Differential Used Intronic
Briana Mittleman
2/23/2020
Last updated: 2020-04-10
Checks: 7 0
Knit directory: Comparative_APA/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190902) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: code/chimp_log/
Ignored: code/human_log/
Ignored: data/.DS_Store
Ignored: data/TrialFiltersMeta.txt.sb-9845453e-R58Y0Q/
Ignored: data/mediation_prot/
Ignored: data/metadata_HCpanel.txt.sb-284518db-RGf0kd/
Ignored: data/metadata_HCpanel.txt.sb-a5794dd2-i594qs/
Ignored: output/.DS_Store
Untracked files:
Untracked: ._.DS_Store
Untracked: Chimp/
Untracked: Human/
Untracked: analysis/CrossChimpThreePrime.Rmd
Untracked: analysis/DiffTransProtvsExpression.Rmd
Untracked: analysis/DiffUsedUTR.Rmd
Untracked: analysis/GvizPlots.Rmd
Untracked: analysis/HandC.TvN
Untracked: analysis/PhenotypeOverlap10.Rmd
Untracked: analysis/annotationBias.Rmd
Untracked: analysis/assessReadQual.Rmd
Untracked: analysis/diffExpressionPantro6.Rmd
Untracked: code/._AlignmentScores.sh
Untracked: code/._BothFCMM.sh
Untracked: code/._BothFCMMPrim.sh
Untracked: code/._BothFCnewOInclusive.sh
Untracked: code/._ChimpStarMM2.sh
Untracked: code/._ClassifyLeafviz.sh
Untracked: code/._ClosestorthoEx.sh
Untracked: code/._Config_chimp.yaml
Untracked: code/._Config_chimp_full.yaml
Untracked: code/._Config_human.yaml
Untracked: code/._ConvertJunc2Bed.sh
Untracked: code/._CountNucleotides.py
Untracked: code/._CrossMapChimpRNA.sh
Untracked: code/._CrossMapThreeprime.sh
Untracked: code/._DiffSplice.sh
Untracked: code/._DiffSplicePlots.sh
Untracked: code/._DiffSplicePlots_gencode.sh
Untracked: code/._DiffSplice_gencode.sh
Untracked: code/._DiffSplice_removebad.sh
Untracked: code/._Filter255MM.sh
Untracked: code/._FilterPrimSec.sh
Untracked: code/._FindIntronForDomPAS.sh
Untracked: code/._FindIntronForDomPAS_DF.sh
Untracked: code/._GetMAPQscore.py
Untracked: code/._GetSecondaryMap.py
Untracked: code/._Lift5perPAS.sh
Untracked: code/._LiftFinalChimpJunc2Human.sh
Untracked: code/._LiftOrthoPAS2chimp.sh
Untracked: code/._MapBadSamples.sh
Untracked: code/._MismatchNumbers.sh
Untracked: code/._PAS_ATTAAA.sh
Untracked: code/._PAS_ATTAAA_df.sh
Untracked: code/._PAS_seqExpanded.sh
Untracked: code/._PASsequences.sh
Untracked: code/._PASsequences_DF.sh
Untracked: code/._PlotNuclearUsagebySpecies.R
Untracked: code/._PlotNuclearUsagebySpecies_DF.R
Untracked: code/._QuantMergedClusters.sh
Untracked: code/._RNATranscriptDTplot.sh
Untracked: code/._ReverseLiftFilter.R
Untracked: code/._RunFixLeafCluster.sh
Untracked: code/._RunNegMCMediation.sh
Untracked: code/._RunNegMCMediationDF.sh
Untracked: code/._RunPosMCMediationDF.err
Untracked: code/._RunPosMCMediationDF.sh
Untracked: code/._SAF2Bed.py
Untracked: code/._Snakefile
Untracked: code/._SnakefilePAS
Untracked: code/._SnakefilePASfilt
Untracked: code/._SortIndexBadSamples.sh
Untracked: code/._StarMM2.sh
Untracked: code/._TestFC.sh
Untracked: code/._assignPeak2Intronicregion
Untracked: code/._assignPeak2Intronicregion.sh
Untracked: code/._bed215upbed.py
Untracked: code/._bed2Bedbothstrand.py
Untracked: code/._bed2SAF_gen.py
Untracked: code/._buildIndecpantro5
Untracked: code/._buildIndecpantro5.sh
Untracked: code/._buildLeafviz.sh
Untracked: code/._buildLeafviz_leadAnno.sh
Untracked: code/._buildStarIndex.sh
Untracked: code/._chimpChromprder.sh
Untracked: code/._chimpMultiCov.sh
Untracked: code/._chimpMultiCov255.sh
Untracked: code/._chimpMultiCovInclusive.sh
Untracked: code/._chooseSignalSite.py
Untracked: code/._cleanbed2saf.py
Untracked: code/._cluster.json
Untracked: code/._cluster2bed.py
Untracked: code/._clusterLiftReverse.sh
Untracked: code/._clusterLiftReverse_removebad.sh
Untracked: code/._clusterLiftprimary.sh
Untracked: code/._clusterLiftprimary_removebad.sh
Untracked: code/._converBam2Junc.sh
Untracked: code/._converBam2Junc_removeBad.sh
Untracked: code/._extraSnakefiltpas
Untracked: code/._extractPhyloReg.py
Untracked: code/._extractPhyloRegGene.py
Untracked: code/._extractPhylopGeneral.ph
Untracked: code/._extractPhylopGeneral.py
Untracked: code/._extractPhylopReg200down.py
Untracked: code/._extractPhylopReg200up.py
Untracked: code/._filter5percPAS.py
Untracked: code/._filterNumChroms.py
Untracked: code/._filterPASforMP.py
Untracked: code/._filterPostLift.py
Untracked: code/._filterPrimaryread.py
Untracked: code/._filterSecondaryread.py
Untracked: code/._fixExonFC.py
Untracked: code/._fixFCheadforExp.py
Untracked: code/._fixLeafCluster.py
Untracked: code/._fixLiftedJunc.py
Untracked: code/._fixUTRexonanno.py
Untracked: code/._formathg38Anno.py
Untracked: code/._formatpantro6Anno.py
Untracked: code/._getRNAseqMapStats.sh
Untracked: code/._hg19MapStats.sh
Untracked: code/._humanChromorder.sh
Untracked: code/._humanMultiCov.sh
Untracked: code/._humanMultiCov255.sh
Untracked: code/._humanMultiCov_inclusive.sh
Untracked: code/._intersectLiftedPAS.sh
Untracked: code/._liftJunctionFiles.sh
Untracked: code/._liftPAS19to38.sh
Untracked: code/._liftedchimpJunc2human.sh
Untracked: code/._makeNuclearDapaplots.sh
Untracked: code/._makeNuclearDapaplots_DF.sh
Untracked: code/._makeSamplyGroupsHuman_TvN.py
Untracked: code/._mapRNAseqhg19.sh
Untracked: code/._mapRNAseqhg19_newPipeline.sh
Untracked: code/._maphg19.sh
Untracked: code/._maphg19_subjunc.sh
Untracked: code/._mediation_test.R
Untracked: code/._mergeChimp3prime_inhg38.sh
Untracked: code/._mergeandBWRNAseq.sh
Untracked: code/._mergedBam2BW.sh
Untracked: code/._nameClusters.py
Untracked: code/._negativeMediation_montecarlo.R
Untracked: code/._negativeMediation_montecarloDF.R
Untracked: code/._numMultimap.py
Untracked: code/._overlapMMandOrthoexon.sh
Untracked: code/._overlapPASandOrthoexon.sh
Untracked: code/._overlapapaQTLPAS.sh
Untracked: code/._parseHg38.py
Untracked: code/._postiveMediation_montecarlo_DF.R
Untracked: code/._prepareCleanLiftedFC_5perc4LC.py
Untracked: code/._prepareLeafvizAnno.sh
Untracked: code/._preparePAS4lift.py
Untracked: code/._primaryLift.sh
Untracked: code/._processhg38exons.py
Untracked: code/._quantJunc.sh
Untracked: code/._quantJunc_TEST.sh
Untracked: code/._quantJunc_removeBad.sh
Untracked: code/._quantLiftedPASPrimary.sh
Untracked: code/._quantMerged_seperatly.sh
Untracked: code/._recLiftchim2human.sh
Untracked: code/._revLiftPAShg38to19.sh
Untracked: code/._reverseLift.sh
Untracked: code/._runCheckReverseLift.sh
Untracked: code/._runChimpDiffIso.sh
Untracked: code/._runCountNucleotides.sh
Untracked: code/._runFilterNumChroms.sh
Untracked: code/._runHumanDiffIso.sh
Untracked: code/._runNuclearDiffIso_DF.sh
Untracked: code/._runNuclearDifffIso.sh
Untracked: code/._runTotalDiffIso.sh
Untracked: code/._run_chimpverifybam.sh
Untracked: code/._run_verifyBam.sh
Untracked: code/._snakemake.batch
Untracked: code/._snakemakePAS.batch
Untracked: code/._snakemakePASchimp.batch
Untracked: code/._snakemakePAShuman.batch
Untracked: code/._snakemake_chimp.batch
Untracked: code/._snakemake_human.batch
Untracked: code/._snakemakefiltPAS.batch
Untracked: code/._snakemakefiltPAS_chimp
Untracked: code/._snakemakefiltPAS_chimp.sh
Untracked: code/._snakemakefiltPAS_human.sh
Untracked: code/._spliceSite2Fasta.py
Untracked: code/._submit-snakemake-chimp.sh
Untracked: code/._submit-snakemake-human.sh
Untracked: code/._submit-snakemakePAS-chimp.sh
Untracked: code/._submit-snakemakePAS-human.sh
Untracked: code/._submit-snakemakefiltPAS-chimp.sh
Untracked: code/._submit-snakemakefiltPAS-human.sh
Untracked: code/._subset_diffisopheno_Nuclear_HvC.py
Untracked: code/._subset_diffisopheno_Nuclear_HvC_DF.py
Untracked: code/._subset_diffisopheno_Total_HvC.py
Untracked: code/._threeprimeOrthoFC.sh
Untracked: code/._transcriptDTplotsNuclear.sh
Untracked: code/._verifyBam4973.sh
Untracked: code/._verifyBam4973inHuman.sh
Untracked: code/._wrap_chimpverifybam.sh
Untracked: code/._wrap_verifyBam.sh
Untracked: code/._writeMergecode.py
Untracked: code/.snakemake/
Untracked: code/ALLPAS_sequenceDF.err
Untracked: code/ALLPAS_sequenceDF.out
Untracked: code/AlignmentScores.err
Untracked: code/AlignmentScores.out
Untracked: code/AlignmentScores.sh
Untracked: code/BothFCMM.err
Untracked: code/BothFCMM.out
Untracked: code/BothFCMM.sh
Untracked: code/BothFCMMPrim.err
Untracked: code/BothFCMMPrim.out
Untracked: code/BothFCMMPrim.sh
Untracked: code/BothFCnewOInclusive.sh
Untracked: code/BothFCnewOInclusive.sh.err
Untracked: code/BothFCnewOInclusive.sh.out
Untracked: code/ChimpStarMM2.err
Untracked: code/ChimpStarMM2.out
Untracked: code/ChimpStarMM2.sh
Untracked: code/ClassifyLeafviz.sh
Untracked: code/ClosestorthoEx.err
Untracked: code/ClosestorthoEx.out
Untracked: code/ClosestorthoEx.sh
Untracked: code/Config_chimp.yaml
Untracked: code/Config_chimp_full.yaml
Untracked: code/Config_human.yaml
Untracked: code/ConvertJunc2Bed.err
Untracked: code/ConvertJunc2Bed.out
Untracked: code/ConvertJunc2Bed.sh
Untracked: code/CountNucleotides.py
Untracked: code/CrossMapChimpRNA.sh
Untracked: code/CrossMapThreeprime.sh
Untracked: code/CrossmapChimp3prime.err
Untracked: code/CrossmapChimp3prime.out
Untracked: code/CrossmapChimpRNA.err
Untracked: code/CrossmapChimpRNA.out
Untracked: code/DiffSplice.err
Untracked: code/DiffSplice.out
Untracked: code/DiffSplice.sh
Untracked: code/DiffSplicePlots.err
Untracked: code/DiffSplicePlots.out
Untracked: code/DiffSplicePlots.sh
Untracked: code/DiffSplicePlots_gencode.sh
Untracked: code/DiffSplice_gencode.sh
Untracked: code/DiffSplice_removebad.err
Untracked: code/DiffSplice_removebad.out
Untracked: code/DiffSplice_removebad.sh
Untracked: code/Filter255.err
Untracked: code/Filter255.out
Untracked: code/Filter255MM.sh
Untracked: code/FilterPrimSec.err
Untracked: code/FilterPrimSec.out
Untracked: code/FilterPrimSec.sh
Untracked: code/FilterReverseLift.err
Untracked: code/FilterReverseLift.out
Untracked: code/FindIntronForDomPAS.err
Untracked: code/FindIntronForDomPAS.out
Untracked: code/FindIntronForDomPAS.sh
Untracked: code/FindIntronForDomPAS_DF.sh
Untracked: code/GencodeDiffSplice.err
Untracked: code/GencodeDiffSplice.out
Untracked: code/GetMAPQscore.py
Untracked: code/GetSecondaryMap.py
Untracked: code/HchromOrder.err
Untracked: code/HchromOrder.out
Untracked: code/IntersectMMandOrtho.err
Untracked: code/IntersectMMandOrtho.out
Untracked: code/IntersectPASandOrtho.err
Untracked: code/IntersectPASandOrtho.out
Untracked: code/JunctionLift.err
Untracked: code/JunctionLift.out
Untracked: code/JunctionLiftFinalChimp.err
Untracked: code/JunctionLiftFinalChimp.out
Untracked: code/Lift5perPAS.sh
Untracked: code/Lift5perPASbed.err
Untracked: code/Lift5perPASbed.out
Untracked: code/LiftClustersFirst.err
Untracked: code/LiftClustersFirst.out
Untracked: code/LiftClustersFirst_remove.err
Untracked: code/LiftClustersFirst_remove.out
Untracked: code/LiftClustersSecond.err
Untracked: code/LiftClustersSecond.out
Untracked: code/LiftClustersSecond_remove.err
Untracked: code/LiftClustersSecond_remove.out
Untracked: code/LiftFinalChimpJunc2Human.sh
Untracked: code/LiftOrthoPAS2chimp.sh
Untracked: code/LiftorthoPAS.err
Untracked: code/LiftorthoPASt.out
Untracked: code/Log.out
Untracked: code/MapBadSamples.err
Untracked: code/MapBadSamples.out
Untracked: code/MapBadSamples.sh
Untracked: code/MapStats.err
Untracked: code/MapStats.out
Untracked: code/MaxEntCode/
Untracked: code/MergeClusters.err
Untracked: code/MergeClusters.out
Untracked: code/MergeClusters.sh
Untracked: code/MismatchNumbers.err
Untracked: code/MismatchNumbers.out
Untracked: code/MismatchNumbers.sh
Untracked: code/PAS_ATTAAA.err
Untracked: code/PAS_ATTAAA.out
Untracked: code/PAS_ATTAAA.sh
Untracked: code/PAS_ATTAAADF.err
Untracked: code/PAS_ATTAAADF.out
Untracked: code/PAS_ATTAAA_df.sh
Untracked: code/PAS_seqExpanded.sh
Untracked: code/PAS_sequence.err
Untracked: code/PAS_sequence.out
Untracked: code/PAS_sequenceDF.err
Untracked: code/PAS_sequenceDF.out
Untracked: code/PASexpanded_sequenceDF.err
Untracked: code/PASexpanded_sequenceDF.out
Untracked: code/PASsequences.sh
Untracked: code/PASsequences_DF.sh
Untracked: code/PlotNuclearUsagebySpecies.R
Untracked: code/PlotNuclearUsagebySpecies_DF.R
Untracked: code/QuantMergeClusters
Untracked: code/QuantMergeClusters.err
Untracked: code/QuantMergeClusters.out
Untracked: code/QuantMergedClusters.sh
Untracked: code/RNATranscriptDTplot.err
Untracked: code/RNATranscriptDTplot.out
Untracked: code/RNATranscriptDTplot.sh
Untracked: code/Rev_liftoverPAShg19to38.err
Untracked: code/Rev_liftoverPAShg19to38.out
Untracked: code/ReverseLiftFilter.R
Untracked: code/RunFixCluster.err
Untracked: code/RunFixCluster.out
Untracked: code/RunFixLeafCluster.sh
Untracked: code/RunNegMCMediation.err
Untracked: code/RunNegMCMediation.sh
Untracked: code/RunNegMCMediationDF.err
Untracked: code/RunNegMCMediationDF.out
Untracked: code/RunNegMCMediationDF.sh
Untracked: code/RunNegMCMediationr.out
Untracked: code/RunPosMCMediation.err
Untracked: code/RunPosMCMediation.sh
Untracked: code/RunPosMCMediationDF.err
Untracked: code/RunPosMCMediationDF.out
Untracked: code/RunPosMCMediationDF.sh
Untracked: code/RunPosMCMediationr.out
Untracked: code/SAF215upbed_gen.py
Untracked: code/SAF2Bed.py
Untracked: code/Snakefile
Untracked: code/SnakefilePAS
Untracked: code/SnakefilePASfilt
Untracked: code/SortIndexBadSamples.err
Untracked: code/SortIndexBadSamples.out
Untracked: code/SortIndexBadSamples.sh
Untracked: code/StarMM2.err
Untracked: code/StarMM2.out
Untracked: code/StarMM2.sh
Untracked: code/TestFC.err
Untracked: code/TestFC.out
Untracked: code/TestFC.sh
Untracked: code/TotalTranscriptDTplot.err
Untracked: code/TotalTranscriptDTplot.out
Untracked: code/Upstream10Bases_general.py
Untracked: code/allPASSeq_df.sh
Untracked: code/apaQTLsnake.err
Untracked: code/apaQTLsnake.out
Untracked: code/apaQTLsnakePAS.err
Untracked: code/apaQTLsnakePAS.out
Untracked: code/apaQTLsnakePAShuman.err
Untracked: code/apaQTLsnakefiltPAS.err
Untracked: code/apaQTLsnakefiltPAS.out
Untracked: code/assignPeak2Intronicregion.err
Untracked: code/assignPeak2Intronicregion.out
Untracked: code/assignPeak2Intronicregion.sh
Untracked: code/bam2junc.err
Untracked: code/bam2junc.out
Untracked: code/bam2junc_remove.err
Untracked: code/bam2junc_remove.out
Untracked: code/bed215upbed.py
Untracked: code/bed2Bedbothstrand.py
Untracked: code/bed2SAF_gen.py
Untracked: code/bed2saf.py
Untracked: code/bg_to_cov.py
Untracked: code/buildIndecpantro5
Untracked: code/buildIndecpantro5.sh
Untracked: code/buildLeafviz.err
Untracked: code/buildLeafviz.out
Untracked: code/buildLeafviz.sh
Untracked: code/buildLeafviz_leadAnno.sh
Untracked: code/buildLeafviz_leafanno.err
Untracked: code/buildLeafviz_leafanno.out
Untracked: code/buildStarIndex.sh
Untracked: code/callPeaksYL.py
Untracked: code/chimpChromprder.sh
Untracked: code/chimpMultiCov.err
Untracked: code/chimpMultiCov.out
Untracked: code/chimpMultiCov.sh
Untracked: code/chimpMultiCov255.sh
Untracked: code/chimpMultiCovInclusive.err
Untracked: code/chimpMultiCovInclusive.out
Untracked: code/chimpMultiCovInclusive.sh
Untracked: code/chooseAnno2Bed.py
Untracked: code/chooseAnno2SAF.py
Untracked: code/chooseSignalSite.py
Untracked: code/chromOrder.err
Untracked: code/chromOrder.out
Untracked: code/classifyLeafviz.err
Untracked: code/classifyLeafviz.out
Untracked: code/cleanbed2saf.py
Untracked: code/cluster.json
Untracked: code/cluster2bed.py
Untracked: code/clusterLiftReverse.sh
Untracked: code/clusterLiftReverse_removebad.sh
Untracked: code/clusterLiftprimary.sh
Untracked: code/clusterLiftprimary_removebad.sh
Untracked: code/clusterPAS.json
Untracked: code/clusterfiltPAS.json
Untracked: code/comands2Mege.sh
Untracked: code/converBam2Junc.sh
Untracked: code/converBam2Junc_removeBad.sh
Untracked: code/convertNumeric.py
Untracked: code/environment.yaml
Untracked: code/extraSnakefiltpas
Untracked: code/extractPhyloReg.py
Untracked: code/extractPhyloRegGene.py
Untracked: code/extractPhylopGeneral.py
Untracked: code/extractPhylopReg200down.py
Untracked: code/extractPhylopReg200up.py
Untracked: code/filter5perc.R
Untracked: code/filter5percPAS.py
Untracked: code/filter5percPheno.py
Untracked: code/filterBamforMP.pysam2_gen.py
Untracked: code/filterJuncChroms.err
Untracked: code/filterJuncChroms.out
Untracked: code/filterMissprimingInNuc10_gen.py
Untracked: code/filterNumChroms.py
Untracked: code/filterPASforMP.py
Untracked: code/filterPostLift.py
Untracked: code/filterPrimaryread.py
Untracked: code/filterSAFforMP_gen.py
Untracked: code/filterSecondaryread.py
Untracked: code/filterSortBedbyCleanedBed_gen.R
Untracked: code/filterpeaks.py
Untracked: code/fixExonFC.py
Untracked: code/fixFChead.py
Untracked: code/fixFChead_bothfrac.py
Untracked: code/fixFCheadforExp.py
Untracked: code/fixLeafCluster.py
Untracked: code/fixLiftedJunc.py
Untracked: code/fixUTRexonanno.py
Untracked: code/formathg38Anno.py
Untracked: code/generateStarIndex.err
Untracked: code/generateStarIndex.out
Untracked: code/generateStarIndexHuman.err
Untracked: code/generateStarIndexHuman.out
Untracked: code/getAlloverlap.py
Untracked: code/getRNAseqMapStats.sh
Untracked: code/hg19MapStats.err
Untracked: code/hg19MapStats.out
Untracked: code/hg19MapStats.sh
Untracked: code/humanChromorder.sh
Untracked: code/humanFiles
Untracked: code/humanMultiCov.err
Untracked: code/humanMultiCov.out
Untracked: code/humanMultiCov.sh
Untracked: code/humanMultiCov255.err
Untracked: code/humanMultiCov255.out
Untracked: code/humanMultiCov255.sh
Untracked: code/humanMultiCovInclusive.err
Untracked: code/humanMultiCovInclusive.out
Untracked: code/humanMultiCov_inclusive.sh
Untracked: code/intersectAnno.err
Untracked: code/intersectAnno.out
Untracked: code/intersectAnnoExt.err
Untracked: code/intersectAnnoExt.out
Untracked: code/intersectLiftedPAS.sh
Untracked: code/leafcutter_merge_regtools_redo.py
Untracked: code/liftJunctionFiles.sh
Untracked: code/liftPAS19to38.sh
Untracked: code/liftoverPAShg19to38.err
Untracked: code/liftoverPAShg19to38.out
Untracked: code/log/
Untracked: code/make5percPeakbed.py
Untracked: code/makeFileID.py
Untracked: code/makeNuclearDapaplots.sh
Untracked: code/makeNuclearDapaplots_DF.sh
Untracked: code/makeNuclearPlots.err
Untracked: code/makeNuclearPlots.out
Untracked: code/makeNuclearPlotsDF.err
Untracked: code/makeNuclearPlotsDF.out
Untracked: code/makePheno.py
Untracked: code/makeSamplyGroupsChimp_TvN.py
Untracked: code/makeSamplyGroupsHuman_TvN.py
Untracked: code/mapRNAseqhg19.sh
Untracked: code/mapRNAseqhg19_newPipeline.sh
Untracked: code/maphg19.err
Untracked: code/maphg19.out
Untracked: code/maphg19.sh
Untracked: code/maphg19_new.err
Untracked: code/maphg19_new.out
Untracked: code/maphg19_sub.err
Untracked: code/maphg19_sub.out
Untracked: code/maphg19_subjunc.sh
Untracked: code/mediation_test.R
Untracked: code/merge.err
Untracked: code/mergeChimp3prime_inhg38.sh
Untracked: code/merge_leafcutter_clusters_redo.py
Untracked: code/mergeandBWRNAseq.sh
Untracked: code/mergeandsort_ChimpinHuman.err
Untracked: code/mergeandsort_ChimpinHuman.out
Untracked: code/mergedBam2BW.sh
Untracked: code/mergedbam2bw.err
Untracked: code/mergedbam2bw.out
Untracked: code/mergedbamRNAand2bw.err
Untracked: code/mergedbamRNAand2bw.out
Untracked: code/nameClusters.py
Untracked: code/namePeaks.py
Untracked: code/negativeMediation_montecarlo.R
Untracked: code/negativeMediation_montecarloDF.R
Untracked: code/nuclearTranscriptDTplot.err
Untracked: code/nuclearTranscriptDTplot.out
Untracked: code/numMultimap.py
Untracked: code/overlapMMandOrthoexon.sh
Untracked: code/overlapPAS.err
Untracked: code/overlapPAS.out
Untracked: code/overlapPASandOrthoexon.sh
Untracked: code/overlapapaQTLPAS.sh
Untracked: code/overlapapaQTLPAS_extended.sh
Untracked: code/overlapapaQTLPAS_samples.sh
Untracked: code/parseHg38.py
Untracked: code/peak2PAS.py
Untracked: code/pheno2countonly.R
Untracked: code/postiveMediation_montecarlo.R
Untracked: code/postiveMediation_montecarlo_DF.R
Untracked: code/prepareAnnoLeafviz.err
Untracked: code/prepareAnnoLeafviz.out
Untracked: code/prepareCleanLiftedFC_5perc4LC.py
Untracked: code/prepareLeafvizAnno.sh
Untracked: code/preparePAS4lift.py
Untracked: code/prepare_phenotype_table.py
Untracked: code/primaryLift.err
Untracked: code/primaryLift.out
Untracked: code/primaryLift.sh
Untracked: code/processhg38exons.py
Untracked: code/quantJunc.sh
Untracked: code/quantJunc_TEST.sh
Untracked: code/quantJunc_removeBad.sh
Untracked: code/quantLiftedPAS.err
Untracked: code/quantLiftedPAS.out
Untracked: code/quantLiftedPAS.sh
Untracked: code/quantLiftedPASPrimary.err
Untracked: code/quantLiftedPASPrimary.out
Untracked: code/quantLiftedPASPrimary.sh
Untracked: code/quatJunc.err
Untracked: code/quatJunc.out
Untracked: code/recChimpback2Human.err
Untracked: code/recChimpback2Human.out
Untracked: code/recLiftchim2human.sh
Untracked: code/revLift.err
Untracked: code/revLift.out
Untracked: code/revLiftPAShg38to19.sh
Untracked: code/reverseLift.sh
Untracked: code/runCheckReverseLift.sh
Untracked: code/runChimpDiffIso.sh
Untracked: code/runChimpDiffIsoDF.sh
Untracked: code/runCountNucleotides.err
Untracked: code/runCountNucleotides.out
Untracked: code/runCountNucleotides.sh
Untracked: code/runCountNucleotidesPantro6.err
Untracked: code/runCountNucleotidesPantro6.out
Untracked: code/runCountNucleotides_pantro6.sh
Untracked: code/runFilterNumChroms.sh
Untracked: code/runHumanDiffIso.sh
Untracked: code/runHumanDiffIsoDF.sh
Untracked: code/runNuclearDiffIso_DF.sh
Untracked: code/runNuclearDifffIso.sh
Untracked: code/runTotalDiffIso.sh
Untracked: code/run_Chimpleafcutter_ds.err
Untracked: code/run_Chimpleafcutter_ds.out
Untracked: code/run_Chimpverifybam.err
Untracked: code/run_Chimpverifybam.out
Untracked: code/run_Humanleafcutter_dF.err
Untracked: code/run_Humanleafcutter_dF.out
Untracked: code/run_Humanleafcutter_ds.err
Untracked: code/run_Humanleafcutter_ds.out
Untracked: code/run_Nuclearleafcutter_ds.err
Untracked: code/run_Nuclearleafcutter_ds.out
Untracked: code/run_Nuclearleafcutter_dsDF.err
Untracked: code/run_Nuclearleafcutter_dsDF.out
Untracked: code/run_Totalleafcutter_ds.err
Untracked: code/run_Totalleafcutter_ds.out
Untracked: code/run_chimpverifybam.sh
Untracked: code/run_verifyBam.sh
Untracked: code/run_verifybam.err
Untracked: code/run_verifybam.out
Untracked: code/slurm-62824013.out
Untracked: code/slurm-62825841.out
Untracked: code/slurm-62826116.out
Untracked: code/slurm-64108209.out
Untracked: code/slurm-64108521.out
Untracked: code/slurm-64108557.out
Untracked: code/snakePASChimp.err
Untracked: code/snakePASChimp.out
Untracked: code/snakePAShuman.out
Untracked: code/snakemake.batch
Untracked: code/snakemakeChimp.err
Untracked: code/snakemakeChimp.out
Untracked: code/snakemakeHuman.err
Untracked: code/snakemakeHuman.out
Untracked: code/snakemakePAS.batch
Untracked: code/snakemakePASFiltChimp.err
Untracked: code/snakemakePASFiltChimp.out
Untracked: code/snakemakePASFiltHuman.err
Untracked: code/snakemakePASFiltHuman.out
Untracked: code/snakemakePAS_Human.batch
Untracked: code/snakemakePASchimp.batch
Untracked: code/snakemakePAShuman.batch
Untracked: code/snakemake_chimp.batch
Untracked: code/snakemake_human.batch
Untracked: code/snakemakefiltPAS.batch
Untracked: code/snakemakefiltPAS_chimp.sh
Untracked: code/snakemakefiltPAS_human.batch
Untracked: code/snakemakefiltPAS_human.sh
Untracked: code/spliceSite2Fasta.py
Untracked: code/submit-snakemake-chimp.sh
Untracked: code/submit-snakemake-human.sh
Untracked: code/submit-snakemakePAS-chimp.sh
Untracked: code/submit-snakemakePAS-human.sh
Untracked: code/submit-snakemakefiltPAS-chimp.sh
Untracked: code/submit-snakemakefiltPAS-human.sh
Untracked: code/subset_diffisopheno.py
Untracked: code/subset_diffisopheno_Chimp_tvN.py
Untracked: code/subset_diffisopheno_Chimp_tvN_DF.py
Untracked: code/subset_diffisopheno_Huma_tvN.py
Untracked: code/subset_diffisopheno_Huma_tvN_DF.py
Untracked: code/subset_diffisopheno_Nuclear_HvC.py
Untracked: code/subset_diffisopheno_Nuclear_HvC_DF.py
Untracked: code/subset_diffisopheno_Total_HvC.py
Untracked: code/test
Untracked: code/test.txt
Untracked: code/threeprimeOrthoFC.out
Untracked: code/threeprimeOrthoFC.sh
Untracked: code/threeprimeOrthoFCcd.err
Untracked: code/transcriptDTplotsNuclear.sh
Untracked: code/transcriptDTplotsTotal.sh
Untracked: code/verifyBam4973.sh
Untracked: code/verifyBam4973inHuman.sh
Untracked: code/verifybam4973.err
Untracked: code/verifybam4973.out
Untracked: code/verifybam4973HumanMap.err
Untracked: code/verifybam4973HumanMap.out
Untracked: code/wrap_Chimpverifybam.err
Untracked: code/wrap_Chimpverifybam.out
Untracked: code/wrap_chimpverifybam.sh
Untracked: code/wrap_verifyBam.sh
Untracked: code/wrap_verifybam.err
Untracked: code/wrap_verifybam.out
Untracked: code/writeMergecode.py
Untracked: data/._.DS_Store
Untracked: data/._HC_filenames.txt
Untracked: data/._HC_filenames.txt.sb-4426323c-IKIs0S
Untracked: data/._HC_filenames.xlsx
Untracked: data/._MapPantro6_meta.txt
Untracked: data/._MapPantro6_meta.txt.sb-a5794dd2-Cskmlm
Untracked: data/._MapPantro6_meta.xlsx
Untracked: data/._OppositeSpeciesMap.txt
Untracked: data/._OppositeSpeciesMap.txt.sb-a5794dd2-mayWJf
Untracked: data/._OppositeSpeciesMap.xlsx
Untracked: data/._RNASEQ_metadata.txt
Untracked: data/._RNASEQ_metadata.txt.sb-4426323c-TE4ns3
Untracked: data/._RNASEQ_metadata.txt.sb-51f67ae1-HXp7Gq
Untracked: data/._RNASEQ_metadata_2Removed.txt
Untracked: data/._RNASEQ_metadata_2Removed.txt.sb-4426323c-a4lBwx
Untracked: data/._RNASEQ_metadata_2Removed.xlsx
Untracked: data/._RNASEQ_metadata_stranded.txt
Untracked: data/._RNASEQ_metadata_stranded.txt.sb-a5794dd2-D659m2
Untracked: data/._RNASEQ_metadata_stranded.txt.sb-a5794dd2-ImNMoY
Untracked: data/._RNASEQ_metadata_stranded.txt.sb-e4bf31f0-ZGnGgl
Untracked: data/._RNASEQ_metadata_stranded.xlsx
Untracked: data/._TrialFiltersMeta.txt
Untracked: data/._TrialFiltersMeta.txt.sb-9845453e-R58Y0Q
Untracked: data/._metadata_HCpanel.txt
Untracked: data/._metadata_HCpanel.txt.sb-a3d92a2d-b9cYoF
Untracked: data/._metadata_HCpanel.txt.sb-a5794dd2-i594qs
Untracked: data/._metadata_HCpanel.txt.sb-f4823d1e-qihGek
Untracked: data/._metadata_HCpanel_frompantro5.xlsx
Untracked: data/._~$RNASEQ_metadata.xlsx
Untracked: data/._~$metadata_HCpanel.xlsx
Untracked: data/._.xlsx
Untracked: data/BaseComp/
Untracked: data/CleanLiftedPeaks_FC_primary/
Untracked: data/CompapaQTLpas/
Untracked: data/DNDS/
Untracked: data/DTmatrix/
Untracked: data/DiffExpression/
Untracked: data/DiffIso_Nuclear/
Untracked: data/DiffIso_Nuclear_DF/
Untracked: data/DiffIso_Total/
Untracked: data/DiffSplice/
Untracked: data/DiffSplice_liftedJunc/
Untracked: data/DiffSplice_removeBad/
Untracked: data/DominantPAS/
Untracked: data/DominantPAS_DF/
Untracked: data/EvalPantro5/
Untracked: data/HC_filenames.txt
Untracked: data/HC_filenames.xlsx
Untracked: data/Khan_prot/
Untracked: data/Li_eqtls/
Untracked: data/MapPantro6_meta.txt
Untracked: data/MapPantro6_meta.xlsx
Untracked: data/MapStats/
Untracked: data/NormalizedClusters/
Untracked: data/NuclearHvC/
Untracked: data/NuclearHvC_DF/
Untracked: data/OppositeSpeciesMap.txt
Untracked: data/OppositeSpeciesMap.xlsx
Untracked: data/OrthoExonBed/
Untracked: data/OverlapBenchmark/
Untracked: data/OverlappingPAS/
Untracked: data/PAS/
Untracked: data/PAS_SAF/
Untracked: data/PAS_doubleFilter/
Untracked: data/Peaks_5perc/
Untracked: data/Pheno_5perc/
Untracked: data/Pheno_5perc_DF_nuclear/
Untracked: data/Pheno_5perc_nuclear/
Untracked: data/Pheno_5perc_nuclear_old/
Untracked: data/Pheno_5perc_total/
Untracked: data/PhyloP/
Untracked: data/Pol2Chip/
Untracked: data/RNASEQ_metadata.txt
Untracked: data/RNASEQ_metadata_2Removed.txt
Untracked: data/RNASEQ_metadata_2Removed.xlsx
Untracked: data/RNASEQ_metadata_stranded.txt
Untracked: data/RNASEQ_metadata_stranded.txt.sb-e4bf31f0-ZGnGgl/
Untracked: data/RNASEQ_metadata_stranded.xlsx
Untracked: data/SignalSites/
Untracked: data/SignalSites_doublefilter/
Untracked: data/SpliceSite/
Untracked: data/TestAnnoBiasOE/
Untracked: data/TestMM2/
Untracked: data/TestMM2_AS/
Untracked: data/TestMM2_PrimaryRead/
Untracked: data/TestMM2_SeondaryRead/
Untracked: data/TestMM2_mismatch/
Untracked: data/TestMM2_quality/
Untracked: data/TestWithinMergePAS/
Untracked: data/Test_FC_methods/
Untracked: data/Threeprime2Ortho/
Untracked: data/TotalFractionPAS/
Untracked: data/TotalHvC/
Untracked: data/TrialFiltersMeta.txt
Untracked: data/TwoBadSampleAnalysis/
Untracked: data/Wang_ribo/
Untracked: data/apaQTLGenes/
Untracked: data/bioGRID/
Untracked: data/chainFiles/
Untracked: data/cleanPeaks_anno/
Untracked: data/cleanPeaks_byspecies/
Untracked: data/cleanPeaks_lifted/
Untracked: data/files4viz_nuclear/
Untracked: data/files4viz_nuclear_DF/
Untracked: data/gviz/
Untracked: data/leafviz/
Untracked: data/liftover_files/
Untracked: data/mediation/
Untracked: data/mediation_DF/
Untracked: data/metadata_HCpanel.txt
Untracked: data/metadata_HCpanel.xlsx
Untracked: data/metadata_HCpanel_extra.txt
Untracked: data/metadata_HCpanel_frompantro5.txt
Untracked: data/metadata_HCpanel_frompantro5.xlsx
Untracked: data/multimap/
Untracked: data/orthoUTR/
Untracked: data/primaryLift/
Untracked: data/reverseLift/
Untracked: data/testQuant/
Untracked: data/~$RNASEQ_metadata.xlsx
Untracked: data/~$metadata_HCpanel.xlsx
Untracked: data/.xlsx
Untracked: output/._.DS_Store
Untracked: output/dtPlots/
Untracked: projectNotes.Rmd
Untracked: proteinModelSet.Rmd
Unstaged changes:
Modified: analysis/ExploredAPA.Rmd
Modified: analysis/MMExpreiment.Rmd
Modified: analysis/OppositeMap.Rmd
Modified: analysis/TotalVNuclearBothSpecies.Rmd
Modified: analysis/annotationInfo.Rmd
Modified: analysis/changeMisprimcut.Rmd
Modified: analysis/comp2apaQTLPAS.Rmd
Modified: analysis/correlationPhenos.Rmd
Modified: analysis/establishCutoffs.Rmd
Modified: analysis/investigatePantro5.Rmd
Modified: analysis/multiMap.Rmd
Modified: analysis/pol2.Rmd
Modified: analysis/speciesSpecific.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the R Markdown and HTML files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view them.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | c2a0778 | brimittleman | 2020-04-06 | Build site. |
| Rmd | 9676d72 | brimittleman | 2020-04-06 | updated anno |
| html | c9c6b6a | brimittleman | 2020-02-29 | Build site. |
| Rmd | 8366a88 | brimittleman | 2020-02-29 | add cowplots |
| html | 0a68177 | brimittleman | 2020-02-27 | Build site. |
| Rmd | 0ef2c6d | brimittleman | 2020-02-27 | add protien res |
| html | 1d56205 | brimittleman | 2020-02-27 | Build site. |
| Rmd | cc9f594 | brimittleman | 2020-02-27 | add more plots for meeting |
| html | 09ad482 | brimittleman | 2020-02-24 | Build site. |
| Rmd | 7385496 | brimittleman | 2020-02-24 | add correlations plotted by location |
| html | 5f821ee | brimittleman | 2020-02-23 | Build site. |
| Rmd | f4ae857 | brimittleman | 2020-02-23 | wflow_publish(c(“analysis/index.Rmd”, “analysis/DiffUsedIntronic.Rmd”)) |
library(workflowr)This is workflowr version 1.6.0
Run ?workflowr for help getting startedlibrary(ggpubr)Loading required package: ggplot2Loading required package: magrittrlibrary(limma)
library(qvalue)
library(tidyverse)── Attaching packages ─────────────────────────────────────────────────────────── tidyverse 1.2.1 ──✔ tibble 2.1.1 ✔ purrr 0.3.2
✔ tidyr 0.8.3 ✔ dplyr 0.8.0.1
✔ readr 1.3.1 ✔ stringr 1.3.1
✔ tibble 2.1.1 ✔ forcats 0.3.0 ── Conflicts ────────────────────────────────────────────────────────────── tidyverse_conflicts() ──
✖ tidyr::extract() masks magrittr::extract()
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
✖ purrr::set_names() masks magrittr::set_names()library(cowplot)
Attaching package: 'cowplot'The following object is masked from 'package:ggpubr':
get_legendThe following object is masked from 'package:ggplot2':
ggsaveFor this analysis I will look at the differentially used PAS in introns and ask if I can used information from DE and dribosome to better understand these. I subset intornic because I believe the intronic and utr mechanisms are different.
Meta=read.table("../data/PAS_doubleFilter/PAS_5perc_either_HumanCoord_BothUsage_meta_doubleFilter.txt", header = T, stringsAsFactors = F) %>% dplyr::select(PAS, chr, start,end, loc)
DiffIso= read.table("../data/DiffIso_Nuclear_DF/AllPAS_withGeneSig.txt", header = T,stringsAsFactors = F) %>% inner_join(Meta, by=c("chr", 'start','end')) %>% filter(loc %in% c("intron","utr3"))
DiffIsoSig= DiffIso %>% filter(SigPAU2=="Yes")I can compare the effect sizes with these genes in the DE.
Compare with expression
nameID=read.table("../../genome_anotation_data/ensemble_to_genename.txt",sep="\t", header = T, stringsAsFactors = F) %>% dplyr::select(Gene_stable_ID, Gene.name)
DE=read.table("../data/DiffExpression/DEtested_allres.txt",stringsAsFactors = F,header = F, col.names = c("Gene_stable_ID" ,"logFC" ,"AveExpr" , "t" , "P.Value" , "adj.P.Val", "B" )) %>% inner_join(nameID,by="Gene_stable_ID") %>% dplyr::rename('gene'=Gene.name) %>% dplyr::select(-Gene_stable_ID)First do all of the genes:
DeandAPA= DiffIso %>% inner_join(DE, by="gene")This pas I will include each PAS
ggplot(DeandAPA,aes(y=deltaPAU, x=logFC,col=loc)) + geom_point(alpha=.3) + geom_smooth(aes(col=loc), method="lm") + labs(title="Intronic and 3' UTR APA v DE") + scale_color_brewer(palette = "Dark2")+ stat_cor(aes(color = loc), label.x = 3)
ggplot(DeandAPA,aes(y=deltaPAU, x=logFC)) + geom_point(alpha=.3) + geom_smooth(method="lm") + labs(title="Intronic and 3' UTR APA v DE") + scale_color_brewer(palette = "Dark2")+ stat_cor( label.x = 3)
Just the genes with significant differences in PAS
DeandAPA_sigAPA= DeandAPA %>% filter(SigPAU2=="Yes")ggplot(DeandAPA_sigAPA,aes(y=deltaPAU, x=logFC, col=loc)) + geom_point(alpha=.3) + geom_smooth(method="lm")+ labs(title="Significant Intronic and 3' UTR APA v DE")+ scale_color_brewer(palette = "Dark2")+ stat_cor(aes(color = loc), label.x = 3)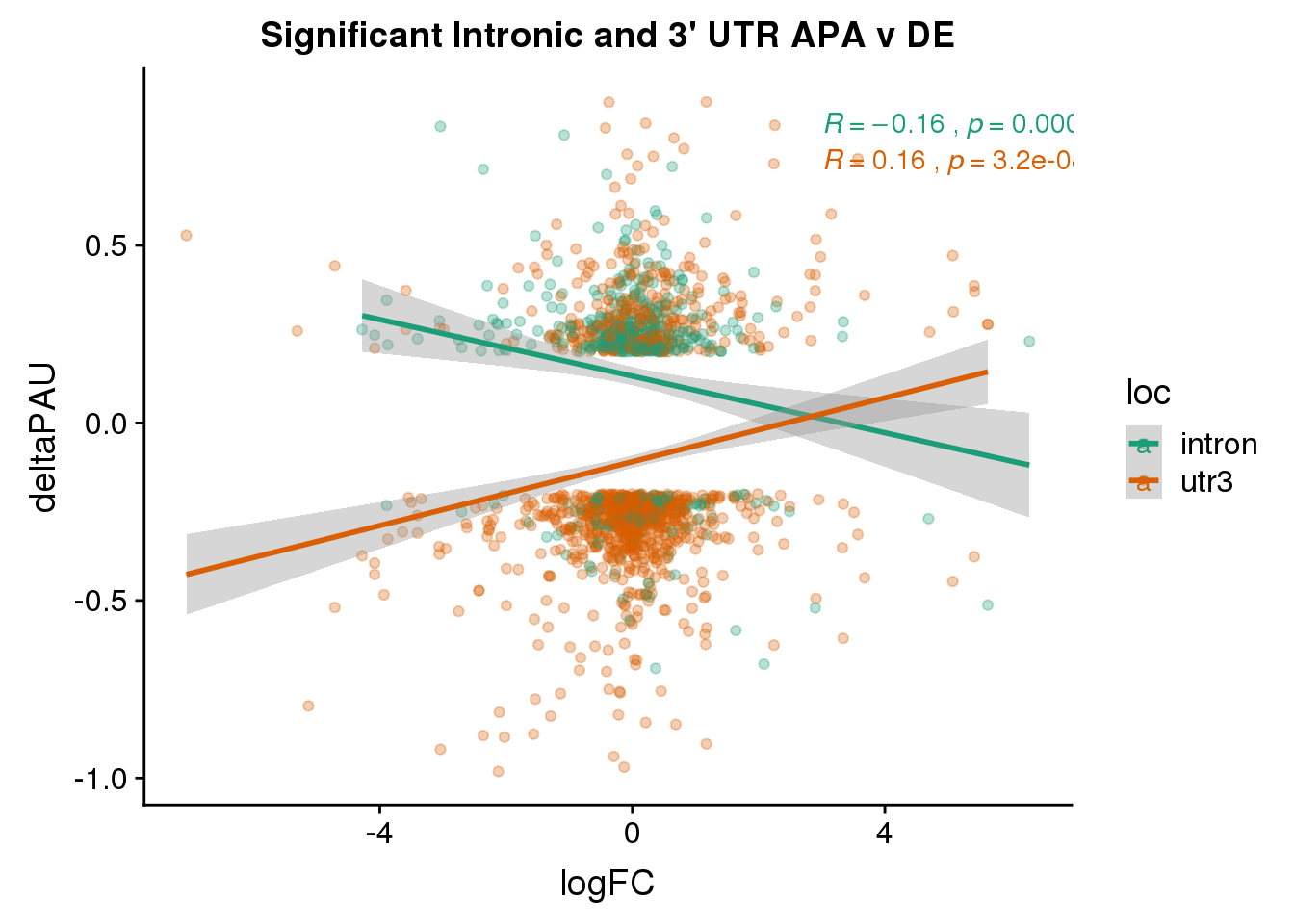
ggplot(DeandAPA_sigAPA,aes(y=deltaPAU, x=logFC)) + geom_point(alpha=.3) + geom_smooth(method="lm")+ labs(title="Significant Intronic and 3' UTR APA v DE")+ scale_color_brewer(palette = "Dark2")+ stat_cor(label.x = 3)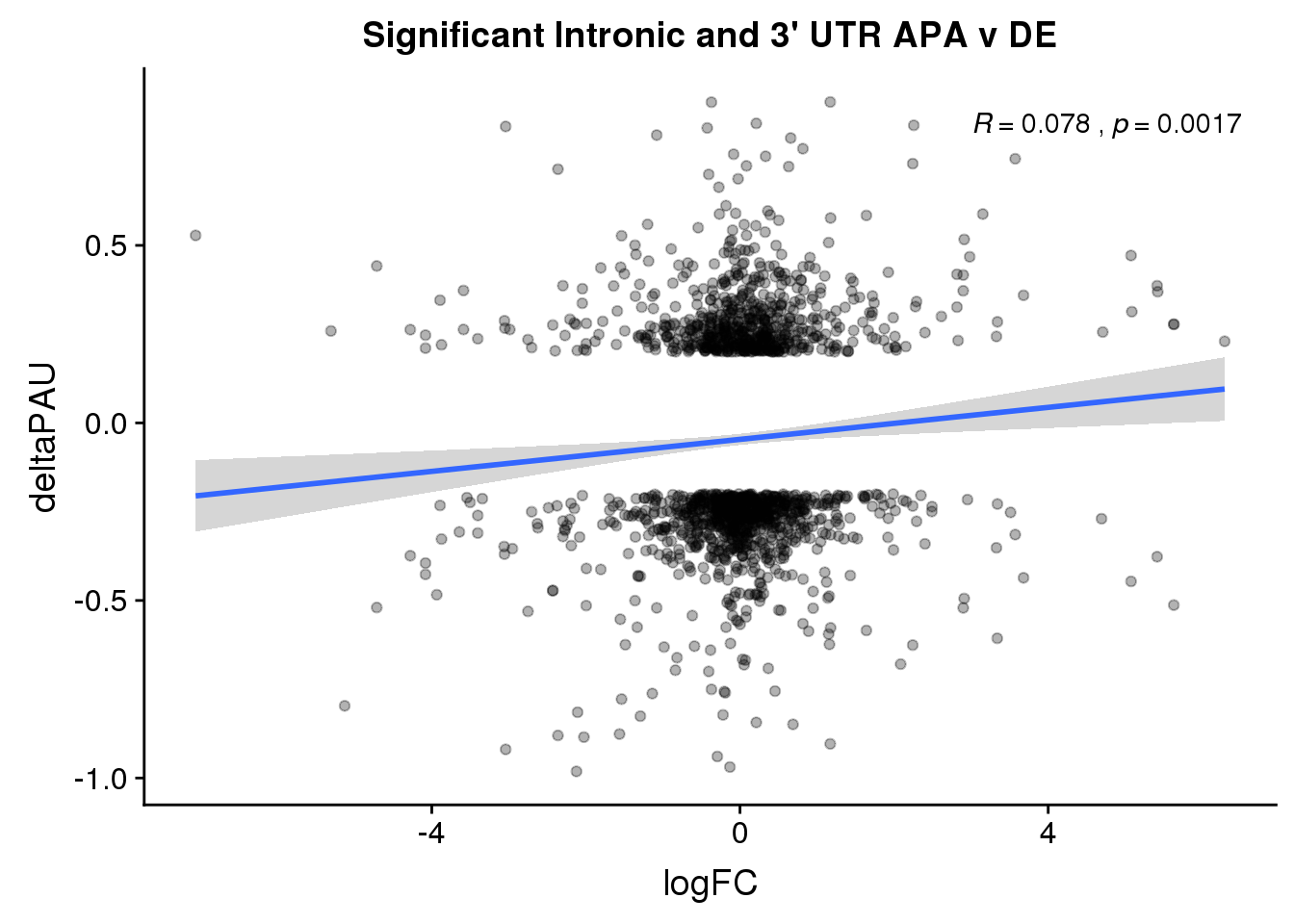
Sig both:
DeandAPA_sigAPAandE= DeandAPA %>% filter(SigPAU2=="Yes", adj.P.Val<.05)ggplot(DeandAPA_sigAPAandE,aes(y=deltaPAU, x=logFC,col=loc)) + geom_point() + geom_smooth(method="lm")+ labs(title="Significant Intronic and 3' UTR APA v Significant DE") + scale_color_brewer(palette = "Dark2")+ stat_cor(aes(color = loc), label.x = 3)
ggplot(DeandAPA_sigAPAandE,aes(y=deltaPAU, x=logFC)) + geom_point() + geom_smooth(method="lm")+ labs(title="Significant Intronic and 3' UTR APA v Significant DE") + scale_color_brewer(palette = "Dark2")+ stat_cor( label.x = 3)
Choose most Sig PAS
To break ties I will use the top average usage. I will not worry about location when chosing top PAS.
DeandAPA_topPAS= DeandAPA %>% mutate(AvgUsageBoth=(Human+Chimp)/2) %>% group_by(gene) %>% arrange(p.adjust,desc(AvgUsageBoth)) %>% slice(1) %>% ungroup()
#intron
nrow(DeandAPA_topPAS %>% filter(loc=="intron"))[1] 1313nrow(DeandAPA_topPAS %>% filter(loc=="intron", SigPAU2=="Yes"))[1] 161#3 utr
nrow(DeandAPA_topPAS %>% filter(loc=="utr3"))[1] 6075nrow(DeandAPA_topPAS %>% filter(loc=="utr3", SigPAU2=="Yes"))[1] 826Plot the correlation in effect size
ggplot(DeandAPA_topPAS,aes(y=deltaPAU, x=logFC, col=loc)) + geom_point(alpha=.3) + geom_smooth(method="lm") + labs(title="Intronic and 3' UTR APA top PAS v DE") + scale_color_brewer(palette = "Dark2") + stat_cor(aes(color = loc), label.x = 3)
ggplot(DeandAPA_topPAS,aes(y=deltaPAU, x=logFC)) + geom_point(alpha=.3) + geom_smooth(method="lm") + stat_cor(label.x = 3)
DeandAPA_topPASsigAPA= DeandAPA_topPAS %>% filter(SigPAU2=="Yes")ggplot(DeandAPA_topPASsigAPA,aes(y=deltaPAU, x=logFC,col=loc)) + geom_point(alpha=.4) + geom_smooth(method="lm") + labs(title="Significant APA, Top PAS v DE ") + scale_color_brewer(palette = "Dark2")+ stat_cor(aes(color = loc), label.x = 3)
ggplot(DeandAPA_topPASsigAPA,aes(y=deltaPAU, x=logFC)) + geom_point(alpha=.4) + geom_smooth(method="lm") + labs(title="Significant APA, Top PAS v DE ") + scale_color_brewer(palette = "Dark2")+ stat_cor( label.x = 3)
Sig both:
DeandAPA_topPASsigAPAandE= DeandAPA_topPASsigAPA %>% filter(SigPAU2=="Yes", adj.P.Val<.05)ggplot(DeandAPA_topPASsigAPAandE,aes(y=deltaPAU, x=logFC,col=loc)) + geom_point(alpha=.3) + geom_smooth(method="lm")+ labs(title="Significant APA, Top PAS v Significant DE") + scale_color_brewer(palette = "Dark2")+ stat_cor(aes(color = loc), label.x = 3)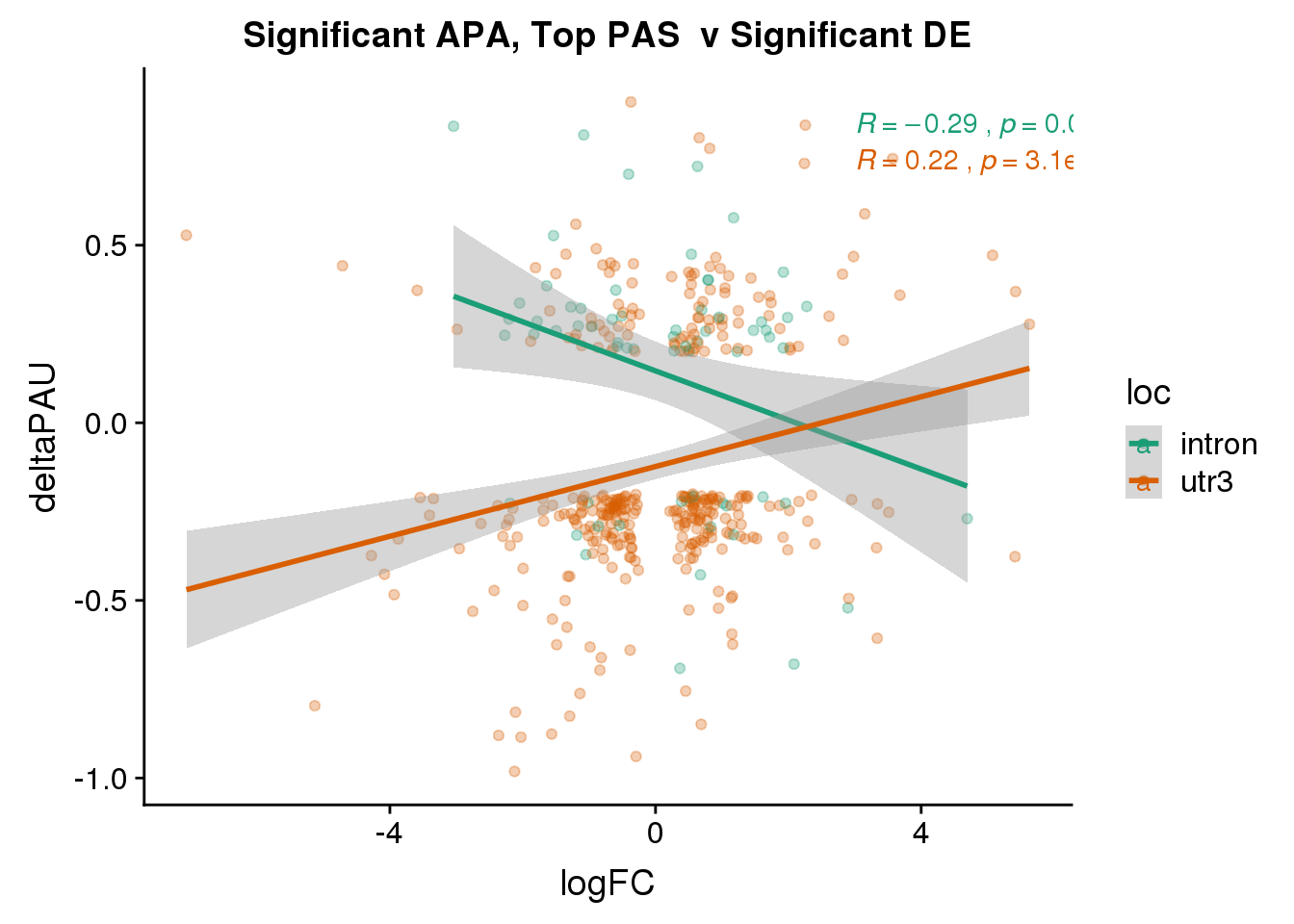
ggplot(DeandAPA_topPASsigAPAandE,aes(y=deltaPAU, x=logFC)) + geom_point(alpha=.3) + geom_smooth(method="lm")+ labs(title="Significant APA, Top PAS v Significant DE") + scale_color_brewer(palette = "Dark2")+ stat_cor(label.x = 3)
Plot together:
allboth=ggplot(DeandAPA,aes(y=deltaPAU, x=logFC)) + geom_point(alpha=.3) + geom_smooth( method="lm") + labs(title="APA v DE") + scale_color_brewer(palette = "Dark2")+ stat_cor(label.x = 1) +theme_classic(base_size = 12)
allSep= ggplot(DeandAPA_topPAS,aes(y=deltaPAU, x=logFC, col=loc)) + geom_point(alpha=.3) + geom_smooth(method="lm") + labs(title="APA, top PAS v DE") + scale_color_brewer(palette = "Dark2") + stat_cor(aes(color = loc), label.x = 1)+theme_classic(base_size = 12)
sigAPAboth=ggplot(DeandAPA_sigAPA,aes(y=deltaPAU, x=logFC)) + geom_point(alpha=.3) + geom_smooth(method="lm")+ labs(title="Significant APA v DE")+ scale_color_brewer(palette = "Dark2")+ stat_cor(label.x = 1)+theme_classic(base_size = 12)
sigAPSep=ggplot(DeandAPA_topPASsigAPA,aes(y=deltaPAU, x=logFC,col=loc)) + geom_point(alpha=.4) + geom_smooth(method="lm") + labs(title="Significant APA, Top PAS v DE ") + scale_color_brewer(palette = "Dark2")+ stat_cor(aes(color = loc), label.x = 1)+theme_classic(base_size = 12)
SigBoth= ggplot(DeandAPA_sigAPAandE,aes(y=deltaPAU, x=logFC)) + geom_point() + geom_smooth(method="lm")+ labs(title="Significant APA v Significant DE") + scale_color_brewer(palette = "Dark2")+ stat_cor( label.x = 1)+theme_classic(base_size = 12)
SigSep=ggplot(DeandAPA_topPASsigAPAandE,aes(y=deltaPAU, x=logFC,col=loc)) + geom_point(alpha=.3) + geom_smooth(method="lm")+ labs(title="Significant APA, Top PAS v Significant DE") + scale_color_brewer(palette = "Dark2")+ stat_cor(aes(color = loc), label.x = 1)+theme_classic(base_size = 12) plot_grid(allboth,allSep,sigAPAboth,sigAPSep,SigBoth,SigSep, ncol=2)
Ribosome occupancy
Ribo=read.table("../data/Wang_ribo/Additionaltable5_translationComparisons.txt",header = T, stringsAsFactors = F) %>% rename("Gene_stable_ID"= ENSG) %>% inner_join(nameID,by="Gene_stable_ID") %>% dplyr::select(Gene.name, HvC.beta, HvC.pvalue, HvC.FDR) %>% rename("gene"=Gene.name)Join with APA
RiboandAPA=DiffIso %>% inner_join(Ribo, by="gene")
RiboandAPA %>% group_by(gene) %>% n_distinct()[1] 23590ggplot(RiboandAPA,aes(y=deltaPAU, x=HvC.beta, col=loc)) + geom_point(alpha=.3) + geom_smooth(method="lm") + labs(title="APA v Ribosome Occupany")+ scale_color_brewer(palette = "Dark2")+ stat_cor(aes(color = loc), label.x = 3)
ggplot(RiboandAPA,aes(y=deltaPAU, x=HvC.beta)) + geom_point(alpha=.3) + geom_smooth(method="lm") + labs(title="APA v Ribosome Occupany")+ scale_color_brewer(palette = "Dark2")+ stat_cor( label.x = 3)
Just the genes with significant differences in PAS
RiboandAPA_sigAPA= RiboandAPA %>% filter(SigPAU2=="Yes")ggplot(RiboandAPA_sigAPA,aes(y=deltaPAU, x=HvC.beta,col=loc)) + geom_point(alpha=.3) + geom_smooth(method="lm")+ labs(title="Significant APA v Ribosome Occupany")+ scale_color_brewer(palette = "Dark2")+ stat_cor(aes(color = loc), label.x = 3)
ggplot(RiboandAPA_sigAPA,aes(y=deltaPAU, x=HvC.beta)) + geom_point(alpha=.3) + geom_smooth(method="lm")+ labs(title="Significant APA v Ribosome Occupany")+ scale_color_brewer(palette = "Dark2")+ stat_cor( label.x = 3)
Sig both:
RiboandAPA_sigAPAandR= RiboandAPA_sigAPA %>% filter(SigPAU2=="Yes", HvC.FDR<.05)ggplot(RiboandAPA_sigAPAandR,aes(y=deltaPAU, x=HvC.beta,col=loc)) + geom_point(alpha=.3) + geom_smooth(method="lm")+ labs(title="Significant APA v Significant Ribosome Occupany") + scale_color_brewer(palette = "Dark2")+ stat_cor(aes(color = loc), label.x = 3)
ggplot(RiboandAPA_sigAPAandR,aes(y=deltaPAU, x=HvC.beta)) + geom_point(alpha=.3) + geom_smooth(method="lm")+ labs(title="Significant APA v Significant Ribosome Occupany") + scale_color_brewer(palette = "Dark2")+ stat_cor(label.x = 3)
The correlation in expression with intronic is not there in ribosome occupancy.
Choose most Sig PAS
To break ties I will use the top average usage. I will not worry about location at first.
RiboandAPA_topPAS= RiboandAPA %>% mutate(AvgUsageBoth=(Human+Chimp)/2) %>% group_by(gene) %>% arrange(p.adjust,desc(AvgUsageBoth)) %>% slice(1) %>% ungroup()
nrow(RiboandAPA %>% filter(loc=="intron"))[1] 9994nrow(RiboandAPA_topPAS %>% filter(loc=="intron"))[1] 1138nrow(RiboandAPA %>% filter(loc=="utr3"))[1] 13596nrow(RiboandAPA_topPAS %>% filter(loc=="utr3"))[1] 5269Plot the correlation in effect size
ggplot(RiboandAPA_topPAS,aes(y=deltaPAU, x=HvC.beta,col=loc)) + geom_point(alpha=.3) + geom_smooth(method="lm") + labs(title="APA, top PAS v Ribosome Occupany")+ scale_color_brewer(palette = "Dark2")+ stat_cor(aes(color = loc), label.x = 3)
ggplot(RiboandAPA_topPAS,aes(y=deltaPAU, x=HvC.beta)) + geom_point(alpha=.3) + geom_smooth(method="lm") + labs(title="APA, top PAS v Ribosome Occupany")+ scale_color_brewer(palette = "Dark2")+ stat_cor(label.x = 3)
Sig APA
RiboandAPA_topPASsigAPA= RiboandAPA_topPAS %>% filter(SigPAU2=="Yes")
nrow(RiboandAPA_topPASsigAPA %>% filter(loc=="intron"))[1] 136nrow(RiboandAPA_topPASsigAPA %>% filter(loc=="utr3"))[1] 675199 intronic significant, 910 significant 3’ utr
ggplot(RiboandAPA_topPASsigAPA,aes(y=deltaPAU, x=HvC.beta,col=loc)) + geom_point(alpha=.3) + geom_smooth(method="lm")+ labs(title="Significant APA, top PAS v Ribosome Occupany") +scale_color_brewer(palette = "Dark2")+ stat_cor(aes(color = loc), label.x = 3)
ggplot(RiboandAPA_topPASsigAPA,aes(y=deltaPAU, x=HvC.beta)) + geom_point(alpha=.3) + geom_smooth(method="lm")+ labs(title="Significant APA, top PAS v Ribosome Occupany") +scale_color_brewer(palette = "Dark2")+ stat_cor(label.x = 3)
Sig both:
RiboandAPA_topPASsigAPAandR= RiboandAPA_topPASsigAPA %>% filter(SigPAU2=="Yes", HvC.FDR<.05)
nrow(RiboandAPA_topPASsigAPAandR %>% filter(loc=="intron"))[1] 37nrow(RiboandAPA_topPASsigAPAandR %>% filter(loc=="utr3"))[1] 17747 PAS for intrnic 229 for 3’ UTR
ggplot(RiboandAPA_topPASsigAPAandR,aes(y=deltaPAU, x=HvC.beta,col=loc)) + geom_point(alpha=.3) + geom_smooth(method="lm")+ labs(title="Significant APA, top PAS v Significant Ribosome Occupany") + scale_color_brewer(palette = "Dark2")+ stat_cor(aes(color = loc), label.x = 3)
ggplot(RiboandAPA_topPASsigAPAandR,aes(y=deltaPAU, x=HvC.beta)) + geom_point(alpha=.3) + geom_smooth(method="lm")+ labs(title="Significant APA, top PAS v Significant Ribosome Occupany") + scale_color_brewer(palette = "Dark2")+ stat_cor(label.x = 3)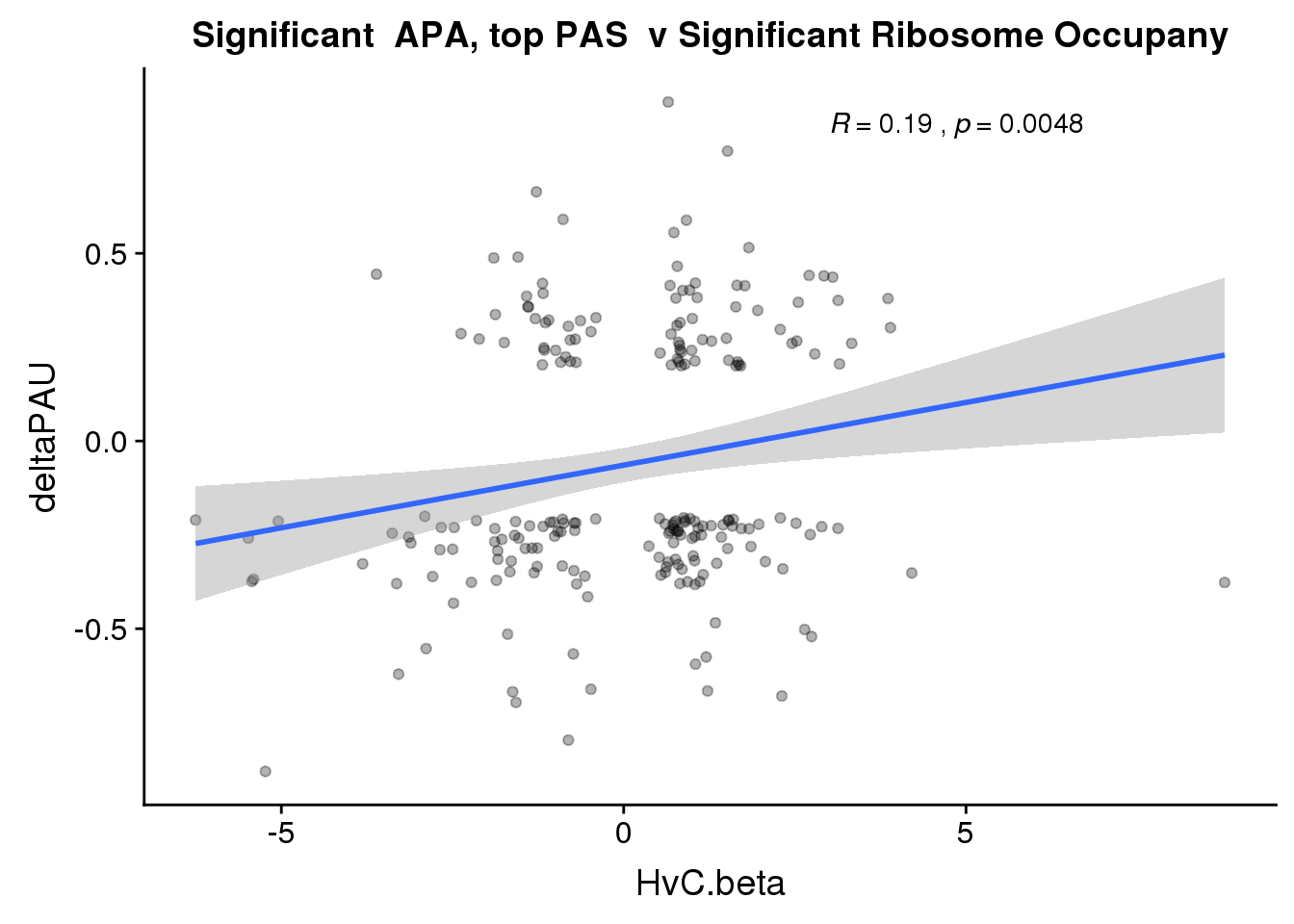
Correlation in UTR but not intronic. Not sure if this is due to the number of PAS.
Plot all together
riboBoth=ggplot(RiboandAPA,aes(y=deltaPAU, x=HvC.beta)) + geom_point(alpha=.3) + geom_smooth(method="lm") + labs(title="APA v Ribosome Occupany")+ scale_color_brewer(palette = "Dark2")+ stat_cor( label.x = 1)+theme_classic(base_size = 12)
riboTop=ggplot(RiboandAPA_topPAS,aes(y=deltaPAU, x=HvC.beta,col=loc)) + geom_point(alpha=.3) + geom_smooth(method="lm") + labs(title="APA, top PAS v Ribosome Occupany")+ scale_color_brewer(palette = "Dark2")+ stat_cor(aes(color = loc), label.x = 1)+theme_classic(base_size = 12)
ribosigapaboth=ggplot(RiboandAPA_sigAPA,aes(y=deltaPAU, x=HvC.beta)) + geom_point(alpha=.3) + geom_smooth(method="lm")+ labs(title="Significant APA v Ribosome Occupany")+ scale_color_brewer(palette = "Dark2")+ stat_cor( label.x = 1)+theme_classic(base_size = 12)
ribosigapasep=ggplot(RiboandAPA_topPASsigAPA,aes(y=deltaPAU, x=HvC.beta,col=loc)) + geom_point(alpha=.3) + geom_smooth(method="lm")+ labs(title="Significant APA, top PAS v Ribosome Occupany") +scale_color_brewer(palette = "Dark2")+ stat_cor(aes(color = loc), label.x = 1)+theme_classic(base_size = 12)
sigapasigriboboth=ggplot(RiboandAPA_sigAPAandR,aes(y=deltaPAU, x=HvC.beta)) + geom_point(alpha=.3) + geom_smooth(method="lm")+ labs(title="Significant APA v Significant Ribosome Occupany") + scale_color_brewer(palette = "Dark2")+ stat_cor(label.x = 1)+theme_classic(base_size = 12)
sigapasigribosep=ggplot(RiboandAPA_topPASsigAPAandR,aes(y=deltaPAU, x=HvC.beta,col=loc)) + geom_point(alpha=.3) + geom_smooth(method="lm")+ labs(title="Significant APA, top PAS v Significant Ribosome Occupany") + scale_color_brewer(palette = "Dark2")+ stat_cor(aes(color = loc), label.x = 1)+theme_classic(base_size = 12) plot_grid(riboBoth,riboTop,ribosigapaboth, ribosigapasep,sigapasigriboboth,sigapasigribosep, ncol=2)
Protein
I will use code from https://github.com/siddisis/project_primate_ribo to fit the linear model again and get effect sizes.
load(“../tables/fileS4.RData”)
load(“../rdas/HCR.protein.TMM.RData”)
Put both of these in ../data/Khan_prot
load("../data/Khan_prot/fileS4.RData")
load("../data/Khan_prot/HCR.protein.TMM.RData")expressed.gene.names <- as.character(HCR.protein.TMM.norm.ESNGlabeled[rownames(HCR.protein.TMM.norm.ESNGlabeled) %in% rownames(protein.expressed.data),16])
names(expressed.gene.names) <- rownames(protein.expressed.data)Use to make design matrix
# HvC
RNA.expressed.data.HC<-RNA.expressed.data[,1:10]
species.label <- substring(colnames(RNA.expressed.data.HC),1,1)
design <- model.matrix(~species.label)
colnames(design)<-c("Chimp","Human")Protien
protein.expressed.data.HC<-protein.expressed.data[,1:10]
protein.fit<-lmFit(protein.expressed.data.HC ,design = design)
HvC.prot<- eBayes(protein.fit)
top.table <- topTable(HvC.prot, n = Inf)
volcanoplot(HvC.prot,coef=2,highlight=2)
| Version | Author | Date |
|---|---|---|
| c9c6b6a | brimittleman | 2020-02-29 |
effectsizeDF= as.data.frame(cbind(Gene_stable_ID=rownames(protein.expressed.data.HC),logEf=HvC.prot$coefficients[,2], pval=top.table$adj.P.Val)) %>% inner_join(nameID,by="Gene_stable_ID") %>% dplyr::rename('gene'=Gene.name) %>% dplyr::select(-Gene_stable_ID)Warning: Column `Gene_stable_ID` joining factor and character vector,
coercing into character vectorwrite.table(effectsizeDF, "../data/Khan_prot/ProtData_effectSize.txt",col.names = T, row.names = F, quote = F)DPandAPA= DiffIso %>% inner_join(effectsizeDF, by="gene")
DPandAPA %>% group_by(gene) %>% summarise(n()) %>% nrow()[1] 2611DPandAPA$logEf= as.numeric(as.character(DPandAPA$logEf))
DPandAPA$pval= as.numeric(as.character(DPandAPA$pval))Looking at 2557 common genes.
ggplot(DPandAPA,aes(y=deltaPAU, x=logEf,col=loc)) + geom_point(alpha=.3) + geom_smooth(aes(col=loc), method="lm") + labs(title="Intronic and 3' UTR APA v DP") + scale_color_brewer(palette = "Dark2")+ stat_cor(aes(color = loc), label.x = 1)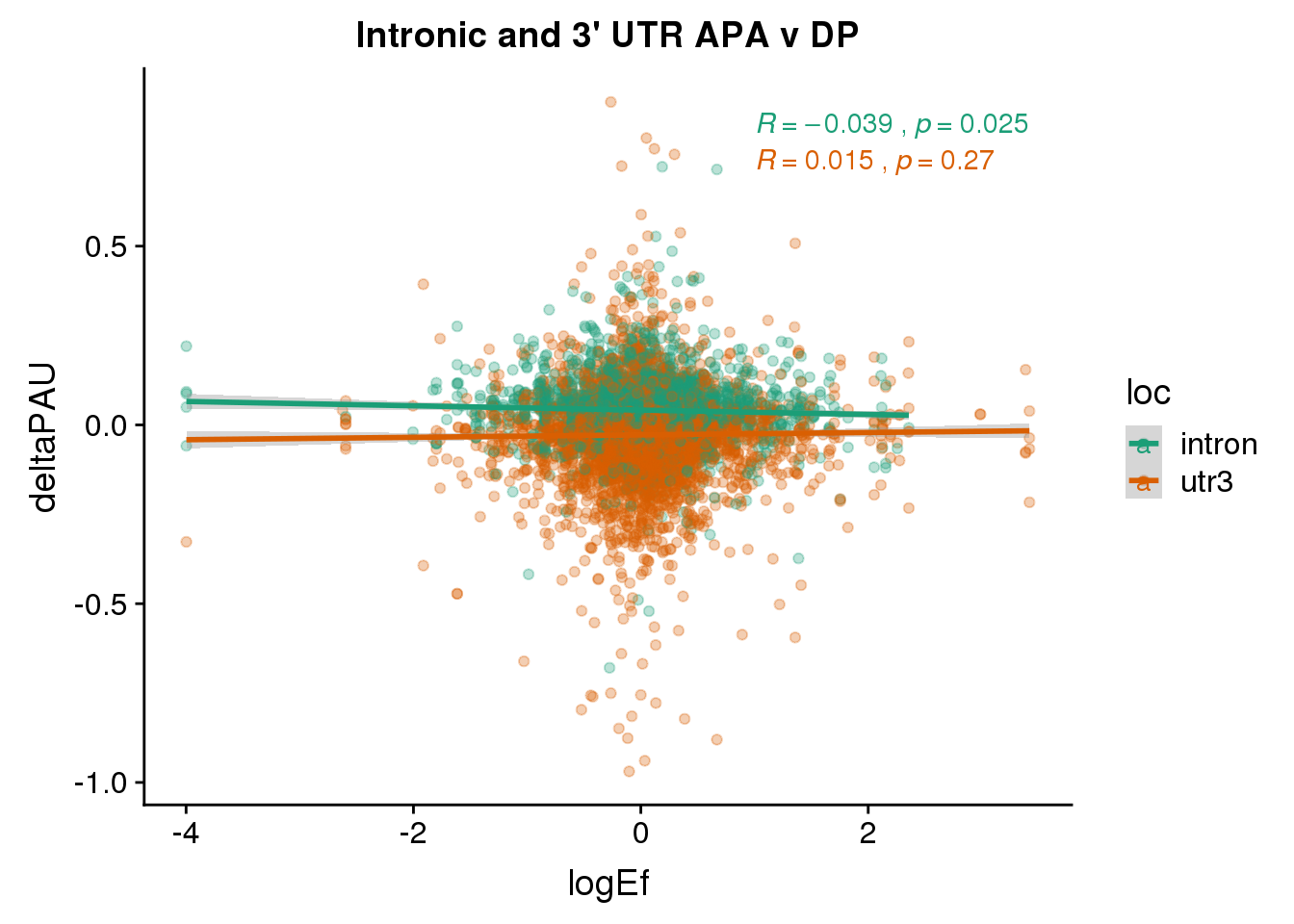
ggplot(DPandAPA,aes(y=deltaPAU, x=logEf)) + geom_point(alpha=.3) + geom_smooth(method="lm") + labs(title="Intronic and 3' UTR APA v DP") + scale_color_brewer(palette = "Dark2")+ stat_cor( label.x = 1)
Just the genes with significant differences in PAS
PandAPA_sigAPA= DPandAPA %>% filter(SigPAU2=="Yes")ggplot(PandAPA_sigAPA,aes(y=deltaPAU, x=logEf,col=loc)) + geom_point(alpha=.3) + geom_smooth(method="lm")+ labs(title="Significant APA v Protein")+ scale_color_brewer(palette = "Dark2")+ stat_cor(aes(color = loc), label.x = 1)
ggplot(PandAPA_sigAPA,aes(y=deltaPAU, x=logEf)) + geom_point(alpha=.3) + geom_smooth(method="lm")+ labs(title="Significant APA v Protein")+ scale_color_brewer(palette = "Dark2")+ stat_cor( label.x = 1)
Sig both:
PandAPA_sigAPAandP= PandAPA_sigAPA %>% filter(SigPAU2=="Yes", pval <.05)ggplot(PandAPA_sigAPAandP,aes(y=deltaPAU, x=logEf,col=loc)) + geom_point(alpha=.3) + geom_smooth(method="lm")+ labs(title="Significant APA v Signficant Protein")+ scale_color_brewer(palette = "Dark2")+ stat_cor(aes(color = loc), label.x = 1)
ggplot(PandAPA_sigAPAandP,aes(y=deltaPAU, x=logEf)) + geom_point(alpha=.3) + geom_smooth(method="lm")+ labs(title="Significant APA v Signficant Protein")+ scale_color_brewer(palette = "Dark2")+ stat_cor( label.x = 1)
Choose most Sig PAS
To break ties I will use the top average usage. I will not worry about location at first.
PandAPA_topPAS= DPandAPA %>% mutate(AvgUsageBoth=(Human+Chimp)/2) %>% group_by(gene) %>% arrange(p.adjust,desc(AvgUsageBoth)) %>% slice(1) %>% ungroup()ggplot(PandAPA_topPAS,aes(y=deltaPAU, x=logEf,col=loc)) + geom_point(alpha=.3) + geom_smooth(aes(col=loc), method="lm") + labs(title="Intronic and 3' UTR APA, top PAS v DP") + scale_color_brewer(palette = "Dark2")+ stat_cor(aes(color = loc), label.x = 1)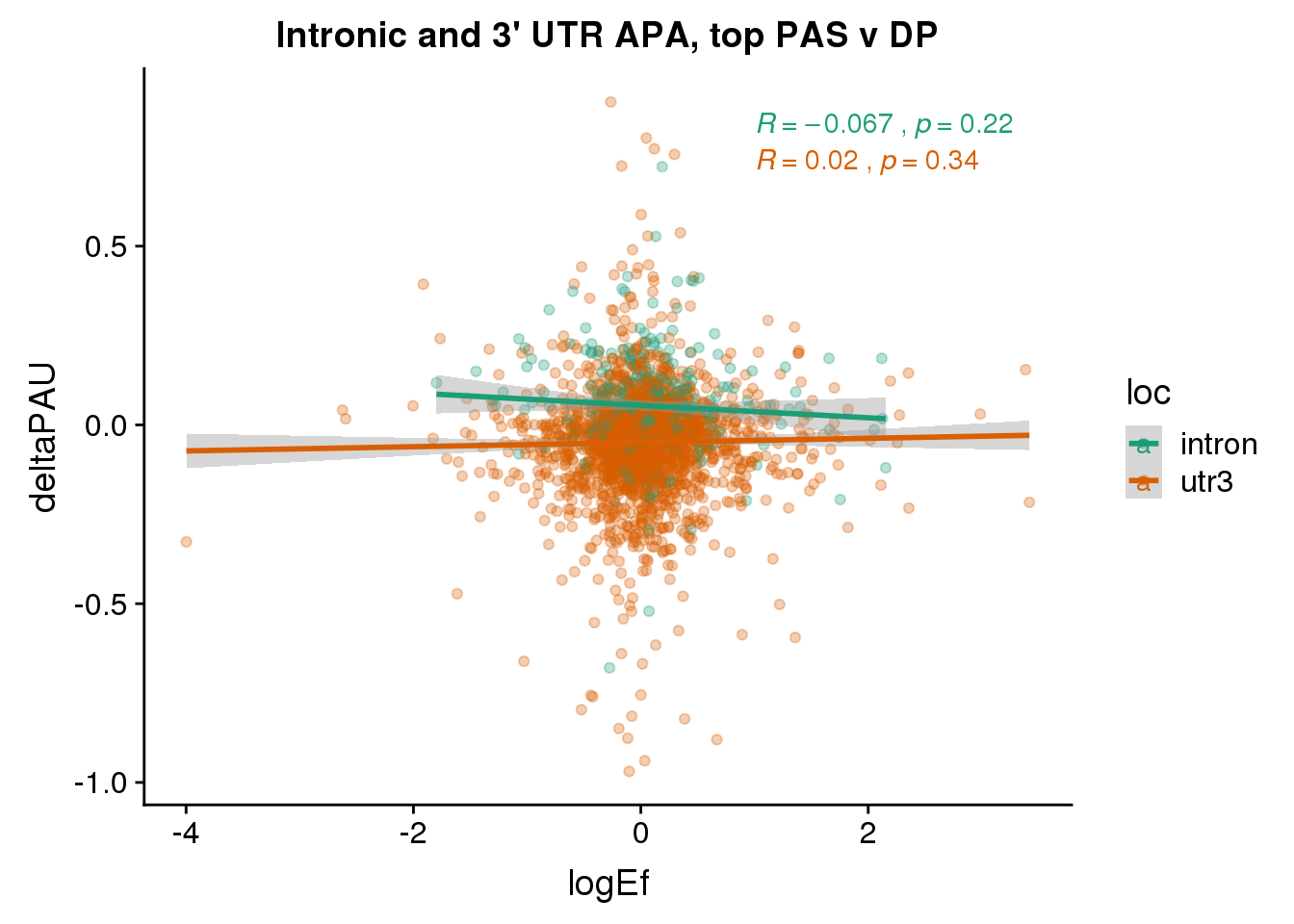
ggplot(PandAPA_topPAS,aes(y=deltaPAU, x=logEf)) + geom_point(alpha=.3) + geom_smooth(method="lm") + labs(title="Intronic and 3' UTR APA v DP") + scale_color_brewer(palette = "Dark2")+ stat_cor( label.x = 1)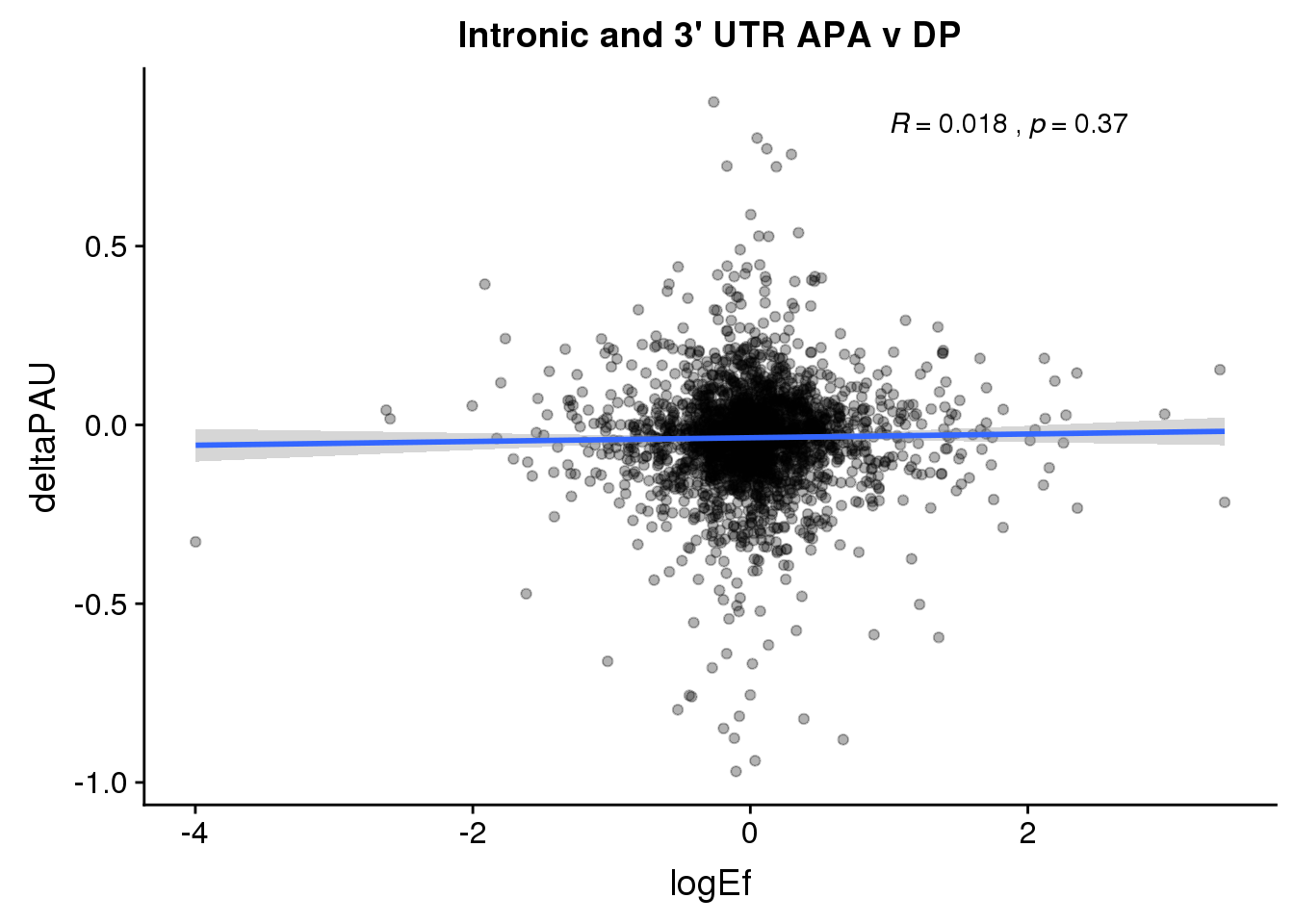
PandAPA_topPAS_sigAPA= PandAPA_topPAS %>% filter(SigPAU2=="Yes")ggplot(PandAPA_topPAS_sigAPA,aes(y=deltaPAU, x=logEf,col=loc)) + geom_point(alpha=.3) + geom_smooth(method="lm")+ labs(title="Significant APA, top PAS v Protein")+ scale_color_brewer(palette = "Dark2")+ stat_cor(aes(color = loc), label.x = 1)
ggplot(PandAPA_topPAS_sigAPA,aes(y=deltaPAU, x=logEf)) + geom_point(alpha=.3) + geom_smooth(method="lm")+ labs(title="Significant APA top PAS v Protein")+ scale_color_brewer(palette = "Dark2")+ stat_cor( label.x = 1)
Sig both:
PandAPA_topPAS_sigAPAandP= PandAPA_topPAS_sigAPA %>% filter(SigPAU2=="Yes", pval <.05)ggplot(PandAPA_topPAS_sigAPAandP,aes(y=deltaPAU, x=logEf,col=loc)) + geom_point(alpha=.3) + geom_smooth(method="lm")+ labs(title="Significant APA top PAS v Signficant Protein")+ scale_color_brewer(palette = "Dark2")+ stat_cor(aes(color = loc), label.x = 1)
ggplot(PandAPA_topPAS_sigAPAandP,aes(y=deltaPAU, x=logEf)) + geom_point(alpha=.3) + geom_smooth(method="lm")+ labs(title="Significant APA top PAS v Signficant Protein")+ scale_color_brewer(palette = "Dark2")+ stat_cor( label.x = 1)
Check pvalues:
protKhan=read.csv("../data/Khan_prot/Khan_TableS4.csv",header = T) %>% rename("Gene_stable_ID"= ENSG) %>% inner_join(nameID,by="Gene_stable_ID") %>% rename("gene"=Gene.name) Warning: Column `Gene_stable_ID` joining factor and character vector,
coercing into character vectorprotKhanwmine= protKhan %>% inner_join(effectsizeDF, by="gene")
protKhanwmine$logEf=as.numeric(as.character(protKhanwmine$logEf))
protKhanwmine$pval=as.numeric(as.character(protKhanwmine$pval))
cor.test(protKhanwmine$pval,protKhanwmine$HC.pvalues.protein )
Pearson's product-moment correlation
data: protKhanwmine$pval and protKhanwmine$HC.pvalues.protein
t = -0.71198, df = 3248, p-value = 0.4765
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
-0.04685410 0.02189991
sample estimates:
cor
-0.01249186 This is not good. Try the difference in means approach:
Chimp-human
protKhanSmall= protKhan %>% select(gene,mean.H.protein,mean.C.protein, HC.qvalues.rna) %>% mutate(Effect=mean.C.protein-mean.H.protein)deltaPandAPA= DiffIso %>% inner_join(protKhanSmall, by="gene")
deltaPandAPA %>% group_by(gene) %>% summarise(n()) %>% nrow()[1] 26662607 genes
ggplot(deltaPandAPA,aes(y=deltaPAU, x=Effect,col=loc)) + geom_point(alpha=.3) + geom_smooth(aes(col=loc), method="lm") + labs(title="Intronic and 3' UTR APA v DP") + scale_color_brewer(palette = "Dark2")+ stat_cor(aes(color = loc), label.x = 2)
ggplot(deltaPandAPA,aes(y=deltaPAU, x=Effect)) + geom_point(alpha=.3) + geom_smooth(method="lm") + labs(title="Intronic and 3' UTR APA v DP") + scale_color_brewer(palette = "Dark2")+ stat_cor( label.x = 2)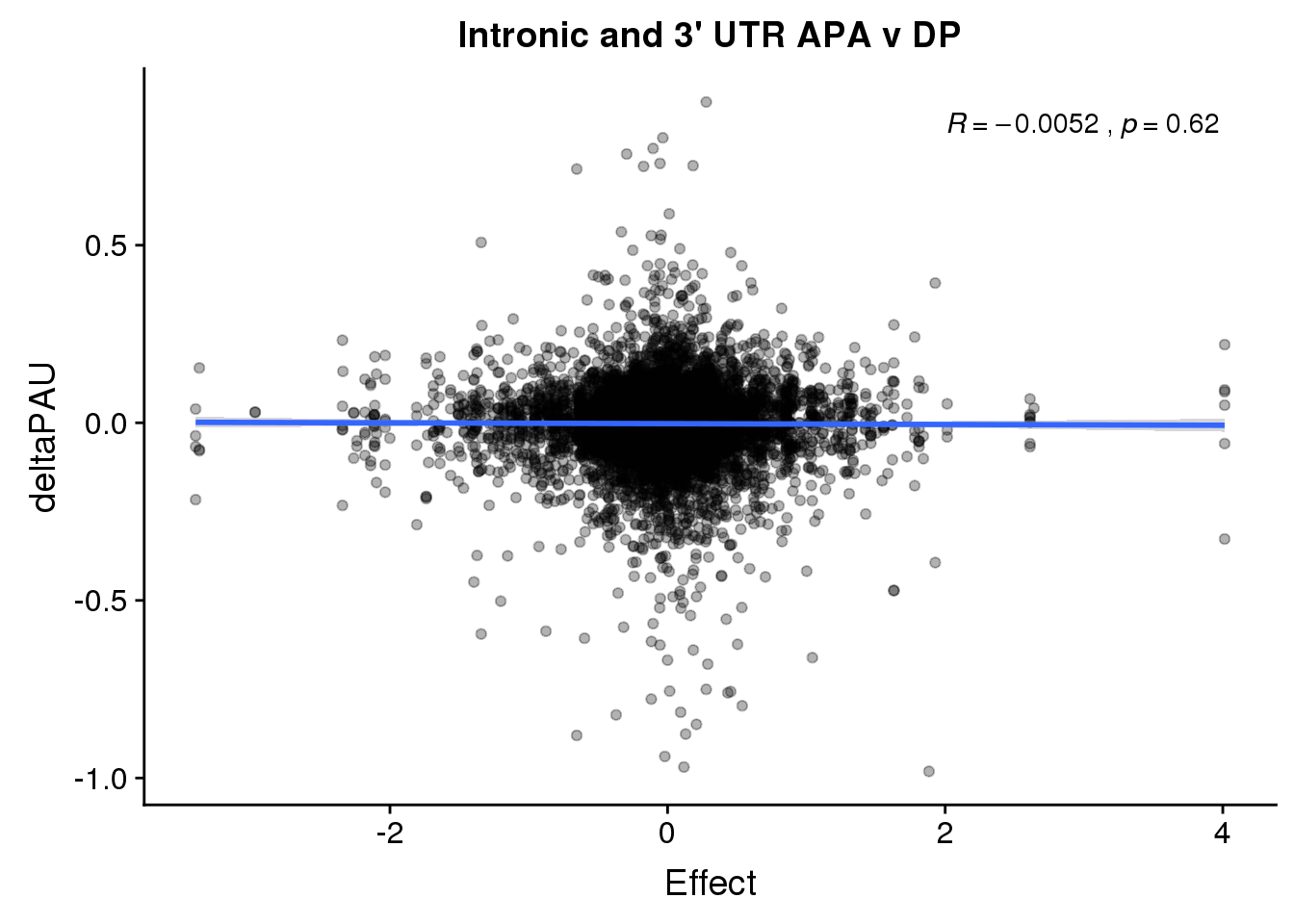
Correlation effect sizes:
protKhanSmall_withmone= protKhanSmall %>% inner_join(effectsizeDF, by="gene")
protKhanSmall_withmone$logEf=as.numeric(as.character(protKhanSmall_withmone$logEf))
cor.test(protKhanSmall_withmone$logEf, protKhanSmall_withmone$Effect)
Pearson's product-moment correlation
data: protKhanSmall_withmone$logEf and protKhanSmall_withmone$Effect
t = -11459, df = 3248, p-value < 2.2e-16
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
-0.9999885 -0.9999868
sample estimates:
cor
-0.9999876 Ok this is equal but opposite. So this is correct.
I need to check the direction of the effects.
Plot together:
protboth=ggplot(DPandAPA,aes(y=deltaPAU, x=logEf)) + geom_point(alpha=.3) + geom_smooth(method="lm") + labs(title="APA v Protein") + scale_color_brewer(palette = "Dark2")+ stat_cor( label.x = 2)+theme_classic(base_size = 12)
protsep=ggplot(PandAPA_topPAS,aes(y=deltaPAU, x=logEf,col=loc)) + geom_point(alpha=.3) + geom_smooth(aes(col=loc), method="lm") + labs(title="APA, top PAS v Protein") + scale_color_brewer(palette = "Dark2")+ stat_cor(aes(color = loc), label.x = 1)+theme_classic(base_size = 12)
protsigapa=ggplot(PandAPA_sigAPA,aes(y=deltaPAU, x=logEf)) + geom_point(alpha=.3) + geom_smooth(method="lm")+ labs(title="Significant APA v Protein")+ scale_color_brewer(palette = "Dark2")+ stat_cor( label.x = 1)+theme_classic(base_size = 12)
protsigapasep=ggplot(PandAPA_topPAS_sigAPA,aes(y=deltaPAU, x=logEf,col=loc)) + geom_point(alpha=.3) + geom_smooth(method="lm")+ labs(title="Significant APA, top PAS v Protein")+ scale_color_brewer(palette = "Dark2")+ stat_cor(aes(color = loc), label.x = 1)+theme_classic(base_size = 12)
protsigall=ggplot(PandAPA_sigAPAandP,aes(y=deltaPAU, x=logEf)) + geom_point(alpha=.3) + geom_smooth(method="lm")+ labs(title="Significant APA v Signficant Protein")+ scale_color_brewer(palette = "Dark2")+ stat_cor( label.x = 1)+theme_classic(base_size = 12)
protsigallsep=ggplot(PandAPA_topPAS_sigAPAandP,aes(y=deltaPAU, x=logEf,col=loc)) + geom_point(alpha=.3) + geom_smooth(method="lm")+ labs(title="Significant APA top PAS v Signficant Protein")+ scale_color_brewer(palette = "Dark2")+ stat_cor(aes(color = loc), label.x = 1)+theme_classic(base_size = 12) plot_grid(protboth,protsep,protsigapa,protsigapasep,protsigall,protsigallsep, ncol=2)
sessionInfo()R version 3.5.1 (2018-07-02)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] cowplot_0.9.4 forcats_0.3.0 stringr_1.3.1 dplyr_0.8.0.1
[5] purrr_0.3.2 readr_1.3.1 tidyr_0.8.3 tibble_2.1.1
[9] tidyverse_1.2.1 qvalue_2.14.0 limma_3.38.2 ggpubr_0.2
[13] magrittr_1.5 ggplot2_3.1.1 workflowr_1.6.0
loaded via a namespace (and not attached):
[1] tidyselect_0.2.5 reshape2_1.4.3 splines_3.5.1
[4] haven_1.1.2 lattice_0.20-38 colorspace_1.3-2
[7] generics_0.0.2 htmltools_0.3.6 yaml_2.2.0
[10] rlang_0.4.0 later_0.7.5 pillar_1.3.1
[13] glue_1.3.0 withr_2.1.2 RColorBrewer_1.1-2
[16] modelr_0.1.2 readxl_1.1.0 plyr_1.8.4
[19] munsell_0.5.0 gtable_0.2.0 cellranger_1.1.0
[22] rvest_0.3.2 evaluate_0.12 labeling_0.3
[25] knitr_1.20 httpuv_1.4.5 broom_0.5.1
[28] Rcpp_1.0.2 promises_1.0.1 scales_1.0.0
[31] backports_1.1.2 jsonlite_1.6 fs_1.3.1
[34] hms_0.4.2 digest_0.6.18 stringi_1.2.4
[37] grid_3.5.1 rprojroot_1.3-2 cli_1.1.0
[40] tools_3.5.1 lazyeval_0.2.1 crayon_1.3.4
[43] whisker_0.3-2 pkgconfig_2.0.2 xml2_1.2.0
[46] lubridate_1.7.4 rstudioapi_0.10 assertthat_0.2.0
[49] rmarkdown_1.10 httr_1.3.1 R6_2.3.0
[52] nlme_3.1-137 git2r_0.26.1 compiler_3.5.1