Inflammation of Paediatric Pulmonary Diseases
Cell type proportions analysis - annotation level 1
Jovana Maksimovic
December 24, 2024
Last updated: 2024-12-24
Checks: 7 0
Knit directory: paed-inflammation-CITEseq/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20240216) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version ff4fd99. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/obsolete/
Ignored: data/C133_Neeland_batch1/
Ignored: data/C133_Neeland_merged/
Ignored: output/dge_analysis/obsolete/
Ignored: renv/library/
Ignored: renv/staging/
Untracked files:
Untracked: analysis/14.0_DGE_analysis_T-cells.Rmd
Untracked: analysis/15.1_proportions_analysis_ann_level_3_non-macrophages.Rmd
Untracked: analysis/15.2_proportions_analysis_ann_level_3_macrophages.Rmd
Untracked: analysis/17.0_Figure_3.Rmd
Untracked: analysis/17.0_Figure_4.Rmd
Untracked: analysis/17.0_Figure_5.Rmd
Untracked: broad_markers_seurat.csv
Untracked: code/background_job.R
Untracked: code/reverse_modifier_severity_comparisons.sh
Untracked: data/intermediate_objects/CD4 T cells.CF_samples.fit.rds
Untracked: data/intermediate_objects/CD4 T cells.all_samples.fit.rds
Untracked: data/intermediate_objects/CD8 T cells.CF_samples.fit.rds
Untracked: data/intermediate_objects/CD8 T cells.all_samples.fit.rds
Untracked: data/intermediate_objects/DC cells.CF_samples.fit.rds
Untracked: data/intermediate_objects/DC cells.all_samples.fit.rds
Untracked: data/intermediate_objects/T cells.CF_samples.fit.rds
Untracked: data/intermediate_objects/T cells.all_samples.fit.rds
Untracked: output/dge_analysis/T cells/
Unstaged changes:
Modified: .gitignore
Modified: analysis/06.0_azimuth_annotation.Rmd
Modified: analysis/09.0_integrate_cluster_macro_cells.Rmd
Deleted: analysis/14.0_proportions_analysis_ann_level_1.Rmd
Deleted: analysis/14.1_proportions_analysis_ann_level_3_non-macrophages.Rmd
Deleted: analysis/14.2_proportions_analysis_ann_level_3_macrophages.Rmd
Modified: analysis/15.0_Figure_1.Rmd
Modified: analysis/16.0_Figure_2.Rmd
Modified: data/cluster_annotations/marker_proteins_TNK_supp.xlsx
Modified: data/cluster_annotations/marker_proteins_macrophages_supp.xlsx
Modified: data/cluster_annotations/marker_proteins_other_supp.xlsx
Modified: data/cluster_annotations/seurat_markers_all_cells.rds
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown
(analysis/15.0_proportions_analysis_ann_level_1.Rmd) and
HTML (docs/15.0_proportions_analysis_ann_level_1.html)
files. If you’ve configured a remote Git repository (see
?wflow_git_remote), click on the hyperlinks in the table
below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | ff4fd99 | Jovana Maksimovic | 2024-12-24 | wflow_publish("analysis/15.0_proportions_analysis_ann_level_1.Rmd") |
Load libraries
suppressPackageStartupMessages({
library(SingleCellExperiment)
library(edgeR)
library(tidyverse)
library(ggplot2)
library(Seurat)
library(glmGamPoi)
library(dittoSeq)
library(clustree)
library(AnnotationDbi)
library(org.Hs.eg.db)
library(glue)
library(speckle)
library(patchwork)
library(paletteer)
library(tidyHeatmap)
library(here)
})
set.seed(42)
options(scipen=999)
options(future.globals.maxSize = 6500 * 1024^2)Load Data
files <- list.files(here("data/C133_Neeland_merged"),
pattern = "C133_Neeland_full_clean.*(macrophages|t_cells|other_cells)_annotated_diet.SEU.rds",
full.names = TRUE)
seuLst <- lapply(files[2:4], function(f) readRDS(f))
seu <- merge(seuLst[[1]],
y = c(seuLst[[2]],
seuLst[[3]]))
seuAn object of class Seurat
21568 features across 194407 samples within 1 assay
Active assay: RNA (21568 features, 0 variable features) used (Mb) gc trigger (Mb) max used (Mb)
Ncells 12078855 645.1 20174672 1077.5 14082721 752.1
Vcells 1354150870 10331.4 3693733670 28181.0 3551484612 27095.7Analyse Cell type proportions
# Differences in cell type proportions
props <- getTransformedProps(clusters = seu$ann_level_1,
sample = seu$sample.id, transform="asin")
props$Proportions %>% knitr::kable()| sample_1.1 | sample_15.1 | sample_16.1 | sample_17.1 | sample_18.1 | sample_19.1 | sample_2.1 | sample_20.1 | sample_21.1 | sample_22.1 | sample_23.1 | sample_24.1 | sample_25.1 | sample_26.1 | sample_27.1 | sample_28.1 | sample_29.1 | sample_3.1 | sample_30.1 | sample_31.1 | sample_32.1 | sample_33.1 | sample_34.1 | sample_34.2 | sample_34.3 | sample_35.1 | sample_35.2 | sample_36.1 | sample_36.2 | sample_37.1 | sample_37.2 | sample_37.3 | sample_38.1 | sample_38.2 | sample_38.3 | sample_39.1 | sample_39.2 | sample_4.1 | sample_40.1 | sample_41.1 | sample_42.1 | sample_43.1 | sample_5.1 | sample_6.1 | sample_7.1 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B cells | 0.0223723 | 0.0071704 | 0.0018883 | 0.0428725 | 0.0015235 | 0.0023866 | 0.0950689 | 0.0017933 | 0.0095134 | 0.0023447 | 0.2436149 | 0.0057254 | 0.0027506 | 0.0122549 | 0.0020268 | 0.0006984 | 0.0918367 | 0.0119505 | 0.0064935 | 0.0041667 | 0.0027000 | 0.0607761 | 0.0026738 | 0.0000000 | 0.0016584 | 0.0181452 | 0.0081239 | 0.0304653 | 0.0502624 | 0.0069589 | 0.0124824 | 0.0105605 | 0.0048374 | 0.0058787 | 0.0356175 | 0.0434431 | 0.0275087 | 0.0096690 | 0.1603532 | 0.0202830 | 0.0107823 | 0.0427193 | 0.0181922 | 0.0046205 | 0.0282528 |
| CD4 T cells | 0.0092866 | 0.0275786 | 0.0129485 | 0.0176849 | 0.0082267 | 0.0023866 | 0.0364283 | 0.0087101 | 0.0192097 | 0.0082063 | 0.1650295 | 0.0262504 | 0.0174205 | 0.0323529 | 0.0133766 | 0.0060064 | 0.0299745 | 0.0061886 | 0.1911977 | 0.0722222 | 0.0155248 | 0.1547452 | 0.0113636 | 0.0051207 | 0.0035537 | 0.0173387 | 0.0164368 | 0.0113827 | 0.0425297 | 0.0153097 | 0.0163076 | 0.0268075 | 0.0056436 | 0.0102104 | 0.0274874 | 0.0250944 | 0.0585045 | 0.0106607 | 0.1119339 | 0.0521226 | 0.0361976 | 0.0390446 | 0.0801592 | 0.0137671 | 0.0381306 |
| CD8 T cells | 0.0063318 | 0.0295091 | 0.0283248 | 0.0353698 | 0.0164534 | 0.0011933 | 0.0510884 | 0.0098629 | 0.0179290 | 0.0095252 | 0.1277014 | 0.0310036 | 0.0134474 | 0.0308824 | 0.0267531 | 0.0153653 | 0.0586735 | 0.0091763 | 0.0977633 | 0.0949074 | 0.0175498 | 0.1280972 | 0.0100267 | 0.0029261 | 0.0052120 | 0.0203629 | 0.0102022 | 0.0033478 | 0.0248550 | 0.0146138 | 0.0074492 | 0.0199025 | 0.0037624 | 0.0049505 | 0.0251645 | 0.0070157 | 0.0730337 | 0.0254122 | 0.0780404 | 0.0662736 | 0.0866982 | 0.1748966 | 0.0949403 | 0.0271570 | 0.0380244 |
| DC cells | 0.0097087 | 0.0184777 | 0.0037766 | 0.0391211 | 0.0173675 | 0.0055688 | 0.0017770 | 0.0067888 | 0.0080498 | 0.0016120 | 0.0628684 | 0.0398617 | 0.0201711 | 0.0230392 | 0.0162140 | 0.0065652 | 0.0440051 | 0.0147247 | 0.0497835 | 0.0157407 | 0.0121498 | 0.0719963 | 0.0788770 | 0.0643745 | 0.0322199 | 0.0219758 | 0.0251275 | 0.0348175 | 0.0544049 | 0.0073069 | 0.0062412 | 0.0048741 | 0.0010750 | 0.0024752 | 0.0081301 | 0.0232056 | 0.0350639 | 0.0126441 | 0.0185132 | 0.0570755 | 0.0064914 | 0.0705099 | 0.0204662 | 0.0045262 | 0.0059480 |
| dividing innate cells | 0.0000000 | 0.0002758 | 0.0002698 | 0.0024116 | 0.0006094 | 0.0003978 | 0.0093292 | 0.0002562 | 0.0000000 | 0.0000000 | 0.0108055 | 0.0012963 | 0.0006112 | 0.0000000 | 0.0000000 | 0.0001397 | 0.0031888 | 0.0006402 | 0.0003608 | 0.0000000 | 0.0003375 | 0.0032726 | 0.0013369 | 0.0000000 | 0.0002369 | 0.0002016 | 0.0001889 | 0.0016739 | 0.0008285 | 0.0000000 | 0.0000000 | 0.0004062 | 0.0000000 | 0.0003094 | 0.0011614 | 0.0035078 | 0.0017435 | 0.0006198 | 0.0034178 | 0.0023585 | 0.0003301 | 0.0026412 | 0.0000000 | 0.0000000 | 0.0001062 |
| epithelial cells | 0.0426340 | 0.0132377 | 0.0005395 | 0.0032154 | 0.0053321 | 0.0019889 | 0.1537095 | 0.0043551 | 0.0104281 | 0.0038101 | 0.2092338 | 0.0034568 | 0.0006112 | 0.0034314 | 0.0275638 | 0.0006984 | 0.0446429 | 0.0017072 | 0.0010823 | 0.0018519 | 0.0000000 | 0.0219729 | 0.0093583 | 0.0087783 | 0.0037906 | 0.0094758 | 0.0094464 | 0.0077000 | 0.0038663 | 0.0076548 | 0.0020133 | 0.0052803 | 0.0016125 | 0.0018564 | 0.0154859 | 0.0029682 | 0.0069740 | 0.0032230 | 0.0287667 | 0.0188679 | 0.0083618 | 0.0096463 | 0.0062536 | 0.0012258 | 0.0011683 |
| gamma delta T cells | 0.0000000 | 0.0005516 | 0.0008093 | 0.0008039 | 0.0003047 | 0.0000000 | 0.0000000 | 0.0003843 | 0.0007318 | 0.0000000 | 0.0000000 | 0.0003241 | 0.0006112 | 0.0004902 | 0.0012161 | 0.0001397 | 0.0000000 | 0.0000000 | 0.0223665 | 0.0064815 | 0.0003375 | 0.0014025 | 0.0000000 | 0.0000000 | 0.0000000 | 0.0002016 | 0.0000000 | 0.0003348 | 0.0000000 | 0.0010438 | 0.0000000 | 0.0000000 | 0.0000000 | 0.0003094 | 0.0038715 | 0.0002698 | 0.0027121 | 0.0000000 | 0.0133865 | 0.0025943 | 0.0027506 | 0.0014929 | 0.0005685 | 0.0000000 | 0.0009559 |
| innate lymphocyte | 0.0029548 | 0.0353006 | 0.0045859 | 0.0077706 | 0.0039610 | 0.0023866 | 0.0075522 | 0.0019214 | 0.0012806 | 0.0038101 | 0.0245580 | 0.0046451 | 0.0058068 | 0.0083333 | 0.0044589 | 0.0006984 | 0.0082908 | 0.0014938 | 0.0339105 | 0.0129630 | 0.0006750 | 0.0144928 | 0.0006684 | 0.0007315 | 0.0016584 | 0.0036290 | 0.0039675 | 0.0006696 | 0.0138083 | 0.0013918 | 0.0018120 | 0.0020309 | 0.0002687 | 0.0003094 | 0.0046458 | 0.0010793 | 0.0048431 | 0.0047105 | 0.0179436 | 0.0158019 | 0.0366377 | 0.0142398 | 0.0136441 | 0.0071664 | 0.0023367 |
| macrophages | 0.8518362 | 0.8019857 | 0.9072026 | 0.7607181 | 0.8740098 | 0.9240255 | 0.6010662 | 0.9126425 | 0.8940724 | 0.9422626 | 0.1070727 | 0.7980987 | 0.8719438 | 0.8401961 | 0.8479935 | 0.9353262 | 0.6696429 | 0.9026889 | 0.5313853 | 0.7277778 | 0.8876139 | 0.4684432 | 0.8288770 | 0.8580834 | 0.8995499 | 0.8252016 | 0.8730399 | 0.7937730 | 0.7321182 | 0.9060543 | 0.8969197 | 0.9102356 | 0.9180328 | 0.9260520 | 0.8180410 | 0.8345926 | 0.7268501 | 0.8098426 | 0.5351752 | 0.7023585 | 0.7680713 | 0.5799265 | 0.7072200 | 0.8938237 | 0.8326075 |
| mast cells | 0.0000000 | 0.0000000 | 0.0000000 | 0.0005359 | 0.0001523 | 0.0000000 | 0.0008885 | 0.0000000 | 0.0000000 | 0.0000000 | 0.0098232 | 0.0001080 | 0.0012225 | 0.0014706 | 0.0008107 | 0.0000000 | 0.0063776 | 0.0000000 | 0.0007215 | 0.0023148 | 0.0000000 | 0.0112202 | 0.0000000 | 0.0000000 | 0.0004738 | 0.0004032 | 0.0003779 | 0.0010044 | 0.0000000 | 0.0000000 | 0.0000000 | 0.0004062 | 0.0002687 | 0.0000000 | 0.0007743 | 0.0002698 | 0.0001937 | 0.0021074 | 0.0000000 | 0.0000000 | 0.0000000 | 0.0000000 | 0.0005685 | 0.0000000 | 0.0000000 |
| monocytes | 0.0113972 | 0.0013789 | 0.0013488 | 0.0200965 | 0.0195003 | 0.0023866 | 0.0035540 | 0.0044832 | 0.0012806 | 0.0013189 | 0.0098232 | 0.0468834 | 0.0097800 | 0.0127451 | 0.0097284 | 0.0108954 | 0.0114796 | 0.0134443 | 0.0119048 | 0.0087963 | 0.0155248 | 0.0107527 | 0.0106952 | 0.0065838 | 0.0073442 | 0.0217742 | 0.0171925 | 0.0133914 | 0.0265120 | 0.0059151 | 0.0094625 | 0.0064988 | 0.0037624 | 0.0046411 | 0.0061943 | 0.0218564 | 0.0240217 | 0.1063592 | 0.0005696 | 0.0073113 | 0.0061613 | 0.0297428 | 0.0187607 | 0.0086752 | 0.0065852 |
| neutrophils | 0.0000000 | 0.0000000 | 0.0000000 | 0.0056270 | 0.0012188 | 0.0000000 | 0.0000000 | 0.0001281 | 0.0000000 | 0.0000000 | 0.0108055 | 0.0006482 | 0.0003056 | 0.0014706 | 0.0000000 | 0.0004191 | 0.0063776 | 0.0004268 | 0.0010823 | 0.0013889 | 0.0006750 | 0.0229079 | 0.0033422 | 0.0051207 | 0.0021322 | 0.0022177 | 0.0005668 | 0.0549046 | 0.0035902 | 0.0003479 | 0.0004027 | 0.0012185 | 0.0000000 | 0.0003094 | 0.0000000 | 0.0089045 | 0.0092987 | 0.0004958 | 0.0079749 | 0.0025943 | 0.0004401 | 0.0008039 | 0.0000000 | 0.0000000 | 0.0000000 |
| NK cells | 0.0046433 | 0.0085494 | 0.0051254 | 0.0058950 | 0.0036563 | 0.0000000 | 0.0026655 | 0.0014090 | 0.0020124 | 0.0011723 | 0.0157171 | 0.0016204 | 0.0015281 | 0.0014706 | 0.0008107 | 0.0000000 | 0.0044643 | 0.0014938 | 0.0266955 | 0.0060185 | 0.0016875 | 0.0079476 | 0.0020053 | 0.0000000 | 0.0002369 | 0.0020161 | 0.0011336 | 0.0010044 | 0.0033140 | 0.0003479 | 0.0004027 | 0.0012185 | 0.0013437 | 0.0006188 | 0.0011614 | 0.0000000 | 0.0019372 | 0.0019834 | 0.0136713 | 0.0089623 | 0.0066014 | 0.0062012 | 0.0068221 | 0.0005658 | 0.0098779 |
| NK-T cells | 0.0000000 | 0.0041368 | 0.0002698 | 0.0005359 | 0.0001523 | 0.0000000 | 0.0013327 | 0.0002562 | 0.0005488 | 0.0001465 | 0.0019646 | 0.0000000 | 0.0003056 | 0.0014706 | 0.0000000 | 0.0001397 | 0.0012755 | 0.0002134 | 0.0014430 | 0.0000000 | 0.0000000 | 0.0014025 | 0.0000000 | 0.0000000 | 0.0000000 | 0.0004032 | 0.0011336 | 0.0000000 | 0.0005523 | 0.0003479 | 0.0000000 | 0.0000000 | 0.0000000 | 0.0000000 | 0.0000000 | 0.0000000 | 0.0007749 | 0.0000000 | 0.0019937 | 0.0002358 | 0.0004401 | 0.0009187 | 0.0005685 | 0.0000943 | 0.0009559 |
| proliferating macrophages | 0.0367244 | 0.0493657 | 0.0321014 | 0.0560021 | 0.0467703 | 0.0572792 | 0.0310973 | 0.0462406 | 0.0332967 | 0.0247655 | 0.0000000 | 0.0392136 | 0.0531785 | 0.0303922 | 0.0490474 | 0.0222098 | 0.0197704 | 0.0354247 | 0.0151515 | 0.0444444 | 0.0445494 | 0.0187003 | 0.0407754 | 0.0475494 | 0.0414594 | 0.0556452 | 0.0317400 | 0.0451958 | 0.0425297 | 0.0320111 | 0.0457016 | 0.0097482 | 0.0588551 | 0.0411510 | 0.0514905 | 0.0275229 | 0.0236343 | 0.0115284 | 0.0065508 | 0.0422170 | 0.0259655 | 0.0264125 | 0.0278567 | 0.0375295 | 0.0333510 |
| proliferating T/NK | 0.0021106 | 0.0024821 | 0.0008093 | 0.0013398 | 0.0007617 | 0.0000000 | 0.0044425 | 0.0007685 | 0.0016465 | 0.0010258 | 0.0009823 | 0.0008642 | 0.0003056 | 0.0000000 | 0.0000000 | 0.0006984 | 0.0000000 | 0.0004268 | 0.0086580 | 0.0009259 | 0.0006750 | 0.0018700 | 0.0000000 | 0.0007315 | 0.0004738 | 0.0010081 | 0.0013225 | 0.0003348 | 0.0008285 | 0.0006959 | 0.0008053 | 0.0008123 | 0.0005375 | 0.0009282 | 0.0007743 | 0.0002698 | 0.0029059 | 0.0007438 | 0.0017089 | 0.0009434 | 0.0040709 | 0.0008039 | 0.0039795 | 0.0008487 | 0.0016994 |
Cell type proportions by sample
props$Proportions %>%
data.frame %>%
inner_join(seu@meta.data %>%
dplyr::select(sample.id,
Disease,
Treatment,
Severity,
Group,
Batch,
Age,
Sex),
by = c("sample" = "sample.id")) %>%
distinct()-> dat
ggplot(dat, aes(x = sample, y = Freq, fill = clusters)) +
geom_bar(stat = "identity", color = "black", size = 0.1) +
theme(axis.text.x = element_text(angle = 90,
vjust = 0.5,
hjust = 1),
legend.text = element_text(size = 8),
legend.position = "bottom") +
labs(y = "Proportion", fill = "Cell Label") +
scale_fill_paletteer_d("miscpalettes::pastel", direction = 1) +
facet_grid(~Group, scales = "free_x", space = "free_x")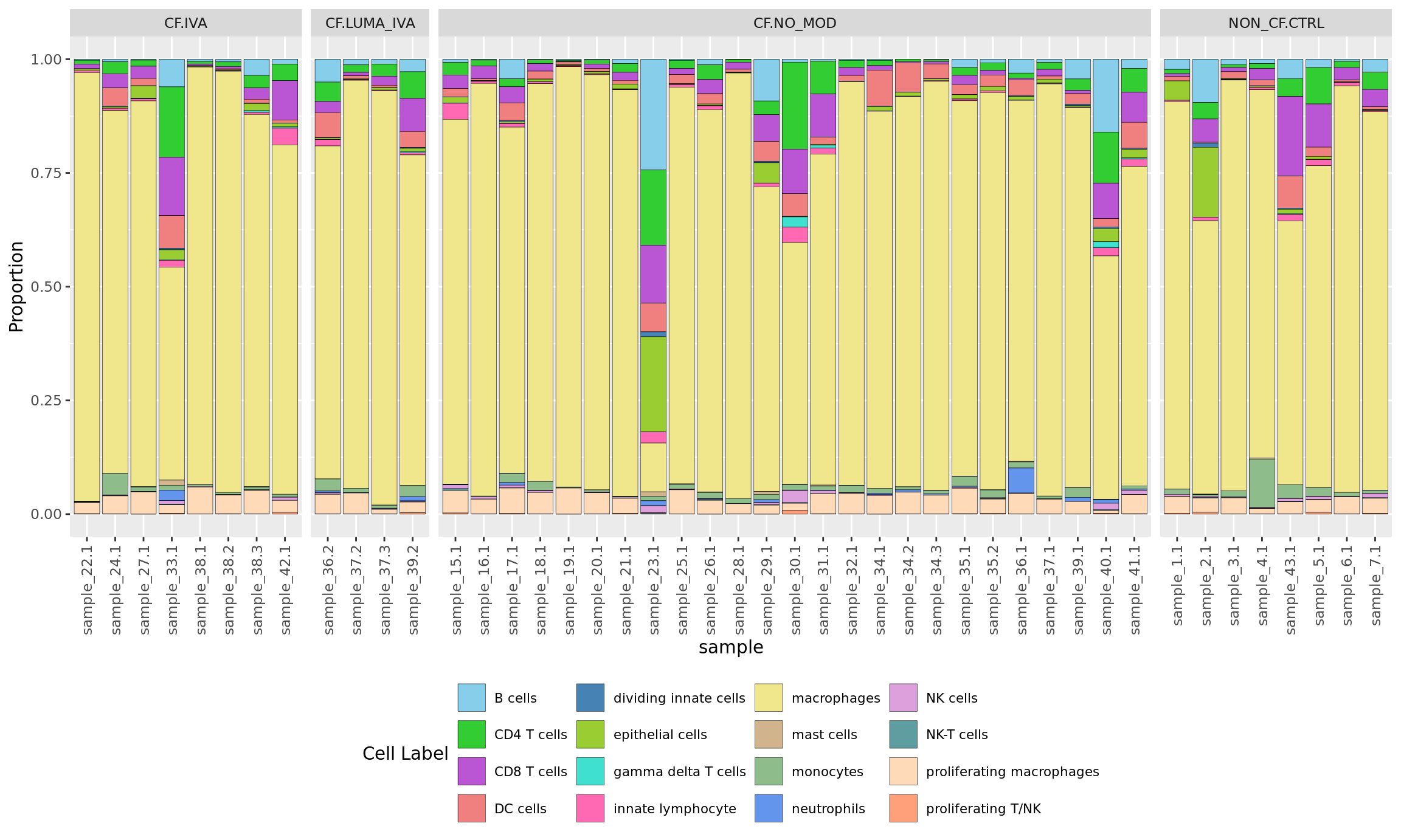
No. cells per sample
props$Counts %>%
data.frame %>%
inner_join(seu@meta.data %>%
dplyr::select(sample.id,
ann_level_1,
Disease,
Treatment,
Severity,
Group,
Group_severity,
Batch,
Age,
Sex),
by = c("sample" = "sample.id", "clusters" = "ann_level_1")) %>%
distinct() -> dat
ggplot(dat, aes(x = sample, y = Freq, fill = Disease)) +
geom_bar(stat = "identity") +
theme(axis.text.x = element_text(angle = 90,
vjust = 0.5,
hjust = 1),
legend.text = element_text(size = 8),
legend.position = "bottom") +
labs(y = "No. cells", fill = "Disease") +
facet_grid(~Group, scales = "free_x", space = "free_x") +
geom_hline(yintercept = 1000, linetype = "dashed")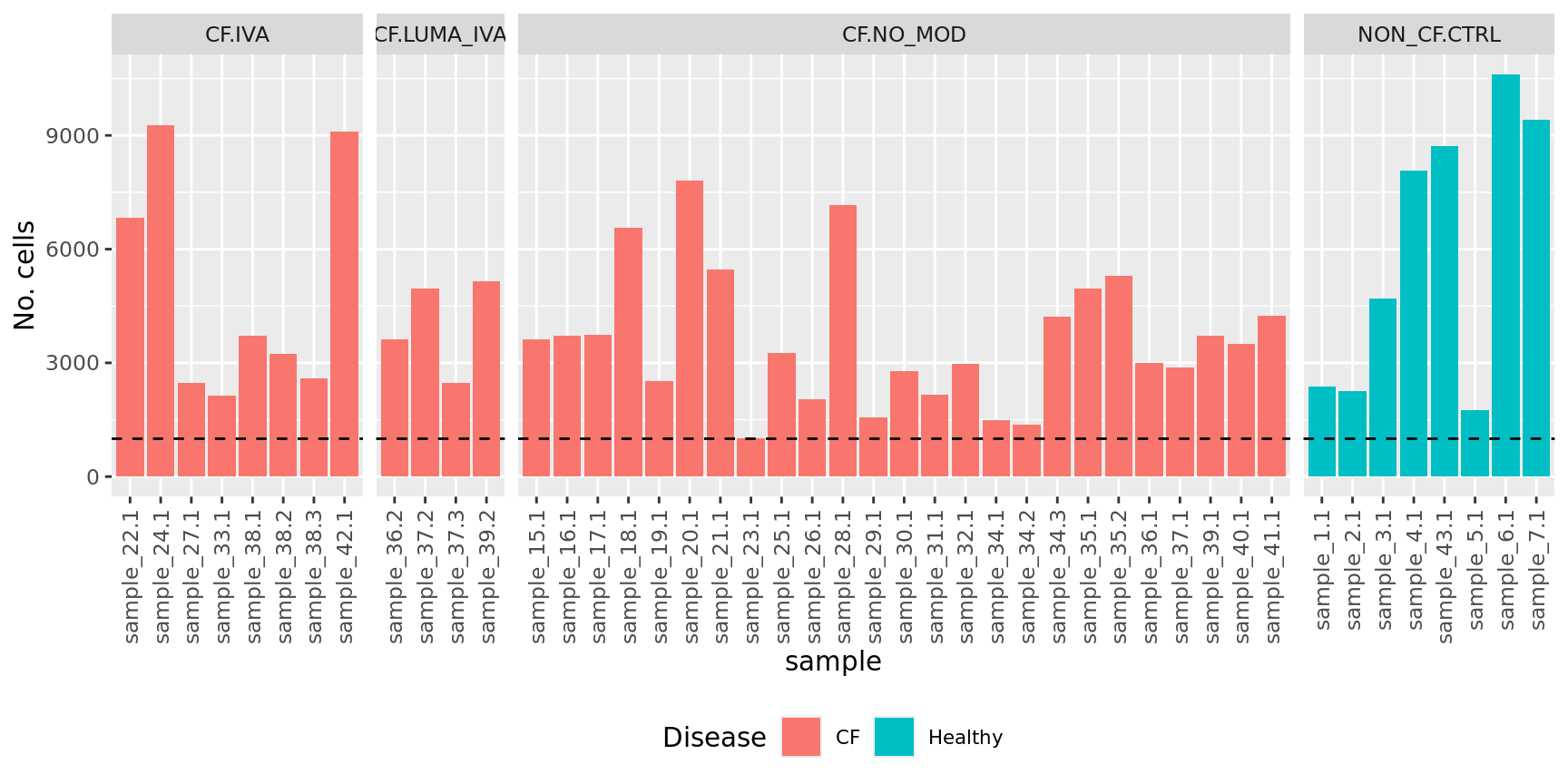
Cell proportions by sample
props$Proportions %>%
data.frame %>%
inner_join(seu@meta.data %>%
dplyr::select(sample.id,
ann_level_1,
Disease,
Treatment,
Severity,
Group,
Group_severity,
Batch,
Age,
Sex),
by = c("sample" = "sample.id", "clusters" = "ann_level_1")) %>%
distinct() -> dat
ggplot(dat, aes(x = sample, y = Freq, fill = clusters)) +
geom_bar(stat = "identity", color = "black", size = 0.1) +
theme(axis.text.x = element_text(angle = 90,
vjust = 0.5,
hjust = 1),
legend.text = element_text(size = 8),
legend.position = "bottom") +
labs(y = "Proportion", fill = "Cell Label") +
scale_fill_paletteer_d("miscpalettes::pastel", direction = 1) +
facet_grid(~Group, scales = "free_x", space = "free_x")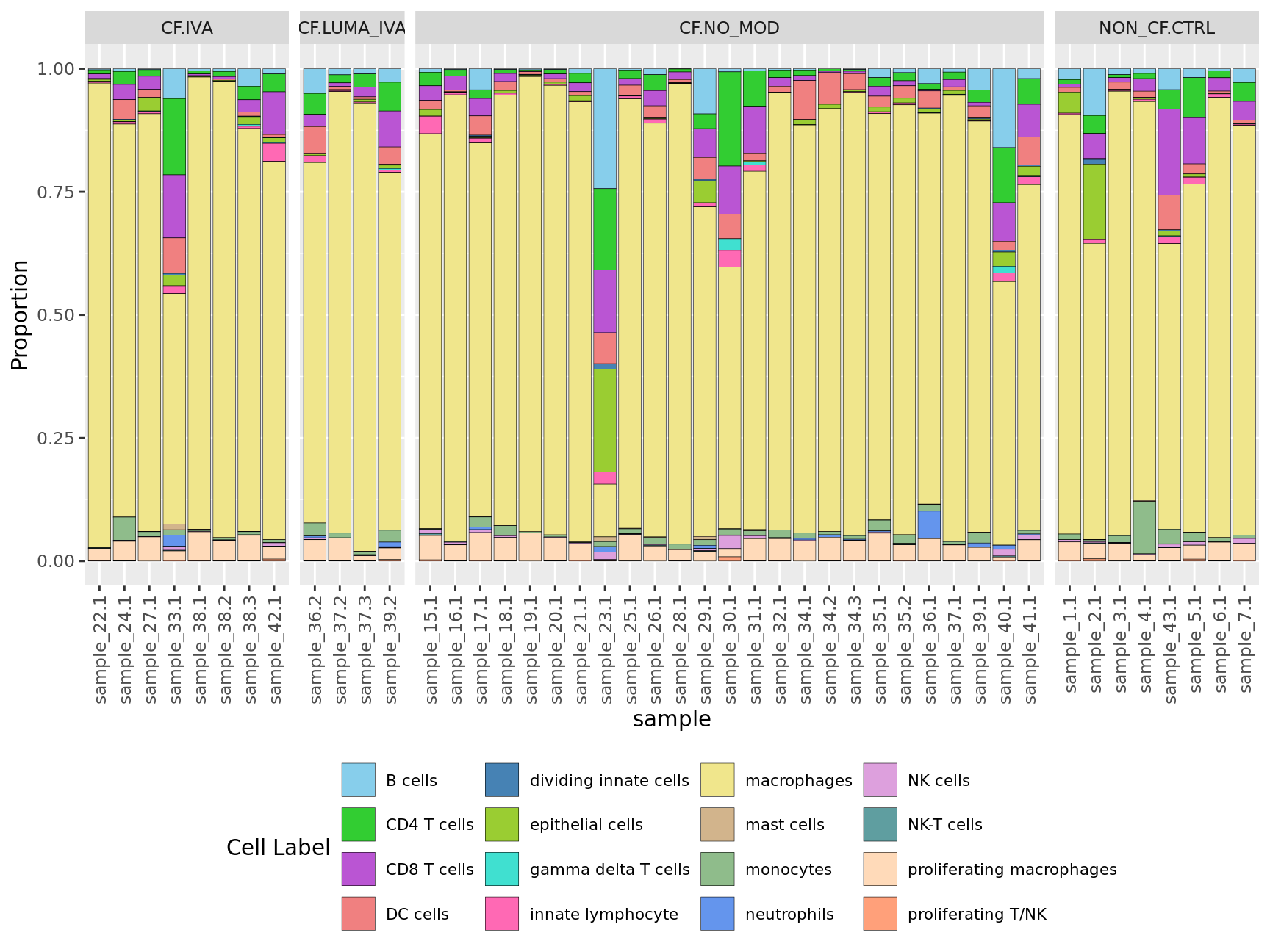
Cell proportions by cell type
props$Proportions %>%
data.frame %>%
inner_join(seu@meta.data %>%
dplyr::select(sample.id,
ann_level_1,
Disease,
Treatment,
Severity,
Group,
Group_severity,
Batch,
Age,
Sex),
by = c("sample" = "sample.id", "clusters" = "ann_level_1")) %>%
distinct() -> dat
ggplot(dat,
aes(x = clusters, y = Freq, fill = clusters)) +
geom_boxplot(outlier.size = 0.1, size = 0.25) +
theme(axis.text.x = element_text(angle = 45,
vjust = 1,
hjust = 1),
legend.text = element_text(size = 8)) +
labs(y = "Proportion") +
scale_fill_paletteer_d("miscpalettes::pastel", direction = 1) +
NoLegend()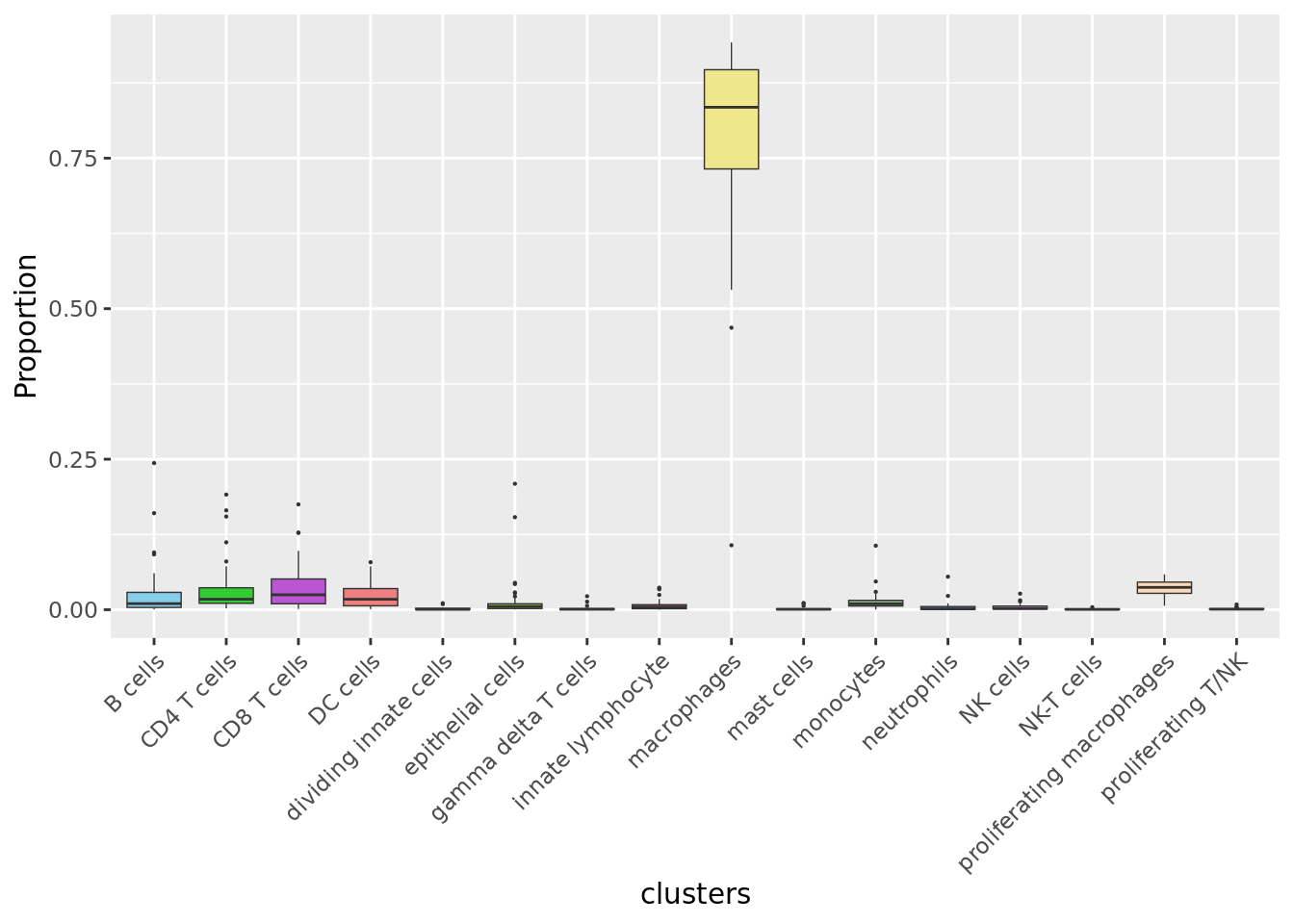
Explore sources of variation
Cell count data
Look at the sources of variation in the raw cell count level data.
dims <- list(c(1,2), c(2:3), c(3,4), c(4,5))
p <- vector("list", length(dims))
for(i in 1:length(dims)){
mds <- plotMDS(props$Counts,
gene.selection = "common",
plot = FALSE, dim.plot = dims[[i]])
data.frame(x = mds$x,
y = mds$y,
sample = rownames(mds$distance.matrix.squared)) %>%
left_join(seu@meta.data %>%
dplyr::select(sample.id,
Disease,
Treatment,
Severity,
Batch,
Age,
Sex),
by = c("sample" = "sample.id")) %>%
distinct() -> dat
p[[i]] <- ggplot(dat, aes(x = x, y = y,
shape = as.factor(Disease),
color = as.factor(Batch))) +
geom_point(size = 3) +
labs(x = glue("Principal Component {dims[[i]][1]}"),
y = glue("Principal Component {dims[[i]][2]}"),
colour = "Batch",
shape = "Disease") +
theme(legend.direction = "horizontal",
legend.text = element_text(size = 8),
legend.title = element_text(size = 9),
axis.text = element_text(size = 8),
axis.title = element_text(size = 9))
}
wrap_plots(p, cols = 2) + plot_layout(guides = "collect") &
theme(legend.position = "bottom") 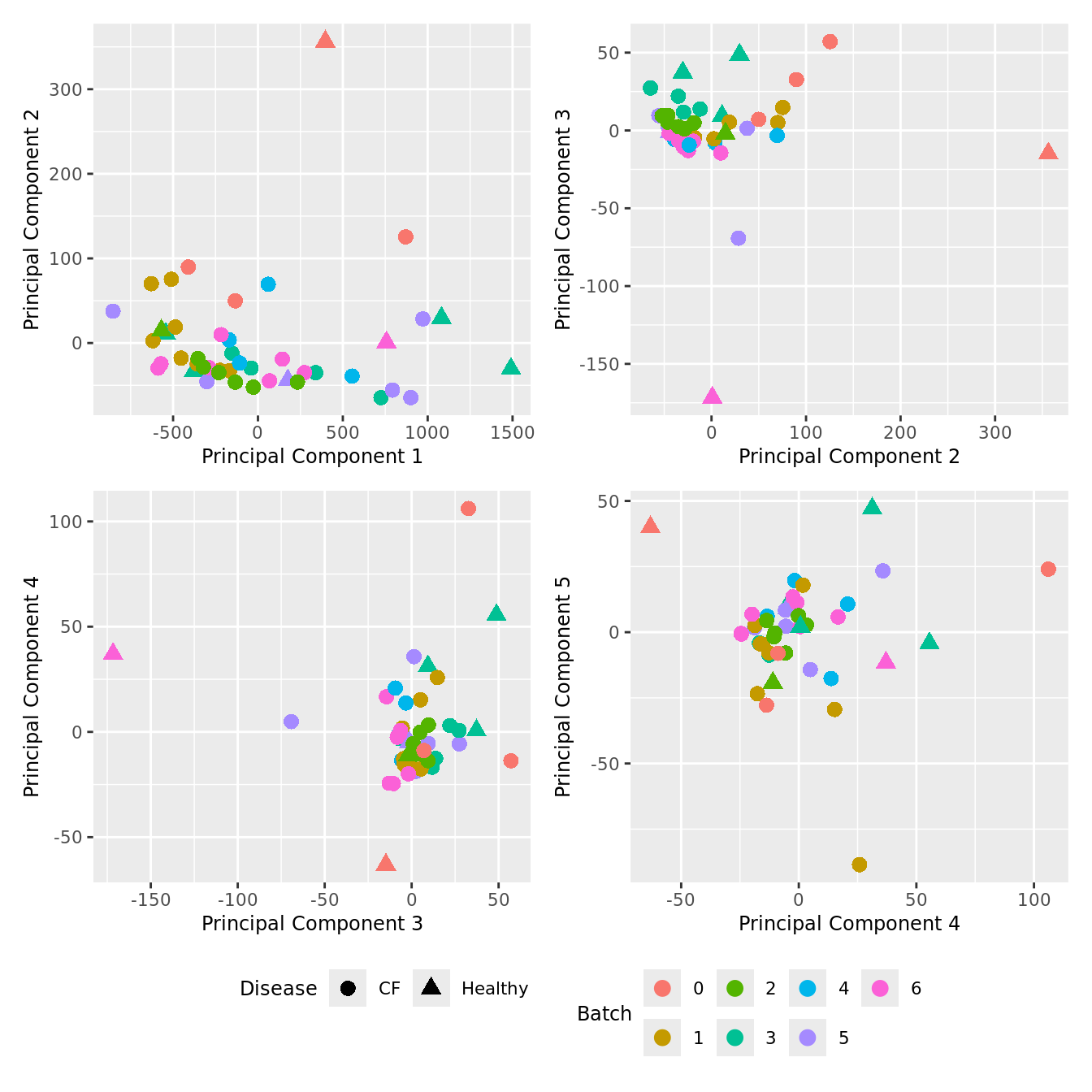
dims <- list(c(1,2), c(2:3), c(3,4), c(4,5))
p <- vector("list", length(dims))
for(i in 1:length(dims)){
mds <- plotMDS(props$Counts,
gene.selection = "common",
plot = FALSE, dim.plot = dims[[i]])
data.frame(x = mds$x,
y = mds$y,
sample = rownames(mds$distance.matrix.squared)) %>%
left_join(seu@meta.data %>%
dplyr::select(sample.id,
Disease,
Treatment,
Severity,
Participant,
Batch,
Age,
Sex),
by = c("sample" = "sample.id")) %>%
group_by(sample) %>%
mutate(ncells = n()) %>%
ungroup() %>%
distinct() -> dat
p[[i]] <- ggplot(dat, aes(x = x, y = y,
colour = log2(ncells)))+
geom_text(aes(label = str_remove_all(sample, "sample_")), size = 2.5) +
labs(x = glue("Principal Component {dims[[i]][1]}"),
y = glue("Principal Component {dims[[i]][2]}"),
colour = "Log2 No. Cells") +
theme(legend.direction = "horizontal",
legend.text = element_text(size = 8),
legend.title = element_text(size = 9),
axis.text = element_text(size = 8),
axis.title = element_text(size = 9)) +
scale_colour_viridis_c(option = "magma")
}
wrap_plots(p, cols = 2) + plot_layout(guides = "collect") &
theme(legend.position = "bottom") 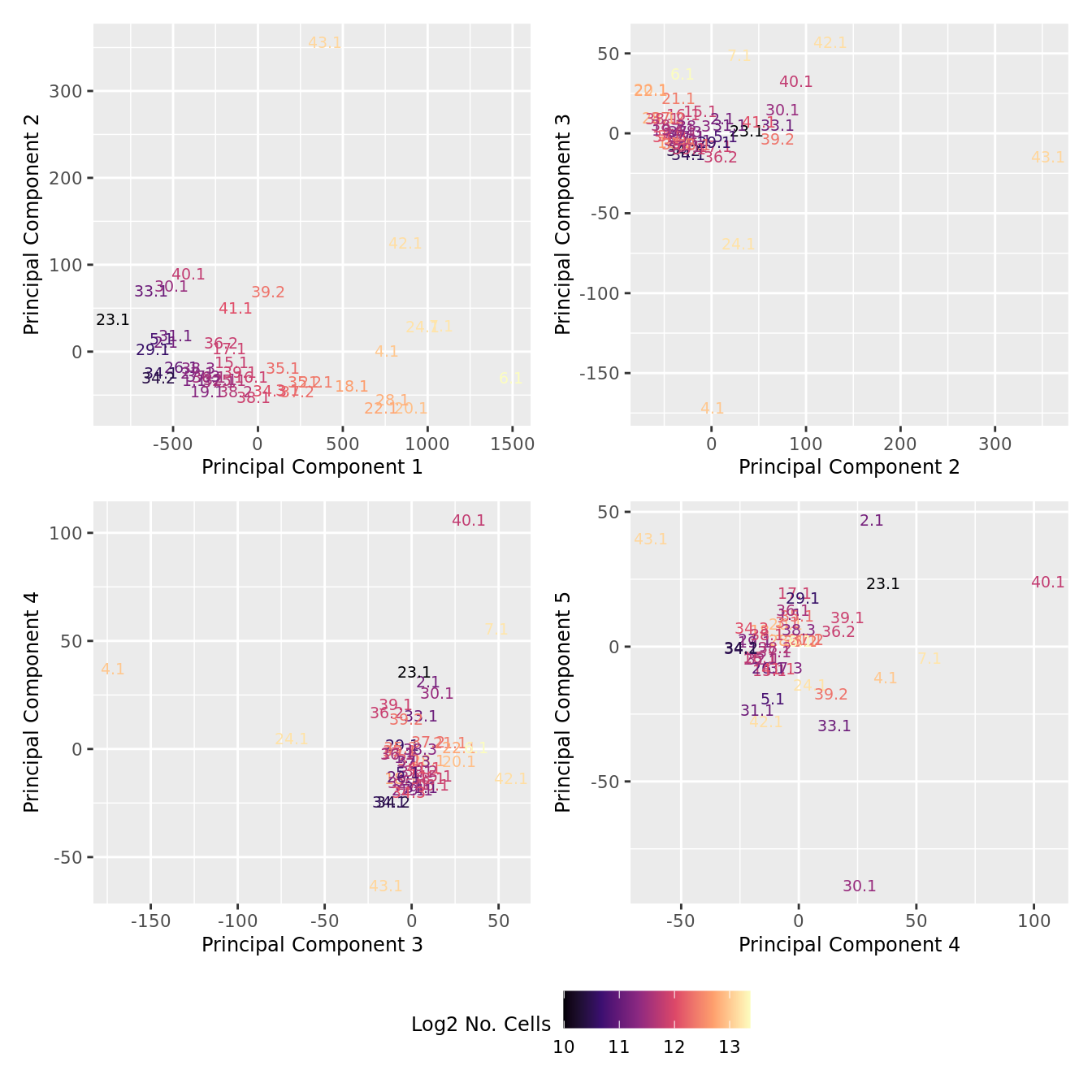
Cell proportion data
Look at the sources of variation in the cell proportions data.
dims <- list(c(1,2), c(2:3), c(3,4), c(4,5))
p <- vector("list", length(dims))
for(i in 1:length(dims)){
mds <- plotMDS(props$TransformedProps,
gene.selection = "common",
plot = FALSE, dim.plot = dims[[i]])
data.frame(x = mds$x,
y = mds$y,
sample = rownames(mds$distance.matrix.squared)) %>%
left_join(seu@meta.data %>%
dplyr::select(sample.id,
Disease,
Treatment,
Severity,
Batch,
Age,
Sex),
by = c("sample" = "sample.id")) %>%
distinct() -> dat
p[[i]] <- ggplot(dat, aes(x = x, y = y,
shape = as.factor(Disease),
color = as.factor(Batch)))+
geom_point(size = 3) +
labs(x = glue("Principal Component {dims[[i]][1]}"),
y = glue("Principal Component {dims[[i]][2]}"),
colour = "Batch",
shape = "Disease") +
theme(legend.direction = "horizontal",
legend.text = element_text(size = 8),
legend.title = element_text(size = 9),
axis.text = element_text(size = 8),
axis.title = element_text(size = 9))
}
wrap_plots(p, cols = 2) + plot_layout(guides = "collect") &
theme(legend.position = "bottom") 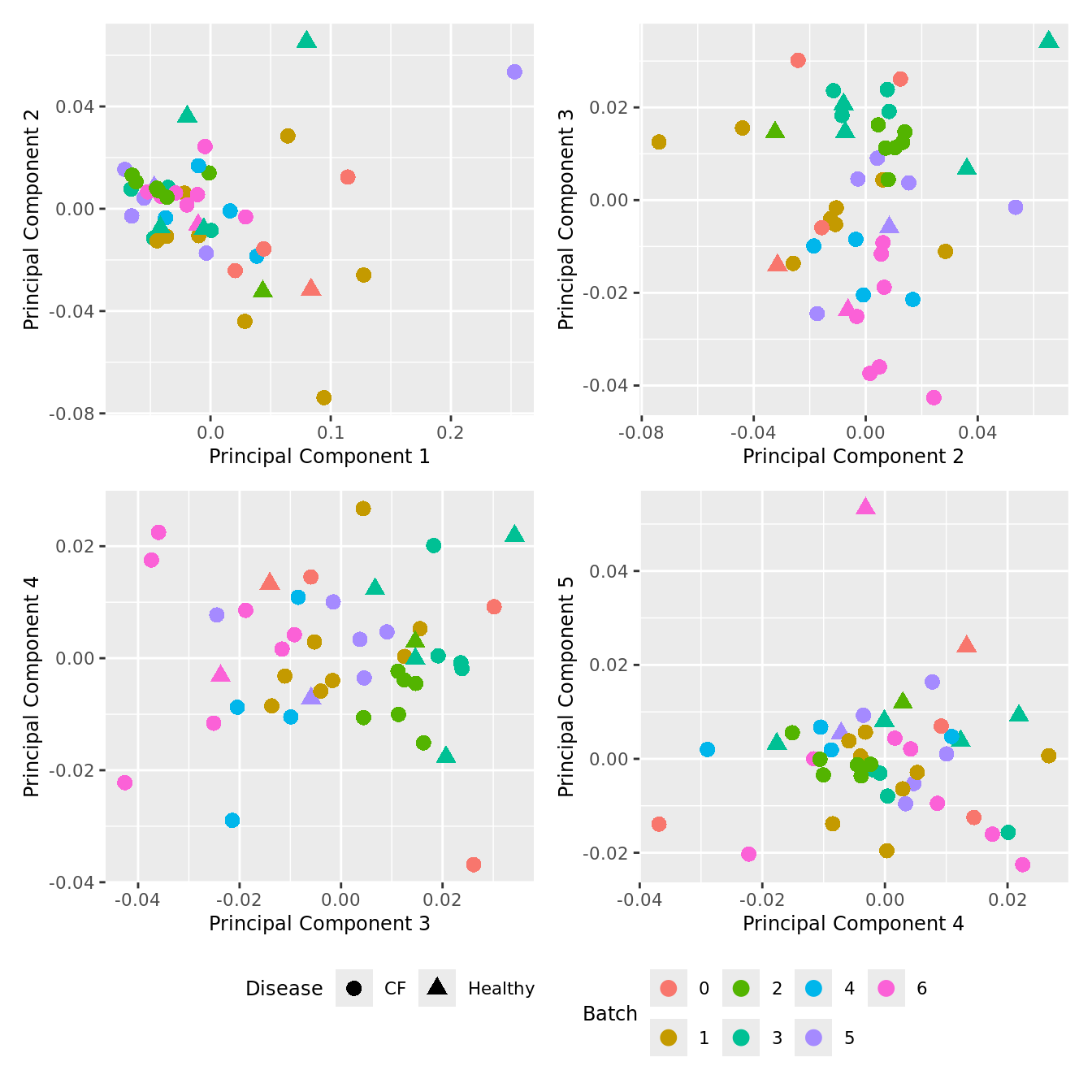
dims <- list(c(1,2), c(2:3), c(3,4), c(4,5))
p <- vector("list", length(dims))
for(i in 1:length(dims)){
mds <- plotMDS(props$TransformedProps,
gene.selection = "common",
plot = FALSE, dim.plot = dims[[i]])
data.frame(x = mds$x,
y = mds$y,
sample = rownames(mds$distance.matrix.squared)) %>%
left_join(seu@meta.data %>%
dplyr::select(sample.id,
Disease,
Treatment,
Severity,
Batch,
Age,
Sex),
by = c("sample" = "sample.id")) %>%
distinct() -> dat
p[[i]] <- ggplot(dat, aes(x = x, y = y,
shape = as.factor(Disease),
color = Sex))+
geom_point(size = 3) +
labs(x = glue("Principal Component {dims[[i]][1]}"),
y = glue("Principal Component {dims[[i]][2]}"),
colour = "Sex",
shape = "Disease") +
theme(legend.direction = "horizontal",
legend.text = element_text(size = 8),
legend.title = element_text(size = 9),
axis.text = element_text(size = 8),
axis.title = element_text(size = 9))
}
wrap_plots(p, cols = 2) + plot_layout(guides = "collect") &
theme(legend.position = "bottom") 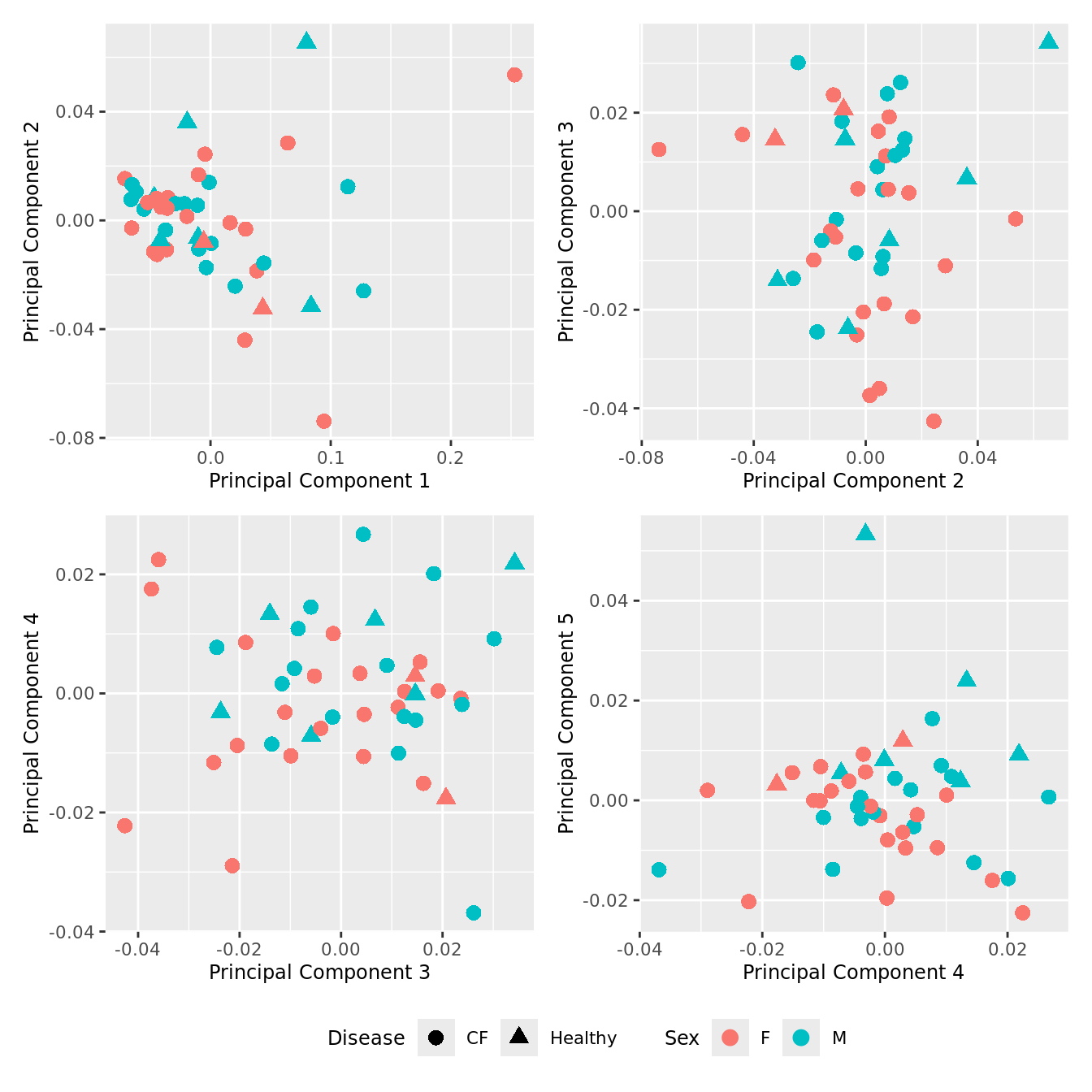
dims <- list(c(1,2), c(2:3), c(3,4), c(4,5))
p <- vector("list", length(dims))
for(i in 1:length(dims)){
mds <- plotMDS(props$TransformedProps,
gene.selection = "common",
plot = FALSE, dim.plot = dims[[i]])
data.frame(x = mds$x,
y = mds$y,
sample = rownames(mds$distance.matrix.squared)) %>%
left_join(seu@meta.data %>%
dplyr::select(sample.id,
Disease,
Treatment,
Severity,
Participant,
Batch,
Age,
Sex),
by = c("sample" = "sample.id")) %>%
group_by(sample) %>%
mutate(ncells = n()) %>%
ungroup() %>%
distinct() -> dat
p[[i]] <- ggplot(dat, aes(x = x, y = y,
colour = log2(Age)))+
geom_text(aes(label = str_remove_all(sample, "sample_")), size = 2.5) +
labs(x = glue("Principal Component {dims[[i]][1]}"),
y = glue("Principal Component {dims[[i]][2]}"),
colour = "Log2 Age") +
theme(legend.direction = "horizontal",
legend.text = element_text(size = 8),
legend.title = element_text(size = 9),
axis.text = element_text(size = 8),
axis.title = element_text(size = 9)) +
scale_colour_viridis_c(option = "magma")
}
wrap_plots(p, cols = 2) + plot_layout(guides = "collect") &
theme(legend.position = "bottom") 
dims <- list(c(1,2), c(2:3), c(3,4), c(4,5))
p <- vector("list", length(dims))
for(i in 1:length(dims)){
mds <- plotMDS(props$TransformedProps,
gene.selection = "common",
plot = FALSE, dim.plot = dims[[i]])
data.frame(x = mds$x,
y = mds$y,
sample = rownames(mds$distance.matrix.squared)) %>%
left_join(seu@meta.data %>%
dplyr::select(sample.id,
Disease,
Treatment,
Severity,
Participant,
Batch,
Age,
Sex),
by = c("sample" = "sample.id")) %>%
group_by(sample) %>%
mutate(ncells = n()) %>%
ungroup() %>%
distinct() -> dat
p[[i]] <- ggplot(dat, aes(x = x, y = y,
colour = log2(ncells)))+
geom_text(aes(label = str_remove_all(sample, "sample_")), size = 2.5) +
labs(x = glue("Principal Component {dims[[i]][1]}"),
y = glue("Principal Component {dims[[i]][2]}"),
colour = "Log2 No. Cells") +
theme(legend.direction = "horizontal",
legend.text = element_text(size = 8),
legend.title = element_text(size = 9),
axis.text = element_text(size = 8),
axis.title = element_text(size = 9)) +
scale_colour_viridis_c(option = "magma")
}
wrap_plots(p, cols = 2) + plot_layout(guides = "collect") &
theme(legend.position = "bottom") 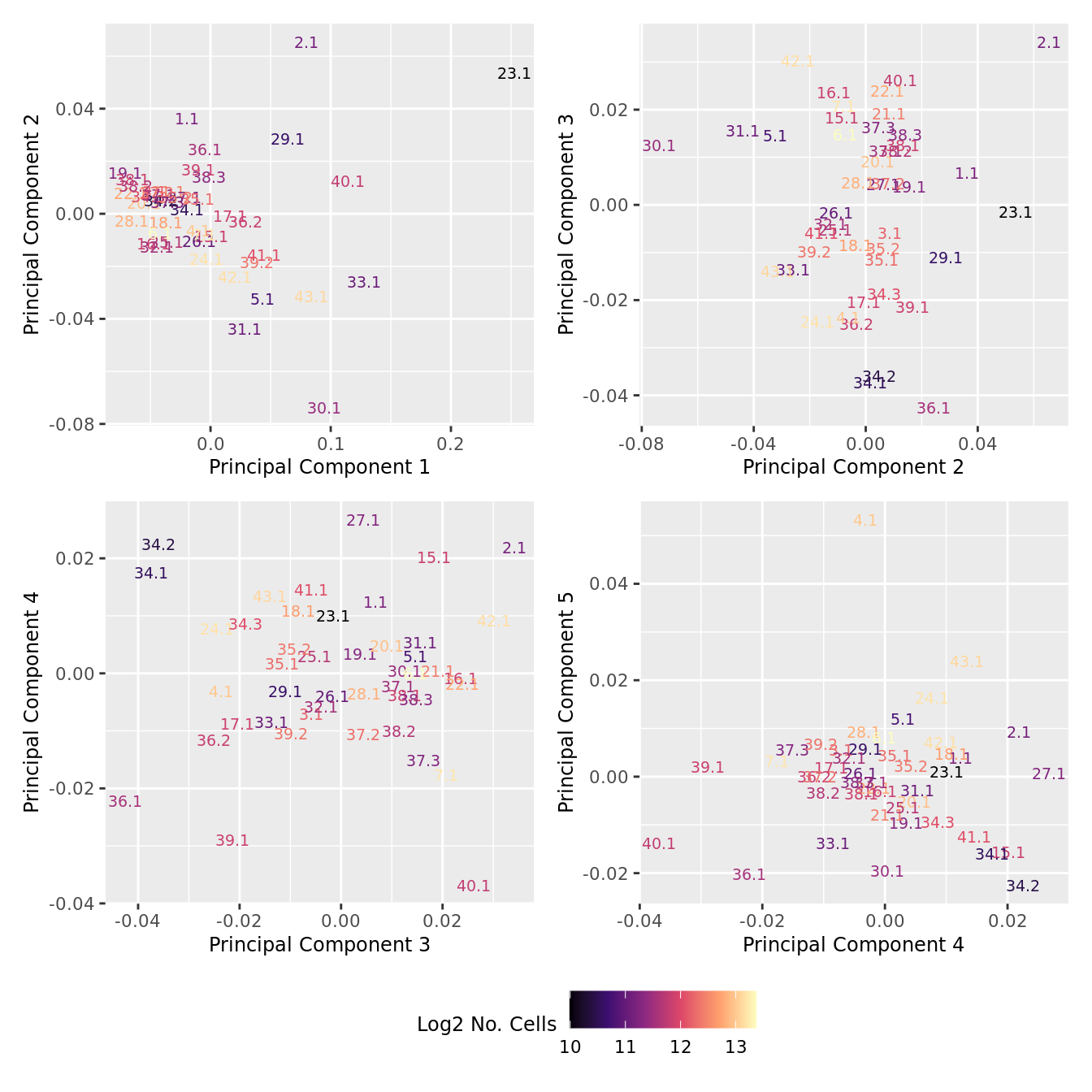
Principal components versus traits
Principal components analysis (PCA) allows us to mathematically determine the sources of variation in the data. We can then investigate whether these correlate with any of the specifed covariates. First, we calculate the principal components. The scree plot belows shows us that most of the variation in this data is captured by the top 7 principal components.
# remove outlier sample
transformed_props <- props$TransformedProps[,colnames(props$TransformedProps) != "sample_23.1"]
PCs <- prcomp(t(transformed_props), center = TRUE,
scale = TRUE, retx = TRUE)
loadings = PCs$x # pc loadings
plot(PCs, type="lines") # scree plot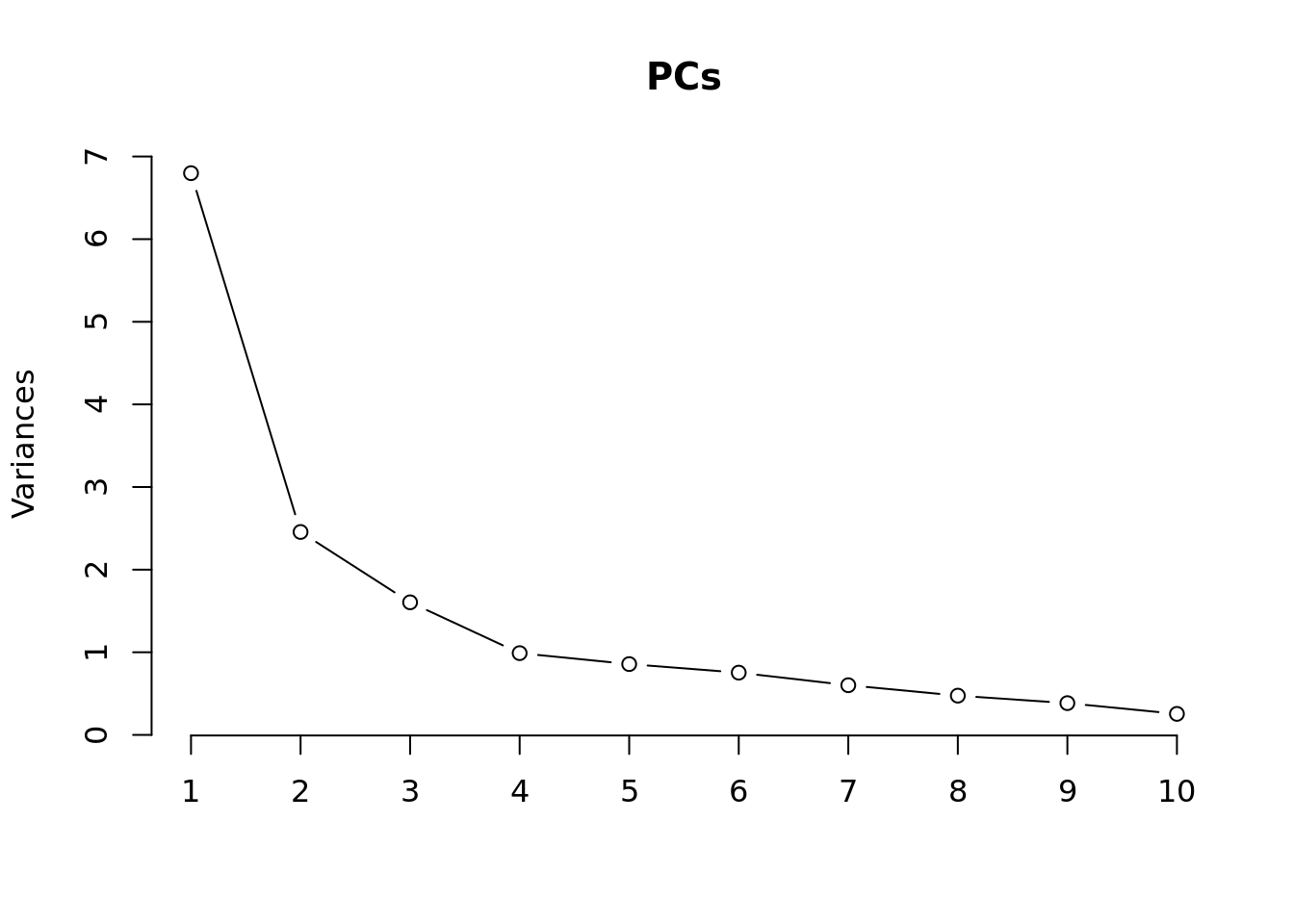
Collect all of the known sample traits.
nGenes = nrow(transformed_props)
nSamples = ncol(transformed_props)
info <- seu@meta.data %>%
dplyr::select(sample.id,
Disease,
Treatment,
Severity,
Participant,
Group_severity,
Group,
Batch,
Age,
Sex) %>%
group_by(sample.id) %>%
mutate(ncells = n()) %>%
ungroup() %>%
distinct()
m <- match(colnames(transformed_props), info$sample.id)
info <- info[m,]
datTraits <- info %>% dplyr::select(Participant, Batch, Disease, Treatment,
Group, Severity, Age, Sex, ncells) %>%
mutate(Age = log2(Age),
ncells = log2(ncells),
Donor = factor(Participant),
Batch = factor(Batch),
Disease = factor(Disease,
labels = 1:length(unique(Disease))),
Group = factor(Group,
labels = 1:length(unique(Group))),
Treatment = factor(Treatment,
labels = 1:length(unique(Treatment))),
Sex = factor(Sex, labels = length(unique(Sex))),
Severity = factor(Severity, labels = length(unique(Severity)))) %>%
mutate(across(everything(), as.numeric)) %>%
dplyr::select(-Participant)
datTraits %>%
knitr::kable()| Batch | Disease | Treatment | Group | Severity | Age | Sex | ncells | Donor |
|---|---|---|---|---|---|---|---|---|
| 4 | 2 | 1 | 4 | 1 | -0.2590872 | 2 | 11.21006 | 1 |
| 4 | 1 | 4 | 3 | 2 | -0.0939001 | 2 | 11.82416 | 2 |
| 4 | 1 | 4 | 3 | 2 | -0.1151479 | 1 | 11.85604 | 3 |
| 5 | 1 | 4 | 3 | 2 | -0.0441471 | 1 | 11.86573 | 4 |
| 5 | 1 | 4 | 3 | 2 | 0.1428834 | 2 | 12.68036 | 5 |
| 6 | 1 | 4 | 3 | 2 | -0.0729608 | 1 | 11.29577 | 6 |
| 4 | 2 | 1 | 4 | 1 | 0.1464588 | 2 | 11.13635 | 7 |
| 6 | 1 | 4 | 3 | 3 | 0.5597097 | 2 | 12.93055 | 8 |
| 4 | 1 | 4 | 3 | 3 | 1.5743836 | 1 | 12.41627 | 9 |
| 4 | 1 | 2 | 1 | 2 | 1.5993830 | 2 | 12.73640 | 10 |
| 6 | 1 | 2 | 1 | 2 | 2.3883594 | 2 | 13.17633 | 11 |
| 2 | 1 | 4 | 3 | 3 | 2.2957230 | 1 | 11.67596 | 12 |
| 2 | 1 | 4 | 3 | 2 | 2.3360877 | 2 | 10.99435 | 13 |
| 2 | 1 | 2 | 1 | 2 | 2.2980155 | 2 | 11.26854 | 14 |
| 6 | 1 | 4 | 3 | 2 | 2.5790214 | 1 | 12.80554 | 15 |
| 2 | 1 | 4 | 3 | 3 | 2.5823250 | 1 | 10.61471 | 16 |
| 6 | 2 | 1 | 4 | 1 | 0.1321035 | 2 | 12.19414 | 17 |
| 2 | 1 | 4 | 3 | 3 | 2.5889097 | 1 | 11.43671 | 18 |
| 2 | 1 | 4 | 3 | 2 | 2.5583683 | 1 | 11.07682 | 19 |
| 2 | 1 | 4 | 3 | 2 | 2.5670653 | 1 | 11.53284 | 20 |
| 2 | 1 | 2 | 1 | 3 | 2.5730557 | 2 | 11.06272 | 21 |
| 7 | 1 | 4 | 3 | 2 | -0.9343238 | 1 | 10.54689 | 22 |
| 7 | 1 | 4 | 3 | 2 | 0.0918737 | 1 | 10.41680 | 22 |
| 7 | 1 | 4 | 3 | 2 | 1.0409164 | 1 | 12.04337 | 22 |
| 7 | 1 | 4 | 3 | 2 | 0.0807044 | 2 | 12.27612 | 23 |
| 7 | 1 | 4 | 3 | 2 | 0.9940589 | 2 | 12.36987 | 23 |
| 7 | 1 | 4 | 3 | 3 | -0.0564254 | 1 | 11.54448 | 24 |
| 7 | 1 | 3 | 2 | 3 | 1.1764977 | 1 | 11.82217 | 24 |
| 3 | 1 | 4 | 3 | 2 | 1.5597097 | 1 | 11.48884 | 25 |
| 3 | 1 | 3 | 2 | 2 | 2.1930156 | 1 | 12.27816 | 25 |
| 3 | 1 | 3 | 2 | 2 | 2.2980155 | 1 | 11.26562 | 25 |
| 3 | 1 | 2 | 1 | 2 | 1.5703964 | 2 | 11.86147 | 26 |
| 3 | 1 | 2 | 1 | 2 | 2.0206033 | 2 | 11.65821 | 26 |
| 3 | 1 | 2 | 1 | 2 | 2.3485584 | 2 | 11.33483 | 26 |
| 5 | 1 | 4 | 3 | 2 | 1.9730702 | 1 | 11.85565 | 27 |
| 5 | 1 | 3 | 2 | 2 | 2.6297159 | 1 | 12.33371 | 27 |
| 7 | 2 | 1 | 4 | 1 | 0.2923784 | 2 | 12.97782 | 28 |
| 1 | 1 | 4 | 3 | 3 | 1.5801455 | 2 | 11.77767 | 29 |
| 1 | 1 | 4 | 3 | 2 | 1.5801455 | 2 | 12.04985 | 30 |
| 1 | 1 | 2 | 1 | 3 | 1.5993178 | 2 | 13.14991 | 31 |
| 1 | 2 | 1 | 4 | 1 | 1.5849625 | 2 | 13.08813 | 32 |
| 3 | 2 | 1 | 4 | 1 | 3.0699187 | 1 | 10.78054 | 33 |
| 4 | 2 | 1 | 4 | 1 | 2.4204621 | 2 | 13.37246 | 34 |
| 4 | 2 | 1 | 4 | 1 | 2.2356012 | 1 | 13.20075 | 35 |
Correlate known sample traits with the top 10 principal components. This can help us determine which traits are potentially contributing to the main sources of variation in the data and should thus be included in our statistical analysis.
moduleTraitCor <- suppressWarnings(cor(loadings[, 1:10], datTraits, use = "p"))
moduleTraitPvalue <- WGCNA::corPvalueStudent(moduleTraitCor, (nSamples - 2))
textMatrix <- paste(signif(moduleTraitCor, 2), "\n(",
signif(moduleTraitPvalue, 1), ")", sep = "")
dim(textMatrix) <- dim(moduleTraitCor)
## Display the correlation values within a heatmap plot
par(cex=0.75, mar = c(6, 8.5, 3, 3))
WGCNA::labeledHeatmap(Matrix = t(moduleTraitCor),
xLabels = colnames(loadings)[1:10],
yLabels = names(datTraits),
colorLabels = FALSE,
colors = WGCNA::blueWhiteRed(6),
textMatrix = t(textMatrix),
setStdMargins = FALSE,
cex.text = 1,
zlim = c(-1,1),
main = paste("PCA-trait relationships: Top 10 PCs"))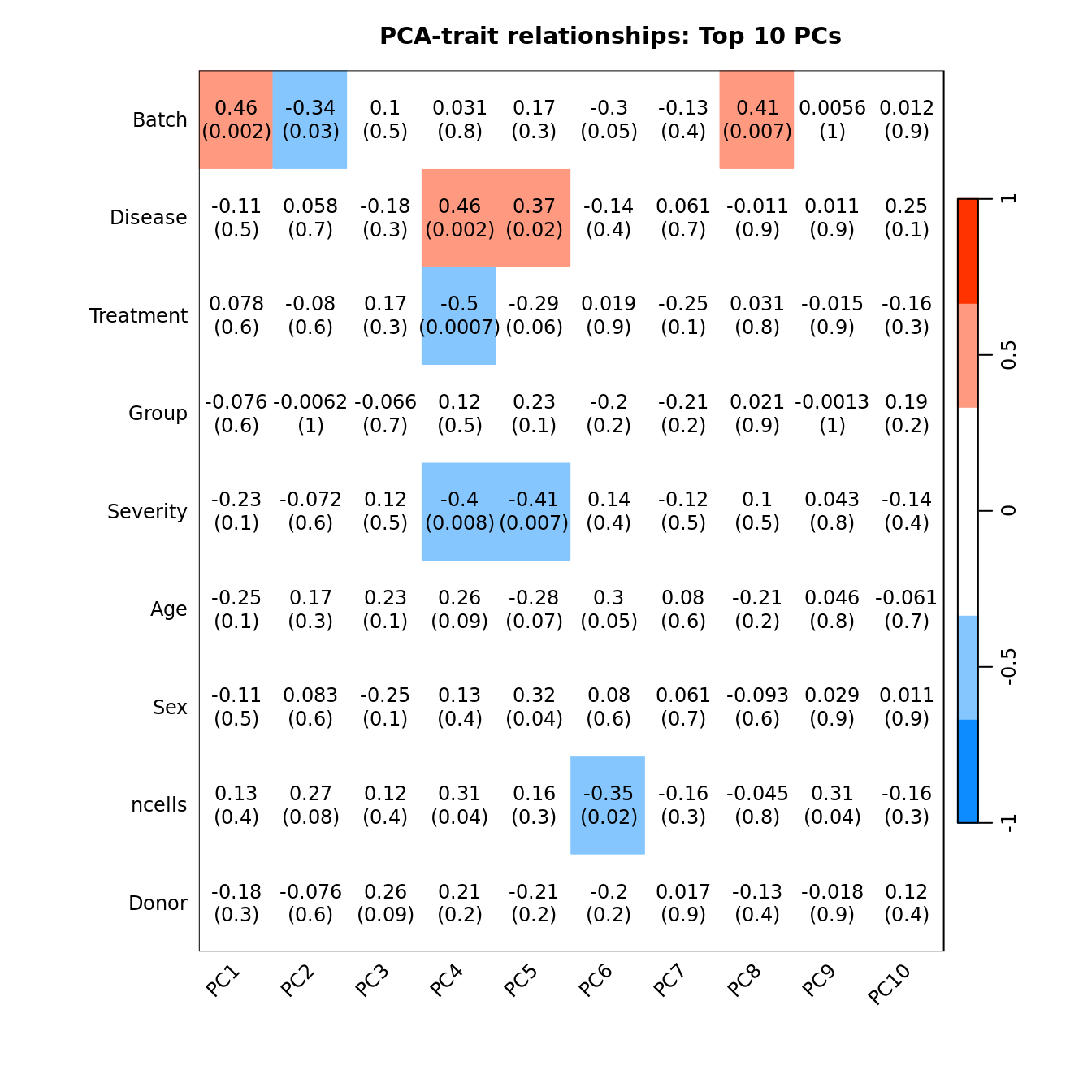
Statistical analysis using propeller and
limma
Create the design matrix.
group <- factor(info$Group_severity)
participant <- factor(info$Participant)
age <- log2(info$Age)
batch <- factor(info$Batch)
sex <- factor(info$Sex)
design <- model.matrix(~ 0 + group + batch + age + sex)
colnames(design)[1:7] <- levels(group)
design CF.IVA.M CF.IVA.S CF.LUMA_IVA.M CF.LUMA_IVA.S CF.NO_MOD.M CF.NO_MOD.S
1 0 0 0 0 0 0
2 0 0 0 0 1 0
3 0 0 0 0 1 0
4 0 0 0 0 1 0
5 0 0 0 0 1 0
6 0 0 0 0 1 0
7 0 0 0 0 0 0
8 0 0 0 0 0 1
9 0 0 0 0 0 1
10 1 0 0 0 0 0
11 1 0 0 0 0 0
12 0 0 0 0 0 1
13 0 0 0 0 1 0
14 1 0 0 0 0 0
15 0 0 0 0 1 0
16 0 0 0 0 0 1
17 0 0 0 0 0 0
18 0 0 0 0 0 1
19 0 0 0 0 1 0
20 0 0 0 0 1 0
21 0 1 0 0 0 0
22 0 0 0 0 1 0
23 0 0 0 0 1 0
24 0 0 0 0 1 0
25 0 0 0 0 1 0
26 0 0 0 0 1 0
27 0 0 0 0 0 1
28 0 0 0 1 0 0
29 0 0 0 0 1 0
30 0 0 1 0 0 0
31 0 0 1 0 0 0
32 1 0 0 0 0 0
33 1 0 0 0 0 0
34 1 0 0 0 0 0
35 0 0 0 0 1 0
36 0 0 1 0 0 0
37 0 0 0 0 0 0
38 0 0 0 0 0 1
39 0 0 0 0 1 0
40 0 1 0 0 0 0
41 0 0 0 0 0 0
42 0 0 0 0 0 0
43 0 0 0 0 0 0
44 0 0 0 0 0 0
NON_CF.CTRL batch1 batch2 batch3 batch4 batch5 batch6 age sexM
1 1 0 0 1 0 0 0 -0.25908722 1
2 0 0 0 1 0 0 0 -0.09390014 1
3 0 0 0 1 0 0 0 -0.11514787 0
4 0 0 0 0 1 0 0 -0.04414710 0
5 0 0 0 0 1 0 0 0.14288337 1
6 0 0 0 0 0 1 0 -0.07296080 0
7 1 0 0 1 0 0 0 0.14645883 1
8 0 0 0 0 0 1 0 0.55970971 1
9 0 0 0 1 0 0 0 1.57438357 0
10 0 0 0 1 0 0 0 1.59938302 1
11 0 0 0 0 0 1 0 2.38835941 1
12 0 1 0 0 0 0 0 2.29572302 0
13 0 1 0 0 0 0 0 2.33608770 1
14 0 1 0 0 0 0 0 2.29801547 1
15 0 0 0 0 0 1 0 2.57902140 0
16 0 1 0 0 0 0 0 2.58232503 0
17 1 0 0 0 0 1 0 0.13210354 1
18 0 1 0 0 0 0 0 2.58890969 0
19 0 1 0 0 0 0 0 2.55836829 0
20 0 1 0 0 0 0 0 2.56706530 0
21 0 1 0 0 0 0 0 2.57305573 1
22 0 0 0 0 0 0 1 -0.93432383 0
23 0 0 0 0 0 0 1 0.09187369 0
24 0 0 0 0 0 0 1 1.04091644 0
25 0 0 0 0 0 0 1 0.08070438 1
26 0 0 0 0 0 0 1 0.99405890 1
27 0 0 0 0 0 0 1 -0.05642543 0
28 0 0 0 0 0 0 1 1.17649766 0
29 0 0 1 0 0 0 0 1.55970971 0
30 0 0 1 0 0 0 0 2.19301559 0
31 0 0 1 0 0 0 0 2.29801547 0
32 0 0 1 0 0 0 0 1.57039639 1
33 0 0 1 0 0 0 0 2.02060327 1
34 0 0 1 0 0 0 0 2.34855840 1
35 0 0 0 0 1 0 0 1.97307024 0
36 0 0 0 0 1 0 0 2.62971590 0
37 1 0 0 0 0 0 1 0.29237837 1
38 0 0 0 0 0 0 0 1.58014548 1
39 0 0 0 0 0 0 0 1.58014548 1
40 0 0 0 0 0 0 0 1.59931779 1
41 1 0 0 0 0 0 0 1.58496250 1
42 1 0 1 0 0 0 0 3.06991870 0
43 1 0 0 1 0 0 0 2.42046210 1
44 1 0 0 1 0 0 0 2.23560118 0
attr(,"assign")
[1] 1 1 1 1 1 1 1 2 2 2 2 2 2 3 4
attr(,"contrasts")
attr(,"contrasts")$group
[1] "contr.treatment"
attr(,"contrasts")$batch
[1] "contr.treatment"
attr(,"contrasts")$sex
[1] "contr.treatment"Create the contrast matrix.
contr <- makeContrasts(CF.NO_MODvNON_CF.CTRL = 0.5*(CF.NO_MOD.M + CF.NO_MOD.S) - NON_CF.CTRL,
CF.IVAvCF.NO_MOD = 0.5*(CF.IVA.S + CF.IVA.M) - 0.5*(CF.NO_MOD.S + CF.NO_MOD.M),
CF.LUMA_IVAvCF.NO_MOD = 0.5*(CF.LUMA_IVA.S + CF.LUMA_IVA.M) - 0.5*(CF.NO_MOD.S + CF.NO_MOD.M),
CF.NO_MOD.SvCF.NO_MOD.M = CF.NO_MOD.S - CF.NO_MOD.M,
levels = design)
contr Contrasts
Levels CF.NO_MODvNON_CF.CTRL CF.IVAvCF.NO_MOD CF.LUMA_IVAvCF.NO_MOD
CF.IVA.M 0.0 0.5 0.0
CF.IVA.S 0.0 0.5 0.0
CF.LUMA_IVA.M 0.0 0.0 0.5
CF.LUMA_IVA.S 0.0 0.0 0.5
CF.NO_MOD.M 0.5 -0.5 -0.5
CF.NO_MOD.S 0.5 -0.5 -0.5
NON_CF.CTRL -1.0 0.0 0.0
batch1 0.0 0.0 0.0
batch2 0.0 0.0 0.0
batch3 0.0 0.0 0.0
batch4 0.0 0.0 0.0
batch5 0.0 0.0 0.0
batch6 0.0 0.0 0.0
age 0.0 0.0 0.0
sexM 0.0 0.0 0.0
Contrasts
Levels CF.NO_MOD.SvCF.NO_MOD.M
CF.IVA.M 0
CF.IVA.S 0
CF.LUMA_IVA.M 0
CF.LUMA_IVA.S 0
CF.NO_MOD.M -1
CF.NO_MOD.S 1
NON_CF.CTRL 0
batch1 0
batch2 0
batch3 0
batch4 0
batch5 0
batch6 0
age 0
sexM 0Add random effect for samples from the same individual.
dupcor <- duplicateCorrelation(transformed_props, design=design,
block=participant)
dupcor$consensus.correlation
[1] 0.6815988
$cor
[1] 0.6815988
$atanh.correlations
[1] 0.91280819 1.07781785 -0.05536504 0.85854344 0.52144441 1.11093619
[7] 0.20770523 1.17122725 0.94099741 0.54921399 1.71581017 1.63069725
[13] 1.14397325 0.51267513 -0.53606034 0.97778570Fit the model.
fit <- lmFit(transformed_props, design=design, block=participant,
correlation=dupcor$consensus)
fit2 <- contrasts.fit(fit, contr)
fit2 <- eBayes(fit2, robust=TRUE, trend=FALSE)
pvalue <- 0.05
summary(decideTests(fit2, p.value = pvalue)) CF.NO_MODvNON_CF.CTRL CF.IVAvCF.NO_MOD CF.LUMA_IVAvCF.NO_MOD
Down 1 0 1
NotSig 15 16 15
Up 0 0 0
CF.NO_MOD.SvCF.NO_MOD.M
Down 0
NotSig 16
Up 0Results
p <- vector("list", ncol(contr))
for(i in 1:ncol(contr)){
print(knitr::kable(topTable(fit2, coef = i),
caption = colnames(contr)[i]))
transformed_props %>% data.frame %>%
left_join(info,
by = c("sample" = "sample.id")) %>%
mutate(Group = Group_severity) %>%
dplyr::filter(Group %in% names(contr[,i])[abs(contr[, i]) > 0]) -> dat
if(length(unique(dat$Group)) > 2) dat$Group <- str_remove(dat$Group, ".(M|S)$")
ggplot(dat, aes(x = Group,
y = Freq,
colour = Group,
group = Group)) +
geom_jitter(stat = "identity",
width = 0.15,
size = 2) +
stat_summary(geom = "point",
fun.y = "mean",
col = "black",
shape = "_",
size = 14) +
theme_classic() +
theme(axis.text.x = element_text(angle = 90,
hjust = 1,
vjust = 0.5),
legend.position = "bottom",
legend.direction = "horizontal") +
labs(x = "Group", y = "Proportion",
colour = "Condition") +
facet_wrap(~clusters, scales = "free_y") +
ggtitle(colnames(contr)[i]) -> p[[i]]
print(p[[i]])
}| logFC | AveExpr | t | P.Value | adj.P.Val | B | |
|---|---|---|---|---|---|---|
| monocytes | -0.0866804 | 0.1046134 | -4.5306909 | 0.0000812 | 0.0012991 | 1.413145 |
| CD8 T cells | -0.0741143 | 0.1671905 | -1.8914421 | 0.0679821 | 0.5438572 | -4.779980 |
| macrophages | 0.0887124 | 1.1311489 | 1.3773538 | 0.1783099 | 0.7581057 | -5.557175 |
| NK-T cells | 0.0121502 | 0.0158486 | 1.3412624 | 0.1895264 | 0.7581057 | -5.603834 |
| gamma delta T cells | 0.0183142 | 0.0242845 | 1.1081569 | 0.2762713 | 0.8449842 | -5.878127 |
| neutrophils | 0.0195569 | 0.0365714 | 1.0017495 | 0.3241777 | 0.8449842 | -5.987338 |
| B cells | -0.0286482 | 0.1209003 | -0.6654712 | 0.5106920 | 0.8449842 | -6.263453 |
| DC cells | -0.0196543 | 0.1376617 | -0.6424225 | 0.5253424 | 0.8449842 | -6.278446 |
| dividing innate cells | -0.0073307 | 0.0232467 | -0.5587345 | 0.5803349 | 0.8449842 | -6.328590 |
| innate lymphocyte | 0.0095538 | 0.0733282 | 0.4797679 | 0.6347417 | 0.8449842 | -6.369579 |
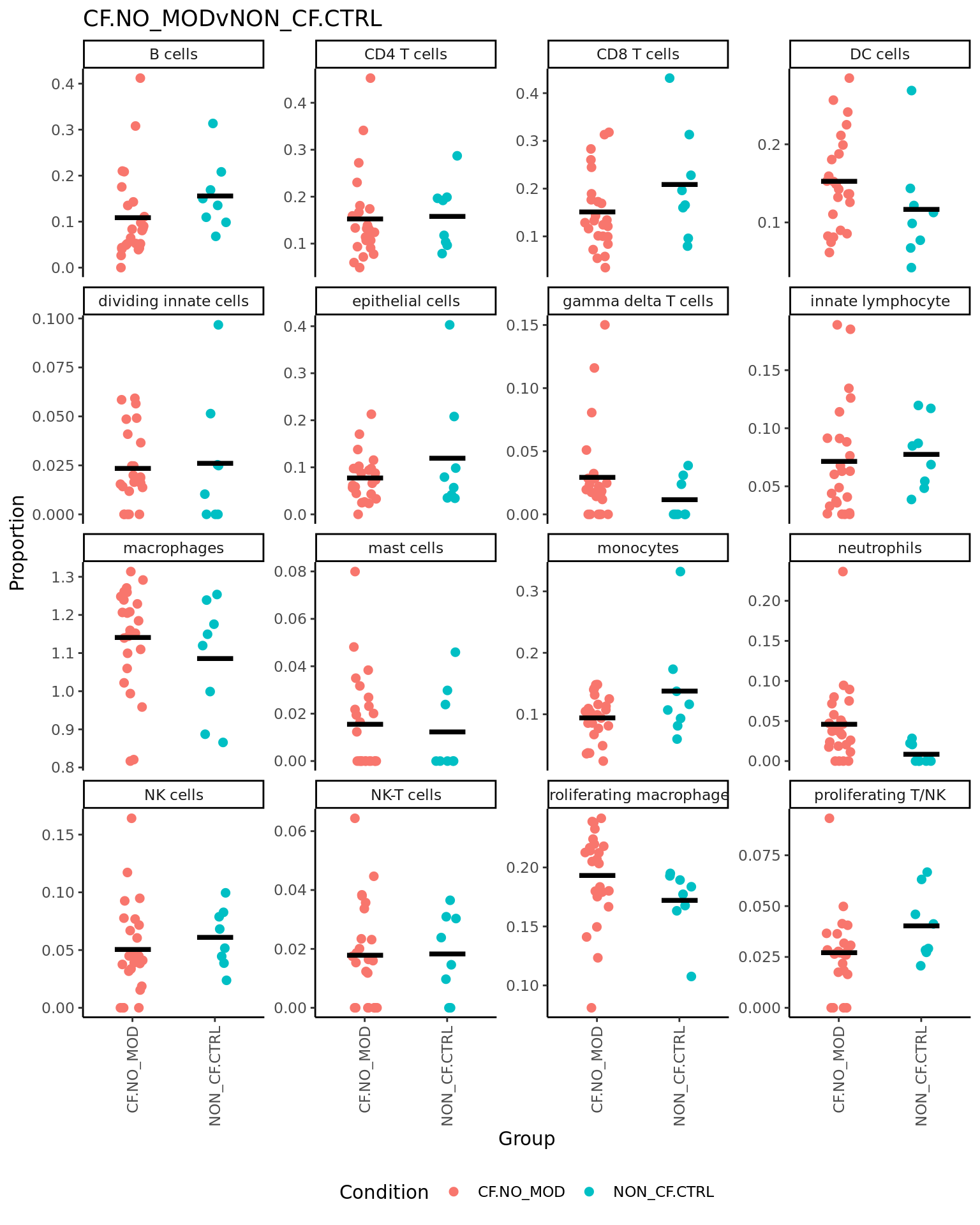
| logFC | AveExpr | t | P.Value | adj.P.Val | B | |
|---|---|---|---|---|---|---|
| NK-T cells | -0.0178018 | 0.0158486 | -1.6771544 | 0.1035136 | 0.9866568 | -4.880636 |
| mast cells | 0.0125230 | 0.0158193 | 1.0766777 | 0.2898866 | 0.9866568 | -5.649573 |
| monocytes | 0.0204331 | 0.1046134 | 0.9115003 | 0.3690271 | 0.9866568 | -5.807150 |
| CD8 T cells | 0.0365180 | 0.1671905 | 0.7953837 | 0.4324648 | 0.9866568 | -5.903003 |
| neutrophils | 0.0180237 | 0.0365714 | 0.7879191 | 0.4366948 | 0.9866568 | -5.908806 |
| gamma delta T cells | -0.0131149 | 0.0242845 | -0.6772627 | 0.5032350 | 0.9866568 | -5.987763 |
| B cells | -0.0232246 | 0.1209003 | -0.4604248 | 0.6484389 | 0.9866568 | -6.108684 |
| macrophages | -0.0324599 | 1.1311489 | -0.4301181 | 0.6700981 | 0.9866568 | -6.121979 |
| DC cells | 0.0105310 | 0.1376617 | 0.2937740 | 0.7708971 | 0.9866568 | -6.170703 |
| proliferating T/NK | 0.0033930 | 0.0310578 | 0.2711860 | 0.7880358 | 0.9866568 | -6.177025 |
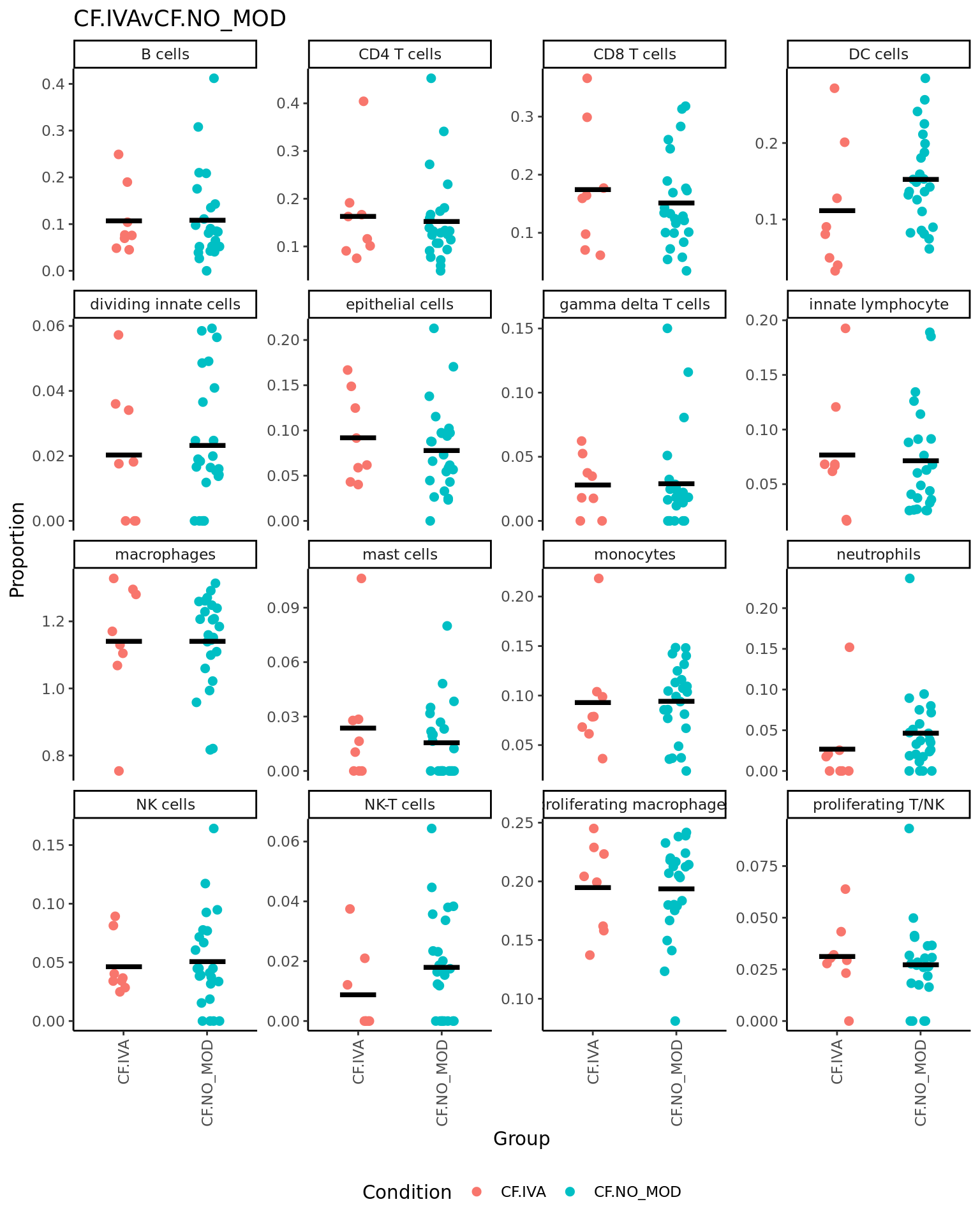
| logFC | AveExpr | t | P.Value | adj.P.Val | B | |
|---|---|---|---|---|---|---|
| neutrophils | -0.0623699 | 0.0365714 | -3.409832 | 0.0018145 | 0.0290321 | -1.482959 |
| innate lymphocyte | 0.0449772 | 0.0733282 | 2.410706 | 0.0220055 | 0.1760439 | -3.756250 |
| mast cells | -0.0165537 | 0.0158193 | -1.779886 | 0.0848459 | 0.4089010 | -4.914571 |
| CD8 T cells | 0.0618318 | 0.1671905 | 1.684230 | 0.1022253 | 0.4089010 | -5.066366 |
| NK cells | 0.0236804 | 0.0505933 | 1.557941 | 0.1293431 | 0.4138978 | -5.255670 |
| CD4 T cells | 0.0514344 | 0.1587382 | 1.414851 | 0.1671225 | 0.4456600 | -5.455042 |
| monocytes | 0.0223527 | 0.1046134 | 1.247018 | 0.2216793 | 0.4552617 | -5.667274 |
| DC cells | 0.0342966 | 0.1376617 | 1.196499 | 0.2406122 | 0.4552617 | -5.726384 |
| macrophages | -0.0697972 | 1.1311489 | -1.156640 | 0.2562936 | 0.4552617 | -5.771489 |
| gamma delta T cells | -0.0168610 | 0.0242845 | -1.088915 | 0.2845386 | 0.4552617 | -5.844982 |
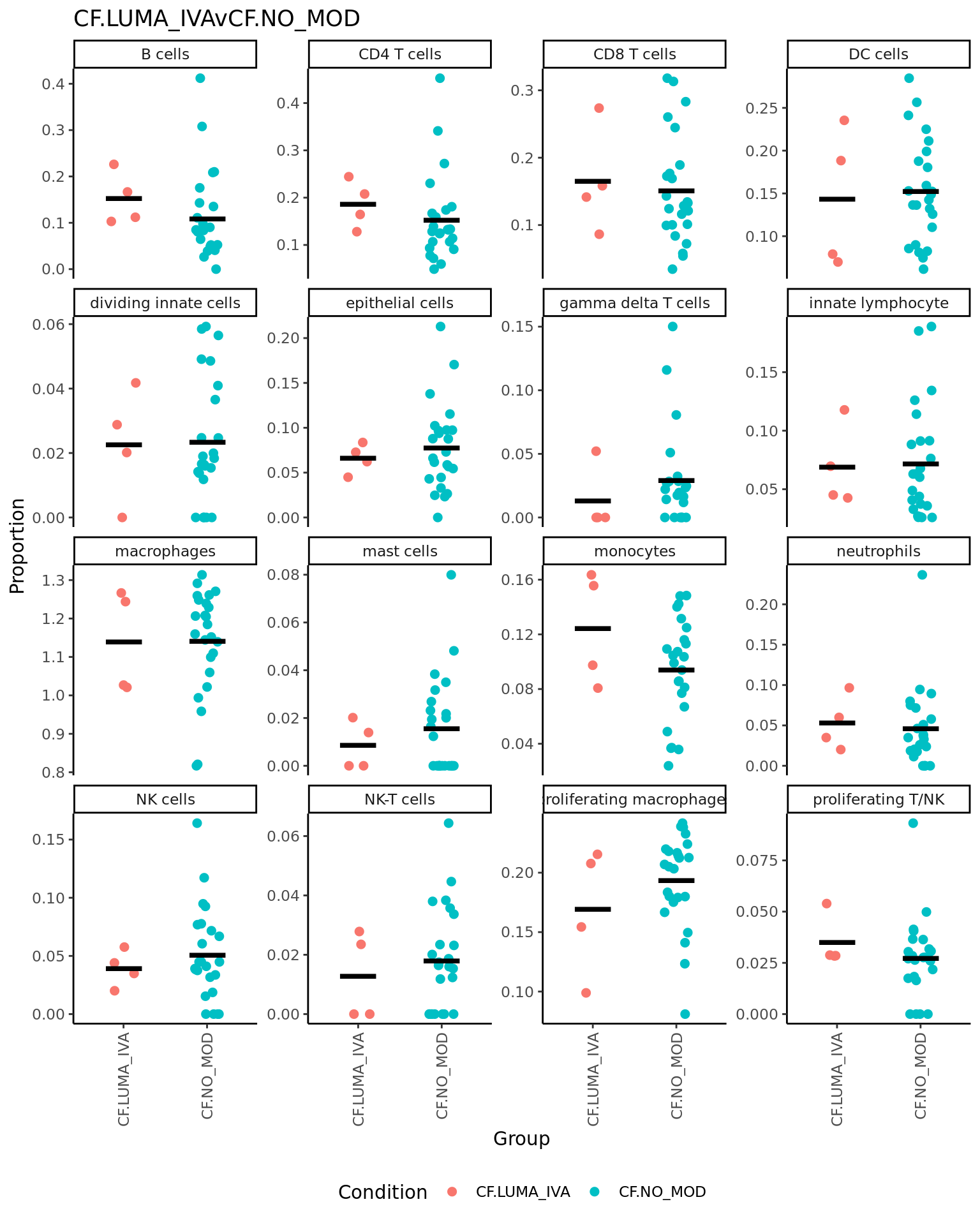
| logFC | AveExpr | t | P.Value | adj.P.Val | B | |
|---|---|---|---|---|---|---|
| neutrophils | 0.0399716 | 0.0365714 | 2.159654 | 0.0386053 | 0.3092130 | -4.238117 |
| B cells | 0.0881572 | 0.1209003 | 2.160049 | 0.0386516 | 0.3092130 | -4.238184 |
| macrophages | -0.0954724 | 1.1311489 | -1.563554 | 0.1281186 | 0.4022276 | -5.236842 |
| NK cells | 0.0222587 | 0.0505933 | 1.447224 | 0.1578166 | 0.4022276 | -5.400533 |
| CD4 T cells | 0.0486446 | 0.1587382 | 1.322411 | 0.1957475 | 0.4022276 | -5.563891 |
| gamma delta T cells | 0.0204081 | 0.0242845 | 1.302533 | 0.2022756 | 0.4022276 | -5.588735 |
| mast cells | 0.0113367 | 0.0158193 | 1.204650 | 0.2374051 | 0.4022276 | -5.705992 |
| proliferating macrophages | -0.0258037 | 0.1876178 | -1.179550 | 0.2471137 | 0.4022276 | -5.734717 |
| proliferating T/NK | 0.0114745 | 0.0310578 | 1.133464 | 0.2656620 | 0.4022276 | -5.786035 |
| dividing innate cells | 0.0139205 | 0.0232467 | 1.119152 | 0.2716253 | 0.4022276 | -5.801588 |
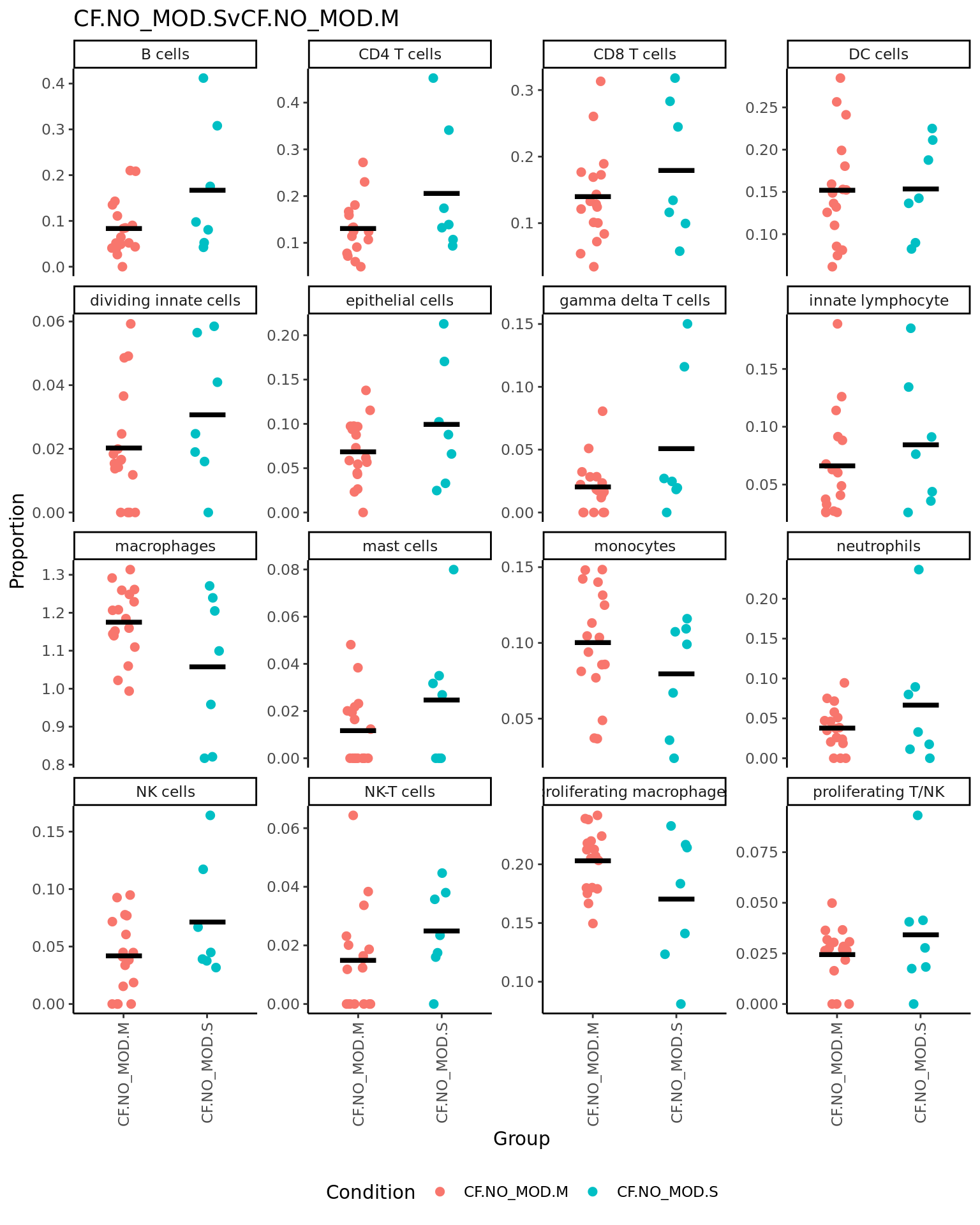
Session info
sessioninfo::session_info()─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.3.3 (2024-02-29)
os Ubuntu 22.04.4 LTS
system x86_64, linux-gnu
ui X11
language (EN)
collate en_AU.UTF-8
ctype en_AU.UTF-8
tz Etc/UTC
date 2024-12-24
pandoc 3.1.1 @ /usr/lib/rstudio-server/bin/quarto/bin/tools/ (via rmarkdown)
─ Packages ───────────────────────────────────────────────────────────────────
! package * version date (UTC) lib source
P abind 1.4-5 2016-07-21 [?] RSPM (R 4.3.0)
P AnnotationDbi * 1.64.1 2023-11-03 [?] Bioconductor
P backports 1.4.1 2021-12-13 [?] RSPM (R 4.3.0)
P base64enc 0.1-3 2015-07-28 [?] RSPM (R 4.3.0)
P Biobase * 2.62.0 2023-10-24 [?] Bioconductor
P BiocGenerics * 0.48.1 2023-11-01 [?] Bioconductor
P BiocManager 1.30.22 2023-08-08 [?] RSPM (R 4.3.0)
P Biostrings 2.70.2 2024-01-28 [?] Bioconductor 3.18 (R 4.3.3)
P bit 4.0.5 2022-11-15 [?] RSPM (R 4.3.0)
P bit64 4.0.5 2020-08-30 [?] RSPM (R 4.3.0)
P bitops 1.0-7 2021-04-24 [?] RSPM (R 4.3.0)
P blob 1.2.4 2023-03-17 [?] RSPM (R 4.3.0)
P bslib 0.6.1 2023-11-28 [?] RSPM (R 4.3.0)
P cachem 1.0.8 2023-05-01 [?] RSPM (R 4.3.0)
P callr 3.7.3 2022-11-02 [?] RSPM (R 4.3.0)
P checkmate 2.3.1 2023-12-04 [?] RSPM (R 4.3.0)
P circlize 0.4.15 2022-05-10 [?] RSPM (R 4.3.0)
P cli 3.6.2 2023-12-11 [?] RSPM (R 4.3.0)
P clue 0.3-65 2023-09-23 [?] RSPM (R 4.3.0)
P cluster 2.1.6 2023-12-01 [?] CRAN (R 4.3.2)
P clustree * 0.5.1 2023-11-05 [?] RSPM (R 4.3.0)
P codetools 0.2-19 2023-02-01 [?] CRAN (R 4.2.2)
P colorspace 2.1-0 2023-01-23 [?] RSPM (R 4.3.0)
P ComplexHeatmap 2.18.0 2023-10-24 [?] Bioconductor
P cowplot 1.1.3 2024-01-22 [?] RSPM (R 4.3.0)
P crayon 1.5.2 2022-09-29 [?] RSPM (R 4.3.0)
P data.table 1.15.0 2024-01-30 [?] RSPM (R 4.3.0)
P DBI 1.2.1 2024-01-12 [?] RSPM (R 4.3.0)
P DelayedArray 0.28.0 2023-10-24 [?] Bioconductor
P deldir 2.0-2 2023-11-23 [?] RSPM (R 4.3.0)
P dendextend 1.17.1 2023-03-25 [?] RSPM (R 4.3.0)
P digest 0.6.34 2024-01-11 [?] RSPM (R 4.3.0)
P dittoSeq * 1.14.2 2024-02-09 [?] Bioconductor 3.18 (R 4.3.3)
P doParallel 1.0.17 2022-02-07 [?] RSPM (R 4.3.0)
P dplyr * 1.1.4 2023-11-17 [?] RSPM (R 4.3.0)
P dynamicTreeCut 1.63-1 2016-03-11 [?] RSPM (R 4.3.0)
P edgeR * 4.0.15 2024-02-11 [?] Bioconductor 3.18 (R 4.3.3)
P ellipsis 0.3.2 2021-04-29 [?] RSPM (R 4.3.0)
P evaluate 0.23 2023-11-01 [?] RSPM (R 4.3.0)
P fansi 1.0.6 2023-12-08 [?] RSPM (R 4.3.0)
P farver 2.1.1 2022-07-06 [?] RSPM (R 4.3.0)
P fastcluster 1.2.6 2024-01-12 [?] RSPM (R 4.3.0)
P fastmap 1.1.1 2023-02-24 [?] RSPM (R 4.3.0)
P fitdistrplus 1.1-11 2023-04-25 [?] RSPM (R 4.3.0)
P forcats * 1.0.0 2023-01-29 [?] RSPM (R 4.3.0)
P foreach 1.5.2 2022-02-02 [?] RSPM (R 4.3.0)
P foreign 0.8-86 2023-11-28 [?] CRAN (R 4.3.2)
P Formula 1.2-5 2023-02-24 [?] RSPM (R 4.3.0)
P fs 1.6.3 2023-07-20 [?] RSPM (R 4.3.0)
P future 1.33.1 2023-12-22 [?] RSPM (R 4.3.0)
P future.apply 1.11.1 2023-12-21 [?] RSPM (R 4.3.0)
P generics 0.1.3 2022-07-05 [?] RSPM (R 4.3.0)
P GenomeInfoDb * 1.38.6 2024-02-08 [?] Bioconductor 3.18 (R 4.3.3)
P GenomeInfoDbData 1.2.11 2024-04-23 [?] Bioconductor
P GenomicRanges * 1.54.1 2023-10-29 [?] Bioconductor
P GetoptLong 1.0.5 2020-12-15 [?] RSPM (R 4.3.0)
P getPass 0.2-4 2023-12-10 [?] RSPM (R 4.3.0)
P ggforce 0.4.2 2024-02-19 [?] RSPM (R 4.3.0)
P ggplot2 * 3.5.0 2024-02-23 [?] RSPM (R 4.3.0)
P ggraph * 2.2.0 2024-02-27 [?] RSPM (R 4.3.0)
P ggrepel 0.9.5 2024-01-10 [?] RSPM (R 4.3.0)
P ggridges 0.5.6 2024-01-23 [?] RSPM (R 4.3.0)
P git2r 0.33.0 2023-11-26 [?] RSPM (R 4.3.0)
P glmGamPoi * 1.14.3 2024-02-11 [?] Bioconductor 3.18 (R 4.3.3)
P GlobalOptions 0.1.2 2020-06-10 [?] RSPM (R 4.3.0)
P globals 0.16.2 2022-11-21 [?] RSPM (R 4.3.0)
P glue * 1.7.0 2024-01-09 [?] RSPM (R 4.3.0)
P GO.db 3.18.0 2024-04-23 [?] Bioconductor
P goftest 1.2-3 2021-10-07 [?] RSPM (R 4.3.0)
P graphlayouts 1.1.0 2024-01-19 [?] RSPM (R 4.3.0)
P gridExtra 2.3 2017-09-09 [?] RSPM (R 4.3.0)
P gtable 0.3.4 2023-08-21 [?] RSPM (R 4.3.0)
P here * 1.0.1 2020-12-13 [?] RSPM (R 4.3.0)
P highr 0.10 2022-12-22 [?] RSPM (R 4.3.0)
P Hmisc 5.1-1 2023-09-12 [?] RSPM (R 4.3.0)
P hms 1.1.3 2023-03-21 [?] RSPM (R 4.3.0)
P htmlTable 2.4.2 2023-10-29 [?] RSPM (R 4.3.0)
P htmltools 0.5.7 2023-11-03 [?] RSPM (R 4.3.0)
P htmlwidgets 1.6.4 2023-12-06 [?] RSPM (R 4.3.0)
P httpuv 1.6.14 2024-01-26 [?] RSPM (R 4.3.0)
P httr 1.4.7 2023-08-15 [?] RSPM (R 4.3.0)
P ica 1.0-3 2022-07-08 [?] RSPM (R 4.3.0)
P igraph 2.0.1.1 2024-01-30 [?] RSPM (R 4.3.0)
P impute 1.76.0 2023-10-24 [?] Bioconductor
P IRanges * 2.36.0 2023-10-24 [?] Bioconductor
P irlba 2.3.5.1 2022-10-03 [?] RSPM (R 4.3.0)
P iterators 1.0.14 2022-02-05 [?] RSPM (R 4.3.0)
P jquerylib 0.1.4 2021-04-26 [?] RSPM (R 4.3.0)
P jsonlite 1.8.8 2023-12-04 [?] RSPM (R 4.3.0)
P KEGGREST 1.42.0 2023-10-24 [?] Bioconductor
P KernSmooth 2.23-24 2024-05-17 [?] RSPM (R 4.3.0)
P knitr 1.45 2023-10-30 [?] RSPM (R 4.3.0)
P labeling 0.4.3 2023-08-29 [?] RSPM (R 4.3.0)
P later 1.3.2 2023-12-06 [?] RSPM (R 4.3.0)
P lattice 0.22-5 2023-10-24 [?] CRAN (R 4.3.1)
P lazyeval 0.2.2 2019-03-15 [?] RSPM (R 4.3.0)
P leiden 0.4.3.1 2023-11-17 [?] RSPM (R 4.3.0)
P lifecycle 1.0.4 2023-11-07 [?] RSPM (R 4.3.0)
P limma * 3.58.1 2023-10-31 [?] Bioconductor
P listenv 0.9.1 2024-01-29 [?] RSPM (R 4.3.0)
P lmtest 0.9-40 2022-03-21 [?] RSPM (R 4.3.0)
P locfit 1.5-9.8 2023-06-11 [?] RSPM (R 4.3.0)
P lubridate * 1.9.3 2023-09-27 [?] RSPM (R 4.3.0)
P magrittr 2.0.3 2022-03-30 [?] RSPM (R 4.3.0)
P MASS 7.3-60.0.1 2024-01-13 [?] RSPM (R 4.3.0)
P Matrix 1.6-5 2024-01-11 [?] CRAN (R 4.3.3)
P MatrixGenerics * 1.14.0 2023-10-24 [?] Bioconductor
P matrixStats * 1.2.0 2023-12-11 [?] RSPM (R 4.3.0)
P memoise 2.0.1 2021-11-26 [?] RSPM (R 4.3.0)
P mime 0.12 2021-09-28 [?] RSPM (R 4.3.0)
P miniUI 0.1.1.1 2018-05-18 [?] RSPM (R 4.3.0)
P munsell 0.5.0 2018-06-12 [?] RSPM (R 4.3.0)
P nlme 3.1-164 2023-11-27 [?] RSPM (R 4.3.0)
P nnet 7.3-19 2023-05-03 [?] CRAN (R 4.3.1)
P org.Hs.eg.db * 3.18.0 2024-04-23 [?] Bioconductor
P paletteer * 1.6.0 2024-01-21 [?] RSPM (R 4.3.0)
P parallelly 1.37.0 2024-02-14 [?] RSPM (R 4.3.0)
P patchwork * 1.2.0 2024-01-08 [?] RSPM (R 4.3.0)
P pbapply 1.7-2 2023-06-27 [?] RSPM (R 4.3.0)
P pheatmap 1.0.12 2019-01-04 [?] RSPM (R 4.3.0)
P pillar 1.9.0 2023-03-22 [?] RSPM (R 4.3.0)
P pkgconfig 2.0.3 2019-09-22 [?] RSPM (R 4.3.0)
P plotly 4.10.4 2024-01-13 [?] RSPM (R 4.3.0)
P plyr 1.8.9 2023-10-02 [?] RSPM (R 4.3.0)
P png 0.1-8 2022-11-29 [?] RSPM (R 4.3.0)
P polyclip 1.10-6 2023-09-27 [?] RSPM (R 4.3.0)
P preprocessCore 1.64.0 2023-10-24 [?] Bioconductor
P prismatic 1.1.1 2022-08-15 [?] RSPM (R 4.3.0)
P processx 3.8.3 2023-12-10 [?] RSPM (R 4.3.0)
P progressr 0.14.0 2023-08-10 [?] RSPM (R 4.3.0)
P promises 1.2.1 2023-08-10 [?] RSPM (R 4.3.0)
P ps 1.7.6 2024-01-18 [?] RSPM (R 4.3.0)
P purrr * 1.0.2 2023-08-10 [?] RSPM (R 4.3.0)
P R6 2.5.1 2021-08-19 [?] RSPM (R 4.3.0)
P RANN 2.6.1 2019-01-08 [?] RSPM (R 4.3.0)
P RColorBrewer 1.1-3 2022-04-03 [?] RSPM (R 4.3.0)
P Rcpp 1.0.12 2024-01-09 [?] RSPM (R 4.3.0)
P RcppAnnoy 0.0.22 2024-01-23 [?] RSPM (R 4.3.0)
P RCurl 1.98-1.14 2024-01-09 [?] RSPM (R 4.3.0)
P readr * 2.1.5 2024-01-10 [?] RSPM (R 4.3.0)
P rematch2 2.1.2 2020-05-01 [?] RSPM (R 4.3.0)
renv 1.0.3 2023-09-19 [1] CRAN (R 4.3.3)
P reshape2 1.4.4 2020-04-09 [?] RSPM (R 4.3.0)
P reticulate 1.35.0 2024-01-31 [?] RSPM (R 4.3.0)
P rjson 0.2.21 2022-01-09 [?] RSPM (R 4.3.0)
P rlang 1.1.3 2024-01-10 [?] RSPM (R 4.3.0)
P rmarkdown 2.25 2023-09-18 [?] RSPM (R 4.3.0)
P ROCR 1.0-11 2020-05-02 [?] RSPM (R 4.3.0)
P rpart 4.1.23 2023-12-05 [?] RSPM (R 4.3.0)
P rprojroot 2.0.4 2023-11-05 [?] RSPM (R 4.3.0)
P RSQLite 2.3.5 2024-01-21 [?] RSPM (R 4.3.0)
P rstudioapi 0.15.0 2023-07-07 [?] RSPM (R 4.3.0)
P Rtsne 0.17 2023-12-07 [?] RSPM (R 4.3.0)
P S4Arrays 1.2.0 2023-10-24 [?] Bioconductor
P S4Vectors * 0.40.2 2023-11-23 [?] Bioconductor 3.18 (R 4.3.3)
P sass 0.4.8 2023-12-06 [?] RSPM (R 4.3.0)
P scales 1.3.0 2023-11-28 [?] RSPM (R 4.3.0)
P scattermore 1.2 2023-06-12 [?] RSPM (R 4.3.0)
sctransform 0.4.1 2023-10-19 [1] RSPM (R 4.3.0)
P sessioninfo 1.2.2 2021-12-06 [?] RSPM (R 4.3.0)
Seurat * 4.4.0 2024-04-25 [1] https://satijalab.r-universe.dev (R 4.3.3)
SeuratObject * 4.1.4 2024-04-25 [1] https://satijalab.r-universe.dev (R 4.3.3)
P shape 1.4.6 2021-05-19 [?] RSPM (R 4.3.0)
P shiny 1.8.0 2023-11-17 [?] RSPM (R 4.3.0)
P SingleCellExperiment * 1.24.0 2023-10-24 [?] Bioconductor
P sp 2.1-3 2024-01-30 [?] RSPM (R 4.3.0)
P SparseArray 1.2.4 2024-02-11 [?] Bioconductor 3.18 (R 4.3.3)
P spatstat.data 3.0-4 2024-01-15 [?] RSPM (R 4.3.0)
P spatstat.explore 3.2-6 2024-02-01 [?] RSPM (R 4.3.0)
P spatstat.geom 3.2-8 2024-01-26 [?] RSPM (R 4.3.0)
P spatstat.random 3.2-2 2023-11-29 [?] RSPM (R 4.3.0)
P spatstat.sparse 3.0-3 2023-10-24 [?] RSPM (R 4.3.0)
P spatstat.utils 3.0-4 2023-10-24 [?] RSPM (R 4.3.0)
P speckle * 1.2.0 2023-10-24 [?] Bioconductor
P statmod 1.5.0 2023-01-06 [?] RSPM (R 4.3.0)
P stringi 1.8.3 2023-12-11 [?] RSPM (R 4.3.0)
P stringr * 1.5.1 2023-11-14 [?] RSPM (R 4.3.0)
P SummarizedExperiment * 1.32.0 2023-10-24 [?] Bioconductor
P survival 3.7-0 2024-06-05 [?] RSPM (R 4.3.0)
P tensor 1.5 2012-05-05 [?] RSPM (R 4.3.0)
P tibble * 3.2.1 2023-03-20 [?] RSPM (R 4.3.0)
P tidygraph 1.3.1 2024-01-30 [?] RSPM (R 4.3.0)
P tidyHeatmap * 1.8.1 2022-05-20 [?] RSPM (R 4.3.3)
P tidyr * 1.3.1 2024-01-24 [?] RSPM (R 4.3.0)
P tidyselect 1.2.0 2022-10-10 [?] RSPM (R 4.3.0)
P tidyverse * 2.0.0 2023-02-22 [?] RSPM (R 4.3.0)
P timechange 0.3.0 2024-01-18 [?] RSPM (R 4.3.0)
P tweenr 2.0.3 2024-02-26 [?] RSPM (R 4.3.0)
P tzdb 0.4.0 2023-05-12 [?] RSPM (R 4.3.0)
P utf8 1.2.4 2023-10-22 [?] RSPM (R 4.3.0)
P uwot 0.1.16 2023-06-29 [?] RSPM (R 4.3.0)
P vctrs 0.6.5 2023-12-01 [?] RSPM (R 4.3.0)
P viridis 0.6.5 2024-01-29 [?] RSPM (R 4.3.0)
P viridisLite 0.4.2 2023-05-02 [?] RSPM (R 4.3.0)
P WGCNA 1.72-5 2023-12-07 [?] RSPM (R 4.3.3)
P whisker 0.4.1 2022-12-05 [?] RSPM (R 4.3.0)
P withr 3.0.0 2024-01-16 [?] RSPM (R 4.3.0)
P workflowr * 1.7.1 2023-08-23 [?] RSPM (R 4.3.0)
P xfun 0.42 2024-02-08 [?] RSPM (R 4.3.0)
P xtable 1.8-4 2019-04-21 [?] RSPM (R 4.3.0)
P XVector 0.42.0 2023-10-24 [?] Bioconductor
P yaml 2.3.8 2023-12-11 [?] RSPM (R 4.3.0)
P zlibbioc 1.48.0 2023-10-24 [?] Bioconductor
P zoo 1.8-12 2023-04-13 [?] RSPM (R 4.3.0)
[1] /mnt/allandata/jovana_data/paed-inflammation-CITEseq/renv/library/R-4.3/x86_64-pc-linux-gnu
[2] /home/jovana/.cache/R/renv/sandbox/R-4.3/x86_64-pc-linux-gnu/9a444a72
P ── Loaded and on-disk path mismatch.
──────────────────────────────────────────────────────────────────────────────
sessionInfo()R version 4.3.3 (2024-02-29)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 22.04.4 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
locale:
[1] LC_CTYPE=en_AU.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_AU.UTF-8 LC_COLLATE=en_AU.UTF-8
[5] LC_MONETARY=en_AU.UTF-8 LC_MESSAGES=en_AU.UTF-8
[7] LC_PAPER=en_AU.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_AU.UTF-8 LC_IDENTIFICATION=C
time zone: Etc/UTC
tzcode source: system (glibc)
attached base packages:
[1] stats4 stats graphics grDevices datasets utils methods
[8] base
other attached packages:
[1] here_1.0.1 tidyHeatmap_1.8.1
[3] paletteer_1.6.0 patchwork_1.2.0
[5] speckle_1.2.0 glue_1.7.0
[7] org.Hs.eg.db_3.18.0 AnnotationDbi_1.64.1
[9] clustree_0.5.1 ggraph_2.2.0
[11] dittoSeq_1.14.2 glmGamPoi_1.14.3
[13] SeuratObject_4.1.4 Seurat_4.4.0
[15] lubridate_1.9.3 forcats_1.0.0
[17] stringr_1.5.1 dplyr_1.1.4
[19] purrr_1.0.2 readr_2.1.5
[21] tidyr_1.3.1 tibble_3.2.1
[23] ggplot2_3.5.0 tidyverse_2.0.0
[25] edgeR_4.0.15 limma_3.58.1
[27] SingleCellExperiment_1.24.0 SummarizedExperiment_1.32.0
[29] Biobase_2.62.0 GenomicRanges_1.54.1
[31] GenomeInfoDb_1.38.6 IRanges_2.36.0
[33] S4Vectors_0.40.2 BiocGenerics_0.48.1
[35] MatrixGenerics_1.14.0 matrixStats_1.2.0
[37] workflowr_1.7.1
loaded via a namespace (and not attached):
[1] fs_1.6.3 spatstat.sparse_3.0-3 bitops_1.0-7
[4] httr_1.4.7 RColorBrewer_1.1-3 doParallel_1.0.17
[7] dynamicTreeCut_1.63-1 backports_1.4.1 tools_4.3.3
[10] sctransform_0.4.1 utf8_1.2.4 R6_2.5.1
[13] lazyeval_0.2.2 uwot_0.1.16 GetoptLong_1.0.5
[16] withr_3.0.0 sp_2.1-3 gridExtra_2.3
[19] preprocessCore_1.64.0 progressr_0.14.0 WGCNA_1.72-5
[22] cli_3.6.2 spatstat.explore_3.2-6 labeling_0.4.3
[25] sass_0.4.8 prismatic_1.1.1 spatstat.data_3.0-4
[28] ggridges_0.5.6 pbapply_1.7-2 foreign_0.8-86
[31] sessioninfo_1.2.2 parallelly_1.37.0 impute_1.76.0
[34] rstudioapi_0.15.0 RSQLite_2.3.5 generics_0.1.3
[37] shape_1.4.6 ica_1.0-3 spatstat.random_3.2-2
[40] dendextend_1.17.1 GO.db_3.18.0 Matrix_1.6-5
[43] fansi_1.0.6 abind_1.4-5 lifecycle_1.0.4
[46] whisker_0.4.1 yaml_2.3.8 SparseArray_1.2.4
[49] Rtsne_0.17 grid_4.3.3 blob_1.2.4
[52] promises_1.2.1 crayon_1.5.2 miniUI_0.1.1.1
[55] lattice_0.22-5 cowplot_1.1.3 KEGGREST_1.42.0
[58] pillar_1.9.0 knitr_1.45 ComplexHeatmap_2.18.0
[61] rjson_0.2.21 future.apply_1.11.1 codetools_0.2-19
[64] leiden_0.4.3.1 getPass_0.2-4 data.table_1.15.0
[67] vctrs_0.6.5 png_0.1-8 gtable_0.3.4
[70] rematch2_2.1.2 cachem_1.0.8 xfun_0.42
[73] S4Arrays_1.2.0 mime_0.12 tidygraph_1.3.1
[76] survival_3.7-0 pheatmap_1.0.12 iterators_1.0.14
[79] statmod_1.5.0 ellipsis_0.3.2 fitdistrplus_1.1-11
[82] ROCR_1.0-11 nlme_3.1-164 bit64_4.0.5
[85] RcppAnnoy_0.0.22 rprojroot_2.0.4 bslib_0.6.1
[88] irlba_2.3.5.1 rpart_4.1.23 KernSmooth_2.23-24
[91] Hmisc_5.1-1 colorspace_2.1-0 DBI_1.2.1
[94] nnet_7.3-19 tidyselect_1.2.0 processx_3.8.3
[97] bit_4.0.5 compiler_4.3.3 git2r_0.33.0
[100] htmlTable_2.4.2 DelayedArray_0.28.0 plotly_4.10.4
[103] checkmate_2.3.1 scales_1.3.0 lmtest_0.9-40
[106] callr_3.7.3 digest_0.6.34 goftest_1.2-3
[109] spatstat.utils_3.0-4 rmarkdown_2.25 XVector_0.42.0
[112] base64enc_0.1-3 htmltools_0.5.7 pkgconfig_2.0.3
[115] highr_0.10 fastmap_1.1.1 rlang_1.1.3
[118] GlobalOptions_0.1.2 htmlwidgets_1.6.4 shiny_1.8.0
[121] farver_2.1.1 jquerylib_0.1.4 zoo_1.8-12
[124] jsonlite_1.8.8 RCurl_1.98-1.14 magrittr_2.0.3
[127] Formula_1.2-5 GenomeInfoDbData_1.2.11 munsell_0.5.0
[130] Rcpp_1.0.12 viridis_0.6.5 reticulate_1.35.0
[133] stringi_1.8.3 zlibbioc_1.48.0 MASS_7.3-60.0.1
[136] plyr_1.8.9 parallel_4.3.3 listenv_0.9.1
[139] ggrepel_0.9.5 deldir_2.0-2 Biostrings_2.70.2
[142] graphlayouts_1.1.0 splines_4.3.3 tensor_1.5
[145] hms_1.1.3 circlize_0.4.15 locfit_1.5-9.8
[148] ps_1.7.6 fastcluster_1.2.6 igraph_2.0.1.1
[151] spatstat.geom_3.2-8 reshape2_1.4.4 evaluate_0.23
[154] renv_1.0.3 BiocManager_1.30.22 tzdb_0.4.0
[157] foreach_1.5.2 tweenr_2.0.3 httpuv_1.6.14
[160] RANN_2.6.1 polyclip_1.10-6 future_1.33.1
[163] clue_0.3-65 scattermore_1.2 ggforce_0.4.2
[166] xtable_1.8-4 later_1.3.2 viridisLite_0.4.2
[169] memoise_2.0.1 cluster_2.1.6 timechange_0.3.0
[172] globals_0.16.2