QTL Analysis - binary [ICI vs EOI]
Belinda Cornes
2022-02-11
Last updated: 2022-02-11
Checks: 6 1
Knit directory: Serreze-T1D_Workflow/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20220210) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /Users/corneb/Documents/MyJax/CS/Projects/Serreze/qc/workflowr/Serreze-T1D_Workflow | . |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version fc17722. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Untracked files:
Untracked: data/GM_covar.csv
Untracked: data/bad_markers_all_4.batches.RData
Untracked: data/covar_cleaned_ici.vs.eoi.csv
Untracked: data/covar_cleaned_ici.vs.pbs.csv
Untracked: data/e.RData
Untracked: data/e_snpg_samqc_4.batches.RData
Untracked: data/e_snpg_samqc_4.batches_bc.RData
Untracked: data/errors_ind_4.batches.RData
Untracked: data/errors_ind_4.batches_bc.RData
Untracked: data/genetic_map.csv
Untracked: data/genotype_errors_marker_4.batches.RData
Untracked: data/genotype_freq_marker_4.batches.RData
Untracked: data/gm_allqc_4.batches.RData
Untracked: data/gm_samqc_3.batches.RData
Untracked: data/gm_samqc_4.batches.RData
Untracked: data/gm_samqc_4.batches_bc.RData
Untracked: data/gm_serreze.192.RData
Untracked: data/percent_missing_id_3.batches.RData
Untracked: data/percent_missing_id_4.batches.RData
Untracked: data/percent_missing_id_4.batches_bc.RData
Untracked: data/percent_missing_marker_4.batches.RData
Untracked: data/pheno.csv
Untracked: data/physical_map.csv
Untracked: data/qc_info_bad_sample_3.batches.RData
Untracked: data/qc_info_bad_sample_4.batches.RData
Untracked: data/qc_info_bad_sample_4.batches_bc.RData
Untracked: data/sample_geno.csv
Untracked: data/sample_geno_bc.csv
Untracked: data/serreze_probs.rds
Untracked: data/serreze_probs_allqc.rds
Untracked: data/summary.cg_3.batches.RData
Untracked: data/summary.cg_4.batches.RData
Untracked: data/summary.cg_4.batches_bc.RData
Untracked: output/Percent_missing_genotype_data_4.batches.pdf
Untracked: output/Percent_missing_genotype_data_per_marker.pdf
Untracked: output/Proportion_matching_genotypes_before_removal_of_bad_samples_4.batches.pdf
Untracked: output/genotype_error_marker.pdf
Untracked: output/genotype_frequency_marker.pdf
Unstaged changes:
Modified: analysis/2.2.1_snp_qc_4.batches.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/4.1.1_qtl.analysis_binary_ici.vs.eoi_snpsqc.Rmd) and HTML (docs/4.1.1_qtl.analysis_binary_ici.vs.eoi_snpsqc.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | fc17722 | Belinda Cornes | 2022-02-11 | QTL Mapping correcting MAF cutoff |
| html | 1218e08 | Belinda Cornes | 2022-02-11 | Build site. |
| Rmd | 8427b21 | Belinda Cornes | 2022-02-11 | QTL Mapping after snp qc |
Loading Data
We will load the data and subset indivials out that are in the groups of interest. We will create a binary phenotype from this (EOI ==0, ICI == 1).
load("data/gm_allqc_4.batches.RData")
#gm_allqc
gm=gm_allqc
gmObject of class cross2 (crosstype "bc")
Total individuals 188
No. genotyped individuals 188
No. phenotyped individuals 188
No. with both geno & pheno 188
No. phenotypes 1
No. covariates 6
No. phenotype covariates 0
No. chromosomes 20
Total markers 131578
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13
9977 10005 7858 7589 7621 7758 7413 6472 6725 6396 7154 6137 6085
14 15 16 17 18 19 X
5981 5346 5019 5093 4607 3564 4778 #pr <- readRDS("data/serreze_probs_allqc.rds")
#pr <- readRDS("data/serreze_probs.rds")
##extracting animals with ici and eoi group status
miceinfo <- gm$covar[gm$covar$group == "EOI" | gm$covar$group == "ICI",]
table(miceinfo$group)
EOI ICI
69 92 mice.ids <- rownames(miceinfo)
gm <- gm[mice.ids]
gmObject of class cross2 (crosstype "bc")
Total individuals 161
No. genotyped individuals 161
No. phenotyped individuals 161
No. with both geno & pheno 161
No. phenotypes 1
No. covariates 6
No. phenotype covariates 0
No. chromosomes 20
Total markers 131578
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13
9977 10005 7858 7589 7621 7758 7413 6472 6725 6396 7154 6137 6085
14 15 16 17 18 19 X
5981 5346 5019 5093 4607 3564 4778 #pr.qc <- pr
#for (i in 1:20){pr.qc[[i]] = pr.qc[[i]][mice.ids,,]}
#bin_pheno <- NULL
#bin_pheno$EOI <- ifelse(gm$covar$group == "EOI", 1, 0)
#bin_pheno$ICI <- ifelse(gm$covar$group == "ICI", 1, 0)
#bin_pheno <- as.data.frame(bin_pheno)
#rownames(bin_pheno) <- rownames(gm$covar)
gm$covar$ICI.vs.EOI <- ifelse(gm$covar$group == "EOI", 0, 1)
##dropping monomorphic markers within the dataset
g <- do.call("cbind", gm$geno)
#g[1,1:ncol(g)]
gf_mar <- t(apply(g, 2, function(a) table(factor(a, 1:2))/sum(a != 0)))
#gn_mar <- t(apply(g, 2, function(a) table(factor(a, 1:2))))
gf_mar <- gf_mar[gf_mar[,2] != "NaN",]
count <- rowSums(gf_mar <=0.05)
low_freq_df <- merge(as.data.frame(gf_mar),as.data.frame(count), by="row.names",all=T)
low_freq_df[is.na(low_freq_df)] <- ''
low_freq_df <- low_freq_df[low_freq_df$count == 1,]
rownames(low_freq_df) <- low_freq_df$Row.names
low_freq <- find_markerpos(gm, rownames(low_freq_df))
low_freq$id <- rownames(low_freq)
nrow(low_freq)[1] 99077low_freq_bad <- merge(low_freq,low_freq_df, by="row.names",all=T)
names(low_freq_bad)[1] <- c("marker")
gf_mar <- gf_mar[gf_mar[,2] != "NaN",]
MAF <- apply(gf_mar, 1, function(x) min(x))
MAF <- as.data.frame(MAF)
MAF$index <- 1:nrow(gf_mar)
gf_mar_maf <- merge(gf_mar,as.data.frame(MAF), by="row.names")
gf_mar_maf <- gf_mar_maf[order(gf_mar_maf$index),]
gfmar <- NULL
gfmar$gfmar_mar_0 <- sum(gf_mar_maf$MAF==0)
gfmar$gfmar_mar_1 <- sum(gf_mar_maf$MAF< 0.01)
gfmar$gfmar_mar_5 <- sum(gf_mar_maf$MAF< 0.05)
gfmar$gfmar_mar_10 <- sum(gf_mar_maf$MAF< 0.10)
gfmar$gfmar_mar_15 <- sum(gf_mar_maf$MAF< 0.15)
gfmar$gfmar_mar_25 <- sum(gf_mar_maf$MAF< 0.25)
gfmar$gfmar_mar_50 <- sum(gf_mar_maf$MAF< 0.50)
gfmar$total_snps <- nrow(as.data.frame(gf_mar_maf))
gfmar <- t(as.data.frame(gfmar))
gfmar <- as.data.frame(gfmar)
gfmar$count <- gfmar$V1
gfmar[c(2)] %>%
kable(escape = F,align = c("ccccccccc"),linesep ="\\hline") %>%
kable_styling(full_width = F) %>%
kable_styling("striped", full_width = F) %>%
row_spec(8 ,bold=T,color= "white",background = "black")| count | |
|---|---|
| gfmar_mar_0 | 89667 |
| gfmar_mar_1 | 92615 |
| gfmar_mar_5 | 99045 |
| gfmar_mar_10 | 99502 |
| gfmar_mar_15 | 99619 |
| gfmar_mar_25 | 100097 |
| gfmar_mar_50 | 131139 |
| total_snps | 131578 |
gm_qc <- drop_markers(gm, low_freq_bad$marker)
gm_qc <- drop_nullmarkers(gm_qc)
gm = gm_qc
gmObject of class cross2 (crosstype "bc")
Total individuals 161
No. genotyped individuals 161
No. phenotyped individuals 161
No. with both geno & pheno 161
No. phenotypes 1
No. covariates 7
No. phenotype covariates 0
No. chromosomes 20
Total markers 32501
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
2940 2853 2033 2045 1931 2041 1858 1677 1991 1208 2057 1387 1617 1685 1194 922
17 18 19 X
382 1073 1052 555 markers <- marker_names(gm)
gmapdf <- read.csv("/Users/corneb/Documents/MyJax/CS/Projects/Serreze/haplotype.reconstruction/output_hh/genetic_map.csv")
pmapdf <- read.csv("/Users/corneb/Documents/MyJax/CS/Projects/Serreze/haplotype.reconstruction/output_hh/physical_map.csv")
mapdf <- merge(gmapdf,pmapdf, by=c("marker","chr"), all=T)
rownames(mapdf) <- mapdf$marker
mapdf <- mapdf[markers,]
names(mapdf) <- c('marker','chr','gmapdf','pmapdf')
mapdf <- mapdf[!duplicated(mapdf[c(2:3)]),]
pr.qc <- calc_genoprob(gm)Genome-wide scan
For each of the phenotype analyzed, permutations were used for each model to obtain genome-wide LOD significance threshold for p < 0.01, p < 0.05, p < 0.10, respectively, separately for X and automsomes (A).
The table shows the estimated significance thresholds from permutation test.
We also looked at the kinship to see how correlated each sample is. Kinship values between pairs of samples range between 0 (no relationship) and 1.0 (completely identical). The darker the colour the more indentical the pairs are.
Xcovar <- get_x_covar(gm)
#addcovar = model.matrix(~Sex, data = covars)[,-1]
#K <- calc_kinship(pr.qc, type = "loco")
#heatmap(K[[1]])
#K.overall <- calc_kinship(pr.qc, type = "overall")
#heatmap(K.overall)
kinship <- calc_kinship(pr.qc)
heatmap(kinship)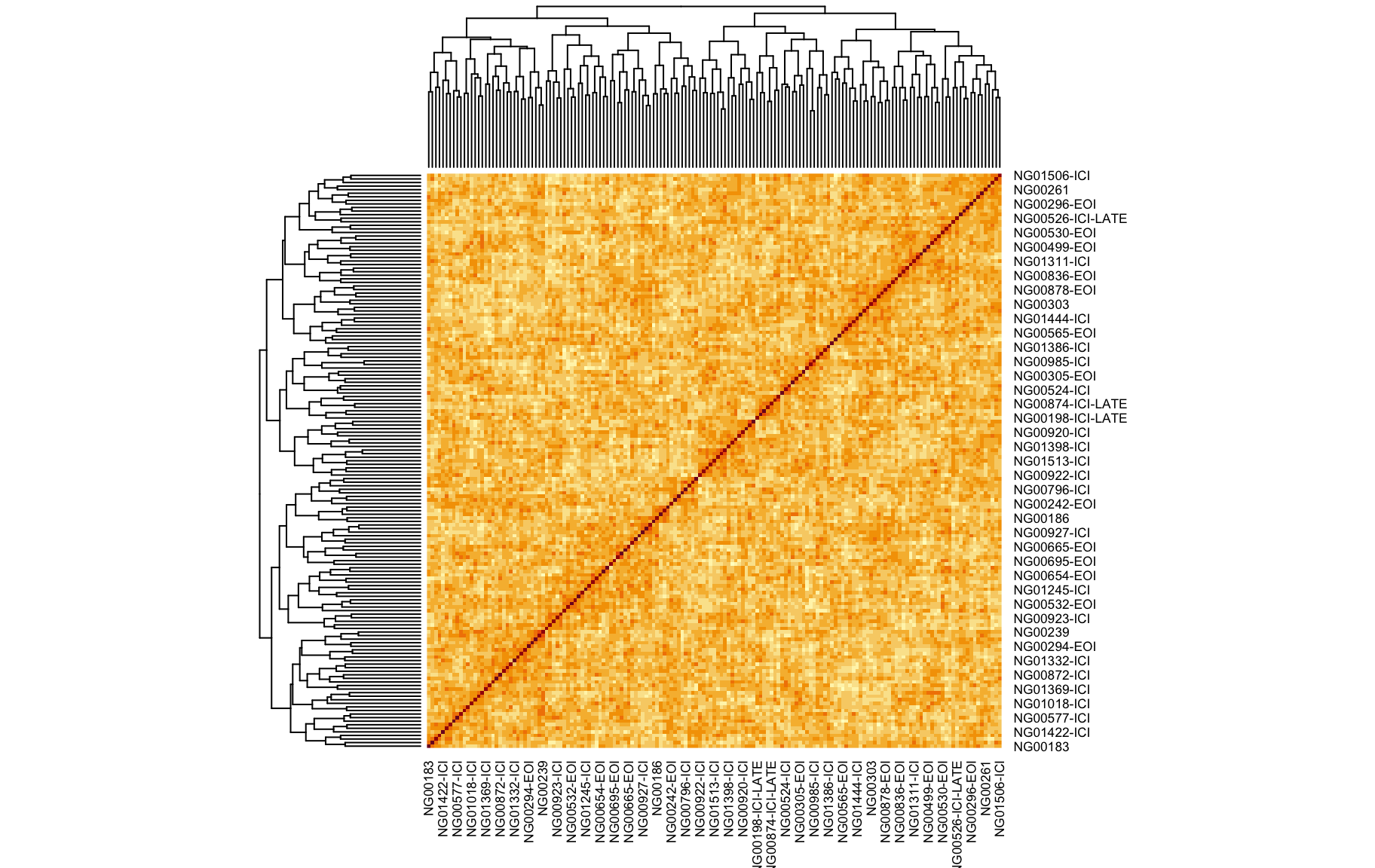
| Version | Author | Date |
|---|---|---|
| 1218e08 | Belinda Cornes | 2022-02-11 |
#operm <- scan1perm(pr.qc, gm$covar$phenos, Xcovar=Xcovar, n_perm=2000)
#operm <- scan1perm(pr.qc, gm$covar$phenos, addcovar = addcovar, n_perm=2000)
#operm <- scan1perm(pr.qc, gm$covar$phenos, n_perm=2000)
operm <- scan1perm(pr.qc, gm$covar["ICI.vs.EOI"], model="binary", n_perm=10, perm_Xsp=TRUE, chr_lengths=chr_lengths(gm$gmap))
summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
names(summary_table) <- c("autosomes","X")
summary_table$significance.level <- rownames(summary_table)
rownames(summary_table) <- NULL
summary_table[c(3,1:2)] %>%
kable(escape = F,align = c("ccc")) %>%
kable_styling("striped", full_width = T) %>%
column_spec(1, bold=TRUE)| significance.level | autosomes | X |
|---|---|---|
| 0.01 | 4.033409 | 2.907967 |
| 0.05 | 3.877426 | 2.847949 |
| 0.1 | 3.681965 | 2.769463 |
The figures below show QTL maps for each phenotype
out <- scan1(pr.qc, gm$covar["ICI.vs.EOI"], Xcovar=Xcovar, model="binary")
summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
plot_lod<-function(out,map){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, gm$gmap, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = colnames(out)[i])
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
}
}
plot_lod(out,gm$gmap)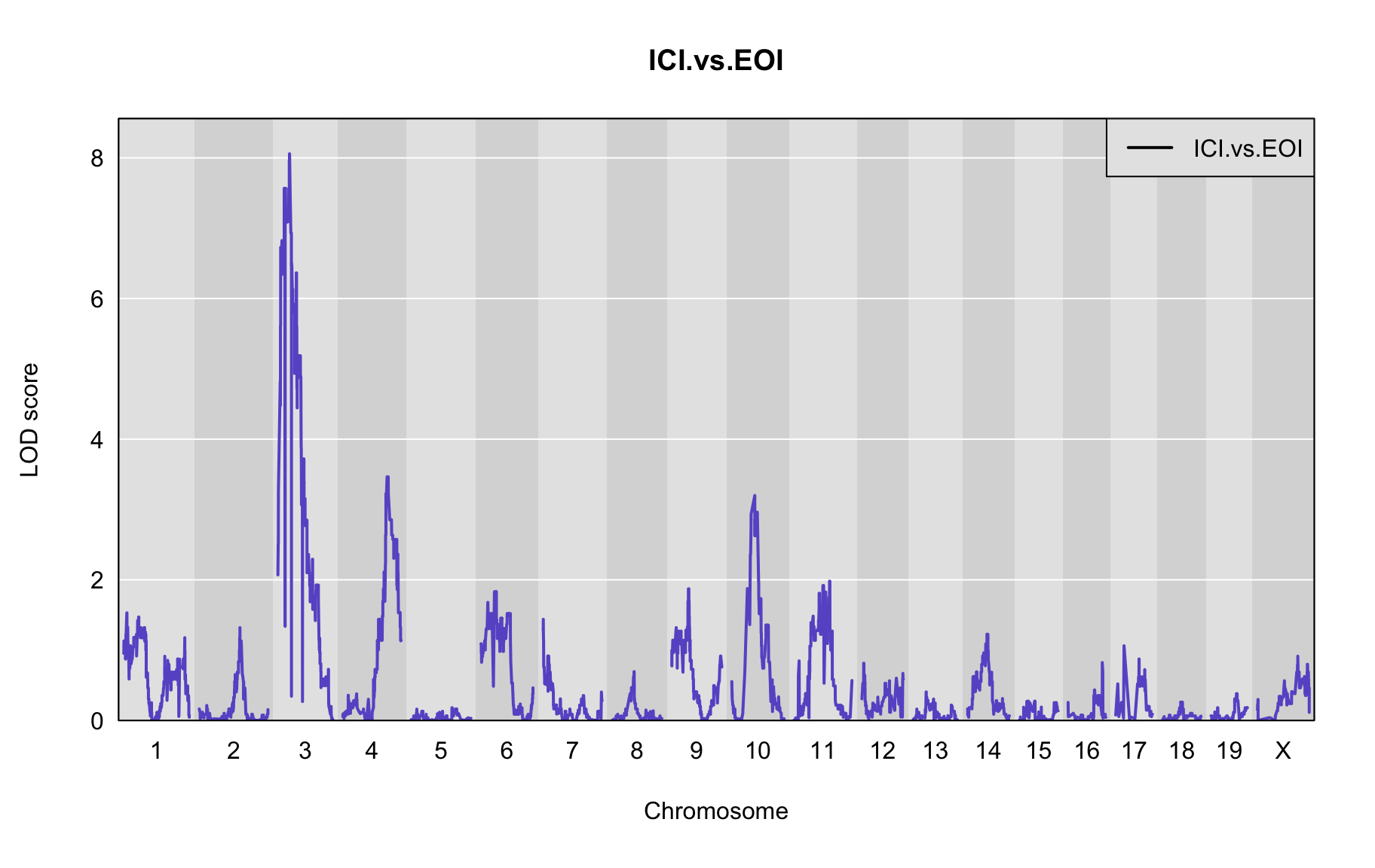
| Version | Author | Date |
|---|---|---|
| 1218e08 | Belinda Cornes | 2022-02-11 |
LOD peaks
The table below shows QTL peaks associated with the phenotype. We use the 95% threshold from the permutations to find peaks.
peaks<-find_peaks(out, gm$gmap, threshold=4, drop=1.5)
rownames(peaks) <- NULL
peaks[] %>%
kable(escape = F,align = c("ccccccc")) %>%
kable_styling("striped", full_width = T) %>%
column_spec(1, bold=TRUE)| lodindex | lodcolumn | chr | pos | lod | ci_lo | ci_hi |
|---|---|---|---|---|---|---|
| 1 | ICI.vs.EOI | 3 | 18.624 | 8.058029 | 5.566 | 21.449 |
peaks_mbl <- list()
#corresponding info in Mb
for(i in 1:nrow(peaks)){
lodindex <- peaks$lodindex[i]
lodcolumn <- peaks$lodcolumn[i]
chr <- peaks$chr[i]
lod <- peaks$lod[i]
pos <- mapdf$pmapdf[which(mapdf$gmapdf == peaks$pos[i] & mapdf$chr == peaks$chr[i])]
ci_lo <- mapdf$pmapdf[which(mapdf$gmapdf == peaks$ci_lo[i] & mapdf$chr == peaks$chr[i])]
ci_hi <- mapdf$pmapdf[which(mapdf$gmapdf == peaks$ci_hi[i] & mapdf$chr == peaks$chr[i])]
peaks_mb=cbind(lodindex, lodcolumn, chr, pos, lod, ci_lo, ci_hi)
peaks_mbl[[i]] <- peaks_mb
}
peaks_mba <- do.call(rbind, peaks_mbl)
peaks_mba <- as.data.frame(peaks_mba)
#peaks_mba[,c("chr", "pos", "lod", "ci_lo", "ci_hi")] <- sapply(peaks_mba[,c("chr", "pos", "lod", "ci_lo", "ci_hi")], as.numeric)
rownames(peaks_mba) <- NULL
peaks_mba[] %>%
kable(escape = F,align = c("ccccccc")) %>%
kable_styling("striped", full_width = T) %>%
column_spec(1, bold=TRUE)| lodindex | lodcolumn | chr | pos | lod | ci_lo | ci_hi |
|---|---|---|---|---|---|---|
| 1 | ICI.vs.EOI | 3 | 38.523543 | 8.05802845906476 | 19.493219 | 48.682283 |
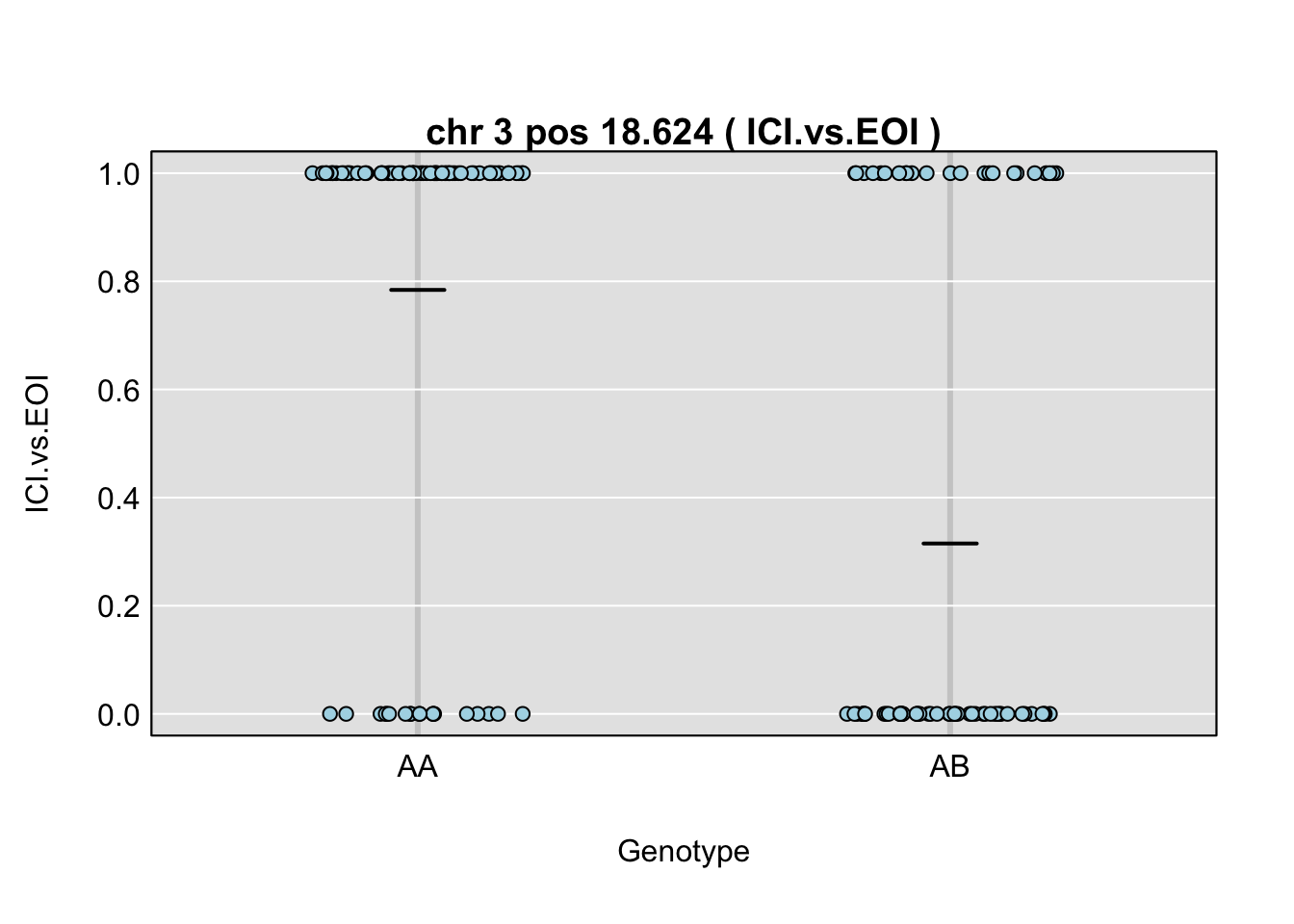
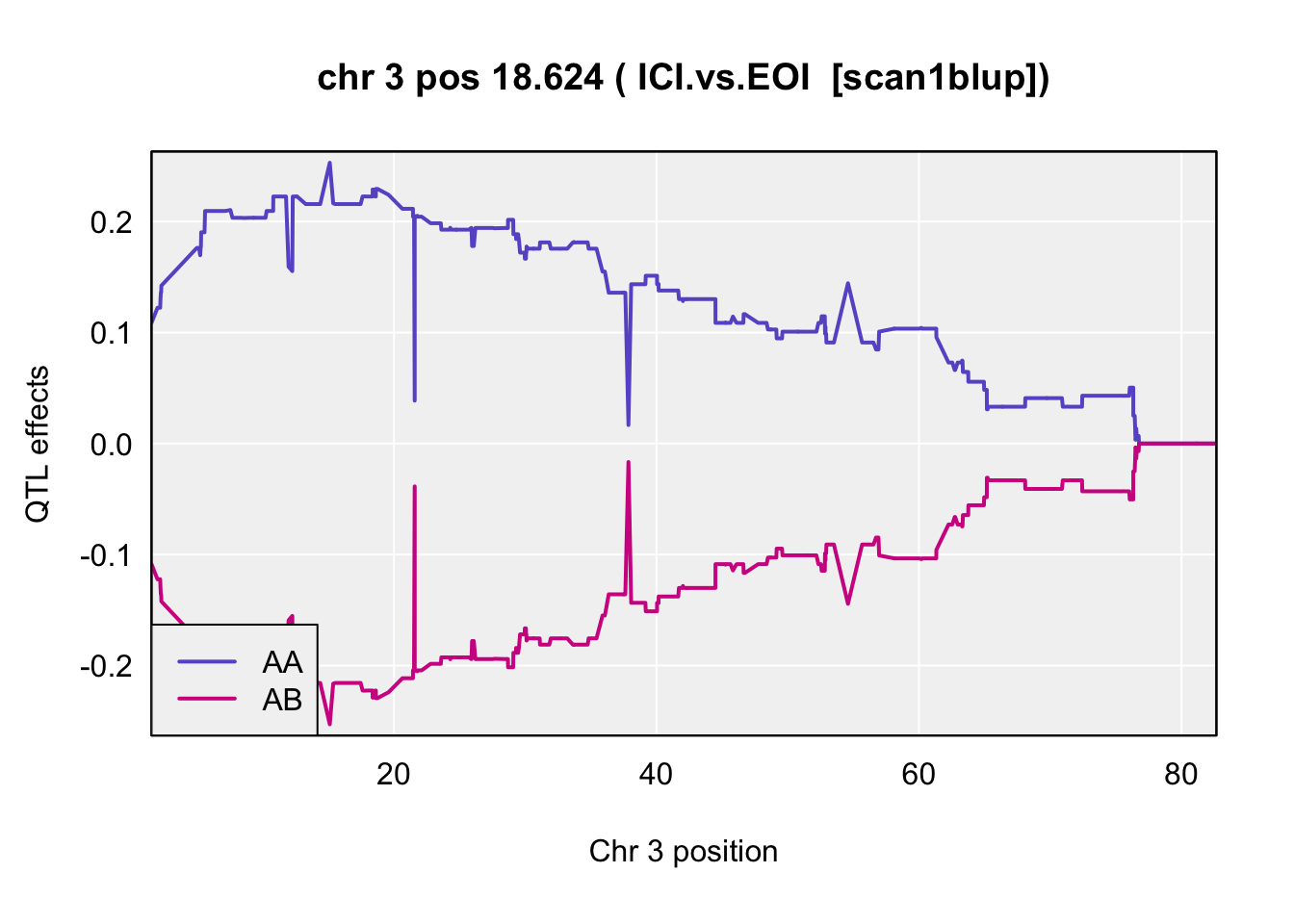
QTL effects
For each peak LOD location we give a list of gene
query_variants <- create_variant_query_func("/Users/corneb/Documents/MyJax/CS/Projects/support.files/qtl2/cc_variants.sqlite")
query_genes <- create_gene_query_func("/Users/corneb/Documents/MyJax/CS/Projects/support.files/qtl2/mouse_genes_mgi.sqlite")
for (i in 1:nrow(peaks)){
#for (i in 1:1){
#Plot 1
marker = find_marker(gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i])
#g <- maxmarg(pr.qc, gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i], return_char=TRUE, minprob = 0.5)
gp <- g[,marker]
gp[gp==1] <- "AA"
gp[gp==2] <- "AB"
gp[gp==0] <- NA
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/","qtl_effect_", i, ".png"))
#par(mar=c(4.1, 4.1, 1.5, 0.6))
plot_pxg(gp, gm$covar[,peaks$lodcolumn[i]], ylab=peaks$lodcolumn[i], sort=FALSE)
title(main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")"), line=0.2)
##dev.off()
chr = peaks$chr[i]
# Plot 2
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar)
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], Xcovar=Xcovar)
coeff <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
blup <- scan1blup(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]])
#plot_coef(coeff,
# gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i]," [scan1coeff])")
# )
plot_coef(blup,
gm$gmap[chr], columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i]," [scan1blup])")
)
#last_coef <- unclass(coeff)[nrow(coeff),1:3]
#for(t in seq(along=last_coef))
#axis(side=4, at=last_coef[t], names(last_coef)[t], tick=FALSE)
# Plot 3
#c2effB <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary", contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
#c2effBb <- scan1blup(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]],Xcovar=Xcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
#plot(c2effB, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
#plot(c2effBb, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
##last_coef <- unclass(c2effB)[nrow(c2effB),2:3] # last two coefficients
##for(t in seq(along=last_coef))
## axis(side=4, at=last_coef[t], names(last_coef)[t], tick=FALSE)
#Table 1
chr = peaks_mba$chr[i]
start=as.numeric(peaks_mba$ci_lo[i])
end=as.numeric(peaks_mba$ci_hi[i])
genesgss = query_genes(chr, start, end)
rownames(genesgss) <- NULL
genesgss$strand_old = genesgss$strand
genesgss$strand[genesgss$strand=="+"] <- "positive"
genesgss$strand[genesgss$strand=="-"] <- "negative"
#genesgss <-
#table <-
#genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")] %>%
#kable(escape = F,align = c("ccccccccccc")) %>%
#kable_styling("striped", full_width = T) #%>%
#cat #%>%
#column_spec(1, bold=TRUE)
#
#print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], escape = F,align = c("ccccccccccc")))
print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], "html") %>% kable_styling("striped", full_width = T))
#table
}
| Version | Author | Date |
|---|---|---|
| 1218e08 | Belinda Cornes | 2022-02-11 |
| Version | Author | Date |
|---|---|---|
| 1218e08 | Belinda Cornes | 2022-02-11 |
| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | gene | 19.50859 | 19.61086 | positive | MGI_C57BL6J_1913576 | Dnajc5b | NCBI_Gene:66326,ENSEMBL:ENSMUSG00000027606 | MGI:1913576 | protein coding gene | DnaJ heat shock protein family (Hsp40) member C5 beta |
| 3 | gene | 19.56025 | 19.56042 | positive | MGI_C57BL6J_5453107 | Gm23330 | ENSEMBL:ENSMUSG00000096211 | MGI:5453107 | snRNA gene | predicted gene, 23330 |
| 3 | pseudogene | 19.58479 | 19.58554 | positive | MGI_C57BL6J_3646012 | Gm6141 | NCBI_Gene:620290,ENSEMBL:ENSMUSG00000084339 | MGI:3646012 | pseudogene | predicted pseudogene 6141 |
| 3 | gene | 19.61037 | 19.62948 | negative | MGI_C57BL6J_1920674 | 1700064H15Rik | NCBI_Gene:102633032,ENSEMBL:ENSMUSG00000082766 | MGI:1920674 | protein coding gene | RIKEN cDNA 1700064H15 gene |
| 3 | gene | 19.62796 | 19.62803 | negative | MGI_C57BL6J_4413716 | n-TRacg3 | NCBI_Gene:102467637 | MGI:4413716 | tRNA gene | nuclear encoded tRNA arginine 3 (anticodon ACG) |
| 3 | gene | 19.62835 | 19.62845 | positive | MGI_C57BL6J_4414074 | n-TYgta1 | NCBI_Gene:102467442 | MGI:4414074 | tRNA gene | nuclear encoded tRNA tyrosine 1 (anticodon GTA) |
| 3 | gene | 19.62878 | 19.62887 | positive | MGI_C57BL6J_4414075 | n-TYgta2 | NCBI_Gene:102467395 | MGI:4414075 | tRNA gene | nuclear encoded tRNA tyrosine 2 (anticodon GTA) |
| 3 | gene | 19.62899 | 19.62907 | positive | MGI_C57BL6J_4413687 | n-TAagc14 | NCBI_Gene:102467233 | MGI:4413687 | tRNA gene | nuclear encoded tRNA alanine 14 (anticodon AGC) |
| 3 | gene | 19.64434 | 19.69242 | positive | MGI_C57BL6J_3036269 | Trim55 | NCBI_Gene:381485,ENSEMBL:ENSMUSG00000060913 | MGI:3036269 | protein coding gene | tripartite motif-containing 55 |
| 3 | gene | 19.68597 | 19.68657 | positive | MGI_C57BL6J_3642300 | BC002189 | ENSEMBL:ENSMUSG00000104465 | MGI:3642300 | lncRNA gene | cDNA sequence BC002189 |
| 3 | gene | 19.69340 | 19.69540 | negative | MGI_C57BL6J_88496 | Crh | NCBI_Gene:12918,ENSEMBL:ENSMUSG00000049796 | MGI:88496 | protein coding gene | corticotropin releasing hormone |
| 3 | gene | 19.69449 | 19.70735 | positive | MGI_C57BL6J_5625074 | Gm42189 | NCBI_Gene:105247005 | MGI:5625074 | lncRNA gene | predicted gene, 42189 |
| 3 | pseudogene | 19.70825 | 19.70932 | negative | MGI_C57BL6J_5011049 | Gm18864 | NCBI_Gene:100417853,ENSEMBL:ENSMUSG00000102261 | MGI:5011049 | pseudogene | predicted gene, 18864 |
| 3 | pseudogene | 19.76883 | 19.76933 | positive | MGI_C57BL6J_3644940 | Gm7442 | NCBI_Gene:665002,ENSEMBL:ENSMUSG00000106182 | MGI:3644940 | pseudogene | predicted gene 7442 |
| 3 | pseudogene | 19.88087 | 19.88175 | negative | MGI_C57BL6J_3780317 | Gm2148 | NCBI_Gene:102633274 | MGI:3780317 | pseudogene | predicted gene 2148 |
| 3 | pseudogene | 19.89487 | 19.90103 | positive | MGI_C57BL6J_1918058 | 4632415L05Rik | NCBI_Gene:70808,ENSEMBL:ENSMUSG00000048106 | MGI:1918058 | pseudogene | RIKEN cDNA 4632415L05 gene |
| 3 | pseudogene | 19.91380 | 19.91436 | positive | MGI_C57BL6J_5610751 | Gm37523 | ENSEMBL:ENSMUSG00000103781 | MGI:5610751 | pseudogene | predicted gene, 37523 |
| 3 | pseudogene | 19.93424 | 19.93539 | positive | MGI_C57BL6J_5009935 | Gm17771 | NCBI_Gene:100046095,ENSEMBL:ENSMUSG00000104031 | MGI:5009935 | pseudogene | predicted gene, 17771 |
| 3 | gene | 19.94515 | 19.95395 | negative | MGI_C57BL6J_5625075 | Gm42190 | NCBI_Gene:105247006 | MGI:5625075 | lncRNA gene | predicted gene, 42190 |
| 3 | gene | 19.95698 | 20.00975 | positive | MGI_C57BL6J_88476 | Cp | NCBI_Gene:12870,ENSEMBL:ENSMUSG00000003617 | MGI:88476 | protein coding gene | ceruloplasmin |
| 3 | gene | 19.99594 | 20.03538 | negative | MGI_C57BL6J_2153839 | Hps3 | NCBI_Gene:12807,ENSEMBL:ENSMUSG00000027615 | MGI:2153839 | protein coding gene | HPS3, biogenesis of lysosomal organelles complex 2 subunit 1 |
| 3 | gene | 20.05657 | 20.05816 | negative | MGI_C57BL6J_2445023 | D830024F11Rik | NA | NA | unclassified gene | RIKEN cDNA D830024F11 gene |
| 3 | gene | 20.05781 | 20.11849 | positive | MGI_C57BL6J_1196437 | Hltf | NCBI_Gene:20585,ENSEMBL:ENSMUSG00000002428 | MGI:1196437 | protein coding gene | helicase-like transcription factor |
| 3 | gene | 20.12208 | 20.15532 | negative | MGI_C57BL6J_1351614 | Gyg | NCBI_Gene:27357,ENSEMBL:ENSMUSG00000019528 | MGI:1351614 | protein coding gene | glycogenin |
| 3 | gene | 20.21562 | 20.24221 | negative | MGI_C57BL6J_88479 | Cpa3 | NCBI_Gene:12873,ENSEMBL:ENSMUSG00000001865 | MGI:88479 | protein coding gene | carboxypeptidase A3, mast cell |
| 3 | gene | 20.22229 | 20.22236 | negative | MGI_C57BL6J_5531279 | Mir7007 | miRBase:MI0022856,NCBI_Gene:102465609,ENSEMBL:ENSMUSG00000099105 | MGI:5531279 | miRNA gene | microRNA 7007 |
| 3 | gene | 20.22229 | 20.22948 | positive | MGI_C57BL6J_5590479 | Gm31320 | NCBI_Gene:102633510 | MGI:5590479 | lncRNA gene | predicted gene, 31320 |
| 3 | pseudogene | 20.22377 | 20.22469 | negative | MGI_C57BL6J_3647720 | Gm7428 | NCBI_Gene:664965 | MGI:3647720 | pseudogene | predicted gene 7428 |
| 3 | gene | 20.24826 | 20.27573 | negative | MGI_C57BL6J_1923953 | Cpb1 | NCBI_Gene:76703,ENSEMBL:ENSMUSG00000011463 | MGI:1923953 | protein coding gene | carboxypeptidase B1 (tissue) |
| 3 | gene | 20.27671 | 20.28605 | negative | MGI_C57BL6J_5826434 | Gm46797 | NCBI_Gene:108168859 | MGI:5826434 | lncRNA gene | predicted gene, 46797 |
| 3 | gene | 20.30841 | 20.30992 | positive | MGI_C57BL6J_5663662 | Gm43525 | ENSEMBL:ENSMUSG00000105208 | MGI:5663662 | lncRNA gene | predicted gene 43525 |
| 3 | gene | 20.31447 | 20.36718 | negative | MGI_C57BL6J_87965 | Agtr1b | NCBI_Gene:11608,ENSEMBL:ENSMUSG00000054988 | MGI:87965 | protein coding gene | angiotensin II receptor, type 1b |
| 3 | pseudogene | 20.39101 | 20.39198 | negative | MGI_C57BL6J_5010611 | Gm18426 | NCBI_Gene:100417153,ENSEMBL:ENSMUSG00000103573 | MGI:5010611 | pseudogene | predicted gene, 18426 |
| 3 | gene | 20.40250 | 20.50621 | negative | MGI_C57BL6J_5590625 | Gm31466 | NCBI_Gene:108168860,ENSEMBL:ENSMUSG00000102636 | MGI:5590625 | lncRNA gene | predicted gene, 31466 |
| 3 | gene | 20.40467 | 20.40491 | positive | MGI_C57BL6J_5662731 | Gm42594 | ENSEMBL:ENSMUSG00000106442 | MGI:5662731 | unclassified gene | predicted gene 42594 |
| 3 | pseudogene | 20.78247 | 20.78407 | negative | MGI_C57BL6J_5010676 | Gm18491 | NCBI_Gene:100417265,ENSEMBL:ENSMUSG00000103460 | MGI:5010676 | pseudogene | predicted gene, 18491 |
| 3 | pseudogene | 20.78794 | 20.78815 | positive | MGI_C57BL6J_5611551 | Gm38323 | ENSEMBL:ENSMUSG00000102986 | MGI:5611551 | pseudogene | predicted gene, 38323 |
| 3 | pseudogene | 20.80448 | 20.80609 | positive | MGI_C57BL6J_5010678 | Gm18493 | NCBI_Gene:100417268,ENSEMBL:ENSMUSG00000103533 | MGI:5010678 | pseudogene | predicted gene, 18493 |
| 3 | gene | 20.81639 | 20.82236 | negative | MGI_C57BL6J_5590711 | Gm31552 | NCBI_Gene:102633820 | MGI:5590711 | lncRNA gene | predicted gene, 31552 |
| 3 | gene | 20.81962 | 20.82369 | positive | MGI_C57BL6J_5610708 | Gm37480 | ENSEMBL:ENSMUSG00000102807 | MGI:5610708 | unclassified gene | predicted gene, 37480 |
| 3 | pseudogene | 21.04945 | 21.05053 | positive | MGI_C57BL6J_3647006 | Gm7488 | NCBI_Gene:665094,ENSEMBL:ENSMUSG00000039617 | MGI:3647006 | pseudogene | predicted gene 7488 |
| 3 | pseudogene | 21.11358 | 21.11400 | negative | MGI_C57BL6J_5663297 | Gm43160 | ENSEMBL:ENSMUSG00000106051 | MGI:5663297 | pseudogene | predicted gene 43160 |
| 3 | gene | 21.26837 | 21.26909 | negative | MGI_C57BL6J_5579843 | Gm29137 | ENSEMBL:ENSMUSG00000101897 | MGI:5579843 | lncRNA gene | predicted gene 29137 |
| 3 | gene | 21.42179 | 21.48966 | negative | MGI_C57BL6J_5590769 | Gm31610 | NCBI_Gene:102633894 | MGI:5590769 | lncRNA gene | predicted gene, 31610 |
| 3 | pseudogene | 21.60381 | 21.60551 | positive | MGI_C57BL6J_5663812 | Gm43675 | ENSEMBL:ENSMUSG00000106085 | MGI:5663812 | pseudogene | predicted gene 43675 |
| 3 | pseudogene | 21.64839 | 21.64918 | positive | MGI_C57BL6J_5010120 | Gm17935 | NCBI_Gene:100416137,ENSEMBL:ENSMUSG00000106460 | MGI:5010120 | pseudogene | predicted gene, 17935 |
| 3 | gene | 21.70282 | 21.71246 | positive | MGI_C57BL6J_5590852 | Gm31693 | NCBI_Gene:102634003,ENSEMBL:ENSMUSG00000105440 | MGI:5590852 | lncRNA gene | predicted gene, 31693 |
| 3 | gene | 21.75353 | 21.80080 | negative | MGI_C57BL6J_2442162 | 7530428D23Rik | NCBI_Gene:319506,ENSEMBL:ENSMUSG00000103441 | MGI:2442162 | lncRNA gene | RIKEN cDNA 7530428D23 gene |
| 3 | gene | 21.86417 | 21.96435 | negative | MGI_C57BL6J_5590972 | Gm31813 | NCBI_Gene:102634161 | MGI:5590972 | lncRNA gene | predicted gene, 31813 |
| 3 | gene | 21.99768 | 21.99847 | negative | MGI_C57BL6J_5663811 | Gm43674 | ENSEMBL:ENSMUSG00000106321 | MGI:5663811 | lncRNA gene | predicted gene 43674 |
| 3 | pseudogene | 22.02120 | 22.02163 | positive | MGI_C57BL6J_5663808 | Gm43671 | ENSEMBL:ENSMUSG00000106527 | MGI:5663808 | pseudogene | predicted gene 43671 |
| 3 | gene | 22.07441 | 22.07614 | negative | MGI_C57BL6J_5663809 | Gm43672 | ENSEMBL:ENSMUSG00000106019 | MGI:5663809 | lncRNA gene | predicted gene 43672 |
| 3 | gene | 22.07441 | 22.07620 | negative | MGI_C57BL6J_1914374 | 2810416G20Rik | NA | NA | protein coding gene | RIKEN cDNA 2810416G20 gene |
| 3 | gene | 22.07659 | 22.21659 | positive | MGI_C57BL6J_2441730 | Tbl1xr1 | NCBI_Gene:81004,ENSEMBL:ENSMUSG00000027630 | MGI:2441730 | protein coding gene | transducin (beta)-like 1X-linked receptor 1 |
| 3 | pseudogene | 22.08040 | 22.08125 | negative | MGI_C57BL6J_3645251 | Gm5842 | NCBI_Gene:545506,ENSEMBL:ENSMUSG00000103293 | MGI:3645251 | pseudogene | predicted gene 5842 |
| 3 | gene | 22.25137 | 22.25166 | positive | MGI_C57BL6J_105104 | Rprl2 | NCBI_Gene:19784,ENSEMBL:ENSMUSG00000087775 | MGI:105104 | RNase P RNA gene | ribonuclease P RNA-like 2 |
| 3 | gene | 22.28508 | 22.29783 | negative | MGI_C57BL6J_5611552 | Gm38324 | ENSEMBL:ENSMUSG00000102987 | MGI:5611552 | lncRNA gene | predicted gene, 38324 |
| 3 | pseudogene | 22.34885 | 22.34961 | negative | MGI_C57BL6J_5610864 | Gm37636 | NCBI_Gene:108168850,ENSEMBL:ENSMUSG00000102866 | MGI:5610864 | pseudogene | predicted gene, 37636 |
| 3 | gene | 22.40992 | 22.41005 | negative | MGI_C57BL6J_5456131 | Gm26354 | ENSEMBL:ENSMUSG00000088846 | MGI:5456131 | snoRNA gene | predicted gene, 26354 |
| 3 | pseudogene | 22.52501 | 22.52624 | positive | MGI_C57BL6J_3780056 | Spin3-ps | NCBI_Gene:675593,ENSEMBL:ENSMUSG00000103801 | MGI:3780056 | pseudogene | spindlin family, member 3, pseudogene |
| 3 | gene | 22.81221 | 22.81246 | positive | MGI_C57BL6J_5611117 | Gm37889 | ENSEMBL:ENSMUSG00000102233 | MGI:5611117 | unclassified gene | predicted gene, 37889 |
| 3 | pseudogene | 23.07242 | 23.07492 | positive | MGI_C57BL6J_5610211 | Gm36983 | ENSEMBL:ENSMUSG00000103275 | MGI:5610211 | pseudogene | predicted gene, 36983 |
| 3 | gene | 23.08598 | 23.08895 | positive | MGI_C57BL6J_5610249 | Gm37021 | ENSEMBL:ENSMUSG00000103607 | MGI:5610249 | lncRNA gene | predicted gene, 37021 |
| 3 | pseudogene | 23.37494 | 23.37532 | negative | MGI_C57BL6J_5610720 | Gm37492 | ENSEMBL:ENSMUSG00000102214 | MGI:5610720 | pseudogene | predicted gene, 37492 |
| 3 | gene | 23.46105 | 23.52140 | positive | MGI_C57BL6J_1924114 | 4930412E21Rik | NCBI_Gene:100039548,ENSEMBL:ENSMUSG00000102203 | MGI:1924114 | lncRNA gene | RIKEN cDNA 4930412E21 gene |
| 3 | gene | 23.79810 | 25.14426 | negative | MGI_C57BL6J_2685867 | Naaladl2 | NCBI_Gene:635702,ENSEMBL:ENSMUSG00000102758 | MGI:2685867 | protein coding gene | N-acetylated alpha-linked acidic dipeptidase-like 2 |
| 3 | pseudogene | 23.93096 | 23.93200 | positive | MGI_C57BL6J_5011009 | Gm18824 | NCBI_Gene:100417784 | MGI:5011009 | pseudogene | predicted gene, 18824 |
| 3 | gene | 23.94002 | 23.94593 | positive | MGI_C57BL6J_5625076 | Gm42191 | NCBI_Gene:105247007 | MGI:5625076 | lncRNA gene | predicted gene, 42191 |
| 3 | gene | 24.16303 | 24.16311 | negative | MGI_C57BL6J_4413931 | n-TLcag1 | NCBI_Gene:102467553 | MGI:4413931 | tRNA gene | nuclear encoded tRNA leucine 1 (anticodon CAG) |
| 3 | pseudogene | 24.33304 | 24.33355 | positive | MGI_C57BL6J_3645137 | Gm7536 | NCBI_Gene:665189,ENSEMBL:ENSMUSG00000057036 | MGI:3645137 | pseudogene | predicted gene 7536 |
| 3 | gene | 24.48671 | 24.48749 | positive | MGI_C57BL6J_1920306 | 2900080J11Rik | NA | NA | unclassified gene | RIKEN cDNA 2900080J11 gene |
| 3 | gene | 24.56207 | 24.56218 | positive | MGI_C57BL6J_5454481 | Gm24704 | ENSEMBL:ENSMUSG00000070227 | MGI:5454481 | snRNA gene | predicted gene, 24704 |
| 3 | gene | 24.71872 | 24.71898 | positive | MGI_C57BL6J_5453839 | Gm24062 | ENSEMBL:ENSMUSG00000088115 | MGI:5453839 | unclassified non-coding RNA gene | predicted gene, 24062 |
| 3 | pseudogene | 25.00742 | 25.00771 | negative | MGI_C57BL6J_5662911 | Gm42774 | ENSEMBL:ENSMUSG00000105009 | MGI:5662911 | pseudogene | predicted gene 42774 |
| 3 | gene | 25.14407 | 25.14426 | positive | MGI_C57BL6J_5610364 | Gm37136 | ENSEMBL:ENSMUSG00000102876 | MGI:5610364 | unclassified gene | predicted gene, 37136 |
| 3 | pseudogene | 25.18480 | 25.18498 | negative | MGI_C57BL6J_5611230 | Gm38002 | ENSEMBL:ENSMUSG00000102855 | MGI:5611230 | pseudogene | predicted gene, 38002 |
| 3 | pseudogene | 25.21076 | 25.21273 | negative | MGI_C57BL6J_3704211 | Gm10259 | NCBI_Gene:100039592,ENSEMBL:ENSMUSG00000069083 | MGI:3704211 | pseudogene | predicted pseudogene 10259 |
| 3 | gene | 25.42621 | 26.33286 | negative | MGI_C57BL6J_2179435 | Nlgn1 | NCBI_Gene:192167,ENSEMBL:ENSMUSG00000063887 | MGI:2179435 | protein coding gene | neuroligin 1 |
| 3 | gene | 25.54602 | 25.54615 | positive | MGI_C57BL6J_5454027 | Gm24250 | ENSEMBL:ENSMUSG00000087945 | MGI:5454027 | rRNA gene | predicted gene, 24250 |
| 3 | gene | 25.64956 | 25.65077 | negative | MGI_C57BL6J_1920377 | 3110039C02Rik | NA | NA | unclassified gene | RIKEN cDNA 3110039C02 gene |
| 3 | gene | 25.69519 | 25.70030 | negative | MGI_C57BL6J_5610801 | Gm37573 | ENSEMBL:ENSMUSG00000102397 | MGI:5610801 | lncRNA gene | predicted gene, 37573 |
| 3 | pseudogene | 25.73123 | 25.73261 | positive | MGI_C57BL6J_3647151 | Gm6197 | NCBI_Gene:621017,ENSEMBL:ENSMUSG00000104295 | MGI:3647151 | pseudogene | predicted gene 6197 |
| 3 | gene | 25.86566 | 25.86752 | negative | MGI_C57BL6J_2443798 | A830010M09Rik | NA | NA | unclassified gene | RIKEN cDNA A830010M09 gene |
| 3 | gene | 26.33115 | 26.33688 | positive | MGI_C57BL6J_3698881 | A830092H15Rik | NCBI_Gene:100379095,ENSEMBL:ENSMUSG00000074664 | MGI:3698881 | lncRNA gene | RIKEN cDNA A830092H15 gene |
| 3 | pseudogene | 26.38044 | 26.38256 | positive | MGI_C57BL6J_3648363 | Gm4855 | NCBI_Gene:229152,ENSEMBL:ENSMUSG00000102274 | MGI:3648363 | pseudogene | predicted gene 4855 |
| 3 | gene | 26.62618 | 26.62631 | negative | MGI_C57BL6J_5455550 | Gm25773 | ENSEMBL:ENSMUSG00000080368 | MGI:5455550 | miRNA gene | predicted gene, 25773 |
| 3 | gene | 26.63762 | 26.98321 | positive | MGI_C57BL6J_1918112 | Spata16 | NCBI_Gene:70862,ENSEMBL:ENSMUSG00000039335 | MGI:1918112 | protein coding gene | spermatogenesis associated 16 |
| 3 | gene | 26.85020 | 26.87914 | negative | MGI_C57BL6J_5591617 | Gm32458 | NCBI_Gene:102635017 | MGI:5591617 | lncRNA gene | predicted gene, 32458 |
| 3 | pseudogene | 27.01276 | 27.01287 | positive | MGI_C57BL6J_5610887 | Gm37659 | ENSEMBL:ENSMUSG00000103712 | MGI:5610887 | pseudogene | predicted gene, 37659 |
| 3 | gene | 27.02642 | 27.02655 | negative | MGI_C57BL6J_5690856 | Gm44464 | ENSEMBL:ENSMUSG00000105670 | MGI:5690856 | miRNA gene | predicted gene, 44464 |
| 3 | pseudogene | 27.04674 | 27.04798 | negative | MGI_C57BL6J_3645515 | Gm7558 | NCBI_Gene:665258,ENSEMBL:ENSMUSG00000102908 | MGI:3645515 | pseudogene | predicted gene 7558 |
| 3 | gene | 27.09722 | 27.15391 | negative | MGI_C57BL6J_95281 | Ect2 | NCBI_Gene:13605,ENSEMBL:ENSMUSG00000027699 | MGI:95281 | protein coding gene | ect2 oncogene |
| 3 | gene | 27.10898 | 27.10909 | positive | MGI_C57BL6J_5454929 | Gm25152 | ENSEMBL:ENSMUSG00000087742 | MGI:5454929 | snoRNA gene | predicted gene, 25152 |
| 3 | gene | 27.15403 | 27.15702 | positive | MGI_C57BL6J_1923907 | 1700125G22Rik | NCBI_Gene:76657,ENSEMBL:ENSMUSG00000097584 | MGI:1923907 | lncRNA gene | RIKEN cDNA 1700125G22 gene |
| 3 | gene | 27.18296 | 27.28461 | positive | MGI_C57BL6J_2443191 | Nceh1 | NCBI_Gene:320024,ENSEMBL:ENSMUSG00000027698 | MGI:2443191 | protein coding gene | neutral cholesterol ester hydrolase 1 |
| 3 | gene | 27.26033 | 27.28446 | positive | MGI_C57BL6J_5625077 | Gm42192 | NCBI_Gene:105247008 | MGI:5625077 | lncRNA gene | predicted gene, 42192 |
| 3 | pseudogene | 27.27806 | 27.27845 | positive | MGI_C57BL6J_5610419 | Gm37191 | ENSEMBL:ENSMUSG00000102186 | MGI:5610419 | pseudogene | predicted gene, 37191 |
| 3 | pseudogene | 27.30841 | 27.30867 | negative | MGI_C57BL6J_5663482 | Gm43345 | ENSEMBL:ENSMUSG00000105806 | MGI:5663482 | pseudogene | predicted gene 43345 |
| 3 | gene | 27.31703 | 27.34243 | positive | MGI_C57BL6J_107414 | Tnfsf10 | NCBI_Gene:22035,ENSEMBL:ENSMUSG00000039304 | MGI:107414 | protein coding gene | tumor necrosis factor (ligand) superfamily, member 10 |
| 3 | gene | 27.37131 | 27.37923 | positive | MGI_C57BL6J_2441906 | Ghsr | NCBI_Gene:208188,ENSEMBL:ENSMUSG00000051136 | MGI:2441906 | protein coding gene | growth hormone secretagogue receptor |
| 3 | gene | 27.41511 | 27.41615 | negative | MGI_C57BL6J_1924904 | C430049A07Rik | NA | NA | unclassified gene | RIKEN cDNA C430049A07 gene |
| 3 | gene | 27.41616 | 27.71311 | negative | MGI_C57BL6J_1919257 | Fndc3b | NCBI_Gene:72007,ENSEMBL:ENSMUSG00000039286 | MGI:1919257 | protein coding gene | fibronectin type III domain containing 3B |
| 3 | gene | 27.44166 | 27.44411 | positive | MGI_C57BL6J_5625078 | Gm42193 | NCBI_Gene:105247009 | MGI:5625078 | lncRNA gene | predicted gene, 42193 |
| 3 | gene | 27.48228 | 27.48424 | positive | MGI_C57BL6J_5591897 | Gm32738 | NCBI_Gene:102635381 | MGI:5591897 | lncRNA gene | predicted gene, 32738 |
| 3 | gene | 27.54664 | 27.54787 | positive | MGI_C57BL6J_5663481 | Gm43344 | ENSEMBL:ENSMUSG00000106625 | MGI:5663481 | unclassified gene | predicted gene 43344 |
| 3 | gene | 27.58490 | 27.58499 | negative | MGI_C57BL6J_4834306 | Mir3092 | miRBase:MI0014085,NCBI_Gene:100526525,ENSEMBL:ENSMUSG00000092695 | MGI:4834306 | miRNA gene | microRNA 3092 |
| 3 | gene | 27.77786 | 27.84092 | positive | MGI_C57BL6J_5625079 | Gm42194 | NCBI_Gene:105247010 | MGI:5625079 | lncRNA gene | predicted gene, 42194 |
| 3 | gene | 27.79788 | 27.81538 | negative | MGI_C57BL6J_5591954 | Gm32795 | NCBI_Gene:102635463 | MGI:5591954 | lncRNA gene | predicted gene, 32795 |
| 3 | gene | 27.86606 | 27.89639 | negative | MGI_C57BL6J_2685410 | Tmem212 | NCBI_Gene:208613,ENSEMBL:ENSMUSG00000043164 | MGI:2685410 | protein coding gene | transmembrane protein 212 |
| 3 | gene | 27.86811 | 27.86827 | positive | MGI_C57BL6J_5455817 | Gm26040 | ENSEMBL:ENSMUSG00000088772 | MGI:5455817 | snoRNA gene | predicted gene, 26040 |
| 3 | gene | 27.89680 | 27.90023 | negative | MGI_C57BL6J_5625080 | Gm42195 | NCBI_Gene:105247011 | MGI:5625080 | lncRNA gene | predicted gene, 42195 |
| 3 | gene | 27.93841 | 28.13336 | positive | MGI_C57BL6J_109585 | Pld1 | NCBI_Gene:18805,ENSEMBL:ENSMUSG00000027695 | MGI:109585 | protein coding gene | phospholipase D1 |
| 3 | gene | 28.17651 | 28.21221 | positive | MGI_C57BL6J_5625081 | Gm42196 | NCBI_Gene:105247012,ENSEMBL:ENSMUSG00000109716 | MGI:5625081 | lncRNA gene | predicted gene, 42196 |
| 3 | pseudogene | 28.24552 | 28.24577 | positive | MGI_C57BL6J_5610326 | Gm37098 | ENSEMBL:ENSMUSG00000103729 | MGI:5610326 | pseudogene | predicted gene, 37098 |
| 3 | gene | 28.26321 | 28.67586 | positive | MGI_C57BL6J_1916264 | Tnik | NCBI_Gene:665113,ENSEMBL:ENSMUSG00000027692 | MGI:1916264 | protein coding gene | TRAF2 and NCK interacting kinase |
| 3 | gene | 28.26928 | 28.26983 | positive | MGI_C57BL6J_2139599 | AA589472 | NA | NA | unclassified gene | expressed sequence AA589472 |
| 3 | gene | 28.32436 | 28.32609 | positive | MGI_C57BL6J_5610765 | Gm37537 | ENSEMBL:ENSMUSG00000103420 | MGI:5610765 | unclassified gene | predicted gene, 37537 |
| 3 | gene | 28.41994 | 28.42002 | positive | MGI_C57BL6J_4950412 | Mir466q | miRBase:MI0018032,NCBI_Gene:100628591,ENSEMBL:ENSMUSG00000092990 | MGI:4950412 | miRNA gene | microRNA 466q |
| 3 | gene | 28.69790 | 28.73136 | positive | MGI_C57BL6J_1095438 | Slc2a2 | NCBI_Gene:20526,ENSEMBL:ENSMUSG00000027690 | MGI:1095438 | protein coding gene | solute carrier family 2 (facilitated glucose transporter), member 2 |
| 3 | gene | 28.70267 | 28.78111 | negative | MGI_C57BL6J_1920861 | 1700112D23Rik | NCBI_Gene:73611,ENSEMBL:ENSMUSG00000091329 | MGI:1920861 | lncRNA gene | RIKEN cDNA 1700112D23 gene |
| 3 | pseudogene | 28.76353 | 28.76539 | negative | MGI_C57BL6J_3648080 | Gm6505 | NCBI_Gene:624446,ENSEMBL:ENSMUSG00000070522 | MGI:3648080 | pseudogene | predicted pseudogene 6505 |
| 3 | gene | 28.78127 | 28.79885 | positive | MGI_C57BL6J_1933735 | Eif5a2 | NCBI_Gene:208691,ENSEMBL:ENSMUSG00000050192 | MGI:1933735 | protein coding gene | eukaryotic translation initiation factor 5A2 |
| 3 | gene | 28.79064 | 28.80542 | negative | MGI_C57BL6J_5592109 | Gm32950 | NCBI_Gene:102635671,ENSEMBL:ENSMUSG00000103827 | MGI:5592109 | lncRNA gene | predicted gene, 32950 |
| 3 | gene | 28.80544 | 28.80742 | positive | MGI_C57BL6J_1915278 | Rpl22l1 | NCBI_Gene:68028,ENSEMBL:ENSMUSG00000039221 | MGI:1915278 | protein coding gene | ribosomal protein L22 like 1 |
| 3 | gene | 28.80720 | 28.81618 | negative | MGI_C57BL6J_5592210 | Gm33051 | NCBI_Gene:102635804,ENSEMBL:ENSMUSG00000104444 | MGI:5592210 | lncRNA gene | predicted gene, 33051 |
| 3 | gene | 28.85203 | 28.85426 | negative | MGI_C57BL6J_5611095 | Gm37867 | ENSEMBL:ENSMUSG00000104275 | MGI:5611095 | lncRNA gene | predicted gene, 37867 |
| 3 | gene | 28.87981 | 28.88346 | positive | MGI_C57BL6J_5611152 | Gm37924 | ENSEMBL:ENSMUSG00000102482 | MGI:5611152 | unclassified gene | predicted gene, 37924 |
| 3 | gene | 28.88346 | 28.89728 | negative | MGI_C57BL6J_5592261 | Gm33102 | NCBI_Gene:102635876 | MGI:5592261 | lncRNA gene | predicted gene, 33102 |
| 3 | gene | 28.89262 | 28.92672 | positive | MGI_C57BL6J_2686373 | Gm1527 | NCBI_Gene:385263,ENSEMBL:ENSMUSG00000074655 | MGI:2686373 | protein coding gene | predicted gene 1527 |
| 3 | gene | 29.04377 | 29.04555 | negative | MGI_C57BL6J_5592313 | Gm33154 | NCBI_Gene:102635945 | MGI:5592313 | lncRNA gene | predicted gene, 33154 |
| 3 | gene | 29.08072 | 29.69121 | positive | MGI_C57BL6J_1922990 | Egfem1 | NCBI_Gene:75740,ENSEMBL:ENSMUSG00000063600 | MGI:1922990 | protein coding gene | EGF-like and EMI domain containing 1 |
| 3 | gene | 29.17484 | 29.17693 | positive | MGI_C57BL6J_5611257 | Gm38029 | ENSEMBL:ENSMUSG00000103992 | MGI:5611257 | unclassified gene | predicted gene, 38029 |
| 3 | gene | 29.20934 | 29.21198 | positive | MGI_C57BL6J_2442122 | D230021J17Rik | NA | NA | unclassified gene | RIKEN cDNA D230021J17 gene |
| 3 | gene | 29.21989 | 29.22125 | positive | MGI_C57BL6J_5611193 | Gm37965 | ENSEMBL:ENSMUSG00000102579 | MGI:5611193 | unclassified gene | predicted gene, 37965 |
| 3 | gene | 29.26628 | 29.26684 | negative | MGI_C57BL6J_2139582 | AA516838 | NA | NA | unclassified gene | expressed sequence AA516838 |
| 3 | gene | 29.41682 | 29.41692 | positive | MGI_C57BL6J_3691605 | Mir551b | miRBase:MI0004131,NCBI_Gene:791072,ENSEMBL:ENSMUSG00000076120 | MGI:3691605 | miRNA gene | microRNA 551b |
| 3 | gene | 29.63674 | 29.63685 | positive | MGI_C57BL6J_5531333 | Mir21b | miRBase:MI0021906,NCBI_Gene:102466637,ENSEMBL:ENSMUSG00000098315 | MGI:5531333 | miRNA gene | microRNA 21b |
| 3 | gene | 29.66251 | 29.69656 | negative | MGI_C57BL6J_5625082 | Gm42197 | NCBI_Gene:105247013 | MGI:5625082 | lncRNA gene | predicted gene, 42197 |
| 3 | gene | 29.76404 | 29.89323 | negative | MGI_C57BL6J_5592365 | Gm33206 | NCBI_Gene:102636014,ENSEMBL:ENSMUSG00000102574 | MGI:5592365 | lncRNA gene | predicted gene, 33206 |
| 3 | pseudogene | 29.77128 | 29.77142 | positive | MGI_C57BL6J_5610785 | Gm37557 | ENSEMBL:ENSMUSG00000103095 | MGI:5610785 | pseudogene | predicted gene, 37557 |
| 3 | gene | 29.89101 | 29.92419 | positive | MGI_C57BL6J_5564803 | Mannr | NCBI_Gene:102636083,ENSEMBL:ENSMUSG00000102590 | MGI:5564803 | lncRNA gene | Mecom adjacent non-protein coding RNA |
| 3 | pseudogene | 29.92654 | 29.92697 | positive | MGI_C57BL6J_5610509 | Gm37281 | ENSEMBL:ENSMUSG00000102884 | MGI:5610509 | pseudogene | predicted gene, 37281 |
| 3 | gene | 29.95130 | 30.54801 | negative | MGI_C57BL6J_95457 | Mecom | NCBI_Gene:14013,ENSEMBL:ENSMUSG00000027684 | MGI:95457 | protein coding gene | MDS1 and EVI1 complex locus |
| 3 | gene | 30.01238 | 30.05481 | positive | MGI_C57BL6J_4439646 | Mecomos | ENSEMBL:ENSMUSG00000085702 | MGI:4439646 | antisense lncRNA gene | MDS1 and EVI1 complex locus, opposite strand |
| 3 | gene | 30.16025 | 30.16189 | negative | MGI_C57BL6J_5611294 | Gm38066 | ENSEMBL:ENSMUSG00000104486 | MGI:5611294 | unclassified gene | predicted gene, 38066 |
| 3 | gene | 30.22557 | 30.22769 | negative | MGI_C57BL6J_5611425 | Gm38197 | ENSEMBL:ENSMUSG00000102460 | MGI:5611425 | unclassified gene | predicted gene, 38197 |
| 3 | gene | 30.26684 | 30.28019 | negative | MGI_C57BL6J_3648282 | Gm10258 | NCBI_Gene:545510,ENSEMBL:ENSMUSG00000069074 | MGI:3648282 | unclassified non-coding RNA gene | predicted gene 10258 |
| 3 | gene | 30.27232 | 30.27438 | negative | MGI_C57BL6J_5611590 | Gm38362 | ENSEMBL:ENSMUSG00000103870 | MGI:5611590 | unclassified gene | predicted gene, 38362 |
| 3 | gene | 30.34998 | 30.35132 | negative | MGI_C57BL6J_1924752 | 9530010C24Rik | NA | NA | unclassified gene | RIKEN cDNA 9530010C24 gene |
| 3 | gene | 30.48843 | 30.48929 | positive | MGI_C57BL6J_5610252 | Gm37024 | ENSEMBL:ENSMUSG00000104079 | MGI:5610252 | unclassified gene | predicted gene, 37024 |
| 3 | gene | 30.50172 | 30.50381 | negative | MGI_C57BL6J_5610880 | Gm37652 | ENSEMBL:ENSMUSG00000103738 | MGI:5610880 | unclassified gene | predicted gene, 37652 |
| 3 | gene | 30.55270 | 30.56424 | negative | MGI_C57BL6J_5611486 | Gm38258 | ENSEMBL:ENSMUSG00000102256 | MGI:5611486 | lncRNA gene | predicted gene, 38258 |
| 3 | pseudogene | 30.56166 | 30.56213 | negative | MGI_C57BL6J_5611381 | Gm38153 | ENSEMBL:ENSMUSG00000104243 | MGI:5611381 | pseudogene | predicted gene, 38153 |
| 3 | gene | 30.59484 | 30.59576 | negative | MGI_C57BL6J_1920499 | 1600017P15Rik | ENSEMBL:ENSMUSG00000102505,ENSEMBL:ENSMUSG00000065361 | MGI:1920499 | unclassified gene | RIKEN cDNA 1600017P15 gene |
| 3 | gene | 30.59707 | 30.59994 | negative | MGI_C57BL6J_1923902 | Actrt3 | NCBI_Gene:76652,ENSEMBL:ENSMUSG00000037737 | MGI:1923902 | protein coding gene | actin related protein T3 |
| 3 | gene | 30.60127 | 30.60134 | positive | MGI_C57BL6J_4414091 | n-TVaac8 | NCBI_Gene:102467402 | MGI:4414091 | tRNA gene | nuclear encoded tRNA valine 8 (anticodon AAC) |
| 3 | gene | 30.60206 | 30.61987 | positive | MGI_C57BL6J_1931415 | Mynn | NCBI_Gene:80732,ENSEMBL:ENSMUSG00000037730 | MGI:1931415 | protein coding gene | myoneurin |
| 3 | gene | 30.62427 | 30.64787 | negative | MGI_C57BL6J_1919077 | Lrrc34 | NCBI_Gene:71827,ENSEMBL:ENSMUSG00000027702 | MGI:1919077 | protein coding gene | leucine rich repeat containing 34 |
| 3 | gene | 30.64451 | 30.67243 | positive | MGI_C57BL6J_1915557 | Lrriq4 | NCBI_Gene:68307,ENSEMBL:ENSMUSG00000027703 | MGI:1915557 | protein coding gene | leucine-rich repeats and IQ motif containing 4 |
| 3 | gene | 30.67855 | 30.69984 | negative | MGI_C57BL6J_2443864 | Lrrc31 | NCBI_Gene:320352,ENSEMBL:ENSMUSG00000074653 | MGI:2443864 | protein coding gene | leucine rich repeat containing 31 |
| 3 | gene | 30.68594 | 30.69301 | positive | MGI_C57BL6J_3705271 | Gm15462 | NCBI_Gene:108168861,ENSEMBL:ENSMUSG00000086189 | MGI:3705271 | lncRNA gene | predicted gene 15462 |
| 3 | gene | 30.74629 | 30.76717 | positive | MGI_C57BL6J_1923203 | Samd7 | NCBI_Gene:75953,ENSEMBL:ENSMUSG00000051860 | MGI:1923203 | protein coding gene | sterile alpha motif domain containing 7 |
| 3 | gene | 30.75231 | 30.75665 | negative | MGI_C57BL6J_5592749 | Gm33590 | NCBI_Gene:102636556 | MGI:5592749 | lncRNA gene | predicted gene, 33590 |
| 3 | gene | 30.79257 | 30.79356 | negative | MGI_C57BL6J_1918530 | 4933429H19Rik | ENSEMBL:ENSMUSG00000097786 | MGI:1918530 | lncRNA gene | RIKEN cDNA 4933429H19 gene |
| 3 | gene | 30.79276 | 30.82126 | positive | MGI_C57BL6J_1916526 | Sec62 | NCBI_Gene:69276,ENSEMBL:ENSMUSG00000027706 | MGI:1916526 | protein coding gene | SEC62 homolog (S. cerevisiae) |
| 3 | pseudogene | 30.82857 | 30.82883 | negative | MGI_C57BL6J_5611308 | Gm38080 | ENSEMBL:ENSMUSG00000104361 | MGI:5611308 | pseudogene | predicted gene, 38080 |
| 3 | gene | 30.84154 | 30.85199 | negative | MGI_C57BL6J_5625083 | Gm42198 | NCBI_Gene:105247015 | MGI:5625083 | lncRNA gene | predicted gene, 42198 |
| 3 | gene | 30.85248 | 30.85586 | negative | MGI_C57BL6J_5625084 | Gm42199 | NCBI_Gene:105247016 | MGI:5625084 | lncRNA gene | predicted gene, 42199 |
| 3 | gene | 30.85587 | 30.89719 | positive | MGI_C57BL6J_1919112 | Gpr160 | NCBI_Gene:71862,ENSEMBL:ENSMUSG00000037661 | MGI:1919112 | protein coding gene | G protein-coupled receptor 160 |
| 3 | gene | 30.89929 | 30.96948 | negative | MGI_C57BL6J_2181434 | Phc3 | NCBI_Gene:241915,ENSEMBL:ENSMUSG00000037652 | MGI:2181434 | protein coding gene | polyhomeotic 3 |
| 3 | gene | 30.95733 | 30.95764 | negative | MGI_C57BL6J_1924590 | 9530022L04Rik | ENSEMBL:ENSMUSG00000102369 | MGI:1924590 | unclassified gene | RIKEN cDNA 9530022L04 gene |
| 3 | pseudogene | 30.97291 | 30.97511 | positive | MGI_C57BL6J_3781157 | Gm2979 | NCBI_Gene:100040809,ENSEMBL:ENSMUSG00000102741 | MGI:3781157 | pseudogene | predicted gene 2979 |
| 3 | gene | 30.99574 | 31.05296 | positive | MGI_C57BL6J_99260 | Prkci | NCBI_Gene:18759,ENSEMBL:ENSMUSG00000037643 | MGI:99260 | protein coding gene | protein kinase C, iota |
| 3 | gene | 31.03897 | 31.03903 | positive | MGI_C57BL6J_5531134 | Mir7008 | miRBase:MI0022857,NCBI_Gene:102465610,ENSEMBL:ENSMUSG00000098890 | MGI:5531134 | miRNA gene | microRNA 7008 |
| 3 | gene | 31.06130 | 31.06669 | positive | MGI_C57BL6J_5592800 | Gm33641 | NCBI_Gene:102636630 | MGI:5592800 | lncRNA gene | predicted gene, 33641 |
| 3 | gene | 31.06243 | 31.07994 | negative | MGI_C57BL6J_5592853 | Gm33694 | NCBI_Gene:102636694 | MGI:5592853 | lncRNA gene | predicted gene, 33694 |
| 3 | gene | 31.09480 | 31.12292 | positive | MGI_C57BL6J_106203 | Skil | NCBI_Gene:20482,ENSEMBL:ENSMUSG00000027660 | MGI:106203 | protein coding gene | SKI-like |
| 3 | gene | 31.14992 | 31.16433 | positive | MGI_C57BL6J_106925 | Cldn11 | NCBI_Gene:18417,ENSEMBL:ENSMUSG00000037625 | MGI:106925 | protein coding gene | claudin 11 |
| 3 | gene | 31.20285 | 31.31057 | negative | MGI_C57BL6J_3040688 | Slc7a14 | NCBI_Gene:241919,ENSEMBL:ENSMUSG00000069072 | MGI:3040688 | protein coding gene | solute carrier family 7 (cationic amino acid transporter, y+ system), member 14 |
| 3 | gene | 31.23262 | 31.23736 | positive | MGI_C57BL6J_5592993 | Gm33834 | NCBI_Gene:102636884 | MGI:5592993 | lncRNA gene | predicted gene, 33834 |
| 3 | gene | 31.24614 | 31.31690 | positive | MGI_C57BL6J_3782942 | Gm15496 | NCBI_Gene:102636803,ENSEMBL:ENSMUSG00000085953 | MGI:3782942 | lncRNA gene | predicted gene 15496 |
| 3 | gene | 31.34101 | 31.34528 | positive | MGI_C57BL6J_5611253 | Gm38025 | ENSEMBL:ENSMUSG00000103998 | MGI:5611253 | lncRNA gene | predicted gene, 38025 |
| 3 | pseudogene | 31.64480 | 31.69058 | negative | MGI_C57BL6J_3779607 | Gm6558 | NCBI_Gene:625125,ENSEMBL:ENSMUSG00000103499 | MGI:3779607 | pseudogene | predicted gene 6558 |
| 3 | gene | 31.68642 | 31.69214 | positive | MGI_C57BL6J_5593045 | Gm33886 | NCBI_Gene:102636961 | MGI:5593045 | lncRNA gene | predicted gene, 33886 |
| 3 | gene | 31.89883 | 31.90212 | negative | MGI_C57BL6J_5593190 | Gm34031 | NCBI_Gene:102637147 | MGI:5593190 | lncRNA gene | predicted gene, 34031 |
| 3 | gene | 31.90213 | 32.20018 | positive | MGI_C57BL6J_1919663 | Kcnmb2 | NCBI_Gene:72413,ENSEMBL:ENSMUSG00000037610 | MGI:1919663 | protein coding gene | potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| 3 | gene | 31.95476 | 31.95780 | positive | MGI_C57BL6J_5611062 | Gm37834 | ENSEMBL:ENSMUSG00000103539 | MGI:5611062 | unclassified gene | predicted gene, 37834 |
| 3 | gene | 32.23753 | 32.32631 | negative | MGI_C57BL6J_5625086 | Gm42201 | NCBI_Gene:105247019 | MGI:5625086 | lncRNA gene | predicted gene, 42201 |
| 3 | pseudogene | 32.26033 | 32.26111 | positive | MGI_C57BL6J_5610998 | Gm37770 | NCBI_Gene:108168862,ENSEMBL:ENSMUSG00000103643 | MGI:5610998 | pseudogene | predicted gene, 37770 |
| 3 | gene | 32.33479 | 32.36601 | negative | MGI_C57BL6J_1195270 | Zmat3 | NCBI_Gene:22401,ENSEMBL:ENSMUSG00000027663 | MGI:1195270 | protein coding gene | zinc finger matrin type 3 |
| 3 | gene | 32.36549 | 32.36744 | positive | MGI_C57BL6J_1914826 | 4930429B21Rik | NCBI_Gene:67576 | MGI:1914826 | lncRNA gene | RIKEN cDNA 4930429B21 gene |
| 3 | gene | 32.37158 | 32.37299 | negative | MGI_C57BL6J_5610218 | Gm36990 | ENSEMBL:ENSMUSG00000102825 | MGI:5610218 | unclassified gene | predicted gene, 36990 |
| 3 | gene | 32.37898 | 32.38470 | negative | MGI_C57BL6J_5625087 | Gm42202 | NCBI_Gene:105247020 | MGI:5625087 | lncRNA gene | predicted gene, 42202 |
| 3 | gene | 32.39707 | 32.46849 | positive | MGI_C57BL6J_1206581 | Pik3ca | NCBI_Gene:18706,ENSEMBL:ENSMUSG00000027665 | MGI:1206581 | protein coding gene | phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| 3 | gene | 32.45459 | 32.45576 | positive | MGI_C57BL6J_5610351 | Gm37123 | ENSEMBL:ENSMUSG00000103039 | MGI:5610351 | unclassified gene | predicted gene, 37123 |
| 3 | gene | 32.46778 | 32.50290 | negative | MGI_C57BL6J_3612244 | Kcnmb3 | NCBI_Gene:100502876,ENSEMBL:ENSMUSG00000091091 | MGI:3612244 | protein coding gene | potassium large conductance calcium-activated channel, subfamily M, beta member 3 |
| 3 | gene | 32.51026 | 32.52083 | positive | MGI_C57BL6J_1915028 | Zfp639 | NCBI_Gene:67778,ENSEMBL:ENSMUSG00000027667 | MGI:1915028 | protein coding gene | zinc finger protein 639 |
| 3 | gene | 32.51180 | 32.51234 | negative | MGI_C57BL6J_5610820 | Gm37592 | ENSEMBL:ENSMUSG00000104231 | MGI:5610820 | lncRNA gene | predicted gene, 37592 |
| 3 | gene | 32.52946 | 32.57924 | positive | MGI_C57BL6J_1914664 | Mfn1 | NCBI_Gene:67414,ENSEMBL:ENSMUSG00000027668 | MGI:1914664 | protein coding gene | mitofusin 1 |
| 3 | gene | 32.58033 | 32.61659 | negative | MGI_C57BL6J_104581 | Gnb4 | NCBI_Gene:14696,ENSEMBL:ENSMUSG00000027669 | MGI:104581 | protein coding gene | guanine nucleotide binding protein (G protein), beta 4 |
| 3 | gene | 32.61086 | 32.61351 | positive | MGI_C57BL6J_5610868 | Gm37640 | ENSEMBL:ENSMUSG00000102862 | MGI:5610868 | lncRNA gene | predicted gene, 37640 |
| 3 | gene | 32.70630 | 32.72697 | positive | MGI_C57BL6J_1861453 | Actl6a | NCBI_Gene:56456,ENSEMBL:ENSMUSG00000027671 | MGI:1861453 | protein coding gene | actin-like 6A |
| 3 | gene | 32.72178 | 32.72209 | negative | MGI_C57BL6J_5610581 | Gm37353 | ENSEMBL:ENSMUSG00000103590 | MGI:5610581 | unclassified gene | predicted gene, 37353 |
| 3 | gene | 32.72540 | 32.73761 | negative | MGI_C57BL6J_1921850 | Mrpl47 | NCBI_Gene:74600,ENSEMBL:ENSMUSG00000037531 | MGI:1921850 | protein coding gene | mitochondrial ribosomal protein L47 |
| 3 | gene | 32.73610 | 32.73761 | negative | MGI_C57BL6J_3704212 | Gm9859 | NA | NA | unclassified gene | predicted gene 9859 |
| 3 | gene | 32.73699 | 32.75157 | positive | MGI_C57BL6J_1913296 | Ndufb5 | NCBI_Gene:66046,ENSEMBL:ENSMUSG00000027673 | MGI:1913296 | protein coding gene | NADH:ubiquinone oxidoreductase subunit B5 |
| 3 | gene | 32.81702 | 32.93807 | positive | MGI_C57BL6J_1919857 | Usp13 | NCBI_Gene:72607,ENSEMBL:ENSMUSG00000056900 | MGI:1919857 | protein coding gene | ubiquitin specific peptidase 13 (isopeptidase T-3) |
| 3 | pseudogene | 32.88001 | 32.88061 | negative | MGI_C57BL6J_3783022 | Gm15574 | ENSEMBL:ENSMUSG00000081883 | MGI:3783022 | pseudogene | predicted gene 15574 |
| 3 | gene | 32.93931 | 32.98330 | positive | MGI_C57BL6J_5610687 | Gm37459 | ENSEMBL:ENSMUSG00000104283 | MGI:5610687 | lncRNA gene | predicted gene, 37459 |
| 3 | gene | 32.94793 | 33.14327 | negative | MGI_C57BL6J_1916672 | Pex5l | NCBI_Gene:58869,ENSEMBL:ENSMUSG00000027674 | MGI:1916672 | protein coding gene | peroxisomal biogenesis factor 5-like |
| 3 | gene | 33.01196 | 33.01211 | positive | MGI_C57BL6J_5452991 | Gm23214 | ENSEMBL:ENSMUSG00000084665 | MGI:5452991 | snRNA gene | predicted gene, 23214 |
| 3 | gene | 33.02478 | 33.08861 | positive | MGI_C57BL6J_1921124 | 4930419G24Rik | NCBI_Gene:73874,ENSEMBL:ENSMUSG00000087625 | MGI:1921124 | lncRNA gene | RIKEN cDNA 4930419G24 gene |
| 3 | gene | 33.03423 | 33.03705 | negative | MGI_C57BL6J_5610671 | Gm37443 | ENSEMBL:ENSMUSG00000102581 | MGI:5610671 | unclassified gene | predicted gene, 37443 |
| 3 | gene | 33.07392 | 33.07636 | negative | MGI_C57BL6J_5011630 | Gm19445 | ENSEMBL:ENSMUSG00000102498 | MGI:5011630 | unclassified gene | predicted gene, 19445 |
| 3 | pseudogene | 33.19080 | 33.19104 | positive | MGI_C57BL6J_5610356 | Gm37128 | ENSEMBL:ENSMUSG00000103896 | MGI:5610356 | pseudogene | predicted gene, 37128 |
| 3 | gene | 33.26072 | 33.26082 | negative | MGI_C57BL6J_5531356 | Gm27974 | ENSEMBL:ENSMUSG00000098622 | MGI:5531356 | miRNA gene | predicted gene, 27974 |
| 3 | gene | 33.28312 | 33.28565 | positive | MGI_C57BL6J_5610259 | Gm37031 | ENSEMBL:ENSMUSG00000104384 | MGI:5610259 | unclassified gene | predicted gene, 37031 |
| 3 | gene | 33.39306 | 33.39320 | negative | MGI_C57BL6J_5454557 | Gm24780 | ENSEMBL:ENSMUSG00000088260 | MGI:5454557 | snoRNA gene | predicted gene, 24780 |
| 3 | gene | 33.39338 | 33.39485 | negative | MGI_C57BL6J_5610626 | Gm37398 | ENSEMBL:ENSMUSG00000104203 | MGI:5610626 | unclassified gene | predicted gene, 37398 |
| 3 | gene | 33.54027 | 33.54342 | positive | MGI_C57BL6J_5593364 | Gm34205 | NCBI_Gene:102637377 | MGI:5593364 | lncRNA gene | predicted gene, 34205 |
| 3 | pseudogene | 33.62159 | 33.62245 | positive | MGI_C57BL6J_3649050 | Gm5845 | NCBI_Gene:545512,ENSEMBL:ENSMUSG00000103423 | MGI:3649050 | pseudogene | predicted gene 5845 |
| 3 | pseudogene | 33.68451 | 33.68487 | negative | MGI_C57BL6J_5662831 | Gm42694 | ENSEMBL:ENSMUSG00000104954 | MGI:5662831 | pseudogene | predicted gene 42694 |
| 3 | gene | 33.68696 | 33.69200 | negative | MGI_C57BL6J_5625088 | Gm42203 | NCBI_Gene:105247021 | MGI:5625088 | lncRNA gene | predicted gene, 42203 |
| 3 | gene | 33.70336 | 33.75962 | negative | MGI_C57BL6J_5663803 | Gm43666 | ENSEMBL:ENSMUSG00000104588 | MGI:5663803 | lncRNA gene | predicted gene 43666 |
| 3 | pseudogene | 33.73060 | 33.73353 | positive | MGI_C57BL6J_5010796 | Gm18611 | NCBI_Gene:100417436,ENSEMBL:ENSMUSG00000106231 | MGI:5010796 | pseudogene | predicted gene, 18611 |
| 3 | gene | 33.79983 | 33.81486 | positive | MGI_C57BL6J_1914370 | Ttc14 | NCBI_Gene:67120,ENSEMBL:ENSMUSG00000027677 | MGI:1914370 | protein coding gene | tetratricopeptide repeat domain 14 |
| 3 | gene | 33.81076 | 33.84432 | negative | MGI_C57BL6J_1289263 | Ccdc39 | NCBI_Gene:51938,ENSEMBL:ENSMUSG00000027676 | MGI:1289263 | protein coding gene | coiled-coil domain containing 39 |
| 3 | gene | 33.81726 | 33.81795 | positive | MGI_C57BL6J_5663277 | Gm43140 | ENSEMBL:ENSMUSG00000105286 | MGI:5663277 | lncRNA gene | predicted gene 43140 |
| 3 | gene | 33.86681 | 33.89758 | negative | MGI_C57BL6J_1921204 | 4930431L21Rik | ENSEMBL:ENSMUSG00000105475 | MGI:1921204 | lncRNA gene | RIKEN cDNA 4930431L21 gene |
| 3 | gene | 33.90684 | 33.90823 | positive | MGI_C57BL6J_5625089 | Gm42204 | NCBI_Gene:105247022 | MGI:5625089 | lncRNA gene | predicted gene, 42204 |
| 3 | gene | 33.95620 | 33.95715 | positive | MGI_C57BL6J_1922225 | 4930502C17Rik | ENSEMBL:ENSMUSG00000106584 | MGI:1922225 | unclassified gene | RIKEN cDNA 4930502C17 gene |
| 3 | gene | 33.95907 | 33.95920 | negative | MGI_C57BL6J_5452952 | Gm23175 | ENSEMBL:ENSMUSG00000077559 | MGI:5452952 | snoRNA gene | predicted gene, 23175 |
| 3 | gene | 33.97218 | 33.97545 | negative | MGI_C57BL6J_5826470 | Gm46833 | NCBI_Gene:108168913 | MGI:5826470 | lncRNA gene | predicted gene, 46833 |
| 3 | pseudogene | 34.00477 | 34.00551 | negative | MGI_C57BL6J_3642317 | Gm9791 | NCBI_Gene:100040232,ENSEMBL:ENSMUSG00000044434 | MGI:3642317 | pseudogene | predicted pseudogene 9791 |
| 3 | gene | 34.00880 | 34.01188 | negative | MGI_C57BL6J_5826435 | Gm46798 | NCBI_Gene:108168863 | MGI:5826435 | lncRNA gene | predicted gene, 46798 |
| 3 | gene | 34.01771 | 34.01988 | negative | MGI_C57BL6J_5663214 | Gm43077 | ENSEMBL:ENSMUSG00000105535 | MGI:5663214 | lncRNA gene | predicted gene 43077 |
| 3 | gene | 34.01994 | 34.07032 | positive | MGI_C57BL6J_104860 | Fxr1 | NCBI_Gene:14359,ENSEMBL:ENSMUSG00000027680 | MGI:104860 | protein coding gene | fragile X mental retardation gene 1, autosomal homolog |
| 3 | gene | 34.05434 | 34.05543 | negative | MGI_C57BL6J_5663804 | Gm43667 | ENSEMBL:ENSMUSG00000105135 | MGI:5663804 | unclassified gene | predicted gene 43667 |
| 3 | gene | 34.05602 | 34.08135 | negative | MGI_C57BL6J_1914963 | Dnajc19 | NCBI_Gene:67713,ENSEMBL:ENSMUSG00000027679 | MGI:1914963 | protein coding gene | DnaJ heat shock protein family (Hsp40) member C19 |
| 3 | gene | 34.07066 | 34.07600 | positive | MGI_C57BL6J_5663805 | Gm43668 | ENSEMBL:ENSMUSG00000105176 | MGI:5663805 | unclassified gene | predicted gene 43668 |
| 3 | gene | 34.10427 | 34.68262 | positive | MGI_C57BL6J_2444112 | Sox2ot | NCBI_Gene:320478,ENSEMBL:ENSMUSG00000105265 | MGI:2444112 | lncRNA gene | SOX2 overlapping transcript (non-protein coding) |
| 3 | pseudogene | 34.18331 | 34.18419 | positive | MGI_C57BL6J_3647478 | Gm5708 | NCBI_Gene:435727,ENSEMBL:ENSMUSG00000105739 | MGI:3647478 | pseudogene | predicted gene 5708 |
| 3 | gene | 34.28133 | 34.35199 | negative | MGI_C57BL6J_5621390 | Gm38505 | NCBI_Gene:102638008,ENSEMBL:ENSMUSG00000092196 | MGI:5621390 | lncRNA gene | predicted gene, 38505 |
| 3 | gene | 34.41544 | 34.41816 | positive | MGI_C57BL6J_5593758 | Gm34599 | NCBI_Gene:102637903,ENSEMBL:ENSMUSG00000105577 | MGI:5593758 | lncRNA gene | predicted gene, 34599 |
| 3 | gene | 34.42737 | 34.43510 | negative | MGI_C57BL6J_5141980 | Gm20515 | ENSEMBL:ENSMUSG00000092242 | MGI:5141980 | lncRNA gene | predicted gene 20515 |
| 3 | gene | 34.48143 | 34.48221 | negative | MGI_C57BL6J_5579841 | Gm29135 | ENSEMBL:ENSMUSG00000099437 | MGI:5579841 | lncRNA gene | predicted gene 29135 |
| 3 | gene | 34.56034 | 34.56055 | positive | MGI_C57BL6J_5530785 | Gm27403 | ENSEMBL:ENSMUSG00000098795 | MGI:5530785 | unclassified non-coding RNA gene | predicted gene, 27403 |
| 3 | gene | 34.56526 | 34.57220 | negative | MGI_C57BL6J_5662832 | Gm42695 | ENSEMBL:ENSMUSG00000106525 | MGI:5662832 | lncRNA gene | predicted gene 42695 |
| 3 | gene | 34.59782 | 34.60488 | negative | MGI_C57BL6J_5625090 | Gm42205 | NCBI_Gene:105247023,ENSEMBL:ENSMUSG00000105222 | MGI:5625090 | lncRNA gene | predicted gene, 42205 |
| 3 | gene | 34.63846 | 34.63854 | positive | MGI_C57BL6J_3811407 | Mir1897 | miRBase:MI0008314,NCBI_Gene:100316679,ENSEMBL:ENSMUSG00000104797 | MGI:3811407 | miRNA gene | microRNA 1897 |
| 3 | gene | 34.64326 | 34.64610 | positive | MGI_C57BL6J_5662829 | Gm42692 | ENSEMBL:ENSMUSG00000104901 | MGI:5662829 | unclassified gene | predicted gene 42692 |
| 3 | gene | 34.64999 | 34.65246 | positive | MGI_C57BL6J_98364 | Sox2 | NCBI_Gene:20674,ENSEMBL:ENSMUSG00000074637 | MGI:98364 | protein coding gene | SRY (sex determining region Y)-box 2 |
| 3 | gene | 34.65318 | 34.65328 | positive | MGI_C57BL6J_5530993 | Gm27611 | ENSEMBL:ENSMUSG00000098254 | MGI:5530993 | unclassified non-coding RNA gene | predicted gene, 27611 |
| 3 | gene | 34.66366 | 34.66429 | negative | MGI_C57BL6J_5662830 | Gm42693 | ENSEMBL:ENSMUSG00000105284 | MGI:5662830 | unclassified gene | predicted gene 42693 |
| 3 | gene | 34.68263 | 34.68275 | positive | MGI_C57BL6J_5663343 | Gm43206 | ENSEMBL:ENSMUSG00000105101 | MGI:5663343 | unclassified gene | predicted gene 43206 |
| 3 | gene | 34.68938 | 34.69292 | positive | MGI_C57BL6J_5594100 | Gm34941 | NCBI_Gene:102638356 | MGI:5594100 | lncRNA gene | predicted gene, 34941 |
| 3 | gene | 34.69964 | 34.71666 | negative | MGI_C57BL6J_3781322 | Gm3143 | NCBI_Gene:100041108,ENSEMBL:ENSMUSG00000105153 | MGI:3781322 | lncRNA gene | predicted gene 3143 |
| 3 | gene | 34.77208 | 34.78235 | positive | MGI_C57BL6J_5621394 | Gm38509 | NCBI_Gene:102638436,ENSEMBL:ENSMUSG00000106519 | MGI:5621394 | lncRNA gene | predicted gene, 38509 |
| 3 | gene | 34.82656 | 34.87928 | positive | MGI_C57BL6J_5434743 | Gm21388 | NCBI_Gene:100861998,ENSEMBL:ENSMUSG00000114832 | MGI:5434743 | lncRNA gene | predicted gene, 21388 |
| 3 | gene | 34.92254 | 34.92265 | negative | MGI_C57BL6J_5531088 | Mir6378 | miRBase:MI0021909,NCBI_Gene:102466155,ENSEMBL:ENSMUSG00000098862 | MGI:5531088 | miRNA gene | microRNA 6378 |
| 3 | gene | 34.95023 | 34.95953 | positive | MGI_C57BL6J_5594414 | Gm35255 | NCBI_Gene:102638771 | MGI:5594414 | lncRNA gene | predicted gene, 35255 |
| 3 | gene | 35.25646 | 35.27613 | negative | MGI_C57BL6J_1923648 | 1700017M07Rik | NCBI_Gene:76398,ENSEMBL:ENSMUSG00000105651 | MGI:1923648 | lncRNA gene | RIKEN cDNA 1700017M07 gene |
| 3 | gene | 35.31844 | 35.33011 | positive | MGI_C57BL6J_5594584 | Gm35425 | NCBI_Gene:102639001 | MGI:5594584 | lncRNA gene | predicted gene, 35425 |
| 3 | gene | 35.32561 | 35.32570 | positive | MGI_C57BL6J_5455219 | Gm25442 | ENSEMBL:ENSMUSG00000093499 | MGI:5455219 | miRNA gene | predicted gene, 25442 |
| 3 | gene | 35.33770 | 35.40595 | negative | MGI_C57BL6J_5621397 | Gm38512 | NCBI_Gene:102638848 | MGI:5621397 | lncRNA gene | predicted gene, 38512 |
| 3 | gene | 35.41383 | 35.41400 | positive | MGI_C57BL6J_5663215 | Gm43078 | ENSEMBL:ENSMUSG00000106512 | MGI:5663215 | unclassified gene | predicted gene 43078 |
| 3 | pseudogene | 35.53688 | 35.53780 | negative | MGI_C57BL6J_3643628 | Gm7733 | NCBI_Gene:665655,ENSEMBL:ENSMUSG00000106496 | MGI:3643628 | pseudogene | predicted gene 7733 |
| 3 | pseudogene | 35.54589 | 35.54633 | positive | MGI_C57BL6J_1347022 | Ube2l3-ps1 | ENSEMBL:ENSMUSG00000105040 | MGI:1347022 | pseudogene | ubiquitin-conjugating enzyme E2L 3, pseudogene 1 |
| 3 | gene | 35.59715 | 35.66616 | negative | MGI_C57BL6J_3779620 | Gm6639 | NCBI_Gene:625963,ENSEMBL:ENSMUSG00000105662 | MGI:3779620 | lncRNA gene | predicted gene 6639 |
| 3 | gene | 35.73537 | 35.73548 | positive | MGI_C57BL6J_5455473 | Gm25696 | ENSEMBL:ENSMUSG00000064834 | MGI:5455473 | snRNA gene | predicted gene, 25696 |
| 3 | gene | 35.75406 | 35.85628 | positive | MGI_C57BL6J_1923545 | Atp11b | NCBI_Gene:76295,ENSEMBL:ENSMUSG00000037400 | MGI:1923545 | protein coding gene | ATPase, class VI, type 11B |
| 3 | gene | 35.84534 | 35.84590 | positive | MGI_C57BL6J_5663340 | Gm43203 | ENSEMBL:ENSMUSG00000104897 | MGI:5663340 | unclassified gene | predicted gene 43203 |
| 3 | gene | 35.86162 | 35.86212 | positive | MGI_C57BL6J_1915850 | 1110021P09Rik | NA | NA | unclassified gene | RIKEN cDNA 1110021P09 gene |
| 3 | pseudogene | 35.86386 | 35.86507 | positive | MGI_C57BL6J_3647088 | Gm7741 | NCBI_Gene:665667,ENSEMBL:ENSMUSG00000097713 | MGI:3647088 | pseudogene | predicted gene 7741 |
| 3 | pseudogene | 35.87565 | 35.88335 | negative | MGI_C57BL6J_5295701 | Gm20595 | NCBI_Gene:100463514 | MGI:5295701 | pseudogene | predicted gene, 20595 |
| 3 | gene | 35.89211 | 35.93744 | negative | MGI_C57BL6J_2150386 | Dcun1d1 | NCBI_Gene:114893,ENSEMBL:ENSMUSG00000027708 | MGI:2150386 | protein coding gene | DCN1, defective in cullin neddylation 1, domain containing 1 (S. cerevisiae) |
| 3 | gene | 35.93304 | 35.99084 | positive | MGI_C57BL6J_3802149 | Gm15952 | NCBI_Gene:102639351,ENSEMBL:ENSMUSG00000085655 | MGI:3802149 | lncRNA gene | predicted gene 15952 |
| 3 | gene | 35.95544 | 35.95626 | negative | MGI_C57BL6J_1925753 | 8430422M14Rik | ENSEMBL:ENSMUSG00000105095 | MGI:1925753 | unclassified gene | RIKEN cDNA 8430422M14 gene |
| 3 | gene | 35.95629 | 36.00068 | negative | MGI_C57BL6J_1919289 | Mccc1 | NCBI_Gene:72039,ENSEMBL:ENSMUSG00000027709 | MGI:1919289 | protein coding gene | methylcrotonoyl-Coenzyme A carboxylase 1 (alpha) |
| 3 | gene | 35.96938 | 35.97203 | negative | MGI_C57BL6J_5663217 | Gm43080 | ENSEMBL:ENSMUSG00000105556 | MGI:5663217 | unclassified gene | predicted gene 43080 |
| 3 | pseudogene | 35.97377 | 35.97409 | positive | MGI_C57BL6J_3704213 | B230207O21Rik | ENSEMBL:ENSMUSG00000106096 | MGI:3704213 | pseudogene | RIKEN cDNA B230207O21 gene |
| 3 | gene | 35.99983 | 36.02511 | positive | MGI_C57BL6J_3590669 | Mccc1os | NCBI_Gene:654424,ENSEMBL:ENSMUSG00000086392 | MGI:3590669 | antisense lncRNA gene | methylcrotonoyl-Coenzyme A carboxylase 1 (alpha), opposite strand |
| 3 | gene | 36.00724 | 36.05355 | negative | MGI_C57BL6J_2681852 | Ccdc144b | NCBI_Gene:241943,ENSEMBL:ENSMUSG00000047696 | MGI:2681852 | protein coding gene | coiled-coil domain containing 144B |
| 3 | gene | 36.06598 | 36.09286 | positive | MGI_C57BL6J_1914272 | Acad9 | NCBI_Gene:229211,ENSEMBL:ENSMUSG00000027710 | MGI:1914272 | protein coding gene | acyl-Coenzyme A dehydrogenase family, member 9 |
| 3 | gene | 36.08886 | 36.09145 | positive | MGI_C57BL6J_5663216 | Gm43079 | ENSEMBL:ENSMUSG00000105055 | MGI:5663216 | lncRNA gene | predicted gene 43079 |
| 3 | gene | 36.09718 | 36.09914 | positive | MGI_C57BL6J_5663342 | Gm43205 | ENSEMBL:ENSMUSG00000105419 | MGI:5663342 | unclassified gene | predicted gene 43205 |
| 3 | gene | 36.15102 | 36.17034 | positive | MGI_C57BL6J_1098769 | Zfp267 | NCBI_Gene:241944,ENSEMBL:ENSMUSG00000033883 | MGI:1098769 | protein coding gene | zinc finger protein 267 |
| 3 | gene | 36.17942 | 36.22231 | negative | MGI_C57BL6J_2677633 | Qrfpr | NCBI_Gene:229214,ENSEMBL:ENSMUSG00000058400 | MGI:2677633 | protein coding gene | pyroglutamylated RFamide peptide receptor |
| 3 | gene | 36.20329 | 36.20562 | positive | MGI_C57BL6J_5663392 | Gm43255 | ENSEMBL:ENSMUSG00000104568 | MGI:5663392 | lncRNA gene | predicted gene 43255 |
| 3 | pseudogene | 36.21381 | 36.21476 | negative | MGI_C57BL6J_5011078 | Gm18893 | NCBI_Gene:100417907,ENSEMBL:ENSMUSG00000106180 | MGI:5011078 | pseudogene | predicted gene, 18893 |
| 3 | gene | 36.27233 | 36.27896 | negative | MGI_C57BL6J_5625093 | Gm42208 | NCBI_Gene:105247026,ENSEMBL:ENSMUSG00000106140 | MGI:5625093 | lncRNA gene | predicted gene, 42208 |
| 3 | gene | 36.35422 | 36.40651 | negative | MGI_C57BL6J_5625094 | Gm42209 | NCBI_Gene:105247027 | MGI:5625094 | lncRNA gene | predicted gene, 42209 |
| 3 | pseudogene | 36.39470 | 36.39520 | negative | MGI_C57BL6J_5010020 | Gm17835 | NCBI_Gene:100415952,ENSEMBL:ENSMUSG00000105281 | MGI:5010020 | pseudogene | predicted gene, 17835 |
| 3 | pseudogene | 36.42158 | 36.42212 | positive | MGI_C57BL6J_5010072 | Gm17887 | NCBI_Gene:100416041,ENSEMBL:ENSMUSG00000105792 | MGI:5010072 | pseudogene | predicted gene, 17887 |
| 3 | gene | 36.44892 | 36.47589 | negative | MGI_C57BL6J_106008 | Anxa5 | NCBI_Gene:11747,ENSEMBL:ENSMUSG00000027712 | MGI:106008 | protein coding gene | annexin A5 |
| 3 | gene | 36.47546 | 36.47552 | negative | MGI_C57BL6J_5530854 | Mir7009 | miRBase:MI0022858,NCBI_Gene:102466786,ENSEMBL:ENSMUSG00000098808 | MGI:5530854 | miRNA gene | microRNA 7009 |
| 3 | gene | 36.47594 | 36.48230 | positive | MGI_C57BL6J_1919532 | 1810062G17Rik | NCBI_Gene:72282,ENSEMBL:ENSMUSG00000027713 | MGI:1919532 | protein coding gene | RIKEN cDNA 1810062G17 gene |
| 3 | gene | 36.49980 | 36.50681 | negative | MGI_C57BL6J_3650340 | Gm11548 | NCBI_Gene:329623,ENSEMBL:ENSMUSG00000085320 | MGI:3650340 | lncRNA gene | predicted gene 11548 |
| 3 | gene | 36.51506 | 36.52181 | negative | MGI_C57BL6J_3650339 | Gm11549 | NCBI_Gene:100503068,ENSEMBL:ENSMUSG00000085007 | MGI:3650339 | lincRNA gene | predicted gene 11549 |
| 3 | gene | 36.52922 | 36.53233 | negative | MGI_C57BL6J_5663674 | Gm43537 | ENSEMBL:ENSMUSG00000105022 | MGI:5663674 | unclassified gene | predicted gene 43537 |
| 3 | pseudogene | 36.53917 | 36.54010 | negative | MGI_C57BL6J_3650891 | Gm11550 | NCBI_Gene:665709,ENSEMBL:ENSMUSG00000083388 | MGI:3650891 | pseudogene | predicted gene 11550 |
| 3 | gene | 36.55261 | 36.56573 | positive | MGI_C57BL6J_1355319 | Exosc9 | NCBI_Gene:50911,ENSEMBL:ENSMUSG00000027714 | MGI:1355319 | protein coding gene | exosome component 9 |
| 3 | gene | 36.56486 | 36.57303 | negative | MGI_C57BL6J_108069 | Ccna2 | NCBI_Gene:12428,ENSEMBL:ENSMUSG00000027715 | MGI:108069 | protein coding gene | cyclin A2 |
| 3 | gene | 36.57314 | 36.61349 | negative | MGI_C57BL6J_1918742 | Bbs7 | NCBI_Gene:71492,ENSEMBL:ENSMUSG00000037325 | MGI:1918742 | protein coding gene | Bardet-Biedl syndrome 7 (human) |
| 3 | gene | 36.62048 | 36.69022 | negative | MGI_C57BL6J_109526 | Trpc3 | NCBI_Gene:22065,ENSEMBL:ENSMUSG00000027716 | MGI:109526 | protein coding gene | transient receptor potential cation channel, subfamily C, member 3 |
| 3 | gene | 36.65407 | 36.65501 | negative | MGI_C57BL6J_5595015 | Gm35856 | NCBI_Gene:102639576 | MGI:5595015 | lncRNA gene | predicted gene, 35856 |
| 3 | pseudogene | 36.69480 | 36.69505 | negative | MGI_C57BL6J_3651607 | Gm12377 | ENSEMBL:ENSMUSG00000082548 | MGI:3651607 | pseudogene | predicted gene 12377 |
| 3 | pseudogene | 36.82435 | 36.82506 | negative | MGI_C57BL6J_3651608 | Gm12376 | NCBI_Gene:100417988,ENSEMBL:ENSMUSG00000084027 | MGI:3651608 | pseudogene | predicted gene 12376 |
| 3 | gene | 36.86306 | 37.05303 | positive | MGI_C57BL6J_2444631 | 4932438A13Rik | NCBI_Gene:229227,ENSEMBL:ENSMUSG00000037270 | MGI:2444631 | protein coding gene | RIKEN cDNA 4932438A13 gene |
| 3 | gene | 37.05221 | 37.06003 | negative | MGI_C57BL6J_3650531 | Gm12531 | ENSEMBL:ENSMUSG00000086090 | MGI:3650531 | lncRNA gene | predicted gene 12531 |
| 3 | gene | 37.06309 | 37.12193 | positive | MGI_C57BL6J_103258 | Adad1 | NCBI_Gene:21744,ENSEMBL:ENSMUSG00000027719 | MGI:103258 | protein coding gene | adenosine deaminase domain containing 1 (testis specific) |
| 3 | pseudogene | 37.07756 | 37.07891 | positive | MGI_C57BL6J_3651489 | Gm12582 | NCBI_Gene:652987,ENSEMBL:ENSMUSG00000081705 | MGI:3651489 | pseudogene | predicted gene 12582 |
| 3 | pseudogene | 37.10808 | 37.10933 | positive | MGI_C57BL6J_3651485 | Gm12583 | NCBI_Gene:100416060,ENSEMBL:ENSMUSG00000082878 | MGI:3651485 | pseudogene | predicted gene 12583 |
| 3 | pseudogene | 37.11673 | 37.11784 | positive | MGI_C57BL6J_3651486 | Gm12584 | ENSEMBL:ENSMUSG00000083857 | MGI:3651486 | pseudogene | predicted gene 12584 |
| 3 | gene | 37.12052 | 37.12596 | negative | MGI_C57BL6J_96548 | Il2 | NCBI_Gene:16183,ENSEMBL:ENSMUSG00000027720 | MGI:96548 | protein coding gene | interleukin 2 |
| 3 | gene | 37.16972 | 37.17665 | negative | MGI_C57BL6J_5595159 | Gm36000 | NCBI_Gene:102639768 | MGI:5595159 | lncRNA gene | predicted gene, 36000 |
| 3 | gene | 37.18218 | 37.18860 | positive | MGI_C57BL6J_3650618 | Gm12532 | NCBI_Gene:102639848,ENSEMBL:ENSMUSG00000084754 | MGI:3650618 | lncRNA gene | predicted gene 12532 |
| 3 | gene | 37.22276 | 37.23264 | negative | MGI_C57BL6J_1890474 | Il21 | NCBI_Gene:60505,ENSEMBL:ENSMUSG00000027718 | MGI:1890474 | protein coding gene | interleukin 21 |
| 3 | gene | 37.22538 | 37.22711 | positive | MGI_C57BL6J_5595327 | Gm36168 | NCBI_Gene:102639985 | MGI:5595327 | lncRNA gene | predicted gene, 36168 |
| 3 | gene | 37.23133 | 37.23247 | positive | MGI_C57BL6J_3650303 | Gm12534 | ENSEMBL:ENSMUSG00000086517 | MGI:3650303 | lncRNA gene | predicted gene 12534 |
| 3 | gene | 37.23287 | 37.23334 | positive | MGI_C57BL6J_5663959 | Gm43822 | ENSEMBL:ENSMUSG00000104998 | MGI:5663959 | unclassified gene | predicted gene 43822 |
| 3 | gene | 37.25678 | 37.25689 | negative | MGI_C57BL6J_5453946 | Gm24169 | ENSEMBL:ENSMUSG00000094007 | MGI:5453946 | miRNA gene | predicted gene, 24169 |
| 3 | gene | 37.30775 | 37.31271 | negative | MGI_C57BL6J_2677454 | Cetn4 | NCBI_Gene:207175,ENSEMBL:ENSMUSG00000045031 | MGI:2677454 | protein coding gene | centrin 4 |
| 3 | gene | 37.31249 | 37.32145 | positive | MGI_C57BL6J_2686651 | Bbs12 | NCBI_Gene:241950,ENSEMBL:ENSMUSG00000051444 | MGI:2686651 | protein coding gene | Bardet-Biedl syndrome 12 (human) |
| 3 | gene | 37.31252 | 37.40487 | positive | MGI_C57BL6J_5663576 | Gm43439 | ENSEMBL:ENSMUSG00000106407 | MGI:5663576 | lncRNA gene | predicted gene 43439 |
| 3 | pseudogene | 37.31253 | 37.32919 | positive | MGI_C57BL6J_3651849 | Gm12540 | NCBI_Gene:102640067,ENSEMBL:ENSMUSG00000083353 | MGI:3651849 | pseudogene | predicted gene 12540 |
| 3 | gene | 37.33329 | 37.35103 | negative | MGI_C57BL6J_3649376 | Fgf2os | NCBI_Gene:329626,ENSEMBL:ENSMUSG00000054779 | MGI:3649376 | antisense lncRNA gene | fibroblast growth factor 2, opposite strand |
| 3 | gene | 37.33902 | 37.34152 | positive | MGI_C57BL6J_5663625 | Gm43488 | ENSEMBL:ENSMUSG00000105063 | MGI:5663625 | unclassified gene | predicted gene 43488 |
| 3 | gene | 37.34835 | 37.41011 | positive | MGI_C57BL6J_95516 | Fgf2 | NCBI_Gene:14173,ENSEMBL:ENSMUSG00000037225 | MGI:95516 | protein coding gene | fibroblast growth factor 2 |
| 3 | gene | 37.39221 | 37.39510 | negative | MGI_C57BL6J_5663060 | Gm42923 | ENSEMBL:ENSMUSG00000105134 | MGI:5663060 | unclassified gene | predicted gene 42923 |
| 3 | gene | 37.40491 | 37.42021 | negative | MGI_C57BL6J_2387618 | Nudt6 | NCBI_Gene:229228,ENSEMBL:ENSMUSG00000050174 | MGI:2387618 | protein coding gene | nudix (nucleoside diphosphate linked moiety X)-type motif 6 |
| 3 | gene | 37.41990 | 37.57910 | positive | MGI_C57BL6J_1927170 | Spata5 | NCBI_Gene:57815,ENSEMBL:ENSMUSG00000027722 | MGI:1927170 | protein coding gene | spermatogenesis associated 5 |
| 3 | gene | 37.48883 | 37.49479 | positive | MGI_C57BL6J_5663621 | Gm43484 | ENSEMBL:ENSMUSG00000106166 | MGI:5663621 | unclassified gene | predicted gene 43484 |
| 3 | gene | 37.54188 | 37.60239 | negative | MGI_C57BL6J_5595571 | Gm36412 | NCBI_Gene:102640314 | MGI:5595571 | lncRNA gene | predicted gene, 36412 |
| 3 | pseudogene | 37.55618 | 37.55650 | negative | MGI_C57BL6J_3652207 | Gm12564 | ENSEMBL:ENSMUSG00000080723 | MGI:3652207 | pseudogene | predicted gene 12564 |
| 3 | pseudogene | 37.59942 | 37.60007 | negative | MGI_C57BL6J_3652206 | Gm12565 | NCBI_Gene:665778,ENSEMBL:ENSMUSG00000081093 | MGI:3652206 | pseudogene | predicted gene 12565 |
| 3 | gene | 37.60989 | 37.61528 | positive | MGI_C57BL6J_1923197 | 4930594O21Rik | NCBI_Gene:75947,ENSEMBL:ENSMUSG00000085677 | MGI:1923197 | lncRNA gene | RIKEN cDNA 4930594O21 gene |
| 3 | gene | 37.63858 | 37.64066 | negative | MGI_C57BL6J_5663059 | Gm42922 | ENSEMBL:ENSMUSG00000104605 | MGI:5663059 | unclassified gene | predicted gene 42922 |
| 3 | gene | 37.63995 | 37.64460 | positive | MGI_C57BL6J_1345139 | Spry1 | NCBI_Gene:24063,ENSEMBL:ENSMUSG00000037211 | MGI:1345139 | protein coding gene | sprouty RTK signaling antagonist 1 |
| 3 | gene | 37.64461 | 37.64795 | positive | MGI_C57BL6J_5663957 | Gm43820 | ENSEMBL:ENSMUSG00000106062 | MGI:5663957 | unclassified gene | predicted gene 43820 |
| 3 | gene | 37.65268 | 37.65652 | negative | MGI_C57BL6J_5826436 | Gm46799 | NCBI_Gene:108168864 | MGI:5826436 | lncRNA gene | predicted gene, 46799 |
| 3 | gene | 37.69046 | 37.69486 | negative | MGI_C57BL6J_5625096 | Gm42211 | NCBI_Gene:105247029 | MGI:5625096 | lncRNA gene | predicted gene, 42211 |
| 3 | gene | 37.70802 | 37.71121 | positive | MGI_C57BL6J_5663551 | Gm43414 | ENSEMBL:ENSMUSG00000104935 | MGI:5663551 | unclassified gene | predicted gene 43414 |
| 3 | gene | 37.71419 | 37.72449 | negative | MGI_C57BL6J_3646006 | Gm5148 | NCBI_Gene:381438,ENSEMBL:ENSMUSG00000058174 | MGI:3646006 | protein coding gene | predicted gene 5148 |
| 3 | pseudogene | 37.71471 | 37.71522 | positive | MGI_C57BL6J_3705429 | Rps23-ps1 | NCBI_Gene:100038875,ENSEMBL:ENSMUSG00000100755 | MGI:3705429 | pseudogene | ribosomal protein S23, pseudogene 1 |
| 3 | gene | 37.72423 | 37.74132 | positive | MGI_C57BL6J_5595779 | Gm36620 | NCBI_Gene:102640592 | MGI:5595779 | lncRNA gene | predicted gene, 36620 |
| 3 | pseudogene | 37.73468 | 37.73514 | positive | MGI_C57BL6J_3643738 | Rpl31-ps15 | NCBI_Gene:626367,ENSEMBL:ENSMUSG00000097304 | MGI:3643738 | pseudogene | ribosomal protein L31, pseudogene 15 |
| 3 | gene | 37.75320 | 37.77440 | positive | MGI_C57BL6J_5625095 | Gm42210 | NCBI_Gene:105247028 | MGI:5625095 | lncRNA gene | predicted gene, 42210 |
| 3 | gene | 37.76104 | 37.76730 | positive | MGI_C57BL6J_5663058 | Gm42921 | ENSEMBL:ENSMUSG00000106441 | MGI:5663058 | lncRNA gene | predicted gene 42921 |
| 3 | gene | 37.80254 | 37.80265 | negative | MGI_C57BL6J_5456181 | Gm26404 | ENSEMBL:ENSMUSG00000089123 | MGI:5456181 | snoRNA gene | predicted gene, 26404 |
| 3 | gene | 37.82822 | 37.84829 | positive | MGI_C57BL6J_1925294 | 4930556A17Rik | NCBI_Gene:102640520 | MGI:1925294 | lncRNA gene | RIKEN cDNA 4930556A17 gene |
| 3 | gene | 37.83847 | 37.83957 | positive | MGI_C57BL6J_5662889 | Gm42752 | ENSEMBL:ENSMUSG00000106621 | MGI:5662889 | unclassified gene | predicted gene 42752 |
| 3 | gene | 37.84180 | 37.84307 | positive | MGI_C57BL6J_5662890 | Gm42753 | ENSEMBL:ENSMUSG00000105252 | MGI:5662890 | unclassified gene | predicted gene 42753 |
| 3 | gene | 37.89881 | 37.99273 | positive | MGI_C57BL6J_5434111 | Gm20755 | NCBI_Gene:626410,ENSEMBL:ENSMUSG00000106461 | MGI:5434111 | lncRNA gene | predicted gene, 20755 |
| 3 | pseudogene | 37.95961 | 37.96009 | negative | MGI_C57BL6J_3643378 | Gm7799 | ENSEMBL:ENSMUSG00000087457 | MGI:3643378 | pseudogene | predicted gene 7799 |
| 3 | gene | 37.98279 | 37.98335 | positive | MGI_C57BL6J_5662888 | Gm42751 | ENSEMBL:ENSMUSG00000105099 | MGI:5662888 | unclassified gene | predicted gene 42751 |
| 3 | gene | 37.99511 | 37.99524 | negative | MGI_C57BL6J_5452676 | Gm22899 | ENSEMBL:ENSMUSG00000077728 | MGI:5452676 | snoRNA gene | predicted gene, 22899 |
| 3 | gene | 38.09463 | 38.13019 | negative | MGI_C57BL6J_5663958 | Gm43821 | ENSEMBL:ENSMUSG00000105219 | MGI:5663958 | lncRNA gene | predicted gene 43821 |
| 3 | pseudogene | 38.10123 | 38.10798 | negative | MGI_C57BL6J_3643358 | Rpl21-ps10 | NCBI_Gene:665821,ENSEMBL:ENSMUSG00000105359 | MGI:3643358 | pseudogene | ribosomal protein L21, pseudogene 10 |
| 3 | gene | 38.15191 | 38.15915 | positive | MGI_C57BL6J_5595839 | Gm36680 | NCBI_Gene:102640666 | MGI:5595839 | lncRNA gene | predicted gene, 36680 |
| 3 | gene | 38.20315 | 38.20812 | negative | MGI_C57BL6J_1918592 | 5430434I15Rik | NCBI_Gene:71342,ENSEMBL:ENSMUSG00000106002 | MGI:1918592 | lncRNA gene | RIKEN cDNA 5430434I15 gene |
| 3 | pseudogene | 38.22345 | 38.22382 | positive | MGI_C57BL6J_3781143 | Gm2965 | NCBI_Gene:100040785,ENSEMBL:ENSMUSG00000105943 | MGI:3781143 | pseudogene | predicted gene 2965 |
| 3 | pseudogene | 38.25402 | 38.25480 | positive | MGI_C57BL6J_5010140 | Gm17955 | NCBI_Gene:100416169 | MGI:5010140 | pseudogene | predicted gene, 17955 |
| 3 | gene | 38.26784 | 38.26808 | negative | MGI_C57BL6J_5455351 | Gm25574 | ENSEMBL:ENSMUSG00000092986 | MGI:5455351 | unclassified non-coding RNA gene | predicted gene, 25574 |
| 3 | gene | 38.36615 | 38.36888 | negative | MGI_C57BL6J_5625098 | Gm42213 | NCBI_Gene:105247031 | MGI:5625098 | lncRNA gene | predicted gene, 42213 |
| 3 | gene | 38.36940 | 38.37364 | positive | MGI_C57BL6J_5663057 | Gm42920 | ENSEMBL:ENSMUSG00000106008 | MGI:5663057 | lncRNA gene | predicted gene 42920 |
| 3 | gene | 38.41897 | 38.42015 | negative | MGI_C57BL6J_1917958 | 3830422I06Rik | ENSEMBL:ENSMUSG00000106485 | MGI:1917958 | unclassified gene | RIKEN cDNA 3830422I06 gene |
| 3 | gene | 38.42655 | 38.44387 | negative | MGI_C57BL6J_5595982 | Gm36823 | NCBI_Gene:102640855,ENSEMBL:ENSMUSG00000105516 | MGI:5595982 | lncRNA gene | predicted gene, 36823 |
| 3 | pseudogene | 38.43895 | 38.43984 | negative | MGI_C57BL6J_3646285 | Gm7815 | NCBI_Gene:665838,ENSEMBL:ENSMUSG00000106201 | MGI:3646285 | pseudogene | predicted gene 7815 |
| 3 | gene | 38.44926 | 38.48486 | negative | MGI_C57BL6J_2139777 | Ankrd50 | NCBI_Gene:99696,ENSEMBL:ENSMUSG00000044864 | MGI:2139777 | protein coding gene | ankyrin repeat domain 50 |
| 3 | pseudogene | 38.55797 | 38.55886 | negative | MGI_C57BL6J_3779766 | Gm7824 | NCBI_Gene:665855,ENSEMBL:ENSMUSG00000105360 | MGI:3779766 | pseudogene | predicted gene 7824 |
| 3 | gene | 38.55811 | 38.55821 | negative | MGI_C57BL6J_5454239 | Gm24462 | ENSEMBL:ENSMUSG00000076385 | MGI:5454239 | miRNA gene | predicted gene, 24462 |
| 3 | gene | 38.61425 | 38.61556 | positive | MGI_C57BL6J_5663675 | Gm43538 | ENSEMBL:ENSMUSG00000106168 | MGI:5663675 | lncRNA gene | predicted gene 43538 |
| 3 | gene | 38.66361 | 38.70922 | positive | MGI_C57BL6J_5589038 | Gm29879 | NCBI_Gene:102631576 | MGI:5589038 | lncRNA gene | predicted gene, 29879 |
| 3 | gene | 38.76048 | 38.76195 | negative | MGI_C57BL6J_5589118 | Gm29959 | NCBI_Gene:102631683 | MGI:5589118 | lncRNA gene | predicted gene, 29959 |
| 3 | gene | 38.87457 | 38.88826 | negative | MGI_C57BL6J_2441892 | C230034O21Rik | NCBI_Gene:102632538,ENSEMBL:ENSMUSG00000086181 | MGI:2441892 | lncRNA gene | RIKEN cDNA C230034O21 gene |
| 3 | gene | 38.88563 | 39.01199 | positive | MGI_C57BL6J_3045256 | Fat4 | NCBI_Gene:329628,ENSEMBL:ENSMUSG00000046743 | MGI:3045256 | protein coding gene | FAT atypical cadherin 4 |
| 3 | gene | 39.00860 | 39.00968 | negative | MGI_C57BL6J_5663676 | Gm43539 | ENSEMBL:ENSMUSG00000105910 | MGI:5663676 | lncRNA gene | predicted gene 43539 |
| 3 | pseudogene | 39.12853 | 39.12891 | negative | MGI_C57BL6J_5663445 | Gm43308 | ENSEMBL:ENSMUSG00000104792 | MGI:5663445 | pseudogene | predicted gene 43308 |
| 3 | gene | 39.15083 | 39.15967 | negative | MGI_C57BL6J_5589170 | Gm30011 | NCBI_Gene:102631750 | MGI:5589170 | lncRNA gene | predicted gene, 30011 |
| 3 | pseudogene | 39.21756 | 39.21785 | negative | MGI_C57BL6J_5663145 | Gm43008 | ENSEMBL:ENSMUSG00000107333 | MGI:5663145 | pseudogene | predicted gene 43008 |
| 3 | gene | 39.27713 | 39.30593 | positive | MGI_C57BL6J_5625099 | Gm42214 | NCBI_Gene:105247032 | MGI:5625099 | lncRNA gene | predicted gene, 42214 |
| 3 | pseudogene | 39.35737 | 39.35913 | negative | MGI_C57BL6J_3704215 | Gm9845 | NCBI_Gene:100040853,ENSEMBL:ENSMUSG00000050533 | MGI:3704215 | pseudogene | predicted pseudogene 9845 |
| 3 | pseudogene | 39.44389 | 39.44435 | positive | MGI_C57BL6J_5295673 | Gm20566 | NCBI_Gene:100047050 | MGI:5295673 | pseudogene | predicted gene, 20566 |
| 3 | gene | 39.46078 | 39.59865 | positive | MGI_C57BL6J_5589222 | Gm30063 | NCBI_Gene:102631823 | MGI:5589222 | lncRNA gene | predicted gene, 30063 |
| 3 | gene | 39.52552 | 39.53415 | negative | MGI_C57BL6J_5625100 | Gm42215 | NCBI_Gene:105247033 | MGI:5625100 | lncRNA gene | predicted gene, 42215 |
| 3 | gene | 39.63596 | 39.63608 | negative | MGI_C57BL6J_4422060 | n-R5s195 | ENSEMBL:ENSMUSG00000093952 | MGI:4422060 | rRNA gene | nuclear encoded rRNA 5S 195 |
| 3 | gene | 39.63606 | 39.63878 | positive | MGI_C57BL6J_5662918 | Gm42781 | ENSEMBL:ENSMUSG00000106786 | MGI:5662918 | unclassified gene | predicted gene 42781 |
| 3 | gene | 39.66629 | 39.66645 | positive | MGI_C57BL6J_5453573 | Gm23796 | ENSEMBL:ENSMUSG00000065945 | MGI:5453573 | snRNA gene | predicted gene, 23796 |
| 3 | gene | 39.71315 | 39.72565 | positive | MGI_C57BL6J_5826471 | Gm46834 | NCBI_Gene:108168914 | MGI:5826471 | lncRNA gene | predicted gene, 46834 |
| 3 | gene | 39.79963 | 39.79972 | negative | MGI_C57BL6J_5453608 | Gm23831 | ENSEMBL:ENSMUSG00000089520 | MGI:5453608 | snoRNA gene | predicted gene, 23831 |
| 3 | pseudogene | 39.82048 | 39.82076 | negative | MGI_C57BL6J_5662923 | Gm42786 | ENSEMBL:ENSMUSG00000106846 | MGI:5662923 | pseudogene | predicted gene 42786 |
| 3 | pseudogene | 39.82358 | 39.82741 | positive | MGI_C57BL6J_5662922 | Gm42785 | ENSEMBL:ENSMUSG00000106948 | MGI:5662922 | pseudogene | predicted gene 42785 |
| 3 | gene | 40.42571 | 40.45380 | negative | MGI_C57BL6J_5589343 | Gm30184 | NCBI_Gene:102632001 | MGI:5589343 | lncRNA gene | predicted gene, 30184 |
| 3 | gene | 40.45843 | 40.46115 | positive | MGI_C57BL6J_5625101 | Gm42216 | NCBI_Gene:105247034 | MGI:5625101 | lncRNA gene | predicted gene, 42216 |
| 3 | gene | 40.48090 | 40.48573 | positive | MGI_C57BL6J_5625102 | Gm42217 | NCBI_Gene:105247035 | MGI:5625102 | lncRNA gene | predicted gene, 42217 |
| 3 | gene | 40.48702 | 40.48993 | positive | MGI_C57BL6J_5826438 | Gm46801 | NCBI_Gene:108168866 | MGI:5826438 | lncRNA gene | predicted gene, 46801 |
| 3 | gene | 40.50487 | 40.52291 | negative | MGI_C57BL6J_1919477 | 1700017G19Rik | NCBI_Gene:100040882,ENSEMBL:ENSMUSG00000037884 | MGI:1919477 | lncRNA gene | RIKEN cDNA 1700017G19 gene |
| 3 | gene | 40.50998 | 40.51183 | positive | MGI_C57BL6J_5826437 | Gm46800 | NCBI_Gene:108168865 | MGI:5826437 | lncRNA gene | predicted gene, 46800 |
| 3 | gene | 40.51882 | 40.51889 | positive | MGI_C57BL6J_5453587 | Gm23810 | ENSEMBL:ENSMUSG00000092891 | MGI:5453587 | miRNA gene | predicted gene, 23810 |
| 3 | gene | 40.53129 | 40.70478 | positive | MGI_C57BL6J_2443752 | Intu | NCBI_Gene:380614,ENSEMBL:ENSMUSG00000060798 | MGI:2443752 | protein coding gene | inturned planar cell polarity protein |
| 3 | pseudogene | 40.65887 | 40.65917 | negative | MGI_C57BL6J_3801939 | Gm16114 | ENSEMBL:ENSMUSG00000089932 | MGI:3801939 | pseudogene | predicted gene 16114 |
| 3 | gene | 40.70885 | 40.72610 | positive | MGI_C57BL6J_1920583 | Slc25a31 | NCBI_Gene:73333,ENSEMBL:ENSMUSG00000069041 | MGI:1920583 | protein coding gene | solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 31 |
| 3 | gene | 40.74440 | 40.79610 | positive | MGI_C57BL6J_107422 | Hspa4l | NCBI_Gene:18415,ENSEMBL:ENSMUSG00000025757 | MGI:107422 | protein coding gene | heat shock protein 4 like |
| 3 | gene | 40.79364 | 40.79846 | positive | MGI_C57BL6J_1918107 | 4921509J17Rik | NA | NA | unclassified gene | RIKEN cDNA 4921509J17 gene |
| 3 | gene | 40.79995 | 40.81688 | positive | MGI_C57BL6J_101783 | Plk4 | NCBI_Gene:20873,ENSEMBL:ENSMUSG00000025758 | MGI:101783 | protein coding gene | polo like kinase 4 |
| 3 | gene | 40.81810 | 40.84689 | negative | MGI_C57BL6J_1919425 | Mfsd8 | NCBI_Gene:72175,ENSEMBL:ENSMUSG00000025759 | MGI:1919425 | protein coding gene | major facilitator superfamily domain containing 8 |
| 3 | pseudogene | 40.84682 | 40.87561 | positive | MGI_C57BL6J_3780180 | Gm2011 | NCBI_Gene:100039027,ENSEMBL:ENSMUSG00000107741 | MGI:3780180 | pseudogene | predicted gene 2011 |
| 3 | gene | 40.84697 | 40.93814 | positive | MGI_C57BL6J_1915468 | Abhd18 | NCBI_Gene:269423,ENSEMBL:ENSMUSG00000037818 | MGI:1915468 | protein coding gene | abhydrolase domain containing 18 |
| 3 | pseudogene | 40.85348 | 40.85529 | negative | MGI_C57BL6J_3795712 | Gm10731 | NCBI_Gene:100039043,ENSEMBL:ENSMUSG00000074620 | MGI:3795712 | pseudogene | predicted gene 10731 |
| 3 | gene | 40.86395 | 40.86505 | negative | MGI_C57BL6J_5690528 | Gm44136 | ENSEMBL:ENSMUSG00000107524 | MGI:5690528 | unclassified gene | predicted gene, 44136 |
| 3 | gene | 40.87665 | 40.89455 | negative | MGI_C57BL6J_3648839 | 6430590A07Rik | NCBI_Gene:619719,ENSEMBL:ENSMUSG00000097583 | MGI:3648839 | lncRNA gene | RIKEN cDNA 6430590A07 gene |
| 3 | pseudogene | 40.87698 | 40.87725 | negative | MGI_C57BL6J_5753100 | Gm44524 | ENSEMBL:ENSMUSG00000107892 | MGI:5753100 | pseudogene | predicted gene 44524 |
| 3 | gene | 40.90020 | 40.90254 | negative | MGI_C57BL6J_1920547 | 1700034I23Rik | NCBI_Gene:73297,ENSEMBL:ENSMUSG00000074619 | MGI:1920547 | protein coding gene | RIKEN cDNA 1700034I23 gene |
| 3 | gene | 40.94869 | 40.94986 | negative | MGI_C57BL6J_5826439 | Gm46802 | NCBI_Gene:108168867 | MGI:5826439 | lncRNA gene | predicted gene, 46802 |
| 3 | gene | 40.94972 | 41.04023 | positive | MGI_C57BL6J_1914604 | Larp1b | NCBI_Gene:214048,ENSEMBL:ENSMUSG00000025762 | MGI:1914604 | protein coding gene | La ribonucleoprotein domain family, member 1B |
| 3 | gene | 40.97850 | 41.00609 | positive | MGI_C57BL6J_5589448 | Gm30289 | NA | NA | protein coding gene | predicted gene%2c 30289 |
| 3 | gene | 40.99269 | 40.99285 | positive | MGI_C57BL6J_5451820 | Gm22043 | ENSEMBL:ENSMUSG00000064927 | MGI:5451820 | snRNA gene | predicted gene, 22043 |
| 3 | gene | 41.00243 | 41.00609 | positive | MGI_C57BL6J_5662573 | Gm42436 | ENSEMBL:ENSMUSG00000105517 | MGI:5662573 | unclassified gene | predicted gene 42436 |
| 3 | gene | 41.00552 | 41.00644 | negative | MGI_C57BL6J_5662574 | Gm42437 | ENSEMBL:ENSMUSG00000105674 | MGI:5662574 | unclassified gene | predicted gene 42437 |
| 3 | gene | 41.06633 | 41.08305 | negative | MGI_C57BL6J_1918054 | Pgrmc2 | NCBI_Gene:70804,ENSEMBL:ENSMUSG00000049940 | MGI:1918054 | protein coding gene | progesterone receptor membrane component 2 |
| 3 | gene | 41.15803 | 41.17346 | negative | MGI_C57BL6J_5622923 | Gm40038 | NCBI_Gene:105244429,ENSEMBL:ENSMUSG00000105258 | MGI:5622923 | lncRNA gene | predicted gene, 40038 |
| 3 | gene | 41.28606 | 41.29059 | negative | MGI_C57BL6J_3642810 | Gm16508 | ENSEMBL:ENSMUSG00000106000 | MGI:3642810 | lncRNA gene | predicted gene 16508 |
| 3 | gene | 41.35415 | 41.40446 | positive | MGI_C57BL6J_1920712 | 1700052H01Rik | NCBI_Gene:73462,ENSEMBL:ENSMUSG00000105297 | MGI:1920712 | lncRNA gene | RIKEN cDNA 1700052H01 gene |
| 3 | gene | 41.44134 | 41.44147 | negative | MGI_C57BL6J_5455264 | Gm25487 | ENSEMBL:ENSMUSG00000089350 | MGI:5455264 | snoRNA gene | predicted gene, 25487 |
| 3 | gene | 41.47491 | 41.48400 | negative | MGI_C57BL6J_5622925 | Gm40040 | NCBI_Gene:105244431,ENSEMBL:ENSMUSG00000105260 | MGI:5622925 | lncRNA gene | predicted gene, 40040 |
| 3 | gene | 41.48402 | 41.49320 | negative | MGI_C57BL6J_1925956 | Platr4 | NCBI_Gene:78706,ENSEMBL:ENSMUSG00000097639 | MGI:1925956 | lncRNA gene | pluripotency associated transcript 4 |
| 3 | pseudogene | 41.48569 | 41.48641 | positive | MGI_C57BL6J_3648422 | Gm6069 | NCBI_Gene:606542,ENSEMBL:ENSMUSG00000104616 | MGI:3648422 | pseudogene | predicted gene 6069 |
| 3 | gene | 41.50093 | 41.52847 | negative | MGI_C57BL6J_1925898 | 2400006E01Rik | NCBI_Gene:78648,ENSEMBL:ENSMUSG00000106223 | MGI:1925898 | lncRNA gene | RIKEN cDNA 2400006E01 gene |
| 3 | gene | 41.55573 | 41.61686 | positive | MGI_C57BL6J_1925835 | Jade1 | NCBI_Gene:269424,ENSEMBL:ENSMUSG00000025764 | MGI:1925835 | protein coding gene | jade family PHD finger 1 |
| 3 | gene | 41.56050 | 41.56370 | negative | MGI_C57BL6J_5590425 | Gm31266 | NCBI_Gene:105244433,ENSEMBL:ENSMUSG00000103843 | MGI:5590425 | lncRNA gene | predicted gene, 31266 |
| 3 | gene | 41.61755 | 41.62371 | negative | MGI_C57BL6J_5622926 | Gm40041 | NCBI_Gene:105244432 | MGI:5622926 | lncRNA gene | predicted gene, 40041 |
| 3 | gene | 41.62671 | 41.74253 | negative | MGI_C57BL6J_1914411 | Sclt1 | NCBI_Gene:67161,ENSEMBL:ENSMUSG00000059834 | MGI:1914411 | protein coding gene | sodium channel and clathrin linker 1 |
| 3 | gene | 41.64812 | 41.64925 | negative | MGI_C57BL6J_5611275 | Gm38047 | ENSEMBL:ENSMUSG00000102436 | MGI:5611275 | unclassified gene | predicted gene, 38047 |
| 3 | gene | 41.74261 | 41.80332 | positive | MGI_C57BL6J_1289213 | D3Ertd751e | NCBI_Gene:73852,ENSEMBL:ENSMUSG00000025766 | MGI:1289213 | protein coding gene | DNA segment, Chr 3, ERATO Doi 751, expressed |
| 3 | gene | 41.77318 | 41.77579 | positive | MGI_C57BL6J_5610496 | Gm37268 | ENSEMBL:ENSMUSG00000103952 | MGI:5610496 | unclassified gene | predicted gene, 37268 |
| 3 | pseudogene | 41.97607 | 41.97812 | negative | MGI_C57BL6J_3645044 | Gm7899 | NCBI_Gene:666031,ENSEMBL:ENSMUSG00000103823 | MGI:3645044 | pseudogene | predicted gene 7899 |
| 3 | pseudogene | 41.97633 | 41.97812 | positive | MGI_C57BL6J_5610576 | Gm37348 | ENSEMBL:ENSMUSG00000102972 | MGI:5610576 | pseudogene | predicted gene, 37348 |
| 3 | gene | 42.16387 | 42.16764 | positive | MGI_C57BL6J_5611074 | Gm37846 | ENSEMBL:ENSMUSG00000102610 | MGI:5611074 | unclassified gene | predicted gene, 37846 |
| 3 | pseudogene | 42.36263 | 42.36278 | negative | MGI_C57BL6J_5611502 | Gm38274 | ENSEMBL:ENSMUSG00000104089 | MGI:5611502 | pseudogene | predicted gene, 38274 |
| 3 | gene | 42.57246 | 42.57300 | positive | MGI_C57BL6J_5610761 | Gm37533 | ENSEMBL:ENSMUSG00000103426 | MGI:5610761 | unclassified gene | predicted gene, 37533 |
| 3 | pseudogene | 42.58511 | 42.58560 | negative | MGI_C57BL6J_5611272 | Gm38044 | ENSEMBL:ENSMUSG00000104294 | MGI:5611272 | pseudogene | predicted gene, 38044 |
| 3 | pseudogene | 43.25039 | 43.25093 | positive | MGI_C57BL6J_3780131 | Gm9723 | NCBI_Gene:677788,ENSEMBL:ENSMUSG00000102149 | MGI:3780131 | pseudogene | predicted gene 9723 |
| 3 | pseudogene | 43.39077 | 43.39194 | negative | MGI_C57BL6J_5011050 | Gm18865 | NCBI_Gene:100417854,ENSEMBL:ENSMUSG00000102334 | MGI:5011050 | pseudogene | predicted gene, 18865 |
| 3 | gene | 43.54717 | 43.54828 | positive | MGI_C57BL6J_5610204 | Gm36976 | ENSEMBL:ENSMUSG00000104303 | MGI:5610204 | unclassified gene | predicted gene, 36976 |
| 3 | pseudogene | 43.76405 | 43.76476 | negative | MGI_C57BL6J_5588898 | Gm29739 | NCBI_Gene:101055891,ENSEMBL:ENSMUSG00000104083 | MGI:5588898 | pseudogene | predicted gene, 29739 |
| 3 | gene | 43.92557 | 43.92586 | positive | MGI_C57BL6J_5455491 | Gm25714 | ENSEMBL:ENSMUSG00000089251 | MGI:5455491 | unclassified non-coding RNA gene | predicted gene, 25714 |
| 3 | pseudogene | 43.92760 | 43.92795 | negative | MGI_C57BL6J_5591334 | Gm32175 | ENSEMBL:ENSMUSG00000102792 | MGI:5591334 | pseudogene | predicted gene, 32175 |
| 3 | gene | 43.95572 | 43.95592 | positive | MGI_C57BL6J_5610974 | Gm37746 | ENSEMBL:ENSMUSG00000103148 | MGI:5610974 | unclassified gene | predicted gene, 37746 |
| 3 | pseudogene | 44.49350 | 44.49390 | negative | MGI_C57BL6J_5610593 | Gm37365 | ENSEMBL:ENSMUSG00000104013 | MGI:5610593 | pseudogene | predicted gene, 37365 |
| 3 | gene | 44.65990 | 44.66315 | positive | MGI_C57BL6J_5611356 | Gm38128 | ENSEMBL:ENSMUSG00000104051 | MGI:5611356 | unclassified gene | predicted gene, 38128 |
| 3 | pseudogene | 44.71813 | 44.71851 | negative | MGI_C57BL6J_5610825 | Gm37597 | ENSEMBL:ENSMUSG00000103353 | MGI:5610825 | pseudogene | predicted gene, 37597 |
| 3 | pseudogene | 44.72516 | 44.72574 | positive | MGI_C57BL6J_5011051 | Gm18866 | NCBI_Gene:100417856 | MGI:5011051 | pseudogene | predicted gene, 18866 |
| 3 | gene | 44.98320 | 44.98330 | positive | MGI_C57BL6J_5454774 | Gm24997 | ENSEMBL:ENSMUSG00000088535 | MGI:5454774 | snRNA gene | predicted gene, 24997 |
| 3 | gene | 45.16069 | 45.16080 | positive | MGI_C57BL6J_5531371 | Gm27989 | ENSEMBL:ENSMUSG00000099285 | MGI:5531371 | miRNA gene | predicted gene, 27989 |
| 3 | pseudogene | 45.17179 | 45.17324 | negative | MGI_C57BL6J_5434370 | Gm21015 | NCBI_Gene:100534291 | MGI:5434370 | pseudogene | predicted gene, 21015 |
| 3 | gene | 45.26999 | 45.27257 | negative | MGI_C57BL6J_5611352 | Gm38124 | ENSEMBL:ENSMUSG00000103082 | MGI:5611352 | unclassified gene | predicted gene, 38124 |
| 3 | gene | 45.27342 | 45.27497 | negative | MGI_C57BL6J_1924316 | 5930409G06Rik | ENSEMBL:ENSMUSG00000103477 | MGI:1924316 | unclassified gene | RIKEN cDNA 5930409G06 gene |
| 3 | gene | 45.28044 | 45.37826 | negative | MGI_C57BL6J_1919761 | 2610316D01Rik | NCBI_Gene:72511,ENSEMBL:ENSMUSG00000097040 | MGI:1919761 | lncRNA gene | RIKEN cDNA 2610316D01 gene |
| 3 | pseudogene | 45.33216 | 45.33241 | negative | MGI_C57BL6J_5610224 | Gm36996 | ENSEMBL:ENSMUSG00000103430 | MGI:5610224 | pseudogene | predicted gene, 36996 |
| 3 | gene | 45.37840 | 45.43562 | positive | MGI_C57BL6J_1338042 | Pcdh10 | NCBI_Gene:18526,ENSEMBL:ENSMUSG00000049100 | MGI:1338042 | protein coding gene | protocadherin 10 |
| 3 | gene | 45.38548 | 45.38960 | positive | MGI_C57BL6J_2442672 | 6330566A10Rik | NA | NA | unclassified gene | RIKEN cDNA 6330566A10 gene |
| 3 | gene | 45.39986 | 45.40200 | positive | MGI_C57BL6J_2444936 | D930002L09Rik | NA | NA | unclassified gene | RIKEN cDNA D930002L09 gene |
| 3 | gene | 45.41658 | 45.43931 | negative | MGI_C57BL6J_1919524 | 1700027H10Rik | NCBI_Gene:72274,ENSEMBL:ENSMUSG00000100799 | MGI:1919524 | lncRNA gene | RIKEN cDNA 1700027H10 gene |
| 3 | gene | 45.43838 | 45.44096 | positive | MGI_C57BL6J_3780305 | Gm2136 | ENSEMBL:ENSMUSG00000103713 | MGI:3780305 | protein coding gene | predicted gene 2136 |
| 3 | gene | 45.44567 | 45.44881 | positive | MGI_C57BL6J_5610573 | Gm37345 | ENSEMBL:ENSMUSG00000103947 | MGI:5610573 | unclassified gene | predicted gene, 37345 |
| 3 | pseudogene | 45.69054 | 45.69090 | negative | MGI_C57BL6J_5611063 | Gm37835 | ENSEMBL:ENSMUSG00000103534 | MGI:5611063 | pseudogene | predicted gene, 37835 |
| 3 | gene | 45.77542 | 45.77589 | negative | MGI_C57BL6J_5611394 | Gm38166 | ENSEMBL:ENSMUSG00000102190 | MGI:5611394 | unclassified gene | predicted gene, 38166 |
| 3 | pseudogene | 45.77587 | 45.86058 | negative | MGI_C57BL6J_5591499 | Gm32340 | NCBI_Gene:102634856,ENSEMBL:ENSMUSG00000104491 | MGI:5591499 | pseudogene | predicted gene, 32340 |
| 3 | gene | 45.81774 | 45.81790 | positive | MGI_C57BL6J_5455573 | Gm25796 | ENSEMBL:ENSMUSG00000064579 | MGI:5455573 | snRNA gene | predicted gene, 25796 |
| 3 | pseudogene | 45.86121 | 45.86995 | negative | MGI_C57BL6J_5591575 | Gm32416 | NCBI_Gene:102634958 | MGI:5591575 | pseudogene | predicted gene, 32416 |
| 3 | gene | 45.97858 | 45.97870 | positive | MGI_C57BL6J_5453114 | Gm23337 | ENSEMBL:ENSMUSG00000089584 | MGI:5453114 | snRNA gene | predicted gene, 23337 |
| 3 | gene | 46.28983 | 46.28996 | positive | MGI_C57BL6J_5455858 | Gm26081 | ENSEMBL:ENSMUSG00000077256 | MGI:5455858 | snoRNA gene | predicted gene, 26081 |
| 3 | pseudogene | 46.41340 | 46.41412 | positive | MGI_C57BL6J_5011028 | Gm18843 | NCBI_Gene:100417811,ENSEMBL:ENSMUSG00000104511 | MGI:5011028 | pseudogene | predicted gene, 18843 |
| 3 | gene | 46.44085 | 46.44822 | negative | MGI_C57BL6J_3643087 | Pabpc4l | NCBI_Gene:241989,ENSEMBL:ENSMUSG00000090919 | MGI:3643087 | protein coding gene | poly(A) binding protein, cytoplasmic 4-like |
| 3 | pseudogene | 46.62112 | 46.62136 | positive | MGI_C57BL6J_5610673 | Gm37445 | ENSEMBL:ENSMUSG00000102589 | MGI:5610673 | pseudogene | predicted gene, 37445 |
| 3 | gene | 46.90067 | 46.90078 | negative | MGI_C57BL6J_5531112 | Mir6379 | miRBase:MI0021910,NCBI_Gene:102465199,ENSEMBL:ENSMUSG00000098417 | MGI:5531112 | miRNA gene | microRNA 6379 |
| 3 | pseudogene | 47.04911 | 47.05178 | negative | MGI_C57BL6J_3641761 | Gm10356 | NCBI_Gene:545517,ENSEMBL:ENSMUSG00000072387 | MGI:3641761 | pseudogene | predicted gene 10356 |
| 3 | pseudogene | 47.08453 | 47.08575 | negative | MGI_C57BL6J_3646161 | Gm5274 | NCBI_Gene:383849,ENSEMBL:ENSMUSG00000097985 | MGI:3646161 | pseudogene | predicted gene 5274 |
| 3 | gene | 47.20063 | 47.20076 | positive | MGI_C57BL6J_5453668 | Gm23891 | ENSEMBL:ENSMUSG00000088739 | MGI:5453668 | snoRNA gene | predicted gene, 23891 |
| 3 | pseudogene | 47.23050 | 47.23100 | negative | MGI_C57BL6J_3643228 | Rpl23a-ps5 | NCBI_Gene:383850,ENSEMBL:ENSMUSG00000102220 | MGI:3643228 | pseudogene | ribosomal protein L23A, pseudogene 5 |
| 3 | gene | 47.26308 | 47.26580 | negative | MGI_C57BL6J_3809922 | Gm10730 | ENSEMBL:ENSMUSG00000103491 | MGI:3809922 | unclassified gene | predicted gene 10730 |
| 3 | pseudogene | 47.45961 | 47.46034 | negative | MGI_C57BL6J_3780399 | Gm2229 | NCBI_Gene:100039426,ENSEMBL:ENSMUSG00000103798 | MGI:3780399 | pseudogene | predicted gene 2229 |
| 3 | pseudogene | 48.02601 | 48.02676 | negative | MGI_C57BL6J_3648898 | Gm7977 | NCBI_Gene:666202,ENSEMBL:ENSMUSG00000103257 | MGI:3648898 | pseudogene | predicted gene 7977 |
| 3 | gene | 48.07834 | 48.07845 | positive | MGI_C57BL6J_5610523 | Gm37295 | ENSEMBL:ENSMUSG00000104110 | MGI:5610523 | unclassified gene | predicted gene, 37295 |
| 3 | gene | 48.54642 | 48.57947 | positive | MGI_C57BL6J_5826472 | Gm46835 | NCBI_Gene:108168915 | MGI:5826472 | lncRNA gene | predicted gene, 46835 |
| 3 | gene | 48.60573 | 48.60910 | negative | MGI_C57BL6J_1913582 | 1700018B24Rik | NCBI_Gene:66332,ENSEMBL:ENSMUSG00000037910 | MGI:1913582 | lncRNA gene | RIKEN cDNA 1700018B24 gene |
R version 3.6.2 (2019-12-12)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS Catalina 10.15.7
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRlapack.dylib
locale:
[1] en_AU.UTF-8/en_AU.UTF-8/en_AU.UTF-8/C/en_AU.UTF-8/en_AU.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] abind_1.4-5 qtl2_0.22 reshape2_1.4.4 ggplot2_3.3.5
[5] tibble_3.1.2 psych_2.0.7 readxl_1.3.1 cluster_2.1.0
[9] dplyr_0.8.5 optparse_1.6.6 rhdf5_2.28.1 mclust_5.4.6
[13] tidyr_1.0.2 data.table_1.14.0 knitr_1.33 kableExtra_1.1.0
[17] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] httr_1.4.1 bit64_4.0.5 viridisLite_0.4.0 assertthat_0.2.1
[5] highr_0.9 blob_1.2.1 cellranger_1.1.0 yaml_2.2.1
[9] pillar_1.6.1 RSQLite_2.2.7 backports_1.2.1 lattice_0.20-38
[13] glue_1.4.2 digest_0.6.27 promises_1.1.0 rvest_0.3.5
[17] colorspace_2.0-2 htmltools_0.5.1.1 httpuv_1.5.2 plyr_1.8.6
[21] pkgconfig_2.0.3 purrr_0.3.4 scales_1.1.1 webshot_0.5.2
[25] whisker_0.4 getopt_1.20.3 later_1.0.0 git2r_0.26.1
[29] ellipsis_0.3.2 cachem_1.0.5 withr_2.4.2 mnormt_1.5-7
[33] magrittr_2.0.1 crayon_1.4.1 memoise_2.0.0 evaluate_0.14
[37] fs_1.4.1 fansi_0.5.0 nlme_3.1-142 xml2_1.3.1
[41] tools_3.6.2 hms_0.5.3 lifecycle_1.0.0 stringr_1.4.0
[45] Rhdf5lib_1.6.3 munsell_0.5.0 compiler_3.6.2 rlang_0.4.11
[49] grid_3.6.2 rstudioapi_0.13 rmarkdown_2.1 gtable_0.3.0
[53] DBI_1.1.1 R6_2.5.0 fastmap_1.1.0 bit_4.0.4
[57] utf8_1.2.1 rprojroot_1.3-2 readr_1.3.1 stringi_1.7.2
[61] parallel_3.6.2 Rcpp_1.0.7 vctrs_0.3.8 tidyselect_1.0.0
[65] xfun_0.24