Data Frames in R
Last updated: 2025-01-02
Checks: 7 0
Knit directory: R_tutorial/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20241223) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version f7832cd. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/Data_Frames.Rmd) and HTML
(docs/Data_Frames.html) files. If you’ve configured a
remote Git repository (see ?wflow_git_remote), click on the
hyperlinks in the table below to view the files as they were in that
past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | f7832cd | Ohm-Np | 2025-01-02 | wflow_publish("analysis/Data_Frames.Rmd") |
| html | d11d577 | Ohm-Np | 2025-01-01 | Build site. |
| Rmd | 1782ac4 | Ohm-Np | 2025-01-01 | wflow_publish("analysis/Data_Frames.Rmd") |
What is a Data Frame?
Data frames are a cornerstone of data manipulation and analysis in R, designed for handling tabular data efficiently. They are two-dimensional, rectangular structures where rows represent observations and columns represent variables. Each column is a vector of equal length, allowing for the storage of mixed data types such as numerical, character, or logical values. Analogous to spreadsheets or SQL tables, data frames are indispensable in tasks like statistical analysis, data visualization, and machine learning.
Key characteristics of data frames:
- Columns can contain different data types (e.g., numeric, character, factor).
- Rows represent individual observations or cases.
- Data frames can hold large datasets efficiently.
Creating Data Frames
We can create a data frame in R using the data.frame()
function from base R. Here’s an example:
# Creating a data frame
students <- data.frame(
Name = c("Amrit", "Ishwor", "Indra", "Rakshya"),
Age = c(27, 24, 21, 25),
Grade = c("A", "B", "A", "B"),
Major = c("Maths", "Science", "Maths", "Science")
)
# Display the data frame
print(students) Name Age Grade Major
1 Amrit 27 A Maths
2 Ishwor 24 B Science
3 Indra 21 A Maths
4 Rakshya 25 B ScienceAccessing and Modifying Data
Selecting Rows and Columns
We can access elements of a data frame using indexing, column names, or logical conditions.
# Access a specific column
students$Name[1] "Amrit" "Ishwor" "Indra" "Rakshya"# Access a specific row
students[1, ] Name Age Grade Major
1 Amrit 27 A Maths# Access a specific value
students[2, "Grade"][1] "B"Adding and Removing Columns
We can add a new column to a data frame dynamically or remove an existing column.
# Adding a new column
students$Passed <- c(TRUE, FALSE, TRUE, FALSE)
# Display the data frame
print(students) Name Age Grade Major Passed
1 Amrit 27 A Maths TRUE
2 Ishwor 24 B Science FALSE
3 Indra 21 A Maths TRUE
4 Rakshya 25 B Science FALSE# Removing a column
students$Grade <- NULL
# Display the data frame
print(students) Name Age Major Passed
1 Amrit 27 Maths TRUE
2 Ishwor 24 Science FALSE
3 Indra 21 Maths TRUE
4 Rakshya 25 Science FALSEData Frame Operations
Filtering and Subsetting
We can filter rows based on conditions using logical operators.
# Filter students with Age > 22
students[students$Age > 22, ] Name Age Major Passed
1 Amrit 27 Maths TRUE
2 Ishwor 24 Science FALSE
4 Rakshya 25 Science FALSESorting and Reordering
Sorting a data frame is straightforward with the order()
function.
# Sort by Age
students <- students[order(students$Age), ]
# Display the data frame
print(students) Name Age Major Passed
3 Indra 21 Maths TRUE
2 Ishwor 24 Science FALSE
4 Rakshya 25 Science FALSE
1 Amrit 27 Maths TRUESummary and Statistical Functions
R provides built-in functions to summarize and analyze data frames.
# Summary statistics
summary(students) Name Age Major Passed
Length:4 Min. :21.00 Length:4 Mode :logical
Class :character 1st Qu.:23.25 Class :character FALSE:2
Mode :character Median :24.50 Mode :character TRUE :2
Mean :24.25
3rd Qu.:25.50
Max. :27.00 # Number of rows and columns
nrow(students)[1] 4ncol(students)[1] 4Aggregation and Grouping
The dplyr package functions enable grouping and
summarization.
# Load dplyr package
library(dplyr)
# grouping
students %>%
group_by(Major) %>%
summarise(Average_Age = mean(Age))# A tibble: 2 × 2
Major Average_Age
<chr> <dbl>
1 Maths 24
2 Science 24.5So far, this chapter has introduced you to the versatility and power of data frames in R, laying the foundation for data manipulation and analysis. Now, we will delve into data frames specifically designed for geospatial data.
Geospatial Data Frames
A Geospatial Data Frame (GDF) extends a standard data frame by including a geometry column, which stores spatial data in formats such as points, lines, and polygons. This integration of spatial and attribute data makes geospatial data frames ideal for spatial analysis and mapping.
The sf package represents geospatial data frames as
sf objects, offering seamless compatibility with other R
packages for geospatial and statistical analysis.
Geometry is central to geospatial data frames, defining the spatial features associated with each observation. Each row corresponds to a spatial feature with related attributes, and the geometry column typically stores spatial data in Well-Known Text (WKT) or binary format.
Creating GDF
You can create a geospatial data frame using the st_as_sf() function from the sf package. Here’s an example:
library(sf)
# Create a data frame with spatial data
data <- data.frame(
Name = c("Point A", "Point B", "Point C"),
Latitude = c(27.7, 28.2, 26.9),
Longitude = c(85.3, 84.9, 83.8)
)
# Convert to an sf object
geo_data <- st_as_sf(
data,
coords = c("Longitude", "Latitude"),
crs = 4326 # WGS 84 Coordinate Reference System
)
print(geo_data)Simple feature collection with 3 features and 1 field
Geometry type: POINT
Dimension: XY
Bounding box: xmin: 83.8 ymin: 26.9 xmax: 85.3 ymax: 28.2
Geodetic CRS: WGS 84
Name geometry
1 Point A POINT (85.3 27.7)
2 Point B POINT (84.9 28.2)
3 Point C POINT (83.8 26.9)Manipulating GDF
Spatial Queries
Geospatial data frames support operations like spatial filtering and proximity analysis.
# Define a bounding box and convert it to an sfc object
bbox <- st_bbox(c(xmin = 84, ymin = 27, xmax = 86, ymax = 29),
crs = st_crs(geo_data))
bbox_polygon <-
st_as_sfc(bbox)
# Filter points within a specific bounding box
filtered_data <-
geo_data[st_within(geo_data,
bbox_polygon,
sparse = FALSE),
]
print(filtered_data)Simple feature collection with 2 features and 1 field
Geometry type: POINT
Dimension: XY
Bounding box: xmin: 84.9 ymin: 27.7 xmax: 85.3 ymax: 28.2
Geodetic CRS: WGS 84
Name geometry
1 Point A POINT (85.3 27.7)
2 Point B POINT (84.9 28.2)Attribute-Based Operations
Just like standard data frames, we can manipulate attributes in geospatial data frames.
# Add a new column
geo_data$Elevation <-
c(1500, 2000, 1700)
print(geo_data)Simple feature collection with 3 features and 2 fields
Geometry type: POINT
Dimension: XY
Bounding box: xmin: 83.8 ymin: 26.9 xmax: 85.3 ymax: 28.2
Geodetic CRS: WGS 84
Name geometry Elevation
1 Point A POINT (85.3 27.7) 1500
2 Point B POINT (84.9 28.2) 2000
3 Point C POINT (83.8 26.9) 1700Visualization of GDF
Geospatial data frames can be visualized using the
plot() function from the sf package or
integrated with advanced visualization libraries such as
ggplot2.
plot(geo_data["Name"],
main = "Geospatial Data Points")
For enhanced visualization, we can use geom_sf() in
ggplot2.
library(ggplot2)
ggplot(data = geo_data) +
geom_sf(aes(color = Name), size = 3) +
labs(title = "Geospatial Data Points") +
theme_minimal()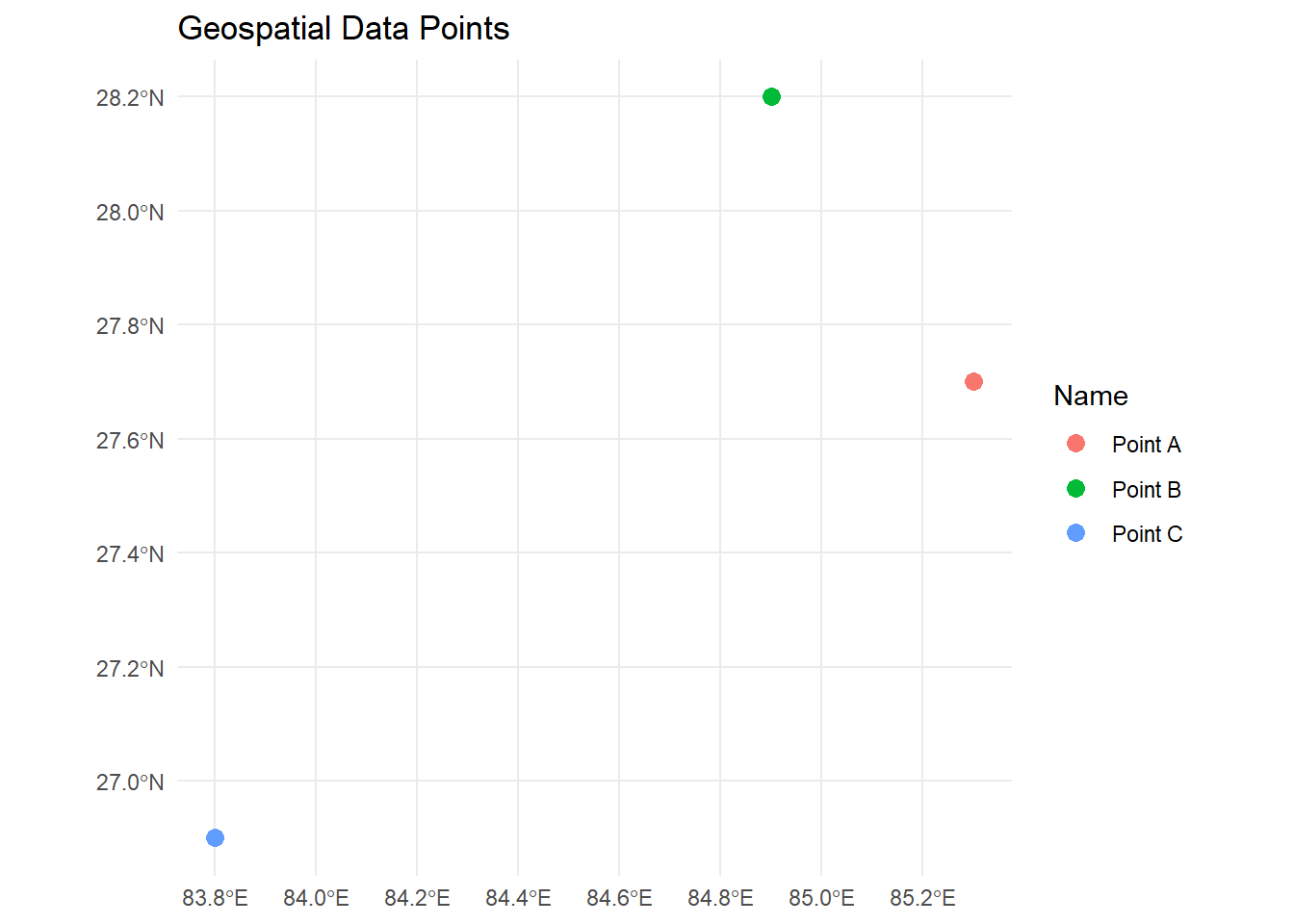
This approach allows you to set individual colors based on the Name attribute or other properties and provides more control over styling.
Geospatial data frames bridge the gap between tabular data and spatial analysis, making them indispensable for modern geospatial workflows. Their versatility, combined with R’s ecosystem of geospatial packages, provides an efficient and intuitive way to analyze and visualize spatial data.
sessionInfo()R version 4.4.0 (2024-04-24 ucrt)
Platform: x86_64-w64-mingw32/x64
Running under: Windows 11 x64 (build 22631)
Matrix products: default
locale:
[1] LC_COLLATE=English_Germany.utf8 LC_CTYPE=English_Germany.utf8
[3] LC_MONETARY=English_Germany.utf8 LC_NUMERIC=C
[5] LC_TIME=English_Germany.utf8
time zone: Europe/Berlin
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] ggplot2_3.5.1 sf_1.0-19 dplyr_1.1.4 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] s2_1.1.7 sass_0.4.9 utf8_1.2.4 generics_0.1.3
[5] class_7.3-22 KernSmooth_2.23-22 stringi_1.8.4 digest_0.6.36
[9] magrittr_2.0.3 evaluate_0.24.0 grid_4.4.0 fastmap_1.2.0
[13] rprojroot_2.0.4 jsonlite_1.8.8 processx_3.8.4 whisker_0.4.1
[17] e1071_1.7-16 DBI_1.2.3 ps_1.8.1 promises_1.3.0
[21] httr_1.4.7 fansi_1.0.6 scales_1.3.0 jquerylib_0.1.4
[25] cli_3.6.3 rlang_1.1.4 units_0.8-5 munsell_0.5.1
[29] withr_3.0.2 cachem_1.1.0 yaml_2.3.10 tools_4.4.0
[33] colorspace_2.1-1 httpuv_1.6.15 vctrs_0.6.5 R6_2.5.1
[37] proxy_0.4-27 lifecycle_1.0.4 classInt_0.4-10 git2r_0.33.0
[41] stringr_1.5.1 fs_1.6.4 pkgconfig_2.0.3 callr_3.7.6
[45] gtable_0.3.5 pillar_1.9.0 bslib_0.8.0 later_1.3.2
[49] glue_1.7.0 Rcpp_1.0.13 highr_0.11 xfun_0.47
[53] tibble_3.2.1 tidyselect_1.2.1 rstudioapi_0.16.0 knitr_1.48
[57] farver_2.1.2 htmltools_0.5.8.1 rmarkdown_2.28 wk_0.9.4
[61] compiler_4.4.0 getPass_0.2-4