RPKM_vs_TPM
Last updated: 2022-09-19
Checks: 6 1
Knit directory: ChromatinSplicingQTLs/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20191126) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version e9163e8. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.Rhistory
Ignored: code/.DS_Store
Ignored: code/.RData
Ignored: code/._.DS_Store
Ignored: code/._README.md
Ignored: code/._report.html
Ignored: code/.ipynb_checkpoints/
Ignored: code/.snakemake/
Ignored: code/APA_Processing/
Ignored: code/Alignments/
Ignored: code/ChromHMM/
Ignored: code/ENCODE/
Ignored: code/ExpressionAnalysis/
Ignored: code/FastqFastp/
Ignored: code/FastqFastpSE/
Ignored: code/Genotypes/
Ignored: code/IntronSlopes/
Ignored: code/Misc/
Ignored: code/MiscCountTables/
Ignored: code/Multiqc/
Ignored: code/Multiqc_chRNA/
Ignored: code/NonCodingRNA_annotation/
Ignored: code/PeakCalling/
Ignored: code/Phenotypes/
Ignored: code/PlotGruberQTLs/
Ignored: code/PlotQTLs/
Ignored: code/ProCapAnalysis/
Ignored: code/QC/
Ignored: code/QTL_SNP_Enrichment/
Ignored: code/QTLs/
Ignored: code/ReferenceGenome/
Ignored: code/Rplots.pdf
Ignored: code/Session.vim
Ignored: code/SplicingAnalysis/
Ignored: code/TODO
Ignored: code/Tehranchi/
Ignored: code/bigwigs/
Ignored: code/bigwigs_FromNonWASPFilteredReads/
Ignored: code/config/.DS_Store
Ignored: code/config/._.DS_Store
Ignored: code/config/.ipynb_checkpoints/
Ignored: code/debug.ipynb
Ignored: code/debug_python.ipynb
Ignored: code/deepTools/
Ignored: code/featureCounts/
Ignored: code/gwas_summary_stats/
Ignored: code/hyprcoloc/
Ignored: code/igv_session.xml
Ignored: code/log
Ignored: code/logs/
Ignored: code/notebooks/.ipynb_checkpoints/
Ignored: code/rules/.QTLTools.smk.swp
Ignored: code/rules/.ipynb_checkpoints/
Ignored: code/rules/OldRules/
Ignored: code/rules/notebooks/
Ignored: code/scratch/
Ignored: code/scripts/.ipynb_checkpoints/
Ignored: code/scripts/GTFtools_0.8.0/
Ignored: code/scripts/__pycache__/
Ignored: code/scripts/liftOverBedpe/liftOverBedpe.py
Ignored: code/snakemake.log
Ignored: code/snakemake.sbatch.log
Ignored: data/.DS_Store
Ignored: data/._.DS_Store
Ignored: data/._20220414203249_JASPAR2022_combined_matrices_25818_jaspar.txt
Ignored: data/GWAS_catalog_summary_stats_sources/._list_gwas_summary_statistics_6_Apr_2022-10.csv
Ignored: data/GWAS_catalog_summary_stats_sources/._list_gwas_summary_statistics_6_Apr_2022-11.csv
Ignored: data/GWAS_catalog_summary_stats_sources/._list_gwas_summary_statistics_6_Apr_2022-2.csv
Ignored: data/GWAS_catalog_summary_stats_sources/._list_gwas_summary_statistics_6_Apr_2022-3.csv
Ignored: data/GWAS_catalog_summary_stats_sources/._list_gwas_summary_statistics_6_Apr_2022-4.csv
Ignored: data/GWAS_catalog_summary_stats_sources/._list_gwas_summary_statistics_6_Apr_2022-5.csv
Ignored: data/GWAS_catalog_summary_stats_sources/._list_gwas_summary_statistics_6_Apr_2022-6.csv
Ignored: data/GWAS_catalog_summary_stats_sources/._list_gwas_summary_statistics_6_Apr_2022-7.csv
Ignored: data/GWAS_catalog_summary_stats_sources/._list_gwas_summary_statistics_6_Apr_2022-8.csv
Ignored: data/GWAS_catalog_summary_stats_sources/._list_gwas_summary_statistics_6_Apr_2022.csv
Untracked files:
Untracked: code/snakemake_profiles/slurm/__pycache__/
Unstaged changes:
Modified: analysis/20220713_RPKM_v_TPM.Rmd
Deleted: code/envs/spliceq.yml
Modified: code/scripts/GenometracksByGenotype
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/20220713_RPKM_v_TPM.Rmd) and HTML (docs/20220713_RPKM_v_TPM.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 5a188df | Benjmain Fair | 2022-07-26 | update coloc results |
Analysis
…Just curious how RPKM and TPM correlate in practice…
library(tidyverse)
library(edgeR)
library(DGEobj.utils)
#Read in data:
dat <- read_tsv("../code/featureCounts/chRNA.Expression/Counts.txt", comment = '#')
genes <- read_tsv("../code/ExpressionAnalysis/polyA/ExpressedGeneList.txt", col_names = c("chrom", "start", "stop", "Geneid", "score", "strand"))
counts <- dat %>%
dplyr::select(-c(2:6)) %>%
inner_join(genes, by="Geneid") %>%
dplyr::select(1, matches("Alignments/STAR_Align/chRNA.Expression.Splicing/(.+?)/1/Filtered.bam")) %>%
rename_at(-1, ~ str_replace(.x, "Alignments/STAR_Align/chRNA.Expression.Splicing/(.+?)/1/Filtered.bam", "\\1")) %>%
column_to_rownames("Geneid") %>%
as.matrix() %>%
DGEList()
geneLengths <- counts$counts %>%
as.data.frame() %>%
rownames_to_column("Geneid") %>%
dplyr::select(Geneid) %>%
left_join(
dat %>% dplyr::select(Geneid, Length)
) %>% pull(Length)
rpkm <- rpkm(counts, gene.length = geneLengths, prior.count=0.25, log=T)
tpm <- convertCounts(counts$counts, unit="TPM", geneLength=geneLengths, log=T, prior.count = 0.25)
rpkm.other <- convertCounts(counts$counts, unit="FPKM", geneLength=geneLengths, log=T, prior.count = 0.25)
plot(rpkm.other[,1], rpkm[,1])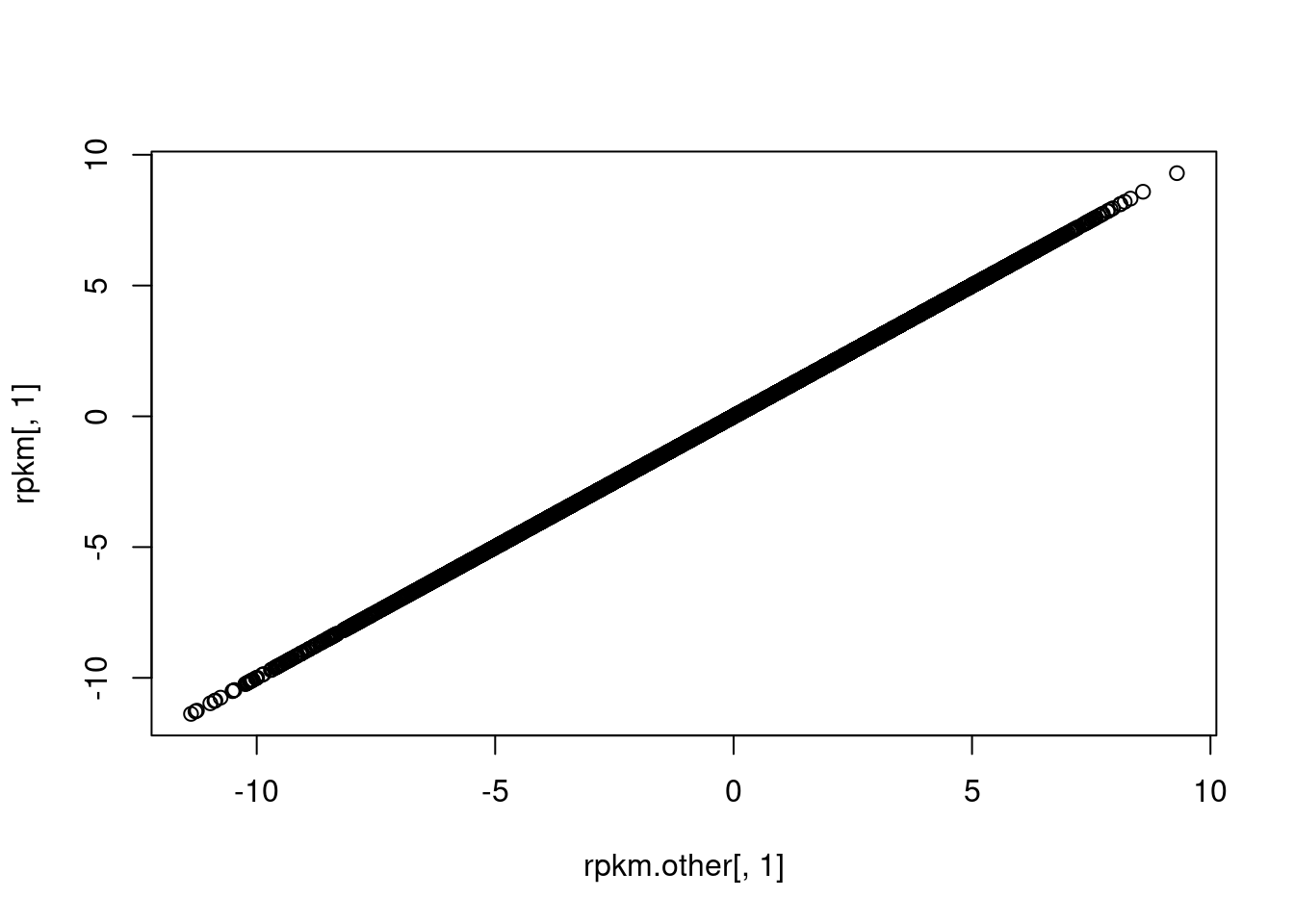
plot(rpkm.other[,1], tpm[,1])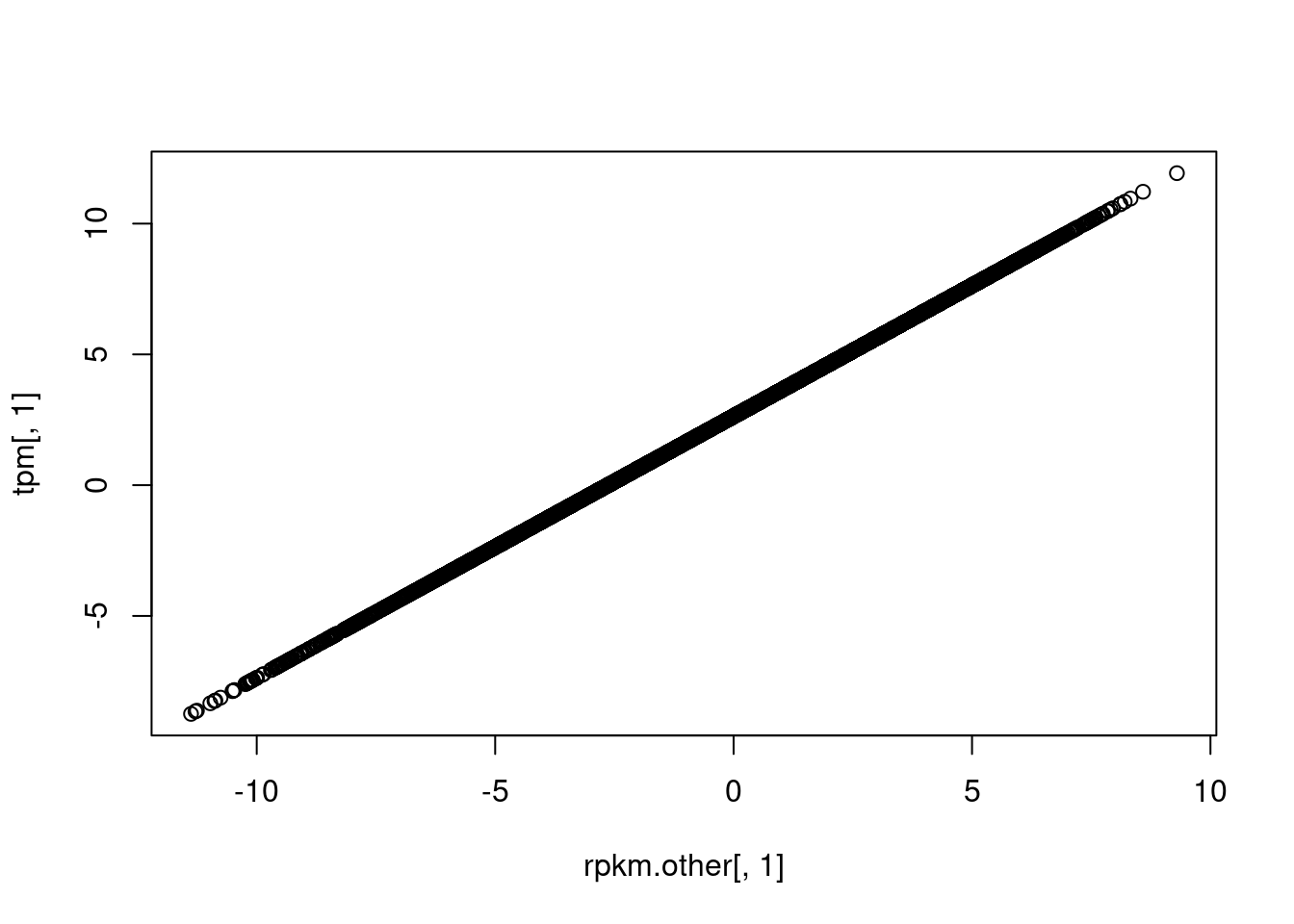
rpkm %>% cor(use="complete.obs") %>% mean()[1] 0.9270265tpm %>% cor(use="complete.obs") %>% mean()[1] 0.9270265data.frame(tpm = tpm[,1], rpkm=rpkm.other[,1]) %>%
ggplot(aes(x=tpm, y=rpkm)) +
geom_point() +
geom_abline(color='red') +
theme_bw()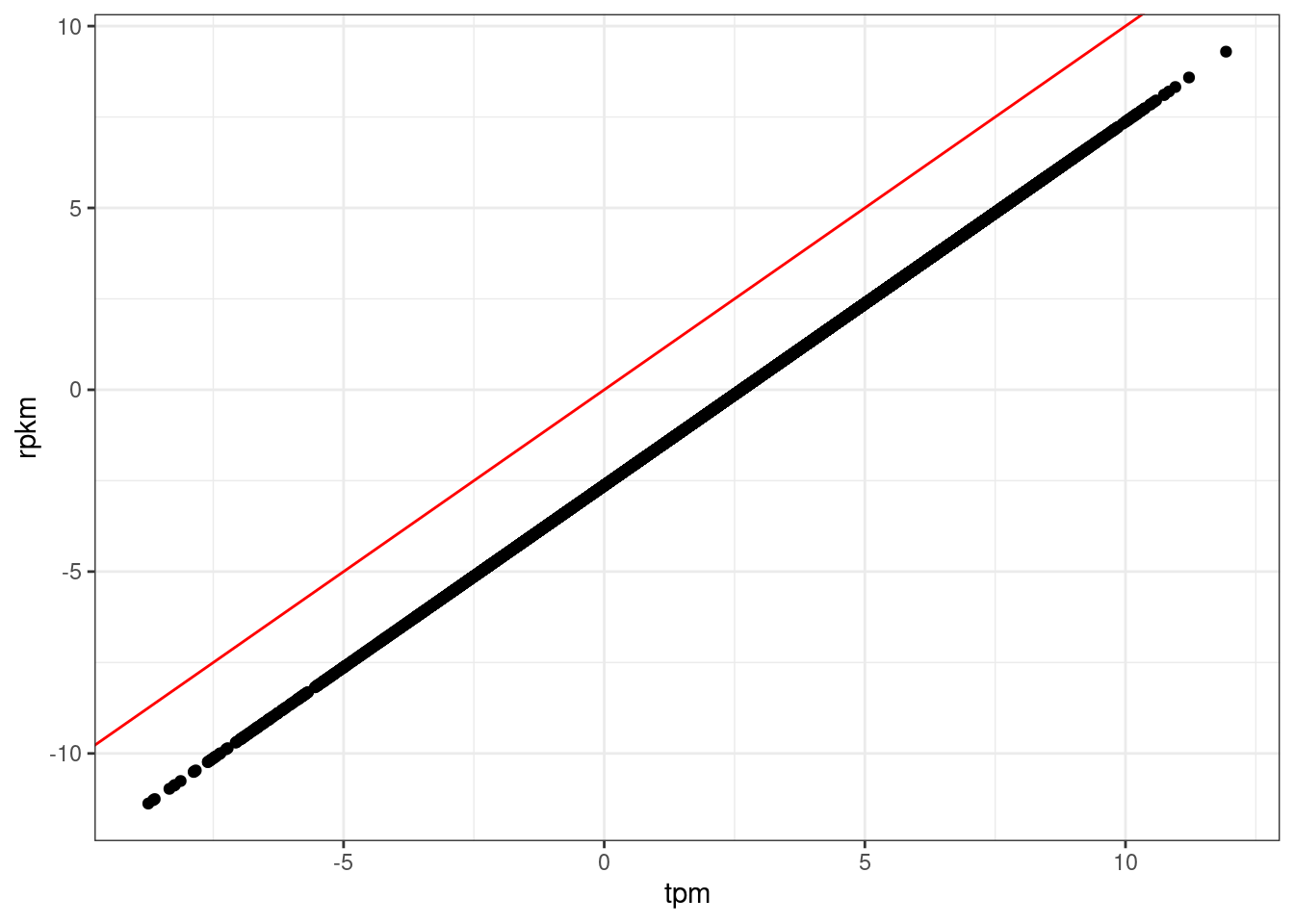
2**median(tpm[,1]-rpkm.other[,1])[1] 6.199072tpm %>%
as.data.frame() %>%
filter_all(any_vars(is.na(.))) [1] NA18853 NA19122 NA18523 NA18499 NA18511 NA19150 NA19098 NA19141 NA18852
[10] NA19210 NA19101 NA18915 NA19102 NA19114 NA18504 NA19190 NA19147 NA19131
[19] NA19138 NA19130 NA19099 NA19239 NA19200 NA19238 NA18881 NA19257 NA18486
[28] NA19137 NA18879 NA18497 NA18923 NA19117 NA19214 NA18520 NA18507 NA19127
[37] NA19152 NA18864 NA18867 NA19119 NA18510 NA19225 NA19184 NA18924 NA19236
[46] NA18868 NA19213 NA19107 NA18877 NA18516 NA19247 NA18855 NA19206 NA19160
[55] NA18913 NA18870 NA19095 NA19093 NA18858 NA19092 NA18522 NA18917 NA18862
[64] NA19198 NA19171 NA19201 NA19096 NA19140 NA19121 NA18508 NA18519 NA19153
[73] NA18910 NA19143 NA19118 NA18934 NA19209 NA18498 NA19207 NA19146 NA18876
[82] NA18909 NA19108 NA18505 NA18502 NA19203 NA19128
<0 rows> (or 0-length row.names)Conclusion:
RPKM, TPM… It really doesn’t matter! They are basically perfectly correlated within a sample and across samples the mean correlation coefficient is basically the same… But maybe TPM is slightly more interpretable units imo… But make sure to add a pseudocount otherwise the convertCounts(method="TPM") function will output NA values.
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=C
[4] LC_COLLATE=C LC_MONETARY=C LC_MESSAGES=C
[7] LC_PAPER=C LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=C LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] DGEobj.utils_1.0.6 edgeR_3.26.5 limma_3.40.6 forcats_0.4.0
[5] stringr_1.4.0 dplyr_1.0.9 purrr_0.3.4 readr_1.3.1
[9] tidyr_1.2.0 tibble_3.1.7 ggplot2_3.3.6 tidyverse_1.3.0
loaded via a namespace (and not attached):
[1] Rcpp_1.0.5 locfit_1.5-9.1 lubridate_1.7.4 lattice_0.20-38
[5] DGEobj_1.1.2 assertthat_0.2.1 rprojroot_2.0.2 digest_0.6.20
[9] utf8_1.1.4 R6_2.4.0 cellranger_1.1.0 backports_1.4.1
[13] reprex_0.3.0 evaluate_0.15 highr_0.9 httr_1.4.4
[17] pillar_1.7.0 rlang_1.0.5 readxl_1.3.1 rstudioapi_0.14
[21] whisker_0.3-2 rmarkdown_1.13 labeling_0.3 munsell_0.5.0
[25] broom_1.0.0 compiler_3.6.1 httpuv_1.5.1 modelr_0.1.8
[29] xfun_0.31 pkgconfig_2.0.2 htmltools_0.5.3 tidyselect_1.1.2
[33] workflowr_1.6.2 fansi_0.4.0 crayon_1.3.4 dbplyr_1.4.2
[37] withr_2.5.0 later_0.8.0 grid_3.6.1 jsonlite_1.6
[41] gtable_0.3.0 lifecycle_1.0.1 DBI_1.1.0 git2r_0.26.1
[45] magrittr_1.5 scales_1.1.0 cli_3.3.0 stringi_1.4.3
[49] farver_2.1.0 fs_1.5.2 promises_1.0.1 xml2_1.3.2
[53] ellipsis_0.3.2 generics_0.1.3 vctrs_0.4.1 tools_3.6.1
[57] glue_1.6.2 hms_0.5.3 fastmap_1.1.0 yaml_2.2.0
[61] colorspace_1.4-1 rvest_0.3.5 knitr_1.39 haven_2.3.1