Ovarian tissue data
George Howitt
2023-05-16
Last updated: 2023-07-05
Checks: 7 0
Knit directory: hashtag-demux-paper/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20230522) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 9e9164b. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/figure/
Ignored: data/BAL_data/.DS_Store
Untracked files:
Untracked: .ipynb_checkpoints/
Untracked: Dear editors.docx
Untracked: NAR.cls
Untracked: analysis/Fscore_MCC_comparison.Rmd
Untracked: analysis/cell_line_analysis.Rmd
Untracked: analysis/gmm_demux.Rmd
Untracked: analysis/hashsolo_prep.Rmd
Untracked: analysis/run_GMM_demux_BAL.sh
Untracked: cover_letter.docx
Untracked: data/BAL_data/batch1_all_methods.SEU.rds
Untracked: data/BAL_data/batch1_c1_donors_original.csv
Untracked: data/BAL_data/batch1_c1_hto_counts_original.csv
Untracked: data/BAL_data/batch1_c1_relabelled.SEU.rds
Untracked: data/BAL_data/batch1_c2_donors_original.csv
Untracked: data/BAL_data/batch1_c2_hto_counts_original.csv
Untracked: data/BAL_data/batch1_c2_relabelled.SEU.rds
Untracked: data/BAL_data/batch1_relabelled.SEU.rds
Untracked: data/BAL_data/batch2_all_methods.SEU.rds
Untracked: data/BAL_data/batch2_c1_donors_original.csv
Untracked: data/BAL_data/batch2_c1_hto_counts_original.csv
Untracked: data/BAL_data/batch2_c1_relabelled.SEU.rds
Untracked: data/BAL_data/batch2_c2_donors_original.csv
Untracked: data/BAL_data/batch2_c2_hto_counts_original.csv
Untracked: data/BAL_data/batch2_c2_relabelled.SEU.rds
Untracked: data/BAL_data/batch2_relabelled.SEU.rds
Untracked: data/BAL_data/batch3_all_methods.SEU.rds
Untracked: data/BAL_data/batch3_c1_donors_original.csv
Untracked: data/BAL_data/batch3_c1_hto_counts_original.csv
Untracked: data/BAL_data/batch3_c1_relabelled.SEU.rds
Untracked: data/BAL_data/batch3_c2_donors_original.csv
Untracked: data/BAL_data/batch3_c2_hto_counts_original.csv
Untracked: data/BAL_data/batch3_c2_relabelled.SEU.rds
Untracked: data/BAL_data/batch3_relabelled.SEU.rds
Untracked: data/adata/batch1_c1_hashsolo.csv
Untracked: data/adata/batch1_c1_hs_n10_d10.csv
Untracked: data/adata/batch1_c1_hs_n10_d20.csv
Untracked: data/adata/batch1_c1_hs_n10_d30.csv
Untracked: data/adata/batch1_c1_hs_n1_d10.csv
Untracked: data/adata/batch1_c1_hs_n1_d20.csv
Untracked: data/adata/batch1_c1_hs_n1_d30.csv
Untracked: data/adata/batch1_c1_hs_n5_d10.csv
Untracked: data/adata/batch1_c1_hs_n5_d30.csv
Untracked: data/adata/batch1_c2_hashsolo.csv
Untracked: data/adata/batch1_c2_hs_n10_d10.csv
Untracked: data/adata/batch1_c2_hs_n10_d20.csv
Untracked: data/adata/batch1_c2_hs_n10_d30.csv
Untracked: data/adata/batch1_c2_hs_n1_d10.csv
Untracked: data/adata/batch1_c2_hs_n1_d20.csv
Untracked: data/adata/batch1_c2_hs_n1_d30.csv
Untracked: data/adata/batch1_c2_hs_n5_d10.csv
Untracked: data/adata/batch1_c2_hs_n5_d30.csv
Untracked: data/adata/batch2_c1_hashsolo.csv
Untracked: data/adata/batch2_c1_hs_n10_d10.csv
Untracked: data/adata/batch2_c1_hs_n10_d20.csv
Untracked: data/adata/batch2_c1_hs_n10_d30.csv
Untracked: data/adata/batch2_c1_hs_n1_d10.csv
Untracked: data/adata/batch2_c1_hs_n1_d20.csv
Untracked: data/adata/batch2_c1_hs_n1_d30.csv
Untracked: data/adata/batch2_c1_hs_n5_d10.csv
Untracked: data/adata/batch2_c1_hs_n5_d30.csv
Untracked: data/adata/batch2_c2_hashsolo.csv
Untracked: data/adata/batch2_c2_hs_n10_d10.csv
Untracked: data/adata/batch2_c2_hs_n10_d20.csv
Untracked: data/adata/batch2_c2_hs_n10_d30.csv
Untracked: data/adata/batch2_c2_hs_n1_d10.csv
Untracked: data/adata/batch2_c2_hs_n1_d20.csv
Untracked: data/adata/batch2_c2_hs_n1_d30.csv
Untracked: data/adata/batch2_c2_hs_n5_d10.csv
Untracked: data/adata/batch2_c2_hs_n5_d30.csv
Untracked: data/adata/batch3_c1_hashsolo.csv
Untracked: data/adata/batch3_c1_hs_n10_d10.csv
Untracked: data/adata/batch3_c1_hs_n10_d20.csv
Untracked: data/adata/batch3_c1_hs_n10_d30.csv
Untracked: data/adata/batch3_c1_hs_n1_d10.csv
Untracked: data/adata/batch3_c1_hs_n1_d20.csv
Untracked: data/adata/batch3_c1_hs_n1_d30.csv
Untracked: data/adata/batch3_c1_hs_n5_d10.csv
Untracked: data/adata/batch3_c1_hs_n5_d30.csv
Untracked: data/adata/batch3_c2_hashsolo.csv
Untracked: data/adata/batch3_c2_hs_n10_d10.csv
Untracked: data/adata/batch3_c2_hs_n10_d20.csv
Untracked: data/adata/batch3_c2_hs_n10_d30.csv
Untracked: data/adata/batch3_c2_hs_n1_d10.csv
Untracked: data/adata/batch3_c2_hs_n1_d20.csv
Untracked: data/adata/batch3_c2_hs_n1_d30.csv
Untracked: data/adata/batch3_c2_hs_n5_d10.csv
Untracked: data/adata/batch3_c2_hs_n5_d30.csv
Untracked: data/adata/solid_tissue_batch1_hashsolo.csv
Untracked: data/adata/solid_tissue_batch2_hashsolo.csv
Untracked: data/solid_tumor_data/
Untracked: figures/QC_plots_new.png
Untracked: figures/Users/
Untracked: filter_wrong_empties.Rmd
Untracked: iscb_long_abstract.docx
Untracked: iscb_long_abstract.pdf
Untracked: oup-authoring-template/
Untracked: output/mean_fscore_mcc.xlsx
Untracked: paper_latex/
Unstaged changes:
Modified: analysis/index.Rmd
Modified: data/.DS_Store
Deleted: data/GMM-Demux/SSD_mtx/barcodes.tsv.gz
Deleted: data/GMM-Demux/SSD_mtx/features.tsv.gz
Deleted: data/GMM-Demux/SSD_mtx/matrix.mtx.gz
Deleted: data/GMM-Demux/batch1_c1_hto_counts_transpose.csv
Deleted: data/GMM-Demux/batch1_c2_hto_counts_transpose.csv
Deleted: data/GMM-Demux/batch2_c1_hto_counts_transpose.csv
Deleted: data/GMM-Demux/batch2_c2_hto_counts_transpose.csv
Deleted: data/GMM-Demux/batch3_c1_hto_counts_transpose.csv
Deleted: data/GMM-Demux/batch3_c2_hto_counts_transpose.csv
Deleted: data/GMM-Demux/gmm_out_LMO_c1/full_report/GMM_full.config
Deleted: data/GMM-Demux/gmm_out_LMO_c1/full_report/GMM_full.csv
Deleted: data/GMM-Demux/gmm_out_LMO_c1/simplified_report/GMM_simplified.config
Deleted: data/GMM-Demux/gmm_out_LMO_c1/simplified_report/GMM_simplified.csv
Deleted: data/GMM-Demux/gmm_out_LMO_c2/full_report/GMM_full.config
Deleted: data/GMM-Demux/gmm_out_LMO_c2/full_report/GMM_full.csv
Deleted: data/GMM-Demux/gmm_out_LMO_c2/simplified_report/GMM_simplified.config
Deleted: data/GMM-Demux/gmm_out_LMO_c2/simplified_report/GMM_simplified.csv
Deleted: data/GMM-Demux/gmm_out_LMO_c3/full_report/GMM_full.config
Deleted: data/GMM-Demux/gmm_out_LMO_c3/full_report/GMM_full.csv
Deleted: data/GMM-Demux/gmm_out_LMO_c3/simplified_report/GMM_simplified.config
Deleted: data/GMM-Demux/gmm_out_LMO_c3/simplified_report/GMM_simplified.csv
Deleted: data/GMM-Demux/gmm_out_batch1_c1/full_report/GMM_full.config
Deleted: data/GMM-Demux/gmm_out_batch1_c1/full_report/GMM_full.csv
Deleted: data/GMM-Demux/gmm_out_batch1_c1/simplified_report/GMM_simplified.config
Deleted: data/GMM-Demux/gmm_out_batch1_c1/simplified_report/GMM_simplified.csv
Deleted: data/GMM-Demux/gmm_out_batch1_c2/full_report/GMM_full.config
Deleted: data/GMM-Demux/gmm_out_batch1_c2/full_report/GMM_full.csv
Deleted: data/GMM-Demux/gmm_out_batch1_c2/simplified_report/GMM_simplified.config
Deleted: data/GMM-Demux/gmm_out_batch1_c2/simplified_report/GMM_simplified.csv
Deleted: data/GMM-Demux/gmm_out_batch2_c1/full_report/GMM_full.config
Deleted: data/GMM-Demux/gmm_out_batch2_c1/full_report/GMM_full.csv
Deleted: data/GMM-Demux/gmm_out_batch2_c1/simplified_report/GMM_simplified.config
Deleted: data/GMM-Demux/gmm_out_batch2_c1/simplified_report/GMM_simplified.csv
Deleted: data/GMM-Demux/gmm_out_batch2_c2/full_report/GMM_full.config
Deleted: data/GMM-Demux/gmm_out_batch2_c2/full_report/GMM_full.csv
Deleted: data/GMM-Demux/gmm_out_batch2_c2/simplified_report/GMM_simplified.config
Deleted: data/GMM-Demux/gmm_out_batch2_c2/simplified_report/GMM_simplified.csv
Deleted: data/GMM-Demux/gmm_out_batch3_c1/full_report/GMM_full.config
Deleted: data/GMM-Demux/gmm_out_batch3_c1/full_report/GMM_full.csv
Deleted: data/GMM-Demux/gmm_out_batch3_c1/simplified_report/GMM_simplified.config
Deleted: data/GMM-Demux/gmm_out_batch3_c1/simplified_report/GMM_simplified.csv
Deleted: data/GMM-Demux/gmm_out_batch3_c2/full_report/GMM_full.config
Deleted: data/GMM-Demux/gmm_out_batch3_c2/full_report/GMM_full.csv
Deleted: data/GMM-Demux/gmm_out_batch3_c2/simplified_report/GMM_simplified.config
Deleted: data/GMM-Demux/gmm_out_batch3_c2/simplified_report/GMM_simplified.csv
Deleted: data/GMM-Demux/lmo_counts_capture1_transpose.csv
Deleted: data/GMM-Demux/lmo_counts_capture2_transpose.csv
Deleted: data/GMM-Demux/lmo_counts_capture3_transpose.csv
Deleted: data/GMM-Demux/run_GMM_demux_BAL.sh
Deleted: data/GMM-Demux/run_GMM_demux_LMO.sh
Modified: data/adata/batch1_HTOs.csv
Modified: data/adata/batch1_c1_barcodes.csv
Modified: data/adata/batch1_c1_counts.mtx
Modified: data/adata/batch1_c2_barcodes.csv
Modified: data/adata/batch1_c2_counts.mtx
Modified: data/adata/batch2_HTOs.csv
Modified: data/adata/batch2_c1_barcodes.csv
Modified: data/adata/batch2_c1_counts.mtx
Modified: data/adata/batch2_c2_barcodes.csv
Modified: data/adata/batch2_c2_counts.mtx
Modified: data/adata/batch3_HTOs.csv
Modified: data/adata/batch3_c1_barcodes.csv
Modified: data/adata/batch3_c1_counts.mtx
Modified: data/adata/batch3_c2_barcodes.csv
Modified: data/adata/batch3_c2_counts.mtx
Deleted: data/batch1_c1_donors.csv
Deleted: data/batch1_c1_hto_counts.csv
Deleted: data/batch1_c2_donors.csv
Deleted: data/batch1_c2_hto_counts.csv
Deleted: data/batch1_hto_counts.csv
Deleted: data/batch2_c1_donors.csv
Deleted: data/batch2_c1_hto_counts.csv
Deleted: data/batch2_c2_donors.csv
Deleted: data/batch2_c2_hto_counts.csv
Deleted: data/batch2_hto_counts.csv
Deleted: data/batch3_c1_donors.csv
Deleted: data/batch3_c1_hto_counts.csv
Deleted: data/batch3_c2_donors.csv
Deleted: data/batch3_c2_hto_counts.csv
Deleted: data/batch3_hto_counts.csv
Deleted: data/lmo_counts.csv
Deleted: data/lmo_counts_capture1.csv
Deleted: data/lmo_counts_capture2.csv
Deleted: data/lmo_counts_capture3.csv
Deleted: data/lmo_donors.csv
Deleted: data/lmo_donors_capture1.csv
Deleted: data/lmo_donors_capture2.csv
Deleted: data/lmo_donors_capture3.csv
Modified: figures/QC_plots.png
Modified: figures/category_fractions.png
Modified: hashsolo_calls.ipynb
Modified: notebook_for_paper.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/solid_tissue_analysis.Rmd)
and HTML (docs/solid_tissue_analysis.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 9e9164b | George Howitt | 2023-07-05 | wflow_publish("analysis/solid_tissue_analysis.Rmd") |
#Load libraries
suppressPackageStartupMessages({
library(here)
library(BiocStyle)
library(dplyr)
library(janitor)
library(ggplot2)
library(cowplot)
library(patchwork)
library(DropletUtils)
library(tidyverse)
library(scuttle)
library(scater)
library(Seurat)
library(pheatmap)
library(speckle)
library(dittoSeq)
library(cellhashR)
library(RColorBrewer)
library(demuxmix)
library(ComplexHeatmap)
library(tidyHeatmap)
library(viridis)
})Ovarian tissue data
This notebook contains the analysis code for all results and figures relating to the cell line data set in the paper “Benchmarking single-cell hashtag oligo demultiplexing methods”.
This data consists of eight samples where scRNA-seq was performed in two batches of four samples each. on ovarian tumours.
Data loading and reduction
Data is available here https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs002262.v2.p1&phv=436020&phd=&pha=&pht=10575&phvf=&phdf=&phaf=&phtf=&dssp=1&consent=&temp=1
hashtag_counts_b1 <- read.csv(here("data", "solid_tumor_data", "ovarian_tumour_b1.csv"),
check.names = FALSE, row.names = 1)
hashtag_counts_b2 <- read.csv(here("data", "solid_tumor_data", "ovarian_tumour_b2.csv"),
check.names = FALSE, row.names = 1)
donors_b1 <- read.csv(here("data", "solid_tumor_data", "ovarian_tumour_donors_b1.csv"),
row.names = 1)
donors_b2 <- read.csv(here("data", "solid_tumor_data", "ovarian_tumour_donors_b2.csv"),
row.names = 1)seu_b1 <- CreateSeuratObject(counts = hashtag_counts_b1, assay = "HTO")
seu_b2 <- CreateSeuratObject(counts = hashtag_counts_b2, assay = "HTO")seu_b1$Barcode <- colnames(seu_b1)
seu_b2$Barcode <- colnames(seu_b2)Add genetic donor information to Seurat objects
seu_b1$genetic_donor <- donors_b1$genetic_donors
seu_b2$genetic_donor <- donors_b2$genetic_donorsUseful lists
hashtag_list_b1 <- c("OT 01",
"OT 02",
"OT 03",
"OT 04")
donor_hashtag_list_b1 <- list("OT A" = "OT 01",
"OT B" = "OT 02",
"OT C" = "OT 03",
"OT D" = "OT 04",
"Doublet" = "Doublet",
"Negative" = "Negative")
hashtag_donor_list_b1 <- list("OT 01" = "OT A",
"OT 02" = "OT B",
"OT 03" = "OT C",
"OT 04" = "OT D",
"Doublet" = "Doublet",
"Negative" = "Negative")
hashtag_list_b2 <- c("OT 05",
"OT 06",
"OT 07",
"OT 08")
donor_hashtag_list_b2 <- list("OT E" = "OT 05",
"OT F" = "OT 06",
"OT G" = "OT 07",
"OT H" = "OT 08",
"Doublet" = "Doublet",
"Negative" = "Negative")
hashtag_donor_list_b2 <- list("OT 05" = "OT E",
"OT 06" = "OT F",
"OT 07" = "OT G",
"OT 08" = "OT H",
"Doublet" = "Doublet",
"Negative" = "Negative")DefaultAssay(seu_b1) <- "HTO"
seu_b1 <- NormalizeData(seu_b1, assay = "HTO", normalization.method = "CLR")Normalizing across featuresseu_b1 <- ScaleData(seu_b1, features = rownames(seu_b1),
verbose = FALSE)
seu_b1 <- RunPCA(seu_b1, features = rownames(seu_b1), approx = FALSE, verbose = FALSE)
seu_b1 <- RunTSNE(seu_b1, dims = 1:3, perplexity = 100, check_duplicates = FALSE, verbose = FALSE)
DefaultAssay(seu_b2) <- "HTO"
seu_b2 <- NormalizeData(seu_b2, assay = "HTO", normalization.method = "CLR")Normalizing across featuresseu_b2 <- ScaleData(seu_b2, features = rownames(seu_b2),
verbose = FALSE)
seu_b2 <- RunPCA(seu_b2, features = rownames(seu_b2), approx = FALSE, verbose = FALSE)
seu_b2 <- RunTSNE(seu_b2, dims = 1:3, perplexity = 100, check_duplicates = FALSE, verbose = FALSE)QC Plots
Density plots per barcode. In ideal conditions the density of the hashtag counts should appear bimodal, with a lower peak corresponding to the background and the higher peak corresponding to the signal.
df <- as.data.frame(t(seu_b1[["HTO"]]@counts))
df %>%
pivot_longer(cols = starts_with("OT")) %>%
mutate(logged = log(value + 1)) %>%
ggplot(aes(x = logged)) +
xlab("log(counts)") +
xlim(3,10) +
geom_density(adjust = 2) +
facet_wrap(~name, scales = "fixed", ncol = 2) -> p1
df <- as.data.frame(t(seu_b2[["HTO"]]@counts))
df %>%
pivot_longer(cols = starts_with("OT")) %>%
mutate(logged = log(value + 1)) %>%
ggplot(aes(x = logged)) +
xlab("log(counts)") +
xlim(3,10) +
geom_density(adjust = 2) +
facet_wrap(~name, scales = "fixed", ncol = 2) -> p2
p1 / p2Warning: Removed 188 rows containing non-finite values (`stat_density()`).Warning: Removed 14885 rows containing non-finite values (`stat_density()`).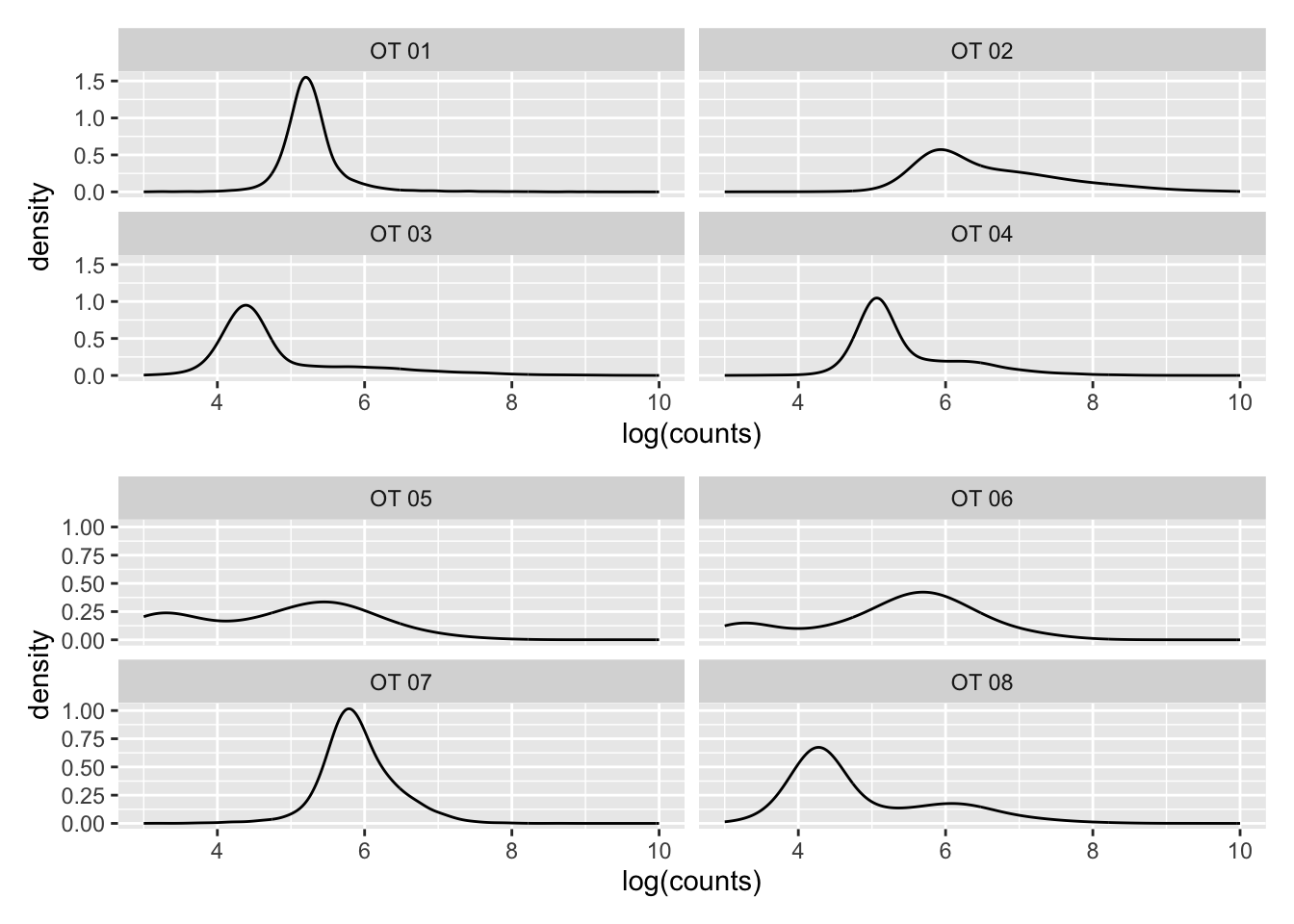 These data aren’t looking so hot. Hard to see a second peak in most
hashtags…
These data aren’t looking so hot. Hard to see a second peak in most
hashtags…
p3 <- DimPlot(seu_b1, group.by = "genetic_donor") +
ggtitle("Batch 1")
p4 <- DimPlot(seu_b2, group.by = "genetic_donor") +
ggtitle("Batch 2")
p3 / p4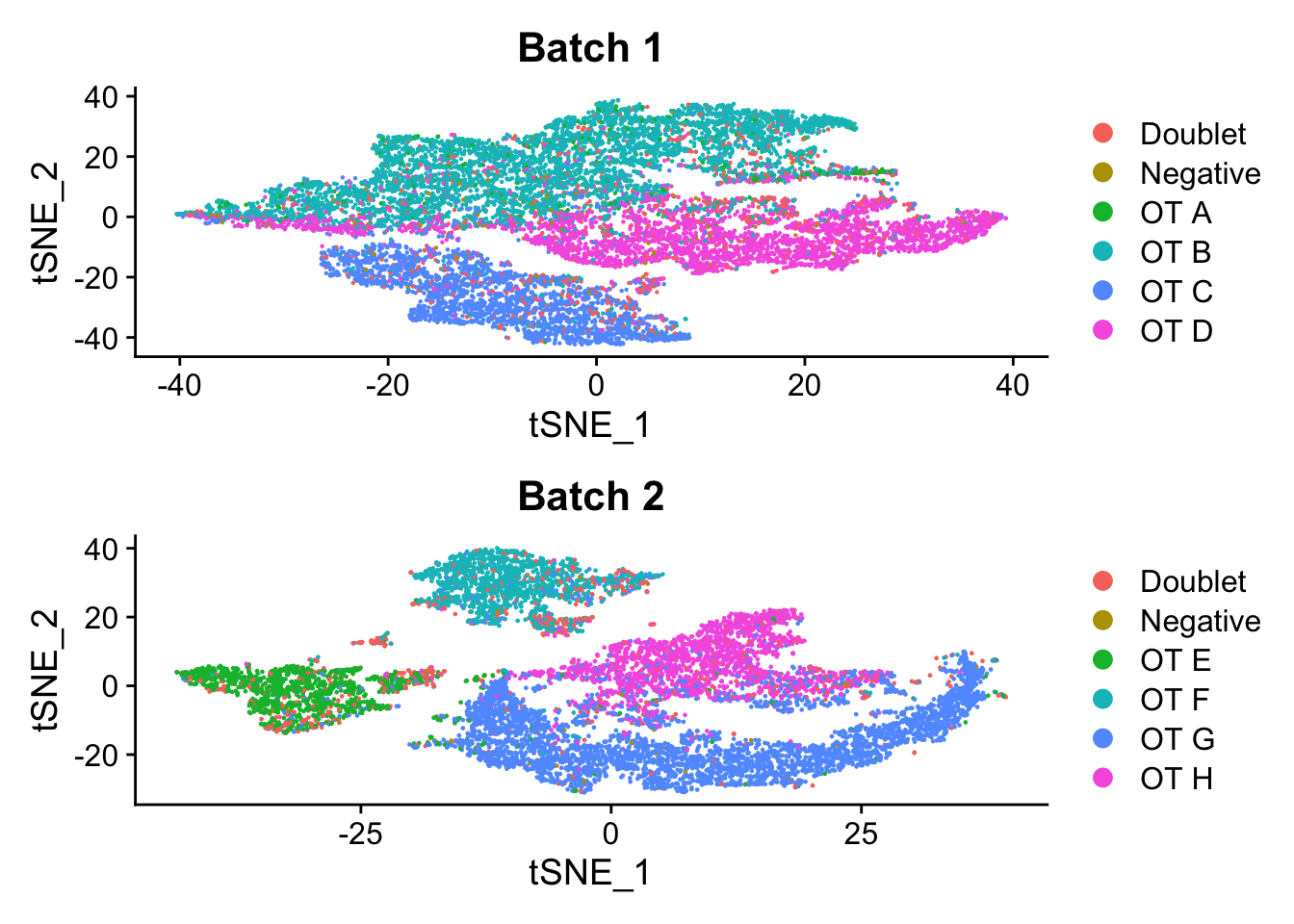
(((p1 / p2 ) | (p3 / p4)) + plot_annotation(tag_levels = 'a')) &
theme(plot.title = element_text(face = "plain", size = 10),
plot.tag = element_text(face = 'plain'))Warning: Removed 188 rows containing non-finite values (`stat_density()`).Warning: Removed 14885 rows containing non-finite values (`stat_density()`).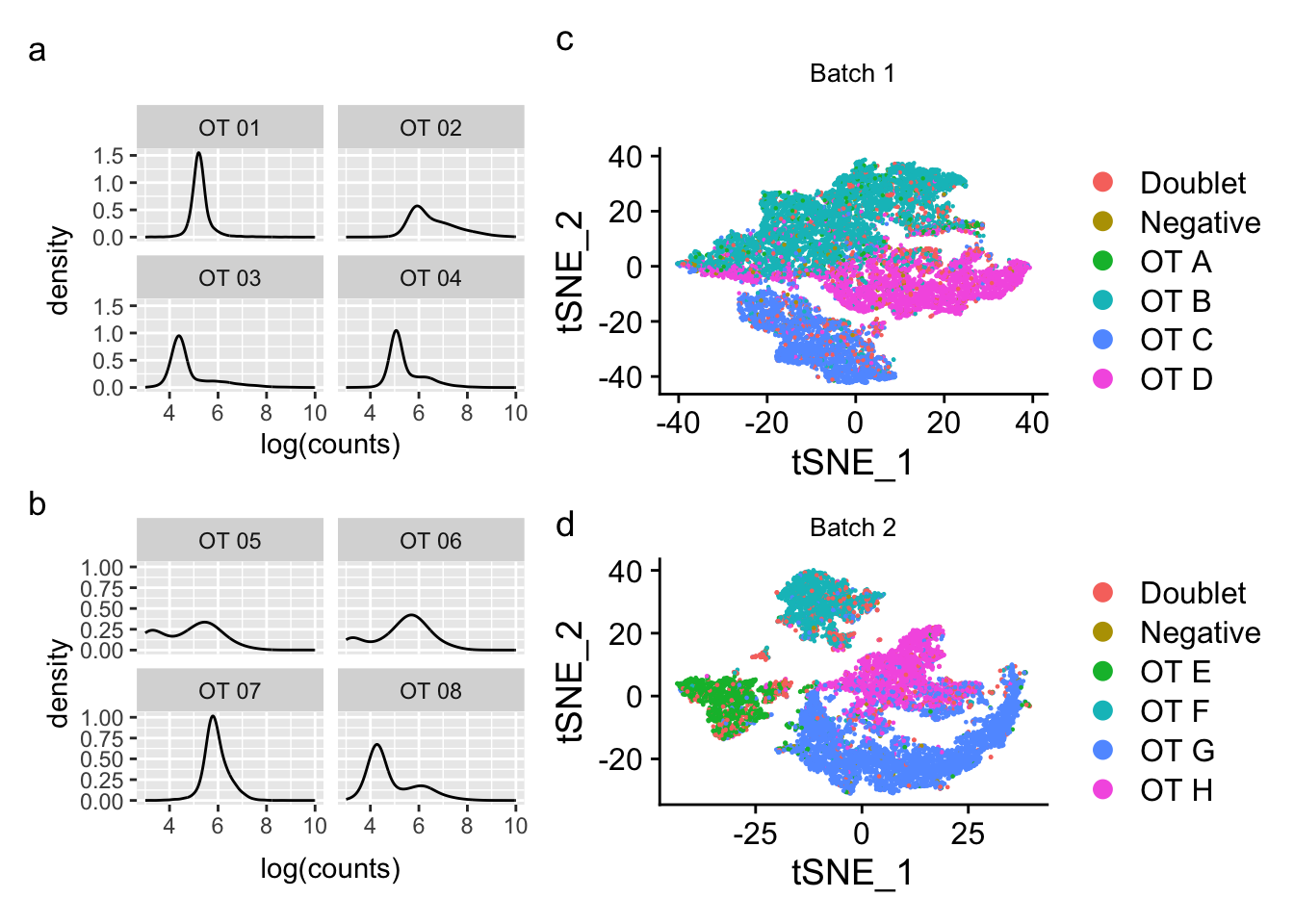
Demultiplexing
hashedDrops
This function creates a list of hashedDrops calls. Its defaults are the same as hashedDrops
create_hashedDrops_factor <- function(seurat_object, confident.min = 2,
doublet.nmads = 3, doublet.min = 2) {
hto_counts <- GetAssayData(seurat_object[["HTO"]], slot = "counts")
hash_stats <- DropletUtils::hashedDrops(hto_counts, confident.min = confident.min,
doublet.nmads = doublet.nmads, doublet.min = doublet.min)
hash_stats$Best <- rownames(seurat_object[["HTO"]])[hash_stats$Best]
hash_stats$Second <- rownames(seurat_object[["HTO"]])[hash_stats$Second]
HTO_assignments <- factor(case_when(
hash_stats$Confident == TRUE ~ hash_stats$Best,
hash_stats$Doublet == TRUE ~ "Doublet",
TRUE ~ "Negative"))
return(HTO_assignments)
}Making factors with best parameters
seu_b1$hashedDrops_calls <- create_hashedDrops_factor(seu_b1, confident.min = 0.5)
seu_b2$hashedDrops_calls <- create_hashedDrops_factor(seu_b2, confident.min = 0.5)Now with default parameters
seu_b1$hashedDrops_default_calls <- create_hashedDrops_factor(seu_b1)
seu_b2$hashedDrops_default_calls <- create_hashedDrops_factor(seu_b2)HashSolo
HashSolo is a scanpy program. Needs a bit of prep Write to anndata compatible files Counts
library(Matrix)
Attaching package: 'Matrix'The following objects are masked from 'package:tidyr':
expand, pack, unpackThe following object is masked from 'package:S4Vectors':
expandwriteMM(seu_b1@assays$HTO@counts, here("data", "solid_tumor_data", "adata", "b1_counts.mtx"))NULLwriteMM(seu_b2@assays$HTO@counts, here("data", "solid_tumor_data", "adata", "b2_counts.mtx"))NULLBarcodes
barcodes <- data.frame(colnames(seu_b1))
colnames(barcodes)<-'Barcode'
write.csv(barcodes, here("data", "solid_tumor_data", "adata", "b1_barcodes.csv"),
quote = FALSE,row.names = FALSE)
barcodes <- data.frame(colnames(seu_b2))
colnames(barcodes)<-'Barcode'
write.csv(barcodes, here("data", "solid_tumor_data", "adata", "b2_barcodes.csv"),
quote = FALSE,row.names = FALSE)Save hashtag names
HTOs_b1 <- data.frame(rownames(seu_b1))
colnames(HTOs_b1) <- 'HTO'
write.csv(HTOs_b1, here("data", "solid_tumor_data", "adata", "HTOs_b1.csv"),
quote = FALSE,row.names = FALSE)
HTOs_b2 <- data.frame(rownames(seu_b2))
colnames(HTOs_b2) <- 'HTO'
write.csv(HTOs_b2, here("data", "solid_tumor_data", "adata", "HTOs_b2.csv"),
quote = FALSE,row.names = FALSE)See hashsolo_calls.ipynb for how we get these assignments
seu_b1$hashsolo_calls <- read.csv(here("data", "solid_tumor_data", "adata", "b1_hashsolo.csv"))$Classification
seu_b2$hashsolo_calls <- read.csv(here("data", "solid_tumor_data", "adata", "b2_hashsolo.csv"))$ClassificationdeMULTIplex
seu_b1$deMULTIplex_calls <- MULTIseqDemux(seu_b1, autoThresh = TRUE)$MULTI_IDNo threshold found for OT 03...
No threshold found for OT 03...
No threshold found for OT 03...
No threshold found for OT 03...
No threshold found for OT 03...
No threshold found for OT 03...
No threshold found for OT 03...
No threshold found for OT 03...
No threshold found for OT 03...
No threshold found for OT 03...
No threshold found for OT 03...
No threshold found for OT 03...
No threshold found for OT 03...
No threshold found for OT 03...
No threshold found for OT 03...
No threshold found for OT 03...
No threshold found for OT 03...Iteration 1Using quantile 0.1No threshold found for OT 03...No threshold found for OT 04...
No threshold found for OT 04...
No threshold found for OT 04...
No threshold found for OT 04...
No threshold found for OT 04...
No threshold found for OT 04...
No threshold found for OT 04...
No threshold found for OT 04...
No threshold found for OT 04...
No threshold found for OT 04...
No threshold found for OT 04...
No threshold found for OT 04...
No threshold found for OT 04...
No threshold found for OT 04...
No threshold found for OT 04...
No threshold found for OT 04...
No threshold found for OT 04...Iteration 2Using quantile 0.1No threshold found for OT 04...Iteration 3Using quantile 0.1Iteration 4Using quantile 0.1Iteration 5Using quantile 0.3seu_b2$deMULTIplex_calls <- MULTIseqDemux(seu_b2, autoThresh = TRUE)$MULTI_IDIteration 1Using quantile 0.2Iteration 2Using quantile 0.2Iteration 3Using quantile 0.2HTODemux
HDmux <- HTODemux(seu_b1)Cutoff for OT 01 : 422 readsCutoff for OT 02 : 910 readsCutoff for OT 03 : 188 readsCutoff for OT 04 : 352 readsseu_b1$HTODemux_calls <- HDmux$hash.ID
HDmux <- HTODemux(seu_b2)Cutoff for OT 05 : 35 readsCutoff for OT 06 : 31 readsCutoff for OT 07 : 876 readsCutoff for OT 08 : 166 readsseu_b2$HTODemux_calls <- HDmux$hash.IDGMM-Demux
GMM-Demux is run on the command line and needs a function to read in the results and format them all properly.
create_gmm_demux_factor <- function(seu, GMM_path, hto_list) {
#Read in output, have to use the "full" report, not the simplified one.
calls <- read.csv(paste0(GMM_path, "/GMM_full.csv"), row.names = 1)
#Read in names of clusters
cluster_names <- read.table(paste0(GMM_path, "/GMM_full.config"), sep = ",")
names(cluster_names) <- c("Cluster_id", "assignment")
#Need to fix the formatting of the assignment names, for some reason there's a leading space.
cluster_names$assignment <- gsub(x = cluster_names$assignment, pattern = '^ ', replacement = '')
#Add cell barcodes
calls$Barcode <- rownames(calls)
calls <- merge(calls, cluster_names, by = "Cluster_id", sort = FALSE)
#Need to re-order after merge for some reason
calls <- calls[order(match(calls$Barcode, names(seu$Barcode))), ]
#Rename the negative cluster for consistency
calls$assignment[calls$assignment == "negative"] <- "Negative"
#Put all the multiplet states into one assignment category
calls$assignment[!calls$assignment %in% c("Negative", hto_list)] <- "Doublet"
return(as.factor(calls$assignment))
}Write the transpose of the counts matrix for GMM-Demux
write.csv(t(hashtag_counts_b1), here("data", "solid_tumor_data", "GMM-Demux", "hashtag_counts_b1_transpose.csv"))
write.csv(t(hashtag_counts_b2), here("data", "solid_tumor_data", "GMM-Demux", "hashtag_counts_b2_transpose.csv"))Run the script run_GMM_demux_solid_tumor.sh in command line.
Add to objects
seu_b1$GMMDemux_calls <- create_gmm_demux_factor(seu_b1, here("data", "solid_tumor_data", "GMM-Demux", "gmm_out_b1", "full_report"), hashtag_list_b1)
seu_b2$GMMDemux_calls <- create_gmm_demux_factor(seu_b2, here("data", "solid_tumor_data", "GMM-Demux", "gmm_out_b2", "full_report"), hashtag_list_b2) BFF
cellhashR_calls <- GenerateCellHashingCalls(barcodeMatrix = hashtag_counts_b1,
methods = c("bff_raw", "bff_cluster"),
doTSNE = FALSE, doHeatmap = FALSE)[1] "Converting input data.frame to a matrix"
[1] "Starting BFF"
[1] "rows dropped for low counts: 0 of 4"
[1] "Running BFF_raw"
[1] "Only one peak found, using max value as cutoff: OT 01"
[1] "Only one peak found, using max value as cutoff: OT 02"
[1] "Only one peak found, using max value as cutoff: OT 03"
[1] "Only one peak found, using max value as cutoff: OT 04"
[1] "Thresholds:"
[1] "OT 04: 182082.316992221"
[1] "OT 03: 112127.839767934"
[1] "OT 02: 214644.500496396"
[1] "OT 01: 31165.7553663721"
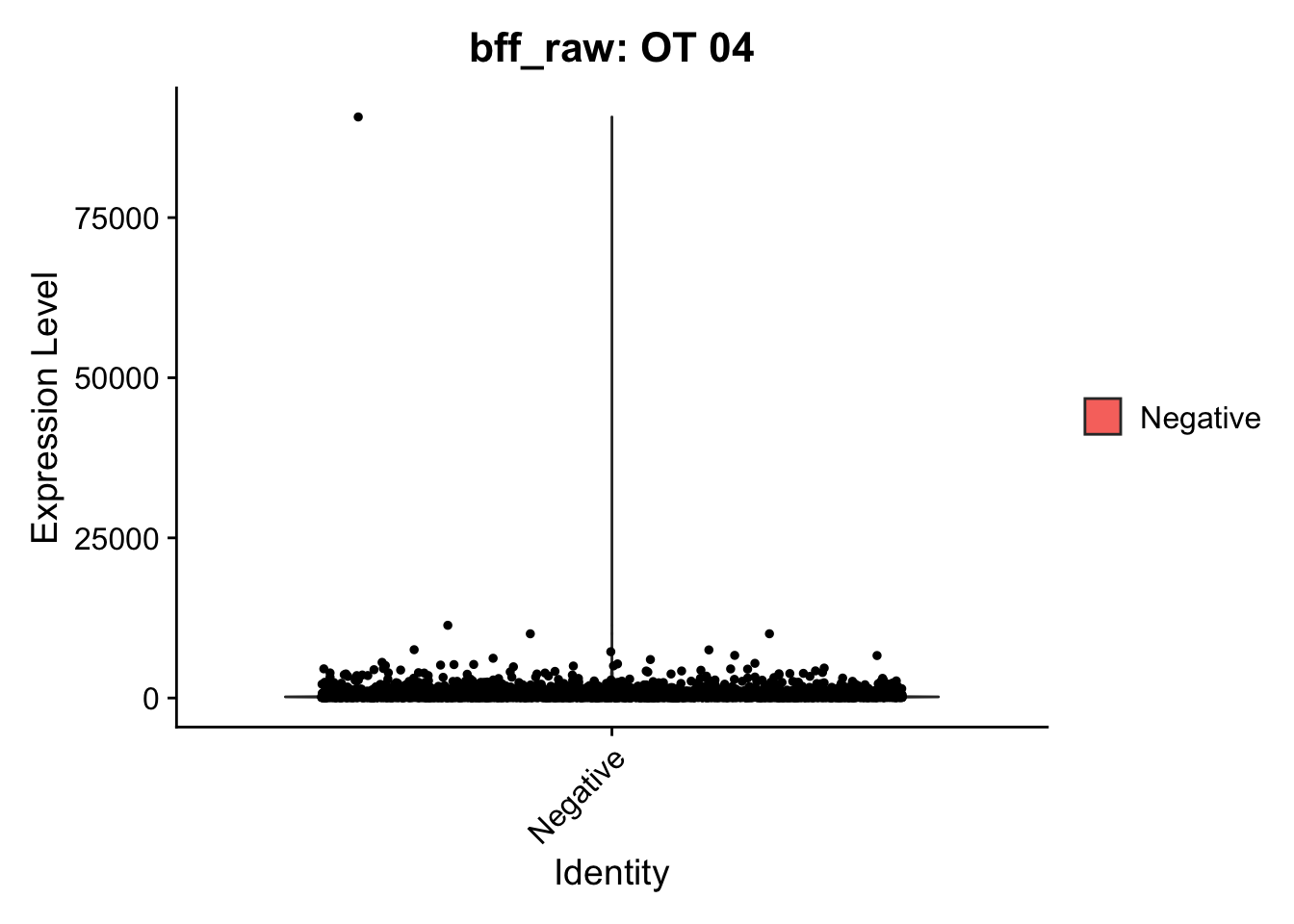
[1] "Starting BFF"
[1] "rows dropped for low counts: 0 of 4"
[1] "Running BFF_cluster"
[1] "Doublet threshold: 0.05"
[1] "Neg threshold: 0.05"
[1] "Min distance as fraction of distance between peaks: 0.1"
[1] "Only one peak found, using max value as cutoff: OT 01"
[1] "Only one peak found, using max value as cutoff: OT 02"
[1] "Only one peak found, using max value as cutoff: OT 03"
[1] "Only one peak found, using max value as cutoff: OT 04"
[1] "Thresholds:"
[1] "OT 04: 182082.316992221"
[1] "OT 03: 112127.839767934"
[1] "OT 02: 214644.500496396"
[1] "OT 01: 31165.7553663721"
[1] "Only one peak found, using max value as cutoff: OT 01"
[1] "Only one peak found, using max value as cutoff: OT 02"
[1] "Only one peak found, using max value as cutoff: OT 03"
[1] "Only one peak found, using max value as cutoff: OT 04"
[1] "Error running BFF"
[1] "Generating consensus calls"
[1] "adding missing method: bff_cluster"
[1] "Consensus calls will be generated using: bff_raw,bff_cluster"
[1] "Total concordant: 12510"
[1] "Total discordant: 0 (0%)"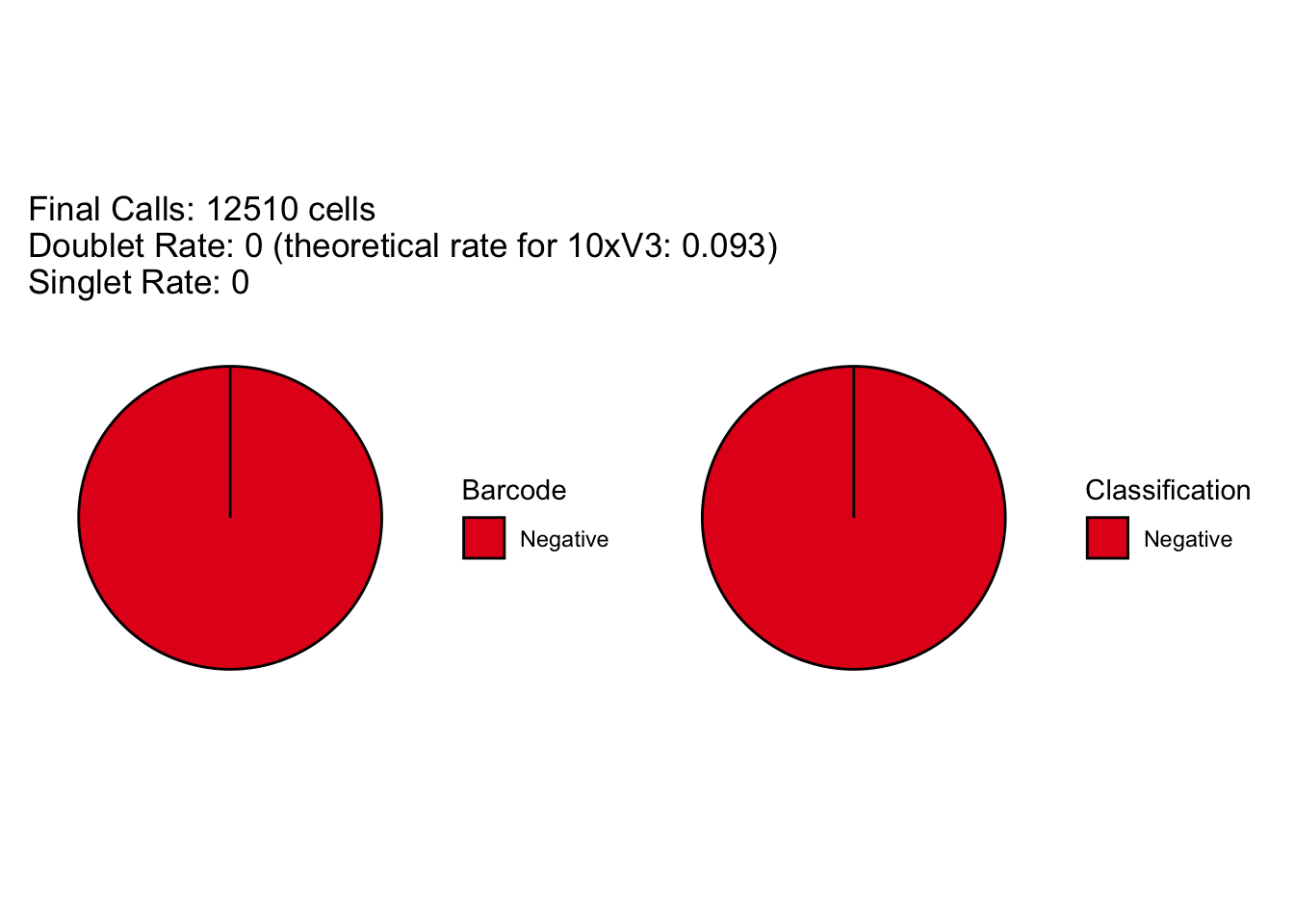
seu_b1$BFF_raw_calls <- cellhashR_calls$bff_raw
seu_b1$BFF_cluster_calls <- cellhashR_calls$bff_cluster
cellhashR_calls <- GenerateCellHashingCalls(barcodeMatrix = hashtag_counts_b2,
methods = c("bff_raw", "bff_cluster"),
doTSNE = FALSE, doHeatmap = FALSE)[1] "Converting input data.frame to a matrix"
[1] "Starting BFF"
[1] "rows dropped for low counts: 0 of 4"
[1] "Running BFF_raw"
[1] "Only one peak found, using max value as cutoff: OT 07"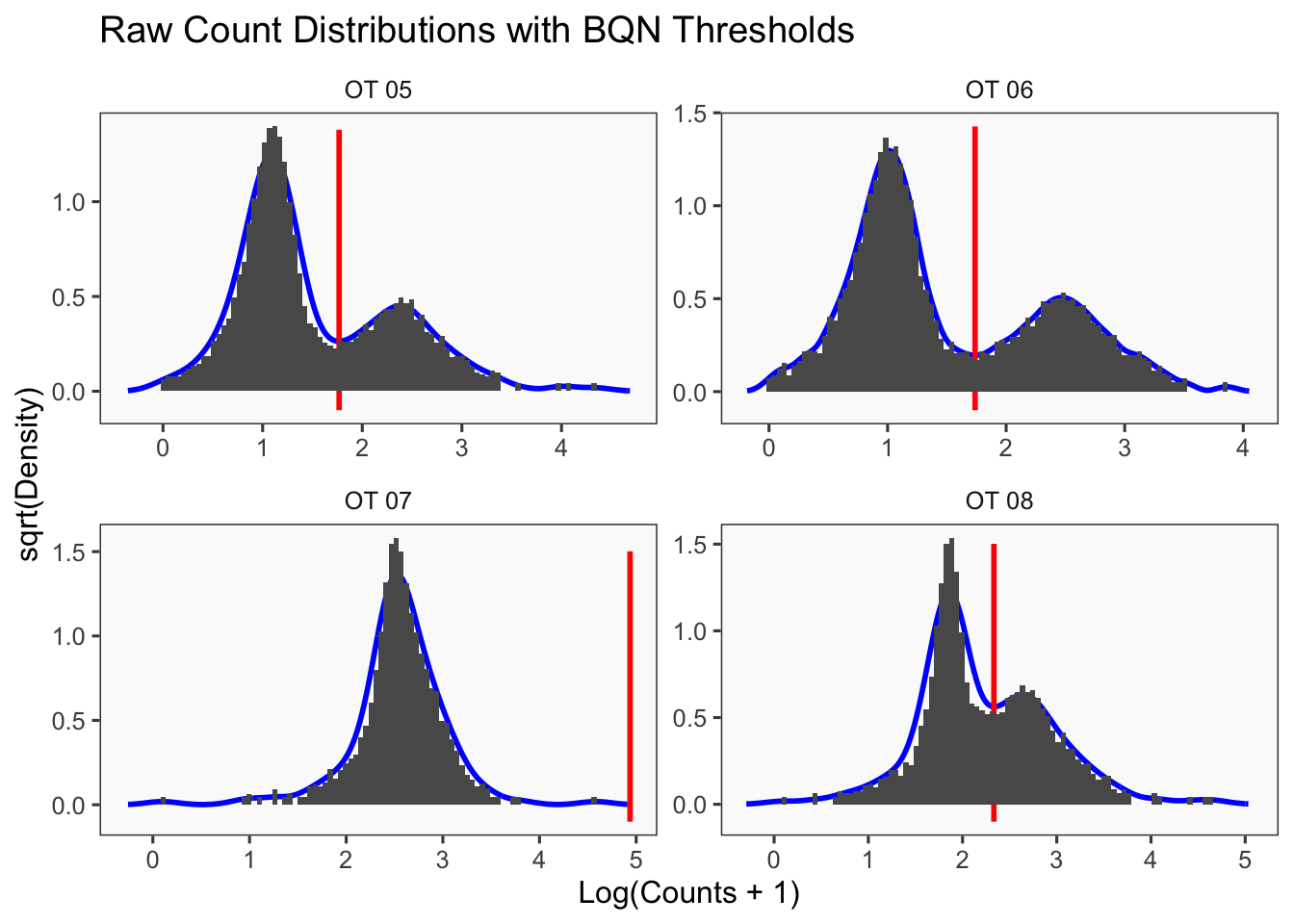
[1] "Thresholds:"
[1] "OT 08: 215.227799018146"
[1] "OT 07: 86433.9145009996"
[1] "OT 06: 54.6952290109095"
[1] "OT 05: 58.5737138510189"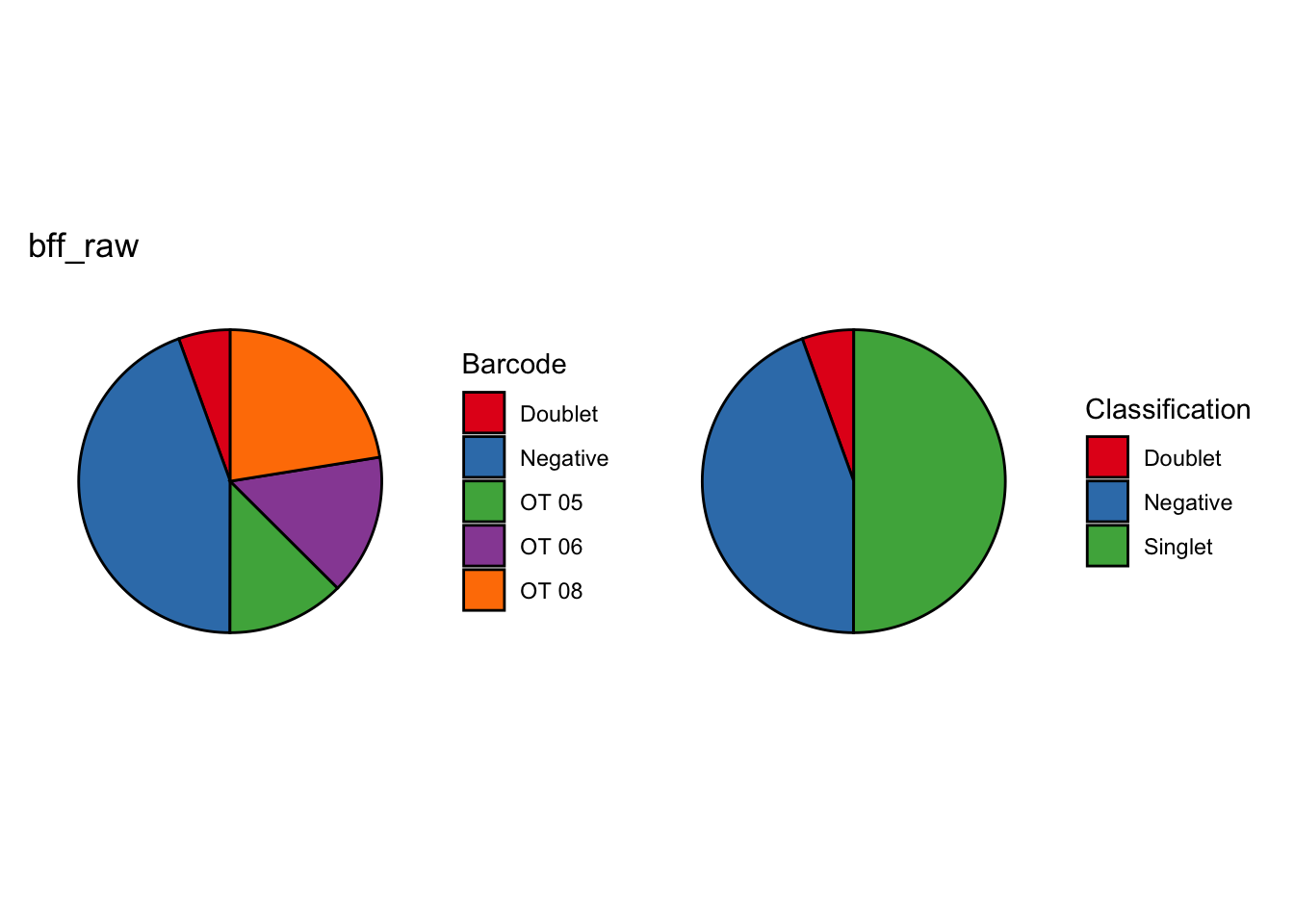
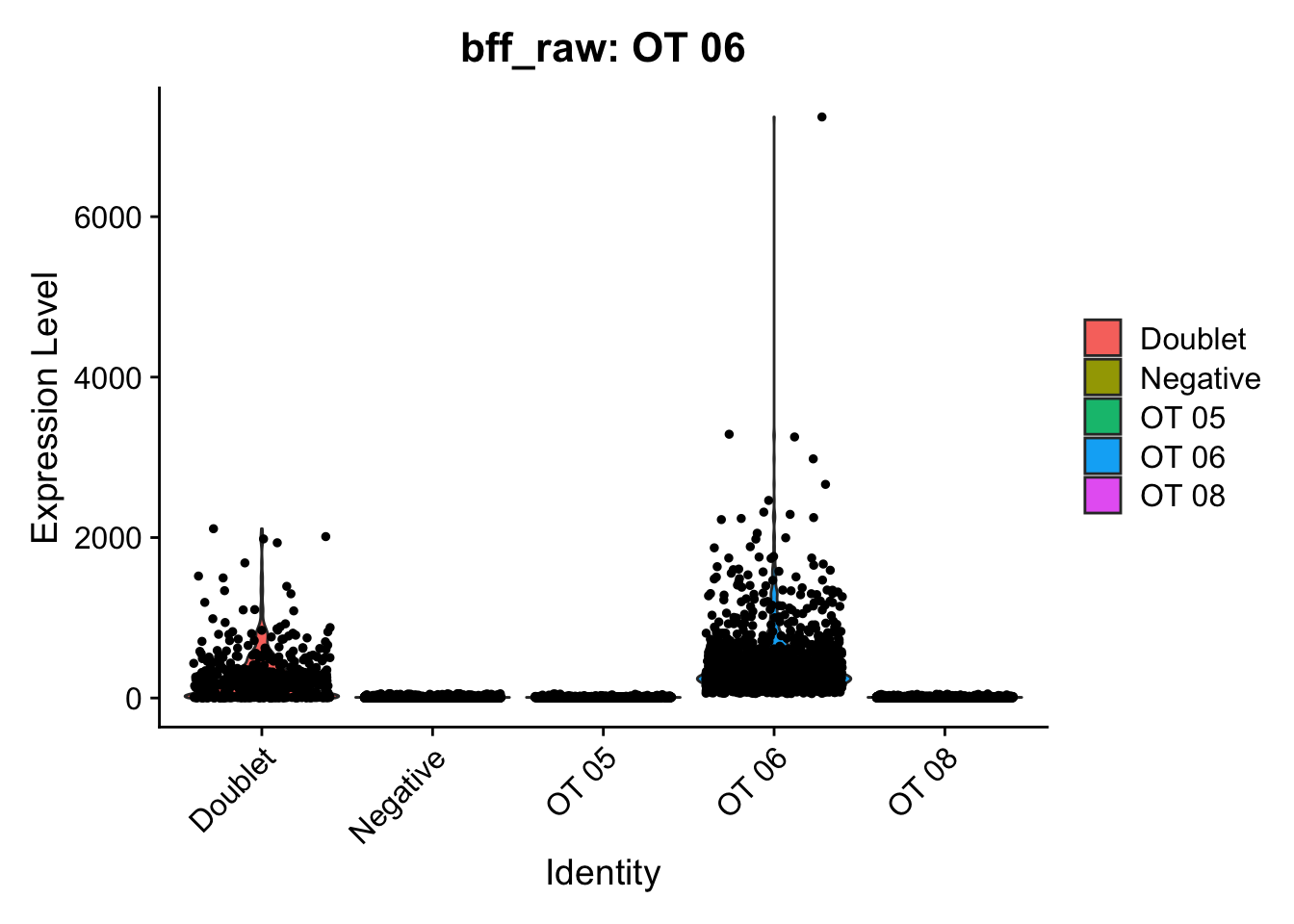
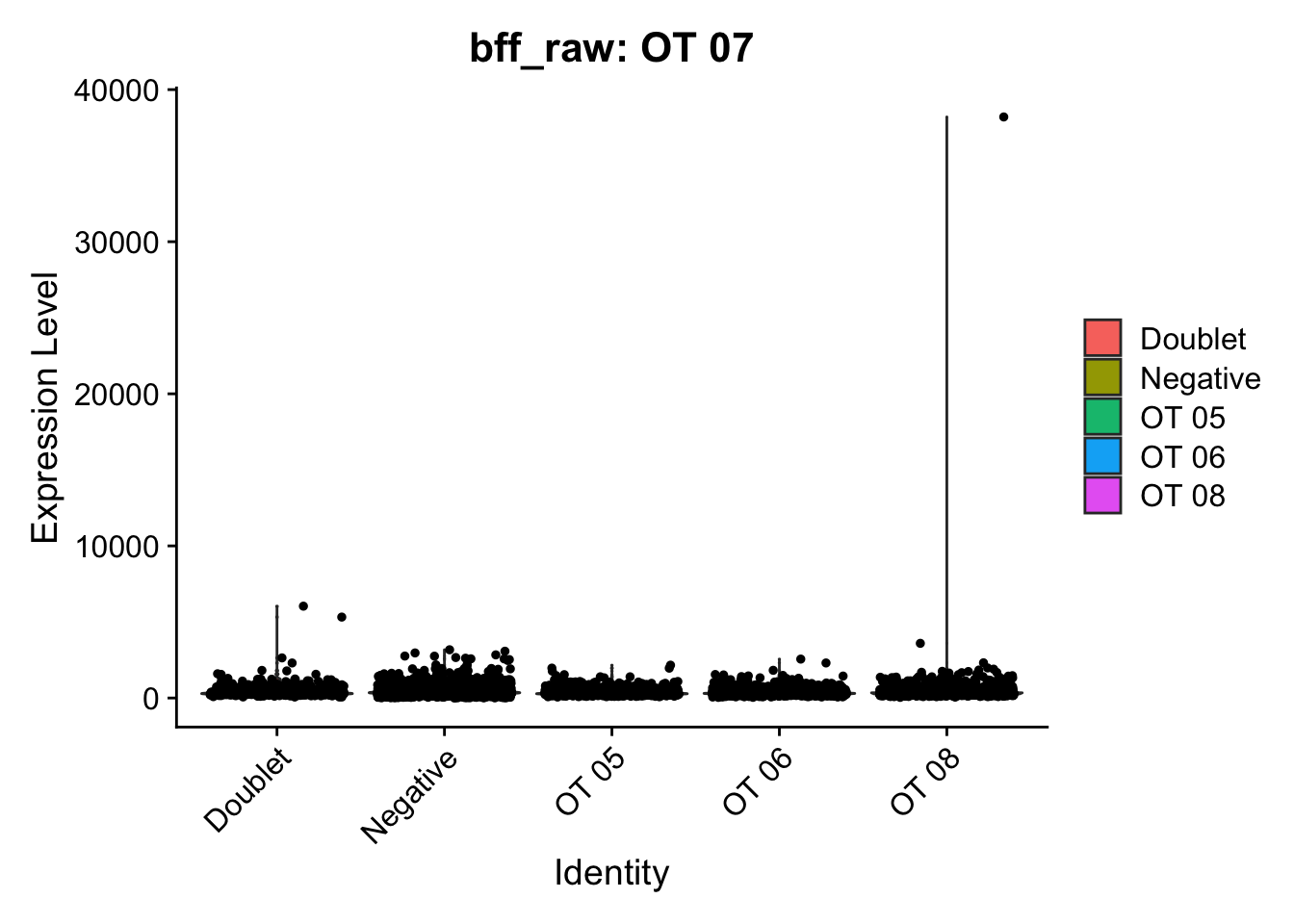
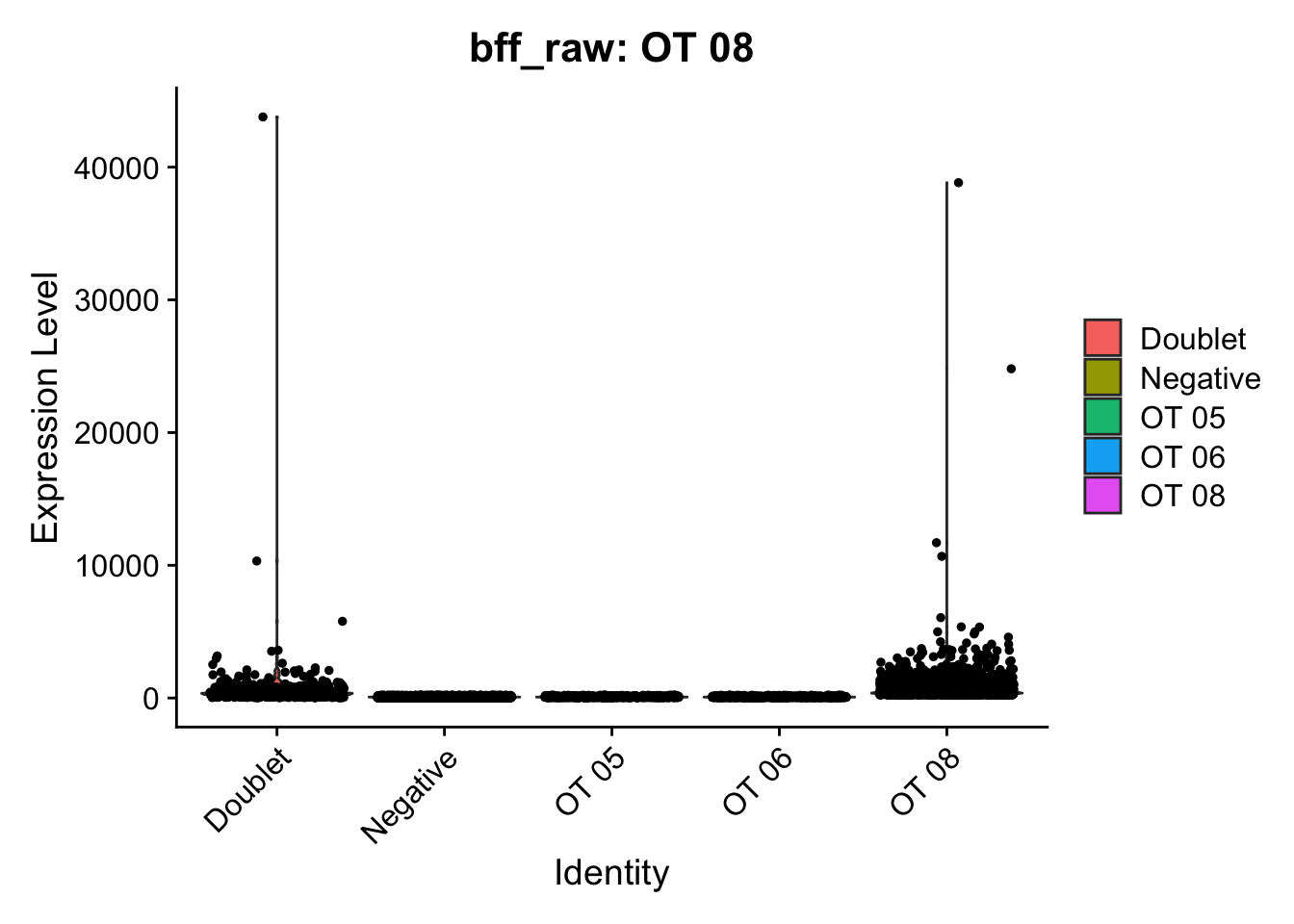
[1] "Starting BFF"
[1] "rows dropped for low counts: 0 of 4"
[1] "Running BFF_cluster"
[1] "Doublet threshold: 0.05"
[1] "Neg threshold: 0.05"
[1] "Min distance as fraction of distance between peaks: 0.1"
[1] "Only one peak found, using max value as cutoff: OT 07"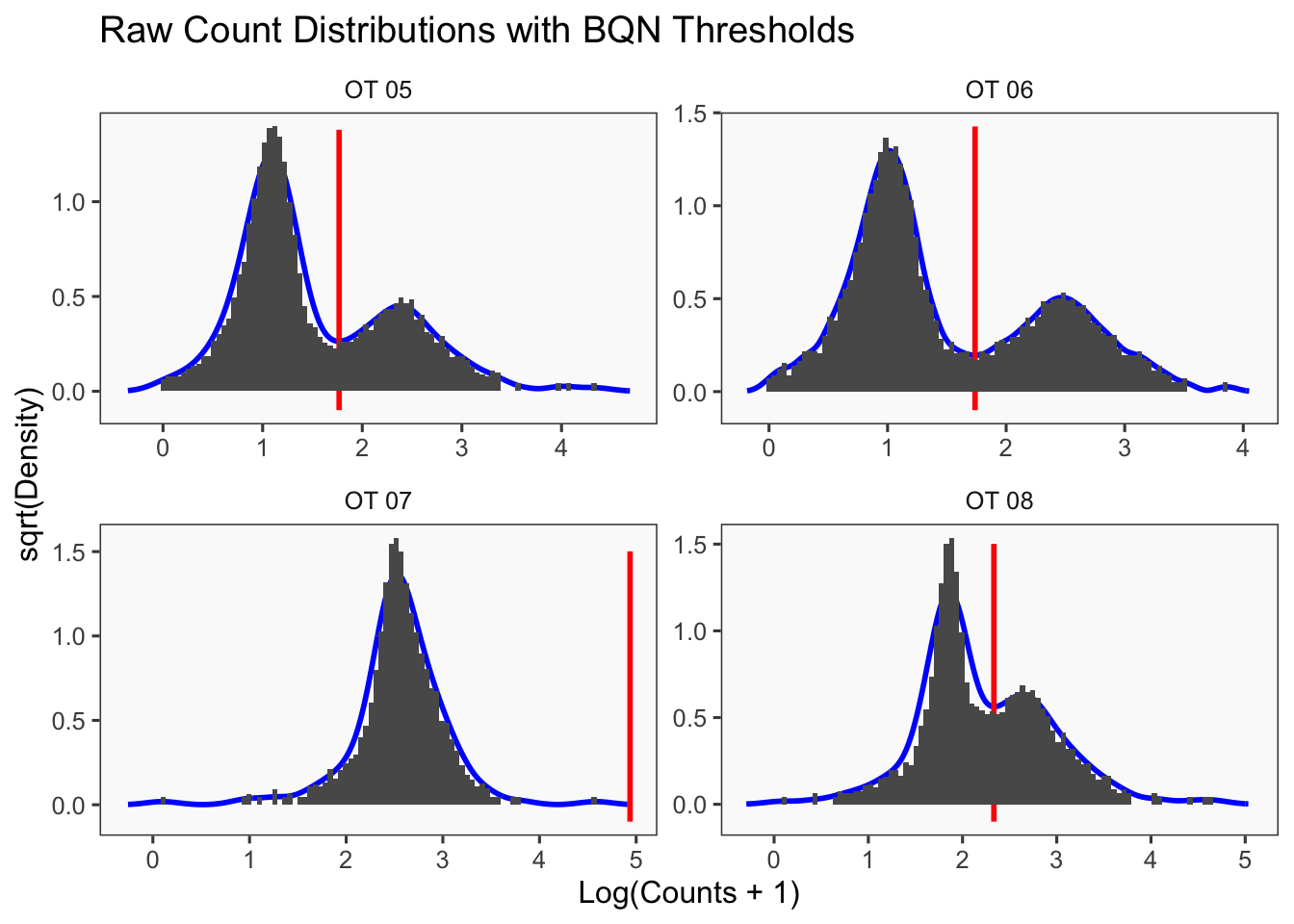
[1] "Thresholds:"
[1] "OT 08: 215.227799018146"
[1] "OT 07: 86433.9145009996"
[1] "OT 06: 54.6952290109095"
[1] "OT 05: 58.5737138510189"
[1] "Only one peak found, using max value as cutoff: OT 07"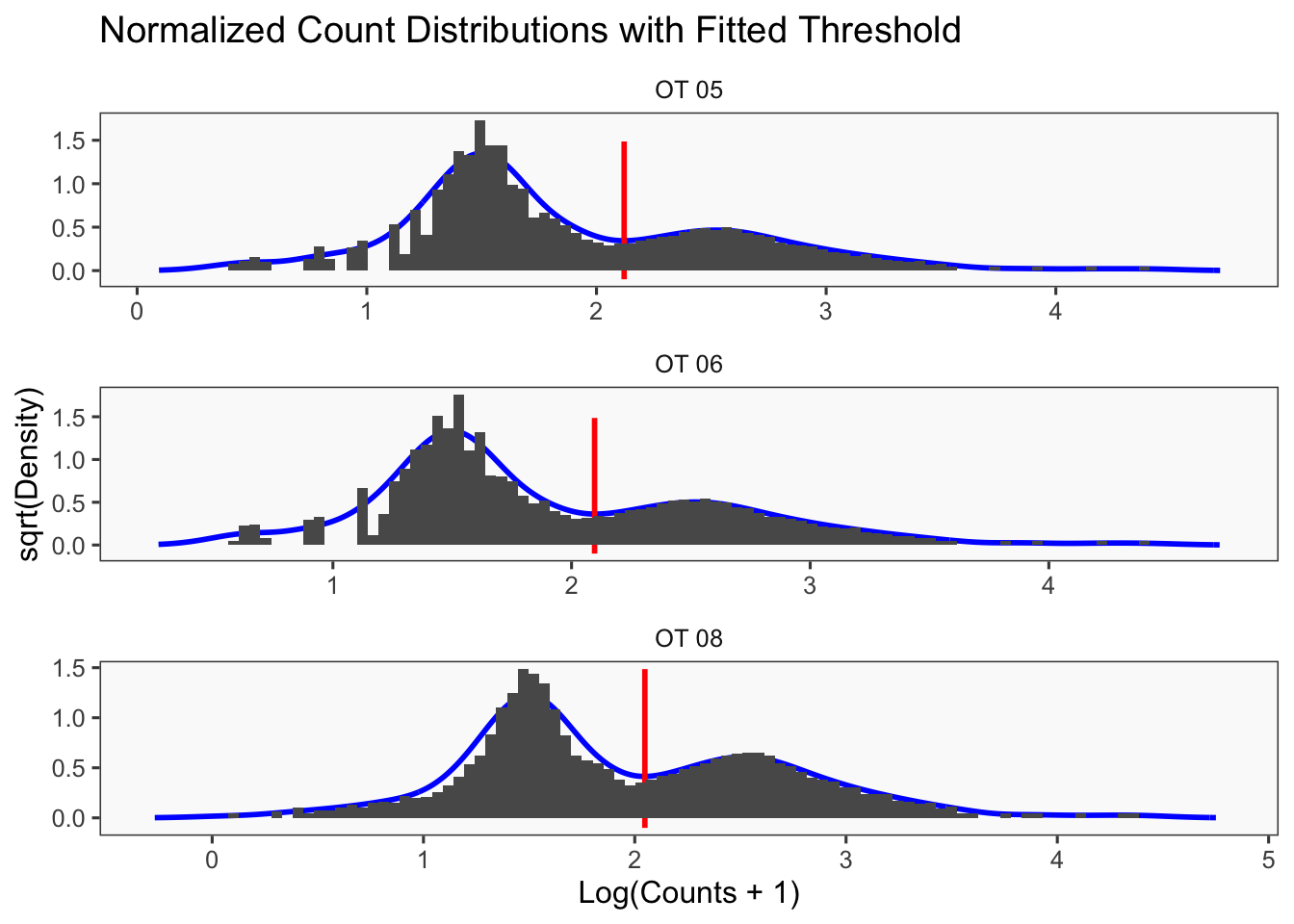
[1] "Smoothing parameter j = 5"
[1] "Smoothing parameter j = 10"
[1] "Smoothing parameter j = 15"
[1] "Smoothing parameter j = 20"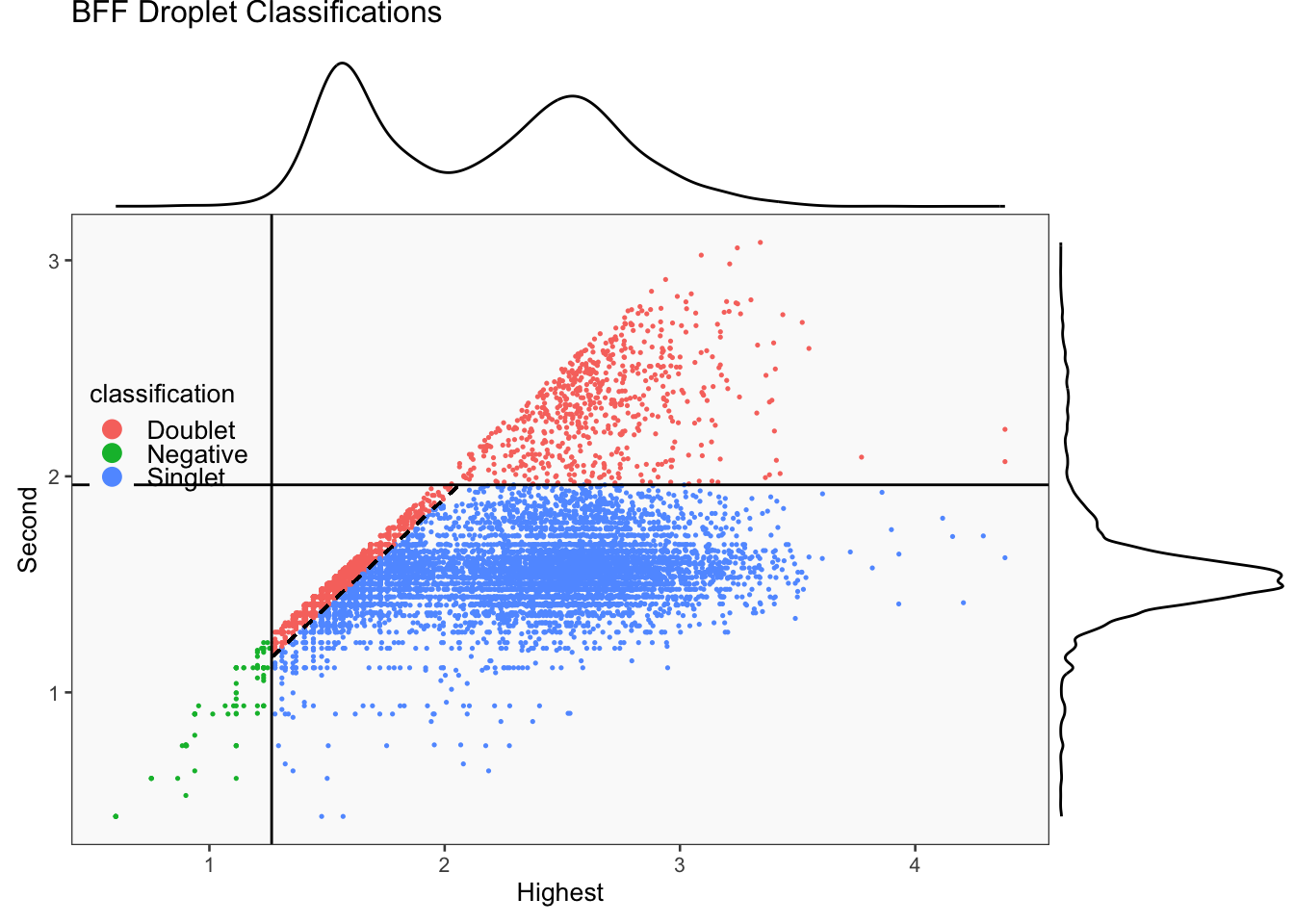
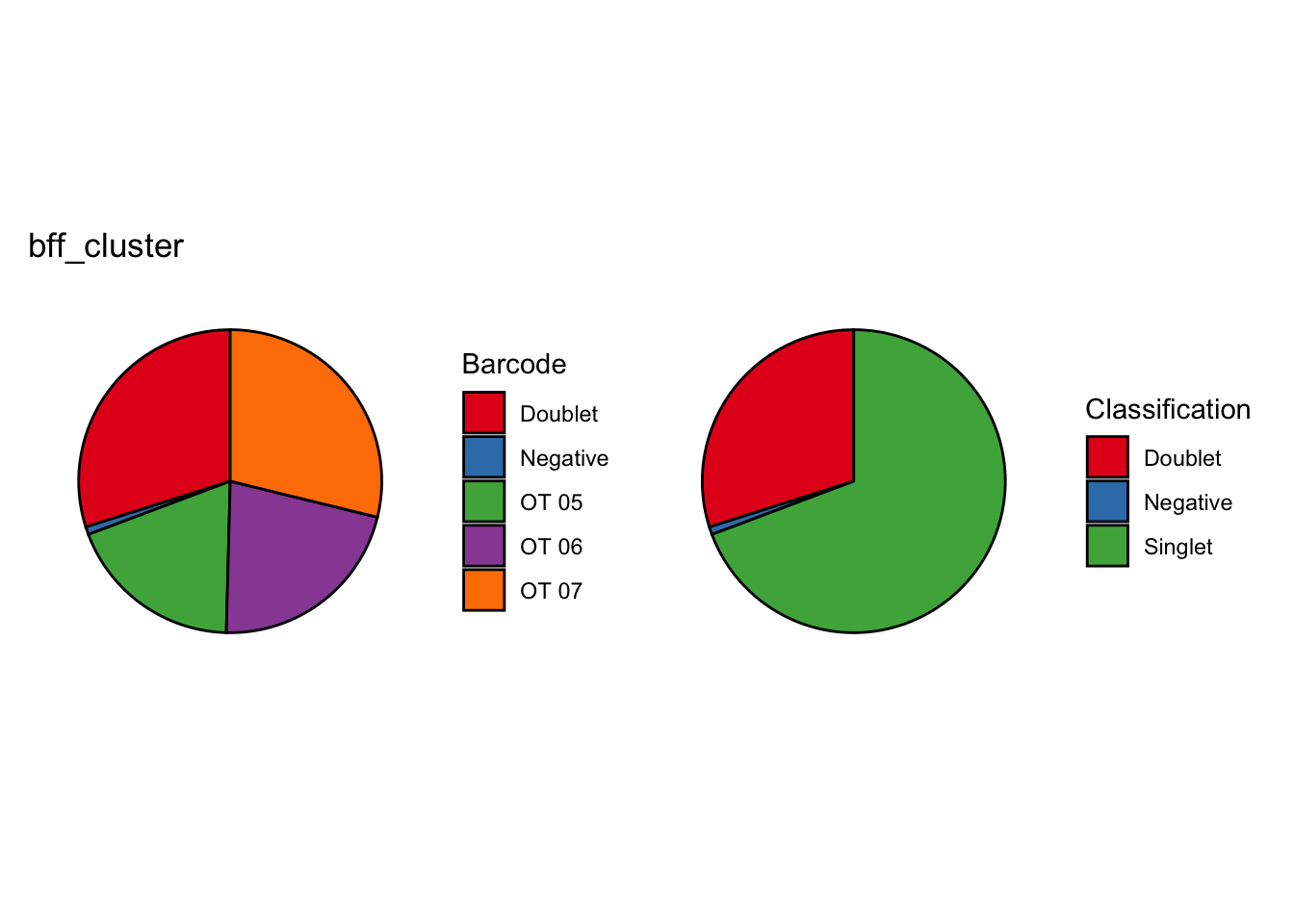
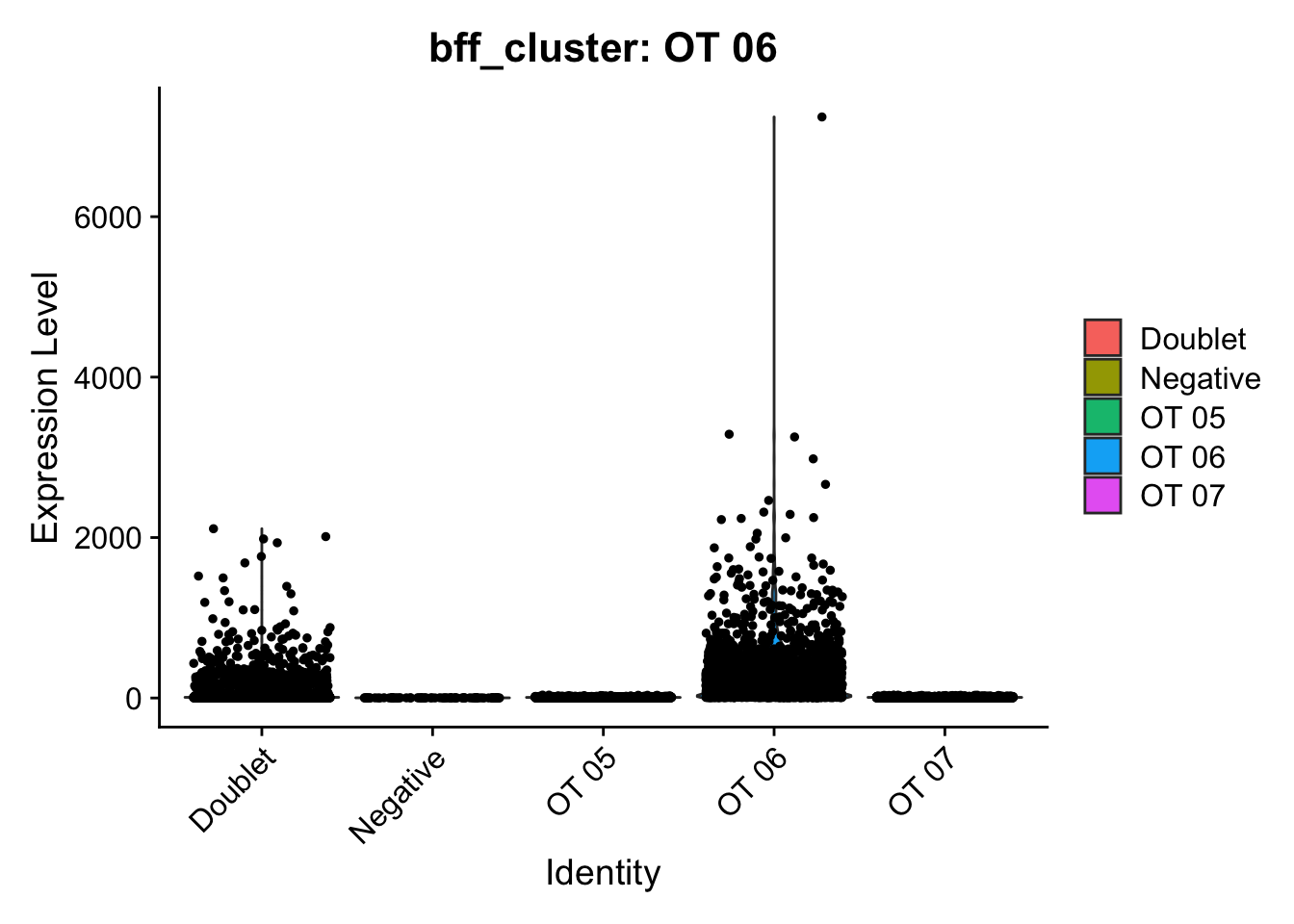
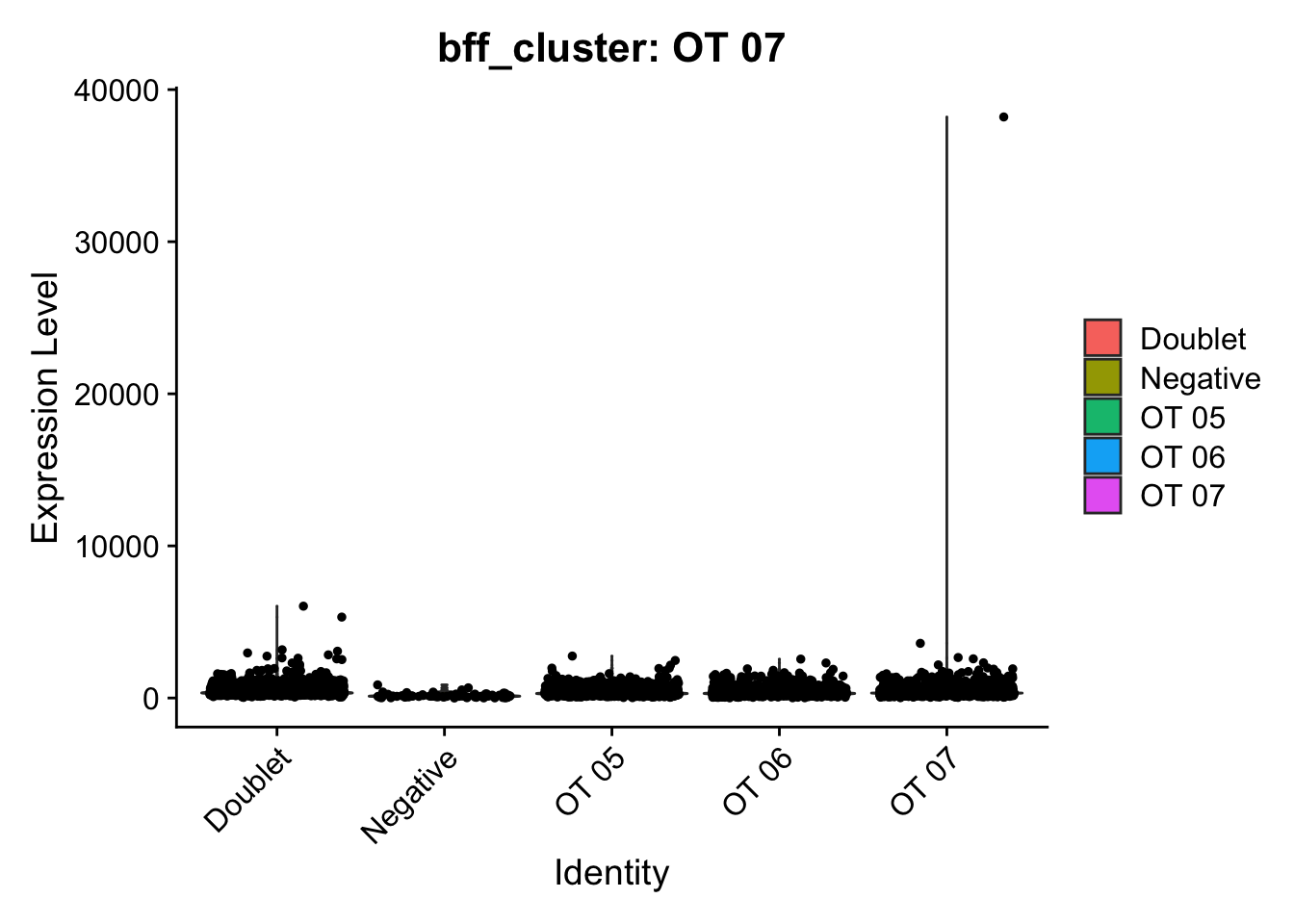
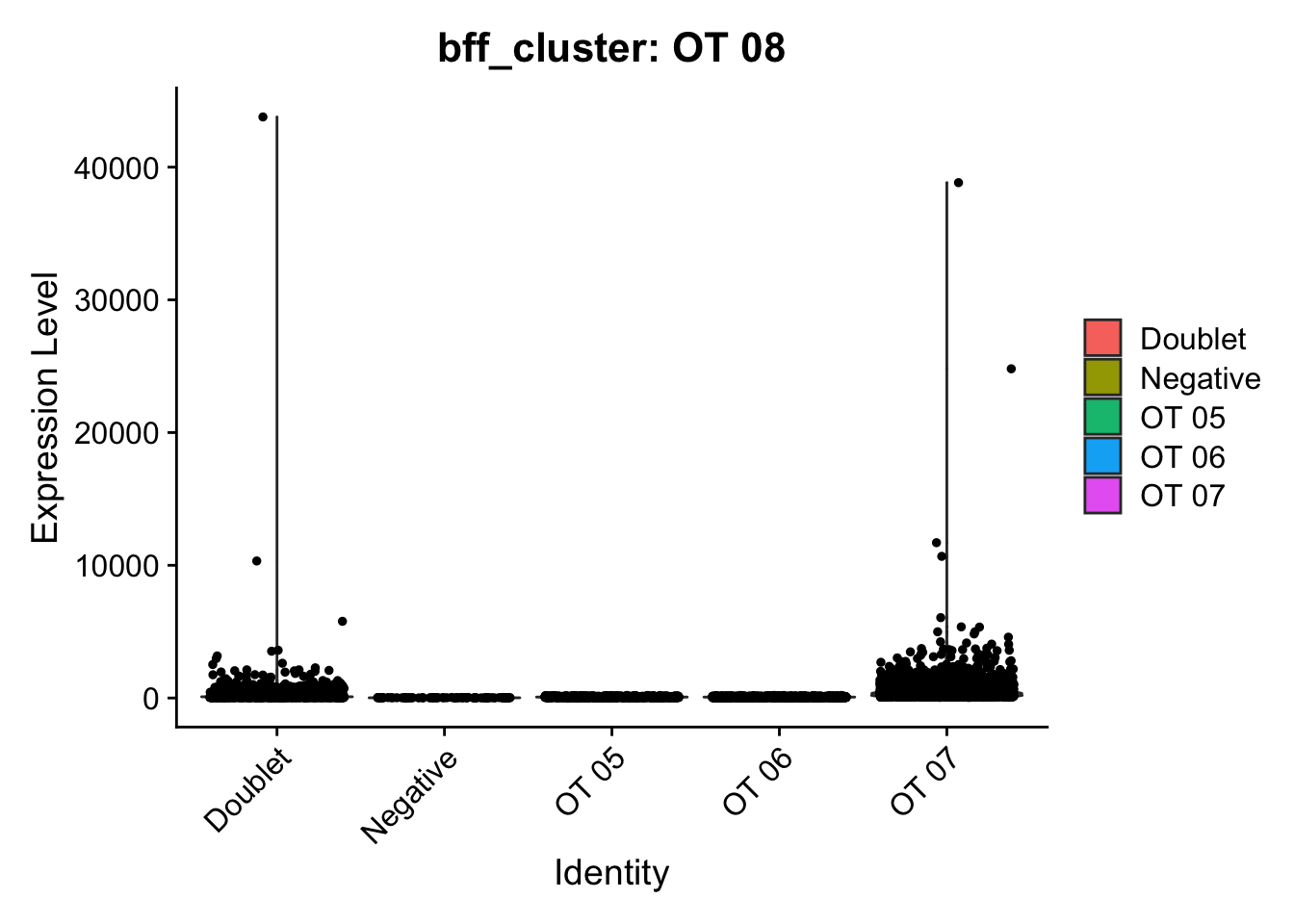
[1] "Generating consensus calls"
[1] "Consensus calls will be generated using: bff_raw,bff_cluster"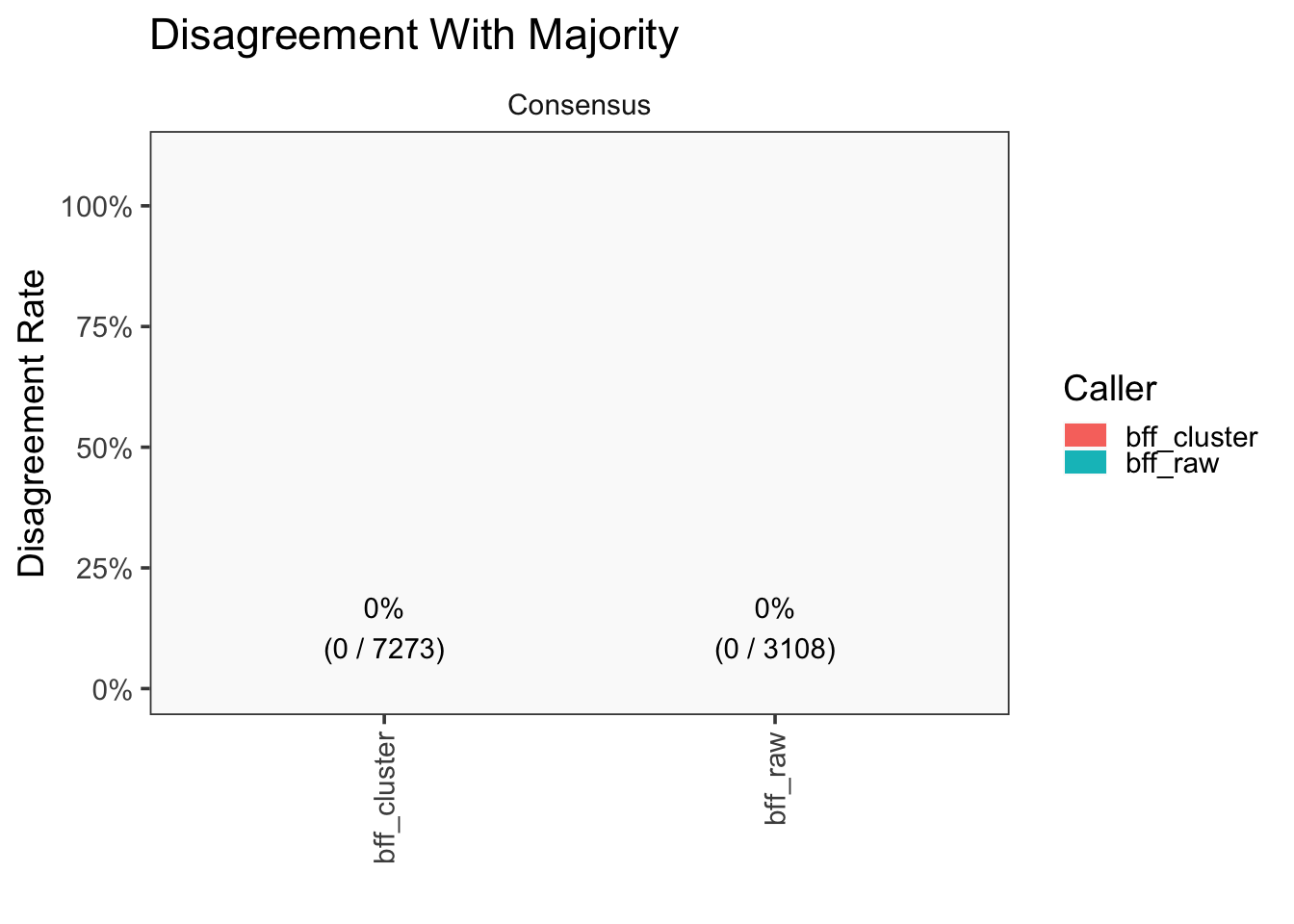
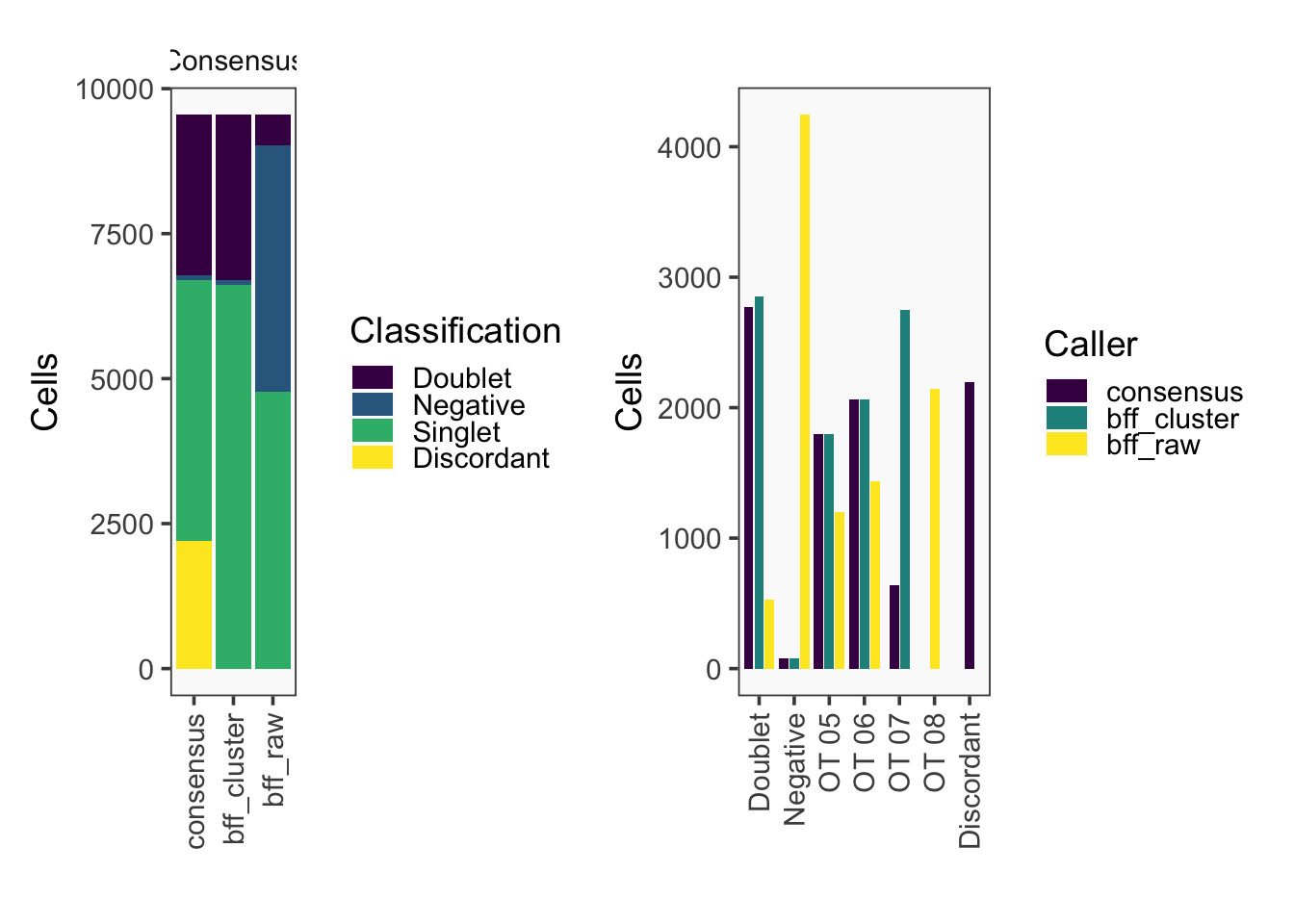
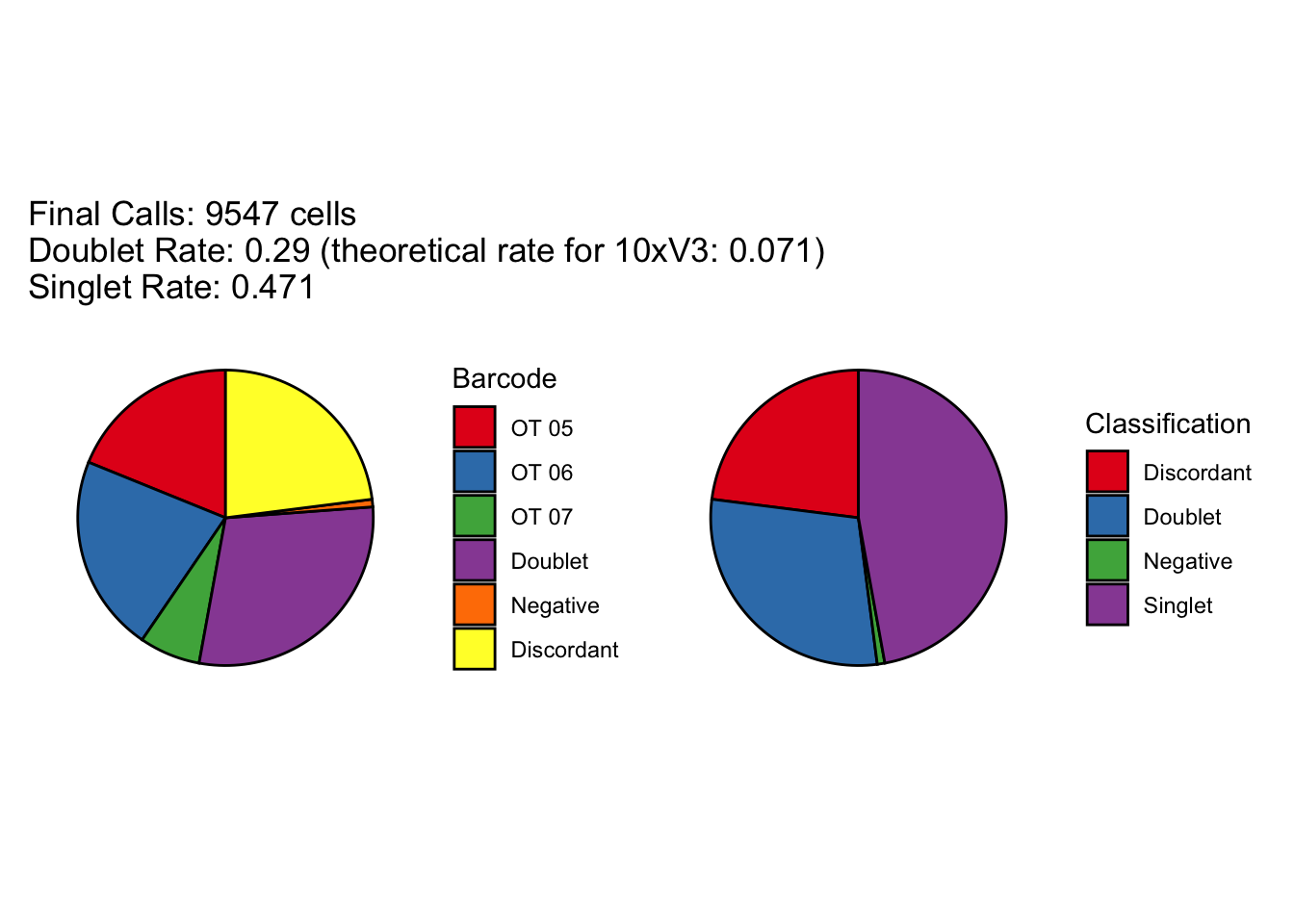
[1] "Total concordant: 7352"
[1] "Total discordant: 2195 (22.99%)"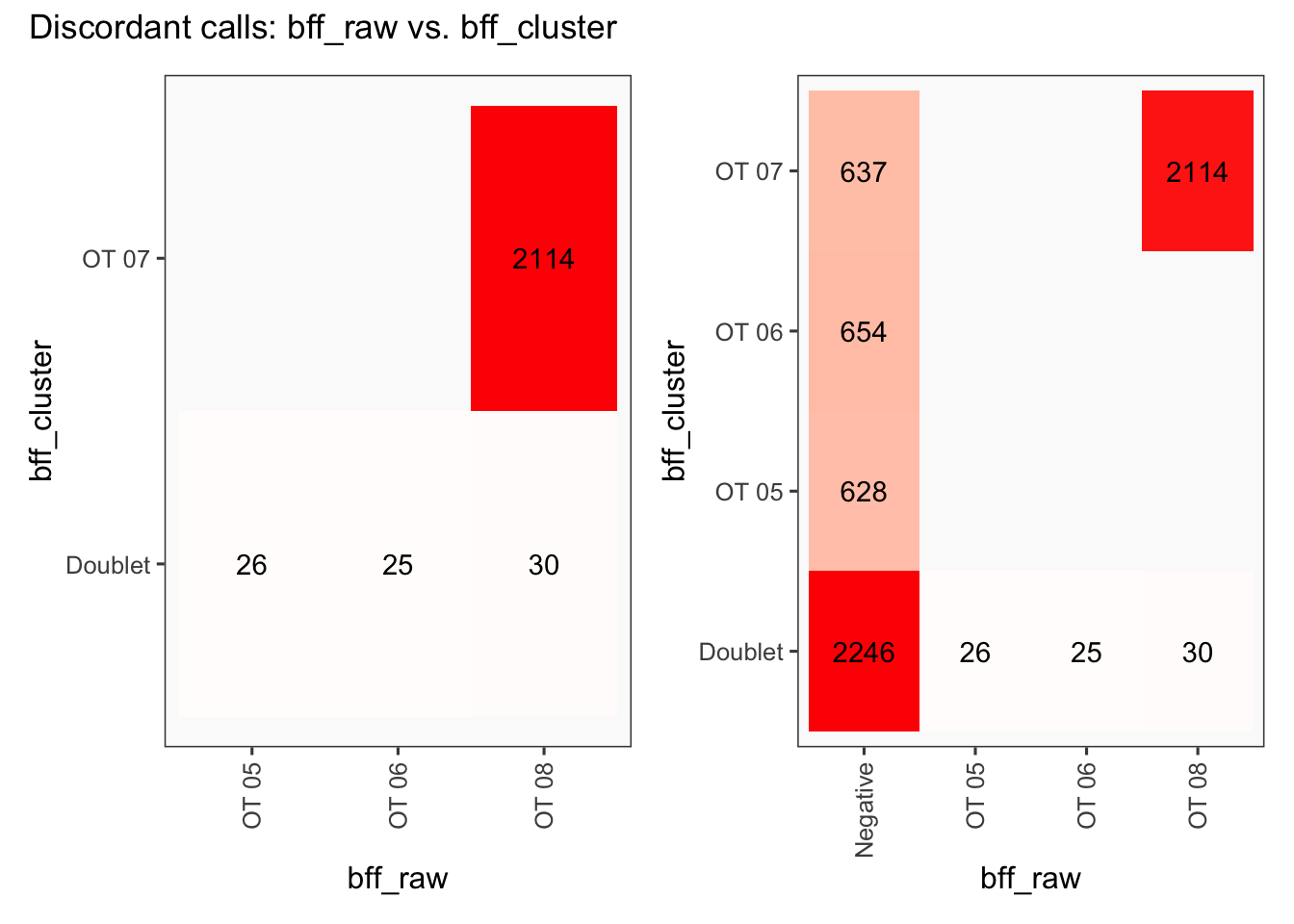
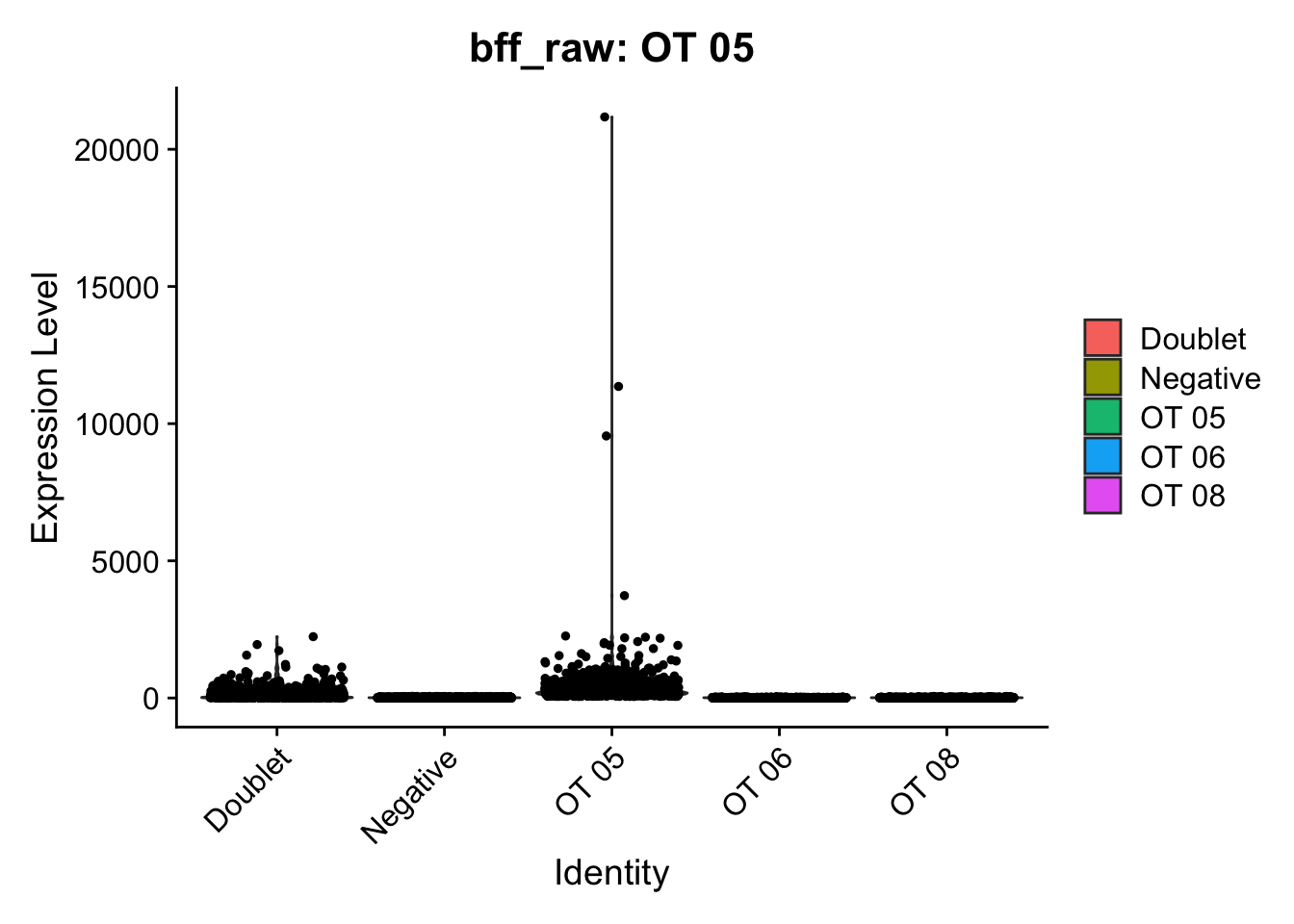
seu_b2$BFF_raw_calls <- cellhashR_calls$bff_raw
seu_b2$BFF_cluster_calls <- cellhashR_calls$bff_clusterseu_b1$BFF_cluster_calls <- case_when(seu_b1$BFF_cluster_calls == "Not Called" ~ "Negative")demuxmix
This function turns the output of demuxmix into something consistent with the other methods
demuxmix_calls_consistent <- function(seurat_object, model = "naive", hto_list) {
hto_counts <- as.matrix(GetAssayData(seurat_object[["HTO"]], slot = "counts"))
dmm <- demuxmix(hto_counts, model = model)
dmm_calls <- dmmClassify(dmm)
calls_out <- case_when(dmm_calls$HTO %in% hto_list ~ dmm_calls$HTO,
!dmm_calls$HTO %in% hto_list ~ case_when(
dmm_calls$Type == "multiplet" ~ "Doublet",
dmm_calls$Type %in% c("negative", "uncertain") ~ "Negative")
)
return(as.factor(calls_out))
}seu_b1$demuxmix_calls <- demuxmix_calls_consistent(seu_b1, hto_list = hashtag_list_b1)
seu_b2$demuxmix_calls <- demuxmix_calls_consistent(seu_b2, hto_list = hashtag_list_b2)Save Seurat objects with all the hashtag assignments
#saveRDS(seu_b1, here("data", "solid_tumor_data", "batch1_all_methods.SEU.rds"))
#saveRDS(seu_b2, here("data", "solid_tumor_data", "batch2_all_methods.SEU.rds"))Making plots
Category plots
Looking at what fraction of droplets are assigned as singlets, doublets or negative by each method
method_calls <- c("hashedDrops_calls",
"hashedDrops_default_calls",
"hashsolo_calls",
"HTODemux_calls",
"GMMDemux_calls",
"deMULTIplex_calls",
"BFF_raw_calls",
"BFF_cluster_calls",
"demuxmix_calls")
for (method in method_calls){
seu_b1[[gsub(method, paste0(method, "_donors"), method)]] <- as.factor(unlist(hashtag_list_b1[unlist(seu_b1[[method]])]))
seu_b2[[gsub(method, paste0(method, "_donors"), method)]] <- as.factor(unlist(hashtag_list_b2[unlist(seu_b2[[method]])]))
}
seu_b1$Batch <- "Batch 1"
seu_b2$Batch <- "Batch 2"
seu <- merge(seu_b1, seu_b2)Warning in CheckDuplicateCellNames(object.list = objects): Some cell names are
duplicated across objects provided. Renaming to enforce unique cell names.seu$Batch <- as.factor(seu$Batch)Need to make a factor which reduces the assignments to one of (“Singlet”, “Doublet”, “Negative”).
sd_or_u_hashtags <- function(seurat_object, method) {
return(case_when(seurat_object[[method]] == "Doublet" ~ "Doublet",
seurat_object[[method]] == "Negative" ~ "Negative",
TRUE ~ "Singlet"))
}
for (method in method_calls) {
seu[[gsub(method, paste0(method, "_category"), method)]] <- sd_or_u_hashtags(seu, method)
}
seu$genetic_donor_category <- case_when(seu$genetic_donor == "Doublet" ~ "Doublet",
seu$genetic_donor == "Negative" ~ "Negative",
TRUE ~ "Singlet")p1 <- dittoBarPlot(seu, var = "genetic_donor_category", group.by = "Batch") + NoLegend() +
ggtitle("vireo (genetics)") +
theme(axis.title.x = element_blank(), axis.title.y = element_blank(),
axis.text.x = element_blank())
p2 <- dittoBarPlot(seu, var = "BFF_cluster_calls_category", group.by = "Batch") + NoLegend() +
ggtitle("BFF_cluster") +
theme(axis.title.x = element_blank(), axis.title.y = element_blank(),
axis.text.x = element_blank(), axis.text.y = element_blank())
p3 <- dittoBarPlot(seu, var = "BFF_raw_calls_category", group.by = "Batch") + NoLegend() +
ggtitle("BFF_raw") +
theme(axis.title.x = element_blank(), axis.title.y = element_blank(),
axis.text.x = element_blank())
p4 <- dittoBarPlot(seu, var = "deMULTIplex_calls_category", group.by = "Batch") +
ggtitle("deMULTIplex") +
theme(axis.title.x = element_blank(), axis.title.y = element_blank(),
axis.text.x = element_blank(), axis.text.y = element_blank())
p5 <- dittoBarPlot(seu, var = "demuxmix_calls_category", group.by = "Batch") + NoLegend() +
ggtitle("demuxmix") +
theme(axis.title.x = element_blank(), axis.title.y = element_blank(),
axis.text.x = element_blank())
p6 <- dittoBarPlot(seu, var = "GMMDemux_calls_category", group.by = "Batch") + NoLegend() +
ggtitle("GMM-Demux") +
theme(axis.title.x = element_blank(), axis.title.y = element_blank(),
axis.text.x = element_blank(), axis.text.y = element_blank())
p7 <- dittoBarPlot(seu, var = "hashedDrops_calls_category", group.by = "Batch") + NoLegend() +
ggtitle("hashedDrops - best") +
theme(axis.title.x = element_blank(), axis.title.y = element_blank(),
axis.text.x = element_blank())
p8 <- dittoBarPlot(seu, var = "hashedDrops_default_calls_category", group.by = "Batch") + NoLegend() +
ggtitle("hashedDrops - default") +
theme(axis.title.x = element_blank(), axis.title.y = element_blank(),
axis.text.x = element_blank(), axis.text.y = element_blank())
p9 <- dittoBarPlot(seu, var = "hashsolo_calls_category", group.by = "Batch") + NoLegend() +
ggtitle("HashSolo") +
theme(axis.title.x = element_blank(), axis.title.y = element_blank(),
axis.text.x = element_text(size = 14))
p10 <- dittoBarPlot(seu, var = "HTODemux_calls_category", group.by = "Batch") + NoLegend() +
ggtitle("HTODemux") +
theme(axis.title.x = element_blank(), axis.title.y = element_blank(),
axis.text.x = element_text(size = 14), axis.text.y = element_blank())
(p1 + p2) / (p3 + p4) / (p5 + p6) / (p7 + p8) / (p9 + p10)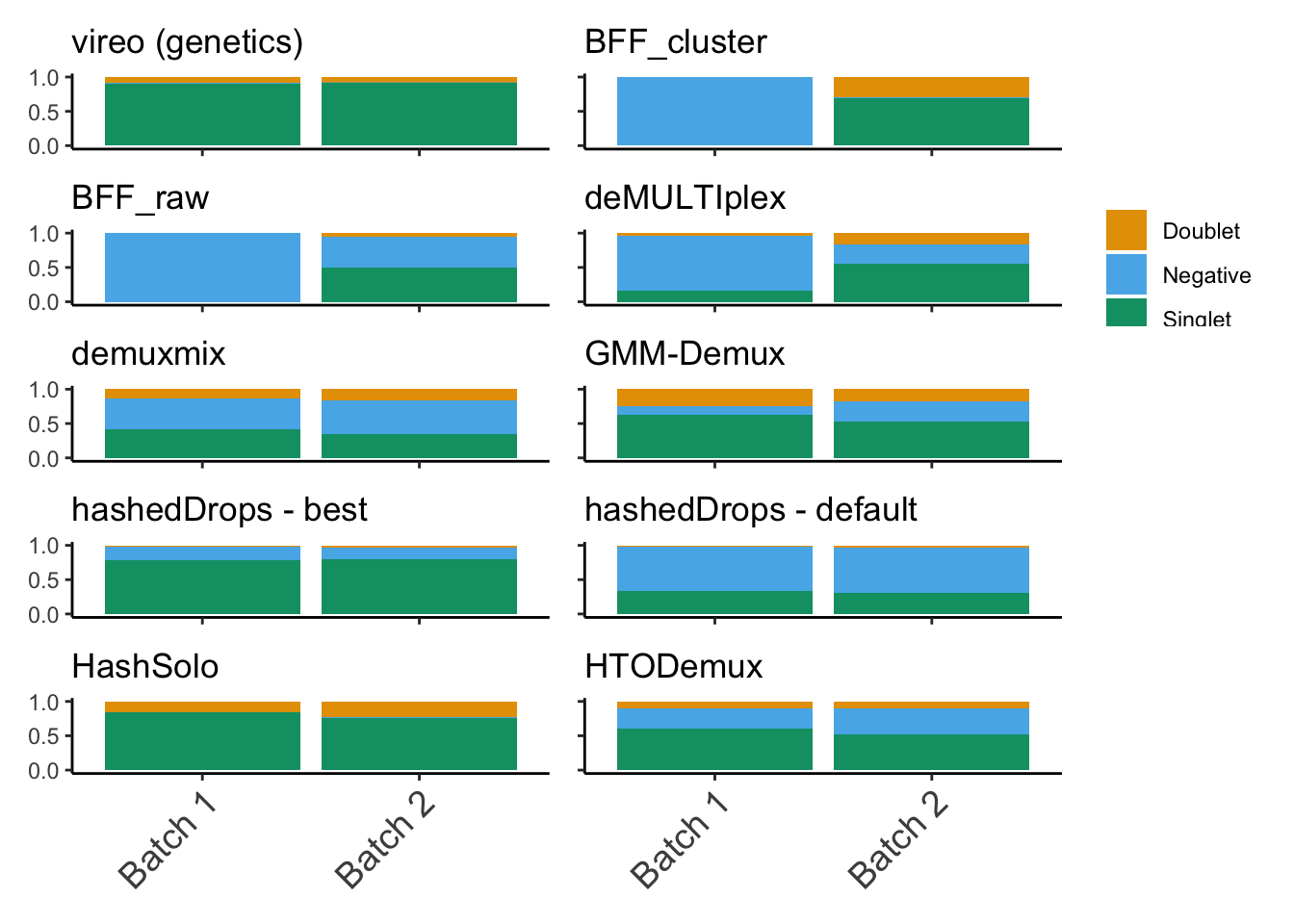
Fscore
calculate_HTO_fscore <- function(seurat_object, donor_hto_list, method) {
calls <- seurat_object[[method]]
f <- NULL
for (HTO in donor_hto_list) {
tp <- sum(calls == HTO & donor_hto_list[seurat_object$genetic_donor] == HTO) #True positive rate
fp <- sum(calls == HTO & donor_hto_list[seurat_object$genetic_donor] != HTO) #False positive rate
fn <- sum(calls != HTO & donor_hto_list[seurat_object$genetic_donor] == HTO) #False negative rate
f <- c(f, tp / (tp + 0.5 * (fp + fn)))
}
# f <- c(f, median(f)) #Add median F score
f <- c(f, mean(f)) #Add mean F score
names(f) <- c(donor_hto_list, "Average")
return(f)
}Fscore_hashedDrops_b1 <- calculate_HTO_fscore(seu_b1, donor_hashtag_list_b1[1:4], "hashedDrops_calls")
Fscore_hashedDrops_b2 <- calculate_HTO_fscore(seu_b2, donor_hashtag_list_b2[1:4], "hashedDrops_calls")
Fscore_hashedDrops_default_b1 <- calculate_HTO_fscore(seu_b1, donor_hashtag_list_b1[1:4], "hashedDrops_default_calls")
Fscore_hashedDrops_default_b2 <- calculate_HTO_fscore(seu_b2, donor_hashtag_list_b2[1:4], "hashedDrops_default_calls")
Fscore_hashsolo_b1 <- calculate_HTO_fscore(seu_b1, donor_hashtag_list_b1[1:4], "hashsolo_calls")
Fscore_hashsolo_b2 <- calculate_HTO_fscore(seu_b2, donor_hashtag_list_b2[1:4], "hashsolo_calls")
Fscore_HTODemux_b1 <- calculate_HTO_fscore(seu_b1, donor_hashtag_list_b1[1:4], "HTODemux_calls")
Fscore_HTODemux_b2 <- calculate_HTO_fscore(seu_b2, donor_hashtag_list_b2[1:4], "HTODemux_calls")
Fscore_GMMDemux_b1 <- calculate_HTO_fscore(seu_b1, donor_hashtag_list_b1[1:4], "GMMDemux_calls")
Fscore_GMMDemux_b2 <- calculate_HTO_fscore(seu_b2, donor_hashtag_list_b2[1:4], "GMMDemux_calls")
Fscore_deMULTIplex_b1 <- calculate_HTO_fscore(seu_b1, donor_hashtag_list_b1[1:4], "deMULTIplex_calls")
Fscore_deMULTIplex_b2 <- calculate_HTO_fscore(seu_b2, donor_hashtag_list_b2[1:4], "deMULTIplex_calls")
Fscore_BFF_raw_b1 <- calculate_HTO_fscore(seu_b1, donor_hashtag_list_b1[1:4], "BFF_raw_calls")
Fscore_BFF_raw_b2 <- calculate_HTO_fscore(seu_b2, donor_hashtag_list_b2[1:4], "BFF_raw_calls")
Fscore_BFF_cluster_b1 <- calculate_HTO_fscore(seu_b1, donor_hashtag_list_b1[1:4], "BFF_cluster_calls")
Fscore_BFF_cluster_b2 <- calculate_HTO_fscore(seu_b2, donor_hashtag_list_b2[1:4], "BFF_cluster_calls")
Fscore_demuxmix_b1 <- calculate_HTO_fscore(seu_b1, donor_hashtag_list_b1[1:4], "demuxmix_calls")
Fscore_demuxmix_b2 <- calculate_HTO_fscore(seu_b2, donor_hashtag_list_b2[1:4], "demuxmix_calls")Fscore_matrix_b1 <- data.frame("Hashtag" = c(hashtag_list_b1[1:4], "Mean"),
"hashedDrops" = Fscore_hashedDrops_b1,
"hashedDrops_default" = Fscore_hashedDrops_default_b1,
"HashSolo" = Fscore_hashsolo_b1,
"HTODemux" = Fscore_HTODemux_b1,
"GMM_Demux" = Fscore_GMMDemux_b1,
"deMULTIplex" = Fscore_deMULTIplex_b1,
"BFF_raw" = Fscore_BFF_raw_b1,
"BFF_cluster" = Fscore_BFF_cluster_b1,
"demuxmix" = Fscore_demuxmix_b1)
Fscore_matrix_b1 Hashtag hashedDrops hashedDrops_default HashSolo HTODemux GMM_Demux
OT 01 OT 01 0.3878205 0.3508772 0.3773585 0.3414634 0.3529412
OT 02 OT 02 0.8148297 0.6387852 0.7554054 0.6926214 0.7661009
OT 03 OT 03 0.7790672 0.3617088 0.6282509 0.7844753 0.7506263
OT 04 OT 04 0.7350051 0.2707768 0.6620463 0.6610695 0.7087034
Average Mean 0.6791806 0.4055370 0.6057653 0.6199074 0.6445929
deMULTIplex BFF_raw BFF_cluster demuxmix
OT 01 0.35600000 0 0 0.3066955
OT 02 0.32425422 0 0 0.6209396
OT 03 0.08955224 0 0 0.5673913
OT 04 0.10065189 0 0 0.5490760
Average 0.21761459 0 0 0.5110256#Removing average information
Fscore_matrix_b1 = Fscore_matrix_b1[1:4,]Fscore_matrix_b2 <- data.frame("Hashtag" = c(hashtag_list_b2[1:4], "Mean"),
"hashedDrops" = Fscore_hashedDrops_b2,
"hashedDrops_default" = Fscore_hashedDrops_default_b2,
"HashSolo" = Fscore_hashsolo_b2,
"HTODemux" = Fscore_HTODemux_b2,
"GMM_Demux" = Fscore_GMMDemux_b2,
"deMULTIplex" = Fscore_deMULTIplex_b2,
"BFF_raw" = Fscore_BFF_raw_b2,
"BFF_cluster" = Fscore_BFF_cluster_b2,
"demuxmix" = Fscore_demuxmix_b2)
Fscore_matrix_b2 Hashtag hashedDrops hashedDrops_default HashSolo HTODemux GMM_Demux
OT 05 OT 05 0.7832734 0.3160055 0.4639805 0.8184874 0.7510431
OT 06 OT 06 0.8346273 0.4904387 0.6341223 0.8396890 0.7891936
OT 07 OT 07 0.8075230 0.3776145 0.8724261 0.1335944 0.2358240
OT 08 OT 08 0.8257874 0.6737542 0.7803532 0.8230288 0.7537453
Average Mean 0.8128028 0.4644532 0.6877205 0.6536999 0.6324515
deMULTIplex BFF_raw BFF_cluster demuxmix
OT 05 0.7888938 0.8201499 0.7037284 0.4491315
OT 06 0.8063492 0.8550619 0.7051852 0.4615385
OT 07 0.3422171 0.0000000 0.2221590 0.4526467
OT 08 0.7998987 0.8186554 0.0000000 0.6608187
Average 0.6843397 0.6234668 0.4077681 0.5060339#Removing average information
Fscore_matrix_b2 = Fscore_matrix_b2[1:4,]Fscore_matrix_b1 %>%
pivot_longer(cols = c("hashedDrops", "hashedDrops_default", "HashSolo",
"HTODemux", "GMM_Demux", "deMULTIplex",
"BFF_raw", "BFF_cluster", "demuxmix"),
names_to = "method",
values_to = "Fscore") -> Fscore_matrix_b1
Fscore_matrix_b2 %>%
pivot_longer(cols = c("hashedDrops", "hashedDrops_default", "HashSolo",
"HTODemux", "GMM_Demux", "deMULTIplex",
"BFF_raw", "BFF_cluster", "demuxmix"),
names_to = "method",
values_to = "Fscore") -> Fscore_matrix_b2Fscore_matrix_b1$Batch <- "Batch 1"
Fscore_matrix_b2$Batch <- "Batch 2"
Fscore_matrix <- bind_rows(Fscore_matrix_b1,
Fscore_matrix_b2)Fscore_matrix %>%
group_by(Batch) %>%
heatmap(.row = method,
.column = Hashtag,
.value = Fscore,
column_title = "F-score - solid tissue data",
cluster_rows = TRUE,
row_names_gp = gpar(fontsize = 10),
show_row_dend = FALSE,
row_names_side = "left",
row_title = "",
cluster_columns = FALSE,
column_names_gp = gpar(fontsize = 10),
palette_value = plasma(3)) -> p1tidyHeatmap says: (once per session) from release 1.7.0 the scaling is set to "none" by default. Please use scale = "row", "column" or "both" to apply scalingp1 <- wrap_heatmap(p1)
p1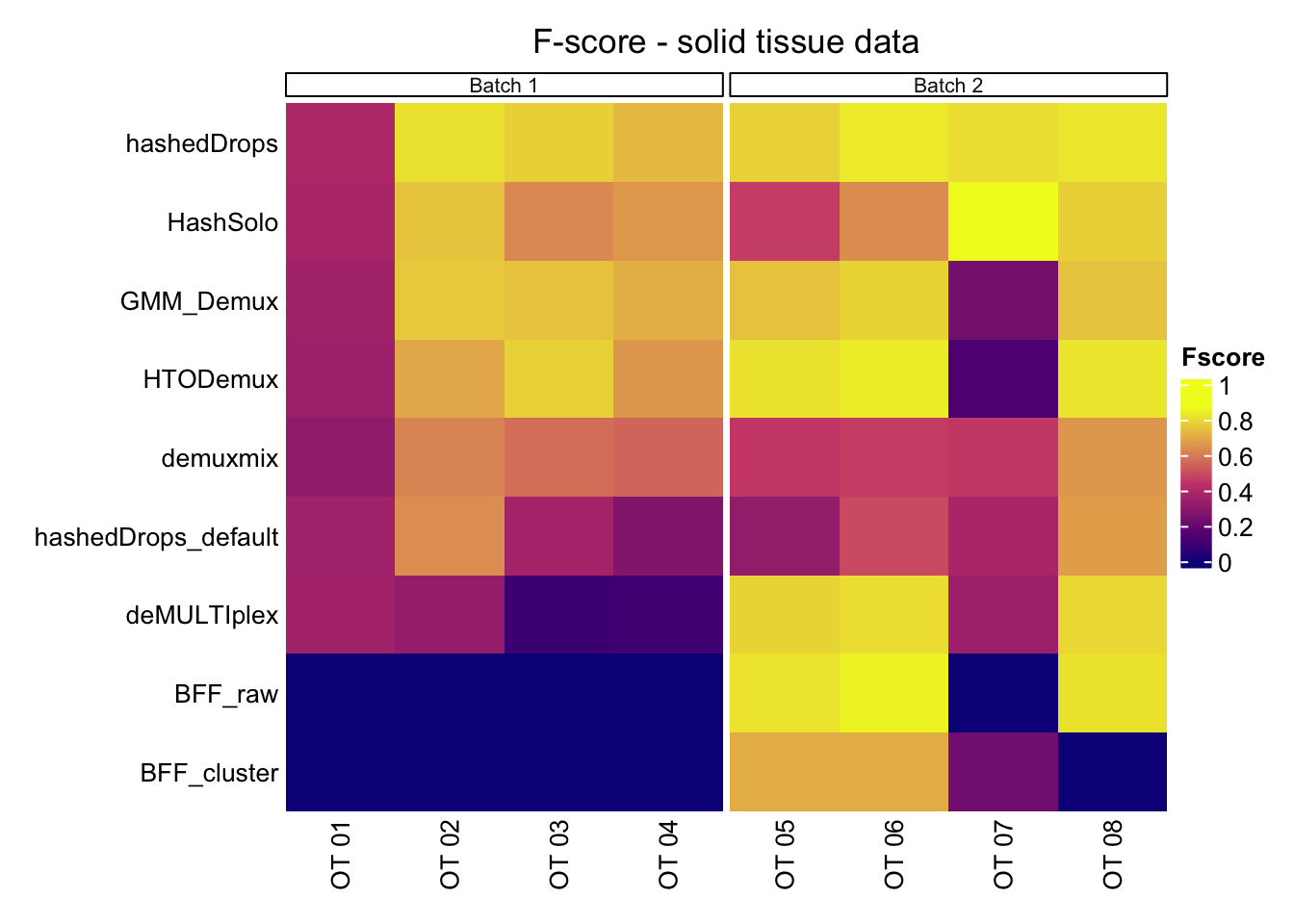
# ggsave(here("paper_latex", "figures", "OT_Fscore.png"),
# p1,
# device = "png",
# width = 8, height = 5,
# units = "in",
# dpi = 350
# )Doublet assignments
Also worried about whether any of these methods are assigning genetic doublets as singlets. Going to look at the fraction of genetic doublets that get assigned to each of the possible HTO categories.
b1_doublets <- seu_b1[, seu_b1$genetic_donor == "Doublet"]
b2_doublets <- seu_b2[, seu_b2$genetic_donor == "Doublet"]
b1_doublet_doublet <- NULL
b1_doublet_negative <- NULL
b1_doublet_singlet <- NULL
b2_doublet_doublet <- NULL
b2_doublet_negative <- NULL
b2_doublet_singlet <- NULL
for (method in method_calls) {
b1_doublet_doublet <- c(b1_doublet_doublet, sum(seu_b1$genetic_donor == "Doublet" & seu_b1[[method]] == "Doublet") / sum(seu_b1$genetic_donor == "Doublet"))
b1_doublet_negative <- c(b1_doublet_negative, sum(seu_b1$genetic_donor == "Doublet" & seu_b1[[method]] == "Negative") / sum(seu_b1$genetic_donor == "Doublet"))
b1_doublet_singlet <- c(b1_doublet_singlet, sum(seu_b1$genetic_donor == "Doublet" & Reduce("|", lapply(hashtag_list_b1[1:4], function(x) seu_b1[[method]] == x))) / sum(seu_b1$genetic_donor == "Doublet"))
b2_doublet_doublet <- c(b2_doublet_doublet, sum(seu_b2$genetic_donor == "Doublet" & seu_b2[[method]] == "Doublet") / sum(seu_b2$genetic_donor == "Doublet"))
b2_doublet_negative <- c(b2_doublet_negative, sum(seu_b2$genetic_donor == "Doublet" & seu_b2[[method]] == "Negative") / sum(seu_b2$genetic_donor == "Doublet"))
b2_doublet_singlet <- c(b2_doublet_singlet, sum(seu_b2$genetic_donor == "Doublet" & Reduce("|", lapply(hashtag_list_b2[1:4], function(x) seu_b2[[method]] == x))) / sum(seu_b2$genetic_donor == "Doublet"))
}
names(b1_doublet_doublet) <- method_calls
names(b1_doublet_negative) <- method_calls
names(b1_doublet_singlet) <- method_calls
names(b2_doublet_doublet) <- method_calls
names(b2_doublet_negative) <- method_calls
names(b2_doublet_singlet) <- method_callsb1_doublet_assignments <- data.frame("method" = c("hashedDrops",
"hashedDrops (default)",
"HashSolo",
"HTODemux",
"GMM-Demux",
"deMULTIplex",
"BFF_raw",
"BFF_cluster",
"demuxmix"),
"Doublet" = b1_doublet_doublet,
"Negative" = b1_doublet_negative,
"Singlet" = b1_doublet_singlet) %>%
pivot_longer(cols = c("Doublet", "Negative", "Singlet"),
names_to = "assignment",
values_to = "fraction")
b2_doublet_assignments <- data.frame("method" = c("hashedDrops",
"hashedDrops (default)",
"HashSolo",
"HTODemux",
"GMM-Demux",
"deMULTIplex",
"BFF_raw",
"BFF_cluster",
"demuxmix"),
"Doublet" = b2_doublet_doublet,
"Negative" = b2_doublet_negative,
"Singlet" = b2_doublet_singlet) %>%
pivot_longer(cols = c("Doublet", "Negative", "Singlet"),
names_to = "assignment",
values_to = "fraction")doublet_colours <- c("black", "gray60", "firebrick1")
p2 <- ggplot(b1_doublet_assignments %>%
mutate(method = factor(method, levels = c("BFF_cluster",
"BFF_raw",
"deMULTIplex",
"hashedDrops (default)",
"demuxmix",
"HTODemux",
"GMM-Demux",
"HashSolo",
"hashedDrops")))) +
geom_bar(aes(x = method, y = fraction, fill = assignment),
stat = "identity") +
ggtitle("Batch 1 (1048 doublets)") +
ylim(0, 1) +
scale_fill_manual(values = doublet_colours) +
theme(axis.ticks.x = element_blank(),
axis.title.y = element_blank(),
axis.text.y = element_text(size = 12)) +
coord_flip() + NoLegend()
p3 <- ggplot(b2_doublet_assignments %>%
mutate(method = factor(method, levels = c("BFF_cluster",
"BFF_raw",
"deMULTIplex",
"hashedDrops (default)",
"demuxmix",
"HTODemux",
"GMM-Demux",
"HashSolo",
"hashedDrops")))) +
geom_bar(aes(x = method, y = fraction, fill = assignment),
stat = "identity") +
ggtitle("Batch 2 (805 doublets)") +
ylim(0, 1) +
scale_fill_manual(values = doublet_colours) +
theme(axis.ticks.x = element_blank(),
axis.ticks.y = element_blank(),
axis.title.y = element_blank(),
axis.text.y = element_blank()
) + coord_flip()
p2 | p3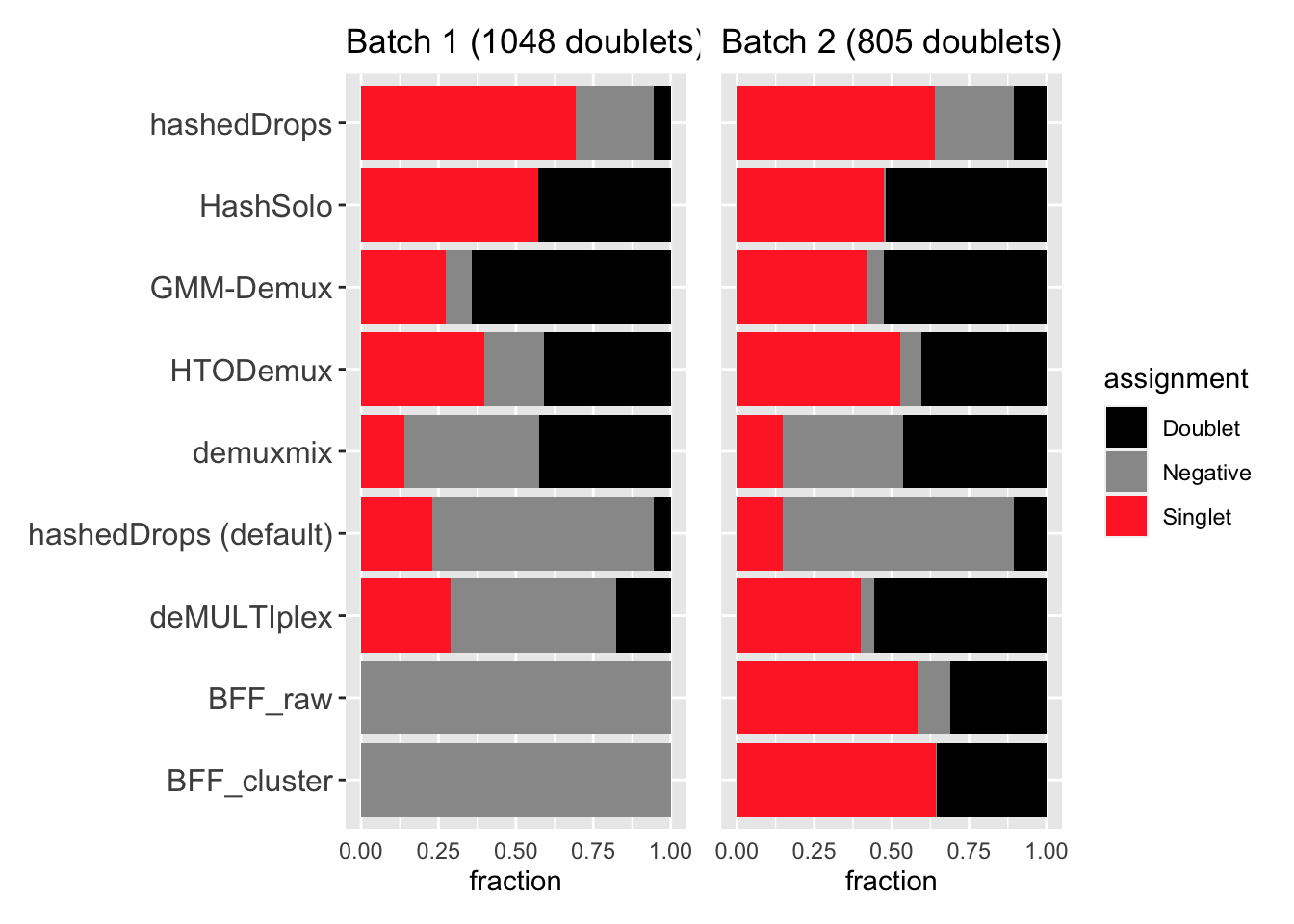
# ggsave(here("paper_latex", "figures", "OT_doublet_assignments.png"),
# p2 | p3,
# device = "png",
# width = 8, height = 5,
# units = "in",
# dpi = 350
# )
sessionInfo()R version 4.2.2 (2022-10-31)
Platform: aarch64-apple-darwin20 (64-bit)
Running under: macOS Ventura 13.0.1
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] grid stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] Matrix_1.5-4 viridis_0.6.3
[3] viridisLite_0.4.2 tidyHeatmap_1.8.1
[5] ComplexHeatmap_2.14.0 demuxmix_1.0.0
[7] RColorBrewer_1.1-3 cellhashR_1.0.3
[9] dittoSeq_1.10.0 speckle_0.99.7
[11] pheatmap_1.0.12 SeuratObject_4.1.3
[13] Seurat_4.3.0 scater_1.26.1
[15] scuttle_1.8.4 lubridate_1.9.2
[17] forcats_1.0.0 stringr_1.5.0
[19] purrr_1.0.1 readr_2.1.4
[21] tidyr_1.3.0 tibble_3.2.1
[23] tidyverse_2.0.0 DropletUtils_1.18.1
[25] SingleCellExperiment_1.20.1 SummarizedExperiment_1.28.0
[27] Biobase_2.58.0 GenomicRanges_1.50.2
[29] GenomeInfoDb_1.34.9 IRanges_2.32.0
[31] S4Vectors_0.36.2 BiocGenerics_0.44.0
[33] MatrixGenerics_1.10.0 matrixStats_0.63.0
[35] patchwork_1.1.2 cowplot_1.1.1
[37] ggplot2_3.4.2 janitor_2.2.0
[39] dplyr_1.1.2 BiocStyle_2.26.0
[41] here_1.0.1 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] scattermore_1.0 R.methodsS3_1.8.2
[3] knitr_1.42 irlba_2.3.5.1
[5] DelayedArray_0.24.0 R.utils_2.12.2
[7] data.table_1.14.8 RCurl_1.98-1.12
[9] doParallel_1.0.17 generics_0.1.3
[11] preprocessCore_1.61.0 ScaledMatrix_1.6.0
[13] callr_3.7.3 RANN_2.6.1
[15] future_1.32.0 tzdb_0.3.0
[17] spatstat.data_3.0-1 httpuv_1.6.11
[19] xfun_0.39 hms_1.1.3
[21] jquerylib_0.1.4 evaluate_0.21
[23] promises_1.2.0.1 fansi_1.0.4
[25] dendextend_1.17.1 igraph_1.4.2
[27] DBI_1.1.3 htmlwidgets_1.6.2
[29] spatstat.geom_3.2-1 ellipsis_0.3.2
[31] bookdown_0.34 deldir_1.0-6
[33] sparseMatrixStats_1.10.0 vctrs_0.6.2
[35] Cairo_1.6-0 ROCR_1.0-11
[37] abind_1.4-5 cachem_1.0.8
[39] withr_2.5.0 ggforce_0.4.1
[41] progressr_0.13.0 sctransform_0.3.5
[43] goftest_1.2-3 cluster_2.1.4
[45] lazyeval_0.2.2 crayon_1.5.2
[47] spatstat.explore_3.2-1 edgeR_3.40.2
[49] pkgconfig_2.0.3 labeling_0.4.2
[51] tweenr_2.0.2 nlme_3.1-162
[53] vipor_0.4.5 rlang_1.1.1
[55] globals_0.16.2 lifecycle_1.0.3
[57] miniUI_0.1.1.1 rsvd_1.0.5
[59] ggrastr_1.0.1 rprojroot_2.0.3
[61] polyclip_1.10-4 lmtest_0.9-40
[63] Rhdf5lib_1.20.0 zoo_1.8-12
[65] beeswarm_0.4.0 whisker_0.4.1
[67] ggridges_0.5.4 GlobalOptions_0.1.2
[69] processx_3.8.1 png_0.1-8
[71] rjson_0.2.21 bitops_1.0-7
[73] getPass_0.2-2 R.oo_1.25.0
[75] KernSmooth_2.23-21 rhdf5filters_1.10.1
[77] ggExtra_0.10.0 DelayedMatrixStats_1.20.0
[79] shape_1.4.6 parallelly_1.35.0
[81] spatstat.random_3.1-5 beachmat_2.14.2
[83] scales_1.2.1 magrittr_2.0.3
[85] plyr_1.8.8 ica_1.0-3
[87] zlibbioc_1.44.0 compiler_4.2.2
[89] dqrng_0.3.0 clue_0.3-64
[91] fitdistrplus_1.1-11 snakecase_0.11.0
[93] cli_3.6.1 XVector_0.38.0
[95] listenv_0.9.0 pbapply_1.7-0
[97] ps_1.7.5 MASS_7.3-60
[99] tidyselect_1.2.0 stringi_1.7.12
[101] highr_0.10 yaml_2.3.7
[103] BiocSingular_1.14.0 locfit_1.5-9.7
[105] ggrepel_0.9.3 sass_0.4.6
[107] tools_4.2.2 timechange_0.2.0
[109] future.apply_1.10.0 parallel_4.2.2
[111] circlize_0.4.15 rstudioapi_0.14
[113] foreach_1.5.2 git2r_0.32.0
[115] gridExtra_2.3 rmdformats_1.0.4
[117] farver_2.1.1 Rtsne_0.16
[119] digest_0.6.31 BiocManager_1.30.20
[121] shiny_1.7.4 Rcpp_1.0.10
[123] egg_0.4.5 later_1.3.1
[125] RcppAnnoy_0.0.20 httr_1.4.6
[127] naturalsort_0.1.3 colorspace_2.1-0
[129] fs_1.6.2 tensor_1.5
[131] reticulate_1.28 splines_4.2.2
[133] uwot_0.1.14 spatstat.utils_3.0-3
[135] sp_1.6-0 plotly_4.10.1
[137] xtable_1.8-4 jsonlite_1.8.4
[139] R6_2.5.1 pillar_1.9.0
[141] htmltools_0.5.5 mime_0.12
[143] glue_1.6.2 fastmap_1.1.1
[145] BiocParallel_1.32.6 BiocNeighbors_1.16.0
[147] codetools_0.2-19 utf8_1.2.3
[149] lattice_0.21-8 bslib_0.4.2
[151] spatstat.sparse_3.0-1 ggbeeswarm_0.7.2
[153] leiden_0.4.3 magick_2.7.4
[155] survival_3.5-5 limma_3.54.2
[157] rmarkdown_2.21 munsell_0.5.0
[159] GetoptLong_1.0.5 rhdf5_2.42.1
[161] GenomeInfoDbData_1.2.9 iterators_1.0.14
[163] HDF5Array_1.26.0 reshape2_1.4.4
[165] gtable_0.3.3