Gene set testing for Illumina HumanMethylation Arrays
Exploring the biases on EPIC arrays
Jovana Maksimovic, Alicia Oshlack and Belinda Phipson
June 23, 2020
Last updated: 2020-06-23
Checks: 7 0
Knit directory: methyl-geneset-testing/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200302) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 8bd9b75. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: code/.job/
Ignored: code/old/
Ignored: data/
Ignored: output/FDR-analysis/
Ignored: output/Fig-1A.pdf
Ignored: output/Fig-1B.pdf
Ignored: output/Fig-1C.pdf
Ignored: output/Fig-1D.pdf
Ignored: output/Fig-3A.pdf
Ignored: output/Fig-3B.pdf
Ignored: output/Fig-3C.pdf
Ignored: output/Fig-3D.pdf
Ignored: output/Fig-4A.pdf
Ignored: output/Fig-4B.pdf
Ignored: output/Fig-4C-G.pdf
Ignored: output/SFig-1A.pdf
Ignored: output/SFig-1B.pdf
Ignored: output/SFig-3A.pdf
Ignored: output/SFig-3B.pdf
Ignored: output/SFig-4B.pdf
Ignored: output/SFig-5B.pdf
Ignored: output/SFig-6A.pdf
Ignored: output/SFig-6B-F.pdf
Ignored: output/SFig-7A.pdf
Ignored: output/SFig-7B-F.pdf
Ignored: output/SFig-8A.pdf
Ignored: output/SFig-8B-F.pdf
Ignored: output/SFig-X1.pdf
Ignored: output/SFig-X2.pdf
Ignored: output/compare-methods/
Ignored: output/methylgsa-params/
Ignored: output/random-cpg-sims/
Unstaged changes:
Modified: analysis/exploreArrayBias450.Rmd
Modified: analysis/exploreData.Rmd
Modified: analysis/fdrAnalysis.Rmd
Modified: code/utility.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/exploreArrayBiasEPIC.Rmd) and HTML (docs/exploreArrayBiasEPIC.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 8bd9b75 | Jovana Maksimovic | 2020-06-23 | wflow_publish(c(“analysis/exploreArrayBiasEPIC.Rmd”)) |
| html | 25a3336 | Jovana Maksimovic | 2020-06-23 | Build site. |
| Rmd | 6d928b5 | Jovana Maksimovic | 2020-06-23 | wflow_publish(c(“analysis/exploreArrayBiasEPIC.Rmd”)) |
| Rmd | 823b793 | Jovana Maksimovic | 2020-06-16 | UPdated analyses with label translation and new colour palette. |
| html | 0f4cf3b | Jovana Maksimovic | 2020-06-16 | Build site. |
| Rmd | 136a9f0 | Jovana Maksimovic | 2020-06-16 | wflow_publish(“analysis/exploreArrayBiasEPIC.Rmd”) |
| html | 22f00e9 | Jovana Maksimovic | 2020-05-19 | Build site. |
| Rmd | 63b0011 | Jovana Maksimovic | 2020-05-19 | wflow_publish(c(“analysis/exploreArrayBias450.Rmd”, “analysis/exploreArrayBiasEPIC.Rmd”, |
| html | 358cd52 | JovMaksimovic | 2020-03-16 | Build site. |
| Rmd | 250d8ae | JovMaksimovic | 2020-03-16 | Comitting local changes. |
| Rmd | 24fb166 | Jovana Maksimovic | 2020-03-16 | Adding Rmd file changes. |
| Rmd | d7cd66e | Jovana Maksimovic | 2020-03-02 | Initial Commit |
library(here)
library(glue)
library(minfi)
library(IlluminaHumanMethylationEPICanno.ilm10b4.hg19)
library(IlluminaHumanMethylation450kanno.ilmn12.hg19)
library(missMethyl)
library(org.Hs.eg.db)
library(GO.db)
library(patchwork)
library(grid)
library(ggplot2)
library(tibble)
library(dplyr)
source(here("code/utility.R"))Array properties
Get the array annotation data.
ann <- loadAnnotation(arrayType="EPIC")Associate CpGs to genes (ENTREZ ID) using the Illumina annotation information.
flatAnn <- loadFlatAnnotation(ann)The number of CpGs annotated to a gene is highly variable.
numCpgsPerGene <- as.vector(table(flatAnn$entrezid))
summary(numCpgsPerGene) Min. 1st Qu. Median Mean 3rd Qu. Max.
1.00 10.00 20.00 27.25 33.00 1485.00 Figure 1A
View
dat <- data.frame(table(table(flatAnn$entrezid)))
numCpgsPerGene <- as.vector(table(flatAnn$entrezid))
med <- median(numCpgsPerGene)
mod <- getMode(numCpgsPerGene)
p <- ggplot(dat, aes(x=Var1, y=Freq)) +
geom_segment(aes(x=Var1, xend=Var1, y=0, yend=Freq), color="darkgrey") +
geom_point( color="black", size=1) +
geom_vline(xintercept = med, linetype = "dashed", color = "red") +
annotate("text", x = med + 2, label=glue("median = {med}"), y=720, colour="red",
size = 3, hjust="left") +
geom_vline(xintercept = mod, linetype = "dashed", color = "red") +
annotate("text", x = mod + 2, label=glue("mode = {mod}"), y=780, colour="red",
size = 3, hjust="left") +
scale_x_discrete(breaks = c(1,10,20,30,45,55,70,80,90,100,120,135,150,160,180,
200,215,250,330,429,1485)) +
theme(axis.text.x=element_text(angle=45, hjust=1)) +
xlab("No. CpGs per gene") +
ylab("Frequency")
p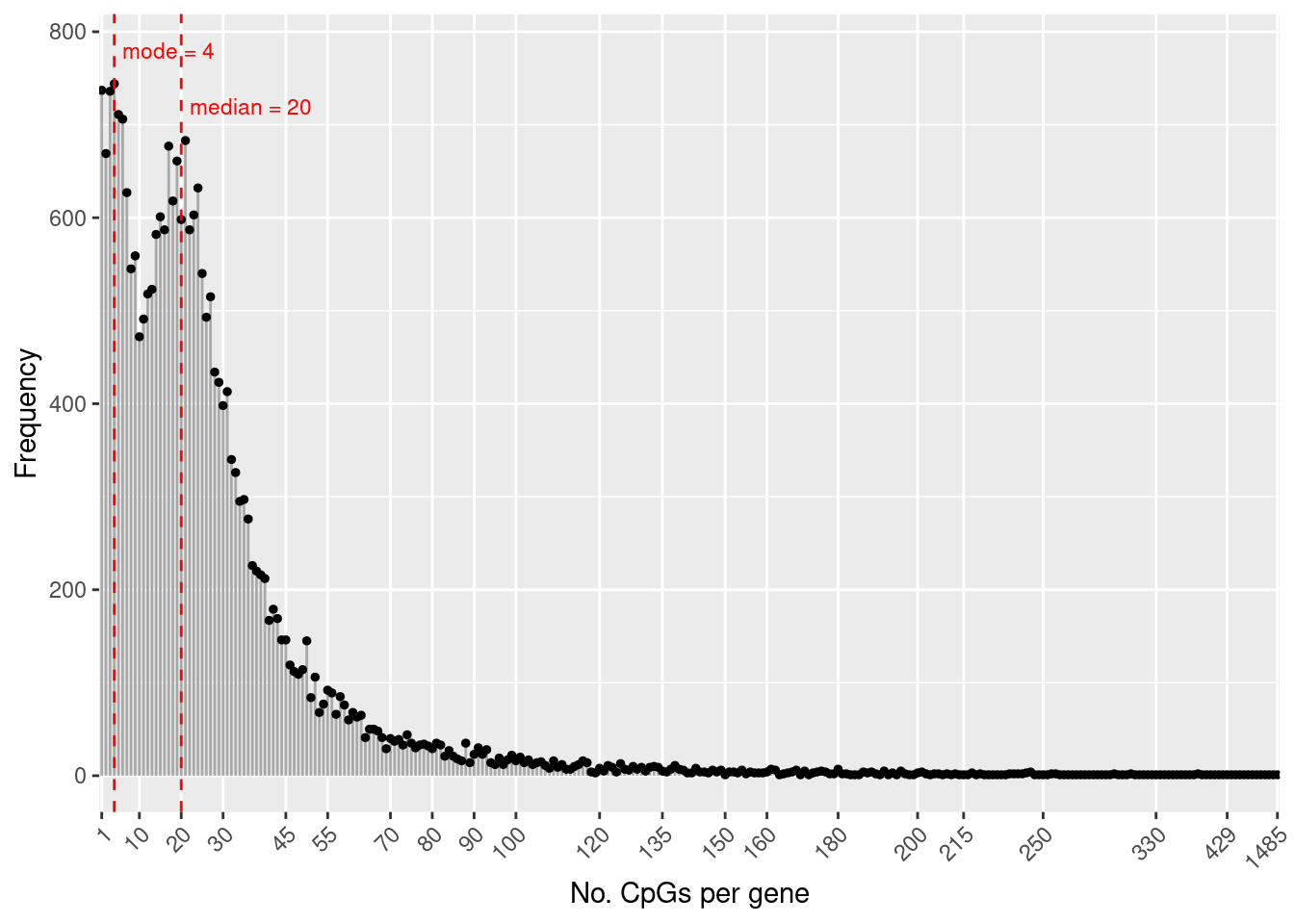
Create PDF
fig <- here("output/Fig-1A.pdf")
pdf(file = fig, width = 9)
p
dev.off()Figure 1D
View
The number of genes a CpG maps to can also vary, although the majority of CpGs only map to one gene.
dat <- data.frame(table(table(flatAnn$cpg)))
dat$Split <- ifelse(dat$Freq > 2500, "A",
ifelse(dat$Freq < 2500 & dat$Freq > 65,"B","C"))
a <- ggplot(dat[dat$Split == "A",], aes(x=Var1, y=Freq)) +
geom_bar(stat = "identity") +
theme(axis.title.x = element_blank(),
axis.text.y=element_text(angle=90, hjust=0.5)) +
ylab("Frequency")
b <- ggplot(dat[dat$Split == "B",], aes(x=Var1, y=Freq)) +
geom_bar(stat = "identity") +
theme(axis.title.x = element_blank(),
axis.title.y = element_blank())
c <- ggplot(dat[dat$Split == "C",], aes(x=Var1, y=Freq)) +
geom_bar(stat = "identity") +
theme(axis.title.x=element_blank(),
axis.title.y = element_blank())
p <- a + b + c + plot_layout(widths = c(1,2,6)) +
plot_annotation(caption = "No. genes mapped to a CpG",
theme = theme(plot.caption = element_text(hjust = 0.5,
size = 12)))
p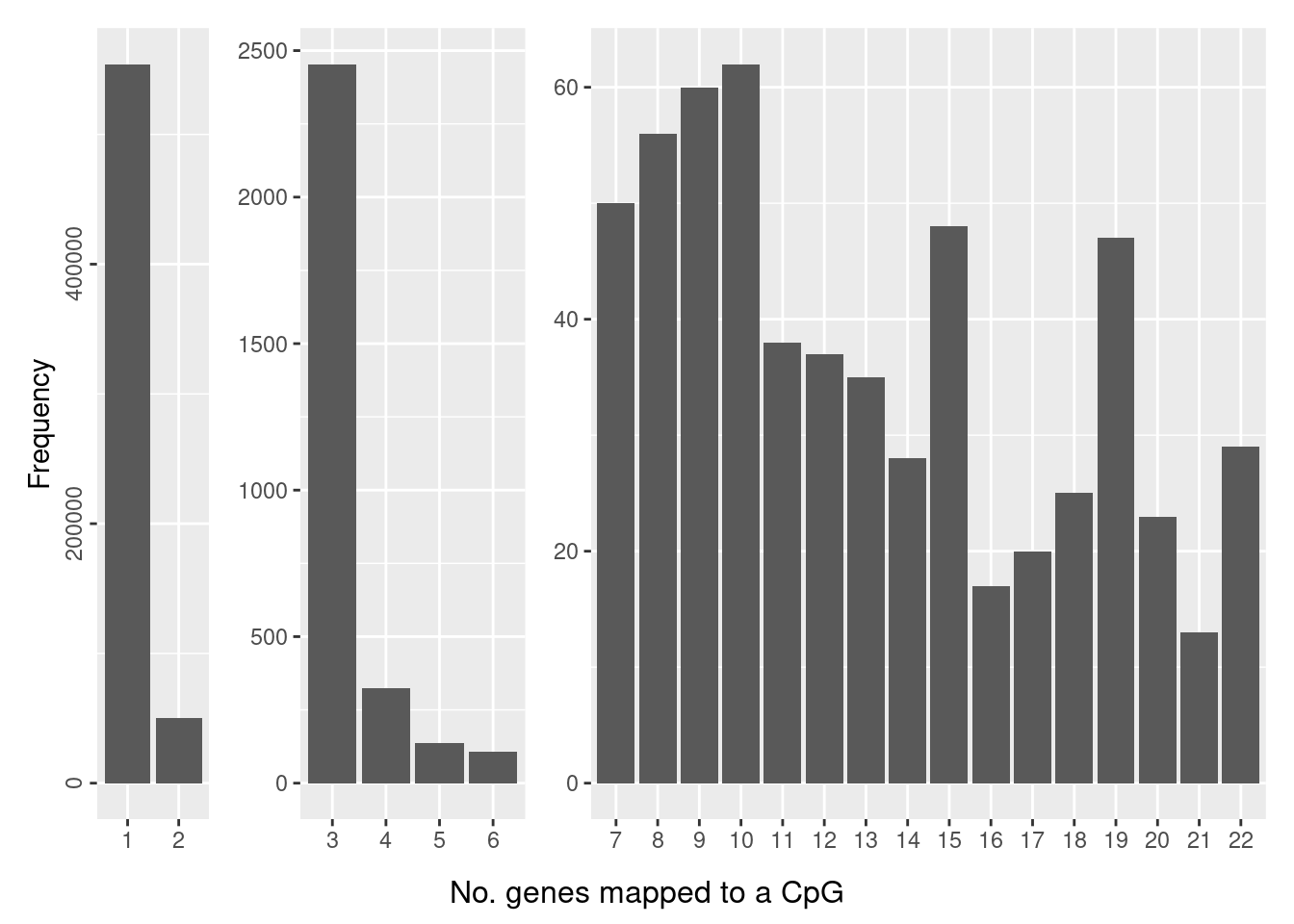
Create PDF
fig <- here("output/Fig-1D.pdf")
pdf(file = fig, width = 9)
p
dev.off()Explore the distribution of the average number of CpGs per gene, per GO category.
Figure 1C
View
ann450 <- loadAnnotation("450k")
flatAnn450 <- loadFlatAnnotation(ann450)
cpgEgGoEPIC <- cpgsEgGoFreqs(flatAnn)
cpgEgGoEPIC$Array <- "EPIC"
cpgEgGo450 <- cpgsEgGoFreqs(flatAnn450)
cpgEgGo450$Array <- "450k"
cpgEgGoEPIC %>% bind_rows(cpgEgGo450) %>%
group_by(Array, GO) %>%
summarise(avg = mean(Freq)) -> datSumWarning in bind_rows_(x, .id): Unequal factor levels: coercing to characterWarning in bind_rows_(x, .id): binding character and factor vector, coercing
into character vector
Warning in bind_rows_(x, .id): binding character and factor vector, coercing
into character vectormaxEPIC <- max(datSum$avg[datSum$Array == "EPIC"])
max450 <- max(datSum$avg[datSum$Array == "450k"])
bw <- 2
pal <- paletteer::paletteer_d("RColorBrewer::Set1", 2)
platform <- c("EPIC" = pal[1], "450k" = pal[2])
p <- ggplot(datSum, aes(x=avg)) +
geom_histogram(dat = subset(datSum, Array == "450k"),
alpha = 0.5, aes(fill = "450k"), binwidth = bw) +
geom_histogram(dat = subset(datSum, Array == "EPIC"),
alpha = 0.5, aes(fill = "EPIC"), binwidth = bw) +
geom_vline(xintercept = maxEPIC, linetype="dashed", colour = pal[1]) +
geom_vline(xintercept = max450, linetype="dashed", colour = pal[2]) +
labs(x="Mean no. CpGs per gene per GO category", y = "Frequency",
fill = "Platform") +
annotate("text", x = maxEPIC - 4, label=glue("Max. EPIC = {maxEPIC}"),
y = 1500, colour=pal[1], size = 2.5, hjust="right") +
annotate("text", x = max450 - 4, label=glue("Max. 450k = {max450}"),
y = 2500, colour=pal[2], size = 2.5, hjust="right") +
scale_fill_manual(values = platform)
p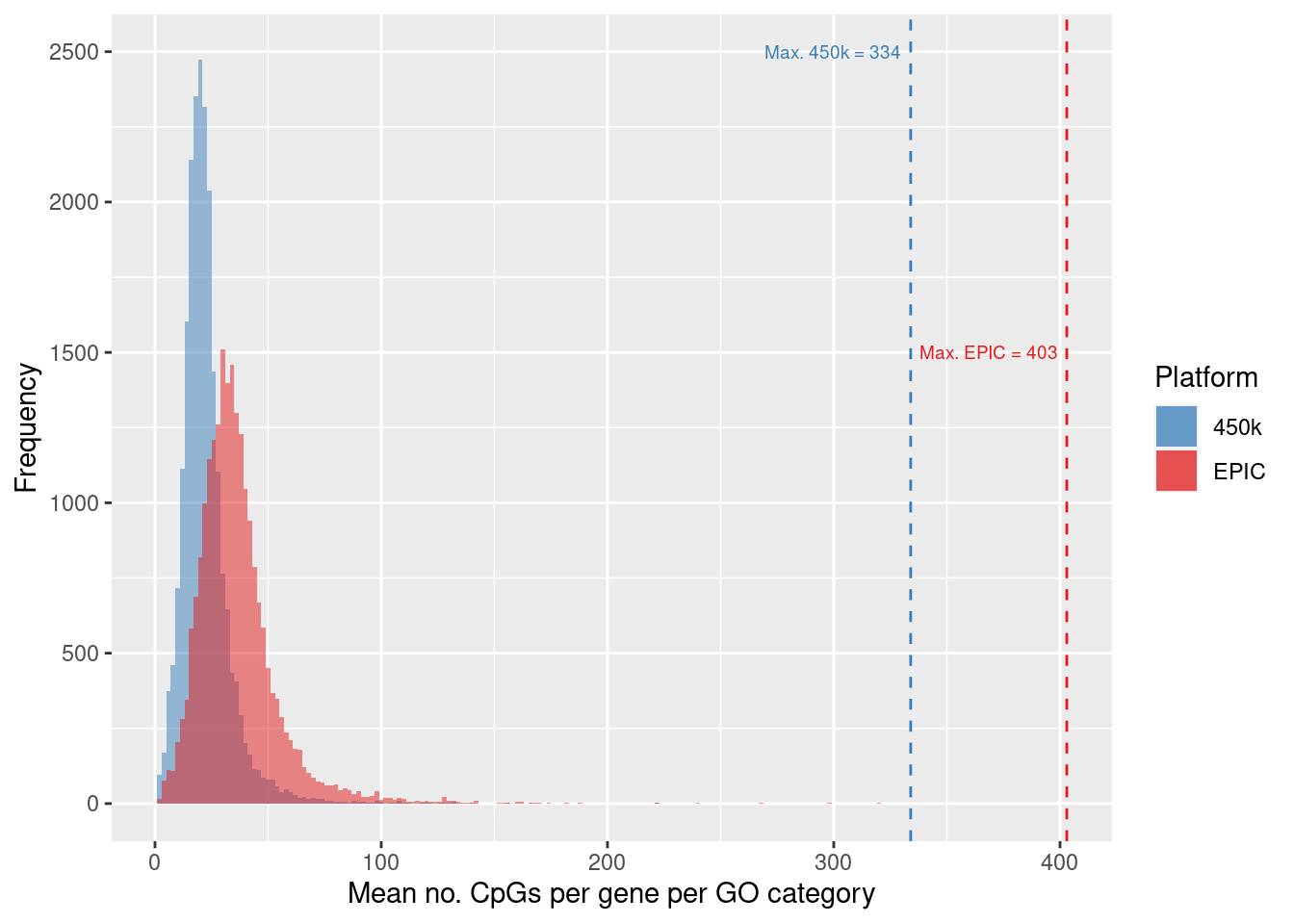
Create PDF
fig <- here("output/Fig-1C.pdf")
pdf(file = fig, width = 9)
p
dev.off()Null simulations
Null simulation strategy: randomly select 50, 100, 500, 1000, 5000 and 10000 sets of CpGs and perform GO testing on each set 100 times, with and without adjusting for the various biases on the array.
The code used for the simulation can be found in /oshlack_lab/jovana.maksimovic/research/methyl-geneset-testing/code/randCpgSim.R.
Figure 3A
View
The following boxplots show what proportion of the 100 simulations, at each level of CpGs sampled, had a raw p-value less than 0.05. This gives us an idea of the false discovery rate with and without adjustment for the number of CpGs annotated to a gene.
dat <- readRDS(here("output/random-cpg-sims/EPIC.rds"))
dat %>% filter(method %in% names(dict)) %>%
mutate(method = unname(dict[method])) %>%
group_by(simNo, noCpgs, method) %>%
summarise(pSig = sum(P.DE < 0.05)/length(P.DE)) -> sigDat
p <- ggplot(sigDat, aes(x=noCpgs, y=pSig, fill=method)) +
geom_violin() +
geom_hline(yintercept=0.05, linetype="dashed", color = "red") +
labs(y="Prop. GO cat. with p-value < 0.05", x="No. randomly sampled CpGs",
fill="Method") +
scale_fill_manual(values = methodCols)
p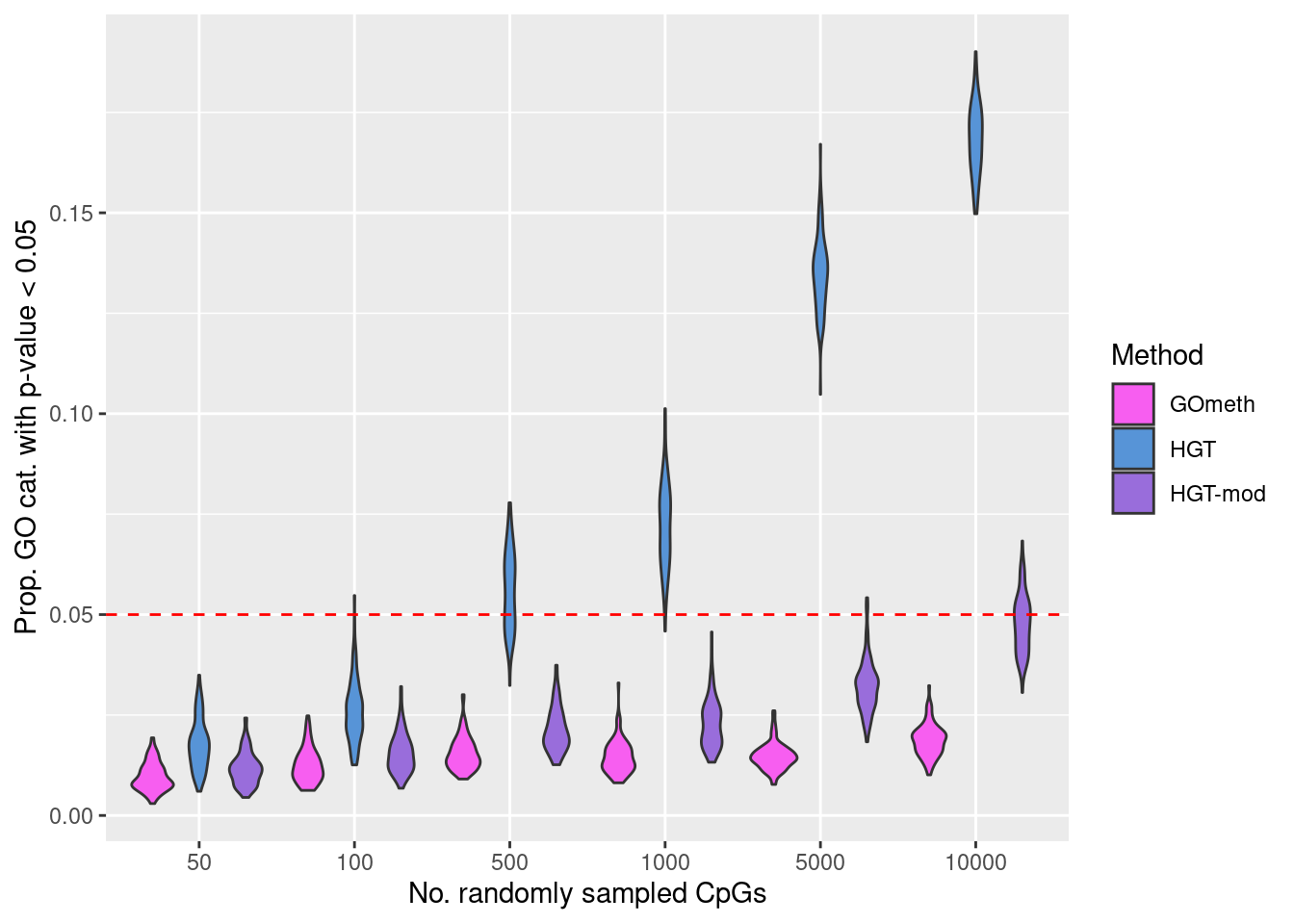
Create PDF
fig <- here("output/Fig-3A.pdf")
pdf(file = fig, width = 9)
p
dev.off()QQ plots of randomly selected simulations at each level of CpGs sampled.
set.seed(42)
s <- sample(1:100, 3)
dat %>% filter(simNo %in% s) %>%
filter(method %in% names(dict)) %>%
mutate(method = unname(dict[method])) %>%
arrange(simNo, noCpgs, method, P.DE) %>%
group_by(simNo, noCpgs, method) %>%
mutate(exp = 1:n()/n()) -> subDat
p <- ggplot(subDat, aes(x=-log10(exp), y=-log10(P.DE), color=method)) +
geom_point(shape = 1, size = 0.5) +
facet_grid(noCpgs ~ simNo, scales = "free_y") +
scale_color_manual(values = methodCols)
p + geom_line(aes(x=-log10(exp), y=-log10(exp)),
linetype="dashed", color = "black") +
labs(y=expression(Observed~~-log[10](italic(p))),
x=expression(Expected~~-log[10](italic(p))),
color="Method") +
theme(legend.position="bottom", strip.text.x = element_blank())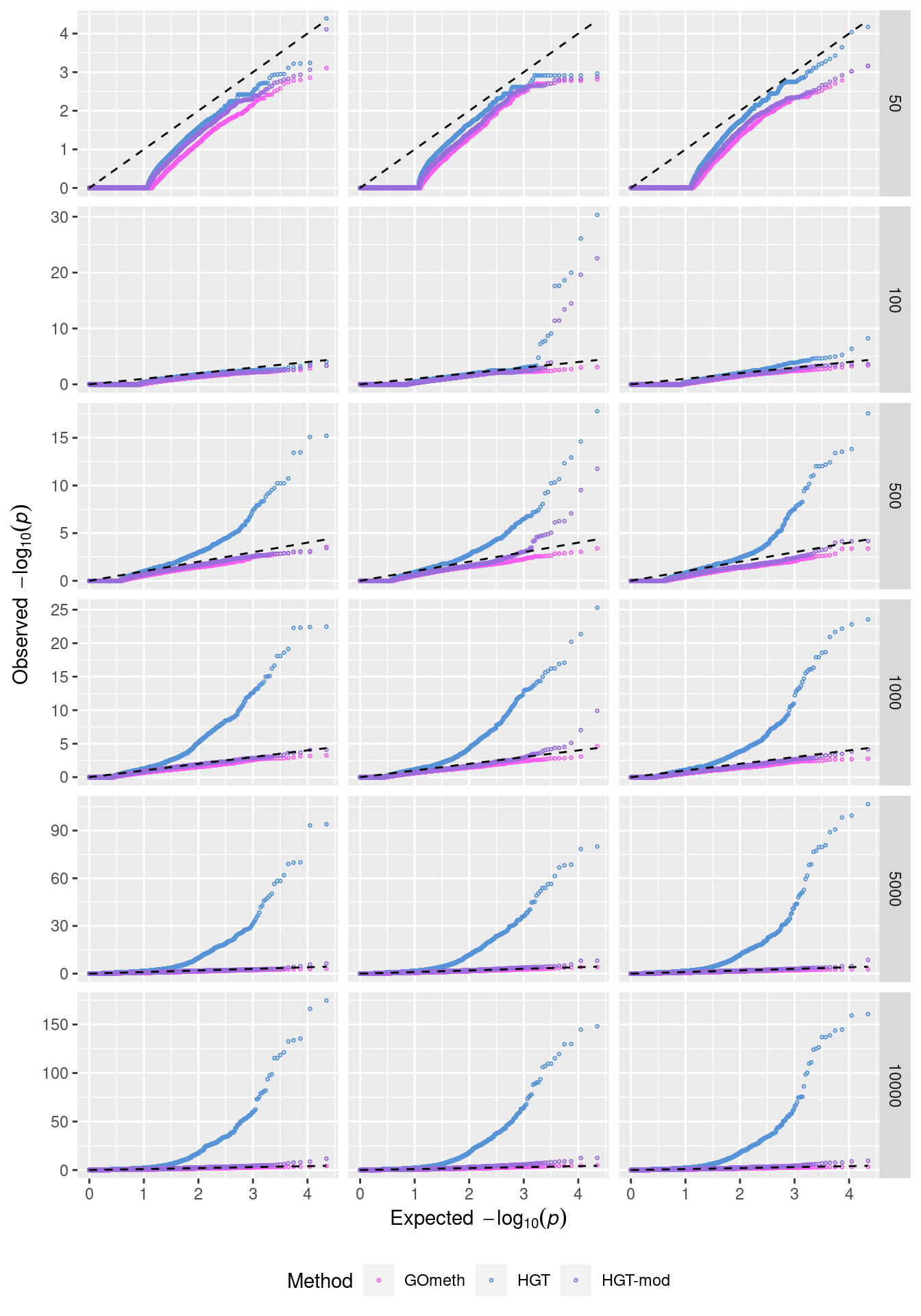
Figure 3A
View
Explore the relationship between the median, average number of CpGs, per gene, per GO category and the various sources of bias on the array.
goFreq <- as_tibble(unique(cpgEgGoEPIC[,c("GO","Freq")]))
dat %>% filter(method %in% names(dict)) %>%
mutate(method = unname(dict[method])) %>%
filter(P.DE < 0.05) %>%
inner_join(goFreq, by=c("GO" = "GO")) %>%
group_by(simNo, noCpgs, method, GO) %>%
summarise(avgFreq=mean(Freq)) %>%
group_by(simNo, noCpgs, method) %>%
summarise(medAvgFreq=median(avgFreq)) -> medAvgDat
p <- ggplot(medAvgDat, aes(x=noCpgs, y=medAvgFreq, fill=method)) +
geom_violin() +
stat_summary(geom="point", size=1, color="white", position = position_dodge(0.9),
show.legend = FALSE, fun = median) +
labs(y="Median avg. no. CpGs/gene/GO cat.",
x="No. randomly sampled CpGs",
fill="Method") +
scale_fill_manual(values = methodCols)
p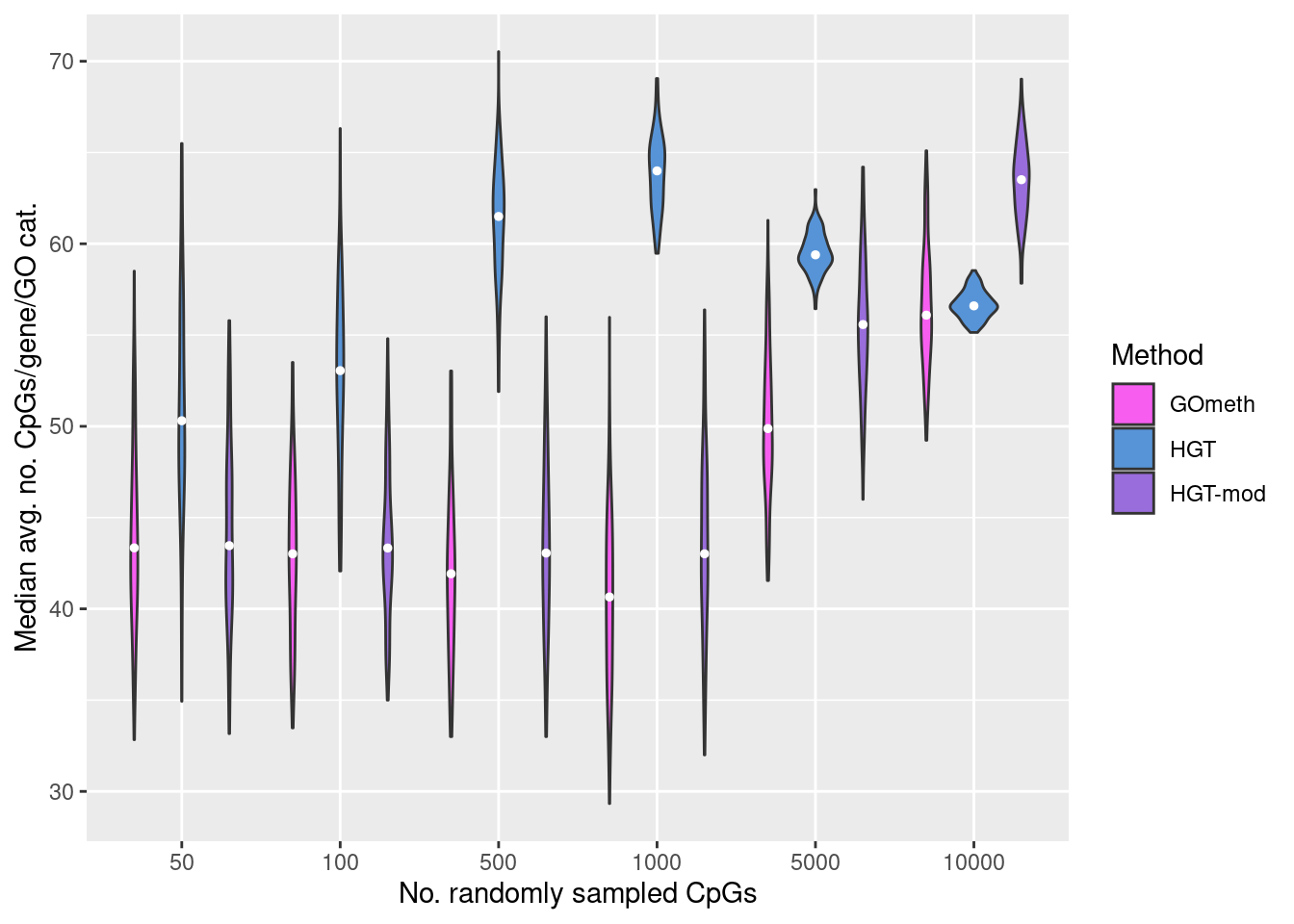
| Version | Author | Date |
|---|---|---|
| 25a3336 | Jovana Maksimovic | 2020-06-23 |
Create PDF
fig <- here("output/Fig-3B.pdf")
pdf(file = fig, width = 9)
p
dev.off()
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS: /config/RStudio/R/3.6.1/lib64/R/lib/libRblas.so
LAPACK: /config/RStudio/R/3.6.1/lib64/R/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_AU.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_AU.UTF-8 LC_COLLATE=en_AU.UTF-8
[5] LC_MONETARY=en_AU.UTF-8 LC_MESSAGES=en_AU.UTF-8
[7] LC_PAPER=en_AU.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_AU.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] grid stats4 parallel stats graphics grDevices utils
[8] datasets methods base
other attached packages:
[1] dplyr_0.8.5
[2] tibble_3.0.1
[3] ggplot2_3.3.0
[4] patchwork_1.0.0
[5] GO.db_3.8.2
[6] org.Hs.eg.db_3.8.2
[7] AnnotationDbi_1.46.1
[8] missMethyl_1.20.4
[9] IlluminaHumanMethylation450kanno.ilmn12.hg19_0.6.0
[10] IlluminaHumanMethylationEPICanno.ilm10b4.hg19_0.6.0
[11] minfi_1.32.0
[12] bumphunter_1.26.0
[13] locfit_1.5-9.1
[14] iterators_1.0.12
[15] foreach_1.5.0
[16] Biostrings_2.54.0
[17] XVector_0.24.0
[18] SummarizedExperiment_1.16.1
[19] DelayedArray_0.12.3
[20] BiocParallel_1.20.1
[21] matrixStats_0.56.0
[22] Biobase_2.46.0
[23] GenomicRanges_1.38.0
[24] GenomeInfoDb_1.22.1
[25] IRanges_2.20.2
[26] S4Vectors_0.24.4
[27] BiocGenerics_0.32.0
[28] glue_1.4.1
[29] here_0.1
[30] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] backports_1.1.7
[2] BiocFileCache_1.10.2
[3] plyr_1.8.6
[4] splines_3.6.1
[5] digest_0.6.25
[6] htmltools_0.4.0
[7] jcolors_0.0.4
[8] magrittr_1.5
[9] memoise_1.1.0
[10] cluster_2.1.0
[11] paletteer_1.1.0
[12] limma_3.42.2
[13] readr_1.3.1
[14] annotate_1.62.0
[15] askpass_1.1
[16] siggenes_1.60.0
[17] prettyunits_1.1.1
[18] colorspace_1.4-1
[19] blob_1.2.0
[20] rappdirs_0.3.1
[21] BiasedUrn_1.07
[22] xfun_0.14
[23] prismatic_0.2.0
[24] crayon_1.3.4
[25] RCurl_1.95-4.12
[26] genefilter_1.68.0
[27] GEOquery_2.54.1
[28] IlluminaHumanMethylationEPICmanifest_0.3.0
[29] survival_2.44-1.1
[30] palr_0.2.0
[31] ruv_0.9.7.1
[32] pals_1.6
[33] gtable_0.3.0
[34] registry_0.5-1
[35] zlibbioc_1.30.0
[36] scico_1.1.0
[37] Rhdf5lib_1.6.1
[38] maps_3.3.0
[39] HDF5Array_1.14.4
[40] scales_1.1.1
[41] DBI_1.0.0
[42] rngtools_1.4
[43] bibtex_0.4.2
[44] Rcpp_1.0.4.6
[45] viridisLite_0.3.0
[46] xtable_1.8-4
[47] progress_1.2.2
[48] mapproj_1.2.6
[49] bit_1.1-14
[50] mclust_5.4.6
[51] preprocessCore_1.48.0
[52] httr_1.4.1
[53] RColorBrewer_1.1-2
[54] ellipsis_0.3.1
[55] farver_2.0.3
[56] pkgconfig_2.0.3
[57] reshape_0.8.8
[58] XML_3.98-1.20
[59] dbplyr_1.4.2
[60] reshape2_1.4.3
[61] labeling_0.3
[62] tidyselect_1.1.0
[63] rlang_0.4.6
[64] later_1.0.0
[65] munsell_0.5.0
[66] tools_3.6.1
[67] RSQLite_2.1.2
[68] evaluate_0.14
[69] stringr_1.4.0
[70] yaml_2.2.1
[71] rematch2_2.1.0
[72] knitr_1.28
[73] bit64_0.9-7
[74] fs_1.4.1
[75] beanplot_1.2
[76] scrime_1.3.5
[77] methylumi_2.30.0
[78] purrr_0.3.4
[79] nlme_3.1-147
[80] doRNG_1.7.1
[81] whisker_0.4
[82] nor1mix_1.3-0
[83] xml2_1.3.2
[84] biomaRt_2.42.1
[85] compiler_3.6.1
[86] curl_4.3
[87] statmod_1.4.32
[88] stringi_1.4.6
[89] GenomicFeatures_1.36.4
[90] lattice_0.20-41
[91] Matrix_1.2-18
[92] IlluminaHumanMethylation450kmanifest_0.4.0
[93] multtest_2.40.0
[94] vctrs_0.3.0
[95] pillar_1.4.4
[96] lifecycle_0.2.0
[97] data.table_1.12.8
[98] bitops_1.0-6
[99] httpuv_1.5.2
[100] rtracklayer_1.44.4
[101] oompaBase_3.2.9
[102] R6_2.4.1
[103] promises_1.1.0
[104] gridExtra_2.3
[105] codetools_0.2-16
[106] dichromat_2.0-0
[107] MASS_7.3-51.6
[108] assertthat_0.2.1
[109] rhdf5_2.30.1
[110] openssl_1.4.1
[111] pkgmaker_0.27
[112] rprojroot_1.3-2
[113] withr_2.2.0
[114] GenomicAlignments_1.20.1
[115] Rsamtools_2.0.1
[116] GenomeInfoDbData_1.2.1
[117] hms_0.5.3
[118] quadprog_1.5-8
[119] tidyr_1.1.0
[120] base64_2.0
[121] rmarkdown_2.1
[122] DelayedMatrixStats_1.8.0
[123] illuminaio_0.28.0
[124] git2r_0.27.1