PEM vs. PAM split point sensitivity
Last updated: 2018-05-22
workflowr checks: (Click a bullet for more information)-
✔ R Markdown file: up-to-date
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
-
✔ Environment: empty
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
-
✔ Seed:
set.seed(20180517)The command
set.seed(20180517)was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible. -
✔ Session information: recorded
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
-
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.✔ Repository version: 14e326b
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can usewflow_publishorwflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.Ignored files: Ignored: analysis/ll_selection_cache/ Ignored: analysis/pem_vs_pam_cache/ Untracked files: Untracked: output/sim-lag-lead-registry/ Untracked: output/sim-pem-vs-pam-registry/ Untracked: sandbox/ Unstaged changes: Modified: analysis/_site.yml Modified: analysis/about.Rmd
Expand here to see past versions:
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 14e326b | adibender | 2018-05-22 | wflow_publish(c(“analysis/pem_vs_pam.Rmd”)) |
| html | 1e706df | adibender | 2018-05-22 | Build site. |
| Rmd | f963aae | adibender | 2018-05-22 | wflow_publish(c(“analysis/pem_vs_pam.Rmd”)) |
library(ggplot2)
theme_set(theme_bw())
library(batchtools)Loading required package: data.tableBreaking change introduced in batchtools v0.9.6: The format of the returned data.table of the functions `reduceResultsDataTable()`, getJobTable()`, `getJobPars()`, and `getJobResources()` has changed. List columns are not unnested automatically anymore. To manually unnest tables, batchtools provides the helper function `unwrap()` now, e.g. `unwrap(getJobPars())`. The previously introduced helper function `flatten()` will be deprecated due to a name clash with `purrr::flatten()`.Function for data simulation (using pammtools::sim_pexp):
## simulation function
sim_wrapper <- function(data, job, n = 250, time_grid = seq(0, 10, by = 0.05)) {
# create data set with covariates
df <- tibble::tibble(x1 = runif(n, -3, 3), x2 = runif(n, 0, 6))
# baseline hazard
f0 <- function(t) {dgamma(t, 8, 2) * 6}
# define function that generates nz exposures z(t_{z,1}), ..., z(t_{z,Q})
sim_pexp(formula = ~ -3.5 + f0(t), data = df, cut = time_grid)
}Function to estimate hazard from simulated data, either by a PEM or PAM
## estimation function
pam_wrapper <- function(data, job, instance,
cut = NA,
bs = "ps",
mod_type = c("pem", "pam") ,
max_time = 10) {
if(is.na(cut)) {
cut <- NULL
} else {
if(cut == "rough") {
cut <- seq(0, max_time, by = 0.5)
} else {
if(cut == "fine") {
cut <- seq(0, max_time, by = 0.2)
}
}
}
ped <- as_ped(data = instance, formula = Surv(time, status) ~ ., cut = cut, id="id")
form <- "ped_status ~ s(tend) + s(x1) + s(x2)"
if(mod_type == "pem") {
form <- ped_status ~ interval
time_var <- "interval"
} else {
form <- ped_status ~ s(tend, bs = bs,k = k)
time_var <- "tend"
}
mod <- gam(formula = form, data = ped, family = poisson(), offset = offset, method = "REML")
# summary(mod)
make_newdata(ped, tend=unique(tend)) %>%
add_hazard(mod, type="link", se_mult = qnorm(0.975), time_var = time_var) %>%
mutate(truth = -3.5 + dgamma(tend, 8, 2) * 6)
}Setup simulation using batchtools:
if(!checkmate::test_directory_exists("output/sim-pem-vs-pam-registry")) {
reg <- makeExperimentRegistry("output/sim-pem-vs-pam-registry",
packages = c("mgcv", "dplyr", "tidyr", "pammtools"),
seed = 20052018)
reg$cluster.functions = makeClusterFunctionsMulticore(ncpus = 2)
addProblem(name = "pem-vs-pam", fun = sim_wrapper)
addAlgorithm(name = "pem-vs-pam", fun = pam_wrapper)
algo_df <- tidyr::crossing(
cut = c(NA, "fine", "rough"),
mod_type = c("pem", "pam"))
addExperiments(algo.design = list("pem-vs-pam" = algo_df), repls = 20)
submitJobs()
waitForJobs()
}Warning: replacing previous import 'dplyr::vars' by 'ggplot2::vars' when
loading 'pammtools'[1] FALSEEvaluate Simulation:
reg <- loadRegistry("output/sim-pem-vs-pam-registry", writeable = TRUE)Reading registry in read-write modeLoading required package: mgcvLoading required package: nlmeThis is mgcv 1.8-23. For overview type 'help("mgcv-package")'.Loading required package: dplyr
Attaching package: 'dplyr'The following object is masked from 'package:nlme':
collapseThe following objects are masked from 'package:data.table':
between, first, lastThe following object is masked from 'package:ggplot2':
varsThe following objects are masked from 'package:stats':
filter, lagThe following objects are masked from 'package:base':
intersect, setdiff, setequal, unionLoading required package: tidyrLoading required package: pammtoolsWarning: replacing previous import 'dplyr::vars' by 'ggplot2::vars' when
loading 'pammtools'
Attaching package: 'pammtools'The following object is masked from 'package:stats':
filterNo configuration file foundids_pam <- findExperiments(prob.name="pem-vs-pam", algo.name="pem-vs-pam")
pars <- unwrap(getJobPars()) %>% as_tibble()
res <- reduceResultsDataTable(ids=findDone(ids_pam)) %>%
as_tibble() %>%
tidyr::unnest() %>%
left_join(pars) %>%
mutate(cut = case_when(is.na(cut) ~ "default", TRUE ~ cut))Warning in bind_rows_(x, .id): Unequal factor levels: coercing to characterWarning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vector
Warning in bind_rows_(x, .id): binding character and factor vector,
coercing into character vectorJoining, by = "job.id"res %>%
mutate(
sq_error = (truth - hazard)^2,
covered = (truth >= ci_lower) & (truth <= ci_upper)) %>%
group_by(job.id, mod_type, cut) %>%
summarize(
RMSE = sqrt(mean(sq_error)),
coverage = mean(covered)) %>%
group_by(mod_type, cut) %>%
summarize(
RMSE = mean(RMSE),
coverage = mean(coverage))# A tibble: 3 x 4
# Groups: mod_type [?]
mod_type cut RMSE coverage
<chr> <chr> <dbl> <dbl>
1 pem default 1.38 0.870
2 pem fine 6.84 0.966
3 pem rough 3.02 0.920ggplot(res, aes(x=tend, y = hazard)) +
geom_step(aes(group = job.id), alpha = 0.3) +
geom_line(aes(y = truth, col = "truth"), lwd = 2) +
facet_grid(cut ~ mod_type) +
coord_cartesian(ylim=c(-5, 0)) +
geom_smooth(aes(col="average estimate"), method="gam", formula = y ~ s(x),
se=FALSE) +
scale_color_brewer("Method", palette = "Dark2") +
xlab("time")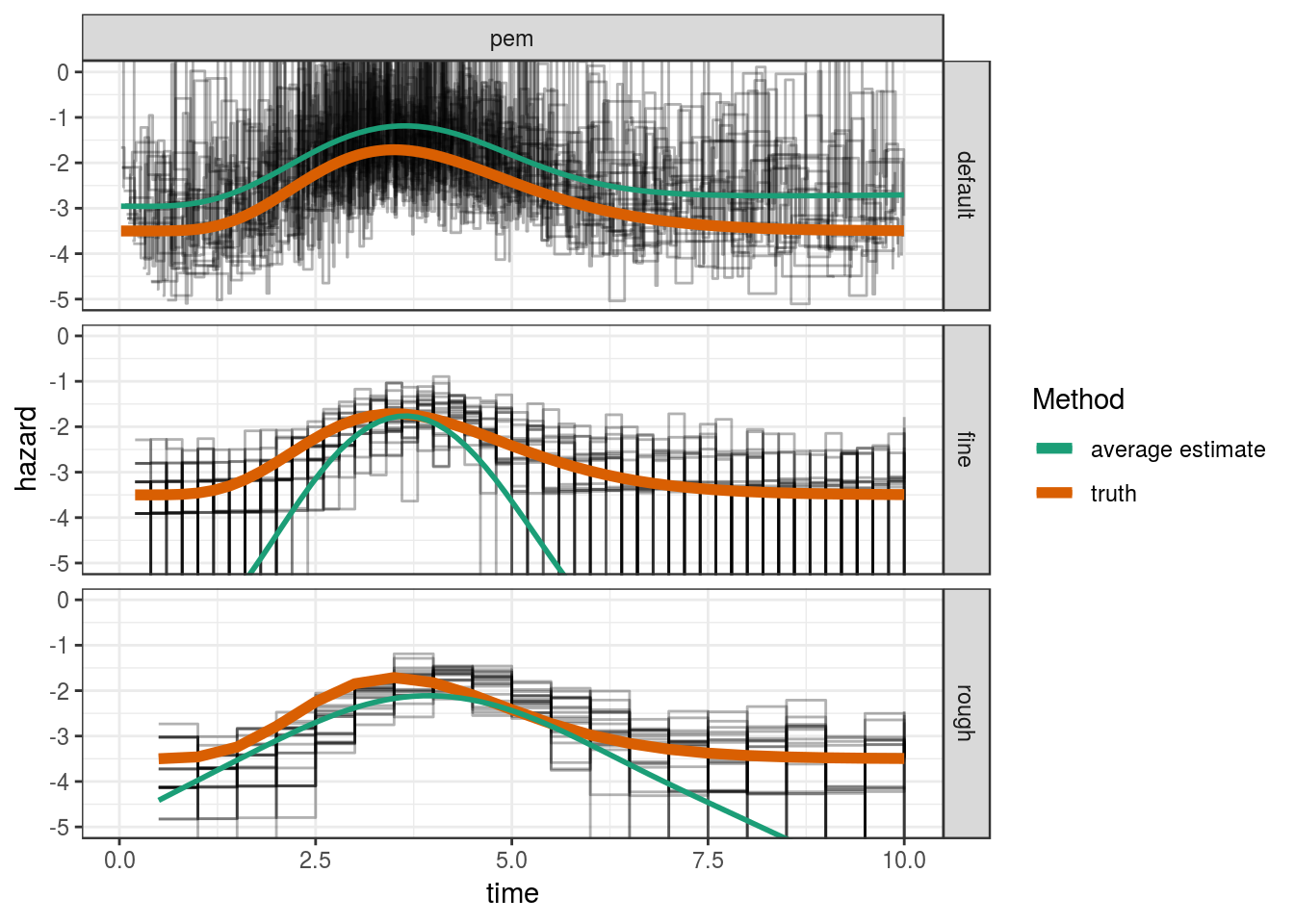
Expand here to see past versions of unnamed-chunk-5-1.png:
| Version | Author | Date |
|---|---|---|
| 1e706df | adibender | 2018-05-22 |
Session information
sessionInfo()R version 3.4.4 (2018-03-15)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 16.04.4 LTS
Matrix products: default
BLAS: /usr/lib/openblas-base/libblas.so.3
LAPACK: /usr/lib/libopenblasp-r0.2.18.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=de_DE.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=de_DE.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=de_DE.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=de_DE.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] bindrcpp_0.2.2 pammtools_0.0.9.9003 tidyr_0.8.0
[4] dplyr_0.7.4 mgcv_1.8-23 nlme_3.1-137
[7] batchtools_0.9.8 data.table_1.10.4-3 ggplot2_2.2.1.9000
loaded via a namespace (and not attached):
[1] progress_1.1.2 tidyselect_0.2.4 reshape2_1.4.3
[4] purrr_0.2.4 splines_3.4.4 lattice_0.20-35
[7] expm_0.999-2 colorspace_1.3-2 htmltools_0.3.6
[10] yaml_2.1.18 utf8_1.1.3 survival_2.42-3
[13] rlang_0.2.0.9001 R.oo_1.21.0 pillar_1.2.1
[16] glue_1.2.0 withr_2.1.2 R.utils_2.6.0
[19] rappdirs_0.3.1 RColorBrewer_1.1-2 plyr_1.8.4
[22] bindr_0.1.1 stringr_1.3.0 munsell_0.4.3
[25] gtable_0.2.0 workflowr_1.0.1 R.methodsS3_1.7.1
[28] mvtnorm_1.0-7 evaluate_0.10.1 labeling_0.3
[31] knitr_1.20 Rcpp_0.12.16 scales_0.5.0.9000
[34] backports_1.1.2 checkmate_1.8.5 debugme_1.1.0
[37] brew_1.0-6 digest_0.6.15 stringi_1.1.7
[40] msm_1.6.6 grid_3.4.4 rprojroot_1.3-2
[43] cli_1.0.0 tools_3.4.4 magrittr_1.5
[46] base64url_1.3 lazyeval_0.2.1 tibble_1.4.2
[49] Formula_1.2-3 crayon_1.3.4 whisker_0.3-2
[52] pkgconfig_2.0.1 Matrix_1.2-13 prettyunits_1.0.2
[55] assertthat_0.2.0 rmarkdown_1.9 R6_2.2.2
[58] git2r_0.21.0 compiler_3.4.4 This reproducible R Markdown analysis was created with workflowr 1.0.1