20190326_Admixture
Ben Fair
3/26/2019
Last updated: 2019-04-03
Checks: 5 1
Knit directory: Comparative_eQTL/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.2.0). The Report tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190319) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Untracked files:
Untracked: analysis/20190321_Check-Kinship-And-PopulationStructure.Rmd
Untracked: analysis/20190325_MergingRNASeqLanes.Rmd
Untracked: analysis/20190326_Admixture.Rmd
Untracked: analysis/20190326_PCA.Rmd
Untracked: analysis/20190327_MakeFamAndCovariateFiles.Rmd
Untracked: analysis/20190327_MakeFamPhenotypeFile.Rmd
Untracked: docs/figure/20190321_Check-Kinship-And-PopulationStructure.Rmd/
Untracked: docs/figure/20190325_MergingRNASeqLanes.Rmd/
Untracked: docs/figure/20190326_PCA.Rmd/
Unstaged changes:
Deleted: ._workflowr.yml.swp
Modified: analysis/20190320_Check-RNAseq-PCs.Rmd
Modified: analysis/index.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with wflow_publish() to start tracking its development.
genotype data from this study (as vcf-files) was merged with that from deManuel et al 2016 (after LiftOver to PanTro5) which contains 65 chimp whole-genome genotype data spanning all of 4 recognized sub-species. Snps were pruned to get variants in approximate equilibrium. Admixture algoroithm was used to examine population substructure and admixture. Results for K=4 is plotted below. See PCA of genotypes for a different look at the same set of samples based on the same set of snps.
library(plyr)
library(tidyverse)
library(knitr)
library(reshape2)AdmixtureResults <- read.table("../output/PopulationStructure/Admixture/MergedForAdmixture.4.Q.labelled") %>%
mutate(id=paste(V1, V2)) %>%
select( fam = 1, ind=2, everything() ) %>%
select(-V3, -V4, -V5, -V6)
kable(head(AdmixtureResults))| fam | ind | V7 | V8 | V9 | V10 | id |
|---|---|---|---|---|---|---|
| Pan_troglodytes_ThisStudy | 549 | 1.0e-05 | 0.000010 | 0.999970 | 1e-05 | Pan_troglodytes_ThisStudy 549 |
| Pan_troglodytes_ThisStudy | 570 | 1.3e-05 | 0.059266 | 0.940711 | 1e-05 | Pan_troglodytes_ThisStudy 570 |
| Pan_troglodytes_ThisStudy | 389 | 1.0e-05 | 0.000010 | 0.999970 | 1e-05 | Pan_troglodytes_ThisStudy 389 |
| Pan_troglodytes_ThisStudy | 456 | 1.1e-05 | 0.000010 | 0.999969 | 1e-05 | Pan_troglodytes_ThisStudy 456 |
| Pan_troglodytes_ThisStudy | 623 | 1.0e-05 | 0.000010 | 0.999970 | 1e-05 | Pan_troglodytes_ThisStudy 623 |
| Pan_troglodytes_ThisStudy | 438 | 1.0e-05 | 0.000010 | 0.999970 | 1e-05 | Pan_troglodytes_ThisStudy 438 |
Columns 3 to end represent admixed membership among K sub-populations.
# Reordering samples makes admixture plots prettier
AdmixtureResults$ind <- reorder(AdmixtureResults$ind, AdmixtureResults$V8)
# Rename species (fam-ID) to common name
AdmixtureResults$fam <- mapvalues(AdmixtureResults$fam, from=c("Pan_troglodytes_schweinfurthii", "Pan_troglodytes_ellioti", "Pan_troglodytes_ThisStudy", "Pan_troglodytes", "Pan_troglodytes_troglodytes", "Pan_troglodytes_verus"), to=c("Eastern", "Nigeria\nCameroon", "This Study", "Eastern", "Central", "Western"))
# plot
melt(AdmixtureResults) %>%
ggplot(aes(x = ind, y = value, fill = variable)) +
geom_bar(stat = "identity") +
scale_y_continuous(limits = c(-0.001,1.001), expand = c(0, 0)) +
facet_grid(~fam, scales="free_x", space="free_x") +
theme(legend.position="none") +
theme(axis.title.y=element_blank(), axis.title.x=element_blank(), axis.ticks.x=element_blank(), axis.text.x=element_text(size=rel(0.5), angle=70, hjust=1))Using fam, ind, id as id variables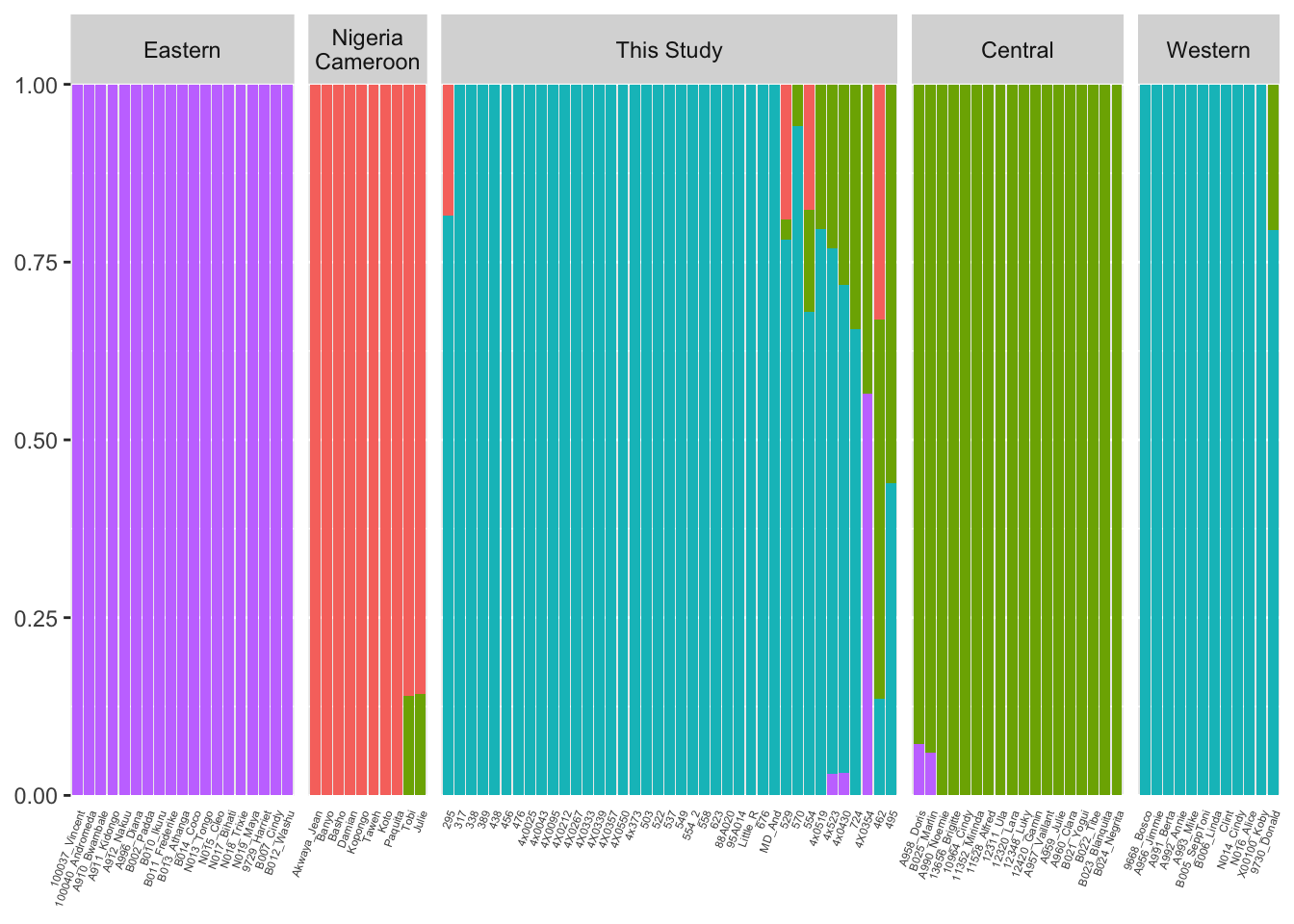
sessionInfo()R version 3.5.1 (2018-07-02)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS 10.14
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] reshape2_1.4.3 knitr_1.22 forcats_0.4.0 stringr_1.4.0
[5] dplyr_0.8.0.1 purrr_0.3.2 readr_1.3.1 tidyr_0.8.2
[9] tibble_2.1.1 ggplot2_3.1.0 tidyverse_1.2.1 plyr_1.8.4
loaded via a namespace (and not attached):
[1] Rcpp_1.0.1 highr_0.8 cellranger_1.1.0 pillar_1.3.1
[5] compiler_3.5.1 git2r_0.24.0 workflowr_1.2.0 tools_3.5.1
[9] digest_0.6.18 lubridate_1.7.4 jsonlite_1.6 evaluate_0.13
[13] nlme_3.1-137 gtable_0.3.0 lattice_0.20-38 pkgconfig_2.0.2
[17] rlang_0.3.3 cli_1.1.0 rstudioapi_0.10 yaml_2.2.0
[21] haven_2.1.0 xfun_0.6 withr_2.1.2 xml2_1.2.0
[25] httr_1.4.0 hms_0.4.2 generics_0.0.2 fs_1.2.6
[29] rprojroot_1.3-2 grid_3.5.1 tidyselect_0.2.5 glue_1.3.1
[33] R6_2.4.0 readxl_1.1.0 rmarkdown_1.11 modelr_0.1.4
[37] magrittr_1.5 backports_1.1.3 scales_1.0.0 htmltools_0.3.6
[41] rvest_0.3.2 assertthat_0.2.1 colorspace_1.4-1 labeling_0.3
[45] stringi_1.4.3 lazyeval_0.2.2 munsell_0.5.0 broom_0.5.1
[49] crayon_1.3.4