QTL Analysis - binary [ICI vs EOI]
Belinda Cornes
2022-03-09
Last updated: 2022-03-09
Checks: 6 1
Knit directory: Serreze-T1D_Workflow/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20220210) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /Users/corneb/Documents/MyJax/CS/Projects/Serreze/qc/workflowr/Serreze-T1D_Workflow | . |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 46d3df1. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: analysis/.DS_Store
Untracked files:
Untracked: data/GM_covar.csv
Untracked: data/bad_markers_all_4.batches.RData
Untracked: data/covar_cleaned_ici.vs.eoi.csv
Untracked: data/covar_cleaned_ici.vs.pbs.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi.csv
Untracked: data/covar_corrected.cleaned_ici.vs.eoi1.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs.csv
Untracked: data/covar_corrected.cleaned_ici.vs.pbs1.csv
Untracked: data/covar_corrected_ici.vs.eoi.csv
Untracked: data/covar_corrected_ici.vs.eoi1.csv
Untracked: data/covar_corrected_ici.vs.pbs.csv
Untracked: data/covar_corrected_ici.vs.pbs1.csv
Untracked: data/e.RData
Untracked: data/e_snpg_samqc_4.batches.RData
Untracked: data/e_snpg_samqc_4.batches_bc.RData
Untracked: data/errors_ind_4.batches.RData
Untracked: data/errors_ind_4.batches_bc.RData
Untracked: data/genetic_map.csv
Untracked: data/genotype_errors_marker_4.batches.RData
Untracked: data/genotype_freq_marker_4.batches.RData
Untracked: data/gm_allqc_4.batches.RData
Untracked: data/gm_samqc_3.batches.RData
Untracked: data/gm_samqc_4.batches.RData
Untracked: data/gm_samqc_4.batches_bc.RData
Untracked: data/gm_serreze.192.RData
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio.csv
Untracked: data/ici.vs.eoi_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_geno.ratio.csv
Untracked: data/ici.vs.eoi_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.geno.freq.removed.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.probs.freq.removed.csv
Untracked: data/ici.vs.eoi_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_geno.ratio.csv
Untracked: data/ici.vs.pbs_marker.freq_low.geno.freq.removed_sample.outliers.removed_geno.ratio.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_geno.ratio.csv
Untracked: data/ici.vs.pbs_marker.freq_low.probs.freq.removed_sample.outliers.removed_geno.ratio.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.geno.freq.removed_sample.outliers.removed.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.probs.freq.removed.csv
Untracked: data/ici.vs.pbs_sample.genos_marker.freq_low.probs.freq.removed_sample.outliers.removed.csv
Untracked: data/percent_missing_id_3.batches.RData
Untracked: data/percent_missing_id_4.batches.RData
Untracked: data/percent_missing_id_4.batches_bc.RData
Untracked: data/percent_missing_marker_4.batches.RData
Untracked: data/pheno.csv
Untracked: data/physical_map.csv
Untracked: data/qc_info_bad_sample_3.batches.RData
Untracked: data/qc_info_bad_sample_4.batches.RData
Untracked: data/qc_info_bad_sample_4.batches_bc.RData
Untracked: data/remaining.markers_geno.freq.xlsx
Untracked: data/sample_geno.csv
Untracked: data/sample_geno_bc.csv
Untracked: data/serreze_probs.rds
Untracked: data/serreze_probs_allqc.rds
Untracked: data/summary.cg_3.batches.RData
Untracked: data/summary.cg_4.batches.RData
Untracked: data/summary.cg_4.batches_bc.RData
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/4.1.1_qtl.analysis_binary_ici.vs.eoi_snpsqc_dis.Rmd) and HTML (docs/4.1.1_qtl.analysis_binary_ici.vs.eoi_snpsqc_dis.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 46d3df1 | Belinda Cornes | 2022-03-09 | no xcovar or kinship |
Data Information
- binary phenotype (therefore no kinship)
- low frequency (< 5% in samples) removed
- disproportionate markers removed
Loading Data
We will load the data and subset indivials out that are in the groups of interest. We will create a binary phenotype from this (EOI ==0, ICI == 1).
load("data/gm_allqc_4.batches.RData")
#gm_allqc
gm=gm_allqc
gmObject of class cross2 (crosstype "bc")
Total individuals 188
No. genotyped individuals 188
No. phenotyped individuals 188
No. with both geno & pheno 188
No. phenotypes 1
No. covariates 6
No. phenotype covariates 0
No. chromosomes 20
Total markers 131578
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13
9977 10005 7858 7589 7621 7758 7413 6472 6725 6396 7154 6137 6085
14 15 16 17 18 19 X
5981 5346 5019 5093 4607 3564 4778 #pr <- readRDS("data/serreze_probs_allqc.rds")
#pr <- readRDS("data/serreze_probs.rds")
##extracting animals with ici and eoi group status
miceinfo <- gm$covar[gm$covar$group == "EOI" | gm$covar$group == "ICI",]
table(miceinfo$group)
EOI ICI
69 92 mice.ids <- rownames(miceinfo)
gm <- gm[mice.ids]
gmObject of class cross2 (crosstype "bc")
Total individuals 161
No. genotyped individuals 161
No. phenotyped individuals 161
No. with both geno & pheno 161
No. phenotypes 1
No. covariates 6
No. phenotype covariates 0
No. chromosomes 20
Total markers 131578
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13
9977 10005 7858 7589 7621 7758 7413 6472 6725 6396 7154 6137 6085
14 15 16 17 18 19 X
5981 5346 5019 5093 4607 3564 4778 table(gm$covar$group)
EOI ICI
69 92 gm$covar$ICI.vs.EOI <- ifelse(gm$covar$group == "EOI", 0, 1)
covars <- read_csv("data/covar_corrected.cleaned_ici.vs.eoi.csv")
#removing any missing info
covars <- subset(covars, covars$ICI.vs.EOI!='')
nrow(covars)[1] 161table(covars$group)
EOI ICI
69 92 #keeping only informative mice
gm <- gm[covars$Mouse.ID]
gmObject of class cross2 (crosstype "bc")
Total individuals 161
No. genotyped individuals 161
No. phenotyped individuals 161
No. with both geno & pheno 161
No. phenotypes 1
No. covariates 7
No. phenotype covariates 0
No. chromosomes 20
Total markers 131578
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13
9977 10005 7858 7589 7621 7758 7413 6472 6725 6396 7154 6137 6085
14 15 16 17 18 19 X
5981 5346 5019 5093 4607 3564 4778 table(gm$covar$group)
EOI ICI
69 92 #pr.qc.ids <- pr
#for (i in 1:20){pr.qc.ids[[i]] = pr.qc.ids[[i]][covars$Mouse.ID,,]}
##dropping monomorphic markers within the dataset
g <- do.call("cbind", gm$geno)
gf_mar <- t(apply(g, 2, function(a) table(factor(a, 1:2))/sum(a != 0)))
#gn_mar <- t(apply(g, 2, function(a) table(factor(a, 1:2))))
gf_mar <- gf_mar[gf_mar[,2] != "NaN",]
count <- rowSums(gf_mar <=0.05)
low_freq_df <- merge(as.data.frame(gf_mar),as.data.frame(count), by="row.names",all=T)
low_freq_df[is.na(low_freq_df)] <- ''
low_freq_df <- low_freq_df[low_freq_df$count == 1,]
rownames(low_freq_df) <- low_freq_df$Row.names
low_freq <- find_markerpos(gm, rownames(low_freq_df))
low_freq$id <- rownames(low_freq)
nrow(low_freq)[1] 99077low_freq_bad <- merge(low_freq,low_freq_df, by="row.names",all=T)
names(low_freq_bad)[1] <- c("marker")
gf_mar <- gf_mar[gf_mar[,2] != "NaN",]
MAF <- apply(gf_mar, 1, function(x) min(x))
MAF <- as.data.frame(MAF)
MAF$index <- 1:nrow(gf_mar)
gf_mar_maf <- merge(gf_mar,as.data.frame(MAF), by="row.names")
gf_mar_maf <- gf_mar_maf[order(gf_mar_maf$index),]
gfmar <- NULL
gfmar$gfmar_mar_0 <- sum(gf_mar_maf$MAF==0)
gfmar$gfmar_mar_1 <- sum(gf_mar_maf$MAF< 0.01)
gfmar$gfmar_mar_5 <- sum(gf_mar_maf$MAF< 0.05)
gfmar$gfmar_mar_10 <- sum(gf_mar_maf$MAF< 0.10)
gfmar$gfmar_mar_15 <- sum(gf_mar_maf$MAF< 0.15)
gfmar$gfmar_mar_25 <- sum(gf_mar_maf$MAF< 0.25)
gfmar$gfmar_mar_50 <- sum(gf_mar_maf$MAF< 0.50)
gfmar$total_snps <- nrow(as.data.frame(gf_mar_maf))
gfmar <- t(as.data.frame(gfmar))
gfmar <- as.data.frame(gfmar)
gfmar$count <- gfmar$V1
gfmar[c(2)] %>%
kable(escape = F,align = c("ccccccccc"),linesep ="\\hline") %>%
kable_styling(full_width = F) %>%
kable_styling("striped", full_width = F) %>%
row_spec(8 ,bold=T,color= "white",background = "black")| count | |
|---|---|
| gfmar_mar_0 | 89667 |
| gfmar_mar_1 | 92615 |
| gfmar_mar_5 | 99045 |
| gfmar_mar_10 | 99502 |
| gfmar_mar_15 | 99619 |
| gfmar_mar_25 | 100097 |
| gfmar_mar_50 | 131139 |
| total_snps | 131578 |
gm_qc <- drop_markers(gm, low_freq_bad$marker)
gm_qc <- drop_nullmarkers(gm_qc)
gm_qcObject of class cross2 (crosstype "bc")
Total individuals 161
No. genotyped individuals 161
No. phenotyped individuals 161
No. with both geno & pheno 161
No. phenotypes 1
No. covariates 7
No. phenotype covariates 0
No. chromosomes 20
Total markers 32501
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
2940 2853 2033 2045 1931 2041 1858 1677 1991 1208 2057 1387 1617 1685 1194 922
17 18 19 X
382 1073 1052 555 ## dropping disproportionate markers
dismark <- read.csv("data/ici.vs.eoi_marker.freq_low.geno.freq.removed_geno.ratio.csv")
nrow(dismark)[1] 32501names(dismark)[1] <- c("marker")
dismark <- dismark[!dismark$Include,]
nrow(dismark)[1] 18736gm_qc_dis <- drop_markers(gm_qc, dismark$marker)
gm_qc_dis <- drop_nullmarkers(gm_qc_dis)
gm = gm_qc_dis
gmObject of class cross2 (crosstype "bc")
Total individuals 161
No. genotyped individuals 161
No. phenotyped individuals 161
No. with both geno & pheno 161
No. phenotypes 1
No. covariates 7
No. phenotype covariates 0
No. chromosomes 19
Total markers 13765
No. markers by chr:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 16 17
2142 1860 1022 570 688 514 1060 294 772 205 718 11 1270 1105 490 291
18 19 X
494 208 51 markers <- marker_names(gm)
gmapdf <- read.csv("/Users/corneb/Documents/MyJax/CS/Projects/Serreze/haplotype.reconstruction/output_hh/genetic_map.csv")
pmapdf <- read.csv("/Users/corneb/Documents/MyJax/CS/Projects/Serreze/haplotype.reconstruction/output_hh/physical_map.csv")
mapdf <- merge(gmapdf,pmapdf, by=c("marker","chr"), all=T)
rownames(mapdf) <- mapdf$marker
mapdf <- mapdf[markers,]
names(mapdf) <- c('marker','chr','gmapdf','pmapdf')
mapdfnd <- mapdf[!duplicated(mapdf[c(2:3)]),]
pr.qc <- calc_genoprob(gm)Genome-wide scan
For each of the phenotype analyzed, permutations were used for each model to obtain genome-wide LOD significance threshold for p < 0.01, p < 0.05, p < 0.10, respectively, separately for X and automsomes (A).
The table shows the estimated significance thresholds from permutation test.
We also looked at the kinship to see how correlated each sample is. Kinship values between pairs of samples range between 0 (no relationship) and 1.0 (completely identical). The darker the colour the more indentical the pairs are.
Xcovar <- get_x_covar(gm)
#addcovar = model.matrix(~Sex, data = covars)[,-1]
#K <- calc_kinship(pr.qc, type = "loco")
#heatmap(K[[1]])
#K.overall <- calc_kinship(pr.qc, type = "overall")
#heatmap(K.overall)
kinship <- calc_kinship(pr.qc)
heatmap(kinship)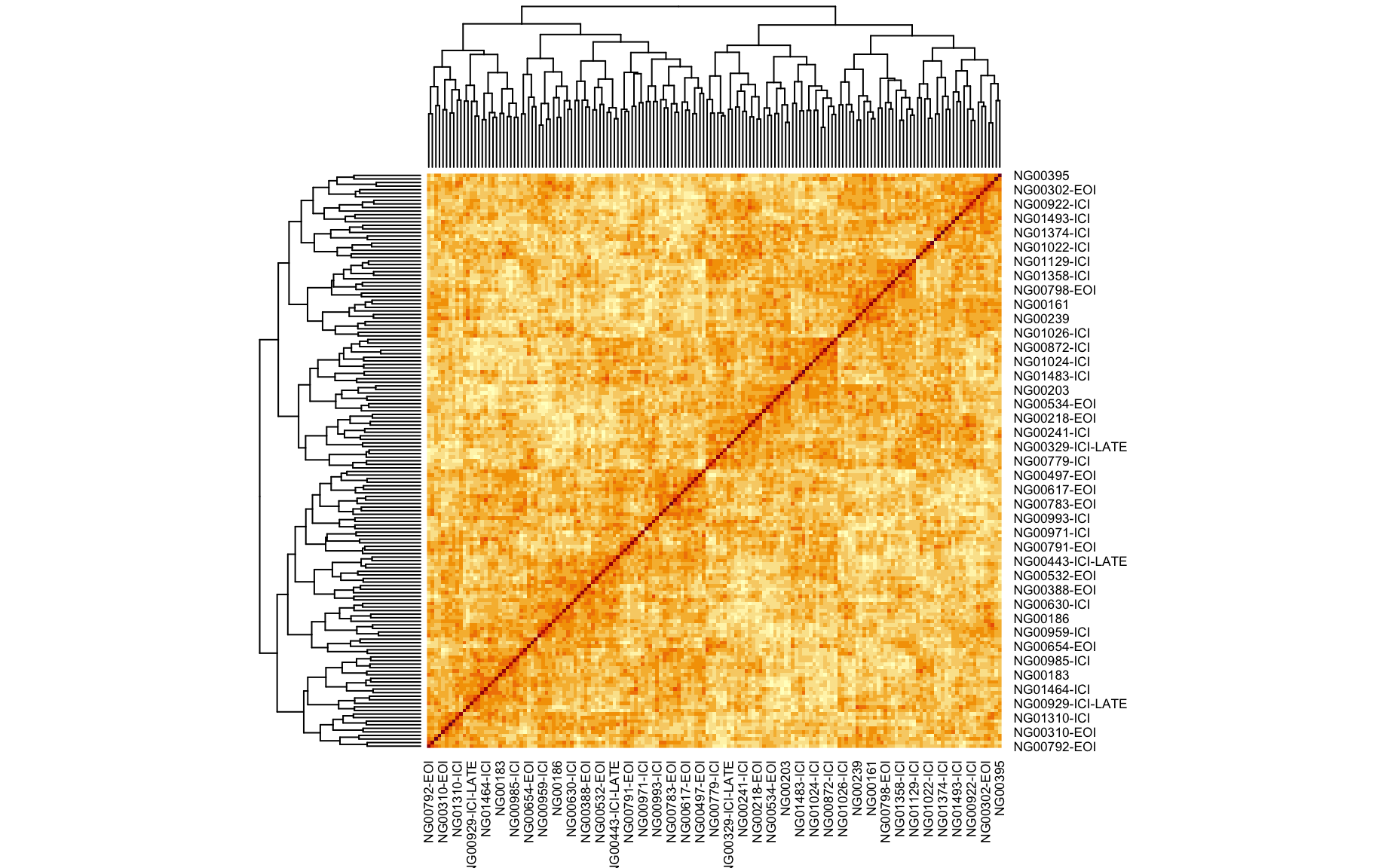
#operm <- scan1perm(pr.qc, gm$covar$phenos, Xcovar=Xcovar, n_perm=2000)
#operm <- scan1perm(pr.qc, gm$covar$phenos, addcovar = addcovar, n_perm=2000)
#operm <- scan1perm(pr.qc, gm$covar$phenos, n_perm=2000)
operm <- scan1perm(pr.qc, gm$covar["ICI.vs.EOI"], model="binary", n_perm=10, perm_Xsp=TRUE, chr_lengths=chr_lengths(gm$gmap))
summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
names(summary_table) <- c("autosomes","X")
summary_table$significance.level <- rownames(summary_table)
rownames(summary_table) <- NULL
summary_table[c(3,1:2)] %>%
kable(escape = F,align = c("ccc")) %>%
kable_styling("striped", full_width = T) %>%
column_spec(1, bold=TRUE)| significance.level | autosomes | X |
|---|---|---|
| 0.01 | 3.934232 | 2.903277 |
| 0.05 | 3.381052 | 2.591771 |
| 0.1 | 2.688646 | 2.196548 |
The figures below show QTL maps for each phenotype
out <- scan1(pr.qc, gm$covar["ICI.vs.EOI"], Xcovar=Xcovar, model="binary")
summary_table<-data.frame(unclass(summary(operm, alpha=c(0.01, 0.05, 0.1))))
plot_lod<-function(out,map){
for (i in 1:dim(out)[2]){
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/",colnames(out)[i], "_lod.png"))
#par(mar=c(5.1, 6.1, 1.1, 1.1))
ymx <- maxlod(out) # overall maximum LOD score
plot(out, gm$gmap, lodcolumn=i, col="slateblue", ylim=c(0, ymx+0.5))
legend("topright", lwd=2, colnames(out)[i], bg="gray90")
title(main = colnames(out)[i])
#for (j in 1: dim(summary_table)[1]){
# abline(h=summary_table[j, i],col="red")
# text(x=400, y =summary_table[j, i]+0.12, labels = paste("p=", row.names(summary_table)[j]))
#}
#dev.off()
}
}
plot_lod(out,gm$gmap)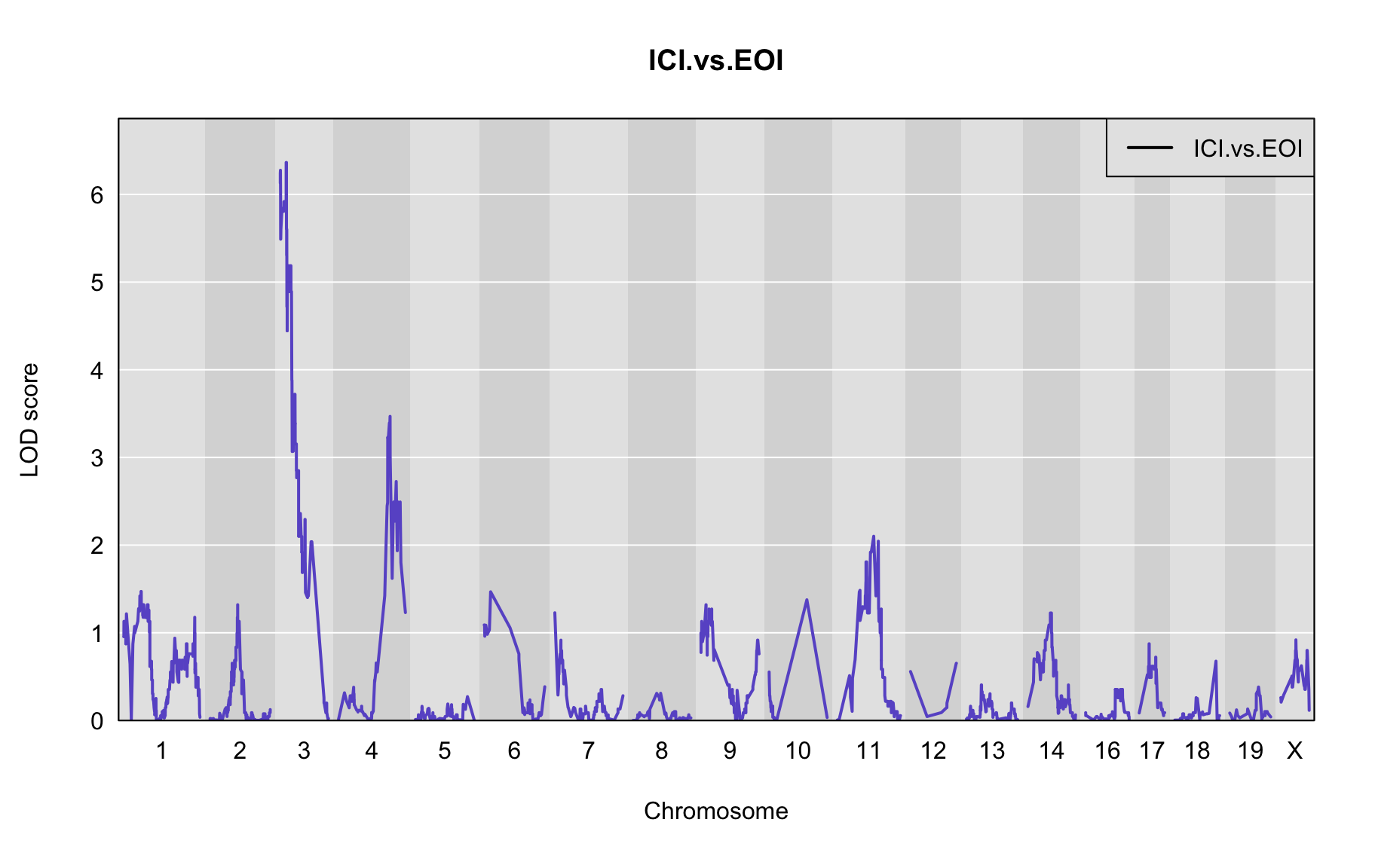
LOD peaks
The table below shows QTL peaks associated with the phenotype. We use the 95% threshold from the permutations to find peaks.
peaks<-find_peaks(out, gm$gmap, threshold=4, drop=1.5)
rownames(peaks) <- NULL
peaks[] %>%
kable(escape = F,align = c("ccccccc")) %>%
kable_styling("striped", full_width = T) %>%
column_spec(1, bold=TRUE)| lodindex | lodcolumn | chr | pos | lod | ci_lo | ci_hi |
|---|---|---|---|---|---|---|
| 1 | ICI.vs.EOI | 3 | 29.038 | 6.364394 | 21.228 | 35.891 |
peaks_mbl <- list()
#corresponding info in Mb
for(i in 1:nrow(peaks)){
lodindex <- peaks$lodindex[i]
lodcolumn <- peaks$lodcolumn[i]
chr <- peaks$chr[i]
lod <- peaks$lod[i]
pos <- mapdf$pmapdf[which(mapdf$gmapdf == peaks$pos[i] & mapdf$chr == peaks$chr[i])]
ci_lo <- mapdf$pmapdf[which(mapdf$gmapdf == peaks$ci_lo[i] & mapdf$chr == peaks$chr[i])]
ci_hi <- mapdf$pmapdf[which(mapdf$gmapdf == peaks$ci_hi[i] & mapdf$chr == peaks$chr[i])]
peaks_mb=cbind(lodindex, lodcolumn, chr, pos, lod, ci_lo, ci_hi)
peaks_mbl[[i]] <- peaks_mb
}
peaks_mba <- do.call(rbind, peaks_mbl)
peaks_mba <- as.data.frame(peaks_mba)
#peaks_mba[,c("chr", "pos", "lod", "ci_lo", "ci_hi")] <- sapply(peaks_mba[,c("chr", "pos", "lod", "ci_lo", "ci_hi")], as.numeric)
rownames(peaks_mba) <- NULL
peaks_mba[] %>%
kable(escape = F,align = c("ccccccc")) %>%
kable_styling("striped", full_width = T) %>%
column_spec(1, bold=TRUE)| lodindex | lodcolumn | chr | pos | lod | ci_lo | ci_hi |
|---|---|---|---|---|---|---|
| 1 | ICI.vs.EOI | 3 | 59.429352 | 6.36439381654824 | 48.232305 | 81.503929 |
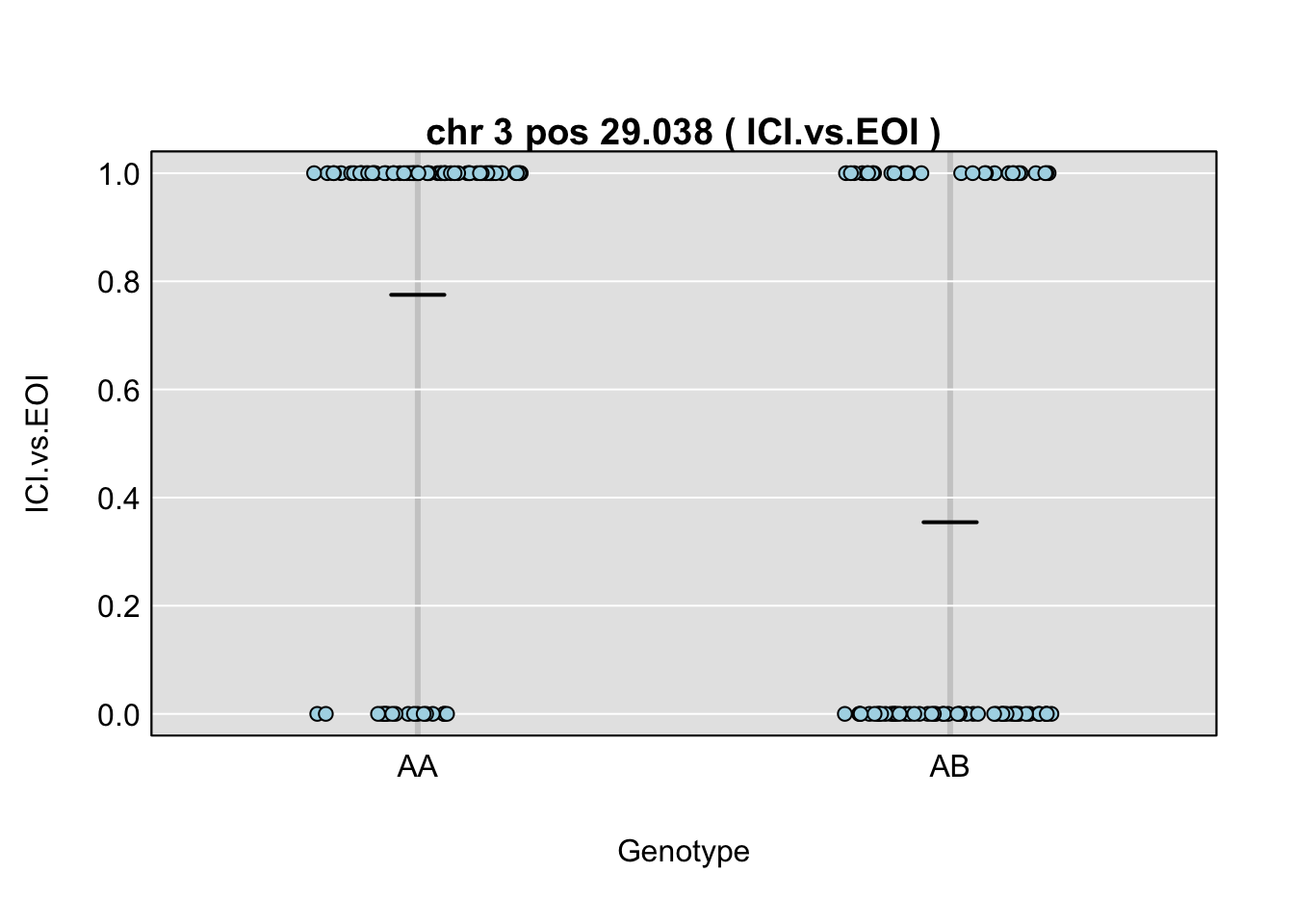
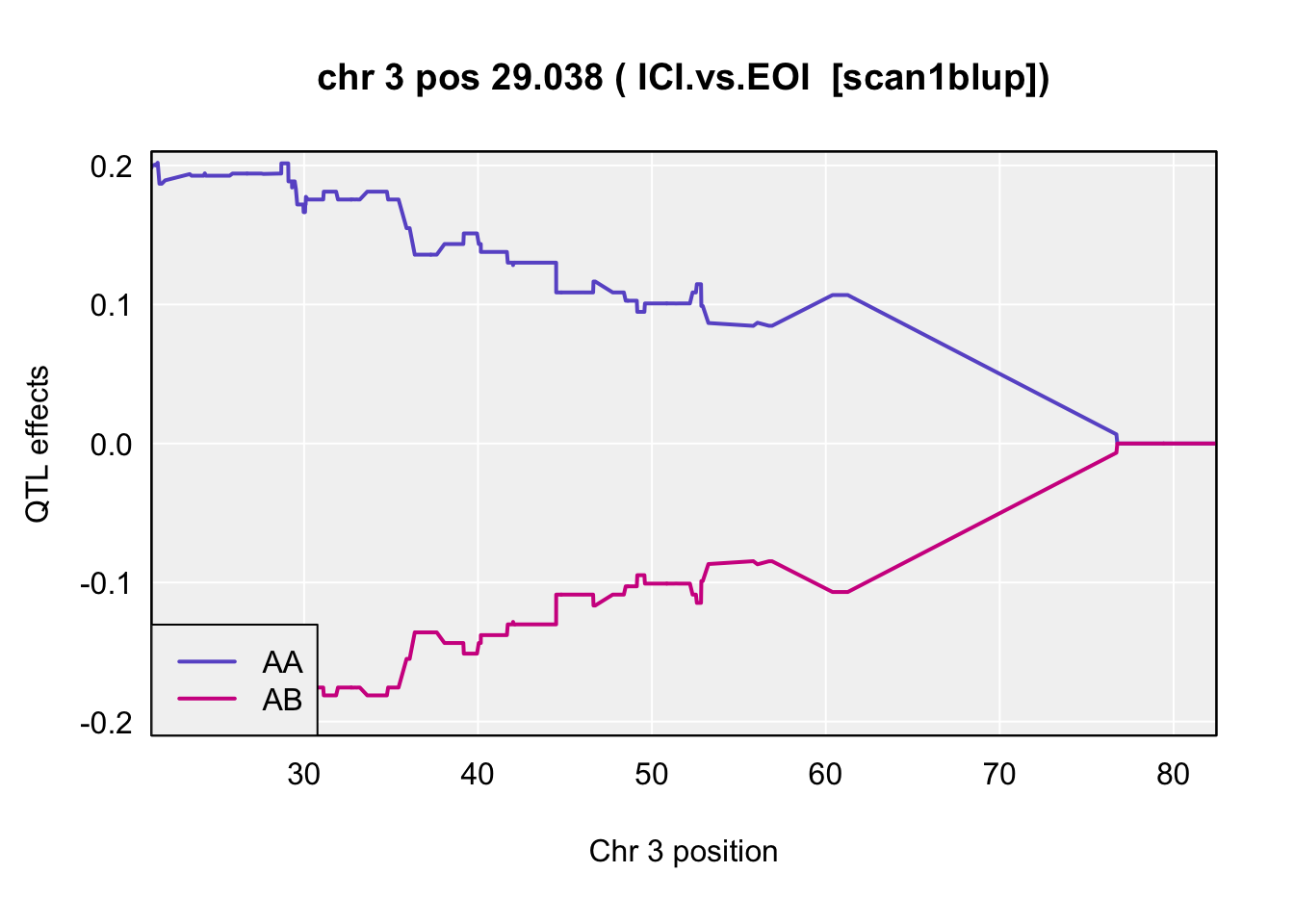
QTL effects
For each peak LOD location we give a list of gene
query_variants <- create_variant_query_func("/Users/corneb/Documents/MyJax/CS/Projects/support.files/qtl2/cc_variants.sqlite")
query_genes <- create_gene_query_func("/Users/corneb/Documents/MyJax/CS/Projects/support.files/qtl2/mouse_genes_mgi.sqlite")
for (i in 1:nrow(peaks)){
#for (i in 1:1){
#Plot 1
marker = find_marker(gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i])
#g <- maxmarg(pr.qc, gm$gmap, chr=peaks$chr[i], pos=peaks$pos[i], return_char=TRUE, minprob = 0.5)
gp <- g[,marker]
gp[gp==1] <- "AA"
gp[gp==2] <- "AB"
gp[gp==0] <- NA
#png(filename=paste0("/Users/chenm/Documents/qtl/Jai/","qtl_effect_", i, ".png"))
#par(mar=c(4.1, 4.1, 1.5, 0.6))
plot_pxg(gp, gm$covar[,peaks$lodcolumn[i]], ylab=peaks$lodcolumn[i], sort=FALSE)
title(main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")"), line=0.2)
##dev.off()
chr = peaks$chr[i]
# Plot 2
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar)
#coeff <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], Xcovar=Xcovar)
coeff <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary")
blup <- scan1blup(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]])
#plot_coef(coeff,
# gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i]," [scan1coeff])")
# )
plot_coef(blup,
gm$gmap[chr], columns=1:2,
bgcolor="gray95", legend="bottomleft",
main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i]," [scan1blup])")
)
#last_coef <- unclass(coeff)[nrow(coeff),1:3]
#for(t in seq(along=last_coef))
#axis(side=4, at=last_coef[t], names(last_coef)[t], tick=FALSE)
# Plot 3
#c2effB <- scan1coef(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], model="binary", contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
#c2effBb <- scan1blup(pr.qc[,chr], gm$covar[peaks$lodcolumn[i]], contrasts=cbind(a=c(-1, 0), d=c(0, -1)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]], addcovar = addcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
##c2effB <- scan1coef(pr[,chr], cross$pheno[,peaks$lodcolumn[i]],Xcovar=Xcovar, contrasts=cbind(mu=c(1,1,1), a=c(-1, 0, 1), d=c(0, 1, 0)))
#plot(c2effB, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
#plot(c2effBb, gm$gmap[chr], columns=1:2,
# bgcolor="gray95", legend="bottomleft",
# main = paste("chr", chr=peaks$chr[i], "pos", peaks$pos[i], "(",peaks$lodcolumn[i],")")
# )
##last_coef <- unclass(c2effB)[nrow(c2effB),2:3] # last two coefficients
##for(t in seq(along=last_coef))
## axis(side=4, at=last_coef[t], names(last_coef)[t], tick=FALSE)
#Table 1
chr = peaks_mba$chr[i]
start=as.numeric(peaks_mba$ci_lo[i])
end=as.numeric(peaks_mba$ci_hi[i])
genesgss = query_genes(chr, start, end)
rownames(genesgss) <- NULL
genesgss$strand_old = genesgss$strand
genesgss$strand[genesgss$strand=="+"] <- "positive"
genesgss$strand[genesgss$strand=="-"] <- "negative"
#genesgss <-
#table <-
#genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")] %>%
#kable(escape = F,align = c("ccccccccccc")) %>%
#kable_styling("striped", full_width = T) #%>%
#cat #%>%
#column_spec(1, bold=TRUE)
#
#print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], escape = F,align = c("ccccccccccc")))
print(kable(genesgss[,c("chr","type","start","stop","strand","ID","Name","Dbxref","gene_id","mgi_type","description")], "html") %>% kable_styling("striped", full_width = T))
#table
}
| chr | type | start | stop | strand | ID | Name | Dbxref | gene_id | mgi_type | description |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | gene | 48.54642 | 48.57947 | positive | MGI_C57BL6J_5826472 | Gm46835 | NCBI_Gene:108168915 | MGI:5826472 | lncRNA gene | predicted gene, 46835 |
| 3 | gene | 48.60573 | 48.60910 | negative | MGI_C57BL6J_1913582 | 1700018B24Rik | NCBI_Gene:66332,ENSEMBL:ENSMUSG00000037910 | MGI:1913582 | lncRNA gene | RIKEN cDNA 1700018B24 gene |
| 3 | gene | 48.74851 | 48.76095 | negative | MGI_C57BL6J_5610418 | Gm37190 | ENSEMBL:ENSMUSG00000102181 | MGI:5610418 | lncRNA gene | predicted gene, 37190 |
| 3 | gene | 48.95472 | 48.95488 | positive | MGI_C57BL6J_5453288 | Gm23511 | ENSEMBL:ENSMUSG00000065018 | MGI:5453288 | snRNA gene | predicted gene, 23511 |
| 3 | gene | 49.06237 | 49.09636 | positive | MGI_C57BL6J_5590574 | Gm31415 | NCBI_Gene:102633632,ENSEMBL:ENSMUSG00000102263 | MGI:5590574 | lncRNA gene | predicted gene, 31415 |
| 3 | gene | 49.13746 | 49.13853 | positive | MGI_C57BL6J_5611047 | Gm37819 | ENSEMBL:ENSMUSG00000104186 | MGI:5611047 | unclassified gene | predicted gene, 37819 |
| 3 | pseudogene | 49.20771 | 49.20807 | negative | MGI_C57BL6J_5610912 | Gm37684 | ENSEMBL:ENSMUSG00000103027 | MGI:5610912 | pseudogene | predicted gene, 37684 |
| 3 | gene | 49.32568 | 49.36736 | positive | MGI_C57BL6J_5295664 | Gm20557 | NCBI_Gene:329633,ENSEMBL:ENSMUSG00000102679 | MGI:5295664 | lncRNA gene | predicted gene, 20557 |
| 3 | pseudogene | 49.38115 | 49.38285 | negative | MGI_C57BL6J_3643624 | Gm5846 | NCBI_Gene:545519,ENSEMBL:ENSMUSG00000104309 | MGI:3643624 | pseudogene | predicted gene 5846 |
| 3 | pseudogene | 49.40082 | 49.46744 | negative | MGI_C57BL6J_3780091 | Gm9683 | NCBI_Gene:676568 | MGI:3780091 | pseudogene | predicted gene 9683 |
| 3 | gene | 49.49135 | 49.49148 | positive | MGI_C57BL6J_5452257 | Gm22480 | ENSEMBL:ENSMUSG00000084517 | MGI:5452257 | snoRNA gene | predicted gene, 22480 |
| 3 | pseudogene | 49.53075 | 49.53131 | positive | MGI_C57BL6J_5610652 | Gm37424 | ENSEMBL:ENSMUSG00000104437 | MGI:5610652 | pseudogene | predicted gene, 37424 |
| 3 | pseudogene | 49.60356 | 49.60388 | positive | MGI_C57BL6J_5611489 | Gm38261 | ENSEMBL:ENSMUSG00000102731 | MGI:5611489 | pseudogene | predicted gene, 38261 |
| 3 | pseudogene | 49.60423 | 49.60447 | positive | MGI_C57BL6J_5611316 | Gm38088 | ENSEMBL:ENSMUSG00000103249 | MGI:5611316 | pseudogene | predicted gene, 38088 |
| 3 | pseudogene | 49.63244 | 49.63297 | negative | MGI_C57BL6J_3780498 | Gm2328 | NCBI_Gene:100039587,ENSEMBL:ENSMUSG00000104134 | MGI:3780498 | pseudogene | predicted gene 2328 |
| 3 | pseudogene | 49.64348 | 49.64413 | positive | MGI_C57BL6J_5588900 | Gm29741 | NCBI_Gene:101055903,ENSEMBL:ENSMUSG00000103568 | MGI:5588900 | pseudogene | predicted gene, 29741 |
| 3 | gene | 49.65876 | 49.65887 | positive | MGI_C57BL6J_5452873 | Gm23096 | ENSEMBL:ENSMUSG00000065381 | MGI:5452873 | snRNA gene | predicted gene, 23096 |
| 3 | gene | 49.74329 | 49.75738 | negative | MGI_C57BL6J_1920423 | Pcdh18 | NCBI_Gene:73173,ENSEMBL:ENSMUSG00000037892 | MGI:1920423 | protein coding gene | protocadherin 18 |
| 3 | pseudogene | 49.78043 | 49.78089 | negative | MGI_C57BL6J_5610778 | Gm37550 | ENSEMBL:ENSMUSG00000102943 | MGI:5610778 | pseudogene | predicted gene, 37550 |
| 3 | gene | 49.89253 | 50.49909 | negative | MGI_C57BL6J_1347355 | Slc7a11 | NCBI_Gene:26570,ENSEMBL:ENSMUSG00000027737 | MGI:1347355 | protein coding gene | solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
| 3 | gene | 50.00197 | 50.01078 | positive | MGI_C57BL6J_5611082 | Gm37854 | ENSEMBL:ENSMUSG00000102892 | MGI:5611082 | lncRNA gene | predicted gene, 37854 |
| 3 | gene | 50.01177 | 50.02624 | negative | MGI_C57BL6J_5622927 | Gm40042 | NCBI_Gene:105244434 | MGI:5622927 | lncRNA gene | predicted gene, 40042 |
| 3 | pseudogene | 50.02967 | 50.02999 | positive | MGI_C57BL6J_5611054 | Gm37826 | ENSEMBL:ENSMUSG00000104457 | MGI:5611054 | pseudogene | predicted gene, 37826 |
| 3 | pseudogene | 50.16666 | 50.16689 | negative | MGI_C57BL6J_5610920 | Gm37692 | ENSEMBL:ENSMUSG00000104349 | MGI:5610920 | pseudogene | predicted gene, 37692 |
| 3 | pseudogene | 50.25485 | 50.25544 | positive | MGI_C57BL6J_3780515 | Gm2345 | NCBI_Gene:100039628,ENSEMBL:ENSMUSG00000102167 | MGI:3780515 | pseudogene | predicted gene 2345 |
| 3 | gene | 50.40028 | 50.40428 | negative | MGI_C57BL6J_5610726 | Gm37498 | ENSEMBL:ENSMUSG00000103907 | MGI:5610726 | unclassified gene | predicted gene, 37498 |
| 3 | gene | 50.51640 | 50.60731 | negative | MGI_C57BL6J_3643374 | Gm6209 | NCBI_Gene:621304,ENSEMBL:ENSMUSG00000102715 | MGI:3643374 | lncRNA gene | predicted gene 6209 |
| 3 | gene | 50.57519 | 50.57652 | negative | MGI_C57BL6J_1923259 | 5830415G21Rik | ENSEMBL:ENSMUSG00000102895 | MGI:1923259 | unclassified gene | RIKEN cDNA 5830415G21 gene |
| 3 | pseudogene | 50.59799 | 50.59850 | negative | MGI_C57BL6J_5611465 | Gm38237 | NCBI_Gene:108168851,ENSEMBL:ENSMUSG00000103745 | MGI:5611465 | pseudogene | predicted gene, 38237 |
| 3 | gene | 50.62435 | 50.62841 | negative | MGI_C57BL6J_5610427 | Gm37199 | ENSEMBL:ENSMUSG00000104068 | MGI:5610427 | unclassified gene | predicted gene, 37199 |
| 3 | gene | 50.67970 | 50.68340 | negative | MGI_C57BL6J_5610689 | Gm37461 | ENSEMBL:ENSMUSG00000104281 | MGI:5610689 | unclassified gene | predicted gene, 37461 |
| 3 | gene | 50.70690 | 50.71189 | negative | MGI_C57BL6J_5622928 | Gm40043 | NCBI_Gene:105244435 | MGI:5622928 | lncRNA gene | predicted gene, 40043 |
| 3 | gene | 50.83858 | 50.83872 | negative | MGI_C57BL6J_5454280 | Gm24503 | ENSEMBL:ENSMUSG00000089079 | MGI:5454280 | snoRNA gene | predicted gene, 24503 |
| 3 | gene | 50.88324 | 50.88889 | negative | MGI_C57BL6J_5610776 | Gm37548 | NCBI_Gene:105244436,ENSEMBL:ENSMUSG00000103012 | MGI:5610776 | lncRNA gene | predicted gene, 37548 |
| 3 | gene | 50.91889 | 50.92153 | negative | MGI_C57BL6J_5611071 | Gm37843 | ENSEMBL:ENSMUSG00000102614 | MGI:5611071 | unclassified gene | predicted gene, 37843 |
| 3 | gene | 50.96656 | 50.96677 | positive | MGI_C57BL6J_2139603 | AA672641 | NA | NA | unclassified gene | expressed sequence AA672641 |
| 3 | gene | 50.97754 | 50.97881 | negative | MGI_C57BL6J_5610437 | Gm37209 | ENSEMBL:ENSMUSG00000103652 | MGI:5610437 | unclassified gene | predicted gene, 37209 |
| 3 | gene | 51.03335 | 51.04284 | negative | MGI_C57BL6J_5594915 | Gm35756 | NCBI_Gene:102639442 | MGI:5594915 | lncRNA gene | predicted gene, 35756 |
| 3 | gene | 51.05891 | 51.08934 | negative | MGI_C57BL6J_5579936 | Gm29230 | NCBI_Gene:102639521,ENSEMBL:ENSMUSG00000102067 | MGI:5579936 | lncRNA gene | predicted gene 29230 |
| 3 | gene | 51.10366 | 51.10517 | positive | MGI_C57BL6J_5611474 | Gm38246 | ENSEMBL:ENSMUSG00000103464 | MGI:5611474 | lncRNA gene | predicted gene, 38246 |
| 3 | gene | 51.12669 | 51.12894 | negative | MGI_C57BL6J_5595050 | Gm35891 | NCBI_Gene:102639626 | MGI:5595050 | lncRNA gene | predicted gene, 35891 |
| 3 | gene | 51.21847 | 51.22033 | negative | MGI_C57BL6J_5622929 | Gm40044 | NCBI_Gene:105244437 | MGI:5622929 | lncRNA gene | predicted gene, 40044 |
| 3 | gene | 51.22445 | 51.25165 | positive | MGI_C57BL6J_109382 | Noct | NCBI_Gene:12457,ENSEMBL:ENSMUSG00000023087 | MGI:109382 | protein coding gene | nocturnin |
| 3 | gene | 51.23192 | 51.23438 | positive | MGI_C57BL6J_5611585 | Gm38357 | ENSEMBL:ENSMUSG00000103313 | MGI:5611585 | unclassified gene | predicted gene, 38357 |
| 3 | gene | 51.25272 | 51.34092 | negative | MGI_C57BL6J_1916507 | Elf2 | NCBI_Gene:69257,ENSEMBL:ENSMUSG00000037174 | MGI:1916507 | protein coding gene | E74-like factor 2 |
| 3 | gene | 51.27676 | 51.27841 | positive | MGI_C57BL6J_1914996 | 4930577N17Rik | NCBI_Gene:67746,ENSEMBL:ENSMUSG00000087440 | MGI:1914996 | lncRNA gene | RIKEN cDNA 4930577N17 gene |
| 3 | gene | 51.29718 | 51.29724 | positive | MGI_C57BL6J_5454059 | Gm24282 | ENSEMBL:ENSMUSG00000077420 | MGI:5454059 | miRNA gene | predicted gene, 24282 |
| 3 | gene | 51.30249 | 51.30290 | negative | MGI_C57BL6J_5610627 | Gm37399 | ENSEMBL:ENSMUSG00000104200 | MGI:5610627 | unclassified gene | predicted gene, 37399 |
| 3 | gene | 51.35835 | 51.35842 | positive | MGI_C57BL6J_4414019 | n-TPtgg7 | NCBI_Gene:102467437 | MGI:4414019 | tRNA gene | nuclear encoded tRNA proline 7 (anticodon TGG) |
| 3 | gene | 51.35941 | 51.35948 | positive | MGI_C57BL6J_4414007 | n-TPagg5 | NCBI_Gene:102467366 | MGI:4414007 | tRNA gene | nuclear encoded tRNA proline 5 (anticodon AGG) |
| 3 | pseudogene | 51.37022 | 51.37061 | positive | MGI_C57BL6J_5610431 | Gm37203 | ENSEMBL:ENSMUSG00000103654 | MGI:5610431 | pseudogene | predicted gene, 37203 |
| 3 | gene | 51.37802 | 51.38632 | positive | MGI_C57BL6J_3779459 | Gm5103 | NCBI_Gene:329636,ENSEMBL:ENSMUSG00000102544 | MGI:3779459 | lncRNA gene | predicted gene 5103 |
| 3 | gene | 51.38841 | 51.39674 | negative | MGI_C57BL6J_1914999 | Mgarp | NCBI_Gene:67749,ENSEMBL:ENSMUSG00000037161 | MGI:1914999 | protein coding gene | mitochondria localized glutamic acid rich protein |
| 3 | pseudogene | 51.40246 | 51.40320 | negative | MGI_C57BL6J_3779852 | Gm9442 | NCBI_Gene:669130,ENSEMBL:ENSMUSG00000104241 | MGI:3779852 | pseudogene | predicted gene 9442 |
| 3 | gene | 51.40468 | 51.40899 | negative | MGI_C57BL6J_1913627 | Ndufc1 | NCBI_Gene:66377,ENSEMBL:ENSMUSG00000037152 | MGI:1913627 | protein coding gene | NADH:ubiquinone oxidoreductase subunit C1 |
| 3 | gene | 51.41515 | 51.47651 | positive | MGI_C57BL6J_1922088 | Naa15 | NCBI_Gene:74838,ENSEMBL:ENSMUSG00000063273 | MGI:1922088 | protein coding gene | N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| 3 | gene | 51.42361 | 51.42372 | positive | MGI_C57BL6J_5610874 | Gm37646 | ENSEMBL:ENSMUSG00000103191 | MGI:5610874 | unclassified gene | predicted gene, 37646 |
| 3 | gene | 51.48233 | 51.48483 | negative | MGI_C57BL6J_5611388 | Gm38160 | ENSEMBL:ENSMUSG00000102775 | MGI:5611388 | unclassified gene | predicted gene, 38160 |
| 3 | gene | 51.48392 | 51.49623 | positive | MGI_C57BL6J_1330805 | Rab33b | NCBI_Gene:19338,ENSEMBL:ENSMUSG00000027739 | MGI:1330805 | protein coding gene | RAB33B, member RAS oncogene family |
| 3 | gene | 51.48962 | 51.49282 | positive | MGI_C57BL6J_5611196 | Gm37968 | ENSEMBL:ENSMUSG00000102570 | MGI:5611196 | unclassified gene | predicted gene, 37968 |
| 3 | gene | 51.51099 | 51.51430 | positive | MGI_C57BL6J_5595167 | Gm36008 | NCBI_Gene:102639777 | MGI:5595167 | lncRNA gene | predicted gene, 36008 |
| 3 | gene | 51.51532 | 51.56088 | negative | MGI_C57BL6J_1920501 | Setd7 | NCBI_Gene:73251,ENSEMBL:ENSMUSG00000037111 | MGI:1920501 | protein coding gene | SET domain containing (lysine methyltransferase) 7 |
| 3 | gene | 51.55754 | 51.55965 | negative | MGI_C57BL6J_5611428 | Gm38200 | ENSEMBL:ENSMUSG00000102466 | MGI:5611428 | unclassified gene | predicted gene, 38200 |
| 3 | gene | 51.55976 | 51.56712 | positive | MGI_C57BL6J_1923230 | 5031434O11Rik | NCBI_Gene:100039684,ENSEMBL:ENSMUSG00000097885 | MGI:1923230 | lncRNA gene | RIKEN cDNA 5031434O11 gene |
| 3 | pseudogene | 51.57111 | 51.57142 | negative | MGI_C57BL6J_5611562 | Gm38334 | ENSEMBL:ENSMUSG00000104001 | MGI:5611562 | pseudogene | predicted gene, 38334 |
| 3 | gene | 51.58231 | 51.58285 | positive | MGI_C57BL6J_1918620 | 5430433H01Rik | ENSEMBL:ENSMUSG00000104075 | MGI:1918620 | unclassified gene | RIKEN cDNA 5430433H01 gene |
| 3 | pseudogene | 51.63649 | 51.63705 | negative | MGI_C57BL6J_5611475 | Gm38247 | NCBI_Gene:108168852,ENSEMBL:ENSMUSG00000104166 | MGI:5611475 | pseudogene | predicted gene, 38247 |
| 3 | gene | 51.66036 | 51.68268 | positive | MGI_C57BL6J_2448481 | Mgst2 | NCBI_Gene:211666,ENSEMBL:ENSMUSG00000074604 | MGI:2448481 | protein coding gene | microsomal glutathione S-transferase 2 |
| 3 | gene | 51.67280 | 51.67390 | positive | MGI_C57BL6J_5610489 | Gm37261 | ENSEMBL:ENSMUSG00000103527 | MGI:5610489 | unclassified gene | predicted gene, 37261 |
| 3 | gene | 51.68591 | 52.10509 | negative | MGI_C57BL6J_2389461 | Maml3 | NCBI_Gene:433586,ENSEMBL:ENSMUSG00000061143 | MGI:2389461 | protein coding gene | mastermind like transcriptional coactivator 3 |
| 3 | gene | 51.69372 | 51.69509 | positive | MGI_C57BL6J_3642905 | Gm10729 | ENSEMBL:ENSMUSG00000074603 | MGI:3642905 | protein coding gene | predicted gene 10729 |
| 3 | gene | 51.70772 | 51.71225 | positive | MGI_C57BL6J_5826440 | Gm46803 | NCBI_Gene:108168869 | MGI:5826440 | lncRNA gene | predicted gene, 46803 |
| 3 | gene | 51.75712 | 51.76209 | negative | MGI_C57BL6J_5610570 | Gm37342 | ENSEMBL:ENSMUSG00000102732 | MGI:5610570 | unclassified gene | predicted gene, 37342 |
| 3 | gene | 51.81295 | 51.81931 | positive | MGI_C57BL6J_5610457 | Gm37229 | ENSEMBL:ENSMUSG00000102539 | MGI:5610457 | lncRNA gene | predicted gene, 37229 |
| 3 | gene | 51.82891 | 51.82903 | positive | MGI_C57BL6J_4422061 | n-R5s196 | ENSEMBL:ENSMUSG00000065070 | MGI:4422061 | rRNA gene | nuclear encoded rRNA 5S 196 |
| 3 | gene | 51.82939 | 51.83040 | negative | MGI_C57BL6J_1920491 | 3110080O07Rik | ENSEMBL:ENSMUSG00000103284 | MGI:1920491 | unclassified gene | RIKEN cDNA 3110080O07 gene |
| 3 | gene | 51.86735 | 51.86856 | negative | MGI_C57BL6J_5611228 | Gm38000 | ENSEMBL:ENSMUSG00000103484 | MGI:5611228 | unclassified gene | predicted gene, 38000 |
| 3 | gene | 51.87299 | 51.87584 | negative | MGI_C57BL6J_5610903 | Gm37675 | ENSEMBL:ENSMUSG00000102620 | MGI:5610903 | unclassified gene | predicted gene, 37675 |
| 3 | gene | 51.88428 | 51.88689 | negative | MGI_C57BL6J_5611480 | Gm38252 | ENSEMBL:ENSMUSG00000102786 | MGI:5611480 | unclassified gene | predicted gene, 38252 |
| 3 | pseudogene | 51.90699 | 51.90882 | negative | MGI_C57BL6J_3708672 | Gm10728 | ENSEMBL:ENSMUSG00000103701 | MGI:3708672 | pseudogene | predicted gene 10728 |
| 3 | gene | 51.98094 | 51.99007 | positive | MGI_C57BL6J_5012274 | Gm20089 | ENSEMBL:ENSMUSG00000104161 | MGI:5012274 | lncRNA gene | predicted gene, 20089 |
| 3 | gene | 51.98763 | 51.98985 | negative | MGI_C57BL6J_5611283 | Gm38055 | ENSEMBL:ENSMUSG00000102147 | MGI:5611283 | unclassified gene | predicted gene, 38055 |
| 3 | gene | 51.99134 | 51.99196 | negative | MGI_C57BL6J_5610754 | Gm37526 | ENSEMBL:ENSMUSG00000103784 | MGI:5610754 | unclassified gene | predicted gene, 37526 |
| 3 | gene | 52.00074 | 52.00402 | negative | MGI_C57BL6J_5610693 | Gm37465 | ENSEMBL:ENSMUSG00000102423 | MGI:5610693 | unclassified gene | predicted gene, 37465 |
| 3 | gene | 52.05729 | 52.06424 | negative | MGI_C57BL6J_2444420 | C130089K02Rik | ENSEMBL:ENSMUSG00000104339 | MGI:2444420 | unclassified gene | RIKEN cDNA C130089K02 gene |
| 3 | gene | 52.07159 | 52.07339 | negative | MGI_C57BL6J_1921741 | 5430420F09Rik | ENSEMBL:ENSMUSG00000102808 | MGI:1921741 | unclassified gene | RIKEN cDNA 5430420F09 gene |
| 3 | gene | 52.13405 | 52.22200 | negative | MGI_C57BL6J_5610430 | Gm37202 | ENSEMBL:ENSMUSG00000103655 | MGI:5610430 | lncRNA gene | predicted gene, 37202 |
| 3 | gene | 52.17108 | 52.18013 | negative | MGI_C57BL6J_5589233 | Gm30074 | NCBI_Gene:102631843,ENSEMBL:ENSMUSG00000103726 | MGI:5589233 | lncRNA gene | predicted gene, 30074 |
| 3 | gene | 52.18657 | 52.18796 | positive | MGI_C57BL6J_5611360 | Gm38132 | ENSEMBL:ENSMUSG00000104055 | MGI:5611360 | lncRNA gene | predicted gene, 38132 |
| 3 | gene | 52.19825 | 52.20049 | positive | MGI_C57BL6J_5610398 | Gm37170 | ENSEMBL:ENSMUSG00000103170 | MGI:5610398 | lncRNA gene | predicted gene, 37170 |
| 3 | pseudogene | 52.23684 | 52.24694 | negative | MGI_C57BL6J_3031232 | Olfr1398-ps1 | NCBI_Gene:404507,ENSEMBL:ENSMUSG00000102510 | MGI:3031232 | pseudogene | olfactory receptor 1398, pseudogene 1 |
| 3 | gene | 52.23745 | 52.25650 | positive | MGI_C57BL6J_5589284 | Gm30125 | NCBI_Gene:102631910 | MGI:5589284 | lncRNA gene | predicted gene, 30125 |
| 3 | gene | 52.26834 | 52.35322 | positive | MGI_C57BL6J_1890077 | Foxo1 | NCBI_Gene:56458,ENSEMBL:ENSMUSG00000044167 | MGI:1890077 | protein coding gene | forkhead box O1 |
| 3 | gene | 52.26843 | 52.26944 | negative | MGI_C57BL6J_5141867 | Gm20402 | ENSEMBL:ENSMUSG00000092405 | MGI:5141867 | lncRNA gene | predicted gene 20402 |
| 3 | gene | 52.28224 | 52.28431 | positive | MGI_C57BL6J_5611262 | Gm38034 | ENSEMBL:ENSMUSG00000102844 | MGI:5611262 | unclassified gene | predicted gene, 38034 |
| 3 | pseudogene | 52.42535 | 52.42573 | positive | MGI_C57BL6J_5593462 | Gm34303 | NCBI_Gene:102637513 | MGI:5593462 | pseudogene | predicted gene, 34303 |
| 3 | pseudogene | 52.42541 | 52.42565 | positive | MGI_C57BL6J_5611326 | Gm38098 | ENSEMBL:ENSMUSG00000103454 | MGI:5611326 | pseudogene | predicted gene, 38098 |
| 3 | gene | 52.46717 | 52.49327 | positive | MGI_C57BL6J_5621422 | Gm38537 | NCBI_Gene:102641152 | MGI:5621422 | lncRNA gene | predicted gene, 38537 |
| 3 | gene | 52.50277 | 52.55022 | positive | MGI_C57BL6J_5589332 | Gm30173 | NCBI_Gene:102631981,ENSEMBL:ENSMUSG00000103965 | MGI:5589332 | lncRNA gene | predicted gene, 30173 |
| 3 | gene | 52.58013 | 52.58907 | positive | MGI_C57BL6J_5622932 | Gm40047 | NCBI_Gene:105244440 | MGI:5622932 | lncRNA gene | predicted gene, 40047 |
| 3 | pseudogene | 52.61282 | 52.61402 | positive | MGI_C57BL6J_3704216 | Gm10293 | NCBI_Gene:100039762,ENSEMBL:ENSMUSG00000070490 | MGI:3704216 | pseudogene | predicted pseudogene 10293 |
| 3 | gene | 52.64989 | 52.70971 | positive | MGI_C57BL6J_5589394 | Gm30235 | NCBI_Gene:102632062 | MGI:5589394 | lncRNA gene | predicted gene, 30235 |
| 3 | gene | 52.72265 | 52.72275 | negative | MGI_C57BL6J_5452575 | Gm22798 | ENSEMBL:ENSMUSG00000065926 | MGI:5452575 | snRNA gene | predicted gene, 22798 |
| 3 | gene | 52.73972 | 52.77666 | positive | MGI_C57BL6J_3780614 | Gm2447 | NCBI_Gene:329639,ENSEMBL:ENSMUSG00000102785 | MGI:3780614 | lncRNA gene | predicted gene 2447 |
| 3 | gene | 52.78900 | 52.79781 | negative | MGI_C57BL6J_5589451 | Gm30292 | NCBI_Gene:102632139,ENSEMBL:ENSMUSG00000104109 | MGI:5589451 | lncRNA gene | predicted gene, 30292 |
| 3 | gene | 52.90676 | 52.92767 | negative | MGI_C57BL6J_5434106 | Gm20750 | NCBI_Gene:622124,ENSEMBL:ENSMUSG00000104401 | MGI:5434106 | lncRNA gene | predicted gene, 20750 |
| 3 | gene | 52.93325 | 52.95628 | positive | MGI_C57BL6J_5589510 | Gm30351 | NCBI_Gene:102632211 | MGI:5589510 | lncRNA gene | predicted gene, 30351 |
| 3 | gene | 52.95618 | 52.95789 | negative | MGI_C57BL6J_5589561 | Gm30402 | NCBI_Gene:102632287 | MGI:5589561 | lncRNA gene | predicted gene, 30402 |
| 3 | pseudogene | 52.96005 | 52.96037 | negative | MGI_C57BL6J_5610235 | Gm37007 | ENSEMBL:ENSMUSG00000103886 | MGI:5610235 | pseudogene | predicted gene, 37007 |
| 3 | gene | 52.98188 | 53.01724 | negative | MGI_C57BL6J_1914792 | Cog6 | NCBI_Gene:67542,ENSEMBL:ENSMUSG00000027742 | MGI:1914792 | protein coding gene | component of oligomeric golgi complex 6 |
| 3 | gene | 53.03858 | 53.04176 | negative | MGI_C57BL6J_5663038 | Gm42901 | ENSEMBL:ENSMUSG00000104850 | MGI:5663038 | lncRNA gene | predicted gene 42901 |
| 3 | gene | 53.04153 | 53.26168 | positive | MGI_C57BL6J_1920048 | Lhfp | NCBI_Gene:108927,ENSEMBL:ENSMUSG00000048332 | MGI:1920048 | protein coding gene | lipoma HMGIC fusion partner |
| 3 | gene | 53.07216 | 53.07524 | negative | MGI_C57BL6J_5663035 | Gm42898 | ENSEMBL:ENSMUSG00000104828 | MGI:5663035 | unclassified gene | predicted gene 42898 |
| 3 | gene | 53.08322 | 53.08588 | positive | MGI_C57BL6J_5663036 | Gm42899 | ENSEMBL:ENSMUSG00000104687 | MGI:5663036 | unclassified gene | predicted gene 42899 |
| 3 | gene | 53.09900 | 53.10318 | positive | MGI_C57BL6J_2443940 | C130075A20Rik | ENSEMBL:ENSMUSG00000104735 | MGI:2443940 | unclassified gene | RIKEN cDNA C130075A20 gene |
| 3 | gene | 53.10967 | 53.11012 | positive | MGI_C57BL6J_5663941 | Gm43804 | ENSEMBL:ENSMUSG00000104615 | MGI:5663941 | unclassified gene | predicted gene 43804 |
| 3 | gene | 53.11040 | 53.11341 | negative | MGI_C57BL6J_5663940 | Gm43803 | ENSEMBL:ENSMUSG00000104968 | MGI:5663940 | lncRNA gene | predicted gene 43803 |
| 3 | gene | 53.11217 | 53.11299 | positive | MGI_C57BL6J_5663942 | Gm43805 | ENSEMBL:ENSMUSG00000105971 | MGI:5663942 | unclassified gene | predicted gene 43805 |
| 3 | pseudogene | 53.34649 | 53.34723 | negative | MGI_C57BL6J_5663609 | Gm43472 | ENSEMBL:ENSMUSG00000106660 | MGI:5663609 | pseudogene | predicted gene 43472 |
| 3 | gene | 53.39575 | 53.39850 | positive | MGI_C57BL6J_5622933 | Gm40048 | NCBI_Gene:105244441 | MGI:5622933 | lncRNA gene | predicted gene, 40048 |
| 3 | pseudogene | 53.40161 | 53.42457 | negative | MGI_C57BL6J_5663608 | Gm43471 | NCBI_Gene:108168853,ENSEMBL:ENSMUSG00000105397 | MGI:5663608 | pseudogene | predicted gene 43471 |
| 3 | gene | 53.44858 | 53.46333 | negative | MGI_C57BL6J_2444520 | Nhlrc3 | NCBI_Gene:212114,ENSEMBL:ENSMUSG00000042997 | MGI:2444520 | protein coding gene | NHL repeat containing 3 |
| 3 | gene | 53.45262 | 53.45805 | positive | MGI_C57BL6J_3801870 | Gm16206 | NCBI_Gene:102632669,ENSEMBL:ENSMUSG00000085174 | MGI:3801870 | lncRNA gene | predicted gene 16206 |
| 3 | gene | 53.46367 | 53.48175 | positive | MGI_C57BL6J_1919933 | Proser1 | NCBI_Gene:212127,ENSEMBL:ENSMUSG00000049504 | MGI:1919933 | protein coding gene | proline and serine rich 1 |
| 3 | gene | 53.48865 | 53.50850 | positive | MGI_C57BL6J_2388072 | Stoml3 | NCBI_Gene:229277,ENSEMBL:ENSMUSG00000027744 | MGI:2388072 | protein coding gene | stomatin (Epb7.2)-like 3 |
| 3 | gene | 53.50893 | 53.51010 | positive | MGI_C57BL6J_1924132 | 6430500D05Rik | ENSEMBL:ENSMUSG00000105481 | MGI:1924132 | unclassified gene | RIKEN cDNA 6430500D05 gene |
| 3 | gene | 53.51200 | 53.51213 | positive | MGI_C57BL6J_5453452 | Gm23675 | ENSEMBL:ENSMUSG00000087936 | MGI:5453452 | snoRNA gene | predicted gene, 23675 |
| 3 | gene | 53.51394 | 53.65736 | negative | MGI_C57BL6J_2444465 | Frem2 | NCBI_Gene:242022,ENSEMBL:ENSMUSG00000037016 | MGI:2444465 | protein coding gene | Fras1 related extracellular matrix protein 2 |
| 3 | pseudogene | 53.66666 | 53.66739 | negative | MGI_C57BL6J_3648425 | Gm6073 | NCBI_Gene:606547,ENSEMBL:ENSMUSG00000105840 | MGI:3648425 | pseudogene | predicted gene 6073 |
| 3 | pseudogene | 53.69237 | 53.69292 | negative | MGI_C57BL6J_3649038 | Gm6204 | NCBI_Gene:621155,ENSEMBL:ENSMUSG00000105879 | MGI:3649038 | pseudogene | predicted gene 6204 |
| 3 | gene | 53.70304 | 53.73576 | positive | MGI_C57BL6J_5589894 | Gm30735 | NCBI_Gene:102632743,ENSEMBL:ENSMUSG00000105168 | MGI:5589894 | lncRNA gene | predicted gene, 30735 |
| 3 | gene | 53.73622 | 53.73783 | positive | MGI_C57BL6J_3642187 | Gm10727 | ENSEMBL:ENSMUSG00000105444 | MGI:3642187 | unclassified gene | predicted gene 10727 |
| 3 | gene | 53.75673 | 53.77295 | positive | MGI_C57BL6J_2443105 | C820005J03Rik | ENSEMBL:ENSMUSG00000106270 | MGI:2443105 | lncRNA gene | RIKEN cDNA C820005J03 gene |
| 3 | gene | 53.84341 | 53.84351 | positive | MGI_C57BL6J_5454628 | Gm24851 | ENSEMBL:ENSMUSG00000088653 | MGI:5454628 | snRNA gene | predicted gene, 24851 |
| 3 | gene | 53.84509 | 53.84528 | positive | MGI_C57BL6J_3779200 | Gm10985 | ENSEMBL:ENSMUSG00000078742 | MGI:3779200 | protein coding gene | predicted gene 10985 |
| 3 | gene | 53.85338 | 53.86383 | negative | MGI_C57BL6J_1915140 | Ufm1 | NCBI_Gene:67890,ENSEMBL:ENSMUSG00000027746 | MGI:1915140 | protein coding gene | ubiquitin-fold modifier 1 |
| 3 | gene | 53.92484 | 53.94353 | positive | MGI_C57BL6J_5590185 | Gm31026 | NCBI_Gene:102633119,ENSEMBL:ENSMUSG00000106577 | MGI:5590185 | lncRNA gene | predicted gene, 31026 |
| 3 | pseudogene | 54.02115 | 54.02195 | positive | MGI_C57BL6J_3645533 | Gm8109 | NCBI_Gene:666444,ENSEMBL:ENSMUSG00000103155 | MGI:3645533 | pseudogene | predicted gene 8109 |
| 3 | pseudogene | 54.02988 | 54.03041 | positive | MGI_C57BL6J_5010203 | Gm18018 | NCBI_Gene:100416282,ENSEMBL:ENSMUSG00000105067 | MGI:5010203 | pseudogene | predicted gene, 18018 |
| 3 | gene | 54.15604 | 54.31847 | positive | MGI_C57BL6J_109525 | Trpc4 | NCBI_Gene:22066,ENSEMBL:ENSMUSG00000027748 | MGI:109525 | protein coding gene | transient receptor potential cation channel, subfamily C, member 4 |
| 3 | gene | 54.35927 | 54.39104 | positive | MGI_C57BL6J_1926321 | Postn | NCBI_Gene:50706,ENSEMBL:ENSMUSG00000027750 | MGI:1926321 | protein coding gene | periostin, osteoblast specific factor |
| 3 | gene | 54.35966 | 54.36101 | negative | MGI_C57BL6J_5590254 | Gm31095 | NCBI_Gene:102633208 | MGI:5590254 | lncRNA gene | predicted gene, 31095 |
| 3 | pseudogene | 54.48141 | 54.48293 | positive | MGI_C57BL6J_3645731 | Gm5641 | NCBI_Gene:434807,ENSEMBL:ENSMUSG00000069014 | MGI:3645731 | pseudogene | predicted gene 5641 |
| 3 | gene | 54.48856 | 54.53489 | negative | MGI_C57BL6J_5590313 | Gm31154 | NCBI_Gene:102633293 | MGI:5590313 | lncRNA gene | predicted gene, 31154 |
| 3 | gene | 54.59055 | 54.60139 | negative | MGI_C57BL6J_5622935 | Gm40050 | NCBI_Gene:105244443 | MGI:5622935 | lncRNA gene | predicted gene, 40050 |
| 3 | gene | 54.63144 | 54.63463 | positive | MGI_C57BL6J_5477172 | Gm26678 | ENSEMBL:ENSMUSG00000097599 | MGI:5477172 | lncRNA gene | predicted gene, 26678 |
| 3 | pseudogene | 54.63288 | 54.63457 | negative | MGI_C57BL6J_3588285 | B020017C02Rik | NCBI_Gene:100503708,ENSEMBL:ENSMUSG00000104831 | MGI:3588285 | pseudogene | RIKEN cDNA B020017C02 gene |
| 3 | gene | 54.69276 | 54.72877 | positive | MGI_C57BL6J_1929651 | Supt20 | NCBI_Gene:56790,ENSEMBL:ENSMUSG00000027751 | MGI:1929651 | protein coding gene | SPT20 SAGA complex component |
| 3 | gene | 54.71318 | 54.71435 | negative | MGI_C57BL6J_5439427 | Gm21958 | ENSEMBL:ENSMUSG00000094487 | MGI:5439427 | protein coding gene | predicted gene, 21958 |
| 3 | gene | 54.72868 | 54.73539 | negative | MGI_C57BL6J_1916889 | Exosc8 | NCBI_Gene:69639,ENSEMBL:ENSMUSG00000027752 | MGI:1916889 | protein coding gene | exosome component 8 |
| 3 | gene | 54.73554 | 54.75132 | positive | MGI_C57BL6J_1913498 | Alg5 | NCBI_Gene:66248,ENSEMBL:ENSMUSG00000036632 | MGI:1913498 | protein coding gene | asparagine-linked glycosylation 5 (dolichyl-phosphate beta-glucosyltransferase) |
| 3 | gene | 54.75545 | 54.80166 | positive | MGI_C57BL6J_1859993 | Smad9 | NCBI_Gene:55994,ENSEMBL:ENSMUSG00000027796 | MGI:1859993 | protein coding gene | SMAD family member 9 |
| 3 | gene | 54.80311 | 54.80779 | negative | MGI_C57BL6J_2180854 | Rfxap | NCBI_Gene:170767,ENSEMBL:ENSMUSG00000036615 | MGI:2180854 | protein coding gene | regulatory factor X-associated protein |
| 3 | pseudogene | 54.80997 | 54.81140 | negative | MGI_C57BL6J_3708673 | Gm10254 | NCBI_Gene:545524,ENSEMBL:ENSMUSG00000069011 | MGI:3708673 | pseudogene | predicted gene 10254 |
| 3 | gene | 54.89707 | 54.91589 | negative | MGI_C57BL6J_3607715 | Sertm1 | NCBI_Gene:329641,ENSEMBL:ENSMUSG00000056306 | MGI:3607715 | protein coding gene | serine rich and transmembrane domain containing 1 |
| 3 | gene | 54.91602 | 54.95326 | positive | MGI_C57BL6J_5590517 | Gm31358 | NCBI_Gene:102633562 | MGI:5590517 | lncRNA gene | predicted gene, 31358 |
| 3 | gene | 55.04547 | 55.05550 | negative | MGI_C57BL6J_108042 | Ccna1 | NCBI_Gene:12427,ENSEMBL:ENSMUSG00000027793 | MGI:108042 | protein coding gene | cyclin A1 |
| 3 | gene | 55.05524 | 55.08400 | positive | MGI_C57BL6J_1918220 | 4931419H13Rik | NCBI_Gene:70970,ENSEMBL:ENSMUSG00000106589 | MGI:1918220 | lncRNA gene | RIKEN cDNA 4931419H13 gene |
| 3 | pseudogene | 55.05735 | 55.05766 | negative | MGI_C57BL6J_5663692 | Gm43555 | ENSEMBL:ENSMUSG00000105977 | MGI:5663692 | pseudogene | predicted gene 43555 |
| 3 | gene | 55.11207 | 55.13733 | positive | MGI_C57BL6J_2139806 | Spg20 | NCBI_Gene:229285,ENSEMBL:ENSMUSG00000036580 | MGI:2139806 | protein coding gene | spastic paraplegia 20, spartin (Troyer syndrome) homolog (human) |
| 3 | gene | 55.13734 | 55.17525 | positive | MGI_C57BL6J_2444356 | Ccdc169 | NCBI_Gene:320604,ENSEMBL:ENSMUSG00000048655 | MGI:2444356 | protein coding gene | coiled-coil domain containing 169 |
| 3 | gene | 55.16231 | 55.16243 | negative | MGI_C57BL6J_5451806 | Gm22029 | ENSEMBL:ENSMUSG00000080590 | MGI:5451806 | miRNA gene | predicted gene, 22029 |
| 3 | gene | 55.18203 | 55.20996 | positive | MGI_C57BL6J_1921684 | Sohlh2 | NCBI_Gene:74434,ENSEMBL:ENSMUSG00000027794 | MGI:1921684 | protein coding gene | spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
| 3 | gene | 55.24236 | 55.53907 | positive | MGI_C57BL6J_1330861 | Dclk1 | NCBI_Gene:13175,ENSEMBL:ENSMUSG00000027797 | MGI:1330861 | protein coding gene | doublecortin-like kinase 1 |
| 3 | gene | 55.29094 | 55.29194 | positive | MGI_C57BL6J_1924494 | 9430012M22Rik | NA | NA | unclassified gene | RIKEN cDNA 9430012M22 gene |
| 3 | gene | 55.30532 | 55.30542 | positive | MGI_C57BL6J_5454909 | Gm25132 | ENSEMBL:ENSMUSG00000064586 | MGI:5454909 | snRNA gene | predicted gene, 25132 |
| 3 | gene | 55.33084 | 55.33297 | positive | MGI_C57BL6J_5012002 | Gm19817 | ENSEMBL:ENSMUSG00000105302 | MGI:5012002 | unclassified gene | predicted gene, 19817 |
| 3 | gene | 55.35240 | 55.38079 | negative | MGI_C57BL6J_5622936 | Gm40051 | NCBI_Gene:105244444 | MGI:5622936 | lncRNA gene | predicted gene, 40051 |
| 3 | gene | 55.37746 | 55.40312 | negative | MGI_C57BL6J_5662746 | Gm42609 | ENSEMBL:ENSMUSG00000105975 | MGI:5662746 | lncRNA gene | predicted gene 42609 |
| 3 | gene | 55.39181 | 55.40420 | negative | MGI_C57BL6J_3642047 | Gm9831 | NCBI_Gene:101056032 | MGI:3642047 | protein coding gene | predicted gene 9831 |
| 3 | gene | 55.46198 | 55.47221 | negative | MGI_C57BL6J_5439423 | Gm21954 | ENSEMBL:ENSMUSG00000094962 | MGI:5439423 | protein coding gene | predicted gene, 21954 |
| 3 | gene | 55.47609 | 55.48162 | negative | MGI_C57BL6J_5590602 | Gm31443 | NCBI_Gene:102633676 | MGI:5590602 | lncRNA gene | predicted gene, 31443 |
| 3 | gene | 55.48379 | 55.48718 | positive | MGI_C57BL6J_5662744 | Gm42607 | ENSEMBL:ENSMUSG00000106354 | MGI:5662744 | unclassified gene | predicted gene 42607 |
| 3 | gene | 55.51757 | 55.51959 | positive | MGI_C57BL6J_5662745 | Gm42608 | ENSEMBL:ENSMUSG00000106651 | MGI:5662745 | unclassified gene | predicted gene 42608 |
| 3 | gene | 55.54045 | 55.54338 | negative | MGI_C57BL6J_5590955 | Gm31796 | NCBI_Gene:102634140 | MGI:5590955 | lncRNA gene | predicted gene, 31796 |
| 3 | gene | 55.54354 | 55.54855 | positive | MGI_C57BL6J_5455181 | Gm25404 | NCBI_Gene:102634218,ENSEMBL:ENSMUSG00000064904 | MGI:5455181 | snRNA gene | predicted gene, 25404 |
| 3 | gene | 55.57492 | 55.57727 | positive | MGI_C57BL6J_5663685 | Gm43548 | ENSEMBL:ENSMUSG00000105627 | MGI:5663685 | unclassified gene | predicted gene 43548 |
| 3 | gene | 55.57811 | 55.58273 | negative | MGI_C57BL6J_5663686 | Gm43549 | NCBI_Gene:108168870,ENSEMBL:ENSMUSG00000106334 | MGI:5663686 | lncRNA gene | predicted gene 43549 |
| 3 | gene | 55.62519 | 56.18427 | negative | MGI_C57BL6J_1347075 | Nbea | NCBI_Gene:26422,ENSEMBL:ENSMUSG00000027799 | MGI:1347075 | protein coding gene | neurobeachin |
| 3 | gene | 55.70589 | 55.71565 | positive | MGI_C57BL6J_5826441 | Gm46804 | NCBI_Gene:108168871 | MGI:5826441 | lncRNA gene | predicted gene, 46804 |
| 3 | gene | 55.72593 | 55.72844 | negative | MGI_C57BL6J_3609650 | D430004P15Rik | NA | NA | unclassified gene | Riken cDNA D430004P15 gene |
| 3 | gene | 55.77534 | 55.78795 | negative | MGI_C57BL6J_3698047 | 9630024C22Rik | NA | NA | unclassified non-coding RNA gene | RIKEN cDNA 9630024C22 gene |
| 3 | gene | 55.78251 | 55.78529 | positive | MGI_C57BL6J_1333773 | Mab21l1 | NCBI_Gene:17116,ENSEMBL:ENSMUSG00000056947 | MGI:1333773 | protein coding gene | mab-21-like 1 |
| 3 | gene | 55.83367 | 55.83446 | negative | MGI_C57BL6J_1924979 | 6720451E02Rik | NA | NA | unclassified gene | RIKEN cDNA 6720451E02 gene |
| 3 | pseudogene | 55.84710 | 55.84840 | positive | MGI_C57BL6J_1918432 | 4933417G07Rik | NCBI_Gene:71182,ENSEMBL:ENSMUSG00000106572 | MGI:1918432 | pseudogene | RIKEN cDNA 4933417G07 gene |
| 3 | gene | 55.85525 | 55.85615 | negative | MGI_C57BL6J_1926125 | B230206L23Rik | NA | NA | unclassified gene | RIKEN cDNA B230206L23 gene |
| 3 | gene | 55.88327 | 56.00032 | positive | MGI_C57BL6J_5591073 | Gm31914 | NCBI_Gene:102634293 | MGI:5591073 | lncRNA gene | predicted gene, 31914 |
| 3 | gene | 55.89031 | 55.93670 | positive | MGI_C57BL6J_5663513 | Gm43376 | ENSEMBL:ENSMUSG00000104677 | MGI:5663513 | lncRNA gene | predicted gene 43376 |
| 3 | gene | 55.95678 | 55.95691 | negative | MGI_C57BL6J_5452561 | Gm22784 | ENSEMBL:ENSMUSG00000077276 | MGI:5452561 | snoRNA gene | predicted gene, 22784 |
| 3 | gene | 56.01492 | 56.01717 | negative | MGI_C57BL6J_5662977 | Gm42840 | ENSEMBL:ENSMUSG00000104938 | MGI:5662977 | unclassified gene | predicted gene 42840 |
| 3 | gene | 56.12316 | 56.12616 | negative | MGI_C57BL6J_5662979 | Gm42842 | ENSEMBL:ENSMUSG00000106211 | MGI:5662979 | unclassified gene | predicted gene 42842 |
| 3 | gene | 56.12860 | 56.13053 | negative | MGI_C57BL6J_5662978 | Gm42841 | ENSEMBL:ENSMUSG00000106528 | MGI:5662978 | unclassified gene | predicted gene 42841 |
| 3 | gene | 56.16395 | 56.16757 | negative | MGI_C57BL6J_5662980 | Gm42843 | ENSEMBL:ENSMUSG00000105293 | MGI:5662980 | unclassified gene | predicted gene 42843 |
| 3 | gene | 56.21791 | 56.22124 | negative | MGI_C57BL6J_5622937 | Gm40052 | NCBI_Gene:105244445 | MGI:5622937 | lncRNA gene | predicted gene, 40052 |
| 3 | gene | 56.25590 | 56.26994 | negative | MGI_C57BL6J_5622938 | Gm40053 | NCBI_Gene:105244446 | MGI:5622938 | lncRNA gene | predicted gene, 40053 |
| 3 | gene | 56.34703 | 56.35755 | negative | MGI_C57BL6J_5622939 | Gm40054 | NCBI_Gene:105244447 | MGI:5622939 | lncRNA gene | predicted gene, 40054 |
| 3 | gene | 56.35224 | 56.35230 | positive | MGI_C57BL6J_102792 | Mcptl | NA | NA | protein coding gene | mast cell protease-like |
| 3 | gene | 56.43600 | 56.43875 | negative | MGI_C57BL6J_5591336 | Gm32177 | NCBI_Gene:102634641 | MGI:5591336 | lncRNA gene | predicted gene, 32177 |
| 3 | gene | 56.44528 | 56.44541 | negative | MGI_C57BL6J_5455504 | Gm25727 | ENSEMBL:ENSMUSG00000077710 | MGI:5455504 | snoRNA gene | predicted gene, 25727 |
| 3 | gene | 56.69842 | 56.70506 | positive | MGI_C57BL6J_5591396 | Gm32237 | NCBI_Gene:102634719 | MGI:5591396 | lncRNA gene | predicted gene, 32237 |
| 3 | pseudogene | 56.82832 | 56.82884 | negative | MGI_C57BL6J_3780790 | Gm2622 | NCBI_Gene:100040139,ENSEMBL:ENSMUSG00000105113 | MGI:3780790 | pseudogene | predicted gene 2622 |
| 3 | gene | 56.88806 | 56.89133 | negative | MGI_C57BL6J_3588185 | F830104G03Rik | NA | NA | unclassified gene | RIKEN cDNA F830104G03 gene |
| 3 | gene | 57.05415 | 57.05428 | positive | MGI_C57BL6J_5452046 | Gm22269 | ENSEMBL:ENSMUSG00000077190 | MGI:5452046 | snoRNA gene | predicted gene, 22269 |
| 3 | pseudogene | 57.13549 | 57.13683 | negative | MGI_C57BL6J_3805962 | Gm8177 | NCBI_Gene:666585,ENSEMBL:ENSMUSG00000097366 | MGI:3805962 | pseudogene | predicted gene 8177 |
| 3 | gene | 57.22772 | 57.22918 | negative | MGI_C57BL6J_1918389 | 4933408K01Rik | NA | NA | unclassified gene | RIKEN cDNA 4933408K01 gene |
| 3 | pseudogene | 57.24925 | 57.24950 | positive | MGI_C57BL6J_5663561 | Gm43424 | ENSEMBL:ENSMUSG00000106286 | MGI:5663561 | pseudogene | predicted gene 43424 |
| 3 | pseudogene | 57.27174 | 57.27237 | negative | MGI_C57BL6J_5010845 | Gm18660 | NCBI_Gene:100417512,ENSEMBL:ENSMUSG00000104534 | MGI:5010845 | pseudogene | predicted gene, 18660 |
| 3 | gene | 57.28561 | 57.30199 | negative | MGI_C57BL6J_104678 | Tm4sf1 | NCBI_Gene:17112,ENSEMBL:ENSMUSG00000027800 | MGI:104678 | protein coding gene | transmembrane 4 superfamily member 1 |
| 3 | pseudogene | 57.36225 | 57.36361 | positive | MGI_C57BL6J_3646382 | Gm5276 | NCBI_Gene:383862,ENSEMBL:ENSMUSG00000105452 | MGI:3646382 | pseudogene | predicted gene 5276 |
| 3 | gene | 57.37929 | 57.39455 | negative | MGI_C57BL6J_5591526 | Gm32367 | NCBI_Gene:102634892 | MGI:5591526 | lncRNA gene | predicted gene, 32367 |
| 3 | gene | 57.42531 | 57.44168 | positive | MGI_C57BL6J_2385173 | Tm4sf4 | NCBI_Gene:229302,ENSEMBL:ENSMUSG00000027801 | MGI:2385173 | protein coding gene | transmembrane 4 superfamily member 4 |
| 3 | gene | 57.45564 | 57.57591 | negative | MGI_C57BL6J_1917649 | Wwtr1 | NCBI_Gene:97064,ENSEMBL:ENSMUSG00000027803 | MGI:1917649 | protein coding gene | WW domain containing transcription regulator 1 |
| 3 | pseudogene | 57.52655 | 57.53048 | positive | MGI_C57BL6J_3802068 | Gm16016 | NCBI_Gene:105244448,ENSEMBL:ENSMUSG00000083538 | MGI:3802068 | pseudogene | predicted gene 16016 |
| 3 | gene | 57.61259 | 57.62736 | negative | MGI_C57BL6J_5477165 | Gm26671 | NCBI_Gene:102635136,ENSEMBL:ENSMUSG00000097339 | MGI:5477165 | lncRNA gene | predicted gene, 26671 |
| 3 | gene | 57.61661 | 57.61672 | positive | MGI_C57BL6J_5530798 | Mir6377 | miRBase:MI0021907,NCBI_Gene:102465197,ENSEMBL:ENSMUSG00000098401 | MGI:5530798 | miRNA gene | microRNA 6377 |
| 3 | gene | 57.63688 | 57.64084 | negative | MGI_C57BL6J_5663574 | Gm43437 | ENSEMBL:ENSMUSG00000104965 | MGI:5663574 | unclassified gene | predicted gene 43437 |
| 3 | gene | 57.63768 | 57.64073 | positive | MGI_C57BL6J_5826442 | Gm46805 | NCBI_Gene:108168872 | MGI:5826442 | lncRNA gene | predicted gene, 46805 |
| 3 | gene | 57.64435 | 57.65172 | negative | MGI_C57BL6J_1098806 | Commd2 | NCBI_Gene:52245,ENSEMBL:ENSMUSG00000036513 | MGI:1098806 | protein coding gene | COMM domain containing 2 |
| 3 | gene | 57.65440 | 57.65558 | negative | MGI_C57BL6J_2140033 | BB187690 | ENSEMBL:ENSMUSG00000105389 | MGI:2140033 | unclassified gene | expressed sequence BB187690 |
| 3 | gene | 57.65739 | 57.69280 | negative | MGI_C57BL6J_2685256 | Ankub1 | NCBI_Gene:242037,ENSEMBL:ENSMUSG00000074591 | MGI:2685256 | protein coding gene | ankrin repeat and ubiquitin domain containing 1 |
| 3 | gene | 57.69618 | 57.69748 | negative | MGI_C57BL6J_5663235 | Gm43098 | ENSEMBL:ENSMUSG00000106635 | MGI:5663235 | unclassified gene | predicted gene 43098 |
| 3 | pseudogene | 57.71777 | 57.71832 | negative | MGI_C57BL6J_3648319 | Gm6394 | NCBI_Gene:623114,ENSEMBL:ENSMUSG00000104623 | MGI:3648319 | pseudogene | predicted gene 6394 |
| 3 | gene | 57.73604 | 57.83543 | positive | MGI_C57BL6J_1346341 | Rnf13 | NCBI_Gene:24017,ENSEMBL:ENSMUSG00000036503 | MGI:1346341 | protein coding gene | ring finger protein 13 |
| 3 | pseudogene | 57.76582 | 57.76667 | negative | MGI_C57BL6J_3780052 | Gm9645 | NCBI_Gene:675537,ENSEMBL:ENSMUSG00000106421 | MGI:3780052 | pseudogene | predicted gene 9645 |
| 3 | pseudogene | 57.80976 | 57.81027 | negative | MGI_C57BL6J_5010136 | Gm17951 | NCBI_Gene:100416165,ENSEMBL:ENSMUSG00000106006 | MGI:5010136 | pseudogene | predicted gene, 17951 |
| 3 | gene | 57.82159 | 57.82240 | positive | MGI_C57BL6J_1925970 | D530035G10Rik | NA | NA | unclassified gene | RIKEN cDNA D530035G10 gene |
| 3 | gene | 57.84190 | 57.84808 | negative | MGI_C57BL6J_97550 | Pfn2 | NCBI_Gene:18645,ENSEMBL:ENSMUSG00000027805 | MGI:97550 | protein coding gene | profilin 2 |
| 3 | gene | 57.92209 | 57.92219 | negative | MGI_C57BL6J_5454308 | Gm24531 | ENSEMBL:ENSMUSG00000088570 | MGI:5454308 | snRNA gene | predicted gene, 24531 |
| 3 | pseudogene | 57.94292 | 57.94315 | positive | MGI_C57BL6J_5662648 | Gm42511 | ENSEMBL:ENSMUSG00000104730 | MGI:5662648 | pseudogene | predicted gene 42511 |
| 3 | gene | 57.94911 | 57.97883 | positive | MGI_C57BL6J_5591912 | Gm32753 | NCBI_Gene:102635402 | MGI:5591912 | lncRNA gene | predicted gene, 32753 |
| 3 | gene | 57.95198 | 58.03171 | negative | MGI_C57BL6J_5622940 | Gm40055 | NCBI_Gene:105244449,ENSEMBL:ENSMUSG00000105003 | MGI:5622940 | lncRNA gene | predicted gene, 40055 |
| 3 | pseudogene | 57.96219 | 57.96337 | positive | MGI_C57BL6J_5011052 | Gm18867 | NCBI_Gene:100417858,ENSEMBL:ENSMUSG00000106246 | MGI:5011052 | pseudogene | predicted gene, 18867 |
| 3 | gene | 58.07972 | 58.08183 | positive | MGI_C57BL6J_5662650 | Gm42513 | ENSEMBL:ENSMUSG00000105065 | MGI:5662650 | unclassified gene | predicted gene 42513 |
| 3 | gene | 58.08960 | 58.16235 | positive | MGI_C57BL6J_5622941 | Gm40056 | NCBI_Gene:105244450 | MGI:5622941 | lncRNA gene | predicted gene, 40056 |
| 3 | pseudogene | 58.10256 | 58.10430 | negative | MGI_C57BL6J_3643179 | Gm5537 | NCBI_Gene:433594,ENSEMBL:ENSMUSG00000069008 | MGI:3643179 | pseudogene | predicted gene 5537 |
| 3 | gene | 58.11705 | 58.16762 | positive | MGI_C57BL6J_1925506 | 4921539H07Rik | ENSEMBL:ENSMUSG00000104586 | MGI:1925506 | lncRNA gene | RIKEN cDNA 4921539H07 gene |
| 3 | gene | 58.14171 | 58.16381 | negative | MGI_C57BL6J_1915100 | 1700007F19Rik | NCBI_Gene:67850,ENSEMBL:ENSMUSG00000100666 | MGI:1915100 | lncRNA gene | RIKEN cDNA 1700007F19 gene |
| 3 | gene | 58.21192 | 58.21337 | positive | MGI_C57BL6J_5622942 | Gm40057 | NCBI_Gene:105244451 | MGI:5622942 | lncRNA gene | predicted gene, 40057 |
| 3 | gene | 58.29025 | 58.29036 | negative | MGI_C57BL6J_5455943 | Gm26166 | ENSEMBL:ENSMUSG00000065722 | MGI:5455943 | snRNA gene | predicted gene, 26166 |
| 3 | gene | 58.30186 | 58.31762 | positive | MGI_C57BL6J_5592006 | Gm32847 | NCBI_Gene:102635539 | MGI:5592006 | lncRNA gene | predicted gene, 32847 |
| 3 | pseudogene | 58.31494 | 58.31531 | negative | MGI_C57BL6J_5663867 | Gm43730 | ENSEMBL:ENSMUSG00000106176 | MGI:5663867 | pseudogene | predicted gene 43730 |
| 3 | pseudogene | 58.40465 | 58.40512 | positive | MGI_C57BL6J_5012313 | Gm20128 | NCBI_Gene:100504238,ENSEMBL:ENSMUSG00000105154 | MGI:5012313 | pseudogene | predicted gene, 20128 |
| 3 | gene | 58.41472 | 58.46679 | positive | MGI_C57BL6J_1919283 | Tsc22d2 | NCBI_Gene:72033,ENSEMBL:ENSMUSG00000027806 | MGI:1919283 | protein coding gene | TSC22 domain family, member 2 |
| 3 | gene | 58.43268 | 58.43301 | positive | MGI_C57BL6J_1338064 | Gdap9 | NA | NA | unclassified gene | ganglioside-induced differentiation-associated-protein 9 |
| 3 | gene | 58.51982 | 58.52589 | negative | MGI_C57BL6J_92638 | Serp1 | NCBI_Gene:28146,ENSEMBL:ENSMUSG00000027808 | MGI:92638 | protein coding gene | stress-associated endoplasmic reticulum protein 1 |
| 3 | gene | 58.52582 | 58.55750 | positive | MGI_C57BL6J_1098684 | Eif2a | NCBI_Gene:229317,ENSEMBL:ENSMUSG00000027810 | MGI:1098684 | protein coding gene | eukaryotic translation initiation factor 2A |
| 3 | gene | 58.52693 | 58.52812 | positive | MGI_C57BL6J_1925148 | 6720482G16Rik | ENSEMBL:ENSMUSG00000106553 | MGI:1925148 | unclassified gene | RIKEN cDNA 6720482G16 gene |
| 3 | gene | 58.57664 | 58.59354 | positive | MGI_C57BL6J_1916477 | Selenot | NCBI_Gene:69227,ENSEMBL:ENSMUSG00000075700 | MGI:1916477 | protein coding gene | selenoprotein T |
| 3 | gene | 58.61630 | 58.63721 | negative | MGI_C57BL6J_3588212 | Erich6 | NCBI_Gene:545527,ENSEMBL:ENSMUSG00000070471 | MGI:3588212 | protein coding gene | glutamate rich 6 |
| 3 | gene | 58.63671 | 58.63749 | positive | MGI_C57BL6J_5826443 | Gm46806 | NCBI_Gene:108168873 | MGI:5826443 | lncRNA gene | predicted gene, 46806 |
| 3 | pseudogene | 58.65204 | 58.65404 | positive | MGI_C57BL6J_3779790 | Gm8234 | NCBI_Gene:666680,ENSEMBL:ENSMUSG00000105021 | MGI:3779790 | pseudogene | predicted gene 8234 |
| 3 | pseudogene | 58.66547 | 58.66664 | negative | MGI_C57BL6J_2139888 | AU022133 | NCBI_Gene:100417266,ENSEMBL:ENSMUSG00000105336 | MGI:2139888 | pseudogene | expressed sequence AU022133 |
| 3 | gene | 58.67494 | 58.69244 | negative | MGI_C57BL6J_108062 | Siah2 | NCBI_Gene:20439,ENSEMBL:ENSMUSG00000036432 | MGI:108062 | protein coding gene | siah E3 ubiquitin protein ligase 2 |
| 3 | gene | 58.69259 | 58.79733 | positive | MGI_C57BL6J_1923132 | 4930593A02Rik | NCBI_Gene:71301,ENSEMBL:ENSMUSG00000097725 | MGI:1923132 | lncRNA gene | RIKEN cDNA 4930593A02 gene |
| 3 | gene | 58.69756 | 58.69867 | positive | MGI_C57BL6J_1925070 | A930028O11Rik | ENSEMBL:ENSMUSG00000105226 | MGI:1925070 | unclassified gene | RIKEN cDNA A930028O11 gene |
| 3 | pseudogene | 58.76149 | 58.76275 | negative | MGI_C57BL6J_5592251 | Gm33092 | NCBI_Gene:102635856 | MGI:5592251 | pseudogene | predicted gene, 33092 |
| 3 | pseudogene | 58.78841 | 58.82231 | negative | MGI_C57BL6J_2140018 | Mindy4b-ps | NCBI_Gene:100040293,ENSEMBL:ENSMUSG00000101860 | MGI:2140018 | pseudogene | MINDY lysine 48 deubiquitinase 4B, pseudogene |
| 3 | gene | 58.82499 | 58.83590 | positive | MGI_C57BL6J_5622944 | Gm40059 | NCBI_Gene:105244453 | MGI:5622944 | lncRNA gene | predicted gene, 40059 |
| 3 | gene | 58.84210 | 58.86675 | positive | MGI_C57BL6J_5622945 | Gm40060 | NCBI_Gene:105244454 | MGI:5622945 | lncRNA gene | predicted gene, 40060 |
| 3 | gene | 58.84403 | 58.88534 | negative | MGI_C57BL6J_2388124 | Clrn1 | NCBI_Gene:229320,ENSEMBL:ENSMUSG00000043850 | MGI:2388124 | protein coding gene | clarin 1 |
| 3 | gene | 58.97563 | 58.97573 | positive | MGI_C57BL6J_5452268 | Gm22491 | ENSEMBL:ENSMUSG00000087815 | MGI:5452268 | snRNA gene | predicted gene, 22491 |
| 3 | gene | 58.97886 | 58.97903 | negative | MGI_C57BL6J_5455349 | Gm25572 | ENSEMBL:ENSMUSG00000088152 | MGI:5455349 | snRNA gene | predicted gene, 25572 |
| 3 | pseudogene | 58.98713 | 58.98786 | positive | MGI_C57BL6J_3642685 | Rpl13-ps6 | NCBI_Gene:100040416,ENSEMBL:ENSMUSG00000059776 | MGI:3642685 | pseudogene | ribosomal protein L13, pseudogene 6 |
| 3 | gene | 59.00162 | 59.00169 | positive | MGI_C57BL6J_5531175 | Gm27793 | ENSEMBL:ENSMUSG00000099052 | MGI:5531175 | unclassified non-coding RNA gene | predicted gene, 27793 |
| 3 | gene | 59.00247 | 59.00616 | negative | MGI_C57BL6J_5663707 | Gm43570 | ENSEMBL:ENSMUSG00000105945 | MGI:5663707 | lncRNA gene | predicted gene 43570 |
| 3 | gene | 59.00559 | 59.31868 | positive | MGI_C57BL6J_2139916 | Med12l | NCBI_Gene:329650,ENSEMBL:ENSMUSG00000056476 | MGI:2139916 | protein coding gene | mediator complex subunit 12-like |
| 3 | gene | 59.09645 | 59.10182 | negative | MGI_C57BL6J_2442043 | Gpr171 | NCBI_Gene:229323,ENSEMBL:ENSMUSG00000050075 | MGI:2442043 | protein coding gene | G protein-coupled receptor 171 |
| 3 | gene | 59.11386 | 59.15362 | negative | MGI_C57BL6J_2155705 | P2ry14 | NCBI_Gene:140795,ENSEMBL:ENSMUSG00000036381 | MGI:2155705 | protein coding gene | purinergic receptor P2Y, G-protein coupled, 14 |
| 3 | gene | 59.14630 | 59.15363 | negative | MGI_C57BL6J_2444074 | F630111L10Rik | NCBI_Gene:320463 | MGI:2444074 | lncRNA gene | RIKEN cDNA F630111L10 gene |
| 3 | gene | 59.17890 | 59.19547 | negative | MGI_C57BL6J_1934133 | Gpr87 | NCBI_Gene:84111,ENSEMBL:ENSMUSG00000051431 | MGI:1934133 | protein coding gene | G protein-coupled receptor 87 |
| 3 | gene | 59.20789 | 59.21088 | negative | MGI_C57BL6J_1921441 | P2ry13 | NCBI_Gene:74191,ENSEMBL:ENSMUSG00000036362 | MGI:1921441 | protein coding gene | purinergic receptor P2Y, G-protein coupled 13 |
| 3 | gene | 59.21627 | 59.26301 | negative | MGI_C57BL6J_1918089 | P2ry12 | NCBI_Gene:70839,ENSEMBL:ENSMUSG00000036353 | MGI:1918089 | protein coding gene | purinergic receptor P2Y, G-protein coupled 12 |
| 3 | gene | 59.26979 | 59.27131 | positive | MGI_C57BL6J_5663726 | Gm43589 | ENSEMBL:ENSMUSG00000104550 | MGI:5663726 | unclassified gene | predicted gene 43589 |
| 3 | gene | 59.31674 | 59.34444 | negative | MGI_C57BL6J_1923481 | Igsf10 | NCBI_Gene:242050,ENSEMBL:ENSMUSG00000036334 | MGI:1923481 | protein coding gene | immunoglobulin superfamily, member 10 |
| 3 | gene | 59.34427 | 59.35117 | positive | MGI_C57BL6J_5611414 | Gm38186 | ENSEMBL:ENSMUSG00000103668 | MGI:5611414 | lncRNA gene | predicted gene, 38186 |
| 3 | pseudogene | 59.42453 | 59.42476 | positive | MGI_C57BL6J_5610943 | Gm37715 | ENSEMBL:ENSMUSG00000102247 | MGI:5610943 | pseudogene | predicted gene, 37715 |
| 3 | pseudogene | 59.43154 | 59.43335 | positive | MGI_C57BL6J_3648019 | Gm8276 | NCBI_Gene:666756,ENSEMBL:ENSMUSG00000102550 | MGI:3648019 | pseudogene | predicted gene 8276 |
| 3 | pseudogene | 59.46967 | 59.47409 | negative | MGI_C57BL6J_5611516 | Gm38288 | ENSEMBL:ENSMUSG00000103211 | MGI:5611516 | pseudogene | predicted gene, 38288 |
| 3 | gene | 59.47493 | 59.47500 | negative | MGI_C57BL6J_4414092 | n-TVcac1 | NCBI_Gene:102467620 | MGI:4414092 | tRNA gene | nuclear encoded tRNA valine 1 (anticodon CAC) |
| 3 | pseudogene | 59.48302 | 59.48329 | positive | MGI_C57BL6J_5610999 | Gm37771 | ENSEMBL:ENSMUSG00000104038 | MGI:5610999 | pseudogene | predicted gene, 37771 |
| 3 | pseudogene | 59.50558 | 59.53523 | negative | MGI_C57BL6J_3644725 | Gm4856 | NCBI_Gene:229330,ENSEMBL:ENSMUSG00000103598 | MGI:3644725 | pseudogene | predicted gene 4856 |
| 3 | pseudogene | 59.60245 | 59.63576 | negative | MGI_C57BL6J_3644280 | Gm5709 | NCBI_Gene:435732,ENSEMBL:ENSMUSG00000095128 | MGI:3644280 | pseudogene | predicted gene 5709 |
| 3 | gene | 59.66286 | 59.66528 | positive | MGI_C57BL6J_5611154 | Gm37926 | ENSEMBL:ENSMUSG00000102480 | MGI:5611154 | unclassified gene | predicted gene, 37926 |
| 3 | gene | 59.72979 | 59.75240 | positive | MGI_C57BL6J_3779495 | Aadacl2fm2 | NCBI_Gene:433597,ENSEMBL:ENSMUSG00000090527 | MGI:3779495 | protein coding gene | AADACL2 family member 2 |
| 3 | pseudogene | 59.73675 | 59.73686 | negative | MGI_C57BL6J_5611436 | Gm38208 | ENSEMBL:ENSMUSG00000104471 | MGI:5611436 | pseudogene | predicted gene, 38208 |
| 3 | pseudogene | 59.73718 | 59.73732 | negative | MGI_C57BL6J_5610807 | Gm37579 | ENSEMBL:ENSMUSG00000102390 | MGI:5610807 | pseudogene | predicted gene, 37579 |
| 3 | pseudogene | 59.76721 | 59.76891 | positive | MGI_C57BL6J_3644451 | Gm5539 | NCBI_Gene:433598,ENSEMBL:ENSMUSG00000103986 | MGI:3644451 | pseudogene | predicted gene 5539 |
| 3 | gene | 59.77781 | 59.84682 | positive | MGI_C57BL6J_1921922 | 4930449A18Rik | ENSEMBL:ENSMUSG00000074589 | MGI:1921922 | protein coding gene | RIKEN cDNA 4930449A18 gene |
| 3 | pseudogene | 59.78322 | 59.78863 | negative | MGI_C57BL6J_3780925 | Gm2756 | NCBI_Gene:100040407,ENSEMBL:ENSMUSG00000102757 | MGI:3780925 | pseudogene | predicted gene 2756 |
| 3 | gene | 59.83837 | 59.84682 | positive | MGI_C57BL6J_4361133 | Gm16527 | NCBI_Gene:669780 | MGI:4361133 | protein coding gene | predicted gene, 16527 |
| 3 | gene | 59.86105 | 59.87731 | positive | MGI_C57BL6J_3643798 | Aadacl2fm3 | NCBI_Gene:666803,ENSEMBL:ENSMUSG00000095522 | MGI:3643798 | protein coding gene | AADACL2 family member 3 |
| 3 | gene | 59.92521 | 59.93795 | positive | MGI_C57BL6J_3028051 | Aadacl2fm1 | NCBI_Gene:229333,ENSEMBL:ENSMUSG00000036951 | MGI:3028051 | protein coding gene | AADACL2 family member 1 |
| 3 | pseudogene | 59.95231 | 59.97359 | positive | MGI_C57BL6J_3819177 | Gm9696 | NCBI_Gene:676914,ENSEMBL:ENSMUSG00000070465 | MGI:3819177 | pseudogene | predicted gene 9696 |
| 3 | gene | 60.00674 | 60.02542 | positive | MGI_C57BL6J_3646333 | Aadacl2 | NCBI_Gene:639634,ENSEMBL:ENSMUSG00000091376 | MGI:3646333 | protein coding gene | arylacetamide deacetylase like 2 |
| 3 | pseudogene | 60.02036 | 60.02087 | negative | MGI_C57BL6J_5010612 | Gm18427 | NCBI_Gene:100417154,ENSEMBL:ENSMUSG00000103177 | MGI:5010612 | pseudogene | predicted gene, 18427 |
| 3 | gene | 60.02572 | 60.04016 | positive | MGI_C57BL6J_1915008 | Aadac | NCBI_Gene:67758,ENSEMBL:ENSMUSG00000027761 | MGI:1915008 | protein coding gene | arylacetamide deacetylase |
| 3 | gene | 60.08187 | 60.08757 | positive | MGI_C57BL6J_1934135 | Sucnr1 | NCBI_Gene:84112,ENSEMBL:ENSMUSG00000027762 | MGI:1934135 | protein coding gene | succinate receptor 1 |
| 3 | gene | 60.12764 | 60.12775 | negative | MGI_C57BL6J_5454159 | Gm24382 | ENSEMBL:ENSMUSG00000084647 | MGI:5454159 | snRNA gene | predicted gene, 24382 |
| 3 | gene | 60.29342 | 60.30941 | negative | MGI_C57BL6J_5592809 | Gm33650 | NCBI_Gene:102636642 | MGI:5592809 | lncRNA gene | predicted gene, 33650 |
| 3 | pseudogene | 60.34219 | 60.34309 | negative | MGI_C57BL6J_3779880 | Tgif1-ps | NCBI_Gene:669823,ENSEMBL:ENSMUSG00000107270 | MGI:3779880 | pseudogene | TGFB-induced factor homeobox 1, pseudogene |
| 3 | gene | 60.47279 | 60.62975 | positive | MGI_C57BL6J_1928482 | Mbnl1 | NCBI_Gene:56758,ENSEMBL:ENSMUSG00000027763 | MGI:1928482 | protein coding gene | muscleblind like splicing factor 1 |
| 3 | gene | 60.53575 | 60.53974 | positive | MGI_C57BL6J_5610817 | Gm37589 | ENSEMBL:ENSMUSG00000104235 | MGI:5610817 | unclassified gene | predicted gene, 37589 |
| 3 | gene | 60.57770 | 60.58136 | positive | MGI_C57BL6J_5012001 | Gm19816 | NA | NA | unclassified gene | predicted gene%2c 19816 |
| 3 | gene | 60.60438 | 60.60776 | positive | MGI_C57BL6J_5610716 | Gm37488 | ENSEMBL:ENSMUSG00000104125 | MGI:5610716 | unclassified gene | predicted gene, 37488 |
| 3 | pseudogene | 60.73763 | 60.73779 | negative | MGI_C57BL6J_5611508 | Gm38280 | ENSEMBL:ENSMUSG00000104337 | MGI:5611508 | pseudogene | predicted gene, 38280 |
| 3 | pseudogene | 60.76958 | 60.77164 | negative | MGI_C57BL6J_5592866 | Gm33707 | NCBI_Gene:102636711,ENSEMBL:ENSMUSG00000102298 | MGI:5592866 | pseudogene | predicted gene, 33707 |
| 3 | gene | 60.78148 | 61.00379 | negative | MGI_C57BL6J_5610263 | Gm37035 | NCBI_Gene:105244455,ENSEMBL:ENSMUSG00000102564 | MGI:5610263 | lncRNA gene | predicted gene, 37035 |
| 3 | gene | 60.78836 | 60.79473 | positive | MGI_C57BL6J_5611554 | Gm38326 | NCBI_Gene:102642162,ENSEMBL:ENSMUSG00000103587 | MGI:5611554 | lncRNA gene | predicted gene, 38326 |
| 3 | pseudogene | 60.86071 | 60.86109 | positive | MGI_C57BL6J_5610213 | Gm36985 | ENSEMBL:ENSMUSG00000102490 | MGI:5610213 | pseudogene | predicted gene, 36985 |
| 3 | pseudogene | 60.87700 | 60.87884 | positive | MGI_C57BL6J_3644852 | Gm8325 | NCBI_Gene:666853,ENSEMBL:ENSMUSG00000027694 | MGI:3644852 | pseudogene | predicted pseudogene 8325 |
| 3 | gene | 61.00279 | 61.00898 | positive | MGI_C57BL6J_105049 | P2ry1 | NCBI_Gene:18441,ENSEMBL:ENSMUSG00000027765 | MGI:105049 | protein coding gene | purinergic receptor P2Y, G-protein coupled 1 |
| 3 | pseudogene | 61.29116 | 61.29145 | negative | MGI_C57BL6J_5610947 | Gm37719 | ENSEMBL:ENSMUSG00000102242 | MGI:5610947 | pseudogene | predicted gene, 37719 |
| 3 | pseudogene | 61.32849 | 61.32923 | negative | MGI_C57BL6J_5012086 | Gm19901 | NCBI_Gene:100503798,ENSEMBL:ENSMUSG00000102667 | MGI:5012086 | pseudogene | predicted gene, 19901 |
| 3 | gene | 61.35855 | 61.36190 | negative | MGI_C57BL6J_5610924 | Gm37696 | ENSEMBL:ENSMUSG00000103473 | MGI:5610924 | lncRNA gene | predicted gene, 37696 |
| 3 | gene | 61.36164 | 61.36870 | positive | MGI_C57BL6J_1921262 | Rap2b | NCBI_Gene:74012,ENSEMBL:ENSMUSG00000036894 | MGI:1921262 | protein coding gene | RAP2B, member of RAS oncogene family |
| 3 | gene | 61.36225 | 61.36595 | negative | MGI_C57BL6J_3697707 | B430305J03Rik | ENSEMBL:ENSMUSG00000053706 | MGI:3697707 | protein coding gene | RIKEN cDNA B430305J03 gene |
| 3 | gene | 61.39215 | 61.41303 | positive | MGI_C57BL6J_5622946 | Gm40061 | NCBI_Gene:105244456 | MGI:5622946 | lncRNA gene | predicted gene, 40061 |
| 3 | pseudogene | 61.46166 | 61.46288 | negative | MGI_C57BL6J_3643135 | Gm8349 | NCBI_Gene:666891,ENSEMBL:ENSMUSG00000098208 | MGI:3643135 | pseudogene | predicted gene 8349 |
| 3 | pseudogene | 62.06754 | 62.06979 | negative | MGI_C57BL6J_3780107 | Gm9700 | NCBI_Gene:676965,ENSEMBL:ENSMUSG00000104489 | MGI:3780107 | pseudogene | predicted gene 9700 |
| 3 | gene | 62.12321 | 62.12356 | positive | MGI_C57BL6J_5610632 | Gm37404 | ENSEMBL:ENSMUSG00000103322 | MGI:5610632 | unclassified gene | predicted gene, 37404 |
| 3 | gene | 62.33752 | 62.34053 | negative | MGI_C57BL6J_3704218 | 9330121J05Rik | ENSEMBL:ENSMUSG00000103502 | MGI:3704218 | lncRNA gene | RIKEN cDNA 9330121J05 gene |
| 3 | gene | 62.33834 | 62.46222 | positive | MGI_C57BL6J_1918053 | Arhgef26 | NCBI_Gene:622434,ENSEMBL:ENSMUSG00000036885 | MGI:1918053 | protein coding gene | Rho guanine nucleotide exchange factor (GEF) 26 |
| 3 | gene | 62.34139 | 62.35052 | negative | MGI_C57BL6J_5593082 | Gm33923 | NCBI_Gene:102637008 | MGI:5593082 | lncRNA gene | predicted gene, 33923 |
| 3 | gene | 62.46801 | 62.50703 | negative | MGI_C57BL6J_1919412 | Dhx36 | NCBI_Gene:72162,ENSEMBL:ENSMUSG00000027770 | MGI:1919412 | protein coding gene | DEAH (Asp-Glu-Ala-His) box polypeptide 36 |
| 3 | gene | 62.52908 | 62.60544 | negative | MGI_C57BL6J_2443628 | Gpr149 | NCBI_Gene:229357,ENSEMBL:ENSMUSG00000043441 | MGI:2443628 | protein coding gene | G protein-coupled receptor 149 |
| 3 | gene | 62.60493 | 62.76393 | positive | MGI_C57BL6J_5593207 | Gm34048 | NCBI_Gene:102637165 | MGI:5593207 | lncRNA gene | predicted gene, 34048 |
| 3 | pseudogene | 62.76144 | 62.76202 | positive | MGI_C57BL6J_5010567 | Gm18382 | NCBI_Gene:100417044,ENSEMBL:ENSMUSG00000103764 | MGI:5010567 | pseudogene | predicted gene, 18382 |
| 3 | gene | 62.77198 | 62.86064 | positive | MGI_C57BL6J_5622947 | Gm40062 | NCBI_Gene:105244457 | MGI:5622947 | lncRNA gene | predicted gene, 40062 |
| 3 | gene | 62.86768 | 62.88900 | positive | MGI_C57BL6J_5622948 | Gm40063 | NCBI_Gene:105244458 | MGI:5622948 | lncRNA gene | predicted gene, 40063 |
| 3 | pseudogene | 62.92376 | 62.92426 | negative | MGI_C57BL6J_3780108 | Gm9701 | NCBI_Gene:676984,ENSEMBL:ENSMUSG00000063001 | MGI:3780108 | pseudogene | predicted gene 9701 |
| 3 | gene | 63.15222 | 63.15236 | positive | MGI_C57BL6J_5452210 | Gm22433 | ENSEMBL:ENSMUSG00000089175 | MGI:5452210 | snoRNA gene | predicted gene, 22433 |
| 3 | pseudogene | 63.17311 | 63.17386 | negative | MGI_C57BL6J_3645001 | Gm8388 | NCBI_Gene:666967,ENSEMBL:ENSMUSG00000102270 | MGI:3645001 | pseudogene | predicted gene 8388 |
| 3 | gene | 63.23282 | 63.24545 | negative | MGI_C57BL6J_5622949 | Gm40064 | NCBI_Gene:105244459 | MGI:5622949 | lncRNA gene | predicted gene, 40064 |
| 3 | gene | 63.24154 | 63.38603 | positive | MGI_C57BL6J_97004 | Mme | NCBI_Gene:17380,ENSEMBL:ENSMUSG00000027820 | MGI:97004 | protein coding gene | membrane metallo endopeptidase |
| 3 | gene | 63.43925 | 63.45790 | positive | MGI_C57BL6J_5593399 | Gm34240 | NCBI_Gene:102637430,ENSEMBL:ENSMUSG00000104336 | MGI:5593399 | lncRNA gene | predicted gene, 34240 |
| 3 | gene | 63.48110 | 63.48391 | negative | MGI_C57BL6J_5593461 | Strit1 | NCBI_Gene:102637511,ENSEMBL:ENSMUSG00000103476 | MGI:5593461 | protein coding gene | small transmembrane regulator of ion transport 1 |
| 3 | gene | 63.48388 | 63.48437 | positive | MGI_C57BL6J_5610507 | Gm37279 | ENSEMBL:ENSMUSG00000102883 | MGI:5610507 | unclassified gene | predicted gene, 37279 |
| 3 | gene | 63.48899 | 63.48910 | positive | MGI_C57BL6J_5454723 | Gm24946 | ENSEMBL:ENSMUSG00000088957 | MGI:5454723 | snRNA gene | predicted gene, 24946 |
| 3 | gene | 63.69620 | 63.89972 | negative | MGI_C57BL6J_2683547 | Plch1 | NCBI_Gene:269437,ENSEMBL:ENSMUSG00000036834 | MGI:2683547 | protein coding gene | phospholipase C, eta 1 |
| 3 | gene | 63.77021 | 63.77031 | positive | MGI_C57BL6J_5453277 | Gm23500 | ENSEMBL:ENSMUSG00000088360 | MGI:5453277 | snRNA gene | predicted gene, 23500 |
| 3 | gene | 63.79610 | 63.83685 | positive | MGI_C57BL6J_5826444 | Gm46807 | NCBI_Gene:108168874 | MGI:5826444 | lncRNA gene | predicted gene, 46807 |
| 3 | pseudogene | 63.82579 | 63.82644 | positive | MGI_C57BL6J_3802047 | Gm16162 | ENSEMBL:ENSMUSG00000090131 | MGI:3802047 | pseudogene | predicted gene 16162 |
| 3 | pseudogene | 63.84312 | 63.84407 | positive | MGI_C57BL6J_3801947 | Gm16129 | ENSEMBL:ENSMUSG00000085290 | MGI:3801947 | pseudogene | predicted gene 16129 |
| 3 | pseudogene | 63.90543 | 63.90573 | positive | MGI_C57BL6J_5593538 | Gm34379 | NCBI_Gene:102637617,ENSEMBL:ENSMUSG00000103905 | MGI:5593538 | pseudogene | predicted gene, 34379 |
| 3 | gene | 63.91468 | 63.92947 | negative | MGI_C57BL6J_3607716 | E130311K13Rik | NCBI_Gene:329659,ENSEMBL:ENSMUSG00000048581 | MGI:3607716 | protein coding gene | RIKEN cDNA E130311K13 gene |
| 3 | gene | 63.93351 | 63.96477 | negative | MGI_C57BL6J_1332247 | Slc33a1 | NCBI_Gene:11416,ENSEMBL:ENSMUSG00000027822 | MGI:1332247 | protein coding gene | solute carrier family 33 (acetyl-CoA transporter), member 1 |
| 3 | gene | 63.96363 | 63.96866 | positive | MGI_C57BL6J_5477344 | Gm26850 | ENSEMBL:ENSMUSG00000097724 | MGI:5477344 | lncRNA gene | predicted gene, 26850 |
| 3 | gene | 63.96482 | 64.02258 | positive | MGI_C57BL6J_2448526 | Gmps | NCBI_Gene:229363,ENSEMBL:ENSMUSG00000027823 | MGI:2448526 | protein coding gene | guanine monophosphate synthetase |
| 3 | pseudogene | 64.05303 | 64.05561 | negative | MGI_C57BL6J_3757653 | Vmn2r-ps3 | NCBI_Gene:100124546,ENSEMBL:ENSMUSG00000090303 | MGI:3757653 | pseudogene | vomeronasal 2, receptor, pseudogene 3 |
| 3 | gene | 64.07977 | 64.22085 | positive | MGI_C57BL6J_3645892 | Vmn2r1 | NCBI_Gene:56544,ENSEMBL:ENSMUSG00000027824 | MGI:3645892 | protein coding gene | vomeronasal 2, receptor 1 |
| 3 | gene | 64.10963 | 64.10974 | positive | MGI_C57BL6J_5451942 | Gm22165 | ENSEMBL:ENSMUSG00000089039 | MGI:5451942 | snRNA gene | predicted gene, 22165 |
| 3 | gene | 64.11528 | 64.17092 | negative | MGI_C57BL6J_3757666 | Vmn2r2 | NCBI_Gene:100125586,ENSEMBL:ENSMUSG00000043897 | MGI:3757666 | protein coding gene | vomeronasal 2, receptor 2 |
| 3 | pseudogene | 64.16339 | 64.16385 | positive | MGI_C57BL6J_3779465 | Gm5139 | NCBI_Gene:380739,ENSEMBL:ENSMUSG00000093447 | MGI:3779465 | pseudogene | predicted gene 5139 |
| 3 | pseudogene | 64.19557 | 64.21368 | negative | MGI_C57BL6J_5010479 | Gm18294 | NCBI_Gene:100416861,ENSEMBL:ENSMUSG00000093416 | MGI:5010479 | pseudogene | predicted gene, 18294 |
| 3 | gene | 64.23373 | 64.23546 | positive | MGI_C57BL6J_5593621 | Gm34462 | NCBI_Gene:102637724 | MGI:5593621 | lncRNA gene | predicted gene, 34462 |
| 3 | gene | 64.25722 | 64.28971 | negative | MGI_C57BL6J_3643995 | Vmn2r3 | NCBI_Gene:637004,ENSEMBL:ENSMUSG00000091572 | MGI:3643995 | protein coding gene | vomeronasal 2, receptor 3 |
| 3 | gene | 64.27582 | 64.27619 | negative | MGI_C57BL6J_1345615 | Ifld3 | NA | NA | unclassified gene | induced in fatty liver dystrophy 3 |
| 3 | gene | 64.31841 | 64.31856 | negative | MGI_C57BL6J_5453674 | Gm23897 | ENSEMBL:ENSMUSG00000088735 | MGI:5453674 | snoRNA gene | predicted gene, 23897 |
| 3 | pseudogene | 64.32866 | 64.33285 | positive | MGI_C57BL6J_3757669 | Vmn2r-ps4 | NCBI_Gene:100124547,ENSEMBL:ENSMUSG00000093503 | MGI:3757669 | pseudogene | vomeronasal 2, receptor, pseudogene 4 |
| 3 | pseudogene | 64.33956 | 64.35772 | negative | MGI_C57BL6J_3645404 | Vmn2r-ps5 | NCBI_Gene:667031,ENSEMBL:ENSMUSG00000091967 | MGI:3645404 | polymorphic pseudogene | vomeronasal 2, receptor, pseudogene 5 |
| 3 | gene | 64.38304 | 64.41537 | negative | MGI_C57BL6J_3648229 | Vmn2r4 | NCBI_Gene:637053,ENSEMBL:ENSMUSG00000092049 | MGI:3648229 | protein coding gene | vomeronasal 2, receptor 4 |
| 3 | pseudogene | 64.43688 | 64.43829 | negative | MGI_C57BL6J_3757678 | Vmn2r-ps6 | NCBI_Gene:100124548,ENSEMBL:ENSMUSG00000093395 | MGI:3757678 | pseudogene | vomeronasal 2, receptor, pseudogene 6 |
| 3 | pseudogene | 64.44260 | 64.44397 | negative | MGI_C57BL6J_3757679 | Vmn2r-ps7 | NCBI_Gene:672951,ENSEMBL:ENSMUSG00000093546 | MGI:3757679 | pseudogene | vomeronasal 2, receptor, pseudogene 7 |
| 3 | pseudogene | 64.44797 | 64.46009 | positive | MGI_C57BL6J_5313103 | Gm20656 | ENSEMBL:ENSMUSG00000093671 | MGI:5313103 | pseudogene | predicted gene 20656 |
| 3 | pseudogene | 64.47702 | 64.47932 | positive | MGI_C57BL6J_3757680 | Vmn2r-ps8 | NCBI_Gene:100124549,ENSEMBL:ENSMUSG00000094482 | MGI:3757680 | pseudogene | vomeronasal 2, receptor, pseudogene 8 |
| 3 | gene | 64.48861 | 64.50984 | negative | MGI_C57BL6J_3649074 | Vmn2r5 | NCBI_Gene:667060,ENSEMBL:ENSMUSG00000068999 | MGI:3649074 | protein coding gene | vomeronasal 2, receptor 5 |
| 3 | gene | 64.53131 | 64.56583 | negative | MGI_C57BL6J_3649068 | Vmn2r6 | NCBI_Gene:667069,ENSEMBL:ENSMUSG00000090581 | MGI:3649068 | protein coding gene | vomeronasal 2, receptor 6 |
| 3 | pseudogene | 64.59152 | 64.59290 | negative | MGI_C57BL6J_3757686 | Vmn2r-ps9 | NCBI_Gene:100124550,ENSEMBL:ENSMUSG00000093699 | MGI:3757686 | pseudogene | vomeronasal 2, receptor, pseudogene 9 |
| 3 | pseudogene | 64.59691 | 64.60577 | positive | MGI_C57BL6J_5313104 | Gm20657 | ENSEMBL:ENSMUSG00000093484 | MGI:5313104 | pseudogene | predicted gene 20657 |
| 3 | pseudogene | 64.60043 | 64.60114 | positive | MGI_C57BL6J_3781218 | Gm3040 | NCBI_Gene:100416170 | MGI:3781218 | pseudogene | predicted gene 3040 |
| 3 | pseudogene | 64.62169 | 64.62399 | positive | MGI_C57BL6J_3757943 | Vmn2r-ps10 | NCBI_Gene:100124554,ENSEMBL:ENSMUSG00000095107 | MGI:3757943 | pseudogene | vomeronasal 2, receptor, pseudogene 10 |
| 3 | pseudogene | 64.63257 | 64.84474 | negative | MGI_C57BL6J_2441682 | Vmn2r-ps11 | NCBI_Gene:319210,ENSEMBL:ENSMUSG00000093434 | MGI:2441682 | pseudogene | vomeronasal 2, receptor, pseudogene 11 |
| 3 | gene | 64.63286 | 64.84474 | negative | MGI_C57BL6J_5504054 | Gm26939 | ENSEMBL:ENSMUSG00000097538 | MGI:5504054 | lncRNA gene | predicted gene, 26939 |
| 3 | gene | 64.68504 | 64.84476 | negative | MGI_C57BL6J_2441693 | Vmn2r7 | NCBI_Gene:319217,ENSEMBL:ENSMUSG00000062200 | MGI:2441693 | protein coding gene | vomeronasal 2, receptor 7 |
| 3 | pseudogene | 64.74475 | 64.74564 | negative | MGI_C57BL6J_3757944 | Vmn2r-ps12 | NCBI_Gene:100125273,ENSEMBL:ENSMUSG00000093474 | MGI:3757944 | pseudogene | vomeronasal 2, receptor, pseudogene 12 |
| 3 | gene | 64.89447 | 64.89460 | negative | MGI_C57BL6J_5454999 | Gm25222 | ENSEMBL:ENSMUSG00000088824 | MGI:5454999 | snoRNA gene | predicted gene, 25222 |
| 3 | pseudogene | 64.89885 | 64.89976 | positive | MGI_C57BL6J_3757946 | Vmn2r-ps13 | NCBI_Gene:670110,ENSEMBL:ENSMUSG00000093767 | MGI:3757946 | pseudogene | vomeronasal 2, receptor, pseudogene 13 |
| 3 | gene | 64.94905 | 65.37823 | positive | MGI_C57BL6J_109155 | Kcnab1 | NCBI_Gene:16497,ENSEMBL:ENSMUSG00000027827 | MGI:109155 | protein coding gene | potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| 3 | gene | 64.94922 | 65.22370 | positive | MGI_C57BL6J_5611276 | Gm38048 | ENSEMBL:ENSMUSG00000102437 | MGI:5611276 | lncRNA gene | predicted gene, 38048 |
| 3 | gene | 65.13306 | 65.30268 | negative | MGI_C57BL6J_2443553 | A330015K06Rik | NCBI_Gene:102637797,ENSEMBL:ENSMUSG00000104093 | MGI:2443553 | lncRNA gene | RIKEN cDNA A330015K06 gene |
| 3 | gene | 65.32402 | 65.34943 | negative | MGI_C57BL6J_2442748 | A730090N16Rik | NCBI_Gene:319798,ENSEMBL:ENSMUSG00000090120 | MGI:2442748 | lncRNA gene | RIKEN cDNA A730090N16 gene |
| 3 | gene | 65.37965 | 65.39262 | negative | MGI_C57BL6J_1914687 | Ssr3 | NCBI_Gene:67437,ENSEMBL:ENSMUSG00000027828 | MGI:1914687 | protein coding gene | signal sequence receptor, gamma |
| 3 | pseudogene | 65.39332 | 65.39629 | positive | MGI_C57BL6J_3644824 | Gm5540 | NCBI_Gene:433604,ENSEMBL:ENSMUSG00000048916 | MGI:3644824 | pseudogene | predicted pseudogene 5540 |
| 3 | gene | 65.44941 | 65.47981 | positive | MGI_C57BL6J_5593735 | Gm34576 | NCBI_Gene:102637876 | MGI:5593735 | lncRNA gene | predicted gene, 34576 |
| 3 | gene | 65.49409 | 65.49563 | negative | MGI_C57BL6J_5593939 | Gm34780 | NCBI_Gene:102638146,ENSEMBL:ENSMUSG00000103513 | MGI:5593939 | lncRNA gene | predicted gene, 34780 |
| 3 | gene | 65.49517 | 65.51729 | positive | MGI_C57BL6J_5622951 | Gm40066 | NCBI_Gene:105244461 | MGI:5622951 | lncRNA gene | predicted gene, 40066 |
| 3 | pseudogene | 65.50628 | 65.50664 | negative | MGI_C57BL6J_5610237 | Gm37009 | ENSEMBL:ENSMUSG00000103880 | MGI:5610237 | pseudogene | predicted gene, 37009 |
| 3 | gene | 65.50646 | 65.52004 | negative | MGI_C57BL6J_5621388 | Gm38503 | NCBI_Gene:102637953 | MGI:5621388 | lncRNA gene | predicted gene, 38503 |
| 3 | gene | 65.50726 | 65.52941 | negative | MGI_C57BL6J_1918254 | 4931440P22Rik | NCBI_Gene:71004,ENSEMBL:ENSMUSG00000074580 | MGI:1918254 | lncRNA gene | RIKEN cDNA 4931440P22 gene |
| 3 | gene | 65.52570 | 65.52579 | positive | MGI_C57BL6J_5452056 | Gm22279 | ENSEMBL:ENSMUSG00000087825 | MGI:5452056 | snRNA gene | predicted gene, 22279 |
| 3 | gene | 65.52841 | 65.55552 | positive | MGI_C57BL6J_2159210 | Tiparp | NCBI_Gene:99929,ENSEMBL:ENSMUSG00000034640 | MGI:2159210 | protein coding gene | TCDD-inducible poly(ADP-ribose) polymerase |
| 3 | gene | 65.57522 | 65.58085 | negative | MGI_C57BL6J_5594083 | Gm34924 | NCBI_Gene:102638334 | MGI:5594083 | lncRNA gene | predicted gene, 34924 |
| 3 | pseudogene | 65.61350 | 65.61532 | positive | MGI_C57BL6J_3648046 | Gm5847 | NCBI_Gene:545530,ENSEMBL:ENSMUSG00000082280 | MGI:3648046 | pseudogene | predicted gene 5847 |
| 3 | pseudogene | 65.61537 | 65.61617 | negative | MGI_C57BL6J_5611461 | Gm38233 | ENSEMBL:ENSMUSG00000103294 | MGI:5611461 | pseudogene | predicted gene, 38233 |
| 3 | gene | 65.65929 | 65.65943 | positive | MGI_C57BL6J_5531019 | Mir8120 | miRBase:MI0026052,NCBI_Gene:102465905,ENSEMBL:ENSMUSG00000098944 | MGI:5531019 | miRNA gene | microRNA 8120 |
| 3 | gene | 65.65954 | 65.83248 | positive | MGI_C57BL6J_3645902 | Lekr1 | NCBI_Gene:624866,ENSEMBL:ENSMUSG00000074579 | MGI:3645902 | protein coding gene | leucine, glutamate and lysine rich 1 |
| 3 | gene | 65.67839 | 65.68088 | positive | MGI_C57BL6J_5611004 | Gm37776 | ENSEMBL:ENSMUSG00000104037 | MGI:5611004 | unclassified gene | predicted gene, 37776 |
| 3 | gene | 65.77151 | 65.77162 | negative | MGI_C57BL6J_5455532 | Gm25755 | ENSEMBL:ENSMUSG00000065330 | MGI:5455532 | snRNA gene | predicted gene, 25755 |
| 3 | gene | 65.82912 | 65.87228 | negative | MGI_C57BL6J_5610587 | Gm37359 | ENSEMBL:ENSMUSG00000103620 | MGI:5610587 | lncRNA gene | predicted gene, 37359 |
| 3 | gene | 65.86327 | 65.86483 | negative | MGI_C57BL6J_5610359 | Gm37131 | ENSEMBL:ENSMUSG00000103440 | MGI:5610359 | unclassified gene | predicted gene, 37131 |
| 3 | gene | 65.92610 | 65.92828 | negative | MGI_C57BL6J_5594198 | Gm35039 | NCBI_Gene:102638487 | MGI:5594198 | lncRNA gene | predicted gene, 35039 |
| 3 | pseudogene | 65.93349 | 65.93391 | negative | MGI_C57BL6J_5610265 | Gm37037 | ENSEMBL:ENSMUSG00000102566 | MGI:5610265 | pseudogene | predicted gene, 37037 |
| 3 | gene | 65.94591 | 65.95839 | negative | MGI_C57BL6J_1922664 | Ccnl1 | NCBI_Gene:56706,ENSEMBL:ENSMUSG00000027829 | MGI:1922664 | protein coding gene | cyclin L1 |
| 3 | gene | 65.95776 | 65.96204 | positive | MGI_C57BL6J_5610533 | Gm37305 | ENSEMBL:ENSMUSG00000103041 | MGI:5610533 | lncRNA gene | predicted gene, 37305 |
| 3 | pseudogene | 65.96873 | 65.97028 | negative | MGI_C57BL6J_5594265 | Gm35106 | NCBI_Gene:105244463,ENSEMBL:ENSMUSG00000102914 | MGI:5594265 | pseudogene | predicted gene, 35106 |
| 3 | gene | 65.96991 | 65.97192 | positive | MGI_C57BL6J_5610306 | Gm37078 | ENSEMBL:ENSMUSG00000102652 | MGI:5610306 | unclassified gene | predicted gene, 37078 |
| 3 | pseudogene | 65.98716 | 65.98975 | negative | MGI_C57BL6J_5011209 | Gm19024 | NCBI_Gene:100418133,ENSEMBL:ENSMUSG00000103641 | MGI:5011209 | pseudogene | predicted gene, 19024 |
| 3 | pseudogene | 66.02950 | 66.03152 | negative | MGI_C57BL6J_3643872 | Gm6546 | NCBI_Gene:625048,ENSEMBL:ENSMUSG00000103369 | MGI:3643872 | pseudogene | predicted gene 6546 |
| 3 | gene | 66.05356 | 66.29707 | negative | MGI_C57BL6J_1920039 | Veph1 | NCBI_Gene:72789,ENSEMBL:ENSMUSG00000027831 | MGI:1920039 | protein coding gene | ventricular zone expressed PH domain-containing 1 |
| 3 | gene | 66.08668 | 66.08779 | positive | MGI_C57BL6J_5611050 | Gm37822 | ENSEMBL:ENSMUSG00000104189 | MGI:5611050 | unclassified gene | predicted gene, 37822 |
| 3 | gene | 66.08811 | 66.08826 | negative | MGI_C57BL6J_5456219 | Gm26442 | ENSEMBL:ENSMUSG00000064522 | MGI:5456219 | snRNA gene | predicted gene, 26442 |
| 3 | pseudogene | 66.09981 | 66.10006 | positive | MGI_C57BL6J_5610201 | Gm36973 | ENSEMBL:ENSMUSG00000104307 | MGI:5610201 | pseudogene | predicted gene, 36973 |
| 3 | pseudogene | 66.12840 | 66.12975 | negative | MGI_C57BL6J_98800 | Tpi-rs2 | NCBI_Gene:111205,ENSEMBL:ENSMUSG00000097968 | MGI:98800 | pseudogene | triosephosphate isomerase related sequence 2 |
| 3 | pseudogene | 66.19109 | 66.19124 | negative | MGI_C57BL6J_5610558 | Gm37330 | ENSEMBL:ENSMUSG00000103208 | MGI:5610558 | pseudogene | predicted gene, 37330 |
| 3 | gene | 66.21989 | 66.22581 | positive | MGI_C57BL6J_104641 | Ptx3 | NCBI_Gene:19288,ENSEMBL:ENSMUSG00000027832 | MGI:104641 | protein coding gene | pentraxin related gene |
| 3 | pseudogene | 66.36454 | 66.36463 | positive | MGI_C57BL6J_6275305 | Gm50014 | ENSEMBL:ENSMUSG00000117623 | MGI:6275305 | pseudogene | predicted gene, 50014 |
| 3 | gene | 66.37430 | 66.38810 | positive | MGI_C57BL6J_5594343 | Gm35184 | NCBI_Gene:102638679 | MGI:5594343 | lncRNA gene | predicted gene, 35184 |
| 3 | gene | 66.40041 | 66.40607 | positive | MGI_C57BL6J_5594395 | Gm35236 | NCBI_Gene:102638746 | MGI:5594395 | lncRNA gene | predicted gene, 35236 |
| 3 | pseudogene | 66.56278 | 66.56355 | negative | MGI_C57BL6J_5010137 | Gm17952 | NCBI_Gene:100416166,ENSEMBL:ENSMUSG00000103600 | MGI:5010137 | pseudogene | predicted gene, 17952 |
| 3 | pseudogene | 66.88235 | 66.88314 | positive | MGI_C57BL6J_3646021 | Gm6555 | NCBI_Gene:625115,ENSEMBL:ENSMUSG00000103700 | MGI:3646021 | pseudogene | predicted gene 6555 |
| 3 | pseudogene | 66.90736 | 66.90777 | negative | MGI_C57BL6J_5611584 | Gm38356 | ENSEMBL:ENSMUSG00000103318 | MGI:5611584 | pseudogene | predicted gene, 38356 |
| 3 | pseudogene | 66.95354 | 66.95386 | positive | MGI_C57BL6J_5663653 | Gm43516 | ENSEMBL:ENSMUSG00000105794 | MGI:5663653 | pseudogene | predicted gene 43516 |
| 3 | gene | 66.97172 | 66.98177 | negative | MGI_C57BL6J_1201673 | Shox2 | NCBI_Gene:20429,ENSEMBL:ENSMUSG00000027833 | MGI:1201673 | protein coding gene | short stature homeobox 2 |
| 3 | gene | 66.98139 | 67.35840 | positive | MGI_C57BL6J_1914130 | Rsrc1 | NCBI_Gene:66880,ENSEMBL:ENSMUSG00000034544 | MGI:1914130 | protein coding gene | arginine/serine-rich coiled-coil 1 |
| 3 | gene | 67.05596 | 67.05606 | positive | MGI_C57BL6J_5452072 | Gm22295 | ENSEMBL:ENSMUSG00000065245 | MGI:5452072 | snRNA gene | predicted gene, 22295 |
| 3 | gene | 67.36546 | 67.37516 | negative | MGI_C57BL6J_4937036 | Gm17402 | ENSEMBL:ENSMUSG00000090408 | MGI:4937036 | protein coding gene | predicted gene, 17402 |
| 3 | gene | 67.37410 | 67.40000 | positive | MGI_C57BL6J_1341819 | Mlf1 | NCBI_Gene:17349,ENSEMBL:ENSMUSG00000048416 | MGI:1341819 | protein coding gene | myeloid leukemia factor 1 |
| 3 | gene | 67.43010 | 67.47653 | positive | MGI_C57BL6J_107339 | Gfm1 | NCBI_Gene:28030,ENSEMBL:ENSMUSG00000027774 | MGI:107339 | protein coding gene | G elongation factor, mitochondrial 1 |
| 3 | gene | 67.45800 | 67.46393 | negative | MGI_C57BL6J_107633 | Lxn | NCBI_Gene:17035,ENSEMBL:ENSMUSG00000047557 | MGI:107633 | protein coding gene | latexin |
| 3 | gene | 67.47698 | 67.47882 | positive | MGI_C57BL6J_5662705 | Gm42568 | ENSEMBL:ENSMUSG00000103703 | MGI:5662705 | unclassified gene | predicted gene 42568 |
| 3 | gene | 67.47889 | 67.51552 | negative | MGI_C57BL6J_1924461 | Rarres1 | NCBI_Gene:109222,ENSEMBL:ENSMUSG00000049404 | MGI:1924461 | protein coding gene | retinoic acid receptor responder (tazarotene induced) 1 |
| 3 | gene | 67.51567 | 67.51966 | positive | MGI_C57BL6J_5622952 | Gm40067 | NCBI_Gene:105244464 | MGI:5622952 | lncRNA gene | predicted gene, 40067 |
| 3 | gene | 67.52266 | 67.55343 | negative | MGI_C57BL6J_5594458 | Gm35299 | NCBI_Gene:102638825,ENSEMBL:ENSMUSG00000103718 | MGI:5594458 | lncRNA gene | predicted gene, 35299 |
| 3 | gene | 67.57057 | 67.57282 | negative | MGI_C57BL6J_1921179 | 4930402C01Rik | ENSEMBL:ENSMUSG00000104382 | MGI:1921179 | lncRNA gene | RIKEN cDNA 4930402C01 gene |
| 3 | gene | 67.57316 | 67.57329 | negative | MGI_C57BL6J_5455696 | Gm25919 | ENSEMBL:ENSMUSG00000077459 | MGI:5455696 | snoRNA gene | predicted gene, 25919 |
| 3 | gene | 67.58274 | 67.60424 | positive | MGI_C57BL6J_1914118 | Mfsd1 | NCBI_Gene:66868,ENSEMBL:ENSMUSG00000027775 | MGI:1914118 | protein coding gene | major facilitator superfamily domain containing 1 |
| 3 | pseudogene | 67.60575 | 67.60785 | negative | MGI_C57BL6J_3648243 | Gm8515 | NCBI_Gene:667207,ENSEMBL:ENSMUSG00000103206 | MGI:3648243 | pseudogene | predicted gene 8515 |
| 3 | pseudogene | 67.61978 | 67.62073 | negative | MGI_C57BL6J_2685258 | Gm412 | NCBI_Gene:208424,ENSEMBL:ENSMUSG00000103606 | MGI:2685258 | pseudogene | predicted gene 412 |
| 3 | pseudogene | 67.68436 | 67.68475 | negative | MGI_C57BL6J_5610829 | Gm37601 | ENSEMBL:ENSMUSG00000102746 | MGI:5610829 | pseudogene | predicted gene, 37601 |
| 3 | pseudogene | 67.74197 | 67.74330 | negative | MGI_C57BL6J_5010902 | Gm18717 | NCBI_Gene:100417618,ENSEMBL:ENSMUSG00000104477 | MGI:5010902 | pseudogene | predicted gene, 18717 |
| 3 | gene | 67.75360 | 67.75565 | positive | MGI_C57BL6J_5594566 | Gm35407 | NCBI_Gene:102638975 | MGI:5594566 | lncRNA gene | predicted gene, 35407 |
| 3 | gene | 67.75566 | 67.76628 | positive | MGI_C57BL6J_5622953 | Gm40068 | NCBI_Gene:105244465 | MGI:5622953 | lncRNA gene | predicted gene, 40068 |
| 3 | gene | 67.89222 | 68.05659 | positive | MGI_C57BL6J_3644166 | Iqcj | NCBI_Gene:208426,ENSEMBL:ENSMUSG00000051777 | MGI:3644166 | protein coding gene | IQ motif containing J |
| 3 | gene | 67.89222 | 68.62648 | positive | MGI_C57BL6J_5439400 | Iqschfp | NCBI_Gene:100505386,ENSEMBL:ENSMUSG00000102422 | MGI:5439400 | protein coding gene | Iqcj and Schip1 fusion protein |
| 3 | gene | 68.06480 | 68.62648 | positive | MGI_C57BL6J_1353557 | Schip1 | NCBI_Gene:30953,ENSEMBL:ENSMUSG00000027777 | MGI:1353557 | protein coding gene | schwannomin interacting protein 1 |
| 3 | pseudogene | 68.30377 | 68.31036 | negative | MGI_C57BL6J_3648047 | Gm5848 | NCBI_Gene:545531 | MGI:3648047 | pseudogene | predicted pseudogene 5848 |
| 3 | pseudogene | 68.33319 | 68.33378 | positive | MGI_C57BL6J_3642851 | Gm10292 | NCBI_Gene:631379,ENSEMBL:ENSMUSG00000102846 | MGI:3642851 | pseudogene | predicted gene 10292 |
| 3 | gene | 68.40778 | 68.41470 | positive | MGI_C57BL6J_3641783 | Gm10723 | ENSEMBL:ENSMUSG00000103345 | MGI:3641783 | unclassified gene | predicted gene 10723 |
| 3 | gene | 68.54752 | 68.54771 | negative | MGI_C57BL6J_5451850 | Gm22073 | ENSEMBL:ENSMUSG00000084617 | MGI:5451850 | snRNA gene | predicted gene, 22073 |
| 3 | gene | 68.69064 | 68.69855 | positive | MGI_C57BL6J_96539 | Il12a | NCBI_Gene:16159,ENSEMBL:ENSMUSG00000027776 | MGI:96539 | protein coding gene | interleukin 12a |
| 3 | pseudogene | 68.73314 | 68.73808 | positive | MGI_C57BL6J_3708676 | Gm10040 | ENSEMBL:ENSMUSG00000058360 | MGI:3708676 | pseudogene | predicted gene 10040 |
| 3 | gene | 68.75991 | 68.76143 | negative | MGI_C57BL6J_1922468 | 4930535E02Rik | ENSEMBL:ENSMUSG00000102677 | MGI:1922468 | lncRNA gene | RIKEN cDNA 4930535E02 gene |
| 3 | gene | 68.76136 | 68.76267 | positive | MGI_C57BL6J_5594743 | Gm35584 | NCBI_Gene:102639225,ENSEMBL:ENSMUSG00000102936 | MGI:5594743 | lncRNA gene | predicted gene, 35584 |
| 3 | pseudogene | 68.84608 | 68.84741 | positive | MGI_C57BL6J_3644317 | Gm7270 | NCBI_Gene:639530,ENSEMBL:ENSMUSG00000103218,ENSEMBL:ENSMUSG00000102853 | MGI:3644317 | pseudogene | predicted gene 7270 |
| 3 | gene | 68.86792 | 68.87027 | negative | MGI_C57BL6J_4937275 | Gm17641 | ENSEMBL:ENSMUSG00000091272 | MGI:4937275 | protein coding gene | predicted gene, 17641 |
| 3 | gene | 68.86959 | 68.87216 | positive | MGI_C57BL6J_1915975 | 1110032F04Rik | NCBI_Gene:68725,ENSEMBL:ENSMUSG00000046999 | MGI:1915975 | protein coding gene | RIKEN cDNA 1110032F04 gene |
| 3 | gene | 68.89250 | 69.00464 | negative | MGI_C57BL6J_1915509 | Ift80 | NCBI_Gene:68259,ENSEMBL:ENSMUSG00000027778 | MGI:1915509 | protein coding gene | intraflagellar transport 80 |
| 3 | pseudogene | 68.90017 | 68.90045 | negative | MGI_C57BL6J_5313136 | Gm20689 | ENSEMBL:ENSMUSG00000093384 | MGI:5313136 | pseudogene | predicted gene 20689 |
| 3 | pseudogene | 68.90181 | 68.90216 | positive | MGI_C57BL6J_5313137 | Gm20690 | ENSEMBL:ENSMUSG00000093730,ENSEMBL:ENSMUSG00000103007 | MGI:5313137 | pseudogene | predicted gene 20690 |
| 3 | pseudogene | 68.90216 | 68.90278 | negative | MGI_C57BL6J_5010773 | Gm18588 | NCBI_Gene:100417394,ENSEMBL:ENSMUSG00000093402 | MGI:5010773 | pseudogene | predicted gene, 18588 |
| 3 | pseudogene | 68.90382 | 68.90415 | negative | MGI_C57BL6J_5313138 | Gm20691 | ENSEMBL:ENSMUSG00000093505 | MGI:5313138 | pseudogene | predicted gene 20691 |
| 3 | gene | 68.91650 | 68.91732 | negative | MGI_C57BL6J_6097561 | Gm48183 | ENSEMBL:ENSMUSG00000111756 | MGI:6097561 | unclassified gene | predicted gene, 48183 |
| 3 | gene | 69.00474 | 69.03462 | positive | MGI_C57BL6J_1917349 | Smc4 | NCBI_Gene:70099,ENSEMBL:ENSMUSG00000034349 | MGI:1917349 | protein coding gene | structural maintenance of chromosomes 4 |
| 3 | gene | 69.00977 | 69.00983 | positive | MGI_C57BL6J_2676842 | Mir15b | miRBase:MI0000140,NCBI_Gene:387175,ENSEMBL:ENSMUSG00000065580 | MGI:2676842 | miRNA gene | microRNA 15b |
| 3 | gene | 69.00990 | 69.01000 | positive | MGI_C57BL6J_3618690 | Mir16-2 | miRBase:MI0000566,NCBI_Gene:723949,ENSEMBL:ENSMUSG00000065606 | MGI:3618690 | miRNA gene | microRNA 16-2 |
| 3 | gene | 69.03529 | 69.04640 | negative | MGI_C57BL6J_1914199 | Trim59 | NCBI_Gene:66949,ENSEMBL:ENSMUSG00000034317 | MGI:1914199 | protein coding gene | tripartite motif-containing 59 |
| 3 | gene | 69.06065 | 69.06525 | positive | MGI_C57BL6J_2442898 | 9630025H16Rik | NCBI_Gene:105244467 | MGI:2442898 | lncRNA gene | RIKEN cDNA 9630025H16 gene |
| 3 | gene | 69.06715 | 69.12711 | negative | MGI_C57BL6J_1100848 | Kpna4 | NCBI_Gene:16649,ENSEMBL:ENSMUSG00000027782 | MGI:1100848 | protein coding gene | karyopherin (importin) alpha 4 |
| 3 | gene | 69.08510 | 69.08545 | negative | MGI_C57BL6J_5451786 | Gm22009 | ENSEMBL:ENSMUSG00000089417 | MGI:5451786 | snoRNA gene | predicted gene, 22009 |
| 3 | gene | 69.11974 | 69.12272 | negative | MGI_C57BL6J_5610786 | Gm37558 | ENSEMBL:ENSMUSG00000103094 | MGI:5610786 | unclassified gene | predicted gene, 37558 |
| 3 | gene | 69.13123 | 69.15718 | positive | MGI_C57BL6J_2686493 | Gm1647 | NCBI_Gene:381452,ENSEMBL:ENSMUSG00000094763 | MGI:2686493 | lncRNA gene | predicted gene 1647 |
| 3 | gene | 69.22242 | 69.22362 | positive | MGI_C57BL6J_1918869 | Arl14 | NCBI_Gene:71619,ENSEMBL:ENSMUSG00000098207 | MGI:1918869 | protein coding gene | ADP-ribosylation factor-like 14 |
| 3 | gene | 69.25461 | 69.26383 | positive | MGI_C57BL6J_5622955 | Gm40070 | NCBI_Gene:105244468 | MGI:5622955 | lncRNA gene | predicted gene, 40070 |
| 3 | gene | 69.31686 | 69.56080 | positive | MGI_C57BL6J_2139740 | Ppm1l | NCBI_Gene:242083,ENSEMBL:ENSMUSG00000027784 | MGI:2139740 | protein coding gene | protein phosphatase 1 (formerly 2C)-like |
| 3 | gene | 69.31816 | 69.31929 | negative | MGI_C57BL6J_1920514 | 1700037N05Rik | NA | NA | unclassified gene | RIKEN cDNA 1700037N05 gene |
| 3 | pseudogene | 69.32001 | 69.32062 | negative | MGI_C57BL6J_4938041 | Gm17214 | NCBI_Gene:100416279,ENSEMBL:ENSMUSG00000091315 | MGI:4938041 | pseudogene | predicted gene 17214 |
| 3 | pseudogene | 69.33519 | 69.33579 | negative | MGI_C57BL6J_4938039 | Gm17212 | NCBI_Gene:100416280,ENSEMBL:ENSMUSG00000090941 | MGI:4938039 | pseudogene | predicted gene 17212 |
| 3 | gene | 69.37640 | 69.38370 | positive | MGI_C57BL6J_2443158 | A330078L11Rik | NA | NA | unclassified gene | RIKEN cDNA A330078L11 gene |
| 3 | pseudogene | 69.43101 | 69.43238 | negative | MGI_C57BL6J_4938040 | Gm17213 | NCBI_Gene:100417908,ENSEMBL:ENSMUSG00000090743 | MGI:4938040 | pseudogene | predicted gene 17213 |
| 3 | pseudogene | 69.56624 | 69.56747 | negative | MGI_C57BL6J_3644126 | Gm8565 | NCBI_Gene:667309,ENSEMBL:ENSMUSG00000098006 | MGI:3644126 | pseudogene | predicted gene 8565 |
| 3 | gene | 69.57392 | 69.59896 | negative | MGI_C57BL6J_1349405 | B3galnt1 | NCBI_Gene:26879,ENSEMBL:ENSMUSG00000043300 | MGI:1349405 | protein coding gene | UDP-GalNAc:betaGlcNAc beta 1,3-galactosaminyltransferase, polypeptide 1 |
| 3 | pseudogene | 69.66177 | 69.66216 | positive | MGI_C57BL6J_5610608 | Gm37380 | ENSEMBL:ENSMUSG00000102342 | MGI:5610608 | pseudogene | predicted gene, 37380 |
| 3 | gene | 69.68547 | 69.68558 | positive | MGI_C57BL6J_5455398 | Gm25621 | ENSEMBL:ENSMUSG00000087848 | MGI:5455398 | rRNA gene | predicted gene, 25621 |
| 3 | pseudogene | 69.71694 | 69.71744 | negative | MGI_C57BL6J_98039 | Rpl32-ps | NCBI_Gene:19952,ENSEMBL:ENSMUSG00000068969 | MGI:98039 | pseudogene | ribosomal protein L32, pseudogene |
| 3 | gene | 69.72199 | 69.75637 | positive | MGI_C57BL6J_2140103 | Nmd3 | NCBI_Gene:97112,ENSEMBL:ENSMUSG00000027787 | MGI:2140103 | protein coding gene | NMD3 ribosome export adaptor |
| 3 | gene | 69.75311 | 69.75637 | positive | MGI_C57BL6J_2442565 | 9530051E23Rik | NA | NA | unclassified gene | RIKEN cDNA 9530051E23 gene |
| 3 | pseudogene | 69.80846 | 69.80902 | negative | MGI_C57BL6J_5011240 | Gm19055 | NCBI_Gene:100418175 | MGI:5011240 | pseudogene | predicted gene, 19055 |
| 3 | gene | 69.81954 | 69.85994 | negative | MGI_C57BL6J_1913433 | Sptssb | NCBI_Gene:66183,ENSEMBL:ENSMUSG00000043461 | MGI:1913433 | protein coding gene | serine palmitoyltransferase, small subunit B |
| 3 | gene | 69.88247 | 69.88411 | positive | MGI_C57BL6J_5594976 | Gm35817 | NCBI_Gene:102639522 | MGI:5594976 | lncRNA gene | predicted gene, 35817 |
| 3 | pseudogene | 69.95481 | 69.95568 | negative | MGI_C57BL6J_5010080 | Gm17895 | NCBI_Gene:100416058,ENSEMBL:ENSMUSG00000103549 | MGI:5010080 | pseudogene | predicted gene, 17895 |
| 3 | gene | 69.96232 | 69.96245 | negative | MGI_C57BL6J_5453261 | Gm23484 | ENSEMBL:ENSMUSG00000077366 | MGI:5453261 | snoRNA gene | predicted gene, 23484 |
| 3 | pseudogene | 69.96328 | 69.96347 | positive | MGI_C57BL6J_5611012 | Gm37784 | ENSEMBL:ENSMUSG00000102525 | MGI:5611012 | pseudogene | predicted gene, 37784 |
| 3 | gene | 70.00761 | 70.02871 | positive | MGI_C57BL6J_2685260 | Otol1 | NCBI_Gene:229389,ENSEMBL:ENSMUSG00000027788 | MGI:2685260 | protein coding gene | otolin 1 |
| 3 | pseudogene | 70.05975 | 70.06047 | positive | MGI_C57BL6J_5610909 | Gm37681 | ENSEMBL:ENSMUSG00000103020 | MGI:5610909 | pseudogene | predicted gene, 37681 |
| 3 | pseudogene | 70.11625 | 70.11653 | negative | MGI_C57BL6J_5610813 | Gm37585 | ENSEMBL:ENSMUSG00000103113 | MGI:5610813 | pseudogene | predicted gene, 37585 |
| 3 | gene | 70.22875 | 70.22887 | negative | MGI_C57BL6J_5453254 | Gm23477 | ENSEMBL:ENSMUSG00000089507 | MGI:5453254 | snoRNA gene | predicted gene, 23477 |
| 3 | pseudogene | 70.41328 | 70.41388 | positive | MGI_C57BL6J_3644892 | Gm6631 | NCBI_Gene:625868,ENSEMBL:ENSMUSG00000102663 | MGI:3644892 | pseudogene | predicted gene 6631 |
| 3 | pseudogene | 70.56626 | 70.56730 | negative | MGI_C57BL6J_5610441 | Gm37213 | ENSEMBL:ENSMUSG00000104023 | MGI:5610441 | pseudogene | predicted gene, 37213 |
| 3 | pseudogene | 70.68909 | 70.68938 | positive | MGI_C57BL6J_3642422 | Gm10780 | ENSEMBL:ENSMUSG00000074877 | MGI:3642422 | pseudogene | predicted gene 10780 |
| 3 | gene | 70.77238 | 70.80729 | negative | MGI_C57BL6J_3643829 | Gm6634 | NCBI_Gene:625901,ENSEMBL:ENSMUSG00000097252 | MGI:3643829 | lncRNA gene | predicted gene 6634 |
| 3 | pseudogene | 71.40104 | 71.40155 | negative | MGI_C57BL6J_3646556 | Gm8599 | NCBI_Gene:108168916,ENSEMBL:ENSMUSG00000102666 | MGI:3646556 | pseudogene | predicted gene 8599 |
| 3 | pseudogene | 71.46523 | 71.46567 | negative | MGI_C57BL6J_5611533 | Gm38305 | ENSEMBL:ENSMUSG00000103396 | MGI:5611533 | pseudogene | predicted gene, 38305 |
| 3 | pseudogene | 71.67252 | 71.67712 | positive | MGI_C57BL6J_5011379 | Gm19194 | NCBI_Gene:100418413,ENSEMBL:ENSMUSG00000103815 | MGI:5011379 | pseudogene | predicted gene, 19194 |
| 3 | pseudogene | 71.85918 | 71.86015 | positive | MGI_C57BL6J_3643186 | Gm8604 | NCBI_Gene:667385,ENSEMBL:ENSMUSG00000098189 | MGI:3643186 | pseudogene | predicted gene 8604 |
| 3 | pseudogene | 71.89484 | 71.89499 | positive | MGI_C57BL6J_5610845 | Gm37617 | ENSEMBL:ENSMUSG00000103465 | MGI:5610845 | pseudogene | predicted gene, 37617 |
| 3 | pseudogene | 72.00992 | 72.01046 | negative | MGI_C57BL6J_5010416 | Gm18231 | NCBI_Gene:100416754 | MGI:5010416 | pseudogene | predicted gene, 18231 |
| 3 | pseudogene | 72.05000 | 72.05062 | negative | MGI_C57BL6J_3781529 | Gm3351 | NCBI_Gene:100041459,ENSEMBL:ENSMUSG00000103279 | MGI:3781529 | pseudogene | predicted gene 3351 |
| 3 | pseudogene | 72.18074 | 72.18095 | positive | MGI_C57BL6J_5610831 | Gm37603 | ENSEMBL:ENSMUSG00000103859 | MGI:5610831 | pseudogene | predicted gene, 37603 |
| 3 | gene | 72.31294 | 72.31303 | positive | MGI_C57BL6J_5530847 | Gm27465 | ENSEMBL:ENSMUSG00000098869 | MGI:5530847 | unclassified non-coding RNA gene | predicted gene, 27465 |
| 3 | pseudogene | 72.60384 | 72.60435 | negative | MGI_C57BL6J_5611129 | Gm37901 | ENSEMBL:ENSMUSG00000103078 | MGI:5611129 | pseudogene | predicted gene, 37901 |
| 3 | gene | 72.73783 | 72.73798 | negative | MGI_C57BL6J_5452228 | Gm22451 | ENSEMBL:ENSMUSG00000084752 | MGI:5452228 | snRNA gene | predicted gene, 22451 |
| 3 | gene | 72.88855 | 72.96786 | negative | MGI_C57BL6J_1917233 | Sis | NCBI_Gene:69983,ENSEMBL:ENSMUSG00000027790 | MGI:1917233 | protein coding gene | sucrase isomaltase (alpha-glucosidase) |
| 3 | gene | 73.04197 | 73.04384 | negative | MGI_C57BL6J_5610284 | Gm37056 | ENSEMBL:ENSMUSG00000102753 | MGI:5610284 | unclassified gene | predicted gene, 37056 |
| 3 | gene | 73.04725 | 73.05788 | negative | MGI_C57BL6J_2679447 | Slitrk3 | NCBI_Gene:386750,ENSEMBL:ENSMUSG00000048304 | MGI:2679447 | protein coding gene | SLIT and NTRK-like family, member 3 |
| 3 | gene | 73.06632 | 73.59483 | positive | MGI_C57BL6J_5434110 | Gm20754 | NCBI_Gene:626082,ENSEMBL:ENSMUSG00000103428 | MGI:5434110 | lncRNA gene | predicted gene, 20754 |
| 3 | gene | 73.14685 | 73.14696 | positive | MGI_C57BL6J_5454321 | Gm24544 | ENSEMBL:ENSMUSG00000077050 | MGI:5454321 | miRNA gene | predicted gene, 24544 |
| 3 | gene | 73.16409 | 73.16621 | positive | MGI_C57BL6J_5610953 | Gm37725 | ENSEMBL:ENSMUSG00000102916 | MGI:5610953 | unclassified gene | predicted gene, 37725 |
| 3 | gene | 73.20770 | 73.20800 | negative | MGI_C57BL6J_5452315 | Gm22538 | ENSEMBL:ENSMUSG00000088794 | MGI:5452315 | unclassified non-coding RNA gene | predicted gene, 22538 |
| 3 | gene | 73.32943 | 73.47001 | positive | MGI_C57BL6J_5611581 | Gm38353 | ENSEMBL:ENSMUSG00000102820 | MGI:5611581 | lncRNA gene | predicted gene, 38353 |
| 3 | gene | 73.35121 | 73.48165 | negative | MGI_C57BL6J_1921943 | 4930509J09Rik | NCBI_Gene:74693,ENSEMBL:ENSMUSG00000102362 | MGI:1921943 | lncRNA gene | RIKEN cDNA 4930509J09 gene |
| 3 | gene | 73.63581 | 73.70844 | negative | MGI_C57BL6J_894278 | Bche | NCBI_Gene:12038,ENSEMBL:ENSMUSG00000027792 | MGI:894278 | protein coding gene | butyrylcholinesterase |
| 3 | pseudogene | 73.93305 | 73.93412 | positive | MGI_C57BL6J_3644686 | Gm8625 | NCBI_Gene:667425,ENSEMBL:ENSMUSG00000102859 | MGI:3644686 | pseudogene | predicted gene 8625 |
| 3 | gene | 74.00922 | 74.01728 | negative | MGI_C57BL6J_5012541 | Gm20356 | NCBI_Gene:100504692,ENSEMBL:ENSMUSG00000104096 | MGI:5012541 | lncRNA gene | predicted gene, 20356 |
| 3 | gene | 74.10541 | 74.12645 | negative | MGI_C57BL6J_5595228 | Gm36069 | NCBI_Gene:102639855 | MGI:5595228 | lncRNA gene | predicted gene, 36069 |
| 3 | pseudogene | 74.24226 | 74.29687 | negative | MGI_C57BL6J_3779552 | Gm6098 | NCBI_Gene:100861784,ENSEMBL:ENSMUSG00000096400 | MGI:3779552 | pseudogene | predicted gene 6098 |
| 3 | pseudogene | 74.32353 | 74.32399 | negative | MGI_C57BL6J_5610278 | Gm37050 | ENSEMBL:ENSMUSG00000104227 | MGI:5610278 | pseudogene | predicted gene, 37050 |
| 3 | pseudogene | 74.81472 | 74.81534 | positive | MGI_C57BL6J_3647458 | Gm8643 | NCBI_Gene:108168917,ENSEMBL:ENSMUSG00000102809 | MGI:3647458 | pseudogene | predicted gene 8643 |
| 3 | gene | 75.02240 | 75.02461 | negative | MGI_C57BL6J_5610692 | Gm37464 | ENSEMBL:ENSMUSG00000102420 | MGI:5610692 | unclassified gene | predicted gene, 37464 |
| 3 | gene | 75.03790 | 75.16505 | negative | MGI_C57BL6J_2674085 | Zbbx | NCBI_Gene:213234,ENSEMBL:ENSMUSG00000034151 | MGI:2674085 | protein coding gene | zinc finger, B-box domain containing |
| 3 | gene | 75.16506 | 75.20808 | positive | MGI_C57BL6J_5622956 | Gm40071 | NCBI_Gene:105244469 | MGI:5622956 | lncRNA gene | predicted gene, 40071 |
| 3 | gene | 75.24236 | 75.27008 | negative | MGI_C57BL6J_1915181 | Serpini2 | NCBI_Gene:67931,ENSEMBL:ENSMUSG00000034139 | MGI:1915181 | protein coding gene | serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| 3 | gene | 75.27393 | 75.48216 | negative | MGI_C57BL6J_3645287 | Wdr49 | NCBI_Gene:213248,ENSEMBL:ENSMUSG00000104301 | MGI:3645287 | protein coding gene | WD repeat domain 49 |
| 3 | pseudogene | 75.39132 | 75.39264 | positive | MGI_C57BL6J_5010021 | Gm17836 | NCBI_Gene:100415953,ENSEMBL:ENSMUSG00000103444 | MGI:5010021 | pseudogene | predicted gene, 17836 |
| 3 | gene | 75.51649 | 75.55686 | negative | MGI_C57BL6J_1928396 | Pdcd10 | NCBI_Gene:56426,ENSEMBL:ENSMUSG00000027835 | MGI:1928396 | protein coding gene | programmed cell death 10 |
| 3 | gene | 75.55753 | 75.64350 | positive | MGI_C57BL6J_1194506 | Serpini1 | NCBI_Gene:20713,ENSEMBL:ENSMUSG00000027834 | MGI:1194506 | protein coding gene | serine (or cysteine) peptidase inhibitor, clade I, member 1 |
| 3 | gene | 75.61935 | 75.62336 | positive | MGI_C57BL6J_5610900 | Gm37672 | ENSEMBL:ENSMUSG00000102625 | MGI:5610900 | unclassified gene | predicted gene, 37672 |
| 3 | gene | 75.64744 | 75.65573 | negative | MGI_C57BL6J_1916443 | Platr10 | NCBI_Gene:69193,ENSEMBL:ENSMUSG00000099370 | MGI:1916443 | lncRNA gene | pluripotency associated transcript 10 |
| 3 | gene | 75.69895 | 75.69946 | negative | MGI_C57BL6J_5610421 | Gm37193 | ENSEMBL:ENSMUSG00000102188 | MGI:5610421 | unclassified gene | predicted gene, 37193 |
| 3 | gene | 75.71479 | 75.71492 | positive | MGI_C57BL6J_5455623 | Gm25846 | ENSEMBL:ENSMUSG00000088638 | MGI:5455623 | snoRNA gene | predicted gene, 25846 |
| 3 | gene | 75.74820 | 75.78765 | positive | MGI_C57BL6J_5579839 | Gm29133 | ENSEMBL:ENSMUSG00000100962 | MGI:5579839 | protein coding gene | predicted gene 29133 |
| 3 | gene | 75.87508 | 75.95695 | negative | MGI_C57BL6J_1920374 | Golim4 | NCBI_Gene:73124,ENSEMBL:ENSMUSG00000034109 | MGI:1920374 | protein coding gene | golgi integral membrane protein 4 |
| 3 | gene | 75.95212 | 75.95430 | positive | MGI_C57BL6J_5610913 | Gm37685 | ENSEMBL:ENSMUSG00000103024 | MGI:5610913 | unclassified gene | predicted gene, 37685 |
| 3 | gene | 75.95610 | 76.00114 | positive | MGI_C57BL6J_5610484 | Gm37256 | ENSEMBL:ENSMUSG00000104448 | MGI:5610484 | lncRNA gene | predicted gene, 37256 |
| 3 | pseudogene | 76.04092 | 76.04172 | negative | MGI_C57BL6J_5010613 | Gm18428 | NCBI_Gene:100417155,ENSEMBL:ENSMUSG00000104223 | MGI:5010613 | pseudogene | predicted gene, 18428 |
| 3 | gene | 76.07427 | 76.71002 | positive | MGI_C57BL6J_2442179 | Fstl5 | NCBI_Gene:213262,ENSEMBL:ENSMUSG00000034098 | MGI:2442179 | protein coding gene | follistatin-like 5 |
| 3 | gene | 76.54669 | 76.55025 | positive | MGI_C57BL6J_5611125 | Gm37897 | ENSEMBL:ENSMUSG00000103920 | MGI:5611125 | unclassified gene | predicted gene, 37897 |
| 3 | pseudogene | 76.82206 | 76.82240 | positive | MGI_C57BL6J_5610518 | Gm37290 | ENSEMBL:ENSMUSG00000103702 | MGI:5610518 | pseudogene | predicted gene, 37290 |
| 3 | pseudogene | 76.88450 | 76.88533 | positive | MGI_C57BL6J_5011416 | Gm19231 | NCBI_Gene:100418471,ENSEMBL:ENSMUSG00000102216 | MGI:5011416 | pseudogene | predicted gene, 19231 |
| 3 | gene | 77.10521 | 77.10528 | negative | MGI_C57BL6J_5453421 | Gm23644 | ENSEMBL:ENSMUSG00000088481 | MGI:5453421 | snoRNA gene | predicted gene, 23644 |
| 3 | pseudogene | 77.56081 | 77.56394 | negative | MGI_C57BL6J_2674154 | Dnmt3a-ps1 | NCBI_Gene:445261,ENSEMBL:ENSMUSG00000102489 | MGI:2674154 | pseudogene | DNA methyltransferase 3A, pseudogene 1 |
| 3 | gene | 77.56496 | 77.57134 | negative | MGI_C57BL6J_5611034 | Gm37806 | ENSEMBL:ENSMUSG00000103849 | MGI:5611034 | unclassified gene | predicted gene, 37806 |
| 3 | pseudogene | 77.69137 | 77.69223 | negative | MGI_C57BL6J_5010480 | Gm18295 | NCBI_Gene:100416866 | MGI:5010480 | pseudogene | predicted gene, 18295 |
| 3 | gene | 77.70289 | 77.70439 | negative | MGI_C57BL6J_5611423 | Gm38195 | ENSEMBL:ENSMUSG00000102872 | MGI:5611423 | unclassified gene | predicted gene, 38195 |
| 3 | pseudogene | 77.87278 | 77.87315 | negative | MGI_C57BL6J_5610804 | Gm37576 | ENSEMBL:ENSMUSG00000102393 | MGI:5610804 | pseudogene | predicted gene, 37576 |
| 3 | pseudogene | 77.96138 | 77.96280 | positive | MGI_C57BL6J_5010904 | Gm18719 | NCBI_Gene:100417620,ENSEMBL:ENSMUSG00000105026 | MGI:5010904 | pseudogene | predicted gene, 18719 |
| 3 | pseudogene | 78.00646 | 78.00698 | positive | MGI_C57BL6J_3648953 | Gm6677 | NCBI_Gene:626461,ENSEMBL:ENSMUSG00000103312 | MGI:3648953 | pseudogene | predicted gene 6677 |
| 3 | pseudogene | 78.08583 | 78.08740 | negative | MGI_C57BL6J_3643677 | Gm8684 | NCBI_Gene:667526,ENSEMBL:ENSMUSG00000102637 | MGI:3643677 | pseudogene | predicted gene 8684 |
| 3 | pseudogene | 78.13446 | 78.13558 | positive | MGI_C57BL6J_3645908 | Gm6680 | NCBI_Gene:626479,ENSEMBL:ENSMUSG00000103817 | MGI:3645908 | pseudogene | predicted gene 6680 |
| 3 | gene | 78.40430 | 78.40594 | negative | MGI_C57BL6J_5663463 | Gm43326 | ENSEMBL:ENSMUSG00000104782 | MGI:5663463 | unclassified gene | predicted gene 43326 |
| 3 | gene | 78.40616 | 78.41077 | negative | MGI_C57BL6J_5595447 | Gm36288 | NCBI_Gene:102640153 | MGI:5595447 | lncRNA gene | predicted gene, 36288 |
| 3 | pseudogene | 78.48658 | 78.48868 | negative | MGI_C57BL6J_3705708 | Gm15442 | NCBI_Gene:100041691,ENSEMBL:ENSMUSG00000084370 | MGI:3705708 | pseudogene | predicted gene 15442 |
| 3 | pseudogene | 78.75862 | 78.75946 | positive | MGI_C57BL6J_5011137 | Gm18952 | NCBI_Gene:100418022,ENSEMBL:ENSMUSG00000102341 | MGI:5011137 | pseudogene | predicted gene, 18952 |
| 3 | gene | 78.78973 | 78.79223 | negative | MGI_C57BL6J_5595500 | Gm36341 | NCBI_Gene:102640221 | MGI:5595500 | lncRNA gene | predicted gene, 36341 |
| 3 | gene | 78.84222 | 78.84257 | positive | MGI_C57BL6J_5454526 | Gm24749 | ENSEMBL:ENSMUSG00000088463 | MGI:5454526 | unclassified non-coding RNA gene | predicted gene, 24749 |
| 3 | pseudogene | 78.86489 | 78.86557 | negative | MGI_C57BL6J_5010138 | Gm17953 | NCBI_Gene:100416167,ENSEMBL:ENSMUSG00000102438 | MGI:5010138 | pseudogene | predicted gene, 17953 |
| 3 | pseudogene | 78.89183 | 78.89247 | negative | MGI_C57BL6J_3646590 | Gm5277 | NCBI_Gene:383883,ENSEMBL:ENSMUSG00000091361 | MGI:3646590 | pseudogene | predicted gene 5277 |
| 3 | pseudogene | 78.91724 | 78.91846 | positive | MGI_C57BL6J_3641638 | Gm10291 | NCBI_Gene:100041748,ENSEMBL:ENSMUSG00000070443 | MGI:3641638 | pseudogene | predicted pseudogene 10291 |
| 3 | gene | 78.91846 | 78.93631 | negative | MGI_C57BL6J_5621427 | Gm38542 | NCBI_Gene:102641233 | MGI:5621427 | lncRNA gene | predicted gene, 38542 |
| 3 | pseudogene | 78.96549 | 78.96715 | negative | MGI_C57BL6J_3704220 | Gm9762 | NCBI_Gene:100041766,ENSEMBL:ENSMUSG00000037096 | MGI:3704220 | pseudogene | predicted pseudogene 9762 |
| 3 | gene | 78.97992 | 78.98005 | positive | MGI_C57BL6J_5453217 | Gm23440 | ENSEMBL:ENSMUSG00000077487 | MGI:5453217 | snoRNA gene | predicted gene, 23440 |
| 3 | gene | 79.06251 | 79.28724 | negative | MGI_C57BL6J_2659071 | Rapgef2 | NCBI_Gene:76089,ENSEMBL:ENSMUSG00000062232 | MGI:2659071 | protein coding gene | Rap guanine nucleotide exchange factor (GEF) 2 |
| 3 | gene | 79.19637 | 79.19648 | positive | MGI_C57BL6J_5453927 | Gm24150 | ENSEMBL:ENSMUSG00000064693 | MGI:5453927 | snRNA gene | predicted gene, 24150 |
| 3 | gene | 79.21292 | 79.25391 | positive | MGI_C57BL6J_1918174 | 4921511C10Rik | NCBI_Gene:70924,ENSEMBL:ENSMUSG00000104585 | MGI:1918174 | lncRNA gene | RIKEN cDNA 4921511C10 gene |
| 3 | gene | 79.23228 | 79.23413 | negative | MGI_C57BL6J_2443462 | B930012P20Rik | NA | NA | unclassified gene | RIKEN cDNA B930012P20 gene |
| 3 | gene | 79.28717 | 79.28985 | positive | MGI_C57BL6J_2686494 | 6430573P05Rik | ENSEMBL:ENSMUSG00000102545 | MGI:2686494 | lncRNA gene | RIKEN cDNA 6430573P05 gene |
| 3 | gene | 79.33420 | 79.33658 | negative | MGI_C57BL6J_5622957 | Gm40072 | NCBI_Gene:105244470 | MGI:5622957 | lncRNA gene | predicted gene, 40072 |
| 3 | gene | 79.33666 | 79.46413 | positive | MGI_C57BL6J_4936993 | Gm17359 | NCBI_Gene:100233207,ENSEMBL:ENSMUSG00000091685 | MGI:4936993 | protein coding gene | predicted gene, 17359 |
| 3 | gene | 79.45597 | 79.56780 | negative | MGI_C57BL6J_2683054 | Fnip2 | NCBI_Gene:329679,ENSEMBL:ENSMUSG00000061175 | MGI:2683054 | protein coding gene | folliculin interacting protein 2 |
| 3 | gene | 79.50979 | 79.51210 | negative | MGI_C57BL6J_5595601 | Gm36442 | NCBI_Gene:102640362 | MGI:5595601 | lncRNA gene | predicted gene, 36442 |
| 3 | gene | 79.55131 | 79.55219 | negative | MGI_C57BL6J_3781690 | Gm3513 | ENSEMBL:ENSMUSG00000089746 | MGI:3781690 | lncRNA gene | predicted gene 3513 |
| 3 | gene | 79.56819 | 79.57332 | positive | MGI_C57BL6J_1923129 | 4930589L23Rik | NCBI_Gene:75879,ENSEMBL:ENSMUSG00000068957 | MGI:1923129 | lncRNA gene | RIKEN cDNA 4930589L23 gene |
| 3 | pseudogene | 79.58259 | 79.58323 | positive | MGI_C57BL6J_5594226 | Gm35067 | ENSEMBL:ENSMUSG00000103564 | MGI:5594226 | pseudogene | predicted gene, 35067 |
| 3 | gene | 79.59134 | 79.60365 | positive | MGI_C57BL6J_1914988 | Ppid | NCBI_Gene:67738,ENSEMBL:ENSMUSG00000027804 | MGI:1914988 | protein coding gene | peptidylprolyl isomerase D (cyclophilin D) |
| 3 | gene | 79.60379 | 79.62950 | negative | MGI_C57BL6J_106100 | Etfdh | NCBI_Gene:66841,ENSEMBL:ENSMUSG00000027809 | MGI:106100 | protein coding gene | electron transferring flavoprotein, dehydrogenase |
| 3 | gene | 79.61054 | 79.61183 | negative | MGI_C57BL6J_5611201 | Gm37973 | ENSEMBL:ENSMUSG00000102370 | MGI:5611201 | unclassified gene | predicted gene, 37973 |
| 3 | gene | 79.62908 | 79.63282 | positive | MGI_C57BL6J_1923189 | 4930579G24Rik | NCBI_Gene:75939,ENSEMBL:ENSMUSG00000027811 | MGI:1923189 | protein coding gene | RIKEN cDNA 4930579G24 gene |
| 3 | gene | 79.64160 | 79.73795 | negative | MGI_C57BL6J_2682211 | Rxfp1 | NCBI_Gene:381489,ENSEMBL:ENSMUSG00000034009 | MGI:2682211 | protein coding gene | relaxin/insulin-like family peptide receptor 1 |
| 3 | gene | 79.67337 | 79.67688 | negative | MGI_C57BL6J_5504099 | Gm26984 | ENSEMBL:ENSMUSG00000098204 | MGI:5504099 | lncRNA gene | predicted gene, 26984 |
| 3 | gene | 79.81256 | 79.85279 | negative | MGI_C57BL6J_1917902 | Tmem144 | NCBI_Gene:70652,ENSEMBL:ENSMUSG00000027956 | MGI:1917902 | protein coding gene | transmembrane protein 144 |
| 3 | gene | 79.83493 | 79.83504 | positive | MGI_C57BL6J_5456197 | Gm26420 | ENSEMBL:ENSMUSG00000077461 | MGI:5456197 | snRNA gene | predicted gene, 26420 |
| 3 | gene | 79.85287 | 79.85666 | positive | MGI_C57BL6J_5622958 | Gm40073 | NCBI_Gene:105244471 | MGI:5622958 | lncRNA gene | predicted gene, 40073 |
| 3 | gene | 79.85929 | 79.88681 | negative | MGI_C57BL6J_5595728 | Gm36569 | NCBI_Gene:102640530,ENSEMBL:ENSMUSG00000102191 | MGI:5595728 | lncRNA gene | predicted gene, 36569 |
| 3 | gene | 79.88447 | 79.94628 | positive | MGI_C57BL6J_1915909 | Gask1b | NCBI_Gene:68659,ENSEMBL:ENSMUSG00000027955 | MGI:1915909 | protein coding gene | golgi associated kinase 1B |
| 3 | gene | 80.05489 | 80.21309 | positive | MGI_C57BL6J_2685464 | A330069K06Rik | NCBI_Gene:213544,ENSEMBL:ENSMUSG00000103181 | MGI:2685464 | lncRNA gene | RIKEN cDNA A330069K06 gene |
| 3 | gene | 80.05987 | 80.10161 | positive | MGI_C57BL6J_5622959 | Gm40074 | NCBI_Gene:105244472 | MGI:5622959 | lncRNA gene | predicted gene, 40074 |
| 3 | pseudogene | 80.08808 | 80.08916 | negative | MGI_C57BL6J_5011251 | Gm19066 | NCBI_Gene:100418193,ENSEMBL:ENSMUSG00000103193 | MGI:5011251 | pseudogene | predicted gene, 19066 |
| 3 | gene | 80.13413 | 80.13646 | positive | MGI_C57BL6J_5663976 | Gm43839 | ENSEMBL:ENSMUSG00000105307 | MGI:5663976 | unclassified gene | predicted gene 43839 |
| 3 | pseudogene | 80.19448 | 80.19463 | negative | MGI_C57BL6J_5611523 | Gm38295 | ENSEMBL:ENSMUSG00000103154 | MGI:5611523 | pseudogene | predicted gene, 38295 |
| 3 | gene | 80.20249 | 80.20560 | negative | MGI_C57BL6J_5611120 | Gm37892 | ENSEMBL:ENSMUSG00000103929 | MGI:5611120 | lncRNA gene | predicted gene, 37892 |
| 3 | gene | 80.28512 | 80.33459 | negative | MGI_C57BL6J_5611199 | Gm37971 | ENSEMBL:ENSMUSG00000102379 | MGI:5611199 | lncRNA gene | predicted gene, 37971 |
| 3 | pseudogene | 80.39935 | 80.39967 | negative | MGI_C57BL6J_3644992 | Gm6706 | ENSEMBL:ENSMUSG00000087123 | MGI:3644992 | pseudogene | predicted gene 6706 |
| 3 | gene | 80.68145 | 80.80320 | negative | MGI_C57BL6J_95809 | Gria2 | NCBI_Gene:14800,ENSEMBL:ENSMUSG00000033981 | MGI:95809 | protein coding gene | glutamate receptor, ionotropic, AMPA2 (alpha 2) |
| 3 | gene | 80.69220 | 80.69231 | negative | MGI_C57BL6J_5455526 | Gm25749 | ENSEMBL:ENSMUSG00000093226 | MGI:5455526 | miRNA gene | predicted gene, 25749 |
| 3 | gene | 80.84315 | 80.84355 | negative | MGI_C57BL6J_1920346 | 2900092N11Rik | NA | NA | unclassified gene | RIKEN cDNA 2900092N11 gene |
| 3 | gene | 80.84360 | 80.91366 | negative | MGI_C57BL6J_95751 | Glrb | NCBI_Gene:14658,ENSEMBL:ENSMUSG00000028020 | MGI:95751 | protein coding gene | glycine receptor, beta subunit |
| 3 | pseudogene | 80.88509 | 80.88699 | positive | MGI_C57BL6J_5010730 | Gm18545 | NCBI_Gene:100417342 | MGI:5010730 | pseudogene | predicted gene, 18545 |
| 3 | gene | 80.96296 | 80.97163 | positive | MGI_C57BL6J_5662605 | Gm42468 | ENSEMBL:ENSMUSG00000105092 | MGI:5662605 | lncRNA gene | predicted gene 42468 |
| 3 | pseudogene | 80.99156 | 81.00159 | positive | MGI_C57BL6J_5588967 | Gm29808 | NCBI_Gene:101056241,ENSEMBL:ENSMUSG00000103419 | MGI:5588967 | pseudogene | predicted gene, 29808 |
| 3 | gene | 81.03295 | 81.03377 | positive | MGI_C57BL6J_5662613 | Gm42476 | ENSEMBL:ENSMUSG00000106293 | MGI:5662613 | lncRNA gene | predicted gene 42476 |
| 3 | gene | 81.03589 | 81.21404 | positive | MGI_C57BL6J_1859631 | Pdgfc | NCBI_Gene:54635,ENSEMBL:ENSMUSG00000028019 | MGI:1859631 | protein coding gene | platelet-derived growth factor, C polypeptide |
| 3 | gene | 81.03741 | 81.04044 | negative | MGI_C57BL6J_3801866 | Gm16000 | ENSEMBL:ENSMUSG00000085106 | MGI:3801866 | lncRNA gene | predicted gene 16000 |
| 3 | pseudogene | 81.40529 | 81.40557 | negative | MGI_C57BL6J_5610528 | Gm37300 | ENSEMBL:ENSMUSG00000102223 | MGI:5610528 | pseudogene | predicted gene, 37300 |
R version 3.6.2 (2019-12-12)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS Catalina 10.15.7
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRlapack.dylib
locale:
[1] en_AU.UTF-8/en_AU.UTF-8/en_AU.UTF-8/C/en_AU.UTF-8/en_AU.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] abind_1.4-5 qtl2_0.22 reshape2_1.4.4 ggplot2_3.3.5
[5] tibble_3.1.2 psych_2.0.7 readxl_1.3.1 cluster_2.1.0
[9] dplyr_0.8.5 optparse_1.6.6 rhdf5_2.28.1 mclust_5.4.6
[13] tidyr_1.0.2 data.table_1.14.0 knitr_1.33 kableExtra_1.1.0
[17] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] httr_1.4.1 bit64_4.0.5 viridisLite_0.4.0 assertthat_0.2.1
[5] highr_0.9 blob_1.2.1 cellranger_1.1.0 yaml_2.2.1
[9] pillar_1.6.1 RSQLite_2.2.7 backports_1.2.1 lattice_0.20-38
[13] glue_1.4.2 digest_0.6.27 promises_1.1.0 rvest_0.3.5
[17] colorspace_2.0-2 htmltools_0.5.1.1 httpuv_1.5.2 plyr_1.8.6
[21] pkgconfig_2.0.3 purrr_0.3.4 scales_1.1.1 webshot_0.5.2
[25] whisker_0.4 getopt_1.20.3 later_1.0.0 git2r_0.26.1
[29] ellipsis_0.3.2 cachem_1.0.5 withr_2.4.2 mnormt_1.5-7
[33] magrittr_2.0.1 crayon_1.4.1 memoise_2.0.0 evaluate_0.14
[37] fs_1.4.1 fansi_0.5.0 nlme_3.1-142 xml2_1.3.1
[41] tools_3.6.2 hms_0.5.3 lifecycle_1.0.0 stringr_1.4.0
[45] Rhdf5lib_1.6.3 munsell_0.5.0 compiler_3.6.2 rlang_0.4.11
[49] grid_3.6.2 rstudioapi_0.13 rmarkdown_2.1 gtable_0.3.0
[53] DBI_1.1.1 R6_2.5.0 fastmap_1.1.0 bit_4.0.4
[57] utf8_1.2.1 rprojroot_1.3-2 readr_1.3.1 stringi_1.7.2
[61] parallel_3.6.2 Rcpp_1.0.7 vctrs_0.3.8 tidyselect_1.0.0
[65] xfun_0.24