integrate data from adult mLN and iLN
A.DeMartin
2025-05-06
Last updated: 2025-07-11
Checks: 6 1
Knit directory: LNdevMouse24.2/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R
Markdown file created these results, you’ll want to first commit it to
the Git repo. If you’re still working on the analysis, you can ignore
this warning. When you’re finished, you can run
wflow_publish to commit the R Markdown file and build the
HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20250625) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version e2b9f79. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.DS_Store
Untracked files:
Untracked: analysis/adult_mLN_iLN.Rmd
Unstaged changes:
Modified: analysis/index.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with
wflow_publish() to start tracking its development.
preprocessing
load packages
library(ExploreSCdataSeurat3)
library(runSeurat3)
library(Seurat)
library(ggpubr)
library(pheatmap)
library(SingleCellExperiment)
library(dplyr)
library(tidyverse)
library(viridis)
library(here)
library(muscat)
library(circlize)
library(destiny)
library(scater)
library(metap)
library(multtest)
library(clusterProfiler)
library(org.Hs.eg.db)
library(msigdbr)
library(enrichplot)
library(DOSE)
library(grid)
library(gridExtra)
library(ggupset)
library(NCmisc)load object all
basedir <- here()
fileNam <- paste0(basedir, "/data/LNmLToRev_allmerged_seurat.rds")
seuratM <- readRDS(fileNam)
table(seuratM$orig.ident)
140291 subset adult
seuratA <- subset(seuratM, timepoint == "8w")
table(seuratM$timepoint)
E18 P7 3w 8w
42711 44836 29577 23167 table(seuratA$timepoint)
8w
23167 #rerun seurat
seuratA <- NormalizeData (object = seuratA)
seuratA <- FindVariableFeatures(object = seuratA)
seuratA <- ScaleData(object = seuratA, verbose = TRUE)
seuratA <- RunPCA(object=seuratA, npcs = 30, verbose = FALSE)
seuratA <- RunTSNE(object=seuratA, reduction="pca", dims = 1:20)
seuratA <- RunUMAP(object=seuratA, reduction="pca", dims = 1:20)
seuratA <- FindNeighbors(object = seuratA, reduction = "pca", dims= 1:20)
res <- c(0.25, 0.6, 0.8, 0.4)
for (i in 1:length(res)) {
seuratA <- FindClusters(object = seuratA, resolution = res[i], random.seed = 1234)
}Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 23167
Number of edges: 747906
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.9267
Number of communities: 12
Elapsed time: 5 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 23167
Number of edges: 747906
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.8790
Number of communities: 17
Elapsed time: 4 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 23167
Number of edges: 747906
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.8611
Number of communities: 20
Elapsed time: 4 seconds
Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 23167
Number of edges: 747906
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.9025
Number of communities: 15
Elapsed time: 5 secondsplot umaps
clustering
colPal <- c("#DAF7A6", "#FFC300", "#FF5733", "#C70039", "#900C3F", "#b66e8d",
"#61a4ba", "#6178ba", "#54a87f", "#25328a", "#b6856e",
"#ba6161", "#20714a", "#0073C2FF", "#EFC000FF", "#868686FF",
"#CD534CFF","#7AA6DCFF", "#003C67FF", "#8F7700FF", "#3B3B3BFF",
"#A73030FF", "#4A6990FF")[1:length(unique(seuratM$RNA_snn_res.0.25))]
names(colPal) <- unique(seuratM$RNA_snn_res.0.25)
DimPlot(seuratA, reduction = "umap", group.by = "RNA_snn_res.0.25" ,
pt.size = 0.1, label = T, shuffle = T, cols = colPal) +
theme_bw() +
theme(axis.text = element_blank(), axis.ticks = element_blank(),
panel.grid.minor = element_blank()) +
xlab("umap1") +
ylab("umap2")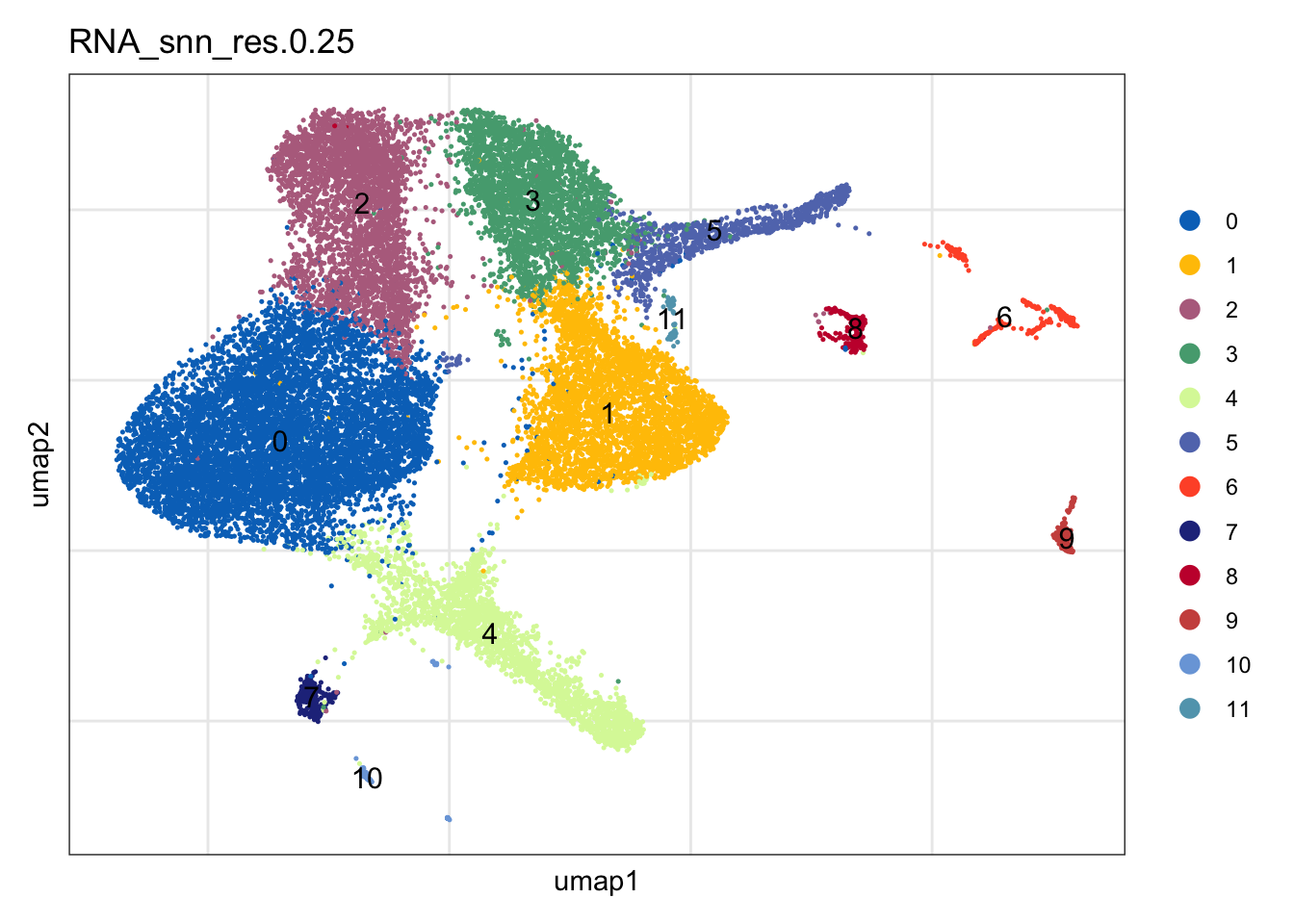
location
collocation <- c("#61baba", "#ba6161")
names(collocation) <- c("iLN", "mLN")
DimPlot(seuratA, reduction = "umap", group.by = "location", cols = collocation,
pt.size = 0.1, label = T, shuffle = T) +
theme_bw() +
theme(axis.text = element_blank(), axis.ticks = element_blank(),
panel.grid.minor = element_blank()) +
xlab("umap1") +
ylab("umap2")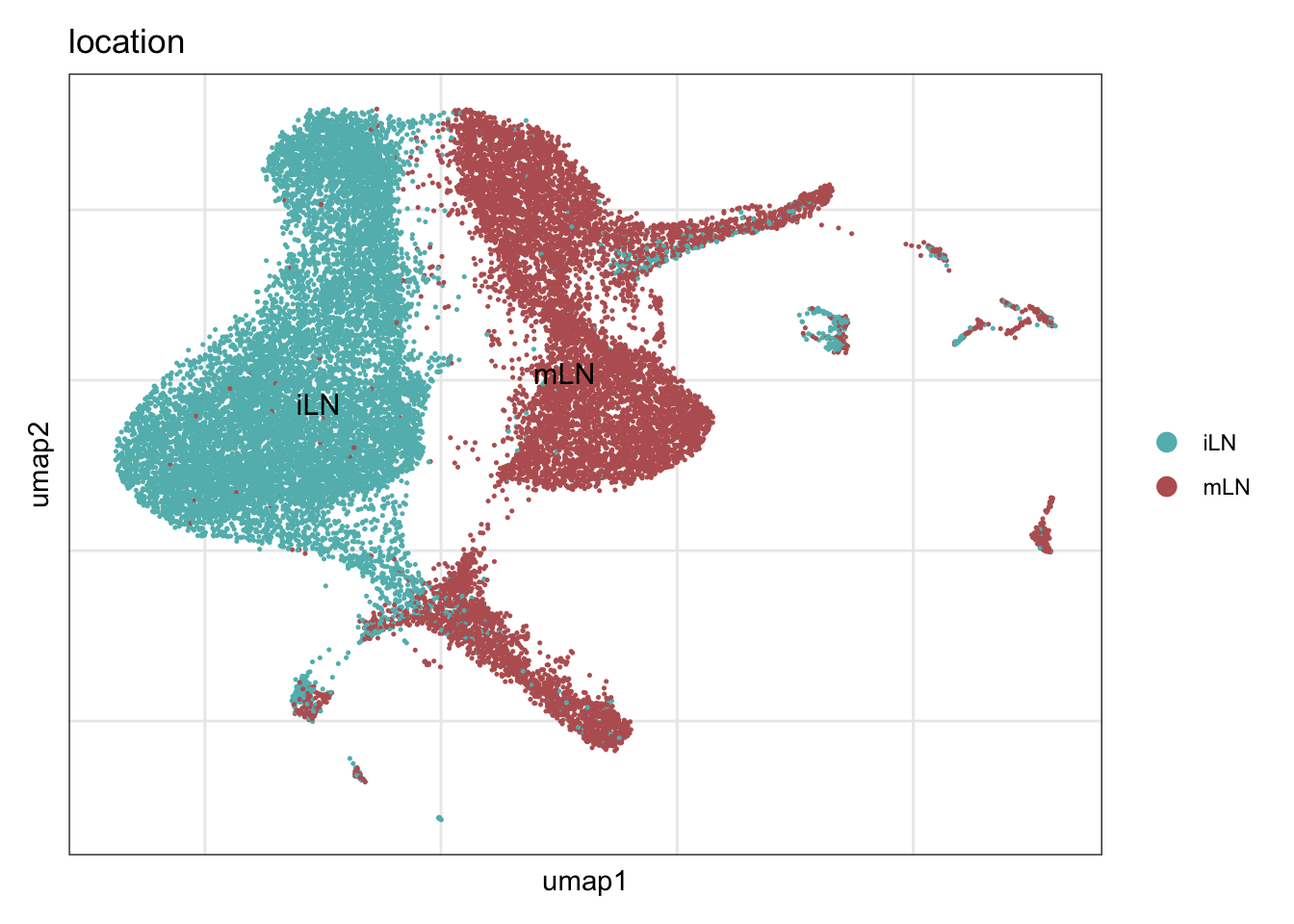
calculate cluster marker genes pre filter
Idents(seuratA) <- seuratA$RNA_snn_res.0.25
levels(seuratA)
markerGenes <- FindAllMarkers(seuratA, only.pos=T) %>%
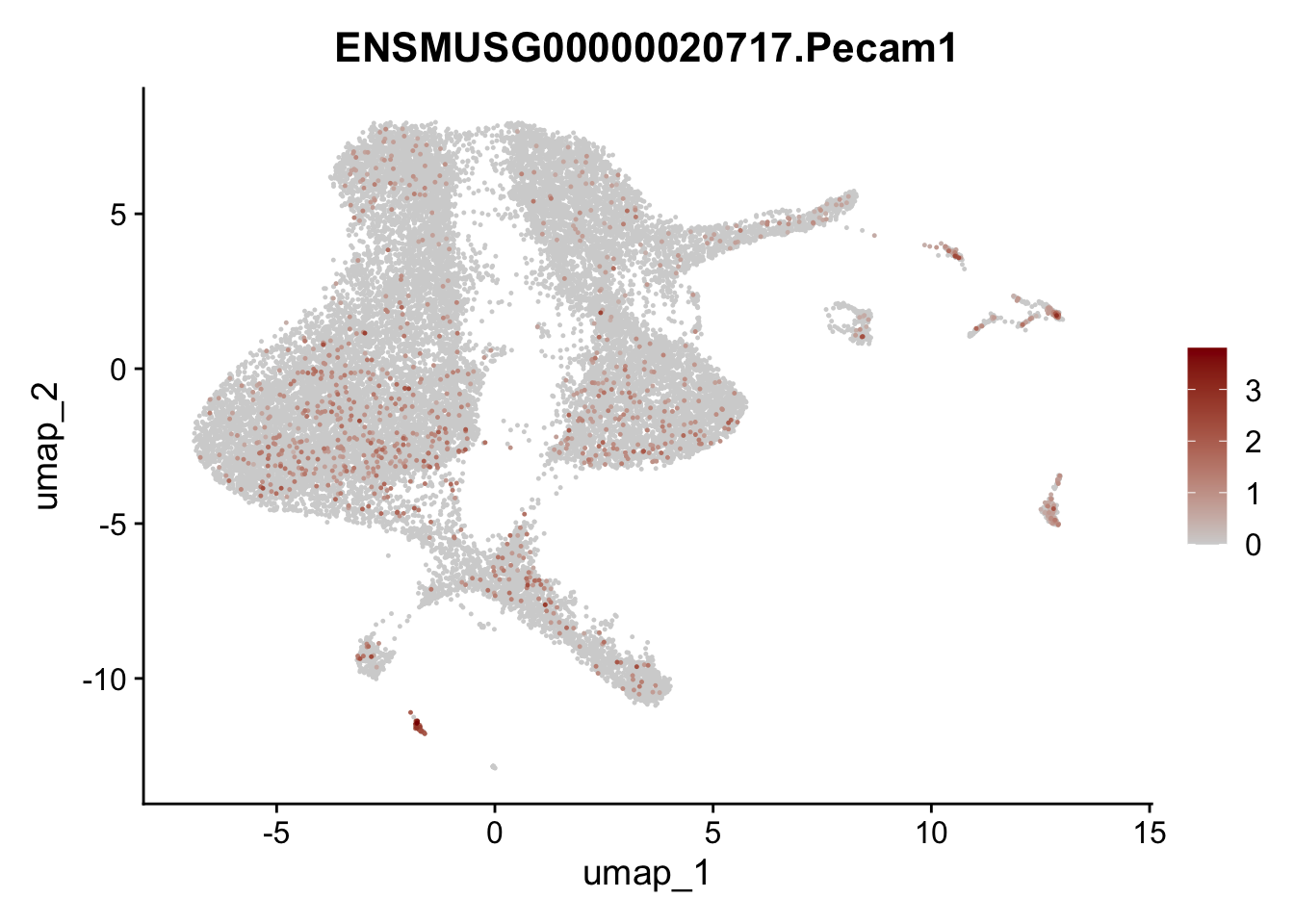
dplyr::filter(p_val_adj < 0.01) features pre filter
selGenesViolin <- c("ENSMUSG00000020717.Pecam1", "ENSMUSG00000026395.Ptprc", "ENSMUSG00000031004.Mki67")
pList <- sapply(selGenesViolin, function(x){
p <- FeaturePlot(seuratA, reduction = "umap",
features = x,
cols=c("lightgrey", "darkred"),
order = T)+
theme(legend.position="right")
plot(p)
})
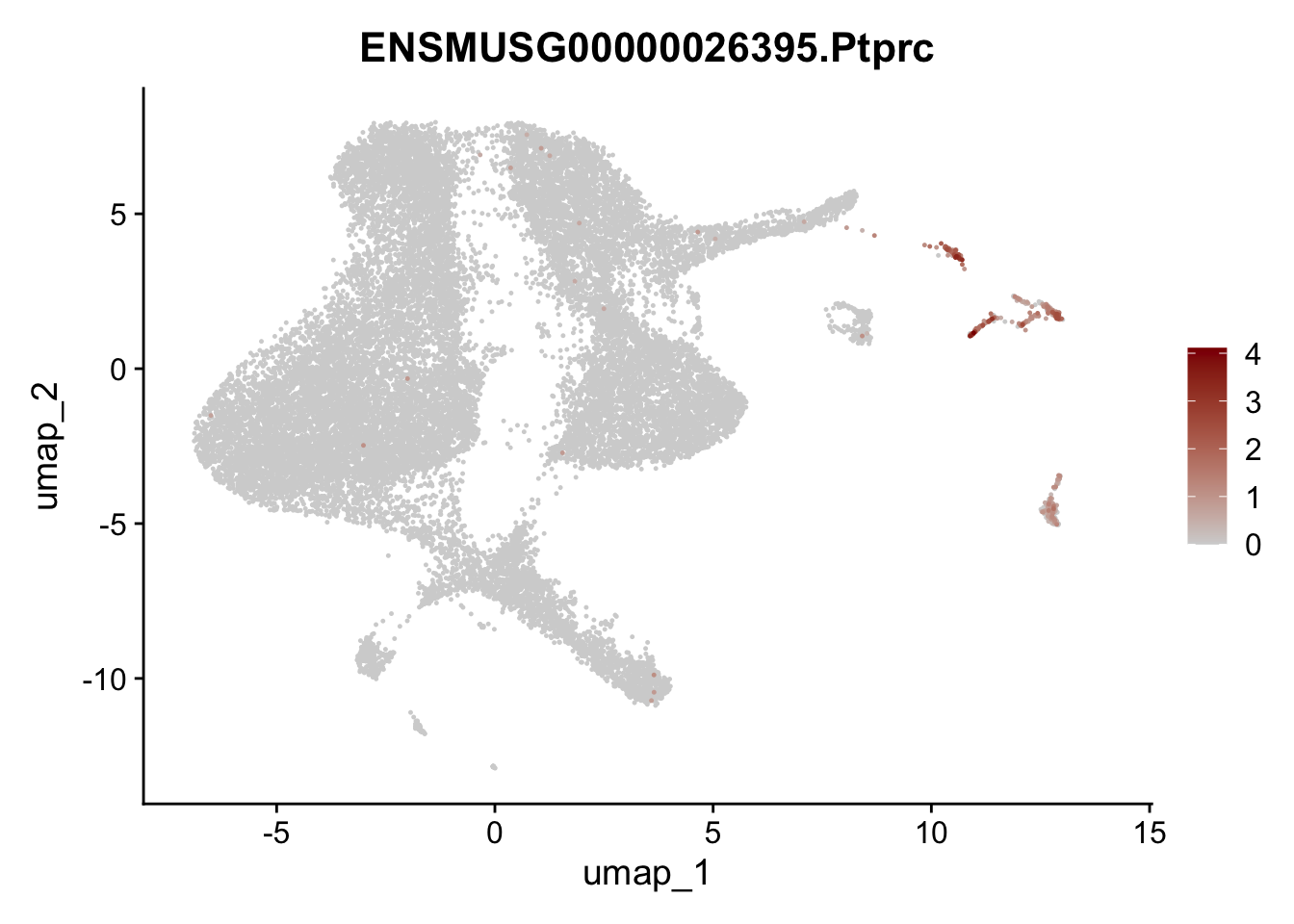
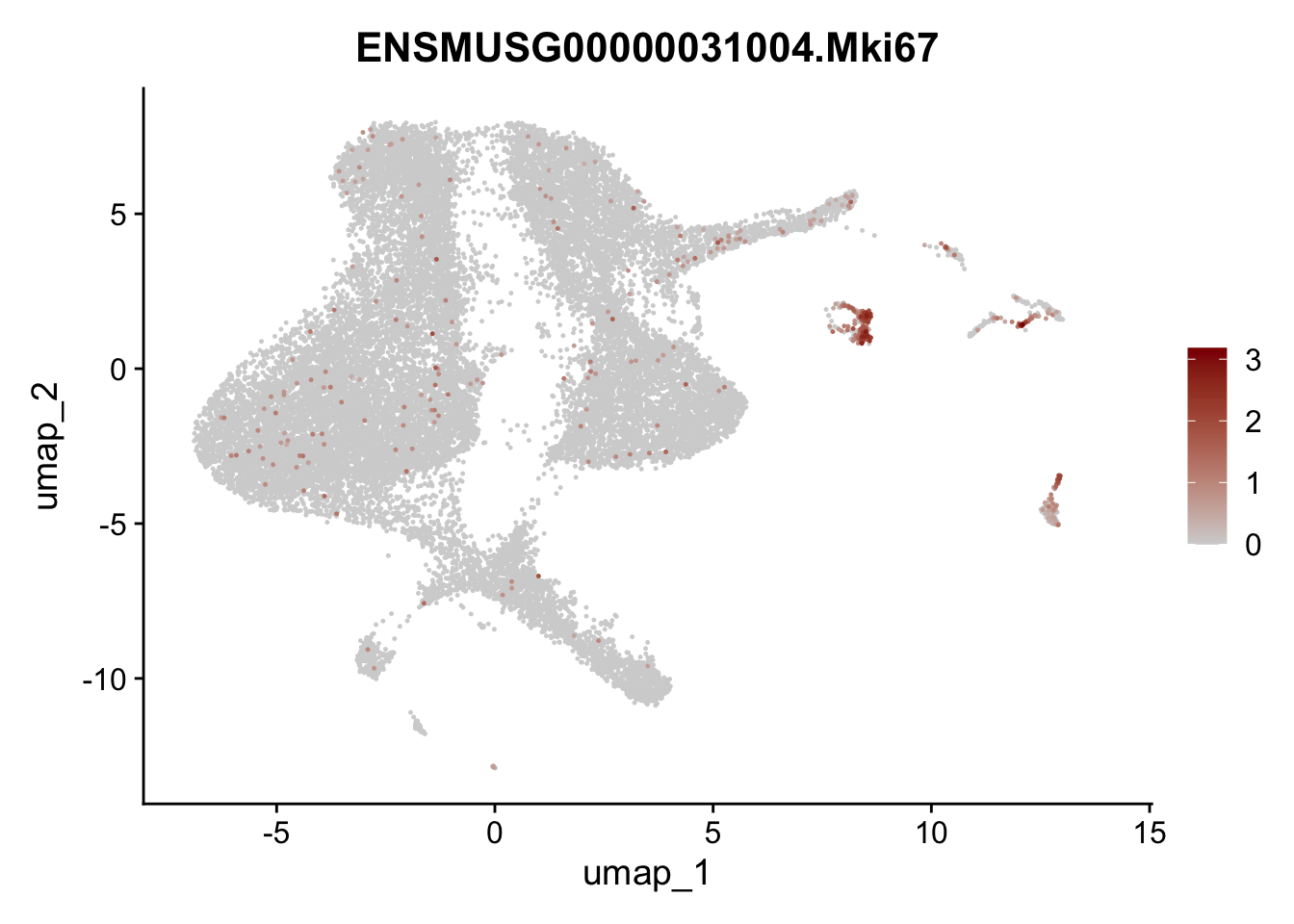
filter object
## filter Pecam1 (cluster #6) and Ptprc (cluster #9 and #10) Mki67 (#8) and pancreatic cells (#11)
table(seuratA$RNA_snn_res.0.25)
seuratF <- subset(seuratA, RNA_snn_res.0.25 %in% c("6", "8" ,"9", "10", "11"), invert = TRUE)
table(seuratF$RNA_snn_res.0.25)
seuratA <- seuratF
remove(seuratF)rerun seurat after filter
seuratA <- NormalizeData (object = seuratA)
seuratA <- FindVariableFeatures(object = seuratA)
seuratA <- ScaleData(object = seuratA, verbose = TRUE)
seuratA <- RunPCA(object=seuratA, npcs = 30, verbose = FALSE)
seuratA <- RunTSNE(object=seuratA, reduction="pca", dims = 1:20)
seuratA <- RunUMAP(object=seuratA, reduction="pca", dims = 1:20)
seuratA <- FindNeighbors(object = seuratA, reduction = "pca", dims= 1:20)
res <- c(0.25, 0.6, 0.8, 0.4)
for (i in 1:length(res)) {
seuratA <- FindClusters(object = seuratA, resolution = res[i], random.seed = 1234)
}## save object
saveRDS(seuratA, file=paste0(basedir,"/data/LNmLToRev_adultonly_seurat.rds"))load object adult
## load object adult only
fileNam <- paste0(basedir, "/data/LNmLToRev_adultonly_seurat.rds")
seuratA <- readRDS(fileNam)
table(seuratA$dataset)
380131_11-11_20250305_Mu_Cxcl13EYFP_Adult_pLN_FRC 380131_12-12_20250305_Mu_Cxcl13EYFP_Adult_mLN_FRC
5253 6419
382581_08-8_20250320_Mu_Cxcl13EYFP_Adult_pLN_FRC 382581_09-9_20250320_Mu_Cxcl13EYFP_Adult_mLN_FRC
7094 3522 set color vectors
colLoc <- c("#61baba", "#ba6161")
names(colLoc) <- unique(seuratA$location)
colLab <- c("#42a071", "#900C3F","#b66e8d", "#8F7700FF", "#61a4ba","#003C67FF",
"#e3953d","#ab5711", "#714542", "#b6856e", "#FFC300")
names(colLab) <- c("FDC", "TRC", "TBRC", "IFRC", "medRC1" , "medRC2",
"PRC1", "PRC2", "Pi16+RC", "PRC3", "VSMC")
coltimepoint <- c("#440154FF", "#3B528BFF", "#21908CFF", "#5DC863FF")
names(coltimepoint) <- c("E18", "P7", "3w", "8w")
collocation <- c("#61baba", "#ba6161")
names(collocation) <- c("iLN", "mLN")plot umaps
clustering
colPal <- c("#DAF7A6", "#FFC300", "#FF5733", "#C70039", "#900C3F", "#b66e8d",
"#61a4ba", "#6178ba", "#54a87f", "#25328a", "#b6856e",
"#ba6161", "#20714a", "#0073C2FF", "#EFC000FF", "#868686FF",
"#CD534CFF","#7AA6DCFF", "#003C67FF", "#8F7700FF", "#3B3B3BFF",
"#A73030FF", "#4A6990FF")[1:length(unique(seuratA$RNA_snn_res.0.25))]
names(colPal) <- unique(seuratA$RNA_snn_res.0.25)
DimPlot(seuratA, reduction = "umap", group.by = "RNA_snn_res.0.25", cols = colPal,
pt.size = 0.1, label = T, shuffle = T) +
theme_bw() +
theme(axis.text = element_blank(), axis.ticks = element_blank(),
panel.grid.minor = element_blank()) +
xlab("umap1") +
ylab("umap2")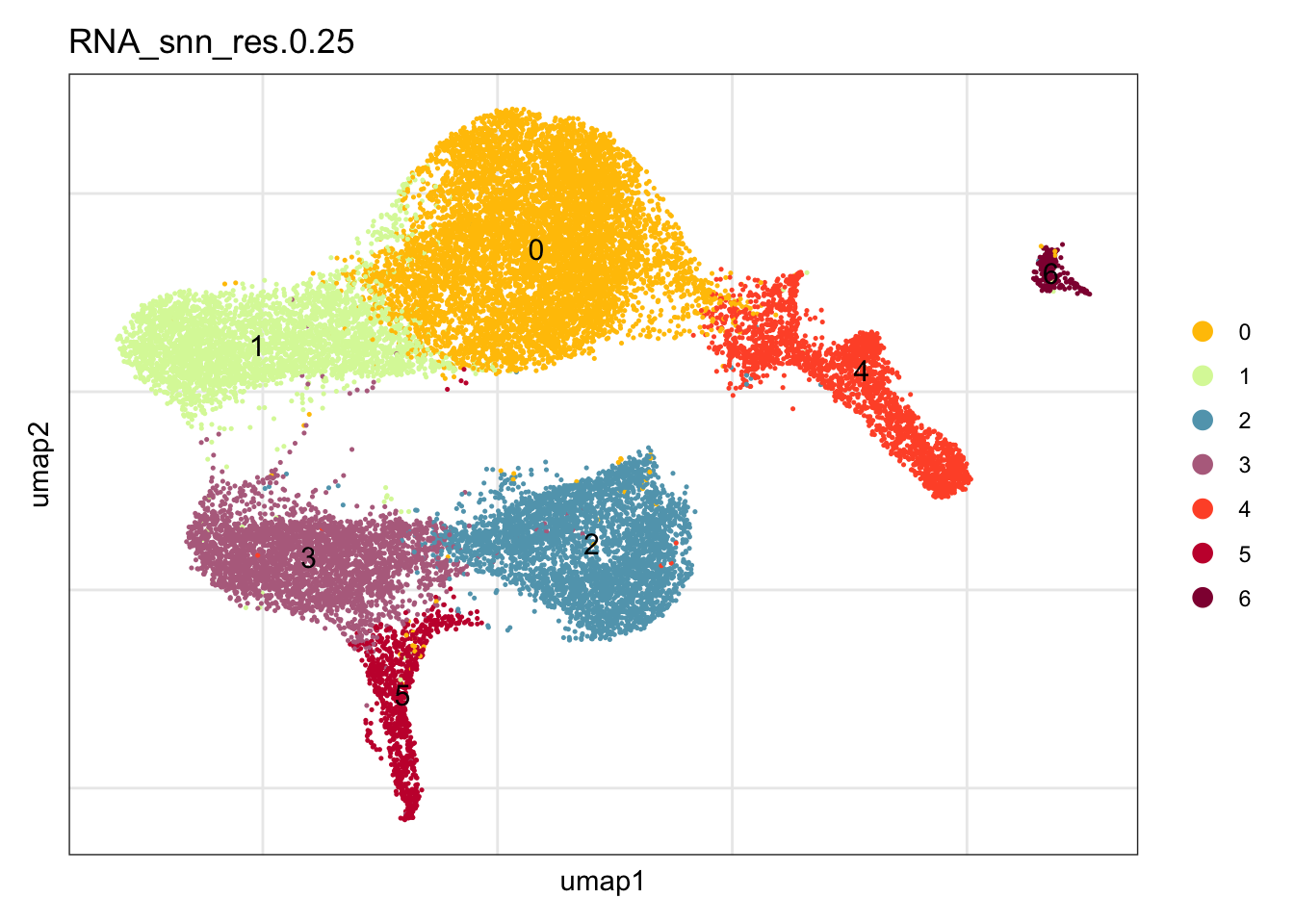
location
DimPlot(seuratA, reduction = "umap", group.by = "location", cols = collocation,
pt.size = 0.1, label = T, shuffle = T) +
theme_bw() +
theme(axis.text = element_blank(), axis.ticks = element_blank(),
panel.grid.minor = element_blank()) +
xlab("umap1") +
ylab("umap2")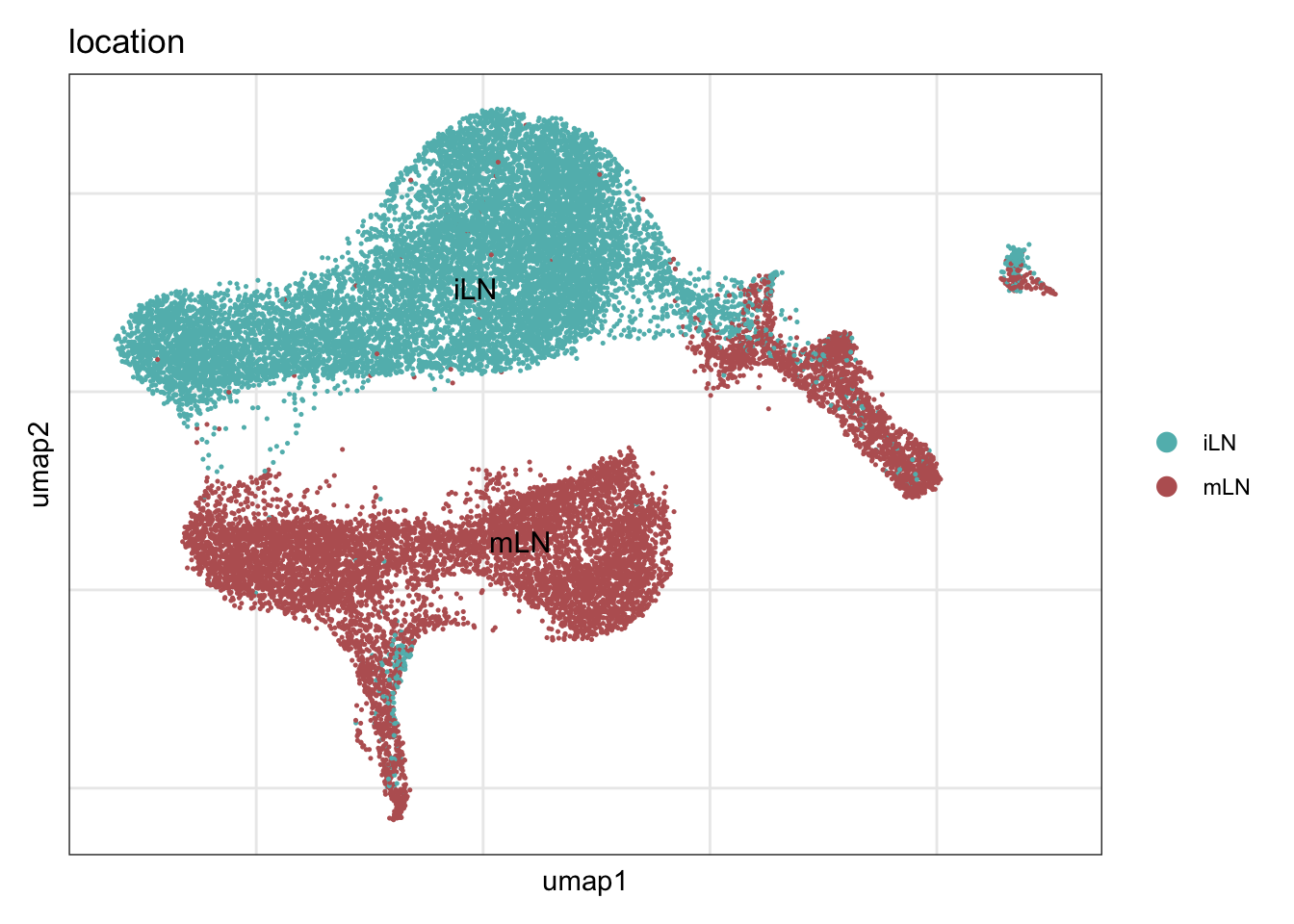
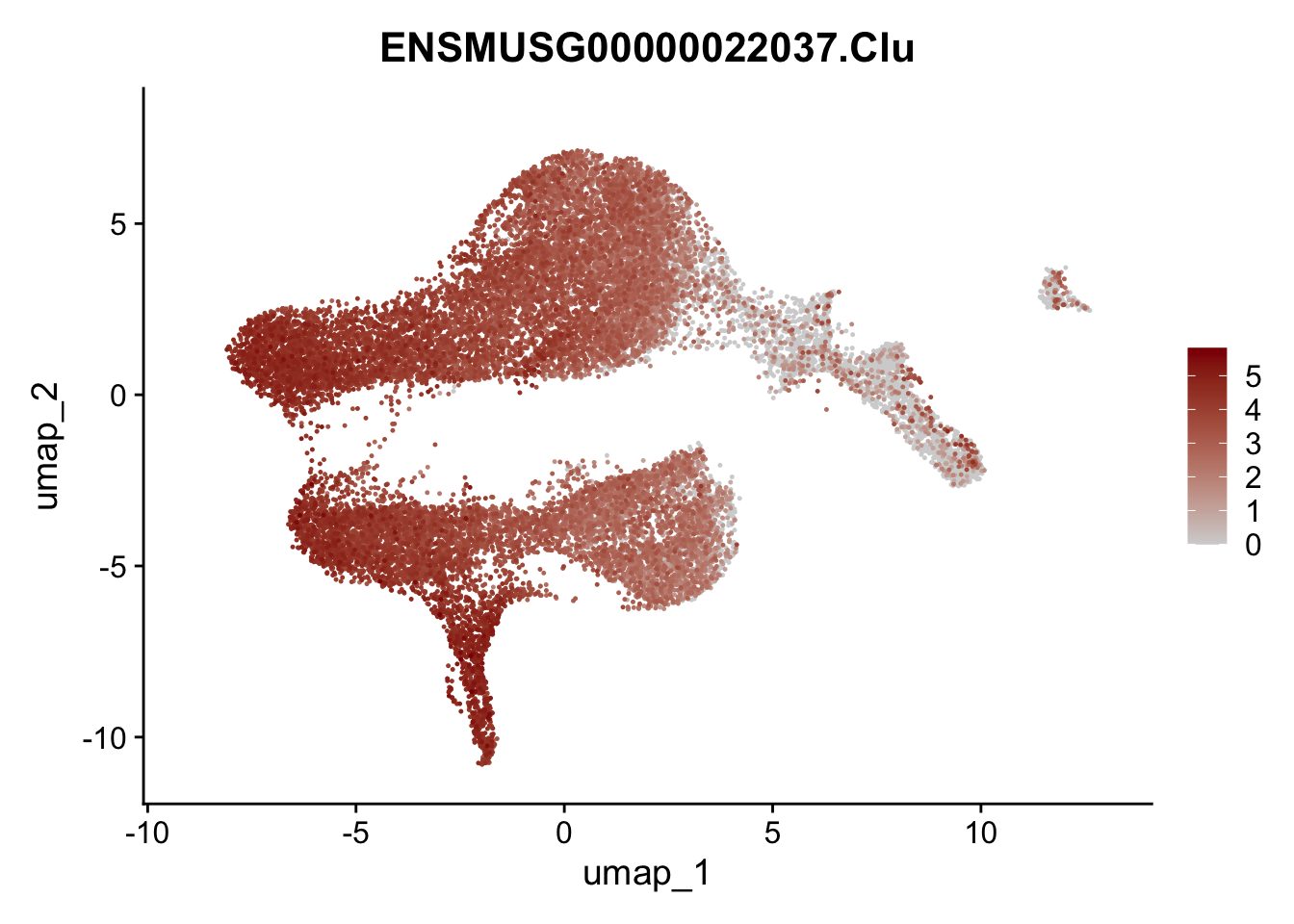
plot features
selGenesViolin <- c("ENSMUSG00000022037.Clu", "ENSMUSG00000094686.Ccl21a",
"ENSMUSG00000074934.Grem1", "ENSMUSG00000050069.Grem2",
"ENSMUSG00000042436.Mfap4", "ENSMUSG00000071005.Ccl19",
"ENSMUSG00000016494.Cd34", "ENSMUSG00000001119.Col6a1",
"ENSMUSG00000020241.Col6a2","Rosa26eyfp.Rosa26eyfp",
"ENSMUSG00000023078.Cxcl13", "ENSMUSG00000032135.Mcam")
pList <- sapply(selGenesViolin, function(x){
p <- FeaturePlot(seuratA, reduction = "umap",
features = x,
cols=c("lightgrey", "darkred"),
order = T)+
theme(legend.position="right")
plot(p)
})
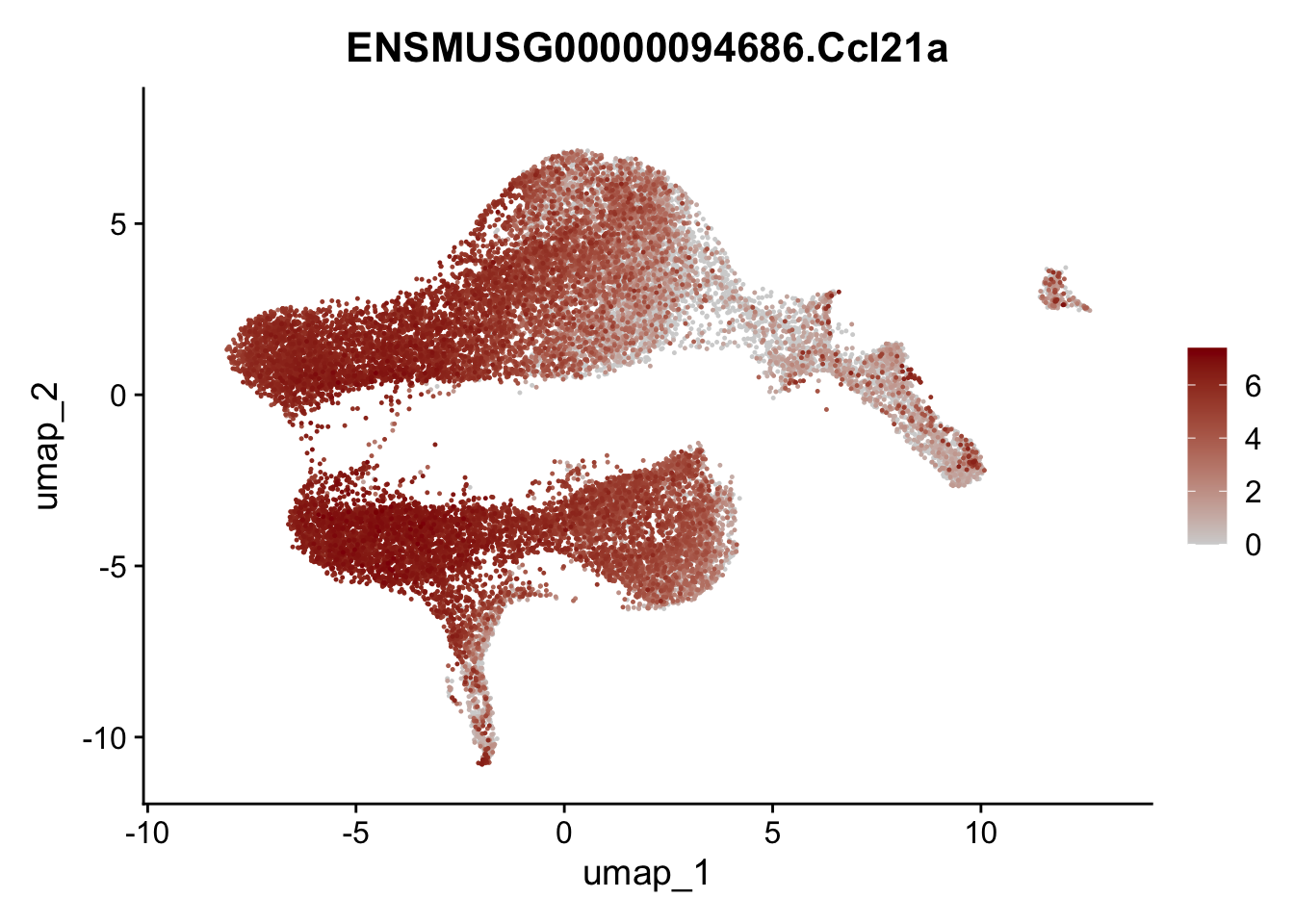
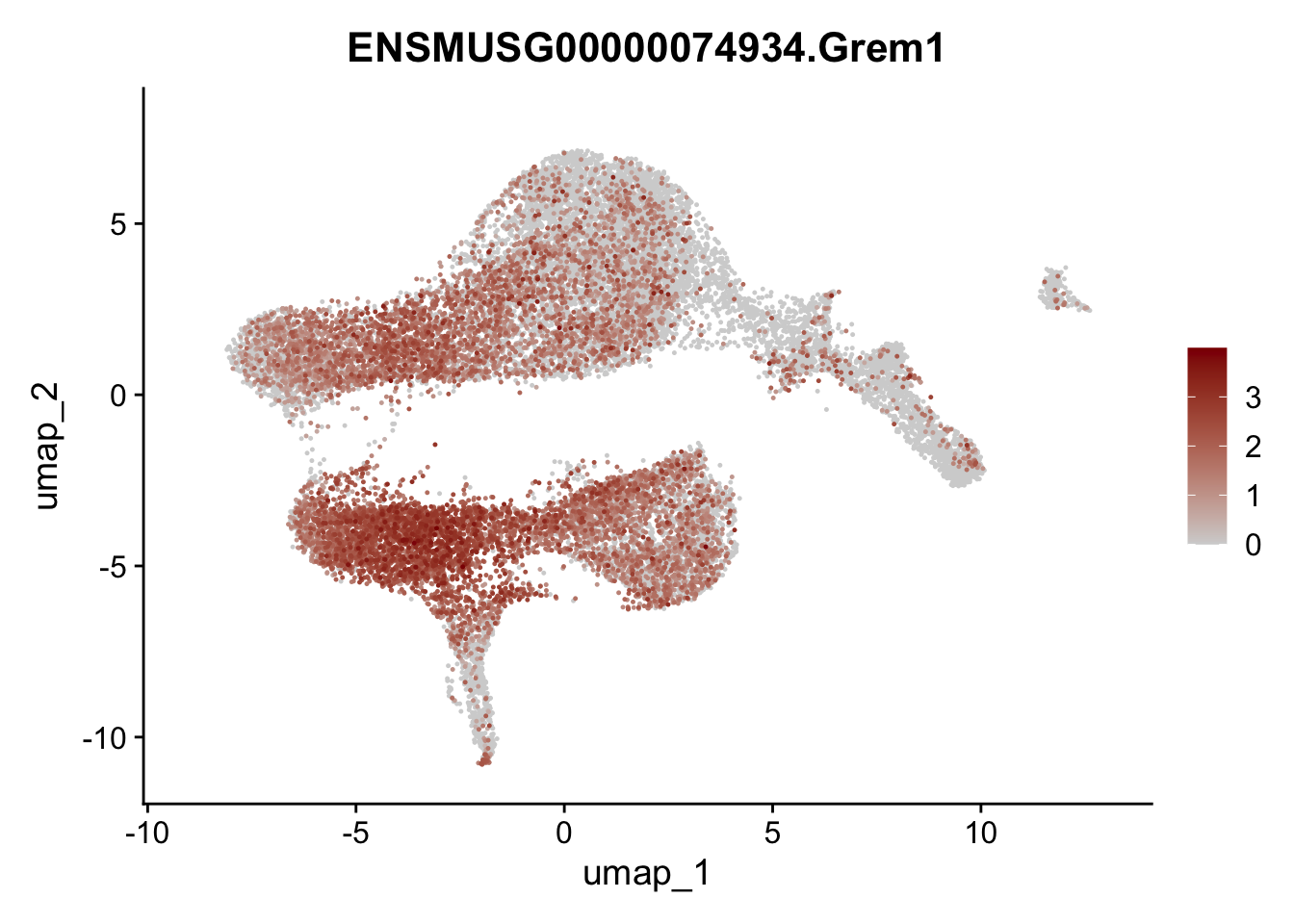
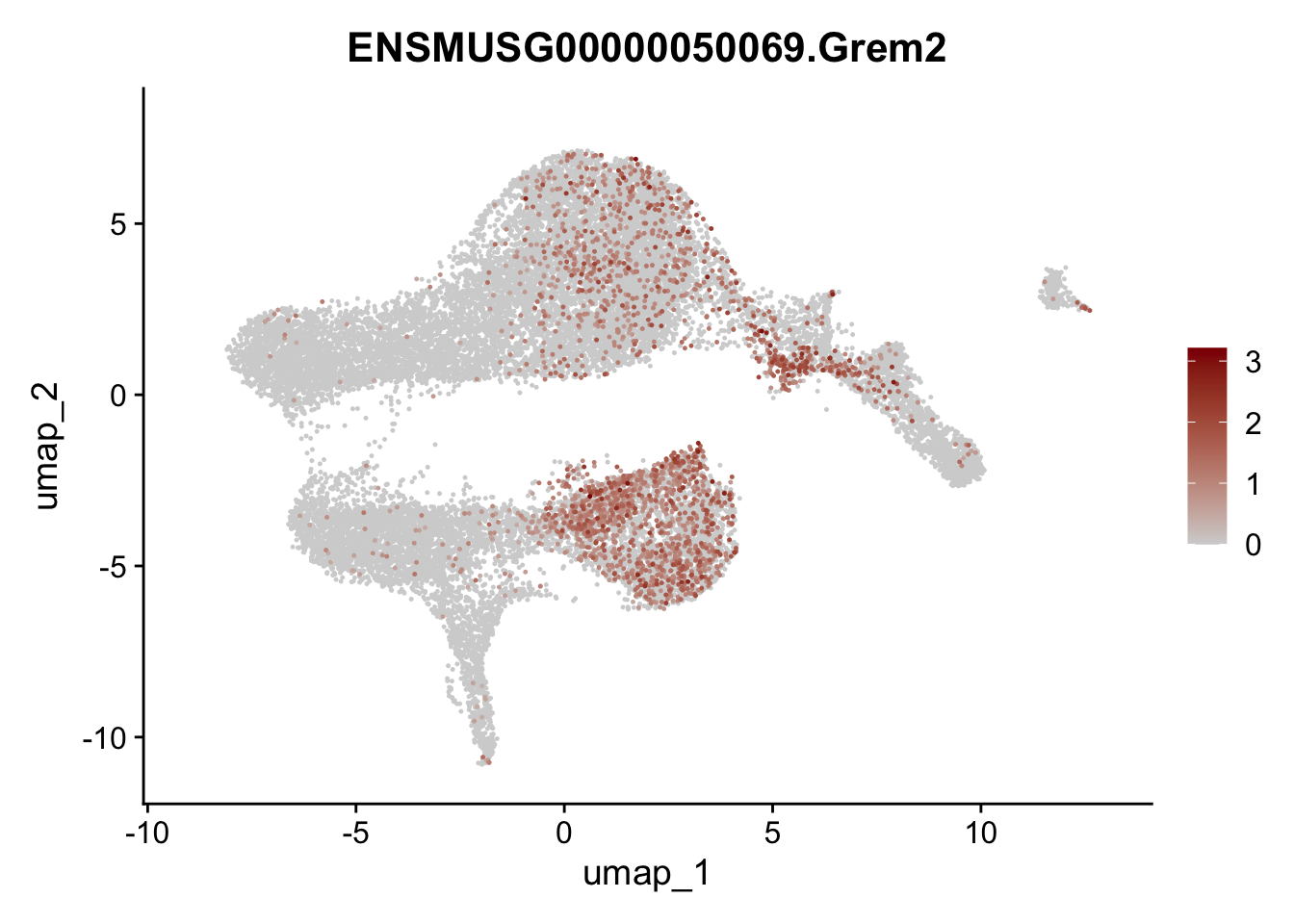
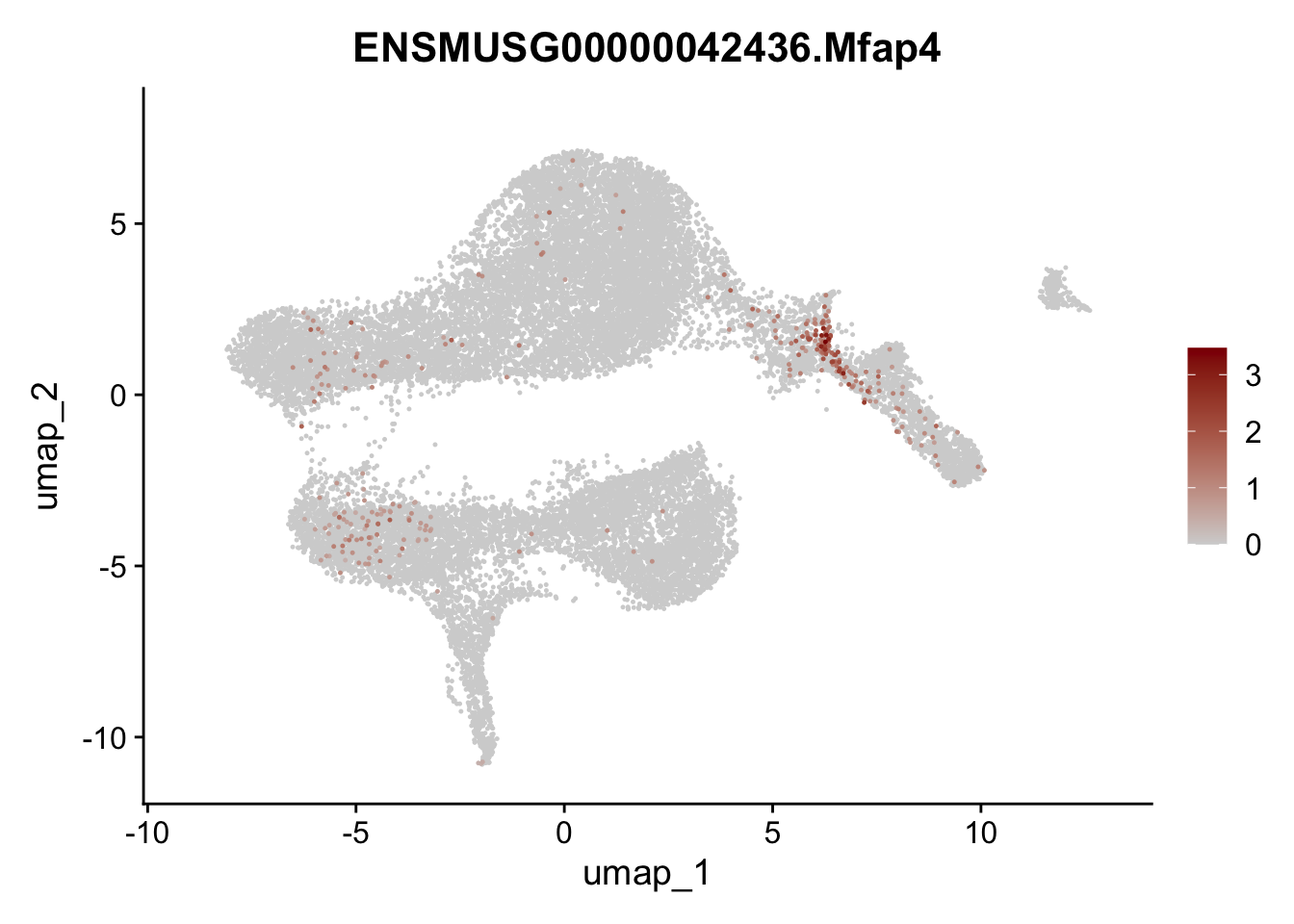
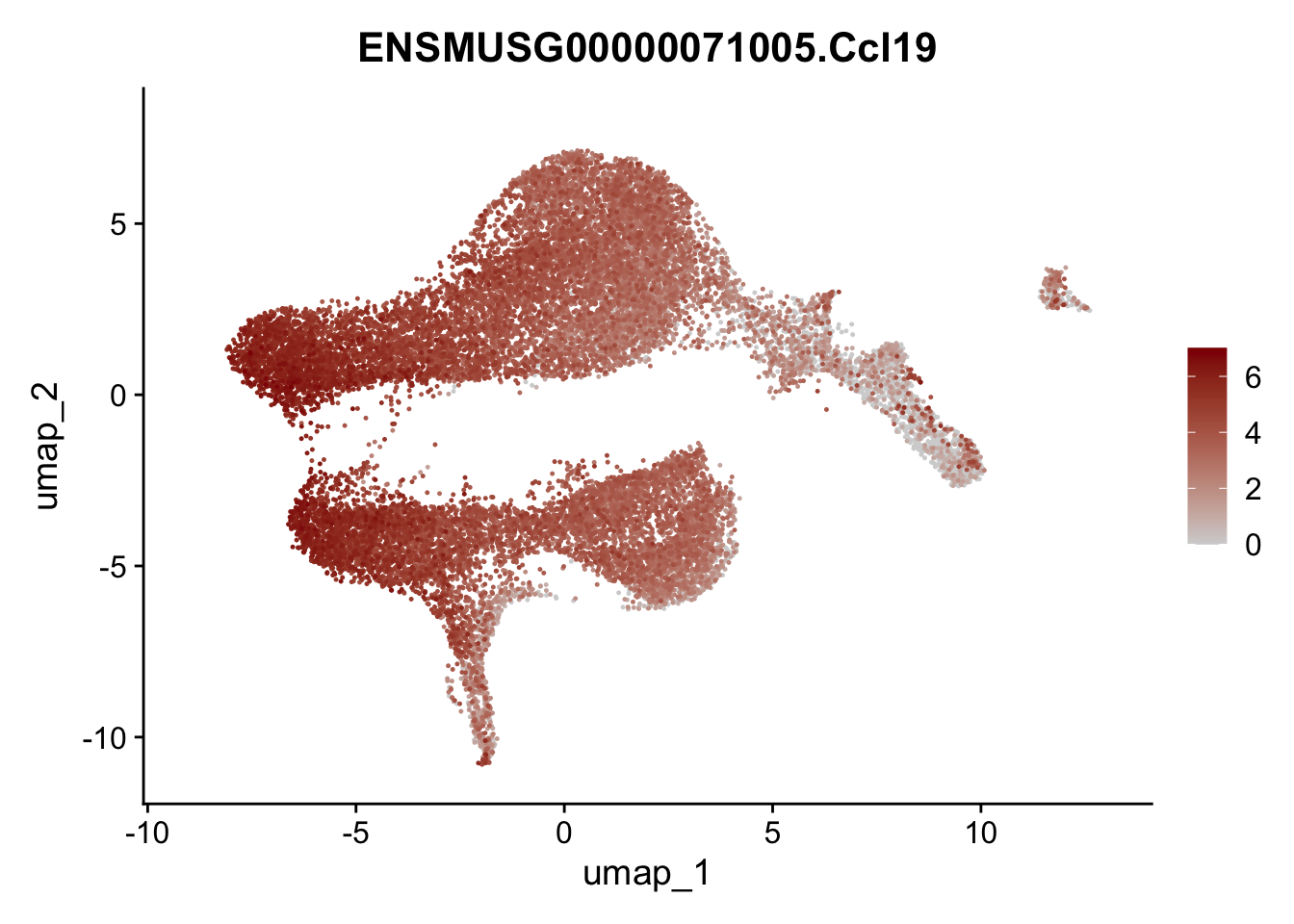
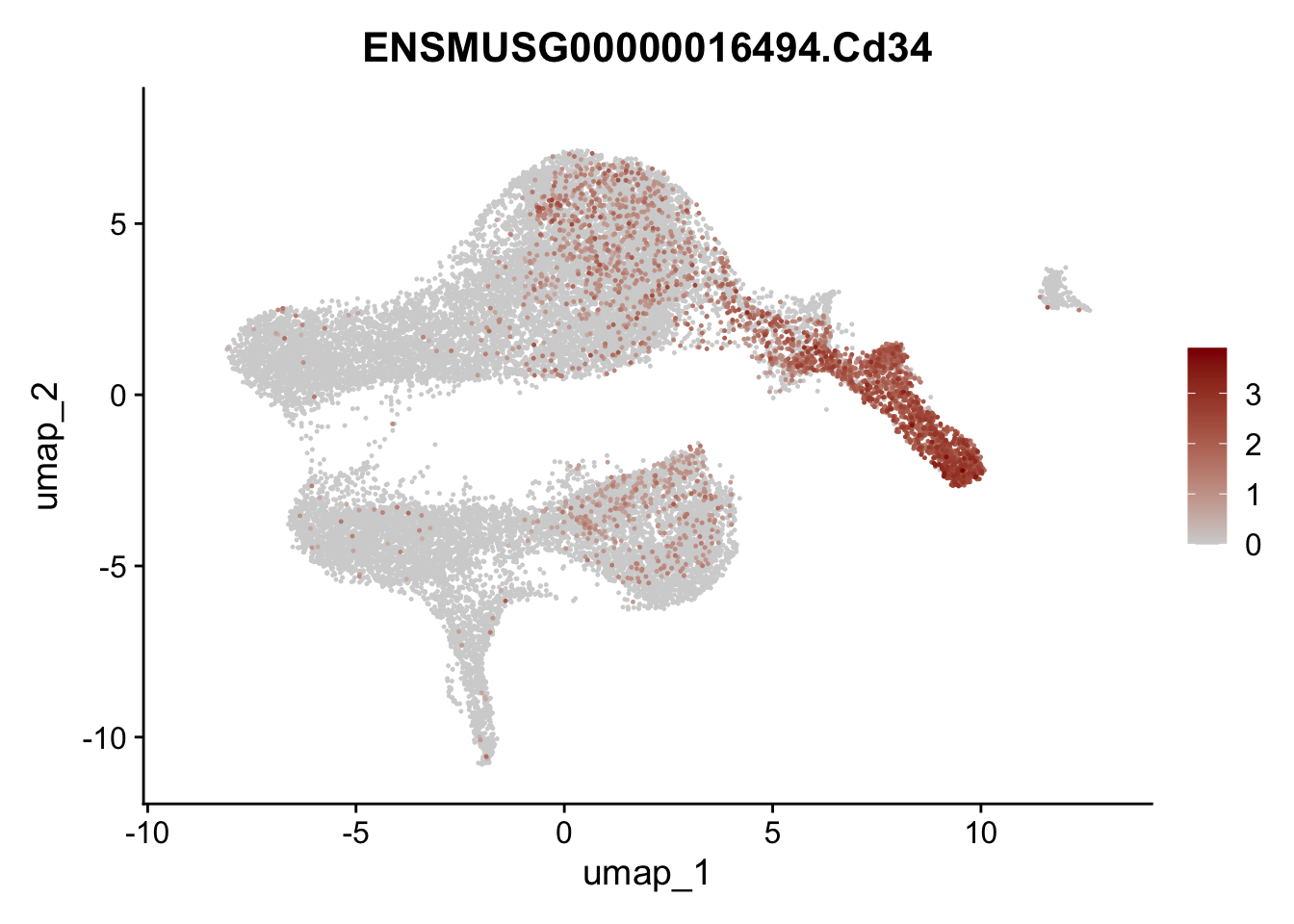
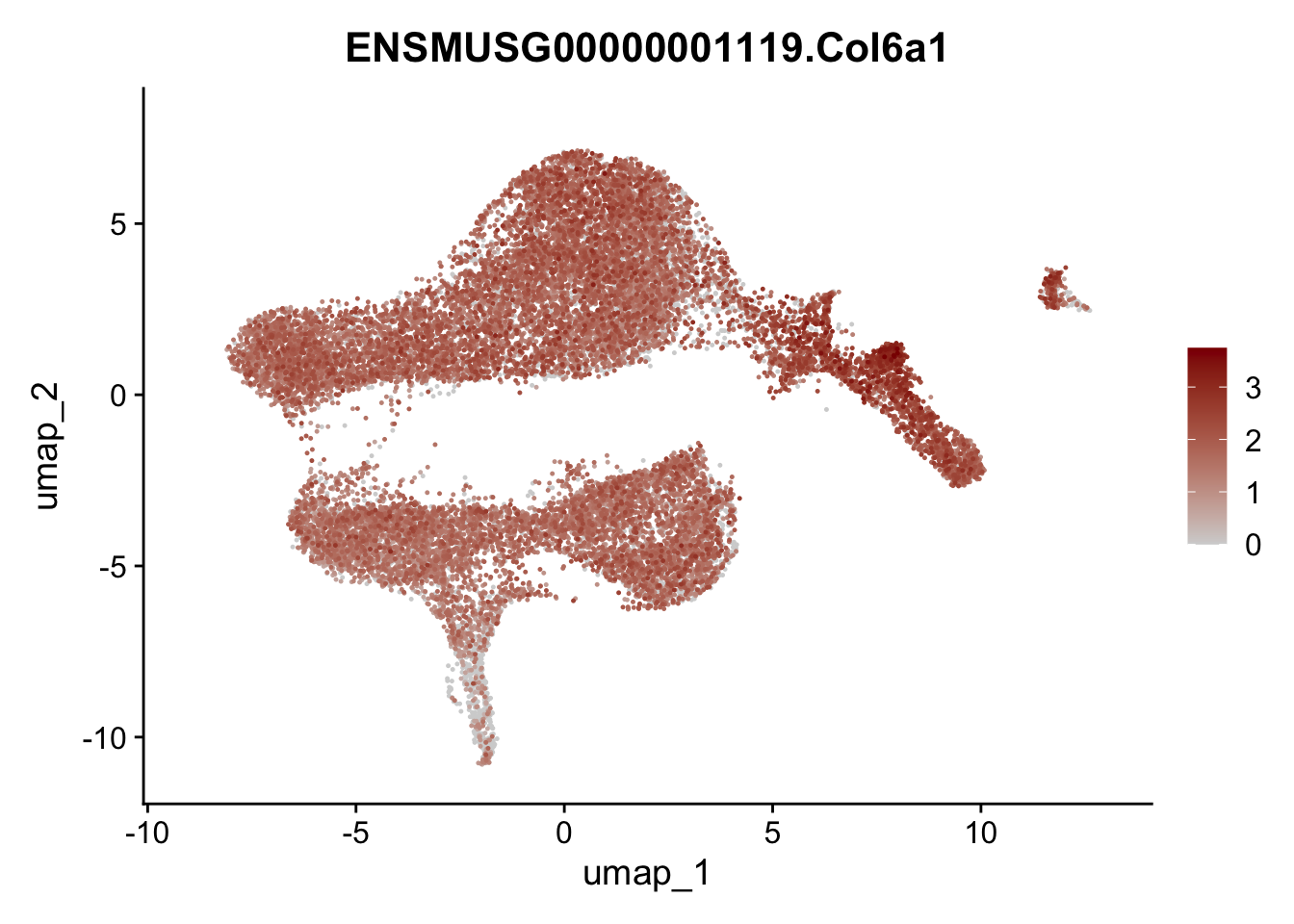
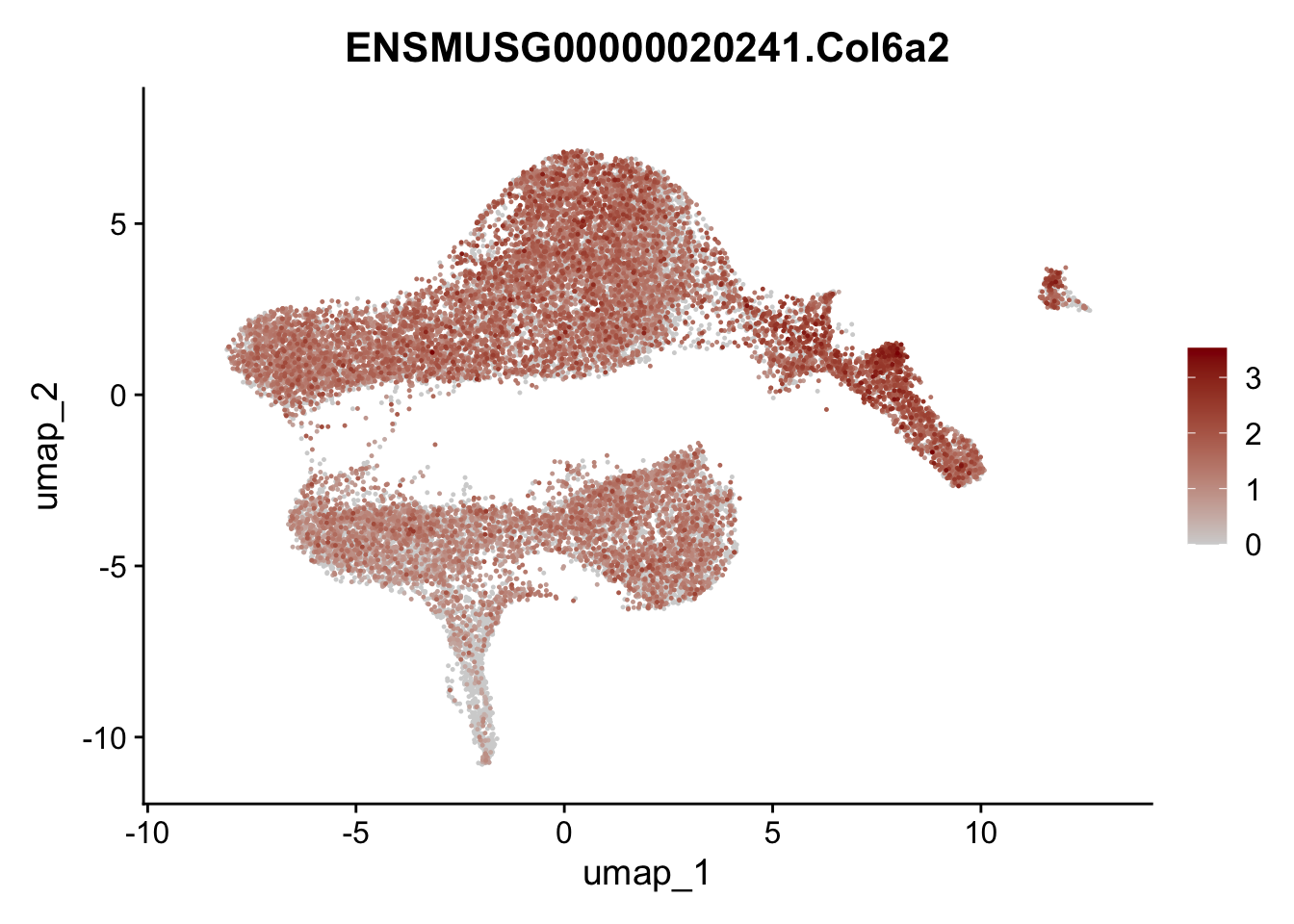
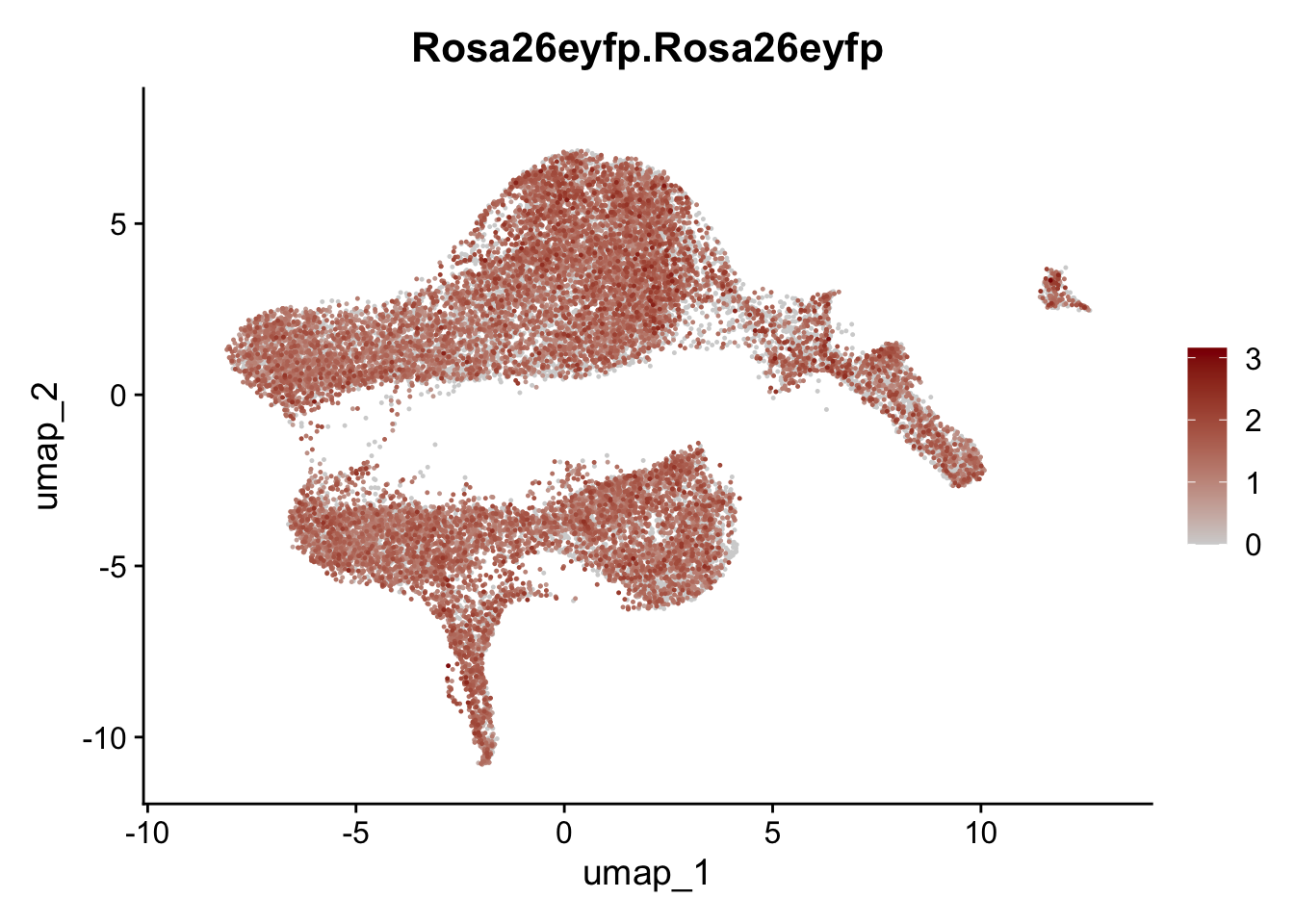
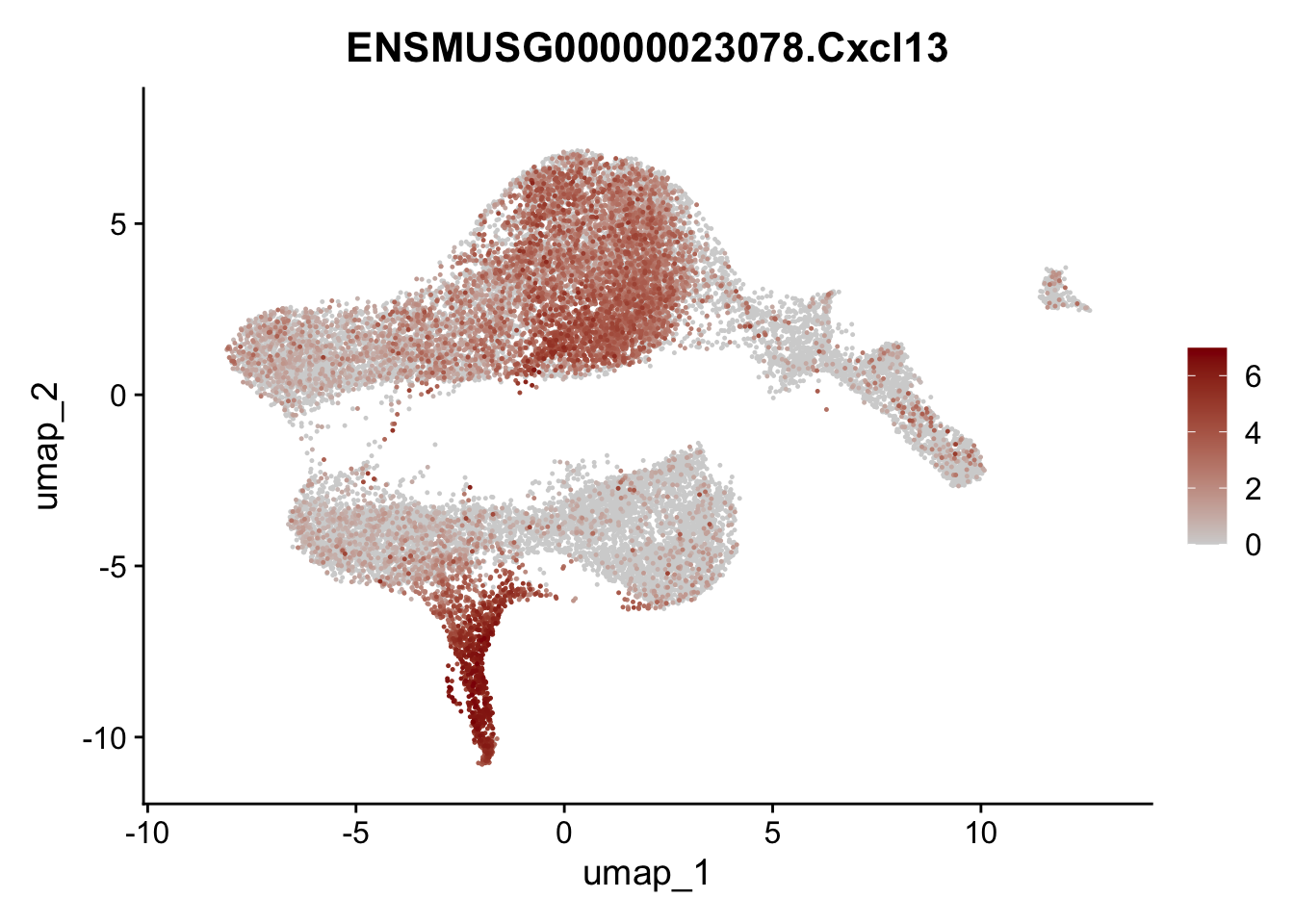
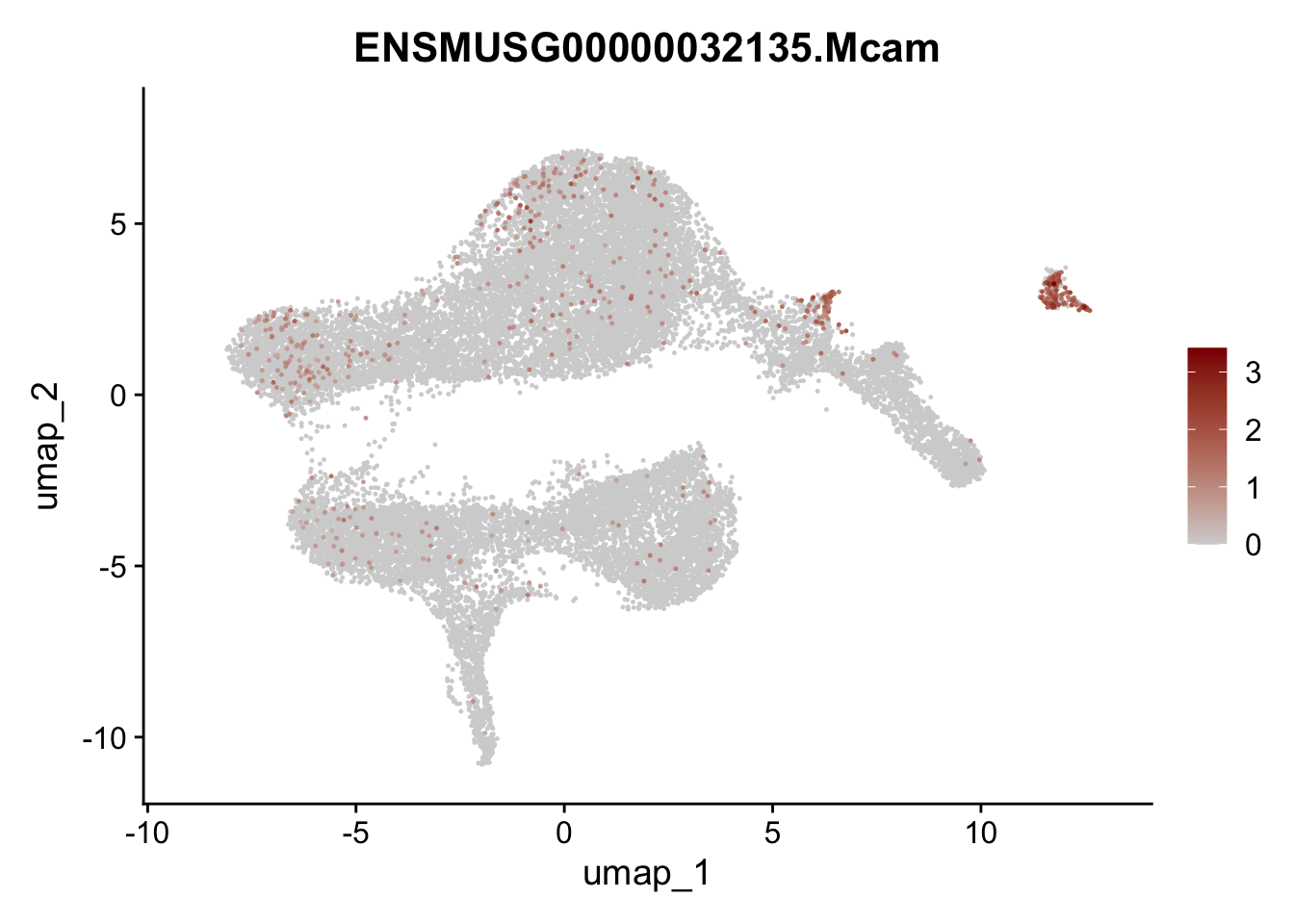
integrate data across location
Idents(seuratA) <- seuratA$location
seurat.list <- SplitObject(object = seuratA, split.by = "location")
for (i in 1:length(x = seurat.list)) {
seurat.list[[i]] <- NormalizeData(object = seurat.list[[i]],
verbose = FALSE)
seurat.list[[i]] <- FindVariableFeatures(object = seurat.list[[i]],
selection.method = "vst", nfeatures = 2000, verbose = FALSE)
}
seurat.anchors <- FindIntegrationAnchors(object.list = seurat.list, dims = 1:20)
seuratA.int <- IntegrateData(anchorset = seurat.anchors, dims = 1:20)
DefaultAssay(object = seuratA.int) <- "integrated"
## rerun seurat
seuratA.int <- ScaleData(object = seuratA.int, verbose = FALSE,
features = rownames(seuratA.int))
seuratA.int <- RunPCA(object = seuratA.int, npcs = 20, verbose = FALSE)
seuratA.int <- RunTSNE(object = seuratA.int, recuction = "pca", dims = 1:20)
seuratA.int <- RunUMAP(object = seuratA.int, recuction = "pca", dims = 1:20)
seuratA.int <- FindNeighbors(object = seuratA.int, reduction = "pca", dims = 1:20)
res <- c(0.6, 0.8, 0.4, 0.25)
for (i in 1:length(res)){
seuratA.int <- FindClusters(object = seuratA.int, resolution = res[i],
random.seed = 1234)
}saveRDS(seuratA.int, file=paste0(basedir,"/data/LNmLToRev_adultonly_seurat.integrated.rds"))load integrated object adult
fileNam <- paste0(basedir, "/data/LNmLToRev_adultonly_seurat.integrated.rds")
seuratA.int <- readRDS(fileNam)DefaultAssay(object = seuratA.int) <- "RNA"
seuratA.int$intCluster <- seuratA.int$integrated_snn_res.0.4
Idents(seuratA.int) <- seuratA.int$intCluster
colPal <- c("#DAF7A6", "#FFC300", "#FF5733", "#C70039", "#900C3F", "#b66e8d",
"#61a4ba", "#6178ba", "#54a87f", "#25328a", "#b6856e",
"#ba6161", "#20714a", "#0073C2FF", "#EFC000FF", "#868686FF",
"#CD534CFF","#7AA6DCFF", "#003C67FF", "#8F7700FF", "#3B3B3BFF",
"#A73030FF", "#4A6990FF")[1:length(unique(seuratA.int$intCluster))]
names(colPal) <- unique(seuratA.int$intCluster)assign label
seuratA.int$label <- "label"
seuratA.int$label[which(seuratA.int$intCluster == "0")] <- "MedRC/IFRC"
seuratA.int$label[which(seuratA.int$intCluster == "1")] <- "actMedRC"
seuratA.int$label[which(seuratA.int$intCluster == "2")] <- "TBRC"
seuratA.int$label[which(seuratA.int$intCluster == "3")] <- "TRC"
seuratA.int$label[which(seuratA.int$intCluster == "4")] <- "MedRC"
seuratA.int$label[which(seuratA.int$intCluster == "5")] <- "PRC"
seuratA.int$label[which(seuratA.int$intCluster == "6")] <- "FDC/MRC"
seuratA.int$label[which(seuratA.int$intCluster == "7")] <- "Pi16+RC"
seuratA.int$label[which(seuratA.int$intCluster == "8")] <- "VSMC"
table(seuratA.int$label)
actMedRC FDC/MRC MedRC MedRC/IFRC Pi16+RC PRC TBRC TRC VSMC
4020 1292 2371 4868 600 2192 3841 2839 265 colLab2 <- colLab
colLab <- c("#42a071", "#900C3F","#b66e8d", "#61a4ba", "#424671", "#003C67FF",
"#e3953d", "#714542", "#b6856e", "#FFC300")
names(colLab) <- c("FDC/MRC", "TRC", "TBRC", "MedRC/IFRC", "MedRC" , "actMedRC",
"PRC", "Pi16+RC", "VSMC")Dimplot int data
clustering
DimPlot(seuratA.int, reduction = "umap",
pt.size = 0.1, label = T, shuffle = T, cols = colPal) +
theme_bw() +
theme(axis.text = element_blank(), axis.ticks = element_blank(),
panel.grid.minor = element_blank()) +
xlab("umap1") +
ylab("umap2")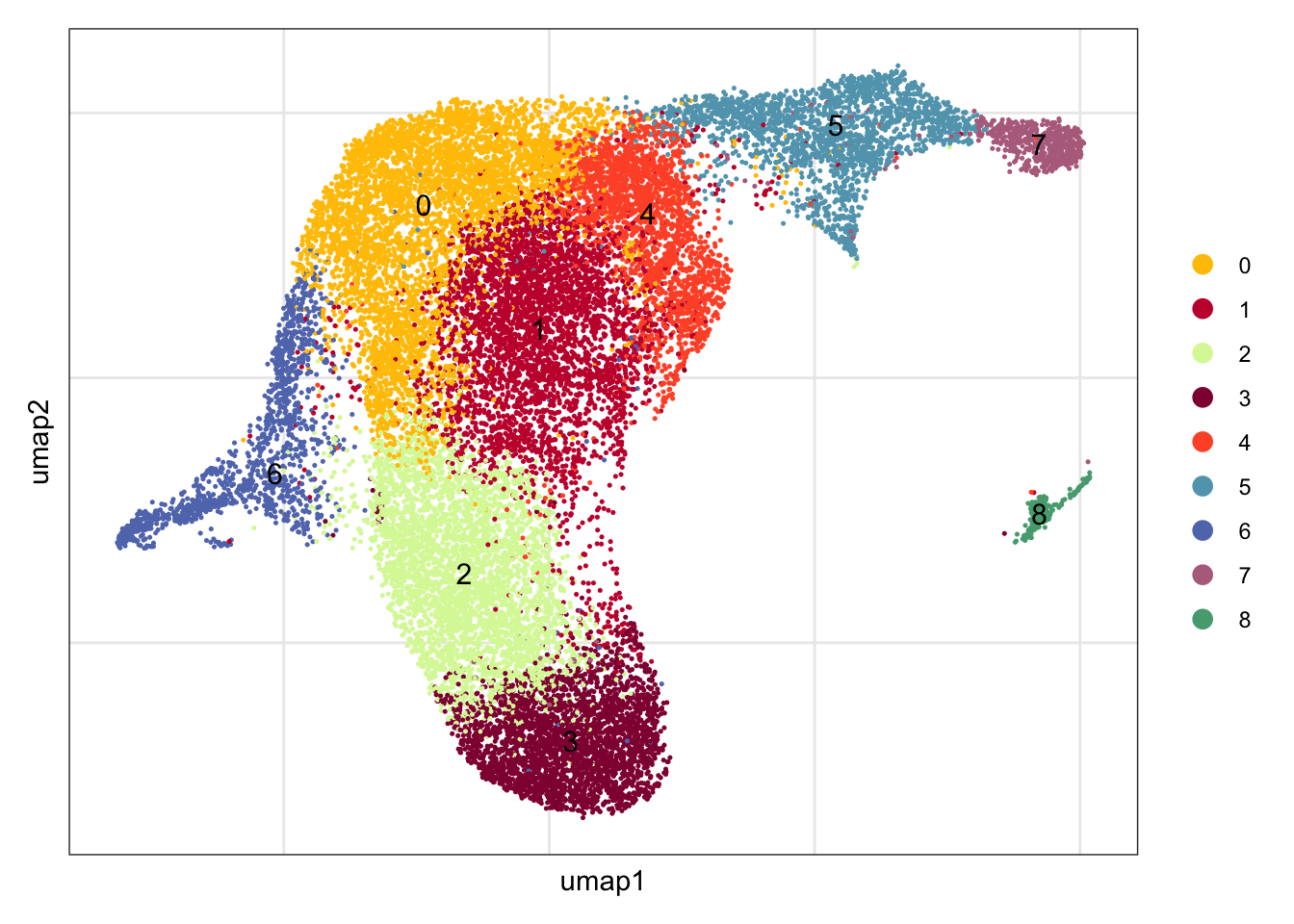
label
DimPlot(seuratA.int, reduction = "umap", group.by = "label", cols = colLab)+
theme_bw() +
theme(axis.text = element_blank(), axis.ticks = element_blank(),
panel.grid.minor = element_blank()) +
xlab("UMAP1") +
ylab("UMAP2")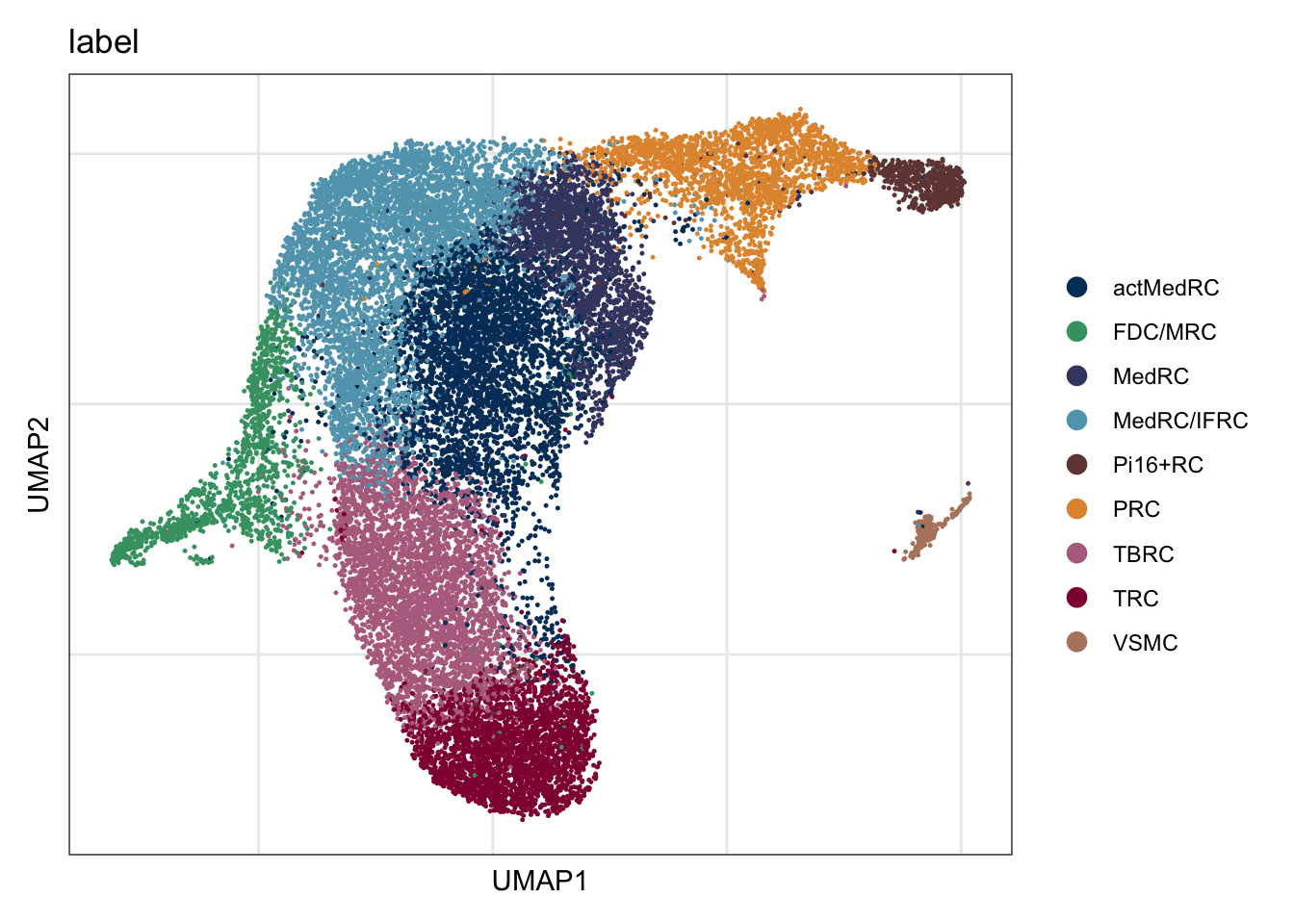
DimPlot(seuratA.int, reduction = "umap", group.by = "label", pt.size=0.5,
cols = colLab, shuffle = T)+
theme_void()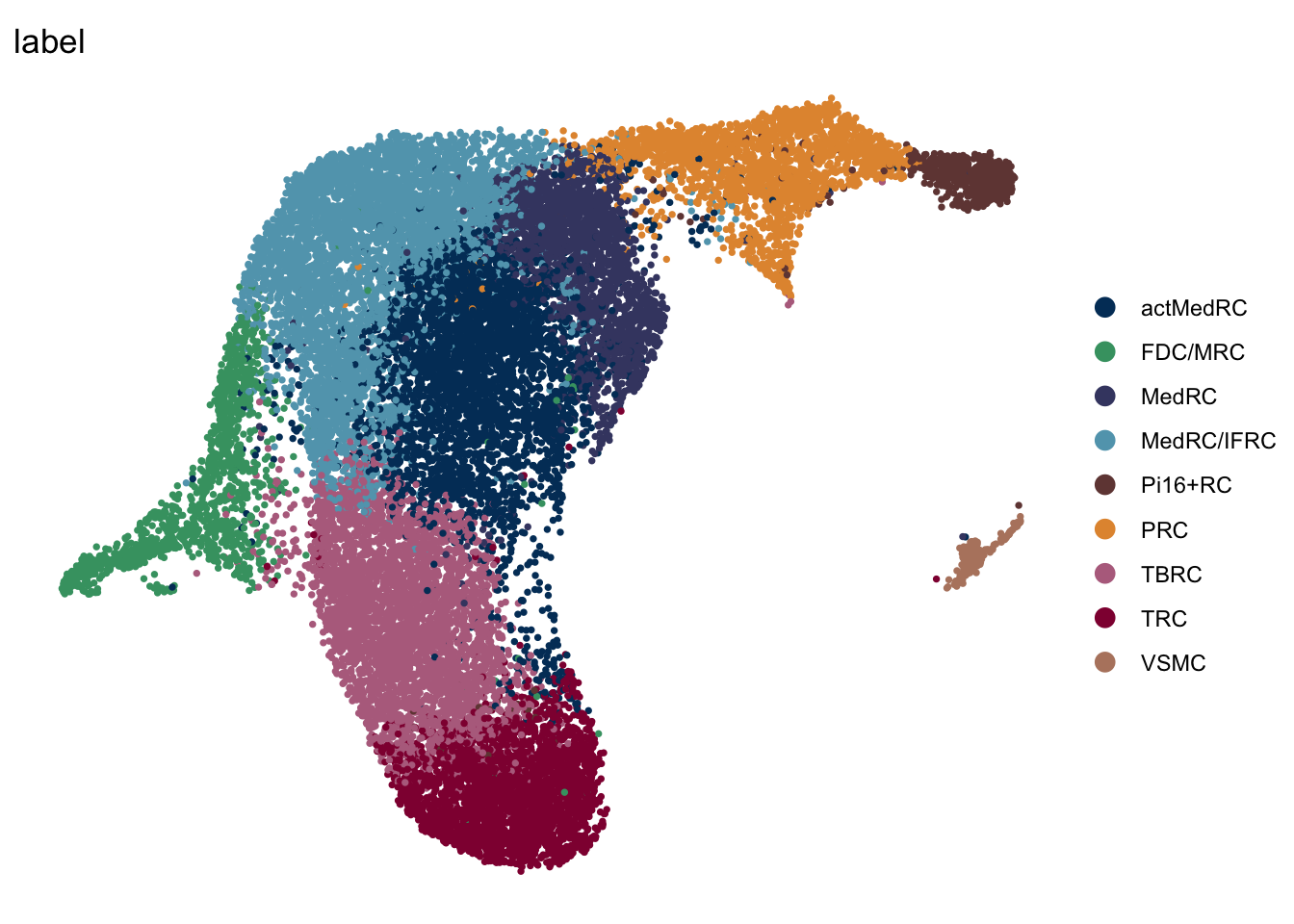
DimPlot(seuratA.int, reduction = "umap", group.by = "label", pt.size=0.5,
cols = colLab, shuffle = T)+
theme_void() +
theme(legend.position = "none") 
label mLN iLN sep
mLN
seuratA.intmLN <- subset(seuratA.int, location == "mLN")
DimPlot(seuratA.intmLN, reduction = "umap", group.by = "label", pt.size=0.5,
cols = colLab, shuffle = T)+
theme_void()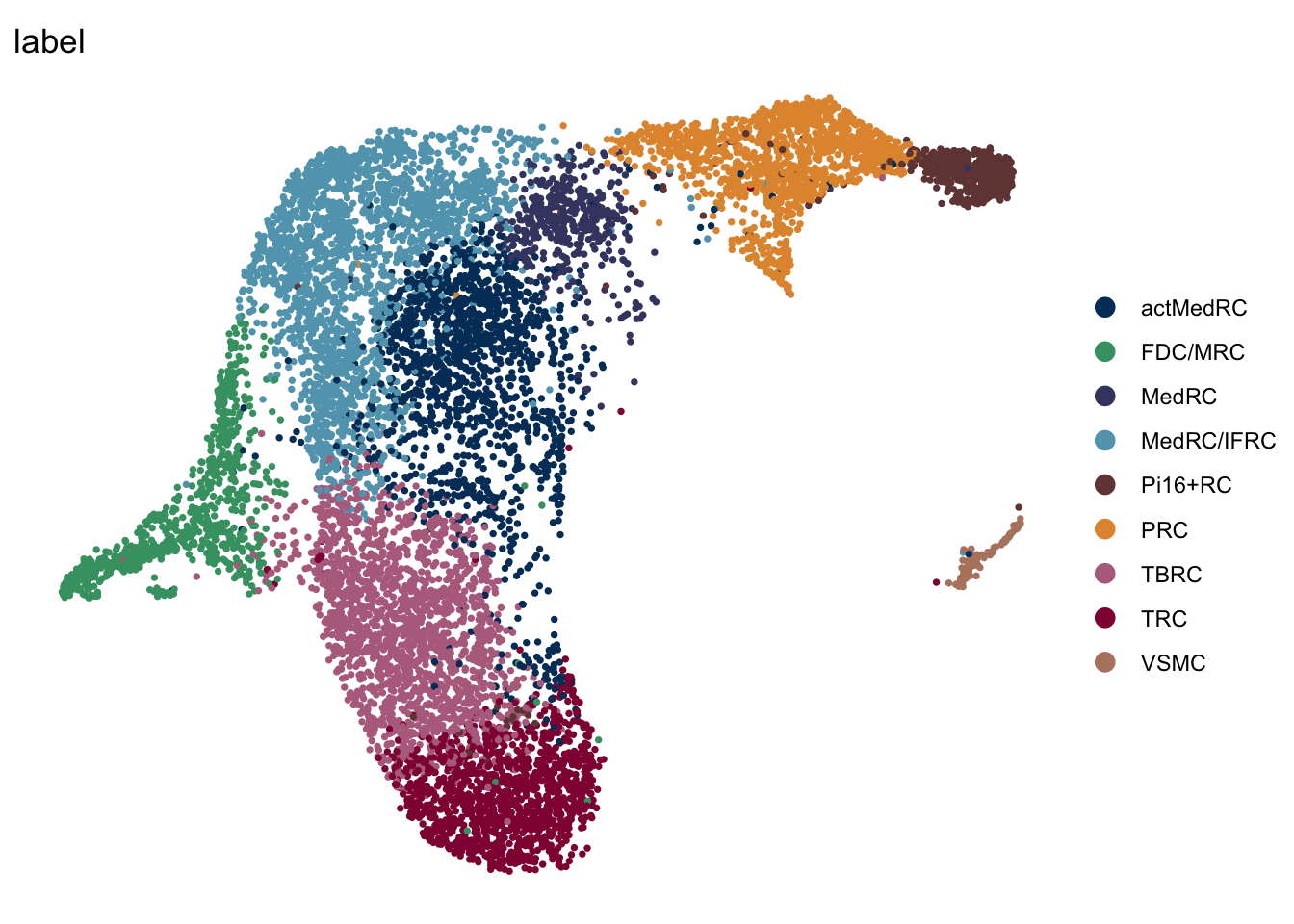
DimPlot(seuratA.intmLN, reduction = "umap", group.by = "label", pt.size=0.5,
cols = colLab, shuffle = T)+
theme_void() +
theme(legend.position = "none") 
iLN
seuratA.intiLN <- subset(seuratA.int, location == "iLN")
DimPlot(seuratA.intiLN, reduction = "umap", group.by = "label", pt.size=0.5,
cols = colLab, shuffle = T)+
theme_void()
DimPlot(seuratA.intiLN, reduction = "umap", group.by = "label", pt.size=0.5,
cols = colLab, shuffle = T)+
theme_void() +
theme(legend.position = "none") 
label split by location
DimPlot(seuratA.int, reduction = "umap", group.by = "label", cols = colLab,
split.by = "location")+
theme_bw() +
theme(axis.text = element_blank(), axis.ticks = element_blank(),
panel.grid.minor = element_blank()) +
xlab("UMAP1") +
ylab("UMAP2")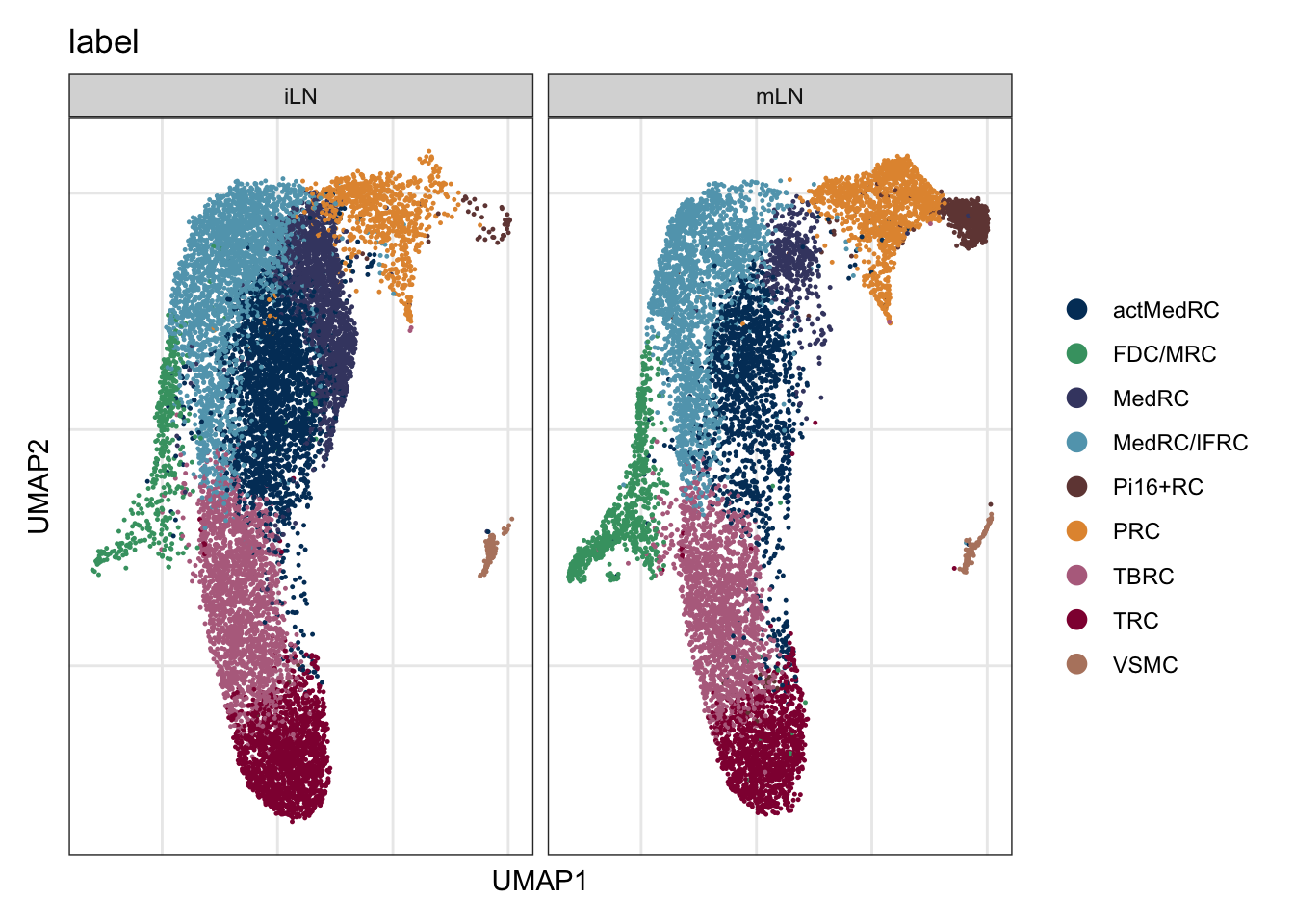
DimPlot(seuratA.int, reduction = "umap", group.by = "label", pt.size=0.5,
cols = colLab, split.by = "location", shuffle = T)+
theme_void()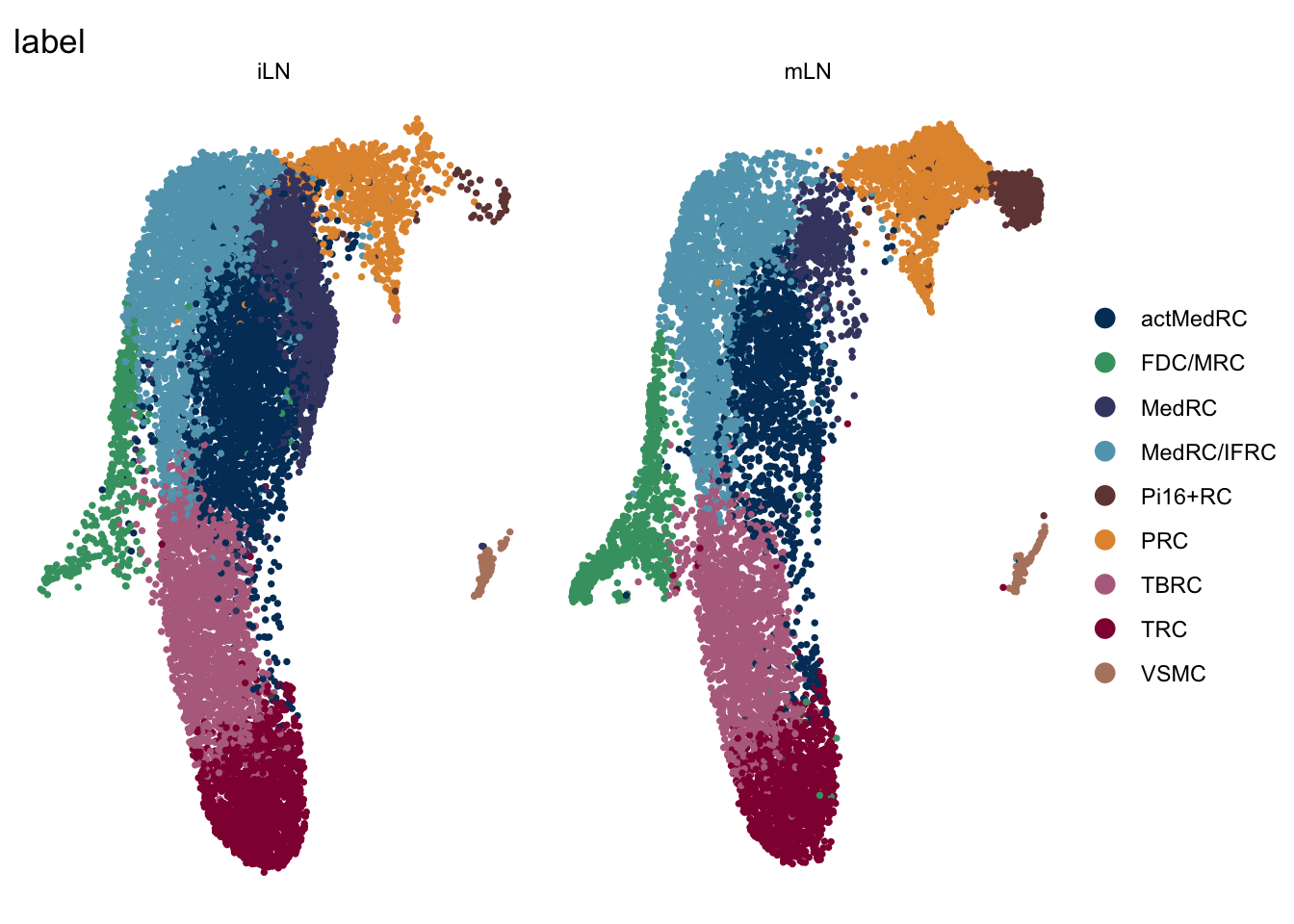
location
DimPlot(seuratA.int, reduction = "umap", group.by = "location", cols = colLoc,
pt.size = 0.1, shuffle = T) +
theme_bw() +
theme(axis.text = element_blank(), axis.ticks = element_blank(),
panel.grid.minor = element_blank()) +
xlab("umap1") +
ylab("umap2")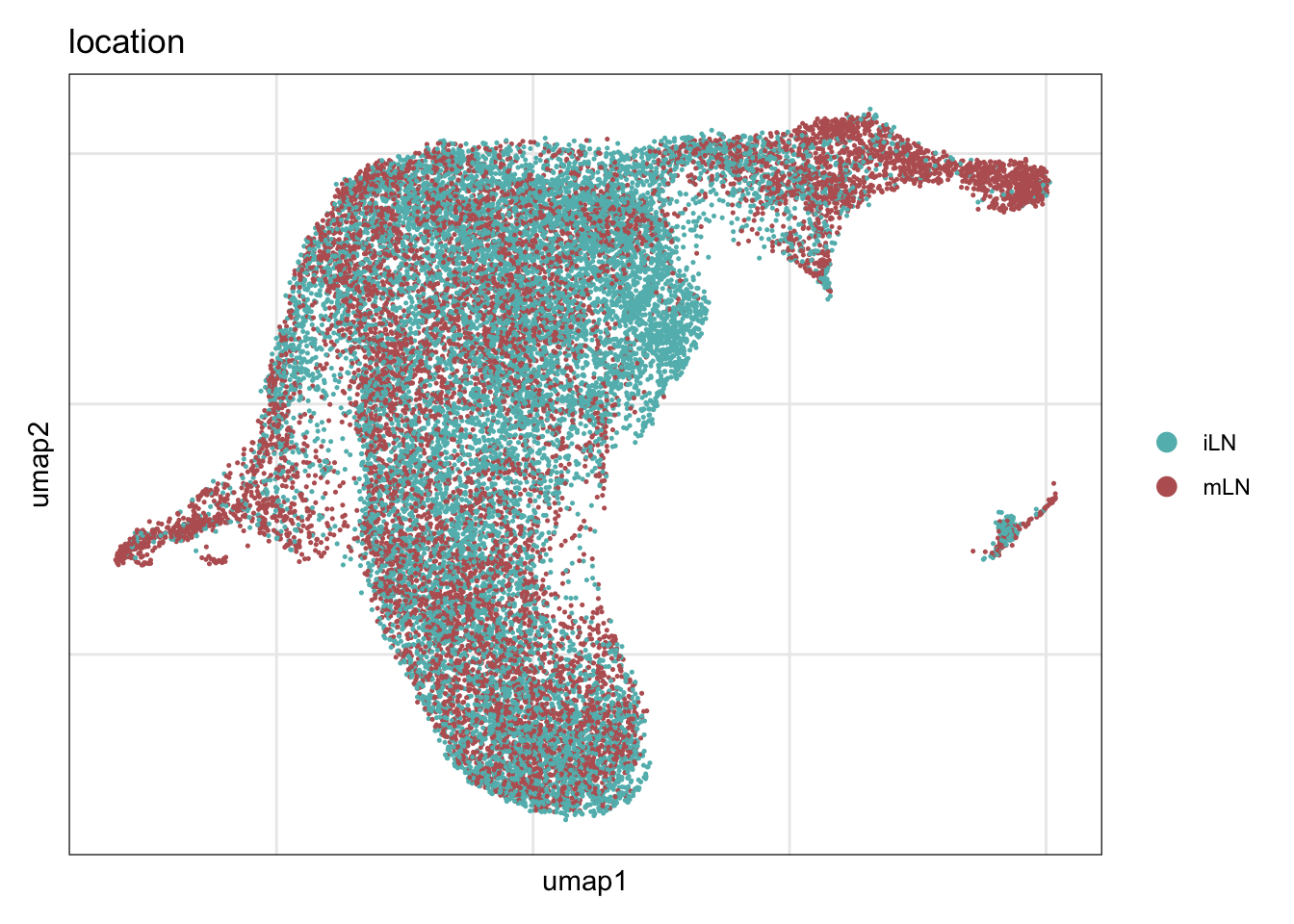
DimPlot(seuratA.int, reduction = "umap", group.by = "location", pt.size=0.5,
cols = colLoc, split.by = "location", shuffle = T)+
theme_void()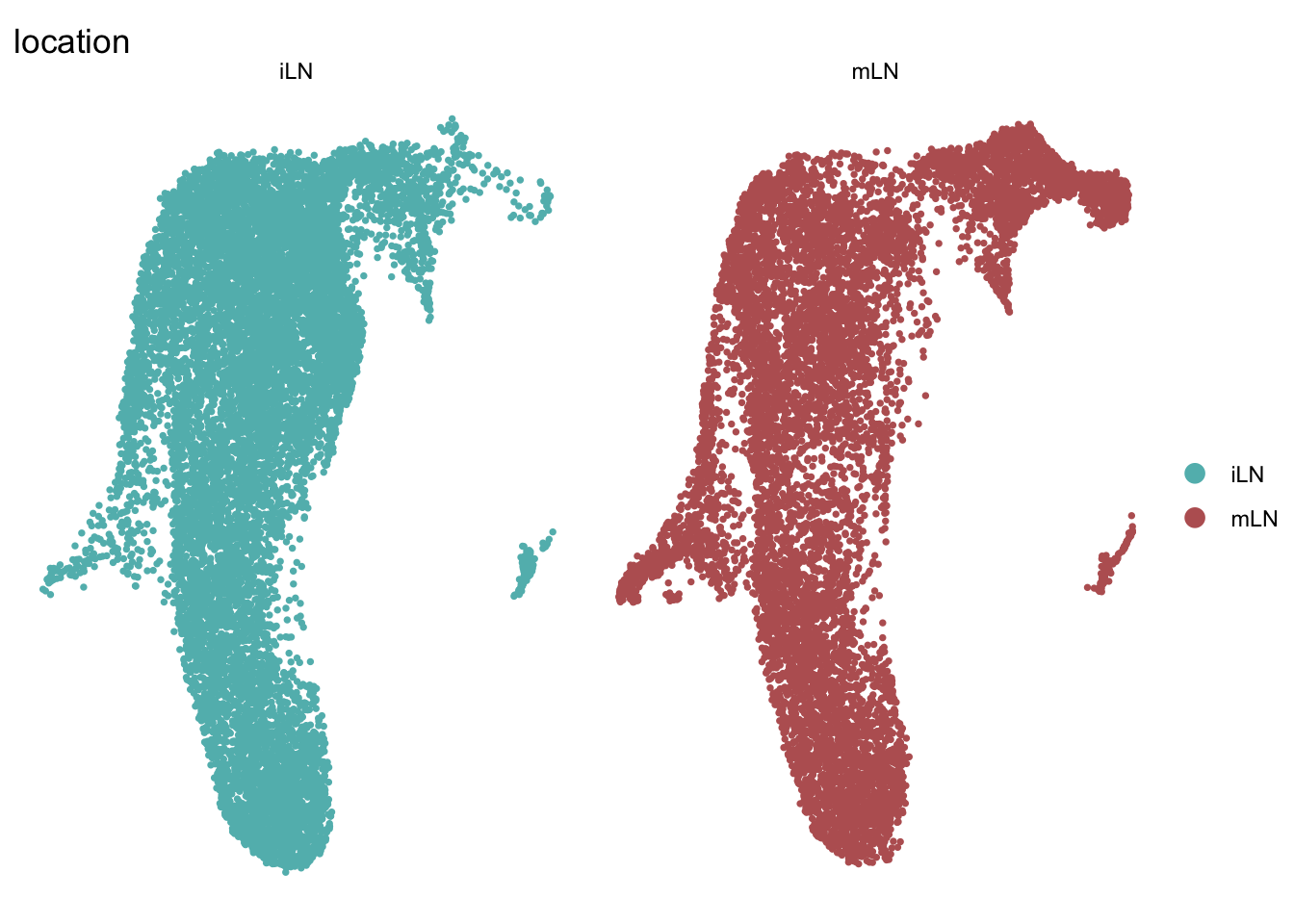
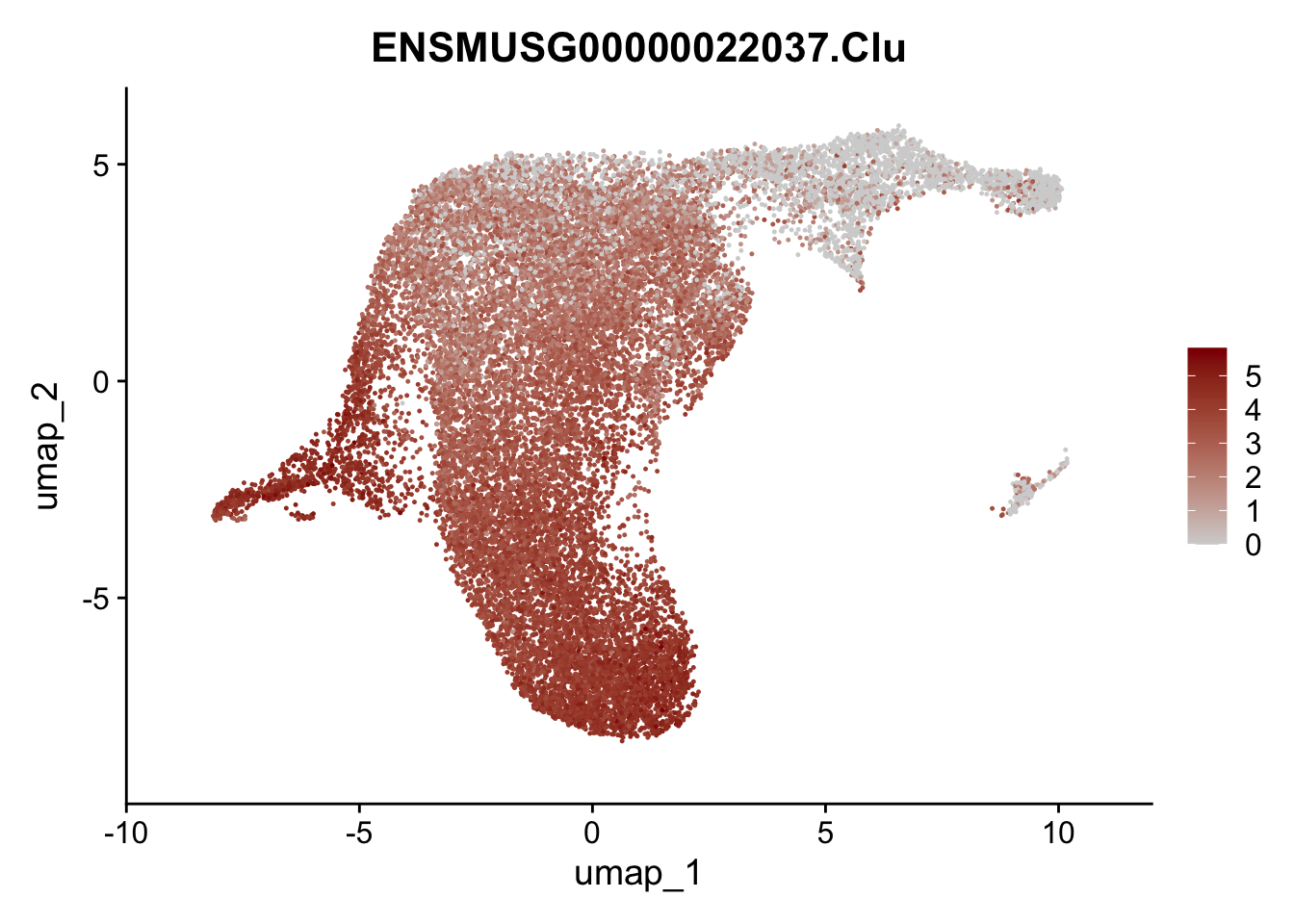
plot features int
selGenesViolin <- c("ENSMUSG00000022037.Clu", "ENSMUSG00000094686.Ccl21a",
"ENSMUSG00000074934.Grem1", "ENSMUSG00000050069.Grem2",
"ENSMUSG00000042436.Mfap4", "ENSMUSG00000071005.Ccl19",
"ENSMUSG00000016494.Cd34", "ENSMUSG00000024011.Pi16",
"ENSMUSG00000001119.Col6a1", "ENSMUSG00000020241.Col6a2",
"Rosa26eyfp.Rosa26eyfp", "ENSMUSG00000023078.Cxcl13",
"ENSMUSG00000032135.Mcam", "ENSMUSG00000023034.Nr4a1")
pList <- sapply(selGenesViolin, function(x){
p <- FeaturePlot(seuratA.int, reduction = "umap",
features = x,
cols=c("lightgrey", "darkred"),
order = F)+
theme(legend.position="right")
plot(p)
})
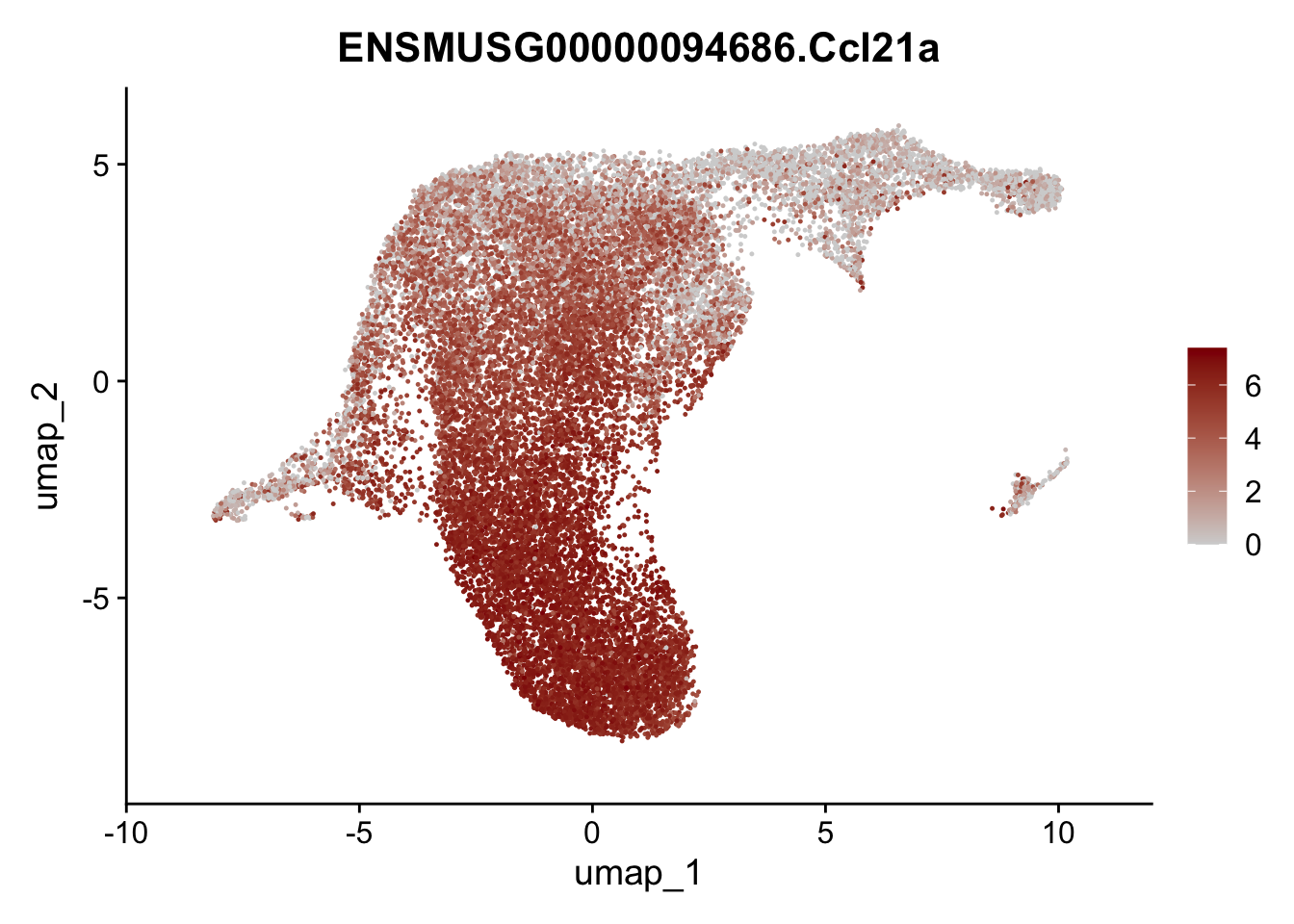
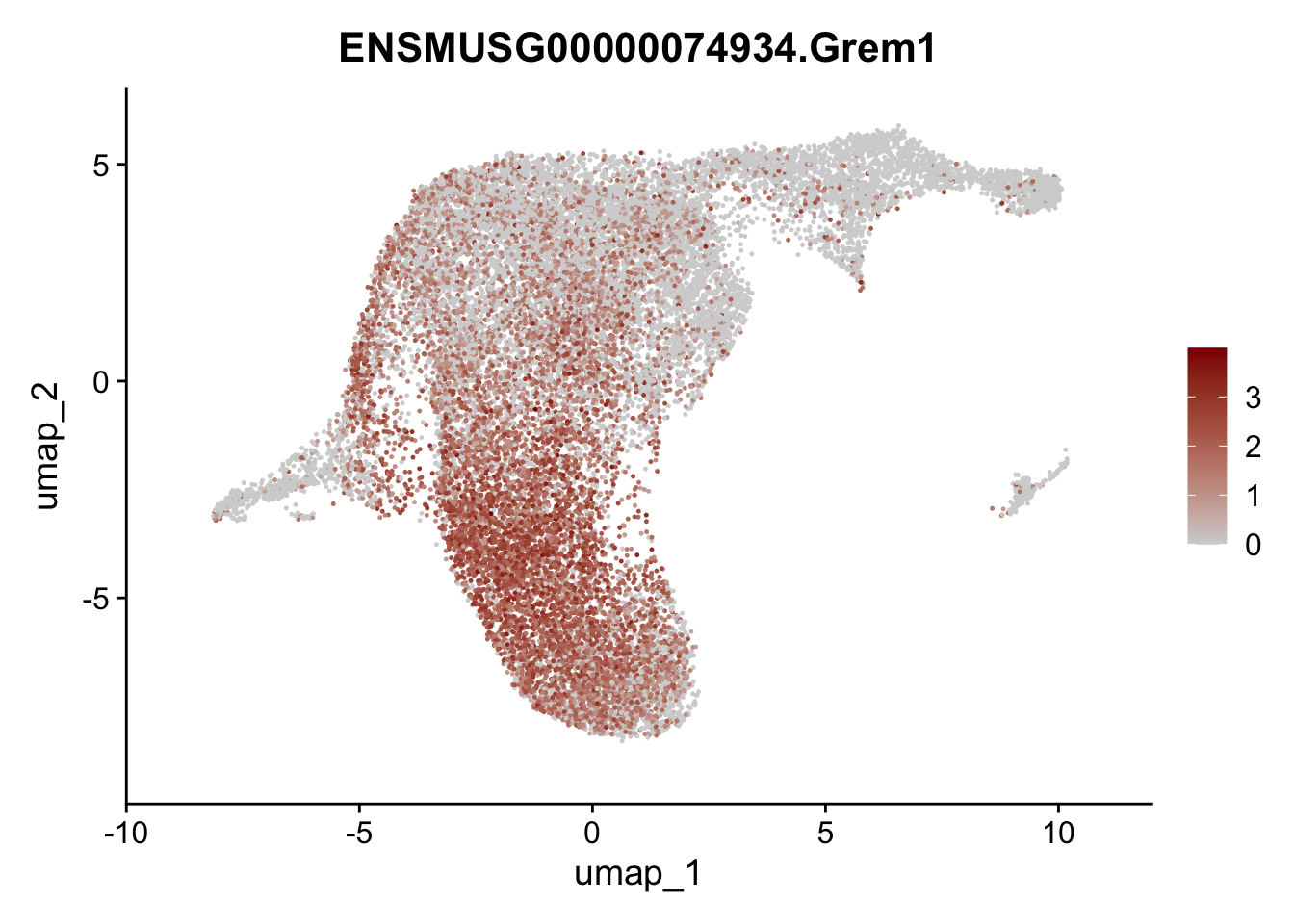
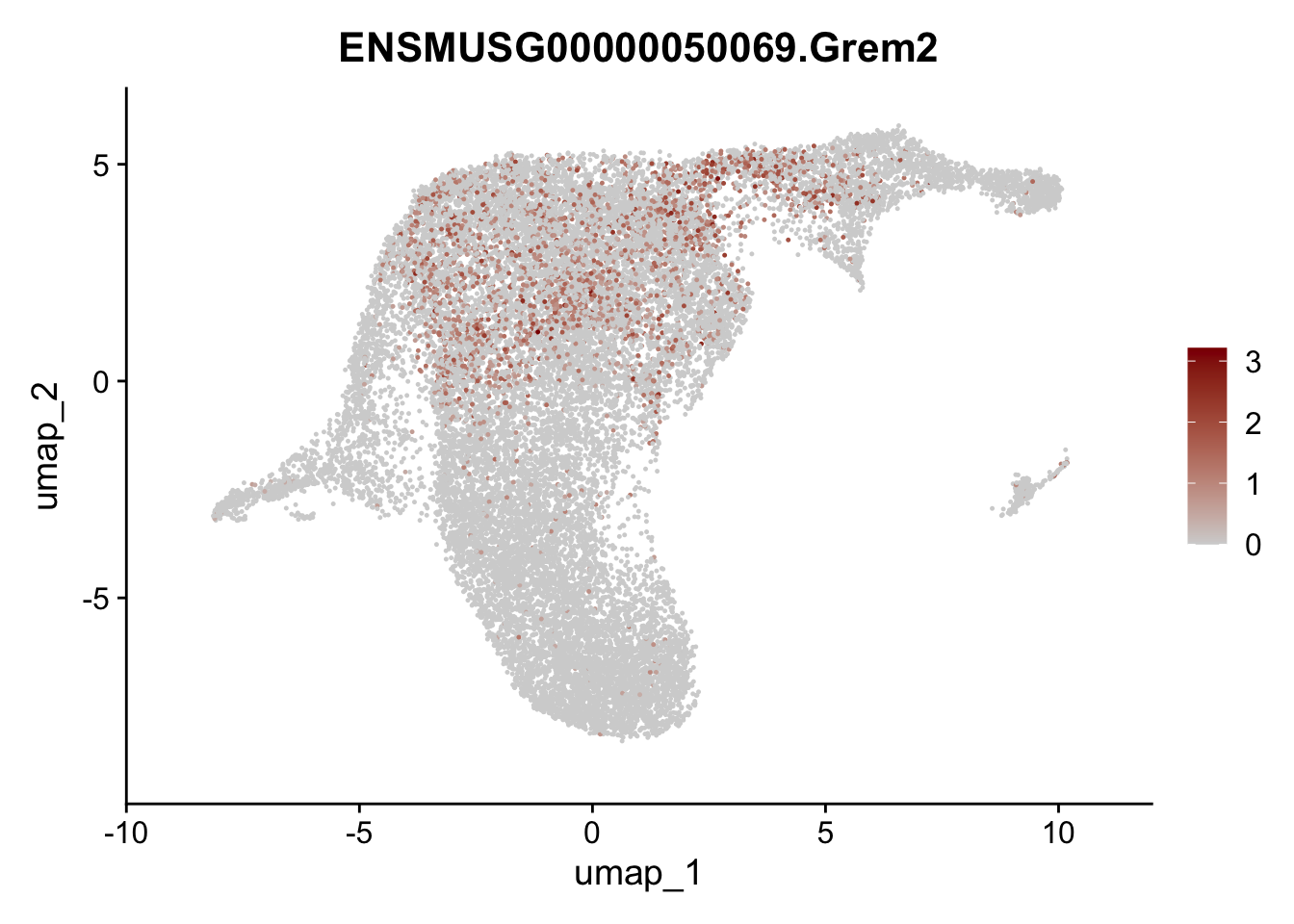
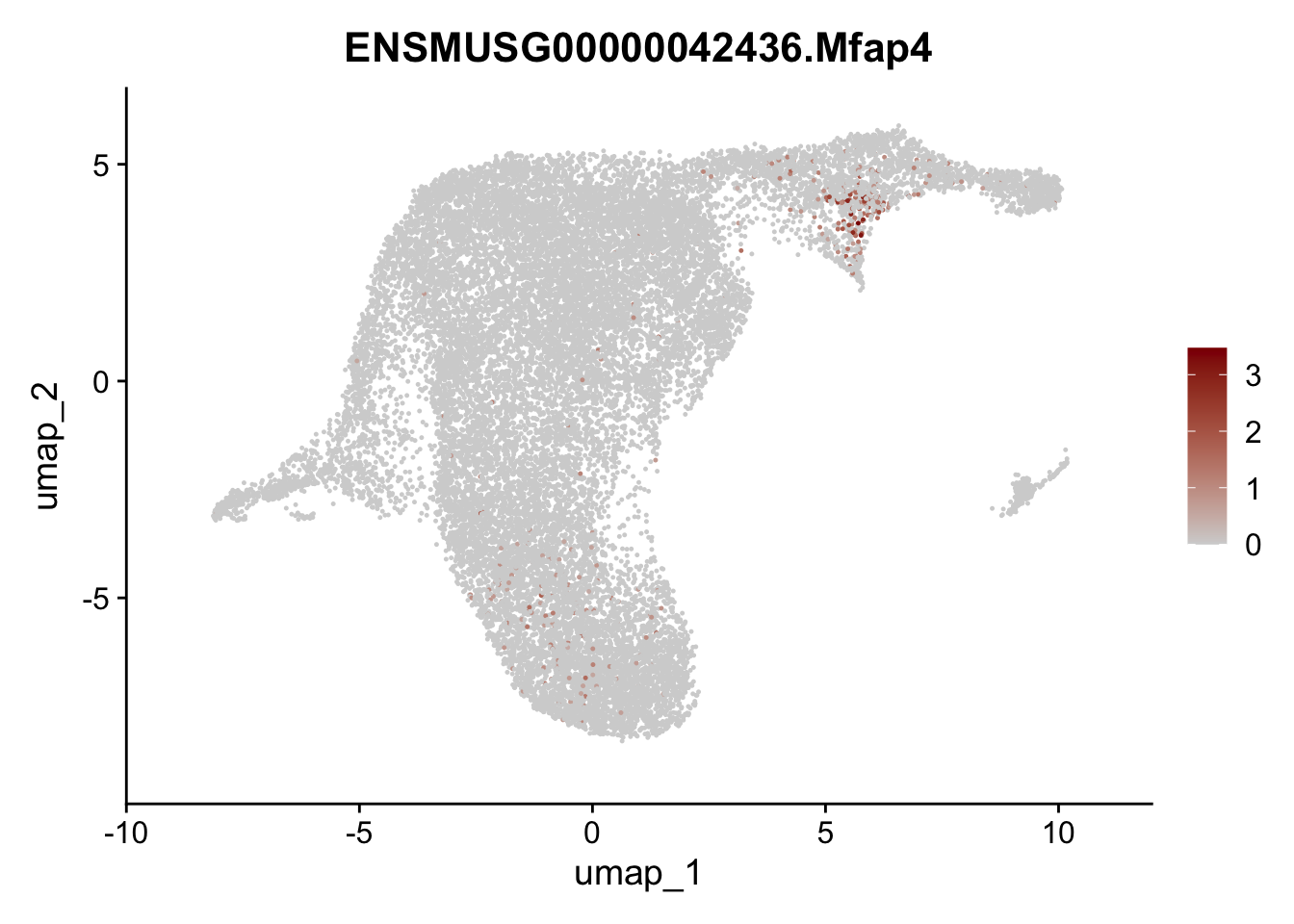
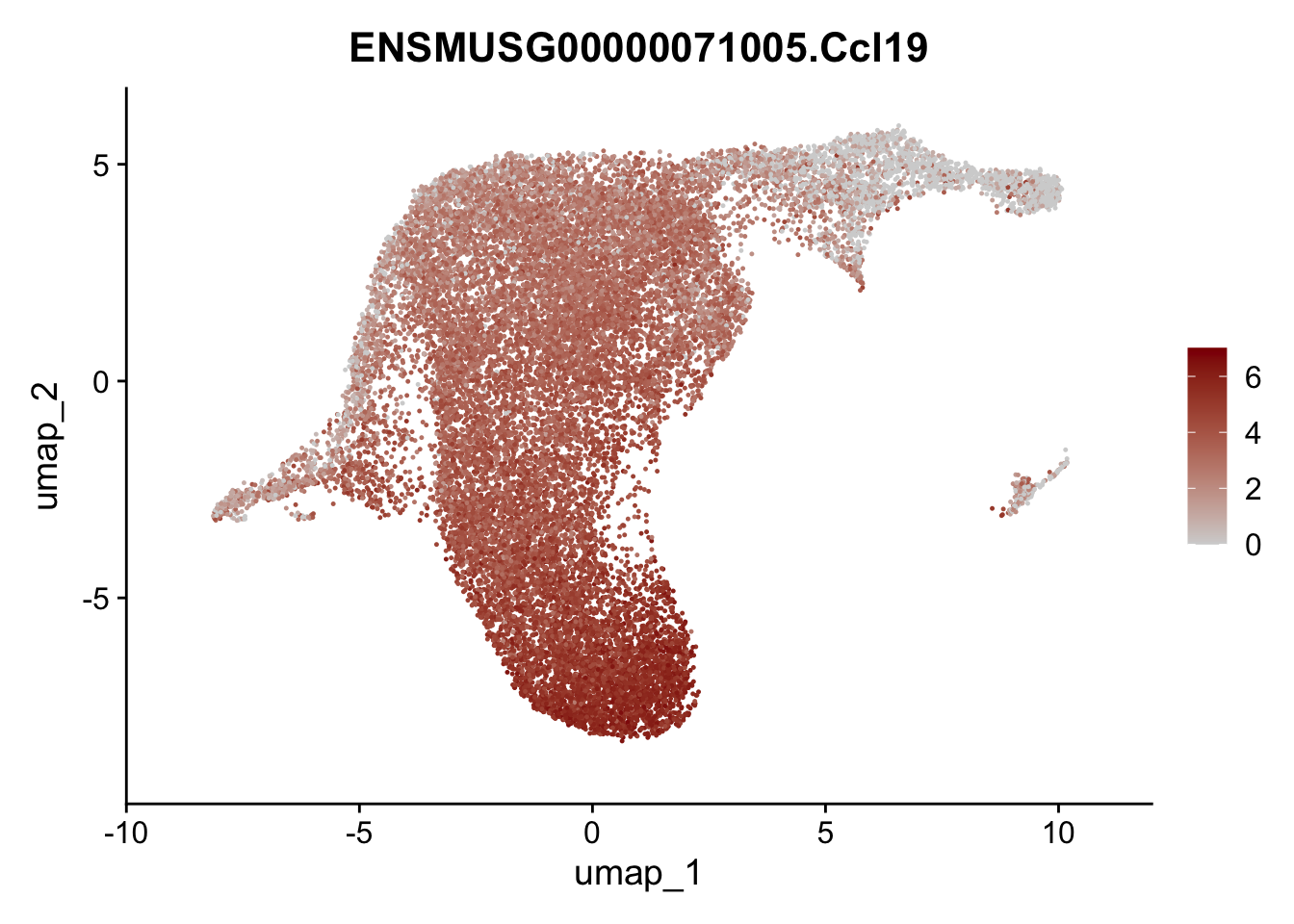
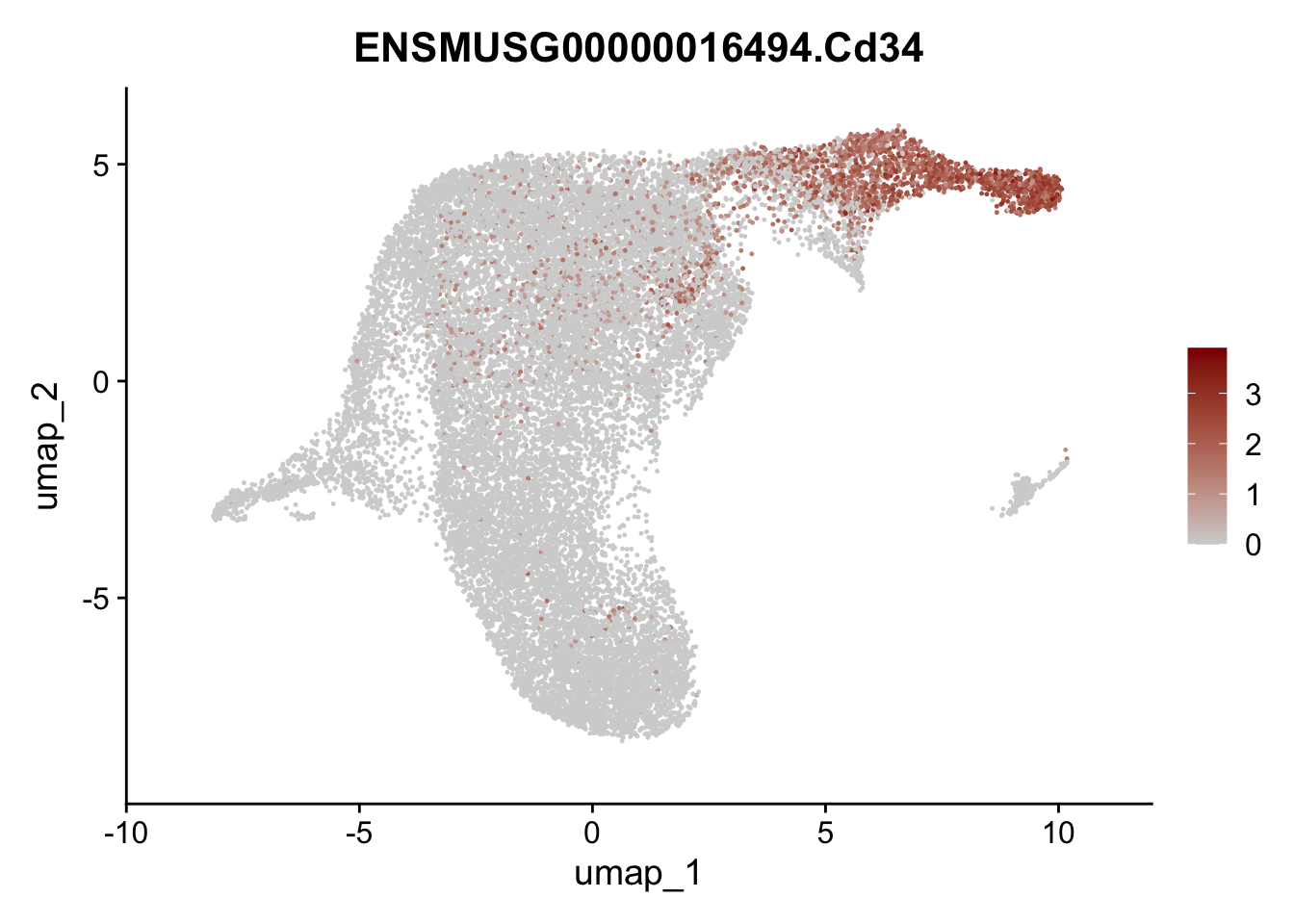
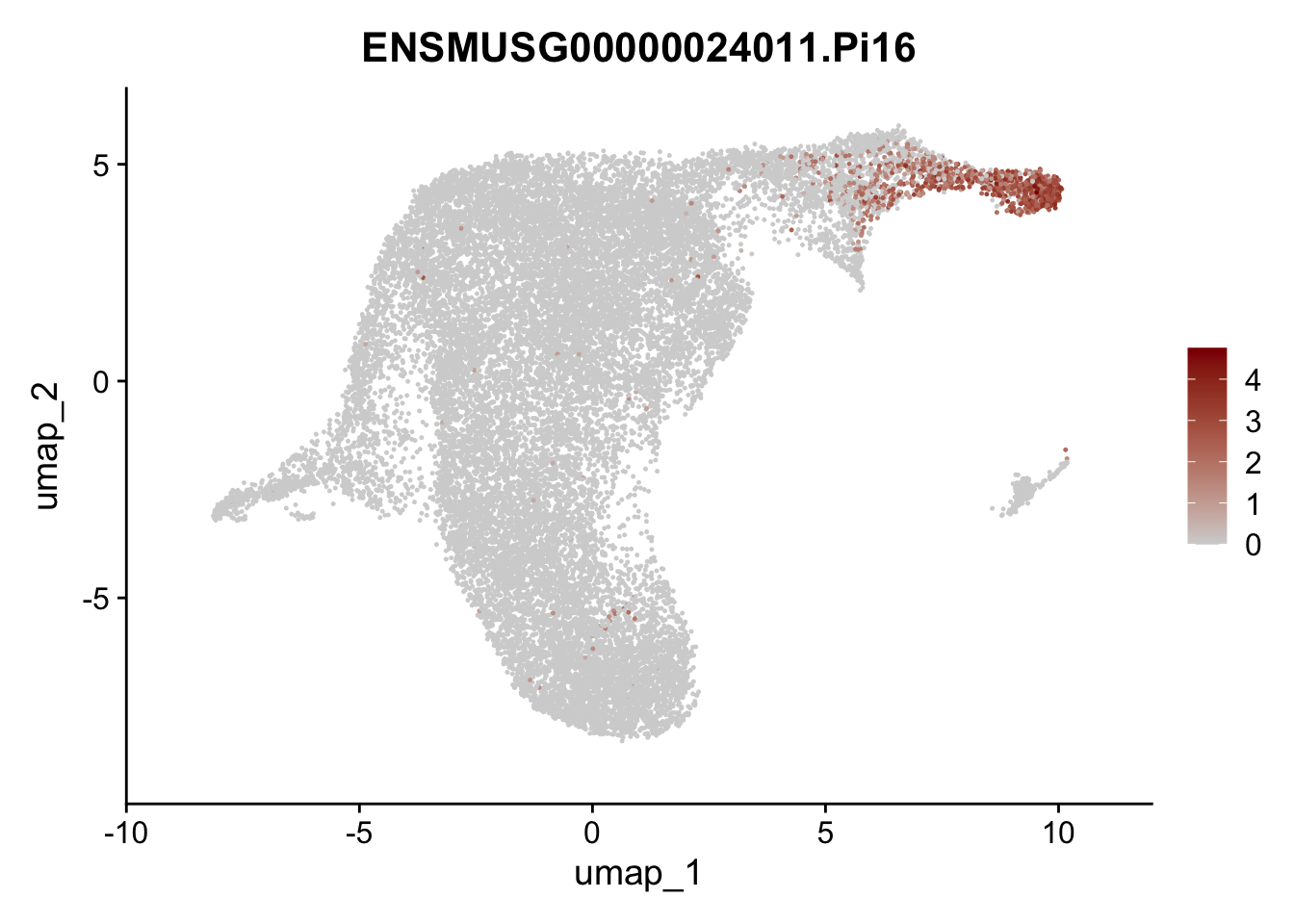
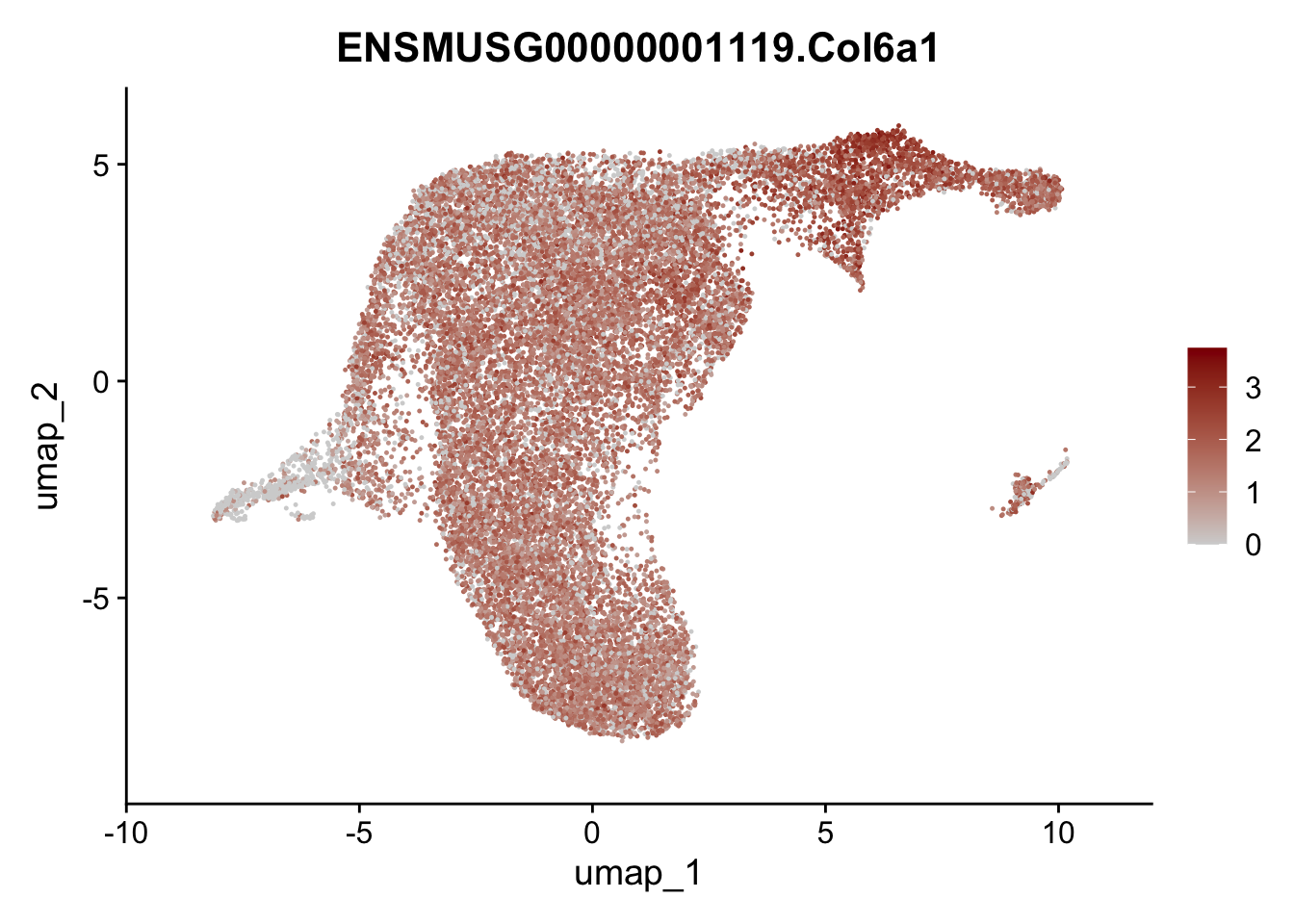
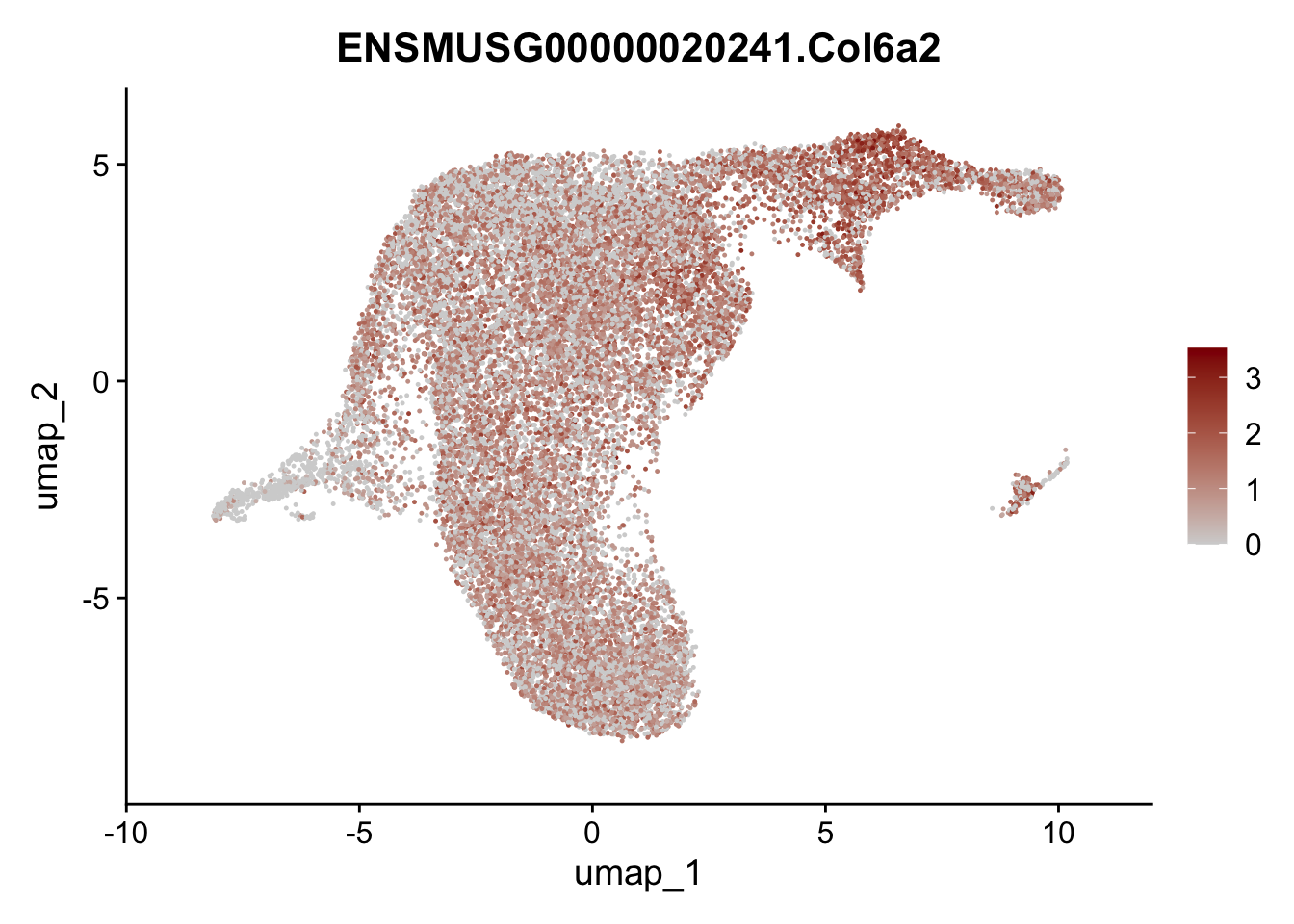
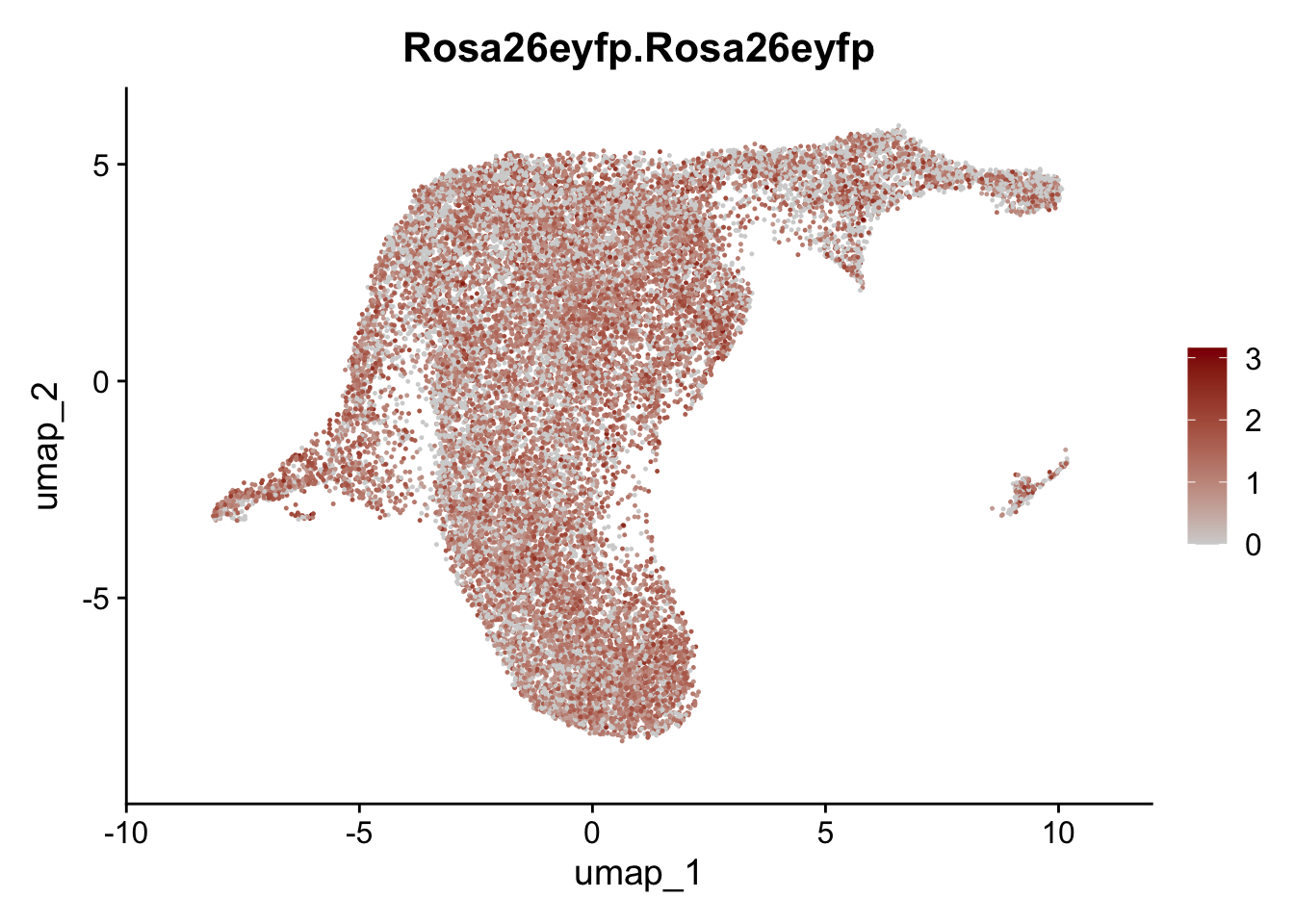
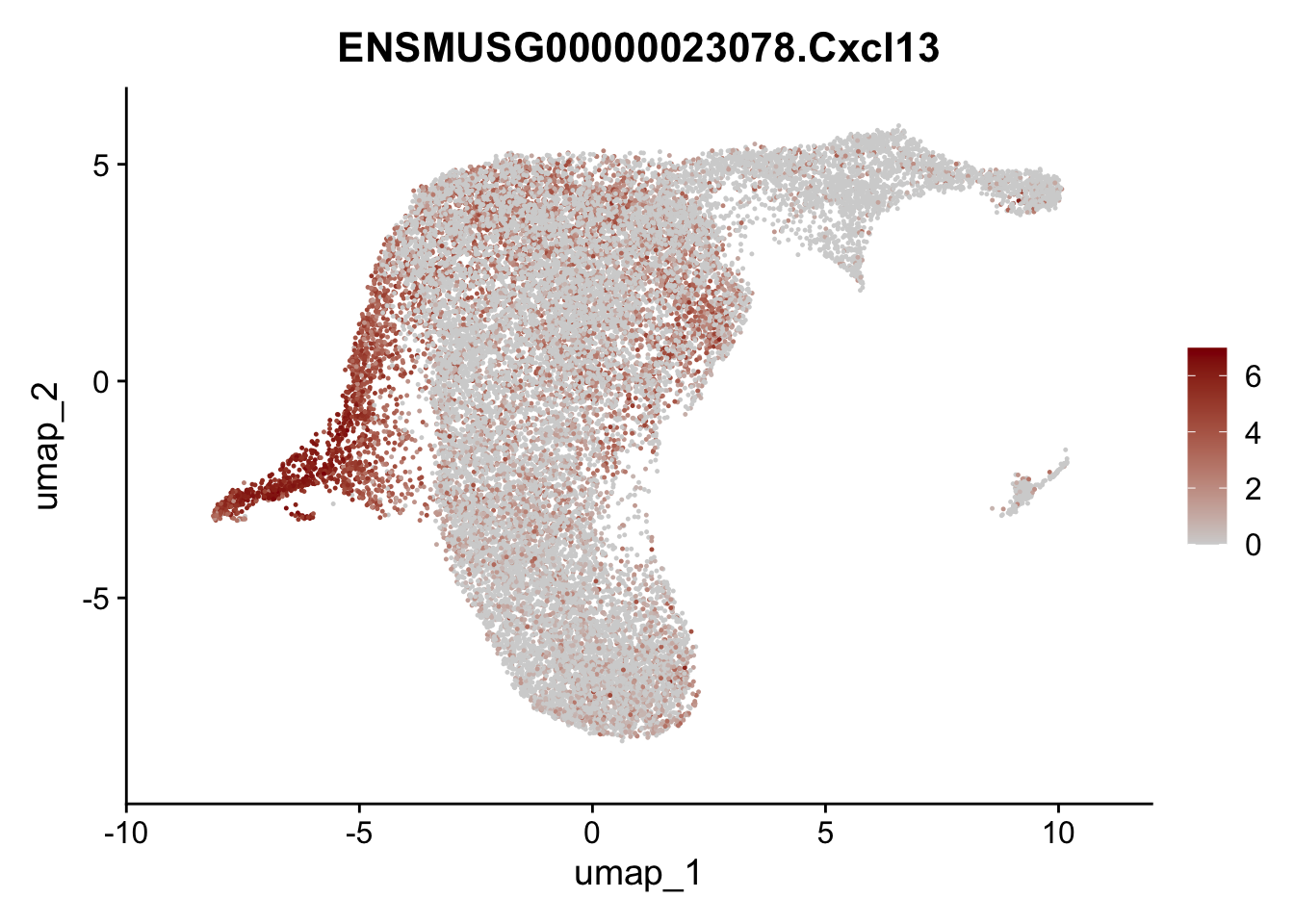
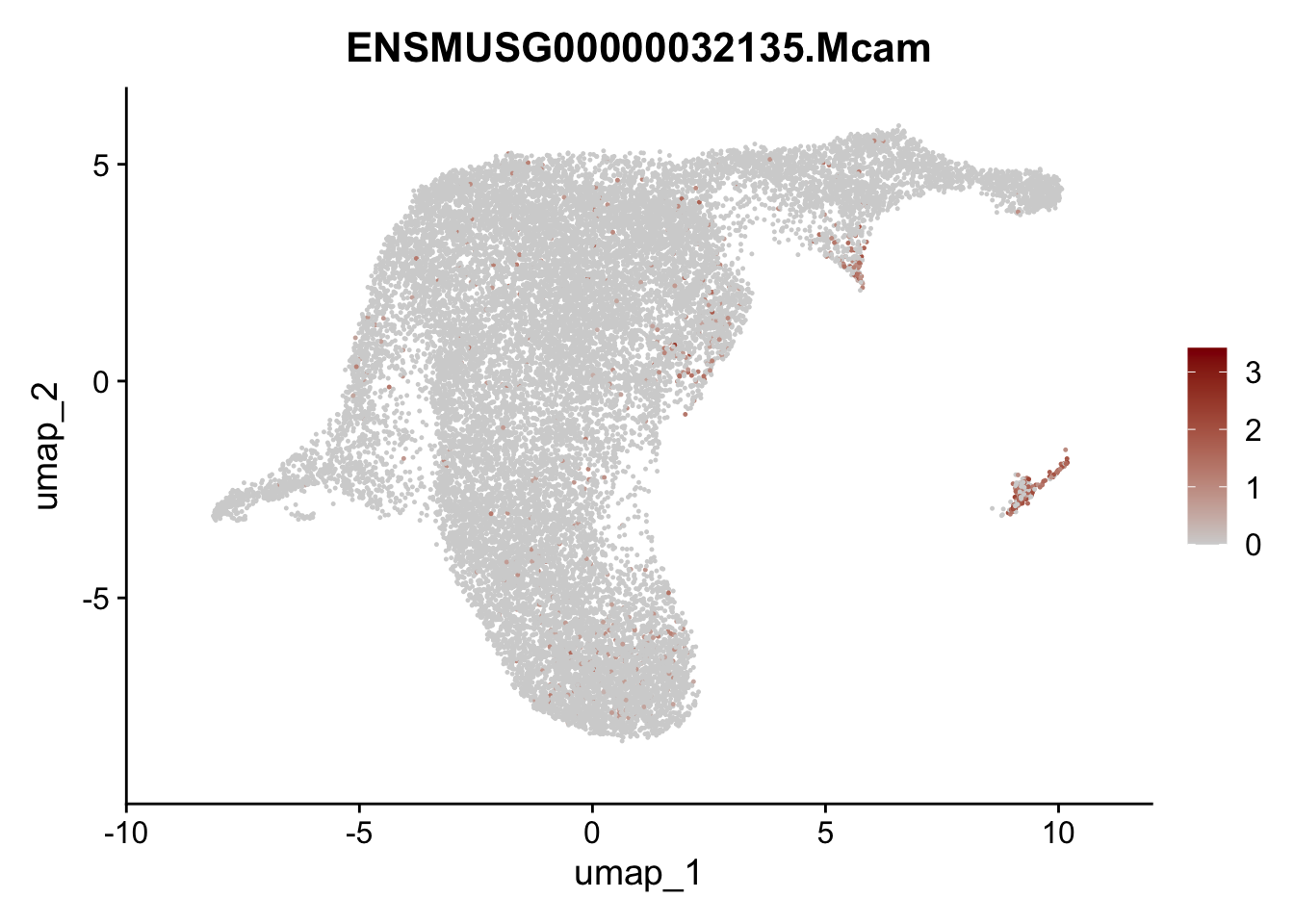
cluster characterization
heatmap function
avgHeatmap <- function(seurat, selGenes, colVecIdent, colVecCond=NULL,
ordVec=NULL, gapVecR=NULL, gapVecC=NULL,cc=FALSE,
cr=FALSE, condCol=FALSE){
selGenes <- selGenes$gene
## assay data
clusterAssigned <- as.data.frame(Idents(seurat)) %>%
dplyr::mutate(cell=rownames(.))
colnames(clusterAssigned)[1] <- "ident"
seuratDat <- GetAssayData(seurat)
## genes of interest
genes <- data.frame(gene=rownames(seurat)) %>%
mutate(geneID=gsub("^.*\\.", "", gene)) %>% filter(geneID %in% selGenes)
## matrix with averaged cnts per ident
logNormExpres <- as.data.frame(t(as.matrix(
seuratDat[which(rownames(seuratDat) %in% genes$gene),])))
logNormExpres <- logNormExpres %>% dplyr::mutate(cell=rownames(.)) %>%
dplyr::left_join(.,clusterAssigned, by=c("cell")) %>%
dplyr::select(-cell) %>% dplyr::group_by(ident) %>%
dplyr::summarise_all(mean)
logNormExpresMa <- logNormExpres %>% dplyr::select(-ident) %>% as.matrix()
rownames(logNormExpresMa) <- logNormExpres$ident
logNormExpresMa <- t(logNormExpresMa)
rownames(logNormExpresMa) <- gsub("^.*?\\.","",rownames(logNormExpresMa))
## remove genes if they are all the same in all groups
ind <- apply(logNormExpresMa, 1, sd) == 0
logNormExpresMa <- logNormExpresMa[!ind,]
genes <- genes[!ind,]
## color columns according to cluster
annotation_col <- as.data.frame(gsub("(^.*?_)","",
colnames(logNormExpresMa)))%>%
dplyr::mutate(celltype=gsub("(_.*$)","",colnames(logNormExpresMa)))
colnames(annotation_col)[1] <- "col1"
annotation_col <- annotation_col %>%
dplyr::mutate(cond = gsub(".*_","",col1)) %>%
dplyr::select(cond, celltype)
rownames(annotation_col) <- colnames(logNormExpresMa)
ann_colors = list(
cond = colVecCond,
celltype=colVecIdent)
if(is.null(ann_colors$cond)){
annotation_col$cond <- NULL
}
## adjust order
logNormExpresMa <- logNormExpresMa[selGenes,]
if(is.null(ordVec)){
ordVec <- levels(seurat)
}
logNormExpresMa <- logNormExpresMa[,ordVec]
## scaled row-wise
pheatmap(logNormExpresMa, scale="row" ,treeheight_row = 0, cluster_rows = cr,
cluster_cols = cc,
color = colorRampPalette(c("#2166AC", "#F7F7F7", "#B2182B"))(50),
annotation_col = annotation_col, cellwidth=15, cellheight=10,
annotation_colors = ann_colors, gaps_row = gapVecR, gaps_col = gapVecC)
}heatmap
seuratA.int <- JoinLayers(seuratA.int)
Idents(seuratA.int) <- seuratA.int$intCluster
seuratAint_markers <- FindAllMarkers(seuratA.int, only.pos = T, logfc.threshold = 0.25)
### plot DE genes top 10 avg logFC
markerAll <- seuratAint_markers %>% group_by(cluster) %>%
mutate(geneID = gene) %>% top_n(10, avg_log2FC) %>%
mutate(gene=gsub(".*\\.", "", geneID)) %>%
filter(nchar(gene)>1)
grpCnt <- markerAll %>% group_by(cluster) %>% summarise(cnt=n())
gapR <- data.frame(cluster=unique(markerAll$cluster)) %>%
left_join(.,grpCnt, by="cluster") %>% mutate(cumSum=cumsum(cnt))
ordVec <- levels(seuratA.int)
pOut <- avgHeatmap(seurat = seuratA.int, selGenes = markerAll,
colVecIdent = colPal,
ordVec=ordVec,
gapVecR=gapR$cumSum, gapVecC=NULL,cc=T,
cr=F, condCol=F)
session info
date()[1] "Fri Jul 11 16:38:30 2025"sessionInfo()R version 4.4.0 (2024-04-24)
Platform: x86_64-apple-darwin20
Running under: macOS Ventura 13.7.6
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.4-x86_64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.4-x86_64/Resources/lib/libRlapack.dylib; LAPACK version 3.12.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: Europe/Zurich
tzcode source: internal
attached base packages:
[1] grid stats4 stats graphics grDevices utils datasets methods base
other attached packages:
[1] future_1.58.0 NCmisc_1.2.0 ggupset_0.4.1
[4] gridExtra_2.3 DOSE_3.30.5 enrichplot_1.24.4
[7] msigdbr_24.1.0 org.Hs.eg.db_3.19.1 AnnotationDbi_1.66.0
[10] clusterProfiler_4.12.6 multtest_2.60.0 metap_1.12
[13] scater_1.32.1 scuttle_1.14.0 destiny_3.18.0
[16] circlize_0.4.16 muscat_1.18.0 here_1.0.1
[19] viridis_0.6.5 viridisLite_0.4.2 lubridate_1.9.4
[22] forcats_1.0.0 stringr_1.5.1 purrr_1.0.4
[25] readr_2.1.5 tidyr_1.3.1 tibble_3.2.1
[28] tidyverse_2.0.0 dplyr_1.1.4 SingleCellExperiment_1.26.0
[31] SummarizedExperiment_1.34.0 Biobase_2.64.0 GenomicRanges_1.56.2
[34] GenomeInfoDb_1.40.1 IRanges_2.38.1 S4Vectors_0.42.1
[37] BiocGenerics_0.50.0 MatrixGenerics_1.16.0 matrixStats_1.5.0
[40] pheatmap_1.0.13 ggpubr_0.6.0 ggplot2_3.5.2
[43] Seurat_5.3.0 SeuratObject_5.1.0 sp_2.2-0
[46] runSeurat3_0.1.0 ExploreSCdataSeurat3_0.1.0
loaded via a namespace (and not attached):
[1] igraph_2.1.4 ica_1.0-3 plotly_4.10.4
[4] Formula_1.2-5 zlibbioc_1.50.0 tidyselect_1.2.1
[7] bit_4.6.0 doParallel_1.0.17 clue_0.3-66
[10] lattice_0.22-7 rjson_0.2.23 blob_1.2.4
[13] S4Arrays_1.4.1 pbkrtest_0.5.4 parallel_4.4.0
[16] png_0.1-8 plotrix_3.8-4 cli_3.6.5
[19] ggplotify_0.1.2 goftest_1.2-3 VIM_6.2.2
[22] variancePartition_1.34.0 BiocNeighbors_1.22.0 shadowtext_0.1.4
[25] uwot_0.2.3 curl_6.2.3 tidytree_0.4.6
[28] mime_0.13 evaluate_1.0.3 ComplexHeatmap_2.20.0
[31] stringi_1.8.7 backports_1.5.0 lmerTest_3.1-3
[34] qqconf_1.3.2 httpuv_1.6.16 magrittr_2.0.3
[37] rappdirs_0.3.3 splines_4.4.0 ggraph_2.2.1
[40] sctransform_0.4.2 ggbeeswarm_0.7.2 DBI_1.2.3
[43] jquerylib_0.1.4 smoother_1.3 withr_3.0.2
[46] git2r_0.36.2 corpcor_1.6.10 reformulas_0.4.1
[49] class_7.3-23 rprojroot_2.0.4 lmtest_0.9-40
[52] tidygraph_1.3.1 colourpicker_1.3.0 htmlwidgets_1.6.4
[55] fs_1.6.6 ggrepel_0.9.6 labeling_0.4.3
[58] fANCOVA_0.6-1 SparseArray_1.4.8 DESeq2_1.44.0
[61] ranger_0.17.0 DEoptimR_1.1-3-1 reticulate_1.42.0
[64] hexbin_1.28.5 zoo_1.8-14 XVector_0.44.0
[67] knitr_1.50 ggplot.multistats_1.0.1 UCSC.utils_1.0.0
[70] RhpcBLASctl_0.23-42 timechange_0.3.0 foreach_1.5.2
[73] patchwork_1.3.0 caTools_1.18.3 ggtree_3.12.0
[76] data.table_1.17.4 R.oo_1.27.1 RSpectra_0.16-2
[79] irlba_2.3.5.1 fastDummies_1.7.5 gridGraphics_0.5-1
[82] lazyeval_0.2.2 yaml_2.3.10 survival_3.8-3
[85] scattermore_1.2 crayon_1.5.3 RcppAnnoy_0.0.22
[88] RColorBrewer_1.1-3 progressr_0.15.1 tweenr_2.0.3
[91] later_1.4.2 ggridges_0.5.6 codetools_0.2-20
[94] GlobalOptions_0.1.2 aod_1.3.3 KEGGREST_1.44.1
[97] Rtsne_0.17 shape_1.4.6.1 limma_3.60.6
[100] pkgconfig_2.0.3 TMB_1.9.17 spatstat.univar_3.1-3
[103] mathjaxr_1.8-0 EnvStats_3.1.0 aplot_0.2.5
[106] scatterplot3d_0.3-44 ape_5.8-1 spatstat.sparse_3.1-0
[109] xtable_1.8-4 car_3.1-3 plyr_1.8.9
[112] httr_1.4.7 rbibutils_2.3 tools_4.4.0
[115] globals_0.18.0 beeswarm_0.4.0 broom_1.0.8
[118] nlme_3.1-168 assertthat_0.2.1 lme4_1.1-37
[121] digest_0.6.37 numDeriv_2016.8-1.1 Matrix_1.7-3
[124] farver_2.1.2 tzdb_0.5.0 remaCor_0.0.18
[127] reshape2_1.4.4 yulab.utils_0.2.0 glue_1.8.0
[130] cachem_1.1.0 polyclip_1.10-7 generics_0.1.4
[133] Biostrings_2.72.1 mvtnorm_1.3-3 presto_1.0.0
[136] parallelly_1.45.0 mnormt_2.1.1 statmod_1.5.0
[139] RcppHNSW_0.6.0 ScaledMatrix_1.12.0 carData_3.0-5
[142] minqa_1.2.8 pbapply_1.7-2 httr2_1.1.2
[145] spam_2.11-1 gson_0.1.0 graphlayouts_1.2.2
[148] gtools_3.9.5 ggsignif_0.6.4 RcppEigen_0.3.4.0.2
[151] shiny_1.10.0 GenomeInfoDbData_1.2.12 glmmTMB_1.1.11
[154] R.utils_2.13.0 memoise_2.0.1 rmarkdown_2.29
[157] scales_1.4.0 R.methodsS3_1.8.2 RANN_2.6.2
[160] Cairo_1.6-2 spatstat.data_3.1-6 rstudioapi_0.17.1
[163] cluster_2.1.8.1 mutoss_0.1-13 spatstat.utils_3.1-4
[166] hms_1.1.3 fitdistrplus_1.2-2 cowplot_1.1.3
[169] colorspace_2.1-1 rlang_1.1.6 DelayedMatrixStats_1.26.0
[172] sparseMatrixStats_1.16.0 xts_0.14.1 dotCall64_1.2
[175] shinydashboard_0.7.3 ggforce_0.4.2 laeken_0.5.3
[178] mgcv_1.9-3 xfun_0.52 e1071_1.7-16
[181] TH.data_1.1-3 iterators_1.0.14 abind_1.4-8
[184] GOSemSim_2.30.2 treeio_1.28.0 bitops_1.0-9
[187] Rdpack_2.6.4 promises_1.3.3 scatterpie_0.2.4
[190] RSQLite_2.4.0 qvalue_2.36.0 sandwich_3.1-1
[193] fgsea_1.30.0 DelayedArray_0.30.1 proxy_0.4-27
[196] GO.db_3.19.1 compiler_4.4.0 prettyunits_1.2.0
[199] boot_1.3-31 beachmat_2.20.0 listenv_0.9.1
[202] Rcpp_1.0.14 edgeR_4.2.2 workflowr_1.7.1
[205] BiocSingular_1.20.0 tensor_1.5 MASS_7.3-65
[208] progress_1.2.3 BiocParallel_1.38.0 babelgene_22.9
[211] spatstat.random_3.4-1 R6_2.6.1 fastmap_1.2.0
[214] multcomp_1.4-28 fastmatch_1.1-6 rstatix_0.7.2
[217] vipor_0.4.7 TTR_0.24.4 ROCR_1.0-11
[220] TFisher_0.2.0 rsvd_1.0.5 vcd_1.4-13
[223] nnet_7.3-20 gtable_0.3.6 KernSmooth_2.23-26
[226] miniUI_0.1.2 deldir_2.0-4 htmltools_0.5.8.1
[229] ggthemes_5.1.0 bit64_4.6.0-1 spatstat.explore_3.4-3
[232] lifecycle_1.0.4 blme_1.0-6 nloptr_2.2.1
[235] sass_0.4.10 vctrs_0.6.5 robustbase_0.99-4-1
[238] spatstat.geom_3.4-1 sn_2.1.1 ggfun_0.1.8
[241] future.apply_1.11.3 bslib_0.9.0 pillar_1.10.2
[244] gplots_3.2.0 pcaMethods_1.96.0 locfit_1.5-9.12
[247] jsonlite_2.0.0 GetoptLong_1.0.5
sessionInfo()R version 4.4.0 (2024-04-24)
Platform: x86_64-apple-darwin20
Running under: macOS Ventura 13.7.6
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.4-x86_64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.4-x86_64/Resources/lib/libRlapack.dylib; LAPACK version 3.12.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: Europe/Zurich
tzcode source: internal
attached base packages:
[1] grid stats4 stats graphics grDevices utils datasets methods base
other attached packages:
[1] future_1.58.0 NCmisc_1.2.0 ggupset_0.4.1
[4] gridExtra_2.3 DOSE_3.30.5 enrichplot_1.24.4
[7] msigdbr_24.1.0 org.Hs.eg.db_3.19.1 AnnotationDbi_1.66.0
[10] clusterProfiler_4.12.6 multtest_2.60.0 metap_1.12
[13] scater_1.32.1 scuttle_1.14.0 destiny_3.18.0
[16] circlize_0.4.16 muscat_1.18.0 here_1.0.1
[19] viridis_0.6.5 viridisLite_0.4.2 lubridate_1.9.4
[22] forcats_1.0.0 stringr_1.5.1 purrr_1.0.4
[25] readr_2.1.5 tidyr_1.3.1 tibble_3.2.1
[28] tidyverse_2.0.0 dplyr_1.1.4 SingleCellExperiment_1.26.0
[31] SummarizedExperiment_1.34.0 Biobase_2.64.0 GenomicRanges_1.56.2
[34] GenomeInfoDb_1.40.1 IRanges_2.38.1 S4Vectors_0.42.1
[37] BiocGenerics_0.50.0 MatrixGenerics_1.16.0 matrixStats_1.5.0
[40] pheatmap_1.0.13 ggpubr_0.6.0 ggplot2_3.5.2
[43] Seurat_5.3.0 SeuratObject_5.1.0 sp_2.2-0
[46] runSeurat3_0.1.0 ExploreSCdataSeurat3_0.1.0
loaded via a namespace (and not attached):
[1] igraph_2.1.4 ica_1.0-3 plotly_4.10.4
[4] Formula_1.2-5 zlibbioc_1.50.0 tidyselect_1.2.1
[7] bit_4.6.0 doParallel_1.0.17 clue_0.3-66
[10] lattice_0.22-7 rjson_0.2.23 blob_1.2.4
[13] S4Arrays_1.4.1 pbkrtest_0.5.4 parallel_4.4.0
[16] png_0.1-8 plotrix_3.8-4 cli_3.6.5
[19] ggplotify_0.1.2 goftest_1.2-3 VIM_6.2.2
[22] variancePartition_1.34.0 BiocNeighbors_1.22.0 shadowtext_0.1.4
[25] uwot_0.2.3 curl_6.2.3 tidytree_0.4.6
[28] mime_0.13 evaluate_1.0.3 ComplexHeatmap_2.20.0
[31] stringi_1.8.7 backports_1.5.0 lmerTest_3.1-3
[34] qqconf_1.3.2 httpuv_1.6.16 magrittr_2.0.3
[37] rappdirs_0.3.3 splines_4.4.0 ggraph_2.2.1
[40] sctransform_0.4.2 ggbeeswarm_0.7.2 DBI_1.2.3
[43] jquerylib_0.1.4 smoother_1.3 withr_3.0.2
[46] git2r_0.36.2 corpcor_1.6.10 reformulas_0.4.1
[49] class_7.3-23 rprojroot_2.0.4 lmtest_0.9-40
[52] tidygraph_1.3.1 colourpicker_1.3.0 htmlwidgets_1.6.4
[55] fs_1.6.6 ggrepel_0.9.6 labeling_0.4.3
[58] fANCOVA_0.6-1 SparseArray_1.4.8 DESeq2_1.44.0
[61] ranger_0.17.0 DEoptimR_1.1-3-1 reticulate_1.42.0
[64] hexbin_1.28.5 zoo_1.8-14 XVector_0.44.0
[67] knitr_1.50 ggplot.multistats_1.0.1 UCSC.utils_1.0.0
[70] RhpcBLASctl_0.23-42 timechange_0.3.0 foreach_1.5.2
[73] patchwork_1.3.0 caTools_1.18.3 ggtree_3.12.0
[76] data.table_1.17.4 R.oo_1.27.1 RSpectra_0.16-2
[79] irlba_2.3.5.1 fastDummies_1.7.5 gridGraphics_0.5-1
[82] lazyeval_0.2.2 yaml_2.3.10 survival_3.8-3
[85] scattermore_1.2 crayon_1.5.3 RcppAnnoy_0.0.22
[88] RColorBrewer_1.1-3 progressr_0.15.1 tweenr_2.0.3
[91] later_1.4.2 ggridges_0.5.6 codetools_0.2-20
[94] GlobalOptions_0.1.2 aod_1.3.3 KEGGREST_1.44.1
[97] Rtsne_0.17 shape_1.4.6.1 limma_3.60.6
[100] pkgconfig_2.0.3 TMB_1.9.17 spatstat.univar_3.1-3
[103] mathjaxr_1.8-0 EnvStats_3.1.0 aplot_0.2.5
[106] scatterplot3d_0.3-44 ape_5.8-1 spatstat.sparse_3.1-0
[109] xtable_1.8-4 car_3.1-3 plyr_1.8.9
[112] httr_1.4.7 rbibutils_2.3 tools_4.4.0
[115] globals_0.18.0 beeswarm_0.4.0 broom_1.0.8
[118] nlme_3.1-168 assertthat_0.2.1 lme4_1.1-37
[121] digest_0.6.37 numDeriv_2016.8-1.1 Matrix_1.7-3
[124] farver_2.1.2 tzdb_0.5.0 remaCor_0.0.18
[127] reshape2_1.4.4 yulab.utils_0.2.0 glue_1.8.0
[130] cachem_1.1.0 polyclip_1.10-7 generics_0.1.4
[133] Biostrings_2.72.1 mvtnorm_1.3-3 presto_1.0.0
[136] parallelly_1.45.0 mnormt_2.1.1 statmod_1.5.0
[139] RcppHNSW_0.6.0 ScaledMatrix_1.12.0 carData_3.0-5
[142] minqa_1.2.8 pbapply_1.7-2 httr2_1.1.2
[145] spam_2.11-1 gson_0.1.0 graphlayouts_1.2.2
[148] gtools_3.9.5 ggsignif_0.6.4 RcppEigen_0.3.4.0.2
[151] shiny_1.10.0 GenomeInfoDbData_1.2.12 glmmTMB_1.1.11
[154] R.utils_2.13.0 memoise_2.0.1 rmarkdown_2.29
[157] scales_1.4.0 R.methodsS3_1.8.2 RANN_2.6.2
[160] Cairo_1.6-2 spatstat.data_3.1-6 rstudioapi_0.17.1
[163] cluster_2.1.8.1 mutoss_0.1-13 spatstat.utils_3.1-4
[166] hms_1.1.3 fitdistrplus_1.2-2 cowplot_1.1.3
[169] colorspace_2.1-1 rlang_1.1.6 DelayedMatrixStats_1.26.0
[172] sparseMatrixStats_1.16.0 xts_0.14.1 dotCall64_1.2
[175] shinydashboard_0.7.3 ggforce_0.4.2 laeken_0.5.3
[178] mgcv_1.9-3 xfun_0.52 e1071_1.7-16
[181] TH.data_1.1-3 iterators_1.0.14 abind_1.4-8
[184] GOSemSim_2.30.2 treeio_1.28.0 bitops_1.0-9
[187] Rdpack_2.6.4 promises_1.3.3 scatterpie_0.2.4
[190] RSQLite_2.4.0 qvalue_2.36.0 sandwich_3.1-1
[193] fgsea_1.30.0 DelayedArray_0.30.1 proxy_0.4-27
[196] GO.db_3.19.1 compiler_4.4.0 prettyunits_1.2.0
[199] boot_1.3-31 beachmat_2.20.0 listenv_0.9.1
[202] Rcpp_1.0.14 edgeR_4.2.2 workflowr_1.7.1
[205] BiocSingular_1.20.0 tensor_1.5 MASS_7.3-65
[208] progress_1.2.3 BiocParallel_1.38.0 babelgene_22.9
[211] spatstat.random_3.4-1 R6_2.6.1 fastmap_1.2.0
[214] multcomp_1.4-28 fastmatch_1.1-6 rstatix_0.7.2
[217] vipor_0.4.7 TTR_0.24.4 ROCR_1.0-11
[220] TFisher_0.2.0 rsvd_1.0.5 vcd_1.4-13
[223] nnet_7.3-20 gtable_0.3.6 KernSmooth_2.23-26
[226] miniUI_0.1.2 deldir_2.0-4 htmltools_0.5.8.1
[229] ggthemes_5.1.0 bit64_4.6.0-1 spatstat.explore_3.4-3
[232] lifecycle_1.0.4 blme_1.0-6 nloptr_2.2.1
[235] sass_0.4.10 vctrs_0.6.5 robustbase_0.99-4-1
[238] spatstat.geom_3.4-1 sn_2.1.1 ggfun_0.1.8
[241] future.apply_1.11.3 bslib_0.9.0 pillar_1.10.2
[244] gplots_3.2.0 pcaMethods_1.96.0 locfit_1.5-9.12
[247] jsonlite_2.0.0 GetoptLong_1.0.5