20230324_MakeFigs
2023-03-24
Last updated: 2023-03-27
Checks: 6 1
Knit directory:
ChromatinSplicingQTLs/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R
Markdown file created these results, you’ll want to first commit it to
the Git repo. If you’re still working on the analysis, you can ignore
this warning. When you’re finished, you can run
wflow_publish to commit the R Markdown file and build the
HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20191126) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version de90abf. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.Rhistory
Ignored: code/.DS_Store
Ignored: code/.RData
Ignored: code/._report.html
Ignored: code/.ipynb_checkpoints/
Ignored: code/.snakemake/
Ignored: code/APA_Processing/
Ignored: code/Alignments/
Ignored: code/ChromHMM/
Ignored: code/ENCODE/
Ignored: code/ExpressionAnalysis/
Ignored: code/FastqFastp/
Ignored: code/FastqFastpSE/
Ignored: code/FastqSE/
Ignored: code/Genotypes/
Ignored: code/H3K36me3_CutAndTag.pdf
Ignored: code/IntronSlopes/
Ignored: code/LR.bed
Ignored: code/LR.seq.bed
Ignored: code/LongReads/
Ignored: code/Metaplots/
Ignored: code/Misc/
Ignored: code/MiscCountTables/
Ignored: code/Multiqc/
Ignored: code/Multiqc_chRNA/
Ignored: code/NonCodingRNA/
Ignored: code/NonCodingRNA_annotation/
Ignored: code/PairwisePi1Traits.P.all.txt.gz
Ignored: code/PeakCalling/
Ignored: code/Phenotypes/
Ignored: code/PlotGruberQTLs/
Ignored: code/PlotQTLs/
Ignored: code/ProCapAnalysis/
Ignored: code/QC/
Ignored: code/QTL_SNP_Enrichment/
Ignored: code/QTLs/
Ignored: code/RPKM_tables/
Ignored: code/ReadLengthMapExperiment/
Ignored: code/ReadLengthMapExperimentResults/
Ignored: code/ReadLengthMapExperimentSpliceCounts/
Ignored: code/ReferenceGenome/
Ignored: code/Rplots.pdf
Ignored: code/Session.vim
Ignored: code/SmallMolecule/
Ignored: code/SplicingAnalysis/
Ignored: code/TODO
Ignored: code/Tehranchi/
Ignored: code/bigwigs/
Ignored: code/bigwigs_FromNonWASPFilteredReads/
Ignored: code/config/.DS_Store
Ignored: code/config/._.DS_Store
Ignored: code/config/.ipynb_checkpoints/
Ignored: code/config/config.local.yaml
Ignored: code/dag.pdf
Ignored: code/dag.png
Ignored: code/dag.svg
Ignored: code/debug.ipynb
Ignored: code/debug_python.ipynb
Ignored: code/deepTools/
Ignored: code/featureCounts/
Ignored: code/featureCountsBasicGtf/
Ignored: code/gwas_summary_stats/
Ignored: code/hyprcoloc/
Ignored: code/igv_session.xml
Ignored: code/isoseqbams/
Ignored: code/log
Ignored: code/logs/
Ignored: code/notebooks/.ipynb_checkpoints/
Ignored: code/pi1/
Ignored: code/rules/.CalculatePi1.smk.swp
Ignored: code/rules/.ipynb_checkpoints/
Ignored: code/rules/OldRules/
Ignored: code/rules/notebooks/
Ignored: code/scratch/
Ignored: code/scripts/.ipynb_checkpoints/
Ignored: code/scripts/GTFtools_0.8.0/
Ignored: code/scripts/__pycache__/
Ignored: code/scripts/liftOverBedpe/liftOverBedpe.py
Ignored: code/snakemake.dryrun.log
Ignored: code/snakemake.log
Ignored: code/snakemake.sbatch.log
Ignored: code/snakemake_profiles/slurm/__pycache__/
Ignored: code/test.introns.bed
Ignored: code/test.introns2.bed
Ignored: code/test.log
Ignored: code/tracks.xml
Ignored: data/.DS_Store
Ignored: data/._.DS_Store
Ignored: data/._20220414203249_JASPAR2022_combined_matrices_25818_jaspar.txt
Ignored: data/GWAS_catalog_summary_stats_sources/._list_gwas_summary_statistics_6_Apr_2022-10.csv
Ignored: data/GWAS_catalog_summary_stats_sources/._list_gwas_summary_statistics_6_Apr_2022-11.csv
Ignored: data/GWAS_catalog_summary_stats_sources/._list_gwas_summary_statistics_6_Apr_2022-2.csv
Ignored: data/GWAS_catalog_summary_stats_sources/._list_gwas_summary_statistics_6_Apr_2022-3.csv
Ignored: data/GWAS_catalog_summary_stats_sources/._list_gwas_summary_statistics_6_Apr_2022-4.csv
Ignored: data/GWAS_catalog_summary_stats_sources/._list_gwas_summary_statistics_6_Apr_2022-5.csv
Ignored: data/GWAS_catalog_summary_stats_sources/._list_gwas_summary_statistics_6_Apr_2022-6.csv
Ignored: data/GWAS_catalog_summary_stats_sources/._list_gwas_summary_statistics_6_Apr_2022-7.csv
Ignored: data/GWAS_catalog_summary_stats_sources/._list_gwas_summary_statistics_6_Apr_2022-8.csv
Ignored: data/GWAS_catalog_summary_stats_sources/._list_gwas_summary_statistics_6_Apr_2022.csv
Ignored: data/Metaplots/.DS_Store
Untracked files:
Untracked: analysis/20230324_MakeFigs.Rmd
Unstaged changes:
Modified: code/scripts/GenometracksByGenotype
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with
wflow_publish() to start tracking its development.
knitr::opts_chunk$set(echo = TRUE, warning = F, message = F)
library(tidyverse)── Attaching packages ─────────────────────────────────────── tidyverse 1.3.1 ──✔ ggplot2 3.3.6 ✔ purrr 0.3.4
✔ tibble 3.1.7 ✔ dplyr 1.0.9
✔ tidyr 1.2.0 ✔ stringr 1.4.0
✔ readr 2.1.2 ✔ forcats 0.5.1── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()library(RColorBrewer)
library(data.table)
Attaching package: 'data.table'The following objects are masked from 'package:dplyr':
between, first, lastThe following object is masked from 'package:purrr':
transposelibrary(edgeR)Loading required package: limma# Set theme
theme_set(
theme_classic() +
theme(text=element_text(size=16, family="Helvetica")))
# I use layer a lot, to rotate long x-axis labels
Rotate_x_labels <- theme(axis.text.x = element_text(angle = 45, vjust = 1, hjust=1))
#test plot
ggplot(mtcars, aes(x=mpg, y=cyl)) +
geom_point()
Intro
I notebook to make figure panels for publication…
Figures to make: 1. Placeholder, Nascent RNA reveals abundance of NMD-destined transcripts 2. sQTLs, eQTLs a. pi heatmap across promoter hQTLs, H3K36me3, chRNA, 4sU, polyA. Number of QTLs in margin b. Enrichment of annotations in transcriptional vs post-transcriptional eQTLs. Classify transcriptional vs post-transcriptional by comparing hQTL to eQTL significance (or effect size). c. QQ-plot of eQTL signal, grouped by hQTLs, sQTLs that affect productive junctions, and sQTLs that affect unproductive junctions d. polyA sQTL beta vs polyA eQTL beta scatter, faceted by stable vs unstable. e. Beta vs beta spearman slope with different lines for chRNA, 4sU, polyA, faceted by stable vs unstable 3. Contribution of splicing-mediated decay to complex trait a. A single GWAS trait (ie MS) QQ plot, colored by splicing-mediated eQTLs, transcription-mediated QTLs, productive-AS sQTLs b. Heatmap of similar enrichment in A, but across many traits c. Example locus showing colocalization eQTL/sQTL/GWAS scatter, and IGV screenshot of the splicing and eQTL effect 4. The effect of splice-modifying drugs on gene expression a. Cartoon showing inset with risdiplam bulge repair, along with dose:response trend showing the tested doses and the increasing response in GA|GT but not AG|GT splicing. b. Scatter that shows splicing vs expression effect, faeted by productive unproductive. Or something like that c. Something that shows the 4B effect is not in chRNA d. An illustrative example effect in IGV.
Supplemental Figures to make 1. Placeholder 2. sQTLs, eQTLs a. Total number of QTLs in each type of type of trait. Bargraph, with multiple lines for different FDR levels b. Correlation of effect sizes across promoter hQTLs, H3K36me3, chRNA, 4sU, polyA i. Need to select set of eQTLs to plot… Previous versions of this plot I tend to just pick colocalized things. That would be easiest. c. sQTL QQ plot, with different colors for post-transcriptional eQTLs to transcriptional eQTLs (like what Carlos has done in red and black) d. eQTL QQ plot, with different colors for H3K27AC QTLs and sQTLs e. lead gene-wise sQTLs are not strongly enriched for eQTLs compared to control SNPs. eQTL QQ-plot with top gene-wise sQTL SNP, compared to some controls. Or do this with a pi1, and can use various thresholds f. enrichment of eQTL signals is explained by both primary and non-primary eQTL signals (FigX). More precisely, we used the SuSiE finemapping approach which does not assume a single causal SNP for each expression trait, and can thus be used to finemap multiple eQTL signals per gene. We find that approximately x% of primary signals could be overlapped with sQTLs, compared to y% of primary signals that we could overlap to hQTLs. Furthermore, x% and y% of non primary signals could be mapped sQTLs and hQTLs, respectively g. More extensive version of 2d, so we can explain the nuances in filtering out transcriptional effects, and there are some lingering non-NMD effects that are degraded in cytoplasm 3. Contribution of splicing-mediated decay to complex trait a. Colocalization framework cartoon b. Version of 3b that uses colocalization. Shown as barplots similar to Phoenix’s paper 4. The effect of splice-modifying drugs on gene expression a. In total, we observe x% of observed non-canonincal GA-GT splice sites are activated with x criteria… ecdf of spearman correlation coefficient, for different groups of 5’ss.
Figure2
A
pi heatmap of gene-feature QTLs. Num QTLs in margin.
Only include hQTLs at promoter, chRNA, 4sU, and polyA.
library(qvalue)
library(viridis)
PeaksToTSS <- Sys.glob("../code/Misc/PeaksClosestToTSS/*_assigned.tsv.gz") %>%
setNames(str_replace(., "../code/Misc/PeaksClosestToTSS/(.+?)_assigned.tsv.gz", "\\1")) %>%
lapply(read_tsv) %>%
bind_rows(.id="ChromatinMark") %>%
mutate(GenePeakPair = paste(gene, peak, sep = ";")) %>%
distinct(ChromatinMark, peak, gene, .keep_all=T)
PC1.filter <- c("Expression.Splicing", "MetabolicLabelled.30min", "MetabolicLabelled.60min", "H3K36ME3", "H3K27AC", "H3K4ME3", "H3K4ME1", "Expression.Splicing.Subset_YRI", "chRNA.Expression.Splicing")
dat <- fread("../code/PairwisePi1Traits.P.all.txt.gz") %>%
filter((PC1 %in% PC1.filter) & (PC2 %in% PC1.filter))
dat$PC1 %>% unique()[1] "Expression.Splicing" "Expression.Splicing.Subset_YRI"
[3] "chRNA.Expression.Splicing" "MetabolicLabelled.30min"
[5] "MetabolicLabelled.60min" "H3K27AC"
[7] "H3K4ME3" "H3K4ME1"
[9] "H3K36ME3" PhenotypeAliases <- read_tsv("../data/Phenotypes_recode_for_Plotting.txt")Many genes have multiple promoters that should all be accounted for as potential transcriptional effects. So calculating pi1 can be tricky. I want to ask the question, “What fraction of eQTLs have a promoter hQTL in at least one of the promoters?”… I have already solved this problem… For each gene, get lowest P value for across tests (ie all the promoters) and use that a a test statistic to come up with a new Pvalue, to calculate pi1. The test statistic distribution under the null can be approximate by drawing from uniform n times.
First we need to approximate that null using runif…
MaxSampleSizeToCreateANull <- 250
NSamplesToEstimateDistribution <- 10000
NullSimulatedTestStats <- matrix(nrow=MaxSampleSizeToCreateANull, ncol=NSamplesToEstimateDistribution)
rownames(NullSimulatedTestStats) <- paste0("runif_samplesize", 1:MaxSampleSizeToCreateANull)
colnames(NullSimulatedTestStats) <- paste0("Sample_", 1:NSamplesToEstimateDistribution)
for (i in 1:MaxSampleSizeToCreateANull){
SampleSizeFromUniform <- i
SampledDat <- matrix(runif(SampleSizeFromUniform*NSamplesToEstimateDistribution), nrow=NSamplesToEstimateDistribution)
SampleDatNullTestStatistics <- -log10(apply(SampledDat, 1, min))
NullSimulatedTestStats[i,] <- SampleDatNullTestStatistics
}
NullSimulatedTestStats %>%
as.data.frame() %>%
rownames_to_column("runif_samplesize") %>%
slice(1:250) %>%
mutate(runif_samplesize = as.numeric(str_remove(runif_samplesize, "runif_samplesize"))) %>%
gather(key="Sample", value="value", -runif_samplesize) %>%
ggplot(aes(x=value, color=runif_samplesize, group=runif_samplesize)) +
geom_density() +
scale_color_viridis_c() +
theme_bw() +
labs(x="Test statistic under null\n-log10(MinP) across n draws from uniform", color="n")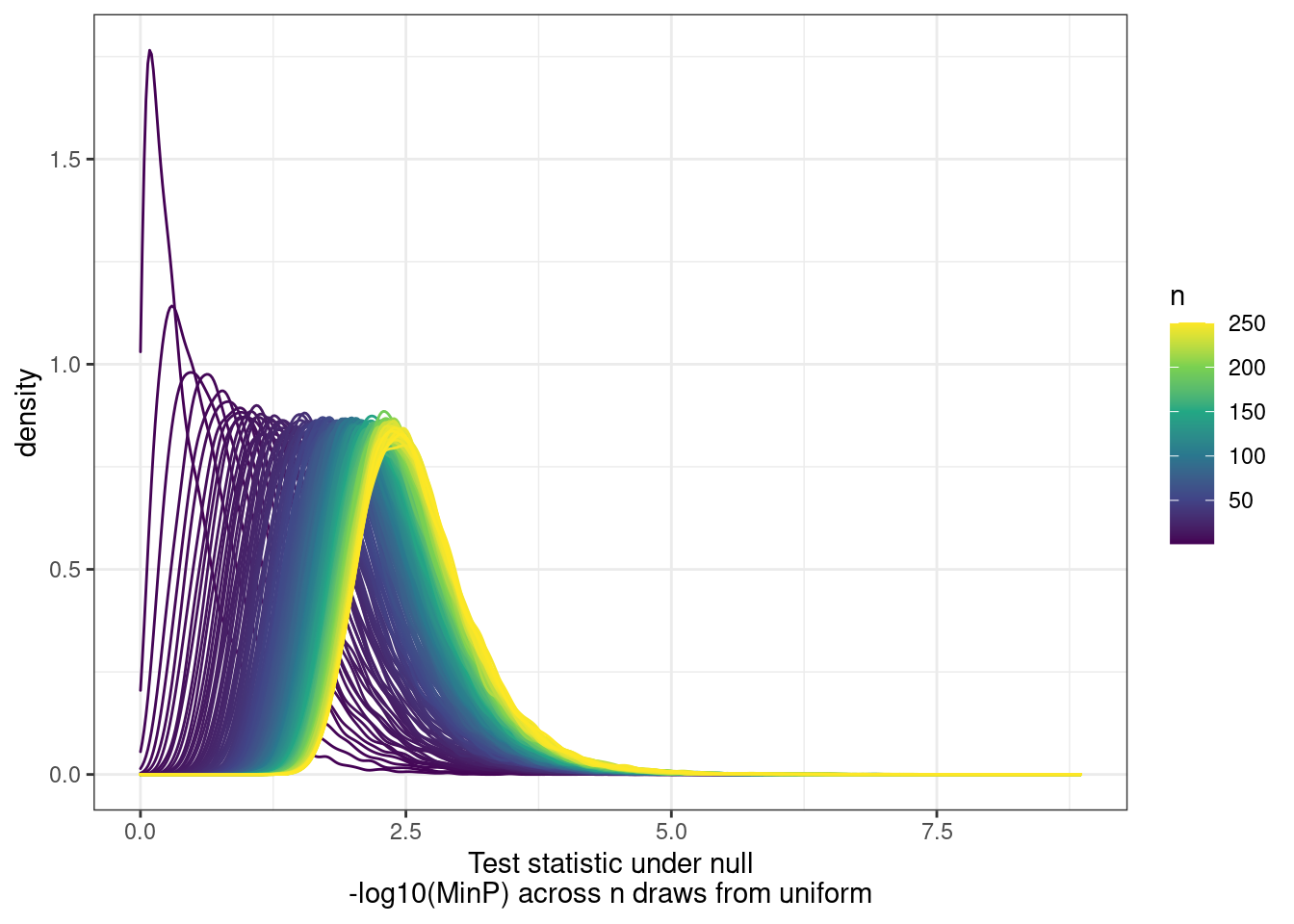
ecdf.functions <- apply(NullSimulatedTestStats, 1, ecdf)
ecdf.functions[[1]](1)[1] 0.9034Make a function to calculate pi1
# pi0est function breaks sometimes. this wrapper function returns 1 when that happens
CalculatePi1 <- function (dat.in, ...) {
return(tryCatch(1-pi0est(dat.in$Pvals.For.Pi1, ...)$pi0, error=function(e) 1))
}And now, recode peaks as promoters, and calculate test statistics.
Recode.PCs = c("ActivatingMark_HostGenePromoter"="Promoter marks*", "H3K36ME3"="H3K36ME3", "chRNA.Expression.Splicing"="chRNA", "MetabolicLabelled.30min"="4sU 30min", "MetabolicLabelled.60min"="4sU 60min", "Expression.Splicing.Subset_YRI"="polyA YRI", "Expression.Splicing"="polyA")
ActivatingMarksToConsider <- c("H3K27AC")
ProcessPi1HeatmapDat <- function(FDR.cutoff=0.1){
dat.split <- dat %>%
filter(FDR.x < FDR.cutoff) %>%
mutate(PC1 = case_when(
paste(GeneLocus, P1, sep=";") %in% PeaksToTSS$GenePeakPair & PC1 %in% ActivatingMarksToConsider ~ "ActivatingMark_HostGenePromoter",
P1 %in% PeaksToTSS$peak ~ "ActivatingMark_OtherGenePromoter",
PC1 %in% c("H3K4ME1", "H3K4ME3", "H3K27AC") ~ "ActivatingMark_Enhancer",
TRUE ~ PC1
)) %>%
mutate(PC2 = case_when(
paste(GeneLocus, P2, sep=";") %in% PeaksToTSS$GenePeakPair & PC2 %in% ActivatingMarksToConsider ~ "ActivatingMark_HostGenePromoter",
P2 %in% PeaksToTSS$peak ~ "ActivatingMark_OtherGenePromoter",
PC2 %in% c("H3K4ME1", "H3K4ME3", "H3K27AC") ~ "ActivatingMark_Enhancer",
TRUE ~ PC2
)) %>%
group_by(PC1, P1, PC2) %>%
mutate(test.stat.obs = -log10(min(trait.x.p.in.y))) %>%
ungroup() %>%
add_count(PC1, P1, PC2) %>%
filter(n<=250) %>%
group_by(PC1, PC2) %>%
rowwise() %>%
mutate(Pvals.For.Pi1 = 1-ecdf.functions[[n]](test.stat.obs)) %>%
ungroup() %>%
select(PC1, PC2, Pvals.For.Pi1) %>%
filter(!PC1==PC2) %>%
split(paste(.$PC1, .$PC2, sep = ";"))
dat.out <- lapply(dat.split, CalculatePi1, pi0.method="bootstrap") %>%
unlist() %>%
data.frame(pi1=.) %>%
rownames_to_column("PC1_PC2") %>%
separate(PC1_PC2, into=c("PC1", "PC2"), sep=';') %>%
filter((PC1%in% c(PC1.filter, "ActivatingMark_HostGenePromoter") & (PC2%in% c(PC1.filter, "ActivatingMark_HostGenePromoter") ))) %>%
mutate(TextColor = case_when(
pi1 >= 0.7 ~ "white",
TRUE ~ "black"
)) %>%
mutate(PC1 = recode(PC1, !!!Recode.PCs)) %>%
mutate(PC2 = recode(PC2, !!!Recode.PCs)) %>%
mutate(PC1 = factor(PC1, levels=Recode.PCs)) %>%
mutate(PC2 = factor(PC2, levels=Recode.PCs))
P <- ggplot(dat.out, aes(x=PC1, y=PC2, fill=pi1, color=TextColor)) +
geom_raster() +
geom_text(aes(label=signif(pi1*100, 2))) +
scale_color_identity() +
scale_fill_viridis(option="A", direction = -1, limits=c(0,1)) +
coord_flip() +
scale_x_discrete(limits=rev) +
theme_classic() +
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1)) +
labs(x="Discovery QTL phenotype", y="Phenotype assessed for overlap", caption=paste("*Promoter marks include", paste(ActivatingMarksToConsider, collapse=",")))
return(list(dat=dat, P=P))
}
ProcessPi1HeatmapDat()$P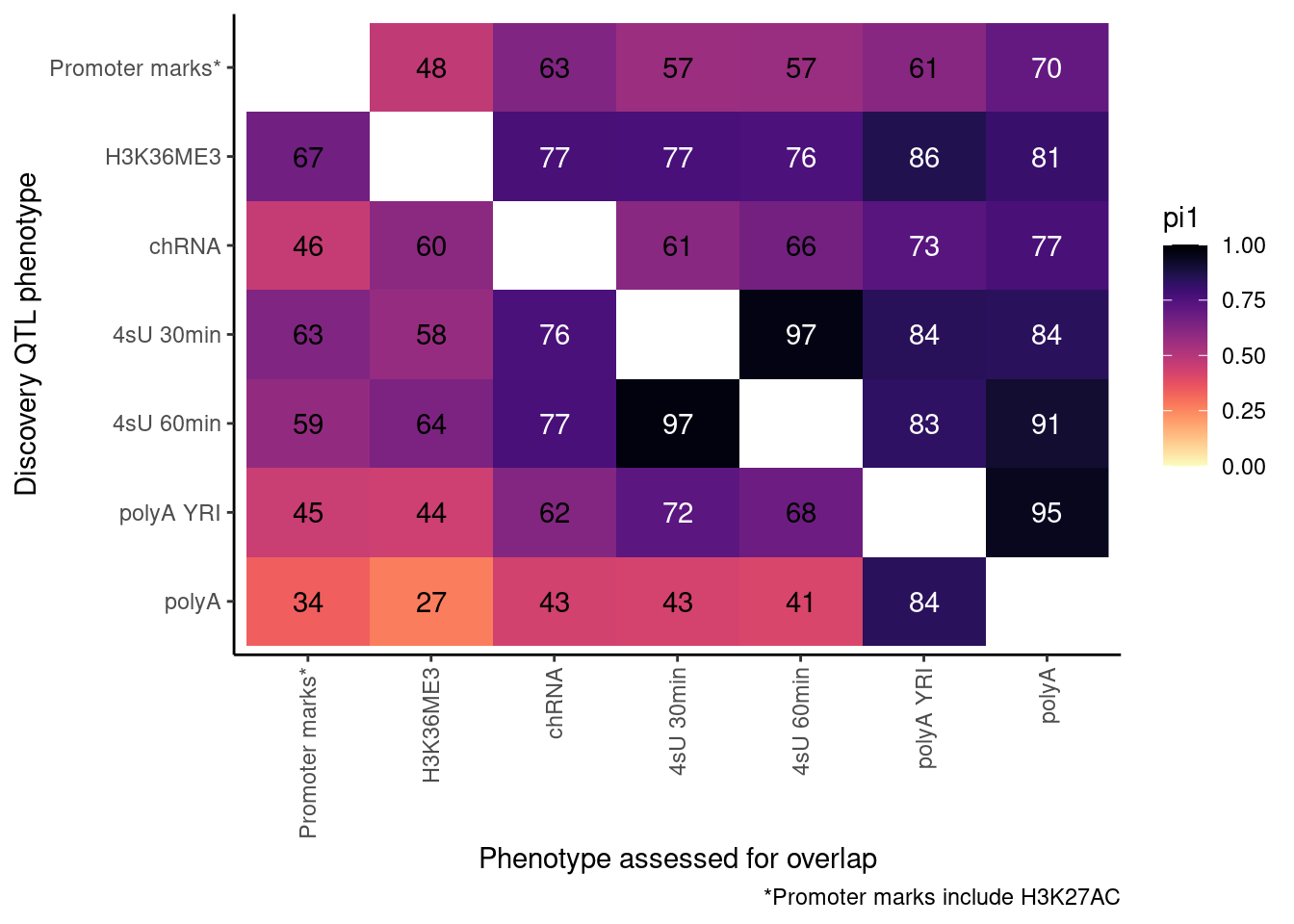
Redo, but also consider both H3K27AC and H3K4ME3 at promoter…
ActivatingMarksToConsider <- c("H3K27AC", "H3K4ME3")
ProcessPi1HeatmapDat()$P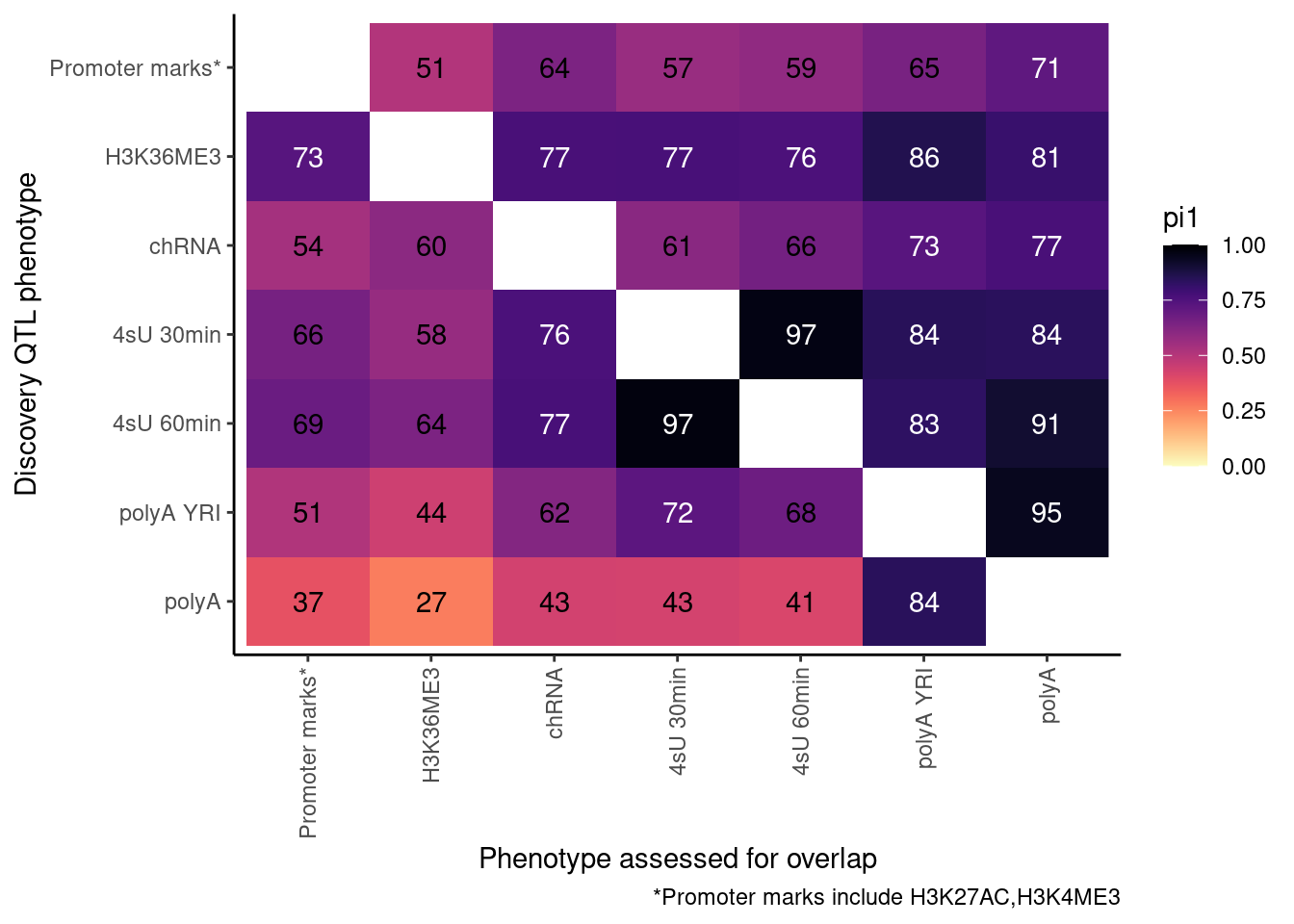
Redo but consider H3K4ME3, H3K4ME1, and H3K27AC
ActivatingMarksToConsider <- c("H3K27AC", "H3K4ME3", "H3K4ME1")
ProcessPi1HeatmapDat()$P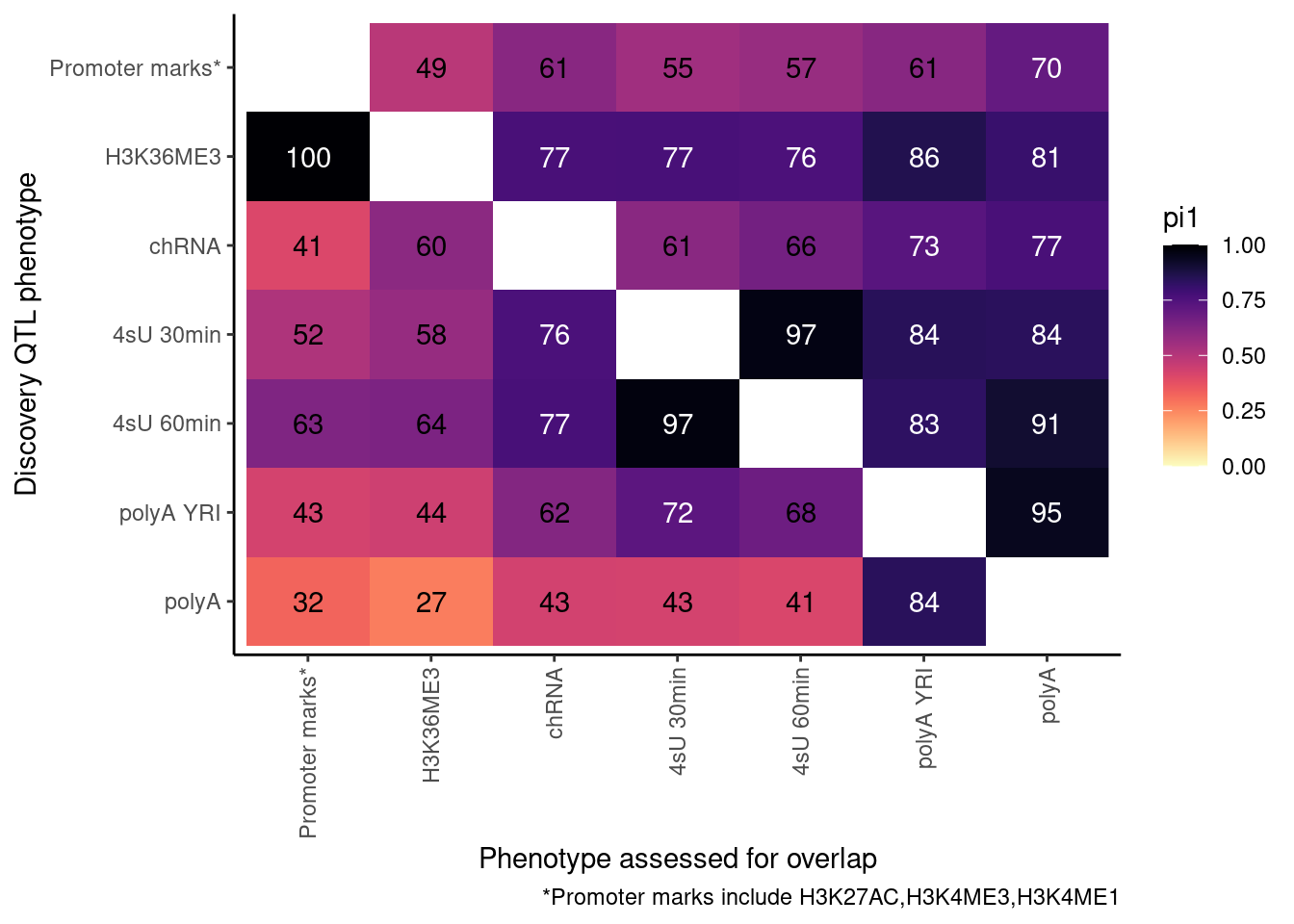 In theory things should only go up as you include H3K4ME1, but that is
not the case. This phenomena is something I’ve noticed before, and I
think it’s because adding non-overlapping phenotypes to promoter marks
only modestly increases the lowest P-value, but more greatly can
increase the n number of features used to determine the null.
In theory things should only go up as you include H3K4ME1, but that is
not the case. This phenomena is something I’ve noticed before, and I
think it’s because adding non-overlapping phenotypes to promoter marks
only modestly increases the lowest P-value, but more greatly can
increase the n number of features used to determine the null.
So, let’s now include H3K4ME1. This is more thought of as an enhancer rather than promoter mark anyway.
So now let’s redo, but only include H3K27AC and H3K4ME3 but use lower FDR cutoffs.
ActivatingMarksToConsider <- c("H3K27AC", "H3K4ME3")
ProcessPi1HeatmapDat(FDR.cutoff = 0.05)$P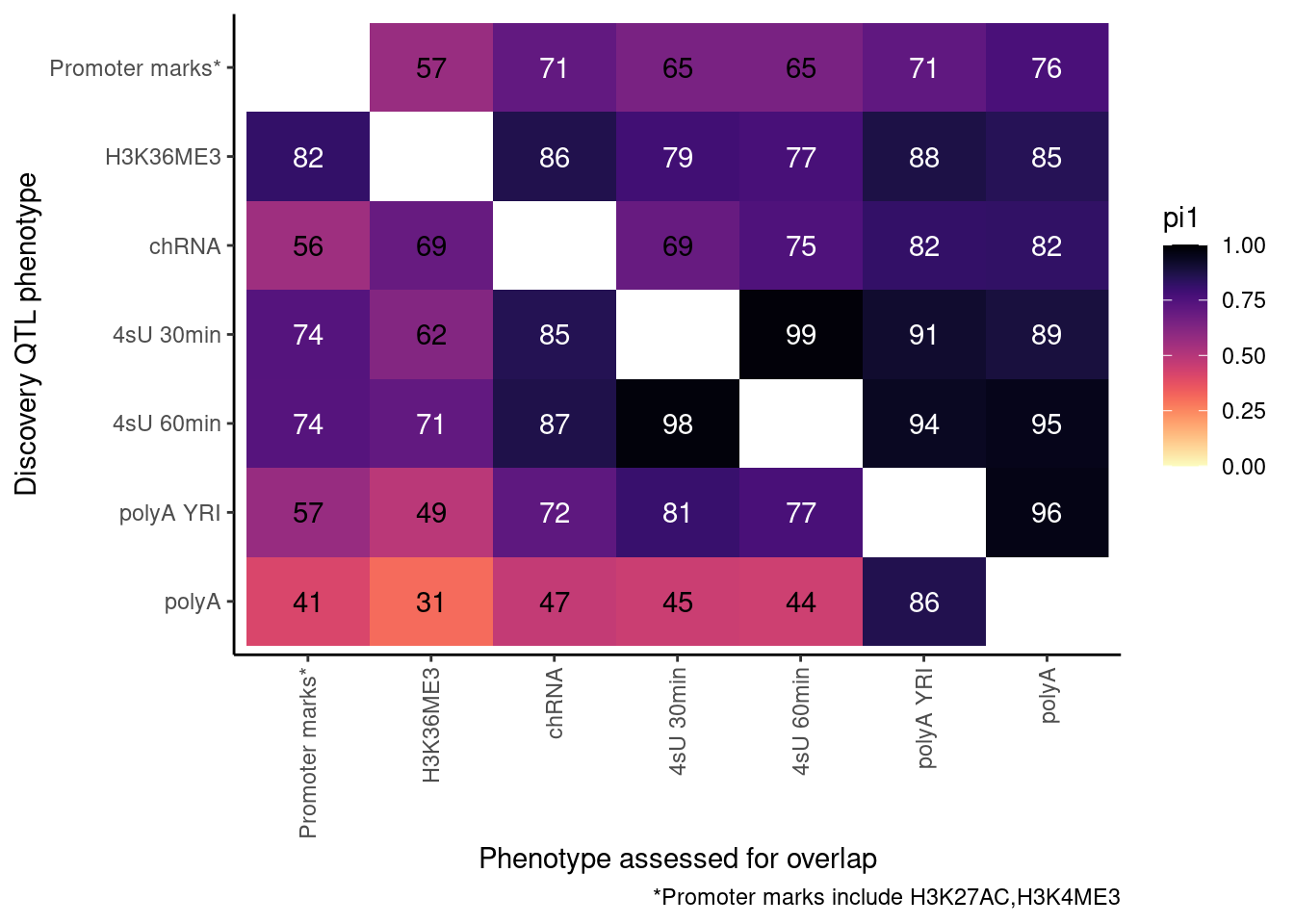
ProcessPi1HeatmapDat(FDR.cutoff = 0.01)$P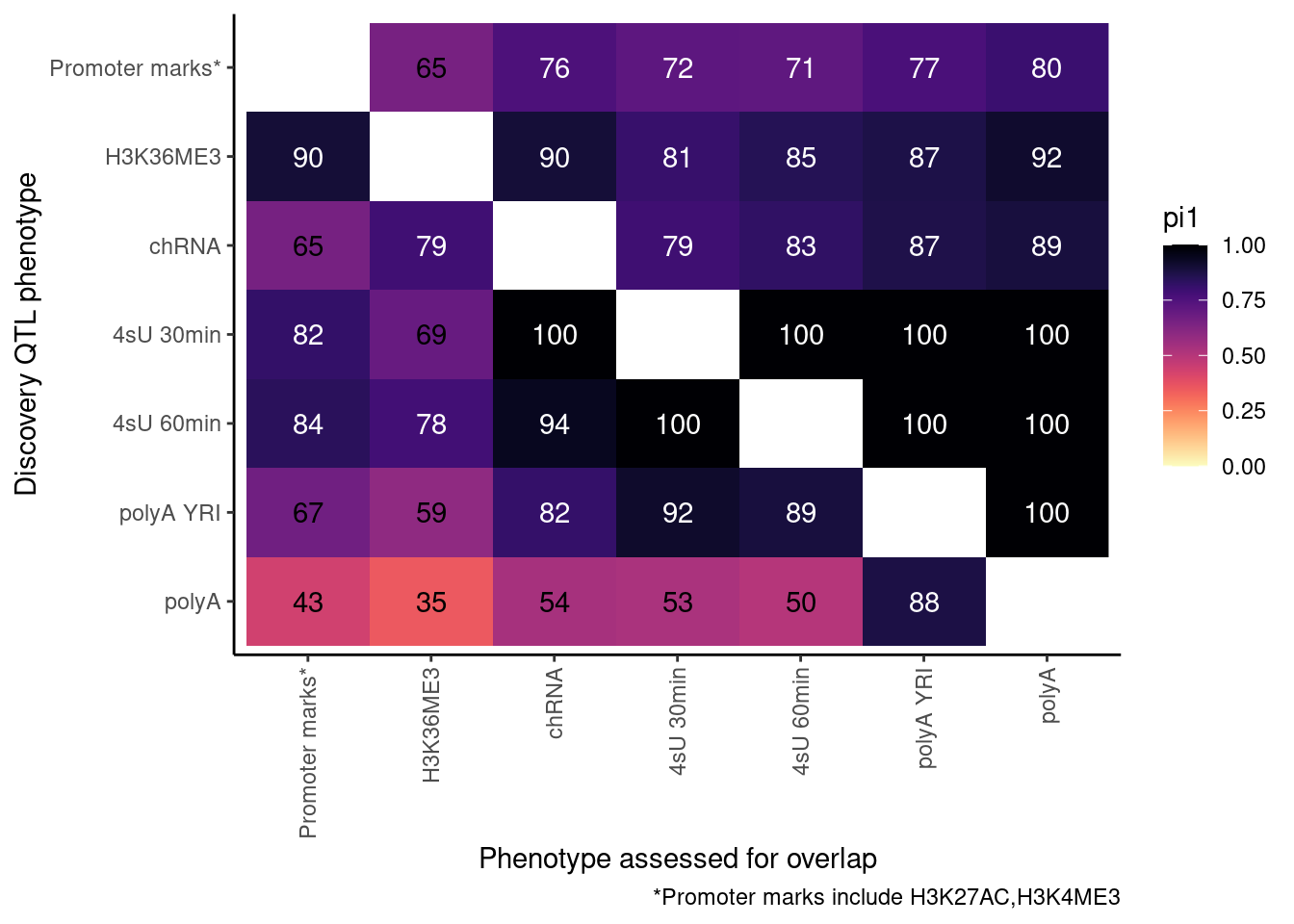
Ok, cool. I mean slightly different versions. I’ll leave it to Yang as to which version should be the main figure, and perhaps we can include alternate versions as supplements. In every case though I notice that the top right corner is dark, suggesting histone effects can be picked up pretty well in downstream assays (ie polyA), but the bottom left corner is light, because of post-transcriptional mechanisms of gene regulation.
sessionInfo()R version 4.2.0 (2022-04-22)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.3.13-el7-x86_64/lib/libopenblas_haswellp-r0.3.13.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=C
[4] LC_COLLATE=C LC_MONETARY=C LC_MESSAGES=C
[7] LC_PAPER=C LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=C LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] viridis_0.6.2 viridisLite_0.4.0 qvalue_2.28.0 edgeR_3.38.4
[5] limma_3.52.4 data.table_1.14.2 RColorBrewer_1.1-3 forcats_0.5.1
[9] stringr_1.4.0 dplyr_1.0.9 purrr_0.3.4 readr_2.1.2
[13] tidyr_1.2.0 tibble_3.1.7 ggplot2_3.3.6 tidyverse_1.3.1
loaded via a namespace (and not attached):
[1] fs_1.5.2 lubridate_1.8.0 bit64_4.0.5 httr_1.4.3
[5] rprojroot_2.0.3 tools_4.2.0 backports_1.4.1 bslib_0.3.1
[9] utf8_1.2.2 R6_2.5.1 DBI_1.1.2 colorspace_2.0-3
[13] withr_2.5.0 tidyselect_1.1.2 gridExtra_2.3 bit_4.0.4
[17] compiler_4.2.0 git2r_0.30.1 cli_3.3.0 rvest_1.0.2
[21] xml2_1.3.3 labeling_0.4.2 sass_0.4.1 scales_1.2.0
[25] digest_0.6.29 rmarkdown_2.14 R.utils_2.11.0 pkgconfig_2.0.3
[29] htmltools_0.5.2 dbplyr_2.1.1 fastmap_1.1.0 highr_0.9
[33] rlang_1.0.2 readxl_1.4.0 rstudioapi_0.13 jquerylib_0.1.4
[37] farver_2.1.0 generics_0.1.2 jsonlite_1.8.0 vroom_1.5.7
[41] R.oo_1.24.0 magrittr_2.0.3 Rcpp_1.0.8.3 munsell_0.5.0
[45] fansi_1.0.3 lifecycle_1.0.1 R.methodsS3_1.8.1 stringi_1.7.6
[49] yaml_2.3.5 plyr_1.8.7 grid_4.2.0 parallel_4.2.0
[53] promises_1.2.0.1 crayon_1.5.1 lattice_0.20-45 haven_2.5.0
[57] splines_4.2.0 hms_1.1.1 locfit_1.5-9.7 knitr_1.39
[61] pillar_1.7.0 reshape2_1.4.4 reprex_2.0.1 glue_1.6.2
[65] evaluate_0.15 modelr_0.1.8 vctrs_0.4.1 tzdb_0.3.0
[69] httpuv_1.6.5 cellranger_1.1.0 gtable_0.3.0 assertthat_0.2.1
[73] xfun_0.30 broom_0.8.0 later_1.3.0 workflowr_1.7.0
[77] ellipsis_0.3.2