Figure 5
Jovana Maksimovic
January 08, 2025
Last updated: 2025-01-08
Checks: 7 0
Knit directory: paed-inflammation-CITEseq/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20240216) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 7da3393. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/obsolete/
Ignored: data/C133_Neeland_batch1/
Ignored: data/C133_Neeland_merged/
Ignored: output/dge_analysis/obsolete/
Ignored: renv/library/
Ignored: renv/staging/
Untracked files:
Untracked: broad_markers_seurat.csv
Untracked: code/background_job.R
Untracked: code/reverse_modifier_severity_comparisons.sh
Untracked: data/intermediate_objects/CD4 T cells.CF_samples.fit.rds
Untracked: data/intermediate_objects/CD4 T cells.all_samples.fit.rds
Untracked: data/intermediate_objects/CD8 T cells.CF_samples.fit.rds
Untracked: data/intermediate_objects/CD8 T cells.all_samples.fit.rds
Untracked: data/intermediate_objects/DC cells.CF_samples.fit.rds
Untracked: data/intermediate_objects/DC cells.all_samples.fit.rds
Unstaged changes:
Modified: .gitignore
Modified: analysis/06.0_azimuth_annotation.Rmd
Modified: analysis/09.0_integrate_cluster_macro_cells.Rmd
Modified: analysis/13.1_DGE_analysis_macro-alveolar.Rmd
Modified: analysis/13.7_DGE_analysis_macro-proliferating.Rmd
Modified: analysis/99.0_Figure_4.Rmd
Modified: code/utility.R
Modified: data/intermediate_objects/macro-APOC2+.CF_samples.fit.rds
Modified: data/intermediate_objects/macro-APOC2+.all_samples.fit.rds
Modified: data/intermediate_objects/macro-CCL.CF_samples.fit.rds
Modified: data/intermediate_objects/macro-CCL.all_samples.fit.rds
Modified: data/intermediate_objects/macro-IFI27.CF_samples.fit.rds
Modified: data/intermediate_objects/macro-IFI27.all_samples.fit.rds
Modified: data/intermediate_objects/macro-alveolar.CF_samples.fit.rds
Modified: data/intermediate_objects/macro-alveolar.all_samples.fit.rds
Modified: data/intermediate_objects/macro-lipid.CF_samples.fit.rds
Modified: data/intermediate_objects/macro-lipid.all_samples.fit.rds
Modified: data/intermediate_objects/macro-monocyte-derived.CF_samples.fit.rds
Modified: data/intermediate_objects/macro-monocyte-derived.all_samples.fit.rds
Modified: data/intermediate_objects/macro-proliferating.CF_samples.fit.rds
Modified: data/intermediate_objects/macro-proliferating.all_samples.fit.rds
Modified: data/intermediate_objects/macrophages.CF_samples.fit.rds
Modified: data/intermediate_objects/macrophages.all_samples.fit.rds
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/99.0_Figure_5.Rmd) and
HTML (docs/99.0_Figure_5.html) files. If you’ve configured
a remote Git repository (see ?wflow_git_remote), click on
the hyperlinks in the table below to view the files as they were in that
past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 7da3393 | Jovana Maksimovic | 2025-01-08 | wflow_publish("analysis/99.0_Figure_5.Rmd") |
| html | bb42dbf | Jovana Maksimovic | 2025-01-06 | Build site. |
| Rmd | a30b829 | Jovana Maksimovic | 2025-01-06 | wflow_publish("analysis/99.0_Figure_5.Rmd") |
Load libraries.
suppressPackageStartupMessages({
library(SingleCellExperiment)
library(edgeR)
library(tidyverse)
library(ggplot2)
library(Seurat)
library(glmGamPoi)
library(dittoSeq)
library(here)
library(clustree)
library(patchwork)
library(AnnotationDbi)
library(org.Hs.eg.db)
library(glue)
library(speckle)
library(tidyHeatmap)
library(paletteer)
library(dsb)
library(ggh4x)
library(readxl)
})
source(here("code/utility.R"))Load data
files <- list.files(here("data/C133_Neeland_merged"),
pattern = "C133_Neeland_full_clean.*(macrophages|t_cells|other_cells)_annotated_diet.SEU.rds",
full.names = TRUE)
seuLst <- lapply(files[2:4], function(f) readRDS(f))
seu <- merge(seuLst[[1]],
y = c(seuLst[[2]],
seuLst[[3]]))
seuAn object of class Seurat
21568 features across 194407 samples within 1 assay
Active assay: RNA (21568 features, 0 variable features) used (Mb) gc trigger (Mb) max used (Mb)
Ncells 12101517 646.3 20150177 1076.2 13844582 739.4
Vcells 1354183553 10331.7 3693778851 28181.3 3551517295 27096.0Create sample meta data table.
props <- getTransformedProps(clusters = seu$ann_level_3,
sample = seu$sample.id, transform="asin")
seu@meta.data %>%
dplyr::select(sample.id,
Participant,
Disease,
Treatment,
Severity,
Group,
Group_severity,
Batch,
Age,
Sex) %>%
left_join(props$Counts %>%
data.frame %>%
group_by(sample) %>%
summarise(ncells = sum(Freq)),
by = c("sample.id" = "sample")) %>%
distinct() -> info
head(info) %>% knitr::kable()| sample.id | Participant | Disease | Treatment | Severity | Group | Group_severity | Batch | Age | Sex | ncells |
|---|---|---|---|---|---|---|---|---|---|---|
| sample_33.1 | sample_33 | CF | treated (ivacaftor) | severe | CF.IVA | CF.IVA.S | 1 | 5.950685 | M | 2139 |
| sample_25.1 | sample_25 | CF | untreated | severe | CF.NO_MOD | CF.NO_MOD.S | 1 | 4.910000 | F | 3272 |
| sample_29.1 | sample_29 | CF | untreated | severe | CF.NO_MOD | CF.NO_MOD.S | 1 | 5.989041 | F | 1568 |
| sample_27.1 | sample_27 | CF | treated (ivacaftor) | mild | CF.IVA | CF.IVA.M | 1 | 4.917808 | M | 2467 |
| sample_32.1 | sample_32 | CF | untreated | mild | CF.NO_MOD | CF.NO_MOD.M | 1 | 5.926027 | F | 2963 |
| sample_26.1 | sample_26 | CF | untreated | mild | CF.NO_MOD | CF.NO_MOD.M | 1 | 5.049315 | M | 2040 |
Prepare figure panels
props <- getTransformedProps(clusters = seu$ann_level_3[!str_detect(seu$ann_level_3, "macro")],
sample = seu$sample.id[!str_detect(seu$ann_level_3, "macro")], transform="asin")
# IVA 5A: NK-T cells, CD4 T-IFN, monocytes
props$Proportions %>% data.frame %>%
left_join(info,
by = c("sample" = "sample.id")) %>%
dplyr::filter(Group %in% c("CF.IVA", "CF.NO_MOD", "NON_CF.CTRL"),
clusters %in% c("NK-T cells",
"CD4 T-IFN",
"monocytes")) -> dat
sig_names <- as_labeller(
c("CD4 T-IFN" = "CD4 T-IFN",
"monocytes" = "monocytes",
"NK-T cells" = "NK-T cells"))
pal <- RColorBrewer::brewer.pal(8, "Set2")[1:4]
names(pal) <- c("CF.IVA","CF.LUMA_IVA","CF.NO_MOD","NON_CF.CTRL")
dat %>%
dplyr::filter(Group %in% c("CF.IVA", "CF.NO_MOD"),
clusters %in% "NK-T cells") %>%
ggplot(aes(x = Group,
y = Freq,
colour = Group)) +
geom_jitter(stat = "identity",
width = 0.15,
size = 1) +
theme_classic() +
theme(axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
axis.title.x = element_blank(),
axis.text.y = element_text(7),
legend.position = "bottom",
legend.direction = "horizontal",
strip.text = element_text(size = 8)) +
labs(x = "Group", y = "Proportion") +
facet_wrap(~clusters, scales = "free_y", ncol = 4,
labeller = sig_names) +
stat_summary(
geom = "point",
fun.y = "mean",
col = "black",
shape = "_",
size = 10) +
scale_color_manual(values = pal) -> p1
dat %>%
dplyr::filter(Group %in% c("CF.IVA", "NON_CF.CTRL"),
clusters %in% c("CD4 T-IFN",
"monocytes")) %>%
ggplot(aes(x = Group,
y = Freq,
colour = Group)) +
geom_jitter(stat = "identity",
width = 0.15,
size = 1) +
theme_classic() +
theme(axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
axis.title.x = element_blank(),
axis.text.y = element_text(7),
legend.position = "bottom",
legend.direction = "horizontal",
strip.text = element_text(size = 8)) +
labs(x = "Group", y = "Proportion") +
facet_wrap(~clusters, scales = "free_y", ncol = 4,
labeller = sig_names) +
stat_summary(
geom = "point",
fun.y = "mean",
col = "black",
shape = "_",
size = 10) +
scale_color_manual(values = pal) -> p2
layout <- "ABB"
iva_props <- (p1 +
(p2 + theme(axis.title.y = element_blank()))) +
plot_layout(design = layout) &
theme(axis.text.y = element_text(size = 6,
angle = 90,
hjust = 0.5),
legend.text = element_text(size = 8),
legend.key.spacing = unit(0, "lines"),
legend.margin = margin(-0.5,0,0,0, unit="lines"),
legend.direction = "horizontal") &
guides(colour = guide_legend(title.position="top", title.hjust = 0.5))
iva_props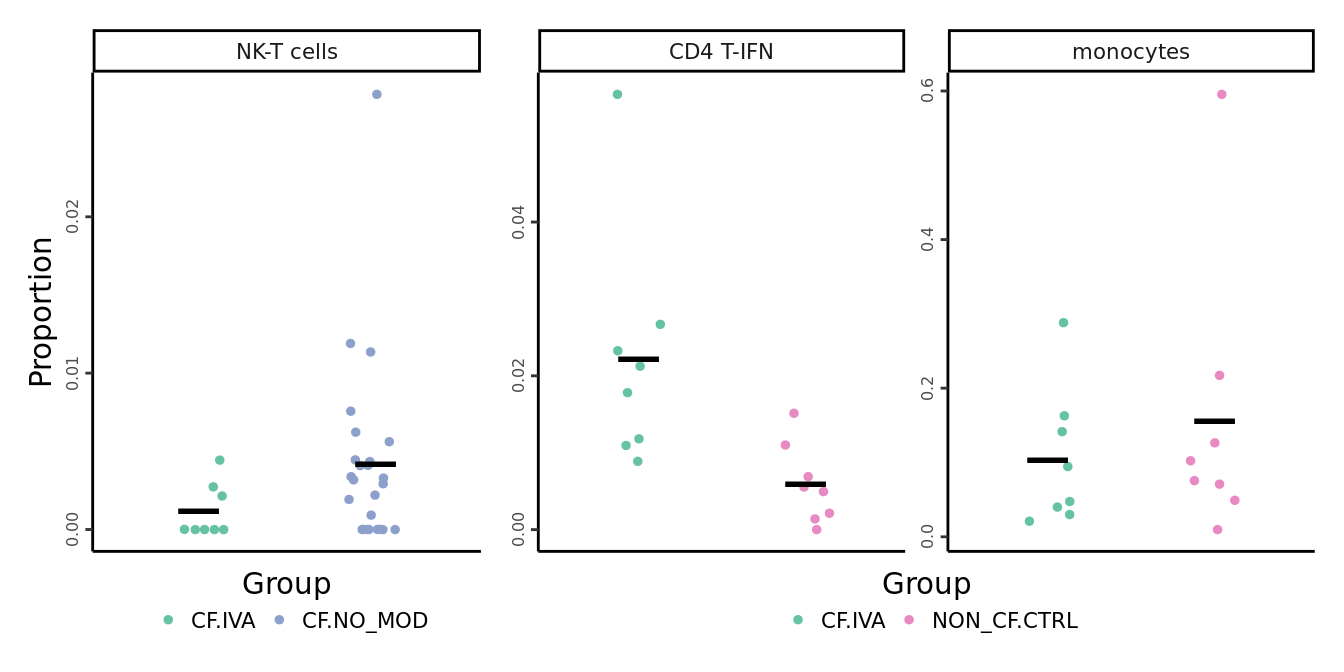
| Version | Author | Date |
|---|---|---|
| bb42dbf | Jovana Maksimovic | 2025-01-06 |
# LUMA-IVA 5B: NK-T cells, CD4 Tregs, monocytes, CD4 T-IFN, macro-CCL18
props <- getTransformedProps(clusters = seu$ann_level_3[!str_detect(seu$ann_level_3, "macro")],
sample = seu$sample.id[!str_detect(seu$ann_level_3, "macro")], transform="asin")
props$Proportions %>% data.frame %>%
left_join(info,
by = c("sample" = "sample.id")) %>%
dplyr::filter(Group %in% c("CF.LUMA_IVA", "CF.NO_MOD", "NON_CF.CTRL"),
clusters %in% c("NK-T cells",
"CD4 T-reg",
"CD4 T-IFN",
"monocytes")) -> dat
sig_names <- as_labeller(
c("CD4 T-IFN" = "CD4 T-IFN",
"CD4 T-reg" = "CD4 T-reg",
"monocytes" = "monocytes",
"macro-CCL18" = "macro-CCL18",
"NK-T cells" = "NK-T cells"))
dat %>%
dplyr::filter(Group %in% c("CF.LUMA_IVA", "CF.NO_MOD"),
clusters %in% "NK-T cells") %>%
ggplot(aes(x = Group,
y = Freq,
colour = Group)) +
geom_jitter(stat = "identity",
width = 0.15,
size = 1,
show.legend = FALSE) +
theme_classic() +
theme(axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
axis.title.x = element_blank(),
axis.text.y = element_text(7),
legend.position = "bottom",
legend.direction = "horizontal",
strip.text = element_text(size = 8)) +
labs(x = "Group", y = "Proportion") +
facet_wrap(~clusters, scales = "free_y", ncol = 4,
labeller = sig_names) +
stat_summary(
geom = "point",
fun.y = "mean",
col = "black",
shape = "_",
size = 10) +
scale_color_manual(values = pal) -> p1
dat %>%
dplyr::filter(Group %in% c("CF.LUMA_IVA", "NON_CF.CTRL"),
clusters %in% c("CD4 T-IFN",
"CD4 T-reg",
"monocytes")) %>%
ggplot(aes(x = Group,
y = Freq,
colour = Group)) +
geom_jitter(stat = "identity",
width = 0.15,
size = 1,
show.legend = FALSE) +
theme_classic() +
theme(axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
axis.title.x = element_blank(),
axis.text.y = element_text(7),
legend.position = "bottom",
legend.direction = "horizontal",
strip.text = element_text(size = 8)) +
labs(x = "Group", y = "Proportion") +
facet_wrap(~clusters, scales = "free_y", ncol = 4,
labeller = sig_names) +
stat_summary(
geom = "point",
fun.y = "mean",
col = "black",
shape = "_",
size = 10) +
scale_color_manual(values = pal) -> p2props <- getTransformedProps(clusters = seu$ann_level_3[str_detect(seu$ann_level_3, "macro")],
sample = seu$sample.id[str_detect(seu$ann_level_3, "macro")], transform="asin")
props$Proportions %>% data.frame %>%
left_join(info,
by = c("sample" = "sample.id")) %>%
dplyr::filter(Group %in% c("CF.LUMA_IVA", "CF.NO_MOD", "NON_CF.CTRL"),
clusters %in% "macro-CCL18") -> dat
dat %>%
dplyr::filter(Group %in% c("CF.LUMA_IVA", "CF.NO_MOD", "NON_CF.CTRL"),
clusters %in% c("macro-CCL18")) %>%
ggplot(aes(x = Group,
y = Freq,
colour = Group)) +
geom_jitter(stat = "identity",
width = 0.15,
size = 1) +
theme_classic() +
theme(axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
axis.title.x = element_blank(),
axis.text.y = element_text(7),
legend.position = "bottom",
legend.direction = "horizontal",
strip.text = element_text(size = 8)) +
labs(x = "Group", y = "Proportion") +
facet_wrap(~clusters, scales = "free_y", ncol = 4,
labeller = sig_names) +
stat_summary(
geom = "point",
fun.y = "mean",
col = "black",
shape = "_",
size = 10) +
scale_color_manual(values = pal) -> p3
layout <- "ABBBC"
lumaiva_props <- (p1 +
(p2 + theme(axis.title.y = element_blank())) +
(p3 + theme(axis.title.y = element_blank()))) +
plot_layout(design = layout, guides = "collect") &
theme(axis.text.y = element_text(size = 6,
angle = 90,
hjust = 0.5),
legend.text = element_text(size = 8),
legend.key.spacing = unit(0, "lines"),
legend.position = "bottom",
legend.margin = margin(-0.5,0,0,0, unit="lines"))
lumaiva_props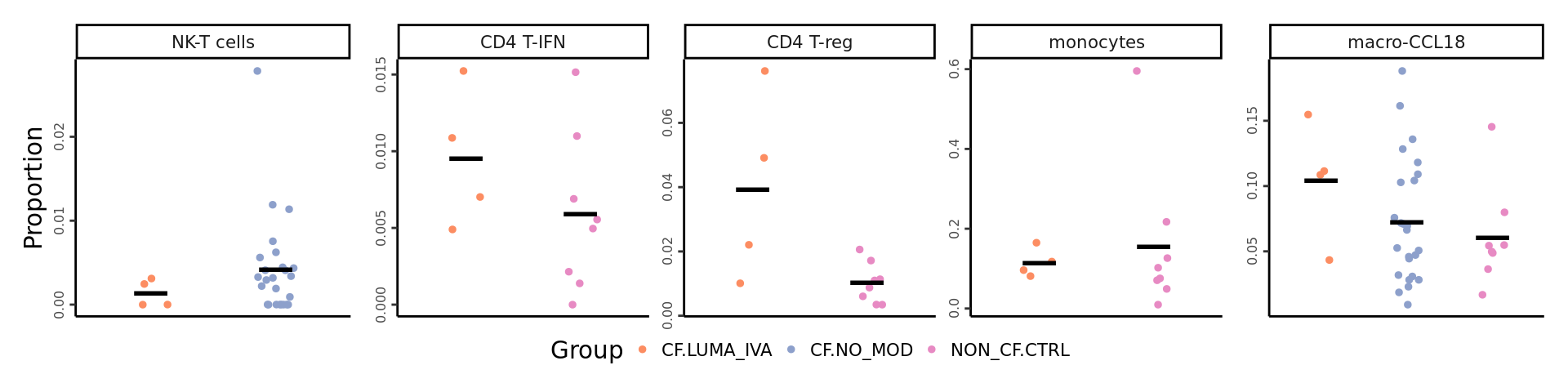
| Version | Author | Date |
|---|---|---|
| bb42dbf | Jovana Maksimovic | 2025-01-06 |
seu@meta.data %>%
data.frame %>%
dplyr::select(ann_level_2) %>%
dplyr::filter(str_detect(ann_level_2, "macro")) %>%
group_by(ann_level_2) %>%
count() %>%
janitor::adorn_totals(name = "macrophages") %>%
arrange(-n) %>%
dplyr::rename(cell = ann_level_2) -> cell_freq
cell_freq cell n
macrophages 165209
macro-alveolar 52563
macro-IFI27 24864
macro-CCL 21246
macro-monocyte-derived 13461
macro-APOC2+ 13354
macro-lipid 12452
macro-IGF1 8229
macro-proliferating 6821
macro-MT 4037
macro-interstitial 3412
macro-T 2722
macro-IFN 2048files <- list.files(here("data/intermediate_objects"),
pattern = "macro.*CF_samples",
full.names = TRUE)
cutoff <- 0.05
cont_name <- "CF.IVAvCF.NO_MOD"
lfc_cutoff <- log2(1.2)
suffix <- ".CF_samples.fit.rds"
get_deg_data(files, cont_name, cell_freq, treat_lfc = lfc_cutoff,
suffix = suffix) -> datdat %>%
dplyr::select(gene, cell, logFC) %>%
distinct() %>%
pivot_wider(
names_from = cell, # Column whose values become new column names
values_from = logFC,
values_fill = list(logFC = NA)) %>%
arrange(across(all_of(cell_freq$cell[cell_freq$cell %in% dat$cell]))) %>%
column_to_rownames(var = "gene") -> dat_lfccol_fun <- circlize::colorRamp2(seq(0, 100000, length.out = 9),
(RColorBrewer::brewer.pal(9, "PiYG")))
col_split <- c(rep("aggregate", 1), rep("sub-type", ncol(dat_lfc) - 1))
pal_dt <- c(paletteer::paletteer_d("RColorBrewer::Set1")[2:1], "grey")
col_lfc_fun <- circlize::colorRamp2(seq(-2, 2, length.out = 3),
c(pal_dt[1], "white", pal_dt[2]))
ComplexHeatmap::HeatmapAnnotation(df = cell_freq %>%
dplyr::filter(cell %in% colnames(dat_lfc)) %>%
column_to_rownames(var = "cell") %>%
dplyr::rename(`No. cells` = n),
which = "column",
show_annotation_name = FALSE,
col = list(`No. cells` = col_fun),
annotation_legend_param = list(
`No. cells` = list(direction = "vertical"))) -> col_ann
ComplexHeatmap::HeatmapAnnotation(df = data.frame(multiple = (rowSums(!is.na(dat_lfc)) > 1)),
which = "row",
col = list(multiple = c("FALSE" = "#fdcce5","TRUE" = "#8bd3c7")),
annotation_legend_param = list(
multiple = list(direction = "vertical",
ncol = 1))) -> row_ann
ComplexHeatmap::Heatmap(dat_lfc,
name = "logFC",
column_split = col_split,
column_title = NULL,
cluster_rows = FALSE,
cluster_columns = FALSE,
rect_gp = grid::gpar(col = "white", lwd = 1),
row_names_gp = grid::gpar(fontsize = 7),
column_names_gp = grid::gpar(fontsize = 7),
col = col_lfc_fun,
top_annotation = col_ann,
right_annotation = row_ann,
heatmap_legend_param = list(direction = "vertical")) -> plot_lfc
ComplexHeatmap::draw(as(list(plot_lfc), "HeatmapList"),
heatmap_legend_side = "right",
annotation_legend_side = "right",
merge_legends = TRUE) -> plot_lfc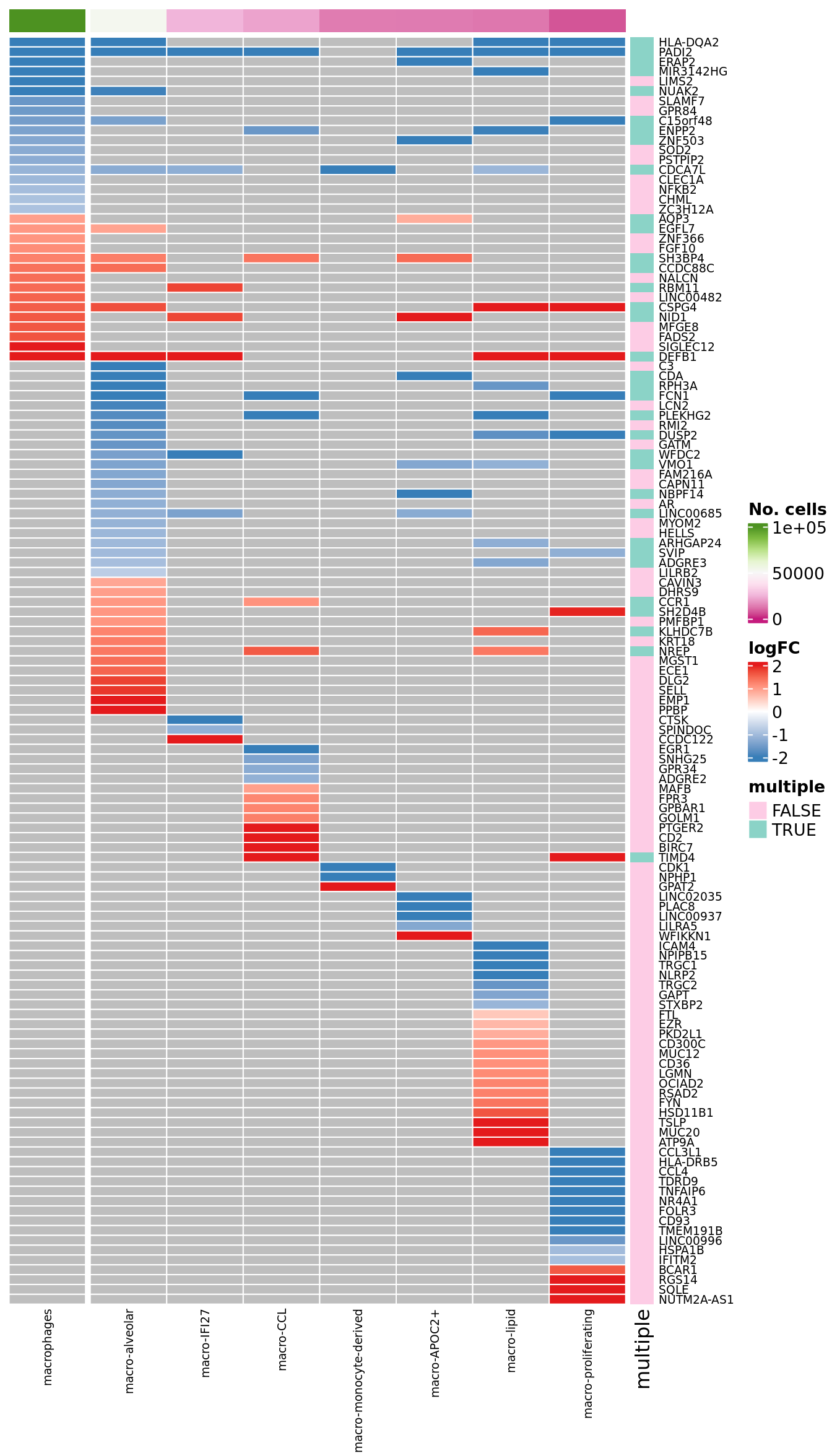
| Version | Author | Date |
|---|---|---|
| bb42dbf | Jovana Maksimovic | 2025-01-06 |
plot_lfc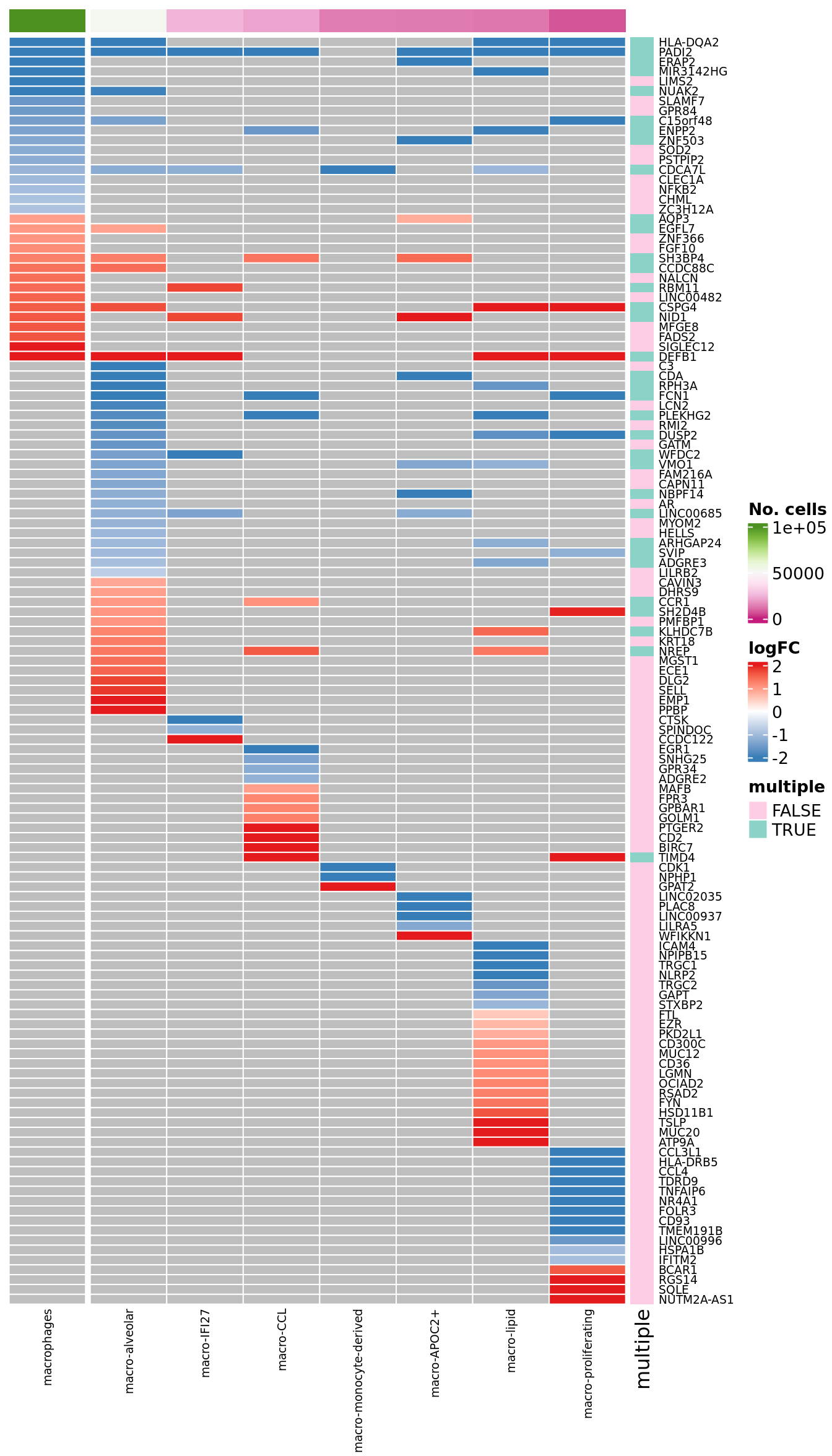
| Version | Author | Date |
|---|---|---|
| bb42dbf | Jovana Maksimovic | 2025-01-06 |
bind_rows(lapply(files, function(f){
deg_results <- readRDS(f)
lrt <- glmLRT(deg_results$fit,
contrast = deg_results$contr[,cont_name])
tmp <- cbind(summary(decideTests(lrt, p.value = cutoff)) %>% data.frame,
cell = unlist(str_split(str_remove(f, ".CF_samples.fit.rds"), "/"))[8])
tmp
})) -> dat_degdat_deg %>%
left_join(cell_freq) -> dat_deg
pal_dt <- c(paletteer::paletteer_d("RColorBrewer::Set1")[2:1], "grey")
dat_deg %>%
dplyr::filter(Var1 != "NotSig") %>%
ggplot(aes(x = fct_reorder(cell, n), y = Freq, fill = Var1)) +
geom_col(position = "dodge") +
scale_fill_manual(values = pal_dt) +
theme_classic() +
theme(axis.text.y = element_text(angle = -45,
hjust = 1,
vjust = 1,
size = 7),
legend.position = "top") +
geom_text(aes(label = Freq),
position = position_dodge(width = 0.9),
vjust = -0.5,
angle = 270,
size = 2.5) +
labs(x = "Cell Type",
y = "No. DEG (FDR < 0.05)",
fill = "Direction") +
coord_flip() -> deg_barplot
deg_barplot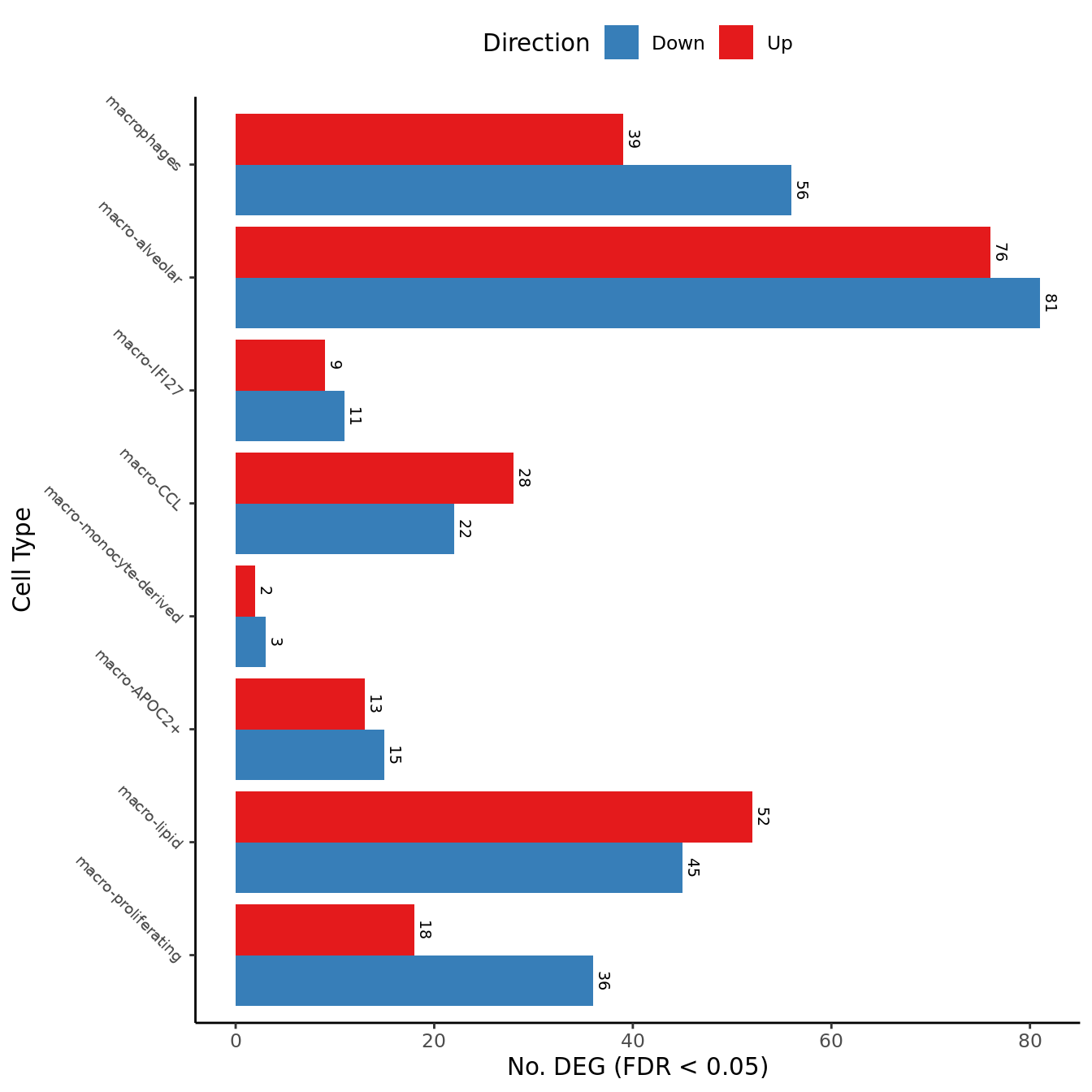
| Version | Author | Date |
|---|---|---|
| bb42dbf | Jovana Maksimovic | 2025-01-06 |
volc_plot <- draw_treat_volcano_plot(cell = "macro-alveolar",
suffix = suffix,
cutoff = cutoff,
lfc_cutoff = lfc_cutoff)
volc_plot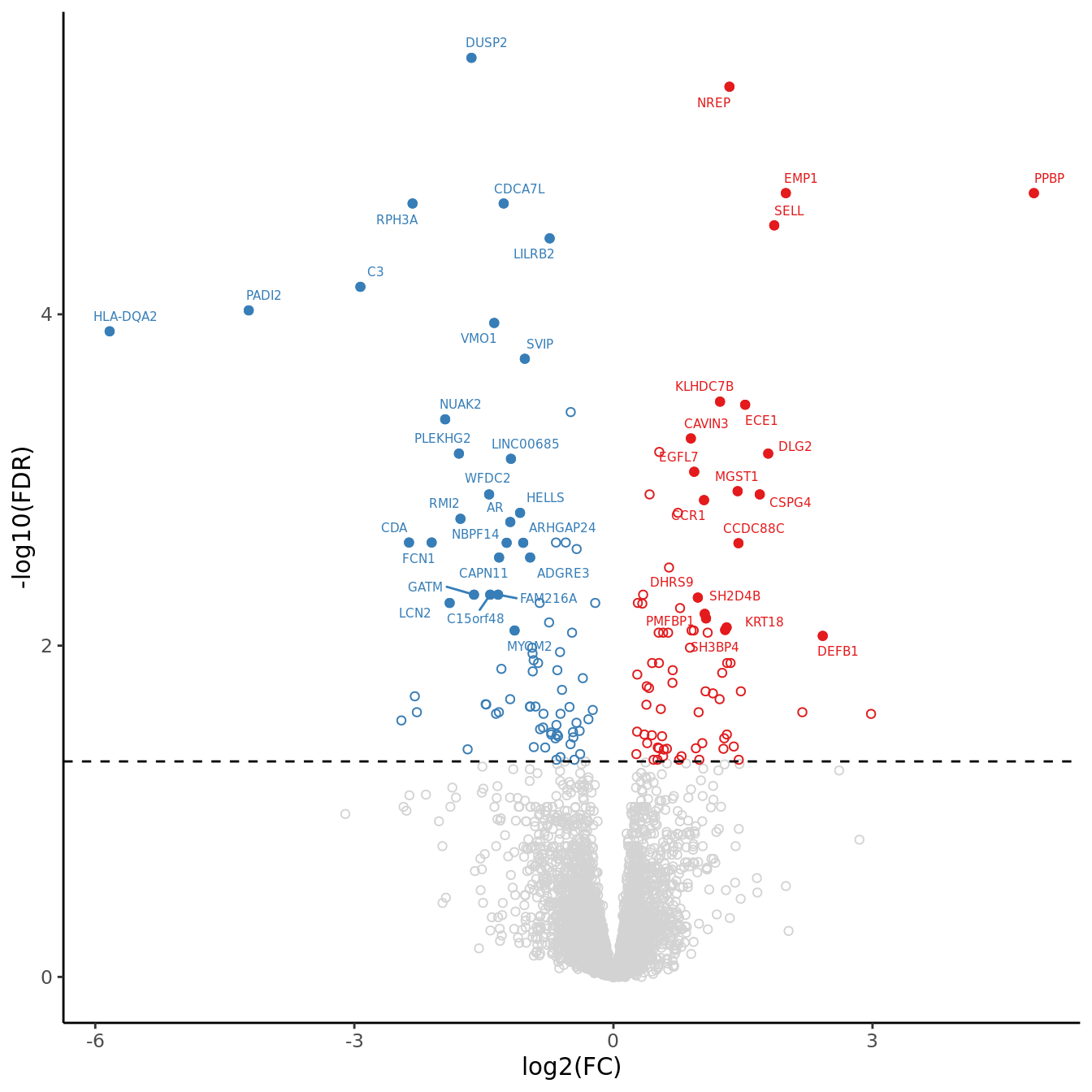
| Version | Author | Date |
|---|---|---|
| bb42dbf | Jovana Maksimovic | 2025-01-06 |
Hs.c2.all <- convert_gmt_to_list(here("data/c2.all.v2024.1.Hs.entrez.gmt"))
Hs.h.all <- convert_gmt_to_list(here("data/h.all.v2024.1.Hs.entrez.gmt"))
Hs.c5.all <- convert_gmt_to_list(here("data/c5.all.v2024.1.Hs.entrez.gmt"))
fibrosis <- create_custom_gene_lists_from_file(here("data/fibrosis_gene_sets.csv"))
# add fibrosis sets from REACTOME and WIKIPATHWAYS
fibrosis <- c(lapply(fibrosis, function(l) l[!is.na(l)]),
Hs.c2.all[str_detect(names(Hs.c2.all), "FIBROSIS")])
gene_sets_list <- list(HALLMARK = Hs.h.all,
GO = Hs.c5.all,
REACTOME = Hs.c2.all[str_detect(names(Hs.c2.all), "REACTOME")],
WP = Hs.c2.all[str_detect(names(Hs.c2.all), "^WP")],
FIBROSIS = fibrosis)hallmark <- read_csv(file = here("output",
"dge_analysis",
"macro-alveolar",
"CAM.HALLMARK.CF.IVAvCF.NO_MOD.csv"))
reactome <- read_csv(file = here("output",
"dge_analysis",
"macro-alveolar",
"CAM.REACTOME.CF.IVAvCF.NO_MOD.csv"))
results_list <- list(HALLMARK = hallmark %>% column_to_rownames(var = "Set"),
REACTOME = reactome %>% column_to_rownames(var = "Set"))
top_camera_sets(results_list, num = 4) +
theme(plot.title = element_blank(),
axis.text.y = element_text(size = 7,
angle = 35)) -> cam_plot
cam_plot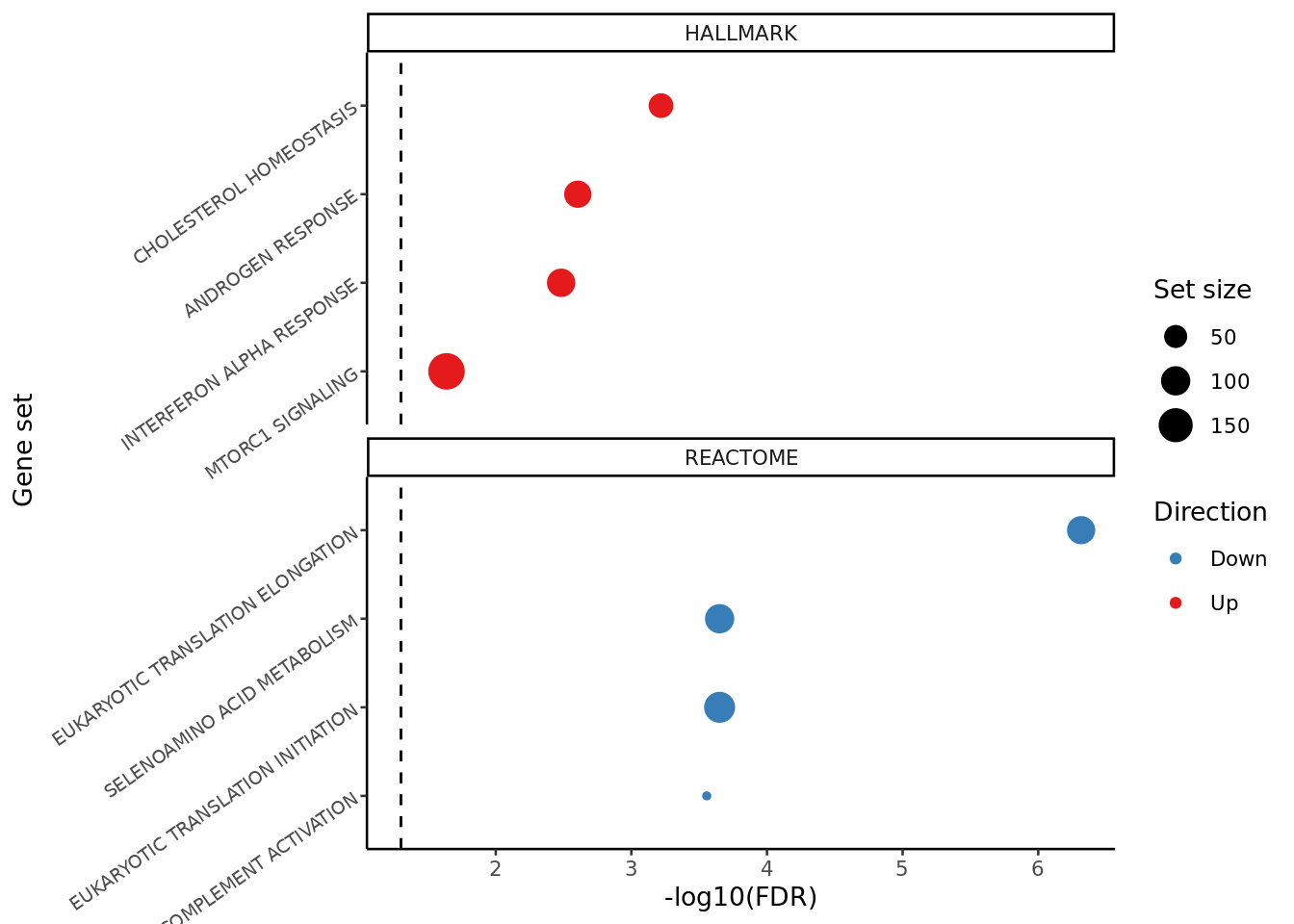
| Version | Author | Date |
|---|---|---|
| bb42dbf | Jovana Maksimovic | 2025-01-06 |
reactome <- read_csv(file = here("output",
"dge_analysis",
"macro-alveolar",
"ORA.REACTOME.CF.IVAvCF.NO_MOD.csv"))
go <- read_csv(file = here("output",
"dge_analysis",
"macro-alveolar",
"ORA.GO.CF.IVAvCF.NO_MOD.csv"))
results_list <- list(REACTOME = reactome %>% column_to_rownames(var = "Set"),
GO = go %>% column_to_rownames(var = "Set"))
top_ora_sets(results_list, num = 3) +
theme(plot.title = element_blank(),
axis.text.y = element_text(size = 7,
angle = 35)) -> ora_plot
ora_plot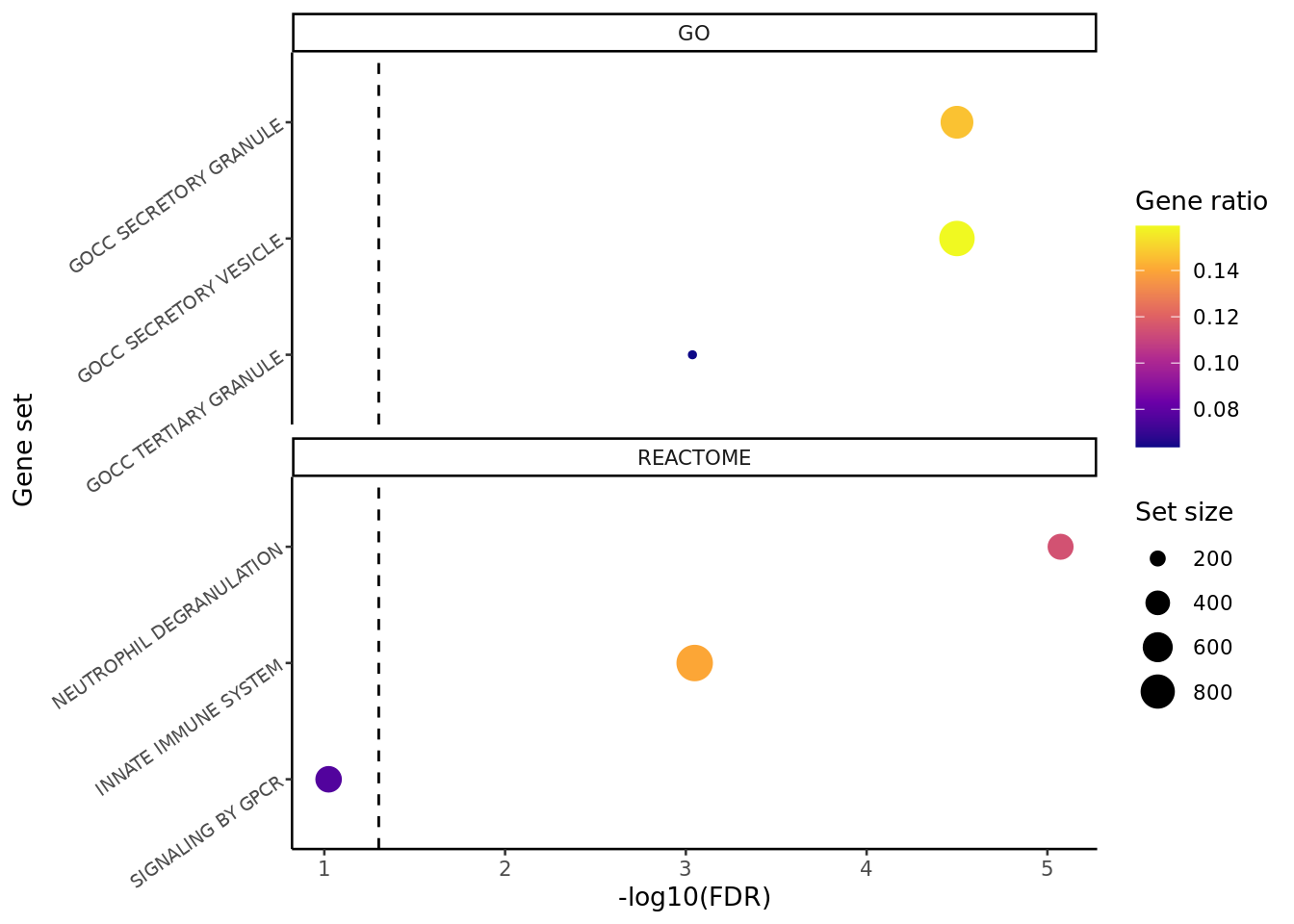
| Version | Author | Date |
|---|---|---|
| bb42dbf | Jovana Maksimovic | 2025-01-06 |
genes <- c("LILRB2", "C3", "PADI2", "SVIP", "FCN1", "CDA", "ADGRE3", "CAPN11", "LCN2", "CLEC12A", "LYZ")
f <- files[1]
deg_results <- readRDS(f)
norm_counts <- deg_results$adj$normalizedCounts
group <- deg_results$fit$samples[,"group", drop = FALSE]
cpm(norm_counts, log = TRUE) %>%
data.frame %>%
rownames_to_column(var = "gene") %>%
pivot_longer(-gene,
names_to = "sample",
values_to = "logCPM") %>%
left_join(group %>%
data.frame %>%
rownames_to_column(var = "sample")) %>%
dplyr::filter(gene %in% genes) %>%
mutate(group = str_remove_all(group, "(.M|.S)$")) %>%
dplyr::filter(group %in% str_split(cont_name, "v")[[1]]) -> dat
ggplot(dat, aes(x = group, y = logCPM, colour = group)) +
# geom_violin() +
geom_jitter(width = 0.15,
size = 1) +
stat_summary(geom = "point",
fun.y = "mean",
col = "black",
shape = "_",
size = 10) +
facet_wrap(~gene, nrow = 1, scales = "free_y") +
theme_classic() +
theme(axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
axis.title.x = element_blank(),
axis.text.y = element_text(7),
legend.position = "bottom",
legend.direction = "horizontal",
strip.text = element_text(size = 8)) +
labs(colour = "Group") +
scale_color_manual(values = pal) -> gene_plot
gene_plot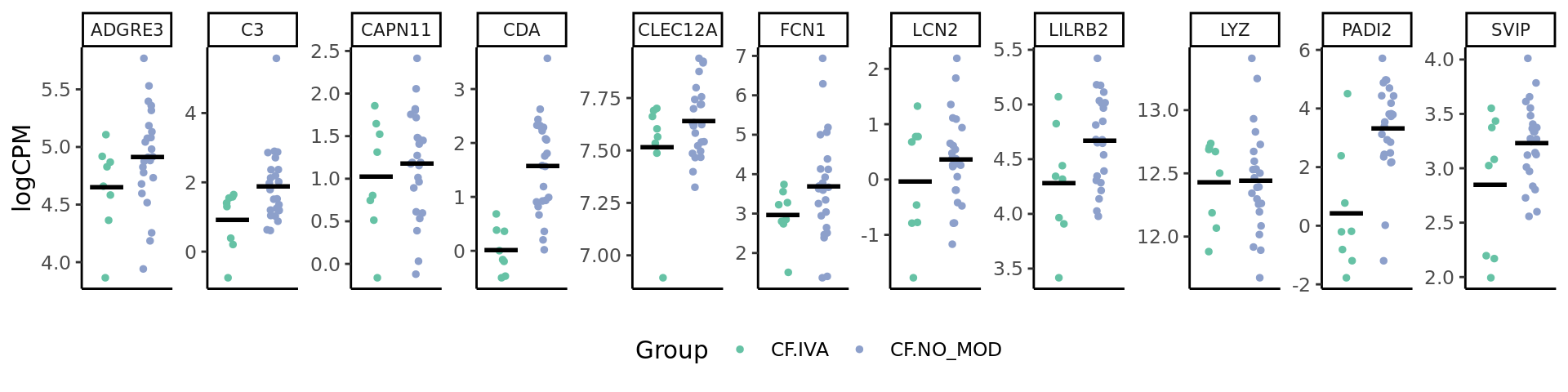
| Version | Author | Date |
|---|---|---|
| bb42dbf | Jovana Maksimovic | 2025-01-06 |
Figure 5
layout <- "
AAABBBB
AAABBBB
CCCDDDD
CCCDDDD
CCCDDDD
CCCDDDD
EEEDDDD
EEEDDDD
EEEDDDD
FFFDDDD
FFFDDDD
FFFDDDD
GGGDDDD
GGGDDDD
GGGDDDD
HHHHHHH
HHHHHHH
"
wrap_elements(iva_props) +
wrap_elements(lumaiva_props) +
wrap_elements(deg_barplot) +
wrap_elements(grid::grid.grabExpr(ComplexHeatmap::draw(plot_lfc))) +
wrap_elements(volc_plot) +
wrap_elements(cam_plot + theme(legend.position = "bottom",
legend.direction = "horizontal",
legend.box = "vertical",
legend.margin = margin(-0.5,0,0,0, unit="lines"),
legend.text = element_text(size = 8))) +
wrap_elements(ora_plot +
theme(legend.position = "bottom",
legend.direction = "horizontal",
legend.box = "vertical",
legend.margin = margin(-0.5,0,0,0, unit="lines"),
legend.text = element_text(size = 7))) +
wrap_elements(gene_plot) +
plot_layout(design = layout) +
plot_annotation(tag_levels = "A") &
theme(plot.tag = element_text(size = 16,
face = "bold",
family = "arial"))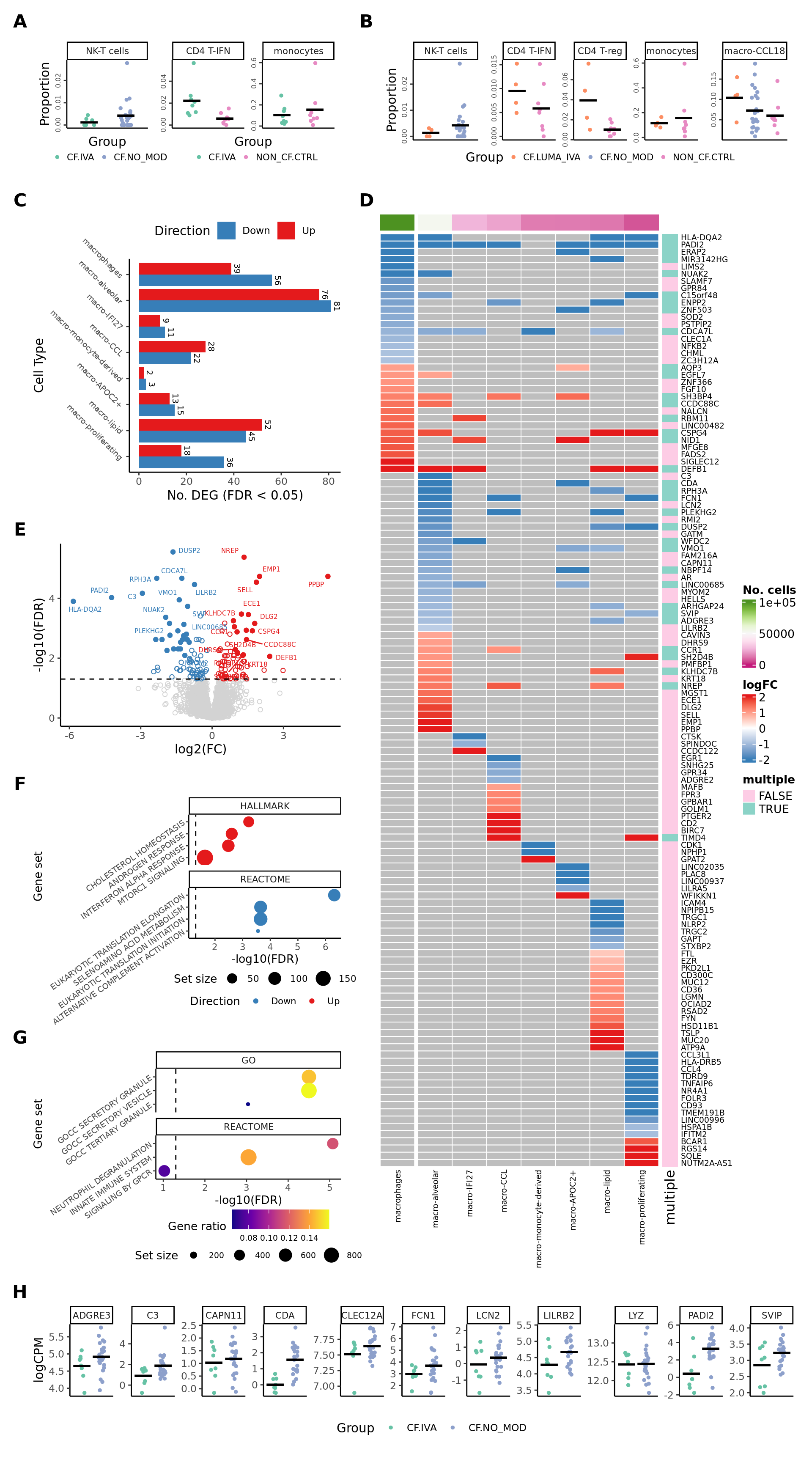
| Version | Author | Date |
|---|---|---|
| bb42dbf | Jovana Maksimovic | 2025-01-06 |
Session info
sessionInfo()R version 4.3.3 (2024-02-29)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 22.04.4 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
locale:
[1] LC_CTYPE=en_AU.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_AU.UTF-8 LC_COLLATE=en_AU.UTF-8
[5] LC_MONETARY=en_AU.UTF-8 LC_MESSAGES=en_AU.UTF-8
[7] LC_PAPER=en_AU.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_AU.UTF-8 LC_IDENTIFICATION=C
time zone: Etc/UTC
tzcode source: system (glibc)
attached base packages:
[1] stats4 stats graphics grDevices datasets utils methods
[8] base
other attached packages:
[1] readxl_1.4.3 ggh4x_0.2.8
[3] dsb_1.0.3 paletteer_1.6.0
[5] tidyHeatmap_1.8.1 speckle_1.2.0
[7] glue_1.8.0 org.Hs.eg.db_3.18.0
[9] AnnotationDbi_1.64.1 patchwork_1.3.0
[11] clustree_0.5.1 ggraph_2.2.0
[13] here_1.0.1 dittoSeq_1.14.2
[15] glmGamPoi_1.14.3 SeuratObject_4.1.4
[17] Seurat_4.4.0 lubridate_1.9.3
[19] forcats_1.0.0 stringr_1.5.1
[21] dplyr_1.1.4 purrr_1.0.2
[23] readr_2.1.5 tidyr_1.3.1
[25] tibble_3.2.1 ggplot2_3.5.0
[27] tidyverse_2.0.0 edgeR_4.0.15
[29] limma_3.58.1 SingleCellExperiment_1.24.0
[31] SummarizedExperiment_1.32.0 Biobase_2.62.0
[33] GenomicRanges_1.54.1 GenomeInfoDb_1.38.6
[35] IRanges_2.36.0 S4Vectors_0.40.2
[37] BiocGenerics_0.48.1 MatrixGenerics_1.14.0
[39] matrixStats_1.2.0 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] fs_1.6.5 spatstat.sparse_3.0-3 bitops_1.0-7
[4] httr_1.4.7 RColorBrewer_1.1-3 doParallel_1.0.17
[7] tools_4.3.3 sctransform_0.4.1 utf8_1.2.4
[10] R6_2.5.1 lazyeval_0.2.2 uwot_0.1.16
[13] GetoptLong_1.0.5 withr_3.0.0 sp_2.1-3
[16] gridExtra_2.3 progressr_0.14.0 cli_3.6.3
[19] Cairo_1.6-2 spatstat.explore_3.2-6 prismatic_1.1.1
[22] labeling_0.4.3 sass_0.4.9 spatstat.data_3.0-4
[25] ggridges_0.5.6 pbapply_1.7-2 parallelly_1.37.0
[28] rstudioapi_0.15.0 RSQLite_2.3.5 generics_0.1.3
[31] shape_1.4.6 vroom_1.6.5 ica_1.0-3
[34] spatstat.random_3.2-2 dendextend_1.17.1 Matrix_1.6-5
[37] fansi_1.0.6 abind_1.4-5 lifecycle_1.0.4
[40] whisker_0.4.1 yaml_2.3.8 snakecase_0.11.1
[43] SparseArray_1.2.4 Rtsne_0.17 grid_4.3.3
[46] blob_1.2.4 promises_1.2.1 crayon_1.5.2
[49] miniUI_0.1.1.1 lattice_0.22-5 cowplot_1.1.3
[52] KEGGREST_1.42.0 pillar_1.9.0 knitr_1.45
[55] ComplexHeatmap_2.18.0 rjson_0.2.21 future.apply_1.11.1
[58] codetools_0.2-19 leiden_0.4.3.1 getPass_0.2-4
[61] data.table_1.15.0 vctrs_0.6.5 png_0.1-8
[64] cellranger_1.1.0 gtable_0.3.4 rematch2_2.1.2
[67] cachem_1.0.8 xfun_0.42 S4Arrays_1.2.0
[70] mime_0.12 tidygraph_1.3.1 survival_3.7-0
[73] pheatmap_1.0.12 iterators_1.0.14 statmod_1.5.0
[76] ellipsis_0.3.2 fitdistrplus_1.1-11 ROCR_1.0-11
[79] nlme_3.1-164 bit64_4.0.5 RcppAnnoy_0.0.22
[82] rprojroot_2.0.4 bslib_0.6.1 irlba_2.3.5.1
[85] KernSmooth_2.23-24 colorspace_2.1-0 DBI_1.2.1
[88] tidyselect_1.2.1 processx_3.8.3 bit_4.0.5
[91] compiler_4.3.3 git2r_0.33.0 DelayedArray_0.28.0
[94] plotly_4.10.4 scales_1.3.0 lmtest_0.9-40
[97] callr_3.7.3 digest_0.6.34 goftest_1.2-3
[100] spatstat.utils_3.0-4 rmarkdown_2.25 XVector_0.42.0
[103] htmltools_0.5.8.1 pkgconfig_2.0.3 highr_0.10
[106] fastmap_1.1.1 rlang_1.1.4 GlobalOptions_0.1.2
[109] htmlwidgets_1.6.4 shiny_1.8.0 farver_2.1.1
[112] jquerylib_0.1.4 zoo_1.8-12 jsonlite_1.8.8
[115] mclust_6.1 RCurl_1.98-1.14 magrittr_2.0.3
[118] GenomeInfoDbData_1.2.11 munsell_0.5.0 Rcpp_1.0.12
[121] viridis_0.6.5 reticulate_1.35.0 stringi_1.8.3
[124] zlibbioc_1.48.0 MASS_7.3-60.0.1 plyr_1.8.9
[127] parallel_4.3.3 listenv_0.9.1 ggrepel_0.9.5
[130] deldir_2.0-2 Biostrings_2.70.2 graphlayouts_1.1.0
[133] splines_4.3.3 tensor_1.5 hms_1.1.3
[136] circlize_0.4.15 locfit_1.5-9.8 ps_1.7.6
[139] igraph_2.0.1.1 spatstat.geom_3.2-8 reshape2_1.4.4
[142] evaluate_0.23 renv_1.0.3 BiocManager_1.30.22
[145] tzdb_0.4.0 foreach_1.5.2 tweenr_2.0.3
[148] httpuv_1.6.14 RANN_2.6.1 polyclip_1.10-6
[151] future_1.33.1 clue_0.3-65 scattermore_1.2
[154] ggforce_0.4.2 janitor_2.2.0 xtable_1.8-4
[157] later_1.3.2 viridisLite_0.4.2 memoise_2.0.1
[160] cluster_2.1.6 timechange_0.3.0 globals_0.16.2