Mediation analysis
Briana Mittleman
1/13/2020
Last updated: 2020-01-15
Checks: 7 0
Knit directory: Comparative_APA/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.5.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190902) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: code/chimp_log/
Ignored: code/human_log/
Ignored: data/.DS_Store
Ignored: data/metadata_HCpanel.txt.sb-a5794dd2-i594qs/
Untracked files:
Untracked: ._.DS_Store
Untracked: Chimp/
Untracked: Human/
Untracked: analysis/CrossChimpThreePrime.Rmd
Untracked: analysis/DiffTransProtvsExpression.Rmd
Untracked: analysis/assessReadQual.Rmd
Untracked: analysis/diffExpressionPantro6.Rmd
Untracked: code/._ClassifyLeafviz.sh
Untracked: code/._Config_chimp.yaml
Untracked: code/._Config_chimp_full.yaml
Untracked: code/._Config_human.yaml
Untracked: code/._ConvertJunc2Bed.sh
Untracked: code/._CountNucleotides.py
Untracked: code/._CrossMapChimpRNA.sh
Untracked: code/._CrossMapThreeprime.sh
Untracked: code/._DiffSplice.sh
Untracked: code/._DiffSplicePlots.sh
Untracked: code/._DiffSplicePlots_gencode.sh
Untracked: code/._DiffSplice_gencode.sh
Untracked: code/._DiffSplice_removebad.sh
Untracked: code/._FindIntronForDomPAS.sh
Untracked: code/._GetMAPQscore.py
Untracked: code/._GetSecondaryMap.py
Untracked: code/._Lift5perPAS.sh
Untracked: code/._LiftFinalChimpJunc2Human.sh
Untracked: code/._LiftOrthoPAS2chimp.sh
Untracked: code/._MapBadSamples.sh
Untracked: code/._PAS_ATTAAA.sh
Untracked: code/._PASsequences.sh
Untracked: code/._PlotNuclearUsagebySpecies.R
Untracked: code/._QuantMergedClusters.sh
Untracked: code/._ReverseLiftFilter.R
Untracked: code/._RunFixLeafCluster.sh
Untracked: code/._RunNegMCMediation.sh
Untracked: code/._Snakefile
Untracked: code/._SnakefilePAS
Untracked: code/._SnakefilePASfilt
Untracked: code/._SortIndexBadSamples.sh
Untracked: code/._bed215upbed.py
Untracked: code/._bed2SAF_gen.py
Untracked: code/._buildIndecpantro5
Untracked: code/._buildIndecpantro5.sh
Untracked: code/._buildLeafviz.sh
Untracked: code/._buildLeafviz_leadAnno.sh
Untracked: code/._buildStarIndex.sh
Untracked: code/._chimpChromprder.sh
Untracked: code/._cleanbed2saf.py
Untracked: code/._cluster.json
Untracked: code/._cluster2bed.py
Untracked: code/._clusterLiftReverse.sh
Untracked: code/._clusterLiftReverse_removebad.sh
Untracked: code/._clusterLiftprimary.sh
Untracked: code/._clusterLiftprimary_removebad.sh
Untracked: code/._converBam2Junc.sh
Untracked: code/._converBam2Junc_removeBad.sh
Untracked: code/._extraSnakefiltpas
Untracked: code/._filter5percPAS.py
Untracked: code/._filterNumChroms.py
Untracked: code/._filterPASforMP.py
Untracked: code/._filterPostLift.py
Untracked: code/._fixExonFC.py
Untracked: code/._fixLeafCluster.py
Untracked: code/._fixLiftedJunc.py
Untracked: code/._fixUTRexonanno.py
Untracked: code/._formathg38Anno.py
Untracked: code/._formatpantro6Anno.py
Untracked: code/._getRNAseqMapStats.sh
Untracked: code/._hg19MapStats.sh
Untracked: code/._humanChromorder.sh
Untracked: code/._intersectLiftedPAS.sh
Untracked: code/._liftJunctionFiles.sh
Untracked: code/._liftPAS19to38.sh
Untracked: code/._liftedchimpJunc2human.sh
Untracked: code/._makeNuclearDapaplots.sh
Untracked: code/._makeSamplyGroupsHuman_TvN.py
Untracked: code/._mapRNAseqhg19.sh
Untracked: code/._mapRNAseqhg19_newPipeline.sh
Untracked: code/._maphg19.sh
Untracked: code/._maphg19_subjunc.sh
Untracked: code/._mediation_test.R
Untracked: code/._mergeChimp3prime_inhg38.sh
Untracked: code/._mergedBam2BW.sh
Untracked: code/._nameClusters.py
Untracked: code/._negativeMediation_montecarlo.R
Untracked: code/._numMultimap.py
Untracked: code/._overlapapaQTLPAS.sh
Untracked: code/._prepareCleanLiftedFC_5perc4LC.py
Untracked: code/._prepareLeafvizAnno.sh
Untracked: code/._preparePAS4lift.py
Untracked: code/._primaryLift.sh
Untracked: code/._processhg38exons.py
Untracked: code/._quantJunc.sh
Untracked: code/._quantJunc_TEST.sh
Untracked: code/._quantJunc_removeBad.sh
Untracked: code/._quantMerged_seperatly.sh
Untracked: code/._recLiftchim2human.sh
Untracked: code/._revLiftPAShg38to19.sh
Untracked: code/._reverseLift.sh
Untracked: code/._runCheckReverseLift.sh
Untracked: code/._runChimpDiffIso.sh
Untracked: code/._runCountNucleotides.sh
Untracked: code/._runFilterNumChroms.sh
Untracked: code/._runHumanDiffIso.sh
Untracked: code/._runNuclearDifffIso.sh
Untracked: code/._runTotalDiffIso.sh
Untracked: code/._run_chimpverifybam.sh
Untracked: code/._run_verifyBam.sh
Untracked: code/._snakemake.batch
Untracked: code/._snakemakePAS.batch
Untracked: code/._snakemakePASchimp.batch
Untracked: code/._snakemakePAShuman.batch
Untracked: code/._snakemake_chimp.batch
Untracked: code/._snakemake_human.batch
Untracked: code/._snakemakefiltPAS.batch
Untracked: code/._snakemakefiltPAS_chimp
Untracked: code/._snakemakefiltPAS_chimp.sh
Untracked: code/._snakemakefiltPAS_human.sh
Untracked: code/._submit-snakemake-chimp.sh
Untracked: code/._submit-snakemake-human.sh
Untracked: code/._submit-snakemakePAS-chimp.sh
Untracked: code/._submit-snakemakePAS-human.sh
Untracked: code/._submit-snakemakefiltPAS-chimp.sh
Untracked: code/._submit-snakemakefiltPAS-human.sh
Untracked: code/._subset_diffisopheno_Nuclear_HvC.py
Untracked: code/._subset_diffisopheno_Total_HvC.py
Untracked: code/._transcriptDTplotsNuclear.sh
Untracked: code/._verifyBam4973.sh
Untracked: code/._verifyBam4973inHuman.sh
Untracked: code/._wrap_chimpverifybam.sh
Untracked: code/._wrap_verifyBam.sh
Untracked: code/._writeMergecode.py
Untracked: code/.snakemake/
Untracked: code/ClassifyLeafviz.sh
Untracked: code/Config_chimp.yaml
Untracked: code/Config_chimp_full.yaml
Untracked: code/Config_human.yaml
Untracked: code/ConvertJunc2Bed.err
Untracked: code/ConvertJunc2Bed.out
Untracked: code/ConvertJunc2Bed.sh
Untracked: code/CountNucleotides.py
Untracked: code/CrossMapChimpRNA.sh
Untracked: code/CrossMapThreeprime.sh
Untracked: code/CrossmapChimp3prime.err
Untracked: code/CrossmapChimp3prime.out
Untracked: code/CrossmapChimpRNA.err
Untracked: code/CrossmapChimpRNA.out
Untracked: code/DiffSplice.err
Untracked: code/DiffSplice.out
Untracked: code/DiffSplice.sh
Untracked: code/DiffSplicePlots.err
Untracked: code/DiffSplicePlots.out
Untracked: code/DiffSplicePlots.sh
Untracked: code/DiffSplicePlots_gencode.sh
Untracked: code/DiffSplice_gencode.sh
Untracked: code/DiffSplice_removebad.err
Untracked: code/DiffSplice_removebad.out
Untracked: code/DiffSplice_removebad.sh
Untracked: code/FilterReverseLift.err
Untracked: code/FilterReverseLift.out
Untracked: code/FindIntronForDomPAS.err
Untracked: code/FindIntronForDomPAS.out
Untracked: code/FindIntronForDomPAS.sh
Untracked: code/GencodeDiffSplice.err
Untracked: code/GencodeDiffSplice.out
Untracked: code/GetMAPQscore.py
Untracked: code/GetSecondaryMap.py
Untracked: code/HchromOrder.err
Untracked: code/HchromOrder.out
Untracked: code/JunctionLift.err
Untracked: code/JunctionLift.out
Untracked: code/JunctionLiftFinalChimp.err
Untracked: code/JunctionLiftFinalChimp.out
Untracked: code/Lift5perPAS.sh
Untracked: code/Lift5perPASbed.err
Untracked: code/Lift5perPASbed.out
Untracked: code/LiftClustersFirst.err
Untracked: code/LiftClustersFirst.out
Untracked: code/LiftClustersFirst_remove.err
Untracked: code/LiftClustersFirst_remove.out
Untracked: code/LiftClustersSecond.err
Untracked: code/LiftClustersSecond.out
Untracked: code/LiftClustersSecond_remove.err
Untracked: code/LiftClustersSecond_remove.out
Untracked: code/LiftFinalChimpJunc2Human.sh
Untracked: code/LiftOrthoPAS2chimp.sh
Untracked: code/LiftorthoPAS.err
Untracked: code/LiftorthoPASt.out
Untracked: code/Log.out
Untracked: code/MapBadSamples.err
Untracked: code/MapBadSamples.out
Untracked: code/MapBadSamples.sh
Untracked: code/MapStats.err
Untracked: code/MapStats.out
Untracked: code/MergeClusters.err
Untracked: code/MergeClusters.out
Untracked: code/MergeClusters.sh
Untracked: code/PAS_ATTAAA.err
Untracked: code/PAS_ATTAAA.out
Untracked: code/PAS_ATTAAA.sh
Untracked: code/PAS_sequence.err
Untracked: code/PAS_sequence.out
Untracked: code/PASsequences.sh
Untracked: code/PlotNuclearUsagebySpecies.R
Untracked: code/QuantMergeClusters
Untracked: code/QuantMergeClusters.err
Untracked: code/QuantMergeClusters.out
Untracked: code/QuantMergedClusters.sh
Untracked: code/Rev_liftoverPAShg19to38.err
Untracked: code/Rev_liftoverPAShg19to38.out
Untracked: code/ReverseLiftFilter.R
Untracked: code/RunFixCluster.err
Untracked: code/RunFixCluster.out
Untracked: code/RunFixLeafCluster.sh
Untracked: code/RunNegMCMediation.err
Untracked: code/RunNegMCMediation.sh
Untracked: code/RunNegMCMediationr.out
Untracked: code/RunPosMCMediation.err
Untracked: code/RunPosMCMediation.sh
Untracked: code/RunPosMCMediationr.out
Untracked: code/SAF215upbed_gen.py
Untracked: code/Snakefile
Untracked: code/SnakefilePAS
Untracked: code/SnakefilePASfilt
Untracked: code/SortIndexBadSamples.err
Untracked: code/SortIndexBadSamples.out
Untracked: code/SortIndexBadSamples.sh
Untracked: code/TotalTranscriptDTplot.err
Untracked: code/TotalTranscriptDTplot.out
Untracked: code/Upstream10Bases_general.py
Untracked: code/apaQTLsnake.err
Untracked: code/apaQTLsnake.out
Untracked: code/apaQTLsnakePAS.err
Untracked: code/apaQTLsnakePAS.out
Untracked: code/apaQTLsnakePAShuman.err
Untracked: code/bam2junc.err
Untracked: code/bam2junc.out
Untracked: code/bam2junc_remove.err
Untracked: code/bam2junc_remove.out
Untracked: code/bed215upbed.py
Untracked: code/bed2SAF_gen.py
Untracked: code/bed2saf.py
Untracked: code/bg_to_cov.py
Untracked: code/buildIndecpantro5
Untracked: code/buildIndecpantro5.sh
Untracked: code/buildLeafviz.err
Untracked: code/buildLeafviz.out
Untracked: code/buildLeafviz.sh
Untracked: code/buildLeafviz_leadAnno.sh
Untracked: code/buildLeafviz_leafanno.err
Untracked: code/buildLeafviz_leafanno.out
Untracked: code/buildStarIndex.sh
Untracked: code/callPeaksYL.py
Untracked: code/chimpChromprder.sh
Untracked: code/chooseAnno2Bed.py
Untracked: code/chooseAnno2SAF.py
Untracked: code/chromOrder.err
Untracked: code/chromOrder.out
Untracked: code/classifyLeafviz.err
Untracked: code/classifyLeafviz.out
Untracked: code/cleanbed2saf.py
Untracked: code/cluster.json
Untracked: code/cluster2bed.py
Untracked: code/clusterLiftReverse.sh
Untracked: code/clusterLiftReverse_removebad.sh
Untracked: code/clusterLiftprimary.sh
Untracked: code/clusterLiftprimary_removebad.sh
Untracked: code/clusterPAS.json
Untracked: code/clusterfiltPAS.json
Untracked: code/comands2Mege.sh
Untracked: code/converBam2Junc.sh
Untracked: code/converBam2Junc_removeBad.sh
Untracked: code/convertNumeric.py
Untracked: code/environment.yaml
Untracked: code/extraSnakefiltpas
Untracked: code/filter5perc.R
Untracked: code/filter5percPAS.py
Untracked: code/filter5percPheno.py
Untracked: code/filterBamforMP.pysam2_gen.py
Untracked: code/filterJuncChroms.err
Untracked: code/filterJuncChroms.out
Untracked: code/filterMissprimingInNuc10_gen.py
Untracked: code/filterNumChroms.py
Untracked: code/filterPASforMP.py
Untracked: code/filterPostLift.py
Untracked: code/filterSAFforMP_gen.py
Untracked: code/filterSortBedbyCleanedBed_gen.R
Untracked: code/filterpeaks.py
Untracked: code/fixExonFC.py
Untracked: code/fixFChead.py
Untracked: code/fixFChead_bothfrac.py
Untracked: code/fixLeafCluster.py
Untracked: code/fixLiftedJunc.py
Untracked: code/fixUTRexonanno.py
Untracked: code/formathg38Anno.py
Untracked: code/generateStarIndex.err
Untracked: code/generateStarIndex.out
Untracked: code/generateStarIndexHuman.err
Untracked: code/generateStarIndexHuman.out
Untracked: code/getRNAseqMapStats.sh
Untracked: code/hg19MapStats.err
Untracked: code/hg19MapStats.out
Untracked: code/hg19MapStats.sh
Untracked: code/humanChromorder.sh
Untracked: code/humanFiles
Untracked: code/intersectAnno.err
Untracked: code/intersectAnno.out
Untracked: code/intersectAnnoExt.err
Untracked: code/intersectAnnoExt.out
Untracked: code/intersectLiftedPAS.sh
Untracked: code/leafcutter_merge_regtools_redo.py
Untracked: code/liftJunctionFiles.sh
Untracked: code/liftPAS19to38.sh
Untracked: code/liftoverPAShg19to38.err
Untracked: code/liftoverPAShg19to38.out
Untracked: code/log/
Untracked: code/make5percPeakbed.py
Untracked: code/makeFileID.py
Untracked: code/makeNuclearDapaplots.sh
Untracked: code/makeNuclearPlots.err
Untracked: code/makeNuclearPlots.out
Untracked: code/makePheno.py
Untracked: code/makeSamplyGroupsChimp_TvN.py
Untracked: code/makeSamplyGroupsHuman_TvN.py
Untracked: code/mapRNAseqhg19.sh
Untracked: code/mapRNAseqhg19_newPipeline.sh
Untracked: code/maphg19.err
Untracked: code/maphg19.out
Untracked: code/maphg19.sh
Untracked: code/maphg19_new.err
Untracked: code/maphg19_new.out
Untracked: code/maphg19_sub.err
Untracked: code/maphg19_sub.out
Untracked: code/maphg19_subjunc.sh
Untracked: code/mediation_test.R
Untracked: code/merge.err
Untracked: code/mergeChimp3prime_inhg38.sh
Untracked: code/merge_leafcutter_clusters_redo.py
Untracked: code/mergeandsort_ChimpinHuman.err
Untracked: code/mergeandsort_ChimpinHuman.out
Untracked: code/mergedBam2BW.sh
Untracked: code/mergedbam2bw.err
Untracked: code/mergedbam2bw.out
Untracked: code/nameClusters.py
Untracked: code/namePeaks.py
Untracked: code/negativeMediation_montecarlo.R
Untracked: code/nuclearTranscriptDTplot.err
Untracked: code/nuclearTranscriptDTplot.out
Untracked: code/numMultimap.py
Untracked: code/overlapPAS.err
Untracked: code/overlapPAS.out
Untracked: code/overlapapaQTLPAS.sh
Untracked: code/overlapapaQTLPAS_extended.sh
Untracked: code/peak2PAS.py
Untracked: code/pheno2countonly.R
Untracked: code/postiveMediation_montecarlo.R
Untracked: code/prepareAnnoLeafviz.err
Untracked: code/prepareAnnoLeafviz.out
Untracked: code/prepareCleanLiftedFC_5perc4LC.py
Untracked: code/prepareLeafvizAnno.sh
Untracked: code/preparePAS4lift.py
Untracked: code/prepare_phenotype_table.py
Untracked: code/primaryLift.err
Untracked: code/primaryLift.out
Untracked: code/primaryLift.sh
Untracked: code/processhg38exons.py
Untracked: code/quantJunc.sh
Untracked: code/quantJunc_TEST.sh
Untracked: code/quantJunc_removeBad.sh
Untracked: code/quantLiftedPAS.err
Untracked: code/quantLiftedPAS.out
Untracked: code/quantLiftedPAS.sh
Untracked: code/quatJunc.err
Untracked: code/quatJunc.out
Untracked: code/recChimpback2Human.err
Untracked: code/recChimpback2Human.out
Untracked: code/recLiftchim2human.sh
Untracked: code/revLift.err
Untracked: code/revLift.out
Untracked: code/revLiftPAShg38to19.sh
Untracked: code/reverseLift.sh
Untracked: code/runCheckReverseLift.sh
Untracked: code/runChimpDiffIso.sh
Untracked: code/runCountNucleotides.err
Untracked: code/runCountNucleotides.out
Untracked: code/runCountNucleotides.sh
Untracked: code/runCountNucleotidesPantro6.err
Untracked: code/runCountNucleotidesPantro6.out
Untracked: code/runCountNucleotides_pantro6.sh
Untracked: code/runFilterNumChroms.sh
Untracked: code/runHumanDiffIso.sh
Untracked: code/runNuclearDifffIso.sh
Untracked: code/runTotalDiffIso.sh
Untracked: code/run_Chimpleafcutter_ds.err
Untracked: code/run_Chimpleafcutter_ds.out
Untracked: code/run_Chimpverifybam.err
Untracked: code/run_Chimpverifybam.out
Untracked: code/run_Humanleafcutter_ds.err
Untracked: code/run_Humanleafcutter_ds.out
Untracked: code/run_Nuclearleafcutter_ds.err
Untracked: code/run_Nuclearleafcutter_ds.out
Untracked: code/run_Totalleafcutter_ds.err
Untracked: code/run_Totalleafcutter_ds.out
Untracked: code/run_chimpverifybam.sh
Untracked: code/run_verifyBam.sh
Untracked: code/run_verifybam.err
Untracked: code/run_verifybam.out
Untracked: code/slurm-62824013.out
Untracked: code/slurm-62825841.out
Untracked: code/slurm-62826116.out
Untracked: code/slurm-64108209.out
Untracked: code/slurm-64108521.out
Untracked: code/slurm-64108557.out
Untracked: code/snakePASChimp.err
Untracked: code/snakePASChimp.out
Untracked: code/snakePAShuman.out
Untracked: code/snakemake.batch
Untracked: code/snakemakeChimp.err
Untracked: code/snakemakeChimp.out
Untracked: code/snakemakeHuman.err
Untracked: code/snakemakeHuman.out
Untracked: code/snakemakePAS.batch
Untracked: code/snakemakePASFiltChimp.err
Untracked: code/snakemakePASFiltChimp.out
Untracked: code/snakemakePASFiltHuman.err
Untracked: code/snakemakePASFiltHuman.out
Untracked: code/snakemakePASchimp.batch
Untracked: code/snakemakePAShuman.batch
Untracked: code/snakemake_chimp.batch
Untracked: code/snakemake_human.batch
Untracked: code/snakemakefiltPAS.batch
Untracked: code/snakemakefiltPAS_chimp.sh
Untracked: code/snakemakefiltPAS_human.sh
Untracked: code/submit-snakemake-chimp.sh
Untracked: code/submit-snakemake-human.sh
Untracked: code/submit-snakemakePAS-chimp.sh
Untracked: code/submit-snakemakePAS-human.sh
Untracked: code/submit-snakemakefiltPAS-chimp.sh
Untracked: code/submit-snakemakefiltPAS-human.sh
Untracked: code/subset_diffisopheno.py
Untracked: code/subset_diffisopheno_Chimp_tvN.py
Untracked: code/subset_diffisopheno_Huma_tvN.py
Untracked: code/subset_diffisopheno_Nuclear_HvC.py
Untracked: code/subset_diffisopheno_Total_HvC.py
Untracked: code/test
Untracked: code/transcriptDTplotsNuclear.sh
Untracked: code/transcriptDTplotsTotal.sh
Untracked: code/verifyBam4973.sh
Untracked: code/verifyBam4973inHuman.sh
Untracked: code/verifybam4973.err
Untracked: code/verifybam4973.out
Untracked: code/verifybam4973HumanMap.err
Untracked: code/verifybam4973HumanMap.out
Untracked: code/wrap_Chimpverifybam.err
Untracked: code/wrap_Chimpverifybam.out
Untracked: code/wrap_chimpverifybam.sh
Untracked: code/wrap_verifyBam.sh
Untracked: code/wrap_verifybam.err
Untracked: code/wrap_verifybam.out
Untracked: code/writeMergecode.py
Untracked: data/._.DS_Store
Untracked: data/._HC_filenames.txt
Untracked: data/._HC_filenames.txt.sb-4426323c-IKIs0S
Untracked: data/._HC_filenames.xlsx
Untracked: data/._MapPantro6_meta.txt
Untracked: data/._MapPantro6_meta.txt.sb-a5794dd2-Cskmlm
Untracked: data/._MapPantro6_meta.xlsx
Untracked: data/._OppositeSpeciesMap.txt
Untracked: data/._OppositeSpeciesMap.txt.sb-a5794dd2-mayWJf
Untracked: data/._OppositeSpeciesMap.xlsx
Untracked: data/._RNASEQ_metadata.txt
Untracked: data/._RNASEQ_metadata.txt.sb-4426323c-TE4ns3
Untracked: data/._RNASEQ_metadata.txt.sb-51f67ae1-HXp7Gq
Untracked: data/._RNASEQ_metadata_2Removed.txt
Untracked: data/._RNASEQ_metadata_2Removed.txt.sb-4426323c-a4lBwx
Untracked: data/._RNASEQ_metadata_2Removed.xlsx
Untracked: data/._RNASEQ_metadata_stranded.txt
Untracked: data/._RNASEQ_metadata_stranded.txt.sb-a5794dd2-D659m2
Untracked: data/._RNASEQ_metadata_stranded.txt.sb-a5794dd2-ImNMoY
Untracked: data/._RNASEQ_metadata_stranded.txt.sb-e4bf31f0-ZGnGgl
Untracked: data/._RNASEQ_metadata_stranded.xlsx
Untracked: data/._metadata_HCpanel.txt
Untracked: data/._metadata_HCpanel.txt.sb-a3d92a2d-b9cYoF
Untracked: data/._metadata_HCpanel.txt.sb-a5794dd2-i594qs
Untracked: data/._metadata_HCpanel.txt.sb-f4823d1e-qihGek
Untracked: data/._metadata_HCpanel.xlsx
Untracked: data/._metadata_HCpanel_frompantro5.xlsx
Untracked: data/._~$RNASEQ_metadata.xlsx
Untracked: data/._~$metadata_HCpanel.xlsx
Untracked: data/._.xlsx
Untracked: data/CompapaQTLpas/
Untracked: data/DTmatrix/
Untracked: data/DiffExpression/
Untracked: data/DiffIso_Nuclear/
Untracked: data/DiffIso_Total/
Untracked: data/DiffSplice/
Untracked: data/DiffSplice_liftedJunc/
Untracked: data/DiffSplice_removeBad/
Untracked: data/DominantPAS/
Untracked: data/EvalPantro5/
Untracked: data/HC_filenames.txt
Untracked: data/HC_filenames.xlsx
Untracked: data/Khan_prot/
Untracked: data/Li_eqtls/
Untracked: data/MapPantro6_meta.txt
Untracked: data/MapPantro6_meta.xlsx
Untracked: data/MapStats/
Untracked: data/NormalizedClusters/
Untracked: data/NuclearHvC/
Untracked: data/OppositeSpeciesMap.txt
Untracked: data/OppositeSpeciesMap.xlsx
Untracked: data/PAS/
Untracked: data/Peaks_5perc/
Untracked: data/Pheno_5perc/
Untracked: data/Pheno_5perc_nuclear/
Untracked: data/Pheno_5perc_nuclear_old/
Untracked: data/Pheno_5perc_total/
Untracked: data/RNASEQ_metadata.txt
Untracked: data/RNASEQ_metadata_2Removed.txt
Untracked: data/RNASEQ_metadata_2Removed.xlsx
Untracked: data/RNASEQ_metadata_stranded.txt
Untracked: data/RNASEQ_metadata_stranded.txt.sb-e4bf31f0-ZGnGgl/
Untracked: data/RNASEQ_metadata_stranded.xlsx
Untracked: data/SignalSites/
Untracked: data/TotalHvC/
Untracked: data/TwoBadSampleAnalysis/
Untracked: data/Wang_ribo/
Untracked: data/chainFiles/
Untracked: data/cleanPeaks_anno/
Untracked: data/cleanPeaks_byspecies/
Untracked: data/cleanPeaks_lifted/
Untracked: data/files4viz_nuclear/
Untracked: data/leafviz/
Untracked: data/liftover_files/
Untracked: data/mediation/
Untracked: data/metadata_HCpanel.txt
Untracked: data/metadata_HCpanel.xlsx
Untracked: data/metadata_HCpanel_frompantro5.txt
Untracked: data/metadata_HCpanel_frompantro5.xlsx
Untracked: data/primaryLift/
Untracked: data/reverseLift/
Untracked: data/~$RNASEQ_metadata.xlsx
Untracked: data/~$metadata_HCpanel.xlsx
Untracked: data/.xlsx
Untracked: output/dtPlots/
Untracked: projectNotes.Rmd
Unstaged changes:
Modified: analysis/OppositeMap.Rmd
Modified: analysis/annotationInfo.Rmd
Modified: analysis/investigatePantro5.Rmd
Modified: analysis/multiMap.Rmd
Modified: analysis/speciesSpecific.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the R Markdown and HTML files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view them.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | a624a7e | brimittleman | 2020-01-16 | normalize splice and start mediation code |
| html | 5b4069c | brimittleman | 2020-01-15 | Build site. |
| Rmd | c7e1c1f | brimittleman | 2020-01-15 | add negative |
| html | 4f5d492 | brimittleman | 2020-01-14 | Build site. |
| Rmd | 3cce3da | brimittleman | 2020-01-14 | start mediation analysis |
library(workflowr)This is workflowr version 1.5.0
Run ?workflowr for help getting startedlibrary(tidyverse)── Attaching packages ──────────────────────────────────────────────────────────────────────────────────────────────────────────── tidyverse 1.2.1 ──✔ ggplot2 3.1.1 ✔ purrr 0.3.2
✔ tibble 2.1.1 ✔ dplyr 0.8.0.1
✔ tidyr 0.8.3 ✔ stringr 1.3.1
✔ readr 1.3.1 ✔ forcats 0.3.0 ── Conflicts ─────────────────────────────────────────────────────────────────────────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()library(limma)
library(MASS)
Attaching package: 'MASS'The following object is masked from 'package:dplyr':
selectIn this analysis, I will ask if differrecnes in APA or splicing are causal for differences in expression. I will use midiation analysis as implemented in (https://lauren-blake.github.io/Regulatory_Evol/analysis/Final_effect_size_human_chimp.html) using Joyces package mediation.
Notes:
need one value for gene, sample, expression values, mol pheno values (normalized and non normalized)
uses ashR,vashr,medinome package
df= sample -2 (4)
run for DE and non DE genes seperatly
Ittai’s version of the analysis: https://ittaieres.github.io/HiCiPSC/gene_expression.html#now,_get_the_appropriate_data_and_actually_run_the_mediation_analysis
For this version I do not need the package. I only need limma. I will try this first.
To deal with multiple phenotypes per gene I will take the highest absolute effect size cluster or PAS for splicing and APA respectively.
mkdir ../data/mediationsource("../code/mediation_test.R") #Obtain necessary functionsNow I need to pull in the expression, apa, and splicing data.
- expression= log2RPKM, and adju.P.val - normalized nuclear apa phenotype
- splicing normalized clusters
Expression and APA
#expression
nameID=read.table("../../genome_anotation_data/ensemble_to_genename.txt",sep="\t", header = T, stringsAsFactors = F) %>% dplyr::select(Gene_stable_ID,Gene.name)
ExpRes=read.table("../data/DiffExpression/DEtested_allres.txt", header = F, stringsAsFactors = F, col.names = c("Gene_stable_ID", "logFC", "AveExpr", "t", "P.Value", "adj.P.Val", "B")) %>% inner_join(nameID,by="Gene_stable_ID") %>% dplyr::select(Gene.name,adj.P.Val )
ExpNorm=read.table("../data/DiffExpression/NormalizedExpressionPassCutoff.txt", header = T, stringsAsFactors = F) %>% inner_join(nameID,by="Gene_stable_ID") %>% dplyr::select(-Gene_stable_ID)
#join indiv with adjust pval
Exp=ExpRes %>% inner_join(ExpNorm, by="Gene.name")%>% rename("gene"= Gene.name) %>% dplyr::select(gene, adj.P.Val, NA18498, NA18499, NA18502, NA18504, NA18510,NA18523,NA18358, NA3622,NA3659,NA4973,NAPT30,NAPT91)
#Chr start end ID NA18498_N NA18499_N NA18502_N NA18504_N NA18510_N NA18523_N NA18358_N NA3622_N NA3659_N NA4973_N NApt30_N NApt91_N
#apa:
apaN= read.table("../data/Pheno_5perc_nuclear/ALLPAS_postLift_LocParsed_bothSpecies_pheno_5perc_Nuclear.txt.gz.phen_AllChrom", col.names = c("chr", "start", "end", "id","NA18498_APA", "NA18499_APA", "NA18502_APA", "NA18504_APA", "NA18510_APA","NA18523_APA", "NA18358_APA", "NA3622_APA","NA3659_APA", "NA4973_APA","NAPT30_APA","NAPT91_APA")) %>% separate(id, into=c("ch", "st", "en","id2"),sep=":") %>% separate(id2, into=c("gene", "strand","id3"),sep="_") %>% separate(id3, into=c("loc", "disc", "PAS"), sep="-") %>% dplyr::select(gene, PAS,contains("NA"))
PASMeta=read.table("../data/PAS/PAS_5perc_either_HumanCoord_BothUsage_meta.txt", header = T, stringsAsFactors = F) %>% dplyr::select(PAS, chr, start,end, gene)
apaRes= read.table("../data/DiffIso_Nuclear/TN_diff_isoform_allChrom.txt_significance.txt",sep="\t" ,col.names = c('status','loglr','df','p','cluster','p.adjust'),stringsAsFactors = F) %>% filter(status=="Success") %>% separate(cluster, into=c("chr","gene"),sep=":")
apaPASres=read.table("../data/DiffIso_Nuclear/TN_diff_isoform_allChrom.txt_effect_sizes.txt", stringsAsFactors = F, col.names=c('intron', 'logef' ,'Human', 'Chimp','deltaPAU')) %>% filter(intron != "intron") %>% separate(intron, into=c("chr","start", "end","gene"), sep=":")
apaPASres$start=as.integer(apaPASres$start)
apaPASres$end=as.integer(apaPASres$end)
apaPASres$deltaPAU=as.numeric(apaPASres$deltaPAU)
apaPASres=apaPASres%>% inner_join(PASMeta,by=c("chr", "start", "end", "gene"))
#problem if there are 2 pas then the are opposite but same value - do with all one direction for
apaPASres_topPos= apaPASres %>% group_by(gene) %>% top_n(1,abs(deltaPAU)) %>% top_n(1,deltaPAU)
apaPASres_topNeg= apaPASres %>% group_by(gene) %>% top_n(1,abs(deltaPAU)) %>% top_n(-1,deltaPAU)
#join with pvalue:
apaPASres_topPos_pval=apaPASres_topPos %>% inner_join(apaRes, by=c("gene")) %>% dplyr::select( PAS, p.adjust) Adding missing grouping variables: `gene`apaPASres_topNeg_pval=apaPASres_topNeg %>% inner_join(apaRes, by=c("gene")) %>% dplyr::select( PAS, p.adjust)Adding missing grouping variables: `gene`#looks like the naming convention is not consistent...
apaPASres_topPos_pval_normAPA=apaPASres_topPos_pval %>% inner_join(apaN,by=c("PAS","gene")) %>% dplyr::select(-PAS,-p.adjust)
apaPASres_topNeg_pval_normAPA=apaPASres_topNeg_pval %>% inner_join(apaN,by=c("PAS","gene"))%>% dplyr::select(-PAS,-p.adjust)
#create a dataframe with expression and apa
APA_posandExp=Exp %>% inner_join(apaPASres_topPos_pval_normAPA, by="gene")
APA_negandExp=Exp %>% inner_join(apaPASres_topNeg_pval_normAPA,by="gene")First do the positive:
Seperate De and not De
is_de <- which(APA_posandExp$adj.P.Val < .05)
isnot_de <- which(APA_posandExp$adj.P.Val >= .05)
gvec <- APA_posandExp$genes
Exp_pos <- APA_posandExp %>% dplyr::select(-gene,-adj.P.Val, -contains("APA"))
APA_pos <- APA_posandExp %>% dplyr::select(contains("APA"))
# metadata label
species <- factor(c("H","H","H","H","H","H","C","C","C","C","C","C"))
batch <- factor(c("A", "A", "B", "A", "B", "B", "B", "B","A", "A","A","B"))
metadata <- data.frame(sample=names(Exp_pos),
species=species,
batch=batch)Compute indirect effects:
fit_de_pos <- test_mediation(exprs = Exp_pos[is_de,],
fixed_covariates = list(species=metadata$species,
batch=metadata$batch),
varying_covariate = APA_pos[is_de,])
fit_node_pos <- test_mediation(exprs = Exp_pos[isnot_de,],
fixed_covariates = list(species=metadata$species,
batch=metadata$batch),
varying_covariate = APA_pos[isnot_de,])
#save as R data object:
save(fit_de_pos, fit_node_pos, is_de, isnot_de, Exp_pos, APA_pos, metadata, gvec, file = "../data/mediation/positive_mediation.rda")Montecarlo:
I will have to run this seperatly as an Rscript.
sbatch RunPosMCMediation.sh
Load results
mc_pos_de <- readRDS(file = "../data/mediation/mc_de_postive.rds")
mc_pos_node <- readRDS(file = "../data/mediation/mc_node_postive.rds")
##In DE genes
ngenes <- ncol(mc_pos_de)
ab <- fit_de_pos$alpha*fit_de_pos$beta
out <- sapply(1:ngenes, function(g) {
x <- unlist(mc_pos_de[,g])
q <- quantile(x, prob = c(.025, .975))
# q <- quantile(x, prob = c(.005, .995))
ifelse(0 > q[1] & 0 < q[2], F, T)
})
table(out)out
FALSE TRUE
2823 85 85/(85+2823) #[1] 0.02922971DEdat <- data.frame(bf=fit_de_pos$tau, af=fit_de_pos$tau_prime,significance=out)
DEdat$color <- ifelse(DEdat$significance==TRUE, "red", "black")
plot(x=DEdat$bf, y=DEdat$af, ylab="Effect Size After Controlling for APA", xlab="Effect Size Before Controlling for APA", main="Effect of APA on Expression Divergence in DE genes", pch=16, cex=0.6, xlim=c(-10, 10), ylim=c(-10,10),col=alpha(DEdat$color, 0.6))
legend("topleft", legend=c("95% CI Significant (n=85)", "95% CI Non significant (n=2823)"), col=c("red", "black"), pch=16:16, cex=0.8)
abline(0, 1)
abline(h=0)
abline(v=0)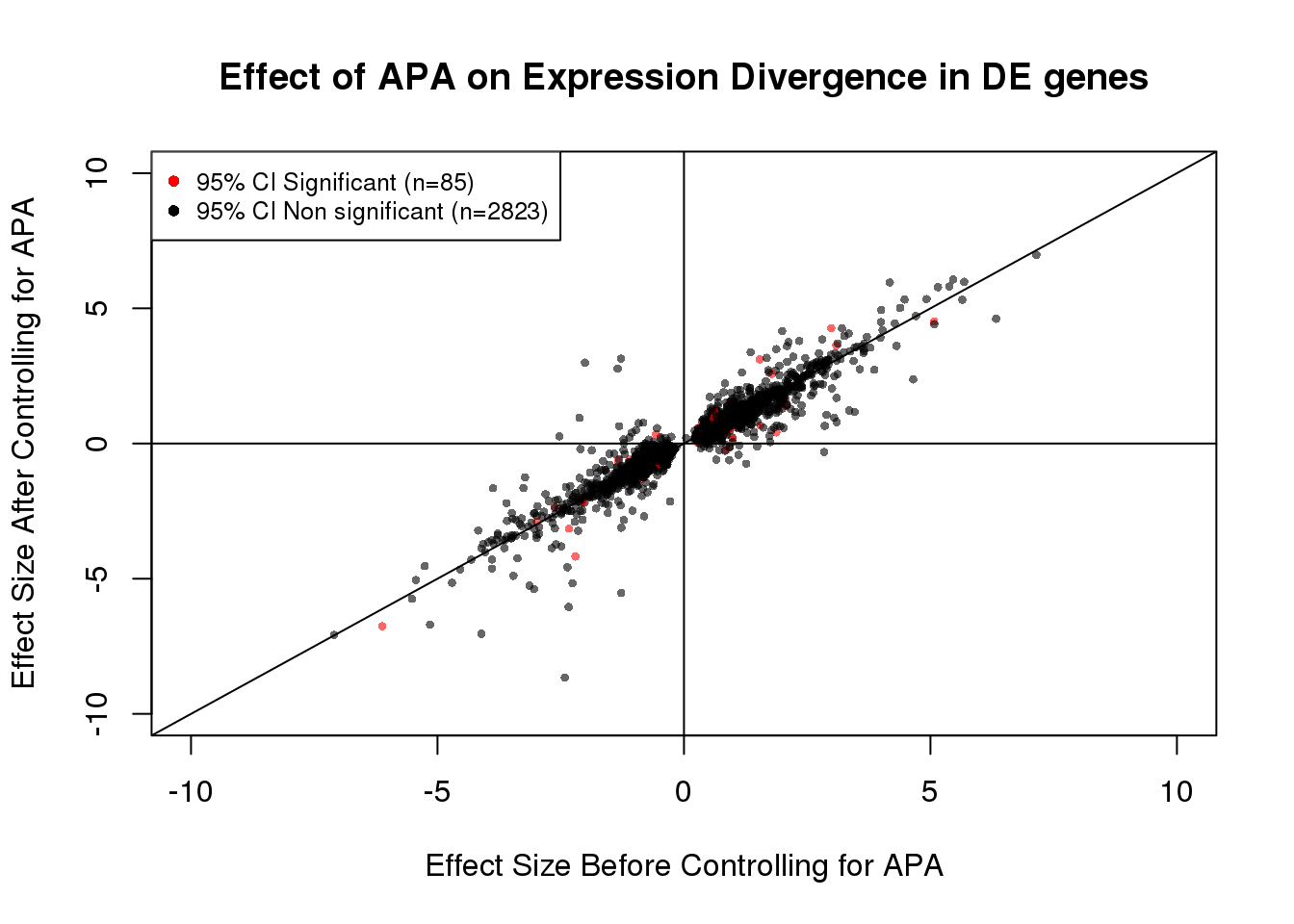
| Version | Author | Date |
|---|---|---|
| 5b4069c | brimittleman | 2020-01-15 |
Not DE:
##In non-DE genes
ngenes <- ncol(mc_pos_node)
ab <- fit_node_pos$alpha*fit_node_pos$beta
out <- sapply(1:ngenes, function(g) {
x <- unlist(mc_pos_node[,g])
q <- quantile(x, prob = c(.025, .975))
# q <- quantile(x, prob = c(.005, .995))
ifelse(0 > q[1] & 0 < q[2], F, T)
})
table(out)out
FALSE TRUE
4852 110 110/(110+4852)[1] 0.02216848#Visualization for non DE genes:
noDEdat <- data.frame(bf=fit_node_pos$tau, af=fit_node_pos$tau_prime,significance=out)
noDEdat$color <- ifelse(noDEdat$significance==TRUE, "red", "black")
plot(x=noDEdat$bf, y=noDEdat$af, ylab="Effect Size After Controlling for APA", xlab="Effect Size Before Controlling for APA", main="Effect of APA on Expression Divergence in non-DE genes", pch=16, cex=0.6, xlim=c(-10, 10), ylim=c(-10,10), adj=0.6,col=alpha(noDEdat$color, 0.6))
legend("topleft", legend=c("95% CI Significant (n=110)", "95% CI Non significant (n=4852)"), col=c("red", "black"), pch=16:16, cex=0.8)
abline(0, 1)
abline(h=0)
abline(v=0)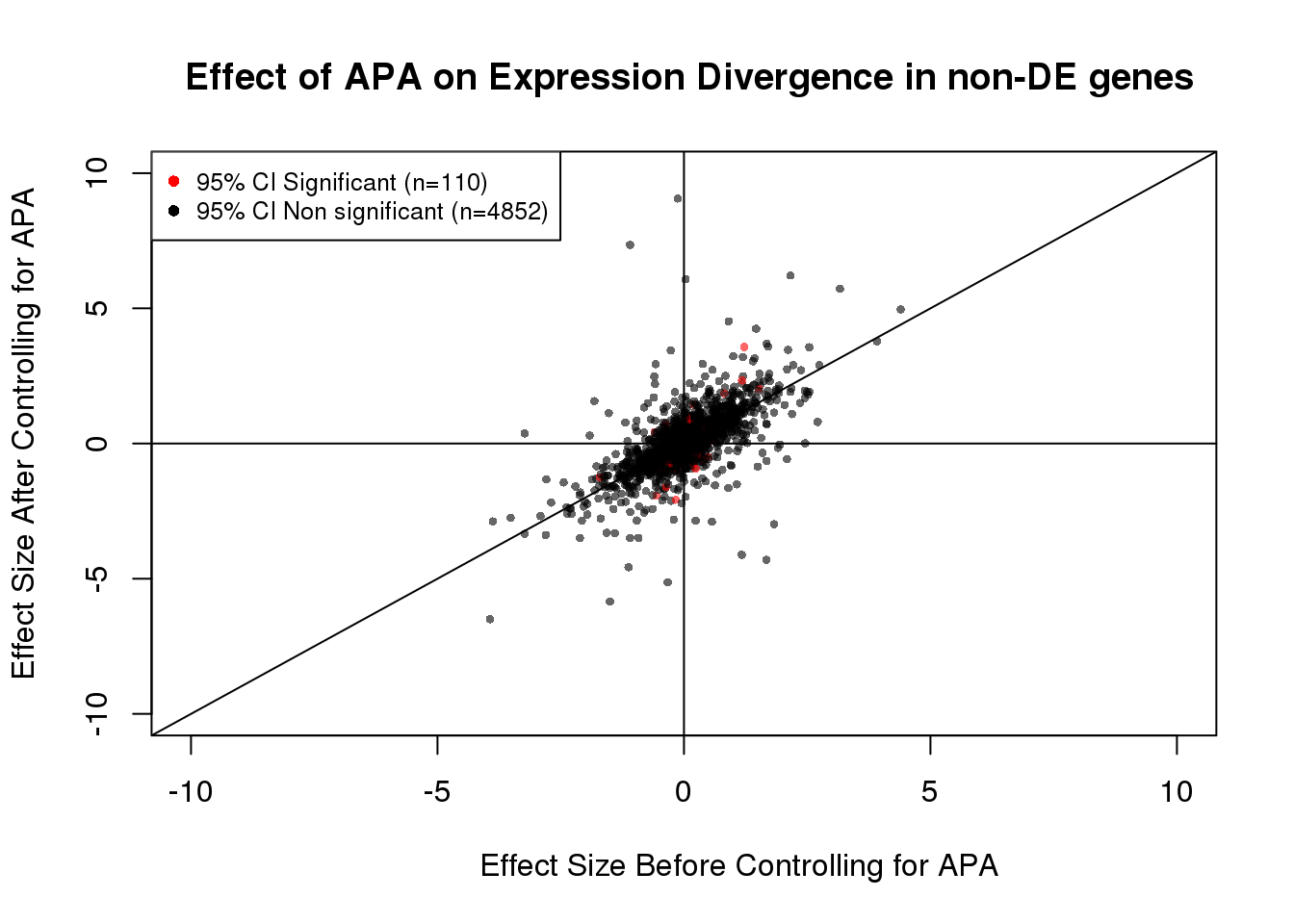
Negative:
Seperate De and not De
is_de_neg <- which(APA_negandExp$adj.P.Val < .05)
isnot_de_gen <- which(APA_negandExp$adj.P.Val >= .05)
gvec_neg <- APA_negandExp$genes
Exp_neg <- APA_negandExp %>% dplyr::select(-gene,-adj.P.Val, -contains("APA"))
APA_neg <- APA_negandExp %>% dplyr::select(contains("APA"))Compute indirect effects:
fit_de_neg<- test_mediation(exprs = Exp_neg[is_de_neg,],
fixed_covariates = list(species=metadata$species,
batch=metadata$batch),
varying_covariate = APA_neg[is_de_neg,])
fit_node_neg <- test_mediation(exprs = Exp_neg[isnot_de_gen,],
fixed_covariates = list(species=metadata$species,
batch=metadata$batch),
varying_covariate = APA_neg[isnot_de_gen,])
#save as R data object:
save(fit_de_neg, fit_node_neg, is_de_neg, isnot_de_gen, Exp_neg, APA_neg, metadata, gvec_neg, file = "../data/mediation/negative_mediation.rda")Montecarlo:
I will have to run this seperatly as an Rscript.
sbatch RunNegMCMediation.sh
Pull in results.
mc_neg_de <- readRDS(file = "../data/mediation/mc_de_negative.rds")
mc_neg_node <- readRDS(file = "../data/mediation/mc_node_negative.rds")
##In DE genes
ngenes <- ncol(mc_neg_de)
ab <- fit_de_neg$alpha*fit_de_neg$beta
out <- sapply(1:ngenes, function(g) {
x <- unlist(mc_neg_de[,g])
q <- quantile(x, prob = c(.025, .975))
# q <- quantile(x, prob = c(.005, .995))
ifelse(0 > q[1] & 0 < q[2], F, T)
})
table(out)out
FALSE TRUE
2815 93 93/(93+2823) #[1] 0.031893DEdat_neg <- data.frame(bf=fit_de_neg$tau, af=fit_de_neg$tau_prime,significance=out)
DEdat_neg$color <- ifelse(DEdat_neg$significance==TRUE, "red", "black")
plot(x=DEdat_neg$bf, y=DEdat_neg$af, ylab="Effect Size After Controlling for APA", xlab="Effect Size Before Controlling for APA", main="Effect of APA on Expression Divergence in DE genes (Negative)", pch=16, cex=0.6, xlim=c(-10, 10), ylim=c(-10,10),col=alpha(DEdat$color, 0.6))
legend("topleft", legend=c("95% CI Significant (n=93)", "95% CI Non significant (n=2823)"), col=c("red", "black"), pch=16:16, cex=0.8)
abline(0, 1)
abline(h=0)
abline(v=0)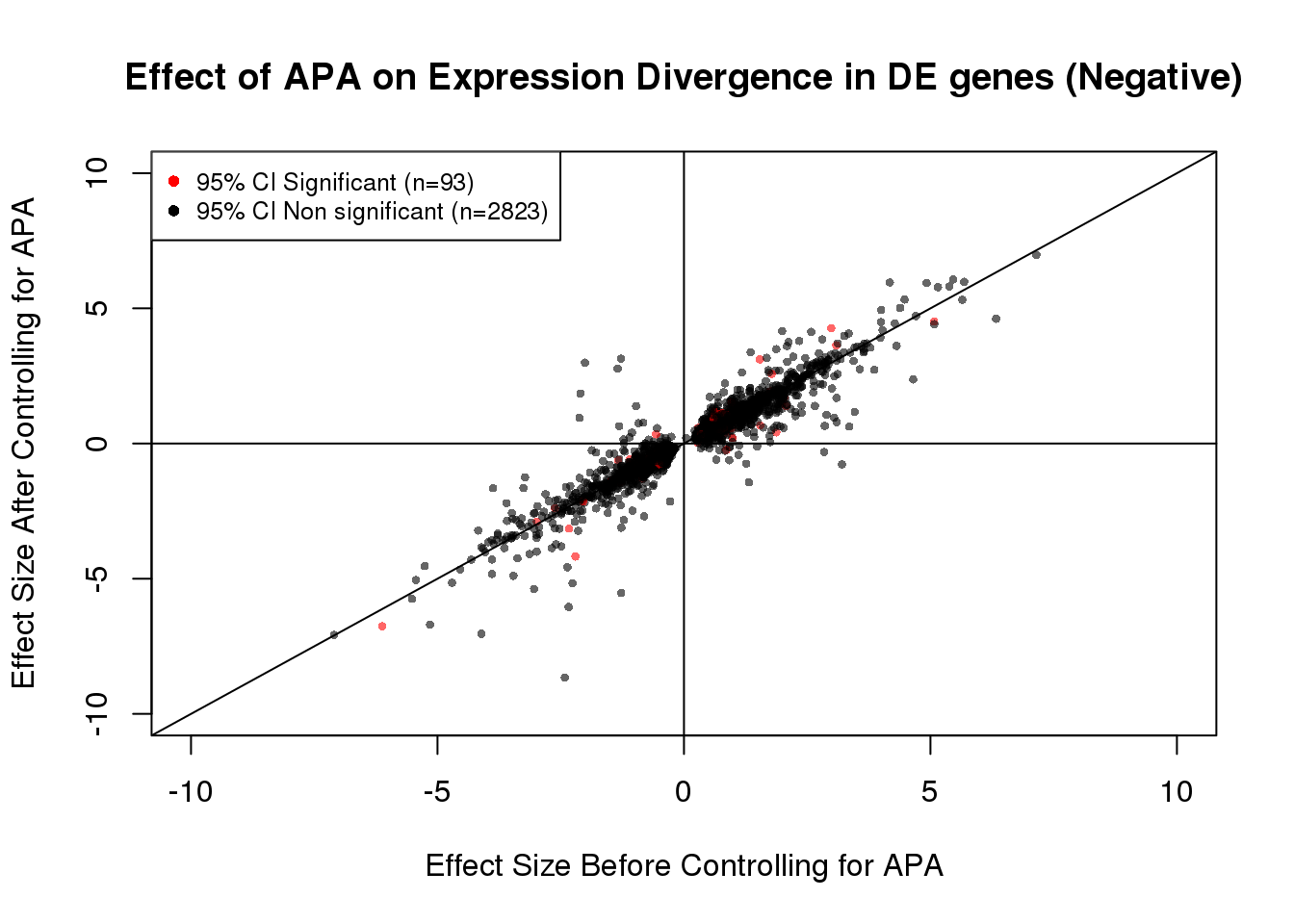
| Version | Author | Date |
|---|---|---|
| 5b4069c | brimittleman | 2020-01-15 |
Not DE:
##In not DE genes
ngenes <- ncol(mc_neg_node)
ab <- fit_node_neg$alpha*fit_node_neg$beta
out <- sapply(1:ngenes, function(g) {
x <- unlist(mc_neg_node[,g])
q <- quantile(x, prob = c(.025, .975))
# q <- quantile(x, prob = c(.005, .995))
ifelse(0 > q[1] & 0 < q[2], F, T)
})
table(out)out
FALSE TRUE
4859 102 102/(102+4859) #[1] 0.02056037#Visualization for non DE genes:
noDEdat_neg <- data.frame(bf=fit_node_neg$tau, af=fit_node_neg$tau_prime,significance=out)
noDEdat$color <- ifelse(noDEdat$significance==TRUE, "red", "black")
plot(x=noDEdat_neg$bf, y=noDEdat_neg$af, ylab="Effect Size After Controlling for APA", xlab="Effect Size Before Controlling for APA", main="Effect of APA on Expression Divergence in non-DE genes (negative)", pch=16, cex=0.6, xlim=c(-10, 10), ylim=c(-10,10), adj=0.6,col=alpha(noDEdat_neg$color, 0.6))
legend("topleft", legend=c("95% CI Significant (n=102)", "95% CI Non significant (n=4859)"), col=c("red", "black"), pch=16:16, cex=0.8)
abline(0, 1)
abline(h=0)
abline(v=0)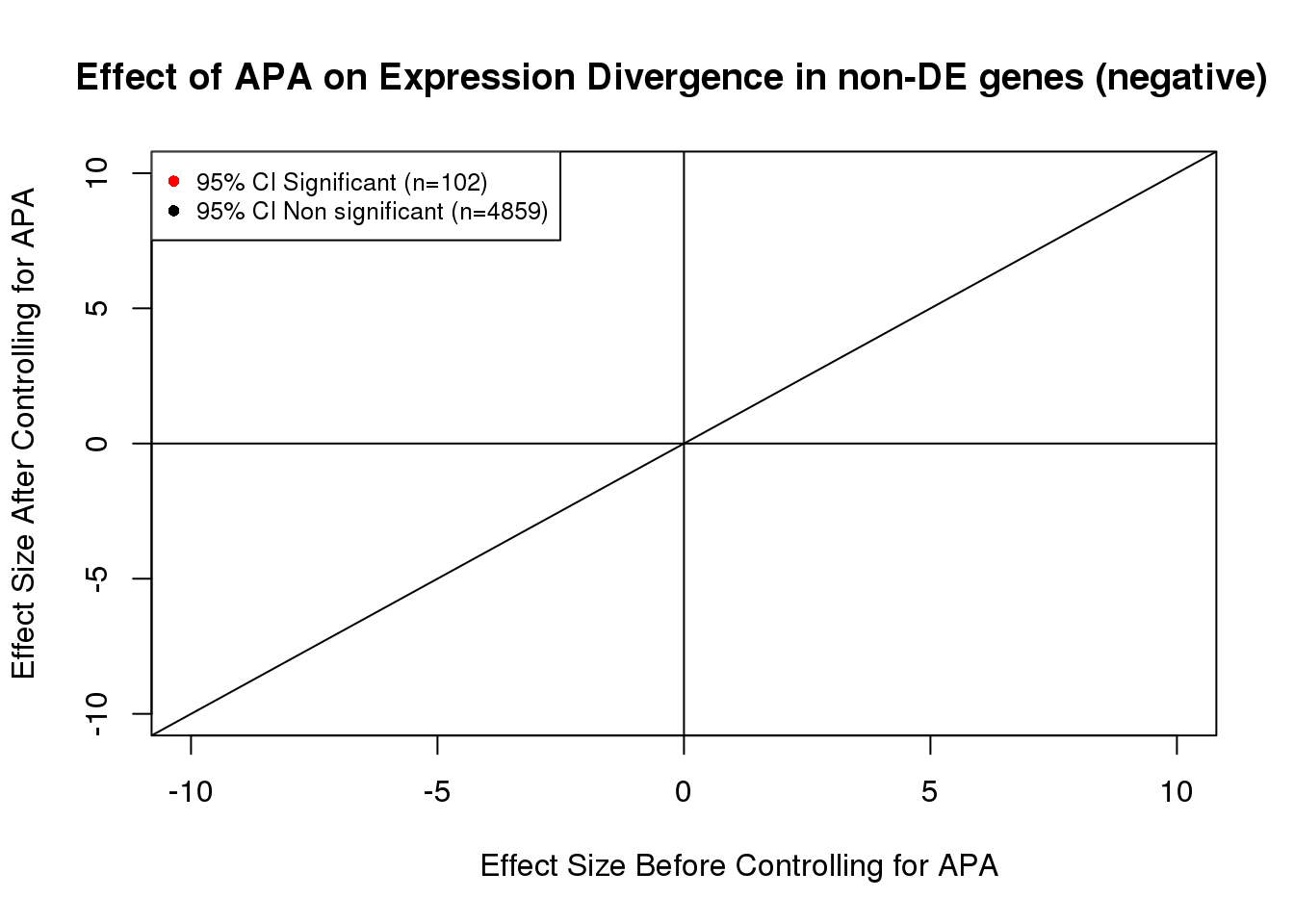
| Version | Author | Date |
|---|---|---|
| 5b4069c | brimittleman | 2020-01-15 |
Expression and Splicing
#YG-BM-S3-3622C-Total_S3_R1_001-sort.bam YG-BM-FIXED-18498H-Total_S5_R1_001-sort.bam YG-BM-S12-18523H-Total_S12_R1_001-sort.baYG-BM-S4-3659C-Total_S4_R1_001-sort.bam YG-BM-S2-PT91C-Total_S2_R1_001-sort.bam YG-BM-S11-18510H-Total_S11_R1_001-sort.bam YG-BM-S9-18502H-Total_S9_R1_001-sort.bam YG-BM-S8-18499H-Total_S8_R1_001-sort.bam YG-BM-S1-PT30C-Total_S1_R1_001-sort.bam YG-BM-FIXED-4973C-Total_S7_R1_001-sort.bam YG-BM-S10-18504H-Total_S10_R1_001-sort.bam YG-BM-S6-18358C-Total_S6_R1_001-sort.bam
Splicing= read.table("../data/NormalizedClusters/MergeCombined_perind.counts.fixed.gz.qqnorm.allchr", col.names = c("chr", "start", "end", "id","NA3622_Splice", "NA18498_Splice", "NA18523_Splice", "NA3659_Splice", "NAPT91_Splice","NA18510_Splice", "NA18502_Splice", "NA18499_Splice","NAPT30_Splice", "NA4973_Splice","NA18504_Splice","NA18358_Splice")) %>% separate(id, into=c("ch", "st", "en","clus"),sep=":")
Sp_res=read.table("../data/DiffSplice_liftedJunc/MergedRes_cluster_significance.txt",stringsAsFactors = F, header = T, sep="\t") %>% separate(cluster, into=c("chrom", "clus"),sep=":") %>% filter(status=="Success") %>% dplyr::select(p.adjust,clus, genes,loglr)
#%>% dplyr::select(genes,clus, p.adjust)
Splicing_andres=Splicing %>% inner_join(Sp_res, by=c("clus") )
#%>% dplyr::select(genes, p.adjust, NA18498_Splice, NA18499_Splice, NA18502_Splice, NA18504_Splice, NA18510_Splice,NA18523_Splice,NA18358_Splice, NA3622_Splice,NA3659_Splice,NA4973_Splice,NAPT30_Splice,NAPT91_Splice) %>% rename("gene"=genes)
#create a dataframe with expression and apa
#SpandExp=Exp %>% inner_join(Splicing_andres, by="gene")Current problem: genes to cluster mapping is not trivial. I do not know how to chose the top DS isoform per gene.
sessionInfo()R version 3.5.1 (2018-07-02)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] MASS_7.3-51.1 limma_3.38.2 forcats_0.3.0 stringr_1.3.1
[5] dplyr_0.8.0.1 purrr_0.3.2 readr_1.3.1 tidyr_0.8.3
[9] tibble_2.1.1 ggplot2_3.1.1 tidyverse_1.2.1 workflowr_1.5.0
loaded via a namespace (and not attached):
[1] tidyselect_0.2.5 haven_1.1.2 lattice_0.20-38 colorspace_1.3-2
[5] generics_0.0.2 htmltools_0.3.6 yaml_2.2.0 rlang_0.4.0
[9] later_0.7.5 pillar_1.3.1 glue_1.3.0 withr_2.1.2
[13] modelr_0.1.2 readxl_1.1.0 plyr_1.8.4 munsell_0.5.0
[17] gtable_0.2.0 cellranger_1.1.0 rvest_0.3.2 evaluate_0.12
[21] knitr_1.20 httpuv_1.4.5 broom_0.5.1 Rcpp_1.0.2
[25] promises_1.0.1 scales_1.0.0 backports_1.1.2 jsonlite_1.6
[29] fs_1.3.1 hms_0.4.2 digest_0.6.18 stringi_1.2.4
[33] grid_3.5.1 rprojroot_1.3-2 cli_1.1.0 tools_3.5.1
[37] magrittr_1.5 lazyeval_0.2.1 crayon_1.3.4 whisker_0.3-2
[41] pkgconfig_2.0.2 xml2_1.2.0 lubridate_1.7.4 assertthat_0.2.0
[45] rmarkdown_1.10 httr_1.3.1 rstudioapi_0.10 R6_2.3.0
[49] nlme_3.1-137 git2r_0.26.1 compiler_3.5.1