Exploratory analysis
Antonio J Perez-Luque
2021-06-30
Last updated: 2021-07-01
Checks: 7 0
Knit directory: fire_alcontar/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210630) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version a2df5c6. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Untracked files:
Untracked: analysis/ecology-letters.csl
Untracked: data/Cobertura.xlsx
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/exploratory_analysis.Rmd) and HTML (docs/exploratory_analysis.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | a2df5c6 | Antonio J Perez-Luque | 2021-07-01 | add exploratory analysis |
Introduction
Evaluar la variación de parámetros relacionados con vegetación tras la realización de quemas prescritas en parcelas con diferentes tratamientos de pastoreo.
Analizar si la fecha de la quema afecta a la velocidad de recuperación. Existen algunos estudios que señalan que la recuperación de la vegetación en las zonas afectadas por fuegos de primavera es rápida (ver referencias en Pereira et al. 2016)
library(here)
library(tidyverse)
library(readxl)
library(plotrix)
library(DT)Preparar los datos
Recodifico los datos para facilidad de manejo.
- ZONA. Se convierte a factor (
zonaCod):- Quemado con pastoreo ~ OP
- Quemado sin pastoreo ~ ONP
- Quemado primavera ~ PP
- RANGO_INFOCA. Se convierte a factor (
rango. Se añade “R” delante de cada rango (i.e. RANGO_INFOCA = 1 ~ R1)
Se añade una variable de tiempo tras el fuego (time). Para ello, previamente establecemos la fecha de fuego en otoño (2018-12-18) y en primavera (2019-05-07). Seguidamente computamos el número de meses tras el fuego. Los muestreos previos al fuego se codifican como -1 en la variable time
Ver código!
df <- read_excel(path=here::here("data/Cobertura.xlsx"))
quema_oto <- as.Date("2018-12-18")
quema_pri <- as.Date("2019-05-07")
cobertura <- df %>%
mutate(zonaCod =
as.factor(
case_when(
ZONA == "Quemado con pastoreo" ~ "OP",
ZONA == "Quemado sin pastoreo" ~ "ONP",
ZONA == "Quemado primavera" ~ "PP")),
rango =
as.factor(
case_when(
RANGO_INFOCA == 1 ~ "R1",
RANGO_INFOCA == 2 ~ "R2",
RANGO_INFOCA == 3 ~ "R3",
RANGO_INFOCA == 4 ~ "R4"))) %>%
mutate(time =
case_when(
ZONA == "Quemado primavera" ~ (lubridate::interval(quema_pri, FECHA_MUESTREOS)) %/% months(1),
TRUE ~ (lubridate::interval(quema_oto, FECHA_MUESTREOS)) %/% months(1),
)) %>%
mutate(time =
case_when(time == 0 ~ -1,
TRUE ~ time),
season = getSeason(FECHA_MUESTREOS),
year = as.factor(lubridate::year(FECHA_MUESTREOS))) %>%
unite("timeSeason", year, season, sep="_", remove=FALSE) %>%
filter(FECHA_MUESTREOS != as.Date("2020-12-21"))Análisis exploratorio
Evolución temporal de la cobertura agrupada por zonas (
OP,ONP,PP) para cada uno de los rangos.Calculamos el promedio de cobertura (
group_byzonaCod y RANGO)Se añaden las fechas de las quemas
Ver código!
cob <- cobertura %>%
group_by(zonaCod, time, rango) %>%
summarise(mean = mean(COB_TOTAL, na.rm=TRUE),
sd = sd(COB_TOTAL, na.rm=TRUE),
se = plotrix::std.error(COB_TOTAL, na.rm=TRUE),
n = length(COB_TOTAL)) %>%
mutate(pastoreo =
case_when(
zonaCod == "OP" ~ "pastoreo",
zonaCod == "ONP" ~ "no pastoreo",
zonaCod == "PP" ~ "pastoreo")
)
cob_season <- cobertura %>%
group_by(zonaCod, timeSeason, rango, FECHA_MUESTREOS) %>%
summarise(mean = mean(COB_TOTAL, na.rm=TRUE),
sd = sd(COB_TOTAL, na.rm=TRUE),
se = plotrix::std.error(COB_TOTAL, na.rm=TRUE),
n = length(COB_TOTAL)) %>%
mutate(pastoreo =
case_when(
zonaCod == "OP" ~ "pastoreo",
zonaCod == "ONP" ~ "no pastoreo",
zonaCod == "PP" ~ "pastoreo")
) %>%
mutate(timeSeason = factor(timeSeason,
levels = c("2018_Autumn",
"2019_Spring","2019_Autumn",
"2020_Spring","2020_Autumn",
"2021_Spring"))) %>%
ungroup()datatable(cob_season) %>% formatRound(c("mean","sd","se"), 2)Ver código!
p <- position_dodge(0.9)
plot_vegcob <- cob_season %>% ggplot(aes(x=FECHA_MUESTREOS, y=mean, colour=zonaCod,
group=zonaCod)) +
geom_line(position = p) +
geom_point(position = p,
aes(shape=pastoreo),
size = 3) +
geom_errorbar(aes(ymin = mean-se,
ymax = mean+se),
position = p) +
facet_wrap(~rango, ncol=1, scales = "free_y") +
theme_bw() +
theme(panel.grid = element_blank(),
strip.background = element_rect(fill="white")) +
ylab("veg. cover (%)") +
xlab("Year") +
scale_shape_manual(values=c(15,16)) +
geom_vline(xintercept = as.POSIXct(quema_oto), linetype="dotted", size = 1) +
geom_vline(xintercept = as.POSIXct(quema_pri), linetype="dotted", colour="#00BFC4", size=1)plot_vegcob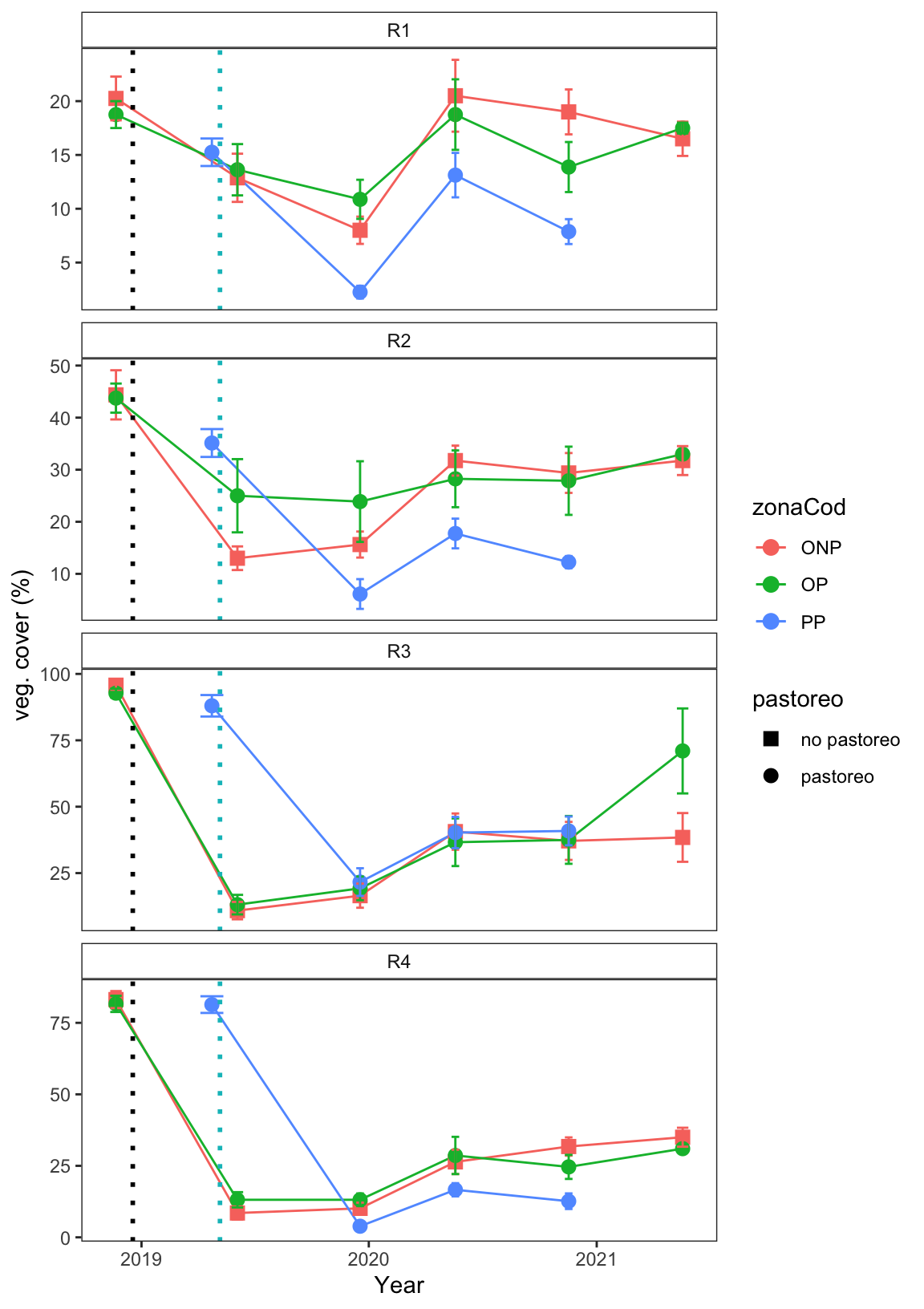
Evolución de la cobertura vegetal (%) tras las quemas prescritas en las tres parcelas de estudio. Se muestran valores medios y error estándar. Los cuadrados corresponden a las parcelas de no pastoreo, los circulos a las parcelas con pastoreo. Las líneas de puntos verticales indican el momento en el que se realizaron las quemas
Notas
Keeley et al. (2005) en un estudio sobre recuperación de la vegetación tras el fuego en matorrales mediterráneos de California, computaron índices de similaridad para la densidad (cobertura) de cada una de las especies antes y después de las quemas. De esta forma podían estimar el comportamiento de las especies tras el fuego. Además podían analizar las posibles relaciones de competencia que se producen tras el fuego entre las especies. Asímismo usando un índice de Jaccard, pudieron determinan cambios a nivel de comunidad.
Existen varios estudios interesantes que presentan un diseño experimental similar, ver Alcañiz et al. (2016) y Alcañiz et al. (2020)
Incluir otras covariables como la precipitación tras el fuego (nº de días sin lluvía, etc); la pendiente, etc. Ver el estudio de Pereira et al. (2016).
References
sessionInfo()R version 4.0.2 (2020-06-22)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS Catalina 10.15.3
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRblas.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] DT_0.17 plotrix_3.8-1 readxl_1.3.1 forcats_0.5.1
[5] stringr_1.4.0 dplyr_1.0.6 purrr_0.3.4 readr_1.4.0
[9] tidyr_1.1.3 tibble_3.1.2 ggplot2_3.3.3 tidyverse_1.3.1
[13] here_1.0.1 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] Rcpp_1.0.6 lubridate_1.7.10 assertthat_0.2.1 rprojroot_2.0.2
[5] digest_0.6.27 utf8_1.1.4 R6_2.5.0 cellranger_1.1.0
[9] backports_1.2.1 reprex_2.0.0 evaluate_0.14 highr_0.8
[13] httr_1.4.2 pillar_1.6.1 rlang_0.4.10 rstudioapi_0.13
[17] whisker_0.4 jquerylib_0.1.3 rmarkdown_2.8 labeling_0.4.2
[21] htmlwidgets_1.5.3 munsell_0.5.0 broom_0.7.6 compiler_4.0.2
[25] httpuv_1.5.5 modelr_0.1.8 xfun_0.23 pkgconfig_2.0.3
[29] htmltools_0.5.1.1 tidyselect_1.1.0 fansi_0.4.2 crayon_1.4.1
[33] dbplyr_2.1.1 withr_2.4.1 later_1.1.0.1 grid_4.0.2
[37] jsonlite_1.7.2 gtable_0.3.0 lifecycle_1.0.0 DBI_1.1.1
[41] git2r_0.28.0 magrittr_2.0.1 scales_1.1.1 cli_2.5.0
[45] stringi_1.5.3 farver_2.0.3 fs_1.5.0 promises_1.2.0.1
[49] xml2_1.3.2 bslib_0.2.4 ellipsis_0.3.2 generics_0.1.0
[53] vctrs_0.3.8 tools_4.0.2 glue_1.4.2 crosstalk_1.1.1
[57] hms_1.0.0 yaml_2.2.1 colorspace_2.0-0 rvest_1.0.0
[61] knitr_1.31 haven_2.3.1 sass_0.3.1