20190821_eGeneTissueCount
Ben Fair
8/21/2019
Last updated: 2019-10-15
Checks: 6 1
Knit directory: Comparative_eQTL/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.4.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has staged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190319) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.DS_Store
Ignored: analysis/20190521_eQTL_CrossSpeciesEnrichment_cache/
Ignored: analysis_temp/.DS_Store
Ignored: code/.DS_Store
Ignored: code/snakemake_workflow/.DS_Store
Ignored: data/.DS_Store
Ignored: data/PastAnalysesDataToKeep/.DS_Store
Ignored: docs/.DS_Store
Ignored: docs/assets/.DS_Store
Untracked files:
Untracked: code/MakeFigures.R
Unstaged changes:
Modified: code/CustomFunctions.R
Staged changes:
Modified: analysis/20190821_eGeneTissueCount.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the R Markdown and HTML files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view them.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | b62b089 | Benjmain Fair | 2019-08-21 | update site |
| html | b62b089 | Benjmain Fair | 2019-08-21 | update site |
library(tidyverse)
library(knitr)
library(matrixStats)
library(dplyr)
library(ggExtra)
library(reshape2)Are eGenes that are shared between humans and chimps more likely to be eGenes in many tissues?
From a table of eGene qvalues across all GTEx tissues (GTEx v7 release), first get distribution of how many tissues each gene has a qval under some threshold
GTEx <- read.table("../data/AllGTExTissues.egenes.txt", header=T, sep='\t')
Threshold=0.1
TissueEgeneCount <- data.frame(TissueCount=rowSums(GTEx[,-1]<=Threshold, na.rm=T), Gene.stable.ID=gsub("\\.\\d+", "", GTEx$gene_id, perl=T))
hist(TissueEgeneCount$TissueCount, breaks=50)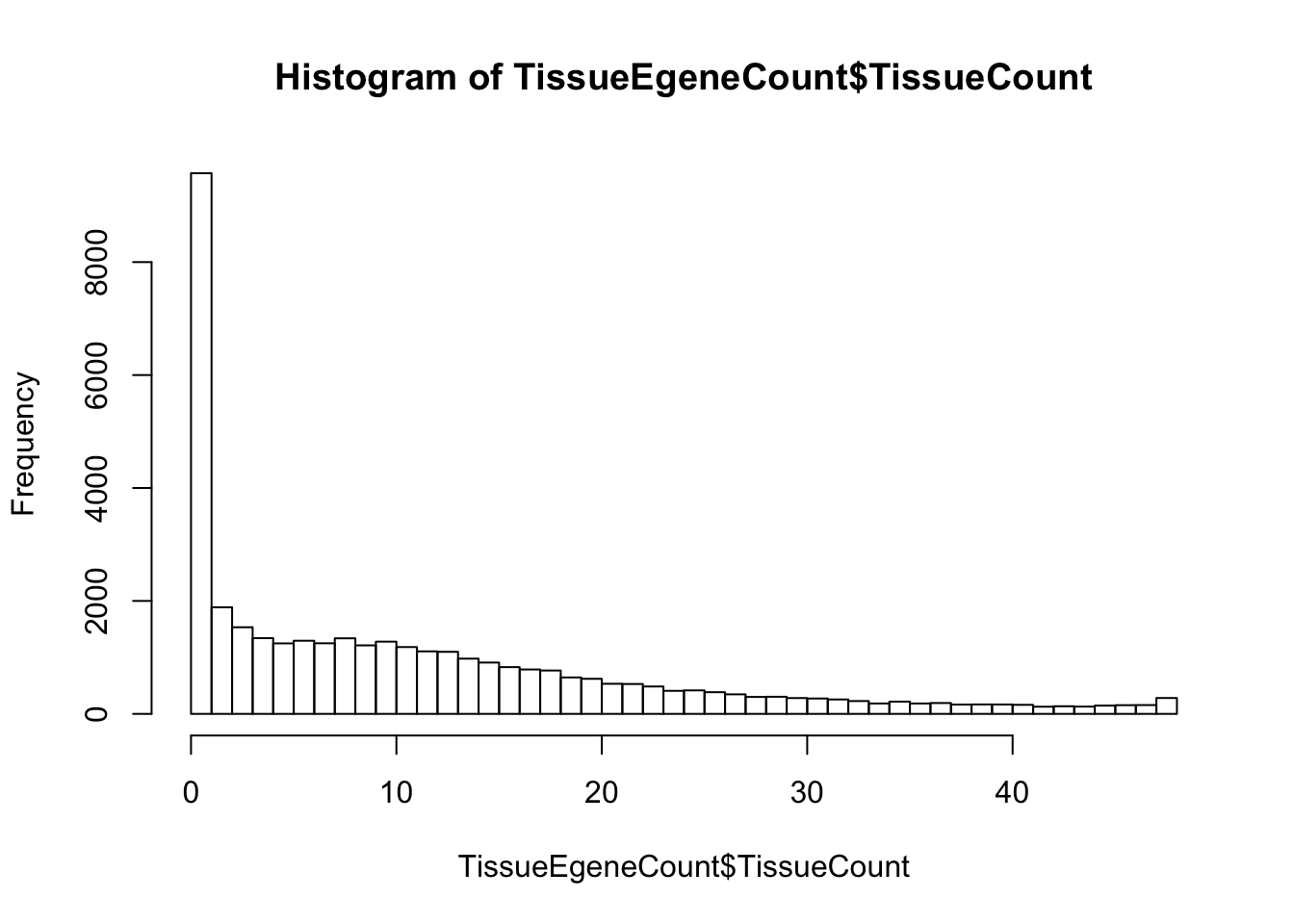
| Version | Author | Date |
|---|---|---|
| b62b089 | Benjmain Fair | 2019-08-21 |
From my data, plot this distribution (as a cumulative dist) after stratifying into human-specific eGenes, versus shared eGenes.
eQTLs <- read.table(gzfile("../data/PastAnalysesDataToKeep/20190521_eQTLs_250kB_10MAF.txt.gz"), header=T)
kable(head(eQTLs))| snps | gene | beta | statistic | pvalue | FDR | qvalue |
|---|---|---|---|---|---|---|
| ID.1.126459696.ACCCTAGTAAG.A | ENSPTRG00000001061 | 3.570039 | 12.50828 | 0 | 3.5e-06 | 3.5e-06 |
| ID.1.126465687.TTGT.A | ENSPTRG00000001061 | 3.570039 | 12.50828 | 0 | 3.5e-06 | 3.5e-06 |
| ID.1.126465750.TG.CT | ENSPTRG00000001061 | 3.570039 | 12.50828 | 0 | 3.5e-06 | 3.5e-06 |
| ID.1.126465756.T.C | ENSPTRG00000001061 | 3.570039 | 12.50828 | 0 | 3.5e-06 | 3.5e-06 |
| ID.1.126465766.C.A | ENSPTRG00000001061 | 3.570039 | 12.50828 | 0 | 3.5e-06 | 3.5e-06 |
| ID.1.126465774.G.A | ENSPTRG00000001061 | 3.570039 | 12.50828 | 0 | 3.5e-06 | 3.5e-06 |
# List of chimp tested genes
ChimpTestedGenes <- rownames(read.table('../output/ExpressionMatrix.un-normalized.txt.gz', header=T, check.names=FALSE, row.names = 1))
ChimpToHumanGeneMap <- read.table("../data/Biomart_export.Hsap.Ptro.orthologs.txt.gz", header=T, sep='\t', stringsAsFactors = F)
kable(head(ChimpToHumanGeneMap))| Gene.stable.ID | Transcript.stable.ID | Chimpanzee.gene.stable.ID | Chimpanzee.gene.name | Chimpanzee.protein.or.transcript.stable.ID | Chimpanzee.homology.type | X.id..target.Chimpanzee.gene.identical.to.query.gene | X.id..query.gene.identical.to.target.Chimpanzee.gene | dN.with.Chimpanzee | dS.with.Chimpanzee | Chimpanzee.orthology.confidence..0.low..1.high. |
|---|---|---|---|---|---|---|---|---|---|---|
| ENSG00000198888 | ENST00000361390 | ENSPTRG00000042641 | MT-ND1 | ENSPTRP00000061407 | ortholog_one2one | 94.6541 | 94.6541 | 0.0267 | 0.5455 | 1 |
| ENSG00000198763 | ENST00000361453 | ENSPTRG00000042626 | MT-ND2 | ENSPTRP00000061406 | ortholog_one2one | 96.2536 | 96.2536 | 0.0185 | 0.7225 | 1 |
| ENSG00000210127 | ENST00000387392 | ENSPTRG00000042642 | MT-TA | ENSPTRT00000076396 | ortholog_one2one | 100.0000 | 100.0000 | NA | NA | NA |
| ENSG00000198804 | ENST00000361624 | ENSPTRG00000042657 | MT-CO1 | ENSPTRP00000061408 | ortholog_one2one | 98.8304 | 98.8304 | 0.0065 | 0.5486 | 1 |
| ENSG00000198712 | ENST00000361739 | ENSPTRG00000042660 | MT-CO2 | ENSPTRP00000061402 | ortholog_one2one | 97.7974 | 97.7974 | 0.0106 | 0.5943 | 1 |
| ENSG00000228253 | ENST00000361851 | ENSPTRG00000042653 | MT-ATP8 | ENSPTRP00000061400 | ortholog_one2one | 94.1176 | 94.1176 | 0.0325 | 0.3331 | 1 |
# Of this ortholog list, how many genes are one2one
table(ChimpToHumanGeneMap$Chimpanzee.homology.type)
ortholog_many2many ortholog_one2many ortholog_one2one
2278 19917 140351 OneToOneMap <- ChimpToHumanGeneMap %>%
filter(Chimpanzee.homology.type=="ortholog_one2one") %>%
distinct(Chimpanzee.gene.stable.ID, .keep_all = TRUE) %>%
left_join(TissueEgeneCount, by="Gene.stable.ID")
# Read gtex heart egene list
# Only consider those that were tested in both species and are one2one orthologs
GtexHeartEgenes <- read.table("../data/Heart_Left_Ventricle.v7.egenes.txt.gz", header=T, sep='\t', stringsAsFactors = F) %>%
mutate(gene_id_stable = gsub(".\\d+$","",gene_id)) %>%
filter(gene_id_stable %in% OneToOneMap$Gene.stable.ID) %>%
mutate(chimp_id = plyr::mapvalues(gene_id_stable, OneToOneMap$Gene.stable.ID, OneToOneMap$Chimpanzee.gene.stable.ID, warn_missing = F)) %>%
filter(chimp_id %in% ChimpTestedGenes)
ChimpToHuman.ID <- function(Chimp.ID){
#function to convert chimp ensembl to human ensembl gene ids
return(
plyr::mapvalues(Chimp.ID, OneToOneMap$Chimpanzee.gene.stable.ID, OneToOneMap$Gene.stable.ID, warn_missing = F)
)}
HumanFDR <- 0.1
ChimpFDR <- 0.1
#Get chimp eQTLs
Chimp_eQTLs <- eQTLs %>%
filter(qvalue<ChimpFDR)
# Count chimp eGenes
length(unique(Chimp_eQTLs$gene))[1] 336# Count human eGenes
length(GtexHeartEgenes %>% filter(qval< HumanFDR) %>% pull(chimp_id))[1] 5410# Count number genes tested in both species (already filtered for 1to1 orthologs)
length(GtexHeartEgenes$gene_id_stable)[1] 11586#Change FDR thresholds or take top N eGenes by qvalue
HumanTopN <- 600
HumanFDR <- 0.1
ChimpFDR <- 0.1
# Filter human eGenes by qval threshold
# HumanSigGenes <- GtexHeartEgenes %>% filter(qval<HumanFDR) %>% pull(chimp_id)
# Filter human eGenes by topN qval
HumanSigGenes <- GtexHeartEgenes %>% top_n(-HumanTopN, qval) %>% pull(chimp_id)
# Filter human eGeness by qval threshold then topN betas
# HumanSigGenes <- GtexHeartEgenes %>% filter(qval<HumanFDR) %>% top_n(1000, abs(slope)) %>% pull(chimp_id)
HumanNonSigGenes <- GtexHeartEgenes %>%
filter(!chimp_id %in% HumanSigGenes) %>%
pull(chimp_id)
ChimpSigGenes <- GtexHeartEgenes %>%
filter(chimp_id %in% Chimp_eQTLs$gene) %>%
pull(chimp_id)
ChimpNonSigGenes <- GtexHeartEgenes %>%
filter(! chimp_id %in% Chimp_eQTLs$gene) %>%
pull(chimp_id)
ContigencyTable <- matrix( c( length(intersect(ChimpSigGenes,HumanSigGenes)),
length(intersect(HumanSigGenes,ChimpNonSigGenes)),
length(intersect(ChimpSigGenes,HumanNonSigGenes)),
length(intersect(ChimpNonSigGenes,HumanNonSigGenes))),
nrow = 2)
rownames(ContigencyTable) <- c("Chimp eGene", "Not Chimp eGene")
colnames(ContigencyTable) <- c("Human eGene", "Not human eGene")
#what is qval threshold for human eGene classification in this contigency table
print(GtexHeartEgenes %>% top_n(-HumanTopN, qval) %>% top_n(1, qval) %>% pull(qval))[1] 5.83223e-12#Contigency table of one to one orthologs tested in both chimps and humans of whether significant in humans, or chimps, or both, or neither
ContigencyTable Human eGene Not human eGene
Chimp eGene 28 252
Not Chimp eGene 572 10734#One-sided Fisher test for greater overlap than expected by chance
fisher.test(ContigencyTable, alternative="greater")
Fisher's Exact Test for Count Data
data: ContigencyTable
p-value = 0.0006395
alternative hypothesis: true odds ratio is greater than 1
95 percent confidence interval:
1.444504 Inf
sample estimates:
odds ratio
2.084996 #Chimp eGenes vs non chimp eGenes
ToPlot <- GtexHeartEgenes %>%
mutate(group = case_when(
chimp_id %in% ChimpSigGenes ~ "chimp.eGene",
!chimp_id %in% ChimpSigGenes ~ "not.chimp.eGene")) %>%
left_join(OneToOneMap, by=c("chimp_id"="Chimpanzee.gene.stable.ID"))
Chimp.tissue.plot <- ggplot(ToPlot, aes(color=group,x=TissueCount)) +
stat_ecdf(geom = "step") +
ylab("Cumulative frequency") +
xlab("TissueCount") +
annotate("text", x = 40, y = 0.4, label = paste("Mann-Whitney\none-sided P =", signif(wilcox.test(data=ToPlot, TissueCount ~ group, alternative="greater")$p.value, 2) )) +
theme_bw() +
theme(legend.position = c(.80, .2), legend.title=element_blank())
#Human eGenes vs non human eGenes
ToPlot <- GtexHeartEgenes %>%
mutate(group = case_when(
chimp_id %in% HumanSigGenes ~ "human.eGene",
!chimp_id %in% HumanSigGenes ~ "not.human.eGene")) %>%
left_join(OneToOneMap, by=c("chimp_id"="Chimpanzee.gene.stable.ID"))
Human.tissue.plot <- ggplot(ToPlot, aes(color=group,x=TissueCount)) +
stat_ecdf(geom = "step") +
ylab("Cumulative frequency") +
xlab("TissueCount") +
annotate("text", x = 40, y = 0.4, label = paste("Mann-Whitney\none-sided P =", signif(wilcox.test(data=ToPlot, TissueCount ~ group, alternative="greater")$p.value, 2) )) +
theme_bw() +
theme(legend.position = c(.80, .2), legend.title=element_blank())
#Shared eGenes vs human-specific eGenes
ToPlot <- GtexHeartEgenes %>%
mutate(group = case_when(
chimp_id %in% intersect(HumanSigGenes, ChimpSigGenes) ~ "shared.eGene",
chimp_id %in% setdiff(HumanSigGenes, ChimpSigGenes) ~ "human.specific.eGene")) %>%
filter(chimp_id %in% union(intersect(HumanSigGenes, ChimpSigGenes), setdiff(HumanSigGenes, ChimpSigGenes)))%>%
left_join(OneToOneMap, by=c("chimp_id"="Chimpanzee.gene.stable.ID"))
Shared.human.tissue.plot <- ggplot(ToPlot, aes(color=group,x=TissueCount)) +
stat_ecdf(geom = "step") +
ylab("Cumulative frequency") +
xlab("TissueCount") +
annotate("text", x = 40, y = 0.4, label = paste("Mann-Whitney\none-sided P =", signif(wilcox.test(data=ToPlot, TissueCount ~ group, alternative="less")$p.value, 2) )) +
theme_bw() +
theme(legend.position = c(.80, .2), legend.title=element_blank())
#Shared eGenes vs chimp-specific eGenes
ToPlot <- GtexHeartEgenes %>%
left_join(OneToOneMap, by=c("chimp_id"="Chimpanzee.gene.stable.ID")) %>%
mutate(dN.dS = dN.with.Chimpanzee/dS.with.Chimpanzee) %>%
mutate(group = case_when(
chimp_id %in% intersect(HumanSigGenes, ChimpSigGenes) ~ "shared.eGene",
chimp_id %in% setdiff(ChimpSigGenes, HumanSigGenes) ~ "chimp.specific.eGene")) %>%
dplyr::filter(chimp_id %in% union(intersect(HumanSigGenes, ChimpSigGenes), setdiff(ChimpSigGenes, HumanSigGenes)))
Shared.chimp.tissue.plot <- ggplot(ToPlot, aes(color=group,x=TissueCount)) +
stat_ecdf(geom = "step") +
ylab("Cumulative frequency") +
xlab("TissueCount") +
annotate("text", x = 40, y = 0.4, label = paste("Mann-Whitney\none-sided P =", signif(wilcox.test(data=ToPlot, TissueCount ~ group, alternative="less")$p.value, 2) )) +
theme_bw() +
theme(legend.position = c(.80, .2), legend.title=element_blank())
Chimp.tissue.plot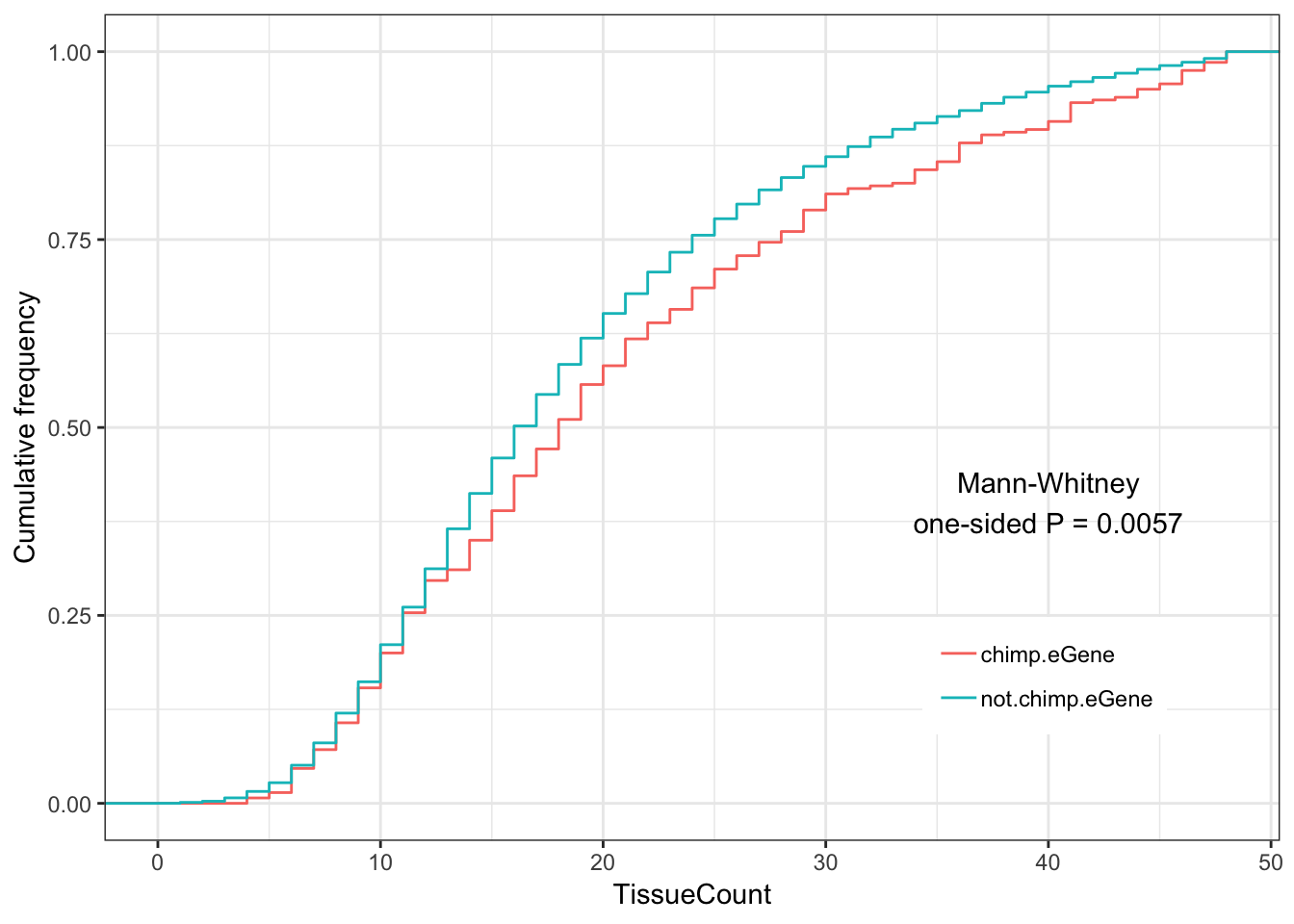
| Version | Author | Date |
|---|---|---|
| b62b089 | Benjmain Fair | 2019-08-21 |
Human.tissue.plot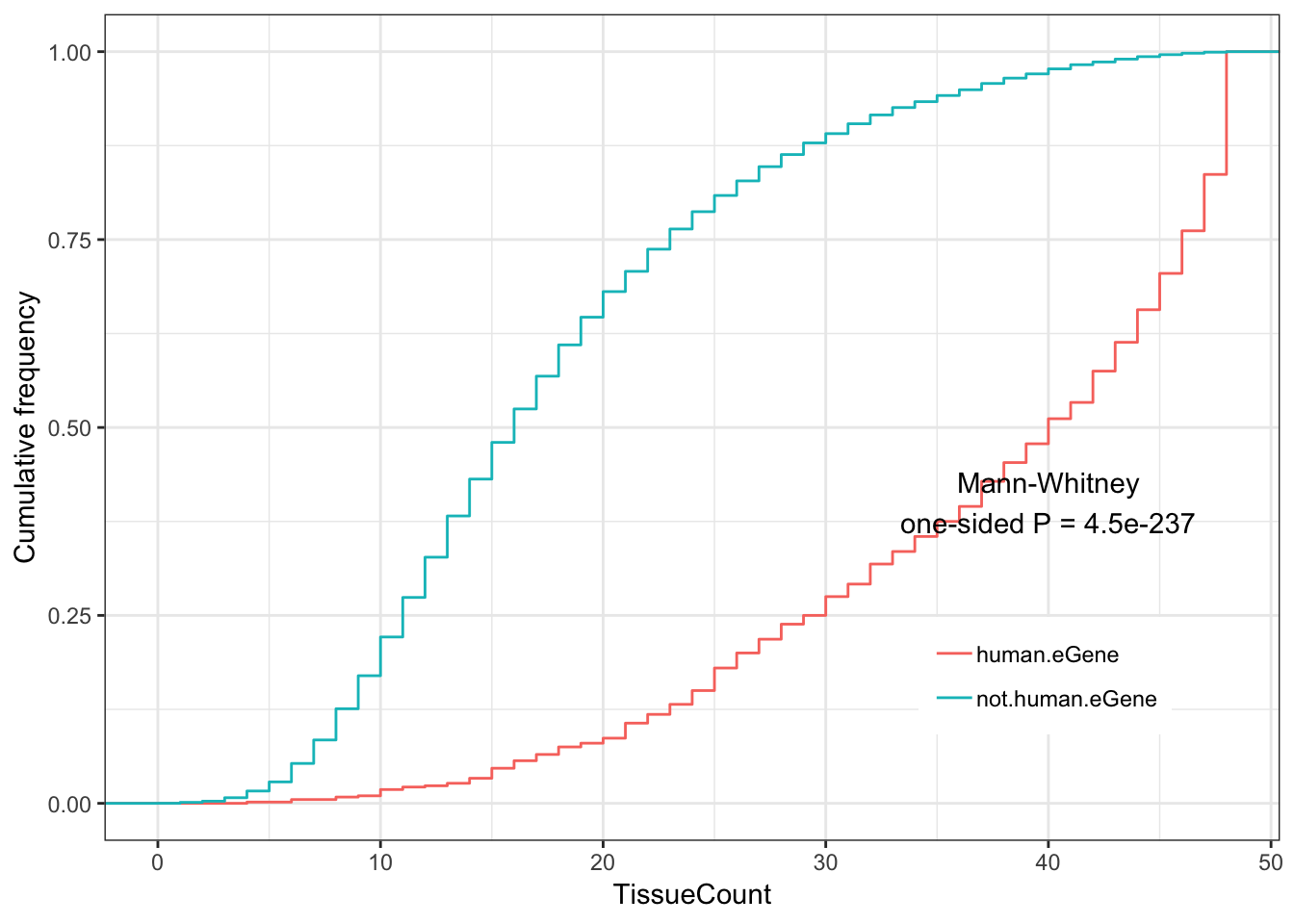
| Version | Author | Date |
|---|---|---|
| b62b089 | Benjmain Fair | 2019-08-21 |
Shared.human.tissue.plot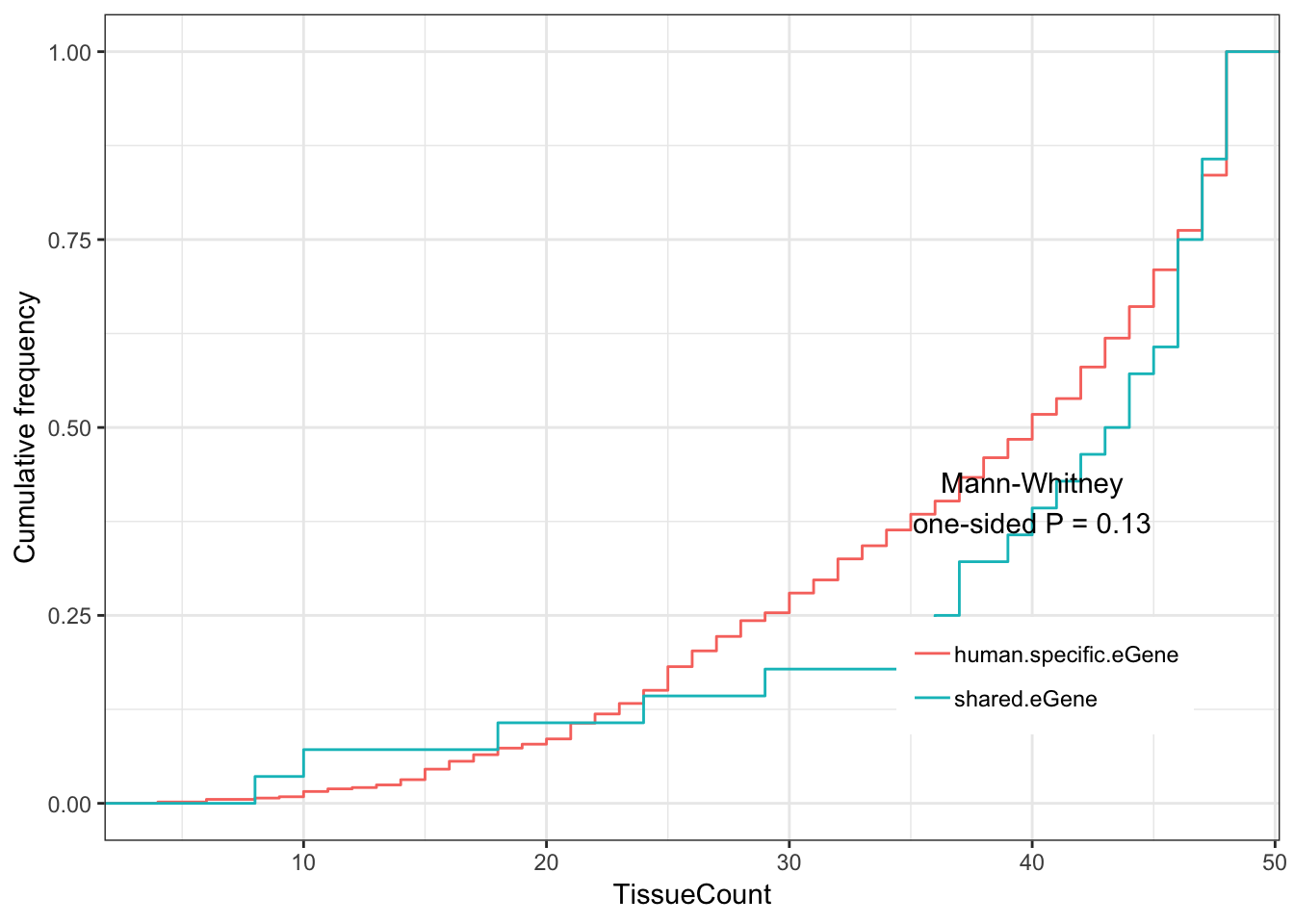
| Version | Author | Date |
|---|---|---|
| b62b089 | Benjmain Fair | 2019-08-21 |
Shared.chimp.tissue.plot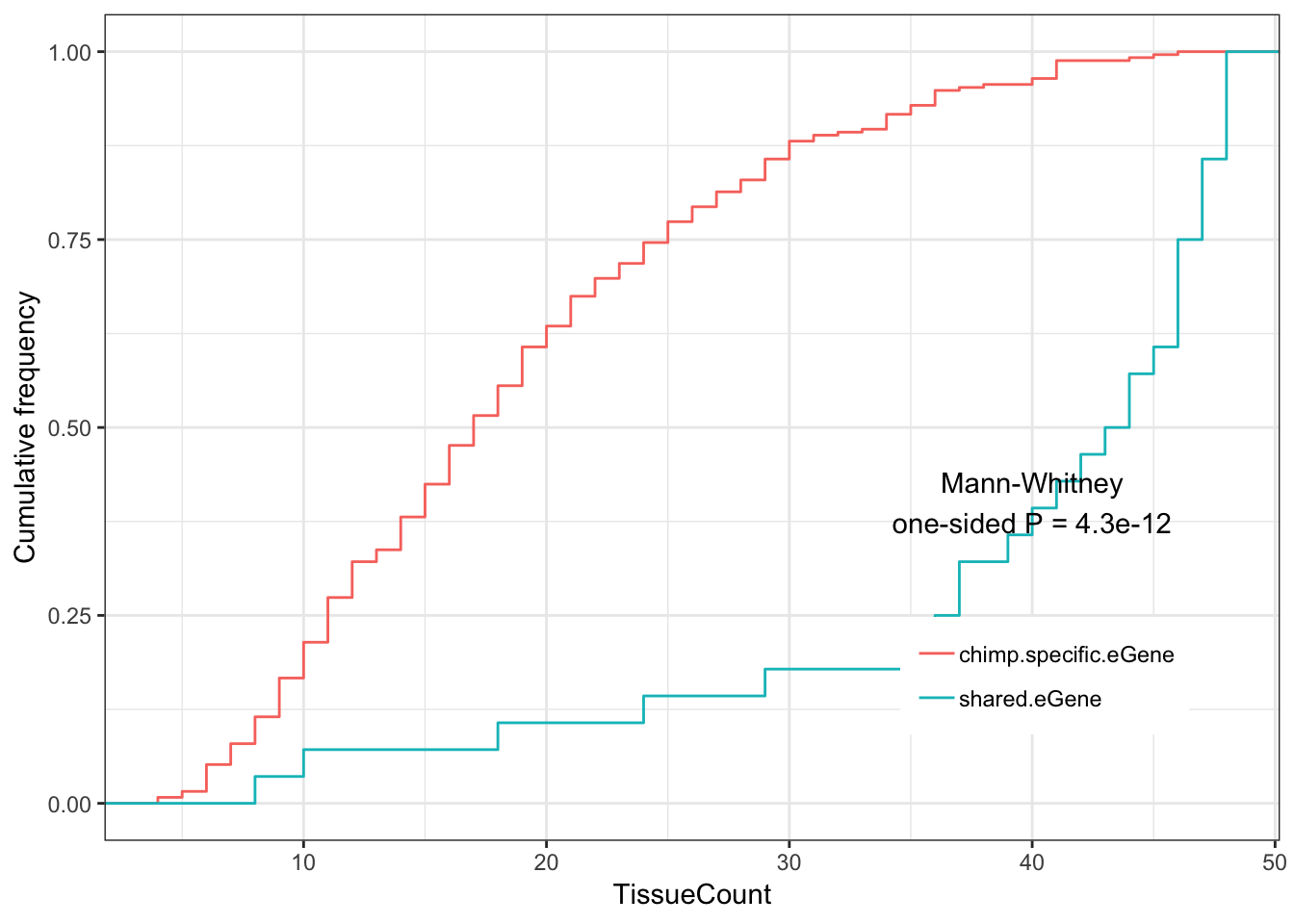
| Version | Author | Date |
|---|---|---|
| b62b089 | Benjmain Fair | 2019-08-21 |
Main finding:
Shared eGenes tend to be eGenes in more GTEx tissues than human specific eGenes
Make same plots, but using top 600 human eGenes to classify heart eGene for purposes of species sharing.
#Change FDR thresholds or take top N eGenes by qvalue
HumanTopN <- 1000
HumanFDR <- 0.1
ChimpFDR <- 0.1
# Filter human eGenes by qval threshold
# HumanSigGenes <- GtexHeartEgenes %>% filter(qval<HumanFDR) %>% pull(chimp_id)
# Filter human eGenes by topN qval
HumanSigGenes <- GtexHeartEgenes %>% top_n(-HumanTopN, qval) %>% pull(chimp_id)
# Filter human eGeness by qval threshold then topN betas
# HumanSigGenes <- GtexHeartEgenes %>% filter(qval<HumanFDR) %>% top_n(1000, abs(slope)) %>% pull(chimp_id)
HumanNonSigGenes <- GtexHeartEgenes %>%
filter(!chimp_id %in% HumanSigGenes) %>%
pull(chimp_id)
ChimpSigGenes <- GtexHeartEgenes %>%
filter(chimp_id %in% Chimp_eQTLs$gene) %>%
pull(chimp_id)
ChimpNonSigGenes <- GtexHeartEgenes %>%
filter(! chimp_id %in% Chimp_eQTLs$gene) %>%
pull(chimp_id)
ContigencyTable <- matrix( c( length(intersect(ChimpSigGenes,HumanSigGenes)),
length(intersect(HumanSigGenes,ChimpNonSigGenes)),
length(intersect(ChimpSigGenes,HumanNonSigGenes)),
length(intersect(ChimpNonSigGenes,HumanNonSigGenes))),
nrow = 2)
rownames(ContigencyTable) <- c("Chimp eGene", "Not Chimp eGene")
colnames(ContigencyTable) <- c("Human eGene", "Not human eGene")
#what is qval threshold for human eGene classification in this contigency table
print(GtexHeartEgenes %>% top_n(-HumanTopN, qval) %>% top_n(1, qval) %>% pull(qval))[1] 3.95822e-08#Contigency table of one to one orthologs tested in both chimps and humans of whether significant in humans, or chimps, or both, or neither
ContigencyTable Human eGene Not human eGene
Chimp eGene 36 244
Not Chimp eGene 964 10342#One-sided Fisher test for greater overlap than expected by chance
fisher.test(ContigencyTable, alternative="greater")
Fisher's Exact Test for Count Data
data: ContigencyTable
p-value = 0.01
alternative hypothesis: true odds ratio is greater than 1
95 percent confidence interval:
1.14537 Inf
sample estimates:
odds ratio
1.582765 #Chimp eGenes vs non chimp eGenes
ToPlot <- GtexHeartEgenes %>%
mutate(group = case_when(
chimp_id %in% ChimpSigGenes ~ "chimp.eGene",
!chimp_id %in% ChimpSigGenes ~ "not.chimp.eGene")) %>%
left_join(OneToOneMap, by=c("chimp_id"="Chimpanzee.gene.stable.ID"))
Chimp.tissue.plot <- ggplot(ToPlot, aes(color=group,x=TissueCount)) +
stat_ecdf(geom = "step") +
ylab("Cumulative frequency") +
xlab("TissueCount") +
annotate("text", x = 40, y = 0.4, label = paste("Mann-Whitney\none-sided P =", signif(wilcox.test(data=ToPlot, TissueCount ~ group, alternative="greater")$p.value, 2) )) +
theme_bw() +
theme(legend.position = c(.80, .2), legend.title=element_blank())
#Human eGenes vs non human eGenes
ToPlot <- GtexHeartEgenes %>%
mutate(group = case_when(
chimp_id %in% HumanSigGenes ~ "human.eGene",
!chimp_id %in% HumanSigGenes ~ "not.human.eGene")) %>%
left_join(OneToOneMap, by=c("chimp_id"="Chimpanzee.gene.stable.ID"))
Human.tissue.plot <- ggplot(ToPlot, aes(color=group,x=TissueCount)) +
stat_ecdf(geom = "step") +
ylab("Cumulative frequency") +
xlab("TissueCount") +
annotate("text", x = 40, y = 0.4, label = paste("Mann-Whitney\none-sided P =", signif(wilcox.test(data=ToPlot, TissueCount ~ group, alternative="greater")$p.value, 2) )) +
theme_bw() +
theme(legend.position = c(.80, .2), legend.title=element_blank())
#Shared eGenes vs human-specific eGenes
ToPlot <- GtexHeartEgenes %>%
mutate(group = case_when(
chimp_id %in% intersect(HumanSigGenes, ChimpSigGenes) ~ "shared.eGene",
chimp_id %in% setdiff(HumanSigGenes, ChimpSigGenes) ~ "human.specific.eGene")) %>%
filter(chimp_id %in% union(intersect(HumanSigGenes, ChimpSigGenes), setdiff(HumanSigGenes, ChimpSigGenes)))%>%
left_join(OneToOneMap, by=c("chimp_id"="Chimpanzee.gene.stable.ID"))
Shared.human.tissue.plot <- ggplot(ToPlot, aes(color=group,x=TissueCount)) +
stat_ecdf(geom = "step") +
ylab("Cumulative frequency") +
xlab("TissueCount") +
annotate("text", x = 40, y = 0.4, label = paste("Mann-Whitney\none-sided P =", signif(wilcox.test(data=ToPlot, TissueCount ~ group, alternative="less")$p.value, 2) )) +
theme_bw() +
theme(legend.position = c(.80, .2), legend.title=element_blank())
#Shared eGenes vs chimp-specific eGenes
ToPlot <- GtexHeartEgenes %>%
left_join(OneToOneMap, by=c("chimp_id"="Chimpanzee.gene.stable.ID")) %>%
mutate(dN.dS = dN.with.Chimpanzee/dS.with.Chimpanzee) %>%
mutate(group = case_when(
chimp_id %in% intersect(HumanSigGenes, ChimpSigGenes) ~ "shared.eGene",
chimp_id %in% setdiff(ChimpSigGenes, HumanSigGenes) ~ "chimp.specific.eGene")) %>%
dplyr::filter(chimp_id %in% union(intersect(HumanSigGenes, ChimpSigGenes), setdiff(ChimpSigGenes, HumanSigGenes)))
Shared.chimp.tissue.plot <- ggplot(ToPlot, aes(color=group,x=TissueCount)) +
stat_ecdf(geom = "step") +
ylab("Cumulative frequency") +
xlab("TissueCount") +
annotate("text", x = 40, y = 0.4, label = paste("Mann-Whitney\none-sided P =", signif(wilcox.test(data=ToPlot, TissueCount ~ group, alternative="less")$p.value, 2) )) +
theme_bw() +
theme(legend.position = c(.80, .2), legend.title=element_blank())
Chimp.tissue.plot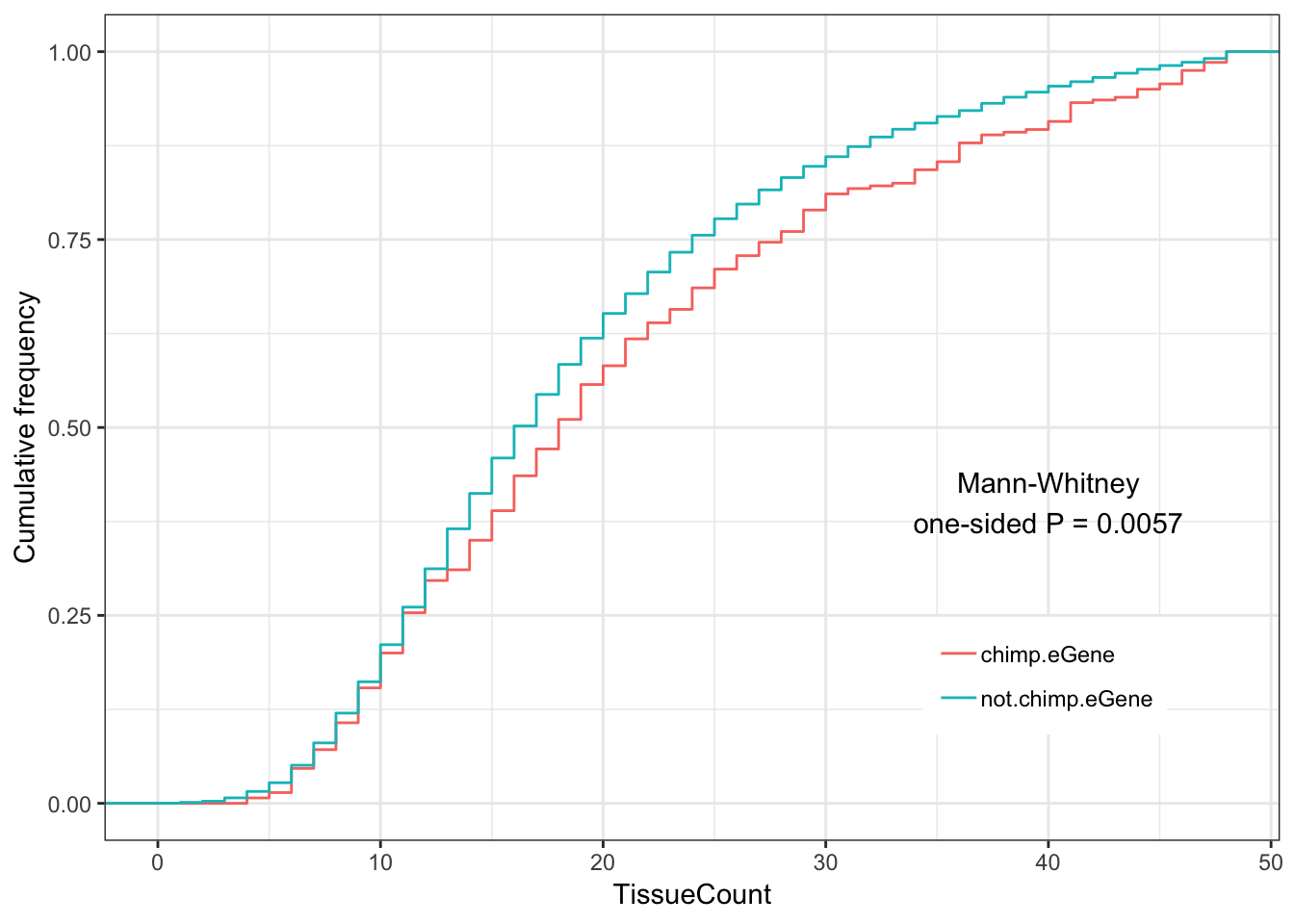
| Version | Author | Date |
|---|---|---|
| b62b089 | Benjmain Fair | 2019-08-21 |
Human.tissue.plot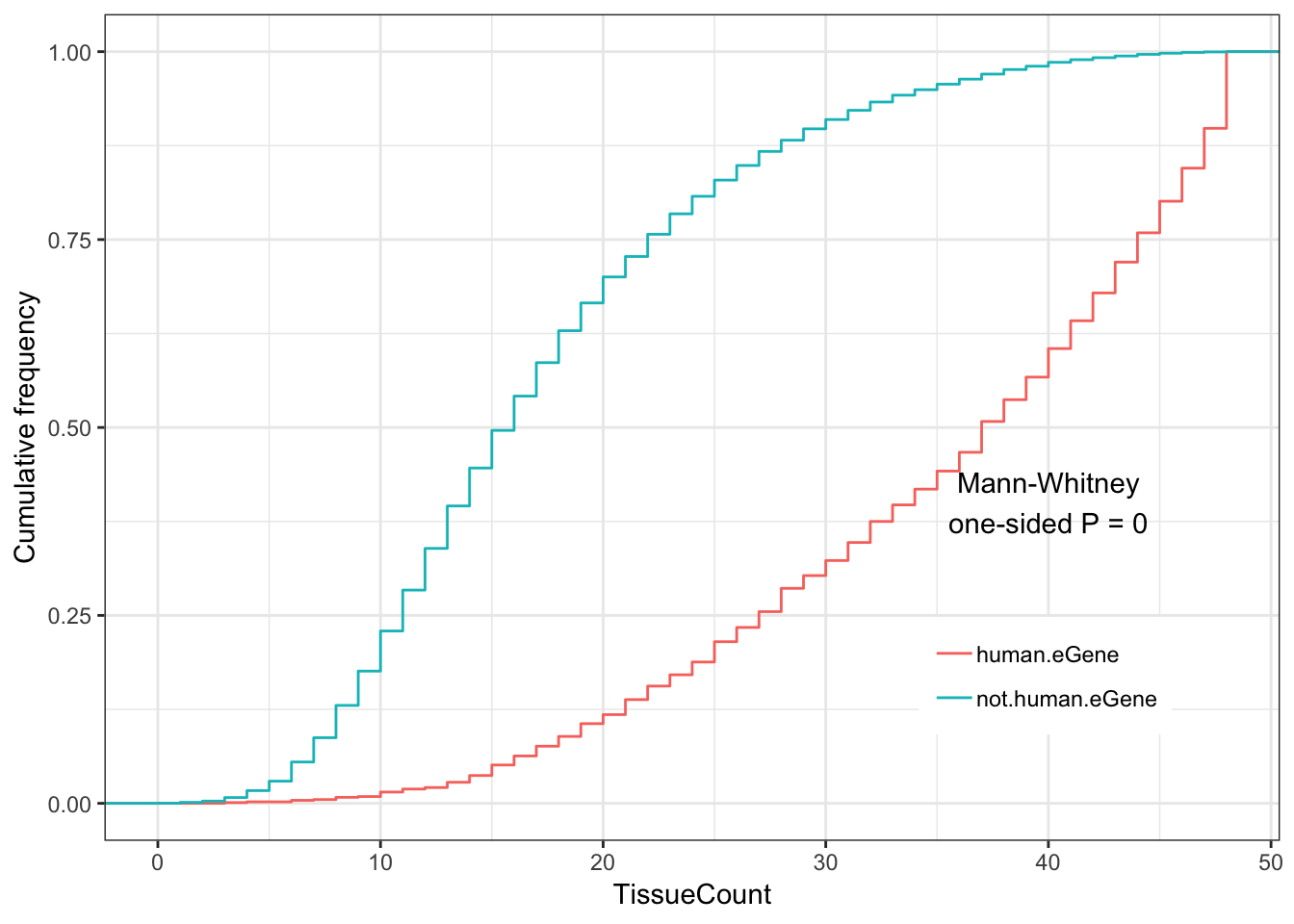
| Version | Author | Date |
|---|---|---|
| b62b089 | Benjmain Fair | 2019-08-21 |
Shared.human.tissue.plot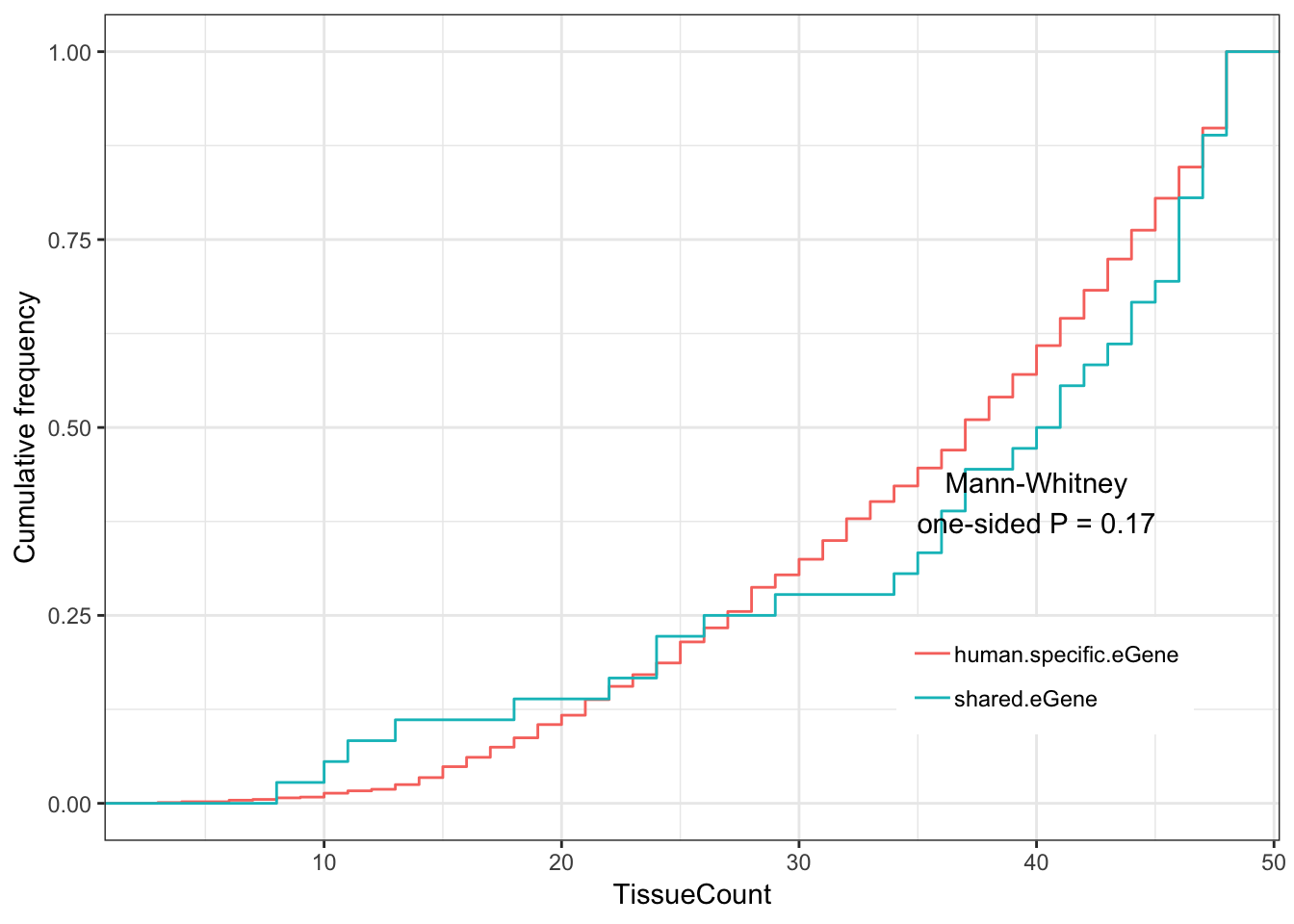
| Version | Author | Date |
|---|---|---|
| b62b089 | Benjmain Fair | 2019-08-21 |
Shared.chimp.tissue.plot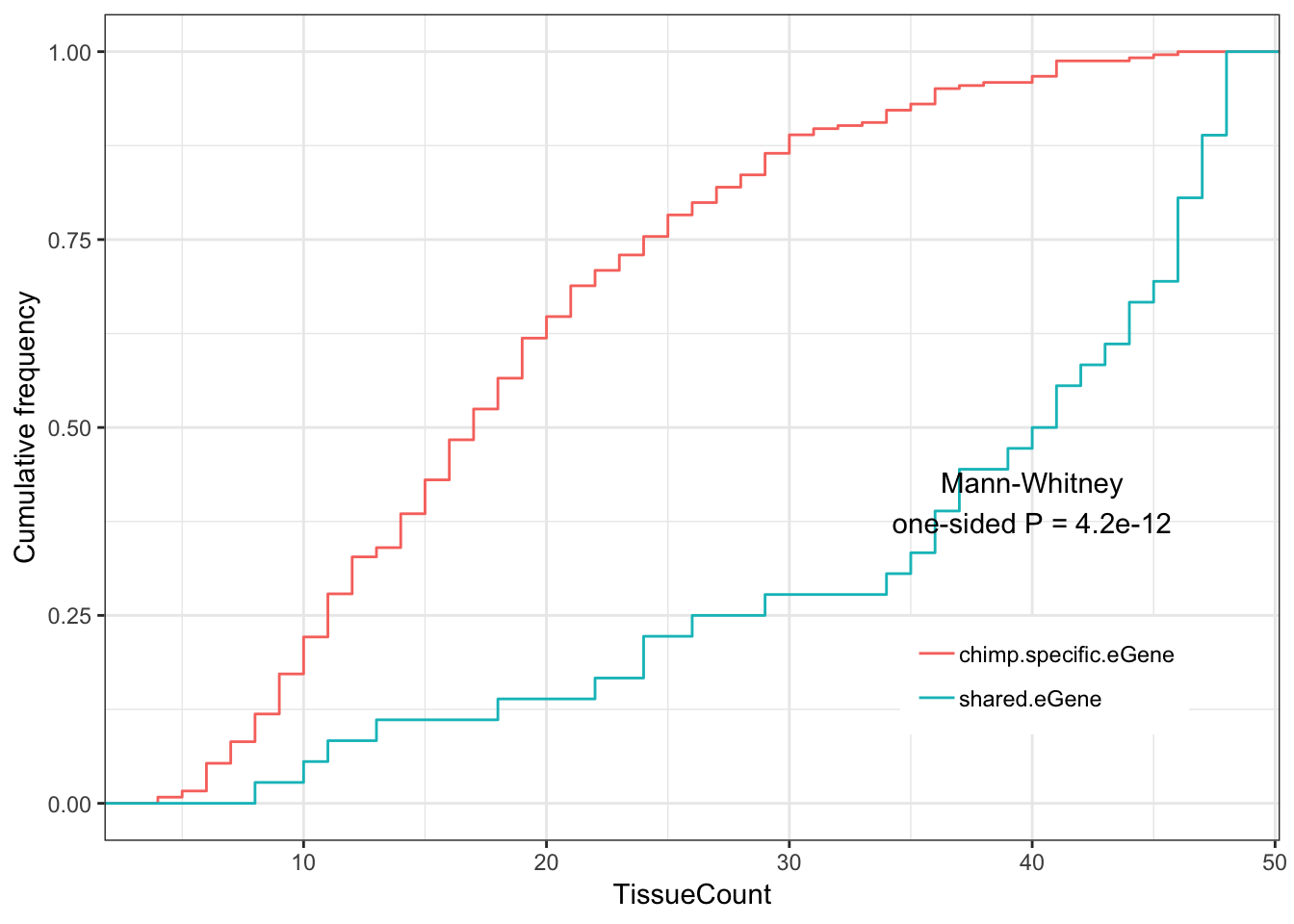
| Version | Author | Date |
|---|---|---|
| b62b089 | Benjmain Fair | 2019-08-21 |
Ok the same main result seems true with this much more stringent cutoff for human eGenes (which only leaves 28 shared eGenes), but could be due to chance (P=0.11). Perhaps for publication it is best to show results at these various thresholds in the supplement, to show these trends generally hold across various reasonable thresholds for classifying human eGenes.
Next I want to see if the shared eGenes are more likely to be expressed in just a few tissues or highly expressed in all tissues. I hypothesize the shared eGenes are more likely to be tissue specific, as tissue specific genes are more likely to be neutral, since deleterious genes need only be deleterious in one tissue for it to be selected against.
First I will look at expression levels (TPM) of shared eGenes vs species specific eGenes
#read in median GTEx TPM for each gene
GTEx.Expression <- read.table("../data/GTEx_Analysis_2016-01-15_v7_RNASeQCv1.1.8_gene_median_tpm.gct.gz", sep='\t', skip=2, header=T,check.names=FALSE)
GTEx.Expression[1:10,1:10] %>% kable()| gene_id | Description | Adipose - Subcutaneous | Adipose - Visceral (Omentum) | Adrenal Gland | Artery - Aorta | Artery - Coronary | Artery - Tibial | Bladder | Brain - Amygdala |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000223972.4 | DDX11L1 | 0.056945 | 0.05054 | 0.074600 | 0.03976 | 0.04386 | 0.04977 | 0.05878 | 0.089315 |
| ENSG00000227232.4 | WASH7P | 11.850000 | 9.75300 | 8.023000 | 12.51000 | 12.30000 | 11.59000 | 14.24000 | 5.743000 |
| ENSG00000243485.2 | MIR1302-11 | 0.061460 | 0.05959 | 0.081790 | 0.04297 | 0.05848 | 0.05184 | 0.06097 | 0.115450 |
| ENSG00000237613.2 | FAM138A | 0.038600 | 0.03245 | 0.040500 | 0.02815 | 0.03678 | 0.03894 | 0.04113 | 0.056265 |
| ENSG00000268020.2 | OR4G4P | 0.035695 | 0.00000 | 0.034790 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.050520 |
| ENSG00000240361.1 | OR4G11P | 0.042680 | 0.03988 | 0.049065 | 0.03399 | 0.00000 | 0.04286 | 0.00000 | 0.062955 |
| ENSG00000186092.4 | OR4F5 | 0.051450 | 0.04558 | 0.061360 | 0.00000 | 0.04069 | 0.04669 | 0.05461 | 0.097705 |
| ENSG00000238009.2 | RP11-34P13.7 | 0.162500 | 0.12020 | 0.087785 | 0.13510 | 0.13690 | 0.14720 | 0.14300 | 0.070360 |
| ENSG00000233750.3 | CICP27 | 0.124400 | 0.13470 | 0.148800 | 0.10260 | 0.11950 | 0.11450 | 0.07610 | 0.166150 |
| ENSG00000237683.5 | AL627309.1 | 5.992500 | 8.38500 | 6.595500 | 6.37800 | 6.06100 | 2.69100 | 4.28200 | 1.679500 |
#Get list of brain subtypes to exclude (let's just include brain cerebellum for this analysis)
#...too many brain subtypes is redundant
BrainsSubtypesToExclude<-colnames(GTEx.Expression) %>% grep("Brain", ., value=T) %>% grep("Cerebellum", ., value=T, invert=T)
GTEx.Expression.filtered <- GTEx.Expression %>%
mutate(human_id=gsub("\\.\\d+$", "", gene_id, perl=T)) %>%
select(-c(gene_id, BrainsSubtypesToExclude, Description)) %>%
filter(human_id %in% GtexHeartEgenes$gene_id_stable) %>%
mutate(Means = rowMedians(as.matrix(.[,-42]))) %>%
mutate(chimp_id=plyr::mapvalues(human_id, OneToOneMap$Gene.stable.ID, OneToOneMap$Chimpanzee.gene.stable.ID, warn_missing = F))
ToPlot <- GTEx.Expression.filtered %>%
mutate(group = case_when(
chimp_id %in% intersect(HumanSigGenes, ChimpSigGenes) ~ "shared.eGene",
chimp_id %in% setdiff(HumanSigGenes, ChimpSigGenes) ~ "human.specific.eGene")) %>%
filter(chimp_id %in% union(intersect(HumanSigGenes, ChimpSigGenes), setdiff(HumanSigGenes, ChimpSigGenes)))%>%
select(human_id, group, Means, 1:41) %>%
melt(value.name="expression")
GtexTissueColors <- read.table("../data/GTEx_Analysis_TissueColorCodes.txt", header=T, sep='\t') %>%
rbind(data.frame(Tissue="Means", Color.code="000000"), .) %>%
filter(Tissue %in% ToPlot$variable) %>%
mutate(HexCode=paste0("#", Color.code))
Human.tissue.expression.plot <- ggplot(ToPlot, aes(color=group,x=variable, y=expression)) +
geom_boxplot(outlier.shape = NA, fatten = 4) +
scale_y_continuous(trans="log10", limits=c(0.01, 5000)) +
ylab("expression\nlog10(TPM)") +
# geom_point(data=GtexTissueColors, aes(y=1,x=Tissue, color=HexCode)) +
theme_bw() +
theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust=0.5),
axis.ticks.x = element_line(colour = GtexTissueColors$HexCode, size = 2),
axis.title.x=element_blank())
Human.tissue.expression.plot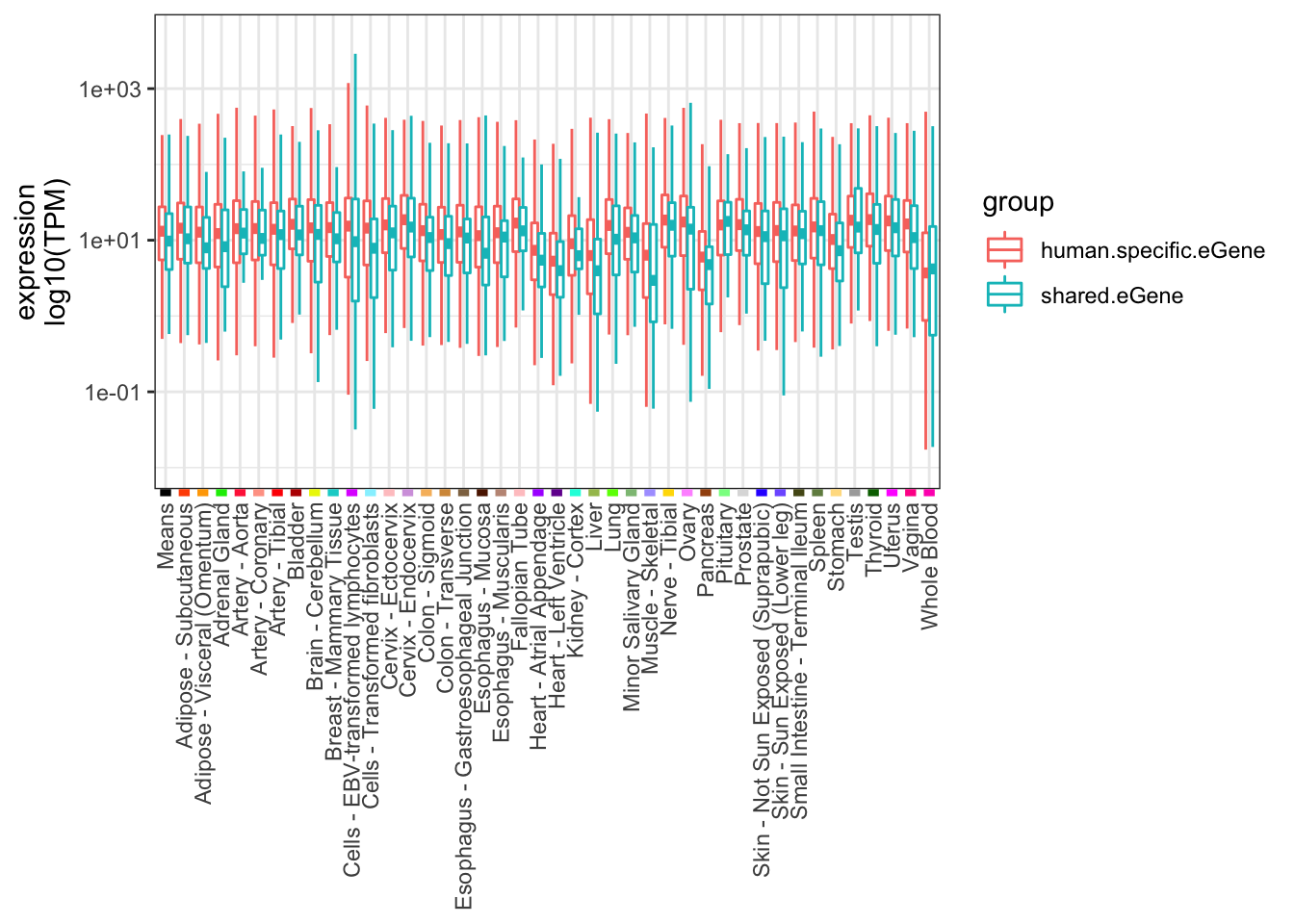
| Version | Author | Date |
|---|---|---|
| 7810819 | Benjmain Fair | 2019-10-01 |
#Are any of the differences in those groups significant... Try muscle for example
ToCompare <- GTEx.Expression.filtered %>%
mutate(group = case_when(
chimp_id %in% intersect(HumanSigGenes, ChimpSigGenes) ~ "shared.eGene",
chimp_id %in% setdiff(HumanSigGenes, ChimpSigGenes) ~ "human.specific.eGene")) %>%
filter(chimp_id %in% union(intersect(HumanSigGenes, ChimpSigGenes), setdiff(HumanSigGenes, ChimpSigGenes))) %>%
select(human_id, group, Means, 1:41)
wilcox.test(data=ToCompare, `Muscle - Skeletal` ~ group, alternative="greater")
Wilcoxon rank sum test with continuity correction
data: Muscle - Skeletal by group
W = 19718, p-value = 0.08218
alternative hypothesis: true location shift is greater than 0Ok so the shared eGenes seem to be slightly lower expressed in all tissues, though it could be just chance.
Ok now I will look at tissue specificity on a per-tissue level (are the shared eGenes specific for any particular tissue?). First I will do this by looking at Z-scores of expression (for each gene, standardize the measurements across tissues into Z-score).
ToPlot <- GTEx.Expression.filtered %>%
select(1:41) %>% as.matrix() %>% log10() %>% t() %>% scale() %>% t() %>% as.data.frame() %>%
mutate(human_id=GTEx.Expression.filtered$human_id, chimp_id=GTEx.Expression.filtered$chimp_id) %>%
mutate(group = case_when(
# chimp_id %in% HumanSigGenes ~ "human.eGene",
# !chimp_id %in% HumanSigGenes ~ "not.human.eGene")) %>%
chimp_id %in% intersect(HumanSigGenes, ChimpSigGenes) ~ "shared.eGene",
chimp_id %in% setdiff(HumanSigGenes, ChimpSigGenes) ~ "human.specific.eGene")) %>%
filter(chimp_id %in% union(intersect(HumanSigGenes, ChimpSigGenes), setdiff(HumanSigGenes, ChimpSigGenes)))%>%
melt(value.name="expression")
GtexTissueColors <- read.table("../data/GTEx_Analysis_TissueColorCodes.txt", header=T, sep='\t') %>%
# rbind(data.frame(Tissue="Means", Color.code="000000"), .) %>%
filter(Tissue %in% ToPlot$variable) %>%
mutate(HexCode=paste0("#", Color.code))
Human.tissue.expression.Z.plot <- ggplot(ToPlot, aes(color=group,x=variable, y=expression)) +
geom_boxplot(outlier.shape = NA, fatten = 4) +
ylab("expression\nZ-score") +
# geom_point(data=GtexTissueColors, aes(y=1,x=Tissue, color=HexCode)) +
theme_bw() +
theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust=0.5),
axis.ticks.x = element_line(colour = GtexTissueColors$HexCode, size = 2),
axis.title.x=element_blank())
Human.tissue.expression.Z.plot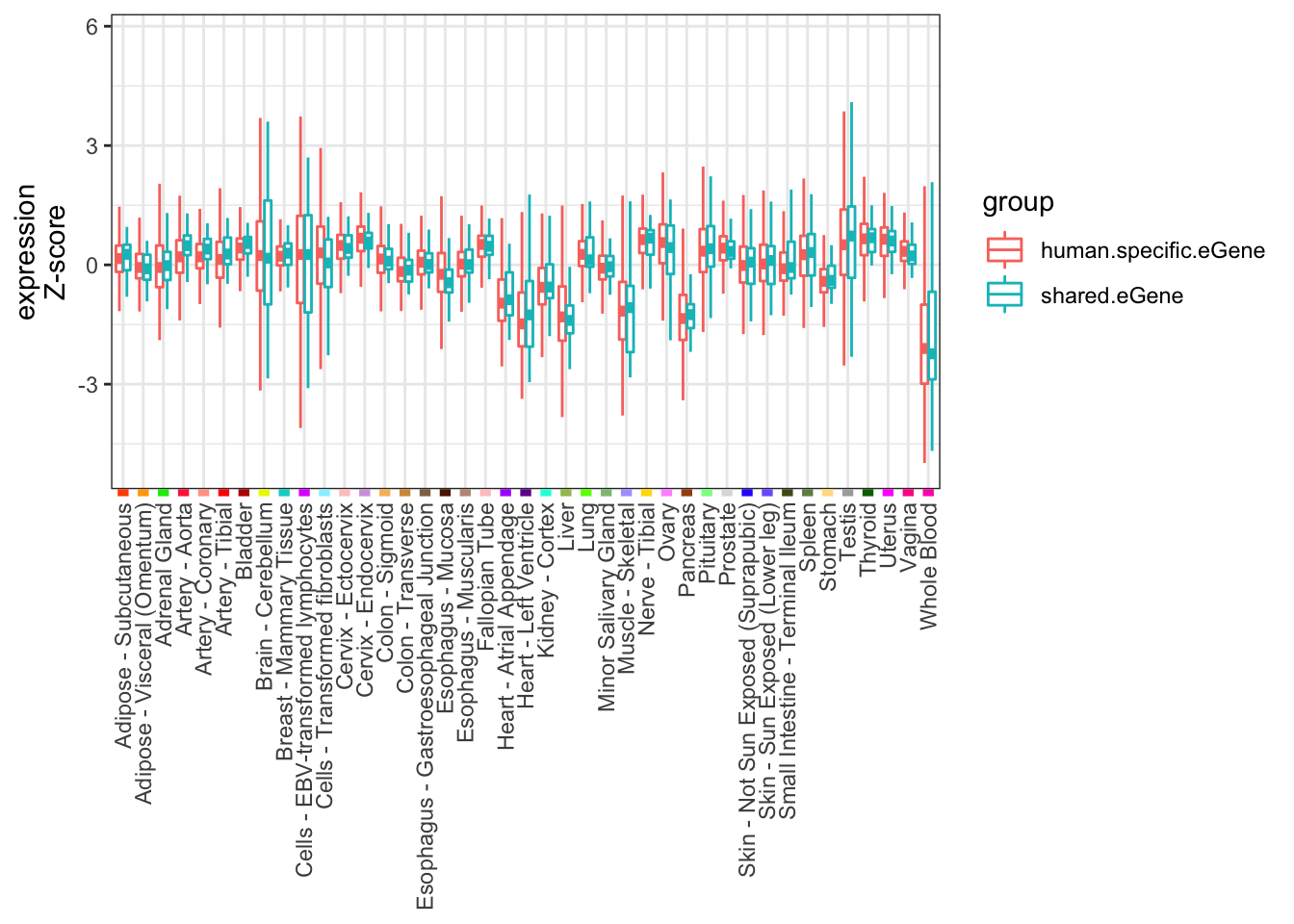
| Version | Author | Date |
|---|---|---|
| 7810819 | Benjmain Fair | 2019-10-01 |
Now look at tissue specificity, as calculated with gini and tau statistics from the GTEx tissue data. Using both slightly different statistics to make sure the general finding is robust to choice of statistic
tau<-read.table("../output/TissueSpecificity/tau.log.txt", col.names =c('gene', 'tau'), sep='\t')
gini<-read.table("../output/TissueSpecificity/gini.log.txt", col.names =c('gene', 'gini'), sep='\t')
tissue.specificity <- merge(tau, gini) %>%
filter(gene %in% GtexHeartEgenes$gene_id_stable) %>%
mutate(chimp_id=plyr::mapvalues(gene, OneToOneMap$Gene.stable.ID, OneToOneMap$Chimpanzee.gene.stable.ID, warn_missing = F))
p<- ggplot(tissue.specificity, aes(x=gini, y=tau)) +
geom_point(alpha=0.05) +
theme_bw()
#scatter plot and histograms of tau and gini over all tested genes
tauVgini <- ggExtra::ggMarginal(p, type = "histogram")
library(grid)
grid.newpage()
grid.draw(tauVgini)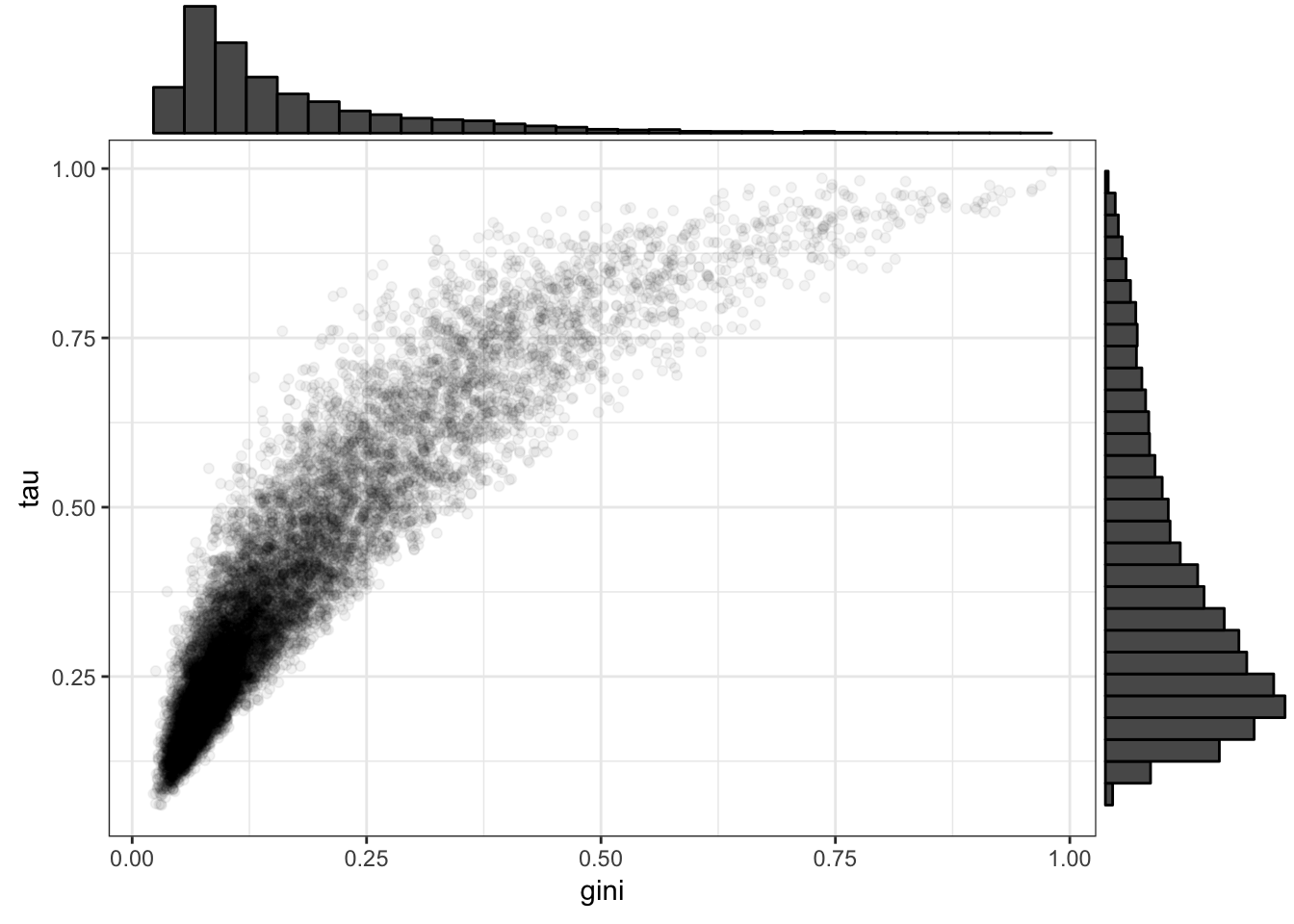
| Version | Author | Date |
|---|---|---|
| 7810819 | Benjmain Fair | 2019-10-01 |
ToPlot <- tissue.specificity %>%
mutate(group = case_when(
chimp_id %in% intersect(HumanSigGenes, ChimpSigGenes) ~ "shared.eGene",
chimp_id %in% setdiff(HumanSigGenes, ChimpSigGenes) ~ "human.specific.eGene")) %>%
filter(chimp_id %in% union(intersect(HumanSigGenes, ChimpSigGenes), setdiff(HumanSigGenes, ChimpSigGenes)))
# chimp_id %in% HumanSigGenes ~ "chimp.eGene",
# !chimp_id %in% HumanSigGenes ~ "not.chimp.eGene"))
Human.tissue.specificity.plot <- ggplot(ToPlot, aes(color=group,x=tau)) +
stat_ecdf(geom = "step") +
ylab("Cumulative frequency") +
xlab("Tissue specificity (tau)") +
annotate("text", x = 0.4, y = 0.4, label = paste("Mann-Whitney\none-sided P =", signif(wilcox.test(data=ToPlot, tau ~ group, alternative="less")$p.value, 2) )) +
theme_bw() +
theme(legend.position = c(.80, .2), legend.title=element_blank())
Human.tissue.specificity.plot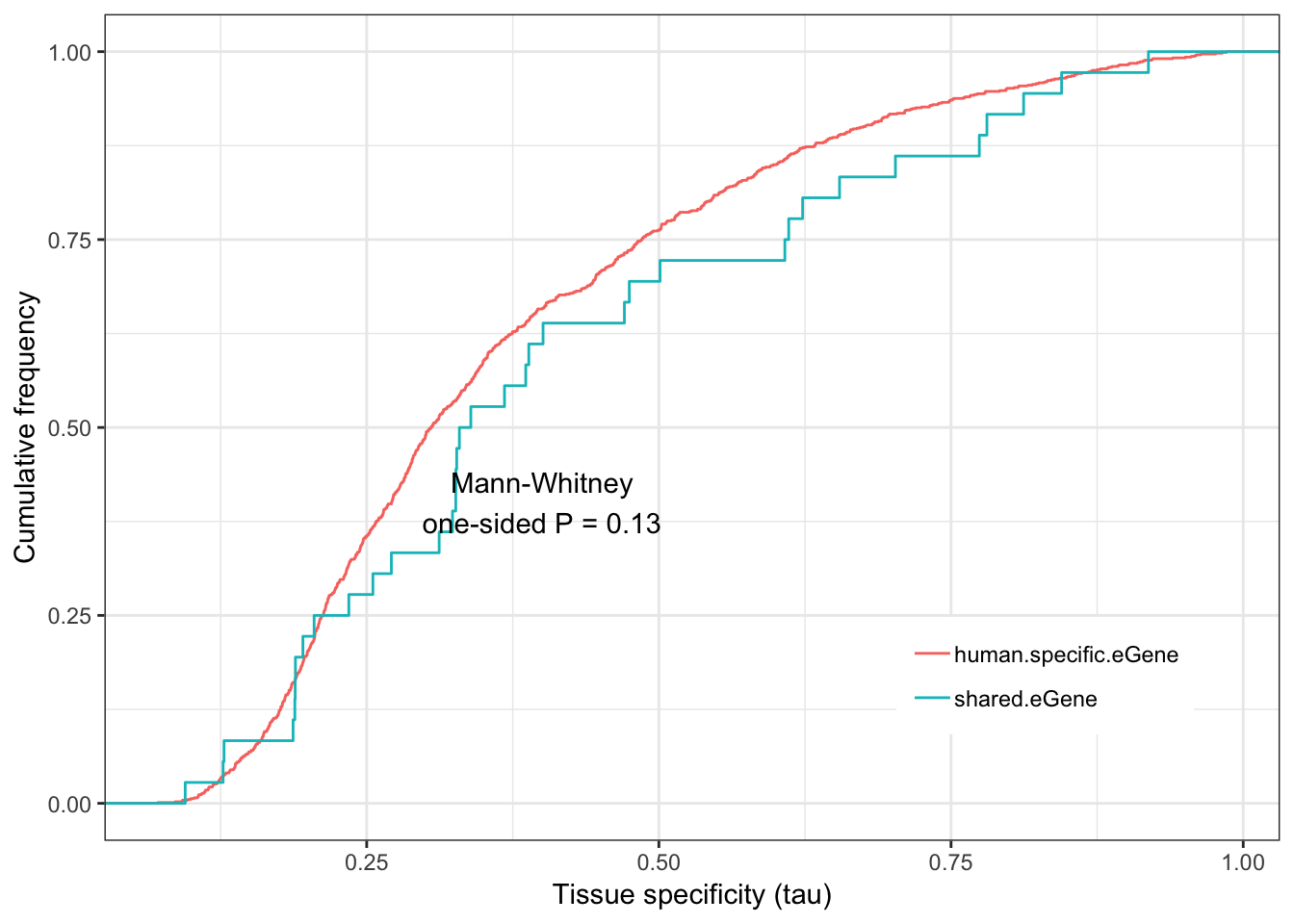
| Version | Author | Date |
|---|---|---|
| 7810819 | Benjmain Fair | 2019-10-01 |
sessionInfo()R version 3.5.1 (2018-07-02)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS 10.14
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] grid stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] reshape2_1.4.3 ggExtra_0.9 matrixStats_0.54.0
[4] knitr_1.23 forcats_0.4.0 stringr_1.4.0
[7] dplyr_0.8.1 purrr_0.3.2 readr_1.3.1
[10] tidyr_0.8.3 tibble_2.1.3 ggplot2_3.1.1
[13] tidyverse_1.2.1
loaded via a namespace (and not attached):
[1] tidyselect_0.2.5 xfun_0.7 haven_2.1.0 lattice_0.20-38
[5] colorspace_1.4-1 generics_0.0.2 miniUI_0.1.1.1 htmltools_0.3.6
[9] yaml_2.2.0 rlang_0.3.4 later_0.8.0 pillar_1.4.1
[13] glue_1.3.1 withr_2.1.2 modelr_0.1.4 readxl_1.3.1
[17] plyr_1.8.4 munsell_0.5.0 gtable_0.3.0 workflowr_1.4.0
[21] cellranger_1.1.0 rvest_0.3.4 evaluate_0.14 labeling_0.3
[25] httpuv_1.5.1 highr_0.8 broom_0.5.2 Rcpp_1.0.1
[29] xtable_1.8-4 promises_1.0.1 scales_1.0.0 backports_1.1.4
[33] jsonlite_1.6 mime_0.7 fs_1.3.1 hms_0.4.2
[37] digest_0.6.19 stringi_1.4.3 shiny_1.3.2 rprojroot_1.3-2
[41] cli_1.1.0 tools_3.5.1 magrittr_1.5 lazyeval_0.2.2
[45] crayon_1.3.4 whisker_0.3-2 pkgconfig_2.0.2 xml2_1.2.0
[49] lubridate_1.7.4 assertthat_0.2.1 rmarkdown_1.13 httr_1.4.0
[53] rstudioapi_0.10 R6_2.4.0 nlme_3.1-140 git2r_0.25.2
[57] compiler_3.5.1