Last updated: 2019-10-20
Checks: 6 1
Knit directory: csna_workflow/
This reproducible R Markdown analysis was created with workflowr (version 1.4.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190918) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /projects/csna/csna_workflow/code/reconst_utils.R | code/reconst_utils.R |
| /projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/Gagnon_gm.RData | data/Jackson_Lab_Gagnon/Gagnon_gm.RData |
| /projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/Percent_missing_genotype_data.pdf | data/Jackson_Lab_Gagnon/Percent_missing_genotype_data.pdf |
| /projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/percent_missing_id.RData | data/Jackson_Lab_Gagnon/percent_missing_id.RData |
| /projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/Jackson_Lab_Gagnon_qtl2_chrXint.csv | data/Jackson_Lab_Gagnon/Jackson_Lab_Gagnon_qtl2_chrXint.csv |
| /projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/Jackson_Lab_Gagnon_qtl2_chrYint.csv | data/Jackson_Lab_Gagnon/Jackson_Lab_Gagnon_qtl2_chrYint.csv |
| /projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/intensities.fst.RData | data/Jackson_Lab_Gagnon/intensities.fst.RData |
| /projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/summary.cg.RData | data/Jackson_Lab_Gagnon/summary.cg.RData |
| /projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/Proportion_matching_genotypes_before_removal_of_bad_samples.pdf | data/Jackson_Lab_Gagnon/Proportion_matching_genotypes_before_removal_of_bad_samples.pdf |
| /projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/Proportion_matching_genotypes_after_removal_of_bad_samples.pdf | data/Jackson_Lab_Gagnon/Proportion_matching_genotypes_after_removal_of_bad_samples.pdf |
| /projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/pr.RData | data/Jackson_Lab_Gagnon/pr.RData |
| /projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/m.RData | data/Jackson_Lab_Gagnon/m.RData |
| /projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/nxo.RData | data/Jackson_Lab_Gagnon/nxo.RData |
| /projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/number_crossover.pdf | data/Jackson_Lab_Gagnon/number_crossover.pdf |
| /projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/e.RData | data/Jackson_Lab_Gagnon/e.RData |
| /projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/errors_ind.RData | data/Jackson_Lab_Gagnon/errors_ind.RData |
| /projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/snpg.RData | data/Jackson_Lab_Gagnon/snpg.RData |
| /projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/Percent_genotype_errors_obs_vs_predicted.pdf | data/Jackson_Lab_Gagnon/Percent_genotype_errors_obs_vs_predicted.pdf |
| /projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/Percent_missing_genotype_data_per_marker.pdf | data/Jackson_Lab_Gagnon/Percent_missing_genotype_data_per_marker.pdf |
| /projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/genotype_frequency_marker.pdf | data/Jackson_Lab_Gagnon/genotype_frequency_marker.pdf |
| /projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/genotype_error_marker.pdf | data/Jackson_Lab_Gagnon/genotype_error_marker.pdf |
| /projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/qc_info_bad_sample.RData | data/Jackson_Lab_Gagnon/qc_info_bad_sample.RData |
| /projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/bad_markers.RData | data/Jackson_Lab_Gagnon/bad_markers.RData |
| /projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/gm_DO2437_qc.RData | data/Jackson_Lab_Gagnon/gm_DO2437_qc.RData |
| /projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/e_g_snpg_qc.RData | data/Jackson_Lab_Gagnon/e_g_snpg_qc.RData |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Untracked files:
Untracked: analysis/.nfs0000000135a08b9b00002448
Untracked: analysis/.nfs0000000135a08ba50000244a
Untracked: analysis/01_geneseek2qtl2.Rout
Untracked: analysis/01_geneseek2qtl2.script
Untracked: analysis/01_geneseek2qtl2.stderr
Untracked: analysis/01_geneseek2qtl2.stdout
Untracked: analysis/02_geneseek2intensity.Rout
Untracked: analysis/02_geneseek2intensity.script
Untracked: analysis/02_geneseek2intensity.stderr
Untracked: analysis/02_geneseek2intensity.stdout
Untracked: analysis/03_firstgm2genoprobs.Rout
Untracked: analysis/03_firstgm2genoprobs.script
Untracked: analysis/03_firstgm2genoprobs.stderr
Untracked: analysis/03_firstgm2genoprobs.stdout
Untracked: analysis/04_diagnosis_qc_gigamuga_nine_batches.Rout
Untracked: analysis/04_diagnosis_qc_gigamuga_nine_batches.script
Untracked: analysis/04_diagnosis_qc_gigamuga_nine_batches.stderr
Untracked: analysis/04_diagnosis_qc_gigamuga_nine_batches.stdout
Untracked: analysis/05_after_diagnosis_qc_gigamuga_nine_batches.Rout
Untracked: analysis/05_after_diagnosis_qc_gigamuga_nine_batches.script
Untracked: analysis/05_after_diagnosis_qc_gigamuga_nine_batches.stderr
Untracked: analysis/05_after_diagnosis_qc_gigamuga_nine_batches.stdout
Untracked: analysis/06_final_pr_apr_69K.Rout
Untracked: analysis/06_final_pr_apr_69K.script
Untracked: analysis/06_final_pr_apr_69K.stderr
Untracked: analysis/06_final_pr_apr_69K.stdout
Untracked: analysis/07.1_html_founder_prop.Rout
Untracked: analysis/07.1_html_founder_prop.script
Untracked: analysis/07.1_html_founder_prop.stderr
Untracked: analysis/07.1_html_founder_prop.stdout
Untracked: analysis/07_recomb_size_founder_prop.Rout
Untracked: analysis/07_recomb_size_founder_prop.script
Untracked: analysis/07_recomb_size_founder_prop.stderr
Untracked: analysis/07_recomb_size_founder_prop.stdout
Untracked: analysis/12_plot_qtl_mapping.R
Untracked: analysis/12_plot_qtl_mapping.Rout
Untracked: analysis/12_plot_qtl_mapping.script
Untracked: analysis/12_plot_qtl_mapping.stderr
Untracked: analysis/12_plot_qtl_mapping.stdout
Untracked: analysis/13_conditional_m2_qtlmapping.R
Untracked: analysis/13_conditional_m2_qtlmapping.Rout
Untracked: analysis/13_conditional_m2_qtlmapping.e10916958
Untracked: analysis/13_conditional_m2_qtlmapping.e10920851
Untracked: analysis/13_conditional_m2_qtlmapping.o10916958
Untracked: analysis/13_conditional_m2_qtlmapping.o10920851
Untracked: analysis/13_conditional_m2_qtlmapping.script
Untracked: analysis/13_plot_conditional_m2_qtlmapping.R
Untracked: analysis/13_plot_conditional_m2_qtlmapping.Rout
Untracked: analysis/13_plot_conditional_m2_qtlmapping.script
Untracked: analysis/14_geneseek2qtl2fst_Gagnon.R
Untracked: analysis/14_geneseek2qtl2fst_Gagnon.Rout
Untracked: analysis/14_geneseek2qtl2fst_Gagnon.script
Untracked: analysis/14_geneseek2qtl2fst_Gagnon.stderr
Untracked: analysis/14_geneseek2qtl2fst_Gagnon.stdout
Untracked: analysis/15_firstgm2genoprobs_Gagnon.R
Untracked: analysis/15_firstgm2genoprobs_Gagnon.Rout
Untracked: analysis/15_firstgm2genoprobs_Gagnon.script
Untracked: analysis/15_firstgm2genoprobs_Gagnon.stderr
Untracked: analysis/15_firstgm2genoprobs_Gagnon.stdout
Untracked: analysis/16_diagnosis_qc_gigamuga_gagnon.R
Untracked: analysis/16_diagnosis_qc_gigamuga_gagnon.Rout
Untracked: analysis/16_diagnosis_qc_gigamuga_gagnon.script
Untracked: analysis/16_diagnosis_qc_gigamuga_gagnon.stderr
Untracked: analysis/16_diagnosis_qc_gigamuga_gagnon.stdout
Untracked: analysis/Novelty_resids_datarelease_07302918.gcta.Rout
Untracked: analysis/Novelty_resids_datarelease_07302918.gcta.script
Untracked: analysis/Novelty_resids_datarelease_07302918.gcta.stderr
Untracked: analysis/Novelty_resids_datarelease_07302918.gcta.stdout
Untracked: analysis/Novelty_resids_datarelease_07302918_m1.Rout
Untracked: analysis/Novelty_resids_datarelease_07302918_m1.script
Untracked: analysis/Novelty_resids_datarelease_07302918_m1.stderr
Untracked: analysis/Novelty_resids_datarelease_07302918_m1.stdout
Untracked: analysis/Novelty_resids_datarelease_07302918_m2.Rout
Untracked: analysis/Novelty_resids_datarelease_07302918_m2.script
Untracked: analysis/Novelty_resids_datarelease_07302918_m2.stderr
Untracked: analysis/Novelty_resids_datarelease_07302918_m2.stdout
Untracked: analysis/Novelty_resids_datarelease_07302918_m3.Rout
Untracked: analysis/Novelty_resids_datarelease_07302918_m3.script
Untracked: analysis/Novelty_resids_datarelease_07302918_m3.stderr
Untracked: analysis/Novelty_resids_datarelease_07302918_m3.stdout
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918.gcta.Rout
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918.gcta.script
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918.gcta.stderr
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918.gcta.stdout
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918_m1.Rout
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918_m1.script
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918_m1.stderr
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918_m1.stdout
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918_m2.Rout
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918_m2.script
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918_m2.stderr
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918_m2.stdout
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918_m3.Rout
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918_m3.script
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918_m3.stderr
Untracked: analysis/Novelty_residuals_RankNormal_datarelease_07302918_m3.stdout
Untracked: analysis/blup_14_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_14_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_14_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_14_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_14_m1_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_14_m1_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_14_m1_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_14_m1_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_14_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_14_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_14_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_14_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_14_m2_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_14_m2_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_14_m2_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_14_m2_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_14_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_14_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_14_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_14_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_14_m3_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_14_m3_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_14_m3_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_14_m3_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_15_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_15_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_15_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_15_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_15_m1_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_15_m1_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_15_m1_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_15_m1_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_15_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_15_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_15_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_15_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_15_m2_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_15_m2_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_15_m2_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_15_m2_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_15_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_15_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_15_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_15_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_15_m3_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_15_m3_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_15_m3_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_15_m3_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_16_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_16_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_16_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_16_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_16_m1_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_16_m1_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_16_m1_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_16_m1_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_16_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_16_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_16_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_16_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_16_m2_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_16_m2_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_16_m2_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_16_m2_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_16_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_16_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_16_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_16_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_16_m3_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_16_m3_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_16_m3_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_16_m3_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_17_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_17_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_17_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_17_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_17_m1_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_17_m1_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_17_m1_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_17_m1_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_17_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_17_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_17_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_17_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_17_m2_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_17_m2_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_17_m2_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_17_m2_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_17_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_17_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_17_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_17_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_17_m3_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_17_m3_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_17_m3_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_17_m3_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_18_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_18_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_18_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_18_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_18_m1_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_18_m1_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_18_m1_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_18_m1_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_18_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_18_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_18_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_18_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_18_m2_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_18_m2_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_18_m2_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_18_m2_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_18_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_18_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_18_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_18_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_18_m3_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_18_m3_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_18_m3_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_18_m3_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_19_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_19_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_19_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_19_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_19_m1_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_19_m1_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_19_m1_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_19_m1_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_19_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_19_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_19_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_19_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_19_m2_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_19_m2_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_19_m2_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_19_m2_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_19_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_19_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_19_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_19_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_19_m3_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_19_m3_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_19_m3_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_19_m3_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_20_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_20_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_20_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_20_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_20_m1_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_20_m1_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_20_m1_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_20_m1_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_20_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_20_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_20_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_20_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_20_m2_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_20_m2_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_20_m2_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_20_m2_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_20_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_20_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_20_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_20_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_20_m3_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_20_m3_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_20_m3_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_20_m3_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_21_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_21_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_21_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_21_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_21_m1_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_21_m1_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_21_m1_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_21_m1_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_21_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_21_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_21_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_21_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_21_m2_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_21_m2_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_21_m2_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_21_m2_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_21_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_21_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_21_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_21_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_21_m3_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_21_m3_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_21_m3_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_21_m3_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_22_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_22_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_22_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_22_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_22_m1_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_22_m1_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_22_m1_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_22_m1_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_22_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_22_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_22_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_22_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_22_m2_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_22_m2_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_22_m2_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_22_m2_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_22_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_22_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_22_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_22_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_22_m3_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_22_m3_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_22_m3_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_22_m3_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_23_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_23_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_23_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_23_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_23_m1_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_23_m1_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_23_m1_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_23_m1_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_23_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_23_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_23_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_23_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_23_m2_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_23_m2_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_23_m2_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_23_m2_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_23_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_23_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_23_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_23_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_23_m3_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_23_m3_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_23_m3_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_23_m3_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_24_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_24_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_24_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_24_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_24_m1_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_24_m1_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_24_m1_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_24_m1_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_24_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_24_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_24_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_24_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_24_m2_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_24_m2_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_24_m2_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_24_m2_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_24_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_24_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_24_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_24_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_24_m3_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_24_m3_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_24_m3_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_24_m3_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_25_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_25_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_25_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_25_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_25_m1_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_25_m1_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_25_m1_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_25_m1_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_25_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_25_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_25_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_25_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_25_m2_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_25_m2_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_25_m2_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_25_m2_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_25_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_25_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_25_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_25_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_25_m3_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_25_m3_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_25_m3_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_25_m3_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_26_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_26_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_26_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_26_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_26_m1_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_26_m1_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_26_m1_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_26_m1_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_26_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_26_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_26_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_26_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_26_m2_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_26_m2_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_26_m2_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_26_m2_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_26_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_26_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_26_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_26_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_26_m3_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_26_m3_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_26_m3_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_26_m3_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_27_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_27_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_27_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_27_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_27_m1_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_27_m1_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_27_m1_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_27_m1_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_27_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_27_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_27_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_27_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_27_m2_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_27_m2_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_27_m2_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_27_m2_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_27_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_27_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_27_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_27_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_27_m3_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_27_m3_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_27_m3_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_27_m3_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_28_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_28_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_28_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_28_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_28_m1_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_28_m1_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_28_m1_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_28_m1_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_28_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_28_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_28_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_28_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_28_m2_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_28_m2_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_28_m2_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_28_m2_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_28_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_28_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_28_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_28_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_28_m3_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_28_m3_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_28_m3_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_28_m3_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_29_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_29_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_29_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_29_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_29_m1_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_29_m1_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_29_m1_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_29_m1_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_29_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_29_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_29_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_29_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_29_m2_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_29_m2_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_29_m2_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_29_m2_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_29_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_29_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_29_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_29_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_29_m3_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_29_m3_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_29_m3_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_29_m3_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_30_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_30_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_30_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_30_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_30_m1_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_30_m1_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_30_m1_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_30_m1_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_30_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_30_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_30_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_30_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_30_m2_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_30_m2_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_30_m2_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_30_m2_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_30_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_30_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_30_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_30_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_30_m3_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_30_m3_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_30_m3_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_30_m3_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_31_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_31_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_31_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_31_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_31_m1_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_31_m1_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_31_m1_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_31_m1_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_31_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_31_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_31_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_31_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_31_m2_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_31_m2_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_31_m2_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_31_m2_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_31_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_31_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_31_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_31_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_31_m3_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_31_m3_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_31_m3_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_31_m3_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_32_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_32_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_32_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_32_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_32_m1_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_32_m1_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_32_m1_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_32_m1_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_32_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_32_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_32_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_32_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_32_m2_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_32_m2_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_32_m2_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_32_m2_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_32_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_32_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_32_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_32_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_32_m3_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_32_m3_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_32_m3_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_32_m3_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_33_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_33_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_33_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_33_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_33_m1_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_33_m1_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_33_m1_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_33_m1_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_33_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_33_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_33_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_33_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_33_m2_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_33_m2_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_33_m2_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_33_m2_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_33_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_33_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_33_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_33_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_33_m3_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_33_m3_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_33_m3_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_33_m3_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_34_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_34_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_34_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_34_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_34_m1_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_34_m1_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_34_m1_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_34_m1_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_34_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_34_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_34_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_34_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_34_m2_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_34_m2_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_34_m2_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_34_m2_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_34_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_34_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_34_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_34_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_34_m3_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_34_m3_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_34_m3_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_34_m3_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_35_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_35_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_35_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_35_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_35_m1_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_35_m1_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_35_m1_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_35_m1_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_35_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_35_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_35_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_35_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_35_m2_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_35_m2_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_35_m2_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_35_m2_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_35_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_35_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_35_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_35_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_35_m3_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_35_m3_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_35_m3_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_35_m3_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_36_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_36_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_36_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_36_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_36_m1_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_36_m1_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_36_m1_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_36_m1_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_36_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_36_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_36_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_36_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_36_m2_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_36_m2_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_36_m2_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_36_m2_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_36_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_36_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_36_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_36_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_36_m3_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_36_m3_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_36_m3_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_36_m3_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_37_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_37_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_37_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_37_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_37_m1_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_37_m1_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_37_m1_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_37_m1_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_37_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_37_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_37_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_37_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_37_m2_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_37_m2_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_37_m2_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_37_m2_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_37_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_37_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_37_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_37_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_37_m3_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_37_m3_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_37_m3_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_37_m3_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_38_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_38_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_38_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_38_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_38_m1_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_38_m1_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_38_m1_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_38_m1_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_38_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_38_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_38_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_38_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_38_m2_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_38_m2_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_38_m2_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_38_m2_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_38_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_38_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_38_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_38_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_38_m3_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_38_m3_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_38_m3_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_38_m3_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_39_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_39_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_39_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_39_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_39_m1_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_39_m1_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_39_m1_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_39_m1_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_39_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_39_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_39_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_39_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_39_m2_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_39_m2_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_39_m2_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_39_m2_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_39_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_39_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_39_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_39_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_39_m3_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_39_m3_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_39_m3_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_39_m3_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_40_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_40_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_40_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_40_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_40_m1_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_40_m1_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_40_m1_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_40_m1_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_40_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_40_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_40_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_40_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_40_m2_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_40_m2_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_40_m2_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_40_m2_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_40_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_40_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_40_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_40_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_40_m3_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_40_m3_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_40_m3_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_40_m3_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_41_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_41_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_41_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_41_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_41_m1_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_41_m1_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_41_m1_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_41_m1_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_41_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_41_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_41_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_41_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_41_m2_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_41_m2_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_41_m2_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_41_m2_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/blup_41_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/blup_41_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/blup_41_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/blup_41_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/blup_41_m3_Novelty_residuals_RankNormal_datarelease_07302918.Rout
Untracked: analysis/blup_41_m3_Novelty_residuals_RankNormal_datarelease_07302918.script
Untracked: analysis/blup_41_m3_Novelty_residuals_RankNormal_datarelease_07302918.stderr
Untracked: analysis/blup_41_m3_Novelty_residuals_RankNormal_datarelease_07302918.stdout
Untracked: analysis/permu_10_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_10_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_10_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_10_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_10_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_10_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_10_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_10_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_10_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_10_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_10_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_10_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_11_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_11_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_11_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_11_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_11_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_11_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_11_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_11_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_11_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_11_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_11_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_11_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_12_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_12_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_12_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_12_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_12_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_12_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_12_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_12_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_12_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_12_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_12_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_12_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_13_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_13_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_13_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_13_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_13_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_13_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_13_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_13_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_13_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_13_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_13_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_13_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_14_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_14_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_14_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_14_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_14_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_14_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_14_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_14_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_14_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_14_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_14_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_14_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_15_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_15_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_15_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_15_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_15_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_15_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_15_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_15_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_15_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_15_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_15_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_15_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_16_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_16_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_16_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_16_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_16_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_16_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_16_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_16_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_16_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_16_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_16_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_16_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_17_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_17_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_17_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_17_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_17_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_17_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_17_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_17_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_17_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_17_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_17_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_17_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_18_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_18_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_18_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_18_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_18_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_18_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_18_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_18_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_18_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_18_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_18_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_18_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_19_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_19_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_19_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_19_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_19_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_19_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_19_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_19_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_19_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_19_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_19_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_19_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_1_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_1_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_1_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_1_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_1_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_1_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_1_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_1_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_1_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_1_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_1_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_1_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_20_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_20_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_20_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_20_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_20_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_20_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_20_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_20_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_20_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_20_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_20_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_20_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_21_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_21_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_21_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_21_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_21_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_21_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_21_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_21_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_21_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_21_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_21_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_21_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_22_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_22_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_22_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_22_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_22_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_22_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_22_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_22_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_22_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_22_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_22_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_22_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_23_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_23_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_23_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_23_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_23_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_23_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_23_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_23_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_23_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_23_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_23_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_23_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_24_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_24_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_24_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_24_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_24_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_24_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_24_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_24_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_24_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_24_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_24_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_24_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_25_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_25_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_25_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_25_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_25_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_25_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_25_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_25_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_25_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_25_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_25_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_25_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_26_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_26_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_26_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_26_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_26_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_26_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_26_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_26_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_26_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_26_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_26_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_26_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_27_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_27_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_27_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_27_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_27_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_27_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_27_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_27_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_27_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_27_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_27_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_27_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_28_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_28_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_28_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_28_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_28_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_28_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_28_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_28_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_28_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_28_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_28_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_28_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_29_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_29_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_29_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_29_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_29_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_29_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_29_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_29_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_29_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_29_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_29_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_29_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_2_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_2_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_2_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_2_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_2_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_2_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_2_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_2_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_2_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_2_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_2_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_2_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_30_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_30_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_30_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_30_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_30_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_30_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_30_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_30_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_30_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_30_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_30_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_30_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_31_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_31_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_31_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_31_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_31_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_31_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_31_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_31_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_31_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_31_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_31_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_31_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_32_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_32_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_32_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_32_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_32_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_32_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_32_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_32_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_32_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_32_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_32_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_32_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_33_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_33_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_33_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_33_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_33_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_33_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_33_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_33_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_33_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_33_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_33_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_33_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_34_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_34_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_34_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_34_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_34_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_34_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_34_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_34_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_34_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_34_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_34_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_34_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_35_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_35_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_35_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_35_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_35_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_35_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_35_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_35_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_35_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_35_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_35_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_35_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_36_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_36_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_36_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_36_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_36_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_36_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_36_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_36_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_36_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_36_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_36_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_36_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_37_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_37_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_37_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_37_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_37_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_37_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_37_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_37_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_37_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_37_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_37_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_37_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_38_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_38_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_38_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_38_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_38_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_38_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_38_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_38_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_38_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_38_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_38_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_38_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_39_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_39_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_39_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_39_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_39_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_39_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_39_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_39_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_39_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_39_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_39_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_39_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_3_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_3_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_3_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_3_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_3_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_3_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_3_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_3_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_3_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_3_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_3_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_3_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_40_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_40_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_40_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_40_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_40_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_40_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_40_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_40_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_40_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_40_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_40_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_40_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_41_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_41_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_41_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_41_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_41_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_41_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_41_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_41_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_41_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_41_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_41_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_41_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_42_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_42_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_42_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_42_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_42_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_42_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_42_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_42_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_42_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_42_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_42_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_42_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_43_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_43_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_43_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_43_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_43_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_43_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_43_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_43_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_43_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_43_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_43_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_43_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_44_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_44_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_44_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_44_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_44_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_44_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_44_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_44_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_44_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_44_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_44_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_44_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_45_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_45_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_45_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_45_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_45_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_45_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_45_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_45_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_45_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_45_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_45_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_45_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_46_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_46_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_46_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_46_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_46_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_46_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_46_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_46_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_46_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_46_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_46_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_46_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_47_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_47_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_47_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_47_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_47_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_47_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_47_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_47_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_47_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_47_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_47_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_47_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_48_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_48_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_48_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_48_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_48_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_48_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_48_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_48_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_48_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_48_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_48_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_48_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_49_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_49_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_49_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_49_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_49_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_49_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_49_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_49_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_49_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_49_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_49_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_49_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_4_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_4_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_4_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_4_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_4_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_4_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_4_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_4_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_4_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_4_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_4_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_4_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_50_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_50_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_50_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_50_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_50_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_50_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_50_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_50_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_50_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_50_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_50_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_50_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_5_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_5_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_5_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_5_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_5_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_5_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_5_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_5_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_5_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_5_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_5_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_5_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_6_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_6_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_6_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_6_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_6_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_6_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_6_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_6_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_6_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_6_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_6_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_6_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_7_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_7_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_7_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_7_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_7_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_7_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_7_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_7_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_7_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_7_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_7_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_7_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_8_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_8_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_8_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_8_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_8_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_8_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_8_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_8_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_8_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_8_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_8_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_8_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_9_m1_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_9_m1_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_9_m1_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_9_m1_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_9_m2_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_9_m2_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_9_m2_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_9_m2_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/permu_9_m3_Novelty_resids_datarelease_07302918.Rout
Untracked: analysis/permu_9_m3_Novelty_resids_datarelease_07302918.script
Untracked: analysis/permu_9_m3_Novelty_resids_datarelease_07302918.stderr
Untracked: analysis/permu_9_m3_Novelty_resids_datarelease_07302918.stdout
Untracked: analysis/run_10_qtl_permu.R.Rout
Untracked: analysis/run_10_qtl_permu.e10855280
Untracked: analysis/run_10_qtl_permu.o10855280
Untracked: analysis/run_11_qtl_blup.R.Rout
Untracked: analysis/run_12_plot_qtl_mapping.R
Untracked: analysis/run_14_geneseek2qtl2fst_Gagnon.R
Untracked: analysis/run_15_firstgm2genoprobs_Gagnon.R
Untracked: analysis/run_16_diagnosis_qc_gigamuga_gagnon.R
Untracked: analysis/run_run_10_qtl_permu.script
Untracked: analysis/run_run_11_qtl_blup.script
Untracked: code/reconst_utils.R
Untracked: data/FinalReport/
Untracked: data/GCTA/
Untracked: data/GM/
Untracked: data/Jackson_Lab_Bubier_MURGIGV01/
Untracked: data/Jackson_Lab_Gagnon/
Untracked: data/cc_variants.sqlite
Untracked: data/marker_grid_0.02cM_plus.txt
Untracked: data/mouse_genes_mgi.sqlite
Untracked: data/pheno/
Untracked: output/AfterQC_Percent_missing_genotype_data.pdf
Untracked: output/AfterQC_Proportion_matching_genotypes_after_removal_of_bad_samples.pdf
Untracked: output/AfterQC_Proportion_matching_genotypes_before_removal_of_bad_samples.pdf
Untracked: output/AfterQC_number_crossover.pdf
Untracked: output/Percent_genotype_errors_obs_vs_predicted.pdf
Untracked: output/Percent_missing_genotype_data.pdf
Untracked: output/Percent_missing_genotype_data_per_marker.pdf
Untracked: output/Proportion_matching_genotypes_after_removal_of_bad_samples.pdf
Untracked: output/Proportion_matching_genotypes_before_removal_of_bad_samples.pdf
Untracked: output/blup/
Untracked: output/condi.m2.qtl.out.RData
Untracked: output/genotype_error_marker.pdf
Untracked: output/genotype_frequency_marker.pdf
Untracked: output/m1.Novelty_resids_datarelease_07302918.RData
Untracked: output/m1.Novelty_resids_datarelease_07302918_coeffgeneplot.pdf
Untracked: output/m1.Novelty_resids_datarelease_07302918_genomescan.pdf
Untracked: output/m1.Novelty_residuals_RankNormal_datarelease_07302918.RData
Untracked: output/m1.Novelty_residuals_RankNormal_datarelease_07302918_genomescan.pdf
Untracked: output/m2.Novelty_resids_datarelease_07302918.RData
Untracked: output/m2.Novelty_resids_datarelease_07302918_genomescan.pdf
Untracked: output/m2.Novelty_residuals_RankNormal_datarelease_07302918.RData
Untracked: output/m2.Novelty_residuals_RankNormal_datarelease_07302918_genomescan.pdf
Untracked: output/m3.Novelty_resids_datarelease_07302918.RData
Untracked: output/m3.Novelty_resids_datarelease_07302918_genomescan.pdf
Untracked: output/m3.Novelty_residuals_RankNormal_datarelease_07302918.RData
Untracked: output/m3.Novelty_residuals_RankNormal_datarelease_07302918_genomescan.pdf
Untracked: output/num.geno.pheno.in.Novelty_resids_datarelease_07302918.csv
Untracked: output/num.geno.pheno.in.Novelty_residuals_RankNormal_datarelease_07302918.csv
Untracked: output/number_crossover.pdf
Untracked: output/permu/
Untracked: output/prop_across_generation_chr_p.RData
Unstaged changes:
Modified: README.md
Modified: _workflowr.yml
Modified: analysis/10_qtl_permu.R
Modified: analysis/run_10_qtl_permu.R
Modified: analysis/run_11_qtl_blup.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the R Markdown and HTML files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view them.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 884447c | xhyuo | 2019-10-20 | 16_diagnosis_qc_gigamuga_gagnon.Rmd |
| html | a6d6886 | xhyuo | 2019-10-20 | Build site. |
| Rmd | 7f95a3a | xhyuo | 2019-10-20 | 16_diagnosis_qc_gigamuga_gagnon.Rmd |
author: “Hao He” date: “2019-10-20” output: html_document —
Genotype diagnostics for diversity outbred mice
We first load the R/qtl2 package and the data. We’ll also load the R/broman package for some utilities and plotting functions, and R/qtlcharts for interactive graphs.
library
library(broman)
library(qtl2)
library(qtlcharts)
library(ggplot2)
library(ggrepel)
library(DOQTL)
library(mclust)
source("/projects/csna/csna_workflow/code/reconst_utils.R")Missing data per sample
load("/projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/Gagnon_gm.RData")
gmObject of class cross2 (crosstype "do")
Total individuals 384
No. genotyped individuals 384
No. phenotyped individuals 384
No. with both geno & pheno 384
No. phenotypes 1
No. covariates 3
No. phenotype covariates 0
No. chromosomes 20
Total markers 112728
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15
8555 8666 6420 6615 6571 6443 6294 5677 5870 5447 6352 5167 5274 5039 4555
16 17 18 19 X
4369 4330 4002 3108 3974 percent_missing <- n_missing(gm, "ind", "prop")*100
setScreenSize(height=100, width=300)Set screen size to height=100 x width=300labels <- paste0(names(percent_missing), " (", round(percent_missing,2), "%)")
iplot(seq_along(percent_missing), percent_missing, indID=labels,
chartOpts=list(xlab="Mouse", ylab="Percent missing genotype data",
ylim=c(0, 100)))#save into pdf
pdf(file = "/projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/Percent_missing_genotype_data.pdf", width = 20, height = 20)
labels <- as.character(do.call(rbind.data.frame, strsplit(ind_ids(gm), "V01_"))[,2])
labels[percent_missing < 5] = ""
# Change point shapes and colors
p <- ggplot(data = data.frame(Mouse=seq_along(percent_missing),
Percent_missing_genotype_data = percent_missing,
batch = factor(as.character(do.call(rbind.data.frame, strsplit(ind_ids(gm), "_"))[,6]))),
aes(x=Mouse, y=Percent_missing_genotype_data, color = batch)) +
geom_point() +
geom_hline(yintercept=5, linetype="solid", color = "red") +
geom_text_repel(aes(label=labels), vjust = 0, nudge_y = 0.01, show.legend = FALSE, size=3) +
theme(text = element_text(size = 20))
p
dev.off()png
2 p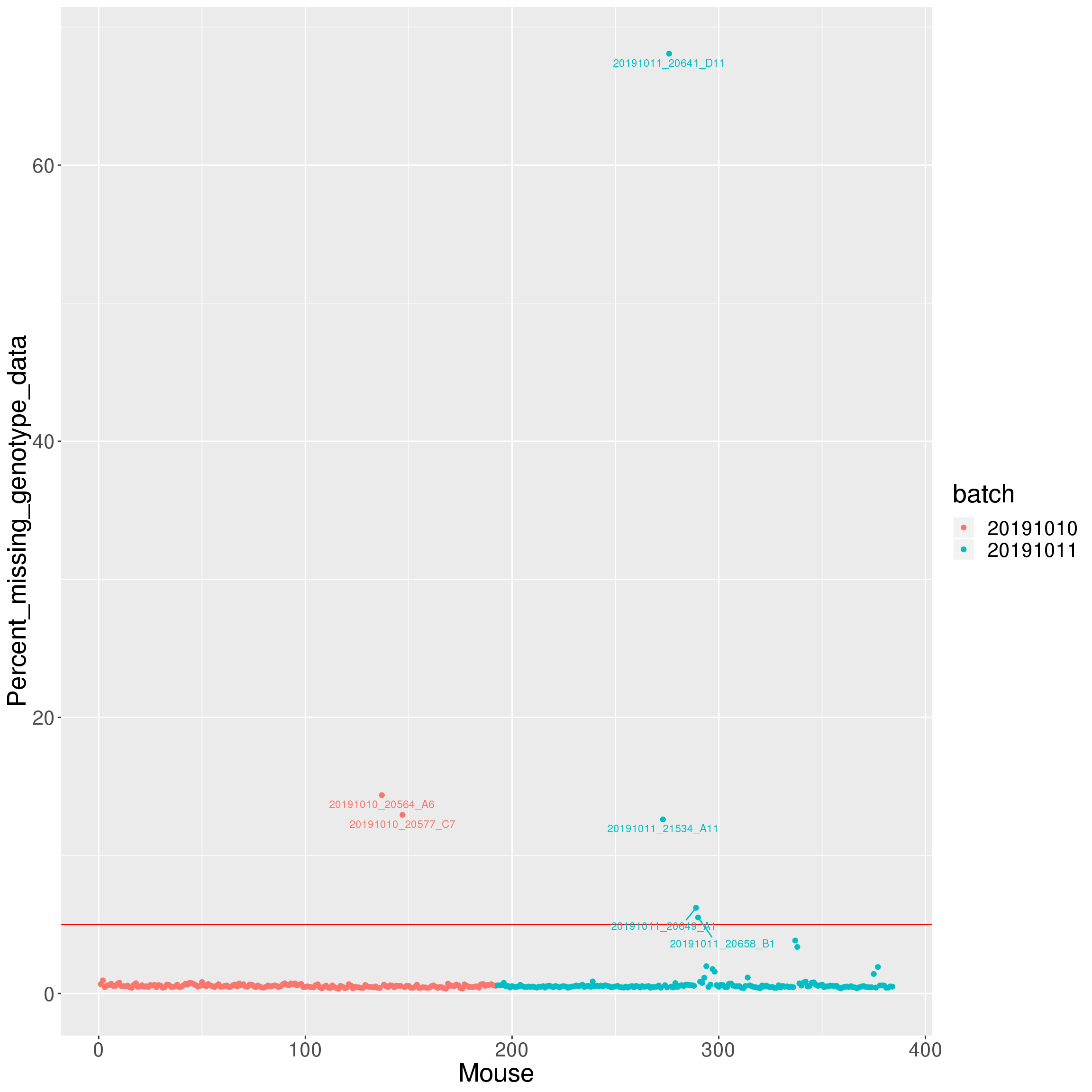
| Version | Author | Date |
|---|---|---|
| a6d6886 | xhyuo | 2019-10-20 |
save(percent_missing,
file = "/projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/percent_missing_id.RData")Sexes
xint <- read_csv_numer("/projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/Jackson_Lab_Gagnon_qtl2_chrXint.csv", transpose=TRUE)
yint <- read_csv_numer("/projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/Jackson_Lab_Gagnon_qtl2_chrYint.csv", transpose=TRUE)
# Gigamuga marker annotation file from UNC.
gm_marker_file = "http://csbio.unc.edu/MUGA/snps.gigamuga.Rdata" #FIXED
# Read in the UNC GigaMUGA SNPs and clusters.
load(url(gm_marker_file))
#subset down to gm
snps$marker = as.character(snps$marker)
#load the intensities.fst.RData
load("/projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/intensities.fst.RData")
#X and Y channel
X <- result[result$channel == "X",]
rownames(X) <- X$snp
X <- X[,c(-1,-2)]
Y <- result[result$channel == "Y",]
rownames(Y) <- Y$snp
Y <- Y[,c(-1,-2)]
#determine sex
sex = determine_sex_chry_m(x = X, y = Y, markers = snps)$sex
gm$covar <- merge(data.frame(id = names(sex),
predict.sex = sex,stringsAsFactors = F),
gm$covar,
by.x = "id",
by.y = "row.names")
rownames(gm$covar) <- gm$covar$id
#sex order
sex <- gm$covar[rownames(xint),"predict.sex"]
x_pval <- apply(xint, 2, function(a) t.test(a ~ sex)$p.value)
y_pval <- apply(yint, 2, function(a) t.test(a ~ sex)$p.value)
xint_ave <- rowMeans(xint[, x_pval < 0.05/length(x_pval)], na.rm=TRUE)
yint_ave <- rowMeans(yint[, y_pval < 0.05/length(y_pval)], na.rm=TRUE)
point_colors <- as.character( brocolors("web")[c("green", "purple")] )
labels <- paste0(names(xint_ave))
iplot(xint_ave, yint_ave, group=sex, indID=labels,
chartOpts=list(pointcolor=point_colors, pointsize=4,
xlab="Average X chr intensity", ylab="Average Y chr intensity"))phetX <- rowSums(gm$geno$X == 2)/rowSums(gm$geno$X != 0)
phetX <- phetX[names(phetX) %in% names(xint_ave)]
iplot(xint_ave, phetX, group=sex, indID=labels,
chartOpts=list(pointcolor=point_colors, pointsize=4,
xlab="Average X chr intensity", ylab="Proportion het on X chr"))Sample duplicates
cg <- compare_geno(gm, cores=10)
summary.cg <- summary(cg)
summary.cg ind1
The_Jackson_Lab_Gagnon_MURGIGV01_20191011_21433_H9
ind2 prop_match n_mismatch
The_Jackson_Lab_Gagnon_MURGIGV01_20191011_21412_A10 1 35
n_typed n_match index1 index2
112027 111992 360 361summary.cg$Name.ind1 <- as.character(do.call(rbind.data.frame, strsplit(as.character(summary.cg$ind1), "_"))[,6])
summary.cg$Name.ind2 <- as.character(do.call(rbind.data.frame, strsplit(as.character(summary.cg$ind2), "_"))[,6])
summary.cg$miss.ind1 <- percent_missing[match(summary.cg$ind1, names(percent_missing))]
summary.cg$miss.ind2 <- percent_missing[match(summary.cg$ind2, names(percent_missing))]
summary.cg$remove.id <- ifelse(summary.cg$miss.ind1 > summary.cg$miss.ind2, summary.cg$ind1, summary.cg$ind2)
summary.cg$remove.id [1] "The_Jackson_Lab_Gagnon_MURGIGV01_20191011_21412_A10"save(summary.cg,
file = "/projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/summary.cg.RData")
pdf(file = "/projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/Proportion_matching_genotypes_before_removal_of_bad_samples.pdf", width = 20, height = 20)
par(mar=c(5.1,0.6,0.6, 0.6))
hist(cg[upper.tri(cg)], breaks=seq(0, 1, length=201),
main="", yaxt="n", ylab="", xlab="Proportion matching genotypes")
rug(cg[upper.tri(cg)])
dev.off()png
2 par(mar=c(5.1,0.6,0.6, 0.6))
hist(cg[upper.tri(cg)], breaks=seq(0, 1, length=201),
main="", yaxt="n", ylab="", xlab="Proportion matching genotypes")
rug(cg[upper.tri(cg)])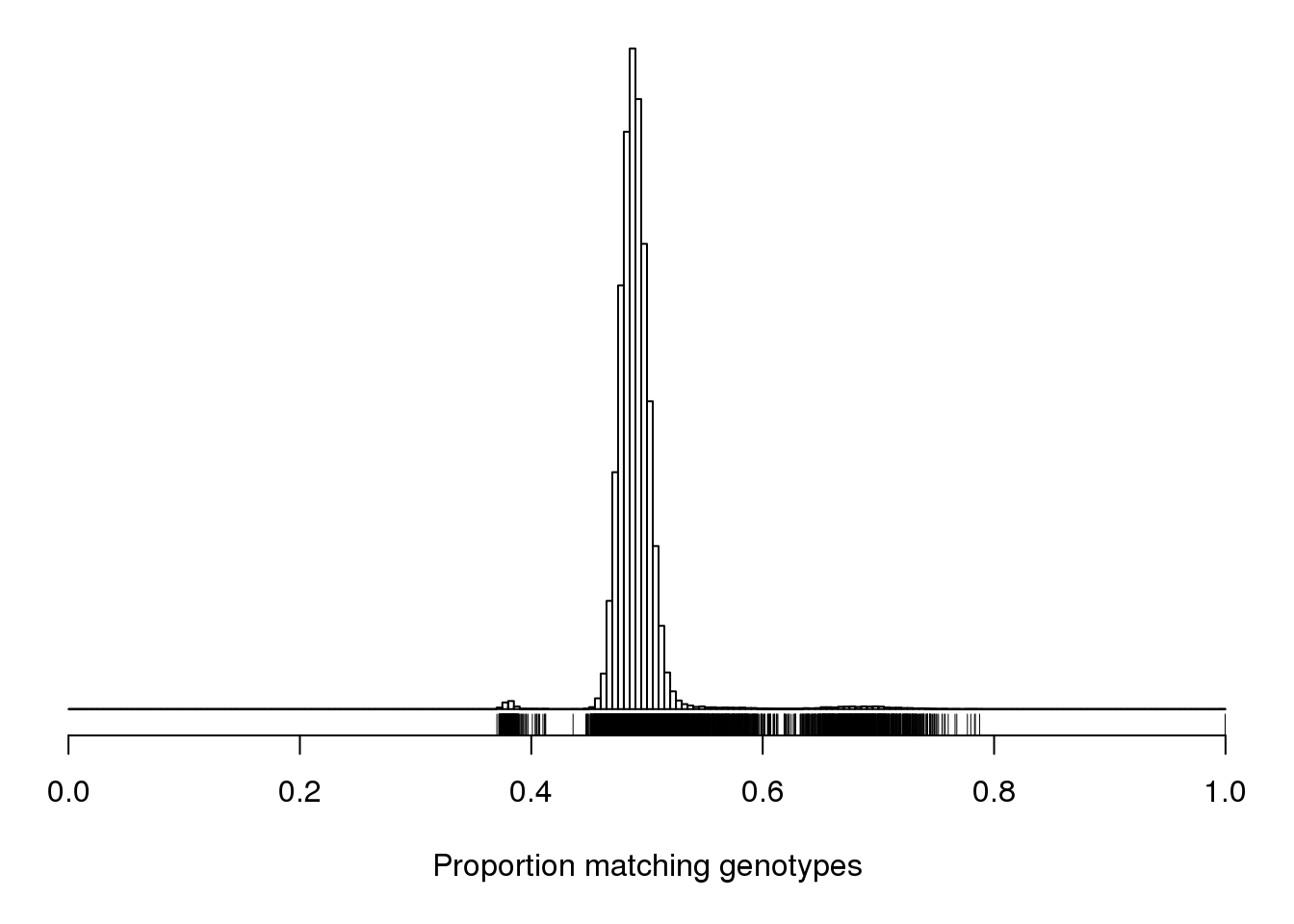
| Version | Author | Date |
|---|---|---|
| a6d6886 | xhyuo | 2019-10-20 |
pdf(file = "/projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/Proportion_matching_genotypes_after_removal_of_bad_samples.pdf",width = 20, height = 20)
cgsub <- cg[percent_missing < 5, percent_missing < 5]
par(mar=c(5.1,0.6,0.6, 0.6))
hist(cgsub[upper.tri(cgsub)], breaks=seq(0, 1, length=201),
main="", yaxt="n", ylab="", xlab="Proportion matching genotypes")
rug(cgsub[upper.tri(cgsub)])
dev.off()png
2 cgsub <- cg[percent_missing < 5, percent_missing < 5]
par(mar=c(5.1,0.6,0.6, 0.6))
hist(cgsub[upper.tri(cgsub)], breaks=seq(0, 1, length=201),
main="", yaxt="n", ylab="", xlab="Proportion matching genotypes")
rug(cgsub[upper.tri(cgsub)])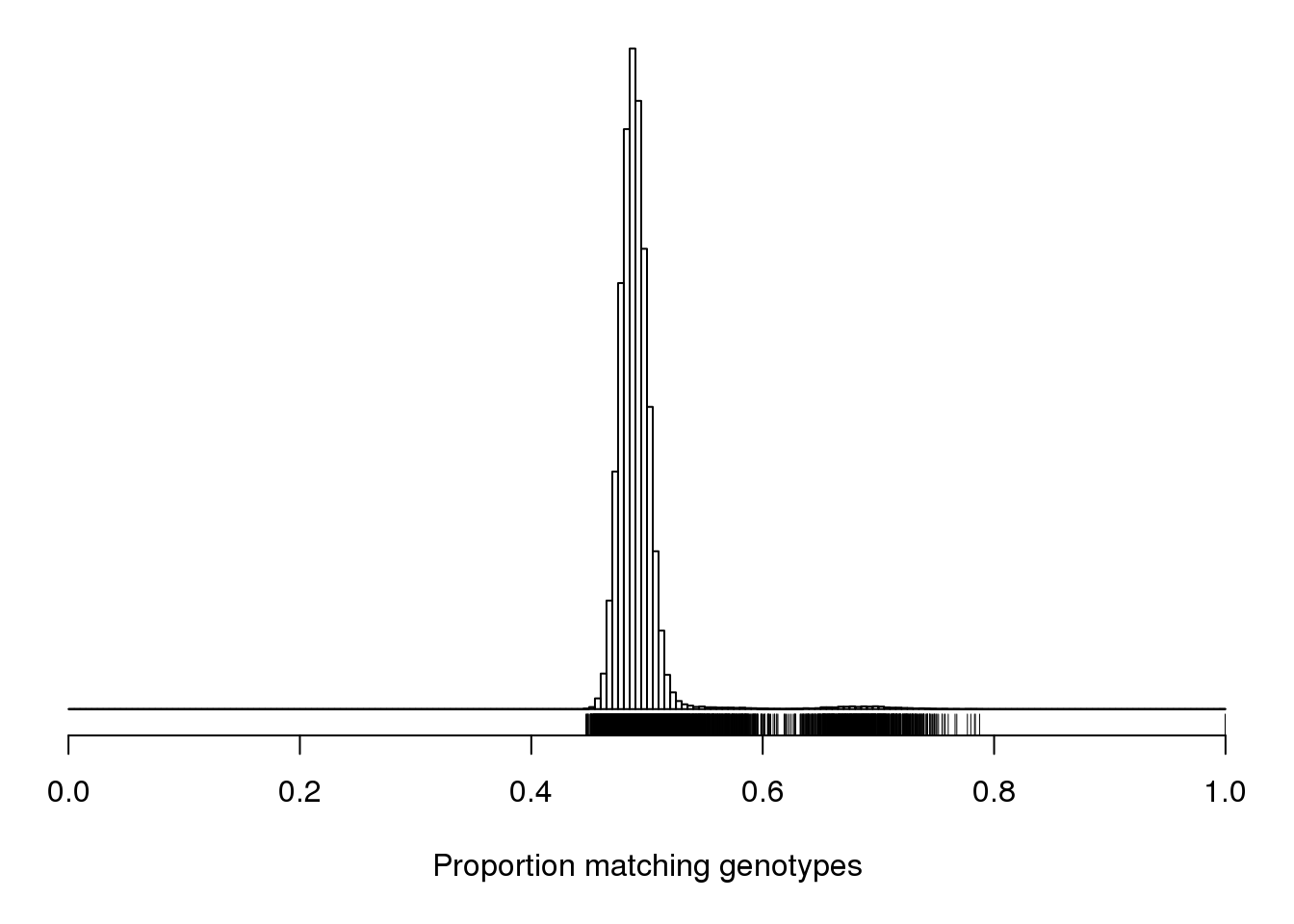
| Version | Author | Date |
|---|---|---|
| a6d6886 | xhyuo | 2019-10-20 |
#show top 20 samples with missing genotypes
percent_missing <- n_missing(gm, "ind", "prop")*100
round(sort(percent_missing, decreasing=TRUE)[1:20], 1)The_Jackson_Lab_Gagnon_MURGIGV01_20191011_20641_D11
68.1
The_Jackson_Lab_Gagnon_MURGIGV01_20191010_20564_A6
14.4
The_Jackson_Lab_Gagnon_MURGIGV01_20191010_20577_C7
12.9
The_Jackson_Lab_Gagnon_MURGIGV01_20191011_21534_A11
12.6
The_Jackson_Lab_Gagnon_MURGIGV01_20191011_20649_A1
6.2
The_Jackson_Lab_Gagnon_MURGIGV01_20191011_20658_B1
5.5
The_Jackson_Lab_Gagnon_MURGIGV01_20191011_21440_A7
3.8
The_Jackson_Lab_Gagnon_MURGIGV01_20191011_20692_B7
3.4
The_Jackson_Lab_Gagnon_MURGIGV01_20191011_20656_F1
2.0
The_Jackson_Lab_Gagnon_MURGIGV01_20191011_22396_A12
1.9
The_Jackson_Lab_Gagnon_MURGIGV01_20191011_20660_A2
1.8
The_Jackson_Lab_Gagnon_MURGIGV01_20191011_21526_B2
1.6
The_Jackson_Lab_Gagnon_MURGIGV01_20191011_22637_G11
1.4
The_Jackson_Lab_Gagnon_MURGIGV01_20191011_21411_B4
1.2
The_Jackson_Lab_Gagnon_MURGIGV01_20191011_21490_E1
1.1
The_Jackson_Lab_Gagnon_MURGIGV01_20191010_20716_B1
0.9
The_Jackson_Lab_Gagnon_MURGIGV01_20191011_21516_G6
0.9
The_Jackson_Lab_Gagnon_MURGIGV01_20191011_21405_F7
0.9
The_Jackson_Lab_Gagnon_MURGIGV01_20191011_21535_C1
0.9
The_Jackson_Lab_Gagnon_MURGIGV01_20191010_22640_B7
0.8 Array intensities and Genotype frequencies
int <- result
rm(result)
int <- int[seq(1, nrow(int), by=2),-(1:2)] + int[-seq(1, nrow(int), by=2),-(1:2)]
int <- int[,intersect(ind_ids(gm), colnames(int))]
n <- names(sort(percent_missing[intersect(ind_ids(gm), colnames(int))], decreasing=TRUE))
iboxplot(log10(t(int[,n])+1), orderByMedian=FALSE, chartOpts=list(ylab="log10(SNP intensity + 1)"))# Genotype frequencies
g <- do.call("cbind", gm$geno[1:19])
fg <- do.call("cbind", gm$founder_geno[1:19])
g <- g[,colSums(fg==0)==0]
fg <- fg[,colSums(fg==0)==0]
fgn <- colSums(fg==3)
gf_ind <- vector("list", 4)
for(i in 1:4) {
gf_ind[[i]] <- t(apply(g[,fgn==i], 1, function(a) table(factor(a, 1:3))/sum(a != 0)))
}
par(mfrow=c(4,1), mar=c(0.6, 0.6, 2.6, 0.6))
for(i in 1:4) {
triplot(c("AA", "AB", "BB"), main=paste0("MAF = ", i, "/8"))
tripoints(gf_ind[[i]], pch=21, bg="lightblue")
tripoints(c((1-i/8)^2, 2*i/8*(1-i/8), (i/8)^2), pch=21, bg="violetred")
if(i>=3) { # label mouse with lowest het
wh <- which(gf_ind[[i]][,2] == min(gf_ind[[i]][,2]))
tritext(gf_ind[[i]][wh,,drop=FALSE] + c(0.02, -0.02, 0),
names(wh), adj=c(0, 1))
}
# label other mice
if(i==1) {
lab <- rownames(gf_ind[[i]])[gf_ind[[i]][,2]>0.3]
}
else if(i==2) {
lab <- rownames(gf_ind[[i]])[gf_ind[[i]][,2]>0.48]
}
else if(i==3) {
lab <- rownames(gf_ind[[i]])[gf_ind[[i]][,2]>0.51]
}
else if(i==4) {
lab <- rownames(gf_ind[[i]])[gf_ind[[i]][,2]>0.6]
}
for(ind in lab) {
if(grepl("^F", ind) && i != 3) {
tritext(gf_ind[[i]][ind,,drop=FALSE] + c(-0.01, 0, +0.01), ind, adj=c(1,0.5))
} else {
tritext(gf_ind[[i]][ind,,drop=FALSE] + c(0.01, 0, -0.01), ind, adj=c(0,0.5))
}
}
}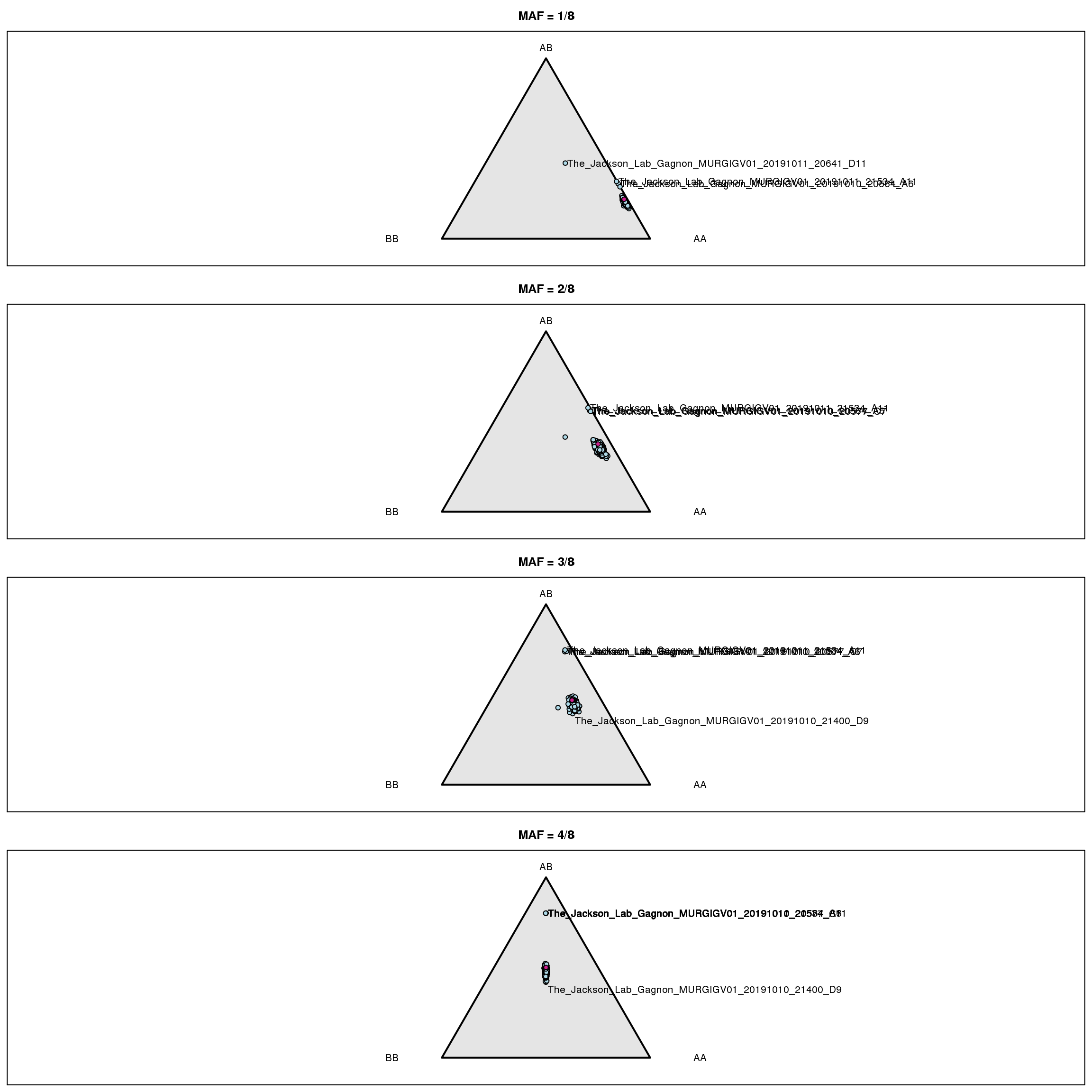
| Version | Author | Date |
|---|---|---|
| a6d6886 | xhyuo | 2019-10-20 |
Crossover counts and Genotyping error LOD scores
# qsub qsub pr_cross_over.sh to get the pr, m and nxo
#load pre-caluated results
load("/projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/pr.RData")
load("/projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/m.RData")
load("/projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/nxo.RData")
#crossover
totxo <- rowSums(nxo)[rownames(gm$covar)]
iplot(seq_along(totxo),
totxo,
group=gm$covar$ngen,
chartOpts=list(xlab="Mouse", ylab="Number of crossovers",
margin=list(left=80,top=40,right=40,bottom=40,inner=5),
axispos=list(xtitle=25,ytitle=50,xlabel=5,ylabel=5)))#save crossover into pdf
pdf(file = "/projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/number_crossover.pdf")
cross_over <- data.frame(Mouse = seq_along(totxo), Number_crossovers = totxo, generation = gm$covar$ngen)
names(totxo) <- as.character(do.call(rbind.data.frame, strsplit(names(totxo), "V01_"))[,2])
names(totxo)[totxo <= 800] = ""
# Change point shapes and colors
p <-ggplot(cross_over, aes(x=Mouse, y=Number_crossovers, fill = generation, color=generation)) +
geom_point() +
geom_text_repel(aes(label=names(totxo),hjust=0,vjust=0), show.legend = FALSE)
p
dev.off()png
2 p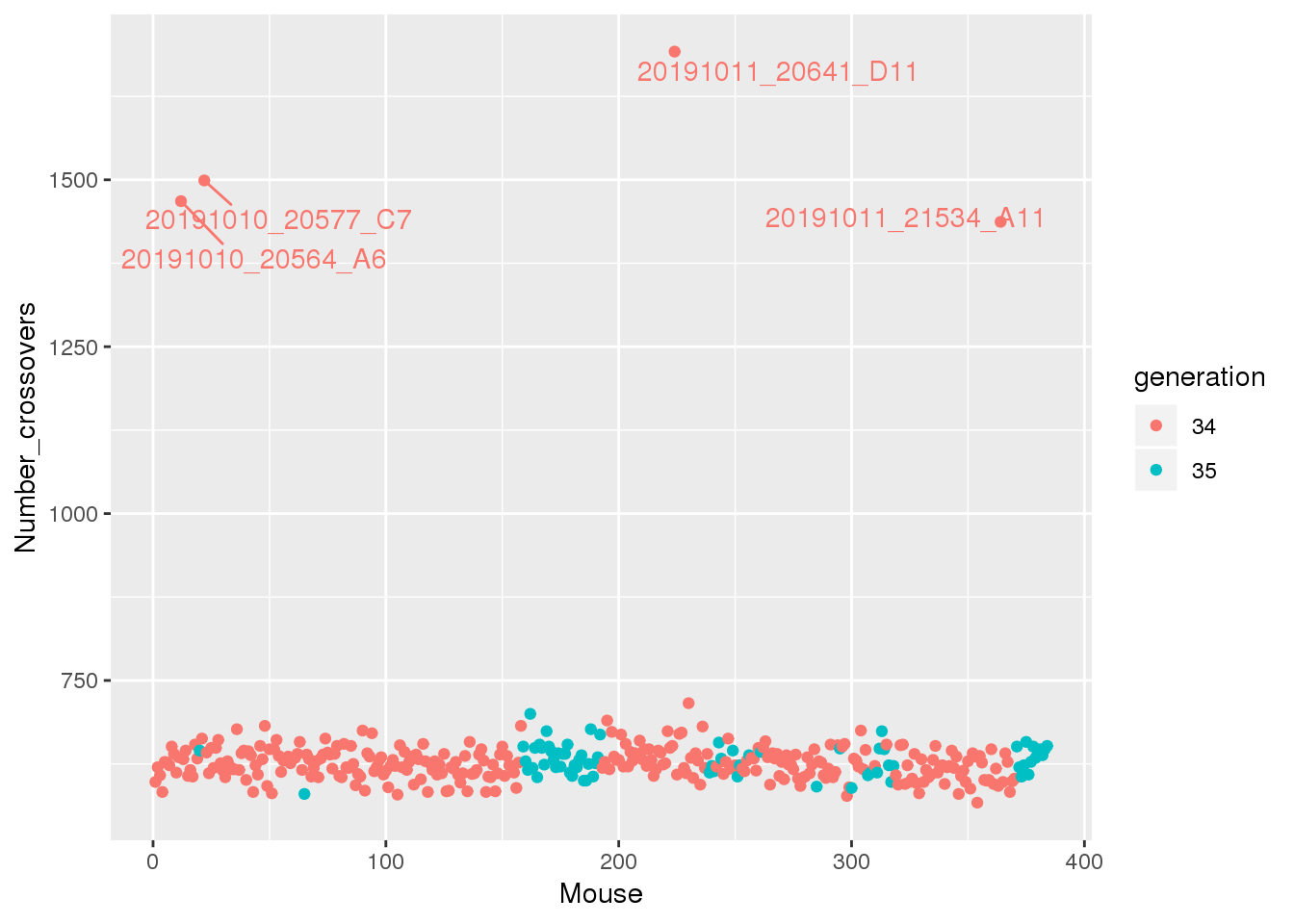
| Version | Author | Date |
|---|---|---|
| a6d6886 | xhyuo | 2019-10-20 |
#Here are the crossover counts for those mice:
tmp <- cbind(percent_missing=round(percent_missing,2), total_xo=totxo)[percent_missing >= 5,]
tmp[order(tmp[,1]),] percent_missing
The_Jackson_Lab_Gagnon_MURGIGV01_20191011_20658_B1 5.51
The_Jackson_Lab_Gagnon_MURGIGV01_20191011_20649_A1 6.20
The_Jackson_Lab_Gagnon_MURGIGV01_20191011_21534_A11 12.61
The_Jackson_Lab_Gagnon_MURGIGV01_20191010_20577_C7 12.94
The_Jackson_Lab_Gagnon_MURGIGV01_20191010_20564_A6 14.37
The_Jackson_Lab_Gagnon_MURGIGV01_20191011_20641_D11 68.07
total_xo
The_Jackson_Lab_Gagnon_MURGIGV01_20191011_20658_B1 618
The_Jackson_Lab_Gagnon_MURGIGV01_20191011_20649_A1 604
The_Jackson_Lab_Gagnon_MURGIGV01_20191011_21534_A11 624
The_Jackson_Lab_Gagnon_MURGIGV01_20191010_20577_C7 584
The_Jackson_Lab_Gagnon_MURGIGV01_20191010_20564_A6 610
The_Jackson_Lab_Gagnon_MURGIGV01_20191011_20641_D11 639# Genotyping error LOD scores
load("/projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/e.RData")
errors_ind <- rowSums(e>2)/n_typed(gm)*100
lab <- paste0(names(errors_ind), " (", myround(percent_missing,1), "%)")
iplot(seq_along(errors_ind), errors_ind, indID=lab,
chartOpts=list(xlab="Mouse", ylab="Percent genotyping errors", ylim=c(0, 8),
axispos=list(xtitle=25, ytitle=50, xlabel=5, ylabel=5)))save(errors_ind, file = "/projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/errors_ind.RData")
# Apparent genotyping errors
load("/projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/snpg.RData")
gobs <- do.call("cbind", gm$geno)
gobs[gobs==0] <- NA
par(pty="s")
err_direct <- rowMeans(snpg != gobs, na.rm=TRUE)*100
errors_ind_0 <- rowSums(e > 0)/n_typed(gm)*100
par(mar=c(4.1,4.1,0.6, 0.6))
grayplot(errors_ind_0, err_direct,
xlab="Percent errors (error LOD > 0)",
ylab="Percent errors (obs vs predicted)",
xlim=c(0, 2), ylim=c(0, 2))
abline(0,1,lty=2, col="gray60")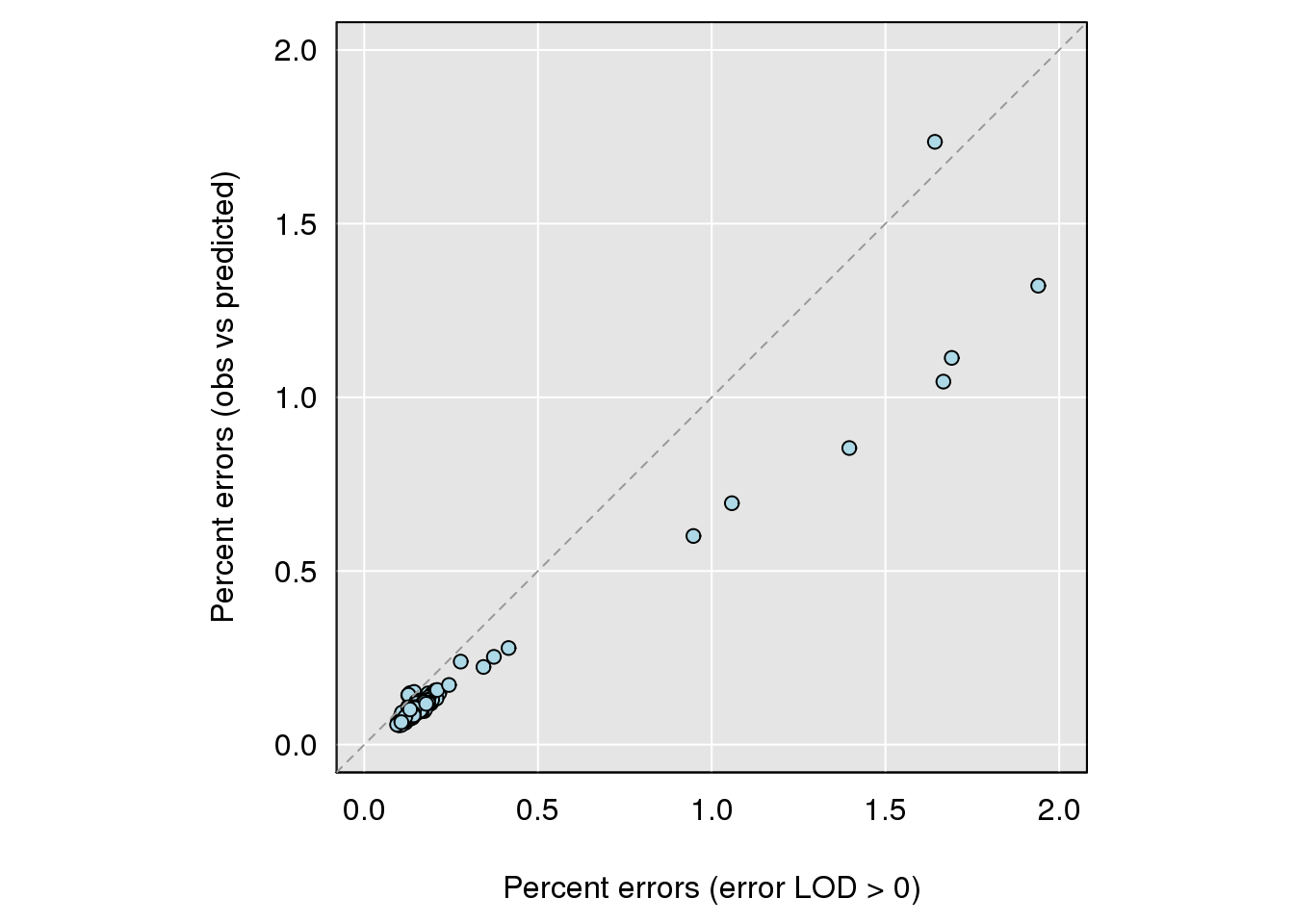
| Version | Author | Date |
|---|---|---|
| a6d6886 | xhyuo | 2019-10-20 |
pdf(file = "/projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/Percent_genotype_errors_obs_vs_predicted.pdf",width = 20, height = 20)
par(pty="s")
err_direct <- rowMeans(snpg != gobs, na.rm=TRUE)*100
errors_ind_0 <- rowSums(e > 0)/n_typed(gm)*100
par(mar=c(4.1,4.1,0.6, 0.6))
grayplot(errors_ind_0, err_direct,
xlab="Percent errors (error LOD > 0)",
ylab="Percent errors (obs vs predicted)",
xlim=c(0, 2), ylim=c(0, 2))
abline(0,1,lty=2, col="gray60")
dev.off()png
2 Missing data in Markers and Genotype frequencies Markers
#It can also be useful to look at the proportion of missing genotypes by marker.
#Markers with a lot of missing data were likely difficult to call, and so the genotypes that were called may contain a lot of errors.
pmis_mar <- n_missing(gm, "marker", "proportion")*100
par(mar=c(5.1,0.6,0.6, 0.6))
hist(pmis_mar, breaks=seq(0, 100, length=201),
main="", yaxt="n", ylab="", xlab="Percent missing genotypes")
rug(pmis_mar)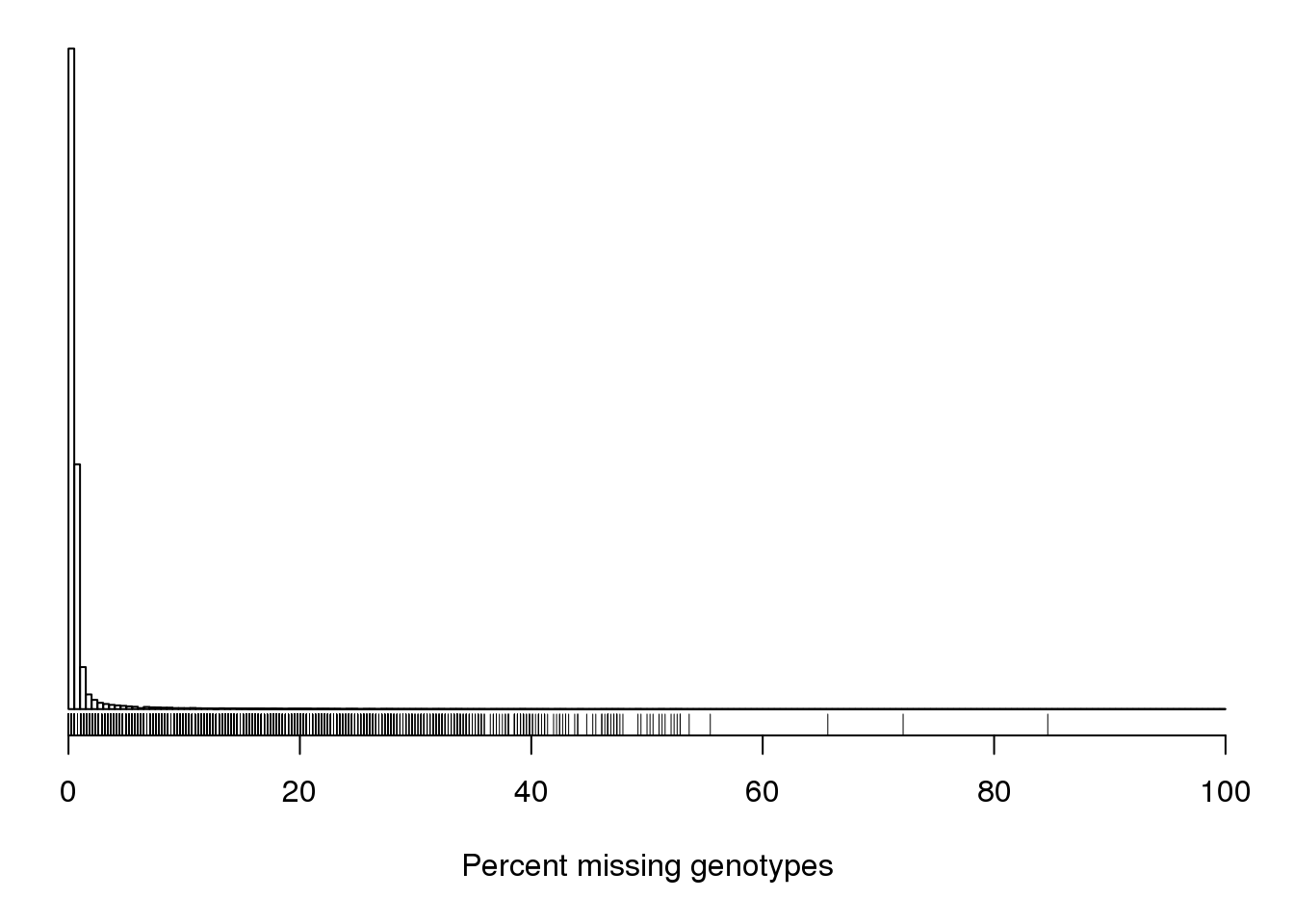
| Version | Author | Date |
|---|---|---|
| a6d6886 | xhyuo | 2019-10-20 |
pdf(file = "/projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/Percent_missing_genotype_data_per_marker.pdf")
par(mar=c(5.1,0.6,0.6, 0.6))
hist(pmis_mar, breaks=seq(0, 100, length=201),
main="", yaxt="n", ylab="", xlab="Percent missing genotypes")
rug(pmis_mar)
dev.off()png
2 # Genotype frequencies Markers
gf_mar <- t(apply(g, 2, function(a) table(factor(a, 1:3))/sum(a != 0)))
gn_mar <- t(apply(g, 2, function(a) table(factor(a, 1:3))))
pdf(file = "/projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/genotype_frequency_marker.pdf")
par(mfrow=c(2,2), mar=c(0.6, 0.6, 2.6, 0.6))
for(i in 1:4) {
triplot(c("AA", "AB", "BB"), main=paste0("MAF = ", i, "/8"))
z <- gf_mar[fgn==i,]
z <- z[rowSums(is.na(z)) < 3,]
tripoints(z, pch=21, bg="gray80", cex=0.6)
tripoints(c((1-i/8)^2, 2*i/8*(1-i/8), (i/8)^2), pch=21, bg="violetred")
}
dev.off()png
2 par(mfrow=c(2,2), mar=c(0.6, 0.6, 2.6, 0.6))
for(i in 1:4) {
triplot(c("AA", "AB", "BB"), main=paste0("MAF = ", i, "/8"))
z <- gf_mar[fgn==i,]
z <- z[rowSums(is.na(z)) < 3,]
tripoints(z, pch=21, bg="gray80", cex=0.6)
tripoints(c((1-i/8)^2, 2*i/8*(1-i/8), (i/8)^2), pch=21, bg="violetred")
}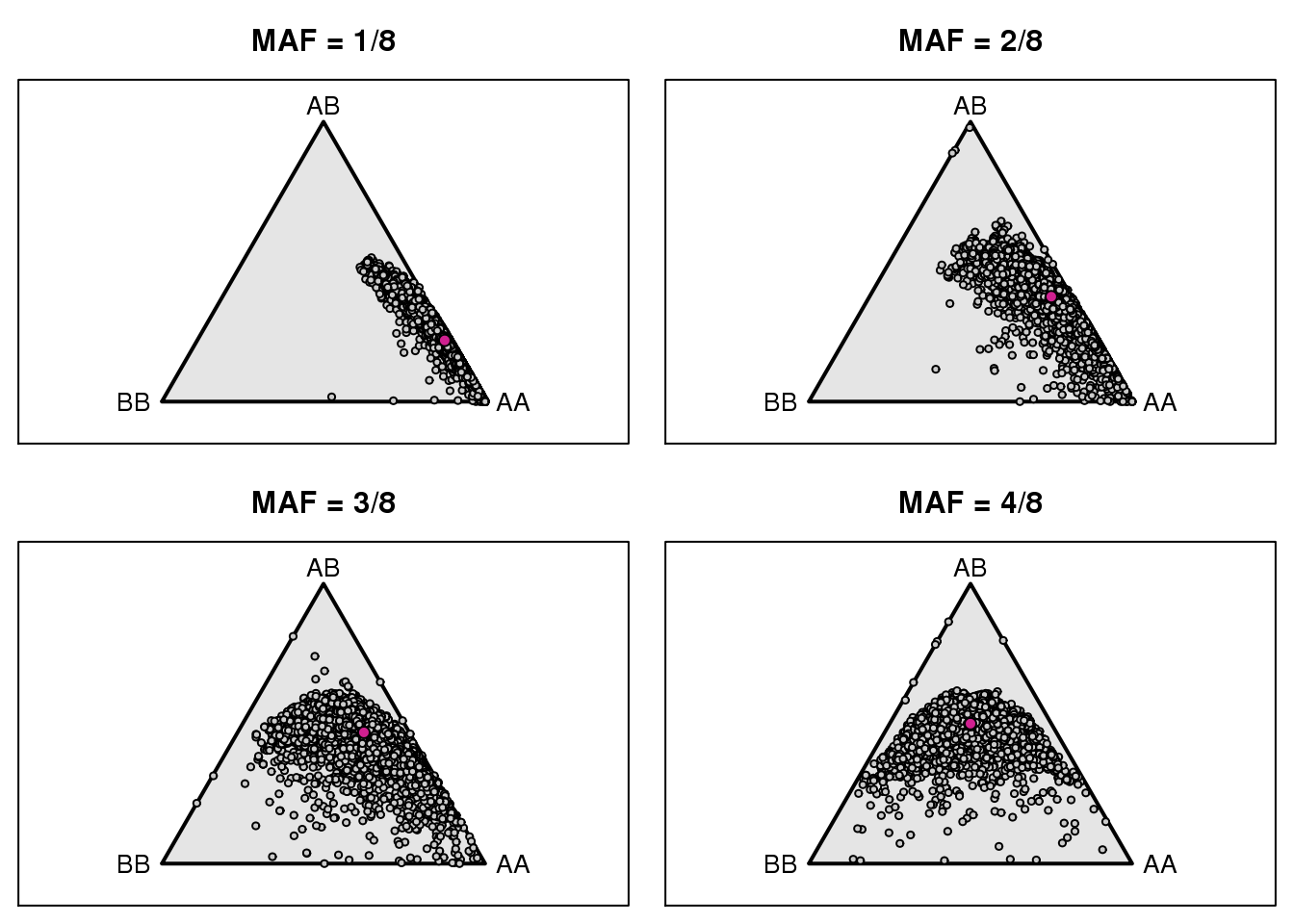
| Version | Author | Date |
|---|---|---|
| a6d6886 | xhyuo | 2019-10-20 |
# Genotype errors Markers
errors_mar <- colSums(e>2)/n_typed(gm, "marker")*100
grayplot(pmis_mar, errors_mar,
xlab="Proportion missing", ylab="Proportion genotyping errors")
pdf(file = "/projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/genotype_error_marker.pdf")
grayplot(pmis_mar, errors_mar,
xlab="Proportion missing", ylab="Proportion genotyping errors")
dev.off()png
2 Remove bad samples
#percent missing
qc_info <- merge(gm$covar,
data.frame(id = names(percent_missing),
percent_missing = percent_missing,stringsAsFactors = F),by = "id")
#cross_over
qc_info <- merge(qc_info,
data.frame(id = rownames(cross_over),
Number_crossovers = cross_over$Number_crossovers,stringsAsFactors = F),by = "id")
#missing sex
qc_info[qc_info$id %in% tot.id.covar.csna.sexna$ID, "sex"] <- NA
qc_info$sex.match <- ifelse(qc_info$predict.sex == qc_info$sex, TRUE, FALSE)
#genotype errors
qc_info <- merge(qc_info,
data.frame(id = names(errors_ind),
genotype_erros = errors_ind,stringsAsFactors = F),by = "id")
#duplicated id to be remove
qc_info$remove.id.duplicated <- ifelse(qc_info$id %in% summary.cg$remove.id, TRUE,FALSE)
bad.sample <- qc_info[qc_info$ngen ==1 | qc_info$Number_crossovers <= 200 | qc_info$Number_crossovers >=1000 | qc_info$percent_missing >= 10 | qc_info$genotype_erros >= 1 | qc_info$remove.id.duplicated == TRUE,]
save(qc_info, bad.sample, file = "/projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/qc_info_bad_sample.RData")
#remove 77 bad samples
gm.no.bad <- gm[paste0("-",as.character(bad.sample$id)),]
#fix sex mismatch
gm.no.bad$covar[gm.no.bad$covar$id %in% names(yint_ave[!names(yint_ave) %in% bad.sample$id][yint_ave[!names(yint_ave) %in% bad.sample$id] <= 0.1]),"predict.sex"] <- "F"
gm.no.bad
#2437 subjects
# update other stuff
e <- e[ind_ids(gm.no.bad),]
g <- g[ind_ids(gm.no.bad),]
snpg <- snpg[ind_ids(gm.no.bad),]
length(errors_mar[errors_mar > 5])
# omit the 273 markers with error rates >5%.
bad_markers <- find_markerpos(gm.no.bad, names(errors_mar[errors_mar > 5]))
save(bad_markers, file = "/projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/bad_markers.RData")
#drop bad markers
gm_DO2437_qc <- drop_markers(gm.no.bad, names(errors_mar)[errors_mar > 5])
gm_DO2437_qc
save(gm_DO2437_qc, file = "/projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/gm_DO2437_qc.RData")
save(e,g,snpg, file = "/projects/csna/csna_workflow/data/Jackson_Lab_Gagnon/e_g_snpg_qc.RData")
sessionInfo()R version 3.3.2 (2016-10-31)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS release 6.5 (Final)
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats4 parallel methods stats graphics grDevices utils
[8] datasets base
other attached packages:
[1] mclust_5.2.1 DOQTL_1.10.0
[3] VariantAnnotation_1.20.3 Rsamtools_1.26.2
[5] SummarizedExperiment_1.4.0 Biobase_2.34.0
[7] BSgenome.Mmusculus.UCSC.mm10_1.4.0 BSgenome_1.42.0
[9] rtracklayer_1.34.2 Biostrings_2.42.1
[11] XVector_0.14.1 GenomicRanges_1.26.4
[13] GenomeInfoDb_1.10.3 IRanges_2.8.2
[15] S4Vectors_0.12.2 BiocGenerics_0.20.0
[17] ggrepel_0.8.1 ggplot2_3.1.0
[19] qtlcharts_0.9-6 qtl2_0.18
[21] broman_0.68-2
loaded via a namespace (and not attached):
[1] bitops_1.0-6 fs_1.2.6
[3] bit64_0.9-7 doParallel_1.0.10
[5] rprojroot_1.3-2 prabclus_2.2-6
[7] regress_1.3-15 tools_3.3.2
[9] backports_1.1.2 R6_2.4.0
[11] DBI_1.0.0 lazyeval_0.2.1
[13] colorspace_1.4-0 trimcluster_0.1-2
[15] annotationTools_1.48.0 nnet_7.3-12
[17] withr_2.1.2 tidyselect_0.2.5
[19] bit_1.1-14 git2r_0.23.0
[21] labeling_0.3 diptest_0.75-7
[23] scales_1.0.0 QTLRel_1.0
[25] DEoptimR_1.0-8 mvtnorm_1.0-5
[27] robustbase_0.92-7 stringr_1.3.1
[29] digest_0.6.18 rmarkdown_1.11
[31] pkgconfig_2.0.1 htmltools_0.3.6
[33] highr_0.6 htmlwidgets_1.3
[35] rlang_0.4.0 RSQLite_2.1.1
[37] jsonlite_1.6 hwriter_1.3.2
[39] gtools_3.5.0 BiocParallel_1.8.2
[41] dplyr_0.8.3 RCurl_1.95-4.12
[43] magrittr_1.5 modeltools_0.2-21
[45] qtl_1.41-6 Matrix_1.2-14
[47] Rcpp_1.0.2 munsell_0.5.0
[49] stringi_1.2.4 whisker_0.3-2
[51] yaml_2.2.0 MASS_7.3-50
[53] zlibbioc_1.20.0 rhdf5_2.18.0
[55] flexmix_2.3-13 plyr_1.8.4
[57] grid_3.3.2 blob_1.1.1
[59] gdata_2.18.0 crayon_1.3.4
[61] lattice_0.20-35 GenomicFeatures_1.26.2
[63] annotate_1.52.1 knitr_1.20
[65] pillar_1.3.1 RUnit_0.4.31
[67] fpc_2.1-10 corpcor_1.6.9
[69] codetools_0.2-15 biomaRt_2.30.0
[71] XML_3.98-1.16 glue_1.3.1
[73] evaluate_0.10 data.table_1.11.4
[75] foreach_1.4.4 gtable_0.2.0
[77] purrr_0.3.2 kernlab_0.9-25
[79] assertthat_0.2.1 xtable_1.8-2
[81] class_7.3-14 tibble_2.1.3
[83] iterators_1.0.10 GenomicAlignments_1.10.1
[85] AnnotationDbi_1.36.2 memoise_1.1.0
[87] workflowr_1.4.0 cluster_2.0.7-1