Additional Enrichments for revisions
Briana Mittleman
12/13/2020
Last updated: 2020-12-14
Checks: 7 0
Knit directory: Comparative_APA/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190902) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version d261561. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: code/chimp_log/
Ignored: code/human_log/
Ignored: data/.DS_Store
Ignored: data/TrialFiltersMeta.txt.sb-9845453e-R58Y0Q/
Ignored: data/mediation_prot/
Ignored: data/metadata_HCpanel.txt.sb-284518db-RGf0kd/
Ignored: data/metadata_HCpanel.txt.sb-a5794dd2-i594qs/
Ignored: output/.DS_Store
Untracked files:
Untracked: ._.DS_Store
Untracked: Chimp/
Untracked: GEO/
Untracked: Human/
Untracked: analysis/ALUelements.Rmd
Untracked: analysis/AREstabilityScores.Rmd
Untracked: analysis/AllLoc_effectSizeCor.Rmd
Untracked: analysis/Conservation_bydAPAset.Rmd
Untracked: analysis/CrossChimpThreePrime.Rmd
Untracked: analysis/DiffTransProtvsExpression.Rmd
Untracked: analysis/DiffUsedUTR.Rmd
Untracked: analysis/GvizPlots.Rmd
Untracked: analysis/HandC.TvN
Untracked: analysis/PhenotypeOverlap10.Rmd
Untracked: analysis/ResultsNoUnlifted.md
Untracked: analysis/SuppTables.Rmd
Untracked: analysis/annotationBias.Rmd
Untracked: analysis/assessReadQual.Rmd
Untracked: analysis/diffExpressionPantro6.Rmd
Untracked: analysis/isoformdivandexp.Rmd
Untracked: code/._AlignmentScores.sh
Untracked: code/._BothFCMM.sh
Untracked: code/._BothFCMMPrim.sh
Untracked: code/._BothFCnewOInclusive.sh
Untracked: code/._ChimpStarMM2.sh
Untracked: code/._ClassifyLeafviz.sh
Untracked: code/._ClosestorthoEx.sh
Untracked: code/._Config_chimp.yaml
Untracked: code/._Config_chimp_full.yaml
Untracked: code/._Config_human.yaml
Untracked: code/._ConvertJunc2Bed.sh
Untracked: code/._CountNucleotides.py
Untracked: code/._CrossMapChimpRNA.sh
Untracked: code/._CrossMapThreeprime.sh
Untracked: code/._DiffSplice.sh
Untracked: code/._DiffSplicePlots.sh
Untracked: code/._DiffSplicePlots_gencode.sh
Untracked: code/._DiffSplice_gencode.sh
Untracked: code/._DiffSplice_removebad.sh
Untracked: code/._Filter255MM.sh
Untracked: code/._FilterPrimSec.sh
Untracked: code/._FindIntronForDomPAS.sh
Untracked: code/._FindIntronForDomPAS_DF.sh
Untracked: code/._GetMAPQscore.py
Untracked: code/._GetSecondaryMap.py
Untracked: code/._Lift5perPAS.sh
Untracked: code/._LiftFinalChimpJunc2Human.sh
Untracked: code/._LiftOrthoPAS2chimp.sh
Untracked: code/._MapBadSamples.sh
Untracked: code/._MismatchNumbers.sh
Untracked: code/._PAS_ATTAAA.sh
Untracked: code/._PAS_ATTAAA_df.sh
Untracked: code/._PAS_seqExpanded.sh
Untracked: code/._PASsequences.sh
Untracked: code/._PASsequences_DF.sh
Untracked: code/._PlotNuclearUsagebySpecies.R
Untracked: code/._PlotNuclearUsagebySpecies_DF.R
Untracked: code/._QuantMergedClusters.sh
Untracked: code/._RNATranscriptDTplot.sh
Untracked: code/._ReverseLiftFilter.R
Untracked: code/._RunFixLeafCluster.sh
Untracked: code/._RunNegMCMediation.sh
Untracked: code/._RunNegMCMediationDF.sh
Untracked: code/._RunPosMCMediationDF.err
Untracked: code/._RunPosMCMediationDF.sh
Untracked: code/._SAF2Bed.py
Untracked: code/._Snakefile
Untracked: code/._SnakefilePAS
Untracked: code/._SnakefilePASfilt
Untracked: code/._SortIndexBadSamples.sh
Untracked: code/._StarMM2.sh
Untracked: code/._TestFC.sh
Untracked: code/._assignPeak2Intronicregion
Untracked: code/._assignPeak2Intronicregion.sh
Untracked: code/._bed215upbed.py
Untracked: code/._bed2Bedbothstrand.py
Untracked: code/._bed2SAF_gen.py
Untracked: code/._buildIndecpantro5
Untracked: code/._buildIndecpantro5.sh
Untracked: code/._buildLeafviz.sh
Untracked: code/._buildLeafviz_leadAnno.sh
Untracked: code/._buildStarIndex.sh
Untracked: code/._chimpChromprder.sh
Untracked: code/._chimpMultiCov.sh
Untracked: code/._chimpMultiCov255.sh
Untracked: code/._chimpMultiCovInclusive.sh
Untracked: code/._chooseSignalSite.py
Untracked: code/._cleanbed2saf.py
Untracked: code/._cluster.json
Untracked: code/._cluster2bed.py
Untracked: code/._clusterLiftReverse.sh
Untracked: code/._clusterLiftReverse_removebad.sh
Untracked: code/._clusterLiftprimary.sh
Untracked: code/._clusterLiftprimary_removebad.sh
Untracked: code/._converBam2Junc.sh
Untracked: code/._converBam2Junc_removeBad.sh
Untracked: code/._extraSnakefiltpas
Untracked: code/._extractPhyloReg.py
Untracked: code/._extractPhyloRegGene.py
Untracked: code/._extractPhylopGeneral.ph
Untracked: code/._extractPhylopGeneral.py
Untracked: code/._extractPhylopReg200down.py
Untracked: code/._extractPhylopReg200up.py
Untracked: code/._filter5percPAS.py
Untracked: code/._filterNumChroms.py
Untracked: code/._filterPASforMP.py
Untracked: code/._filterPostLift.py
Untracked: code/._filterPrimaryread.py
Untracked: code/._filterSecondaryread.py
Untracked: code/._fixExonFC.py
Untracked: code/._fixFCheadforExp.py
Untracked: code/._fixLeafCluster.py
Untracked: code/._fixLiftedJunc.py
Untracked: code/._fixUTRexonanno.py
Untracked: code/._formathg38Anno.py
Untracked: code/._formatpantro6Anno.py
Untracked: code/._getRNAseqMapStats.sh
Untracked: code/._hg19MapStats.sh
Untracked: code/._humanChromorder.sh
Untracked: code/._humanMultiCov.sh
Untracked: code/._humanMultiCov255.sh
Untracked: code/._humanMultiCov_inclusive.sh
Untracked: code/._intersectLiftedPAS.sh
Untracked: code/._liftJunctionFiles.sh
Untracked: code/._liftPAS19to38.sh
Untracked: code/._liftedchimpJunc2human.sh
Untracked: code/._makeNuclearDapaplots.sh
Untracked: code/._makeNuclearDapaplots_DF.sh
Untracked: code/._makeSamplyGroupsHuman_TvN.py
Untracked: code/._mapRNAseqhg19.sh
Untracked: code/._mapRNAseqhg19_newPipeline.sh
Untracked: code/._maphg19.sh
Untracked: code/._maphg19_subjunc.sh
Untracked: code/._mediation_test.R
Untracked: code/._mergeChimp3prime_inhg38.sh
Untracked: code/._mergeandBWRNAseq.sh
Untracked: code/._mergedBam2BW.sh
Untracked: code/._nameClusters.py
Untracked: code/._negativeMediation_montecarlo.R
Untracked: code/._negativeMediation_montecarloDF.R
Untracked: code/._numMultimap.py
Untracked: code/._overlapMMandOrthoexon.sh
Untracked: code/._overlapPASandOrthoexon.sh
Untracked: code/._overlapapaQTLPAS.sh
Untracked: code/._parseHg38.py
Untracked: code/._postiveMediation_montecarlo_DF.R
Untracked: code/._prepareCleanLiftedFC_5perc4LC.py
Untracked: code/._prepareLeafvizAnno.sh
Untracked: code/._preparePAS4lift.py
Untracked: code/._primaryLift.sh
Untracked: code/._processhg38exons.py
Untracked: code/._quantJunc.sh
Untracked: code/._quantJunc_TEST.sh
Untracked: code/._quantJunc_removeBad.sh
Untracked: code/._quantLiftedPASPrimary.sh
Untracked: code/._quantMerged_seperatly.sh
Untracked: code/._recLiftchim2human.sh
Untracked: code/._revLiftPAShg38to19.sh
Untracked: code/._reverseLift.sh
Untracked: code/._runCheckReverseLift.sh
Untracked: code/._runChimpDiffIso.sh
Untracked: code/._runCountNucleotides.sh
Untracked: code/._runFilterNumChroms.sh
Untracked: code/._runHumanDiffIso.sh
Untracked: code/._runNuclearDiffIso_DF.sh
Untracked: code/._runNuclearDifffIso.sh
Untracked: code/._runTotalDiffIso.sh
Untracked: code/._run_chimpverifybam.sh
Untracked: code/._run_verifyBam.sh
Untracked: code/._snakemake.batch
Untracked: code/._snakemakePAS.batch
Untracked: code/._snakemakePASchimp.batch
Untracked: code/._snakemakePAShuman.batch
Untracked: code/._snakemake_chimp.batch
Untracked: code/._snakemake_human.batch
Untracked: code/._snakemakefiltPAS.batch
Untracked: code/._snakemakefiltPAS_chimp
Untracked: code/._snakemakefiltPAS_chimp.sh
Untracked: code/._snakemakefiltPAS_human.sh
Untracked: code/._spliceSite2Fasta.py
Untracked: code/._submit-snakemake-chimp.sh
Untracked: code/._submit-snakemake-human.sh
Untracked: code/._submit-snakemakePAS-chimp.sh
Untracked: code/._submit-snakemakePAS-human.sh
Untracked: code/._submit-snakemakefiltPAS-chimp.sh
Untracked: code/._submit-snakemakefiltPAS-human.sh
Untracked: code/._subset_diffisopheno_Nuclear_HvC.py
Untracked: code/._subset_diffisopheno_Nuclear_HvC_DF.py
Untracked: code/._subset_diffisopheno_Total_HvC.py
Untracked: code/._threeprimeOrthoFC.sh
Untracked: code/._transcriptDTplotsNuclear.sh
Untracked: code/._verifyBam4973.sh
Untracked: code/._verifyBam4973inHuman.sh
Untracked: code/._wrap_chimpverifybam.sh
Untracked: code/._wrap_verifyBam.sh
Untracked: code/._writeMergecode.py
Untracked: code/.snakemake/
Untracked: code/ALLPAS_sequenceDF.err
Untracked: code/ALLPAS_sequenceDF.out
Untracked: code/AlignmentScores.err
Untracked: code/AlignmentScores.out
Untracked: code/AlignmentScores.sh
Untracked: code/BothFCMM.err
Untracked: code/BothFCMM.out
Untracked: code/BothFCMM.sh
Untracked: code/BothFCMMPrim.err
Untracked: code/BothFCMMPrim.out
Untracked: code/BothFCMMPrim.sh
Untracked: code/BothFCnewOInclusive.sh
Untracked: code/BothFCnewOInclusive.sh.err
Untracked: code/BothFCnewOInclusive.sh.out
Untracked: code/ChimpStarMM2.err
Untracked: code/ChimpStarMM2.out
Untracked: code/ChimpStarMM2.sh
Untracked: code/ClassifyLeafviz.sh
Untracked: code/ClosestorthoEx.err
Untracked: code/ClosestorthoEx.out
Untracked: code/ClosestorthoEx.sh
Untracked: code/Config_chimp.yaml
Untracked: code/Config_chimp_full.yaml
Untracked: code/Config_human.yaml
Untracked: code/ConvertJunc2Bed.err
Untracked: code/ConvertJunc2Bed.out
Untracked: code/ConvertJunc2Bed.sh
Untracked: code/CountNucleotides.py
Untracked: code/CrossMapChimpRNA.sh
Untracked: code/CrossMapThreeprime.sh
Untracked: code/CrossmapChimp3prime.err
Untracked: code/CrossmapChimp3prime.out
Untracked: code/CrossmapChimpRNA.err
Untracked: code/CrossmapChimpRNA.out
Untracked: code/DTUTR.sh
Untracked: code/DiffDom_RNAmotif_4.err
Untracked: code/DiffDom_RNAmotif_4.out
Untracked: code/DiffDom_RNAmotif_4.sh
Untracked: code/DiffDom_RNAmotif_4_splitDE.err
Untracked: code/DiffDom_RNAmotif_4_splitDE.out
Untracked: code/DiffDom_RNAmotif_4_splitDE.sh
Untracked: code/DiffSplice.err
Untracked: code/DiffSplice.out
Untracked: code/DiffSplice.sh
Untracked: code/DiffSplicePlots.err
Untracked: code/DiffSplicePlots.out
Untracked: code/DiffSplicePlots.sh
Untracked: code/DiffSplicePlots_gencode.sh
Untracked: code/DiffSplice_gencode.sh
Untracked: code/DiffSplice_removebad.err
Untracked: code/DiffSplice_removebad.out
Untracked: code/DiffSplice_removebad.sh
Untracked: code/Filter255.err
Untracked: code/Filter255.out
Untracked: code/Filter255MM.sh
Untracked: code/FilterPrimSec.err
Untracked: code/FilterPrimSec.out
Untracked: code/FilterPrimSec.sh
Untracked: code/FilterReverseLift.err
Untracked: code/FilterReverseLift.out
Untracked: code/FindDomXCutoff.py
Untracked: code/FindIntronForDomPAS.err
Untracked: code/FindIntronForDomPAS.out
Untracked: code/FindIntronForDomPAS.sh
Untracked: code/FindIntronForDomPAS_DF.sh
Untracked: code/GencodeDiffSplice.err
Untracked: code/GencodeDiffSplice.out
Untracked: code/GetMAPQscore.py
Untracked: code/GetSecondaryMap.py
Untracked: code/GetTopminus2Usage.py
Untracked: code/H3K36me3DTplot.err
Untracked: code/H3K36me3DTplot.out
Untracked: code/H3K36me3DTplot.sh
Untracked: code/H3K36me3DTplot_DiffIso.err
Untracked: code/H3K36me3DTplot_DiffIso.out
Untracked: code/H3K36me3DTplot_DiffIso.sh
Untracked: code/H3K36me3DTplot_Specific.err
Untracked: code/H3K36me3DTplot_Specific.out
Untracked: code/H3K36me3DTplot_Specific.sh
Untracked: code/H3K36me3DTplot_distalPAS.err
Untracked: code/H3K36me3DTplot_distalPAS.out
Untracked: code/H3K36me3DTplot_distalPAS.sh
Untracked: code/H3K36me3DTplot_transcript.err
Untracked: code/H3K36me3DTplot_transcript.out
Untracked: code/H3K36me3DTplot_transcript.sh
Untracked: code/H3K36me3DTplotwide.err
Untracked: code/H3K36me3DTplotwide.out
Untracked: code/H3K36me3DTplotwide.sh
Untracked: code/H3K9me3DTplot_transcript.err
Untracked: code/H3K9me3DTplot_transcript.out
Untracked: code/H3K9me3DTplot_transcript.sh
Untracked: code/H3K9me3_processandDT.sh
Untracked: code/HchromOrder.err
Untracked: code/HchromOrder.out
Untracked: code/InfoContentShannon.py
Untracked: code/InfoContentbyInd.py
Untracked: code/IntersectMMandOrtho.err
Untracked: code/IntersectMMandOrtho.out
Untracked: code/IntersectPASandOrtho.err
Untracked: code/IntersectPASandOrtho.out
Untracked: code/JunctionLift.err
Untracked: code/JunctionLift.out
Untracked: code/JunctionLiftFinalChimp.err
Untracked: code/JunctionLiftFinalChimp.out
Untracked: code/Lift5perPAS.sh
Untracked: code/Lift5perPASbed.err
Untracked: code/Lift5perPASbed.out
Untracked: code/LiftClustersFirst.err
Untracked: code/LiftClustersFirst.out
Untracked: code/LiftClustersFirst_remove.err
Untracked: code/LiftClustersFirst_remove.out
Untracked: code/LiftClustersSecond.err
Untracked: code/LiftClustersSecond.out
Untracked: code/LiftClustersSecond_remove.err
Untracked: code/LiftClustersSecond_remove.out
Untracked: code/LiftFinalChimpJunc2Human.sh
Untracked: code/LiftOrthoPAS2chimp.sh
Untracked: code/LiftorthoPAS.err
Untracked: code/LiftorthoPASt.out
Untracked: code/Log.out
Untracked: code/MapBadSamples.err
Untracked: code/MapBadSamples.out
Untracked: code/MapBadSamples.sh
Untracked: code/MapStats.err
Untracked: code/MapStats.out
Untracked: code/MaxEntCode/
Untracked: code/MergeClusters.err
Untracked: code/MergeClusters.out
Untracked: code/MergeClusters.sh
Untracked: code/MismatchNumbers.err
Untracked: code/MismatchNumbers.out
Untracked: code/MismatchNumbers.sh
Untracked: code/NuclearDTUTR.err
Untracked: code/NuclearDTUTRt.out
Untracked: code/NuclearPlotsDEandDiffDom_4.err
Untracked: code/NuclearPlotsDEandDiffDom_4.out
Untracked: code/NuclearPlotsDEandDiffDom_4.sh
Untracked: code/PAS_ATTAAA.err
Untracked: code/PAS_ATTAAA.out
Untracked: code/PAS_ATTAAA.sh
Untracked: code/PAS_ATTAAADF.err
Untracked: code/PAS_ATTAAADF.out
Untracked: code/PAS_ATTAAA_df.sh
Untracked: code/PAS_seqExpanded.sh
Untracked: code/PAS_sequence.err
Untracked: code/PAS_sequence.out
Untracked: code/PAS_sequenceDF.err
Untracked: code/PAS_sequenceDF.out
Untracked: code/PASexpanded_sequenceDF.err
Untracked: code/PASexpanded_sequenceDF.out
Untracked: code/PASsequences.sh
Untracked: code/PASsequences_DF.sh
Untracked: code/PlotNuclearUsagebySpecies.R
Untracked: code/PlotNuclearUsagebySpecies_DF.R
Untracked: code/PlotNuclearUsagebySpecies_DF_4DIC.R
Untracked: code/PlotNuclearUsagebySpecies_DF_DEout.R
Untracked: code/QuantMergeClusters
Untracked: code/QuantMergeClusters.err
Untracked: code/QuantMergeClusters.out
Untracked: code/QuantMergedClusters.sh
Untracked: code/RNATranscriptDTplot.err
Untracked: code/RNATranscriptDTplot.out
Untracked: code/RNATranscriptDTplot.sh
Untracked: code/RNAmotif_PAS.err
Untracked: code/RNAmotif_PAS.out
Untracked: code/RNAmotif_PAS.sh
Untracked: code/RNAmotif_PAS_chimp.err
Untracked: code/RNAmotif_PAS_chimp.out
Untracked: code/RNAmotif_PAS_chimp.sh
Untracked: code/Rev_liftoverPAShg19to38.err
Untracked: code/Rev_liftoverPAShg19to38.out
Untracked: code/ReverseLiftFilter.R
Untracked: code/RunFixCluster.err
Untracked: code/RunFixCluster.out
Untracked: code/RunFixLeafCluster.sh
Untracked: code/RunNegMCMediation.err
Untracked: code/RunNegMCMediation.sh
Untracked: code/RunNegMCMediationDF.err
Untracked: code/RunNegMCMediationDF.out
Untracked: code/RunNegMCMediationDF.sh
Untracked: code/RunNegMCMediationr.out
Untracked: code/RunNewDom.err
Untracked: code/RunNewDom.out
Untracked: code/RunPosMCMediation.err
Untracked: code/RunPosMCMediation.sh
Untracked: code/RunPosMCMediationDF.err
Untracked: code/RunPosMCMediationDF.out
Untracked: code/RunPosMCMediationDF.sh
Untracked: code/RunPosMCMediationr.out
Untracked: code/SAF215upbed_gen.py
Untracked: code/SAF2Bed.py
Untracked: code/Snakefile
Untracked: code/SnakefilePAS
Untracked: code/SnakefilePASfilt
Untracked: code/SortIndexBadSamples.err
Untracked: code/SortIndexBadSamples.out
Untracked: code/SortIndexBadSamples.sh
Untracked: code/StarMM2.err
Untracked: code/StarMM2.out
Untracked: code/StarMM2.sh
Untracked: code/TestFC.err
Untracked: code/TestFC.out
Untracked: code/TestFC.sh
Untracked: code/TotalTranscriptDTplot.err
Untracked: code/TotalTranscriptDTplot.out
Untracked: code/UTR2FASTA.py
Untracked: code/Upstream10Bases_general.py
Untracked: code/allPASSeq_df.sh
Untracked: code/apaQTLsnake.err
Untracked: code/apaQTLsnake.out
Untracked: code/apaQTLsnakePAS.err
Untracked: code/apaQTLsnakePAS.out
Untracked: code/apaQTLsnakePAShuman.err
Untracked: code/apaQTLsnakefiltPAS.err
Untracked: code/apaQTLsnakefiltPAS.out
Untracked: code/assignPeak2Intronicregion.err
Untracked: code/assignPeak2Intronicregion.out
Untracked: code/assignPeak2Intronicregion.sh
Untracked: code/bam2junc.err
Untracked: code/bam2junc.out
Untracked: code/bam2junc_remove.err
Untracked: code/bam2junc_remove.out
Untracked: code/bed215upbed.py
Untracked: code/bed2Bedbothstrand.py
Untracked: code/bed2SAF_gen.py
Untracked: code/bed2saf.py
Untracked: code/bg_to_cov.py
Untracked: code/buildIndecpantro5
Untracked: code/buildIndecpantro5.sh
Untracked: code/buildLeafviz.err
Untracked: code/buildLeafviz.out
Untracked: code/buildLeafviz.sh
Untracked: code/buildLeafviz_leadAnno.sh
Untracked: code/buildLeafviz_leafanno.err
Untracked: code/buildLeafviz_leafanno.out
Untracked: code/buildStarIndex.sh
Untracked: code/callPeaksYL.py
Untracked: code/chimpChromprder.sh
Untracked: code/chimpMultiCov.err
Untracked: code/chimpMultiCov.out
Untracked: code/chimpMultiCov.sh
Untracked: code/chimpMultiCov255.sh
Untracked: code/chimpMultiCovInclusive.err
Untracked: code/chimpMultiCovInclusive.out
Untracked: code/chimpMultiCovInclusive.sh
Untracked: code/chooseAnno2Bed.py
Untracked: code/chooseAnno2SAF.py
Untracked: code/chooseSignalSite.py
Untracked: code/chromOrder.err
Untracked: code/chromOrder.out
Untracked: code/classifyLeafviz.err
Untracked: code/classifyLeafviz.out
Untracked: code/cleanbed2saf.py
Untracked: code/cluster.json
Untracked: code/cluster2bed.py
Untracked: code/clusterLiftReverse.sh
Untracked: code/clusterLiftReverse_removebad.sh
Untracked: code/clusterLiftprimary.sh
Untracked: code/clusterLiftprimary_removebad.sh
Untracked: code/clusterPAS.json
Untracked: code/clusterfiltPAS.json
Untracked: code/comands2Mege.sh
Untracked: code/converBam2Junc.sh
Untracked: code/converBam2Junc_removeBad.sh
Untracked: code/convertNumeric.py
Untracked: code/extraSnakefiltpas
Untracked: code/extractPhaastConGeneral.py
Untracked: code/extractPhyloReg.py
Untracked: code/extractPhyloRegGene.py
Untracked: code/extractPhylopGeneral.py
Untracked: code/extractPhylopGeneral_20way.py
Untracked: code/extractPhylopGeneral_noName.py
Untracked: code/extractPhylopReg200down.py
Untracked: code/extractPhylopReg200up.py
Untracked: code/filter5perc.R
Untracked: code/filter5percPAS.py
Untracked: code/filter5percPheno.py
Untracked: code/filterBamforMP.pysam2_gen.py
Untracked: code/filterJuncChroms.err
Untracked: code/filterJuncChroms.out
Untracked: code/filterMissprimingInNuc10_gen.py
Untracked: code/filterNumChroms.py
Untracked: code/filterPASforMP.py
Untracked: code/filterPostLift.py
Untracked: code/filterPrimaryread.py
Untracked: code/filterSAFforMP_gen.py
Untracked: code/filterSecondaryread.py
Untracked: code/filterSortBedbyCleanedBed_gen.R
Untracked: code/filterpeaks.py
Untracked: code/fixExonFC.py
Untracked: code/fixFChead.py
Untracked: code/fixFChead_bothfrac.py
Untracked: code/fixFCheadforExp.py
Untracked: code/fixLeafCluster.py
Untracked: code/fixLiftedJunc.py
Untracked: code/fixUTRexonanno.py
Untracked: code/formathg38Anno.py
Untracked: code/generateStarIndex.err
Untracked: code/generateStarIndex.out
Untracked: code/generateStarIndexHuman.err
Untracked: code/generateStarIndexHuman.out
Untracked: code/getAlloverlap.py
Untracked: code/getRNAseqMapStats.sh
Untracked: code/hg19MapStats.err
Untracked: code/hg19MapStats.out
Untracked: code/hg19MapStats.sh
Untracked: code/humanChromorder.sh
Untracked: code/humanFiles
Untracked: code/humanMultiCov.err
Untracked: code/humanMultiCov.out
Untracked: code/humanMultiCov.sh
Untracked: code/humanMultiCov255.err
Untracked: code/humanMultiCov255.out
Untracked: code/humanMultiCov255.sh
Untracked: code/humanMultiCovInclusive.err
Untracked: code/humanMultiCovInclusive.out
Untracked: code/humanMultiCov_inclusive.sh
Untracked: code/infoContentSimpson.py
Untracked: code/intersectAnno.err
Untracked: code/intersectAnno.out
Untracked: code/intersectAnnoExt.err
Untracked: code/intersectAnnoExt.out
Untracked: code/intersectLiftedPAS.sh
Untracked: code/leafcutter_merge_regtools_redo.py
Untracked: code/liftJunctionFiles.sh
Untracked: code/liftPAS19to38.sh
Untracked: code/liftVCF.out
Untracked: code/liftVCF.sh
Untracked: code/liftoverPAShg19to38.err
Untracked: code/liftoverPAShg19to38.out
Untracked: code/lliftVCF.err
Untracked: code/log/
Untracked: code/make5percPeakbed.py
Untracked: code/makeDIC.err
Untracked: code/makeDIC.out
Untracked: code/makeFileID.py
Untracked: code/makeNuclearDapaplots.sh
Untracked: code/makeNuclearDapaplots_DF.sh
Untracked: code/makeNuclearPlots.err
Untracked: code/makeNuclearPlots.out
Untracked: code/makeNuclearPlotsDF.err
Untracked: code/makeNuclearPlotsDF.out
Untracked: code/makePheno.py
Untracked: code/makeSamplyGroupsChimp_TvN.py
Untracked: code/makeSamplyGroupsHuman_TvN.py
Untracked: code/makedICPlots_DF.sh
Untracked: code/mapRNAseqhg19.sh
Untracked: code/mapRNAseqhg19_newPipeline.sh
Untracked: code/maphg19.err
Untracked: code/maphg19.out
Untracked: code/maphg19.sh
Untracked: code/maphg19_new.err
Untracked: code/maphg19_new.out
Untracked: code/maphg19_sub.err
Untracked: code/maphg19_sub.out
Untracked: code/maphg19_subjunc.sh
Untracked: code/mediation_test.R
Untracked: code/merge.err
Untracked: code/mergeChimp3prime_inhg38.sh
Untracked: code/mergeChimpRNA.sh
Untracked: code/merge_leafcutter_clusters_redo.py
Untracked: code/mergeandBWRNAseq.sh
Untracked: code/mergeandsort_ChimpinHuman.err
Untracked: code/mergeandsort_ChimpinHuman.out
Untracked: code/mergeandsort_H3K9me3
Untracked: code/mergeandsort_h3k36me3
Untracked: code/mergeandsorth3k36me3.sh
Untracked: code/mergedBam2BW.sh
Untracked: code/mergedbam2bw.err
Untracked: code/mergedbam2bw.out
Untracked: code/mergedbamRNAand2bw.err
Untracked: code/mergedbamRNAand2bw.out
Untracked: code/nameClusters.py
Untracked: code/namePeaks.py
Untracked: code/negativeMediation_montecarlo.R
Untracked: code/negativeMediation_montecarloDF.R
Untracked: code/nuclearTranscriptDTplot.err
Untracked: code/nuclearTranscriptDTplot.out
Untracked: code/numMultimap.py
Untracked: code/overlapMMandOrthoexon.sh
Untracked: code/overlapPAS.err
Untracked: code/overlapPAS.out
Untracked: code/overlapPASandOrthoexon.sh
Untracked: code/overlapapaQTLPAS.sh
Untracked: code/overlapapaQTLPAS_extended.sh
Untracked: code/overlapapaQTLPAS_samples.sh
Untracked: code/parseHg38.py
Untracked: code/peak2PAS.py
Untracked: code/pheno2countonly.R
Untracked: code/postiveMediation_montecarlo.R
Untracked: code/postiveMediation_montecarlo_DF.R
Untracked: code/prepareAnnoLeafviz.err
Untracked: code/prepareAnnoLeafviz.out
Untracked: code/prepareCleanLiftedFC_5perc4LC.py
Untracked: code/prepareLeafvizAnno.sh
Untracked: code/preparePAS4lift.py
Untracked: code/prepare_phenotype_table.py
Untracked: code/primaryLift.err
Untracked: code/primaryLift.out
Untracked: code/primaryLift.sh
Untracked: code/processhg38exons.py
Untracked: code/quantJunc.sh
Untracked: code/quantJunc_TEST.sh
Untracked: code/quantJunc_removeBad.sh
Untracked: code/quantLiftedPAS.err
Untracked: code/quantLiftedPAS.out
Untracked: code/quantLiftedPAS.sh
Untracked: code/quantLiftedPASPrimary.err
Untracked: code/quantLiftedPASPrimary.out
Untracked: code/quantLiftedPASPrimary.sh
Untracked: code/quatJunc.err
Untracked: code/quatJunc.out
Untracked: code/recChimpback2Human.err
Untracked: code/recChimpback2Human.out
Untracked: code/recLiftchim2human.sh
Untracked: code/revLift.err
Untracked: code/revLift.out
Untracked: code/revLiftPAShg38to19.sh
Untracked: code/reverseLift.sh
Untracked: code/runCheckReverseLift.sh
Untracked: code/runChimpDiffIso.sh
Untracked: code/runChimpDiffIsoDF.sh
Untracked: code/runCountNucleotides.err
Untracked: code/runCountNucleotides.out
Untracked: code/runCountNucleotides.sh
Untracked: code/runCountNucleotidesPantro6.err
Untracked: code/runCountNucleotidesPantro6.out
Untracked: code/runCountNucleotides_pantro6.sh
Untracked: code/runFilterNumChroms.sh
Untracked: code/runHumanDiffIso.sh
Untracked: code/runHumanDiffIsoDF.sh
Untracked: code/runNewDom.sh
Untracked: code/runNuclearDiffIso_DF.sh
Untracked: code/runNuclearDifffIso.sh
Untracked: code/runTotalDiffIso.sh
Untracked: code/run_Chimpleafcutter_ds.err
Untracked: code/run_Chimpleafcutter_ds.out
Untracked: code/run_Chimpverifybam.err
Untracked: code/run_Chimpverifybam.out
Untracked: code/run_Humanleafcutter_dF.err
Untracked: code/run_Humanleafcutter_dF.out
Untracked: code/run_Humanleafcutter_ds.err
Untracked: code/run_Humanleafcutter_ds.out
Untracked: code/run_Nuclearleafcutter_ds.err
Untracked: code/run_Nuclearleafcutter_ds.out
Untracked: code/run_Nuclearleafcutter_dsDF.err
Untracked: code/run_Nuclearleafcutter_dsDF.out
Untracked: code/run_Totalleafcutter_ds.err
Untracked: code/run_Totalleafcutter_ds.out
Untracked: code/run_chimpverifybam.sh
Untracked: code/run_verifyBam.sh
Untracked: code/run_verifybam.err
Untracked: code/run_verifybam.out
Untracked: code/slurm-62824013.out
Untracked: code/slurm-62825841.out
Untracked: code/slurm-62826116.out
Untracked: code/slurm-64108209.out
Untracked: code/slurm-64108521.out
Untracked: code/slurm-64108557.out
Untracked: code/snakePASChimp.err
Untracked: code/snakePASChimp.out
Untracked: code/snakePAShuman.out
Untracked: code/snakemake.batch
Untracked: code/snakemakeChimp.err
Untracked: code/snakemakeChimp.out
Untracked: code/snakemakeHuman.err
Untracked: code/snakemakeHuman.out
Untracked: code/snakemakePAS.batch
Untracked: code/snakemakePASFiltChimp.err
Untracked: code/snakemakePASFiltChimp.out
Untracked: code/snakemakePASFiltHuman.err
Untracked: code/snakemakePASFiltHuman.out
Untracked: code/snakemakePAS_Human.batch
Untracked: code/snakemakePASchimp.batch
Untracked: code/snakemakePAShuman.batch
Untracked: code/snakemake_chimp.batch
Untracked: code/snakemake_human.batch
Untracked: code/snakemakefiltPAS.batch
Untracked: code/snakemakefiltPAS_chimp.sh
Untracked: code/snakemakefiltPAS_human.batch
Untracked: code/snakemakefiltPAS_human.sh
Untracked: code/spliceSite2Fasta.py
Untracked: code/submit-snakemake-chimp.sh
Untracked: code/submit-snakemake-human.sh
Untracked: code/submit-snakemakePAS-chimp.sh
Untracked: code/submit-snakemakePAS-human.sh
Untracked: code/submit-snakemakefiltPAS-chimp.sh
Untracked: code/submit-snakemakefiltPAS-human.sh
Untracked: code/subset_diffisopheno.py
Untracked: code/subset_diffisopheno_Chimp_tvN.py
Untracked: code/subset_diffisopheno_Chimp_tvN_DF.py
Untracked: code/subset_diffisopheno_Huma_tvN.py
Untracked: code/subset_diffisopheno_Huma_tvN_DF.py
Untracked: code/subset_diffisopheno_Nuclear_HvC.py
Untracked: code/subset_diffisopheno_Nuclear_HvC_DF.py
Untracked: code/subset_diffisopheno_Total_HvC.py
Untracked: code/test
Untracked: code/test.txt
Untracked: code/threeprimeOrthoFC.out
Untracked: code/threeprimeOrthoFC.sh
Untracked: code/threeprimeOrthoFCcd.err
Untracked: code/transcriptDTplotsNuclear.sh
Untracked: code/transcriptDTplotsTotal.sh
Untracked: code/tripseq-analysis/
Untracked: code/verifyBam4973.sh
Untracked: code/verifyBam4973inHuman.sh
Untracked: code/verifybam4973.err
Untracked: code/verifybam4973.out
Untracked: code/verifybam4973HumanMap.err
Untracked: code/verifybam4973HumanMap.out
Untracked: code/wrap_Chimpverifybam.err
Untracked: code/wrap_Chimpverifybam.out
Untracked: code/wrap_chimpverifybam.sh
Untracked: code/wrap_verifyBam.sh
Untracked: code/wrap_verifybam.err
Untracked: code/wrap_verifybam.out
Untracked: code/writeMergecode.py
Untracked: data/._.DS_Store
Untracked: data/._HC_filenames.txt
Untracked: data/._HC_filenames.txt.sb-4426323c-IKIs0S
Untracked: data/._HC_filenames.xlsx
Untracked: data/._MapPantro6_meta.txt
Untracked: data/._MapPantro6_meta.txt.sb-a5794dd2-Cskmlm
Untracked: data/._MapPantro6_meta.xlsx
Untracked: data/._OppositeSpeciesMap.txt
Untracked: data/._OppositeSpeciesMap.txt.sb-a5794dd2-mayWJf
Untracked: data/._OppositeSpeciesMap.xlsx
Untracked: data/._RNASEQ_metadata.txt
Untracked: data/._RNASEQ_metadata.txt.sb-4426323c-TE4ns3
Untracked: data/._RNASEQ_metadata.txt.sb-51f67ae1-HXp7Gq
Untracked: data/._RNASEQ_metadata_2Removed.txt
Untracked: data/._RNASEQ_metadata_2Removed.txt.sb-4426323c-a4lBwx
Untracked: data/._RNASEQ_metadata_2Removed.xlsx
Untracked: data/._RNASEQ_metadata_stranded.txt
Untracked: data/._RNASEQ_metadata_stranded.txt.sb-a5794dd2-D659m2
Untracked: data/._RNASEQ_metadata_stranded.txt.sb-a5794dd2-ImNMoY
Untracked: data/._RNASEQ_metadata_stranded.txt.sb-e4bf31f0-ZGnGgl
Untracked: data/._RNASEQ_metadata_stranded.xlsx
Untracked: data/._TrialFiltersMeta.txt
Untracked: data/._TrialFiltersMeta.txt.sb-9845453e-R58Y0Q
Untracked: data/._metadata_HCpanel.txt
Untracked: data/._metadata_HCpanel.txt.sb-a3d92a2d-b9cYoF
Untracked: data/._metadata_HCpanel.txt.sb-a5794dd2-i594qs
Untracked: data/._metadata_HCpanel.txt.sb-f4823d1e-qihGek
Untracked: data/._metadata_HCpanel_frompantro5.xlsx
Untracked: data/._~$RNASEQ_metadata.xlsx
Untracked: data/._~$metadata_HCpanel.xlsx
Untracked: data/._.xlsx
Untracked: data/ALU/
Untracked: data/AREelements/
Untracked: data/BaseComp/
Untracked: data/CleanLiftedPeaks_FC_primary/
Untracked: data/CompapaQTLpas/
Untracked: data/DIC_Viz/
Untracked: data/DNDS/
Untracked: data/DTmatrix/
Untracked: data/DiffDomandDE_example/
Untracked: data/DiffExpression/
Untracked: data/DiffIso_Nuclear/
Untracked: data/DiffIso_Nuclear_DF/
Untracked: data/DiffIso_Total/
Untracked: data/DiffSplice/
Untracked: data/DiffSplice_liftedJunc/
Untracked: data/DiffSplice_removeBad/
Untracked: data/DifferentialPASUsageResults.txt
Untracked: data/DistTwoDom/
Untracked: data/DomDefGreaterX/
Untracked: data/DomStructure_4/
Untracked: data/DominantPAS/
Untracked: data/DominantPAS_DF/
Untracked: data/DoubleFilterUsageNumeric/
Untracked: data/EvalPantro5/
Untracked: data/H3K36me3/
Untracked: data/HC_filenames.txt
Untracked: data/HC_filenames.xlsx
Untracked: data/HumanMolPheno/
Untracked: data/HumanUTR/
Untracked: data/IndInfoContent/
Untracked: data/InfoContent/
Untracked: data/Khan_prot/
Untracked: data/Li_eqtls/
Untracked: data/MapPantro6_meta.txt
Untracked: data/MapPantro6_meta.xlsx
Untracked: data/MapStats/
Untracked: data/NormalizedClusters/
Untracked: data/NuclearHvC/
Untracked: data/NuclearHvC_DF/
Untracked: data/OppositeSpeciesMap.txt
Untracked: data/OppositeSpeciesMap.xlsx
Untracked: data/OrthoExonBed/
Untracked: data/OverlapBenchmark/
Untracked: data/OverlappingPAS/
Untracked: data/PAS/
Untracked: data/PAS_SAF/
Untracked: data/PAS_doubleFilter/
Untracked: data/PTM/
Untracked: data/Peaks_5perc/
Untracked: data/PhastCon/
Untracked: data/Pheno_5perc/
Untracked: data/Pheno_5perc_DF_nuclear/
Untracked: data/Pheno_5perc_nuclear/
Untracked: data/Pheno_5perc_nuclear_old/
Untracked: data/Pheno_5perc_total/
Untracked: data/PhyloP/
Untracked: data/Pol2Chip/
Untracked: data/QTLPASoverlap/
Untracked: data/RNASEQ_metadata.txt
Untracked: data/RNASEQ_metadata_2Removed.txt
Untracked: data/RNASEQ_metadata_2Removed.xlsx
Untracked: data/RNASEQ_metadata_stranded.txt
Untracked: data/RNASEQ_metadata_stranded.txt.sb-e4bf31f0-ZGnGgl/
Untracked: data/RNASEQ_metadata_stranded.xlsx
Untracked: data/SignalSites/
Untracked: data/SignalSites_doublefilter/
Untracked: data/SpliceSite/
Untracked: data/TestAnnoBiasOE/
Untracked: data/TestMM2/
Untracked: data/TestMM2_AS/
Untracked: data/TestMM2_PrimaryRead/
Untracked: data/TestMM2_SeondaryRead/
Untracked: data/TestMM2_mismatch/
Untracked: data/TestMM2_quality/
Untracked: data/TestWithinMergePAS/
Untracked: data/Test_FC_methods/
Untracked: data/Threeprime2Ortho/
Untracked: data/TotalFractionPAS/
Untracked: data/TotalHvC/
Untracked: data/TrialFiltersMeta.txt
Untracked: data/TwoBadSampleAnalysis/
Untracked: data/UnliftedSites/
Untracked: data/UrichElements/
Untracked: data/Wang_ribo/
Untracked: data/apaQTLGenes/
Untracked: data/bioGRID/
Untracked: data/chainFiles/
Untracked: data/cleanPeaks_anno/
Untracked: data/cleanPeaks_byspecies/
Untracked: data/cleanPeaks_lifted/
Untracked: data/files4viz_nuclear/
Untracked: data/files4viz_nuclear_DF/
Untracked: data/gsea/
Untracked: data/gviz/
Untracked: data/hypoxia/
Untracked: data/leafviz/
Untracked: data/liftover_files/
Untracked: data/mediation/
Untracked: data/mediation_DF/
Untracked: data/metadata_HCpanel.txt
Untracked: data/metadata_HCpanel.xlsx
Untracked: data/metadata_HCpanel_extra.txt
Untracked: data/metadata_HCpanel_frompantro5.txt
Untracked: data/metadata_HCpanel_frompantro5.xlsx
Untracked: data/miRNA/
Untracked: data/multimap/
Untracked: data/orthoUTR/
Untracked: data/paiDecay/
Untracked: data/primaryLift/
Untracked: data/randomIntronCons/
Untracked: data/reverseLift/
Untracked: data/testQuant/
Untracked: data/utrDB/
Untracked: data/~$RNASEQ_metadata.xlsx
Untracked: data/~$metadata_HCpanel.xlsx
Untracked: data/.xlsx
Untracked: output/._.DS_Store
Untracked: output/AUcount_density.pdf
Untracked: output/AUcount_density_sm.pdf
Untracked: output/DEandAPA.txt
Untracked: output/DEandAPA_sig.txt
Untracked: output/DEandTEeffectsize
Untracked: output/DEandTEeffectsize.pdf
Untracked: output/DEeffectsize
Untracked: output/DEeffectsize.pdf
Untracked: output/FigureDF/
Untracked: output/FigurePresColors/
Untracked: output/PropSamesdom
Untracked: output/PropSamesdom.pdf
Untracked: output/SupplementaryTable/
Untracked: output/TEeffectsize
Untracked: output/TEeffectsize.pdf
Untracked: output/Total_DEeffectsize
Untracked: output/Total_DEeffectsize.pdf
Untracked: output/Total_DEeffectsizeNotJusttop.pdf
Untracked: output/Total_DErelationship.pdf
Untracked: output/Total_TEeffectsize
Untracked: output/Total_TEeffectsize.pdf
Untracked: output/Ubiqplot
Untracked: output/Ubiqplot.pdf
Untracked: output/dAPAandDomEnrich.png
Untracked: output/dEandDomEnrich.png
Untracked: output/dediffdom.pdf
Untracked: output/dpnotDE
Untracked: output/dtPlots/
Untracked: output/exandte
Untracked: output/fig1.pdf
Untracked: output/fig2.pdf
Untracked: output/fig3.pdf
Untracked: output/fig4.pdf
Untracked: output/fig5.pdf
Untracked: output/fig6.pdf
Untracked: output/piecharts
Untracked: output/piecharts.pdf
Untracked: output/removeUnilift_fig6 copy.pdf
Untracked: output/removeUnilift_fig6.pdf
Untracked: output/removeUnlift_fig3.pdf
Untracked: output/removeUnlift_fig4.pdf
Untracked: output/removeUnlift_fig5.pdf
Untracked: output/simpson.pdf
Untracked: output/simpson_alTogether.pdf
Untracked: output/supplement/
Untracked: output/whichSiteplot.pdf
Untracked: projectNotes.Rmd
Untracked: proteinModelSet.Rmd
Unstaged changes:
Modified: analysis/DeandNumPAS.Rmd
Modified: analysis/DirSelectionKhan.Rmd
Modified: analysis/ExploredAPA.Rmd
Modified: analysis/ExploredAPA_DF.Rmd
Modified: analysis/MMExpreiment.Rmd
Modified: analysis/OppositeMap.Rmd
Modified: analysis/PTM_analysis.Rmd
Modified: analysis/ResultsNoUnlifted.Rmd
Modified: analysis/TotalDomStructure.Rmd
Modified: analysis/TotalVNuclearBothSpecies.Rmd
Modified: analysis/annotationInfo.Rmd
Modified: analysis/changeMisprimcut.Rmd
Modified: analysis/comp2apaQTLPAS.Rmd
Modified: analysis/correlationPhenos.Rmd
Modified: analysis/dInforContent.Rmd
Modified: analysis/df_QC.Rmd
Modified: analysis/diffExpression.Rmd
Modified: analysis/establishCutoffs.Rmd
Modified: analysis/incorporateQTLsAncestral.Rmd
Modified: analysis/infoContent.Rmd
Modified: analysis/investigatePantro5.Rmd
Modified: analysis/mRNADecay.Rmd
Modified: analysis/miRNAanalysis.Rmd
Modified: analysis/multiMap.Rmd
Modified: analysis/phastCon.Rmd
Modified: analysis/pol2.Rmd
Modified: analysis/speciesSpecific.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/additionalGSEA.Rmd) and HTML (docs/additionalGSEA.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | d261561 | brimittleman | 2020-12-13 | add GOrilla results |
| html | e1bb6e8 | brimittleman | 2020-12-13 | Build site. |
| Rmd | e57e4b5 | brimittleman | 2020-12-13 | add analysis for FGSEA |
In this analysis I will look for gene set enrichments for categories requested by reviewers.
library(tidyverse)── Attaching packages ────────────────────────────────── tidyverse 1.3.0 ──✓ ggplot2 3.2.1 ✓ purrr 0.3.4
✓ tibble 2.1.3 ✓ dplyr 0.8.3
✓ tidyr 1.1.0 ✓ stringr 1.4.0
✓ readr 1.3.1 ✓ forcats 0.4.0── Conflicts ───────────────────────────────────── tidyverse_conflicts() ──
x dplyr::filter() masks stats::filter()
x dplyr::lag() masks stats::lag()library(workflowr)This is workflowr version 1.6.2
Run ?workflowr for help getting startedlibrary(fgsea)Gene sets:
Hset=gmtPathways("../data/gsea/h.all.v7.1.symbols.gmt")
C3=gmtPathways("../data/gsea/c3.all.v7.1.symbols.gmt")
C5=gmtPathways("../data/gsea/c5.all.v7.1.symbols.gmt")DE genes withing dAPA
First I will look at the differential APA genes that are differentially expressed. I will ask the question if DE genes are enriched within dAPA genes.
#load DE genes
nameID=read.table("../../genome_anotation_data/ensemble_to_genename.txt",sep="\t", header = T, stringsAsFactors = F)
DE= read.table("../data/DiffExpression/DEtested_allres.txt",header=F, stringsAsFactors = F,col.names = c('Gene_stable_ID', 'logFC' ,'AveExpr', 't', 'P.Value', 'adj.P.Val', 'B')) %>% inner_join(nameID, by="Gene_stable_ID") %>% dplyr::select(-Gene_stable_ID, -Source_of_gene_name) %>% rename("gene"=Gene.name)
#loaddAPA
dAPA=read.table("../data/DiffIso_Nuclear_DF/AllPAS_withGeneSig.txt", header = T, stringsAsFactors = F) %>% filter(SigPAU2=='Yes') %>% select(gene) %>% unique()
DEwithdAPA=DE %>% inner_join(dAPA, by="gene") %>% arrange(t)
DEwithdAPAVec= setNames(DEwithdAPA$t, DEwithdAPA$gene)Run FGSEA
FGSEA_Funtion=function(path,stat){
fgseaResDiffUsed <- fgsea(pathways = path,
stats = stat,
minSize = 15,
maxSize = 500,
nperm = 100000)
fgseaResTidy <- fgseaResDiffUsed %>%
as_tibble() %>%
arrange(desc(NES)) %>%
select(-leadingEdge, -ES, -nMoreExtreme) %>%
arrange(padj)
return(fgseaResTidy)
}
DE_halmark=FGSEA_Funtion(Hset,DEwithdAPAVec)Warning in fgsea(pathways = path, stats = stat, minSize = 15, maxSize
= 500, : You are trying to run fgseaSimple. It is recommended to use
fgseaMultilevel. To run fgseaMultilevel, you need to remove the nperm
argument in the fgsea function call.ggplot(DE_halmark, aes(reorder(pathway, NES), NES)) +
geom_col(aes(fill=padj<0.05)) +
coord_flip() +
labs(x="Pathway", y="Normalized Enrichment Score",
title="Hallmark pathways NES from GSEA") +
theme_minimal()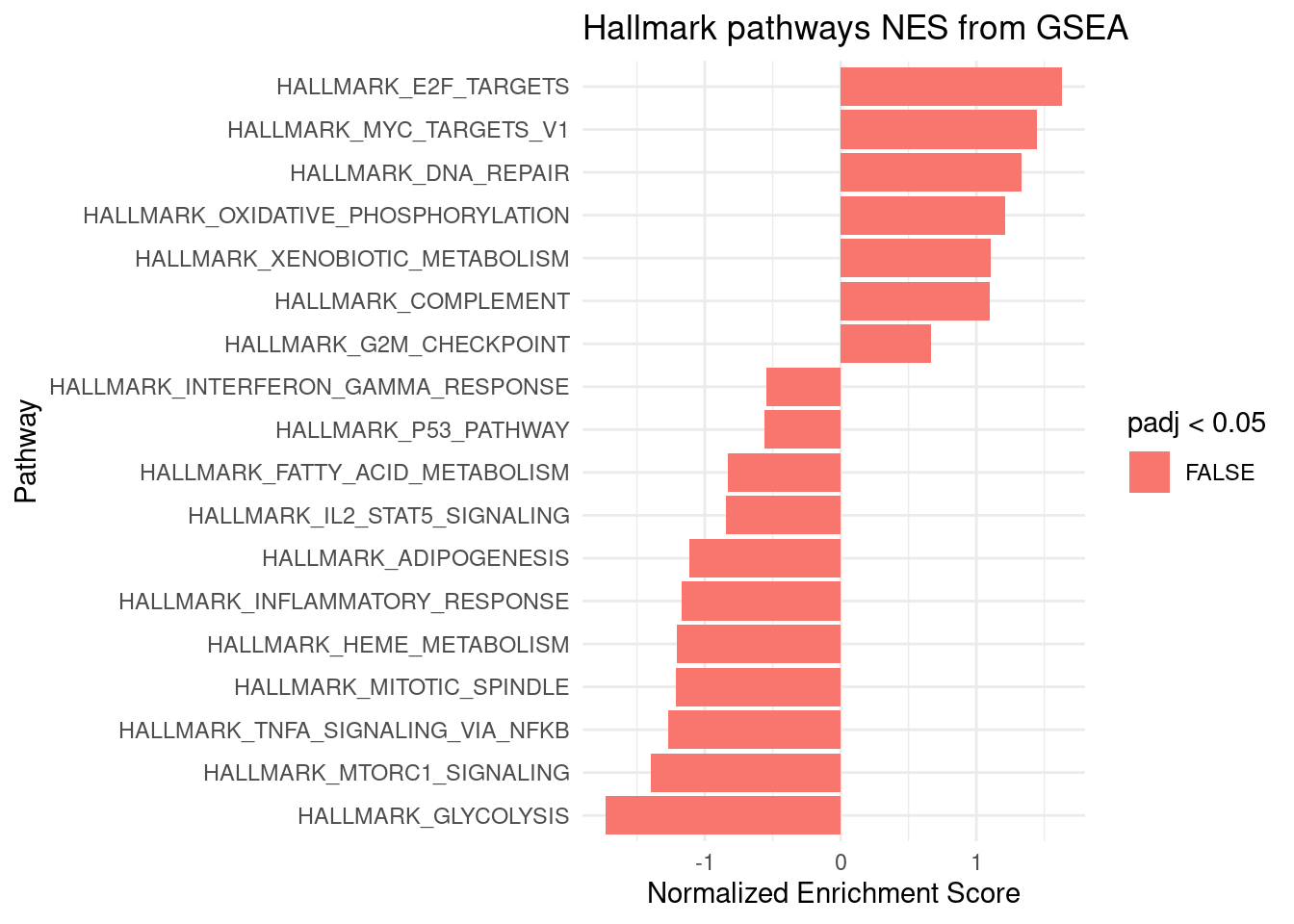
| Version | Author | Date |
|---|---|---|
| e1bb6e8 | brimittleman | 2020-12-13 |
DE_C3=FGSEA_Funtion(C3,DEwithdAPAVec)Warning in fgsea(pathways = path, stats = stat, minSize = 15, maxSize
= 500, : You are trying to run fgseaSimple. It is recommended to use
fgseaMultilevel. To run fgseaMultilevel, you need to remove the nperm
argument in the fgsea function call.head(DE_C3)# A tibble: 6 x 5
pathway pval padj NES size
<chr> <dbl> <dbl> <dbl> <int>
1 AUTS2_TARGET_GENES 0.000253 0.162 1.90 70
2 TAF9B_TARGET_GENES 0.000401 0.162 1.84 78
3 ZFHX3_TARGET_GENES 0.000305 0.162 1.64 195
4 THAP1_TARGET_GENES 0.000269 0.162 1.60 229
5 MTA1_TARGET_GENES 0.000542 0.176 1.60 215
6 ACCATTT_MIR522 0.000945 0.255 -1.92 17DE_C5=FGSEA_Funtion(C5,DEwithdAPAVec)Warning in fgsea(pathways = path, stats = stat, minSize = 15, maxSize
= 500, : You are trying to run fgseaSimple. It is recommended to use
fgseaMultilevel. To run fgseaMultilevel, you need to remove the nperm
argument in the fgsea function call.DE_C5_sig <- DE_C5 %>%
filter(padj < 0.05) %>%
select(pathway, padj, NES)
DE_C5# A tibble: 1,056 x 5
pathway pval padj NES size
<chr> <dbl> <dbl> <dbl> <int>
1 GO_RIBONUCLEOPROTEIN_COMPLEX 1.66e-5 0.00876 1.95 88
2 GO_MRNA_METABOLIC_PROCESS 1.62e-5 0.00876 1.91 112
3 GO_RNA_CATABOLIC_PROCESS 3.43e-5 0.0121 1.98 58
4 GO_NUCLEUS_ORGANIZATION 5.49e-5 0.0145 2.07 19
5 GO_RIBOSOME 1.23e-4 0.0222 2.04 40
6 GO_NUCLEAR_TRANSCRIBED_MRNA_CATABOLIC_PROC… 1.59e-4 0.0222 2.01 38
7 GO_RIBOSOMAL_SUBUNIT 1.42e-4 0.0222 1.98 34
8 GO_STRUCTURAL_MOLECULE_ACTIVITY 1.89e-4 0.0222 1.94 54
9 GO_RNA_BINDING 1.81e-4 0.0222 1.64 214
10 GO_NUCLEAR_TRANSCRIBED_MRNA_CATABOLIC_PROC… 2.16e-4 0.0228 2.03 29
# … with 1,046 more rowsdAPA and TE
For this I will have to use GOrilla because I only have FDRs. I will run the same analysis but with the differentially translated:
nameID=read.table("../../genome_anotation_data/ensemble_to_genename.txt",sep="\t", header = T, stringsAsFactors = F)
colnames(nameID)=c("ENSG", "gene", "source")
translation=read.table("../data/Wang_ribo/Additionaltable5_translationComparisons.txt",stringsAsFactors = F, header = T) %>% inner_join(nameID,by="ENSG") %>% select(gene,HvC.pvalue, HvC.FDR)
translationandAPA=translation %>% inner_join(dAPA,by="gene") %>% arrange(HvC.FDR)
TEgenes=as.vector(translationandAPA$gene)
write.table(TEgenes, "../data/gsea/TEgenes.txt", quote = F, sep="/n",row.names = F, col.names = F)Results:
TE_GO=read.table("../data/gsea/GO_termsTE.txt", sep="\t", header = T) %>% arrange(FDR.q.value)
TE_GO_sig=TE_GO %>% filter(FDR.q.value<=0.05)
TE_GO_sig GO.Term Description P.value FDR.q.value Enrichment N
1 GO:0006413 translational initiation 1.35e-06 0.00908 32.08 1155
B n b
1 20 9 5
Genes
1 [EIF4H - eukaryotic translation initiation factor 4h, RPL22 - ribosomal protein l22, RPL3 - ribosomal protein l3, EIF3F - eukaryotic translation initiation factor 3, subunit f, EIF1 - eukaryotic translation initiation factor 1]dAPA and DP
ProtComp=read.csv("../data/Khan_prot/Khan_TableS4.csv", stringsAsFactors = F, header = T)%>% inner_join(nameID,by="ENSG") %>% select(gene.symbol ,HC.qvalues.protein) %>% rename("gene"=gene.symbol)
protandAPA=ProtComp %>% inner_join(dAPA,by="gene") %>% arrange(HC.qvalues.protein)
PEgenes=as.vector(protandAPA$gene)
write.table(PEgenes, "../data/gsea/Pgenes.txt", quote = F, sep="/n",row.names = F, col.names = F)Results:
dP_GO=read.table("../data/gsea/Goterms_dP.txt", sep="\t", header = T) %>% arrange(FDR.q.value)
dP_GO_sig=dP_GO %>% filter(FDR.q.value<=0.05)
dP_GO_sig [1] GO.Term Description P.value FDR.q.value Enrichment
[6] N B n b Genes
<0 rows> (or 0-length row.names)dAPA, DP not DE
I will use the two set analysis in GORilla. I will compare the DP not DE genes with all of the dAPA genes.
DE_sig=DE %>% filter(adj.P.Val <0.05)
protandAPA_notDE= protandAPA %>% anti_join(DE_sig,by="gene") %>% filter(HC.qvalues.protein<0.05)
PnotDEgenes=as.vector(protandAPA$gene)
write.table(PnotDEgenes, "../data/gsea/PnotEgenes.txt", quote = F, sep="/n", row.names = F, col.names = F)
#write dAPA genes as background
dAPAgeness=as.vector(dAPA$gene)
write.table(dAPAgeness, "../data/gsea/dAPAgenes.txt", quote = F, sep="/n", row.names = F, col.names = F)Results:
dPnotDE_GO=read.table("../data/gsea/GO_termsDPnotDE.txt", sep="\t", header = T) %>% arrange(FDR.q.value)
dPnotDE_GO_sig=dPnotDE_GO %>% filter(FDR.q.value<=0.05)
dPnotDE_GO_sig GO.Term
1 GO:0044444
2 GO:0003723
3 GO:0005829
4 GO:0070062
5 GO:1903561
6 GO:0043230
7 GO:0009987
8 GO:0031982
9 GO:0044421
10 GO:0032991
11 GO:0044237
12 GO:0031974
13 GO:0070013
14 GO:0002252
15 GO:0043233
16 GO:0008152
17 GO:0044424
18 GO:0002376
19 GO:0043299
20 GO:0002275
21 GO:0042119
22 GO:0051704
23 GO:0036230
24 GO:0002283
25 GO:0045055
26 GO:0043312
27 GO:0044419
28 GO:0006810
29 GO:0006887
30 GO:0044403
31 GO:0016032
32 GO:0002263
33 GO:0002366
34 GO:0051234
35 GO:0071704
36 GO:0034641
37 GO:0046907
38 GO:0002274
39 GO:1904813
40 GO:0044249
41 GO:0006886
42 GO:0016020
43 GO:0016192
44 GO:0005515
45 GO:0044429
46 GO:0044238
47 GO:1901566
48 GO:1901576
49 GO:0006807
50 GO:0051649
51 GO:0009058
52 GO:0046903
53 GO:0044271
54 GO:0032940
55 GO:0044446
56 GO:0043226
57 GO:0015031
58 GO:0045321
59 GO:0019693
60 GO:0043043
61 GO:0045184
62 GO:1901564
63 GO:0051179
64 GO:0043603
65 GO:0006518
66 GO:0015833
67 GO:1990904
68 GO:0006139
69 GO:0046483
70 GO:0006725
71 GO:0006412
72 GO:0051641
73 GO:1901360
74 GO:0042221
75 GO:0071705
76 GO:0060205
77 GO:0044248
78 GO:0031983
79 GO:0044422
80 GO:0034774
81 GO:0042886
82 GO:0006163
83 GO:0010033
84 GO:0009259
85 GO:0043604
86 GO:0001775
87 GO:0044433
88 GO:0072594
89 GO:0044464
90 GO:0044391
91 GO:0006413
92 GO:0031966
93 GO:0009117
94 GO:0006753
95 GO:0005575
96 GO:1901575
97 GO:0022411
98 GO:0003735
99 GO:0043227
100 GO:0055086
101 GO:0009150
102 GO:0008150
103 GO:0005488
104 GO:0009124
105 GO:0045296
106 GO:0009123
107 GO:0005198
108 GO:0009056
109 GO:0010608
110 GO:0009165
111 GO:1901293
112 GO:0043933
113 GO:0003674
114 GO:0043229
115 GO:0005739
116 GO:0072521
117 GO:0046390
118 GO:0048193
119 GO:0034645
120 GO:0032984
121 GO:0071840
122 GO:0071702
123 GO:0044281
124 GO:0005737
125 GO:0098798
126 GO:0009156
127 GO:0009141
128 GO:0019637
129 GO:0009260
130 GO:0005743
131 GO:0009161
132 GO:0072522
133 GO:0008104
134 GO:0009059
135 GO:0006164
136 GO:0005840
137 GO:0015934
138 GO:0070887
139 GO:0016050
140 GO:0002757
141 GO:1901698
142 GO:0033036
143 GO:1901363
144 GO:0050896
145 GO:0005759
146 GO:0097159
147 GO:0009057
148 GO:0031090
149 GO:0006793
150 GO:0003743
151 GO:0002429
152 GO:0009199
153 GO:0016043
154 GO:0009126
155 GO:0043488
156 GO:0033365
157 GO:0061024
158 GO:0043487
159 GO:0009127
160 GO:0019866
161 GO:0030054
162 GO:0071310
163 GO:0002253
164 GO:0010243
165 GO:0050839
166 GO:0070125
167 GO:0006796
168 GO:0006091
169 GO:0016071
170 GO:0009152
171 GO:0009144
172 GO:0009205
173 GO:0006414
174 GO:0090407
175 GO:0043170
176 GO:0055114
177 GO:0061013
178 GO:0043624
179 GO:0009167
180 GO:0044265
181 GO:1903311
182 GO:0003824
183 GO:0051716
184 GO:0019899
185 GO:0030667
186 GO:0098796
187 GO:0009168
188 GO:0009132
189 GO:0009142
190 GO:0003729
191 GO:0009185
192 GO:1901068
193 GO:0031329
194 GO:0002764
195 GO:0036094
196 GO:0009719
197 GO:0019439
198 GO:0006520
199 GO:0016874
200 GO:0009894
201 GO:0034654
202 GO:0042493
203 GO:0008135
204 GO:1901699
205 GO:0009119
206 GO:0006415
207 GO:1901361
208 GO:0009116
209 GO:0034404
210 GO:0099023
211 GO:0006996
212 GO:1905369
213 GO:1905368
214 GO:0000502
215 GO:0031410
216 GO:1901362
217 GO:0034655
218 GO:0009135
219 GO:0003725
220 GO:0009179
221 GO:0060071
222 GO:0006605
223 GO:0000166
224 GO:1901135
225 GO:0046700
226 GO:0044270
227 GO:0007006
228 GO:0006396
229 GO:0002768
230 GO:0032555
231 GO:0019438
232 GO:0018130
233 GO:1901265
234 GO:1901137
235 GO:0042802
236 GO:0031145
237 GO:0071345
238 GO:0006906
239 GO:0006614
240 GO:0007007
241 GO:0046039
242 GO:0034248
243 GO:0046031
244 GO:0032784
245 GO:0016052
246 GO:0032727
247 GO:0032481
248 GO:0048284
249 GO:0043039
250 GO:0061025
251 GO:0090174
252 GO:0071826
253 GO:0043038
254 GO:0006888
255 GO:0070126
256 GO:0032647
257 GO:0034613
258 GO:0051169
259 GO:0007005
260 GO:0006913
261 GO:0017076
262 GO:0035639
263 GO:0044877
264 GO:0032553
Description
1 cytoplasmic part
2 RNA binding
3 cytosol
4 extracellular exosome
5 extracellular vesicle
6 extracellular organelle
7 cellular process
8 vesicle
9 extracellular region part
10 protein-containing complex
11 cellular metabolic process
12 membrane-enclosed lumen
13 intracellular organelle lumen
14 immune effector process
15 organelle lumen
16 metabolic process
17 intracellular part
18 immune system process
19 leukocyte degranulation
20 myeloid cell activation involved in immune response
21 neutrophil activation
22 multi-organism process
23 granulocyte activation
24 neutrophil activation involved in immune response
25 regulated exocytosis
26 neutrophil degranulation
27 interspecies interaction between organisms
28 transport
29 exocytosis
30 symbiont process
31 viral process
32 cell activation involved in immune response
33 leukocyte activation involved in immune response
34 establishment of localization
35 organic substance metabolic process
36 cellular nitrogen compound metabolic process
37 intracellular transport
38 myeloid leukocyte activation
39 ficolin-1-rich granule lumen
40 cellular biosynthetic process
41 intracellular protein transport
42 membrane
43 vesicle-mediated transport
44 protein binding
45 mitochondrial part
46 primary metabolic process
47 organonitrogen compound biosynthetic process
48 organic substance biosynthetic process
49 nitrogen compound metabolic process
50 establishment of localization in cell
51 biosynthetic process
52 secretion
53 cellular nitrogen compound biosynthetic process
54 secretion by cell
55 intracellular organelle part
56 organelle
57 protein transport
58 leukocyte activation
59 ribose phosphate metabolic process
60 peptide biosynthetic process
61 establishment of protein localization
62 organonitrogen compound metabolic process
63 localization
64 cellular amide metabolic process
65 peptide metabolic process
66 peptide transport
67 ribonucleoprotein complex
68 nucleobase-containing compound metabolic process
69 heterocycle metabolic process
70 cellular aromatic compound metabolic process
71 translation
72 cellular localization
73 organic cyclic compound metabolic process
74 response to chemical
75 nitrogen compound transport
76 cytoplasmic vesicle lumen
77 cellular catabolic process
78 vesicle lumen
79 organelle part
80 secretory granule lumen
81 amide transport
82 purine nucleotide metabolic process
83 response to organic substance
84 ribonucleotide metabolic process
85 amide biosynthetic process
86 cell activation
87 cytoplasmic vesicle part
88 establishment of protein localization to organelle
89 cell part
90 ribosomal subunit
91 translational initiation
92 mitochondrial membrane
93 nucleotide metabolic process
94 nucleoside phosphate metabolic process
95 cellular_component
96 organic substance catabolic process
97 cellular component disassembly
98 structural constituent of ribosome
99 membrane-bounded organelle
100 nucleobase-containing small molecule metabolic process
101 purine ribonucleotide metabolic process
102 biological_process
103 binding
104 nucleoside monophosphate biosynthetic process
105 cadherin binding
106 nucleoside monophosphate metabolic process
107 structural molecule activity
108 catabolic process
109 posttranscriptional regulation of gene expression
110 nucleotide biosynthetic process
111 nucleoside phosphate biosynthetic process
112 protein-containing complex subunit organization
113 molecular_function
114 intracellular organelle
115 mitochondrion
116 purine-containing compound metabolic process
117 ribose phosphate biosynthetic process
118 Golgi vesicle transport
119 cellular macromolecule biosynthetic process
120 protein-containing complex disassembly
121 cellular component organization or biogenesis
122 organic substance transport
123 small molecule metabolic process
124 cytoplasm
125 mitochondrial protein complex
126 ribonucleoside monophosphate biosynthetic process
127 nucleoside triphosphate metabolic process
128 organophosphate metabolic process
129 ribonucleotide biosynthetic process
130 mitochondrial inner membrane
131 ribonucleoside monophosphate metabolic process
132 purine-containing compound biosynthetic process
133 protein localization
134 macromolecule biosynthetic process
135 purine nucleotide biosynthetic process
136 ribosome
137 large ribosomal subunit
138 cellular response to chemical stimulus
139 vesicle organization
140 immune response-activating signal transduction
141 response to nitrogen compound
142 macromolecule localization
143 heterocyclic compound binding
144 response to stimulus
145 mitochondrial matrix
146 organic cyclic compound binding
147 macromolecule catabolic process
148 organelle membrane
149 phosphorus metabolic process
150 translation initiation factor activity
151 immune response-activating cell surface receptor signaling pathway
152 ribonucleoside triphosphate metabolic process
153 cellular component organization
154 purine nucleoside monophosphate metabolic process
155 regulation of mRNA stability
156 protein localization to organelle
157 membrane organization
158 regulation of RNA stability
159 purine nucleoside monophosphate biosynthetic process
160 organelle inner membrane
161 cell junction
162 cellular response to organic substance
163 activation of immune response
164 response to organonitrogen compound
165 cell adhesion molecule binding
166 mitochondrial translational elongation
167 phosphate-containing compound metabolic process
168 generation of precursor metabolites and energy
169 mRNA metabolic process
170 purine ribonucleotide biosynthetic process
171 purine nucleoside triphosphate metabolic process
172 purine ribonucleoside triphosphate metabolic process
173 translational elongation
174 organophosphate biosynthetic process
175 macromolecule metabolic process
176 oxidation-reduction process
177 regulation of mRNA catabolic process
178 cellular protein complex disassembly
179 purine ribonucleoside monophosphate metabolic process
180 cellular macromolecule catabolic process
181 regulation of mRNA metabolic process
182 catalytic activity
183 cellular response to stimulus
184 enzyme binding
185 secretory granule membrane
186 membrane protein complex
187 purine ribonucleoside monophosphate biosynthetic process
188 nucleoside diphosphate metabolic process
189 nucleoside triphosphate biosynthetic process
190 mRNA binding
191 ribonucleoside diphosphate metabolic process
192 guanosine-containing compound metabolic process
193 regulation of cellular catabolic process
194 immune response-regulating signaling pathway
195 small molecule binding
196 response to endogenous stimulus
197 aromatic compound catabolic process
198 cellular amino acid metabolic process
199 ligase activity
200 regulation of catabolic process
201 nucleobase-containing compound biosynthetic process
202 response to drug
203 translation factor activity, RNA binding
204 cellular response to nitrogen compound
205 ribonucleoside metabolic process
206 translational termination
207 organic cyclic compound catabolic process
208 nucleoside metabolic process
209 nucleobase-containing small molecule biosynthetic process
210 tethering complex
211 organelle organization
212 endopeptidase complex
213 peptidase complex
214 proteasome complex
215 cytoplasmic vesicle
216 organic cyclic compound biosynthetic process
217 nucleobase-containing compound catabolic process
218 purine nucleoside diphosphate metabolic process
219 double-stranded RNA binding
220 purine ribonucleoside diphosphate metabolic process
221 Wnt signaling pathway, planar cell polarity pathway
222 protein targeting
223 nucleotide binding
224 carbohydrate derivative metabolic process
225 heterocycle catabolic process
226 cellular nitrogen compound catabolic process
227 mitochondrial membrane organization
228 RNA processing
229 immune response-regulating cell surface receptor signaling pathway
230 purine ribonucleotide binding
231 aromatic compound biosynthetic process
232 heterocycle biosynthetic process
233 nucleoside phosphate binding
234 carbohydrate derivative biosynthetic process
235 identical protein binding
236 anaphase-promoting complex-dependent catabolic process
237 cellular response to cytokine stimulus
238 vesicle fusion
239 SRP-dependent cotranslational protein targeting to membrane
240 inner mitochondrial membrane organization
241 GTP metabolic process
242 regulation of cellular amide metabolic process
243 ADP metabolic process
244 regulation of DNA-templated transcription, elongation
245 carbohydrate catabolic process
246 positive regulation of interferon-alpha production
247 positive regulation of type I interferon production
248 organelle fusion
249 tRNA aminoacylation
250 membrane fusion
251 organelle membrane fusion
252 ribonucleoprotein complex subunit organization
253 amino acid activation
254 ER to Golgi vesicle-mediated transport
255 mitochondrial translational termination
256 regulation of interferon-alpha production
257 cellular protein localization
258 nuclear transport
259 mitochondrion organization
260 nucleocytoplasmic transport
261 purine nucleotide binding
262 purine ribonucleoside triphosphate binding
263 protein-containing complex binding
264 ribonucleotide binding
P.value FDR.q.value Enrichment N B n b
1 3.69e-22 3.95e-19 1.27 1474 959 455 375
2 1.86e-21 3.22e-18 1.92 1474 219 455 130
3 6.45e-21 3.46e-18 1.46 1474 578 455 260
4 4.35e-18 9.32e-16 1.99 1474 166 455 102
5 4.35e-18 1.16e-15 1.99 1474 166 455 102
6 4.35e-18 1.55e-15 1.99 1474 166 455 102
7 7.07e-19 5.23e-15 1.16 1474 1181 455 423
8 5.58e-16 9.97e-14 1.69 1474 266 455 139
9 2.78e-13 4.26e-11 1.72 1474 211 455 112
10 1.12e-12 1.50e-10 1.37 1474 541 455 228
11 1.65e-13 6.11e-10 1.28 1474 756 455 298
12 1.11e-11 1.08e-09 1.93 1474 119 455 71
13 1.11e-11 1.19e-09 1.93 1474 119 455 71
14 5.39e-13 1.33e-09 2.16 1474 90 455 60
15 1.11e-11 1.33e-09 1.93 1474 119 455 71
16 2.19e-12 4.04e-09 1.25 1474 793 455 306
17 5.72e-11 5.11e-09 1.08 1474 1325 455 441
18 5.13e-12 7.59e-09 1.77 1474 170 455 93
19 8.35e-12 1.03e-08 2.55 1474 47 455 37
20 9.82e-12 1.04e-08 2.51 1474 49 455 38
21 2.25e-11 1.51e-08 2.54 1474 46 455 36
22 2.53e-11 1.56e-08 1.74 1474 171 455 92
23 2.25e-11 1.66e-08 2.54 1474 46 455 36
24 2.25e-11 1.85e-08 2.54 1474 46 455 36
25 3.56e-11 2.03e-08 2.37 1474 56 455 41
26 2.25e-11 2.08e-08 2.54 1474 46 455 36
27 4.93e-11 2.60e-08 1.88 1474 124 455 72
28 7.89e-11 3.89e-08 1.44 1474 379 455 168
29 8.68e-11 4.01e-08 2.26 1474 63 455 44
30 9.95e-11 4.09e-08 1.93 1474 109 455 65
31 9.95e-11 4.33e-08 1.93 1474 109 455 65
32 2.24e-10 7.91e-08 2.31 1474 56 455 40
33 2.24e-10 8.30e-08 2.31 1474 56 455 40
34 2.15e-10 8.35e-08 1.41 1474 397 455 173
35 3.53e-10 1.19e-07 1.24 1474 742 455 284
36 3.81e-10 1.22e-07 1.42 1474 376 455 165
37 4.96e-10 1.53e-07 1.67 1474 178 455 92
38 5.50e-10 1.63e-07 2.30 1474 55 455 39
39 2.19e-09 1.81e-07 3.08 1474 20 455 19
40 6.67e-10 1.90e-07 1.54 1474 255 455 121
41 7.50e-10 2.05e-07 1.89 1474 108 455 63
42 3.15e-09 2.41e-07 1.27 1474 614 455 241
43 1.23e-09 3.25e-07 1.70 1474 160 455 84
44 4.60e-10 3.99e-07 1.11 1474 1199 455 411
45 6.52e-09 4.66e-07 1.72 1474 139 455 74
46 1.94e-09 4.94e-07 1.24 1474 714 455 273
47 2.25e-09 5.55e-07 1.81 1474 122 455 68
48 3.55e-09 8.47e-07 1.50 1474 263 455 122
49 4.26e-09 9.86e-07 1.25 1474 679 455 261
50 4.53e-09 1.02e-06 1.59 1474 202 455 99
51 4.82e-09 1.05e-06 1.50 1474 264 455 122
52 6.53e-09 1.38e-06 1.98 1474 82 455 50
53 8.66e-09 1.78e-06 1.64 1474 170 455 86
54 1.02e-08 2.03e-06 2.00 1474 76 455 47
55 5.53e-08 3.49e-06 1.15 1474 981 455 347
56 5.45e-08 3.65e-06 1.14 1474 1020 455 358
57 2.12e-08 4.12e-06 1.65 1474 157 455 80
58 2.45e-08 4.65e-06 1.94 1474 82 455 49
59 4.63e-08 8.57e-06 2.39 1474 38 455 28
60 4.79e-08 8.64e-06 2.35 1474 40 455 29
61 4.93e-08 8.68e-06 1.61 1474 167 455 83
62 7.76e-08 1.34e-05 1.29 1474 493 455 197
63 8.27e-08 1.39e-05 1.32 1474 445 455 181
64 8.73e-08 1.43e-05 1.90 1474 82 455 48
65 9.05e-08 1.46e-05 2.16 1474 51 455 34
66 9.26e-08 1.46e-05 1.61 1474 161 455 80
67 2.59e-07 1.54e-05 1.70 1474 118 455 62
68 1.06e-07 1.57e-05 1.39 1474 323 455 139
69 1.03e-07 1.59e-05 1.38 1474 343 455 146
70 1.06e-07 1.60e-05 1.38 1474 346 455 147
71 1.17e-07 1.70e-05 2.33 1474 39 455 28
72 1.37e-07 1.94e-05 1.46 1474 248 455 112
73 1.47e-07 2.06e-05 1.36 1474 359 455 151
74 1.80e-07 2.47e-05 1.51 1474 208 455 97
75 1.86e-07 2.50e-05 1.54 1474 192 455 91
76 5.44e-07 2.78e-05 2.52 1474 27 455 21
77 2.15e-07 2.84e-05 1.50 1474 214 455 99
78 5.44e-07 2.92e-05 2.52 1474 27 455 21
79 6.22e-07 3.03e-05 1.13 1474 994 455 347
80 5.44e-07 3.07e-05 2.52 1474 27 455 21
81 2.62e-07 3.40e-05 1.58 1474 164 455 80
82 2.85e-07 3.45e-05 2.30 1474 38 455 27
83 2.74e-07 3.49e-05 1.56 1474 172 455 83
84 2.84e-07 3.50e-05 2.34 1474 36 455 26
85 2.94e-07 3.51e-05 2.05 1474 57 455 36
86 2.83e-07 3.55e-05 1.82 1474 89 455 50
87 8.26e-07 3.85e-05 1.65 1474 126 455 64
88 3.63e-07 4.26e-05 1.97 1474 64 455 39
89 1.70e-06 7.29e-05 1.05 1474 1375 455 444
90 1.66e-06 7.43e-05 2.29 1474 34 455 24
91 6.84e-07 7.91e-05 2.36 1474 33 455 24
92 1.93e-06 7.96e-05 1.84 1474 74 455 42
93 7.19e-07 8.05e-05 2.04 1474 54 455 34
94 7.19e-07 8.18e-05 2.04 1474 54 455 34
95 2.12e-06 8.40e-05 1.03 1474 1418 455 452
96 9.16e-07 1.01e-04 1.49 1474 200 455 92
97 1.35e-06 1.47e-04 2.16 1474 42 455 28
98 2.61e-07 1.51e-04 2.43 1474 32 455 24
99 3.98e-06 1.52e-04 1.14 1474 898 455 316
100 1.56e-06 1.67e-04 1.93 1474 62 455 37
101 1.66e-06 1.76e-04 2.29 1474 34 455 24
102 1.70e-06 1.77e-04 1.05 1474 1375 455 444
103 4.54e-07 1.97e-04 1.06 1474 1342 455 438
104 2.06e-06 2.12e-04 3.01 1474 14 455 13
105 6.30e-07 2.19e-04 2.21 1474 41 455 28
106 3.24e-06 3.28e-04 2.54 1474 23 455 18
107 1.17e-06 3.39e-04 1.99 1474 57 455 35
108 3.54e-06 3.54e-04 1.43 1474 227 455 100
109 3.75e-06 3.70e-04 1.82 1474 73 455 41
110 4.04e-06 3.88e-04 2.30 1474 31 455 22
111 4.04e-06 3.93e-04 2.30 1474 31 455 22
112 4.75e-06 4.51e-04 1.49 1474 176 455 81
113 1.83e-06 4.54e-04 1.04 1474 1406 455 450
114 1.51e-05 5.39e-04 1.12 1474 961 455 332
115 1.49e-05 5.51e-04 1.47 1474 172 455 78
116 6.09e-06 5.70e-04 2.08 1474 42 455 27
117 7.23e-06 6.68e-04 2.59 1474 20 455 16
118 7.45e-06 6.80e-04 2.13 1474 38 455 25
119 8.75e-06 7.80e-04 1.58 1474 125 455 61
120 8.73e-06 7.88e-04 2.19 1474 34 455 23
121 9.38e-06 8.26e-04 1.24 1474 506 455 193
122 1.14e-05 9.91e-04 1.42 1474 215 455 94
123 1.38e-05 1.19e-03 1.48 1474 169 455 77
124 3.67e-05 1.27e-03 1.23 1474 465 455 177
125 4.86e-05 1.58e-03 2.16 1474 30 455 20
126 1.93e-05 1.59e-03 2.97 1474 12 455 11
127 1.93e-05 1.60e-03 2.56 1474 19 455 15
128 1.89e-05 1.61e-03 1.64 1474 99 455 50
129 1.93e-05 1.62e-03 2.56 1474 19 455 15
130 4.84e-05 1.62e-03 1.88 1474 50 455 29
131 2.18e-05 1.70e-03 2.47 1474 21 455 16
132 2.18e-05 1.71e-03 2.47 1474 21 455 16
133 2.12e-05 1.72e-03 1.41 1474 212 455 92
134 2.14e-05 1.72e-03 1.52 1474 141 455 66
135 2.18e-05 1.73e-03 2.47 1474 21 455 16
136 5.53e-05 1.74e-03 2.43 1474 20 455 15
137 5.71e-05 1.75e-03 2.29 1474 24 455 17
138 2.35e-05 1.81e-03 1.52 1474 136 455 64
139 2.48e-05 1.87e-03 2.01 1474 42 455 26
140 2.48e-05 1.89e-03 2.01 1474 42 455 26
141 2.56e-05 1.91e-03 1.76 1474 70 455 38
142 2.81e-05 2.08e-03 1.39 1474 216 455 93
143 1.32e-05 2.54e-03 1.20 1474 608 455 225
144 3.56e-05 2.60e-03 1.26 1474 395 455 154
145 9.09e-05 2.71e-03 1.79 1474 56 455 31
146 1.25e-05 2.71e-03 1.20 1474 614 455 227
147 3.81e-05 2.76e-03 1.50 1474 143 455 66
148 1.10e-04 3.18e-03 1.31 1474 274 455 111
149 4.48e-05 3.22e-03 1.40 1474 204 455 88
150 1.93e-05 3.35e-03 2.97 1474 12 455 11
151 4.86e-05 3.46e-03 2.16 1474 30 455 20
152 5.06e-05 3.56e-03 2.52 1474 18 455 14
153 5.24e-05 3.66e-03 1.22 1474 498 455 187
154 5.53e-05 3.83e-03 2.43 1474 20 455 15
155 5.71e-05 3.84e-03 2.29 1474 24 455 17
156 5.63e-05 3.86e-03 1.60 1474 97 455 48
157 5.85e-05 3.86e-03 1.62 1474 92 455 46
158 5.71e-05 3.88e-03 2.29 1474 24 455 17
159 5.84e-05 3.89e-03 2.95 1474 11 455 10
160 1.71e-04 4.81e-03 1.77 1474 55 455 30
161 1.78e-04 4.88e-03 1.60 1474 85 455 42
162 7.89e-05 5.12e-03 1.55 1474 111 455 53
163 7.84e-05 5.13e-03 1.91 1474 44 455 26
164 8.24e-05 5.30e-03 1.75 1474 63 455 34
165 3.47e-05 5.47e-03 1.86 1474 54 455 31
166 9.06e-05 5.78e-03 2.74 1474 13 455 11
167 1.02e-04 6.43e-03 1.38 1474 202 455 86
168 1.09e-04 6.81e-03 2.12 1474 29 455 19
169 1.21e-04 7.54e-03 1.60 1474 89 455 44
170 1.31e-04 7.56e-03 2.48 1474 17 455 13
171 1.31e-04 7.62e-03 2.48 1474 17 455 13
172 1.31e-04 7.68e-03 2.48 1474 17 455 13
173 1.31e-04 7.74e-03 2.48 1474 17 455 13
174 1.29e-04 7.77e-03 1.69 1474 69 455 36
175 1.30e-04 7.78e-03 1.18 1474 598 455 217
176 1.29e-04 7.82e-03 1.64 1474 79 455 40
177 1.28e-04 7.83e-03 2.20 1474 25 455 17
178 1.28e-04 7.90e-03 2.20 1474 25 455 17
179 1.38e-04 7.93e-03 2.39 1474 19 455 14
180 1.44e-04 8.12e-03 1.48 1474 129 455 59
181 1.43e-04 8.13e-03 1.98 1474 36 455 22
182 5.68e-05 8.21e-03 1.21 1474 514 455 192
183 1.52e-04 8.53e-03 1.31 1474 267 455 108
184 6.39e-05 8.53e-03 1.36 1474 231 455 97
185 3.32e-04 8.89e-03 2.27 1474 20 455 14
186 3.61e-04 9.43e-03 1.53 1474 95 455 45
187 1.75e-04 9.44e-03 2.92 1474 10 455 9
188 1.75e-04 9.51e-03 2.92 1474 10 455 9
189 1.75e-04 9.58e-03 2.92 1474 10 455 9
190 7.76e-05 9.61e-03 2.04 1474 35 455 22
191 1.75e-04 9.65e-03 2.92 1474 10 455 9
192 1.75e-04 9.72e-03 2.92 1474 10 455 9
193 2.10e-04 1.13e-02 1.52 1474 109 455 51
194 2.18e-04 1.16e-02 1.83 1474 46 455 26
195 1.00e-04 1.16e-02 1.35 1474 236 455 98
196 2.23e-04 1.18e-02 1.62 1474 78 455 39
197 2.29e-04 1.20e-02 1.67 1474 68 455 35
198 2.39e-04 1.24e-02 2.08 1474 28 455 18
199 1.15e-04 1.25e-02 2.59 1474 15 455 12
200 2.52e-04 1.30e-02 1.47 1474 123 455 56
201 2.54e-04 1.30e-02 1.51 1474 107 455 50
202 2.73e-04 1.39e-02 1.70 1474 61 455 32
203 1.38e-04 1.41e-02 2.39 1474 19 455 14
204 2.94e-04 1.49e-02 1.94 1474 35 455 21
205 3.06e-04 1.54e-02 2.55 1474 14 455 11
206 3.33e-04 1.65e-02 2.43 1474 16 455 12
207 3.33e-04 1.66e-02 1.64 1474 69 455 35
208 3.40e-04 1.66e-02 2.34 1474 18 455 13
209 3.40e-04 1.67e-02 2.34 1474 18 455 13
210 6.99e-04 1.78e-02 2.65 1474 11 455 9
211 3.67e-04 1.79e-02 1.29 1474 266 455 106
212 8.46e-04 2.01e-02 3.24 1474 6 455 6
213 8.46e-04 2.06e-02 3.24 1474 6 455 6
214 8.46e-04 2.11e-02 3.24 1474 6 455 6
215 9.39e-04 2.19e-02 1.41 1474 131 455 57
216 4.56e-04 2.20e-02 1.44 1474 128 455 57
217 4.85e-04 2.33e-02 1.64 1474 65 455 33
218 5.18e-04 2.44e-02 2.88 1474 9 455 8
219 2.54e-04 2.44e-02 2.70 1474 12 455 10
220 5.18e-04 2.46e-02 2.88 1474 9 455 8
221 5.18e-04 2.47e-02 2.88 1474 9 455 8
222 5.45e-04 2.55e-02 1.75 1474 48 455 26
223 3.02e-04 2.62e-02 1.34 1474 210 455 87
224 5.65e-04 2.63e-02 1.54 1474 86 455 41
225 5.74e-04 2.64e-02 1.62 1474 68 455 34
226 5.74e-04 2.65e-02 1.62 1474 68 455 34
227 5.82e-04 2.66e-02 2.07 1474 25 455 16
228 5.90e-04 2.68e-02 1.49 1474 102 455 47
229 5.94e-04 2.68e-02 1.91 1474 34 455 20
230 3.25e-04 2.68e-02 1.37 1474 182 455 77
231 6.11e-04 2.72e-02 1.45 1474 121 455 54
232 6.11e-04 2.74e-02 1.45 1474 121 455 54
233 3.02e-04 2.75e-02 1.34 1474 210 455 87
234 6.54e-04 2.90e-02 1.76 1474 46 455 25
235 3.75e-04 2.96e-02 1.35 1474 194 455 81
236 6.99e-04 3.06e-02 2.65 1474 11 455 9
237 7.05e-04 3.07e-02 1.83 1474 39 455 22
238 6.99e-04 3.08e-02 2.65 1474 11 455 9
239 7.17e-04 3.10e-02 2.16 1474 21 455 14
240 8.46e-04 3.34e-02 3.24 1474 6 455 6
241 8.46e-04 3.36e-02 3.24 1474 6 455 6
242 8.53e-04 3.36e-02 1.65 1474 59 455 30
243 8.46e-04 3.38e-02 3.24 1474 6 455 6
244 8.46e-04 3.40e-02 3.24 1474 6 455 6
245 7.97e-04 3.41e-02 2.49 1474 13 455 10
246 8.46e-04 3.42e-02 3.24 1474 6 455 6
247 7.97e-04 3.43e-02 2.49 1474 13 455 10
248 8.17e-04 3.43e-02 2.29 1474 17 455 12
249 8.46e-04 3.44e-02 3.24 1474 6 455 6
250 8.17e-04 3.45e-02 2.29 1474 17 455 12
251 8.30e-04 3.45e-02 2.38 1474 15 455 11
252 8.37e-04 3.46e-02 1.84 1474 37 455 21
253 8.46e-04 3.46e-02 3.24 1474 6 455 6
254 8.17e-04 3.47e-02 2.29 1474 17 455 12
255 8.30e-04 3.47e-02 2.38 1474 15 455 11
256 8.46e-04 3.47e-02 3.24 1474 6 455 6
257 8.87e-04 3.47e-02 1.42 1474 128 455 56
258 9.42e-04 3.63e-02 1.97 1474 28 455 17
259 9.38e-04 3.65e-02 1.77 1474 42 455 23
260 9.42e-04 3.65e-02 1.97 1474 28 455 17
261 4.94e-04 3.73e-02 1.36 1474 184 455 77
262 5.46e-04 3.79e-02 1.36 1474 176 455 74
263 5.27e-04 3.81e-02 1.46 1474 115 455 52
264 6.06e-04 4.04e-02 1.35 1474 185 455 77
Genes
1 [DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, DST - dystonin, NCBP2 - nuclear cap binding protein subunit 2, 20kda, EXOSC5 - exosome component 5, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, SACM1L - sac1 suppressor of actin mutations 1-like (yeast), SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, NDUFA5 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, GFM2 - g elongation factor, mitochondrial 2, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, TMED2 - transmembrane emp24 domain trafficking protein 2, HOOK3 - hook microtubule-tethering protein 3, DERL1 - derlin 1, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, BTF3 - basic transcription factor 3, DOCK8 - dedicator of cytokinesis 8, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, SPC24 - spc24, ndc80 kinetochore complex component, ACAT1 - acetyl-coa acetyltransferase 1, EIF4B - eukaryotic translation initiation factor 4b, CPOX - coproporphyrinogen oxidase, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), SUGT1 - sgt1, suppressor of g2 allele of skp1 (s. cerevisiae), EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, GOSR2 - golgi snap receptor complex member 2, USP25 - ubiquitin specific peptidase 25, SORD - sorbitol dehydrogenase, OTUB1 - otu domain, ubiquitin aldehyde binding 1, PLCG1 - phospholipase c, gamma 1, LIMS1 - lim and senescent cell antigen-like domains 1, WDR77 - wd repeat domain 77, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, CARHSP1 - calcium regulated heat stable protein 1, 24kda, CREB1 - camp responsive element binding protein 1, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, MYO6 - myosin vi, SNX1 - sorting nexin 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, SGPL1 - sphingosine-1-phosphate lyase 1, EIF2D - eukaryotic translation initiation factor 2d, TPM3 - tropomyosin 3, SNRPE - small nuclear ribonucleoprotein polypeptide e, STK38L - serine/threonine kinase 38 like, ANAPC7 - anaphase promoting complex subunit 7, SEH1L - seh1-like (s. cerevisiae), EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, GLS - glutaminase, IARS - isoleucyl-trna synthetase, GLO1 - glyoxalase i, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), GLG1 - golgi glycoprotein 1, NIP7 - nip7, nucleolar pre-rrna processing protein, GOSR1 - golgi snap receptor complex member 1, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), POLR1C - polymerase (rna) i polypeptide c, 30kda, DCAF7 - ddb1 and cul4 associated factor 7, EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, ADPGK - adp-dependent glucokinase, PPA1 - pyrophosphatase (inorganic) 1, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, EIF2S2 - eukaryotic translation initiation factor 2, subunit 2 beta, 38kda, SNX6 - sorting nexin 6, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, TSFM - ts translation elongation factor, mitochondrial, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), MVD - mevalonate (diphospho) decarboxylase, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, DENND4A - denn/madd domain containing 4a, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, COMT - catechol-o-methyltransferase, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, KYNU - kynureninase, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, STX8 - syntaxin 8, ERP44 - endoplasmic reticulum protein 44, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), NAMPT - nicotinamide phosphoribosyltransferase, HSPA4 - heat shock 70kda protein 4, PARP4 - poly (adp-ribose) polymerase family, member 4, SCPEP1 - serine carboxypeptidase 1, TMEM30A - transmembrane protein 30a, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, UQCRB - ubiquinol-cytochrome c reductase binding protein, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, CAPN2 - calpain 2, (m/ii) large subunit, WNK1 - wnk lysine deficient protein kinase 1, ZWILCH - zwilch kinetochore protein, OSTF1 - osteoclast stimulating factor 1, UBE2Z - ubiquitin-conjugating enzyme e2z, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, HMGB2 - high mobility group box 2, AGK - acylglycerol kinase, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), TBC1D2B - tbc1 domain family, member 2b, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), PTCD3 - pentatricopeptide repeat domain 3, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, HOMER1 - homer homolog 1 (drosophila), EIF4G3 - eukaryotic translation initiation factor 4 gamma, 3, FTH1 - ferritin, heavy polypeptide 1, UCK2 - uridine-cytidine kinase 2, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, UMPS - uridine monophosphate synthetase, USP48 - ubiquitin specific peptidase 48, PPP5C - protein phosphatase 5, catalytic subunit, RPL3 - ribosomal protein l3, CARS - cysteinyl-trna synthetase, SURF4 - surfeit 4, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), IRAK4 - interleukin-1 receptor-associated kinase 4, EIF3F - eukaryotic translation initiation factor 3, subunit f, DNAJC11 - dnaj (hsp40) homolog, subfamily c, member 11, MRPS35 - mitochondrial ribosomal protein s35, MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, RPN1 - ribophorin i, TNPO1 - transportin 1, AK4 - adenylate kinase 4, STX17 - syntaxin 17, HK2 - hexokinase 2, KPNB1 - karyopherin (importin) beta 1, TRIM38 - tripartite motif containing 38, EXOC2 - exocyst complex component 2, CDC23 - cell division cycle 23, EXOC1 - exocyst complex component 1, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, IFIT5 - interferon-induced protein with tetratricopeptide repeats 5, TJP2 - tight junction protein 2, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, VDAC2 - voltage-dependent anion channel 2, MRPL13 - mitochondrial ribosomal protein l13, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, NIPBL - nipped-b homolog (drosophila), SCYL2 - scy1-like 2 (s. cerevisiae), GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, GALNT2 - udp-n-acetyl-alpha-d-galactosamine:polypeptide n-acetylgalactosaminyltransferase 2 (galnac-t2), TMEM70 - transmembrane protein 70, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), MFN1 - mitofusin 1, STAT6 - signal transducer and activator of transcription 6, interleukin-4 induced, WDR1 - wd repeat domain 1, LARP4B - la ribonucleoprotein domain family, member 4b, RPS8 - ribosomal protein s8, ESD - esterase d, MRPL42 - mitochondrial ribosomal protein l42, RPS13 - ribosomal protein s13, RINT1 - rad50 interactor 1, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, STOM - stomatin, UCHL3 - ubiquitin carboxyl-terminal esterase l3 (ubiquitin thiolesterase), SUMO1 - small ubiquitin-like modifier 1, SRP72 - signal recognition particle 72kda, GCDH - glutaryl-coa dehydrogenase, CISD2 - cdgsh iron sulfur domain 2, CAT - catalase, USP15 - ubiquitin specific peptidase 15, UGDH - udp-glucose 6-dehydrogenase, XPO6 - exportin 6, NDUFAF4 - nadh dehydrogenase (ubiquinone) complex i, assembly factor 4, SSBP1 - single-stranded dna binding protein 1, mitochondrial, SAMM50 - samm50 sorting and assembly machinery component, MRPL47 - mitochondrial ribosomal protein l47, ANXA4 - annexin a4, TUBGCP3 - tubulin, gamma complex associated protein 3, UBE2I - ubiquitin-conjugating enzyme e2i, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, PRKCD - protein kinase c, delta, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, CCDC6 - coiled-coil domain containing 6, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, RRBP1 - ribosome binding protein 1, EPS15 - epidermal growth factor receptor pathway substrate 15, RRM1 - ribonucleotide reductase m1, DDX31 - dead (asp-glu-ala-asp) box polypeptide 31, ERAP1 - endoplasmic reticulum aminopeptidase 1, DMXL1 - dmx-like 1, DDX6 - dead (asp-glu-ala-asp) box helicase 6, TCOF1 - treacher collins-franceschetti syndrome 1, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, ZMYM2 - zinc finger, mym-type 2, F11R - f11 receptor, GSPT1 - g1 to s phase transition 1, STAMBP - stam binding protein, SCFD1 - sec1 family domain containing 1, GSN - gelsolin, CISD1 - cdgsh iron sulfur domain 1, RAB30 - rab30, member ras oncogene family, FLNB - filamin b, beta, PPFIA1 - protein tyrosine phosphatase, receptor type, f polypeptide (ptprf), interacting protein (liprin), alpha 1, ECI1 - enoyl-coa delta isomerase 1, DCK - deoxycytidine kinase, PPFIBP1 - ptprf interacting protein, binding protein 1 (liprin beta 1), IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, DBN1 - drebrin 1, TRIM25 - tripartite motif containing 25, ILF3 - interleukin enhancer binding factor 3, 90kda, XPOT - exportin, trna, ANKRD28 - ankyrin repeat domain 28, NUDCD2 - nudc domain containing 2, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, CYCS - cytochrome c, somatic, VTI1A - vesicle transport through interaction with t-snares 1a, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, CCDC51 - coiled-coil domain containing 51, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), MMS22L - mms22-like, dna repair protein, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, EHBP1 - eh domain binding protein 1, ARCN1 - archain 1, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, LRRK1 - leucine-rich repeat kinase 1, PA2G4 - proliferation-associated 2g4, 38kda, AGPS - alkylglycerone phosphate synthase, CAB39 - calcium binding protein 39, TBC1D4 - tbc1 domain family, member 4, TMED5 - transmembrane emp24 protein transport domain containing 5, RAB23 - rab23, member ras oncogene family, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, ANAPC1 - anaphase promoting complex subunit 1, MRPL3 - mitochondrial ribosomal protein l3, EXOSC1 - exosome component 1, TELO2 - telomere maintenance 2, PITRM1 - pitrilysin metallopeptidase 1, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), RNF31 - ring finger protein 31, MCM7 - minichromosome maintenance complex component 7, ATG7 - autophagy related 7, MCU - mitochondrial calcium uniporter, ARL1 - adp-ribosylation factor-like 1, TUBGCP4 - tubulin, gamma complex associated protein 4, PRRC1 - proline-rich coiled-coil 1, KDELC2 - kdel (lys-asp-glu-leu) containing 2, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), SYNE1 - spectrin repeat containing, nuclear envelope 1, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), NUP88 - nucleoporin 88kda, SCO1 - sco1 cytochrome c oxidase assembly protein, NFU1 - nfu1 iron-sulfur cluster scaffold homolog (s. cerevisiae), HADH - hydroxyacyl-coa dehydrogenase, OSBPL3 - oxysterol binding protein-like 3, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), FECH - ferrochelatase, CDK6 - cyclin-dependent kinase 6, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, CDK13 - cyclin-dependent kinase 13, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, CCT4 - chaperonin containing tcp1, subunit 4 (delta), GNPDA2 - glucosamine-6-phosphate deaminase 2, CUTC - cutc copper transporter, TBCA - tubulin folding cofactor a, INPP5B - inositol polyphosphate-5-phosphatase, 75kda, RPL7L1 - ribosomal protein l7-like 1, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, IRF3 - interferon regulatory factor 3, ARL6IP5 - adp-ribosylation-like factor 6 interacting protein 5, STX18 - syntaxin 18, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, LARP1 - la ribonucleoprotein domain family, member 1, IRF5 - interferon regulatory factor 5, DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), RPTOR - regulatory associated protein of mtor, complex 1, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, EIF3E - eukaryotic translation initiation factor 3, subunit e, QSOX2 - quiescin q6 sulfhydryl oxidase 2, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, LYRM7 - lyr motif containing 7, VTA1 - vesicle (multivesicular body) trafficking 1, SERBP1 - serpine1 mrna binding protein 1, PURA - purine-rich element binding protein a, GAPVD1 - gtpase activating protein and vps9 domains 1, DENND3 - denn/madd domain containing 3, SPG11 - spastic paraplegia 11 (autosomal recessive), ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, TDRD7 - tudor domain containing 7, PGM1 - phosphoglucomutase 1, NEK6 - nima-related kinase 6, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, NOC3L - nucleolar complex associated 3 homolog (s. cerevisiae), PYGB - phosphorylase, glycogen; brain, PCYT2 - phosphate cytidylyltransferase 2, ethanolamine, MAP4 - microtubule-associated protein 4, RAB14 - rab14, member ras oncogene family, PHKB - phosphorylase kinase, beta, MANBA - mannosidase, beta a, lysosomal, VKORC1L1 - vitamin k epoxide reductase complex, subunit 1-like 1, MAN2A1 - mannosidase, alpha, class 2a, member 1, USP8 - ubiquitin specific peptidase 8, TRAPPC10 - trafficking protein particle complex 10, MBNL1 - muscleblind-like splicing regulator 1, EIF4H - eukaryotic translation initiation factor 4h, METTL15 - methyltransferase like 15, CCDC58 - coiled-coil domain containing 58, DUT - deoxyuridine triphosphatase, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, MAT2A - methionine adenosyltransferase ii, alpha, MCM3 - minichromosome maintenance complex component 3, IDE - insulin-degrading enzyme, CHMP5 - charged multivesicular body protein 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, USP34 - ubiquitin specific peptidase 34, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), IFIH1 - interferon induced with helicase c domain 1, MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, MAD1L1 - mad1 mitotic arrest deficient-like 1 (yeast), CTSC - cathepsin c, WIPF1 - was/wasl interacting protein family, member 1, LSS - lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase), UBA6 - ubiquitin-like modifier activating enzyme 6, RANBP1 - ran binding protein 1, DLG1 - discs, large homolog 1 (drosophila), RAP1A - rap1a, member of ras oncogene family, PUS1 - pseudouridylate synthase 1, RPL14 - ribosomal protein l14, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, NSMAF - neutral sphingomyelinase (n-smase) activation associated factor, GNPAT - glyceronephosphate o-acyltransferase, NUB1 - negative regulator of ubiquitin-like proteins 1, CYFIP2 - cytoplasmic fmr1 interacting protein 2, DGUOK - deoxyguanosine kinase, TRAPPC9 - trafficking protein particle complex 9, CLNS1A - chloride channel, nucleotide-sensitive, 1a, UBA3 - ubiquitin-like modifier activating enzyme 3, RAP1B - rap1b, member of ras oncogene family, GOLPH3 - golgi phosphoprotein 3 (coat-protein), TMX3 - thioredoxin-related transmembrane protein 3, DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, ACOT7 - acyl-coa thioesterase 7, PDAP1 - pdgfa associated protein 1, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, FAM49B - family with sequence similarity 49, member b, ADI1 - acireductone dioxygenase 1, CAPRIN1 - cell cycle associated protein 1, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), MRPS14 - mitochondrial ribosomal protein s14, RHOT1 - ras homolog family member t1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, CUL1 - cullin 1, BCL2L1 - bcl2-like 1, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, PGM2 - phosphoglucomutase 2, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
2 [DDX31 - dead (asp-glu-ala-asp) box polypeptide 31, ZC3H7B - zinc finger ccch-type containing 7b, NCBP2 - nuclear cap binding protein subunit 2, 20kda, EXOSC5 - exosome component 5, DDX6 - dead (asp-glu-ala-asp) box helicase 6, TCOF1 - treacher collins-franceschetti syndrome 1, ZNF638 - zinc finger protein 638, SF3A3 - splicing factor 3a, subunit 3, 60kda, GSPT1 - g1 to s phase transition 1, URB1 - urb1 ribosome biogenesis 1 homolog (s. cerevisiae), FAM98A - family with sequence similarity 98, member a, FLNB - filamin b, beta, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, BTF3 - basic transcription factor 3, ILF3 - interleukin enhancer binding factor 3, 90kda, TRIM25 - tripartite motif containing 25, XPOT - exportin, trna, EIF4B - eukaryotic translation initiation factor 4b, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, RBM19 - rna binding motif protein 19, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), ARCN1 - archain 1, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), PA2G4 - proliferation-associated 2g4, 38kda, EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, ZNF326 - zinc finger protein 326, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, CARHSP1 - calcium regulated heat stable protein 1, 24kda, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, GAR1 - gar1 ribonucleoprotein, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, MRPL3 - mitochondrial ribosomal protein l3, THUMPD1 - thump domain containing 1, EXOSC1 - exosome component 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, EIF2D - eukaryotic translation initiation factor 2d, SNRPE - small nuclear ribonucleoprotein polypeptide e, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, SYNE1 - spectrin repeat containing, nuclear envelope 1, IARS - isoleucyl-trna synthetase, NIP7 - nip7, nucleolar pre-rrna processing protein, NOM1 - nucleolar protein with mif4g domain 1, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, EIF2S2 - eukaryotic translation initiation factor 2, subunit 2 beta, 38kda, THOC5 - tho complex 5, MRPL2 - mitochondrial ribosomal protein l2, TSFM - ts translation elongation factor, mitochondrial, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, CDK13 - cyclin-dependent kinase 13, HSPD1 - heat shock 60kda protein 1 (chaperonin), CCT4 - chaperonin containing tcp1, subunit 4 (delta), FCF1 - fcf1 rrna-processing protein, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, TBCA - tubulin folding cofactor a, RPL7L1 - ribosomal protein l7-like 1, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, LARP1 - la ribonucleoprotein domain family, member 1, NEMF - nuclear export mediator factor, DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), EIF3E - eukaryotic translation initiation factor 3, subunit e, PURA - purine-rich element binding protein a, SERBP1 - serpine1 mrna binding protein 1, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, QKI - qki, kh domain containing, rna binding, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, TDRD7 - tudor domain containing 7, HMGB2 - high mobility group box 2, NOC3L - nucleolar complex associated 3 homolog (s. cerevisiae), HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, PTCD3 - pentatricopeptide repeat domain 3, RPL6 - ribosomal protein l6, MAP4 - microtubule-associated protein 4, RPL7 - ribosomal protein l7, EIF4G3 - eukaryotic translation initiation factor 4 gamma, 3, PPP5C - protein phosphatase 5, catalytic subunit, RPL3 - ribosomal protein l3, CARS - cysteinyl-trna synthetase, EIF3F - eukaryotic translation initiation factor 3, subunit f, EIF4H - eukaryotic translation initiation factor 4h, MBNL1 - muscleblind-like splicing regulator 1, MRPS35 - mitochondrial ribosomal protein s35, DUT - deoxyuridine triphosphatase, MRPL1 - mitochondrial ribosomal protein l1, RPN1 - ribophorin i, MRPL9 - mitochondrial ribosomal protein l9, TNPO1 - transportin 1, KPNB1 - karyopherin (importin) beta 1, IFIH1 - interferon induced with helicase c domain 1, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, RPL31 - ribosomal protein l31, IFIT5 - interferon-induced protein with tetratricopeptide repeats 5, RPL32 - ribosomal protein l32, SART3 - squamous cell carcinoma antigen recognized by t cells 3, SCAF8 - sr-related ctd-associated factor 8, MRPL13 - mitochondrial ribosomal protein l13, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, PUS1 - pseudouridylate synthase 1, RPL14 - ribosomal protein l14, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), AKAP8L - a kinase (prka) anchor protein 8-like, RNMT - rna (guanine-7-) methyltransferase, RBM26 - rna binding motif protein 26, CLNS1A - chloride channel, nucleotide-sensitive, 1a, EIF1AD - eukaryotic translation initiation factor 1a domain containing, LARP4B - la ribonucleoprotein domain family, member 4b, RPS8 - ribosomal protein s8, MRPL42 - mitochondrial ribosomal protein l42, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, NHP2 - nhp2 ribonucleoprotein, RPS7 - ribosomal protein s7, SUMO1 - small ubiquitin-like modifier 1, BZW1 - basic leucine zipper and w2 domains 1, SRP72 - signal recognition particle 72kda, CISD2 - cdgsh iron sulfur domain 2, PDAP1 - pdgfa associated protein 1, CAPRIN1 - cell cycle associated protein 1, SSBP1 - single-stranded dna binding protein 1, mitochondrial, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), MRPS14 - mitochondrial ribosomal protein s14, UBE2I - ubiquitin-conjugating enzyme e2i, RSL1D1 - ribosomal l1 domain containing 1, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, RRBP1 - ribosome binding protein 1]
3 [DST - dystonin, NCBP2 - nuclear cap binding protein subunit 2, 20kda, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, EXOSC5 - exosome component 5, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, HOOK3 - hook microtubule-tethering protein 3, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, BTF3 - basic transcription factor 3, DOCK8 - dedicator of cytokinesis 8, SPC24 - spc24, ndc80 kinetochore complex component, EIF4B - eukaryotic translation initiation factor 4b, CPOX - coproporphyrinogen oxidase, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), SUGT1 - sgt1, suppressor of g2 allele of skp1 (s. cerevisiae), EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, GOSR2 - golgi snap receptor complex member 2, USP25 - ubiquitin specific peptidase 25, SORD - sorbitol dehydrogenase, OTUB1 - otu domain, ubiquitin aldehyde binding 1, PLCG1 - phospholipase c, gamma 1, LIMS1 - lim and senescent cell antigen-like domains 1, WDR77 - wd repeat domain 77, CARHSP1 - calcium regulated heat stable protein 1, 24kda, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, SNX1 - sorting nexin 1, MYO6 - myosin vi, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, EIF2D - eukaryotic translation initiation factor 2d, SNRPE - small nuclear ribonucleoprotein polypeptide e, TPM3 - tropomyosin 3, ANAPC7 - anaphase promoting complex subunit 7, STK38L - serine/threonine kinase 38 like, SEH1L - seh1-like (s. cerevisiae), PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, GLS - glutaminase, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, GLO1 - glyoxalase i, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), GOSR1 - golgi snap receptor complex member 1, NIP7 - nip7, nucleolar pre-rrna processing protein, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), POLR1C - polymerase (rna) i polypeptide c, 30kda, EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, DCAF7 - ddb1 and cul4 associated factor 7, PPA1 - pyrophosphatase (inorganic) 1, ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, EIF2S2 - eukaryotic translation initiation factor 2, subunit 2 beta, 38kda, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, SNX6 - sorting nexin 6, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, HSPD1 - heat shock 60kda protein 1 (chaperonin), MVD - mevalonate (diphospho) decarboxylase, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, DENND4A - denn/madd domain containing 4a, COMT - catechol-o-methyltransferase, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, KYNU - kynureninase, STX8 - syntaxin 8, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), NAMPT - nicotinamide phosphoribosyltransferase, HSPA4 - heat shock 70kda protein 4, PARP4 - poly (adp-ribose) polymerase family, member 4, SCPEP1 - serine carboxypeptidase 1, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, CAPN2 - calpain 2, (m/ii) large subunit, WNK1 - wnk lysine deficient protein kinase 1, ZWILCH - zwilch kinetochore protein, UBE2Z - ubiquitin-conjugating enzyme e2z, AGK - acylglycerol kinase, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PTCD3 - pentatricopeptide repeat domain 3, TBC1D2B - tbc1 domain family, member 2b, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, HOMER1 - homer homolog 1 (drosophila), EIF4G3 - eukaryotic translation initiation factor 4 gamma, 3, FTH1 - ferritin, heavy polypeptide 1, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, UCK2 - uridine-cytidine kinase 2, USP48 - ubiquitin specific peptidase 48, PPP5C - protein phosphatase 5, catalytic subunit, RPL3 - ribosomal protein l3, UMPS - uridine monophosphate synthetase, CARS - cysteinyl-trna synthetase, SURF4 - surfeit 4, IRAK4 - interleukin-1 receptor-associated kinase 4, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), EIF3F - eukaryotic translation initiation factor 3, subunit f, RPN1 - ribophorin i, TNPO1 - transportin 1, STX17 - syntaxin 17, HK2 - hexokinase 2, KPNB1 - karyopherin (importin) beta 1, TRIM38 - tripartite motif containing 38, EXOC2 - exocyst complex component 2, CDC23 - cell division cycle 23, EXOC1 - exocyst complex component 1, IFIT5 - interferon-induced protein with tetratricopeptide repeats 5, TJP2 - tight junction protein 2, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, NIPBL - nipped-b homolog (drosophila), STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), STAT6 - signal transducer and activator of transcription 6, interleukin-4 induced, WDR1 - wd repeat domain 1, LARP4B - la ribonucleoprotein domain family, member 4b, RPS8 - ribosomal protein s8, ESD - esterase d, RPS13 - ribosomal protein s13, RINT1 - rad50 interactor 1, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, UCHL3 - ubiquitin carboxyl-terminal esterase l3 (ubiquitin thiolesterase), SUMO1 - small ubiquitin-like modifier 1, SRP72 - signal recognition particle 72kda, CAT - catalase, USP15 - ubiquitin specific peptidase 15, UGDH - udp-glucose 6-dehydrogenase, XPO6 - exportin 6, TUBGCP3 - tubulin, gamma complex associated protein 3, UBE2I - ubiquitin-conjugating enzyme e2i, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, PRKCD - protein kinase c, delta, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, CCDC6 - coiled-coil domain containing 6, EPS15 - epidermal growth factor receptor pathway substrate 15, RRM1 - ribonucleotide reductase m1, ERAP1 - endoplasmic reticulum aminopeptidase 1, DDX6 - dead (asp-glu-ala-asp) box helicase 6, TCOF1 - treacher collins-franceschetti syndrome 1, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, ZMYM2 - zinc finger, mym-type 2, STAMBP - stam binding protein, GSPT1 - g1 to s phase transition 1, SCFD1 - sec1 family domain containing 1, GSN - gelsolin, FLNB - filamin b, beta, PPFIA1 - protein tyrosine phosphatase, receptor type, f polypeptide (ptprf), interacting protein (liprin), alpha 1, DCK - deoxycytidine kinase, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, PPFIBP1 - ptprf interacting protein, binding protein 1 (liprin beta 1), DBN1 - drebrin 1, TRIM25 - tripartite motif containing 25, XPOT - exportin, trna, ANKRD28 - ankyrin repeat domain 28, NUDCD2 - nudc domain containing 2, CYCS - cytochrome c, somatic, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, VTI1A - vesicle transport through interaction with t-snares 1a, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, MMS22L - mms22-like, dna repair protein, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, EHBP1 - eh domain binding protein 1, ARCN1 - archain 1, LRRK1 - leucine-rich repeat kinase 1, AGPS - alkylglycerone phosphate synthase, CAB39 - calcium binding protein 39, TBC1D4 - tbc1 domain family, member 4, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, RAB23 - rab23, member ras oncogene family, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, ANAPC1 - anaphase promoting complex subunit 1, EXOSC1 - exosome component 1, TELO2 - telomere maintenance 2, RNF31 - ring finger protein 31, ATG7 - autophagy related 7, MCM7 - minichromosome maintenance complex component 7, TUBGCP4 - tubulin, gamma complex associated protein 4, ARL1 - adp-ribosylation factor-like 1, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), NUP88 - nucleoporin 88kda, NFU1 - nfu1 iron-sulfur cluster scaffold homolog (s. cerevisiae), OSBPL3 - oxysterol binding protein-like 3, CDK6 - cyclin-dependent kinase 6, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, CDK13 - cyclin-dependent kinase 13, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, GNPDA2 - glucosamine-6-phosphate deaminase 2, CCT4 - chaperonin containing tcp1, subunit 4 (delta), NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, CUTC - cutc copper transporter, TBCA - tubulin folding cofactor a, INPP5B - inositol polyphosphate-5-phosphatase, 75kda, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, IRF3 - interferon regulatory factor 3, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, IRF5 - interferon regulatory factor 5, DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), EIF3E - eukaryotic translation initiation factor 3, subunit e, RPTOR - regulatory associated protein of mtor, complex 1, VTA1 - vesicle (multivesicular body) trafficking 1, SERBP1 - serpine1 mrna binding protein 1, DENND3 - denn/madd domain containing 3, GAPVD1 - gtpase activating protein and vps9 domains 1, SPG11 - spastic paraplegia 11 (autosomal recessive), NEK6 - nima-related kinase 6, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), MAP4 - microtubule-associated protein 4, RAB14 - rab14, member ras oncogene family, PHKB - phosphorylase kinase, beta, USP8 - ubiquitin specific peptidase 8, TRAPPC10 - trafficking protein particle complex 10, MBNL1 - muscleblind-like splicing regulator 1, EIF4H - eukaryotic translation initiation factor 4h, MAT2A - methionine adenosyltransferase ii, alpha, IDE - insulin-degrading enzyme, CHMP5 - charged multivesicular body protein 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, IFIH1 - interferon induced with helicase c domain 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), USP34 - ubiquitin specific peptidase 34, MAD1L1 - mad1 mitotic arrest deficient-like 1 (yeast), WIPF1 - was/wasl interacting protein family, member 1, UBA6 - ubiquitin-like modifier activating enzyme 6, RANBP1 - ran binding protein 1, DLG1 - discs, large homolog 1 (drosophila), RAP1A - rap1a, member of ras oncogene family, NSMAF - neutral sphingomyelinase (n-smase) activation associated factor, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, RPL14 - ribosomal protein l14, GNPAT - glyceronephosphate o-acyltransferase, CYFIP2 - cytoplasmic fmr1 interacting protein 2, NUB1 - negative regulator of ubiquitin-like proteins 1, CLNS1A - chloride channel, nucleotide-sensitive, 1a, TRAPPC9 - trafficking protein particle complex 9, DGUOK - deoxyguanosine kinase, UBA3 - ubiquitin-like modifier activating enzyme 3, RAP1B - rap1b, member of ras oncogene family, GOLPH3 - golgi phosphoprotein 3 (coat-protein), DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, ACOT7 - acyl-coa thioesterase 7, PDAP1 - pdgfa associated protein 1, ADI1 - acireductone dioxygenase 1, CAPRIN1 - cell cycle associated protein 1, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), RHOT1 - ras homolog family member t1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, CUL1 - cullin 1, BCL2L1 - bcl2-like 1, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, PGM2 - phosphoglucomutase 2]
4 [ERAP1 - endoplasmic reticulum aminopeptidase 1, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, F11R - f11 receptor, STAMBP - stam binding protein, GSN - gelsolin, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, FLNB - filamin b, beta, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, ACAT1 - acetyl-coa acetyltransferase 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), CD48 - cd48 molecule, PA2G4 - proliferation-associated 2g4, 38kda, CAB39 - calcium binding protein 39, OTUB1 - otu domain, ubiquitin aldehyde binding 1, SORD - sorbitol dehydrogenase, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, MYO6 - myosin vi, LRRC57 - leucine rich repeat containing 57, TPM3 - tropomyosin 3, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, GLO1 - glyoxalase i, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), GLG1 - golgi glycoprotein 1, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), MBLAC2 - metallo-beta-lactamase domain containing 2, PPA1 - pyrophosphatase (inorganic) 1, ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, HSPD1 - heat shock 60kda protein 1 (chaperonin), CCT4 - chaperonin containing tcp1, subunit 4 (delta), NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, COMT - catechol-o-methyltransferase, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, ERP44 - endoplasmic reticulum protein 44, NAMPT - nicotinamide phosphoribosyltransferase, EIF3E - eukaryotic translation initiation factor 3, subunit e, HSPA4 - heat shock 70kda protein 4, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, PARP4 - poly (adp-ribose) polymerase family, member 4, SCPEP1 - serine carboxypeptidase 1, VTA1 - vesicle (multivesicular body) trafficking 1, SERBP1 - serpine1 mrna binding protein 1, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, CAPN2 - calpain 2, (m/ii) large subunit, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), FABP5 - fatty acid binding protein 5 (psoriasis-associated), MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, PYGB - phosphorylase, glycogen; brain, RAB14 - rab14, member ras oncogene family, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, FTH1 - ferritin, heavy polypeptide 1, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, RPL3 - ribosomal protein l3, MAN2A1 - mannosidase, alpha, class 2a, member 1, SLAMF1 - signaling lymphocytic activation molecule family member 1, DUT - deoxyuridine triphosphatase, TNPO1 - transportin 1, KPNB1 - karyopherin (importin) beta 1, IDE - insulin-degrading enzyme, CHMP5 - charged multivesicular body protein 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), RPL31 - ribosomal protein l31, CTSC - cathepsin c, NIPBL - nipped-b homolog (drosophila), DLG1 - discs, large homolog 1 (drosophila), RAP1A - rap1a, member of ras oncogene family, RPL14 - ribosomal protein l14, CYFIP2 - cytoplasmic fmr1 interacting protein 2, WDR1 - wd repeat domain 1, RPS8 - ribosomal protein s8, ESD - esterase d, RPS13 - ribosomal protein s13, RAP1B - rap1b, member of ras oncogene family, DSTN - destrin (actin depolymerizing factor), STOM - stomatin, ACOT7 - acyl-coa thioesterase 7, CAT - catalase, UGDH - udp-glucose 6-dehydrogenase, FAM49B - family with sequence similarity 49, member b, SSBP1 - single-stranded dna binding protein 1, mitochondrial, SAMM50 - samm50 sorting and assembly machinery component, ANXA4 - annexin a4, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, PRKCD - protein kinase c, delta, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, PGM2 - phosphoglucomutase 2]
5 [ERAP1 - endoplasmic reticulum aminopeptidase 1, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, F11R - f11 receptor, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, STAMBP - stam binding protein, GSN - gelsolin, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, FLNB - filamin b, beta, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, ACAT1 - acetyl-coa acetyltransferase 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), CD48 - cd48 molecule, PA2G4 - proliferation-associated 2g4, 38kda, CAB39 - calcium binding protein 39, SORD - sorbitol dehydrogenase, OTUB1 - otu domain, ubiquitin aldehyde binding 1, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, MYO6 - myosin vi, LRRC57 - leucine rich repeat containing 57, TPM3 - tropomyosin 3, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, GLO1 - glyoxalase i, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), GLG1 - golgi glycoprotein 1, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), MBLAC2 - metallo-beta-lactamase domain containing 2, PPA1 - pyrophosphatase (inorganic) 1, ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, HSPD1 - heat shock 60kda protein 1 (chaperonin), CCT4 - chaperonin containing tcp1, subunit 4 (delta), NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, COMT - catechol-o-methyltransferase, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, ERP44 - endoplasmic reticulum protein 44, NAMPT - nicotinamide phosphoribosyltransferase, EIF3E - eukaryotic translation initiation factor 3, subunit e, HSPA4 - heat shock 70kda protein 4, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, PARP4 - poly (adp-ribose) polymerase family, member 4, SCPEP1 - serine carboxypeptidase 1, VTA1 - vesicle (multivesicular body) trafficking 1, SERBP1 - serpine1 mrna binding protein 1, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, CAPN2 - calpain 2, (m/ii) large subunit, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), FABP5 - fatty acid binding protein 5 (psoriasis-associated), MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, PYGB - phosphorylase, glycogen; brain, RAB14 - rab14, member ras oncogene family, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, FTH1 - ferritin, heavy polypeptide 1, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, RPL3 - ribosomal protein l3, MAN2A1 - mannosidase, alpha, class 2a, member 1, SLAMF1 - signaling lymphocytic activation molecule family member 1, DUT - deoxyuridine triphosphatase, TNPO1 - transportin 1, KPNB1 - karyopherin (importin) beta 1, IDE - insulin-degrading enzyme, CHMP5 - charged multivesicular body protein 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), RPL31 - ribosomal protein l31, CTSC - cathepsin c, DLG1 - discs, large homolog 1 (drosophila), NIPBL - nipped-b homolog (drosophila), RAP1A - rap1a, member of ras oncogene family, RPL14 - ribosomal protein l14, CYFIP2 - cytoplasmic fmr1 interacting protein 2, WDR1 - wd repeat domain 1, RPS8 - ribosomal protein s8, ESD - esterase d, RPS13 - ribosomal protein s13, RAP1B - rap1b, member of ras oncogene family, DSTN - destrin (actin depolymerizing factor), STOM - stomatin, ACOT7 - acyl-coa thioesterase 7, CAT - catalase, UGDH - udp-glucose 6-dehydrogenase, FAM49B - family with sequence similarity 49, member b, SSBP1 - single-stranded dna binding protein 1, mitochondrial, SAMM50 - samm50 sorting and assembly machinery component, ANXA4 - annexin a4, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, PRKCD - protein kinase c, delta, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, PGM2 - phosphoglucomutase 2]
6 [ERAP1 - endoplasmic reticulum aminopeptidase 1, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, F11R - f11 receptor, STAMBP - stam binding protein, GSN - gelsolin, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, FLNB - filamin b, beta, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, ACAT1 - acetyl-coa acetyltransferase 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), CD48 - cd48 molecule, PA2G4 - proliferation-associated 2g4, 38kda, CAB39 - calcium binding protein 39, OTUB1 - otu domain, ubiquitin aldehyde binding 1, SORD - sorbitol dehydrogenase, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, MYO6 - myosin vi, LRRC57 - leucine rich repeat containing 57, TPM3 - tropomyosin 3, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, GLO1 - glyoxalase i, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), GLG1 - golgi glycoprotein 1, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), MBLAC2 - metallo-beta-lactamase domain containing 2, PPA1 - pyrophosphatase (inorganic) 1, ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, HSPD1 - heat shock 60kda protein 1 (chaperonin), CCT4 - chaperonin containing tcp1, subunit 4 (delta), NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, COMT - catechol-o-methyltransferase, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, ERP44 - endoplasmic reticulum protein 44, NAMPT - nicotinamide phosphoribosyltransferase, EIF3E - eukaryotic translation initiation factor 3, subunit e, HSPA4 - heat shock 70kda protein 4, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, PARP4 - poly (adp-ribose) polymerase family, member 4, SCPEP1 - serine carboxypeptidase 1, VTA1 - vesicle (multivesicular body) trafficking 1, SERBP1 - serpine1 mrna binding protein 1, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, CAPN2 - calpain 2, (m/ii) large subunit, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), FABP5 - fatty acid binding protein 5 (psoriasis-associated), MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, PYGB - phosphorylase, glycogen; brain, RAB14 - rab14, member ras oncogene family, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, FTH1 - ferritin, heavy polypeptide 1, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, RPL3 - ribosomal protein l3, MAN2A1 - mannosidase, alpha, class 2a, member 1, SLAMF1 - signaling lymphocytic activation molecule family member 1, DUT - deoxyuridine triphosphatase, TNPO1 - transportin 1, KPNB1 - karyopherin (importin) beta 1, IDE - insulin-degrading enzyme, CHMP5 - charged multivesicular body protein 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), RPL31 - ribosomal protein l31, CTSC - cathepsin c, NIPBL - nipped-b homolog (drosophila), DLG1 - discs, large homolog 1 (drosophila), RAP1A - rap1a, member of ras oncogene family, RPL14 - ribosomal protein l14, CYFIP2 - cytoplasmic fmr1 interacting protein 2, WDR1 - wd repeat domain 1, RPS8 - ribosomal protein s8, ESD - esterase d, RPS13 - ribosomal protein s13, RAP1B - rap1b, member of ras oncogene family, DSTN - destrin (actin depolymerizing factor), STOM - stomatin, ACOT7 - acyl-coa thioesterase 7, CAT - catalase, UGDH - udp-glucose 6-dehydrogenase, FAM49B - family with sequence similarity 49, member b, SSBP1 - single-stranded dna binding protein 1, mitochondrial, SAMM50 - samm50 sorting and assembly machinery component, ANXA4 - annexin a4, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, PRKCD - protein kinase c, delta, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, PGM2 - phosphoglucomutase 2]
7 [GFPT1 - glutamine--fructose-6-phosphate transaminase 1, SF3A3 - splicing factor 3a, subunit 3, 60kda, GFM2 - g elongation factor, mitochondrial 2, FKBP11 - fk506 binding protein 11, 19 kda, TMED2 - transmembrane emp24 domain trafficking protein 2, HOOK3 - hook microtubule-tethering protein 3, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, ACAT1 - acetyl-coa acetyltransferase 1, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, CPOX - coproporphyrinogen oxidase, LRRC40 - leucine rich repeat containing 40, SUGT1 - sgt1, suppressor of g2 allele of skp1 (s. cerevisiae), GOSR2 - golgi snap receptor complex member 2, OTUB1 - otu domain, ubiquitin aldehyde binding 1, SORD - sorbitol dehydrogenase, PLCG1 - phospholipase c, gamma 1, LIMS1 - lim and senescent cell antigen-like domains 1, MORF4L1 - mortality factor 4 like 1, CARHSP1 - calcium regulated heat stable protein 1, 24kda, CREB1 - camp responsive element binding protein 1, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, SNX1 - sorting nexin 1, THUMPD1 - thump domain containing 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, SNRPE - small nuclear ribonucleoprotein polypeptide e, ANAPC7 - anaphase promoting complex subunit 7, PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, GLS - glutaminase, GLO1 - glyoxalase i, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), GLG1 - golgi glycoprotein 1, NIP7 - nip7, nucleolar pre-rrna processing protein, GOSR1 - golgi snap receptor complex member 1, POLR1C - polymerase (rna) i polypeptide c, 30kda, PPA1 - pyrophosphatase (inorganic) 1, SUMO3 - small ubiquitin-like modifier 3, SNX6 - sorting nexin 6, SMARCD1 - swi/snf related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, COMT - catechol-o-methyltransferase, STX8 - syntaxin 8, TAOK3 - tao kinase 3, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), PARP4 - poly (adp-ribose) polymerase family, member 4, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, TMEM30A - transmembrane protein 30a, SCPEP1 - serine carboxypeptidase 1, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, QKI - qki, kh domain containing, rna binding, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, AGK - acylglycerol kinase, HOMER1 - homer homolog 1 (drosophila), FTH1 - ferritin, heavy polypeptide 1, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, PPP5C - protein phosphatase 5, catalytic subunit, USP48 - ubiquitin specific peptidase 48, SURF4 - surfeit 4, IRAK4 - interleukin-1 receptor-associated kinase 4, DNAJC11 - dnaj (hsp40) homolog, subfamily c, member 11, MRPS35 - mitochondrial ribosomal protein s35, AK4 - adenylate kinase 4, TNPO1 - transportin 1, KPNB1 - karyopherin (importin) beta 1, EXOC2 - exocyst complex component 2, EXOC1 - exocyst complex component 1, TJP2 - tight junction protein 2, DCAKD - dephospho-coa kinase domain containing, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, SCYL2 - scy1-like 2 (s. cerevisiae), NIPBL - nipped-b homolog (drosophila), GALNT2 - udp-n-acetyl-alpha-d-galactosamine:polypeptide n-acetylgalactosaminyltransferase 2 (galnac-t2), STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), SNRNP27 - small nuclear ribonucleoprotein 27kda (u4/u6.u5), MFN1 - mitofusin 1, STAT6 - signal transducer and activator of transcription 6, interleukin-4 induced, FCRLA - fc receptor-like a, EIF1AD - eukaryotic translation initiation factor 1a domain containing, RINT1 - rad50 interactor 1, NHP2 - nhp2 ribonucleoprotein, DSTN - destrin (actin depolymerizing factor), RIC8A - ric8 guanine nucleotide exchange factor a, GCDH - glutaryl-coa dehydrogenase, CISD2 - cdgsh iron sulfur domain 2, SSBP1 - single-stranded dna binding protein 1, mitochondrial, SAMM50 - samm50 sorting and assembly machinery component, ANXA4 - annexin a4, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, TNFAIP8 - tumor necrosis factor, alpha-induced protein 8, PRKCD - protein kinase c, delta, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, CCDC6 - coiled-coil domain containing 6, DMXL1 - dmx-like 1, ERAP1 - endoplasmic reticulum aminopeptidase 1, DDX6 - dead (asp-glu-ala-asp) box helicase 6, SDAD1 - sda1 domain containing 1, GSPT1 - g1 to s phase transition 1, GSN - gelsolin, FAM98A - family with sequence similarity 98, member a, ECI1 - enoyl-coa delta isomerase 1, DCK - deoxycytidine kinase, DBN1 - drebrin 1, TRIM25 - tripartite motif containing 25, RIF1 - rap1 interacting factor homolog (yeast), VTI1A - vesicle transport through interaction with t-snares 1a, CCDC51 - coiled-coil domain containing 51, MMS22L - mms22-like, dna repair protein, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, PA2G4 - proliferation-associated 2g4, 38kda, LRRK1 - leucine-rich repeat kinase 1, CAB39 - calcium binding protein 39, GATAD2B - gata zinc finger domain containing 2b, RAB23 - rab23, member ras oncogene family, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, MRPL3 - mitochondrial ribosomal protein l3, RNF31 - ring finger protein 31, MCM7 - minichromosome maintenance complex component 7, MCU - mitochondrial calcium uniporter, ARL1 - adp-ribosylation factor-like 1, KDELC2 - kdel (lys-asp-glu-leu) containing 2, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, PELO - pelota homolog (drosophila), SCO1 - sco1 cytochrome c oxidase assembly protein, WDHD1 - wd repeat and hmg-box dna binding protein 1, HADH - hydroxyacyl-coa dehydrogenase, PPIL3 - peptidylprolyl isomerase (cyclophilin)-like 3, OSBPL3 - oxysterol binding protein-like 3, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, RPL7L1 - ribosomal protein l7-like 1, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, STX18 - syntaxin 18, NEMF - nuclear export mediator factor, RPTOR - regulatory associated protein of mtor, complex 1, QSOX2 - quiescin q6 sulfhydryl oxidase 2, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, LYRM7 - lyr motif containing 7, SERBP1 - serpine1 mrna binding protein 1, VTA1 - vesicle (multivesicular body) trafficking 1, DENND3 - denn/madd domain containing 3, GAPVD1 - gtpase activating protein and vps9 domains 1, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, MAP4 - microtubule-associated protein 4, RAB14 - rab14, member ras oncogene family, PHKB - phosphorylase kinase, beta, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, MANBA - mannosidase, beta a, lysosomal, MAN2A1 - mannosidase, alpha, class 2a, member 1, USP8 - ubiquitin specific peptidase 8, EIF4H - eukaryotic translation initiation factor 4h, MBNL1 - muscleblind-like splicing regulator 1, METTL15 - methyltransferase like 15, SLAMF1 - signaling lymphocytic activation molecule family member 1, DUT - deoxyuridine triphosphatase, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, MAT2A - methionine adenosyltransferase ii, alpha, MCM3 - minichromosome maintenance complex component 3, CHMP5 - charged multivesicular body protein 5, DOCK7 - dedicator of cytokinesis 7, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, WIPF1 - was/wasl interacting protein family, member 1, SCAF8 - sr-related ctd-associated factor 8, UBA6 - ubiquitin-like modifier activating enzyme 6, GRK6 - g protein-coupled receptor kinase 6, LSS - lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase), DLG1 - discs, large homolog 1 (drosophila), PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, RPL14 - ribosomal protein l14, NUB1 - negative regulator of ubiquitin-like proteins 1, TRAPPC9 - trafficking protein particle complex 9, DGUOK - deoxyguanosine kinase, UBA3 - ubiquitin-like modifier activating enzyme 3, DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, ACOT7 - acyl-coa thioesterase 7, PDAP1 - pdgfa associated protein 1, CBX3 - chromobox homolog 3, ADI1 - acireductone dioxygenase 1, FAM49B - family with sequence similarity 49, member b, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), RHOT1 - ras homolog family member t1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, RSL1D1 - ribosomal l1 domain containing 1, BCL2L1 - bcl2-like 1, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, PGM2 - phosphoglucomutase 2, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1, CCNK - cyclin k, DST - dystonin, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, NCBP2 - nuclear cap binding protein subunit 2, 20kda, EXOSC5 - exosome component 5, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, RFC3 - replication factor c (activator 1) 3, 38kda, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, SACM1L - sac1 suppressor of actin mutations 1-like (yeast), NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, NDUFA5 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, RECQL - recq protein-like (dna helicase q1-like), DERL1 - derlin 1, BTF3 - basic transcription factor 3, DOCK8 - dedicator of cytokinesis 8, SPC24 - spc24, ndc80 kinetochore complex component, EIF4B - eukaryotic translation initiation factor 4b, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, USP25 - ubiquitin specific peptidase 25, ZNF326 - zinc finger protein 326, WDR77 - wd repeat domain 77, RCSD1 - rcsd domain containing 1, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, MYO6 - myosin vi, SGPL1 - sphingosine-1-phosphate lyase 1, EIF2D - eukaryotic translation initiation factor 2d, TPM3 - tropomyosin 3, STK38L - serine/threonine kinase 38 like, SEH1L - seh1-like (s. cerevisiae), EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, DCAF7 - ddb1 and cul4 associated factor 7, ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, ADPGK - adp-dependent glucokinase, EIF2S2 - eukaryotic translation initiation factor 2, subunit 2 beta, 38kda, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, TSFM - ts translation elongation factor, mitochondrial, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), MVD - mevalonate (diphospho) decarboxylase, KYNU - kynureninase, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, ERP44 - endoplasmic reticulum protein 44, NAMPT - nicotinamide phosphoribosyltransferase, HSPA4 - heat shock 70kda protein 4, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, UQCRB - ubiquinol-cytochrome c reductase binding protein, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, CAPN2 - calpain 2, (m/ii) large subunit, WNK1 - wnk lysine deficient protein kinase 1, ZWILCH - zwilch kinetochore protein, OSTF1 - osteoclast stimulating factor 1, UBE2Z - ubiquitin-conjugating enzyme e2z, HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PTCD3 - pentatricopeptide repeat domain 3, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, EIF4G3 - eukaryotic translation initiation factor 4 gamma, 3, UCK2 - uridine-cytidine kinase 2, RPL3 - ribosomal protein l3, UMPS - uridine monophosphate synthetase, CARS - cysteinyl-trna synthetase, EIF3F - eukaryotic translation initiation factor 3, subunit f, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, RPN1 - ribophorin i, HK2 - hexokinase 2, STX17 - syntaxin 17, TRIM38 - tripartite motif containing 38, CDC23 - cell division cycle 23, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, VDAC2 - voltage-dependent anion channel 2, TTLL12 - tubulin tyrosine ligase-like family, member 12, MRPL13 - mitochondrial ribosomal protein l13, GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, TMEM70 - transmembrane protein 70, RNMT - rna (guanine-7-) methyltransferase, WDR1 - wd repeat domain 1, RPS8 - ribosomal protein s8, ESD - esterase d, MRPL42 - mitochondrial ribosomal protein l42, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, STOM - stomatin, UCHL3 - ubiquitin carboxyl-terminal esterase l3 (ubiquitin thiolesterase), SUMO1 - small ubiquitin-like modifier 1, CAT - catalase, USP15 - ubiquitin specific peptidase 15, UGDH - udp-glucose 6-dehydrogenase, NDUFAF4 - nadh dehydrogenase (ubiquinone) complex i, assembly factor 4, TUBGCP3 - tubulin, gamma complex associated protein 3, MRPL47 - mitochondrial ribosomal protein l47, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, UBE2I - ubiquitin-conjugating enzyme e2i, RRBP1 - ribosome binding protein 1, EPS15 - epidermal growth factor receptor pathway substrate 15, RRM1 - ribonucleotide reductase m1, ZC3H7B - zinc finger ccch-type containing 7b, TCOF1 - treacher collins-franceschetti syndrome 1, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, ZNF638 - zinc finger protein 638, F11R - f11 receptor, STAMBP - stam binding protein, SCFD1 - sec1 family domain containing 1, URB1 - urb1 ribosome biogenesis 1 homolog (s. cerevisiae), RAB30 - rab30, member ras oncogene family, FLNB - filamin b, beta, PPFIA1 - protein tyrosine phosphatase, receptor type, f polypeptide (ptprf), interacting protein (liprin), alpha 1, PPP4R2 - protein phosphatase 4, regulatory subunit 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, PPFIBP1 - ptprf interacting protein, binding protein 1 (liprin beta 1), ILF3 - interleukin enhancer binding factor 3, 90kda, ANKRD28 - ankyrin repeat domain 28, NUDCD2 - nudc domain containing 2, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, CYCS - cytochrome c, somatic, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, RBM19 - rna binding motif protein 19, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, EHBP1 - eh domain binding protein 1, CD48 - cd48 molecule, AGPS - alkylglycerone phosphate synthase, TBC1D4 - tbc1 domain family, member 4, TMED5 - transmembrane emp24 protein transport domain containing 5, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, GAR1 - gar1 ribonucleoprotein, KDM3A - lysine (k)-specific demethylase 3a, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, ANAPC1 - anaphase promoting complex subunit 1, TELO2 - telomere maintenance 2, EXOSC1 - exosome component 1, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), ATG7 - autophagy related 7, TUBGCP4 - tubulin, gamma complex associated protein 4, HDDC2 - hd domain containing 2, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), SYNE1 - spectrin repeat containing, nuclear envelope 1, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), NUP88 - nucleoporin 88kda, ZBTB8OS - zinc finger and btb domain containing 8 opposite strand, NFU1 - nfu1 iron-sulfur cluster scaffold homolog (s. cerevisiae), ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), THOC5 - tho complex 5, FECH - ferrochelatase, CDK6 - cyclin-dependent kinase 6, ADNP - activity-dependent neuroprotector homeobox, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, CDK13 - cyclin-dependent kinase 13, TBCD - tubulin folding cofactor d, FCF1 - fcf1 rrna-processing protein, GNPDA2 - glucosamine-6-phosphate deaminase 2, CCT4 - chaperonin containing tcp1, subunit 4 (delta), TBCA - tubulin folding cofactor a, CUTC - cutc copper transporter, INPP5B - inositol polyphosphate-5-phosphatase, 75kda, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, IRF3 - interferon regulatory factor 3, ARL6IP5 - adp-ribosylation-like factor 6 interacting protein 5, LARP1 - la ribonucleoprotein domain family, member 1, IRF5 - interferon regulatory factor 5, DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), EIF3E - eukaryotic translation initiation factor 3, subunit e, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, TCEA1 - transcription elongation factor a (sii), 1, PURA - purine-rich element binding protein a, SPG11 - spastic paraplegia 11 (autosomal recessive), TDRD7 - tudor domain containing 7, NEK6 - nima-related kinase 6, NOC3L - nucleolar complex associated 3 homolog (s. cerevisiae), PYGB - phosphorylase, glycogen; brain, PCYT2 - phosphate cytidylyltransferase 2, ethanolamine, NUP50 - nucleoporin 50kda, VKORC1L1 - vitamin k epoxide reductase complex, subunit 1-like 1, TRAPPC10 - trafficking protein particle complex 10, IDE - insulin-degrading enzyme, USP34 - ubiquitin specific peptidase 34, IFIH1 - interferon induced with helicase c domain 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, SART3 - squamous cell carcinoma antigen recognized by t cells 3, MAD1L1 - mad1 mitotic arrest deficient-like 1 (yeast), CTSC - cathepsin c, RANBP1 - ran binding protein 1, RAP1A - rap1a, member of ras oncogene family, PARVG - parvin, gamma, PUS1 - pseudouridylate synthase 1, NSMAF - neutral sphingomyelinase (n-smase) activation associated factor, GNPAT - glyceronephosphate o-acyltransferase, AKAP8L - a kinase (prka) anchor protein 8-like, CYFIP2 - cytoplasmic fmr1 interacting protein 2, RBM26 - rna binding motif protein 26, CLNS1A - chloride channel, nucleotide-sensitive, 1a, RAP1B - rap1b, member of ras oncogene family, GOLPH3 - golgi phosphoprotein 3 (coat-protein), TMX3 - thioredoxin-related transmembrane protein 3, RB1 - retinoblastoma 1, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), MRPS14 - mitochondrial ribosomal protein s14, CUL1 - cullin 1, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3]
8 [DST - dystonin, ERAP1 - endoplasmic reticulum aminopeptidase 1, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, F11R - f11 receptor, STAMBP - stam binding protein, GSN - gelsolin, SCFD1 - sec1 family domain containing 1, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, TMED2 - transmembrane emp24 domain trafficking protein 2, FLNB - filamin b, beta, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, DERL1 - derlin 1, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, ACAT1 - acetyl-coa acetyltransferase 1, VTI1A - vesicle transport through interaction with t-snares 1a, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, EHBP1 - eh domain binding protein 1, ARCN1 - archain 1, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), CD48 - cd48 molecule, PA2G4 - proliferation-associated 2g4, 38kda, CAB39 - calcium binding protein 39, TBC1D4 - tbc1 domain family, member 4, OTUB1 - otu domain, ubiquitin aldehyde binding 1, SORD - sorbitol dehydrogenase, TMED5 - transmembrane emp24 protein transport domain containing 5, RAB23 - rab23, member ras oncogene family, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, SNX1 - sorting nexin 1, MYO6 - myosin vi, LRRC57 - leucine rich repeat containing 57, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), TUBGCP4 - tubulin, gamma complex associated protein 4, TPM3 - tropomyosin 3, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, GLO1 - glyoxalase i, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), GLG1 - golgi glycoprotein 1, GOSR1 - golgi snap receptor complex member 1, MBLAC2 - metallo-beta-lactamase domain containing 2, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, PPA1 - pyrophosphatase (inorganic) 1, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, SNX6 - sorting nexin 6, HSPD1 - heat shock 60kda protein 1 (chaperonin), NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, CCT4 - chaperonin containing tcp1, subunit 4 (delta), ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, DENND4A - denn/madd domain containing 4a, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, COMT - catechol-o-methyltransferase, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, STX8 - syntaxin 8, ERP44 - endoplasmic reticulum protein 44, NAMPT - nicotinamide phosphoribosyltransferase, EIF3E - eukaryotic translation initiation factor 3, subunit e, HSPA4 - heat shock 70kda protein 4, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, PARP4 - poly (adp-ribose) polymerase family, member 4, SCPEP1 - serine carboxypeptidase 1, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, SERBP1 - serpine1 mrna binding protein 1, VTA1 - vesicle (multivesicular body) trafficking 1, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, DENND3 - denn/madd domain containing 3, GAPVD1 - gtpase activating protein and vps9 domains 1, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, SPG11 - spastic paraplegia 11 (autosomal recessive), RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, CAPN2 - calpain 2, (m/ii) large subunit, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), HMGB1 - high mobility group box 1, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PYGB - phosphorylase, glycogen; brain, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RAB14 - rab14, member ras oncogene family, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, FTH1 - ferritin, heavy polypeptide 1, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, RPL3 - ribosomal protein l3, SURF4 - surfeit 4, MAN2A1 - mannosidase, alpha, class 2a, member 1, USP8 - ubiquitin specific peptidase 8, SLAMF1 - signaling lymphocytic activation molecule family member 1, DUT - deoxyuridine triphosphatase, RPN1 - ribophorin i, TNPO1 - transportin 1, STX17 - syntaxin 17, KPNB1 - karyopherin (importin) beta 1, IDE - insulin-degrading enzyme, CHMP5 - charged multivesicular body protein 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), RPL31 - ribosomal protein l31, VDAC2 - voltage-dependent anion channel 2, CTSC - cathepsin c, WIPF1 - was/wasl interacting protein family, member 1, DLG1 - discs, large homolog 1 (drosophila), NIPBL - nipped-b homolog (drosophila), SCYL2 - scy1-like 2 (s. cerevisiae), RAP1A - rap1a, member of ras oncogene family, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, RPL14 - ribosomal protein l14, CYFIP2 - cytoplasmic fmr1 interacting protein 2, WDR1 - wd repeat domain 1, RPS8 - ribosomal protein s8, ESD - esterase d, RPS13 - ribosomal protein s13, RAP1B - rap1b, member of ras oncogene family, GOLPH3 - golgi phosphoprotein 3 (coat-protein), DSTN - destrin (actin depolymerizing factor), STOM - stomatin, DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, ACOT7 - acyl-coa thioesterase 7, CAT - catalase, UGDH - udp-glucose 6-dehydrogenase, FAM49B - family with sequence similarity 49, member b, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, SSBP1 - single-stranded dna binding protein 1, mitochondrial, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), SAMM50 - samm50 sorting and assembly machinery component, ANXA4 - annexin a4, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, PRKCD - protein kinase c, delta, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, PGM2 - phosphoglucomutase 2]
9 [DST - dystonin, ERAP1 - endoplasmic reticulum aminopeptidase 1, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, F11R - f11 receptor, STAMBP - stam binding protein, GSN - gelsolin, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, FLNB - filamin b, beta, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, NUDCD2 - nudc domain containing 2, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, ACAT1 - acetyl-coa acetyltransferase 1, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), CD48 - cd48 molecule, PA2G4 - proliferation-associated 2g4, 38kda, CAB39 - calcium binding protein 39, SORD - sorbitol dehydrogenase, OTUB1 - otu domain, ubiquitin aldehyde binding 1, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, MYO6 - myosin vi, LRRC57 - leucine rich repeat containing 57, TPM3 - tropomyosin 3, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, GLO1 - glyoxalase i, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), GLG1 - golgi glycoprotein 1, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), MBLAC2 - metallo-beta-lactamase domain containing 2, PPA1 - pyrophosphatase (inorganic) 1, ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, ADNP - activity-dependent neuroprotector homeobox, CDK13 - cyclin-dependent kinase 13, HSPD1 - heat shock 60kda protein 1 (chaperonin), CCT4 - chaperonin containing tcp1, subunit 4 (delta), NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, COMT - catechol-o-methyltransferase, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, ERP44 - endoplasmic reticulum protein 44, NAMPT - nicotinamide phosphoribosyltransferase, EIF3E - eukaryotic translation initiation factor 3, subunit e, HSPA4 - heat shock 70kda protein 4, QSOX2 - quiescin q6 sulfhydryl oxidase 2, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, PARP4 - poly (adp-ribose) polymerase family, member 4, SCPEP1 - serine carboxypeptidase 1, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, SERBP1 - serpine1 mrna binding protein 1, VTA1 - vesicle (multivesicular body) trafficking 1, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, CAPN2 - calpain 2, (m/ii) large subunit, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, FABP5 - fatty acid binding protein 5 (psoriasis-associated), MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, PYGB - phosphorylase, glycogen; brain, RAB14 - rab14, member ras oncogene family, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, FTH1 - ferritin, heavy polypeptide 1, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, RPL3 - ribosomal protein l3, IRAK4 - interleukin-1 receptor-associated kinase 4, MAN2A1 - mannosidase, alpha, class 2a, member 1, SLAMF1 - signaling lymphocytic activation molecule family member 1, DUT - deoxyuridine triphosphatase, TNPO1 - transportin 1, KPNB1 - karyopherin (importin) beta 1, IDE - insulin-degrading enzyme, CHMP5 - charged multivesicular body protein 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, RPL31 - ribosomal protein l31, CTSC - cathepsin c, DLG1 - discs, large homolog 1 (drosophila), NIPBL - nipped-b homolog (drosophila), RAP1A - rap1a, member of ras oncogene family, RPL14 - ribosomal protein l14, CYFIP2 - cytoplasmic fmr1 interacting protein 2, WDR1 - wd repeat domain 1, RPS8 - ribosomal protein s8, ESD - esterase d, RPS13 - ribosomal protein s13, RAP1B - rap1b, member of ras oncogene family, DSTN - destrin (actin depolymerizing factor), STOM - stomatin, ACOT7 - acyl-coa thioesterase 7, CAT - catalase, UGDH - udp-glucose 6-dehydrogenase, FAM49B - family with sequence similarity 49, member b, SSBP1 - single-stranded dna binding protein 1, mitochondrial, SAMM50 - samm50 sorting and assembly machinery component, ANXA4 - annexin a4, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, PRKCD - protein kinase c, delta, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, PGM2 - phosphoglucomutase 2]
10 [CCNK - cyclin k, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, NCBP2 - nuclear cap binding protein subunit 2, 20kda, EXOSC5 - exosome component 5, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, RFC3 - replication factor c (activator 1) 3, 38kda, SF3A3 - splicing factor 3a, subunit 3, 60kda, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, SACM1L - sac1 suppressor of actin mutations 1-like (yeast), NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, NDUFA5 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, HOOK3 - hook microtubule-tethering protein 3, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, DERL1 - derlin 1, BTF3 - basic transcription factor 3, SPC24 - spc24, ndc80 kinetochore complex component, EIF4B - eukaryotic translation initiation factor 4b, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), SUGT1 - sgt1, suppressor of g2 allele of skp1 (s. cerevisiae), EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, GOSR2 - golgi snap receptor complex member 2, ZNF326 - zinc finger protein 326, PLCG1 - phospholipase c, gamma 1, WDR77 - wd repeat domain 77, CARHSP1 - calcium regulated heat stable protein 1, 24kda, MORF4L1 - mortality factor 4 like 1, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, CREB1 - camp responsive element binding protein 1, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, SNX1 - sorting nexin 1, MYO6 - myosin vi, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, EIF2D - eukaryotic translation initiation factor 2d, SNRPE - small nuclear ribonucleoprotein polypeptide e, ANAPC7 - anaphase promoting complex subunit 7, SEH1L - seh1-like (s. cerevisiae), EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, NIP7 - nip7, nucleolar pre-rrna processing protein, GOSR1 - golgi snap receptor complex member 1, POLR1C - polymerase (rna) i polypeptide c, 30kda, EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, DCAF7 - ddb1 and cul4 associated factor 7, ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, SUMO3 - small ubiquitin-like modifier 3, EIF2S2 - eukaryotic translation initiation factor 2, subunit 2 beta, 38kda, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, SNX6 - sorting nexin 6, SMARCD1 - swi/snf related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1, HSPD1 - heat shock 60kda protein 1 (chaperonin), ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, STX8 - syntaxin 8, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), PARP4 - poly (adp-ribose) polymerase family, member 4, TMEM30A - transmembrane protein 30a, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, UQCRB - ubiquinol-cytochrome c reductase binding protein, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, ZWILCH - zwilch kinetochore protein, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, HMGB2 - high mobility group box 2, AGK - acylglycerol kinase, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), PTCD3 - pentatricopeptide repeat domain 3, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, EIF4G3 - eukaryotic translation initiation factor 4 gamma, 3, FTH1 - ferritin, heavy polypeptide 1, PPP5C - protein phosphatase 5, catalytic subunit, RPL3 - ribosomal protein l3, EIF3F - eukaryotic translation initiation factor 3, subunit f, DNAJC11 - dnaj (hsp40) homolog, subfamily c, member 11, MRPS35 - mitochondrial ribosomal protein s35, MRPL1 - mitochondrial ribosomal protein l1, RPN1 - ribophorin i, MRPL9 - mitochondrial ribosomal protein l9, STX17 - syntaxin 17, KPNB1 - karyopherin (importin) beta 1, EXOC2 - exocyst complex component 2, CDC23 - cell division cycle 23, EXOC1 - exocyst complex component 1, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, IFIT5 - interferon-induced protein with tetratricopeptide repeats 5, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, VDAC2 - voltage-dependent anion channel 2, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, MRPL13 - mitochondrial ribosomal protein l13, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, NIPBL - nipped-b homolog (drosophila), STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), MFN1 - mitofusin 1, RNMT - rna (guanine-7-) methyltransferase, WDR1 - wd repeat domain 1, LARP4B - la ribonucleoprotein domain family, member 4b, RPS8 - ribosomal protein s8, MRPL42 - mitochondrial ribosomal protein l42, RPS13 - ribosomal protein s13, RINT1 - rad50 interactor 1, RPS6 - ribosomal protein s6, NHP2 - nhp2 ribonucleoprotein, RPS7 - ribosomal protein s7, SUMO1 - small ubiquitin-like modifier 1, SRP72 - signal recognition particle 72kda, CISD2 - cdgsh iron sulfur domain 2, CAT - catalase, XPO6 - exportin 6, SAMM50 - samm50 sorting and assembly machinery component, MRPL47 - mitochondrial ribosomal protein l47, TUBGCP3 - tubulin, gamma complex associated protein 3, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, UBE2I - ubiquitin-conjugating enzyme e2i, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, RRBP1 - ribosome binding protein 1, EPS15 - epidermal growth factor receptor pathway substrate 15, RRM1 - ribonucleotide reductase m1, DMXL1 - dmx-like 1, DDX6 - dead (asp-glu-ala-asp) box helicase 6, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, F11R - f11 receptor, GSPT1 - g1 to s phase transition 1, GSN - gelsolin, FAM98A - family with sequence similarity 98, member a, PPP4R2 - protein phosphatase 4, regulatory subunit 2, TRIM25 - tripartite motif containing 25, ILF3 - interleukin enhancer binding factor 3, 90kda, XPOT - exportin, trna, NUDCD2 - nudc domain containing 2, CYCS - cytochrome c, somatic, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, VTI1A - vesicle transport through interaction with t-snares 1a, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, MMS22L - mms22-like, dna repair protein, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, ARCN1 - archain 1, EHBP1 - eh domain binding protein 1, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, CAB39 - calcium binding protein 39, GATAD2B - gata zinc finger domain containing 2b, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, GAR1 - gar1 ribonucleoprotein, KDM3A - lysine (k)-specific demethylase 3a, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, ANAPC1 - anaphase promoting complex subunit 1, MRPL3 - mitochondrial ribosomal protein l3, EXOSC1 - exosome component 1, TELO2 - telomere maintenance 2, RNF31 - ring finger protein 31, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), MCM7 - minichromosome maintenance complex component 7, MCU - mitochondrial calcium uniporter, TUBGCP4 - tubulin, gamma complex associated protein 4, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, SYNE1 - spectrin repeat containing, nuclear envelope 1, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), ZBTB8OS - zinc finger and btb domain containing 8 opposite strand, NUP88 - nucleoporin 88kda, PPIL3 - peptidylprolyl isomerase (cyclophilin)-like 3, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), THOC5 - tho complex 5, CDK6 - cyclin-dependent kinase 6, ADNP - activity-dependent neuroprotector homeobox, MRPL2 - mitochondrial ribosomal protein l2, CDK13 - cyclin-dependent kinase 13, FCF1 - fcf1 rrna-processing protein, CCT4 - chaperonin containing tcp1, subunit 4 (delta), RPL7L1 - ribosomal protein l7-like 1, STX18 - syntaxin 18, LARP1 - la ribonucleoprotein domain family, member 1, NEMF - nuclear export mediator factor, EIF3E - eukaryotic translation initiation factor 3, subunit e, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, RPTOR - regulatory associated protein of mtor, complex 1, TCEA1 - transcription elongation factor a (sii), 1, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, TDRD7 - tudor domain containing 7, NEK6 - nima-related kinase 6, NUP50 - nucleoporin 50kda, MAP4 - microtubule-associated protein 4, PHKB - phosphorylase kinase, beta, TRAPPC10 - trafficking protein particle complex 10, MBNL1 - muscleblind-like splicing regulator 1, EIF4H - eukaryotic translation initiation factor 4h, MAT2A - methionine adenosyltransferase ii, alpha, IDE - insulin-degrading enzyme, MCM3 - minichromosome maintenance complex component 3, DOCK7 - dedicator of cytokinesis 7, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), MAD1L1 - mad1 mitotic arrest deficient-like 1 (yeast), SART3 - squamous cell carcinoma antigen recognized by t cells 3, SCAF8 - sr-related ctd-associated factor 8, RANBP1 - ran binding protein 1, DLG1 - discs, large homolog 1 (drosophila), RAP1A - rap1a, member of ras oncogene family, RPL14 - ribosomal protein l14, AKAP8L - a kinase (prka) anchor protein 8-like, CLNS1A - chloride channel, nucleotide-sensitive, 1a, TRAPPC9 - trafficking protein particle complex 9, UBA3 - ubiquitin-like modifier activating enzyme 3, DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, RB1 - retinoblastoma 1, CBX3 - chromobox homolog 3, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, CAPRIN1 - cell cycle associated protein 1, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), MRPS14 - mitochondrial ribosomal protein s14, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, CUL1 - cullin 1, RSL1D1 - ribosomal l1 domain containing 1, BCL2L1 - bcl2-like 1, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
11 [CCNK - cyclin k, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, NCBP2 - nuclear cap binding protein subunit 2, 20kda, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, EXOSC5 - exosome component 5, RFC3 - replication factor c (activator 1) 3, 38kda, SF3A3 - splicing factor 3a, subunit 3, 60kda, SACM1L - sac1 suppressor of actin mutations 1-like (yeast), SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, GFM2 - g elongation factor, mitochondrial 2, NDUFA5 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, FKBP11 - fk506 binding protein 11, 19 kda, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, RECQL - recq protein-like (dna helicase q1-like), DERL1 - derlin 1, BTF3 - basic transcription factor 3, ACAT1 - acetyl-coa acetyltransferase 1, EIF4B - eukaryotic translation initiation factor 4b, CPOX - coproporphyrinogen oxidase, LRRC40 - leucine rich repeat containing 40, EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, USP25 - ubiquitin specific peptidase 25, SORD - sorbitol dehydrogenase, OTUB1 - otu domain, ubiquitin aldehyde binding 1, ZNF326 - zinc finger protein 326, PLCG1 - phospholipase c, gamma 1, MORF4L1 - mortality factor 4 like 1, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, CREB1 - camp responsive element binding protein 1, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, SNX1 - sorting nexin 1, THUMPD1 - thump domain containing 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, SGPL1 - sphingosine-1-phosphate lyase 1, SNRPE - small nuclear ribonucleoprotein polypeptide e, ANAPC7 - anaphase promoting complex subunit 7, STK38L - serine/threonine kinase 38 like, SEH1L - seh1-like (s. cerevisiae), PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, GLS - glutaminase, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, GLO1 - glyoxalase i, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), POLR1C - polymerase (rna) i polypeptide c, 30kda, DCAF7 - ddb1 and cul4 associated factor 7, EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, PPA1 - pyrophosphatase (inorganic) 1, ADPGK - adp-dependent glucokinase, SUMO3 - small ubiquitin-like modifier 3, EIF2S2 - eukaryotic translation initiation factor 2, subunit 2 beta, 38kda, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, TSFM - ts translation elongation factor, mitochondrial, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), MVD - mevalonate (diphospho) decarboxylase, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, COMT - catechol-o-methyltransferase, KYNU - kynureninase, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, TAOK3 - tao kinase 3, ERP44 - endoplasmic reticulum protein 44, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), NAMPT - nicotinamide phosphoribosyltransferase, PARP4 - poly (adp-ribose) polymerase family, member 4, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, SCPEP1 - serine carboxypeptidase 1, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, QKI - qki, kh domain containing, rna binding, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, UQCRB - ubiquinol-cytochrome c reductase binding protein, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, WNK1 - wnk lysine deficient protein kinase 1, UBE2Z - ubiquitin-conjugating enzyme e2z, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, HMGB2 - high mobility group box 2, AGK - acylglycerol kinase, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PTCD3 - pentatricopeptide repeat domain 3, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, EIF4G3 - eukaryotic translation initiation factor 4 gamma, 3, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, UCK2 - uridine-cytidine kinase 2, USP48 - ubiquitin specific peptidase 48, PPP5C - protein phosphatase 5, catalytic subunit, UMPS - uridine monophosphate synthetase, RPL3 - ribosomal protein l3, CARS - cysteinyl-trna synthetase, EIF3F - eukaryotic translation initiation factor 3, subunit f, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), IRAK4 - interleukin-1 receptor-associated kinase 4, MRPS35 - mitochondrial ribosomal protein s35, MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, RPN1 - ribophorin i, HK2 - hexokinase 2, AK4 - adenylate kinase 4, KPNB1 - karyopherin (importin) beta 1, TRIM38 - tripartite motif containing 38, CDC23 - cell division cycle 23, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, RPL31 - ribosomal protein l31, TJP2 - tight junction protein 2, RPL32 - ribosomal protein l32, DCAKD - dephospho-coa kinase domain containing, TTLL12 - tubulin tyrosine ligase-like family, member 12, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, MRPL13 - mitochondrial ribosomal protein l13, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, NIPBL - nipped-b homolog (drosophila), SCYL2 - scy1-like 2 (s. cerevisiae), GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, GALNT2 - udp-n-acetyl-alpha-d-galactosamine:polypeptide n-acetylgalactosaminyltransferase 2 (galnac-t2), STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), RNMT - rna (guanine-7-) methyltransferase, SNRNP27 - small nuclear ribonucleoprotein 27kda (u4/u6.u5), MFN1 - mitofusin 1, RPS8 - ribosomal protein s8, EIF1AD - eukaryotic translation initiation factor 1a domain containing, ESD - esterase d, MRPL42 - mitochondrial ribosomal protein l42, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, NHP2 - nhp2 ribonucleoprotein, UCHL3 - ubiquitin carboxyl-terminal esterase l3 (ubiquitin thiolesterase), SUMO1 - small ubiquitin-like modifier 1, GCDH - glutaryl-coa dehydrogenase, CISD2 - cdgsh iron sulfur domain 2, CAT - catalase, USP15 - ubiquitin specific peptidase 15, UGDH - udp-glucose 6-dehydrogenase, SSBP1 - single-stranded dna binding protein 1, mitochondrial, MRPL47 - mitochondrial ribosomal protein l47, UBE2I - ubiquitin-conjugating enzyme e2i, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, PRKCD - protein kinase c, delta, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), RRBP1 - ribosome binding protein 1, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, RRM1 - ribonucleotide reductase m1, ZC3H7B - zinc finger ccch-type containing 7b, ERAP1 - endoplasmic reticulum aminopeptidase 1, DDX6 - dead (asp-glu-ala-asp) box helicase 6, TCOF1 - treacher collins-franceschetti syndrome 1, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, ZNF638 - zinc finger protein 638, STAMBP - stam binding protein, GSPT1 - g1 to s phase transition 1, GSN - gelsolin, URB1 - urb1 ribosome biogenesis 1 homolog (s. cerevisiae), FAM98A - family with sequence similarity 98, member a, ECI1 - enoyl-coa delta isomerase 1, DCK - deoxycytidine kinase, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, PPP4R2 - protein phosphatase 4, regulatory subunit 2, ILF3 - interleukin enhancer binding factor 3, 90kda, TRIM25 - tripartite motif containing 25, CYCS - cytochrome c, somatic, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, RIF1 - rap1 interacting factor homolog (yeast), PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, VTI1A - vesicle transport through interaction with t-snares 1a, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, RBM19 - rna binding motif protein 19, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, MMS22L - mms22-like, dna repair protein, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, LRRK1 - leucine-rich repeat kinase 1, PA2G4 - proliferation-associated 2g4, 38kda, AGPS - alkylglycerone phosphate synthase, CAB39 - calcium binding protein 39, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, RAB23 - rab23, member ras oncogene family, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, GAR1 - gar1 ribonucleoprotein, KDM3A - lysine (k)-specific demethylase 3a, ANAPC1 - anaphase promoting complex subunit 1, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, MRPL3 - mitochondrial ribosomal protein l3, EXOSC1 - exosome component 1, TELO2 - telomere maintenance 2, RNF31 - ring finger protein 31, ATG7 - autophagy related 7, MCM7 - minichromosome maintenance complex component 7, ARL1 - adp-ribosylation factor-like 1, HDDC2 - hd domain containing 2, KDELC2 - kdel (lys-asp-glu-leu) containing 2, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), PELO - pelota homolog (drosophila), ZBTB8OS - zinc finger and btb domain containing 8 opposite strand, NUP88 - nucleoporin 88kda, NFU1 - nfu1 iron-sulfur cluster scaffold homolog (s. cerevisiae), WDHD1 - wd repeat and hmg-box dna binding protein 1, PPIL3 - peptidylprolyl isomerase (cyclophilin)-like 3, HADH - hydroxyacyl-coa dehydrogenase, OSBPL3 - oxysterol binding protein-like 3, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), THOC5 - tho complex 5, FECH - ferrochelatase, CDK6 - cyclin-dependent kinase 6, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, CDK13 - cyclin-dependent kinase 13, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, GNPDA2 - glucosamine-6-phosphate deaminase 2, FCF1 - fcf1 rrna-processing protein, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, INPP5B - inositol polyphosphate-5-phosphatase, 75kda, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, RPL7L1 - ribosomal protein l7-like 1, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, LARP1 - la ribonucleoprotein domain family, member 1, NEMF - nuclear export mediator factor, DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, EIF3E - eukaryotic translation initiation factor 3, subunit e, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, TCEA1 - transcription elongation factor a (sii), 1, LYRM7 - lyr motif containing 7, PURA - purine-rich element binding protein a, VTA1 - vesicle (multivesicular body) trafficking 1, DENND3 - denn/madd domain containing 3, NEK6 - nima-related kinase 6, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, NOC3L - nucleolar complex associated 3 homolog (s. cerevisiae), PYGB - phosphorylase, glycogen; brain, PCYT2 - phosphate cytidylyltransferase 2, ethanolamine, NUP50 - nucleoporin 50kda, RAB14 - rab14, member ras oncogene family, PHKB - phosphorylase kinase, beta, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, MANBA - mannosidase, beta a, lysosomal, VKORC1L1 - vitamin k epoxide reductase complex, subunit 1-like 1, MAN2A1 - mannosidase, alpha, class 2a, member 1, USP8 - ubiquitin specific peptidase 8, MBNL1 - muscleblind-like splicing regulator 1, EIF4H - eukaryotic translation initiation factor 4h, METTL15 - methyltransferase like 15, DUT - deoxyuridine triphosphatase, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, MAT2A - methionine adenosyltransferase ii, alpha, IDE - insulin-degrading enzyme, MCM3 - minichromosome maintenance complex component 3, USP34 - ubiquitin specific peptidase 34, IFIH1 - interferon induced with helicase c domain 1, MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, SART3 - squamous cell carcinoma antigen recognized by t cells 3, SCAF8 - sr-related ctd-associated factor 8, LSS - lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase), UBA6 - ubiquitin-like modifier activating enzyme 6, GRK6 - g protein-coupled receptor kinase 6, DLG1 - discs, large homolog 1 (drosophila), PUS1 - pseudouridylate synthase 1, NSMAF - neutral sphingomyelinase (n-smase) activation associated factor, RPL14 - ribosomal protein l14, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, GNPAT - glyceronephosphate o-acyltransferase, AKAP8L - a kinase (prka) anchor protein 8-like, NUB1 - negative regulator of ubiquitin-like proteins 1, RBM26 - rna binding motif protein 26, DGUOK - deoxyguanosine kinase, UBA3 - ubiquitin-like modifier activating enzyme 3, GOLPH3 - golgi phosphoprotein 3 (coat-protein), TMX3 - thioredoxin-related transmembrane protein 3, ACOT7 - acyl-coa thioesterase 7, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, ADI1 - acireductone dioxygenase 1, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), MRPS14 - mitochondrial ribosomal protein s14, RHOT1 - ras homolog family member t1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, CUL1 - cullin 1, RSL1D1 - ribosomal l1 domain containing 1, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, PGM2 - phosphoglucomutase 2, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
12 [ERAP1 - endoplasmic reticulum aminopeptidase 1, CAPN2 - calpain 2, (m/ii) large subunit, PGM1 - phosphoglucomutase 1, TDRD7 - tudor domain containing 7, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, OSTF1 - osteoclast stimulating factor 1, PGAM1 - phosphoglycerate mutase 1 (brain), GSN - gelsolin, AGK - acylglycerol kinase, HMGB1 - high mobility group box 1, GFM2 - g elongation factor, mitochondrial 2, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PYGB - phosphorylase, glycogen; brain, ECI1 - enoyl-coa delta isomerase 1, FTH1 - ferritin, heavy polypeptide 1, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, MANBA - mannosidase, beta a, lysosomal, CYCS - cytochrome c, somatic, METTL15 - methyltransferase like 15, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, ACAT1 - acetyl-coa acetyltransferase 1, CPOX - coproporphyrinogen oxidase, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, AK4 - adenylate kinase 4, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, PA2G4 - proliferation-associated 2g4, 38kda, KPNB1 - karyopherin (importin) beta 1, AGPS - alkylglycerone phosphate synthase, CAB39 - calcium binding protein 39, IDE - insulin-degrading enzyme, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, CREB1 - camp responsive element binding protein 1, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, CTSC - cathepsin c, PITRM1 - pitrilysin metallopeptidase 1, ATG7 - autophagy related 7, KDELC2 - kdel (lys-asp-glu-leu) containing 2, PUS1 - pseudouridylate synthase 1, GLS - glutaminase, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, GNPAT - glyceronephosphate o-acyltransferase, DGUOK - deoxyguanosine kinase, ESD - esterase d, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), HADH - hydroxyacyl-coa dehydrogenase, GOLPH3 - golgi phosphoprotein 3 (coat-protein), FECH - ferrochelatase, TSFM - ts translation elongation factor, mitochondrial, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, GCDH - glutaryl-coa dehydrogenase, PDAP1 - pdgfa associated protein 1, CAT - catalase, CDK13 - cyclin-dependent kinase 13, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, FAM49B - family with sequence similarity 49, member b, SSBP1 - single-stranded dna binding protein 1, mitochondrial, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, ERP44 - endoplasmic reticulum protein 44, PRKCD - protein kinase c, delta, BCL2L1 - bcl2-like 1, DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3, PGM2 - phosphoglucomutase 2, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, LYRM7 - lyr motif containing 7]
13 [ERAP1 - endoplasmic reticulum aminopeptidase 1, CAPN2 - calpain 2, (m/ii) large subunit, PGM1 - phosphoglucomutase 1, TDRD7 - tudor domain containing 7, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, OSTF1 - osteoclast stimulating factor 1, PGAM1 - phosphoglycerate mutase 1 (brain), GSN - gelsolin, AGK - acylglycerol kinase, HMGB1 - high mobility group box 1, GFM2 - g elongation factor, mitochondrial 2, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PYGB - phosphorylase, glycogen; brain, ECI1 - enoyl-coa delta isomerase 1, FTH1 - ferritin, heavy polypeptide 1, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, MANBA - mannosidase, beta a, lysosomal, CYCS - cytochrome c, somatic, METTL15 - methyltransferase like 15, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, ACAT1 - acetyl-coa acetyltransferase 1, CPOX - coproporphyrinogen oxidase, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, AK4 - adenylate kinase 4, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, PA2G4 - proliferation-associated 2g4, 38kda, KPNB1 - karyopherin (importin) beta 1, AGPS - alkylglycerone phosphate synthase, CAB39 - calcium binding protein 39, IDE - insulin-degrading enzyme, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, CREB1 - camp responsive element binding protein 1, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, CTSC - cathepsin c, PITRM1 - pitrilysin metallopeptidase 1, ATG7 - autophagy related 7, KDELC2 - kdel (lys-asp-glu-leu) containing 2, PUS1 - pseudouridylate synthase 1, GLS - glutaminase, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, GNPAT - glyceronephosphate o-acyltransferase, DGUOK - deoxyguanosine kinase, ESD - esterase d, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), HADH - hydroxyacyl-coa dehydrogenase, GOLPH3 - golgi phosphoprotein 3 (coat-protein), FECH - ferrochelatase, TSFM - ts translation elongation factor, mitochondrial, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, GCDH - glutaryl-coa dehydrogenase, PDAP1 - pdgfa associated protein 1, CAT - catalase, CDK13 - cyclin-dependent kinase 13, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, FAM49B - family with sequence similarity 49, member b, SSBP1 - single-stranded dna binding protein 1, mitochondrial, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, ERP44 - endoplasmic reticulum protein 44, PRKCD - protein kinase c, delta, BCL2L1 - bcl2-like 1, DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3, PGM2 - phosphoglucomutase 2, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, LYRM7 - lyr motif containing 7]
14 [TMEM30A - transmembrane protein 30a, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, EXOSC5 - exosome component 5, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, PGM1 - phosphoglucomutase 1, OSTF1 - osteoclast stimulating factor 1, PGAM1 - phosphoglycerate mutase 1 (brain), GSN - gelsolin, HMGB1 - high mobility group box 1, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PYGB - phosphorylase, glycogen; brain, RAB14 - rab14, member ras oncogene family, FTH1 - ferritin, heavy polypeptide 1, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, MANBA - mannosidase, beta a, lysosomal, SURF4 - surfeit 4, ILF3 - interleukin enhancer binding factor 3, 90kda, IRAK4 - interleukin-1 receptor-associated kinase 4, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, SLAMF1 - signaling lymphocytic activation molecule family member 1, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PA2G4 - proliferation-associated 2g4, 38kda, KPNB1 - karyopherin (importin) beta 1, CAB39 - calcium binding protein 39, PLCG1 - phospholipase c, gamma 1, EXOC1 - exocyst complex component 1, IFIH1 - interferon induced with helicase c domain 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), IFIT5 - interferon-induced protein with tetratricopeptide repeats 5, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, CTSC - cathepsin c, WIPF1 - was/wasl interacting protein family, member 1, ATG7 - autophagy related 7, DLG1 - discs, large homolog 1 (drosophila), RAP1A - rap1a, member of ras oncogene family, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, CYFIP2 - cytoplasmic fmr1 interacting protein 2, WDR1 - wd repeat domain 1, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), RAP1B - rap1b, member of ras oncogene family, RPS6 - ribosomal protein s6, ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, STOM - stomatin, PDAP1 - pdgfa associated protein 1, CDK13 - cyclin-dependent kinase 13, CAT - catalase, HSPD1 - heat shock 60kda protein 1 (chaperonin), ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, IRF3 - interferon regulatory factor 3, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, ERP44 - endoplasmic reticulum protein 44, PRKCD - protein kinase c, delta, BCL2L1 - bcl2-like 1, IRF5 - interferon regulatory factor 5, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, PGM2 - phosphoglucomutase 2]
15 [ERAP1 - endoplasmic reticulum aminopeptidase 1, CAPN2 - calpain 2, (m/ii) large subunit, PGM1 - phosphoglucomutase 1, TDRD7 - tudor domain containing 7, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, OSTF1 - osteoclast stimulating factor 1, PGAM1 - phosphoglycerate mutase 1 (brain), GSN - gelsolin, AGK - acylglycerol kinase, HMGB1 - high mobility group box 1, GFM2 - g elongation factor, mitochondrial 2, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PYGB - phosphorylase, glycogen; brain, ECI1 - enoyl-coa delta isomerase 1, FTH1 - ferritin, heavy polypeptide 1, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, MANBA - mannosidase, beta a, lysosomal, CYCS - cytochrome c, somatic, METTL15 - methyltransferase like 15, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, ACAT1 - acetyl-coa acetyltransferase 1, CPOX - coproporphyrinogen oxidase, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, AK4 - adenylate kinase 4, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, PA2G4 - proliferation-associated 2g4, 38kda, KPNB1 - karyopherin (importin) beta 1, AGPS - alkylglycerone phosphate synthase, CAB39 - calcium binding protein 39, IDE - insulin-degrading enzyme, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, CREB1 - camp responsive element binding protein 1, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, CTSC - cathepsin c, PITRM1 - pitrilysin metallopeptidase 1, ATG7 - autophagy related 7, KDELC2 - kdel (lys-asp-glu-leu) containing 2, PUS1 - pseudouridylate synthase 1, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, GLS - glutaminase, GNPAT - glyceronephosphate o-acyltransferase, DGUOK - deoxyguanosine kinase, ESD - esterase d, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), HADH - hydroxyacyl-coa dehydrogenase, GOLPH3 - golgi phosphoprotein 3 (coat-protein), FECH - ferrochelatase, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, TSFM - ts translation elongation factor, mitochondrial, GCDH - glutaryl-coa dehydrogenase, CAT - catalase, PDAP1 - pdgfa associated protein 1, CDK13 - cyclin-dependent kinase 13, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), FAM49B - family with sequence similarity 49, member b, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, SSBP1 - single-stranded dna binding protein 1, mitochondrial, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, ERP44 - endoplasmic reticulum protein 44, BCL2L1 - bcl2-like 1, PRKCD - protein kinase c, delta, DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3, PGM2 - phosphoglucomutase 2, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, LYRM7 - lyr motif containing 7]
16 [CCNK - cyclin k, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, NCBP2 - nuclear cap binding protein subunit 2, 20kda, EXOSC5 - exosome component 5, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, RFC3 - replication factor c (activator 1) 3, 38kda, SF3A3 - splicing factor 3a, subunit 3, 60kda, SACM1L - sac1 suppressor of actin mutations 1-like (yeast), SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, NDUFA5 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, GFM2 - g elongation factor, mitochondrial 2, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, FKBP11 - fk506 binding protein 11, 19 kda, RECQL - recq protein-like (dna helicase q1-like), DERL1 - derlin 1, BTF3 - basic transcription factor 3, ACAT1 - acetyl-coa acetyltransferase 1, EIF4B - eukaryotic translation initiation factor 4b, CPOX - coproporphyrinogen oxidase, LRRC40 - leucine rich repeat containing 40, EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, USP25 - ubiquitin specific peptidase 25, OTUB1 - otu domain, ubiquitin aldehyde binding 1, SORD - sorbitol dehydrogenase, ZNF326 - zinc finger protein 326, PLCG1 - phospholipase c, gamma 1, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, MORF4L1 - mortality factor 4 like 1, CREB1 - camp responsive element binding protein 1, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, SNX1 - sorting nexin 1, THUMPD1 - thump domain containing 1, SGPL1 - sphingosine-1-phosphate lyase 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, SNRPE - small nuclear ribonucleoprotein polypeptide e, STK38L - serine/threonine kinase 38 like, ANAPC7 - anaphase promoting complex subunit 7, SEH1L - seh1-like (s. cerevisiae), EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, GLS - glutaminase, IARS - isoleucyl-trna synthetase, GLO1 - glyoxalase i, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), POLR1C - polymerase (rna) i polypeptide c, 30kda, EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, DCAF7 - ddb1 and cul4 associated factor 7, ADPGK - adp-dependent glucokinase, PPA1 - pyrophosphatase (inorganic) 1, EIF2S2 - eukaryotic translation initiation factor 2, subunit 2 beta, 38kda, SUMO3 - small ubiquitin-like modifier 3, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, TSFM - ts translation elongation factor, mitochondrial, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), MVD - mevalonate (diphospho) decarboxylase, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, COMT - catechol-o-methyltransferase, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, KYNU - kynureninase, ERP44 - endoplasmic reticulum protein 44, TAOK3 - tao kinase 3, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), NAMPT - nicotinamide phosphoribosyltransferase, PARP4 - poly (adp-ribose) polymerase family, member 4, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, SCPEP1 - serine carboxypeptidase 1, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, UQCRB - ubiquinol-cytochrome c reductase binding protein, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, QKI - qki, kh domain containing, rna binding, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, CAPN2 - calpain 2, (m/ii) large subunit, WNK1 - wnk lysine deficient protein kinase 1, UBE2Z - ubiquitin-conjugating enzyme e2z, HMGB2 - high mobility group box 2, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, HMGB1 - high mobility group box 1, AGK - acylglycerol kinase, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PTCD3 - pentatricopeptide repeat domain 3, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, EIF4G3 - eukaryotic translation initiation factor 4 gamma, 3, FTH1 - ferritin, heavy polypeptide 1, UCK2 - uridine-cytidine kinase 2, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, UMPS - uridine monophosphate synthetase, PPP5C - protein phosphatase 5, catalytic subunit, RPL3 - ribosomal protein l3, USP48 - ubiquitin specific peptidase 48, CARS - cysteinyl-trna synthetase, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), EIF3F - eukaryotic translation initiation factor 3, subunit f, IRAK4 - interleukin-1 receptor-associated kinase 4, MRPS35 - mitochondrial ribosomal protein s35, MRPL1 - mitochondrial ribosomal protein l1, RPN1 - ribophorin i, MRPL9 - mitochondrial ribosomal protein l9, AK4 - adenylate kinase 4, HK2 - hexokinase 2, KPNB1 - karyopherin (importin) beta 1, TRIM38 - tripartite motif containing 38, CDC23 - cell division cycle 23, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, TJP2 - tight junction protein 2, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, DCAKD - dephospho-coa kinase domain containing, TTLL12 - tubulin tyrosine ligase-like family, member 12, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, MRPL13 - mitochondrial ribosomal protein l13, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, NIPBL - nipped-b homolog (drosophila), SCYL2 - scy1-like 2 (s. cerevisiae), GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, GALNT2 - udp-n-acetyl-alpha-d-galactosamine:polypeptide n-acetylgalactosaminyltransferase 2 (galnac-t2), STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), SNRNP27 - small nuclear ribonucleoprotein 27kda (u4/u6.u5), MFN1 - mitofusin 1, RNMT - rna (guanine-7-) methyltransferase, EIF1AD - eukaryotic translation initiation factor 1a domain containing, RPS8 - ribosomal protein s8, MRPL42 - mitochondrial ribosomal protein l42, ESD - esterase d, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, NHP2 - nhp2 ribonucleoprotein, RPS7 - ribosomal protein s7, UCHL3 - ubiquitin carboxyl-terminal esterase l3 (ubiquitin thiolesterase), SUMO1 - small ubiquitin-like modifier 1, CISD2 - cdgsh iron sulfur domain 2, GCDH - glutaryl-coa dehydrogenase, CAT - catalase, USP15 - ubiquitin specific peptidase 15, UGDH - udp-glucose 6-dehydrogenase, SSBP1 - single-stranded dna binding protein 1, mitochondrial, MRPL47 - mitochondrial ribosomal protein l47, UBE2I - ubiquitin-conjugating enzyme e2i, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, PRKCD - protein kinase c, delta, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, RRBP1 - ribosome binding protein 1, RRM1 - ribonucleotide reductase m1, ZC3H7B - zinc finger ccch-type containing 7b, ERAP1 - endoplasmic reticulum aminopeptidase 1, DDX6 - dead (asp-glu-ala-asp) box helicase 6, TCOF1 - treacher collins-franceschetti syndrome 1, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, ZNF638 - zinc finger protein 638, GSPT1 - g1 to s phase transition 1, STAMBP - stam binding protein, GSN - gelsolin, URB1 - urb1 ribosome biogenesis 1 homolog (s. cerevisiae), FAM98A - family with sequence similarity 98, member a, ECI1 - enoyl-coa delta isomerase 1, DCK - deoxycytidine kinase, PPP4R2 - protein phosphatase 4, regulatory subunit 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, TRIM25 - tripartite motif containing 25, ILF3 - interleukin enhancer binding factor 3, 90kda, RIF1 - rap1 interacting factor homolog (yeast), CYCS - cytochrome c, somatic, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, VTI1A - vesicle transport through interaction with t-snares 1a, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, RBM19 - rna binding motif protein 19, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), MMS22L - mms22-like, dna repair protein, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, LRRK1 - leucine-rich repeat kinase 1, PA2G4 - proliferation-associated 2g4, 38kda, AGPS - alkylglycerone phosphate synthase, CAB39 - calcium binding protein 39, RAB23 - rab23, member ras oncogene family, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, GAR1 - gar1 ribonucleoprotein, KDM3A - lysine (k)-specific demethylase 3a, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, ANAPC1 - anaphase promoting complex subunit 1, MRPL3 - mitochondrial ribosomal protein l3, EXOSC1 - exosome component 1, TELO2 - telomere maintenance 2, PITRM1 - pitrilysin metallopeptidase 1, RNF31 - ring finger protein 31, MCM7 - minichromosome maintenance complex component 7, ATG7 - autophagy related 7, ARL1 - adp-ribosylation factor-like 1, HDDC2 - hd domain containing 2, KDELC2 - kdel (lys-asp-glu-leu) containing 2, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), PELO - pelota homolog (drosophila), NUP88 - nucleoporin 88kda, ZBTB8OS - zinc finger and btb domain containing 8 opposite strand, WDHD1 - wd repeat and hmg-box dna binding protein 1, NFU1 - nfu1 iron-sulfur cluster scaffold homolog (s. cerevisiae), PPIL3 - peptidylprolyl isomerase (cyclophilin)-like 3, HADH - hydroxyacyl-coa dehydrogenase, OSBPL3 - oxysterol binding protein-like 3, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), THOC5 - tho complex 5, FECH - ferrochelatase, CDK6 - cyclin-dependent kinase 6, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, CDK13 - cyclin-dependent kinase 13, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, FCF1 - fcf1 rrna-processing protein, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, GNPDA2 - glucosamine-6-phosphate deaminase 2, INPP5B - inositol polyphosphate-5-phosphatase, 75kda, RPL7L1 - ribosomal protein l7-like 1, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, LARP1 - la ribonucleoprotein domain family, member 1, NEMF - nuclear export mediator factor, DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, EIF3E - eukaryotic translation initiation factor 3, subunit e, QSOX2 - quiescin q6 sulfhydryl oxidase 2, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, TCEA1 - transcription elongation factor a (sii), 1, LYRM7 - lyr motif containing 7, VTA1 - vesicle (multivesicular body) trafficking 1, PURA - purine-rich element binding protein a, DENND3 - denn/madd domain containing 3, PGM1 - phosphoglucomutase 1, NEK6 - nima-related kinase 6, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, NOC3L - nucleolar complex associated 3 homolog (s. cerevisiae), PYGB - phosphorylase, glycogen; brain, PCYT2 - phosphate cytidylyltransferase 2, ethanolamine, NUP50 - nucleoporin 50kda, RAB14 - rab14, member ras oncogene family, PHKB - phosphorylase kinase, beta, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, MANBA - mannosidase, beta a, lysosomal, VKORC1L1 - vitamin k epoxide reductase complex, subunit 1-like 1, MAN2A1 - mannosidase, alpha, class 2a, member 1, USP8 - ubiquitin specific peptidase 8, MBNL1 - muscleblind-like splicing regulator 1, EIF4H - eukaryotic translation initiation factor 4h, METTL15 - methyltransferase like 15, DUT - deoxyuridine triphosphatase, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, MAT2A - methionine adenosyltransferase ii, alpha, MCM3 - minichromosome maintenance complex component 3, IDE - insulin-degrading enzyme, USP34 - ubiquitin specific peptidase 34, IFIH1 - interferon induced with helicase c domain 1, MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, SART3 - squamous cell carcinoma antigen recognized by t cells 3, CTSC - cathepsin c, SCAF8 - sr-related ctd-associated factor 8, LSS - lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase), GRK6 - g protein-coupled receptor kinase 6, UBA6 - ubiquitin-like modifier activating enzyme 6, DLG1 - discs, large homolog 1 (drosophila), PUS1 - pseudouridylate synthase 1, RPL14 - ribosomal protein l14, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, NSMAF - neutral sphingomyelinase (n-smase) activation associated factor, DHRS7 - dehydrogenase/reductase (sdr family) member 7, GNPAT - glyceronephosphate o-acyltransferase, AKAP8L - a kinase (prka) anchor protein 8-like, NUB1 - negative regulator of ubiquitin-like proteins 1, CYFIP2 - cytoplasmic fmr1 interacting protein 2, RBM26 - rna binding motif protein 26, DGUOK - deoxyguanosine kinase, UBA3 - ubiquitin-like modifier activating enzyme 3, GOLPH3 - golgi phosphoprotein 3 (coat-protein), TMX3 - thioredoxin-related transmembrane protein 3, ACOT7 - acyl-coa thioesterase 7, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, ADI1 - acireductone dioxygenase 1, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), RHOT1 - ras homolog family member t1, MRPS14 - mitochondrial ribosomal protein s14, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, CUL1 - cullin 1, RSL1D1 - ribosomal l1 domain containing 1, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, METTL13 - methyltransferase like 13, PGM2 - phosphoglucomutase 2, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
17 [FAM136A - family with sequence similarity 136, member a, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, SF3A3 - splicing factor 3a, subunit 3, 60kda, GFM2 - g elongation factor, mitochondrial 2, TMED2 - transmembrane emp24 domain trafficking protein 2, HOOK3 - hook microtubule-tethering protein 3, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, ACAT1 - acetyl-coa acetyltransferase 1, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, CPOX - coproporphyrinogen oxidase, LRRC40 - leucine rich repeat containing 40, SUGT1 - sgt1, suppressor of g2 allele of skp1 (s. cerevisiae), GOSR2 - golgi snap receptor complex member 2, OTUB1 - otu domain, ubiquitin aldehyde binding 1, SORD - sorbitol dehydrogenase, PLCG1 - phospholipase c, gamma 1, LIMS1 - lim and senescent cell antigen-like domains 1, MORF4L1 - mortality factor 4 like 1, CARHSP1 - calcium regulated heat stable protein 1, 24kda, CREB1 - camp responsive element binding protein 1, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, SNX1 - sorting nexin 1, THUMPD1 - thump domain containing 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, SNRPE - small nuclear ribonucleoprotein polypeptide e, ANAPC7 - anaphase promoting complex subunit 7, PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, GLS - glutaminase, GLO1 - glyoxalase i, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), GLG1 - golgi glycoprotein 1, GOSR1 - golgi snap receptor complex member 1, NIP7 - nip7, nucleolar pre-rrna processing protein, COMMD10 - comm domain containing 10, POLR1C - polymerase (rna) i polypeptide c, 30kda, PPA1 - pyrophosphatase (inorganic) 1, SUMO3 - small ubiquitin-like modifier 3, SNX6 - sorting nexin 6, SMARCD1 - swi/snf related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, COMT - catechol-o-methyltransferase, STX8 - syntaxin 8, TAOK3 - tao kinase 3, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), PARP4 - poly (adp-ribose) polymerase family, member 4, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, TMEM30A - transmembrane protein 30a, SCPEP1 - serine carboxypeptidase 1, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, QKI - qki, kh domain containing, rna binding, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, AGK - acylglycerol kinase, HOMER1 - homer homolog 1 (drosophila), FTH1 - ferritin, heavy polypeptide 1, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, PPP5C - protein phosphatase 5, catalytic subunit, USP48 - ubiquitin specific peptidase 48, SURF4 - surfeit 4, IRAK4 - interleukin-1 receptor-associated kinase 4, DNAJC11 - dnaj (hsp40) homolog, subfamily c, member 11, MRPS35 - mitochondrial ribosomal protein s35, AK4 - adenylate kinase 4, TNPO1 - transportin 1, KPNB1 - karyopherin (importin) beta 1, EXOC2 - exocyst complex component 2, EXOC1 - exocyst complex component 1, TJP2 - tight junction protein 2, IFIT5 - interferon-induced protein with tetratricopeptide repeats 5, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, SCYL2 - scy1-like 2 (s. cerevisiae), NIPBL - nipped-b homolog (drosophila), GALNT2 - udp-n-acetyl-alpha-d-galactosamine:polypeptide n-acetylgalactosaminyltransferase 2 (galnac-t2), STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), MFN1 - mitofusin 1, SNRNP27 - small nuclear ribonucleoprotein 27kda (u4/u6.u5), STAT6 - signal transducer and activator of transcription 6, interleukin-4 induced, EIF1AD - eukaryotic translation initiation factor 1a domain containing, FCRLA - fc receptor-like a, RINT1 - rad50 interactor 1, NHP2 - nhp2 ribonucleoprotein, DSTN - destrin (actin depolymerizing factor), RIC8A - ric8 guanine nucleotide exchange factor a, SRP72 - signal recognition particle 72kda, GCDH - glutaryl-coa dehydrogenase, CISD2 - cdgsh iron sulfur domain 2, SSBP1 - single-stranded dna binding protein 1, mitochondrial, SAMM50 - samm50 sorting and assembly machinery component, ANXA4 - annexin a4, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, TNFAIP8 - tumor necrosis factor, alpha-induced protein 8, PRKCD - protein kinase c, delta, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), CCDC6 - coiled-coil domain containing 6, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, DMXL1 - dmx-like 1, ERAP1 - endoplasmic reticulum aminopeptidase 1, DDX6 - dead (asp-glu-ala-asp) box helicase 6, SDAD1 - sda1 domain containing 1, ZMYM2 - zinc finger, mym-type 2, GSPT1 - g1 to s phase transition 1, GSN - gelsolin, FAM98A - family with sequence similarity 98, member a, ECI1 - enoyl-coa delta isomerase 1, DCK - deoxycytidine kinase, DBN1 - drebrin 1, TRIM25 - tripartite motif containing 25, XPOT - exportin, trna, RIF1 - rap1 interacting factor homolog (yeast), VTI1A - vesicle transport through interaction with t-snares 1a, CCDC51 - coiled-coil domain containing 51, MMS22L - mms22-like, dna repair protein, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), ARCN1 - archain 1, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, LRRK1 - leucine-rich repeat kinase 1, PA2G4 - proliferation-associated 2g4, 38kda, CAB39 - calcium binding protein 39, GATAD2B - gata zinc finger domain containing 2b, RAB23 - rab23, member ras oncogene family, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, MRPL3 - mitochondrial ribosomal protein l3, RNF31 - ring finger protein 31, MCM7 - minichromosome maintenance complex component 7, MCU - mitochondrial calcium uniporter, ARL1 - adp-ribosylation factor-like 1, KDELC2 - kdel (lys-asp-glu-leu) containing 2, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, PELO - pelota homolog (drosophila), SCO1 - sco1 cytochrome c oxidase assembly protein, WDHD1 - wd repeat and hmg-box dna binding protein 1, HADH - hydroxyacyl-coa dehydrogenase, PPIL3 - peptidylprolyl isomerase (cyclophilin)-like 3, OSBPL3 - oxysterol binding protein-like 3, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, C15orf40 - chromosome 15 open reading frame 40, RPL7L1 - ribosomal protein l7-like 1, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, STX18 - syntaxin 18, NEMF - nuclear export mediator factor, RPTOR - regulatory associated protein of mtor, complex 1, QSOX2 - quiescin q6 sulfhydryl oxidase 2, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, LYRM7 - lyr motif containing 7, VTA1 - vesicle (multivesicular body) trafficking 1, SERBP1 - serpine1 mrna binding protein 1, DENND3 - denn/madd domain containing 3, GAPVD1 - gtpase activating protein and vps9 domains 1, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, MAP4 - microtubule-associated protein 4, RAB14 - rab14, member ras oncogene family, PHKB - phosphorylase kinase, beta, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, MANBA - mannosidase, beta a, lysosomal, MAN2A1 - mannosidase, alpha, class 2a, member 1, USP8 - ubiquitin specific peptidase 8, EIF4H - eukaryotic translation initiation factor 4h, MBNL1 - muscleblind-like splicing regulator 1, METTL15 - methyltransferase like 15, DUT - deoxyuridine triphosphatase, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, MAT2A - methionine adenosyltransferase ii, alpha, MCM3 - minichromosome maintenance complex component 3, CHMP5 - charged multivesicular body protein 5, DOCK7 - dedicator of cytokinesis 7, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, WIPF1 - was/wasl interacting protein family, member 1, SCAF8 - sr-related ctd-associated factor 8, UBA6 - ubiquitin-like modifier activating enzyme 6, LSS - lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase), DLG1 - discs, large homolog 1 (drosophila), PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, RPL14 - ribosomal protein l14, NUB1 - negative regulator of ubiquitin-like proteins 1, TRAPPC9 - trafficking protein particle complex 9, DGUOK - deoxyguanosine kinase, UBA3 - ubiquitin-like modifier activating enzyme 3, DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, ACOT7 - acyl-coa thioesterase 7, PDAP1 - pdgfa associated protein 1, CBX3 - chromobox homolog 3, ADI1 - acireductone dioxygenase 1, FAM49B - family with sequence similarity 49, member b, CAPRIN1 - cell cycle associated protein 1, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), RHOT1 - ras homolog family member t1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, RSL1D1 - ribosomal l1 domain containing 1, BCL2L1 - bcl2-like 1, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, PGM2 - phosphoglucomutase 2, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1, CCNK - cyclin k, DST - dystonin, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, NCBP2 - nuclear cap binding protein subunit 2, 20kda, EXOSC5 - exosome component 5, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, RFC3 - replication factor c (activator 1) 3, 38kda, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, SACM1L - sac1 suppressor of actin mutations 1-like (yeast), NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, NDUFA5 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, RECQL - recq protein-like (dna helicase q1-like), DERL1 - derlin 1, BTF3 - basic transcription factor 3, DOCK8 - dedicator of cytokinesis 8, SPC24 - spc24, ndc80 kinetochore complex component, EIF4B - eukaryotic translation initiation factor 4b, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, USP25 - ubiquitin specific peptidase 25, ZNF326 - zinc finger protein 326, WDR77 - wd repeat domain 77, RCSD1 - rcsd domain containing 1, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, MYO6 - myosin vi, SGPL1 - sphingosine-1-phosphate lyase 1, EIF2D - eukaryotic translation initiation factor 2d, TPM3 - tropomyosin 3, STK38L - serine/threonine kinase 38 like, SEH1L - seh1-like (s. cerevisiae), EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, CC2D1B - coiled-coil and c2 domain containing 1b, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, DCAF7 - ddb1 and cul4 associated factor 7, ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, ADPGK - adp-dependent glucokinase, EIF2S2 - eukaryotic translation initiation factor 2, subunit 2 beta, 38kda, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, TSFM - ts translation elongation factor, mitochondrial, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), MVD - mevalonate (diphospho) decarboxylase, DENND4A - denn/madd domain containing 4a, KYNU - kynureninase, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, ERP44 - endoplasmic reticulum protein 44, NAMPT - nicotinamide phosphoribosyltransferase, HSPA4 - heat shock 70kda protein 4, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, UQCRB - ubiquinol-cytochrome c reductase binding protein, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, CAPN2 - calpain 2, (m/ii) large subunit, ZWILCH - zwilch kinetochore protein, WNK1 - wnk lysine deficient protein kinase 1, OSTF1 - osteoclast stimulating factor 1, UBE2Z - ubiquitin-conjugating enzyme e2z, HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PTCD3 - pentatricopeptide repeat domain 3, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), TBC1D2B - tbc1 domain family, member 2b, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, EIF4G3 - eukaryotic translation initiation factor 4 gamma, 3, UCK2 - uridine-cytidine kinase 2, RPL3 - ribosomal protein l3, UMPS - uridine monophosphate synthetase, CARS - cysteinyl-trna synthetase, EIF3F - eukaryotic translation initiation factor 3, subunit f, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), MRPL1 - mitochondrial ribosomal protein l1, RPN1 - ribophorin i, MRPL9 - mitochondrial ribosomal protein l9, STX17 - syntaxin 17, HK2 - hexokinase 2, TRIM38 - tripartite motif containing 38, CDC23 - cell division cycle 23, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, VDAC2 - voltage-dependent anion channel 2, TTLL12 - tubulin tyrosine ligase-like family, member 12, MRPL13 - mitochondrial ribosomal protein l13, GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, TMEM70 - transmembrane protein 70, RNMT - rna (guanine-7-) methyltransferase, WDR1 - wd repeat domain 1, RPS8 - ribosomal protein s8, LARP4B - la ribonucleoprotein domain family, member 4b, MRPL42 - mitochondrial ribosomal protein l42, ESD - esterase d, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, STOM - stomatin, UCHL3 - ubiquitin carboxyl-terminal esterase l3 (ubiquitin thiolesterase), SUMO1 - small ubiquitin-like modifier 1, CAT - catalase, USP15 - ubiquitin specific peptidase 15, XPO6 - exportin 6, UGDH - udp-glucose 6-dehydrogenase, NDUFAF4 - nadh dehydrogenase (ubiquinone) complex i, assembly factor 4, MRPL47 - mitochondrial ribosomal protein l47, TUBGCP3 - tubulin, gamma complex associated protein 3, UBE2I - ubiquitin-conjugating enzyme e2i, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, AIDA - axin interactor, dorsalization associated, RRBP1 - ribosome binding protein 1, EPS15 - epidermal growth factor receptor pathway substrate 15, RRM1 - ribonucleotide reductase m1, ZC3H7B - zinc finger ccch-type containing 7b, DDX31 - dead (asp-glu-ala-asp) box polypeptide 31, TCOF1 - treacher collins-franceschetti syndrome 1, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, ZNF638 - zinc finger protein 638, F11R - f11 receptor, STAMBP - stam binding protein, SCFD1 - sec1 family domain containing 1, CISD1 - cdgsh iron sulfur domain 1, URB1 - urb1 ribosome biogenesis 1 homolog (s. cerevisiae), RAB30 - rab30, member ras oncogene family, FLNB - filamin b, beta, PPFIA1 - protein tyrosine phosphatase, receptor type, f polypeptide (ptprf), interacting protein (liprin), alpha 1, PPP4R2 - protein phosphatase 4, regulatory subunit 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, PPFIBP1 - ptprf interacting protein, binding protein 1 (liprin beta 1), ILF3 - interleukin enhancer binding factor 3, 90kda, ANKRD28 - ankyrin repeat domain 28, NUDCD2 - nudc domain containing 2, CYCS - cytochrome c, somatic, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, RBM19 - rna binding motif protein 19, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, EHBP1 - eh domain binding protein 1, AGPS - alkylglycerone phosphate synthase, TBC1D4 - tbc1 domain family, member 4, TMED5 - transmembrane emp24 protein transport domain containing 5, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, GAR1 - gar1 ribonucleoprotein, KDM3A - lysine (k)-specific demethylase 3a, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, ANAPC1 - anaphase promoting complex subunit 1, TELO2 - telomere maintenance 2, EXOSC1 - exosome component 1, PITRM1 - pitrilysin metallopeptidase 1, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), ATG7 - autophagy related 7, TUBGCP4 - tubulin, gamma complex associated protein 4, HDDC2 - hd domain containing 2, PRRC1 - proline-rich coiled-coil 1, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), SYNE1 - spectrin repeat containing, nuclear envelope 1, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), NUP88 - nucleoporin 88kda, ZBTB8OS - zinc finger and btb domain containing 8 opposite strand, NOM1 - nucleolar protein with mif4g domain 1, NFU1 - nfu1 iron-sulfur cluster scaffold homolog (s. cerevisiae), ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), THOC5 - tho complex 5, FECH - ferrochelatase, CDK6 - cyclin-dependent kinase 6, ADNP - activity-dependent neuroprotector homeobox, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, CDK13 - cyclin-dependent kinase 13, TBCD - tubulin folding cofactor d, FCF1 - fcf1 rrna-processing protein, GNPDA2 - glucosamine-6-phosphate deaminase 2, CCT4 - chaperonin containing tcp1, subunit 4 (delta), TBCA - tubulin folding cofactor a, CUTC - cutc copper transporter, INPP5B - inositol polyphosphate-5-phosphatase, 75kda, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, IRF3 - interferon regulatory factor 3, ARL6IP5 - adp-ribosylation-like factor 6 interacting protein 5, LARP1 - la ribonucleoprotein domain family, member 1, IRF5 - interferon regulatory factor 5, DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, EIF3E - eukaryotic translation initiation factor 3, subunit e, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, TCEA1 - transcription elongation factor a (sii), 1, PURA - purine-rich element binding protein a, SPG11 - spastic paraplegia 11 (autosomal recessive), TDRD7 - tudor domain containing 7, NEK6 - nima-related kinase 6, NOC3L - nucleolar complex associated 3 homolog (s. cerevisiae), PYGB - phosphorylase, glycogen; brain, PCYT2 - phosphate cytidylyltransferase 2, ethanolamine, NUP50 - nucleoporin 50kda, VKORC1L1 - vitamin k epoxide reductase complex, subunit 1-like 1, TRAPPC10 - trafficking protein particle complex 10, CCDC58 - coiled-coil domain containing 58, IDE - insulin-degrading enzyme, USP34 - ubiquitin specific peptidase 34, IFIH1 - interferon induced with helicase c domain 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, SART3 - squamous cell carcinoma antigen recognized by t cells 3, MAD1L1 - mad1 mitotic arrest deficient-like 1 (yeast), CTSC - cathepsin c, RANBP1 - ran binding protein 1, RAP1A - rap1a, member of ras oncogene family, PARVG - parvin, gamma, PUS1 - pseudouridylate synthase 1, NSMAF - neutral sphingomyelinase (n-smase) activation associated factor, GNPAT - glyceronephosphate o-acyltransferase, AKAP8L - a kinase (prka) anchor protein 8-like, ZNF592 - zinc finger protein 592, CYFIP2 - cytoplasmic fmr1 interacting protein 2, RBM26 - rna binding motif protein 26, CLNS1A - chloride channel, nucleotide-sensitive, 1a, RAP1B - rap1b, member of ras oncogene family, GOLPH3 - golgi phosphoprotein 3 (coat-protein), TMX3 - thioredoxin-related transmembrane protein 3, BZW1 - basic leucine zipper and w2 domains 1, RB1 - retinoblastoma 1, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), MRPS14 - mitochondrial ribosomal protein s14, CUL1 - cullin 1, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3]
18 [TMEM30A - transmembrane protein 30a, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, PURA - purine-rich element binding protein a, ERAP1 - endoplasmic reticulum aminopeptidase 1, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, EXOSC5 - exosome component 5, RPL22 - ribosomal protein l22, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, WNK1 - wnk lysine deficient protein kinase 1, PGM1 - phosphoglucomutase 1, OSTF1 - osteoclast stimulating factor 1, F11R - f11 receptor, PGAM1 - phosphoglycerate mutase 1 (brain), GSN - gelsolin, HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PYGB - phosphorylase, glycogen; brain, RAB14 - rab14, member ras oncogene family, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, FTH1 - ferritin, heavy polypeptide 1, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, MANBA - mannosidase, beta a, lysosomal, SURF4 - surfeit 4, TRIM25 - tripartite motif containing 25, ILF3 - interleukin enhancer binding factor 3, 90kda, IRAK4 - interleukin-1 receptor-associated kinase 4, DOCK8 - dedicator of cytokinesis 8, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, SLAMF1 - signaling lymphocytic activation molecule family member 1, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, CD48 - cd48 molecule, PA2G4 - proliferation-associated 2g4, 38kda, KPNB1 - karyopherin (importin) beta 1, TRIM38 - tripartite motif containing 38, EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, CAB39 - calcium binding protein 39, IDE - insulin-degrading enzyme, OTUB1 - otu domain, ubiquitin aldehyde binding 1, PLCG1 - phospholipase c, gamma 1, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), EXOC1 - exocyst complex component 1, IFIH1 - interferon induced with helicase c domain 1, IFIT5 - interferon-induced protein with tetratricopeptide repeats 5, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, CTSC - cathepsin c, WIPF1 - was/wasl interacting protein family, member 1, TTLL12 - tubulin tyrosine ligase-like family, member 12, RNF31 - ring finger protein 31, ATG7 - autophagy related 7, DLG1 - discs, large homolog 1 (drosophila), RAP1A - rap1a, member of ras oncogene family, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, GALNT2 - udp-n-acetyl-alpha-d-galactosamine:polypeptide n-acetylgalactosaminyltransferase 2 (galnac-t2), EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), CYFIP2 - cytoplasmic fmr1 interacting protein 2, WDR1 - wd repeat domain 1, GLG1 - golgi glycoprotein 1, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), RAP1B - rap1b, member of ras oncogene family, EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, RPS6 - ribosomal protein s6, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, STOM - stomatin, CDK6 - cyclin-dependent kinase 6, DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, CAT - catalase, PDAP1 - pdgfa associated protein 1, CDK13 - cyclin-dependent kinase 13, HSPD1 - heat shock 60kda protein 1 (chaperonin), ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, IRF3 - interferon regulatory factor 3, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, CUL1 - cullin 1, ERP44 - endoplasmic reticulum protein 44, PRKCD - protein kinase c, delta, BCL2L1 - bcl2-like 1, IRF5 - interferon regulatory factor 5, NAMPT - nicotinamide phosphoribosyltransferase, PGM2 - phosphoglucomutase 2, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1]
19 [TMEM30A - transmembrane protein 30a, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, ATG7 - autophagy related 7, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, RAP1A - rap1a, member of ras oncogene family, PGM1 - phosphoglucomutase 1, OSTF1 - osteoclast stimulating factor 1, PGAM1 - phosphoglycerate mutase 1 (brain), GSN - gelsolin, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, HMGB1 - high mobility group box 1, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PYGB - phosphorylase, glycogen; brain, RAB14 - rab14, member ras oncogene family, RAP1B - rap1b, member of ras oncogene family, FTH1 - ferritin, heavy polypeptide 1, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, MANBA - mannosidase, beta a, lysosomal, SURF4 - surfeit 4, STOM - stomatin, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, CAT - catalase, CDK13 - cyclin-dependent kinase 13, PDAP1 - pdgfa associated protein 1, PA2G4 - proliferation-associated 2g4, 38kda, KPNB1 - karyopherin (importin) beta 1, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, CAB39 - calcium binding protein 39, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, ERP44 - endoplasmic reticulum protein 44, PRKCD - protein kinase c, delta, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), PGM2 - phosphoglucomutase 2, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, CTSC - cathepsin c]
20 [TMEM30A - transmembrane protein 30a, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, ATG7 - autophagy related 7, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, PGM1 - phosphoglucomutase 1, RAP1A - rap1a, member of ras oncogene family, OSTF1 - osteoclast stimulating factor 1, PGAM1 - phosphoglycerate mutase 1 (brain), EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, GSN - gelsolin, HMGB1 - high mobility group box 1, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PYGB - phosphorylase, glycogen; brain, RAB14 - rab14, member ras oncogene family, RAP1B - rap1b, member of ras oncogene family, FTH1 - ferritin, heavy polypeptide 1, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, MANBA - mannosidase, beta a, lysosomal, SURF4 - surfeit 4, STOM - stomatin, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, SLAMF1 - signaling lymphocytic activation molecule family member 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PDAP1 - pdgfa associated protein 1, CDK13 - cyclin-dependent kinase 13, CAT - catalase, PA2G4 - proliferation-associated 2g4, 38kda, KPNB1 - karyopherin (importin) beta 1, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, CAB39 - calcium binding protein 39, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, ERP44 - endoplasmic reticulum protein 44, PRKCD - protein kinase c, delta, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), PGM2 - phosphoglucomutase 2, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, CTSC - cathepsin c]
21 [AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, TMEM30A - transmembrane protein 30a, ATG7 - autophagy related 7, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, PGM1 - phosphoglucomutase 1, RAP1A - rap1a, member of ras oncogene family, OSTF1 - osteoclast stimulating factor 1, PGAM1 - phosphoglycerate mutase 1 (brain), EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, GSN - gelsolin, HMGB1 - high mobility group box 1, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PYGB - phosphorylase, glycogen; brain, RAB14 - rab14, member ras oncogene family, RAP1B - rap1b, member of ras oncogene family, FTH1 - ferritin, heavy polypeptide 1, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, MANBA - mannosidase, beta a, lysosomal, SURF4 - surfeit 4, STOM - stomatin, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PDAP1 - pdgfa associated protein 1, CDK13 - cyclin-dependent kinase 13, CAT - catalase, PA2G4 - proliferation-associated 2g4, 38kda, KPNB1 - karyopherin (importin) beta 1, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, CAB39 - calcium binding protein 39, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, ERP44 - endoplasmic reticulum protein 44, PRKCD - protein kinase c, delta, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, PGM2 - phosphoglucomutase 2, CTSC - cathepsin c]
22 [ZC3H7B - zinc finger ccch-type containing 7b, CCNK - cyclin k, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, VTA1 - vesicle (multivesicular body) trafficking 1, ERAP1 - endoplasmic reticulum aminopeptidase 1, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, EXOSC5 - exosome component 5, DDX6 - dead (asp-glu-ala-asp) box helicase 6, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, CAPN2 - calpain 2, (m/ii) large subunit, F11R - f11 receptor, HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, NUP50 - nucleoporin 50kda, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, RAB14 - rab14, member ras oncogene family, RPL3 - ribosomal protein l3, UMPS - uridine monophosphate synthetase, DERL1 - derlin 1, ILF3 - interleukin enhancer binding factor 3, 90kda, TRIM25 - tripartite motif containing 25, EIF3F - eukaryotic translation initiation factor 3, subunit f, EIF4H - eukaryotic translation initiation factor 4h, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, SLAMF1 - signaling lymphocytic activation molecule family member 1, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, TNPO1 - transportin 1, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), SUGT1 - sgt1, suppressor of g2 allele of skp1 (s. cerevisiae), KPNB1 - karyopherin (importin) beta 1, IDE - insulin-degrading enzyme, CHMP5 - charged multivesicular body protein 5, PLCG1 - phospholipase c, gamma 1, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, EXOC1 - exocyst complex component 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), IFIH1 - interferon induced with helicase c domain 1, RPL31 - ribosomal protein l31, IFIT5 - interferon-induced protein with tetratricopeptide repeats 5, RPL32 - ribosomal protein l32, CREB1 - camp responsive element binding protein 1, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, WIPF1 - was/wasl interacting protein family, member 1, EIF2D - eukaryotic translation initiation factor 2d, ATG7 - autophagy related 7, RANBP1 - ran binding protein 1, DLG1 - discs, large homolog 1 (drosophila), SEH1L - seh1-like (s. cerevisiae), PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, RPL14 - ribosomal protein l14, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, GLS - glutaminase, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), AKAP8L - a kinase (prka) anchor protein 8-like, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), NUP88 - nucleoporin 88kda, RPS8 - ribosomal protein s8, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), RPS7 - ribosomal protein s7, THOC5 - tho complex 5, STOM - stomatin, CDK6 - cyclin-dependent kinase 6, DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, SUMO1 - small ubiquitin-like modifier 1, RB1 - retinoblastoma 1, CDK13 - cyclin-dependent kinase 13, USP15 - ubiquitin specific peptidase 15, HSPD1 - heat shock 60kda protein 1 (chaperonin), LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, COMT - catechol-o-methyltransferase, IRF3 - interferon regulatory factor 3, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, UBE2I - ubiquitin-conjugating enzyme e2i, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, CUL1 - cullin 1, BCL2L1 - bcl2-like 1, PRKCD - protein kinase c, delta, IRF5 - interferon regulatory factor 5, NAMPT - nicotinamide phosphoribosyltransferase, RPTOR - regulatory associated protein of mtor, complex 1, EPS15 - epidermal growth factor receptor pathway substrate 15]
23 [TMEM30A - transmembrane protein 30a, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, ATG7 - autophagy related 7, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, RAP1A - rap1a, member of ras oncogene family, PGM1 - phosphoglucomutase 1, OSTF1 - osteoclast stimulating factor 1, PGAM1 - phosphoglycerate mutase 1 (brain), GSN - gelsolin, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, HMGB1 - high mobility group box 1, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PYGB - phosphorylase, glycogen; brain, RAB14 - rab14, member ras oncogene family, RAP1B - rap1b, member of ras oncogene family, FTH1 - ferritin, heavy polypeptide 1, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, MANBA - mannosidase, beta a, lysosomal, SURF4 - surfeit 4, STOM - stomatin, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, CAT - catalase, CDK13 - cyclin-dependent kinase 13, PDAP1 - pdgfa associated protein 1, PA2G4 - proliferation-associated 2g4, 38kda, KPNB1 - karyopherin (importin) beta 1, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, CAB39 - calcium binding protein 39, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, ERP44 - endoplasmic reticulum protein 44, PRKCD - protein kinase c, delta, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), PGM2 - phosphoglucomutase 2, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, CTSC - cathepsin c]
24 [TMEM30A - transmembrane protein 30a, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, ATG7 - autophagy related 7, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, RAP1A - rap1a, member of ras oncogene family, PGM1 - phosphoglucomutase 1, OSTF1 - osteoclast stimulating factor 1, PGAM1 - phosphoglycerate mutase 1 (brain), GSN - gelsolin, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, HMGB1 - high mobility group box 1, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PYGB - phosphorylase, glycogen; brain, RAB14 - rab14, member ras oncogene family, RAP1B - rap1b, member of ras oncogene family, FTH1 - ferritin, heavy polypeptide 1, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, MANBA - mannosidase, beta a, lysosomal, SURF4 - surfeit 4, STOM - stomatin, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, CAT - catalase, CDK13 - cyclin-dependent kinase 13, PDAP1 - pdgfa associated protein 1, PA2G4 - proliferation-associated 2g4, 38kda, KPNB1 - karyopherin (importin) beta 1, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, CAB39 - calcium binding protein 39, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, ERP44 - endoplasmic reticulum protein 44, PRKCD - protein kinase c, delta, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), PGM2 - phosphoglucomutase 2, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, CTSC - cathepsin c]
25 [TMEM30A - transmembrane protein 30a, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, ATG7 - autophagy related 7, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, PGM1 - phosphoglucomutase 1, RAP1A - rap1a, member of ras oncogene family, OSTF1 - osteoclast stimulating factor 1, PGAM1 - phosphoglycerate mutase 1 (brain), GSN - gelsolin, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, HMGB1 - high mobility group box 1, FABP5 - fatty acid binding protein 5 (psoriasis-associated), WDR1 - wd repeat domain 1, PYGB - phosphorylase, glycogen; brain, RAB14 - rab14, member ras oncogene family, RAP1B - rap1b, member of ras oncogene family, FTH1 - ferritin, heavy polypeptide 1, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, MANBA - mannosidase, beta a, lysosomal, SURF4 - surfeit 4, STOM - stomatin, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, TMX3 - thioredoxin-related transmembrane protein 3, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PDAP1 - pdgfa associated protein 1, CDK13 - cyclin-dependent kinase 13, CAT - catalase, PA2G4 - proliferation-associated 2g4, 38kda, KPNB1 - karyopherin (importin) beta 1, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, FAM49B - family with sequence similarity 49, member b, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), CAB39 - calcium binding protein 39, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, ERP44 - endoplasmic reticulum protein 44, PRKCD - protein kinase c, delta, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, PGM2 - phosphoglucomutase 2, CTSC - cathepsin c]
26 [AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, TMEM30A - transmembrane protein 30a, ATG7 - autophagy related 7, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, PGM1 - phosphoglucomutase 1, RAP1A - rap1a, member of ras oncogene family, OSTF1 - osteoclast stimulating factor 1, PGAM1 - phosphoglycerate mutase 1 (brain), EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, GSN - gelsolin, HMGB1 - high mobility group box 1, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PYGB - phosphorylase, glycogen; brain, RAB14 - rab14, member ras oncogene family, RAP1B - rap1b, member of ras oncogene family, FTH1 - ferritin, heavy polypeptide 1, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, MANBA - mannosidase, beta a, lysosomal, SURF4 - surfeit 4, STOM - stomatin, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PDAP1 - pdgfa associated protein 1, CDK13 - cyclin-dependent kinase 13, CAT - catalase, PA2G4 - proliferation-associated 2g4, 38kda, KPNB1 - karyopherin (importin) beta 1, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, CAB39 - calcium binding protein 39, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, ERP44 - endoplasmic reticulum protein 44, PRKCD - protein kinase c, delta, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, PGM2 - phosphoglucomutase 2, CTSC - cathepsin c]
27 [CCNK - cyclin k, ZC3H7B - zinc finger ccch-type containing 7b, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, VTA1 - vesicle (multivesicular body) trafficking 1, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, DDX6 - dead (asp-glu-ala-asp) box helicase 6, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, F11R - f11 receptor, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, RPL6 - ribosomal protein l6, NUP50 - nucleoporin 50kda, RPL7 - ribosomal protein l7, RPL3 - ribosomal protein l3, DERL1 - derlin 1, TRIM25 - tripartite motif containing 25, EIF3F - eukaryotic translation initiation factor 3, subunit f, EIF4H - eukaryotic translation initiation factor 4h, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, SLAMF1 - signaling lymphocytic activation molecule family member 1, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, TNPO1 - transportin 1, SUGT1 - sgt1, suppressor of g2 allele of skp1 (s. cerevisiae), KPNB1 - karyopherin (importin) beta 1, IDE - insulin-degrading enzyme, CHMP5 - charged multivesicular body protein 5, PLCG1 - phospholipase c, gamma 1, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, EXOC1 - exocyst complex component 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), IFIH1 - interferon induced with helicase c domain 1, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, CREB1 - camp responsive element binding protein 1, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, ATG7 - autophagy related 7, EIF2D - eukaryotic translation initiation factor 2d, RANBP1 - ran binding protein 1, DLG1 - discs, large homolog 1 (drosophila), PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, SEH1L - seh1-like (s. cerevisiae), PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, RPL14 - ribosomal protein l14, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), AKAP8L - a kinase (prka) anchor protein 8-like, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), NUP88 - nucleoporin 88kda, RPS8 - ribosomal protein s8, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), RPS7 - ribosomal protein s7, THOC5 - tho complex 5, STOM - stomatin, DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, SUMO1 - small ubiquitin-like modifier 1, RB1 - retinoblastoma 1, CDK13 - cyclin-dependent kinase 13, USP15 - ubiquitin specific peptidase 15, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, HSPD1 - heat shock 60kda protein 1 (chaperonin), IRF3 - interferon regulatory factor 3, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, UBE2I - ubiquitin-conjugating enzyme e2i, CUL1 - cullin 1, BCL2L1 - bcl2-like 1, RPTOR - regulatory associated protein of mtor, complex 1, EPS15 - epidermal growth factor receptor pathway substrate 15]
28 [DST - dystonin, NCBP2 - nuclear cap binding protein subunit 2, 20kda, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, SDAD1 - sda1 domain containing 1, F11R - f11 receptor, GSN - gelsolin, SCFD1 - sec1 family domain containing 1, TMED2 - transmembrane emp24 domain trafficking protein 2, HOOK3 - hook microtubule-tethering protein 3, PPFIA1 - protein tyrosine phosphatase, receptor type, f polypeptide (ptprf), interacting protein (liprin), alpha 1, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, DERL1 - derlin 1, BTF3 - basic transcription factor 3, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, XPOT - exportin, trna, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, VTI1A - vesicle transport through interaction with t-snares 1a, CCDC51 - coiled-coil domain containing 51, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, EHBP1 - eh domain binding protein 1, ARCN1 - archain 1, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), PA2G4 - proliferation-associated 2g4, 38kda, AGPS - alkylglycerone phosphate synthase, CAB39 - calcium binding protein 39, GOSR2 - golgi snap receptor complex member 2, TBC1D4 - tbc1 domain family, member 4, TMED5 - transmembrane emp24 protein transport domain containing 5, RAB23 - rab23, member ras oncogene family, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, CREB1 - camp responsive element binding protein 1, SNX1 - sorting nexin 1, MYO6 - myosin vi, PITRM1 - pitrilysin metallopeptidase 1, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, ATG7 - autophagy related 7, EIF2D - eukaryotic translation initiation factor 2d, MCU - mitochondrial calcium uniporter, ARL1 - adp-ribosylation factor-like 1, SNRPE - small nuclear ribonucleoprotein polypeptide e, SEH1L - seh1-like (s. cerevisiae), EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, GLS - glutaminase, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), NUP88 - nucleoporin 88kda, SCO1 - sco1 cytochrome c oxidase assembly protein, GOSR1 - golgi snap receptor complex member 1, OSBPL3 - oxysterol binding protein-like 3, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), THOC5 - tho complex 5, EIF2S2 - eukaryotic translation initiation factor 2, subunit 2 beta, 38kda, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, SNX6 - sorting nexin 6, CDK13 - cyclin-dependent kinase 13, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, HSPD1 - heat shock 60kda protein 1 (chaperonin), CCT4 - chaperonin containing tcp1, subunit 4 (delta), CUTC - cutc copper transporter, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, ARL6IP5 - adp-ribosylation-like factor 6 interacting protein 5, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, STX8 - syntaxin 8, STX18 - syntaxin 18, ERP44 - endoplasmic reticulum protein 44, NEMF - nuclear export mediator factor, HSPA4 - heat shock 70kda protein 4, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, TMEM30A - transmembrane protein 30a, VTA1 - vesicle (multivesicular body) trafficking 1, PURA - purine-rich element binding protein a, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, GAPVD1 - gtpase activating protein and vps9 domains 1, DENND3 - denn/madd domain containing 3, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, QKI - qki, kh domain containing, rna binding, SPG11 - spastic paraplegia 11 (autosomal recessive), RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, PGM1 - phosphoglucomutase 1, WNK1 - wnk lysine deficient protein kinase 1, OSTF1 - osteoclast stimulating factor 1, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, AGK - acylglycerol kinase, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), TBC1D2B - tbc1 domain family, member 2b, PYGB - phosphorylase, glycogen; brain, NUP50 - nucleoporin 50kda, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, RAB14 - rab14, member ras oncogene family, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, FTH1 - ferritin, heavy polypeptide 1, UMPS - uridine monophosphate synthetase, RPL3 - ribosomal protein l3, MANBA - mannosidase, beta a, lysosomal, SURF4 - surfeit 4, TRAPPC10 - trafficking protein particle complex 10, SLAMF1 - signaling lymphocytic activation molecule family member 1, HK2 - hexokinase 2, STX17 - syntaxin 17, TNPO1 - transportin 1, KPNB1 - karyopherin (importin) beta 1, IDE - insulin-degrading enzyme, CHMP5 - charged multivesicular body protein 5, EXOC2 - exocyst complex component 2, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, DOCK7 - dedicator of cytokinesis 7, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), EXOC1 - exocyst complex component 1, TJP2 - tight junction protein 2, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, VDAC2 - voltage-dependent anion channel 2, CTSC - cathepsin c, RANBP1 - ran binding protein 1, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, SCYL2 - scy1-like 2 (s. cerevisiae), RAP1A - rap1a, member of ras oncogene family, GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, RPL14 - ribosomal protein l14, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), GNPAT - glyceronephosphate o-acyltransferase, WDR1 - wd repeat domain 1, CLNS1A - chloride channel, nucleotide-sensitive, 1a, RPS8 - ribosomal protein s8, RPS13 - ribosomal protein s13, RAP1B - rap1b, member of ras oncogene family, RINT1 - rad50 interactor 1, RPS6 - ribosomal protein s6, GOLPH3 - golgi phosphoprotein 3 (coat-protein), RPS7 - ribosomal protein s7, STOM - stomatin, TMX3 - thioredoxin-related transmembrane protein 3, DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, SRP72 - signal recognition particle 72kda, PDAP1 - pdgfa associated protein 1, CAT - catalase, XPO6 - exportin 6, FAM49B - family with sequence similarity 49, member b, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), SAMM50 - samm50 sorting and assembly machinery component, RHOT1 - ras homolog family member t1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, BCL2L1 - bcl2-like 1, PRKCD - protein kinase c, delta, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, RRBP1 - ribosome binding protein 1, PGM2 - phosphoglucomutase 2, EPS15 - epidermal growth factor receptor pathway substrate 15]
29 [TMEM30A - transmembrane protein 30a, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, ATG7 - autophagy related 7, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, RAP1A - rap1a, member of ras oncogene family, PGM1 - phosphoglucomutase 1, OSTF1 - osteoclast stimulating factor 1, PGAM1 - phosphoglycerate mutase 1 (brain), EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, GSN - gelsolin, HMGB1 - high mobility group box 1, FABP5 - fatty acid binding protein 5 (psoriasis-associated), WDR1 - wd repeat domain 1, PYGB - phosphorylase, glycogen; brain, RAB14 - rab14, member ras oncogene family, RAP1B - rap1b, member of ras oncogene family, FTH1 - ferritin, heavy polypeptide 1, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, MANBA - mannosidase, beta a, lysosomal, SURF4 - surfeit 4, STOM - stomatin, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, TMX3 - thioredoxin-related transmembrane protein 3, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, STX17 - syntaxin 17, PDAP1 - pdgfa associated protein 1, CDK13 - cyclin-dependent kinase 13, CAT - catalase, PA2G4 - proliferation-associated 2g4, 38kda, KPNB1 - karyopherin (importin) beta 1, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, FAM49B - family with sequence similarity 49, member b, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), CAB39 - calcium binding protein 39, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, EXOC2 - exocyst complex component 2, ERP44 - endoplasmic reticulum protein 44, PRKCD - protein kinase c, delta, EXOC1 - exocyst complex component 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), PGM2 - phosphoglucomutase 2, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, CTSC - cathepsin c]
30 [ZC3H7B - zinc finger ccch-type containing 7b, CCNK - cyclin k, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, VTA1 - vesicle (multivesicular body) trafficking 1, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, DDX6 - dead (asp-glu-ala-asp) box helicase 6, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, RPL6 - ribosomal protein l6, NUP50 - nucleoporin 50kda, RPL7 - ribosomal protein l7, RPL3 - ribosomal protein l3, DERL1 - derlin 1, TRIM25 - tripartite motif containing 25, EIF3F - eukaryotic translation initiation factor 3, subunit f, EIF4H - eukaryotic translation initiation factor 4h, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, TNPO1 - transportin 1, KPNB1 - karyopherin (importin) beta 1, CHMP5 - charged multivesicular body protein 5, PLCG1 - phospholipase c, gamma 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), IFIH1 - interferon induced with helicase c domain 1, EXOC1 - exocyst complex component 1, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, RPL31 - ribosomal protein l31, CREB1 - camp responsive element binding protein 1, RPL32 - ribosomal protein l32, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, EIF2D - eukaryotic translation initiation factor 2d, ATG7 - autophagy related 7, RANBP1 - ran binding protein 1, DLG1 - discs, large homolog 1 (drosophila), SEH1L - seh1-like (s. cerevisiae), PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, RPL14 - ribosomal protein l14, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), AKAP8L - a kinase (prka) anchor protein 8-like, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), NUP88 - nucleoporin 88kda, RPS8 - ribosomal protein s8, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, SUMO1 - small ubiquitin-like modifier 1, RB1 - retinoblastoma 1, CDK13 - cyclin-dependent kinase 13, USP15 - ubiquitin specific peptidase 15, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, HSPD1 - heat shock 60kda protein 1 (chaperonin), IRF3 - interferon regulatory factor 3, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, UBE2I - ubiquitin-conjugating enzyme e2i, CUL1 - cullin 1, BCL2L1 - bcl2-like 1, RPTOR - regulatory associated protein of mtor, complex 1, EPS15 - epidermal growth factor receptor pathway substrate 15]
31 [CCNK - cyclin k, ZC3H7B - zinc finger ccch-type containing 7b, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, VTA1 - vesicle (multivesicular body) trafficking 1, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, DDX6 - dead (asp-glu-ala-asp) box helicase 6, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, RPL6 - ribosomal protein l6, NUP50 - nucleoporin 50kda, RPL7 - ribosomal protein l7, RPL3 - ribosomal protein l3, DERL1 - derlin 1, TRIM25 - tripartite motif containing 25, EIF3F - eukaryotic translation initiation factor 3, subunit f, EIF4H - eukaryotic translation initiation factor 4h, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, TNPO1 - transportin 1, KPNB1 - karyopherin (importin) beta 1, CHMP5 - charged multivesicular body protein 5, PLCG1 - phospholipase c, gamma 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), IFIH1 - interferon induced with helicase c domain 1, EXOC1 - exocyst complex component 1, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, RPL31 - ribosomal protein l31, CREB1 - camp responsive element binding protein 1, RPL32 - ribosomal protein l32, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, ATG7 - autophagy related 7, EIF2D - eukaryotic translation initiation factor 2d, RANBP1 - ran binding protein 1, DLG1 - discs, large homolog 1 (drosophila), PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, SEH1L - seh1-like (s. cerevisiae), PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, RPL14 - ribosomal protein l14, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), AKAP8L - a kinase (prka) anchor protein 8-like, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), NUP88 - nucleoporin 88kda, RPS8 - ribosomal protein s8, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, SUMO1 - small ubiquitin-like modifier 1, RB1 - retinoblastoma 1, CDK13 - cyclin-dependent kinase 13, USP15 - ubiquitin specific peptidase 15, HSPD1 - heat shock 60kda protein 1 (chaperonin), LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, IRF3 - interferon regulatory factor 3, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, UBE2I - ubiquitin-conjugating enzyme e2i, CUL1 - cullin 1, BCL2L1 - bcl2-like 1, RPTOR - regulatory associated protein of mtor, complex 1, EPS15 - epidermal growth factor receptor pathway substrate 15]
32 [TMEM30A - transmembrane protein 30a, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, ATG7 - autophagy related 7, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, PGM1 - phosphoglucomutase 1, RAP1A - rap1a, member of ras oncogene family, OSTF1 - osteoclast stimulating factor 1, PGAM1 - phosphoglycerate mutase 1 (brain), EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, GSN - gelsolin, HMGB1 - high mobility group box 1, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PYGB - phosphorylase, glycogen; brain, RAB14 - rab14, member ras oncogene family, RAP1B - rap1b, member of ras oncogene family, FTH1 - ferritin, heavy polypeptide 1, RPS6 - ribosomal protein s6, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, MANBA - mannosidase, beta a, lysosomal, SURF4 - surfeit 4, STOM - stomatin, SLAMF1 - signaling lymphocytic activation molecule family member 1, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PDAP1 - pdgfa associated protein 1, CDK13 - cyclin-dependent kinase 13, CAT - catalase, HSPD1 - heat shock 60kda protein 1 (chaperonin), PA2G4 - proliferation-associated 2g4, 38kda, KPNB1 - karyopherin (importin) beta 1, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, CAB39 - calcium binding protein 39, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, ERP44 - endoplasmic reticulum protein 44, PRKCD - protein kinase c, delta, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), PGM2 - phosphoglucomutase 2, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, CTSC - cathepsin c]
33 [TMEM30A - transmembrane protein 30a, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, ATG7 - autophagy related 7, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, PGM1 - phosphoglucomutase 1, RAP1A - rap1a, member of ras oncogene family, OSTF1 - osteoclast stimulating factor 1, PGAM1 - phosphoglycerate mutase 1 (brain), EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, GSN - gelsolin, HMGB1 - high mobility group box 1, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PYGB - phosphorylase, glycogen; brain, RAB14 - rab14, member ras oncogene family, RAP1B - rap1b, member of ras oncogene family, FTH1 - ferritin, heavy polypeptide 1, RPS6 - ribosomal protein s6, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, MANBA - mannosidase, beta a, lysosomal, SURF4 - surfeit 4, STOM - stomatin, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, SLAMF1 - signaling lymphocytic activation molecule family member 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PDAP1 - pdgfa associated protein 1, CDK13 - cyclin-dependent kinase 13, CAT - catalase, HSPD1 - heat shock 60kda protein 1 (chaperonin), PA2G4 - proliferation-associated 2g4, 38kda, KPNB1 - karyopherin (importin) beta 1, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, CAB39 - calcium binding protein 39, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, ERP44 - endoplasmic reticulum protein 44, PRKCD - protein kinase c, delta, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, PGM2 - phosphoglucomutase 2, CTSC - cathepsin c]
34 [DST - dystonin, NCBP2 - nuclear cap binding protein subunit 2, 20kda, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, SDAD1 - sda1 domain containing 1, F11R - f11 receptor, SCFD1 - sec1 family domain containing 1, GSN - gelsolin, TMED2 - transmembrane emp24 domain trafficking protein 2, HOOK3 - hook microtubule-tethering protein 3, PPFIA1 - protein tyrosine phosphatase, receptor type, f polypeptide (ptprf), interacting protein (liprin), alpha 1, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, DERL1 - derlin 1, BTF3 - basic transcription factor 3, XPOT - exportin, trna, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, VTI1A - vesicle transport through interaction with t-snares 1a, CCDC51 - coiled-coil domain containing 51, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, EHBP1 - eh domain binding protein 1, ARCN1 - archain 1, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), PA2G4 - proliferation-associated 2g4, 38kda, AGPS - alkylglycerone phosphate synthase, CAB39 - calcium binding protein 39, GOSR2 - golgi snap receptor complex member 2, TBC1D4 - tbc1 domain family, member 4, TMED5 - transmembrane emp24 protein transport domain containing 5, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, RAB23 - rab23, member ras oncogene family, LIMS1 - lim and senescent cell antigen-like domains 1, CREB1 - camp responsive element binding protein 1, SNX1 - sorting nexin 1, MYO6 - myosin vi, PITRM1 - pitrilysin metallopeptidase 1, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, ATG7 - autophagy related 7, EIF2D - eukaryotic translation initiation factor 2d, MCU - mitochondrial calcium uniporter, ARL1 - adp-ribosylation factor-like 1, SNRPE - small nuclear ribonucleoprotein polypeptide e, SEH1L - seh1-like (s. cerevisiae), GLS - glutaminase, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), NUP88 - nucleoporin 88kda, SCO1 - sco1 cytochrome c oxidase assembly protein, GOSR1 - golgi snap receptor complex member 1, OSBPL3 - oxysterol binding protein-like 3, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), THOC5 - tho complex 5, EIF2S2 - eukaryotic translation initiation factor 2, subunit 2 beta, 38kda, SNX6 - sorting nexin 6, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, CDK13 - cyclin-dependent kinase 13, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, HSPD1 - heat shock 60kda protein 1 (chaperonin), CCT4 - chaperonin containing tcp1, subunit 4 (delta), CUTC - cutc copper transporter, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, ARL6IP5 - adp-ribosylation-like factor 6 interacting protein 5, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, STX8 - syntaxin 8, STX18 - syntaxin 18, ERP44 - endoplasmic reticulum protein 44, NEMF - nuclear export mediator factor, HSPA4 - heat shock 70kda protein 4, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, TMEM30A - transmembrane protein 30a, PURA - purine-rich element binding protein a, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, VTA1 - vesicle (multivesicular body) trafficking 1, QKI - qki, kh domain containing, rna binding, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, DENND3 - denn/madd domain containing 3, GAPVD1 - gtpase activating protein and vps9 domains 1, SPG11 - spastic paraplegia 11 (autosomal recessive), RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, WNK1 - wnk lysine deficient protein kinase 1, PGM1 - phosphoglucomutase 1, OSTF1 - osteoclast stimulating factor 1, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, HMGB1 - high mobility group box 1, AGK - acylglycerol kinase, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), TBC1D2B - tbc1 domain family, member 2b, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), PYGB - phosphorylase, glycogen; brain, NUP50 - nucleoporin 50kda, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, MAP4 - microtubule-associated protein 4, RAB14 - rab14, member ras oncogene family, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, FTH1 - ferritin, heavy polypeptide 1, UMPS - uridine monophosphate synthetase, RPL3 - ribosomal protein l3, MANBA - mannosidase, beta a, lysosomal, SURF4 - surfeit 4, TRAPPC10 - trafficking protein particle complex 10, SLAMF1 - signaling lymphocytic activation molecule family member 1, HK2 - hexokinase 2, STX17 - syntaxin 17, TNPO1 - transportin 1, KPNB1 - karyopherin (importin) beta 1, IDE - insulin-degrading enzyme, EXOC2 - exocyst complex component 2, CHMP5 - charged multivesicular body protein 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, DOCK7 - dedicator of cytokinesis 7, CDC23 - cell division cycle 23, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), EXOC1 - exocyst complex component 1, RPL31 - ribosomal protein l31, TJP2 - tight junction protein 2, RPL32 - ribosomal protein l32, VDAC2 - voltage-dependent anion channel 2, CTSC - cathepsin c, RANBP1 - ran binding protein 1, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, NIPBL - nipped-b homolog (drosophila), SCYL2 - scy1-like 2 (s. cerevisiae), DLG1 - discs, large homolog 1 (drosophila), RAP1A - rap1a, member of ras oncogene family, GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, RPL14 - ribosomal protein l14, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), GNPAT - glyceronephosphate o-acyltransferase, WDR1 - wd repeat domain 1, CLNS1A - chloride channel, nucleotide-sensitive, 1a, RPS8 - ribosomal protein s8, RPS13 - ribosomal protein s13, RAP1B - rap1b, member of ras oncogene family, RPS6 - ribosomal protein s6, RINT1 - rad50 interactor 1, GOLPH3 - golgi phosphoprotein 3 (coat-protein), RPS7 - ribosomal protein s7, STOM - stomatin, TMX3 - thioredoxin-related transmembrane protein 3, DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, SRP72 - signal recognition particle 72kda, CAT - catalase, PDAP1 - pdgfa associated protein 1, XPO6 - exportin 6, FAM49B - family with sequence similarity 49, member b, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), SAMM50 - samm50 sorting and assembly machinery component, RHOT1 - ras homolog family member t1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, PRKCD - protein kinase c, delta, BCL2L1 - bcl2-like 1, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, RRBP1 - ribosome binding protein 1, EPS15 - epidermal growth factor receptor pathway substrate 15, PGM2 - phosphoglucomutase 2]
35 [CCNK - cyclin k, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, NCBP2 - nuclear cap binding protein subunit 2, 20kda, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, EXOSC5 - exosome component 5, RFC3 - replication factor c (activator 1) 3, 38kda, SF3A3 - splicing factor 3a, subunit 3, 60kda, SACM1L - sac1 suppressor of actin mutations 1-like (yeast), SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, GFM2 - g elongation factor, mitochondrial 2, FKBP11 - fk506 binding protein 11, 19 kda, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, RECQL - recq protein-like (dna helicase q1-like), DERL1 - derlin 1, BTF3 - basic transcription factor 3, ACAT1 - acetyl-coa acetyltransferase 1, CPOX - coproporphyrinogen oxidase, LRRC40 - leucine rich repeat containing 40, USP25 - ubiquitin specific peptidase 25, SORD - sorbitol dehydrogenase, OTUB1 - otu domain, ubiquitin aldehyde binding 1, ZNF326 - zinc finger protein 326, PLCG1 - phospholipase c, gamma 1, MORF4L1 - mortality factor 4 like 1, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, CREB1 - camp responsive element binding protein 1, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, SNX1 - sorting nexin 1, THUMPD1 - thump domain containing 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, SGPL1 - sphingosine-1-phosphate lyase 1, SNRPE - small nuclear ribonucleoprotein polypeptide e, ANAPC7 - anaphase promoting complex subunit 7, STK38L - serine/threonine kinase 38 like, SEH1L - seh1-like (s. cerevisiae), PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, GLS - glutaminase, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, GLO1 - glyoxalase i, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), POLR1C - polymerase (rna) i polypeptide c, 30kda, DCAF7 - ddb1 and cul4 associated factor 7, PPA1 - pyrophosphatase (inorganic) 1, ADPGK - adp-dependent glucokinase, SUMO3 - small ubiquitin-like modifier 3, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, TSFM - ts translation elongation factor, mitochondrial, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), MVD - mevalonate (diphospho) decarboxylase, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, COMT - catechol-o-methyltransferase, KYNU - kynureninase, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, TAOK3 - tao kinase 3, ERP44 - endoplasmic reticulum protein 44, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), NAMPT - nicotinamide phosphoribosyltransferase, PARP4 - poly (adp-ribose) polymerase family, member 4, SCPEP1 - serine carboxypeptidase 1, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, QKI - qki, kh domain containing, rna binding, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, UQCRB - ubiquinol-cytochrome c reductase binding protein, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, CAPN2 - calpain 2, (m/ii) large subunit, WNK1 - wnk lysine deficient protein kinase 1, UBE2Z - ubiquitin-conjugating enzyme e2z, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, HMGB2 - high mobility group box 2, AGK - acylglycerol kinase, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PTCD3 - pentatricopeptide repeat domain 3, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, UCK2 - uridine-cytidine kinase 2, PPP5C - protein phosphatase 5, catalytic subunit, USP48 - ubiquitin specific peptidase 48, UMPS - uridine monophosphate synthetase, RPL3 - ribosomal protein l3, CARS - cysteinyl-trna synthetase, IRAK4 - interleukin-1 receptor-associated kinase 4, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), EIF3F - eukaryotic translation initiation factor 3, subunit f, MRPS35 - mitochondrial ribosomal protein s35, MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, RPN1 - ribophorin i, AK4 - adenylate kinase 4, HK2 - hexokinase 2, KPNB1 - karyopherin (importin) beta 1, TRIM38 - tripartite motif containing 38, CDC23 - cell division cycle 23, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, TJP2 - tight junction protein 2, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, DCAKD - dephospho-coa kinase domain containing, TTLL12 - tubulin tyrosine ligase-like family, member 12, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, MRPL13 - mitochondrial ribosomal protein l13, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, SCYL2 - scy1-like 2 (s. cerevisiae), NIPBL - nipped-b homolog (drosophila), GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, GALNT2 - udp-n-acetyl-alpha-d-galactosamine:polypeptide n-acetylgalactosaminyltransferase 2 (galnac-t2), STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), RNMT - rna (guanine-7-) methyltransferase, SNRNP27 - small nuclear ribonucleoprotein 27kda (u4/u6.u5), MFN1 - mitofusin 1, RPS8 - ribosomal protein s8, MRPL42 - mitochondrial ribosomal protein l42, ESD - esterase d, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, NHP2 - nhp2 ribonucleoprotein, UCHL3 - ubiquitin carboxyl-terminal esterase l3 (ubiquitin thiolesterase), SUMO1 - small ubiquitin-like modifier 1, GCDH - glutaryl-coa dehydrogenase, CAT - catalase, USP15 - ubiquitin specific peptidase 15, UGDH - udp-glucose 6-dehydrogenase, SSBP1 - single-stranded dna binding protein 1, mitochondrial, MRPL47 - mitochondrial ribosomal protein l47, UBE2I - ubiquitin-conjugating enzyme e2i, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, PRKCD - protein kinase c, delta, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), RRBP1 - ribosome binding protein 1, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, RRM1 - ribonucleotide reductase m1, ZC3H7B - zinc finger ccch-type containing 7b, ERAP1 - endoplasmic reticulum aminopeptidase 1, DDX6 - dead (asp-glu-ala-asp) box helicase 6, TCOF1 - treacher collins-franceschetti syndrome 1, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, ZNF638 - zinc finger protein 638, STAMBP - stam binding protein, GSPT1 - g1 to s phase transition 1, GSN - gelsolin, URB1 - urb1 ribosome biogenesis 1 homolog (s. cerevisiae), FAM98A - family with sequence similarity 98, member a, ECI1 - enoyl-coa delta isomerase 1, DCK - deoxycytidine kinase, PPP4R2 - protein phosphatase 4, regulatory subunit 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, ILF3 - interleukin enhancer binding factor 3, 90kda, TRIM25 - tripartite motif containing 25, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, CYCS - cytochrome c, somatic, RIF1 - rap1 interacting factor homolog (yeast), PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, RBM19 - rna binding motif protein 19, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, MMS22L - mms22-like, dna repair protein, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, PA2G4 - proliferation-associated 2g4, 38kda, LRRK1 - leucine-rich repeat kinase 1, AGPS - alkylglycerone phosphate synthase, CAB39 - calcium binding protein 39, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, RAB23 - rab23, member ras oncogene family, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, GAR1 - gar1 ribonucleoprotein, KDM3A - lysine (k)-specific demethylase 3a, ANAPC1 - anaphase promoting complex subunit 1, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, MRPL3 - mitochondrial ribosomal protein l3, TELO2 - telomere maintenance 2, EXOSC1 - exosome component 1, PITRM1 - pitrilysin metallopeptidase 1, RNF31 - ring finger protein 31, ATG7 - autophagy related 7, MCM7 - minichromosome maintenance complex component 7, KDELC2 - kdel (lys-asp-glu-leu) containing 2, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, PELO - pelota homolog (drosophila), ZBTB8OS - zinc finger and btb domain containing 8 opposite strand, NUP88 - nucleoporin 88kda, NFU1 - nfu1 iron-sulfur cluster scaffold homolog (s. cerevisiae), WDHD1 - wd repeat and hmg-box dna binding protein 1, PPIL3 - peptidylprolyl isomerase (cyclophilin)-like 3, HADH - hydroxyacyl-coa dehydrogenase, OSBPL3 - oxysterol binding protein-like 3, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), THOC5 - tho complex 5, FECH - ferrochelatase, CDK6 - cyclin-dependent kinase 6, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, CDK13 - cyclin-dependent kinase 13, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, FCF1 - fcf1 rrna-processing protein, GNPDA2 - glucosamine-6-phosphate deaminase 2, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, INPP5B - inositol polyphosphate-5-phosphatase, 75kda, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, RPL7L1 - ribosomal protein l7-like 1, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, NEMF - nuclear export mediator factor, DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), EIF3E - eukaryotic translation initiation factor 3, subunit e, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, TCEA1 - transcription elongation factor a (sii), 1, PURA - purine-rich element binding protein a, DENND3 - denn/madd domain containing 3, NEK6 - nima-related kinase 6, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, NOC3L - nucleolar complex associated 3 homolog (s. cerevisiae), PYGB - phosphorylase, glycogen; brain, PCYT2 - phosphate cytidylyltransferase 2, ethanolamine, NUP50 - nucleoporin 50kda, RAB14 - rab14, member ras oncogene family, PHKB - phosphorylase kinase, beta, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, MANBA - mannosidase, beta a, lysosomal, VKORC1L1 - vitamin k epoxide reductase complex, subunit 1-like 1, MAN2A1 - mannosidase, alpha, class 2a, member 1, USP8 - ubiquitin specific peptidase 8, MBNL1 - muscleblind-like splicing regulator 1, METTL15 - methyltransferase like 15, DUT - deoxyuridine triphosphatase, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, MAT2A - methionine adenosyltransferase ii, alpha, IDE - insulin-degrading enzyme, MCM3 - minichromosome maintenance complex component 3, IFIH1 - interferon induced with helicase c domain 1, USP34 - ubiquitin specific peptidase 34, MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, SART3 - squamous cell carcinoma antigen recognized by t cells 3, CTSC - cathepsin c, SCAF8 - sr-related ctd-associated factor 8, LSS - lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase), UBA6 - ubiquitin-like modifier activating enzyme 6, GRK6 - g protein-coupled receptor kinase 6, DLG1 - discs, large homolog 1 (drosophila), PUS1 - pseudouridylate synthase 1, NSMAF - neutral sphingomyelinase (n-smase) activation associated factor, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, RPL14 - ribosomal protein l14, AKAP8L - a kinase (prka) anchor protein 8-like, GNPAT - glyceronephosphate o-acyltransferase, CYFIP2 - cytoplasmic fmr1 interacting protein 2, NUB1 - negative regulator of ubiquitin-like proteins 1, RBM26 - rna binding motif protein 26, DGUOK - deoxyguanosine kinase, UBA3 - ubiquitin-like modifier activating enzyme 3, GOLPH3 - golgi phosphoprotein 3 (coat-protein), TMX3 - thioredoxin-related transmembrane protein 3, ACOT7 - acyl-coa thioesterase 7, ADI1 - acireductone dioxygenase 1, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), MRPS14 - mitochondrial ribosomal protein s14, RHOT1 - ras homolog family member t1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, CUL1 - cullin 1, RSL1D1 - ribosomal l1 domain containing 1, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, PGM2 - phosphoglucomutase 2, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
36 [CCNK - cyclin k, ZC3H7B - zinc finger ccch-type containing 7b, NCBP2 - nuclear cap binding protein subunit 2, 20kda, ERAP1 - endoplasmic reticulum aminopeptidase 1, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, EXOSC5 - exosome component 5, DDX6 - dead (asp-glu-ala-asp) box helicase 6, TCOF1 - treacher collins-franceschetti syndrome 1, RFC3 - replication factor c (activator 1) 3, 38kda, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, ZNF638 - zinc finger protein 638, SF3A3 - splicing factor 3a, subunit 3, 60kda, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, GSPT1 - g1 to s phase transition 1, URB1 - urb1 ribosome biogenesis 1 homolog (s. cerevisiae), NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, GFM2 - g elongation factor, mitochondrial 2, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, RECQL - recq protein-like (dna helicase q1-like), DCK - deoxycytidine kinase, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, PPP4R2 - protein phosphatase 4, regulatory subunit 2, TRIM25 - tripartite motif containing 25, BTF3 - basic transcription factor 3, RIF1 - rap1 interacting factor homolog (yeast), ACAT1 - acetyl-coa acetyltransferase 1, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, RBM19 - rna binding motif protein 19, CPOX - coproporphyrinogen oxidase, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), MMS22L - mms22-like, dna repair protein, PA2G4 - proliferation-associated 2g4, 38kda, OTUB1 - otu domain, ubiquitin aldehyde binding 1, ZNF326 - zinc finger protein 326, RAB23 - rab23, member ras oncogene family, MORF4L1 - mortality factor 4 like 1, GAR1 - gar1 ribonucleoprotein, CREB1 - camp responsive element binding protein 1, MRPL3 - mitochondrial ribosomal protein l3, TELO2 - telomere maintenance 2, EXOSC1 - exosome component 1, THUMPD1 - thump domain containing 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, SGPL1 - sphingosine-1-phosphate lyase 1, ATG7 - autophagy related 7, MCM7 - minichromosome maintenance complex component 7, SNRPE - small nuclear ribonucleoprotein polypeptide e, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, GLO1 - glyoxalase i, PELO - pelota homolog (drosophila), ZBTB8OS - zinc finger and btb domain containing 8 opposite strand, WDHD1 - wd repeat and hmg-box dna binding protein 1, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), PPIL3 - peptidylprolyl isomerase (cyclophilin)-like 3, POLR1C - polymerase (rna) i polypeptide c, 30kda, ADPGK - adp-dependent glucokinase, PPA1 - pyrophosphatase (inorganic) 1, THOC5 - tho complex 5, FECH - ferrochelatase, MRPL2 - mitochondrial ribosomal protein l2, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, CDK13 - cyclin-dependent kinase 13, HSPD1 - heat shock 60kda protein 1 (chaperonin), GNPDA2 - glucosamine-6-phosphate deaminase 2, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, FCF1 - fcf1 rrna-processing protein, MVD - mevalonate (diphospho) decarboxylase, RPL7L1 - ribosomal protein l7-like 1, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, KYNU - kynureninase, TAOK3 - tao kinase 3, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), NAMPT - nicotinamide phosphoribosyltransferase, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, EIF3E - eukaryotic translation initiation factor 3, subunit e, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, TCEA1 - transcription elongation factor a (sii), 1, PARP4 - poly (adp-ribose) polymerase family, member 4, PURA - purine-rich element binding protein a, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, UQCRB - ubiquinol-cytochrome c reductase binding protein, QKI - qki, kh domain containing, rna binding, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, HMGB2 - high mobility group box 2, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, NOC3L - nucleolar complex associated 3 homolog (s. cerevisiae), AGK - acylglycerol kinase, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, PTCD3 - pentatricopeptide repeat domain 3, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, UCK2 - uridine-cytidine kinase 2, PPP5C - protein phosphatase 5, catalytic subunit, RPL3 - ribosomal protein l3, UMPS - uridine monophosphate synthetase, CARS - cysteinyl-trna synthetase, MBNL1 - muscleblind-like splicing regulator 1, METTL15 - methyltransferase like 15, DUT - deoxyuridine triphosphatase, MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, HK2 - hexokinase 2, AK4 - adenylate kinase 4, KPNB1 - karyopherin (importin) beta 1, IDE - insulin-degrading enzyme, MCM3 - minichromosome maintenance complex component 3, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, TJP2 - tight junction protein 2, RPL31 - ribosomal protein l31, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, RPL32 - ribosomal protein l32, SART3 - squamous cell carcinoma antigen recognized by t cells 3, DCAKD - dephospho-coa kinase domain containing, SCAF8 - sr-related ctd-associated factor 8, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, MRPL13 - mitochondrial ribosomal protein l13, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, DLG1 - discs, large homolog 1 (drosophila), NIPBL - nipped-b homolog (drosophila), PUS1 - pseudouridylate synthase 1, NSMAF - neutral sphingomyelinase (n-smase) activation associated factor, RPL14 - ribosomal protein l14, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), AKAP8L - a kinase (prka) anchor protein 8-like, SNRNP27 - small nuclear ribonucleoprotein 27kda (u4/u6.u5), MFN1 - mitofusin 1, RNMT - rna (guanine-7-) methyltransferase, RBM26 - rna binding motif protein 26, DGUOK - deoxyguanosine kinase, RPS8 - ribosomal protein s8, MRPL42 - mitochondrial ribosomal protein l42, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, NHP2 - nhp2 ribonucleoprotein, RPS7 - ribosomal protein s7, SUMO1 - small ubiquitin-like modifier 1, ACOT7 - acyl-coa thioesterase 7, GCDH - glutaryl-coa dehydrogenase, UGDH - udp-glucose 6-dehydrogenase, SSBP1 - single-stranded dna binding protein 1, mitochondrial, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), MRPS14 - mitochondrial ribosomal protein s14, MRPL47 - mitochondrial ribosomal protein l47, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, RSL1D1 - ribosomal l1 domain containing 1, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, RRBP1 - ribosome binding protein 1, PGM2 - phosphoglucomutase 2, RRM1 - ribonucleotide reductase m1, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
37 [AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, DST - dystonin, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, VTA1 - vesicle (multivesicular body) trafficking 1, PURA - purine-rich element binding protein a, NCBP2 - nuclear cap binding protein subunit 2, 20kda, DENND3 - denn/madd domain containing 3, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, SPG11 - spastic paraplegia 11 (autosomal recessive), RPL23A - ribosomal protein l23a, SDAD1 - sda1 domain containing 1, RPL22 - ribosomal protein l22, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, SCFD1 - sec1 family domain containing 1, AGK - acylglycerol kinase, RPL13 - ribosomal protein l13, TBC1D2B - tbc1 domain family, member 2b, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), TMED2 - transmembrane emp24 domain trafficking protein 2, NUP50 - nucleoporin 50kda, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, HOOK3 - hook microtubule-tethering protein 3, RAB14 - rab14, member ras oncogene family, RPL3 - ribosomal protein l3, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, DERL1 - derlin 1, XPOT - exportin, trna, TRAPPC10 - trafficking protein particle complex 10, VTI1A - vesicle transport through interaction with t-snares 1a, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, TNPO1 - transportin 1, STX17 - syntaxin 17, ARCN1 - archain 1, KPNB1 - karyopherin (importin) beta 1, AGPS - alkylglycerone phosphate synthase, GOSR2 - golgi snap receptor complex member 2, IDE - insulin-degrading enzyme, TBC1D4 - tbc1 domain family, member 4, CHMP5 - charged multivesicular body protein 5, TMED5 - transmembrane emp24 protein transport domain containing 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, DOCK7 - dedicator of cytokinesis 7, RAB23 - rab23, member ras oncogene family, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, MYO6 - myosin vi, SNX1 - sorting nexin 1, CTSC - cathepsin c, PITRM1 - pitrilysin metallopeptidase 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, EIF2D - eukaryotic translation initiation factor 2d, RANBP1 - ran binding protein 1, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, ARL1 - adp-ribosylation factor-like 1, SNRPE - small nuclear ribonucleoprotein polypeptide e, SCYL2 - scy1-like 2 (s. cerevisiae), GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, SEH1L - seh1-like (s. cerevisiae), RPL14 - ribosomal protein l14, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), GNPAT - glyceronephosphate o-acyltransferase, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), NUP88 - nucleoporin 88kda, RPS8 - ribosomal protein s8, GOSR1 - golgi snap receptor complex member 1, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, THOC5 - tho complex 5, SNX6 - sorting nexin 6, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, SRP72 - signal recognition particle 72kda, CAT - catalase, HSPD1 - heat shock 60kda protein 1 (chaperonin), XPO6 - exportin 6, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), SAMM50 - samm50 sorting and assembly machinery component, RHOT1 - ras homolog family member t1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, STX8 - syntaxin 8, STX18 - syntaxin 18, NEMF - nuclear export mediator factor, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, HSPA4 - heat shock 70kda protein 4, EPS15 - epidermal growth factor receptor pathway substrate 15]
38 [TMEM30A - transmembrane protein 30a, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, ATG7 - autophagy related 7, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, PGM1 - phosphoglucomutase 1, RAP1A - rap1a, member of ras oncogene family, OSTF1 - osteoclast stimulating factor 1, PGAM1 - phosphoglycerate mutase 1 (brain), EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, GSN - gelsolin, HMGB1 - high mobility group box 1, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PYGB - phosphorylase, glycogen; brain, RAB14 - rab14, member ras oncogene family, RAP1B - rap1b, member of ras oncogene family, FTH1 - ferritin, heavy polypeptide 1, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, MANBA - mannosidase, beta a, lysosomal, SURF4 - surfeit 4, STOM - stomatin, SLAMF1 - signaling lymphocytic activation molecule family member 1, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PDAP1 - pdgfa associated protein 1, CDK13 - cyclin-dependent kinase 13, CAT - catalase, PA2G4 - proliferation-associated 2g4, 38kda, KPNB1 - karyopherin (importin) beta 1, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, CAB39 - calcium binding protein 39, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, ERP44 - endoplasmic reticulum protein 44, PRKCD - protein kinase c, delta, NAMPT - nicotinamide phosphoribosyltransferase, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, PGM2 - phosphoglucomutase 2, CTSC - cathepsin c]
39 [PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, ATG7 - autophagy related 7, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PGM1 - phosphoglucomutase 1, OSTF1 - osteoclast stimulating factor 1, PDAP1 - pdgfa associated protein 1, CDK13 - cyclin-dependent kinase 13, CAT - catalase, PGAM1 - phosphoglycerate mutase 1 (brain), EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, GSN - gelsolin, HMGB1 - high mobility group box 1, KPNB1 - karyopherin (importin) beta 1, CAB39 - calcium binding protein 39, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, FTH1 - ferritin, heavy polypeptide 1, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, PGM2 - phosphoglucomutase 2]
40 [CCNK - cyclin k, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, NCBP2 - nuclear cap binding protein subunit 2, 20kda, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, TCOF1 - treacher collins-franceschetti syndrome 1, RFC3 - replication factor c (activator 1) 3, 38kda, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, SACM1L - sac1 suppressor of actin mutations 1-like (yeast), GSPT1 - g1 to s phase transition 1, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, GFM2 - g elongation factor, mitochondrial 2, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, DCK - deoxycytidine kinase, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, BTF3 - basic transcription factor 3, TRIM25 - tripartite motif containing 25, ACAT1 - acetyl-coa acetyltransferase 1, CPOX - coproporphyrinogen oxidase, AGPS - alkylglycerone phosphate synthase, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, GAR1 - gar1 ribonucleoprotein, CREB1 - camp responsive element binding protein 1, KDM3A - lysine (k)-specific demethylase 3a, MRPL3 - mitochondrial ribosomal protein l3, TELO2 - telomere maintenance 2, SGPL1 - sphingosine-1-phosphate lyase 1, MCM7 - minichromosome maintenance complex component 7, SNRPE - small nuclear ribonucleoprotein polypeptide e, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, GLS - glutaminase, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, WDHD1 - wd repeat and hmg-box dna binding protein 1, OSBPL3 - oxysterol binding protein-like 3, POLR1C - polymerase (rna) i polypeptide c, 30kda, ADPGK - adp-dependent glucokinase, FECH - ferrochelatase, MRPL2 - mitochondrial ribosomal protein l2, TSFM - ts translation elongation factor, mitochondrial, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, CDK13 - cyclin-dependent kinase 13, MVD - mevalonate (diphospho) decarboxylase, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, GNPDA2 - glucosamine-6-phosphate deaminase 2, KYNU - kynureninase, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, NEMF - nuclear export mediator factor, NAMPT - nicotinamide phosphoribosyltransferase, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, TCEA1 - transcription elongation factor a (sii), 1, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, QKI - qki, kh domain containing, rna binding, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, AGK - acylglycerol kinase, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PTCD3 - pentatricopeptide repeat domain 3, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, PCYT2 - phosphate cytidylyltransferase 2, ethanolamine, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, RAB14 - rab14, member ras oncogene family, UCK2 - uridine-cytidine kinase 2, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, UMPS - uridine monophosphate synthetase, RPL3 - ribosomal protein l3, PPP5C - protein phosphatase 5, catalytic subunit, MRPS35 - mitochondrial ribosomal protein s35, DUT - deoxyuridine triphosphatase, MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, AK4 - adenylate kinase 4, HK2 - hexokinase 2, MAT2A - methionine adenosyltransferase ii, alpha, MCM3 - minichromosome maintenance complex component 3, RPL31 - ribosomal protein l31, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, RPL32 - ribosomal protein l32, DCAKD - dephospho-coa kinase domain containing, SCAF8 - sr-related ctd-associated factor 8, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, LSS - lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase), MRPL13 - mitochondrial ribosomal protein l13, RPL14 - ribosomal protein l14, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), GNPAT - glyceronephosphate o-acyltransferase, RNMT - rna (guanine-7-) methyltransferase, DGUOK - deoxyguanosine kinase, RPS8 - ribosomal protein s8, ESD - esterase d, MRPL42 - mitochondrial ribosomal protein l42, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, GOLPH3 - golgi phosphoprotein 3 (coat-protein), NHP2 - nhp2 ribonucleoprotein, RPS7 - ribosomal protein s7, ACOT7 - acyl-coa thioesterase 7, GCDH - glutaryl-coa dehydrogenase, UGDH - udp-glucose 6-dehydrogenase, ADI1 - acireductone dioxygenase 1, SSBP1 - single-stranded dna binding protein 1, mitochondrial, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), MRPS14 - mitochondrial ribosomal protein s14, MRPL47 - mitochondrial ribosomal protein l47, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, AK3 - adenylate kinase 3, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, RRBP1 - ribosome binding protein 1, PGM2 - phosphoglucomutase 2, RRM1 - ribonucleotide reductase m1, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
41 [AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, PURA - purine-rich element binding protein a, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, RPL23A - ribosomal protein l23a, SDAD1 - sda1 domain containing 1, RPL22 - ribosomal protein l22, SCFD1 - sec1 family domain containing 1, AGK - acylglycerol kinase, RPL13 - ribosomal protein l13, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), TBC1D2B - tbc1 domain family, member 2b, TMED2 - transmembrane emp24 domain trafficking protein 2, RPL6 - ribosomal protein l6, NUP50 - nucleoporin 50kda, RPL7 - ribosomal protein l7, RAB14 - rab14, member ras oncogene family, RPL3 - ribosomal protein l3, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, DERL1 - derlin 1, VTI1A - vesicle transport through interaction with t-snares 1a, STX17 - syntaxin 17, TNPO1 - transportin 1, ARCN1 - archain 1, KPNB1 - karyopherin (importin) beta 1, AGPS - alkylglycerone phosphate synthase, GOSR2 - golgi snap receptor complex member 2, TBC1D4 - tbc1 domain family, member 4, IDE - insulin-degrading enzyme, TMED5 - transmembrane emp24 protein transport domain containing 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, RAB23 - rab23, member ras oncogene family, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, SNX1 - sorting nexin 1, MYO6 - myosin vi, PITRM1 - pitrilysin metallopeptidase 1, EIF2D - eukaryotic translation initiation factor 2d, ARL1 - adp-ribosylation factor-like 1, GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, RPL14 - ribosomal protein l14, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), GNPAT - glyceronephosphate o-acyltransferase, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), NUP88 - nucleoporin 88kda, RPS8 - ribosomal protein s8, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, SNX6 - sorting nexin 6, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, SRP72 - signal recognition particle 72kda, CAT - catalase, HSPD1 - heat shock 60kda protein 1 (chaperonin), XPO6 - exportin 6, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, SAMM50 - samm50 sorting and assembly machinery component, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, STX8 - syntaxin 8, STX18 - syntaxin 18, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, HSPA4 - heat shock 70kda protein 4]
42 [DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, DST - dystonin, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, SACM1L - sac1 suppressor of actin mutations 1-like (yeast), NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, NDUFA5 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, FKBP11 - fk506 binding protein 11, 19 kda, TMED2 - transmembrane emp24 domain trafficking protein 2, RECQL - recq protein-like (dna helicase q1-like), YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, DERL1 - derlin 1, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, DOCK8 - dedicator of cytokinesis 8, CPOX - coproporphyrinogen oxidase, LRRC40 - leucine rich repeat containing 40, EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, GOSR2 - golgi snap receptor complex member 2, SORD - sorbitol dehydrogenase, PLCG1 - phospholipase c, gamma 1, LIMS1 - lim and senescent cell antigen-like domains 1, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, SNX1 - sorting nexin 1, MYO6 - myosin vi, SGPL1 - sphingosine-1-phosphate lyase 1, STK38L - serine/threonine kinase 38 like, SEH1L - seh1-like (s. cerevisiae), PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, GLO1 - glyoxalase i, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), GLG1 - golgi glycoprotein 1, GOSR1 - golgi snap receptor complex member 1, ADPGK - adp-dependent glucokinase, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, SNX6 - sorting nexin 6, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, HSPD1 - heat shock 60kda protein 1 (chaperonin), ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, COMT - catechol-o-methyltransferase, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, TAOK3 - tao kinase 3, ERP44 - endoplasmic reticulum protein 44, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), NAMPT - nicotinamide phosphoribosyltransferase, PARP4 - poly (adp-ribose) polymerase family, member 4, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, TMEM30A - transmembrane protein 30a, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, UQCRB - ubiquinol-cytochrome c reductase binding protein, CAPN2 - calpain 2, (m/ii) large subunit, WNK1 - wnk lysine deficient protein kinase 1, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, AGK - acylglycerol kinase, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), PTCD3 - pentatricopeptide repeat domain 3, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, HOMER1 - homer homolog 1 (drosophila), PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, PPP5C - protein phosphatase 5, catalytic subunit, SURF4 - surfeit 4, IRAK4 - interleukin-1 receptor-associated kinase 4, EIF3F - eukaryotic translation initiation factor 3, subunit f, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), MRPS35 - mitochondrial ribosomal protein s35, MRPL1 - mitochondrial ribosomal protein l1, RPN1 - ribophorin i, MRPL9 - mitochondrial ribosomal protein l9, STX17 - syntaxin 17, HK2 - hexokinase 2, KPNB1 - karyopherin (importin) beta 1, EXOC2 - exocyst complex component 2, ABCF2 - atp-binding cassette, sub-family f (gcn20), member 2, EXOC1 - exocyst complex component 1, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, IFIT5 - interferon-induced protein with tetratricopeptide repeats 5, TJP2 - tight junction protein 2, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, DCAKD - dephospho-coa kinase domain containing, VDAC2 - voltage-dependent anion channel 2, MRPL13 - mitochondrial ribosomal protein l13, SCYL2 - scy1-like 2 (s. cerevisiae), GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, GALNT2 - udp-n-acetyl-alpha-d-galactosamine:polypeptide n-acetylgalactosaminyltransferase 2 (galnac-t2), TMEM70 - transmembrane protein 70, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), MFN1 - mitofusin 1, STAT6 - signal transducer and activator of transcription 6, interleukin-4 induced, WDR1 - wd repeat domain 1, LARP4B - la ribonucleoprotein domain family, member 4b, RPS8 - ribosomal protein s8, MRPL42 - mitochondrial ribosomal protein l42, RPS13 - ribosomal protein s13, RINT1 - rad50 interactor 1, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, RIC8A - ric8 guanine nucleotide exchange factor a, STOM - stomatin, SUMO1 - small ubiquitin-like modifier 1, CISD2 - cdgsh iron sulfur domain 2, CAT - catalase, XPO6 - exportin 6, NDUFAF4 - nadh dehydrogenase (ubiquinone) complex i, assembly factor 4, SAMM50 - samm50 sorting and assembly machinery component, ANXA4 - annexin a4, TUBGCP3 - tubulin, gamma complex associated protein 3, MRPL47 - mitochondrial ribosomal protein l47, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, AIDA - axin interactor, dorsalization associated, PRKCD - protein kinase c, delta, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, RRBP1 - ribosome binding protein 1, EPS15 - epidermal growth factor receptor pathway substrate 15, ERAP1 - endoplasmic reticulum aminopeptidase 1, DDX6 - dead (asp-glu-ala-asp) box helicase 6, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, F11R - f11 receptor, STAMBP - stam binding protein, GSN - gelsolin, SCFD1 - sec1 family domain containing 1, CISD1 - cdgsh iron sulfur domain 1, RAB30 - rab30, member ras oncogene family, FLNB - filamin b, beta, PPFIBP1 - ptprf interacting protein, binding protein 1 (liprin beta 1), IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, DBN1 - drebrin 1, ILF3 - interleukin enhancer binding factor 3, 90kda, ANKRD28 - ankyrin repeat domain 28, RIF1 - rap1 interacting factor homolog (yeast), PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, CYCS - cytochrome c, somatic, VTI1A - vesicle transport through interaction with t-snares 1a, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, RBM19 - rna binding motif protein 19, EHBP1 - eh domain binding protein 1, ARCN1 - archain 1, CD48 - cd48 molecule, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, PA2G4 - proliferation-associated 2g4, 38kda, AGPS - alkylglycerone phosphate synthase, TMED5 - transmembrane emp24 protein transport domain containing 5, KIAA2013 - kiaa2013, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, RAB23 - rab23, member ras oncogene family, KDM3A - lysine (k)-specific demethylase 3a, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, MRPL3 - mitochondrial ribosomal protein l3, TELO2 - telomere maintenance 2, LRRC57 - leucine rich repeat containing 57, MCM7 - minichromosome maintenance complex component 7, MCU - mitochondrial calcium uniporter, TUBGCP4 - tubulin, gamma complex associated protein 4, ARL1 - adp-ribosylation factor-like 1, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, SYNE1 - spectrin repeat containing, nuclear envelope 1, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), OSBPL3 - oxysterol binding protein-like 3, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), FECH - ferrochelatase, MRPL2 - mitochondrial ribosomal protein l2, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, INPP5B - inositol polyphosphate-5-phosphatase, 75kda, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, ARL6IP5 - adp-ribosylation-like factor 6 interacting protein 5, STX18 - syntaxin 18, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, LARP1 - la ribonucleoprotein domain family, member 1, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, EIF3E - eukaryotic translation initiation factor 3, subunit e, RPTOR - regulatory associated protein of mtor, complex 1, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, QSOX2 - quiescin q6 sulfhydryl oxidase 2, LYRM7 - lyr motif containing 7, VTA1 - vesicle (multivesicular body) trafficking 1, SERBP1 - serpine1 mrna binding protein 1, GAPVD1 - gtpase activating protein and vps9 domains 1, SPG11 - spastic paraplegia 11 (autosomal recessive), ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, PYGB - phosphorylase, glycogen; brain, PCYT2 - phosphate cytidylyltransferase 2, ethanolamine, NUP50 - nucleoporin 50kda, MAP4 - microtubule-associated protein 4, RAB14 - rab14, member ras oncogene family, PHKB - phosphorylase kinase, beta, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, MANBA - mannosidase, beta a, lysosomal, VKORC1L1 - vitamin k epoxide reductase complex, subunit 1-like 1, MAN2A1 - mannosidase, alpha, class 2a, member 1, TRAPPC10 - trafficking protein particle complex 10, EIF4H - eukaryotic translation initiation factor 4h, SLAMF1 - signaling lymphocytic activation molecule family member 1, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, IDE - insulin-degrading enzyme, MCM3 - minichromosome maintenance complex component 3, CHMP5 - charged multivesicular body protein 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), CTSC - cathepsin c, LSS - lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase), GRK6 - g protein-coupled receptor kinase 6, DLG1 - discs, large homolog 1 (drosophila), RAP1A - rap1a, member of ras oncogene family, PARVG - parvin, gamma, RPL14 - ribosomal protein l14, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, GNPAT - glyceronephosphate o-acyltransferase, DHRS7 - dehydrogenase/reductase (sdr family) member 7, CYFIP2 - cytoplasmic fmr1 interacting protein 2, CLNS1A - chloride channel, nucleotide-sensitive, 1a, TRAPPC9 - trafficking protein particle complex 9, RAP1B - rap1b, member of ras oncogene family, GOLPH3 - golgi phosphoprotein 3 (coat-protein), TMX3 - thioredoxin-related transmembrane protein 3, BZW1 - basic leucine zipper and w2 domains 1, PDAP1 - pdgfa associated protein 1, CBX3 - chromobox homolog 3, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, FAM49B - family with sequence similarity 49, member b, ADI1 - acireductone dioxygenase 1, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, CAPRIN1 - cell cycle associated protein 1, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), MRPS14 - mitochondrial ribosomal protein s14, RHOT1 - ras homolog family member t1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, CUL1 - cullin 1, RSL1D1 - ribosomal l1 domain containing 1, BCL2L1 - bcl2-like 1]
43 [TMEM30A - transmembrane protein 30a, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, VTA1 - vesicle (multivesicular body) trafficking 1, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, DENND3 - denn/madd domain containing 3, GAPVD1 - gtpase activating protein and vps9 domains 1, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, SPG11 - spastic paraplegia 11 (autosomal recessive), BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, PGM1 - phosphoglucomutase 1, OSTF1 - osteoclast stimulating factor 1, PGAM1 - phosphoglycerate mutase 1 (brain), GSN - gelsolin, SCFD1 - sec1 family domain containing 1, HMGB1 - high mobility group box 1, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PYGB - phosphorylase, glycogen; brain, TMED2 - transmembrane emp24 domain trafficking protein 2, HOOK3 - hook microtubule-tethering protein 3, RAB14 - rab14, member ras oncogene family, FTH1 - ferritin, heavy polypeptide 1, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, MANBA - mannosidase, beta a, lysosomal, SURF4 - surfeit 4, TRAPPC10 - trafficking protein particle complex 10, SLAMF1 - signaling lymphocytic activation molecule family member 1, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, VTI1A - vesicle transport through interaction with t-snares 1a, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, STX17 - syntaxin 17, ARCN1 - archain 1, EHBP1 - eh domain binding protein 1, PA2G4 - proliferation-associated 2g4, 38kda, KPNB1 - karyopherin (importin) beta 1, GOSR2 - golgi snap receptor complex member 2, CAB39 - calcium binding protein 39, TBC1D4 - tbc1 domain family, member 4, CHMP5 - charged multivesicular body protein 5, EXOC2 - exocyst complex component 2, TMED5 - transmembrane emp24 protein transport domain containing 5, EXOC1 - exocyst complex component 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), SNX1 - sorting nexin 1, MYO6 - myosin vi, CTSC - cathepsin c, ATG7 - autophagy related 7, ARL1 - adp-ribosylation factor-like 1, SCYL2 - scy1-like 2 (s. cerevisiae), GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, RAP1A - rap1a, member of ras oncogene family, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), WDR1 - wd repeat domain 1, GOSR1 - golgi snap receptor complex member 1, RAP1B - rap1b, member of ras oncogene family, RINT1 - rad50 interactor 1, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), GOLPH3 - golgi phosphoprotein 3 (coat-protein), STOM - stomatin, TMX3 - thioredoxin-related transmembrane protein 3, SNX6 - sorting nexin 6, DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, CAT - catalase, PDAP1 - pdgfa associated protein 1, CDK13 - cyclin-dependent kinase 13, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, FAM49B - family with sequence similarity 49, member b, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, STX8 - syntaxin 8, ERP44 - endoplasmic reticulum protein 44, STX18 - syntaxin 18, BCL2L1 - bcl2-like 1, PRKCD - protein kinase c, delta, EPS15 - epidermal growth factor receptor pathway substrate 15, PGM2 - phosphoglucomutase 2, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1]
44 [FAM136A - family with sequence similarity 136, member a, SF3A3 - splicing factor 3a, subunit 3, 60kda, GFM2 - g elongation factor, mitochondrial 2, TMED2 - transmembrane emp24 domain trafficking protein 2, HOOK3 - hook microtubule-tethering protein 3, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, ACAT1 - acetyl-coa acetyltransferase 1, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, CPOX - coproporphyrinogen oxidase, LRRC40 - leucine rich repeat containing 40, SUGT1 - sgt1, suppressor of g2 allele of skp1 (s. cerevisiae), GOSR2 - golgi snap receptor complex member 2, OTUB1 - otu domain, ubiquitin aldehyde binding 1, SORD - sorbitol dehydrogenase, PLCG1 - phospholipase c, gamma 1, LIMS1 - lim and senescent cell antigen-like domains 1, TTC27 - tetratricopeptide repeat domain 27, MORF4L1 - mortality factor 4 like 1, CARHSP1 - calcium regulated heat stable protein 1, 24kda, CREB1 - camp responsive element binding protein 1, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, SNX1 - sorting nexin 1, THUMPD1 - thump domain containing 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, SNRPE - small nuclear ribonucleoprotein polypeptide e, ANAPC7 - anaphase promoting complex subunit 7, PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, GLS - glutaminase, GLO1 - glyoxalase i, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), GLG1 - golgi glycoprotein 1, NIP7 - nip7, nucleolar pre-rrna processing protein, GOSR1 - golgi snap receptor complex member 1, COMMD10 - comm domain containing 10, POLR1C - polymerase (rna) i polypeptide c, 30kda, SUMO3 - small ubiquitin-like modifier 3, SNX6 - sorting nexin 6, SMARCD1 - swi/snf related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, COMT - catechol-o-methyltransferase, STX8 - syntaxin 8, TAOK3 - tao kinase 3, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), PARP4 - poly (adp-ribose) polymerase family, member 4, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, TMEM30A - transmembrane protein 30a, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, QKI - qki, kh domain containing, rna binding, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, AGK - acylglycerol kinase, HOMER1 - homer homolog 1 (drosophila), FTH1 - ferritin, heavy polypeptide 1, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, PPP5C - protein phosphatase 5, catalytic subunit, USP48 - ubiquitin specific peptidase 48, SURF4 - surfeit 4, IRAK4 - interleukin-1 receptor-associated kinase 4, DNAJC11 - dnaj (hsp40) homolog, subfamily c, member 11, AK4 - adenylate kinase 4, TNPO1 - transportin 1, KPNB1 - karyopherin (importin) beta 1, EXOC2 - exocyst complex component 2, EXOC1 - exocyst complex component 1, IFIT5 - interferon-induced protein with tetratricopeptide repeats 5, TJP2 - tight junction protein 2, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, SCYL2 - scy1-like 2 (s. cerevisiae), NIPBL - nipped-b homolog (drosophila), GALNT2 - udp-n-acetyl-alpha-d-galactosamine:polypeptide n-acetylgalactosaminyltransferase 2 (galnac-t2), STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), SNRNP27 - small nuclear ribonucleoprotein 27kda (u4/u6.u5), MFN1 - mitofusin 1, STAT6 - signal transducer and activator of transcription 6, interleukin-4 induced, EIF1AD - eukaryotic translation initiation factor 1a domain containing, RINT1 - rad50 interactor 1, NHP2 - nhp2 ribonucleoprotein, DSTN - destrin (actin depolymerizing factor), RIC8A - ric8 guanine nucleotide exchange factor a, SRP72 - signal recognition particle 72kda, CISD2 - cdgsh iron sulfur domain 2, SSBP1 - single-stranded dna binding protein 1, mitochondrial, SAMM50 - samm50 sorting and assembly machinery component, ANXA4 - annexin a4, TNFAIP8 - tumor necrosis factor, alpha-induced protein 8, PRKCD - protein kinase c, delta, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), CCDC6 - coiled-coil domain containing 6, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, ERAP1 - endoplasmic reticulum aminopeptidase 1, DDX6 - dead (asp-glu-ala-asp) box helicase 6, ZMYM2 - zinc finger, mym-type 2, GSPT1 - g1 to s phase transition 1, GSN - gelsolin, FAM98A - family with sequence similarity 98, member a, DCK - deoxycytidine kinase, DBN1 - drebrin 1, TRIM25 - tripartite motif containing 25, XPOT - exportin, trna, RIF1 - rap1 interacting factor homolog (yeast), VTI1A - vesicle transport through interaction with t-snares 1a, CCDC51 - coiled-coil domain containing 51, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), MMS22L - mms22-like, dna repair protein, ARCN1 - archain 1, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, PA2G4 - proliferation-associated 2g4, 38kda, LRRK1 - leucine-rich repeat kinase 1, CAB39 - calcium binding protein 39, GATAD2B - gata zinc finger domain containing 2b, KIAA2013 - kiaa2013, RAB23 - rab23, member ras oncogene family, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, RNF31 - ring finger protein 31, MCM7 - minichromosome maintenance complex component 7, MCU - mitochondrial calcium uniporter, ARL1 - adp-ribosylation factor-like 1, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, PELO - pelota homolog (drosophila), SCO1 - sco1 cytochrome c oxidase assembly protein, WDHD1 - wd repeat and hmg-box dna binding protein 1, HADH - hydroxyacyl-coa dehydrogenase, PPIL3 - peptidylprolyl isomerase (cyclophilin)-like 3, OSBPL3 - oxysterol binding protein-like 3, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, RPL7L1 - ribosomal protein l7-like 1, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, STX18 - syntaxin 18, RPTOR - regulatory associated protein of mtor, complex 1, LYRM7 - lyr motif containing 7, VTA1 - vesicle (multivesicular body) trafficking 1, SERBP1 - serpine1 mrna binding protein 1, DENND3 - denn/madd domain containing 3, GAPVD1 - gtpase activating protein and vps9 domains 1, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, MAP4 - microtubule-associated protein 4, RAB14 - rab14, member ras oncogene family, PHKB - phosphorylase kinase, beta, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, MAN2A1 - mannosidase, alpha, class 2a, member 1, USP8 - ubiquitin specific peptidase 8, EIF4H - eukaryotic translation initiation factor 4h, MBNL1 - muscleblind-like splicing regulator 1, SLAMF1 - signaling lymphocytic activation molecule family member 1, DUT - deoxyuridine triphosphatase, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, MAT2A - methionine adenosyltransferase ii, alpha, MCM3 - minichromosome maintenance complex component 3, CHMP5 - charged multivesicular body protein 5, DOCK7 - dedicator of cytokinesis 7, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, WIPF1 - was/wasl interacting protein family, member 1, SCAF8 - sr-related ctd-associated factor 8, GRK6 - g protein-coupled receptor kinase 6, UBA6 - ubiquitin-like modifier activating enzyme 6, LSS - lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase), DLG1 - discs, large homolog 1 (drosophila), PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, RPL14 - ribosomal protein l14, DHRS7 - dehydrogenase/reductase (sdr family) member 7, LRRC20 - leucine rich repeat containing 20, NUB1 - negative regulator of ubiquitin-like proteins 1, TRAPPC9 - trafficking protein particle complex 9, UBA3 - ubiquitin-like modifier activating enzyme 3, DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, ACOT7 - acyl-coa thioesterase 7, PDAP1 - pdgfa associated protein 1, CBX3 - chromobox homolog 3, ADI1 - acireductone dioxygenase 1, FAM49B - family with sequence similarity 49, member b, CAPRIN1 - cell cycle associated protein 1, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), RHOT1 - ras homolog family member t1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, RSL1D1 - ribosomal l1 domain containing 1, BCL2L1 - bcl2-like 1, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, METTL13 - methyltransferase like 13, PGM2 - phosphoglucomutase 2, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1, CCNK - cyclin k, DST - dystonin, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, NCBP2 - nuclear cap binding protein subunit 2, 20kda, EXOSC5 - exosome component 5, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, RFC3 - replication factor c (activator 1) 3, 38kda, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, SACM1L - sac1 suppressor of actin mutations 1-like (yeast), NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, NDUFA5 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, RECQL - recq protein-like (dna helicase q1-like), DERL1 - derlin 1, BTF3 - basic transcription factor 3, DOCK8 - dedicator of cytokinesis 8, SPC24 - spc24, ndc80 kinetochore complex component, EIF4B - eukaryotic translation initiation factor 4b, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, USP25 - ubiquitin specific peptidase 25, ZNF326 - zinc finger protein 326, WDR77 - wd repeat domain 77, RCSD1 - rcsd domain containing 1, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, MYO6 - myosin vi, SGPL1 - sphingosine-1-phosphate lyase 1, TPM3 - tropomyosin 3, STK38L - serine/threonine kinase 38 like, SEH1L - seh1-like (s. cerevisiae), EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, CC2D1B - coiled-coil and c2 domain containing 1b, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, DCAF7 - ddb1 and cul4 associated factor 7, ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, EIF2S2 - eukaryotic translation initiation factor 2, subunit 2 beta, 38kda, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, TSFM - ts translation elongation factor, mitochondrial, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), MVD - mevalonate (diphospho) decarboxylase, DENND4A - denn/madd domain containing 4a, KYNU - kynureninase, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, ERP44 - endoplasmic reticulum protein 44, NAMPT - nicotinamide phosphoribosyltransferase, HSPA4 - heat shock 70kda protein 4, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, UQCRB - ubiquinol-cytochrome c reductase binding protein, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, CAPN2 - calpain 2, (m/ii) large subunit, WNK1 - wnk lysine deficient protein kinase 1, ZWILCH - zwilch kinetochore protein, OSTF1 - osteoclast stimulating factor 1, UBE2Z - ubiquitin-conjugating enzyme e2z, HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), PTCD3 - pentatricopeptide repeat domain 3, TBC1D2B - tbc1 domain family, member 2b, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, UCK2 - uridine-cytidine kinase 2, RPL3 - ribosomal protein l3, BTF3L4 - basic transcription factor 3-like 4, CARS - cysteinyl-trna synthetase, EIF3F - eukaryotic translation initiation factor 3, subunit f, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, RPN1 - ribophorin i, STX17 - syntaxin 17, HK2 - hexokinase 2, TRIM38 - tripartite motif containing 38, CDC23 - cell division cycle 23, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, RPL31 - ribosomal protein l31, VDAC2 - voltage-dependent anion channel 2, TTLL12 - tubulin tyrosine ligase-like family, member 12, MRPL13 - mitochondrial ribosomal protein l13, GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, TMEM70 - transmembrane protein 70, RNMT - rna (guanine-7-) methyltransferase, WDR1 - wd repeat domain 1, RPS8 - ribosomal protein s8, LARP4B - la ribonucleoprotein domain family, member 4b, ESD - esterase d, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, STOM - stomatin, UCHL3 - ubiquitin carboxyl-terminal esterase l3 (ubiquitin thiolesterase), SUMO1 - small ubiquitin-like modifier 1, CAT - catalase, USP15 - ubiquitin specific peptidase 15, XPO6 - exportin 6, UGDH - udp-glucose 6-dehydrogenase, NDUFAF4 - nadh dehydrogenase (ubiquinone) complex i, assembly factor 4, TUBGCP3 - tubulin, gamma complex associated protein 3, MRPL47 - mitochondrial ribosomal protein l47, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, UBE2I - ubiquitin-conjugating enzyme e2i, AIDA - axin interactor, dorsalization associated, EPS15 - epidermal growth factor receptor pathway substrate 15, RRM1 - ribonucleotide reductase m1, DDX31 - dead (asp-glu-ala-asp) box polypeptide 31, ZC3H7B - zinc finger ccch-type containing 7b, TCOF1 - treacher collins-franceschetti syndrome 1, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, F11R - f11 receptor, STAMBP - stam binding protein, SCFD1 - sec1 family domain containing 1, CISD1 - cdgsh iron sulfur domain 1, RAB30 - rab30, member ras oncogene family, FLNB - filamin b, beta, PPFIA1 - protein tyrosine phosphatase, receptor type, f polypeptide (ptprf), interacting protein (liprin), alpha 1, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, PPFIBP1 - ptprf interacting protein, binding protein 1 (liprin beta 1), PPP4R2 - protein phosphatase 4, regulatory subunit 2, ILF3 - interleukin enhancer binding factor 3, 90kda, ANKRD28 - ankyrin repeat domain 28, NUDCD2 - nudc domain containing 2, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, CYCS - cytochrome c, somatic, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, CD48 - cd48 molecule, AGPS - alkylglycerone phosphate synthase, TBC1D4 - tbc1 domain family, member 4, TMED5 - transmembrane emp24 protein transport domain containing 5, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, GAR1 - gar1 ribonucleoprotein, KDM3A - lysine (k)-specific demethylase 3a, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, EXOSC1 - exosome component 1, TELO2 - telomere maintenance 2, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), ATG7 - autophagy related 7, TUBGCP4 - tubulin, gamma complex associated protein 4, HDDC2 - hd domain containing 2, PRRC1 - proline-rich coiled-coil 1, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, SYNE1 - spectrin repeat containing, nuclear envelope 1, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), NUP88 - nucleoporin 88kda, ZBTB8OS - zinc finger and btb domain containing 8 opposite strand, NOM1 - nucleolar protein with mif4g domain 1, NFU1 - nfu1 iron-sulfur cluster scaffold homolog (s. cerevisiae), MBLAC2 - metallo-beta-lactamase domain containing 2, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), THOC5 - tho complex 5, FECH - ferrochelatase, CDK6 - cyclin-dependent kinase 6, ADNP - activity-dependent neuroprotector homeobox, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, CDK13 - cyclin-dependent kinase 13, TBCD - tubulin folding cofactor d, FCF1 - fcf1 rrna-processing protein, GNPDA2 - glucosamine-6-phosphate deaminase 2, CCT4 - chaperonin containing tcp1, subunit 4 (delta), TBCA - tubulin folding cofactor a, CUTC - cutc copper transporter, INPP5B - inositol polyphosphate-5-phosphatase, 75kda, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, IRF3 - interferon regulatory factor 3, ARL6IP5 - adp-ribosylation-like factor 6 interacting protein 5, LARP1 - la ribonucleoprotein domain family, member 1, IRF5 - interferon regulatory factor 5, EIF3E - eukaryotic translation initiation factor 3, subunit e, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, TCEA1 - transcription elongation factor a (sii), 1, PURA - purine-rich element binding protein a, SPG11 - spastic paraplegia 11 (autosomal recessive), TDRD7 - tudor domain containing 7, NEK6 - nima-related kinase 6, PYGB - phosphorylase, glycogen; brain, PCYT2 - phosphate cytidylyltransferase 2, ethanolamine, NUP50 - nucleoporin 50kda, VKORC1L1 - vitamin k epoxide reductase complex, subunit 1-like 1, TRAPPC10 - trafficking protein particle complex 10, IDE - insulin-degrading enzyme, USP34 - ubiquitin specific peptidase 34, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), IFIH1 - interferon induced with helicase c domain 1, MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, SART3 - squamous cell carcinoma antigen recognized by t cells 3, MAD1L1 - mad1 mitotic arrest deficient-like 1 (yeast), CTSC - cathepsin c, RANBP1 - ran binding protein 1, RAP1A - rap1a, member of ras oncogene family, PARVG - parvin, gamma, NSMAF - neutral sphingomyelinase (n-smase) activation associated factor, AKAP8L - a kinase (prka) anchor protein 8-like, ZNF592 - zinc finger protein 592, CYFIP2 - cytoplasmic fmr1 interacting protein 2, RBM26 - rna binding motif protein 26, CLNS1A - chloride channel, nucleotide-sensitive, 1a, RAP1B - rap1b, member of ras oncogene family, GOLPH3 - golgi phosphoprotein 3 (coat-protein), TMX3 - thioredoxin-related transmembrane protein 3, BZW1 - basic leucine zipper and w2 domains 1, RB1 - retinoblastoma 1, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), MRPS14 - mitochondrial ribosomal protein s14, CUL1 - cullin 1, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3]
45 [UQCRB - ubiquinol-cytochrome c reductase binding protein, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, CAPN2 - calpain 2, (m/ii) large subunit, TDRD7 - tudor domain containing 7, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, TMEM14C - transmembrane protein 14c, CISD1 - cdgsh iron sulfur domain 1, AGK - acylglycerol kinase, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, GFM2 - g elongation factor, mitochondrial 2, NDUFA5 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, PTCD3 - pentatricopeptide repeat domain 3, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), ECI1 - enoyl-coa delta isomerase 1, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, DNAJC11 - dnaj (hsp40) homolog, subfamily c, member 11, CYCS - cytochrome c, somatic, MRPS35 - mitochondrial ribosomal protein s35, METTL15 - methyltransferase like 15, ACAT1 - acetyl-coa acetyltransferase 1, CCDC51 - coiled-coil domain containing 51, CPOX - coproporphyrinogen oxidase, MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, HK2 - hexokinase 2, AK4 - adenylate kinase 4, SORD - sorbitol dehydrogenase, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, CREB1 - camp responsive element binding protein 1, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, VDAC2 - voltage-dependent anion channel 2, MRPL3 - mitochondrial ribosomal protein l3, PITRM1 - pitrilysin metallopeptidase 1, MRPL13 - mitochondrial ribosomal protein l13, MCU - mitochondrial calcium uniporter, PUS1 - pseudouridylate synthase 1, TMEM70 - transmembrane protein 70, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, GLS - glutaminase, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), GNPAT - glyceronephosphate o-acyltransferase, MFN1 - mitofusin 1, SCO1 - sco1 cytochrome c oxidase assembly protein, DGUOK - deoxyguanosine kinase, MRPL42 - mitochondrial ribosomal protein l42, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), HADH - hydroxyacyl-coa dehydrogenase, GOLPH3 - golgi phosphoprotein 3 (coat-protein), FECH - ferrochelatase, MRPL2 - mitochondrial ribosomal protein l2, TSFM - ts translation elongation factor, mitochondrial, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, GCDH - glutaryl-coa dehydrogenase, CISD2 - cdgsh iron sulfur domain 2, CAT - catalase, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), NDUFAF4 - nadh dehydrogenase (ubiquinone) complex i, assembly factor 4, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, SSBP1 - single-stranded dna binding protein 1, mitochondrial, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), SAMM50 - samm50 sorting and assembly machinery component, RHOT1 - ras homolog family member t1, MRPL47 - mitochondrial ribosomal protein l47, MRPS14 - mitochondrial ribosomal protein s14, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, BCL2L1 - bcl2-like 1, DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, LYRM7 - lyr motif containing 7]
46 [CCNK - cyclin k, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, NCBP2 - nuclear cap binding protein subunit 2, 20kda, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, EXOSC5 - exosome component 5, RFC3 - replication factor c (activator 1) 3, 38kda, SF3A3 - splicing factor 3a, subunit 3, 60kda, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, SACM1L - sac1 suppressor of actin mutations 1-like (yeast), NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, GFM2 - g elongation factor, mitochondrial 2, FKBP11 - fk506 binding protein 11, 19 kda, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, RECQL - recq protein-like (dna helicase q1-like), DERL1 - derlin 1, BTF3 - basic transcription factor 3, ACAT1 - acetyl-coa acetyltransferase 1, LRRC40 - leucine rich repeat containing 40, USP25 - ubiquitin specific peptidase 25, OTUB1 - otu domain, ubiquitin aldehyde binding 1, SORD - sorbitol dehydrogenase, ZNF326 - zinc finger protein 326, PLCG1 - phospholipase c, gamma 1, MORF4L1 - mortality factor 4 like 1, CREB1 - camp responsive element binding protein 1, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, THUMPD1 - thump domain containing 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, SGPL1 - sphingosine-1-phosphate lyase 1, SNRPE - small nuclear ribonucleoprotein polypeptide e, ANAPC7 - anaphase promoting complex subunit 7, STK38L - serine/threonine kinase 38 like, SEH1L - seh1-like (s. cerevisiae), GLS - glutaminase, PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, GLO1 - glyoxalase i, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), POLR1C - polymerase (rna) i polypeptide c, 30kda, DCAF7 - ddb1 and cul4 associated factor 7, PPA1 - pyrophosphatase (inorganic) 1, ADPGK - adp-dependent glucokinase, SUMO3 - small ubiquitin-like modifier 3, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), MVD - mevalonate (diphospho) decarboxylase, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, COMT - catechol-o-methyltransferase, KYNU - kynureninase, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, TAOK3 - tao kinase 3, ERP44 - endoplasmic reticulum protein 44, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), NAMPT - nicotinamide phosphoribosyltransferase, PARP4 - poly (adp-ribose) polymerase family, member 4, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, SCPEP1 - serine carboxypeptidase 1, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, QKI - qki, kh domain containing, rna binding, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, UQCRB - ubiquinol-cytochrome c reductase binding protein, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, CAPN2 - calpain 2, (m/ii) large subunit, WNK1 - wnk lysine deficient protein kinase 1, UBE2Z - ubiquitin-conjugating enzyme e2z, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, HMGB2 - high mobility group box 2, AGK - acylglycerol kinase, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PTCD3 - pentatricopeptide repeat domain 3, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, UCK2 - uridine-cytidine kinase 2, USP48 - ubiquitin specific peptidase 48, PPP5C - protein phosphatase 5, catalytic subunit, UMPS - uridine monophosphate synthetase, RPL3 - ribosomal protein l3, CARS - cysteinyl-trna synthetase, IRAK4 - interleukin-1 receptor-associated kinase 4, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), EIF3F - eukaryotic translation initiation factor 3, subunit f, MRPL1 - mitochondrial ribosomal protein l1, RPN1 - ribophorin i, MRPL9 - mitochondrial ribosomal protein l9, AK4 - adenylate kinase 4, HK2 - hexokinase 2, KPNB1 - karyopherin (importin) beta 1, TRIM38 - tripartite motif containing 38, CDC23 - cell division cycle 23, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, TJP2 - tight junction protein 2, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, DCAKD - dephospho-coa kinase domain containing, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, TTLL12 - tubulin tyrosine ligase-like family, member 12, MRPL13 - mitochondrial ribosomal protein l13, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, SCYL2 - scy1-like 2 (s. cerevisiae), NIPBL - nipped-b homolog (drosophila), GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, GALNT2 - udp-n-acetyl-alpha-d-galactosamine:polypeptide n-acetylgalactosaminyltransferase 2 (galnac-t2), STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), SNRNP27 - small nuclear ribonucleoprotein 27kda (u4/u6.u5), MFN1 - mitofusin 1, RNMT - rna (guanine-7-) methyltransferase, RPS8 - ribosomal protein s8, MRPL42 - mitochondrial ribosomal protein l42, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, NHP2 - nhp2 ribonucleoprotein, UCHL3 - ubiquitin carboxyl-terminal esterase l3 (ubiquitin thiolesterase), SUMO1 - small ubiquitin-like modifier 1, GCDH - glutaryl-coa dehydrogenase, CAT - catalase, USP15 - ubiquitin specific peptidase 15, UGDH - udp-glucose 6-dehydrogenase, SSBP1 - single-stranded dna binding protein 1, mitochondrial, MRPL47 - mitochondrial ribosomal protein l47, UBE2I - ubiquitin-conjugating enzyme e2i, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, PRKCD - protein kinase c, delta, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), RRBP1 - ribosome binding protein 1, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, RRM1 - ribonucleotide reductase m1, ZC3H7B - zinc finger ccch-type containing 7b, ERAP1 - endoplasmic reticulum aminopeptidase 1, DDX6 - dead (asp-glu-ala-asp) box helicase 6, TCOF1 - treacher collins-franceschetti syndrome 1, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, ZNF638 - zinc finger protein 638, STAMBP - stam binding protein, GSPT1 - g1 to s phase transition 1, GSN - gelsolin, URB1 - urb1 ribosome biogenesis 1 homolog (s. cerevisiae), FAM98A - family with sequence similarity 98, member a, ECI1 - enoyl-coa delta isomerase 1, DCK - deoxycytidine kinase, PPP4R2 - protein phosphatase 4, regulatory subunit 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, ILF3 - interleukin enhancer binding factor 3, 90kda, TRIM25 - tripartite motif containing 25, CYCS - cytochrome c, somatic, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, RIF1 - rap1 interacting factor homolog (yeast), PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, RBM19 - rna binding motif protein 19, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), MMS22L - mms22-like, dna repair protein, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, PA2G4 - proliferation-associated 2g4, 38kda, LRRK1 - leucine-rich repeat kinase 1, AGPS - alkylglycerone phosphate synthase, CAB39 - calcium binding protein 39, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, RAB23 - rab23, member ras oncogene family, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, GAR1 - gar1 ribonucleoprotein, KDM3A - lysine (k)-specific demethylase 3a, ANAPC1 - anaphase promoting complex subunit 1, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, MRPL3 - mitochondrial ribosomal protein l3, EXOSC1 - exosome component 1, TELO2 - telomere maintenance 2, PITRM1 - pitrilysin metallopeptidase 1, RNF31 - ring finger protein 31, ATG7 - autophagy related 7, MCM7 - minichromosome maintenance complex component 7, KDELC2 - kdel (lys-asp-glu-leu) containing 2, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, PELO - pelota homolog (drosophila), ZBTB8OS - zinc finger and btb domain containing 8 opposite strand, NUP88 - nucleoporin 88kda, NFU1 - nfu1 iron-sulfur cluster scaffold homolog (s. cerevisiae), WDHD1 - wd repeat and hmg-box dna binding protein 1, PPIL3 - peptidylprolyl isomerase (cyclophilin)-like 3, HADH - hydroxyacyl-coa dehydrogenase, OSBPL3 - oxysterol binding protein-like 3, THOC5 - tho complex 5, CDK6 - cyclin-dependent kinase 6, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, CDK13 - cyclin-dependent kinase 13, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, FCF1 - fcf1 rrna-processing protein, GNPDA2 - glucosamine-6-phosphate deaminase 2, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, INPP5B - inositol polyphosphate-5-phosphatase, 75kda, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, RPL7L1 - ribosomal protein l7-like 1, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, NEMF - nuclear export mediator factor, DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), EIF3E - eukaryotic translation initiation factor 3, subunit e, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, TCEA1 - transcription elongation factor a (sii), 1, PURA - purine-rich element binding protein a, DENND3 - denn/madd domain containing 3, NEK6 - nima-related kinase 6, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), NOC3L - nucleolar complex associated 3 homolog (s. cerevisiae), PYGB - phosphorylase, glycogen; brain, PCYT2 - phosphate cytidylyltransferase 2, ethanolamine, NUP50 - nucleoporin 50kda, RAB14 - rab14, member ras oncogene family, PHKB - phosphorylase kinase, beta, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, MANBA - mannosidase, beta a, lysosomal, VKORC1L1 - vitamin k epoxide reductase complex, subunit 1-like 1, MAN2A1 - mannosidase, alpha, class 2a, member 1, USP8 - ubiquitin specific peptidase 8, MBNL1 - muscleblind-like splicing regulator 1, METTL15 - methyltransferase like 15, DUT - deoxyuridine triphosphatase, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, IDE - insulin-degrading enzyme, MCM3 - minichromosome maintenance complex component 3, IFIH1 - interferon induced with helicase c domain 1, USP34 - ubiquitin specific peptidase 34, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, SART3 - squamous cell carcinoma antigen recognized by t cells 3, CTSC - cathepsin c, SCAF8 - sr-related ctd-associated factor 8, UBA6 - ubiquitin-like modifier activating enzyme 6, GRK6 - g protein-coupled receptor kinase 6, LSS - lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase), DLG1 - discs, large homolog 1 (drosophila), PUS1 - pseudouridylate synthase 1, NSMAF - neutral sphingomyelinase (n-smase) activation associated factor, RPL14 - ribosomal protein l14, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, GNPAT - glyceronephosphate o-acyltransferase, AKAP8L - a kinase (prka) anchor protein 8-like, CYFIP2 - cytoplasmic fmr1 interacting protein 2, NUB1 - negative regulator of ubiquitin-like proteins 1, RBM26 - rna binding motif protein 26, DGUOK - deoxyguanosine kinase, UBA3 - ubiquitin-like modifier activating enzyme 3, GOLPH3 - golgi phosphoprotein 3 (coat-protein), TMX3 - thioredoxin-related transmembrane protein 3, ACOT7 - acyl-coa thioesterase 7, ADI1 - acireductone dioxygenase 1, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), MRPS14 - mitochondrial ribosomal protein s14, RHOT1 - ras homolog family member t1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, CUL1 - cullin 1, RSL1D1 - ribosomal l1 domain containing 1, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, PGM2 - phosphoglucomutase 2, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
47 [MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, PGM1 - phosphoglucomutase 1, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, GSPT1 - g1 to s phase transition 1, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, AGK - acylglycerol kinase, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, RPL13 - ribosomal protein l13, GFM2 - g elongation factor, mitochondrial 2, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PTCD3 - pentatricopeptide repeat domain 3, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, DCK - deoxycytidine kinase, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, UCK2 - uridine-cytidine kinase 2, RPL3 - ribosomal protein l3, UMPS - uridine monophosphate synthetase, ACAT1 - acetyl-coa acetyltransferase 1, DUT - deoxyuridine triphosphatase, CPOX - coproporphyrinogen oxidase, MRPL9 - mitochondrial ribosomal protein l9, HK2 - hexokinase 2, AK4 - adenylate kinase 4, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, DCAKD - dephospho-coa kinase domain containing, MRPL3 - mitochondrial ribosomal protein l3, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, SGPL1 - sphingosine-1-phosphate lyase 1, MRPL13 - mitochondrial ribosomal protein l13, RPL14 - ribosomal protein l14, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, GLS - glutaminase, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, DGUOK - deoxyguanosine kinase, RPS8 - ribosomal protein s8, ESD - esterase d, MRPL42 - mitochondrial ribosomal protein l42, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, GOLPH3 - golgi phosphoprotein 3 (coat-protein), RPS7 - ribosomal protein s7, ADPGK - adp-dependent glucokinase, FECH - ferrochelatase, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, ACOT7 - acyl-coa thioesterase 7, GCDH - glutaryl-coa dehydrogenase, UGDH - udp-glucose 6-dehydrogenase, ADI1 - acireductone dioxygenase 1, MRPS14 - mitochondrial ribosomal protein s14, MRPL47 - mitochondrial ribosomal protein l47, KYNU - kynureninase, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, NAMPT - nicotinamide phosphoribosyltransferase, AK3 - adenylate kinase 3, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, RRBP1 - ribosome binding protein 1, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
48 [CCNK - cyclin k, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, NCBP2 - nuclear cap binding protein subunit 2, 20kda, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, TCOF1 - treacher collins-franceschetti syndrome 1, RFC3 - replication factor c (activator 1) 3, 38kda, SACM1L - sac1 suppressor of actin mutations 1-like (yeast), SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, GSPT1 - g1 to s phase transition 1, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, GFM2 - g elongation factor, mitochondrial 2, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, DCK - deoxycytidine kinase, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, BTF3 - basic transcription factor 3, TRIM25 - tripartite motif containing 25, ACAT1 - acetyl-coa acetyltransferase 1, CPOX - coproporphyrinogen oxidase, AGPS - alkylglycerone phosphate synthase, SORD - sorbitol dehydrogenase, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, GAR1 - gar1 ribonucleoprotein, CREB1 - camp responsive element binding protein 1, KDM3A - lysine (k)-specific demethylase 3a, MRPL3 - mitochondrial ribosomal protein l3, TELO2 - telomere maintenance 2, SGPL1 - sphingosine-1-phosphate lyase 1, MCM7 - minichromosome maintenance complex component 7, SNRPE - small nuclear ribonucleoprotein polypeptide e, GLS - glutaminase, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, WDHD1 - wd repeat and hmg-box dna binding protein 1, OSBPL3 - oxysterol binding protein-like 3, POLR1C - polymerase (rna) i polypeptide c, 30kda, ADPGK - adp-dependent glucokinase, FECH - ferrochelatase, MRPL2 - mitochondrial ribosomal protein l2, TSFM - ts translation elongation factor, mitochondrial, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, CDK13 - cyclin-dependent kinase 13, MVD - mevalonate (diphospho) decarboxylase, GNPDA2 - glucosamine-6-phosphate deaminase 2, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, KYNU - kynureninase, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, NEMF - nuclear export mediator factor, NAMPT - nicotinamide phosphoribosyltransferase, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, TCEA1 - transcription elongation factor a (sii), 1, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, QKI - qki, kh domain containing, rna binding, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, AGK - acylglycerol kinase, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PTCD3 - pentatricopeptide repeat domain 3, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, PCYT2 - phosphate cytidylyltransferase 2, ethanolamine, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, RAB14 - rab14, member ras oncogene family, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, UCK2 - uridine-cytidine kinase 2, PPP5C - protein phosphatase 5, catalytic subunit, RPL3 - ribosomal protein l3, UMPS - uridine monophosphate synthetase, MRPS35 - mitochondrial ribosomal protein s35, DUT - deoxyuridine triphosphatase, MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, HK2 - hexokinase 2, AK4 - adenylate kinase 4, MAT2A - methionine adenosyltransferase ii, alpha, MCM3 - minichromosome maintenance complex component 3, RPL31 - ribosomal protein l31, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, RPL32 - ribosomal protein l32, DCAKD - dephospho-coa kinase domain containing, SCAF8 - sr-related ctd-associated factor 8, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, LSS - lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase), MRPL13 - mitochondrial ribosomal protein l13, RPL14 - ribosomal protein l14, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), GNPAT - glyceronephosphate o-acyltransferase, RNMT - rna (guanine-7-) methyltransferase, DGUOK - deoxyguanosine kinase, RPS8 - ribosomal protein s8, ESD - esterase d, MRPL42 - mitochondrial ribosomal protein l42, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, GOLPH3 - golgi phosphoprotein 3 (coat-protein), NHP2 - nhp2 ribonucleoprotein, RPS7 - ribosomal protein s7, ACOT7 - acyl-coa thioesterase 7, GCDH - glutaryl-coa dehydrogenase, UGDH - udp-glucose 6-dehydrogenase, ADI1 - acireductone dioxygenase 1, SSBP1 - single-stranded dna binding protein 1, mitochondrial, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), MRPS14 - mitochondrial ribosomal protein s14, MRPL47 - mitochondrial ribosomal protein l47, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, AK3 - adenylate kinase 3, RRBP1 - ribosome binding protein 1, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, PGM2 - phosphoglucomutase 2, RRM1 - ribonucleotide reductase m1, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
49 [CCNK - cyclin k, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, NCBP2 - nuclear cap binding protein subunit 2, 20kda, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, EXOSC5 - exosome component 5, RFC3 - replication factor c (activator 1) 3, 38kda, SF3A3 - splicing factor 3a, subunit 3, 60kda, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, GFM2 - g elongation factor, mitochondrial 2, FKBP11 - fk506 binding protein 11, 19 kda, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, RECQL - recq protein-like (dna helicase q1-like), DERL1 - derlin 1, BTF3 - basic transcription factor 3, ACAT1 - acetyl-coa acetyltransferase 1, CPOX - coproporphyrinogen oxidase, LRRC40 - leucine rich repeat containing 40, USP25 - ubiquitin specific peptidase 25, OTUB1 - otu domain, ubiquitin aldehyde binding 1, ZNF326 - zinc finger protein 326, MORF4L1 - mortality factor 4 like 1, CREB1 - camp responsive element binding protein 1, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, THUMPD1 - thump domain containing 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, SGPL1 - sphingosine-1-phosphate lyase 1, SNRPE - small nuclear ribonucleoprotein polypeptide e, ANAPC7 - anaphase promoting complex subunit 7, STK38L - serine/threonine kinase 38 like, SEH1L - seh1-like (s. cerevisiae), GLS - glutaminase, PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, GLO1 - glyoxalase i, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), POLR1C - polymerase (rna) i polypeptide c, 30kda, DCAF7 - ddb1 and cul4 associated factor 7, PPA1 - pyrophosphatase (inorganic) 1, ADPGK - adp-dependent glucokinase, SUMO3 - small ubiquitin-like modifier 3, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), MVD - mevalonate (diphospho) decarboxylase, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, COMT - catechol-o-methyltransferase, KYNU - kynureninase, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, TAOK3 - tao kinase 3, ERP44 - endoplasmic reticulum protein 44, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), NAMPT - nicotinamide phosphoribosyltransferase, PARP4 - poly (adp-ribose) polymerase family, member 4, SCPEP1 - serine carboxypeptidase 1, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, QKI - qki, kh domain containing, rna binding, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, UQCRB - ubiquinol-cytochrome c reductase binding protein, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, CAPN2 - calpain 2, (m/ii) large subunit, WNK1 - wnk lysine deficient protein kinase 1, UBE2Z - ubiquitin-conjugating enzyme e2z, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, HMGB2 - high mobility group box 2, AGK - acylglycerol kinase, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PTCD3 - pentatricopeptide repeat domain 3, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, UCK2 - uridine-cytidine kinase 2, USP48 - ubiquitin specific peptidase 48, PPP5C - protein phosphatase 5, catalytic subunit, UMPS - uridine monophosphate synthetase, RPL3 - ribosomal protein l3, CARS - cysteinyl-trna synthetase, IRAK4 - interleukin-1 receptor-associated kinase 4, EIF3F - eukaryotic translation initiation factor 3, subunit f, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), MRPL1 - mitochondrial ribosomal protein l1, RPN1 - ribophorin i, MRPL9 - mitochondrial ribosomal protein l9, AK4 - adenylate kinase 4, HK2 - hexokinase 2, KPNB1 - karyopherin (importin) beta 1, TRIM38 - tripartite motif containing 38, CDC23 - cell division cycle 23, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, TJP2 - tight junction protein 2, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, DCAKD - dephospho-coa kinase domain containing, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, TTLL12 - tubulin tyrosine ligase-like family, member 12, MRPL13 - mitochondrial ribosomal protein l13, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, SCYL2 - scy1-like 2 (s. cerevisiae), NIPBL - nipped-b homolog (drosophila), GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, GALNT2 - udp-n-acetyl-alpha-d-galactosamine:polypeptide n-acetylgalactosaminyltransferase 2 (galnac-t2), STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), MFN1 - mitofusin 1, SNRNP27 - small nuclear ribonucleoprotein 27kda (u4/u6.u5), RNMT - rna (guanine-7-) methyltransferase, RPS8 - ribosomal protein s8, ESD - esterase d, MRPL42 - mitochondrial ribosomal protein l42, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, NHP2 - nhp2 ribonucleoprotein, RPS7 - ribosomal protein s7, UCHL3 - ubiquitin carboxyl-terminal esterase l3 (ubiquitin thiolesterase), SUMO1 - small ubiquitin-like modifier 1, GCDH - glutaryl-coa dehydrogenase, CAT - catalase, USP15 - ubiquitin specific peptidase 15, UGDH - udp-glucose 6-dehydrogenase, SSBP1 - single-stranded dna binding protein 1, mitochondrial, MRPL47 - mitochondrial ribosomal protein l47, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, UBE2I - ubiquitin-conjugating enzyme e2i, PRKCD - protein kinase c, delta, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), RRBP1 - ribosome binding protein 1, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, RRM1 - ribonucleotide reductase m1, ZC3H7B - zinc finger ccch-type containing 7b, ERAP1 - endoplasmic reticulum aminopeptidase 1, DDX6 - dead (asp-glu-ala-asp) box helicase 6, TCOF1 - treacher collins-franceschetti syndrome 1, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, ZNF638 - zinc finger protein 638, STAMBP - stam binding protein, GSPT1 - g1 to s phase transition 1, GSN - gelsolin, URB1 - urb1 ribosome biogenesis 1 homolog (s. cerevisiae), FAM98A - family with sequence similarity 98, member a, DCK - deoxycytidine kinase, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, PPP4R2 - protein phosphatase 4, regulatory subunit 2, ILF3 - interleukin enhancer binding factor 3, 90kda, TRIM25 - tripartite motif containing 25, CYCS - cytochrome c, somatic, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, RIF1 - rap1 interacting factor homolog (yeast), PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, RBM19 - rna binding motif protein 19, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), MMS22L - mms22-like, dna repair protein, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, PA2G4 - proliferation-associated 2g4, 38kda, LRRK1 - leucine-rich repeat kinase 1, CAB39 - calcium binding protein 39, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, RAB23 - rab23, member ras oncogene family, GAR1 - gar1 ribonucleoprotein, KDM3A - lysine (k)-specific demethylase 3a, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, ANAPC1 - anaphase promoting complex subunit 1, MRPL3 - mitochondrial ribosomal protein l3, TELO2 - telomere maintenance 2, EXOSC1 - exosome component 1, PITRM1 - pitrilysin metallopeptidase 1, RNF31 - ring finger protein 31, ATG7 - autophagy related 7, MCM7 - minichromosome maintenance complex component 7, KDELC2 - kdel (lys-asp-glu-leu) containing 2, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, PELO - pelota homolog (drosophila), ZBTB8OS - zinc finger and btb domain containing 8 opposite strand, NUP88 - nucleoporin 88kda, NFU1 - nfu1 iron-sulfur cluster scaffold homolog (s. cerevisiae), WDHD1 - wd repeat and hmg-box dna binding protein 1, PPIL3 - peptidylprolyl isomerase (cyclophilin)-like 3, THOC5 - tho complex 5, FECH - ferrochelatase, CDK6 - cyclin-dependent kinase 6, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, CDK13 - cyclin-dependent kinase 13, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, GNPDA2 - glucosamine-6-phosphate deaminase 2, FCF1 - fcf1 rrna-processing protein, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, RPL7L1 - ribosomal protein l7-like 1, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, NEMF - nuclear export mediator factor, DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, EIF3E - eukaryotic translation initiation factor 3, subunit e, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, TCEA1 - transcription elongation factor a (sii), 1, PURA - purine-rich element binding protein a, DENND3 - denn/madd domain containing 3, NEK6 - nima-related kinase 6, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, NOC3L - nucleolar complex associated 3 homolog (s. cerevisiae), NUP50 - nucleoporin 50kda, PHKB - phosphorylase kinase, beta, MANBA - mannosidase, beta a, lysosomal, VKORC1L1 - vitamin k epoxide reductase complex, subunit 1-like 1, MAN2A1 - mannosidase, alpha, class 2a, member 1, USP8 - ubiquitin specific peptidase 8, MBNL1 - muscleblind-like splicing regulator 1, METTL15 - methyltransferase like 15, DUT - deoxyuridine triphosphatase, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, IDE - insulin-degrading enzyme, MCM3 - minichromosome maintenance complex component 3, USP34 - ubiquitin specific peptidase 34, IFIH1 - interferon induced with helicase c domain 1, MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, SART3 - squamous cell carcinoma antigen recognized by t cells 3, CTSC - cathepsin c, SCAF8 - sr-related ctd-associated factor 8, UBA6 - ubiquitin-like modifier activating enzyme 6, GRK6 - g protein-coupled receptor kinase 6, DLG1 - discs, large homolog 1 (drosophila), PUS1 - pseudouridylate synthase 1, NSMAF - neutral sphingomyelinase (n-smase) activation associated factor, RPL14 - ribosomal protein l14, AKAP8L - a kinase (prka) anchor protein 8-like, CYFIP2 - cytoplasmic fmr1 interacting protein 2, NUB1 - negative regulator of ubiquitin-like proteins 1, RBM26 - rna binding motif protein 26, DGUOK - deoxyguanosine kinase, UBA3 - ubiquitin-like modifier activating enzyme 3, GOLPH3 - golgi phosphoprotein 3 (coat-protein), TMX3 - thioredoxin-related transmembrane protein 3, ACOT7 - acyl-coa thioesterase 7, ADI1 - acireductone dioxygenase 1, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), MRPS14 - mitochondrial ribosomal protein s14, RHOT1 - ras homolog family member t1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, CUL1 - cullin 1, RSL1D1 - ribosomal l1 domain containing 1, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, PGM2 - phosphoglucomutase 2, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
50 [DST - dystonin, NCBP2 - nuclear cap binding protein subunit 2, 20kda, SDAD1 - sda1 domain containing 1, SCFD1 - sec1 family domain containing 1, TMED2 - transmembrane emp24 domain trafficking protein 2, HOOK3 - hook microtubule-tethering protein 3, PPFIA1 - protein tyrosine phosphatase, receptor type, f polypeptide (ptprf), interacting protein (liprin), alpha 1, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, DERL1 - derlin 1, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, XPOT - exportin, trna, VTI1A - vesicle transport through interaction with t-snares 1a, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, ARCN1 - archain 1, AGPS - alkylglycerone phosphate synthase, GOSR2 - golgi snap receptor complex member 2, TBC1D4 - tbc1 domain family, member 4, TMED5 - transmembrane emp24 protein transport domain containing 5, RAB23 - rab23, member ras oncogene family, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, MYO6 - myosin vi, SNX1 - sorting nexin 1, PITRM1 - pitrilysin metallopeptidase 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, EIF2D - eukaryotic translation initiation factor 2d, SNRPE - small nuclear ribonucleoprotein polypeptide e, ARL1 - adp-ribosylation factor-like 1, SEH1L - seh1-like (s. cerevisiae), TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), NUP88 - nucleoporin 88kda, GOSR1 - golgi snap receptor complex member 1, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), THOC5 - tho complex 5, SNX6 - sorting nexin 6, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, HSPD1 - heat shock 60kda protein 1 (chaperonin), HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, STX8 - syntaxin 8, STX18 - syntaxin 18, NEMF - nuclear export mediator factor, HSPA4 - heat shock 70kda protein 4, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, VTA1 - vesicle (multivesicular body) trafficking 1, PURA - purine-rich element binding protein a, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, DENND3 - denn/madd domain containing 3, SPG11 - spastic paraplegia 11 (autosomal recessive), RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, AGK - acylglycerol kinase, RPL13 - ribosomal protein l13, TBC1D2B - tbc1 domain family, member 2b, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), RPL6 - ribosomal protein l6, NUP50 - nucleoporin 50kda, RPL7 - ribosomal protein l7, MAP4 - microtubule-associated protein 4, RAB14 - rab14, member ras oncogene family, RPL3 - ribosomal protein l3, TRAPPC10 - trafficking protein particle complex 10, TNPO1 - transportin 1, STX17 - syntaxin 17, KPNB1 - karyopherin (importin) beta 1, IDE - insulin-degrading enzyme, CHMP5 - charged multivesicular body protein 5, DOCK7 - dedicator of cytokinesis 7, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, CDC23 - cell division cycle 23, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, CTSC - cathepsin c, RANBP1 - ran binding protein 1, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, SCYL2 - scy1-like 2 (s. cerevisiae), DLG1 - discs, large homolog 1 (drosophila), GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, RPL14 - ribosomal protein l14, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), GNPAT - glyceronephosphate o-acyltransferase, RPS8 - ribosomal protein s8, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, SRP72 - signal recognition particle 72kda, CAT - catalase, XPO6 - exportin 6, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, SAMM50 - samm50 sorting and assembly machinery component, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), RHOT1 - ras homolog family member t1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, EPS15 - epidermal growth factor receptor pathway substrate 15]
51 [CCNK - cyclin k, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, NCBP2 - nuclear cap binding protein subunit 2, 20kda, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, TCOF1 - treacher collins-franceschetti syndrome 1, RFC3 - replication factor c (activator 1) 3, 38kda, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, SACM1L - sac1 suppressor of actin mutations 1-like (yeast), GSPT1 - g1 to s phase transition 1, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, GFM2 - g elongation factor, mitochondrial 2, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, DCK - deoxycytidine kinase, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, BTF3 - basic transcription factor 3, TRIM25 - tripartite motif containing 25, ACAT1 - acetyl-coa acetyltransferase 1, CPOX - coproporphyrinogen oxidase, AGPS - alkylglycerone phosphate synthase, SORD - sorbitol dehydrogenase, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, GAR1 - gar1 ribonucleoprotein, CREB1 - camp responsive element binding protein 1, KDM3A - lysine (k)-specific demethylase 3a, MRPL3 - mitochondrial ribosomal protein l3, TELO2 - telomere maintenance 2, SGPL1 - sphingosine-1-phosphate lyase 1, MCM7 - minichromosome maintenance complex component 7, SNRPE - small nuclear ribonucleoprotein polypeptide e, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, GLS - glutaminase, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, WDHD1 - wd repeat and hmg-box dna binding protein 1, OSBPL3 - oxysterol binding protein-like 3, POLR1C - polymerase (rna) i polypeptide c, 30kda, ADPGK - adp-dependent glucokinase, FECH - ferrochelatase, MRPL2 - mitochondrial ribosomal protein l2, TSFM - ts translation elongation factor, mitochondrial, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, CDK13 - cyclin-dependent kinase 13, MVD - mevalonate (diphospho) decarboxylase, GNPDA2 - glucosamine-6-phosphate deaminase 2, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, KYNU - kynureninase, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, NEMF - nuclear export mediator factor, NAMPT - nicotinamide phosphoribosyltransferase, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, TCEA1 - transcription elongation factor a (sii), 1, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, QKI - qki, kh domain containing, rna binding, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, AGK - acylglycerol kinase, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PTCD3 - pentatricopeptide repeat domain 3, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, PCYT2 - phosphate cytidylyltransferase 2, ethanolamine, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, RAB14 - rab14, member ras oncogene family, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, UCK2 - uridine-cytidine kinase 2, PPP5C - protein phosphatase 5, catalytic subunit, RPL3 - ribosomal protein l3, UMPS - uridine monophosphate synthetase, MRPS35 - mitochondrial ribosomal protein s35, DUT - deoxyuridine triphosphatase, MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, AK4 - adenylate kinase 4, HK2 - hexokinase 2, MAT2A - methionine adenosyltransferase ii, alpha, MCM3 - minichromosome maintenance complex component 3, RPL31 - ribosomal protein l31, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, RPL32 - ribosomal protein l32, DCAKD - dephospho-coa kinase domain containing, SCAF8 - sr-related ctd-associated factor 8, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, LSS - lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase), MRPL13 - mitochondrial ribosomal protein l13, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, RPL14 - ribosomal protein l14, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), GNPAT - glyceronephosphate o-acyltransferase, RNMT - rna (guanine-7-) methyltransferase, DGUOK - deoxyguanosine kinase, RPS8 - ribosomal protein s8, ESD - esterase d, MRPL42 - mitochondrial ribosomal protein l42, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, GOLPH3 - golgi phosphoprotein 3 (coat-protein), NHP2 - nhp2 ribonucleoprotein, RPS7 - ribosomal protein s7, ACOT7 - acyl-coa thioesterase 7, GCDH - glutaryl-coa dehydrogenase, UGDH - udp-glucose 6-dehydrogenase, ADI1 - acireductone dioxygenase 1, SSBP1 - single-stranded dna binding protein 1, mitochondrial, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), MRPS14 - mitochondrial ribosomal protein s14, MRPL47 - mitochondrial ribosomal protein l47, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, AK3 - adenylate kinase 3, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, RRBP1 - ribosome binding protein 1, PGM2 - phosphoglucomutase 2, RRM1 - ribonucleotide reductase m1, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
52 [TMEM30A - transmembrane protein 30a, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, PGM1 - phosphoglucomutase 1, OSTF1 - osteoclast stimulating factor 1, PGAM1 - phosphoglycerate mutase 1 (brain), GSN - gelsolin, HMGB1 - high mobility group box 1, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PYGB - phosphorylase, glycogen; brain, RAB14 - rab14, member ras oncogene family, PPFIA1 - protein tyrosine phosphatase, receptor type, f polypeptide (ptprf), interacting protein (liprin), alpha 1, FTH1 - ferritin, heavy polypeptide 1, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, UMPS - uridine monophosphate synthetase, MANBA - mannosidase, beta a, lysosomal, SURF4 - surfeit 4, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, STX17 - syntaxin 17, HK2 - hexokinase 2, PA2G4 - proliferation-associated 2g4, 38kda, KPNB1 - karyopherin (importin) beta 1, CAB39 - calcium binding protein 39, EXOC2 - exocyst complex component 2, EXOC1 - exocyst complex component 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), CREB1 - camp responsive element binding protein 1, CTSC - cathepsin c, ATG7 - autophagy related 7, RAP1A - rap1a, member of ras oncogene family, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, GLS - glutaminase, WDR1 - wd repeat domain 1, RAP1B - rap1b, member of ras oncogene family, GOLPH3 - golgi phosphoprotein 3 (coat-protein), STOM - stomatin, TMX3 - thioredoxin-related transmembrane protein 3, CAT - catalase, CDK13 - cyclin-dependent kinase 13, PDAP1 - pdgfa associated protein 1, FAM49B - family with sequence similarity 49, member b, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, ERP44 - endoplasmic reticulum protein 44, PRKCD - protein kinase c, delta, PGM2 - phosphoglucomutase 2, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1]
53 [CCNK - cyclin k, NCBP2 - nuclear cap binding protein subunit 2, 20kda, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, TCOF1 - treacher collins-franceschetti syndrome 1, RPL23A - ribosomal protein l23a, RFC3 - replication factor c (activator 1) 3, 38kda, RPL22 - ribosomal protein l22, PGM1 - phosphoglucomutase 1, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, GSPT1 - g1 to s phase transition 1, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, AGK - acylglycerol kinase, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, RPL13 - ribosomal protein l13, GFM2 - g elongation factor, mitochondrial 2, PTCD3 - pentatricopeptide repeat domain 3, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, DCK - deoxycytidine kinase, UCK2 - uridine-cytidine kinase 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, UMPS - uridine monophosphate synthetase, RPL3 - ribosomal protein l3, PPP5C - protein phosphatase 5, catalytic subunit, BTF3 - basic transcription factor 3, TRIM25 - tripartite motif containing 25, ACAT1 - acetyl-coa acetyltransferase 1, DUT - deoxyuridine triphosphatase, CPOX - coproporphyrinogen oxidase, MRPL9 - mitochondrial ribosomal protein l9, AK4 - adenylate kinase 4, HK2 - hexokinase 2, RPL31 - ribosomal protein l31, GAR1 - gar1 ribonucleoprotein, RPL32 - ribosomal protein l32, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, CREB1 - camp responsive element binding protein 1, DCAKD - dephospho-coa kinase domain containing, MRPL3 - mitochondrial ribosomal protein l3, TELO2 - telomere maintenance 2, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, SCAF8 - sr-related ctd-associated factor 8, MRPL13 - mitochondrial ribosomal protein l13, SNRPE - small nuclear ribonucleoprotein polypeptide e, RPL14 - ribosomal protein l14, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), RNMT - rna (guanine-7-) methyltransferase, DGUOK - deoxyguanosine kinase, RPS8 - ribosomal protein s8, MRPL42 - mitochondrial ribosomal protein l42, RPS13 - ribosomal protein s13, POLR1C - polymerase (rna) i polypeptide c, 30kda, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, NHP2 - nhp2 ribonucleoprotein, ADPGK - adp-dependent glucokinase, FECH - ferrochelatase, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, ACOT7 - acyl-coa thioesterase 7, GCDH - glutaryl-coa dehydrogenase, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, CDK13 - cyclin-dependent kinase 13, UGDH - udp-glucose 6-dehydrogenase, GNPDA2 - glucosamine-6-phosphate deaminase 2, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), MRPL47 - mitochondrial ribosomal protein l47, MRPS14 - mitochondrial ribosomal protein s14, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, KYNU - kynureninase, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, NAMPT - nicotinamide phosphoribosyltransferase, AK3 - adenylate kinase 3, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, RRBP1 - ribosome binding protein 1, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, RRM1 - ribonucleotide reductase m1, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1, TCEA1 - transcription elongation factor a (sii), 1]
54 [AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, TMEM30A - transmembrane protein 30a, ATG7 - autophagy related 7, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, RAP1A - rap1a, member of ras oncogene family, PGM1 - phosphoglucomutase 1, OSTF1 - osteoclast stimulating factor 1, PGAM1 - phosphoglycerate mutase 1 (brain), GLS - glutaminase, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, GSN - gelsolin, HMGB1 - high mobility group box 1, FABP5 - fatty acid binding protein 5 (psoriasis-associated), WDR1 - wd repeat domain 1, PYGB - phosphorylase, glycogen; brain, RAB14 - rab14, member ras oncogene family, PPFIA1 - protein tyrosine phosphatase, receptor type, f polypeptide (ptprf), interacting protein (liprin), alpha 1, RAP1B - rap1b, member of ras oncogene family, FTH1 - ferritin, heavy polypeptide 1, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, GOLPH3 - golgi phosphoprotein 3 (coat-protein), MANBA - mannosidase, beta a, lysosomal, SURF4 - surfeit 4, STOM - stomatin, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, TMX3 - thioredoxin-related transmembrane protein 3, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, STX17 - syntaxin 17, CAT - catalase, CDK13 - cyclin-dependent kinase 13, PDAP1 - pdgfa associated protein 1, PA2G4 - proliferation-associated 2g4, 38kda, KPNB1 - karyopherin (importin) beta 1, FAM49B - family with sequence similarity 49, member b, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), CAB39 - calcium binding protein 39, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, EXOC2 - exocyst complex component 2, ERP44 - endoplasmic reticulum protein 44, PRKCD - protein kinase c, delta, EXOC1 - exocyst complex component 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, PGM2 - phosphoglucomutase 2, CTSC - cathepsin c]
55 [CCNK - cyclin k, DST - dystonin, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, NCBP2 - nuclear cap binding protein subunit 2, 20kda, EXOSC5 - exosome component 5, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, RFC3 - replication factor c (activator 1) 3, 38kda, SF3A3 - splicing factor 3a, subunit 3, 60kda, SACM1L - sac1 suppressor of actin mutations 1-like (yeast), SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, NDUFA5 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, GFM2 - g elongation factor, mitochondrial 2, TMED2 - transmembrane emp24 domain trafficking protein 2, RECQL - recq protein-like (dna helicase q1-like), HOOK3 - hook microtubule-tethering protein 3, DERL1 - derlin 1, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, SPC24 - spc24, ndc80 kinetochore complex component, ACAT1 - acetyl-coa acetyltransferase 1, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, CPOX - coproporphyrinogen oxidase, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), SUGT1 - sgt1, suppressor of g2 allele of skp1 (s. cerevisiae), GOSR2 - golgi snap receptor complex member 2, OTUB1 - otu domain, ubiquitin aldehyde binding 1, SORD - sorbitol dehydrogenase, ZNF326 - zinc finger protein 326, PLCG1 - phospholipase c, gamma 1, WDR77 - wd repeat domain 77, RCSD1 - rcsd domain containing 1, MORF4L1 - mortality factor 4 like 1, CREB1 - camp responsive element binding protein 1, MYO6 - myosin vi, SNX1 - sorting nexin 1, THUMPD1 - thump domain containing 1, SGPL1 - sphingosine-1-phosphate lyase 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, EIF2D - eukaryotic translation initiation factor 2d, TPM3 - tropomyosin 3, SNRPE - small nuclear ribonucleoprotein polypeptide e, ANAPC7 - anaphase promoting complex subunit 7, SEH1L - seh1-like (s. cerevisiae), EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, GLS - glutaminase, PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, GLO1 - glyoxalase i, CC2D1B - coiled-coil and c2 domain containing 1b, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), GLG1 - golgi glycoprotein 1, NIP7 - nip7, nucleolar pre-rrna processing protein, GOSR1 - golgi snap receptor complex member 1, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), COMMD10 - comm domain containing 10, POLR1C - polymerase (rna) i polypeptide c, 30kda, DCAF7 - ddb1 and cul4 associated factor 7, ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, ADPGK - adp-dependent glucokinase, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, SUMO3 - small ubiquitin-like modifier 3, SNX6 - sorting nexin 6, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, TSFM - ts translation elongation factor, mitochondrial, SMARCD1 - swi/snf related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, KYNU - kynureninase, STX8 - syntaxin 8, ERP44 - endoplasmic reticulum protein 44, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), NAMPT - nicotinamide phosphoribosyltransferase, PARP4 - poly (adp-ribose) polymerase family, member 4, TMEM30A - transmembrane protein 30a, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, UQCRB - ubiquinol-cytochrome c reductase binding protein, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, CAPN2 - calpain 2, (m/ii) large subunit, ZWILCH - zwilch kinetochore protein, OSTF1 - osteoclast stimulating factor 1, UBE2Z - ubiquitin-conjugating enzyme e2z, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, AGK - acylglycerol kinase, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), PTCD3 - pentatricopeptide repeat domain 3, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, FTH1 - ferritin, heavy polypeptide 1, USP48 - ubiquitin specific peptidase 48, PPP5C - protein phosphatase 5, catalytic subunit, RPL3 - ribosomal protein l3, SURF4 - surfeit 4, IRAK4 - interleukin-1 receptor-associated kinase 4, DNAJC11 - dnaj (hsp40) homolog, subfamily c, member 11, MRPS35 - mitochondrial ribosomal protein s35, MRPL1 - mitochondrial ribosomal protein l1, RPN1 - ribophorin i, MRPL9 - mitochondrial ribosomal protein l9, AK4 - adenylate kinase 4, STX17 - syntaxin 17, HK2 - hexokinase 2, KPNB1 - karyopherin (importin) beta 1, TRIM38 - tripartite motif containing 38, CDC23 - cell division cycle 23, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, TJP2 - tight junction protein 2, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, VDAC2 - voltage-dependent anion channel 2, TTLL12 - tubulin tyrosine ligase-like family, member 12, MRPL13 - mitochondrial ribosomal protein l13, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, NIPBL - nipped-b homolog (drosophila), SCYL2 - scy1-like 2 (s. cerevisiae), GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, GALNT2 - udp-n-acetyl-alpha-d-galactosamine:polypeptide n-acetylgalactosaminyltransferase 2 (galnac-t2), TMEM70 - transmembrane protein 70, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), MFN1 - mitofusin 1, SNRNP27 - small nuclear ribonucleoprotein 27kda (u4/u6.u5), RNMT - rna (guanine-7-) methyltransferase, STAT6 - signal transducer and activator of transcription 6, interleukin-4 induced, WDR1 - wd repeat domain 1, LARP4B - la ribonucleoprotein domain family, member 4b, RPS8 - ribosomal protein s8, MRPL42 - mitochondrial ribosomal protein l42, ESD - esterase d, RPS13 - ribosomal protein s13, RINT1 - rad50 interactor 1, RPS6 - ribosomal protein s6, NHP2 - nhp2 ribonucleoprotein, RPS7 - ribosomal protein s7, STOM - stomatin, UCHL3 - ubiquitin carboxyl-terminal esterase l3 (ubiquitin thiolesterase), SUMO1 - small ubiquitin-like modifier 1, CISD2 - cdgsh iron sulfur domain 2, GCDH - glutaryl-coa dehydrogenase, CAT - catalase, UGDH - udp-glucose 6-dehydrogenase, XPO6 - exportin 6, NDUFAF4 - nadh dehydrogenase (ubiquinone) complex i, assembly factor 4, SSBP1 - single-stranded dna binding protein 1, mitochondrial, SAMM50 - samm50 sorting and assembly machinery component, MRPL47 - mitochondrial ribosomal protein l47, ANXA4 - annexin a4, TUBGCP3 - tubulin, gamma complex associated protein 3, UBE2I - ubiquitin-conjugating enzyme e2i, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, TNFAIP8 - tumor necrosis factor, alpha-induced protein 8, PRKCD - protein kinase c, delta, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, RRBP1 - ribosome binding protein 1, EPS15 - epidermal growth factor receptor pathway substrate 15, RRM1 - ribonucleotide reductase m1, DDX31 - dead (asp-glu-ala-asp) box polypeptide 31, ERAP1 - endoplasmic reticulum aminopeptidase 1, DDX6 - dead (asp-glu-ala-asp) box helicase 6, TCOF1 - treacher collins-franceschetti syndrome 1, SDAD1 - sda1 domain containing 1, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, ZMYM2 - zinc finger, mym-type 2, ZNF638 - zinc finger protein 638, STAMBP - stam binding protein, SCFD1 - sec1 family domain containing 1, GSN - gelsolin, CISD1 - cdgsh iron sulfur domain 1, URB1 - urb1 ribosome biogenesis 1 homolog (s. cerevisiae), RAB30 - rab30, member ras oncogene family, FLNB - filamin b, beta, ECI1 - enoyl-coa delta isomerase 1, DCK - deoxycytidine kinase, PPP4R2 - protein phosphatase 4, regulatory subunit 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, DBN1 - drebrin 1, TRIM25 - tripartite motif containing 25, ILF3 - interleukin enhancer binding factor 3, 90kda, XPOT - exportin, trna, ANKRD28 - ankyrin repeat domain 28, NUDCD2 - nudc domain containing 2, RIF1 - rap1 interacting factor homolog (yeast), PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, CYCS - cytochrome c, somatic, VTI1A - vesicle transport through interaction with t-snares 1a, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, CCDC51 - coiled-coil domain containing 51, RBM19 - rna binding motif protein 19, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), MMS22L - mms22-like, dna repair protein, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, EHBP1 - eh domain binding protein 1, ARCN1 - archain 1, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, PA2G4 - proliferation-associated 2g4, 38kda, AGPS - alkylglycerone phosphate synthase, CAB39 - calcium binding protein 39, TMED5 - transmembrane emp24 protein transport domain containing 5, GATAD2B - gata zinc finger domain containing 2b, RAB23 - rab23, member ras oncogene family, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, GAR1 - gar1 ribonucleoprotein, KDM3A - lysine (k)-specific demethylase 3a, ANAPC1 - anaphase promoting complex subunit 1, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, MRPL3 - mitochondrial ribosomal protein l3, TELO2 - telomere maintenance 2, EXOSC1 - exosome component 1, PITRM1 - pitrilysin metallopeptidase 1, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), MCM7 - minichromosome maintenance complex component 7, ATG7 - autophagy related 7, MCU - mitochondrial calcium uniporter, ARL1 - adp-ribosylation factor-like 1, TUBGCP4 - tubulin, gamma complex associated protein 4, KDELC2 - kdel (lys-asp-glu-leu) containing 2, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), SYNE1 - spectrin repeat containing, nuclear envelope 1, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), NUP88 - nucleoporin 88kda, SCO1 - sco1 cytochrome c oxidase assembly protein, ZBTB8OS - zinc finger and btb domain containing 8 opposite strand, NOM1 - nucleolar protein with mif4g domain 1, WDHD1 - wd repeat and hmg-box dna binding protein 1, NFU1 - nfu1 iron-sulfur cluster scaffold homolog (s. cerevisiae), PPIL3 - peptidylprolyl isomerase (cyclophilin)-like 3, HADH - hydroxyacyl-coa dehydrogenase, OSBPL3 - oxysterol binding protein-like 3, THOC5 - tho complex 5, FECH - ferrochelatase, CDK6 - cyclin-dependent kinase 6, ADNP - activity-dependent neuroprotector homeobox, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, CDK13 - cyclin-dependent kinase 13, TBCD - tubulin folding cofactor d, FCF1 - fcf1 rrna-processing protein, CCT4 - chaperonin containing tcp1, subunit 4 (delta), TBCA - tubulin folding cofactor a, CUTC - cutc copper transporter, INPP5B - inositol polyphosphate-5-phosphatase, 75kda, RPL7L1 - ribosomal protein l7-like 1, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, IRF3 - interferon regulatory factor 3, ARL6IP5 - adp-ribosylation-like factor 6 interacting protein 5, STX18 - syntaxin 18, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, IRF5 - interferon regulatory factor 5, DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), RPTOR - regulatory associated protein of mtor, complex 1, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, EIF3E - eukaryotic translation initiation factor 3, subunit e, QSOX2 - quiescin q6 sulfhydryl oxidase 2, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, TCEA1 - transcription elongation factor a (sii), 1, LYRM7 - lyr motif containing 7, VTA1 - vesicle (multivesicular body) trafficking 1, PURA - purine-rich element binding protein a, SPG11 - spastic paraplegia 11 (autosomal recessive), ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, TDRD7 - tudor domain containing 7, PGM1 - phosphoglucomutase 1, NEK6 - nima-related kinase 6, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, NOC3L - nucleolar complex associated 3 homolog (s. cerevisiae), PYGB - phosphorylase, glycogen; brain, PCYT2 - phosphate cytidylyltransferase 2, ethanolamine, NUP50 - nucleoporin 50kda, MAP4 - microtubule-associated protein 4, RAB14 - rab14, member ras oncogene family, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, MANBA - mannosidase, beta a, lysosomal, VKORC1L1 - vitamin k epoxide reductase complex, subunit 1-like 1, MAN2A1 - mannosidase, alpha, class 2a, member 1, USP8 - ubiquitin specific peptidase 8, TRAPPC10 - trafficking protein particle complex 10, MBNL1 - muscleblind-like splicing regulator 1, METTL15 - methyltransferase like 15, DUT - deoxyuridine triphosphatase, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, MCM3 - minichromosome maintenance complex component 3, IDE - insulin-degrading enzyme, CHMP5 - charged multivesicular body protein 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, DOCK7 - dedicator of cytokinesis 7, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, SART3 - squamous cell carcinoma antigen recognized by t cells 3, MAD1L1 - mad1 mitotic arrest deficient-like 1 (yeast), CTSC - cathepsin c, WIPF1 - was/wasl interacting protein family, member 1, SCAF8 - sr-related ctd-associated factor 8, LSS - lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase), RANBP1 - ran binding protein 1, DLG1 - discs, large homolog 1 (drosophila), RAP1A - rap1a, member of ras oncogene family, PUS1 - pseudouridylate synthase 1, RPL14 - ribosomal protein l14, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, GNPAT - glyceronephosphate o-acyltransferase, AKAP8L - a kinase (prka) anchor protein 8-like, NUB1 - negative regulator of ubiquitin-like proteins 1, TRAPPC9 - trafficking protein particle complex 9, DGUOK - deoxyguanosine kinase, CLNS1A - chloride channel, nucleotide-sensitive, 1a, RAP1B - rap1b, member of ras oncogene family, GOLPH3 - golgi phosphoprotein 3 (coat-protein), TMX3 - thioredoxin-related transmembrane protein 3, DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, ACOT7 - acyl-coa thioesterase 7, RB1 - retinoblastoma 1, PDAP1 - pdgfa associated protein 1, CBX3 - chromobox homolog 3, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, FAM49B - family with sequence similarity 49, member b, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), RHOT1 - ras homolog family member t1, MRPS14 - mitochondrial ribosomal protein s14, CUL1 - cullin 1, RSL1D1 - ribosomal l1 domain containing 1, BCL2L1 - bcl2-like 1, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, PGM2 - phosphoglucomutase 2]
56 [CCNK - cyclin k, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, DST - dystonin, NCBP2 - nuclear cap binding protein subunit 2, 20kda, EXOSC5 - exosome component 5, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, RFC3 - replication factor c (activator 1) 3, 38kda, SF3A3 - splicing factor 3a, subunit 3, 60kda, SACM1L - sac1 suppressor of actin mutations 1-like (yeast), SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, GFM2 - g elongation factor, mitochondrial 2, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, TMED2 - transmembrane emp24 domain trafficking protein 2, RECQL - recq protein-like (dna helicase q1-like), HOOK3 - hook microtubule-tethering protein 3, DERL1 - derlin 1, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, BTF3 - basic transcription factor 3, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, SPC24 - spc24, ndc80 kinetochore complex component, ACAT1 - acetyl-coa acetyltransferase 1, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, CPOX - coproporphyrinogen oxidase, LRRC40 - leucine rich repeat containing 40, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), SUGT1 - sgt1, suppressor of g2 allele of skp1 (s. cerevisiae), GOSR2 - golgi snap receptor complex member 2, USP25 - ubiquitin specific peptidase 25, OTUB1 - otu domain, ubiquitin aldehyde binding 1, SORD - sorbitol dehydrogenase, ZNF326 - zinc finger protein 326, WDR77 - wd repeat domain 77, CARHSP1 - calcium regulated heat stable protein 1, 24kda, CREB1 - camp responsive element binding protein 1, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, MYO6 - myosin vi, SNX1 - sorting nexin 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, SGPL1 - sphingosine-1-phosphate lyase 1, TPM3 - tropomyosin 3, SNRPE - small nuclear ribonucleoprotein polypeptide e, STK38L - serine/threonine kinase 38 like, ANAPC7 - anaphase promoting complex subunit 7, SEH1L - seh1-like (s. cerevisiae), EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, GLS - glutaminase, PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, IARS - isoleucyl-trna synthetase, GLO1 - glyoxalase i, CC2D1B - coiled-coil and c2 domain containing 1b, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), GLG1 - golgi glycoprotein 1, GOSR1 - golgi snap receptor complex member 1, NIP7 - nip7, nucleolar pre-rrna processing protein, DCAF7 - ddb1 and cul4 associated factor 7, ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, ADPGK - adp-dependent glucokinase, PPA1 - pyrophosphatase (inorganic) 1, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, SUMO3 - small ubiquitin-like modifier 3, SNX6 - sorting nexin 6, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, TSFM - ts translation elongation factor, mitochondrial, SMARCD1 - swi/snf related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, DENND4A - denn/madd domain containing 4a, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, COMT - catechol-o-methyltransferase, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, KYNU - kynureninase, STX8 - syntaxin 8, ERP44 - endoplasmic reticulum protein 44, NAMPT - nicotinamide phosphoribosyltransferase, HSPA4 - heat shock 70kda protein 4, PARP4 - poly (adp-ribose) polymerase family, member 4, SCPEP1 - serine carboxypeptidase 1, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, TMEM30A - transmembrane protein 30a, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, QKI - qki, kh domain containing, rna binding, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, CAPN2 - calpain 2, (m/ii) large subunit, ZWILCH - zwilch kinetochore protein, UBE2Z - ubiquitin-conjugating enzyme e2z, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, HMGB2 - high mobility group box 2, AGK - acylglycerol kinase, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PTCD3 - pentatricopeptide repeat domain 3, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, HOMER1 - homer homolog 1 (drosophila), FTH1 - ferritin, heavy polypeptide 1, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, UMPS - uridine monophosphate synthetase, USP48 - ubiquitin specific peptidase 48, PPP5C - protein phosphatase 5, catalytic subunit, RPL3 - ribosomal protein l3, SURF4 - surfeit 4, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), IRAK4 - interleukin-1 receptor-associated kinase 4, DNAJC11 - dnaj (hsp40) homolog, subfamily c, member 11, MRPL1 - mitochondrial ribosomal protein l1, RPN1 - ribophorin i, MRPL9 - mitochondrial ribosomal protein l9, TNPO1 - transportin 1, AK4 - adenylate kinase 4, STX17 - syntaxin 17, HK2 - hexokinase 2, KPNB1 - karyopherin (importin) beta 1, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, TJP2 - tight junction protein 2, IFIT5 - interferon-induced protein with tetratricopeptide repeats 5, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, VDAC2 - voltage-dependent anion channel 2, TTLL12 - tubulin tyrosine ligase-like family, member 12, MRPL13 - mitochondrial ribosomal protein l13, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, NIPBL - nipped-b homolog (drosophila), SCYL2 - scy1-like 2 (s. cerevisiae), GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, GALNT2 - udp-n-acetyl-alpha-d-galactosamine:polypeptide n-acetylgalactosaminyltransferase 2 (galnac-t2), TMEM70 - transmembrane protein 70, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), MFN1 - mitofusin 1, RNMT - rna (guanine-7-) methyltransferase, WDR1 - wd repeat domain 1, LARP4B - la ribonucleoprotein domain family, member 4b, EIF1AD - eukaryotic translation initiation factor 1a domain containing, RPS8 - ribosomal protein s8, ESD - esterase d, MRPL42 - mitochondrial ribosomal protein l42, RPS13 - ribosomal protein s13, RINT1 - rad50 interactor 1, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, DSTN - destrin (actin depolymerizing factor), STOM - stomatin, SUMO1 - small ubiquitin-like modifier 1, SRP72 - signal recognition particle 72kda, GCDH - glutaryl-coa dehydrogenase, CISD2 - cdgsh iron sulfur domain 2, CAT - catalase, USP15 - ubiquitin specific peptidase 15, UGDH - udp-glucose 6-dehydrogenase, XPO6 - exportin 6, NDUFAF4 - nadh dehydrogenase (ubiquinone) complex i, assembly factor 4, SSBP1 - single-stranded dna binding protein 1, mitochondrial, SAMM50 - samm50 sorting and assembly machinery component, MRPL47 - mitochondrial ribosomal protein l47, ANXA4 - annexin a4, TUBGCP3 - tubulin, gamma complex associated protein 3, UBE2I - ubiquitin-conjugating enzyme e2i, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, PRKCD - protein kinase c, delta, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), CCDC6 - coiled-coil domain containing 6, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, RRBP1 - ribosome binding protein 1, EPS15 - epidermal growth factor receptor pathway substrate 15, ZC3H7B - zinc finger ccch-type containing 7b, DDX31 - dead (asp-glu-ala-asp) box polypeptide 31, ERAP1 - endoplasmic reticulum aminopeptidase 1, DDX6 - dead (asp-glu-ala-asp) box helicase 6, TCOF1 - treacher collins-franceschetti syndrome 1, SDAD1 - sda1 domain containing 1, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, ZNF638 - zinc finger protein 638, F11R - f11 receptor, STAMBP - stam binding protein, SCFD1 - sec1 family domain containing 1, GSN - gelsolin, CISD1 - cdgsh iron sulfur domain 1, URB1 - urb1 ribosome biogenesis 1 homolog (s. cerevisiae), RAB30 - rab30, member ras oncogene family, FLNB - filamin b, beta, ECI1 - enoyl-coa delta isomerase 1, DCK - deoxycytidine kinase, PPP4R2 - protein phosphatase 4, regulatory subunit 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, DBN1 - drebrin 1, TRIM25 - tripartite motif containing 25, ILF3 - interleukin enhancer binding factor 3, 90kda, NUDCD2 - nudc domain containing 2, RIF1 - rap1 interacting factor homolog (yeast), CYCS - cytochrome c, somatic, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, VTI1A - vesicle transport through interaction with t-snares 1a, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, RBM19 - rna binding motif protein 19, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, EHBP1 - eh domain binding protein 1, ARCN1 - archain 1, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, CD48 - cd48 molecule, LRRK1 - leucine-rich repeat kinase 1, PA2G4 - proliferation-associated 2g4, 38kda, AGPS - alkylglycerone phosphate synthase, CAB39 - calcium binding protein 39, TBC1D4 - tbc1 domain family, member 4, TMED5 - transmembrane emp24 protein transport domain containing 5, RAB23 - rab23, member ras oncogene family, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, KDM3A - lysine (k)-specific demethylase 3a, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, MRPL3 - mitochondrial ribosomal protein l3, TELO2 - telomere maintenance 2, EXOSC1 - exosome component 1, LRRC57 - leucine rich repeat containing 57, PITRM1 - pitrilysin metallopeptidase 1, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), MCM7 - minichromosome maintenance complex component 7, MCU - mitochondrial calcium uniporter, ARL1 - adp-ribosylation factor-like 1, TUBGCP4 - tubulin, gamma complex associated protein 4, PRRC1 - proline-rich coiled-coil 1, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), SYNE1 - spectrin repeat containing, nuclear envelope 1, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), PELO - pelota homolog (drosophila), SCO1 - sco1 cytochrome c oxidase assembly protein, NOM1 - nucleolar protein with mif4g domain 1, NFU1 - nfu1 iron-sulfur cluster scaffold homolog (s. cerevisiae), HADH - hydroxyacyl-coa dehydrogenase, OSBPL3 - oxysterol binding protein-like 3, MBLAC2 - metallo-beta-lactamase domain containing 2, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), THOC5 - tho complex 5, FECH - ferrochelatase, CDK6 - cyclin-dependent kinase 6, ADNP - activity-dependent neuroprotector homeobox, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, CDK13 - cyclin-dependent kinase 13, FCF1 - fcf1 rrna-processing protein, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, CCT4 - chaperonin containing tcp1, subunit 4 (delta), GNPDA2 - glucosamine-6-phosphate deaminase 2, CUTC - cutc copper transporter, TBCA - tubulin folding cofactor a, INPP5B - inositol polyphosphate-5-phosphatase, 75kda, RPL7L1 - ribosomal protein l7-like 1, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, IRF3 - interferon regulatory factor 3, ARL6IP5 - adp-ribosylation-like factor 6 interacting protein 5, STX18 - syntaxin 18, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, LARP1 - la ribonucleoprotein domain family, member 1, NEMF - nuclear export mediator factor, IRF5 - interferon regulatory factor 5, DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), RPTOR - regulatory associated protein of mtor, complex 1, EIF3E - eukaryotic translation initiation factor 3, subunit e, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, QSOX2 - quiescin q6 sulfhydryl oxidase 2, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, TCEA1 - transcription elongation factor a (sii), 1, VTA1 - vesicle (multivesicular body) trafficking 1, SERBP1 - serpine1 mrna binding protein 1, PURA - purine-rich element binding protein a, GAPVD1 - gtpase activating protein and vps9 domains 1, DENND3 - denn/madd domain containing 3, SPG11 - spastic paraplegia 11 (autosomal recessive), ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, TDRD7 - tudor domain containing 7, PGM1 - phosphoglucomutase 1, NEK6 - nima-related kinase 6, PGAM1 - phosphoglycerate mutase 1 (brain), NOC3L - nucleolar complex associated 3 homolog (s. cerevisiae), PYGB - phosphorylase, glycogen; brain, NUP50 - nucleoporin 50kda, MAP4 - microtubule-associated protein 4, RAB14 - rab14, member ras oncogene family, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, MANBA - mannosidase, beta a, lysosomal, VKORC1L1 - vitamin k epoxide reductase complex, subunit 1-like 1, MAN2A1 - mannosidase, alpha, class 2a, member 1, USP8 - ubiquitin specific peptidase 8, MBNL1 - muscleblind-like splicing regulator 1, CCDC58 - coiled-coil domain containing 58, SLAMF1 - signaling lymphocytic activation molecule family member 1, DUT - deoxyuridine triphosphatase, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, MCM3 - minichromosome maintenance complex component 3, IDE - insulin-degrading enzyme, CHMP5 - charged multivesicular body protein 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, USP34 - ubiquitin specific peptidase 34, IFIH1 - interferon induced with helicase c domain 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, SART3 - squamous cell carcinoma antigen recognized by t cells 3, MAD1L1 - mad1 mitotic arrest deficient-like 1 (yeast), CTSC - cathepsin c, WIPF1 - was/wasl interacting protein family, member 1, SCAF8 - sr-related ctd-associated factor 8, LSS - lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase), UBA6 - ubiquitin-like modifier activating enzyme 6, RANBP1 - ran binding protein 1, DLG1 - discs, large homolog 1 (drosophila), RAP1A - rap1a, member of ras oncogene family, PARVG - parvin, gamma, PUS1 - pseudouridylate synthase 1, RPL14 - ribosomal protein l14, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, GNPAT - glyceronephosphate o-acyltransferase, AKAP8L - a kinase (prka) anchor protein 8-like, ZNF592 - zinc finger protein 592, NUB1 - negative regulator of ubiquitin-like proteins 1, CYFIP2 - cytoplasmic fmr1 interacting protein 2, RBM26 - rna binding motif protein 26, DGUOK - deoxyguanosine kinase, TRAPPC9 - trafficking protein particle complex 9, CLNS1A - chloride channel, nucleotide-sensitive, 1a, UBA3 - ubiquitin-like modifier activating enzyme 3, RAP1B - rap1b, member of ras oncogene family, GOLPH3 - golgi phosphoprotein 3 (coat-protein), DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, ACOT7 - acyl-coa thioesterase 7, RB1 - retinoblastoma 1, CBX3 - chromobox homolog 3, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, FAM49B - family with sequence similarity 49, member b, ADI1 - acireductone dioxygenase 1, CAPRIN1 - cell cycle associated protein 1, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), RHOT1 - ras homolog family member t1, MRPS14 - mitochondrial ribosomal protein s14, RSL1D1 - ribosomal l1 domain containing 1, BCL2L1 - bcl2-like 1, AK3 - adenylate kinase 3, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, PGM2 - phosphoglucomutase 2, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
57 [AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, VTA1 - vesicle (multivesicular body) trafficking 1, PURA - purine-rich element binding protein a, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, RPL23A - ribosomal protein l23a, SDAD1 - sda1 domain containing 1, RPL22 - ribosomal protein l22, SCFD1 - sec1 family domain containing 1, AGK - acylglycerol kinase, RPL13 - ribosomal protein l13, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), TBC1D2B - tbc1 domain family, member 2b, TMED2 - transmembrane emp24 domain trafficking protein 2, RPL6 - ribosomal protein l6, NUP50 - nucleoporin 50kda, HOOK3 - hook microtubule-tethering protein 3, RPL7 - ribosomal protein l7, RAB14 - rab14, member ras oncogene family, RPL3 - ribosomal protein l3, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, DERL1 - derlin 1, BTF3 - basic transcription factor 3, VTI1A - vesicle transport through interaction with t-snares 1a, TNPO1 - transportin 1, STX17 - syntaxin 17, EHBP1 - eh domain binding protein 1, ARCN1 - archain 1, KPNB1 - karyopherin (importin) beta 1, AGPS - alkylglycerone phosphate synthase, GOSR2 - golgi snap receptor complex member 2, IDE - insulin-degrading enzyme, TBC1D4 - tbc1 domain family, member 4, CHMP5 - charged multivesicular body protein 5, EXOC2 - exocyst complex component 2, TMED5 - transmembrane emp24 protein transport domain containing 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, RAB23 - rab23, member ras oncogene family, EXOC1 - exocyst complex component 1, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, MYO6 - myosin vi, SNX1 - sorting nexin 1, PITRM1 - pitrilysin metallopeptidase 1, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), EIF2D - eukaryotic translation initiation factor 2d, ATG7 - autophagy related 7, ARL1 - adp-ribosylation factor-like 1, GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, RPL14 - ribosomal protein l14, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), GNPAT - glyceronephosphate o-acyltransferase, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), NUP88 - nucleoporin 88kda, RPS8 - ribosomal protein s8, GOSR1 - golgi snap receptor complex member 1, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RINT1 - rad50 interactor 1, GOLPH3 - golgi phosphoprotein 3 (coat-protein), RPS7 - ribosomal protein s7, SNX6 - sorting nexin 6, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, SRP72 - signal recognition particle 72kda, CAT - catalase, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, HSPD1 - heat shock 60kda protein 1 (chaperonin), XPO6 - exportin 6, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), SAMM50 - samm50 sorting and assembly machinery component, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, STX8 - syntaxin 8, STX18 - syntaxin 18, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, HSPA4 - heat shock 70kda protein 4, RRBP1 - ribosome binding protein 1, EPS15 - epidermal growth factor receptor pathway substrate 15]
58 [AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, TMEM30A - transmembrane protein 30a, PURA - purine-rich element binding protein a, RPL22 - ribosomal protein l22, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, PGM1 - phosphoglucomutase 1, OSTF1 - osteoclast stimulating factor 1, PGAM1 - phosphoglycerate mutase 1 (brain), GSN - gelsolin, HMGB1 - high mobility group box 1, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PYGB - phosphorylase, glycogen; brain, RAB14 - rab14, member ras oncogene family, FTH1 - ferritin, heavy polypeptide 1, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, MANBA - mannosidase, beta a, lysosomal, SURF4 - surfeit 4, DOCK8 - dedicator of cytokinesis 8, SLAMF1 - signaling lymphocytic activation molecule family member 1, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PA2G4 - proliferation-associated 2g4, 38kda, KPNB1 - karyopherin (importin) beta 1, CAB39 - calcium binding protein 39, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), CTSC - cathepsin c, ATG7 - autophagy related 7, DLG1 - discs, large homolog 1 (drosophila), RAP1A - rap1a, member of ras oncogene family, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, RAP1B - rap1b, member of ras oncogene family, RPS6 - ribosomal protein s6, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), STOM - stomatin, CDK6 - cyclin-dependent kinase 6, PDAP1 - pdgfa associated protein 1, CDK13 - cyclin-dependent kinase 13, CAT - catalase, HSPD1 - heat shock 60kda protein 1 (chaperonin), ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, ERP44 - endoplasmic reticulum protein 44, PRKCD - protein kinase c, delta, NAMPT - nicotinamide phosphoribosyltransferase, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, PGM2 - phosphoglucomutase 2]
59 [PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, UQCRB - ubiquinol-cytochrome c reductase binding protein, DLG1 - discs, large homolog 1 (drosophila), PGM1 - phosphoglucomutase 1, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, PGAM1 - phosphoglycerate mutase 1 (brain), MFN1 - mitofusin 1, DGUOK - deoxyguanosine kinase, UCK2 - uridine-cytidine kinase 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, UMPS - uridine monophosphate synthetase, ADPGK - adp-dependent glucokinase, ACAT1 - acetyl-coa acetyltransferase 1, HK2 - hexokinase 2, AK4 - adenylate kinase 4, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, ACOT7 - acyl-coa thioesterase 7, GCDH - glutaryl-coa dehydrogenase, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, MVD - mevalonate (diphospho) decarboxylase, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, KYNU - kynureninase, RAB23 - rab23, member ras oncogene family, TJP2 - tight junction protein 2, AK3 - adenylate kinase 3, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, DCAKD - dephospho-coa kinase domain containing, PGM2 - phosphoglucomutase 2]
60 [MRPL13 - mitochondrial ribosomal protein l13, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, GSPT1 - g1 to s phase transition 1, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, RPL14 - ribosomal protein l14, RPL13 - ribosomal protein l13, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, GFM2 - g elongation factor, mitochondrial 2, PTCD3 - pentatricopeptide repeat domain 3, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, RPS8 - ribosomal protein s8, MRPL42 - mitochondrial ribosomal protein l42, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPL3 - ribosomal protein l3, RPS7 - ribosomal protein s7, MRPL2 - mitochondrial ribosomal protein l2, MRPL9 - mitochondrial ribosomal protein l9, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, MRPS14 - mitochondrial ribosomal protein s14, MRPL47 - mitochondrial ribosomal protein l47, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, RRBP1 - ribosome binding protein 1, MRPL3 - mitochondrial ribosomal protein l3, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
61 [AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, PURA - purine-rich element binding protein a, VTA1 - vesicle (multivesicular body) trafficking 1, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, RPL23A - ribosomal protein l23a, SDAD1 - sda1 domain containing 1, RPL22 - ribosomal protein l22, SCFD1 - sec1 family domain containing 1, AGK - acylglycerol kinase, RPL13 - ribosomal protein l13, TBC1D2B - tbc1 domain family, member 2b, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), TMED2 - transmembrane emp24 domain trafficking protein 2, NUP50 - nucleoporin 50kda, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, HOOK3 - hook microtubule-tethering protein 3, RAB14 - rab14, member ras oncogene family, RPL3 - ribosomal protein l3, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, DERL1 - derlin 1, BTF3 - basic transcription factor 3, VTI1A - vesicle transport through interaction with t-snares 1a, TNPO1 - transportin 1, HK2 - hexokinase 2, STX17 - syntaxin 17, EHBP1 - eh domain binding protein 1, ARCN1 - archain 1, KPNB1 - karyopherin (importin) beta 1, AGPS - alkylglycerone phosphate synthase, GOSR2 - golgi snap receptor complex member 2, IDE - insulin-degrading enzyme, TBC1D4 - tbc1 domain family, member 4, EXOC2 - exocyst complex component 2, CHMP5 - charged multivesicular body protein 5, TMED5 - transmembrane emp24 protein transport domain containing 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, RAB23 - rab23, member ras oncogene family, LIMS1 - lim and senescent cell antigen-like domains 1, EXOC1 - exocyst complex component 1, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, SNX1 - sorting nexin 1, MYO6 - myosin vi, PITRM1 - pitrilysin metallopeptidase 1, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), ATG7 - autophagy related 7, EIF2D - eukaryotic translation initiation factor 2d, ARL1 - adp-ribosylation factor-like 1, NIPBL - nipped-b homolog (drosophila), GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, RPL14 - ribosomal protein l14, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), GNPAT - glyceronephosphate o-acyltransferase, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), NUP88 - nucleoporin 88kda, RPS8 - ribosomal protein s8, GOSR1 - golgi snap receptor complex member 1, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RINT1 - rad50 interactor 1, GOLPH3 - golgi phosphoprotein 3 (coat-protein), RPS7 - ribosomal protein s7, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, SNX6 - sorting nexin 6, SRP72 - signal recognition particle 72kda, CAT - catalase, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, HSPD1 - heat shock 60kda protein 1 (chaperonin), XPO6 - exportin 6, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), SAMM50 - samm50 sorting and assembly machinery component, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, STX8 - syntaxin 8, STX18 - syntaxin 18, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, HSPA4 - heat shock 70kda protein 4, RRBP1 - ribosome binding protein 1, EPS15 - epidermal growth factor receptor pathway substrate 15]
62 [CCNK - cyclin k, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, GFM2 - g elongation factor, mitochondrial 2, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, FKBP11 - fk506 binding protein 11, 19 kda, DERL1 - derlin 1, ACAT1 - acetyl-coa acetyltransferase 1, CPOX - coproporphyrinogen oxidase, LRRC40 - leucine rich repeat containing 40, USP25 - ubiquitin specific peptidase 25, OTUB1 - otu domain, ubiquitin aldehyde binding 1, MORF4L1 - mortality factor 4 like 1, CREB1 - camp responsive element binding protein 1, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, SGPL1 - sphingosine-1-phosphate lyase 1, STK38L - serine/threonine kinase 38 like, ANAPC7 - anaphase promoting complex subunit 7, SEH1L - seh1-like (s. cerevisiae), EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, GLS - glutaminase, PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, IARS - isoleucyl-trna synthetase, GLO1 - glyoxalase i, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), DCAF7 - ddb1 and cul4 associated factor 7, PPA1 - pyrophosphatase (inorganic) 1, ADPGK - adp-dependent glucokinase, SUMO3 - small ubiquitin-like modifier 3, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), MVD - mevalonate (diphospho) decarboxylase, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, COMT - catechol-o-methyltransferase, KYNU - kynureninase, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, TAOK3 - tao kinase 3, ERP44 - endoplasmic reticulum protein 44, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), NAMPT - nicotinamide phosphoribosyltransferase, PARP4 - poly (adp-ribose) polymerase family, member 4, SCPEP1 - serine carboxypeptidase 1, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, UQCRB - ubiquinol-cytochrome c reductase binding protein, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, CAPN2 - calpain 2, (m/ii) large subunit, WNK1 - wnk lysine deficient protein kinase 1, UBE2Z - ubiquitin-conjugating enzyme e2z, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, AGK - acylglycerol kinase, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PTCD3 - pentatricopeptide repeat domain 3, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, UCK2 - uridine-cytidine kinase 2, PPP5C - protein phosphatase 5, catalytic subunit, USP48 - ubiquitin specific peptidase 48, UMPS - uridine monophosphate synthetase, RPL3 - ribosomal protein l3, CARS - cysteinyl-trna synthetase, IRAK4 - interleukin-1 receptor-associated kinase 4, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), EIF3F - eukaryotic translation initiation factor 3, subunit f, RPN1 - ribophorin i, MRPL9 - mitochondrial ribosomal protein l9, AK4 - adenylate kinase 4, HK2 - hexokinase 2, TRIM38 - tripartite motif containing 38, CDC23 - cell division cycle 23, TJP2 - tight junction protein 2, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, DCAKD - dephospho-coa kinase domain containing, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, TTLL12 - tubulin tyrosine ligase-like family, member 12, MRPL13 - mitochondrial ribosomal protein l13, SCYL2 - scy1-like 2 (s. cerevisiae), GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, GALNT2 - udp-n-acetyl-alpha-d-galactosamine:polypeptide n-acetylgalactosaminyltransferase 2 (galnac-t2), MFN1 - mitofusin 1, RPS8 - ribosomal protein s8, MRPL42 - mitochondrial ribosomal protein l42, ESD - esterase d, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, UCHL3 - ubiquitin carboxyl-terminal esterase l3 (ubiquitin thiolesterase), SUMO1 - small ubiquitin-like modifier 1, GCDH - glutaryl-coa dehydrogenase, CAT - catalase, USP15 - ubiquitin specific peptidase 15, UGDH - udp-glucose 6-dehydrogenase, MRPL47 - mitochondrial ribosomal protein l47, UBE2I - ubiquitin-conjugating enzyme e2i, PRKCD - protein kinase c, delta, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, RRBP1 - ribosome binding protein 1, RRM1 - ribonucleotide reductase m1, ERAP1 - endoplasmic reticulum aminopeptidase 1, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, GSPT1 - g1 to s phase transition 1, STAMBP - stam binding protein, GSN - gelsolin, FAM98A - family with sequence similarity 98, member a, DCK - deoxycytidine kinase, PPP4R2 - protein phosphatase 4, regulatory subunit 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, TRIM25 - tripartite motif containing 25, ILF3 - interleukin enhancer binding factor 3, 90kda, CYCS - cytochrome c, somatic, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, LRRK1 - leucine-rich repeat kinase 1, CAB39 - calcium binding protein 39, RAB23 - rab23, member ras oncogene family, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, KDM3A - lysine (k)-specific demethylase 3a, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, ANAPC1 - anaphase promoting complex subunit 1, MRPL3 - mitochondrial ribosomal protein l3, PITRM1 - pitrilysin metallopeptidase 1, RNF31 - ring finger protein 31, ATG7 - autophagy related 7, KDELC2 - kdel (lys-asp-glu-leu) containing 2, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, NUP88 - nucleoporin 88kda, NFU1 - nfu1 iron-sulfur cluster scaffold homolog (s. cerevisiae), PPIL3 - peptidylprolyl isomerase (cyclophilin)-like 3, FECH - ferrochelatase, CDK6 - cyclin-dependent kinase 6, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, CDK13 - cyclin-dependent kinase 13, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, NEMF - nuclear export mediator factor, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, DENND3 - denn/madd domain containing 3, PGM1 - phosphoglucomutase 1, NEK6 - nima-related kinase 6, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, NUP50 - nucleoporin 50kda, PHKB - phosphorylase kinase, beta, MANBA - mannosidase, beta a, lysosomal, MAN2A1 - mannosidase, alpha, class 2a, member 1, VKORC1L1 - vitamin k epoxide reductase complex, subunit 1-like 1, USP8 - ubiquitin specific peptidase 8, DUT - deoxyuridine triphosphatase, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, IDE - insulin-degrading enzyme, USP34 - ubiquitin specific peptidase 34, IFIH1 - interferon induced with helicase c domain 1, MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, CTSC - cathepsin c, UBA6 - ubiquitin-like modifier activating enzyme 6, GRK6 - g protein-coupled receptor kinase 6, DLG1 - discs, large homolog 1 (drosophila), RPL14 - ribosomal protein l14, NSMAF - neutral sphingomyelinase (n-smase) activation associated factor, NUB1 - negative regulator of ubiquitin-like proteins 1, CYFIP2 - cytoplasmic fmr1 interacting protein 2, DGUOK - deoxyguanosine kinase, UBA3 - ubiquitin-like modifier activating enzyme 3, GOLPH3 - golgi phosphoprotein 3 (coat-protein), TMX3 - thioredoxin-related transmembrane protein 3, ACOT7 - acyl-coa thioesterase 7, ADI1 - acireductone dioxygenase 1, MRPS14 - mitochondrial ribosomal protein s14, RHOT1 - ras homolog family member t1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, CUL1 - cullin 1, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, PGM2 - phosphoglucomutase 2, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
63 [DST - dystonin, NCBP2 - nuclear cap binding protein subunit 2, 20kda, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, SDAD1 - sda1 domain containing 1, F11R - f11 receptor, SCFD1 - sec1 family domain containing 1, GSN - gelsolin, FAM98A - family with sequence similarity 98, member a, TMED2 - transmembrane emp24 domain trafficking protein 2, HOOK3 - hook microtubule-tethering protein 3, PPFIA1 - protein tyrosine phosphatase, receptor type, f polypeptide (ptprf), interacting protein (liprin), alpha 1, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, DBN1 - drebrin 1, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, DERL1 - derlin 1, BTF3 - basic transcription factor 3, XPOT - exportin, trna, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, VTI1A - vesicle transport through interaction with t-snares 1a, CCDC51 - coiled-coil domain containing 51, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, EHBP1 - eh domain binding protein 1, ARCN1 - archain 1, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), PA2G4 - proliferation-associated 2g4, 38kda, AGPS - alkylglycerone phosphate synthase, CAB39 - calcium binding protein 39, GOSR2 - golgi snap receptor complex member 2, TBC1D4 - tbc1 domain family, member 4, TMED5 - transmembrane emp24 protein transport domain containing 5, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, LIMS1 - lim and senescent cell antigen-like domains 1, RAB23 - rab23, member ras oncogene family, CREB1 - camp responsive element binding protein 1, SNX1 - sorting nexin 1, MYO6 - myosin vi, PITRM1 - pitrilysin metallopeptidase 1, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, ATG7 - autophagy related 7, EIF2D - eukaryotic translation initiation factor 2d, MCU - mitochondrial calcium uniporter, SNRPE - small nuclear ribonucleoprotein polypeptide e, ARL1 - adp-ribosylation factor-like 1, SEH1L - seh1-like (s. cerevisiae), GLS - glutaminase, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), SYNE1 - spectrin repeat containing, nuclear envelope 1, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), NUP88 - nucleoporin 88kda, SCO1 - sco1 cytochrome c oxidase assembly protein, GOSR1 - golgi snap receptor complex member 1, OSBPL3 - oxysterol binding protein-like 3, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), THOC5 - tho complex 5, EIF2S2 - eukaryotic translation initiation factor 2, subunit 2 beta, 38kda, SNX6 - sorting nexin 6, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, CDK13 - cyclin-dependent kinase 13, HSPD1 - heat shock 60kda protein 1 (chaperonin), LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, CCT4 - chaperonin containing tcp1, subunit 4 (delta), CUTC - cutc copper transporter, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, ARL6IP5 - adp-ribosylation-like factor 6 interacting protein 5, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, STX8 - syntaxin 8, ERP44 - endoplasmic reticulum protein 44, STX18 - syntaxin 18, NEMF - nuclear export mediator factor, HSPA4 - heat shock 70kda protein 4, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, TMEM30A - transmembrane protein 30a, PURA - purine-rich element binding protein a, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, VTA1 - vesicle (multivesicular body) trafficking 1, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, QKI - qki, kh domain containing, rna binding, DENND3 - denn/madd domain containing 3, GAPVD1 - gtpase activating protein and vps9 domains 1, SPG11 - spastic paraplegia 11 (autosomal recessive), RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, WNK1 - wnk lysine deficient protein kinase 1, PGM1 - phosphoglucomutase 1, OSTF1 - osteoclast stimulating factor 1, PGAM1 - phosphoglycerate mutase 1 (brain), PPP1R9B - protein phosphatase 1, regulatory subunit 9b, TMEM14C - transmembrane protein 14c, HMGB1 - high mobility group box 1, AGK - acylglycerol kinase, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), TBC1D2B - tbc1 domain family, member 2b, PYGB - phosphorylase, glycogen; brain, NUP50 - nucleoporin 50kda, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, MAP4 - microtubule-associated protein 4, RAB14 - rab14, member ras oncogene family, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, FTH1 - ferritin, heavy polypeptide 1, UMPS - uridine monophosphate synthetase, RPL3 - ribosomal protein l3, MANBA - mannosidase, beta a, lysosomal, SURF4 - surfeit 4, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), TRAPPC10 - trafficking protein particle complex 10, SLAMF1 - signaling lymphocytic activation molecule family member 1, HK2 - hexokinase 2, STX17 - syntaxin 17, TNPO1 - transportin 1, KPNB1 - karyopherin (importin) beta 1, IDE - insulin-degrading enzyme, EXOC2 - exocyst complex component 2, CHMP5 - charged multivesicular body protein 5, DOCK7 - dedicator of cytokinesis 7, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, CDC23 - cell division cycle 23, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), EXOC1 - exocyst complex component 1, RPL31 - ribosomal protein l31, TJP2 - tight junction protein 2, RPL32 - ribosomal protein l32, VDAC2 - voltage-dependent anion channel 2, CTSC - cathepsin c, RANBP1 - ran binding protein 1, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, SCYL2 - scy1-like 2 (s. cerevisiae), NIPBL - nipped-b homolog (drosophila), DLG1 - discs, large homolog 1 (drosophila), RAP1A - rap1a, member of ras oncogene family, GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, RPL14 - ribosomal protein l14, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), GNPAT - glyceronephosphate o-acyltransferase, MFN1 - mitofusin 1, WDR1 - wd repeat domain 1, CLNS1A - chloride channel, nucleotide-sensitive, 1a, RPS8 - ribosomal protein s8, RPS13 - ribosomal protein s13, RAP1B - rap1b, member of ras oncogene family, RPS6 - ribosomal protein s6, RINT1 - rad50 interactor 1, GOLPH3 - golgi phosphoprotein 3 (coat-protein), RPS7 - ribosomal protein s7, STOM - stomatin, TMX3 - thioredoxin-related transmembrane protein 3, DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, SUMO1 - small ubiquitin-like modifier 1, SRP72 - signal recognition particle 72kda, RB1 - retinoblastoma 1, CAT - catalase, PDAP1 - pdgfa associated protein 1, XPO6 - exportin 6, FAM49B - family with sequence similarity 49, member b, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), SAMM50 - samm50 sorting and assembly machinery component, RHOT1 - ras homolog family member t1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, PRKCD - protein kinase c, delta, BCL2L1 - bcl2-like 1, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, RRBP1 - ribosome binding protein 1, EPS15 - epidermal growth factor receptor pathway substrate 15, PGM2 - phosphoglucomutase 2]
64 [ERAP1 - endoplasmic reticulum aminopeptidase 1, SGPL1 - sphingosine-1-phosphate lyase 1, ATG7 - autophagy related 7, RPL23A - ribosomal protein l23a, MRPL13 - mitochondrial ribosomal protein l13, RPL22 - ribosomal protein l22, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, GSPT1 - g1 to s phase transition 1, RPL14 - ribosomal protein l14, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, NSMAF - neutral sphingomyelinase (n-smase) activation associated factor, AGK - acylglycerol kinase, GLO1 - glyoxalase i, RPL13 - ribosomal protein l13, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, GFM2 - g elongation factor, mitochondrial 2, PTCD3 - pentatricopeptide repeat domain 3, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, RPS8 - ribosomal protein s8, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, MRPL42 - mitochondrial ribosomal protein l42, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPL3 - ribosomal protein l3, RPS7 - ribosomal protein s7, ACAT1 - acetyl-coa acetyltransferase 1, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, MRPL9 - mitochondrial ribosomal protein l9, GCDH - glutaryl-coa dehydrogenase, ACOT7 - acyl-coa thioesterase 7, MVD - mevalonate (diphospho) decarboxylase, MRPL47 - mitochondrial ribosomal protein l47, MRPS14 - mitochondrial ribosomal protein s14, IDE - insulin-degrading enzyme, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, KYNU - kynureninase, MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, RPL31 - ribosomal protein l31, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, RPL32 - ribosomal protein l32, RRBP1 - ribosome binding protein 1, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, MRPL3 - mitochondrial ribosomal protein l3, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
65 [ERAP1 - endoplasmic reticulum aminopeptidase 1, ATG7 - autophagy related 7, RPL23A - ribosomal protein l23a, MRPL13 - mitochondrial ribosomal protein l13, RPL22 - ribosomal protein l22, GSPT1 - g1 to s phase transition 1, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, RPL14 - ribosomal protein l14, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, RPL13 - ribosomal protein l13, GLO1 - glyoxalase i, GFM2 - g elongation factor, mitochondrial 2, PTCD3 - pentatricopeptide repeat domain 3, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, RPS8 - ribosomal protein s8, RPL6 - ribosomal protein l6, MRPL42 - mitochondrial ribosomal protein l42, RPL7 - ribosomal protein l7, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPL3 - ribosomal protein l3, RPS7 - ribosomal protein s7, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, MRPL9 - mitochondrial ribosomal protein l9, MRPL47 - mitochondrial ribosomal protein l47, MRPS14 - mitochondrial ribosomal protein s14, IDE - insulin-degrading enzyme, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, RRBP1 - ribosome binding protein 1, MRPL3 - mitochondrial ribosomal protein l3, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
66 [AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, VTA1 - vesicle (multivesicular body) trafficking 1, PURA - purine-rich element binding protein a, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, RPL23A - ribosomal protein l23a, SDAD1 - sda1 domain containing 1, RPL22 - ribosomal protein l22, SCFD1 - sec1 family domain containing 1, AGK - acylglycerol kinase, RPL13 - ribosomal protein l13, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), TBC1D2B - tbc1 domain family, member 2b, TMED2 - transmembrane emp24 domain trafficking protein 2, RPL6 - ribosomal protein l6, NUP50 - nucleoporin 50kda, HOOK3 - hook microtubule-tethering protein 3, RPL7 - ribosomal protein l7, RAB14 - rab14, member ras oncogene family, RPL3 - ribosomal protein l3, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, DERL1 - derlin 1, BTF3 - basic transcription factor 3, VTI1A - vesicle transport through interaction with t-snares 1a, STX17 - syntaxin 17, TNPO1 - transportin 1, ARCN1 - archain 1, EHBP1 - eh domain binding protein 1, KPNB1 - karyopherin (importin) beta 1, AGPS - alkylglycerone phosphate synthase, GOSR2 - golgi snap receptor complex member 2, IDE - insulin-degrading enzyme, TBC1D4 - tbc1 domain family, member 4, EXOC2 - exocyst complex component 2, CHMP5 - charged multivesicular body protein 5, TMED5 - transmembrane emp24 protein transport domain containing 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, RAB23 - rab23, member ras oncogene family, EXOC1 - exocyst complex component 1, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, SNX1 - sorting nexin 1, MYO6 - myosin vi, PITRM1 - pitrilysin metallopeptidase 1, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), EIF2D - eukaryotic translation initiation factor 2d, ATG7 - autophagy related 7, ARL1 - adp-ribosylation factor-like 1, GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, RPL14 - ribosomal protein l14, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), GNPAT - glyceronephosphate o-acyltransferase, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), NUP88 - nucleoporin 88kda, RPS8 - ribosomal protein s8, GOSR1 - golgi snap receptor complex member 1, RPS13 - ribosomal protein s13, RINT1 - rad50 interactor 1, RPS6 - ribosomal protein s6, GOLPH3 - golgi phosphoprotein 3 (coat-protein), RPS7 - ribosomal protein s7, SNX6 - sorting nexin 6, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, SRP72 - signal recognition particle 72kda, CAT - catalase, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, HSPD1 - heat shock 60kda protein 1 (chaperonin), XPO6 - exportin 6, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, SAMM50 - samm50 sorting and assembly machinery component, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, STX8 - syntaxin 8, STX18 - syntaxin 18, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, HSPA4 - heat shock 70kda protein 4, RRBP1 - ribosome binding protein 1, EPS15 - epidermal growth factor receptor pathway substrate 15]
67 [DDX6 - dead (asp-glu-ala-asp) box helicase 6, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, SF3A3 - splicing factor 3a, subunit 3, 60kda, TDRD7 - tudor domain containing 7, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, RPL13 - ribosomal protein l13, PTCD3 - pentatricopeptide repeat domain 3, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, RPL3 - ribosomal protein l3, DERL1 - derlin 1, ILF3 - interleukin enhancer binding factor 3, 90kda, BTF3 - basic transcription factor 3, TRIM25 - tripartite motif containing 25, EIF3F - eukaryotic translation initiation factor 3, subunit f, MBNL1 - muscleblind-like splicing regulator 1, MRPS35 - mitochondrial ribosomal protein s35, MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), KPNB1 - karyopherin (importin) beta 1, CARHSP1 - calcium regulated heat stable protein 1, 24kda, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, GAR1 - gar1 ribonucleoprotein, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, SART3 - squamous cell carcinoma antigen recognized by t cells 3, MRPL3 - mitochondrial ribosomal protein l3, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, EIF2D - eukaryotic translation initiation factor 2d, MRPL13 - mitochondrial ribosomal protein l13, SNRPE - small nuclear ribonucleoprotein polypeptide e, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, RPL14 - ribosomal protein l14, SYNE1 - spectrin repeat containing, nuclear envelope 1, AKAP8L - a kinase (prka) anchor protein 8-like, RPS8 - ribosomal protein s8, NIP7 - nip7, nucleolar pre-rrna processing protein, LARP4B - la ribonucleoprotein domain family, member 4b, MRPL42 - mitochondrial ribosomal protein l42, PPIL3 - peptidylprolyl isomerase (cyclophilin)-like 3, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, NHP2 - nhp2 ribonucleoprotein, RPS7 - ribosomal protein s7, MRPL2 - mitochondrial ribosomal protein l2, SUMO1 - small ubiquitin-like modifier 1, SRP72 - signal recognition particle 72kda, FCF1 - fcf1 rrna-processing protein, CAPRIN1 - cell cycle associated protein 1, RPL7L1 - ribosomal protein l7-like 1, MRPL47 - mitochondrial ribosomal protein l47, MRPS14 - mitochondrial ribosomal protein s14, RSL1D1 - ribosomal l1 domain containing 1, LARP1 - la ribonucleoprotein domain family, member 1, EIF3E - eukaryotic translation initiation factor 3, subunit e, RPTOR - regulatory associated protein of mtor, complex 1, RRBP1 - ribosome binding protein 1, PARP4 - poly (adp-ribose) polymerase family, member 4]
68 [CCNK - cyclin k, ZC3H7B - zinc finger ccch-type containing 7b, NCBP2 - nuclear cap binding protein subunit 2, 20kda, EXOSC5 - exosome component 5, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, DDX6 - dead (asp-glu-ala-asp) box helicase 6, TCOF1 - treacher collins-franceschetti syndrome 1, RFC3 - replication factor c (activator 1) 3, 38kda, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, ZNF638 - zinc finger protein 638, SF3A3 - splicing factor 3a, subunit 3, 60kda, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, GSPT1 - g1 to s phase transition 1, URB1 - urb1 ribosome biogenesis 1 homolog (s. cerevisiae), RECQL - recq protein-like (dna helicase q1-like), DCK - deoxycytidine kinase, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, PPP4R2 - protein phosphatase 4, regulatory subunit 2, BTF3 - basic transcription factor 3, TRIM25 - tripartite motif containing 25, RIF1 - rap1 interacting factor homolog (yeast), ACAT1 - acetyl-coa acetyltransferase 1, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, RBM19 - rna binding motif protein 19, MMS22L - mms22-like, dna repair protein, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), PA2G4 - proliferation-associated 2g4, 38kda, OTUB1 - otu domain, ubiquitin aldehyde binding 1, ZNF326 - zinc finger protein 326, RAB23 - rab23, member ras oncogene family, MORF4L1 - mortality factor 4 like 1, GAR1 - gar1 ribonucleoprotein, CREB1 - camp responsive element binding protein 1, EXOSC1 - exosome component 1, THUMPD1 - thump domain containing 1, TELO2 - telomere maintenance 2, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, MCM7 - minichromosome maintenance complex component 7, SNRPE - small nuclear ribonucleoprotein polypeptide e, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, IARS - isoleucyl-trna synthetase, PELO - pelota homolog (drosophila), ZBTB8OS - zinc finger and btb domain containing 8 opposite strand, WDHD1 - wd repeat and hmg-box dna binding protein 1, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), PPIL3 - peptidylprolyl isomerase (cyclophilin)-like 3, POLR1C - polymerase (rna) i polypeptide c, 30kda, ADPGK - adp-dependent glucokinase, PPA1 - pyrophosphatase (inorganic) 1, THOC5 - tho complex 5, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, CDK13 - cyclin-dependent kinase 13, HSPD1 - heat shock 60kda protein 1 (chaperonin), FCF1 - fcf1 rrna-processing protein, GNPDA2 - glucosamine-6-phosphate deaminase 2, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, MVD - mevalonate (diphospho) decarboxylase, RPL7L1 - ribosomal protein l7-like 1, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, KYNU - kynureninase, TAOK3 - tao kinase 3, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), NAMPT - nicotinamide phosphoribosyltransferase, EIF3E - eukaryotic translation initiation factor 3, subunit e, TCEA1 - transcription elongation factor a (sii), 1, PARP4 - poly (adp-ribose) polymerase family, member 4, PURA - purine-rich element binding protein a, QKI - qki, kh domain containing, rna binding, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, UQCRB - ubiquinol-cytochrome c reductase binding protein, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), HMGB2 - high mobility group box 2, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, NOC3L - nucleolar complex associated 3 homolog (s. cerevisiae), HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, UCK2 - uridine-cytidine kinase 2, PPP5C - protein phosphatase 5, catalytic subunit, RPL3 - ribosomal protein l3, UMPS - uridine monophosphate synthetase, CARS - cysteinyl-trna synthetase, MBNL1 - muscleblind-like splicing regulator 1, METTL15 - methyltransferase like 15, DUT - deoxyuridine triphosphatase, MRPL1 - mitochondrial ribosomal protein l1, HK2 - hexokinase 2, AK4 - adenylate kinase 4, KPNB1 - karyopherin (importin) beta 1, MCM3 - minichromosome maintenance complex component 3, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, TJP2 - tight junction protein 2, RPL31 - ribosomal protein l31, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, RPL32 - ribosomal protein l32, SART3 - squamous cell carcinoma antigen recognized by t cells 3, DCAKD - dephospho-coa kinase domain containing, SCAF8 - sr-related ctd-associated factor 8, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, DLG1 - discs, large homolog 1 (drosophila), NIPBL - nipped-b homolog (drosophila), PUS1 - pseudouridylate synthase 1, RPL14 - ribosomal protein l14, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), AKAP8L - a kinase (prka) anchor protein 8-like, MFN1 - mitofusin 1, RNMT - rna (guanine-7-) methyltransferase, SNRNP27 - small nuclear ribonucleoprotein 27kda (u4/u6.u5), RBM26 - rna binding motif protein 26, DGUOK - deoxyguanosine kinase, RPS8 - ribosomal protein s8, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, NHP2 - nhp2 ribonucleoprotein, SUMO1 - small ubiquitin-like modifier 1, ACOT7 - acyl-coa thioesterase 7, GCDH - glutaryl-coa dehydrogenase, UGDH - udp-glucose 6-dehydrogenase, SSBP1 - single-stranded dna binding protein 1, mitochondrial, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), POLR3B - polymerase (rna) iii (dna directed) polypeptide b, RSL1D1 - ribosomal l1 domain containing 1, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, PGM2 - phosphoglucomutase 2, RRM1 - ribonucleotide reductase m1, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
69 [CCNK - cyclin k, ZC3H7B - zinc finger ccch-type containing 7b, NCBP2 - nuclear cap binding protein subunit 2, 20kda, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, EXOSC5 - exosome component 5, DDX6 - dead (asp-glu-ala-asp) box helicase 6, TCOF1 - treacher collins-franceschetti syndrome 1, RFC3 - replication factor c (activator 1) 3, 38kda, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, ZNF638 - zinc finger protein 638, SF3A3 - splicing factor 3a, subunit 3, 60kda, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, GSPT1 - g1 to s phase transition 1, URB1 - urb1 ribosome biogenesis 1 homolog (s. cerevisiae), BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, RECQL - recq protein-like (dna helicase q1-like), DCK - deoxycytidine kinase, PPP4R2 - protein phosphatase 4, regulatory subunit 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, BTF3 - basic transcription factor 3, TRIM25 - tripartite motif containing 25, RIF1 - rap1 interacting factor homolog (yeast), ACAT1 - acetyl-coa acetyltransferase 1, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, CPOX - coproporphyrinogen oxidase, RBM19 - rna binding motif protein 19, MMS22L - mms22-like, dna repair protein, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), PA2G4 - proliferation-associated 2g4, 38kda, OTUB1 - otu domain, ubiquitin aldehyde binding 1, ZNF326 - zinc finger protein 326, RAB23 - rab23, member ras oncogene family, MORF4L1 - mortality factor 4 like 1, GAR1 - gar1 ribonucleoprotein, CREB1 - camp responsive element binding protein 1, TELO2 - telomere maintenance 2, EXOSC1 - exosome component 1, THUMPD1 - thump domain containing 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, MCM7 - minichromosome maintenance complex component 7, SNRPE - small nuclear ribonucleoprotein polypeptide e, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, IARS - isoleucyl-trna synthetase, PELO - pelota homolog (drosophila), ZBTB8OS - zinc finger and btb domain containing 8 opposite strand, WDHD1 - wd repeat and hmg-box dna binding protein 1, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), PPIL3 - peptidylprolyl isomerase (cyclophilin)-like 3, POLR1C - polymerase (rna) i polypeptide c, 30kda, PPA1 - pyrophosphatase (inorganic) 1, ADPGK - adp-dependent glucokinase, THOC5 - tho complex 5, FECH - ferrochelatase, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, CDK13 - cyclin-dependent kinase 13, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), GNPDA2 - glucosamine-6-phosphate deaminase 2, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, FCF1 - fcf1 rrna-processing protein, MVD - mevalonate (diphospho) decarboxylase, RPL7L1 - ribosomal protein l7-like 1, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, KYNU - kynureninase, TAOK3 - tao kinase 3, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), NAMPT - nicotinamide phosphoribosyltransferase, EIF3E - eukaryotic translation initiation factor 3, subunit e, TCEA1 - transcription elongation factor a (sii), 1, PARP4 - poly (adp-ribose) polymerase family, member 4, PURA - purine-rich element binding protein a, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, UQCRB - ubiquinol-cytochrome c reductase binding protein, QKI - qki, kh domain containing, rna binding, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, HMGB2 - high mobility group box 2, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, NOC3L - nucleolar complex associated 3 homolog (s. cerevisiae), HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, UCK2 - uridine-cytidine kinase 2, RPL3 - ribosomal protein l3, UMPS - uridine monophosphate synthetase, PPP5C - protein phosphatase 5, catalytic subunit, CARS - cysteinyl-trna synthetase, MBNL1 - muscleblind-like splicing regulator 1, METTL15 - methyltransferase like 15, DUT - deoxyuridine triphosphatase, MRPL1 - mitochondrial ribosomal protein l1, HK2 - hexokinase 2, AK4 - adenylate kinase 4, KPNB1 - karyopherin (importin) beta 1, MCM3 - minichromosome maintenance complex component 3, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, RPL31 - ribosomal protein l31, TJP2 - tight junction protein 2, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, RPL32 - ribosomal protein l32, SART3 - squamous cell carcinoma antigen recognized by t cells 3, DCAKD - dephospho-coa kinase domain containing, SCAF8 - sr-related ctd-associated factor 8, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, DLG1 - discs, large homolog 1 (drosophila), NIPBL - nipped-b homolog (drosophila), PUS1 - pseudouridylate synthase 1, RPL14 - ribosomal protein l14, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), AKAP8L - a kinase (prka) anchor protein 8-like, SNRNP27 - small nuclear ribonucleoprotein 27kda (u4/u6.u5), MFN1 - mitofusin 1, RNMT - rna (guanine-7-) methyltransferase, RBM26 - rna binding motif protein 26, DGUOK - deoxyguanosine kinase, RPS8 - ribosomal protein s8, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, NHP2 - nhp2 ribonucleoprotein, SUMO1 - small ubiquitin-like modifier 1, ACOT7 - acyl-coa thioesterase 7, GCDH - glutaryl-coa dehydrogenase, UGDH - udp-glucose 6-dehydrogenase, SSBP1 - single-stranded dna binding protein 1, mitochondrial, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), POLR3B - polymerase (rna) iii (dna directed) polypeptide b, RSL1D1 - ribosomal l1 domain containing 1, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, PGM2 - phosphoglucomutase 2, RRM1 - ribonucleotide reductase m1, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
70 [CCNK - cyclin k, ZC3H7B - zinc finger ccch-type containing 7b, NCBP2 - nuclear cap binding protein subunit 2, 20kda, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, EXOSC5 - exosome component 5, DDX6 - dead (asp-glu-ala-asp) box helicase 6, TCOF1 - treacher collins-franceschetti syndrome 1, RFC3 - replication factor c (activator 1) 3, 38kda, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, ZNF638 - zinc finger protein 638, SF3A3 - splicing factor 3a, subunit 3, 60kda, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, GSPT1 - g1 to s phase transition 1, URB1 - urb1 ribosome biogenesis 1 homolog (s. cerevisiae), BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, RECQL - recq protein-like (dna helicase q1-like), DCK - deoxycytidine kinase, PPP4R2 - protein phosphatase 4, regulatory subunit 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, BTF3 - basic transcription factor 3, TRIM25 - tripartite motif containing 25, RIF1 - rap1 interacting factor homolog (yeast), ACAT1 - acetyl-coa acetyltransferase 1, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, CPOX - coproporphyrinogen oxidase, RBM19 - rna binding motif protein 19, MMS22L - mms22-like, dna repair protein, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), PA2G4 - proliferation-associated 2g4, 38kda, OTUB1 - otu domain, ubiquitin aldehyde binding 1, ZNF326 - zinc finger protein 326, RAB23 - rab23, member ras oncogene family, MORF4L1 - mortality factor 4 like 1, GAR1 - gar1 ribonucleoprotein, CREB1 - camp responsive element binding protein 1, TELO2 - telomere maintenance 2, EXOSC1 - exosome component 1, THUMPD1 - thump domain containing 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, MCM7 - minichromosome maintenance complex component 7, SNRPE - small nuclear ribonucleoprotein polypeptide e, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, IARS - isoleucyl-trna synthetase, PELO - pelota homolog (drosophila), ZBTB8OS - zinc finger and btb domain containing 8 opposite strand, WDHD1 - wd repeat and hmg-box dna binding protein 1, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), PPIL3 - peptidylprolyl isomerase (cyclophilin)-like 3, POLR1C - polymerase (rna) i polypeptide c, 30kda, ADPGK - adp-dependent glucokinase, PPA1 - pyrophosphatase (inorganic) 1, THOC5 - tho complex 5, FECH - ferrochelatase, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, CDK13 - cyclin-dependent kinase 13, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, GNPDA2 - glucosamine-6-phosphate deaminase 2, FCF1 - fcf1 rrna-processing protein, MVD - mevalonate (diphospho) decarboxylase, RPL7L1 - ribosomal protein l7-like 1, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, COMT - catechol-o-methyltransferase, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, KYNU - kynureninase, TAOK3 - tao kinase 3, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), NAMPT - nicotinamide phosphoribosyltransferase, EIF3E - eukaryotic translation initiation factor 3, subunit e, TCEA1 - transcription elongation factor a (sii), 1, PARP4 - poly (adp-ribose) polymerase family, member 4, PURA - purine-rich element binding protein a, QKI - qki, kh domain containing, rna binding, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, UQCRB - ubiquinol-cytochrome c reductase binding protein, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, HMGB2 - high mobility group box 2, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, NOC3L - nucleolar complex associated 3 homolog (s. cerevisiae), HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, UCK2 - uridine-cytidine kinase 2, UMPS - uridine monophosphate synthetase, RPL3 - ribosomal protein l3, PPP5C - protein phosphatase 5, catalytic subunit, CARS - cysteinyl-trna synthetase, MBNL1 - muscleblind-like splicing regulator 1, METTL15 - methyltransferase like 15, DUT - deoxyuridine triphosphatase, MRPL1 - mitochondrial ribosomal protein l1, HK2 - hexokinase 2, AK4 - adenylate kinase 4, KPNB1 - karyopherin (importin) beta 1, MCM3 - minichromosome maintenance complex component 3, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, TJP2 - tight junction protein 2, RPL31 - ribosomal protein l31, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, RPL32 - ribosomal protein l32, SART3 - squamous cell carcinoma antigen recognized by t cells 3, DCAKD - dephospho-coa kinase domain containing, SCAF8 - sr-related ctd-associated factor 8, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, DLG1 - discs, large homolog 1 (drosophila), NIPBL - nipped-b homolog (drosophila), PUS1 - pseudouridylate synthase 1, RPL14 - ribosomal protein l14, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), AKAP8L - a kinase (prka) anchor protein 8-like, MFN1 - mitofusin 1, RNMT - rna (guanine-7-) methyltransferase, SNRNP27 - small nuclear ribonucleoprotein 27kda (u4/u6.u5), RBM26 - rna binding motif protein 26, DGUOK - deoxyguanosine kinase, RPS8 - ribosomal protein s8, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, NHP2 - nhp2 ribonucleoprotein, SUMO1 - small ubiquitin-like modifier 1, ACOT7 - acyl-coa thioesterase 7, GCDH - glutaryl-coa dehydrogenase, UGDH - udp-glucose 6-dehydrogenase, SSBP1 - single-stranded dna binding protein 1, mitochondrial, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), POLR3B - polymerase (rna) iii (dna directed) polypeptide b, RSL1D1 - ribosomal l1 domain containing 1, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, PGM2 - phosphoglucomutase 2, RRM1 - ribonucleotide reductase m1, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
71 [RPL23A - ribosomal protein l23a, MRPL13 - mitochondrial ribosomal protein l13, RPL22 - ribosomal protein l22, GSPT1 - g1 to s phase transition 1, RPL14 - ribosomal protein l14, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, RPL13 - ribosomal protein l13, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, GFM2 - g elongation factor, mitochondrial 2, PTCD3 - pentatricopeptide repeat domain 3, RPS8 - ribosomal protein s8, RPL6 - ribosomal protein l6, MRPL42 - mitochondrial ribosomal protein l42, RPL7 - ribosomal protein l7, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPL3 - ribosomal protein l3, RPS7 - ribosomal protein s7, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, MRPL9 - mitochondrial ribosomal protein l9, MRPL47 - mitochondrial ribosomal protein l47, MRPS14 - mitochondrial ribosomal protein s14, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, RRBP1 - ribosome binding protein 1, MRPL3 - mitochondrial ribosomal protein l3, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
72 [DST - dystonin, NCBP2 - nuclear cap binding protein subunit 2, 20kda, SDAD1 - sda1 domain containing 1, F11R - f11 receptor, SCFD1 - sec1 family domain containing 1, FAM98A - family with sequence similarity 98, member a, TMED2 - transmembrane emp24 domain trafficking protein 2, HOOK3 - hook microtubule-tethering protein 3, DERL1 - derlin 1, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, XPOT - exportin, trna, VTI1A - vesicle transport through interaction with t-snares 1a, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, ARCN1 - archain 1, AGPS - alkylglycerone phosphate synthase, GOSR2 - golgi snap receptor complex member 2, TBC1D4 - tbc1 domain family, member 4, TMED5 - transmembrane emp24 protein transport domain containing 5, RAB23 - rab23, member ras oncogene family, MYO6 - myosin vi, SNX1 - sorting nexin 1, PITRM1 - pitrilysin metallopeptidase 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, EIF2D - eukaryotic translation initiation factor 2d, SNRPE - small nuclear ribonucleoprotein polypeptide e, ARL1 - adp-ribosylation factor-like 1, SEH1L - seh1-like (s. cerevisiae), TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), NUP88 - nucleoporin 88kda, GOSR1 - golgi snap receptor complex member 1, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), THOC5 - tho complex 5, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, SNX6 - sorting nexin 6, HSPD1 - heat shock 60kda protein 1 (chaperonin), LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, STX8 - syntaxin 8, STX18 - syntaxin 18, NEMF - nuclear export mediator factor, HSPA4 - heat shock 70kda protein 4, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, TMEM30A - transmembrane protein 30a, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, PURA - purine-rich element binding protein a, VTA1 - vesicle (multivesicular body) trafficking 1, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, DENND3 - denn/madd domain containing 3, SPG11 - spastic paraplegia 11 (autosomal recessive), RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, AGK - acylglycerol kinase, RPL13 - ribosomal protein l13, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), TBC1D2B - tbc1 domain family, member 2b, RPL6 - ribosomal protein l6, NUP50 - nucleoporin 50kda, RPL7 - ribosomal protein l7, MAP4 - microtubule-associated protein 4, RAB14 - rab14, member ras oncogene family, RPL3 - ribosomal protein l3, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), TRAPPC10 - trafficking protein particle complex 10, TNPO1 - transportin 1, HK2 - hexokinase 2, STX17 - syntaxin 17, KPNB1 - karyopherin (importin) beta 1, IDE - insulin-degrading enzyme, CHMP5 - charged multivesicular body protein 5, DOCK7 - dedicator of cytokinesis 7, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, CDC23 - cell division cycle 23, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), EXOC1 - exocyst complex component 1, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, CTSC - cathepsin c, RANBP1 - ran binding protein 1, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, DLG1 - discs, large homolog 1 (drosophila), NIPBL - nipped-b homolog (drosophila), SCYL2 - scy1-like 2 (s. cerevisiae), RAP1A - rap1a, member of ras oncogene family, GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, RPL14 - ribosomal protein l14, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), GNPAT - glyceronephosphate o-acyltransferase, MFN1 - mitofusin 1, RPS8 - ribosomal protein s8, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, GOLPH3 - golgi phosphoprotein 3 (coat-protein), RPS7 - ribosomal protein s7, DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, SUMO1 - small ubiquitin-like modifier 1, SRP72 - signal recognition particle 72kda, RB1 - retinoblastoma 1, CAT - catalase, XPO6 - exportin 6, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), SAMM50 - samm50 sorting and assembly machinery component, RHOT1 - ras homolog family member t1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, EPS15 - epidermal growth factor receptor pathway substrate 15]
73 [CCNK - cyclin k, ZC3H7B - zinc finger ccch-type containing 7b, NCBP2 - nuclear cap binding protein subunit 2, 20kda, EXOSC5 - exosome component 5, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, DDX6 - dead (asp-glu-ala-asp) box helicase 6, TCOF1 - treacher collins-franceschetti syndrome 1, RFC3 - replication factor c (activator 1) 3, 38kda, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, ZNF638 - zinc finger protein 638, SF3A3 - splicing factor 3a, subunit 3, 60kda, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, GSPT1 - g1 to s phase transition 1, URB1 - urb1 ribosome biogenesis 1 homolog (s. cerevisiae), BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, RECQL - recq protein-like (dna helicase q1-like), DCK - deoxycytidine kinase, PPP4R2 - protein phosphatase 4, regulatory subunit 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, BTF3 - basic transcription factor 3, TRIM25 - tripartite motif containing 25, RIF1 - rap1 interacting factor homolog (yeast), ACAT1 - acetyl-coa acetyltransferase 1, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, RBM19 - rna binding motif protein 19, CPOX - coproporphyrinogen oxidase, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), MMS22L - mms22-like, dna repair protein, PA2G4 - proliferation-associated 2g4, 38kda, OTUB1 - otu domain, ubiquitin aldehyde binding 1, ZNF326 - zinc finger protein 326, RAB23 - rab23, member ras oncogene family, MORF4L1 - mortality factor 4 like 1, GAR1 - gar1 ribonucleoprotein, CREB1 - camp responsive element binding protein 1, EXOSC1 - exosome component 1, TELO2 - telomere maintenance 2, THUMPD1 - thump domain containing 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, SGPL1 - sphingosine-1-phosphate lyase 1, MCM7 - minichromosome maintenance complex component 7, SNRPE - small nuclear ribonucleoprotein polypeptide e, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, IARS - isoleucyl-trna synthetase, PELO - pelota homolog (drosophila), ZBTB8OS - zinc finger and btb domain containing 8 opposite strand, WDHD1 - wd repeat and hmg-box dna binding protein 1, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), PPIL3 - peptidylprolyl isomerase (cyclophilin)-like 3, OSBPL3 - oxysterol binding protein-like 3, POLR1C - polymerase (rna) i polypeptide c, 30kda, PPA1 - pyrophosphatase (inorganic) 1, ADPGK - adp-dependent glucokinase, THOC5 - tho complex 5, FECH - ferrochelatase, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, CDK13 - cyclin-dependent kinase 13, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, GNPDA2 - glucosamine-6-phosphate deaminase 2, FCF1 - fcf1 rrna-processing protein, MVD - mevalonate (diphospho) decarboxylase, RPL7L1 - ribosomal protein l7-like 1, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, COMT - catechol-o-methyltransferase, KYNU - kynureninase, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, TAOK3 - tao kinase 3, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), NAMPT - nicotinamide phosphoribosyltransferase, EIF3E - eukaryotic translation initiation factor 3, subunit e, TCEA1 - transcription elongation factor a (sii), 1, PARP4 - poly (adp-ribose) polymerase family, member 4, PURA - purine-rich element binding protein a, UQCRB - ubiquinol-cytochrome c reductase binding protein, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, QKI - qki, kh domain containing, rna binding, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, HMGB2 - high mobility group box 2, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, NOC3L - nucleolar complex associated 3 homolog (s. cerevisiae), HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, UCK2 - uridine-cytidine kinase 2, PPP5C - protein phosphatase 5, catalytic subunit, UMPS - uridine monophosphate synthetase, RPL3 - ribosomal protein l3, CARS - cysteinyl-trna synthetase, MBNL1 - muscleblind-like splicing regulator 1, METTL15 - methyltransferase like 15, DUT - deoxyuridine triphosphatase, MRPL1 - mitochondrial ribosomal protein l1, AK4 - adenylate kinase 4, HK2 - hexokinase 2, KPNB1 - karyopherin (importin) beta 1, MCM3 - minichromosome maintenance complex component 3, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, TJP2 - tight junction protein 2, RPL31 - ribosomal protein l31, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, RPL32 - ribosomal protein l32, SART3 - squamous cell carcinoma antigen recognized by t cells 3, DCAKD - dephospho-coa kinase domain containing, SCAF8 - sr-related ctd-associated factor 8, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, LSS - lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase), POLDIP3 - polymerase (dna-directed), delta interacting protein 3, DLG1 - discs, large homolog 1 (drosophila), NIPBL - nipped-b homolog (drosophila), PUS1 - pseudouridylate synthase 1, RPL14 - ribosomal protein l14, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), AKAP8L - a kinase (prka) anchor protein 8-like, SNRNP27 - small nuclear ribonucleoprotein 27kda (u4/u6.u5), RNMT - rna (guanine-7-) methyltransferase, MFN1 - mitofusin 1, RBM26 - rna binding motif protein 26, DGUOK - deoxyguanosine kinase, RPS8 - ribosomal protein s8, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, NHP2 - nhp2 ribonucleoprotein, RPS7 - ribosomal protein s7, SUMO1 - small ubiquitin-like modifier 1, ACOT7 - acyl-coa thioesterase 7, GCDH - glutaryl-coa dehydrogenase, CAT - catalase, UGDH - udp-glucose 6-dehydrogenase, SSBP1 - single-stranded dna binding protein 1, mitochondrial, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), POLR3B - polymerase (rna) iii (dna directed) polypeptide b, RSL1D1 - ribosomal l1 domain containing 1, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, PGM2 - phosphoglucomutase 2, RRM1 - ribonucleotide reductase m1, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
74 [RFC3 - replication factor c (activator 1) 3, 38kda, SCFD1 - sec1 family domain containing 1, GSN - gelsolin, FLNB - filamin b, beta, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, DERL1 - derlin 1, TRIM25 - tripartite motif containing 25, DOCK8 - dedicator of cytokinesis 8, RIF1 - rap1 interacting factor homolog (yeast), CYCS - cytochrome c, somatic, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, ACAT1 - acetyl-coa acetyltransferase 1, CPOX - coproporphyrinogen oxidase, EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, TBC1D4 - tbc1 domain family, member 4, SORD - sorbitol dehydrogenase, OTUB1 - otu domain, ubiquitin aldehyde binding 1, PLCG1 - phospholipase c, gamma 1, LIMS1 - lim and senescent cell antigen-like domains 1, CREB1 - camp responsive element binding protein 1, KDM3A - lysine (k)-specific demethylase 3a, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, MYO6 - myosin vi, ATG7 - autophagy related 7, MCM7 - minichromosome maintenance complex component 7, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), HADH - hydroxyacyl-coa dehydrogenase, EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, FECH - ferrochelatase, ADNP - activity-dependent neuroprotector homeobox, SNX6 - sorting nexin 6, SMARCD1 - swi/snf related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1, HSPD1 - heat shock 60kda protein 1 (chaperonin), PTPN2 - protein tyrosine phosphatase, non-receptor type 2, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, COMT - catechol-o-methyltransferase, IRF3 - interferon regulatory factor 3, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, KYNU - kynureninase, STX8 - syntaxin 8, ERP44 - endoplasmic reticulum protein 44, LARP1 - la ribonucleoprotein domain family, member 1, IRF5 - interferon regulatory factor 5, NAMPT - nicotinamide phosphoribosyltransferase, RPTOR - regulatory associated protein of mtor, complex 1, HSPA4 - heat shock 70kda protein 4, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, PARP4 - poly (adp-ribose) polymerase family, member 4, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, CAPN2 - calpain 2, (m/ii) large subunit, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, WNK1 - wnk lysine deficient protein kinase 1, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, HOMER1 - homer homolog 1 (drosophila), SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, UMPS - uridine monophosphate synthetase, RPL3 - ribosomal protein l3, PPP5C - protein phosphatase 5, catalytic subunit, VKORC1L1 - vitamin k epoxide reductase complex, subunit 1-like 1, USP8 - ubiquitin specific peptidase 8, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, AK4 - adenylate kinase 4, HK2 - hexokinase 2, MAT2A - methionine adenosyltransferase ii, alpha, CHMP5 - charged multivesicular body protein 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, IFIH1 - interferon induced with helicase c domain 1, CTSC - cathepsin c, RAP1A - rap1a, member of ras oncogene family, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), GNPAT - glyceronephosphate o-acyltransferase, RNMT - rna (guanine-7-) methyltransferase, STAT6 - signal transducer and activator of transcription 6, interleukin-4 induced, NUB1 - negative regulator of ubiquitin-like proteins 1, RAP1B - rap1b, member of ras oncogene family, GOLPH3 - golgi phosphoprotein 3 (coat-protein), SUMO1 - small ubiquitin-like modifier 1, RB1 - retinoblastoma 1, CAT - catalase, FAM49B - family with sequence similarity 49, member b, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), PRKCD - protein kinase c, delta, BCL2L1 - bcl2-like 1, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha]
75 [TMEM30A - transmembrane protein 30a, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, VTA1 - vesicle (multivesicular body) trafficking 1, PURA - purine-rich element binding protein a, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, NCBP2 - nuclear cap binding protein subunit 2, 20kda, QKI - qki, kh domain containing, rna binding, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, SDAD1 - sda1 domain containing 1, SCFD1 - sec1 family domain containing 1, AGK - acylglycerol kinase, RPL13 - ribosomal protein l13, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), TBC1D2B - tbc1 domain family, member 2b, TMED2 - transmembrane emp24 domain trafficking protein 2, RPL6 - ribosomal protein l6, NUP50 - nucleoporin 50kda, RPL7 - ribosomal protein l7, HOOK3 - hook microtubule-tethering protein 3, RAB14 - rab14, member ras oncogene family, PPFIA1 - protein tyrosine phosphatase, receptor type, f polypeptide (ptprf), interacting protein (liprin), alpha 1, RPL3 - ribosomal protein l3, DERL1 - derlin 1, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, BTF3 - basic transcription factor 3, XPOT - exportin, trna, VTI1A - vesicle transport through interaction with t-snares 1a, STX17 - syntaxin 17, TNPO1 - transportin 1, EHBP1 - eh domain binding protein 1, ARCN1 - archain 1, KPNB1 - karyopherin (importin) beta 1, AGPS - alkylglycerone phosphate synthase, GOSR2 - golgi snap receptor complex member 2, IDE - insulin-degrading enzyme, TBC1D4 - tbc1 domain family, member 4, EXOC2 - exocyst complex component 2, CHMP5 - charged multivesicular body protein 5, TMED5 - transmembrane emp24 protein transport domain containing 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, RAB23 - rab23, member ras oncogene family, EXOC1 - exocyst complex component 1, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, MYO6 - myosin vi, SNX1 - sorting nexin 1, PITRM1 - pitrilysin metallopeptidase 1, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), ATG7 - autophagy related 7, EIF2D - eukaryotic translation initiation factor 2d, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, ARL1 - adp-ribosylation factor-like 1, GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, SEH1L - seh1-like (s. cerevisiae), RPL14 - ribosomal protein l14, GLS - glutaminase, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), GNPAT - glyceronephosphate o-acyltransferase, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), NUP88 - nucleoporin 88kda, RPS8 - ribosomal protein s8, GOSR1 - golgi snap receptor complex member 1, RPS13 - ribosomal protein s13, RINT1 - rad50 interactor 1, RPS6 - ribosomal protein s6, GOLPH3 - golgi phosphoprotein 3 (coat-protein), RPS7 - ribosomal protein s7, THOC5 - tho complex 5, SNX6 - sorting nexin 6, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, SRP72 - signal recognition particle 72kda, CAT - catalase, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, HSPD1 - heat shock 60kda protein 1 (chaperonin), XPO6 - exportin 6, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, SAMM50 - samm50 sorting and assembly machinery component, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), ARL6IP5 - adp-ribosylation-like factor 6 interacting protein 5, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, STX8 - syntaxin 8, STX18 - syntaxin 18, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, HSPA4 - heat shock 70kda protein 4, RRBP1 - ribosome binding protein 1, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, EPS15 - epidermal growth factor receptor pathway substrate 15]
76 [PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, ATG7 - autophagy related 7, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, OSTF1 - osteoclast stimulating factor 1, PGAM1 - phosphoglycerate mutase 1 (brain), CAT - catalase, GSN - gelsolin, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, PA2G4 - proliferation-associated 2g4, 38kda, KPNB1 - karyopherin (importin) beta 1, HMGB1 - high mobility group box 1, FAM49B - family with sequence similarity 49, member b, FABP5 - fatty acid binding protein 5 (psoriasis-associated), CAB39 - calcium binding protein 39, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, PYGB - phosphorylase, glycogen; brain, ERP44 - endoplasmic reticulum protein 44, PRKCD - protein kinase c, delta, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, PGM2 - phosphoglucomutase 2, CTSC - cathepsin c]
77 [NCBP2 - nuclear cap binding protein subunit 2, 20kda, ERAP1 - endoplasmic reticulum aminopeptidase 1, EXOSC5 - exosome component 5, DDX6 - dead (asp-glu-ala-asp) box helicase 6, GSPT1 - g1 to s phase transition 1, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, ECI1 - enoyl-coa delta isomerase 1, DERL1 - derlin 1, TRIM25 - tripartite motif containing 25, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, ACAT1 - acetyl-coa acetyltransferase 1, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, VTI1A - vesicle transport through interaction with t-snares 1a, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, USP25 - ubiquitin specific peptidase 25, SORD - sorbitol dehydrogenase, PLCG1 - phospholipase c, gamma 1, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, ANAPC1 - anaphase promoting complex subunit 1, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, EXOSC1 - exosome component 1, SGPL1 - sphingosine-1-phosphate lyase 1, ATG7 - autophagy related 7, ANAPC7 - anaphase promoting complex subunit 7, GLS - glutaminase, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), PELO - pelota homolog (drosophila), HADH - hydroxyacyl-coa dehydrogenase, ADPGK - adp-dependent glucokinase, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, MUT - methylmalonyl coa mutase, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, GNPDA2 - glucosamine-6-phosphate deaminase 2, COMT - catechol-o-methyltransferase, KYNU - kynureninase, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), NEMF - nuclear export mediator factor, EIF3E - eukaryotic translation initiation factor 3, subunit e, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, VTA1 - vesicle (multivesicular body) trafficking 1, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, DENND3 - denn/madd domain containing 3, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), UBE2Z - ubiquitin-conjugating enzyme e2z, HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PYGB - phosphorylase, glycogen; brain, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, PHKB - phosphorylase kinase, beta, RPL3 - ribosomal protein l3, USP48 - ubiquitin specific peptidase 48, MANBA - mannosidase, beta a, lysosomal, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), USP8 - ubiquitin specific peptidase 8, DUT - deoxyuridine triphosphatase, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, HK2 - hexokinase 2, KPNB1 - karyopherin (importin) beta 1, TRIM38 - tripartite motif containing 38, IDE - insulin-degrading enzyme, CDC23 - cell division cycle 23, USP34 - ubiquitin specific peptidase 34, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, UBA6 - ubiquitin-like modifier activating enzyme 6, RPL14 - ribosomal protein l14, MFN1 - mitofusin 1, NUB1 - negative regulator of ubiquitin-like proteins 1, RPS8 - ribosomal protein s8, ESD - esterase d, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, UCHL3 - ubiquitin carboxyl-terminal esterase l3 (ubiquitin thiolesterase), CISD2 - cdgsh iron sulfur domain 2, GCDH - glutaryl-coa dehydrogenase, ACOT7 - acyl-coa thioesterase 7, CAT - catalase, USP15 - ubiquitin specific peptidase 15, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), UBE2I - ubiquitin-conjugating enzyme e2i, CUL1 - cullin 1, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), PGM2 - phosphoglucomutase 2]
78 [PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, ATG7 - autophagy related 7, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, OSTF1 - osteoclast stimulating factor 1, PGAM1 - phosphoglycerate mutase 1 (brain), CAT - catalase, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, GSN - gelsolin, PA2G4 - proliferation-associated 2g4, 38kda, KPNB1 - karyopherin (importin) beta 1, HMGB1 - high mobility group box 1, FAM49B - family with sequence similarity 49, member b, FABP5 - fatty acid binding protein 5 (psoriasis-associated), CAB39 - calcium binding protein 39, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, PYGB - phosphorylase, glycogen; brain, ERP44 - endoplasmic reticulum protein 44, PRKCD - protein kinase c, delta, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, PGM2 - phosphoglucomutase 2, CTSC - cathepsin c]
79 [CCNK - cyclin k, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, DST - dystonin, NCBP2 - nuclear cap binding protein subunit 2, 20kda, EXOSC5 - exosome component 5, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, RFC3 - replication factor c (activator 1) 3, 38kda, SF3A3 - splicing factor 3a, subunit 3, 60kda, SACM1L - sac1 suppressor of actin mutations 1-like (yeast), SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, NDUFA5 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, GFM2 - g elongation factor, mitochondrial 2, TMED2 - transmembrane emp24 domain trafficking protein 2, RECQL - recq protein-like (dna helicase q1-like), HOOK3 - hook microtubule-tethering protein 3, DERL1 - derlin 1, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, SPC24 - spc24, ndc80 kinetochore complex component, ACAT1 - acetyl-coa acetyltransferase 1, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, CPOX - coproporphyrinogen oxidase, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), SUGT1 - sgt1, suppressor of g2 allele of skp1 (s. cerevisiae), GOSR2 - golgi snap receptor complex member 2, OTUB1 - otu domain, ubiquitin aldehyde binding 1, SORD - sorbitol dehydrogenase, ZNF326 - zinc finger protein 326, PLCG1 - phospholipase c, gamma 1, WDR77 - wd repeat domain 77, RCSD1 - rcsd domain containing 1, MORF4L1 - mortality factor 4 like 1, CREB1 - camp responsive element binding protein 1, MYO6 - myosin vi, SNX1 - sorting nexin 1, THUMPD1 - thump domain containing 1, SGPL1 - sphingosine-1-phosphate lyase 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, EIF2D - eukaryotic translation initiation factor 2d, TPM3 - tropomyosin 3, SNRPE - small nuclear ribonucleoprotein polypeptide e, ANAPC7 - anaphase promoting complex subunit 7, SEH1L - seh1-like (s. cerevisiae), EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, GLS - glutaminase, PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, GLO1 - glyoxalase i, CC2D1B - coiled-coil and c2 domain containing 1b, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), GLG1 - golgi glycoprotein 1, NIP7 - nip7, nucleolar pre-rrna processing protein, GOSR1 - golgi snap receptor complex member 1, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), COMMD10 - comm domain containing 10, POLR1C - polymerase (rna) i polypeptide c, 30kda, DCAF7 - ddb1 and cul4 associated factor 7, ADPGK - adp-dependent glucokinase, ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, SUMO3 - small ubiquitin-like modifier 3, SNX6 - sorting nexin 6, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, TSFM - ts translation elongation factor, mitochondrial, SMARCD1 - swi/snf related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, KYNU - kynureninase, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, STX8 - syntaxin 8, ERP44 - endoplasmic reticulum protein 44, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), NAMPT - nicotinamide phosphoribosyltransferase, PARP4 - poly (adp-ribose) polymerase family, member 4, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, TMEM30A - transmembrane protein 30a, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, UQCRB - ubiquinol-cytochrome c reductase binding protein, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, CAPN2 - calpain 2, (m/ii) large subunit, ZWILCH - zwilch kinetochore protein, OSTF1 - osteoclast stimulating factor 1, UBE2Z - ubiquitin-conjugating enzyme e2z, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, AGK - acylglycerol kinase, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PTCD3 - pentatricopeptide repeat domain 3, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, FTH1 - ferritin, heavy polypeptide 1, USP48 - ubiquitin specific peptidase 48, PPP5C - protein phosphatase 5, catalytic subunit, RPL3 - ribosomal protein l3, SURF4 - surfeit 4, IRAK4 - interleukin-1 receptor-associated kinase 4, DNAJC11 - dnaj (hsp40) homolog, subfamily c, member 11, MRPS35 - mitochondrial ribosomal protein s35, MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, RPN1 - ribophorin i, AK4 - adenylate kinase 4, STX17 - syntaxin 17, HK2 - hexokinase 2, KPNB1 - karyopherin (importin) beta 1, TRIM38 - tripartite motif containing 38, CDC23 - cell division cycle 23, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, TJP2 - tight junction protein 2, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, VDAC2 - voltage-dependent anion channel 2, TTLL12 - tubulin tyrosine ligase-like family, member 12, MRPL13 - mitochondrial ribosomal protein l13, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, NIPBL - nipped-b homolog (drosophila), SCYL2 - scy1-like 2 (s. cerevisiae), GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, GALNT2 - udp-n-acetyl-alpha-d-galactosamine:polypeptide n-acetylgalactosaminyltransferase 2 (galnac-t2), TMEM70 - transmembrane protein 70, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), MFN1 - mitofusin 1, SNRNP27 - small nuclear ribonucleoprotein 27kda (u4/u6.u5), RNMT - rna (guanine-7-) methyltransferase, STAT6 - signal transducer and activator of transcription 6, interleukin-4 induced, WDR1 - wd repeat domain 1, LARP4B - la ribonucleoprotein domain family, member 4b, RPS8 - ribosomal protein s8, ESD - esterase d, MRPL42 - mitochondrial ribosomal protein l42, RPS13 - ribosomal protein s13, RINT1 - rad50 interactor 1, RPS6 - ribosomal protein s6, NHP2 - nhp2 ribonucleoprotein, RPS7 - ribosomal protein s7, STOM - stomatin, UCHL3 - ubiquitin carboxyl-terminal esterase l3 (ubiquitin thiolesterase), SUMO1 - small ubiquitin-like modifier 1, CISD2 - cdgsh iron sulfur domain 2, GCDH - glutaryl-coa dehydrogenase, CAT - catalase, XPO6 - exportin 6, UGDH - udp-glucose 6-dehydrogenase, NDUFAF4 - nadh dehydrogenase (ubiquinone) complex i, assembly factor 4, SSBP1 - single-stranded dna binding protein 1, mitochondrial, SAMM50 - samm50 sorting and assembly machinery component, MRPL47 - mitochondrial ribosomal protein l47, ANXA4 - annexin a4, TUBGCP3 - tubulin, gamma complex associated protein 3, UBE2I - ubiquitin-conjugating enzyme e2i, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, TNFAIP8 - tumor necrosis factor, alpha-induced protein 8, PRKCD - protein kinase c, delta, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, RRBP1 - ribosome binding protein 1, EPS15 - epidermal growth factor receptor pathway substrate 15, RRM1 - ribonucleotide reductase m1, DDX31 - dead (asp-glu-ala-asp) box polypeptide 31, ERAP1 - endoplasmic reticulum aminopeptidase 1, DDX6 - dead (asp-glu-ala-asp) box helicase 6, TCOF1 - treacher collins-franceschetti syndrome 1, SDAD1 - sda1 domain containing 1, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, ZMYM2 - zinc finger, mym-type 2, ZNF638 - zinc finger protein 638, STAMBP - stam binding protein, SCFD1 - sec1 family domain containing 1, GSN - gelsolin, CISD1 - cdgsh iron sulfur domain 1, URB1 - urb1 ribosome biogenesis 1 homolog (s. cerevisiae), RAB30 - rab30, member ras oncogene family, FLNB - filamin b, beta, ECI1 - enoyl-coa delta isomerase 1, DCK - deoxycytidine kinase, PPP4R2 - protein phosphatase 4, regulatory subunit 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, DBN1 - drebrin 1, TRIM25 - tripartite motif containing 25, ILF3 - interleukin enhancer binding factor 3, 90kda, XPOT - exportin, trna, ANKRD28 - ankyrin repeat domain 28, NUDCD2 - nudc domain containing 2, RIF1 - rap1 interacting factor homolog (yeast), CYCS - cytochrome c, somatic, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, VTI1A - vesicle transport through interaction with t-snares 1a, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, CCDC51 - coiled-coil domain containing 51, RBM19 - rna binding motif protein 19, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, MMS22L - mms22-like, dna repair protein, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, EHBP1 - eh domain binding protein 1, ARCN1 - archain 1, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, PA2G4 - proliferation-associated 2g4, 38kda, AGPS - alkylglycerone phosphate synthase, CAB39 - calcium binding protein 39, TMED5 - transmembrane emp24 protein transport domain containing 5, GATAD2B - gata zinc finger domain containing 2b, RAB23 - rab23, member ras oncogene family, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, GAR1 - gar1 ribonucleoprotein, KDM3A - lysine (k)-specific demethylase 3a, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, ANAPC1 - anaphase promoting complex subunit 1, MRPL3 - mitochondrial ribosomal protein l3, EXOSC1 - exosome component 1, TELO2 - telomere maintenance 2, PITRM1 - pitrilysin metallopeptidase 1, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), MCM7 - minichromosome maintenance complex component 7, ATG7 - autophagy related 7, MCU - mitochondrial calcium uniporter, ARL1 - adp-ribosylation factor-like 1, TUBGCP4 - tubulin, gamma complex associated protein 4, KDELC2 - kdel (lys-asp-glu-leu) containing 2, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), SYNE1 - spectrin repeat containing, nuclear envelope 1, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), NUP88 - nucleoporin 88kda, SCO1 - sco1 cytochrome c oxidase assembly protein, ZBTB8OS - zinc finger and btb domain containing 8 opposite strand, NOM1 - nucleolar protein with mif4g domain 1, WDHD1 - wd repeat and hmg-box dna binding protein 1, NFU1 - nfu1 iron-sulfur cluster scaffold homolog (s. cerevisiae), PPIL3 - peptidylprolyl isomerase (cyclophilin)-like 3, HADH - hydroxyacyl-coa dehydrogenase, OSBPL3 - oxysterol binding protein-like 3, THOC5 - tho complex 5, FECH - ferrochelatase, CDK6 - cyclin-dependent kinase 6, ADNP - activity-dependent neuroprotector homeobox, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, CDK13 - cyclin-dependent kinase 13, TBCD - tubulin folding cofactor d, FCF1 - fcf1 rrna-processing protein, CCT4 - chaperonin containing tcp1, subunit 4 (delta), CUTC - cutc copper transporter, TBCA - tubulin folding cofactor a, INPP5B - inositol polyphosphate-5-phosphatase, 75kda, RPL7L1 - ribosomal protein l7-like 1, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, IRF3 - interferon regulatory factor 3, ARL6IP5 - adp-ribosylation-like factor 6 interacting protein 5, STX18 - syntaxin 18, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, IRF5 - interferon regulatory factor 5, DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), RPTOR - regulatory associated protein of mtor, complex 1, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, EIF3E - eukaryotic translation initiation factor 3, subunit e, QSOX2 - quiescin q6 sulfhydryl oxidase 2, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, TCEA1 - transcription elongation factor a (sii), 1, LYRM7 - lyr motif containing 7, VTA1 - vesicle (multivesicular body) trafficking 1, PURA - purine-rich element binding protein a, SPG11 - spastic paraplegia 11 (autosomal recessive), ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, TDRD7 - tudor domain containing 7, PGM1 - phosphoglucomutase 1, NEK6 - nima-related kinase 6, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, NOC3L - nucleolar complex associated 3 homolog (s. cerevisiae), PYGB - phosphorylase, glycogen; brain, PCYT2 - phosphate cytidylyltransferase 2, ethanolamine, NUP50 - nucleoporin 50kda, MAP4 - microtubule-associated protein 4, RAB14 - rab14, member ras oncogene family, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, MANBA - mannosidase, beta a, lysosomal, VKORC1L1 - vitamin k epoxide reductase complex, subunit 1-like 1, MAN2A1 - mannosidase, alpha, class 2a, member 1, USP8 - ubiquitin specific peptidase 8, TRAPPC10 - trafficking protein particle complex 10, MBNL1 - muscleblind-like splicing regulator 1, METTL15 - methyltransferase like 15, DUT - deoxyuridine triphosphatase, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, MCM3 - minichromosome maintenance complex component 3, IDE - insulin-degrading enzyme, CHMP5 - charged multivesicular body protein 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, DOCK7 - dedicator of cytokinesis 7, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, SART3 - squamous cell carcinoma antigen recognized by t cells 3, MAD1L1 - mad1 mitotic arrest deficient-like 1 (yeast), CTSC - cathepsin c, WIPF1 - was/wasl interacting protein family, member 1, SCAF8 - sr-related ctd-associated factor 8, LSS - lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase), RANBP1 - ran binding protein 1, DLG1 - discs, large homolog 1 (drosophila), RAP1A - rap1a, member of ras oncogene family, PUS1 - pseudouridylate synthase 1, RPL14 - ribosomal protein l14, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, GNPAT - glyceronephosphate o-acyltransferase, AKAP8L - a kinase (prka) anchor protein 8-like, NUB1 - negative regulator of ubiquitin-like proteins 1, DGUOK - deoxyguanosine kinase, TRAPPC9 - trafficking protein particle complex 9, CLNS1A - chloride channel, nucleotide-sensitive, 1a, RAP1B - rap1b, member of ras oncogene family, GOLPH3 - golgi phosphoprotein 3 (coat-protein), TMX3 - thioredoxin-related transmembrane protein 3, DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, ACOT7 - acyl-coa thioesterase 7, RB1 - retinoblastoma 1, PDAP1 - pdgfa associated protein 1, CBX3 - chromobox homolog 3, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, FAM49B - family with sequence similarity 49, member b, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), RHOT1 - ras homolog family member t1, MRPS14 - mitochondrial ribosomal protein s14, CUL1 - cullin 1, RSL1D1 - ribosomal l1 domain containing 1, BCL2L1 - bcl2-like 1, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, PGM2 - phosphoglucomutase 2]
80 [PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, ATG7 - autophagy related 7, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, OSTF1 - osteoclast stimulating factor 1, PGAM1 - phosphoglycerate mutase 1 (brain), CAT - catalase, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, GSN - gelsolin, PA2G4 - proliferation-associated 2g4, 38kda, KPNB1 - karyopherin (importin) beta 1, HMGB1 - high mobility group box 1, FAM49B - family with sequence similarity 49, member b, FABP5 - fatty acid binding protein 5 (psoriasis-associated), CAB39 - calcium binding protein 39, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, PYGB - phosphorylase, glycogen; brain, ERP44 - endoplasmic reticulum protein 44, PRKCD - protein kinase c, delta, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, PGM2 - phosphoglucomutase 2, CTSC - cathepsin c]
81 [AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, VTA1 - vesicle (multivesicular body) trafficking 1, PURA - purine-rich element binding protein a, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, RPL23A - ribosomal protein l23a, SDAD1 - sda1 domain containing 1, RPL22 - ribosomal protein l22, SCFD1 - sec1 family domain containing 1, AGK - acylglycerol kinase, RPL13 - ribosomal protein l13, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), TBC1D2B - tbc1 domain family, member 2b, TMED2 - transmembrane emp24 domain trafficking protein 2, RPL6 - ribosomal protein l6, NUP50 - nucleoporin 50kda, HOOK3 - hook microtubule-tethering protein 3, RPL7 - ribosomal protein l7, RAB14 - rab14, member ras oncogene family, RPL3 - ribosomal protein l3, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, DERL1 - derlin 1, BTF3 - basic transcription factor 3, VTI1A - vesicle transport through interaction with t-snares 1a, TNPO1 - transportin 1, STX17 - syntaxin 17, ARCN1 - archain 1, EHBP1 - eh domain binding protein 1, KPNB1 - karyopherin (importin) beta 1, AGPS - alkylglycerone phosphate synthase, GOSR2 - golgi snap receptor complex member 2, IDE - insulin-degrading enzyme, TBC1D4 - tbc1 domain family, member 4, CHMP5 - charged multivesicular body protein 5, EXOC2 - exocyst complex component 2, TMED5 - transmembrane emp24 protein transport domain containing 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, RAB23 - rab23, member ras oncogene family, EXOC1 - exocyst complex component 1, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, MYO6 - myosin vi, SNX1 - sorting nexin 1, PITRM1 - pitrilysin metallopeptidase 1, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), ATG7 - autophagy related 7, EIF2D - eukaryotic translation initiation factor 2d, ARL1 - adp-ribosylation factor-like 1, GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, RPL14 - ribosomal protein l14, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), GNPAT - glyceronephosphate o-acyltransferase, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), NUP88 - nucleoporin 88kda, RPS8 - ribosomal protein s8, GOSR1 - golgi snap receptor complex member 1, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RINT1 - rad50 interactor 1, GOLPH3 - golgi phosphoprotein 3 (coat-protein), RPS7 - ribosomal protein s7, SNX6 - sorting nexin 6, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, SRP72 - signal recognition particle 72kda, CAT - catalase, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, HSPD1 - heat shock 60kda protein 1 (chaperonin), XPO6 - exportin 6, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), SAMM50 - samm50 sorting and assembly machinery component, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, STX8 - syntaxin 8, STX18 - syntaxin 18, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, HSPA4 - heat shock 70kda protein 4, RRBP1 - ribosome binding protein 1, EPS15 - epidermal growth factor receptor pathway substrate 15]
82 [PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, UQCRB - ubiquinol-cytochrome c reductase binding protein, DLG1 - discs, large homolog 1 (drosophila), PGM1 - phosphoglucomutase 1, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, PGAM1 - phosphoglycerate mutase 1 (brain), MFN1 - mitofusin 1, DGUOK - deoxyguanosine kinase, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, DCK - deoxycytidine kinase, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, ADPGK - adp-dependent glucokinase, ACAT1 - acetyl-coa acetyltransferase 1, HK2 - hexokinase 2, AK4 - adenylate kinase 4, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, ACOT7 - acyl-coa thioesterase 7, GCDH - glutaryl-coa dehydrogenase, MVD - mevalonate (diphospho) decarboxylase, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, KYNU - kynureninase, RAB23 - rab23, member ras oncogene family, TJP2 - tight junction protein 2, AK3 - adenylate kinase 3, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, DCAKD - dephospho-coa kinase domain containing]
83 [RFC3 - replication factor c (activator 1) 3, 38kda, CAPN2 - calpain 2, (m/ii) large subunit, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, WNK1 - wnk lysine deficient protein kinase 1, GSN - gelsolin, HMGB2 - high mobility group box 2, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, HMGB1 - high mobility group box 1, FLNB - filamin b, beta, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, RPL3 - ribosomal protein l3, PPP5C - protein phosphatase 5, catalytic subunit, DERL1 - derlin 1, TRIM25 - tripartite motif containing 25, DOCK8 - dedicator of cytokinesis 8, USP8 - ubiquitin specific peptidase 8, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, RIF1 - rap1 interacting factor homolog (yeast), ACAT1 - acetyl-coa acetyltransferase 1, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, CPOX - coproporphyrinogen oxidase, HK2 - hexokinase 2, MAT2A - methionine adenosyltransferase ii, alpha, EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, TBC1D4 - tbc1 domain family, member 4, CHMP5 - charged multivesicular body protein 5, OTUB1 - otu domain, ubiquitin aldehyde binding 1, SORD - sorbitol dehydrogenase, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, PLCG1 - phospholipase c, gamma 1, LIMS1 - lim and senescent cell antigen-like domains 1, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, IFIH1 - interferon induced with helicase c domain 1, KDM3A - lysine (k)-specific demethylase 3a, CREB1 - camp responsive element binding protein 1, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, CTSC - cathepsin c, MCM7 - minichromosome maintenance complex component 7, ATG7 - autophagy related 7, RAP1A - rap1a, member of ras oncogene family, PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), GNPAT - glyceronephosphate o-acyltransferase, RNMT - rna (guanine-7-) methyltransferase, STAT6 - signal transducer and activator of transcription 6, interleukin-4 induced, NUB1 - negative regulator of ubiquitin-like proteins 1, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), HADH - hydroxyacyl-coa dehydrogenase, RAP1B - rap1b, member of ras oncogene family, EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), GOLPH3 - golgi phosphoprotein 3 (coat-protein), ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, FECH - ferrochelatase, ADNP - activity-dependent neuroprotector homeobox, SNX6 - sorting nexin 6, SMARCD1 - swi/snf related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1, CAT - catalase, HSPD1 - heat shock 60kda protein 1 (chaperonin), FAM49B - family with sequence similarity 49, member b, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), COMT - catechol-o-methyltransferase, IRF3 - interferon regulatory factor 3, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, KYNU - kynureninase, STX8 - syntaxin 8, ERP44 - endoplasmic reticulum protein 44, BCL2L1 - bcl2-like 1, PRKCD - protein kinase c, delta, IRF5 - interferon regulatory factor 5, LARP1 - la ribonucleoprotein domain family, member 1, NAMPT - nicotinamide phosphoribosyltransferase, RPTOR - regulatory associated protein of mtor, complex 1, HSPA4 - heat shock 70kda protein 4, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1]
84 [UQCRB - ubiquinol-cytochrome c reductase binding protein, DLG1 - discs, large homolog 1 (drosophila), PGM1 - phosphoglucomutase 1, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, PGAM1 - phosphoglycerate mutase 1 (brain), MFN1 - mitofusin 1, DGUOK - deoxyguanosine kinase, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, UCK2 - uridine-cytidine kinase 2, UMPS - uridine monophosphate synthetase, ADPGK - adp-dependent glucokinase, ACAT1 - acetyl-coa acetyltransferase 1, HK2 - hexokinase 2, AK4 - adenylate kinase 4, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, ACOT7 - acyl-coa thioesterase 7, GCDH - glutaryl-coa dehydrogenase, MVD - mevalonate (diphospho) decarboxylase, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, KYNU - kynureninase, RAB23 - rab23, member ras oncogene family, TJP2 - tight junction protein 2, AK3 - adenylate kinase 3, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, DCAKD - dephospho-coa kinase domain containing]
85 [RPL23A - ribosomal protein l23a, MRPL13 - mitochondrial ribosomal protein l13, RPL22 - ribosomal protein l22, GSPT1 - g1 to s phase transition 1, RPL14 - ribosomal protein l14, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, AGK - acylglycerol kinase, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, RPL13 - ribosomal protein l13, GFM2 - g elongation factor, mitochondrial 2, PTCD3 - pentatricopeptide repeat domain 3, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RPS8 - ribosomal protein s8, RPL6 - ribosomal protein l6, MRPL42 - mitochondrial ribosomal protein l42, RPL7 - ribosomal protein l7, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPL3 - ribosomal protein l3, RPS7 - ribosomal protein s7, ACAT1 - acetyl-coa acetyltransferase 1, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, MRPL9 - mitochondrial ribosomal protein l9, GCDH - glutaryl-coa dehydrogenase, MRPL47 - mitochondrial ribosomal protein l47, MRPS14 - mitochondrial ribosomal protein s14, RPL31 - ribosomal protein l31, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, RPL32 - ribosomal protein l32, RRBP1 - ribosome binding protein 1, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, MRPL3 - mitochondrial ribosomal protein l3, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
86 [AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, TMEM30A - transmembrane protein 30a, PURA - purine-rich element binding protein a, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, RPL22 - ribosomal protein l22, PGM1 - phosphoglucomutase 1, OSTF1 - osteoclast stimulating factor 1, PGAM1 - phosphoglycerate mutase 1 (brain), GSN - gelsolin, HMGB1 - high mobility group box 1, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PYGB - phosphorylase, glycogen; brain, RAB14 - rab14, member ras oncogene family, FTH1 - ferritin, heavy polypeptide 1, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, MANBA - mannosidase, beta a, lysosomal, SURF4 - surfeit 4, DOCK8 - dedicator of cytokinesis 8, SLAMF1 - signaling lymphocytic activation molecule family member 1, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PA2G4 - proliferation-associated 2g4, 38kda, KPNB1 - karyopherin (importin) beta 1, CAB39 - calcium binding protein 39, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), CTSC - cathepsin c, ATG7 - autophagy related 7, DLG1 - discs, large homolog 1 (drosophila), RAP1A - rap1a, member of ras oncogene family, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, RAP1B - rap1b, member of ras oncogene family, RPS6 - ribosomal protein s6, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), CDK6 - cyclin-dependent kinase 6, STOM - stomatin, CAT - catalase, CDK13 - cyclin-dependent kinase 13, PDAP1 - pdgfa associated protein 1, HSPD1 - heat shock 60kda protein 1 (chaperonin), ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, ERP44 - endoplasmic reticulum protein 44, PRKCD - protein kinase c, delta, NAMPT - nicotinamide phosphoribosyltransferase, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, PGM2 - phosphoglucomutase 2]
87 [TMEM30A - transmembrane protein 30a, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, VTA1 - vesicle (multivesicular body) trafficking 1, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, PGM1 - phosphoglucomutase 1, OSTF1 - osteoclast stimulating factor 1, PGAM1 - phosphoglycerate mutase 1 (brain), GSN - gelsolin, HMGB1 - high mobility group box 1, FABP5 - fatty acid binding protein 5 (psoriasis-associated), TMED2 - transmembrane emp24 domain trafficking protein 2, PYGB - phosphorylase, glycogen; brain, RAB14 - rab14, member ras oncogene family, FTH1 - ferritin, heavy polypeptide 1, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, MANBA - mannosidase, beta a, lysosomal, SURF4 - surfeit 4, IRAK4 - interleukin-1 receptor-associated kinase 4, USP8 - ubiquitin specific peptidase 8, TRAPPC10 - trafficking protein particle complex 10, VTI1A - vesicle transport through interaction with t-snares 1a, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, STX17 - syntaxin 17, ARCN1 - archain 1, PA2G4 - proliferation-associated 2g4, 38kda, KPNB1 - karyopherin (importin) beta 1, CAB39 - calcium binding protein 39, GOSR2 - golgi snap receptor complex member 2, CHMP5 - charged multivesicular body protein 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, RAB23 - rab23, member ras oncogene family, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), MYO6 - myosin vi, SNX1 - sorting nexin 1, CTSC - cathepsin c, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), ATG7 - autophagy related 7, SCYL2 - scy1-like 2 (s. cerevisiae), RAP1A - rap1a, member of ras oncogene family, GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), RAP1B - rap1b, member of ras oncogene family, STOM - stomatin, TMX3 - thioredoxin-related transmembrane protein 3, SNX6 - sorting nexin 6, CAT - catalase, PDAP1 - pdgfa associated protein 1, CDK13 - cyclin-dependent kinase 13, FAM49B - family with sequence similarity 49, member b, INPP5B - inositol polyphosphate-5-phosphatase, 75kda, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, ERP44 - endoplasmic reticulum protein 44, BCL2L1 - bcl2-like 1, PRKCD - protein kinase c, delta, EPS15 - epidermal growth factor receptor pathway substrate 15, PGM2 - phosphoglucomutase 2, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1]
88 [PITRM1 - pitrilysin metallopeptidase 1, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, NIPBL - nipped-b homolog (drosophila), RPL14 - ribosomal protein l14, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), AGK - acylglycerol kinase, GNPAT - glyceronephosphate o-acyltransferase, RPL13 - ribosomal protein l13, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), NUP88 - nucleoporin 88kda, RPS8 - ribosomal protein s8, NUP50 - nucleoporin 50kda, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPL3 - ribosomal protein l3, RPS7 - ribosomal protein s7, VTI1A - vesicle transport through interaction with t-snares 1a, SRP72 - signal recognition particle 72kda, TNPO1 - transportin 1, HK2 - hexokinase 2, CAT - catalase, HSPD1 - heat shock 60kda protein 1 (chaperonin), LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, KPNB1 - karyopherin (importin) beta 1, AGPS - alkylglycerone phosphate synthase, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), SAMM50 - samm50 sorting and assembly machinery component, GOSR2 - golgi snap receptor complex member 2, IDE - insulin-degrading enzyme, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, HSPA4 - heat shock 70kda protein 4]
89 [FAM136A - family with sequence similarity 136, member a, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, SF3A3 - splicing factor 3a, subunit 3, 60kda, GFM2 - g elongation factor, mitochondrial 2, TMED2 - transmembrane emp24 domain trafficking protein 2, HOOK3 - hook microtubule-tethering protein 3, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, ACAT1 - acetyl-coa acetyltransferase 1, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, CPOX - coproporphyrinogen oxidase, LRRC40 - leucine rich repeat containing 40, SUGT1 - sgt1, suppressor of g2 allele of skp1 (s. cerevisiae), GOSR2 - golgi snap receptor complex member 2, OTUB1 - otu domain, ubiquitin aldehyde binding 1, SORD - sorbitol dehydrogenase, PLCG1 - phospholipase c, gamma 1, LIMS1 - lim and senescent cell antigen-like domains 1, MORF4L1 - mortality factor 4 like 1, CARHSP1 - calcium regulated heat stable protein 1, 24kda, CREB1 - camp responsive element binding protein 1, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, SNX1 - sorting nexin 1, THUMPD1 - thump domain containing 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, SNRPE - small nuclear ribonucleoprotein polypeptide e, ANAPC7 - anaphase promoting complex subunit 7, GLS - glutaminase, PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, GLO1 - glyoxalase i, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), GLG1 - golgi glycoprotein 1, NIP7 - nip7, nucleolar pre-rrna processing protein, GOSR1 - golgi snap receptor complex member 1, COMMD10 - comm domain containing 10, POLR1C - polymerase (rna) i polypeptide c, 30kda, PPA1 - pyrophosphatase (inorganic) 1, SUMO3 - small ubiquitin-like modifier 3, SNX6 - sorting nexin 6, SMARCD1 - swi/snf related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, COMT - catechol-o-methyltransferase, STX8 - syntaxin 8, TAOK3 - tao kinase 3, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), PARP4 - poly (adp-ribose) polymerase family, member 4, TMEM30A - transmembrane protein 30a, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, SCPEP1 - serine carboxypeptidase 1, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, QKI - qki, kh domain containing, rna binding, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, AGK - acylglycerol kinase, HOMER1 - homer homolog 1 (drosophila), FTH1 - ferritin, heavy polypeptide 1, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, PPP5C - protein phosphatase 5, catalytic subunit, USP48 - ubiquitin specific peptidase 48, SURF4 - surfeit 4, IRAK4 - interleukin-1 receptor-associated kinase 4, DNAJC11 - dnaj (hsp40) homolog, subfamily c, member 11, MRPS35 - mitochondrial ribosomal protein s35, AK4 - adenylate kinase 4, TNPO1 - transportin 1, KPNB1 - karyopherin (importin) beta 1, EXOC2 - exocyst complex component 2, EXOC1 - exocyst complex component 1, TJP2 - tight junction protein 2, IFIT5 - interferon-induced protein with tetratricopeptide repeats 5, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, SCYL2 - scy1-like 2 (s. cerevisiae), NIPBL - nipped-b homolog (drosophila), GALNT2 - udp-n-acetyl-alpha-d-galactosamine:polypeptide n-acetylgalactosaminyltransferase 2 (galnac-t2), STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), SNRNP27 - small nuclear ribonucleoprotein 27kda (u4/u6.u5), MFN1 - mitofusin 1, STAT6 - signal transducer and activator of transcription 6, interleukin-4 induced, EIF1AD - eukaryotic translation initiation factor 1a domain containing, FCRLA - fc receptor-like a, RINT1 - rad50 interactor 1, NHP2 - nhp2 ribonucleoprotein, DSTN - destrin (actin depolymerizing factor), RIC8A - ric8 guanine nucleotide exchange factor a, SRP72 - signal recognition particle 72kda, CISD2 - cdgsh iron sulfur domain 2, GCDH - glutaryl-coa dehydrogenase, SSBP1 - single-stranded dna binding protein 1, mitochondrial, SAMM50 - samm50 sorting and assembly machinery component, ANXA4 - annexin a4, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, TNFAIP8 - tumor necrosis factor, alpha-induced protein 8, PRKCD - protein kinase c, delta, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, CCDC6 - coiled-coil domain containing 6, ERAP1 - endoplasmic reticulum aminopeptidase 1, DMXL1 - dmx-like 1, DDX6 - dead (asp-glu-ala-asp) box helicase 6, SDAD1 - sda1 domain containing 1, ZMYM2 - zinc finger, mym-type 2, GSPT1 - g1 to s phase transition 1, GSN - gelsolin, FAM98A - family with sequence similarity 98, member a, ECI1 - enoyl-coa delta isomerase 1, DCK - deoxycytidine kinase, DBN1 - drebrin 1, TRIM25 - tripartite motif containing 25, XPOT - exportin, trna, RIF1 - rap1 interacting factor homolog (yeast), VTI1A - vesicle transport through interaction with t-snares 1a, CCDC51 - coiled-coil domain containing 51, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), MMS22L - mms22-like, dna repair protein, ARCN1 - archain 1, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, LRRK1 - leucine-rich repeat kinase 1, PA2G4 - proliferation-associated 2g4, 38kda, CAB39 - calcium binding protein 39, GATAD2B - gata zinc finger domain containing 2b, RAB23 - rab23, member ras oncogene family, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, MRPL3 - mitochondrial ribosomal protein l3, RNF31 - ring finger protein 31, MCM7 - minichromosome maintenance complex component 7, MCU - mitochondrial calcium uniporter, ARL1 - adp-ribosylation factor-like 1, KDELC2 - kdel (lys-asp-glu-leu) containing 2, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, PELO - pelota homolog (drosophila), SCO1 - sco1 cytochrome c oxidase assembly protein, WDHD1 - wd repeat and hmg-box dna binding protein 1, HADH - hydroxyacyl-coa dehydrogenase, PPIL3 - peptidylprolyl isomerase (cyclophilin)-like 3, OSBPL3 - oxysterol binding protein-like 3, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, C15orf40 - chromosome 15 open reading frame 40, RPL7L1 - ribosomal protein l7-like 1, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, STX18 - syntaxin 18, NEMF - nuclear export mediator factor, RPTOR - regulatory associated protein of mtor, complex 1, QSOX2 - quiescin q6 sulfhydryl oxidase 2, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, LYRM7 - lyr motif containing 7, SERBP1 - serpine1 mrna binding protein 1, VTA1 - vesicle (multivesicular body) trafficking 1, DENND3 - denn/madd domain containing 3, GAPVD1 - gtpase activating protein and vps9 domains 1, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, MAP4 - microtubule-associated protein 4, RAB14 - rab14, member ras oncogene family, PHKB - phosphorylase kinase, beta, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, MANBA - mannosidase, beta a, lysosomal, MAN2A1 - mannosidase, alpha, class 2a, member 1, USP8 - ubiquitin specific peptidase 8, EIF4H - eukaryotic translation initiation factor 4h, MBNL1 - muscleblind-like splicing regulator 1, METTL15 - methyltransferase like 15, SLAMF1 - signaling lymphocytic activation molecule family member 1, DUT - deoxyuridine triphosphatase, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, MAT2A - methionine adenosyltransferase ii, alpha, MCM3 - minichromosome maintenance complex component 3, CHMP5 - charged multivesicular body protein 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, DOCK7 - dedicator of cytokinesis 7, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, WIPF1 - was/wasl interacting protein family, member 1, SCAF8 - sr-related ctd-associated factor 8, GRK6 - g protein-coupled receptor kinase 6, UBA6 - ubiquitin-like modifier activating enzyme 6, LSS - lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase), DLG1 - discs, large homolog 1 (drosophila), PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, RPL14 - ribosomal protein l14, NUB1 - negative regulator of ubiquitin-like proteins 1, DGUOK - deoxyguanosine kinase, TRAPPC9 - trafficking protein particle complex 9, UBA3 - ubiquitin-like modifier activating enzyme 3, DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, ACOT7 - acyl-coa thioesterase 7, PDAP1 - pdgfa associated protein 1, CBX3 - chromobox homolog 3, ADI1 - acireductone dioxygenase 1, FAM49B - family with sequence similarity 49, member b, CAPRIN1 - cell cycle associated protein 1, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), RHOT1 - ras homolog family member t1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, RSL1D1 - ribosomal l1 domain containing 1, BCL2L1 - bcl2-like 1, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, PGM2 - phosphoglucomutase 2, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1, CCNK - cyclin k, DST - dystonin, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, NCBP2 - nuclear cap binding protein subunit 2, 20kda, EXOSC5 - exosome component 5, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, RFC3 - replication factor c (activator 1) 3, 38kda, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, SACM1L - sac1 suppressor of actin mutations 1-like (yeast), NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, NDUFA5 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, RECQL - recq protein-like (dna helicase q1-like), DERL1 - derlin 1, BTF3 - basic transcription factor 3, DOCK8 - dedicator of cytokinesis 8, SPC24 - spc24, ndc80 kinetochore complex component, EIF4B - eukaryotic translation initiation factor 4b, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, USP25 - ubiquitin specific peptidase 25, ZNF326 - zinc finger protein 326, WDR77 - wd repeat domain 77, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, RCSD1 - rcsd domain containing 1, MYO6 - myosin vi, SGPL1 - sphingosine-1-phosphate lyase 1, EIF2D - eukaryotic translation initiation factor 2d, TPM3 - tropomyosin 3, STK38L - serine/threonine kinase 38 like, SEH1L - seh1-like (s. cerevisiae), EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, CC2D1B - coiled-coil and c2 domain containing 1b, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, DCAF7 - ddb1 and cul4 associated factor 7, ADPGK - adp-dependent glucokinase, ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, EIF2S2 - eukaryotic translation initiation factor 2, subunit 2 beta, 38kda, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, TSFM - ts translation elongation factor, mitochondrial, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), MVD - mevalonate (diphospho) decarboxylase, DENND4A - denn/madd domain containing 4a, KYNU - kynureninase, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, ERP44 - endoplasmic reticulum protein 44, NAMPT - nicotinamide phosphoribosyltransferase, HSPA4 - heat shock 70kda protein 4, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, UQCRB - ubiquinol-cytochrome c reductase binding protein, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, CAPN2 - calpain 2, (m/ii) large subunit, ZWILCH - zwilch kinetochore protein, WNK1 - wnk lysine deficient protein kinase 1, OSTF1 - osteoclast stimulating factor 1, UBE2Z - ubiquitin-conjugating enzyme e2z, HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PTCD3 - pentatricopeptide repeat domain 3, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), TBC1D2B - tbc1 domain family, member 2b, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, EIF4G3 - eukaryotic translation initiation factor 4 gamma, 3, UCK2 - uridine-cytidine kinase 2, RPL3 - ribosomal protein l3, UMPS - uridine monophosphate synthetase, CARS - cysteinyl-trna synthetase, EIF3F - eukaryotic translation initiation factor 3, subunit f, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), MRPL1 - mitochondrial ribosomal protein l1, RPN1 - ribophorin i, MRPL9 - mitochondrial ribosomal protein l9, STX17 - syntaxin 17, HK2 - hexokinase 2, TRIM38 - tripartite motif containing 38, CDC23 - cell division cycle 23, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, VDAC2 - voltage-dependent anion channel 2, TTLL12 - tubulin tyrosine ligase-like family, member 12, MRPL13 - mitochondrial ribosomal protein l13, GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, TMEM70 - transmembrane protein 70, RNMT - rna (guanine-7-) methyltransferase, WDR1 - wd repeat domain 1, LARP4B - la ribonucleoprotein domain family, member 4b, RPS8 - ribosomal protein s8, ESD - esterase d, MRPL42 - mitochondrial ribosomal protein l42, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, STOM - stomatin, UCHL3 - ubiquitin carboxyl-terminal esterase l3 (ubiquitin thiolesterase), SUMO1 - small ubiquitin-like modifier 1, CAT - catalase, USP15 - ubiquitin specific peptidase 15, UGDH - udp-glucose 6-dehydrogenase, XPO6 - exportin 6, NDUFAF4 - nadh dehydrogenase (ubiquinone) complex i, assembly factor 4, MRPL47 - mitochondrial ribosomal protein l47, TUBGCP3 - tubulin, gamma complex associated protein 3, UBE2I - ubiquitin-conjugating enzyme e2i, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, AIDA - axin interactor, dorsalization associated, RRBP1 - ribosome binding protein 1, EPS15 - epidermal growth factor receptor pathway substrate 15, RRM1 - ribonucleotide reductase m1, ZC3H7B - zinc finger ccch-type containing 7b, DDX31 - dead (asp-glu-ala-asp) box polypeptide 31, TCOF1 - treacher collins-franceschetti syndrome 1, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, ZNF638 - zinc finger protein 638, F11R - f11 receptor, STAMBP - stam binding protein, SCFD1 - sec1 family domain containing 1, URB1 - urb1 ribosome biogenesis 1 homolog (s. cerevisiae), CISD1 - cdgsh iron sulfur domain 1, RAB30 - rab30, member ras oncogene family, FLNB - filamin b, beta, PPFIA1 - protein tyrosine phosphatase, receptor type, f polypeptide (ptprf), interacting protein (liprin), alpha 1, PPP4R2 - protein phosphatase 4, regulatory subunit 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, PPFIBP1 - ptprf interacting protein, binding protein 1 (liprin beta 1), ILF3 - interleukin enhancer binding factor 3, 90kda, ANKRD28 - ankyrin repeat domain 28, NUDCD2 - nudc domain containing 2, CYCS - cytochrome c, somatic, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, RBM19 - rna binding motif protein 19, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, EHBP1 - eh domain binding protein 1, CD48 - cd48 molecule, AGPS - alkylglycerone phosphate synthase, TBC1D4 - tbc1 domain family, member 4, TMED5 - transmembrane emp24 protein transport domain containing 5, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, GAR1 - gar1 ribonucleoprotein, KDM3A - lysine (k)-specific demethylase 3a, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, ANAPC1 - anaphase promoting complex subunit 1, EXOSC1 - exosome component 1, TELO2 - telomere maintenance 2, PITRM1 - pitrilysin metallopeptidase 1, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), ATG7 - autophagy related 7, TUBGCP4 - tubulin, gamma complex associated protein 4, HDDC2 - hd domain containing 2, PRRC1 - proline-rich coiled-coil 1, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, SYNE1 - spectrin repeat containing, nuclear envelope 1, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), NUP88 - nucleoporin 88kda, ZBTB8OS - zinc finger and btb domain containing 8 opposite strand, NOM1 - nucleolar protein with mif4g domain 1, NFU1 - nfu1 iron-sulfur cluster scaffold homolog (s. cerevisiae), ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), THOC5 - tho complex 5, FECH - ferrochelatase, CDK6 - cyclin-dependent kinase 6, ADNP - activity-dependent neuroprotector homeobox, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, CDK13 - cyclin-dependent kinase 13, TBCD - tubulin folding cofactor d, FCF1 - fcf1 rrna-processing protein, GNPDA2 - glucosamine-6-phosphate deaminase 2, CCT4 - chaperonin containing tcp1, subunit 4 (delta), CUTC - cutc copper transporter, TBCA - tubulin folding cofactor a, INPP5B - inositol polyphosphate-5-phosphatase, 75kda, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, IRF3 - interferon regulatory factor 3, ARL6IP5 - adp-ribosylation-like factor 6 interacting protein 5, LARP1 - la ribonucleoprotein domain family, member 1, IRF5 - interferon regulatory factor 5, DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, EIF3E - eukaryotic translation initiation factor 3, subunit e, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, TCEA1 - transcription elongation factor a (sii), 1, PURA - purine-rich element binding protein a, SPG11 - spastic paraplegia 11 (autosomal recessive), TDRD7 - tudor domain containing 7, NEK6 - nima-related kinase 6, NOC3L - nucleolar complex associated 3 homolog (s. cerevisiae), PYGB - phosphorylase, glycogen; brain, PCYT2 - phosphate cytidylyltransferase 2, ethanolamine, NUP50 - nucleoporin 50kda, VKORC1L1 - vitamin k epoxide reductase complex, subunit 1-like 1, TRAPPC10 - trafficking protein particle complex 10, CCDC58 - coiled-coil domain containing 58, IDE - insulin-degrading enzyme, USP34 - ubiquitin specific peptidase 34, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), IFIH1 - interferon induced with helicase c domain 1, MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, SART3 - squamous cell carcinoma antigen recognized by t cells 3, MAD1L1 - mad1 mitotic arrest deficient-like 1 (yeast), CTSC - cathepsin c, RANBP1 - ran binding protein 1, RAP1A - rap1a, member of ras oncogene family, PARVG - parvin, gamma, PUS1 - pseudouridylate synthase 1, NSMAF - neutral sphingomyelinase (n-smase) activation associated factor, AKAP8L - a kinase (prka) anchor protein 8-like, GNPAT - glyceronephosphate o-acyltransferase, ZNF592 - zinc finger protein 592, CYFIP2 - cytoplasmic fmr1 interacting protein 2, RBM26 - rna binding motif protein 26, CLNS1A - chloride channel, nucleotide-sensitive, 1a, RAP1B - rap1b, member of ras oncogene family, GOLPH3 - golgi phosphoprotein 3 (coat-protein), TMX3 - thioredoxin-related transmembrane protein 3, BZW1 - basic leucine zipper and w2 domains 1, RB1 - retinoblastoma 1, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), MRPS14 - mitochondrial ribosomal protein s14, CUL1 - cullin 1, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3]
90 [MRPS35 - mitochondrial ribosomal protein s35, MRPL2 - mitochondrial ribosomal protein l2, EIF2D - eukaryotic translation initiation factor 2d, MRPL1 - mitochondrial ribosomal protein l1, RPL23A - ribosomal protein l23a, MRPL13 - mitochondrial ribosomal protein l13, MRPL9 - mitochondrial ribosomal protein l9, RPL22 - ribosomal protein l22, RPL14 - ribosomal protein l14, RPL13 - ribosomal protein l13, RPL7L1 - ribosomal protein l7-like 1, MRPS14 - mitochondrial ribosomal protein s14, MRPL47 - mitochondrial ribosomal protein l47, RPS8 - ribosomal protein s8, RPL6 - ribosomal protein l6, MRPL42 - mitochondrial ribosomal protein l42, RPL7 - ribosomal protein l7, RPS13 - ribosomal protein s13, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, RPS6 - ribosomal protein s6, RPL3 - ribosomal protein l3, RPS7 - ribosomal protein s7, MRPL3 - mitochondrial ribosomal protein l3]
91 [EIF4H - eukaryotic translation initiation factor 4h, EIF4B - eukaryotic translation initiation factor 4b, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, RPL14 - ribosomal protein l14, RPL13 - ribosomal protein l13, EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, RPS8 - ribosomal protein s8, EIF1AD - eukaryotic translation initiation factor 1a domain containing, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, LARP1 - la ribonucleoprotein domain family, member 1, RPS13 - ribosomal protein s13, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, EIF4G3 - eukaryotic translation initiation factor 4 gamma, 3, RPL31 - ribosomal protein l31, EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, EIF3E - eukaryotic translation initiation factor 3, subunit e, RPL32 - ribosomal protein l32, RPS6 - ribosomal protein s6, RPL3 - ribosomal protein l3, RPS7 - ribosomal protein s7, EIF3F - eukaryotic translation initiation factor 3, subunit f, EIF2S2 - eukaryotic translation initiation factor 2, subunit 2 beta, 38kda]
92 [UQCRB - ubiquinol-cytochrome c reductase binding protein, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, MRPL13 - mitochondrial ribosomal protein l13, MCU - mitochondrial calcium uniporter, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, TMEM70 - transmembrane protein 70, TMEM14C - transmembrane protein 14c, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), GNPAT - glyceronephosphate o-acyltransferase, AGK - acylglycerol kinase, CISD1 - cdgsh iron sulfur domain 1, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, MFN1 - mitofusin 1, NDUFA5 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, PTCD3 - pentatricopeptide repeat domain 3, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), MRPL42 - mitochondrial ribosomal protein l42, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, FECH - ferrochelatase, CYCS - cytochrome c, somatic, MRPS35 - mitochondrial ribosomal protein s35, MRPL2 - mitochondrial ribosomal protein l2, MRPL1 - mitochondrial ribosomal protein l1, CPOX - coproporphyrinogen oxidase, MRPL9 - mitochondrial ribosomal protein l9, HK2 - hexokinase 2, CISD2 - cdgsh iron sulfur domain 2, HSPD1 - heat shock 60kda protein 1 (chaperonin), NDUFAF4 - nadh dehydrogenase (ubiquinone) complex i, assembly factor 4, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, SAMM50 - samm50 sorting and assembly machinery component, MRPS14 - mitochondrial ribosomal protein s14, MRPL47 - mitochondrial ribosomal protein l47, RHOT1 - ras homolog family member t1, SORD - sorbitol dehydrogenase, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, BCL2L1 - bcl2-like 1, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, VDAC2 - voltage-dependent anion channel 2, MRPL3 - mitochondrial ribosomal protein l3, LYRM7 - lyr motif containing 7]
93 [PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, UQCRB - ubiquinol-cytochrome c reductase binding protein, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, DLG1 - discs, large homolog 1 (drosophila), PGM1 - phosphoglucomutase 1, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, PGAM1 - phosphoglycerate mutase 1 (brain), MFN1 - mitofusin 1, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, DGUOK - deoxyguanosine kinase, DCK - deoxycytidine kinase, UCK2 - uridine-cytidine kinase 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, UMPS - uridine monophosphate synthetase, ADPGK - adp-dependent glucokinase, ACAT1 - acetyl-coa acetyltransferase 1, DUT - deoxyuridine triphosphatase, AK4 - adenylate kinase 4, HK2 - hexokinase 2, ACOT7 - acyl-coa thioesterase 7, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, GCDH - glutaryl-coa dehydrogenase, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, MVD - mevalonate (diphospho) decarboxylase, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, KYNU - kynureninase, RAB23 - rab23, member ras oncogene family, NAMPT - nicotinamide phosphoribosyltransferase, TJP2 - tight junction protein 2, AK3 - adenylate kinase 3, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, PGM2 - phosphoglucomutase 2, DCAKD - dephospho-coa kinase domain containing, RRM1 - ribonucleotide reductase m1]
94 [PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, UQCRB - ubiquinol-cytochrome c reductase binding protein, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, DLG1 - discs, large homolog 1 (drosophila), PGM1 - phosphoglucomutase 1, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, PGAM1 - phosphoglycerate mutase 1 (brain), MFN1 - mitofusin 1, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, DGUOK - deoxyguanosine kinase, DCK - deoxycytidine kinase, UCK2 - uridine-cytidine kinase 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, UMPS - uridine monophosphate synthetase, ADPGK - adp-dependent glucokinase, ACAT1 - acetyl-coa acetyltransferase 1, DUT - deoxyuridine triphosphatase, HK2 - hexokinase 2, AK4 - adenylate kinase 4, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, GCDH - glutaryl-coa dehydrogenase, ACOT7 - acyl-coa thioesterase 7, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, MVD - mevalonate (diphospho) decarboxylase, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, KYNU - kynureninase, RAB23 - rab23, member ras oncogene family, NAMPT - nicotinamide phosphoribosyltransferase, TJP2 - tight junction protein 2, AK3 - adenylate kinase 3, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, PGM2 - phosphoglucomutase 2, DCAKD - dephospho-coa kinase domain containing, RRM1 - ribonucleotide reductase m1]
95 [FAM136A - family with sequence similarity 136, member a, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, SF3A3 - splicing factor 3a, subunit 3, 60kda, GFM2 - g elongation factor, mitochondrial 2, FKBP11 - fk506 binding protein 11, 19 kda, TMED2 - transmembrane emp24 domain trafficking protein 2, HOOK3 - hook microtubule-tethering protein 3, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, ACAT1 - acetyl-coa acetyltransferase 1, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, CPOX - coproporphyrinogen oxidase, LRRC40 - leucine rich repeat containing 40, SUGT1 - sgt1, suppressor of g2 allele of skp1 (s. cerevisiae), GOSR2 - golgi snap receptor complex member 2, OTUB1 - otu domain, ubiquitin aldehyde binding 1, SORD - sorbitol dehydrogenase, PLCG1 - phospholipase c, gamma 1, LIMS1 - lim and senescent cell antigen-like domains 1, MORF4L1 - mortality factor 4 like 1, TTC27 - tetratricopeptide repeat domain 27, CARHSP1 - calcium regulated heat stable protein 1, 24kda, CREB1 - camp responsive element binding protein 1, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, SNX1 - sorting nexin 1, THUMPD1 - thump domain containing 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, SNRPE - small nuclear ribonucleoprotein polypeptide e, ANAPC7 - anaphase promoting complex subunit 7, PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, GLS - glutaminase, GLO1 - glyoxalase i, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), GLG1 - golgi glycoprotein 1, NIP7 - nip7, nucleolar pre-rrna processing protein, GOSR1 - golgi snap receptor complex member 1, COMMD10 - comm domain containing 10, POLR1C - polymerase (rna) i polypeptide c, 30kda, PPA1 - pyrophosphatase (inorganic) 1, SUMO3 - small ubiquitin-like modifier 3, SNX6 - sorting nexin 6, SMARCD1 - swi/snf related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, COMT - catechol-o-methyltransferase, STX8 - syntaxin 8, TAOK3 - tao kinase 3, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), PARP4 - poly (adp-ribose) polymerase family, member 4, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, TMEM30A - transmembrane protein 30a, SCPEP1 - serine carboxypeptidase 1, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, QKI - qki, kh domain containing, rna binding, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, AGK - acylglycerol kinase, HOMER1 - homer homolog 1 (drosophila), FTH1 - ferritin, heavy polypeptide 1, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, PPP5C - protein phosphatase 5, catalytic subunit, USP48 - ubiquitin specific peptidase 48, SURF4 - surfeit 4, IRAK4 - interleukin-1 receptor-associated kinase 4, DNAJC11 - dnaj (hsp40) homolog, subfamily c, member 11, MRPS35 - mitochondrial ribosomal protein s35, AK4 - adenylate kinase 4, TNPO1 - transportin 1, KPNB1 - karyopherin (importin) beta 1, EXOC2 - exocyst complex component 2, EXOC1 - exocyst complex component 1, IFIT5 - interferon-induced protein with tetratricopeptide repeats 5, TJP2 - tight junction protein 2, DCAKD - dephospho-coa kinase domain containing, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, SCYL2 - scy1-like 2 (s. cerevisiae), NIPBL - nipped-b homolog (drosophila), GALNT2 - udp-n-acetyl-alpha-d-galactosamine:polypeptide n-acetylgalactosaminyltransferase 2 (galnac-t2), STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), SNRNP27 - small nuclear ribonucleoprotein 27kda (u4/u6.u5), MFN1 - mitofusin 1, STAT6 - signal transducer and activator of transcription 6, interleukin-4 induced, FCRLA - fc receptor-like a, EIF1AD - eukaryotic translation initiation factor 1a domain containing, RINT1 - rad50 interactor 1, NHP2 - nhp2 ribonucleoprotein, DSTN - destrin (actin depolymerizing factor), RIC8A - ric8 guanine nucleotide exchange factor a, SRP72 - signal recognition particle 72kda, GCDH - glutaryl-coa dehydrogenase, CISD2 - cdgsh iron sulfur domain 2, SSBP1 - single-stranded dna binding protein 1, mitochondrial, SAMM50 - samm50 sorting and assembly machinery component, ANXA4 - annexin a4, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, TNFAIP8 - tumor necrosis factor, alpha-induced protein 8, PRKCD - protein kinase c, delta, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, CCDC6 - coiled-coil domain containing 6, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, DMXL1 - dmx-like 1, ERAP1 - endoplasmic reticulum aminopeptidase 1, DDX6 - dead (asp-glu-ala-asp) box helicase 6, SDAD1 - sda1 domain containing 1, ZMYM2 - zinc finger, mym-type 2, GSPT1 - g1 to s phase transition 1, GSN - gelsolin, FAM98A - family with sequence similarity 98, member a, ECI1 - enoyl-coa delta isomerase 1, DCK - deoxycytidine kinase, DBN1 - drebrin 1, TRIM25 - tripartite motif containing 25, XPOT - exportin, trna, RIF1 - rap1 interacting factor homolog (yeast), VTI1A - vesicle transport through interaction with t-snares 1a, CCDC51 - coiled-coil domain containing 51, MMS22L - mms22-like, dna repair protein, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), ARCN1 - archain 1, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, LRRK1 - leucine-rich repeat kinase 1, PA2G4 - proliferation-associated 2g4, 38kda, CAB39 - calcium binding protein 39, GATAD2B - gata zinc finger domain containing 2b, KIAA2013 - kiaa2013, RAB23 - rab23, member ras oncogene family, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, MRPL3 - mitochondrial ribosomal protein l3, LRRC57 - leucine rich repeat containing 57, RNF31 - ring finger protein 31, MCM7 - minichromosome maintenance complex component 7, MCU - mitochondrial calcium uniporter, ARL1 - adp-ribosylation factor-like 1, KDELC2 - kdel (lys-asp-glu-leu) containing 2, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, PELO - pelota homolog (drosophila), SCO1 - sco1 cytochrome c oxidase assembly protein, WDHD1 - wd repeat and hmg-box dna binding protein 1, HADH - hydroxyacyl-coa dehydrogenase, PPIL3 - peptidylprolyl isomerase (cyclophilin)-like 3, OSBPL3 - oxysterol binding protein-like 3, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, RPL7L1 - ribosomal protein l7-like 1, C15orf40 - chromosome 15 open reading frame 40, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, STX18 - syntaxin 18, NEMF - nuclear export mediator factor, RPTOR - regulatory associated protein of mtor, complex 1, QSOX2 - quiescin q6 sulfhydryl oxidase 2, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, LYRM7 - lyr motif containing 7, VTA1 - vesicle (multivesicular body) trafficking 1, SERBP1 - serpine1 mrna binding protein 1, DENND3 - denn/madd domain containing 3, GAPVD1 - gtpase activating protein and vps9 domains 1, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, MAP4 - microtubule-associated protein 4, RAB14 - rab14, member ras oncogene family, PHKB - phosphorylase kinase, beta, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, MANBA - mannosidase, beta a, lysosomal, MAN2A1 - mannosidase, alpha, class 2a, member 1, USP8 - ubiquitin specific peptidase 8, EIF4H - eukaryotic translation initiation factor 4h, MBNL1 - muscleblind-like splicing regulator 1, METTL15 - methyltransferase like 15, SLAMF1 - signaling lymphocytic activation molecule family member 1, DUT - deoxyuridine triphosphatase, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, MAT2A - methionine adenosyltransferase ii, alpha, MCM3 - minichromosome maintenance complex component 3, CHMP5 - charged multivesicular body protein 5, DOCK7 - dedicator of cytokinesis 7, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, WIPF1 - was/wasl interacting protein family, member 1, SCAF8 - sr-related ctd-associated factor 8, UBA6 - ubiquitin-like modifier activating enzyme 6, GRK6 - g protein-coupled receptor kinase 6, LSS - lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase), DLG1 - discs, large homolog 1 (drosophila), PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, RPL14 - ribosomal protein l14, DHRS7 - dehydrogenase/reductase (sdr family) member 7, NUB1 - negative regulator of ubiquitin-like proteins 1, TRAPPC9 - trafficking protein particle complex 9, DGUOK - deoxyguanosine kinase, UBA3 - ubiquitin-like modifier activating enzyme 3, DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, ACOT7 - acyl-coa thioesterase 7, PDAP1 - pdgfa associated protein 1, CBX3 - chromobox homolog 3, ADI1 - acireductone dioxygenase 1, FAM49B - family with sequence similarity 49, member b, CAPRIN1 - cell cycle associated protein 1, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), RHOT1 - ras homolog family member t1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, RSL1D1 - ribosomal l1 domain containing 1, BCL2L1 - bcl2-like 1, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, PGM2 - phosphoglucomutase 2, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1, CCNK - cyclin k, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, DST - dystonin, NCBP2 - nuclear cap binding protein subunit 2, 20kda, EXOSC5 - exosome component 5, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, RFC3 - replication factor c (activator 1) 3, 38kda, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, SACM1L - sac1 suppressor of actin mutations 1-like (yeast), NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, NDUFA5 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, RECQL - recq protein-like (dna helicase q1-like), DERL1 - derlin 1, BTF3 - basic transcription factor 3, DOCK8 - dedicator of cytokinesis 8, SPC24 - spc24, ndc80 kinetochore complex component, EIF4B - eukaryotic translation initiation factor 4b, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, USP25 - ubiquitin specific peptidase 25, ZNF326 - zinc finger protein 326, WDR77 - wd repeat domain 77, RCSD1 - rcsd domain containing 1, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, MYO6 - myosin vi, SGPL1 - sphingosine-1-phosphate lyase 1, EIF2D - eukaryotic translation initiation factor 2d, TPM3 - tropomyosin 3, STK38L - serine/threonine kinase 38 like, SEH1L - seh1-like (s. cerevisiae), EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, CC2D1B - coiled-coil and c2 domain containing 1b, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), DCAF7 - ddb1 and cul4 associated factor 7, EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, ADPGK - adp-dependent glucokinase, EIF2S2 - eukaryotic translation initiation factor 2, subunit 2 beta, 38kda, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, TSFM - ts translation elongation factor, mitochondrial, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), MVD - mevalonate (diphospho) decarboxylase, DENND4A - denn/madd domain containing 4a, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, KYNU - kynureninase, ERP44 - endoplasmic reticulum protein 44, NAMPT - nicotinamide phosphoribosyltransferase, HSPA4 - heat shock 70kda protein 4, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, UQCRB - ubiquinol-cytochrome c reductase binding protein, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, CAPN2 - calpain 2, (m/ii) large subunit, WNK1 - wnk lysine deficient protein kinase 1, ZWILCH - zwilch kinetochore protein, OSTF1 - osteoclast stimulating factor 1, UBE2Z - ubiquitin-conjugating enzyme e2z, HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PTCD3 - pentatricopeptide repeat domain 3, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), TBC1D2B - tbc1 domain family, member 2b, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, EIF4G3 - eukaryotic translation initiation factor 4 gamma, 3, UCK2 - uridine-cytidine kinase 2, UMPS - uridine monophosphate synthetase, RPL3 - ribosomal protein l3, CARS - cysteinyl-trna synthetase, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), EIF3F - eukaryotic translation initiation factor 3, subunit f, MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, RPN1 - ribophorin i, HK2 - hexokinase 2, STX17 - syntaxin 17, TRIM38 - tripartite motif containing 38, ABCF2 - atp-binding cassette, sub-family f (gcn20), member 2, CDC23 - cell division cycle 23, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, VDAC2 - voltage-dependent anion channel 2, TTLL12 - tubulin tyrosine ligase-like family, member 12, MRPL13 - mitochondrial ribosomal protein l13, GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, TMEM70 - transmembrane protein 70, RNMT - rna (guanine-7-) methyltransferase, WDR1 - wd repeat domain 1, LARP4B - la ribonucleoprotein domain family, member 4b, RPS8 - ribosomal protein s8, ESD - esterase d, MRPL42 - mitochondrial ribosomal protein l42, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, STOM - stomatin, UCHL3 - ubiquitin carboxyl-terminal esterase l3 (ubiquitin thiolesterase), SUMO1 - small ubiquitin-like modifier 1, CAT - catalase, USP15 - ubiquitin specific peptidase 15, XPO6 - exportin 6, UGDH - udp-glucose 6-dehydrogenase, NDUFAF4 - nadh dehydrogenase (ubiquinone) complex i, assembly factor 4, MRPL47 - mitochondrial ribosomal protein l47, TUBGCP3 - tubulin, gamma complex associated protein 3, UBE2I - ubiquitin-conjugating enzyme e2i, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, AIDA - axin interactor, dorsalization associated, RRBP1 - ribosome binding protein 1, EPS15 - epidermal growth factor receptor pathway substrate 15, RRM1 - ribonucleotide reductase m1, ZC3H7B - zinc finger ccch-type containing 7b, DDX31 - dead (asp-glu-ala-asp) box polypeptide 31, TCOF1 - treacher collins-franceschetti syndrome 1, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, ZNF638 - zinc finger protein 638, F11R - f11 receptor, STAMBP - stam binding protein, SCFD1 - sec1 family domain containing 1, CISD1 - cdgsh iron sulfur domain 1, URB1 - urb1 ribosome biogenesis 1 homolog (s. cerevisiae), RAB30 - rab30, member ras oncogene family, FLNB - filamin b, beta, PPFIA1 - protein tyrosine phosphatase, receptor type, f polypeptide (ptprf), interacting protein (liprin), alpha 1, PPP4R2 - protein phosphatase 4, regulatory subunit 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, PPFIBP1 - ptprf interacting protein, binding protein 1 (liprin beta 1), ILF3 - interleukin enhancer binding factor 3, 90kda, ANKRD28 - ankyrin repeat domain 28, NUDCD2 - nudc domain containing 2, CYCS - cytochrome c, somatic, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, RBM19 - rna binding motif protein 19, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, EHBP1 - eh domain binding protein 1, CD48 - cd48 molecule, AGPS - alkylglycerone phosphate synthase, TBC1D4 - tbc1 domain family, member 4, TMED5 - transmembrane emp24 protein transport domain containing 5, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, GAR1 - gar1 ribonucleoprotein, KDM3A - lysine (k)-specific demethylase 3a, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, ANAPC1 - anaphase promoting complex subunit 1, TELO2 - telomere maintenance 2, EXOSC1 - exosome component 1, PITRM1 - pitrilysin metallopeptidase 1, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), ATG7 - autophagy related 7, TUBGCP4 - tubulin, gamma complex associated protein 4, HDDC2 - hd domain containing 2, PRRC1 - proline-rich coiled-coil 1, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, SYNE1 - spectrin repeat containing, nuclear envelope 1, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), NUP88 - nucleoporin 88kda, ZBTB8OS - zinc finger and btb domain containing 8 opposite strand, NOM1 - nucleolar protein with mif4g domain 1, NFU1 - nfu1 iron-sulfur cluster scaffold homolog (s. cerevisiae), MBLAC2 - metallo-beta-lactamase domain containing 2, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), THOC5 - tho complex 5, FECH - ferrochelatase, CDK6 - cyclin-dependent kinase 6, ADNP - activity-dependent neuroprotector homeobox, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, CDK13 - cyclin-dependent kinase 13, TBCD - tubulin folding cofactor d, FCF1 - fcf1 rrna-processing protein, GNPDA2 - glucosamine-6-phosphate deaminase 2, CCT4 - chaperonin containing tcp1, subunit 4 (delta), TBCA - tubulin folding cofactor a, CUTC - cutc copper transporter, INPP5B - inositol polyphosphate-5-phosphatase, 75kda, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, IRF3 - interferon regulatory factor 3, ARL6IP5 - adp-ribosylation-like factor 6 interacting protein 5, LARP1 - la ribonucleoprotein domain family, member 1, IRF5 - interferon regulatory factor 5, DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), EIF3E - eukaryotic translation initiation factor 3, subunit e, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, TCEA1 - transcription elongation factor a (sii), 1, PURA - purine-rich element binding protein a, SPG11 - spastic paraplegia 11 (autosomal recessive), TDRD7 - tudor domain containing 7, NEK6 - nima-related kinase 6, NOC3L - nucleolar complex associated 3 homolog (s. cerevisiae), PYGB - phosphorylase, glycogen; brain, PCYT2 - phosphate cytidylyltransferase 2, ethanolamine, NUP50 - nucleoporin 50kda, VKORC1L1 - vitamin k epoxide reductase complex, subunit 1-like 1, TRAPPC10 - trafficking protein particle complex 10, CCDC58 - coiled-coil domain containing 58, IDE - insulin-degrading enzyme, USP34 - ubiquitin specific peptidase 34, IFIH1 - interferon induced with helicase c domain 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, SART3 - squamous cell carcinoma antigen recognized by t cells 3, MAD1L1 - mad1 mitotic arrest deficient-like 1 (yeast), CTSC - cathepsin c, RANBP1 - ran binding protein 1, RAP1A - rap1a, member of ras oncogene family, PARVG - parvin, gamma, PUS1 - pseudouridylate synthase 1, NSMAF - neutral sphingomyelinase (n-smase) activation associated factor, AKAP8L - a kinase (prka) anchor protein 8-like, GNPAT - glyceronephosphate o-acyltransferase, ZNF592 - zinc finger protein 592, CYFIP2 - cytoplasmic fmr1 interacting protein 2, RBM26 - rna binding motif protein 26, CLNS1A - chloride channel, nucleotide-sensitive, 1a, RAP1B - rap1b, member of ras oncogene family, GOLPH3 - golgi phosphoprotein 3 (coat-protein), TMX3 - thioredoxin-related transmembrane protein 3, BZW1 - basic leucine zipper and w2 domains 1, RB1 - retinoblastoma 1, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), MRPS14 - mitochondrial ribosomal protein s14, CUL1 - cullin 1, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3]
96 [AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, DENND3 - denn/madd domain containing 3, ERAP1 - endoplasmic reticulum aminopeptidase 1, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, NCBP2 - nuclear cap binding protein subunit 2, 20kda, DDX6 - dead (asp-glu-ala-asp) box helicase 6, EXOSC5 - exosome component 5, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), GSPT1 - g1 to s phase transition 1, UBE2Z - ubiquitin-conjugating enzyme e2z, HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, PYGB - phosphorylase, glycogen; brain, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, PHKB - phosphorylase kinase, beta, ECI1 - enoyl-coa delta isomerase 1, RPL3 - ribosomal protein l3, USP48 - ubiquitin specific peptidase 48, MANBA - mannosidase, beta a, lysosomal, DERL1 - derlin 1, TRIM25 - tripartite motif containing 25, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), USP8 - ubiquitin specific peptidase 8, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, ACAT1 - acetyl-coa acetyltransferase 1, DUT - deoxyuridine triphosphatase, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, HK2 - hexokinase 2, KPNB1 - karyopherin (importin) beta 1, TRIM38 - tripartite motif containing 38, IDE - insulin-degrading enzyme, USP25 - ubiquitin specific peptidase 25, SORD - sorbitol dehydrogenase, PLCG1 - phospholipase c, gamma 1, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, CDC23 - cell division cycle 23, USP34 - ubiquitin specific peptidase 34, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, ANAPC1 - anaphase promoting complex subunit 1, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, EXOSC1 - exosome component 1, UBA6 - ubiquitin-like modifier activating enzyme 6, SGPL1 - sphingosine-1-phosphate lyase 1, ATG7 - autophagy related 7, ANAPC7 - anaphase promoting complex subunit 7, RPL14 - ribosomal protein l14, GLS - glutaminase, NUB1 - negative regulator of ubiquitin-like proteins 1, PELO - pelota homolog (drosophila), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), RPS8 - ribosomal protein s8, ESD - esterase d, HADH - hydroxyacyl-coa dehydrogenase, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, ADPGK - adp-dependent glucokinase, RPS7 - ribosomal protein s7, UCHL3 - ubiquitin carboxyl-terminal esterase l3 (ubiquitin thiolesterase), HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, ACOT7 - acyl-coa thioesterase 7, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, GCDH - glutaryl-coa dehydrogenase, MUT - methylmalonyl coa mutase, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, USP15 - ubiquitin specific peptidase 15, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, GNPDA2 - glucosamine-6-phosphate deaminase 2, INPP5B - inositol polyphosphate-5-phosphatase, 75kda, COMT - catechol-o-methyltransferase, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, KYNU - kynureninase, UBE2I - ubiquitin-conjugating enzyme e2i, CUL1 - cullin 1, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), NEMF - nuclear export mediator factor, EIF3E - eukaryotic translation initiation factor 3, subunit e, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), PGM2 - phosphoglucomutase 2, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1]
97 [VTA1 - vesicle (multivesicular body) trafficking 1, EIF2D - eukaryotic translation initiation factor 2d, ATG7 - autophagy related 7, MRPL13 - mitochondrial ribosomal protein l13, CAPN2 - calpain 2, (m/ii) large subunit, NEK6 - nima-related kinase 6, GSPT1 - g1 to s phase transition 1, GSN - gelsolin, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, AKAP8L - a kinase (prka) anchor protein 8-like, GFM2 - g elongation factor, mitochondrial 2, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), PTCD3 - pentatricopeptide repeat domain 3, PELO - pelota homolog (drosophila), WDR1 - wd repeat domain 1, MRPL42 - mitochondrial ribosomal protein l42, DSTN - destrin (actin depolymerizing factor), MRPS35 - mitochondrial ribosomal protein s35, MRPL2 - mitochondrial ribosomal protein l2, MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, STX17 - syntaxin 17, SMARCD1 - swi/snf related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1, CISD2 - cdgsh iron sulfur domain 2, MRPL47 - mitochondrial ribosomal protein l47, MRPS14 - mitochondrial ribosomal protein s14, CHMP5 - charged multivesicular body protein 5, MRPL3 - mitochondrial ribosomal protein l3]
98 [MRPS35 - mitochondrial ribosomal protein s35, MRPL2 - mitochondrial ribosomal protein l2, MRPL1 - mitochondrial ribosomal protein l1, RPL23A - ribosomal protein l23a, MRPL13 - mitochondrial ribosomal protein l13, MRPL9 - mitochondrial ribosomal protein l9, RPL22 - ribosomal protein l22, RPL14 - ribosomal protein l14, RPL13 - ribosomal protein l13, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, RPL7L1 - ribosomal protein l7-like 1, MRPL47 - mitochondrial ribosomal protein l47, MRPS14 - mitochondrial ribosomal protein s14, RPS8 - ribosomal protein s8, RPL6 - ribosomal protein l6, MRPL42 - mitochondrial ribosomal protein l42, RPL7 - ribosomal protein l7, RPS13 - ribosomal protein s13, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, RPS6 - ribosomal protein s6, RPL3 - ribosomal protein l3, RPS7 - ribosomal protein s7, MRPL3 - mitochondrial ribosomal protein l3]
99 [CCNK - cyclin k, DST - dystonin, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, NCBP2 - nuclear cap binding protein subunit 2, 20kda, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, RFC3 - replication factor c (activator 1) 3, 38kda, SF3A3 - splicing factor 3a, subunit 3, 60kda, SACM1L - sac1 suppressor of actin mutations 1-like (yeast), SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, GFM2 - g elongation factor, mitochondrial 2, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, TMED2 - transmembrane emp24 domain trafficking protein 2, RECQL - recq protein-like (dna helicase q1-like), HOOK3 - hook microtubule-tethering protein 3, DERL1 - derlin 1, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, BTF3 - basic transcription factor 3, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, ACAT1 - acetyl-coa acetyltransferase 1, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, CPOX - coproporphyrinogen oxidase, LRRC40 - leucine rich repeat containing 40, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), SUGT1 - sgt1, suppressor of g2 allele of skp1 (s. cerevisiae), GOSR2 - golgi snap receptor complex member 2, USP25 - ubiquitin specific peptidase 25, OTUB1 - otu domain, ubiquitin aldehyde binding 1, SORD - sorbitol dehydrogenase, ZNF326 - zinc finger protein 326, WDR77 - wd repeat domain 77, CREB1 - camp responsive element binding protein 1, MYO6 - myosin vi, SNX1 - sorting nexin 1, SGPL1 - sphingosine-1-phosphate lyase 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, TPM3 - tropomyosin 3, SNRPE - small nuclear ribonucleoprotein polypeptide e, ANAPC7 - anaphase promoting complex subunit 7, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, GLS - glutaminase, IARS - isoleucyl-trna synthetase, GLO1 - glyoxalase i, CC2D1B - coiled-coil and c2 domain containing 1b, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), GLG1 - golgi glycoprotein 1, GOSR1 - golgi snap receptor complex member 1, DCAF7 - ddb1 and cul4 associated factor 7, ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, ADPGK - adp-dependent glucokinase, PPA1 - pyrophosphatase (inorganic) 1, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, SUMO3 - small ubiquitin-like modifier 3, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, SNX6 - sorting nexin 6, TSFM - ts translation elongation factor, mitochondrial, SMARCD1 - swi/snf related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, DENND4A - denn/madd domain containing 4a, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, COMT - catechol-o-methyltransferase, KYNU - kynureninase, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, STX8 - syntaxin 8, ERP44 - endoplasmic reticulum protein 44, NAMPT - nicotinamide phosphoribosyltransferase, HSPA4 - heat shock 70kda protein 4, PARP4 - poly (adp-ribose) polymerase family, member 4, TMEM30A - transmembrane protein 30a, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, SCPEP1 - serine carboxypeptidase 1, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, QKI - qki, kh domain containing, rna binding, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, CAPN2 - calpain 2, (m/ii) large subunit, UBE2Z - ubiquitin-conjugating enzyme e2z, HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, AGK - acylglycerol kinase, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PTCD3 - pentatricopeptide repeat domain 3, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, FTH1 - ferritin, heavy polypeptide 1, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, UMPS - uridine monophosphate synthetase, USP48 - ubiquitin specific peptidase 48, PPP5C - protein phosphatase 5, catalytic subunit, RPL3 - ribosomal protein l3, SURF4 - surfeit 4, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), IRAK4 - interleukin-1 receptor-associated kinase 4, DNAJC11 - dnaj (hsp40) homolog, subfamily c, member 11, MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, RPN1 - ribophorin i, TNPO1 - transportin 1, AK4 - adenylate kinase 4, HK2 - hexokinase 2, STX17 - syntaxin 17, KPNB1 - karyopherin (importin) beta 1, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, TJP2 - tight junction protein 2, RPL31 - ribosomal protein l31, VDAC2 - voltage-dependent anion channel 2, TTLL12 - tubulin tyrosine ligase-like family, member 12, MRPL13 - mitochondrial ribosomal protein l13, NIPBL - nipped-b homolog (drosophila), SCYL2 - scy1-like 2 (s. cerevisiae), GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, GALNT2 - udp-n-acetyl-alpha-d-galactosamine:polypeptide n-acetylgalactosaminyltransferase 2 (galnac-t2), TMEM70 - transmembrane protein 70, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), MFN1 - mitofusin 1, RNMT - rna (guanine-7-) methyltransferase, WDR1 - wd repeat domain 1, EIF1AD - eukaryotic translation initiation factor 1a domain containing, RPS8 - ribosomal protein s8, MRPL42 - mitochondrial ribosomal protein l42, ESD - esterase d, RPS13 - ribosomal protein s13, RINT1 - rad50 interactor 1, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, DSTN - destrin (actin depolymerizing factor), STOM - stomatin, SUMO1 - small ubiquitin-like modifier 1, SRP72 - signal recognition particle 72kda, GCDH - glutaryl-coa dehydrogenase, CISD2 - cdgsh iron sulfur domain 2, CAT - catalase, USP15 - ubiquitin specific peptidase 15, XPO6 - exportin 6, UGDH - udp-glucose 6-dehydrogenase, NDUFAF4 - nadh dehydrogenase (ubiquinone) complex i, assembly factor 4, SSBP1 - single-stranded dna binding protein 1, mitochondrial, SAMM50 - samm50 sorting and assembly machinery component, MRPL47 - mitochondrial ribosomal protein l47, ANXA4 - annexin a4, UBE2I - ubiquitin-conjugating enzyme e2i, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, PRKCD - protein kinase c, delta, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, RRBP1 - ribosome binding protein 1, EPS15 - epidermal growth factor receptor pathway substrate 15, ZC3H7B - zinc finger ccch-type containing 7b, DDX31 - dead (asp-glu-ala-asp) box polypeptide 31, ERAP1 - endoplasmic reticulum aminopeptidase 1, DDX6 - dead (asp-glu-ala-asp) box helicase 6, ZNF638 - zinc finger protein 638, F11R - f11 receptor, STAMBP - stam binding protein, SCFD1 - sec1 family domain containing 1, GSN - gelsolin, CISD1 - cdgsh iron sulfur domain 1, RAB30 - rab30, member ras oncogene family, FLNB - filamin b, beta, ECI1 - enoyl-coa delta isomerase 1, DCK - deoxycytidine kinase, PPP4R2 - protein phosphatase 4, regulatory subunit 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, ILF3 - interleukin enhancer binding factor 3, 90kda, RIF1 - rap1 interacting factor homolog (yeast), PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, CYCS - cytochrome c, somatic, VTI1A - vesicle transport through interaction with t-snares 1a, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, EHBP1 - eh domain binding protein 1, ARCN1 - archain 1, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, CD48 - cd48 molecule, LRRK1 - leucine-rich repeat kinase 1, PA2G4 - proliferation-associated 2g4, 38kda, AGPS - alkylglycerone phosphate synthase, CAB39 - calcium binding protein 39, TBC1D4 - tbc1 domain family, member 4, TMED5 - transmembrane emp24 protein transport domain containing 5, RAB23 - rab23, member ras oncogene family, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, KDM3A - lysine (k)-specific demethylase 3a, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, MRPL3 - mitochondrial ribosomal protein l3, TELO2 - telomere maintenance 2, LRRC57 - leucine rich repeat containing 57, PITRM1 - pitrilysin metallopeptidase 1, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), MCM7 - minichromosome maintenance complex component 7, MCU - mitochondrial calcium uniporter, ARL1 - adp-ribosylation factor-like 1, TUBGCP4 - tubulin, gamma complex associated protein 4, PRRC1 - proline-rich coiled-coil 1, SYNE1 - spectrin repeat containing, nuclear envelope 1, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), PELO - pelota homolog (drosophila), SCO1 - sco1 cytochrome c oxidase assembly protein, NFU1 - nfu1 iron-sulfur cluster scaffold homolog (s. cerevisiae), HADH - hydroxyacyl-coa dehydrogenase, OSBPL3 - oxysterol binding protein-like 3, MBLAC2 - metallo-beta-lactamase domain containing 2, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), THOC5 - tho complex 5, FECH - ferrochelatase, CDK6 - cyclin-dependent kinase 6, ADNP - activity-dependent neuroprotector homeobox, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, CDK13 - cyclin-dependent kinase 13, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, CCT4 - chaperonin containing tcp1, subunit 4 (delta), GNPDA2 - glucosamine-6-phosphate deaminase 2, CUTC - cutc copper transporter, INPP5B - inositol polyphosphate-5-phosphatase, 75kda, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, IRF3 - interferon regulatory factor 3, STX18 - syntaxin 18, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, NEMF - nuclear export mediator factor, IRF5 - interferon regulatory factor 5, RPTOR - regulatory associated protein of mtor, complex 1, EIF3E - eukaryotic translation initiation factor 3, subunit e, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, QSOX2 - quiescin q6 sulfhydryl oxidase 2, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, VTA1 - vesicle (multivesicular body) trafficking 1, SERBP1 - serpine1 mrna binding protein 1, PURA - purine-rich element binding protein a, GAPVD1 - gtpase activating protein and vps9 domains 1, DENND3 - denn/madd domain containing 3, SPG11 - spastic paraplegia 11 (autosomal recessive), ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, PGM1 - phosphoglucomutase 1, NEK6 - nima-related kinase 6, PGAM1 - phosphoglycerate mutase 1 (brain), NOC3L - nucleolar complex associated 3 homolog (s. cerevisiae), PYGB - phosphorylase, glycogen; brain, NUP50 - nucleoporin 50kda, RAB14 - rab14, member ras oncogene family, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, MANBA - mannosidase, beta a, lysosomal, VKORC1L1 - vitamin k epoxide reductase complex, subunit 1-like 1, MAN2A1 - mannosidase, alpha, class 2a, member 1, USP8 - ubiquitin specific peptidase 8, MBNL1 - muscleblind-like splicing regulator 1, CCDC58 - coiled-coil domain containing 58, SLAMF1 - signaling lymphocytic activation molecule family member 1, DUT - deoxyuridine triphosphatase, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, MCM3 - minichromosome maintenance complex component 3, IDE - insulin-degrading enzyme, CHMP5 - charged multivesicular body protein 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, USP34 - ubiquitin specific peptidase 34, IFIH1 - interferon induced with helicase c domain 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, SART3 - squamous cell carcinoma antigen recognized by t cells 3, CTSC - cathepsin c, WIPF1 - was/wasl interacting protein family, member 1, SCAF8 - sr-related ctd-associated factor 8, UBA6 - ubiquitin-like modifier activating enzyme 6, RANBP1 - ran binding protein 1, DLG1 - discs, large homolog 1 (drosophila), RAP1A - rap1a, member of ras oncogene family, PUS1 - pseudouridylate synthase 1, RPL14 - ribosomal protein l14, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, GNPAT - glyceronephosphate o-acyltransferase, AKAP8L - a kinase (prka) anchor protein 8-like, ZNF592 - zinc finger protein 592, CYFIP2 - cytoplasmic fmr1 interacting protein 2, RBM26 - rna binding motif protein 26, DGUOK - deoxyguanosine kinase, TRAPPC9 - trafficking protein particle complex 9, CLNS1A - chloride channel, nucleotide-sensitive, 1a, UBA3 - ubiquitin-like modifier activating enzyme 3, RAP1B - rap1b, member of ras oncogene family, GOLPH3 - golgi phosphoprotein 3 (coat-protein), DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, ACOT7 - acyl-coa thioesterase 7, RB1 - retinoblastoma 1, CBX3 - chromobox homolog 3, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, FAM49B - family with sequence similarity 49, member b, ADI1 - acireductone dioxygenase 1, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), RHOT1 - ras homolog family member t1, BCL2L1 - bcl2-like 1, AK3 - adenylate kinase 3, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, PGM2 - phosphoglucomutase 2, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
100 [PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, UQCRB - ubiquinol-cytochrome c reductase binding protein, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, DLG1 - discs, large homolog 1 (drosophila), PGM1 - phosphoglucomutase 1, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, PGAM1 - phosphoglycerate mutase 1 (brain), MFN1 - mitofusin 1, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, DGUOK - deoxyguanosine kinase, DCK - deoxycytidine kinase, UCK2 - uridine-cytidine kinase 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, UMPS - uridine monophosphate synthetase, ADPGK - adp-dependent glucokinase, ACAT1 - acetyl-coa acetyltransferase 1, DUT - deoxyuridine triphosphatase, AK4 - adenylate kinase 4, HK2 - hexokinase 2, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, GCDH - glutaryl-coa dehydrogenase, ACOT7 - acyl-coa thioesterase 7, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, UGDH - udp-glucose 6-dehydrogenase, GNPDA2 - glucosamine-6-phosphate deaminase 2, MVD - mevalonate (diphospho) decarboxylase, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, KYNU - kynureninase, RAB23 - rab23, member ras oncogene family, NAMPT - nicotinamide phosphoribosyltransferase, TJP2 - tight junction protein 2, AK3 - adenylate kinase 3, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, PGM2 - phosphoglucomutase 2, DCAKD - dephospho-coa kinase domain containing, RRM1 - ribonucleotide reductase m1]
101 [UQCRB - ubiquinol-cytochrome c reductase binding protein, ACAT1 - acetyl-coa acetyltransferase 1, DLG1 - discs, large homolog 1 (drosophila), PGM1 - phosphoglucomutase 1, HK2 - hexokinase 2, AK4 - adenylate kinase 4, GCDH - glutaryl-coa dehydrogenase, ACOT7 - acyl-coa thioesterase 7, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, PGAM1 - phosphoglycerate mutase 1 (brain), NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, MVD - mevalonate (diphospho) decarboxylase, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, MFN1 - mitofusin 1, KYNU - kynureninase, DGUOK - deoxyguanosine kinase, RAB23 - rab23, member ras oncogene family, TJP2 - tight junction protein 2, AK3 - adenylate kinase 3, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, DCAKD - dephospho-coa kinase domain containing, ADPGK - adp-dependent glucokinase]
102 [GFPT1 - glutamine--fructose-6-phosphate transaminase 1, SF3A3 - splicing factor 3a, subunit 3, 60kda, GFM2 - g elongation factor, mitochondrial 2, FKBP11 - fk506 binding protein 11, 19 kda, TMED2 - transmembrane emp24 domain trafficking protein 2, HOOK3 - hook microtubule-tethering protein 3, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, ACAT1 - acetyl-coa acetyltransferase 1, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, CPOX - coproporphyrinogen oxidase, LRRC40 - leucine rich repeat containing 40, SUGT1 - sgt1, suppressor of g2 allele of skp1 (s. cerevisiae), GOSR2 - golgi snap receptor complex member 2, OTUB1 - otu domain, ubiquitin aldehyde binding 1, SORD - sorbitol dehydrogenase, PLCG1 - phospholipase c, gamma 1, LIMS1 - lim and senescent cell antigen-like domains 1, MORF4L1 - mortality factor 4 like 1, TTC27 - tetratricopeptide repeat domain 27, CARHSP1 - calcium regulated heat stable protein 1, 24kda, CREB1 - camp responsive element binding protein 1, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, SNX1 - sorting nexin 1, THUMPD1 - thump domain containing 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, SNRPE - small nuclear ribonucleoprotein polypeptide e, ANAPC7 - anaphase promoting complex subunit 7, PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, GLS - glutaminase, GLO1 - glyoxalase i, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), GLG1 - golgi glycoprotein 1, NIP7 - nip7, nucleolar pre-rrna processing protein, GOSR1 - golgi snap receptor complex member 1, POLR1C - polymerase (rna) i polypeptide c, 30kda, PPA1 - pyrophosphatase (inorganic) 1, SUMO3 - small ubiquitin-like modifier 3, SNX6 - sorting nexin 6, SMARCD1 - swi/snf related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, COMT - catechol-o-methyltransferase, STX8 - syntaxin 8, TAOK3 - tao kinase 3, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), PARP4 - poly (adp-ribose) polymerase family, member 4, TMEM30A - transmembrane protein 30a, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, SCPEP1 - serine carboxypeptidase 1, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, QKI - qki, kh domain containing, rna binding, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, AGK - acylglycerol kinase, HOMER1 - homer homolog 1 (drosophila), FTH1 - ferritin, heavy polypeptide 1, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, PPP5C - protein phosphatase 5, catalytic subunit, USP48 - ubiquitin specific peptidase 48, SURF4 - surfeit 4, IRAK4 - interleukin-1 receptor-associated kinase 4, DNAJC11 - dnaj (hsp40) homolog, subfamily c, member 11, MRPS35 - mitochondrial ribosomal protein s35, AK4 - adenylate kinase 4, TNPO1 - transportin 1, KPNB1 - karyopherin (importin) beta 1, EXOC2 - exocyst complex component 2, EXOC1 - exocyst complex component 1, IFIT5 - interferon-induced protein with tetratricopeptide repeats 5, TJP2 - tight junction protein 2, DCAKD - dephospho-coa kinase domain containing, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, SCYL2 - scy1-like 2 (s. cerevisiae), NIPBL - nipped-b homolog (drosophila), GALNT2 - udp-n-acetyl-alpha-d-galactosamine:polypeptide n-acetylgalactosaminyltransferase 2 (galnac-t2), STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), SNRNP27 - small nuclear ribonucleoprotein 27kda (u4/u6.u5), MFN1 - mitofusin 1, STAT6 - signal transducer and activator of transcription 6, interleukin-4 induced, FCRLA - fc receptor-like a, EIF1AD - eukaryotic translation initiation factor 1a domain containing, RINT1 - rad50 interactor 1, NHP2 - nhp2 ribonucleoprotein, DSTN - destrin (actin depolymerizing factor), RIC8A - ric8 guanine nucleotide exchange factor a, SRP72 - signal recognition particle 72kda, GCDH - glutaryl-coa dehydrogenase, CISD2 - cdgsh iron sulfur domain 2, SSBP1 - single-stranded dna binding protein 1, mitochondrial, SAMM50 - samm50 sorting and assembly machinery component, ANXA4 - annexin a4, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, TNFAIP8 - tumor necrosis factor, alpha-induced protein 8, PRKCD - protein kinase c, delta, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, CCDC6 - coiled-coil domain containing 6, DMXL1 - dmx-like 1, ERAP1 - endoplasmic reticulum aminopeptidase 1, DDX6 - dead (asp-glu-ala-asp) box helicase 6, SDAD1 - sda1 domain containing 1, ZMYM2 - zinc finger, mym-type 2, GSPT1 - g1 to s phase transition 1, GSN - gelsolin, FAM98A - family with sequence similarity 98, member a, ECI1 - enoyl-coa delta isomerase 1, DCK - deoxycytidine kinase, DBN1 - drebrin 1, TRIM25 - tripartite motif containing 25, XPOT - exportin, trna, RIF1 - rap1 interacting factor homolog (yeast), VTI1A - vesicle transport through interaction with t-snares 1a, CCDC51 - coiled-coil domain containing 51, MMS22L - mms22-like, dna repair protein, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), ARCN1 - archain 1, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, PA2G4 - proliferation-associated 2g4, 38kda, LRRK1 - leucine-rich repeat kinase 1, CAB39 - calcium binding protein 39, GATAD2B - gata zinc finger domain containing 2b, RAB23 - rab23, member ras oncogene family, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, MRPL3 - mitochondrial ribosomal protein l3, RNF31 - ring finger protein 31, MCM7 - minichromosome maintenance complex component 7, MCU - mitochondrial calcium uniporter, ARL1 - adp-ribosylation factor-like 1, KDELC2 - kdel (lys-asp-glu-leu) containing 2, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, PELO - pelota homolog (drosophila), SCO1 - sco1 cytochrome c oxidase assembly protein, WDHD1 - wd repeat and hmg-box dna binding protein 1, HADH - hydroxyacyl-coa dehydrogenase, PPIL3 - peptidylprolyl isomerase (cyclophilin)-like 3, OSBPL3 - oxysterol binding protein-like 3, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, RPL7L1 - ribosomal protein l7-like 1, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, STX18 - syntaxin 18, NEMF - nuclear export mediator factor, RPTOR - regulatory associated protein of mtor, complex 1, QSOX2 - quiescin q6 sulfhydryl oxidase 2, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, LYRM7 - lyr motif containing 7, VTA1 - vesicle (multivesicular body) trafficking 1, SERBP1 - serpine1 mrna binding protein 1, DENND3 - denn/madd domain containing 3, GAPVD1 - gtpase activating protein and vps9 domains 1, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, MAP4 - microtubule-associated protein 4, RAB14 - rab14, member ras oncogene family, PHKB - phosphorylase kinase, beta, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, MANBA - mannosidase, beta a, lysosomal, MAN2A1 - mannosidase, alpha, class 2a, member 1, USP8 - ubiquitin specific peptidase 8, EIF4H - eukaryotic translation initiation factor 4h, MBNL1 - muscleblind-like splicing regulator 1, METTL15 - methyltransferase like 15, SLAMF1 - signaling lymphocytic activation molecule family member 1, DUT - deoxyuridine triphosphatase, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, MAT2A - methionine adenosyltransferase ii, alpha, MCM3 - minichromosome maintenance complex component 3, CHMP5 - charged multivesicular body protein 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, DOCK7 - dedicator of cytokinesis 7, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, WIPF1 - was/wasl interacting protein family, member 1, SCAF8 - sr-related ctd-associated factor 8, GRK6 - g protein-coupled receptor kinase 6, UBA6 - ubiquitin-like modifier activating enzyme 6, LSS - lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase), DLG1 - discs, large homolog 1 (drosophila), PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, RPL14 - ribosomal protein l14, DHRS7 - dehydrogenase/reductase (sdr family) member 7, NUB1 - negative regulator of ubiquitin-like proteins 1, DGUOK - deoxyguanosine kinase, TRAPPC9 - trafficking protein particle complex 9, UBA3 - ubiquitin-like modifier activating enzyme 3, DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, ACOT7 - acyl-coa thioesterase 7, PDAP1 - pdgfa associated protein 1, CBX3 - chromobox homolog 3, ADI1 - acireductone dioxygenase 1, FAM49B - family with sequence similarity 49, member b, CAPRIN1 - cell cycle associated protein 1, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), RHOT1 - ras homolog family member t1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, RSL1D1 - ribosomal l1 domain containing 1, BCL2L1 - bcl2-like 1, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, METTL13 - methyltransferase like 13, PGM2 - phosphoglucomutase 2, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1, CCNK - cyclin k, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, DST - dystonin, NCBP2 - nuclear cap binding protein subunit 2, 20kda, EXOSC5 - exosome component 5, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, RFC3 - replication factor c (activator 1) 3, 38kda, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, SACM1L - sac1 suppressor of actin mutations 1-like (yeast), NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, NDUFA5 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, RECQL - recq protein-like (dna helicase q1-like), DERL1 - derlin 1, BTF3 - basic transcription factor 3, DOCK8 - dedicator of cytokinesis 8, SPC24 - spc24, ndc80 kinetochore complex component, EIF4B - eukaryotic translation initiation factor 4b, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, USP25 - ubiquitin specific peptidase 25, ZNF326 - zinc finger protein 326, WDR77 - wd repeat domain 77, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, RCSD1 - rcsd domain containing 1, MYO6 - myosin vi, SGPL1 - sphingosine-1-phosphate lyase 1, EIF2D - eukaryotic translation initiation factor 2d, TPM3 - tropomyosin 3, STK38L - serine/threonine kinase 38 like, SEH1L - seh1-like (s. cerevisiae), EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, CC2D1B - coiled-coil and c2 domain containing 1b, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), DCAF7 - ddb1 and cul4 associated factor 7, EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, ADPGK - adp-dependent glucokinase, ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, EIF2S2 - eukaryotic translation initiation factor 2, subunit 2 beta, 38kda, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, TSFM - ts translation elongation factor, mitochondrial, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), MVD - mevalonate (diphospho) decarboxylase, DENND4A - denn/madd domain containing 4a, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, KYNU - kynureninase, ERP44 - endoplasmic reticulum protein 44, NAMPT - nicotinamide phosphoribosyltransferase, HSPA4 - heat shock 70kda protein 4, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, UQCRB - ubiquinol-cytochrome c reductase binding protein, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, CAPN2 - calpain 2, (m/ii) large subunit, ZWILCH - zwilch kinetochore protein, WNK1 - wnk lysine deficient protein kinase 1, OSTF1 - osteoclast stimulating factor 1, UBE2Z - ubiquitin-conjugating enzyme e2z, HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), PTCD3 - pentatricopeptide repeat domain 3, TBC1D2B - tbc1 domain family, member 2b, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, EIF4G3 - eukaryotic translation initiation factor 4 gamma, 3, UCK2 - uridine-cytidine kinase 2, RPL3 - ribosomal protein l3, UMPS - uridine monophosphate synthetase, CARS - cysteinyl-trna synthetase, EIF3F - eukaryotic translation initiation factor 3, subunit f, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), MRPL1 - mitochondrial ribosomal protein l1, RPN1 - ribophorin i, MRPL9 - mitochondrial ribosomal protein l9, STX17 - syntaxin 17, HK2 - hexokinase 2, TRIM38 - tripartite motif containing 38, CDC23 - cell division cycle 23, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, VDAC2 - voltage-dependent anion channel 2, TTLL12 - tubulin tyrosine ligase-like family, member 12, MRPL13 - mitochondrial ribosomal protein l13, GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, TMEM70 - transmembrane protein 70, RNMT - rna (guanine-7-) methyltransferase, WDR1 - wd repeat domain 1, LARP4B - la ribonucleoprotein domain family, member 4b, RPS8 - ribosomal protein s8, ESD - esterase d, MRPL42 - mitochondrial ribosomal protein l42, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, STOM - stomatin, UCHL3 - ubiquitin carboxyl-terminal esterase l3 (ubiquitin thiolesterase), SUMO1 - small ubiquitin-like modifier 1, CAT - catalase, USP15 - ubiquitin specific peptidase 15, UGDH - udp-glucose 6-dehydrogenase, XPO6 - exportin 6, NDUFAF4 - nadh dehydrogenase (ubiquinone) complex i, assembly factor 4, MRPL47 - mitochondrial ribosomal protein l47, TUBGCP3 - tubulin, gamma complex associated protein 3, UBE2I - ubiquitin-conjugating enzyme e2i, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, AIDA - axin interactor, dorsalization associated, RRBP1 - ribosome binding protein 1, EPS15 - epidermal growth factor receptor pathway substrate 15, RRM1 - ribonucleotide reductase m1, ZC3H7B - zinc finger ccch-type containing 7b, DDX31 - dead (asp-glu-ala-asp) box polypeptide 31, TCOF1 - treacher collins-franceschetti syndrome 1, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, ZNF638 - zinc finger protein 638, F11R - f11 receptor, STAMBP - stam binding protein, SCFD1 - sec1 family domain containing 1, URB1 - urb1 ribosome biogenesis 1 homolog (s. cerevisiae), CISD1 - cdgsh iron sulfur domain 1, RAB30 - rab30, member ras oncogene family, FLNB - filamin b, beta, PPFIA1 - protein tyrosine phosphatase, receptor type, f polypeptide (ptprf), interacting protein (liprin), alpha 1, PPP4R2 - protein phosphatase 4, regulatory subunit 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, PPFIBP1 - ptprf interacting protein, binding protein 1 (liprin beta 1), ILF3 - interleukin enhancer binding factor 3, 90kda, ANKRD28 - ankyrin repeat domain 28, NUDCD2 - nudc domain containing 2, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, CYCS - cytochrome c, somatic, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, RBM19 - rna binding motif protein 19, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, EHBP1 - eh domain binding protein 1, CD48 - cd48 molecule, AGPS - alkylglycerone phosphate synthase, TBC1D4 - tbc1 domain family, member 4, TMED5 - transmembrane emp24 protein transport domain containing 5, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, GAR1 - gar1 ribonucleoprotein, KDM3A - lysine (k)-specific demethylase 3a, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, ANAPC1 - anaphase promoting complex subunit 1, EXOSC1 - exosome component 1, TELO2 - telomere maintenance 2, PITRM1 - pitrilysin metallopeptidase 1, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), ATG7 - autophagy related 7, TUBGCP4 - tubulin, gamma complex associated protein 4, HDDC2 - hd domain containing 2, PRRC1 - proline-rich coiled-coil 1, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, SYNE1 - spectrin repeat containing, nuclear envelope 1, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), NUP88 - nucleoporin 88kda, ZBTB8OS - zinc finger and btb domain containing 8 opposite strand, NOM1 - nucleolar protein with mif4g domain 1, NFU1 - nfu1 iron-sulfur cluster scaffold homolog (s. cerevisiae), MBLAC2 - metallo-beta-lactamase domain containing 2, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), THOC5 - tho complex 5, FECH - ferrochelatase, CDK6 - cyclin-dependent kinase 6, ADNP - activity-dependent neuroprotector homeobox, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, CDK13 - cyclin-dependent kinase 13, TBCD - tubulin folding cofactor d, FCF1 - fcf1 rrna-processing protein, CCT4 - chaperonin containing tcp1, subunit 4 (delta), GNPDA2 - glucosamine-6-phosphate deaminase 2, CUTC - cutc copper transporter, TBCA - tubulin folding cofactor a, INPP5B - inositol polyphosphate-5-phosphatase, 75kda, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, IRF3 - interferon regulatory factor 3, ARL6IP5 - adp-ribosylation-like factor 6 interacting protein 5, LARP1 - la ribonucleoprotein domain family, member 1, IRF5 - interferon regulatory factor 5, DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, EIF3E - eukaryotic translation initiation factor 3, subunit e, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, TCEA1 - transcription elongation factor a (sii), 1, PURA - purine-rich element binding protein a, SPG11 - spastic paraplegia 11 (autosomal recessive), TDRD7 - tudor domain containing 7, NEK6 - nima-related kinase 6, NOC3L - nucleolar complex associated 3 homolog (s. cerevisiae), PYGB - phosphorylase, glycogen; brain, PCYT2 - phosphate cytidylyltransferase 2, ethanolamine, NUP50 - nucleoporin 50kda, VKORC1L1 - vitamin k epoxide reductase complex, subunit 1-like 1, TRAPPC10 - trafficking protein particle complex 10, IDE - insulin-degrading enzyme, USP34 - ubiquitin specific peptidase 34, IFIH1 - interferon induced with helicase c domain 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, SART3 - squamous cell carcinoma antigen recognized by t cells 3, MAD1L1 - mad1 mitotic arrest deficient-like 1 (yeast), CTSC - cathepsin c, RANBP1 - ran binding protein 1, RAP1A - rap1a, member of ras oncogene family, PARVG - parvin, gamma, PUS1 - pseudouridylate synthase 1, NSMAF - neutral sphingomyelinase (n-smase) activation associated factor, GNPAT - glyceronephosphate o-acyltransferase, AKAP8L - a kinase (prka) anchor protein 8-like, CYFIP2 - cytoplasmic fmr1 interacting protein 2, RBM26 - rna binding motif protein 26, CLNS1A - chloride channel, nucleotide-sensitive, 1a, RAP1B - rap1b, member of ras oncogene family, GOLPH3 - golgi phosphoprotein 3 (coat-protein), TMX3 - thioredoxin-related transmembrane protein 3, RB1 - retinoblastoma 1, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), MRPS14 - mitochondrial ribosomal protein s14, CUL1 - cullin 1, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3]
103 [FAM136A - family with sequence similarity 136, member a, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, SF3A3 - splicing factor 3a, subunit 3, 60kda, GFM2 - g elongation factor, mitochondrial 2, TMED2 - transmembrane emp24 domain trafficking protein 2, HOOK3 - hook microtubule-tethering protein 3, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, ACAT1 - acetyl-coa acetyltransferase 1, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, CPOX - coproporphyrinogen oxidase, LRRC40 - leucine rich repeat containing 40, SUGT1 - sgt1, suppressor of g2 allele of skp1 (s. cerevisiae), GOSR2 - golgi snap receptor complex member 2, OTUB1 - otu domain, ubiquitin aldehyde binding 1, SORD - sorbitol dehydrogenase, PLCG1 - phospholipase c, gamma 1, LIMS1 - lim and senescent cell antigen-like domains 1, TTC27 - tetratricopeptide repeat domain 27, MORF4L1 - mortality factor 4 like 1, CARHSP1 - calcium regulated heat stable protein 1, 24kda, CREB1 - camp responsive element binding protein 1, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, SNX1 - sorting nexin 1, THUMPD1 - thump domain containing 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, SNRPE - small nuclear ribonucleoprotein polypeptide e, ANAPC7 - anaphase promoting complex subunit 7, PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, GLS - glutaminase, GLO1 - glyoxalase i, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), GLG1 - golgi glycoprotein 1, NIP7 - nip7, nucleolar pre-rrna processing protein, GOSR1 - golgi snap receptor complex member 1, COMMD10 - comm domain containing 10, POLR1C - polymerase (rna) i polypeptide c, 30kda, PPA1 - pyrophosphatase (inorganic) 1, SUMO3 - small ubiquitin-like modifier 3, SNX6 - sorting nexin 6, SMARCD1 - swi/snf related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, COMT - catechol-o-methyltransferase, STX8 - syntaxin 8, TAOK3 - tao kinase 3, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), PARP4 - poly (adp-ribose) polymerase family, member 4, TMEM30A - transmembrane protein 30a, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, QKI - qki, kh domain containing, rna binding, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, AGK - acylglycerol kinase, HOMER1 - homer homolog 1 (drosophila), FTH1 - ferritin, heavy polypeptide 1, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, PPP5C - protein phosphatase 5, catalytic subunit, USP48 - ubiquitin specific peptidase 48, SURF4 - surfeit 4, IRAK4 - interleukin-1 receptor-associated kinase 4, DNAJC11 - dnaj (hsp40) homolog, subfamily c, member 11, MRPS35 - mitochondrial ribosomal protein s35, AK4 - adenylate kinase 4, TNPO1 - transportin 1, KPNB1 - karyopherin (importin) beta 1, EXOC2 - exocyst complex component 2, EXOC1 - exocyst complex component 1, IFIT5 - interferon-induced protein with tetratricopeptide repeats 5, TJP2 - tight junction protein 2, DCAKD - dephospho-coa kinase domain containing, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, SCYL2 - scy1-like 2 (s. cerevisiae), NIPBL - nipped-b homolog (drosophila), GALNT2 - udp-n-acetyl-alpha-d-galactosamine:polypeptide n-acetylgalactosaminyltransferase 2 (galnac-t2), STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), SNRNP27 - small nuclear ribonucleoprotein 27kda (u4/u6.u5), MFN1 - mitofusin 1, STAT6 - signal transducer and activator of transcription 6, interleukin-4 induced, EIF1AD - eukaryotic translation initiation factor 1a domain containing, RINT1 - rad50 interactor 1, NHP2 - nhp2 ribonucleoprotein, DSTN - destrin (actin depolymerizing factor), RIC8A - ric8 guanine nucleotide exchange factor a, SRP72 - signal recognition particle 72kda, CISD2 - cdgsh iron sulfur domain 2, GCDH - glutaryl-coa dehydrogenase, SSBP1 - single-stranded dna binding protein 1, mitochondrial, SAMM50 - samm50 sorting and assembly machinery component, ANXA4 - annexin a4, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, TNFAIP8 - tumor necrosis factor, alpha-induced protein 8, PRKCD - protein kinase c, delta, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), CCDC6 - coiled-coil domain containing 6, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, ERAP1 - endoplasmic reticulum aminopeptidase 1, DDX6 - dead (asp-glu-ala-asp) box helicase 6, ZMYM2 - zinc finger, mym-type 2, GSPT1 - g1 to s phase transition 1, GSN - gelsolin, FAM98A - family with sequence similarity 98, member a, DCK - deoxycytidine kinase, DBN1 - drebrin 1, TRIM25 - tripartite motif containing 25, XPOT - exportin, trna, RIF1 - rap1 interacting factor homolog (yeast), VTI1A - vesicle transport through interaction with t-snares 1a, CCDC51 - coiled-coil domain containing 51, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), MMS22L - mms22-like, dna repair protein, ARCN1 - archain 1, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, PA2G4 - proliferation-associated 2g4, 38kda, LRRK1 - leucine-rich repeat kinase 1, CAB39 - calcium binding protein 39, GATAD2B - gata zinc finger domain containing 2b, KIAA2013 - kiaa2013, RAB23 - rab23, member ras oncogene family, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, MRPL3 - mitochondrial ribosomal protein l3, RNF31 - ring finger protein 31, MCM7 - minichromosome maintenance complex component 7, MCU - mitochondrial calcium uniporter, ARL1 - adp-ribosylation factor-like 1, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, PELO - pelota homolog (drosophila), SCO1 - sco1 cytochrome c oxidase assembly protein, WDHD1 - wd repeat and hmg-box dna binding protein 1, HADH - hydroxyacyl-coa dehydrogenase, PPIL3 - peptidylprolyl isomerase (cyclophilin)-like 3, OSBPL3 - oxysterol binding protein-like 3, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, RPL7L1 - ribosomal protein l7-like 1, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, STX18 - syntaxin 18, NEMF - nuclear export mediator factor, RPTOR - regulatory associated protein of mtor, complex 1, LYRM7 - lyr motif containing 7, SERBP1 - serpine1 mrna binding protein 1, VTA1 - vesicle (multivesicular body) trafficking 1, DENND3 - denn/madd domain containing 3, GAPVD1 - gtpase activating protein and vps9 domains 1, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, MAP4 - microtubule-associated protein 4, RAB14 - rab14, member ras oncogene family, PHKB - phosphorylase kinase, beta, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, MANBA - mannosidase, beta a, lysosomal, MAN2A1 - mannosidase, alpha, class 2a, member 1, USP8 - ubiquitin specific peptidase 8, EIF4H - eukaryotic translation initiation factor 4h, MBNL1 - muscleblind-like splicing regulator 1, SLAMF1 - signaling lymphocytic activation molecule family member 1, DUT - deoxyuridine triphosphatase, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, MAT2A - methionine adenosyltransferase ii, alpha, MCM3 - minichromosome maintenance complex component 3, CHMP5 - charged multivesicular body protein 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, DOCK7 - dedicator of cytokinesis 7, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, WIPF1 - was/wasl interacting protein family, member 1, SCAF8 - sr-related ctd-associated factor 8, UBA6 - ubiquitin-like modifier activating enzyme 6, GRK6 - g protein-coupled receptor kinase 6, LSS - lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase), DLG1 - discs, large homolog 1 (drosophila), PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, RPL14 - ribosomal protein l14, DHRS7 - dehydrogenase/reductase (sdr family) member 7, LRRC20 - leucine rich repeat containing 20, NUB1 - negative regulator of ubiquitin-like proteins 1, DGUOK - deoxyguanosine kinase, TRAPPC9 - trafficking protein particle complex 9, UBA3 - ubiquitin-like modifier activating enzyme 3, DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, ACOT7 - acyl-coa thioesterase 7, PDAP1 - pdgfa associated protein 1, CBX3 - chromobox homolog 3, ADI1 - acireductone dioxygenase 1, FAM49B - family with sequence similarity 49, member b, CAPRIN1 - cell cycle associated protein 1, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), RHOT1 - ras homolog family member t1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, RSL1D1 - ribosomal l1 domain containing 1, BCL2L1 - bcl2-like 1, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, METTL13 - methyltransferase like 13, PGM2 - phosphoglucomutase 2, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1, CCNK - cyclin k, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, DST - dystonin, NCBP2 - nuclear cap binding protein subunit 2, 20kda, EXOSC5 - exosome component 5, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, RFC3 - replication factor c (activator 1) 3, 38kda, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, SACM1L - sac1 suppressor of actin mutations 1-like (yeast), NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, NDUFA5 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, RECQL - recq protein-like (dna helicase q1-like), DERL1 - derlin 1, BTF3 - basic transcription factor 3, DOCK8 - dedicator of cytokinesis 8, SPC24 - spc24, ndc80 kinetochore complex component, EIF4B - eukaryotic translation initiation factor 4b, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, USP25 - ubiquitin specific peptidase 25, ZNF326 - zinc finger protein 326, WDR77 - wd repeat domain 77, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, RCSD1 - rcsd domain containing 1, MYO6 - myosin vi, SGPL1 - sphingosine-1-phosphate lyase 1, EIF2D - eukaryotic translation initiation factor 2d, TPM3 - tropomyosin 3, STK38L - serine/threonine kinase 38 like, SEH1L - seh1-like (s. cerevisiae), EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, CC2D1B - coiled-coil and c2 domain containing 1b, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), DCAF7 - ddb1 and cul4 associated factor 7, EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, ADPGK - adp-dependent glucokinase, ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, EIF2S2 - eukaryotic translation initiation factor 2, subunit 2 beta, 38kda, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, TSFM - ts translation elongation factor, mitochondrial, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), MVD - mevalonate (diphospho) decarboxylase, DENND4A - denn/madd domain containing 4a, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, KYNU - kynureninase, ERP44 - endoplasmic reticulum protein 44, NAMPT - nicotinamide phosphoribosyltransferase, HSPA4 - heat shock 70kda protein 4, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, UQCRB - ubiquinol-cytochrome c reductase binding protein, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, CAPN2 - calpain 2, (m/ii) large subunit, ZWILCH - zwilch kinetochore protein, WNK1 - wnk lysine deficient protein kinase 1, OSTF1 - osteoclast stimulating factor 1, UBE2Z - ubiquitin-conjugating enzyme e2z, HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PTCD3 - pentatricopeptide repeat domain 3, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), TBC1D2B - tbc1 domain family, member 2b, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, EIF4G3 - eukaryotic translation initiation factor 4 gamma, 3, UCK2 - uridine-cytidine kinase 2, RPL3 - ribosomal protein l3, BTF3L4 - basic transcription factor 3-like 4, CARS - cysteinyl-trna synthetase, EIF3F - eukaryotic translation initiation factor 3, subunit f, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), MRPL1 - mitochondrial ribosomal protein l1, RPN1 - ribophorin i, MRPL9 - mitochondrial ribosomal protein l9, HK2 - hexokinase 2, STX17 - syntaxin 17, TRIM38 - tripartite motif containing 38, CDC23 - cell division cycle 23, ABCF2 - atp-binding cassette, sub-family f (gcn20), member 2, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, VDAC2 - voltage-dependent anion channel 2, TTLL12 - tubulin tyrosine ligase-like family, member 12, MRPL13 - mitochondrial ribosomal protein l13, GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, TMEM70 - transmembrane protein 70, RNMT - rna (guanine-7-) methyltransferase, WDR1 - wd repeat domain 1, RPS8 - ribosomal protein s8, LARP4B - la ribonucleoprotein domain family, member 4b, MRPL42 - mitochondrial ribosomal protein l42, ESD - esterase d, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, STOM - stomatin, UCHL3 - ubiquitin carboxyl-terminal esterase l3 (ubiquitin thiolesterase), SUMO1 - small ubiquitin-like modifier 1, CAT - catalase, USP15 - ubiquitin specific peptidase 15, XPO6 - exportin 6, UGDH - udp-glucose 6-dehydrogenase, NDUFAF4 - nadh dehydrogenase (ubiquinone) complex i, assembly factor 4, MRPL47 - mitochondrial ribosomal protein l47, TUBGCP3 - tubulin, gamma complex associated protein 3, UBE2I - ubiquitin-conjugating enzyme e2i, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, AIDA - axin interactor, dorsalization associated, RRBP1 - ribosome binding protein 1, EPS15 - epidermal growth factor receptor pathway substrate 15, RRM1 - ribonucleotide reductase m1, ZC3H7B - zinc finger ccch-type containing 7b, DDX31 - dead (asp-glu-ala-asp) box polypeptide 31, TCOF1 - treacher collins-franceschetti syndrome 1, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, ZNF638 - zinc finger protein 638, F11R - f11 receptor, STAMBP - stam binding protein, SCFD1 - sec1 family domain containing 1, CISD1 - cdgsh iron sulfur domain 1, URB1 - urb1 ribosome biogenesis 1 homolog (s. cerevisiae), RAB30 - rab30, member ras oncogene family, FLNB - filamin b, beta, PPFIA1 - protein tyrosine phosphatase, receptor type, f polypeptide (ptprf), interacting protein (liprin), alpha 1, PPP4R2 - protein phosphatase 4, regulatory subunit 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, PPFIBP1 - ptprf interacting protein, binding protein 1 (liprin beta 1), ILF3 - interleukin enhancer binding factor 3, 90kda, ANKRD28 - ankyrin repeat domain 28, NUDCD2 - nudc domain containing 2, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, CYCS - cytochrome c, somatic, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, RBM19 - rna binding motif protein 19, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, CD48 - cd48 molecule, AGPS - alkylglycerone phosphate synthase, TBC1D4 - tbc1 domain family, member 4, TMED5 - transmembrane emp24 protein transport domain containing 5, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, GAR1 - gar1 ribonucleoprotein, KDM3A - lysine (k)-specific demethylase 3a, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, ANAPC1 - anaphase promoting complex subunit 1, EXOSC1 - exosome component 1, TELO2 - telomere maintenance 2, PITRM1 - pitrilysin metallopeptidase 1, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), ATG7 - autophagy related 7, TUBGCP4 - tubulin, gamma complex associated protein 4, HDDC2 - hd domain containing 2, PRRC1 - proline-rich coiled-coil 1, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), SYNE1 - spectrin repeat containing, nuclear envelope 1, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), NUP88 - nucleoporin 88kda, ZBTB8OS - zinc finger and btb domain containing 8 opposite strand, NOM1 - nucleolar protein with mif4g domain 1, NFU1 - nfu1 iron-sulfur cluster scaffold homolog (s. cerevisiae), MBLAC2 - metallo-beta-lactamase domain containing 2, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), THOC5 - tho complex 5, FECH - ferrochelatase, CDK6 - cyclin-dependent kinase 6, ADNP - activity-dependent neuroprotector homeobox, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, CDK13 - cyclin-dependent kinase 13, TBCD - tubulin folding cofactor d, FCF1 - fcf1 rrna-processing protein, GNPDA2 - glucosamine-6-phosphate deaminase 2, CCT4 - chaperonin containing tcp1, subunit 4 (delta), CUTC - cutc copper transporter, TBCA - tubulin folding cofactor a, INPP5B - inositol polyphosphate-5-phosphatase, 75kda, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, IRF3 - interferon regulatory factor 3, ARL6IP5 - adp-ribosylation-like factor 6 interacting protein 5, LARP1 - la ribonucleoprotein domain family, member 1, IRF5 - interferon regulatory factor 5, DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, EIF3E - eukaryotic translation initiation factor 3, subunit e, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, TCEA1 - transcription elongation factor a (sii), 1, PURA - purine-rich element binding protein a, SPG11 - spastic paraplegia 11 (autosomal recessive), TDRD7 - tudor domain containing 7, NEK6 - nima-related kinase 6, NOC3L - nucleolar complex associated 3 homolog (s. cerevisiae), PYGB - phosphorylase, glycogen; brain, PCYT2 - phosphate cytidylyltransferase 2, ethanolamine, NUP50 - nucleoporin 50kda, VKORC1L1 - vitamin k epoxide reductase complex, subunit 1-like 1, TRAPPC10 - trafficking protein particle complex 10, IDE - insulin-degrading enzyme, USP34 - ubiquitin specific peptidase 34, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), IFIH1 - interferon induced with helicase c domain 1, MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, SART3 - squamous cell carcinoma antigen recognized by t cells 3, MAD1L1 - mad1 mitotic arrest deficient-like 1 (yeast), CTSC - cathepsin c, RANBP1 - ran binding protein 1, RAP1A - rap1a, member of ras oncogene family, PARVG - parvin, gamma, PUS1 - pseudouridylate synthase 1, NSMAF - neutral sphingomyelinase (n-smase) activation associated factor, AKAP8L - a kinase (prka) anchor protein 8-like, ZNF592 - zinc finger protein 592, CYFIP2 - cytoplasmic fmr1 interacting protein 2, RBM26 - rna binding motif protein 26, CLNS1A - chloride channel, nucleotide-sensitive, 1a, RAP1B - rap1b, member of ras oncogene family, GOLPH3 - golgi phosphoprotein 3 (coat-protein), TMX3 - thioredoxin-related transmembrane protein 3, BZW1 - basic leucine zipper and w2 domains 1, RB1 - retinoblastoma 1, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), MRPS14 - mitochondrial ribosomal protein s14, CUL1 - cullin 1, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3]
104 [DUT - deoxyuridine triphosphatase, HK2 - hexokinase 2, PGM1 - phosphoglucomutase 1, AK4 - adenylate kinase 4, PGAM1 - phosphoglycerate mutase 1 (brain), DGUOK - deoxyguanosine kinase, DCK - deoxycytidine kinase, UCK2 - uridine-cytidine kinase 2, AK3 - adenylate kinase 3, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, UMPS - uridine monophosphate synthetase, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, ADPGK - adp-dependent glucokinase]
105 [SERBP1 - serpine1 mrna binding protein 1, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, GAPVD1 - gtpase activating protein and vps9 domains 1, DDX6 - dead (asp-glu-ala-asp) box helicase 6, RANBP1 - ran binding protein 1, RPL23A - ribosomal protein l23a, DLG1 - discs, large homolog 1 (drosophila), F11R - f11 receptor, RPL14 - ribosomal protein l14, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, RPL6 - ribosomal protein l6, FLNB - filamin b, beta, PPFIBP1 - ptprf interacting protein, binding protein 1 (liprin beta 1), ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), DBN1 - drebrin 1, TRIM25 - tripartite motif containing 25, USP8 - ubiquitin specific peptidase 8, EIF4H - eukaryotic translation initiation factor 4h, BZW1 - basic leucine zipper and w2 domains 1, CHMP5 - charged multivesicular body protein 5, RSL1D1 - ribosomal l1 domain containing 1, LARP1 - la ribonucleoprotein domain family, member 1, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, TJP2 - tight junction protein 2, EIF3E - eukaryotic translation initiation factor 3, subunit e, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, SNX1 - sorting nexin 1, EPS15 - epidermal growth factor receptor pathway substrate 15]
106 [UQCRB - ubiquinol-cytochrome c reductase binding protein, DUT - deoxyuridine triphosphatase, DLG1 - discs, large homolog 1 (drosophila), AK4 - adenylate kinase 4, PGM1 - phosphoglucomutase 1, HK2 - hexokinase 2, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, PGAM1 - phosphoglycerate mutase 1 (brain), NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, DGUOK - deoxyguanosine kinase, DCK - deoxycytidine kinase, TJP2 - tight junction protein 2, UCK2 - uridine-cytidine kinase 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, AK3 - adenylate kinase 3, UMPS - uridine monophosphate synthetase, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, ADPGK - adp-dependent glucokinase]
107 [DST - dystonin, RPL23A - ribosomal protein l23a, MRPL13 - mitochondrial ribosomal protein l13, RPL22 - ribosomal protein l22, TUBGCP4 - tubulin, gamma complex associated protein 4, DLG1 - discs, large homolog 1 (drosophila), SEH1L - seh1-like (s. cerevisiae), RPL14 - ribosomal protein l14, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, RPL13 - ribosomal protein l13, NUP88 - nucleoporin 88kda, RPS8 - ribosomal protein s8, RPL6 - ribosomal protein l6, MRPL42 - mitochondrial ribosomal protein l42, RPL7 - ribosomal protein l7, MAP4 - microtubule-associated protein 4, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPL3 - ribosomal protein l3, ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, RPS7 - ribosomal protein s7, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, MRPS35 - mitochondrial ribosomal protein s35, MRPL2 - mitochondrial ribosomal protein l2, CPOX - coproporphyrinogen oxidase, MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, RPL7L1 - ribosomal protein l7-like 1, TUBGCP3 - tubulin, gamma complex associated protein 3, MRPL47 - mitochondrial ribosomal protein l47, MRPS14 - mitochondrial ribosomal protein s14, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, CCDC6 - coiled-coil domain containing 6, MRPL3 - mitochondrial ribosomal protein l3]
108 [NCBP2 - nuclear cap binding protein subunit 2, 20kda, ERAP1 - endoplasmic reticulum aminopeptidase 1, EXOSC5 - exosome component 5, DDX6 - dead (asp-glu-ala-asp) box helicase 6, GSPT1 - g1 to s phase transition 1, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, ECI1 - enoyl-coa delta isomerase 1, DERL1 - derlin 1, TRIM25 - tripartite motif containing 25, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, ACAT1 - acetyl-coa acetyltransferase 1, VTI1A - vesicle transport through interaction with t-snares 1a, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, USP25 - ubiquitin specific peptidase 25, SORD - sorbitol dehydrogenase, PLCG1 - phospholipase c, gamma 1, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, ANAPC1 - anaphase promoting complex subunit 1, EXOSC1 - exosome component 1, SGPL1 - sphingosine-1-phosphate lyase 1, ATG7 - autophagy related 7, ANAPC7 - anaphase promoting complex subunit 7, GLS - glutaminase, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), PELO - pelota homolog (drosophila), HADH - hydroxyacyl-coa dehydrogenase, ADPGK - adp-dependent glucokinase, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, MUT - methylmalonyl coa mutase, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, GNPDA2 - glucosamine-6-phosphate deaminase 2, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, INPP5B - inositol polyphosphate-5-phosphatase, 75kda, COMT - catechol-o-methyltransferase, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, KYNU - kynureninase, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), NEMF - nuclear export mediator factor, EIF3E - eukaryotic translation initiation factor 3, subunit e, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, VTA1 - vesicle (multivesicular body) trafficking 1, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, DENND3 - denn/madd domain containing 3, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), UBE2Z - ubiquitin-conjugating enzyme e2z, HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PYGB - phosphorylase, glycogen; brain, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, PHKB - phosphorylase kinase, beta, RPL3 - ribosomal protein l3, USP48 - ubiquitin specific peptidase 48, MANBA - mannosidase, beta a, lysosomal, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), USP8 - ubiquitin specific peptidase 8, DUT - deoxyuridine triphosphatase, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, HK2 - hexokinase 2, KPNB1 - karyopherin (importin) beta 1, TRIM38 - tripartite motif containing 38, IDE - insulin-degrading enzyme, CDC23 - cell division cycle 23, USP34 - ubiquitin specific peptidase 34, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, UBA6 - ubiquitin-like modifier activating enzyme 6, RPL14 - ribosomal protein l14, MFN1 - mitofusin 1, NUB1 - negative regulator of ubiquitin-like proteins 1, RPS8 - ribosomal protein s8, ESD - esterase d, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, UCHL3 - ubiquitin carboxyl-terminal esterase l3 (ubiquitin thiolesterase), GCDH - glutaryl-coa dehydrogenase, CISD2 - cdgsh iron sulfur domain 2, ACOT7 - acyl-coa thioesterase 7, CAT - catalase, USP15 - ubiquitin specific peptidase 15, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), UBE2I - ubiquitin-conjugating enzyme e2i, CUL1 - cullin 1, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), PGM2 - phosphoglucomutase 2]
109 [ZC3H7B - zinc finger ccch-type containing 7b, PURA - purine-rich element binding protein a, SERBP1 - serpine1 mrna binding protein 1, QKI - qki, kh domain containing, rna binding, NCBP2 - nuclear cap binding protein subunit 2, 20kda, DDX6 - dead (asp-glu-ala-asp) box helicase 6, EXOSC5 - exosome component 5, MRPL13 - mitochondrial ribosomal protein l13, TCOF1 - treacher collins-franceschetti syndrome 1, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, TDRD7 - tudor domain containing 7, GSPT1 - g1 to s phase transition 1, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), PTCD3 - pentatricopeptide repeat domain 3, LARP4B - la ribonucleoprotein domain family, member 4b, TMED2 - transmembrane emp24 domain trafficking protein 2, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), EIF4G3 - eukaryotic translation initiation factor 4 gamma, 3, ILF3 - interleukin enhancer binding factor 3, 90kda, EIF4H - eukaryotic translation initiation factor 4h, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, EIF4B - eukaryotic translation initiation factor 4b, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, TSFM - ts translation elongation factor, mitochondrial, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, TNPO1 - transportin 1, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, PA2G4 - proliferation-associated 2g4, 38kda, CAPRIN1 - cell cycle associated protein 1, EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, NEMF - nuclear export mediator factor, LARP1 - la ribonucleoprotein domain family, member 1, PRKCD - protein kinase c, delta, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, CARHSP1 - calcium regulated heat stable protein 1, 24kda, EIF3E - eukaryotic translation initiation factor 3, subunit e, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, EXOSC1 - exosome component 1]
110 [PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, ACAT1 - acetyl-coa acetyltransferase 1, DUT - deoxyuridine triphosphatase, AK4 - adenylate kinase 4, PGM1 - phosphoglucomutase 1, HK2 - hexokinase 2, GCDH - glutaryl-coa dehydrogenase, ACOT7 - acyl-coa thioesterase 7, PGAM1 - phosphoglycerate mutase 1 (brain), KYNU - kynureninase, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, DGUOK - deoxyguanosine kinase, NAMPT - nicotinamide phosphoribosyltransferase, DCK - deoxycytidine kinase, AK3 - adenylate kinase 3, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, UCK2 - uridine-cytidine kinase 2, UMPS - uridine monophosphate synthetase, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, DCAKD - dephospho-coa kinase domain containing, ADPGK - adp-dependent glucokinase, RRM1 - ribonucleotide reductase m1]
111 [PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, ACAT1 - acetyl-coa acetyltransferase 1, DUT - deoxyuridine triphosphatase, AK4 - adenylate kinase 4, PGM1 - phosphoglucomutase 1, HK2 - hexokinase 2, GCDH - glutaryl-coa dehydrogenase, ACOT7 - acyl-coa thioesterase 7, PGAM1 - phosphoglycerate mutase 1 (brain), KYNU - kynureninase, DGUOK - deoxyguanosine kinase, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, NAMPT - nicotinamide phosphoribosyltransferase, DCK - deoxycytidine kinase, AK3 - adenylate kinase 3, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, UCK2 - uridine-cytidine kinase 2, UMPS - uridine monophosphate synthetase, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, DCAKD - dephospho-coa kinase domain containing, ADPGK - adp-dependent glucokinase, RRM1 - ribonucleotide reductase m1]
112 [CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, VTA1 - vesicle (multivesicular body) trafficking 1, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, DDX6 - dead (asp-glu-ala-asp) box helicase 6, RPL23A - ribosomal protein l23a, SF3A3 - splicing factor 3a, subunit 3, 60kda, GSPT1 - g1 to s phase transition 1, SCFD1 - sec1 family domain containing 1, GSN - gelsolin, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, HMGB2 - high mobility group box 2, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, NDUFA5 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, GFM2 - g elongation factor, mitochondrial 2, PTCD3 - pentatricopeptide repeat domain 3, TMED2 - transmembrane emp24 domain trafficking protein 2, RPL6 - ribosomal protein l6, HOMER1 - homer homolog 1 (drosophila), RPL3 - ribosomal protein l3, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), EIF3F - eukaryotic translation initiation factor 3, subunit f, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, ANKRD28 - ankyrin repeat domain 28, EIF4H - eukaryotic translation initiation factor 4h, TRAPPC10 - trafficking protein particle complex 10, MRPS35 - mitochondrial ribosomal protein s35, EIF4B - eukaryotic translation initiation factor 4b, MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, STX17 - syntaxin 17, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, MAT2A - methionine adenosyltransferase ii, alpha, GOSR2 - golgi snap receptor complex member 2, MCM3 - minichromosome maintenance complex component 3, CHMP5 - charged multivesicular body protein 5, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, WDR77 - wd repeat domain 77, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), SART3 - squamous cell carcinoma antigen recognized by t cells 3, CTSC - cathepsin c, MRPL3 - mitochondrial ribosomal protein l3, WIPF1 - was/wasl interacting protein family, member 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, MCM7 - minichromosome maintenance complex component 7, EIF2D - eukaryotic translation initiation factor 2d, MRPL13 - mitochondrial ribosomal protein l13, MCU - mitochondrial calcium uniporter, SNRPE - small nuclear ribonucleoprotein polypeptide e, TUBGCP4 - tubulin, gamma complex associated protein 4, SEH1L - seh1-like (s. cerevisiae), TMEM70 - transmembrane protein 70, GLS - glutaminase, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), PELO - pelota homolog (drosophila), WDR1 - wd repeat domain 1, SCO1 - sco1 cytochrome c oxidase assembly protein, CLNS1A - chloride channel, nucleotide-sensitive, 1a, TRAPPC9 - trafficking protein particle complex 9, NIP7 - nip7, nucleolar pre-rrna processing protein, MRPL42 - mitochondrial ribosomal protein l42, DSTN - destrin (actin depolymerizing factor), EIF2S2 - eukaryotic translation initiation factor 2, subunit 2 beta, 38kda, MRPL2 - mitochondrial ribosomal protein l2, SMARCD1 - swi/snf related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1, TBCD - tubulin folding cofactor d, HSPD1 - heat shock 60kda protein 1 (chaperonin), UGDH - udp-glucose 6-dehydrogenase, NDUFAF4 - nadh dehydrogenase (ubiquinone) complex i, assembly factor 4, CUTC - cutc copper transporter, TBCA - tubulin folding cofactor a, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, SAMM50 - samm50 sorting and assembly machinery component, MRPS14 - mitochondrial ribosomal protein s14, MRPL47 - mitochondrial ribosomal protein l47, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, CUL1 - cullin 1, EIF3E - eukaryotic translation initiation factor 3, subunit e, HSPA4 - heat shock 70kda protein 4, EPS15 - epidermal growth factor receptor pathway substrate 15, RRM1 - ribonucleotide reductase m1, LYRM7 - lyr motif containing 7]
113 [FAM136A - family with sequence similarity 136, member a, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, SF3A3 - splicing factor 3a, subunit 3, 60kda, GFM2 - g elongation factor, mitochondrial 2, FKBP11 - fk506 binding protein 11, 19 kda, TMED2 - transmembrane emp24 domain trafficking protein 2, HOOK3 - hook microtubule-tethering protein 3, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, ACAT1 - acetyl-coa acetyltransferase 1, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, CPOX - coproporphyrinogen oxidase, LRRC40 - leucine rich repeat containing 40, SUGT1 - sgt1, suppressor of g2 allele of skp1 (s. cerevisiae), GOSR2 - golgi snap receptor complex member 2, OTUB1 - otu domain, ubiquitin aldehyde binding 1, SORD - sorbitol dehydrogenase, PLCG1 - phospholipase c, gamma 1, LIMS1 - lim and senescent cell antigen-like domains 1, MORF4L1 - mortality factor 4 like 1, TTC27 - tetratricopeptide repeat domain 27, CARHSP1 - calcium regulated heat stable protein 1, 24kda, CREB1 - camp responsive element binding protein 1, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, SNX1 - sorting nexin 1, THUMPD1 - thump domain containing 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, SNRPE - small nuclear ribonucleoprotein polypeptide e, ANAPC7 - anaphase promoting complex subunit 7, GLS - glutaminase, PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, GLO1 - glyoxalase i, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), GLG1 - golgi glycoprotein 1, GOSR1 - golgi snap receptor complex member 1, NIP7 - nip7, nucleolar pre-rrna processing protein, COMMD10 - comm domain containing 10, POLR1C - polymerase (rna) i polypeptide c, 30kda, PPA1 - pyrophosphatase (inorganic) 1, SUMO3 - small ubiquitin-like modifier 3, SNX6 - sorting nexin 6, SMARCD1 - swi/snf related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, COMT - catechol-o-methyltransferase, STX8 - syntaxin 8, TAOK3 - tao kinase 3, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), PARP4 - poly (adp-ribose) polymerase family, member 4, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, TMEM30A - transmembrane protein 30a, SCPEP1 - serine carboxypeptidase 1, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, QKI - qki, kh domain containing, rna binding, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, AGK - acylglycerol kinase, HOMER1 - homer homolog 1 (drosophila), FTH1 - ferritin, heavy polypeptide 1, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, PPP5C - protein phosphatase 5, catalytic subunit, USP48 - ubiquitin specific peptidase 48, SURF4 - surfeit 4, IRAK4 - interleukin-1 receptor-associated kinase 4, DNAJC11 - dnaj (hsp40) homolog, subfamily c, member 11, MRPS35 - mitochondrial ribosomal protein s35, AK4 - adenylate kinase 4, TNPO1 - transportin 1, KPNB1 - karyopherin (importin) beta 1, EXOC2 - exocyst complex component 2, EXOC1 - exocyst complex component 1, TJP2 - tight junction protein 2, IFIT5 - interferon-induced protein with tetratricopeptide repeats 5, DCAKD - dephospho-coa kinase domain containing, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, SCYL2 - scy1-like 2 (s. cerevisiae), NIPBL - nipped-b homolog (drosophila), GALNT2 - udp-n-acetyl-alpha-d-galactosamine:polypeptide n-acetylgalactosaminyltransferase 2 (galnac-t2), STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), SNRNP27 - small nuclear ribonucleoprotein 27kda (u4/u6.u5), MFN1 - mitofusin 1, STAT6 - signal transducer and activator of transcription 6, interleukin-4 induced, EIF1AD - eukaryotic translation initiation factor 1a domain containing, FCRLA - fc receptor-like a, RINT1 - rad50 interactor 1, NHP2 - nhp2 ribonucleoprotein, DSTN - destrin (actin depolymerizing factor), RIC8A - ric8 guanine nucleotide exchange factor a, SRP72 - signal recognition particle 72kda, CISD2 - cdgsh iron sulfur domain 2, GCDH - glutaryl-coa dehydrogenase, SSBP1 - single-stranded dna binding protein 1, mitochondrial, SAMM50 - samm50 sorting and assembly machinery component, ANXA4 - annexin a4, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, TNFAIP8 - tumor necrosis factor, alpha-induced protein 8, PRKCD - protein kinase c, delta, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, CCDC6 - coiled-coil domain containing 6, ERAP1 - endoplasmic reticulum aminopeptidase 1, DDX6 - dead (asp-glu-ala-asp) box helicase 6, SDAD1 - sda1 domain containing 1, ZMYM2 - zinc finger, mym-type 2, GSPT1 - g1 to s phase transition 1, GSN - gelsolin, FAM98A - family with sequence similarity 98, member a, ECI1 - enoyl-coa delta isomerase 1, DCK - deoxycytidine kinase, DBN1 - drebrin 1, TRIM25 - tripartite motif containing 25, XPOT - exportin, trna, RIF1 - rap1 interacting factor homolog (yeast), VTI1A - vesicle transport through interaction with t-snares 1a, CCDC51 - coiled-coil domain containing 51, MMS22L - mms22-like, dna repair protein, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), ARCN1 - archain 1, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, PA2G4 - proliferation-associated 2g4, 38kda, LRRK1 - leucine-rich repeat kinase 1, CAB39 - calcium binding protein 39, GATAD2B - gata zinc finger domain containing 2b, KIAA2013 - kiaa2013, RAB23 - rab23, member ras oncogene family, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, MRPL3 - mitochondrial ribosomal protein l3, RNF31 - ring finger protein 31, MCM7 - minichromosome maintenance complex component 7, MCU - mitochondrial calcium uniporter, ARL1 - adp-ribosylation factor-like 1, KDELC2 - kdel (lys-asp-glu-leu) containing 2, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, PELO - pelota homolog (drosophila), SCO1 - sco1 cytochrome c oxidase assembly protein, WDHD1 - wd repeat and hmg-box dna binding protein 1, HADH - hydroxyacyl-coa dehydrogenase, PPIL3 - peptidylprolyl isomerase (cyclophilin)-like 3, OSBPL3 - oxysterol binding protein-like 3, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, RPL7L1 - ribosomal protein l7-like 1, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, STX18 - syntaxin 18, NEMF - nuclear export mediator factor, RPTOR - regulatory associated protein of mtor, complex 1, QSOX2 - quiescin q6 sulfhydryl oxidase 2, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, LYRM7 - lyr motif containing 7, VTA1 - vesicle (multivesicular body) trafficking 1, SERBP1 - serpine1 mrna binding protein 1, DENND3 - denn/madd domain containing 3, GAPVD1 - gtpase activating protein and vps9 domains 1, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, MAP4 - microtubule-associated protein 4, RAB14 - rab14, member ras oncogene family, PHKB - phosphorylase kinase, beta, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, MANBA - mannosidase, beta a, lysosomal, MAN2A1 - mannosidase, alpha, class 2a, member 1, USP8 - ubiquitin specific peptidase 8, EIF4H - eukaryotic translation initiation factor 4h, MBNL1 - muscleblind-like splicing regulator 1, METTL15 - methyltransferase like 15, SLAMF1 - signaling lymphocytic activation molecule family member 1, DUT - deoxyuridine triphosphatase, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, MAT2A - methionine adenosyltransferase ii, alpha, MCM3 - minichromosome maintenance complex component 3, CHMP5 - charged multivesicular body protein 5, DOCK7 - dedicator of cytokinesis 7, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, WIPF1 - was/wasl interacting protein family, member 1, SCAF8 - sr-related ctd-associated factor 8, UBA6 - ubiquitin-like modifier activating enzyme 6, GRK6 - g protein-coupled receptor kinase 6, LSS - lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase), DLG1 - discs, large homolog 1 (drosophila), PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, RPL14 - ribosomal protein l14, DHRS7 - dehydrogenase/reductase (sdr family) member 7, LRRC20 - leucine rich repeat containing 20, NUB1 - negative regulator of ubiquitin-like proteins 1, TRAPPC9 - trafficking protein particle complex 9, DGUOK - deoxyguanosine kinase, UBA3 - ubiquitin-like modifier activating enzyme 3, DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, ACOT7 - acyl-coa thioesterase 7, PDAP1 - pdgfa associated protein 1, CBX3 - chromobox homolog 3, ADI1 - acireductone dioxygenase 1, FAM49B - family with sequence similarity 49, member b, CAPRIN1 - cell cycle associated protein 1, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), RHOT1 - ras homolog family member t1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, RSL1D1 - ribosomal l1 domain containing 1, BCL2L1 - bcl2-like 1, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, METTL13 - methyltransferase like 13, PGM2 - phosphoglucomutase 2, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1, CCNK - cyclin k, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, DST - dystonin, NCBP2 - nuclear cap binding protein subunit 2, 20kda, EXOSC5 - exosome component 5, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, RFC3 - replication factor c (activator 1) 3, 38kda, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, SACM1L - sac1 suppressor of actin mutations 1-like (yeast), NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, NDUFA5 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, RECQL - recq protein-like (dna helicase q1-like), DERL1 - derlin 1, BTF3 - basic transcription factor 3, DOCK8 - dedicator of cytokinesis 8, SPC24 - spc24, ndc80 kinetochore complex component, EIF4B - eukaryotic translation initiation factor 4b, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, USP25 - ubiquitin specific peptidase 25, ZNF326 - zinc finger protein 326, WDR77 - wd repeat domain 77, RCSD1 - rcsd domain containing 1, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, MYO6 - myosin vi, SGPL1 - sphingosine-1-phosphate lyase 1, EIF2D - eukaryotic translation initiation factor 2d, TPM3 - tropomyosin 3, STK38L - serine/threonine kinase 38 like, SEH1L - seh1-like (s. cerevisiae), EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, CC2D1B - coiled-coil and c2 domain containing 1b, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), DCAF7 - ddb1 and cul4 associated factor 7, EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, ADPGK - adp-dependent glucokinase, ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, EIF2S2 - eukaryotic translation initiation factor 2, subunit 2 beta, 38kda, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, TSFM - ts translation elongation factor, mitochondrial, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), MVD - mevalonate (diphospho) decarboxylase, DENND4A - denn/madd domain containing 4a, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, KYNU - kynureninase, ERP44 - endoplasmic reticulum protein 44, NAMPT - nicotinamide phosphoribosyltransferase, HSPA4 - heat shock 70kda protein 4, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, UQCRB - ubiquinol-cytochrome c reductase binding protein, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, CAPN2 - calpain 2, (m/ii) large subunit, ZWILCH - zwilch kinetochore protein, WNK1 - wnk lysine deficient protein kinase 1, OSTF1 - osteoclast stimulating factor 1, UBE2Z - ubiquitin-conjugating enzyme e2z, HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PTCD3 - pentatricopeptide repeat domain 3, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), TBC1D2B - tbc1 domain family, member 2b, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, EIF4G3 - eukaryotic translation initiation factor 4 gamma, 3, UCK2 - uridine-cytidine kinase 2, RPL3 - ribosomal protein l3, UMPS - uridine monophosphate synthetase, BTF3L4 - basic transcription factor 3-like 4, CARS - cysteinyl-trna synthetase, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), EIF3F - eukaryotic translation initiation factor 3, subunit f, MRPL1 - mitochondrial ribosomal protein l1, RPN1 - ribophorin i, MRPL9 - mitochondrial ribosomal protein l9, HK2 - hexokinase 2, STX17 - syntaxin 17, TRIM38 - tripartite motif containing 38, ABCF2 - atp-binding cassette, sub-family f (gcn20), member 2, CDC23 - cell division cycle 23, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, VDAC2 - voltage-dependent anion channel 2, TTLL12 - tubulin tyrosine ligase-like family, member 12, MRPL13 - mitochondrial ribosomal protein l13, GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, TMEM70 - transmembrane protein 70, RNMT - rna (guanine-7-) methyltransferase, WDR1 - wd repeat domain 1, LARP4B - la ribonucleoprotein domain family, member 4b, RPS8 - ribosomal protein s8, MRPL42 - mitochondrial ribosomal protein l42, ESD - esterase d, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, STOM - stomatin, UCHL3 - ubiquitin carboxyl-terminal esterase l3 (ubiquitin thiolesterase), SUMO1 - small ubiquitin-like modifier 1, CAT - catalase, USP15 - ubiquitin specific peptidase 15, UGDH - udp-glucose 6-dehydrogenase, XPO6 - exportin 6, NDUFAF4 - nadh dehydrogenase (ubiquinone) complex i, assembly factor 4, MRPL47 - mitochondrial ribosomal protein l47, TUBGCP3 - tubulin, gamma complex associated protein 3, UBE2I - ubiquitin-conjugating enzyme e2i, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, AIDA - axin interactor, dorsalization associated, RRBP1 - ribosome binding protein 1, EPS15 - epidermal growth factor receptor pathway substrate 15, RRM1 - ribonucleotide reductase m1, ZC3H7B - zinc finger ccch-type containing 7b, DDX31 - dead (asp-glu-ala-asp) box polypeptide 31, TCOF1 - treacher collins-franceschetti syndrome 1, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, ZNF638 - zinc finger protein 638, F11R - f11 receptor, STAMBP - stam binding protein, SCFD1 - sec1 family domain containing 1, URB1 - urb1 ribosome biogenesis 1 homolog (s. cerevisiae), CISD1 - cdgsh iron sulfur domain 1, RAB30 - rab30, member ras oncogene family, FLNB - filamin b, beta, PPFIA1 - protein tyrosine phosphatase, receptor type, f polypeptide (ptprf), interacting protein (liprin), alpha 1, PPP4R2 - protein phosphatase 4, regulatory subunit 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, PPFIBP1 - ptprf interacting protein, binding protein 1 (liprin beta 1), ILF3 - interleukin enhancer binding factor 3, 90kda, ANKRD28 - ankyrin repeat domain 28, NUDCD2 - nudc domain containing 2, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, CYCS - cytochrome c, somatic, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, RBM19 - rna binding motif protein 19, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, CD48 - cd48 molecule, AGPS - alkylglycerone phosphate synthase, TBC1D4 - tbc1 domain family, member 4, TMED5 - transmembrane emp24 protein transport domain containing 5, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, GAR1 - gar1 ribonucleoprotein, KDM3A - lysine (k)-specific demethylase 3a, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, ANAPC1 - anaphase promoting complex subunit 1, TELO2 - telomere maintenance 2, EXOSC1 - exosome component 1, PITRM1 - pitrilysin metallopeptidase 1, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), ATG7 - autophagy related 7, TUBGCP4 - tubulin, gamma complex associated protein 4, HDDC2 - hd domain containing 2, PRRC1 - proline-rich coiled-coil 1, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), SYNE1 - spectrin repeat containing, nuclear envelope 1, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), NUP88 - nucleoporin 88kda, ZBTB8OS - zinc finger and btb domain containing 8 opposite strand, NOM1 - nucleolar protein with mif4g domain 1, NFU1 - nfu1 iron-sulfur cluster scaffold homolog (s. cerevisiae), MBLAC2 - metallo-beta-lactamase domain containing 2, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), THOC5 - tho complex 5, FECH - ferrochelatase, CDK6 - cyclin-dependent kinase 6, ADNP - activity-dependent neuroprotector homeobox, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, CDK13 - cyclin-dependent kinase 13, TBCD - tubulin folding cofactor d, FCF1 - fcf1 rrna-processing protein, CCT4 - chaperonin containing tcp1, subunit 4 (delta), GNPDA2 - glucosamine-6-phosphate deaminase 2, TBCA - tubulin folding cofactor a, CUTC - cutc copper transporter, INPP5B - inositol polyphosphate-5-phosphatase, 75kda, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, IRF3 - interferon regulatory factor 3, ARL6IP5 - adp-ribosylation-like factor 6 interacting protein 5, LARP1 - la ribonucleoprotein domain family, member 1, IRF5 - interferon regulatory factor 5, DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, EIF3E - eukaryotic translation initiation factor 3, subunit e, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, TCEA1 - transcription elongation factor a (sii), 1, PURA - purine-rich element binding protein a, SPG11 - spastic paraplegia 11 (autosomal recessive), TDRD7 - tudor domain containing 7, NEK6 - nima-related kinase 6, NOC3L - nucleolar complex associated 3 homolog (s. cerevisiae), PYGB - phosphorylase, glycogen; brain, PCYT2 - phosphate cytidylyltransferase 2, ethanolamine, NUP50 - nucleoporin 50kda, VKORC1L1 - vitamin k epoxide reductase complex, subunit 1-like 1, TRAPPC10 - trafficking protein particle complex 10, IDE - insulin-degrading enzyme, USP34 - ubiquitin specific peptidase 34, IFIH1 - interferon induced with helicase c domain 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, SART3 - squamous cell carcinoma antigen recognized by t cells 3, MAD1L1 - mad1 mitotic arrest deficient-like 1 (yeast), CTSC - cathepsin c, RANBP1 - ran binding protein 1, RAP1A - rap1a, member of ras oncogene family, PARVG - parvin, gamma, PUS1 - pseudouridylate synthase 1, NSMAF - neutral sphingomyelinase (n-smase) activation associated factor, GNPAT - glyceronephosphate o-acyltransferase, AKAP8L - a kinase (prka) anchor protein 8-like, ZNF592 - zinc finger protein 592, CYFIP2 - cytoplasmic fmr1 interacting protein 2, RBM26 - rna binding motif protein 26, CLNS1A - chloride channel, nucleotide-sensitive, 1a, RAP1B - rap1b, member of ras oncogene family, GOLPH3 - golgi phosphoprotein 3 (coat-protein), TMX3 - thioredoxin-related transmembrane protein 3, BZW1 - basic leucine zipper and w2 domains 1, RB1 - retinoblastoma 1, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), MRPS14 - mitochondrial ribosomal protein s14, CUL1 - cullin 1, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3]
114 [CCNK - cyclin k, DST - dystonin, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, NCBP2 - nuclear cap binding protein subunit 2, 20kda, EXOSC5 - exosome component 5, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, RFC3 - replication factor c (activator 1) 3, 38kda, SF3A3 - splicing factor 3a, subunit 3, 60kda, SACM1L - sac1 suppressor of actin mutations 1-like (yeast), SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, GFM2 - g elongation factor, mitochondrial 2, TMED2 - transmembrane emp24 domain trafficking protein 2, RECQL - recq protein-like (dna helicase q1-like), HOOK3 - hook microtubule-tethering protein 3, DERL1 - derlin 1, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, BTF3 - basic transcription factor 3, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, SPC24 - spc24, ndc80 kinetochore complex component, ACAT1 - acetyl-coa acetyltransferase 1, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, CPOX - coproporphyrinogen oxidase, LRRC40 - leucine rich repeat containing 40, SUGT1 - sgt1, suppressor of g2 allele of skp1 (s. cerevisiae), GOSR2 - golgi snap receptor complex member 2, USP25 - ubiquitin specific peptidase 25, OTUB1 - otu domain, ubiquitin aldehyde binding 1, ZNF326 - zinc finger protein 326, WDR77 - wd repeat domain 77, CARHSP1 - calcium regulated heat stable protein 1, 24kda, CREB1 - camp responsive element binding protein 1, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, MYO6 - myosin vi, SNX1 - sorting nexin 1, SGPL1 - sphingosine-1-phosphate lyase 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, TPM3 - tropomyosin 3, SNRPE - small nuclear ribonucleoprotein polypeptide e, STK38L - serine/threonine kinase 38 like, ANAPC7 - anaphase promoting complex subunit 7, SEH1L - seh1-like (s. cerevisiae), EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, GLS - glutaminase, CC2D1B - coiled-coil and c2 domain containing 1b, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), GLG1 - golgi glycoprotein 1, NIP7 - nip7, nucleolar pre-rrna processing protein, GOSR1 - golgi snap receptor complex member 1, DCAF7 - ddb1 and cul4 associated factor 7, ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, ADPGK - adp-dependent glucokinase, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, SUMO3 - small ubiquitin-like modifier 3, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, SNX6 - sorting nexin 6, TSFM - ts translation elongation factor, mitochondrial, SMARCD1 - swi/snf related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), DENND4A - denn/madd domain containing 4a, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, COMT - catechol-o-methyltransferase, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, KYNU - kynureninase, STX8 - syntaxin 8, ERP44 - endoplasmic reticulum protein 44, HSPA4 - heat shock 70kda protein 4, PARP4 - poly (adp-ribose) polymerase family, member 4, TMEM30A - transmembrane protein 30a, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, QKI - qki, kh domain containing, rna binding, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, CAPN2 - calpain 2, (m/ii) large subunit, ZWILCH - zwilch kinetochore protein, UBE2Z - ubiquitin-conjugating enzyme e2z, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, AGK - acylglycerol kinase, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PTCD3 - pentatricopeptide repeat domain 3, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, FTH1 - ferritin, heavy polypeptide 1, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, UMPS - uridine monophosphate synthetase, USP48 - ubiquitin specific peptidase 48, RPL3 - ribosomal protein l3, PPP5C - protein phosphatase 5, catalytic subunit, SURF4 - surfeit 4, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), IRAK4 - interleukin-1 receptor-associated kinase 4, DNAJC11 - dnaj (hsp40) homolog, subfamily c, member 11, MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, RPN1 - ribophorin i, TNPO1 - transportin 1, AK4 - adenylate kinase 4, HK2 - hexokinase 2, STX17 - syntaxin 17, KPNB1 - karyopherin (importin) beta 1, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, TJP2 - tight junction protein 2, IFIT5 - interferon-induced protein with tetratricopeptide repeats 5, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, VDAC2 - voltage-dependent anion channel 2, TTLL12 - tubulin tyrosine ligase-like family, member 12, MRPL13 - mitochondrial ribosomal protein l13, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, NIPBL - nipped-b homolog (drosophila), SCYL2 - scy1-like 2 (s. cerevisiae), GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, GALNT2 - udp-n-acetyl-alpha-d-galactosamine:polypeptide n-acetylgalactosaminyltransferase 2 (galnac-t2), TMEM70 - transmembrane protein 70, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), MFN1 - mitofusin 1, RNMT - rna (guanine-7-) methyltransferase, WDR1 - wd repeat domain 1, LARP4B - la ribonucleoprotein domain family, member 4b, EIF1AD - eukaryotic translation initiation factor 1a domain containing, RPS8 - ribosomal protein s8, MRPL42 - mitochondrial ribosomal protein l42, ESD - esterase d, RPS13 - ribosomal protein s13, RINT1 - rad50 interactor 1, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, DSTN - destrin (actin depolymerizing factor), STOM - stomatin, SUMO1 - small ubiquitin-like modifier 1, SRP72 - signal recognition particle 72kda, GCDH - glutaryl-coa dehydrogenase, CISD2 - cdgsh iron sulfur domain 2, CAT - catalase, USP15 - ubiquitin specific peptidase 15, XPO6 - exportin 6, UGDH - udp-glucose 6-dehydrogenase, NDUFAF4 - nadh dehydrogenase (ubiquinone) complex i, assembly factor 4, SSBP1 - single-stranded dna binding protein 1, mitochondrial, SAMM50 - samm50 sorting and assembly machinery component, MRPL47 - mitochondrial ribosomal protein l47, ANXA4 - annexin a4, TUBGCP3 - tubulin, gamma complex associated protein 3, UBE2I - ubiquitin-conjugating enzyme e2i, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, PRKCD - protein kinase c, delta, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), CCDC6 - coiled-coil domain containing 6, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, RRBP1 - ribosome binding protein 1, EPS15 - epidermal growth factor receptor pathway substrate 15, ZC3H7B - zinc finger ccch-type containing 7b, DDX31 - dead (asp-glu-ala-asp) box polypeptide 31, ERAP1 - endoplasmic reticulum aminopeptidase 1, DDX6 - dead (asp-glu-ala-asp) box helicase 6, TCOF1 - treacher collins-franceschetti syndrome 1, SDAD1 - sda1 domain containing 1, ZNF638 - zinc finger protein 638, F11R - f11 receptor, STAMBP - stam binding protein, SCFD1 - sec1 family domain containing 1, GSN - gelsolin, CISD1 - cdgsh iron sulfur domain 1, URB1 - urb1 ribosome biogenesis 1 homolog (s. cerevisiae), RAB30 - rab30, member ras oncogene family, FLNB - filamin b, beta, ECI1 - enoyl-coa delta isomerase 1, DCK - deoxycytidine kinase, PPP4R2 - protein phosphatase 4, regulatory subunit 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, DBN1 - drebrin 1, TRIM25 - tripartite motif containing 25, ILF3 - interleukin enhancer binding factor 3, 90kda, NUDCD2 - nudc domain containing 2, RIF1 - rap1 interacting factor homolog (yeast), CYCS - cytochrome c, somatic, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, VTI1A - vesicle transport through interaction with t-snares 1a, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, RBM19 - rna binding motif protein 19, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, EHBP1 - eh domain binding protein 1, ARCN1 - archain 1, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, PA2G4 - proliferation-associated 2g4, 38kda, LRRK1 - leucine-rich repeat kinase 1, AGPS - alkylglycerone phosphate synthase, TMED5 - transmembrane emp24 protein transport domain containing 5, RAB23 - rab23, member ras oncogene family, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, KDM3A - lysine (k)-specific demethylase 3a, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, MRPL3 - mitochondrial ribosomal protein l3, TELO2 - telomere maintenance 2, EXOSC1 - exosome component 1, PITRM1 - pitrilysin metallopeptidase 1, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), MCM7 - minichromosome maintenance complex component 7, MCU - mitochondrial calcium uniporter, ARL1 - adp-ribosylation factor-like 1, TUBGCP4 - tubulin, gamma complex associated protein 4, PRRC1 - proline-rich coiled-coil 1, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), SYNE1 - spectrin repeat containing, nuclear envelope 1, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), PELO - pelota homolog (drosophila), SCO1 - sco1 cytochrome c oxidase assembly protein, NOM1 - nucleolar protein with mif4g domain 1, NFU1 - nfu1 iron-sulfur cluster scaffold homolog (s. cerevisiae), HADH - hydroxyacyl-coa dehydrogenase, OSBPL3 - oxysterol binding protein-like 3, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), THOC5 - tho complex 5, FECH - ferrochelatase, CDK6 - cyclin-dependent kinase 6, ADNP - activity-dependent neuroprotector homeobox, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, CDK13 - cyclin-dependent kinase 13, FCF1 - fcf1 rrna-processing protein, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, CCT4 - chaperonin containing tcp1, subunit 4 (delta), GNPDA2 - glucosamine-6-phosphate deaminase 2, CUTC - cutc copper transporter, TBCA - tubulin folding cofactor a, INPP5B - inositol polyphosphate-5-phosphatase, 75kda, RPL7L1 - ribosomal protein l7-like 1, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, IRF3 - interferon regulatory factor 3, ARL6IP5 - adp-ribosylation-like factor 6 interacting protein 5, STX18 - syntaxin 18, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, LARP1 - la ribonucleoprotein domain family, member 1, NEMF - nuclear export mediator factor, IRF5 - interferon regulatory factor 5, DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), RPTOR - regulatory associated protein of mtor, complex 1, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, EIF3E - eukaryotic translation initiation factor 3, subunit e, QSOX2 - quiescin q6 sulfhydryl oxidase 2, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, TCEA1 - transcription elongation factor a (sii), 1, SERBP1 - serpine1 mrna binding protein 1, VTA1 - vesicle (multivesicular body) trafficking 1, PURA - purine-rich element binding protein a, GAPVD1 - gtpase activating protein and vps9 domains 1, DENND3 - denn/madd domain containing 3, SPG11 - spastic paraplegia 11 (autosomal recessive), ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, TDRD7 - tudor domain containing 7, NEK6 - nima-related kinase 6, PGAM1 - phosphoglycerate mutase 1 (brain), NOC3L - nucleolar complex associated 3 homolog (s. cerevisiae), NUP50 - nucleoporin 50kda, MAP4 - microtubule-associated protein 4, RAB14 - rab14, member ras oncogene family, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, MANBA - mannosidase, beta a, lysosomal, VKORC1L1 - vitamin k epoxide reductase complex, subunit 1-like 1, MAN2A1 - mannosidase, alpha, class 2a, member 1, USP8 - ubiquitin specific peptidase 8, MBNL1 - muscleblind-like splicing regulator 1, CCDC58 - coiled-coil domain containing 58, DUT - deoxyuridine triphosphatase, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, MCM3 - minichromosome maintenance complex component 3, IDE - insulin-degrading enzyme, CHMP5 - charged multivesicular body protein 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, USP34 - ubiquitin specific peptidase 34, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), IFIH1 - interferon induced with helicase c domain 1, MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, SART3 - squamous cell carcinoma antigen recognized by t cells 3, MAD1L1 - mad1 mitotic arrest deficient-like 1 (yeast), CTSC - cathepsin c, WIPF1 - was/wasl interacting protein family, member 1, SCAF8 - sr-related ctd-associated factor 8, LSS - lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase), UBA6 - ubiquitin-like modifier activating enzyme 6, RANBP1 - ran binding protein 1, DLG1 - discs, large homolog 1 (drosophila), RAP1A - rap1a, member of ras oncogene family, PARVG - parvin, gamma, PUS1 - pseudouridylate synthase 1, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, GNPAT - glyceronephosphate o-acyltransferase, AKAP8L - a kinase (prka) anchor protein 8-like, ZNF592 - zinc finger protein 592, NUB1 - negative regulator of ubiquitin-like proteins 1, CYFIP2 - cytoplasmic fmr1 interacting protein 2, RBM26 - rna binding motif protein 26, DGUOK - deoxyguanosine kinase, TRAPPC9 - trafficking protein particle complex 9, CLNS1A - chloride channel, nucleotide-sensitive, 1a, UBA3 - ubiquitin-like modifier activating enzyme 3, RAP1B - rap1b, member of ras oncogene family, GOLPH3 - golgi phosphoprotein 3 (coat-protein), ACOT7 - acyl-coa thioesterase 7, RB1 - retinoblastoma 1, CBX3 - chromobox homolog 3, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, FAM49B - family with sequence similarity 49, member b, ADI1 - acireductone dioxygenase 1, CAPRIN1 - cell cycle associated protein 1, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), RHOT1 - ras homolog family member t1, MRPS14 - mitochondrial ribosomal protein s14, RSL1D1 - ribosomal l1 domain containing 1, BCL2L1 - bcl2-like 1, AK3 - adenylate kinase 3, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
115 [DDX6 - dead (asp-glu-ala-asp) box helicase 6, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, CISD1 - cdgsh iron sulfur domain 1, NOC3L - nucleolar complex associated 3 homolog (s. cerevisiae), AGK - acylglycerol kinase, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, GFM2 - g elongation factor, mitochondrial 2, PTCD3 - pentatricopeptide repeat domain 3, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, ECI1 - enoyl-coa delta isomerase 1, DCK - deoxycytidine kinase, USP48 - ubiquitin specific peptidase 48, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, ILF3 - interleukin enhancer binding factor 3, 90kda, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, DNAJC11 - dnaj (hsp40) homolog, subfamily c, member 11, CYCS - cytochrome c, somatic, CCDC58 - coiled-coil domain containing 58, ACAT1 - acetyl-coa acetyltransferase 1, DUT - deoxyuridine triphosphatase, CPOX - coproporphyrinogen oxidase, MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, STX17 - syntaxin 17, HK2 - hexokinase 2, AK4 - adenylate kinase 4, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, LRRK1 - leucine-rich repeat kinase 1, AGPS - alkylglycerone phosphate synthase, IDE - insulin-degrading enzyme, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, VDAC2 - voltage-dependent anion channel 2, MRPL3 - mitochondrial ribosomal protein l3, PITRM1 - pitrilysin metallopeptidase 1, MRPL13 - mitochondrial ribosomal protein l13, MCU - mitochondrial calcium uniporter, PUS1 - pseudouridylate synthase 1, TMEM70 - transmembrane protein 70, GLS - glutaminase, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), MFN1 - mitofusin 1, SCO1 - sco1 cytochrome c oxidase assembly protein, DGUOK - deoxyguanosine kinase, MRPL42 - mitochondrial ribosomal protein l42, NFU1 - nfu1 iron-sulfur cluster scaffold homolog (s. cerevisiae), HADH - hydroxyacyl-coa dehydrogenase, GOLPH3 - golgi phosphoprotein 3 (coat-protein), FECH - ferrochelatase, STOM - stomatin, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, TSFM - ts translation elongation factor, mitochondrial, ACOT7 - acyl-coa thioesterase 7, GCDH - glutaryl-coa dehydrogenase, CAT - catalase, MUT - methylmalonyl coa mutase, USP15 - ubiquitin specific peptidase 15, HSPD1 - heat shock 60kda protein 1 (chaperonin), NDUFAF4 - nadh dehydrogenase (ubiquinone) complex i, assembly factor 4, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, FAM49B - family with sequence similarity 49, member b, SSBP1 - single-stranded dna binding protein 1, mitochondrial, SAMM50 - samm50 sorting and assembly machinery component, RHOT1 - ras homolog family member t1, MRPL47 - mitochondrial ribosomal protein l47, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, KYNU - kynureninase, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, BCL2L1 - bcl2-like 1, PRKCD - protein kinase c, delta, AK3 - adenylate kinase 3, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, HSPA4 - heat shock 70kda protein 4, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
116 [PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, UQCRB - ubiquinol-cytochrome c reductase binding protein, DLG1 - discs, large homolog 1 (drosophila), PGM1 - phosphoglucomutase 1, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, PGAM1 - phosphoglycerate mutase 1 (brain), MFN1 - mitofusin 1, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, DGUOK - deoxyguanosine kinase, DCK - deoxycytidine kinase, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, ADPGK - adp-dependent glucokinase, ACAT1 - acetyl-coa acetyltransferase 1, HK2 - hexokinase 2, AK4 - adenylate kinase 4, GCDH - glutaryl-coa dehydrogenase, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, ACOT7 - acyl-coa thioesterase 7, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, MVD - mevalonate (diphospho) decarboxylase, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, KYNU - kynureninase, RAB23 - rab23, member ras oncogene family, TJP2 - tight junction protein 2, AK3 - adenylate kinase 3, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, DCAKD - dephospho-coa kinase domain containing]
117 [PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, ACAT1 - acetyl-coa acetyltransferase 1, AK4 - adenylate kinase 4, HK2 - hexokinase 2, PGM1 - phosphoglucomutase 1, GCDH - glutaryl-coa dehydrogenase, ACOT7 - acyl-coa thioesterase 7, PGAM1 - phosphoglycerate mutase 1 (brain), DGUOK - deoxyguanosine kinase, UCK2 - uridine-cytidine kinase 2, AK3 - adenylate kinase 3, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, UMPS - uridine monophosphate synthetase, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, ADPGK - adp-dependent glucokinase, DCAKD - dephospho-coa kinase domain containing]
118 [CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, ARL1 - adp-ribosylation factor-like 1, GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, SCFD1 - sec1 family domain containing 1, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, TMED2 - transmembrane emp24 domain trafficking protein 2, GOSR1 - golgi snap receptor complex member 1, RAB14 - rab14, member ras oncogene family, RINT1 - rad50 interactor 1, GOLPH3 - golgi phosphoprotein 3 (coat-protein), SURF4 - surfeit 4, TRAPPC10 - trafficking protein particle complex 10, VTI1A - vesicle transport through interaction with t-snares 1a, DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, STX17 - syntaxin 17, ARCN1 - archain 1, GOSR2 - golgi snap receptor complex member 2, TMED5 - transmembrane emp24 protein transport domain containing 5, EXOC2 - exocyst complex component 2, STX18 - syntaxin 18, EXOC1 - exocyst complex component 1, SNX1 - sorting nexin 1, EPS15 - epidermal growth factor receptor pathway substrate 15, CTSC - cathepsin c]
119 [CCNK - cyclin k, NCBP2 - nuclear cap binding protein subunit 2, 20kda, RPL23A - ribosomal protein l23a, TCOF1 - treacher collins-franceschetti syndrome 1, RFC3 - replication factor c (activator 1) 3, 38kda, RPL22 - ribosomal protein l22, PGM1 - phosphoglucomutase 1, GSPT1 - g1 to s phase transition 1, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, RPL13 - ribosomal protein l13, GFM2 - g elongation factor, mitochondrial 2, PTCD3 - pentatricopeptide repeat domain 3, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, PPP5C - protein phosphatase 5, catalytic subunit, RPL3 - ribosomal protein l3, TRIM25 - tripartite motif containing 25, BTF3 - basic transcription factor 3, MRPS35 - mitochondrial ribosomal protein s35, DUT - deoxyuridine triphosphatase, MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, MCM3 - minichromosome maintenance complex component 3, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, GAR1 - gar1 ribonucleoprotein, RPL31 - ribosomal protein l31, CREB1 - camp responsive element binding protein 1, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, RPL32 - ribosomal protein l32, MRPL3 - mitochondrial ribosomal protein l3, TELO2 - telomere maintenance 2, MCM7 - minichromosome maintenance complex component 7, MRPL13 - mitochondrial ribosomal protein l13, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, RPL14 - ribosomal protein l14, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), RNMT - rna (guanine-7-) methyltransferase, RPS8 - ribosomal protein s8, MRPL42 - mitochondrial ribosomal protein l42, WDHD1 - wd repeat and hmg-box dna binding protein 1, RPS13 - ribosomal protein s13, POLR1C - polymerase (rna) i polypeptide c, 30kda, RPS6 - ribosomal protein s6, GOLPH3 - golgi phosphoprotein 3 (coat-protein), RPS7 - ribosomal protein s7, NHP2 - nhp2 ribonucleoprotein, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, TSFM - ts translation elongation factor, mitochondrial, UGDH - udp-glucose 6-dehydrogenase, SSBP1 - single-stranded dna binding protein 1, mitochondrial, MRPS14 - mitochondrial ribosomal protein s14, MRPL47 - mitochondrial ribosomal protein l47, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, NEMF - nuclear export mediator factor, RRBP1 - ribosome binding protein 1, PGM2 - phosphoglucomutase 2, RRM1 - ribonucleotide reductase m1, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1, TCEA1 - transcription elongation factor a (sii), 1]
120 [MRPS35 - mitochondrial ribosomal protein s35, VTA1 - vesicle (multivesicular body) trafficking 1, MRPL2 - mitochondrial ribosomal protein l2, EIF2D - eukaryotic translation initiation factor 2d, MRPL1 - mitochondrial ribosomal protein l1, MRPL13 - mitochondrial ribosomal protein l13, MRPL9 - mitochondrial ribosomal protein l9, STX17 - syntaxin 17, SMARCD1 - swi/snf related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1, GSPT1 - g1 to s phase transition 1, GSN - gelsolin, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, GFM2 - g elongation factor, mitochondrial 2, PELO - pelota homolog (drosophila), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), PTCD3 - pentatricopeptide repeat domain 3, MRPS14 - mitochondrial ribosomal protein s14, MRPL47 - mitochondrial ribosomal protein l47, WDR1 - wd repeat domain 1, CHMP5 - charged multivesicular body protein 5, MRPL42 - mitochondrial ribosomal protein l42, MRPL3 - mitochondrial ribosomal protein l3, DSTN - destrin (actin depolymerizing factor)]
121 [DST - dystonin, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, RFC3 - replication factor c (activator 1) 3, 38kda, SF3A3 - splicing factor 3a, subunit 3, 60kda, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, NDUFA5 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, GFM2 - g elongation factor, mitochondrial 2, TMED2 - transmembrane emp24 domain trafficking protein 2, HOOK3 - hook microtubule-tethering protein 3, RECQL - recq protein-like (dna helicase q1-like), YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, EIF4B - eukaryotic translation initiation factor 4b, GOSR2 - golgi snap receptor complex member 2, LIMS1 - lim and senescent cell antigen-like domains 1, WDR77 - wd repeat domain 77, MORF4L1 - mortality factor 4 like 1, CREB1 - camp responsive element binding protein 1, SNX1 - sorting nexin 1, MYO6 - myosin vi, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, EIF2D - eukaryotic translation initiation factor 2d, SNRPE - small nuclear ribonucleoprotein polypeptide e, TPM3 - tropomyosin 3, SEH1L - seh1-like (s. cerevisiae), GLS - glutaminase, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), GOSR1 - golgi snap receptor complex member 1, NIP7 - nip7, nucleolar pre-rrna processing protein, ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, EIF2S2 - eukaryotic translation initiation factor 2, subunit 2 beta, 38kda, SMARCD1 - swi/snf related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1, HSPD1 - heat shock 60kda protein 1 (chaperonin), ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, STX8 - syntaxin 8, TAOK3 - tao kinase 3, HSPA4 - heat shock 70kda protein 4, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, TMEM30A - transmembrane protein 30a, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, RPL23A - ribosomal protein l23a, CAPN2 - calpain 2, (m/ii) large subunit, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, HMGB2 - high mobility group box 2, AGK - acylglycerol kinase, HMGB1 - high mobility group box 1, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), PTCD3 - pentatricopeptide repeat domain 3, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, HOMER1 - homer homolog 1 (drosophila), PPP5C - protein phosphatase 5, catalytic subunit, RPL3 - ribosomal protein l3, SURF4 - surfeit 4, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), EIF3F - eukaryotic translation initiation factor 3, subunit f, DNAJC11 - dnaj (hsp40) homolog, subfamily c, member 11, MRPS35 - mitochondrial ribosomal protein s35, MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, HK2 - hexokinase 2, STX17 - syntaxin 17, KPNB1 - karyopherin (importin) beta 1, TJP2 - tight junction protein 2, VDAC2 - voltage-dependent anion channel 2, TTLL12 - tubulin tyrosine ligase-like family, member 12, MRPL13 - mitochondrial ribosomal protein l13, NIPBL - nipped-b homolog (drosophila), TMEM70 - transmembrane protein 70, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), MFN1 - mitofusin 1, WDR1 - wd repeat domain 1, MRPL42 - mitochondrial ribosomal protein l42, RPS6 - ribosomal protein s6, NHP2 - nhp2 ribonucleoprotein, RPS7 - ribosomal protein s7, DSTN - destrin (actin depolymerizing factor), RIC8A - ric8 guanine nucleotide exchange factor a, SUMO1 - small ubiquitin-like modifier 1, CISD2 - cdgsh iron sulfur domain 2, CAT - catalase, USP15 - ubiquitin specific peptidase 15, UGDH - udp-glucose 6-dehydrogenase, NDUFAF4 - nadh dehydrogenase (ubiquinone) complex i, assembly factor 4, SSBP1 - single-stranded dna binding protein 1, mitochondrial, SAMM50 - samm50 sorting and assembly machinery component, MRPL47 - mitochondrial ribosomal protein l47, TUBGCP3 - tubulin, gamma complex associated protein 3, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, PRKCD - protein kinase c, delta, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, CCDC6 - coiled-coil domain containing 6, EPS15 - epidermal growth factor receptor pathway substrate 15, RRM1 - ribonucleotide reductase m1, DDX31 - dead (asp-glu-ala-asp) box polypeptide 31, DDX6 - dead (asp-glu-ala-asp) box helicase 6, SDAD1 - sda1 domain containing 1, F11R - f11 receptor, GSPT1 - g1 to s phase transition 1, GSN - gelsolin, SCFD1 - sec1 family domain containing 1, RAB30 - rab30, member ras oncogene family, FLNB - filamin b, beta, PPFIA1 - protein tyrosine phosphatase, receptor type, f polypeptide (ptprf), interacting protein (liprin), alpha 1, PPFIBP1 - ptprf interacting protein, binding protein 1 (liprin beta 1), DBN1 - drebrin 1, ANKRD28 - ankyrin repeat domain 28, RIF1 - rap1 interacting factor homolog (yeast), CYCS - cytochrome c, somatic, VTI1A - vesicle transport through interaction with t-snares 1a, EHBP1 - eh domain binding protein 1, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, AGPS - alkylglycerone phosphate synthase, GATAD2B - gata zinc finger domain containing 2b, TMED5 - transmembrane emp24 protein transport domain containing 5, RAB23 - rab23, member ras oncogene family, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, GAR1 - gar1 ribonucleoprotein, KDM3A - lysine (k)-specific demethylase 3a, MRPL3 - mitochondrial ribosomal protein l3, TELO2 - telomere maintenance 2, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), MCM7 - minichromosome maintenance complex component 7, ATG7 - autophagy related 7, MCU - mitochondrial calcium uniporter, ARL1 - adp-ribosylation factor-like 1, TUBGCP4 - tubulin, gamma complex associated protein 4, SYNE1 - spectrin repeat containing, nuclear envelope 1, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), PELO - pelota homolog (drosophila), SCO1 - sco1 cytochrome c oxidase assembly protein, NOM1 - nucleolar protein with mif4g domain 1, NFU1 - nfu1 iron-sulfur cluster scaffold homolog (s. cerevisiae), ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), MRPL2 - mitochondrial ribosomal protein l2, TBCD - tubulin folding cofactor d, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, CUTC - cutc copper transporter, TBCA - tubulin folding cofactor a, STX18 - syntaxin 18, RPTOR - regulatory associated protein of mtor, complex 1, EIF3E - eukaryotic translation initiation factor 3, subunit e, LYRM7 - lyr motif containing 7, SERBP1 - serpine1 mrna binding protein 1, VTA1 - vesicle (multivesicular body) trafficking 1, PURA - purine-rich element binding protein a, GAPVD1 - gtpase activating protein and vps9 domains 1, SPG11 - spastic paraplegia 11 (autosomal recessive), ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, NEK6 - nima-related kinase 6, MAP4 - microtubule-associated protein 4, RAB14 - rab14, member ras oncogene family, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, MAN2A1 - mannosidase, alpha, class 2a, member 1, USP8 - ubiquitin specific peptidase 8, EIF4H - eukaryotic translation initiation factor 4h, TRAPPC10 - trafficking protein particle complex 10, MAT2A - methionine adenosyltransferase ii, alpha, MCM3 - minichromosome maintenance complex component 3, IDE - insulin-degrading enzyme, CHMP5 - charged multivesicular body protein 5, DOCK7 - dedicator of cytokinesis 7, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), POLDIP2 - polymerase (dna-directed), delta interacting protein 2, SART3 - squamous cell carcinoma antigen recognized by t cells 3, CTSC - cathepsin c, WIPF1 - was/wasl interacting protein family, member 1, DLG1 - discs, large homolog 1 (drosophila), PARVG - parvin, gamma, RPL14 - ribosomal protein l14, AKAP8L - a kinase (prka) anchor protein 8-like, GNPAT - glyceronephosphate o-acyltransferase, CYFIP2 - cytoplasmic fmr1 interacting protein 2, TRAPPC9 - trafficking protein particle complex 9, CLNS1A - chloride channel, nucleotide-sensitive, 1a, GOLPH3 - golgi phosphoprotein 3 (coat-protein), RB1 - retinoblastoma 1, CBX3 - chromobox homolog 3, FAM49B - family with sequence similarity 49, member b, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), RHOT1 - ras homolog family member t1, MRPS14 - mitochondrial ribosomal protein s14, CUL1 - cullin 1, BCL2L1 - bcl2-like 1, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide]
122 [AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, TMEM30A - transmembrane protein 30a, VTA1 - vesicle (multivesicular body) trafficking 1, PURA - purine-rich element binding protein a, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, QKI - qki, kh domain containing, rna binding, NCBP2 - nuclear cap binding protein subunit 2, 20kda, RPL23A - ribosomal protein l23a, SDAD1 - sda1 domain containing 1, RPL22 - ribosomal protein l22, SCFD1 - sec1 family domain containing 1, AGK - acylglycerol kinase, RPL13 - ribosomal protein l13, FABP5 - fatty acid binding protein 5 (psoriasis-associated), TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), TBC1D2B - tbc1 domain family, member 2b, TMED2 - transmembrane emp24 domain trafficking protein 2, NUP50 - nucleoporin 50kda, RPL6 - ribosomal protein l6, HOOK3 - hook microtubule-tethering protein 3, RPL7 - ribosomal protein l7, RAB14 - rab14, member ras oncogene family, PPFIA1 - protein tyrosine phosphatase, receptor type, f polypeptide (ptprf), interacting protein (liprin), alpha 1, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, RPL3 - ribosomal protein l3, DERL1 - derlin 1, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, BTF3 - basic transcription factor 3, XPOT - exportin, trna, VTI1A - vesicle transport through interaction with t-snares 1a, TNPO1 - transportin 1, STX17 - syntaxin 17, ARCN1 - archain 1, EHBP1 - eh domain binding protein 1, KPNB1 - karyopherin (importin) beta 1, AGPS - alkylglycerone phosphate synthase, GOSR2 - golgi snap receptor complex member 2, IDE - insulin-degrading enzyme, TBC1D4 - tbc1 domain family, member 4, CHMP5 - charged multivesicular body protein 5, TMED5 - transmembrane emp24 protein transport domain containing 5, EXOC2 - exocyst complex component 2, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, RAB23 - rab23, member ras oncogene family, EXOC1 - exocyst complex component 1, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, MYO6 - myosin vi, SNX1 - sorting nexin 1, PITRM1 - pitrilysin metallopeptidase 1, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), ATG7 - autophagy related 7, EIF2D - eukaryotic translation initiation factor 2d, ARL1 - adp-ribosylation factor-like 1, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, SEH1L - seh1-like (s. cerevisiae), RPL14 - ribosomal protein l14, GLS - glutaminase, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), GNPAT - glyceronephosphate o-acyltransferase, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), NUP88 - nucleoporin 88kda, GOSR1 - golgi snap receptor complex member 1, RPS8 - ribosomal protein s8, OSBPL3 - oxysterol binding protein-like 3, RPS13 - ribosomal protein s13, RINT1 - rad50 interactor 1, RPS6 - ribosomal protein s6, GOLPH3 - golgi phosphoprotein 3 (coat-protein), RPS7 - ribosomal protein s7, THOC5 - tho complex 5, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, SNX6 - sorting nexin 6, SRP72 - signal recognition particle 72kda, CAT - catalase, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, HSPD1 - heat shock 60kda protein 1 (chaperonin), XPO6 - exportin 6, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), SAMM50 - samm50 sorting and assembly machinery component, ARL6IP5 - adp-ribosylation-like factor 6 interacting protein 5, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, STX8 - syntaxin 8, STX18 - syntaxin 18, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, HSPA4 - heat shock 70kda protein 4, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, RRBP1 - ribosome binding protein 1, EPS15 - epidermal growth factor receptor pathway substrate 15]
123 [SCPEP1 - serine carboxypeptidase 1, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, UQCRB - ubiquinol-cytochrome c reductase binding protein, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, QKI - qki, kh domain containing, rna binding, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, PGM1 - phosphoglucomutase 1, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, PGAM1 - phosphoglycerate mutase 1 (brain), AGK - acylglycerol kinase, FABP5 - fatty acid binding protein 5 (psoriasis-associated), BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, ECI1 - enoyl-coa delta isomerase 1, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, DCK - deoxycytidine kinase, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, UCK2 - uridine-cytidine kinase 2, UMPS - uridine monophosphate synthetase, CARS - cysteinyl-trna synthetase, VKORC1L1 - vitamin k epoxide reductase complex, subunit 1-like 1, MAN2A1 - mannosidase, alpha, class 2a, member 1, ACAT1 - acetyl-coa acetyltransferase 1, DUT - deoxyuridine triphosphatase, HK2 - hexokinase 2, AK4 - adenylate kinase 4, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, MAT2A - methionine adenosyltransferase ii, alpha, AGPS - alkylglycerone phosphate synthase, SORD - sorbitol dehydrogenase, PLCG1 - phospholipase c, gamma 1, RAB23 - rab23, member ras oncogene family, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, TJP2 - tight junction protein 2, KDM3A - lysine (k)-specific demethylase 3a, DCAKD - dephospho-coa kinase domain containing, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, LSS - lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase), SGPL1 - sphingosine-1-phosphate lyase 1, DLG1 - discs, large homolog 1 (drosophila), GLS - glutaminase, GNPAT - glyceronephosphate o-acyltransferase, IARS - isoleucyl-trna synthetase, GLO1 - glyoxalase i, MFN1 - mitofusin 1, DGUOK - deoxyguanosine kinase, ESD - esterase d, HADH - hydroxyacyl-coa dehydrogenase, OSBPL3 - oxysterol binding protein-like 3, ADPGK - adp-dependent glucokinase, PPA1 - pyrophosphatase (inorganic) 1, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, GCDH - glutaryl-coa dehydrogenase, ACOT7 - acyl-coa thioesterase 7, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, CAT - catalase, MUT - methylmalonyl coa mutase, UGDH - udp-glucose 6-dehydrogenase, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, GNPDA2 - glucosamine-6-phosphate deaminase 2, MVD - mevalonate (diphospho) decarboxylase, ADI1 - acireductone dioxygenase 1, INPP5B - inositol polyphosphate-5-phosphatase, 75kda, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, KYNU - kynureninase, NAMPT - nicotinamide phosphoribosyltransferase, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, PGM2 - phosphoglucomutase 2, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, RRM1 - ribonucleotide reductase m1, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
124 [FAM136A - family with sequence similarity 136, member a, DST - dystonin, ERAP1 - endoplasmic reticulum aminopeptidase 1, NCBP2 - nuclear cap binding protein subunit 2, 20kda, DDX6 - dead (asp-glu-ala-asp) box helicase 6, EXOSC5 - exosome component 5, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, ZNF638 - zinc finger protein 638, GSN - gelsolin, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, FLNB - filamin b, beta, HOOK3 - hook microtubule-tethering protein 3, RECQL - recq protein-like (dna helicase q1-like), PPFIA1 - protein tyrosine phosphatase, receptor type, f polypeptide (ptprf), interacting protein (liprin), alpha 1, DCK - deoxycytidine kinase, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, PPP4R2 - protein phosphatase 4, regulatory subunit 2, DBN1 - drebrin 1, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, ILF3 - interleukin enhancer binding factor 3, 90kda, XPOT - exportin, trna, DOCK8 - dedicator of cytokinesis 8, NUDCD2 - nudc domain containing 2, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, RIF1 - rap1 interacting factor homolog (yeast), PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, RBM19 - rna binding motif protein 19, CPOX - coproporphyrinogen oxidase, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, LRRC40 - leucine rich repeat containing 40, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, LRRK1 - leucine-rich repeat kinase 1, PA2G4 - proliferation-associated 2g4, 38kda, EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, PLCG1 - phospholipase c, gamma 1, LIMS1 - lim and senescent cell antigen-like domains 1, RAB23 - rab23, member ras oncogene family, WDR77 - wd repeat domain 77, CARHSP1 - calcium regulated heat stable protein 1, 24kda, KDM3A - lysine (k)-specific demethylase 3a, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, SNX1 - sorting nexin 1, MYO6 - myosin vi, EXOSC1 - exosome component 1, TELO2 - telomere maintenance 2, ATG7 - autophagy related 7, EIF2D - eukaryotic translation initiation factor 2d, ARL1 - adp-ribosylation factor-like 1, HDDC2 - hd domain containing 2, PRRC1 - proline-rich coiled-coil 1, ANAPC7 - anaphase promoting complex subunit 7, STK38L - serine/threonine kinase 38 like, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, SYNE1 - spectrin repeat containing, nuclear envelope 1, IARS - isoleucyl-trna synthetase, GLO1 - glyoxalase i, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), PELO - pelota homolog (drosophila), WDHD1 - wd repeat and hmg-box dna binding protein 1, COMMD10 - comm domain containing 10, HADH - hydroxyacyl-coa dehydrogenase, EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, DCAF7 - ddb1 and cul4 associated factor 7, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), PPA1 - pyrophosphatase (inorganic) 1, THOC5 - tho complex 5, EIF2S2 - eukaryotic translation initiation factor 2, subunit 2 beta, 38kda, SUMO3 - small ubiquitin-like modifier 3, CDK6 - cyclin-dependent kinase 6, ADNP - activity-dependent neuroprotector homeobox, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, SNX6 - sorting nexin 6, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, MUT - methylmalonyl coa mutase, TBCD - tubulin folding cofactor d, HSPD1 - heat shock 60kda protein 1 (chaperonin), LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, GNPDA2 - glucosamine-6-phosphate deaminase 2, TBCA - tubulin folding cofactor a, CUTC - cutc copper transporter, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, C15orf40 - chromosome 15 open reading frame 40, IRF3 - interferon regulatory factor 3, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, KYNU - kynureninase, TAOK3 - tao kinase 3, IRF5 - interferon regulatory factor 5, LARP1 - la ribonucleoprotein domain family, member 1, EIF3E - eukaryotic translation initiation factor 3, subunit e, RPTOR - regulatory associated protein of mtor, complex 1, PARP4 - poly (adp-ribose) polymerase family, member 4, PURA - purine-rich element binding protein a, SERBP1 - serpine1 mrna binding protein 1, QKI - qki, kh domain containing, rna binding, SPG11 - spastic paraplegia 11 (autosomal recessive), RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, CAPN2 - calpain 2, (m/ii) large subunit, NEK6 - nima-related kinase 6, TDRD7 - tudor domain containing 7, WNK1 - wnk lysine deficient protein kinase 1, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), HMGB2 - high mobility group box 2, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, AGK - acylglycerol kinase, FABP5 - fatty acid binding protein 5 (psoriasis-associated), PYGB - phosphorylase, glycogen; brain, RPL7 - ribosomal protein l7, HOMER1 - homer homolog 1 (drosophila), FTH1 - ferritin, heavy polypeptide 1, RPL3 - ribosomal protein l3, UMPS - uridine monophosphate synthetase, CARS - cysteinyl-trna synthetase, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), IRAK4 - interleukin-1 receptor-associated kinase 4, USP8 - ubiquitin specific peptidase 8, MBNL1 - muscleblind-like splicing regulator 1, TNPO1 - transportin 1, AK4 - adenylate kinase 4, KPNB1 - karyopherin (importin) beta 1, TRIM38 - tripartite motif containing 38, IDE - insulin-degrading enzyme, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, CDC23 - cell division cycle 23, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), IFIH1 - interferon induced with helicase c domain 1, EXOC1 - exocyst complex component 1, SART3 - squamous cell carcinoma antigen recognized by t cells 3, TTLL12 - tubulin tyrosine ligase-like family, member 12, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, SCAF8 - sr-related ctd-associated factor 8, UBA6 - ubiquitin-like modifier activating enzyme 6, RANBP1 - ran binding protein 1, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, DLG1 - discs, large homolog 1 (drosophila), RAP1A - rap1a, member of ras oncogene family, PARVG - parvin, gamma, NSMAF - neutral sphingomyelinase (n-smase) activation associated factor, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), AKAP8L - a kinase (prka) anchor protein 8-like, CYFIP2 - cytoplasmic fmr1 interacting protein 2, FCRLA - fc receptor-like a, DGUOK - deoxyguanosine kinase, UBA3 - ubiquitin-like modifier activating enzyme 3, DSTN - destrin (actin depolymerizing factor), RIC8A - ric8 guanine nucleotide exchange factor a, UCHL3 - ubiquitin carboxyl-terminal esterase l3 (ubiquitin thiolesterase), DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, BZW1 - basic leucine zipper and w2 domains 1, CAT - catalase, USP15 - ubiquitin specific peptidase 15, XPO6 - exportin 6, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, ADI1 - acireductone dioxygenase 1, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, CAPRIN1 - cell cycle associated protein 1, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), TUBGCP3 - tubulin, gamma complex associated protein 3, ANXA4 - annexin a4, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, UBE2I - ubiquitin-conjugating enzyme e2i, AIDA - axin interactor, dorsalization associated, TNFAIP8 - tumor necrosis factor, alpha-induced protein 8, PRKCD - protein kinase c, delta, BCL2L1 - bcl2-like 1, AK3 - adenylate kinase 3, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, EPS15 - epidermal growth factor receptor pathway substrate 15]
125 [MRPS35 - mitochondrial ribosomal protein s35, UQCRB - ubiquinol-cytochrome c reductase binding protein, MRPL2 - mitochondrial ribosomal protein l2, MRPL13 - mitochondrial ribosomal protein l13, MCU - mitochondrial calcium uniporter, MRPL9 - mitochondrial ribosomal protein l9, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), AGK - acylglycerol kinase, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, NDUFA5 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, MFN1 - mitofusin 1, SAMM50 - samm50 sorting and assembly machinery component, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), MRPS14 - mitochondrial ribosomal protein s14, MRPL47 - mitochondrial ribosomal protein l47, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, MRPL42 - mitochondrial ribosomal protein l42, MRPL3 - mitochondrial ribosomal protein l3, DNAJC11 - dnaj (hsp40) homolog, subfamily c, member 11]
126 [DGUOK - deoxyguanosine kinase, AK4 - adenylate kinase 4, PGM1 - phosphoglucomutase 1, HK2 - hexokinase 2, PGAM1 - phosphoglycerate mutase 1 (brain), IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, AK3 - adenylate kinase 3, UCK2 - uridine-cytidine kinase 2, UMPS - uridine monophosphate synthetase, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, ADPGK - adp-dependent glucokinase]
127 [UQCRB - ubiquinol-cytochrome c reductase binding protein, DUT - deoxyuridine triphosphatase, AK4 - adenylate kinase 4, PGM1 - phosphoglucomutase 1, HK2 - hexokinase 2, PGAM1 - phosphoglycerate mutase 1 (brain), NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, MFN1 - mitofusin 1, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, DGUOK - deoxyguanosine kinase, RAB23 - rab23, member ras oncogene family, AK3 - adenylate kinase 3, UCK2 - uridine-cytidine kinase 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, ADPGK - adp-dependent glucokinase]
128 [DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, UQCRB - ubiquinol-cytochrome c reductase binding protein, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, PGM1 - phosphoglucomutase 1, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, SACM1L - sac1 suppressor of actin mutations 1-like (yeast), PGAM1 - phosphoglycerate mutase 1 (brain), AGK - acylglycerol kinase, FABP5 - fatty acid binding protein 5 (psoriasis-associated), MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, PCYT2 - phosphate cytidylyltransferase 2, ethanolamine, RAB14 - rab14, member ras oncogene family, DCK - deoxycytidine kinase, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, UCK2 - uridine-cytidine kinase 2, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, UMPS - uridine monophosphate synthetase, ACAT1 - acetyl-coa acetyltransferase 1, DUT - deoxyuridine triphosphatase, AK4 - adenylate kinase 4, HK2 - hexokinase 2, SORD - sorbitol dehydrogenase, PLCG1 - phospholipase c, gamma 1, RAB23 - rab23, member ras oncogene family, TJP2 - tight junction protein 2, DCAKD - dephospho-coa kinase domain containing, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, DLG1 - discs, large homolog 1 (drosophila), PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, GNPAT - glyceronephosphate o-acyltransferase, MFN1 - mitofusin 1, DGUOK - deoxyguanosine kinase, ADPGK - adp-dependent glucokinase, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, GCDH - glutaryl-coa dehydrogenase, ACOT7 - acyl-coa thioesterase 7, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, MVD - mevalonate (diphospho) decarboxylase, INPP5B - inositol polyphosphate-5-phosphatase, 75kda, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, KYNU - kynureninase, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, NAMPT - nicotinamide phosphoribosyltransferase, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, AK3 - adenylate kinase 3, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, PGM2 - phosphoglucomutase 2, RRM1 - ribonucleotide reductase m1]
129 [ACAT1 - acetyl-coa acetyltransferase 1, AK4 - adenylate kinase 4, HK2 - hexokinase 2, PGM1 - phosphoglucomutase 1, GCDH - glutaryl-coa dehydrogenase, ACOT7 - acyl-coa thioesterase 7, PGAM1 - phosphoglycerate mutase 1 (brain), DGUOK - deoxyguanosine kinase, UCK2 - uridine-cytidine kinase 2, AK3 - adenylate kinase 3, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, UMPS - uridine monophosphate synthetase, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, DCAKD - dephospho-coa kinase domain containing, ADPGK - adp-dependent glucokinase]
130 [UQCRB - ubiquinol-cytochrome c reductase binding protein, MRPL13 - mitochondrial ribosomal protein l13, MCU - mitochondrial calcium uniporter, TMEM70 - transmembrane protein 70, TMEM14C - transmembrane protein 14c, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), AGK - acylglycerol kinase, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, NDUFA5 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, PTCD3 - pentatricopeptide repeat domain 3, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), MRPL42 - mitochondrial ribosomal protein l42, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, FECH - ferrochelatase, MRPS35 - mitochondrial ribosomal protein s35, CYCS - cytochrome c, somatic, MRPL2 - mitochondrial ribosomal protein l2, MRPL1 - mitochondrial ribosomal protein l1, CPOX - coproporphyrinogen oxidase, MRPL9 - mitochondrial ribosomal protein l9, HSPD1 - heat shock 60kda protein 1 (chaperonin), NDUFAF4 - nadh dehydrogenase (ubiquinone) complex i, assembly factor 4, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, MRPS14 - mitochondrial ribosomal protein s14, MRPL47 - mitochondrial ribosomal protein l47, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, BCL2L1 - bcl2-like 1, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, MRPL3 - mitochondrial ribosomal protein l3]
131 [UQCRB - ubiquinol-cytochrome c reductase binding protein, DLG1 - discs, large homolog 1 (drosophila), AK4 - adenylate kinase 4, PGM1 - phosphoglucomutase 1, HK2 - hexokinase 2, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, PGAM1 - phosphoglycerate mutase 1 (brain), NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, DGUOK - deoxyguanosine kinase, TJP2 - tight junction protein 2, UCK2 - uridine-cytidine kinase 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, AK3 - adenylate kinase 3, UMPS - uridine monophosphate synthetase, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, ADPGK - adp-dependent glucokinase]
132 [PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, ACAT1 - acetyl-coa acetyltransferase 1, AK4 - adenylate kinase 4, HK2 - hexokinase 2, PGM1 - phosphoglucomutase 1, GCDH - glutaryl-coa dehydrogenase, ACOT7 - acyl-coa thioesterase 7, PGAM1 - phosphoglycerate mutase 1 (brain), MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, DGUOK - deoxyguanosine kinase, DCK - deoxycytidine kinase, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, AK3 - adenylate kinase 3, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, ADPGK - adp-dependent glucokinase, DCAKD - dephospho-coa kinase domain containing]
133 [TMEM30A - transmembrane protein 30a, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, PURA - purine-rich element binding protein a, VTA1 - vesicle (multivesicular body) trafficking 1, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, RPL23A - ribosomal protein l23a, SDAD1 - sda1 domain containing 1, RPL22 - ribosomal protein l22, F11R - f11 receptor, SCFD1 - sec1 family domain containing 1, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, AGK - acylglycerol kinase, RPL13 - ribosomal protein l13, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), TBC1D2B - tbc1 domain family, member 2b, TMED2 - transmembrane emp24 domain trafficking protein 2, NUP50 - nucleoporin 50kda, RPL6 - ribosomal protein l6, HOOK3 - hook microtubule-tethering protein 3, RPL7 - ribosomal protein l7, RAB14 - rab14, member ras oncogene family, RPL3 - ribosomal protein l3, DERL1 - derlin 1, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, BTF3 - basic transcription factor 3, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), VTI1A - vesicle transport through interaction with t-snares 1a, STX17 - syntaxin 17, HK2 - hexokinase 2, TNPO1 - transportin 1, ARCN1 - archain 1, EHBP1 - eh domain binding protein 1, KPNB1 - karyopherin (importin) beta 1, AGPS - alkylglycerone phosphate synthase, GOSR2 - golgi snap receptor complex member 2, IDE - insulin-degrading enzyme, TBC1D4 - tbc1 domain family, member 4, CHMP5 - charged multivesicular body protein 5, TMED5 - transmembrane emp24 protein transport domain containing 5, EXOC2 - exocyst complex component 2, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, RAB23 - rab23, member ras oncogene family, LIMS1 - lim and senescent cell antigen-like domains 1, EXOC1 - exocyst complex component 1, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, MYO6 - myosin vi, SNX1 - sorting nexin 1, PITRM1 - pitrilysin metallopeptidase 1, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), ATG7 - autophagy related 7, EIF2D - eukaryotic translation initiation factor 2d, ARL1 - adp-ribosylation factor-like 1, DLG1 - discs, large homolog 1 (drosophila), NIPBL - nipped-b homolog (drosophila), RAP1A - rap1a, member of ras oncogene family, GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, SEH1L - seh1-like (s. cerevisiae), RPL14 - ribosomal protein l14, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), GNPAT - glyceronephosphate o-acyltransferase, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), NUP88 - nucleoporin 88kda, GOSR1 - golgi snap receptor complex member 1, RPS8 - ribosomal protein s8, RPS13 - ribosomal protein s13, RINT1 - rad50 interactor 1, RPS6 - ribosomal protein s6, GOLPH3 - golgi phosphoprotein 3 (coat-protein), RPS7 - ribosomal protein s7, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, SNX6 - sorting nexin 6, SUMO1 - small ubiquitin-like modifier 1, SRP72 - signal recognition particle 72kda, RB1 - retinoblastoma 1, CAT - catalase, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, HSPD1 - heat shock 60kda protein 1 (chaperonin), XPO6 - exportin 6, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), SAMM50 - samm50 sorting and assembly machinery component, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, STX8 - syntaxin 8, STX18 - syntaxin 18, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, HSPA4 - heat shock 70kda protein 4, RRBP1 - ribosome binding protein 1, EPS15 - epidermal growth factor receptor pathway substrate 15]
134 [CCNK - cyclin k, NCBP2 - nuclear cap binding protein subunit 2, 20kda, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, TCOF1 - treacher collins-franceschetti syndrome 1, RPL23A - ribosomal protein l23a, RFC3 - replication factor c (activator 1) 3, 38kda, RPL22 - ribosomal protein l22, PGM1 - phosphoglucomutase 1, GSPT1 - g1 to s phase transition 1, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, RPL13 - ribosomal protein l13, GFM2 - g elongation factor, mitochondrial 2, PTCD3 - pentatricopeptide repeat domain 3, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, RPL3 - ribosomal protein l3, PPP5C - protein phosphatase 5, catalytic subunit, BTF3 - basic transcription factor 3, TRIM25 - tripartite motif containing 25, MRPS35 - mitochondrial ribosomal protein s35, DUT - deoxyuridine triphosphatase, MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, MCM3 - minichromosome maintenance complex component 3, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, GAR1 - gar1 ribonucleoprotein, RPL31 - ribosomal protein l31, CREB1 - camp responsive element binding protein 1, RPL32 - ribosomal protein l32, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, MRPL3 - mitochondrial ribosomal protein l3, TELO2 - telomere maintenance 2, SCAF8 - sr-related ctd-associated factor 8, MCM7 - minichromosome maintenance complex component 7, MRPL13 - mitochondrial ribosomal protein l13, SNRPE - small nuclear ribonucleoprotein polypeptide e, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, RPL14 - ribosomal protein l14, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), RNMT - rna (guanine-7-) methyltransferase, RPS8 - ribosomal protein s8, MRPL42 - mitochondrial ribosomal protein l42, WDHD1 - wd repeat and hmg-box dna binding protein 1, RPS13 - ribosomal protein s13, POLR1C - polymerase (rna) i polypeptide c, 30kda, RPS6 - ribosomal protein s6, GOLPH3 - golgi phosphoprotein 3 (coat-protein), NHP2 - nhp2 ribonucleoprotein, RPS7 - ribosomal protein s7, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, TSFM - ts translation elongation factor, mitochondrial, CDK13 - cyclin-dependent kinase 13, UGDH - udp-glucose 6-dehydrogenase, SSBP1 - single-stranded dna binding protein 1, mitochondrial, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), MRPL47 - mitochondrial ribosomal protein l47, MRPS14 - mitochondrial ribosomal protein s14, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, NEMF - nuclear export mediator factor, RRBP1 - ribosome binding protein 1, PGM2 - phosphoglucomutase 2, RRM1 - ribonucleotide reductase m1, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1, TCEA1 - transcription elongation factor a (sii), 1]
135 [PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, ACAT1 - acetyl-coa acetyltransferase 1, AK4 - adenylate kinase 4, HK2 - hexokinase 2, PGM1 - phosphoglucomutase 1, GCDH - glutaryl-coa dehydrogenase, ACOT7 - acyl-coa thioesterase 7, PGAM1 - phosphoglycerate mutase 1 (brain), MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, DGUOK - deoxyguanosine kinase, DCK - deoxycytidine kinase, AK3 - adenylate kinase 3, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, DCAKD - dephospho-coa kinase domain containing, ADPGK - adp-dependent glucokinase]
136 [MRPL13 - mitochondrial ribosomal protein l13, MRPL9 - mitochondrial ribosomal protein l9, RPL13 - ribosomal protein l13, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, PTCD3 - pentatricopeptide repeat domain 3, MRPS14 - mitochondrial ribosomal protein s14, LARP4B - la ribonucleoprotein domain family, member 4b, RPL6 - ribosomal protein l6, RPS13 - ribosomal protein s13, LARP1 - la ribonucleoprotein domain family, member 1, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, RRBP1 - ribosome binding protein 1, BTF3 - basic transcription factor 3, RPS7 - ribosomal protein s7]
137 [MRPL2 - mitochondrial ribosomal protein l2, MRPL13 - mitochondrial ribosomal protein l13, MRPL1 - mitochondrial ribosomal protein l1, RPL23A - ribosomal protein l23a, MRPL9 - mitochondrial ribosomal protein l9, RPL22 - ribosomal protein l22, RPL14 - ribosomal protein l14, RPL13 - ribosomal protein l13, RPL7L1 - ribosomal protein l7-like 1, MRPL47 - mitochondrial ribosomal protein l47, RPL6 - ribosomal protein l6, MRPL42 - mitochondrial ribosomal protein l42, RPL7 - ribosomal protein l7, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, RPL3 - ribosomal protein l3, MRPL3 - mitochondrial ribosomal protein l3]
138 [MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, CAPN2 - calpain 2, (m/ii) large subunit, WNK1 - wnk lysine deficient protein kinase 1, GSN - gelsolin, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, FLNB - filamin b, beta, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, UMPS - uridine monophosphate synthetase, RPL3 - ribosomal protein l3, PPP5C - protein phosphatase 5, catalytic subunit, TRIM25 - tripartite motif containing 25, DOCK8 - dedicator of cytokinesis 8, VKORC1L1 - vitamin k epoxide reductase complex, subunit 1-like 1, USP8 - ubiquitin specific peptidase 8, CYCS - cytochrome c, somatic, RIF1 - rap1 interacting factor homolog (yeast), AK4 - adenylate kinase 4, HK2 - hexokinase 2, MAT2A - methionine adenosyltransferase ii, alpha, TBC1D4 - tbc1 domain family, member 4, OTUB1 - otu domain, ubiquitin aldehyde binding 1, CHMP5 - charged multivesicular body protein 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, PLCG1 - phospholipase c, gamma 1, LIMS1 - lim and senescent cell antigen-like domains 1, IFIH1 - interferon induced with helicase c domain 1, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, CREB1 - camp responsive element binding protein 1, KDM3A - lysine (k)-specific demethylase 3a, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, MCM7 - minichromosome maintenance complex component 7, ATG7 - autophagy related 7, RAP1A - rap1a, member of ras oncogene family, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), RNMT - rna (guanine-7-) methyltransferase, STAT6 - signal transducer and activator of transcription 6, interleukin-4 induced, RAP1B - rap1b, member of ras oncogene family, GOLPH3 - golgi phosphoprotein 3 (coat-protein), ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, FECH - ferrochelatase, SNX6 - sorting nexin 6, SUMO1 - small ubiquitin-like modifier 1, SMARCD1 - swi/snf related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1, RB1 - retinoblastoma 1, CAT - catalase, HSPD1 - heat shock 60kda protein 1 (chaperonin), PTPN2 - protein tyrosine phosphatase, non-receptor type 2, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), IRF3 - interferon regulatory factor 3, STX8 - syntaxin 8, PRKCD - protein kinase c, delta, BCL2L1 - bcl2-like 1, LARP1 - la ribonucleoprotein domain family, member 1, NAMPT - nicotinamide phosphoribosyltransferase, RPTOR - regulatory associated protein of mtor, complex 1, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1]
139 [VTA1 - vesicle (multivesicular body) trafficking 1, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), SPG11 - spastic paraplegia 11 (autosomal recessive), SCFD1 - sec1 family domain containing 1, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), TMED2 - transmembrane emp24 domain trafficking protein 2, TRAPPC9 - trafficking protein particle complex 9, GOSR1 - golgi snap receptor complex member 1, HOOK3 - hook microtubule-tethering protein 3, RAB14 - rab14, member ras oncogene family, GOLPH3 - golgi phosphoprotein 3 (coat-protein), USP8 - ubiquitin specific peptidase 8, ANKRD28 - ankyrin repeat domain 28, TRAPPC10 - trafficking protein particle complex 10, VTI1A - vesicle transport through interaction with t-snares 1a, STX17 - syntaxin 17, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, GOSR2 - golgi snap receptor complex member 2, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), CHMP5 - charged multivesicular body protein 5, STX8 - syntaxin 8, CREB1 - camp responsive element binding protein 1, EPS15 - epidermal growth factor receptor pathway substrate 15, CTSC - cathepsin c]
140 [RNF31 - ring finger protein 31, WNK1 - wnk lysine deficient protein kinase 1, HMGB1 - high mobility group box 1, CYFIP2 - cytoplasmic fmr1 interacting protein 2, EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, IRAK4 - interleukin-1 receptor-associated kinase 4, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, HSPD1 - heat shock 60kda protein 1 (chaperonin), EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, IRF3 - interferon regulatory factor 3, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, CUL1 - cullin 1, PLCG1 - phospholipase c, gamma 1, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, PRKCD - protein kinase c, delta, IFIH1 - interferon induced with helicase c domain 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, WIPF1 - was/wasl interacting protein family, member 1]
141 [MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, ATG7 - autophagy related 7, CAPN2 - calpain 2, (m/ii) large subunit, RAP1A - rap1a, member of ras oncogene family, GSN - gelsolin, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), STAT6 - signal transducer and activator of transcription 6, interleukin-4 induced, HADH - hydroxyacyl-coa dehydrogenase, RAP1B - rap1b, member of ras oncogene family, EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, PPP5C - protein phosphatase 5, catalytic subunit, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), GOLPH3 - golgi phosphoprotein 3 (coat-protein), DERL1 - derlin 1, TRIM25 - tripartite motif containing 25, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, SNX6 - sorting nexin 6, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, CAT - catalase, HSPD1 - heat shock 60kda protein 1 (chaperonin), EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, IRF3 - interferon regulatory factor 3, TBC1D4 - tbc1 domain family, member 4, KYNU - kynureninase, CHMP5 - charged multivesicular body protein 5, LARP1 - la ribonucleoprotein domain family, member 1, PRKCD - protein kinase c, delta, BCL2L1 - bcl2-like 1, IRF5 - interferon regulatory factor 5, NAMPT - nicotinamide phosphoribosyltransferase, IFIH1 - interferon induced with helicase c domain 1, RPTOR - regulatory associated protein of mtor, complex 1, CREB1 - camp responsive element binding protein 1, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha]
142 [AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, TMEM30A - transmembrane protein 30a, VTA1 - vesicle (multivesicular body) trafficking 1, PURA - purine-rich element binding protein a, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, RPL23A - ribosomal protein l23a, SDAD1 - sda1 domain containing 1, RPL22 - ribosomal protein l22, F11R - f11 receptor, SCFD1 - sec1 family domain containing 1, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, AGK - acylglycerol kinase, RPL13 - ribosomal protein l13, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), TBC1D2B - tbc1 domain family, member 2b, TMED2 - transmembrane emp24 domain trafficking protein 2, NUP50 - nucleoporin 50kda, RPL6 - ribosomal protein l6, HOOK3 - hook microtubule-tethering protein 3, RPL7 - ribosomal protein l7, RAB14 - rab14, member ras oncogene family, RPL3 - ribosomal protein l3, DERL1 - derlin 1, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, BTF3 - basic transcription factor 3, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), VTI1A - vesicle transport through interaction with t-snares 1a, STX17 - syntaxin 17, TNPO1 - transportin 1, HK2 - hexokinase 2, ARCN1 - archain 1, EHBP1 - eh domain binding protein 1, KPNB1 - karyopherin (importin) beta 1, AGPS - alkylglycerone phosphate synthase, GOSR2 - golgi snap receptor complex member 2, IDE - insulin-degrading enzyme, TBC1D4 - tbc1 domain family, member 4, CHMP5 - charged multivesicular body protein 5, TMED5 - transmembrane emp24 protein transport domain containing 5, EXOC2 - exocyst complex component 2, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, RAB23 - rab23, member ras oncogene family, LIMS1 - lim and senescent cell antigen-like domains 1, EXOC1 - exocyst complex component 1, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, MYO6 - myosin vi, SNX1 - sorting nexin 1, PITRM1 - pitrilysin metallopeptidase 1, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), ATG7 - autophagy related 7, EIF2D - eukaryotic translation initiation factor 2d, ARL1 - adp-ribosylation factor-like 1, DLG1 - discs, large homolog 1 (drosophila), NIPBL - nipped-b homolog (drosophila), RAP1A - rap1a, member of ras oncogene family, GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, SEH1L - seh1-like (s. cerevisiae), RPL14 - ribosomal protein l14, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), GNPAT - glyceronephosphate o-acyltransferase, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), NUP88 - nucleoporin 88kda, GOSR1 - golgi snap receptor complex member 1, RPS8 - ribosomal protein s8, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RINT1 - rad50 interactor 1, GOLPH3 - golgi phosphoprotein 3 (coat-protein), RPS7 - ribosomal protein s7, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, SNX6 - sorting nexin 6, SUMO1 - small ubiquitin-like modifier 1, SRP72 - signal recognition particle 72kda, RB1 - retinoblastoma 1, CAT - catalase, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, HSPD1 - heat shock 60kda protein 1 (chaperonin), CCT4 - chaperonin containing tcp1, subunit 4 (delta), XPO6 - exportin 6, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), SAMM50 - samm50 sorting and assembly machinery component, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, STX8 - syntaxin 8, STX18 - syntaxin 18, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, HSPA4 - heat shock 70kda protein 4, RRBP1 - ribosome binding protein 1, EPS15 - epidermal growth factor receptor pathway substrate 15]
143 [NCBP2 - nuclear cap binding protein subunit 2, 20kda, EXOSC5 - exosome component 5, RFC3 - replication factor c (activator 1) 3, 38kda, SF3A3 - splicing factor 3a, subunit 3, 60kda, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, GFM2 - g elongation factor, mitochondrial 2, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, RECQL - recq protein-like (dna helicase q1-like), BTF3 - basic transcription factor 3, EIF4B - eukaryotic translation initiation factor 4b, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, SORD - sorbitol dehydrogenase, ZNF326 - zinc finger protein 326, WDR77 - wd repeat domain 77, CARHSP1 - calcium regulated heat stable protein 1, 24kda, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, CREB1 - camp responsive element binding protein 1, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, MYO6 - myosin vi, THUMPD1 - thump domain containing 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, SGPL1 - sphingosine-1-phosphate lyase 1, EIF2D - eukaryotic translation initiation factor 2d, SNRPE - small nuclear ribonucleoprotein polypeptide e, STK38L - serine/threonine kinase 38 like, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, CC2D1B - coiled-coil and c2 domain containing 1b, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), NIP7 - nip7, nucleolar pre-rrna processing protein, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), POLR1C - polymerase (rna) i polypeptide c, 30kda, EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, EIF2S2 - eukaryotic translation initiation factor 2, subunit 2 beta, 38kda, TSFM - ts translation elongation factor, mitochondrial, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), MVD - mevalonate (diphospho) decarboxylase, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, DENND4A - denn/madd domain containing 4a, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, KYNU - kynureninase, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, TAOK3 - tao kinase 3, HSPA4 - heat shock 70kda protein 4, PARP4 - poly (adp-ribose) polymerase family, member 4, QKI - qki, kh domain containing, rna binding, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, WNK1 - wnk lysine deficient protein kinase 1, UBE2Z - ubiquitin-conjugating enzyme e2z, HMGB2 - high mobility group box 2, AGK - acylglycerol kinase, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, PTCD3 - pentatricopeptide repeat domain 3, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, EIF4G3 - eukaryotic translation initiation factor 4 gamma, 3, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, UCK2 - uridine-cytidine kinase 2, PPP5C - protein phosphatase 5, catalytic subunit, RPL3 - ribosomal protein l3, CARS - cysteinyl-trna synthetase, IRAK4 - interleukin-1 receptor-associated kinase 4, EIF3F - eukaryotic translation initiation factor 3, subunit f, MRPS35 - mitochondrial ribosomal protein s35, MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, RPN1 - ribophorin i, AK4 - adenylate kinase 4, TNPO1 - transportin 1, HK2 - hexokinase 2, KPNB1 - karyopherin (importin) beta 1, ABCF2 - atp-binding cassette, sub-family f (gcn20), member 2, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, IFIT5 - interferon-induced protein with tetratricopeptide repeats 5, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, DCAKD - dephospho-coa kinase domain containing, VDAC2 - voltage-dependent anion channel 2, TTLL12 - tubulin tyrosine ligase-like family, member 12, MRPL13 - mitochondrial ribosomal protein l13, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, SCYL2 - scy1-like 2 (s. cerevisiae), STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), MFN1 - mitofusin 1, SNRNP27 - small nuclear ribonucleoprotein 27kda (u4/u6.u5), RNMT - rna (guanine-7-) methyltransferase, STAT6 - signal transducer and activator of transcription 6, interleukin-4 induced, EIF1AD - eukaryotic translation initiation factor 1a domain containing, LARP4B - la ribonucleoprotein domain family, member 4b, RPS8 - ribosomal protein s8, MRPL42 - mitochondrial ribosomal protein l42, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, NHP2 - nhp2 ribonucleoprotein, RPS7 - ribosomal protein s7, SUMO1 - small ubiquitin-like modifier 1, SRP72 - signal recognition particle 72kda, GCDH - glutaryl-coa dehydrogenase, CISD2 - cdgsh iron sulfur domain 2, CAT - catalase, UGDH - udp-glucose 6-dehydrogenase, SSBP1 - single-stranded dna binding protein 1, mitochondrial, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, UBE2I - ubiquitin-conjugating enzyme e2i, PRKCD - protein kinase c, delta, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, RRBP1 - ribosome binding protein 1, RRM1 - ribonucleotide reductase m1, DDX31 - dead (asp-glu-ala-asp) box polypeptide 31, ZC3H7B - zinc finger ccch-type containing 7b, DDX6 - dead (asp-glu-ala-asp) box helicase 6, TCOF1 - treacher collins-franceschetti syndrome 1, ZNF638 - zinc finger protein 638, GSPT1 - g1 to s phase transition 1, URB1 - urb1 ribosome biogenesis 1 homolog (s. cerevisiae), RAB30 - rab30, member ras oncogene family, FAM98A - family with sequence similarity 98, member a, FLNB - filamin b, beta, DCK - deoxycytidine kinase, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, TRIM25 - tripartite motif containing 25, ILF3 - interleukin enhancer binding factor 3, 90kda, XPOT - exportin, trna, CYCS - cytochrome c, somatic, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, RBM19 - rna binding motif protein 19, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), ARCN1 - archain 1, PA2G4 - proliferation-associated 2g4, 38kda, LRRK1 - leucine-rich repeat kinase 1, AGPS - alkylglycerone phosphate synthase, GATAD2B - gata zinc finger domain containing 2b, RAB23 - rab23, member ras oncogene family, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, GAR1 - gar1 ribonucleoprotein, KDM3A - lysine (k)-specific demethylase 3a, MRPL3 - mitochondrial ribosomal protein l3, TELO2 - telomere maintenance 2, EXOSC1 - exosome component 1, MCM7 - minichromosome maintenance complex component 7, ARL1 - adp-ribosylation factor-like 1, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, SYNE1 - spectrin repeat containing, nuclear envelope 1, NOM1 - nucleolar protein with mif4g domain 1, WDHD1 - wd repeat and hmg-box dna binding protein 1, HADH - hydroxyacyl-coa dehydrogenase, THOC5 - tho complex 5, CDK6 - cyclin-dependent kinase 6, ADNP - activity-dependent neuroprotector homeobox, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, CDK13 - cyclin-dependent kinase 13, CCT4 - chaperonin containing tcp1, subunit 4 (delta), FCF1 - fcf1 rrna-processing protein, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, TBCA - tubulin folding cofactor a, RPL7L1 - ribosomal protein l7-like 1, IRF3 - interferon regulatory factor 3, IRF5 - interferon regulatory factor 5, LARP1 - la ribonucleoprotein domain family, member 1, NEMF - nuclear export mediator factor, DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), EIF3E - eukaryotic translation initiation factor 3, subunit e, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, RPTOR - regulatory associated protein of mtor, complex 1, TCEA1 - transcription elongation factor a (sii), 1, PURA - purine-rich element binding protein a, SERBP1 - serpine1 mrna binding protein 1, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, TDRD7 - tudor domain containing 7, NEK6 - nima-related kinase 6, NOC3L - nucleolar complex associated 3 homolog (s. cerevisiae), PYGB - phosphorylase, glycogen; brain, MAP4 - microtubule-associated protein 4, RAB14 - rab14, member ras oncogene family, MBNL1 - muscleblind-like splicing regulator 1, EIF4H - eukaryotic translation initiation factor 4h, DUT - deoxyuridine triphosphatase, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, MAT2A - methionine adenosyltransferase ii, alpha, IDE - insulin-degrading enzyme, MCM3 - minichromosome maintenance complex component 3, IFIH1 - interferon induced with helicase c domain 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), POLDIP2 - polymerase (dna-directed), delta interacting protein 2, SART3 - squamous cell carcinoma antigen recognized by t cells 3, SCAF8 - sr-related ctd-associated factor 8, UBA6 - ubiquitin-like modifier activating enzyme 6, GRK6 - g protein-coupled receptor kinase 6, RAP1A - rap1a, member of ras oncogene family, PUS1 - pseudouridylate synthase 1, RPL14 - ribosomal protein l14, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, AKAP8L - a kinase (prka) anchor protein 8-like, ZNF592 - zinc finger protein 592, RBM26 - rna binding motif protein 26, CLNS1A - chloride channel, nucleotide-sensitive, 1a, DGUOK - deoxyguanosine kinase, UBA3 - ubiquitin-like modifier activating enzyme 3, RAP1B - rap1b, member of ras oncogene family, BZW1 - basic leucine zipper and w2 domains 1, RB1 - retinoblastoma 1, ACOT7 - acyl-coa thioesterase 7, PDAP1 - pdgfa associated protein 1, CAPRIN1 - cell cycle associated protein 1, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), MRPS14 - mitochondrial ribosomal protein s14, RHOT1 - ras homolog family member t1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, RSL1D1 - ribosomal l1 domain containing 1, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
144 [CCNK - cyclin k, DST - dystonin, ERAP1 - endoplasmic reticulum aminopeptidase 1, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, EXOSC5 - exosome component 5, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, RFC3 - replication factor c (activator 1) 3, 38kda, F11R - f11 receptor, SCFD1 - sec1 family domain containing 1, GSN - gelsolin, TMED2 - transmembrane emp24 domain trafficking protein 2, FLNB - filamin b, beta, RECQL - recq protein-like (dna helicase q1-like), IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, DERL1 - derlin 1, ILF3 - interleukin enhancer binding factor 3, 90kda, TRIM25 - tripartite motif containing 25, DOCK8 - dedicator of cytokinesis 8, CYCS - cytochrome c, somatic, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, RIF1 - rap1 interacting factor homolog (yeast), PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, ACAT1 - acetyl-coa acetyltransferase 1, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, CPOX - coproporphyrinogen oxidase, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, MMS22L - mms22-like, dna repair protein, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, CD48 - cd48 molecule, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, CAB39 - calcium binding protein 39, GOSR2 - golgi snap receptor complex member 2, TBC1D4 - tbc1 domain family, member 4, OTUB1 - otu domain, ubiquitin aldehyde binding 1, SORD - sorbitol dehydrogenase, PLCG1 - phospholipase c, gamma 1, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, RAB23 - rab23, member ras oncogene family, LIMS1 - lim and senescent cell antigen-like domains 1, MORF4L1 - mortality factor 4 like 1, RCSD1 - rcsd domain containing 1, KDM3A - lysine (k)-specific demethylase 3a, CREB1 - camp responsive element binding protein 1, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, MYO6 - myosin vi, ATG7 - autophagy related 7, MCM7 - minichromosome maintenance complex component 7, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, SEH1L - seh1-like (s. cerevisiae), PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), WDHD1 - wd repeat and hmg-box dna binding protein 1, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), HADH - hydroxyacyl-coa dehydrogenase, EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, FECH - ferrochelatase, CDK6 - cyclin-dependent kinase 6, ADNP - activity-dependent neuroprotector homeobox, SNX6 - sorting nexin 6, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, SMARCD1 - swi/snf related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1, HSPD1 - heat shock 60kda protein 1 (chaperonin), PTPN2 - protein tyrosine phosphatase, non-receptor type 2, IRF3 - interferon regulatory factor 3, COMT - catechol-o-methyltransferase, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, KYNU - kynureninase, STX8 - syntaxin 8, TAOK3 - tao kinase 3, ERP44 - endoplasmic reticulum protein 44, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), IRF5 - interferon regulatory factor 5, LARP1 - la ribonucleoprotein domain family, member 1, NAMPT - nicotinamide phosphoribosyltransferase, RPTOR - regulatory associated protein of mtor, complex 1, HSPA4 - heat shock 70kda protein 4, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, TCEA1 - transcription elongation factor a (sii), 1, PARP4 - poly (adp-ribose) polymerase family, member 4, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, CAPN2 - calpain 2, (m/ii) large subunit, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, WNK1 - wnk lysine deficient protein kinase 1, HMGB2 - high mobility group box 2, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, HMGB1 - high mobility group box 1, HOMER1 - homer homolog 1 (drosophila), RAB14 - rab14, member ras oncogene family, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, FTH1 - ferritin, heavy polypeptide 1, RPL3 - ribosomal protein l3, UMPS - uridine monophosphate synthetase, PPP5C - protein phosphatase 5, catalytic subunit, VKORC1L1 - vitamin k epoxide reductase complex, subunit 1-like 1, IRAK4 - interleukin-1 receptor-associated kinase 4, USP8 - ubiquitin specific peptidase 8, MRPS35 - mitochondrial ribosomal protein s35, SLAMF1 - signaling lymphocytic activation molecule family member 1, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, HK2 - hexokinase 2, AK4 - adenylate kinase 4, TRIM38 - tripartite motif containing 38, MAT2A - methionine adenosyltransferase ii, alpha, MCM3 - minichromosome maintenance complex component 3, CHMP5 - charged multivesicular body protein 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, IFIH1 - interferon induced with helicase c domain 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, EXOC1 - exocyst complex component 1, IFIT5 - interferon-induced protein with tetratricopeptide repeats 5, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, CTSC - cathepsin c, WIPF1 - was/wasl interacting protein family, member 1, TTLL12 - tubulin tyrosine ligase-like family, member 12, UBA6 - ubiquitin-like modifier activating enzyme 6, NIPBL - nipped-b homolog (drosophila), DLG1 - discs, large homolog 1 (drosophila), RAP1A - rap1a, member of ras oncogene family, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), GNPAT - glyceronephosphate o-acyltransferase, RNMT - rna (guanine-7-) methyltransferase, STAT6 - signal transducer and activator of transcription 6, interleukin-4 induced, NUB1 - negative regulator of ubiquitin-like proteins 1, RAP1B - rap1b, member of ras oncogene family, GOLPH3 - golgi phosphoprotein 3 (coat-protein), RIC8A - ric8 guanine nucleotide exchange factor a, SUMO1 - small ubiquitin-like modifier 1, RB1 - retinoblastoma 1, CAT - catalase, CBX3 - chromobox homolog 3, FAM49B - family with sequence similarity 49, member b, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, CUL1 - cullin 1, PRKCD - protein kinase c, delta, BCL2L1 - bcl2-like 1, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, RRM1 - ribonucleotide reductase m1]
145 [PITRM1 - pitrilysin metallopeptidase 1, TDRD7 - tudor domain containing 7, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, PUS1 - pseudouridylate synthase 1, GLS - glutaminase, GFM2 - g elongation factor, mitochondrial 2, DGUOK - deoxyguanosine kinase, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), HADH - hydroxyacyl-coa dehydrogenase, ECI1 - enoyl-coa delta isomerase 1, FECH - ferrochelatase, METTL15 - methyltransferase like 15, ACAT1 - acetyl-coa acetyltransferase 1, TSFM - ts translation elongation factor, mitochondrial, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, AK4 - adenylate kinase 4, GCDH - glutaryl-coa dehydrogenase, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, SSBP1 - single-stranded dna binding protein 1, mitochondrial, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), BCL2L1 - bcl2-like 1, DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, AK3 - adenylate kinase 3, CREB1 - camp responsive element binding protein 1, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, LYRM7 - lyr motif containing 7]
146 [NCBP2 - nuclear cap binding protein subunit 2, 20kda, EXOSC5 - exosome component 5, RFC3 - replication factor c (activator 1) 3, 38kda, SF3A3 - splicing factor 3a, subunit 3, 60kda, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, GFM2 - g elongation factor, mitochondrial 2, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, RECQL - recq protein-like (dna helicase q1-like), BTF3 - basic transcription factor 3, EIF4B - eukaryotic translation initiation factor 4b, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, SORD - sorbitol dehydrogenase, ZNF326 - zinc finger protein 326, WDR77 - wd repeat domain 77, CARHSP1 - calcium regulated heat stable protein 1, 24kda, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, CREB1 - camp responsive element binding protein 1, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, MYO6 - myosin vi, THUMPD1 - thump domain containing 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, SGPL1 - sphingosine-1-phosphate lyase 1, EIF2D - eukaryotic translation initiation factor 2d, SNRPE - small nuclear ribonucleoprotein polypeptide e, STK38L - serine/threonine kinase 38 like, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, CC2D1B - coiled-coil and c2 domain containing 1b, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), NIP7 - nip7, nucleolar pre-rrna processing protein, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), POLR1C - polymerase (rna) i polypeptide c, 30kda, EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, EIF2S2 - eukaryotic translation initiation factor 2, subunit 2 beta, 38kda, TSFM - ts translation elongation factor, mitochondrial, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), MVD - mevalonate (diphospho) decarboxylase, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, DENND4A - denn/madd domain containing 4a, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, KYNU - kynureninase, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, TAOK3 - tao kinase 3, HSPA4 - heat shock 70kda protein 4, PARP4 - poly (adp-ribose) polymerase family, member 4, QKI - qki, kh domain containing, rna binding, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, WNK1 - wnk lysine deficient protein kinase 1, UBE2Z - ubiquitin-conjugating enzyme e2z, HMGB2 - high mobility group box 2, AGK - acylglycerol kinase, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, PTCD3 - pentatricopeptide repeat domain 3, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, EIF4G3 - eukaryotic translation initiation factor 4 gamma, 3, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, UCK2 - uridine-cytidine kinase 2, PPP5C - protein phosphatase 5, catalytic subunit, RPL3 - ribosomal protein l3, CARS - cysteinyl-trna synthetase, IRAK4 - interleukin-1 receptor-associated kinase 4, EIF3F - eukaryotic translation initiation factor 3, subunit f, MRPS35 - mitochondrial ribosomal protein s35, MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, RPN1 - ribophorin i, AK4 - adenylate kinase 4, TNPO1 - transportin 1, HK2 - hexokinase 2, KPNB1 - karyopherin (importin) beta 1, ABCF2 - atp-binding cassette, sub-family f (gcn20), member 2, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, IFIT5 - interferon-induced protein with tetratricopeptide repeats 5, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, DCAKD - dephospho-coa kinase domain containing, VDAC2 - voltage-dependent anion channel 2, TTLL12 - tubulin tyrosine ligase-like family, member 12, MRPL13 - mitochondrial ribosomal protein l13, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, SCYL2 - scy1-like 2 (s. cerevisiae), STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), MFN1 - mitofusin 1, SNRNP27 - small nuclear ribonucleoprotein 27kda (u4/u6.u5), RNMT - rna (guanine-7-) methyltransferase, STAT6 - signal transducer and activator of transcription 6, interleukin-4 induced, EIF1AD - eukaryotic translation initiation factor 1a domain containing, LARP4B - la ribonucleoprotein domain family, member 4b, RPS8 - ribosomal protein s8, MRPL42 - mitochondrial ribosomal protein l42, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, NHP2 - nhp2 ribonucleoprotein, RPS7 - ribosomal protein s7, SUMO1 - small ubiquitin-like modifier 1, SRP72 - signal recognition particle 72kda, GCDH - glutaryl-coa dehydrogenase, CISD2 - cdgsh iron sulfur domain 2, CAT - catalase, UGDH - udp-glucose 6-dehydrogenase, SSBP1 - single-stranded dna binding protein 1, mitochondrial, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, UBE2I - ubiquitin-conjugating enzyme e2i, PRKCD - protein kinase c, delta, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, RRBP1 - ribosome binding protein 1, RRM1 - ribonucleotide reductase m1, DDX31 - dead (asp-glu-ala-asp) box polypeptide 31, ZC3H7B - zinc finger ccch-type containing 7b, DDX6 - dead (asp-glu-ala-asp) box helicase 6, TCOF1 - treacher collins-franceschetti syndrome 1, ZNF638 - zinc finger protein 638, GSPT1 - g1 to s phase transition 1, URB1 - urb1 ribosome biogenesis 1 homolog (s. cerevisiae), RAB30 - rab30, member ras oncogene family, FAM98A - family with sequence similarity 98, member a, FLNB - filamin b, beta, DCK - deoxycytidine kinase, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, TRIM25 - tripartite motif containing 25, ILF3 - interleukin enhancer binding factor 3, 90kda, XPOT - exportin, trna, CYCS - cytochrome c, somatic, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, RBM19 - rna binding motif protein 19, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), ARCN1 - archain 1, PA2G4 - proliferation-associated 2g4, 38kda, LRRK1 - leucine-rich repeat kinase 1, AGPS - alkylglycerone phosphate synthase, GATAD2B - gata zinc finger domain containing 2b, RAB23 - rab23, member ras oncogene family, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, GAR1 - gar1 ribonucleoprotein, KDM3A - lysine (k)-specific demethylase 3a, MRPL3 - mitochondrial ribosomal protein l3, TELO2 - telomere maintenance 2, EXOSC1 - exosome component 1, MCM7 - minichromosome maintenance complex component 7, ARL1 - adp-ribosylation factor-like 1, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, SYNE1 - spectrin repeat containing, nuclear envelope 1, NOM1 - nucleolar protein with mif4g domain 1, WDHD1 - wd repeat and hmg-box dna binding protein 1, HADH - hydroxyacyl-coa dehydrogenase, OSBPL3 - oxysterol binding protein-like 3, THOC5 - tho complex 5, CDK6 - cyclin-dependent kinase 6, ADNP - activity-dependent neuroprotector homeobox, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, CDK13 - cyclin-dependent kinase 13, CCT4 - chaperonin containing tcp1, subunit 4 (delta), FCF1 - fcf1 rrna-processing protein, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, TBCA - tubulin folding cofactor a, RPL7L1 - ribosomal protein l7-like 1, IRF3 - interferon regulatory factor 3, IRF5 - interferon regulatory factor 5, LARP1 - la ribonucleoprotein domain family, member 1, NEMF - nuclear export mediator factor, DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), EIF3E - eukaryotic translation initiation factor 3, subunit e, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, RPTOR - regulatory associated protein of mtor, complex 1, TCEA1 - transcription elongation factor a (sii), 1, PURA - purine-rich element binding protein a, SERBP1 - serpine1 mrna binding protein 1, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, TDRD7 - tudor domain containing 7, NEK6 - nima-related kinase 6, NOC3L - nucleolar complex associated 3 homolog (s. cerevisiae), PYGB - phosphorylase, glycogen; brain, MAP4 - microtubule-associated protein 4, RAB14 - rab14, member ras oncogene family, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, MBNL1 - muscleblind-like splicing regulator 1, EIF4H - eukaryotic translation initiation factor 4h, DUT - deoxyuridine triphosphatase, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, MAT2A - methionine adenosyltransferase ii, alpha, IDE - insulin-degrading enzyme, MCM3 - minichromosome maintenance complex component 3, IFIH1 - interferon induced with helicase c domain 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), POLDIP2 - polymerase (dna-directed), delta interacting protein 2, SART3 - squamous cell carcinoma antigen recognized by t cells 3, SCAF8 - sr-related ctd-associated factor 8, UBA6 - ubiquitin-like modifier activating enzyme 6, GRK6 - g protein-coupled receptor kinase 6, RAP1A - rap1a, member of ras oncogene family, PUS1 - pseudouridylate synthase 1, RPL14 - ribosomal protein l14, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, AKAP8L - a kinase (prka) anchor protein 8-like, ZNF592 - zinc finger protein 592, RBM26 - rna binding motif protein 26, CLNS1A - chloride channel, nucleotide-sensitive, 1a, DGUOK - deoxyguanosine kinase, UBA3 - ubiquitin-like modifier activating enzyme 3, RAP1B - rap1b, member of ras oncogene family, BZW1 - basic leucine zipper and w2 domains 1, RB1 - retinoblastoma 1, ACOT7 - acyl-coa thioesterase 7, PDAP1 - pdgfa associated protein 1, CAPRIN1 - cell cycle associated protein 1, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), MRPS14 - mitochondrial ribosomal protein s14, RHOT1 - ras homolog family member t1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, RSL1D1 - ribosomal l1 domain containing 1, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
147 [AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, NCBP2 - nuclear cap binding protein subunit 2, 20kda, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, DENND3 - denn/madd domain containing 3, EXOSC5 - exosome component 5, DDX6 - dead (asp-glu-ala-asp) box helicase 6, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, PGM1 - phosphoglucomutase 1, GSPT1 - g1 to s phase transition 1, UBE2Z - ubiquitin-conjugating enzyme e2z, HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, PYGB - phosphorylase, glycogen; brain, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, PHKB - phosphorylase kinase, beta, RPL3 - ribosomal protein l3, USP48 - ubiquitin specific peptidase 48, DERL1 - derlin 1, MANBA - mannosidase, beta a, lysosomal, TRIM25 - tripartite motif containing 25, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), USP8 - ubiquitin specific peptidase 8, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, KPNB1 - karyopherin (importin) beta 1, TRIM38 - tripartite motif containing 38, USP25 - ubiquitin specific peptidase 25, IDE - insulin-degrading enzyme, CDC23 - cell division cycle 23, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, USP34 - ubiquitin specific peptidase 34, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, ANAPC1 - anaphase promoting complex subunit 1, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, EXOSC1 - exosome component 1, UBA6 - ubiquitin-like modifier activating enzyme 6, ATG7 - autophagy related 7, ANAPC7 - anaphase promoting complex subunit 7, RPL14 - ribosomal protein l14, NUB1 - negative regulator of ubiquitin-like proteins 1, PELO - pelota homolog (drosophila), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), RPS8 - ribosomal protein s8, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, UCHL3 - ubiquitin carboxyl-terminal esterase l3 (ubiquitin thiolesterase), HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, USP15 - ubiquitin specific peptidase 15, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, UBE2I - ubiquitin-conjugating enzyme e2i, CUL1 - cullin 1, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), NEMF - nuclear export mediator factor, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), EIF3E - eukaryotic translation initiation factor 3, subunit e, PGM2 - phosphoglucomutase 2]
148 [BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, SACM1L - sac1 suppressor of actin mutations 1-like (yeast), SCFD1 - sec1 family domain containing 1, CISD1 - cdgsh iron sulfur domain 1, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, RAB30 - rab30, member ras oncogene family, NDUFA5 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, TMED2 - transmembrane emp24 domain trafficking protein 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, ANKRD28 - ankyrin repeat domain 28, CYCS - cytochrome c, somatic, RIF1 - rap1 interacting factor homolog (yeast), VTI1A - vesicle transport through interaction with t-snares 1a, CPOX - coproporphyrinogen oxidase, ARCN1 - archain 1, AGPS - alkylglycerone phosphate synthase, GOSR2 - golgi snap receptor complex member 2, SORD - sorbitol dehydrogenase, TMED5 - transmembrane emp24 protein transport domain containing 5, RAB23 - rab23, member ras oncogene family, SNX1 - sorting nexin 1, MYO6 - myosin vi, MRPL3 - mitochondrial ribosomal protein l3, MCU - mitochondrial calcium uniporter, ARL1 - adp-ribosylation factor-like 1, SEH1L - seh1-like (s. cerevisiae), SYNE1 - spectrin repeat containing, nuclear envelope 1, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), GLG1 - golgi glycoprotein 1, GOSR1 - golgi snap receptor complex member 1, OSBPL3 - oxysterol binding protein-like 3, FECH - ferrochelatase, SNX6 - sorting nexin 6, MRPL2 - mitochondrial ribosomal protein l2, HSPD1 - heat shock 60kda protein 1 (chaperonin), ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, INPP5B - inositol polyphosphate-5-phosphatase, 75kda, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, STX18 - syntaxin 18, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), RPTOR - regulatory associated protein of mtor, complex 1, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, QSOX2 - quiescin q6 sulfhydryl oxidase 2, LYRM7 - lyr motif containing 7, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, TMEM30A - transmembrane protein 30a, VTA1 - vesicle (multivesicular body) trafficking 1, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, UQCRB - ubiquinol-cytochrome c reductase binding protein, SPG11 - spastic paraplegia 11 (autosomal recessive), ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, TMEM14C - transmembrane protein 14c, AGK - acylglycerol kinase, FABP5 - fatty acid binding protein 5 (psoriasis-associated), TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), PTCD3 - pentatricopeptide repeat domain 3, NUP50 - nucleoporin 50kda, RAB14 - rab14, member ras oncogene family, MANBA - mannosidase, beta a, lysosomal, SURF4 - surfeit 4, IRAK4 - interleukin-1 receptor-associated kinase 4, MAN2A1 - mannosidase, alpha, class 2a, member 1, TRAPPC10 - trafficking protein particle complex 10, MRPS35 - mitochondrial ribosomal protein s35, MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, STX17 - syntaxin 17, HK2 - hexokinase 2, KPNB1 - karyopherin (importin) beta 1, CHMP5 - charged multivesicular body protein 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), VDAC2 - voltage-dependent anion channel 2, CTSC - cathepsin c, MRPL13 - mitochondrial ribosomal protein l13, SCYL2 - scy1-like 2 (s. cerevisiae), RAP1A - rap1a, member of ras oncogene family, GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, GALNT2 - udp-n-acetyl-alpha-d-galactosamine:polypeptide n-acetylgalactosaminyltransferase 2 (galnac-t2), PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, TMEM70 - transmembrane protein 70, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), GNPAT - glyceronephosphate o-acyltransferase, MFN1 - mitofusin 1, TRAPPC9 - trafficking protein particle complex 9, MRPL42 - mitochondrial ribosomal protein l42, RAP1B - rap1b, member of ras oncogene family, GOLPH3 - golgi phosphoprotein 3 (coat-protein), STOM - stomatin, TMX3 - thioredoxin-related transmembrane protein 3, SUMO1 - small ubiquitin-like modifier 1, CISD2 - cdgsh iron sulfur domain 2, CAT - catalase, CBX3 - chromobox homolog 3, NDUFAF4 - nadh dehydrogenase (ubiquinone) complex i, assembly factor 4, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), SAMM50 - samm50 sorting and assembly machinery component, MRPS14 - mitochondrial ribosomal protein s14, RHOT1 - ras homolog family member t1, MRPL47 - mitochondrial ribosomal protein l47, ANXA4 - annexin a4, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, BCL2L1 - bcl2-like 1, EPS15 - epidermal growth factor receptor pathway substrate 15]
149 [CCNK - cyclin k, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, UQCRB - ubiquinol-cytochrome c reductase binding protein, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, PGM1 - phosphoglucomutase 1, NEK6 - nima-related kinase 6, WNK1 - wnk lysine deficient protein kinase 1, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, SACM1L - sac1 suppressor of actin mutations 1-like (yeast), PGAM1 - phosphoglycerate mutase 1 (brain), PPP1R9B - protein phosphatase 1, regulatory subunit 9b, AGK - acylglycerol kinase, FABP5 - fatty acid binding protein 5 (psoriasis-associated), MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, PCYT2 - phosphate cytidylyltransferase 2, ethanolamine, RAB14 - rab14, member ras oncogene family, PHKB - phosphorylase kinase, beta, DCK - deoxycytidine kinase, UCK2 - uridine-cytidine kinase 2, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, PPP4R2 - protein phosphatase 4, regulatory subunit 2, UMPS - uridine monophosphate synthetase, PPP5C - protein phosphatase 5, catalytic subunit, ILF3 - interleukin enhancer binding factor 3, 90kda, IRAK4 - interleukin-1 receptor-associated kinase 4, CYCS - cytochrome c, somatic, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, ACAT1 - acetyl-coa acetyltransferase 1, DUT - deoxyuridine triphosphatase, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, LRRC40 - leucine rich repeat containing 40, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, HK2 - hexokinase 2, AK4 - adenylate kinase 4, LRRK1 - leucine-rich repeat kinase 1, CAB39 - calcium binding protein 39, SORD - sorbitol dehydrogenase, PLCG1 - phospholipase c, gamma 1, RAB23 - rab23, member ras oncogene family, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, TJP2 - tight junction protein 2, CREB1 - camp responsive element binding protein 1, DCAKD - dephospho-coa kinase domain containing, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, GRK6 - g protein-coupled receptor kinase 6, DLG1 - discs, large homolog 1 (drosophila), HDDC2 - hd domain containing 2, SCYL2 - scy1-like 2 (s. cerevisiae), STK38L - serine/threonine kinase 38 like, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), GNPAT - glyceronephosphate o-acyltransferase, MFN1 - mitofusin 1, DGUOK - deoxyguanosine kinase, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), ADPGK - adp-dependent glucokinase, PPA1 - pyrophosphatase (inorganic) 1, CDK6 - cyclin-dependent kinase 6, ACOT7 - acyl-coa thioesterase 7, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, GCDH - glutaryl-coa dehydrogenase, CDK13 - cyclin-dependent kinase 13, USP15 - ubiquitin specific peptidase 15, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, GNPDA2 - glucosamine-6-phosphate deaminase 2, MVD - mevalonate (diphospho) decarboxylase, UGDH - udp-glucose 6-dehydrogenase, INPP5B - inositol polyphosphate-5-phosphatase, 75kda, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, KYNU - kynureninase, CUL1 - cullin 1, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, TAOK3 - tao kinase 3, PRKCD - protein kinase c, delta, NAMPT - nicotinamide phosphoribosyltransferase, AK3 - adenylate kinase 3, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, PGM2 - phosphoglucomutase 2, RRM1 - ribonucleotide reductase m1]
150 [EIF4H - eukaryotic translation initiation factor 4h, EIF1AD - eukaryotic translation initiation factor 1a domain containing, EIF2D - eukaryotic translation initiation factor 2d, EIF4B - eukaryotic translation initiation factor 4b, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, EIF4G3 - eukaryotic translation initiation factor 4 gamma, 3, EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, EIF3E - eukaryotic translation initiation factor 3, subunit e, EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, EIF3F - eukaryotic translation initiation factor 3, subunit f, EIF2S2 - eukaryotic translation initiation factor 2, subunit 2 beta, 38kda]
151 [PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, RNF31 - ring finger protein 31, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, WNK1 - wnk lysine deficient protein kinase 1, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, CYFIP2 - cytoplasmic fmr1 interacting protein 2, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, CUL1 - cullin 1, PLCG1 - phospholipase c, gamma 1, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, PRKCD - protein kinase c, delta, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, WIPF1 - was/wasl interacting protein family, member 1]
152 [UQCRB - ubiquinol-cytochrome c reductase binding protein, AK4 - adenylate kinase 4, PGM1 - phosphoglucomutase 1, HK2 - hexokinase 2, PGAM1 - phosphoglycerate mutase 1 (brain), NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, MFN1 - mitofusin 1, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, DGUOK - deoxyguanosine kinase, RAB23 - rab23, member ras oncogene family, UCK2 - uridine-cytidine kinase 2, AK3 - adenylate kinase 3, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, ADPGK - adp-dependent glucokinase]
153 [DST - dystonin, DDX6 - dead (asp-glu-ala-asp) box helicase 6, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, SDAD1 - sda1 domain containing 1, RFC3 - replication factor c (activator 1) 3, 38kda, SF3A3 - splicing factor 3a, subunit 3, 60kda, F11R - f11 receptor, GSPT1 - g1 to s phase transition 1, SCFD1 - sec1 family domain containing 1, GSN - gelsolin, RAB30 - rab30, member ras oncogene family, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, GFM2 - g elongation factor, mitochondrial 2, NDUFA5 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, TMED2 - transmembrane emp24 domain trafficking protein 2, FLNB - filamin b, beta, HOOK3 - hook microtubule-tethering protein 3, RECQL - recq protein-like (dna helicase q1-like), PPFIA1 - protein tyrosine phosphatase, receptor type, f polypeptide (ptprf), interacting protein (liprin), alpha 1, PPFIBP1 - ptprf interacting protein, binding protein 1 (liprin beta 1), YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, DBN1 - drebrin 1, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, ANKRD28 - ankyrin repeat domain 28, CYCS - cytochrome c, somatic, RIF1 - rap1 interacting factor homolog (yeast), VTI1A - vesicle transport through interaction with t-snares 1a, EIF4B - eukaryotic translation initiation factor 4b, EHBP1 - eh domain binding protein 1, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, AGPS - alkylglycerone phosphate synthase, GOSR2 - golgi snap receptor complex member 2, TMED5 - transmembrane emp24 protein transport domain containing 5, GATAD2B - gata zinc finger domain containing 2b, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, LIMS1 - lim and senescent cell antigen-like domains 1, RAB23 - rab23, member ras oncogene family, WDR77 - wd repeat domain 77, MORF4L1 - mortality factor 4 like 1, GAR1 - gar1 ribonucleoprotein, KDM3A - lysine (k)-specific demethylase 3a, CREB1 - camp responsive element binding protein 1, MYO6 - myosin vi, SNX1 - sorting nexin 1, MRPL3 - mitochondrial ribosomal protein l3, TELO2 - telomere maintenance 2, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, ATG7 - autophagy related 7, EIF2D - eukaryotic translation initiation factor 2d, MCM7 - minichromosome maintenance complex component 7, MCU - mitochondrial calcium uniporter, TUBGCP4 - tubulin, gamma complex associated protein 4, TPM3 - tropomyosin 3, SNRPE - small nuclear ribonucleoprotein polypeptide e, ARL1 - adp-ribosylation factor-like 1, SEH1L - seh1-like (s. cerevisiae), GLS - glutaminase, SYNE1 - spectrin repeat containing, nuclear envelope 1, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), PELO - pelota homolog (drosophila), SCO1 - sco1 cytochrome c oxidase assembly protein, GOSR1 - golgi snap receptor complex member 1, NIP7 - nip7, nucleolar pre-rrna processing protein, NFU1 - nfu1 iron-sulfur cluster scaffold homolog (s. cerevisiae), ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, EIF2S2 - eukaryotic translation initiation factor 2, subunit 2 beta, 38kda, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, MRPL2 - mitochondrial ribosomal protein l2, SMARCD1 - swi/snf related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1, TBCD - tubulin folding cofactor d, HSPD1 - heat shock 60kda protein 1 (chaperonin), NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, CUTC - cutc copper transporter, TBCA - tubulin folding cofactor a, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, STX8 - syntaxin 8, TAOK3 - tao kinase 3, STX18 - syntaxin 18, EIF3E - eukaryotic translation initiation factor 3, subunit e, RPTOR - regulatory associated protein of mtor, complex 1, HSPA4 - heat shock 70kda protein 4, LYRM7 - lyr motif containing 7, TMEM30A - transmembrane protein 30a, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, PURA - purine-rich element binding protein a, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, SERBP1 - serpine1 mrna binding protein 1, VTA1 - vesicle (multivesicular body) trafficking 1, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, GAPVD1 - gtpase activating protein and vps9 domains 1, SPG11 - spastic paraplegia 11 (autosomal recessive), RPL23A - ribosomal protein l23a, CAPN2 - calpain 2, (m/ii) large subunit, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, NEK6 - nima-related kinase 6, HMGB2 - high mobility group box 2, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, HMGB1 - high mobility group box 1, AGK - acylglycerol kinase, PTCD3 - pentatricopeptide repeat domain 3, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), RPL6 - ribosomal protein l6, MAP4 - microtubule-associated protein 4, HOMER1 - homer homolog 1 (drosophila), RAB14 - rab14, member ras oncogene family, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, RPL3 - ribosomal protein l3, PPP5C - protein phosphatase 5, catalytic subunit, SURF4 - surfeit 4, EIF3F - eukaryotic translation initiation factor 3, subunit f, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), MAN2A1 - mannosidase, alpha, class 2a, member 1, DNAJC11 - dnaj (hsp40) homolog, subfamily c, member 11, USP8 - ubiquitin specific peptidase 8, TRAPPC10 - trafficking protein particle complex 10, EIF4H - eukaryotic translation initiation factor 4h, MRPS35 - mitochondrial ribosomal protein s35, MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, STX17 - syntaxin 17, HK2 - hexokinase 2, KPNB1 - karyopherin (importin) beta 1, MAT2A - methionine adenosyltransferase ii, alpha, IDE - insulin-degrading enzyme, MCM3 - minichromosome maintenance complex component 3, CHMP5 - charged multivesicular body protein 5, DOCK7 - dedicator of cytokinesis 7, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), TJP2 - tight junction protein 2, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, SART3 - squamous cell carcinoma antigen recognized by t cells 3, VDAC2 - voltage-dependent anion channel 2, CTSC - cathepsin c, WIPF1 - was/wasl interacting protein family, member 1, TTLL12 - tubulin tyrosine ligase-like family, member 12, MRPL13 - mitochondrial ribosomal protein l13, NIPBL - nipped-b homolog (drosophila), DLG1 - discs, large homolog 1 (drosophila), PARVG - parvin, gamma, TMEM70 - transmembrane protein 70, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), AKAP8L - a kinase (prka) anchor protein 8-like, GNPAT - glyceronephosphate o-acyltransferase, MFN1 - mitofusin 1, CYFIP2 - cytoplasmic fmr1 interacting protein 2, WDR1 - wd repeat domain 1, CLNS1A - chloride channel, nucleotide-sensitive, 1a, TRAPPC9 - trafficking protein particle complex 9, MRPL42 - mitochondrial ribosomal protein l42, GOLPH3 - golgi phosphoprotein 3 (coat-protein), NHP2 - nhp2 ribonucleoprotein, DSTN - destrin (actin depolymerizing factor), RIC8A - ric8 guanine nucleotide exchange factor a, SUMO1 - small ubiquitin-like modifier 1, RB1 - retinoblastoma 1, CISD2 - cdgsh iron sulfur domain 2, CAT - catalase, USP15 - ubiquitin specific peptidase 15, UGDH - udp-glucose 6-dehydrogenase, CBX3 - chromobox homolog 3, NDUFAF4 - nadh dehydrogenase (ubiquinone) complex i, assembly factor 4, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, FAM49B - family with sequence similarity 49, member b, SSBP1 - single-stranded dna binding protein 1, mitochondrial, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), SAMM50 - samm50 sorting and assembly machinery component, MRPS14 - mitochondrial ribosomal protein s14, TUBGCP3 - tubulin, gamma complex associated protein 3, MRPL47 - mitochondrial ribosomal protein l47, RHOT1 - ras homolog family member t1, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, CUL1 - cullin 1, PRKCD - protein kinase c, delta, BCL2L1 - bcl2-like 1, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, CCDC6 - coiled-coil domain containing 6, EPS15 - epidermal growth factor receptor pathway substrate 15, RRM1 - ribonucleotide reductase m1]
154 [UQCRB - ubiquinol-cytochrome c reductase binding protein, DLG1 - discs, large homolog 1 (drosophila), AK4 - adenylate kinase 4, PGM1 - phosphoglucomutase 1, HK2 - hexokinase 2, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, PGAM1 - phosphoglycerate mutase 1 (brain), NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, DGUOK - deoxyguanosine kinase, DCK - deoxycytidine kinase, TJP2 - tight junction protein 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, AK3 - adenylate kinase 3, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, ADPGK - adp-dependent glucokinase]
155 [SERBP1 - serpine1 mrna binding protein 1, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, EXOSC5 - exosome component 5, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, TNPO1 - transportin 1, LARP4B - la ribonucleoprotein domain family, member 4b, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, PRKCD - protein kinase c, delta, LARP1 - la ribonucleoprotein domain family, member 1, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, CARHSP1 - calcium regulated heat stable protein 1, 24kda, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, EXOSC1 - exosome component 1]
156 [PITRM1 - pitrilysin metallopeptidase 1, TMEM30A - transmembrane protein 30a, RPL23A - ribosomal protein l23a, ARL1 - adp-ribosylation factor-like 1, RPL22 - ribosomal protein l22, NIPBL - nipped-b homolog (drosophila), DLG1 - discs, large homolog 1 (drosophila), GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, RPL14 - ribosomal protein l14, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), PPP1R9B - protein phosphatase 1, regulatory subunit 9b, GNPAT - glyceronephosphate o-acyltransferase, AGK - acylglycerol kinase, RPL13 - ribosomal protein l13, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), NUP88 - nucleoporin 88kda, RPS8 - ribosomal protein s8, NUP50 - nucleoporin 50kda, RPL6 - ribosomal protein l6, HOOK3 - hook microtubule-tethering protein 3, RPL7 - ribosomal protein l7, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPL3 - ribosomal protein l3, RPS7 - ribosomal protein s7, VTI1A - vesicle transport through interaction with t-snares 1a, SUMO1 - small ubiquitin-like modifier 1, SRP72 - signal recognition particle 72kda, HK2 - hexokinase 2, TNPO1 - transportin 1, RB1 - retinoblastoma 1, CAT - catalase, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, HSPD1 - heat shock 60kda protein 1 (chaperonin), KPNB1 - karyopherin (importin) beta 1, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, AGPS - alkylglycerone phosphate synthase, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), SAMM50 - samm50 sorting and assembly machinery component, GOSR2 - golgi snap receptor complex member 2, IDE - insulin-degrading enzyme, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, HSPA4 - heat shock 70kda protein 4]
157 [TMEM30A - transmembrane protein 30a, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, GAPVD1 - gtpase activating protein and vps9 domains 1, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), SPG11 - spastic paraplegia 11 (autosomal recessive), BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, DLG1 - discs, large homolog 1 (drosophila), NEK6 - nima-related kinase 6, GSN - gelsolin, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), AKAP8L - a kinase (prka) anchor protein 8-like, GNPAT - glyceronephosphate o-acyltransferase, AGK - acylglycerol kinase, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, MFN1 - mitofusin 1, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), GOSR1 - golgi snap receptor complex member 1, TMED2 - transmembrane emp24 domain trafficking protein 2, GOLPH3 - golgi phosphoprotein 3 (coat-protein), YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, DNAJC11 - dnaj (hsp40) homolog, subfamily c, member 11, VTI1A - vesicle transport through interaction with t-snares 1a, HK2 - hexokinase 2, STX17 - syntaxin 17, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), SAMM50 - samm50 sorting and assembly machinery component, GOSR2 - golgi snap receptor complex member 2, RHOT1 - ras homolog family member t1, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, CHMP5 - charged multivesicular body protein 5, STX8 - syntaxin 8, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, STX18 - syntaxin 18, PRKCD - protein kinase c, delta, BCL2L1 - bcl2-like 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), HSPA4 - heat shock 70kda protein 4, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, EPS15 - epidermal growth factor receptor pathway substrate 15, VDAC2 - voltage-dependent anion channel 2]
158 [SERBP1 - serpine1 mrna binding protein 1, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, EXOSC5 - exosome component 5, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, TNPO1 - transportin 1, LARP4B - la ribonucleoprotein domain family, member 4b, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, LARP1 - la ribonucleoprotein domain family, member 1, PRKCD - protein kinase c, delta, CARHSP1 - calcium regulated heat stable protein 1, 24kda, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, EXOSC1 - exosome component 1]
159 [DGUOK - deoxyguanosine kinase, AK4 - adenylate kinase 4, PGM1 - phosphoglucomutase 1, HK2 - hexokinase 2, PGAM1 - phosphoglycerate mutase 1 (brain), DCK - deoxycytidine kinase, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, AK3 - adenylate kinase 3, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, ADPGK - adp-dependent glucokinase]
160 [UQCRB - ubiquinol-cytochrome c reductase binding protein, MRPL13 - mitochondrial ribosomal protein l13, MCU - mitochondrial calcium uniporter, TMEM70 - transmembrane protein 70, TMEM14C - transmembrane protein 14c, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), AGK - acylglycerol kinase, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, NDUFA5 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, PTCD3 - pentatricopeptide repeat domain 3, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), MRPL42 - mitochondrial ribosomal protein l42, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, FECH - ferrochelatase, CYCS - cytochrome c, somatic, MRPS35 - mitochondrial ribosomal protein s35, MRPL2 - mitochondrial ribosomal protein l2, CPOX - coproporphyrinogen oxidase, MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, HSPD1 - heat shock 60kda protein 1 (chaperonin), CBX3 - chromobox homolog 3, NDUFAF4 - nadh dehydrogenase (ubiquinone) complex i, assembly factor 4, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, MRPS14 - mitochondrial ribosomal protein s14, MRPL47 - mitochondrial ribosomal protein l47, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, BCL2L1 - bcl2-like 1, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, MRPL3 - mitochondrial ribosomal protein l3]
161 [DST - dystonin, DDX6 - dead (asp-glu-ala-asp) box helicase 6, RPL22 - ribosomal protein l22, DLG1 - discs, large homolog 1 (drosophila), CAPN2 - calpain 2, (m/ii) large subunit, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, RAP1A - rap1a, member of ras oncogene family, PARVG - parvin, gamma, F11R - f11 receptor, GSN - gelsolin, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, WDR1 - wd repeat domain 1, RPS8 - ribosomal protein s8, RPL6 - ribosomal protein l6, FLNB - filamin b, beta, RPL7 - ribosomal protein l7, PPFIA1 - protein tyrosine phosphatase, receptor type, f polypeptide (ptprf), interacting protein (liprin), alpha 1, RPS13 - ribosomal protein s13, RAP1B - rap1b, member of ras oncogene family, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, PPFIBP1 - ptprf interacting protein, binding protein 1 (liprin beta 1), ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), RPL3 - ribosomal protein l3, DBN1 - drebrin 1, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, RPS7 - ribosomal protein s7, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, CAT - catalase, TBCD - tubulin folding cofactor d, DOCK7 - dedicator of cytokinesis 7, PLCG1 - phospholipase c, gamma 1, RAB23 - rab23, member ras oncogene family, LIMS1 - lim and senescent cell antigen-like domains 1, PRKCD - protein kinase c, delta, NAMPT - nicotinamide phosphoribosyltransferase, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), TJP2 - tight junction protein 2, RPL31 - ribosomal protein l31, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58]
162 [CAPN2 - calpain 2, (m/ii) large subunit, WNK1 - wnk lysine deficient protein kinase 1, GSN - gelsolin, HMGB2 - high mobility group box 2, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, HMGB1 - high mobility group box 1, FLNB - filamin b, beta, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, RPL3 - ribosomal protein l3, TRIM25 - tripartite motif containing 25, DOCK8 - dedicator of cytokinesis 8, USP8 - ubiquitin specific peptidase 8, RIF1 - rap1 interacting factor homolog (yeast), HK2 - hexokinase 2, MAT2A - methionine adenosyltransferase ii, alpha, TBC1D4 - tbc1 domain family, member 4, OTUB1 - otu domain, ubiquitin aldehyde binding 1, CHMP5 - charged multivesicular body protein 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, PLCG1 - phospholipase c, gamma 1, LIMS1 - lim and senescent cell antigen-like domains 1, IFIH1 - interferon induced with helicase c domain 1, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, KDM3A - lysine (k)-specific demethylase 3a, CREB1 - camp responsive element binding protein 1, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, MCM7 - minichromosome maintenance complex component 7, ATG7 - autophagy related 7, RAP1A - rap1a, member of ras oncogene family, PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), RNMT - rna (guanine-7-) methyltransferase, RAP1B - rap1b, member of ras oncogene family, GOLPH3 - golgi phosphoprotein 3 (coat-protein), ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, FECH - ferrochelatase, SNX6 - sorting nexin 6, SMARCD1 - swi/snf related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1, CAT - catalase, HSPD1 - heat shock 60kda protein 1 (chaperonin), PTPN2 - protein tyrosine phosphatase, non-receptor type 2, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), IRF3 - interferon regulatory factor 3, STX8 - syntaxin 8, BCL2L1 - bcl2-like 1, PRKCD - protein kinase c, delta, LARP1 - la ribonucleoprotein domain family, member 1, NAMPT - nicotinamide phosphoribosyltransferase, RPTOR - regulatory associated protein of mtor, complex 1, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1]
163 [RNF31 - ring finger protein 31, WNK1 - wnk lysine deficient protein kinase 1, HMGB1 - high mobility group box 1, CYFIP2 - cytoplasmic fmr1 interacting protein 2, EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, IRAK4 - interleukin-1 receptor-associated kinase 4, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, HSPD1 - heat shock 60kda protein 1 (chaperonin), EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, IRF3 - interferon regulatory factor 3, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, CUL1 - cullin 1, PLCG1 - phospholipase c, gamma 1, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, PRKCD - protein kinase c, delta, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), IFIH1 - interferon induced with helicase c domain 1, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, WIPF1 - was/wasl interacting protein family, member 1]
164 [ATG7 - autophagy related 7, CAPN2 - calpain 2, (m/ii) large subunit, RAP1A - rap1a, member of ras oncogene family, GSN - gelsolin, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), STAT6 - signal transducer and activator of transcription 6, interleukin-4 induced, HADH - hydroxyacyl-coa dehydrogenase, RAP1B - rap1b, member of ras oncogene family, EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, PPP5C - protein phosphatase 5, catalytic subunit, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), DERL1 - derlin 1, GOLPH3 - golgi phosphoprotein 3 (coat-protein), TRIM25 - tripartite motif containing 25, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, SNX6 - sorting nexin 6, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, CAT - catalase, HSPD1 - heat shock 60kda protein 1 (chaperonin), EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, TBC1D4 - tbc1 domain family, member 4, KYNU - kynureninase, CHMP5 - charged multivesicular body protein 5, PRKCD - protein kinase c, delta, IRF5 - interferon regulatory factor 5, BCL2L1 - bcl2-like 1, LARP1 - la ribonucleoprotein domain family, member 1, NAMPT - nicotinamide phosphoribosyltransferase, RPTOR - regulatory associated protein of mtor, complex 1, CREB1 - camp responsive element binding protein 1, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha]
165 [DST - dystonin, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, SERBP1 - serpine1 mrna binding protein 1, GAPVD1 - gtpase activating protein and vps9 domains 1, DDX6 - dead (asp-glu-ala-asp) box helicase 6, RPL23A - ribosomal protein l23a, RANBP1 - ran binding protein 1, DLG1 - discs, large homolog 1 (drosophila), F11R - f11 receptor, RPL14 - ribosomal protein l14, HMGB1 - high mobility group box 1, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, RPL6 - ribosomal protein l6, FLNB - filamin b, beta, PPFIBP1 - ptprf interacting protein, binding protein 1 (liprin beta 1), ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), DBN1 - drebrin 1, TRIM25 - tripartite motif containing 25, USP8 - ubiquitin specific peptidase 8, EIF4H - eukaryotic translation initiation factor 4h, BZW1 - basic leucine zipper and w2 domains 1, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, CHMP5 - charged multivesicular body protein 5, RSL1D1 - ribosomal l1 domain containing 1, LARP1 - la ribonucleoprotein domain family, member 1, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, TJP2 - tight junction protein 2, EIF3E - eukaryotic translation initiation factor 3, subunit e, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, SNX1 - sorting nexin 1, EPS15 - epidermal growth factor receptor pathway substrate 15]
166 [PTCD3 - pentatricopeptide repeat domain 3, MRPS14 - mitochondrial ribosomal protein s14, MRPL47 - mitochondrial ribosomal protein l47, MRPS35 - mitochondrial ribosomal protein s35, MRPL2 - mitochondrial ribosomal protein l2, MRPL42 - mitochondrial ribosomal protein l42, MRPL1 - mitochondrial ribosomal protein l1, MRPL13 - mitochondrial ribosomal protein l13, MRPL9 - mitochondrial ribosomal protein l9, TSFM - ts translation elongation factor, mitochondrial, MRPL3 - mitochondrial ribosomal protein l3]
167 [CCNK - cyclin k, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, UQCRB - ubiquinol-cytochrome c reductase binding protein, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, PGM1 - phosphoglucomutase 1, NEK6 - nima-related kinase 6, WNK1 - wnk lysine deficient protein kinase 1, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, SACM1L - sac1 suppressor of actin mutations 1-like (yeast), PGAM1 - phosphoglycerate mutase 1 (brain), PPP1R9B - protein phosphatase 1, regulatory subunit 9b, AGK - acylglycerol kinase, FABP5 - fatty acid binding protein 5 (psoriasis-associated), MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, PCYT2 - phosphate cytidylyltransferase 2, ethanolamine, RAB14 - rab14, member ras oncogene family, PHKB - phosphorylase kinase, beta, DCK - deoxycytidine kinase, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, UCK2 - uridine-cytidine kinase 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, PPP4R2 - protein phosphatase 4, regulatory subunit 2, PPP5C - protein phosphatase 5, catalytic subunit, UMPS - uridine monophosphate synthetase, ILF3 - interleukin enhancer binding factor 3, 90kda, IRAK4 - interleukin-1 receptor-associated kinase 4, CYCS - cytochrome c, somatic, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, ACAT1 - acetyl-coa acetyltransferase 1, DUT - deoxyuridine triphosphatase, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, LRRC40 - leucine rich repeat containing 40, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, AK4 - adenylate kinase 4, HK2 - hexokinase 2, LRRK1 - leucine-rich repeat kinase 1, CAB39 - calcium binding protein 39, SORD - sorbitol dehydrogenase, PLCG1 - phospholipase c, gamma 1, RAB23 - rab23, member ras oncogene family, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, TJP2 - tight junction protein 2, CREB1 - camp responsive element binding protein 1, DCAKD - dephospho-coa kinase domain containing, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, GRK6 - g protein-coupled receptor kinase 6, DLG1 - discs, large homolog 1 (drosophila), HDDC2 - hd domain containing 2, SCYL2 - scy1-like 2 (s. cerevisiae), STK38L - serine/threonine kinase 38 like, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), GNPAT - glyceronephosphate o-acyltransferase, MFN1 - mitofusin 1, DGUOK - deoxyguanosine kinase, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), ADPGK - adp-dependent glucokinase, PPA1 - pyrophosphatase (inorganic) 1, CDK6 - cyclin-dependent kinase 6, ACOT7 - acyl-coa thioesterase 7, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, GCDH - glutaryl-coa dehydrogenase, CDK13 - cyclin-dependent kinase 13, USP15 - ubiquitin specific peptidase 15, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, MVD - mevalonate (diphospho) decarboxylase, INPP5B - inositol polyphosphate-5-phosphatase, 75kda, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, KYNU - kynureninase, CUL1 - cullin 1, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, TAOK3 - tao kinase 3, PRKCD - protein kinase c, delta, NAMPT - nicotinamide phosphoribosyltransferase, AK3 - adenylate kinase 3, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, PGM2 - phosphoglucomutase 2, RRM1 - ribonucleotide reductase m1]
168 [CYCS - cytochrome c, somatic, ACAT1 - acetyl-coa acetyltransferase 1, UQCRB - ubiquinol-cytochrome c reductase binding protein, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, HK2 - hexokinase 2, PGM1 - phosphoglucomutase 1, CAT - catalase, PGAM1 - phosphoglycerate mutase 1 (brain), NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, NDUFA5 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, DGUOK - deoxyguanosine kinase, PYGB - phosphorylase, glycogen; brain, PHKB - phosphorylase kinase, beta, PGM2 - phosphoglucomutase 2, ADPGK - adp-dependent glucokinase, LYRM7 - lyr motif containing 7, FECH - ferrochelatase]
169 [SCAF8 - sr-related ctd-associated factor 8, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, NCBP2 - nuclear cap binding protein subunit 2, 20kda, QKI - qki, kh domain containing, rna binding, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, DDX6 - dead (asp-glu-ala-asp) box helicase 6, EXOSC5 - exosome component 5, RPL23A - ribosomal protein l23a, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, SNRPE - small nuclear ribonucleoprotein polypeptide e, RPL22 - ribosomal protein l22, SF3A3 - splicing factor 3a, subunit 3, 60kda, PUS1 - pseudouridylate synthase 1, GSPT1 - g1 to s phase transition 1, RPL14 - ribosomal protein l14, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), AKAP8L - a kinase (prka) anchor protein 8-like, RPL13 - ribosomal protein l13, RNMT - rna (guanine-7-) methyltransferase, SNRNP27 - small nuclear ribonucleoprotein 27kda (u4/u6.u5), PELO - pelota homolog (drosophila), RBM26 - rna binding motif protein 26, RPS8 - ribosomal protein s8, RPL6 - ribosomal protein l6, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), RPL7 - ribosomal protein l7, PPIL3 - peptidylprolyl isomerase (cyclophilin)-like 3, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, PPP4R2 - protein phosphatase 4, regulatory subunit 2, RPL3 - ribosomal protein l3, RPS7 - ribosomal protein s7, THOC5 - tho complex 5, MBNL1 - muscleblind-like splicing regulator 1, RBM19 - rna binding motif protein 19, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), CDK13 - cyclin-dependent kinase 13, ZNF326 - zinc finger protein 326, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, RPL31 - ribosomal protein l31, EIF3E - eukaryotic translation initiation factor 3, subunit e, RPL32 - ribosomal protein l32, SART3 - squamous cell carcinoma antigen recognized by t cells 3, EXOSC1 - exosome component 1]
170 [ACAT1 - acetyl-coa acetyltransferase 1, AK4 - adenylate kinase 4, HK2 - hexokinase 2, PGM1 - phosphoglucomutase 1, GCDH - glutaryl-coa dehydrogenase, ACOT7 - acyl-coa thioesterase 7, PGAM1 - phosphoglycerate mutase 1 (brain), DGUOK - deoxyguanosine kinase, AK3 - adenylate kinase 3, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, DCAKD - dephospho-coa kinase domain containing, ADPGK - adp-dependent glucokinase]
171 [UQCRB - ubiquinol-cytochrome c reductase binding protein, AK4 - adenylate kinase 4, PGM1 - phosphoglucomutase 1, HK2 - hexokinase 2, PGAM1 - phosphoglycerate mutase 1 (brain), NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, MFN1 - mitofusin 1, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, DGUOK - deoxyguanosine kinase, RAB23 - rab23, member ras oncogene family, AK3 - adenylate kinase 3, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, ADPGK - adp-dependent glucokinase]
172 [UQCRB - ubiquinol-cytochrome c reductase binding protein, AK4 - adenylate kinase 4, PGM1 - phosphoglucomutase 1, HK2 - hexokinase 2, PGAM1 - phosphoglycerate mutase 1 (brain), NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, MFN1 - mitofusin 1, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, DGUOK - deoxyguanosine kinase, RAB23 - rab23, member ras oncogene family, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, AK3 - adenylate kinase 3, ADPGK - adp-dependent glucokinase]
173 [MRPS35 - mitochondrial ribosomal protein s35, MRPL2 - mitochondrial ribosomal protein l2, MRPL13 - mitochondrial ribosomal protein l13, MRPL1 - mitochondrial ribosomal protein l1, TSFM - ts translation elongation factor, mitochondrial, MRPL9 - mitochondrial ribosomal protein l9, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, PTCD3 - pentatricopeptide repeat domain 3, MRPL47 - mitochondrial ribosomal protein l47, MRPS14 - mitochondrial ribosomal protein s14, MRPL42 - mitochondrial ribosomal protein l42, NEMF - nuclear export mediator factor, MRPL3 - mitochondrial ribosomal protein l3]
174 [PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, PGM1 - phosphoglucomutase 1, SACM1L - sac1 suppressor of actin mutations 1-like (yeast), PGAM1 - phosphoglycerate mutase 1 (brain), PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, AGK - acylglycerol kinase, GNPAT - glyceronephosphate o-acyltransferase, FABP5 - fatty acid binding protein 5 (psoriasis-associated), MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, DGUOK - deoxyguanosine kinase, PCYT2 - phosphate cytidylyltransferase 2, ethanolamine, RAB14 - rab14, member ras oncogene family, DCK - deoxycytidine kinase, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, UCK2 - uridine-cytidine kinase 2, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, UMPS - uridine monophosphate synthetase, ADPGK - adp-dependent glucokinase, ACAT1 - acetyl-coa acetyltransferase 1, DUT - deoxyuridine triphosphatase, AK4 - adenylate kinase 4, HK2 - hexokinase 2, ACOT7 - acyl-coa thioesterase 7, GCDH - glutaryl-coa dehydrogenase, MVD - mevalonate (diphospho) decarboxylase, KYNU - kynureninase, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, SORD - sorbitol dehydrogenase, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, NAMPT - nicotinamide phosphoribosyltransferase, AK3 - adenylate kinase 3, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, DCAKD - dephospho-coa kinase domain containing, RRM1 - ribonucleotide reductase m1]
175 [CCNK - cyclin k, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, NCBP2 - nuclear cap binding protein subunit 2, 20kda, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, EXOSC5 - exosome component 5, RFC3 - replication factor c (activator 1) 3, 38kda, SF3A3 - splicing factor 3a, subunit 3, 60kda, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, GFM2 - g elongation factor, mitochondrial 2, FKBP11 - fk506 binding protein 11, 19 kda, RECQL - recq protein-like (dna helicase q1-like), DERL1 - derlin 1, BTF3 - basic transcription factor 3, LRRC40 - leucine rich repeat containing 40, USP25 - ubiquitin specific peptidase 25, OTUB1 - otu domain, ubiquitin aldehyde binding 1, ZNF326 - zinc finger protein 326, MORF4L1 - mortality factor 4 like 1, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, CREB1 - camp responsive element binding protein 1, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, SNX1 - sorting nexin 1, THUMPD1 - thump domain containing 1, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, SNRPE - small nuclear ribonucleoprotein polypeptide e, ANAPC7 - anaphase promoting complex subunit 7, STK38L - serine/threonine kinase 38 like, SEH1L - seh1-like (s. cerevisiae), PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), POLR1C - polymerase (rna) i polypeptide c, 30kda, DCAF7 - ddb1 and cul4 associated factor 7, PPA1 - pyrophosphatase (inorganic) 1, SUMO3 - small ubiquitin-like modifier 3, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, TSFM - ts translation elongation factor, mitochondrial, HSPD1 - heat shock 60kda protein 1 (chaperonin), HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, TAOK3 - tao kinase 3, ERP44 - endoplasmic reticulum protein 44, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), PARP4 - poly (adp-ribose) polymerase family, member 4, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, SCPEP1 - serine carboxypeptidase 1, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, QKI - qki, kh domain containing, rna binding, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, CAPN2 - calpain 2, (m/ii) large subunit, WNK1 - wnk lysine deficient protein kinase 1, UBE2Z - ubiquitin-conjugating enzyme e2z, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, PTCD3 - pentatricopeptide repeat domain 3, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, PPP5C - protein phosphatase 5, catalytic subunit, USP48 - ubiquitin specific peptidase 48, RPL3 - ribosomal protein l3, CARS - cysteinyl-trna synthetase, IRAK4 - interleukin-1 receptor-associated kinase 4, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), EIF3F - eukaryotic translation initiation factor 3, subunit f, MRPS35 - mitochondrial ribosomal protein s35, MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, RPN1 - ribophorin i, KPNB1 - karyopherin (importin) beta 1, TRIM38 - tripartite motif containing 38, CDC23 - cell division cycle 23, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, TTLL12 - tubulin tyrosine ligase-like family, member 12, MRPL13 - mitochondrial ribosomal protein l13, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, NIPBL - nipped-b homolog (drosophila), SCYL2 - scy1-like 2 (s. cerevisiae), GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, GALNT2 - udp-n-acetyl-alpha-d-galactosamine:polypeptide n-acetylgalactosaminyltransferase 2 (galnac-t2), STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), SNRNP27 - small nuclear ribonucleoprotein 27kda (u4/u6.u5), RNMT - rna (guanine-7-) methyltransferase, RPS8 - ribosomal protein s8, MRPL42 - mitochondrial ribosomal protein l42, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, NHP2 - nhp2 ribonucleoprotein, RPS7 - ribosomal protein s7, UCHL3 - ubiquitin carboxyl-terminal esterase l3 (ubiquitin thiolesterase), SUMO1 - small ubiquitin-like modifier 1, CAT - catalase, USP15 - ubiquitin specific peptidase 15, UGDH - udp-glucose 6-dehydrogenase, SSBP1 - single-stranded dna binding protein 1, mitochondrial, MRPL47 - mitochondrial ribosomal protein l47, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, UBE2I - ubiquitin-conjugating enzyme e2i, PRKCD - protein kinase c, delta, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), RRBP1 - ribosome binding protein 1, RRM1 - ribonucleotide reductase m1, ZC3H7B - zinc finger ccch-type containing 7b, ERAP1 - endoplasmic reticulum aminopeptidase 1, DDX6 - dead (asp-glu-ala-asp) box helicase 6, TCOF1 - treacher collins-franceschetti syndrome 1, ZNF638 - zinc finger protein 638, GSPT1 - g1 to s phase transition 1, STAMBP - stam binding protein, GSN - gelsolin, URB1 - urb1 ribosome biogenesis 1 homolog (s. cerevisiae), FAM98A - family with sequence similarity 98, member a, PPP4R2 - protein phosphatase 4, regulatory subunit 2, TRIM25 - tripartite motif containing 25, ILF3 - interleukin enhancer binding factor 3, 90kda, RIF1 - rap1 interacting factor homolog (yeast), PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, CYCS - cytochrome c, somatic, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, RBM19 - rna binding motif protein 19, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), MMS22L - mms22-like, dna repair protein, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, LRRK1 - leucine-rich repeat kinase 1, PA2G4 - proliferation-associated 2g4, 38kda, CAB39 - calcium binding protein 39, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, GAR1 - gar1 ribonucleoprotein, KDM3A - lysine (k)-specific demethylase 3a, ANAPC1 - anaphase promoting complex subunit 1, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, MRPL3 - mitochondrial ribosomal protein l3, TELO2 - telomere maintenance 2, EXOSC1 - exosome component 1, PITRM1 - pitrilysin metallopeptidase 1, RNF31 - ring finger protein 31, MCM7 - minichromosome maintenance complex component 7, ATG7 - autophagy related 7, KDELC2 - kdel (lys-asp-glu-leu) containing 2, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), PELO - pelota homolog (drosophila), ZBTB8OS - zinc finger and btb domain containing 8 opposite strand, NUP88 - nucleoporin 88kda, WDHD1 - wd repeat and hmg-box dna binding protein 1, NFU1 - nfu1 iron-sulfur cluster scaffold homolog (s. cerevisiae), PPIL3 - peptidylprolyl isomerase (cyclophilin)-like 3, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), THOC5 - tho complex 5, CDK6 - cyclin-dependent kinase 6, MRPL2 - mitochondrial ribosomal protein l2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, CDK13 - cyclin-dependent kinase 13, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, FCF1 - fcf1 rrna-processing protein, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, RPL7L1 - ribosomal protein l7-like 1, NEMF - nuclear export mediator factor, DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), EIF3E - eukaryotic translation initiation factor 3, subunit e, TCEA1 - transcription elongation factor a (sii), 1, PURA - purine-rich element binding protein a, DENND3 - denn/madd domain containing 3, NEK6 - nima-related kinase 6, PGM1 - phosphoglucomutase 1, NOC3L - nucleolar complex associated 3 homolog (s. cerevisiae), PYGB - phosphorylase, glycogen; brain, NUP50 - nucleoporin 50kda, PHKB - phosphorylase kinase, beta, MANBA - mannosidase, beta a, lysosomal, VKORC1L1 - vitamin k epoxide reductase complex, subunit 1-like 1, MAN2A1 - mannosidase, alpha, class 2a, member 1, USP8 - ubiquitin specific peptidase 8, MBNL1 - muscleblind-like splicing regulator 1, METTL15 - methyltransferase like 15, DUT - deoxyuridine triphosphatase, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, IDE - insulin-degrading enzyme, MCM3 - minichromosome maintenance complex component 3, USP34 - ubiquitin specific peptidase 34, IFIH1 - interferon induced with helicase c domain 1, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, SART3 - squamous cell carcinoma antigen recognized by t cells 3, CTSC - cathepsin c, SCAF8 - sr-related ctd-associated factor 8, GRK6 - g protein-coupled receptor kinase 6, UBA6 - ubiquitin-like modifier activating enzyme 6, DLG1 - discs, large homolog 1 (drosophila), PUS1 - pseudouridylate synthase 1, RPL14 - ribosomal protein l14, AKAP8L - a kinase (prka) anchor protein 8-like, CYFIP2 - cytoplasmic fmr1 interacting protein 2, NUB1 - negative regulator of ubiquitin-like proteins 1, RBM26 - rna binding motif protein 26, DGUOK - deoxyguanosine kinase, UBA3 - ubiquitin-like modifier activating enzyme 3, GOLPH3 - golgi phosphoprotein 3 (coat-protein), TMX3 - thioredoxin-related transmembrane protein 3, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), MRPS14 - mitochondrial ribosomal protein s14, RHOT1 - ras homolog family member t1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, CUL1 - cullin 1, RSL1D1 - ribosomal l1 domain containing 1, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, PGM2 - phosphoglucomutase 2, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
176 [UQCRB - ubiquinol-cytochrome c reductase binding protein, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), DHRS7 - dehydrogenase/reductase (sdr family) member 7, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, NDUFA5 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, DGUOK - deoxyguanosine kinase, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, PYGB - phosphorylase, glycogen; brain, HADH - hydroxyacyl-coa dehydrogenase, PHKB - phosphorylase kinase, beta, ECI1 - enoyl-coa delta isomerase 1, FTH1 - ferritin, heavy polypeptide 1, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, ADPGK - adp-dependent glucokinase, VKORC1L1 - vitamin k epoxide reductase complex, subunit 1-like 1, CYCS - cytochrome c, somatic, ACAT1 - acetyl-coa acetyltransferase 1, TMX3 - thioredoxin-related transmembrane protein 3, CPOX - coproporphyrinogen oxidase, HK2 - hexokinase 2, GCDH - glutaryl-coa dehydrogenase, CAT - catalase, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, UGDH - udp-glucose 6-dehydrogenase, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, ADI1 - acireductone dioxygenase 1, AGPS - alkylglycerone phosphate synthase, SORD - sorbitol dehydrogenase, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, KDM3A - lysine (k)-specific demethylase 3a, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, QSOX2 - quiescin q6 sulfhydryl oxidase 2, PGM2 - phosphoglucomutase 2, RRM1 - ribonucleotide reductase m1, LYRM7 - lyr motif containing 7]
177 [SERBP1 - serpine1 mrna binding protein 1, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, EXOSC5 - exosome component 5, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, TNPO1 - transportin 1, LARP4B - la ribonucleoprotein domain family, member 4b, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, LARP1 - la ribonucleoprotein domain family, member 1, PRKCD - protein kinase c, delta, CARHSP1 - calcium regulated heat stable protein 1, 24kda, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, EXOSC1 - exosome component 1]
178 [MRPS35 - mitochondrial ribosomal protein s35, MRPL2 - mitochondrial ribosomal protein l2, MRPL13 - mitochondrial ribosomal protein l13, MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, GSPT1 - g1 to s phase transition 1, GSN - gelsolin, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, GFM2 - g elongation factor, mitochondrial 2, PTCD3 - pentatricopeptide repeat domain 3, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), MRPL47 - mitochondrial ribosomal protein l47, MRPS14 - mitochondrial ribosomal protein s14, WDR1 - wd repeat domain 1, MRPL42 - mitochondrial ribosomal protein l42, MRPL3 - mitochondrial ribosomal protein l3, DSTN - destrin (actin depolymerizing factor)]
179 [UQCRB - ubiquinol-cytochrome c reductase binding protein, DLG1 - discs, large homolog 1 (drosophila), AK4 - adenylate kinase 4, PGM1 - phosphoglucomutase 1, HK2 - hexokinase 2, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, PGAM1 - phosphoglycerate mutase 1 (brain), NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, DGUOK - deoxyguanosine kinase, TJP2 - tight junction protein 2, AK3 - adenylate kinase 3, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, ADPGK - adp-dependent glucokinase]
180 [AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, NCBP2 - nuclear cap binding protein subunit 2, 20kda, DENND3 - denn/madd domain containing 3, DDX6 - dead (asp-glu-ala-asp) box helicase 6, EXOSC5 - exosome component 5, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, GSPT1 - g1 to s phase transition 1, UBE2Z - ubiquitin-conjugating enzyme e2z, HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, RPL3 - ribosomal protein l3, USP48 - ubiquitin specific peptidase 48, DERL1 - derlin 1, MANBA - mannosidase, beta a, lysosomal, TRIM25 - tripartite motif containing 25, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), USP8 - ubiquitin specific peptidase 8, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, KPNB1 - karyopherin (importin) beta 1, TRIM38 - tripartite motif containing 38, USP25 - ubiquitin specific peptidase 25, IDE - insulin-degrading enzyme, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, CDC23 - cell division cycle 23, USP34 - ubiquitin specific peptidase 34, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, ANAPC1 - anaphase promoting complex subunit 1, EXOSC1 - exosome component 1, UBA6 - ubiquitin-like modifier activating enzyme 6, ANAPC7 - anaphase promoting complex subunit 7, RPL14 - ribosomal protein l14, NUB1 - negative regulator of ubiquitin-like proteins 1, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), PELO - pelota homolog (drosophila), RPS8 - ribosomal protein s8, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPS7 - ribosomal protein s7, UCHL3 - ubiquitin carboxyl-terminal esterase l3 (ubiquitin thiolesterase), HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, USP15 - ubiquitin specific peptidase 15, UBE2I - ubiquitin-conjugating enzyme e2i, CUL1 - cullin 1, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), NEMF - nuclear export mediator factor, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), EIF3E - eukaryotic translation initiation factor 3, subunit e]
181 [MBNL1 - muscleblind-like splicing regulator 1, SERBP1 - serpine1 mrna binding protein 1, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, QKI - qki, kh domain containing, rna binding, NCBP2 - nuclear cap binding protein subunit 2, 20kda, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, EXOSC5 - exosome component 5, RBM19 - rna binding motif protein 19, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, TNPO1 - transportin 1, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, LARP4B - la ribonucleoprotein domain family, member 4b, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, PRKCD - protein kinase c, delta, LARP1 - la ribonucleoprotein domain family, member 1, CARHSP1 - calcium regulated heat stable protein 1, 24kda, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, SART3 - squamous cell carcinoma antigen recognized by t cells 3, EXOSC1 - exosome component 1]
182 [DDX31 - dead (asp-glu-ala-asp) box polypeptide 31, CCNK - cyclin k, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, ERAP1 - endoplasmic reticulum aminopeptidase 1, DDX6 - dead (asp-glu-ala-asp) box helicase 6, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, EXOSC5 - exosome component 5, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, RFC3 - replication factor c (activator 1) 3, 38kda, SACM1L - sac1 suppressor of actin mutations 1-like (yeast), SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, STAMBP - stam binding protein, GSPT1 - g1 to s phase transition 1, RAB30 - rab30, member ras oncogene family, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, NDUFA5 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, FAM98A - family with sequence similarity 98, member a, GFM2 - g elongation factor, mitochondrial 2, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, FKBP11 - fk506 binding protein 11, 19 kda, RECQL - recq protein-like (dna helicase q1-like), ECI1 - enoyl-coa delta isomerase 1, DCK - deoxycytidine kinase, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, TRIM25 - tripartite motif containing 25, CYCS - cytochrome c, somatic, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, ACAT1 - acetyl-coa acetyltransferase 1, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, CPOX - coproporphyrinogen oxidase, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, LRRC40 - leucine rich repeat containing 40, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, LRRK1 - leucine-rich repeat kinase 1, AGPS - alkylglycerone phosphate synthase, USP25 - ubiquitin specific peptidase 25, OTUB1 - otu domain, ubiquitin aldehyde binding 1, SORD - sorbitol dehydrogenase, PLCG1 - phospholipase c, gamma 1, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, RAB23 - rab23, member ras oncogene family, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, KDM3A - lysine (k)-specific demethylase 3a, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, ANAPC1 - anaphase promoting complex subunit 1, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, MYO6 - myosin vi, PITRM1 - pitrilysin metallopeptidase 1, SGPL1 - sphingosine-1-phosphate lyase 1, RNF31 - ring finger protein 31, ATG7 - autophagy related 7, MCM7 - minichromosome maintenance complex component 7, ARL1 - adp-ribosylation factor-like 1, HDDC2 - hd domain containing 2, STK38L - serine/threonine kinase 38 like, KDELC2 - kdel (lys-asp-glu-leu) containing 2, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, GLS - glutaminase, IARS - isoleucyl-trna synthetase, GLO1 - glyoxalase i, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), PELO - pelota homolog (drosophila), RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), HADH - hydroxyacyl-coa dehydrogenase, PPIL3 - peptidylprolyl isomerase (cyclophilin)-like 3, POLR1C - polymerase (rna) i polypeptide c, 30kda, MBLAC2 - metallo-beta-lactamase domain containing 2, ADPGK - adp-dependent glucokinase, PPA1 - pyrophosphatase (inorganic) 1, FECH - ferrochelatase, CDK6 - cyclin-dependent kinase 6, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, CDK13 - cyclin-dependent kinase 13, MUT - methylmalonyl coa mutase, HSPD1 - heat shock 60kda protein 1 (chaperonin), LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, GNPDA2 - glucosamine-6-phosphate deaminase 2, MVD - mevalonate (diphospho) decarboxylase, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, INPP5B - inositol polyphosphate-5-phosphatase, 75kda, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, COMT - catechol-o-methyltransferase, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, KYNU - kynureninase, ERP44 - endoplasmic reticulum protein 44, TAOK3 - tao kinase 3, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), NAMPT - nicotinamide phosphoribosyltransferase, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, QSOX2 - quiescin q6 sulfhydryl oxidase 2, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, PARP4 - poly (adp-ribose) polymerase family, member 4, SCPEP1 - serine carboxypeptidase 1, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, CAPN2 - calpain 2, (m/ii) large subunit, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, NEK6 - nima-related kinase 6, WNK1 - wnk lysine deficient protein kinase 1, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), UBE2Z - ubiquitin-conjugating enzyme e2z, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, HMGB1 - high mobility group box 1, AGK - acylglycerol kinase, PYGB - phosphorylase, glycogen; brain, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, PCYT2 - phosphate cytidylyltransferase 2, ethanolamine, RAB14 - rab14, member ras oncogene family, FTH1 - ferritin, heavy polypeptide 1, UCK2 - uridine-cytidine kinase 2, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, UMPS - uridine monophosphate synthetase, USP48 - ubiquitin specific peptidase 48, PPP5C - protein phosphatase 5, catalytic subunit, CARS - cysteinyl-trna synthetase, MANBA - mannosidase, beta a, lysosomal, VKORC1L1 - vitamin k epoxide reductase complex, subunit 1-like 1, EIF3F - eukaryotic translation initiation factor 3, subunit f, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), IRAK4 - interleukin-1 receptor-associated kinase 4, MAN2A1 - mannosidase, alpha, class 2a, member 1, USP8 - ubiquitin specific peptidase 8, METTL15 - methyltransferase like 15, DUT - deoxyuridine triphosphatase, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, RPN1 - ribophorin i, HK2 - hexokinase 2, AK4 - adenylate kinase 4, TRIM38 - tripartite motif containing 38, MAT2A - methionine adenosyltransferase ii, alpha, IDE - insulin-degrading enzyme, MCM3 - minichromosome maintenance complex component 3, CDC23 - cell division cycle 23, ABCF2 - atp-binding cassette, sub-family f (gcn20), member 2, IFIH1 - interferon induced with helicase c domain 1, USP34 - ubiquitin specific peptidase 34, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), MTHFD2 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 2, methenyltetrahydrofolate cyclohydrolase, TJP2 - tight junction protein 2, DCAKD - dephospho-coa kinase domain containing, CTSC - cathepsin c, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, UBA6 - ubiquitin-like modifier activating enzyme 6, GRK6 - g protein-coupled receptor kinase 6, LSS - lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase), SCYL2 - scy1-like 2 (s. cerevisiae), DLG1 - discs, large homolog 1 (drosophila), RAP1A - rap1a, member of ras oncogene family, PUS1 - pseudouridylate synthase 1, GALNT2 - udp-n-acetyl-alpha-d-galactosamine:polypeptide n-acetylgalactosaminyltransferase 2 (galnac-t2), PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, GNPAT - glyceronephosphate o-acyltransferase, DHRS7 - dehydrogenase/reductase (sdr family) member 7, RNMT - rna (guanine-7-) methyltransferase, MFN1 - mitofusin 1, DGUOK - deoxyguanosine kinase, ESD - esterase d, UBA3 - ubiquitin-like modifier activating enzyme 3, RAP1B - rap1b, member of ras oncogene family, UCHL3 - ubiquitin carboxyl-terminal esterase l3 (ubiquitin thiolesterase), TMX3 - thioredoxin-related transmembrane protein 3, DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, GCDH - glutaryl-coa dehydrogenase, ACOT7 - acyl-coa thioesterase 7, CAT - catalase, USP15 - ubiquitin specific peptidase 15, UGDH - udp-glucose 6-dehydrogenase, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, ADI1 - acireductone dioxygenase 1, RHOT1 - ras homolog family member t1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, UBE2I - ubiquitin-conjugating enzyme e2i, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, CUL1 - cullin 1, PRKCD - protein kinase c, delta, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, METTL13 - methyltransferase like 13, PGM2 - phosphoglucomutase 2, RRM1 - ribonucleotide reductase m1, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
183 [CCNK - cyclin k, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, RFC3 - replication factor c (activator 1) 3, 38kda, F11R - f11 receptor, GSN - gelsolin, TMED2 - transmembrane emp24 domain trafficking protein 2, RECQL - recq protein-like (dna helicase q1-like), FLNB - filamin b, beta, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, DERL1 - derlin 1, TRIM25 - tripartite motif containing 25, DOCK8 - dedicator of cytokinesis 8, RIF1 - rap1 interacting factor homolog (yeast), CYCS - cytochrome c, somatic, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, MMS22L - mms22-like, dna repair protein, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, GOSR2 - golgi snap receptor complex member 2, CAB39 - calcium binding protein 39, TBC1D4 - tbc1 domain family, member 4, OTUB1 - otu domain, ubiquitin aldehyde binding 1, PLCG1 - phospholipase c, gamma 1, LIMS1 - lim and senescent cell antigen-like domains 1, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, MORF4L1 - mortality factor 4 like 1, RCSD1 - rcsd domain containing 1, CREB1 - camp responsive element binding protein 1, KDM3A - lysine (k)-specific demethylase 3a, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, MYO6 - myosin vi, ATG7 - autophagy related 7, MCM7 - minichromosome maintenance complex component 7, SEH1L - seh1-like (s. cerevisiae), PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, WDHD1 - wd repeat and hmg-box dna binding protein 1, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, FECH - ferrochelatase, ADNP - activity-dependent neuroprotector homeobox, SNX6 - sorting nexin 6, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, SMARCD1 - swi/snf related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1, HSPD1 - heat shock 60kda protein 1 (chaperonin), PTPN2 - protein tyrosine phosphatase, non-receptor type 2, COMT - catechol-o-methyltransferase, IRF3 - interferon regulatory factor 3, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, STX8 - syntaxin 8, ERP44 - endoplasmic reticulum protein 44, TAOK3 - tao kinase 3, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), LARP1 - la ribonucleoprotein domain family, member 1, NAMPT - nicotinamide phosphoribosyltransferase, RPTOR - regulatory associated protein of mtor, complex 1, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, PARP4 - poly (adp-ribose) polymerase family, member 4, TCEA1 - transcription elongation factor a (sii), 1, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, CAPN2 - calpain 2, (m/ii) large subunit, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, WNK1 - wnk lysine deficient protein kinase 1, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, UMPS - uridine monophosphate synthetase, RPL3 - ribosomal protein l3, PPP5C - protein phosphatase 5, catalytic subunit, IRAK4 - interleukin-1 receptor-associated kinase 4, VKORC1L1 - vitamin k epoxide reductase complex, subunit 1-like 1, USP8 - ubiquitin specific peptidase 8, MRPS35 - mitochondrial ribosomal protein s35, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, AK4 - adenylate kinase 4, HK2 - hexokinase 2, MAT2A - methionine adenosyltransferase ii, alpha, MCM3 - minichromosome maintenance complex component 3, CHMP5 - charged multivesicular body protein 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), IFIH1 - interferon induced with helicase c domain 1, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, UBA6 - ubiquitin-like modifier activating enzyme 6, NIPBL - nipped-b homolog (drosophila), RAP1A - rap1a, member of ras oncogene family, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), RNMT - rna (guanine-7-) methyltransferase, STAT6 - signal transducer and activator of transcription 6, interleukin-4 induced, RAP1B - rap1b, member of ras oncogene family, GOLPH3 - golgi phosphoprotein 3 (coat-protein), SUMO1 - small ubiquitin-like modifier 1, RB1 - retinoblastoma 1, CAT - catalase, CBX3 - chromobox homolog 3, FAM49B - family with sequence similarity 49, member b, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), CUL1 - cullin 1, BCL2L1 - bcl2-like 1, PRKCD - protein kinase c, delta, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha]
184 [CCNK - cyclin k, ZMYM2 - zinc finger, mym-type 2, GSN - gelsolin, DERL1 - derlin 1, DOCK8 - dedicator of cytokinesis 8, XPOT - exportin, trna, ACAT1 - acetyl-coa acetyltransferase 1, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, PA2G4 - proliferation-associated 2g4, 38kda, EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, CAB39 - calcium binding protein 39, USP25 - ubiquitin specific peptidase 25, TBC1D4 - tbc1 domain family, member 4, OTUB1 - otu domain, ubiquitin aldehyde binding 1, ZNF326 - zinc finger protein 326, PLCG1 - phospholipase c, gamma 1, LIMS1 - lim and senescent cell antigen-like domains 1, CARHSP1 - calcium regulated heat stable protein 1, 24kda, CREB1 - camp responsive element binding protein 1, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, DNAJC10 - dnaj (hsp40) homolog, subfamily c, member 10, TELO2 - telomere maintenance 2, RNF31 - ring finger protein 31, ANAPC7 - anaphase promoting complex subunit 7, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, SYNE1 - spectrin repeat containing, nuclear envelope 1, IARS - isoleucyl-trna synthetase, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), SUMO3 - small ubiquitin-like modifier 3, HERC2 - hect and rld domain containing e3 ubiquitin protein ligase 2, CDK13 - cyclin-dependent kinase 13, HSPD1 - heat shock 60kda protein 1 (chaperonin), DENND4A - denn/madd domain containing 4a, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, STX8 - syntaxin 8, IRF5 - interferon regulatory factor 5, RPTOR - regulatory associated protein of mtor, complex 1, PARP4 - poly (adp-ribose) polymerase family, member 4, AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, GAPVD1 - gtpase activating protein and vps9 domains 1, DENND3 - denn/madd domain containing 3, CAPN2 - calpain 2, (m/ii) large subunit, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, WNK1 - wnk lysine deficient protein kinase 1, NEK6 - nima-related kinase 6, PGAM1 - phosphoglycerate mutase 1 (brain), PPP1R9B - protein phosphatase 1, regulatory subunit 9b, HMGB1 - high mobility group box 1, TBC1D2B - tbc1 domain family, member 2b, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, TNPO1 - transportin 1, STX17 - syntaxin 17, KPNB1 - karyopherin (importin) beta 1, TRIM38 - tripartite motif containing 38, EXOC2 - exocyst complex component 2, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, DOCK7 - dedicator of cytokinesis 7, EXOC1 - exocyst complex component 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), TJP2 - tight junction protein 2, SART3 - squamous cell carcinoma antigen recognized by t cells 3, CTSC - cathepsin c, SCAF8 - sr-related ctd-associated factor 8, RANBP1 - ran binding protein 1, NIPBL - nipped-b homolog (drosophila), DLG1 - discs, large homolog 1 (drosophila), GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, RAP1A - rap1a, member of ras oncogene family, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), AKAP8L - a kinase (prka) anchor protein 8-like, STAT6 - signal transducer and activator of transcription 6, interleukin-4 induced, CYFIP2 - cytoplasmic fmr1 interacting protein 2, RPS6 - ribosomal protein s6, GOLPH3 - golgi phosphoprotein 3 (coat-protein), RPS7 - ribosomal protein s7, RIC8A - ric8 guanine nucleotide exchange factor a, STOM - stomatin, SUMO1 - small ubiquitin-like modifier 1, RB1 - retinoblastoma 1, CAT - catalase, XPO6 - exportin 6, CBX3 - chromobox homolog 3, FAM49B - family with sequence similarity 49, member b, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, UBE2I - ubiquitin-conjugating enzyme e2i, CUL1 - cullin 1, PRKCD - protein kinase c, delta, BCL2L1 - bcl2-like 1, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha]
185 [AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, TMEM30A - transmembrane protein 30a, STOM - stomatin, TMX3 - thioredoxin-related transmembrane protein 3, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, RAP1A - rap1a, member of ras oncogene family, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, FABP5 - fatty acid binding protein 5 (psoriasis-associated), TMED2 - transmembrane emp24 domain trafficking protein 2, RAB14 - rab14, member ras oncogene family, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), RAP1B - rap1b, member of ras oncogene family, MANBA - mannosidase, beta a, lysosomal, SURF4 - surfeit 4]
186 [AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, UQCRB - ubiquinol-cytochrome c reductase binding protein, RNF31 - ring finger protein 31, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, MCU - mitochondrial calcium uniporter, KCNAB2 - potassium voltage-gated channel, shaker-related subfamily, beta member 2, DLG1 - discs, large homolog 1 (drosophila), SACM1L - sac1 suppressor of actin mutations 1-like (yeast), SYNE1 - spectrin repeat containing, nuclear envelope 1, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), HMGB1 - high mobility group box 1, AGK - acylglycerol kinase, NDUFA7 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kda, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), MFN1 - mitofusin 1, NDUFA5 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, GOSR1 - golgi snap receptor complex member 1, HOOK3 - hook microtubule-tethering protein 3, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), DERL1 - derlin 1, DNAJC11 - dnaj (hsp40) homolog, subfamily c, member 11, VTI1A - vesicle transport through interaction with t-snares 1a, SNX6 - sorting nexin 6, SUMO1 - small ubiquitin-like modifier 1, RPN1 - ribophorin i, STX17 - syntaxin 17, ARCN1 - archain 1, HSPD1 - heat shock 60kda protein 1 (chaperonin), ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, NDUFA10 - nadh dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kda, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), SAMM50 - samm50 sorting and assembly machinery component, GOSR2 - golgi snap receptor complex member 2, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, STX8 - syntaxin 8, STX18 - syntaxin 18, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, SNX1 - sorting nexin 1, EPS15 - epidermal growth factor receptor pathway substrate 15, VDAC2 - voltage-dependent anion channel 2]
187 [DGUOK - deoxyguanosine kinase, HK2 - hexokinase 2, PGM1 - phosphoglucomutase 1, AK4 - adenylate kinase 4, PGAM1 - phosphoglycerate mutase 1 (brain), AK3 - adenylate kinase 3, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, ADPGK - adp-dependent glucokinase]
188 [DLG1 - discs, large homolog 1 (drosophila), AK4 - adenylate kinase 4, PGM1 - phosphoglucomutase 1, HK2 - hexokinase 2, PGAM1 - phosphoglycerate mutase 1 (brain), TJP2 - tight junction protein 2, AK3 - adenylate kinase 3, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, ADPGK - adp-dependent glucokinase]
189 [DGUOK - deoxyguanosine kinase, HK2 - hexokinase 2, PGM1 - phosphoglucomutase 1, AK4 - adenylate kinase 4, PGAM1 - phosphoglycerate mutase 1 (brain), UCK2 - uridine-cytidine kinase 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, AK3 - adenylate kinase 3, ADPGK - adp-dependent glucokinase]
190 [SCAF8 - sr-related ctd-associated factor 8, SERBP1 - serpine1 mrna binding protein 1, NCBP2 - nuclear cap binding protein subunit 2, 20kda, QKI - qki, kh domain containing, rna binding, DDX6 - dead (asp-glu-ala-asp) box helicase 6, MRPL13 - mitochondrial ribosomal protein l13, TDRD7 - tudor domain containing 7, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), RBM26 - rna binding motif protein 26, LARP4B - la ribonucleoprotein domain family, member 4b, RPL7 - ribosomal protein l7, RSL1D1 - ribosomal l1 domain containing 1, LARP1 - la ribonucleoprotein domain family, member 1, RPS13 - ribosomal protein s13, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, CARHSP1 - calcium regulated heat stable protein 1, 24kda, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, EIF4G3 - eukaryotic translation initiation factor 4 gamma, 3, ILF3 - interleukin enhancer binding factor 3, 90kda, RPS7 - ribosomal protein s7, THOC5 - tho complex 5, EIF2S2 - eukaryotic translation initiation factor 2, subunit 2 beta, 38kda]
191 [DLG1 - discs, large homolog 1 (drosophila), HK2 - hexokinase 2, PGM1 - phosphoglucomutase 1, AK4 - adenylate kinase 4, TJP2 - tight junction protein 2, PGAM1 - phosphoglycerate mutase 1 (brain), AK3 - adenylate kinase 3, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, ADPGK - adp-dependent glucokinase]
192 [DGUOK - deoxyguanosine kinase, RAB23 - rab23, member ras oncogene family, DLG1 - discs, large homolog 1 (drosophila), AK4 - adenylate kinase 4, TJP2 - tight junction protein 2, AK3 - adenylate kinase 3, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, MFN1 - mitofusin 1]
193 [SERBP1 - serpine1 mrna binding protein 1, EXOSC5 - exosome component 5, PGAM1 - phosphoglycerate mutase 1 (brain), SCFD1 - sec1 family domain containing 1, HMGB1 - high mobility group box 1, NUP50 - nucleoporin 50kda, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, TNPO1 - transportin 1, HK2 - hexokinase 2, TRIM38 - tripartite motif containing 38, USP25 - ubiquitin specific peptidase 25, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, CARHSP1 - calcium regulated heat stable protein 1, 24kda, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, EXOC1 - exocyst complex component 1, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, CTSC - cathepsin c, EXOSC1 - exosome component 1, ATG7 - autophagy related 7, SEH1L - seh1-like (s. cerevisiae), EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), NUB1 - negative regulator of ubiquitin-like proteins 1, NUP88 - nucleoporin 88kda, SCO1 - sco1 cytochrome c oxidase assembly protein, LARP4B - la ribonucleoprotein domain family, member 4b, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), RPS7 - ribosomal protein s7, SNX6 - sorting nexin 6, SUMO1 - small ubiquitin-like modifier 1, CISD2 - cdgsh iron sulfur domain 2, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), LARP1 - la ribonucleoprotein domain family, member 1, PRKCD - protein kinase c, delta, NAMPT - nicotinamide phosphoribosyltransferase, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, RPTOR - regulatory associated protein of mtor, complex 1, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12]
194 [RNF31 - ring finger protein 31, WNK1 - wnk lysine deficient protein kinase 1, HMGB1 - high mobility group box 1, CYFIP2 - cytoplasmic fmr1 interacting protein 2, EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, IRAK4 - interleukin-1 receptor-associated kinase 4, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, HSPD1 - heat shock 60kda protein 1 (chaperonin), EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, IRF3 - interferon regulatory factor 3, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, CUL1 - cullin 1, PLCG1 - phospholipase c, gamma 1, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, PRKCD - protein kinase c, delta, IFIH1 - interferon induced with helicase c domain 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, WIPF1 - was/wasl interacting protein family, member 1]
195 [DDX31 - dead (asp-glu-ala-asp) box polypeptide 31, DDX6 - dead (asp-glu-ala-asp) box helicase 6, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, GSPT1 - g1 to s phase transition 1, RAB30 - rab30, member ras oncogene family, GFM2 - g elongation factor, mitochondrial 2, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, RECQL - recq protein-like (dna helicase q1-like), DCK - deoxycytidine kinase, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, LRRK1 - leucine-rich repeat kinase 1, AGPS - alkylglycerone phosphate synthase, SORD - sorbitol dehydrogenase, RAB23 - rab23, member ras oncogene family, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, WDR77 - wd repeat domain 77, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, MYO6 - myosin vi, SGPL1 - sphingosine-1-phosphate lyase 1, MCM7 - minichromosome maintenance complex component 7, ARL1 - adp-ribosylation factor-like 1, STK38L - serine/threonine kinase 38 like, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), HADH - hydroxyacyl-coa dehydrogenase, OSBPL3 - oxysterol binding protein-like 3, CDK6 - cyclin-dependent kinase 6, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, MUT - methylmalonyl coa mutase, CDK13 - cyclin-dependent kinase 13, HSPD1 - heat shock 60kda protein 1 (chaperonin), MVD - mevalonate (diphospho) decarboxylase, CCT4 - chaperonin containing tcp1, subunit 4 (delta), GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, KYNU - kynureninase, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, TAOK3 - tao kinase 3, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, HSPA4 - heat shock 70kda protein 4, MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, WNK1 - wnk lysine deficient protein kinase 1, NEK6 - nima-related kinase 6, UBE2Z - ubiquitin-conjugating enzyme e2z, AGK - acylglycerol kinase, FABP5 - fatty acid binding protein 5 (psoriasis-associated), MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, PYGB - phosphorylase, glycogen; brain, RAB14 - rab14, member ras oncogene family, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, UCK2 - uridine-cytidine kinase 2, PPP5C - protein phosphatase 5, catalytic subunit, CARS - cysteinyl-trna synthetase, MANBA - mannosidase, beta a, lysosomal, IRAK4 - interleukin-1 receptor-associated kinase 4, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, HK2 - hexokinase 2, AK4 - adenylate kinase 4, MAT2A - methionine adenosyltransferase ii, alpha, IDE - insulin-degrading enzyme, MCM3 - minichromosome maintenance complex component 3, ABCF2 - atp-binding cassette, sub-family f (gcn20), member 2, IFIH1 - interferon induced with helicase c domain 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), DCAKD - dephospho-coa kinase domain containing, VDAC2 - voltage-dependent anion channel 2, TTLL12 - tubulin tyrosine ligase-like family, member 12, UBA6 - ubiquitin-like modifier activating enzyme 6, GRK6 - g protein-coupled receptor kinase 6, SCYL2 - scy1-like 2 (s. cerevisiae), RAP1A - rap1a, member of ras oncogene family, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, MFN1 - mitofusin 1, DGUOK - deoxyguanosine kinase, UBA3 - ubiquitin-like modifier activating enzyme 3, RAP1B - rap1b, member of ras oncogene family, GCDH - glutaryl-coa dehydrogenase, ACOT7 - acyl-coa thioesterase 7, CAT - catalase, UGDH - udp-glucose 6-dehydrogenase, RHOT1 - ras homolog family member t1, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, UBE2I - ubiquitin-conjugating enzyme e2i, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, PRKCD - protein kinase c, delta, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, RRM1 - ribonucleotide reductase m1, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
196 [MCM7 - minichromosome maintenance complex component 7, ATG7 - autophagy related 7, CAPN2 - calpain 2, (m/ii) large subunit, RAP1A - rap1a, member of ras oncogene family, PPM1A - protein phosphatase, mg2+/mn2+ dependent, 1a, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, HMGB2 - high mobility group box 2, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), HMGB1 - high mobility group box 1, STAT6 - signal transducer and activator of transcription 6, interleukin-4 induced, HADH - hydroxyacyl-coa dehydrogenase, RAP1B - rap1b, member of ras oncogene family, EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), GOLPH3 - golgi phosphoprotein 3 (coat-protein), ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, FECH - ferrochelatase, USP8 - ubiquitin specific peptidase 8, ACAT1 - acetyl-coa acetyltransferase 1, SNX6 - sorting nexin 6, CAT - catalase, HSPD1 - heat shock 60kda protein 1 (chaperonin), EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), TBC1D4 - tbc1 domain family, member 4, CHMP5 - charged multivesicular body protein 5, SORD - sorbitol dehydrogenase, PLCG1 - phospholipase c, gamma 1, LIMS1 - lim and senescent cell antigen-like domains 1, BCL2L1 - bcl2-like 1, LARP1 - la ribonucleoprotein domain family, member 1, PRKCD - protein kinase c, delta, NAMPT - nicotinamide phosphoribosyltransferase, RPTOR - regulatory associated protein of mtor, complex 1, CREB1 - camp responsive element binding protein 1, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha]
197 [NCBP2 - nuclear cap binding protein subunit 2, 20kda, DDX6 - dead (asp-glu-ala-asp) box helicase 6, EXOSC5 - exosome component 5, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), GSPT1 - g1 to s phase transition 1, RPL14 - ribosomal protein l14, HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, PELO - pelota homolog (drosophila), RPS8 - ribosomal protein s8, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPL3 - ribosomal protein l3, ADPGK - adp-dependent glucokinase, RPS7 - ribosomal protein s7, ACAT1 - acetyl-coa acetyltransferase 1, DUT - deoxyuridine triphosphatase, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), HK2 - hexokinase 2, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, ACOT7 - acyl-coa thioesterase 7, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, KPNB1 - karyopherin (importin) beta 1, COMT - catechol-o-methyltransferase, KYNU - kynureninase, RPL31 - ribosomal protein l31, EIF3E - eukaryotic translation initiation factor 3, subunit e, RPL32 - ribosomal protein l32, EXOSC1 - exosome component 1]
198 [MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, ACAT1 - acetyl-coa acetyltransferase 1, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, GCDH - glutaryl-coa dehydrogenase, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, MUT - methylmalonyl coa mutase, GLS - glutaminase, IARS - isoleucyl-trna synthetase, ADI1 - acireductone dioxygenase 1, KYNU - kynureninase, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, CARS - cysteinyl-trna synthetase, PPA1 - pyrophosphatase (inorganic) 1, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
199 [UBA6 - ubiquitin-like modifier activating enzyme 6, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, ATG7 - autophagy related 7, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, UBA3 - ubiquitin-like modifier activating enzyme 3, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, CARS - cysteinyl-trna synthetase, IARS - isoleucyl-trna synthetase, TRIM25 - tripartite motif containing 25, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
200 [SERBP1 - serpine1 mrna binding protein 1, EXOSC5 - exosome component 5, PGAM1 - phosphoglycerate mutase 1 (brain), SCFD1 - sec1 family domain containing 1, HMGB1 - high mobility group box 1, NUP50 - nucleoporin 50kda, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, USP8 - ubiquitin specific peptidase 8, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, TNPO1 - transportin 1, HK2 - hexokinase 2, TRIM38 - tripartite motif containing 38, USP25 - ubiquitin specific peptidase 25, IDE - insulin-degrading enzyme, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, CARHSP1 - calcium regulated heat stable protein 1, 24kda, EXOC1 - exocyst complex component 1, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, SNX1 - sorting nexin 1, CTSC - cathepsin c, EXOSC1 - exosome component 1, ATG7 - autophagy related 7, GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, SEH1L - seh1-like (s. cerevisiae), EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), NUB1 - negative regulator of ubiquitin-like proteins 1, SCO1 - sco1 cytochrome c oxidase assembly protein, NUP88 - nucleoporin 88kda, LARP4B - la ribonucleoprotein domain family, member 4b, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), RPS7 - ribosomal protein s7, SNX6 - sorting nexin 6, SUMO1 - small ubiquitin-like modifier 1, CISD2 - cdgsh iron sulfur domain 2, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), PRKCD - protein kinase c, delta, LARP1 - la ribonucleoprotein domain family, member 1, NAMPT - nicotinamide phosphoribosyltransferase, ARIH1 - ariadne homolog, ubiquitin-conjugating enzyme e2 binding protein, 1 (drosophila), RPTOR - regulatory associated protein of mtor, complex 1, SPTLC1 - serine palmitoyltransferase, long chain base subunit 1, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12]
201 [CCNK - cyclin k, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, NCBP2 - nuclear cap binding protein subunit 2, 20kda, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, TCOF1 - treacher collins-franceschetti syndrome 1, RFC3 - replication factor c (activator 1) 3, 38kda, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, DCK - deoxycytidine kinase, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, UCK2 - uridine-cytidine kinase 2, PPP5C - protein phosphatase 5, catalytic subunit, UMPS - uridine monophosphate synthetase, BTF3 - basic transcription factor 3, TRIM25 - tripartite motif containing 25, ACAT1 - acetyl-coa acetyltransferase 1, DUT - deoxyuridine triphosphatase, HK2 - hexokinase 2, AK4 - adenylate kinase 4, GAR1 - gar1 ribonucleoprotein, CREB1 - camp responsive element binding protein 1, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, DCAKD - dephospho-coa kinase domain containing, TELO2 - telomere maintenance 2, SCAF8 - sr-related ctd-associated factor 8, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, SNRPE - small nuclear ribonucleoprotein polypeptide e, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), RNMT - rna (guanine-7-) methyltransferase, DGUOK - deoxyguanosine kinase, POLR1C - polymerase (rna) i polypeptide c, 30kda, ADPGK - adp-dependent glucokinase, NHP2 - nhp2 ribonucleoprotein, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, ACOT7 - acyl-coa thioesterase 7, GCDH - glutaryl-coa dehydrogenase, CDK13 - cyclin-dependent kinase 13, GNPDA2 - glucosamine-6-phosphate deaminase 2, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, UGDH - udp-glucose 6-dehydrogenase, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), KYNU - kynureninase, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, NAMPT - nicotinamide phosphoribosyltransferase, AK3 - adenylate kinase 3, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, RRM1 - ribonucleotide reductase m1, TCEA1 - transcription elongation factor a (sii), 1]
202 [MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, ATG7 - autophagy related 7, MCM7 - minichromosome maintenance complex component 7, CAPN2 - calpain 2, (m/ii) large subunit, RAP1A - rap1a, member of ras oncogene family, GSN - gelsolin, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), GNPAT - glyceronephosphate o-acyltransferase, STAT6 - signal transducer and activator of transcription 6, interleukin-4 induced, HADH - hydroxyacyl-coa dehydrogenase, RAP1B - rap1b, member of ras oncogene family, UMPS - uridine monophosphate synthetase, PPP5C - protein phosphatase 5, catalytic subunit, GOLPH3 - golgi phosphoprotein 3 (coat-protein), USP8 - ubiquitin specific peptidase 8, FECH - ferrochelatase, AK4 - adenylate kinase 4, CAT - catalase, HSPD1 - heat shock 60kda protein 1 (chaperonin), NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), COMT - catechol-o-methyltransferase, KYNU - kynureninase, CHMP5 - charged multivesicular body protein 5, SORD - sorbitol dehydrogenase, IRF5 - interferon regulatory factor 5, PRKCD - protein kinase c, delta, BCL2L1 - bcl2-like 1, LARP1 - la ribonucleoprotein domain family, member 1, CREB1 - camp responsive element binding protein 1, MYO6 - myosin vi, PARP4 - poly (adp-ribose) polymerase family, member 4]
203 [EIF4H - eukaryotic translation initiation factor 4h, EIF4B - eukaryotic translation initiation factor 4b, EIF2D - eukaryotic translation initiation factor 2d, TSFM - ts translation elongation factor, mitochondrial, GSPT1 - g1 to s phase transition 1, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, EIF1AD - eukaryotic translation initiation factor 1a domain containing, EIF4G3 - eukaryotic translation initiation factor 4 gamma, 3, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, EIF3E - eukaryotic translation initiation factor 3, subunit e, EIF3F - eukaryotic translation initiation factor 3, subunit f, EIF2S2 - eukaryotic translation initiation factor 2, subunit 2 beta, 38kda]
204 [MTR - 5-methyltetrahydrofolate-homocysteine methyltransferase, ATG7 - autophagy related 7, SNX6 - sorting nexin 6, CAPN2 - calpain 2, (m/ii) large subunit, RAP1A - rap1a, member of ras oncogene family, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, STAT6 - signal transducer and activator of transcription 6, interleukin-4 induced, IRF3 - interferon regulatory factor 3, TBC1D4 - tbc1 domain family, member 4, CHMP5 - charged multivesicular body protein 5, LARP1 - la ribonucleoprotein domain family, member 1, PRKCD - protein kinase c, delta, BCL2L1 - bcl2-like 1, NAMPT - nicotinamide phosphoribosyltransferase, IFIH1 - interferon induced with helicase c domain 1, RAP1B - rap1b, member of ras oncogene family, CREB1 - camp responsive element binding protein 1, RPTOR - regulatory associated protein of mtor, complex 1, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, GOLPH3 - golgi phosphoprotein 3 (coat-protein)]
205 [DGUOK - deoxyguanosine kinase, RAB23 - rab23, member ras oncogene family, DLG1 - discs, large homolog 1 (drosophila), AK4 - adenylate kinase 4, TJP2 - tight junction protein 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, UCK2 - uridine-cytidine kinase 2, AK3 - adenylate kinase 3, UMPS - uridine monophosphate synthetase, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, MFN1 - mitofusin 1]
206 [PTCD3 - pentatricopeptide repeat domain 3, MRPS14 - mitochondrial ribosomal protein s14, MRPL47 - mitochondrial ribosomal protein l47, MRPS35 - mitochondrial ribosomal protein s35, MRPL2 - mitochondrial ribosomal protein l2, MRPL42 - mitochondrial ribosomal protein l42, MRPL13 - mitochondrial ribosomal protein l13, MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, GSPT1 - g1 to s phase transition 1, MRPL3 - mitochondrial ribosomal protein l3, GFM2 - g elongation factor, mitochondrial 2]
207 [NCBP2 - nuclear cap binding protein subunit 2, 20kda, DDX6 - dead (asp-glu-ala-asp) box helicase 6, EXOSC5 - exosome component 5, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), GSPT1 - g1 to s phase transition 1, RPL14 - ribosomal protein l14, HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, PELO - pelota homolog (drosophila), RPS8 - ribosomal protein s8, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPL3 - ribosomal protein l3, ADPGK - adp-dependent glucokinase, RPS7 - ribosomal protein s7, ACAT1 - acetyl-coa acetyltransferase 1, DUT - deoxyuridine triphosphatase, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), HK2 - hexokinase 2, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, ACOT7 - acyl-coa thioesterase 7, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, KPNB1 - karyopherin (importin) beta 1, COMT - catechol-o-methyltransferase, KYNU - kynureninase, RPL31 - ribosomal protein l31, EIF3E - eukaryotic translation initiation factor 3, subunit e, RPL32 - ribosomal protein l32, EXOSC1 - exosome component 1]
208 [PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, DLG1 - discs, large homolog 1 (drosophila), AK4 - adenylate kinase 4, MFN1 - mitofusin 1, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, DGUOK - deoxyguanosine kinase, RAB23 - rab23, member ras oncogene family, DCK - deoxycytidine kinase, TJP2 - tight junction protein 2, AK3 - adenylate kinase 3, UCK2 - uridine-cytidine kinase 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, UMPS - uridine monophosphate synthetase]
209 [ACAT1 - acetyl-coa acetyltransferase 1, DUT - deoxyuridine triphosphatase, PGM1 - phosphoglucomutase 1, HK2 - hexokinase 2, ACOT7 - acyl-coa thioesterase 7, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, PGAM1 - phosphoglycerate mutase 1 (brain), NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, DCK - deoxycytidine kinase, UCK2 - uridine-cytidine kinase 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, UMPS - uridine monophosphate synthetase, ADPGK - adp-dependent glucokinase]
210 [TRAPPC10 - trafficking protein particle complex 10, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), TRAPPC9 - trafficking protein particle complex 9, EXOC2 - exocyst complex component 2, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), HOOK3 - hook microtubule-tethering protein 3, STX17 - syntaxin 17, EXOC1 - exocyst complex component 1, RINT1 - rad50 interactor 1]
211 [DST - dystonin, DDX6 - dead (asp-glu-ala-asp) box helicase 6, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, RFC3 - replication factor c (activator 1) 3, 38kda, SDAD1 - sda1 domain containing 1, F11R - f11 receptor, SCFD1 - sec1 family domain containing 1, GSN - gelsolin, RAB30 - rab30, member ras oncogene family, GFM2 - g elongation factor, mitochondrial 2, TMED2 - transmembrane emp24 domain trafficking protein 2, HOOK3 - hook microtubule-tethering protein 3, RECQL - recq protein-like (dna helicase q1-like), FLNB - filamin b, beta, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, DBN1 - drebrin 1, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, ANKRD28 - ankyrin repeat domain 28, CYCS - cytochrome c, somatic, RIF1 - rap1 interacting factor homolog (yeast), VTI1A - vesicle transport through interaction with t-snares 1a, EHBP1 - eh domain binding protein 1, AGPS - alkylglycerone phosphate synthase, GOSR2 - golgi snap receptor complex member 2, TMED5 - transmembrane emp24 protein transport domain containing 5, RAB23 - rab23, member ras oncogene family, GAR1 - gar1 ribonucleoprotein, CREB1 - camp responsive element binding protein 1, KDM3A - lysine (k)-specific demethylase 3a, TELO2 - telomere maintenance 2, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), EIF2D - eukaryotic translation initiation factor 2d, MCM7 - minichromosome maintenance complex component 7, ATG7 - autophagy related 7, TUBGCP4 - tubulin, gamma complex associated protein 4, ARL1 - adp-ribosylation factor-like 1, SEH1L - seh1-like (s. cerevisiae), SYNE1 - spectrin repeat containing, nuclear envelope 1, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), PELO - pelota homolog (drosophila), GOSR1 - golgi snap receptor complex member 1, NIP7 - nip7, nucleolar pre-rrna processing protein, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), HSPD1 - heat shock 60kda protein 1 (chaperonin), TBCD - tubulin folding cofactor d, ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, STX8 - syntaxin 8, HSPA4 - heat shock 70kda protein 4, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, SERBP1 - serpine1 mrna binding protein 1, PURA - purine-rich element binding protein a, VTA1 - vesicle (multivesicular body) trafficking 1, SPG11 - spastic paraplegia 11 (autosomal recessive), ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, NEK6 - nima-related kinase 6, HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, AGK - acylglycerol kinase, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), MAP4 - microtubule-associated protein 4, RAB14 - rab14, member ras oncogene family, SURF4 - surfeit 4, MAN2A1 - mannosidase, alpha, class 2a, member 1, DNAJC11 - dnaj (hsp40) homolog, subfamily c, member 11, USP8 - ubiquitin specific peptidase 8, TRAPPC10 - trafficking protein particle complex 10, STX17 - syntaxin 17, HK2 - hexokinase 2, KPNB1 - karyopherin (importin) beta 1, IDE - insulin-degrading enzyme, MCM3 - minichromosome maintenance complex component 3, CHMP5 - charged multivesicular body protein 5, DOCK7 - dedicator of cytokinesis 7, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), POLDIP2 - polymerase (dna-directed), delta interacting protein 2, VDAC2 - voltage-dependent anion channel 2, CTSC - cathepsin c, NIPBL - nipped-b homolog (drosophila), DLG1 - discs, large homolog 1 (drosophila), PARVG - parvin, gamma, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), AKAP8L - a kinase (prka) anchor protein 8-like, GNPAT - glyceronephosphate o-acyltransferase, MFN1 - mitofusin 1, WDR1 - wd repeat domain 1, TRAPPC9 - trafficking protein particle complex 9, GOLPH3 - golgi phosphoprotein 3 (coat-protein), NHP2 - nhp2 ribonucleoprotein, SUMO1 - small ubiquitin-like modifier 1, CISD2 - cdgsh iron sulfur domain 2, CAT - catalase, SSBP1 - single-stranded dna binding protein 1, mitochondrial, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), SAMM50 - samm50 sorting and assembly machinery component, RHOT1 - ras homolog family member t1, TUBGCP3 - tubulin, gamma complex associated protein 3, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, BCL2L1 - bcl2-like 1, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, CCDC6 - coiled-coil domain containing 6, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, EPS15 - epidermal growth factor receptor pathway substrate 15]
212 [PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, IDE - insulin-degrading enzyme, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3]
213 [IDE - insulin-degrading enzyme, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3]
214 [IDE - insulin-degrading enzyme, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3]
215 [AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, DST - dystonin, VTA1 - vesicle (multivesicular body) trafficking 1, DENND3 - denn/madd domain containing 3, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, GAPVD1 - gtpase activating protein and vps9 domains 1, SPG11 - spastic paraplegia 11 (autosomal recessive), BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, F11R - f11 receptor, STAMBP - stam binding protein, GSN - gelsolin, SCFD1 - sec1 family domain containing 1, HMGB1 - high mobility group box 1, TMED2 - transmembrane emp24 domain trafficking protein 2, FLNB - filamin b, beta, RAB14 - rab14, member ras oncogene family, DERL1 - derlin 1, SURF4 - surfeit 4, USP8 - ubiquitin specific peptidase 8, VTI1A - vesicle transport through interaction with t-snares 1a, RPN1 - ribophorin i, STX17 - syntaxin 17, ARCN1 - archain 1, EHBP1 - eh domain binding protein 1, TMED5 - transmembrane emp24 protein transport domain containing 5, CHMP5 - charged multivesicular body protein 5, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, RAB23 - rab23, member ras oncogene family, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), SNX1 - sorting nexin 1, MYO6 - myosin vi, VDAC2 - voltage-dependent anion channel 2, CTSC - cathepsin c, WIPF1 - was/wasl interacting protein family, member 1, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), TUBGCP4 - tubulin, gamma complex associated protein 4, SCYL2 - scy1-like 2 (s. cerevisiae), RAP1A - rap1a, member of ras oncogene family, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), GOSR1 - golgi snap receptor complex member 1, ESD - esterase d, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), GOLPH3 - golgi phosphoprotein 3 (coat-protein), STOM - stomatin, SNX6 - sorting nexin 6, HSPD1 - heat shock 60kda protein 1 (chaperonin), CCT4 - chaperonin containing tcp1, subunit 4 (delta), AP3S1 - adaptor-related protein complex 3, sigma 1 subunit, DENND4A - denn/madd domain containing 4a, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, STX8 - syntaxin 8, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha]
216 [CCNK - cyclin k, NCBP2 - nuclear cap binding protein subunit 2, 20kda, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, TCOF1 - treacher collins-franceschetti syndrome 1, RFC3 - replication factor c (activator 1) 3, 38kda, PGM1 - phosphoglucomutase 1, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, DCK - deoxycytidine kinase, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, UCK2 - uridine-cytidine kinase 2, UMPS - uridine monophosphate synthetase, PPP5C - protein phosphatase 5, catalytic subunit, TRIM25 - tripartite motif containing 25, BTF3 - basic transcription factor 3, ACAT1 - acetyl-coa acetyltransferase 1, DUT - deoxyuridine triphosphatase, CPOX - coproporphyrinogen oxidase, HK2 - hexokinase 2, AK4 - adenylate kinase 4, GAR1 - gar1 ribonucleoprotein, CREB1 - camp responsive element binding protein 1, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, DCAKD - dephospho-coa kinase domain containing, TELO2 - telomere maintenance 2, SCAF8 - sr-related ctd-associated factor 8, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, LSS - lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase), SNRPE - small nuclear ribonucleoprotein polypeptide e, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), RNMT - rna (guanine-7-) methyltransferase, DGUOK - deoxyguanosine kinase, OSBPL3 - oxysterol binding protein-like 3, POLR1C - polymerase (rna) i polypeptide c, 30kda, ADPGK - adp-dependent glucokinase, NHP2 - nhp2 ribonucleoprotein, FECH - ferrochelatase, GCDH - glutaryl-coa dehydrogenase, ACOT7 - acyl-coa thioesterase 7, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, CDK13 - cyclin-dependent kinase 13, UGDH - udp-glucose 6-dehydrogenase, GNPDA2 - glucosamine-6-phosphate deaminase 2, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, MVD - mevalonate (diphospho) decarboxylase, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), KYNU - kynureninase, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, NAMPT - nicotinamide phosphoribosyltransferase, AK3 - adenylate kinase 3, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, RRM1 - ribonucleotide reductase m1, TCEA1 - transcription elongation factor a (sii), 1]
217 [NCBP2 - nuclear cap binding protein subunit 2, 20kda, DDX6 - dead (asp-glu-ala-asp) box helicase 6, EXOSC5 - exosome component 5, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, PGM1 - phosphoglucomutase 1, GSPT1 - g1 to s phase transition 1, PGAM1 - phosphoglycerate mutase 1 (brain), RPL14 - ribosomal protein l14, HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, PELO - pelota homolog (drosophila), RPS8 - ribosomal protein s8, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPL3 - ribosomal protein l3, RPS7 - ribosomal protein s7, ADPGK - adp-dependent glucokinase, ACAT1 - acetyl-coa acetyltransferase 1, DUT - deoxyuridine triphosphatase, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), HK2 - hexokinase 2, ACOT7 - acyl-coa thioesterase 7, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, KPNB1 - karyopherin (importin) beta 1, RPL31 - ribosomal protein l31, EIF3E - eukaryotic translation initiation factor 3, subunit e, RPL32 - ribosomal protein l32, EXOSC1 - exosome component 1]
218 [DLG1 - discs, large homolog 1 (drosophila), HK2 - hexokinase 2, PGM1 - phosphoglucomutase 1, AK4 - adenylate kinase 4, TJP2 - tight junction protein 2, PGAM1 - phosphoglycerate mutase 1 (brain), AK3 - adenylate kinase 3, ADPGK - adp-dependent glucokinase]
219 [EIF4H - eukaryotic translation initiation factor 4h, MBNL1 - muscleblind-like splicing regulator 1, EIF4B - eukaryotic translation initiation factor 4b, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, IFIH1 - interferon induced with helicase c domain 1, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, HSPD1 - heat shock 60kda protein 1 (chaperonin), DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, HMGB1 - high mobility group box 1, ILF3 - interleukin enhancer binding factor 3, 90kda]
220 [DLG1 - discs, large homolog 1 (drosophila), AK4 - adenylate kinase 4, PGM1 - phosphoglucomutase 1, HK2 - hexokinase 2, PGAM1 - phosphoglycerate mutase 1 (brain), TJP2 - tight junction protein 2, AK3 - adenylate kinase 3, ADPGK - adp-dependent glucokinase]
221 [AP2A2 - adaptor-related protein complex 2, alpha 2 subunit, PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1)]
222 [PITRM1 - pitrilysin metallopeptidase 1, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, RPL14 - ribosomal protein l14, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), GNPAT - glyceronephosphate o-acyltransferase, RPL13 - ribosomal protein l13, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), RPS8 - ribosomal protein s8, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPL3 - ribosomal protein l3, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, RPS7 - ribosomal protein s7, VTI1A - vesicle transport through interaction with t-snares 1a, SRP72 - signal recognition particle 72kda, CAT - catalase, AGPS - alkylglycerone phosphate synthase, GOSR2 - golgi snap receptor complex member 2, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), IDE - insulin-degrading enzyme, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide]
223 [DDX31 - dead (asp-glu-ala-asp) box polypeptide 31, DDX6 - dead (asp-glu-ala-asp) box helicase 6, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, NEK6 - nima-related kinase 6, WNK1 - wnk lysine deficient protein kinase 1, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, GSPT1 - g1 to s phase transition 1, UBE2Z - ubiquitin-conjugating enzyme e2z, AGK - acylglycerol kinase, RAB30 - rab30, member ras oncogene family, GFM2 - g elongation factor, mitochondrial 2, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RECQL - recq protein-like (dna helicase q1-like), RAB14 - rab14, member ras oncogene family, DCK - deoxycytidine kinase, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, UCK2 - uridine-cytidine kinase 2, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, PPP5C - protein phosphatase 5, catalytic subunit, CARS - cysteinyl-trna synthetase, IRAK4 - interleukin-1 receptor-associated kinase 4, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, AK4 - adenylate kinase 4, HK2 - hexokinase 2, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), LRRK1 - leucine-rich repeat kinase 1, MAT2A - methionine adenosyltransferase ii, alpha, AGPS - alkylglycerone phosphate synthase, IDE - insulin-degrading enzyme, MCM3 - minichromosome maintenance complex component 3, SORD - sorbitol dehydrogenase, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, RAB23 - rab23, member ras oncogene family, ABCF2 - atp-binding cassette, sub-family f (gcn20), member 2, WDR77 - wd repeat domain 77, IFIH1 - interferon induced with helicase c domain 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, MYO6 - myosin vi, VDAC2 - voltage-dependent anion channel 2, DCAKD - dephospho-coa kinase domain containing, TTLL12 - tubulin tyrosine ligase-like family, member 12, UBA6 - ubiquitin-like modifier activating enzyme 6, GRK6 - g protein-coupled receptor kinase 6, MCM7 - minichromosome maintenance complex component 7, ARL1 - adp-ribosylation factor-like 1, SCYL2 - scy1-like 2 (s. cerevisiae), RAP1A - rap1a, member of ras oncogene family, STK38L - serine/threonine kinase 38 like, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, MFN1 - mitofusin 1, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), DGUOK - deoxyguanosine kinase, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), HADH - hydroxyacyl-coa dehydrogenase, UBA3 - ubiquitin-like modifier activating enzyme 3, RAP1B - rap1b, member of ras oncogene family, CDK6 - cyclin-dependent kinase 6, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, ACOT7 - acyl-coa thioesterase 7, GCDH - glutaryl-coa dehydrogenase, CDK13 - cyclin-dependent kinase 13, CAT - catalase, HSPD1 - heat shock 60kda protein 1 (chaperonin), CCT4 - chaperonin containing tcp1, subunit 4 (delta), MVD - mevalonate (diphospho) decarboxylase, UGDH - udp-glucose 6-dehydrogenase, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, RHOT1 - ras homolog family member t1, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, UBE2I - ubiquitin-conjugating enzyme e2i, TAOK3 - tao kinase 3, PRKCD - protein kinase c, delta, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3, HSPA4 - heat shock 70kda protein 4, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, RRM1 - ribonucleotide reductase m1, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
224 [PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, UQCRB - ubiquinol-cytochrome c reductase binding protein, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, DLG1 - discs, large homolog 1 (drosophila), PGM1 - phosphoglucomutase 1, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, PGAM1 - phosphoglycerate mutase 1 (brain), MFN1 - mitofusin 1, DGUOK - deoxyguanosine kinase, DCK - deoxycytidine kinase, UCK2 - uridine-cytidine kinase 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, UMPS - uridine monophosphate synthetase, MANBA - mannosidase, beta a, lysosomal, GOLPH3 - golgi phosphoprotein 3 (coat-protein), ADPGK - adp-dependent glucokinase, MAN2A1 - mannosidase, alpha, class 2a, member 1, ACAT1 - acetyl-coa acetyltransferase 1, DUT - deoxyuridine triphosphatase, HK2 - hexokinase 2, AK4 - adenylate kinase 4, GCDH - glutaryl-coa dehydrogenase, ACOT7 - acyl-coa thioesterase 7, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, MVD - mevalonate (diphospho) decarboxylase, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, GNPDA2 - glucosamine-6-phosphate deaminase 2, UGDH - udp-glucose 6-dehydrogenase, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, KYNU - kynureninase, SORD - sorbitol dehydrogenase, ERP44 - endoplasmic reticulum protein 44, RAB23 - rab23, member ras oncogene family, TJP2 - tight junction protein 2, AK3 - adenylate kinase 3, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, PGM2 - phosphoglucomutase 2, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, DCAKD - dephospho-coa kinase domain containing, RRM1 - ribonucleotide reductase m1]
225 [NCBP2 - nuclear cap binding protein subunit 2, 20kda, DDX6 - dead (asp-glu-ala-asp) box helicase 6, EXOSC5 - exosome component 5, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), GSPT1 - g1 to s phase transition 1, RPL14 - ribosomal protein l14, HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, PELO - pelota homolog (drosophila), RPS8 - ribosomal protein s8, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPL3 - ribosomal protein l3, ADPGK - adp-dependent glucokinase, RPS7 - ribosomal protein s7, ACAT1 - acetyl-coa acetyltransferase 1, DUT - deoxyuridine triphosphatase, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), HK2 - hexokinase 2, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, ACOT7 - acyl-coa thioesterase 7, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, KPNB1 - karyopherin (importin) beta 1, KYNU - kynureninase, RPL31 - ribosomal protein l31, EIF3E - eukaryotic translation initiation factor 3, subunit e, RPL32 - ribosomal protein l32, EXOSC1 - exosome component 1]
226 [NCBP2 - nuclear cap binding protein subunit 2, 20kda, DDX6 - dead (asp-glu-ala-asp) box helicase 6, EXOSC5 - exosome component 5, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), GSPT1 - g1 to s phase transition 1, RPL14 - ribosomal protein l14, HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, RPL13 - ribosomal protein l13, PELO - pelota homolog (drosophila), RPS8 - ribosomal protein s8, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, RPL3 - ribosomal protein l3, ADPGK - adp-dependent glucokinase, RPS7 - ribosomal protein s7, ACAT1 - acetyl-coa acetyltransferase 1, DUT - deoxyuridine triphosphatase, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), HK2 - hexokinase 2, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, ACOT7 - acyl-coa thioesterase 7, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, KPNB1 - karyopherin (importin) beta 1, KYNU - kynureninase, RPL31 - ribosomal protein l31, EIF3E - eukaryotic translation initiation factor 3, subunit e, RPL32 - ribosomal protein l32, EXOSC1 - exosome component 1]
227 [HK2 - hexokinase 2, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), AGK - acylglycerol kinase, MFN1 - mitofusin 1, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), SAMM50 - samm50 sorting and assembly machinery component, RHOT1 - ras homolog family member t1, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, BCL2L1 - bcl2-like 1, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, HSPA4 - heat shock 70kda protein 4, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, VDAC2 - voltage-dependent anion channel 2, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, DNAJC11 - dnaj (hsp40) homolog, subfamily c, member 11]
228 [SCAF8 - sr-related ctd-associated factor 8, ZC3H7B - zinc finger ccch-type containing 7b, QKI - qki, kh domain containing, rna binding, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, NCBP2 - nuclear cap binding protein subunit 2, 20kda, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, EXOSC5 - exosome component 5, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, SNRPE - small nuclear ribonucleoprotein polypeptide e, ZNF638 - zinc finger protein 638, SF3A3 - splicing factor 3a, subunit 3, 60kda, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, PUS1 - pseudouridylate synthase 1, RPL14 - ribosomal protein l14, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, AKAP8L - a kinase (prka) anchor protein 8-like, URB1 - urb1 ribosome biogenesis 1 homolog (s. cerevisiae), SNRNP27 - small nuclear ribonucleoprotein 27kda (u4/u6.u5), RNMT - rna (guanine-7-) methyltransferase, RBM26 - rna binding motif protein 26, ZBTB8OS - zinc finger and btb domain containing 8 opposite strand, RPS8 - ribosomal protein s8, RPL7 - ribosomal protein l7, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), PPIL3 - peptidylprolyl isomerase (cyclophilin)-like 3, RPS6 - ribosomal protein s6, PPP4R2 - protein phosphatase 4, regulatory subunit 2, RPS7 - ribosomal protein s7, NHP2 - nhp2 ribonucleoprotein, THOC5 - tho complex 5, MBNL1 - muscleblind-like splicing regulator 1, METTL15 - methyltransferase like 15, MRPL1 - mitochondrial ribosomal protein l1, RBM19 - rna binding motif protein 19, LSM7 - lsm7 homolog, u6 small nuclear rna associated (s. cerevisiae), CDK13 - cyclin-dependent kinase 13, PA2G4 - proliferation-associated 2g4, 38kda, FCF1 - fcf1 rrna-processing protein, RPL7L1 - ribosomal protein l7-like 1, ZNF326 - zinc finger protein 326, RSL1D1 - ribosomal l1 domain containing 1, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, DIMT1 - dim1 dimethyladenosine transferase 1 homolog (s. cerevisiae), GAR1 - gar1 ribonucleoprotein, SART3 - squamous cell carcinoma antigen recognized by t cells 3, EXOSC1 - exosome component 1, THUMPD1 - thump domain containing 1]
229 [PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, RNF31 - ring finger protein 31, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, WNK1 - wnk lysine deficient protein kinase 1, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, CYFIP2 - cytoplasmic fmr1 interacting protein 2, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, CUL1 - cullin 1, PLCG1 - phospholipase c, gamma 1, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, PRKCD - protein kinase c, delta, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), EIF2B3 - eukaryotic translation initiation factor 2b, subunit 3 gamma, 58kda, ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, WIPF1 - was/wasl interacting protein family, member 1]
230 [DDX31 - dead (asp-glu-ala-asp) box polypeptide 31, DDX6 - dead (asp-glu-ala-asp) box helicase 6, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, WNK1 - wnk lysine deficient protein kinase 1, NEK6 - nima-related kinase 6, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, GSPT1 - g1 to s phase transition 1, UBE2Z - ubiquitin-conjugating enzyme e2z, AGK - acylglycerol kinase, RAB30 - rab30, member ras oncogene family, GFM2 - g elongation factor, mitochondrial 2, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RECQL - recq protein-like (dna helicase q1-like), RAB14 - rab14, member ras oncogene family, DCK - deoxycytidine kinase, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, UCK2 - uridine-cytidine kinase 2, PPP5C - protein phosphatase 5, catalytic subunit, CARS - cysteinyl-trna synthetase, IRAK4 - interleukin-1 receptor-associated kinase 4, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, AK4 - adenylate kinase 4, HK2 - hexokinase 2, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), LRRK1 - leucine-rich repeat kinase 1, MAT2A - methionine adenosyltransferase ii, alpha, IDE - insulin-degrading enzyme, MCM3 - minichromosome maintenance complex component 3, ABCF2 - atp-binding cassette, sub-family f (gcn20), member 2, RAB23 - rab23, member ras oncogene family, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, IFIH1 - interferon induced with helicase c domain 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, MYO6 - myosin vi, DCAKD - dephospho-coa kinase domain containing, TTLL12 - tubulin tyrosine ligase-like family, member 12, UBA6 - ubiquitin-like modifier activating enzyme 6, GRK6 - g protein-coupled receptor kinase 6, MCM7 - minichromosome maintenance complex component 7, ARL1 - adp-ribosylation factor-like 1, SCYL2 - scy1-like 2 (s. cerevisiae), RAP1A - rap1a, member of ras oncogene family, STK38L - serine/threonine kinase 38 like, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, MFN1 - mitofusin 1, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), DGUOK - deoxyguanosine kinase, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), UBA3 - ubiquitin-like modifier activating enzyme 3, RAP1B - rap1b, member of ras oncogene family, CDK6 - cyclin-dependent kinase 6, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, ACOT7 - acyl-coa thioesterase 7, GCDH - glutaryl-coa dehydrogenase, CDK13 - cyclin-dependent kinase 13, HSPD1 - heat shock 60kda protein 1 (chaperonin), MVD - mevalonate (diphospho) decarboxylase, CCT4 - chaperonin containing tcp1, subunit 4 (delta), GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, RHOT1 - ras homolog family member t1, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, UBE2I - ubiquitin-conjugating enzyme e2i, TAOK3 - tao kinase 3, PRKCD - protein kinase c, delta, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3, HSPA4 - heat shock 70kda protein 4, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, RRM1 - ribonucleotide reductase m1, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
231 [CCNK - cyclin k, NCBP2 - nuclear cap binding protein subunit 2, 20kda, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, TCOF1 - treacher collins-franceschetti syndrome 1, RFC3 - replication factor c (activator 1) 3, 38kda, PGM1 - phosphoglucomutase 1, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, DCK - deoxycytidine kinase, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, UCK2 - uridine-cytidine kinase 2, PPP5C - protein phosphatase 5, catalytic subunit, UMPS - uridine monophosphate synthetase, BTF3 - basic transcription factor 3, TRIM25 - tripartite motif containing 25, ACAT1 - acetyl-coa acetyltransferase 1, DUT - deoxyuridine triphosphatase, CPOX - coproporphyrinogen oxidase, HK2 - hexokinase 2, AK4 - adenylate kinase 4, GAR1 - gar1 ribonucleoprotein, CREB1 - camp responsive element binding protein 1, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, DCAKD - dephospho-coa kinase domain containing, TELO2 - telomere maintenance 2, SCAF8 - sr-related ctd-associated factor 8, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, SNRPE - small nuclear ribonucleoprotein polypeptide e, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), RNMT - rna (guanine-7-) methyltransferase, DGUOK - deoxyguanosine kinase, POLR1C - polymerase (rna) i polypeptide c, 30kda, ADPGK - adp-dependent glucokinase, NHP2 - nhp2 ribonucleoprotein, FECH - ferrochelatase, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, ACOT7 - acyl-coa thioesterase 7, GCDH - glutaryl-coa dehydrogenase, CDK13 - cyclin-dependent kinase 13, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, GNPDA2 - glucosamine-6-phosphate deaminase 2, UGDH - udp-glucose 6-dehydrogenase, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), KYNU - kynureninase, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, NAMPT - nicotinamide phosphoribosyltransferase, AK3 - adenylate kinase 3, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, RRM1 - ribonucleotide reductase m1, TCEA1 - transcription elongation factor a (sii), 1]
232 [CCNK - cyclin k, NCBP2 - nuclear cap binding protein subunit 2, 20kda, CSTF1 - cleavage stimulation factor, 3' pre-rna, subunit 1, 50kda, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, TCOF1 - treacher collins-franceschetti syndrome 1, RFC3 - replication factor c (activator 1) 3, 38kda, PGM1 - phosphoglucomutase 1, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, PGAM1 - phosphoglycerate mutase 1 (brain), TMEM14C - transmembrane protein 14c, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, DCK - deoxycytidine kinase, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, UCK2 - uridine-cytidine kinase 2, UMPS - uridine monophosphate synthetase, PPP5C - protein phosphatase 5, catalytic subunit, TRIM25 - tripartite motif containing 25, BTF3 - basic transcription factor 3, ACAT1 - acetyl-coa acetyltransferase 1, DUT - deoxyuridine triphosphatase, CPOX - coproporphyrinogen oxidase, HK2 - hexokinase 2, AK4 - adenylate kinase 4, GAR1 - gar1 ribonucleoprotein, CREB1 - camp responsive element binding protein 1, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, DCAKD - dephospho-coa kinase domain containing, TELO2 - telomere maintenance 2, SCAF8 - sr-related ctd-associated factor 8, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, SNRPE - small nuclear ribonucleoprotein polypeptide e, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), RNMT - rna (guanine-7-) methyltransferase, DGUOK - deoxyguanosine kinase, POLR1C - polymerase (rna) i polypeptide c, 30kda, ADPGK - adp-dependent glucokinase, NHP2 - nhp2 ribonucleoprotein, FECH - ferrochelatase, NUDT4 - nudix (nucleoside diphosphate linked moiety x)-type motif 4, ACOT7 - acyl-coa thioesterase 7, GCDH - glutaryl-coa dehydrogenase, CDK13 - cyclin-dependent kinase 13, GNPDA2 - glucosamine-6-phosphate deaminase 2, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, UGDH - udp-glucose 6-dehydrogenase, NR3C1 - nuclear receptor subfamily 3, group c, member 1 (glucocorticoid receptor), HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, KYNU - kynureninase, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, NAMPT - nicotinamide phosphoribosyltransferase, AK3 - adenylate kinase 3, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, RRM1 - ribonucleotide reductase m1, TCEA1 - transcription elongation factor a (sii), 1]
233 [DDX31 - dead (asp-glu-ala-asp) box polypeptide 31, DDX6 - dead (asp-glu-ala-asp) box helicase 6, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, NEK6 - nima-related kinase 6, WNK1 - wnk lysine deficient protein kinase 1, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, GSPT1 - g1 to s phase transition 1, UBE2Z - ubiquitin-conjugating enzyme e2z, AGK - acylglycerol kinase, RAB30 - rab30, member ras oncogene family, GFM2 - g elongation factor, mitochondrial 2, BDH2 - 3-hydroxybutyrate dehydrogenase, type 2, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RECQL - recq protein-like (dna helicase q1-like), RAB14 - rab14, member ras oncogene family, DCK - deoxycytidine kinase, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, UCK2 - uridine-cytidine kinase 2, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, PPP5C - protein phosphatase 5, catalytic subunit, CARS - cysteinyl-trna synthetase, IRAK4 - interleukin-1 receptor-associated kinase 4, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, AK4 - adenylate kinase 4, HK2 - hexokinase 2, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), LRRK1 - leucine-rich repeat kinase 1, MAT2A - methionine adenosyltransferase ii, alpha, AGPS - alkylglycerone phosphate synthase, IDE - insulin-degrading enzyme, MCM3 - minichromosome maintenance complex component 3, SORD - sorbitol dehydrogenase, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, RAB23 - rab23, member ras oncogene family, ABCF2 - atp-binding cassette, sub-family f (gcn20), member 2, WDR77 - wd repeat domain 77, IFIH1 - interferon induced with helicase c domain 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, MYO6 - myosin vi, DCAKD - dephospho-coa kinase domain containing, VDAC2 - voltage-dependent anion channel 2, TTLL12 - tubulin tyrosine ligase-like family, member 12, GRK6 - g protein-coupled receptor kinase 6, UBA6 - ubiquitin-like modifier activating enzyme 6, MCM7 - minichromosome maintenance complex component 7, ARL1 - adp-ribosylation factor-like 1, SCYL2 - scy1-like 2 (s. cerevisiae), RAP1A - rap1a, member of ras oncogene family, STK38L - serine/threonine kinase 38 like, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, MFN1 - mitofusin 1, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), DGUOK - deoxyguanosine kinase, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), HADH - hydroxyacyl-coa dehydrogenase, UBA3 - ubiquitin-like modifier activating enzyme 3, RAP1B - rap1b, member of ras oncogene family, CDK6 - cyclin-dependent kinase 6, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, ACOT7 - acyl-coa thioesterase 7, GCDH - glutaryl-coa dehydrogenase, CDK13 - cyclin-dependent kinase 13, CAT - catalase, HSPD1 - heat shock 60kda protein 1 (chaperonin), CCT4 - chaperonin containing tcp1, subunit 4 (delta), MVD - mevalonate (diphospho) decarboxylase, UGDH - udp-glucose 6-dehydrogenase, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, RHOT1 - ras homolog family member t1, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, UBE2I - ubiquitin-conjugating enzyme e2i, TAOK3 - tao kinase 3, PRKCD - protein kinase c, delta, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3, HSPA4 - heat shock 70kda protein 4, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, RRM1 - ribonucleotide reductase m1, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
234 [PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, DPM1 - dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit, GFPT1 - glutamine--fructose-6-phosphate transaminase 1, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), DGUOK - deoxyguanosine kinase, DCK - deoxycytidine kinase, UCK2 - uridine-cytidine kinase 2, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, UMPS - uridine monophosphate synthetase, GOLPH3 - golgi phosphoprotein 3 (coat-protein), ADPGK - adp-dependent glucokinase, ACAT1 - acetyl-coa acetyltransferase 1, DUT - deoxyuridine triphosphatase, AK4 - adenylate kinase 4, HK2 - hexokinase 2, GCDH - glutaryl-coa dehydrogenase, ACOT7 - acyl-coa thioesterase 7, GNPDA2 - glucosamine-6-phosphate deaminase 2, UGDH - udp-glucose 6-dehydrogenase, SORD - sorbitol dehydrogenase, AK3 - adenylate kinase 3, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, DCAKD - dephospho-coa kinase domain containing, RRM1 - ribonucleotide reductase m1]
235 [RPL22 - ribosomal protein l22, F11R - f11 receptor, CISD1 - cdgsh iron sulfur domain 1, FABP5 - fatty acid binding protein 5 (psoriasis-associated), RPL7 - ribosomal protein l7, HOOK3 - hook microtubule-tethering protein 3, FLNB - filamin b, beta, SLC16A1 - solute carrier family 16 (monocarboxylate transporter), member 1, DCK - deoxycytidine kinase, UCK2 - uridine-cytidine kinase 2, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, PPP5C - protein phosphatase 5, catalytic subunit, CARS - cysteinyl-trna synthetase, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, DERL1 - derlin 1, MAN2A1 - mannosidase, alpha, class 2a, member 1, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), EIF3F - eukaryotic translation initiation factor 3, subunit f, SLAMF1 - signaling lymphocytic activation molecule family member 1, ACAT1 - acetyl-coa acetyltransferase 1, CPOX - coproporphyrinogen oxidase, P4HA1 - prolyl 4-hydroxylase, alpha polypeptide i, LRRK1 - leucine-rich repeat kinase 1, MAT2A - methionine adenosyltransferase ii, alpha, EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, TRIM38 - tripartite motif containing 38, IDE - insulin-degrading enzyme, SORD - sorbitol dehydrogenase, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, IFIH1 - interferon induced with helicase c domain 1, DECR1 - 2,4-dienoyl coa reductase 1, mitochondrial, CREB1 - camp responsive element binding protein 1, MAD1L1 - mad1 mitotic arrest deficient-like 1 (yeast), DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, SNX1 - sorting nexin 1, CTSC - cathepsin c, PRPSAP2 - phosphoribosyl pyrophosphate synthetase-associated protein 2, RNF31 - ring finger protein 31, ATG7 - autophagy related 7, MCU - mitochondrial calcium uniporter, PRRC1 - proline-rich coiled-coil 1, PRKRA - protein kinase, interferon-inducible double stranded rna dependent activator, SYNE1 - spectrin repeat containing, nuclear envelope 1, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, MFN1 - mitofusin 1, STAT6 - signal transducer and activator of transcription 6, interleukin-4 induced, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), ESD - esterase d, HADH - hydroxyacyl-coa dehydrogenase, UBA3 - ubiquitin-like modifier activating enzyme 3, STOM - stomatin, SNX6 - sorting nexin 6, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, RB1 - retinoblastoma 1, ACOT7 - acyl-coa thioesterase 7, CISD2 - cdgsh iron sulfur domain 2, MUT - methylmalonyl coa mutase, CAT - catalase, USP15 - ubiquitin specific peptidase 15, CBX3 - chromobox homolog 3, GNPDA2 - glucosamine-6-phosphate deaminase 2, NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, MVD - mevalonate (diphospho) decarboxylase, UGDH - udp-glucose 6-dehydrogenase, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), IRF3 - interferon regulatory factor 3, ANXA4 - annexin a4, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, KYNU - kynureninase, PCYT1A - phosphate cytidylyltransferase 1, choline, alpha, BCL2L1 - bcl2-like 1, IRF5 - interferon regulatory factor 5, NAMPT - nicotinamide phosphoribosyltransferase, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, SLC25A12 - solute carrier family 25 (aspartate/glutamate carrier), member 12, CCDC6 - coiled-coil domain containing 6, EPS15 - epidermal growth factor receptor pathway substrate 15, RRM1 - ribonucleotide reductase m1]
236 [PSMB2 - proteasome (prosome, macropain) subunit, beta type, 2, PSMB1 - proteasome (prosome, macropain) subunit, beta type, 1, PSMA5 - proteasome (prosome, macropain) subunit, alpha type, 5, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, CDC23 - cell division cycle 23, PSMA3 - proteasome (prosome, macropain) subunit, alpha type, 3, ANAPC7 - anaphase promoting complex subunit 7, ANAPC1 - anaphase promoting complex subunit 1]
237 [RIF1 - rap1 interacting factor homolog (yeast), CAPN2 - calpain 2, (m/ii) large subunit, WNK1 - wnk lysine deficient protein kinase 1, HK2 - hexokinase 2, HSPD1 - heat shock 60kda protein 1 (chaperonin), GSN - gelsolin, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), HMGB1 - high mobility group box 1, MAT2A - methionine adenosyltransferase ii, alpha, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, RNMT - rna (guanine-7-) methyltransferase, OTUB1 - otu domain, ubiquitin aldehyde binding 1, STX8 - syntaxin 8, FLNB - filamin b, beta, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, CREB1 - camp responsive element binding protein 1, KDM3A - lysine (k)-specific demethylase 3a, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, RPL3 - ribosomal protein l3, ASAH1 - n-acylsphingosine amidohydrolase (acid ceramidase) 1, TRIM25 - tripartite motif containing 25, DOCK8 - dedicator of cytokinesis 8]
238 [GOSR2 - golgi snap receptor complex member 2, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), VTI1A - vesicle transport through interaction with t-snares 1a, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), GOSR1 - golgi snap receptor complex member 1, SPG11 - spastic paraplegia 11 (autosomal recessive), STX8 - syntaxin 8, STX17 - syntaxin 17, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3]
239 [RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, SRP72 - signal recognition particle 72kda, RPL14 - ribosomal protein l14, RPL13 - ribosomal protein l13, RPS8 - ribosomal protein s8, RPL6 - ribosomal protein l6, RPL7 - ribosomal protein l7, RPS13 - ribosomal protein s13, RPL31 - ribosomal protein l31, RPS6 - ribosomal protein s6, RPL32 - ribosomal protein l32, RPL3 - ribosomal protein l3, RPS7 - ribosomal protein s7]
240 [TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), SAMM50 - samm50 sorting and assembly machinery component, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, AGK - acylglycerol kinase, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, DNAJC11 - dnaj (hsp40) homolog, subfamily c, member 11]
241 [RAB23 - rab23, member ras oncogene family, AK4 - adenylate kinase 4, IMPDH2 - imp (inosine 5'-monophosphate) dehydrogenase 2, AK3 - adenylate kinase 3, MFN1 - mitofusin 1, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3]
242 [PURA - purine-rich element binding protein a, QKI - qki, kh domain containing, rna binding, NCBP2 - nuclear cap binding protein subunit 2, 20kda, DDX6 - dead (asp-glu-ala-asp) box helicase 6, TCOF1 - treacher collins-franceschetti syndrome 1, MRPL13 - mitochondrial ribosomal protein l13, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, GSPT1 - g1 to s phase transition 1, PGAM1 - phosphoglycerate mutase 1 (brain), NSMAF - neutral sphingomyelinase (n-smase) activation associated factor, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), PTCD3 - pentatricopeptide repeat domain 3, TMED2 - transmembrane emp24 domain trafficking protein 2, LARP4B - la ribonucleoprotein domain family, member 4b, EIF4G3 - eukaryotic translation initiation factor 4 gamma, 3, ILF3 - interleukin enhancer binding factor 3, 90kda, EIF4H - eukaryotic translation initiation factor 4h, EIF4B - eukaryotic translation initiation factor 4b, TSFM - ts translation elongation factor, mitochondrial, PA2G4 - proliferation-associated 2g4, 38kda, CAPRIN1 - cell cycle associated protein 1, EIF2B1 - eukaryotic translation initiation factor 2b, subunit 1 alpha, 26kda, PRKCD - protein kinase c, delta, LARP1 - la ribonucleoprotein domain family, member 1, NEMF - nuclear export mediator factor, EIF4G2 - eukaryotic translation initiation factor 4 gamma, 2, SYNCRIP - synaptotagmin binding, cytoplasmic rna interacting protein, EIF3E - eukaryotic translation initiation factor 3, subunit e]
243 [HK2 - hexokinase 2, PGM1 - phosphoglucomutase 1, AK4 - adenylate kinase 4, PGAM1 - phosphoglycerate mutase 1 (brain), AK3 - adenylate kinase 3, ADPGK - adp-dependent glucokinase]
244 [CCNK - cyclin k, SCAF8 - sr-related ctd-associated factor 8, ZNF326 - zinc finger protein 326, TSFM - ts translation elongation factor, mitochondrial, CDK13 - cyclin-dependent kinase 13, THOC5 - tho complex 5]
245 [PYGB - phosphorylase, glycogen; brain, SORD - sorbitol dehydrogenase, PHKB - phosphorylase kinase, beta, HK2 - hexokinase 2, PGM1 - phosphoglucomutase 1, PGAM1 - phosphoglycerate mutase 1 (brain), NUDT5 - nudix (nucleoside diphosphate linked moiety x)-type motif 5, MANBA - mannosidase, beta a, lysosomal, ADPGK - adp-dependent glucokinase, PGM2 - phosphoglucomutase 2]
246 [IRF3 - interferon regulatory factor 3, IRF5 - interferon regulatory factor 5, IFIH1 - interferon induced with helicase c domain 1, HSPD1 - heat shock 60kda protein 1 (chaperonin), DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, HMGB1 - high mobility group box 1]
247 [IRF3 - interferon regulatory factor 3, POLR3B - polymerase (rna) iii (dna directed) polypeptide b, IRF5 - interferon regulatory factor 5, POLR1C - polymerase (rna) i polypeptide c, 30kda, IFIH1 - interferon induced with helicase c domain 1, HSPD1 - heat shock 60kda protein 1 (chaperonin), DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, HMGB2 - high mobility group box 2, HMGB1 - high mobility group box 1, STAT6 - signal transducer and activator of transcription 6, interleukin-4 induced]
248 [GOSR2 - golgi snap receptor complex member 2, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), GOSR1 - golgi snap receptor complex member 1, VTI1A - vesicle transport through interaction with t-snares 1a, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, STX8 - syntaxin 8, SPG11 - spastic paraplegia 11 (autosomal recessive), BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, STX17 - syntaxin 17, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, MFN1 - mitofusin 1]
249 [YARS2 - tyrosyl-trna synthetase 2, mitochondrial, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), IARS - isoleucyl-trna synthetase, CARS - cysteinyl-trna synthetase, PPA1 - pyrophosphatase (inorganic) 1, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
250 [GOSR2 - golgi snap receptor complex member 2, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), GOSR1 - golgi snap receptor complex member 1, VTI1A - vesicle transport through interaction with t-snares 1a, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), STX8 - syntaxin 8, SPG11 - spastic paraplegia 11 (autosomal recessive), STX18 - syntaxin 18, BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, STX17 - syntaxin 17, MFN1 - mitofusin 1, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3]
251 [GOSR2 - golgi snap receptor complex member 2, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), GOSR1 - golgi snap receptor complex member 1, VTI1A - vesicle transport through interaction with t-snares 1a, VPS8 - vacuolar protein sorting 8 homolog (s. cerevisiae), STX8 - syntaxin 8, SPG11 - spastic paraplegia 11 (autosomal recessive), BNIP1 - bcl2/adenovirus e1b 19kda interacting protein 1, STX17 - syntaxin 17, MFN1 - mitofusin 1, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3]
252 [EIF4H - eukaryotic translation initiation factor 4h, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, EIF4B - eukaryotic translation initiation factor 4b, EIF2D - eukaryotic translation initiation factor 2d, DDX6 - dead (asp-glu-ala-asp) box helicase 6, RPL23A - ribosomal protein l23a, SNRPE - small nuclear ribonucleoprotein polypeptide e, SF3A3 - splicing factor 3a, subunit 3, 60kda, GFM2 - g elongation factor, mitochondrial 2, PELO - pelota homolog (drosophila), HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, CLNS1A - chloride channel, nucleotide-sensitive, 1a, NIP7 - nip7, nucleolar pre-rrna processing protein, RPL6 - ribosomal protein l6, WDR77 - wd repeat domain 77, EIF3E - eukaryotic translation initiation factor 3, subunit e, SART3 - squamous cell carcinoma antigen recognized by t cells 3, RPL3 - ribosomal protein l3, EIF2S2 - eukaryotic translation initiation factor 2, subunit 2 beta, 38kda, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), EIF3F - eukaryotic translation initiation factor 3, subunit f]
253 [YARS2 - tyrosyl-trna synthetase 2, mitochondrial, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), CARS - cysteinyl-trna synthetase, IARS - isoleucyl-trna synthetase, PPA1 - pyrophosphatase (inorganic) 1, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
254 [GOSR2 - golgi snap receptor complex member 2, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, TMED5 - transmembrane emp24 protein transport domain containing 5, TMED2 - transmembrane emp24 domain trafficking protein 2, GOSR1 - golgi snap receptor complex member 1, VTI1A - vesicle transport through interaction with t-snares 1a, DYNC1I2 - dynein, cytoplasmic 1, intermediate chain 2, STX17 - syntaxin 17, ARCN1 - archain 1, SCFD1 - sec1 family domain containing 1, VAPB - vamp (vesicle-associated membrane protein)-associated protein b and c, CTSC - cathepsin c]
255 [PTCD3 - pentatricopeptide repeat domain 3, MRPS14 - mitochondrial ribosomal protein s14, MRPL47 - mitochondrial ribosomal protein l47, MRPS35 - mitochondrial ribosomal protein s35, MRPL2 - mitochondrial ribosomal protein l2, MRPL42 - mitochondrial ribosomal protein l42, MRPL13 - mitochondrial ribosomal protein l13, MRPL1 - mitochondrial ribosomal protein l1, MRPL9 - mitochondrial ribosomal protein l9, MRPL3 - mitochondrial ribosomal protein l3, GFM2 - g elongation factor, mitochondrial 2]
256 [IRF3 - interferon regulatory factor 3, IRF5 - interferon regulatory factor 5, IFIH1 - interferon induced with helicase c domain 1, HSPD1 - heat shock 60kda protein 1 (chaperonin), DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, HMGB1 - high mobility group box 1]
257 [TMEM30A - transmembrane protein 30a, RPL23A - ribosomal protein l23a, RPL22 - ribosomal protein l22, F11R - f11 receptor, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, AGK - acylglycerol kinase, RPL13 - ribosomal protein l13, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), TMED2 - transmembrane emp24 domain trafficking protein 2, NUP50 - nucleoporin 50kda, RPL6 - ribosomal protein l6, HOOK3 - hook microtubule-tethering protein 3, RPL7 - ribosomal protein l7, RPL3 - ribosomal protein l3, USP4 - ubiquitin specific peptidase 4 (proto-oncogene), VTI1A - vesicle transport through interaction with t-snares 1a, HK2 - hexokinase 2, TNPO1 - transportin 1, STX17 - syntaxin 17, KPNB1 - karyopherin (importin) beta 1, AGPS - alkylglycerone phosphate synthase, GOSR2 - golgi snap receptor complex member 2, IDE - insulin-degrading enzyme, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, EXOC1 - exocyst complex component 1, RPL31 - ribosomal protein l31, RPL32 - ribosomal protein l32, PITRM1 - pitrilysin metallopeptidase 1, ARL1 - adp-ribosylation factor-like 1, NIPBL - nipped-b homolog (drosophila), DLG1 - discs, large homolog 1 (drosophila), RAP1A - rap1a, member of ras oncogene family, GGA3 - golgi-associated, gamma adaptin ear containing, arf binding protein 3, SEH1L - seh1-like (s. cerevisiae), RPL14 - ribosomal protein l14, TOMM20 - translocase of outer mitochondrial membrane 20 homolog (yeast), STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), GNPAT - glyceronephosphate o-acyltransferase, VPS36 - vacuolar protein sorting 36 homolog (s. cerevisiae), NUP88 - nucleoporin 88kda, RPS8 - ribosomal protein s8, RPS13 - ribosomal protein s13, RPS6 - ribosomal protein s6, GOLPH3 - golgi phosphoprotein 3 (coat-protein), RPS7 - ribosomal protein s7, SUMO1 - small ubiquitin-like modifier 1, SRP72 - signal recognition particle 72kda, RB1 - retinoblastoma 1, CAT - catalase, LRSAM1 - leucine rich repeat and sterile alpha motif containing 1, HSPD1 - heat shock 60kda protein 1 (chaperonin), ATP6V1D - atpase, h+ transporting, lysosomal 34kda, v1 subunit d, SAMM50 - samm50 sorting and assembly machinery component, VPS41 - vacuolar protein sorting 41 homolog (s. cerevisiae), HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, HSPA4 - heat shock 70kda protein 4]
258 [SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, NCBP2 - nuclear cap binding protein subunit 2, 20kda, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, SNRPE - small nuclear ribonucleoprotein polypeptide e, SDAD1 - sda1 domain containing 1, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, TNPO1 - transportin 1, SEH1L - seh1-like (s. cerevisiae), XPO6 - exportin 6, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), KPNB1 - karyopherin (importin) beta 1, NUP88 - nucleoporin 88kda, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, NUP50 - nucleoporin 50kda, NEMF - nuclear export mediator factor, THOC5 - tho complex 5, XPOT - exportin, trna]
259 [CYCS - cytochrome c, somatic, ATG7 - autophagy related 7, HK2 - hexokinase 2, CISD2 - cdgsh iron sulfur domain 2, HSPD1 - heat shock 60kda protein 1 (chaperonin), STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), AGK - acylglycerol kinase, SSBP1 - single-stranded dna binding protein 1, mitochondrial, MFN1 - mitofusin 1, SAMM50 - samm50 sorting and assembly machinery component, TIMM13 - translocase of inner mitochondrial membrane 13 homolog (yeast), RHOT1 - ras homolog family member t1, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, CHCHD3 - coiled-coil-helix-coiled-coil-helix domain containing 3, BCL2L1 - bcl2-like 1, POLDIP2 - polymerase (dna-directed), delta interacting protein 2, HSPA4 - heat shock 70kda protein 4, YWHAZ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, YWHAQ - tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide, VDAC2 - voltage-dependent anion channel 2, MAN2A1 - mannosidase, alpha, class 2a, member 1, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, DNAJC11 - dnaj (hsp40) homolog, subfamily c, member 11]
260 [NCBP2 - nuclear cap binding protein subunit 2, 20kda, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, ANP32A - acidic (leucine-rich) nuclear phosphoprotein 32 family, member a, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, SDAD1 - sda1 domain containing 1, SNRPE - small nuclear ribonucleoprotein polypeptide e, TNPO1 - transportin 1, SEH1L - seh1-like (s. cerevisiae), XPO6 - exportin 6, STAT3 - signal transducer and activator of transcription 3 (acute-phase response factor), KPNB1 - karyopherin (importin) beta 1, NUP88 - nucleoporin 88kda, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, NUP50 - nucleoporin 50kda, NEMF - nuclear export mediator factor, XPOT - exportin, trna, THOC5 - tho complex 5]
261 [DDX31 - dead (asp-glu-ala-asp) box polypeptide 31, DDX6 - dead (asp-glu-ala-asp) box helicase 6, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, WNK1 - wnk lysine deficient protein kinase 1, NEK6 - nima-related kinase 6, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, GSPT1 - g1 to s phase transition 1, UBE2Z - ubiquitin-conjugating enzyme e2z, AGK - acylglycerol kinase, RAB30 - rab30, member ras oncogene family, GFM2 - g elongation factor, mitochondrial 2, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RECQL - recq protein-like (dna helicase q1-like), RAB14 - rab14, member ras oncogene family, DCK - deoxycytidine kinase, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, UCK2 - uridine-cytidine kinase 2, PPP5C - protein phosphatase 5, catalytic subunit, CARS - cysteinyl-trna synthetase, IRAK4 - interleukin-1 receptor-associated kinase 4, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, AK4 - adenylate kinase 4, HK2 - hexokinase 2, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), LRRK1 - leucine-rich repeat kinase 1, MAT2A - methionine adenosyltransferase ii, alpha, IDE - insulin-degrading enzyme, MCM3 - minichromosome maintenance complex component 3, ABCF2 - atp-binding cassette, sub-family f (gcn20), member 2, RAB23 - rab23, member ras oncogene family, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, IFIH1 - interferon induced with helicase c domain 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, MYO6 - myosin vi, DCAKD - dephospho-coa kinase domain containing, TTLL12 - tubulin tyrosine ligase-like family, member 12, UBA6 - ubiquitin-like modifier activating enzyme 6, GRK6 - g protein-coupled receptor kinase 6, MCM7 - minichromosome maintenance complex component 7, ARL1 - adp-ribosylation factor-like 1, SCYL2 - scy1-like 2 (s. cerevisiae), RAP1A - rap1a, member of ras oncogene family, STK38L - serine/threonine kinase 38 like, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, MFN1 - mitofusin 1, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), DGUOK - deoxyguanosine kinase, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), UBA3 - ubiquitin-like modifier activating enzyme 3, RAP1B - rap1b, member of ras oncogene family, CDK6 - cyclin-dependent kinase 6, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, GCDH - glutaryl-coa dehydrogenase, ACOT7 - acyl-coa thioesterase 7, CDK13 - cyclin-dependent kinase 13, HSPD1 - heat shock 60kda protein 1 (chaperonin), CCT4 - chaperonin containing tcp1, subunit 4 (delta), MVD - mevalonate (diphospho) decarboxylase, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, RHOT1 - ras homolog family member t1, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, UBE2I - ubiquitin-conjugating enzyme e2i, TAOK3 - tao kinase 3, PRKCD - protein kinase c, delta, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3, HSPA4 - heat shock 70kda protein 4, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, RRM1 - ribonucleotide reductase m1, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
262 [DDX31 - dead (asp-glu-ala-asp) box polypeptide 31, DDX6 - dead (asp-glu-ala-asp) box helicase 6, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, WNK1 - wnk lysine deficient protein kinase 1, NEK6 - nima-related kinase 6, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, GSPT1 - g1 to s phase transition 1, UBE2Z - ubiquitin-conjugating enzyme e2z, AGK - acylglycerol kinase, RAB30 - rab30, member ras oncogene family, GFM2 - g elongation factor, mitochondrial 2, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RECQL - recq protein-like (dna helicase q1-like), RAB14 - rab14, member ras oncogene family, DCK - deoxycytidine kinase, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, UCK2 - uridine-cytidine kinase 2, PPP5C - protein phosphatase 5, catalytic subunit, CARS - cysteinyl-trna synthetase, IRAK4 - interleukin-1 receptor-associated kinase 4, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, AK4 - adenylate kinase 4, HK2 - hexokinase 2, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), LRRK1 - leucine-rich repeat kinase 1, MAT2A - methionine adenosyltransferase ii, alpha, IDE - insulin-degrading enzyme, MCM3 - minichromosome maintenance complex component 3, ABCF2 - atp-binding cassette, sub-family f (gcn20), member 2, RAB23 - rab23, member ras oncogene family, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, IFIH1 - interferon induced with helicase c domain 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, MYO6 - myosin vi, DCAKD - dephospho-coa kinase domain containing, TTLL12 - tubulin tyrosine ligase-like family, member 12, GRK6 - g protein-coupled receptor kinase 6, UBA6 - ubiquitin-like modifier activating enzyme 6, MCM7 - minichromosome maintenance complex component 7, ARL1 - adp-ribosylation factor-like 1, SCYL2 - scy1-like 2 (s. cerevisiae), RAP1A - rap1a, member of ras oncogene family, STK38L - serine/threonine kinase 38 like, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, MFN1 - mitofusin 1, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), DGUOK - deoxyguanosine kinase, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), UBA3 - ubiquitin-like modifier activating enzyme 3, RAP1B - rap1b, member of ras oncogene family, CDK6 - cyclin-dependent kinase 6, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, CDK13 - cyclin-dependent kinase 13, HSPD1 - heat shock 60kda protein 1 (chaperonin), CCT4 - chaperonin containing tcp1, subunit 4 (delta), MVD - mevalonate (diphospho) decarboxylase, GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, RHOT1 - ras homolog family member t1, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, UBE2I - ubiquitin-conjugating enzyme e2i, TAOK3 - tao kinase 3, PRKCD - protein kinase c, delta, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3, HSPA4 - heat shock 70kda protein 4, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, RRM1 - ribonucleotide reductase m1, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
263 [DST - dystonin, CAPZA1 - capping protein (actin filament) muscle z-line, alpha 1, AP2B1 - adaptor-related protein complex 2, beta 1 subunit, RPL23A - ribosomal protein l23a, CAPN2 - calpain 2, (m/ii) large subunit, F11R - f11 receptor, GSN - gelsolin, SCFD1 - sec1 family domain containing 1, PPP1R9B - protein phosphatase 1, regulatory subunit 9b, HMGB1 - high mobility group box 1, PTCD3 - pentatricopeptide repeat domain 3, PPP5C - protein phosphatase 5, catalytic subunit, DBN1 - drebrin 1, LETM1 - leucine zipper-ef-hand containing transmembrane protein 1, EIF4H - eukaryotic translation initiation factor 4h, EIF4B - eukaryotic translation initiation factor 4b, IDE - insulin-degrading enzyme, GATAD2B - gata zinc finger domain containing 2b, APPL1 - adaptor protein, phosphotyrosine interaction, ph domain and leucine zipper containing 1, MS4A1 - membrane-spanning 4-domains, subfamily a, member 1, RCSD1 - rcsd domain containing 1, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), IFIH1 - interferon induced with helicase c domain 1, MAD1L1 - mad1 mitotic arrest deficient-like 1 (yeast), SNX1 - sorting nexin 1, MYO6 - myosin vi, TELO2 - telomere maintenance 2, SNRPD1 - small nuclear ribonucleoprotein d1 polypeptide 16kda, TPM3 - tropomyosin 3, POLDIP3 - polymerase (dna-directed), delta interacting protein 3, NIPBL - nipped-b homolog (drosophila), RAP1A - rap1a, member of ras oncogene family, SYNE1 - spectrin repeat containing, nuclear envelope 1, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), WDR1 - wd repeat domain 1, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), UBA3 - ubiquitin-like modifier activating enzyme 3, RAP1B - rap1b, member of ras oncogene family, ITGB1 - integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen cd29 includes mdf2, msk12), ARPC1B - actin related protein 2/3 complex, subunit 1b, 41kda, DSTN - destrin (actin depolymerizing factor), ARPC3 - actin related protein 2/3 complex, subunit 3, 21kda, SRP72 - signal recognition particle 72kda, HSPD1 - heat shock 60kda protein 1 (chaperonin), GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, PTPN2 - protein tyrosine phosphatase, non-receptor type 2, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, FZR1 - fizzy/cell division cycle 20 related 1 (drosophila), LARP1 - la ribonucleoprotein domain family, member 1, NEMF - nuclear export mediator factor, RPTOR - regulatory associated protein of mtor, complex 1]
264 [DDX31 - dead (asp-glu-ala-asp) box polypeptide 31, DDX6 - dead (asp-glu-ala-asp) box helicase 6, ATP2A2 - atpase, ca++ transporting, cardiac muscle, slow twitch 2, WNK1 - wnk lysine deficient protein kinase 1, NEK6 - nima-related kinase 6, SUCLA2 - succinate-coa ligase, adp-forming, beta subunit, GSPT1 - g1 to s phase transition 1, UBE2Z - ubiquitin-conjugating enzyme e2z, AGK - acylglycerol kinase, RAB30 - rab30, member ras oncogene family, GFM2 - g elongation factor, mitochondrial 2, MTHFD1 - methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase, RECQL - recq protein-like (dna helicase q1-like), RAB14 - rab14, member ras oncogene family, DCK - deoxycytidine kinase, PIP4K2C - phosphatidylinositol-5-phosphate 4-kinase, type ii, gamma, UCK2 - uridine-cytidine kinase 2, PPP5C - protein phosphatase 5, catalytic subunit, CARS - cysteinyl-trna synthetase, IRAK4 - interleukin-1 receptor-associated kinase 4, OAS3 - 2'-5'-oligoadenylate synthetase 3, 100kda, UBE2J1 - ubiquitin-conjugating enzyme e2, j1, HK2 - hexokinase 2, AK4 - adenylate kinase 4, CCT5 - chaperonin containing tcp1, subunit 5 (epsilon), LRRK1 - leucine-rich repeat kinase 1, MAT2A - methionine adenosyltransferase ii, alpha, MCM3 - minichromosome maintenance complex component 3, IDE - insulin-degrading enzyme, ABCF2 - atp-binding cassette, sub-family f (gcn20), member 2, PSMC1 - proteasome (prosome, macropain) 26s subunit, atpase, 1, RAB23 - rab23, member ras oncogene family, RAC1 - ras-related c3 botulinum toxin substrate 1 (rho family, small gtp binding protein rac1), IFIH1 - interferon induced with helicase c domain 1, DDX58 - dead (asp-glu-ala-asp) box polypeptide 58, MYO6 - myosin vi, DCAKD - dephospho-coa kinase domain containing, TTLL12 - tubulin tyrosine ligase-like family, member 12, GRK6 - g protein-coupled receptor kinase 6, UBA6 - ubiquitin-like modifier activating enzyme 6, MCM7 - minichromosome maintenance complex component 7, ARL1 - adp-ribosylation factor-like 1, SCYL2 - scy1-like 2 (s. cerevisiae), RAP1A - rap1a, member of ras oncogene family, STK38L - serine/threonine kinase 38 like, PI4KB - phosphatidylinositol 4-kinase, catalytic, beta, EEF1A1 - eukaryotic translation elongation factor 1 alpha 1, IARS - isoleucyl-trna synthetase, MFN1 - mitofusin 1, VPS4B - vacuolar protein sorting 4 homolog b (s. cerevisiae), DGUOK - deoxyguanosine kinase, RNASEL - ribonuclease l (2',5'-oligoisoadenylate synthetase-dependent), UBA3 - ubiquitin-like modifier activating enzyme 3, RAP1B - rap1b, member of ras oncogene family, CDK6 - cyclin-dependent kinase 6, YARS2 - tyrosyl-trna synthetase 2, mitochondrial, GCDH - glutaryl-coa dehydrogenase, ACOT7 - acyl-coa thioesterase 7, CDK13 - cyclin-dependent kinase 13, HSPD1 - heat shock 60kda protein 1 (chaperonin), MVD - mevalonate (diphospho) decarboxylase, CCT4 - chaperonin containing tcp1, subunit 4 (delta), GNAI3 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 3, RHOT1 - ras homolog family member t1, GNAI2 - guanine nucleotide binding protein (g protein), alpha inhibiting activity polypeptide 2, PIK3CD - phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta, HSP90AA1 - heat shock protein 90kda alpha (cytosolic), class a member 1, UBE2I - ubiquitin-conjugating enzyme e2i, TAOK3 - tao kinase 3, PRKCD - protein kinase c, delta, TARS2 - threonyl-trna synthetase 2, mitochondrial (putative), AK3 - adenylate kinase 3, HSPA4 - heat shock 70kda protein 4, GART - phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase, PRKAR1A - protein kinase, camp-dependent, regulatory, type i, alpha, RRM1 - ribonucleotide reductase m1, QRSL1 - glutaminyl-trna synthase (glutamine-hydrolyzing)-like 1]
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] fgsea_1.15.1 workflowr_1.6.2 forcats_0.4.0 stringr_1.4.0
[5] dplyr_0.8.3 purrr_0.3.4 readr_1.3.1 tidyr_1.1.0
[9] tibble_2.1.3 ggplot2_3.2.1 tidyverse_1.3.0
loaded via a namespace (and not attached):
[1] Rcpp_1.0.5 lubridate_1.7.4 lattice_0.20-38
[4] utf8_1.1.4 assertthat_0.2.1 rprojroot_1.3-2
[7] digest_0.6.20 R6_2.4.0 cellranger_1.1.0
[10] backports_1.1.4 reprex_0.3.0 evaluate_0.14
[13] httr_1.4.1 pillar_1.4.2 rlang_0.4.6
[16] lazyeval_0.2.2 readxl_1.3.1 rstudioapi_0.10
[19] data.table_1.13.2 whisker_0.3-2 Matrix_1.2-18
[22] rmarkdown_1.13 labeling_0.3 BiocParallel_1.18.0
[25] munsell_0.5.0 broom_0.5.2 compiler_3.6.1
[28] httpuv_1.5.1 modelr_0.1.8 xfun_0.8
[31] pkgconfig_2.0.2 htmltools_0.3.6 tidyselect_1.1.0
[34] gridExtra_2.3 fansi_0.4.0 crayon_1.3.4
[37] dbplyr_1.4.2 withr_2.1.2 later_0.8.0
[40] grid_3.6.1 nlme_3.1-140 jsonlite_1.6
[43] gtable_0.3.0 lifecycle_0.1.0 DBI_1.1.0
[46] git2r_0.26.1 magrittr_1.5 scales_1.1.0
[49] cli_2.2.0 stringi_1.4.3 farver_2.0.1
[52] fs_1.3.1 promises_1.0.1 xml2_1.3.2
[55] generics_0.0.2 vctrs_0.3.0 fastmatch_1.1-0
[58] tools_3.6.1 glue_1.3.1 hms_0.5.3
[61] parallel_3.6.1 yaml_2.2.0 colorspace_1.4-1
[64] rvest_0.3.5 knitr_1.23 haven_2.3.1