QTL Analysis - binary [ICI vs PBS]
Belinda Cornes
2022-02-11
Last updated: 2022-02-11
Checks: 6 1
Knit directory: Serreze-T1D_Workflow/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20220210) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /Users/corneb/Documents/MyJax/CS/Projects/Serreze/qc/workflowr/Serreze-T1D_Workflow | . |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 8427b21. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Untracked files:
Untracked: analysis/4.1.2_qtl.analysis_cont_rzage_ici.vs.eoi_snpsqc.Rmd
Untracked: analysis/4.1.2_qtl.analysis_cont_rzage_ici.vs.pbs_snpsqc.Rmd
Untracked: data/GM_covar.csv
Untracked: data/bad_markers_all_4.batches.RData
Untracked: data/covar_cleaned_ici.vs.eoi.csv
Untracked: data/covar_cleaned_ici.vs.pbs.csv
Untracked: data/e.RData
Untracked: data/e_snpg_samqc_4.batches.RData
Untracked: data/e_snpg_samqc_4.batches_bc.RData
Untracked: data/errors_ind_4.batches.RData
Untracked: data/errors_ind_4.batches_bc.RData
Untracked: data/genetic_map.csv
Untracked: data/genotype_errors_marker_4.batches.RData
Untracked: data/genotype_freq_marker_4.batches.RData
Untracked: data/gm_allqc_4.batches.RData
Untracked: data/gm_samqc_3.batches.RData
Untracked: data/gm_samqc_4.batches.RData
Untracked: data/gm_samqc_4.batches_bc.RData
Untracked: data/gm_serreze.192.RData
Untracked: data/percent_missing_id_3.batches.RData
Untracked: data/percent_missing_id_4.batches.RData
Untracked: data/percent_missing_id_4.batches_bc.RData
Untracked: data/percent_missing_marker_4.batches.RData
Untracked: data/pheno.csv
Untracked: data/physical_map.csv
Untracked: data/qc_info_bad_sample_3.batches.RData
Untracked: data/qc_info_bad_sample_4.batches.RData
Untracked: data/qc_info_bad_sample_4.batches_bc.RData
Untracked: data/sample_geno.csv
Untracked: data/sample_geno_bc.csv
Untracked: data/serreze_probs.rds
Untracked: data/serreze_probs_allqc.rds
Untracked: data/summary.cg_3.batches.RData
Untracked: data/summary.cg_4.batches.RData
Untracked: data/summary.cg_4.batches_bc.RData
Untracked: output/Percent_missing_genotype_data_4.batches.pdf
Untracked: output/Percent_missing_genotype_data_per_marker.pdf
Untracked: output/Proportion_matching_genotypes_before_removal_of_bad_samples_4.batches.pdf
Untracked: output/genotype_error_marker.pdf
Untracked: output/genotype_frequency_marker.pdf
Unstaged changes:
Modified: analysis/2.2.1_snp_qc_4.batches.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc.Rmd) and HTML (docs/4.1.1_qtl.analysis_binary_ici.vs.pbs_snpsqc.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 8427b21 | Belinda Cornes | 2022-02-11 | QTL Mapping after snp qc |
Loading Data
We will load the data and subset indivials out that are in the groups of interest. We will create a binary phenotype from this (PBS ==0, ICI == 1).
load("data/gm_allqc_4.batches.RData")
#gm_allqc
gm=gm_allqc
gmObject of class cross2 (crosstype "bc")
Total individuals 188
No. genotyped individuals 188
No. phenotyped individuals 188
No. with both geno & pheno 188
No. phenotypes 1
No. covariates 6
No. phenotype covariates 0
No. chromosomes 20
Total markers 131578
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13
9977 10005 7858 7589 7621 7758 7413 6472 6725 6396 7154 6137 6085
14 15 16 17 18 19 X
5981 5346 5019 5093 4607 3564 4778 #pr <- readRDS("data/serreze_probs_allqc.rds")
#pr <- readRDS("data/serreze_probs.rds")
##extracting animals with ici and eoi group status
miceinfo <- gm$covar[gm$covar$group == "PBS" | gm$covar$group == "ICI",]
table(miceinfo$group)
ICI PBS
92 21 mice.ids <- rownames(miceinfo)
gm <- gm[mice.ids]
gmObject of class cross2 (crosstype "bc")
Total individuals 113
No. genotyped individuals 113
No. phenotyped individuals 113
No. with both geno & pheno 113
No. phenotypes 1
No. covariates 6
No. phenotype covariates 0
No. chromosomes 20
Total markers 131578
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13
9977 10005 7858 7589 7621 7758 7413 6472 6725 6396 7154 6137 6085
14 15 16 17 18 19 X
5981 5346 5019 5093 4607 3564 4778 #pr.qc <- pr
#for (i in 1:20){pr.qc[[i]] = pr.qc[[i]][mice.ids,,]}
#bin_pheno <- NULL
#bin_pheno$PBS <- ifelse(gm$covar$group == "PBS", 1, 0)
#bin_pheno$ICI <- ifelse(gm$covar$group == "ICI", 1, 0)
#bin_pheno <- as.data.frame(bin_pheno)
#rownames(bin_pheno) <- rownames(gm$covar)
gm$covar$ICI.vs.PBS <- ifelse(gm$covar$group == "PBS", 0, 1)
##dropping monomorphic markers within the dataset
g <- do.call("cbind", gm$geno)
gf_mar <- t(apply(g, 2, function(a) table(factor(a, 1:2))/sum(a != 0)))
#gn_mar <- t(apply(g, 2, function(a) table(factor(a, 1:2))))
gf_mar <- gf_mar[gf_mar[,2] != "NaN",]
count <- rowSums(gf_mar ==0)
low_freq_df <- merge(as.data.frame(gf_mar),as.data.frame(count), by="row.names",all=T)
low_freq_df[is.na(low_freq_df)] <- ''
low_freq_df <- low_freq_df[low_freq_df$count == 1,]
rownames(low_freq_df) <- low_freq_df$Row.names
low_freq <- find_markerpos(gm, rownames(low_freq_df))
low_freq$id <- rownames(low_freq)
nrow(low_freq)[1] 89064low_freq_bad <- merge(low_freq,low_freq_df, by="row.names",all=T)
names(low_freq_bad)[1] <- c("marker")
gf_mar <- gf_mar[gf_mar[,2] != "NaN",]
MAF <- apply(gf_mar, 1, function(x) min(x))
MAF <- as.data.frame(MAF)
MAF$index <- 1:nrow(gf_mar)
gf_mar_maf <- merge(gf_mar,as.data.frame(MAF), by="row.names")
gf_mar_maf <- gf_mar_maf[order(gf_mar_maf$index),]
gfmar <- NULL
gfmar$gfmar_mar_0 <- sum(gf_mar_maf$MAF==0)
gfmar$gfmar_mar_1 <- sum(gf_mar_maf$MAF< 0.01)
gfmar$gfmar_mar_5 <- sum(gf_mar_maf$MAF< 0.05)
gfmar$gfmar_mar_10 <- sum(gf_mar_maf$MAF< 0.10)
gfmar$gfmar_mar_15 <- sum(gf_mar_maf$MAF< 0.15)
gfmar$gfmar_mar_25 <- sum(gf_mar_maf$MAF< 0.25)
gfmar$gfmar_mar_50 <- sum(gf_mar_maf$MAF< 0.50)
gfmar$total_snps <- nrow(as.data.frame(gf_mar_maf))
gfmar <- t(as.data.frame(gfmar))
gfmar <- as.data.frame(gfmar)
gfmar$count <- gfmar$V1
gfmar[c(2)] %>%
kable(escape = F,align = c("ccccccccc"),linesep ="\\hline") %>%
kable_styling(full_width = F) %>%
kable_styling("striped", full_width = F) %>%
row_spec(8 ,bold=T,color= "white",background = "black")| count | |
|---|---|
| gfmar_mar_0 | 89064 |
| gfmar_mar_1 | 92616 |
| gfmar_mar_5 | 98209 |
| gfmar_mar_10 | 99489 |
| gfmar_mar_15 | 99575 |
| gfmar_mar_25 | 100325 |
| gfmar_mar_50 | 131283 |
| total_snps | 131578 |
gm_qc <- drop_markers(gm, low_freq_bad$marker)
gm_qc <- drop_nullmarkers(gm_qc)
gm = gm_qc
gmObject of class cross2 (crosstype "bc")
Total individuals 113
No. genotyped individuals 113
No. phenotyped individuals 113
No. with both geno & pheno 113
No. phenotypes 1
No. covariates 7
No. phenotype covariates 0
No. chromosomes 20
Total markers 42514
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
3669 3548 2703 2593 2484 2667 2346 2134 2498 1749 2527 1864 2047 2153 1679 1349
17 18 19 X
841 1445 1288 930 markers <- marker_names(gm)
gmapdf <- read.csv("/Users/corneb/Documents/MyJax/CS/Projects/Serreze/haplotype.reconstruction/output_hh/genetic_map.csv")
pmapdf <- read.csv("/Users/corneb/Documents/MyJax/CS/Projects/Serreze/haplotype.reconstruction/output_hh/physical_map.csv")
mapdf <- merge(gmapdf,pmapdf, by=c("marker","chr"), all=T)
rownames(mapdf) <- mapdf$marker
mapdf <- mapdf[markers,]
names(mapdf) <- c('marker','chr','gmapdf','pmapdf')
mapdf <- mapdf[!duplicated(mapdf[c(2:3)]),]
pr.qc <- calc_genoprob(gm)Genome-wide scan
For each of the phenotype analyzed, permutations were used for each model to obtain genome-wide LOD significance threshold for p < 0.01, p < 0.05, p < 0.10, respectively, separately for X and automsomes (A).
The table shows the estimated significance thresholds from permutation test.
We also looked at the kinship to see how correlated each sample is. Kinship values between pairs of samples range between 0 (no relationship) and 1.0 (completely identical). The darker the colour the more indentical the pairs are.
Xcovar <- get_x_covar(gm)
#addcovar = model.matrix(~Sex, data = covars)[,-1]
#K <- calc_kinship(pr.qc, type = "loco")
#heatmap(K[[1]])
#K.overall <- calc_kinship(pr.qc, type = "overall")
#heatmap(K.overall)
kinship <- calc_kinship(pr.qc)
heatmap(kinship)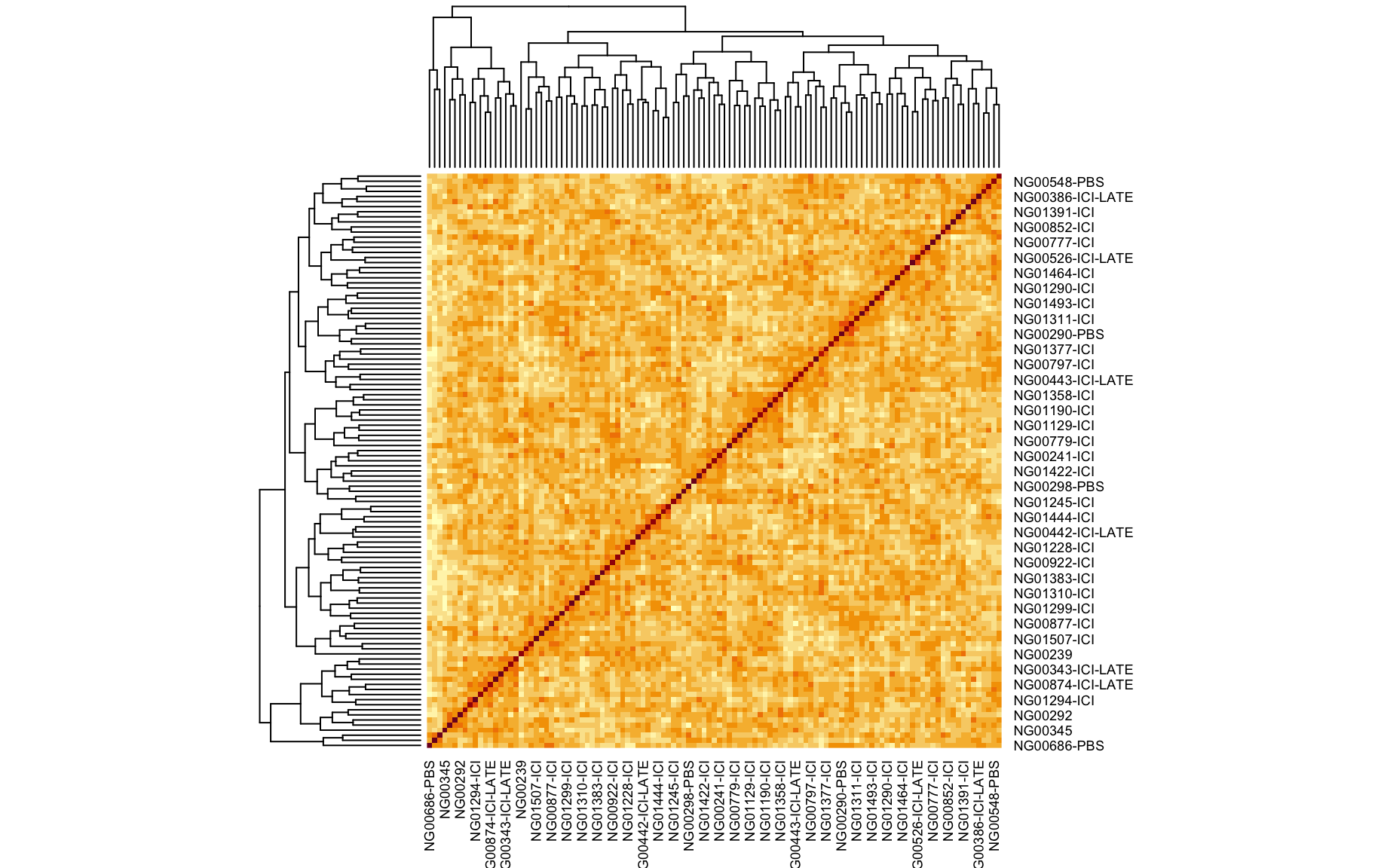
#operm <- scan1perm(pr.qc, gm$covar$phenos, Xcovar=Xcovar, n_perm=2000)
#operm <- scan1perm(pr.qc, gm$covar$phenos, addcovar = addcovar, n_perm=2000)
#operm <- scan1perm(pr.qc, gm$covar$phenos, n_perm=2000)
operm <- scan1perm(pr.qc, gm$covar["ICI.vs.PBS"], model="binary", n_perm=10, perm_Xsp=TRUE, chr_lengths=chr_lengths(gm$gmap))
summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
names(summary_table) <- c("autosomes","X")
summary_table$significance.level <- rownames(summary_table)
rownames(summary_table) <- NULL
summary_table[c(3,1:2)] %>%
kable(escape = F,align = c("ccc")) %>%
kable_styling("striped", full_width = T) %>%
column_spec(1, bold=TRUE)| significance.level | autosomes | X |
|---|---|---|
| 0.01 | 4.501880 | 4.483706 |
| 0.05 | 4.202418 | 4.232069 |
| 0.1 | 3.827168 | 3.902999 |
The figures below show QTL maps for each phenotype
out <- scan1(pr.qc, gm$covar["ICI.vs.PBS"], Xcovar=Xcovar, model="binary")
#summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
plot_lod<-function(out,map){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, gm$gmap, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = colnames(out)[i])
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
}
}
plot_lod(out,gm$gmap)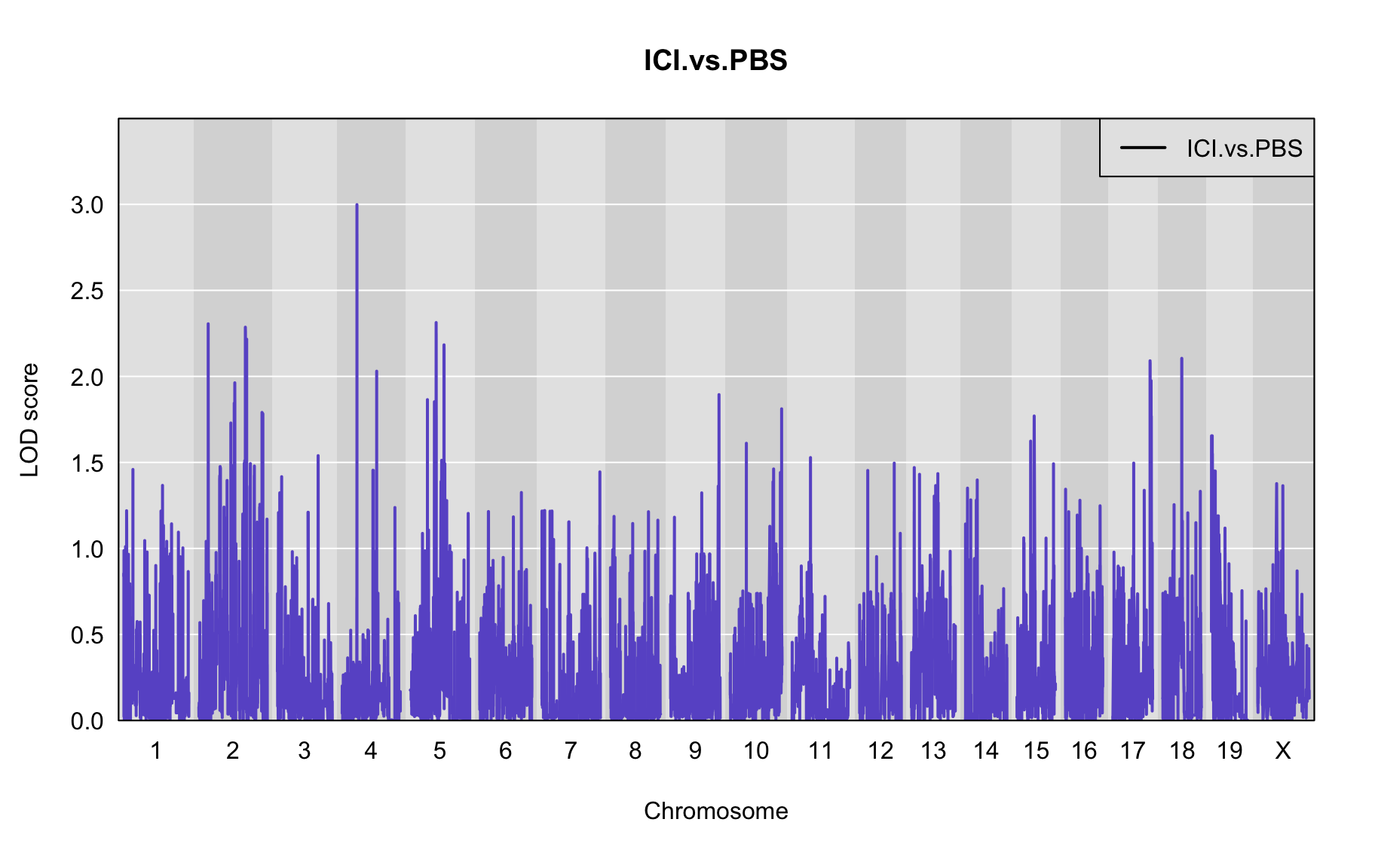
LOD peaks
The table below shows QTL peaks associated with the phenotype. We use the 95% threshold from the permutations to find peaks.
peaks<-find_peaks(out, gm$gmap, threshold=2.5, drop=1.5)
rownames(peaks) <- NULL
peaks[] %>%
kable(escape = F,align = c("ccccccc")) %>%
kable_styling("striped", full_width = T) %>%
column_spec(1, bold=TRUE)| lodindex | lodcolumn | chr | pos | lod | ci_lo | ci_hi |
|---|---|---|---|---|---|---|
| 1 | ICI.vs.PBS | 4 | 24.124 | 2.998475 | 24.106 | 53.597 |
peaks_mbl <- list()
#corresponding info in Mb
for(i in 1:nrow(peaks)){
lodindex <- peaks$lodindex[i]
lodcolumn <- peaks$lodcolumn[i]
chr <- peaks$chr[i]
lod <- peaks$lod[i]
pos <- mapdf$pmapdf[which(mapdf$gmapdf == peaks$pos[i] & mapdf$chr == peaks$chr[i])]
ci_lo <- mapdf$pmapdf[which(mapdf$gmapdf == peaks$ci_lo[i] & mapdf$chr == peaks$chr[i])]
ci_hi <- mapdf$pmapdf[which(mapdf$gmapdf == peaks$ci_hi[i] & mapdf$chr == peaks$chr[i])]
peaks_mb=cbind(lodindex, lodcolumn, chr, pos, lod, ci_lo, ci_hi)
peaks_mbl[[i]] <- peaks_mb
}
peaks_mba <- do.call(rbind, peaks_mbl)
peaks_mba <- as.data.frame(peaks_mba)
#peaks_mba[,c("chr", "pos", "lod", "ci_lo", "ci_hi")] <- sapply(peaks_mba[,c("chr", "pos", "lod", "ci_lo", "ci_hi")], as.numeric)
rownames(peaks_mba) <- NULL
peaks_mba[] %>%
kable(escape = F,align = c("ccccccc")) %>%
kable_styling("striped", full_width = T) %>%
column_spec(1, bold=TRUE)| lodindex | lodcolumn | chr | pos | lod | ci_lo | ci_hi |
|---|---|---|---|---|---|---|
| 1 | ICI.vs.PBS | 4 | 45.577851 | 2.99847470094299 | 45.561949 | 117.678975 |
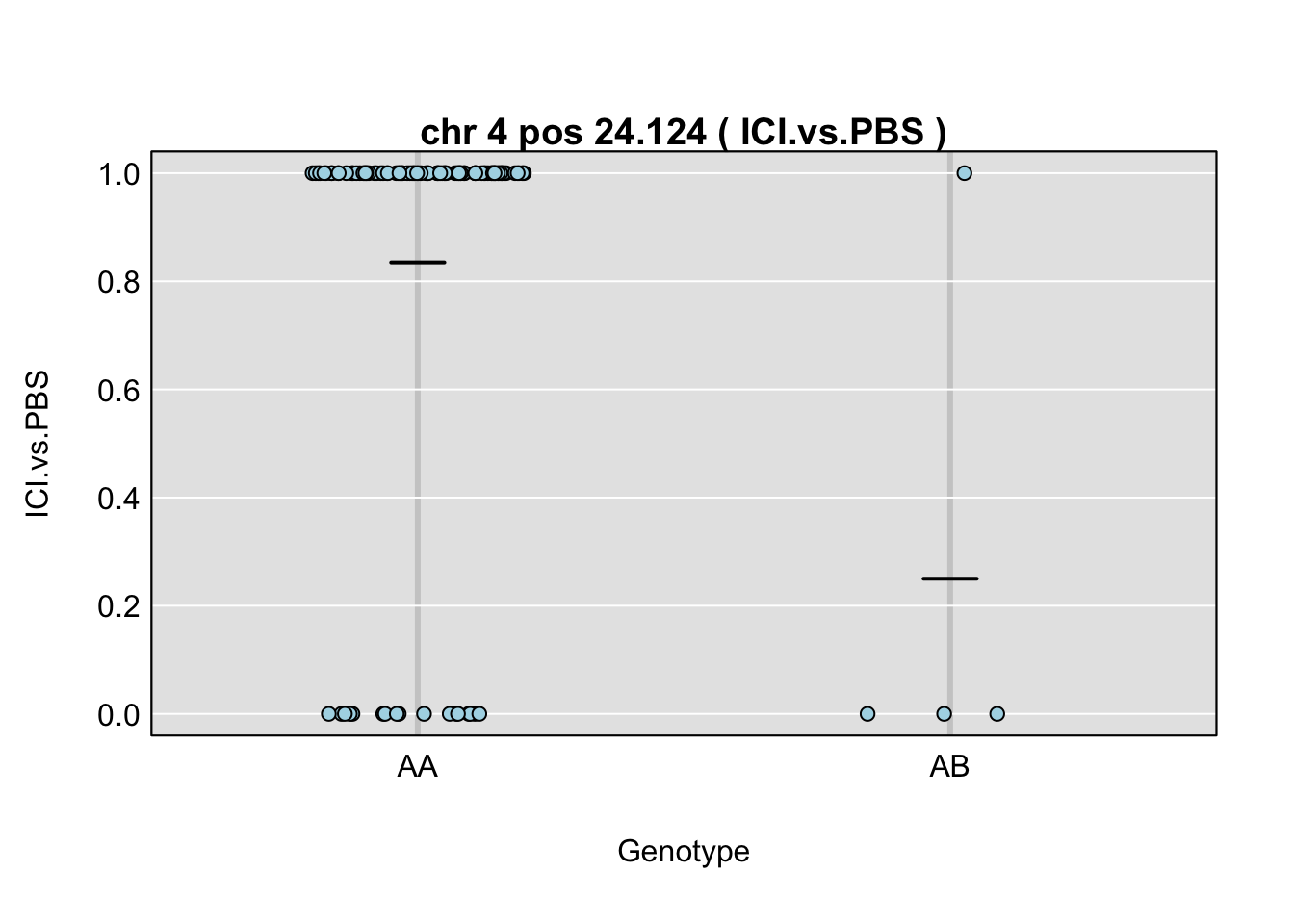
QTL effects
For each peak LOD location we give a list of gene
query_variants <- create_variant_query_func("/Users/corneb/Documents/MyJax/CS/Projects/support.files/qtl2/cc_variants.sqlite")
query_genes <- create_gene_query_func("/Users/corneb/Documents/MyJax/CS/Projects/support.files/qtl2/mouse_genes_mgi.sqlite")
for (i in 1:nrow(peaks)){
#for (i in 1:1){
#Plot 1
marker = find_marker(gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i])
#g <- maxmarg(pr.qc, gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i], return_char=TRUE, minprob = 0.5)
gp <- g[,marker]
gp[gp==1] <- "AA"
gp[gp==2] <- "AB"
gp[gp==0] <- NA
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/","qtl_effect_", i, ".png"))
#par(mar=c(4.1, 4.1, 1.5, 0.6))
plot_pxg(gp, gm$covar[,peaks$lodcolumn[i]], ylab=peaks$lodcolumn[i], sort=FALSE)
title(main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")"), line=0.2)
##dev.off()
chr = peaks$chr[i]
# Plot 2
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar)
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], Xcovar=Xcovar)
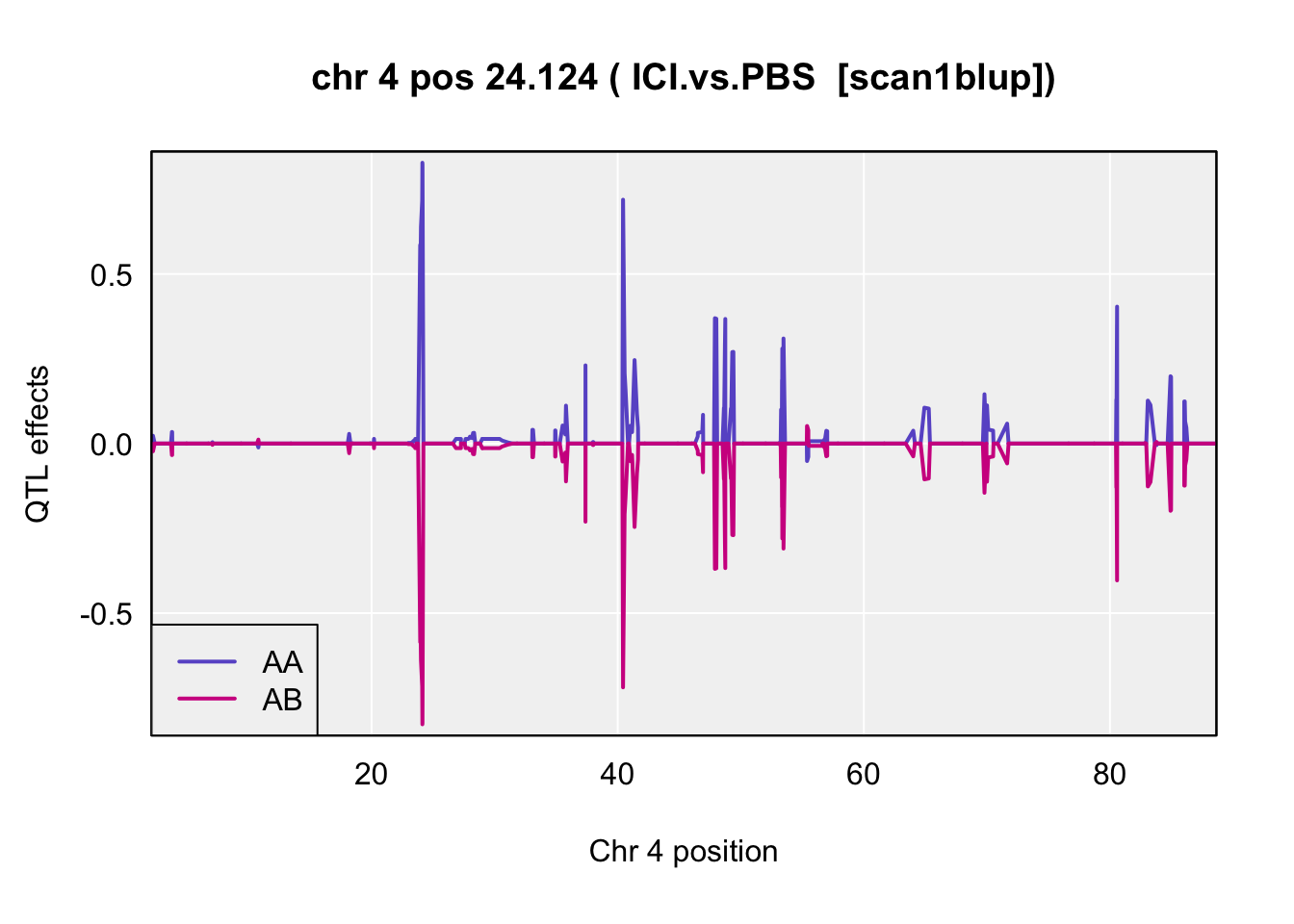
coeff <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
blup <- scan1blup(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]])
#plot_coef(coeff,
# gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i]," [scan1coeff])")
# )
plot_coef(blup,
gm$gmap[chr], columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i]," [scan1blup])")
)
#last_coef <- unclass(coeff)[nrow(coeff),1:3]
#for(t in seq(along=last_coef))
#axis(side=4, at=last_coef[t], names(last_coef)[t], tick=FALSE)
# Plot 3
#c2effB <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary", contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
#c2effBb <- scan1blup(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]],Xcovar=Xcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
#plot(c2effB, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
#plot(c2effBb, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
##last_coef <- unclass(c2effB)[nrow(c2effB),2:3] # last two coefficients
##for(t in seq(along=last_coef))
## axis(side=4, at=last_coef[t], names(last_coef)[t], tick=FALSE)
#Table 1
chr = peaks_mba$chr[i]
start=as.numeric(peaks_mba$ci_lo[i])
end=as.numeric(peaks_mba$ci_hi[i])
genesgss = query_genes(chr, start, end)
rownames(genesgss) <- NULL
genesgss$strand_old = genesgss$strand
genesgss$strand[genesgss$strand=="+"] <- "positive"
genesgss$strand[genesgss$strand=="-"] <- "negative"
#genesgss <-
#table <-
#genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")] %>%
#kable(escape = F,align = c("ccccccccccc")) %>%
#kable_styling("striped", full_width = T) #%>%
#cat #%>%
#column_spec(1, bold=TRUE)
#
#print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], escape = F,align = c("ccccccccccc")))
print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], "html") %>% kable_styling("striped", full_width = T))
#table
}
| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 4 | gene | 45.63937 | 45.64611 | positive | MGI_C57BL6J_3649921 | Gm12408 | NCBI_Gene:100040557,ENSEMBL:ENSMUSG00000085980 | MGI:3649921 | lncRNA gene | predicted gene 12408 |
| 4 | gene | 45.68359 | 45.72371 | positive | MGI_C57BL6J_2685675 | Gm829 | NCBI_Gene:329839,ENSEMBL:ENSMUSG00000073860 | MGI:2685675 | lncRNA gene | predicted gene 829 |
| 4 | gene | 45.71517 | 45.71642 | positive | MGI_C57BL6J_3650364 | Gm12410 | ENSEMBL:ENSMUSG00000085305 | MGI:3650364 | lncRNA gene | predicted gene 12410 |
| 4 | gene | 45.72377 | 45.74030 | negative | MGI_C57BL6J_5590722 | Gm31563 | NCBI_Gene:102633832 | MGI:5590722 | lncRNA gene | predicted gene, 31563 |
| 4 | gene | 45.76238 | 45.76422 | negative | MGI_C57BL6J_3649923 | Gm12409 | NCBI_Gene:100040567,ENSEMBL:ENSMUSG00000085876 | MGI:3649923 | lncRNA gene | predicted gene 12409 |
| 4 | pseudogene | 45.77688 | 45.77721 | negative | MGI_C57BL6J_3649919 | Gm12412 | ENSEMBL:ENSMUSG00000083685 | MGI:3649919 | pseudogene | predicted gene 12412 |
| 4 | gene | 45.79902 | 45.80461 | positive | MGI_C57BL6J_1919785 | Aldh1b1 | NCBI_Gene:72535,ENSEMBL:ENSMUSG00000035561 | MGI:1919785 | protein coding gene | aldehyde dehydrogenase 1 family, member B1 |
| 4 | gene | 45.80947 | 45.82692 | negative | MGI_C57BL6J_1933198 | Igfbpl1 | NCBI_Gene:75426,ENSEMBL:ENSMUSG00000035551 | MGI:1933198 | protein coding gene | insulin-like growth factor binding protein-like 1 |
| 4 | gene | 45.82256 | 45.88701 | positive | MGI_C57BL6J_1921402 | Stra6l | NCBI_Gene:74152,ENSEMBL:ENSMUSG00000028327 | MGI:1921402 | protein coding gene | STRA6-like |
| 4 | gene | 45.89030 | 45.95077 | positive | MGI_C57BL6J_2685871 | Ccdc180 | NCBI_Gene:381522,ENSEMBL:ENSMUSG00000035539 | MGI:2685871 | protein coding gene | coiled-coil domain containing 180 |
| 4 | gene | 45.96503 | 46.03477 | positive | MGI_C57BL6J_2140279 | Tdrd7 | NCBI_Gene:100121,ENSEMBL:ENSMUSG00000035517 | MGI:2140279 | protein coding gene | tudor domain containing 7 |
| 4 | gene | 46.03894 | 46.11603 | positive | MGI_C57BL6J_98775 | Tmod1 | NCBI_Gene:21916,ENSEMBL:ENSMUSG00000028328 | MGI:98775 | protein coding gene | tropomodulin 1 |
| 4 | gene | 46.11475 | 46.13869 | negative | MGI_C57BL6J_3039624 | Tstd2 | NCBI_Gene:272027,ENSEMBL:ENSMUSG00000035495 | MGI:3039624 | protein coding gene | thiosulfate sulfurtransferase (rhodanese)-like domain containing 2 |
| 4 | gene | 46.11818 | 46.11845 | positive | MGI_C57BL6J_5453027 | Gm23250 | ENSEMBL:ENSMUSG00000088069 | MGI:5453027 | unclassified non-coding RNA gene | predicted gene, 23250 |
| 4 | gene | 46.13851 | 46.17240 | positive | MGI_C57BL6J_1891840 | Ncbp1 | NCBI_Gene:433702,ENSEMBL:ENSMUSG00000028330 | MGI:1891840 | protein coding gene | nuclear cap binding protein subunit 1 |
| 4 | gene | 46.15535 | 46.19635 | negative | MGI_C57BL6J_99135 | Xpa | NCBI_Gene:22590,ENSEMBL:ENSMUSG00000028329 | MGI:99135 | protein coding gene | xeroderma pigmentosum, complementation group A |
| 4 | gene | 46.20244 | 46.20725 | positive | MGI_C57BL6J_5590776 | Gm31617 | NCBI_Gene:102633902 | MGI:5590776 | lncRNA gene | predicted gene, 31617 |
| 4 | pseudogene | 46.24299 | 46.24393 | negative | MGI_C57BL6J_3650375 | Gm12444 | NCBI_Gene:624430,ENSEMBL:ENSMUSG00000082194 | MGI:3650375 | pseudogene | predicted gene 12444 |
| 4 | pseudogene | 46.27382 | 46.27494 | negative | MGI_C57BL6J_3650256 | Gm12445 | NCBI_Gene:666379,ENSEMBL:ENSMUSG00000081483 | MGI:3650256 | pseudogene | predicted gene 12445 |
| 4 | gene | 46.30001 | 46.34551 | negative | MGI_C57BL6J_3651433 | Gm12446 | ENSEMBL:ENSMUSG00000087383 | MGI:3651433 | lncRNA gene | predicted gene 12446 |
| 4 | pseudogene | 46.32289 | 46.32777 | positive | MGI_C57BL6J_3650247 | Gm12447 | NCBI_Gene:666389,ENSEMBL:ENSMUSG00000080816 | MGI:3650247 | pseudogene | predicted gene 12447 |
| 4 | gene | 46.32790 | 46.33066 | positive | MGI_C57BL6J_5625165 | Gm42280 | NCBI_Gene:105247123 | MGI:5625165 | lncRNA gene | predicted gene, 42280 |
| 4 | gene | 46.34361 | 46.34641 | positive | MGI_C57BL6J_1353500 | Foxe1 | NCBI_Gene:110805,ENSEMBL:ENSMUSG00000070990 | MGI:1353500 | protein coding gene | forkhead box E1 |
| 4 | gene | 46.37650 | 46.39356 | negative | MGI_C57BL6J_1922003 | Trmo | NCBI_Gene:74753,ENSEMBL:ENSMUSG00000028331 | MGI:1922003 | protein coding gene | tRNA methyltransferase O |
| 4 | gene | 46.39399 | 46.41351 | negative | MGI_C57BL6J_2136910 | Hemgn | NCBI_Gene:93966,ENSEMBL:ENSMUSG00000028332 | MGI:2136910 | protein coding gene | hemogen |
| 4 | pseudogene | 46.41998 | 46.42030 | negative | MGI_C57BL6J_3651437 | Gm12443 | ENSEMBL:ENSMUSG00000082242 | MGI:3651437 | pseudogene | predicted gene 12443 |
| 4 | gene | 46.45090 | 46.47266 | positive | MGI_C57BL6J_1914878 | Anp32b | NCBI_Gene:67628,ENSEMBL:ENSMUSG00000028333 | MGI:1914878 | protein coding gene | acidic (leucine-rich) nuclear phosphoprotein 32 family, member B |
| 4 | gene | 46.48925 | 46.50363 | positive | MGI_C57BL6J_2149820 | Nans | NCBI_Gene:94181,ENSEMBL:ENSMUSG00000028334 | MGI:2149820 | protein coding gene | N-acetylneuraminic acid synthase (sialic acid synthase) |
| 4 | gene | 46.49378 | 46.53615 | negative | MGI_C57BL6J_1921985 | Trim14 | NCBI_Gene:74735,ENSEMBL:ENSMUSG00000039853 | MGI:1921985 | protein coding gene | tripartite motif-containing 14 |
| 4 | gene | 46.49515 | 46.49529 | positive | MGI_C57BL6J_5452024 | Gm22247 | ENSEMBL:ENSMUSG00000064410 | MGI:5452024 | snoRNA gene | predicted gene, 22247 |
| 4 | gene | 46.53590 | 46.53988 | positive | MGI_C57BL6J_4439655 | Gm16731 | NCBI_Gene:105247126,ENSEMBL:ENSMUSG00000085990 | MGI:4439655 | lncRNA gene | predicted gene, 16731 |
| 4 | gene | 46.53694 | 46.60220 | negative | MGI_C57BL6J_1345966 | Coro2a | NCBI_Gene:107684,ENSEMBL:ENSMUSG00000028337 | MGI:1345966 | protein coding gene | coronin, actin binding protein 2A |
| 4 | gene | 46.56714 | 46.56791 | positive | MGI_C57BL6J_1923098 | 4930566K23Rik | NA | NA | unclassified gene | RIKEN cDNA 4930566K23 gene |
| 4 | gene | 46.57721 | 46.57879 | positive | MGI_C57BL6J_2655157 | BC005685 | NA | NA | protein coding gene | cDNA sequence BC005685 |
| 4 | gene | 46.60439 | 46.65088 | negative | MGI_C57BL6J_2652885 | Tbc1d2 | NCBI_Gene:381605,ENSEMBL:ENSMUSG00000039813 | MGI:2652885 | protein coding gene | TBC1 domain family, member 2 |
| 4 | gene | 46.60715 | 46.61013 | positive | MGI_C57BL6J_5591009 | Gm31850 | NCBI_Gene:102634210 | MGI:5591009 | lncRNA gene | predicted gene, 31850 |
| 4 | gene | 46.62160 | 46.62833 | positive | MGI_C57BL6J_5625166 | Gm42281 | NCBI_Gene:105247124 | MGI:5625166 | lncRNA gene | predicted gene, 42281 |
| 4 | gene | 46.66231 | 46.99187 | negative | MGI_C57BL6J_2386030 | Gabbr2 | NCBI_Gene:242425,ENSEMBL:ENSMUSG00000039809 | MGI:2386030 | protein coding gene | gamma-aminobutyric acid (GABA) B receptor, 2 |
| 4 | gene | 46.86017 | 46.87770 | positive | MGI_C57BL6J_5591367 | Gm32208 | NCBI_Gene:102634676 | MGI:5591367 | lncRNA gene | predicted gene, 32208 |
| 4 | gene | 46.88407 | 46.89762 | positive | MGI_C57BL6J_5591294 | Gm32135 | NCBI_Gene:102634594 | MGI:5591294 | lncRNA gene | predicted gene, 32135 |
| 4 | gene | 47.00832 | 47.01078 | negative | MGI_C57BL6J_2685414 | Gm568 | ENSEMBL:ENSMUSG00000048824 | MGI:2685414 | lncRNA gene | predicted gene 568 |
| 4 | gene | 47.01537 | 47.05743 | negative | MGI_C57BL6J_1922941 | Anks6 | NCBI_Gene:75691,ENSEMBL:ENSMUSG00000066191 | MGI:1922941 | protein coding gene | ankyrin repeat and sterile alpha motif domain containing 6 |
| 4 | pseudogene | 47.04160 | 47.04196 | positive | MGI_C57BL6J_3650990 | Gm12425 | ENSEMBL:ENSMUSG00000081561 | MGI:3650990 | pseudogene | predicted gene 12425 |
| 4 | pseudogene | 47.07165 | 47.07195 | negative | MGI_C57BL6J_3651148 | Gm12422 | ENSEMBL:ENSMUSG00000084199 | MGI:3651148 | pseudogene | predicted gene 12422 |
| 4 | gene | 47.09191 | 47.12307 | positive | MGI_C57BL6J_2444664 | Galnt12 | NCBI_Gene:230145,ENSEMBL:ENSMUSG00000039774 | MGI:2444664 | protein coding gene | polypeptide N-acetylgalactosaminyltransferase 12 |
| 4 | pseudogene | 47.10103 | 47.10148 | positive | MGI_C57BL6J_3650991 | Gm12424 | ENSEMBL:ENSMUSG00000094286 | MGI:3650991 | pseudogene | predicted gene 12424 |
| 4 | pseudogene | 47.17167 | 47.17180 | positive | MGI_C57BL6J_3650594 | Gm12423 | ENSEMBL:ENSMUSG00000083491 | MGI:3650594 | pseudogene | predicted gene 12423 |
| 4 | gene | 47.19749 | 47.20931 | negative | MGI_C57BL6J_3650989 | Gm12426 | NCBI_Gene:102634926,ENSEMBL:ENSMUSG00000087500 | MGI:3650989 | lncRNA gene | predicted gene 12426 |
| 4 | gene | 47.20763 | 47.31317 | positive | MGI_C57BL6J_88449 | Col15a1 | NCBI_Gene:12819,ENSEMBL:ENSMUSG00000028339 | MGI:88449 | protein coding gene | collagen, type XV, alpha 1 |
| 4 | gene | 47.22516 | 47.24534 | negative | MGI_C57BL6J_5591476 | Gm32317 | NCBI_Gene:102634823 | MGI:5591476 | lncRNA gene | predicted gene, 32317 |
| 4 | gene | 47.33402 | 47.34660 | negative | MGI_C57BL6J_5625167 | Gm42282 | NCBI_Gene:105247127 | MGI:5625167 | lncRNA gene | predicted gene, 42282 |
| 4 | gene | 47.35322 | 47.41493 | positive | MGI_C57BL6J_98728 | Tgfbr1 | NCBI_Gene:21812,ENSEMBL:ENSMUSG00000007613 | MGI:98728 | protein coding gene | transforming growth factor, beta receptor I |
| 4 | gene | 47.35386 | 47.38614 | negative | MGI_C57BL6J_5591594 | Gm32435 | NCBI_Gene:102634986 | MGI:5591594 | lncRNA gene | predicted gene, 32435 |
| 4 | pseudogene | 47.45356 | 47.45540 | positive | MGI_C57BL6J_3651061 | Gm12430 | NCBI_Gene:666473,ENSEMBL:ENSMUSG00000081152 | MGI:3651061 | pseudogene | predicted gene 12430 |
| 4 | gene | 47.46507 | 47.47437 | negative | MGI_C57BL6J_1914731 | Alg2 | NCBI_Gene:56737,ENSEMBL:ENSMUSG00000039740 | MGI:1914731 | protein coding gene | asparagine-linked glycosylation 2 (alpha-1,3-mannosyltransferase) |
| 4 | gene | 47.47466 | 47.48324 | positive | MGI_C57BL6J_1913462 | Sec61b | NCBI_Gene:66212,ENSEMBL:ENSMUSG00000053317 | MGI:1913462 | protein coding gene | Sec61 beta subunit |
| 4 | gene | 47.48924 | 47.49486 | negative | MGI_C57BL6J_5625168 | Gm42283 | NCBI_Gene:105247128 | MGI:5625168 | lncRNA gene | predicted gene, 42283 |
| 4 | gene | 47.59334 | 47.59346 | positive | MGI_C57BL6J_5530983 | Gm27601 | ENSEMBL:ENSMUSG00000098999 | MGI:5530983 | unclassified non-coding RNA gene | predicted gene, 27601 |
| 4 | gene | 47.59526 | 47.60605 | negative | MGI_C57BL6J_5625169 | Gm42284 | NCBI_Gene:105247129 | MGI:5625169 | lncRNA gene | predicted gene, 42284 |
| 4 | gene | 47.60342 | 47.60829 | positive | MGI_C57BL6J_5591646 | Gm32487 | NCBI_Gene:102635054 | MGI:5591646 | lncRNA gene | predicted gene, 32487 |
| 4 | gene | 47.60389 | 47.60401 | positive | MGI_C57BL6J_5531050 | Gm27668 | ENSEMBL:ENSMUSG00000098518 | MGI:5531050 | unclassified non-coding RNA gene | predicted gene, 27668 |
| 4 | gene | 47.62602 | 47.63888 | negative | MGI_C57BL6J_5591703 | Gm32544 | NCBI_Gene:102635125 | MGI:5591703 | lncRNA gene | predicted gene, 32544 |
| 4 | gene | 47.82722 | 47.92737 | negative | MGI_C57BL6J_5611571 | Gm38343 | ENSEMBL:ENSMUSG00000102450 | MGI:5611571 | unclassified gene | predicted gene, 38343 |
| 4 | gene | 47.89977 | 47.91313 | positive | MGI_C57BL6J_5826520 | Gm46883 | NCBI_Gene:108168995 | MGI:5826520 | lncRNA gene | predicted gene, 46883 |
| 4 | gene | 47.92075 | 47.92090 | positive | MGI_C57BL6J_5452447 | Gm22670 | ENSEMBL:ENSMUSG00000089349 | MGI:5452447 | snoRNA gene | predicted gene, 22670 |
| 4 | gene | 47.94618 | 47.95158 | negative | MGI_C57BL6J_5826490 | Gm46853 | NCBI_Gene:108168941 | MGI:5826490 | lncRNA gene | predicted gene, 46853 |
| 4 | gene | 47.99646 | 47.99826 | positive | MGI_C57BL6J_3649453 | Gm12434 | ENSEMBL:ENSMUSG00000085660 | MGI:3649453 | lncRNA gene | predicted gene 12434 |
| 4 | gene | 48.04489 | 48.08645 | positive | MGI_C57BL6J_1352457 | Nr4a3 | NCBI_Gene:18124,ENSEMBL:ENSMUSG00000028341 | MGI:1352457 | protein coding gene | nuclear receptor subfamily 4, group A, member 3 |
| 4 | gene | 48.12492 | 48.18651 | positive | MGI_C57BL6J_1914977 | Stx17 | NCBI_Gene:67727,ENSEMBL:ENSMUSG00000061455 | MGI:1914977 | protein coding gene | syntaxin 17 |
| 4 | gene | 48.19332 | 48.27960 | negative | MGI_C57BL6J_1923549 | Erp44 | NCBI_Gene:76299,ENSEMBL:ENSMUSG00000028343 | MGI:1923549 | protein coding gene | endoplasmic reticulum protein 44 |
| 4 | gene | 48.21312 | 48.21321 | negative | MGI_C57BL6J_3837123 | Mir1958 | miRBase:MI0009955,NCBI_Gene:100316708,ENSEMBL:ENSMUSG00000087844 | MGI:3837123 | miRNA gene | microRNA 1958 |
| 4 | gene | 48.27976 | 48.43196 | positive | MGI_C57BL6J_1335082 | Invs | NCBI_Gene:16348,ENSEMBL:ENSMUSG00000028344 | MGI:1335082 | protein coding gene | inversin |
| 4 | pseudogene | 48.34421 | 48.34529 | negative | MGI_C57BL6J_3651950 | Gm12435 | NCBI_Gene:100417846,ENSEMBL:ENSMUSG00000081953 | MGI:3651950 | pseudogene | predicted gene 12435 |
| 4 | gene | 48.42773 | 48.42923 | negative | MGI_C57BL6J_3028047 | C030004N09Rik | NA | NA | unclassified gene | RIKEN cDNA C030004N09 gene |
| 4 | gene | 48.43086 | 48.47346 | negative | MGI_C57BL6J_1344413 | Tex10 | NCBI_Gene:269536,ENSEMBL:ENSMUSG00000028345 | MGI:1344413 | protein coding gene | testis expressed gene 10 |
| 4 | gene | 48.53983 | 48.56589 | positive | MGI_C57BL6J_1913915 | Msantd3 | NCBI_Gene:66665,ENSEMBL:ENSMUSG00000039693 | MGI:1913915 | protein coding gene | Myb/SANT-like DNA-binding domain containing 3 |
| 4 | gene | 48.58511 | 48.66313 | positive | MGI_C57BL6J_1926810 | Tmeff1 | NCBI_Gene:230157,ENSEMBL:ENSMUSG00000028347 | MGI:1926810 | protein coding gene | transmembrane protein with EGF-like and two follistatin-like domains 1 |
| 4 | gene | 48.66351 | 48.67350 | positive | MGI_C57BL6J_1915266 | Cavin4 | NCBI_Gene:68016,ENSEMBL:ENSMUSG00000028348 | MGI:1915266 | protein coding gene | caveolae associated 4 |
| 4 | pseudogene | 48.67516 | 48.67552 | positive | MGI_C57BL6J_3651708 | Gm12439 | ENSEMBL:ENSMUSG00000080850 | MGI:3651708 | pseudogene | predicted gene 12439 |
| 4 | pseudogene | 48.74551 | 48.74585 | negative | MGI_C57BL6J_3651707 | Gm12438 | ENSEMBL:ENSMUSG00000083091 | MGI:3651707 | pseudogene | predicted gene 12438 |
| 4 | gene | 48.76285 | 48.76296 | negative | MGI_C57BL6J_5454350 | Gm24573 | ENSEMBL:ENSMUSG00000093240 | MGI:5454350 | rRNA gene | predicted gene, 24573 |
| 4 | gene | 48.89650 | 48.89662 | positive | MGI_C57BL6J_5453523 | Gm23746 | ENSEMBL:ENSMUSG00000077417 | MGI:5453523 | snoRNA gene | predicted gene, 23746 |
| 4 | gene | 48.89859 | 48.96324 | positive | MGI_C57BL6J_3651705 | Gm12436 | NCBI_Gene:102635202,ENSEMBL:ENSMUSG00000084787 | MGI:3651705 | lncRNA gene | predicted gene 12436 |
| 4 | gene | 48.97070 | 48.97216 | negative | MGI_C57BL6J_1914031 | 4933437F24Rik | NA | NA | unclassified gene | RIKEN cDNA 4933437F24 gene |
| 4 | pseudogene | 48.97298 | 48.97331 | positive | MGI_C57BL6J_3649531 | Gm12442 | ENSEMBL:ENSMUSG00000083869 | MGI:3649531 | pseudogene | predicted gene 12442 |
| 4 | gene | 49.05927 | 49.34026 | positive | MGI_C57BL6J_2445015 | Plppr1 | NCBI_Gene:272031,ENSEMBL:ENSMUSG00000063446 | MGI:2445015 | protein coding gene | phospholipid phosphatase related 1 |
| 4 | gene | 49.22744 | 49.24472 | negative | MGI_C57BL6J_5591816 | Gm32657 | NCBI_Gene:102635276 | MGI:5591816 | lncRNA gene | predicted gene, 32657 |
| 4 | gene | 49.28930 | 49.28940 | positive | MGI_C57BL6J_5452462 | Gm22685 | ENSEMBL:ENSMUSG00000064782 | MGI:5452462 | snRNA gene | predicted gene, 22685 |
| 4 | pseudogene | 49.34193 | 49.34271 | positive | MGI_C57BL6J_3650226 | Gm12453 | NCBI_Gene:102635389,ENSEMBL:ENSMUSG00000096476 | MGI:3650226 | pseudogene | predicted gene 12453 |
| 4 | pseudogene | 49.35049 | 49.37464 | positive | MGI_C57BL6J_96523 | Rbpj-ps1 | NCBI_Gene:19665,ENSEMBL:ENSMUSG00000081770 | MGI:96523 | pseudogene | recombination signal binding protein for immunoglobulin kappa J region, pseudogene 1 |
| 4 | gene | 49.37534 | 49.40815 | negative | MGI_C57BL6J_2444345 | Acnat2 | NCBI_Gene:209186,ENSEMBL:ENSMUSG00000060317 | MGI:2444345 | protein coding gene | acyl-coenzyme A amino acid N-acyltransferase 2 |
| 4 | pseudogene | 49.39611 | 49.41031 | positive | MGI_C57BL6J_3649963 | Gm12451 | NCBI_Gene:100418419,ENSEMBL:ENSMUSG00000082725 | MGI:3649963 | pseudogene | predicted gene 12451 |
| 4 | pseudogene | 49.41222 | 49.41277 | positive | MGI_C57BL6J_3650619 | Gm12452 | NCBI_Gene:666617,ENSEMBL:ENSMUSG00000094082 | MGI:3650619 | pseudogene | predicted gene 12452 |
| 4 | pseudogene | 49.43023 | 49.44408 | positive | MGI_C57BL6J_96524 | Rbpj-ps2 | NCBI_Gene:19666 | MGI:96524 | pseudogene | recombination signal binding protein for immunoglobulin kappa J region, pseudogene 2 |
| 4 | gene | 49.44353 | 49.48132 | negative | MGI_C57BL6J_2140197 | Acnat1 | NCBI_Gene:230161,ENSEMBL:ENSMUSG00000070985 | MGI:2140197 | protein coding gene | acyl-coenzyme A amino acid N-acyltransferase 1 |
| 4 | gene | 49.47431 | 49.47441 | positive | MGI_C57BL6J_5456201 | Gm26424 | ENSEMBL:ENSMUSG00000084479 | MGI:5456201 | snRNA gene | predicted gene, 26424 |
| 4 | gene | 49.47471 | 49.48727 | positive | MGI_C57BL6J_5592168 | Gm33009 | NCBI_Gene:102635749 | MGI:5592168 | lncRNA gene | predicted gene, 33009 |
| 4 | gene | 49.47839 | 49.48104 | negative | MGI_C57BL6J_1921795 | 9030417H13Rik | NA | NA | unclassified gene | RIKEN cDNA 9030417H13 gene |
| 4 | gene | 49.48848 | 49.50447 | positive | MGI_C57BL6J_5592053 | Gm32894 | NCBI_Gene:102635603 | MGI:5592053 | lncRNA gene | predicted gene, 32894 |
| 4 | gene | 49.48942 | 49.50792 | negative | MGI_C57BL6J_106642 | Baat | NCBI_Gene:12012,ENSEMBL:ENSMUSG00000039653 | MGI:106642 | protein coding gene | bile acid-Coenzyme A: amino acid N-acyltransferase |
| 4 | gene | 49.51260 | 49.52109 | negative | MGI_C57BL6J_107329 | Mrpl50 | NCBI_Gene:28028,ENSEMBL:ENSMUSG00000044018 | MGI:107329 | protein coding gene | mitochondrial ribosomal protein L50 |
| 4 | gene | 49.52118 | 49.53156 | positive | MGI_C57BL6J_2444707 | Zfp189 | NCBI_Gene:230162,ENSEMBL:ENSMUSG00000039634 | MGI:2444707 | protein coding gene | zinc finger protein 189 |
| 4 | gene | 49.53218 | 49.53379 | positive | MGI_C57BL6J_3026937 | 1110065F06Rik | NA | NA | unclassified gene | RIKEN cDNA 1110065F06 gene |
| 4 | gene | 49.53599 | 49.54955 | negative | MGI_C57BL6J_87995 | Aldob | NCBI_Gene:230163,ENSEMBL:ENSMUSG00000028307 | MGI:87995 | protein coding gene | aldolase B, fructose-bisphosphate |
| 4 | gene | 49.58451 | 49.59788 | negative | MGI_C57BL6J_1914313 | Tmem246 | NCBI_Gene:67063,ENSEMBL:ENSMUSG00000039611 | MGI:1914313 | protein coding gene | transmembrane protein 246 |
| 4 | gene | 49.59799 | 49.59977 | positive | MGI_C57BL6J_5592265 | Gm33106 | NCBI_Gene:102635882 | MGI:5592265 | lncRNA gene | predicted gene, 33106 |
| 4 | gene | 49.60803 | 49.61780 | negative | MGI_C57BL6J_5592318 | Gm33159 | NCBI_Gene:102635952 | MGI:5592318 | lncRNA gene | predicted gene, 33159 |
| 4 | gene | 49.63066 | 49.63195 | negative | MGI_C57BL6J_5825589 | Gm45952 | NCBI_Gene:108167345 | MGI:5825589 | lncRNA gene | predicted gene, 45952 |
| 4 | gene | 49.63201 | 49.65689 | positive | MGI_C57BL6J_1925927 | Rnf20 | NCBI_Gene:109331,ENSEMBL:ENSMUSG00000028309 | MGI:1925927 | protein coding gene | ring finger protein 20 |
| 4 | gene | 49.66161 | 49.84577 | negative | MGI_C57BL6J_1933206 | Grin3a | NCBI_Gene:242443,ENSEMBL:ENSMUSG00000039579 | MGI:1933206 | protein coding gene | glutamate receptor ionotropic, NMDA3A |
| 4 | gene | 49.67875 | 49.68202 | negative | MGI_C57BL6J_107171 | Ppp3r2 | NCBI_Gene:19059,ENSEMBL:ENSMUSG00000028310 | MGI:107171 | protein coding gene | protein phosphatase 3, regulatory subunit B, alpha isoform (calcineurin B, type II) |
| 4 | gene | 49.68200 | 49.69279 | positive | MGI_C57BL6J_5625171 | Gm42286 | NCBI_Gene:105247131 | MGI:5625171 | lncRNA gene | predicted gene, 42286 |
| 4 | pseudogene | 49.74673 | 49.74725 | negative | MGI_C57BL6J_3651071 | Gm12459 | ENSEMBL:ENSMUSG00000083153 | MGI:3651071 | pseudogene | predicted gene 12459 |
| 4 | pseudogene | 50.05091 | 50.05140 | negative | MGI_C57BL6J_3650856 | Gm12458 | ENSEMBL:ENSMUSG00000081120 | MGI:3650856 | pseudogene | predicted gene 12458 |
| 4 | gene | 50.28675 | 50.28688 | positive | MGI_C57BL6J_5455262 | Gm25485 | ENSEMBL:ENSMUSG00000095394 | MGI:5455262 | snoRNA gene | predicted gene, 25485 |
| 4 | gene | 50.78823 | 50.82700 | negative | MGI_C57BL6J_5625173 | Gm42288 | NCBI_Gene:105247133 | MGI:5625173 | lncRNA gene | predicted gene, 42288 |
| 4 | gene | 50.78962 | 50.90373 | positive | MGI_C57BL6J_5625172 | Gm42287 | NCBI_Gene:105247132 | MGI:5625172 | lncRNA gene | predicted gene, 42287 |
| 4 | pseudogene | 51.19009 | 51.19046 | positive | MGI_C57BL6J_3650440 | Gm12460 | NCBI_Gene:666681,ENSEMBL:ENSMUSG00000084116 | MGI:3650440 | pseudogene | predicted gene 12460 |
| 4 | gene | 51.21667 | 51.25062 | positive | MGI_C57BL6J_1922164 | Cylc2 | NCBI_Gene:74914,ENSEMBL:ENSMUSG00000039555 | MGI:1922164 | protein coding gene | cylicin, basic protein of sperm head cytoskeleton 2 |
| 4 | pseudogene | 51.25247 | 51.25436 | negative | MGI_C57BL6J_3650439 | Amd-ps4 | NCBI_Gene:100040814,ENSEMBL:ENSMUSG00000019836 | MGI:3650439 | pseudogene | S-adenosylmethionine decarboxylase, pseudogene 4 |
| 4 | gene | 51.54075 | 51.54102 | positive | MGI_C57BL6J_5452340 | Gm22563 | ENSEMBL:ENSMUSG00000087958 | MGI:5452340 | unclassified non-coding RNA gene | predicted gene, 22563 |
| 4 | gene | 51.65577 | 51.65588 | positive | MGI_C57BL6J_5452820 | Gm23043 | ENSEMBL:ENSMUSG00000088515 | MGI:5452820 | snoRNA gene | predicted gene, 23043 |
| 4 | gene | 51.79992 | 51.80035 | positive | MGI_C57BL6J_5454720 | Gm24943 | ENSEMBL:ENSMUSG00000088954 | MGI:5454720 | ribozyme gene | predicted gene, 24943 |
| 4 | gene | 51.81248 | 51.81265 | negative | MGI_C57BL6J_5451819 | Gm22042 | ENSEMBL:ENSMUSG00000064923 | MGI:5451819 | snRNA gene | predicted gene, 22042 |
| 4 | gene | 51.87665 | 51.87743 | positive | MGI_C57BL6J_5826491 | Gm46854 | NCBI_Gene:108168942 | MGI:5826491 | lncRNA gene | predicted gene, 46854 |
| 4 | pseudogene | 51.89285 | 51.89421 | positive | MGI_C57BL6J_3650438 | Gm12461 | NCBI_Gene:384015,ENSEMBL:ENSMUSG00000081050 | MGI:3650438 | pseudogene | predicted gene 12461 |
| 4 | gene | 51.96809 | 52.05111 | negative | MGI_C57BL6J_3045376 | C630028M04Rik | NCBI_Gene:329844,ENSEMBL:ENSMUSG00000085515 | MGI:3045376 | lncRNA gene | RIKEN cDNA C630028M04 gene |
| 4 | pseudogene | 52.07717 | 52.07747 | negative | MGI_C57BL6J_3649306 | Gm12465 | ENSEMBL:ENSMUSG00000082963 | MGI:3649306 | pseudogene | predicted gene 12465 |
| 4 | pseudogene | 52.24087 | 52.24157 | positive | MGI_C57BL6J_3651763 | Gm12466 | NCBI_Gene:100041026,ENSEMBL:ENSMUSG00000083539 | MGI:3651763 | pseudogene | predicted gene 12466 |
| 4 | pseudogene | 52.24171 | 52.32313 | positive | MGI_C57BL6J_3651688 | Gm12467 | NCBI_Gene:100041038,ENSEMBL:ENSMUSG00000082267 | MGI:3651688 | pseudogene | predicted gene 12467 |
| 4 | pseudogene | 52.36082 | 52.36221 | negative | MGI_C57BL6J_3649584 | Gm12475 | NCBI_Gene:666724,ENSEMBL:ENSMUSG00000083338 | MGI:3649584 | pseudogene | predicted gene 12475 |
| 4 | gene | 52.43028 | 52.43897 | negative | MGI_C57BL6J_1919898 | Smc2os | NCBI_Gene:72648,ENSEMBL:ENSMUSG00000085763 | MGI:1919898 | antisense lncRNA gene | structural maintenance of chromosomes 2, opposite strand |
| 4 | gene | 52.43922 | 52.48837 | positive | MGI_C57BL6J_106067 | Smc2 | NCBI_Gene:14211,ENSEMBL:ENSMUSG00000028312 | MGI:106067 | protein coding gene | structural maintenance of chromosomes 2 |
| 4 | pseudogene | 52.44375 | 52.44593 | negative | MGI_C57BL6J_3651628 | Tmc4-ps | NCBI_Gene:102636091,ENSEMBL:ENSMUSG00000084380 | MGI:3651628 | pseudogene | Tmc4 retrotransposed pseudogene |
| 4 | pseudogene | 52.49495 | 52.49732 | negative | MGI_C57BL6J_3643145 | Vma21-ps | NCBI_Gene:100040883,ENSEMBL:ENSMUSG00000061619 | MGI:3643145 | pseudogene | VMA21 vacuolar H+-ATPase homolog (S. cerevisiae), pseudogene |
| 4 | gene | 52.59627 | 52.61243 | positive | MGI_C57BL6J_1915524 | Toporsl | NCBI_Gene:68274,ENSEMBL:ENSMUSG00000028314 | MGI:1915524 | protein coding gene | topoisomerase I binding, arginine/serine-rich like |
| 4 | gene | 52.61545 | 52.61788 | negative | MGI_C57BL6J_5592594 | Gm33435 | NCBI_Gene:102636342 | MGI:5592594 | lncRNA gene | predicted gene, 33435 |
| 4 | gene | 52.81460 | 52.82827 | positive | MGI_C57BL6J_1922617 | 4930552N02Rik | NCBI_Gene:75367 | MGI:1922617 | lncRNA gene | RIKEN cDNA 4930552N02 gene |
| 4 | gene | 52.81460 | 52.82827 | positive | MGI_C57BL6J_3030109 | Olfr275 | NCBI_Gene:258857,ENSEMBL:ENSMUSG00000089717 | MGI:3030109 | protein coding gene | olfactory receptor 275 |
| 4 | pseudogene | 52.82145 | 52.82224 | positive | MGI_C57BL6J_3651627 | Gm12482 | NCBI_Gene:666761,ENSEMBL:ENSMUSG00000081975 | MGI:3651627 | pseudogene | predicted gene 12482 |
| 4 | pseudogene | 52.84220 | 52.84305 | negative | MGI_C57BL6J_3030108 | Olfr274-ps1 | NCBI_Gene:404368,ENSEMBL:ENSMUSG00000110940 | MGI:3030108 | pseudogene | olfactory receptor 274, pseudogene 1 |
| 4 | gene | 52.85215 | 52.85924 | negative | MGI_C57BL6J_3030107 | Olfr273 | NCBI_Gene:258821,ENSEMBL:ENSMUSG00000049648 | MGI:3030107 | protein coding gene | olfactory receptor 273 |
| 4 | pseudogene | 52.87801 | 52.87870 | positive | MGI_C57BL6J_6096954 | Gm47786 | ENSEMBL:ENSMUSG00000110756 | MGI:6096954 | pseudogene | predicted gene, 47786 |
| 4 | pseudogene | 52.90055 | 52.90288 | positive | MGI_C57BL6J_109293 | Cct3-ps1 | NCBI_Gene:632982,ENSEMBL:ENSMUSG00000083937 | MGI:109293 | pseudogene | chaperonin containing Tcp1, subunit 3 (gamma) pseudogene 1 |
| 4 | gene | 52.91021 | 52.91918 | negative | MGI_C57BL6J_3030106 | Olfr272 | NCBI_Gene:258836,ENSEMBL:ENSMUSG00000051593 | MGI:3030106 | protein coding gene | olfactory receptor 272 |
| 4 | pseudogene | 52.93543 | 52.93638 | negative | MGI_C57BL6J_3030105 | Olfr271-ps1 | NCBI_Gene:258856,ENSEMBL:ENSMUSG00000082216 | MGI:3030105 | polymorphic pseudogene | olfactory receptor 271, pseudogene 1 |
| 4 | gene | 52.94434 | 52.95877 | negative | MGI_C57BL6J_5592638 | Gm33479 | NCBI_Gene:102636404 | MGI:5592638 | lncRNA gene | predicted gene, 33479 |
| 4 | gene | 52.96455 | 52.97247 | positive | MGI_C57BL6J_3030104 | Olfr270 | NCBI_Gene:258600,ENSEMBL:ENSMUSG00000070983 | MGI:3030104 | protein coding gene | olfactory receptor 270 |
| 4 | gene | 52.98927 | 53.00116 | positive | MGI_C57BL6J_1920648 | Nipsnap3a | NCBI_Gene:73398,ENSEMBL:ENSMUSG00000015242 | MGI:1920648 | protein coding gene | nipsnap homolog 3A |
| 4 | gene | 53.01187 | 53.02206 | positive | MGI_C57BL6J_1913786 | Nipsnap3b | NCBI_Gene:66536,ENSEMBL:ENSMUSG00000015247 | MGI:1913786 | protein coding gene | nipsnap homolog 3B |
| 4 | gene | 53.03079 | 53.15999 | negative | MGI_C57BL6J_99607 | Abca1 | NCBI_Gene:11303,ENSEMBL:ENSMUSG00000015243 | MGI:99607 | protein coding gene | ATP-binding cassette, sub-family A (ABC1), member 1 |
| 4 | gene | 53.20152 | 53.22001 | negative | MGI_C57BL6J_1921191 | 4930412L05Rik | NCBI_Gene:73941,ENSEMBL:ENSMUSG00000087192 | MGI:1921191 | lncRNA gene | RIKEN cDNA 4930412L05 gene |
| 4 | gene | 53.24620 | 53.25112 | positive | MGI_C57BL6J_1925430 | 4930522O17Rik | NCBI_Gene:78180,ENSEMBL:ENSMUSG00000086947 | MGI:1925430 | lncRNA gene | RIKEN cDNA 4930522O17 gene |
| 4 | gene | 53.26136 | 53.27023 | negative | MGI_C57BL6J_2140270 | AI427809 | NCBI_Gene:381524,ENSEMBL:ENSMUSG00000086712 | MGI:2140270 | lncRNA gene | expressed sequence AI427809 |
| 4 | pseudogene | 53.30823 | 53.30867 | positive | MGI_C57BL6J_3650574 | Gm12495 | ENSEMBL:ENSMUSG00000083735 | MGI:3650574 | pseudogene | predicted gene 12495 |
| 4 | gene | 53.34842 | 53.40403 | negative | MGI_C57BL6J_3650573 | Gm12496 | NCBI_Gene:102636602,ENSEMBL:ENSMUSG00000086267 | MGI:3650573 | lncRNA gene | predicted gene 12496 |
| 4 | gene | 53.43801 | 53.44069 | negative | MGI_C57BL6J_5826495 | Gm46858 | NCBI_Gene:108168953 | MGI:5826495 | protein coding gene | predicted gene, 46858 |
| 4 | gene | 53.44041 | 53.62248 | positive | MGI_C57BL6J_2140592 | Slc44a1 | NCBI_Gene:100434,ENSEMBL:ENSMUSG00000028412 | MGI:2140592 | protein coding gene | solute carrier family 44, member 1 |
| 4 | gene | 53.56298 | 53.56470 | negative | MGI_C57BL6J_1920641 | 1700060J05Rik | ENSEMBL:ENSMUSG00000084970 | MGI:1920641 | lncRNA gene | RIKEN cDNA 1700060J05 gene |
| 4 | gene | 53.57244 | 53.57303 | positive | MGI_C57BL6J_1924958 | 9130223C08Rik | NA | NA | unclassified gene | RIKEN cDNA 9130223C08 gene |
| 4 | gene | 53.58320 | 53.62987 | negative | MGI_C57BL6J_5592888 | Gm33729 | NCBI_Gene:102636739 | MGI:5592888 | lncRNA gene | predicted gene, 33729 |
| 4 | gene | 53.62993 | 53.70701 | positive | MGI_C57BL6J_2442443 | Fsd1l | NCBI_Gene:319636,ENSEMBL:ENSMUSG00000054752 | MGI:2442443 | protein coding gene | fibronectin type III and SPRY domain containing 1-like |
| 4 | pseudogene | 53.66230 | 53.66334 | positive | MGI_C57BL6J_3652113 | Gm12470 | NCBI_Gene:100534358,ENSEMBL:ENSMUSG00000081516 | MGI:3652113 | pseudogene | predicted gene 12470 |
| 4 | gene | 53.71273 | 53.77789 | positive | MGI_C57BL6J_2179507 | Fktn | NCBI_Gene:246179,ENSEMBL:ENSMUSG00000028414 | MGI:2179507 | protein coding gene | fukutin |
| 4 | gene | 53.72090 | 53.72120 | negative | MGI_C57BL6J_5455222 | Gm25445 | ENSEMBL:ENSMUSG00000089621 | MGI:5455222 | unclassified non-coding RNA gene | predicted gene, 25445 |
| 4 | gene | 53.77970 | 53.78871 | positive | MGI_C57BL6J_99540 | Tal2 | NCBI_Gene:21350,ENSEMBL:ENSMUSG00000028417 | MGI:99540 | protein coding gene | T cell acute lymphocytic leukemia 2 |
| 4 | pseudogene | 53.79103 | 53.79147 | negative | MGI_C57BL6J_3652112 | Rps15a-ps8 | NCBI_Gene:435784,ENSEMBL:ENSMUSG00000084403 | MGI:3652112 | pseudogene | ribosomal protein S15A, pseudogene 8 |
| 4 | gene | 53.82605 | 53.86202 | positive | MGI_C57BL6J_1098718 | Tmem38b | NCBI_Gene:52076,ENSEMBL:ENSMUSG00000028420 | MGI:1098718 | protein coding gene | transmembrane protein 38B |
| 4 | gene | 53.86065 | 53.86077 | positive | MGI_C57BL6J_4422050 | n-R5s185 | ENSEMBL:ENSMUSG00000065887 | MGI:4422050 | rRNA gene | nuclear encoded rRNA 5S 185 |
| 4 | pseudogene | 53.87659 | 53.87843 | positive | MGI_C57BL6J_3652115 | Gm12468 | NCBI_Gene:670124,ENSEMBL:ENSMUSG00000083985 | MGI:3652115 | pseudogene | predicted gene 12468 |
| 4 | gene | 53.90570 | 53.90582 | positive | MGI_C57BL6J_5453522 | Gm23745 | ENSEMBL:ENSMUSG00000077418 | MGI:5453522 | snoRNA gene | predicted gene, 23745 |
| 4 | gene | 53.94826 | 53.95131 | negative | MGI_C57BL6J_5592999 | Gm33840 | NCBI_Gene:102636891 | MGI:5592999 | lncRNA gene | predicted gene, 33840 |
| 4 | gene | 54.00406 | 54.00419 | positive | MGI_C57BL6J_5453954 | Gm24177 | ENSEMBL:ENSMUSG00000089552 | MGI:5453954 | snoRNA gene | predicted gene, 24177 |
| 4 | gene | 54.02972 | 54.07069 | positive | MGI_C57BL6J_5593052 | Gm33893 | NCBI_Gene:102636969 | MGI:5593052 | lncRNA gene | predicted gene, 33893 |
| 4 | gene | 54.15183 | 54.16368 | negative | MGI_C57BL6J_3651626 | Gm12484 | ENSEMBL:ENSMUSG00000086987 | MGI:3651626 | lncRNA gene | predicted gene 12484 |
| 4 | gene | 54.18322 | 54.19183 | positive | MGI_C57BL6J_5593167 | Gm34008 | NCBI_Gene:102637114 | MGI:5593167 | lncRNA gene | predicted gene, 34008 |
| 4 | pseudogene | 54.23567 | 54.23588 | negative | MGI_C57BL6J_3652114 | Gm12469 | ENSEMBL:ENSMUSG00000083624 | MGI:3652114 | pseudogene | predicted gene 12469 |
| 4 | gene | 54.46835 | 54.46859 | positive | MGI_C57BL6J_5455464 | Gm25687 | ENSEMBL:ENSMUSG00000093183 | MGI:5455464 | unclassified non-coding RNA gene | predicted gene, 25687 |
| 4 | gene | 54.55032 | 54.55148 | negative | MGI_C57BL6J_5593231 | Gm34072 | NCBI_Gene:102637198 | MGI:5593231 | lncRNA gene | predicted gene, 34072 |
| 4 | pseudogene | 54.56457 | 54.56534 | positive | MGI_C57BL6J_2664996 | Tpt1-ps2 | NCBI_Gene:666901,ENSEMBL:ENSMUSG00000081428 | MGI:2664996 | pseudogene | tumor protein, translationally-controlled, pseudogene 2 |
| 4 | gene | 54.64581 | 54.64628 | negative | MGI_C57BL6J_5579773 | Gm29067 | ENSEMBL:ENSMUSG00000099421 | MGI:5579773 | lncRNA gene | predicted gene 29067 |
| 4 | gene | 54.64654 | 54.68945 | positive | MGI_C57BL6J_3651221 | Gm12478 | ENSEMBL:ENSMUSG00000086371 | MGI:3651221 | lncRNA gene | predicted gene 12478 |
| 4 | gene | 54.65190 | 54.65608 | positive | MGI_C57BL6J_3651952 | Gm12480 | ENSEMBL:ENSMUSG00000087077 | MGI:3651952 | lncRNA gene | predicted gene 12480 |
| 4 | pseudogene | 54.66962 | 54.67019 | negative | MGI_C57BL6J_3651220 | Gm12477 | NCBI_Gene:100416401,ENSEMBL:ENSMUSG00000083868 | MGI:3651220 | pseudogene | predicted gene 12477 |
| 4 | gene | 54.71062 | 54.71091 | positive | MGI_C57BL6J_5454405 | Gm24628 | ENSEMBL:ENSMUSG00000084710 | MGI:5454405 | unclassified non-coding RNA gene | predicted gene, 24628 |
| 4 | gene | 54.85124 | 54.89394 | negative | MGI_C57BL6J_5593292 | Gm34133 | NCBI_Gene:102637273 | MGI:5593292 | lncRNA gene | predicted gene, 34133 |
| 4 | pseudogene | 54.85249 | 54.85471 | positive | MGI_C57BL6J_3651218 | Gm12479 | NCBI_Gene:384018,ENSEMBL:ENSMUSG00000082111 | MGI:3651218 | pseudogene | predicted gene 12479 |
| 4 | gene | 54.94386 | 55.08356 | positive | MGI_C57BL6J_107690 | Zfp462 | NCBI_Gene:242466,ENSEMBL:ENSMUSG00000060206 | MGI:107690 | protein coding gene | zinc finger protein 462 |
| 4 | gene | 54.94447 | 54.94794 | negative | MGI_C57BL6J_5826496 | Gm46859 | NCBI_Gene:108168957 | MGI:5826496 | lncRNA gene | predicted gene, 46859 |
| 4 | gene | 54.94449 | 54.94586 | negative | MGI_C57BL6J_3645200 | Gm8364 | NA | NA | unclassified gene | predicted gene 8364 |
| 4 | gene | 55.02091 | 55.02127 | positive | MGI_C57BL6J_1924603 | 9430095M17Rik | NA | NA | unclassified gene | RIKEN cDNA 9430095M17 gene |
| 4 | gene | 55.04319 | 55.04481 | negative | MGI_C57BL6J_3641952 | Gm10588 | NA | NA | unclassified gene | predicted gene 10588 |
| 4 | gene | 55.06052 | 55.06927 | negative | MGI_C57BL6J_5625174 | Gm42289 | NCBI_Gene:105247134 | MGI:5625174 | lncRNA gene | predicted gene, 42289 |
| 4 | pseudogene | 55.11656 | 55.11692 | positive | MGI_C57BL6J_3651354 | Gm12515 | ENSEMBL:ENSMUSG00000082704 | MGI:3651354 | pseudogene | predicted gene 12515 |
| 4 | gene | 55.12971 | 55.27948 | negative | MGI_C57BL6J_3651364 | Gm12514 | NCBI_Gene:105247135,ENSEMBL:ENSMUSG00000087614 | MGI:3651364 | lncRNA gene | predicted gene 12514 |
| 4 | pseudogene | 55.16301 | 55.16362 | positive | MGI_C57BL6J_3651361 | Gm12509 | NCBI_Gene:100417778,ENSEMBL:ENSMUSG00000081177 | MGI:3651361 | pseudogene | predicted gene 12509 |
| 4 | gene | 55.20786 | 55.23894 | positive | MGI_C57BL6J_5593390 | Gm34231 | NCBI_Gene:102637421 | MGI:5593390 | lncRNA gene | predicted gene, 34231 |
| 4 | pseudogene | 55.24367 | 55.25522 | positive | MGI_C57BL6J_3651359 | Gm12508 | NCBI_Gene:670211,ENSEMBL:ENSMUSG00000081504 | MGI:3651359 | pseudogene | predicted gene 12508 |
| 4 | pseudogene | 55.28206 | 55.28261 | negative | MGI_C57BL6J_3651362 | Gm12512 | NCBI_Gene:108168954,ENSEMBL:ENSMUSG00000081542 | MGI:3651362 | pseudogene | predicted gene 12512 |
| 4 | gene | 55.29157 | 55.29181 | positive | MGI_C57BL6J_1918945 | 0910001E24Rik | NA | NA | unclassified gene | RIKEN cDNA 0910001E24 gene |
| 4 | gene | 55.29966 | 55.29984 | negative | MGI_C57BL6J_5455196 | Gm25419 | ENSEMBL:ENSMUSG00000077403 | MGI:5455196 | snoRNA gene | predicted gene, 25419 |
| 4 | gene | 55.34123 | 55.34962 | negative | MGI_C57BL6J_5625175 | Gm42290 | NCBI_Gene:105247136 | MGI:5625175 | lncRNA gene | predicted gene, 42290 |
| 4 | gene | 55.35004 | 55.39224 | positive | MGI_C57BL6J_105128 | Rad23b | NCBI_Gene:19359,ENSEMBL:ENSMUSG00000028426 | MGI:105128 | protein coding gene | RAD23 homolog B, nucleotide excision repair protein |
| 4 | pseudogene | 55.39227 | 55.39292 | negative | MGI_C57BL6J_3651353 | Gm12516 | NCBI_Gene:108168955,ENSEMBL:ENSMUSG00000084144 | MGI:3651353 | pseudogene | predicted gene 12516 |
| 4 | gene | 55.39380 | 55.39939 | negative | MGI_C57BL6J_5593449 | Gm34290 | NCBI_Gene:102637498 | MGI:5593449 | lncRNA gene | predicted gene, 34290 |
| 4 | pseudogene | 55.40896 | 55.40931 | negative | MGI_C57BL6J_3649753 | Gm12513 | ENSEMBL:ENSMUSG00000084404 | MGI:3649753 | pseudogene | predicted gene 12513 |
| 4 | gene | 55.41044 | 55.41884 | negative | MGI_C57BL6J_3651173 | Gm12505 | NCBI_Gene:100415914,ENSEMBL:ENSMUSG00000087070 | MGI:3651173 | lncRNA gene | predicted gene 12505 |
| 4 | gene | 55.45068 | 55.46581 | negative | MGI_C57BL6J_3651360 | Gm12510 | ENSEMBL:ENSMUSG00000086346 | MGI:3651360 | lncRNA gene | predicted gene 12510 |
| 4 | gene | 55.46903 | 55.46916 | negative | MGI_C57BL6J_5451927 | Gm22150 | ENSEMBL:ENSMUSG00000077635 | MGI:5451927 | snoRNA gene | predicted gene, 22150 |
| 4 | pseudogene | 55.50829 | 55.50871 | positive | MGI_C57BL6J_3651171 | Gm12507 | ENSEMBL:ENSMUSG00000082952 | MGI:3651171 | pseudogene | predicted gene 12507 |
| 4 | gene | 55.52714 | 55.53247 | negative | MGI_C57BL6J_1342287 | Klf4 | NCBI_Gene:16600,ENSEMBL:ENSMUSG00000003032 | MGI:1342287 | protein coding gene | Kruppel-like factor 4 (gut) |
| 4 | gene | 55.53287 | 55.63919 | positive | MGI_C57BL6J_3651170 | Gm12506 | NCBI_Gene:102637640,ENSEMBL:ENSMUSG00000085375 | MGI:3651170 | lncRNA gene | predicted gene 12506 |
| 4 | gene | 55.53888 | 55.54174 | positive | MGI_C57BL6J_5593502 | Gm34343 | NCBI_Gene:102637567 | MGI:5593502 | lncRNA gene | predicted gene, 34343 |
| 4 | gene | 55.56327 | 55.59930 | positive | MGI_C57BL6J_3651363 | Gm12511 | NCBI_Gene:791419,ENSEMBL:ENSMUSG00000066176 | MGI:3651363 | lncRNA gene | predicted gene 12511 |
| 4 | gene | 55.94800 | 55.95341 | negative | MGI_C57BL6J_5593641 | Gm34482 | NCBI_Gene:102637751 | MGI:5593641 | lncRNA gene | predicted gene, 34482 |
| 4 | gene | 55.96654 | 56.00082 | negative | MGI_C57BL6J_3652272 | Gm12519 | NCBI_Gene:102637828,ENSEMBL:ENSMUSG00000085643 | MGI:3652272 | lncRNA gene | predicted gene 12519 |
| 4 | pseudogene | 56.16294 | 56.16325 | positive | MGI_C57BL6J_3652271 | Gm12520 | ENSEMBL:ENSMUSG00000081290 | MGI:3652271 | pseudogene | predicted gene 12520 |
| 4 | gene | 56.22062 | 56.22452 | negative | MGI_C57BL6J_1919656 | 2310081O03Rik | ENSEMBL:ENSMUSG00000085020 | MGI:1919656 | lncRNA gene | RIKEN cDNA 2310081O03 gene |
| 4 | gene | 56.26732 | 56.31597 | negative | MGI_C57BL6J_5625176 | Gm42291 | NCBI_Gene:105247137 | MGI:5625176 | lncRNA gene | predicted gene, 42291 |
| 4 | pseudogene | 56.38780 | 56.38858 | positive | MGI_C57BL6J_3652273 | Gm12518 | NCBI_Gene:100416258,ENSEMBL:ENSMUSG00000082613 | MGI:3652273 | pseudogene | predicted gene 12518 |
| 4 | gene | 56.45591 | 56.47840 | negative | MGI_C57BL6J_5593787 | Gm34628 | NCBI_Gene:102637940 | MGI:5593787 | lncRNA gene | predicted gene, 34628 |
| 4 | gene | 56.62295 | 56.71442 | negative | MGI_C57BL6J_5593846 | Gm34687 | NCBI_Gene:102638019 | MGI:5593846 | lncRNA gene | predicted gene, 34687 |
| 4 | gene | 56.67890 | 56.67903 | negative | MGI_C57BL6J_5455921 | Gm26144 | ENSEMBL:ENSMUSG00000077663 | MGI:5455921 | snoRNA gene | predicted gene, 26144 |
| 4 | gene | 56.74000 | 56.74144 | negative | MGI_C57BL6J_1343053 | Actl7b | NCBI_Gene:11471,ENSEMBL:ENSMUSG00000070980 | MGI:1343053 | protein coding gene | actin-like 7b |
| 4 | gene | 56.74007 | 56.74138 | positive | MGI_C57BL6J_5477151 | Gm26657 | ENSEMBL:ENSMUSG00000096930 | MGI:5477151 | protein coding gene | predicted gene, 26657 |
| 4 | gene | 56.74341 | 56.74493 | positive | MGI_C57BL6J_1343051 | Actl7a | NCBI_Gene:11470,ENSEMBL:ENSMUSG00000070979 | MGI:1343051 | protein coding gene | actin-like 7a |
| 4 | gene | 56.74968 | 56.80233 | negative | MGI_C57BL6J_1914544 | Elp1 | NCBI_Gene:230233,ENSEMBL:ENSMUSG00000028431 | MGI:1914544 | protein coding gene | elongator complex protein 1 |
| 4 | gene | 56.76192 | 56.76204 | negative | MGI_C57BL6J_5452729 | Gm22952 | ENSEMBL:ENSMUSG00000080370 | MGI:5452729 | snoRNA gene | predicted gene, 22952 |
| 4 | gene | 56.80233 | 56.80960 | positive | MGI_C57BL6J_2677850 | Abitram | NCBI_Gene:230234,ENSEMBL:ENSMUSG00000038827 | MGI:2677850 | protein coding gene | actin binding transcription modulator |
| 4 | gene | 56.81094 | 56.86538 | negative | MGI_C57BL6J_1859649 | Ctnnal1 | NCBI_Gene:54366,ENSEMBL:ENSMUSG00000038816 | MGI:1859649 | protein coding gene | catenin (cadherin associated protein), alpha-like 1 |
| 4 | gene | 56.86692 | 56.94756 | negative | MGI_C57BL6J_2445107 | Tmem245 | NCBI_Gene:242474,ENSEMBL:ENSMUSG00000055296 | MGI:2445107 | protein coding gene | transmembrane protein 245 |
| 4 | gene | 56.89523 | 56.89530 | negative | MGI_C57BL6J_3619331 | Mir32 | miRBase:MI0000691,NCBI_Gene:723837,ENSEMBL:ENSMUSG00000065544 | MGI:3619331 | miRNA gene | microRNA 32 |
| 4 | gene | 56.89762 | 56.89776 | negative | MGI_C57BL6J_5454830 | Gm25053 | ENSEMBL:ENSMUSG00000106585,ENSEMBL:ENSMUSG00000099002 | MGI:5454830 | snoRNA gene | predicted gene, 25053 |
| 4 | gene | 56.91364 | 56.91377 | positive | MGI_C57BL6J_5452077 | Gm22300 | ENSEMBL:ENSMUSG00000064595 | MGI:5452077 | snoRNA gene | predicted gene, 22300 |
| 4 | gene | 56.95717 | 56.95787 | positive | MGI_C57BL6J_2140381 | 9630041G16Rik | NA | NA | unclassified gene | RIKEN cDNA 9630041G16 gene |
| 4 | gene | 56.95717 | 56.99039 | negative | MGI_C57BL6J_2442704 | Frrs1l | NCBI_Gene:230235,ENSEMBL:ENSMUSG00000045589 | MGI:2442704 | protein coding gene | ferric-chelate reductase 1 like |
| 4 | gene | 56.99197 | 57.14344 | negative | MGI_C57BL6J_1859149 | Epb41l4b | NCBI_Gene:54357,ENSEMBL:ENSMUSG00000028434 | MGI:1859149 | protein coding gene | erythrocyte membrane protein band 4.1 like 4b |
| 4 | gene | 57.05275 | 57.05411 | positive | MGI_C57BL6J_5593897 | Gm34738 | NCBI_Gene:102638093 | MGI:5593897 | lncRNA gene | predicted gene, 34738 |
| 4 | gene | 57.16066 | 57.16415 | positive | MGI_C57BL6J_5593961 | Gm34802 | NCBI_Gene:102638176 | MGI:5593961 | lncRNA gene | predicted gene, 34802 |
| 4 | gene | 57.17207 | 57.17655 | positive | MGI_C57BL6J_3651546 | Gm12530 | NCBI_Gene:329851,ENSEMBL:ENSMUSG00000085967 | MGI:3651546 | lncRNA gene | predicted gene 12530 |
| 4 | gene | 57.19084 | 57.33904 | negative | MGI_C57BL6J_105307 | Ptpn3 | NCBI_Gene:545622,ENSEMBL:ENSMUSG00000038764 | MGI:105307 | protein coding gene | protein tyrosine phosphatase, non-receptor type 3 |
| 4 | gene | 57.29979 | 57.30859 | positive | MGI_C57BL6J_3650719 | Gm12536 | NCBI_Gene:102638251,ENSEMBL:ENSMUSG00000085654 | MGI:3650719 | lncRNA gene | predicted gene 12536 |
| 4 | gene | 57.35979 | 57.36429 | negative | MGI_C57BL6J_1920599 | 1700042G15Rik | NCBI_Gene:73349,ENSEMBL:ENSMUSG00000087044 | MGI:1920599 | lncRNA gene | RIKEN cDNA 1700042G15 gene |
| 4 | pseudogene | 57.37077 | 57.37197 | negative | MGI_C57BL6J_3650720 | Gm12537 | NCBI_Gene:100041342,ENSEMBL:ENSMUSG00000095042 | MGI:3650720 | pseudogene | predicted gene 12537 |
| 4 | pseudogene | 57.41367 | 57.41427 | negative | MGI_C57BL6J_3650715 | Gm12538 | NCBI_Gene:619973,ENSEMBL:ENSMUSG00000081540 | MGI:3650715 | pseudogene | predicted gene 12538 |
| 4 | gene | 57.43425 | 57.89698 | positive | MGI_C57BL6J_5141924 | Pakap | NCBI_Gene:677884,NCBI_Gene:11641,NCBI_Gene:242481,ENSEMBL:ENSMUSG00000038729,ENSEMBL:ENSMUSG00000089945,ENSEMBL:ENSMUSG00000090053 | MGI:5141924 | protein coding gene | paralemmin A kinase anchor protein |
| 4 | pseudogene | 57.45366 | 57.45759 | negative | MGI_C57BL6J_3650716 | Gm12539 | NCBI_Gene:667049,ENSEMBL:ENSMUSG00000081615 | MGI:3650716 | pseudogene | predicted gene 12539 |
| 4 | gene | 57.61154 | 57.61187 | positive | MGI_C57BL6J_1919814 | 2700063P09Rik | NA | NA | unclassified gene | RIKEN cDNA 2700063P09 gene |
| 4 | gene | 57.72777 | 57.73042 | negative | MGI_C57BL6J_1924878 | 4930544N03Rik | NA | NA | unclassified gene | RIKEN cDNA 4930544N03 gene |
| 4 | gene | 57.73008 | 57.75602 | negative | MGI_C57BL6J_5594258 | Gm35099 | NCBI_Gene:102638565 | MGI:5594258 | lncRNA gene | predicted gene, 35099 |
| 4 | gene | 57.75774 | 57.76390 | negative | MGI_C57BL6J_3651778 | Gm12526 | ENSEMBL:ENSMUSG00000073846 | MGI:3651778 | lncRNA gene | predicted gene 12526 |
| 4 | gene | 57.76261 | 57.76271 | positive | MGI_C57BL6J_5454054 | Gm24277 | ENSEMBL:ENSMUSG00000089286 | MGI:5454054 | snRNA gene | predicted gene, 24277 |
| 4 | gene | 57.87739 | 57.87750 | negative | MGI_C57BL6J_5455915 | Gm26138 | ENSEMBL:ENSMUSG00000096195 | MGI:5455915 | snRNA gene | predicted gene, 26138 |
| 4 | gene | 57.90838 | 57.91630 | negative | MGI_C57BL6J_2442889 | D630039A03Rik | NCBI_Gene:242484,ENSEMBL:ENSMUSG00000052117 | MGI:2442889 | protein coding gene | RIKEN cDNA D630039A03 gene |
| 4 | gene | 57.91664 | 57.91842 | positive | MGI_C57BL6J_5625178 | Gm42293 | NCBI_Gene:105247139 | MGI:5625178 | lncRNA gene | predicted gene, 42293 |
| 4 | gene | 57.92142 | 57.92343 | negative | MGI_C57BL6J_5625177 | Gm42292 | NCBI_Gene:105247138 | MGI:5625177 | lncRNA gene | predicted gene, 42292 |
| 4 | gene | 57.94337 | 57.95641 | negative | MGI_C57BL6J_98874 | Txn1 | NCBI_Gene:22166,ENSEMBL:ENSMUSG00000028367 | MGI:98874 | protein coding gene | thioredoxin 1 |
| 4 | gene | 57.95651 | 57.96815 | positive | MGI_C57BL6J_5594503 | Gm35344 | NCBI_Gene:102638892 | MGI:5594503 | lncRNA gene | predicted gene, 35344 |
| 4 | gene | 57.98402 | 58.00914 | negative | MGI_C57BL6J_1914652 | Txndc8 | NCBI_Gene:67402,ENSEMBL:ENSMUSG00000038709 | MGI:1914652 | protein coding gene | thioredoxin domain containing 8 |
| 4 | gene | 58.03451 | 58.06582 | positive | MGI_C57BL6J_5594561 | Gm35402 | NCBI_Gene:102638969 | MGI:5594561 | lncRNA gene | predicted gene, 35402 |
| 4 | gene | 58.04244 | 58.20686 | negative | MGI_C57BL6J_1928849 | Svep1 | NCBI_Gene:64817,ENSEMBL:ENSMUSG00000028369 | MGI:1928849 | protein coding gene | sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| 4 | gene | 58.17825 | 58.20663 | positive | MGI_C57BL6J_5477383 | Gm26889 | ENSEMBL:ENSMUSG00000097249 | MGI:5477383 | lncRNA gene | predicted gene, 26889 |
| 4 | pseudogene | 58.26802 | 58.26820 | negative | MGI_C57BL6J_5590066 | Gm30907 | NCBI_Gene:102632965 | MGI:5590066 | pseudogene | predicted gene, 30907 |
| 4 | gene | 58.28572 | 58.37785 | positive | MGI_C57BL6J_103581 | Musk | NCBI_Gene:18198,ENSEMBL:ENSMUSG00000057280 | MGI:103581 | protein coding gene | muscle, skeletal, receptor tyrosine kinase |
| 4 | gene | 58.43525 | 58.55403 | negative | MGI_C57BL6J_108429 | Lpar1 | NCBI_Gene:14745,ENSEMBL:ENSMUSG00000038668 | MGI:108429 | protein coding gene | lysophosphatidic acid receptor 1 |
| 4 | gene | 58.44101 | 58.44110 | negative | MGI_C57BL6J_4834310 | Mir3095 | miRBase:MI0014088,NCBI_Gene:100526502,ENSEMBL:ENSMUSG00000093293 | MGI:4834310 | miRNA gene | microRNA 3095 |
| 4 | pseudogene | 58.48274 | 58.48383 | negative | MGI_C57BL6J_3651713 | Gm12577 | NCBI_Gene:100417556,ENSEMBL:ENSMUSG00000083834 | MGI:3651713 | pseudogene | predicted gene 12577 |
| 4 | pseudogene | 58.58976 | 58.59018 | positive | MGI_C57BL6J_3651712 | Gm12578 | ENSEMBL:ENSMUSG00000081186 | MGI:3651712 | pseudogene | predicted gene 12578 |
| 4 | pseudogene | 58.61106 | 58.61186 | positive | MGI_C57BL6J_3651711 | Gm12579 | NCBI_Gene:675306,ENSEMBL:ENSMUSG00000083145 | MGI:3651711 | pseudogene | predicted gene 12579 |
| 4 | gene | 58.64839 | 58.65859 | negative | MGI_C57BL6J_3651872 | Gm12580 | ENSEMBL:ENSMUSG00000086470 | MGI:3651872 | lncRNA gene | predicted gene 12580 |
| 4 | gene | 58.78270 | 58.78772 | negative | MGI_C57BL6J_3030101 | Olfr267 | NCBI_Gene:258922,ENSEMBL:ENSMUSG00000043385 | MGI:3030101 | protein coding gene | olfactory receptor 267 |
| 4 | gene | 58.79723 | 58.91311 | negative | MGI_C57BL6J_2140220 | Ecpas | NCBI_Gene:230249,ENSEMBL:ENSMUSG00000050812 | MGI:2140220 | protein coding gene | Ecm29 proteasome adaptor and scaffold |
| 4 | gene | 58.93803 | 58.93811 | positive | MGI_C57BL6J_5456177 | Gm26400 | ENSEMBL:ENSMUSG00000092753 | MGI:5456177 | rRNA gene | predicted gene, 26400 |
| 4 | gene | 58.94357 | 58.96262 | positive | MGI_C57BL6J_3510405 | Zkscan16 | NCBI_Gene:100041581,ENSEMBL:ENSMUSG00000038630 | MGI:3510405 | protein coding gene | zinc finger with KRAB and SCAN domains 16 |
| 4 | gene | 58.96544 | 58.98713 | negative | MGI_C57BL6J_1914353 | Ptgr1 | NCBI_Gene:67103,ENSEMBL:ENSMUSG00000028378 | MGI:1914353 | protein coding gene | prostaglandin reductase 1 |
| 4 | gene | 58.99480 | 59.02557 | positive | MGI_C57BL6J_1919679 | Dnajc25 | NCBI_Gene:72429,ENSEMBL:ENSMUSG00000070972 | MGI:1919679 | protein coding gene | DnaJ heat shock protein family (Hsp40) member C25 |
| 4 | gene | 59.00234 | 59.00308 | negative | MGI_C57BL6J_1921126 | 4930432F03Rik | NA | NA | unclassified gene | RIKEN cDNA 4930432F03 gene |
| 4 | gene | 59.00321 | 59.04189 | positive | MGI_C57BL6J_5141968 | Gm20503 | ENSEMBL:ENSMUSG00000092345 | MGI:5141968 | protein coding gene | predicted gene 20503 |
| 4 | gene | 59.03509 | 59.04190 | positive | MGI_C57BL6J_1336169 | Gng10 | NCBI_Gene:14700,ENSEMBL:ENSMUSG00000038607 | MGI:1336169 | protein coding gene | guanine nucleotide binding protein (G protein), gamma 10 |
| 4 | gene | 59.04198 | 59.13898 | negative | MGI_C57BL6J_2140313 | Shoc1 | NCBI_Gene:100155,ENSEMBL:ENSMUSG00000038598 | MGI:2140313 | protein coding gene | shortage in chiasmata 1 |
| 4 | pseudogene | 59.09019 | 59.09093 | positive | MGI_C57BL6J_3649249 | Gm12593 | ENSEMBL:ENSMUSG00000081432 | MGI:3649249 | pseudogene | predicted gene 12593 |
| 4 | gene | 59.11797 | 59.13581 | positive | MGI_C57BL6J_5594686 | Gm35527 | NCBI_Gene:102639146 | MGI:5594686 | lncRNA gene | predicted gene, 35527 |
| 4 | gene | 59.13507 | 59.13745 | negative | MGI_C57BL6J_3649447 | Gm12594 | ENSEMBL:ENSMUSG00000086660 | MGI:3649447 | lncRNA gene | predicted gene 12594 |
| 4 | pseudogene | 59.16220 | 59.16323 | negative | MGI_C57BL6J_3649246 | Rpsa-ps11 | NCBI_Gene:433712,ENSEMBL:ENSMUSG00000082978 | MGI:3649246 | pseudogene | ribosomal protein SA, pseudogene 11 |
| 4 | gene | 59.18917 | 59.22283 | positive | MGI_C57BL6J_1332243 | Ugcg | NCBI_Gene:22234,ENSEMBL:ENSMUSG00000028381 | MGI:1332243 | protein coding gene | UDP-glucose ceramide glucosyltransferase |
| 4 | gene | 59.26005 | 59.26914 | positive | MGI_C57BL6J_3649247 | Gm12596 | NCBI_Gene:101055769,ENSEMBL:ENSMUSG00000086952 | MGI:3649247 | lncRNA gene | predicted gene 12596 |
| 4 | gene | 59.26920 | 59.28171 | negative | MGI_C57BL6J_5594737 | Gm35578 | NCBI_Gene:102639218 | MGI:5594737 | lncRNA gene | predicted gene, 35578 |
| 4 | gene | 59.31420 | 59.43929 | negative | MGI_C57BL6J_3651543 | Susd1 | NCBI_Gene:634731,ENSEMBL:ENSMUSG00000038578 | MGI:3651543 | protein coding gene | sushi domain containing 1 |
| 4 | pseudogene | 59.36811 | 59.36887 | negative | MGI_C57BL6J_3651542 | Gm12527 | NCBI_Gene:666719,ENSEMBL:ENSMUSG00000083979 | MGI:3651542 | pseudogene | predicted gene 12527 |
| 4 | gene | 59.47187 | 59.54938 | negative | MGI_C57BL6J_1923334 | Ptbp3 | NCBI_Gene:230257,ENSEMBL:ENSMUSG00000028382 | MGI:1923334 | protein coding gene | polypyrimidine tract binding protein 3 |
| 4 | pseudogene | 59.53275 | 59.53301 | positive | MGI_C57BL6J_3651544 | Gm12529 | ENSEMBL:ENSMUSG00000080831 | MGI:3651544 | pseudogene | predicted gene 12529 |
| 4 | pseudogene | 59.54915 | 59.54962 | positive | MGI_C57BL6J_1919495 | 1700018M17Rik | ENSEMBL:ENSMUSG00000046145 | MGI:1919495 | pseudogene | RIKEN cDNA 1700018M17 gene |
| 4 | gene | 59.58133 | 59.61869 | positive | MGI_C57BL6J_1919729 | Hsdl2 | NCBI_Gene:72479,ENSEMBL:ENSMUSG00000028383 | MGI:1919729 | protein coding gene | hydroxysteroid dehydrogenase like 2 |
| 4 | gene | 59.62620 | 59.76144 | positive | MGI_C57BL6J_2442164 | E130308A19Rik | NCBI_Gene:230259,ENSEMBL:ENSMUSG00000045071 | MGI:2442164 | protein coding gene | RIKEN cDNA E130308A19 gene |
| 4 | gene | 59.76912 | 59.76939 | negative | MGI_C57BL6J_1919846 | 2700054B07Rik | NA | NA | unclassified gene | RIKEN cDNA 2700054B07 gene |
| 4 | gene | 59.76964 | 59.80159 | negative | MGI_C57BL6J_1913459 | Inip | NCBI_Gene:66209,ENSEMBL:ENSMUSG00000038544 | MGI:1913459 | protein coding gene | INTS3 and NABP interacting protein |
| 4 | gene | 59.80513 | 59.90474 | positive | MGI_C57BL6J_2443882 | Snx30 | NCBI_Gene:209131,ENSEMBL:ENSMUSG00000028385 | MGI:2443882 | protein coding gene | sorting nexin family member 30 |
| 4 | pseudogene | 59.83367 | 59.83422 | positive | MGI_C57BL6J_3649591 | Gm12543 | NCBI_Gene:100416087,ENSEMBL:ENSMUSG00000080772 | MGI:3649591 | pseudogene | predicted gene 12543 |
| 4 | gene | 59.88999 | 59.89492 | negative | MGI_C57BL6J_3651424 | Gm12542 | ENSEMBL:ENSMUSG00000073843 | MGI:3651424 | lncRNA gene | predicted gene 12542 |
| 4 | gene | 59.90513 | 59.91506 | negative | MGI_C57BL6J_1353616 | Slc46a2 | NCBI_Gene:30936,ENSEMBL:ENSMUSG00000028386 | MGI:1353616 | protein coding gene | solute carrier family 46, member 2 |
| 4 | gene | 59.95680 | 59.96071 | negative | MGI_C57BL6J_97236 | Mup4 | NCBI_Gene:17843,ENSEMBL:ENSMUSG00000041333 | MGI:97236 | protein coding gene | major urinary protein 4 |
| 4 | gene | 59.96429 | 60.00727 | positive | MGI_C57BL6J_3650962 | Mup6 | NCBI_Gene:620807,ENSEMBL:ENSMUSG00000078689 | MGI:3650962 | protein coding gene | major urinary protein 6 |
| 4 | pseudogene | 60.04131 | 60.10684 | positive | MGI_C57BL6J_3651069 | Mup-ps1 | NCBI_Gene:100417331,ENSEMBL:ENSMUSG00000083239 | MGI:3651069 | pseudogene | major urinary protein, pseudogene 1 |
| 4 | gene | 60.06647 | 60.07048 | negative | MGI_C57BL6J_3709615 | Mup7 | NCBI_Gene:100041658,ENSEMBL:ENSMUSG00000073842 | MGI:3709615 | protein coding gene | major urinary protein 7 |
| 4 | gene | 60.13591 | 60.15429 | negative | MGI_C57BL6J_97234 | Mup2 | NCBI_Gene:17841,ENSEMBL:ENSMUSG00000078688 | MGI:97234 | protein coding gene | major urinary protein 2 |
| 4 | pseudogene | 60.14075 | 60.15950 | positive | MGI_C57BL6J_3651073 | Mup-ps2 | NCBI_Gene:620882,ENSEMBL:ENSMUSG00000073840 | MGI:3651073 | pseudogene | major urinary protein, pseudogene 2 |
| 4 | gene | 60.21862 | 60.22260 | negative | MGI_C57BL6J_3709619 | Mup8 | NCBI_Gene:100041687,ENSEMBL:ENSMUSG00000078687 | MGI:3709619 | protein coding gene | major urinary protein 8 |
| 4 | pseudogene | 60.23734 | 60.24489 | positive | MGI_C57BL6J_3650796 | Mup-ps3 | NCBI_Gene:102639433,ENSEMBL:ENSMUSG00000095455 | MGI:3650796 | pseudogene | major urinary protein, pseudogene 3 |
| 4 | pseudogene | 60.38449 | 60.38470 | positive | MGI_C57BL6J_3783596 | Mup-ps29 | ENSEMBL:ENSMUSG00000096654 | MGI:3783596 | pseudogene | major urinary protein, pseudogene 29 |
| 4 | pseudogene | 60.38986 | 60.39015 | positive | MGI_C57BL6J_3782925 | Mup-ps28 | ENSEMBL:ENSMUSG00000083293 | MGI:3782925 | pseudogene | major urinary protein, pseudogene 28 |
| 4 | gene | 60.41805 | 60.42196 | negative | MGI_C57BL6J_3782918 | Mup9 | NCBI_Gene:100038948,ENSEMBL:ENSMUSG00000078686 | MGI:3782918 | protein coding gene | major urinary protein 9 |
| 4 | pseudogene | 60.43736 | 60.44006 | positive | MGI_C57BL6J_3782916 | Mup-ps4 | NCBI_Gene:620997,ENSEMBL:ENSMUSG00000078685 | MGI:3782916 | pseudogene | major urinary protein, pseudogene 4 |
| 4 | pseudogene | 60.47941 | 60.47961 | positive | MGI_C57BL6J_3705793 | Mup-ps26 | ENSEMBL:ENSMUSG00000094202 | MGI:3705793 | pseudogene | major urinary protein, pseudogene 26 |
| 4 | gene | 60.49801 | 60.50196 | negative | MGI_C57BL6J_97233 | Mup1 | NCBI_Gene:17840,ENSEMBL:ENSMUSG00000078683 | MGI:97233 | protein coding gene | major urinary protein 1 |
| 4 | gene | 60.50284 | 60.52123 | positive | MGI_C57BL6J_3712316 | Gm13773 | NCBI_Gene:102639616,ENSEMBL:ENSMUSG00000086253 | MGI:3712316 | lncRNA gene | predicted gene 13773 |
| 4 | pseudogene | 60.51746 | 60.52018 | positive | MGI_C57BL6J_3647161 | Mup-ps6 | ENSEMBL:ENSMUSG00000094222 | MGI:3647161 | pseudogene | major urinary protein, pseudogene 6 |
| 4 | pseudogene | 60.55757 | 60.55786 | positive | MGI_C57BL6J_3650405 | Mup-ps24 | ENSEMBL:ENSMUSG00000094235 | MGI:3650405 | pseudogene | major urinary protein, pseudogene 24 |
| 4 | gene | 60.57826 | 60.58220 | negative | MGI_C57BL6J_1924164 | Mup10 | NCBI_Gene:100039008,ENSEMBL:ENSMUSG00000078680 | MGI:1924164 | protein coding gene | major urinary protein 10 |
| 4 | pseudogene | 60.59760 | 60.59770 | positive | MGI_C57BL6J_3650601 | Mup-ps5 | ENSEMBL:ENSMUSG00000082868 | MGI:3650601 | pseudogene | major urinary protein, pseudogene 5 |
| 4 | gene | 60.65847 | 60.66241 | negative | MGI_C57BL6J_3709617 | Mup11 | NCBI_Gene:100039028,ENSEMBL:ENSMUSG00000073834 | MGI:3709617 | protein coding gene | major urinary protein 11 |
| 4 | pseudogene | 60.67768 | 60.68148 | positive | MGI_C57BL6J_3651245 | Mup-ps7 | NCBI_Gene:620982,ENSEMBL:ENSMUSG00000094743 | MGI:3651245 | pseudogene | major urinary protein, pseudogene 7 |
| 4 | gene | 60.73738 | 60.74133 | negative | MGI_C57BL6J_3780193 | Mup12 | NCBI_Gene:100039054,ENSEMBL:ENSMUSG00000094793 | MGI:3780193 | protein coding gene | major urinary protein 12 |
| 4 | pseudogene | 60.74239 | 60.76058 | positive | MGI_C57BL6J_3780197 | Mup-ps8 | NCBI_Gene:100039063,ENSEMBL:ENSMUSG00000096891 | MGI:3780197 | pseudogene | major urinary protein, pseudogene 8 |
| 4 | gene | 60.81759 | 60.82159 | negative | MGI_C57BL6J_5434675 | Mup22 | NCBI_Gene:100861909,ENSEMBL:ENSMUSG00000110439 | MGI:5434675 | protein coding gene | major urinary protein 22 |
| 4 | pseudogene | 60.83691 | 60.84055 | positive | MGI_C57BL6J_5434641 | Gm21286 | NCBI_Gene:102640145,ENSEMBL:ENSMUSG00000094948 | MGI:5434641 | pseudogene | predicted gene, 21286 |
| 4 | pseudogene | 61.16306 | 61.16670 | positive | MGI_C57BL6J_3649622 | Mup-ps9 | NCBI_Gene:667234,ENSEMBL:ENSMUSG00000096652 | MGI:3649622 | pseudogene | major urinary protein, pseudogene 9 |
| 4 | gene | 61.22431 | 61.22829 | negative | MGI_C57BL6J_3702003 | Mup13 | NCBI_Gene:100039089,ENSEMBL:ENSMUSG00000089873 | MGI:3702003 | protein coding gene | major urinary protein 13 |
| 4 | pseudogene | 61.22909 | 61.24734 | positive | MGI_C57BL6J_3649623 | Mup-ps10 | NCBI_Gene:108168721,ENSEMBL:ENSMUSG00000082173 | MGI:3649623 | pseudogene | major urinary protein, pseudogene 10 |
| 4 | gene | 61.30002 | 61.30400 | negative | MGI_C57BL6J_3702005 | Mup14 | NCBI_Gene:100039116,ENSEMBL:ENSMUSG00000073830 | MGI:3702005 | protein coding gene | major urinary protein 14 |
| 4 | pseudogene | 61.31873 | 61.32638 | positive | MGI_C57BL6J_3649621 | Mup-ps11 | NCBI_Gene:102640754,ENSEMBL:ENSMUSG00000095978 | MGI:3649621 | pseudogene | major urinary protein, pseudogene 11 |
| 4 | gene | 61.43579 | 61.43974 | negative | MGI_C57BL6J_3780235 | Mup15 | NCBI_Gene:100039150,ENSEMBL:ENSMUSG00000096674 | MGI:3780235 | protein coding gene | major urinary protein 15 |
| 4 | pseudogene | 61.44059 | 61.53848 | positive | MGI_C57BL6J_3783148 | Mup-ps12 | NCBI_Gene:100039019,ENSEMBL:ENSMUSG00000073835 | MGI:3783148 | pseudogene | major urinary protein, pseudogene 12 |
| 4 | gene | 61.51559 | 61.51953 | negative | MGI_C57BL6J_3780250 | Mup16 | NCBI_Gene:100039177,ENSEMBL:ENSMUSG00000078675 | MGI:3780250 | protein coding gene | major urinary protein 16 |
| 4 | pseudogene | 61.53423 | 61.53697 | positive | MGI_C57BL6J_3802118 | Mup-ps13 | ENSEMBL:ENSMUSG00000095532 | MGI:3802118 | pseudogene | major urinary protein, pseudogene 13 |
| 4 | gene | 61.59193 | 61.59587 | negative | MGI_C57BL6J_3705217 | Mup17 | NCBI_Gene:100039206,ENSEMBL:ENSMUSG00000096688 | MGI:3705217 | protein coding gene | major urinary protein 17 |
| 4 | pseudogene | 61.60828 | 61.61203 | positive | MGI_C57BL6J_3651980 | Mup-ps14 | NCBI_Gene:621127,ENSEMBL:ENSMUSG00000082065 | MGI:3651980 | pseudogene | major urinary protein, pseudogene 14 |
| 4 | gene | 61.67018 | 61.67414 | negative | MGI_C57BL6J_3705220 | Mup18 | NCBI_Gene:100048884,ENSEMBL:ENSMUSG00000078674 | MGI:3705220 | protein coding gene | major urinary protein 18 |
| 4 | pseudogene | 61.67516 | 61.70243 | positive | MGI_C57BL6J_3652152 | Mup-ps15 | NCBI_Gene:102631971,ENSEMBL:ENSMUSG00000083304 | MGI:3652152 | pseudogene | major urinary protein, pseudogene 15 |
| 4 | pseudogene | 61.75008 | 61.75036 | positive | MGI_C57BL6J_3649353 | Gm12573 | ENSEMBL:ENSMUSG00000095307 | MGI:3649353 | pseudogene | predicted gene 12573 |
| 4 | gene | 61.77832 | 61.78227 | negative | MGI_C57BL6J_3705235 | Mup19 | NCBI_Gene:100189605,ENSEMBL:ENSMUSG00000078673 | MGI:3705235 | protein coding gene | major urinary protein 19 |
| 4 | pseudogene | 61.78306 | 61.80123 | positive | MGI_C57BL6J_3645603 | Mup-ps16 | NCBI_Gene:667278,ENSEMBL:ENSMUSG00000090090 | MGI:3645603 | pseudogene | major urinary protein, pseudogene 16 |
| 4 | gene | 61.78312 | 61.80129 | positive | MGI_C57BL6J_3712466 | Gm13775 | ENSEMBL:ENSMUSG00000081207 | MGI:3712466 | lncRNA gene | predicted gene 13775 |
| 4 | gene | 61.83132 | 61.85804 | negative | MGI_C57BL6J_104974 | Mup5 | NCBI_Gene:17844,ENSEMBL:ENSMUSG00000058523 | MGI:104974 | protein coding gene | major urinary protein 5 |
| 4 | pseudogene | 61.85540 | 61.85655 | negative | MGI_C57BL6J_3650815 | Mup-ps23 | ENSEMBL:ENSMUSG00000081755 | MGI:3650815 | pseudogene | major urinary protein, pseudogene 23 |
| 4 | pseudogene | 61.85732 | 61.85788 | negative | MGI_C57BL6J_3705843 | Mup-ps17 | ENSEMBL:ENSMUSG00000081053 | MGI:3705843 | pseudogene | major urinary protein, pseudogene 17 |
| 4 | pseudogene | 61.87470 | 61.88332 | negative | MGI_C57BL6J_3651979 | Mup-ps18 | NCBI_Gene:667295,ENSEMBL:ENSMUSG00000082363 | MGI:3651979 | pseudogene | major urinary protein, pseudogene 18 |
| 4 | pseudogene | 61.94899 | 61.95998 | negative | MGI_C57BL6J_3652149 | Mup-ps19 | NCBI_Gene:100039305,ENSEMBL:ENSMUSG00000082644 | MGI:3652149 | pseudogene | major urinary protein, pseudogene 19 |
| 4 | gene | 62.01141 | 62.05416 | negative | MGI_C57BL6J_3651981 | Mup20 | NCBI_Gene:381530,ENSEMBL:ENSMUSG00000078672 | MGI:3651981 | protein coding gene | major urinary protein 20 |
| 4 | pseudogene | 62.01241 | 62.01486 | negative | MGI_C57BL6J_3651976 | Mup-ps20 | ENSEMBL:ENSMUSG00000084309 | MGI:3651976 | pseudogene | major urinary protein, pseudogene 20 |
| 4 | pseudogene | 62.02840 | 62.03167 | negative | MGI_C57BL6J_3652151 | Mup-ps21 | NCBI_Gene:546832,ENSEMBL:ENSMUSG00000083524 | MGI:3652151 | pseudogene | major urinary protein, pseudogene 21 |
| 4 | gene | 62.08348 | 62.08737 | negative | MGI_C57BL6J_97235 | Mup3 | NCBI_Gene:17842,ENSEMBL:ENSMUSG00000066154 | MGI:97235 | protein coding gene | major urinary protein 3 |
| 4 | gene | 62.08828 | 62.09844 | positive | MGI_C57BL6J_3651135 | Gm12909 | NCBI_Gene:105243851,ENSEMBL:ENSMUSG00000085404 | MGI:3651135 | lncRNA gene | predicted gene 12909 |
| 4 | gene | 62.11293 | 62.12093 | positive | MGI_C57BL6J_3651134 | Gm12910 | ENSEMBL:ENSMUSG00000086530 | MGI:3651134 | lncRNA gene | predicted gene 12910 |
| 4 | gene | 62.14783 | 62.15086 | negative | MGI_C57BL6J_3650630 | Mup21 | NCBI_Gene:381531,ENSEMBL:ENSMUSG00000066153 | MGI:3650630 | protein coding gene | major urinary protein 21 |
| 4 | gene | 62.18954 | 62.23105 | negative | MGI_C57BL6J_99181 | Zfp37 | NCBI_Gene:22696,ENSEMBL:ENSMUSG00000028389 | MGI:99181 | protein coding gene | zinc finger protein 37 |
| 4 | gene | 62.20892 | 62.20940 | positive | MGI_C57BL6J_5589610 | Gm30451 | NCBI_Gene:102632358 | MGI:5589610 | lncRNA gene | predicted gene, 30451 |
| 4 | gene | 62.22579 | 62.22655 | negative | MGI_C57BL6J_5589670 | Gm30511 | NA | NA | unclassified non-coding RNA gene | predicted gene%2c 30511 |
| 4 | gene | 62.26256 | 62.29841 | positive | MGI_C57BL6J_1333844 | Slc31a2 | NCBI_Gene:20530,ENSEMBL:ENSMUSG00000066152 | MGI:1333844 | protein coding gene | solute carrier family 31, member 2 |
| 4 | gene | 62.26431 | 62.26442 | positive | MGI_C57BL6J_5452925 | Gm23148 | ENSEMBL:ENSMUSG00000096713 | MGI:5452925 | snRNA gene | predicted gene, 23148 |
| 4 | gene | 62.30034 | 62.36059 | negative | MGI_C57BL6J_2444782 | Fkbp15 | NCBI_Gene:338355,ENSEMBL:ENSMUSG00000066151 | MGI:2444782 | protein coding gene | FK506 binding protein 15 |
| 4 | gene | 62.36058 | 62.39177 | positive | MGI_C57BL6J_1333843 | Slc31a1 | NCBI_Gene:20529,ENSEMBL:ENSMUSG00000066150 | MGI:1333843 | protein coding gene | solute carrier family 31, member 1 |
| 4 | gene | 62.38365 | 62.40864 | negative | MGI_C57BL6J_1913690 | Cdc26 | NCBI_Gene:66440,ENSEMBL:ENSMUSG00000066149 | MGI:1913690 | protein coding gene | cell division cycle 26 |
| 4 | gene | 62.40877 | 62.42699 | positive | MGI_C57BL6J_1917302 | Prpf4 | NCBI_Gene:70052,ENSEMBL:ENSMUSG00000066148 | MGI:1917302 | protein coding gene | pre-mRNA processing factor 4 |
| 4 | gene | 62.42754 | 62.43525 | negative | MGI_C57BL6J_1923322 | Rnf183 | NCBI_Gene:76072,ENSEMBL:ENSMUSG00000063851 | MGI:1923322 | protein coding gene | ring finger protein 183 |
| 4 | gene | 62.43460 | 62.44385 | positive | MGI_C57BL6J_5589748 | Gm30589 | NCBI_Gene:102632542 | MGI:5589748 | lncRNA gene | predicted gene, 30589 |
| 4 | gene | 62.44865 | 62.47096 | negative | MGI_C57BL6J_1918604 | Wdr31 | NCBI_Gene:71354,ENSEMBL:ENSMUSG00000028391 | MGI:1918604 | protein coding gene | WD repeat domain 31 |
| 4 | gene | 62.48000 | 62.49730 | positive | MGI_C57BL6J_2177191 | Bspry | NCBI_Gene:192120,ENSEMBL:ENSMUSG00000028392 | MGI:2177191 | protein coding gene | B-box and SPRY domain containing |
| 4 | gene | 62.49901 | 62.50347 | negative | MGI_C57BL6J_1919998 | Hdhd3 | NCBI_Gene:72748,ENSEMBL:ENSMUSG00000038422 | MGI:1919998 | protein coding gene | haloacid dehalogenase-like hydrolase domain containing 3 |
| 4 | gene | 62.50598 | 62.52006 | negative | MGI_C57BL6J_96853 | Alad | NCBI_Gene:17025,ENSEMBL:ENSMUSG00000028393 | MGI:96853 | protein coding gene | aminolevulinate, delta-, dehydratase |
| 4 | gene | 62.52265 | 62.52507 | negative | MGI_C57BL6J_1933378 | Pole3 | NCBI_Gene:59001,ENSEMBL:ENSMUSG00000028394 | MGI:1933378 | protein coding gene | polymerase (DNA directed), epsilon 3 (p17 subunit) |
| 4 | gene | 62.52511 | 62.55303 | positive | MGI_C57BL6J_3045314 | 4933430I17Rik | NCBI_Gene:214106,ENSEMBL:ENSMUSG00000058046 | MGI:3045314 | protein coding gene | RIKEN cDNA 4933430I17 gene |
| 4 | gene | 62.55983 | 62.70400 | positive | MGI_C57BL6J_1354734 | Rgs3 | NCBI_Gene:50780,ENSEMBL:ENSMUSG00000059810 | MGI:1354734 | protein coding gene | regulator of G-protein signaling 3 |
| 4 | gene | 62.62768 | 62.63371 | negative | MGI_C57BL6J_3650632 | Gm11209 | ENSEMBL:ENSMUSG00000085177 | MGI:3650632 | lncRNA gene | predicted gene 11209 |
| 4 | gene | 62.70598 | 62.74715 | positive | MGI_C57BL6J_3650633 | Gm11210 | NCBI_Gene:105247140,ENSEMBL:ENSMUSG00000085223 | MGI:3650633 | lncRNA gene | predicted gene 11210 |
| 4 | gene | 62.72351 | 62.72790 | negative | MGI_C57BL6J_3651244 | Gm11211 | NCBI_Gene:102632611,ENSEMBL:ENSMUSG00000086139 | MGI:3651244 | lncRNA gene | predicted gene 11211 |
| 4 | gene | 62.76521 | 62.77558 | negative | MGI_C57BL6J_5589858 | Gm30699 | NCBI_Gene:102632694 | MGI:5589858 | lncRNA gene | predicted gene, 30699 |
| 4 | gene | 62.81243 | 62.81256 | positive | MGI_C57BL6J_5453894 | Gm24117 | ENSEMBL:ENSMUSG00000077161 | MGI:5453894 | snoRNA gene | predicted gene, 24117 |
| 4 | gene | 62.93934 | 62.93947 | positive | MGI_C57BL6J_5455168 | Gm25391 | ENSEMBL:ENSMUSG00000087967 | MGI:5455168 | snoRNA gene | predicted gene, 25391 |
| 4 | pseudogene | 62.93952 | 62.93977 | positive | MGI_C57BL6J_3652240 | Gm11481 | ENSEMBL:ENSMUSG00000082048 | MGI:3652240 | pseudogene | predicted gene 11481 |
| 4 | pseudogene | 62.93991 | 62.94057 | positive | MGI_C57BL6J_3652010 | Gm11480 | ENSEMBL:ENSMUSG00000083635 | MGI:3652010 | pseudogene | predicted gene 11480 |
| 4 | gene | 62.96556 | 63.13971 | positive | MGI_C57BL6J_1919950 | Zfp618 | NCBI_Gene:72701,ENSEMBL:ENSMUSG00000028358 | MGI:1919950 | protein coding gene | zinc finger protein 618 |
| 4 | gene | 62.97867 | 62.97962 | negative | MGI_C57BL6J_5589923 | Gm30764 | NCBI_Gene:102632780 | MGI:5589923 | lncRNA gene | predicted gene, 30764 |
| 4 | gene | 63.14328 | 63.15480 | negative | MGI_C57BL6J_88002 | Ambp | NCBI_Gene:11699,ENSEMBL:ENSMUSG00000028356 | MGI:88002 | protein coding gene | alpha 1 microglobulin/bikunin precursor |
| 4 | gene | 63.15795 | 63.17215 | negative | MGI_C57BL6J_1098232 | Kif12 | NCBI_Gene:16552,ENSEMBL:ENSMUSG00000028357 | MGI:1098232 | protein coding gene | kinesin family member 12 |
| 4 | gene | 63.21400 | 63.33499 | positive | MGI_C57BL6J_2672118 | Col27a1 | NCBI_Gene:373864,ENSEMBL:ENSMUSG00000045672 | MGI:2672118 | protein coding gene | collagen, type XXVII, alpha 1 |
| 4 | gene | 63.25685 | 63.25693 | positive | MGI_C57BL6J_3629649 | Mir455 | miRBase:MI0004679,NCBI_Gene:735262,ENSEMBL:ENSMUSG00000070102 | MGI:3629649 | miRNA gene | microRNA 455 |
| 4 | gene | 63.34456 | 63.34816 | positive | MGI_C57BL6J_97443 | Orm1 | NCBI_Gene:18405,ENSEMBL:ENSMUSG00000039196 | MGI:97443 | protein coding gene | orosomucoid 1 |
| 4 | pseudogene | 63.35037 | 63.35396 | positive | MGI_C57BL6J_3651243 | Gm11212 | NCBI_Gene:435786,ENSEMBL:ENSMUSG00000082141 | MGI:3651243 | pseudogene | predicted gene 11212 |
| 4 | gene | 63.35528 | 63.35951 | positive | MGI_C57BL6J_97445 | Orm3 | NCBI_Gene:18407,ENSEMBL:ENSMUSG00000028359 | MGI:97445 | protein coding gene | orosomucoid 3 |
| 4 | gene | 63.36245 | 63.36588 | positive | MGI_C57BL6J_97444 | Orm2 | NCBI_Gene:18406,ENSEMBL:ENSMUSG00000061540 | MGI:97444 | protein coding gene | orosomucoid 2 |
| 4 | gene | 63.36712 | 63.41076 | negative | MGI_C57BL6J_2140340 | Akna | NCBI_Gene:100182,ENSEMBL:ENSMUSG00000039158 | MGI:2140340 | protein coding gene | AT-hook transcription factor |
| 4 | gene | 63.37706 | 63.39402 | positive | MGI_C57BL6J_1914033 | Aknaos | NCBI_Gene:66783,ENSEMBL:ENSMUSG00000086953 | MGI:1914033 | antisense lncRNA gene | AT-hook transcription factor, opposite strand |
| 4 | gene | 63.41491 | 63.49608 | negative | MGI_C57BL6J_2682003 | Whrn | NCBI_Gene:73750,ENSEMBL:ENSMUSG00000039137 | MGI:2682003 | protein coding gene | whirlin |
| 4 | gene | 63.54476 | 63.55075 | positive | MGI_C57BL6J_1913540 | Atp6v1g1 | NCBI_Gene:66290,ENSEMBL:ENSMUSG00000039105 | MGI:1913540 | protein coding gene | ATPase, H+ transporting, lysosomal V1 subunit G1 |
| 4 | gene | 63.54529 | 63.54786 | positive | MGI_C57BL6J_5621477 | Gm38592 | NCBI_Gene:102642011 | MGI:5621477 | lncRNA gene | predicted gene, 38592 |
| 4 | gene | 63.55842 | 63.58636 | positive | MGI_C57BL6J_1913920 | Tmem268 | NCBI_Gene:230279,ENSEMBL:ENSMUSG00000045917 | MGI:1913920 | protein coding gene | transmembrane protein 268 |
| 4 | pseudogene | 63.56741 | 63.56766 | negative | MGI_C57BL6J_3652237 | Gm11483 | ENSEMBL:ENSMUSG00000083943 | MGI:3652237 | pseudogene | predicted gene 11483 |
| 4 | gene | 63.56766 | 63.56777 | negative | MGI_C57BL6J_5454225 | Gm24448 | ENSEMBL:ENSMUSG00000065913 | MGI:5454225 | snRNA gene | predicted gene, 24448 |
| 4 | gene | 63.59690 | 63.60526 | negative | MGI_C57BL6J_3701774 | Gm11213 | NCBI_Gene:670833,ENSEMBL:ENSMUSG00000084782 | MGI:3701774 | lncRNA gene | predicted gene 11213 |
| 4 | gene | 63.60708 | 63.62245 | negative | MGI_C57BL6J_1922774 | Tex48 | NCBI_Gene:75524,ENSEMBL:ENSMUSG00000058935 | MGI:1922774 | protein coding gene | testis expressed 48 |
| 4 | gene | 63.63168 | 63.63179 | negative | MGI_C57BL6J_5454011 | Gm24234 | ENSEMBL:ENSMUSG00000077504 | MGI:5454011 | snRNA gene | predicted gene, 24234 |
| 4 | gene | 63.63441 | 63.64198 | positive | MGI_C57BL6J_3652238 | Gm11482 | NCBI_Gene:102632940,ENSEMBL:ENSMUSG00000086817 | MGI:3652238 | lncRNA gene | predicted gene 11482 |
| 4 | gene | 63.66216 | 63.66339 | negative | MGI_C57BL6J_3651045 | Gm11214 | NCBI_Gene:667451,ENSEMBL:ENSMUSG00000099294 | MGI:3651045 | protein coding gene | predicted gene 11214 |
| 4 | gene | 63.69638 | 63.74487 | positive | MGI_C57BL6J_5625180 | Gm42295 | NCBI_Gene:105247142 | MGI:5625180 | lncRNA gene | predicted gene, 42295 |
| 4 | gene | 63.72460 | 63.74511 | negative | MGI_C57BL6J_2180140 | Tnfsf15 | NCBI_Gene:326623,ENSEMBL:ENSMUSG00000050395 | MGI:2180140 | protein coding gene | tumor necrosis factor (ligand) superfamily, member 15 |
| 4 | gene | 63.74503 | 63.77318 | positive | MGI_C57BL6J_5625179 | Gm42294 | NCBI_Gene:105247141 | MGI:5625179 | lncRNA gene | predicted gene, 42294 |
| 4 | gene | 63.83131 | 63.86151 | negative | MGI_C57BL6J_88328 | Tnfsf8 | NCBI_Gene:21949,ENSEMBL:ENSMUSG00000028362 | MGI:88328 | protein coding gene | tumor necrosis factor (ligand) superfamily, member 8 |
| 4 | pseudogene | 63.89551 | 63.89612 | negative | MGI_C57BL6J_3651043 | Rpl17-ps4 | NCBI_Gene:435787,ENSEMBL:ENSMUSG00000084345 | MGI:3651043 | pseudogene | ribosomal protein L17, pseudogene 4 |
| 4 | gene | 63.95978 | 64.04702 | negative | MGI_C57BL6J_101922 | Tnc | NCBI_Gene:21923,ENSEMBL:ENSMUSG00000028364 | MGI:101922 | protein coding gene | tenascin C |
| 4 | gene | 63.97985 | 64.14992 | positive | MGI_C57BL6J_1915353 | 8030451A03Rik | NCBI_Gene:100504061,ENSEMBL:ENSMUSG00000073821 | MGI:1915353 | lncRNA gene | RIKEN cDNA 8030451A03 gene |
| 4 | gene | 64.07553 | 64.08381 | positive | MGI_C57BL6J_3651044 | Gm11216 | ENSEMBL:ENSMUSG00000086755 | MGI:3651044 | lncRNA gene | predicted gene 11216 |
| 4 | gene | 64.15428 | 64.28612 | positive | MGI_C57BL6J_5590274 | Gm31115 | NCBI_Gene:102633240 | MGI:5590274 | lncRNA gene | predicted gene, 31115 |
| 4 | gene | 64.25405 | 64.27660 | negative | MGI_C57BL6J_3651042 | Gm11217 | NCBI_Gene:102633436,ENSEMBL:ENSMUSG00000085032 | MGI:3651042 | lncRNA gene | predicted gene 11217 |
| 4 | gene | 64.44473 | 64.44486 | positive | MGI_C57BL6J_5453727 | Gm23950 | ENSEMBL:ENSMUSG00000077214 | MGI:5453727 | snoRNA gene | predicted gene, 23950 |
| 4 | pseudogene | 64.85629 | 64.85814 | negative | MGI_C57BL6J_3651636 | Gm11218 | NCBI_Gene:100502689,ENSEMBL:ENSMUSG00000081941 | MGI:3651636 | pseudogene | predicted gene 11218 |
| 4 | pseudogene | 65.01123 | 65.01159 | positive | MGI_C57BL6J_3651487 | Gm11219 | ENSEMBL:ENSMUSG00000080805 | MGI:3651487 | pseudogene | predicted gene 11219 |
| 4 | gene | 65.12417 | 65.35751 | positive | MGI_C57BL6J_97479 | Pappa | NCBI_Gene:18491,ENSEMBL:ENSMUSG00000028370 | MGI:97479 | protein coding gene | pregnancy-associated plasma protein A |
| 4 | gene | 65.38080 | 66.40461 | negative | MGI_C57BL6J_1889277 | Astn2 | NCBI_Gene:56079,ENSEMBL:ENSMUSG00000028373 | MGI:1889277 | protein coding gene | astrotactin 2 |
| 4 | gene | 65.60499 | 65.61624 | positive | MGI_C57BL6J_1917057 | Trim32 | NCBI_Gene:69807,ENSEMBL:ENSMUSG00000051675 | MGI:1917057 | protein coding gene | tripartite motif-containing 32 |
| 4 | gene | 65.62320 | 65.62606 | positive | MGI_C57BL6J_2441665 | 3632413A11Rik | NA | NA | unclassified gene | RIKEN cDNA 3632413A11 gene |
| 4 | gene | 65.62895 | 65.63480 | positive | MGI_C57BL6J_5590692 | Gm31533 | NCBI_Gene:102633792 | MGI:5590692 | lncRNA gene | predicted gene, 31533 |
| 4 | pseudogene | 66.35385 | 66.35419 | positive | MGI_C57BL6J_3651701 | Gm11484 | ENSEMBL:ENSMUSG00000083080 | MGI:3651701 | pseudogene | predicted gene 11484 |
| 4 | gene | 66.54406 | 66.54419 | positive | MGI_C57BL6J_5455257 | Gm25480 | ENSEMBL:ENSMUSG00000065151 | MGI:5455257 | snoRNA gene | predicted gene, 25480 |
| 4 | pseudogene | 66.72507 | 66.72554 | negative | MGI_C57BL6J_3651695 | Gm11220 | NCBI_Gene:667499,ENSEMBL:ENSMUSG00000082053 | MGI:3651695 | pseudogene | predicted gene 11220 |
| 4 | gene | 66.82755 | 66.93028 | positive | MGI_C57BL6J_96824 | Tlr4 | NCBI_Gene:21898,ENSEMBL:ENSMUSG00000039005 | MGI:96824 | protein coding gene | toll-like receptor 4 |
| 4 | pseudogene | 67.16548 | 67.16585 | positive | MGI_C57BL6J_3651007 | Gm11403 | ENSEMBL:ENSMUSG00000081435 | MGI:3651007 | pseudogene | predicted gene 11403 |
| 4 | pseudogene | 67.51831 | 67.51899 | negative | MGI_C57BL6J_104763 | Hmgb1-rs18 | NCBI_Gene:15298,ENSEMBL:ENSMUSG00000080815 | MGI:104763 | pseudogene | high mobility group box 1, related sequence 18 |
| 4 | gene | 67.63315 | 67.63323 | positive | MGI_C57BL6J_5690889 | Gm44497 | ENSEMBL:ENSMUSG00000106523 | MGI:5690889 | miRNA gene | predicted gene, 44497 |
| 4 | pseudogene | 67.93085 | 67.93178 | positive | MGI_C57BL6J_3650834 | Gm11249 | NCBI_Gene:433715,ENSEMBL:ENSMUSG00000083087 | MGI:3650834 | pseudogene | predicted gene 11249 |
| 4 | gene | 68.02303 | 68.13711 | positive | MGI_C57BL6J_3650096 | Gm11751 | ENSEMBL:ENSMUSG00000086364 | MGI:3650096 | lncRNA gene | predicted gene 11751 |
| 4 | gene | 68.18253 | 68.18270 | positive | MGI_C57BL6J_5453515 | Gm23738 | ENSEMBL:ENSMUSG00000064762 | MGI:5453515 | snRNA gene | predicted gene, 23738 |
| 4 | pseudogene | 68.30124 | 68.30338 | negative | MGI_C57BL6J_98536 | Tcp1-ps1 | NCBI_Gene:21455,ENSEMBL:ENSMUSG00000081231 | MGI:98536 | pseudogene | t-complex protein 1, pseudogene 1 |
| 4 | gene | 68.31247 | 68.32196 | negative | MGI_C57BL6J_3652009 | Gm12911 | NCBI_Gene:102633975,ENSEMBL:ENSMUSG00000086790 | MGI:3652009 | lncRNA gene | predicted gene 12911 |
| 4 | gene | 68.76137 | 68.95440 | negative | MGI_C57BL6J_1928478 | Brinp1 | NCBI_Gene:56710,ENSEMBL:ENSMUSG00000028351 | MGI:1928478 | protein coding gene | bone morphogenic protein/retinoic acid inducible neural specific 1 |
| 4 | gene | 68.80055 | 68.80163 | negative | MGI_C57BL6J_1924804 | 9330154F10Rik | NA | NA | unclassified gene | RIKEN cDNA 9330154F10 gene |
| 4 | gene | 68.95791 | 68.95967 | positive | MGI_C57BL6J_5625181 | Gm42296 | NCBI_Gene:105247144 | MGI:5625181 | lncRNA gene | predicted gene, 42296 |
| 4 | gene | 69.11067 | 69.11078 | positive | MGI_C57BL6J_5454474 | Gm24697 | ENSEMBL:ENSMUSG00000065815 | MGI:5454474 | snRNA gene | predicted gene, 24697 |
| 4 | gene | 69.16754 | 69.19983 | negative | MGI_C57BL6J_5826521 | Gm46884 | NCBI_Gene:108168996 | MGI:5826521 | lncRNA gene | predicted gene, 46884 |
| 4 | gene | 69.25930 | 69.26203 | negative | MGI_C57BL6J_5591064 | Gm31905 | NCBI_Gene:102634283 | MGI:5591064 | lncRNA gene | predicted gene, 31905 |
| 4 | pseudogene | 69.29161 | 69.29512 | negative | MGI_C57BL6J_3651011 | Gm11404 | NCBI_Gene:100418187,ENSEMBL:ENSMUSG00000082553 | MGI:3651011 | pseudogene | predicted gene 11404 |
| 4 | pseudogene | 69.69195 | 69.69265 | positive | MGI_C57BL6J_3651633 | Gm11221 | NCBI_Gene:622491,ENSEMBL:ENSMUSG00000082998 | MGI:3651633 | pseudogene | predicted gene 11221 |
| 4 | pseudogene | 69.85395 | 69.85468 | positive | MGI_C57BL6J_3651632 | Gm11222 | NCBI_Gene:433716,ENSEMBL:ENSMUSG00000085627 | MGI:3651632 | pseudogene | predicted gene 11222 |
| 4 | pseudogene | 69.92520 | 69.92617 | negative | MGI_C57BL6J_3651631 | Gm11223 | NCBI_Gene:100039888,ENSEMBL:ENSMUSG00000046341 | MGI:3651631 | pseudogene | predicted gene 11223 |
| 4 | pseudogene | 69.95417 | 69.95528 | negative | MGI_C57BL6J_3651640 | Gm11224 | NCBI_Gene:242497,ENSEMBL:ENSMUSG00000084342 | MGI:3651640 | pseudogene | predicted gene 11224 |
| 4 | pseudogene | 70.04716 | 70.04820 | positive | MGI_C57BL6J_3651505 | Gm11225 | NCBI_Gene:671064,ENSEMBL:ENSMUSG00000082368 | MGI:3651505 | pseudogene | predicted gene 11225 |
| 4 | gene | 70.21685 | 70.41044 | negative | MGI_C57BL6J_2384875 | Cdk5rap2 | NCBI_Gene:214444,ENSEMBL:ENSMUSG00000039298 | MGI:2384875 | protein coding gene | CDK5 regulatory subunit associated protein 2 |
| 4 | pseudogene | 70.24795 | 70.24944 | negative | MGI_C57BL6J_3650241 | Ywhaq-ps3 | NCBI_Gene:102634437,ENSEMBL:ENSMUSG00000080902 | MGI:3650241 | pseudogene | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein theta, pseudogene 3 |
| 4 | gene | 70.41262 | 70.41292 | negative | MGI_C57BL6J_5456097 | Gm26320 | ENSEMBL:ENSMUSG00000088367 | MGI:5456097 | unclassified non-coding RNA gene | predicted gene, 26320 |
| 4 | gene | 70.42706 | 70.53499 | negative | MGI_C57BL6J_1918264 | Megf9 | NCBI_Gene:230316,ENSEMBL:ENSMUSG00000039270 | MGI:1918264 | protein coding gene | multiple EGF-like-domains 9 |
| 4 | gene | 70.55368 | 70.66501 | positive | MGI_C57BL6J_5591368 | Gm32209 | NCBI_Gene:102634677 | MGI:5591368 | lncRNA gene | predicted gene, 32209 |
| 4 | gene | 70.67923 | 70.69171 | positive | MGI_C57BL6J_5625182 | Gm42297 | NCBI_Gene:105247145 | MGI:5625182 | lncRNA gene | predicted gene, 42297 |
| 4 | pseudogene | 71.07263 | 71.07477 | positive | MGI_C57BL6J_3651434 | Gm11227 | NCBI_Gene:100039565,ENSEMBL:ENSMUSG00000083549 | MGI:3651434 | pseudogene | predicted gene 11227 |
| 4 | gene | 71.07511 | 71.13113 | negative | MGI_C57BL6J_3651435 | Gm11228 | NCBI_Gene:102634783,ENSEMBL:ENSMUSG00000084937 | MGI:3651435 | lncRNA gene | predicted gene 11228 |
| 4 | pseudogene | 71.09460 | 71.09590 | negative | MGI_C57BL6J_5010719 | Gm18534 | NCBI_Gene:100417328 | MGI:5010719 | pseudogene | predicted gene, 18534 |
| 4 | pseudogene | 71.28594 | 71.28638 | positive | MGI_C57BL6J_3651948 | Gm11229 | ENSEMBL:ENSMUSG00000082017 | MGI:3651948 | pseudogene | predicted gene 11229 |
| 4 | pseudogene | 71.37069 | 71.37122 | negative | MGI_C57BL6J_3650249 | Rps18-ps1 | NCBI_Gene:100039979,ENSEMBL:ENSMUSG00000083914 | MGI:3650249 | pseudogene | ribosomal protein S18, pseudogene 1 |
| 4 | gene | 71.38197 | 71.39723 | positive | MGI_C57BL6J_5625183 | Gm42298 | NCBI_Gene:105247146 | MGI:5625183 | lncRNA gene | predicted gene, 42298 |
| 4 | pseudogene | 71.67747 | 71.67981 | positive | MGI_C57BL6J_3650253 | Gm11231 | NCBI_Gene:667638,ENSEMBL:ENSMUSG00000082432 | MGI:3650253 | pseudogene | predicted gene 11231 |
| 4 | gene | 71.75564 | 71.75796 | negative | MGI_C57BL6J_3651439 | Gm11232 | NCBI_Gene:102634852,ENSEMBL:ENSMUSG00000066141 | MGI:3651439 | protein coding gene | predicted gene 11232 |
| 4 | pseudogene | 71.86799 | 71.86898 | positive | MGI_C57BL6J_3650258 | Gm11233 | ENSEMBL:ENSMUSG00000081117 | MGI:3650258 | pseudogene | predicted gene 11233 |
| 4 | gene | 71.98048 | 71.99956 | positive | MGI_C57BL6J_3651441 | Gm11234 | ENSEMBL:ENSMUSG00000086696 | MGI:3651441 | lncRNA gene | predicted gene 11234 |
| 4 | gene | 72.10994 | 72.20105 | negative | MGI_C57BL6J_104636 | Tle1 | NCBI_Gene:21885,ENSEMBL:ENSMUSG00000008305 | MGI:104636 | protein coding gene | transducin-like enhancer of split 1 |
| 4 | gene | 72.16877 | 72.16945 | positive | MGI_C57BL6J_3651181 | Gm11250 | ENSEMBL:ENSMUSG00000086027 | MGI:3651181 | lncRNA gene | predicted gene 11250 |
| 4 | gene | 72.20124 | 72.21535 | positive | MGI_C57BL6J_1915535 | C630043F03Rik | NCBI_Gene:68285,ENSEMBL:ENSMUSG00000084910 | MGI:1915535 | lncRNA gene | RIKEN cDNA C630043F03 gene |
| 4 | pseudogene | 72.54267 | 72.54300 | positive | MGI_C57BL6J_3651442 | Gm11235 | ENSEMBL:ENSMUSG00000083372 | MGI:3651442 | pseudogene | predicted gene 11235 |
| 4 | gene | 72.85058 | 72.85263 | negative | MGI_C57BL6J_2447811 | Aldoart1 | NCBI_Gene:353204,ENSEMBL:ENSMUSG00000059343 | MGI:2447811 | protein coding gene | aldolase 1 A, retrogene 1 |
| 4 | pseudogene | 72.89378 | 72.89438 | positive | MGI_C57BL6J_3648217 | Gm5287 | NCBI_Gene:384113,ENSEMBL:ENSMUSG00000084173 | MGI:3648217 | pseudogene | predicted gene 5287 |
| 4 | gene | 72.90620 | 72.90634 | negative | MGI_C57BL6J_5455430 | Gm25653 | ENSEMBL:ENSMUSG00000089098 | MGI:5455430 | snoRNA gene | predicted gene, 25653 |
| 4 | pseudogene | 73.02982 | 73.03607 | positive | MGI_C57BL6J_3651054 | Gm11251 | NCBI_Gene:667713,ENSEMBL:ENSMUSG00000084162 | MGI:3651054 | pseudogene | predicted gene 11251 |
| 4 | gene | 73.11466 | 73.11478 | positive | MGI_C57BL6J_5455546 | Gm25769 | ENSEMBL:ENSMUSG00000089194 | MGI:5455546 | snoRNA gene | predicted gene, 25769 |
| 4 | gene | 73.39165 | 73.39193 | positive | MGI_C57BL6J_1100493 | Gtayu1 | NA | NA | unclassified gene | gene trap AYU 1 |
| 4 | gene | 73.40103 | 73.40740 | negative | MGI_C57BL6J_3650704 | Gm11487 | NCBI_Gene:433719,ENSEMBL:ENSMUSG00000066137 | MGI:3650704 | protein coding gene | predicted gene 11487 |
| 4 | pseudogene | 73.41530 | 73.41869 | negative | MGI_C57BL6J_3650706 | Gm11486 | ENSEMBL:ENSMUSG00000080222 | MGI:3650706 | pseudogene | predicted gene 11486 |
| 4 | pseudogene | 73.42215 | 73.42332 | negative | MGI_C57BL6J_3705569 | Gm11485 | NCBI_Gene:667722,ENSEMBL:ENSMUSG00000082566 | MGI:3705569 | pseudogene | predicted gene 11485 |
| 4 | pseudogene | 73.49761 | 73.49852 | negative | MGI_C57BL6J_3650705 | Gm11488 | NCBI_Gene:623115,ENSEMBL:ENSMUSG00000081871 | MGI:3650705 | pseudogene | predicted gene 11488 |
| 4 | gene | 73.58908 | 73.58921 | negative | MGI_C57BL6J_5453947 | Gm24170 | ENSEMBL:ENSMUSG00000077312 | MGI:5453947 | snoRNA gene | predicted gene, 24170 |
| 4 | gene | 73.60554 | 73.61192 | positive | MGI_C57BL6J_5621308 | Gm38423 | NCBI_Gene:100502924 | MGI:5621308 | protein coding gene | predicted gene, 38423 |
| 4 | gene | 73.60788 | 73.65729 | positive | MGI_C57BL6J_3702316 | Gm11238 | NCBI_Gene:100502896,ENSEMBL:ENSMUSG00000095935 | MGI:3702316 | protein coding gene | predicted gene 11238 |
| 4 | gene | 73.62065 | 73.62703 | positive | MGI_C57BL6J_3702292 | Gm11237 | NCBI_Gene:623197,ENSEMBL:ENSMUSG00000094116 | MGI:3702292 | protein coding gene | predicted gene 11237 |
| 4 | gene | 73.63578 | 73.64216 | positive | MGI_C57BL6J_3702277 | Gm11236 | NCBI_Gene:100862015,ENSEMBL:ENSMUSG00000096700 | MGI:3702277 | protein coding gene | predicted gene 11236 |
| 4 | gene | 73.66602 | 73.67240 | positive | MGI_C57BL6J_3702319 | Gm11239 | NCBI_Gene:101055863,ENSEMBL:ENSMUSG00000095048 | MGI:3702319 | protein coding gene | predicted gene 11239 |
| 4 | gene | 73.68115 | 73.68756 | positive | MGI_C57BL6J_2685274 | Gm428 | NCBI_Gene:242502,ENSEMBL:ENSMUSG00000095341 | MGI:2685274 | protein coding gene | predicted gene 428 |
| 4 | gene | 73.71458 | 73.79131 | negative | MGI_C57BL6J_2448565 | Rasef | NCBI_Gene:242505,ENSEMBL:ENSMUSG00000043003 | MGI:2448565 | protein coding gene | RAS and EF hand domain containing |
| 4 | gene | 73.79124 | 73.79838 | positive | MGI_C57BL6J_3651058 | Gm11240 | NCBI_Gene:667768,ENSEMBL:ENSMUSG00000085034 | MGI:3651058 | lncRNA gene | predicted gene 11240 |
| 4 | pseudogene | 73.86440 | 73.86509 | positive | MGI_C57BL6J_3650139 | Gm13867 | ENSEMBL:ENSMUSG00000083530 | MGI:3650139 | pseudogene | predicted gene 13867 |
| 4 | gene | 73.87222 | 73.87627 | negative | MGI_C57BL6J_3702099 | Gm11758 | NCBI_Gene:545637,ENSEMBL:ENSMUSG00000096530 | MGI:3702099 | protein coding gene | predicted gene 11758 |
| 4 | gene | 73.88707 | 73.89111 | negative | MGI_C57BL6J_3702097 | Gm11757 | NCBI_Gene:623272,ENSEMBL:ENSMUSG00000096750 | MGI:3702097 | protein coding gene | predicted gene 11757 |
| 4 | gene | 73.90191 | 73.90595 | negative | MGI_C57BL6J_3702126 | Gm13871 | NCBI_Gene:667780,ENSEMBL:ENSMUSG00000096333 | MGI:3702126 | protein coding gene | predicted gene 13871 |
| 4 | gene | 73.91673 | 73.92076 | negative | MGI_C57BL6J_3702095 | Gm11756 | NCBI_Gene:623281,ENSEMBL:ENSMUSG00000093962 | MGI:3702095 | protein coding gene | predicted gene 11756 |
| 4 | gene | 73.93937 | 73.95086 | negative | MGI_C57BL6J_1916780 | 2310002L09Rik | NCBI_Gene:71886,ENSEMBL:ENSMUSG00000028396 | MGI:1916780 | protein coding gene | RIKEN cDNA 2310002L09 gene |
| 4 | gene | 74.01344 | 74.20221 | positive | MGI_C57BL6J_2442466 | Frmd3 | NCBI_Gene:242506,ENSEMBL:ENSMUSG00000049122 | MGI:2442466 | protein coding gene | FERM domain containing 3 |
| 4 | pseudogene | 74.04437 | 74.04522 | positive | MGI_C57BL6J_3651479 | Gm11760 | NCBI_Gene:100384873,ENSEMBL:ENSMUSG00000081048 | MGI:3651479 | pseudogene | predicted gene 11760 |
| 4 | pseudogene | 74.22820 | 74.22975 | positive | MGI_C57BL6J_3651226 | Gm11405 | NCBI_Gene:100039774,ENSEMBL:ENSMUSG00000083767 | MGI:3651226 | pseudogene | predicted gene 11405 |
| 4 | gene | 74.24250 | 74.40586 | positive | MGI_C57BL6J_1924054 | Kdm4c | NCBI_Gene:76804,ENSEMBL:ENSMUSG00000028397 | MGI:1924054 | protein coding gene | lysine (K)-specific demethylase 4C |
| 4 | gene | 74.25032 | 74.25171 | negative | MGI_C57BL6J_3698879 | A230083N12Rik | ENSEMBL:ENSMUSG00000085482 | MGI:3698879 | lncRNA gene | RIKEN cDNA A230083N12 gene |
| 4 | gene | 74.36470 | 74.36501 | positive | MGI_C57BL6J_1919791 | 2700003L17Rik | NA | NA | unclassified gene | RIKEN cDNA 2700003L17 gene |
| 4 | gene | 74.63521 | 74.63535 | positive | MGI_C57BL6J_5456156 | Gm26379 | ENSEMBL:ENSMUSG00000099334,ENSEMBL:ENSMUSG00000104700 | MGI:5456156 | snoRNA gene | predicted gene, 26379 |
| 4 | pseudogene | 74.66367 | 74.66453 | positive | MGI_C57BL6J_3649979 | Gm11254 | NCBI_Gene:100417943,ENSEMBL:ENSMUSG00000081438 | MGI:3649979 | pseudogene | predicted gene 11254 |
| 4 | pseudogene | 74.68766 | 74.68934 | negative | MGI_C57BL6J_3649983 | Gm11258 | NCBI_Gene:100039814,ENSEMBL:ENSMUSG00000083659 | MGI:3649983 | pseudogene | predicted gene 11258 |
| 4 | pseudogene | 74.90396 | 74.90492 | negative | MGI_C57BL6J_5010929 | Gm18744 | NCBI_Gene:100417656 | MGI:5010929 | pseudogene | predicted gene, 18744 |
| 4 | pseudogene | 75.06801 | 75.07197 | positive | MGI_C57BL6J_5826499 | Gm46862 | NCBI_Gene:108168965 | MGI:5826499 | pseudogene | predicted gene, 46862 |
| 4 | pseudogene | 75.08150 | 75.08170 | negative | MGI_C57BL6J_3649981 | Gm11253 | ENSEMBL:ENSMUSG00000082533 | MGI:3649981 | pseudogene | predicted gene 11253 |
| 4 | pseudogene | 75.12871 | 75.12964 | positive | MGI_C57BL6J_3649985 | Gm11257 | NCBI_Gene:100384870,ENSEMBL:ENSMUSG00000084311 | MGI:3649985 | pseudogene | predicted gene 11257 |
| 4 | pseudogene | 75.14754 | 75.14931 | positive | MGI_C57BL6J_3649320 | Gm12912 | NCBI_Gene:636847,ENSEMBL:ENSMUSG00000081067 | MGI:3649320 | pseudogene | predicted gene 12912 |
| 4 | gene | 75.27735 | 75.27831 | negative | MGI_C57BL6J_1914178 | Dmac1 | NCBI_Gene:66928,ENSEMBL:ENSMUSG00000028398 | MGI:1914178 | protein coding gene | distal membrane arm assembly complex 1 |
| 4 | gene | 75.28063 | 75.28075 | negative | MGI_C57BL6J_5452641 | Gm22864 | ENSEMBL:ENSMUSG00000065884 | MGI:5452641 | snoRNA gene | predicted gene, 22864 |
| 4 | pseudogene | 75.32450 | 75.32568 | positive | MGI_C57BL6J_3649873 | Gm11255 | NCBI_Gene:623529,ENSEMBL:ENSMUSG00000083224 | MGI:3649873 | pseudogene | predicted gene 11255 |
| 4 | pseudogene | 75.33090 | 75.33235 | positive | MGI_C57BL6J_5010839 | Gm18654 | NCBI_Gene:100417504 | MGI:5010839 | pseudogene | predicted gene, 18654 |
| 4 | pseudogene | 75.38076 | 75.38189 | positive | MGI_C57BL6J_3649975 | Gm11259 | NCBI_Gene:100039833,ENSEMBL:ENSMUSG00000084026 | MGI:3649975 | pseudogene | predicted gene 11259 |
| 4 | gene | 75.55963 | 75.58876 | negative | MGI_C57BL6J_5625185 | Gm42300 | NCBI_Gene:105247148 | MGI:5625185 | lncRNA gene | predicted gene, 42300 |
| 4 | gene | 75.56729 | 75.58372 | negative | MGI_C57BL6J_5625184 | Gm42299 | NCBI_Gene:105247147 | MGI:5625184 | lncRNA gene | predicted gene, 42299 |
| 4 | pseudogene | 75.85452 | 75.85539 | positive | MGI_C57BL6J_3650655 | Gm11256 | NCBI_Gene:100039884,ENSEMBL:ENSMUSG00000084253 | MGI:3650655 | pseudogene | predicted gene 11256 |
| 4 | pseudogene | 75.90226 | 75.90315 | negative | MGI_C57BL6J_3652121 | Gm11241 | NCBI_Gene:667845,ENSEMBL:ENSMUSG00000081844 | MGI:3652121 | pseudogene | predicted gene 11241 |
| 4 | gene | 75.94124 | 78.21196 | negative | MGI_C57BL6J_97812 | Ptprd | NCBI_Gene:19266,ENSEMBL:ENSMUSG00000028399 | MGI:97812 | protein coding gene | protein tyrosine phosphatase, receptor type, D |
| 4 | gene | 76.19251 | 76.19663 | negative | MGI_C57BL6J_2442984 | A230006I23Rik | NA | NA | unclassified gene | RIKEN cDNA A230006I23 gene |
| 4 | gene | 76.33816 | 76.34539 | positive | MGI_C57BL6J_5826530 | Gm46893 | NCBI_Gene:108169012 | MGI:5826530 | lncRNA gene | predicted gene, 46893 |
| 4 | pseudogene | 76.39315 | 76.39409 | positive | MGI_C57BL6J_3651309 | Gm11252 | ENSEMBL:ENSMUSG00000080780 | MGI:3651309 | pseudogene | predicted gene 11252 |
| 4 | gene | 76.45077 | 76.49011 | positive | MGI_C57BL6J_5625188 | Gm42303 | NCBI_Gene:105247151,ENSEMBL:ENSMUSG00000110373 | MGI:5625188 | lncRNA gene | predicted gene, 42303 |
| 4 | pseudogene | 76.69214 | 76.69463 | positive | MGI_C57BL6J_3652117 | Gm11242 | NCBI_Gene:100422786,ENSEMBL:ENSMUSG00000083050 | MGI:3652117 | pseudogene | predicted gene 11242 |
| 4 | gene | 76.79119 | 76.81372 | positive | MGI_C57BL6J_5625187 | Gm42302 | NCBI_Gene:105247150 | MGI:5625187 | lncRNA gene | predicted gene, 42302 |
| 4 | pseudogene | 76.79373 | 76.79420 | positive | MGI_C57BL6J_3652116 | Gm11243 | NCBI_Gene:667858,ENSEMBL:ENSMUSG00000081890 | MGI:3652116 | pseudogene | predicted gene 11243 |
| 4 | pseudogene | 76.81155 | 76.81179 | negative | MGI_C57BL6J_3651883 | Gm11244 | ENSEMBL:ENSMUSG00000081936 | MGI:3651883 | pseudogene | predicted gene 11244 |
| 4 | pseudogene | 76.83777 | 76.83881 | positive | MGI_C57BL6J_3651884 | Gm11245 | NCBI_Gene:667862,ENSEMBL:ENSMUSG00000084090 | MGI:3651884 | pseudogene | predicted gene 11245 |
| 4 | pseudogene | 76.92731 | 76.92766 | positive | MGI_C57BL6J_3651881 | Gm11246 | ENSEMBL:ENSMUSG00000081656 | MGI:3651881 | pseudogene | predicted gene 11246 |
| 4 | gene | 76.95842 | 76.95850 | negative | MGI_C57BL6J_5452936 | Gm23159 | ENSEMBL:ENSMUSG00000092829 | MGI:5452936 | miRNA gene | predicted gene, 23159 |
| 4 | gene | 77.31082 | 77.31461 | positive | MGI_C57BL6J_5621531 | Gm38646 | NCBI_Gene:102642962 | MGI:5621531 | lncRNA gene | predicted gene, 38646 |
| 4 | pseudogene | 78.01143 | 78.01267 | negative | MGI_C57BL6J_5585257 | Fkbp1a-ps4 | NCBI_Gene:103926449 | MGI:5585257 | pseudogene | FK506 binding protein 1a, pseudogene 4 |
| 4 | gene | 78.15989 | 78.16154 | negative | MGI_C57BL6J_1923471 | 6430598H11Rik | NA | NA | unclassified gene | RIKEN cDNA 6430598H11 gene |
| 4 | gene | 78.20585 | 78.22104 | positive | MGI_C57BL6J_2442149 | B230208B08Rik | NCBI_Gene:102634132,ENSEMBL:ENSMUSG00000092492 | MGI:2442149 | lncRNA gene | RIKEN cDNA B230208B08 gene |
| 4 | pseudogene | 78.23399 | 78.23493 | positive | MGI_C57BL6J_3651928 | Gm11260 | NCBI_Gene:100039996,ENSEMBL:ENSMUSG00000082351 | MGI:3651928 | pseudogene | predicted gene 11260 |
| 4 | gene | 78.26787 | 78.38271 | positive | MGI_C57BL6J_5591704 | Gm32545 | NCBI_Gene:102635126 | MGI:5591704 | lncRNA gene | predicted gene, 32545 |
| 4 | gene | 78.59405 | 78.59748 | positive | MGI_C57BL6J_3647836 | A430085M09Rik | NCBI_Gene:623726 | MGI:3647836 | lncRNA gene | RIKEN cDNA A430085M09 gene |
| 4 | gene | 78.65986 | 78.66159 | negative | MGI_C57BL6J_3650239 | Gm11261 | ENSEMBL:ENSMUSG00000085840 | MGI:3650239 | lncRNA gene | predicted gene 11261 |
| 4 | gene | 78.67015 | 78.67023 | positive | MGI_C57BL6J_5452039 | Gm22262 | ENSEMBL:ENSMUSG00000089338 | MGI:5452039 | unclassified non-coding RNA gene | predicted gene, 22262 |
| 4 | pseudogene | 78.69765 | 78.69893 | positive | MGI_C57BL6J_3650238 | Gm11262 | ENSEMBL:ENSMUSG00000083629 | MGI:3650238 | pseudogene | predicted gene 11262 |
| 4 | gene | 79.24459 | 79.24471 | negative | MGI_C57BL6J_5455613 | Gm25836 | ENSEMBL:ENSMUSG00000089543 | MGI:5455613 | snoRNA gene | predicted gene, 25836 |
| 4 | pseudogene | 79.32771 | 79.32846 | positive | MGI_C57BL6J_3651899 | Gm11263 | NCBI_Gene:667922,ENSEMBL:ENSMUSG00000083496 | MGI:3651899 | pseudogene | predicted gene 11263 |
| 4 | pseudogene | 79.87435 | 79.87475 | positive | MGI_C57BL6J_3652024 | Gm11409 | ENSEMBL:ENSMUSG00000084248 | MGI:3652024 | pseudogene | predicted gene 11409 |
| 4 | pseudogene | 80.00233 | 80.00339 | positive | MGI_C57BL6J_3651131 | Gm11407 | ENSEMBL:ENSMUSG00000082179 | MGI:3651131 | pseudogene | predicted gene 11407 |
| 4 | pseudogene | 80.00340 | 80.00391 | negative | MGI_C57BL6J_3651130 | Gm11408 | ENSEMBL:ENSMUSG00000083076 | MGI:3651130 | pseudogene | predicted gene 11408 |
| 4 | pseudogene | 80.00399 | 80.00513 | positive | MGI_C57BL6J_3652015 | Gm11410 | ENSEMBL:ENSMUSG00000083219 | MGI:3652015 | pseudogene | predicted gene 11410 |
| 4 | pseudogene | 80.44040 | 80.44088 | positive | MGI_C57BL6J_3651132 | Gm11406 | ENSEMBL:ENSMUSG00000082555 | MGI:3651132 | pseudogene | predicted gene 11406 |
| 4 | gene | 80.50934 | 80.63026 | negative | MGI_C57BL6J_5591817 | Gm32658 | NCBI_Gene:102635277 | MGI:5591817 | lncRNA gene | predicted gene, 32658 |
| 4 | gene | 80.70365 | 80.72613 | positive | MGI_C57BL6J_5826522 | Gm46885 | NCBI_Gene:108168997 | MGI:5826522 | lncRNA gene | predicted gene, 46885 |
| 4 | gene | 80.74030 | 80.74413 | positive | MGI_C57BL6J_5625189 | Gm42304 | NCBI_Gene:105247152 | MGI:5625189 | lncRNA gene | predicted gene, 42304 |
| 4 | gene | 80.83412 | 80.85174 | positive | MGI_C57BL6J_98881 | Tyrp1 | NCBI_Gene:22178,ENSEMBL:ENSMUSG00000005994 | MGI:98881 | protein coding gene | tyrosinase-related protein 1 |
| 4 | gene | 80.91065 | 80.95563 | positive | MGI_C57BL6J_106510 | Lurap1l | NCBI_Gene:52829,ENSEMBL:ENSMUSG00000048706 | MGI:106510 | protein coding gene | leucine rich adaptor protein 1-like |
| 4 | gene | 80.91435 | 80.91481 | positive | MGI_C57BL6J_1923975 | 2010003D24Rik | NA | NA | unclassified gene | RIKEN cDNA 2010003D24 gene |
| 4 | gene | 80.91739 | 80.91876 | positive | MGI_C57BL6J_1924589 | 9430051O21Rik | NA | NA | unclassified gene | RIKEN cDNA 9430051O21 gene |
| 4 | gene | 80.96744 | 80.96754 | positive | MGI_C57BL6J_5530834 | Gm27452 | ENSEMBL:ENSMUSG00000099273 | MGI:5530834 | miRNA gene | predicted gene, 27452 |
| 4 | gene | 81.27850 | 81.44291 | negative | MGI_C57BL6J_1343489 | Mpdz | NCBI_Gene:17475,ENSEMBL:ENSMUSG00000028402 | MGI:1343489 | protein coding gene | multiple PDZ domain protein |
| 4 | gene | 81.33827 | 81.33840 | negative | MGI_C57BL6J_5455580 | Gm25803 | ENSEMBL:ENSMUSG00000077607 | MGI:5455580 | snoRNA gene | predicted gene, 25803 |
| 4 | pseudogene | 81.45594 | 81.48478 | negative | MGI_C57BL6J_3651002 | Gm11765 | NCBI_Gene:100040131,ENSEMBL:ENSMUSG00000081391 | MGI:3651002 | pseudogene | predicted gene 11765 |
| 4 | pseudogene | 81.69644 | 81.69716 | positive | MGI_C57BL6J_3650904 | Gm11411 | ENSEMBL:ENSMUSG00000083385 | MGI:3650904 | pseudogene | predicted gene 11411 |
| 4 | gene | 81.74315 | 81.78910 | positive | MGI_C57BL6J_3650905 | Gm11412 | NCBI_Gene:102635467,ENSEMBL:ENSMUSG00000087540 | MGI:3650905 | lncRNA gene | predicted gene 11412 |
| 4 | gene | 81.91851 | 81.91862 | positive | MGI_C57BL6J_4422052 | n-R5s187 | ENSEMBL:ENSMUSG00000084718 | MGI:4422052 | rRNA gene | nuclear encoded rRNA 5S 187 |
| 4 | gene | 81.93048 | 81.93237 | negative | MGI_C57BL6J_3650483 | Gm12914 | ENSEMBL:ENSMUSG00000087608 | MGI:3650483 | lncRNA gene | predicted gene 12914 |
| 4 | gene | 81.93416 | 81.93615 | positive | MGI_C57BL6J_5826523 | Gm46886 | NCBI_Gene:108168998 | MGI:5826523 | lncRNA gene | predicted gene, 46886 |
| 4 | gene | 82.03331 | 82.06589 | negative | MGI_C57BL6J_3649480 | Gm11264 | NCBI_Gene:102635575,ENSEMBL:ENSMUSG00000087110 | MGI:3649480 | lncRNA gene | predicted gene 11264 |
| 4 | gene | 82.06111 | 82.06122 | negative | MGI_C57BL6J_5452179 | Gm22402 | ENSEMBL:ENSMUSG00000092661 | MGI:5452179 | miRNA gene | predicted gene, 22402 |
| 4 | gene | 82.06538 | 82.10281 | positive | MGI_C57BL6J_3646172 | Gm5860 | NCBI_Gene:545641,ENSEMBL:ENSMUSG00000085022 | MGI:3646172 | lncRNA gene | predicted gene 5860 |
| 4 | gene | 82.29017 | 82.70575 | negative | MGI_C57BL6J_103188 | Nfib | NCBI_Gene:18028,ENSEMBL:ENSMUSG00000008575 | MGI:103188 | protein coding gene | nuclear factor I/B |
| 4 | gene | 82.31559 | 82.31990 | negative | MGI_C57BL6J_2444640 | D830012I16Rik | NA | NA | unclassified gene | RIKEN cDNA D830012I16 gene |
| 4 | gene | 82.43941 | 82.43951 | positive | MGI_C57BL6J_4422053 | n-R5s188 | ENSEMBL:ENSMUSG00000084595 | MGI:4422053 | rRNA gene | nuclear encoded rRNA 5S 188 |
| 4 | gene | 82.45006 | 82.45809 | negative | MGI_C57BL6J_1923705 | 2310067E19Rik | NCBI_Gene:76455 | MGI:1923705 | unclassified gene | RIKEN cDNA 2310067E19 gene |
| 4 | gene | 82.50802 | 82.54730 | positive | MGI_C57BL6J_3649705 | Gm11266 | NCBI_Gene:102635783,ENSEMBL:ENSMUSG00000087413 | MGI:3649705 | lncRNA gene | predicted gene 11266 |
| 4 | gene | 82.58505 | 82.58732 | negative | MGI_C57BL6J_3649847 | Gm11267 | ENSEMBL:ENSMUSG00000084851 | MGI:3649847 | lncRNA gene | predicted gene 11267 |
| 4 | gene | 82.60550 | 82.60692 | positive | MGI_C57BL6J_5625190 | Gm42305 | NCBI_Gene:105247154 | MGI:5625190 | lncRNA gene | predicted gene, 42305 |
| 4 | gene | 82.61962 | 82.61969 | negative | MGI_C57BL6J_4413900 | n-THgtg10 | NCBI_Gene:108169019 | MGI:4413900 | tRNA gene | nuclear encoded tRNA histidine 10 (anticodon GTG) |
| 4 | pseudogene | 82.64240 | 82.64267 | positive | MGI_C57BL6J_3649856 | Gm11268 | ENSEMBL:ENSMUSG00000084357 | MGI:3649856 | pseudogene | predicted gene 11268 |
| 4 | gene | 82.69174 | 82.69488 | positive | MGI_C57BL6J_3649855 | Gm11269 | NCBI_Gene:102635985,ENSEMBL:ENSMUSG00000086293 | MGI:3649855 | lncRNA gene | predicted gene 11269 |
| 4 | gene | 82.79874 | 82.85996 | negative | MGI_C57BL6J_1915518 | Zdhhc21 | NCBI_Gene:68268,ENSEMBL:ENSMUSG00000028403 | MGI:1915518 | protein coding gene | zinc finger, DHHC domain containing 21 |
| 4 | gene | 82.88175 | 82.88515 | negative | MGI_C57BL6J_1201414 | Cer1 | NCBI_Gene:12622,ENSEMBL:ENSMUSG00000038192 | MGI:1201414 | protein coding gene | cerberus 1, DAN family BMP antagonist |
| 4 | gene | 82.89792 | 83.05241 | negative | MGI_C57BL6J_2670972 | Frem1 | NCBI_Gene:329872,ENSEMBL:ENSMUSG00000059049 | MGI:2670972 | protein coding gene | Fras1 related extracellular matrix protein 1 |
| 4 | gene | 82.92231 | 82.94031 | positive | MGI_C57BL6J_5592419 | Gm33260 | NCBI_Gene:102636092 | MGI:5592419 | lncRNA gene | predicted gene, 33260 |
| 4 | gene | 83.02380 | 83.02424 | positive | MGI_C57BL6J_1925414 | 4930441N18Rik | NA | NA | unclassified gene | RIKEN cDNA 4930441N18 gene |
| 4 | gene | 83.07851 | 83.07863 | positive | MGI_C57BL6J_5454661 | Gm24884 | ENSEMBL:ENSMUSG00000089409 | MGI:5454661 | snoRNA gene | predicted gene, 24884 |
| 4 | pseudogene | 83.12154 | 83.12174 | positive | MGI_C57BL6J_3651451 | Gm11248 | ENSEMBL:ENSMUSG00000082637 | MGI:3651451 | pseudogene | predicted gene 11248 |
| 4 | gene | 83.17840 | 83.18654 | negative | MGI_C57BL6J_5826524 | Gm46887 | NCBI_Gene:108168999 | MGI:5826524 | lncRNA gene | predicted gene, 46887 |
| 4 | pseudogene | 83.19397 | 83.19519 | positive | MGI_C57BL6J_3650877 | Gm11185 | NCBI_Gene:667985,ENSEMBL:ENSMUSG00000083245 | MGI:3650877 | pseudogene | predicted gene 11185 |
| 4 | gene | 83.22030 | 83.32427 | negative | MGI_C57BL6J_1917113 | Ttc39b | NCBI_Gene:69863,ENSEMBL:ENSMUSG00000038172 | MGI:1917113 | protein coding gene | tetratricopeptide repeat domain 39B |
| 4 | gene | 83.28309 | 83.28782 | positive | MGI_C57BL6J_5625192 | Gm42307 | NCBI_Gene:105247156 | MGI:5625192 | lncRNA gene | predicted gene, 42307 |
| 4 | gene | 83.28580 | 83.28591 | negative | MGI_C57BL6J_5453189 | Gm23412 | ENSEMBL:ENSMUSG00000096801 | MGI:5453189 | snRNA gene | predicted gene, 23412 |
| 4 | gene | 83.32442 | 83.33055 | positive | MGI_C57BL6J_5625191 | Gm42306 | NCBI_Gene:105247155 | MGI:5625191 | lncRNA gene | predicted gene, 42306 |
| 4 | gene | 83.37832 | 83.39067 | negative | MGI_C57BL6J_3650701 | Gm11413 | NCBI_Gene:790913,ENSEMBL:ENSMUSG00000086321 | MGI:3650701 | lncRNA gene | predicted gene 11413 |
| 4 | gene | 83.38241 | 83.39232 | positive | MGI_C57BL6J_3651087 | Gm11184 | NCBI_Gene:102636232,ENSEMBL:ENSMUSG00000087592 | MGI:3651087 | lncRNA gene | predicted gene 11184 |
| 4 | gene | 83.41770 | 83.47302 | positive | MGI_C57BL6J_1916338 | Snapc3 | NCBI_Gene:77634,ENSEMBL:ENSMUSG00000028483 | MGI:1916338 | protein coding gene | small nuclear RNA activating complex, polypeptide 3 |
| 4 | gene | 83.45568 | 83.48651 | negative | MGI_C57BL6J_2142116 | Psip1 | NCBI_Gene:101739,ENSEMBL:ENSMUSG00000028484 | MGI:2142116 | protein coding gene | PC4 and SFRS1 interacting protein 1 |
| 4 | gene | 83.47174 | 83.47302 | positive | MGI_C57BL6J_1925039 | A930007D18Rik | NA | NA | unclassified gene | RIKEN cDNA A930007D18 gene |
| 4 | pseudogene | 83.50050 | 83.50076 | negative | MGI_C57BL6J_3650503 | Gm11414 | ENSEMBL:ENSMUSG00000082416 | MGI:3650503 | pseudogene | predicted gene 11414 |
| 4 | pseudogene | 83.50514 | 83.50557 | negative | MGI_C57BL6J_3642271 | Gm10154 | NCBI_Gene:100040611,ENSEMBL:ENSMUSG00000066116 | MGI:3642271 | pseudogene | predicted gene 10154 |
| 4 | gene | 83.50828 | 83.51129 | negative | MGI_C57BL6J_5625193 | Gm42308 | NCBI_Gene:105247157 | MGI:5625193 | lncRNA gene | predicted gene, 42308 |
| 4 | gene | 83.52540 | 83.86614 | positive | MGI_C57BL6J_1922152 | Ccdc171 | NCBI_Gene:320226,ENSEMBL:ENSMUSG00000052407 | MGI:1922152 | protein coding gene | coiled-coil domain containing 171 |
| 4 | gene | 83.52763 | 83.52777 | negative | MGI_C57BL6J_5455676 | Gm25899 | ENSEMBL:ENSMUSG00000064606 | MGI:5455676 | snRNA gene | predicted gene, 25899 |
| 4 | pseudogene | 83.59730 | 83.59776 | positive | MGI_C57BL6J_5504161 | Gm27046 | ENSEMBL:ENSMUSG00000098217 | MGI:5504161 | pseudogene | predicted gene, 27046 |
| 4 | pseudogene | 83.61102 | 83.61222 | negative | MGI_C57BL6J_5010872 | Gm18687 | NCBI_Gene:100417557 | MGI:5010872 | pseudogene | predicted gene, 18687 |
| 4 | pseudogene | 83.70098 | 83.70449 | positive | MGI_C57BL6J_1890540 | Cbx3-ps3 | NCBI_Gene:102641235,ENSEMBL:ENSMUSG00000080793 | MGI:1890540 | pseudogene | chromobox 3, pseudogene 3 |
| 4 | gene | 83.74368 | 83.74416 | positive | MGI_C57BL6J_5504083 | Gm26968 | ENSEMBL:ENSMUSG00000097942 | MGI:5504083 | lncRNA gene | predicted gene, 26968 |
| 4 | gene | 83.82613 | 83.84205 | negative | MGI_C57BL6J_5625194 | Gm42309 | NCBI_Gene:105247158 | MGI:5625194 | lncRNA gene | predicted gene, 42309 |
| 4 | gene | 83.87042 | 83.88886 | negative | MGI_C57BL6J_3650502 | Gm11415 | NCBI_Gene:105247159,ENSEMBL:ENSMUSG00000085378 | MGI:3650502 | lncRNA gene | predicted gene 11415 |
| 4 | gene | 84.04204 | 84.04488 | negative | MGI_C57BL6J_1925488 | 6030471H07Rik | NCBI_Gene:78238,ENSEMBL:ENSMUSG00000085217 | MGI:1925488 | lncRNA gene | RIKEN cDNA 6030471H07 gene |
| 4 | gene | 84.07240 | 84.08192 | negative | MGI_C57BL6J_5592639 | Gm33480 | NCBI_Gene:102636405 | MGI:5592639 | lncRNA gene | predicted gene, 33480 |
| 4 | gene | 84.07262 | 84.10143 | positive | MGI_C57BL6J_5592569 | Gm33410 | NCBI_Gene:102636305 | MGI:5592569 | lncRNA gene | predicted gene, 33410 |
| 4 | pseudogene | 84.10488 | 84.10611 | positive | MGI_C57BL6J_3650588 | Gm12416 | NCBI_Gene:668010,ENSEMBL:ENSMUSG00000083044 | MGI:3650588 | pseudogene | predicted gene 12416 |
| 4 | gene | 84.11035 | 84.11335 | positive | MGI_C57BL6J_5625195 | Gm42310 | NCBI_Gene:105247160 | MGI:5625195 | lncRNA gene | predicted gene, 42310 |
| 4 | gene | 84.12573 | 84.13176 | negative | MGI_C57BL6J_3650374 | Gm12414 | NCBI_Gene:102636485,ENSEMBL:ENSMUSG00000087378 | MGI:3650374 | lncRNA gene | predicted gene 12414 |
| 4 | gene | 84.13326 | 84.22073 | negative | MGI_C57BL6J_5592756 | Gm33597 | NCBI_Gene:102636567 | MGI:5592756 | lncRNA gene | predicted gene, 33597 |
| 4 | gene | 84.26680 | 84.67635 | negative | MGI_C57BL6J_2443805 | Bnc2 | NCBI_Gene:242509,ENSEMBL:ENSMUSG00000028487 | MGI:2443805 | protein coding gene | basonuclin 2 |
| 4 | pseudogene | 84.42882 | 84.42912 | negative | MGI_C57BL6J_3650591 | Gm12421 | ENSEMBL:ENSMUSG00000083743 | MGI:3650591 | pseudogene | predicted gene 12421 |
| 4 | pseudogene | 84.69786 | 84.69894 | positive | MGI_C57BL6J_3652148 | Gm12420 | NCBI_Gene:624465,ENSEMBL:ENSMUSG00000081775 | MGI:3652148 | pseudogene | predicted gene 12420 |
| 4 | gene | 84.74147 | 84.74452 | positive | MGI_C57BL6J_3650585 | Gm12415 | NCBI_Gene:105247161,ENSEMBL:ENSMUSG00000086413 | MGI:3650585 | lncRNA gene | predicted gene 12415 |
| 4 | gene | 84.88351 | 85.13192 | positive | MGI_C57BL6J_2443104 | Cntln | NCBI_Gene:338349,ENSEMBL:ENSMUSG00000038070 | MGI:2443104 | protein coding gene | centlein, centrosomal protein |
| 4 | pseudogene | 84.91584 | 84.91775 | positive | MGI_C57BL6J_3650582 | Gm12418 | NCBI_Gene:622811,ENSEMBL:ENSMUSG00000083139 | MGI:3650582 | pseudogene | predicted gene 12418 |
| 4 | pseudogene | 84.92355 | 84.92420 | negative | MGI_C57BL6J_3650581 | Gm12419 | NCBI_Gene:100417804,ENSEMBL:ENSMUSG00000080303 | MGI:3650581 | pseudogene | predicted gene 12419 |
| 4 | pseudogene | 85.17653 | 85.17703 | negative | MGI_C57BL6J_3780870 | Gm2700 | NCBI_Gene:100040303 | MGI:3780870 | pseudogene | predicted gene 2700 |
| 4 | pseudogene | 85.17653 | 85.17693 | positive | MGI_C57BL6J_3650587 | Gm12417 | ENSEMBL:ENSMUSG00000084267 | MGI:3650587 | pseudogene | predicted gene 12417 |
| 4 | gene | 85.20513 | 85.63920 | positive | MGI_C57BL6J_700009 | Sh3gl2 | NCBI_Gene:20404,ENSEMBL:ENSMUSG00000028488 | MGI:700009 | protein coding gene | SH3-domain GRB2-like 2 |
| 4 | pseudogene | 85.24545 | 85.24619 | negative | MGI_C57BL6J_3650373 | Gm12413 | NCBI_Gene:100416925,ENSEMBL:ENSMUSG00000081862 | MGI:3650373 | pseudogene | predicted gene 12413 |
| 4 | gene | 85.51323 | 86.42839 | positive | MGI_C57BL6J_1924989 | Adamtsl1 | NCBI_Gene:77739,ENSEMBL:ENSMUSG00000066113 | MGI:1924989 | protein coding gene | ADAMTS-like 1 |
| 4 | gene | 85.51433 | 85.78313 | positive | MGI_C57BL6J_3650846 | Gm12680 | NA | NA | unclassified non-coding RNA gene | predicted gene 12680 |
| 4 | gene | 85.90991 | 85.91019 | positive | MGI_C57BL6J_5455846 | Gm26069 | ENSEMBL:ENSMUSG00000088122 | MGI:5455846 | unclassified non-coding RNA gene | predicted gene, 26069 |
| 4 | pseudogene | 85.92111 | 85.92194 | negative | MGI_C57BL6J_3779825 | Gm8949 | NCBI_Gene:668052 | MGI:3779825 | pseudogene | predicted gene 8949 |
| 4 | gene | 86.04339 | 86.04349 | negative | MGI_C57BL6J_5455588 | Gm25811 | ENSEMBL:ENSMUSG00000064687 | MGI:5455588 | snRNA gene | predicted gene, 25811 |
| 4 | gene | 86.42125 | 86.44095 | negative | MGI_C57BL6J_5592889 | Gm33730 | NCBI_Gene:102636740 | MGI:5592889 | lncRNA gene | predicted gene, 33730 |
| 4 | pseudogene | 86.44201 | 86.44234 | negative | MGI_C57BL6J_3651460 | Gm12550 | ENSEMBL:ENSMUSG00000083272 | MGI:3651460 | pseudogene | predicted gene 12550 |
| 4 | gene | 86.44442 | 86.55833 | negative | MGI_C57BL6J_1923061 | Saxo1 | NCBI_Gene:75811,ENSEMBL:ENSMUSG00000028492 | MGI:1923061 | protein coding gene | stabilizer of axonemal microtubules 1 |
| 4 | gene | 86.50915 | 86.50923 | positive | MGI_C57BL6J_5455794 | Gm26017 | ENSEMBL:ENSMUSG00000077597 | MGI:5455794 | snRNA gene | predicted gene, 26017 |
| 4 | gene | 86.55844 | 86.55980 | positive | MGI_C57BL6J_1922138 | Saxo1os | NCBI_Gene:74888,ENSEMBL:ENSMUSG00000085108 | MGI:1922138 | antisense lncRNA gene | stabilizer of axonemal microtubules 1, opposite strand |
| 4 | gene | 86.57567 | 86.57729 | positive | MGI_C57BL6J_1915691 | Rraga | NCBI_Gene:68441,ENSEMBL:ENSMUSG00000070934 | MGI:1915691 | protein coding gene | Ras-related GTP binding A |
| 4 | gene | 86.57886 | 86.61208 | negative | MGI_C57BL6J_1923389 | Haus6 | NCBI_Gene:230376,ENSEMBL:ENSMUSG00000038047 | MGI:1923389 | protein coding gene | HAUS augmin-like complex, subunit 6 |
| 4 | gene | 86.58645 | 86.58658 | negative | MGI_C57BL6J_3819488 | Scarna8 | NCBI_Gene:100217448,ENSEMBL:ENSMUSG00000088958 | MGI:3819488 | snoRNA gene | small Cajal body-specific RNA 8 |
| 4 | pseudogene | 86.62685 | 86.63321 | negative | MGI_C57BL6J_3651664 | Gm12551 | NCBI_Gene:101055843,ENSEMBL:ENSMUSG00000081169 | MGI:3651664 | pseudogene | predicted gene 12551 |
| 4 | gene | 86.62691 | 86.67006 | negative | MGI_C57BL6J_87920 | Plin2 | NCBI_Gene:11520,ENSEMBL:ENSMUSG00000028494 | MGI:87920 | protein coding gene | perilipin 2 |
| 4 | gene | 86.71097 | 86.74381 | positive | MGI_C57BL6J_5625197 | Gm42312 | NCBI_Gene:105247163 | MGI:5625197 | lncRNA gene | predicted gene, 42312 |
| 4 | gene | 86.74753 | 86.74946 | negative | MGI_C57BL6J_3645185 | Gm6521 | NA | NA | unclassified gene | predicted gene 6521 |
| 4 | gene | 86.74840 | 86.85060 | positive | MGI_C57BL6J_1914769 | Dennd4c | NCBI_Gene:329877,ENSEMBL:ENSMUSG00000038024 | MGI:1914769 | protein coding gene | DENN/MADD domain containing 4C |
| 4 | gene | 86.76849 | 86.76980 | positive | MGI_C57BL6J_1921836 | 4833420L08Rik | NA | NA | unclassified gene | RIKEN cDNA 4833420L08 gene |
| 4 | pseudogene | 86.81775 | 86.81860 | negative | MGI_C57BL6J_3650968 | Gm12630 | NCBI_Gene:100384871,ENSEMBL:ENSMUSG00000082969 | MGI:3650968 | pseudogene | predicted gene 12630 |
| 4 | gene | 86.85410 | 86.85741 | negative | MGI_C57BL6J_98159 | Rps6 | NCBI_Gene:20104,ENSEMBL:ENSMUSG00000028495 | MGI:98159 | protein coding gene | ribosomal protein S6 |
| 4 | gene | 86.87434 | 86.93482 | positive | MGI_C57BL6J_1920932 | Acer2 | NCBI_Gene:230379,ENSEMBL:ENSMUSG00000038007 | MGI:1920932 | protein coding gene | alkaline ceramidase 2 |
| 4 | gene | 86.98312 | 87.23055 | negative | MGI_C57BL6J_1923626 | Slc24a2 | NCBI_Gene:76376,ENSEMBL:ENSMUSG00000037996 | MGI:1923626 | protein coding gene | solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
| 4 | gene | 87.02385 | 87.02564 | negative | MGI_C57BL6J_1925538 | 5330421F21Rik | NA | NA | unclassified gene | RIKEN cDNA 5330421F21 gene |
| 4 | pseudogene | 87.10040 | 87.10493 | positive | MGI_C57BL6J_5593198 | Gm34039 | NCBI_Gene:108168966 | MGI:5593198 | pseudogene | predicted gene, 34039 |
| 4 | gene | 87.29836 | 87.30127 | positive | MGI_C57BL6J_3651471 | Gm12600 | ENSEMBL:ENSMUSG00000086512 | MGI:3651471 | lncRNA gene | predicted gene 12600 |
| 4 | gene | 87.43227 | 87.43234 | positive | MGI_C57BL6J_5452931 | Gm23154 | ENSEMBL:ENSMUSG00000093065 | MGI:5452931 | miRNA gene | predicted gene, 23154 |
| 4 | gene | 87.53089 | 87.66852 | negative | MGI_C57BL6J_5593339 | Gm34180 | NCBI_Gene:102637345 | MGI:5593339 | lncRNA gene | predicted gene, 34180 |
| 4 | pseudogene | 87.58078 | 87.58156 | positive | MGI_C57BL6J_3651368 | Gm12604 | NCBI_Gene:100418044,ENSEMBL:ENSMUSG00000082443 | MGI:3651368 | pseudogene | predicted gene 12604 |
| 4 | gene | 87.76993 | 88.03341 | negative | MGI_C57BL6J_1917372 | Mllt3 | NCBI_Gene:70122,ENSEMBL:ENSMUSG00000028496 | MGI:1917372 | protein coding gene | myeloid/lymphoid or mixed-lineage leukemia, translocated to, 3 |
| 4 | gene | 87.79709 | 87.79869 | negative | MGI_C57BL6J_1924111 | 4832441B07Rik | NA | NA | unclassified gene | RIKEN cDNA 4832441B07 gene |
| 4 | gene | 87.97462 | 87.98563 | negative | MGI_C57BL6J_5593391 | Gm34232 | NCBI_Gene:102637422 | MGI:5593391 | lncRNA gene | predicted gene, 34232 |
| 4 | gene | 88.03350 | 88.03500 | positive | MGI_C57BL6J_3650969 | Gm12631 | ENSEMBL:ENSMUSG00000084809 | MGI:3650969 | lncRNA gene | predicted gene 12631 |
| 4 | pseudogene | 88.04380 | 88.04404 | positive | MGI_C57BL6J_3649679 | Gm12646 | ENSEMBL:ENSMUSG00000081530 | MGI:3649679 | pseudogene | predicted gene 12646 |
| 4 | gene | 88.09463 | 88.41101 | positive | MGI_C57BL6J_2676921 | Focad | NCBI_Gene:230393,ENSEMBL:ENSMUSG00000038368 | MGI:2676921 | protein coding gene | focadhesin |
| 4 | gene | 88.12204 | 88.12212 | positive | MGI_C57BL6J_3629651 | Mir491 | miRBase:MI0004680,NCBI_Gene:735282,ENSEMBL:ENSMUSG00000070077 | MGI:3629651 | miRNA gene | microRNA 491 |
| 4 | pseudogene | 88.15170 | 88.15197 | positive | MGI_C57BL6J_3649678 | Gm12645 | ENSEMBL:ENSMUSG00000082597 | MGI:3649678 | pseudogene | predicted gene 12645 |
| 4 | gene | 88.39614 | 88.43894 | negative | MGI_C57BL6J_1914025 | Hacd4 | NCBI_Gene:66775,ENSEMBL:ENSMUSG00000028497 | MGI:1914025 | protein coding gene | 3-hydroxyacyl-CoA dehydratase 4 |
| 4 | gene | 88.43173 | 88.43293 | negative | MGI_C57BL6J_1337997 | Gdap6 | NA | NA | unclassified gene | ganglioside-induced differentiation-associated-protein 6 |
| 4 | gene | 88.52202 | 88.52279 | negative | MGI_C57BL6J_107657 | Ifnb1 | NCBI_Gene:15977,ENSEMBL:ENSMUSG00000048806 | MGI:107657 | protein coding gene | interferon beta 1, fibroblast |
| 4 | gene | 88.55767 | 88.55824 | negative | MGI_C57BL6J_3649418 | Ifna15 | NCBI_Gene:242517,ENSEMBL:ENSMUSG00000096011 | MGI:3649418 | protein coding gene | interferon alpha 15 |
| 4 | gene | 88.57123 | 88.57180 | negative | MGI_C57BL6J_3641425 | Ifna14 | NCBI_Gene:404549,ENSEMBL:ENSMUSG00000095896 | MGI:3641425 | protein coding gene | interferon alpha 14 |
| 4 | pseudogene | 88.57889 | 88.58057 | negative | MGI_C57BL6J_107669 | Ifna-ps1 | NCBI_Gene:100416153,ENSEMBL:ENSMUSG00000080964 | MGI:107669 | pseudogene | interferon alpha gene, pseudogene 1 |
| 4 | gene | 88.59181 | 88.59238 | negative | MGI_C57BL6J_107659 | Ifna9 | NCBI_Gene:15972,ENSEMBL:ENSMUSG00000095270 | MGI:107659 | protein coding gene | interferon alpha 9 |
| 4 | pseudogene | 88.59898 | 88.59987 | negative | MGI_C57BL6J_3050089 | Mrpl48-ps | NCBI_Gene:269553,ENSEMBL:ENSMUSG00000078480 | MGI:3050089 | pseudogene | mitochondrial ribosomal protein L48 pseudogene |
| 4 | gene | 88.60258 | 88.60338 | negative | MGI_C57BL6J_2676324 | Ifna12 | NCBI_Gene:242519,ENSEMBL:ENSMUSG00000073811 | MGI:2676324 | protein coding gene | interferon alpha 12 |
| 4 | pseudogene | 88.61265 | 88.61404 | negative | MGI_C57BL6J_3650053 | Gm12601 | ENSEMBL:ENSMUSG00000083958 | MGI:3650053 | pseudogene | predicted gene 12601 |
| 4 | gene | 88.62732 | 88.63441 | negative | MGI_C57BL6J_2140706 | C87499 | NCBI_Gene:381590,ENSEMBL:ENSMUSG00000038330 | MGI:2140706 | protein coding gene | expressed sequence C87499 |
| 4 | gene | 88.64364 | 88.64446 | negative | MGI_C57BL6J_2667155 | Ifna13 | NCBI_Gene:230396,ENSEMBL:ENSMUSG00000063376 | MGI:2667155 | protein coding gene | interferon alpha 13 |
| 4 | pseudogene | 88.64859 | 88.64937 | negative | MGI_C57BL6J_3649513 | Gm13281 | NCBI_Gene:546834,ENSEMBL:ENSMUSG00000081784 | MGI:3649513 | pseudogene | predicted gene 13281 |
| 4 | gene | 88.67592 | 88.67692 | negative | MGI_C57BL6J_3649260 | Ifna16 | NCBI_Gene:230398,ENSEMBL:ENSMUSG00000078355 | MGI:3649260 | protein coding gene | interferon alpha 16 |
| 4 | gene | 88.68288 | 88.68391 | negative | MGI_C57BL6J_107666 | Ifna2 | NCBI_Gene:15965,ENSEMBL:ENSMUSG00000078354 | MGI:107666 | protein coding gene | interferon alpha 2 |
| 4 | gene | 88.69030 | 88.69136 | negative | MGI_C57BL6J_1097683 | Ifnab | NCBI_Gene:15974,ENSEMBL:ENSMUSG00000100079 | MGI:1097683 | protein coding gene | interferon alpha B |
| 4 | pseudogene | 88.69619 | 88.69670 | negative | MGI_C57BL6J_3650885 | Gm13273 | ENSEMBL:ENSMUSG00000081893 | MGI:3650885 | pseudogene | predicted gene 13273 |
| 4 | pseudogene | 88.70789 | 88.70845 | negative | MGI_C57BL6J_3649258 | Gm13282 | NCBI_Gene:546835,ENSEMBL:ENSMUSG00000084192 | MGI:3649258 | pseudogene | predicted gene 13282 |
| 4 | gene | 88.71829 | 88.72251 | negative | MGI_C57BL6J_2180122 | Klhl9 | NCBI_Gene:242521,ENSEMBL:ENSMUSG00000070923 | MGI:2180122 | protein coding gene | kelch-like 9 |
| 4 | gene | 88.72181 | 88.72284 | positive | MGI_C57BL6J_5477060 | Gm26566 | ENSEMBL:ENSMUSG00000097078 | MGI:5477060 | protein coding gene | predicted gene, 26566 |
| 4 | gene | 88.72276 | 88.75758 | negative | MGI_C57BL6J_3649443 | Gm13274 | NCBI_Gene:100037413,ENSEMBL:ENSMUSG00000086074 | MGI:3649443 | lncRNA gene | predicted gene 13274 |
| 4 | pseudogene | 88.75447 | 88.75468 | positive | MGI_C57BL6J_6270567 | Gm49884 | ENSEMBL:ENSMUSG00000117268 | MGI:6270567 | pseudogene | predicted gene, 49884 |
| 4 | gene | 88.75487 | 88.75542 | positive | MGI_C57BL6J_3701966 | Gm13271 | NCBI_Gene:435791,ENSEMBL:ENSMUSG00000094618 | MGI:3701966 | protein coding gene | predicted gene 13271 |
| 4 | pseudogene | 88.75784 | 88.75839 | positive | MGI_C57BL6J_3650855 | Gm13286 | NCBI_Gene:100041062,ENSEMBL:ENSMUSG00000081501 | MGI:3650855 | pseudogene | predicted gene 13286 |
| 4 | pseudogene | 88.75817 | 88.75857 | negative | MGI_C57BL6J_3708693 | Gm10583 | ENSEMBL:ENSMUSG00000073809 | MGI:3708693 | pseudogene | predicted gene 10583 |
| 4 | gene | 88.76012 | 88.76158 | positive | MGI_C57BL6J_3649573 | Gm13283 | NCBI_Gene:545645,ENSEMBL:ENSMUSG00000100505 | MGI:3649573 | protein coding gene | predicted gene 13283 |
| 4 | pseudogene | 88.76110 | 88.76150 | negative | MGI_C57BL6J_3708694 | Gm10582 | ENSEMBL:ENSMUSG00000094498 | MGI:3708694 | pseudogene | predicted gene 10582 |
| 4 | pseudogene | 88.76305 | 88.77186 | positive | MGI_C57BL6J_3701980 | Gm13284 | NCBI_Gene:668147,ENSEMBL:ENSMUSG00000082789 | MGI:3701980 | pseudogene | predicted gene 13284 |
| 4 | pseudogene | 88.77155 | 88.77178 | negative | MGI_C57BL6J_3708695 | Gm10581 | ENSEMBL:ENSMUSG00000073807 | MGI:3708695 | pseudogene | predicted gene 10581 |
| 4 | gene | 88.77383 | 88.77480 | positive | MGI_C57BL6J_3701990 | Gm13290 | NCBI_Gene:545646,ENSEMBL:ENSMUSG00000094271 | MGI:3701990 | protein coding gene | predicted gene 13290 |
| 4 | pseudogene | 88.77432 | 88.77472 | negative | MGI_C57BL6J_3708696 | Gm10580 | ENSEMBL:ENSMUSG00000094167 | MGI:3708696 | pseudogene | predicted gene 10580 |
| 4 | gene | 88.77676 | 88.77773 | positive | MGI_C57BL6J_3701987 | Gm13289 | NCBI_Gene:545647,ENSEMBL:ENSMUSG00000096582 | MGI:3701987 | protein coding gene | predicted gene 13289 |
| 4 | gene | 88.77985 | 88.78066 | positive | MGI_C57BL6J_3701968 | Gm13272 | NCBI_Gene:545648,ENSEMBL:ENSMUSG00000096591 | MGI:3701968 | protein coding gene | predicted gene 13272 |
| 4 | gene | 88.78213 | 88.78359 | positive | MGI_C57BL6J_2448469 | Ifnz | NCBI_Gene:319146,ENSEMBL:ENSMUSG00000096854 | MGI:2448469 | protein coding gene | interferon zeta |
| 4 | gene | 88.78445 | 88.79439 | negative | MGI_C57BL6J_5477019 | Gm26525 | ENSEMBL:ENSMUSG00000096987 | MGI:5477019 | lncRNA gene | predicted gene, 26525 |
| 4 | gene | 88.78505 | 88.78651 | positive | MGI_C57BL6J_3701972 | Gm13276 | NCBI_Gene:545649,ENSEMBL:ENSMUSG00000099545 | MGI:3701972 | protein coding gene | predicted gene 13276 |
| 4 | gene | 88.78798 | 88.78944 | positive | MGI_C57BL6J_3701974 | Gm13277 | NCBI_Gene:545650,ENSEMBL:ENSMUSG00000100234 | MGI:3701974 | protein coding gene | predicted gene 13277 |
| 4 | gene | 88.79090 | 88.79236 | positive | MGI_C57BL6J_3701976 | Gm13278 | NCBI_Gene:545651,ENSEMBL:ENSMUSG00000101163 | MGI:3701976 | protein coding gene | predicted gene 13278 |
| 4 | gene | 88.79374 | 88.79530 | positive | MGI_C57BL6J_3701970 | Gm13275 | NCBI_Gene:545652,ENSEMBL:ENSMUSG00000099518 | MGI:3701970 | protein coding gene | predicted gene 13275 |
| 4 | gene | 88.79614 | 88.80022 | negative | MGI_C57BL6J_5477361 | Gm26867 | ENSEMBL:ENSMUSG00000097873 | MGI:5477361 | lncRNA gene | predicted gene, 26867 |
| 4 | gene | 88.79724 | 88.79820 | positive | MGI_C57BL6J_3701978 | Gm13279 | NCBI_Gene:545653,ENSEMBL:ENSMUSG00000099420 | MGI:3701978 | protein coding gene | predicted gene 13279 |
| 4 | gene | 88.79967 | 88.80113 | positive | MGI_C57BL6J_3701981 | Gm13285 | NCBI_Gene:545654,ENSEMBL:ENSMUSG00000095101 | MGI:3701981 | protein coding gene | predicted gene 13285 |
| 4 | gene | 88.80325 | 88.80380 | positive | MGI_C57BL6J_3701983 | Gm13287 | NCBI_Gene:545655,ENSEMBL:ENSMUSG00000094648 | MGI:3701983 | protein coding gene | predicted gene 13287 |
| 4 | gene | 88.80553 | 88.80838 | positive | MGI_C57BL6J_3701985 | Gm13288 | NCBI_Gene:668208,ENSEMBL:ENSMUSG00000070908 | MGI:3701985 | protein coding gene | predicted gene 13288 |
| 4 | gene | 88.81623 | 88.81680 | positive | MGI_C57BL6J_107661 | Ifna7 | NCBI_Gene:15970,ENSEMBL:ENSMUSG00000100713 | MGI:107661 | protein coding gene | interferon alpha 7 |
| 4 | gene | 88.81992 | 88.82057 | positive | MGI_C57BL6J_109210 | Ifna11 | NCBI_Gene:15964,ENSEMBL:ENSMUSG00000100549 | MGI:109210 | protein coding gene | interferon alpha 11 |
| 4 | gene | 88.82742 | 88.82798 | positive | MGI_C57BL6J_107662 | Ifna6 | NCBI_Gene:15969,ENSEMBL:ENSMUSG00000101252 | MGI:107662 | protein coding gene | interferon alpha 6 |
| 4 | gene | 88.83553 | 88.83609 | positive | MGI_C57BL6J_107663 | Ifna5 | NCBI_Gene:15968,ENSEMBL:ENSMUSG00000096682 | MGI:107663 | protein coding gene | interferon alpha 5 |
| 4 | gene | 88.84182 | 88.84246 | positive | MGI_C57BL6J_107664 | Ifna4 | NCBI_Gene:15967,ENSEMBL:ENSMUSG00000070904 | MGI:107664 | protein coding gene | interferon alpha 4 |
| 4 | gene | 88.85009 | 88.85066 | positive | MGI_C57BL6J_107668 | Ifna1 | NCBI_Gene:15962,ENSEMBL:ENSMUSG00000095498 | MGI:107668 | protein coding gene | interferon alpha 1 |
| 4 | gene | 88.86723 | 88.86838 | negative | MGI_C57BL6J_1922496 | 4930553M12Rik | ENSEMBL:ENSMUSG00000054351 | MGI:1922496 | lncRNA gene | RIKEN cDNA 4930553M12 gene |
| 4 | gene | 88.87174 | 88.87489 | positive | MGI_C57BL6J_5593616 | Gm34457 | NCBI_Gene:102637717 | MGI:5593616 | lncRNA gene | predicted gene, 34457 |
| 4 | gene | 88.87954 | 88.88020 | negative | MGI_C57BL6J_2667156 | Ifne | NCBI_Gene:230405,ENSEMBL:ENSMUSG00000045364 | MGI:2667156 | protein coding gene | interferon epsilon |
| 4 | gene | 88.90985 | 88.93852 | negative | MGI_C57BL6J_6270578 | Gm49890 | ENSEMBL:ENSMUSG00000117123 | MGI:6270578 | lncRNA gene | predicted gene, 49890 |
| 4 | gene | 88.91056 | 88.91066 | negative | MGI_C57BL6J_3619330 | Mir31 | miRBase:MI0000579,NCBI_Gene:723895,ENSEMBL:ENSMUSG00000065408 | MGI:3619330 | miRNA gene | microRNA 31 |
| 4 | gene | 88.95642 | 89.03130 | positive | MGI_C57BL6J_5593727 | Gm34568 | NCBI_Gene:102637866 | MGI:5593727 | lncRNA gene | predicted gene, 34568 |
| 4 | gene | 88.99512 | 89.03919 | negative | MGI_C57BL6J_6270580 | Gm49891 | ENSEMBL:ENSMUSG00000117251 | MGI:6270580 | lncRNA gene | predicted gene, 49891 |
| 4 | gene | 89.05031 | 89.08461 | positive | MGI_C57BL6J_3649757 | Wincr1 | NCBI_Gene:100040617,ENSEMBL:ENSMUSG00000085183 | MGI:3649757 | lncRNA gene | WNT induced non-coding RNA 1 |
| 4 | gene | 89.09987 | 89.12876 | negative | MGI_C57BL6J_3651757 | Gm12602 | NCBI_Gene:102637941,ENSEMBL:ENSMUSG00000085569 | MGI:3651757 | lncRNA gene | predicted gene 12602 |
| 4 | gene | 89.13712 | 89.18109 | positive | MGI_C57BL6J_1914152 | Mtap | NCBI_Gene:66902,ENSEMBL:ENSMUSG00000062937 | MGI:1914152 | protein coding gene | methylthioadenosine phosphorylase |
| 4 | pseudogene | 89.16170 | 89.16222 | negative | MGI_C57BL6J_3649224 | Gm12607 | NCBI_Gene:100416402,ENSEMBL:ENSMUSG00000083989 | MGI:3649224 | pseudogene | predicted gene 12607 |
| 4 | gene | 89.18298 | 89.18312 | negative | MGI_C57BL6J_5456267 | Gm26490 | ENSEMBL:ENSMUSG00000088504 | MGI:5456267 | snoRNA gene | predicted gene, 26490 |
| 4 | pseudogene | 89.21671 | 89.21849 | positive | MGI_C57BL6J_3649223 | Tgif2-ps1 | NCBI_Gene:625574,ENSEMBL:ENSMUSG00000107513 | MGI:3649223 | pseudogene | TGFB-induced factor homeobox 2, pseudogene 1 |
| 4 | gene | 89.23570 | 89.27340 | negative | MGI_C57BL6J_3649222 | Gm12606 | ENSEMBL:ENSMUSG00000087659 | MGI:3649222 | lncRNA gene | predicted gene 12606 |
| 4 | gene | 89.27447 | 89.29465 | negative | MGI_C57BL6J_104738 | Cdkn2a | NCBI_Gene:12578,ENSEMBL:ENSMUSG00000044303 | MGI:104738 | protein coding gene | cyclin dependent kinase inhibitor 2A |
| 4 | gene | 89.29518 | 89.51862 | positive | MGI_C57BL6J_3652143 | Gm12610 | NCBI_Gene:102633081,ENSEMBL:ENSMUSG00000086035 | MGI:3652143 | lncRNA gene | predicted gene 12610 |
| 4 | gene | 89.30629 | 89.31104 | negative | MGI_C57BL6J_104737 | Cdkn2b | NCBI_Gene:12579,ENSEMBL:ENSMUSG00000073802 | MGI:104737 | protein coding gene | cyclin dependent kinase inhibitor 2B |
| 4 | gene | 89.38196 | 89.39988 | positive | MGI_C57BL6J_3652142 | Gm12609 | ENSEMBL:ENSMUSG00000085147 | MGI:3652142 | lncRNA gene | predicted gene 12609 |
| 4 | pseudogene | 89.44222 | 89.44464 | negative | MGI_C57BL6J_3652144 | Gm12608 | NCBI_Gene:664785,ENSEMBL:ENSMUSG00000080808 | MGI:3652144 | pseudogene | predicted gene 12608 |
| 4 | gene | 89.44522 | 89.44537 | positive | MGI_C57BL6J_5530962 | Gm27580 | ENSEMBL:ENSMUSG00000098669 | MGI:5530962 | unclassified non-coding RNA gene | predicted gene, 27580 |
| 4 | gene | 89.67944 | 89.69477 | positive | MGI_C57BL6J_2653627 | Dmrta1 | NCBI_Gene:242523,ENSEMBL:ENSMUSG00000043753 | MGI:2653627 | protein coding gene | doublesex and mab-3 related transcription factor like family A1 |
| 4 | gene | 90.18553 | 90.19001 | positive | MGI_C57BL6J_3650349 | Gm12629 | ENSEMBL:ENSMUSG00000085690 | MGI:3650349 | lncRNA gene | predicted gene 12629 |
| 4 | gene | 90.21882 | 90.22570 | positive | MGI_C57BL6J_2387418 | Zfp352 | NCBI_Gene:236537,ENSEMBL:ENSMUSG00000070902 | MGI:2387418 | protein coding gene | zinc finger protein 352 |
| 4 | pseudogene | 90.35962 | 90.36133 | negative | MGI_C57BL6J_3650768 | Gm12633 | NCBI_Gene:619842,ENSEMBL:ENSMUSG00000080756 | MGI:3650768 | pseudogene | predicted gene 12633 |
| 4 | pseudogene | 90.36871 | 90.36991 | positive | MGI_C57BL6J_3650769 | Gm12634 | NCBI_Gene:435792,ENSEMBL:ENSMUSG00000082569 | MGI:3650769 | pseudogene | predicted gene 12634 |
| 4 | pseudogene | 90.45221 | 90.45363 | positive | MGI_C57BL6J_3650766 | Gm12635 | NCBI_Gene:100417247,ENSEMBL:ENSMUSG00000081563 | MGI:3650766 | pseudogene | predicted gene 12635 |
| 4 | pseudogene | 90.54374 | 90.54438 | negative | MGI_C57BL6J_3650767 | Gm12636 | NCBI_Gene:100417387,ENSEMBL:ENSMUSG00000082502 | MGI:3650767 | pseudogene | predicted gene 12636 |
| 4 | pseudogene | 90.85684 | 90.85757 | positive | MGI_C57BL6J_3650967 | Gm12632 | NCBI_Gene:606536,ENSEMBL:ENSMUSG00000082622 | MGI:3650967 | pseudogene | predicted gene 12632 |
| 4 | gene | 90.99786 | 90.99799 | negative | MGI_C57BL6J_5455021 | Gm25244 | ENSEMBL:ENSMUSG00000096896 | MGI:5455021 | snoRNA gene | predicted gene, 25244 |
| 4 | pseudogene | 91.03788 | 91.03827 | positive | MGI_C57BL6J_3649676 | Gm12643 | NCBI_Gene:108169000,ENSEMBL:ENSMUSG00000079962 | MGI:3649676 | pseudogene | predicted gene 12643 |
| 4 | pseudogene | 91.15667 | 91.15774 | positive | MGI_C57BL6J_3649677 | Gm12644 | NCBI_Gene:664848,ENSEMBL:ENSMUSG00000081876 | MGI:3649677 | pseudogene | predicted gene 12644 |
| 4 | gene | 91.21153 | 91.24604 | negative | MGI_C57BL6J_2443380 | A730080H06Rik | NCBI_Gene:102633497 | MGI:2443380 | lncRNA gene | RIKEN cDNA A730080H06 gene |
| 4 | gene | 91.25076 | 91.40078 | negative | MGI_C57BL6J_1100887 | Elavl2 | NCBI_Gene:15569,ENSEMBL:ENSMUSG00000008489 | MGI:1100887 | protein coding gene | ELAV like RNA binding protein 1 |
| 4 | pseudogene | 91.36538 | 91.36594 | positive | MGI_C57BL6J_3650062 | Gm12653 | NCBI_Gene:100418436,ENSEMBL:ENSMUSG00000083377 | MGI:3650062 | pseudogene | predicted gene 12653 |
| 4 | gene | 91.37328 | 91.37336 | negative | MGI_C57BL6J_5531146 | Mir6402 | miRBase:MI0021938,NCBI_Gene:102466157,ENSEMBL:ENSMUSG00000098563 | MGI:5531146 | miRNA gene | microRNA 6402 |
| 4 | gene | 91.49662 | 91.53649 | positive | MGI_C57BL6J_5623087 | Gm40202 | NCBI_Gene:105244619 | MGI:5623087 | lncRNA gene | predicted gene, 40202 |
| 4 | gene | 91.62659 | 91.63014 | positive | MGI_C57BL6J_3651936 | Gm12668 | ENSEMBL:ENSMUSG00000085817 | MGI:3651936 | lncRNA gene | predicted gene 12668 |
| 4 | gene | 91.64632 | 91.65115 | positive | MGI_C57BL6J_3651682 | Gm12670 | ENSEMBL:ENSMUSG00000086042 | MGI:3651682 | lncRNA gene | predicted gene 12670 |
| 4 | pseudogene | 91.70863 | 91.70997 | positive | MGI_C57BL6J_3651935 | Gm12667 | NCBI_Gene:100417505,ENSEMBL:ENSMUSG00000082523 | MGI:3651935 | pseudogene | predicted gene 12667 |
| 4 | pseudogene | 91.80555 | 91.80656 | negative | MGI_C57BL6J_3651683 | Gm12669 | NCBI_Gene:620016,ENSEMBL:ENSMUSG00000100863 | MGI:3651683 | pseudogene | predicted gene 12669 |
| 4 | pseudogene | 91.85149 | 91.85269 | negative | MGI_C57BL6J_3651684 | Gm12671 | NCBI_Gene:654475,ENSEMBL:ENSMUSG00000095937 | MGI:3651684 | pseudogene | predicted gene 12671 |
| 4 | gene | 91.93885 | 91.93995 | negative | MGI_C57BL6J_1925473 | 4930577H14Rik | ENSEMBL:ENSMUSG00000087517 | MGI:1925473 | lncRNA gene | RIKEN cDNA 4930577H14 gene |
| 4 | gene | 92.14432 | 92.14723 | negative | MGI_C57BL6J_1916564 | Izumo3 | NCBI_Gene:69314,ENSEMBL:ENSMUSG00000028533 | MGI:1916564 | protein coding gene | IZUMO family member 3 |
| 4 | gene | 92.14728 | 92.15095 | positive | MGI_C57BL6J_5623088 | Gm40203 | NCBI_Gene:105244620 | MGI:5623088 | lncRNA gene | predicted gene, 40203 |
| 4 | gene | 92.19069 | 92.19175 | negative | MGI_C57BL6J_3650864 | Gm12666 | NCBI_Gene:68280,ENSEMBL:ENSMUSG00000066107 | MGI:3650864 | protein coding gene | predicted gene 12666 |
| 4 | gene | 92.24176 | 92.26624 | positive | MGI_C57BL6J_5623089 | Gm40204 | NCBI_Gene:105244621 | MGI:5623089 | lncRNA gene | predicted gene, 40204 |
| 4 | pseudogene | 92.37305 | 92.37335 | negative | MGI_C57BL6J_3650765 | Gm12638 | NCBI_Gene:100502690,ENSEMBL:ENSMUSG00000083296 | MGI:3650765 | pseudogene | predicted gene 12638 |
| 4 | gene | 92.48461 | 92.54883 | positive | MGI_C57BL6J_3650764 | Gm12637 | NCBI_Gene:108169002,ENSEMBL:ENSMUSG00000085209 | MGI:3650764 | lncRNA gene | predicted gene 12637 |
| 4 | gene | 92.86647 | 92.86658 | negative | MGI_C57BL6J_5452382 | Gm22605 | ENSEMBL:ENSMUSG00000088914 | MGI:5452382 | snRNA gene | predicted gene, 22605 |
| 4 | pseudogene | 92.97024 | 92.97074 | negative | MGI_C57BL6J_3649270 | Gm12639 | ENSEMBL:ENSMUSG00000081987 | MGI:3649270 | pseudogene | predicted gene 12639 |
| 4 | pseudogene | 93.10697 | 93.11012 | positive | MGI_C57BL6J_3649567 | Gm12641 | NCBI_Gene:620269,ENSEMBL:ENSMUSG00000082919 | MGI:3649567 | pseudogene | predicted gene 12641 |
| 4 | pseudogene | 93.26541 | 93.26629 | negative | MGI_C57BL6J_3649275 | Gm12642 | NCBI_Gene:384125,ENSEMBL:ENSMUSG00000082325 | MGI:3649275 | pseudogene | predicted gene 12642 |
| 4 | gene | 93.33414 | 93.33551 | negative | MGI_C57BL6J_2684283 | Tusc1 | NCBI_Gene:69136,ENSEMBL:ENSMUSG00000054000 | MGI:2684283 | protein coding gene | tumor suppressor candidate 1 |
| 4 | gene | 93.37041 | 93.48140 | negative | MGI_C57BL6J_5590987 | Gm31828 | NCBI_Gene:102634183 | MGI:5590987 | lncRNA gene | predicted gene, 31828 |
| 4 | gene | 93.45326 | 93.67107 | negative | MGI_C57BL6J_3649947 | Gm12649 | ENSEMBL:ENSMUSG00000084833 | MGI:3649947 | lncRNA gene | predicted gene 12649 |
| 4 | gene | 93.54515 | 93.54528 | negative | MGI_C57BL6J_5453220 | Gm23443 | ENSEMBL:ENSMUSG00000064850 | MGI:5453220 | snoRNA gene | predicted gene, 23443 |
| 4 | pseudogene | 93.59919 | 93.59950 | positive | MGI_C57BL6J_3649267 | Gm12640 | ENSEMBL:ENSMUSG00000083607 | MGI:3649267 | pseudogene | predicted gene 12640 |
| 4 | gene | 93.60098 | 93.60108 | positive | MGI_C57BL6J_5454573 | Gm24796 | ENSEMBL:ENSMUSG00000095898 | MGI:5454573 | snRNA gene | predicted gene, 24796 |
| 4 | gene | 93.74589 | 93.74600 | negative | MGI_C57BL6J_5452396 | Gm22619 | ENSEMBL:ENSMUSG00000095856 | MGI:5452396 | miRNA gene | predicted gene, 22619 |
| 4 | pseudogene | 93.80233 | 93.80262 | positive | MGI_C57BL6J_3649946 | Gm12650 | NCBI_Gene:102634336,ENSEMBL:ENSMUSG00000073797 | MGI:3649946 | pseudogene | predicted gene 12650 |
| 4 | gene | 93.83042 | 93.86423 | negative | MGI_C57BL6J_5591194 | Gm32035 | NCBI_Gene:102634455 | MGI:5591194 | lncRNA gene | predicted gene, 32035 |
| 4 | pseudogene | 94.06114 | 94.06165 | positive | MGI_C57BL6J_3649949 | Gm12647 | NCBI_Gene:100417559,ENSEMBL:ENSMUSG00000081685 | MGI:3649949 | pseudogene | predicted gene 12647 |
| 4 | gene | 94.08814 | 94.42559 | negative | MGI_C57BL6J_3649948 | Gm12648 | NCBI_Gene:108168967,ENSEMBL:ENSMUSG00000085931 | MGI:3649948 | lncRNA gene | predicted gene 12648 |
| 4 | gene | 94.09829 | 94.09842 | positive | MGI_C57BL6J_5453613 | Gm23836 | ENSEMBL:ENSMUSG00000077622 | MGI:5453613 | snoRNA gene | predicted gene, 23836 |
| 4 | pseudogene | 94.10404 | 94.10484 | negative | MGI_C57BL6J_5010190 | Gm18005 | NCBI_Gene:100416254 | MGI:5010190 | pseudogene | predicted gene, 18005 |
| 4 | pseudogene | 94.39562 | 94.39624 | positive | MGI_C57BL6J_3651710 | Gm12654 | NCBI_Gene:100416259,ENSEMBL:ENSMUSG00000081508 | MGI:3651710 | pseudogene | predicted gene 12654 |
| 4 | gene | 94.43422 | 94.43448 | negative | MGI_C57BL6J_5456187 | Gm26410 | ENSEMBL:ENSMUSG00000088295 | MGI:5456187 | unclassified non-coding RNA gene | predicted gene, 26410 |
| 4 | gene | 94.46028 | 94.55708 | negative | MGI_C57BL6J_1915020 | Caap1 | NCBI_Gene:67770,ENSEMBL:ENSMUSG00000028578 | MGI:1915020 | protein coding gene | caspase activity and apoptosis inhibitor 1 |
| 4 | gene | 94.46093 | 94.47718 | negative | MGI_C57BL6J_3651648 | Gm12655 | ENSEMBL:ENSMUSG00000087128 | MGI:3651648 | lncRNA gene | predicted gene 12655 |
| 4 | gene | 94.48788 | 94.48798 | positive | MGI_C57BL6J_5452581 | Gm22804 | ENSEMBL:ENSMUSG00000088677 | MGI:5452581 | snoRNA gene | predicted gene, 22804 |
| 4 | pseudogene | 94.49699 | 94.49761 | positive | MGI_C57BL6J_3651715 | Gm12656 | NCBI_Gene:100418440,ENSEMBL:ENSMUSG00000083359 | MGI:3651715 | pseudogene | predicted gene 12656 |
| 4 | gene | 94.55655 | 94.55708 | positive | MGI_C57BL6J_3704234 | Gm10306 | NA | NA | protein coding gene | predicted gene 10306 |
| 4 | gene | 94.56514 | 94.60325 | negative | MGI_C57BL6J_104810 | Plaa | NCBI_Gene:18786,ENSEMBL:ENSMUSG00000028577 | MGI:104810 | protein coding gene | phospholipase A2, activating protein |
| 4 | pseudogene | 94.59365 | 94.60032 | positive | MGI_C57BL6J_3651714 | Gm12657 | NCBI_Gene:667250,ENSEMBL:ENSMUSG00000063314 | MGI:3651714 | pseudogene | predicted gene 12657 |
| 4 | gene | 94.60326 | 94.69323 | positive | MGI_C57BL6J_1914944 | Ift74 | NCBI_Gene:67694,ENSEMBL:ENSMUSG00000028576 | MGI:1914944 | protein coding gene | intraflagellar transport 74 |
| 4 | gene | 94.63665 | 94.65014 | negative | MGI_C57BL6J_2140219 | Lrrc19 | NCBI_Gene:100061,ENSEMBL:ENSMUSG00000049799 | MGI:2140219 | protein coding gene | leucine rich repeat containing 19 |
| 4 | gene | 94.66516 | 94.66524 | positive | MGI_C57BL6J_3718562 | Mir872 | miRBase:MI0005549,NCBI_Gene:100124456,ENSEMBL:ENSMUSG00000077921 | MGI:3718562 | miRNA gene | microRNA 872 |
| 4 | gene | 94.72160 | 94.73256 | positive | MGI_C57BL6J_5623090 | Gm40205 | NCBI_Gene:105244622 | MGI:5623090 | lncRNA gene | predicted gene, 40205 |
| 4 | gene | 94.73929 | 94.87498 | positive | MGI_C57BL6J_98664 | Tek | NCBI_Gene:21687,ENSEMBL:ENSMUSG00000006386 | MGI:98664 | protein coding gene | TEK receptor tyrosine kinase |
| 4 | gene | 94.87570 | 94.92995 | negative | MGI_C57BL6J_1915003 | Eqtn | NCBI_Gene:67753,ENSEMBL:ENSMUSG00000028575 | MGI:1915003 | protein coding gene | equatorin, sperm acrosome associated |
| 4 | pseudogene | 94.93327 | 94.93629 | positive | MGI_C57BL6J_3650224 | Gm12693 | NCBI_Gene:100039252,ENSEMBL:ENSMUSG00000083179 | MGI:3650224 | pseudogene | predicted gene 12693 |
| 4 | gene | 94.94204 | 94.97911 | negative | MGI_C57BL6J_2444584 | Mysm1 | NCBI_Gene:320713,ENSEMBL:ENSMUSG00000062627 | MGI:2444584 | protein coding gene | myb-like, SWIRM and MPN domains 1 |
| 4 | gene | 94.97920 | 95.02854 | positive | MGI_C57BL6J_3651291 | Gm12694 | NCBI_Gene:105244623,ENSEMBL:ENSMUSG00000086251 | MGI:3651291 | lncRNA gene | predicted gene 12694 |
| 4 | gene | 95.02996 | 95.04837 | negative | MGI_C57BL6J_3650667 | Gm12703 | ENSEMBL:ENSMUSG00000052767 | MGI:3650667 | lncRNA gene | predicted gene 12703 |
| 4 | pseudogene | 95.03916 | 95.03963 | negative | MGI_C57BL6J_3650671 | Gm12704 | ENSEMBL:ENSMUSG00000081030 | MGI:3650671 | pseudogene | predicted gene 12704 |
| 4 | gene | 95.04903 | 95.05222 | negative | MGI_C57BL6J_96646 | Jun | NCBI_Gene:16476,ENSEMBL:ENSMUSG00000052684 | MGI:96646 | protein coding gene | jun proto-oncogene |
| 4 | gene | 95.05153 | 95.16904 | positive | MGI_C57BL6J_2652837 | Junos | NCBI_Gene:230451,ENSEMBL:ENSMUSG00000087366 | MGI:2652837 | antisense lncRNA gene | jun proto-oncogene, opposite strand |
| 4 | gene | 95.06265 | 95.06276 | positive | MGI_C57BL6J_4422054 | n-R5s189 | ENSEMBL:ENSMUSG00000084624 | MGI:4422054 | rRNA gene | nuclear encoded rRNA 5S 189 |
| 4 | gene | 95.08908 | 95.09142 | negative | MGI_C57BL6J_1922511 | 4930551L18Rik | ENSEMBL:ENSMUSG00000085377 | MGI:1922511 | lncRNA gene | RIKEN cDNA 4930551L18 gene |
| 4 | gene | 95.15174 | 95.15750 | positive | MGI_C57BL6J_5578802 | Gm28096 | ENSEMBL:ENSMUSG00000100189 | MGI:5578802 | lncRNA gene | predicted gene 28096 |
| 4 | gene | 95.19327 | 95.20973 | positive | MGI_C57BL6J_3652178 | Gm12708 | NCBI_Gene:105244624,ENSEMBL:ENSMUSG00000084918 | MGI:3652178 | lncRNA gene | predicted gene 12708 |
| 4 | gene | 95.19691 | 95.20808 | negative | MGI_C57BL6J_5591399 | Gm32240 | NCBI_Gene:102634722 | MGI:5591399 | lncRNA gene | predicted gene, 32240 |
| 4 | pseudogene | 95.25483 | 95.25916 | positive | MGI_C57BL6J_5826500 | Gm46863 | NCBI_Gene:108168968 | MGI:5826500 | pseudogene | predicted gene, 46863 |
| 4 | gene | 95.27219 | 95.27686 | positive | MGI_C57BL6J_3782003 | Gm3831 | NA | NA | protein coding gene | predicted gene 3831 |
| 4 | gene | 95.40198 | 95.40279 | negative | MGI_C57BL6J_5579770 | Gm29064 | ENSEMBL:ENSMUSG00000100129 | MGI:5579770 | lncRNA gene | predicted gene 29064 |
| 4 | gene | 95.46472 | 95.48739 | negative | MGI_C57BL6J_2685676 | Gm830 | NCBI_Gene:329892,ENSEMBL:ENSMUSG00000084939 | MGI:2685676 | lncRNA gene | predicted gene 830 |
| 4 | gene | 95.48396 | 95.48763 | positive | MGI_C57BL6J_3652172 | Gm12707 | NCBI_Gene:102636934,ENSEMBL:ENSMUSG00000085835 | MGI:3652172 | lncRNA gene | predicted gene 12707 |
| 4 | gene | 95.52404 | 95.52420 | positive | MGI_C57BL6J_5454781 | Gm25004 | ENSEMBL:ENSMUSG00000065745 | MGI:5454781 | snRNA gene | predicted gene, 25004 |
| 4 | gene | 95.55342 | 95.55737 | negative | MGI_C57BL6J_5623091 | Gm40206 | NCBI_Gene:105244625 | MGI:5623091 | lncRNA gene | predicted gene, 40206 |
| 4 | gene | 95.55747 | 95.92694 | positive | MGI_C57BL6J_1922828 | Fggy | NCBI_Gene:75578,ENSEMBL:ENSMUSG00000028573 | MGI:1922828 | protein coding gene | FGGY carbohydrate kinase domain containing |
| 4 | pseudogene | 95.56183 | 95.56216 | negative | MGI_C57BL6J_5313098 | Gm20651 | ENSEMBL:ENSMUSG00000093492 | MGI:5313098 | pseudogene | predicted gene 20651 |
| 4 | gene | 95.63474 | 95.63523 | positive | MGI_C57BL6J_1922327 | 4930509A12Rik | NA | NA | unclassified gene | RIKEN cDNA 4930509A12 gene |
| 4 | gene | 95.95971 | 95.96765 | negative | MGI_C57BL6J_2441751 | 9530080O11Rik | NCBI_Gene:319247,ENSEMBL:ENSMUSG00000044125 | MGI:2441751 | lncRNA gene | RIKEN cDNA 9530080O11 gene |
| 4 | gene | 95.96720 | 96.02541 | positive | MGI_C57BL6J_1925213 | Hook1 | NCBI_Gene:77963,ENSEMBL:ENSMUSG00000028572 | MGI:1925213 | protein coding gene | hook microtubule tethering protein 1 |
| 4 | gene | 96.02753 | 96.07757 | negative | MGI_C57BL6J_2385197 | Cyp2j13 | NCBI_Gene:230459,ENSEMBL:ENSMUSG00000028571 | MGI:2385197 | protein coding gene | cytochrome P450, family 2, subfamily j, polypeptide 13 |
| 4 | gene | 96.02976 | 96.03075 | positive | MGI_C57BL6J_1920262 | 2900072D07Rik | NA | NA | unclassified gene | RIKEN cDNA 2900072D07 gene |
| 4 | pseudogene | 96.07231 | 96.07271 | negative | MGI_C57BL6J_1935167 | Hspe1-ps6 | NCBI_Gene:93753,ENSEMBL:ENSMUSG00000083880 | MGI:1935167 | pseudogene | heat shock protein 1 (chaperonin 10), pseudogene 6 |
| 4 | gene | 96.09931 | 96.14127 | negative | MGI_C57BL6J_3717097 | Cyp2j12 | NCBI_Gene:242546,ENSEMBL:ENSMUSG00000081225 | MGI:3717097 | protein coding gene | cytochrome P450, family 2, subfamily j, polypeptide 12 |
| 4 | pseudogene | 96.15165 | 96.15186 | negative | MGI_C57BL6J_3651269 | Cyp2j7-ps3 | ENSEMBL:ENSMUSG00000080884 | MGI:3651269 | pseudogene | cytochrome P450, family 2, subfamily j, polypeptide 7, pseudogene 3 |
| 4 | pseudogene | 96.17021 | 96.17038 | negative | MGI_C57BL6J_3651264 | Cyp2j7-ps2 | ENSEMBL:ENSMUSG00000083801 | MGI:3651264 | pseudogene | cytochrome P450, family 2, subfamily j, polypeptide 7, pseudogene 2 |
| 4 | pseudogene | 96.19007 | 96.19024 | negative | MGI_C57BL6J_3651268 | Cyp2j7-ps1 | ENSEMBL:ENSMUSG00000082681 | MGI:3651268 | pseudogene | cytochrome P450, family 2, subfamily j, polypeptide 7, pseudogene 1 |
| 4 | gene | 96.19519 | 96.23666 | negative | MGI_C57BL6J_2449816 | Cyp2j7 | NCBI_Gene:546837,ENSEMBL:ENSMUSG00000081362 | MGI:2449816 | protein coding gene | cytochrome P450, family 2, subfamily j, polypeptide 7 |
| 4 | pseudogene | 96.24556 | 96.27427 | negative | MGI_C57BL6J_3652069 | Cyp2j15-ps | NCBI_Gene:100125600,ENSEMBL:ENSMUSG00000084286 | MGI:3652069 | pseudogene | cytochrome P450, family 2, subfamily j, member 15, pseudogene |
| 4 | gene | 96.29451 | 96.34870 | negative | MGI_C57BL6J_2140224 | Cyp2j11 | NCBI_Gene:100066,ENSEMBL:ENSMUSG00000066097 | MGI:2140224 | protein coding gene | cytochrome P450, family 2, subfamily j, polypeptide 11 |
| 4 | pseudogene | 96.35625 | 96.38022 | negative | MGI_C57BL6J_3651954 | Cyp2j14-ps | NCBI_Gene:100125599,ENSEMBL:ENSMUSG00000082119 | MGI:3651954 | pseudogene | cytochrome P450, family 2, subfamily j, member 14, pseudogene |
| 4 | gene | 96.44399 | 96.50794 | negative | MGI_C57BL6J_2449817 | Cyp2j8 | NCBI_Gene:665095,ENSEMBL:ENSMUSG00000082932 | MGI:2449817 | protein coding gene | cytochrome P450, family 2, subfamily j, polypeptide 8 |
| 4 | gene | 96.51614 | 96.56431 | negative | MGI_C57BL6J_1270148 | Cyp2j6 | NCBI_Gene:13110,ENSEMBL:ENSMUSG00000052914 | MGI:1270148 | protein coding gene | cytochrome P450, family 2, subfamily j, polypeptide 6 |
| 4 | gene | 96.56843 | 96.59172 | negative | MGI_C57BL6J_1921769 | Cyp2j9 | NCBI_Gene:74519,ENSEMBL:ENSMUSG00000015224 | MGI:1921769 | protein coding gene | cytochrome P450, family 2, subfamily j, polypeptide 9 |
| 4 | gene | 96.62743 | 96.66417 | negative | MGI_C57BL6J_1270149 | Cyp2j5 | NCBI_Gene:13109,ENSEMBL:ENSMUSG00000052520 | MGI:1270149 | protein coding gene | cytochrome P450, family 2, subfamily j, polypeptide 5 |
| 4 | gene | 96.72335 | 96.78683 | negative | MGI_C57BL6J_3650206 | Gm12695 | NCBI_Gene:620779,ENSEMBL:ENSMUSG00000078639 | MGI:3650206 | protein coding gene | predicted gene 12695 |
| 4 | gene | 96.78732 | 96.79490 | positive | MGI_C57BL6J_5593212 | Gm34053 | NCBI_Gene:102637171 | MGI:5593212 | lncRNA gene | predicted gene, 34053 |
| 4 | gene | 96.91694 | 96.91701 | positive | MGI_C57BL6J_5530961 | Gm27579 | ENSEMBL:ENSMUSG00000088738 | MGI:5530961 | snRNA gene | predicted gene, 27579 |
| 4 | gene | 96.91702 | 96.91708 | positive | MGI_C57BL6J_5530903 | Gm27521 | ENSEMBL:ENSMUSG00000098261 | MGI:5530903 | snRNA gene | predicted gene, 27521 |
| 4 | gene | 97.18108 | 97.18317 | negative | MGI_C57BL6J_3642867 | Gm10192 | NA | NA | protein coding gene | predicted gene 10192 |
| 4 | gene | 97.41542 | 97.41663 | negative | MGI_C57BL6J_5623092 | Gm40207 | NCBI_Gene:105244626 | MGI:5623092 | lncRNA gene | predicted gene, 40207 |
| 4 | pseudogene | 97.52127 | 97.52255 | negative | MGI_C57BL6J_3649288 | Gm12696 | NCBI_Gene:620883,ENSEMBL:ENSMUSG00000084159 | MGI:3649288 | pseudogene | predicted gene 12696 |
| 4 | pseudogene | 97.55640 | 97.55706 | positive | MGI_C57BL6J_3643458 | Gm7248 | NCBI_Gene:638698 | MGI:3643458 | pseudogene | predicted gene 7248 |
| 4 | pseudogene | 97.55640 | 97.55707 | negative | MGI_C57BL6J_3651292 | Gm12697 | ENSEMBL:ENSMUSG00000082683 | MGI:3651292 | pseudogene | predicted gene 12697 |
| 4 | gene | 97.56788 | 97.77808 | negative | MGI_C57BL6J_2442873 | E130114P18Rik | NCBI_Gene:319865,ENSEMBL:ENSMUSG00000048747 | MGI:2442873 | protein coding gene | RIKEN cDNA E130114P18 gene |
| 4 | gene | 97.58172 | 98.11888 | positive | MGI_C57BL6J_108056 | Nfia | NCBI_Gene:18027,ENSEMBL:ENSMUSG00000028565 | MGI:108056 | protein coding gene | nuclear factor I/A |
| 4 | gene | 97.58233 | 97.58462 | negative | MGI_C57BL6J_2444134 | D130039L10Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA D130039L10 gene |
| 4 | gene | 97.74710 | 97.76101 | negative | MGI_C57BL6J_3651064 | Gm12676 | NCBI_Gene:102637577,ENSEMBL:ENSMUSG00000085710 | MGI:3651064 | lncRNA gene | predicted gene 12676 |
| 4 | gene | 97.77369 | 97.77743 | negative | MGI_C57BL6J_5621594 | Gm38709 | NCBI_Gene:105242444 | MGI:5621594 | lncRNA gene | predicted gene, 38709 |
| 4 | gene | 97.77452 | 97.77644 | negative | MGI_C57BL6J_2443783 | A130013F12Rik | NA | NA | unclassified gene | RIKEN cDNA A130013F12 gene |
| 4 | gene | 97.86241 | 97.86923 | negative | MGI_C57BL6J_5011602 | Gm19417 | NCBI_Gene:102637515 | MGI:5011602 | lncRNA gene | predicted gene, 19417 |
| 4 | gene | 97.86726 | 97.87008 | positive | MGI_C57BL6J_1914791 | 4932416J16Rik | NA | NA | unclassified gene | RIKEN cDNA 4932416J16 gene |
| 4 | gene | 97.93042 | 97.94099 | positive | MGI_C57BL6J_5623093 | Gm40208 | NCBI_Gene:105244627 | MGI:5623093 | lncRNA gene | predicted gene, 40208 |
| 4 | gene | 98.01856 | 98.02477 | negative | MGI_C57BL6J_5593377 | Gm34218 | NCBI_Gene:102637398 | MGI:5593377 | lncRNA gene | predicted gene, 34218 |
| 4 | gene | 98.12565 | 98.13127 | positive | MGI_C57BL6J_5593943 | Gm34784 | NCBI_Gene:102638152 | MGI:5593943 | lncRNA gene | predicted gene, 34784 |
| 4 | pseudogene | 98.14527 | 98.24446 | negative | MGI_C57BL6J_3650220 | Gm12691 | NCBI_Gene:100039459,ENSEMBL:ENSMUSG00000081096 | MGI:3650220 | pseudogene | predicted gene 12691 |
| 4 | pseudogene | 98.23475 | 98.23529 | negative | MGI_C57BL6J_3651859 | Gm12692 | NCBI_Gene:384106,ENSEMBL:ENSMUSG00000082388 | MGI:3651859 | pseudogene | predicted gene 12692 |
| 4 | pseudogene | 98.26808 | 98.27129 | negative | MGI_C57BL6J_3650112 | Gm12787 | NCBI_Gene:100502616,ENSEMBL:ENSMUSG00000083644 | MGI:3650112 | pseudogene | predicted gene 12787 |
| 4 | gene | 98.27572 | 98.27584 | positive | MGI_C57BL6J_5453980 | Gm24203 | ENSEMBL:ENSMUSG00000064883 | MGI:5453980 | unclassified non-coding RNA gene | predicted gene, 24203 |
| 4 | gene | 98.32134 | 98.33196 | positive | MGI_C57BL6J_1915618 | 0610025J13Rik | NCBI_Gene:78687,ENSEMBL:ENSMUSG00000046683 | MGI:1915618 | lncRNA gene | RIKEN cDNA 0610025J13 gene |
| 4 | pseudogene | 98.35243 | 98.35273 | positive | MGI_C57BL6J_3649859 | Gm12788 | ENSEMBL:ENSMUSG00000082144 | MGI:3649859 | pseudogene | predicted gene 12788 |
| 4 | gene | 98.35537 | 98.38331 | negative | MGI_C57BL6J_2137022 | Tm2d1 | NCBI_Gene:94043,ENSEMBL:ENSMUSG00000028563 | MGI:2137022 | protein coding gene | TM2 domain containing 1 |
| 4 | gene | 98.38340 | 98.38349 | negative | MGI_C57BL6J_5454737 | Gm24960 | ENSEMBL:ENSMUSG00000088548 | MGI:5454737 | snRNA gene | predicted gene, 24960 |
| 4 | gene | 98.39562 | 98.71960 | positive | MGI_C57BL6J_1277960 | Patj | NCBI_Gene:12695,ENSEMBL:ENSMUSG00000061859 | MGI:1277960 | protein coding gene | PATJ, crumbs cell polarity complex component |
| 4 | gene | 98.55774 | 98.55787 | negative | MGI_C57BL6J_5452402 | Gm22625 | ENSEMBL:ENSMUSG00000094456 | MGI:5452402 | miRNA gene | predicted gene, 22625 |
| 4 | gene | 98.57953 | 98.58801 | positive | MGI_C57BL6J_5623095 | Gm40210 | NCBI_Gene:105244629 | MGI:5623095 | lncRNA gene | predicted gene, 40210 |
| 4 | gene | 98.66164 | 98.66173 | positive | MGI_C57BL6J_5455931 | Gm26154 | ENSEMBL:ENSMUSG00000076294 | MGI:5455931 | miRNA gene | predicted gene, 26154 |
| 4 | pseudogene | 98.66437 | 98.66614 | positive | MGI_C57BL6J_3651461 | Gm12846 | NCBI_Gene:100417687,ENSEMBL:ENSMUSG00000081046 | MGI:3651461 | pseudogene | predicted gene 12846 |
| 4 | gene | 98.69689 | 98.69978 | negative | MGI_C57BL6J_5594296 | Gm35137 | NCBI_Gene:102638614 | MGI:5594296 | lncRNA gene | predicted gene, 35137 |
| 4 | gene | 98.70045 | 98.70076 | negative | MGI_C57BL6J_5594232 | Gm35073 | NCBI_Gene:102638531 | MGI:5594232 | lncRNA gene | predicted gene, 35073 |
| 4 | gene | 98.72673 | 98.73849 | positive | MGI_C57BL6J_3578435 | L1td1 | NCBI_Gene:381591,ENSEMBL:ENSMUSG00000087166 | MGI:3578435 | protein coding gene | LINE-1 type transposase domain containing 1 |
| 4 | pseudogene | 98.74573 | 98.74582 | positive | MGI_C57BL6J_4937010 | Gm17376 | ENSEMBL:ENSMUSG00000090718 | MGI:4937010 | pseudogene | predicted gene, 17376 |
| 4 | gene | 98.75489 | 98.81780 | negative | MGI_C57BL6J_3043381 | Kank4 | NCBI_Gene:242553,ENSEMBL:ENSMUSG00000035407 | MGI:3043381 | protein coding gene | KN motif and ankyrin repeat domains 4 |
| 4 | gene | 98.79377 | 98.79904 | positive | MGI_C57BL6J_3026921 | Kank4os | NCBI_Gene:399570,ENSEMBL:ENSMUSG00000085843 | MGI:3026921 | antisense lncRNA gene | KN motif and ankyrin repeat domains 4, opposite strand |
| 4 | gene | 98.81848 | 98.82384 | negative | MGI_C57BL6J_3588230 | I0C0044D17Rik | NCBI_Gene:545660,ENSEMBL:ENSMUSG00000073794 | MGI:3588230 | protein coding gene | RIKEN cDNA I0C0044D17 gene |
| 4 | pseudogene | 98.85274 | 98.85606 | positive | MGI_C57BL6J_3649254 | Gm12851 | NCBI_Gene:102639194,ENSEMBL:ENSMUSG00000084188 | MGI:3649254 | pseudogene | predicted gene 12851 |
| 4 | gene | 98.92362 | 98.93554 | positive | MGI_C57BL6J_2385198 | Usp1 | NCBI_Gene:230484,ENSEMBL:ENSMUSG00000028560 | MGI:2385198 | protein coding gene | ubiquitin specific peptidase 1 |
| 4 | gene | 98.93666 | 99.12179 | negative | MGI_C57BL6J_1914549 | Dock7 | NCBI_Gene:67299,ENSEMBL:ENSMUSG00000028556 | MGI:1914549 | protein coding gene | dedicator of cytokinesis 7 |
| 4 | gene | 99.03095 | 99.04611 | positive | MGI_C57BL6J_1353627 | Angptl3 | NCBI_Gene:30924,ENSEMBL:ENSMUSG00000028553 | MGI:1353627 | protein coding gene | angiopoietin-like 3 |
| 4 | pseudogene | 99.10295 | 99.10383 | negative | MGI_C57BL6J_3651942 | Gm12852 | NCBI_Gene:100417329,ENSEMBL:ENSMUSG00000081733 | MGI:3651942 | pseudogene | predicted gene 12852 |
| 4 | gene | 99.11641 | 99.11830 | positive | MGI_C57BL6J_3650670 | Gm12705 | ENSEMBL:ENSMUSG00000087395 | MGI:3650670 | lncRNA gene | predicted gene 12705 |
| 4 | gene | 99.14691 | 99.16164 | negative | MGI_C57BL6J_3650649 | Gm12681 | ENSEMBL:ENSMUSG00000086498 | MGI:3650649 | lncRNA gene | predicted gene 12681 |
| 4 | gene | 99.18916 | 99.29726 | positive | MGI_C57BL6J_2651854 | Atg4c | NCBI_Gene:242557,ENSEMBL:ENSMUSG00000028550 | MGI:2651854 | protein coding gene | autophagy related 4C, cysteine peptidase |
| 4 | gene | 99.27267 | 99.27514 | positive | MGI_C57BL6J_3642542 | Gm10305 | ENSEMBL:ENSMUSG00000104479 | MGI:3642542 | unclassified gene | predicted gene 10305 |
| 4 | gene | 99.29590 | 99.29726 | positive | MGI_C57BL6J_3650433 | Gm12689 | ENSEMBL:ENSMUSG00000070891 | MGI:3650433 | protein coding gene | predicted gene 12689 |
| 4 | gene | 99.31463 | 99.33291 | positive | MGI_C57BL6J_5594538 | Gm35379 | NCBI_Gene:102638939 | MGI:5594538 | lncRNA gene | predicted gene, 35379 |
| 4 | gene | 99.34218 | 99.34281 | negative | MGI_C57BL6J_5594696 | Gm35537 | NCBI_Gene:102639160 | MGI:5594696 | lncRNA gene | predicted gene, 35537 |
| 4 | gene | 99.37607 | 99.38571 | negative | MGI_C57BL6J_5610784 | Gm37556 | ENSEMBL:ENSMUSG00000103097 | MGI:5610784 | unclassified gene | predicted gene, 37556 |
| 4 | pseudogene | 99.41850 | 99.41871 | negative | MGI_C57BL6J_3650437 | Gm12685 | ENSEMBL:ENSMUSG00000081184 | MGI:3650437 | pseudogene | predicted gene 12685 |
| 4 | pseudogene | 99.41877 | 99.41917 | positive | MGI_C57BL6J_3652197 | Gm12686 | ENSEMBL:ENSMUSG00000082442 | MGI:3652197 | pseudogene | predicted gene 12686 |
| 4 | pseudogene | 99.43006 | 99.43127 | negative | MGI_C57BL6J_3650091 | Gm12684 | NCBI_Gene:665212,ENSEMBL:ENSMUSG00000081150 | MGI:3650091 | pseudogene | predicted gene 12684 |
| 4 | pseudogene | 99.52005 | 99.52047 | negative | MGI_C57BL6J_3650644 | Gm12683 | ENSEMBL:ENSMUSG00000080803 | MGI:3650644 | pseudogene | predicted gene 12683 |
| 4 | gene | 99.52593 | 99.53066 | positive | MGI_C57BL6J_5610345 | Gm37117 | ENSEMBL:ENSMUSG00000103575 | MGI:5610345 | unclassified gene | predicted gene, 37117 |
| 4 | gene | 99.55479 | 99.56935 | negative | MGI_C57BL6J_5623096 | Gm40211 | NCBI_Gene:105244631 | MGI:5623096 | lncRNA gene | predicted gene, 40211 |
| 4 | gene | 99.56950 | 99.57301 | positive | MGI_C57BL6J_3650444 | Gm12690 | ENSEMBL:ENSMUSG00000087332 | MGI:3650444 | lncRNA gene | predicted gene 12690 |
| 4 | pseudogene | 99.61580 | 99.61656 | negative | MGI_C57BL6J_3650435 | Gm12687 | NCBI_Gene:100417847,ENSEMBL:ENSMUSG00000083698 | MGI:3650435 | pseudogene | predicted gene 12687 |
| 4 | gene | 99.65361 | 99.65498 | negative | MGI_C57BL6J_3650434 | Gm12688 | ENSEMBL:ENSMUSG00000086096 | MGI:3650434 | lncRNA gene | predicted gene 12688 |
| 4 | gene | 99.65572 | 99.65601 | negative | MGI_C57BL6J_1920013 | 2810439L12Rik | NA | NA | unclassified gene | RIKEN cDNA 2810439L12 gene |
| 4 | gene | 99.65630 | 99.65867 | positive | MGI_C57BL6J_1347473 | Foxd3 | NCBI_Gene:15221,ENSEMBL:ENSMUSG00000067261 | MGI:1347473 | protein coding gene | forkhead box D3 |
| 4 | gene | 99.65774 | 99.65783 | positive | MGI_C57BL6J_5453143 | Gm23366 | ENSEMBL:ENSMUSG00000092763 | MGI:5453143 | miRNA gene | predicted gene, 23366 |
| 4 | gene | 99.66254 | 99.71297 | negative | MGI_C57BL6J_3650643 | Gm12682 | ENSEMBL:ENSMUSG00000086592 | MGI:3650643 | lncRNA gene | predicted gene 12682 |
| 4 | gene | 99.71560 | 99.76346 | positive | MGI_C57BL6J_2444031 | Alg6 | NCBI_Gene:320438,ENSEMBL:ENSMUSG00000073792 | MGI:2444031 | protein coding gene | asparagine-linked glycosylation 6 (alpha-1,3,-glucosyltransferase) |
| 4 | gene | 99.76540 | 99.92981 | negative | MGI_C57BL6J_1914983 | Itgb3bp | NCBI_Gene:67733,ENSEMBL:ENSMUSG00000028549 | MGI:1914983 | protein coding gene | integrin beta 3 binding protein (beta3-endonexin) |
| 4 | gene | 99.82920 | 99.91279 | positive | MGI_C57BL6J_2385199 | Efcab7 | NCBI_Gene:230500,ENSEMBL:ENSMUSG00000073791 | MGI:2385199 | protein coding gene | EF-hand calcium binding domain 7 |
| 4 | gene | 99.90980 | 99.90988 | positive | MGI_C57BL6J_5456202 | Gm26425 | ENSEMBL:ENSMUSG00000084478 | MGI:5456202 | snRNA gene | predicted gene, 26425 |
| 4 | gene | 99.92941 | 99.98729 | positive | MGI_C57BL6J_97565 | Pgm1 | NCBI_Gene:72157,ENSEMBL:ENSMUSG00000025791 | MGI:97565 | protein coding gene | phosphoglucomutase 1 |
| 4 | gene | 99.92982 | 99.93233 | positive | MGI_C57BL6J_3040690 | BC062258 | NCBI_Gene:102643247 | MGI:3040690 | lncRNA gene | cDNA sequence BC062258 |
| 4 | gene | 100.05252 | 100.06598 | positive | MGI_C57BL6J_3650874 | Gm12702 | ENSEMBL:ENSMUSG00000085142 | MGI:3650874 | lncRNA gene | predicted gene 12702 |
| 4 | gene | 100.09276 | 100.09564 | negative | MGI_C57BL6J_5594860 | Gm35701 | NCBI_Gene:102639371 | MGI:5594860 | lncRNA gene | predicted gene, 35701 |
| 4 | gene | 100.09579 | 100.44481 | positive | MGI_C57BL6J_1347520 | Ror1 | NCBI_Gene:26563,ENSEMBL:ENSMUSG00000035305 | MGI:1347520 | protein coding gene | receptor tyrosine kinase-like orphan receptor 1 |
| 4 | gene | 100.11962 | 100.12278 | positive | MGI_C57BL6J_3650875 | Gm12701 | NCBI_Gene:105244634,ENSEMBL:ENSMUSG00000086385 | MGI:3650875 | lncRNA gene | predicted gene 12701 |
| 4 | pseudogene | 100.36136 | 100.36206 | negative | MGI_C57BL6J_3650672 | Gm12706 | ENSEMBL:ENSMUSG00000080967 | MGI:3650672 | pseudogene | predicted gene 12706 |
| 4 | gene | 100.37658 | 100.39321 | negative | MGI_C57BL6J_5594919 | Gm35760 | NCBI_Gene:102639448 | MGI:5594919 | lncRNA gene | predicted gene, 35760 |
| 4 | pseudogene | 100.46587 | 100.46714 | positive | MGI_C57BL6J_5010473 | Gm18288 | NCBI_Gene:100416841 | MGI:5010473 | pseudogene | predicted gene, 18288 |
| 4 | gene | 100.47885 | 100.55015 | positive | MGI_C57BL6J_3588216 | Ube2u | NCBI_Gene:381534,ENSEMBL:ENSMUSG00000069733 | MGI:3588216 | protein coding gene | ubiquitin-conjugating enzyme E2U (putative) |
| 4 | gene | 100.52323 | 100.52803 | negative | MGI_C57BL6J_3651101 | Ube2uos | NCBI_Gene:105244635,ENSEMBL:ENSMUSG00000085871 | MGI:3651101 | antisense lncRNA gene | ubiquitin-conjugating enzyme E2U (putative), opposite strand |
| 4 | pseudogene | 100.56657 | 100.56816 | negative | MGI_C57BL6J_3651098 | Ccdc50-ps | NCBI_Gene:277707,ENSEMBL:ENSMUSG00000081553 | MGI:3651098 | pseudogene | Ccdc50 retrotransposed pseudogene |
| 4 | pseudogene | 100.57935 | 100.58173 | positive | MGI_C57BL6J_3651090 | Gm12700 | NCBI_Gene:621573,ENSEMBL:ENSMUSG00000082601 | MGI:3651090 | pseudogene | predicted gene 12700 |
| 4 | gene | 100.68767 | 100.68901 | negative | MGI_C57BL6J_5826525 | Gm46888 | NCBI_Gene:108169003 | MGI:5826525 | lncRNA gene | predicted gene, 46888 |
| 4 | gene | 100.77667 | 101.02922 | positive | MGI_C57BL6J_2444177 | Cachd1 | NCBI_Gene:320508,ENSEMBL:ENSMUSG00000028532 | MGI:2444177 | protein coding gene | cache domain containing 1 |
| 4 | gene | 101.01915 | 101.02070 | negative | MGI_C57BL6J_3642534 | Gm10577 | ENSEMBL:ENSMUSG00000073789 | MGI:3642534 | protein coding gene | predicted gene 10577 |
| 4 | gene | 101.05444 | 101.05526 | negative | MGI_C57BL6J_3642535 | Gm10576 | ENSEMBL:ENSMUSG00000102578 | MGI:3642535 | protein coding gene | predicted gene 10576 |
| 4 | gene | 101.06898 | 101.15237 | positive | MGI_C57BL6J_2443623 | Raver2 | NCBI_Gene:242570,ENSEMBL:ENSMUSG00000035275 | MGI:2443623 | protein coding gene | ribonucleoprotein, PTB-binding 2 |
| 4 | gene | 101.07403 | 101.07670 | positive | MGI_C57BL6J_5595115 | Gm35956 | NCBI_Gene:102639705 | MGI:5595115 | lncRNA gene | predicted gene, 35956 |
| 4 | gene | 101.10359 | 101.10365 | negative | MGI_C57BL6J_5451877 | Gm22100 | ENSEMBL:ENSMUSG00000088551 | MGI:5451877 | snRNA gene | predicted gene, 22100 |
| 4 | gene | 101.15197 | 101.26529 | negative | MGI_C57BL6J_96628 | Jak1 | NCBI_Gene:16451,ENSEMBL:ENSMUSG00000028530 | MGI:96628 | protein coding gene | Janus kinase 1 |
| 4 | gene | 101.18676 | 101.18687 | positive | MGI_C57BL6J_5454245 | Gm24468 | ENSEMBL:ENSMUSG00000077677 | MGI:5454245 | snRNA gene | predicted gene, 24468 |
| 4 | pseudogene | 101.21042 | 101.21059 | negative | MGI_C57BL6J_3650357 | Gm12785 | ENSEMBL:ENSMUSG00000082928 | MGI:3650357 | pseudogene | predicted gene 12785 |
| 4 | pseudogene | 101.21976 | 101.22028 | positive | MGI_C57BL6J_5011248 | Gm19063 | NCBI_Gene:100418189 | MGI:5011248 | pseudogene | predicted gene, 19063 |
| 4 | gene | 101.27114 | 101.27131 | positive | MGI_C57BL6J_5454901 | Gm25124 | ENSEMBL:ENSMUSG00000065279 | MGI:5454901 | snRNA gene | predicted gene, 25124 |
| 4 | gene | 101.28422 | 101.28520 | positive | MGI_C57BL6J_3652119 | Gm12801 | ENSEMBL:ENSMUSG00000085863 | MGI:3652119 | lncRNA gene | predicted gene 12801 |
| 4 | gene | 101.29241 | 101.32175 | negative | MGI_C57BL6J_3649207 | Gm12796 | ENSEMBL:ENSMUSG00000085721 | MGI:3649207 | lncRNA gene | predicted gene 12796 |
| 4 | pseudogene | 101.34136 | 101.34165 | positive | MGI_C57BL6J_3649401 | Gm12793 | ENSEMBL:ENSMUSG00000081621 | MGI:3649401 | pseudogene | predicted gene 12793 |
| 4 | gene | 101.34652 | 101.35674 | negative | MGI_C57BL6J_1925116 | E130102H24Rik | NCBI_Gene:77866,ENSEMBL:ENSMUSG00000086782 | MGI:1925116 | lncRNA gene | RIKEN cDNA E130102H24 gene |
| 4 | gene | 101.34695 | 101.34703 | negative | MGI_C57BL6J_2676803 | Mir101a | miRBase:MI0000148,NCBI_Gene:387143,ENSEMBL:ENSMUSG00000065451 | MGI:2676803 | miRNA gene | microRNA 101a |
| 4 | gene | 101.35378 | 101.39919 | positive | MGI_C57BL6J_1915650 | 0610043K17Rik | NCBI_Gene:68400,ENSEMBL:ENSMUSG00000087361 | MGI:1915650 | lncRNA gene | RIKEN cDNA 0610043K17 gene |
| 4 | pseudogene | 101.36218 | 101.36289 | negative | MGI_C57BL6J_3649205 | Gm12795 | NCBI_Gene:100416255,ENSEMBL:ENSMUSG00000083614 | MGI:3649205 | pseudogene | predicted gene 12795 |
| 4 | gene | 101.40308 | 101.40786 | positive | MGI_C57BL6J_3651483 | Gm12798 | ENSEMBL:ENSMUSG00000084758 | MGI:3651483 | lncRNA gene | predicted gene 12798 |
| 4 | gene | 101.41885 | 101.46777 | positive | MGI_C57BL6J_87979 | Ak4 | NCBI_Gene:11639,ENSEMBL:ENSMUSG00000028527 | MGI:87979 | protein coding gene | adenylate kinase 4 |
| 4 | gene | 101.49602 | 101.64280 | positive | MGI_C57BL6J_1919935 | Dnajc6 | NCBI_Gene:72685,ENSEMBL:ENSMUSG00000028528 | MGI:1919935 | protein coding gene | DnaJ heat shock protein family (Hsp40) member C6 |
| 4 | gene | 101.54330 | 101.55681 | negative | MGI_C57BL6J_5595171 | Gm36012 | NCBI_Gene:102639781 | MGI:5595171 | lncRNA gene | predicted gene, 36012 |
| 4 | gene | 101.55509 | 101.55537 | positive | MGI_C57BL6J_5452310 | Gm22533 | ENSEMBL:ENSMUSG00000089446 | MGI:5452310 | unclassified non-coding RNA gene | predicted gene, 22533 |
| 4 | gene | 101.64772 | 101.65936 | positive | MGI_C57BL6J_2687005 | Leprot | NCBI_Gene:230514,ENSEMBL:ENSMUSG00000035212 | MGI:2687005 | protein coding gene | leptin receptor overlapping transcript |
| 4 | gene | 101.71713 | 101.81535 | positive | MGI_C57BL6J_104993 | Lepr | NCBI_Gene:16847,ENSEMBL:ENSMUSG00000057722 | MGI:104993 | protein coding gene | leptin receptor |
| 4 | gene | 101.83497 | 101.84402 | negative | MGI_C57BL6J_3588238 | B020004J07Rik | NCBI_Gene:545662,ENSEMBL:ENSMUSG00000035201 | MGI:3588238 | protein coding gene | RIKEN cDNA B020004J07 gene |
| 4 | pseudogene | 101.87532 | 101.87611 | positive | MGI_C57BL6J_3649397 | Gm12791 | NCBI_Gene:665362,ENSEMBL:ENSMUSG00000081121 | MGI:3649397 | pseudogene | predicted gene 12791 |
| 4 | pseudogene | 101.88404 | 101.88431 | positive | MGI_C57BL6J_3649203 | Gm12797 | ENSEMBL:ENSMUSG00000081045 | MGI:3649203 | pseudogene | predicted gene 12797 |
| 4 | gene | 101.89002 | 101.89379 | negative | MGI_C57BL6J_3045359 | C130073F10Rik | NCBI_Gene:242574,ENSEMBL:ENSMUSG00000046133 | MGI:3045359 | protein coding gene | RIKEN cDNA C130073F10 gene |
| 4 | pseudogene | 101.89596 | 101.89698 | negative | MGI_C57BL6J_3649402 | Gm12792 | NCBI_Gene:384033,ENSEMBL:ENSMUSG00000084208 | MGI:3649402 | pseudogene | predicted gene 12792 |
| 4 | gene | 101.90912 | 101.91191 | positive | MGI_C57BL6J_3652120 | Gm12800 | NCBI_Gene:381535,ENSEMBL:ENSMUSG00000037028 | MGI:3652120 | protein coding gene | predicted gene 12800 |
| 4 | gene | 101.94041 | 101.94318 | positive | MGI_C57BL6J_3795847 | Gm12794 | NCBI_Gene:332923,ENSEMBL:ENSMUSG00000070890 | MGI:3795847 | protein coding gene | predicted gene 12794 |
| 4 | gene | 101.96745 | 101.98105 | negative | MGI_C57BL6J_3649398 | Gm12790 | ENSEMBL:ENSMUSG00000078626 | MGI:3649398 | protein coding gene | predicted gene 12790 |
| 4 | gene | 101.98663 | 101.99023 | positive | MGI_C57BL6J_3649399 | Gm12789 | NCBI_Gene:381536,ENSEMBL:ENSMUSG00000078625 | MGI:3649399 | protein coding gene | predicted gene 12789 |
| 4 | pseudogene | 102.04638 | 102.04979 | negative | MGI_C57BL6J_3652118 | Gm12799 | NCBI_Gene:433739,ENSEMBL:ENSMUSG00000082264 | MGI:3652118 | pseudogene | predicted gene 12799 |
| 4 | gene | 102.08753 | 102.60726 | positive | MGI_C57BL6J_99557 | Pde4b | NCBI_Gene:18578,ENSEMBL:ENSMUSG00000028525 | MGI:99557 | protein coding gene | phosphodiesterase 4B, cAMP specific |
| 4 | pseudogene | 102.32735 | 102.32802 | positive | MGI_C57BL6J_5010404 | Gm18219 | NCBI_Gene:100416738 | MGI:5010404 | pseudogene | predicted gene, 18219 |
| 4 | pseudogene | 102.33305 | 102.33372 | positive | MGI_C57BL6J_5010405 | Gm18220 | NCBI_Gene:100416739 | MGI:5010405 | pseudogene | predicted gene, 18220 |
| 4 | gene | 102.73681 | 102.73694 | positive | MGI_C57BL6J_5454646 | Gm24869 | ENSEMBL:ENSMUSG00000077260 | MGI:5454646 | snoRNA gene | predicted gene, 24869 |
| 4 | gene | 102.74130 | 102.97743 | positive | MGI_C57BL6J_1920344 | Sgip1 | NCBI_Gene:73094,ENSEMBL:ENSMUSG00000028524 | MGI:1920344 | protein coding gene | SH3-domain GRB2-like (endophilin) interacting protein 1 |
| 4 | gene | 102.96727 | 102.98976 | negative | MGI_C57BL6J_3651946 | Gm12709 | NCBI_Gene:100504717,ENSEMBL:ENSMUSG00000085181 | MGI:3651946 | lncRNA gene | predicted gene 12709 |
| 4 | gene | 102.97839 | 103.00559 | positive | MGI_C57BL6J_1914594 | Tctex1d1 | NCBI_Gene:67344,ENSEMBL:ENSMUSG00000028523 | MGI:1914594 | protein coding gene | Tctex1 domain containing 1 |
| 4 | gene | 103.00411 | 103.01197 | negative | MGI_C57BL6J_2140631 | BB031773 | NCBI_Gene:100473,ENSEMBL:ENSMUSG00000085041 | MGI:2140631 | lncRNA gene | expressed sequence BB031773 |
| 4 | gene | 103.01787 | 103.02684 | negative | MGI_C57BL6J_1346085 | Insl5 | NCBI_Gene:23919,ENSEMBL:ENSMUSG00000066090 | MGI:1346085 | protein coding gene | insulin-like 5 |
| 4 | gene | 103.03476 | 103.11455 | negative | MGI_C57BL6J_2385328 | Wdr78 | NCBI_Gene:242584,ENSEMBL:ENSMUSG00000035126 | MGI:2385328 | protein coding gene | WD repeat domain 78 |
| 4 | pseudogene | 103.10353 | 103.10374 | positive | MGI_C57BL6J_3651493 | Gm12710 | ENSEMBL:ENSMUSG00000080744 | MGI:3651493 | pseudogene | predicted gene 12710 |
| 4 | gene | 103.11144 | 103.11461 | negative | MGI_C57BL6J_2443919 | E030030F06Rik | NA | NA | protein coding gene | RIKEN cDNA E030030F06 gene |
| 4 | gene | 103.11439 | 103.16575 | positive | MGI_C57BL6J_1918398 | Mier1 | NCBI_Gene:71148,ENSEMBL:ENSMUSG00000028522 | MGI:1918398 | protein coding gene | MEIR1 treanscription regulator |
| 4 | gene | 103.17065 | 103.21537 | negative | MGI_C57BL6J_2140361 | Slc35d1 | NCBI_Gene:242585,ENSEMBL:ENSMUSG00000028521 | MGI:2140361 | protein coding gene | solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1 |
| 4 | gene | 103.21494 | 103.22312 | positive | MGI_C57BL6J_5826501 | Gm46864 | NCBI_Gene:108168969 | MGI:5826501 | lncRNA gene | predicted gene, 46864 |
| 4 | gene | 103.21770 | 103.21959 | positive | MGI_C57BL6J_5690429 | Gm44037 | ENSEMBL:ENSMUSG00000107726 | MGI:5690429 | lncRNA gene | predicted gene, 44037 |
| 4 | gene | 103.23045 | 103.29095 | negative | MGI_C57BL6J_1918191 | 4921539E11Rik | NCBI_Gene:70941,ENSEMBL:ENSMUSG00000028520 | MGI:1918191 | protein coding gene | RIKEN cDNA 4921539E11 gene |
| 4 | gene | 103.29096 | 103.31183 | positive | MGI_C57BL6J_1918900 | 4930456L15Rik | NCBI_Gene:71650,ENSEMBL:ENSMUSG00000078618 | MGI:1918900 | lncRNA gene | RIKEN cDNA 4930456L15 gene |
| 4 | gene | 103.30284 | 103.31355 | negative | MGI_C57BL6J_5595682 | Gm36523 | NCBI_Gene:102640470 | MGI:5595682 | lncRNA gene | predicted gene, 36523 |
| 4 | gene | 103.31321 | 103.37187 | positive | MGI_C57BL6J_1914263 | Oma1 | NCBI_Gene:67013,ENSEMBL:ENSMUSG00000035069 | MGI:1914263 | protein coding gene | OMA1 zinc metallopeptidase |
| 4 | gene | 103.38130 | 103.38246 | positive | MGI_C57BL6J_1918233 | 4931409D07Rik | NA | NA | unclassified gene | RIKEN cDNA 4931409D07 gene |
| 4 | gene | 103.38250 | 103.49219 | positive | MGI_C57BL6J_3651516 | Gm12718 | NCBI_Gene:622167,ENSEMBL:ENSMUSG00000086136 | MGI:3651516 | lncRNA gene | predicted gene 12718 |
| 4 | gene | 103.47186 | 103.47196 | positive | MGI_C57BL6J_5452841 | Gm23064 | ENSEMBL:ENSMUSG00000094477 | MGI:5452841 | miRNA gene | predicted gene, 23064 |
| 4 | gene | 103.53884 | 103.55068 | negative | MGI_C57BL6J_3651748 | Gm12720 | ENSEMBL:ENSMUSG00000087482 | MGI:3651748 | lncRNA gene | predicted gene 12720 |
| 4 | pseudogene | 103.54318 | 103.54476 | negative | MGI_C57BL6J_3650738 | Gm12716 | NCBI_Gene:665451,ENSEMBL:ENSMUSG00000081584 | MGI:3650738 | pseudogene | predicted gene 12716 |
| 4 | pseudogene | 103.56333 | 103.56523 | positive | MGI_C57BL6J_3650759 | Gm12715 | NCBI_Gene:100039940,ENSEMBL:ENSMUSG00000059195 | MGI:3650759 | pseudogene | predicted gene 12715 |
| 4 | gene | 103.56420 | 103.56429 | positive | MGI_C57BL6J_5452578 | Gm22801 | ENSEMBL:ENSMUSG00000093365 | MGI:5452578 | miRNA gene | predicted gene, 22801 |
| 4 | gene | 103.58627 | 103.58640 | negative | MGI_C57BL6J_5455654 | Gm25877 | ENSEMBL:ENSMUSG00000077514 | MGI:5455654 | snoRNA gene | predicted gene, 25877 |
| 4 | gene | 103.61042 | 103.61915 | negative | MGI_C57BL6J_5595763 | Gm36604 | NCBI_Gene:102640570 | MGI:5595763 | lncRNA gene | predicted gene, 36604 |
| 4 | gene | 103.61936 | 104.74484 | positive | MGI_C57BL6J_108554 | Dab1 | NCBI_Gene:13131,ENSEMBL:ENSMUSG00000028519 | MGI:108554 | protein coding gene | disabled 1 |
| 4 | gene | 103.71166 | 103.71290 | positive | MGI_C57BL6J_1920104 | 2900034C19Rik | NA | NA | unclassified gene | RIKEN cDNA 2900034C19 gene |
| 4 | pseudogene | 103.93930 | 103.93954 | negative | MGI_C57BL6J_3651747 | Gm12719 | ENSEMBL:ENSMUSG00000080913 | MGI:3651747 | pseudogene | predicted gene 12719 |
| 4 | pseudogene | 103.94096 | 103.94212 | positive | MGI_C57BL6J_3650774 | Gm12717 | NCBI_Gene:622315,ENSEMBL:ENSMUSG00000081696 | MGI:3650774 | pseudogene | predicted gene 12717 |
| 4 | gene | 103.99097 | 104.02872 | negative | MGI_C57BL6J_5595820 | Gm36661 | NCBI_Gene:102640646 | MGI:5595820 | lncRNA gene | predicted gene, 36661 |
| 4 | gene | 104.32825 | 104.33056 | positive | MGI_C57BL6J_3642623 | Gm10304 | NA | NA | unclassified gene | predicted gene 10304 |
| 4 | gene | 104.52840 | 104.53225 | positive | MGI_C57BL6J_3641719 | AY512949 | NA | NA | unclassified gene | cDNA sequence AY512949 |
| 4 | gene | 104.76632 | 104.80455 | positive | MGI_C57BL6J_88236 | C8b | NCBI_Gene:110382,ENSEMBL:ENSMUSG00000029656 | MGI:88236 | protein coding gene | complement component 8, beta polypeptide |
| 4 | gene | 104.81568 | 104.87649 | negative | MGI_C57BL6J_2668347 | C8a | NCBI_Gene:230558,ENSEMBL:ENSMUSG00000035031 | MGI:2668347 | protein coding gene | complement component 8, alpha polypeptide |
| 4 | gene | 104.83383 | 105.01686 | positive | MGI_C57BL6J_2685466 | Fyb2 | NCBI_Gene:242594,ENSEMBL:ENSMUSG00000078612 | MGI:2685466 | protein coding gene | FYN binding protein 2 |
| 4 | gene | 104.85733 | 104.85914 | positive | MGI_C57BL6J_4937296 | Gm17662 | ENSEMBL:ENSMUSG00000095386 | MGI:4937296 | protein coding gene | predicted gene, 17662 |
| 4 | gene | 104.88721 | 104.89008 | negative | MGI_C57BL6J_5623101 | Gm40216 | NCBI_Gene:105244639 | MGI:5623101 | lncRNA gene | predicted gene, 40216 |
| 4 | pseudogene | 104.96944 | 104.97092 | negative | MGI_C57BL6J_3650292 | Gm12721 | NCBI_Gene:108168943,ENSEMBL:ENSMUSG00000083791 | MGI:3650292 | pseudogene | predicted gene 12721 |
| 4 | gene | 104.97120 | 104.97264 | negative | MGI_C57BL6J_5595994 | Gm36835 | NCBI_Gene:102640873 | MGI:5595994 | lncRNA gene | predicted gene, 36835 |
| 4 | gene | 105.02916 | 105.10990 | negative | MGI_C57BL6J_1336173 | Prkaa2 | NCBI_Gene:108079,ENSEMBL:ENSMUSG00000028518 | MGI:1336173 | protein coding gene | protein kinase, AMP-activated, alpha 2 catalytic subunit |
| 4 | gene | 105.10998 | 105.11232 | positive | MGI_C57BL6J_5596052 | Gm36893 | NCBI_Gene:102640948 | MGI:5596052 | lncRNA gene | predicted gene, 36893 |
| 4 | gene | 105.15312 | 105.15603 | negative | MGI_C57BL6J_5588997 | Gm29838 | NCBI_Gene:102631520 | MGI:5588997 | lncRNA gene | predicted gene, 29838 |
| 4 | gene | 105.15735 | 105.23277 | positive | MGI_C57BL6J_1915166 | Plpp3 | NCBI_Gene:67916,ENSEMBL:ENSMUSG00000028517 | MGI:1915166 | protein coding gene | phospholipid phosphatase 3 |
| 4 | gene | 105.20074 | 105.20191 | positive | MGI_C57BL6J_1924365 | 6030451C04Rik | NA | NA | unclassified gene | RIKEN cDNA 6030451C04 gene |
| 4 | gene | 105.31038 | 105.31915 | negative | MGI_C57BL6J_5623102 | Gm40217 | NCBI_Gene:105244640 | MGI:5623102 | lncRNA gene | predicted gene, 40217 |
| 4 | pseudogene | 105.37439 | 105.37495 | negative | MGI_C57BL6J_3652209 | Gm12722 | NCBI_Gene:433742,ENSEMBL:ENSMUSG00000081819 | MGI:3652209 | pseudogene | predicted gene 12722 |
| 4 | pseudogene | 105.51123 | 105.51168 | positive | MGI_C57BL6J_3652213 | Gm12723 | ENSEMBL:ENSMUSG00000083128 | MGI:3652213 | pseudogene | predicted gene 12723 |
| 4 | pseudogene | 105.57831 | 105.57899 | positive | MGI_C57BL6J_3649291 | Gm12726 | NCBI_Gene:108169004,ENSEMBL:ENSMUSG00000082061 | MGI:3649291 | pseudogene | predicted gene 12726 |
| 4 | gene | 105.78987 | 105.79491 | positive | MGI_C57BL6J_3649486 | Gm12728 | NCBI_Gene:269569,ENSEMBL:ENSMUSG00000083780 | MGI:3649486 | protein coding gene | predicted gene 12728 |
| 4 | gene | 105.84122 | 105.84571 | negative | MGI_C57BL6J_5589081 | Gm29922 | NCBI_Gene:102631627 | MGI:5589081 | lncRNA gene | predicted gene, 29922 |
| 4 | gene | 105.97558 | 105.97589 | negative | MGI_C57BL6J_5454140 | Gm24363 | ENSEMBL:ENSMUSG00000089065 | MGI:5454140 | unclassified non-coding RNA gene | predicted gene, 24363 |
| 4 | gene | 106.03732 | 106.04880 | negative | MGI_C57BL6J_5589425 | Gm30266 | NCBI_Gene:102632106 | MGI:5589425 | lncRNA gene | predicted gene, 30266 |
| 4 | gene | 106.05400 | 106.05780 | negative | MGI_C57BL6J_5589358 | Gm30199 | NCBI_Gene:102632021 | MGI:5589358 | lncRNA gene | predicted gene, 30199 |
| 4 | gene | 106.08871 | 106.10507 | negative | MGI_C57BL6J_3649488 | Gm12729 | ENSEMBL:ENSMUSG00000085790 | MGI:3649488 | lncRNA gene | predicted gene 12729 |
| 4 | pseudogene | 106.09883 | 106.09953 | positive | MGI_C57BL6J_3649747 | Gm12727 | NCBI_Gene:384038,ENSEMBL:ENSMUSG00000082768 | MGI:3649747 | pseudogene | predicted gene 12727 |
| 4 | gene | 106.11897 | 106.11908 | negative | MGI_C57BL6J_5455824 | Gm26047 | ENSEMBL:ENSMUSG00000106163,ENSEMBL:ENSMUSG00000087933 | MGI:5455824 | snoRNA gene | predicted gene, 26047 |
| 4 | gene | 106.12425 | 106.12718 | positive | MGI_C57BL6J_5589285 | Gm30126 | NCBI_Gene:102631914 | MGI:5589285 | lncRNA gene | predicted gene, 30126 |
| 4 | gene | 106.12625 | 106.14584 | negative | MGI_C57BL6J_5589206 | Gm30047 | NCBI_Gene:102631801 | MGI:5589206 | lncRNA gene | predicted gene, 30047 |
| 4 | pseudogene | 106.13571 | 106.13783 | negative | MGI_C57BL6J_3649750 | Hspd1-ps4 | NCBI_Gene:384039,ENSEMBL:ENSMUSG00000081189 | MGI:3649750 | pseudogene | heat shock protein 1 (chaperonin), pseudogene 4 |
| 4 | gene | 106.23712 | 106.29100 | positive | MGI_C57BL6J_3649755 | Gm12724 | NCBI_Gene:105244641,ENSEMBL:ENSMUSG00000086249 | MGI:3649755 | lncRNA gene | predicted gene 12724 |
| 4 | pseudogene | 106.25703 | 106.25730 | positive | MGI_C57BL6J_3649483 | Gm12731 | ENSEMBL:ENSMUSG00000082958 | MGI:3649483 | pseudogene | predicted gene 12731 |
| 4 | gene | 106.29127 | 106.29225 | positive | MGI_C57BL6J_3651116 | Gm12730 | ENSEMBL:ENSMUSG00000087051 | MGI:3651116 | lncRNA gene | predicted gene 12730 |
| 4 | gene | 106.29335 | 106.29439 | negative | MGI_C57BL6J_5610481 | Gm37253 | ENSEMBL:ENSMUSG00000104192 | MGI:5610481 | unclassified gene | predicted gene, 37253 |
| 4 | gene | 106.31464 | 106.31615 | negative | MGI_C57BL6J_5589588 | Gm30429 | NCBI_Gene:102632328 | MGI:5589588 | lncRNA gene | predicted gene, 30429 |
| 4 | gene | 106.31621 | 106.44657 | positive | MGI_C57BL6J_1919936 | Usp24 | NCBI_Gene:329908,ENSEMBL:ENSMUSG00000028514 | MGI:1919936 | protein coding gene | ubiquitin specific peptidase 24 |
| 4 | gene | 106.32443 | 106.32636 | positive | MGI_C57BL6J_5623103 | Gm40218 | NCBI_Gene:105244642 | MGI:5623103 | lncRNA gene | predicted gene, 40218 |
| 4 | gene | 106.44233 | 106.46433 | negative | MGI_C57BL6J_2140260 | Pcsk9 | NCBI_Gene:100102,ENSEMBL:ENSMUSG00000044254 | MGI:2140260 | protein coding gene | proprotein convertase subtilisin/kexin type 9 |
| 4 | gene | 106.46463 | 106.47739 | positive | MGI_C57BL6J_5589646 | Gm30487 | NCBI_Gene:102632408 | MGI:5589646 | lncRNA gene | predicted gene, 30487 |
| 4 | gene | 106.48346 | 106.49228 | negative | MGI_C57BL6J_2153465 | Bsnd | NCBI_Gene:140475,ENSEMBL:ENSMUSG00000025418 | MGI:2153465 | protein coding gene | barttin CLCNK type accessory beta subunit |
| 4 | gene | 106.49881 | 106.50634 | negative | MGI_C57BL6J_3041156 | Tmem61 | NCBI_Gene:329909,ENSEMBL:ENSMUSG00000085933 | MGI:3041156 | lincRNA gene | transmembrane protein 61 |
| 4 | gene | 106.55467 | 106.55790 | negative | MGI_C57BL6J_1919813 | 2700068H02Rik | ENSEMBL:ENSMUSG00000086172 | MGI:1919813 | lncRNA gene | RIKEN cDNA 2700068H02 gene |
| 4 | gene | 106.56104 | 106.58911 | positive | MGI_C57BL6J_1922004 | Dhcr24 | NCBI_Gene:74754,ENSEMBL:ENSMUSG00000034926 | MGI:1922004 | protein coding gene | 24-dehydrocholesterol reductase |
| 4 | gene | 106.58710 | 106.58911 | negative | MGI_C57BL6J_3702539 | Gm12744 | ENSEMBL:ENSMUSG00000085023 | MGI:3702539 | lncRNA gene | predicted gene 12744 |
| 4 | gene | 106.59091 | 106.61750 | negative | MGI_C57BL6J_2681853 | Lexm | NCBI_Gene:242602,ENSEMBL:ENSMUSG00000054362 | MGI:2681853 | protein coding gene | lymphocyte expansion molecule |
| 4 | gene | 106.62243 | 106.64020 | positive | MGI_C57BL6J_3045307 | Ttc22 | NCBI_Gene:230576,ENSEMBL:ENSMUSG00000034919 | MGI:3045307 | protein coding gene | tetratricopeptide repeat domain 22 |
| 4 | gene | 106.65102 | 106.65528 | positive | MGI_C57BL6J_2386296 | Pars2 | NCBI_Gene:230577,ENSEMBL:ENSMUSG00000043572 | MGI:2386296 | protein coding gene | prolyl-tRNA synthetase (mitochondrial)(putative) |
| 4 | gene | 106.65820 | 106.67897 | negative | MGI_C57BL6J_1919604 | Ttc4 | NCBI_Gene:72354,ENSEMBL:ENSMUSG00000025413 | MGI:1919604 | protein coding gene | tetratricopeptide repeat domain 4 |
| 4 | gene | 106.68042 | 106.73662 | negative | MGI_C57BL6J_2685873 | Mroh7 | NCBI_Gene:381538,ENSEMBL:ENSMUSG00000047502 | MGI:2685873 | protein coding gene | maestro heat-like repeat family member 7 |
| 4 | gene | 106.73389 | 106.74829 | positive | MGI_C57BL6J_2657115 | Fam151a | NCBI_Gene:230579,ENSEMBL:ENSMUSG00000034871 | MGI:2657115 | protein coding gene | family with sequence simliarity 151, member A |
| 4 | gene | 106.74456 | 106.80609 | negative | MGI_C57BL6J_1913736 | Acot11 | NCBI_Gene:329910,ENSEMBL:ENSMUSG00000034853 | MGI:1913736 | protein coding gene | acyl-CoA thioesterase 11 |
| 4 | pseudogene | 106.77482 | 106.77503 | positive | MGI_C57BL6J_3651452 | Gm12745 | ENSEMBL:ENSMUSG00000081110 | MGI:3651452 | pseudogene | predicted gene 12745 |
| 4 | gene | 106.81491 | 106.82834 | negative | MGI_C57BL6J_5590224 | Gm31065 | NCBI_Gene:102633170 | MGI:5590224 | lncRNA gene | predicted gene, 31065 |
| 4 | gene | 106.83928 | 106.84016 | negative | MGI_C57BL6J_5590284 | Gm31125 | NCBI_Gene:102633257 | MGI:5590284 | lncRNA gene | predicted gene, 31125 |
| 4 | gene | 106.84176 | 106.84792 | negative | MGI_C57BL6J_3652135 | Gm12746 | ENSEMBL:ENSMUSG00000086940 | MGI:3652135 | lncRNA gene | predicted gene 12746 |
| 4 | gene | 106.90021 | 106.91290 | negative | MGI_C57BL6J_5826502 | Gm46865 | NCBI_Gene:108168970 | MGI:5826502 | lncRNA gene | predicted gene, 46865 |
| 4 | gene | 106.91070 | 107.04969 | positive | MGI_C57BL6J_1919725 | Ssbp3 | NCBI_Gene:72475,ENSEMBL:ENSMUSG00000061887 | MGI:1919725 | protein coding gene | single-stranded DNA binding protein 3 |
| 4 | gene | 107.01793 | 107.02250 | negative | MGI_C57BL6J_3650114 | Gm12786 | ENSEMBL:ENSMUSG00000085581 | MGI:3650114 | lncRNA gene | predicted gene 12786 |
| 4 | gene | 107.05587 | 107.06687 | negative | MGI_C57BL6J_1926268 | Mrpl37 | NCBI_Gene:56280,ENSEMBL:ENSMUSG00000028622 | MGI:1926268 | protein coding gene | mitochondrial ribosomal protein L37 |
| 4 | gene | 107.06691 | 107.08827 | positive | MGI_C57BL6J_1919657 | Cyb5rl | NCBI_Gene:230582,ENSEMBL:ENSMUSG00000028621 | MGI:1919657 | protein coding gene | cytochrome b5 reductase-like |
| 4 | gene | 107.09689 | 107.11312 | positive | MGI_C57BL6J_3045328 | Cdcp2 | NCBI_Gene:242603,ENSEMBL:ENSMUSG00000047636 | MGI:3045328 | protein coding gene | CUB domain containing protein 2 |
| 4 | gene | 107.12661 | 107.13625 | positive | MGI_C57BL6J_3649683 | Gm12802 | ENSEMBL:ENSMUSG00000085304 | MGI:3649683 | lncRNA gene | predicted gene 12802 |
| 4 | gene | 107.13416 | 107.17912 | negative | MGI_C57BL6J_1913776 | Tceanc2 | NCBI_Gene:66526,ENSEMBL:ENSMUSG00000028619 | MGI:1913776 | protein coding gene | transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| 4 | gene | 107.17840 | 107.20100 | positive | MGI_C57BL6J_1929278 | Tmem59 | NCBI_Gene:56374,ENSEMBL:ENSMUSG00000028618 | MGI:1929278 | protein coding gene | transmembrane protein 59 |
| 4 | gene | 107.20908 | 107.22110 | positive | MGI_C57BL6J_3652166 | Ldlrad1 | NCBI_Gene:546840,ENSEMBL:ENSMUSG00000070877 | MGI:3652166 | protein coding gene | low density lipoprotein receptor class A domain containing 1 |
| 4 | gene | 107.23351 | 107.25353 | negative | MGI_C57BL6J_1925059 | Lrrc42 | NCBI_Gene:77809,ENSEMBL:ENSMUSG00000028617 | MGI:1925059 | protein coding gene | leucine rich repeat containing 42 |
| 4 | gene | 107.25359 | 107.28450 | positive | MGI_C57BL6J_1920188 | Hspb11 | NCBI_Gene:72938,ENSEMBL:ENSMUSG00000063172 | MGI:1920188 | protein coding gene | heat shock protein family B (small), member 11 |
| 4 | pseudogene | 107.27043 | 107.27087 | positive | MGI_C57BL6J_3651240 | Gm12850 | ENSEMBL:ENSMUSG00000082941 | MGI:3651240 | pseudogene | predicted gene 12850 |
| 4 | gene | 107.29147 | 107.30717 | negative | MGI_C57BL6J_94896 | Dio1 | NCBI_Gene:13370,ENSEMBL:ENSMUSG00000034785 | MGI:94896 | protein coding gene | deiodinase, iodothyronine, type I |
| 4 | gene | 107.31422 | 107.35982 | positive | MGI_C57BL6J_1915532 | Yipf1 | NCBI_Gene:230584,ENSEMBL:ENSMUSG00000057375 | MGI:1915532 | protein coding gene | Yip1 domain family, member 1 |
| 4 | pseudogene | 107.32682 | 107.32853 | negative | MGI_C57BL6J_3649408 | Gm12870 | NCBI_Gene:652920,ENSEMBL:ENSMUSG00000082279 | MGI:3649408 | pseudogene | predicted gene 12870 |
| 4 | gene | 107.34904 | 107.35135 | negative | MGI_C57BL6J_3649409 | Gm12869 | ENSEMBL:ENSMUSG00000086041 | MGI:3649409 | lncRNA gene | predicted gene 12869 |
| 4 | gene | 107.36778 | 107.41635 | positive | MGI_C57BL6J_1920037 | Ndc1 | NCBI_Gene:72787,ENSEMBL:ENSMUSG00000028614 | MGI:1920037 | protein coding gene | NDC1 transmembrane nucleoporin |
| 4 | gene | 107.42664 | 107.63506 | positive | MGI_C57BL6J_2386723 | Glis1 | NCBI_Gene:230587,ENSEMBL:ENSMUSG00000034762 | MGI:2386723 | protein coding gene | GLIS family zinc finger 1 |
| 4 | gene | 107.43378 | 107.43449 | negative | MGI_C57BL6J_5826503 | Gm46866 | NCBI_Gene:108168971 | MGI:5826503 | lncRNA gene | predicted gene, 46866 |
| 4 | gene | 107.43457 | 107.43466 | positive | MGI_C57BL6J_5454581 | Gm24804 | ENSEMBL:ENSMUSG00000092723 | MGI:5454581 | miRNA gene | predicted gene, 24804 |
| 4 | gene | 107.59185 | 107.60126 | negative | MGI_C57BL6J_1925456 | 4930552P06Rik | NCBI_Gene:102633643 | MGI:1925456 | lncRNA gene | RIKEN cDNA 4930552P06 gene |
| 4 | gene | 107.67629 | 107.68425 | negative | MGI_C57BL6J_1927125 | Dmrtb1 | NCBI_Gene:56296,ENSEMBL:ENSMUSG00000028610 | MGI:1927125 | protein coding gene | DMRT-like family B with proline-rich C-terminal, 1 |
| 4 | pseudogene | 107.68813 | 107.68871 | positive | MGI_C57BL6J_3650162 | Gm12899 | NCBI_Gene:100417388,ENSEMBL:ENSMUSG00000081916 | MGI:3650162 | pseudogene | predicted gene 12899 |
| 4 | gene | 107.68960 | 107.75898 | positive | MGI_C57BL6J_1920570 | 1700047F07Rik | NCBI_Gene:102633724,ENSEMBL:ENSMUSG00000085549 | MGI:1920570 | lncRNA gene | RIKEN cDNA 1700047F07 gene |
| 4 | gene | 107.69783 | 107.69795 | negative | MGI_C57BL6J_5454900 | Gm25123 | ENSEMBL:ENSMUSG00000094267 | MGI:5454900 | miRNA gene | predicted gene, 25123 |
| 4 | gene | 107.69816 | 107.73654 | negative | MGI_C57BL6J_5590841 | Gm31682 | NCBI_Gene:102633989 | MGI:5590841 | lncRNA gene | predicted gene, 31682 |
| 4 | pseudogene | 107.75944 | 107.76047 | negative | MGI_C57BL6J_3650884 | Gm12906 | NCBI_Gene:433743,ENSEMBL:ENSMUSG00000084874 | MGI:3650884 | pseudogene | predicted gene 12906 |
| 4 | gene | 107.75951 | 107.77099 | positive | MGI_C57BL6J_3650882 | Gm12907 | NCBI_Gene:102634068,ENSEMBL:ENSMUSG00000087195 | MGI:3650882 | lncRNA gene | predicted gene 12907 |
| 4 | gene | 107.78371 | 107.78992 | positive | MGI_C57BL6J_1914798 | 4933424M12Rik | NCBI_Gene:67548,ENSEMBL:ENSMUSG00000087289 | MGI:1914798 | lncRNA gene | RIKEN cDNA 4933424M12 gene |
| 4 | gene | 107.79315 | 107.80217 | negative | MGI_C57BL6J_3651133 | Lrp8os3 | NCBI_Gene:105244644,ENSEMBL:ENSMUSG00000087200 | MGI:3651133 | bidirectional promoter lncRNA gene | low density lipoprotein receptor-related protein 8, apolipoprotein e receptor, opposite strand 3 |
| 4 | gene | 107.80187 | 107.87684 | positive | MGI_C57BL6J_1340044 | Lrp8 | NCBI_Gene:16975,ENSEMBL:ENSMUSG00000028613 | MGI:1340044 | protein coding gene | low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| 4 | gene | 107.80437 | 107.82319 | negative | MGI_C57BL6J_3588233 | Lrp8os2 | NCBI_Gene:619295,ENSEMBL:ENSMUSG00000073779 | MGI:3588233 | antisense lncRNA gene | low density lipoprotein receptor-related protein 8, apolipoprotein e receptor, opposite strand 2 |
| 4 | gene | 107.82790 | 107.83084 | negative | MGI_C57BL6J_1925359 | Lrp8os1 | ENSEMBL:ENSMUSG00000028612 | MGI:1925359 | antisense lncRNA gene | low density lipoprotein receptor-related protein 8, apolipoprotein e receptor, opposite strand 1 |
| 4 | gene | 107.87976 | 107.88742 | positive | MGI_C57BL6J_1330312 | Magoh | NCBI_Gene:17149,ENSEMBL:ENSMUSG00000028609 | MGI:1330312 | protein coding gene | mago homolog, exon junction complex core component |
| 4 | gene | 107.88979 | 107.89938 | positive | MGI_C57BL6J_1921348 | Czib | NCBI_Gene:74098,ENSEMBL:ENSMUSG00000028608 | MGI:1921348 | protein coding gene | CXXC motif containing zinc binding protein |
| 4 | gene | 107.90398 | 107.92361 | negative | MGI_C57BL6J_109176 | Cpt2 | NCBI_Gene:12896,ENSEMBL:ENSMUSG00000028607 | MGI:109176 | protein coding gene | carnitine palmitoyltransferase 2 |
| 4 | gene | 107.93747 | 107.94737 | positive | MGI_C57BL6J_5623106 | Gm40221 | NCBI_Gene:105244646 | MGI:5623106 | lncRNA gene | predicted gene, 40221 |
| 4 | gene | 107.95393 | 107.96072 | positive | MGI_C57BL6J_1924473 | 4930407G08Rik | NCBI_Gene:77223,ENSEMBL:ENSMUSG00000086713 | MGI:1924473 | lncRNA gene | RIKEN cDNA 4930407G08 gene |
| 4 | gene | 107.96833 | 108.01353 | positive | MGI_C57BL6J_2444087 | Slc1a7 | NCBI_Gene:242607,ENSEMBL:ENSMUSG00000008932 | MGI:2444087 | protein coding gene | solute carrier family 1 (glutamate transporter), member 7 |
| 4 | gene | 108.01479 | 108.09645 | negative | MGI_C57BL6J_2674939 | Podn | NCBI_Gene:242608,ENSEMBL:ENSMUSG00000028600 | MGI:2674939 | protein coding gene | podocan |
| 4 | gene | 108.04170 | 108.04180 | negative | MGI_C57BL6J_5453131 | Gm23354 | ENSEMBL:ENSMUSG00000089051 | MGI:5453131 | snRNA gene | predicted gene, 23354 |
| 4 | gene | 108.04383 | 108.14500 | negative | MGI_C57BL6J_98254 | Scp2 | NCBI_Gene:20280,ENSEMBL:ENSMUSG00000028603 | MGI:98254 | protein coding gene | sterol carrier protein 2, liver |
| 4 | gene | 108.08736 | 108.08745 | positive | MGI_C57BL6J_5531386 | Mir6397 | miRBase:MI0021933,NCBI_Gene:102465213,ENSEMBL:ENSMUSG00000098980 | MGI:5531386 | miRNA gene | microRNA 6397 |
| 4 | gene | 108.16544 | 108.17931 | positive | MGI_C57BL6J_1289238 | Echdc2 | NCBI_Gene:52430,ENSEMBL:ENSMUSG00000028601 | MGI:1289238 | protein coding gene | enoyl Coenzyme A hydratase domain containing 2 |
| 4 | gene | 108.18024 | 108.21815 | negative | MGI_C57BL6J_2446208 | Zyg11a | NCBI_Gene:230590,ENSEMBL:ENSMUSG00000034645 | MGI:2446208 | protein coding gene | zyg-11 family member A, cell cycle regulator |
| 4 | gene | 108.21798 | 108.22046 | positive | MGI_C57BL6J_5623107 | Gm40222 | NCBI_Gene:105244647 | MGI:5623107 | lncRNA gene | predicted gene, 40222 |
| 4 | gene | 108.22776 | 108.30114 | negative | MGI_C57BL6J_2685277 | Zyg11b | NCBI_Gene:414872,ENSEMBL:ENSMUSG00000034636 | MGI:2685277 | protein coding gene | zyg-ll family member B, cell cycle regulator |
| 4 | pseudogene | 108.31036 | 108.31115 | negative | MGI_C57BL6J_3651646 | Gm12742 | ENSEMBL:ENSMUSG00000083957 | MGI:3651646 | pseudogene | predicted gene 12742 |
| 4 | pseudogene | 108.32234 | 108.32264 | positive | MGI_C57BL6J_3651641 | Gm12740 | ENSEMBL:ENSMUSG00000081734 | MGI:3651641 | pseudogene | predicted gene 12740 |
| 4 | gene | 108.32814 | 108.34154 | positive | MGI_C57BL6J_1917143 | Coa7 | NCBI_Gene:69893,ENSEMBL:ENSMUSG00000048351 | MGI:1917143 | protein coding gene | cytochrome c oxidase assembly factor 7 |
| 4 | gene | 108.34549 | 108.35120 | negative | MGI_C57BL6J_5826526 | Gm46889 | NCBI_Gene:108169005 | MGI:5826526 | lncRNA gene | predicted gene, 46889 |
| 4 | gene | 108.36778 | 108.38578 | negative | MGI_C57BL6J_3651644 | Shisal2a | NCBI_Gene:545667,ENSEMBL:ENSMUSG00000059816 | MGI:3651644 | protein coding gene | shisa like 2A |
| 4 | gene | 108.38330 | 108.40661 | positive | MGI_C57BL6J_5591456 | Gm32297 | NCBI_Gene:105244648 | MGI:5591456 | lncRNA gene | predicted gene, 32297 |
| 4 | gene | 108.40022 | 108.40696 | negative | MGI_C57BL6J_1914555 | Gpx7 | NCBI_Gene:67305,ENSEMBL:ENSMUSG00000028597 | MGI:1914555 | protein coding gene | glutathione peroxidase 7 |
| 4 | gene | 108.43064 | 108.45417 | positive | MGI_C57BL6J_5623108 | Gm40223 | NCBI_Gene:105244650 | MGI:5623108 | lncRNA gene | predicted gene, 40223 |
| 4 | pseudogene | 108.45071 | 108.45137 | negative | MGI_C57BL6J_3651217 | Gm12738 | NCBI_Gene:665728,ENSEMBL:ENSMUSG00000083255 | MGI:3651217 | pseudogene | predicted gene 12738 |
| 4 | gene | 108.45943 | 108.56073 | positive | MGI_C57BL6J_2445126 | Tut4 | NCBI_Gene:230594,ENSEMBL:ENSMUSG00000034610 | MGI:2445126 | protein coding gene | terminal uridylyl transferase 4 |
| 4 | gene | 108.46694 | 108.46705 | negative | MGI_C57BL6J_5454744 | Gm24967 | ENSEMBL:ENSMUSG00000095361 | MGI:5454744 | snRNA gene | predicted gene, 24967 |
| 4 | pseudogene | 108.48406 | 108.48487 | negative | MGI_C57BL6J_3651018 | Gm12739 | NCBI_Gene:105244651,ENSEMBL:ENSMUSG00000083783 | MGI:3651018 | pseudogene | predicted gene 12739 |
| 4 | gene | 108.49804 | 108.49815 | positive | MGI_C57BL6J_5452915 | Gm23138 | ENSEMBL:ENSMUSG00000065299 | MGI:5452915 | snRNA gene | predicted gene, 23138 |
| 4 | gene | 108.51378 | 108.51682 | positive | MGI_C57BL6J_3651021 | Gm12737 | ENSEMBL:ENSMUSG00000085093 | MGI:3651021 | lncRNA gene | predicted gene 12737 |
| 4 | gene | 108.52192 | 108.52203 | negative | MGI_C57BL6J_5455604 | Gm25827 | ENSEMBL:ENSMUSG00000087714 | MGI:5455604 | snRNA gene | predicted gene, 25827 |
| 4 | gene | 108.53749 | 108.53760 | negative | MGI_C57BL6J_5452122 | Gm22345 | ENSEMBL:ENSMUSG00000084593 | MGI:5452122 | snRNA gene | predicted gene, 22345 |
| 4 | gene | 108.56317 | 108.57934 | negative | MGI_C57BL6J_1916962 | Prpf38a | NCBI_Gene:230596,ENSEMBL:ENSMUSG00000063800 | MGI:1916962 | protein coding gene | PRP38 pre-mRNA processing factor 38 (yeast) domain containing A |
| 4 | gene | 108.57562 | 108.61603 | positive | MGI_C57BL6J_1328337 | Orc1 | NCBI_Gene:18392,ENSEMBL:ENSMUSG00000028587 | MGI:1328337 | protein coding gene | origin recognition complex, subunit 1 |
| 4 | gene | 108.59243 | 108.61985 | negative | MGI_C57BL6J_3651449 | Gm12743 | ENSEMBL:ENSMUSG00000084968 | MGI:3651449 | lncRNA gene | predicted gene 12743 |
| 4 | gene | 108.61977 | 108.63549 | positive | MGI_C57BL6J_2443076 | Cc2d1b | NCBI_Gene:319965,ENSEMBL:ENSMUSG00000028582 | MGI:2443076 | protein coding gene | coiled-coil and C2 domain containing 1B |
| 4 | gene | 108.62293 | 108.62522 | negative | MGI_C57BL6J_5313040 | Gm20731 | ENSEMBL:ENSMUSG00000102912 | MGI:5313040 | protein coding gene | predicted gene, 20731 |
| 4 | gene | 108.63384 | 108.63744 | negative | MGI_C57BL6J_4936988 | Gm17354 | ENSEMBL:ENSMUSG00000091985 | MGI:4936988 | lncRNA gene | predicted gene, 17354 |
| 4 | gene | 108.63747 | 108.78225 | negative | MGI_C57BL6J_2652838 | Zfyve9 | NCBI_Gene:230597,ENSEMBL:ENSMUSG00000034557 | MGI:2652838 | protein coding gene | zinc finger, FYVE domain containing 9 |
| 4 | gene | 108.71965 | 108.78190 | positive | MGI_C57BL6J_1920383 | 3110021N24Rik | ENSEMBL:ENSMUSG00000094958 | MGI:1920383 | protein coding gene | RIKEN cDNA 3110021N24 gene |
| 4 | pseudogene | 108.73171 | 108.73240 | positive | MGI_C57BL6J_3651642 | Gm12741 | NCBI_Gene:100040326,ENSEMBL:ENSMUSG00000082374 | MGI:3651642 | pseudogene | predicted gene 12741 |
| 4 | gene | 108.81430 | 108.83430 | negative | MGI_C57BL6J_1915312 | Btf3l4 | NCBI_Gene:70533,ENSEMBL:ENSMUSG00000028568 | MGI:1915312 | protein coding gene | basic transcription factor 3-like 4 |
| 4 | gene | 108.83460 | 108.86213 | positive | MGI_C57BL6J_1913323 | Txndc12 | NCBI_Gene:66073,ENSEMBL:ENSMUSG00000028567 | MGI:1913323 | protein coding gene | thioredoxin domain containing 12 (endoplasmic reticulum) |
| 4 | gene | 108.84740 | 108.84862 | negative | MGI_C57BL6J_3704235 | A730015C16Rik | NA | NA | lncRNA gene | RIKEN cDNA A730015C16 gene |
| 4 | gene | 108.84779 | 108.84941 | positive | MGI_C57BL6J_1923547 | Kti12 | NCBI_Gene:100087,ENSEMBL:ENSMUSG00000073775 | MGI:1923547 | protein coding gene | KTI12 homolog, chromatin associated |
| 4 | gene | 108.85087 | 108.85202 | positive | MGI_C57BL6J_1924896 | C030043A13Rik | NA | NA | unclassified gene | RIKEN cDNA C030043A13 gene |
| 4 | gene | 108.87906 | 108.94332 | positive | MGI_C57BL6J_1917158 | Rab3b | NCBI_Gene:69908,ENSEMBL:ENSMUSG00000003411 | MGI:1917158 | protein coding gene | RAB3B, member RAS oncogene family |
| 4 | pseudogene | 108.93357 | 108.93381 | negative | MGI_C57BL6J_3650723 | Gm12736 | ENSEMBL:ENSMUSG00000082699 | MGI:3650723 | pseudogene | predicted gene 12736 |
| 4 | gene | 108.94284 | 108.97208 | negative | MGI_C57BL6J_1924423 | 8030443G20Rik | NCBI_Gene:77173,ENSEMBL:ENSMUSG00000086483 | MGI:1924423 | lncRNA gene | RIKEN cDNA 8030443G20 gene |
| 4 | gene | 108.99860 | 108.99871 | negative | MGI_C57BL6J_5453366 | Gm23589 | ENSEMBL:ENSMUSG00000088285 | MGI:5453366 | snRNA gene | predicted gene, 23589 |
| 4 | gene | 109.00061 | 109.06178 | positive | MGI_C57BL6J_1201386 | Nrd1 | NCBI_Gene:230598,ENSEMBL:ENSMUSG00000053510 | MGI:1201386 | protein coding gene | nardilysin, N-arginine dibasic convertase, NRD convertase 1 |
| 4 | gene | 109.01766 | 109.01773 | positive | MGI_C57BL6J_3691609 | Mir761 | miRBase:MI0004306,NCBI_Gene:791075,ENSEMBL:ENSMUSG00000076444 | MGI:3691609 | miRNA gene | microRNA 761 |
| 4 | gene | 109.06114 | 109.20227 | negative | MGI_C57BL6J_1923784 | Osbpl9 | NCBI_Gene:100273,ENSEMBL:ENSMUSG00000028559 | MGI:1923784 | protein coding gene | oxysterol binding protein-like 9 |
| 4 | gene | 109.06981 | 109.07017 | negative | MGI_C57BL6J_1925486 | 5330417P21Rik | NA | NA | unclassified gene | RIKEN cDNA 5330417P21 gene |
| 4 | gene | 109.07849 | 109.08122 | positive | MGI_C57BL6J_5591740 | Gm32581 | NCBI_Gene:102635178 | MGI:5591740 | lncRNA gene | predicted gene, 32581 |
| 4 | gene | 109.10297 | 109.10353 | negative | MGI_C57BL6J_1924670 | C030032G21Rik | NA | NA | unclassified gene | RIKEN cDNA C030032G21 gene |
| 4 | gene | 109.16092 | 109.16120 | negative | MGI_C57BL6J_5453520 | Gm23743 | ENSEMBL:ENSMUSG00000088014 | MGI:5453520 | unclassified non-coding RNA gene | predicted gene, 23743 |
| 4 | gene | 109.20219 | 109.20850 | positive | MGI_C57BL6J_5826504 | Gm46867 | NCBI_Gene:108168972 | MGI:5826504 | lncRNA gene | predicted gene, 46867 |
| 4 | gene | 109.21841 | 109.25513 | positive | MGI_C57BL6J_2140435 | Calr4 | NCBI_Gene:108802,ENSEMBL:ENSMUSG00000028558 | MGI:2140435 | protein coding gene | calreticulin 4 |
| 4 | gene | 109.22409 | 109.28011 | negative | MGI_C57BL6J_5623110 | Gm40225 | NCBI_Gene:105244653 | MGI:5623110 | lncRNA gene | predicted gene, 40225 |
| 4 | gene | 109.28026 | 109.38782 | positive | MGI_C57BL6J_104583 | Eps15 | NCBI_Gene:13858,ENSEMBL:ENSMUSG00000028552 | MGI:104583 | protein coding gene | epidermal growth factor receptor pathway substrate 15 |
| 4 | gene | 109.35544 | 109.36002 | negative | MGI_C57BL6J_5623109 | Gm40224 | NCBI_Gene:105244652 | MGI:5623109 | lncRNA gene | predicted gene, 40224 |
| 4 | pseudogene | 109.37370 | 109.37494 | negative | MGI_C57BL6J_3652196 | Gm12749 | NCBI_Gene:666144,ENSEMBL:ENSMUSG00000083512 | MGI:3652196 | pseudogene | predicted gene 12749 |
| 4 | gene | 109.39415 | 109.44474 | positive | MGI_C57BL6J_2444350 | Ttc39a | NCBI_Gene:230603,ENSEMBL:ENSMUSG00000028555 | MGI:2444350 | protein coding gene | tetratricopeptide repeat domain 39A |
| 4 | gene | 109.40228 | 109.40626 | negative | MGI_C57BL6J_3651956 | Ttc39aos1 | NCBI_Gene:102635290,ENSEMBL:ENSMUSG00000085873 | MGI:3651956 | antisense lncRNA gene | Ttc39a opposite strand RNA 1 |
| 4 | gene | 109.45110 | 109.48473 | negative | MGI_C57BL6J_1352759 | Rnf11 | NCBI_Gene:29864,ENSEMBL:ENSMUSG00000028557 | MGI:1352759 | protein coding gene | ring finger protein 11 |
| 4 | gene | 109.50533 | 109.53130 | negative | MGI_C57BL6J_1914896 | 4930522H14Rik | NCBI_Gene:67646,ENSEMBL:ENSMUSG00000060491 | MGI:1914896 | protein coding gene | RIKEN cDNA 4930522H14 gene |
| 4 | gene | 109.53704 | 109.53717 | positive | MGI_C57BL6J_5452274 | Gm22497 | ENSEMBL:ENSMUSG00000077687 | MGI:5452274 | snoRNA gene | predicted gene, 22497 |
| 4 | gene | 109.55096 | 109.55405 | positive | MGI_C57BL6J_5623111 | Gm40226 | NCBI_Gene:105244654 | MGI:5623111 | lncRNA gene | predicted gene, 40226 |
| 4 | gene | 109.56921 | 109.65679 | positive | MGI_C57BL6J_3650795 | Gm12811 | ENSEMBL:ENSMUSG00000087641 | MGI:3650795 | lncRNA gene | predicted gene 12811 |
| 4 | pseudogene | 109.59937 | 109.60005 | negative | MGI_C57BL6J_5011297 | Gm19112 | NCBI_Gene:100418274 | MGI:5011297 | pseudogene | predicted gene, 19112 |
| 4 | pseudogene | 109.62958 | 109.63086 | positive | MGI_C57BL6J_3651634 | Gm12803 | NCBI_Gene:100418450,ENSEMBL:ENSMUSG00000082771 | MGI:3651634 | pseudogene | predicted gene 12803 |
| 4 | gene | 109.63088 | 109.64910 | negative | MGI_C57BL6J_5623112 | Gm40227 | NCBI_Gene:105244655 | MGI:5623112 | lncRNA gene | predicted gene, 40227 |
| 4 | gene | 109.65153 | 109.65785 | negative | MGI_C57BL6J_2442649 | 9630013D21Rik | NCBI_Gene:319743,ENSEMBL:ENSMUSG00000059027 | MGI:2442649 | lncRNA gene | RIKEN cDNA 9630013D21 gene |
| 4 | gene | 109.66088 | 109.66719 | negative | MGI_C57BL6J_105388 | Cdkn2c | NCBI_Gene:12580,ENSEMBL:ENSMUSG00000028551 | MGI:105388 | protein coding gene | cyclin dependent kinase inhibitor 2C |
| 4 | gene | 109.66687 | 109.66825 | negative | MGI_C57BL6J_3826683 | Gm2703 | NA | NA | protein coding gene | predicted gene 2703 |
| 4 | gene | 109.67659 | 109.96396 | positive | MGI_C57BL6J_109419 | Faf1 | NCBI_Gene:14084,ENSEMBL:ENSMUSG00000010517 | MGI:109419 | protein coding gene | Fas-associated factor 1 |
| 4 | pseudogene | 109.75441 | 109.75482 | negative | MGI_C57BL6J_3650597 | Gm12808 | ENSEMBL:ENSMUSG00000083995 | MGI:3650597 | pseudogene | predicted gene 12808 |
| 4 | gene | 109.97318 | 109.97781 | negative | MGI_C57BL6J_1924703 | Dmrta2os | NCBI_Gene:77453,ENSEMBL:ENSMUSG00000085256 | MGI:1924703 | antisense lncRNA gene | doublesex and mab-3 related transcription factor like family A2, opposite strand |
| 4 | gene | 109.97778 | 109.98370 | positive | MGI_C57BL6J_2653629 | Dmrta2 | NCBI_Gene:242620,ENSEMBL:ENSMUSG00000047143 | MGI:2653629 | protein coding gene | doublesex and mab-3 related transcription factor like family A2 |
| 4 | gene | 110.15267 | 110.15671 | positive | MGI_C57BL6J_5592229 | Gm33070 | NCBI_Gene:102635827 | MGI:5592229 | lncRNA gene | predicted gene, 33070 |
| 4 | gene | 110.20372 | 110.35191 | negative | MGI_C57BL6J_107427 | Elavl4 | NCBI_Gene:15572,ENSEMBL:ENSMUSG00000028546 | MGI:107427 | protein coding gene | ELAV like RNA binding protein 4 |
| 4 | gene | 110.39764 | 111.66432 | positive | MGI_C57BL6J_1918244 | Agbl4 | NCBI_Gene:78933,ENSEMBL:ENSMUSG00000061298 | MGI:1918244 | protein coding gene | ATP/GTP binding protein-like 4 |
| 4 | gene | 110.43480 | 110.43599 | positive | MGI_C57BL6J_1924569 | C030007D22Rik | NA | NA | unclassified gene | RIKEN cDNA C030007D22 gene |
| 4 | pseudogene | 110.51004 | 110.51030 | positive | MGI_C57BL6J_3651080 | Gm12806 | ENSEMBL:ENSMUSG00000081813 | MGI:3651080 | pseudogene | predicted gene 12806 |
| 4 | pseudogene | 110.51276 | 110.51426 | negative | MGI_C57BL6J_3650599 | Gm12807 | NCBI_Gene:102635892,ENSEMBL:ENSMUSG00000082348 | MGI:3650599 | pseudogene | predicted gene 12807 |
| 4 | pseudogene | 111.29012 | 111.29112 | positive | MGI_C57BL6J_3651639 | Gm12804 | NCBI_Gene:100417659,ENSEMBL:ENSMUSG00000081068 | MGI:3651639 | pseudogene | predicted gene 12804 |
| 4 | pseudogene | 111.31955 | 111.32063 | negative | MGI_C57BL6J_3650124 | Gm12805 | NCBI_Gene:100417245,ENSEMBL:ENSMUSG00000083904 | MGI:3650124 | pseudogene | predicted gene 12805 |
| 4 | gene | 111.41379 | 111.46030 | positive | MGI_C57BL6J_1914871 | Bend5 | NCBI_Gene:67621,ENSEMBL:ENSMUSG00000028545 | MGI:1914871 | protein coding gene | BEN domain containing 5 |
| 4 | gene | 111.42782 | 111.42793 | negative | MGI_C57BL6J_4422056 | n-R5s191 | ENSEMBL:ENSMUSG00000084673 | MGI:4422056 | rRNA gene | nuclear encoded rRNA 5S 191 |
| 4 | gene | 111.69561 | 111.72399 | negative | MGI_C57BL6J_5592351 | Gm33192 | NCBI_Gene:102635996 | MGI:5592351 | lncRNA gene | predicted gene, 33192 |
| 4 | gene | 111.71995 | 111.82918 | positive | MGI_C57BL6J_1915196 | Spata6 | NCBI_Gene:67946,ENSEMBL:ENSMUSG00000034401 | MGI:1915196 | protein coding gene | spermatogenesis associated 6 |
| 4 | pseudogene | 111.79062 | 111.79167 | negative | MGI_C57BL6J_3650787 | Gm12961 | NCBI_Gene:100417901,ENSEMBL:ENSMUSG00000082686 | MGI:3650787 | pseudogene | predicted gene 12961 |
| 4 | pseudogene | 111.80700 | 111.80848 | negative | MGI_C57BL6J_3651525 | Gm12960 | NCBI_Gene:105244657,ENSEMBL:ENSMUSG00000100514 | MGI:3651525 | pseudogene | predicted gene 12960 |
| 4 | gene | 111.87537 | 111.90292 | negative | MGI_C57BL6J_2140201 | Slc5a9 | NCBI_Gene:230612,ENSEMBL:ENSMUSG00000028544 | MGI:2140201 | protein coding gene | solute carrier family 5 (sodium/glucose cotransporter), member 9 |
| 4 | gene | 111.91939 | 111.95036 | positive | MGI_C57BL6J_3651523 | Skint8 | NCBI_Gene:639774,ENSEMBL:ENSMUSG00000078599 | MGI:3651523 | protein coding gene | selection and upkeep of intraepithelial T cells 8 |
| 4 | gene | 111.97292 | 111.98826 | positive | MGI_C57BL6J_3041190 | Skint7 | NCBI_Gene:328505,ENSEMBL:ENSMUSG00000049214 | MGI:3041190 | protein coding gene | selection and upkeep of intraepithelial T cells 7 |
| 4 | gene | 111.98865 | 111.98896 | negative | MGI_C57BL6J_5455621 | Gm25844 | ENSEMBL:ENSMUSG00000084536 | MGI:5455621 | unclassified non-coding RNA gene | predicted gene, 25844 |
| 4 | gene | 112.00627 | 112.02954 | positive | MGI_C57BL6J_3649627 | Skint1 | NCBI_Gene:639781,ENSEMBL:ENSMUSG00000089773 | MGI:3649627 | protein coding gene | selection and upkeep of intraepithelial T cells 1 |
| 4 | gene | 112.07202 | 112.16808 | positive | MGI_C57BL6J_2444425 | Skint4 | NCBI_Gene:320640,ENSEMBL:ENSMUSG00000055960 | MGI:2444425 | protein coding gene | selection and upkeep of intraepithelial T cells 4 |
| 4 | pseudogene | 112.12100 | 112.12134 | negative | MGI_C57BL6J_3652018 | Gm12820 | ENSEMBL:ENSMUSG00000084326 | MGI:3652018 | pseudogene | predicted gene 12820 |
| 4 | pseudogene | 112.14610 | 112.14623 | negative | MGI_C57BL6J_3652016 | Gm12815 | ENSEMBL:ENSMUSG00000082638 | MGI:3652016 | pseudogene | predicted gene 12815 |
| 4 | gene | 112.23225 | 112.30047 | positive | MGI_C57BL6J_3045331 | Skint3 | NCBI_Gene:195564,ENSEMBL:ENSMUSG00000070868 | MGI:3045331 | protein coding gene | selection and upkeep of intraepithelial T cells 3 |
| 4 | gene | 112.38597 | 112.43399 | negative | MGI_C57BL6J_3045341 | Skint9 | NCBI_Gene:329918,ENSEMBL:ENSMUSG00000049972 | MGI:3045341 | protein coding gene | selection and upkeep of intraepithelial T cells 9 |
| 4 | pseudogene | 112.47383 | 112.52153 | positive | MGI_C57BL6J_3652017 | Gm12819 | NCBI_Gene:100504555,ENSEMBL:ENSMUSG00000081166 | MGI:3652017 | pseudogene | predicted gene 12819 |
| 4 | pseudogene | 112.53706 | 112.53771 | positive | MGI_C57BL6J_3649625 | Gm12821 | NCBI_Gene:102636060,ENSEMBL:ENSMUSG00000083599 | MGI:3649625 | pseudogene | predicted gene 12821 |
| 4 | gene | 112.55719 | 112.65225 | positive | MGI_C57BL6J_3649629 | Skint2 | NCBI_Gene:329919,ENSEMBL:ENSMUSG00000034359 | MGI:3649629 | protein coding gene | selection and upkeep of intraepithelial T cells 2 |
| 4 | pseudogene | 112.55975 | 112.59330 | positive | MGI_C57BL6J_3651208 | Gm12814 | NCBI_Gene:630887,ENSEMBL:ENSMUSG00000083753 | MGI:3651208 | pseudogene | predicted gene 12814 |
| 4 | gene | 112.59077 | 112.59088 | negative | MGI_C57BL6J_5453876 | Gm24099 | ENSEMBL:ENSMUSG00000096564 | MGI:5453876 | snRNA gene | predicted gene, 24099 |
| 4 | pseudogene | 112.70514 | 112.70624 | positive | MGI_C57BL6J_3649633 | Gm12817 | NCBI_Gene:100040581,ENSEMBL:ENSMUSG00000081066 | MGI:3649633 | pseudogene | predicted gene 12817 |
| 4 | gene | 112.71083 | 112.77487 | negative | MGI_C57BL6J_2685416 | Skint10 | NCBI_Gene:230613,ENSEMBL:ENSMUSG00000048766 | MGI:2685416 | protein coding gene | selection and upkeep of intraepithelial T cells 10 |
| 4 | pseudogene | 112.77209 | 112.77327 | positive | MGI_C57BL6J_5010585 | Gm18400 | NCBI_Gene:100417108 | MGI:5010585 | pseudogene | predicted gene, 18400 |
| 4 | gene | 112.80450 | 113.28697 | negative | MGI_C57BL6J_3649262 | Skint6 | NCBI_Gene:230622,ENSEMBL:ENSMUSG00000087194 | MGI:3649262 | protein coding gene | selection and upkeep of intraepithelial T cells 6 |
| 4 | gene | 112.92370 | 112.94532 | positive | MGI_C57BL6J_3651665 | Gm12823 | ENSEMBL:ENSMUSG00000085878 | MGI:3651665 | lncRNA gene | predicted gene 12823 |
| 4 | pseudogene | 112.95501 | 112.95629 | negative | MGI_C57BL6J_3649628 | Gm12816 | NCBI_Gene:433745,ENSEMBL:ENSMUSG00000083011 | MGI:3649628 | pseudogene | predicted gene 12816 |
| 4 | pseudogene | 112.97846 | 112.97984 | negative | MGI_C57BL6J_3649634 | Gm12818 | NCBI_Gene:100534383,ENSEMBL:ENSMUSG00000082678 | MGI:3649634 | pseudogene | predicted gene 12818 |
| 4 | pseudogene | 113.36819 | 113.39450 | positive | MGI_C57BL6J_3650999 | Gm12813 | NCBI_Gene:100040607,ENSEMBL:ENSMUSG00000083384 | MGI:3650999 | pseudogene | predicted gene 12813 |
| 4 | gene | 113.47786 | 113.99951 | negative | MGI_C57BL6J_3650151 | Skint5 | NCBI_Gene:242627,ENSEMBL:ENSMUSG00000078598 | MGI:3650151 | protein coding gene | selection and upkeep of intraepithelial T cells 5 |
| 4 | pseudogene | 114.09632 | 114.09744 | negative | MGI_C57BL6J_3650153 | Gm12825 | NCBI_Gene:100418129,ENSEMBL:ENSMUSG00000080696 | MGI:3650153 | pseudogene | predicted gene 12825 |
| 4 | gene | 114.16338 | 114.24503 | positive | MGI_C57BL6J_2685415 | Skint11 | NCBI_Gene:230623,ENSEMBL:ENSMUSG00000057977 | MGI:2685415 | protein coding gene | selection and upkeep of intraepithelial T cells 11 |
| 4 | pseudogene | 114.16511 | 114.16591 | negative | MGI_C57BL6J_3783183 | Gm15741 | NCBI_Gene:100417107,ENSEMBL:ENSMUSG00000082825 | MGI:3783183 | pseudogene | predicted gene 15741 |
| 4 | gene | 114.32991 | 114.34662 | negative | MGI_C57BL6J_5623113 | Gm40228 | NCBI_Gene:105244658 | MGI:5623113 | lncRNA gene | predicted gene, 40228 |
| 4 | gene | 114.40620 | 114.61510 | positive | MGI_C57BL6J_3650152 | Trabd2b | NCBI_Gene:666048,ENSEMBL:ENSMUSG00000070867 | MGI:3650152 | protein coding gene | TraB domain containing 2B |
| 4 | gene | 114.41397 | 114.41495 | negative | MGI_C57BL6J_5592869 | Gm33710 | NCBI_Gene:102636714 | MGI:5592869 | lncRNA gene | predicted gene, 33710 |
| 4 | gene | 114.57692 | 114.59222 | negative | MGI_C57BL6J_5592814 | Gm33655 | NCBI_Gene:102636647 | MGI:5592814 | lncRNA gene | predicted gene, 33655 |
| 4 | gene | 114.67270 | 114.67334 | negative | MGI_C57BL6J_5579570 | Gm28864 | ENSEMBL:ENSMUSG00000100998 | MGI:5579570 | lncRNA gene | predicted gene 28864 |
| 4 | gene | 114.68034 | 114.68760 | positive | MGI_C57BL6J_3650003 | Gm12829 | NCBI_Gene:102636500,ENSEMBL:ENSMUSG00000085870 | MGI:3650003 | lncRNA gene | predicted gene 12829 |
| 4 | gene | 114.81609 | 114.90286 | negative | MGI_C57BL6J_1918862 | 9130410C08Rik | NCBI_Gene:105244659,ENSEMBL:ENSMUSG00000085323 | MGI:1918862 | lncRNA gene | RIKEN cDNA 9130410C08 gene |
| 4 | gene | 114.82172 | 114.85617 | positive | MGI_C57BL6J_3649998 | Gm12830 | NCBI_Gene:433746,ENSEMBL:ENSMUSG00000055198 | MGI:3649998 | protein coding gene | predicted gene 12830 |
| 4 | gene | 114.84498 | 114.84512 | negative | MGI_C57BL6J_5453007 | Gm23230 | ENSEMBL:ENSMUSG00000087923 | MGI:5453007 | snRNA gene | predicted gene, 23230 |
| 4 | gene | 114.90628 | 114.90890 | negative | MGI_C57BL6J_1347471 | Foxd2 | NCBI_Gene:17301,ENSEMBL:ENSMUSG00000055210 | MGI:1347471 | protein coding gene | forkhead box D2 |
| 4 | gene | 114.90926 | 114.92197 | positive | MGI_C57BL6J_2444065 | Foxd2os | NCBI_Gene:100040736,ENSEMBL:ENSMUSG00000085399 | MGI:2444065 | lncRNA gene | forkhead box D2, opposite strand |
| 4 | gene | 114.92515 | 114.92601 | negative | MGI_C57BL6J_1353569 | Foxe3 | NCBI_Gene:30923,ENSEMBL:ENSMUSG00000044518 | MGI:1353569 | protein coding gene | forkhead box E3 |
| 4 | gene | 114.94681 | 114.95085 | negative | MGI_C57BL6J_5623114 | Gm40229 | NCBI_Gene:105244660 | MGI:5623114 | lncRNA gene | predicted gene, 40229 |
| 4 | gene | 114.95934 | 114.98724 | negative | MGI_C57BL6J_1913838 | Cmpk1 | NCBI_Gene:66588,ENSEMBL:ENSMUSG00000028719 | MGI:1913838 | protein coding gene | cytidine monophosphate (UMP-CMP) kinase 1 |
| 4 | gene | 114.98796 | 114.98806 | positive | MGI_C57BL6J_5453822 | Gm24045 | ENSEMBL:ENSMUSG00000065047 | MGI:5453822 | unclassified non-coding RNA gene | predicted gene, 24045 |
| 4 | gene | 115.00011 | 115.04320 | positive | MGI_C57BL6J_107477 | Stil | NCBI_Gene:20460,ENSEMBL:ENSMUSG00000028718 | MGI:107477 | protein coding gene | Scl/Tal1 interrupting locus |
| 4 | gene | 115.05643 | 115.07175 | positive | MGI_C57BL6J_98480 | Tal1 | NCBI_Gene:21349,ENSEMBL:ENSMUSG00000028717 | MGI:98480 | protein coding gene | T cell acute lymphocytic leukemia 1 |
| 4 | gene | 115.05762 | 115.05822 | negative | MGI_C57BL6J_5592945 | Gm33786 | NCBI_Gene:102636820 | MGI:5592945 | lncRNA gene | predicted gene, 33786 |
| 4 | gene | 115.08871 | 115.09390 | positive | MGI_C57BL6J_1914432 | Pdzk1ip1 | NCBI_Gene:67182,ENSEMBL:ENSMUSG00000028716 | MGI:1914432 | protein coding gene | PDZK1 interacting protein 1 |
| 4 | gene | 115.10632 | 115.13428 | negative | MGI_C57BL6J_1932403 | Cyp4x1 | NCBI_Gene:81906,ENSEMBL:ENSMUSG00000047155 | MGI:1932403 | protein coding gene | cytochrome P450, family 4, subfamily x, polypeptide 1 |
| 4 | gene | 115.13383 | 115.13786 | positive | MGI_C57BL6J_3649427 | Cyp4x1os | NCBI_Gene:105244662,ENSEMBL:ENSMUSG00000086896 | MGI:3649427 | antisense lncRNA gene | cytochrome P450, family 4, subfamily x, polypeptide 1, opposite strand |
| 4 | pseudogene | 115.13856 | 115.15651 | negative | MGI_C57BL6J_1932405 | Cyp4a28-ps | NCBI_Gene:547220,ENSEMBL:ENSMUSG00000081823 | MGI:1932405 | pseudogene | cytochrome P450, family 4, subfamily a, polypeptide 28, pseudogene |
| 4 | pseudogene | 115.17580 | 115.17684 | positive | MGI_C57BL6J_3651920 | Cyp4a29-ps1 | ENSEMBL:ENSMUSG00000082889 | MGI:3651920 | pseudogene | cytochrome P450, family 4, subfamily a, polypeptide 29, pseudogene 1 |
| 4 | pseudogene | 115.23875 | 115.24158 | negative | MGI_C57BL6J_3649428 | Gm12834 | NCBI_Gene:384042,ENSEMBL:ENSMUSG00000081138 | MGI:3649428 | pseudogene | predicted gene 12834 |
| 4 | gene | 115.24208 | 115.25456 | positive | MGI_C57BL6J_3717143 | Cyp4a29 | NCBI_Gene:230639,ENSEMBL:ENSMUSG00000083138 | MGI:3717143 | protein coding gene | cytochrome P450, family 4, subfamily a, polypeptide 29 |
| 4 | gene | 115.29905 | 115.33281 | positive | MGI_C57BL6J_88612 | Cyp4a12a | NCBI_Gene:277753,ENSEMBL:ENSMUSG00000066071 | MGI:88612 | protein coding gene | cytochrome P450, family 4, subfamily a, polypeptide 12a |
| 4 | pseudogene | 115.34379 | 115.36311 | positive | MGI_C57BL6J_3649426 | Cyp4a30-ps | NCBI_Gene:546842,ENSEMBL:ENSMUSG00000083190 | MGI:3649426 | pseudogene | cytochrome P450, family 4, subfamily a, member 30, pseudogene |
| 4 | gene | 115.41162 | 115.43903 | positive | MGI_C57BL6J_3611747 | Cyp4a12b | NCBI_Gene:13118,ENSEMBL:ENSMUSG00000078597 | MGI:3611747 | protein coding gene | cytochrome P450, family 4, subfamily a, polypeptide 12B |
| 4 | pseudogene | 115.41608 | 115.41645 | negative | MGI_C57BL6J_3649425 | Gm12837 | ENSEMBL:ENSMUSG00000084415 | MGI:3649425 | pseudogene | predicted gene 12837 |
| 4 | gene | 115.45249 | 115.47142 | positive | MGI_C57BL6J_3717145 | Cyp4a30b | NCBI_Gene:435802,ENSEMBL:ENSMUSG00000084346 | MGI:3717145 | protein coding gene | cytochrome P450, family 4, subfamily a, polypeptide 30b |
| 4 | pseudogene | 115.45379 | 115.45412 | negative | MGI_C57BL6J_3649431 | Gm12838 | ENSEMBL:ENSMUSG00000080142 | MGI:3649431 | pseudogene | predicted gene 12838 |
| 4 | pseudogene | 115.47954 | 115.48128 | positive | MGI_C57BL6J_3651919 | Gm12833 | NCBI_Gene:230641,ENSEMBL:ENSMUSG00000086724 | MGI:3651919 | pseudogene | predicted gene 12833 |
| 4 | gene | 115.48620 | 115.49616 | negative | MGI_C57BL6J_1096550 | Cyp4a14 | NCBI_Gene:13119,ENSEMBL:ENSMUSG00000028715 | MGI:1096550 | protein coding gene | cytochrome P450, family 4, subfamily a, polypeptide 14 |
| 4 | gene | 115.51826 | 115.53365 | positive | MGI_C57BL6J_88611 | Cyp4a10 | NCBI_Gene:13117,ENSEMBL:ENSMUSG00000066072 | MGI:88611 | protein coding gene | cytochrome P450, family 4, subfamily a, polypeptide 10 |
| 4 | pseudogene | 115.54338 | 115.54507 | negative | MGI_C57BL6J_3651921 | Cyp4b1-ps1 | NCBI_Gene:100418216,ENSEMBL:ENSMUSG00000081006 | MGI:3651921 | pseudogene | cytochrome P450, family 4, subfamily b, polypeptide 1, pseudogene 1 |
| 4 | gene | 115.56363 | 115.57901 | positive | MGI_C57BL6J_3028580 | Cyp4a31 | NCBI_Gene:666168,ENSEMBL:ENSMUSG00000028712 | MGI:3028580 | protein coding gene | cytochrome P450, family 4, subfamily a, polypeptide 31 |
| 4 | pseudogene | 115.58204 | 115.58314 | negative | MGI_C57BL6J_3649236 | Cyp4b1-ps2 | NCBI_Gene:631037,ENSEMBL:ENSMUSG00000083457 | MGI:3649236 | pseudogene | cytochrome P450, family 4, subfamily b, polypeptide 1, pseudogene 2 |
| 4 | gene | 115.60093 | 115.62251 | positive | MGI_C57BL6J_3717148 | Cyp4a32 | NCBI_Gene:100040843,ENSEMBL:ENSMUSG00000063929 | MGI:3717148 | protein coding gene | cytochrome P450, family 4, subfamily a, polypeptide 32 |
| 4 | gene | 115.62472 | 115.64772 | negative | MGI_C57BL6J_103225 | Cyp4b1 | NCBI_Gene:13120,ENSEMBL:ENSMUSG00000028713 | MGI:103225 | protein coding gene | cytochrome P450, family 4, subfamily b, polypeptide 1 |
| 4 | pseudogene | 115.65239 | 115.65348 | positive | MGI_C57BL6J_3650820 | Gm12848 | NCBI_Gene:546843,ENSEMBL:ENSMUSG00000082043 | MGI:3650820 | pseudogene | predicted gene 12848 |
| 4 | pseudogene | 115.70725 | 115.70830 | positive | MGI_C57BL6J_3650623 | Gm12849 | ENSEMBL:ENSMUSG00000081172 | MGI:3650623 | pseudogene | predicted gene 12849 |
| 4 | gene | 115.73774 | 115.77733 | positive | MGI_C57BL6J_2442397 | Efcab14 | NCBI_Gene:230648,ENSEMBL:ENSMUSG00000034210 | MGI:2442397 | protein coding gene | EF-hand calcium binding domain 14 |
| 4 | gene | 115.77983 | 115.78275 | negative | MGI_C57BL6J_1922423 | Tex38 | NCBI_Gene:75173,ENSEMBL:ENSMUSG00000044556 | MGI:1922423 | protein coding gene | testis expressed 38 |
| 4 | gene | 115.78481 | 115.81883 | positive | MGI_C57BL6J_2180560 | Atpaf1 | NCBI_Gene:230649,ENSEMBL:ENSMUSG00000028710 | MGI:2180560 | protein coding gene | ATP synthase mitochondrial F1 complex assembly factor 1 |
| 4 | gene | 115.81886 | 115.82265 | positive | MGI_C57BL6J_3650822 | Gm12847 | ENSEMBL:ENSMUSG00000085249 | MGI:3650822 | lncRNA gene | predicted gene 12847 |
| 4 | gene | 115.82789 | 115.83619 | positive | MGI_C57BL6J_2140623 | Mob3c | NCBI_Gene:100465,ENSEMBL:ENSMUSG00000028709 | MGI:2140623 | protein coding gene | MOB kinase activator 3C |
| 4 | gene | 115.83899 | 115.87926 | positive | MGI_C57BL6J_894316 | Mknk1 | NCBI_Gene:17346,ENSEMBL:ENSMUSG00000028708 | MGI:894316 | protein coding gene | MAP kinase-interacting serine/threonine kinase 1 |
| 4 | gene | 115.87832 | 115.87858 | negative | MGI_C57BL6J_5580140 | Gm29434 | ENSEMBL:ENSMUSG00000100081 | MGI:5580140 | lncRNA gene | predicted gene 29434 |
| 4 | gene | 115.88440 | 115.88953 | positive | MGI_C57BL6J_3614952 | Kncn | NCBI_Gene:654462,ENSEMBL:ENSMUSG00000073774 | MGI:3614952 | protein coding gene | kinocilin |
| 4 | gene | 115.89066 | 115.89923 | positive | MGI_C57BL6J_1923487 | 6430628N08Rik | NCBI_Gene:76237,ENSEMBL:ENSMUSG00000034185 | MGI:1923487 | protein coding gene | RIKEN cDNA 6430628N08 gene |
| 4 | gene | 115.91512 | 115.94060 | negative | MGI_C57BL6J_2153518 | Dmbx1 | NCBI_Gene:140477,ENSEMBL:ENSMUSG00000028707 | MGI:2153518 | protein coding gene | diencephalon/mesencephalon homeobox 1 |
| 4 | gene | 115.96714 | 115.97092 | negative | MGI_C57BL6J_5621377 | Gm38492 | NCBI_Gene:102637048 | MGI:5621377 | lncRNA gene | predicted gene, 38492 |
| 4 | gene | 115.96715 | 116.01869 | negative | MGI_C57BL6J_109609 | Faah | NCBI_Gene:14073,ENSEMBL:ENSMUSG00000034171 | MGI:109609 | protein coding gene | fatty acid amide hydrolase |
| 4 | gene | 116.03177 | 116.05388 | negative | MGI_C57BL6J_1919431 | Nsun4 | NCBI_Gene:72181,ENSEMBL:ENSMUSG00000028706 | MGI:1919431 | protein coding gene | NOL1/NOP2/Sun domain family, member 4 |
| 4 | gene | 116.06696 | 116.07507 | negative | MGI_C57BL6J_1913826 | Uqcrh | NCBI_Gene:66576,ENSEMBL:ENSMUSG00000063882 | MGI:1913826 | protein coding gene | ubiquinol-cytochrome c reductase hinge protein |
| 4 | pseudogene | 116.06718 | 116.06770 | positive | MGI_C57BL6J_3650410 | Gm12854 | NCBI_Gene:102637129,ENSEMBL:ENSMUSG00000078238 | MGI:3650410 | pseudogene | predicted gene 12854 |
| 4 | gene | 116.07527 | 116.09711 | positive | MGI_C57BL6J_2441984 | Lrrc41 | NCBI_Gene:230654,ENSEMBL:ENSMUSG00000028703 | MGI:2441984 | protein coding gene | leucine rich repeat containing 41 |
| 4 | gene | 116.09426 | 116.12369 | negative | MGI_C57BL6J_894697 | Rad54l | NCBI_Gene:19366,ENSEMBL:ENSMUSG00000028702 | MGI:894697 | protein coding gene | RAD54 like (S. cerevisiae) |
| 4 | gene | 116.12384 | 116.13884 | positive | MGI_C57BL6J_1924036 | 2510003B16Rik | NCBI_Gene:76786 | MGI:1924036 | lncRNA gene | RIKEN cDNA 2510003B16 gene |
| 4 | gene | 116.12384 | 116.15985 | positive | MGI_C57BL6J_1915523 | Pomgnt1 | NCBI_Gene:68273,ENSEMBL:ENSMUSG00000028700 | MGI:1915523 | protein coding gene | protein O-linked mannose beta 1,2-N-acetylglucosaminyltransferase |
| 4 | gene | 116.13241 | 116.15134 | negative | MGI_C57BL6J_1915325 | Lurap1 | NCBI_Gene:68075,ENSEMBL:ENSMUSG00000028701 | MGI:1915325 | protein coding gene | leucine rich adaptor protein 1 |
| 4 | gene | 116.16187 | 116.16762 | negative | MGI_C57BL6J_1914055 | Tspan1 | NCBI_Gene:66805,ENSEMBL:ENSMUSG00000028699 | MGI:1914055 | protein coding gene | tetraspanin 1 |
| 4 | gene | 116.17337 | 116.17430 | positive | MGI_C57BL6J_1914573 | 1700042G07Rik | NCBI_Gene:67323,ENSEMBL:ENSMUSG00000078593 | MGI:1914573 | protein coding gene | RIKEN cDNA 1700042G07 gene |
| 4 | gene | 116.17338 | 116.29984 | positive | MGI_C57BL6J_6121526 | Gm49337 | ENSEMBL:ENSMUSG00000111410 | MGI:6121526 | protein coding gene | predicted gene, 49337 |
| 4 | gene | 116.17625 | 116.17773 | positive | MGI_C57BL6J_3649505 | Gm12951 | ENSEMBL:ENSMUSG00000085920 | MGI:3649505 | lncRNA gene | predicted gene 12951 |
| 4 | pseudogene | 116.20041 | 116.20099 | negative | MGI_C57BL6J_3649506 | Llph-ps1 | NCBI_Gene:433748,ENSEMBL:ENSMUSG00000081459 | MGI:3649506 | pseudogene | LLP homolog, pseudogene 1 |
| 4 | gene | 116.22142 | 116.30306 | positive | MGI_C57BL6J_109277 | Pik3r3 | NCBI_Gene:18710,ENSEMBL:ENSMUSG00000028698 | MGI:109277 | protein coding gene | phosphoinositide-3-kinase regulatory subunit 3 |
| 4 | pseudogene | 116.25002 | 116.25111 | negative | MGI_C57BL6J_5010720 | Gm18535 | NCBI_Gene:100417330 | MGI:5010720 | pseudogene | predicted gene, 18535 |
| 4 | gene | 116.30676 | 116.46425 | negative | MGI_C57BL6J_894676 | Mast2 | NCBI_Gene:17776,ENSEMBL:ENSMUSG00000003810 | MGI:894676 | protein coding gene | microtubule associated serine/threonine kinase 2 |
| 4 | gene | 116.38232 | 116.38575 | negative | MGI_C57BL6J_2444793 | A630078A22Rik | NA | NA | unclassified gene | RIKEN cDNA A630078A22 gene |
| 4 | pseudogene | 116.44879 | 116.44920 | positive | MGI_C57BL6J_3649504 | Gm12950 | ENSEMBL:ENSMUSG00000082592 | MGI:3649504 | pseudogene | predicted gene 12950 |
| 4 | gene | 116.50751 | 116.54256 | positive | MGI_C57BL6J_96581 | Ipp | NCBI_Gene:16351,ENSEMBL:ENSMUSG00000028696 | MGI:96581 | protein coding gene | IAP promoted placental gene |
| 4 | gene | 116.55153 | 116.55704 | negative | MGI_C57BL6J_3045357 | Tmem69 | NCBI_Gene:230657,ENSEMBL:ENSMUSG00000055900 | MGI:3045357 | protein coding gene | transmembrane protein 69 |
| 4 | gene | 116.55174 | 116.55263 | positive | MGI_C57BL6J_3649497 | Gm12953 | ENSEMBL:ENSMUSG00000087299 | MGI:3649497 | lncRNA gene | predicted gene 12953 |
| 4 | gene | 116.55715 | 116.59388 | positive | MGI_C57BL6J_1924360 | Gpbp1l1 | NCBI_Gene:77110,ENSEMBL:ENSMUSG00000034042 | MGI:1924360 | protein coding gene | GC-rich promoter binding protein 1-like 1 |
| 4 | gene | 116.58973 | 116.59763 | negative | MGI_C57BL6J_3612454 | C530005A16Rik | NCBI_Gene:654318,ENSEMBL:ENSMUSG00000085408 | MGI:3612454 | lncRNA gene | RIKEN cDNA C530005A16 gene |
| 4 | gene | 116.59665 | 116.60027 | positive | MGI_C57BL6J_1915667 | Ccdc17 | NCBI_Gene:622665,ENSEMBL:ENSMUSG00000034035 | MGI:1915667 | protein coding gene | coiled-coil domain containing 17 |
| 4 | gene | 116.60105 | 116.62795 | negative | MGI_C57BL6J_1355328 | Nasp | NCBI_Gene:50927,ENSEMBL:ENSMUSG00000028693 | MGI:1355328 | protein coding gene | nuclear autoantigenic sperm protein (histone-binding) |
| 4 | gene | 116.62684 | 116.62869 | positive | MGI_C57BL6J_2441802 | A430091L06Rik | NA | NA | unclassified gene | RIKEN cDNA A430091L06 gene |
| 4 | gene | 116.63651 | 116.65168 | negative | MGI_C57BL6J_1929955 | Akr1a1 | NCBI_Gene:58810,ENSEMBL:ENSMUSG00000028692 | MGI:1929955 | protein coding gene | aldo-keto reductase family 1, member A1 (aldehyde reductase) |
| 4 | gene | 116.67415 | 116.67426 | positive | MGI_C57BL6J_5454914 | Gm25137 | ENSEMBL:ENSMUSG00000065185 | MGI:5454914 | snRNA gene | predicted gene, 25137 |
| 4 | gene | 116.68554 | 116.70082 | positive | MGI_C57BL6J_99523 | Prdx1 | NCBI_Gene:18477,ENSEMBL:ENSMUSG00000028691 | MGI:99523 | protein coding gene | peroxiredoxin 1 |
| 4 | gene | 116.69708 | 116.69751 | positive | MGI_C57BL6J_1924686 | 9530001N24Rik | NA | NA | unclassified gene | RIKEN cDNA 9530001N24 gene |
| 4 | gene | 116.70228 | 116.70841 | negative | MGI_C57BL6J_1914346 | Mmachc | NCBI_Gene:67096,ENSEMBL:ENSMUSG00000028690 | MGI:1914346 | protein coding gene | methylmalonic aciduria cblC type, with homocystinuria |
| 4 | gene | 116.70812 | 116.70843 | positive | MGI_C57BL6J_1925287 | 4930565M07Rik | NA | NA | unclassified gene | RIKEN cDNA 4930565M07 gene |
| 4 | gene | 116.70851 | 116.75121 | positive | MGI_C57BL6J_1915644 | Ccdc163 | NCBI_Gene:68394,ENSEMBL:ENSMUSG00000028689 | MGI:1915644 | protein coding gene | coiled-coil domain containing 163 |
| 4 | gene | 116.72091 | 116.80596 | positive | MGI_C57BL6J_2385204 | Tesk2 | NCBI_Gene:230661,ENSEMBL:ENSMUSG00000033985 | MGI:2385204 | protein coding gene | testis-specific kinase 2 |
| 4 | gene | 116.72590 | 116.72599 | positive | MGI_C57BL6J_5531070 | Gm27688 | ENSEMBL:ENSMUSG00000098491 | MGI:5531070 | unclassified non-coding RNA gene | predicted gene, 27688 |
| 4 | gene | 116.79431 | 116.80764 | negative | MGI_C57BL6J_1915526 | Toe1 | NCBI_Gene:68276,ENSEMBL:ENSMUSG00000028688 | MGI:1915526 | protein coding gene | target of EGR1, member 1 (nuclear) |
| 4 | gene | 116.80768 | 116.81944 | positive | MGI_C57BL6J_1917853 | Mutyh | NCBI_Gene:70603,ENSEMBL:ENSMUSG00000028687 | MGI:1917853 | protein coding gene | mutY DNA glycosylase |
| 4 | gene | 116.81990 | 116.82171 | negative | MGI_C57BL6J_2444646 | Hpdl | NCBI_Gene:242642,ENSEMBL:ENSMUSG00000043155 | MGI:2444646 | protein coding gene | 4-hydroxyphenylpyruvate dioxygenase-like |
| 4 | gene | 116.82426 | 116.83061 | positive | MGI_C57BL6J_3650481 | Gm12996 | NCBI_Gene:100502973,ENSEMBL:ENSMUSG00000086417 | MGI:3650481 | lncRNA gene | predicted gene 12996 |
| 4 | pseudogene | 116.83988 | 116.84024 | negative | MGI_C57BL6J_3650480 | Rpl36-ps8 | NCBI_Gene:625336,ENSEMBL:ENSMUSG00000075390 | MGI:3650480 | pseudogene | ribosomal protein L36, pseudogene 8 |
| 4 | pseudogene | 116.85266 | 116.85311 | positive | MGI_C57BL6J_3650479 | Gm12994 | ENSEMBL:ENSMUSG00000082841 | MGI:3650479 | pseudogene | predicted gene 12994 |
| 4 | pseudogene | 116.87459 | 116.87547 | negative | MGI_C57BL6J_3650477 | Gm12993 | ENSEMBL:ENSMUSG00000082063 | MGI:3650477 | pseudogene | predicted gene 12993 |
| 4 | gene | 116.87738 | 116.98926 | positive | MGI_C57BL6J_1921714 | Zswim5 | NCBI_Gene:74464,ENSEMBL:ENSMUSG00000033948 | MGI:1921714 | protein coding gene | zinc finger SWIM-type containing 5 |
| 4 | gene | 116.90464 | 116.90474 | positive | MGI_C57BL6J_5456132 | Gm26355 | ENSEMBL:ENSMUSG00000088845 | MGI:5456132 | snRNA gene | predicted gene, 26355 |
| 4 | gene | 116.91523 | 116.91549 | positive | MGI_C57BL6J_5455212 | Gm25435 | ENSEMBL:ENSMUSG00000088902 | MGI:5455212 | unclassified non-coding RNA gene | predicted gene, 25435 |
| 4 | gene | 116.92238 | 116.92249 | negative | MGI_C57BL6J_5451869 | Gm22092 | ENSEMBL:ENSMUSG00000095805 | MGI:5451869 | snRNA gene | predicted gene, 22092 |
| 4 | pseudogene | 116.94573 | 116.94653 | negative | MGI_C57BL6J_3651168 | Gm12998 | NCBI_Gene:100417429,ENSEMBL:ENSMUSG00000082207 | MGI:3651168 | pseudogene | predicted gene 12998 |
| 4 | gene | 116.98996 | 116.99444 | negative | MGI_C57BL6J_98916 | Urod | NCBI_Gene:22275,ENSEMBL:ENSMUSG00000028684 | MGI:98916 | protein coding gene | uroporphyrinogen decarboxylase |
| 4 | gene | 116.99529 | 117.00528 | positive | MGI_C57BL6J_1923858 | Hectd3 | NCBI_Gene:76608,ENSEMBL:ENSMUSG00000046861 | MGI:1923858 | protein coding gene | HECT domain E3 ubiquitin protein ligase 3 |
| 4 | gene | 117.00217 | 117.00301 | negative | MGI_C57BL6J_1919485 | 1700021J08Rik | ENSEMBL:ENSMUSG00000086890 | MGI:1919485 | lncRNA gene | RIKEN cDNA 1700021J08 gene |
| 4 | pseudogene | 117.01513 | 117.01577 | negative | MGI_C57BL6J_3650026 | Gm12997 | NCBI_Gene:100416056,ENSEMBL:ENSMUSG00000082249 | MGI:3650026 | pseudogene | predicted gene 12997 |
| 4 | gene | 117.01940 | 117.08731 | positive | MGI_C57BL6J_1313286 | Eif2b3 | NCBI_Gene:108067,ENSEMBL:ENSMUSG00000028683 | MGI:1313286 | protein coding gene | eukaryotic translation initiation factor 2B, subunit 3 |
| 4 | gene | 117.02721 | 117.02731 | positive | MGI_C57BL6J_5452112 | Gm22335 | ENSEMBL:ENSMUSG00000088349 | MGI:5452112 | snRNA gene | predicted gene, 22335 |
| 4 | gene | 117.09578 | 117.11610 | positive | MGI_C57BL6J_1095405 | Ptch2 | NCBI_Gene:19207,ENSEMBL:ENSMUSG00000028681 | MGI:1095405 | protein coding gene | patched 2 |
| 4 | gene | 117.11922 | 117.12573 | negative | MGI_C57BL6J_1925861 | Btbd19 | NCBI_Gene:78611,ENSEMBL:ENSMUSG00000073771 | MGI:1925861 | protein coding gene | BTB (POZ) domain containing 19 |
| 4 | gene | 117.12455 | 117.12874 | positive | MGI_C57BL6J_3045358 | Tctex1d4 | NCBI_Gene:242646,ENSEMBL:ENSMUSG00000047671 | MGI:3045358 | protein coding gene | Tctex1 domain containing 4 |
| 4 | gene | 117.12865 | 117.13396 | negative | MGI_C57BL6J_109604 | Plk3 | NCBI_Gene:12795,ENSEMBL:ENSMUSG00000028680 | MGI:109604 | protein coding gene | polo like kinase 3 |
| 4 | pseudogene | 117.14526 | 117.14864 | positive | MGI_C57BL6J_3663793 | Best4-ps | NCBI_Gene:230664,ENSEMBL:ENSMUSG00000033872 | MGI:3663793 | pseudogene | bestrophin 4, pseudogene |
| 4 | pseudogene | 117.14890 | 117.14930 | positive | MGI_C57BL6J_3651767 | Gm13015 | NCBI_Gene:625405,ENSEMBL:ENSMUSG00000070834 | MGI:3651767 | pseudogene | predicted gene 13015 |
| 4 | gene | 117.15383 | 117.15624 | negative | MGI_C57BL6J_98166 | Rps8 | NCBI_Gene:20116,ENSEMBL:ENSMUSG00000047675 | MGI:98166 | protein coding gene | ribosomal protein S8 |
| 4 | gene | 117.15409 | 117.15416 | negative | MGI_C57BL6J_5452757 | Gm22980 | ENSEMBL:ENSMUSG00000064542 | MGI:5452757 | snoRNA gene | predicted gene, 22980 |
| 4 | gene | 117.15451 | 117.15458 | negative | MGI_C57BL6J_3819531 | Snord38a | NCBI_Gene:100217424,ENSEMBL:ENSMUSG00000065680 | MGI:3819531 | snoRNA gene | small nucleolar RNA, C/D box 38A |
| 4 | gene | 117.15519 | 117.15529 | negative | MGI_C57BL6J_5456107 | Gm26330 | ENSEMBL:ENSMUSG00000064751 | MGI:5456107 | snoRNA gene | predicted gene, 26330 |
| 4 | gene | 117.15577 | 117.15585 | negative | MGI_C57BL6J_3819543 | Snord55 | NCBI_Gene:100216533,ENSEMBL:ENSMUSG00000092680 | MGI:3819543 | snoRNA gene | small nucleolar RNA, C/D box 55 |
| 4 | gene | 117.15831 | 117.15843 | negative | MGI_C57BL6J_5452175 | Gm22398 | ENSEMBL:ENSMUSG00000095209 | MGI:5452175 | miRNA gene | predicted gene, 22398 |
| 4 | gene | 117.15963 | 117.18264 | negative | MGI_C57BL6J_1921054 | Kif2c | NCBI_Gene:73804,ENSEMBL:ENSMUSG00000028678 | MGI:1921054 | protein coding gene | kinesin family member 2C |
| 4 | gene | 117.18994 | 117.19006 | positive | MGI_C57BL6J_5454876 | Gm25099 | ENSEMBL:ENSMUSG00000095676 | MGI:5454876 | snRNA gene | predicted gene, 25099 |
| 4 | gene | 117.20253 | 117.20264 | positive | MGI_C57BL6J_5452920 | Gm23143 | ENSEMBL:ENSMUSG00000094405 | MGI:5452920 | snRNA gene | predicted gene, 23143 |
| 4 | gene | 117.20499 | 117.20536 | positive | MGI_C57BL6J_3629729 | Gt(pU21)107Imeg | NA | NA | unclassified gene | gene trap 107%2c Institute of Molecular Embryology and Genetics |
| 4 | gene | 117.21084 | 117.21096 | positive | MGI_C57BL6J_5453064 | Gm23287 | ENSEMBL:ENSMUSG00000096280 | MGI:5453064 | snRNA gene | predicted gene, 23287 |
| 4 | gene | 117.21333 | 117.25213 | negative | MGI_C57BL6J_2686507 | Armh1 | NCBI_Gene:381544,ENSEMBL:ENSMUSG00000060268 | MGI:2686507 | protein coding gene | armadillo-like helical domain containing 1 |
| 4 | gene | 117.21333 | 117.21378 | negative | MGI_C57BL6J_1923635 | 1700012C08Rik | ENSEMBL:ENSMUSG00000086482 | MGI:1923635 | lncRNA gene | RIKEN cDNA 1700012C08 gene |
| 4 | gene | 117.25195 | 117.26859 | positive | MGI_C57BL6J_1916027 | Tmem53 | NCBI_Gene:68777,ENSEMBL:ENSMUSG00000048772 | MGI:1916027 | protein coding gene | transmembrane protein 53 |
| 4 | gene | 117.27146 | 117.49705 | negative | MGI_C57BL6J_1913993 | Rnf220 | NCBI_Gene:66743,ENSEMBL:ENSMUSG00000028677 | MGI:1913993 | protein coding gene | ring finger protein 220 |
| 4 | gene | 117.28939 | 117.30542 | positive | MGI_C57BL6J_3651051 | Gm12843 | NCBI_Gene:102637763,ENSEMBL:ENSMUSG00000085749 | MGI:3651051 | lncRNA gene | predicted gene 12843 |
| 4 | gene | 117.31174 | 117.31943 | negative | MGI_C57BL6J_5623116 | Gm40231 | NCBI_Gene:105244664 | MGI:5623116 | lncRNA gene | predicted gene, 40231 |
| 4 | gene | 117.32809 | 117.32902 | negative | MGI_C57BL6J_1924582 | C030012D19Rik | NA | NA | unclassified gene | RIKEN cDNA C030012D19 gene |
| 4 | gene | 117.44173 | 117.45504 | positive | MGI_C57BL6J_3650145 | Gm12828 | NCBI_Gene:108168973,ENSEMBL:ENSMUSG00000086876 | MGI:3650145 | lncRNA gene | predicted gene 12828 |
| 4 | gene | 117.49593 | 117.50932 | positive | MGI_C57BL6J_3651358 | Gm12827 | NCBI_Gene:102637578,ENSEMBL:ENSMUSG00000087228 | MGI:3651358 | lncRNA gene | predicted gene 12827 |
| 4 | gene | 117.50936 | 117.51960 | positive | MGI_C57BL6J_5593828 | Gm34669 | NCBI_Gene:102637993 | MGI:5593828 | lncRNA gene | predicted gene, 34669 |
| 4 | pseudogene | 117.51382 | 117.51420 | negative | MGI_C57BL6J_3650150 | Gm12826 | ENSEMBL:ENSMUSG00000083446 | MGI:3650150 | pseudogene | predicted gene 12826 |
| 4 | gene | 117.55029 | 117.67430 | positive | MGI_C57BL6J_2153887 | Eri3 | NCBI_Gene:140546,ENSEMBL:ENSMUSG00000033423 | MGI:2153887 | protein coding gene | exoribonuclease 3 |
| 4 | gene | 117.57013 | 117.57021 | positive | MGI_C57BL6J_5455336 | Gm25559 | ENSEMBL:ENSMUSG00000087807 | MGI:5455336 | miRNA gene | predicted gene, 25559 |
| 4 | pseudogene | 117.59884 | 117.59939 | positive | MGI_C57BL6J_5011377 | Gm19192 | NCBI_Gene:100418411 | MGI:5011377 | pseudogene | predicted gene, 19192 |
| 4 | gene | 117.67468 | 117.68232 | negative | MGI_C57BL6J_1913483 | Dmap1 | NCBI_Gene:66233,ENSEMBL:ENSMUSG00000009640 | MGI:1913483 | protein coding gene | DNA methyltransferase 1-associated protein 1 |
R version 3.6.2 (2019-12-12)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS Catalina 10.15.7
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRlapack.dylib
locale:
[1] en_AU.UTF-8/en_AU.UTF-8/en_AU.UTF-8/C/en_AU.UTF-8/en_AU.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] abind_1.4-5 qtl2_0.22 reshape2_1.4.4 ggplot2_3.3.5
[5] tibble_3.1.2 psych_2.0.7 readxl_1.3.1 cluster_2.1.0
[9] dplyr_0.8.5 optparse_1.6.6 rhdf5_2.28.1 mclust_5.4.6
[13] tidyr_1.0.2 data.table_1.14.0 knitr_1.33 kableExtra_1.1.0
[17] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] httr_1.4.1 bit64_4.0.5 viridisLite_0.4.0 assertthat_0.2.1
[5] highr_0.9 blob_1.2.1 cellranger_1.1.0 yaml_2.2.1
[9] pillar_1.6.1 RSQLite_2.2.7 backports_1.2.1 lattice_0.20-38
[13] glue_1.4.2 digest_0.6.27 promises_1.1.0 rvest_0.3.5
[17] colorspace_2.0-2 htmltools_0.5.1.1 httpuv_1.5.2 plyr_1.8.6
[21] pkgconfig_2.0.3 purrr_0.3.4 scales_1.1.1 webshot_0.5.2
[25] whisker_0.4 getopt_1.20.3 later_1.0.0 git2r_0.26.1
[29] ellipsis_0.3.2 cachem_1.0.5 withr_2.4.2 mnormt_1.5-7
[33] magrittr_2.0.1 crayon_1.4.1 memoise_2.0.0 evaluate_0.14
[37] fs_1.4.1 fansi_0.5.0 nlme_3.1-142 xml2_1.3.1
[41] tools_3.6.2 hms_0.5.3 lifecycle_1.0.0 stringr_1.4.0
[45] Rhdf5lib_1.6.3 munsell_0.5.0 compiler_3.6.2 rlang_0.4.11
[49] grid_3.6.2 rstudioapi_0.13 rmarkdown_2.1 gtable_0.3.0
[53] DBI_1.1.1 R6_2.5.0 fastmap_1.1.0 bit_4.0.4
[57] utf8_1.2.1 rprojroot_1.3-2 readr_1.3.1 stringi_1.7.2
[61] parallel_3.6.2 Rcpp_1.0.7 vctrs_0.3.8 tidyselect_1.0.0
[65] xfun_0.24