RefSubIntegrationComparison
Last updated: 2021-07-05
Checks: 5 2
Knit directory: Embryoid_Body_Pilot_Workflowr/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R Markdown file created these results, you'll want to first commit it to the Git repo. If you're still working on the analysis, you can ignore this warning. When you're finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it's best to always run the code in an empty environment.
The command set.seed(20200804) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /project2/gilad/katie/Pilot_HumanEBs/Embryoid_Body_Pilot_Workflowr/output/TranferredAnnotations_ReferenceInt_JustPluripotent.csv | ../output/TranferredAnnotations_ReferenceInt_JustPluripotent.csv |
| /project2/gilad/katie/Pilot_HumanEBs/Embryoid_Body_Pilot_Workflowr/output/TranferredAnnotations_ReferenceInt_JustEndoderm.csv | ../output/TranferredAnnotations_ReferenceInt_JustEndoderm.csv |
| /project2/gilad/katie/Pilot_HumanEBs/Embryoid_Body_Pilot_Workflowr/output/TranferredAnnotations_ReferenceInt_JustMeso.csv | ../output/TranferredAnnotations_ReferenceInt_JustMeso.csv |
| /project2/gilad/katie/Pilot_HumanEBs/Embryoid_Body_Pilot_Workflowr/output/TranferredAnnotations_ReferenceInt_JustEarlyEcto.csv | ../output/TranferredAnnotations_ReferenceInt_JustEarlyEcto.csv |
| /project2/gilad/katie/Pilot_HumanEBs/Embryoid_Body_Pilot_Workflowr/output/TranferredAnnotations_ReferenceInt_JustNeuron.csv | ../output/TranferredAnnotations_ReferenceInt_JustNeuron.csv |
| /project2/gilad/katie/Pilot_HumanEBs/Embryoid_Body_Pilot_Workflowr/output/TranferredAnnotations_ReferenceInt_JustNeuralCrest.csv | ../output/TranferredAnnotations_ReferenceInt_JustNeuralCrest.csv |
| /project2/gilad/katie/Pilot_HumanEBs/Embryoid_Body_Pilot_Workflowr/output/MostCommonAnnotation.FiveNearestRefCells.csv | ../output/MostCommonAnnotation.FiveNearestRefCells.csv |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version c8767ac. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.Rhistory
Ignored: output/.Rhistory
Untracked files:
Untracked: GSE122380_raw_counts.txt.gz
Untracked: UTF1_plots.Rmd
Untracked: analysis/IntegrateReference_SCTregressCaoPlusScHCL_JustEarlyEcto.Rmd
Untracked: analysis/IntegrateReference_SCTregressCaoPlusScHCL_JustEndo.Rmd
Untracked: analysis/IntegrateReference_SCTregressCaoPlusScHCL_JustMeso.Rmd
Untracked: analysis/IntegrateReference_SCTregressCaoPlusScHCL_JustNeuralCrest.Rmd
Untracked: analysis/IntegrateReference_SCTregressCaoPlusScHCL_JustNeuron.Rmd
Untracked: analysis/IntegrateReference_SCTregressCaoPlusScHCL_JustPluri.Rmd
Untracked: analysis/OLD/
Untracked: analysis/Pseudobulk_Limma_Harmony.BatchIndividual_ClusterRes0.8_minPCT0.2.Rmd
Untracked: analysis/Pseudobulk_Limma_Harmony.BatchIndividual_ClusterRes1_minPCT0.2.Rmd
Untracked: analysis/Pseudobulk_VariancePartition_Harmony.Batchindividual_ClusterRes0.1_byCluster.Rmd
Untracked: analysis/RefInt_ComparingFulltoPartialIntegrationAnnotations.Rmd
Untracked: analysis/ReferenceAnn_DE.Rmd
Untracked: analysis/SingleCell_HierarchicalClustering_NoGeneFilter.Rmd
Untracked: analysis/SingleCell_VariancePartitionByCluster_Harmony.Batchindividual_ClusterRes0.1_minPCT0.2.Rmd
Untracked: analysis/VarPartPlots_res0.1_SCT.Rmd
Untracked: analysis/VarPart_SC_res0.1_SCT.Rmd
Untracked: analysis/child/
Untracked: analysis/k10topics_Explore.Rmd
Untracked: analysis/k6topics_Explore.Rmd
Untracked: build_refint_scale.R
Untracked: build_refint_sct.R
Untracked: build_stuff.R
Untracked: build_varpart_sc.R
Untracked: code/.ipynb_checkpoints/
Untracked: code/CellRangerPreprocess.Rmd
Untracked: code/ConvertToDGE.Rmd
Untracked: code/ConvertToDGE_PseudoBulk.Rmd
Untracked: code/ConvertToDGE_SingleCellRes_minPCT0.2.Rmd
Untracked: code/EB.getHumanMetadata.Rmd
Untracked: code/GEO_processed_data.Rmd
Untracked: code/PowerAnalysis_NoiseRatio.ipynb
Untracked: code/Untitled.ipynb
Untracked: code/Untitled1.ipynb
Untracked: code/compile_fits.Rmd
Untracked: code/fit_all_models.sh
Untracked: code/fit_poisson_nmf.R
Untracked: code/fit_poisson_nmf.sbatch
Untracked: code/functions_for_fit_comparison.Rmd
Untracked: code/get_genelist_byPCTthresh.Rmd
Untracked: code/prefit_poisson_nmf.R
Untracked: code/prefit_poisson_nmf.sbatch
Untracked: code/prepare_data_for_fastTopics.Rmd
Untracked: data/HCL_Fig1_adata.h5ad
Untracked: data/HCL_Fig1_adata.h5seurat
Untracked: data/dge/
Untracked: data/dge_raw_data.tar.gz
Untracked: data/ref.expr.rda
Untracked: figure/
Untracked: output/CR_sampleQCrds/
Untracked: output/CaoEtAl.Obj.CellsOfAllClusters.ProteinCodingGenes.rds
Untracked: output/CaoEtAl.Obj.rds
Untracked: output/ClusterInfo_res0.1.csv
Untracked: output/DGELists/
Untracked: output/DownSampleVarPart.rds
Untracked: output/Frequency.MostCommonAnnotation.FiveNearestRefCells.csv
Untracked: output/GEOsubmissionProcessedFiles/
Untracked: output/GeneLists_by_minPCT/
Untracked: output/MostCommonAnnotation.FiveNearestRefCells.csv
Untracked: output/NearestReferenceCell.Cao.hESC.EuclideanDistanceinHarmonySpace.csv
Untracked: output/NearestReferenceCell.Cao.hESC.FrequencyofEachAnnotation.csv
Untracked: output/NearestReferenceCell.SCTregressRNAassay.Cao.hESC.EuclideanDistanceinHarmonySpace.csv
Untracked: output/NearestReferenceCell.SCTregressRNAassay.Cao.hESC.FrequencyofEachAnnotation.csv
Untracked: output/Pseudobulk_Limma_res0.1_OnevAllTopTables.csv
Untracked: output/Pseudobulk_Limma_res0.1_OnevAll_top10Upregby_adjP.csv
Untracked: output/Pseudobulk_Limma_res0.1_OnevAll_top10Upregby_logFC.csv
Untracked: output/Pseudobulk_Limma_res0.5_OnevAllTopTables.csv
Untracked: output/Pseudobulk_Limma_res0.8_OnevAllTopTables.csv
Untracked: output/Pseudobulk_Limma_res1_OnevAllTopTables.csv
Untracked: output/Pseudobulk_VarPart.ByCluster.Res0.1.rds
Untracked: output/ResidualVariances_fromDownSampAnalysis.csv
Untracked: output/SingleCell_VariancePartition_RNA_Res0.1_minPCT0.2.rds
Untracked: output/SingleCell_VariancePartition_Res0.1_minPCT0.2.rds
Untracked: output/SingleCell_VariancePartition_SCT_Res0.1_minPCT0.2.rds
Untracked: output/TopicModelling_k10_top10drivergenes.byBeta.csv
Untracked: output/TopicModelling_k6_top10drivergenes.byBeta.csv
Untracked: output/TopicModelling_k6_top15drivergenes.byZ.csv
Untracked: output/TranferredAnnotations_ReferenceInt_JustEarlyEcto.csv
Untracked: output/TranferredAnnotations_ReferenceInt_JustEndoderm.csv
Untracked: output/TranferredAnnotations_ReferenceInt_JustMeso.csv
Untracked: output/TranferredAnnotations_ReferenceInt_JustNeuralCrest.csv
Untracked: output/TranferredAnnotations_ReferenceInt_JustNeuron.csv
Untracked: output/TranferredAnnotations_ReferenceInt_JustPluripotent.csv
Untracked: output/VarPart.ByCluster.Res0.1.rds
Untracked: output/azimuth/
Untracked: output/downsamp_10800cells_10subreps_medianexplainedbyresiduals_varpart_PsB.rds
Untracked: output/downsamp_16200cells_10subreps_medianexplainedbyresiduals_varpart_PsB.rds
Untracked: output/downsamp_21600cells_10subreps_medianexplainedbyresiduals_varpart_PsB.rds
Untracked: output/downsamp_2700cells_10subreps_medianexplainedbyresiduals_varpart_PsB.rds
Untracked: output/downsamp_2700cells_10subreps_medianexplainedbyresiduals_varpart_scres.rds
Untracked: output/downsamp_5400cells_10subreps_medianexplainedbyresiduals_varpart_PsB.rds
Untracked: output/downsamp_7200cells_10subreps_medianexplainedbyresiduals_varpart_PsB.rds
Untracked: output/fasttopics/
Untracked: output/figs/
Untracked: output/merge.Cao.SCTwRegressOrigIdent.rds
Untracked: output/merge.all.SCTwRegressOrigIdent.Harmony.rds
Untracked: output/merged.SCT.counts.matrix.rds
Untracked: output/merged.raw.counts.matrix.rds
Untracked: output/mergedObjects/
Untracked: output/pdfs/
Untracked: output/sampleQCrds/
Untracked: output/splitgpm_gsea_results/
Untracked: slurm-12005914.out
Untracked: slurm-12005923.out
Unstaged changes:
Deleted: analysis/IntegrateAnalysis.afterFilter.HarmonyBatch.Rmd
Deleted: analysis/IntegrateAnalysis.afterFilter.HarmonyBatchSampleIDindividual.Rmd
Modified: analysis/IntegrateAnalysis.afterFilter.HarmonyBatchindividual.Rmd
Deleted: analysis/IntegrateAnalysis.afterFilter.NOHARMONYjustmerge.Rmd
Deleted: analysis/IntegrateAnalysis.afterFilter.SCTregressBatchIndividual.Rmd
Deleted: analysis/IntegrateAnalysis.afterFilter.SCTregressBatchIndividualHarmonyBatchindividual.Rmd
Modified: analysis/Pseudobulk_HierarchicalClustering_Harmony.Batchindividual_ClusterRes0.1_minPCT0.2.Rmd
Modified: analysis/Pseudobulk_HierarchicalClustering_Harmony.Batchindividual_ClusterRes0.5_minPCT0.2.Rmd
Modified: analysis/Pseudobulk_HierarchicalClustering_Harmony.Batchindividual_ClusterRes0.8_minPCT0.2.Rmd
Modified: analysis/Pseudobulk_HierarchicalClustering_Harmony.Batchindividual_ClusterRes1_minPCT0.2.Rmd
Modified: analysis/Pseudobulk_Limma_Harmony.BatchIndividual_ClusterRes0.1_minPCT0.2.Rmd
Modified: analysis/Pseudobulk_Limma_Harmony.BatchIndividual_ClusterRes0.5_minPCT0.2.Rmd
Modified: analysis/Pseudobulk_VariancePartition_Harmony.Batchindividual_ClusterRes0.1_minPCT0.2.Rmd
Modified: analysis/Pseudobulk_VariancePartition_Harmony.Batchindividual_ClusterRes0.5_minPCT0.2.Rmd
Modified: analysis/Pseudobulk_VariancePartition_Harmony.Batchindividual_ClusterRes0.8_minPCT0.2.Rmd
Modified: analysis/Pseudobulk_VariancePartition_Harmony.Batchindividual_ClusterRes1_minPCT0.2.Rmd
Deleted: analysis/RunscHCL_HarmonyBatchInd.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with wflow_publish() to start tracking its development.
library(dplyr)
Attaching package: 'dplyr'The following objects are masked from 'package:stats':
filter, lagThe following objects are masked from 'package:base':
intersect, setdiff, setequal, unionplur<- read.csv("/project2/gilad/katie/Pilot_HumanEBs/Embryoid_Body_Pilot_Workflowr/output/TranferredAnnotations_ReferenceInt_JustPluripotent.csv")
end<- read.csv("/project2/gilad/katie/Pilot_HumanEBs/Embryoid_Body_Pilot_Workflowr/output/TranferredAnnotations_ReferenceInt_JustEndoderm.csv")
mes<- read.csv("/project2/gilad/katie/Pilot_HumanEBs/Embryoid_Body_Pilot_Workflowr/output/TranferredAnnotations_ReferenceInt_JustMeso.csv")
ee<- read.csv("/project2/gilad/katie/Pilot_HumanEBs/Embryoid_Body_Pilot_Workflowr/output/TranferredAnnotations_ReferenceInt_JustEarlyEcto.csv")
neur<- read.csv("/project2/gilad/katie/Pilot_HumanEBs/Embryoid_Body_Pilot_Workflowr/output/TranferredAnnotations_ReferenceInt_JustNeuron.csv")
nc<- read.csv("/project2/gilad/katie/Pilot_HumanEBs/Embryoid_Body_Pilot_Workflowr/output/TranferredAnnotations_ReferenceInt_JustNeuralCrest.csv")sepint<- rbind(plur, end, mes, ee, neur, nc)
colnames(sepint)<- c("X.sep", "EB.cell.id", "Annotation.sep", "NofFive.sep")all<- read.csv("/project2/gilad/katie/Pilot_HumanEBs/Embryoid_Body_Pilot_Workflowr/output/MostCommonAnnotation.FiveNearestRefCells.csv")comb<- full_join(all, sepint, by = "EB.cell.id")
dim(comb)[1] 42488 7sum(as.character(comb$Annotation) == as.character(comb$Annotation.sep))[1] 3371433714/42488[1] 0.793494679% of cells are assigned to the same cell type between full integration and partial integration
Let's see if we see any patterns in the cell types that disagree between integrations
dis<- comb[as.character(comb$Annotation) != as.character(comb$Annotation.sep),]barplot(table(dis$Annotation), las=2, cex.names = 0.4)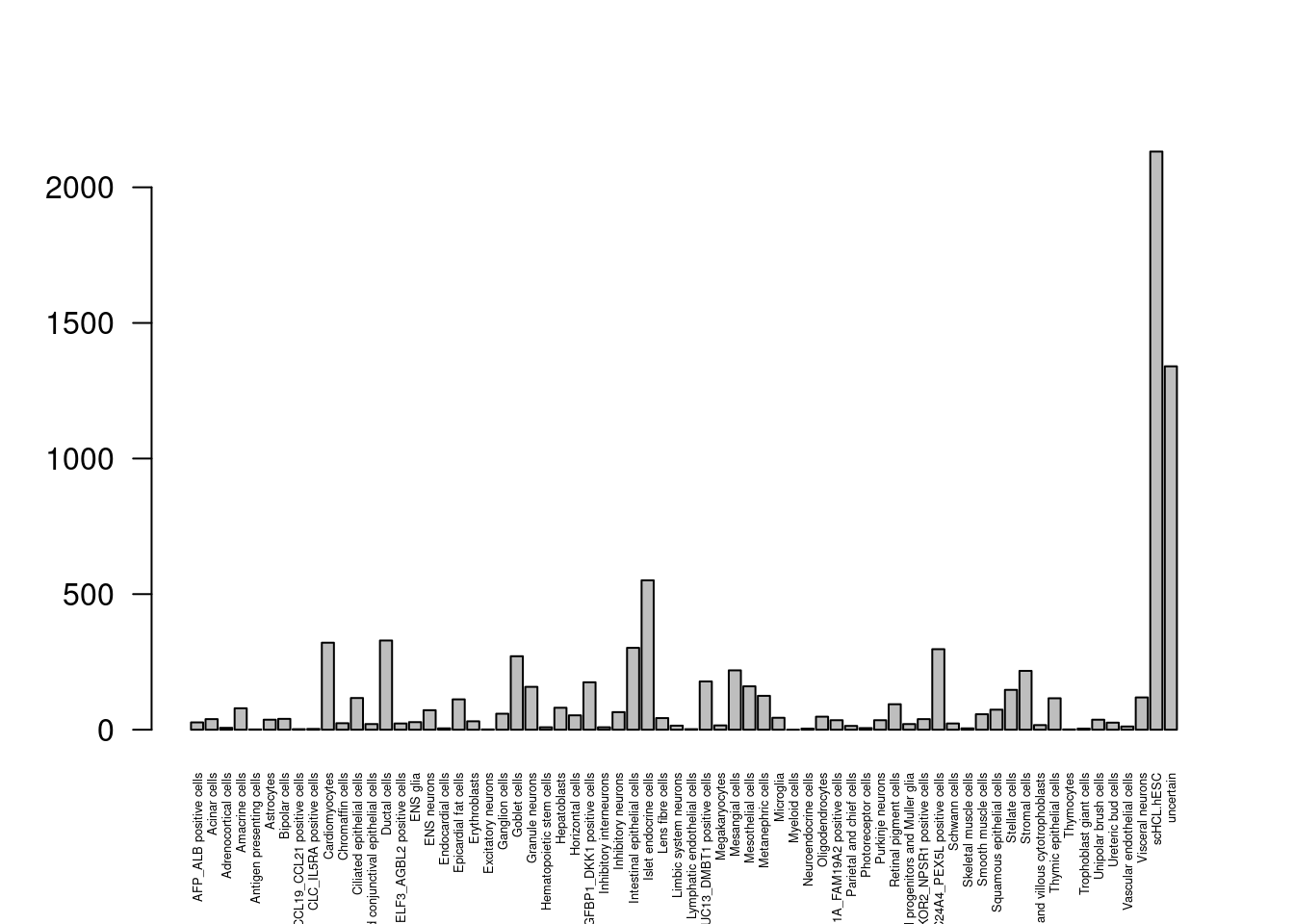
barplot(table(dis$Annotation.sep), las=2, cex.names = 0.4)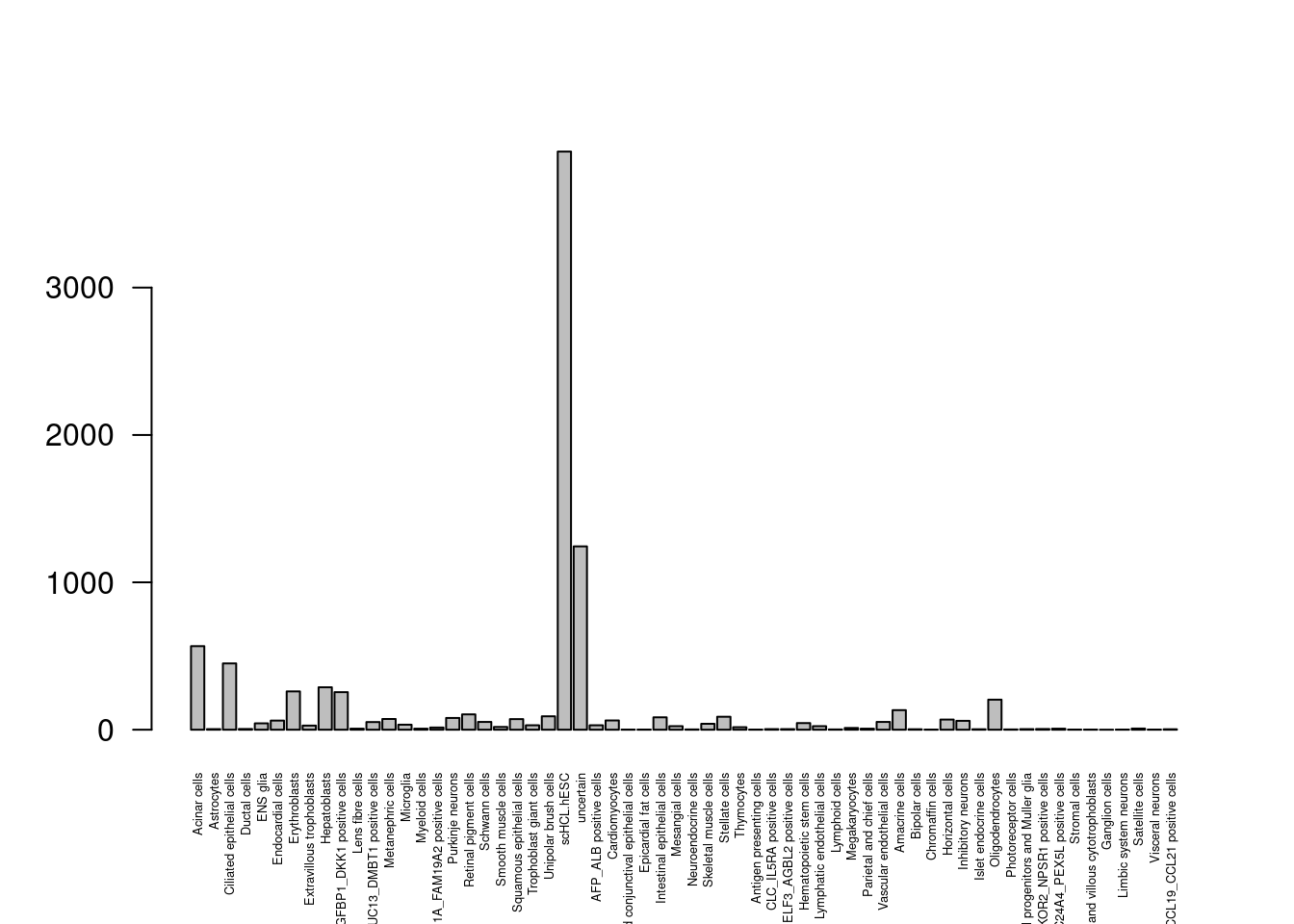
Lots of cells flipping to and from hESC, uncertain.
How many unique cell types total in the separate integrations?
length(unique(sepint$Annotation.sep))[1] 62how many in full integration?
length(unique(all$Annotation))[1] 68Both include uncertain. so its 66 fetal cell types + hESC in total, 60 fetal cell types + hESC from sep int.
Which cell types do we lose?
septypes<- as.character(unique(sepint$Annotation.sep))
alltypes<- as.character(unique(all$Annotation))
'%notin%' <- Negate('%in%')
lost<- alltypes[(alltypes %notin% septypes)]
lost[1] "ENS neurons" "Mesothelial cells"
[3] "Goblet cells" "Thymic epithelial cells"
[5] "Ureteric bud cells" "Granule neurons"
[7] "Inhibitory interneurons" "Adrenocortical cells"
[9] "Excitatory neurons" How many of these cell types were there in the full integration? were they rare?
taball<- table(all$Annotation)
taball[names(taball) %in% lost]
Adrenocortical cells ENS neurons Excitatory neurons
7 72 1
Goblet cells Granule neurons Inhibitory interneurons
271 158 9
Mesothelial cells Thymic epithelial cells Ureteric bud cells
160 116 26 looks like there are some cell types in the separate integration that are not in the full
gain<- septypes[septypes %notin% alltypes]
gain[1] "Extravillous trophoblasts" "Lymphoid cells"
[3] "Satellite cells" tabsep<- table(sepint$Annotation.sep)
tabsep[names(tabsep) %in% gain]
Extravillous trophoblasts Lymphoid cells Satellite cells
28 2 8 see most common label transitions
trans<- paste(dis$Annotation, "_", dis$Annotation.sep)
length(unique(trans))[1] 407switch<- sort(table(trans), decreasing = T)
switchtrans
uncertain _ scHCL.hESC
858
scHCL.hESC _ uncertain
591
Islet endocrine cells _ scHCL.hESC
511
scHCL.hESC _ Ciliated epithelial cells
288
Ductal cells _ scHCL.hESC
259
Intestinal epithelial cells _ scHCL.hESC
244
scHCL.hESC _ Acinar cells
238
scHCL.hESC _ Hepatoblasts
182
Cardiomyocytes _ scHCL.hESC
159
Goblet cells _ scHCL.hESC
155
Stromal cells _ scHCL.hESC
139
scHCL.hESC _ Amacrine cells
129
MUC13_DMBT1 positive cells _ scHCL.hESC
128
Stellate cells _ scHCL.hESC
127
SLC24A4_PEX5L positive cells _ scHCL.hESC
114
Ciliated epithelial cells _ scHCL.hESC
112
SLC24A4_PEX5L positive cells _ uncertain
106
Cardiomyocytes _ Erythroblasts
105
Mesothelial cells _ scHCL.hESC
104
Thymic epithelial cells _ Erythroblasts
95
uncertain _ IGFBP1_DKK1 positive cells
92
scHCL.hESC _ Retinal pigment cells
89
uncertain _ Oligodendrocytes
88
Granule neurons _ scHCL.hESC
86
Epicardial fat cells _ scHCL.hESC
84
Retinal pigment cells _ scHCL.hESC
82
IGFBP1_DKK1 positive cells _ Acinar cells
80
Goblet cells _ Unipolar brush cells
68
uncertain _ Acinar cells
62
Mesangial cells _ Acinar cells
61
Stromal cells _ IGFBP1_DKK1 positive cells
57
scHCL.hESC _ Vascular endothelial cells
52
scHCL.hESC _ Horizontal cells
51
ENS neurons _ Oligodendrocytes
50
Granule neurons _ Ciliated epithelial cells
50
Mesangial cells _ scHCL.hESC
50
scHCL.hESC _ Endocardial cells
50
Cardiomyocytes _ uncertain
49
Mesangial cells _ uncertain
48
Mesangial cells _ Metanephric cells
47
Metanephric cells _ scHCL.hESC
46
scHCL.hESC _ Squamous epithelial cells
46
Visceral neurons _ scHCL.hESC
45
Hepatoblasts _ Intestinal epithelial cells
43
IGFBP1_DKK1 positive cells _ scHCL.hESC
42
SLC24A4_PEX5L positive cells _ Purkinje neurons
41
Metanephric cells _ uncertain
40
Smooth muscle cells _ scHCL.hESC
40
Bipolar cells _ scHCL.hESC
39
Lens fibre cells _ scHCL.hESC
39
Ductal cells _ Acinar cells
37
Oligodendrocytes _ scHCL.hESC
37
SKOR2_NPSR1 positive cells _ scHCL.hESC
36
scHCL.hESC _ Schwann cells
36
Amacrine cells _ uncertain
35
scHCL.hESC _ Erythroblasts
35
uncertain _ Ciliated epithelial cells
34
Microglia _ scHCL.hESC
33
scHCL.hESC _ Stellate cells
33
uncertain _ Cardiomyocytes
33
Acinar cells _ scHCL.hESC
31
Visceral neurons _ Oligodendrocytes
31
Purkinje neurons _ scHCL.hESC
29
uncertain _ Hepatoblasts
29
Goblet cells _ Acinar cells
28
PDE11A_FAM19A2 positive cells _ scHCL.hESC
28
Squamous epithelial cells _ Stellate cells
28
scHCL.hESC _ Hematopoietic stem cells
27
Erythroblasts _ scHCL.hESC
26
Horizontal cells _ uncertain
26
Mesothelial cells _ uncertain
26
SLC24A4_PEX5L positive cells _ Inhibitory neurons
26
Amacrine cells _ Hepatoblasts
25
Astrocytes _ scHCL.hESC
25
scHCL.hESC _ ENS glia
25
Islet endocrine cells _ Ciliated epithelial cells
24
scHCL.hESC _ IGFBP1_DKK1 positive cells
24
Inhibitory neurons _ scHCL.hESC
23
Intestinal epithelial cells _ MUC13_DMBT1 positive cells
23
MUC13_DMBT1 positive cells _ Acinar cells
23
AFP_ALB positive cells _ Hepatoblasts
22
Ganglion cells _ scHCL.hESC
22
Hepatoblasts _ AFP_ALB positive cells
22
Inhibitory neurons _ IGFBP1_DKK1 positive cells
22
Intestinal epithelial cells _ uncertain
22
scHCL.hESC _ Extravillous trophoblasts
22
Corneal and conjunctival epithelial cells _ Squamous epithelial cells
21
scHCL.hESC _ Metanephric cells
21
scHCL.hESC _ Lymphatic endothelial cells
20
MUC13_DMBT1 positive cells _ Intestinal epithelial cells
19
Stromal cells _ uncertain
19
Ganglion cells _ Oligodendrocytes
18
IGFBP1_DKK1 positive cells _ uncertain
18
Mesothelial cells _ MUC13_DMBT1 positive cells
18
Retinal progenitors and Muller glia _ scHCL.hESC
18
Squamous epithelial cells _ IGFBP1_DKK1 positive cells
18
Visceral neurons _ uncertain
18
Epicardial fat cells _ uncertain
17
IGFBP1_DKK1 positive cells _ Mesangial cells
17
Metanephric cells _ Acinar cells
17
Unipolar brush cells _ Ciliated epithelial cells
17
scHCL.hESC _ Trophoblast giant cells
17
Ductal cells _ uncertain
16
ENS glia _ scHCL.hESC
16
Goblet cells _ uncertain
16
Granule neurons _ uncertain
16
Metanephric cells _ Inhibitory neurons
16
scHCL.hESC _ Microglia
16
Horizontal cells _ Hepatoblasts
15
Unipolar brush cells _ uncertain
15
scHCL.hESC _ Purkinje neurons
15
scHCL.hESC _ Unipolar brush cells
14
uncertain _ Purkinje neurons
14
Chromaffin cells _ scHCL.hESC
13
Visceral neurons _ IGFBP1_DKK1 positive cells
13
uncertain _ Erythroblasts
13
uncertain _ Skeletal muscle cells
13
Syncytiotrophoblasts and villous cytotrophoblasts _ scHCL.hESC
12
Ureteric bud cells _ Stellate cells
12
scHCL.hESC _ Skeletal muscle cells
12
scHCL.hESC _ Thymocytes
12
ENS neurons _ scHCL.hESC
11
Ganglion cells _ uncertain
11
Smooth muscle cells _ uncertain
11
Chromaffin cells _ uncertain
10
Islet endocrine cells _ uncertain
10
Schwann cells _ scHCL.hESC
10
Squamous epithelial cells _ scHCL.hESC
10
Stellate cells _ IGFBP1_DKK1 positive cells
10
Thymic epithelial cells _ scHCL.hESC
10
scHCL.hESC _ Inhibitory neurons
10
uncertain _ Horizontal cells
10
uncertain _ Retinal pigment cells
10
Megakaryocytes _ scHCL.hESC
9
Squamous epithelial cells _ uncertain
9
Stellate cells _ uncertain
9
scHCL.hESC _ Megakaryocytes
9
uncertain _ Intestinal epithelial cells
9
uncertain _ PDE11A_FAM19A2 positive cells
9
Ductal cells _ Ciliated epithelial cells
8
ELF3_AGBL2 positive cells _ Cardiomyocytes
8
ELF3_AGBL2 positive cells _ Erythroblasts
8
ENS glia _ Schwann cells
8
IGFBP1_DKK1 positive cells _ Smooth muscle cells
8
Retinal pigment cells _ uncertain
8
uncertain _ Microglia
8
uncertain _ Schwann cells
8
Astrocytes _ uncertain
7
Ganglion cells _ Purkinje neurons
7
Hepatoblasts _ uncertain
7
Limbic system neurons _ uncertain
7
Oligodendrocytes _ uncertain
7
Parietal and chief cells _ scHCL.hESC
7
Thymic epithelial cells _ Unipolar brush cells
7
scHCL.hESC _ Lens fibre cells
7
scHCL.hESC _ Myeloid cells
7
scHCL.hESC _ SLC24A4_PEX5L positive cells
7
uncertain _ AFP_ALB positive cells
7
uncertain _ Hematopoietic stem cells
7
Acinar cells _ Ciliated epithelial cells
6
Amacrine cells _ Trophoblast giant cells
6
Ductal cells _ Cardiomyocytes
6
Inhibitory neurons _ Oligodendrocytes
6
Inhibitory neurons _ Skeletal muscle cells
6
Inhibitory neurons _ uncertain
6
Limbic system neurons _ Inhibitory neurons
6
MUC13_DMBT1 positive cells _ uncertain
6
Mesothelial cells _ Stellate cells
6
Parietal and chief cells _ Acinar cells
6
Schwann cells _ ENS glia
6
Schwann cells _ uncertain
6
scHCL.hESC _ Cardiomyocytes
6
Adrenocortical cells _ scHCL.hESC
5
Epicardial fat cells _ MUC13_DMBT1 positive cells
5
Granule neurons _ IGFBP1_DKK1 positive cells
5
Hepatoblasts _ scHCL.hESC
5
Horizontal cells _ Trophoblast giant cells
5
Intestinal epithelial cells _ Cardiomyocytes
5
Mesothelial cells _ Intestinal epithelial cells
5
Unipolar brush cells _ scHCL.hESC
5
Ureteric bud cells _ scHCL.hESC
5
Visceral neurons _ Skeletal muscle cells
5
Amacrine cells _ ENS glia
4
Amacrine cells _ scHCL.hESC
4
ELF3_AGBL2 positive cells _ uncertain
4
ENS neurons _ uncertain
4
Endocardial cells _ Hematopoietic stem cells
4
Hematopoietic stem cells _ scHCL.hESC
4
Hepatoblasts _ MUC13_DMBT1 positive cells
4
Inhibitory interneurons _ scHCL.hESC
4
Megakaryocytes _ Hematopoietic stem cells
4
Mesangial cells _ Parietal and chief cells
4
PDE11A_FAM19A2 positive cells _ uncertain
4
Photoreceptor cells _ scHCL.hESC
4
SLC24A4_PEX5L positive cells _ Ciliated epithelial cells
4
SLC24A4_PEX5L positive cells _ Oligodendrocytes
4
Squamous epithelial cells _ Ciliated epithelial cells
4
Vascular endothelial cells _ Lymphatic endothelial cells
4
scHCL.hESC _ Oligodendrocytes
4
scHCL.hESC _ Satellite cells
4
uncertain _ ENS glia
4
uncertain _ Stellate cells
4
AFP_ALB positive cells _ Intestinal epithelial cells
3
Cardiomyocytes _ ELF3_AGBL2 positive cells
3
Ciliated epithelial cells _ uncertain
3
ENS glia _ uncertain
3
ENS neurons _ IGFBP1_DKK1 positive cells
3
ENS neurons _ Purkinje neurons
3
Erythroblasts _ uncertain
3
Horizontal cells _ scHCL.hESC
3
IGFBP1_DKK1 positive cells _ Ciliated epithelial cells
3
IGFBP1_DKK1 positive cells _ Stellate cells
3
Intestinal epithelial cells _ Acinar cells
3
Intestinal epithelial cells _ Thymocytes
3
Lens fibre cells _ Retinal pigment cells
3
Megakaryocytes _ uncertain
3
Mesangial cells _ IGFBP1_DKK1 positive cells
3
Mesangial cells _ Smooth muscle cells
3
Purkinje neurons _ Horizontal cells
3
Skeletal muscle cells _ scHCL.hESC
3
Smooth muscle cells _ IGFBP1_DKK1 positive cells
3
Squamous epithelial cells _ Extravillous trophoblasts
3
Trophoblast giant cells _ scHCL.hESC
3
Ureteric bud cells _ IGFBP1_DKK1 positive cells
3
Ureteric bud cells _ uncertain
3
Vascular endothelial cells _ Endocardial cells
3
Vascular endothelial cells _ uncertain
3
Visceral neurons _ Hepatoblasts
3
scHCL.hESC _ Astrocytes
3
scHCL.hESC _ Ductal cells
3
scHCL.hESC _ Retinal progenitors and Muller glia
3
scHCL.hESC _ Smooth muscle cells
3
Acinar cells _ uncertain
2
Adrenocortical cells _ uncertain
2
Amacrine cells _ Ciliated epithelial cells
2
Amacrine cells _ Extravillous trophoblasts
2
Astrocytes _ Oligodendrocytes
2
CLC_IL5RA positive cells _ Cardiomyocytes
2
Ductal cells _ Intestinal epithelial cells
2
Epicardial fat cells _ Intestinal epithelial cells
2
Epicardial fat cells _ Stellate cells
2
Goblet cells _ Neuroendocrine cells
2
Hematopoietic stem cells _ Acinar cells
2
Horizontal cells _ ENS glia
2
IGFBP1_DKK1 positive cells _ Endocardial cells
2
Inhibitory interneurons _ IGFBP1_DKK1 positive cells
2
Inhibitory interneurons _ uncertain
2
Intestinal epithelial cells _ Hepatoblasts
2
MUC13_DMBT1 positive cells _ Cardiomyocytes
2
Metanephric cells _ Microglia
2
Metanephric cells _ Squamous epithelial cells
2
Microglia _ Acinar cells
2
Microglia _ Endocardial cells
2
Neuroendocrine cells _ scHCL.hESC
2
Purkinje neurons _ Microglia
2
Retinal pigment cells _ Photoreceptor cells
2
Retinal progenitors and Muller glia _ uncertain
2
SKOR2_NPSR1 positive cells _ Ciliated epithelial cells
2
Smooth muscle cells _ Acinar cells
2
Stromal cells _ Smooth muscle cells
2
Syncytiotrophoblasts and villous cytotrophoblasts _ Ciliated epithelial cells
2
Syncytiotrophoblasts and villous cytotrophoblasts _ uncertain
2
Thymic epithelial cells _ Hepatoblasts
2
Visceral neurons _ Satellite cells
2
scHCL.hESC _ Bipolar cells
2
scHCL.hESC _ CCL19_CCL21 positive cells
2
scHCL.hESC _ Islet endocrine cells
2
scHCL.hESC _ Mesangial cells
2
scHCL.hESC _ PDE11A_FAM19A2 positive cells
2
scHCL.hESC _ SKOR2_NPSR1 positive cells
2
uncertain _ Ductal cells
2
uncertain _ Endocardial cells
2
uncertain _ Inhibitory neurons
2
uncertain _ Mesangial cells
2
uncertain _ Metanephric cells
2
uncertain _ Trophoblast giant cells
2
uncertain _ Unipolar brush cells
2
AFP_ALB positive cells _ scHCL.hESC
1
AFP_ALB positive cells _ uncertain
1
Amacrine cells _ Satellite cells
1
Antigen presenting cells _ scHCL.hESC
1
Astrocytes _ Amacrine cells
1
Astrocytes _ Endocardial cells
1
Astrocytes _ Microglia
1
Bipolar cells _ uncertain
1
CCL19_CCL21 positive cells _ scHCL.hESC
1
CCL19_CCL21 positive cells _ uncertain
1
CLC_IL5RA positive cells _ scHCL.hESC
1
Cardiomyocytes _ Antigen presenting cells
1
Cardiomyocytes _ Lymphoid cells
1
Cardiomyocytes _ Microglia
1
Cardiomyocytes _ Schwann cells
1
Cardiomyocytes _ Thymocytes
1
Chromaffin cells _ PDE11A_FAM19A2 positive cells
1
Ciliated epithelial cells _ Hepatoblasts
1
Ciliated epithelial cells _ Microglia
1
Ductal cells _ Extravillous trophoblasts
1
ELF3_AGBL2 positive cells _ CLC_IL5RA positive cells
1
ELF3_AGBL2 positive cells _ Microglia
1
ELF3_AGBL2 positive cells _ scHCL.hESC
1
ENS glia _ Oligodendrocytes
1
ENS neurons _ PDE11A_FAM19A2 positive cells
1
Endocardial cells _ CLC_IL5RA positive cells
1
Epicardial fat cells _ Acinar cells
1
Epicardial fat cells _ Smooth muscle cells
1
Erythroblasts _ Megakaryocytes
1
Erythroblasts _ Metanephric cells
1
Excitatory neurons _ SKOR2_NPSR1 positive cells
1
Ganglion cells _ Ciliated epithelial cells
1
Goblet cells _ Cardiomyocytes
1
Goblet cells _ Erythroblasts
1
Granule neurons _ Skeletal muscle cells
1
Hematopoietic stem cells _ Hepatoblasts
1
Hematopoietic stem cells _ MUC13_DMBT1 positive cells
1
Hematopoietic stem cells _ uncertain
1
Horizontal cells _ Satellite cells
1
Horizontal cells _ Skeletal muscle cells
1
IGFBP1_DKK1 positive cells _ Hepatoblasts
1
IGFBP1_DKK1 positive cells _ Metanephric cells
1
Inhibitory interneurons _ Skeletal muscle cells
1
Inhibitory neurons _ Horizontal cells
1
Inhibitory neurons _ PDE11A_FAM19A2 positive cells
1
Islet endocrine cells _ Amacrine cells
1
Islet endocrine cells _ ENS glia
1
Islet endocrine cells _ Endocardial cells
1
Islet endocrine cells _ Hepatoblasts
1
Islet endocrine cells _ Horizontal cells
1
Islet endocrine cells _ Smooth muscle cells
1
Lens fibre cells _ uncertain
1
Limbic system neurons _ Ciliated epithelial cells
1
Limbic system neurons _ scHCL.hESC
1
Lymphatic endothelial cells _ Hematopoietic stem cells
1
Lymphatic endothelial cells _ Megakaryocytes
1
Mesangial cells _ Ciliated epithelial cells
1
Mesangial cells _ ENS glia
1
Mesangial cells _ Hepatoblasts
1
Mesothelial cells _ Acinar cells
1
Metanephric cells _ Hematopoietic stem cells
1
Metanephric cells _ Horizontal cells
1
Microglia _ CCL19_CCL21 positive cells
1
Microglia _ Ciliated epithelial cells
1
Microglia _ Horizontal cells
1
Microglia _ Mesangial cells
1
Microglia _ Retinal pigment cells
1
Microglia _ Squamous epithelial cells
1
Microglia _ uncertain
1
Neuroendocrine cells _ Islet endocrine cells
1
Neuroendocrine cells _ uncertain
1
Oligodendrocytes _ Endocardial cells
1
Oligodendrocytes _ Erythroblasts
1
Oligodendrocytes _ Horizontal cells
1
Oligodendrocytes _ Microglia
1
PDE11A_FAM19A2 positive cells _ Amacrine cells
1
PDE11A_FAM19A2 positive cells _ Erythroblasts
1
PDE11A_FAM19A2 positive cells _ Hepatoblasts
1
Parietal and chief cells _ uncertain
1
Photoreceptor cells _ Ciliated epithelial cells
1
Photoreceptor cells _ Squamous epithelial cells
1
Purkinje neurons _ uncertain
1
Retinal pigment cells _ Acinar cells
1
Retinal pigment cells _ Hepatoblasts
1
Retinal progenitors and Muller glia _ Retinal pigment cells
1
SKOR2_NPSR1 positive cells _ Parietal and chief cells
1
SLC24A4_PEX5L positive cells _ Limbic system neurons
1
SLC24A4_PEX5L positive cells _ SKOR2_NPSR1 positive cells
1
Schwann cells _ Vascular endothelial cells
1
Skeletal muscle cells _ Acinar cells
1
Skeletal muscle cells _ Amacrine cells
1
Smooth muscle cells _ Metanephric cells
1
Squamous epithelial cells _ Hepatoblasts
1
Squamous epithelial cells _ Mesangial cells
1
Stellate cells _ Erythroblasts
1
Syncytiotrophoblasts and villous cytotrophoblasts _ Acinar cells
1
Thymic epithelial cells _ Skeletal muscle cells
1
Thymic epithelial cells _ uncertain
1
Thymocytes _ uncertain
1
Trophoblast giant cells _ uncertain
1
Ureteric bud cells _ Acinar cells
1
Ureteric bud cells _ Mesangial cells
1
Ureteric bud cells _ Microglia
1
Vascular endothelial cells _ Hematopoietic stem cells
1
Vascular endothelial cells _ scHCL.hESC
1
Visceral neurons _ Ciliated epithelial cells
1
Visceral neurons _ PDE11A_FAM19A2 positive cells
1
scHCL.hESC _ AFP_ALB positive cells
1
scHCL.hESC _ CLC_IL5RA positive cells
1
scHCL.hESC _ Corneal and conjunctival epithelial cells
1
scHCL.hESC _ Epicardial fat cells
1
scHCL.hESC _ Intestinal epithelial cells
1
scHCL.hESC _ Lymphoid cells
1
scHCL.hESC _ Parietal and chief cells
1
scHCL.hESC _ Stromal cells
1
scHCL.hESC _ Syncytiotrophoblasts and villous cytotrophoblasts
1
uncertain _ Astrocytes
1
uncertain _ Bipolar cells
1
uncertain _ CLC_IL5RA positive cells
1
uncertain _ Chromaffin cells
1
uncertain _ ELF3_AGBL2 positive cells
1
uncertain _ MUC13_DMBT1 positive cells
1
uncertain _ Megakaryocytes
1
uncertain _ Parietal and chief cells
1
uncertain _ Retinal progenitors and Muller glia
1
uncertain _ SKOR2_NPSR1 positive cells
1
uncertain _ Smooth muscle cells
1
uncertain _ Squamous epithelial cells
1
uncertain _ Thymocytes
1
uncertain _ Visceral neurons
1 how many are switching to or from hESC or uncertain?
length(grep("hESC|uncertain", trans))[1] 71907190/length(trans)[1] 0.8194666Take a closer look at the other mismatches
mis<- trans[-grep("hESC|uncertain", trans)]
mistab<- sort(table(mis), decreasing=T)
mistabmis
Cardiomyocytes _ Erythroblasts
105
Thymic epithelial cells _ Erythroblasts
95
IGFBP1_DKK1 positive cells _ Acinar cells
80
Goblet cells _ Unipolar brush cells
68
Mesangial cells _ Acinar cells
61
Stromal cells _ IGFBP1_DKK1 positive cells
57
ENS neurons _ Oligodendrocytes
50
Granule neurons _ Ciliated epithelial cells
50
Mesangial cells _ Metanephric cells
47
Hepatoblasts _ Intestinal epithelial cells
43
SLC24A4_PEX5L positive cells _ Purkinje neurons
41
Ductal cells _ Acinar cells
37
Visceral neurons _ Oligodendrocytes
31
Goblet cells _ Acinar cells
28
Squamous epithelial cells _ Stellate cells
28
SLC24A4_PEX5L positive cells _ Inhibitory neurons
26
Amacrine cells _ Hepatoblasts
25
Islet endocrine cells _ Ciliated epithelial cells
24
Intestinal epithelial cells _ MUC13_DMBT1 positive cells
23
MUC13_DMBT1 positive cells _ Acinar cells
23
AFP_ALB positive cells _ Hepatoblasts
22
Hepatoblasts _ AFP_ALB positive cells
22
Inhibitory neurons _ IGFBP1_DKK1 positive cells
22
Corneal and conjunctival epithelial cells _ Squamous epithelial cells
21
MUC13_DMBT1 positive cells _ Intestinal epithelial cells
19
Ganglion cells _ Oligodendrocytes
18
Mesothelial cells _ MUC13_DMBT1 positive cells
18
Squamous epithelial cells _ IGFBP1_DKK1 positive cells
18
IGFBP1_DKK1 positive cells _ Mesangial cells
17
Metanephric cells _ Acinar cells
17
Unipolar brush cells _ Ciliated epithelial cells
17
Metanephric cells _ Inhibitory neurons
16
Horizontal cells _ Hepatoblasts
15
Visceral neurons _ IGFBP1_DKK1 positive cells
13
Ureteric bud cells _ Stellate cells
12
Stellate cells _ IGFBP1_DKK1 positive cells
10
Ductal cells _ Ciliated epithelial cells
8
ELF3_AGBL2 positive cells _ Cardiomyocytes
8
ELF3_AGBL2 positive cells _ Erythroblasts
8
ENS glia _ Schwann cells
8
IGFBP1_DKK1 positive cells _ Smooth muscle cells
8
Ganglion cells _ Purkinje neurons
7
Thymic epithelial cells _ Unipolar brush cells
7
Acinar cells _ Ciliated epithelial cells
6
Amacrine cells _ Trophoblast giant cells
6
Ductal cells _ Cardiomyocytes
6
Inhibitory neurons _ Oligodendrocytes
6
Inhibitory neurons _ Skeletal muscle cells
6
Limbic system neurons _ Inhibitory neurons
6
Mesothelial cells _ Stellate cells
6
Parietal and chief cells _ Acinar cells
6
Schwann cells _ ENS glia
6
Epicardial fat cells _ MUC13_DMBT1 positive cells
5
Granule neurons _ IGFBP1_DKK1 positive cells
5
Horizontal cells _ Trophoblast giant cells
5
Intestinal epithelial cells _ Cardiomyocytes
5
Mesothelial cells _ Intestinal epithelial cells
5
Visceral neurons _ Skeletal muscle cells
5
Amacrine cells _ ENS glia
4
Endocardial cells _ Hematopoietic stem cells
4
Hepatoblasts _ MUC13_DMBT1 positive cells
4
Megakaryocytes _ Hematopoietic stem cells
4
Mesangial cells _ Parietal and chief cells
4
SLC24A4_PEX5L positive cells _ Ciliated epithelial cells
4
SLC24A4_PEX5L positive cells _ Oligodendrocytes
4
Squamous epithelial cells _ Ciliated epithelial cells
4
Vascular endothelial cells _ Lymphatic endothelial cells
4
AFP_ALB positive cells _ Intestinal epithelial cells
3
Cardiomyocytes _ ELF3_AGBL2 positive cells
3
ENS neurons _ IGFBP1_DKK1 positive cells
3
ENS neurons _ Purkinje neurons
3
IGFBP1_DKK1 positive cells _ Ciliated epithelial cells
3
IGFBP1_DKK1 positive cells _ Stellate cells
3
Intestinal epithelial cells _ Acinar cells
3
Intestinal epithelial cells _ Thymocytes
3
Lens fibre cells _ Retinal pigment cells
3
Mesangial cells _ IGFBP1_DKK1 positive cells
3
Mesangial cells _ Smooth muscle cells
3
Purkinje neurons _ Horizontal cells
3
Smooth muscle cells _ IGFBP1_DKK1 positive cells
3
Squamous epithelial cells _ Extravillous trophoblasts
3
Ureteric bud cells _ IGFBP1_DKK1 positive cells
3
Vascular endothelial cells _ Endocardial cells
3
Visceral neurons _ Hepatoblasts
3
Amacrine cells _ Ciliated epithelial cells
2
Amacrine cells _ Extravillous trophoblasts
2
Astrocytes _ Oligodendrocytes
2
CLC_IL5RA positive cells _ Cardiomyocytes
2
Ductal cells _ Intestinal epithelial cells
2
Epicardial fat cells _ Intestinal epithelial cells
2
Epicardial fat cells _ Stellate cells
2
Goblet cells _ Neuroendocrine cells
2
Hematopoietic stem cells _ Acinar cells
2
Horizontal cells _ ENS glia
2
IGFBP1_DKK1 positive cells _ Endocardial cells
2
Inhibitory interneurons _ IGFBP1_DKK1 positive cells
2
Intestinal epithelial cells _ Hepatoblasts
2
MUC13_DMBT1 positive cells _ Cardiomyocytes
2
Metanephric cells _ Microglia
2
Metanephric cells _ Squamous epithelial cells
2
Microglia _ Acinar cells
2
Microglia _ Endocardial cells
2
Purkinje neurons _ Microglia
2
Retinal pigment cells _ Photoreceptor cells
2
SKOR2_NPSR1 positive cells _ Ciliated epithelial cells
2
Smooth muscle cells _ Acinar cells
2
Stromal cells _ Smooth muscle cells
2
Syncytiotrophoblasts and villous cytotrophoblasts _ Ciliated epithelial cells
2
Thymic epithelial cells _ Hepatoblasts
2
Visceral neurons _ Satellite cells
2
Amacrine cells _ Satellite cells
1
Astrocytes _ Amacrine cells
1
Astrocytes _ Endocardial cells
1
Astrocytes _ Microglia
1
Cardiomyocytes _ Antigen presenting cells
1
Cardiomyocytes _ Lymphoid cells
1
Cardiomyocytes _ Microglia
1
Cardiomyocytes _ Schwann cells
1
Cardiomyocytes _ Thymocytes
1
Chromaffin cells _ PDE11A_FAM19A2 positive cells
1
Ciliated epithelial cells _ Hepatoblasts
1
Ciliated epithelial cells _ Microglia
1
Ductal cells _ Extravillous trophoblasts
1
ELF3_AGBL2 positive cells _ CLC_IL5RA positive cells
1
ELF3_AGBL2 positive cells _ Microglia
1
ENS glia _ Oligodendrocytes
1
ENS neurons _ PDE11A_FAM19A2 positive cells
1
Endocardial cells _ CLC_IL5RA positive cells
1
Epicardial fat cells _ Acinar cells
1
Epicardial fat cells _ Smooth muscle cells
1
Erythroblasts _ Megakaryocytes
1
Erythroblasts _ Metanephric cells
1
Excitatory neurons _ SKOR2_NPSR1 positive cells
1
Ganglion cells _ Ciliated epithelial cells
1
Goblet cells _ Cardiomyocytes
1
Goblet cells _ Erythroblasts
1
Granule neurons _ Skeletal muscle cells
1
Hematopoietic stem cells _ Hepatoblasts
1
Hematopoietic stem cells _ MUC13_DMBT1 positive cells
1
Horizontal cells _ Satellite cells
1
Horizontal cells _ Skeletal muscle cells
1
IGFBP1_DKK1 positive cells _ Hepatoblasts
1
IGFBP1_DKK1 positive cells _ Metanephric cells
1
Inhibitory interneurons _ Skeletal muscle cells
1
Inhibitory neurons _ Horizontal cells
1
Inhibitory neurons _ PDE11A_FAM19A2 positive cells
1
Islet endocrine cells _ Amacrine cells
1
Islet endocrine cells _ ENS glia
1
Islet endocrine cells _ Endocardial cells
1
Islet endocrine cells _ Hepatoblasts
1
Islet endocrine cells _ Horizontal cells
1
Islet endocrine cells _ Smooth muscle cells
1
Limbic system neurons _ Ciliated epithelial cells
1
Lymphatic endothelial cells _ Hematopoietic stem cells
1
Lymphatic endothelial cells _ Megakaryocytes
1
Mesangial cells _ Ciliated epithelial cells
1
Mesangial cells _ ENS glia
1
Mesangial cells _ Hepatoblasts
1
Mesothelial cells _ Acinar cells
1
Metanephric cells _ Hematopoietic stem cells
1
Metanephric cells _ Horizontal cells
1
Microglia _ CCL19_CCL21 positive cells
1
Microglia _ Ciliated epithelial cells
1
Microglia _ Horizontal cells
1
Microglia _ Mesangial cells
1
Microglia _ Retinal pigment cells
1
Microglia _ Squamous epithelial cells
1
Neuroendocrine cells _ Islet endocrine cells
1
Oligodendrocytes _ Endocardial cells
1
Oligodendrocytes _ Erythroblasts
1
Oligodendrocytes _ Horizontal cells
1
Oligodendrocytes _ Microglia
1
PDE11A_FAM19A2 positive cells _ Amacrine cells
1
PDE11A_FAM19A2 positive cells _ Erythroblasts
1
PDE11A_FAM19A2 positive cells _ Hepatoblasts
1
Photoreceptor cells _ Ciliated epithelial cells
1
Photoreceptor cells _ Squamous epithelial cells
1
Retinal pigment cells _ Acinar cells
1
Retinal pigment cells _ Hepatoblasts
1
Retinal progenitors and Muller glia _ Retinal pigment cells
1
SKOR2_NPSR1 positive cells _ Parietal and chief cells
1
SLC24A4_PEX5L positive cells _ Limbic system neurons
1
SLC24A4_PEX5L positive cells _ SKOR2_NPSR1 positive cells
1
Schwann cells _ Vascular endothelial cells
1
Skeletal muscle cells _ Acinar cells
1
Skeletal muscle cells _ Amacrine cells
1
Smooth muscle cells _ Metanephric cells
1
Squamous epithelial cells _ Hepatoblasts
1
Squamous epithelial cells _ Mesangial cells
1
Stellate cells _ Erythroblasts
1
Syncytiotrophoblasts and villous cytotrophoblasts _ Acinar cells
1
Thymic epithelial cells _ Skeletal muscle cells
1
Ureteric bud cells _ Acinar cells
1
Ureteric bud cells _ Mesangial cells
1
Ureteric bud cells _ Microglia
1
Vascular endothelial cells _ Hematopoietic stem cells
1
Visceral neurons _ Ciliated epithelial cells
1
Visceral neurons _ PDE11A_FAM19A2 positive cells
1 sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] dplyr_1.0.2 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] Rcpp_1.0.6 knitr_1.29 magrittr_2.0.1 tidyselect_1.1.0
[5] R6_2.5.0 rlang_0.4.10 highr_0.8 stringr_1.4.0
[9] tools_3.6.1 xfun_0.16 git2r_0.26.1 htmltools_0.5.0
[13] ellipsis_0.3.1 yaml_2.2.1 digest_0.6.27 rprojroot_2.0.2
[17] tibble_3.0.4 lifecycle_0.2.0 crayon_1.3.4 purrr_0.3.4
[21] later_1.1.0.1 vctrs_0.3.6 promises_1.1.1 fs_1.4.2
[25] glue_1.4.2 evaluate_0.14 rmarkdown_2.3 stringi_1.5.3
[29] compiler_3.6.1 pillar_1.4.7 generics_0.1.0 httpuv_1.5.4
[33] pkgconfig_2.0.3
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] dplyr_1.0.2 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] Rcpp_1.0.6 knitr_1.29 magrittr_2.0.1 tidyselect_1.1.0
[5] R6_2.5.0 rlang_0.4.10 highr_0.8 stringr_1.4.0
[9] tools_3.6.1 xfun_0.16 git2r_0.26.1 htmltools_0.5.0
[13] ellipsis_0.3.1 yaml_2.2.1 digest_0.6.27 rprojroot_2.0.2
[17] tibble_3.0.4 lifecycle_0.2.0 crayon_1.3.4 purrr_0.3.4
[21] later_1.1.0.1 vctrs_0.3.6 promises_1.1.1 fs_1.4.2
[25] glue_1.4.2 evaluate_0.14 rmarkdown_2.3 stringi_1.5.3
[29] compiler_3.6.1 pillar_1.4.7 generics_0.1.0 httpuv_1.5.4
[33] pkgconfig_2.0.3