20190521_eQTL_enrichmentCrossSpecies
Ben Fair
5/21/2019
Last updated: 2019-06-11
Checks: 4 2
Knit directory: Comparative_eQTL/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.3.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190319) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
- GO-2,
- GO-3
- GO-analysis
To ensure reproducibility of the results, delete the cache directory 20190521_eQTL_CrossSpeciesEnrichment_cache and re-run the analysis. To have workflowr automatically delete the cache directory prior to building the file, set delete_cache = TRUE when running wflow_build() or wflow_publish().
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.DS_Store
Ignored: analysis/20190521_eQTL_CrossSpeciesEnrichment_cache/
Ignored: analysis/figure/
Ignored: analysis_temp/.DS_Store
Ignored: code/.DS_Store
Ignored: code/snakemake_workflow/.DS_Store
Ignored: data/.DS_Store
Ignored: data/PastAnalysesDataToKeep/.DS_Store
Ignored: docs/.DS_Store
Ignored: docs/assets/.DS_Store
Untracked files:
Untracked: analysis/20190502_Check_GemmaKinshipMatrix.Rmd
Untracked: analysis/20190502_Check_eQTLs.Rmd
Untracked: analysis/20190521_eQTL_CrossSpeciesEnrichment.Rmd
Untracked: analysis/20190606_eGene_Conservation.Rmd
Untracked: analysis_temp/20190412_Check_eQTLs.Rmd
Untracked: analysis_temp/PlayingWithGOAnalysis.R
Untracked: data/PastAnalysesDataToKeep/20190428_log10TPM.txt.gz
Untracked: docs/figure/20190502_Check_GemmaKinshipMatrix.Rmd/
Untracked: docs/figure/20190521_eQTL_CrossSpeciesEnrichment.Rmd/
Untracked: docs/figure/20190606_eGene_Conservation.Rmd/
Unstaged changes:
Deleted: analysis/20190412_Check_eQTLs.Rmd
Modified: analysis/20190426_CheckRNASeqPCs_2.Rmd
Modified: analysis/20190428_Check_eQTLs.Rmd
Modified: analysis/index.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with wflow_publish() to start tracking its development.
library(plyr)
library(tidyverse)
library(knitr)
library(data.table)
library(ggpmisc)
library("clusterProfiler")
library("org.Hs.eg.db")
library(gridExtra)Here the dataset is eQTLs called from a model with the following pre-testing filters/transformations/checks:
- cis-window=250kB
- MAF>10%
- genes tested must have >6 reads in 80% of samples
- lmm with genetic relatedness matrix produced by gemma
- Gene expression is standardized and normalized
- 10PCs added as covariates
- FDR estimated by Storey’s qvalue
- Pvalues well calibrated under a permutated null
First, Read in the data…
eQTLs <- read.table(gzfile("../data/PastAnalysesDataToKeep/20190521_eQTLs_250kB_10MAF.txt.gz"), header=T)
kable(head(eQTLs))| snps | gene | beta | statistic | pvalue | FDR | qvalue |
|---|---|---|---|---|---|---|
| ID.1.126459696.ACCCTAGTAAG.A | ENSPTRG00000001061 | 3.570039 | 12.50828 | 0 | 3.5e-06 | 3.5e-06 |
| ID.1.126465687.TTGT.A | ENSPTRG00000001061 | 3.570039 | 12.50828 | 0 | 3.5e-06 | 3.5e-06 |
| ID.1.126465750.TG.CT | ENSPTRG00000001061 | 3.570039 | 12.50828 | 0 | 3.5e-06 | 3.5e-06 |
| ID.1.126465756.T.C | ENSPTRG00000001061 | 3.570039 | 12.50828 | 0 | 3.5e-06 | 3.5e-06 |
| ID.1.126465766.C.A | ENSPTRG00000001061 | 3.570039 | 12.50828 | 0 | 3.5e-06 | 3.5e-06 |
| ID.1.126465774.G.A | ENSPTRG00000001061 | 3.570039 | 12.50828 | 0 | 3.5e-06 | 3.5e-06 |
# List of chimp tested genes
ChimpTestedGenes <- rownames(read.table('../output/ExpressionMatrix.un-normalized.txt.gz', header=T, check.names=FALSE, row.names = 1))
ChimpToHumanGeneMap <- read.table("../data/Biomart_export.Hsap.Ptro.orthologs.txt.gz", header=T, sep='\t', stringsAsFactors = F)
kable(head(ChimpToHumanGeneMap))| Gene.stable.ID | Transcript.stable.ID | Chimpanzee.gene.stable.ID | Chimpanzee.gene.name | Chimpanzee.protein.or.transcript.stable.ID | Chimpanzee.homology.type | X.id..target.Chimpanzee.gene.identical.to.query.gene | X.id..query.gene.identical.to.target.Chimpanzee.gene | dN.with.Chimpanzee | dS.with.Chimpanzee | Chimpanzee.orthology.confidence..0.low..1.high. |
|---|---|---|---|---|---|---|---|---|---|---|
| ENSG00000198888 | ENST00000361390 | ENSPTRG00000042641 | MT-ND1 | ENSPTRP00000061407 | ortholog_one2one | 94.6541 | 94.6541 | 0.0267 | 0.5455 | 1 |
| ENSG00000198763 | ENST00000361453 | ENSPTRG00000042626 | MT-ND2 | ENSPTRP00000061406 | ortholog_one2one | 96.2536 | 96.2536 | 0.0185 | 0.7225 | 1 |
| ENSG00000210127 | ENST00000387392 | ENSPTRG00000042642 | MT-TA | ENSPTRT00000076396 | ortholog_one2one | 100.0000 | 100.0000 | NA | NA | NA |
| ENSG00000198804 | ENST00000361624 | ENSPTRG00000042657 | MT-CO1 | ENSPTRP00000061408 | ortholog_one2one | 98.8304 | 98.8304 | 0.0065 | 0.5486 | 1 |
| ENSG00000198712 | ENST00000361739 | ENSPTRG00000042660 | MT-CO2 | ENSPTRP00000061402 | ortholog_one2one | 97.7974 | 97.7974 | 0.0106 | 0.5943 | 1 |
| ENSG00000228253 | ENST00000361851 | ENSPTRG00000042653 | MT-ATP8 | ENSPTRP00000061400 | ortholog_one2one | 94.1176 | 94.1176 | 0.0325 | 0.3331 | 1 |
# Of this ortholog list, how many genes are one2one
table(ChimpToHumanGeneMap$Chimpanzee.homology.type)
ortholog_many2many ortholog_one2many ortholog_one2one
2278 19917 140351 OneToOneMap <- ChimpToHumanGeneMap %>%
filter(Chimpanzee.homology.type=="ortholog_one2one")
# Read gtex heart egene list
# Only consider those that were tested in both species and are one2one orthologs
GtexHeartEgenes <- read.table("../data/Heart_Left_Ventricle.v7.egenes.txt.gz", header=T, sep='\t', stringsAsFactors = F) %>%
mutate(gene_id_stable = gsub(".\\d+$","",gene_id)) %>%
filter(gene_id_stable %in% OneToOneMap$Gene.stable.ID) %>%
mutate(chimp_id = plyr::mapvalues(gene_id_stable, OneToOneMap$Gene.stable.ID, OneToOneMap$Chimpanzee.gene.stable.ID, warn_missing = F)) %>%
filter(chimp_id %in% ChimpTestedGenes)
ChimpToHuman.ID <- function(Chimp.ID){
#function to convert chimp ensembl to human ensembl gene ids
return(
plyr::mapvalues(Chimp.ID, OneToOneMap$Chimpanzee.gene.stable.ID, OneToOneMap$Gene.stable.ID, warn_missing = F)
)}Now compare with GTEx by making 2x2 contigency table (eGene/not-eGene in Chimp/human). The odds ratio from this table is symetrical.
HumanFDR <- 0.1
ChimpFDR <- 0.1
#Get chimp eQTLs
Chimp_eQTLs <- eQTLs %>%
filter(qvalue<ChimpFDR)
# Count chimp eGenes
length(unique(Chimp_eQTLs$gene))[1] 336# Count human eGenes
length(GtexHeartEgenes %>% filter(qval< HumanFDR) %>% pull(chimp_id))[1] 5410# Count number genes tested in both species (already filtered for 1to1 orthologs)
length(GtexHeartEgenes$gene_id_stable)[1] 11586The number of human eGenes is huge (about half of all tested genes) and GTEx over-powered compared to chimp. With huge power, everything is an eGene and the eGene classification becomes devoid of meaningful information. So I will play with different ways to classify human eGenes.
#Change FDR thresholds or take top N eGenes by qvalue
HumanTopN <- 600
HumanFDR <- 0.1
ChimpFDR <- 0.1
# Filter human eGenes by qval threshold
# HumanSigGenes <- GtexHeartEgenes %>% filter(qval<HumanFDR) %>% pull(chimp_id)
# Filter human eGenes by topN qval
HumanSigGenes <- GtexHeartEgenes %>% top_n(-HumanTopN, qval) %>% pull(chimp_id)
# Filter human eGeness by qval threshold then topN betas
# HumanSigGenes <- GtexHeartEgenes %>% filter(qval<HumanFDR) %>% top_n(1000, abs(slope)) %>% pull(chimp_id)
HumanNonSigGenes <- GtexHeartEgenes %>%
filter(!chimp_id %in% HumanSigGenes) %>%
pull(chimp_id)
ChimpSigGenes <- GtexHeartEgenes %>%
filter(chimp_id %in% Chimp_eQTLs$gene) %>%
pull(chimp_id)
ChimpNonSigGenes <- GtexHeartEgenes %>%
filter(! chimp_id %in% Chimp_eQTLs$gene) %>%
pull(chimp_id)
ContigencyTable <- matrix( c( length(intersect(ChimpSigGenes,HumanSigGenes)),
length(intersect(HumanSigGenes,ChimpNonSigGenes)),
length(intersect(ChimpSigGenes,HumanNonSigGenes)),
length(intersect(ChimpNonSigGenes,HumanNonSigGenes))),
nrow = 2)
rownames(ContigencyTable) <- c("Chimp eGene", "Not Chimp eGene")
colnames(ContigencyTable) <- c("Human eGene", "Not human eGene")
#what is qval threshold for human eGene classification in this contigency table
print(GtexHeartEgenes %>% top_n(-HumanTopN, qval) %>% top_n(1, qval) %>% pull(qval))[1] 5.83223e-12#Contigency table of one to one orthologs tested in both chimps and humans of whether significant in humans, or chimps, or both, or neither
ContigencyTable Human eGene Not human eGene
Chimp eGene 28 252
Not Chimp eGene 572 10734#One-sided Fisher test for greater overlap than expected by chance
fisher.test(ContigencyTable, alternative="greater")
Fisher's Exact Test for Count Data
data: ContigencyTable
p-value = 0.0006395
alternative hypothesis: true odds ratio is greater than 1
95 percent confidence interval:
1.444504 Inf
sample estimates:
odds ratio
2.084996 The above contingency table and one-sided fisher test indicated a greater-than-chance overlap between the sets of eGenes in chimp and human.
A few plots might give me a better intuition for thinking about the data…
# Plot betas vs qval for GTEx for qval<0.1
GtexHeartEgenes %>% filter(qval<0.1) %>%
ggplot(aes(x=slope, y=-log10(qval))) +
geom_point(alpha=0.05)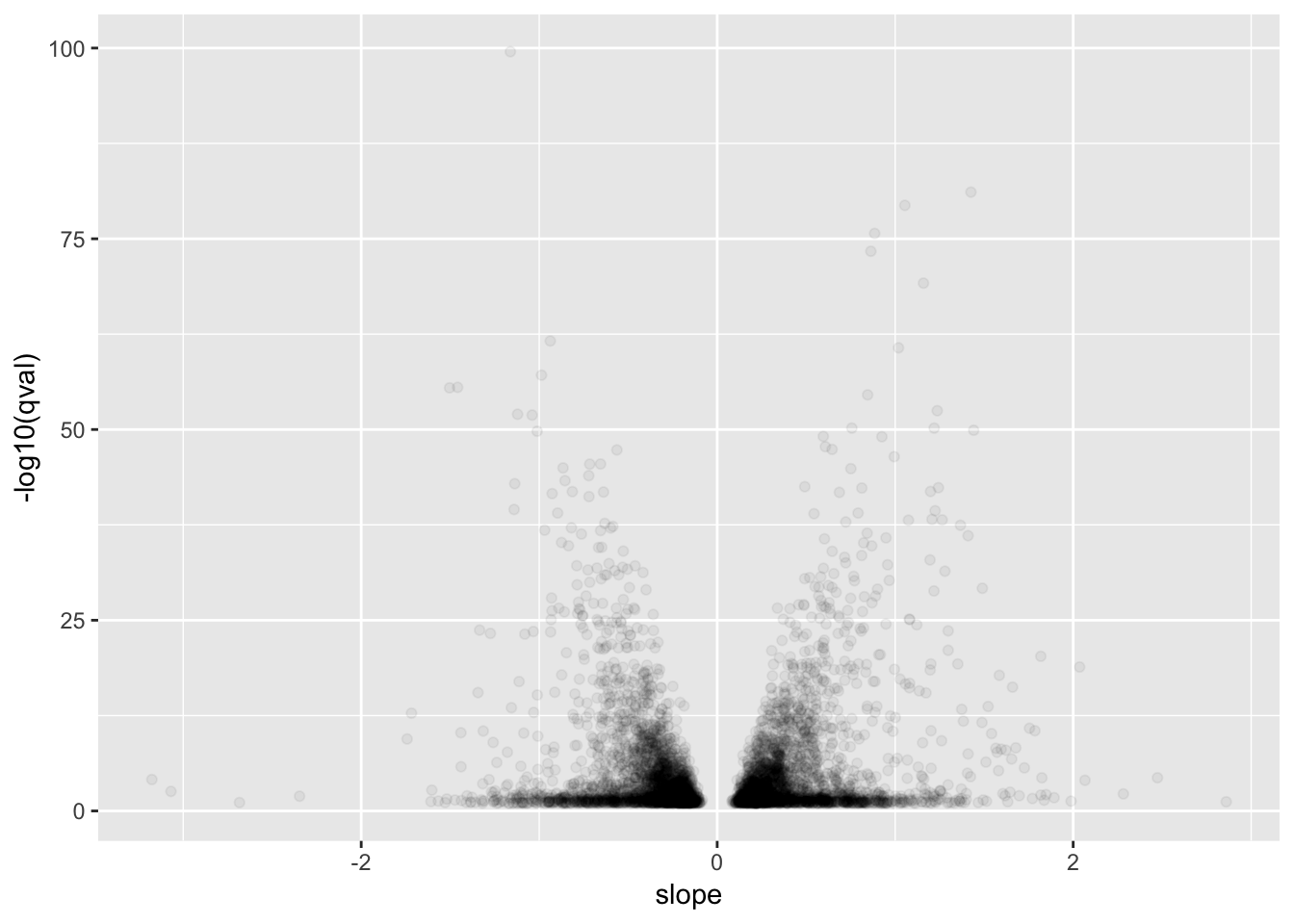
HumanAndChimpEffectSizes <- Chimp_eQTLs %>%
group_by(gene) %>%
dplyr::slice(which.min(qvalue)) %>%
ungroup() %>%
left_join((GtexHeartEgenes %>% filter(qval<0.1)), by=c("gene"="chimp_id")) %>%
dplyr::select(gene, slope, beta)Warning: Column `gene`/`chimp_id` joining factor and character vector,
coercing into character vector# For shared eGenes, are scatter plot of absolute value of betas
ggplot(HumanAndChimpEffectSizes, aes(x=log10(abs(slope)), y=log10(abs(beta)))) +
geom_point() +
geom_smooth(method = "lm") +
theme_bw() +
xlim(c(-1,1)) +
ylim(c(-1,1)) +
xlab("Human |beta|") +
ylab("Chimp |beta|")Warning: Removed 191 rows containing non-finite values (stat_smooth).Warning: Removed 191 rows containing missing values (geom_point).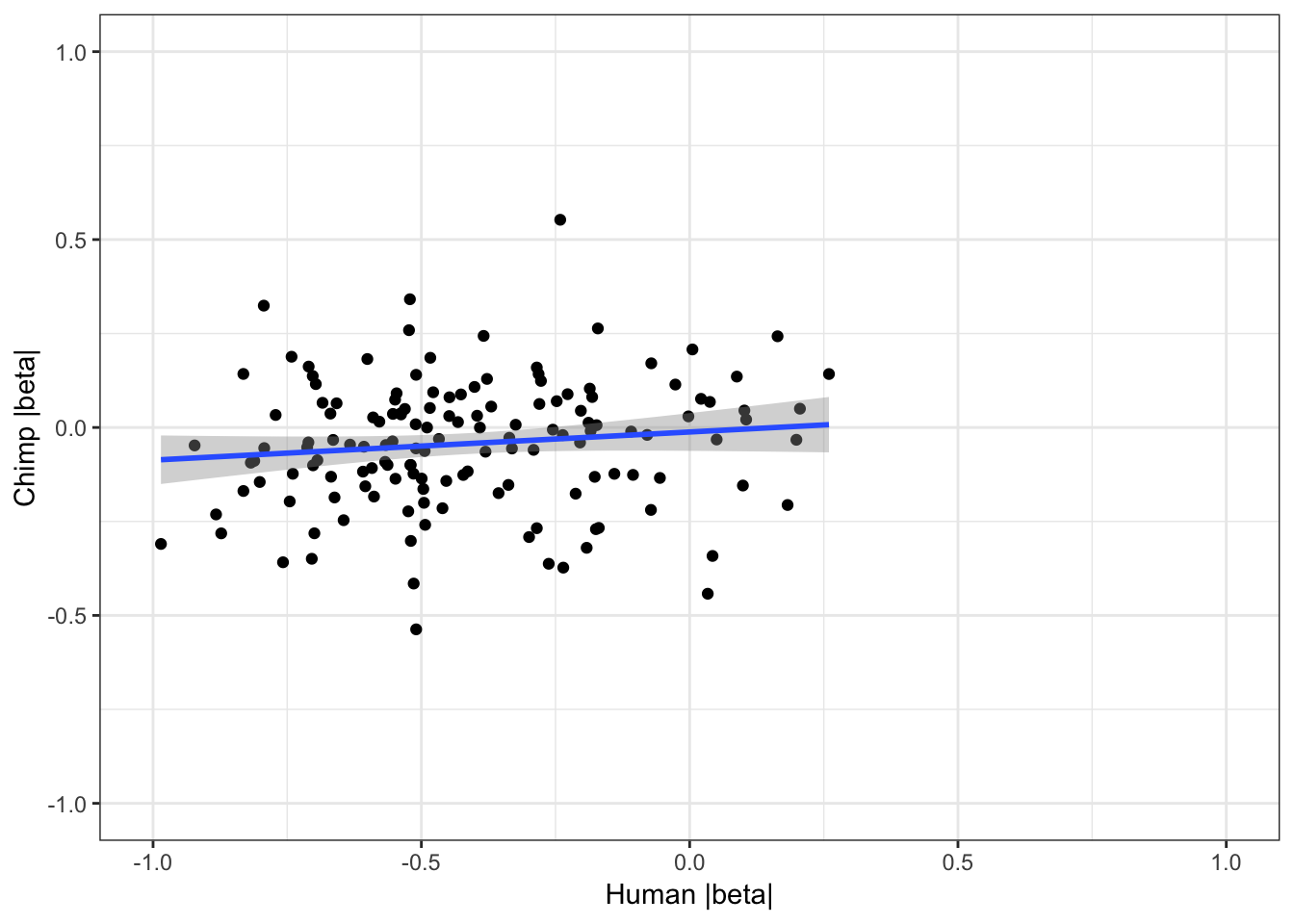
cor.test(HumanAndChimpEffectSizes$slope, HumanAndChimpEffectSizes$beta,method='spearman')
Spearman's rank correlation rho
data: HumanAndChimpEffectSizes$slope and HumanAndChimpEffectSizes$beta
S = 476920, p-value = 0.4633
alternative hypothesis: true rho is not equal to 0
sample estimates:
rho
0.06132499 Effect sizes for cross-species eGenes appear uncorrelated. This might at first appear at odds with the greater-than-chance overlap for the same set of eGenes across sepecies. However, one explanation is that the above effect sizes analysis didn’t consider non eGenes for which the null of effect-size=0 could not be rejected.
OK, so now I will perform the same Odds ratio estimate and Fisher exact test for different thresholds/quantiles of the human data ordered by significance.
ChimpFDR <- 0.1
HumanFDR <- 0.1
Chimp_eQTLs <- eQTLs %>%
filter(qvalue<ChimpFDR)
ChimpSigGenes <- GtexHeartEgenes %>%
filter(chimp_id %in% Chimp_eQTLs$gene) %>%
pull(chimp_id)
ChimpNonSigGenes <- GtexHeartEgenes %>%
filter(! chimp_id %in% Chimp_eQTLs$gene) %>%
pull(chimp_id)
LengthOut=200
TopHumanEgeneCountList <- round(seq(100,5410, length.out=LengthOut))
OddsRatiosList <- rep(0,LengthOut)
CI.lower <- rep(0,LengthOut)
CI.upper <- rep(0,LengthOut)
for (i in seq_along(TopHumanEgeneCountList)){
HumanTopN <- TopHumanEgeneCountList[i]
# Filter human eGenes by qval threshold
# HumanSigGenes <- GtexHeartEgenes %>% filter(qval<HumanFDR) %>% pull(chimp_id)
# Filter human eGenes by topN qval
HumanSigGenes <- GtexHeartEgenes %>% top_n(-HumanTopN, qval) %>% pull(chimp_id)
# Filter human eGeness by qval threshold then topN betas
# HumanSigGenes <- GtexHeartEgenes %>% filter(qval<HumanFDR) %>% top_n(1000, abs(slope)) %>% pull(chimp_id)
HumanNonSigGenes <- GtexHeartEgenes %>%
filter(!chimp_id %in% HumanSigGenes) %>%
pull(chimp_id)
ContigencyTable <- matrix( c( length(intersect(ChimpSigGenes,HumanSigGenes)),
length(intersect(HumanSigGenes,ChimpNonSigGenes)),
length(intersect(ChimpSigGenes,HumanNonSigGenes)),
length(intersect(ChimpNonSigGenes,HumanNonSigGenes))),
nrow = 2)
#Contigency table of one to one orthologs tested in both chimps and humans of whether significant in humans, or chimps, or both, or neither
ContigencyTable
A<-fisher.test(ContigencyTable, conf.int=T)
OddsRatiosList[i]<- as.numeric(A$estimate)
CI.lower[i] <- as.numeric(A$conf.int[1])
CI.upper[i] <- as.numeric(A$conf.int[2])
}
HeartOddsRatios <- data.frame(TopHumanEgeneCountList=TopHumanEgeneCountList, OddsRatio=OddsRatiosList, GTEx.Tissue="Heart_LeftVentricle", CI.lower=CI.lower, CI.upper=CI.upper)
ggplot(HeartOddsRatios, aes(x=TopHumanEgeneCountList, y=OddsRatiosList)) +
geom_point() +
geom_ribbon(aes(ymin=CI.lower, ymax=CI.upper), alpha=0.2) +
theme_bw() +
ylab("Odds Ratio") +
xlab("Considering only top X human eGenes") +
geom_hline(yintercept=1, linetype="dashed")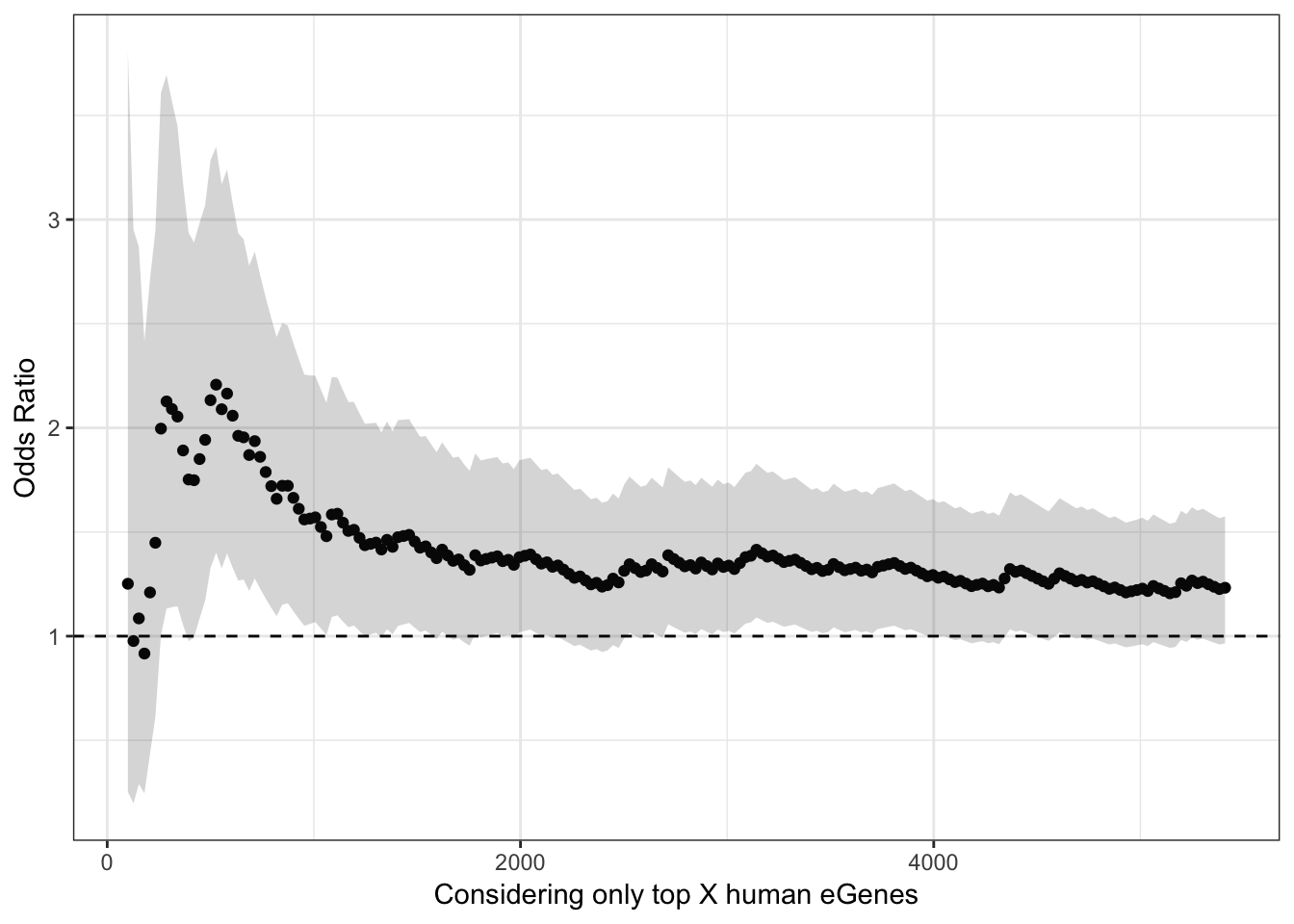
Now do the same for a couple of GTEx tissues
GTEx.eGene.Files <- c("../data/Heart_Left_Ventricle.v7.egenes.txt.gz",
"../data/Adipose_Subcutaneous.v7.egenes.txt.gz",
# "../data/Cells_EBV-transformed_lymphocytes.v7.egenes.txt.gz",
"../data/Thyroid.v7.egenes.txt.gz",
"../data/Lung.v7.egenes.txt.gz")
GTEx.tissue.labels <- c("Heart_Left_Ventricle",
"Adipose_Subcutaneous",
# "LCL",
"Thyroid",
"Lung")
OddsRatios.rbinded <- data.frame()
for (j in seq_along(GTEx.eGene.Files)){
paste0("processing")
GtexEgenes <- read.table(GTEx.eGene.Files[j], header=T, sep='\t', stringsAsFactors = F) %>%
mutate(gene_id_stable = gsub(".\\d+$","",gene_id)) %>%
filter(gene_id_stable %in% OneToOneMap$Gene.stable.ID) %>%
mutate(chimp_id = plyr::mapvalues(gene_id_stable, OneToOneMap$Gene.stable.ID, OneToOneMap$Chimpanzee.gene.stable.ID, warn_missing = F)) %>%
filter(chimp_id %in% ChimpTestedGenes)
LengthOut=200
TopHumanEgeneCountList <- round(seq(100,5410, length.out=LengthOut))
OddsRatiosList <- rep(0,LengthOut)
CI.lower <- rep(0,LengthOut)
CI.upper <- rep(0,LengthOut)
for (i in seq_along(TopHumanEgeneCountList)){
HumanTopN <- TopHumanEgeneCountList[i]
# Filter human eGenes by qval threshold
# HumanSigGenes <- GtexHeartEgenes %>% filter(qval<HumanFDR) %>% pull(chimp_id)
# Filter human eGenes by topN qval
HumanSigGenes <- GtexEgenes %>% top_n(-HumanTopN, qval) %>% pull(chimp_id)
HumanNonSigGenes <- GtexEgenes %>%
filter(!chimp_id %in% HumanSigGenes) %>%
pull(chimp_id)
ContigencyTable <- matrix( c( length(intersect(ChimpSigGenes,HumanSigGenes)),
length(intersect(HumanSigGenes,ChimpNonSigGenes)),
length(intersect(ChimpSigGenes,HumanNonSigGenes)),
length(intersect(ChimpNonSigGenes,HumanNonSigGenes))),
nrow = 2)
#Contigency table of one to one orthologs tested in both chimps and humans of whether significant in humans, or chimps, or both, or neither
A<-fisher.test(ContigencyTable, conf.int=T)
OddsRatiosList[i]<- as.numeric(A$estimate)
CI.lower[i] <- as.numeric(A$conf.int[1])
CI.upper[i] <- as.numeric(A$conf.int[2])
}
OddsRatios <- data.frame(HumanEgeneThreshold=TopHumanEgeneCountList, OR=OddsRatiosList, GTEx.Tissue=GTEx.tissue.labels[j], CI.lower=CI.lower, CI.upper=CI.upper)
OddsRatios.rbinded <- rbind(OddsRatios.rbinded, OddsRatios)
}
ggplot(OddsRatios.rbinded, aes(x=HumanEgeneThreshold, y=OR, color=GTEx.Tissue)) +
geom_line(size=2) +
geom_ribbon(aes(ymin=CI.lower, ymax=CI.upper, fill = GTEx.Tissue), alpha=0.1,linetype = 3) +
theme_bw() +
ylab("Odds Ratio") +
xlab("Considering only top X human eGenes") +
geom_hline(yintercept=1, linetype="dashed") +
ylim(c(0,4)) +
theme(legend.position="top")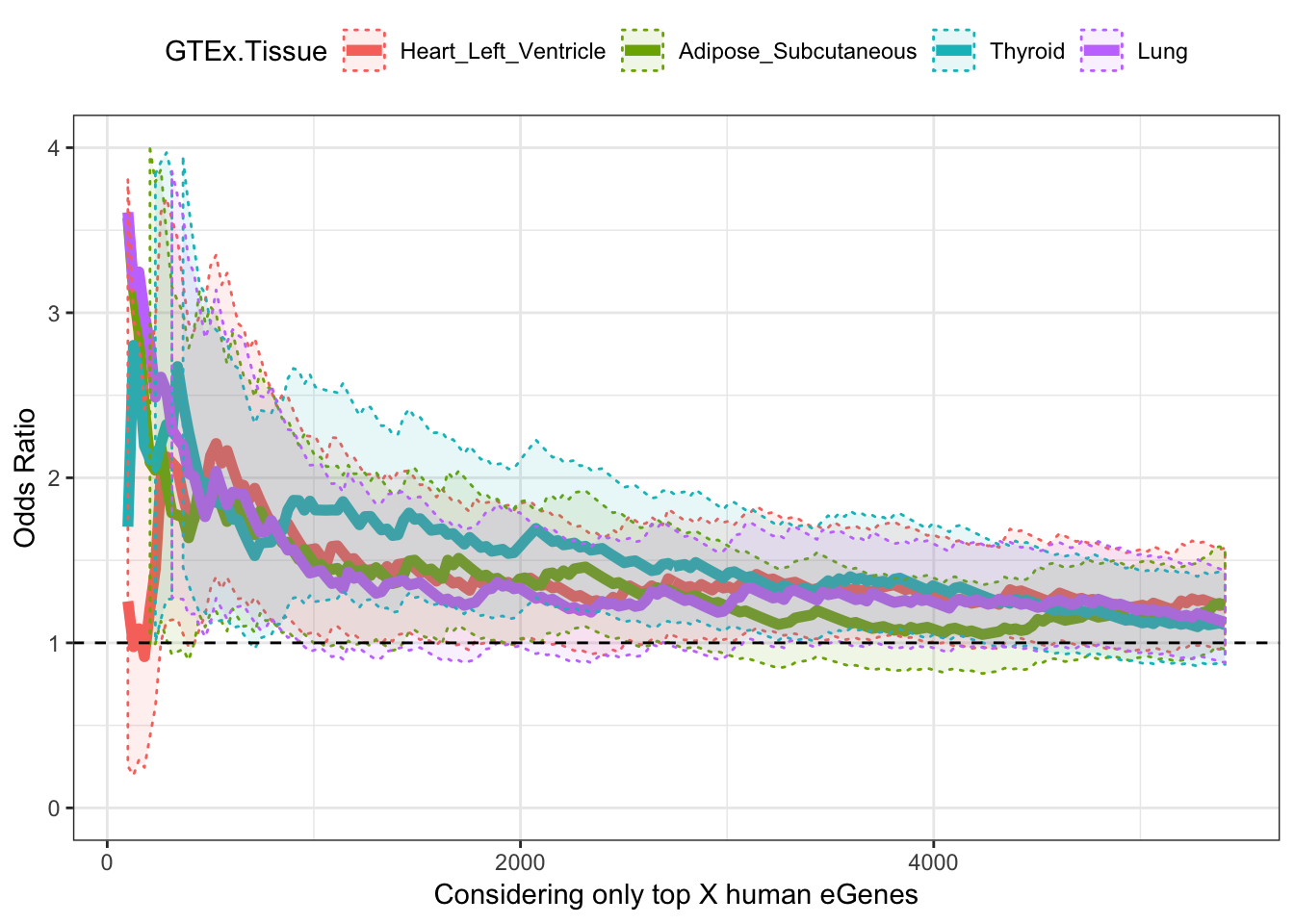
Try one alternative way to present these odds ratios as a function of eGene stringency:
# Get qval ordered ordered list of Chimp genes (that were tested in both)
Chimp_OrderedGenes <- eQTLs %>%
group_by(gene) %>%
dplyr::slice(which.min(qvalue)) %>%
filter(gene %in% GtexHeartEgenes$chimp_id) %>%
arrange(qvalue) %>%
as.data.frame()
# Get qval ordered list of human genes
Human_OrderedGenes <- GtexHeartEgenes %>%
arrange(qval)
LengthOut=60
ChimpEgeneCount <- Chimp_OrderedGenes %>% filter(qvalue<0.2) %>% pull(qvalue) %>% length()
HumanEgeneCount <- Human_OrderedGenes %>% filter(qval<0.2) %>% pull(qval) %>% length()
TopHumanEgeneCountList <- round(seq(1,HumanEgeneCount, by=50))[-1]
TopChimpEgeneCountList <- round(seq(1,ChimpEgeneCount, by=25))[-1]
OddsRatioMatrix <- matrix(nrow = length(TopHumanEgeneCountList), ncol = length(TopChimpEgeneCountList))
for (i in seq_along(TopHumanEgeneCountList)){
HumanTopN <- TopHumanEgeneCountList[i]
# Filter human eGenes by topN qval
HumanSigGenes <- Human_OrderedGenes %>% top_n(-HumanTopN, qval) %>% pull(chimp_id)
HumanNonSigGenes <- Human_OrderedGenes %>%
filter(!chimp_id %in% HumanSigGenes) %>%
pull(chimp_id)
for (j in seq_along(TopChimpEgeneCountList)){
TopChimpN <- TopChimpEgeneCountList[j]
ChimpSigGenes <- Chimp_OrderedGenes %>% top_n(-TopChimpN, qvalue) %>% pull(gene)
ChimpNonSigGenes <- GtexHeartEgenes %>%
filter(!chimp_id %in% ChimpSigGenes) %>%
pull(chimp_id)
ContigencyTable <- matrix( c( length(intersect(ChimpSigGenes,HumanSigGenes)),
length(intersect(HumanSigGenes,ChimpNonSigGenes)),
length(intersect(ChimpSigGenes,HumanNonSigGenes)),
length(intersect(ChimpNonSigGenes,HumanNonSigGenes))),
nrow = 2)
A<-fisher.test(ContigencyTable, conf.int=F)
OddsRatioMatrix[i,j] <- (as.numeric(A$estimate))
}
}
row.names(OddsRatioMatrix) <- TopHumanEgeneCountList
colnames(OddsRatioMatrix) <- TopChimpEgeneCountList
melt(OddsRatioMatrix) %>%
dplyr::rename(OddsRatio=value) %>%
ggplot(aes(x=as.numeric(Var1),y=as.numeric(Var2), fill=OddsRatio)) +
geom_raster(interpolate=F) +
xlab("Top N human eGenes") +
ylab("Top N chimp eGenes") +
scale_y_continuous(expand = c(0,0)) +
scale_x_continuous(expand = c(0,0)) +
scale_fill_distiller(palette = "Spectral") +
# coord_fixed(ratio = 1, xlim = NULL, ylim = NULL, expand = F, clip = "on") +
geom_vline(xintercept=7555, colour="grey") +
geom_text(aes(x=7555, label="FDR=0.2", y=400), angle=90, vjust = 0, color="grey", size=4)+
geom_vline(xintercept=5410, colour="grey") +
geom_text(aes(x=5410, label="FDR=0.1", y=400), angle=90, vjust = 0, color="grey", size=4)+
geom_vline(xintercept=4240, colour="grey") +
geom_text(aes(x=4240, label="FDR=0.05", y=400), angle=90, vjust = 0, color="grey", size=4)+
geom_hline(yintercept=731, colour="grey") +
geom_text(aes(x=2000, label="FDR=0.2", y=731-50), vjust = 0, color="grey", size=4)+
geom_hline(yintercept=280, colour="grey") +
geom_text(aes(x=2000, label="FDR=0.1", y=280-50), vjust = 0, color="grey", size=4)+
geom_hline(yintercept=98, colour="grey") +
geom_text(aes(x=2000, label="FDR=0.05", y=98-50), vjust = 0, color="grey", size=4)+
theme_bw() +
theme(legend.position="bottom")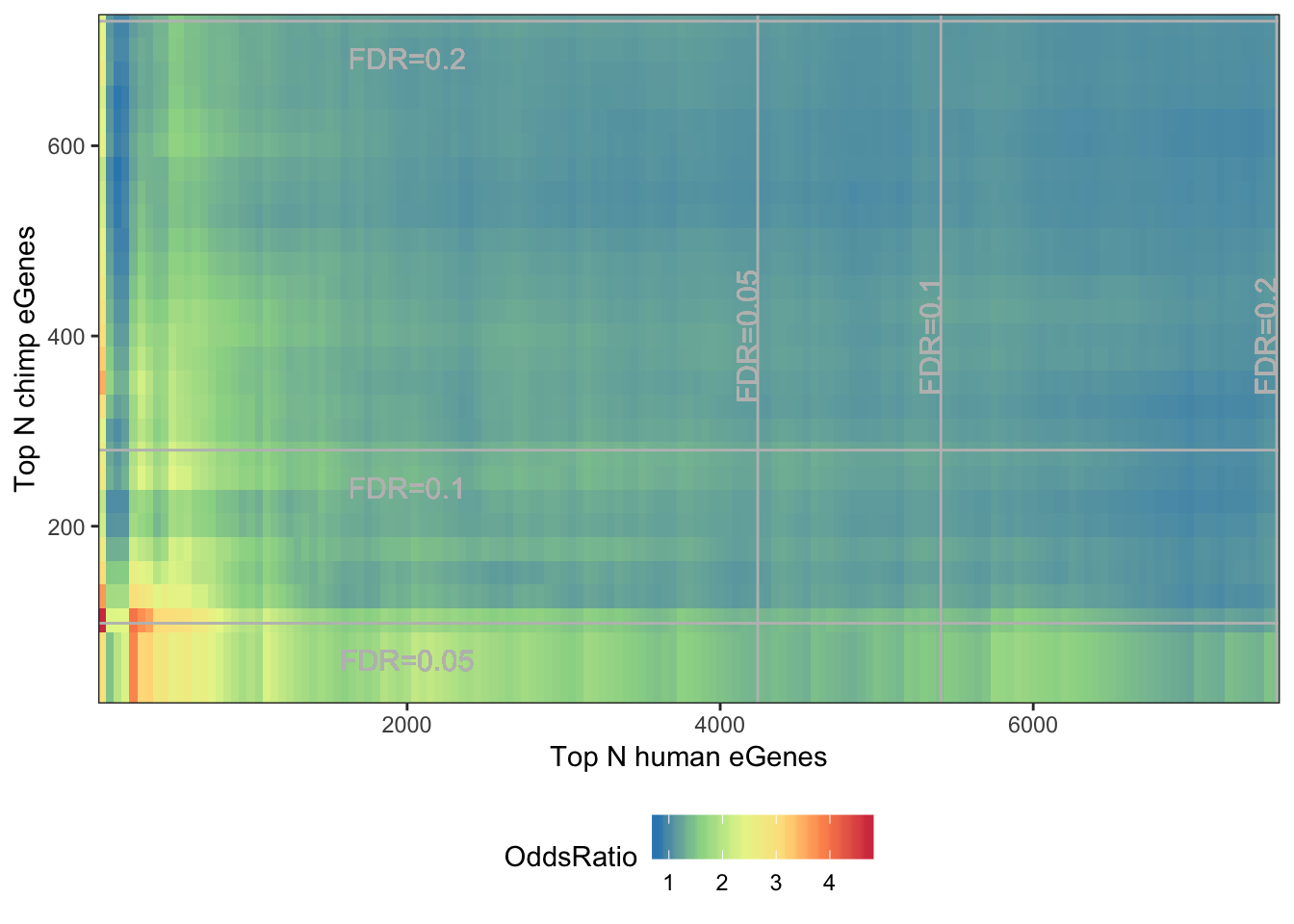
Gene ontology overlap analysis of genes that are Chimp eGenes but not human eGenes. The interpretation is that this is a somewhat prioritized list of candidate genes under stabilizing selection in the human lineage.
#Change FDR thresholds or take top N eGenes by qvalue
HumanFDR <- 0.1
ChimpFDR <- 0.1
HumanTopN <- 600
Chimp_eQTLs <- eQTLs %>%
filter(qvalue<ChimpFDR)
# Filter human eGenes by qval threshold
HumanSigGenes <- GtexHeartEgenes %>% filter(qval<HumanFDR) %>% pull(chimp_id)
# HumanSigGenes <- GtexHeartEgenes %>% top_n(-HumanTopN, qval) %>% pull(chimp_id)
HumanNonSigGenes <- GtexHeartEgenes %>%
filter(!chimp_id %in% HumanSigGenes) %>%
pull(chimp_id)
ChimpSigGenes <- GtexHeartEgenes %>%
filter(chimp_id %in% Chimp_eQTLs$gene) %>%
pull(chimp_id)
ChimpNonSigGenes <- GtexHeartEgenes %>%
filter(! chimp_id %in% Chimp_eQTLs$gene) %>%
pull(chimp_id)
ContigencyTable <- matrix( c( length(intersect(ChimpSigGenes,HumanSigGenes)),
length(intersect(HumanSigGenes,ChimpNonSigGenes)),
length(intersect(ChimpSigGenes,HumanNonSigGenes)),
length(intersect(ChimpNonSigGenes,HumanNonSigGenes))),
nrow = 2)
rownames(ContigencyTable) <- c("Chimp eGene", "Not Chimp eGene")
colnames(ContigencyTable) <- c("Human eGene", "Not human eGene")
#Contigency table of one to one orthologs tested in both chimps and humans of whether significant in humans, or chimps, or both, or neither
ContigencyTable Human eGene Not human eGene
Chimp eGene 145 135
Not Chimp eGene 5265 6041Now that we have a contingency table of eGenes between chimp and human, lets do gene set enrichment analysis. This analysis uses clusterProfiler package to look for gene set enrichment in a foreground set of genes over a background set. For each foreground/background comparison, there will be a gene ontology enrichment for the GO subcategories of - biological process (bp) - cellular component (cc) - molecular function (mf)
Perhaps first most intuitive comparison to make is:
Foreground = Chimp eGene & not human eGene
background = All genes tested in both species
foreground = ChimpToHuman.ID(intersect(ChimpSigGenes,HumanNonSigGenes))
background = ChimpToHuman.ID(GtexHeartEgenes$chimp_id)
length(foreground)[1] 135length(background)[1] 11586ego.mf <- enrichGO(gene = foreground,
universe = background,
OrgDb = org.Hs.eg.db,
keyType = 'ENSEMBL',
ont = "MF",
pAdjustMethod = "BH",
pvalueCutoff = 0.01,
qvalueCutoff = 0.05)
kable(summary(ego.mf))Warning in summary(ego.mf): summary method to convert the object to
data.frame is deprecated, please use as.data.frame instead.| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count |
|---|
ego.bp <- enrichGO(gene = foreground,
universe = background,
OrgDb = org.Hs.eg.db,
keyType = 'ENSEMBL',
ont = "BP",
pAdjustMethod = "BH",
pvalueCutoff = 0.01,
qvalueCutoff = 0.05)
kable(summary(ego.bp))Warning in summary(ego.bp): summary method to convert the object to
data.frame is deprecated, please use as.data.frame instead.| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count |
|---|
ego.cc <- enrichGO(gene = foreground,
universe = background,
OrgDb = org.Hs.eg.db,
keyType = 'ENSEMBL',
ont = "CC",
pAdjustMethod = "BH",
pvalueCutoff = 0.01,
qvalueCutoff = 0.05)
kable(summary(ego.cc))Warning in summary(ego.cc): summary method to convert the object to
data.frame is deprecated, please use as.data.frame instead.| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count |
|---|
Nothing enriched by this comparison of gene sets. Perhaps a more appropriate foreground/background set is:
foreground = chimp specific eGenes
background = all chimp eGenes
foreground = ChimpToHuman.ID(intersect(ChimpSigGenes,HumanNonSigGenes))
background = ChimpToHuman.ID(ChimpSigGenes)
length(foreground)[1] 135length(background)[1] 280ego.mf <- enrichGO(gene = foreground,
universe = background,
OrgDb = org.Hs.eg.db,
keyType = 'ENSEMBL',
ont = "MF",
pAdjustMethod = "BH",
pvalueCutoff = 0.01,
qvalueCutoff = 0.05)
kable(summary(ego.mf))Warning in summary(ego.mf): summary method to convert the object to
data.frame is deprecated, please use as.data.frame instead.| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count | |
|---|---|---|---|---|---|---|---|---|---|
| GO:0003676 | GO:0003676 | nucleic acid binding | 40/127 | 54/256 | 3.91e-05 | 0.0026993 | 0.0025943 | ENSG00000159208/ENSG00000158796/ENSG00000153187/ENSG00000184898/ENSG00000163602/ENSG00000114127/ENSG00000072274/ENSG00000114503/ENSG00000151623/ENSG00000164342/ENSG00000124279/ENSG00000113643/ENSG00000037241/ENSG00000106261/ENSG00000165061/ENSG00000185122/ENSG00000196150/ENSG00000147854/ENSG00000107371/ENSG00000125484/ENSG00000150593/ENSG00000211450/ENSG00000170325/ENSG00000134283/ENSG00000075856/ENSG00000139718/ENSG00000061936/ENSG00000173575/ENSG00000178952/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000125457/ENSG00000185504/ENSG00000141646/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454 | 40 |
ego.bp <- enrichGO(gene = foreground,
universe = background,
OrgDb = org.Hs.eg.db,
keyType = 'ENSEMBL',
ont = "BP",
pAdjustMethod = "BH",
pvalueCutoff = 0.01,
qvalueCutoff = 0.05)
kable(summary(ego.bp))Warning in summary(ego.bp): summary method to convert the object to
data.frame is deprecated, please use as.data.frame instead.| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count | |
|---|---|---|---|---|---|---|---|---|---|
| GO:0051252 | GO:0051252 | regulation of RNA metabolic process | 39/122 | 50/255 | 0.0000014 | 0.0005097 | 0.0003559 | ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000123609/ENSG00000163602/ENSG00000114127/ENSG00000114503/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000107371/ENSG00000171862/ENSG00000150593/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000075856/ENSG00000061936/ENSG00000173575/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000141646/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454 | 39 |
| GO:0019219 | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 41/122 | 54/255 | 0.0000025 | 0.0005097 | 0.0003559 | ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000123609/ENSG00000163602/ENSG00000114127/ENSG00000072274/ENSG00000114503/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000107371/ENSG00000170312/ENSG00000171862/ENSG00000150593/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000075856/ENSG00000061936/ENSG00000173575/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000141646/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454 | 41 |
| GO:0051171 | GO:0051171 | regulation of nitrogen compound metabolic process | 57/122 | 83/255 | 0.0000030 | 0.0005097 | 0.0003559 | ENSG00000121769/ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000172985/ENSG00000163083/ENSG00000123609/ENSG00000163359/ENSG00000163376/ENSG00000163602/ENSG00000114127/ENSG00000072274/ENSG00000114503/ENSG00000163975/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000055163/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000107140/ENSG00000107371/ENSG00000170312/ENSG00000171862/ENSG00000138166/ENSG00000150593/ENSG00000154127/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000075856/ENSG00000061936/ENSG00000133116/ENSG00000173575/ENSG00000150281/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000109062/ENSG00000125457/ENSG00000141646/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454/ENSG00000100985/ENSG00000099958 | 57 |
| GO:0010556 | GO:0010556 | regulation of macromolecule biosynthetic process | 41/122 | 55/255 | 0.0000060 | 0.0006226 | 0.0004347 | ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000163083/ENSG00000123609/ENSG00000163376/ENSG00000163602/ENSG00000114127/ENSG00000114503/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000164219/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000170312/ENSG00000171862/ENSG00000150593/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000173575/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000125457/ENSG00000141646/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454 | 41 |
| GO:0009889 | GO:0009889 | regulation of biosynthetic process | 42/122 | 57/255 | 0.0000074 | 0.0006226 | 0.0004347 | ENSG00000121769/ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000163083/ENSG00000123609/ENSG00000163376/ENSG00000163602/ENSG00000114127/ENSG00000114503/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000164219/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000170312/ENSG00000171862/ENSG00000150593/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000173575/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000125457/ENSG00000141646/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454 | 42 |
| GO:0010468 | GO:0010468 | regulation of gene expression | 42/122 | 57/255 | 0.0000074 | 0.0006226 | 0.0004347 | ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000163376/ENSG00000163602/ENSG00000114127/ENSG00000114503/ENSG00000163975/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000107371/ENSG00000170312/ENSG00000171862/ENSG00000150593/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000075856/ENSG00000061936/ENSG00000173575/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000125457/ENSG00000141646/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454 | 42 |
| GO:0010628 | GO:0010628 | positive regulation of gene expression | 20/122 | 22/255 | 0.0000130 | 0.0007531 | 0.0005259 | ENSG00000153187/ENSG00000163602/ENSG00000114503/ENSG00000163975/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000164342/ENSG00000124279/ENSG00000078401/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000170312/ENSG00000171862/ENSG00000102974/ENSG00000102984/ENSG00000125457/ENSG00000141646/ENSG00000090971 | 20 |
| GO:0031326 | GO:0031326 | regulation of cellular biosynthetic process | 41/122 | 56/255 | 0.0000135 | 0.0007531 | 0.0005259 | ENSG00000121769/ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000163083/ENSG00000123609/ENSG00000163376/ENSG00000163602/ENSG00000114127/ENSG00000114503/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000170312/ENSG00000171862/ENSG00000150593/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000173575/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000125457/ENSG00000141646/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454 | 41 |
| GO:2000112 | GO:2000112 | regulation of cellular macromolecule biosynthetic process | 38/122 | 51/255 | 0.0000161 | 0.0007531 | 0.0005259 | ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000163376/ENSG00000163602/ENSG00000114127/ENSG00000114503/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000170312/ENSG00000171862/ENSG00000150593/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000173575/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000125457/ENSG00000141646/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454 | 38 |
| GO:0006351 | GO:0006351 | transcription, DNA-templated | 35/122 | 46/255 | 0.0000179 | 0.0007531 | 0.0005259 | ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000123609/ENSG00000163602/ENSG00000114503/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000152409/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000125484/ENSG00000171862/ENSG00000150593/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000173575/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000141646/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454 | 35 |
| GO:0032774 | GO:0032774 | RNA biosynthetic process | 35/122 | 46/255 | 0.0000179 | 0.0007531 | 0.0005259 | ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000123609/ENSG00000163602/ENSG00000114503/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000152409/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000125484/ENSG00000171862/ENSG00000150593/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000173575/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000141646/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454 | 35 |
| GO:0097659 | GO:0097659 | nucleic acid-templated transcription | 35/122 | 46/255 | 0.0000179 | 0.0007531 | 0.0005259 | ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000123609/ENSG00000163602/ENSG00000114503/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000152409/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000125484/ENSG00000171862/ENSG00000150593/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000173575/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000141646/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454 | 35 |
| GO:0080090 | GO:0080090 | regulation of primary metabolic process | 58/122 | 88/255 | 0.0000224 | 0.0008703 | 0.0006077 | ENSG00000121769/ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000172985/ENSG00000163083/ENSG00000123609/ENSG00000163359/ENSG00000163376/ENSG00000163602/ENSG00000114127/ENSG00000072274/ENSG00000114503/ENSG00000163975/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000055163/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000155158/ENSG00000107140/ENSG00000107371/ENSG00000170312/ENSG00000171862/ENSG00000138166/ENSG00000150593/ENSG00000154127/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000075856/ENSG00000061936/ENSG00000133116/ENSG00000173575/ENSG00000150281/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000109062/ENSG00000125457/ENSG00000141646/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454/ENSG00000100985/ENSG00000099958 | 58 |
| GO:0016070 | GO:0016070 | RNA metabolic process | 41/122 | 57/255 | 0.0000292 | 0.0010017 | 0.0006995 | ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000123609/ENSG00000163602/ENSG00000114127/ENSG00000114503/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000113643/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000107371/ENSG00000125484/ENSG00000171862/ENSG00000150593/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000075856/ENSG00000061936/ENSG00000173575/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000141646/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454 | 41 |
| GO:0090304 | GO:0090304 | nucleic acid metabolic process | 46/122 | 66/255 | 0.0000298 | 0.0010017 | 0.0006995 | ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000123609/ENSG00000163602/ENSG00000114127/ENSG00000072274/ENSG00000114503/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000113643/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000147854/ENSG00000107371/ENSG00000125484/ENSG00000170312/ENSG00000171862/ENSG00000150593/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000075856/ENSG00000061936/ENSG00000128731/ENSG00000173575/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000185504/ENSG00000141646/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454 | 46 |
| GO:0044260 | GO:0044260 | cellular macromolecule metabolic process | 77/122 | 127/255 | 0.0000366 | 0.0010941 | 0.0007640 | ENSG00000162434/ENSG00000159208/ENSG00000116183/ENSG00000159388/ENSG00000153187/ENSG00000138081/ENSG00000172985/ENSG00000163083/ENSG00000123609/ENSG00000168542/ENSG00000163359/ENSG00000163376/ENSG00000163602/ENSG00000163833/ENSG00000114127/ENSG00000163923/ENSG00000072274/ENSG00000114503/ENSG00000163975/ENSG00000157404/ENSG00000118785/ENSG00000138641/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000164219/ENSG00000055163/ENSG00000113643/ENSG00000037241/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000147854/ENSG00000107140/ENSG00000107371/ENSG00000136930/ENSG00000136840/ENSG00000125484/ENSG00000170312/ENSG00000171862/ENSG00000138166/ENSG00000150593/ENSG00000154127/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000075856/ENSG00000139718/ENSG00000180776/ENSG00000133116/ENSG00000100626/ENSG00000128731/ENSG00000173575/ENSG00000178952/ENSG00000150281/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000109062/ENSG00000125457/ENSG00000185504/ENSG00000141646/ENSG00000166479/ENSG00000167461/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454/ENSG00000100985/ENSG00000099958 | 77 |
| GO:0060255 | GO:0060255 | regulation of macromolecule metabolic process | 57/122 | 87/255 | 0.0000387 | 0.0010941 | 0.0007640 | ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000172985/ENSG00000163083/ENSG00000123609/ENSG00000163359/ENSG00000163376/ENSG00000163602/ENSG00000114127/ENSG00000072274/ENSG00000114503/ENSG00000163975/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000164219/ENSG00000055163/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000107140/ENSG00000107371/ENSG00000170312/ENSG00000171862/ENSG00000138166/ENSG00000150593/ENSG00000154127/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000075856/ENSG00000061936/ENSG00000133116/ENSG00000173575/ENSG00000150281/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000109062/ENSG00000125457/ENSG00000141646/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454/ENSG00000100985/ENSG00000099958 | 57 |
| GO:0044238 | GO:0044238 | primary metabolic process | 89/122 | 153/255 | 0.0000402 | 0.0010941 | 0.0007640 | ENSG00000121769/ENSG00000162434/ENSG00000117600/ENSG00000159208/ENSG00000116183/ENSG00000159388/ENSG00000153187/ENSG00000143797/ENSG00000138081/ENSG00000172985/ENSG00000163083/ENSG00000123609/ENSG00000168542/ENSG00000163359/ENSG00000163376/ENSG00000163602/ENSG00000163833/ENSG00000114127/ENSG00000163923/ENSG00000072274/ENSG00000114503/ENSG00000163975/ENSG00000157404/ENSG00000118785/ENSG00000138641/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000164219/ENSG00000055163/ENSG00000113643/ENSG00000037241/ENSG00000078401/ENSG00000127995/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000147854/ENSG00000155158/ENSG00000107140/ENSG00000107371/ENSG00000136930/ENSG00000136840/ENSG00000125484/ENSG00000160339/ENSG00000170312/ENSG00000171862/ENSG00000138166/ENSG00000150593/ENSG00000152402/ENSG00000154127/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000075856/ENSG00000139718/ENSG00000061936/ENSG00000180776/ENSG00000133116/ENSG00000100626/ENSG00000128731/ENSG00000187720/ENSG00000140526/ENSG00000173575/ENSG00000178952/ENSG00000150281/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000074660/ENSG00000109062/ENSG00000125457/ENSG00000185504/ENSG00000141646/ENSG00000166479/ENSG00000167461/ENSG00000130313/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454/ENSG00000100985/ENSG00000099958 | 89 |
| GO:0010467 | GO:0010467 | gene expression | 49/122 | 72/255 | 0.0000412 | 0.0010941 | 0.0007640 | ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000123609/ENSG00000163376/ENSG00000163602/ENSG00000114127/ENSG00000163923/ENSG00000114503/ENSG00000163975/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000055163/ENSG00000113643/ENSG00000037241/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000107371/ENSG00000125484/ENSG00000170312/ENSG00000171862/ENSG00000150593/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000075856/ENSG00000061936/ENSG00000173575/ENSG00000178952/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000125457/ENSG00000141646/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454 | 49 |
| GO:0009059 | GO:0009059 | macromolecule biosynthetic process | 50/122 | 74/255 | 0.0000450 | 0.0010996 | 0.0007679 | ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000163083/ENSG00000123609/ENSG00000163376/ENSG00000163602/ENSG00000114127/ENSG00000163923/ENSG00000114503/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000164219/ENSG00000113643/ENSG00000037241/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000136840/ENSG00000125484/ENSG00000170312/ENSG00000171862/ENSG00000150593/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000180776/ENSG00000100626/ENSG00000173575/ENSG00000178952/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000125457/ENSG00000141646/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454/ENSG00000099958 | 50 |
| GO:0006355 | GO:0006355 | regulation of transcription, DNA-templated | 32/122 | 42/255 | 0.0000457 | 0.0010996 | 0.0007679 | ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000163602/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000152409/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000171862/ENSG00000150593/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000173575/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000141646/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454 | 32 |
| GO:1903506 | GO:1903506 | regulation of nucleic acid-templated transcription | 33/122 | 44/255 | 0.0000595 | 0.0013053 | 0.0009115 | ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000123609/ENSG00000163602/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000152409/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000171862/ENSG00000150593/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000173575/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000141646/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454 | 33 |
| GO:2001141 | GO:2001141 | regulation of RNA biosynthetic process | 33/122 | 44/255 | 0.0000595 | 0.0013053 | 0.0009115 | ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000123609/ENSG00000163602/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000152409/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000171862/ENSG00000150593/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000173575/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000141646/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454 | 33 |
| GO:0044271 | GO:0044271 | cellular nitrogen compound biosynthetic process | 44/122 | 64/255 | 0.0000891 | 0.0018740 | 0.0013086 | ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000123609/ENSG00000163376/ENSG00000163602/ENSG00000114127/ENSG00000163923/ENSG00000114503/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000113643/ENSG00000037241/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000125484/ENSG00000171862/ENSG00000150593/ENSG00000152402/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000173575/ENSG00000178952/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000125457/ENSG00000141646/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454 | 44 |
| GO:0031323 | GO:0031323 | regulation of cellular metabolic process | 58/122 | 91/255 | 0.0001225 | 0.0023947 | 0.0016721 | ENSG00000121769/ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000172985/ENSG00000163083/ENSG00000123609/ENSG00000163359/ENSG00000163376/ENSG00000163602/ENSG00000114127/ENSG00000072274/ENSG00000114503/ENSG00000163975/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000055163/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000107140/ENSG00000107371/ENSG00000170312/ENSG00000171862/ENSG00000138166/ENSG00000150593/ENSG00000154127/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000075856/ENSG00000061936/ENSG00000133116/ENSG00000173575/ENSG00000150281/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000109062/ENSG00000125457/ENSG00000141646/ENSG00000167461/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454/ENSG00000100985/ENSG00000099958 | 58 |
| GO:0006139 | GO:0006139 | nucleobase-containing compound metabolic process | 48/122 | 72/255 | 0.0001295 | 0.0023947 | 0.0016721 | ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000123609/ENSG00000163602/ENSG00000114127/ENSG00000072274/ENSG00000114503/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000113643/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000147854/ENSG00000107371/ENSG00000125484/ENSG00000170312/ENSG00000171862/ENSG00000150593/ENSG00000152402/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000075856/ENSG00000061936/ENSG00000128731/ENSG00000173575/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000185504/ENSG00000141646/ENSG00000130313/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454 | 48 |
| GO:0034645 | GO:0034645 | cellular macromolecule biosynthetic process | 48/122 | 72/255 | 0.0001295 | 0.0023947 | 0.0016721 | ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000123609/ENSG00000163376/ENSG00000163602/ENSG00000114127/ENSG00000163923/ENSG00000114503/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000113643/ENSG00000037241/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000136840/ENSG00000125484/ENSG00000170312/ENSG00000171862/ENSG00000150593/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000180776/ENSG00000100626/ENSG00000173575/ENSG00000178952/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000125457/ENSG00000141646/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454/ENSG00000099958 | 48 |
| GO:0034654 | GO:0034654 | nucleobase-containing compound biosynthetic process | 37/122 | 52/255 | 0.0001328 | 0.0023947 | 0.0016721 | ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000123609/ENSG00000163602/ENSG00000114127/ENSG00000114503/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000152409/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000125484/ENSG00000171862/ENSG00000150593/ENSG00000152402/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000173575/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000141646/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454 | 37 |
| GO:0071704 | GO:0071704 | organic substance metabolic process | 90/122 | 158/255 | 0.0001503 | 0.0026165 | 0.0018270 | ENSG00000121769/ENSG00000162434/ENSG00000117600/ENSG00000159208/ENSG00000116183/ENSG00000159388/ENSG00000153187/ENSG00000143797/ENSG00000138081/ENSG00000172985/ENSG00000163083/ENSG00000123609/ENSG00000168542/ENSG00000163359/ENSG00000163376/ENSG00000163602/ENSG00000163833/ENSG00000114127/ENSG00000163923/ENSG00000072274/ENSG00000114503/ENSG00000163975/ENSG00000157404/ENSG00000118785/ENSG00000138641/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000164219/ENSG00000055163/ENSG00000113643/ENSG00000037241/ENSG00000078401/ENSG00000127995/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000147854/ENSG00000155158/ENSG00000107140/ENSG00000107371/ENSG00000136930/ENSG00000136840/ENSG00000125484/ENSG00000160339/ENSG00000170312/ENSG00000171862/ENSG00000138166/ENSG00000150593/ENSG00000152402/ENSG00000154127/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000111058/ENSG00000075856/ENSG00000139718/ENSG00000061936/ENSG00000180776/ENSG00000133116/ENSG00000100626/ENSG00000128731/ENSG00000187720/ENSG00000140526/ENSG00000173575/ENSG00000178952/ENSG00000150281/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000074660/ENSG00000109062/ENSG00000125457/ENSG00000185504/ENSG00000141646/ENSG00000166479/ENSG00000167461/ENSG00000130313/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454/ENSG00000100985/ENSG00000099958 | 90 |
| GO:0006807 | GO:0006807 | nitrogen compound metabolic process | 84/122 | 145/255 | 0.0001626 | 0.0027378 | 0.0019117 | ENSG00000121769/ENSG00000162434/ENSG00000159208/ENSG00000116183/ENSG00000159388/ENSG00000153187/ENSG00000143797/ENSG00000138081/ENSG00000172985/ENSG00000163083/ENSG00000123609/ENSG00000168542/ENSG00000163359/ENSG00000163376/ENSG00000163602/ENSG00000163833/ENSG00000114127/ENSG00000163923/ENSG00000072274/ENSG00000114503/ENSG00000163975/ENSG00000157404/ENSG00000118785/ENSG00000138641/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000164219/ENSG00000055163/ENSG00000113643/ENSG00000037241/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000147854/ENSG00000107140/ENSG00000107371/ENSG00000136930/ENSG00000136840/ENSG00000125484/ENSG00000160339/ENSG00000170312/ENSG00000171862/ENSG00000138166/ENSG00000150593/ENSG00000152402/ENSG00000154127/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000075856/ENSG00000139718/ENSG00000061936/ENSG00000180776/ENSG00000133116/ENSG00000100626/ENSG00000128731/ENSG00000187720/ENSG00000173575/ENSG00000178952/ENSG00000150281/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000109062/ENSG00000125457/ENSG00000185504/ENSG00000141646/ENSG00000166479/ENSG00000167461/ENSG00000130313/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454/ENSG00000100985/ENSG00000099958 | 84 |
| GO:0045935 | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 20/122 | 24/255 | 0.0002035 | 0.0032688 | 0.0022825 | ENSG00000159388/ENSG00000153187/ENSG00000163602/ENSG00000072274/ENSG00000114503/ENSG00000118785/ENSG00000168743/ENSG00000164342/ENSG00000152409/ENSG00000078401/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000107371/ENSG00000170312/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000141646/ENSG00000090971 | 20 |
| GO:0043170 | GO:0043170 | macromolecule metabolic process | 80/122 | 137/255 | 0.0002127 | 0.0032688 | 0.0022825 | ENSG00000162434/ENSG00000159208/ENSG00000116183/ENSG00000159388/ENSG00000153187/ENSG00000138081/ENSG00000172985/ENSG00000163083/ENSG00000123609/ENSG00000168542/ENSG00000163359/ENSG00000163376/ENSG00000163602/ENSG00000163833/ENSG00000114127/ENSG00000163923/ENSG00000072274/ENSG00000114503/ENSG00000163975/ENSG00000157404/ENSG00000118785/ENSG00000138641/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000164219/ENSG00000055163/ENSG00000113643/ENSG00000037241/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000147854/ENSG00000107140/ENSG00000107371/ENSG00000136930/ENSG00000136840/ENSG00000125484/ENSG00000160339/ENSG00000170312/ENSG00000171862/ENSG00000138166/ENSG00000150593/ENSG00000154127/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000075856/ENSG00000139718/ENSG00000061936/ENSG00000180776/ENSG00000133116/ENSG00000100626/ENSG00000128731/ENSG00000187720/ENSG00000173575/ENSG00000178952/ENSG00000150281/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000109062/ENSG00000125457/ENSG00000185504/ENSG00000141646/ENSG00000166479/ENSG00000167461/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454/ENSG00000100985/ENSG00000099958 | 80 |
| GO:0019222 | GO:0019222 | regulation of metabolic process | 60/122 | 96/255 | 0.0002136 | 0.0032688 | 0.0022825 | ENSG00000121769/ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000172985/ENSG00000163083/ENSG00000123609/ENSG00000163359/ENSG00000163376/ENSG00000163602/ENSG00000114127/ENSG00000072274/ENSG00000114503/ENSG00000163975/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000164219/ENSG00000055163/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000155158/ENSG00000107140/ENSG00000107371/ENSG00000170312/ENSG00000171862/ENSG00000138166/ENSG00000150593/ENSG00000154127/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000075856/ENSG00000061936/ENSG00000133116/ENSG00000173575/ENSG00000150281/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000109062/ENSG00000125457/ENSG00000141646/ENSG00000167461/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454/ENSG00000100985/ENSG00000099958 | 60 |
| GO:0051254 | GO:0051254 | positive regulation of RNA metabolic process | 16/122 | 18/255 | 0.0002261 | 0.0033268 | 0.0023231 | ENSG00000159388/ENSG00000153187/ENSG00000163602/ENSG00000114503/ENSG00000118785/ENSG00000168743/ENSG00000164342/ENSG00000152409/ENSG00000078401/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000102974/ENSG00000102984/ENSG00000141646/ENSG00000090971 | 16 |
| GO:0046483 | GO:0046483 | heterocycle metabolic process | 48/122 | 73/255 | 0.0002306 | 0.0033268 | 0.0023231 | ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000123609/ENSG00000163602/ENSG00000114127/ENSG00000072274/ENSG00000114503/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000113643/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000147854/ENSG00000107371/ENSG00000125484/ENSG00000170312/ENSG00000171862/ENSG00000150593/ENSG00000152402/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000075856/ENSG00000061936/ENSG00000128731/ENSG00000173575/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000185504/ENSG00000141646/ENSG00000130313/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454 | 48 |
| GO:0018130 | GO:0018130 | heterocycle biosynthetic process | 37/122 | 53/255 | 0.0002628 | 0.0035872 | 0.0025049 | ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000123609/ENSG00000163602/ENSG00000114127/ENSG00000114503/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000152409/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000125484/ENSG00000171862/ENSG00000150593/ENSG00000152402/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000173575/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000141646/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454 | 37 |
| GO:0019438 | GO:0019438 | aromatic compound biosynthetic process | 37/122 | 53/255 | 0.0002628 | 0.0035872 | 0.0025049 | ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000123609/ENSG00000163602/ENSG00000114127/ENSG00000114503/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000152409/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000125484/ENSG00000171862/ENSG00000150593/ENSG00000152402/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000173575/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000141646/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454 | 37 |
| GO:0034641 | GO:0034641 | cellular nitrogen compound metabolic process | 53/122 | 83/255 | 0.0002978 | 0.0039570 | 0.0027631 | ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000123609/ENSG00000163376/ENSG00000163602/ENSG00000114127/ENSG00000163923/ENSG00000072274/ENSG00000114503/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000113643/ENSG00000037241/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000147854/ENSG00000107371/ENSG00000125484/ENSG00000170312/ENSG00000171862/ENSG00000150593/ENSG00000152402/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000075856/ENSG00000061936/ENSG00000128731/ENSG00000173575/ENSG00000178952/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000125457/ENSG00000185504/ENSG00000141646/ENSG00000130313/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454 | 53 |
| GO:0010604 | GO:0010604 | positive regulation of macromolecule metabolic process | 34/122 | 48/255 | 0.0003297 | 0.0041623 | 0.0029065 | ENSG00000159388/ENSG00000153187/ENSG00000172985/ENSG00000163083/ENSG00000123609/ENSG00000163602/ENSG00000072274/ENSG00000114503/ENSG00000163975/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000055163/ENSG00000078401/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000107371/ENSG00000170312/ENSG00000171862/ENSG00000138166/ENSG00000075856/ENSG00000133116/ENSG00000150281/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000125457/ENSG00000141646/ENSG00000090971/ENSG00000100985 | 34 |
| GO:0051173 | GO:0051173 | positive regulation of nitrogen compound metabolic process | 34/122 | 48/255 | 0.0003297 | 0.0041623 | 0.0029065 | ENSG00000159388/ENSG00000153187/ENSG00000172985/ENSG00000163083/ENSG00000123609/ENSG00000163602/ENSG00000072274/ENSG00000114503/ENSG00000163975/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000055163/ENSG00000078401/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000107371/ENSG00000170312/ENSG00000171862/ENSG00000138166/ENSG00000075856/ENSG00000133116/ENSG00000150281/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000125457/ENSG00000141646/ENSG00000090971/ENSG00000100985 | 34 |
| GO:0006366 | GO:0006366 | transcription by RNA polymerase II | 27/122 | 36/255 | 0.0003592 | 0.0044238 | 0.0030890 | ENSG00000159388/ENSG00000153187/ENSG00000123609/ENSG00000163602/ENSG00000114503/ENSG00000157404/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000152409/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000150593/ENSG00000170325/ENSG00000094916/ENSG00000173575/ENSG00000102974/ENSG00000102984/ENSG00000141646/ENSG00000181007/ENSG00000204519/ENSG00000152454 | 27 |
| GO:0009893 | GO:0009893 | positive regulation of metabolic process | 35/122 | 50/255 | 0.0003836 | 0.0044769 | 0.0031261 | ENSG00000121769/ENSG00000159388/ENSG00000153187/ENSG00000172985/ENSG00000163083/ENSG00000123609/ENSG00000163602/ENSG00000072274/ENSG00000114503/ENSG00000163975/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000055163/ENSG00000078401/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000107371/ENSG00000170312/ENSG00000171862/ENSG00000138166/ENSG00000075856/ENSG00000133116/ENSG00000150281/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000125457/ENSG00000141646/ENSG00000090971/ENSG00000100985 | 35 |
| GO:0031325 | GO:0031325 | positive regulation of cellular metabolic process | 35/122 | 50/255 | 0.0003836 | 0.0044769 | 0.0031261 | ENSG00000121769/ENSG00000159388/ENSG00000153187/ENSG00000172985/ENSG00000163083/ENSG00000123609/ENSG00000163602/ENSG00000072274/ENSG00000114503/ENSG00000163975/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000055163/ENSG00000078401/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000107371/ENSG00000170312/ENSG00000171862/ENSG00000138166/ENSG00000075856/ENSG00000133116/ENSG00000150281/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000125457/ENSG00000141646/ENSG00000090971/ENSG00000100985 | 35 |
| GO:0045893 | GO:0045893 | positive regulation of transcription, DNA-templated | 13/122 | 14/255 | 0.0003944 | 0.0044769 | 0.0031261 | ENSG00000153187/ENSG00000163602/ENSG00000118785/ENSG00000168743/ENSG00000164342/ENSG00000078401/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000102974/ENSG00000102984/ENSG00000141646/ENSG00000090971 | 13 |
| GO:0006725 | GO:0006725 | cellular aromatic compound metabolic process | 48/122 | 74/255 | 0.0003989 | 0.0044769 | 0.0031261 | ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000123609/ENSG00000163602/ENSG00000114127/ENSG00000072274/ENSG00000114503/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000113643/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000147854/ENSG00000107371/ENSG00000125484/ENSG00000170312/ENSG00000171862/ENSG00000150593/ENSG00000152402/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000075856/ENSG00000061936/ENSG00000128731/ENSG00000173575/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000185504/ENSG00000141646/ENSG00000130313/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454 | 48 |
| GO:1901362 | GO:1901362 | organic cyclic compound biosynthetic process | 37/122 | 54/255 | 0.0004977 | 0.0054451 | 0.0038022 | ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000123609/ENSG00000163602/ENSG00000114127/ENSG00000114503/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000152409/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000125484/ENSG00000171862/ENSG00000150593/ENSG00000152402/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000173575/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000141646/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454 | 37 |
| GO:0008152 | GO:0008152 | metabolic process | 91/122 | 163/255 | 0.0005068 | 0.0054451 | 0.0038022 | ENSG00000121769/ENSG00000162434/ENSG00000117600/ENSG00000159208/ENSG00000116183/ENSG00000159388/ENSG00000153187/ENSG00000143797/ENSG00000138081/ENSG00000172985/ENSG00000163083/ENSG00000123609/ENSG00000168542/ENSG00000163359/ENSG00000163376/ENSG00000163602/ENSG00000163833/ENSG00000114127/ENSG00000163923/ENSG00000072274/ENSG00000114503/ENSG00000163975/ENSG00000157404/ENSG00000118785/ENSG00000138641/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000164219/ENSG00000055163/ENSG00000113643/ENSG00000037241/ENSG00000078401/ENSG00000127995/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000147854/ENSG00000155158/ENSG00000107140/ENSG00000107371/ENSG00000136930/ENSG00000136840/ENSG00000125484/ENSG00000160339/ENSG00000170312/ENSG00000171862/ENSG00000138166/ENSG00000150593/ENSG00000152402/ENSG00000154127/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000111058/ENSG00000075856/ENSG00000139718/ENSG00000061936/ENSG00000180776/ENSG00000133116/ENSG00000100612/ENSG00000100626/ENSG00000128731/ENSG00000187720/ENSG00000140526/ENSG00000173575/ENSG00000178952/ENSG00000150281/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000074660/ENSG00000109062/ENSG00000125457/ENSG00000185504/ENSG00000141646/ENSG00000166479/ENSG00000167461/ENSG00000130313/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454/ENSG00000100985/ENSG00000099958 | 91 |
| GO:0032268 | GO:0032268 | regulation of cellular protein metabolic process | 33/122 | 47/255 | 0.0005542 | 0.0056520 | 0.0039467 | ENSG00000159388/ENSG00000172985/ENSG00000163083/ENSG00000123609/ENSG00000163359/ENSG00000163376/ENSG00000163602/ENSG00000114127/ENSG00000114503/ENSG00000163975/ENSG00000157404/ENSG00000168743/ENSG00000164342/ENSG00000124279/ENSG00000055163/ENSG00000078401/ENSG00000161057/ENSG00000185122/ENSG00000107140/ENSG00000170312/ENSG00000171862/ENSG00000138166/ENSG00000150593/ENSG00000154127/ENSG00000075856/ENSG00000133116/ENSG00000150281/ENSG00000102974/ENSG00000109062/ENSG00000125457/ENSG00000141646/ENSG00000100985/ENSG00000099958 | 33 |
| GO:0051246 | GO:0051246 | regulation of protein metabolic process | 33/122 | 47/255 | 0.0005542 | 0.0056520 | 0.0039467 | ENSG00000159388/ENSG00000172985/ENSG00000163083/ENSG00000123609/ENSG00000163359/ENSG00000163376/ENSG00000163602/ENSG00000114127/ENSG00000114503/ENSG00000163975/ENSG00000157404/ENSG00000168743/ENSG00000164342/ENSG00000124279/ENSG00000055163/ENSG00000078401/ENSG00000161057/ENSG00000185122/ENSG00000107140/ENSG00000170312/ENSG00000171862/ENSG00000138166/ENSG00000150593/ENSG00000154127/ENSG00000075856/ENSG00000133116/ENSG00000150281/ENSG00000102974/ENSG00000109062/ENSG00000125457/ENSG00000141646/ENSG00000100985/ENSG00000099958 | 33 |
| GO:0010605 | GO:0010605 | negative regulation of macromolecule metabolic process | 29/122 | 40/255 | 0.0005596 | 0.0056520 | 0.0039467 | ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000163083/ENSG00000123609/ENSG00000163359/ENSG00000163602/ENSG00000114127/ENSG00000114503/ENSG00000151718/ENSG00000124279/ENSG00000164219/ENSG00000078401/ENSG00000185122/ENSG00000107140/ENSG00000107371/ENSG00000171862/ENSG00000138166/ENSG00000150593/ENSG00000154127/ENSG00000134283/ENSG00000094916/ENSG00000061936/ENSG00000102974/ENSG00000109062/ENSG00000141646/ENSG00000152454/ENSG00000100985/ENSG00000099958 | 29 |
| GO:0010557 | GO:0010557 | positive regulation of macromolecule biosynthetic process | 18/122 | 22/255 | 0.0007214 | 0.0070980 | 0.0049564 | ENSG00000153187/ENSG00000163602/ENSG00000118785/ENSG00000168743/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000078401/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000170312/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000125457/ENSG00000141646/ENSG00000090971 | 18 |
| GO:1901360 | GO:1901360 | organic cyclic compound metabolic process | 50/122 | 79/255 | 0.0007309 | 0.0070980 | 0.0049564 | ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000123609/ENSG00000163602/ENSG00000114127/ENSG00000072274/ENSG00000114503/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000113643/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000147854/ENSG00000155158/ENSG00000107371/ENSG00000125484/ENSG00000170312/ENSG00000171862/ENSG00000150593/ENSG00000152402/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000075856/ENSG00000061936/ENSG00000128731/ENSG00000173575/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000074660/ENSG00000185504/ENSG00000141646/ENSG00000130313/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454 | 50 |
| GO:0006357 | GO:0006357 | regulation of transcription by RNA polymerase II | 24/122 | 32/255 | 0.0008502 | 0.0079561 | 0.0055556 | ENSG00000159388/ENSG00000153187/ENSG00000163602/ENSG00000157404/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000152409/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000185122/ENSG00000196150/ENSG00000150593/ENSG00000170325/ENSG00000094916/ENSG00000173575/ENSG00000102974/ENSG00000102984/ENSG00000141646/ENSG00000181007/ENSG00000204519/ENSG00000152454 | 24 |
| GO:1901576 | GO:1901576 | organic substance biosynthetic process | 56/122 | 91/255 | 0.0008508 | 0.0079561 | 0.0055556 | ENSG00000121769/ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000143797/ENSG00000163083/ENSG00000123609/ENSG00000163376/ENSG00000163602/ENSG00000114127/ENSG00000163923/ENSG00000114503/ENSG00000157404/ENSG00000118785/ENSG00000168743/ENSG00000151623/ENSG00000151718/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000164219/ENSG00000113643/ENSG00000037241/ENSG00000078401/ENSG00000106261/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000196150/ENSG00000136840/ENSG00000125484/ENSG00000170312/ENSG00000171862/ENSG00000150593/ENSG00000152402/ENSG00000170325/ENSG00000134283/ENSG00000094916/ENSG00000111058/ENSG00000180776/ENSG00000133116/ENSG00000100626/ENSG00000140526/ENSG00000173575/ENSG00000178952/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000125457/ENSG00000141646/ENSG00000181007/ENSG00000128000/ENSG00000090971/ENSG00000204519/ENSG00000152454/ENSG00000099958 | 56 |
| GO:1902680 | GO:1902680 | positive regulation of RNA biosynthetic process | 14/122 | 16/255 | 0.0008854 | 0.0079848 | 0.0055756 | ENSG00000153187/ENSG00000163602/ENSG00000118785/ENSG00000168743/ENSG00000164342/ENSG00000152409/ENSG00000078401/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000102974/ENSG00000102984/ENSG00000141646/ENSG00000090971 | 14 |
| GO:1903508 | GO:1903508 | positive regulation of nucleic acid-templated transcription | 14/122 | 16/255 | 0.0008854 | 0.0079848 | 0.0055756 | ENSG00000153187/ENSG00000163602/ENSG00000118785/ENSG00000168743/ENSG00000164342/ENSG00000152409/ENSG00000078401/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000102974/ENSG00000102984/ENSG00000141646/ENSG00000090971 | 14 |
| GO:0043412 | GO:0043412 | macromolecule modification | 47/122 | 74/255 | 0.0010572 | 0.0090721 | 0.0063349 | ENSG00000162434/ENSG00000159388/ENSG00000138081/ENSG00000172985/ENSG00000163083/ENSG00000123609/ENSG00000168542/ENSG00000163376/ENSG00000163602/ENSG00000163833/ENSG00000163975/ENSG00000157404/ENSG00000118785/ENSG00000138641/ENSG00000168743/ENSG00000164342/ENSG00000124279/ENSG00000164219/ENSG00000078401/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000147854/ENSG00000107140/ENSG00000107371/ENSG00000136930/ENSG00000136840/ENSG00000125484/ENSG00000170312/ENSG00000171862/ENSG00000138166/ENSG00000150593/ENSG00000154127/ENSG00000075856/ENSG00000139718/ENSG00000180776/ENSG00000133116/ENSG00000100626/ENSG00000128731/ENSG00000150281/ENSG00000102974/ENSG00000109062/ENSG00000141646/ENSG00000166479/ENSG00000167461/ENSG00000100985/ENSG00000099958 | 47 |
| GO:0051172 | GO:0051172 | negative regulation of nitrogen compound metabolic process | 25/122 | 34/255 | 0.0010753 | 0.0090721 | 0.0063349 | ENSG00000159208/ENSG00000159388/ENSG00000153187/ENSG00000163083/ENSG00000123609/ENSG00000163359/ENSG00000163602/ENSG00000114127/ENSG00000151718/ENSG00000078401/ENSG00000185122/ENSG00000107140/ENSG00000171862/ENSG00000138166/ENSG00000150593/ENSG00000154127/ENSG00000134283/ENSG00000094916/ENSG00000061936/ENSG00000102974/ENSG00000109062/ENSG00000141646/ENSG00000152454/ENSG00000100985/ENSG00000099958 | 25 |
| GO:0009891 | GO:0009891 | positive regulation of biosynthetic process | 19/122 | 24/255 | 0.0010779 | 0.0090721 | 0.0063349 | ENSG00000121769/ENSG00000153187/ENSG00000163602/ENSG00000118785/ENSG00000168743/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000078401/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000170312/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000125457/ENSG00000141646/ENSG00000090971 | 19 |
| GO:0031328 | GO:0031328 | positive regulation of cellular biosynthetic process | 19/122 | 24/255 | 0.0010779 | 0.0090721 | 0.0063349 | ENSG00000121769/ENSG00000153187/ENSG00000163602/ENSG00000118785/ENSG00000168743/ENSG00000164342/ENSG00000124279/ENSG00000152409/ENSG00000078401/ENSG00000161057/ENSG00000164758/ENSG00000185122/ENSG00000170312/ENSG00000102974/ENSG00000102984/ENSG00000167513/ENSG00000125457/ENSG00000141646/ENSG00000090971 | 19 |
ego.cc <- enrichGO(gene = foreground,
universe = background,
OrgDb = org.Hs.eg.db,
keyType = 'ENSEMBL',
ont = "CC",
pAdjustMethod = "BH",
pvalueCutoff = 0.01,
qvalueCutoff = 0.05)
kable(summary(ego.cc))Warning in summary(ego.cc): summary method to convert the object to
data.frame is deprecated, please use as.data.frame instead.| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count |
|---|
dotplot(ego.mf, font.size=8)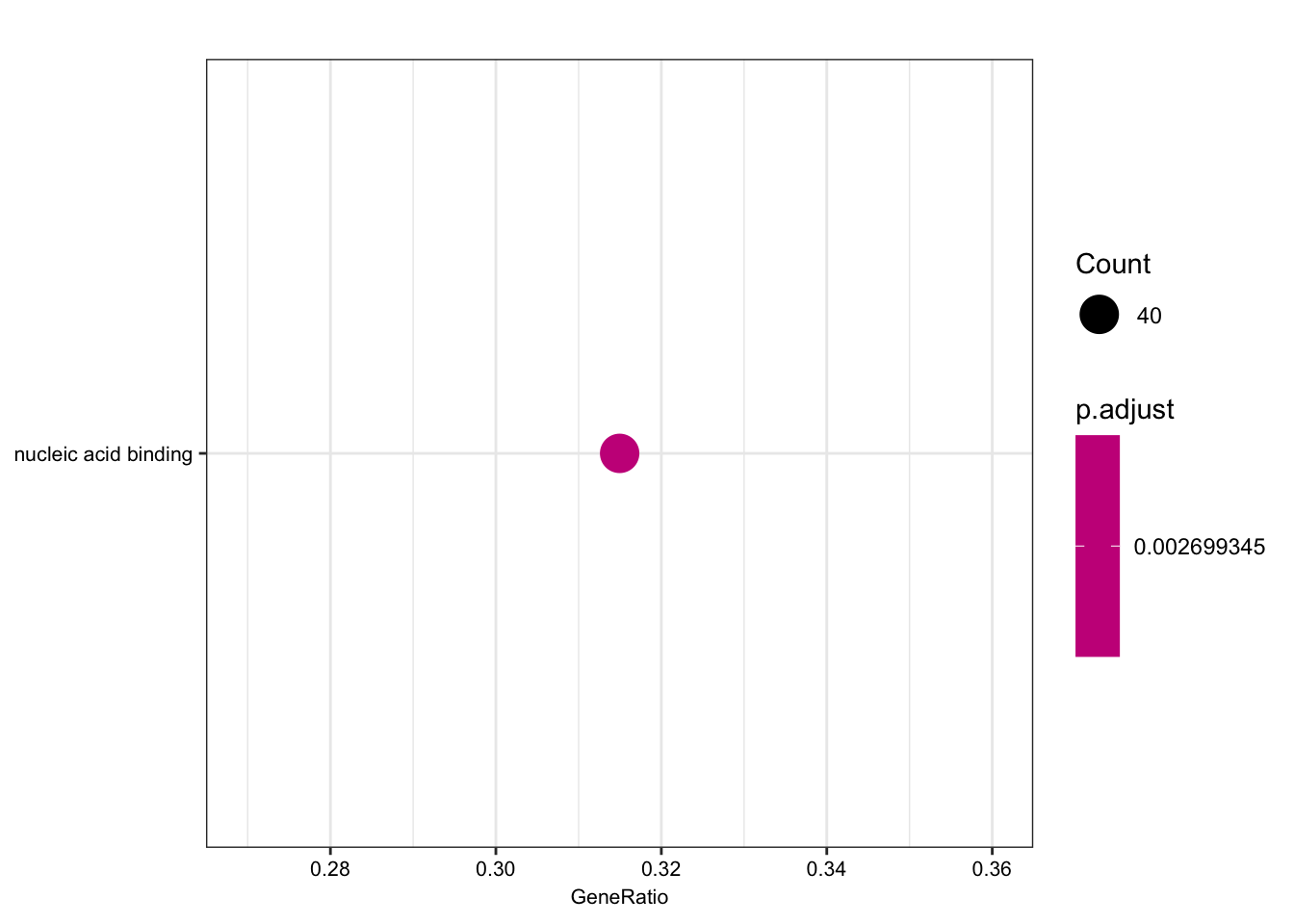
dotplot(ego.bp, font.size=8)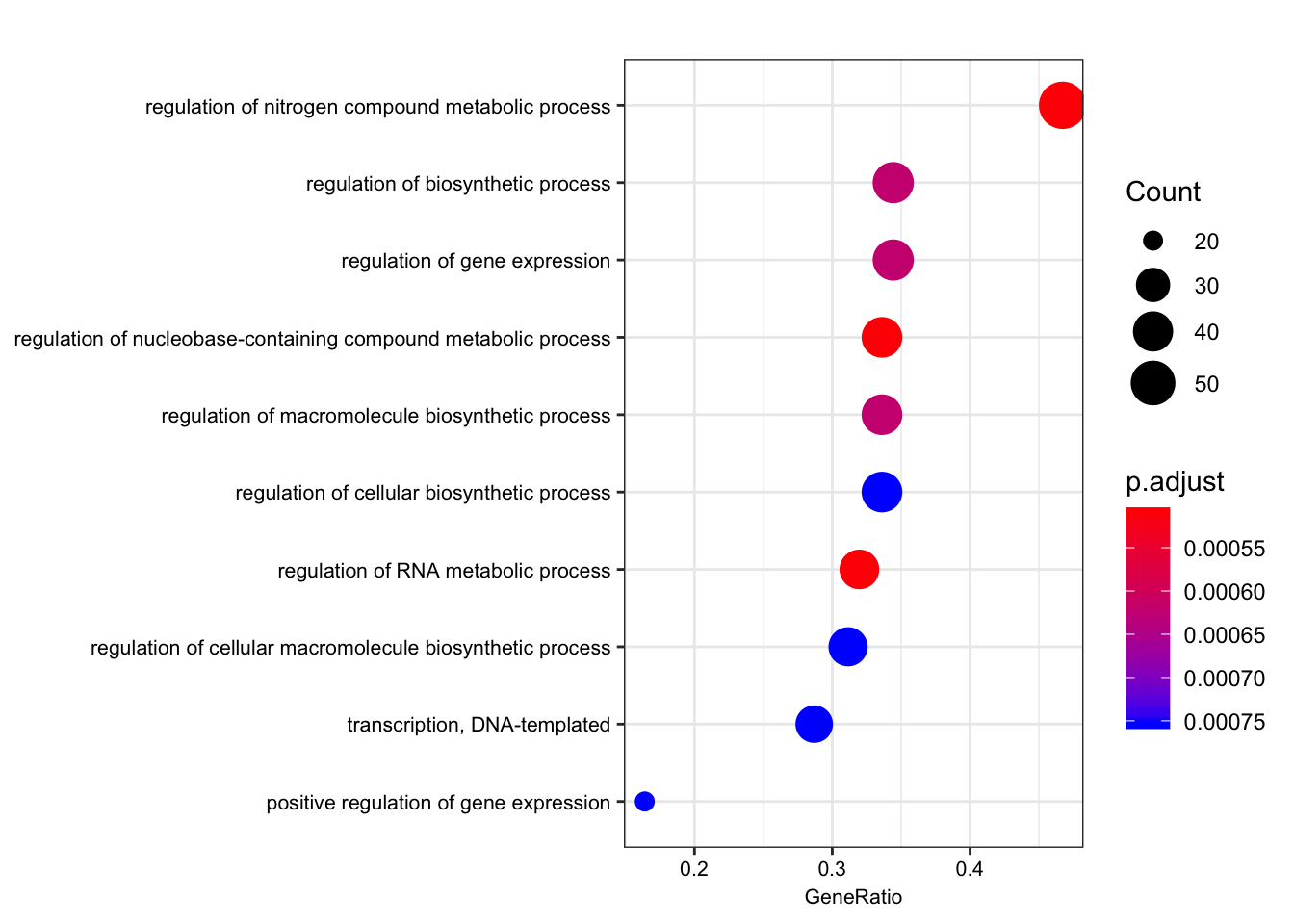
dotplot(ego.cc, font.size=8)
Compare those that are shared species eGenes, with the interpretation that they would be enriched for things not under stabilizing selection in either, perhaps things like relatively useless pseudogenes.
foreground = shared eGenes
background = all genes tested in both species
foreground = ChimpToHuman.ID(intersect(ChimpSigGenes,HumanSigGenes))
background = ChimpToHuman.ID(GtexHeartEgenes$chimp_id)
length(foreground)[1] 145length(background)[1] 11586# foreground = ChimpToHuman.ID(intersect(ChimpSigGenes,(GtexHeartEgenes %>% top_n(-600, qval) %>% pull(chimp_id))))
# Could also use the more stringent intersection of chimp eGenes and top600 human eGenes. You get mostly the sae GO terms, including MHC complex
ego.mf <- enrichGO(gene = foreground,
universe = background,
OrgDb = org.Hs.eg.db,
keyType = 'ENSEMBL',
ont = "MF",
pAdjustMethod = "BH",
pvalueCutoff = 0.01,
qvalueCutoff = 0.05)
kable(summary(ego.mf))Warning in summary(ego.mf): summary method to convert the object to
data.frame is deprecated, please use as.data.frame instead.| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count | |
|---|---|---|---|---|---|---|---|---|---|
| GO:0042605 | GO:0042605 | peptide antigen binding | 7/129 | 19/10605 | 0.00e+00 | 0.0000004 | 0.0000004 | ENSG00000206503/ENSG00000234745/ENSG00000198502/ENSG00000196126/ENSG00000196735/ENSG00000237541/ENSG00000231389 | 7 |
| GO:0003823 | GO:0003823 | antigen binding | 8/129 | 37/10605 | 0.00e+00 | 0.0000016 | 0.0000016 | ENSG00000158473/ENSG00000206503/ENSG00000234745/ENSG00000198502/ENSG00000196126/ENSG00000196735/ENSG00000237541/ENSG00000231389 | 8 |
| GO:0042277 | GO:0042277 | peptide binding | 11/129 | 172/10605 | 7.50e-06 | 0.0007191 | 0.0007172 | ENSG00000134184/ENSG00000158473/ENSG00000169760/ENSG00000163697/ENSG00000206503/ENSG00000234745/ENSG00000198502/ENSG00000196126/ENSG00000196735/ENSG00000237541/ENSG00000231389 | 11 |
| GO:0033218 | GO:0033218 | amide binding | 11/129 | 221/10605 | 7.74e-05 | 0.0055310 | 0.0055167 | ENSG00000134184/ENSG00000158473/ENSG00000169760/ENSG00000163697/ENSG00000206503/ENSG00000234745/ENSG00000198502/ENSG00000196126/ENSG00000196735/ENSG00000237541/ENSG00000231389 | 11 |
ego.bp <- enrichGO(gene = foreground,
universe = background,
OrgDb = org.Hs.eg.db,
keyType = 'ENSEMBL',
ont = "BP",
pAdjustMethod = "BH",
pvalueCutoff = 0.01,
qvalueCutoff = 0.05)
kable(summary(ego.bp))Warning in summary(ego.bp): summary method to convert the object to
data.frame is deprecated, please use as.data.frame instead.| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count | |
|---|---|---|---|---|---|---|---|---|---|
| GO:0019884 | GO:0019884 | antigen processing and presentation of exogenous antigen | 9/133 | 96/10512 | 3.2e-06 | 0.0065215 | 0.0064726 | ENSG00000158473/ENSG00000206503/ENSG00000234745/ENSG00000198502/ENSG00000196126/ENSG00000196735/ENSG00000237541/ENSG00000231389/ENSG00000161800 | 9 |
ego.cc <- enrichGO(gene = foreground,
universe = background,
OrgDb = org.Hs.eg.db,
keyType = 'ENSEMBL',
ont = "CC",
pAdjustMethod = "BH",
pvalueCutoff = 0.01,
qvalueCutoff = 0.05)
kable(summary(ego.cc))Warning in summary(ego.cc): summary method to convert the object to
data.frame is deprecated, please use as.data.frame instead.| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count | |
|---|---|---|---|---|---|---|---|---|---|
| GO:0071556 | GO:0071556 | integral component of lumenal side of endoplasmic reticulum membrane | 8/136 | 21/10999 | 0.0000000 | 0.0000000 | 0.0000000 | ENSG00000206503/ENSG00000234745/ENSG00000198502/ENSG00000196126/ENSG00000196735/ENSG00000237541/ENSG00000231389/ENSG00000005206 | 8 |
| GO:0098553 | GO:0098553 | lumenal side of endoplasmic reticulum membrane | 8/136 | 21/10999 | 0.0000000 | 0.0000000 | 0.0000000 | ENSG00000206503/ENSG00000234745/ENSG00000198502/ENSG00000196126/ENSG00000196735/ENSG00000237541/ENSG00000231389/ENSG00000005206 | 8 |
| GO:0042611 | GO:0042611 | MHC protein complex | 7/136 | 18/10999 | 0.0000000 | 0.0000001 | 0.0000001 | ENSG00000206503/ENSG00000234745/ENSG00000198502/ENSG00000196126/ENSG00000196735/ENSG00000237541/ENSG00000231389 | 7 |
| GO:0042613 | GO:0042613 | MHC class II protein complex | 5/136 | 12/10999 | 0.0000002 | 0.0000131 | 0.0000117 | ENSG00000198502/ENSG00000196126/ENSG00000196735/ENSG00000237541/ENSG00000231389 | 5 |
| GO:0030660 | GO:0030660 | Golgi-associated vesicle membrane | 9/136 | 84/10999 | 0.0000009 | 0.0000461 | 0.0000410 | ENSG00000206503/ENSG00000234745/ENSG00000198502/ENSG00000196126/ENSG00000196735/ENSG00000237541/ENSG00000231389/ENSG00000005206/ENSG00000242247 | 9 |
| GO:0012507 | GO:0012507 | ER to Golgi transport vesicle membrane | 7/136 | 47/10999 | 0.0000016 | 0.0000696 | 0.0000619 | ENSG00000206503/ENSG00000234745/ENSG00000198502/ENSG00000196126/ENSG00000196735/ENSG00000237541/ENSG00000231389 | 7 |
| GO:0030134 | GO:0030134 | COPII-coated ER to Golgi transport vesicle | 7/136 | 71/10999 | 0.0000260 | 0.0009860 | 0.0008773 | ENSG00000206503/ENSG00000234745/ENSG00000198502/ENSG00000196126/ENSG00000196735/ENSG00000237541/ENSG00000231389 | 7 |
| GO:0030658 | GO:0030658 | transport vesicle membrane | 9/136 | 129/10999 | 0.0000303 | 0.0010037 | 0.0008930 | ENSG00000135823/ENSG00000115194/ENSG00000206503/ENSG00000234745/ENSG00000198502/ENSG00000196126/ENSG00000196735/ENSG00000237541/ENSG00000231389 | 9 |
| GO:0005798 | GO:0005798 | Golgi-associated vesicle | 9/136 | 131/10999 | 0.0000342 | 0.0010080 | 0.0008969 | ENSG00000206503/ENSG00000234745/ENSG00000198502/ENSG00000196126/ENSG00000196735/ENSG00000237541/ENSG00000231389/ENSG00000005206/ENSG00000242247 | 9 |
| GO:0030666 | GO:0030666 | endocytic vesicle membrane | 8/136 | 108/10999 | 0.0000554 | 0.0014679 | 0.0013061 | ENSG00000206503/ENSG00000234745/ENSG00000198502/ENSG00000196126/ENSG00000196735/ENSG00000237541/ENSG00000231389/ENSG00000185344 | 8 |
| GO:0030669 | GO:0030669 | clathrin-coated endocytic vesicle membrane | 5/136 | 36/10999 | 0.0000744 | 0.0017305 | 0.0015398 | ENSG00000198502/ENSG00000196126/ENSG00000196735/ENSG00000237541/ENSG00000231389 | 5 |
| GO:0030133 | GO:0030133 | transport vesicle | 12/136 | 259/10999 | 0.0000858 | 0.0017305 | 0.0015398 | ENSG00000135823/ENSG00000115194/ENSG00000127418/ENSG00000206503/ENSG00000234745/ENSG00000198502/ENSG00000196126/ENSG00000196735/ENSG00000237541/ENSG00000231389/ENSG00000019505/ENSG00000100815 | 12 |
| GO:0098552 | GO:0098552 | side of membrane | 13/136 | 302/10999 | 0.0000913 | 0.0017305 | 0.0015398 | ENSG00000169760/ENSG00000112149/ENSG00000206503/ENSG00000234745/ENSG00000198502/ENSG00000196126/ENSG00000196735/ENSG00000237541/ENSG00000231389/ENSG00000120217/ENSG00000161800/ENSG00000134760/ENSG00000005206 | 13 |
| GO:0010008 | GO:0010008 | endosome membrane | 14/136 | 345/10999 | 0.0000914 | 0.0017305 | 0.0015398 | ENSG00000158473/ENSG00000115194/ENSG00000206503/ENSG00000234745/ENSG00000198502/ENSG00000196126/ENSG00000196735/ENSG00000237541/ENSG00000231389/ENSG00000086300/ENSG00000120217/ENSG00000185344/ENSG00000005206/ENSG00000054793 | 14 |
| GO:0030176 | GO:0030176 | integral component of endoplasmic reticulum membrane | 8/136 | 127/10999 | 0.0001733 | 0.0030619 | 0.0027244 | ENSG00000206503/ENSG00000234745/ENSG00000198502/ENSG00000196126/ENSG00000196735/ENSG00000237541/ENSG00000231389/ENSG00000005206 | 8 |
| GO:0044440 | GO:0044440 | endosomal part | 14/136 | 373/10999 | 0.0002073 | 0.0034330 | 0.0030546 | ENSG00000158473/ENSG00000115194/ENSG00000206503/ENSG00000234745/ENSG00000198502/ENSG00000196126/ENSG00000196735/ENSG00000237541/ENSG00000231389/ENSG00000086300/ENSG00000120217/ENSG00000185344/ENSG00000005206/ENSG00000054793 | 14 |
| GO:0031227 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane | 8/136 | 134/10999 | 0.0002506 | 0.0038677 | 0.0034414 | ENSG00000206503/ENSG00000234745/ENSG00000198502/ENSG00000196126/ENSG00000196735/ENSG00000237541/ENSG00000231389/ENSG00000005206 | 8 |
| GO:0045334 | GO:0045334 | clathrin-coated endocytic vesicle | 5/136 | 47/10999 | 0.0002714 | 0.0038677 | 0.0034414 | ENSG00000198502/ENSG00000196126/ENSG00000196735/ENSG00000237541/ENSG00000231389 | 5 |
| GO:0030662 | GO:0030662 | coated vesicle membrane | 8/136 | 136/10999 | 0.0002773 | 0.0038677 | 0.0034414 | ENSG00000206503/ENSG00000234745/ENSG00000198502/ENSG00000196126/ENSG00000196735/ENSG00000237541/ENSG00000231389/ENSG00000242247 | 8 |
| GO:0032588 | GO:0032588 | trans-Golgi network membrane | 6/136 | 74/10999 | 0.0002975 | 0.0039420 | 0.0035075 | ENSG00000135823/ENSG00000198502/ENSG00000196126/ENSG00000196735/ENSG00000237541/ENSG00000231389 | 6 |
| GO:0030139 | GO:0030139 | endocytic vesicle | 9/136 | 197/10999 | 0.0007464 | 0.0094184 | 0.0083802 | ENSG00000135823/ENSG00000206503/ENSG00000234745/ENSG00000198502/ENSG00000196126/ENSG00000196735/ENSG00000237541/ENSG00000231389/ENSG00000185344 | 9 |
dotplot(ego.mf, font.size=8)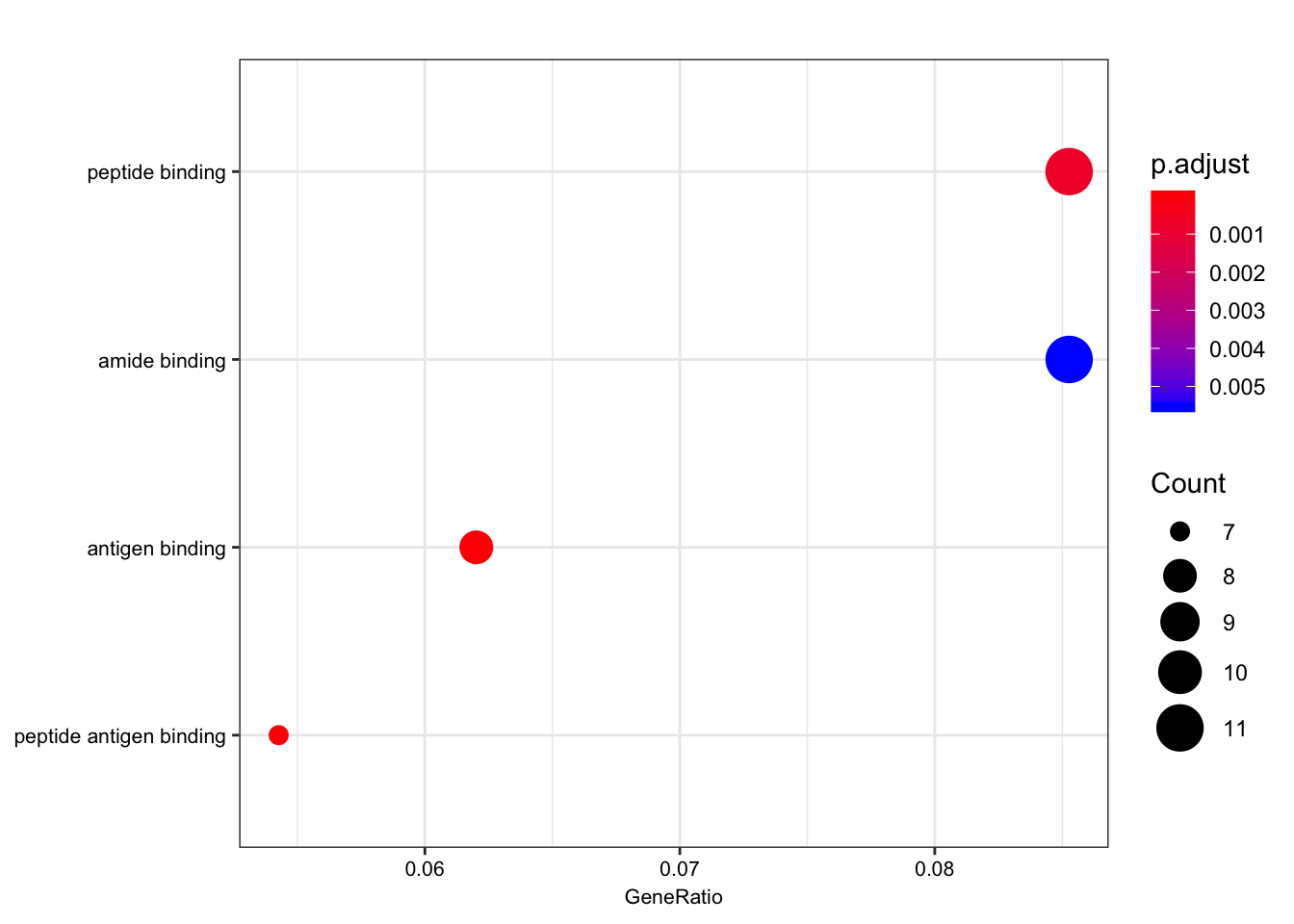
dotplot(ego.bp, font.size=8)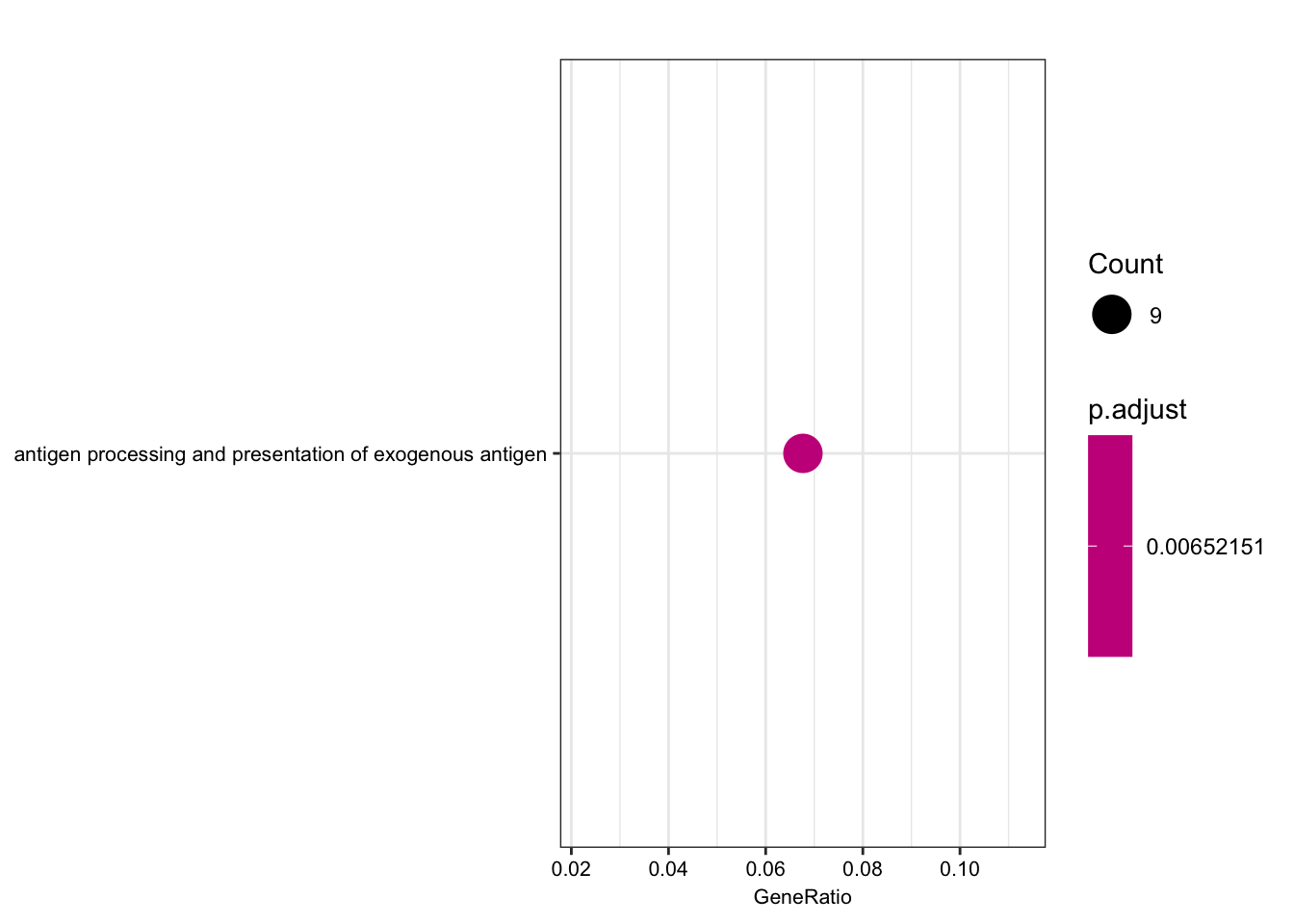
dotplot(ego.cc, font.size=8)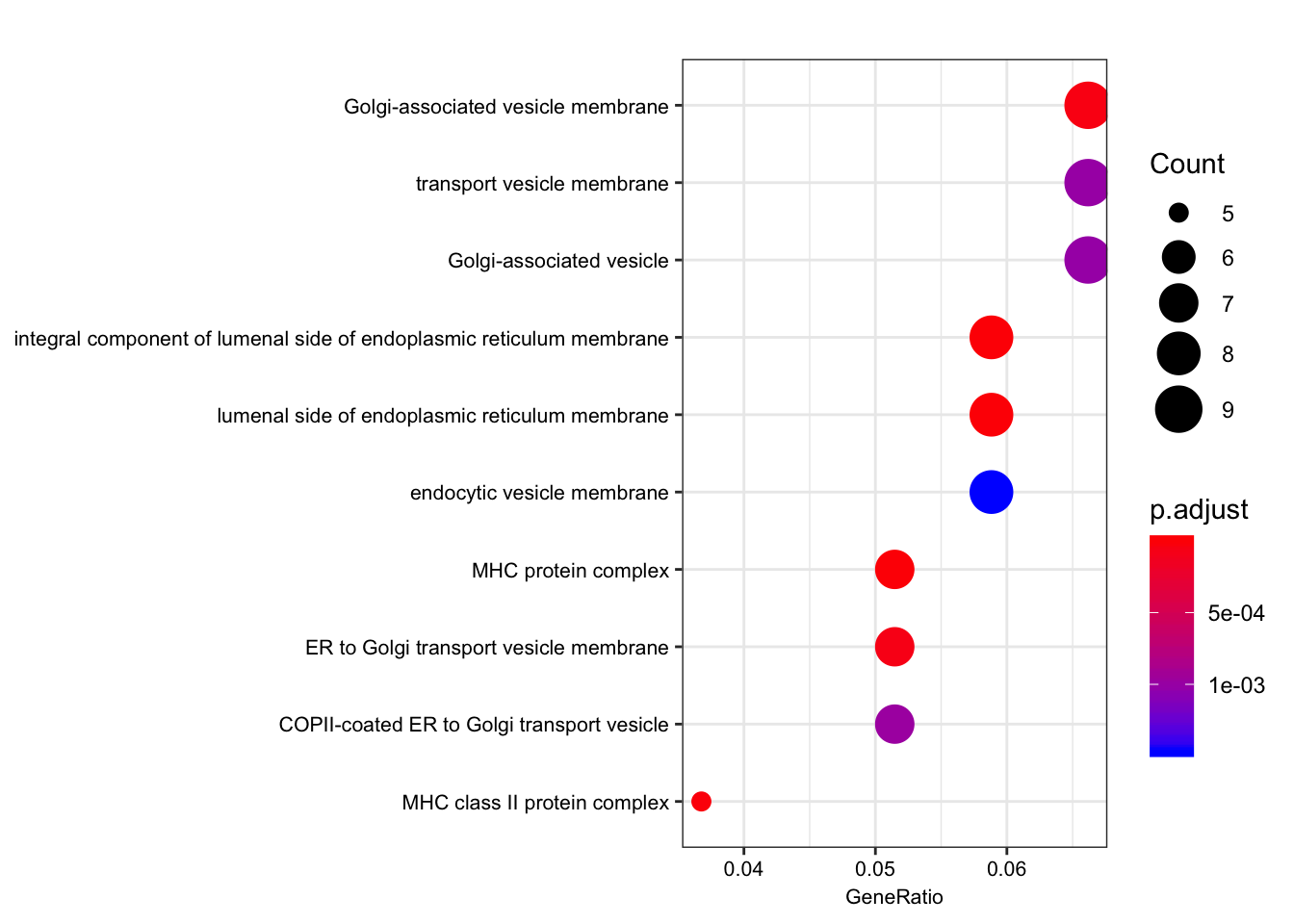
Interestingly, the MHC complex comes out of this analysis as enriched among the genes that are eGenes in both species, consistent with these genes being rapidly evolving targets of selection in between human/chimp, with high diversity in both species.
A complementart analysis to GO overlap is gene set enrichment analysis. Here I will perform GSEA on the chimp qvalue orded list of chimp/human shared eGenes and chimp specific eGenes based on FDR<0.1 in the human dataset…
Actually, maybe a more sensible thing to do is consider the human qvalue ordered list and then separate based on if chimp eGene or not. This is more similar to what the previous odds-ratio plots are.
HumanOrderedGenes <- GtexHeartEgenes %>%
arrange(qval) %>%
filter(chimp_id %in% ChimpSigGenes)
GeneList <- log10(HumanOrderedGenes$qval) * -1
names(GeneList) <- as.character(ChimpToHuman.ID(HumanOrderedGenes$chimp_id))
gsego.cc.shared <- gseGO(gene = GeneList,
OrgDb = org.Hs.eg.db,
keyType = 'ENSEMBL',
maxGSSize = 500,
# pvalueCutoff=0.8,
ont = "CC")
as.data.frame(gsego.cc.shared)
dotplot(gsego.cc.shared, font.size=8, showCategory=15)
gseaplot(gsego.cc.shared, geneSetID = "GO:0042611", pvalue_table = TRUE)# HumanOrderedGenes <- GtexHeartEgenes %>%
# arrange(qval) %>%
# filter(!chimp_id %in% ChimpSigGenes)
# GeneList <- log10(HumanOrderedGenes$qval) * -1
# names(GeneList) <- as.character(ChimpToHuman.ID(HumanOrderedGenes$chimp_id))
#Create chimp-qval-ordered gene list for genes that are human eGenes
#I expect this to return similar results as gene overlap analysis with forground as shared eGenes and background as all all genes tested. So this should return MHC complex among other things.
Chimp_OrderedGenes <- eQTLs %>%
group_by(gene) %>%
dplyr::slice(which.min(qvalue)) %>%
filter(gene %in% GtexHeartEgenes$chimp_id) %>%
arrange(qvalue) %>%
filter(gene %in% HumanSigGenes)
GeneList <- log10(Chimp_OrderedGenes$qvalue) * -1
names(GeneList) <- as.character(ChimpToHuman.ID(Chimp_OrderedGenes$gene))
gsego.cc.shared <- gseGO(gene = GeneList,
OrgDb = org.Hs.eg.db,
keyType = 'ENSEMBL',
maxGSSize = 500,
# pvalueCutoff=0.8,
ont = "CC")preparing geneSet collections...GSEA analysis...Warning in fgsea(pathways = geneSets, stats = geneList, nperm = nPerm, minSize = minGSSize, : There are ties in the preranked stats (38.71% of the list).
The order of those tied genes will be arbitrary, which may produce unexpected results.leading edge analysis...done...as.data.frame(gsego.cc.shared) ID
GO:0005768 GO:0005768
GO:0005802 GO:0005802
GO:0010008 GO:0010008
GO:0030660 GO:0030660
GO:0032588 GO:0032588
GO:0044440 GO:0044440
GO:0012507 GO:0012507
GO:0030669 GO:0030669
GO:0071556 GO:0071556
GO:0098553 GO:0098553
GO:0042611 GO:0042611
GO:0042613 GO:0042613
Description
GO:0005768 endosome
GO:0005802 trans-Golgi network
GO:0010008 endosome membrane
GO:0030660 Golgi-associated vesicle membrane
GO:0032588 trans-Golgi network membrane
GO:0044440 endosomal part
GO:0012507 ER to Golgi transport vesicle membrane
GO:0030669 clathrin-coated endocytic vesicle membrane
GO:0071556 integral component of lumenal side of endoplasmic reticulum membrane
GO:0098553 lumenal side of endoplasmic reticulum membrane
GO:0042611 MHC protein complex
GO:0042613 MHC class II protein complex
setSize enrichmentScore NES pvalue p.adjust
GO:0005768 291 0.4157062 1.311359 0.000999001 0.04211663
GO:0005802 83 0.5198493 1.551804 0.000999001 0.04211663
GO:0010008 159 0.4505316 1.388673 0.000999001 0.04211663
GO:0030660 39 0.6492226 1.826185 0.000999001 0.04211663
GO:0032588 41 0.6042965 1.704040 0.000999001 0.04211663
GO:0044440 172 0.4495002 1.390874 0.000999001 0.04211663
GO:0012507 28 0.7104864 1.920442 0.001007049 0.04211663
GO:0030669 19 0.7044431 1.818832 0.001028807 0.04211663
GO:0071556 16 0.8600123 2.164377 0.001037344 0.04211663
GO:0098553 16 0.8600123 2.164377 0.001037344 0.04211663
GO:0042611 14 0.8498340 2.089347 0.001053741 0.04211663
GO:0042613 10 0.8404787 1.952444 0.001079914 0.04211663
qvalues rank leading_edge
GO:0005768 0.04016521 976 tags=26%, list=19%, signal=22%
GO:0005802 0.04016521 972 tags=36%, list=19%, signal=30%
GO:0010008 0.04016521 976 tags=27%, list=19%, signal=23%
GO:0030660 0.04016521 436 tags=33%, list=8%, signal=31%
GO:0032588 0.04016521 972 tags=44%, list=19%, signal=36%
GO:0044440 0.04016521 976 tags=27%, list=19%, signal=23%
GO:0012507 0.04016521 436 tags=39%, list=8%, signal=36%
GO:0030669 0.04016521 567 tags=47%, list=11%, signal=42%
GO:0071556 0.04016521 436 tags=69%, list=8%, signal=63%
GO:0098553 0.04016521 436 tags=69%, list=8%, signal=63%
GO:0042611 0.04016521 436 tags=71%, list=8%, signal=66%
GO:0042613 0.04016521 436 tags=70%, list=8%, signal=64%
core_enrichment
GO:0005768 ENSG00000158473/ENSG00000196735/ENSG00000196126/ENSG00000145214/ENSG00000054793/ENSG00000010322/ENSG00000206503/ENSG00000198502/ENSG00000135823/ENSG00000120217/ENSG00000120860/ENSG00000237541/ENSG00000005206/ENSG00000234745/ENSG00000231389/ENSG00000185344/ENSG00000086300/ENSG00000115194/ENSG00000100478/ENSG00000204525/ENSG00000064687/ENSG00000131242/ENSG00000177169/ENSG00000110723/ENSG00000164038/ENSG00000197043/ENSG00000155099/ENSG00000182240/ENSG00000179344/ENSG00000055732/ENSG00000108774/ENSG00000204130/ENSG00000125633/ENSG00000129204/ENSG00000185339/ENSG00000099999/ENSG00000223865/ENSG00000117533/ENSG00000103111/ENSG00000129515/ENSG00000142089/ENSG00000167671/ENSG00000083097/ENSG00000006125/ENSG00000136933/ENSG00000171130/ENSG00000168824/ENSG00000177951/ENSG00000136051/ENSG00000140557/ENSG00000161011/ENSG00000155876/ENSG00000085365/ENSG00000196396/ENSG00000079691/ENSG00000175348/ENSG00000122490/ENSG00000175471/ENSG00000140157/ENSG00000176783/ENSG00000136813/ENSG00000102780/ENSG00000010810/ENSG00000091317/ENSG00000181804/ENSG00000049759/ENSG00000070371/ENSG00000070540/ENSG00000105270/ENSG00000136235/ENSG00000089486/ENSG00000087053/ENSG00000145416/ENSG00000197535/ENSG00000142192/ENSG00000215305
GO:0005802 ENSG00000196735/ENSG00000196126/ENSG00000054793/ENSG00000198502/ENSG00000135823/ENSG00000237541/ENSG00000231389/ENSG00000242247/ENSG00000100478/ENSG00000078328/ENSG00000182240/ENSG00000179344/ENSG00000223865/ENSG00000117533/ENSG00000083097/ENSG00000036054/ENSG00000136933/ENSG00000168824/ENSG00000135968/ENSG00000137502/ENSG00000072958/ENSG00000085365/ENSG00000108666/ENSG00000122490/ENSG00000133103/ENSG00000070371/ENSG00000070540/ENSG00000105270/ENSG00000145416/ENSG00000142192
GO:0010008 ENSG00000158473/ENSG00000196735/ENSG00000196126/ENSG00000054793/ENSG00000206503/ENSG00000198502/ENSG00000120217/ENSG00000237541/ENSG00000005206/ENSG00000234745/ENSG00000231389/ENSG00000185344/ENSG00000086300/ENSG00000115194/ENSG00000204525/ENSG00000064687/ENSG00000131242/ENSG00000164038/ENSG00000197043/ENSG00000155099/ENSG00000179344/ENSG00000055732/ENSG00000108774/ENSG00000223865/ENSG00000129515/ENSG00000142089/ENSG00000167671/ENSG00000006125/ENSG00000136933/ENSG00000171130/ENSG00000168824/ENSG00000085365/ENSG00000175348/ENSG00000176783/ENSG00000091317/ENSG00000181804/ENSG00000070540/ENSG00000105270/ENSG00000136235/ENSG00000089486/ENSG00000087053/ENSG00000145416/ENSG00000215305
GO:0030660 ENSG00000196735/ENSG00000196126/ENSG00000206503/ENSG00000198502/ENSG00000237541/ENSG00000005206/ENSG00000234745/ENSG00000231389/ENSG00000242247/ENSG00000204525/ENSG00000148396/ENSG00000179344/ENSG00000223865
GO:0032588 ENSG00000196735/ENSG00000196126/ENSG00000198502/ENSG00000135823/ENSG00000237541/ENSG00000231389/ENSG00000100478/ENSG00000179344/ENSG00000223865/ENSG00000117533/ENSG00000136933/ENSG00000168824/ENSG00000072958/ENSG00000085365/ENSG00000133103/ENSG00000105270/ENSG00000145416/ENSG00000142192
GO:0044440 ENSG00000158473/ENSG00000196735/ENSG00000196126/ENSG00000054793/ENSG00000206503/ENSG00000198502/ENSG00000120217/ENSG00000237541/ENSG00000005206/ENSG00000234745/ENSG00000231389/ENSG00000185344/ENSG00000086300/ENSG00000115194/ENSG00000100478/ENSG00000204525/ENSG00000064687/ENSG00000131242/ENSG00000164038/ENSG00000197043/ENSG00000155099/ENSG00000179344/ENSG00000055732/ENSG00000108774/ENSG00000223865/ENSG00000103111/ENSG00000129515/ENSG00000142089/ENSG00000167671/ENSG00000006125/ENSG00000136933/ENSG00000171130/ENSG00000168824/ENSG00000155876/ENSG00000085365/ENSG00000175348/ENSG00000176783/ENSG00000091317/ENSG00000181804/ENSG00000070540/ENSG00000105270/ENSG00000136235/ENSG00000089486/ENSG00000087053/ENSG00000145416/ENSG00000142192/ENSG00000215305
GO:0012507 ENSG00000196735/ENSG00000196126/ENSG00000206503/ENSG00000198502/ENSG00000237541/ENSG00000234745/ENSG00000231389/ENSG00000204525/ENSG00000148396/ENSG00000179344/ENSG00000223865
GO:0030669 ENSG00000196735/ENSG00000196126/ENSG00000198502/ENSG00000237541/ENSG00000231389/ENSG00000179344/ENSG00000223865/ENSG00000174804/ENSG00000006125
GO:0071556 ENSG00000196735/ENSG00000196126/ENSG00000206503/ENSG00000198502/ENSG00000237541/ENSG00000005206/ENSG00000234745/ENSG00000231389/ENSG00000204525/ENSG00000179344/ENSG00000223865
GO:0098553 ENSG00000196735/ENSG00000196126/ENSG00000206503/ENSG00000198502/ENSG00000237541/ENSG00000005206/ENSG00000234745/ENSG00000231389/ENSG00000204525/ENSG00000179344/ENSG00000223865
GO:0042611 ENSG00000196735/ENSG00000196126/ENSG00000206503/ENSG00000198502/ENSG00000237541/ENSG00000234745/ENSG00000231389/ENSG00000204525/ENSG00000179344/ENSG00000223865
GO:0042613 ENSG00000196735/ENSG00000196126/ENSG00000198502/ENSG00000237541/ENSG00000231389/ENSG00000179344/ENSG00000223865dotplot(gsego.cc.shared, font.size=8, showCategory=15)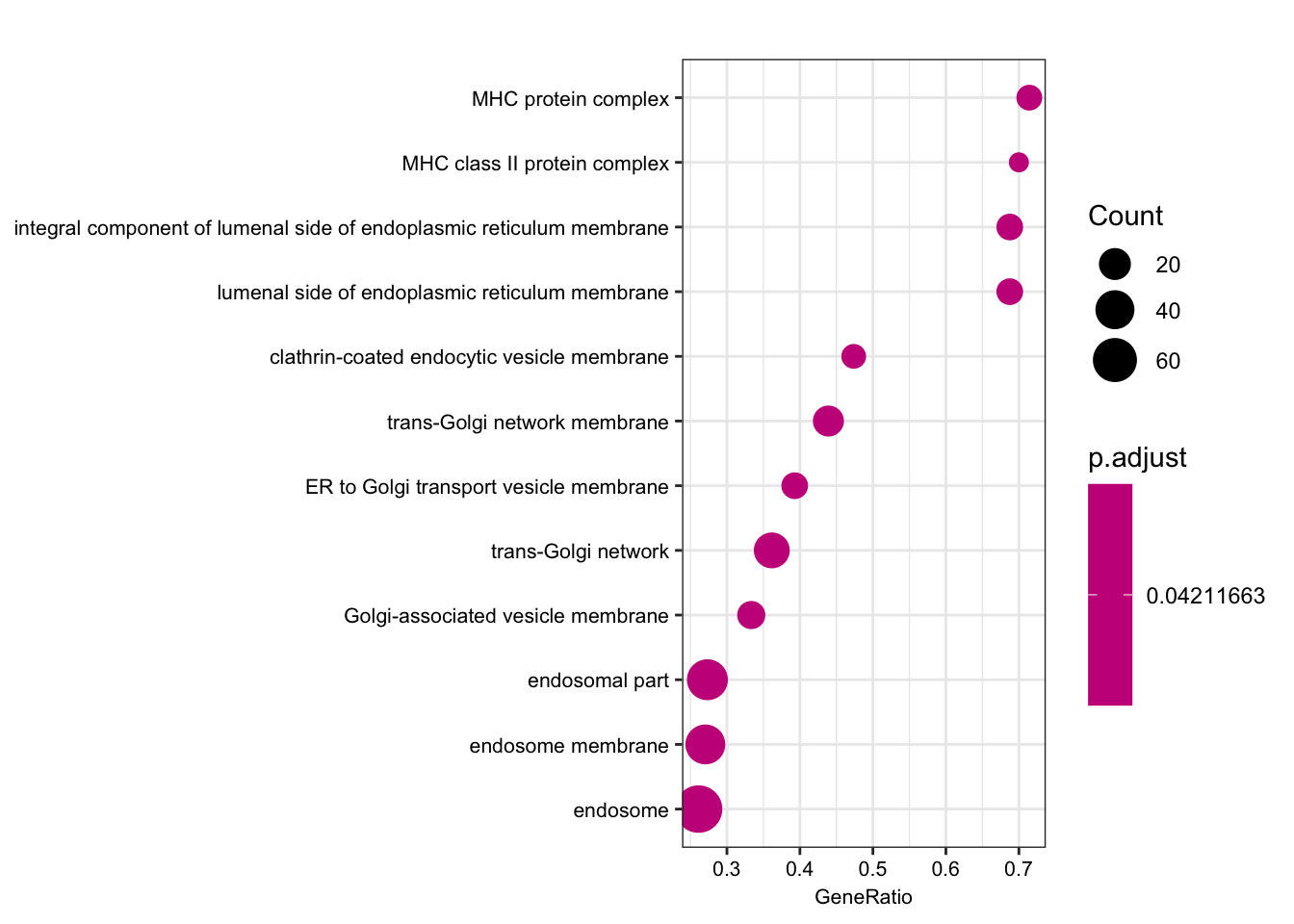
gseaplot(gsego.cc.shared, geneSetID = "GO:0042611", pvalue_table = TRUE)
#Create chimp-qval-ordered gene list for genes that are not human eGenes.
#I expect this to return similar results as gene overlap analysis with forground as chimp-specific eGenes and background as all chimp-eGenes.
Chimp_OrderedGenes <- eQTLs %>%
group_by(gene) %>%
dplyr::slice(which.min(qvalue)) %>%
filter(gene %in% GtexHeartEgenes$chimp_id) %>%
arrange(qvalue) %>%
filter(!gene %in% HumanSigGenes)
GeneList <- log10(Chimp_OrderedGenes$qvalue) * -1
names(GeneList) <- as.character(ChimpToHuman.ID(Chimp_OrderedGenes$gene))
gsego.cc.chimpspecific <- gseGO(gene = GeneList,
OrgDb = org.Hs.eg.db,
keyType = 'ENSEMBL',
maxGSSize = 500,
pvalueCutoff=0.8,
ont = "CC")preparing geneSet collections...GSEA analysis...Warning in fgsea(pathways = geneSets, stats = geneList, nperm = nPerm, minSize = minGSSize, : There are ties in the preranked stats (41.14% of the list).
The order of those tied genes will be arbitrary, which may produce unexpected results.leading edge analysis...done...as.data.frame(gsego.cc.chimpspecific) ID
GO:0015934 GO:0015934
GO:0030173 GO:0030173
GO:0031228 GO:0031228
GO:0000315 GO:0000315
GO:0005762 GO:0005762
GO:0099522 GO:0099522
GO:0043197 GO:0043197
GO:0044309 GO:0044309
GO:0000178 GO:0000178
GO:1905354 GO:1905354
GO:0000779 GO:0000779
GO:0000777 GO:0000777
GO:0000313 GO:0000313
GO:0005761 GO:0005761
GO:0048471 GO:0048471
GO:0022625 GO:0022625
GO:0005913 GO:0005913
GO:0000776 GO:0000776
GO:0005581 GO:0005581
GO:0034704 GO:0034704
GO:0031304 GO:0031304
GO:0031305 GO:0031305
GO:0005743 GO:0005743
GO:0030496 GO:0030496
GO:0005891 GO:0005891
GO:0005795 GO:0005795
GO:0005834 GO:0005834
GO:1905360 GO:1905360
GO:0031985 GO:0031985
GO:0000922 GO:0000922
GO:0044391 GO:0044391
GO:0016342 GO:0016342
GO:0043292 GO:0043292
GO:0098687 GO:0098687
GO:0031514 GO:0031514
GO:0044429 GO:0044429
GO:0005911 GO:0005911
GO:0071005 GO:0071005
GO:0071011 GO:0071011
GO:0030134 GO:0030134
GO:0005740 GO:0005740
GO:0031966 GO:0031966
GO:0005776 GO:0005776
GO:0031258 GO:0031258
GO:0000775 GO:0000775
GO:1905368 GO:1905368
GO:0033017 GO:0033017
GO:0016605 GO:0016605
GO:0019866 GO:0019866
GO:0012507 GO:0012507
GO:0031301 GO:0031301
GO:0042641 GO:0042641
GO:0005840 GO:0005840
GO:0005778 GO:0005778
GO:0031903 GO:0031903
GO:0022626 GO:0022626
GO:0032432 GO:0032432
GO:0032592 GO:0032592
GO:0098573 GO:0098573
GO:0000780 GO:0000780
GO:0016592 GO:0016592
GO:0016324 GO:0016324
GO:0016529 GO:0016529
GO:0043296 GO:0043296
GO:0015629 GO:0015629
GO:0032839 GO:0032839
GO:0098794 GO:0098794
GO:0005640 GO:0005640
GO:0005912 GO:0005912
GO:0030135 GO:0030135
GO:0030315 GO:0030315
GO:0032580 GO:0032580
GO:0005798 GO:0005798
GO:0031012 GO:0031012
GO:0045171 GO:0045171
GO:0005874 GO:0005874
GO:0030054 GO:0030054
GO:0030136 GO:0030136
GO:0031967 GO:0031967
GO:0031975 GO:0031975
GO:0000502 GO:0000502
GO:1905369 GO:1905369
GO:0019005 GO:0019005
GO:0044449 GO:0044449
GO:0016528 GO:0016528
GO:0099081 GO:0099081
GO:0005838 GO:0005838
GO:0030016 GO:0030016
GO:0030426 GO:0030426
GO:0099512 GO:0099512
GO:0000793 GO:0000793
GO:0044445 GO:0044445
GO:0005814 GO:0005814
GO:0099080 GO:0099080
GO:0044420 GO:0044420
GO:0036126 GO:0036126
GO:0030125 GO:0030125
GO:0030427 GO:0030427
GO:0043209 GO:0043209
GO:0036477 GO:0036477
GO:0097223 GO:0097223
GO:0005819 GO:0005819
GO:0014069 GO:0014069
GO:0032279 GO:0032279
GO:0001669 GO:0001669
GO:0009986 GO:0009986
GO:0035327 GO:0035327
GO:0008305 GO:0008305
GO:0098636 GO:0098636
GO:0009897 GO:0009897
GO:0098798 GO:0098798
GO:0043025 GO:0043025
GO:0031941 GO:0031941
GO:0005667 GO:0005667
GO:0150034 GO:0150034
GO:0005923 GO:0005923
GO:0031461 GO:0031461
GO:0000784 GO:0000784
GO:0005791 GO:0005791
GO:0070160 GO:0070160
GO:0097729 GO:0097729
GO:0005876 GO:0005876
GO:0070161 GO:0070161
GO:0030660 GO:0030660
GO:0030120 GO:0030120
GO:0005790 GO:0005790
GO:0043005 GO:0043005
GO:0044798 GO:0044798
GO:0005905 GO:0005905
GO:0042470 GO:0042470
GO:0048770 GO:0048770
GO:0000794 GO:0000794
GO:0030425 GO:0030425
GO:0009295 GO:0009295
GO:0042645 GO:0042645
GO:0030662 GO:0030662
GO:0097546 GO:0097546
GO:0099572 GO:0099572
GO:0045211 GO:0045211
GO:0051233 GO:0051233
GO:0031902 GO:0031902
GO:0031594 GO:0031594
GO:0000151 GO:0000151
GO:0001725 GO:0001725
GO:0097517 GO:0097517
GO:0070461 GO:0070461
GO:0031300 GO:0031300
GO:0097447 GO:0097447
GO:0031526 GO:0031526
GO:0030017 GO:0030017
GO:0032154 GO:0032154
GO:0044455 GO:0044455
GO:0014704 GO:0014704
GO:0030118 GO:0030118
GO:0005901 GO:0005901
GO:0005938 GO:0005938
GO:0000795 GO:0000795
GO:0099086 GO:0099086
GO:0072562 GO:0072562
GO:0005773 GO:0005773
GO:1990391 GO:1990391
GO:0043204 GO:0043204
GO:0035770 GO:0035770
GO:0098984 GO:0098984
GO:0045202 GO:0045202
GO:0062023 GO:0062023
GO:0022624 GO:0022624
GO:0045178 GO:0045178
GO:0005759 GO:0005759
GO:0005758 GO:0005758
GO:0042383 GO:0042383
GO:0099568 GO:0099568
GO:0030055 GO:0030055
GO:0030140 GO:0030140
GO:0035097 GO:0035097
GO:0098590 GO:0098590
GO:0044463 GO:0044463
GO:0120038 GO:0120038
GO:0030992 GO:0030992
GO:0000781 GO:0000781
GO:0005903 GO:0005903
GO:0045177 GO:0045177
GO:0030117 GO:0030117
GO:0048475 GO:0048475
GO:0005925 GO:0005925
GO:0033116 GO:0033116
GO:0098563 GO:0098563
GO:0032155 GO:0032155
GO:0090575 GO:0090575
GO:0005789 GO:0005789
GO:0005747 GO:0005747
GO:0030964 GO:0030964
GO:0045271 GO:0045271
GO:0005924 GO:0005924
GO:0031970 GO:0031970
GO:0036464 GO:0036464
GO:0031463 GO:0031463
GO:0044215 GO:0044215
GO:0044216 GO:0044216
GO:0044217 GO:0044217
Description
GO:0015934 large ribosomal subunit
GO:0030173 integral component of Golgi membrane
GO:0031228 intrinsic component of Golgi membrane
GO:0000315 organellar large ribosomal subunit
GO:0005762 mitochondrial large ribosomal subunit
GO:0099522 region of cytosol
GO:0043197 dendritic spine
GO:0044309 neuron spine
GO:0000178 exosome (RNase complex)
GO:1905354 exoribonuclease complex
GO:0000779 condensed chromosome, centromeric region
GO:0000777 condensed chromosome kinetochore
GO:0000313 organellar ribosome
GO:0005761 mitochondrial ribosome
GO:0048471 perinuclear region of cytoplasm
GO:0022625 cytosolic large ribosomal subunit
GO:0005913 cell-cell adherens junction
GO:0000776 kinetochore
GO:0005581 collagen trimer
GO:0034704 calcium channel complex
GO:0031304 intrinsic component of mitochondrial inner membrane
GO:0031305 integral component of mitochondrial inner membrane
GO:0005743 mitochondrial inner membrane
GO:0030496 midbody
GO:0005891 voltage-gated calcium channel complex
GO:0005795 Golgi stack
GO:0005834 heterotrimeric G-protein complex
GO:1905360 GTPase complex
GO:0031985 Golgi cisterna
GO:0000922 spindle pole
GO:0044391 ribosomal subunit
GO:0016342 catenin complex
GO:0043292 contractile fiber
GO:0098687 chromosomal region
GO:0031514 motile cilium
GO:0044429 mitochondrial part
GO:0005911 cell-cell junction
GO:0071005 U2-type precatalytic spliceosome
GO:0071011 precatalytic spliceosome
GO:0030134 COPII-coated ER to Golgi transport vesicle
GO:0005740 mitochondrial envelope
GO:0031966 mitochondrial membrane
GO:0005776 autophagosome
GO:0031258 lamellipodium membrane
GO:0000775 chromosome, centromeric region
GO:1905368 peptidase complex
GO:0033017 sarcoplasmic reticulum membrane
GO:0016605 PML body
GO:0019866 organelle inner membrane
GO:0012507 ER to Golgi transport vesicle membrane
GO:0031301 integral component of organelle membrane
GO:0042641 actomyosin
GO:0005840 ribosome
GO:0005778 peroxisomal membrane
GO:0031903 microbody membrane
GO:0022626 cytosolic ribosome
GO:0032432 actin filament bundle
GO:0032592 integral component of mitochondrial membrane
GO:0098573 intrinsic component of mitochondrial membrane
GO:0000780 condensed nuclear chromosome, centromeric region
GO:0016592 mediator complex
GO:0016324 apical plasma membrane
GO:0016529 sarcoplasmic reticulum
GO:0043296 apical junction complex
GO:0015629 actin cytoskeleton
GO:0032839 dendrite cytoplasm
GO:0098794 postsynapse
GO:0005640 nuclear outer membrane
GO:0005912 adherens junction
GO:0030135 coated vesicle
GO:0030315 T-tubule
GO:0032580 Golgi cisterna membrane
GO:0005798 Golgi-associated vesicle
GO:0031012 extracellular matrix
GO:0045171 intercellular bridge
GO:0005874 microtubule
GO:0030054 cell junction
GO:0030136 clathrin-coated vesicle
GO:0031967 organelle envelope
GO:0031975 envelope
GO:0000502 proteasome complex
GO:1905369 endopeptidase complex
GO:0019005 SCF ubiquitin ligase complex
GO:0044449 contractile fiber part
GO:0016528 sarcoplasm
GO:0099081 supramolecular polymer
GO:0005838 proteasome regulatory particle
GO:0030016 myofibril
GO:0030426 growth cone
GO:0099512 supramolecular fiber
GO:0000793 condensed chromosome
GO:0044445 cytosolic part
GO:0005814 centriole
GO:0099080 supramolecular complex
GO:0044420 extracellular matrix component
GO:0036126 sperm flagellum
GO:0030125 clathrin vesicle coat
GO:0030427 site of polarized growth
GO:0043209 myelin sheath
GO:0036477 somatodendritic compartment
GO:0097223 sperm part
GO:0005819 spindle
GO:0014069 postsynaptic density
GO:0032279 asymmetric synapse
GO:0001669 acrosomal vesicle
GO:0009986 cell surface
GO:0035327 transcriptionally active chromatin
GO:0008305 integrin complex
GO:0098636 protein complex involved in cell adhesion
GO:0009897 external side of plasma membrane
GO:0098798 mitochondrial protein complex
GO:0043025 neuronal cell body
GO:0031941 filamentous actin
GO:0005667 transcription factor complex
GO:0150034 distal axon
GO:0005923 bicellular tight junction
GO:0031461 cullin-RING ubiquitin ligase complex
GO:0000784 nuclear chromosome, telomeric region
GO:0005791 rough endoplasmic reticulum
GO:0070160 tight junction
GO:0097729 9+2 motile cilium
GO:0005876 spindle microtubule
GO:0070161 anchoring junction
GO:0030660 Golgi-associated vesicle membrane
GO:0030120 vesicle coat
GO:0005790 smooth endoplasmic reticulum
GO:0043005 neuron projection
GO:0044798 nuclear transcription factor complex
GO:0005905 clathrin-coated pit
GO:0042470 melanosome
GO:0048770 pigment granule
GO:0000794 condensed nuclear chromosome
GO:0030425 dendrite
GO:0009295 nucleoid
GO:0042645 mitochondrial nucleoid
GO:0030662 coated vesicle membrane
GO:0097546 ciliary base
GO:0099572 postsynaptic specialization
GO:0045211 postsynaptic membrane
GO:0051233 spindle midzone
GO:0031902 late endosome membrane
GO:0031594 neuromuscular junction
GO:0000151 ubiquitin ligase complex
GO:0001725 stress fiber
GO:0097517 contractile actin filament bundle
GO:0070461 SAGA-type complex
GO:0031300 intrinsic component of organelle membrane
GO:0097447 dendritic tree
GO:0031526 brush border membrane
GO:0030017 sarcomere
GO:0032154 cleavage furrow
GO:0044455 mitochondrial membrane part
GO:0014704 intercalated disc
GO:0030118 clathrin coat
GO:0005901 caveola
GO:0005938 cell cortex
GO:0000795 synaptonemal complex
GO:0099086 synaptonemal structure
GO:0072562 blood microparticle
GO:0005773 vacuole
GO:1990391 DNA repair complex
GO:0043204 perikaryon
GO:0035770 ribonucleoprotein granule
GO:0098984 neuron to neuron synapse
GO:0045202 synapse
GO:0062023 collagen-containing extracellular matrix
GO:0022624 proteasome accessory complex
GO:0045178 basal part of cell
GO:0005759 mitochondrial matrix
GO:0005758 mitochondrial intermembrane space
GO:0042383 sarcolemma
GO:0099568 cytoplasmic region
GO:0030055 cell-substrate junction
GO:0030140 trans-Golgi network transport vesicle
GO:0035097 histone methyltransferase complex
GO:0098590 plasma membrane region
GO:0044463 cell projection part
GO:0120038 plasma membrane bounded cell projection part
GO:0030992 intraciliary transport particle B
GO:0000781 chromosome, telomeric region
GO:0005903 brush border
GO:0045177 apical part of cell
GO:0030117 membrane coat
GO:0048475 coated membrane
GO:0005925 focal adhesion
GO:0033116 endoplasmic reticulum-Golgi intermediate compartment membrane
GO:0098563 intrinsic component of synaptic vesicle membrane
GO:0032155 cell division site part
GO:0090575 RNA polymerase II transcription factor complex
GO:0005789 endoplasmic reticulum membrane
GO:0005747 mitochondrial respiratory chain complex I
GO:0030964 NADH dehydrogenase complex
GO:0045271 respiratory chain complex I
GO:0005924 cell-substrate adherens junction
GO:0031970 organelle envelope lumen
GO:0036464 cytoplasmic ribonucleoprotein granule
GO:0031463 Cul3-RING ubiquitin ligase complex
GO:0044215 other organism
GO:0044216 other organism cell
GO:0044217 other organism part
setSize enrichmentScore NES pvalue p.adjust
GO:0015934 42 0.5337794 1.540643 0.002997003 0.3667954
GO:0030173 20 0.6152928 1.644906 0.003070624 0.3667954
GO:0031228 20 0.6152928 1.644906 0.003070624 0.3667954
GO:0000315 23 0.5958199 1.617803 0.004048583 0.3667954
GO:0005762 23 0.5958199 1.617803 0.004048583 0.3667954
GO:0099522 12 0.6866644 1.674718 0.005313496 0.3667954
GO:0043197 60 0.4886757 1.453480 0.005994006 0.3667954
GO:0044309 61 0.4844915 1.442991 0.006993007 0.3667954
GO:0000178 10 0.7091309 1.673265 0.008705114 0.3667954
GO:1905354 10 0.7091309 1.673265 0.008705114 0.3667954
GO:0000779 45 0.5037988 1.457870 0.008991009 0.3667954
GO:0000777 40 0.5176551 1.486811 0.009990010 0.3667954
GO:0000313 31 0.5437738 1.520908 0.010010010 0.3667954
GO:0005761 31 0.5437738 1.520908 0.010010010 0.3667954
GO:0048471 263 0.3830875 1.223264 0.015984016 0.5245399
GO:0022625 19 0.5787490 1.537684 0.016359918 0.5245399
GO:0005913 46 0.4884025 1.417255 0.018981019 0.5694306
GO:0000776 57 0.4687597 1.386632 0.019980020 0.5694306
GO:0005581 28 0.5321308 1.468014 0.023092369 0.6089623
GO:0034704 20 0.5531787 1.478852 0.026612078 0.6089623
GO:0031304 17 0.5795260 1.509305 0.029896907 0.6089623
GO:0031305 17 0.5795260 1.509305 0.029896907 0.6089623
GO:0005743 140 0.3989638 1.248069 0.030969031 0.6089623
GO:0030496 79 0.4316682 1.309564 0.032967033 0.6089623
GO:0005891 12 0.6157776 1.501831 0.034006376 0.6089623
GO:0005795 48 0.4712130 1.372934 0.034965035 0.6089623
GO:0005834 10 -0.5061665 -1.805022 0.036144578 0.6089623
GO:1905360 10 -0.5061665 -1.805022 0.036144578 0.6089623
GO:0031985 35 0.4968350 1.405189 0.039000000 0.6089623
GO:0000922 66 0.4412074 1.323130 0.039960040 0.6089623
GO:0044391 59 0.4471486 1.326557 0.039960040 0.6089623
GO:0016342 12 0.6054430 1.476625 0.041445271 0.6089623
GO:0043292 97 0.4199546 1.286391 0.041958042 0.6089623
GO:0098687 145 0.3894535 1.219572 0.043956044 0.6089623
GO:0031514 44 0.4682848 1.353743 0.045954046 0.6089623
GO:0044429 315 0.3615502 1.159190 0.045954046 0.6089623
GO:0005911 168 0.3828228 1.206536 0.047952048 0.6089623
GO:0071005 10 -0.4119561 -1.469062 0.048192771 0.6089623
GO:0071011 10 -0.4119561 -1.469062 0.048192771 0.6089623
GO:0030134 28 0.4993601 1.377608 0.049196787 0.6089623
GO:0005740 229 0.3683272 1.171146 0.051948052 0.6089623
GO:0031966 217 0.3713507 1.179597 0.051948052 0.6089623
GO:0005776 29 0.4997835 1.383744 0.052208835 0.6089623
GO:0031258 10 0.6185298 1.459483 0.052230686 0.6089623
GO:0000775 85 0.4190203 1.276363 0.053946054 0.6149850
GO:1905368 47 0.4535813 1.320034 0.056943057 0.6275357
GO:0033017 14 0.5688568 1.418439 0.057591623 0.6275357
GO:0016605 45 0.4577110 1.324503 0.058941059 0.6275357
GO:0019866 156 0.3784873 1.188608 0.059940060 0.6275357
GO:0012507 18 0.5322100 1.401116 0.065573770 0.6406900
GO:0031301 70 0.4224190 1.270701 0.065934066 0.6406900
GO:0042641 31 0.4843835 1.354796 0.066066066 0.6406900
GO:0005840 78 0.4131743 1.252547 0.069930070 0.6406900
GO:0005778 23 -0.2335613 -1.277783 0.071428571 0.6406900
GO:0031903 23 -0.2335613 -1.277783 0.071428571 0.6406900
GO:0022626 32 0.4771270 1.339652 0.072000000 0.6406900
GO:0032432 29 0.4810304 1.331822 0.075301205 0.6406900
GO:0032592 27 0.4886576 1.341874 0.077620968 0.6406900
GO:0098573 27 0.4886576 1.341874 0.077620968 0.6406900
GO:0000780 11 0.5864354 1.405487 0.082073434 0.6406900
GO:0016592 13 0.5616247 1.385760 0.082627119 0.6406900
GO:0016324 94 0.3994289 1.222679 0.084915085 0.6406900
GO:0016529 23 0.4917873 1.335328 0.087044534 0.6406900
GO:0043296 52 0.4341577 1.271727 0.090909091 0.6406900
GO:0015629 190 0.3654037 1.154921 0.091908092 0.6406900
GO:0032839 16 0.5350076 1.375901 0.093360996 0.6406900
GO:0098794 207 0.3609852 1.144723 0.094905095 0.6406900
GO:0005640 11 0.5772499 1.383472 0.095032397 0.6406900
GO:0005912 239 0.3576162 1.137649 0.095904096 0.6406900
GO:0030135 108 0.3903280 1.202281 0.095904096 0.6406900
GO:0030315 20 0.4996057 1.335631 0.098259980 0.6406900
GO:0032580 27 0.4759681 1.307028 0.101814516 0.6406900
GO:0005798 66 0.4081749 1.224069 0.101898102 0.6406900
GO:0031012 153 0.3713800 1.166428 0.101898102 0.6406900
GO:0045171 21 0.4897177 1.313559 0.107252298 0.6406900
GO:0005874 157 0.3668637 1.152535 0.107892108 0.6406900
GO:0030054 449 0.3412679 1.099477 0.107892108 0.6406900
GO:0030136 69 0.4057311 1.218660 0.107892108 0.6406900
GO:0031967 394 0.3433489 1.105549 0.107892108 0.6406900
GO:0031975 394 0.3433489 1.105549 0.107892108 0.6406900
GO:0000502 32 0.4553423 1.278486 0.108000000 0.6406900
GO:1905369 32 0.4553423 1.278486 0.108000000 0.6406900
GO:0019005 24 0.4796853 1.306287 0.111223458 0.6406900
GO:0044449 90 0.3921845 1.196806 0.114885115 0.6406900
GO:0016528 28 0.4651481 1.283226 0.117469880 0.6406900
GO:0099081 314 0.3466858 1.111572 0.117882118 0.6406900
GO:0005838 10 0.5715634 1.348661 0.118607182 0.6406900
GO:0030016 92 0.3899206 1.191865 0.119880120 0.6406900
GO:0030426 59 0.4142325 1.228905 0.120879121 0.6406900
GO:0099512 310 0.3467658 1.111672 0.120879121 0.6406900
GO:0000793 82 0.3933878 1.195823 0.121878122 0.6406900
GO:0044445 87 0.3903979 1.191090 0.122877123 0.6406900
GO:0005814 43 0.4309189 1.245915 0.124875125 0.6406900
GO:0099080 315 0.3454327 1.107515 0.124875125 0.6406900
GO:0044420 21 0.4815141 1.291555 0.125638407 0.6406900
GO:0036126 22 0.4765404 1.284943 0.127291242 0.6406900
GO:0030125 14 0.5322712 1.327213 0.127748691 0.6406900
GO:0030427 60 0.4092049 1.217108 0.127872128 0.6406900
GO:0043209 70 0.3982868 1.198108 0.127872128 0.6406900
GO:0036477 284 0.3468505 1.109523 0.128871129 0.6406900
GO:0097223 48 0.4237332 1.234596 0.128871129 0.6406900
GO:0005819 136 0.3708813 1.157757 0.129870130 0.6406900
GO:0014069 116 0.3765061 1.164662 0.130869131 0.6406900
GO:0032279 116 0.3765061 1.164662 0.130869131 0.6406900
GO:0001669 29 0.4537136 1.256190 0.133534137 0.6406900
GO:0009986 229 0.3514304 1.117420 0.135864136 0.6406900
GO:0035327 12 0.5403870 1.317959 0.136025505 0.6406900
GO:0008305 20 0.4817895 1.288002 0.136131013 0.6406900
GO:0098636 20 0.4817895 1.288002 0.136131013 0.6406900
GO:0009897 84 0.3884309 1.182493 0.138861139 0.6417636
GO:0098798 86 0.3860764 1.178187 0.138861139 0.6417636
GO:0043025 165 0.3587359 1.131044 0.146853147 0.6701760
GO:0031941 10 0.5501879 1.298223 0.149075082 0.6701760
GO:0005667 158 0.3604046 1.132819 0.150849151 0.6701760
GO:0150034 81 0.3859639 1.172535 0.150849151 0.6701760
GO:0005923 43 0.4220205 1.220188 0.151848152 0.6701760
GO:0031461 74 0.3915287 1.183030 0.152847153 0.6701760
GO:0000784 45 0.4177432 1.208846 0.160839161 0.6961289
GO:0005791 28 0.4465985 1.232052 0.162650602 0.6961289
GO:0070160 44 0.4186055 1.210128 0.162837163 0.6961289
GO:0097729 23 0.4625336 1.255897 0.164979757 0.6994596
GO:0005876 23 0.4605103 1.250403 0.171052632 0.7086247
GO:0070161 245 0.3448021 1.097272 0.171828172 0.7086247
GO:0030660 42 0.4143415 1.195910 0.174825175 0.7086247
GO:0030120 26 0.4499199 1.234936 0.177062374 0.7086247
GO:0005790 12 0.5170475 1.261036 0.178533475 0.7086247
GO:0043005 406 0.3350011 1.079356 0.178821179 0.7086247
GO:0044798 97 0.3762601 1.152548 0.178821179 0.7086247
GO:0005905 28 0.4416070 1.218282 0.182730924 0.7086247
GO:0042470 42 0.4129608 1.191925 0.182817183 0.7086247
GO:0048770 42 0.4129608 1.191925 0.182817183 0.7086247
GO:0000794 32 0.4285969 1.203392 0.186000000 0.7086247
GO:0030425 209 0.3478976 1.104005 0.187812188 0.7086247
GO:0009295 12 0.5102941 1.244565 0.188097768 0.7086247
GO:0042645 12 0.5102941 1.244565 0.188097768 0.7086247
GO:0030662 69 0.3816044 1.146193 0.188811189 0.7086247
GO:0097546 16 0.4898827 1.259851 0.196058091 0.7086247
GO:0099572 121 0.3636387 1.127274 0.197802198 0.7086247
GO:0045211 85 0.3743196 1.140202 0.200799201 0.7086247
GO:0051233 16 0.4829030 1.241901 0.205394191 0.7086247
GO:0031902 42 0.4058872 1.171508 0.205794206 0.7086247
GO:0031594 21 0.4552528 1.221115 0.207354443 0.7086247
GO:0000151 126 0.3597339 1.117388 0.207792208 0.7086247
GO:0001725 27 0.4368168 1.199517 0.208669355 0.7086247
GO:0097517 27 0.4368168 1.199517 0.208669355 0.7086247
GO:0070461 13 0.4995723 1.232651 0.208686441 0.7086247
GO:0031300 78 0.3765987 1.141668 0.208791209 0.7086247
GO:0097447 210 0.3455520 1.096829 0.209790210 0.7086247
GO:0031526 13 0.4993482 1.232098 0.210805085 0.7086247
GO:0030017 84 0.3719220 1.132235 0.211788212 0.7086247
GO:0032154 28 0.4349692 1.199970 0.211847390 0.7086247
GO:0044455 73 0.3780167 1.141028 0.215784216 0.7086247
GO:0014704 21 0.4512231 1.210306 0.218590398 0.7086247
GO:0030118 21 0.4508850 1.209399 0.218590398 0.7086247
GO:0005901 28 0.4324979 1.193152 0.218875502 0.7086247
GO:0005938 114 0.3632178 1.122355 0.219780220 0.7086247
GO:0000795 11 0.5091949 1.220368 0.220302376 0.7086247
GO:0099086 11 0.5091949 1.220368 0.220302376 0.7086247
GO:0072562 27 0.4328264 1.188559 0.221774194 0.7086247
GO:0005773 241 0.3394707 1.080158 0.222777223 0.7086247
GO:1990391 12 0.5006701 1.221093 0.223166844 0.7086247
GO:0043204 42 0.4025818 1.161968 0.223776224 0.7086247
GO:0035770 89 0.3677233 1.121786 0.228771229 0.7156076
GO:0098984 121 0.3586891 1.111931 0.228771229 0.7156076
GO:0045202 376 0.3299240 1.061984 0.232767233 0.7193349
GO:0062023 113 0.3613978 1.116738 0.232767233 0.7193349
GO:0022624 12 0.4913673 1.198404 0.244420829 0.7419151
GO:0045178 20 0.4472400 1.195638 0.244626407 0.7419151
GO:0005759 133 0.3525289 1.098412 0.244755245 0.7419151
GO:0005758 19 0.4529371 1.203413 0.248466258 0.7419151
GO:0042383 51 0.3871320 1.132740 0.248751249 0.7419151
GO:0099568 177 0.3436930 1.084551 0.248751249 0.7419151
GO:0030055 186 0.3417898 1.079153 0.252747253 0.7463209
GO:0030140 15 0.4745819 1.202520 0.253138075 0.7463209
GO:0035097 38 0.4016677 1.148879 0.255744256 0.7496960
GO:0098590 348 0.3291236 1.058068 0.258741259 0.7520197
GO:0044463 462 0.3258449 1.051264 0.261738262 0.7520197
GO:0120038 462 0.3258449 1.051264 0.261738262 0.7520197
GO:0030992 10 0.5004139 1.180777 0.265505985 0.7520197
GO:0000781 60 0.3755677 1.117061 0.266733267 0.7520197
GO:0005903 32 0.4075783 1.144377 0.267000000 0.7520197
GO:0045177 113 0.3552323 1.097686 0.267732268 0.7520197
GO:0030117 44 0.3902444 1.128140 0.269730270 0.7520197
GO:0048475 44 0.3902444 1.128140 0.269730270 0.7520197
GO:0005925 184 0.3407568 1.075885 0.272727273 0.7553938
GO:0033116 26 0.4210002 1.155557 0.275653924 0.7553938
GO:0098563 14 -0.2691121 -1.143646 0.276595745 0.7553938
GO:0032155 30 0.4087457 1.139586 0.276830491 0.7553938
GO:0090575 76 0.3648113 1.103385 0.278721279 0.7565292
GO:0005789 369 0.3266385 1.051477 0.287712288 0.7669856
GO:0005747 15 0.4603296 1.166407 0.290794979 0.7669856
GO:0030964 15 0.4603296 1.166407 0.290794979 0.7669856
GO:0045271 15 0.4603296 1.166407 0.290794979 0.7669856
GO:0005924 185 0.3386868 1.069134 0.292707293 0.7669856
GO:0031970 22 0.4263315 1.149559 0.293279022 0.7669856
GO:0036464 83 0.3595010 1.094368 0.293706294 0.7669856
GO:0031463 23 0.4215947 1.144737 0.294534413 0.7669856
GO:0044215 16 0.4446502 1.143525 0.310165975 0.7955757
GO:0044216 16 0.4446502 1.143525 0.310165975 0.7955757
GO:0044217 16 0.4446502 1.143525 0.310165975 0.7955757
qvalues rank leading_edge
GO:0015934 0.3559951 1298 tags=43%, list=22%, signal=34%
GO:0030173 0.3559951 1107 tags=50%, list=19%, signal=41%
GO:0031228 0.3559951 1107 tags=50%, list=19%, signal=41%
GO:0000315 0.3559951 1298 tags=57%, list=22%, signal=44%
GO:0005762 0.3559951 1298 tags=57%, list=22%, signal=44%
GO:0099522 0.3559951 662 tags=42%, list=11%, signal=37%
GO:0043197 0.3559951 1075 tags=33%, list=18%, signal=27%
GO:0044309 0.3559951 1075 tags=33%, list=18%, signal=27%
GO:0000178 0.3559951 559 tags=40%, list=10%, signal=36%
GO:1905354 0.3559951 559 tags=40%, list=10%, signal=36%
GO:0000779 0.3559951 1326 tags=38%, list=23%, signal=29%
GO:0000777 0.3559951 1326 tags=38%, list=23%, signal=29%
GO:0000313 0.3559951 1298 tags=48%, list=22%, signal=38%
GO:0005761 0.3559951 1298 tags=48%, list=22%, signal=38%
GO:0048471 0.5090948 1383 tags=30%, list=24%, signal=24%
GO:0022625 0.5090948 285 tags=21%, list=5%, signal=20%
GO:0005913 0.5526637 1716 tags=50%, list=29%, signal=36%
GO:0000776 0.5526637 1691 tags=40%, list=29%, signal=29%
GO:0005581 0.5910314 542 tags=21%, list=9%, signal=20%
GO:0034704 0.5910314 1409 tags=50%, list=24%, signal=38%
GO:0031304 0.5910314 1334 tags=47%, list=23%, signal=36%
GO:0031305 0.5910314 1334 tags=47%, list=23%, signal=36%
GO:0005743 0.5910314 1788 tags=39%, list=31%, signal=28%
GO:0030496 0.5910314 1546 tags=41%, list=26%, signal=30%
GO:0005891 0.5910314 1409 tags=58%, list=24%, signal=44%
GO:0005795 0.5910314 2023 tags=54%, list=35%, signal=36%
GO:0005834 0.5910314 2894 tags=100%, list=49%, signal=51%
GO:1905360 0.5910314 2894 tags=100%, list=49%, signal=51%
GO:0031985 0.5910314 2023 tags=57%, list=35%, signal=38%
GO:0000922 0.5910314 1365 tags=38%, list=23%, signal=29%
GO:0044391 0.5910314 1298 tags=34%, list=22%, signal=27%
GO:0016342 0.5910314 543 tags=42%, list=9%, signal=38%
GO:0043292 0.5910314 1586 tags=36%, list=27%, signal=27%
GO:0098687 0.5910314 2161 tags=44%, list=37%, signal=29%
GO:0031514 0.5910314 1680 tags=41%, list=29%, signal=29%
GO:0044429 0.5910314 1874 tags=38%, list=32%, signal=27%
GO:0005911 0.5910314 1629 tags=36%, list=28%, signal=27%
GO:0071005 0.5910314 3444 tags=100%, list=59%, signal=41%
GO:0071011 0.5910314 3444 tags=100%, list=59%, signal=41%
GO:0030134 0.5910314 980 tags=32%, list=17%, signal=27%
GO:0005740 0.5910314 1868 tags=39%, list=32%, signal=28%
GO:0031966 0.5910314 1868 tags=39%, list=32%, signal=27%
GO:0005776 0.5910314 1647 tags=45%, list=28%, signal=32%
GO:0031258 0.5910314 504 tags=30%, list=9%, signal=27%
GO:0000775 0.5968768 1991 tags=42%, list=34%, signal=28%
GO:1905368 0.6090580 1887 tags=49%, list=32%, signal=33%
GO:0033017 0.6090580 909 tags=43%, list=16%, signal=36%
GO:0016605 0.6090580 698 tags=22%, list=12%, signal=20%
GO:0019866 0.6090580 1788 tags=37%, list=31%, signal=27%
GO:0012507 0.6218249 927 tags=33%, list=16%, signal=28%
GO:0031301 0.6218249 1189 tags=29%, list=20%, signal=23%
GO:0042641 0.6218249 1070 tags=35%, list=18%, signal=29%
GO:0005840 0.6218249 1298 tags=31%, list=22%, signal=24%
GO:0005778 0.6218249 2835 tags=91%, list=48%, signal=47%
GO:0031903 0.6218249 2835 tags=91%, list=48%, signal=47%
GO:0022626 0.6218249 285 tags=16%, list=5%, signal=15%
GO:0032432 0.6218249 1070 tags=34%, list=18%, signal=28%
GO:0032592 0.6218249 1334 tags=37%, list=23%, signal=29%
GO:0098573 0.6218249 1334 tags=37%, list=23%, signal=29%
GO:0000780 0.6218249 1253 tags=55%, list=21%, signal=43%
GO:0016592 0.6218249 1742 tags=62%, list=30%, signal=43%
GO:0016324 0.6218249 2311 tags=51%, list=40%, signal=31%
GO:0016529 0.6218249 1409 tags=43%, list=24%, signal=33%
GO:0043296 0.6218249 1214 tags=33%, list=21%, signal=26%
GO:0015629 0.6218249 1586 tags=33%, list=27%, signal=25%
GO:0032839 0.6218249 879 tags=31%, list=15%, signal=27%
GO:0098794 0.6218249 1091 tags=23%, list=19%, signal=20%
GO:0005640 0.6218249 2013 tags=73%, list=34%, signal=48%
GO:0005912 0.6218249 1644 tags=35%, list=28%, signal=26%
GO:0030135 0.6218249 1801 tags=35%, list=31%, signal=25%
GO:0030315 0.6218249 1877 tags=55%, list=32%, signal=37%
GO:0032580 0.6218249 1717 tags=44%, list=29%, signal=32%
GO:0005798 0.6218249 980 tags=23%, list=17%, signal=19%
GO:0031012 0.6218249 1278 tags=27%, list=22%, signal=22%
GO:0045171 0.6218249 1442 tags=48%, list=25%, signal=36%
GO:0005874 0.6218249 1365 tags=31%, list=23%, signal=24%
GO:0030054 0.6218249 1644 tags=32%, list=28%, signal=25%
GO:0030136 0.6218249 354 tags=14%, list=6%, signal=14%
GO:0031967 0.6218249 2052 tags=40%, list=35%, signal=28%
GO:0031975 0.6218249 2052 tags=40%, list=35%, signal=28%
GO:0000502 0.6218249 1887 tags=50%, list=32%, signal=34%
GO:1905369 0.6218249 1887 tags=50%, list=32%, signal=34%
GO:0019005 0.6218249 1962 tags=58%, list=34%, signal=39%
GO:0044449 0.6218249 1586 tags=34%, list=27%, signal=25%
GO:0016528 0.6218249 1533 tags=43%, list=26%, signal=32%
GO:0099081 0.6218249 1628 tags=32%, list=28%, signal=24%
GO:0005838 0.6218249 1342 tags=50%, list=23%, signal=39%
GO:0030016 0.6218249 1586 tags=35%, list=27%, signal=26%
GO:0030426 0.6218249 2026 tags=51%, list=35%, signal=34%
GO:0099512 0.6218249 1628 tags=32%, list=28%, signal=24%
GO:0000793 0.6218249 1326 tags=28%, list=23%, signal=22%
GO:0044445 0.6218249 889 tags=20%, list=15%, signal=17%
GO:0005814 0.6218249 334 tags=16%, list=6%, signal=15%
GO:0099080 0.6218249 1628 tags=32%, list=28%, signal=24%
GO:0044420 0.6218249 492 tags=19%, list=8%, signal=18%
GO:0036126 0.6218249 400 tags=18%, list=7%, signal=17%
GO:0030125 0.6218249 354 tags=21%, list=6%, signal=20%
GO:0030427 0.6218249 2026 tags=50%, list=35%, signal=33%
GO:0043209 0.6218249 1567 tags=37%, list=27%, signal=28%
GO:0036477 0.6218249 2311 tags=46%, list=40%, signal=29%
GO:0097223 0.6218249 400 tags=17%, list=7%, signal=16%
GO:0005819 0.6218249 1385 tags=30%, list=24%, signal=24%
GO:0014069 0.6218249 677 tags=18%, list=12%, signal=16%
GO:0032279 0.6218249 677 tags=18%, list=12%, signal=16%
GO:0001669 0.6218249 1571 tags=38%, list=27%, signal=28%
GO:0009986 0.6218249 2227 tags=43%, list=38%, signal=28%
GO:0035327 0.6218249 1241 tags=42%, list=21%, signal=33%
GO:0008305 0.6218249 1256 tags=40%, list=21%, signal=32%
GO:0098636 0.6218249 1256 tags=40%, list=21%, signal=32%
GO:0009897 0.6228669 2221 tags=49%, list=38%, signal=31%
GO:0098798 0.6228669 1788 tags=41%, list=31%, signal=29%
GO:0043025 0.6504427 2137 tags=44%, list=37%, signal=29%
GO:0031941 0.6504427 1500 tags=60%, list=26%, signal=45%
GO:0005667 0.6504427 2394 tags=49%, list=41%, signal=30%
GO:0150034 0.6504427 2026 tags=47%, list=35%, signal=31%
GO:0005923 0.6504427 1176 tags=30%, list=20%, signal=24%
GO:0031461 0.6504427 1962 tags=46%, list=34%, signal=31%
GO:0000784 0.6756314 1738 tags=42%, list=30%, signal=30%
GO:0005791 0.6756314 1644 tags=39%, list=28%, signal=28%
GO:0070160 0.6756314 1176 tags=30%, list=20%, signal=24%
GO:0097729 0.6788641 400 tags=17%, list=7%, signal=16%
GO:0005876 0.6877593 1780 tags=43%, list=30%, signal=30%
GO:0070161 0.6877593 1644 tags=34%, list=28%, signal=25%
GO:0030660 0.6877593 255 tags=14%, list=4%, signal=14%
GO:0030120 0.6877593 354 tags=15%, list=6%, signal=15%
GO:0005790 0.6877593 1239 tags=42%, list=21%, signal=33%
GO:0043005 0.6877593 1721 tags=33%, list=29%, signal=25%
GO:0044798 0.6877593 1084 tags=25%, list=19%, signal=20%
GO:0005905 0.6877593 1306 tags=36%, list=22%, signal=28%
GO:0042470 0.6877593 254 tags=17%, list=4%, signal=16%
GO:0048770 0.6877593 254 tags=17%, list=4%, signal=16%
GO:0000794 0.6877593 1253 tags=34%, list=21%, signal=27%
GO:0030425 0.6877593 1075 tags=23%, list=18%, signal=20%
GO:0009295 0.6877593 375 tags=17%, list=6%, signal=16%
GO:0042645 0.6877593 375 tags=17%, list=6%, signal=16%
GO:0030662 0.6877593 354 tags=13%, list=6%, signal=12%
GO:0097546 0.6877593 757 tags=25%, list=13%, signal=22%
GO:0099572 0.6877593 677 tags=17%, list=12%, signal=16%
GO:0045211 0.6877593 1075 tags=24%, list=18%, signal=19%
GO:0051233 0.6877593 1253 tags=44%, list=21%, signal=34%
GO:0031902 0.6877593 1081 tags=31%, list=18%, signal=25%
GO:0031594 0.6877593 1687 tags=43%, list=29%, signal=31%
GO:0000151 0.6877593 1962 tags=41%, list=34%, signal=28%
GO:0001725 0.6877593 1070 tags=33%, list=18%, signal=27%
GO:0097517 0.6877593 1070 tags=33%, list=18%, signal=27%
GO:0070461 0.6877593 1029 tags=38%, list=18%, signal=32%
GO:0031300 0.6877593 1189 tags=26%, list=20%, signal=21%
GO:0097447 0.6877593 1075 tags=23%, list=18%, signal=20%
GO:0031526 0.6877593 434 tags=23%, list=7%, signal=21%
GO:0030017 0.6877593 1586 tags=33%, list=27%, signal=25%
GO:0032154 0.6877593 1521 tags=39%, list=26%, signal=29%
GO:0044455 0.6877593 1788 tags=38%, list=31%, signal=27%
GO:0014704 0.6877593 887 tags=29%, list=15%, signal=24%
GO:0030118 0.6877593 1306 tags=33%, list=22%, signal=26%
GO:0005901 0.6877593 1482 tags=36%, list=25%, signal=27%
GO:0005938 0.6877593 1960 tags=41%, list=34%, signal=28%
GO:0000795 0.6877593 1105 tags=36%, list=19%, signal=30%
GO:0099086 0.6877593 1105 tags=36%, list=19%, signal=30%
GO:0072562 0.6877593 471 tags=19%, list=8%, signal=17%
GO:0005773 0.6877593 1603 tags=31%, list=27%, signal=23%
GO:1990391 0.6877593 1606 tags=50%, list=27%, signal=36%
GO:0043204 0.6877593 1222 tags=31%, list=21%, signal=25%
GO:0035770 0.6945365 698 tags=18%, list=12%, signal=16%
GO:0098984 0.6945365 677 tags=17%, list=12%, signal=16%
GO:0045202 0.6981541 1911 tags=35%, list=33%, signal=25%
GO:0062023 0.6981541 1239 tags=25%, list=21%, signal=20%
GO:0022624 0.7200694 859 tags=33%, list=15%, signal=28%
GO:0045178 0.7200694 676 tags=25%, list=12%, signal=22%
GO:0005759 0.7200694 1352 tags=29%, list=23%, signal=23%
GO:0005758 0.7200694 2039 tags=58%, list=35%, signal=38%
GO:0042383 0.7200694 1409 tags=29%, list=24%, signal=23%
GO:0099568 0.7200694 1960 tags=38%, list=34%, signal=26%
GO:0030055 0.7243455 2253 tags=45%, list=39%, signal=28%
GO:0030140 0.7243455 120 tags=13%, list=2%, signal=13%
GO:0035097 0.7276213 440 tags=16%, list=8%, signal=15%
GO:0098590 0.7298765 1521 tags=28%, list=26%, signal=22%
GO:0044463 0.7298765 2342 tags=44%, list=40%, signal=29%
GO:0120038 0.7298765 2342 tags=44%, list=40%, signal=29%
GO:0030992 0.7298765 524 tags=20%, list=9%, signal=18%
GO:0000781 0.7298765 2425 tags=50%, list=41%, signal=30%
GO:0005903 0.7298765 2423 tags=56%, list=41%, signal=33%
GO:0045177 0.7298765 2269 tags=45%, list=39%, signal=28%
GO:0030117 0.7298765 1306 tags=27%, list=22%, signal=21%
GO:0048475 0.7298765 1306 tags=27%, list=22%, signal=21%
GO:0005925 0.7331513 2253 tags=45%, list=39%, signal=28%
GO:0033116 0.7331513 1649 tags=38%, list=28%, signal=28%
GO:0098563 0.7331513 4279 tags=100%, list=73%, signal=27%
GO:0032155 0.7331513 1521 tags=37%, list=26%, signal=27%
GO:0090575 0.7342532 1084 tags=24%, list=19%, signal=20%
GO:0005789 0.7444017 992 tags=20%, list=17%, signal=18%
GO:0005747 0.7444017 1936 tags=53%, list=33%, signal=36%
GO:0030964 0.7444017 1936 tags=53%, list=33%, signal=36%
GO:0045271 0.7444017 1936 tags=53%, list=33%, signal=36%
GO:0005924 0.7444017 2253 tags=44%, list=39%, signal=28%
GO:0031970 0.7444017 2039 tags=55%, list=35%, signal=36%
GO:0036464 0.7444017 698 tags=18%, list=12%, signal=16%
GO:0031463 0.7444017 1946 tags=48%, list=33%, signal=32%
GO:0044215 0.7721500 1691 tags=44%, list=29%, signal=31%
GO:0044216 0.7721500 1691 tags=44%, list=29%, signal=31%
GO:0044217 0.7721500 1691 tags=44%, list=29%, signal=31%
core_enrichment
GO:0015934 ENSG00000163923/ENSG00000037241/ENSG00000173545/ENSG00000259494/ENSG00000135900/ENSG00000082516/ENSG00000156928/ENSG00000082515/ENSG00000143314/ENSG00000149792/ENSG00000172590/ENSG00000166902/ENSG00000111639/ENSG00000243147/ENSG00000105364/ENSG00000136522/ENSG00000174444/ENSG00000204822
GO:0030173 ENSG00000127995/ENSG00000180776/ENSG00000198964/ENSG00000120942/ENSG00000157593/ENSG00000204070/ENSG00000156671/ENSG00000160613/ENSG00000167645/ENSG00000100060
GO:0031228 ENSG00000127995/ENSG00000180776/ENSG00000198964/ENSG00000120942/ENSG00000157593/ENSG00000204070/ENSG00000156671/ENSG00000160613/ENSG00000167645/ENSG00000100060
GO:0000315 ENSG00000259494/ENSG00000135900/ENSG00000156928/ENSG00000082515/ENSG00000143314/ENSG00000149792/ENSG00000172590/ENSG00000166902/ENSG00000111639/ENSG00000243147/ENSG00000105364/ENSG00000136522/ENSG00000204822
GO:0005762 ENSG00000259494/ENSG00000135900/ENSG00000156928/ENSG00000082515/ENSG00000143314/ENSG00000149792/ENSG00000172590/ENSG00000166902/ENSG00000111639/ENSG00000243147/ENSG00000105364/ENSG00000136522/ENSG00000204822
GO:0099522 ENSG00000171862/ENSG00000113758/ENSG00000197386/ENSG00000141543/ENSG00000166501
GO:0043197 ENSG00000161057/ENSG00000167461/ENSG00000171862/ENSG00000176749/ENSG00000183023/ENSG00000080709/ENSG00000101040/ENSG00000107404/ENSG00000152767/ENSG00000118160/ENSG00000134909/ENSG00000137845/ENSG00000133026/ENSG00000123700/ENSG00000181790/ENSG00000166313/ENSG00000077063/ENSG00000099260/ENSG00000171992/ENSG00000197555
GO:0044309 ENSG00000161057/ENSG00000167461/ENSG00000171862/ENSG00000176749/ENSG00000183023/ENSG00000080709/ENSG00000101040/ENSG00000107404/ENSG00000152767/ENSG00000118160/ENSG00000134909/ENSG00000137845/ENSG00000133026/ENSG00000123700/ENSG00000181790/ENSG00000166313/ENSG00000077063/ENSG00000099260/ENSG00000171992/ENSG00000197555
GO:0000178 ENSG00000107371/ENSG00000197223/ENSG00000171824/ENSG00000144535
GO:1905354 ENSG00000107371/ENSG00000197223/ENSG00000171824/ENSG00000144535
GO:0000779 ENSG00000167513/ENSG00000185122/ENSG00000153187/ENSG00000166851/ENSG00000122545/ENSG00000085415/ENSG00000101265/ENSG00000134057/ENSG00000117724/ENSG00000069248/ENSG00000080986/ENSG00000076382/ENSG00000105146/ENSG00000143228/ENSG00000166579/ENSG00000087586/ENSG00000145919
GO:0000777 ENSG00000167513/ENSG00000185122/ENSG00000153187/ENSG00000166851/ENSG00000122545/ENSG00000085415/ENSG00000101265/ENSG00000134057/ENSG00000117724/ENSG00000069248/ENSG00000080986/ENSG00000076382/ENSG00000143228/ENSG00000166579/ENSG00000145919
GO:0000313 ENSG00000259494/ENSG00000135900/ENSG00000120333/ENSG00000144029/ENSG00000156928/ENSG00000082515/ENSG00000143314/ENSG00000149792/ENSG00000172590/ENSG00000166902/ENSG00000111639/ENSG00000243147/ENSG00000105364/ENSG00000136522/ENSG00000204822
GO:0005761 ENSG00000259494/ENSG00000135900/ENSG00000120333/ENSG00000144029/ENSG00000156928/ENSG00000082515/ENSG00000143314/ENSG00000149792/ENSG00000172590/ENSG00000166902/ENSG00000111639/ENSG00000243147/ENSG00000105364/ENSG00000136522/ENSG00000204822
GO:0048471 ENSG00000109062/ENSG00000154645/ENSG00000185122/ENSG00000152102/ENSG00000072274/ENSG00000118785/ENSG00000180776/ENSG00000055163/ENSG00000163083/ENSG00000173482/ENSG00000176749/ENSG00000138768/ENSG00000061938/ENSG00000113282/ENSG00000172071/ENSG00000006025/ENSG00000197386/ENSG00000182718/ENSG00000151461/ENSG00000151532/ENSG00000174989/ENSG00000115266/ENSG00000086991/ENSG00000105538/ENSG00000143740/ENSG00000105887/ENSG00000105974/ENSG00000167333/ENSG00000174059/ENSG00000110031/ENSG00000147649/ENSG00000060339/ENSG00000167434/ENSG00000115020/ENSG00000142867/ENSG00000051180/ENSG00000135018/ENSG00000131188/ENSG00000140299/ENSG00000134982/ENSG00000187391/ENSG00000165029/ENSG00000144476/ENSG00000055917/ENSG00000160191/ENSG00000184371/ENSG00000137845/ENSG00000117724/ENSG00000087470/ENSG00000139998/ENSG00000155903/ENSG00000132376/ENSG00000181790/ENSG00000065243/ENSG00000181588/ENSG00000171132/ENSG00000136854/ENSG00000134278/ENSG00000166313/ENSG00000169696/ENSG00000165795/ENSG00000128641/ENSG00000173786/ENSG00000239697/ENSG00000166135/ENSG00000091513/ENSG00000171368/ENSG00000066629/ENSG00000179218/ENSG00000087586/ENSG00000124762/ENSG00000118523/ENSG00000164300/ENSG00000021574/ENSG00000140350/ENSG00000069399/ENSG00000165861/ENSG00000116473
GO:0022625 ENSG00000163923/ENSG00000037241/ENSG00000173545/ENSG00000082516
GO:0005913 ENSG00000173482/ENSG00000044115/ENSG00000138594/ENSG00000182718/ENSG00000179242/ENSG00000169862/ENSG00000163110/ENSG00000196923/ENSG00000173801/ENSG00000134982/ENSG00000100345/ENSG00000110321/ENSG00000113361/ENSG00000148498/ENSG00000166689/ENSG00000172531/ENSG00000084733/ENSG00000183230/ENSG00000107438/ENSG00000198561/ENSG00000111057/ENSG00000149115/ENSG00000144283
GO:0000776 ENSG00000167513/ENSG00000094916/ENSG00000185122/ENSG00000153187/ENSG00000166851/ENSG00000110713/ENSG00000122545/ENSG00000085415/ENSG00000134982/ENSG00000101265/ENSG00000134057/ENSG00000117724/ENSG00000069248/ENSG00000080986/ENSG00000132912/ENSG00000076382/ENSG00000143228/ENSG00000166579/ENSG00000145919/ENSG00000153044/ENSG00000074054/ENSG00000088986/ENSG00000082898
GO:0005581 ENSG00000163359/ENSG00000160339/ENSG00000168542/ENSG00000139329/ENSG00000134871/ENSG00000164932
GO:0034704 ENSG00000151062/ENSG00000151067/ENSG00000113448/ENSG00000119782/ENSG00000198668/ENSG00000141837/ENSG00000179397/ENSG00000051108/ENSG00000165995/ENSG00000153956
GO:0031304 ENSG00000144120/ENSG00000125454/ENSG00000050393/ENSG00000133028/ENSG00000175701/ENSG00000183978/ENSG00000178307/ENSG00000007923
GO:0031305 ENSG00000144120/ENSG00000125454/ENSG00000050393/ENSG00000133028/ENSG00000175701/ENSG00000183978/ENSG00000178307/ENSG00000007923
GO:0005743 ENSG00000169682/ENSG00000130159/ENSG00000144120/ENSG00000174032/ENSG00000136521/ENSG00000259494/ENSG00000135900/ENSG00000108528/ENSG00000125454/ENSG00000120333/ENSG00000099795/ENSG00000050393/ENSG00000133028/ENSG00000144029/ENSG00000175701/ENSG00000101290/ENSG00000082515/ENSG00000174886/ENSG00000131143/ENSG00000143314/ENSG00000100575/ENSG00000198836/ENSG00000004864/ENSG00000149792/ENSG00000172590/ENSG00000115159/ENSG00000166902/ENSG00000111639/ENSG00000183648/ENSG00000243147/ENSG00000115468/ENSG00000163050/ENSG00000183978/ENSG00000215021/ENSG00000105364/ENSG00000167283/ENSG00000173786/ENSG00000136522/ENSG00000178307/ENSG00000150779/ENSG00000204822/ENSG00000007923/ENSG00000075336/ENSG00000167863/ENSG00000084092/ENSG00000023228/ENSG00000168924/ENSG00000131981/ENSG00000185633/ENSG00000176340/ENSG00000140990/ENSG00000179091/ENSG00000167136/ENSG00000197912/ENSG00000145494
GO:0030496 ENSG00000167461/ENSG00000153187/ENSG00000170312/ENSG00000044574/ENSG00000182718/ENSG00000166851/ENSG00000115266/ENSG00000072121/ENSG00000122545/ENSG00000180096/ENSG00000110697/ENSG00000119541/ENSG00000122966/ENSG00000133026/ENSG00000117724/ENSG00000065243/ENSG00000134108/ENSG00000100154/ENSG00000076382/ENSG00000105146/ENSG00000135763/ENSG00000087586/ENSG00000021574/ENSG00000090565/ENSG00000165660/ENSG00000112984/ENSG00000069399/ENSG00000140854/ENSG00000123143/ENSG00000127445/ENSG00000084112/ENSG00000198561
GO:0005891 ENSG00000151062/ENSG00000151067/ENSG00000113448/ENSG00000141837/ENSG00000179397/ENSG00000165995/ENSG00000153956
GO:0005795 ENSG00000166557/ENSG00000151693/ENSG00000092108/ENSG00000138768/ENSG00000158850/ENSG00000198964/ENSG00000158470/ENSG00000112893/ENSG00000105538/ENSG00000139266/ENSG00000110697/ENSG00000058799/ENSG00000122966/ENSG00000171302/ENSG00000117411/ENSG00000165861/ENSG00000196562/ENSG00000103056/ENSG00000086598/ENSG00000205155/ENSG00000134243/ENSG00000115540/ENSG00000011009/ENSG00000103978/ENSG00000137642/ENSG00000108587
GO:0005834 ENSG00000186469/ENSG00000114353/ENSG00000078369/ENSG00000088256/ENSG00000060558/ENSG00000120063/ENSG00000128266/ENSG00000065135/ENSG00000156049
GO:1905360 ENSG00000186469/ENSG00000114353/ENSG00000078369/ENSG00000088256/ENSG00000060558/ENSG00000120063/ENSG00000128266/ENSG00000065135/ENSG00000156049
GO:0031985 ENSG00000166557/ENSG00000151693/ENSG00000092108/ENSG00000158850/ENSG00000198964/ENSG00000158470/ENSG00000112893/ENSG00000110697/ENSG00000058799/ENSG00000122966/ENSG00000171302/ENSG00000117411/ENSG00000103056/ENSG00000086598/ENSG00000205155/ENSG00000134243/ENSG00000115540/ENSG00000103978/ENSG00000137642/ENSG00000108587
GO:0000922 ENSG00000185122/ENSG00000153187/ENSG00000166851/ENSG00000099910/ENSG00000115841/ENSG00000258890/ENSG00000119541/ENSG00000109103/ENSG00000073910/ENSG00000134057/ENSG00000117724/ENSG00000198668/ENSG00000125898/ENSG00000121957/ENSG00000132912/ENSG00000100154/ENSG00000188807/ENSG00000076382/ENSG00000105146/ENSG00000096093/ENSG00000066629/ENSG00000087586/ENSG00000145919/ENSG00000165660/ENSG00000140854
GO:0044391 ENSG00000163923/ENSG00000037241/ENSG00000173545/ENSG00000259494/ENSG00000135900/ENSG00000082516/ENSG00000120333/ENSG00000144029/ENSG00000156928/ENSG00000082515/ENSG00000143314/ENSG00000149792/ENSG00000172590/ENSG00000166902/ENSG00000111639/ENSG00000243147/ENSG00000105364/ENSG00000136522/ENSG00000174444/ENSG00000204822
GO:0016342 ENSG00000044115/ENSG00000179242/ENSG00000115266/ENSG00000173801/ENSG00000134982
GO:0043292 ENSG00000133392/ENSG00000168743/ENSG00000074695/ENSG00000176749/ENSG00000184009/ENSG00000138594/ENSG00000183023/ENSG00000122786/ENSG00000080709/ENSG00000143318/ENSG00000151067/ENSG00000073712/ENSG00000133454/ENSG00000143632/ENSG00000173801/ENSG00000133028/ENSG00000239474/ENSG00000119782/ENSG00000198668/ENSG00000197702/ENSG00000138347/ENSG00000120907/ENSG00000173991/ENSG00000132912/ENSG00000171992/ENSG00000155657/ENSG00000077549/ENSG00000158022/ENSG00000124942/ENSG00000118194/ENSG00000156804/ENSG00000172403/ENSG00000107438/ENSG00000130956/ENSG00000134775
GO:0098687 ENSG00000167513/ENSG00000102974/ENSG00000094916/ENSG00000185122/ENSG00000153187/ENSG00000170312/ENSG00000183765/ENSG00000083642/ENSG00000166851/ENSG00000110713/ENSG00000099284/ENSG00000104885/ENSG00000160957/ENSG00000122545/ENSG00000085415/ENSG00000051180/ENSG00000010803/ENSG00000134982/ENSG00000101265/ENSG00000173559/ENSG00000134057/ENSG00000117724/ENSG00000069248/ENSG00000080986/ENSG00000109534/ENSG00000139579/ENSG00000152457/ENSG00000132912/ENSG00000076382/ENSG00000100109/ENSG00000139351/ENSG00000105146/ENSG00000143228/ENSG00000172531/ENSG00000166579/ENSG00000087586/ENSG00000102977/ENSG00000145919/ENSG00000175054/ENSG00000167986/ENSG00000204569/ENSG00000153044/ENSG00000138385/ENSG00000092330/ENSG00000062822/ENSG00000074054/ENSG00000164091/ENSG00000088986/ENSG00000082898/ENSG00000132475/ENSG00000107077/ENSG00000203760/ENSG00000113575/ENSG00000130779/ENSG00000070366/ENSG00000133247/ENSG00000124216/ENSG00000146670/ENSG00000030066/ENSG00000132646/ENSG00000156831/ENSG00000062650/ENSG00000107672/ENSG00000179051
GO:0031514 ENSG00000154040/ENSG00000109062/ENSG00000140526/ENSG00000135951/ENSG00000158023/ENSG00000061656/ENSG00000122545/ENSG00000105948/ENSG00000166578/ENSG00000163001/ENSG00000144061/ENSG00000179397/ENSG00000132321/ENSG00000133393/ENSG00000169379/ENSG00000111319/ENSG00000137075/ENSG00000225697
GO:0044429 ENSG00000178952/ENSG00000111058/ENSG00000170312/ENSG00000169682/ENSG00000151498/ENSG00000130159/ENSG00000150991/ENSG00000144120/ENSG00000068615/ENSG00000129003/ENSG00000143318/ENSG00000174032/ENSG00000136521/ENSG00000259494/ENSG00000135900/ENSG00000108528/ENSG00000125454/ENSG00000162851/ENSG00000120942/ENSG00000120333/ENSG00000099795/ENSG00000050393/ENSG00000133028/ENSG00000187838/ENSG00000051180/ENSG00000144029/ENSG00000183486/ENSG00000175701/ENSG00000156928/ENSG00000174840/ENSG00000101290/ENSG00000133943/ENSG00000082515/ENSG00000174886/ENSG00000131143/ENSG00000143314/ENSG00000165672/ENSG00000100575/ENSG00000104936/ENSG00000198836/ENSG00000104765/ENSG00000135423/ENSG00000104381/ENSG00000026652/ENSG00000161653/ENSG00000188266/ENSG00000004864/ENSG00000134057/ENSG00000149792/ENSG00000153094/ENSG00000172590/ENSG00000087470/ENSG00000115159/ENSG00000198668/ENSG00000166902/ENSG00000111639/ENSG00000183648/ENSG00000131844/ENSG00000142168/ENSG00000107819/ENSG00000177192/ENSG00000162396/ENSG00000243147/ENSG00000115468/ENSG00000177034/ENSG00000163050/ENSG00000075240/ENSG00000166575/ENSG00000183978/ENSG00000120254/ENSG00000162433/ENSG00000215021/ENSG00000127554/ENSG00000105364/ENSG00000103018/ENSG00000132155/ENSG00000167283/ENSG00000173786/ENSG00000233276/ENSG00000136522/ENSG00000143384/ENSG00000178307/ENSG00000150779/ENSG00000140374/ENSG00000204822/ENSG00000137393/ENSG00000007923/ENSG00000213995/ENSG00000075336/ENSG00000080819/ENSG00000148187/ENSG00000167863/ENSG00000091483/ENSG00000170791/ENSG00000084092/ENSG00000151576/ENSG00000023228/ENSG00000168924/ENSG00000130707/ENSG00000131981/ENSG00000213585/ENSG00000135537/ENSG00000185633/ENSG00000176340/ENSG00000140990/ENSG00000179091/ENSG00000167136/ENSG00000197912/ENSG00000213339/ENSG00000160691/ENSG00000145982/ENSG00000145494/ENSG00000168393/ENSG00000164306/ENSG00000151376/ENSG00000108561/ENSG00000118777/ENSG00000155189/ENSG00000130304/ENSG00000170502
GO:0005911 ENSG00000113758/ENSG00000158769/ENSG00000157404/ENSG00000173482/ENSG00000044115/ENSG00000138594/ENSG00000183023/ENSG00000163399/ENSG00000182718/ENSG00000179242/ENSG00000082397/ENSG00000179820/ENSG00000169862/ENSG00000163110/ENSG00000196923/ENSG00000147649/ENSG00000115020/ENSG00000173801/ENSG00000114019/ENSG00000158092/ENSG00000134982/ENSG00000187391/ENSG00000160593/ENSG00000124171/ENSG00000156453/ENSG00000100345/ENSG00000116539/ENSG00000144061/ENSG00000114796/ENSG00000100266/ENSG00000043462/ENSG00000123700/ENSG00000110321/ENSG00000181790/ENSG00000160460/ENSG00000143776/ENSG00000065243/ENSG00000103494/ENSG00000105707/ENSG00000128298/ENSG00000105443/ENSG00000171992/ENSG00000113361/ENSG00000148498/ENSG00000166689/ENSG00000172531/ENSG00000124942/ENSG00000084733/ENSG00000163531/ENSG00000157483/ENSG00000183230/ENSG00000113263/ENSG00000107438/ENSG00000060656/ENSG00000198561/ENSG00000116584/ENSG00000127314/ENSG00000111057/ENSG00000149115/ENSG00000089041
GO:0071005 ENSG00000125870/ENSG00000113141/ENSG00000174231/ENSG00000169976/ENSG00000115524/ENSG00000175467/ENSG00000146007/ENSG00000117360/ENSG00000130520
GO:0071011 ENSG00000125870/ENSG00000113141/ENSG00000174231/ENSG00000169976/ENSG00000115524/ENSG00000175467/ENSG00000146007/ENSG00000117360/ENSG00000130520
GO:0030134 ENSG00000166557/ENSG00000204642/ENSG00000074695/ENSG00000138768/ENSG00000151532/ENSG00000100528/ENSG00000099203/ENSG00000113615/ENSG00000167645
GO:0005740 ENSG00000169682/ENSG00000130159/ENSG00000150991/ENSG00000144120/ENSG00000068615/ENSG00000129003/ENSG00000174032/ENSG00000136521/ENSG00000259494/ENSG00000135900/ENSG00000108528/ENSG00000125454/ENSG00000120942/ENSG00000120333/ENSG00000099795/ENSG00000050393/ENSG00000133028/ENSG00000187838/ENSG00000144029/ENSG00000183486/ENSG00000175701/ENSG00000101290/ENSG00000082515/ENSG00000174886/ENSG00000131143/ENSG00000143314/ENSG00000100575/ENSG00000104936/ENSG00000198836/ENSG00000104765/ENSG00000104381/ENSG00000026652/ENSG00000004864/ENSG00000149792/ENSG00000153094/ENSG00000172590/ENSG00000087470/ENSG00000115159/ENSG00000198668/ENSG00000166902/ENSG00000111639/ENSG00000183648/ENSG00000142168/ENSG00000107819/ENSG00000243147/ENSG00000115468/ENSG00000177034/ENSG00000163050/ENSG00000075240/ENSG00000166575/ENSG00000183978/ENSG00000215021/ENSG00000127554/ENSG00000105364/ENSG00000103018/ENSG00000132155/ENSG00000167283/ENSG00000173786/ENSG00000136522/ENSG00000143384/ENSG00000178307/ENSG00000150779/ENSG00000204822/ENSG00000137393/ENSG00000007923/ENSG00000075336/ENSG00000080819/ENSG00000167863/ENSG00000170791/ENSG00000084092/ENSG00000151576/ENSG00000023228/ENSG00000168924/ENSG00000130707/ENSG00000131981/ENSG00000213585/ENSG00000135537/ENSG00000185633/ENSG00000176340/ENSG00000140990/ENSG00000179091/ENSG00000167136/ENSG00000197912/ENSG00000213339/ENSG00000145494/ENSG00000168393/ENSG00000118777/ENSG00000155189/ENSG00000130304
GO:0031966 ENSG00000169682/ENSG00000130159/ENSG00000150991/ENSG00000144120/ENSG00000068615/ENSG00000129003/ENSG00000174032/ENSG00000136521/ENSG00000259494/ENSG00000135900/ENSG00000108528/ENSG00000125454/ENSG00000120942/ENSG00000120333/ENSG00000099795/ENSG00000050393/ENSG00000133028/ENSG00000187838/ENSG00000144029/ENSG00000183486/ENSG00000175701/ENSG00000101290/ENSG00000082515/ENSG00000174886/ENSG00000131143/ENSG00000143314/ENSG00000100575/ENSG00000104936/ENSG00000198836/ENSG00000104765/ENSG00000104381/ENSG00000026652/ENSG00000004864/ENSG00000149792/ENSG00000153094/ENSG00000172590/ENSG00000087470/ENSG00000115159/ENSG00000198668/ENSG00000166902/ENSG00000111639/ENSG00000183648/ENSG00000107819/ENSG00000243147/ENSG00000115468/ENSG00000177034/ENSG00000163050/ENSG00000075240/ENSG00000166575/ENSG00000183978/ENSG00000215021/ENSG00000105364/ENSG00000103018/ENSG00000132155/ENSG00000167283/ENSG00000173786/ENSG00000136522/ENSG00000143384/ENSG00000178307/ENSG00000150779/ENSG00000204822/ENSG00000137393/ENSG00000007923/ENSG00000075336/ENSG00000167863/ENSG00000084092/ENSG00000151576/ENSG00000023228/ENSG00000168924/ENSG00000130707/ENSG00000131981/ENSG00000213585/ENSG00000135537/ENSG00000185633/ENSG00000176340/ENSG00000140990/ENSG00000179091/ENSG00000167136/ENSG00000197912/ENSG00000213339/ENSG00000145494/ENSG00000118777/ENSG00000155189/ENSG00000130304
GO:0005776 ENSG00000126775/ENSG00000006025/ENSG00000197386/ENSG00000151532/ENSG00000131374/ENSG00000132405/ENSG00000135018/ENSG00000104142/ENSG00000165861/ENSG00000132109/ENSG00000118705/ENSG00000167996/ENSG00000078804
GO:0031258 ENSG00000073712/ENSG00000115266/ENSG00000141424
GO:0000775 ENSG00000167513/ENSG00000102974/ENSG00000094916/ENSG00000185122/ENSG00000153187/ENSG00000083642/ENSG00000166851/ENSG00000110713/ENSG00000122545/ENSG00000085415/ENSG00000134982/ENSG00000101265/ENSG00000134057/ENSG00000117724/ENSG00000069248/ENSG00000080986/ENSG00000132912/ENSG00000076382/ENSG00000139351/ENSG00000105146/ENSG00000143228/ENSG00000166579/ENSG00000087586/ENSG00000145919/ENSG00000153044/ENSG00000074054/ENSG00000088986/ENSG00000082898/ENSG00000107077/ENSG00000203760/ENSG00000113575/ENSG00000130779/ENSG00000133247/ENSG00000124216/ENSG00000146670/ENSG00000030066
GO:1905368 ENSG00000161057/ENSG00000136930/ENSG00000106290/ENSG00000119318/ENSG00000135018/ENSG00000176476/ENSG00000114062/ENSG00000101557/ENSG00000100519/ENSG00000108671/ENSG00000173692/ENSG00000142507/ENSG00000162227/ENSG00000197170/ENSG00000129128/ENSG00000179262/ENSG00000068878/ENSG00000013275/ENSG00000197912/ENSG00000101182/ENSG00000114902/ENSG00000087152/ENSG00000100567
GO:0033017 ENSG00000143318/ENSG00000239474/ENSG00000104936/ENSG00000198363/ENSG00000119782/ENSG00000110108
GO:0016605 ENSG00000171862/ENSG00000094916/ENSG00000185122/ENSG00000159208/ENSG00000183765/ENSG00000109906/ENSG00000051180/ENSG00000148737/ENSG00000188612/ENSG00000166889
GO:0019866 ENSG00000169682/ENSG00000130159/ENSG00000144120/ENSG00000174032/ENSG00000136521/ENSG00000259494/ENSG00000061656/ENSG00000135900/ENSG00000108528/ENSG00000125454/ENSG00000120333/ENSG00000099795/ENSG00000050393/ENSG00000133028/ENSG00000144029/ENSG00000175701/ENSG00000101290/ENSG00000082515/ENSG00000174886/ENSG00000131143/ENSG00000143314/ENSG00000100575/ENSG00000198836/ENSG00000004864/ENSG00000149792/ENSG00000172590/ENSG00000115159/ENSG00000166902/ENSG00000111639/ENSG00000183648/ENSG00000243147/ENSG00000115468/ENSG00000163050/ENSG00000183978/ENSG00000188807/ENSG00000215021/ENSG00000105364/ENSG00000167283/ENSG00000173786/ENSG00000136522/ENSG00000178307/ENSG00000150779/ENSG00000204822/ENSG00000007923/ENSG00000075336/ENSG00000167863/ENSG00000084092/ENSG00000023228/ENSG00000168924/ENSG00000131981/ENSG00000089041/ENSG00000185633/ENSG00000176340/ENSG00000140990/ENSG00000179091/ENSG00000167136/ENSG00000197912/ENSG00000145494
GO:0012507 ENSG00000204642/ENSG00000074695/ENSG00000138768/ENSG00000151532/ENSG00000100528/ENSG00000113615
GO:0031301 ENSG00000127995/ENSG00000180776/ENSG00000144120/ENSG00000198964/ENSG00000125454/ENSG00000120942/ENSG00000050393/ENSG00000133028/ENSG00000157593/ENSG00000175701/ENSG00000104936/ENSG00000104381/ENSG00000204070/ENSG00000156671/ENSG00000160613/ENSG00000167645/ENSG00000183978/ENSG00000188807/ENSG00000100060/ENSG00000178307
GO:0042641 ENSG00000113758/ENSG00000073712/ENSG00000086991/ENSG00000122545/ENSG00000196923/ENSG00000143632/ENSG00000100345/ENSG00000133026/ENSG00000143776/ENSG00000132912/ENSG00000171992
GO:0005840 ENSG00000163923/ENSG00000037241/ENSG00000172071/ENSG00000173545/ENSG00000259494/ENSG00000135900/ENSG00000082516/ENSG00000120333/ENSG00000144029/ENSG00000156928/ENSG00000158092/ENSG00000082515/ENSG00000143314/ENSG00000198498/ENSG00000149792/ENSG00000172590/ENSG00000166902/ENSG00000111639/ENSG00000243147/ENSG00000105364/ENSG00000136522/ENSG00000174444/ENSG00000198208/ENSG00000204822
GO:0005778 ENSG00000115204/ENSG00000162735/ENSG00000160097/ENSG00000121680/ENSG00000133835/ENSG00000126934/ENSG00000113161/ENSG00000108733/ENSG00000138138/ENSG00000164398/ENSG00000107897/ENSG00000164751/ENSG00000116906/ENSG00000166128/ENSG00000178035/ENSG00000064763/ENSG00000166821/ENSG00000119688/ENSG00000162928/ENSG00000131779
GO:0031903 ENSG00000115204/ENSG00000162735/ENSG00000160097/ENSG00000121680/ENSG00000133835/ENSG00000126934/ENSG00000113161/ENSG00000108733/ENSG00000138138/ENSG00000164398/ENSG00000107897/ENSG00000164751/ENSG00000116906/ENSG00000166128/ENSG00000178035/ENSG00000064763/ENSG00000166821/ENSG00000119688/ENSG00000162928/ENSG00000131779
GO:0022626 ENSG00000163923/ENSG00000037241/ENSG00000172071/ENSG00000173545/ENSG00000082516
GO:0032432 ENSG00000183688/ENSG00000073712/ENSG00000086991/ENSG00000122545/ENSG00000196923/ENSG00000143632/ENSG00000100345/ENSG00000133026/ENSG00000132912/ENSG00000171992
GO:0032592 ENSG00000144120/ENSG00000125454/ENSG00000050393/ENSG00000133028/ENSG00000175701/ENSG00000104936/ENSG00000104381/ENSG00000183978/ENSG00000178307/ENSG00000007923
GO:0098573 ENSG00000144120/ENSG00000125454/ENSG00000050393/ENSG00000133028/ENSG00000175701/ENSG00000104936/ENSG00000104381/ENSG00000183978/ENSG00000178307/ENSG00000007923
GO:0000780 ENSG00000166851/ENSG00000134057/ENSG00000080986/ENSG00000105146/ENSG00000143228/ENSG00000087586
GO:0016592 ENSG00000164758/ENSG00000108590/ENSG00000108510/ENSG00000112237/ENSG00000135077/ENSG00000155868/ENSG00000239306/ENSG00000155846
GO:0016324 ENSG00000116183/ENSG00000133116/ENSG00000109062/ENSG00000171862/ENSG00000130303/ENSG00000163399/ENSG00000086991/ENSG00000163803/ENSG00000080031/ENSG00000122545/ENSG00000113448/ENSG00000174059/ENSG00000147649/ENSG00000167434/ENSG00000232434/ENSG00000114019/ENSG00000124171/ENSG00000136153/ENSG00000065361/ENSG00000171631/ENSG00000154096/ENSG00000105707/ENSG00000101384/ENSG00000148498/ENSG00000091513/ENSG00000145623/ENSG00000164930/ENSG00000124839/ENSG00000111319/ENSG00000174827/ENSG00000131620/ENSG00000110719/ENSG00000225697/ENSG00000154310/ENSG00000162738/ENSG00000188931/ENSG00000118777/ENSG00000092820/ENSG00000010278/ENSG00000116266/ENSG00000067606/ENSG00000196961/ENSG00000116039/ENSG00000085563/ENSG00000110911/ENSG00000111913/ENSG00000197375/ENSG00000169398
GO:0016529 ENSG00000143318/ENSG00000137801/ENSG00000239474/ENSG00000104936/ENSG00000198363/ENSG00000119782/ENSG00000140939/ENSG00000110108/ENSG00000179218/ENSG00000153956
GO:0043296 ENSG00000158769/ENSG00000044115/ENSG00000147649/ENSG00000173801/ENSG00000114019/ENSG00000134982/ENSG00000187391/ENSG00000160593/ENSG00000124171/ENSG00000116539/ENSG00000144061/ENSG00000065243/ENSG00000103494/ENSG00000105443/ENSG00000171992/ENSG00000148498/ENSG00000166689
GO:0015629 ENSG00000183688/ENSG00000109062/ENSG00000133392/ENSG00000113758/ENSG00000044115/ENSG00000088727/ENSG00000184009/ENSG00000138594/ENSG00000122786/ENSG00000073712/ENSG00000179820/ENSG00000115266/ENSG00000163110/ENSG00000086991/ENSG00000105887/ENSG00000198771/ENSG00000103343/ENSG00000122545/ENSG00000110031/ENSG00000196923/ENSG00000133454/ENSG00000143632/ENSG00000152818/ENSG00000173801/ENSG00000119729/ENSG00000134909/ENSG00000100345/ENSG00000066933/ENSG00000164741/ENSG00000133026/ENSG00000113407/ENSG00000104142/ENSG00000149930/ENSG00000197702/ENSG00000160460/ENSG00000143776/ENSG00000128298/ENSG00000111229/ENSG00000163171/ENSG00000132912/ENSG00000077063/ENSG00000128641/ENSG00000171992/ENSG00000132613/ENSG00000211455/ENSG00000155657/ENSG00000128487/ENSG00000136950/ENSG00000077549/ENSG00000167193/ENSG00000113108/ENSG00000124942/ENSG00000118194/ENSG00000167306/ENSG00000145555/ENSG00000157483/ENSG00000119523/ENSG00000124160/ENSG00000172403/ENSG00000112679/ENSG00000116679/ENSG00000116584/ENSG00000134775
GO:0032839 ENSG00000153187/ENSG00000164418/ENSG00000139826/ENSG00000196839/ENSG00000142168
GO:0098794 ENSG00000161057/ENSG00000167461/ENSG00000171862/ENSG00000113758/ENSG00000171208/ENSG00000117600/ENSG00000052126/ENSG00000176749/ENSG00000183023/ENSG00000163399/ENSG00000080709/ENSG00000197386/ENSG00000151067/ENSG00000082397/ENSG00000101040/ENSG00000162946/ENSG00000131409/ENSG00000115266/ENSG00000107404/ENSG00000169862/ENSG00000163110/ENSG00000164418/ENSG00000152767/ENSG00000118160/ENSG00000140992/ENSG00000066044/ENSG00000141543/ENSG00000152818/ENSG00000131188/ENSG00000187391/ENSG00000134909/ENSG00000112851/ENSG00000165802/ENSG00000137845/ENSG00000133026/ENSG00000087470/ENSG00000123700/ENSG00000181790/ENSG00000136854/ENSG00000096968/ENSG00000183783/ENSG00000120907/ENSG00000166313/ENSG00000077063/ENSG00000099260/ENSG00000171992/ENSG00000197555/ENSG00000034677
GO:0005640 ENSG00000104936/ENSG00000110108/ENSG00000165959/ENSG00000067798/ENSG00000176438/ENSG00000224051/ENSG00000070081/ENSG00000131018
GO:0005912 ENSG00000162434/ENSG00000173482/ENSG00000044115/ENSG00000044574/ENSG00000061938/ENSG00000184009/ENSG00000079308/ENSG00000138594/ENSG00000182718/ENSG00000179242/ENSG00000073712/ENSG00000132470/ENSG00000169862/ENSG00000163110/ENSG00000086991/ENSG00000131016/ENSG00000213949/ENSG00000105974/ENSG00000140992/ENSG00000110031/ENSG00000196923/ENSG00000110841/ENSG00000136205/ENSG00000173801/ENSG00000109270/ENSG00000232434/ENSG00000134982/ENSG00000100345/ENSG00000136153/ENSG00000115232/ENSG00000164741/ENSG00000137845/ENSG00000159176/ENSG00000144061/ENSG00000114796/ENSG00000100266/ENSG00000197702/ENSG00000110321/ENSG00000181790/ENSG00000160460/ENSG00000154096/ENSG00000150764/ENSG00000076706/ENSG00000111229/ENSG00000105443/ENSG00000096968/ENSG00000137809/ENSG00000101384/ENSG00000135269/ENSG00000132912/ENSG00000113361/ENSG00000074696/ENSG00000136950/ENSG00000148498/ENSG00000140691/ENSG00000174444/ENSG00000162736/ENSG00000133818/ENSG00000166689/ENSG00000149428/ENSG00000172531/ENSG00000179218/ENSG00000082781/ENSG00000124942/ENSG00000170017/ENSG00000084733/ENSG00000163531/ENSG00000081237/ENSG00000157483/ENSG00000145819/ENSG00000183230/ENSG00000172403/ENSG00000107438/ENSG00000198561/ENSG00000244486/ENSG00000116584/ENSG00000167004/ENSG00000111057/ENSG00000108518/ENSG00000149115/ENSG00000074054/ENSG00000185621/ENSG00000213741
GO:0030135 ENSG00000078401/ENSG00000089818/ENSG00000167461/ENSG00000166557/ENSG00000204642/ENSG00000072274/ENSG00000119844/ENSG00000074695/ENSG00000138768/ENSG00000061938/ENSG00000113282/ENSG00000151532/ENSG00000107404/ENSG00000131374/ENSG00000104142/ENSG00000100528/ENSG00000099203/ENSG00000130147/ENSG00000113615/ENSG00000092929/ENSG00000167645/ENSG00000091513/ENSG00000100280/ENSG00000189195/ENSG00000157483/ENSG00000138798/ENSG00000095139/ENSG00000091622/ENSG00000105649/ENSG00000213047/ENSG00000105438/ENSG00000148219/ENSG00000086598/ENSG00000134243/ENSG00000143858/ENSG00000130203/ENSG00000141367/ENSG00000162738
GO:0030315 ENSG00000183023/ENSG00000163399/ENSG00000143318/ENSG00000151067/ENSG00000123700/ENSG00000120907/ENSG00000124942/ENSG00000165995/ENSG00000153956/ENSG00000119699/ENSG00000092820
GO:0032580 ENSG00000166557/ENSG00000151693/ENSG00000092108/ENSG00000158850/ENSG00000158470/ENSG00000110697/ENSG00000171302/ENSG00000117411/ENSG00000086598/ENSG00000205155/ENSG00000134243/ENSG00000115540
GO:0005798 ENSG00000167461/ENSG00000166557/ENSG00000204642/ENSG00000119844/ENSG00000074695/ENSG00000092108/ENSG00000138768/ENSG00000151532/ENSG00000109220/ENSG00000119729/ENSG00000137845/ENSG00000100528/ENSG00000099203/ENSG00000113615/ENSG00000167645
GO:0031012 ENSG00000163359/ENSG00000100985/ENSG00000164342/ENSG00000168056/ENSG00000168743/ENSG00000168542/ENSG00000187720/ENSG00000074695/ENSG00000166833/ENSG00000139329/ENSG00000123243/ENSG00000182718/ENSG00000133110/ENSG00000137801/ENSG00000127083/ENSG00000151388/ENSG00000159674/ENSG00000134871/ENSG00000106366/ENSG00000164932/ENSG00000112851/ENSG00000132561/ENSG00000168140/ENSG00000105825/ENSG00000038427/ENSG00000142168/ENSG00000142910/ENSG00000174807/ENSG00000170382/ENSG00000092969/ENSG00000064309/ENSG00000082293/ENSG00000163661/ENSG00000154175/ENSG00000214274/ENSG00000100109/ENSG00000163638/ENSG00000049323/ENSG00000173868/ENSG00000179218/ENSG00000118523
GO:0045171 ENSG00000115266/ENSG00000174059/ENSG00000109103/ENSG00000173153/ENSG00000214367/ENSG00000168286/ENSG00000104881/ENSG00000090565/ENSG00000112984/ENSG00000142875
GO:0005874 ENSG00000066735/ENSG00000153187/ENSG00000170312/ENSG00000176749/ENSG00000088727/ENSG00000183023/ENSG00000166851/ENSG00000162946/ENSG00000099910/ENSG00000115266/ENSG00000107404/ENSG00000070761/ENSG00000155324/ENSG00000115841/ENSG00000141376/ENSG00000142867/ENSG00000040341/ENSG00000198718/ENSG00000134982/ENSG00000101162/ENSG00000087470/ENSG00000120694/ENSG00000198668/ENSG00000119661/ENSG00000117632/ENSG00000103494/ENSG00000100271/ENSG00000151849/ENSG00000076382/ENSG00000214367/ENSG00000105146/ENSG00000249115/ENSG00000067798/ENSG00000171368/ENSG00000158022/ENSG00000149557/ENSG00000188229/ENSG00000162849/ENSG00000066629/ENSG00000166579/ENSG00000087586/ENSG00000197892/ENSG00000021574/ENSG00000145919/ENSG00000107581/ENSG00000165660/ENSG00000112984/ENSG00000140854
GO:0030054 ENSG00000162434/ENSG00000113758/ENSG00000158769/ENSG00000157404/ENSG00000055163/ENSG00000173482/ENSG00000044115/ENSG00000044574/ENSG00000061938/ENSG00000184009/ENSG00000079308/ENSG00000138594/ENSG00000183023/ENSG00000163399/ENSG00000182718/ENSG00000151067/ENSG00000179242/ENSG00000082397/ENSG00000073712/ENSG00000259494/ENSG00000179820/ENSG00000162946/ENSG00000131409/ENSG00000132470/ENSG00000169862/ENSG00000163110/ENSG00000086991/ENSG00000164418/ENSG00000152767/ENSG00000131016/ENSG00000213949/ENSG00000105974/ENSG00000140992/ENSG00000110031/ENSG00000196923/ENSG00000147649/ENSG00000110841/ENSG00000115020/ENSG00000136205/ENSG00000152818/ENSG00000173801/ENSG00000131188/ENSG00000109270/ENSG00000232434/ENSG00000175920/ENSG00000114019/ENSG00000112137/ENSG00000158092/ENSG00000134982/ENSG00000187391/ENSG00000160593/ENSG00000124171/ENSG00000196839/ENSG00000216490/ENSG00000119899/ENSG00000134909/ENSG00000112851/ENSG00000156453/ENSG00000100345/ENSG00000136153/ENSG00000165802/ENSG00000115232/ENSG00000164741/ENSG00000137845/ENSG00000116539/ENSG00000159176/ENSG00000144061/ENSG00000087470/ENSG00000114796/ENSG00000100266/ENSG00000043462/ENSG00000134769/ENSG00000069188/ENSG00000123700/ENSG00000197702/ENSG00000110321/ENSG00000181790/ENSG00000135077/ENSG00000160460/ENSG00000154096/ENSG00000150764/ENSG00000143776/ENSG00000065243/ENSG00000103494/ENSG00000105707/ENSG00000128298/ENSG00000076706/ENSG00000111229/ENSG00000105443/ENSG00000096968/ENSG00000183783/ENSG00000137809/ENSG00000101384/ENSG00000135269/ENSG00000132912/ENSG00000130558/ENSG00000125651/ENSG00000171992/ENSG00000197555/ENSG00000113361/ENSG00000074696/ENSG00000136950/ENSG00000148498/ENSG00000160469/ENSG00000140691/ENSG00000174444/ENSG00000162736/ENSG00000133818/ENSG00000166689/ENSG00000149428/ENSG00000172531/ENSG00000179218/ENSG00000006712/ENSG00000104881/ENSG00000082781/ENSG00000189195/ENSG00000124942/ENSG00000170017/ENSG00000162951/ENSG00000116473/ENSG00000084733/ENSG00000163531/ENSG00000081237/ENSG00000157483/ENSG00000145819/ENSG00000183230/ENSG00000172403/ENSG00000183454/ENSG00000113263/ENSG00000107438/ENSG00000060656/ENSG00000198561/ENSG00000244486/ENSG00000116584/ENSG00000167004/ENSG00000127314/ENSG00000111057/ENSG00000108518/ENSG00000149115/ENSG00000074054/ENSG00000108639/ENSG00000089041/ENSG00000124785/ENSG00000185621/ENSG00000213741
GO:0030136 ENSG00000078401/ENSG00000089818/ENSG00000167461/ENSG00000072274/ENSG00000119844/ENSG00000061938/ENSG00000113282/ENSG00000151532/ENSG00000107404/ENSG00000131374
GO:0031967 ENSG00000094916/ENSG00000153132/ENSG00000169682/ENSG00000177932/ENSG00000130159/ENSG00000135951/ENSG00000150991/ENSG00000144120/ENSG00000139496/ENSG00000068615/ENSG00000129003/ENSG00000174032/ENSG00000136521/ENSG00000110713/ENSG00000259494/ENSG00000061656/ENSG00000135900/ENSG00000108528/ENSG00000172292/ENSG00000125454/ENSG00000120942/ENSG00000113448/ENSG00000120333/ENSG00000147649/ENSG00000149547/ENSG00000162231/ENSG00000099795/ENSG00000050393/ENSG00000085415/ENSG00000133028/ENSG00000187838/ENSG00000205339/ENSG00000144029/ENSG00000140299/ENSG00000183486/ENSG00000175701/ENSG00000108506/ENSG00000101290/ENSG00000082515/ENSG00000174886/ENSG00000112851/ENSG00000055917/ENSG00000136153/ENSG00000131143/ENSG00000143314/ENSG00000165802/ENSG00000147457/ENSG00000100575/ENSG00000104936/ENSG00000198836/ENSG00000104765/ENSG00000174574/ENSG00000108559/ENSG00000198700/ENSG00000104381/ENSG00000026652/ENSG00000004864/ENSG00000117724/ENSG00000149792/ENSG00000153094/ENSG00000172590/ENSG00000087470/ENSG00000115159/ENSG00000069248/ENSG00000198668/ENSG00000166902/ENSG00000111639/ENSG00000183648/ENSG00000142168/ENSG00000107819/ENSG00000143418/ENSG00000110108/ENSG00000105707/ENSG00000243147/ENSG00000115468/ENSG00000177034/ENSG00000163050/ENSG00000163577/ENSG00000075240/ENSG00000166575/ENSG00000120907/ENSG00000183978/ENSG00000188807/ENSG00000215021/ENSG00000127554/ENSG00000105364/ENSG00000165959/ENSG00000103018/ENSG00000132155/ENSG00000167283/ENSG00000173786/ENSG00000074696/ENSG00000067798/ENSG00000136522/ENSG00000143384/ENSG00000178307/ENSG00000166579/ENSG00000179218/ENSG00000150779/ENSG00000091732/ENSG00000176438/ENSG00000204822/ENSG00000021574/ENSG00000137393/ENSG00000007923/ENSG00000075336/ENSG00000080819/ENSG00000109133/ENSG00000167863/ENSG00000267680/ENSG00000170214/ENSG00000170791/ENSG00000084092/ENSG00000185085/ENSG00000151576/ENSG00000023228/ENSG00000141448/ENSG00000168924/ENSG00000114030/ENSG00000130707/ENSG00000136770/ENSG00000131981/ENSG00000089041/ENSG00000155561/ENSG00000213585/ENSG00000135537/ENSG00000185633/ENSG00000176340/ENSG00000082898/ENSG00000134243/ENSG00000140990/ENSG00000161217/ENSG00000179091/ENSG00000167136/ENSG00000197912/ENSG00000213339/ENSG00000145494/ENSG00000168393/ENSG00000108349/ENSG00000118777/ENSG00000155189/ENSG00000224051/ENSG00000130304/ENSG00000070081/ENSG00000130779/ENSG00000123358/ENSG00000184060/ENSG00000196497/ENSG00000137038/ENSG00000137642/ENSG00000089163/ENSG00000030066/ENSG00000163904/ENSG00000131018/ENSG00000028116/ENSG00000004455/ENSG00000132591/ENSG00000102738
GO:0031975 ENSG00000094916/ENSG00000153132/ENSG00000169682/ENSG00000177932/ENSG00000130159/ENSG00000135951/ENSG00000150991/ENSG00000144120/ENSG00000139496/ENSG00000068615/ENSG00000129003/ENSG00000174032/ENSG00000136521/ENSG00000110713/ENSG00000259494/ENSG00000061656/ENSG00000135900/ENSG00000108528/ENSG00000172292/ENSG00000125454/ENSG00000120942/ENSG00000113448/ENSG00000120333/ENSG00000147649/ENSG00000149547/ENSG00000162231/ENSG00000099795/ENSG00000050393/ENSG00000085415/ENSG00000133028/ENSG00000187838/ENSG00000205339/ENSG00000144029/ENSG00000140299/ENSG00000183486/ENSG00000175701/ENSG00000108506/ENSG00000101290/ENSG00000082515/ENSG00000174886/ENSG00000112851/ENSG00000055917/ENSG00000136153/ENSG00000131143/ENSG00000143314/ENSG00000165802/ENSG00000147457/ENSG00000100575/ENSG00000104936/ENSG00000198836/ENSG00000104765/ENSG00000174574/ENSG00000108559/ENSG00000198700/ENSG00000104381/ENSG00000026652/ENSG00000004864/ENSG00000117724/ENSG00000149792/ENSG00000153094/ENSG00000172590/ENSG00000087470/ENSG00000115159/ENSG00000069248/ENSG00000198668/ENSG00000166902/ENSG00000111639/ENSG00000183648/ENSG00000142168/ENSG00000107819/ENSG00000143418/ENSG00000110108/ENSG00000105707/ENSG00000243147/ENSG00000115468/ENSG00000177034/ENSG00000163050/ENSG00000163577/ENSG00000075240/ENSG00000166575/ENSG00000120907/ENSG00000183978/ENSG00000188807/ENSG00000215021/ENSG00000127554/ENSG00000105364/ENSG00000165959/ENSG00000103018/ENSG00000132155/ENSG00000167283/ENSG00000173786/ENSG00000074696/ENSG00000067798/ENSG00000136522/ENSG00000143384/ENSG00000178307/ENSG00000166579/ENSG00000179218/ENSG00000150779/ENSG00000091732/ENSG00000176438/ENSG00000204822/ENSG00000021574/ENSG00000137393/ENSG00000007923/ENSG00000075336/ENSG00000080819/ENSG00000109133/ENSG00000167863/ENSG00000267680/ENSG00000170214/ENSG00000170791/ENSG00000084092/ENSG00000185085/ENSG00000151576/ENSG00000023228/ENSG00000141448/ENSG00000168924/ENSG00000114030/ENSG00000130707/ENSG00000136770/ENSG00000131981/ENSG00000089041/ENSG00000155561/ENSG00000213585/ENSG00000135537/ENSG00000185633/ENSG00000176340/ENSG00000082898/ENSG00000134243/ENSG00000140990/ENSG00000161217/ENSG00000179091/ENSG00000167136/ENSG00000197912/ENSG00000213339/ENSG00000145494/ENSG00000168393/ENSG00000108349/ENSG00000118777/ENSG00000155189/ENSG00000224051/ENSG00000130304/ENSG00000070081/ENSG00000130779/ENSG00000123358/ENSG00000184060/ENSG00000196497/ENSG00000137038/ENSG00000137642/ENSG00000089163/ENSG00000030066/ENSG00000163904/ENSG00000131018/ENSG00000028116/ENSG00000004455/ENSG00000132591/ENSG00000102738
GO:0000502 ENSG00000161057/ENSG00000136930/ENSG00000119318/ENSG00000135018/ENSG00000114062/ENSG00000101557/ENSG00000100519/ENSG00000108671/ENSG00000173692/ENSG00000142507/ENSG00000197170/ENSG00000179262/ENSG00000068878/ENSG00000013275/ENSG00000101182/ENSG00000100567
GO:1905369 ENSG00000161057/ENSG00000136930/ENSG00000119318/ENSG00000135018/ENSG00000114062/ENSG00000101557/ENSG00000100519/ENSG00000108671/ENSG00000173692/ENSG00000142507/ENSG00000197170/ENSG00000179262/ENSG00000068878/ENSG00000013275/ENSG00000101182/ENSG00000100567
GO:0019005 ENSG00000174989/ENSG00000107872/ENSG00000162032/ENSG00000159069/ENSG00000127452/ENSG00000145743/ENSG00000175093/ENSG00000132109/ENSG00000156804/ENSG00000153558/ENSG00000100387/ENSG00000112146/ENSG00000166233/ENSG00000151743
GO:0044449 ENSG00000133392/ENSG00000168743/ENSG00000074695/ENSG00000138594/ENSG00000183023/ENSG00000080709/ENSG00000143318/ENSG00000151067/ENSG00000073712/ENSG00000133454/ENSG00000143632/ENSG00000173801/ENSG00000239474/ENSG00000119782/ENSG00000198668/ENSG00000197702/ENSG00000138347/ENSG00000120907/ENSG00000173991/ENSG00000132912/ENSG00000171992/ENSG00000155657/ENSG00000077549/ENSG00000158022/ENSG00000124942/ENSG00000118194/ENSG00000156804/ENSG00000172403/ENSG00000107438/ENSG00000130956/ENSG00000134775
GO:0016528 ENSG00000121769/ENSG00000143318/ENSG00000137801/ENSG00000239474/ENSG00000104936/ENSG00000198363/ENSG00000119782/ENSG00000140939/ENSG00000110108/ENSG00000179218/ENSG00000153956/ENSG00000130956
GO:0099081 ENSG00000163359/ENSG00000066735/ENSG00000133392/ENSG00000168743/ENSG00000168542/ENSG00000153187/ENSG00000187720/ENSG00000074695/ENSG00000170312/ENSG00000139329/ENSG00000176749/ENSG00000088727/ENSG00000184009/ENSG00000138594/ENSG00000183023/ENSG00000122786/ENSG00000080709/ENSG00000143318/ENSG00000166851/ENSG00000151067/ENSG00000073712/ENSG00000162946/ENSG00000099910/ENSG00000115266/ENSG00000107404/ENSG00000070761/ENSG00000198771/ENSG00000155324/ENSG00000115841/ENSG00000141376/ENSG00000133454/ENSG00000143632/ENSG00000142867/ENSG00000173801/ENSG00000133028/ENSG00000040341/ENSG00000198718/ENSG00000134871/ENSG00000134982/ENSG00000119729/ENSG00000239474/ENSG00000101162/ENSG00000108244/ENSG00000087470/ENSG00000119782/ENSG00000120694/ENSG00000198668/ENSG00000119661/ENSG00000104142/ENSG00000117632/ENSG00000197702/ENSG00000103494/ENSG00000111229/ENSG00000100271/ENSG00000138347/ENSG00000120907/ENSG00000173991/ENSG00000132912/ENSG00000128641/ENSG00000151849/ENSG00000171992/ENSG00000076382/ENSG00000214367/ENSG00000105146/ENSG00000249115/ENSG00000155657/ENSG00000128487/ENSG00000067798/ENSG00000077549/ENSG00000171368/ENSG00000158022/ENSG00000149557/ENSG00000049323/ENSG00000188229/ENSG00000162849/ENSG00000066629/ENSG00000166579/ENSG00000087586/ENSG00000197892/ENSG00000021574/ENSG00000124942/ENSG00000145919/ENSG00000118194/ENSG00000107581/ENSG00000165660/ENSG00000112984/ENSG00000140854/ENSG00000125864/ENSG00000156804/ENSG00000172403/ENSG00000112679/ENSG00000107438/ENSG00000130956/ENSG00000116584/ENSG00000134775/ENSG00000165476/ENSG00000111057/ENSG00000141380/ENSG00000074054/ENSG00000204262
GO:0005838 ENSG00000161057/ENSG00000100519/ENSG00000108671/ENSG00000173692/ENSG00000197170
GO:0030016 ENSG00000074695/ENSG00000184009/ENSG00000138594/ENSG00000183023/ENSG00000122786/ENSG00000080709/ENSG00000143318/ENSG00000151067/ENSG00000073712/ENSG00000133454/ENSG00000143632/ENSG00000173801/ENSG00000133028/ENSG00000239474/ENSG00000119782/ENSG00000198668/ENSG00000197702/ENSG00000138347/ENSG00000120907/ENSG00000173991/ENSG00000132912/ENSG00000171992/ENSG00000155657/ENSG00000077549/ENSG00000158022/ENSG00000124942/ENSG00000118194/ENSG00000156804/ENSG00000172403/ENSG00000107438/ENSG00000130956/ENSG00000134775
GO:0030426 ENSG00000113758/ENSG00000176749/ENSG00000107404/ENSG00000100697/ENSG00000152818/ENSG00000144857/ENSG00000115232/ENSG00000121297/ENSG00000133026/ENSG00000198668/ENSG00000149930/ENSG00000154096/ENSG00000111229/ENSG00000105443/ENSG00000166313/ENSG00000130558/ENSG00000165795/ENSG00000214274/ENSG00000148498/ENSG00000166579/ENSG00000162951/ENSG00000140854/ENSG00000084112/ENSG00000198561/ENSG00000124126/ENSG00000099250/ENSG00000106078/ENSG00000151491/ENSG00000119328/ENSG00000079805
GO:0099512 ENSG00000163359/ENSG00000066735/ENSG00000133392/ENSG00000168743/ENSG00000168542/ENSG00000153187/ENSG00000187720/ENSG00000074695/ENSG00000170312/ENSG00000139329/ENSG00000176749/ENSG00000088727/ENSG00000184009/ENSG00000138594/ENSG00000183023/ENSG00000122786/ENSG00000080709/ENSG00000143318/ENSG00000166851/ENSG00000151067/ENSG00000073712/ENSG00000162946/ENSG00000099910/ENSG00000115266/ENSG00000107404/ENSG00000070761/ENSG00000198771/ENSG00000155324/ENSG00000115841/ENSG00000141376/ENSG00000133454/ENSG00000143632/ENSG00000142867/ENSG00000173801/ENSG00000133028/ENSG00000040341/ENSG00000198718/ENSG00000134982/ENSG00000119729/ENSG00000239474/ENSG00000101162/ENSG00000108244/ENSG00000087470/ENSG00000119782/ENSG00000120694/ENSG00000198668/ENSG00000119661/ENSG00000104142/ENSG00000117632/ENSG00000197702/ENSG00000103494/ENSG00000111229/ENSG00000100271/ENSG00000138347/ENSG00000120907/ENSG00000173991/ENSG00000132912/ENSG00000128641/ENSG00000151849/ENSG00000171992/ENSG00000076382/ENSG00000214367/ENSG00000105146/ENSG00000249115/ENSG00000155657/ENSG00000128487/ENSG00000067798/ENSG00000077549/ENSG00000171368/ENSG00000158022/ENSG00000149557/ENSG00000049323/ENSG00000188229/ENSG00000162849/ENSG00000066629/ENSG00000166579/ENSG00000087586/ENSG00000197892/ENSG00000021574/ENSG00000124942/ENSG00000145919/ENSG00000118194/ENSG00000107581/ENSG00000165660/ENSG00000112984/ENSG00000140854/ENSG00000125864/ENSG00000156804/ENSG00000172403/ENSG00000112679/ENSG00000107438/ENSG00000130956/ENSG00000116584/ENSG00000134775/ENSG00000165476/ENSG00000111057/ENSG00000141380/ENSG00000074054/ENSG00000204262
GO:0000793 ENSG00000167513/ENSG00000102974/ENSG00000185122/ENSG00000153187/ENSG00000166851/ENSG00000196074/ENSG00000122545/ENSG00000085415/ENSG00000051180/ENSG00000101265/ENSG00000134057/ENSG00000117724/ENSG00000069248/ENSG00000080986/ENSG00000076242/ENSG00000076382/ENSG00000139351/ENSG00000105146/ENSG00000155657/ENSG00000143228/ENSG00000166579/ENSG00000087586/ENSG00000145919
GO:0044445 ENSG00000163923/ENSG00000171862/ENSG00000113758/ENSG00000037241/ENSG00000108515/ENSG00000172071/ENSG00000173545/ENSG00000197386/ENSG00000082516/ENSG00000143740/ENSG00000162711/ENSG00000141543/ENSG00000166501/ENSG00000100519/ENSG00000166189/ENSG00000173692/ENSG00000164116
GO:0005814 ENSG00000167461/ENSG00000128731/ENSG00000176225/ENSG00000103540/ENSG00000197386/ENSG00000166851/ENSG00000070761
GO:0099080 ENSG00000163359/ENSG00000066735/ENSG00000133392/ENSG00000168743/ENSG00000168542/ENSG00000153187/ENSG00000187720/ENSG00000074695/ENSG00000170312/ENSG00000139329/ENSG00000176749/ENSG00000088727/ENSG00000184009/ENSG00000138594/ENSG00000183023/ENSG00000122786/ENSG00000080709/ENSG00000143318/ENSG00000166851/ENSG00000151067/ENSG00000073712/ENSG00000162946/ENSG00000099910/ENSG00000115266/ENSG00000107404/ENSG00000070761/ENSG00000198771/ENSG00000155324/ENSG00000115841/ENSG00000141376/ENSG00000133454/ENSG00000143632/ENSG00000142867/ENSG00000173801/ENSG00000133028/ENSG00000040341/ENSG00000198718/ENSG00000134871/ENSG00000134982/ENSG00000119729/ENSG00000239474/ENSG00000101162/ENSG00000108244/ENSG00000087470/ENSG00000119782/ENSG00000120694/ENSG00000198668/ENSG00000119661/ENSG00000104142/ENSG00000117632/ENSG00000197702/ENSG00000103494/ENSG00000111229/ENSG00000100271/ENSG00000138347/ENSG00000120907/ENSG00000173991/ENSG00000132912/ENSG00000128641/ENSG00000151849/ENSG00000171992/ENSG00000076382/ENSG00000214367/ENSG00000105146/ENSG00000249115/ENSG00000155657/ENSG00000128487/ENSG00000067798/ENSG00000077549/ENSG00000171368/ENSG00000158022/ENSG00000149557/ENSG00000049323/ENSG00000188229/ENSG00000162849/ENSG00000066629/ENSG00000166579/ENSG00000087586/ENSG00000197892/ENSG00000021574/ENSG00000124942/ENSG00000145919/ENSG00000118194/ENSG00000107581/ENSG00000165660/ENSG00000112984/ENSG00000140854/ENSG00000125864/ENSG00000156804/ENSG00000172403/ENSG00000112679/ENSG00000107438/ENSG00000130956/ENSG00000116584/ENSG00000134775/ENSG00000165476/ENSG00000111057/ENSG00000141380/ENSG00000074054/ENSG00000204262
GO:0044420 ENSG00000168542/ENSG00000187720/ENSG00000139329/ENSG00000134871
GO:0036126 ENSG00000154040/ENSG00000109062/ENSG00000140526/ENSG00000122545
GO:0030125 ENSG00000089818/ENSG00000119844/ENSG00000131374
GO:0030427 ENSG00000113758/ENSG00000176749/ENSG00000107404/ENSG00000100697/ENSG00000152818/ENSG00000144857/ENSG00000115232/ENSG00000121297/ENSG00000133026/ENSG00000198668/ENSG00000149930/ENSG00000154096/ENSG00000111229/ENSG00000105443/ENSG00000166313/ENSG00000130558/ENSG00000165795/ENSG00000214274/ENSG00000148498/ENSG00000166579/ENSG00000162951/ENSG00000140854/ENSG00000084112/ENSG00000198561/ENSG00000124126/ENSG00000099250/ENSG00000106078/ENSG00000151491/ENSG00000119328/ENSG00000079805
GO:0043209 ENSG00000178952/ENSG00000171862/ENSG00000150991/ENSG00000044574/ENSG00000184009/ENSG00000163399/ENSG00000182718/ENSG00000174238/ENSG00000158887/ENSG00000165672/ENSG00000198668/ENSG00000142168/ENSG00000154096/ENSG00000136854/ENSG00000173786/ENSG00000171368/ENSG00000188229/ENSG00000164300/ENSG00000144834/ENSG00000167863/ENSG00000116473/ENSG00000102934/ENSG00000163531/ENSG00000023228/ENSG00000130707/ENSG00000167004
GO:0036477 ENSG00000161057/ENSG00000167461/ENSG00000171862/ENSG00000113758/ENSG00000114127/ENSG00000153187/ENSG00000145911/ENSG00000176749/ENSG00000183023/ENSG00000080709/ENSG00000197386/ENSG00000151067/ENSG00000151532/ENSG00000101040/ENSG00000075413/ENSG00000107404/ENSG00000169862/ENSG00000164418/ENSG00000152767/ENSG00000118160/ENSG00000143740/ENSG00000135317/ENSG00000213949/ENSG00000100697/ENSG00000140992/ENSG00000139826/ENSG00000139567/ENSG00000141543/ENSG00000131188/ENSG00000104133/ENSG00000141564/ENSG00000187391/ENSG00000171617/ENSG00000196839/ENSG00000134909/ENSG00000115252/ENSG00000165802/ENSG00000115232/ENSG00000160191/ENSG00000137845/ENSG00000122966/ENSG00000198836/ENSG00000133026/ENSG00000164961/ENSG00000078725/ENSG00000198844/ENSG00000114796/ENSG00000149930/ENSG00000123700/ENSG00000141837/ENSG00000181790/ENSG00000142168/ENSG00000224470/ENSG00000164902/ENSG00000160460/ENSG00000154096/ENSG00000105707/ENSG00000125851/ENSG00000092969/ENSG00000166313/ENSG00000077063/ENSG00000130558/ENSG00000099260/ENSG00000171992/ENSG00000197555/ENSG00000214274/ENSG00000096093/ENSG00000197283/ENSG00000148498/ENSG00000135387/ENSG00000172531/ENSG00000166579/ENSG00000087586/ENSG00000170017/ENSG00000140854/ENSG00000115041/ENSG00000163531/ENSG00000145555/ENSG00000124839/ENSG00000131788/ENSG00000114030/ENSG00000130707/ENSG00000198561/ENSG00000136848/ENSG00000116584/ENSG00000184792/ENSG00000125149/ENSG00000089041/ENSG00000148219/ENSG00000134243/ENSG00000130203/ENSG00000138592/ENSG00000176658/ENSG00000108852/ENSG00000124126/ENSG00000099250/ENSG00000119699/ENSG00000167136/ENSG00000106078/ENSG00000108349/ENSG00000179632/ENSG00000123908/ENSG00000076321/ENSG00000101210/ENSG00000079215/ENSG00000116729/ENSG00000020129/ENSG00000168329/ENSG00000115904/ENSG00000182324/ENSG00000119328/ENSG00000079805/ENSG00000145632/ENSG00000145545/ENSG00000185896/ENSG00000117400/ENSG00000067606/ENSG00000162409/ENSG00000251322/ENSG00000129460/ENSG00000196639/ENSG00000112541/ENSG00000136868/ENSG00000100722/ENSG00000198835/ENSG00000171867/ENSG00000137497/ENSG00000137880/ENSG00000137825/ENSG00000169398
GO:0097223 ENSG00000154040/ENSG00000109062/ENSG00000140526/ENSG00000157404/ENSG00000044115/ENSG00000213949/ENSG00000105974/ENSG00000122545
GO:0005819 ENSG00000163376/ENSG00000185122/ENSG00000153187/ENSG00000170312/ENSG00000163872/ENSG00000166851/ENSG00000099910/ENSG00000167967/ENSG00000122545/ENSG00000115841/ENSG00000258890/ENSG00000119541/ENSG00000100345/ENSG00000133026/ENSG00000109103/ENSG00000073910/ENSG00000112245/ENSG00000134057/ENSG00000117724/ENSG00000198668/ENSG00000125898/ENSG00000121957/ENSG00000134108/ENSG00000132912/ENSG00000100154/ENSG00000188807/ENSG00000076382/ENSG00000214367/ENSG00000105146/ENSG00000249115/ENSG00000096093/ENSG00000138411/ENSG00000066629/ENSG00000166579/ENSG00000087586/ENSG00000021574/ENSG00000145919/ENSG00000165660/ENSG00000112984/ENSG00000140854/ENSG00000135476
GO:0014069 ENSG00000167461/ENSG00000113758/ENSG00000171208/ENSG00000117600/ENSG00000052126/ENSG00000176749/ENSG00000163399/ENSG00000151067/ENSG00000082397/ENSG00000162946/ENSG00000131409/ENSG00000107404/ENSG00000169862/ENSG00000163110/ENSG00000164418/ENSG00000140992/ENSG00000131188/ENSG00000187391/ENSG00000134909/ENSG00000165802/ENSG00000137845
GO:0032279 ENSG00000167461/ENSG00000113758/ENSG00000171208/ENSG00000117600/ENSG00000052126/ENSG00000176749/ENSG00000163399/ENSG00000151067/ENSG00000082397/ENSG00000162946/ENSG00000131409/ENSG00000107404/ENSG00000169862/ENSG00000163110/ENSG00000164418/ENSG00000140992/ENSG00000131188/ENSG00000187391/ENSG00000134909/ENSG00000165802/ENSG00000137845
GO:0001669 ENSG00000140526/ENSG00000157404/ENSG00000044115/ENSG00000213949/ENSG00000105974/ENSG00000006638/ENSG00000144837/ENSG00000179218/ENSG00000070770/ENSG00000111319/ENSG00000105649
GO:0009986 ENSG00000129048/ENSG00000204642/ENSG00000157404/ENSG00000166479/ENSG00000072274/ENSG00000153187/ENSG00000163975/ENSG00000044574/ENSG00000130303/ENSG00000079308/ENSG00000080709/ENSG00000182718/ENSG00000179242/ENSG00000117090/ENSG00000073712/ENSG00000137801/ENSG00000132470/ENSG00000006016/ENSG00000213949/ENSG00000174059/ENSG00000167434/ENSG00000139567/ENSG00000158869/ENSG00000141424/ENSG00000165029/ENSG00000177575/ENSG00000144476/ENSG00000196839/ENSG00000116824/ENSG00000135678/ENSG00000113594/ENSG00000136153/ENSG00000115232/ENSG00000101557/ENSG00000137845/ENSG00000156886/ENSG00000130770/ENSG00000168140/ENSG00000135424/ENSG00000012223/ENSG00000135077/ENSG00000154096/ENSG00000105707/ENSG00000174807/ENSG00000076706/ENSG00000215021/ENSG00000113361/ENSG00000163638/ENSG00000196776/ENSG00000091513/ENSG00000145623/ENSG00000179218/ENSG00000104853/ENSG00000082781/ENSG00000139193/ENSG00000186827/ENSG00000115884/ENSG00000164930/ENSG00000170017/ENSG00000072571/ENSG00000081237/ENSG00000196562/ENSG00000125845/ENSG00000163513/ENSG00000183454/ENSG00000197442/ENSG00000167004/ENSG00000173535/ENSG00000177119/ENSG00000131981/ENSG00000181284/ENSG00000089041/ENSG00000119535/ENSG00000134243/ENSG00000099250/ENSG00000119699/ENSG00000170989/ENSG00000108561/ENSG00000100385/ENSG00000112715/ENSG00000079215/ENSG00000168329/ENSG00000119912/ENSG00000167191/ENSG00000185896/ENSG00000010278/ENSG00000124145/ENSG00000117400/ENSG00000153563/ENSG00000170458/ENSG00000187688/ENSG00000106483/ENSG00000005844/ENSG00000169604/ENSG00000139626/ENSG00000146013/ENSG00000005884/ENSG00000085563/ENSG00000110911
GO:0035327 ENSG00000107371/ENSG00000171824/ENSG00000141376/ENSG00000005483/ENSG00000006712
GO:0008305 ENSG00000168743/ENSG00000132470/ENSG00000213949/ENSG00000115232/ENSG00000156886/ENSG00000135424/ENSG00000137809/ENSG00000082781
GO:0098636 ENSG00000168743/ENSG00000132470/ENSG00000213949/ENSG00000115232/ENSG00000156886/ENSG00000135424/ENSG00000137809/ENSG00000082781
GO:0009897 ENSG00000129048/ENSG00000157404/ENSG00000072274/ENSG00000137801/ENSG00000006016/ENSG00000213949/ENSG00000174059/ENSG00000167434/ENSG00000158869/ENSG00000165029/ENSG00000177575/ENSG00000144476/ENSG00000196839/ENSG00000116824/ENSG00000113594/ENSG00000154096/ENSG00000174807/ENSG00000076706/ENSG00000091513/ENSG00000145623/ENSG00000179218/ENSG00000104853/ENSG00000139193/ENSG00000186827/ENSG00000115884/ENSG00000170017/ENSG00000081237/ENSG00000163513/ENSG00000197442/ENSG00000089041/ENSG00000119535/ENSG00000170989/ENSG00000100385/ENSG00000168329/ENSG00000185896/ENSG00000010278/ENSG00000153563/ENSG00000170458/ENSG00000169604/ENSG00000146013/ENSG00000005884
GO:0098798 ENSG00000136521/ENSG00000259494/ENSG00000135900/ENSG00000120333/ENSG00000099795/ENSG00000144029/ENSG00000156928/ENSG00000082515/ENSG00000174886/ENSG00000131143/ENSG00000143314/ENSG00000100575/ENSG00000149792/ENSG00000172590/ENSG00000166902/ENSG00000111639/ENSG00000183648/ENSG00000131844/ENSG00000243147/ENSG00000177034/ENSG00000105364/ENSG00000167283/ENSG00000136522/ENSG00000150779/ENSG00000204822/ENSG00000007923/ENSG00000075336/ENSG00000167863/ENSG00000023228/ENSG00000213585/ENSG00000185633/ENSG00000140990/ENSG00000179091/ENSG00000197912/ENSG00000145494
GO:0043025 ENSG00000167461/ENSG00000114127/ENSG00000176749/ENSG00000080709/ENSG00000151067/ENSG00000151532/ENSG00000107404/ENSG00000169862/ENSG00000164418/ENSG00000118160/ENSG00000143740/ENSG00000213949/ENSG00000140992/ENSG00000139567/ENSG00000141543/ENSG00000141564/ENSG00000171617/ENSG00000196839/ENSG00000115252/ENSG00000165802/ENSG00000115232/ENSG00000160191/ENSG00000137845/ENSG00000122966/ENSG00000133026/ENSG00000164961/ENSG00000078725/ENSG00000114796/ENSG00000123700/ENSG00000141837/ENSG00000142168/ENSG00000164902/ENSG00000160460/ENSG00000154096/ENSG00000105707/ENSG00000125851/ENSG00000092969/ENSG00000166313/ENSG00000130558/ENSG00000171992/ENSG00000214274/ENSG00000096093/ENSG00000148498/ENSG00000172531/ENSG00000166579/ENSG00000087586/ENSG00000170017/ENSG00000140854/ENSG00000145555/ENSG00000124839/ENSG00000130707/ENSG00000136848/ENSG00000116584/ENSG00000089041/ENSG00000148219/ENSG00000134243/ENSG00000130203/ENSG00000176658/ENSG00000099250/ENSG00000119699/ENSG00000167136/ENSG00000106078/ENSG00000101210/ENSG00000079215/ENSG00000020129/ENSG00000168329/ENSG00000115904/ENSG00000182324/ENSG00000145545/ENSG00000185896/ENSG00000117400/ENSG00000067606/ENSG00000162409
GO:0031941 ENSG00000184009/ENSG00000115266/ENSG00000133454/ENSG00000111229/ENSG00000128487/ENSG00000112679
GO:0005667 ENSG00000125484/ENSG00000141646/ENSG00000153187/ENSG00000129173/ENSG00000102908/ENSG00000106290/ENSG00000077235/ENSG00000126351/ENSG00000082516/ENSG00000140332/ENSG00000134954/ENSG00000177045/ENSG00000077092/ENSG00000080298/ENSG00000108590/ENSG00000148737/ENSG00000100968/ENSG00000143257/ENSG00000125878/ENSG00000187079/ENSG00000167771/ENSG00000137834/ENSG00000104221/ENSG00000143190/ENSG00000185669/ENSG00000113387/ENSG00000162227/ENSG00000170577/ENSG00000213999/ENSG00000048052/ENSG00000064313/ENSG00000119508/ENSG00000125651/ENSG00000136826/ENSG00000112033/ENSG00000188342/ENSG00000068305/ENSG00000155868/ENSG00000118156/ENSG00000171056/ENSG00000159692/ENSG00000168214/ENSG00000164134/ENSG00000143995/ENSG00000130726/ENSG00000007866/ENSG00000141448/ENSG00000107438/ENSG00000116679/ENSG00000005513/ENSG00000198728/ENSG00000239306/ENSG00000142453/ENSG00000131408/ENSG00000111358/ENSG00000169744/ENSG00000123358/ENSG00000129194/ENSG00000158773/ENSG00000170854/ENSG00000143013/ENSG00000170485/ENSG00000186350/ENSG00000168036/ENSG00000148308/ENSG00000108773/ENSG00000166337/ENSG00000104973/ENSG00000177732/ENSG00000023608/ENSG00000112592/ENSG00000183072/ENSG00000163348/ENSG00000089902/ENSG00000198176/ENSG00000166949/ENSG00000101096
GO:0150034 ENSG00000113758/ENSG00000176749/ENSG00000184009/ENSG00000107404/ENSG00000164418/ENSG00000100697/ENSG00000152818/ENSG00000144857/ENSG00000166501/ENSG00000115232/ENSG00000121297/ENSG00000133026/ENSG00000198668/ENSG00000149930/ENSG00000154096/ENSG00000111229/ENSG00000105443/ENSG00000166313/ENSG00000130558/ENSG00000165795/ENSG00000214274/ENSG00000148498/ENSG00000160469/ENSG00000166579/ENSG00000162951/ENSG00000140854/ENSG00000115041/ENSG00000084112/ENSG00000198561/ENSG00000105649/ENSG00000184160/ENSG00000124126/ENSG00000099250/ENSG00000106078/ENSG00000106367/ENSG00000151491/ENSG00000119328/ENSG00000079805
GO:0005923 ENSG00000158769/ENSG00000147649/ENSG00000114019/ENSG00000134982/ENSG00000187391/ENSG00000160593/ENSG00000124171/ENSG00000116539/ENSG00000144061/ENSG00000103494/ENSG00000105443/ENSG00000171992/ENSG00000148498
GO:0031461 ENSG00000163376/ENSG00000174989/ENSG00000099910/ENSG00000107872/ENSG00000162032/ENSG00000145332/ENSG00000171617/ENSG00000239474/ENSG00000159069/ENSG00000127452/ENSG00000145743/ENSG00000114796/ENSG00000111276/ENSG00000176248/ENSG00000175093/ENSG00000196510/ENSG00000089053/ENSG00000180901/ENSG00000132109/ENSG00000156804/ENSG00000167986/ENSG00000163257/ENSG00000261609/ENSG00000153558/ENSG00000100387/ENSG00000166266/ENSG00000111530/ENSG00000103363/ENSG00000112146/ENSG00000173163/ENSG00000076321/ENSG00000166233/ENSG00000132003/ENSG00000151743
GO:0000784 ENSG00000094916/ENSG00000170312/ENSG00000099284/ENSG00000051180/ENSG00000173559/ENSG00000109534/ENSG00000139579/ENSG00000152457/ENSG00000100109/ENSG00000172531/ENSG00000102977/ENSG00000175054/ENSG00000167986/ENSG00000204569/ENSG00000138385/ENSG00000092330/ENSG00000062822/ENSG00000164091/ENSG00000132475
GO:0005791 ENSG00000078401/ENSG00000099958/ENSG00000158887/ENSG00000167434/ENSG00000083444/ENSG00000123700/ENSG00000174444/ENSG00000176438/ENSG00000065665/ENSG00000118705/ENSG00000213741
GO:0070160 ENSG00000158769/ENSG00000147649/ENSG00000114019/ENSG00000134982/ENSG00000187391/ENSG00000160593/ENSG00000124171/ENSG00000116539/ENSG00000144061/ENSG00000103494/ENSG00000105443/ENSG00000171992/ENSG00000148498
GO:0097729 ENSG00000154040/ENSG00000109062/ENSG00000140526/ENSG00000122545
GO:0005876 ENSG00000153187/ENSG00000170312/ENSG00000166851/ENSG00000099910/ENSG00000198668/ENSG00000105146/ENSG00000087586/ENSG00000145919/ENSG00000074054/ENSG00000141367
GO:0070161 ENSG00000162434/ENSG00000173482/ENSG00000044115/ENSG00000044574/ENSG00000061938/ENSG00000184009/ENSG00000079308/ENSG00000138594/ENSG00000182718/ENSG00000179242/ENSG00000073712/ENSG00000132470/ENSG00000169862/ENSG00000163110/ENSG00000086991/ENSG00000131016/ENSG00000213949/ENSG00000105974/ENSG00000140992/ENSG00000110031/ENSG00000196923/ENSG00000110841/ENSG00000136205/ENSG00000173801/ENSG00000109270/ENSG00000232434/ENSG00000134982/ENSG00000100345/ENSG00000136153/ENSG00000115232/ENSG00000164741/ENSG00000137845/ENSG00000159176/ENSG00000144061/ENSG00000114796/ENSG00000100266/ENSG00000197702/ENSG00000110321/ENSG00000181790/ENSG00000160460/ENSG00000154096/ENSG00000150764/ENSG00000076706/ENSG00000111229/ENSG00000105443/ENSG00000096968/ENSG00000137809/ENSG00000101384/ENSG00000135269/ENSG00000132912/ENSG00000113361/ENSG00000074696/ENSG00000136950/ENSG00000148498/ENSG00000140691/ENSG00000174444/ENSG00000162736/ENSG00000133818/ENSG00000166689/ENSG00000149428/ENSG00000172531/ENSG00000179218/ENSG00000082781/ENSG00000124942/ENSG00000170017/ENSG00000084733/ENSG00000163531/ENSG00000081237/ENSG00000157483/ENSG00000145819/ENSG00000183230/ENSG00000172403/ENSG00000107438/ENSG00000198561/ENSG00000244486/ENSG00000116584/ENSG00000167004/ENSG00000111057/ENSG00000108518/ENSG00000149115/ENSG00000074054/ENSG00000185621/ENSG00000213741
GO:0030660 ENSG00000166557/ENSG00000204642/ENSG00000119844/ENSG00000074695/ENSG00000138768/ENSG00000151532
GO:0030120 ENSG00000089818/ENSG00000166557/ENSG00000119844/ENSG00000131374
GO:0005790 ENSG00000044574/ENSG00000143318/ENSG00000123700/ENSG00000149428/ENSG00000179218
GO:0043005 ENSG00000161057/ENSG00000167461/ENSG00000109062/ENSG00000171862/ENSG00000113758/ENSG00000152102/ENSG00000114127/ENSG00000153187/ENSG00000055163/ENSG00000135951/ENSG00000145911/ENSG00000176749/ENSG00000184009/ENSG00000105186/ENSG00000183023/ENSG00000080709/ENSG00000197386/ENSG00000151067/ENSG00000082397/ENSG00000151532/ENSG00000101040/ENSG00000131409/ENSG00000075413/ENSG00000107404/ENSG00000169862/ENSG00000163110/ENSG00000070404/ENSG00000164418/ENSG00000152767/ENSG00000118160/ENSG00000143740/ENSG00000105887/ENSG00000135317/ENSG00000213949/ENSG00000044524/ENSG00000100697/ENSG00000139826/ENSG00000139567/ENSG00000064999/ENSG00000141543/ENSG00000152818/ENSG00000131188/ENSG00000104133/ENSG00000141564/ENSG00000187391/ENSG00000196839/ENSG00000134909/ENSG00000144857/ENSG00000165802/ENSG00000166501/ENSG00000115232/ENSG00000122591/ENSG00000137845/ENSG00000121297/ENSG00000198836/ENSG00000133026/ENSG00000164961/ENSG00000147862/ENSG00000078725/ENSG00000144061/ENSG00000198844/ENSG00000114796/ENSG00000198668/ENSG00000134769/ENSG00000149930/ENSG00000132376/ENSG00000123700/ENSG00000117632/ENSG00000181790/ENSG00000142168/ENSG00000224470/ENSG00000160460/ENSG00000154096/ENSG00000103494/ENSG00000125851/ENSG00000092969/ENSG00000111229/ENSG00000105443/ENSG00000136854/ENSG00000138347/ENSG00000166313/ENSG00000077063/ENSG00000130558/ENSG00000099260/ENSG00000165795/ENSG00000171992/ENSG00000197555/ENSG00000214274/ENSG00000197283/ENSG00000148498/ENSG00000160469/ENSG00000149557/ENSG00000135387/ENSG00000172531/ENSG00000166579/ENSG00000150540/ENSG00000153253/ENSG00000087586/ENSG00000197892/ENSG00000021574/ENSG00000189195/ENSG00000170017/ENSG00000113248/ENSG00000162951/ENSG00000144619/ENSG00000140854/ENSG00000115041/ENSG00000116473/ENSG00000163531/ENSG00000145555/ENSG00000124839/ENSG00000128512/ENSG00000127445/ENSG00000131788/ENSG00000183454/ENSG00000114030/ENSG00000084112/ENSG00000130707/ENSG00000198561/ENSG00000136848/ENSG00000146072/ENSG00000136381/ENSG00000116584/ENSG00000105649/ENSG00000184792/ENSG00000125149/ENSG00000184160/ENSG00000088986/ENSG00000134243/ENSG00000143858/ENSG00000130203/ENSG00000138592/ENSG00000176658/ENSG00000108852/ENSG00000124126
GO:0044798 ENSG00000125484/ENSG00000141646/ENSG00000153187/ENSG00000129173/ENSG00000102908/ENSG00000106290/ENSG00000077235/ENSG00000126351/ENSG00000082516/ENSG00000140332/ENSG00000077092/ENSG00000108590/ENSG00000148737/ENSG00000100968/ENSG00000143257/ENSG00000125878/ENSG00000187079/ENSG00000104221/ENSG00000143190/ENSG00000162227/ENSG00000064313/ENSG00000125651/ENSG00000136826/ENSG00000112033
GO:0005905 ENSG00000089818/ENSG00000072274/ENSG00000061938/ENSG00000131374/ENSG00000144476/ENSG00000127527/ENSG00000087470/ENSG00000130147/ENSG00000091513/ENSG00000189195
GO:0042470 ENSG00000133392/ENSG00000072274/ENSG00000044574/ENSG00000105186/ENSG00000163399/ENSG00000182718/ENSG00000141258
GO:0048770 ENSG00000133392/ENSG00000072274/ENSG00000044574/ENSG00000105186/ENSG00000163399/ENSG00000182718/ENSG00000141258
GO:0000794 ENSG00000166851/ENSG00000196074/ENSG00000051180/ENSG00000134057/ENSG00000080986/ENSG00000076242/ENSG00000139351/ENSG00000105146/ENSG00000155657/ENSG00000143228/ENSG00000087586
GO:0030425 ENSG00000161057/ENSG00000167461/ENSG00000171862/ENSG00000113758/ENSG00000114127/ENSG00000153187/ENSG00000145911/ENSG00000176749/ENSG00000183023/ENSG00000080709/ENSG00000197386/ENSG00000151067/ENSG00000101040/ENSG00000075413/ENSG00000107404/ENSG00000169862/ENSG00000164418/ENSG00000152767/ENSG00000118160/ENSG00000143740/ENSG00000135317/ENSG00000100697/ENSG00000139826/ENSG00000139567/ENSG00000141543/ENSG00000131188/ENSG00000104133/ENSG00000141564/ENSG00000187391/ENSG00000196839/ENSG00000134909/ENSG00000165802/ENSG00000137845/ENSG00000198836/ENSG00000133026/ENSG00000078725/ENSG00000198844/ENSG00000149930/ENSG00000123700/ENSG00000181790/ENSG00000142168/ENSG00000224470/ENSG00000154096/ENSG00000125851/ENSG00000166313/ENSG00000077063/ENSG00000099260/ENSG00000171992/ENSG00000197555
GO:0009295 ENSG00000178952/ENSG00000162851
GO:0042645 ENSG00000178952/ENSG00000162851
GO:0030662 ENSG00000089818/ENSG00000166557/ENSG00000204642/ENSG00000072274/ENSG00000119844/ENSG00000074695/ENSG00000138768/ENSG00000151532/ENSG00000131374
GO:0097546 ENSG00000167461/ENSG00000162946/ENSG00000105948/ENSG00000163001
GO:0099572 ENSG00000167461/ENSG00000113758/ENSG00000171208/ENSG00000117600/ENSG00000052126/ENSG00000176749/ENSG00000163399/ENSG00000151067/ENSG00000082397/ENSG00000162946/ENSG00000131409/ENSG00000107404/ENSG00000169862/ENSG00000163110/ENSG00000164418/ENSG00000140992/ENSG00000131188/ENSG00000187391/ENSG00000134909/ENSG00000165802/ENSG00000137845
GO:0045211 ENSG00000171862/ENSG00000113758/ENSG00000117600/ENSG00000183023/ENSG00000151067/ENSG00000162946/ENSG00000131409/ENSG00000163110/ENSG00000164418/ENSG00000152818/ENSG00000131188/ENSG00000134909/ENSG00000165802/ENSG00000137845/ENSG00000181790/ENSG00000183783/ENSG00000120907/ENSG00000166313/ENSG00000171992/ENSG00000197555
GO:0051233 ENSG00000153187/ENSG00000166851/ENSG00000109103/ENSG00000134108/ENSG00000105146/ENSG00000066629/ENSG00000087586
GO:0031902 ENSG00000182718/ENSG00000165782/ENSG00000151532/ENSG00000135317/ENSG00000115020/ENSG00000109270/ENSG00000058799/ENSG00000119541/ENSG00000177058/ENSG00000104142/ENSG00000134108/ENSG00000162144/ENSG00000144909
GO:0031594 ENSG00000176749/ENSG00000133110/ENSG00000152818/ENSG00000100345/ENSG00000133026/ENSG00000137478/ENSG00000108639/ENSG00000089041/ENSG00000030304
GO:0000151 ENSG00000164758/ENSG00000163376/ENSG00000138081/ENSG00000174989/ENSG00000099910/ENSG00000114098/ENSG00000107872/ENSG00000118420/ENSG00000162032/ENSG00000145332/ENSG00000108590/ENSG00000141034/ENSG00000110344/ENSG00000171617/ENSG00000136153/ENSG00000239474/ENSG00000159069/ENSG00000127452/ENSG00000145743/ENSG00000114796/ENSG00000134086/ENSG00000111276/ENSG00000176248/ENSG00000175093/ENSG00000115307/ENSG00000034677/ENSG00000196510/ENSG00000155868/ENSG00000089053/ENSG00000137393/ENSG00000180901/ENSG00000132109/ENSG00000156804/ENSG00000167986/ENSG00000163257/ENSG00000261609/ENSG00000108854/ENSG00000153558/ENSG00000177889/ENSG00000100387/ENSG00000166266/ENSG00000111530/ENSG00000048140/ENSG00000103363/ENSG00000112146/ENSG00000156587/ENSG00000173163/ENSG00000164117/ENSG00000076321/ENSG00000166233/ENSG00000132003/ENSG00000151743
GO:0001725 ENSG00000073712/ENSG00000086991/ENSG00000122545/ENSG00000196923/ENSG00000143632/ENSG00000100345/ENSG00000133026/ENSG00000132912/ENSG00000171992
GO:0097517 ENSG00000073712/ENSG00000086991/ENSG00000122545/ENSG00000196923/ENSG00000143632/ENSG00000100345/ENSG00000133026/ENSG00000132912/ENSG00000171992
GO:0070461 ENSG00000106290/ENSG00000176476/ENSG00000173011/ENSG00000162227/ENSG00000064313
GO:0031300 ENSG00000127995/ENSG00000180776/ENSG00000144120/ENSG00000198964/ENSG00000125454/ENSG00000120942/ENSG00000050393/ENSG00000133028/ENSG00000157593/ENSG00000175701/ENSG00000104936/ENSG00000104381/ENSG00000204070/ENSG00000156671/ENSG00000160613/ENSG00000167645/ENSG00000183978/ENSG00000188807/ENSG00000100060/ENSG00000178307
GO:0097447 ENSG00000161057/ENSG00000167461/ENSG00000171862/ENSG00000113758/ENSG00000114127/ENSG00000153187/ENSG00000145911/ENSG00000176749/ENSG00000183023/ENSG00000080709/ENSG00000197386/ENSG00000151067/ENSG00000101040/ENSG00000075413/ENSG00000107404/ENSG00000169862/ENSG00000164418/ENSG00000152767/ENSG00000118160/ENSG00000143740/ENSG00000135317/ENSG00000100697/ENSG00000139826/ENSG00000139567/ENSG00000141543/ENSG00000131188/ENSG00000104133/ENSG00000141564/ENSG00000187391/ENSG00000196839/ENSG00000134909/ENSG00000165802/ENSG00000137845/ENSG00000198836/ENSG00000133026/ENSG00000078725/ENSG00000198844/ENSG00000149930/ENSG00000123700/ENSG00000181790/ENSG00000142168/ENSG00000224470/ENSG00000154096/ENSG00000125851/ENSG00000166313/ENSG00000077063/ENSG00000099260/ENSG00000171992/ENSG00000197555
GO:0031526 ENSG00000109062/ENSG00000163803/ENSG00000167434
GO:0030017 ENSG00000074695/ENSG00000138594/ENSG00000183023/ENSG00000080709/ENSG00000143318/ENSG00000151067/ENSG00000073712/ENSG00000133454/ENSG00000143632/ENSG00000173801/ENSG00000239474/ENSG00000119782/ENSG00000198668/ENSG00000197702/ENSG00000138347/ENSG00000120907/ENSG00000173991/ENSG00000132912/ENSG00000171992/ENSG00000155657/ENSG00000077549/ENSG00000158022/ENSG00000118194/ENSG00000156804/ENSG00000172403/ENSG00000107438/ENSG00000130956/ENSG00000134775
GO:0032154 ENSG00000122545/ENSG00000110697/ENSG00000100345/ENSG00000122966/ENSG00000133026/ENSG00000065243/ENSG00000134278/ENSG00000090565/ENSG00000112984/ENSG00000123143/ENSG00000084112
GO:0044455 ENSG00000144120/ENSG00000136521/ENSG00000125454/ENSG00000099795/ENSG00000050393/ENSG00000133028/ENSG00000175701/ENSG00000174886/ENSG00000131143/ENSG00000104936/ENSG00000198836/ENSG00000104381/ENSG00000183648/ENSG00000177034/ENSG00000163050/ENSG00000183978/ENSG00000167283/ENSG00000178307/ENSG00000007923/ENSG00000075336/ENSG00000167863/ENSG00000084092/ENSG00000023228/ENSG00000185633/ENSG00000140990/ENSG00000179091/ENSG00000197912/ENSG00000145494
GO:0014704 ENSG00000044115/ENSG00000183023/ENSG00000163399/ENSG00000173801/ENSG00000123700/ENSG00000160460
GO:0030118 ENSG00000089818/ENSG00000119844/ENSG00000131374/ENSG00000127527/ENSG00000128298/ENSG00000100280/ENSG00000189195
GO:0005901 ENSG00000044115/ENSG00000163399/ENSG00000105974/ENSG00000079435/ENSG00000164741/ENSG00000100266/ENSG00000096968/ENSG00000120907/ENSG00000170214/ENSG00000163513
GO:0005938 ENSG00000113758/ENSG00000122786/ENSG00000182718/ENSG00000073712/ENSG00000179820/ENSG00000132254/ENSG00000131016/ENSG00000105974/ENSG00000122545/ENSG00000180096/ENSG00000152818/ENSG00000124171/ENSG00000119729/ENSG00000105122/ENSG00000134909/ENSG00000100345/ENSG00000165802/ENSG00000164741/ENSG00000133026/ENSG00000073910/ENSG00000198363/ENSG00000160460/ENSG00000121957/ENSG00000134278/ENSG00000132912/ENSG00000077063/ENSG00000132613/ENSG00000148498/ENSG00000118523/ENSG00000159692/ENSG00000084733/ENSG00000167306/ENSG00000145555/ENSG00000125864/ENSG00000198561/ENSG00000108518/ENSG00000074054/ENSG00000175104/ENSG00000148219/ENSG00000106078/ENSG00000105221/ENSG00000075539/ENSG00000092820/ENSG00000151491/ENSG00000115963/ENSG00000148180/ENSG00000126785
GO:0000795 ENSG00000166851/ENSG00000196074/ENSG00000051180/ENSG00000139351
GO:0099086 ENSG00000166851/ENSG00000196074/ENSG00000051180/ENSG00000139351
GO:0072562 ENSG00000160339/ENSG00000072274/ENSG00000184009/ENSG00000182326/ENSG00000143632
GO:0005773 ENSG00000164342/ENSG00000157404/ENSG00000169682/ENSG00000139329/ENSG00000126775/ENSG00000130303/ENSG00000105186/ENSG00000006025/ENSG00000103222/ENSG00000197386/ENSG00000182718/ENSG00000165782/ENSG00000151532/ENSG00000104324/ENSG00000158887/ENSG00000099910/ENSG00000116005/ENSG00000105851/ENSG00000127083/ENSG00000072121/ENSG00000115363/ENSG00000131374/ENSG00000135317/ENSG00000132405/ENSG00000174059/ENSG00000142867/ENSG00000085415/ENSG00000135018/ENSG00000104133/ENSG00000139266/ENSG00000109270/ENSG00000141564/ENSG00000196839/ENSG00000216490/ENSG00000103381/ENSG00000119899/ENSG00000140950/ENSG00000100365/ENSG00000166189/ENSG00000177058/ENSG00000168140/ENSG00000038427/ENSG00000104142/ENSG00000173692/ENSG00000142168/ENSG00000092929/ENSG00000134108/ENSG00000119508/ENSG00000174206/ENSG00000162144/ENSG00000185127/ENSG00000139370/ENSG00000168907/ENSG00000254470/ENSG00000188229/ENSG00000162736/ENSG00000005381/ENSG00000100280/ENSG00000115884/ENSG00000124942/ENSG00000165861/ENSG00000127580/ENSG00000180448/ENSG00000172575/ENSG00000132109/ENSG00000125730/ENSG00000118705/ENSG00000100099/ENSG00000138798/ENSG00000130707/ENSG00000167996/ENSG00000127314/ENSG00000113441/ENSG00000110719
GO:1990391 ENSG00000119318/ENSG00000076242/ENSG00000152457/ENSG00000116062/ENSG00000152422/ENSG00000062822
GO:0043204 ENSG00000151067/ENSG00000169862/ENSG00000164418/ENSG00000118160/ENSG00000213949/ENSG00000140992/ENSG00000165802/ENSG00000160191/ENSG00000114796/ENSG00000125851/ENSG00000130558/ENSG00000171992/ENSG00000172531
GO:0035770 ENSG00000161057/ENSG00000185122/ENSG00000114127/ENSG00000153187/ENSG00000095627/ENSG00000183496/ENSG00000151461/ENSG00000066044/ENSG00000110697/ENSG00000141564/ENSG00000144535/ENSG00000162639/ENSG00000205323/ENSG00000055917/ENSG00000152520/ENSG00000166889
GO:0098984 ENSG00000167461/ENSG00000113758/ENSG00000171208/ENSG00000117600/ENSG00000052126/ENSG00000176749/ENSG00000163399/ENSG00000151067/ENSG00000082397/ENSG00000162946/ENSG00000131409/ENSG00000107404/ENSG00000169862/ENSG00000163110/ENSG00000164418/ENSG00000140992/ENSG00000131188/ENSG00000187391/ENSG00000134909/ENSG00000165802/ENSG00000137845
GO:0045202 ENSG00000161057/ENSG00000178952/ENSG00000167461/ENSG00000171862/ENSG00000113758/ENSG00000171208/ENSG00000114127/ENSG00000117600/ENSG00000055163/ENSG00000052126/ENSG00000176749/ENSG00000184009/ENSG00000183023/ENSG00000163399/ENSG00000080709/ENSG00000197386/ENSG00000151067/ENSG00000082397/ENSG00000133110/ENSG00000151532/ENSG00000101040/ENSG00000162946/ENSG00000131409/ENSG00000115266/ENSG00000107404/ENSG00000169862/ENSG00000163110/ENSG00000164418/ENSG00000152767/ENSG00000118160/ENSG00000131016/ENSG00000143740/ENSG00000153933/ENSG00000099246/ENSG00000140992/ENSG00000066044/ENSG00000180096/ENSG00000141543/ENSG00000152818/ENSG00000131188/ENSG00000104133/ENSG00000175920/ENSG00000112137/ENSG00000187391/ENSG00000119899/ENSG00000134909/ENSG00000112851/ENSG00000100345/ENSG00000165802/ENSG00000166501/ENSG00000101557/ENSG00000137845/ENSG00000133026/ENSG00000147799/ENSG00000087470/ENSG00000198668/ENSG00000139998/ENSG00000100266/ENSG00000134769/ENSG00000069188/ENSG00000104142/ENSG00000123700/ENSG00000181790/ENSG00000164116/ENSG00000091428/ENSG00000137478/ENSG00000136854/ENSG00000096968/ENSG00000183783/ENSG00000120907/ENSG00000166313/ENSG00000132670/ENSG00000077063/ENSG00000130558/ENSG00000099260/ENSG00000171992/ENSG00000197555/ENSG00000034677/ENSG00000136950/ENSG00000197283/ENSG00000160469/ENSG00000162736/ENSG00000135387/ENSG00000172531/ENSG00000166579/ENSG00000113360/ENSG00000189195/ENSG00000162951/ENSG00000159692/ENSG00000107581/ENSG00000165995/ENSG00000115041/ENSG00000116473/ENSG00000084733/ENSG00000127445/ENSG00000131788/ENSG00000183454/ENSG00000145592/ENSG00000114030/ENSG00000111110/ENSG00000198561/ENSG00000116584/ENSG00000105649/ENSG00000125149/ENSG00000108518/ENSG00000108639/ENSG00000089041/ENSG00000124785/ENSG00000184160/ENSG00000221914/ENSG00000030304/ENSG00000143858/ENSG00000130203/ENSG00000138592/ENSG00000108852/ENSG00000013275/ENSG00000144283/ENSG00000099250/ENSG00000164318/ENSG00000043591/ENSG00000154310/ENSG00000110076/ENSG00000113575/ENSG00000171914/ENSG00000108561/ENSG00000101182/ENSG00000179632/ENSG00000100567/ENSG00000168135/ENSG00000106367/ENSG00000151491
GO:0062023 ENSG00000163359/ENSG00000100985/ENSG00000168743/ENSG00000168542/ENSG00000187720/ENSG00000074695/ENSG00000166833/ENSG00000139329/ENSG00000123243/ENSG00000182718/ENSG00000133110/ENSG00000137801/ENSG00000127083/ENSG00000151388/ENSG00000134871/ENSG00000106366/ENSG00000164932/ENSG00000112851/ENSG00000132561/ENSG00000038427/ENSG00000142910/ENSG00000092969/ENSG00000064309/ENSG00000154175/ENSG00000214274/ENSG00000163638/ENSG00000049323/ENSG00000179218
GO:0022624 ENSG00000161057/ENSG00000100519/ENSG00000108671/ENSG00000173692
GO:0045178 ENSG00000078401/ENSG00000213949/ENSG00000174059/ENSG00000112851/ENSG00000065361
GO:0005759 ENSG00000178952/ENSG00000111058/ENSG00000170312/ENSG00000151498/ENSG00000143318/ENSG00000259494/ENSG00000135900/ENSG00000162851/ENSG00000120333/ENSG00000051180/ENSG00000144029/ENSG00000156928/ENSG00000174840/ENSG00000133943/ENSG00000082515/ENSG00000143314/ENSG00000165672/ENSG00000135423/ENSG00000161653/ENSG00000188266/ENSG00000134057/ENSG00000149792/ENSG00000172590/ENSG00000166902/ENSG00000111639/ENSG00000131844/ENSG00000142168/ENSG00000177192/ENSG00000162396/ENSG00000243147/ENSG00000120254/ENSG00000162433/ENSG00000105364/ENSG00000233276/ENSG00000136522/ENSG00000140374/ENSG00000204822/ENSG00000213995/ENSG00000148187
GO:0005758 ENSG00000099795/ENSG00000100575/ENSG00000198836/ENSG00000142168/ENSG00000127554/ENSG00000150779/ENSG00000080819/ENSG00000170791/ENSG00000023228/ENSG00000168393/ENSG00000004455
GO:0042383 ENSG00000163359/ENSG00000183023/ENSG00000163399/ENSG00000182718/ENSG00000143318/ENSG00000151067/ENSG00000105974/ENSG00000152818/ENSG00000160191/ENSG00000134769/ENSG00000123700/ENSG00000120907/ENSG00000124942/ENSG00000165995/ENSG00000153956
GO:0099568 ENSG00000113758/ENSG00000153187/ENSG00000126775/ENSG00000122786/ENSG00000182718/ENSG00000158023/ENSG00000073712/ENSG00000179820/ENSG00000132254/ENSG00000164418/ENSG00000131016/ENSG00000105974/ENSG00000122545/ENSG00000139826/ENSG00000180096/ENSG00000152818/ENSG00000124171/ENSG00000119729/ENSG00000196839/ENSG00000105122/ENSG00000134909/ENSG00000100345/ENSG00000165802/ENSG00000164741/ENSG00000198836/ENSG00000133026/ENSG00000073910/ENSG00000117724/ENSG00000198363/ENSG00000119661/ENSG00000142168/ENSG00000160460/ENSG00000103494/ENSG00000121957/ENSG00000134278/ENSG00000132912/ENSG00000077063/ENSG00000132613/ENSG00000096093/ENSG00000148498/ENSG00000166579/ENSG00000118523/ENSG00000021574/ENSG00000159692/ENSG00000084733/ENSG00000169379/ENSG00000167306/ENSG00000145555/ENSG00000125864/ENSG00000198561/ENSG00000108518/ENSG00000074054/ENSG00000175104/ENSG00000148219/ENSG00000088986/ENSG00000197912/ENSG00000106078/ENSG00000105221/ENSG00000189114/ENSG00000075539/ENSG00000144451/ENSG00000092820/ENSG00000151491/ENSG00000115963/ENSG00000148180/ENSG00000116729/ENSG00000126785
GO:0030055 ENSG00000162434/ENSG00000044115/ENSG00000044574/ENSG00000184009/ENSG00000079308/ENSG00000073712/ENSG00000132470/ENSG00000086991/ENSG00000131016/ENSG00000213949/ENSG00000105974/ENSG00000140992/ENSG00000110031/ENSG00000196923/ENSG00000110841/ENSG00000136205/ENSG00000173801/ENSG00000109270/ENSG00000112851/ENSG00000100345/ENSG00000136153/ENSG00000115232/ENSG00000164741/ENSG00000137845/ENSG00000159176/ENSG00000100266/ENSG00000197702/ENSG00000181790/ENSG00000154096/ENSG00000150764/ENSG00000076706/ENSG00000111229/ENSG00000096968/ENSG00000137809/ENSG00000135269/ENSG00000132912/ENSG00000074696/ENSG00000136950/ENSG00000140691/ENSG00000174444/ENSG00000162736/ENSG00000133818/ENSG00000149428/ENSG00000179218/ENSG00000082781/ENSG00000124942/ENSG00000170017/ENSG00000084733/ENSG00000163531/ENSG00000081237/ENSG00000145819/ENSG00000172403/ENSG00000107438/ENSG00000244486/ENSG00000116584/ENSG00000167004/ENSG00000108518/ENSG00000074054/ENSG00000185621/ENSG00000213741/ENSG00000099250/ENSG00000141367/ENSG00000171914/ENSG00000092820/ENSG00000182774/ENSG00000128591/ENSG00000115963/ENSG00000148180/ENSG00000063177/ENSG00000079805/ENSG00000168036/ENSG00000070756/ENSG00000138639/ENSG00000010278/ENSG00000124145/ENSG00000168310/ENSG00000139626/ENSG00000112655/ENSG00000005884/ENSG00000130429/ENSG00000151914/ENSG00000100711/ENSG00000197321
GO:0030140 ENSG00000167461/ENSG00000119844
GO:0035097 ENSG00000139718/ENSG00000094916/ENSG00000170325/ENSG00000119403/ENSG00000106290/ENSG00000099381
GO:0098590 ENSG00000116183/ENSG00000133116/ENSG00000109062/ENSG00000171862/ENSG00000113758/ENSG00000072274/ENSG00000117600/ENSG00000044115/ENSG00000130303/ENSG00000183023/ENSG00000103222/ENSG00000163399/ENSG00000182718/ENSG00000151067/ENSG00000082397/ENSG00000073712/ENSG00000158887/ENSG00000162946/ENSG00000131409/ENSG00000115266/ENSG00000088836/ENSG00000163110/ENSG00000086991/ENSG00000164418/ENSG00000118160/ENSG00000163803/ENSG00000141068/ENSG00000080031/ENSG00000105974/ENSG00000122545/ENSG00000113448/ENSG00000174059/ENSG00000147649/ENSG00000167434/ENSG00000152818/ENSG00000173801/ENSG00000131188/ENSG00000141424/ENSG00000110697/ENSG00000232434/ENSG00000114019/ENSG00000134982/ENSG00000124171/ENSG00000134909/ENSG00000079435/ENSG00000112851/ENSG00000100345/ENSG00000136153/ENSG00000165802/ENSG00000160191/ENSG00000164741/ENSG00000065361/ENSG00000137845/ENSG00000122966/ENSG00000171631/ENSG00000133026/ENSG00000087470/ENSG00000100266/ENSG00000043462/ENSG00000132376/ENSG00000181790/ENSG00000154096/ENSG00000065243/ENSG00000105707/ENSG00000136854/ENSG00000096968/ENSG00000183783/ENSG00000134278/ENSG00000101384/ENSG00000120907/ENSG00000166313/ENSG00000132670/ENSG00000171992/ENSG00000197555/ENSG00000132613/ENSG00000168907/ENSG00000146192/ENSG00000148498/ENSG00000091513/ENSG00000145623/ENSG00000162736/ENSG00000164930/ENSG00000189195/ENSG00000162951/ENSG00000090565/ENSG00000112984/ENSG00000169379/ENSG00000145555/ENSG00000123143/ENSG00000180448/ENSG00000124839/ENSG00000170214/ENSG00000103569/ENSG00000163513/ENSG00000183454/ENSG00000111319/ENSG00000174827/ENSG00000084112
GO:0044463 ENSG00000161057/ENSG00000154040/ENSG00000167461/ENSG00000109062/ENSG00000171862/ENSG00000113758/ENSG00000114127/ENSG00000153187/ENSG00000126775/ENSG00000145911/ENSG00000176749/ENSG00000184009/ENSG00000176225/ENSG00000183023/ENSG00000080709/ENSG00000197386/ENSG00000151067/ENSG00000082397/ENSG00000158023/ENSG00000151532/ENSG00000101040/ENSG00000073712/ENSG00000162946/ENSG00000075413/ENSG00000115266/ENSG00000107404/ENSG00000169862/ENSG00000070404/ENSG00000164418/ENSG00000152767/ENSG00000070761/ENSG00000138443/ENSG00000118160/ENSG00000143740/ENSG00000135317/ENSG00000163803/ENSG00000141068/ENSG00000080031/ENSG00000100697/ENSG00000122545/ENSG00000139826/ENSG00000167434/ENSG00000105948/ENSG00000173947/ENSG00000139567/ENSG00000141543/ENSG00000152818/ENSG00000198718/ENSG00000131188/ENSG00000104133/ENSG00000141424/ENSG00000141564/ENSG00000196659/ENSG00000134982/ENSG00000187391/ENSG00000196839/ENSG00000134909/ENSG00000144857/ENSG00000165802/ENSG00000166501/ENSG00000115232/ENSG00000160191/ENSG00000164741/ENSG00000137845/ENSG00000121297/ENSG00000198836/ENSG00000133026/ENSG00000117724/ENSG00000078725/ENSG00000163001/ENSG00000144061/ENSG00000198844/ENSG00000198668/ENSG00000100266/ENSG00000119661/ENSG00000149930/ENSG00000132376/ENSG00000123700/ENSG00000181790/ENSG00000142168/ENSG00000224470/ENSG00000160460/ENSG00000154096/ENSG00000179397/ENSG00000103494/ENSG00000125851/ENSG00000111229/ENSG00000105443/ENSG00000166313/ENSG00000077063/ENSG00000130558/ENSG00000099260/ENSG00000165795/ENSG00000151849/ENSG00000171992/ENSG00000197555/ENSG00000132613/ENSG00000214274/ENSG00000168907/ENSG00000096093/ENSG00000146192/ENSG00000197283/ENSG00000148498/ENSG00000160469/ENSG00000135387/ENSG00000172531/ENSG00000133393/ENSG00000166579/ENSG00000087586/ENSG00000148019/ENSG00000197892/ENSG00000021574/ENSG00000189195/ENSG00000170017/ENSG00000113248/ENSG00000162951/ENSG00000128881/ENSG00000140854/ENSG00000115041/ENSG00000169379/ENSG00000163531/ENSG00000145555/ENSG00000180448/ENSG00000124839/ENSG00000142875/ENSG00000112679/ENSG00000131788/ENSG00000111319/ENSG00000114030/ENSG00000174827/ENSG00000084112/ENSG00000198561/ENSG00000136848/ENSG00000116584/ENSG00000105649/ENSG00000107679/ENSG00000184792/ENSG00000188026/ENSG00000125149/ENSG00000184160/ENSG00000088986/ENSG00000225697/ENSG00000134243/ENSG00000130203/ENSG00000138592/ENSG00000176658/ENSG00000108852/ENSG00000124126/ENSG00000099250/ENSG00000176101/ENSG00000197912/ENSG00000123607/ENSG00000106078/ENSG00000108349/ENSG00000188931/ENSG00000105221/ENSG00000189114/ENSG00000179632/ENSG00000175463/ENSG00000144451/ENSG00000092820/ENSG00000123908/ENSG00000106367/ENSG00000151491/ENSG00000076321/ENSG00000116729/ENSG00000020129/ENSG00000182324/ENSG00000119328/ENSG00000079805/ENSG00000168036/ENSG00000004455/ENSG00000145632/ENSG00000185896/ENSG00000115355/ENSG00000067606/ENSG00000125971/ENSG00000187688/ENSG00000162409/ENSG00000165698/ENSG00000048342/ENSG00000169604/ENSG00000107882/ENSG00000251322/ENSG00000129460/ENSG00000196961/ENSG00000196639/ENSG00000005884/ENSG00000110911/ENSG00000151914/ENSG00000111913/ENSG00000127603/ENSG00000100722/ENSG00000197375/ENSG00000198835/ENSG00000171867/ENSG00000137497/ENSG00000137880/ENSG00000137825/ENSG00000169398/ENSG00000197879/ENSG00000115415/ENSG00000110367
GO:0120038 ENSG00000161057/ENSG00000154040/ENSG00000167461/ENSG00000109062/ENSG00000171862/ENSG00000113758/ENSG00000114127/ENSG00000153187/ENSG00000126775/ENSG00000145911/ENSG00000176749/ENSG00000184009/ENSG00000176225/ENSG00000183023/ENSG00000080709/ENSG00000197386/ENSG00000151067/ENSG00000082397/ENSG00000158023/ENSG00000151532/ENSG00000101040/ENSG00000073712/ENSG00000162946/ENSG00000075413/ENSG00000115266/ENSG00000107404/ENSG00000169862/ENSG00000070404/ENSG00000164418/ENSG00000152767/ENSG00000070761/ENSG00000138443/ENSG00000118160/ENSG00000143740/ENSG00000135317/ENSG00000163803/ENSG00000141068/ENSG00000080031/ENSG00000100697/ENSG00000122545/ENSG00000139826/ENSG00000167434/ENSG00000105948/ENSG00000173947/ENSG00000139567/ENSG00000141543/ENSG00000152818/ENSG00000198718/ENSG00000131188/ENSG00000104133/ENSG00000141424/ENSG00000141564/ENSG00000196659/ENSG00000134982/ENSG00000187391/ENSG00000196839/ENSG00000134909/ENSG00000144857/ENSG00000165802/ENSG00000166501/ENSG00000115232/ENSG00000160191/ENSG00000164741/ENSG00000137845/ENSG00000121297/ENSG00000198836/ENSG00000133026/ENSG00000117724/ENSG00000078725/ENSG00000163001/ENSG00000144061/ENSG00000198844/ENSG00000198668/ENSG00000100266/ENSG00000119661/ENSG00000149930/ENSG00000132376/ENSG00000123700/ENSG00000181790/ENSG00000142168/ENSG00000224470/ENSG00000160460/ENSG00000154096/ENSG00000179397/ENSG00000103494/ENSG00000125851/ENSG00000111229/ENSG00000105443/ENSG00000166313/ENSG00000077063/ENSG00000130558/ENSG00000099260/ENSG00000165795/ENSG00000151849/ENSG00000171992/ENSG00000197555/ENSG00000132613/ENSG00000214274/ENSG00000168907/ENSG00000096093/ENSG00000146192/ENSG00000197283/ENSG00000148498/ENSG00000160469/ENSG00000135387/ENSG00000172531/ENSG00000133393/ENSG00000166579/ENSG00000087586/ENSG00000148019/ENSG00000197892/ENSG00000021574/ENSG00000189195/ENSG00000170017/ENSG00000113248/ENSG00000162951/ENSG00000128881/ENSG00000140854/ENSG00000115041/ENSG00000169379/ENSG00000163531/ENSG00000145555/ENSG00000180448/ENSG00000124839/ENSG00000142875/ENSG00000112679/ENSG00000131788/ENSG00000111319/ENSG00000114030/ENSG00000174827/ENSG00000084112/ENSG00000198561/ENSG00000136848/ENSG00000116584/ENSG00000105649/ENSG00000107679/ENSG00000184792/ENSG00000188026/ENSG00000125149/ENSG00000184160/ENSG00000088986/ENSG00000225697/ENSG00000134243/ENSG00000130203/ENSG00000138592/ENSG00000176658/ENSG00000108852/ENSG00000124126/ENSG00000099250/ENSG00000176101/ENSG00000197912/ENSG00000123607/ENSG00000106078/ENSG00000108349/ENSG00000188931/ENSG00000105221/ENSG00000189114/ENSG00000179632/ENSG00000175463/ENSG00000144451/ENSG00000092820/ENSG00000123908/ENSG00000106367/ENSG00000151491/ENSG00000076321/ENSG00000116729/ENSG00000020129/ENSG00000182324/ENSG00000119328/ENSG00000079805/ENSG00000168036/ENSG00000004455/ENSG00000145632/ENSG00000185896/ENSG00000115355/ENSG00000067606/ENSG00000125971/ENSG00000187688/ENSG00000162409/ENSG00000165698/ENSG00000048342/ENSG00000169604/ENSG00000107882/ENSG00000251322/ENSG00000129460/ENSG00000196961/ENSG00000196639/ENSG00000005884/ENSG00000110911/ENSG00000151914/ENSG00000111913/ENSG00000127603/ENSG00000100722/ENSG00000197375/ENSG00000198835/ENSG00000171867/ENSG00000137497/ENSG00000137880/ENSG00000137825/ENSG00000169398/ENSG00000197879/ENSG00000115415/ENSG00000110367
GO:0030992 ENSG00000105948/ENSG00000196659
GO:0000781 ENSG00000094916/ENSG00000170312/ENSG00000183765/ENSG00000099284/ENSG00000104885/ENSG00000160957/ENSG00000051180/ENSG00000173559/ENSG00000109534/ENSG00000139579/ENSG00000152457/ENSG00000100109/ENSG00000172531/ENSG00000102977/ENSG00000175054/ENSG00000167986/ENSG00000204569/ENSG00000138385/ENSG00000092330/ENSG00000062822/ENSG00000164091/ENSG00000132475/ENSG00000070366/ENSG00000132646/ENSG00000156831/ENSG00000107672/ENSG00000121931/ENSG00000085840/ENSG00000185379/ENSG00000145912
GO:0005903 ENSG00000109062/ENSG00000163803/ENSG00000167434/ENSG00000100345/ENSG00000133026/ENSG00000087470/ENSG00000128641/ENSG00000089006/ENSG00000157483/ENSG00000174827/ENSG00000225697/ENSG00000176658/ENSG00000092820/ENSG00000151491/ENSG00000110911/ENSG00000197375/ENSG00000197879/ENSG00000105357
GO:0045177 ENSG00000116183/ENSG00000133116/ENSG00000109062/ENSG00000171862/ENSG00000130303/ENSG00000163399/ENSG00000086991/ENSG00000163803/ENSG00000080031/ENSG00000122545/ENSG00000113448/ENSG00000174059/ENSG00000147649/ENSG00000167434/ENSG00000232434/ENSG00000114019/ENSG00000124171/ENSG00000136153/ENSG00000065361/ENSG00000171631/ENSG00000154096/ENSG00000105707/ENSG00000101384/ENSG00000128641/ENSG00000148498/ENSG00000091513/ENSG00000145623/ENSG00000164930/ENSG00000167306/ENSG00000124839/ENSG00000111319/ENSG00000174827/ENSG00000131620/ENSG00000110719/ENSG00000225697/ENSG00000154310/ENSG00000162738/ENSG00000188931/ENSG00000118777/ENSG00000092820/ENSG00000168036/ENSG00000010278/ENSG00000116266/ENSG00000067606/ENSG00000196961/ENSG00000116039/ENSG00000085563/ENSG00000110911/ENSG00000134830/ENSG00000111913/ENSG00000197375
GO:0030117 ENSG00000089818/ENSG00000166557/ENSG00000119844/ENSG00000131374/ENSG00000127527/ENSG00000242802/ENSG00000104142/ENSG00000128298/ENSG00000113615/ENSG00000254470/ENSG00000100280/ENSG00000189195
GO:0048475 ENSG00000089818/ENSG00000166557/ENSG00000119844/ENSG00000131374/ENSG00000127527/ENSG00000242802/ENSG00000104142/ENSG00000128298/ENSG00000113615/ENSG00000254470/ENSG00000100280/ENSG00000189195
GO:0005925 ENSG00000162434/ENSG00000044115/ENSG00000044574/ENSG00000184009/ENSG00000079308/ENSG00000073712/ENSG00000132470/ENSG00000086991/ENSG00000131016/ENSG00000213949/ENSG00000105974/ENSG00000140992/ENSG00000110031/ENSG00000196923/ENSG00000110841/ENSG00000136205/ENSG00000173801/ENSG00000109270/ENSG00000100345/ENSG00000136153/ENSG00000115232/ENSG00000164741/ENSG00000137845/ENSG00000159176/ENSG00000100266/ENSG00000197702/ENSG00000181790/ENSG00000154096/ENSG00000150764/ENSG00000076706/ENSG00000111229/ENSG00000096968/ENSG00000137809/ENSG00000135269/ENSG00000132912/ENSG00000074696/ENSG00000136950/ENSG00000140691/ENSG00000174444/ENSG00000162736/ENSG00000133818/ENSG00000149428/ENSG00000179218/ENSG00000082781/ENSG00000124942/ENSG00000170017/ENSG00000084733/ENSG00000163531/ENSG00000081237/ENSG00000145819/ENSG00000172403/ENSG00000107438/ENSG00000244486/ENSG00000116584/ENSG00000167004/ENSG00000108518/ENSG00000074054/ENSG00000185621/ENSG00000213741/ENSG00000099250/ENSG00000141367/ENSG00000171914/ENSG00000092820/ENSG00000182774/ENSG00000128591/ENSG00000115963/ENSG00000148180/ENSG00000063177/ENSG00000079805/ENSG00000168036/ENSG00000070756/ENSG00000138639/ENSG00000010278/ENSG00000124145/ENSG00000168310/ENSG00000139626/ENSG00000112655/ENSG00000005884/ENSG00000130429/ENSG00000151914/ENSG00000100711/ENSG00000197321
GO:0033116 ENSG00000166557/ENSG00000074695/ENSG00000080189/ENSG00000100528/ENSG00000099203/ENSG00000169696/ENSG00000179218/ENSG00000114745/ENSG00000105438/ENSG00000086598
GO:0098563 ENSG00000185896/ENSG00000197106/ENSG00000168118/ENSG00000144566/ENSG00000111737/ENSG00000111540/ENSG00000117758/ENSG00000101152/ENSG00000104888/ENSG00000132718/ENSG00000196517/ENSG00000104915/ENSG00000167964
GO:0032155 ENSG00000122545/ENSG00000110697/ENSG00000100345/ENSG00000122966/ENSG00000133026/ENSG00000065243/ENSG00000134278/ENSG00000090565/ENSG00000112984/ENSG00000123143/ENSG00000084112
GO:0090575 ENSG00000141646/ENSG00000153187/ENSG00000129173/ENSG00000106290/ENSG00000126351/ENSG00000082516/ENSG00000140332/ENSG00000077092/ENSG00000108590/ENSG00000148737/ENSG00000143257/ENSG00000125878/ENSG00000187079/ENSG00000143190/ENSG00000162227/ENSG00000064313/ENSG00000125651/ENSG00000112033
GO:0005789 ENSG00000125037/ENSG00000166557/ENSG00000164342/ENSG00000154645/ENSG00000099958/ENSG00000204642/ENSG00000143797/ENSG00000166479/ENSG00000151623/ENSG00000153132/ENSG00000074695/ENSG00000170312/ENSG00000092108/ENSG00000126775/ENSG00000150991/ENSG00000044574/ENSG00000172296/ENSG00000172071/ENSG00000006025/ENSG00000068615/ENSG00000143318/ENSG00000151532/ENSG00000198964/ENSG00000105223/ENSG00000172046/ENSG00000172292/ENSG00000143570/ENSG00000007541/ENSG00000086991/ENSG00000115363/ENSG00000141068/ENSG00000138678/ENSG00000099246/ENSG00000120942/ENSG00000105974/ENSG00000147649/ENSG00000149547/ENSG00000157593/ENSG00000110697/ENSG00000120697/ENSG00000101290/ENSG00000165029/ENSG00000133872/ENSG00000054392/ENSG00000083444/ENSG00000115970/ENSG00000134909/ENSG00000185813/ENSG00000178467/ENSG00000176454/ENSG00000139914/ENSG00000136908/ENSG00000239474/ENSG00000104936/ENSG00000026652/ENSG00000185000/ENSG00000198363/ENSG00000087470/ENSG00000119782/ENSG00000143195/ENSG00000129667/ENSG00000107566/ENSG00000156671/ENSG00000100528/ENSG00000052802/ENSG00000143418/ENSG00000110108/ENSG00000099203/ENSG00000105707/ENSG00000113615/ENSG00000163577/ENSG00000033011/ENSG00000167645/ENSG00000115307
GO:0005747 ENSG00000136521/ENSG00000099795/ENSG00000174886/ENSG00000183648/ENSG00000023228/ENSG00000140990/ENSG00000145494/ENSG00000137038
GO:0030964 ENSG00000136521/ENSG00000099795/ENSG00000174886/ENSG00000183648/ENSG00000023228/ENSG00000140990/ENSG00000145494/ENSG00000137038
GO:0045271 ENSG00000136521/ENSG00000099795/ENSG00000174886/ENSG00000183648/ENSG00000023228/ENSG00000140990/ENSG00000145494/ENSG00000137038
GO:0005924 ENSG00000162434/ENSG00000044115/ENSG00000044574/ENSG00000184009/ENSG00000079308/ENSG00000073712/ENSG00000132470/ENSG00000086991/ENSG00000131016/ENSG00000213949/ENSG00000105974/ENSG00000140992/ENSG00000110031/ENSG00000196923/ENSG00000110841/ENSG00000136205/ENSG00000173801/ENSG00000109270/ENSG00000100345/ENSG00000136153/ENSG00000115232/ENSG00000164741/ENSG00000137845/ENSG00000159176/ENSG00000100266/ENSG00000197702/ENSG00000181790/ENSG00000154096/ENSG00000150764/ENSG00000076706/ENSG00000111229/ENSG00000096968/ENSG00000137809/ENSG00000135269/ENSG00000132912/ENSG00000074696/ENSG00000136950/ENSG00000140691/ENSG00000174444/ENSG00000162736/ENSG00000133818/ENSG00000149428/ENSG00000179218/ENSG00000082781/ENSG00000124942/ENSG00000170017/ENSG00000084733/ENSG00000163531/ENSG00000081237/ENSG00000145819/ENSG00000172403/ENSG00000107438/ENSG00000244486/ENSG00000116584/ENSG00000167004/ENSG00000108518/ENSG00000074054/ENSG00000185621/ENSG00000213741/ENSG00000099250/ENSG00000141367/ENSG00000171914/ENSG00000092820/ENSG00000182774/ENSG00000128591/ENSG00000115963/ENSG00000148180/ENSG00000063177/ENSG00000079805/ENSG00000168036/ENSG00000070756/ENSG00000138639/ENSG00000010278/ENSG00000124145/ENSG00000168310/ENSG00000139626/ENSG00000112655/ENSG00000005884/ENSG00000130429/ENSG00000151914/ENSG00000100711/ENSG00000197321
GO:0031970 ENSG00000099795/ENSG00000100575/ENSG00000198836/ENSG00000142168/ENSG00000127554/ENSG00000150779/ENSG00000080819/ENSG00000170791/ENSG00000023228/ENSG00000168393/ENSG00000137642/ENSG00000004455
GO:0036464 ENSG00000161057/ENSG00000114127/ENSG00000153187/ENSG00000095627/ENSG00000183496/ENSG00000151461/ENSG00000066044/ENSG00000110697/ENSG00000141564/ENSG00000144535/ENSG00000162639/ENSG00000205323/ENSG00000055917/ENSG00000152520/ENSG00000166889
GO:0031463 ENSG00000163376/ENSG00000099910/ENSG00000145332/ENSG00000171617/ENSG00000239474/ENSG00000114796/ENSG00000180901/ENSG00000261609/ENSG00000100387/ENSG00000076321/ENSG00000166233
GO:0044215 ENSG00000074695/ENSG00000150991/ENSG00000133028/ENSG00000012223/ENSG00000185745/ENSG00000114030/ENSG00000082898
GO:0044216 ENSG00000074695/ENSG00000150991/ENSG00000133028/ENSG00000012223/ENSG00000185745/ENSG00000114030/ENSG00000082898
GO:0044217 ENSG00000074695/ENSG00000150991/ENSG00000133028/ENSG00000012223/ENSG00000185745/ENSG00000114030/ENSG00000082898gseaplot(gsego.cc.chimpspecific, geneSetID = "GO:0042611", pvalue_table = TRUE)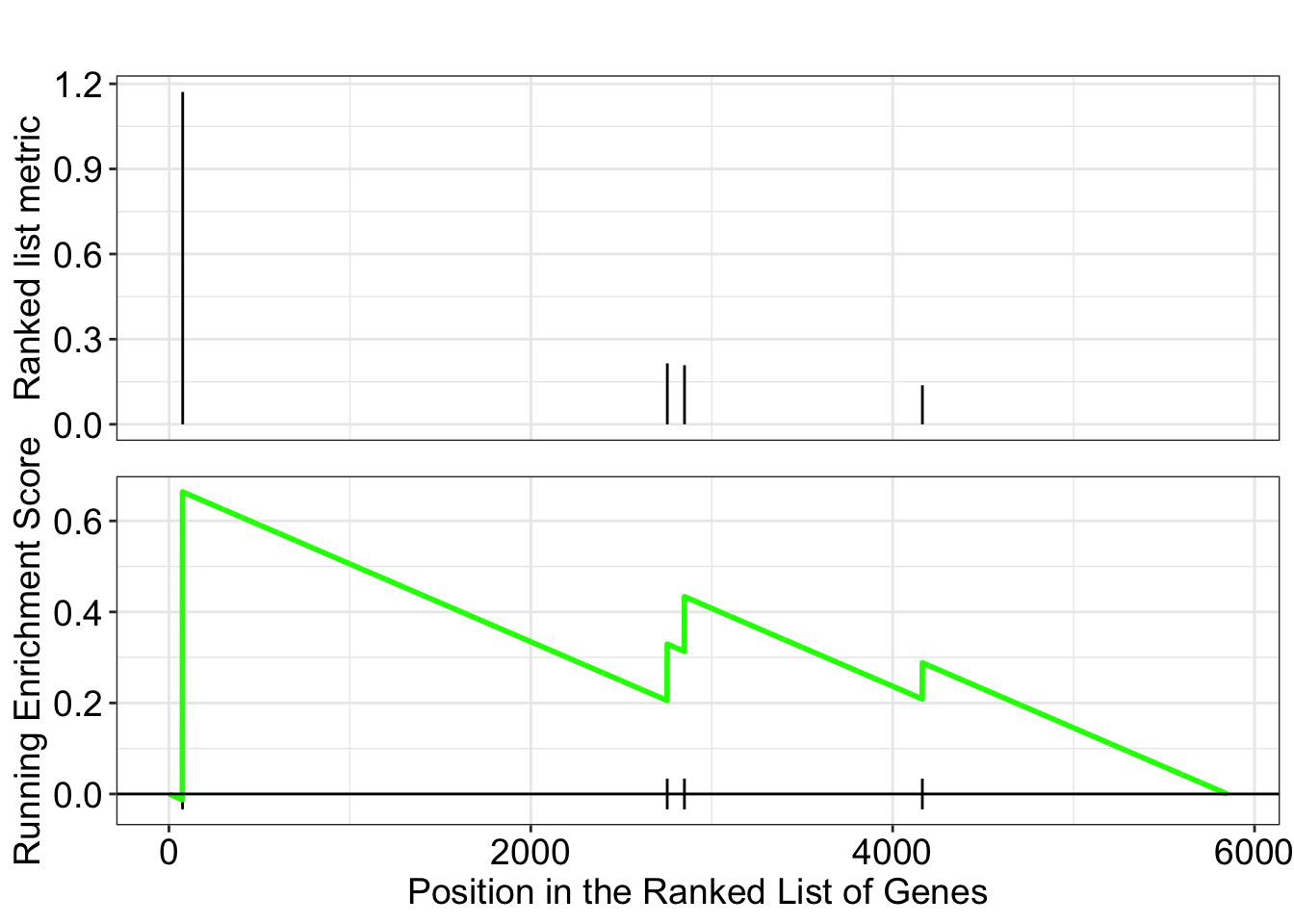
gseaplot(gsego.cc.chimpspecific, geneSetID = "GO:0042611", pvalue_table = TRUE)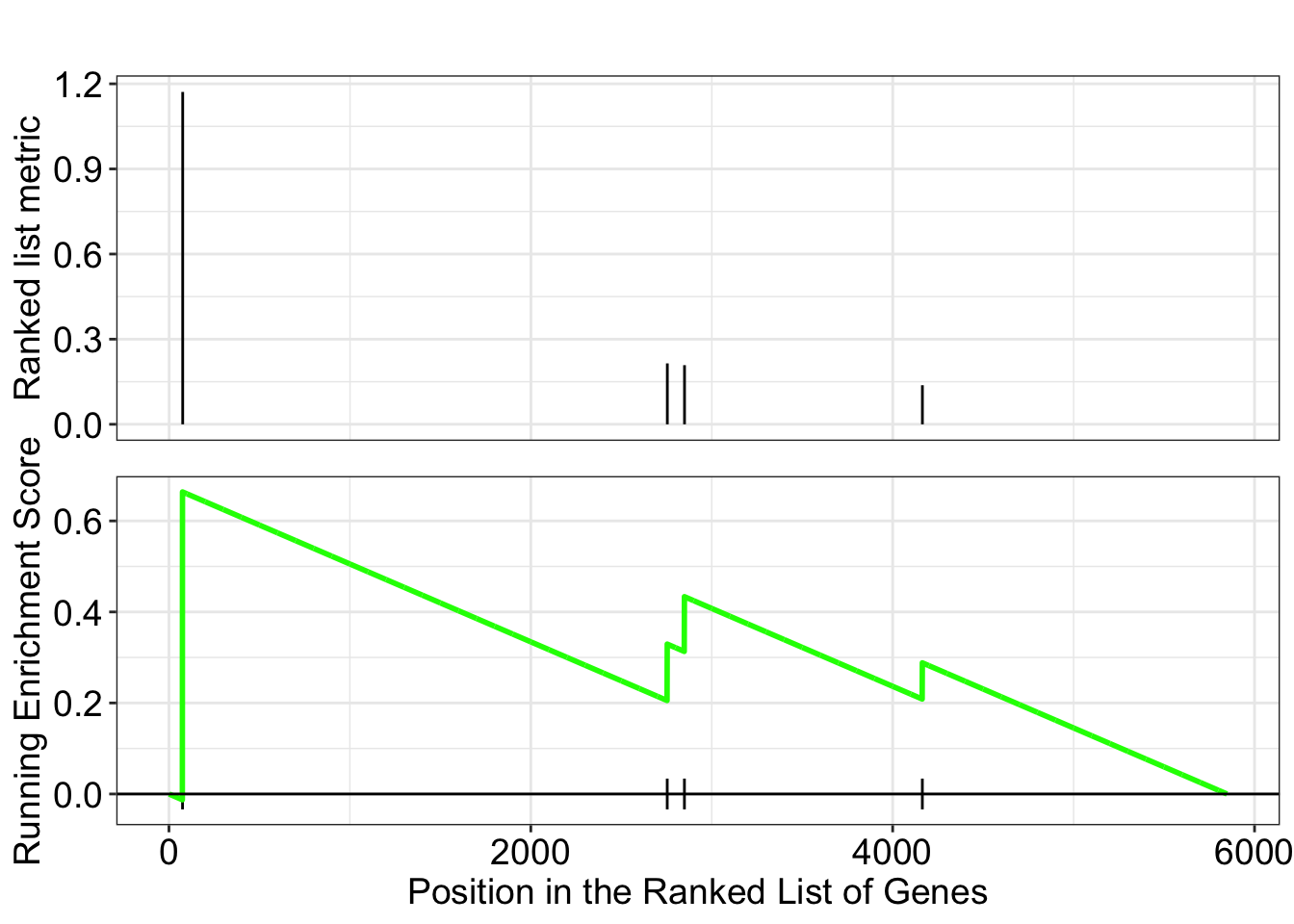
dotplot(gsego.cc.chimpspecific, font.size=8, showCategory=15)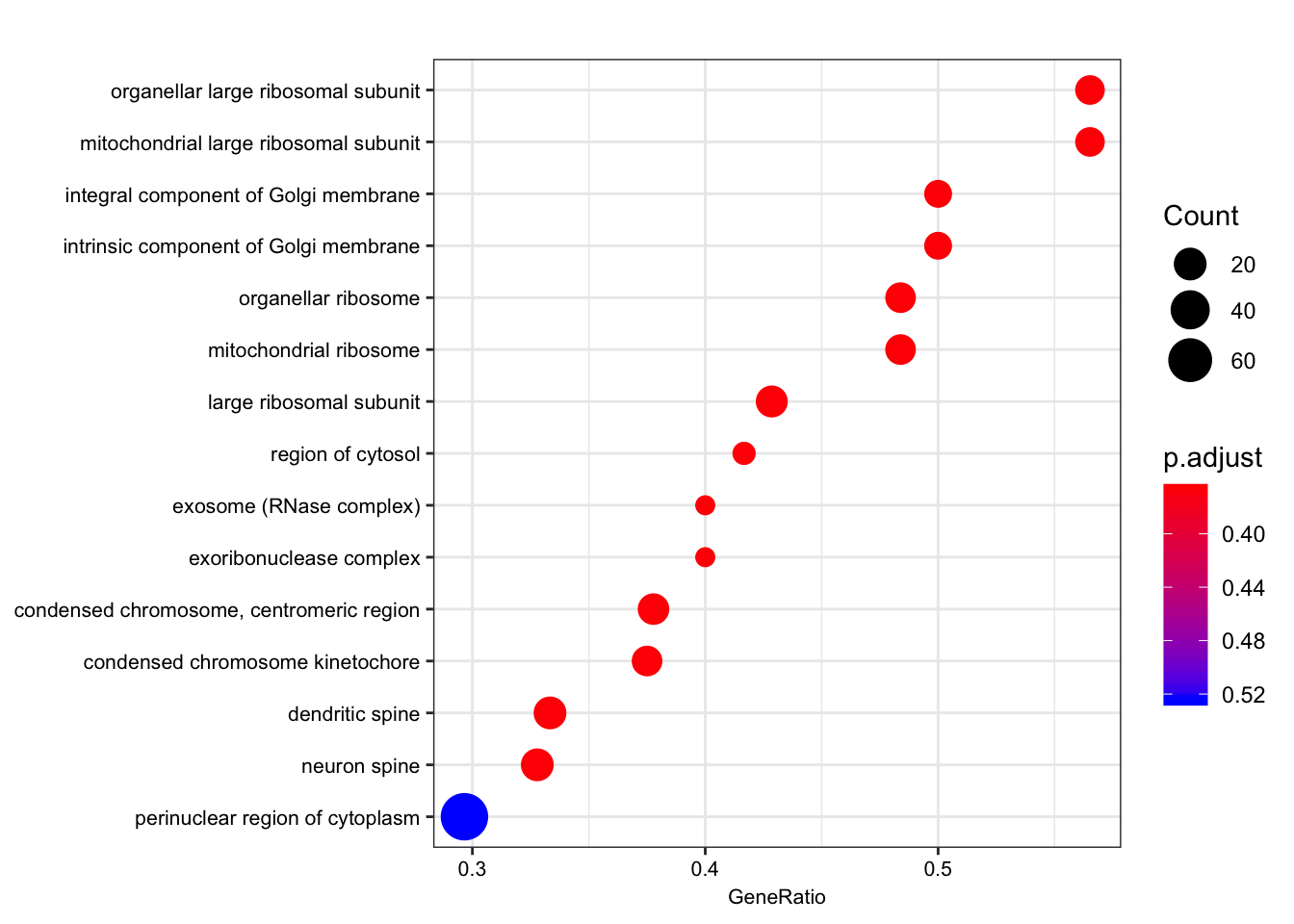
# Chimp_OrderedGenes <- eQTLs %>%
# group_by(gene) %>%
# dplyr::slice(which.min(qvalue)) %>%
# filter(gene %in% GtexHeartEgenes$chimp_id) %>%
# arrange(qvalue) %>%
# filter(!gene %in% HumanSigGenes)
# GeneList <- log10(Chimp_OrderedGenes$qvalue) * -1
# names(GeneList) <- as.character(ChimpToHuman.ID(Chimp_OrderedGenes$gene))
#
# gsego.cc.shared <- gseGO(gene = GeneList,
# OrgDb = org.Hs.eg.db,
# keyType = 'ENSEMBL',
# maxGSSize = 500,
# pvalueCutoff=0.8,
# ont = "CC")
# as.data.frame(gsego.cc.shared)
#
# gseaplot(gsego.cc.shared, geneSetID = "GO:0042611", pvalue_table = TRUE)
sessionInfo()R version 3.5.1 (2018-07-02)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS 10.14
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] parallel stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] gridExtra_2.3 org.Hs.eg.db_3.7.0 AnnotationDbi_1.44.0
[4] IRanges_2.16.0 S4Vectors_0.20.1 Biobase_2.42.0
[7] BiocGenerics_0.28.0 clusterProfiler_3.10.1 ggpmisc_0.3.1
[10] data.table_1.12.2 knitr_1.23 forcats_0.4.0
[13] stringr_1.4.0 dplyr_0.8.1 purrr_0.3.2
[16] readr_1.3.1 tidyr_0.8.3 tibble_2.1.1
[19] ggplot2_3.1.1 tidyverse_1.2.1 plyr_1.8.4
loaded via a namespace (and not attached):
[1] nlme_3.1-140 fs_1.3.1 enrichplot_1.2.0
[4] lubridate_1.7.4 bit64_0.9-7 progress_1.2.2
[7] RColorBrewer_1.1-2 httr_1.4.0 UpSetR_1.4.0
[10] rprojroot_1.3-2 tools_3.5.1 backports_1.1.4
[13] R6_2.4.0 DBI_1.0.0 lazyeval_0.2.2
[16] colorspace_1.4-1 withr_2.1.2 prettyunits_1.0.2
[19] tidyselect_0.2.5 bit_1.1-14 compiler_3.5.1
[22] git2r_0.25.2 cli_1.1.0 rvest_0.3.4
[25] xml2_1.2.0 labeling_0.3 triebeard_0.3.0
[28] scales_1.0.0 ggridges_0.5.1 digest_0.6.19
[31] rmarkdown_1.13 DOSE_3.8.2 pkgconfig_2.0.2
[34] htmltools_0.3.6 highr_0.8 rlang_0.3.4
[37] readxl_1.3.1 rstudioapi_0.10 RSQLite_2.1.1
[40] gridGraphics_0.4-1 generics_0.0.2 farver_1.1.0
[43] jsonlite_1.6 BiocParallel_1.16.6 GOSemSim_2.8.0
[46] magrittr_1.5 ggplotify_0.0.3 GO.db_3.7.0
[49] Matrix_1.2-17 Rcpp_1.0.1 munsell_0.5.0
[52] viridis_0.5.1 stringi_1.4.3 yaml_2.2.0
[55] ggraph_1.0.2 MASS_7.3-51.4 qvalue_2.14.1
[58] grid_3.5.1 blob_1.1.1 ggrepel_0.8.1
[61] DO.db_2.9 crayon_1.3.4 lattice_0.20-38
[64] cowplot_0.9.4 haven_2.1.0 splines_3.5.1
[67] hms_0.4.2 pillar_1.4.0 fgsea_1.8.0
[70] igraph_1.2.4.1 reshape2_1.4.3 fastmatch_1.1-0
[73] glue_1.3.1 evaluate_0.13 modelr_0.1.4
[76] urltools_1.7.3 tweenr_1.0.1 cellranger_1.1.0
[79] gtable_0.3.0 polyclip_1.10-0 assertthat_0.2.1
[82] xfun_0.7 ggforce_0.2.2 europepmc_0.3
[85] broom_0.5.2 viridisLite_0.3.0 rvcheck_0.1.3
[88] memoise_1.1.0 workflowr_1.3.0