20190708_HumanLeadSnps
Ben Fair
7/9/2019
Last updated: 2019-07-11
Checks: 6 1
Knit directory: Comparative_eQTL/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.4.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190319) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.DS_Store
Ignored: analysis/20190521_eQTL_CrossSpeciesEnrichment_cache/
Ignored: analysis_temp/.DS_Store
Ignored: code/.DS_Store
Ignored: code/snakemake_workflow/.DS_Store
Ignored: data/.DS_Store
Ignored: data/PastAnalysesDataToKeep/.DS_Store
Ignored: docs/.DS_Store
Ignored: docs/assets/.DS_Store
Untracked files:
Untracked: analysis/20190627_DiffContactsEgenes.Rmd
Untracked: analysis/20190708_DiffContactsEgenes_CisWindowControlled.Rmd
Untracked: analysis/20190708_SpeciesSharedPolymorphisms_InitialQQPlot.Rmd
Untracked: analysis/20190808_HumanLeadSnps.Rmd
Untracked: docs/figure/20190627_DiffContactsEgenes.Rmd/
Untracked: docs/figure/20190708_DiffContactsEgenes_CisWindowControlled.Rmd/
Untracked: docs/figure/20190708_SpeciesSharedPolymorphisms_InitialQQPlot.Rmd/
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with wflow_publish() to start tracking its development.
The Gtex summary statistics I had been using previously are per-gene qvalues based on permutation test using lead snp to ask if a gene has an eQTL. This was all based on the GTEx v7 pipeline which tests for cis-eQTLs in 1MB window from TSS. However, since my chimp eQTL calling pipeline is based on a 250kB cis-window, this may bias the results of my previous analysis where I concluded that Chimp eGenes have more DNA contacts in the 250kb cis-window in chimp than in human. To address this possible bias, I want to repeat the analysis with human eGene significance values based on a matched cis-window size (250kb). Therefore, I downloaded the full summary statistic table for human left-ventrical eQTLs (all snp-gene pairs) and obtained the lead snp for each gene. this will be used as my-gene level significance value in human. Note that my subsequent analysis is based on ranks of pvalues, so having pvalues that are not well calibrated isn’t as important as getting the ranks of genes right.
Below, I just want to check that there is some rank-correlation (as I would expect) between the lead-snp pvalues (when snps confined to a 250kb window), and the gene-level qvalue based on permutation test.
library(tidyverse)
HumanLeadSnps <- read.table(gzfile('../data/Heart_Left_Ventricle.v7.250kB.leadsnps.txt.gz'), col.names = c("gene", "snp", "tss.dist", "ma_samples", "ma_count", "maf", "p", "slope", "slope_se"))
GtexHeartEgenes <- read.table("../data/Heart_Left_Ventricle.v7.egenes.txt.gz", header=T, sep='\t', stringsAsFactors = F)
Merged <- left_join(GtexHeartEgenes, HumanLeadSnps, by=c("gene_id"="gene"))
ggplot(Merged, aes(x=-log10(qval), y=-log10(p))) +
geom_point(alpha=0.5)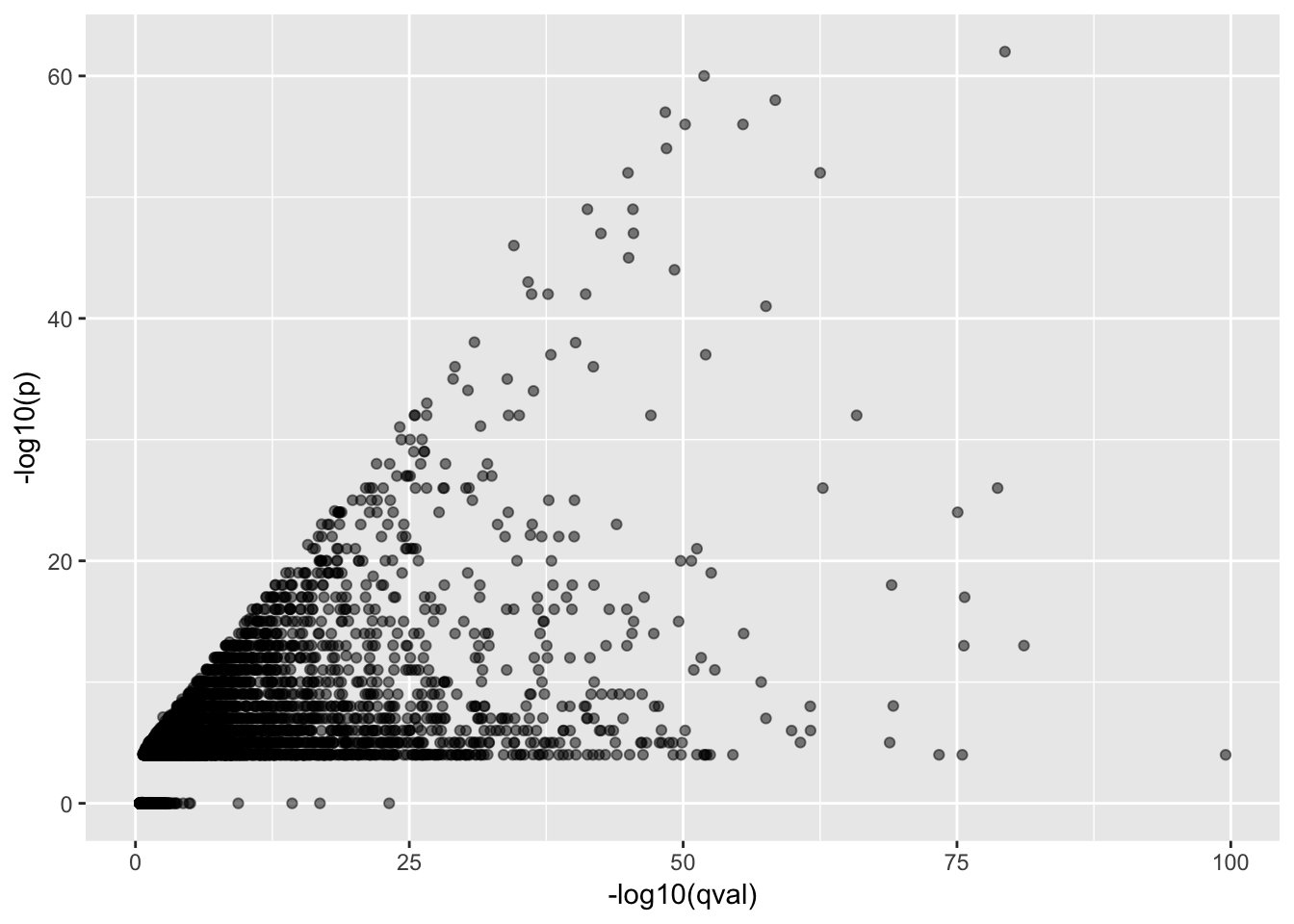
cor.test(Merged$p, Merged$qval, method='spearman')
Spearman's rank correlation rho
data: Merged$p and Merged$qval
S = 3.5902e+11, p-value < 2.2e-16
alternative hypothesis: true rho is not equal to 0
sample estimates:
rho
0.7616891 The correlation is reassuring… I will move on with a repeat of my previous analysis based on these 250kb window lead snp pvalues in humans.
sessionInfo()R version 3.5.1 (2018-07-02)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS 10.14
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] forcats_0.4.0 stringr_1.4.0 dplyr_0.8.1 purrr_0.3.2
[5] readr_1.3.1 tidyr_0.8.3 tibble_2.1.3 ggplot2_3.1.1
[9] tidyverse_1.2.1
loaded via a namespace (and not attached):
[1] Rcpp_1.0.1 cellranger_1.1.0 plyr_1.8.4 pillar_1.4.1
[5] compiler_3.5.1 git2r_0.25.2 workflowr_1.4.0 tools_3.5.1
[9] digest_0.6.19 lubridate_1.7.4 jsonlite_1.6 evaluate_0.14
[13] nlme_3.1-140 gtable_0.3.0 lattice_0.20-38 pkgconfig_2.0.2
[17] rlang_0.3.4 cli_1.1.0 rstudioapi_0.10 yaml_2.2.0
[21] haven_2.1.0 xfun_0.7 withr_2.1.2 xml2_1.2.0
[25] httr_1.4.0 knitr_1.23 hms_0.4.2 generics_0.0.2
[29] fs_1.3.1 rprojroot_1.3-2 grid_3.5.1 tidyselect_0.2.5
[33] glue_1.3.1 R6_2.4.0 readxl_1.3.1 rmarkdown_1.13
[37] modelr_0.1.4 magrittr_1.5 backports_1.1.4 scales_1.0.0
[41] htmltools_0.3.6 rvest_0.3.4 assertthat_0.2.1 colorspace_1.4-1
[45] labeling_0.3 stringi_1.4.3 lazyeval_0.2.2 munsell_0.5.0
[49] broom_0.5.2 crayon_1.3.4