |
| 0.0e+00 | 0.4376333 | 0.969 | 0.699 | 0.0000001 | 14 | Sox5 |
| 0.0e+00 | 0.4347482 | 1.000 | 0.819 | 0.0000000 | 14 | Kcnip4 |
| 0.0e+00 | 0.4342838 | 1.000 | 0.895 | 0.0000000 | 14 | Cadm2 |
| 0.0e+00 | 0.4232970 | 1.000 | 0.768 | 0.0000000 | 14 | Lrrc7 |
| 0.0e+00 | 1.3569120 | 0.960 | 0.040 | 0.0000000 | 3 | Cfap299 |
| 0.0e+00 | 1.3376229 | 0.968 | 0.027 | 0.0000000 | 3 | Dnah12 |
| 0.0e+00 | 1.2419726 | 0.952 | 0.102 | 0.0000000 | 3 | Adamts20 |
| 0.0e+00 | 1.1665717 | 0.960 | 0.040 | 0.0000000 | 3 | Dnah6 |
| 0.0e+00 | 1.1267706 | 0.968 | 0.098 | 0.0000000 | 3 | Cfap54 |
| 0.0e+00 | 1.1096854 | 0.968 | 0.117 | 0.0000000 | 3 | Spag16 |
| 0.0e+00 | 1.0928477 | 0.960 | 0.037 | 0.0000000 | 3 | Hydin |
| 0.0e+00 | 1.0834740 | 0.960 | 0.076 | 0.0000000 | 3 | Ccdc162 |
| 0.0e+00 | 1.0808259 | 0.960 | 0.059 | 0.0000000 | 3 | Rgs22 |
| 0.0e+00 | 1.0784167 | 0.968 | 0.056 | 0.0000000 | 3 | Cfap61 |
| 0.0e+00 | 1.0729439 | 0.935 | 0.074 | 0.0000000 | 3 | Cfap44 |
| 0.0e+00 | 1.0546277 | 0.992 | 0.865 | 0.0000000 | 3 | Syne1 |
| 0.0e+00 | 1.0507064 | 0.952 | 0.114 | 0.0000000 | 3 | Gm973 |
| 0.0e+00 | 1.0437030 | 0.968 | 0.052 | 0.0000000 | 3 | Spef2 |
| 0.0e+00 | 1.0400440 | 0.968 | 0.051 | 0.0000000 | 3 | Ak7 |
| 0.0e+00 | 1.0318951 | 0.976 | 0.131 | 0.0000000 | 3 | Dnah9 |
| 0.0e+00 | 1.0267513 | 0.976 | 0.060 | 0.0000000 | 3 | Kif6 |
| 0.0e+00 | 1.0247695 | 0.952 | 0.032 | 0.0000000 | 3 | Ak9 |
| 0.0e+00 | 1.0199062 | 0.952 | 0.043 | 0.0000000 | 3 | Dnah11 |
| 0.0e+00 | 0.9844545 | 0.968 | 0.027 | 0.0000000 | 3 | Cfap65 |
| 0.0e+00 | 1.6589135 | 0.875 | 0.167 | 0.0000000 | 15 | Htr2c |
| 0.0e+00 | 1.4673531 | 1.000 | 0.087 | 0.0000000 | 15 | Ttr |
| 0.0e+00 | 0.9553699 | 1.000 | 0.511 | 0.0000000 | 15 | Wdr17 |
| 0.0e+00 | 0.9178587 | 0.781 | 0.024 | 0.0000000 | 15 | Gmnc |
| 0.0e+00 | 0.8962658 | 0.875 | 0.181 | 0.0000000 | 15 | Enpp2 |
| 0.0e+00 | 0.8825383 | 0.969 | 0.064 | 0.0000000 | 15 | Rbm47 |
| 0.0e+00 | 0.8818117 | 0.875 | 0.233 | 0.0000000 | 15 | Vat1l |
| 0.0e+00 | 0.8731853 | 0.938 | 0.179 | 0.0000000 | 15 | Otx2os1 |
| 0.0e+00 | 0.8621759 | 0.906 | 0.158 | 0.0000000 | 15 | Sulf1 |
| 0.0e+00 | 0.8325165 | 0.938 | 0.637 | 0.0000000 | 15 | Stk39 |
| 0.0e+00 | 0.8169684 | 1.000 | 0.861 | 0.0000000 | 15 | Trpm3 |
| 0.0e+00 | 0.7779916 | 0.969 | 0.219 | 0.0000000 | 15 | C330002G04Rik |
| 0.0e+00 | 0.7569651 | 0.875 | 0.316 | 0.0000000 | 15 | Nhsl2 |
| 0.0e+00 | 0.7382019 | 1.000 | 0.673 | 0.0000000 | 15 | Frmpd4 |
| 0.0e+00 | 0.7310722 | 0.938 | 0.367 | 0.0000000 | 15 | Itpr1 |
| 0.0e+00 | 0.6973879 | 0.781 | 0.207 | 0.0000000 | 15 | Atp2b3 |
| 0.0e+00 | 0.6920626 | 0.875 | 0.169 | 0.0000000 | 15 | Prdm16 |
| 0.0e+00 | 0.6890443 | 0.812 | 0.245 | 0.0000000 | 15 | Cab39l |
| 0.0e+00 | 0.6844702 | 0.969 | 0.567 | 0.0000000 | 15 | Rfx3 |
| 0.0e+00 | 0.6690855 | 0.938 | 0.627 | 0.0000000 | 15 | Tmem108 |
| 0.0e+00 | 0.8211787 | 0.973 | 0.591 | 0.0000000 | 10 | Zfp804b |
| 0.0e+00 | 0.8144616 | 1.000 | 0.813 | 0.0000000 | 10 | Kcnip4 |
| 0.0e+00 | 0.8054150 | 0.644 | 0.074 | 0.0000000 | 10 | Npsr1 |
| 0.0e+00 | 0.6461324 | 1.000 | 0.707 | 0.0000000 | 10 | Cntn5 |
| 0.0e+00 | 0.6316225 | 1.000 | 0.759 | 0.0000000 | 10 | Tenm2 |
| 0.0e+00 | 0.6087797 | 1.000 | 0.578 | 0.0000000 | 10 | Gm20754 |
| 0.0e+00 | 0.6024773 | 0.973 | 0.566 | 0.0000000 | 10 | Kcnq5 |
| 0.0e+00 | 0.5777072 | 0.973 | 0.458 | 0.0000000 | 10 | St6galnac5 |
| 0.0e+00 | 0.5522407 | 0.575 | 0.207 | 0.0000000 | 10 | Pcsk5 |
| 0.0e+00 | 0.5460744 | 0.945 | 0.740 | 0.0000000 | 10 | Mgat4c |
| 0.0e+00 | 0.5258432 | 0.986 | 0.690 | 0.0000000 | 10 | Sox5 |
| 0.0e+00 | 0.5065918 | 0.986 | 0.474 | 0.0000000 | 10 | Cdh8 |
| 0.0e+00 | 0.4842790 | 0.699 | 0.319 | 0.0000000 | 10 | Cntnap5c |
| 0.0e+00 | 0.4796889 | 0.658 | 0.182 | 0.0000000 | 10 | Prox1 |
| 0.0e+00 | 0.4747579 | 0.836 | 0.661 | 0.0000428 | 10 | Brinp3 |
| 0.0e+00 | 0.4657125 | 1.000 | 0.867 | 0.0000000 | 10 | Kcnma1 |
| 0.0e+00 | 0.4578456 | 0.699 | 0.380 | 0.0000000 | 10 | 4930509J09Rik |
| 0.0e+00 | 0.4474035 | 0.808 | 0.326 | 0.0000000 | 10 | Prr16 |
| 0.0e+00 | 0.4269801 | 0.973 | 0.649 | 0.0000000 | 10 | Rasgef1b |
| 0.0e+00 | 0.4182427 | 0.781 | 0.354 | 0.0000000 | 10 | Tafa2 |
| 0.0e+00 | 0.9804342 | 0.950 | 0.265 | 0.0000000 | 11 | 6330411D24Rik |
| 0.0e+00 | 0.8601788 | 0.967 | 0.304 | 0.0000000 | 11 | Pex5l |
| 0.0e+00 | 0.6614258 | 1.000 | 0.488 | 0.0000000 | 11 | Spock1 |
| 0.0e+00 | 0.6572678 | 0.983 | 0.481 | 0.0000000 | 11 | Ryr3 |
| 0.0e+00 | 0.6184067 | 1.000 | 0.756 | 0.0000000 | 11 | Hs6st3 |
| 0.0e+00 | 0.6148524 | 0.917 | 0.381 | 0.0000000 | 11 | Adarb1 |
| 0.0e+00 | 0.6014458 | 0.867 | 0.151 | 0.0000000 | 11 | 4930419G24Rik |
| 0.0e+00 | 0.5629957 | 0.983 | 0.477 | 0.0000000 | 11 | Edil3 |
| 0.0e+00 | 0.5581578 | 1.000 | 0.666 | 0.0000000 | 11 | Frmpd4 |
| 0.0e+00 | 0.5554781 | 0.933 | 0.383 | 0.0000000 | 11 | Cntn3 |
| 0.0e+00 | 0.5453907 | 0.933 | 0.225 | 0.0000000 | 11 | Ptpn3 |
| 0.0e+00 | 0.5336730 | 1.000 | 0.481 | 0.0000000 | 11 | Zmat4 |
| 0.0e+00 | 0.5239501 | 1.000 | 0.856 | 0.0000000 | 11 | Prickle2 |
| 0.0e+00 | 0.5223944 | 0.983 | 0.634 | 0.0000000 | 11 | Cacnb4 |
| 0.0e+00 | 0.5198736 | 1.000 | 0.593 | 0.0000000 | 11 | Zfp804b |
| 0.0e+00 | 0.5068030 | 1.000 | 0.620 | 0.0000000 | 11 | Unc13c |
| 0.0e+00 | 0.5063109 | 0.967 | 0.390 | 0.0000000 | 11 | Epb41l4b |
| 0.0e+00 | 0.5059856 | 0.933 | 0.370 | 0.0000000 | 11 | Zfp385b |
| 0.0e+00 | 0.5027454 | 0.967 | 0.463 | 0.0000000 | 11 | Camk4 |
| 0.0e+00 | 0.5009991 | 0.950 | 0.547 | 0.0000000 | 11 | Pak7 |
markers.logreg %>%
group_by(cluster) %>%
top_n(n = 10, wt = avg_log10FC) -> top10
DoHeatmap(combined.srt, features = top10$gene) + NoLegend()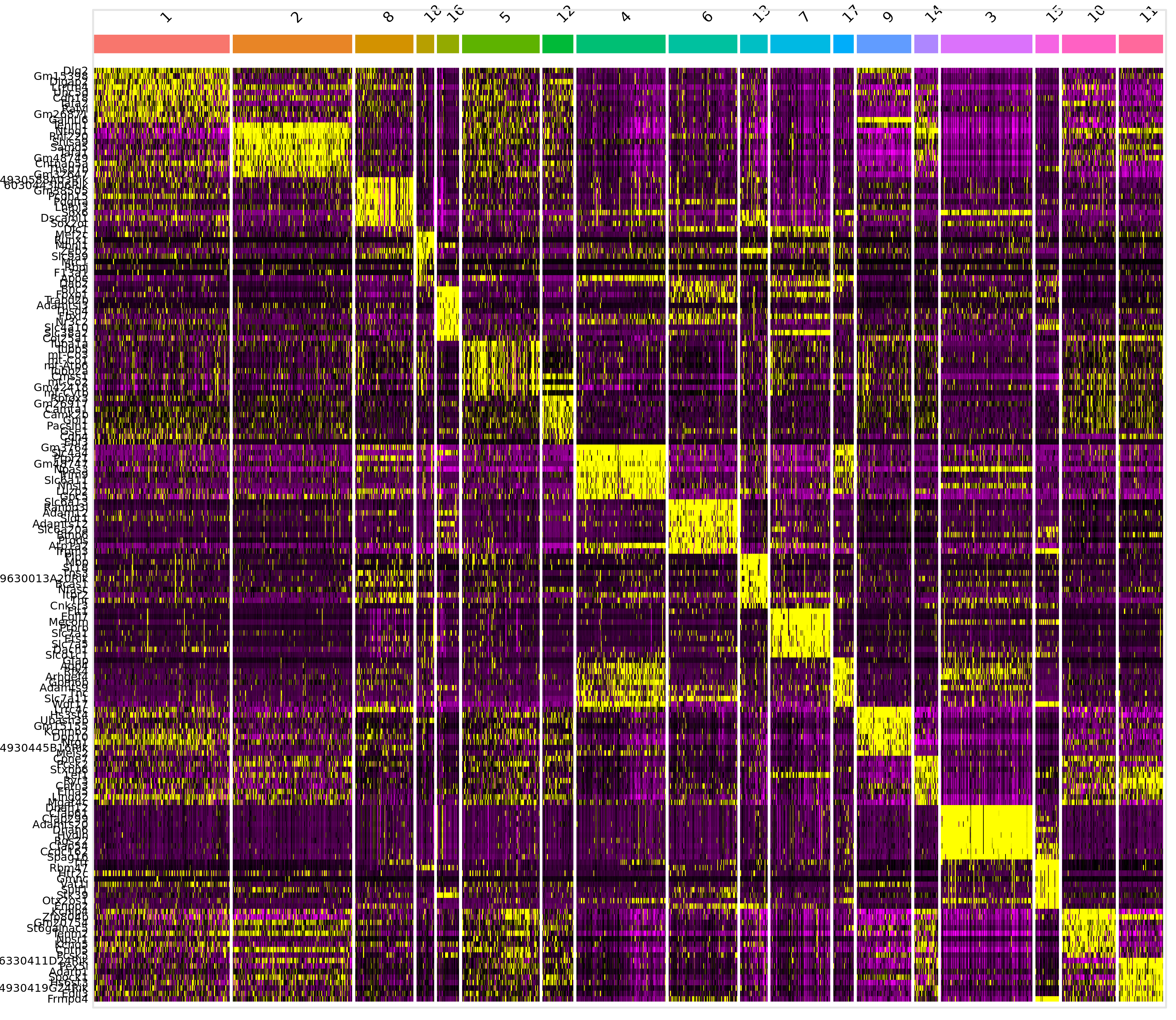
It seems that we should split datasets despite the fact that they’re derived from the same experiment. It should help us to get better embeddings and clusters.
Separate analysis
DefaultAssay(combined.srt) <- "RNA"
srt.list <- SplitObject(combined.srt,
split.by = "orig.ident")
c(THP7, Pr5P7) %<-% srt.listVentrobasal thalamus
n_pcs <- 100
c(THP7, THP7.markers.logreg, THP7.markers.mast) %<-% DeriveKTree(THP7)
|
|======================================================================| 100%
|
| | 0%
|
|= | 1%
|
|= | 2%
|
|== | 2%
|
|== | 3%
|
|== | 4%
|
|=== | 4%
|
|=== | 5%
|
|==== | 5%
|
|==== | 6%
|
|===== | 7%
|
|===== | 8%
|
|====== | 8%
|
|====== | 9%
|
|======= | 10%
|
|======= | 11%
|
|======== | 11%
|
|======== | 12%
|
|========= | 12%
|
|========= | 13%
|
|========== | 14%
|
|=========== | 15%
|
|============ | 17%
|
|============ | 18%
|
|============= | 19%
|
|============== | 20%
|
|=============== | 21%
|
|=============== | 22%
|
|================ | 23%
|
|================= | 24%
|
|================= | 25%
|
|================== | 26%
|
|=================== | 27%
|
|==================== | 28%
|
|==================== | 29%
|
|===================== | 30%
|
|===================== | 31%
|
|====================== | 31%
|
|====================== | 32%
|
|======================= | 32%
|
|======================== | 35%
|
|========================= | 35%
|
|========================= | 36%
|
|========================== | 37%
|
|========================== | 38%
|
|=========================== | 38%
|
|=========================== | 39%
|
|============================ | 39%
|
|============================ | 40%
|
|============================ | 41%
|
|============================= | 41%
|
|============================= | 42%
|
|============================== | 42%
|
|============================== | 43%
|
|================================= | 46%
|
|================================= | 47%
|
|================================= | 48%
|
|================================== | 48%
|
|================================== | 49%
|
|=================================== | 50%
|
|==================================== | 51%
|
|===================================== | 52%
|
|===================================== | 53%
|
|======================================== | 57%
|
|======================================== | 58%
|
|========================================= | 58%
|
|========================================= | 59%
|
|========================================== | 59%
|
|========================================== | 60%
|
|========================================== | 61%
|
|=========================================== | 61%
|
|=========================================== | 62%
|
|============================================ | 62%
|
|============================================ | 63%
|
|============================================= | 64%
|
|============================================= | 65%
|
|============================================== | 65%
|
|============================================== | 66%
|
|=============================================== | 67%
|
|=============================================== | 68%
|
|================================================ | 69%
|
|================================================= | 69%
|
|================================================= | 70%
|
|================================================== | 71%
|
|================================================== | 72%
|
|=================================================== | 73%
|
|==================================================== | 74%
|
|==================================================== | 75%
|
|===================================================== | 75%
|
|===================================================== | 76%
|
|====================================================== | 77%
|
|======================================================= | 78%
|
|======================================================= | 79%
|
|========================================================= | 81%
|
|========================================================= | 82%
|
|========================================================== | 83%
|
|=========================================================== | 84%
|
|=========================================================== | 85%
|
|============================================================ | 85%
|
|============================================================ | 86%
|
|============================================================= | 87%
|
|============================================================== | 88%
|
|============================================================== | 89%
|
|=============================================================== | 89%
|
|=============================================================== | 90%
|
|=============================================================== | 91%
|
|================================================================ | 91%
|
|================================================================ | 92%
|
|================================================================= | 92%
|
|================================================================= | 93%
|
|================================================================== | 94%
|
|================================================================== | 95%
|
|=================================================================== | 95%
|
|=================================================================== | 96%
|
|==================================================================== | 96%
|
|==================================================================== | 97%
|
|==================================================================== | 98%
|
|===================================================================== | 98%
|
|===================================================================== | 99%
|
|======================================================================| 100%THP7.markers.logreg %>%
group_by(cluster) %>%
slice_max(n = 20, order_by = avg_log10FC) %>%
kable("html") %>%
kable_material(
bootstrap_options = c("bordered",
"condensed",
"responsive",
"striped"),
position = "left",
font_size = 14
)| p_val | avg_log10FC | pct.1 | pct.2 | p_val_adj | cluster | gene |
|---|---|---|---|---|---|---|
| 0.0000001 | 0.4921556 | 0.720 | 0.419 | 0.0024499 | 14 | Zfp804b |
| 0.0000004 | 0.4251261 | 0.500 | 0.246 | 0.0095144 | 14 | D030068K23Rik |
| 0.0000000 | 0.3988595 | 0.980 | 0.691 | 0.0000000 | 14 | Ube3a |
| 0.0000000 | 0.3762109 | 0.660 | 0.352 | 0.0000057 | 14 | Stxbp5l |
| 0.0000000 | 0.3660018 | 0.740 | 0.424 | 0.0000503 | 14 | Rmst |
| 0.0000000 | 0.3495472 | 0.860 | 0.688 | 0.0000679 | 14 | Ralyl |
| 0.0000000 | 0.3489561 | 0.940 | 0.684 | 0.0000000 | 14 | Ahi1 |
| 0.0000000 | 0.3433597 | 0.820 | 0.509 | 0.0000003 | 14 | Nrg3os |
| 0.0024840 | 0.3417350 | 0.320 | 0.192 | 1.0000000 | 14 | Meis2 |
| 0.0000000 | 0.3408464 | 0.720 | 0.491 | 0.0004494 | 14 | Gm26871 |
| 0.0000000 | 0.3364576 | 0.900 | 0.575 | 0.0000063 | 14 | Dpp10 |
| 0.0000000 | 0.3350022 | 0.960 | 0.694 | 0.0000000 | 14 | Dlg2 |
| 0.0000000 | 0.3326104 | 0.860 | 0.556 | 0.0000207 | 14 | Cdh18 |
| 0.0000000 | 0.3269318 | 0.980 | 0.666 | 0.0000000 | 14 | Snhg11 |
| 0.0002663 | 0.3248276 | 0.120 | 0.044 | 1.0000000 | 14 | Gm27016 |
| 0.0000000 | 0.3221719 | 0.620 | 0.323 | 0.0006288 | 14 | Chrm3 |
| 0.0000000 | 0.3217724 | 0.760 | 0.372 | 0.0001203 | 14 | Tafa2 |
| 0.0000000 | 0.3165664 | 0.700 | 0.378 | 0.0000164 | 14 | Zfp385b |
| 0.0000911 | 0.3161474 | 0.580 | 0.405 | 1.0000000 | 14 | Sorcs1 |
| 0.0000002 | 0.3146110 | 0.540 | 0.231 | 0.0045314 | 14 | B230217J21Rik |
| 0.0000000 | 0.6262359 | 0.847 | 0.540 | 0.0000000 | 8 | Galntl6 |
| 0.0000000 | 0.5807242 | 0.847 | 0.371 | 0.0000000 | 8 | Gm15398 |
| 0.0000000 | 0.5482611 | 0.847 | 0.361 | 0.0000000 | 8 | Unc5d |
| 0.0000000 | 0.5424823 | 0.917 | 0.340 | 0.0000000 | 8 | Tafa2 |
| 0.0000000 | 0.4523585 | 1.000 | 0.646 | 0.0000000 | 8 | Nrg1 |
| 0.0000000 | 0.4513830 | 0.625 | 0.185 | 0.0000000 | 8 | Klhl1 |
| 0.0000000 | 0.4473121 | 0.972 | 0.554 | 0.0000000 | 8 | Dpp10 |
| 0.0000000 | 0.4396150 | 0.875 | 0.358 | 0.0000000 | 8 | Spock3 |
| 0.0000000 | 0.4330138 | 0.944 | 0.535 | 0.0000039 | 8 | Cdh18 |
| 0.0000000 | 0.4278080 | 0.792 | 0.387 | 0.0000008 | 8 | Cdh8 |
| 0.0000000 | 0.4257400 | 0.806 | 0.405 | 0.0000001 | 8 | Rmst |
| 0.0000000 | 0.4233159 | 0.736 | 0.176 | 0.0000000 | 8 | Vwc2l |
| 0.0000000 | 0.4124411 | 0.931 | 0.315 | 0.0000000 | 8 | Dlgap2 |
| 0.0000000 | 0.4116109 | 0.806 | 0.189 | 0.0000000 | 8 | B230217J21Rik |
| 0.0000000 | 0.4106594 | 0.944 | 0.608 | 0.0000000 | 8 | Asic2 |
| 0.0000000 | 0.4016633 | 0.889 | 0.332 | 0.0000000 | 8 | Grin2a |
| 0.0000000 | 0.3945054 | 0.972 | 0.621 | 0.0000000 | 8 | Lingo2 |
| 0.0000000 | 0.3917071 | 1.000 | 0.634 | 0.0000000 | 8 | Sgcz |
| 0.0000000 | 0.3906737 | 0.903 | 0.393 | 0.0000000 | 8 | Esrrg |
| 0.0000000 | 0.3891959 | 0.917 | 0.475 | 0.0000000 | 8 | Galnt13 |
| 0.0000012 | 0.4458096 | 0.222 | 0.055 | 0.0262961 | 7 | Gm42397 |
| 0.0000002 | 0.4123405 | 0.889 | 0.398 | 0.0048643 | 7 | Kif26b |
| 0.0000000 | 0.4099479 | 1.000 | 0.670 | 0.0000805 | 7 | Nrg1 |
| 0.0000448 | 0.3853220 | 0.852 | 0.394 | 0.9490102 | 7 | Unc5d |
| 0.0042007 | 0.3580457 | 0.556 | 0.167 | 1.0000000 | 7 | Adamts18 |
| 0.0000605 | 0.3493007 | 0.815 | 0.326 | 1.0000000 | 7 | Chrm3 |
| 0.0000619 | 0.3140704 | 0.778 | 0.333 | 1.0000000 | 7 | Kcnmb2 |
| 0.0000037 | 0.3128392 | 0.926 | 0.482 | 0.0792079 | 7 | Pcdh15 |
| 0.0006535 | 0.2916253 | 0.815 | 0.383 | 1.0000000 | 7 | Tafa2 |
| 0.0003387 | 0.2912035 | 0.852 | 0.403 | 1.0000000 | 7 | Gm15398 |
| 0.0000770 | 0.2876327 | 0.926 | 0.483 | 1.0000000 | 7 | Cdr1os |
| 0.0008917 | 0.2833309 | 0.889 | 0.633 | 1.0000000 | 7 | Asic2 |
| 0.0019770 | 0.2819749 | 0.815 | 0.352 | 1.0000000 | 7 | Cntnap4 |
| 0.0000037 | 0.2805227 | 0.926 | 0.352 | 0.0775085 | 7 | Stxbp5l |
| 0.0017438 | 0.2789436 | 0.704 | 0.367 | 1.0000000 | 7 | Ebf1 |
| 0.0072863 | 0.2754964 | 0.481 | 0.173 | 1.0000000 | 7 | 4930419G24Rik |
| 0.0000022 | 0.2722620 | 1.000 | 0.827 | 0.0475139 | 7 | Fgf14 |
| 0.0006842 | 0.2712338 | 0.889 | 0.568 | 1.0000000 | 7 | Kcnq5 |
| 0.0003553 | 0.2676954 | 0.926 | 0.576 | 1.0000000 | 7 | Pde4dip |
| 0.0009787 | 0.2652826 | 0.778 | 0.374 | 1.0000000 | 7 | Grin2a |
| 0.0000000 | 0.7242156 | 1.000 | 0.526 | 0.0000000 | 1 | Ntng1 |
| 0.0000000 | 0.6576643 | 0.992 | 0.490 | 0.0000000 | 1 | Rnf220 |
| 0.0000000 | 0.6574206 | 1.000 | 0.524 | 0.0000000 | 1 | Tenm1 |
| 0.0000000 | 0.6349539 | 0.992 | 0.400 | 0.0000000 | 1 | Samd5 |
| 0.0000000 | 0.6175479 | 0.962 | 0.544 | 0.0000000 | 1 | Shisa9 |
| 0.0000000 | 0.6069198 | 0.977 | 0.497 | 0.0000000 | 1 | Tafa1 |
| 0.0000000 | 0.5998934 | 0.879 | 0.306 | 0.0000000 | 1 | Gm48749 |
| 0.0000000 | 0.5487415 | 0.780 | 0.295 | 0.0000000 | 1 | 6330411D24Rik |
| 0.0000000 | 0.5436846 | 0.841 | 0.319 | 0.0000000 | 1 | Zmat4 |
| 0.0000000 | 0.5429387 | 0.970 | 0.510 | 0.0000000 | 1 | Egfem1 |
| 0.0000000 | 0.5371066 | 0.879 | 0.350 | 0.0000000 | 1 | Cdh6 |
| 0.0000000 | 0.5180337 | 0.924 | 0.395 | 0.0000000 | 1 | Nell1 |
| 0.0000000 | 0.5174248 | 1.000 | 0.546 | 0.0000000 | 1 | Syt1 |
| 0.0000000 | 0.5077017 | 0.902 | 0.420 | 0.0000000 | 1 | Thsd7b |
| 0.0000000 | 0.5036508 | 0.992 | 0.505 | 0.0000000 | 1 | Tox |
| 0.0000000 | 0.5006324 | 0.932 | 0.414 | 0.0000000 | 1 | Nell2 |
| 0.0000000 | 0.4791969 | 0.992 | 0.580 | 0.0000000 | 1 | Rims1 |
| 0.0000000 | 0.4772918 | 1.000 | 0.798 | 0.0000000 | 1 | Ptprd |
| 0.0000000 | 0.4757209 | 0.886 | 0.315 | 0.0000000 | 1 | Kcnh8 |
| 0.0000000 | 0.4727737 | 0.939 | 0.497 | 0.0000000 | 1 | Hs3st5 |
| 0.0000000 | 0.7071187 | 0.444 | 0.167 | 0.0000000 | 6 | mt-Co3 |
| 0.0000000 | 0.5993005 | 0.841 | 0.516 | 0.0000000 | 6 | Tuba1a |
| 0.0000000 | 0.5847852 | 0.444 | 0.117 | 0.0000000 | 6 | mt-Atp6 |
| 0.0000000 | 0.5390167 | 0.413 | 0.114 | 0.0000313 | 6 | mt-Co2 |
| 0.0000000 | 0.5275187 | 0.540 | 0.204 | 0.0000001 | 6 | mt-Co1 |
| 0.0000118 | 0.5169106 | 0.857 | 0.718 | 0.2492124 | 6 | Cmss1 |
| 0.0000000 | 0.5099136 | 0.508 | 0.287 | 0.0000006 | 6 | Actg1 |
| 0.0000000 | 0.5036783 | 0.698 | 0.327 | 0.0000000 | 6 | Tubb5 |
| 0.0000000 | 0.4942592 | 0.603 | 0.325 | 0.0000000 | 6 | Tubb2b |
| 0.0000000 | 0.4800841 | 0.619 | 0.277 | 0.0000000 | 6 | Atp6v0c |
| 0.0000000 | 0.4781357 | 0.952 | 0.837 | 0.0010250 | 6 | Gm42418 |
| 0.0000000 | 0.4762985 | 0.794 | 0.492 | 0.0000000 | 6 | Calm1 |
| 0.0000000 | 0.4712774 | 0.619 | 0.365 | 0.0000002 | 6 | Hspa8 |
| 0.0000000 | 0.4675408 | 0.667 | 0.377 | 0.0000000 | 6 | Tubb2a |
| 0.0000000 | 0.4546950 | 0.476 | 0.200 | 0.0000320 | 6 | Tmsb10 |
| 0.0000000 | 0.4368420 | 0.333 | 0.085 | 0.0007012 | 6 | mt-Nd2 |
| 0.0000002 | 0.4167998 | 0.460 | 0.178 | 0.0042058 | 6 | Tmsb4x |
| 0.0000000 | 0.4095981 | 0.841 | 0.556 | 0.0000000 | 6 | Gap43 |
| 0.0000000 | 0.3995666 | 0.492 | 0.163 | 0.0000000 | 6 | Dynll2 |
| 0.0000000 | 0.3891660 | 0.333 | 0.088 | 0.0000074 | 6 | Tuba1b |
| 0.0000000 | 1.2395835 | 0.755 | 0.054 | 0.0000000 | 4 | Flt1 |
| 0.0000000 | 1.0596388 | 0.679 | 0.057 | 0.0000000 | 4 | Mecom |
| 0.0000000 | 1.0042969 | 0.736 | 0.226 | 0.0000000 | 4 | Dach1 |
| 0.0000000 | 0.9449069 | 0.868 | 0.055 | 0.0000000 | 4 | Fli1 |
| 0.0000000 | 0.9401421 | 0.660 | 0.079 | 0.0000000 | 4 | Slc7a5 |
| 0.0000000 | 0.8640453 | 0.660 | 0.181 | 0.0000000 | 4 | Slc7a1 |
| 0.0000000 | 0.8515687 | 0.623 | 0.025 | 0.0000000 | 4 | Egfl7 |
| 0.0000000 | 0.8413374 | 0.642 | 0.058 | 0.0000000 | 4 | Slco1c1 |
| 0.0000000 | 0.8347802 | 0.717 | 0.159 | 0.0000000 | 4 | Ccdc141 |
| 0.0000000 | 0.8246317 | 0.623 | 0.068 | 0.0000000 | 4 | Ets1 |
| 0.0000000 | 0.8175285 | 0.698 | 0.011 | 0.0000000 | 4 | Cldn5 |
| 0.0000007 | 0.8075258 | 0.170 | 0.063 | 0.0142100 | 4 | F13a1 |
| 0.0000000 | 0.7978844 | 0.623 | 0.035 | 0.0000000 | 4 | Ptprb |
| 0.0000000 | 0.7969139 | 0.151 | 0.019 | 0.0005690 | 4 | Mrc1 |
| 0.0000006 | 0.7737853 | 0.491 | 0.289 | 0.0117230 | 4 | Rbpj |
| 0.0000000 | 0.7697953 | 0.623 | 0.035 | 0.0000000 | 4 | Adgrl4 |
| 0.0000000 | 0.7664865 | 0.755 | 0.229 | 0.0000000 | 4 | Bsg |
| 0.0000000 | 0.7630175 | 0.604 | 0.068 | 0.0000000 | 4 | Apcdd1 |
| 0.0000000 | 0.7608081 | 0.868 | 0.405 | 0.0000000 | 4 | Dlc1 |
| 0.0000000 | 0.7469235 | 0.604 | 0.077 | 0.0000000 | 4 | Itga1 |
| 0.0000000 | 0.8289105 | 0.971 | 0.698 | 0.0000000 | 18 | Gm26917 |
| 0.0000022 | 0.7021306 | 0.286 | 0.077 | 0.0466847 | 18 | Ebf3 |
| 0.0000000 | 0.6773262 | 0.886 | 0.529 | 0.0000000 | 18 | Rbfox3 |
| 0.0000000 | 0.5832218 | 0.800 | 0.347 | 0.0000000 | 18 | Dgkz |
| 0.0000000 | 0.5730225 | 0.771 | 0.540 | 0.0000003 | 18 | Gpi1 |
| 0.0000000 | 0.5687922 | 0.743 | 0.494 | 0.0000000 | 18 | Atp2b2 |
| 0.0000000 | 0.5547723 | 0.629 | 0.479 | 0.0000192 | 18 | Cdh4 |
| 0.0000000 | 0.5455832 | 0.571 | 0.328 | 0.0003714 | 18 | Matk |
| 0.0000000 | 0.5342803 | 0.943 | 0.753 | 0.0000000 | 18 | Camta1 |
| 0.0000000 | 0.5330145 | 0.943 | 0.842 | 0.0000000 | 18 | Gm42418 |
| 0.0000000 | 0.5300448 | 0.857 | 0.454 | 0.0000000 | 18 | Camk2b |
| 0.0000000 | 0.5269742 | 0.657 | 0.311 | 0.0000026 | 18 | Plec |
| 0.0000000 | 0.5268009 | 0.743 | 0.402 | 0.0000002 | 18 | Ank1 |
| 0.0000000 | 0.5197668 | 0.657 | 0.275 | 0.0000000 | 18 | Pde4a |
| 0.0000000 | 0.5107268 | 0.857 | 0.411 | 0.0000000 | 18 | Tspoap1 |
| 0.0000000 | 0.5055357 | 0.657 | 0.374 | 0.0000000 | 18 | Cacna2d2 |
| 0.0000000 | 0.5029930 | 0.771 | 0.515 | 0.0000000 | 18 | Xkr6 |
| 0.0000000 | 0.4993049 | 0.743 | 0.505 | 0.0000009 | 18 | Slc4a3 |
| 0.0000000 | 0.4960239 | 0.829 | 0.600 | 0.0000000 | 18 | Kif1a |
| 0.0000000 | 0.4903034 | 0.657 | 0.373 | 0.0000067 | 18 | Caskin1 |
| 0.0066465 | 0.9080376 | 0.800 | 0.584 | 1.0000000 | 17 | Ptpn4 |
| 0.0018383 | 0.7928303 | 0.800 | 0.479 | 1.0000000 | 17 | Ryr3 |
| 0.0002136 | 0.7275093 | 0.600 | 0.065 | 1.0000000 | 17 | Ttn |
| 0.0030286 | 0.6985545 | 0.800 | 0.519 | 1.0000000 | 17 | 1700025G04Rik |
| 0.0000119 | 0.6800666 | 0.800 | 0.323 | 0.2522807 | 17 | Cdh7 |
| 0.0038616 | 0.6619007 | 0.800 | 0.449 | 1.0000000 | 17 | Kcnab1 |
| 0.0043203 | 0.6615400 | 0.800 | 0.292 | 1.0000000 | 17 | Lef1 |
| 0.0010939 | 0.6294186 | 0.600 | 0.260 | 1.0000000 | 17 | Dnajc11 |
| 0.0000008 | 0.6293353 | 1.000 | 0.740 | 0.0172589 | 17 | Agap1 |
| 0.0001495 | 0.5725820 | 1.000 | 0.614 | 1.0000000 | 17 | Ntng1 |
| 0.0082341 | 0.5679669 | 1.000 | 0.601 | 1.0000000 | 17 | Cntnap5a |
| 0.0047902 | 0.5551751 | 1.000 | 0.827 | 1.0000000 | 17 | Ntm |
| 0.0008758 | 0.5421623 | 1.000 | 0.600 | 1.0000000 | 17 | Mgat5 |
| 0.0090303 | 0.5300646 | 0.200 | 0.018 | 1.0000000 | 17 | Car12 |
| 0.0038889 | 0.5294053 | 0.800 | 0.302 | 1.0000000 | 17 | Cfap36 |
| 0.0029751 | 0.5292644 | 0.600 | 0.229 | 1.0000000 | 17 | Ptdss1 |
| 0.0000115 | 0.5254941 | 0.600 | 0.117 | 0.2444946 | 17 | Spns1 |
| 0.0046333 | 0.5212267 | 1.000 | 0.570 | 1.0000000 | 17 | Khdrbs2 |
| 0.0004305 | 0.5060974 | 1.000 | 0.343 | 1.0000000 | 17 | Coro2b |
| 0.0013712 | 0.5031579 | 1.000 | 0.543 | 1.0000000 | 17 | Unc80 |
| 0.0000000 | 1.5120877 | 0.967 | 0.123 | 0.0000000 | 20 | Bnc2 |
| 0.0000000 | 1.1812980 | 0.833 | 0.097 | 0.0000000 | 20 | Adamtsl3 |
| 0.0000000 | 1.1224331 | 0.900 | 0.166 | 0.0000000 | 20 | Thsd4 |
| 0.0000000 | 1.0866637 | 0.800 | 0.329 | 0.0000000 | 20 | Col25a1 |
| 0.0000000 | 1.0770827 | 0.867 | 0.507 | 0.0000000 | 20 | Slc4a10 |
| 0.0000000 | 1.0668271 | 0.900 | 0.058 | 0.0000000 | 20 | Trabd2b |
| 0.0000000 | 1.0417482 | 0.967 | 0.414 | 0.0000000 | 20 | Slc38a2 |
| 0.0000000 | 1.0247902 | 0.967 | 0.598 | 0.0000000 | 20 | Foxp1 |
| 0.0000000 | 1.0145583 | 0.967 | 0.473 | 0.0000000 | 20 | Fbxl7 |
| 0.0000000 | 0.9948796 | 0.700 | 0.003 | 0.0000000 | 20 | Slc47a1 |
| 0.0000000 | 0.9929258 | 0.800 | 0.247 | 0.0000000 | 20 | Itgbl1 |
| 0.0000000 | 0.9687133 | 0.900 | 0.374 | 0.0000000 | 20 | Nr3c2 |
| 0.0000000 | 0.9654554 | 0.800 | 0.084 | 0.0000000 | 20 | Dock5 |
| 0.0000000 | 0.9386882 | 0.900 | 0.362 | 0.0000000 | 20 | Gulp1 |
| 0.0000000 | 0.9205507 | 0.567 | 0.105 | 0.0000000 | 20 | Crispld1 |
| 0.0000000 | 0.9158102 | 0.867 | 0.315 | 0.0000000 | 20 | Sh3pxd2a |
| 0.0000000 | 0.9059161 | 0.933 | 0.218 | 0.0000000 | 20 | Hmcn1 |
| 0.0000000 | 0.8957423 | 0.867 | 0.282 | 0.0000000 | 20 | Slit2 |
| 0.0000000 | 0.8901833 | 0.900 | 0.152 | 0.0000000 | 20 | Eya2 |
| 0.0000000 | 0.8734660 | 0.900 | 0.473 | 0.0000000 | 20 | Tmtc1 |
| 0.0000000 | 1.1186122 | 1.000 | 0.313 | 0.0000000 | 3 | Gm3764 |
| 0.0000000 | 0.9860336 | 0.914 | 0.420 | 0.0000000 | 3 | Wdr17 |
| 0.0000000 | 0.9623464 | 0.983 | 0.405 | 0.0000000 | 3 | Slc4a4 |
| 0.0000000 | 0.9476649 | 1.000 | 0.609 | 0.0000000 | 3 | Npas3 |
| 0.0000000 | 0.9266210 | 1.000 | 0.525 | 0.0000000 | 3 | Ptprz1 |
| 0.0000000 | 0.8986149 | 1.000 | 0.417 | 0.0000000 | 3 | Gm48747 |
| 0.0000000 | 0.8928927 | 0.862 | 0.237 | 0.0000000 | 3 | Slc6a11 |
| 0.0000000 | 0.8832488 | 0.931 | 0.315 | 0.0000000 | 3 | Nhsl1 |
| 0.0000000 | 0.8102532 | 0.983 | 0.607 | 0.0000000 | 3 | Luzp2 |
| 0.0000000 | 0.7795485 | 0.862 | 0.210 | 0.0000000 | 3 | Lrig1 |
| 0.0000000 | 0.7663200 | 0.724 | 0.172 | 0.0000000 | 3 | Tnfaip8 |
| 0.0000000 | 0.7654431 | 0.810 | 0.211 | 0.0000000 | 3 | Bmpr1b |
| 0.0000000 | 0.7609677 | 0.879 | 0.424 | 0.0000000 | 3 | Slc1a2 |
| 0.0000000 | 0.7403526 | 0.569 | 0.135 | 0.0000000 | 3 | Tnc |
| 0.0000000 | 0.7354705 | 0.931 | 0.572 | 0.0000000 | 3 | Grm3 |
| 0.0000000 | 0.7120208 | 0.897 | 0.339 | 0.0000000 | 3 | Mir9-3hg |
| 0.0000000 | 0.7103010 | 0.897 | 0.482 | 0.0000000 | 3 | Trim9 |
| 0.0000000 | 0.7035664 | 0.776 | 0.369 | 0.0000000 | 3 | Sfxn5 |
| 0.0000000 | 0.7020843 | 0.845 | 0.277 | 0.0000000 | 3 | Sparcl1 |
| 0.0000000 | 0.6743951 | 0.603 | 0.075 | 0.0000000 | 3 | Acsbg1 |
| 0.0000000 | 1.4989001 | 0.673 | 0.094 | 0.0000000 | 9 | Ptgds |
| 0.0000000 | 1.3620210 | 0.962 | 0.076 | 0.0000000 | 9 | Ranbp3l |
| 0.0000000 | 1.2480266 | 0.904 | 0.072 | 0.0000000 | 9 | Slc6a20a |
| 0.0000000 | 1.1642027 | 0.904 | 0.235 | 0.0000000 | 9 | Sidt1 |
| 0.0000000 | 1.1632862 | 0.827 | 0.806 | 0.0000000 | 9 | Trpm3 |
| 0.0000000 | 1.1025386 | 0.846 | 0.107 | 0.0000000 | 9 | Bmp6 |
| 0.0000000 | 1.0917575 | 0.942 | 0.044 | 0.0000000 | 9 | Slc6a13 |
| 0.0000000 | 1.0891028 | 0.923 | 0.304 | 0.0000000 | 9 | Adam12 |
| 0.0000000 | 1.0529576 | 0.923 | 0.243 | 0.0000000 | 9 | Bicc1 |
| 0.0000000 | 1.0292238 | 0.788 | 0.162 | 0.0000000 | 9 | Adamts12 |
| 0.0000000 | 1.0070606 | 0.962 | 0.332 | 0.0000000 | 9 | Atp1a2 |
| 0.0000000 | 0.9796699 | 0.808 | 0.184 | 0.0000000 | 9 | Lrmda |
| 0.0000000 | 0.9779893 | 0.750 | 0.191 | 0.0000000 | 9 | Slc7a11 |
| 0.0000000 | 0.9732712 | 0.827 | 0.235 | 0.0000000 | 9 | Sned1 |
| 0.0000000 | 0.9700557 | 0.904 | 0.235 | 0.0000000 | 9 | Tmtc4 |
| 0.0000000 | 0.9605894 | 0.750 | 0.649 | 0.0000000 | 9 | Nnat |
| 0.0000000 | 0.9415528 | 0.962 | 0.126 | 0.0000000 | 9 | Arhgap29 |
| 0.0000000 | 0.9385549 | 0.885 | 0.225 | 0.0000000 | 9 | Pdzrn3 |
| 0.0000000 | 0.8939220 | 0.788 | 0.099 | 0.0000000 | 9 | Eya1 |
| 0.0000000 | 0.8631456 | 0.865 | 0.137 | 0.0000000 | 9 | Colec12 |
| 0.0000001 | 0.2059569 | 0.714 | 0.173 | 0.0013968 | 21 | Enpp2 |
| 0.0000000 | 1.0896584 | 0.829 | 0.133 | 0.0000000 | 11 | Gm38505 |
| 0.0000000 | 1.0376620 | 0.971 | 0.284 | 0.0000000 | 11 | Sox6 |
| 0.0000000 | 1.0090739 | 0.829 | 0.229 | 0.0000000 | 11 | 4930588A03Rik |
| 0.0000000 | 0.8441985 | 0.886 | 0.393 | 0.0000000 | 11 | Sox2ot |
| 0.0000000 | 0.8221941 | 0.943 | 0.475 | 0.0000000 | 11 | Pcdh15 |
| 0.0000000 | 0.8131103 | 0.686 | 0.153 | 0.0000000 | 11 | Pdgfra |
| 0.0000000 | 0.7985575 | 0.714 | 0.100 | 0.0000000 | 11 | Sox10 |
| 0.0000000 | 0.7558943 | 0.914 | 0.311 | 0.0000000 | 11 | Dscaml1 |
| 0.0000000 | 0.7476119 | 0.486 | 0.118 | 0.0000004 | 11 | Bcas1 |
| 0.0000000 | 0.7460156 | 0.857 | 0.509 | 0.0000000 | 11 | Lhfpl3 |
| 0.0000000 | 0.7265405 | 0.829 | 0.511 | 0.0000000 | 11 | 6030443J06Rik |
| 0.0000000 | 0.7215268 | 0.857 | 0.416 | 0.0000000 | 11 | Tnr |
| 0.0000000 | 0.6465059 | 0.600 | 0.224 | 0.0000000 | 11 | B9d1 |
| 0.0000000 | 0.6443401 | 0.714 | 0.472 | 0.0000000 | 11 | Epn2 |
| 0.0000000 | 0.6434330 | 0.771 | 0.244 | 0.0000000 | 11 | Megf11 |
| 0.0000000 | 0.6422749 | 0.771 | 0.235 | 0.0000000 | 11 | Arhgef3 |
| 0.0000000 | 0.6271295 | 0.914 | 0.480 | 0.0000000 | 11 | Xylt1 |
| 0.0000000 | 0.6162602 | 0.771 | 0.337 | 0.0000000 | 11 | Gm4876 |
| 0.0000000 | 0.6036030 | 0.829 | 0.546 | 0.0000000 | 11 | Pcdh11x |
| 0.0000000 | 0.5953035 | 0.629 | 0.097 | 0.0000000 | 11 | Prkcq |
| 0.0000707 | 0.4417175 | 1.000 | 0.405 | 1.0000000 | 5 | Gm48749 |
| 0.0000847 | 0.3532611 | 1.000 | 0.608 | 1.0000000 | 5 | Tenm1 |
| 0.0000038 | 0.3519949 | 1.000 | 0.724 | 0.0796981 | 5 | Grik2 |
| 0.0001373 | 0.3508967 | 1.000 | 0.496 | 1.0000000 | 5 | Spock1 |
| 0.0000451 | 0.3380302 | 1.000 | 0.579 | 0.9550882 | 5 | Rnf220 |
| 0.0001381 | 0.3282852 | 1.000 | 0.620 | 1.0000000 | 5 | Cntn4 |
| 0.0018579 | 0.3278412 | 0.846 | 0.280 | 1.0000000 | 5 | Gm48727 |
| 0.0034638 | 0.3276071 | 0.769 | 0.298 | 1.0000000 | 5 | Nell1os |
| 0.0001026 | 0.3273768 | 1.000 | 0.322 | 1.0000000 | 5 | Gm42303 |
| 0.0001856 | 0.3237748 | 1.000 | 0.610 | 1.0000000 | 5 | Ntng1 |
| 0.0002390 | 0.3208237 | 1.000 | 0.550 | 1.0000000 | 5 | Gm20754 |
| 0.0002175 | 0.3081936 | 0.846 | 0.251 | 1.0000000 | 5 | Gm15414 |
| 0.0010475 | 0.3052291 | 1.000 | 0.487 | 1.0000000 | 5 | Nell1 |
| 0.0002409 | 0.2996319 | 1.000 | 0.464 | 1.0000000 | 5 | Srrm4 |
| 0.0078936 | 0.2985181 | 0.846 | 0.285 | 1.0000000 | 5 | Lef1 |
| 0.0030178 | 0.2867084 | 0.846 | 0.374 | 1.0000000 | 5 | Kitl |
| 0.0052808 | 0.2844606 | 1.000 | 0.582 | 1.0000000 | 5 | Tafa1 |
| 0.0066094 | 0.2830246 | 0.923 | 0.409 | 1.0000000 | 5 | Zmat4 |
| 0.0009849 | 0.2700701 | 1.000 | 0.504 | 1.0000000 | 5 | Samd5 |
| 0.0019040 | 0.2684313 | 1.000 | 0.521 | 1.0000000 | 5 | Kcnc2 |
| 0.0000000 | 1.1856605 | 1.000 | 0.047 | 0.0000000 | 15 | Gfap |
| 0.0000000 | 1.0664641 | 0.900 | 0.047 | 0.0000000 | 15 | Mamdc2 |
| 0.0000000 | 1.0243340 | 0.900 | 0.137 | 0.0000000 | 15 | Aqp4 |
| 0.0000000 | 0.9577662 | 0.800 | 0.009 | 0.0000000 | 15 | Myoc |
| 0.0000000 | 0.9200572 | 1.000 | 0.160 | 0.0000084 | 15 | Tnc |
| 0.0000000 | 0.9134502 | 0.900 | 0.272 | 0.0000000 | 15 | Arhgef4 |
| 0.0000000 | 0.9045121 | 0.900 | 0.199 | 0.0000157 | 15 | Adamts9 |
| 0.0000000 | 0.8344536 | 1.000 | 0.188 | 0.0000011 | 15 | Rfx4 |
| 0.0000000 | 0.8197856 | 0.900 | 0.013 | 0.0000000 | 15 | Hopx |
| 0.0000000 | 0.8077459 | 1.000 | 0.130 | 0.0000002 | 15 | Ednrb |
| 0.0000111 | 0.7891101 | 0.800 | 0.312 | 0.2349650 | 15 | Apoe |
| 0.0000000 | 0.7735977 | 1.000 | 0.569 | 0.0000000 | 15 | Gpm6b |
| 0.0000144 | 0.7581740 | 0.600 | 0.151 | 0.3045947 | 15 | Dbi |
| 0.0000000 | 0.7523044 | 0.800 | 0.155 | 0.0001182 | 15 | Glis3 |
| 0.0000035 | 0.7371467 | 0.900 | 0.455 | 0.0745124 | 15 | Wdr17 |
| 0.0000000 | 0.7209024 | 0.800 | 0.173 | 0.0003054 | 15 | Igsf11 |
| 0.0000000 | 0.7156398 | 0.900 | 0.316 | 0.0009103 | 15 | Sparcl1 |
| 0.0000000 | 0.7138396 | 0.900 | 0.440 | 0.0000007 | 15 | Ablim2 |
| 0.0000029 | 0.7127229 | 0.800 | 0.205 | 0.0621272 | 15 | Gm29683 |
| 0.0000000 | 0.7071319 | 0.900 | 0.090 | 0.0000307 | 15 | Cd9 |
| 0.0000000 | 1.5949065 | 1.000 | 0.220 | 0.0000000 | 16 | Mbp |
| 0.0000000 | 1.4115742 | 1.000 | 0.092 | 0.0000000 | 16 | Plp1 |
| 0.0000000 | 1.3342487 | 0.786 | 0.184 | 0.0000000 | 16 | 9630013A20Rik |
| 0.0000000 | 1.3102788 | 0.929 | 0.059 | 0.0000000 | 16 | St18 |
| 0.0000000 | 1.1614454 | 0.429 | 0.018 | 0.0000024 | 16 | Mobp |
| 0.0000000 | 1.1103252 | 0.643 | 0.095 | 0.0000000 | 16 | Cnksr3 |
| 0.0000000 | 1.0875635 | 0.857 | 0.215 | 0.0000000 | 16 | Tns3 |
| 0.0000000 | 1.0856082 | 0.857 | 0.122 | 0.0000000 | 16 | Bcas1 |
| 0.0000000 | 1.0665966 | 0.857 | 0.059 | 0.0000000 | 16 | Bcas1os2 |
| 0.0000000 | 1.0493521 | 0.643 | 0.024 | 0.0000000 | 16 | Mag |
| 0.0000000 | 1.0269588 | 1.000 | 0.361 | 0.0000000 | 16 | Pacrg |
| 0.0000000 | 1.0085917 | 0.857 | 0.597 | 0.0000000 | 16 | Nfasc |
| 0.0000001 | 0.9895753 | 0.714 | 0.432 | 0.0017416 | 16 | Tnr |
| 0.0000000 | 0.9767866 | 0.786 | 0.036 | 0.0000000 | 16 | Enpp6 |
| 0.0000000 | 0.9584172 | 0.786 | 0.193 | 0.0000000 | 16 | Dusp15 |
| 0.0000000 | 0.9226141 | 0.929 | 0.322 | 0.0000000 | 16 | Itpr2 |
| 0.0000000 | 0.9079627 | 0.786 | 0.025 | 0.0000000 | 16 | 1700047M11Rik |
| 0.0000000 | 0.9038872 | 0.857 | 0.288 | 0.0000000 | 16 | Sirt2 |
| 0.0000000 | 0.8956121 | 0.857 | 0.116 | 0.0000000 | 16 | Sox10 |
| 0.0000000 | 0.8394800 | 0.929 | 0.639 | 0.0000000 | 16 | Fyn |
| 0.0026080 | 0.5572294 | 1.000 | 0.329 | 1.0000000 | 10 | Lars2 |
| 0.0000771 | 0.5442207 | 1.000 | 0.452 | 1.0000000 | 10 | Slc4a4 |
| 0.0004410 | 0.5373239 | 1.000 | 0.136 | 1.0000000 | 10 | C030005K06Rik |
| 0.0008089 | 0.5003299 | 1.000 | 0.640 | 1.0000000 | 10 | Npas3 |
| 0.0004744 | 0.5003187 | 1.000 | 0.563 | 1.0000000 | 10 | Ptprz1 |
| 0.0002506 | 0.4944531 | 1.000 | 0.364 | 1.0000000 | 10 | Nhsl1 |
| 0.0004323 | 0.4938599 | 1.000 | 0.287 | 1.0000000 | 10 | Slc6a11 |
| 0.0059256 | 0.4824386 | 1.000 | 0.442 | 1.0000000 | 10 | Rtl4 |
| 0.0020579 | 0.4777815 | 1.000 | 0.547 | 1.0000000 | 10 | Cdh10 |
| 0.0086567 | 0.4755130 | 0.667 | 0.156 | 1.0000000 | 10 | Kcnh1 |
| 0.0032977 | 0.4565816 | 1.000 | 0.583 | 1.0000000 | 10 | Erbb4 |
| 0.0001304 | 0.4560045 | 1.000 | 0.177 | 1.0000000 | 10 | Nwd1 |
| 0.0000531 | 0.4556571 | 1.000 | 0.459 | 1.0000000 | 10 | Eps8 |
| 0.0038748 | 0.4550326 | 1.000 | 0.174 | 1.0000000 | 10 | Car8 |
| 0.0027591 | 0.4492483 | 1.000 | 0.500 | 1.0000000 | 10 | Xylt1 |
| 0.0001509 | 0.4431093 | 1.000 | 0.115 | 1.0000000 | 10 | Acsbg1 |
| 0.0000700 | 0.4363668 | 1.000 | 0.066 | 1.0000000 | 10 | Frem2 |
| 0.0013194 | 0.4354670 | 1.000 | 0.121 | 1.0000000 | 10 | Amot |
| 0.0007347 | 0.4320210 | 1.000 | 0.463 | 1.0000000 | 10 | Gm48747 |
| 0.0008066 | 0.4317512 | 1.000 | 0.515 | 1.0000000 | 10 | Trim9 |
THP7.markers.mast %>%
group_by(cluster) %>%
slice_max(n = 20, order_by = avg_log10FC) %>%
kable("html") %>%
kable_material(
bootstrap_options = c("bordered",
"condensed",
"responsive",
"striped"),
position = "left",
font_size = 14
)| p_val | avg_log10FC | pct.1 | pct.2 | p_val_adj | cluster | gene |
|---|---|---|---|---|---|---|
| 0.0000028 | 0.4921556 | 0.720 | 0.419 | 0.0584517 | 14 | Zfp804b |
| 0.0000015 | 0.4251261 | 0.500 | 0.246 | 0.0315524 | 14 | D030068K23Rik |
| 0.0000000 | 0.3988595 | 0.980 | 0.691 | 0.0000000 | 14 | Ube3a |
| 0.0000000 | 0.3762109 | 0.660 | 0.352 | 0.0005931 | 14 | Stxbp5l |
| 0.0000000 | 0.3660018 | 0.740 | 0.424 | 0.0009416 | 14 | Rmst |
| 0.0000002 | 0.3495472 | 0.860 | 0.688 | 0.0035211 | 14 | Ralyl |
| 0.0000000 | 0.3489561 | 0.940 | 0.684 | 0.0000019 | 14 | Ahi1 |
| 0.0000003 | 0.3433597 | 0.820 | 0.509 | 0.0061480 | 14 | Nrg3os |
| 0.0000031 | 0.3408464 | 0.720 | 0.491 | 0.0652623 | 14 | Gm26871 |
| 0.0000000 | 0.3364576 | 0.900 | 0.575 | 0.0005510 | 14 | Dpp10 |
| 0.0000000 | 0.3350022 | 0.960 | 0.694 | 0.0000001 | 14 | Dlg2 |
| 0.0000001 | 0.3326104 | 0.860 | 0.556 | 0.0030495 | 14 | Cdh18 |
| 0.0000000 | 0.3269318 | 0.980 | 0.666 | 0.0000000 | 14 | Snhg11 |
| 0.0084473 | 0.3248276 | 0.120 | 0.044 | 1.0000000 | 14 | Gm27016 |
| 0.0000009 | 0.3221719 | 0.620 | 0.323 | 0.0191148 | 14 | Chrm3 |
| 0.0000000 | 0.3217724 | 0.760 | 0.372 | 0.0003891 | 14 | Tafa2 |
| 0.0000000 | 0.3165664 | 0.700 | 0.378 | 0.0006410 | 14 | Zfp385b |
| 0.0008374 | 0.3161474 | 0.580 | 0.405 | 1.0000000 | 14 | Sorcs1 |
| 0.0000021 | 0.3146110 | 0.540 | 0.231 | 0.0455510 | 14 | B230217J21Rik |
| 0.0000077 | 0.3065923 | 0.700 | 0.399 | 0.1627184 | 14 | Gm15398 |
| 0.0000000 | 0.6262359 | 0.847 | 0.540 | 0.0000000 | 8 | Galntl6 |
| 0.0000000 | 0.5807242 | 0.847 | 0.371 | 0.0000000 | 8 | Gm15398 |
| 0.0000000 | 0.5482611 | 0.847 | 0.361 | 0.0000000 | 8 | Unc5d |
| 0.0000000 | 0.5424823 | 0.917 | 0.340 | 0.0000000 | 8 | Tafa2 |
| 0.0000000 | 0.4523585 | 1.000 | 0.646 | 0.0000000 | 8 | Nrg1 |
| 0.0000000 | 0.4513830 | 0.625 | 0.185 | 0.0000000 | 8 | Klhl1 |
| 0.0000000 | 0.4473121 | 0.972 | 0.554 | 0.0000000 | 8 | Dpp10 |
| 0.0000000 | 0.4396150 | 0.875 | 0.358 | 0.0000000 | 8 | Spock3 |
| 0.0000000 | 0.4330138 | 0.944 | 0.535 | 0.0001727 | 8 | Cdh18 |
| 0.0000000 | 0.4278080 | 0.792 | 0.387 | 0.0000016 | 8 | Cdh8 |
| 0.0000000 | 0.4257400 | 0.806 | 0.405 | 0.0000191 | 8 | Rmst |
| 0.0000000 | 0.4233159 | 0.736 | 0.176 | 0.0000000 | 8 | Vwc2l |
| 0.0000000 | 0.4124411 | 0.931 | 0.315 | 0.0000000 | 8 | Dlgap2 |
| 0.0000000 | 0.4116109 | 0.806 | 0.189 | 0.0000000 | 8 | B230217J21Rik |
| 0.0000000 | 0.4106594 | 0.944 | 0.608 | 0.0000001 | 8 | Asic2 |
| 0.0000000 | 0.4016633 | 0.889 | 0.332 | 0.0000000 | 8 | Grin2a |
| 0.0000000 | 0.3945054 | 0.972 | 0.621 | 0.0000754 | 8 | Lingo2 |
| 0.0000000 | 0.3917071 | 1.000 | 0.634 | 0.0000048 | 8 | Sgcz |
| 0.0000000 | 0.3906737 | 0.903 | 0.393 | 0.0000000 | 8 | Esrrg |
| 0.0000000 | 0.3891959 | 0.917 | 0.475 | 0.0000000 | 8 | Galnt13 |
| 0.0000001 | 0.4123405 | 0.889 | 0.398 | 0.0023999 | 7 | Kif26b |
| 0.0000005 | 0.4099479 | 1.000 | 0.670 | 0.0109247 | 7 | Nrg1 |
| 0.0002265 | 0.3853220 | 0.852 | 0.394 | 1.0000000 | 7 | Unc5d |
| 0.0003056 | 0.3493007 | 0.815 | 0.326 | 1.0000000 | 7 | Chrm3 |
| 0.0028767 | 0.3480289 | 0.667 | 0.517 | 1.0000000 | 7 | Rgs6 |
| 0.0004741 | 0.3140704 | 0.778 | 0.333 | 1.0000000 | 7 | Kcnmb2 |
| 0.0000503 | 0.3128392 | 0.926 | 0.482 | 1.0000000 | 7 | Pcdh15 |
| 0.0021469 | 0.2916253 | 0.815 | 0.383 | 1.0000000 | 7 | Tafa2 |
| 0.0023705 | 0.2912035 | 0.852 | 0.403 | 1.0000000 | 7 | Gm15398 |
| 0.0001941 | 0.2876327 | 0.926 | 0.483 | 1.0000000 | 7 | Cdr1os |
| 0.0011563 | 0.2833309 | 0.889 | 0.633 | 1.0000000 | 7 | Asic2 |
| 0.0045694 | 0.2819749 | 0.815 | 0.352 | 1.0000000 | 7 | Cntnap4 |
| 0.0000020 | 0.2805227 | 0.926 | 0.352 | 0.0433772 | 7 | Stxbp5l |
| 0.0067925 | 0.2789436 | 0.704 | 0.367 | 1.0000000 | 7 | Ebf1 |
| 0.0049380 | 0.2746294 | 0.852 | 0.420 | 1.0000000 | 7 | 4930555F03Rik |
| 0.0018564 | 0.2722620 | 1.000 | 0.827 | 1.0000000 | 7 | Fgf14 |
| 0.0045964 | 0.2712338 | 0.889 | 0.568 | 1.0000000 | 7 | Kcnq5 |
| 0.0006517 | 0.2676954 | 0.926 | 0.576 | 1.0000000 | 7 | Pde4dip |
| 0.0055659 | 0.2652826 | 0.778 | 0.374 | 1.0000000 | 7 | Grin2a |
| 0.0025406 | 0.2621642 | 0.778 | 0.285 | 1.0000000 | 7 | Usp13 |
| 0.0000000 | 0.7242156 | 1.000 | 0.526 | 0.0000000 | 1 | Ntng1 |
| 0.0000000 | 0.6576643 | 0.992 | 0.490 | 0.0000000 | 1 | Rnf220 |
| 0.0000000 | 0.6574206 | 1.000 | 0.524 | 0.0000000 | 1 | Tenm1 |
| 0.0000000 | 0.6349539 | 0.992 | 0.400 | 0.0000000 | 1 | Samd5 |
| 0.0000000 | 0.6175479 | 0.962 | 0.544 | 0.0000000 | 1 | Shisa9 |
| 0.0000000 | 0.6069198 | 0.977 | 0.497 | 0.0000000 | 1 | Tafa1 |
| 0.0000000 | 0.5998934 | 0.879 | 0.306 | 0.0000000 | 1 | Gm48749 |
| 0.0000000 | 0.5487415 | 0.780 | 0.295 | 0.0000000 | 1 | 6330411D24Rik |
| 0.0000000 | 0.5436846 | 0.841 | 0.319 | 0.0000000 | 1 | Zmat4 |
| 0.0000000 | 0.5429387 | 0.970 | 0.510 | 0.0000000 | 1 | Egfem1 |
| 0.0000000 | 0.5371066 | 0.879 | 0.350 | 0.0000000 | 1 | Cdh6 |
| 0.0000000 | 0.5180337 | 0.924 | 0.395 | 0.0000000 | 1 | Nell1 |
| 0.0000000 | 0.5174248 | 1.000 | 0.546 | 0.0000000 | 1 | Syt1 |
| 0.0000000 | 0.5077017 | 0.902 | 0.420 | 0.0000000 | 1 | Thsd7b |
| 0.0000000 | 0.5036508 | 0.992 | 0.505 | 0.0000000 | 1 | Tox |
| 0.0000000 | 0.5006324 | 0.932 | 0.414 | 0.0000000 | 1 | Nell2 |
| 0.0000000 | 0.4791969 | 0.992 | 0.580 | 0.0000000 | 1 | Rims1 |
| 0.0000000 | 0.4772918 | 1.000 | 0.798 | 0.0000000 | 1 | Ptprd |
| 0.0000000 | 0.4757209 | 0.886 | 0.315 | 0.0000000 | 1 | Kcnh8 |
| 0.0000000 | 0.4727737 | 0.939 | 0.497 | 0.0000000 | 1 | Hs3st5 |
| 0.0000000 | 0.7071187 | 0.444 | 0.167 | 0.0000002 | 6 | mt-Co3 |
| 0.0000000 | 0.5993005 | 0.841 | 0.516 | 0.0000000 | 6 | Tuba1a |
| 0.0000000 | 0.5847852 | 0.444 | 0.117 | 0.0000087 | 6 | mt-Atp6 |
| 0.0000004 | 0.5390167 | 0.413 | 0.114 | 0.0080001 | 6 | mt-Co2 |
| 0.0000000 | 0.5275187 | 0.540 | 0.204 | 0.0000750 | 6 | mt-Co1 |
| 0.0000267 | 0.5169106 | 0.857 | 0.718 | 0.5658952 | 6 | Cmss1 |
| 0.0000000 | 0.5099136 | 0.508 | 0.287 | 0.0000000 | 6 | Actg1 |
| 0.0000000 | 0.5036783 | 0.698 | 0.327 | 0.0000000 | 6 | Tubb5 |
| 0.0000000 | 0.4942592 | 0.603 | 0.325 | 0.0000000 | 6 | Tubb2b |
| 0.0000000 | 0.4800841 | 0.619 | 0.277 | 0.0000000 | 6 | Atp6v0c |
| 0.0000000 | 0.4781357 | 0.952 | 0.837 | 0.0007858 | 6 | Gm42418 |
| 0.0000000 | 0.4762985 | 0.794 | 0.492 | 0.0000000 | 6 | Calm1 |
| 0.0000000 | 0.4712774 | 0.619 | 0.365 | 0.0000000 | 6 | Hspa8 |
| 0.0000000 | 0.4675408 | 0.667 | 0.377 | 0.0000000 | 6 | Tubb2a |
| 0.0000000 | 0.4546950 | 0.476 | 0.200 | 0.0001449 | 6 | Tmsb10 |
| 0.0000037 | 0.4368420 | 0.333 | 0.085 | 0.0783737 | 6 | mt-Nd2 |
| 0.0000051 | 0.4167998 | 0.460 | 0.178 | 0.1076967 | 6 | Tmsb4x |
| 0.0000000 | 0.4095981 | 0.841 | 0.556 | 0.0000000 | 6 | Gap43 |
| 0.0000000 | 0.3995666 | 0.492 | 0.163 | 0.0000005 | 6 | Dynll2 |
| 0.0000009 | 0.3891660 | 0.333 | 0.088 | 0.0199444 | 6 | Tuba1b |
| 0.0000000 | 1.2395835 | 0.755 | 0.054 | 0.0000000 | 4 | Flt1 |
| 0.0000000 | 1.0596388 | 0.679 | 0.057 | 0.0000000 | 4 | Mecom |
| 0.0000000 | 1.0042969 | 0.736 | 0.226 | 0.0000000 | 4 | Dach1 |
| 0.0000000 | 0.9449069 | 0.868 | 0.055 | 0.0000000 | 4 | Fli1 |
| 0.0000000 | 0.9401421 | 0.660 | 0.079 | 0.0000000 | 4 | Slc7a5 |
| 0.0000000 | 0.8640453 | 0.660 | 0.181 | 0.0000000 | 4 | Slc7a1 |
| 0.0000000 | 0.8515687 | 0.623 | 0.025 | 0.0000000 | 4 | Egfl7 |
| 0.0000000 | 0.8413374 | 0.642 | 0.058 | 0.0000000 | 4 | Slco1c1 |
| 0.0000000 | 0.8347802 | 0.717 | 0.159 | 0.0000000 | 4 | Ccdc141 |
| 0.0000000 | 0.8246317 | 0.623 | 0.068 | 0.0000000 | 4 | Ets1 |
| 0.0000000 | 0.8175285 | 0.698 | 0.011 | 0.0000000 | 4 | Cldn5 |
| 0.0000002 | 0.8075258 | 0.170 | 0.063 | 0.0032462 | 4 | F13a1 |
| 0.0000000 | 0.7978844 | 0.623 | 0.035 | 0.0000000 | 4 | Ptprb |
| 0.0000011 | 0.7969139 | 0.151 | 0.019 | 0.0223034 | 4 | Mrc1 |
| 0.0000000 | 0.7737853 | 0.491 | 0.289 | 0.0005863 | 4 | Rbpj |
| 0.0000000 | 0.7697953 | 0.623 | 0.035 | 0.0000000 | 4 | Adgrl4 |
| 0.0000000 | 0.7664865 | 0.755 | 0.229 | 0.0000000 | 4 | Bsg |
| 0.0000000 | 0.7630175 | 0.604 | 0.068 | 0.0000000 | 4 | Apcdd1 |
| 0.0000000 | 0.7608081 | 0.868 | 0.405 | 0.0000000 | 4 | Dlc1 |
| 0.0000000 | 0.7469235 | 0.604 | 0.077 | 0.0000000 | 4 | Itga1 |
| 0.0000000 | 0.8289105 | 0.971 | 0.698 | 0.0000000 | 18 | Gm26917 |
| 0.0000024 | 0.7021306 | 0.286 | 0.077 | 0.0513245 | 18 | Ebf3 |
| 0.0000000 | 0.6773262 | 0.886 | 0.529 | 0.0000000 | 18 | Rbfox3 |
| 0.0000000 | 0.5832218 | 0.800 | 0.347 | 0.0000000 | 18 | Dgkz |
| 0.0000000 | 0.5730225 | 0.771 | 0.540 | 0.0000000 | 18 | Gpi1 |
| 0.0000000 | 0.5687922 | 0.743 | 0.494 | 0.0000000 | 18 | Atp2b2 |
| 0.0000000 | 0.5547723 | 0.629 | 0.479 | 0.0000056 | 18 | Cdh4 |
| 0.0000000 | 0.5455832 | 0.571 | 0.328 | 0.0000023 | 18 | Matk |
| 0.0000000 | 0.5342803 | 0.943 | 0.753 | 0.0000000 | 18 | Camta1 |
| 0.0000000 | 0.5330145 | 0.943 | 0.842 | 0.0000000 | 18 | Gm42418 |
| 0.0000000 | 0.5300448 | 0.857 | 0.454 | 0.0000000 | 18 | Camk2b |
| 0.0000000 | 0.5269742 | 0.657 | 0.311 | 0.0000003 | 18 | Plec |
| 0.0000000 | 0.5268009 | 0.743 | 0.402 | 0.0000000 | 18 | Ank1 |
| 0.0000000 | 0.5197668 | 0.657 | 0.275 | 0.0000000 | 18 | Pde4a |
| 0.0000000 | 0.5107268 | 0.857 | 0.411 | 0.0000000 | 18 | Tspoap1 |
| 0.0000000 | 0.5055357 | 0.657 | 0.374 | 0.0000000 | 18 | Cacna2d2 |
| 0.0000000 | 0.5029930 | 0.771 | 0.515 | 0.0000000 | 18 | Xkr6 |
| 0.0000000 | 0.4993049 | 0.743 | 0.505 | 0.0000000 | 18 | Slc4a3 |
| 0.0000000 | 0.4960239 | 0.829 | 0.600 | 0.0000000 | 18 | Kif1a |
| 0.0000000 | 0.4903034 | 0.657 | 0.373 | 0.0000010 | 18 | Caskin1 |
| 0.0000894 | 0.9080376 | 0.800 | 0.584 | 1.0000000 | 17 | Ptpn4 |
| 0.0002315 | 0.7928303 | 0.800 | 0.479 | 1.0000000 | 17 | Ryr3 |
| 0.0000001 | 0.7918593 | 0.200 | 0.085 | 0.0016911 | 17 | Tex261 |
| 0.0002012 | 0.7275093 | 0.600 | 0.065 | 1.0000000 | 17 | Ttn |
| 0.0001204 | 0.6985545 | 0.800 | 0.519 | 1.0000000 | 17 | 1700025G04Rik |
| 0.0000216 | 0.6800666 | 0.800 | 0.323 | 0.4567981 | 17 | Cdh7 |
| 0.0040947 | 0.6619007 | 0.800 | 0.449 | 1.0000000 | 17 | Kcnab1 |
| 0.0026983 | 0.6553041 | 0.600 | 0.261 | 1.0000000 | 17 | Prmt8 |
| 0.0000737 | 0.6294186 | 0.600 | 0.260 | 1.0000000 | 17 | Dnajc11 |
| 0.0000012 | 0.6293353 | 1.000 | 0.740 | 0.0250651 | 17 | Agap1 |
| 0.0000557 | 0.5793077 | 0.400 | 0.359 | 1.0000000 | 17 | Ripor2 |
| 0.0042225 | 0.5668952 | 0.400 | 0.273 | 1.0000000 | 17 | Galnt10 |
| 0.0041209 | 0.5421623 | 1.000 | 0.600 | 1.0000000 | 17 | Mgat5 |
| 0.0086744 | 0.5386761 | 0.600 | 0.218 | 1.0000000 | 17 | Synpo2 |
| 0.0000036 | 0.5377447 | 0.400 | 0.362 | 0.0759692 | 17 | Rab11fip4 |
| 0.0079792 | 0.5294053 | 0.800 | 0.302 | 1.0000000 | 17 | Cfap36 |
| 0.0045500 | 0.5292644 | 0.600 | 0.229 | 1.0000000 | 17 | Ptdss1 |
| 0.0002339 | 0.5254941 | 0.600 | 0.117 | 1.0000000 | 17 | Spns1 |
| 0.0016045 | 0.5060974 | 1.000 | 0.343 | 1.0000000 | 17 | Coro2b |
| 0.0000524 | 0.5004997 | 0.600 | 0.254 | 1.0000000 | 17 | Ndufaf2 |
| 0.0000000 | 1.5120877 | 0.967 | 0.123 | 0.0000000 | 20 | Bnc2 |
| 0.0000000 | 1.1812980 | 0.833 | 0.097 | 0.0000000 | 20 | Adamtsl3 |
| 0.0000000 | 1.1224331 | 0.900 | 0.166 | 0.0000000 | 20 | Thsd4 |
| 0.0000000 | 1.0866637 | 0.800 | 0.329 | 0.0000000 | 20 | Col25a1 |
| 0.0000000 | 1.0770827 | 0.867 | 0.507 | 0.0000000 | 20 | Slc4a10 |
| 0.0000000 | 1.0668271 | 0.900 | 0.058 | 0.0000000 | 20 | Trabd2b |
| 0.0000000 | 1.0417482 | 0.967 | 0.414 | 0.0000000 | 20 | Slc38a2 |
| 0.0000000 | 1.0247902 | 0.967 | 0.598 | 0.0000000 | 20 | Foxp1 |
| 0.0000000 | 1.0145583 | 0.967 | 0.473 | 0.0000000 | 20 | Fbxl7 |
| 0.0000000 | 0.9948796 | 0.700 | 0.003 | 0.0000000 | 20 | Slc47a1 |
| 0.0000000 | 0.9929258 | 0.800 | 0.247 | 0.0000000 | 20 | Itgbl1 |
| 0.0000000 | 0.9687133 | 0.900 | 0.374 | 0.0000000 | 20 | Nr3c2 |
| 0.0000000 | 0.9654554 | 0.800 | 0.084 | 0.0000000 | 20 | Dock5 |
| 0.0000000 | 0.9386882 | 0.900 | 0.362 | 0.0000000 | 20 | Gulp1 |
| 0.0000000 | 0.9205507 | 0.567 | 0.105 | 0.0000000 | 20 | Crispld1 |
| 0.0000000 | 0.9158102 | 0.867 | 0.315 | 0.0000000 | 20 | Sh3pxd2a |
| 0.0000000 | 0.9059161 | 0.933 | 0.218 | 0.0000000 | 20 | Hmcn1 |
| 0.0000000 | 0.8957423 | 0.867 | 0.282 | 0.0000000 | 20 | Slit2 |
| 0.0000000 | 0.8901833 | 0.900 | 0.152 | 0.0000000 | 20 | Eya2 |
| 0.0000000 | 0.8734660 | 0.900 | 0.473 | 0.0000000 | 20 | Tmtc1 |
| 0.0000000 | 1.1186122 | 1.000 | 0.313 | 0.0000000 | 3 | Gm3764 |
| 0.0000000 | 0.9860336 | 0.914 | 0.420 | 0.0000000 | 3 | Wdr17 |
| 0.0000000 | 0.9623464 | 0.983 | 0.405 | 0.0000000 | 3 | Slc4a4 |
| 0.0000000 | 0.9476649 | 1.000 | 0.609 | 0.0000000 | 3 | Npas3 |
| 0.0000000 | 0.9266210 | 1.000 | 0.525 | 0.0000000 | 3 | Ptprz1 |
| 0.0000000 | 0.8986149 | 1.000 | 0.417 | 0.0000000 | 3 | Gm48747 |
| 0.0000000 | 0.8928927 | 0.862 | 0.237 | 0.0000000 | 3 | Slc6a11 |
| 0.0000000 | 0.8832488 | 0.931 | 0.315 | 0.0000000 | 3 | Nhsl1 |
| 0.0000000 | 0.8102532 | 0.983 | 0.607 | 0.0000000 | 3 | Luzp2 |
| 0.0000000 | 0.7795485 | 0.862 | 0.210 | 0.0000000 | 3 | Lrig1 |
| 0.0000000 | 0.7663200 | 0.724 | 0.172 | 0.0000000 | 3 | Tnfaip8 |
| 0.0000000 | 0.7654431 | 0.810 | 0.211 | 0.0000000 | 3 | Bmpr1b |
| 0.0000000 | 0.7609677 | 0.879 | 0.424 | 0.0000000 | 3 | Slc1a2 |
| 0.0000000 | 0.7403526 | 0.569 | 0.135 | 0.0000000 | 3 | Tnc |
| 0.0000000 | 0.7354705 | 0.931 | 0.572 | 0.0000000 | 3 | Grm3 |
| 0.0000000 | 0.7120208 | 0.897 | 0.339 | 0.0000000 | 3 | Mir9-3hg |
| 0.0000000 | 0.7103010 | 0.897 | 0.482 | 0.0000000 | 3 | Trim9 |
| 0.0000000 | 0.7035664 | 0.776 | 0.369 | 0.0000000 | 3 | Sfxn5 |
| 0.0000000 | 0.7020843 | 0.845 | 0.277 | 0.0000000 | 3 | Sparcl1 |
| 0.0000000 | 0.6743951 | 0.603 | 0.075 | 0.0000000 | 3 | Acsbg1 |
| 0.0000000 | 1.4989001 | 0.673 | 0.094 | 0.0000000 | 9 | Ptgds |
| 0.0000000 | 1.3620210 | 0.962 | 0.076 | 0.0000000 | 9 | Ranbp3l |
| 0.0000000 | 1.2480266 | 0.904 | 0.072 | 0.0000000 | 9 | Slc6a20a |
| 0.0000000 | 1.1642027 | 0.904 | 0.235 | 0.0000000 | 9 | Sidt1 |
| 0.0000000 | 1.1632862 | 0.827 | 0.806 | 0.0000000 | 9 | Trpm3 |
| 0.0000000 | 1.1025386 | 0.846 | 0.107 | 0.0000000 | 9 | Bmp6 |
| 0.0000000 | 1.0917575 | 0.942 | 0.044 | 0.0000000 | 9 | Slc6a13 |
| 0.0000000 | 1.0891028 | 0.923 | 0.304 | 0.0000000 | 9 | Adam12 |
| 0.0000000 | 1.0529576 | 0.923 | 0.243 | 0.0000000 | 9 | Bicc1 |
| 0.0000000 | 1.0292238 | 0.788 | 0.162 | 0.0000000 | 9 | Adamts12 |
| 0.0000000 | 1.0070606 | 0.962 | 0.332 | 0.0000000 | 9 | Atp1a2 |
| 0.0000000 | 0.9796699 | 0.808 | 0.184 | 0.0000000 | 9 | Lrmda |
| 0.0000000 | 0.9779893 | 0.750 | 0.191 | 0.0000000 | 9 | Slc7a11 |
| 0.0000000 | 0.9732712 | 0.827 | 0.235 | 0.0000000 | 9 | Sned1 |
| 0.0000000 | 0.9700557 | 0.904 | 0.235 | 0.0000000 | 9 | Tmtc4 |
| 0.0000000 | 0.9605894 | 0.750 | 0.649 | 0.0000000 | 9 | Nnat |
| 0.0000000 | 0.9415528 | 0.962 | 0.126 | 0.0000000 | 9 | Arhgap29 |
| 0.0000000 | 0.9385549 | 0.885 | 0.225 | 0.0000000 | 9 | Pdzrn3 |
| 0.0000000 | 0.8939220 | 0.788 | 0.099 | 0.0000000 | 9 | Eya1 |
| 0.0000000 | 0.8631456 | 0.865 | 0.137 | 0.0000000 | 9 | Colec12 |
| 0.0000000 | 0.2059569 | 0.714 | 0.173 | 0.0000873 | 21 | Enpp2 |
| 0.0000000 | 1.0896584 | 0.829 | 0.133 | 0.0000000 | 11 | Gm38505 |
| 0.0000000 | 1.0376620 | 0.971 | 0.284 | 0.0000000 | 11 | Sox6 |
| 0.0000000 | 1.0090739 | 0.829 | 0.229 | 0.0000000 | 11 | 4930588A03Rik |
| 0.0000000 | 0.8441985 | 0.886 | 0.393 | 0.0000000 | 11 | Sox2ot |
| 0.0000000 | 0.8221941 | 0.943 | 0.475 | 0.0000000 | 11 | Pcdh15 |
| 0.0000000 | 0.8131103 | 0.686 | 0.153 | 0.0000000 | 11 | Pdgfra |
| 0.0000000 | 0.7985575 | 0.714 | 0.100 | 0.0000000 | 11 | Sox10 |
| 0.0000000 | 0.7558943 | 0.914 | 0.311 | 0.0000000 | 11 | Dscaml1 |
| 0.0000000 | 0.7476119 | 0.486 | 0.118 | 0.0000005 | 11 | Bcas1 |
| 0.0000000 | 0.7460156 | 0.857 | 0.509 | 0.0000000 | 11 | Lhfpl3 |
| 0.0000000 | 0.7265405 | 0.829 | 0.511 | 0.0000000 | 11 | 6030443J06Rik |
| 0.0000000 | 0.7215268 | 0.857 | 0.416 | 0.0000000 | 11 | Tnr |
| 0.0000000 | 0.6465059 | 0.600 | 0.224 | 0.0000000 | 11 | B9d1 |
| 0.0000000 | 0.6443401 | 0.714 | 0.472 | 0.0000000 | 11 | Epn2 |
| 0.0000000 | 0.6434330 | 0.771 | 0.244 | 0.0000000 | 11 | Megf11 |
| 0.0000000 | 0.6422749 | 0.771 | 0.235 | 0.0000000 | 11 | Arhgef3 |
| 0.0000000 | 0.6271295 | 0.914 | 0.480 | 0.0000000 | 11 | Xylt1 |
| 0.0000000 | 0.6162602 | 0.771 | 0.337 | 0.0000000 | 11 | Gm4876 |
| 0.0000000 | 0.6036030 | 0.829 | 0.546 | 0.0000000 | 11 | Pcdh11x |
| 0.0000000 | 0.5953035 | 0.629 | 0.097 | 0.0000000 | 11 | Prkcq |
| 0.0021467 | 0.4417175 | 1.000 | 0.405 | 1.0000000 | 5 | Gm48749 |
| 0.0042610 | 0.3519949 | 1.000 | 0.724 | 1.0000000 | 5 | Grik2 |
| 0.0020658 | 0.3508967 | 1.000 | 0.496 | 1.0000000 | 5 | Spock1 |
| 0.0018035 | 0.3380302 | 1.000 | 0.579 | 1.0000000 | 5 | Rnf220 |
| 0.0003696 | 0.3273768 | 1.000 | 0.322 | 1.0000000 | 5 | Gm42303 |
| 0.0072834 | 0.3081936 | 0.846 | 0.251 | 1.0000000 | 5 | Gm15414 |
| 0.0028187 | 0.2996319 | 1.000 | 0.464 | 1.0000000 | 5 | Srrm4 |
| 0.0088547 | 0.2985181 | 0.846 | 0.285 | 1.0000000 | 5 | Lef1 |
| 0.0090276 | 0.2700701 | 1.000 | 0.504 | 1.0000000 | 5 | Samd5 |
| 0.0008151 | 0.2665697 | 0.615 | 0.093 | 1.0000000 | 5 | Gm2912 |
| 0.0017114 | 0.2605020 | 1.000 | 0.433 | 1.0000000 | 5 | St6galnac5 |
| 0.0041365 | 0.2578710 | 1.000 | 0.441 | 1.0000000 | 5 | Cdh6 |
| 0.0036836 | 0.2520009 | 1.000 | 0.573 | 1.0000000 | 5 | Hsp90ab1 |
| 0.0041946 | 0.2457113 | 1.000 | 0.442 | 1.0000000 | 5 | Kcnj6 |
| 0.0038357 | 0.2445412 | 0.846 | 0.267 | 1.0000000 | 5 | Adamts17 |
| 0.0094895 | 0.2415859 | 0.923 | 0.322 | 1.0000000 | 5 | Gm49003 |
| 0.0043400 | 0.2256511 | 1.000 | 0.481 | 1.0000000 | 5 | Syt7 |
| 0.0088944 | 0.2212197 | 0.846 | 0.267 | 1.0000000 | 5 | Gm12709 |
| 0.0080790 | 0.2133374 | 1.000 | 0.441 | 1.0000000 | 5 | Tro |
| 0.0068962 | 0.2072306 | 0.846 | 0.279 | 1.0000000 | 5 | Tub |
| 0.0000000 | 1.1856605 | 1.000 | 0.047 | 0.0000000 | 15 | Gfap |
| 0.0000000 | 1.0664641 | 0.900 | 0.047 | 0.0000000 | 15 | Mamdc2 |
| 0.0000000 | 1.0243340 | 0.900 | 0.137 | 0.0000001 | 15 | Aqp4 |
| 0.0000000 | 0.9577662 | 0.800 | 0.009 | 0.0000000 | 15 | Myoc |
| 0.0000000 | 0.9200572 | 1.000 | 0.160 | 0.0000699 | 15 | Tnc |
| 0.0000000 | 0.9134502 | 0.900 | 0.272 | 0.0000000 | 15 | Arhgef4 |
| 0.0000000 | 0.9045121 | 0.900 | 0.199 | 0.0000229 | 15 | Adamts9 |
| 0.0000000 | 0.8344536 | 1.000 | 0.188 | 0.0000108 | 15 | Rfx4 |
| 0.0000000 | 0.8197856 | 0.900 | 0.013 | 0.0000000 | 15 | Hopx |
| 0.0000000 | 0.8077459 | 1.000 | 0.130 | 0.0000019 | 15 | Ednrb |
| 0.0000057 | 0.7891101 | 0.800 | 0.312 | 0.1213906 | 15 | Apoe |
| 0.0000000 | 0.7735977 | 1.000 | 0.569 | 0.0000000 | 15 | Gpm6b |
| 0.0000014 | 0.7581740 | 0.600 | 0.151 | 0.0297774 | 15 | Dbi |
| 0.0000000 | 0.7523044 | 0.800 | 0.155 | 0.0007812 | 15 | Glis3 |
| 0.0000026 | 0.7371467 | 0.900 | 0.455 | 0.0547285 | 15 | Wdr17 |
| 0.0000003 | 0.7209024 | 0.800 | 0.173 | 0.0058418 | 15 | Igsf11 |
| 0.0000000 | 0.7156398 | 0.900 | 0.316 | 0.0009902 | 15 | Sparcl1 |
| 0.0000000 | 0.7138396 | 0.900 | 0.440 | 0.0000000 | 15 | Ablim2 |
| 0.0000046 | 0.7127229 | 0.800 | 0.205 | 0.0980518 | 15 | Gm29683 |
| 0.0000000 | 0.7071319 | 0.900 | 0.090 | 0.0000722 | 15 | Cd9 |
| 0.0000000 | 1.5949065 | 1.000 | 0.220 | 0.0000000 | 16 | Mbp |
| 0.0000000 | 1.4115742 | 1.000 | 0.092 | 0.0000000 | 16 | Plp1 |
| 0.0000000 | 1.3342487 | 0.786 | 0.184 | 0.0000000 | 16 | 9630013A20Rik |
| 0.0000000 | 1.3102788 | 0.929 | 0.059 | 0.0000000 | 16 | St18 |
| 0.0000000 | 1.1614454 | 0.429 | 0.018 | 0.0000041 | 16 | Mobp |
| 0.0000000 | 1.1103252 | 0.643 | 0.095 | 0.0000000 | 16 | Cnksr3 |
| 0.0000000 | 1.0875635 | 0.857 | 0.215 | 0.0000000 | 16 | Tns3 |
| 0.0000000 | 1.0856082 | 0.857 | 0.122 | 0.0000000 | 16 | Bcas1 |
| 0.0000000 | 1.0665966 | 0.857 | 0.059 | 0.0000000 | 16 | Bcas1os2 |
| 0.0000000 | 1.0493521 | 0.643 | 0.024 | 0.0000000 | 16 | Mag |
| 0.0000000 | 1.0269588 | 1.000 | 0.361 | 0.0000000 | 16 | Pacrg |
| 0.0000000 | 1.0085917 | 0.857 | 0.597 | 0.0000000 | 16 | Nfasc |
| 0.0000000 | 0.9895753 | 0.714 | 0.432 | 0.0000020 | 16 | Tnr |
| 0.0000000 | 0.9767866 | 0.786 | 0.036 | 0.0000000 | 16 | Enpp6 |
| 0.0000000 | 0.9584172 | 0.786 | 0.193 | 0.0000000 | 16 | Dusp15 |
| 0.0000000 | 0.9226141 | 0.929 | 0.322 | 0.0000000 | 16 | Itpr2 |
| 0.0000000 | 0.9079627 | 0.786 | 0.025 | 0.0000000 | 16 | 1700047M11Rik |
| 0.0000000 | 0.9038872 | 0.857 | 0.288 | 0.0000000 | 16 | Sirt2 |
| 0.0000000 | 0.8956121 | 0.857 | 0.116 | 0.0000000 | 16 | Sox10 |
| 0.0000000 | 0.8394800 | 0.929 | 0.639 | 0.0000000 | 16 | Fyn |
| 0.0075008 | 0.5572294 | 1.000 | 0.329 | 1.0000000 | 10 | Lars2 |
| 0.0028114 | 0.5442207 | 1.000 | 0.452 | 1.0000000 | 10 | Slc4a4 |
| 0.0044067 | 0.5373239 | 1.000 | 0.136 | 1.0000000 | 10 | C030005K06Rik |
| 0.0099641 | 0.5003187 | 1.000 | 0.563 | 1.0000000 | 10 | Ptprz1 |
| 0.0061664 | 0.4944531 | 1.000 | 0.364 | 1.0000000 | 10 | Nhsl1 |
| 0.0065948 | 0.4938599 | 1.000 | 0.287 | 1.0000000 | 10 | Slc6a11 |
| 0.0021117 | 0.4560045 | 1.000 | 0.177 | 1.0000000 | 10 | Nwd1 |
| 0.0018577 | 0.4556571 | 1.000 | 0.459 | 1.0000000 | 10 | Eps8 |
| 0.0015974 | 0.4431093 | 1.000 | 0.115 | 1.0000000 | 10 | Acsbg1 |
| 0.0006432 | 0.4363668 | 1.000 | 0.066 | 1.0000000 | 10 | Frem2 |
| 0.0047228 | 0.4354670 | 1.000 | 0.121 | 1.0000000 | 10 | Amot |
| 0.0058295 | 0.4250269 | 1.000 | 0.268 | 1.0000000 | 10 | Kank1 |
| 0.0051470 | 0.4215182 | 1.000 | 0.199 | 1.0000000 | 10 | Prex1 |
| 0.0029669 | 0.4199876 | 1.000 | 0.256 | 1.0000000 | 10 | Rhoq |
| 0.0090750 | 0.4187313 | 1.000 | 0.241 | 1.0000000 | 10 | Fads2 |
| 0.0044944 | 0.4055842 | 1.000 | 0.510 | 1.0000000 | 10 | Zfyve9 |
| 0.0059925 | 0.3957603 | 1.000 | 0.545 | 1.0000000 | 10 | 1110051M20Rik |
| 0.0027831 | 0.3899032 | 1.000 | 0.111 | 1.0000000 | 10 | Gm11266 |
| 0.0036697 | 0.3883996 | 1.000 | 0.126 | 1.0000000 | 10 | Hepacam |
| 0.0002862 | 0.3704361 | 1.000 | 0.042 | 1.0000000 | 10 | Mlc1 |
THP7.markers.mast %>%
group_by(cluster) %>%
top_n(n = 10, wt = avg_log10FC) -> top10
DoHeatmap(combined.srt, features = top10$gene) + NoLegend()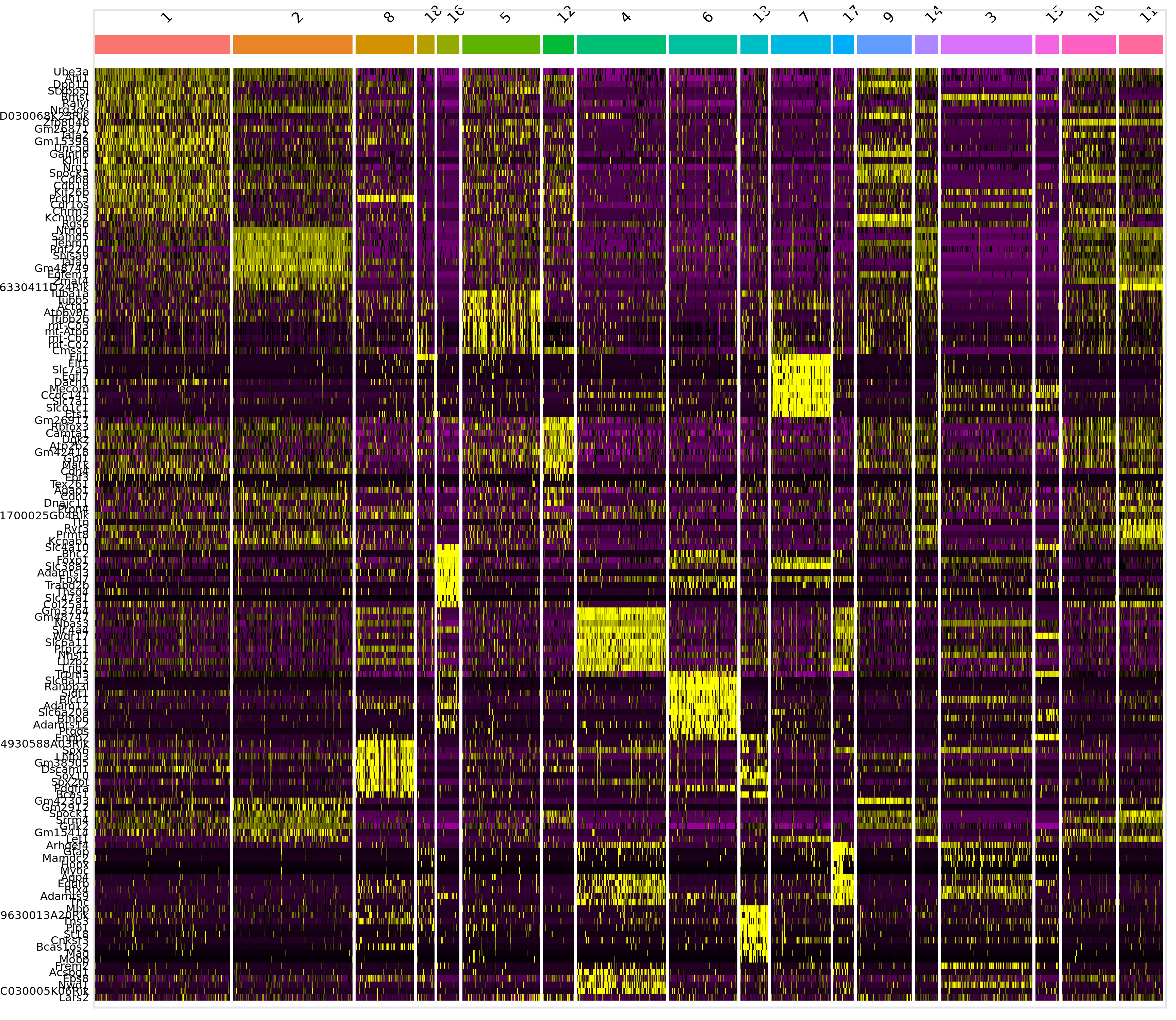
FeaturePlot(THP7, "Galr1", pt.size = 3, order = T) +
scale_colour_gradientn(colours = rev(brewer.pal(n = 11, name = "Spectral"))) + ggtitle("Galr1: ") + theme(plot.title = element_text(size = 24))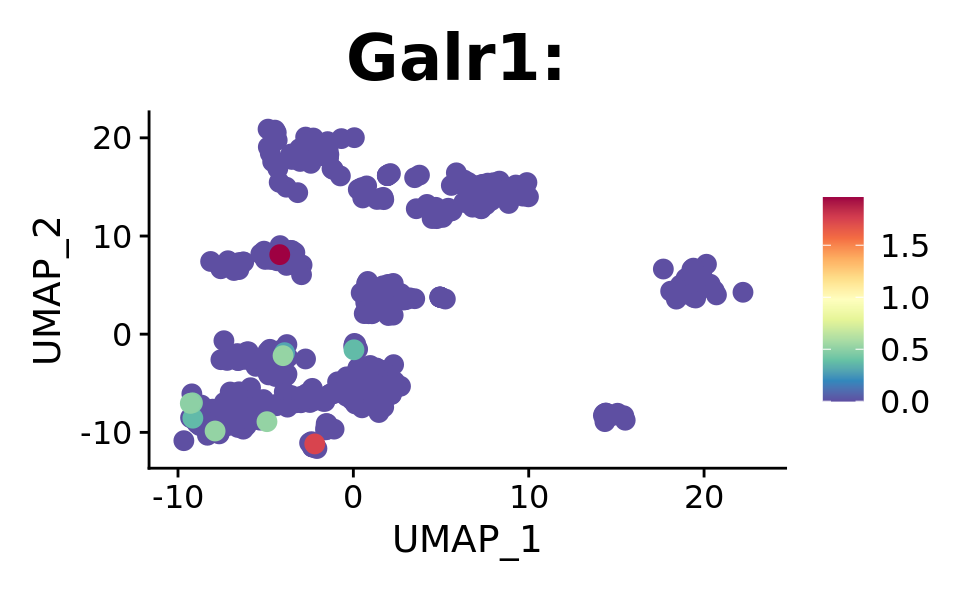
FeaturePlot(THP7, "Gal", pt.size = 3, order = T) +
scale_colour_gradientn(colours = rev(brewer.pal(n = 11, name = "Spectral"))) + ggtitle("Gal: ") + theme(plot.title = element_text(size = 24))
SaveH5Seurat(THP7,
filename = here(data_dir,
"THP7_clusters.h5Seurat"))
Convert(here(data_dir,
"THP7_clusters.h5Seurat"),
dest = "h5ad")Principal sensory trigeminal nucleus
n_pcs <- 100
c(Pr5P7, Pr5P7.markers.logreg, Pr5P7.markers.mast) %<-% DeriveKTree(Pr5P7)
|
|======================================================================| 100%
|
| | 1%
|
|= | 1%
|
|= | 2%
|
|== | 3%
|
|== | 4%
|
|=== | 4%
|
|=== | 5%
|
|==== | 5%
|
|==== | 6%
|
|===== | 7%
|
|===== | 8%
|
|====== | 8%
|
|====== | 9%
|
|======= | 10%
|
|======== | 11%
|
|======== | 12%
|
|========= | 12%
|
|========= | 13%
|
|========== | 14%
|
|========== | 15%
|
|=========== | 15%
|
|=========== | 16%
|
|============ | 17%
|
|============ | 18%
|
|============= | 18%
|
|============= | 19%
|
|============== | 20%
|
|=============== | 21%
|
|=============== | 22%
|
|================ | 22%
|
|================ | 23%
|
|================= | 24%
|
|================== | 25%
|
|================== | 26%
|
|=================== | 27%
|
|=================== | 28%
|
|==================== | 28%
|
|==================== | 29%
|
|===================== | 30%
|
|===================== | 31%
|
|====================== | 31%
|
|========================= | 36%
|
|========================== | 37%
|
|========================== | 38%
|
|=========================== | 38%
|
|=========================== | 39%
|
|============================ | 39%
|
|============================ | 40%
|
|============================= | 41%
|
|============================= | 42%
|
|============================== | 43%
|
|=============================== | 44%
|
|=============================== | 45%
|
|================================ | 45%
|
|================================ | 46%
|
|================================= | 47%
|
|================================== | 49%
|
|=================================== | 49%
|
|=================================== | 50%
|
|=================================== | 51%
|
|==================================== | 51%
|
|==================================== | 52%
|
|===================================== | 53%
|
|======================================= | 56%
|
|======================================== | 57%
|
|======================================== | 58%
|
|========================================= | 58%
|
|========================================= | 59%
|
|=========================================== | 62%
|
|============================================ | 63%
|
|============================================= | 64%
|
|============================================= | 65%
|
|============================================== | 65%
|
|============================================== | 66%
|
|=============================================== | 67%
|
|================================================ | 68%
|
|================================================= | 69%
|
|================================================== | 71%
|
|=================================================== | 73%
|
|==================================================== | 75%
|
|===================================================== | 76%
|
|====================================================== | 77%
|
|====================================================== | 78%
|
|======================================================= | 78%
|
|======================================================= | 79%
|
|======================================================== | 80%
|
|======================================================== | 81%
|
|========================================================== | 82%
|
|========================================================== | 83%
|
|=========================================================== | 84%
|
|=========================================================== | 85%
|
|============================================================ | 85%
|
|============================================================ | 86%
|
|============================================================= | 87%
|
|============================================================= | 88%
|
|============================================================== | 88%
|
|=============================================================== | 90%
|
|================================================================= | 93%
|
|=================================================================== | 96%
|
|==================================================================== | 96%
|
|==================================================================== | 97%
|
|===================================================================== | 98%
|
|===================================================================== | 99%
|
|======================================================================| 99%
|
|======================================================================| 100%Pr5P7.markers.logreg %>%
group_by(cluster) %>%
slice_max(n = 20, order_by = avg_log10FC) %>%
kable("html") %>%
kable_material(
bootstrap_options = c("bordered",
"condensed",
"responsive",
"striped"),
position = "left",
font_size = 14
)| p_val | avg_log10FC | pct.1 | pct.2 | p_val_adj | cluster | gene |
|---|---|---|---|---|---|---|
| 0.0000000 | 0.5887299 | 0.750 | 0.316 | 0.0000034 | 6 | Adarb2 |
| 0.0000000 | 0.5665524 | 0.864 | 0.484 | 0.0000000 | 6 | Ntng1 |
| 0.0000000 | 0.5366819 | 0.886 | 0.551 | 0.0000010 | 6 | Cntnap2 |
| 0.0000000 | 0.5225962 | 0.727 | 0.292 | 0.0000001 | 6 | Stxbp6 |
| 0.0000000 | 0.5084117 | 0.795 | 0.379 | 0.0000001 | 6 | Pcsk2 |
| 0.0000000 | 0.4997253 | 0.795 | 0.392 | 0.0000037 | 6 | Tafa1 |
| 0.0000000 | 0.4737184 | 0.864 | 0.389 | 0.0000000 | 6 | Epha4 |
| 0.0000000 | 0.4732716 | 0.818 | 0.624 | 0.0000528 | 6 | Rnf220 |
| 0.0000000 | 0.4702533 | 0.886 | 0.570 | 0.0000781 | 6 | Mgat4c |
| 0.0000000 | 0.4659155 | 0.773 | 0.303 | 0.0000000 | 6 | Cpne7 |
| 0.0000184 | 0.4633764 | 0.500 | 0.172 | 0.3901997 | 6 | 4930555F03Rik |
| 0.0000000 | 0.4527734 | 0.818 | 0.516 | 0.0005901 | 6 | Tenm1 |
| 0.0000004 | 0.4444636 | 0.864 | 0.512 | 0.0079404 | 6 | Lingo2 |
| 0.0000015 | 0.4426924 | 0.614 | 0.240 | 0.0323020 | 6 | Sv2b |
| 0.0000010 | 0.4301088 | 0.636 | 0.227 | 0.0205300 | 6 | C130073E24Rik |
| 0.0000000 | 0.4285888 | 0.841 | 0.422 | 0.0000000 | 6 | Lef1 |
| 0.0000000 | 0.4187708 | 0.864 | 0.556 | 0.0004753 | 6 | Syt1 |
| 0.0000000 | 0.4160329 | 0.909 | 0.654 | 0.0000069 | 6 | Robo1 |
| 0.0003173 | 0.4087367 | 0.500 | 0.229 | 1.0000000 | 6 | Tmem132cos |
| 0.0000000 | 0.4078876 | 0.841 | 0.464 | 0.0002954 | 6 | Arpp21 |
| 0.0000000 | 1.4223595 | 0.958 | 0.101 | 0.0000000 | 1 | Cfap299 |
| 0.0000000 | 1.4023446 | 0.958 | 0.061 | 0.0000000 | 1 | Dnah12 |
| 0.0000000 | 1.3281690 | 0.950 | 0.234 | 0.0000000 | 1 | Adamts20 |
| 0.0000000 | 1.2144258 | 0.958 | 0.115 | 0.0000000 | 1 | Dnah6 |
| 0.0000000 | 1.1978385 | 0.958 | 0.090 | 0.0000000 | 1 | Hydin |
| 0.0000000 | 1.1972084 | 0.958 | 0.111 | 0.0000000 | 1 | Rgs22 |
| 0.0000000 | 1.1773520 | 0.958 | 0.097 | 0.0000000 | 1 | Ak7 |
| 0.0000000 | 1.1660189 | 0.958 | 0.153 | 0.0000000 | 1 | Cfap61 |
| 0.0000000 | 1.1652030 | 0.966 | 0.217 | 0.0000000 | 1 | Cfap54 |
| 0.0000000 | 1.1617392 | 0.950 | 0.116 | 0.0000000 | 1 | Cfap44 |
| 0.0000000 | 1.1482262 | 0.958 | 0.198 | 0.0000000 | 1 | Ccdc162 |
| 0.0000000 | 1.1393814 | 0.966 | 0.076 | 0.0000000 | 1 | Ak9 |
| 0.0000000 | 1.1355603 | 0.966 | 0.148 | 0.0000000 | 1 | Kif6 |
| 0.0000000 | 1.1291271 | 0.958 | 0.219 | 0.0000000 | 1 | Gm973 |
| 0.0000000 | 1.1285197 | 0.958 | 0.137 | 0.0000000 | 1 | Spef2 |
| 0.0000000 | 1.1106756 | 0.966 | 0.102 | 0.0000000 | 1 | Dnah11 |
| 0.0000000 | 1.1053018 | 0.924 | 0.049 | 0.0000000 | 1 | Spag17 |
| 0.0000000 | 1.1039693 | 0.950 | 0.267 | 0.0000000 | 1 | Spag16 |
| 0.0000000 | 1.0912348 | 0.966 | 0.075 | 0.0000000 | 1 | Cfap65 |
| 0.0000000 | 1.0898457 | 0.958 | 0.226 | 0.0000000 | 1 | Dnah9 |
| 0.0000000 | 1.0539035 | 0.958 | 0.300 | 0.0000000 | 3 | Hs3st4 |
| 0.0000000 | 0.9621409 | 1.000 | 0.660 | 0.0000000 | 3 | Lrrc4c |
| 0.0000000 | 0.9518426 | 0.986 | 0.509 | 0.0000000 | 3 | Dpp10 |
| 0.0000000 | 0.9441776 | 0.986 | 0.488 | 0.0000000 | 3 | Galntl6 |
| 0.0000000 | 0.8927704 | 0.764 | 0.177 | 0.0000000 | 3 | 4930445B16Rik |
| 0.0000000 | 0.8338233 | 0.972 | 0.586 | 0.0000000 | 3 | Nrg1 |
| 0.0000000 | 0.7804064 | 0.917 | 0.291 | 0.0000000 | 3 | Kcnmb2 |
| 0.0000000 | 0.7509803 | 0.958 | 0.714 | 0.0000000 | 3 | Asic2 |
| 0.0000000 | 0.7491942 | 0.903 | 0.159 | 0.0000000 | 3 | Gm15155 |
| 0.0000000 | 0.7438089 | 0.944 | 0.313 | 0.0000000 | 3 | Ubash3b |
| 0.0000000 | 0.7304727 | 0.958 | 0.353 | 0.0000000 | 3 | Fign |
| 0.0000000 | 0.7219656 | 1.000 | 0.449 | 0.0000000 | 3 | Sgcz |
| 0.0000000 | 0.7026695 | 0.889 | 0.234 | 0.0000000 | 3 | Meis2 |
| 0.0000000 | 0.6875301 | 0.958 | 0.387 | 0.0000000 | 3 | Galnt13 |
| 0.0000000 | 0.6777728 | 0.972 | 0.430 | 0.0000000 | 3 | Zfp804a |
| 0.0000000 | 0.6660402 | 0.944 | 0.098 | 0.0000000 | 3 | Gad2 |
| 0.0000000 | 0.6657465 | 0.917 | 0.329 | 0.0000000 | 3 | Ak5 |
| 0.0000000 | 0.6651989 | 0.903 | 0.157 | 0.0000000 | 3 | Ptchd1 |
| 0.0000000 | 0.6568510 | 0.764 | 0.149 | 0.0000000 | 3 | D030068K23Rik |
| 0.0000000 | 0.6533722 | 0.958 | 0.369 | 0.0000000 | 3 | Kctd16 |
| 0.0000000 | 0.7045880 | 0.933 | 0.452 | 0.0000000 | 7 | Gria1 |
| 0.0000000 | 0.6371582 | 0.689 | 0.223 | 0.0000000 | 7 | C130073E24Rik |
| 0.0000000 | 0.6365187 | 0.644 | 0.162 | 0.0000016 | 7 | 4930555F03Rik |
| 0.0000000 | 0.6099695 | 0.800 | 0.237 | 0.0000000 | 7 | Dlgap2 |
| 0.0000000 | 0.6019430 | 0.844 | 0.268 | 0.0000000 | 7 | Gm26871 |
| 0.0000000 | 0.5888953 | 0.689 | 0.377 | 0.0000001 | 7 | Cdh18 |
| 0.0000035 | 0.5845365 | 0.511 | 0.380 | 0.0748373 | 7 | Foxp2 |
| 0.0000000 | 0.5359904 | 0.956 | 0.635 | 0.0000000 | 7 | Nkain2 |
| 0.0000001 | 0.5148576 | 0.422 | 0.135 | 0.0022600 | 7 | Gm45321 |
| 0.0000000 | 0.5040488 | 0.800 | 0.392 | 0.0000017 | 7 | Cntnap5a |
| 0.0000000 | 0.4979773 | 0.644 | 0.345 | 0.0008762 | 7 | Unc5d |
| 0.0000000 | 0.4715143 | 0.933 | 0.540 | 0.0000000 | 7 | Lrrtm4 |
| 0.0000000 | 0.4423627 | 0.689 | 0.360 | 0.0000482 | 7 | Cpne4 |
| 0.0000000 | 0.4420399 | 0.978 | 0.586 | 0.0000000 | 7 | Dlg2 |
| 0.0000000 | 0.4347987 | 0.600 | 0.263 | 0.0000010 | 7 | Pde1a |
| 0.0000000 | 0.4307543 | 0.911 | 0.732 | 0.0005728 | 7 | Tenm3 |
| 0.0000238 | 0.4232842 | 0.133 | 0.018 | 0.5051245 | 7 | Ebf2 |
| 0.0000000 | 0.4231530 | 0.578 | 0.111 | 0.0000000 | 7 | Gm45323 |
| 0.0000000 | 0.4208482 | 0.844 | 0.300 | 0.0000000 | 7 | Stxbp5l |
| 0.0000000 | 0.4167395 | 0.822 | 0.429 | 0.0000001 | 7 | 5730522E02Rik |
| 0.0000000 | 1.9695983 | 0.967 | 0.122 | 0.0000000 | 12 | Htr2c |
| 0.0000000 | 1.5298282 | 1.000 | 0.182 | 0.0000000 | 12 | Ttr |
| 0.0000000 | 1.2141402 | 0.867 | 0.044 | 0.0000000 | 12 | Gmnc |
| 0.0000000 | 1.1683576 | 1.000 | 0.735 | 0.0000000 | 12 | Trpm3 |
| 0.0000000 | 1.1348079 | 0.967 | 0.241 | 0.0000000 | 12 | Vat1l |
| 0.0000000 | 1.1209476 | 0.967 | 0.125 | 0.0000000 | 12 | Sulf1 |
| 0.0000000 | 1.1174114 | 1.000 | 0.084 | 0.0000000 | 12 | Rbm47 |
| 0.0000000 | 1.0869176 | 0.933 | 0.235 | 0.0000000 | 12 | Enpp2 |
| 0.0000000 | 1.0298495 | 1.000 | 0.293 | 0.0000000 | 12 | C330002G04Rik |
| 0.0000000 | 1.0217912 | 0.967 | 0.292 | 0.0000000 | 12 | Otx2os1 |
| 0.0000000 | 1.0157289 | 0.967 | 0.626 | 0.0000000 | 12 | Stk39 |
| 0.0000000 | 1.0104769 | 0.967 | 0.350 | 0.0000000 | 12 | Nhsl2 |
| 0.0000000 | 0.9805817 | 1.000 | 0.591 | 0.0000000 | 12 | Wdr17 |
| 0.0000000 | 0.9593404 | 0.867 | 0.290 | 0.0000000 | 12 | Atp2b3 |
| 0.0000000 | 0.9300492 | 0.800 | 0.038 | 0.0000000 | 12 | Col8a1 |
| 0.0000000 | 0.9014157 | 0.967 | 0.179 | 0.0000000 | 12 | Slc16a2 |
| 0.0000000 | 0.8640925 | 0.833 | 0.411 | 0.0000000 | 12 | Cab39l |
| 0.0000000 | 0.8484966 | 0.867 | 0.531 | 0.0000000 | 12 | Slc4a10 |
| 0.0000000 | 0.8386755 | 0.700 | 0.174 | 0.0000000 | 12 | Gm28376 |
| 0.0000000 | 0.8350280 | 1.000 | 0.564 | 0.0000000 | 12 | Frmpd4 |
| 0.0000000 | 0.8784196 | 0.983 | 0.681 | 0.0000000 | 11 | Kcnip4 |
| 0.0000000 | 0.7869666 | 0.975 | 0.451 | 0.0000000 | 11 | Cntn5 |
| 0.0000000 | 0.7691387 | 0.992 | 0.495 | 0.0000000 | 11 | Tenm2 |
| 0.0000000 | 0.7526718 | 0.967 | 0.530 | 0.0000000 | 11 | Zfp804b |
| 0.0000000 | 0.7454201 | 0.504 | 0.075 | 0.0000000 | 11 | Npsr1 |
| 0.0000000 | 0.6808152 | 0.917 | 0.355 | 0.0000000 | 11 | Gm20754 |
| 0.0000000 | 0.6222545 | 0.950 | 0.514 | 0.0000000 | 11 | Mgat4c |
| 0.0000000 | 0.6040689 | 0.818 | 0.437 | 0.0000000 | 11 | Kcnq5 |
| 0.0000000 | 0.6022047 | 0.851 | 0.326 | 0.0000000 | 11 | Prr16 |
| 0.0000000 | 0.5655895 | 0.826 | 0.463 | 0.0000000 | 11 | Brinp3 |
| 0.0000000 | 0.5504580 | 0.785 | 0.274 | 0.0000000 | 11 | Tafa2 |
| 0.0000000 | 0.5418817 | 0.653 | 0.261 | 0.0000000 | 11 | Pcsk5 |
| 0.0000000 | 0.5250264 | 0.669 | 0.263 | 0.0000000 | 11 | 4930509J09Rik |
| 0.0000000 | 0.5066042 | 0.818 | 0.409 | 0.0000000 | 11 | St6galnac5 |
| 0.0000000 | 0.5062330 | 0.893 | 0.406 | 0.0000000 | 11 | Cntn6 |
| 0.0000000 | 0.4948317 | 0.595 | 0.294 | 0.0000000 | 11 | Cntnap5c |
| 0.0000000 | 0.4844845 | 0.826 | 0.317 | 0.0000000 | 11 | Pcsk2 |
| 0.0000000 | 0.4788825 | 0.942 | 0.613 | 0.0000000 | 11 | Robo1 |
| 0.0000000 | 0.4706050 | 0.959 | 0.462 | 0.0000000 | 11 | Cntn4 |
| 0.0000000 | 0.4663570 | 0.537 | 0.166 | 0.0000000 | 11 | Gm32647 |
| 0.0000000 | 1.3603113 | 0.879 | 0.057 | 0.0000000 | 5 | Flt1 |
| 0.0000000 | 1.1377646 | 0.931 | 0.422 | 0.0000000 | 5 | Dlc1 |
| 0.0000000 | 1.1338448 | 0.914 | 0.141 | 0.0000000 | 5 | Ebf1 |
| 0.0000000 | 1.0823496 | 0.569 | 0.064 | 0.0000000 | 5 | Atp13a5 |
| 0.0000000 | 1.0650516 | 0.741 | 0.166 | 0.0000000 | 5 | Mecom |
| 0.0000000 | 1.0391397 | 0.879 | 0.049 | 0.0000000 | 5 | Itga1 |
| 0.0000000 | 1.0349977 | 0.931 | 0.050 | 0.0000000 | 5 | Ets1 |
| 0.0000000 | 0.9960415 | 0.810 | 0.215 | 0.0000000 | 5 | Slc7a1 |
| 0.0000000 | 0.9843588 | 0.724 | 0.022 | 0.0000000 | 5 | Egfl7 |
| 0.0000000 | 0.9660674 | 0.948 | 0.345 | 0.0000000 | 5 | Rapgef5 |
| 0.0000000 | 0.9567065 | 0.845 | 0.063 | 0.0000000 | 5 | Fli1 |
| 0.0000000 | 0.9387221 | 0.845 | 0.496 | 0.0000000 | 5 | Slc38a2 |
| 0.0000000 | 0.9332474 | 0.793 | 0.024 | 0.0000000 | 5 | Rgs5 |
| 0.0000000 | 0.9239473 | 0.759 | 0.044 | 0.0000000 | 5 | Ptprb |
| 0.0000000 | 0.9204071 | 0.810 | 0.251 | 0.0000000 | 5 | Dach1 |
| 0.0000000 | 0.9175331 | 0.724 | 0.234 | 0.0000000 | 5 | Hmcn1 |
| 0.0000000 | 0.9163048 | 0.741 | 0.097 | 0.0000000 | 5 | Slc7a5 |
| 0.0000000 | 0.9007020 | 0.810 | 0.030 | 0.0000000 | 5 | Morrbid |
| 0.0000000 | 0.8916247 | 0.862 | 0.082 | 0.0000000 | 5 | Lama4 |
| 0.0000000 | 0.8766677 | 0.690 | 0.041 | 0.0000000 | 5 | Adgrl4 |
| 0.0000000 | 1.0422162 | 0.929 | 0.300 | 0.0000000 | 2 | Gm3764 |
| 0.0000000 | 1.0070704 | 0.941 | 0.466 | 0.0000000 | 2 | Slc4a4 |
| 0.0000000 | 0.8575195 | 0.894 | 0.415 | 0.0000000 | 2 | Gm48747 |
| 0.0000000 | 0.8463038 | 0.941 | 0.628 | 0.0000000 | 2 | Ptprz1 |
| 0.0000000 | 0.7881937 | 0.894 | 0.152 | 0.0000000 | 2 | Bmpr1b |
| 0.0000000 | 0.7745003 | 0.765 | 0.174 | 0.0000000 | 2 | Slc6a11 |
| 0.0000000 | 0.7419631 | 0.765 | 0.451 | 0.0000000 | 2 | Gpc5 |
| 0.0000000 | 0.7406946 | 0.929 | 0.289 | 0.0000000 | 2 | Plpp3 |
| 0.0000000 | 0.7340348 | 0.812 | 0.539 | 0.0000000 | 2 | Luzp2 |
| 0.0000000 | 0.7330892 | 0.824 | 0.197 | 0.0000000 | 2 | Pla2g7 |
| 0.0000000 | 0.7305223 | 0.706 | 0.136 | 0.0000000 | 2 | Tnc |
| 0.0000000 | 0.7193021 | 0.894 | 0.316 | 0.0000000 | 2 | Apoe |
| 0.0000000 | 0.7134415 | 0.871 | 0.574 | 0.0000000 | 2 | Trim9 |
| 0.0000000 | 0.7109505 | 0.859 | 0.233 | 0.0000000 | 2 | Lrig1 |
| 0.0000000 | 0.6843853 | 0.859 | 0.228 | 0.0000000 | 2 | Palld |
| 0.0000000 | 0.6781358 | 0.941 | 0.620 | 0.0000000 | 2 | Gpm6b |
| 0.0000000 | 0.6760270 | 0.776 | 0.136 | 0.0000000 | 2 | Egfr |
| 0.0000000 | 0.6756797 | 0.812 | 0.190 | 0.0000000 | 2 | Cdh20 |
| 0.0000000 | 0.6737712 | 0.965 | 0.695 | 0.0000000 | 2 | Npas3 |
| 0.0000000 | 0.6668370 | 0.929 | 0.582 | 0.0000000 | 2 | Ptn |
| 0.0000000 | 1.1536760 | 0.966 | 0.182 | 0.0000000 | 4 | 6330411D24Rik |
| 0.0000000 | 0.8713080 | 1.000 | 0.320 | 0.0000000 | 4 | Pex5l |
| 0.0000000 | 0.7573003 | 1.000 | 0.344 | 0.0000000 | 4 | Spock1 |
| 0.0000000 | 0.7044603 | 1.000 | 0.414 | 0.0000000 | 4 | Ryr3 |
| 0.0000000 | 0.6954763 | 1.000 | 0.650 | 0.0000000 | 4 | Hs6st3 |
| 0.0000000 | 0.6895827 | 1.000 | 0.441 | 0.0000000 | 4 | Arpp21 |
| 0.0000000 | 0.6778960 | 0.983 | 0.386 | 0.0000000 | 4 | Edil3 |
| 0.0000000 | 0.6528223 | 0.914 | 0.171 | 0.0000000 | 4 | 4930419G24Rik |
| 0.0000000 | 0.6316032 | 0.966 | 0.485 | 0.0000000 | 4 | Adarb1 |
| 0.0000000 | 0.6239836 | 1.000 | 0.463 | 0.0000000 | 4 | Ntng1 |
| 0.0000000 | 0.6026214 | 0.983 | 0.408 | 0.0000000 | 4 | Camk4 |
| 0.0000000 | 0.5882924 | 0.948 | 0.281 | 0.0000000 | 4 | Grm1 |
| 0.0000000 | 0.5851017 | 1.000 | 0.551 | 0.0000000 | 4 | Gria4 |
| 0.0000000 | 0.5819572 | 0.845 | 0.138 | 0.0000000 | 4 | Pappa |
| 0.0000000 | 0.5682423 | 1.000 | 0.416 | 0.0000000 | 4 | Zmat4 |
| 0.0000000 | 0.5650541 | 0.983 | 0.429 | 0.0000000 | 4 | Hs3st5 |
| 0.0000000 | 0.5621780 | 0.966 | 0.378 | 0.0000000 | 4 | Epb41l4b |
| 0.0000000 | 0.5534059 | 0.983 | 0.317 | 0.0000000 | 4 | Ptpn3 |
| 0.0000000 | 0.5524678 | 1.000 | 0.548 | 0.0000000 | 4 | Lrrc7 |
| 0.0000000 | 0.5481946 | 0.948 | 0.350 | 0.0000000 | 4 | Cntn3 |
| 0.0000000 | 0.9735890 | 0.854 | 0.367 | 0.0000000 | 9 | Pcdh15 |
| 0.0000000 | 0.9602308 | 0.415 | 0.222 | 0.0000000 | 9 | Mbp |
| 0.0000000 | 0.9524441 | 0.829 | 0.110 | 0.0000000 | 9 | 4930588A03Rik |
| 0.0000000 | 0.9147655 | 0.951 | 0.408 | 0.0000000 | 9 | Tnr |
| 0.0000000 | 0.8757874 | 0.293 | 0.130 | 0.0000001 | 9 | Plp1 |
| 0.0000000 | 0.8633900 | 0.829 | 0.205 | 0.0000000 | 9 | Dscaml1 |
| 0.0000000 | 0.8515065 | 0.854 | 0.492 | 0.0000000 | 9 | 6030443J06Rik |
| 0.0000000 | 0.8470640 | 0.902 | 0.061 | 0.0000000 | 9 | Sox10 |
| 0.0000000 | 0.8440775 | 0.829 | 0.124 | 0.0000000 | 9 | Gm38505 |
| 0.0000000 | 0.8367428 | 0.512 | 0.183 | 0.0000000 | 9 | 9630013A20Rik |
| 0.0000000 | 0.8333975 | 0.756 | 0.170 | 0.0000000 | 9 | Bcas1 |
| 0.0000000 | 0.8047986 | 0.659 | 0.116 | 0.0000000 | 9 | Pdgfra |
| 0.0000000 | 0.7785127 | 0.878 | 0.385 | 0.0000000 | 9 | Lhfpl3 |
| 0.0000000 | 0.7383577 | 0.244 | 0.052 | 0.0000000 | 9 | St18 |
| 0.0000000 | 0.7251675 | 0.902 | 0.598 | 0.0000000 | 9 | Sox2ot |
| 0.0000000 | 0.6847430 | 0.902 | 0.477 | 0.0000000 | 9 | Sox6 |
| 0.0000000 | 0.6739720 | 0.878 | 0.401 | 0.0000000 | 9 | Nxph1 |
| 0.0000000 | 0.6728349 | 0.829 | 0.274 | 0.0000000 | 9 | Tns3 |
| 0.0000000 | 0.6329265 | 0.707 | 0.098 | 0.0000000 | 9 | Vxn |
| 0.0000000 | 0.6283993 | 0.976 | 0.641 | 0.0000000 | 9 | Opcml |
| 0.0000000 | 1.3307978 | 0.955 | 0.135 | 0.0000000 | 13 | Adamts12 |
| 0.0000000 | 1.2983823 | 0.955 | 0.248 | 0.0000000 | 13 | Adam12 |
| 0.0000000 | 1.2333967 | 0.955 | 0.135 | 0.0000000 | 13 | Cped1 |
| 0.0000000 | 1.1018416 | 0.864 | 0.022 | 0.0000000 | 13 | Slc6a13 |
| 0.0000000 | 1.0967793 | 0.773 | 0.065 | 0.0000000 | 13 | Ranbp3l |
| 0.0000000 | 1.0930283 | 0.455 | 0.025 | 0.0000017 | 13 | Ptgds |
| 0.0000000 | 1.0826431 | 0.864 | 0.184 | 0.0000000 | 13 | Sidt1 |
| 0.0000000 | 1.0723408 | 0.773 | 0.224 | 0.0000000 | 13 | Bmp6 |
| 0.0000000 | 1.0451808 | 1.000 | 0.422 | 0.0000000 | 13 | Bicc1 |
| 0.0000000 | 1.0180167 | 0.773 | 0.042 | 0.0000000 | 13 | Aox3 |
| 0.0000000 | 0.9944784 | 0.864 | 0.230 | 0.0000000 | 13 | Slc7a11 |
| 0.0000000 | 0.9472832 | 0.909 | 0.048 | 0.0000000 | 13 | Col3a1 |
| 0.0000000 | 0.9046290 | 0.773 | 0.218 | 0.0000000 | 13 | Tmtc4 |
| 0.0000000 | 0.8772338 | 0.636 | 0.056 | 0.0000002 | 13 | Col1a2 |
| 0.0000000 | 0.8680208 | 0.864 | 0.446 | 0.0000001 | 13 | Svil |
| 0.0000000 | 0.8660678 | 0.727 | 0.046 | 0.0000000 | 13 | Cfh |
| 0.0000000 | 0.8561243 | 0.818 | 0.166 | 0.0000000 | 13 | Col4a5 |
| 0.0000000 | 0.8513968 | 0.545 | 0.088 | 0.0000009 | 13 | Itga8 |
| 0.0000000 | 0.8347594 | 0.864 | 0.155 | 0.0000000 | 13 | Arhgap29 |
| 0.0000000 | 0.8295859 | 0.818 | 0.068 | 0.0000000 | 13 | Col4a6 |
Pr5P7.markers.mast %>%
group_by(cluster) %>%
slice_max(n = 20, order_by = avg_log10FC) %>%
kable("html") %>%
kable_material(
bootstrap_options = c("bordered",
"condensed",
"responsive",
"striped"),
position = "left",
font_size = 14
)| p_val | avg_log10FC | pct.1 | pct.2 | p_val_adj | cluster | gene |
|---|---|---|---|---|---|---|
| 0.0000000 | 0.5887299 | 0.750 | 0.316 | 0.0000105 | 6 | Adarb2 |
| 0.0000000 | 0.5665524 | 0.864 | 0.484 | 0.0000009 | 6 | Ntng1 |
| 0.0000000 | 0.5366819 | 0.886 | 0.551 | 0.0000176 | 6 | Cntnap2 |
| 0.0000000 | 0.5225962 | 0.727 | 0.292 | 0.0000000 | 6 | Stxbp6 |
| 0.0000000 | 0.5084117 | 0.795 | 0.379 | 0.0000000 | 6 | Pcsk2 |
| 0.0000000 | 0.4997253 | 0.795 | 0.392 | 0.0000153 | 6 | Tafa1 |
| 0.0000000 | 0.4737184 | 0.864 | 0.389 | 0.0000000 | 6 | Epha4 |
| 0.0000000 | 0.4732716 | 0.818 | 0.624 | 0.0000007 | 6 | Rnf220 |
| 0.0000000 | 0.4702533 | 0.886 | 0.570 | 0.0000907 | 6 | Mgat4c |
| 0.0000000 | 0.4659155 | 0.773 | 0.303 | 0.0000000 | 6 | Cpne7 |
| 0.0000526 | 0.4633764 | 0.500 | 0.172 | 1.0000000 | 6 | 4930555F03Rik |
| 0.0000000 | 0.4527734 | 0.818 | 0.516 | 0.0002443 | 6 | Tenm1 |
| 0.0000242 | 0.4444636 | 0.864 | 0.512 | 0.5134938 | 6 | Lingo2 |
| 0.0000342 | 0.4426924 | 0.614 | 0.240 | 0.7241354 | 6 | Sv2b |
| 0.0000016 | 0.4301088 | 0.636 | 0.227 | 0.0341635 | 6 | C130073E24Rik |
| 0.0000000 | 0.4285888 | 0.841 | 0.422 | 0.0000000 | 6 | Lef1 |
| 0.0000002 | 0.4187708 | 0.864 | 0.556 | 0.0041521 | 6 | Syt1 |
| 0.0000000 | 0.4160329 | 0.909 | 0.654 | 0.0000023 | 6 | Robo1 |
| 0.0027379 | 0.4087367 | 0.500 | 0.229 | 1.0000000 | 6 | Tmem132cos |
| 0.0000006 | 0.4078876 | 0.841 | 0.464 | 0.0128281 | 6 | Arpp21 |
| 0.0000000 | 1.4223595 | 0.958 | 0.101 | 0.0000000 | 1 | Cfap299 |
| 0.0000000 | 1.4023446 | 0.958 | 0.061 | 0.0000000 | 1 | Dnah12 |
| 0.0000000 | 1.3281690 | 0.950 | 0.234 | 0.0000000 | 1 | Adamts20 |
| 0.0000000 | 1.2144258 | 0.958 | 0.115 | 0.0000000 | 1 | Dnah6 |
| 0.0000000 | 1.1978385 | 0.958 | 0.090 | 0.0000000 | 1 | Hydin |
| 0.0000000 | 1.1972084 | 0.958 | 0.111 | 0.0000000 | 1 | Rgs22 |
| 0.0000000 | 1.1773520 | 0.958 | 0.097 | 0.0000000 | 1 | Ak7 |
| 0.0000000 | 1.1660189 | 0.958 | 0.153 | 0.0000000 | 1 | Cfap61 |
| 0.0000000 | 1.1652030 | 0.966 | 0.217 | 0.0000000 | 1 | Cfap54 |
| 0.0000000 | 1.1617392 | 0.950 | 0.116 | 0.0000000 | 1 | Cfap44 |
| 0.0000000 | 1.1482262 | 0.958 | 0.198 | 0.0000000 | 1 | Ccdc162 |
| 0.0000000 | 1.1393814 | 0.966 | 0.076 | 0.0000000 | 1 | Ak9 |
| 0.0000000 | 1.1355603 | 0.966 | 0.148 | 0.0000000 | 1 | Kif6 |
| 0.0000000 | 1.1291271 | 0.958 | 0.219 | 0.0000000 | 1 | Gm973 |
| 0.0000000 | 1.1285197 | 0.958 | 0.137 | 0.0000000 | 1 | Spef2 |
| 0.0000000 | 1.1106756 | 0.966 | 0.102 | 0.0000000 | 1 | Dnah11 |
| 0.0000000 | 1.1053018 | 0.924 | 0.049 | 0.0000000 | 1 | Spag17 |
| 0.0000000 | 1.1039693 | 0.950 | 0.267 | 0.0000000 | 1 | Spag16 |
| 0.0000000 | 1.0912348 | 0.966 | 0.075 | 0.0000000 | 1 | Cfap65 |
| 0.0000000 | 1.0898457 | 0.958 | 0.226 | 0.0000000 | 1 | Dnah9 |
| 0.0000000 | 1.0539035 | 0.958 | 0.300 | 0.0000000 | 3 | Hs3st4 |
| 0.0000000 | 0.9621409 | 1.000 | 0.660 | 0.0000000 | 3 | Lrrc4c |
| 0.0000000 | 0.9518426 | 0.986 | 0.509 | 0.0000000 | 3 | Dpp10 |
| 0.0000000 | 0.9441776 | 0.986 | 0.488 | 0.0000000 | 3 | Galntl6 |
| 0.0000000 | 0.8927704 | 0.764 | 0.177 | 0.0000000 | 3 | 4930445B16Rik |
| 0.0000000 | 0.8338233 | 0.972 | 0.586 | 0.0000000 | 3 | Nrg1 |
| 0.0000000 | 0.7804064 | 0.917 | 0.291 | 0.0000000 | 3 | Kcnmb2 |
| 0.0000000 | 0.7509803 | 0.958 | 0.714 | 0.0000000 | 3 | Asic2 |
| 0.0000000 | 0.7491942 | 0.903 | 0.159 | 0.0000000 | 3 | Gm15155 |
| 0.0000000 | 0.7438089 | 0.944 | 0.313 | 0.0000000 | 3 | Ubash3b |
| 0.0000000 | 0.7304727 | 0.958 | 0.353 | 0.0000000 | 3 | Fign |
| 0.0000000 | 0.7219656 | 1.000 | 0.449 | 0.0000000 | 3 | Sgcz |
| 0.0000000 | 0.7026695 | 0.889 | 0.234 | 0.0000000 | 3 | Meis2 |
| 0.0000000 | 0.6875301 | 0.958 | 0.387 | 0.0000000 | 3 | Galnt13 |
| 0.0000000 | 0.6777728 | 0.972 | 0.430 | 0.0000000 | 3 | Zfp804a |
| 0.0000000 | 0.6660402 | 0.944 | 0.098 | 0.0000000 | 3 | Gad2 |
| 0.0000000 | 0.6657465 | 0.917 | 0.329 | 0.0000000 | 3 | Ak5 |
| 0.0000000 | 0.6651989 | 0.903 | 0.157 | 0.0000000 | 3 | Ptchd1 |
| 0.0000000 | 0.6568510 | 0.764 | 0.149 | 0.0000000 | 3 | D030068K23Rik |
| 0.0000000 | 0.6533722 | 0.958 | 0.369 | 0.0000000 | 3 | Kctd16 |
| 0.0000000 | 0.7045880 | 0.933 | 0.452 | 0.0000000 | 7 | Gria1 |
| 0.0000000 | 0.6371582 | 0.689 | 0.223 | 0.0000001 | 7 | C130073E24Rik |
| 0.0000000 | 0.6365187 | 0.644 | 0.162 | 0.0000001 | 7 | 4930555F03Rik |
| 0.0000000 | 0.6099695 | 0.800 | 0.237 | 0.0000000 | 7 | Dlgap2 |
| 0.0000000 | 0.6019430 | 0.844 | 0.268 | 0.0000000 | 7 | Gm26871 |
| 0.0000000 | 0.5888953 | 0.689 | 0.377 | 0.0000000 | 7 | Cdh18 |
| 0.0000000 | 0.5845365 | 0.511 | 0.380 | 0.0000009 | 7 | Foxp2 |
| 0.0000000 | 0.5359904 | 0.956 | 0.635 | 0.0000000 | 7 | Nkain2 |
| 0.0000000 | 0.5148576 | 0.422 | 0.135 | 0.0006125 | 7 | Gm45321 |
| 0.0000000 | 0.5040488 | 0.800 | 0.392 | 0.0000404 | 7 | Cntnap5a |
| 0.0030477 | 0.5020671 | 0.333 | 0.262 | 1.0000000 | 7 | Nwd2 |
| 0.0000004 | 0.4979773 | 0.644 | 0.345 | 0.0089892 | 7 | Unc5d |
| 0.0000000 | 0.4715143 | 0.933 | 0.540 | 0.0000010 | 7 | Lrrtm4 |
| 0.0000000 | 0.4423627 | 0.689 | 0.360 | 0.0000602 | 7 | Cpne4 |
| 0.0000000 | 0.4420399 | 0.978 | 0.586 | 0.0000000 | 7 | Dlg2 |
| 0.0000000 | 0.4347987 | 0.600 | 0.263 | 0.0000000 | 7 | Pde1a |
| 0.0000002 | 0.4307543 | 0.911 | 0.732 | 0.0037332 | 7 | Tenm3 |
| 0.0001691 | 0.4232842 | 0.133 | 0.018 | 1.0000000 | 7 | Ebf2 |
| 0.0000000 | 0.4231530 | 0.578 | 0.111 | 0.0000000 | 7 | Gm45323 |
| 0.0000000 | 0.4208482 | 0.844 | 0.300 | 0.0000000 | 7 | Stxbp5l |
| 0.0000000 | 1.9695983 | 0.967 | 0.122 | 0.0000000 | 12 | Htr2c |
| 0.0000000 | 1.5298282 | 1.000 | 0.182 | 0.0000000 | 12 | Ttr |
| 0.0000000 | 1.2141402 | 0.867 | 0.044 | 0.0000000 | 12 | Gmnc |
| 0.0000000 | 1.1683576 | 1.000 | 0.735 | 0.0000000 | 12 | Trpm3 |
| 0.0000000 | 1.1348079 | 0.967 | 0.241 | 0.0000000 | 12 | Vat1l |
| 0.0000000 | 1.1209476 | 0.967 | 0.125 | 0.0000000 | 12 | Sulf1 |
| 0.0000000 | 1.1174114 | 1.000 | 0.084 | 0.0000000 | 12 | Rbm47 |
| 0.0000000 | 1.0869176 | 0.933 | 0.235 | 0.0000000 | 12 | Enpp2 |
| 0.0000000 | 1.0298495 | 1.000 | 0.293 | 0.0000000 | 12 | C330002G04Rik |
| 0.0000000 | 1.0217912 | 0.967 | 0.292 | 0.0000000 | 12 | Otx2os1 |
| 0.0000000 | 1.0157289 | 0.967 | 0.626 | 0.0000000 | 12 | Stk39 |
| 0.0000000 | 1.0104769 | 0.967 | 0.350 | 0.0000000 | 12 | Nhsl2 |
| 0.0000000 | 0.9805817 | 1.000 | 0.591 | 0.0000000 | 12 | Wdr17 |
| 0.0000000 | 0.9593404 | 0.867 | 0.290 | 0.0000000 | 12 | Atp2b3 |
| 0.0000000 | 0.9300492 | 0.800 | 0.038 | 0.0000000 | 12 | Col8a1 |
| 0.0000000 | 0.9014157 | 0.967 | 0.179 | 0.0000000 | 12 | Slc16a2 |
| 0.0000000 | 0.8640925 | 0.833 | 0.411 | 0.0000000 | 12 | Cab39l |
| 0.0000000 | 0.8484966 | 0.867 | 0.531 | 0.0000000 | 12 | Slc4a10 |
| 0.0000000 | 0.8386755 | 0.700 | 0.174 | 0.0000000 | 12 | Gm28376 |
| 0.0000000 | 0.8350280 | 1.000 | 0.564 | 0.0000000 | 12 | Frmpd4 |
| 0.0000000 | 0.8784196 | 0.983 | 0.681 | 0.0000000 | 11 | Kcnip4 |
| 0.0000000 | 0.7869666 | 0.975 | 0.451 | 0.0000000 | 11 | Cntn5 |
| 0.0000000 | 0.7691387 | 0.992 | 0.495 | 0.0000000 | 11 | Tenm2 |
| 0.0000000 | 0.7526718 | 0.967 | 0.530 | 0.0000000 | 11 | Zfp804b |
| 0.0000000 | 0.7454201 | 0.504 | 0.075 | 0.0000000 | 11 | Npsr1 |
| 0.0000000 | 0.6808152 | 0.917 | 0.355 | 0.0000000 | 11 | Gm20754 |
| 0.0000000 | 0.6222545 | 0.950 | 0.514 | 0.0000000 | 11 | Mgat4c |
| 0.0000000 | 0.6040689 | 0.818 | 0.437 | 0.0000000 | 11 | Kcnq5 |
| 0.0000000 | 0.6022047 | 0.851 | 0.326 | 0.0000000 | 11 | Prr16 |
| 0.0000000 | 0.5655895 | 0.826 | 0.463 | 0.0000000 | 11 | Brinp3 |
| 0.0000000 | 0.5504580 | 0.785 | 0.274 | 0.0000000 | 11 | Tafa2 |
| 0.0000000 | 0.5418817 | 0.653 | 0.261 | 0.0000000 | 11 | Pcsk5 |
| 0.0000000 | 0.5250264 | 0.669 | 0.263 | 0.0000000 | 11 | 4930509J09Rik |
| 0.0000000 | 0.5066042 | 0.818 | 0.409 | 0.0000000 | 11 | St6galnac5 |
| 0.0000000 | 0.5062330 | 0.893 | 0.406 | 0.0000000 | 11 | Cntn6 |
| 0.0000000 | 0.4948317 | 0.595 | 0.294 | 0.0000000 | 11 | Cntnap5c |
| 0.0000000 | 0.4844845 | 0.826 | 0.317 | 0.0000000 | 11 | Pcsk2 |
| 0.0000000 | 0.4788825 | 0.942 | 0.613 | 0.0000000 | 11 | Robo1 |
| 0.0000000 | 0.4706050 | 0.959 | 0.462 | 0.0000000 | 11 | Cntn4 |
| 0.0000000 | 0.4663570 | 0.537 | 0.166 | 0.0000000 | 11 | Gm32647 |
| 0.0000000 | 1.3603113 | 0.879 | 0.057 | 0.0000000 | 5 | Flt1 |
| 0.0000000 | 1.1377646 | 0.931 | 0.422 | 0.0000000 | 5 | Dlc1 |
| 0.0000000 | 1.1338448 | 0.914 | 0.141 | 0.0000000 | 5 | Ebf1 |
| 0.0000000 | 1.0823496 | 0.569 | 0.064 | 0.0000000 | 5 | Atp13a5 |
| 0.0000000 | 1.0650516 | 0.741 | 0.166 | 0.0000000 | 5 | Mecom |
| 0.0000000 | 1.0391397 | 0.879 | 0.049 | 0.0000000 | 5 | Itga1 |
| 0.0000000 | 1.0349977 | 0.931 | 0.050 | 0.0000000 | 5 | Ets1 |
| 0.0000000 | 0.9960415 | 0.810 | 0.215 | 0.0000000 | 5 | Slc7a1 |
| 0.0000000 | 0.9843588 | 0.724 | 0.022 | 0.0000000 | 5 | Egfl7 |
| 0.0000000 | 0.9660674 | 0.948 | 0.345 | 0.0000000 | 5 | Rapgef5 |
| 0.0000000 | 0.9567065 | 0.845 | 0.063 | 0.0000000 | 5 | Fli1 |
| 0.0000000 | 0.9387221 | 0.845 | 0.496 | 0.0000000 | 5 | Slc38a2 |
| 0.0000000 | 0.9332474 | 0.793 | 0.024 | 0.0000000 | 5 | Rgs5 |
| 0.0000000 | 0.9239473 | 0.759 | 0.044 | 0.0000000 | 5 | Ptprb |
| 0.0000000 | 0.9204071 | 0.810 | 0.251 | 0.0000000 | 5 | Dach1 |
| 0.0000000 | 0.9175331 | 0.724 | 0.234 | 0.0000000 | 5 | Hmcn1 |
| 0.0000000 | 0.9163048 | 0.741 | 0.097 | 0.0000000 | 5 | Slc7a5 |
| 0.0000000 | 0.9007020 | 0.810 | 0.030 | 0.0000000 | 5 | Morrbid |
| 0.0000000 | 0.8916247 | 0.862 | 0.082 | 0.0000000 | 5 | Lama4 |
| 0.0000000 | 0.8766677 | 0.690 | 0.041 | 0.0000000 | 5 | Adgrl4 |
| 0.0000000 | 1.0422162 | 0.929 | 0.300 | 0.0000000 | 2 | Gm3764 |
| 0.0000000 | 1.0070704 | 0.941 | 0.466 | 0.0000000 | 2 | Slc4a4 |
| 0.0000000 | 0.8575195 | 0.894 | 0.415 | 0.0000000 | 2 | Gm48747 |
| 0.0000000 | 0.8463038 | 0.941 | 0.628 | 0.0000000 | 2 | Ptprz1 |
| 0.0000000 | 0.7881937 | 0.894 | 0.152 | 0.0000000 | 2 | Bmpr1b |
| 0.0000000 | 0.7745003 | 0.765 | 0.174 | 0.0000000 | 2 | Slc6a11 |
| 0.0000000 | 0.7419631 | 0.765 | 0.451 | 0.0000000 | 2 | Gpc5 |
| 0.0000000 | 0.7406946 | 0.929 | 0.289 | 0.0000000 | 2 | Plpp3 |
| 0.0000000 | 0.7340348 | 0.812 | 0.539 | 0.0000000 | 2 | Luzp2 |
| 0.0000000 | 0.7330892 | 0.824 | 0.197 | 0.0000000 | 2 | Pla2g7 |
| 0.0000000 | 0.7305223 | 0.706 | 0.136 | 0.0000000 | 2 | Tnc |
| 0.0000000 | 0.7193021 | 0.894 | 0.316 | 0.0000000 | 2 | Apoe |
| 0.0000000 | 0.7134415 | 0.871 | 0.574 | 0.0000000 | 2 | Trim9 |
| 0.0000000 | 0.7109505 | 0.859 | 0.233 | 0.0000000 | 2 | Lrig1 |
| 0.0000000 | 0.6843853 | 0.859 | 0.228 | 0.0000000 | 2 | Palld |
| 0.0000000 | 0.6781358 | 0.941 | 0.620 | 0.0000000 | 2 | Gpm6b |
| 0.0000000 | 0.6760270 | 0.776 | 0.136 | 0.0000000 | 2 | Egfr |
| 0.0000000 | 0.6756797 | 0.812 | 0.190 | 0.0000000 | 2 | Cdh20 |
| 0.0000000 | 0.6737712 | 0.965 | 0.695 | 0.0000000 | 2 | Npas3 |
| 0.0000000 | 0.6668370 | 0.929 | 0.582 | 0.0000000 | 2 | Ptn |
| 0.0000000 | 1.1536760 | 0.966 | 0.182 | 0.0000000 | 4 | 6330411D24Rik |
| 0.0000000 | 0.8713080 | 1.000 | 0.320 | 0.0000000 | 4 | Pex5l |
| 0.0000000 | 0.7573003 | 1.000 | 0.344 | 0.0000000 | 4 | Spock1 |
| 0.0000000 | 0.7044603 | 1.000 | 0.414 | 0.0000000 | 4 | Ryr3 |
| 0.0000000 | 0.6954763 | 1.000 | 0.650 | 0.0000000 | 4 | Hs6st3 |
| 0.0000000 | 0.6895827 | 1.000 | 0.441 | 0.0000000 | 4 | Arpp21 |
| 0.0000000 | 0.6778960 | 0.983 | 0.386 | 0.0000000 | 4 | Edil3 |
| 0.0000000 | 0.6528223 | 0.914 | 0.171 | 0.0000000 | 4 | 4930419G24Rik |
| 0.0000000 | 0.6316032 | 0.966 | 0.485 | 0.0000000 | 4 | Adarb1 |
| 0.0000000 | 0.6239836 | 1.000 | 0.463 | 0.0000000 | 4 | Ntng1 |
| 0.0000000 | 0.6026214 | 0.983 | 0.408 | 0.0000000 | 4 | Camk4 |
| 0.0000000 | 0.5882924 | 0.948 | 0.281 | 0.0000000 | 4 | Grm1 |
| 0.0000000 | 0.5851017 | 1.000 | 0.551 | 0.0000000 | 4 | Gria4 |
| 0.0000000 | 0.5819572 | 0.845 | 0.138 | 0.0000000 | 4 | Pappa |
| 0.0000000 | 0.5682423 | 1.000 | 0.416 | 0.0000000 | 4 | Zmat4 |
| 0.0000000 | 0.5650541 | 0.983 | 0.429 | 0.0000000 | 4 | Hs3st5 |
| 0.0000000 | 0.5621780 | 0.966 | 0.378 | 0.0000000 | 4 | Epb41l4b |
| 0.0000000 | 0.5534059 | 0.983 | 0.317 | 0.0000000 | 4 | Ptpn3 |
| 0.0000000 | 0.5524678 | 1.000 | 0.548 | 0.0000000 | 4 | Lrrc7 |
| 0.0000000 | 0.5481946 | 0.948 | 0.350 | 0.0000000 | 4 | Cntn3 |
| 0.0000000 | 0.9735890 | 0.854 | 0.367 | 0.0000000 | 9 | Pcdh15 |
| 0.0000000 | 0.9602308 | 0.415 | 0.222 | 0.0000000 | 9 | Mbp |
| 0.0000000 | 0.9524441 | 0.829 | 0.110 | 0.0000000 | 9 | 4930588A03Rik |
| 0.0000000 | 0.9147655 | 0.951 | 0.408 | 0.0000000 | 9 | Tnr |
| 0.0000000 | 0.8757874 | 0.293 | 0.130 | 0.0000000 | 9 | Plp1 |
| 0.0000000 | 0.8633900 | 0.829 | 0.205 | 0.0000000 | 9 | Dscaml1 |
| 0.0000000 | 0.8515065 | 0.854 | 0.492 | 0.0000000 | 9 | 6030443J06Rik |
| 0.0000000 | 0.8470640 | 0.902 | 0.061 | 0.0000000 | 9 | Sox10 |
| 0.0000000 | 0.8440775 | 0.829 | 0.124 | 0.0000000 | 9 | Gm38505 |
| 0.0000000 | 0.8367428 | 0.512 | 0.183 | 0.0000000 | 9 | 9630013A20Rik |
| 0.0000000 | 0.8333975 | 0.756 | 0.170 | 0.0000000 | 9 | Bcas1 |
| 0.0000000 | 0.8047986 | 0.659 | 0.116 | 0.0000000 | 9 | Pdgfra |
| 0.0000000 | 0.7785127 | 0.878 | 0.385 | 0.0000000 | 9 | Lhfpl3 |
| 0.0000000 | 0.7383577 | 0.244 | 0.052 | 0.0000000 | 9 | St18 |
| 0.0000000 | 0.7251675 | 0.902 | 0.598 | 0.0000000 | 9 | Sox2ot |
| 0.0000000 | 0.6847430 | 0.902 | 0.477 | 0.0000000 | 9 | Sox6 |
| 0.0000000 | 0.6739720 | 0.878 | 0.401 | 0.0000000 | 9 | Nxph1 |
| 0.0000000 | 0.6728349 | 0.829 | 0.274 | 0.0000000 | 9 | Tns3 |
| 0.0000000 | 0.6329265 | 0.707 | 0.098 | 0.0000000 | 9 | Vxn |
| 0.0000000 | 0.6283993 | 0.976 | 0.641 | 0.0000000 | 9 | Opcml |
| 0.0000000 | 1.3307978 | 0.955 | 0.135 | 0.0000000 | 13 | Adamts12 |
| 0.0000000 | 1.2983823 | 0.955 | 0.248 | 0.0000000 | 13 | Adam12 |
| 0.0000000 | 1.2333967 | 0.955 | 0.135 | 0.0000000 | 13 | Cped1 |
| 0.0000000 | 1.1018416 | 0.864 | 0.022 | 0.0000000 | 13 | Slc6a13 |
| 0.0000000 | 1.0967793 | 0.773 | 0.065 | 0.0000000 | 13 | Ranbp3l |
| 0.0000000 | 1.0930283 | 0.455 | 0.025 | 0.0000121 | 13 | Ptgds |
| 0.0000000 | 1.0826431 | 0.864 | 0.184 | 0.0000000 | 13 | Sidt1 |
| 0.0000000 | 1.0723408 | 0.773 | 0.224 | 0.0000000 | 13 | Bmp6 |
| 0.0000000 | 1.0451808 | 1.000 | 0.422 | 0.0000000 | 13 | Bicc1 |
| 0.0000000 | 1.0180167 | 0.773 | 0.042 | 0.0000000 | 13 | Aox3 |
| 0.0000000 | 0.9944784 | 0.864 | 0.230 | 0.0000000 | 13 | Slc7a11 |
| 0.0000000 | 0.9472832 | 0.909 | 0.048 | 0.0000000 | 13 | Col3a1 |
| 0.0000000 | 0.9046290 | 0.773 | 0.218 | 0.0000000 | 13 | Tmtc4 |
| 0.0000000 | 0.8772338 | 0.636 | 0.056 | 0.0000002 | 13 | Col1a2 |
| 0.0000000 | 0.8680208 | 0.864 | 0.446 | 0.0000000 | 13 | Svil |
| 0.0000000 | 0.8660678 | 0.727 | 0.046 | 0.0000000 | 13 | Cfh |
| 0.0000000 | 0.8561243 | 0.818 | 0.166 | 0.0000000 | 13 | Col4a5 |
| 0.0000000 | 0.8513968 | 0.545 | 0.088 | 0.0000000 | 13 | Itga8 |
| 0.0000000 | 0.8347594 | 0.864 | 0.155 | 0.0000000 | 13 | Arhgap29 |
| 0.0000000 | 0.8295859 | 0.818 | 0.068 | 0.0000000 | 13 | Col4a6 |
Pr5P7.markers.mast %>%
group_by(cluster) %>%
top_n(n = 10, wt = avg_log10FC) -> top10
DoHeatmap(combined.srt, features = top10$gene) + NoLegend()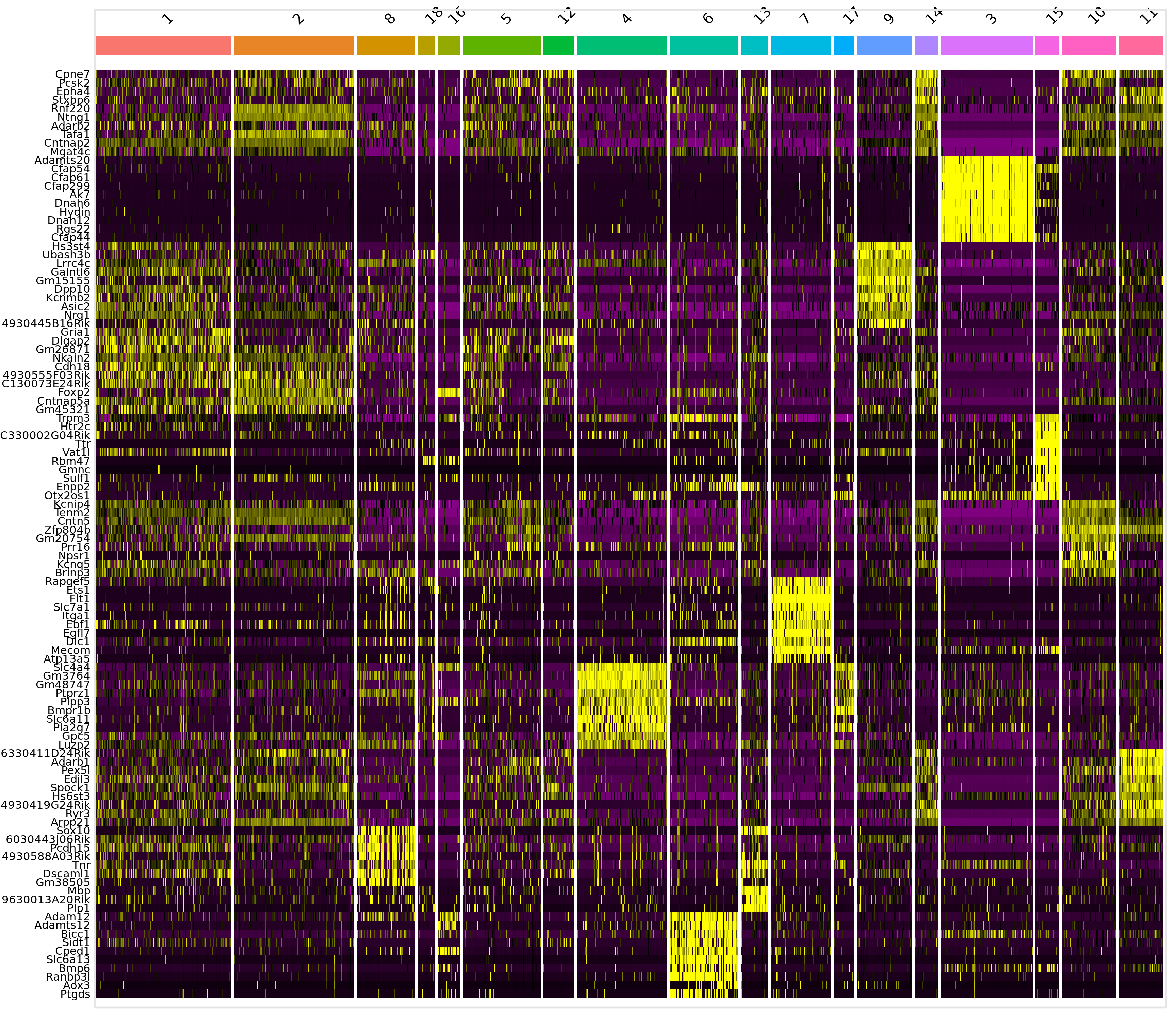
FeaturePlot(Pr5P7, "Galr1", pt.size = 3, order = T) +
scale_colour_gradientn(colours = rev(brewer.pal(n = 11, name = "Spectral"))) + ggtitle("Galr1: ") + theme(plot.title = element_text(size = 24))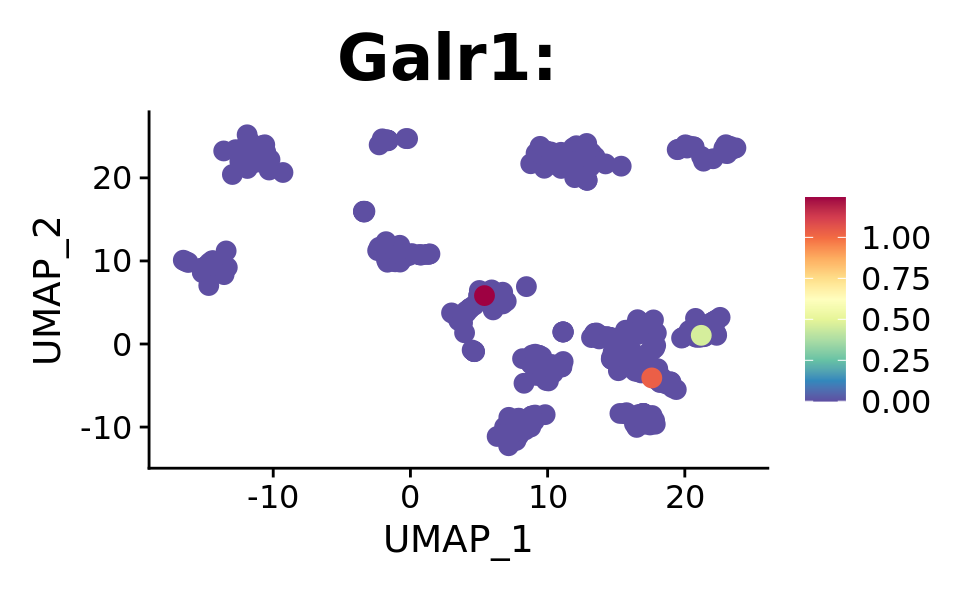
FeaturePlot(Pr5P7, "Gal", pt.size = 3, order = T) +
scale_colour_gradientn(colours = rev(brewer.pal(n = 11, name = "Spectral"))) + ggtitle("Gal: ") + theme(plot.title = element_text(size = 24))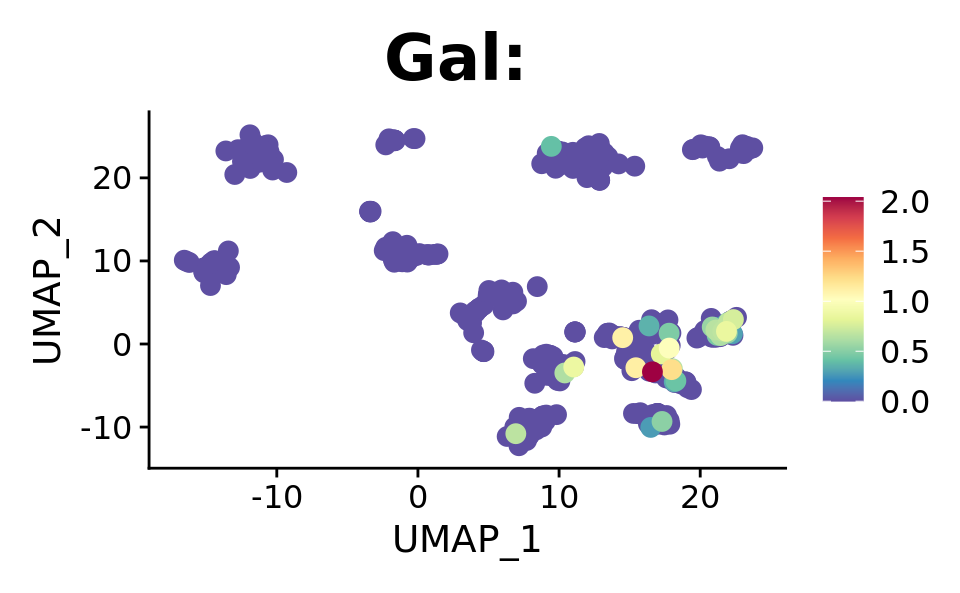
SaveH5Seurat(Pr5P7,
filename = here(data_dir,
"Pr5P7_clusters.h5Seurat"))
Convert(here(data_dir,
"Pr5P7_clusters.h5Seurat"),
dest = "h5ad")
sessionInfo()R version 4.2.2 (2022-10-31)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 22.04.1 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] ggraph_2.1.0.9000 gprofiler2_0.2.1 mrtree_0.0.0.9000
[4] Nebulosa_1.8.0 scCustomize_1.1.0 Scillus_0.5.0
[7] qs_0.25.4 patchwork_1.1.2.9000 glmGamPoi_1.10.1
[10] sctransform_0.3.5 SeuratDisk_0.0.0.9020 SeuratWrappers_0.3.1
[13] SeuratObject_4.1.3 Seurat_4.3.0 reticulate_1.27
[16] kableExtra_1.3.4 zeallot_0.1.0 future_1.30.0
[19] skimr_2.1.5 magrittr_2.0.3 lubridate_1.9.0
[22] timechange_0.1.1 forcats_0.5.2 stringr_1.5.0
[25] dplyr_1.0.10 purrr_1.0.1 readr_2.1.3
[28] tidyr_1.2.1.9001 tibble_3.1.8 ggplot2_3.4.0
[31] tidyverse_1.3.2.9000 viridis_0.6.2 viridisLite_0.4.1
[34] RColorBrewer_1.1-3 here_1.0.1 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] rsvd_1.0.5 ica_1.0-3
[3] svglite_2.1.0 ps_1.7.2
[5] foreach_1.5.2 lmtest_0.9-40
[7] rprojroot_2.0.3 crayon_1.5.2
[9] MASS_7.3-58.1 MAST_1.24.0
[11] nlme_3.1-161 backports_1.4.1
[13] rlang_1.0.6 XVector_0.38.0
[15] ROCR_1.0-11 irlba_2.3.5.1
[17] callr_3.7.3 limma_3.54.0
[19] stringfish_0.15.7 data.tree_1.0.0
[21] rjson_0.2.21 bit64_4.0.5
[23] glue_1.6.2 parallel_4.2.2
[25] processx_3.8.0 vipor_0.4.5
[27] spatstat.sparse_3.0-0 BiocGenerics_0.44.0
[29] spatstat.geom_3.0-3 tidyselect_1.2.0
[31] SummarizedExperiment_1.28.0 fitdistrplus_1.1-8
[33] zoo_1.8-11 xtable_1.8-4
[35] formattable_0.2.1 evaluate_0.19
[37] cli_3.6.0 zlibbioc_1.44.0
[39] rstudioapi_0.14 miniUI_0.1.1.1
[41] sp_1.5-1 whisker_0.4.1
[43] bslib_0.4.2 fastmatch_1.1-3
[45] treeio_1.23.0 maps_3.4.1
[47] shiny_1.7.4 xfun_0.36
[49] clue_0.3-63 cluster_2.1.4
[51] tidygraph_1.2.2 clusterGeneration_1.3.7
[53] expm_0.999-6 SymSim_0.0.0.9000
[55] ggrepel_0.9.2.9999 ape_5.6-2
[57] listenv_0.9.0 dendextend_1.16.0
[59] png_0.1-8 withr_2.5.0
[61] bitops_1.0-7 ggforce_0.4.1.9000
[63] plyr_1.8.8 pracma_2.4.2
[65] coda_0.19-4 pillar_1.8.1
[67] RcppParallel_5.1.5 GlobalOptions_0.1.2
[69] cachem_1.0.6 fs_1.5.2
[71] scatterplot3d_0.3-42 hdf5r_1.3.7
[73] GetoptLong_1.0.5 paletteer_1.5.0
[75] vctrs_0.5.1 ellipsis_0.3.2
[77] generics_0.1.3 RApiSerialize_0.1.2
[79] tools_4.2.2 beeswarm_0.4.0
[81] munsell_0.5.0 tweenr_2.0.2
[83] DelayedArray_0.24.0 fastmap_1.1.0
[85] compiler_4.2.2 abind_1.4-5
[87] httpuv_1.6.7 ggimage_0.3.1
[89] plotly_4.10.1 GenomeInfoDbData_1.2.9
[91] gridExtra_2.3 lattice_0.20-45
[93] deldir_1.0-6 utf8_1.2.2
[95] later_1.3.0 jsonlite_1.8.4
[97] scales_1.2.1 tidytree_0.4.2
[99] pbapply_1.6-0 lazyeval_0.2.2
[101] promises_1.2.0.1 doParallel_1.0.17
[103] R.utils_2.12.2 goftest_1.2-3
[105] spatstat.utils_3.0-1 checkmate_2.1.0
[107] rmarkdown_2.19 cowplot_1.1.1
[109] webshot_0.5.4 Rtsne_0.16
[111] Biobase_2.58.0 uwot_0.1.14
[113] igraph_1.3.5 survival_3.4-0
[115] numDeriv_2016.8-1.1 yaml_2.3.6
[117] plotrix_3.8-2 systemfonts_1.0.4
[119] htmltools_0.5.4 graphlayouts_0.8.4
[121] IRanges_2.32.0 quadprog_1.5-8
[123] digest_0.6.31 assertthat_0.2.1
[125] mime_0.12 repr_1.1.4
[127] yulab.utils_0.0.6 future.apply_1.10.0
[129] ggmin_0.0.0.9000 remotes_2.4.2
[131] data.table_1.14.6 S4Vectors_0.36.1
[133] R.oo_1.25.0 labeling_0.4.2
[135] splines_4.2.2 rematch2_2.1.2
[137] RCurl_1.98-1.9 ks_1.14.0
[139] hms_1.1.2 colorspace_2.0-3
[141] base64enc_0.1-3 BiocManager_1.30.19
[143] mnormt_2.1.1 ggbeeswarm_0.7.1
[145] GenomicRanges_1.50.2 shape_1.4.6
[147] aplot_0.1.9 ggrastr_1.0.1
[149] sass_0.4.4 Rcpp_1.0.9
[151] mclust_6.0.0 RANN_2.6.1
[153] mvtnorm_1.1-3 circlize_0.4.15
[155] fansi_1.0.3 tzdb_0.3.0
[157] parallelly_1.33.0 R6_2.5.1
[159] grid_4.2.2 ggridges_0.5.4
[161] lifecycle_1.0.3 phytools_1.2-0
[163] leiden_0.4.3 phangorn_2.10.0
[165] jquerylib_0.1.4 snakecase_0.11.0
[167] Matrix_1.5-3 RcppAnnoy_0.0.20
[169] iterators_1.0.14 spatstat.explore_3.0-5
[171] htmlwidgets_1.6.1 polyclip_1.10-4
[173] gridGraphics_0.5-1 optimParallel_1.0-2
[175] rvest_1.0.3 ComplexHeatmap_2.14.0
[177] globals_0.16.2 spatstat.random_3.0-1
[179] progressr_0.13.0 codetools_0.2-18
[181] matrixStats_0.63.0 prettyunits_1.1.1
[183] getPass_0.2-2 SingleCellExperiment_1.20.0
[185] RSpectra_0.16-1 R.methodsS3_1.8.2
[187] GenomeInfoDb_1.34.6 gtable_0.3.1
[189] DBI_1.1.3 git2r_0.30.1
[191] stats4_4.2.2 ggfun_0.0.9
[193] tensor_1.5 httr_1.4.4
[195] highr_0.10 KernSmooth_2.23-20
[197] progress_1.2.2 stringi_1.7.8
[199] vroom_1.6.0 reshape2_1.4.4
[201] farver_2.1.1 magick_2.7.3
[203] ggtree_3.7.1.002 xml2_1.3.3
[205] combinat_0.0-8 ggplotify_0.1.0
[207] scattermore_0.8 bit_4.0.5
[209] clustree_0.5.0 MatrixGenerics_1.10.0
[211] spatstat.data_3.0-0 janitor_2.1.0.9000
[213] pkgconfig_2.0.3 ggprism_1.0.4
[215] knitr_1.41