Are Most used PAS dAPA
Briana Mittleman
3/11/2020
Last updated: 2020-03-18
Checks: 7 0
Knit directory: Comparative_APA/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190902) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: code/chimp_log/
Ignored: code/human_log/
Ignored: data/.DS_Store
Ignored: data/TrialFiltersMeta.txt.sb-9845453e-R58Y0Q/
Ignored: data/mediation_prot/
Ignored: data/metadata_HCpanel.txt.sb-a5794dd2-i594qs/
Ignored: output/.DS_Store
Untracked files:
Untracked: ._.DS_Store
Untracked: Chimp/
Untracked: Human/
Untracked: analysis/CrossChimpThreePrime.Rmd
Untracked: analysis/DiffTransProtvsExpression.Rmd
Untracked: analysis/DiffUsedUTR.Rmd
Untracked: analysis/GvizPlots.Rmd
Untracked: analysis/HandC.TvN
Untracked: analysis/PhenotypeOverlap10.Rmd
Untracked: analysis/annotationBias.Rmd
Untracked: analysis/assessReadQual.Rmd
Untracked: analysis/diffExpressionPantro6.Rmd
Untracked: analysis/orthoexonAnno.Rmd
Untracked: analysis/pol2.Rmd
Untracked: code/._ClassifyLeafviz.sh
Untracked: code/._Config_chimp.yaml
Untracked: code/._Config_chimp_full.yaml
Untracked: code/._Config_human.yaml
Untracked: code/._ConvertJunc2Bed.sh
Untracked: code/._CountNucleotides.py
Untracked: code/._CrossMapChimpRNA.sh
Untracked: code/._CrossMapThreeprime.sh
Untracked: code/._DiffSplice.sh
Untracked: code/._DiffSplicePlots.sh
Untracked: code/._DiffSplicePlots_gencode.sh
Untracked: code/._DiffSplice_gencode.sh
Untracked: code/._DiffSplice_removebad.sh
Untracked: code/._FindIntronForDomPAS.sh
Untracked: code/._FindIntronForDomPAS_DF.sh
Untracked: code/._GetMAPQscore.py
Untracked: code/._GetSecondaryMap.py
Untracked: code/._Lift5perPAS.sh
Untracked: code/._LiftFinalChimpJunc2Human.sh
Untracked: code/._LiftOrthoPAS2chimp.sh
Untracked: code/._MapBadSamples.sh
Untracked: code/._PAS_ATTAAA.sh
Untracked: code/._PAS_ATTAAA_df.sh
Untracked: code/._PAS_seqExpanded.sh
Untracked: code/._PASsequences.sh
Untracked: code/._PASsequences_DF.sh
Untracked: code/._PlotNuclearUsagebySpecies.R
Untracked: code/._PlotNuclearUsagebySpecies_DF.R
Untracked: code/._QuantMergedClusters.sh
Untracked: code/._RNATranscriptDTplot.sh
Untracked: code/._ReverseLiftFilter.R
Untracked: code/._RunFixLeafCluster.sh
Untracked: code/._RunNegMCMediation.sh
Untracked: code/._RunNegMCMediationDF.sh
Untracked: code/._RunPosMCMediationDF.err
Untracked: code/._RunPosMCMediationDF.sh
Untracked: code/._SAF2Bed.py
Untracked: code/._Snakefile
Untracked: code/._SnakefilePAS
Untracked: code/._SnakefilePASfilt
Untracked: code/._SortIndexBadSamples.sh
Untracked: code/._assignPeak2Intronicregion
Untracked: code/._assignPeak2Intronicregion.sh
Untracked: code/._bed215upbed.py
Untracked: code/._bed2SAF_gen.py
Untracked: code/._buildIndecpantro5
Untracked: code/._buildIndecpantro5.sh
Untracked: code/._buildLeafviz.sh
Untracked: code/._buildLeafviz_leadAnno.sh
Untracked: code/._buildStarIndex.sh
Untracked: code/._chimpChromprder.sh
Untracked: code/._chooseSignalSite.py
Untracked: code/._cleanbed2saf.py
Untracked: code/._cluster.json
Untracked: code/._cluster2bed.py
Untracked: code/._clusterLiftReverse.sh
Untracked: code/._clusterLiftReverse_removebad.sh
Untracked: code/._clusterLiftprimary.sh
Untracked: code/._clusterLiftprimary_removebad.sh
Untracked: code/._converBam2Junc.sh
Untracked: code/._converBam2Junc_removeBad.sh
Untracked: code/._extraSnakefiltpas
Untracked: code/._extractPhyloReg.py
Untracked: code/._extractPhyloRegGene.py
Untracked: code/._extractPhylopGeneral.ph
Untracked: code/._extractPhylopGeneral.py
Untracked: code/._extractPhylopReg200down.py
Untracked: code/._extractPhylopReg200up.py
Untracked: code/._filter5percPAS.py
Untracked: code/._filterNumChroms.py
Untracked: code/._filterPASforMP.py
Untracked: code/._filterPostLift.py
Untracked: code/._fixExonFC.py
Untracked: code/._fixLeafCluster.py
Untracked: code/._fixLiftedJunc.py
Untracked: code/._fixUTRexonanno.py
Untracked: code/._formathg38Anno.py
Untracked: code/._formatpantro6Anno.py
Untracked: code/._getRNAseqMapStats.sh
Untracked: code/._hg19MapStats.sh
Untracked: code/._humanChromorder.sh
Untracked: code/._intersectLiftedPAS.sh
Untracked: code/._liftJunctionFiles.sh
Untracked: code/._liftPAS19to38.sh
Untracked: code/._liftedchimpJunc2human.sh
Untracked: code/._makeNuclearDapaplots.sh
Untracked: code/._makeNuclearDapaplots_DF.sh
Untracked: code/._makeSamplyGroupsHuman_TvN.py
Untracked: code/._mapRNAseqhg19.sh
Untracked: code/._mapRNAseqhg19_newPipeline.sh
Untracked: code/._maphg19.sh
Untracked: code/._maphg19_subjunc.sh
Untracked: code/._mediation_test.R
Untracked: code/._mergeChimp3prime_inhg38.sh
Untracked: code/._mergeandBWRNAseq.sh
Untracked: code/._mergedBam2BW.sh
Untracked: code/._nameClusters.py
Untracked: code/._negativeMediation_montecarlo.R
Untracked: code/._negativeMediation_montecarloDF.R
Untracked: code/._numMultimap.py
Untracked: code/._overlapapaQTLPAS.sh
Untracked: code/._parseHg38.py
Untracked: code/._postiveMediation_montecarlo_DF.R
Untracked: code/._prepareCleanLiftedFC_5perc4LC.py
Untracked: code/._prepareLeafvizAnno.sh
Untracked: code/._preparePAS4lift.py
Untracked: code/._primaryLift.sh
Untracked: code/._processhg38exons.py
Untracked: code/._quantJunc.sh
Untracked: code/._quantJunc_TEST.sh
Untracked: code/._quantJunc_removeBad.sh
Untracked: code/._quantMerged_seperatly.sh
Untracked: code/._recLiftchim2human.sh
Untracked: code/._revLiftPAShg38to19.sh
Untracked: code/._reverseLift.sh
Untracked: code/._runCheckReverseLift.sh
Untracked: code/._runChimpDiffIso.sh
Untracked: code/._runCountNucleotides.sh
Untracked: code/._runFilterNumChroms.sh
Untracked: code/._runHumanDiffIso.sh
Untracked: code/._runNuclearDiffIso_DF.sh
Untracked: code/._runNuclearDifffIso.sh
Untracked: code/._runTotalDiffIso.sh
Untracked: code/._run_chimpverifybam.sh
Untracked: code/._run_verifyBam.sh
Untracked: code/._snakemake.batch
Untracked: code/._snakemakePAS.batch
Untracked: code/._snakemakePASchimp.batch
Untracked: code/._snakemakePAShuman.batch
Untracked: code/._snakemake_chimp.batch
Untracked: code/._snakemake_human.batch
Untracked: code/._snakemakefiltPAS.batch
Untracked: code/._snakemakefiltPAS_chimp
Untracked: code/._snakemakefiltPAS_chimp.sh
Untracked: code/._snakemakefiltPAS_human.sh
Untracked: code/._spliceSite2Fasta.py
Untracked: code/._submit-snakemake-chimp.sh
Untracked: code/._submit-snakemake-human.sh
Untracked: code/._submit-snakemakePAS-chimp.sh
Untracked: code/._submit-snakemakePAS-human.sh
Untracked: code/._submit-snakemakefiltPAS-chimp.sh
Untracked: code/._submit-snakemakefiltPAS-human.sh
Untracked: code/._subset_diffisopheno_Nuclear_HvC.py
Untracked: code/._subset_diffisopheno_Nuclear_HvC_DF.py
Untracked: code/._subset_diffisopheno_Total_HvC.py
Untracked: code/._threeprimeOrthoFC.sh
Untracked: code/._transcriptDTplotsNuclear.sh
Untracked: code/._verifyBam4973.sh
Untracked: code/._verifyBam4973inHuman.sh
Untracked: code/._wrap_chimpverifybam.sh
Untracked: code/._wrap_verifyBam.sh
Untracked: code/._writeMergecode.py
Untracked: code/.snakemake/
Untracked: code/ClassifyLeafviz.sh
Untracked: code/Config_chimp.yaml
Untracked: code/Config_chimp_full.yaml
Untracked: code/Config_human.yaml
Untracked: code/ConvertJunc2Bed.err
Untracked: code/ConvertJunc2Bed.out
Untracked: code/ConvertJunc2Bed.sh
Untracked: code/CountNucleotides.py
Untracked: code/CrossMapChimpRNA.sh
Untracked: code/CrossMapThreeprime.sh
Untracked: code/CrossmapChimp3prime.err
Untracked: code/CrossmapChimp3prime.out
Untracked: code/CrossmapChimpRNA.err
Untracked: code/CrossmapChimpRNA.out
Untracked: code/DiffSplice.err
Untracked: code/DiffSplice.out
Untracked: code/DiffSplice.sh
Untracked: code/DiffSplicePlots.err
Untracked: code/DiffSplicePlots.out
Untracked: code/DiffSplicePlots.sh
Untracked: code/DiffSplicePlots_gencode.sh
Untracked: code/DiffSplice_gencode.sh
Untracked: code/DiffSplice_removebad.err
Untracked: code/DiffSplice_removebad.out
Untracked: code/DiffSplice_removebad.sh
Untracked: code/FilterReverseLift.err
Untracked: code/FilterReverseLift.out
Untracked: code/FindIntronForDomPAS.err
Untracked: code/FindIntronForDomPAS.out
Untracked: code/FindIntronForDomPAS.sh
Untracked: code/FindIntronForDomPAS_DF.sh
Untracked: code/GencodeDiffSplice.err
Untracked: code/GencodeDiffSplice.out
Untracked: code/GetMAPQscore.py
Untracked: code/GetSecondaryMap.py
Untracked: code/HchromOrder.err
Untracked: code/HchromOrder.out
Untracked: code/JunctionLift.err
Untracked: code/JunctionLift.out
Untracked: code/JunctionLiftFinalChimp.err
Untracked: code/JunctionLiftFinalChimp.out
Untracked: code/Lift5perPAS.sh
Untracked: code/Lift5perPASbed.err
Untracked: code/Lift5perPASbed.out
Untracked: code/LiftClustersFirst.err
Untracked: code/LiftClustersFirst.out
Untracked: code/LiftClustersFirst_remove.err
Untracked: code/LiftClustersFirst_remove.out
Untracked: code/LiftClustersSecond.err
Untracked: code/LiftClustersSecond.out
Untracked: code/LiftClustersSecond_remove.err
Untracked: code/LiftClustersSecond_remove.out
Untracked: code/LiftFinalChimpJunc2Human.sh
Untracked: code/LiftOrthoPAS2chimp.sh
Untracked: code/LiftorthoPAS.err
Untracked: code/LiftorthoPASt.out
Untracked: code/Log.out
Untracked: code/MapBadSamples.err
Untracked: code/MapBadSamples.out
Untracked: code/MapBadSamples.sh
Untracked: code/MapStats.err
Untracked: code/MapStats.out
Untracked: code/MaxEntCode/
Untracked: code/MergeClusters.err
Untracked: code/MergeClusters.out
Untracked: code/MergeClusters.sh
Untracked: code/PAS_ATTAAA.err
Untracked: code/PAS_ATTAAA.out
Untracked: code/PAS_ATTAAA.sh
Untracked: code/PAS_ATTAAADF.err
Untracked: code/PAS_ATTAAADF.out
Untracked: code/PAS_ATTAAA_df.sh
Untracked: code/PAS_seqExpanded.sh
Untracked: code/PAS_sequence.err
Untracked: code/PAS_sequence.out
Untracked: code/PAS_sequenceDF.err
Untracked: code/PAS_sequenceDF.out
Untracked: code/PASexpanded_sequenceDF.err
Untracked: code/PASexpanded_sequenceDF.out
Untracked: code/PASsequences.sh
Untracked: code/PASsequences_DF.sh
Untracked: code/PlotNuclearUsagebySpecies.R
Untracked: code/PlotNuclearUsagebySpecies_DF.R
Untracked: code/QuantMergeClusters
Untracked: code/QuantMergeClusters.err
Untracked: code/QuantMergeClusters.out
Untracked: code/QuantMergedClusters.sh
Untracked: code/RNATranscriptDTplot.err
Untracked: code/RNATranscriptDTplot.out
Untracked: code/RNATranscriptDTplot.sh
Untracked: code/Rev_liftoverPAShg19to38.err
Untracked: code/Rev_liftoverPAShg19to38.out
Untracked: code/ReverseLiftFilter.R
Untracked: code/RunFixCluster.err
Untracked: code/RunFixCluster.out
Untracked: code/RunFixLeafCluster.sh
Untracked: code/RunNegMCMediation.err
Untracked: code/RunNegMCMediation.sh
Untracked: code/RunNegMCMediationDF.err
Untracked: code/RunNegMCMediationDF.out
Untracked: code/RunNegMCMediationDF.sh
Untracked: code/RunNegMCMediationr.out
Untracked: code/RunPosMCMediation.err
Untracked: code/RunPosMCMediation.sh
Untracked: code/RunPosMCMediationDF.err
Untracked: code/RunPosMCMediationDF.out
Untracked: code/RunPosMCMediationDF.sh
Untracked: code/RunPosMCMediationr.out
Untracked: code/SAF215upbed_gen.py
Untracked: code/SAF2Bed.py
Untracked: code/Snakefile
Untracked: code/SnakefilePAS
Untracked: code/SnakefilePASfilt
Untracked: code/SortIndexBadSamples.err
Untracked: code/SortIndexBadSamples.out
Untracked: code/SortIndexBadSamples.sh
Untracked: code/TotalTranscriptDTplot.err
Untracked: code/TotalTranscriptDTplot.out
Untracked: code/Upstream10Bases_general.py
Untracked: code/apaQTLsnake.err
Untracked: code/apaQTLsnake.out
Untracked: code/apaQTLsnakePAS.err
Untracked: code/apaQTLsnakePAS.out
Untracked: code/apaQTLsnakePAShuman.err
Untracked: code/assignPeak2Intronicregion.err
Untracked: code/assignPeak2Intronicregion.out
Untracked: code/assignPeak2Intronicregion.sh
Untracked: code/bam2junc.err
Untracked: code/bam2junc.out
Untracked: code/bam2junc_remove.err
Untracked: code/bam2junc_remove.out
Untracked: code/bed215upbed.py
Untracked: code/bed2SAF_gen.py
Untracked: code/bed2saf.py
Untracked: code/bg_to_cov.py
Untracked: code/buildIndecpantro5
Untracked: code/buildIndecpantro5.sh
Untracked: code/buildLeafviz.err
Untracked: code/buildLeafviz.out
Untracked: code/buildLeafviz.sh
Untracked: code/buildLeafviz_leadAnno.sh
Untracked: code/buildLeafviz_leafanno.err
Untracked: code/buildLeafviz_leafanno.out
Untracked: code/buildStarIndex.sh
Untracked: code/callPeaksYL.py
Untracked: code/chimpChromprder.sh
Untracked: code/chooseAnno2Bed.py
Untracked: code/chooseAnno2SAF.py
Untracked: code/chooseSignalSite.py
Untracked: code/chromOrder.err
Untracked: code/chromOrder.out
Untracked: code/classifyLeafviz.err
Untracked: code/classifyLeafviz.out
Untracked: code/cleanbed2saf.py
Untracked: code/cluster.json
Untracked: code/cluster2bed.py
Untracked: code/clusterLiftReverse.sh
Untracked: code/clusterLiftReverse_removebad.sh
Untracked: code/clusterLiftprimary.sh
Untracked: code/clusterLiftprimary_removebad.sh
Untracked: code/clusterPAS.json
Untracked: code/clusterfiltPAS.json
Untracked: code/comands2Mege.sh
Untracked: code/converBam2Junc.sh
Untracked: code/converBam2Junc_removeBad.sh
Untracked: code/convertNumeric.py
Untracked: code/environment.yaml
Untracked: code/extraSnakefiltpas
Untracked: code/extractPhyloReg.py
Untracked: code/extractPhyloRegGene.py
Untracked: code/extractPhylopGeneral.py
Untracked: code/extractPhylopReg200down.py
Untracked: code/extractPhylopReg200up.py
Untracked: code/filter5perc.R
Untracked: code/filter5percPAS.py
Untracked: code/filter5percPheno.py
Untracked: code/filterBamforMP.pysam2_gen.py
Untracked: code/filterJuncChroms.err
Untracked: code/filterJuncChroms.out
Untracked: code/filterMissprimingInNuc10_gen.py
Untracked: code/filterNumChroms.py
Untracked: code/filterPASforMP.py
Untracked: code/filterPostLift.py
Untracked: code/filterSAFforMP_gen.py
Untracked: code/filterSortBedbyCleanedBed_gen.R
Untracked: code/filterpeaks.py
Untracked: code/fixExonFC.py
Untracked: code/fixFChead.py
Untracked: code/fixFChead_bothfrac.py
Untracked: code/fixLeafCluster.py
Untracked: code/fixLiftedJunc.py
Untracked: code/fixUTRexonanno.py
Untracked: code/formathg38Anno.py
Untracked: code/generateStarIndex.err
Untracked: code/generateStarIndex.out
Untracked: code/generateStarIndexHuman.err
Untracked: code/generateStarIndexHuman.out
Untracked: code/getRNAseqMapStats.sh
Untracked: code/hg19MapStats.err
Untracked: code/hg19MapStats.out
Untracked: code/hg19MapStats.sh
Untracked: code/humanChromorder.sh
Untracked: code/humanFiles
Untracked: code/intersectAnno.err
Untracked: code/intersectAnno.out
Untracked: code/intersectAnnoExt.err
Untracked: code/intersectAnnoExt.out
Untracked: code/intersectLiftedPAS.sh
Untracked: code/leafcutter_merge_regtools_redo.py
Untracked: code/liftJunctionFiles.sh
Untracked: code/liftPAS19to38.sh
Untracked: code/liftoverPAShg19to38.err
Untracked: code/liftoverPAShg19to38.out
Untracked: code/log/
Untracked: code/make5percPeakbed.py
Untracked: code/makeFileID.py
Untracked: code/makeNuclearDapaplots.sh
Untracked: code/makeNuclearDapaplots_DF.sh
Untracked: code/makeNuclearPlots.err
Untracked: code/makeNuclearPlots.out
Untracked: code/makeNuclearPlotsDF.err
Untracked: code/makeNuclearPlotsDF.out
Untracked: code/makePheno.py
Untracked: code/makeSamplyGroupsChimp_TvN.py
Untracked: code/makeSamplyGroupsHuman_TvN.py
Untracked: code/mapRNAseqhg19.sh
Untracked: code/mapRNAseqhg19_newPipeline.sh
Untracked: code/maphg19.err
Untracked: code/maphg19.out
Untracked: code/maphg19.sh
Untracked: code/maphg19_new.err
Untracked: code/maphg19_new.out
Untracked: code/maphg19_sub.err
Untracked: code/maphg19_sub.out
Untracked: code/maphg19_subjunc.sh
Untracked: code/mediation_test.R
Untracked: code/merge.err
Untracked: code/mergeChimp3prime_inhg38.sh
Untracked: code/merge_leafcutter_clusters_redo.py
Untracked: code/mergeandBWRNAseq.sh
Untracked: code/mergeandsort_ChimpinHuman.err
Untracked: code/mergeandsort_ChimpinHuman.out
Untracked: code/mergedBam2BW.sh
Untracked: code/mergedbam2bw.err
Untracked: code/mergedbam2bw.out
Untracked: code/mergedbamRNAand2bw.err
Untracked: code/mergedbamRNAand2bw.out
Untracked: code/nameClusters.py
Untracked: code/namePeaks.py
Untracked: code/negativeMediation_montecarlo.R
Untracked: code/negativeMediation_montecarloDF.R
Untracked: code/nuclearTranscriptDTplot.err
Untracked: code/nuclearTranscriptDTplot.out
Untracked: code/numMultimap.py
Untracked: code/overlapPAS.err
Untracked: code/overlapPAS.out
Untracked: code/overlapapaQTLPAS.sh
Untracked: code/overlapapaQTLPAS_extended.sh
Untracked: code/overlapapaQTLPAS_samples.sh
Untracked: code/parseHg38.py
Untracked: code/peak2PAS.py
Untracked: code/pheno2countonly.R
Untracked: code/postiveMediation_montecarlo.R
Untracked: code/postiveMediation_montecarlo_DF.R
Untracked: code/prepareAnnoLeafviz.err
Untracked: code/prepareAnnoLeafviz.out
Untracked: code/prepareCleanLiftedFC_5perc4LC.py
Untracked: code/prepareLeafvizAnno.sh
Untracked: code/preparePAS4lift.py
Untracked: code/prepare_phenotype_table.py
Untracked: code/primaryLift.err
Untracked: code/primaryLift.out
Untracked: code/primaryLift.sh
Untracked: code/processhg38exons.py
Untracked: code/quantJunc.sh
Untracked: code/quantJunc_TEST.sh
Untracked: code/quantJunc_removeBad.sh
Untracked: code/quantLiftedPAS.err
Untracked: code/quantLiftedPAS.out
Untracked: code/quantLiftedPAS.sh
Untracked: code/quatJunc.err
Untracked: code/quatJunc.out
Untracked: code/recChimpback2Human.err
Untracked: code/recChimpback2Human.out
Untracked: code/recLiftchim2human.sh
Untracked: code/revLift.err
Untracked: code/revLift.out
Untracked: code/revLiftPAShg38to19.sh
Untracked: code/reverseLift.sh
Untracked: code/runCheckReverseLift.sh
Untracked: code/runChimpDiffIso.sh
Untracked: code/runChimpDiffIsoDF.sh
Untracked: code/runCountNucleotides.err
Untracked: code/runCountNucleotides.out
Untracked: code/runCountNucleotides.sh
Untracked: code/runCountNucleotidesPantro6.err
Untracked: code/runCountNucleotidesPantro6.out
Untracked: code/runCountNucleotides_pantro6.sh
Untracked: code/runFilterNumChroms.sh
Untracked: code/runHumanDiffIso.sh
Untracked: code/runHumanDiffIsoDF.sh
Untracked: code/runNuclearDiffIso_DF.sh
Untracked: code/runNuclearDifffIso.sh
Untracked: code/runTotalDiffIso.sh
Untracked: code/run_Chimpleafcutter_ds.err
Untracked: code/run_Chimpleafcutter_ds.out
Untracked: code/run_Chimpverifybam.err
Untracked: code/run_Chimpverifybam.out
Untracked: code/run_Humanleafcutter_dF.err
Untracked: code/run_Humanleafcutter_dF.out
Untracked: code/run_Humanleafcutter_ds.err
Untracked: code/run_Humanleafcutter_ds.out
Untracked: code/run_Nuclearleafcutter_ds.err
Untracked: code/run_Nuclearleafcutter_ds.out
Untracked: code/run_Nuclearleafcutter_dsDF.err
Untracked: code/run_Nuclearleafcutter_dsDF.out
Untracked: code/run_Totalleafcutter_ds.err
Untracked: code/run_Totalleafcutter_ds.out
Untracked: code/run_chimpverifybam.sh
Untracked: code/run_verifyBam.sh
Untracked: code/run_verifybam.err
Untracked: code/run_verifybam.out
Untracked: code/slurm-62824013.out
Untracked: code/slurm-62825841.out
Untracked: code/slurm-62826116.out
Untracked: code/slurm-64108209.out
Untracked: code/slurm-64108521.out
Untracked: code/slurm-64108557.out
Untracked: code/snakePASChimp.err
Untracked: code/snakePASChimp.out
Untracked: code/snakePAShuman.out
Untracked: code/snakemake.batch
Untracked: code/snakemakeChimp.err
Untracked: code/snakemakeChimp.out
Untracked: code/snakemakeHuman.err
Untracked: code/snakemakeHuman.out
Untracked: code/snakemakePAS.batch
Untracked: code/snakemakePASFiltChimp.err
Untracked: code/snakemakePASFiltChimp.out
Untracked: code/snakemakePASFiltHuman.err
Untracked: code/snakemakePASFiltHuman.out
Untracked: code/snakemakePASchimp.batch
Untracked: code/snakemakePAShuman.batch
Untracked: code/snakemake_chimp.batch
Untracked: code/snakemake_human.batch
Untracked: code/snakemakefiltPAS.batch
Untracked: code/snakemakefiltPAS_chimp.sh
Untracked: code/snakemakefiltPAS_human.sh
Untracked: code/spliceSite2Fasta.py
Untracked: code/submit-snakemake-chimp.sh
Untracked: code/submit-snakemake-human.sh
Untracked: code/submit-snakemakePAS-chimp.sh
Untracked: code/submit-snakemakePAS-human.sh
Untracked: code/submit-snakemakefiltPAS-chimp.sh
Untracked: code/submit-snakemakefiltPAS-human.sh
Untracked: code/subset_diffisopheno.py
Untracked: code/subset_diffisopheno_Chimp_tvN.py
Untracked: code/subset_diffisopheno_Chimp_tvN_DF.py
Untracked: code/subset_diffisopheno_Huma_tvN.py
Untracked: code/subset_diffisopheno_Huma_tvN_DF.py
Untracked: code/subset_diffisopheno_Nuclear_HvC.py
Untracked: code/subset_diffisopheno_Nuclear_HvC_DF.py
Untracked: code/subset_diffisopheno_Total_HvC.py
Untracked: code/test
Untracked: code/test.txt
Untracked: code/threeprimeOrthoFC.out
Untracked: code/threeprimeOrthoFC.sh
Untracked: code/threeprimeOrthoFCcd.err
Untracked: code/transcriptDTplotsNuclear.sh
Untracked: code/transcriptDTplotsTotal.sh
Untracked: code/verifyBam4973.sh
Untracked: code/verifyBam4973inHuman.sh
Untracked: code/verifybam4973.err
Untracked: code/verifybam4973.out
Untracked: code/verifybam4973HumanMap.err
Untracked: code/verifybam4973HumanMap.out
Untracked: code/wrap_Chimpverifybam.err
Untracked: code/wrap_Chimpverifybam.out
Untracked: code/wrap_chimpverifybam.sh
Untracked: code/wrap_verifyBam.sh
Untracked: code/wrap_verifybam.err
Untracked: code/wrap_verifybam.out
Untracked: code/writeMergecode.py
Untracked: data/._.DS_Store
Untracked: data/._HC_filenames.txt
Untracked: data/._HC_filenames.txt.sb-4426323c-IKIs0S
Untracked: data/._HC_filenames.xlsx
Untracked: data/._MapPantro6_meta.txt
Untracked: data/._MapPantro6_meta.txt.sb-a5794dd2-Cskmlm
Untracked: data/._MapPantro6_meta.xlsx
Untracked: data/._OppositeSpeciesMap.txt
Untracked: data/._OppositeSpeciesMap.txt.sb-a5794dd2-mayWJf
Untracked: data/._OppositeSpeciesMap.xlsx
Untracked: data/._RNASEQ_metadata.txt
Untracked: data/._RNASEQ_metadata.txt.sb-4426323c-TE4ns3
Untracked: data/._RNASEQ_metadata.txt.sb-51f67ae1-HXp7Gq
Untracked: data/._RNASEQ_metadata_2Removed.txt
Untracked: data/._RNASEQ_metadata_2Removed.txt.sb-4426323c-a4lBwx
Untracked: data/._RNASEQ_metadata_2Removed.xlsx
Untracked: data/._RNASEQ_metadata_stranded.txt
Untracked: data/._RNASEQ_metadata_stranded.txt.sb-a5794dd2-D659m2
Untracked: data/._RNASEQ_metadata_stranded.txt.sb-a5794dd2-ImNMoY
Untracked: data/._RNASEQ_metadata_stranded.txt.sb-e4bf31f0-ZGnGgl
Untracked: data/._RNASEQ_metadata_stranded.xlsx
Untracked: data/._TrialFiltersMeta.txt
Untracked: data/._TrialFiltersMeta.txt.sb-9845453e-R58Y0Q
Untracked: data/._metadata_HCpanel.txt
Untracked: data/._metadata_HCpanel.txt.sb-a3d92a2d-b9cYoF
Untracked: data/._metadata_HCpanel.txt.sb-a5794dd2-i594qs
Untracked: data/._metadata_HCpanel.txt.sb-f4823d1e-qihGek
Untracked: data/._metadata_HCpanel.xlsx
Untracked: data/._metadata_HCpanel_frompantro5.xlsx
Untracked: data/._~$RNASEQ_metadata.xlsx
Untracked: data/._~$metadata_HCpanel.xlsx
Untracked: data/._.xlsx
Untracked: data/BaseComp/
Untracked: data/CompapaQTLpas/
Untracked: data/DNDS/
Untracked: data/DTmatrix/
Untracked: data/DiffExpression/
Untracked: data/DiffIso_Nuclear/
Untracked: data/DiffIso_Nuclear_DF/
Untracked: data/DiffIso_Total/
Untracked: data/DiffSplice/
Untracked: data/DiffSplice_liftedJunc/
Untracked: data/DiffSplice_removeBad/
Untracked: data/DominantPAS/
Untracked: data/DominantPAS_DF/
Untracked: data/EvalPantro5/
Untracked: data/HC_filenames.txt
Untracked: data/HC_filenames.xlsx
Untracked: data/Khan_prot/
Untracked: data/Li_eqtls/
Untracked: data/MapPantro6_meta.txt
Untracked: data/MapPantro6_meta.xlsx
Untracked: data/MapStats/
Untracked: data/NormalizedClusters/
Untracked: data/NuclearHvC/
Untracked: data/NuclearHvC_DF/
Untracked: data/OppositeSpeciesMap.txt
Untracked: data/OppositeSpeciesMap.xlsx
Untracked: data/OrthoExonBed/
Untracked: data/OverlapBenchmark/
Untracked: data/OverlappingPAS/
Untracked: data/PAS/
Untracked: data/PAS_doubleFilter/
Untracked: data/Peaks_5perc/
Untracked: data/Pheno_5perc/
Untracked: data/Pheno_5perc_DF_nuclear/
Untracked: data/Pheno_5perc_nuclear/
Untracked: data/Pheno_5perc_nuclear_old/
Untracked: data/Pheno_5perc_total/
Untracked: data/PhyloP/
Untracked: data/RNASEQ_metadata.txt
Untracked: data/RNASEQ_metadata_2Removed.txt
Untracked: data/RNASEQ_metadata_2Removed.xlsx
Untracked: data/RNASEQ_metadata_stranded.txt
Untracked: data/RNASEQ_metadata_stranded.txt.sb-e4bf31f0-ZGnGgl/
Untracked: data/RNASEQ_metadata_stranded.xlsx
Untracked: data/SignalSites/
Untracked: data/SignalSites_doublefilter/
Untracked: data/SpliceSite/
Untracked: data/Threeprime2Ortho/
Untracked: data/TotalHvC/
Untracked: data/TrialFiltersMeta.txt
Untracked: data/TwoBadSampleAnalysis/
Untracked: data/Wang_ribo/
Untracked: data/apaQTLGenes/
Untracked: data/bioGRID/
Untracked: data/chainFiles/
Untracked: data/cleanPeaks_anno/
Untracked: data/cleanPeaks_byspecies/
Untracked: data/cleanPeaks_lifted/
Untracked: data/files4viz_nuclear/
Untracked: data/files4viz_nuclear_DF/
Untracked: data/gviz/
Untracked: data/leafviz/
Untracked: data/liftover_files/
Untracked: data/mediation/
Untracked: data/mediation_DF/
Untracked: data/metadata_HCpanel.txt
Untracked: data/metadata_HCpanel.xlsx
Untracked: data/metadata_HCpanel_frompantro5.txt
Untracked: data/metadata_HCpanel_frompantro5.xlsx
Untracked: data/primaryLift/
Untracked: data/reverseLift/
Untracked: data/~$RNASEQ_metadata.xlsx
Untracked: data/~$metadata_HCpanel.xlsx
Untracked: data/.xlsx
Untracked: output/._.DS_Store
Untracked: output/dtPlots/
Untracked: projectNotes.Rmd
Untracked: proteinModelSet.Rmd
Unstaged changes:
Modified: analysis/DiffUsedIntronic.Rmd
Modified: analysis/ExploredAPA.Rmd
Modified: analysis/ExploredAPA_DF.Rmd
Modified: analysis/OppositeMap.Rmd
Modified: analysis/SpliceSiteStrength.Rmd
Modified: analysis/TotalVNuclearBothSpecies.Rmd
Modified: analysis/annotationInfo.Rmd
Modified: analysis/changeMisprimcut.Rmd
Modified: analysis/comp2apaQTLPAS.Rmd
Modified: analysis/correlationPhenos.Rmd
Modified: analysis/dAPAandapaQTL_DF.Rmd
Modified: analysis/establishCutoffs.Rmd
Modified: analysis/investigatePantro5.Rmd
Modified: analysis/multiMap.Rmd
Modified: analysis/speciesSpecific.Rmd
Modified: analysis/speciesSpecific_DF.Rmd
Modified: analysis/upsetter_DF.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the R Markdown and HTML files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view them.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 565dd6a | brimittleman | 2020-03-18 | show dom enrich not factor of usage |
| html | 86238eb | brimittleman | 2020-03-16 | Build site. |
| Rmd | 164c237 | brimittleman | 2020-03-16 | ask about anno bias |
I want to see if the most used PAS are also the differentially used.
library(tidyverse)── Attaching packages ─────────────────────────────────────────────────────────── tidyverse 1.2.1 ──✔ ggplot2 3.1.1 ✔ purrr 0.3.2
✔ tibble 2.1.1 ✔ dplyr 0.8.0.1
✔ tidyr 0.8.3 ✔ stringr 1.3.1
✔ readr 1.3.1 ✔ forcats 0.3.0 ── Conflicts ────────────────────────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()library(workflowr)This is workflowr version 1.6.0
Run ?workflowr for help getting startedlibrary(cowplot)
Attaching package: 'cowplot'The following object is masked from 'package:ggplot2':
ggsaveCaveat- ties.
Dominant PAS
allPAS= read.table("../data/PAS_doubleFilter/PAS_10perc_either_HumanCoord_BothUsage_meta_doubleFilter.txt", header = T)
ChimpPASwMean =allPAS %>% dplyr::select(-Human)
HumanPASwMean =allPAS %>% dplyr::select(-Chimp)
Chimp_Dom= ChimpPASwMean %>%
group_by(gene) %>%
arrange(desc(Chimp)) %>%
slice(1) %>%
group_by(gene) %>%
mutate(npas=n()) %>%
dplyr::select(gene,loc,PAS,Chimp) %>%
rename(ChimpLoc=loc, ChimpPAS=PAS)
Chimp_Dom2= ChimpPASwMean %>%
group_by(gene) %>%
top_n(1,Chimp) %>%
mutate(nPer=n())
nrow(Chimp_Dom2%>% filter(nPer>1) )[1] 198Human_Dom= HumanPASwMean %>%
group_by(gene) %>%
arrange(desc(Human)) %>%
slice(1) %>%
group_by(gene) %>%
mutate(npas=n()) %>%
dplyr::select(gene,loc,PAS,Human) %>%
rename(HumanLoc=loc, HumanPAS=PAS)
Human_Dom2= HumanPASwMean %>%
group_by(gene) %>%
top_n(1,Human) %>%
mutate(nPer=n())
nrow(Human_Dom2 %>% filter(nPer>1) )[1] 161198 genes with a tie in chimp and 161 with a tie in human. Picks top for both for analysis.
No PAS usage cutoff for dominant
BothDom= Chimp_Dom %>% inner_join(Human_Dom,by="gene")
SameDom= BothDom %>% filter(ChimpPAS==HumanPAS,HumanLoc!="008559")
nrow(SameDom)[1] 7718SameDom_g= SameDom %>% select(gene, ChimpLoc, HumanLoc) %>% gather("Species", "Location", -gene)
ggplot(SameDom_g, aes(x=Location, by=Species, fill=Species))+ geom_bar(stat="count",position = "Dodge") + labs(x="Location", y="Number of Genes", title="Dominant PAS for genes with matching by species")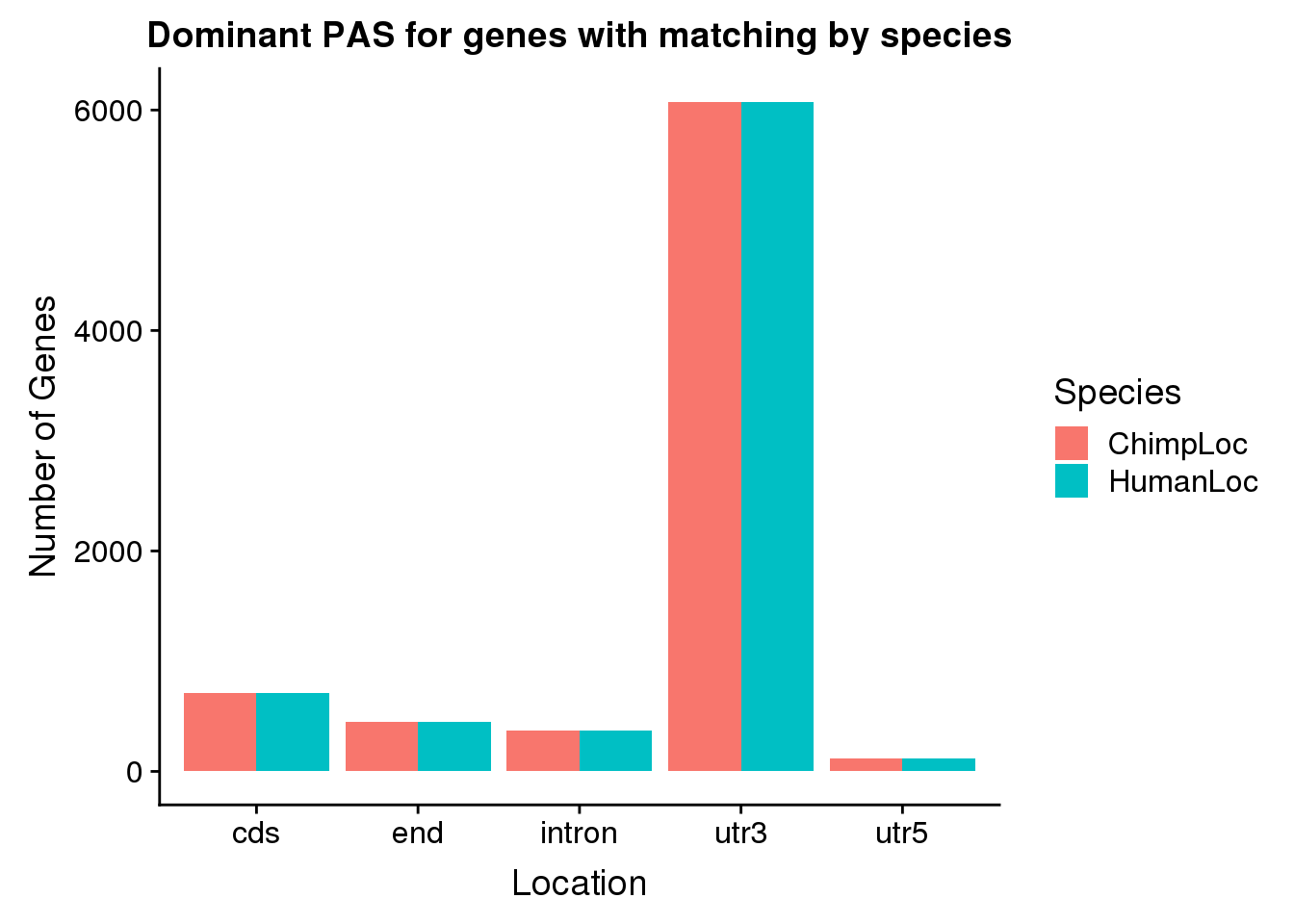
| Version | Author | Date |
|---|---|---|
| 86238eb | brimittleman | 2020-03-16 |
ggplot(SameDom, aes(x=HumanLoc))+ geom_bar(stat="count") + labs(x="Location", y="Number of Genes", title="Dominant PAS for genes with matching by species")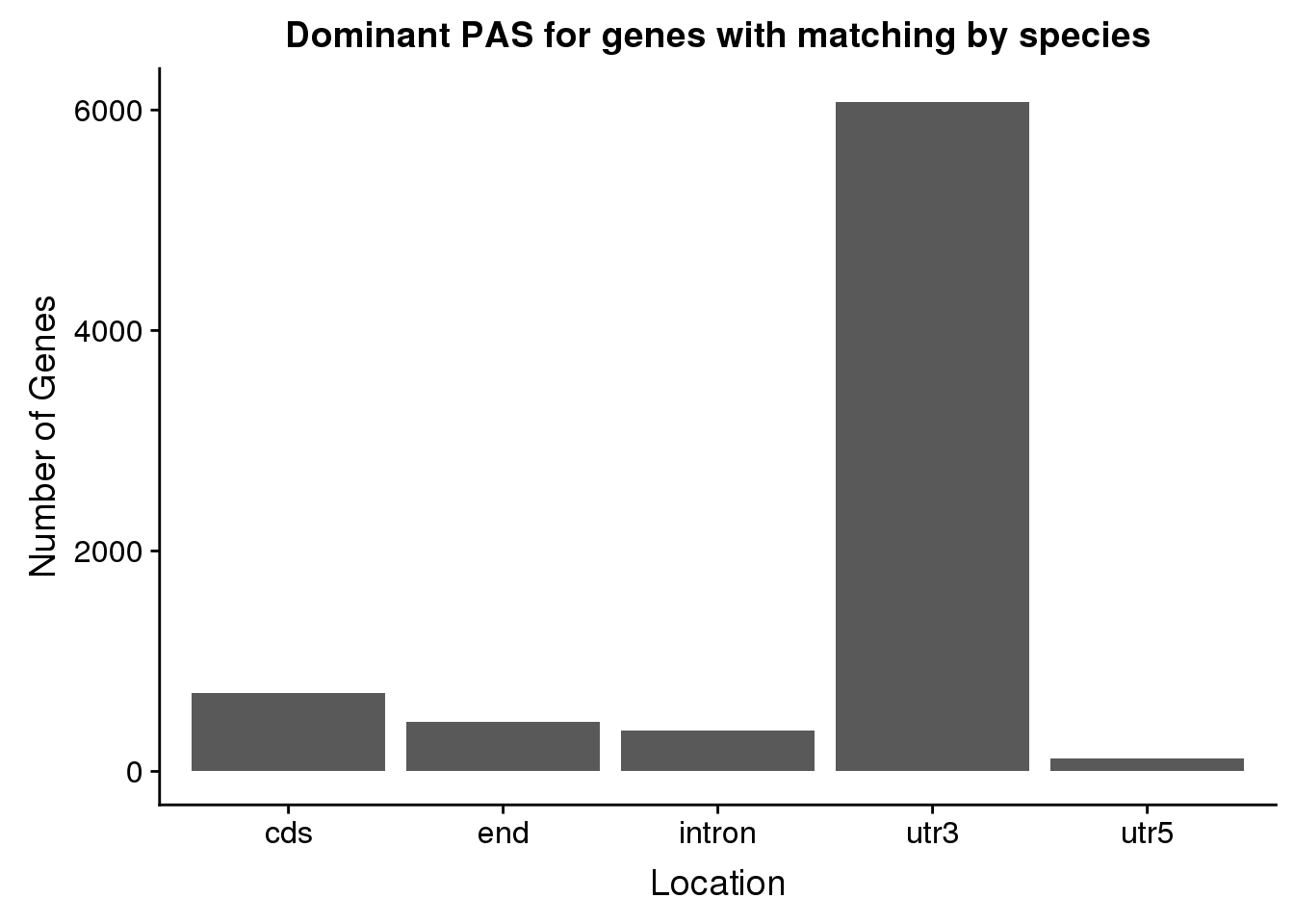
| Version | Author | Date |
|---|---|---|
| 86238eb | brimittleman | 2020-03-16 |
DiffDom=BothDom %>% filter(ChimpPAS!=HumanPAS,HumanLoc!="008559")
nrow(DiffDom)[1] 2092DiffDom_g= DiffDom %>% select(gene, ChimpLoc, HumanLoc) %>% gather("Species", "Location", -gene)
plotnones=ggplot(DiffDom_g,aes(by=Species, x=Location, fill=Species))+ geom_histogram(stat="count",position = "dodge") + labs(x="Location", y="Number of Genes", title="Different Dominant PAS") + scale_fill_brewer(palette = "Dark2")Warning: Ignoring unknown parameters: binwidth, bins, padDiff dominant different locations:
DiffDomDiffLoc=BothDom %>% filter(ChimpPAS!=HumanPAS,HumanLoc!="008559", ChimpLoc!=HumanLoc)
nrow(DiffDomDiffLoc)[1] 1095DiffDomDiffLoc_g= DiffDomDiffLoc %>% select(gene, ChimpLoc, HumanLoc) %>% gather("Species", "Location", -gene)
plotnone=ggplot(DiffDomDiffLoc_g,aes(by=Species, x=Location, fill=Species))+ geom_bar(stat="count",position = "dodge") + labs(x="Location", y="Number of Genes", title="Dominant PAS in different locations") + scale_fill_brewer(palette = "Dark2")Implement filters
I want to filter these and see if these distribution changes. First I will say the most used has to be over 50% in both species.
BothDom_50= BothDom %>% filter(Chimp>=.5, Human>.5)
nrow(BothDom_50)[1] 3526BothDom_40= BothDom %>% filter(Chimp>=.4, Human>.4)
nrow(BothDom_40)[1] 4555BothDom_30= BothDom %>% filter(Chimp>=.3, Human>.3)
nrow(BothDom_30)[1] 5879BothDom_20= BothDom %>% filter(Chimp>=.2, Human>.2)
nrow(BothDom_20)[1] 7584BothDom_10= BothDom %>% filter(Chimp>=.1, Human>.1)
nrow(BothDom_10)[1] 9324cutoffs=c(".5",'.4','.3','.2','.1', "0")
nPAS=c(nrow(BothDom_50),nrow(BothDom_40),nrow(BothDom_30),nrow(BothDom_20),nrow(BothDom_10), nrow(BothDom))
nPAS[1] 3526 4555 5879 7584 9324 9810Different Dominant, different Location
BothDom_50_diff=BothDom_50%>% filter(ChimpPAS!=HumanPAS,HumanLoc!="008559", ChimpLoc!=HumanLoc)
nrow(BothDom_50_diff)[1] 33BothDom_50_diff_g= BothDom_50_diff %>% select(gene, ChimpLoc, HumanLoc) %>% gather("Species", "Location", -gene)
plot50=ggplot(BothDom_50_diff_g,aes(by=Species, x=Location, fill=Species))+ geom_bar(stat="count",position = "dodge") + labs(x="Location", y="Number of Genes", title="Dominant PAS in different locations (50%)") + scale_fill_brewer(palette = "Dark2")BothDom_40_diff=BothDom_40%>% filter(ChimpPAS!=HumanPAS,HumanLoc!="008559", ChimpLoc!=HumanLoc)
nrow(BothDom_40_diff)[1] 85BothDom_40_diff_g= BothDom_40_diff %>% select(gene, ChimpLoc, HumanLoc) %>% gather("Species", "Location", -gene)
plot40=ggplot(BothDom_40_diff_g,aes(by=Species, x=Location, fill=Species))+ geom_bar(stat="count",position = "dodge") + labs(x="Location", y="Number of Genes", title="Dominant PAS in different locations (40%)") + scale_fill_brewer(palette = "Dark2")BothDom_30_diff=BothDom_30%>% filter(ChimpPAS!=HumanPAS,HumanLoc!="008559", ChimpLoc!=HumanLoc)
nrow(BothDom_30_diff)[1] 211BothDom_30_diff_g= BothDom_30_diff %>% select(gene, ChimpLoc, HumanLoc) %>% gather("Species", "Location", -gene)
plot30=ggplot(BothDom_30_diff_g,aes(by=Species, x=Location, fill=Species))+ geom_bar(stat="count",position = "dodge") + labs(x="Location", y="Number of Genes", title="Dominant PAS in different locations (30%)") + scale_fill_brewer(palette = "Dark2")BothDom_20_diff=BothDom_20%>% filter(ChimpPAS!=HumanPAS,HumanLoc!="008559", ChimpLoc!=HumanLoc)
nrow(BothDom_20_diff)[1] 481BothDom_20_diff_g= BothDom_20_diff %>% select(gene, ChimpLoc, HumanLoc) %>% gather("Species", "Location", -gene)
plot20=ggplot(BothDom_20_diff_g,aes(by=Species, x=Location, fill=Species))+ geom_bar(stat="count",position = "dodge") + labs(x="Location", y="Number of Genes", title="Dominant PAS in different locations (20%)") + scale_fill_brewer(palette = "Dark2")BothDom_10_diff=BothDom_10%>% filter(ChimpPAS!=HumanPAS,HumanLoc!="008559", ChimpLoc!=HumanLoc)
nrow(BothDom_10_diff)[1] 931BothDom_10_diff_g= BothDom_10_diff %>% select(gene, ChimpLoc, HumanLoc) %>% gather("Species", "Location", -gene)
plot10=ggplot(BothDom_10_diff_g,aes(by=Species, x=Location, fill=Species))+ geom_bar(stat="count",position = "dodge") + labs(x="Location", y="Number of Genes", title="Dominant PAS in different locations (10%)") + scale_fill_brewer(palette = "Dark2")plot_grid(plot50, plot40, plot30, plot20, plot10,plotnone)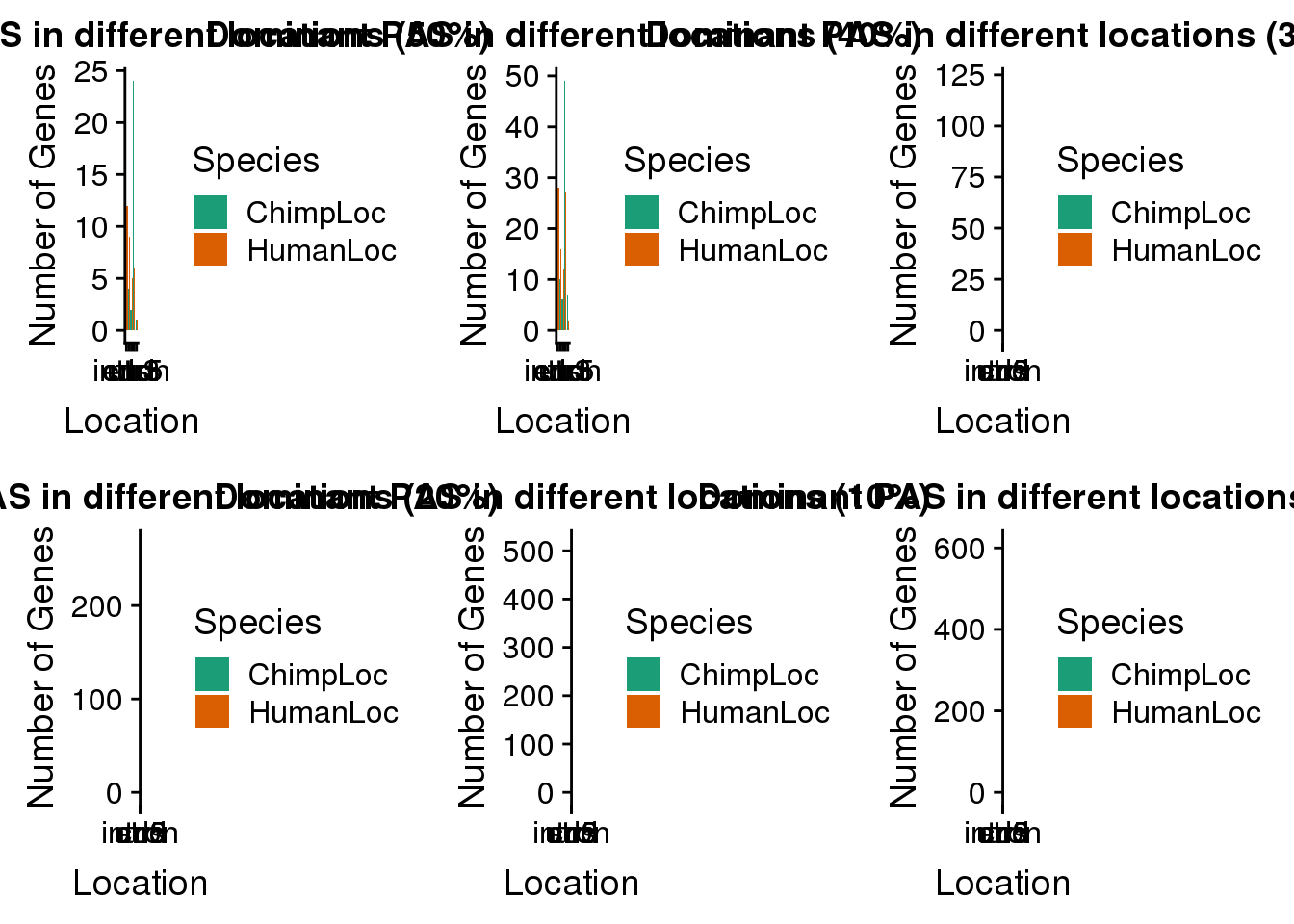
| Version | Author | Date |
|---|---|---|
| 86238eb | brimittleman | 2020-03-16 |
Filters in same loc ok
BothDom_50_same=BothDom_50%>% filter(ChimpPAS!=HumanPAS,HumanLoc!="008559")
nrow(BothDom_50_same)[1] 74BothDom_50_same_g= BothDom_50_same %>% select(gene, ChimpLoc, HumanLoc) %>% gather("Species", "Location", -gene)
plot50s=ggplot(BothDom_50_same_g,aes(by=Species, x=Location, fill=Species))+ geom_bar(stat="count",position = "dodge") + labs(x="Location", y="Number of Genes", title="Different Dominant (50%)") + scale_fill_brewer(palette = "Dark2")BothDom_40_same=BothDom_40%>% filter(ChimpPAS!=HumanPAS,HumanLoc!="008559")
nrow(BothDom_40_same)[1] 223BothDom_40_same_g= BothDom_40_same %>% select(gene, ChimpLoc, HumanLoc) %>% gather("Species", "Location", -gene)
plot40s=ggplot(BothDom_40_same_g,aes(by=Species, x=Location, fill=Species))+ geom_bar(stat="count",position = "dodge") + labs(x="Location", y="Number of Genes", title="Different Dominant (40%)") + scale_fill_brewer(palette = "Dark2")BothDom_30_same=BothDom_30%>% filter(ChimpPAS!=HumanPAS,HumanLoc!="008559")
nrow(BothDom_30_same)[1] 480BothDom_30_same_g= BothDom_30_same %>% select(gene, ChimpLoc, HumanLoc) %>% gather("Species", "Location", -gene)
plot30s=ggplot(BothDom_30_same_g,aes(by=Species, x=Location, fill=Species))+ geom_bar(stat="count",position = "dodge") + labs(x="Location", y="Number of Genes", title="Different Dominant (30%)") + scale_fill_brewer(palette = "Dark2")BothDom_20_same=BothDom_20%>% filter(ChimpPAS!=HumanPAS,HumanLoc!="008559")
nrow(BothDom_20_same)[1] 1011BothDom_20_same_g= BothDom_20_same %>% select(gene, ChimpLoc, HumanLoc) %>% gather("Species", "Location", -gene)
plot20s=ggplot(BothDom_20_same_g,aes(by=Species, x=Location, fill=Species))+ geom_bar(stat="count",position = "dodge") + labs(x="Location", y="Number of Genes", title="Different Dominant (20%)") + scale_fill_brewer(palette = "Dark2")BothDom_10_same=BothDom_10%>% filter(ChimpPAS!=HumanPAS,HumanLoc!="008559")
nrow(BothDom_10_same)[1] 1810BothDom_10_same_g= BothDom_10_same %>% select(gene, ChimpLoc, HumanLoc) %>% gather("Species", "Location", -gene)
plot10s=ggplot(BothDom_10_same_g,aes(by=Species, x=Location, fill=Species))+ geom_bar(stat="count",position = "dodge") + labs(x="Location", y="Number of Genes", title="Different Dominant (10%)") + scale_fill_brewer(palette = "Dark2")plot_grid(plot50s, plot40s, plot30s, plot20s, plot10s,plotnones)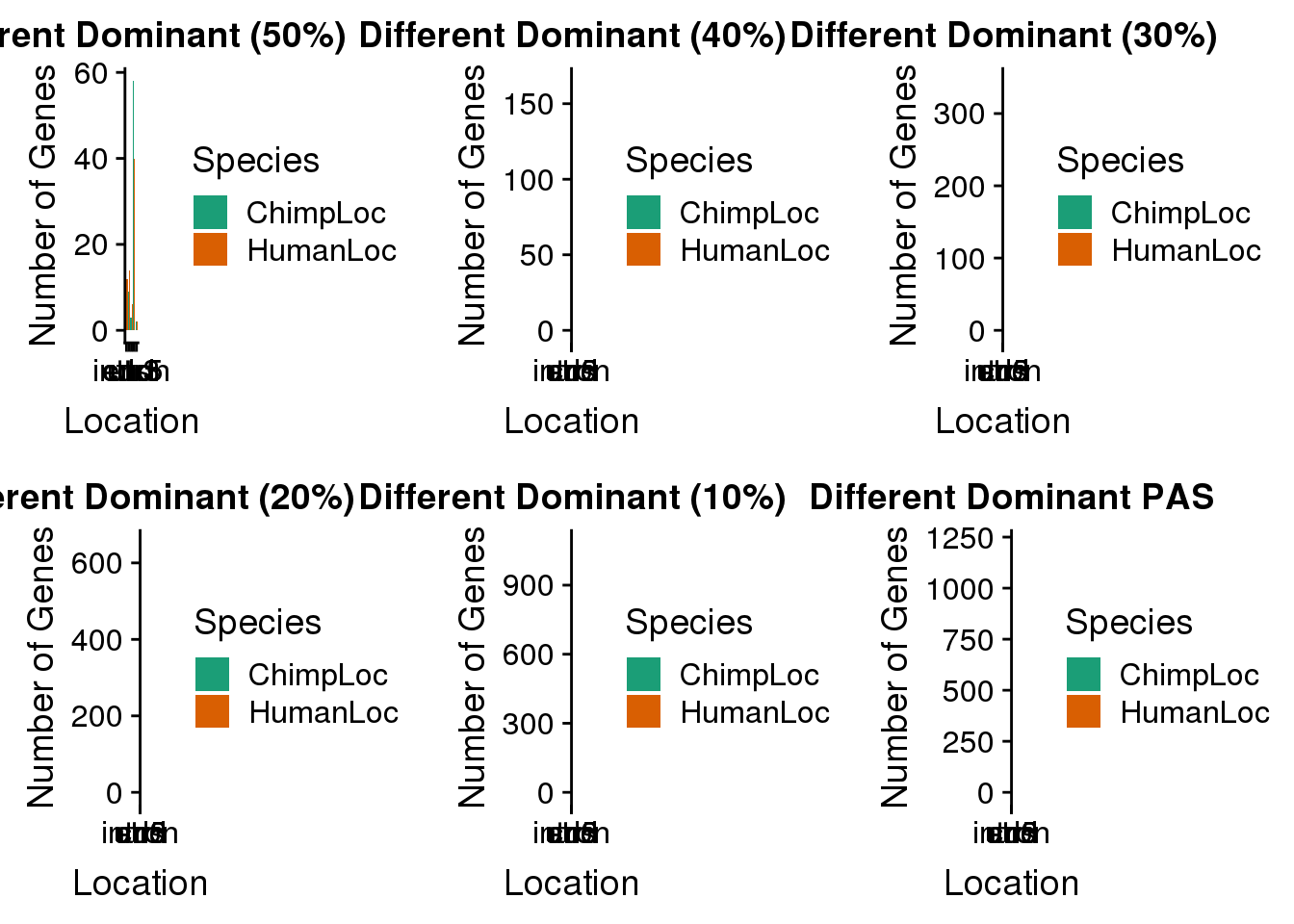
| Version | Author | Date |
|---|---|---|
| 86238eb | brimittleman | 2020-03-16 |
Plot numbers:
cutoffs=c("0.5",'0.4','0.3','0.2','0.1', "0")
#nPAS=c(nrow(BothDom_50),nrow(BothDom_40),nrow(BothDom_30),nrow(BothDom_20),nrow(BothDom_10), nrow(BothDom))
nPASDiff=c(nrow(BothDom_50_diff),nrow(BothDom_40_diff),nrow(BothDom_30_diff),nrow(BothDom_20_diff),nrow(BothDom_10_diff), nrow(DiffDomDiffLoc))
nPASSameLoc=c(nrow(BothDom_50_same),nrow(BothDom_40_same),nrow(BothDom_30_same),nrow(BothDom_20_same),nrow(BothDom_10_same), nrow(DiffDom))
NumberDF= as.data.frame(cbind(cutoffs, Genes=nPAS, DifferentPASandLoc=nPASDiff, DifferentPAS=nPASSameLoc)) %>% gather("Set", "count",-cutoffs )Warning: attributes are not identical across measure variables;
they will be droppedNumberDF$count=as.numeric(NumberDF$count)
ggplot(NumberDF,aes(x=cutoffs, y=count, by=Set, fill=Set)) + geom_bar(stat="identity",position = "dodge" ) +geom_text(stat='identity', aes(label=count), vjust=0,position = position_dodge(width = 1)) + labs(x="Cutoff for PAS to be dominant (Both Species)", y="Number of genes")+ scale_fill_brewer(palette = "Dark2", labels=c("Different PAS Same Location", "Different PAS and Location", "All Genes"),name="") + theme(legend.position = "top")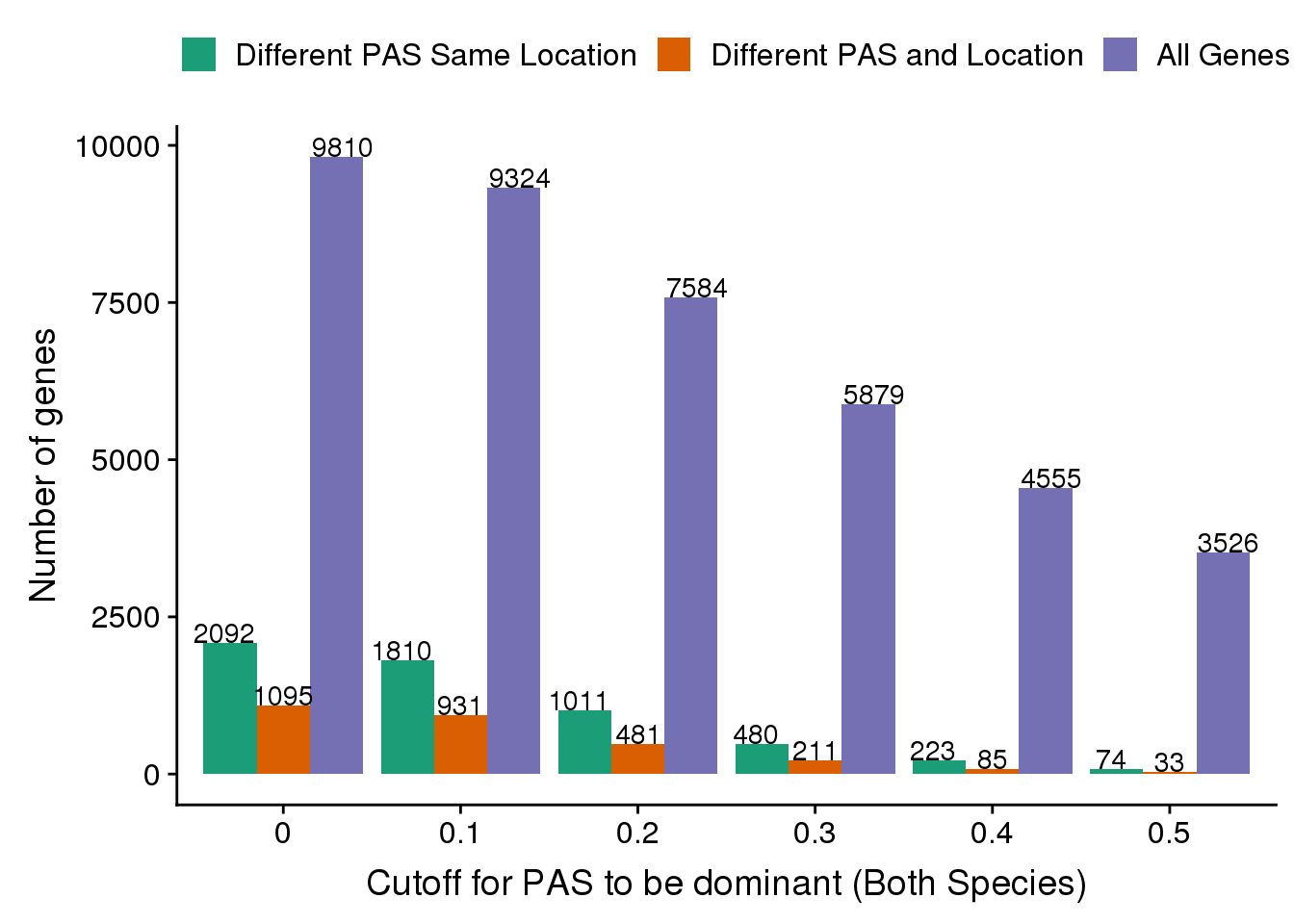
| Version | Author | Date |
|---|---|---|
| 86238eb | brimittleman | 2020-03-16 |
Compare to differencially used
PASMeta=read.table("../data/PAS_doubleFilter/PAS_10perc_either_HumanCoord_BothUsage_meta_doubleFilter.txt", header = T, stringsAsFactors = F) %>% select(PAS, gene, chr, start,end,loc)
DiffIso=read.table("../data/DiffIso_Nuclear_DF/AllPAS_withGeneSig.txt",header = T, stringsAsFactors = F) %>% inner_join(PASMeta, by=c("chr",'start','end',"gene"))
DiffIso_sig= DiffIso %>% filter(SigPAU2=="Yes")
DiffIsowDom=DiffIso %>% mutate(ChimpDom=ifelse(PAS %in% Chimp_Dom2$PAS, "Yes", "No"),HumanDom=ifelse(PAS %in% Human_Dom2$PAS, "Yes", "No") )
ggplot(DiffIsowDom,aes(x=loc,fill=ChimpDom)) + geom_bar(stat = "count") + facet_grid(~SigPAU2) +labs(x="", y="Number of PAS", title="Differentially used PAS colored by Dominant Status in Chimp")+ scale_fill_brewer(palette = "Dark2",name="Is PAS Dominant in Chimp") + theme(legend.position = "top")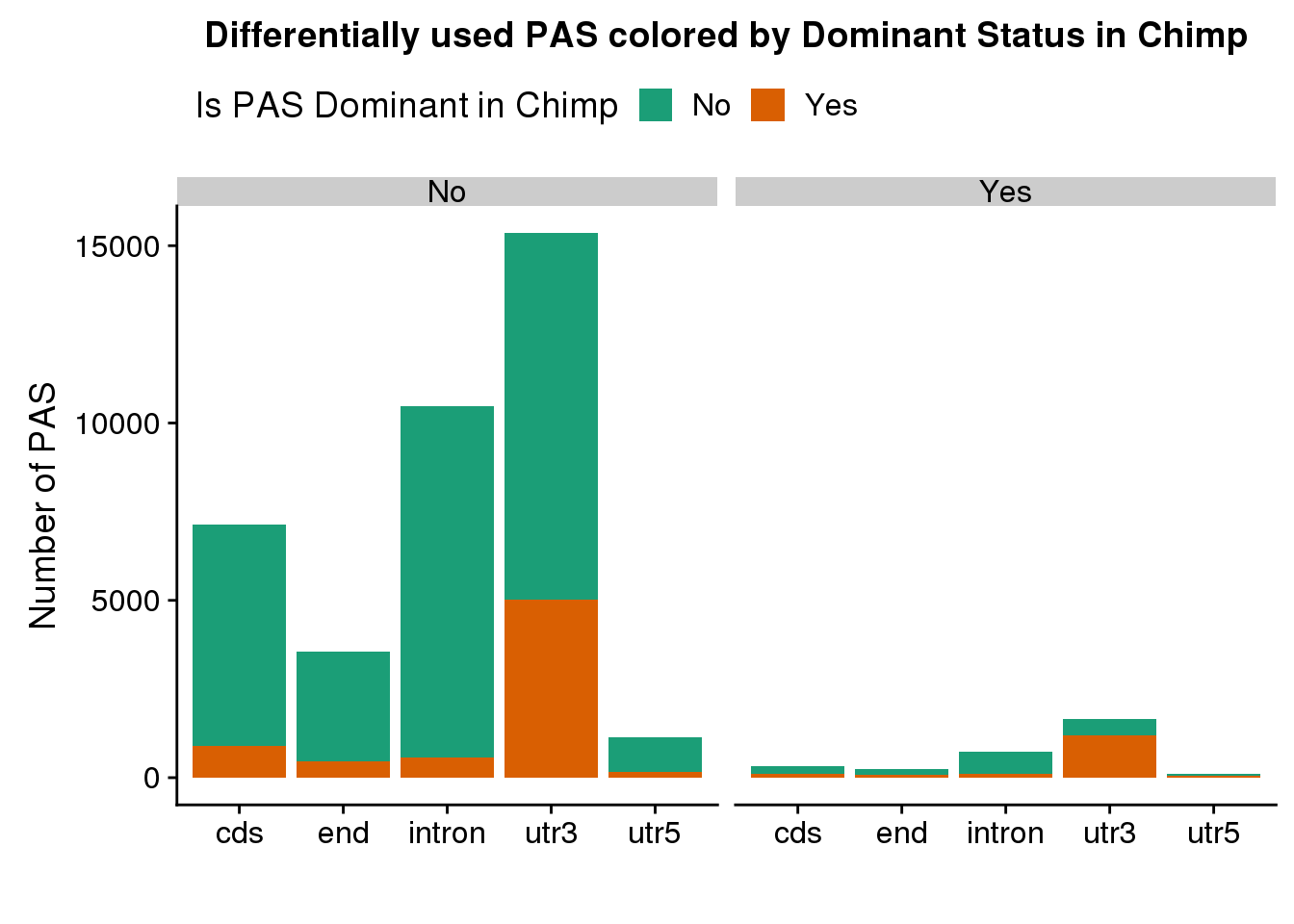
| Version | Author | Date |
|---|---|---|
| 86238eb | brimittleman | 2020-03-16 |
ggplot(DiffIsowDom,aes(x=loc,fill=HumanDom)) + geom_bar(stat = "count") + facet_grid(~SigPAU2) +labs(x="", y="Number of PAS", title="Differentially used PAS colored by Dominant Status in Human")+ scale_fill_brewer(palette = "Dark2",name="Is PAS Dominant in Chimp") + theme(legend.position = "top")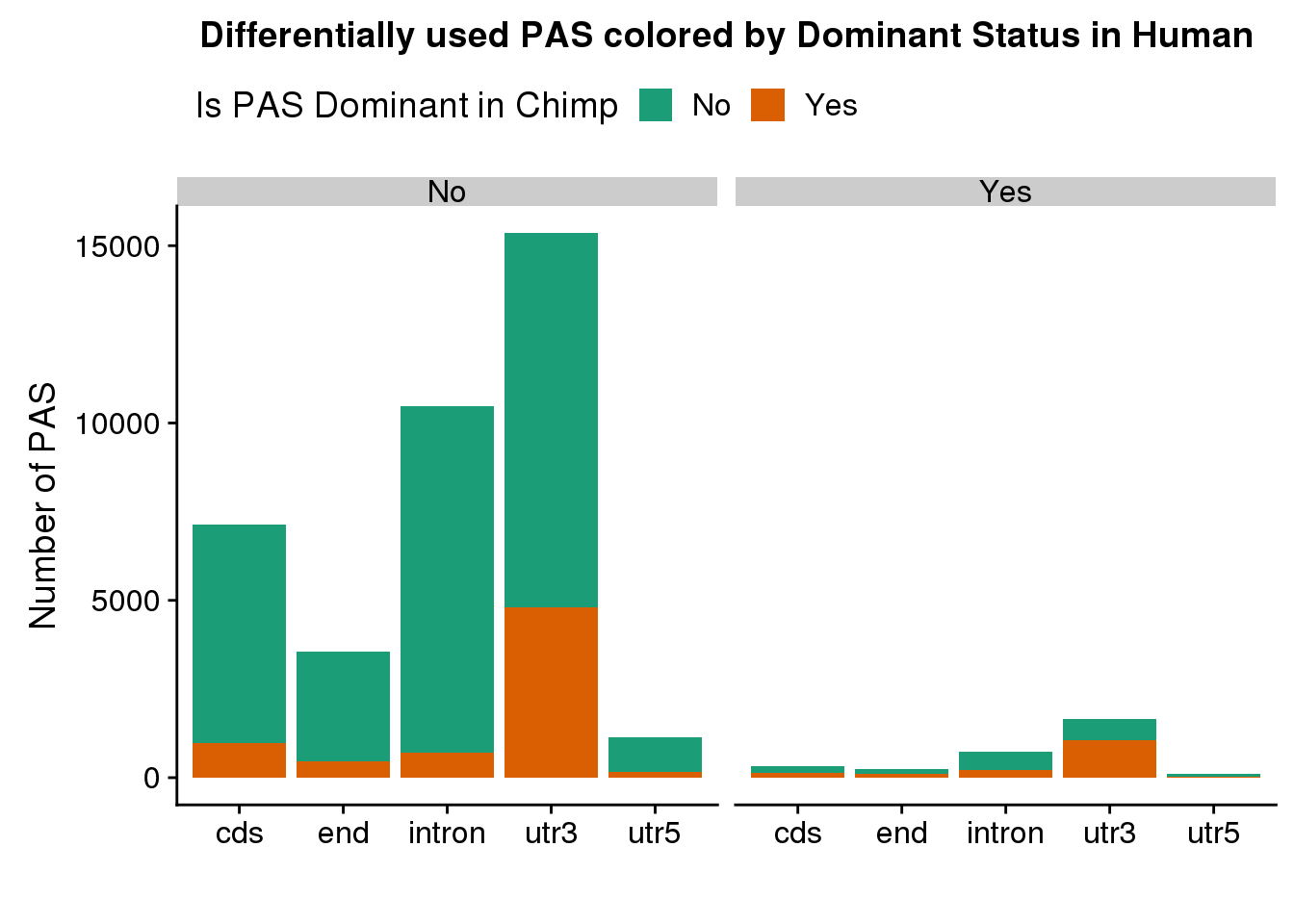
| Version | Author | Date |
|---|---|---|
| 86238eb | brimittleman | 2020-03-16 |
Of the 3076 how many are in the dominant set.
I need to use the set that includes ties here:
SiginChimp_Dom2= Chimp_Dom2 %>% ungroup %>% select(PAS) %>% inner_join(DiffIso_sig, by="PAS")Warning: Column `PAS` joining factor and character vector, coercing into
character vectornrow(SiginChimp_Dom2)[1] 1549SiginHuman_Dom2= Human_Dom2 %>% ungroup %>% select(PAS) %>% inner_join(DiffIso_sig, by="PAS")Warning: Column `PAS` joining factor and character vector, coercing into
character vectornrow(SiginHuman_Dom2)[1] 1514Enrichemnt:
EnrichChimp=c()
PvalueChimp=c()
for (i in c('cds', 'end', 'intron', 'utr3', 'utr5')){
x=nrow(DiffIsowDom %>% filter(ChimpDom=="Yes", SigPAU2=="Yes", loc==i))
m=nrow(DiffIsowDom %>% filter(SigPAU2=="Yes", loc==i))
n=nrow(DiffIsowDom %>% filter(SigPAU2=="No", loc==i))
k=nrow(DiffIsowDom %>% filter(loc==i,ChimpDom=="Yes"))
N=nrow(DiffIsowDom %>% filter(loc==i))
PvalueChimp=c(PvalueChimp, phyper(x,m,n,k,lower.tail=F))
enrich=(x/k)/(m/N)
EnrichChimp=c(EnrichChimp, round(enrich,2))
}
EnrichHuman=c()
PvalueHuman=c()
for (i in c('cds', 'end', 'intron', 'utr3', 'utr5')){
x=nrow(DiffIsowDom %>% filter(HumanDom=="Yes", SigPAU2=="Yes", loc==i))
m=nrow(DiffIsowDom %>% filter(SigPAU2=="Yes", loc==i))
n=nrow(DiffIsowDom %>% filter(SigPAU2=="No", loc==i))
k=nrow(DiffIsowDom %>% filter(loc==i,HumanDom=="Yes"))
N=nrow(DiffIsowDom %>% filter(loc==i))
PvalueHuman=c(PvalueHuman, phyper(x,m,n,k,lower.tail=F))
enrich=(x/k)/(m/N)
EnrichHuman=c(EnrichHuman, round(enrich,2))
}
locations=c('cds', 'end', 'intron', 'utr3', 'utr5')
EnrichDF=as.data.frame(cbind(locations, EnrichChimp,EnrichHuman ))
colnames(EnrichDF)=c("GenicLoc", "Chimp", "Human")
EnrichDF GenicLoc Chimp Human
1 cds 2.64 2.5
2 end 2.51 2.92
3 intron 2.62 3.41
4 utr3 1.95 1.83
5 utr5 2.79 2.4Compare this result to a set of PAS by usage:
For this I will use dominant at 30%, I will then use, used at 30%
SiginChimp_Dom2_30= Chimp_Dom2 %>% ungroup%>% filter(Chimp>=.3) %>% select(PAS) %>% inner_join(DiffIso_sig, by="PAS")Warning: Column `PAS` joining factor and character vector, coercing into
character vectornrow(SiginChimp_Dom2)[1] 1549SiginHuman_Dom2_30= Human_Dom2 %>% ungroup %>% filter(Human>=.3) %>% select(PAS) %>% inner_join(DiffIso_sig, by="PAS")Warning: Column `PAS` joining factor and character vector, coercing into
character vectornrow(SiginHuman_Dom2)[1] 1514HumanUsed30= allPAS %>% filter(Human>=.3)
nrow(HumanUsed30)[1] 6896ChimpUsed30= allPAS %>% filter(Chimp>=.3)
nrow(ChimpUsed30)[1] 7456allPAS_anno= allPAS %>% select(PAS,loc) %>% mutate(SigDPAU=ifelse(PAS %in% DiffIso_sig$PAS, "Yes","No"), HumanUse30=ifelse(PAS %in% HumanUsed30$PAS, "Yes", "No"), ChimpUse30=ifelse(PAS %in% ChimpUsed30$PAS, "Yes","No"), DomHuman=ifelse(PAS %in% SiginHuman_Dom2_30$PAS, "Yes", "No"), DomChimp=ifelse(PAS %in%SiginChimp_Dom2_30$PAS, "Yes", "No" ))Enrichment of dominant in diff used human:
x=nrow(allPAS_anno %>% filter(DomHuman=="Yes", SigDPAU=="Yes"))
m=nrow(allPAS_anno %>% filter(SigDPAU=="Yes"))
n=nrow(allPAS_anno %>% filter(SigDPAU=="No"))
k=nrow(allPAS_anno %>% filter(DomHuman=="Yes"))
N=nrow(allPAS_anno)
phyper(x,m,n,k,lower.tail=F)[1] 0enrich=(x/k)/(m/N)
enrich[1] 13.75748Compare to human just used at 30%
x=nrow(allPAS_anno %>% filter(HumanUse30=="Yes", SigDPAU=="Yes"))
m=nrow(allPAS_anno %>% filter(SigDPAU=="Yes"))
n=nrow(allPAS_anno %>% filter(SigDPAU=="No"))
k=nrow(allPAS_anno %>% filter(HumanUse30=="Yes"))
N=nrow(allPAS_anno)
phyper(x,m,n,k,lower.tail=F)[1] 4.329756e-180enrich=(x/k)/(m/N)
enrich[1] 2.248358Enrichment of dominant in diff used chimp:
x=nrow(allPAS_anno %>% filter(DomChimp=="Yes", SigDPAU=="Yes"))
m=nrow(allPAS_anno %>% filter(SigDPAU=="Yes"))
n=nrow(allPAS_anno %>% filter(SigDPAU=="No"))
k=nrow(allPAS_anno %>% filter(DomChimp=="Yes"))
N=nrow(allPAS_anno)
phyper(x,m,n,k,lower.tail=F)[1] 0enrich=(x/k)/(m/N)
enrich[1] 13.75748Enrichment of used at 30%
x=nrow(allPAS_anno %>% filter(ChimpUse30=="Yes", SigDPAU=="Yes"))
m=nrow(allPAS_anno %>% filter(SigDPAU=="Yes"))
n=nrow(allPAS_anno %>% filter(SigDPAU=="No"))
k=nrow(allPAS_anno %>% filter(ChimpUse30=="Yes"))
N=nrow(allPAS_anno)
phyper(x,m,n,k,lower.tail=F)[1] 4.169482e-274enrich=(x/k)/(m/N)
enrich[1] 2.485424At the 30% cutoff in the domiant PAS has 13.8X enrichement and 30% used has 2.49X enrichment. Is this based on the dominant at 30% always being in the other set?
Do by location:
EnrichChimpDom=c()
PvalueChimpDom=c()
for (i in c('cds', 'end', 'intron', 'utr3', 'utr5')){
x=nrow(allPAS_anno %>% filter(DomChimp=="Yes", SigDPAU=="Yes", loc==i))
m=nrow(allPAS_anno %>% filter(SigDPAU=="Yes", loc==i))
n=nrow(allPAS_anno %>% filter(SigDPAU=="No", loc==i))
k=nrow(allPAS_anno %>% filter(loc==i,DomChimp=="Yes"))
N=nrow(allPAS_anno %>% filter(loc==i))
PvalueChimpDom=c(PvalueChimpDom, phyper(x,m,n,k,lower.tail=F))
enrich=(x/k)/(m/N)
EnrichChimpDom=c(EnrichChimpDom, round(enrich,2))
}
EnrichChimp30=c()
PvalueChimp30=c()
for (i in c('cds', 'end', 'intron', 'utr3', 'utr5')){
x=nrow(allPAS_anno %>% filter(ChimpUse30=="Yes", SigDPAU=="Yes", loc==i))
m=nrow(allPAS_anno %>% filter(SigDPAU=="Yes", loc==i))
n=nrow(allPAS_anno %>% filter(SigDPAU=="No", loc==i))
k=nrow(allPAS_anno %>% filter(loc==i,ChimpUse30=="Yes"))
N=nrow(allPAS_anno %>% filter(loc==i))
PvalueChimp30=c(PvalueChimp30, phyper(x,m,n,k,lower.tail=F))
enrich=(x/k)/(m/N)
EnrichChimp30=c(EnrichChimp30, round(enrich,2))
}EnrichHumanDom=c()
PvalueHumanDom=c()
for (i in c('cds', 'end', 'intron', 'utr3', 'utr5')){
x=nrow(allPAS_anno %>% filter(DomHuman=="Yes", SigDPAU=="Yes", loc==i))
m=nrow(allPAS_anno %>% filter(SigDPAU=="Yes", loc==i))
n=nrow(allPAS_anno %>% filter(SigDPAU=="No", loc==i))
k=nrow(allPAS_anno %>% filter(loc==i,DomHuman=="Yes"))
N=nrow(allPAS_anno %>% filter(loc==i))
PvalueHumanDom=c(PvalueHumanDom, phyper(x,m,n,k,lower.tail=F))
enrich=(x/k)/(m/N)
EnrichHumanDom=c(EnrichHumanDom, round(enrich,2))
}
EnrichHuman30=c()
PvalueHuman30=c()
for (i in c('cds', 'end', 'intron', 'utr3', 'utr5')){
x=nrow(allPAS_anno %>% filter(HumanUse30=="Yes", SigDPAU=="Yes", loc==i))
m=nrow(allPAS_anno %>% filter(SigDPAU=="Yes", loc==i))
n=nrow(allPAS_anno %>% filter(SigDPAU=="No", loc==i))
k=nrow(allPAS_anno %>% filter(loc==i,HumanUse30=="Yes"))
N=nrow(allPAS_anno %>% filter(loc==i))
PvalueHuman30=c(PvalueHuman30, phyper(x,m,n,k,lower.tail=F))
enrich=(x/k)/(m/N)
EnrichHuman30=c(EnrichHuman30, round(enrich,2))
}EnrichDataDFHuman=as.data.frame(cbind(loc=c('cds', 'end', 'intron', 'utr3', 'utr5'), Used30= EnrichHuman30,Dominant=EnrichHumanDom)) %>% gather("Set", "Enrichment", -loc)%>% mutate(species="Human")Warning: attributes are not identical across measure variables;
they will be droppedEnrichDataDFChimp=as.data.frame(cbind(loc=c('cds', 'end', 'intron', 'utr3', 'utr5'), Used30= EnrichChimp30,Dominant=EnrichChimpDom)) %>% gather("Set", "Enrichment", -loc) %>% mutate(species="Chimp")Warning: attributes are not identical across measure variables;
they will be droppedEnrichDataDFBoth= EnrichDataDFHuman %>% bind_rows(EnrichDataDFChimp)
EnrichDataDFBoth$Enrichment=as.numeric(EnrichDataDFBoth$Enrichment)
EnrichDataDFBoth$Set=as.factor(EnrichDataDFBoth$Set)
ggplot(EnrichDataDFBoth, aes(x=loc, y=Enrichment, by=Set, fill=Set)) + geom_bar(stat="identity", position="dodge") + geom_hline(yintercept = 1) + facet_grid(~species)+ scale_fill_brewer(palette = "Dark2",name="") + theme(legend.position = "bottom") + labs(x="Genic Location", title="Dominant PAS are more enriched for dPAS \nthan all PAS used at same level") 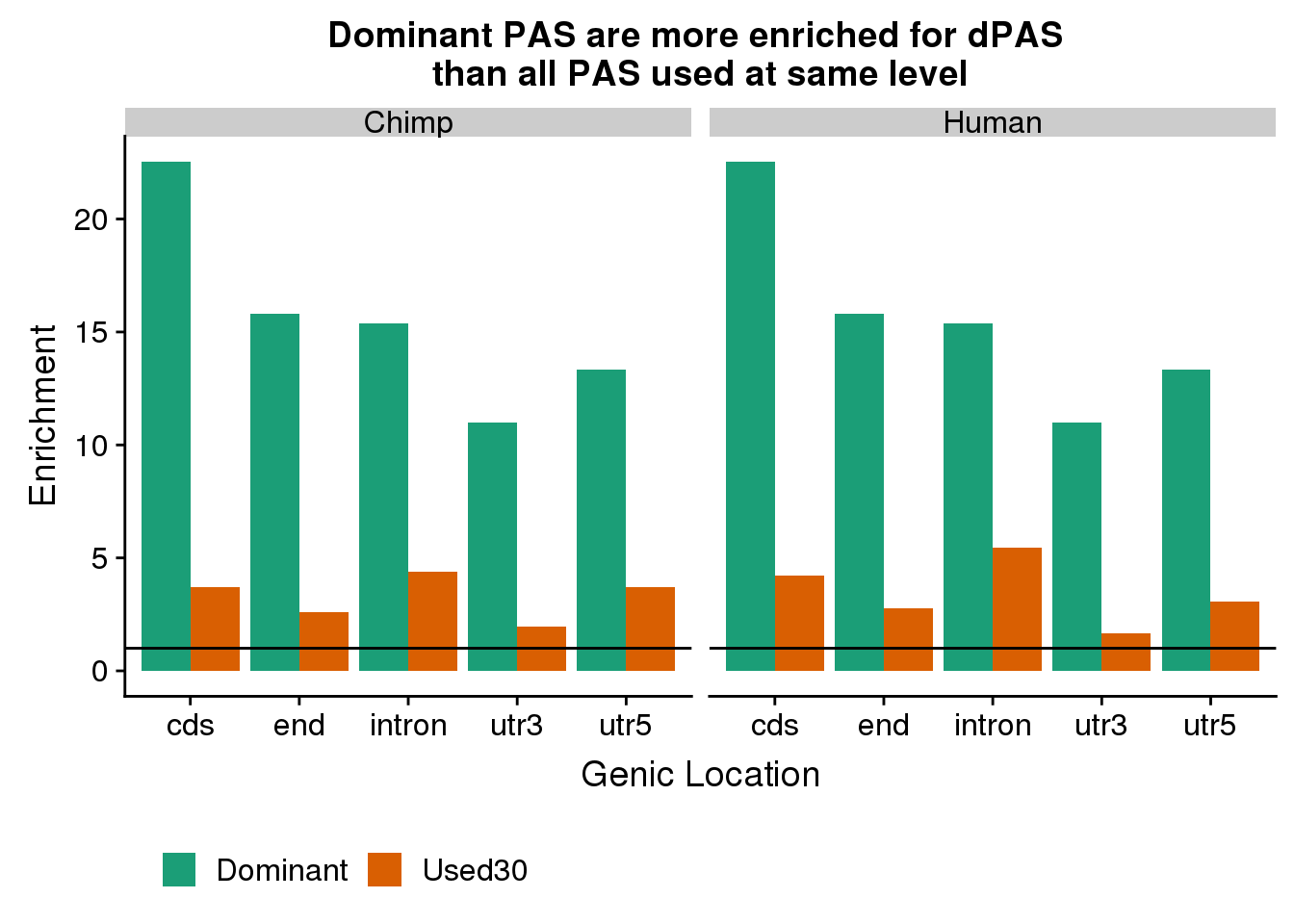
test opposite
EnrichChimpDomOpp=c()
for (i in c('cds', 'end', 'intron', 'utr3', 'utr5')){
x=nrow(allPAS_anno %>% filter(DomChimp=="Yes", SigDPAU=="Yes", loc==i))
m=nrow(allPAS_anno %>% filter(DomChimp=="Yes", loc==i))
n=nrow(allPAS_anno %>% filter(DomChimp=="No", loc==i))
k=nrow(allPAS_anno %>% filter(loc==i,SigDPAU=="Yes"))
N=nrow(allPAS_anno %>% filter(loc==i))
enrich=(x/k)/(m/N)
EnrichChimpDomOpp=c(EnrichChimpDomOpp, round(enrich,2))
}
EnrichChimpDomOpp[1] 22.54 15.80 15.40 10.98 13.32EnrichChimpDom[1] 22.54 15.80 15.40 10.98 13.32ok test. its the same.
##diff dominance and discovery
DiffDom
SamDomLoc=SameDom %>% ungroup() %>% select(ChimpPAS) %>% rename("PAS"=ChimpPAS) %>% inner_join(allPAS, by="PAS")
ggplot(SamDomLoc, aes(x=disc, fill=disc)) +geom_bar(stat="count") + labs(title="Same dominant")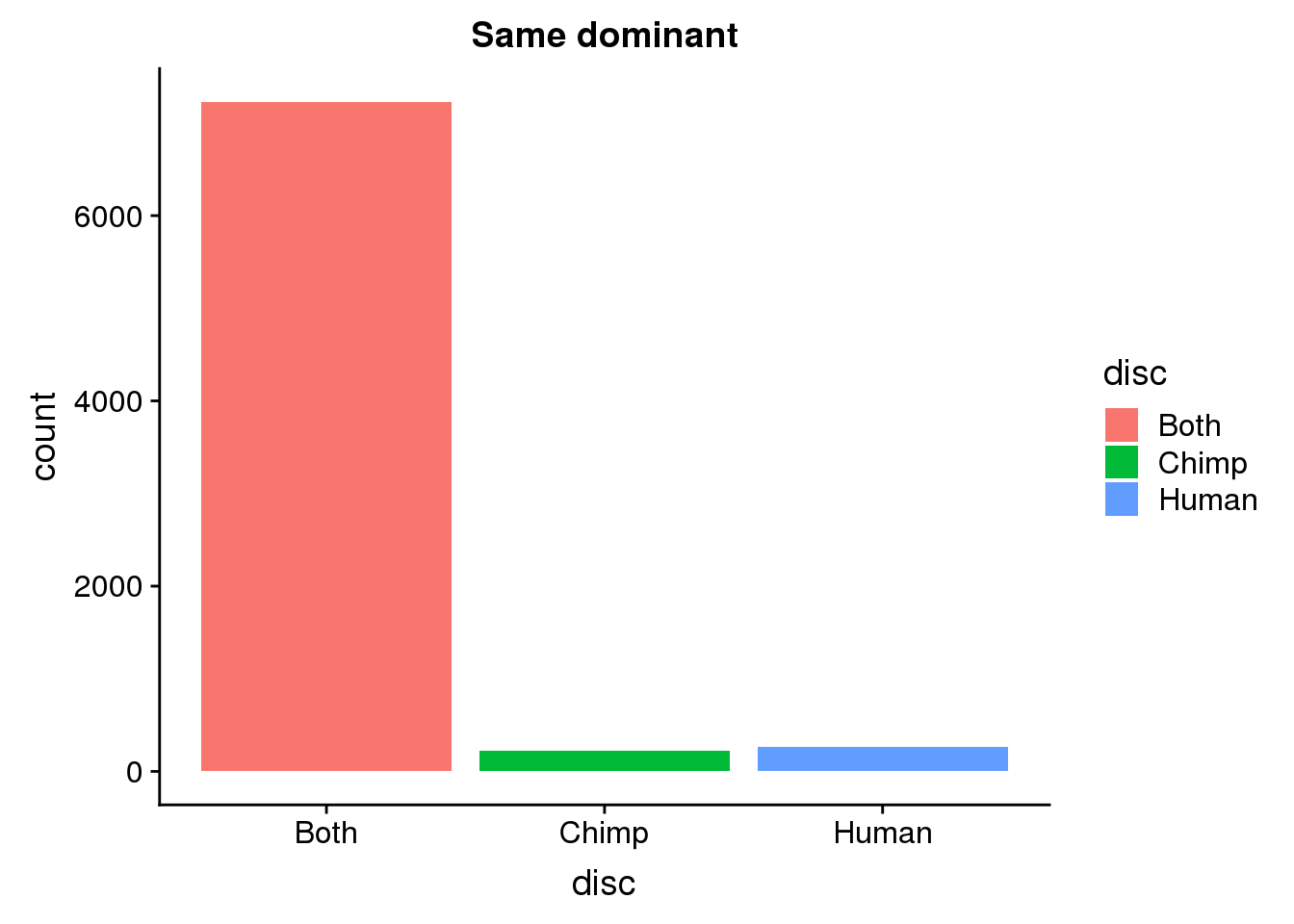
DiffDomChimp= DiffDom %>% ungroup() %>% select(ChimpPAS) %>% rename("PAS"=ChimpPAS) %>% inner_join(allPAS, by="PAS")
ggplot(DiffDomChimp, aes(x=disc, fill=disc)) +geom_bar(stat="count") + labs(title="Different dominant, Chimp PAS")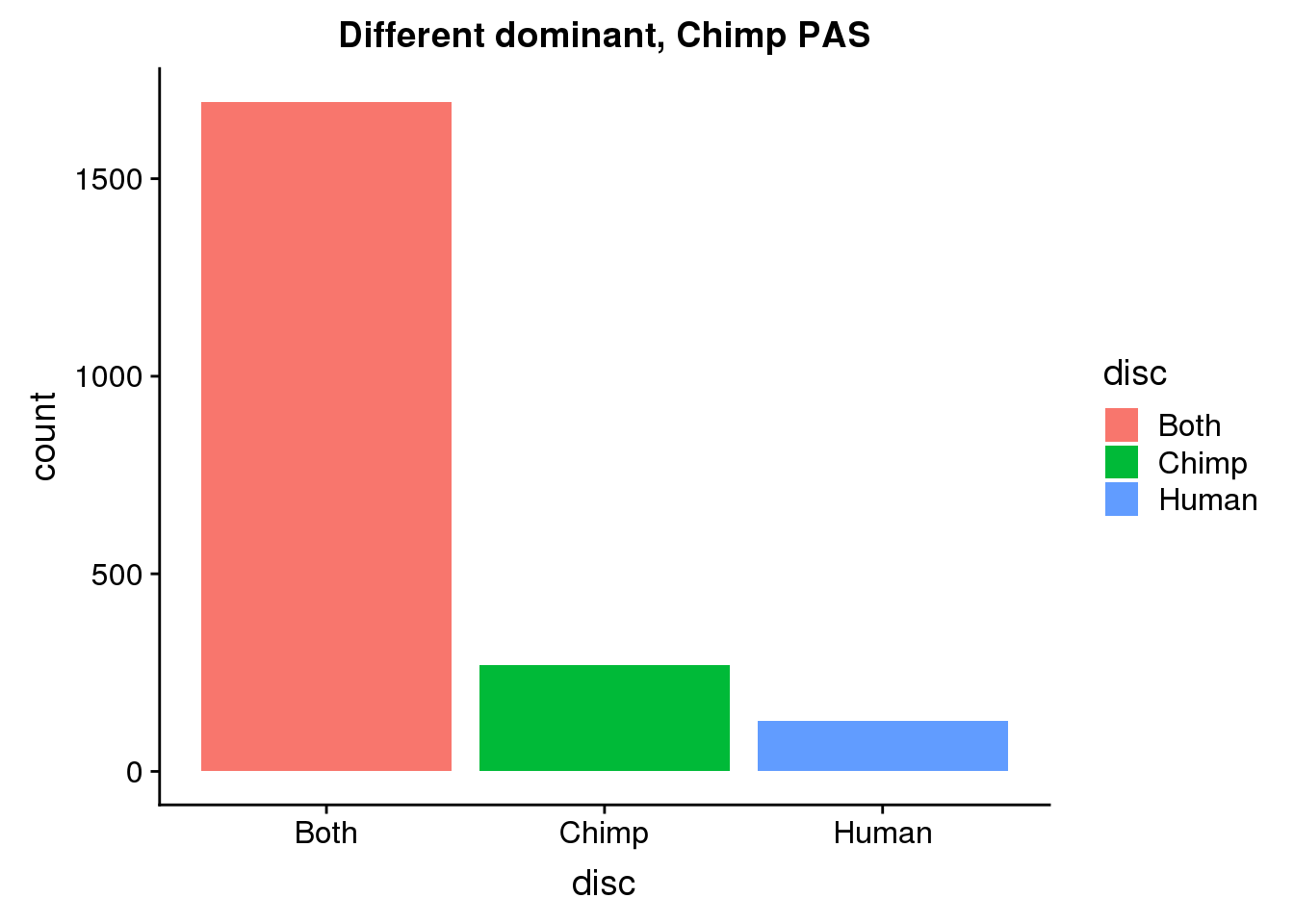
DiffDomHuman= DiffDom %>% ungroup() %>% select(HumanPAS) %>% rename("PAS"=HumanPAS) %>% inner_join(allPAS, by="PAS")
ggplot(DiffDomHuman, aes(x=disc, fill=disc)) +geom_bar(stat="count") + labs(title="Different dominant, Human PAS")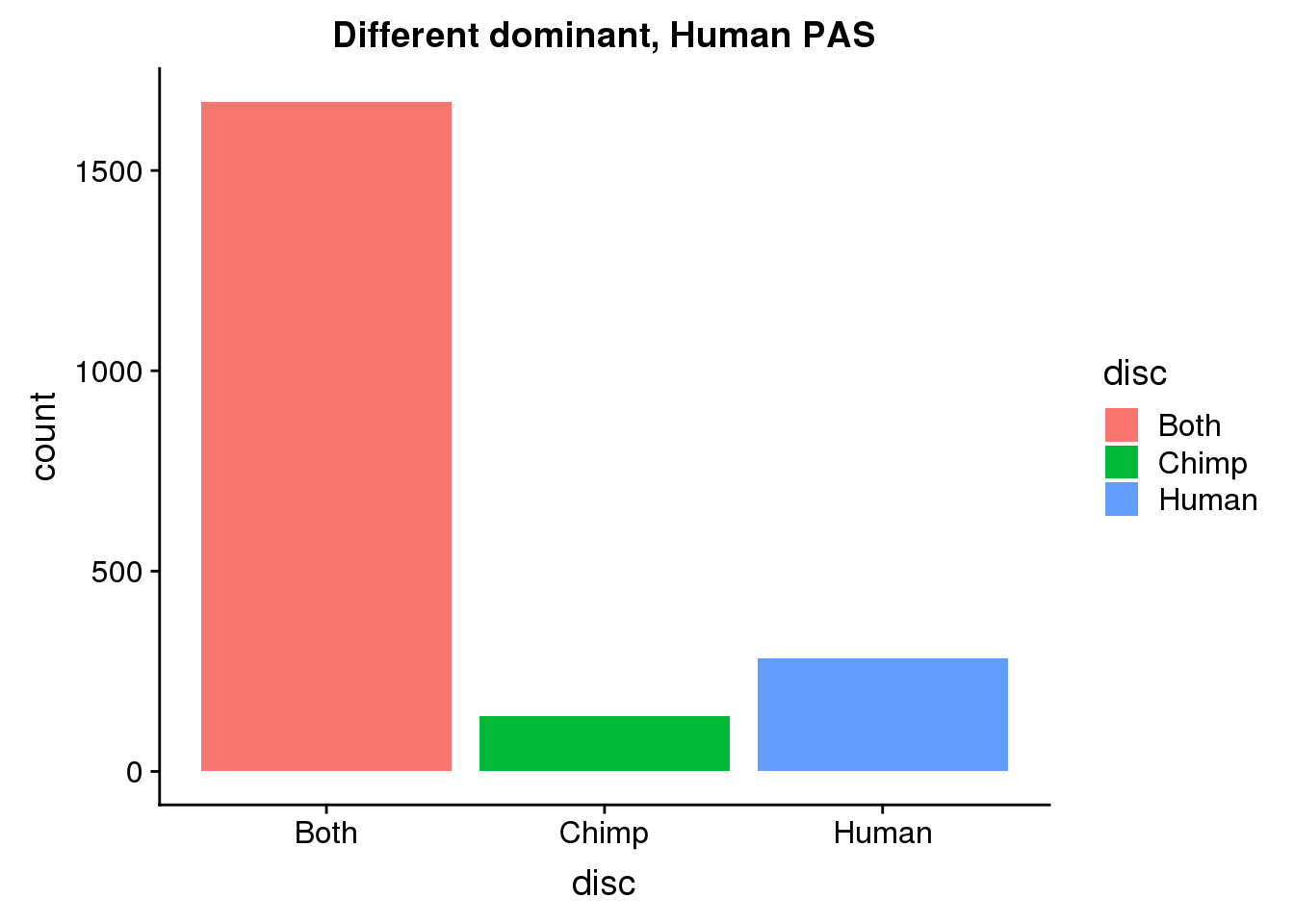
#DiffDomDiffLoc
DiffDomDiffLocChimp= DiffDomDiffLoc %>% ungroup() %>% select(ChimpPAS) %>% rename("PAS"=ChimpPAS) %>% inner_join(allPAS, by="PAS")
ggplot(DiffDomDiffLocChimp, aes(x=disc, fill=disc)) +geom_bar(stat="count") + labs(title="Different dominant, different Loc, Chimp PAS")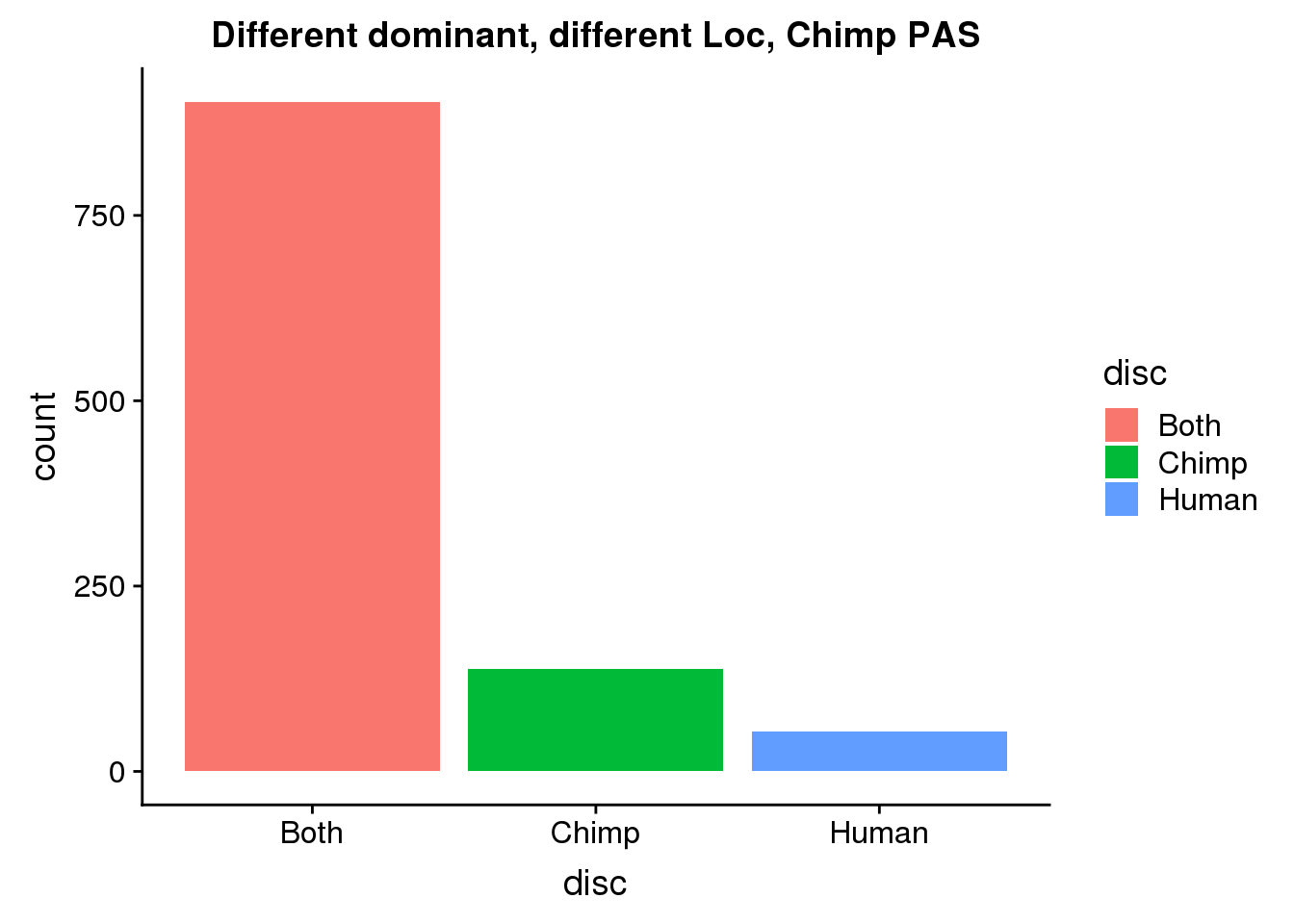
DiffDomDiffLocHuman= DiffDomDiffLoc %>% ungroup() %>% select(HumanPAS) %>% rename("PAS"=HumanPAS) %>% inner_join(allPAS, by="PAS")
ggplot(DiffDomDiffLocHuman, aes(x=disc, fill=disc)) +geom_bar(stat="count") + labs(title="Different dominant, different Loc, Human PAS")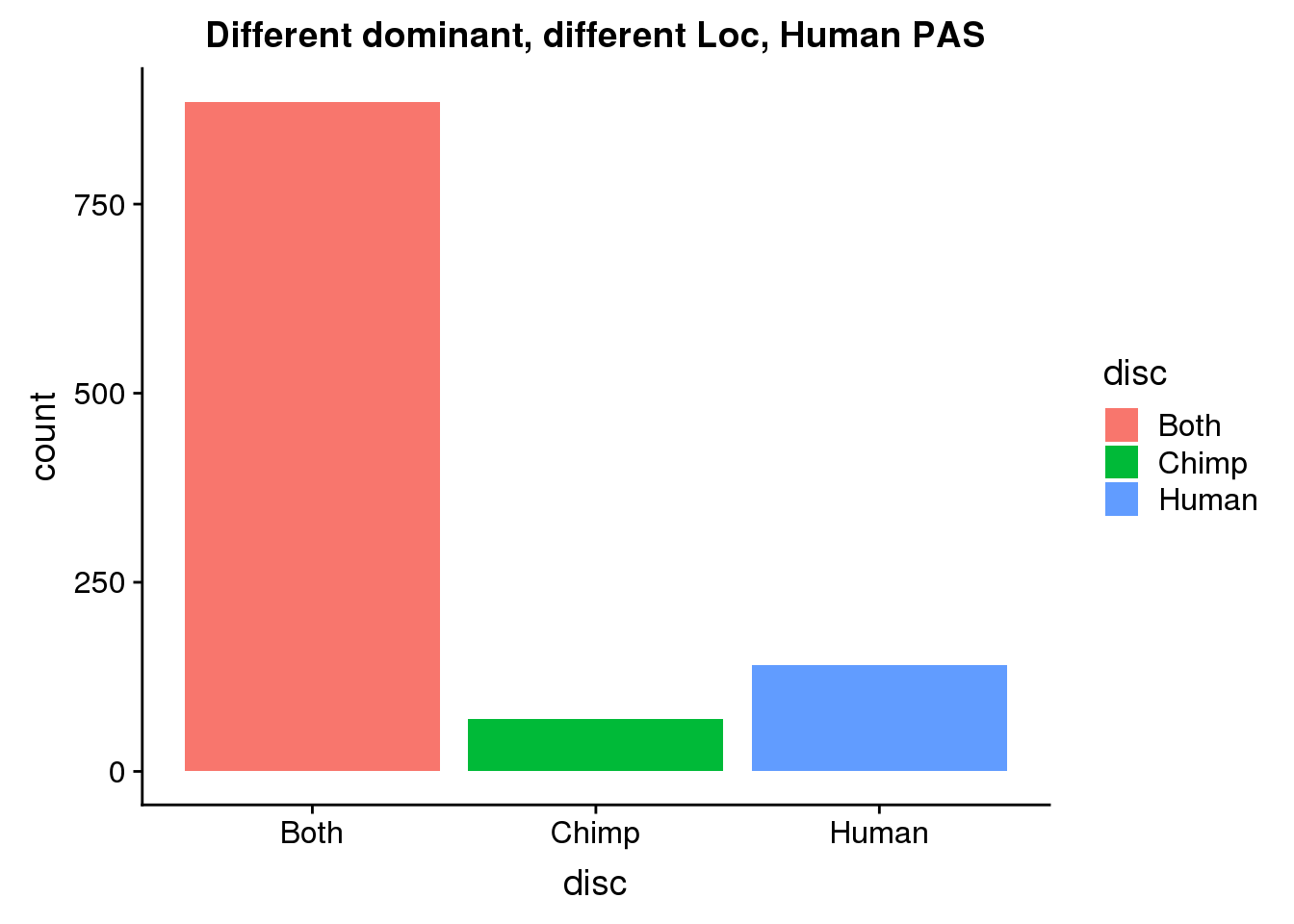
Does this go away if we say has to be 50%
DiffDomChimp50= BothDom_50_same %>% ungroup() %>% select(ChimpPAS) %>% rename("PAS"=ChimpPAS) %>% inner_join(allPAS, by="PAS")
ggplot(DiffDomChimp50, aes(x=disc, fill=disc)) +geom_bar(stat="count") + labs(title="Different dominant, Chimp PAS")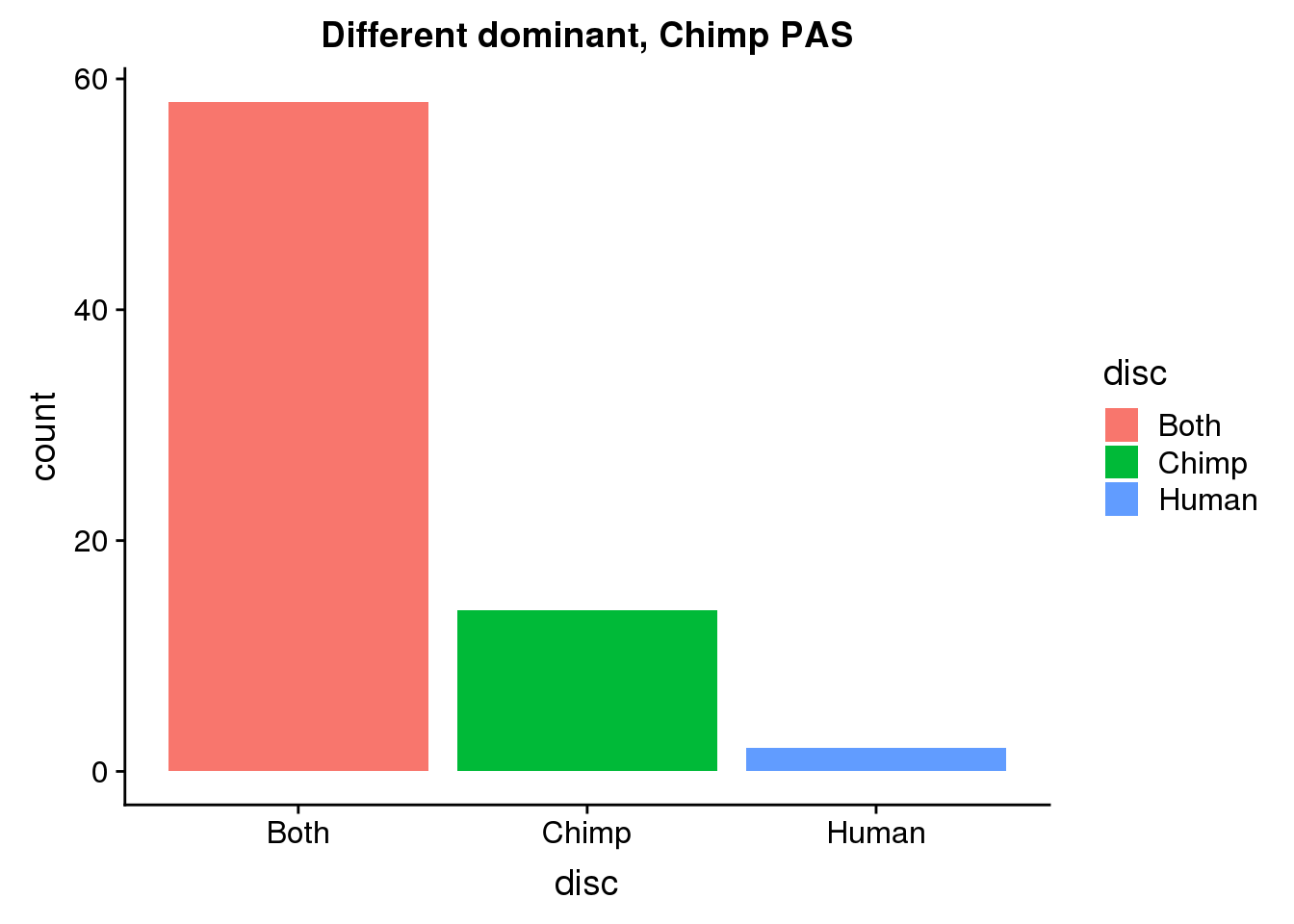
DiffDomHuman50= BothDom_50_same %>% ungroup() %>% select(HumanPAS) %>% rename("PAS"=HumanPAS) %>% inner_join(allPAS, by="PAS")
ggplot(DiffDomHuman50, aes(x=disc, fill=disc)) +geom_bar(stat="count") + labs(title="Different dominant, Human PAS")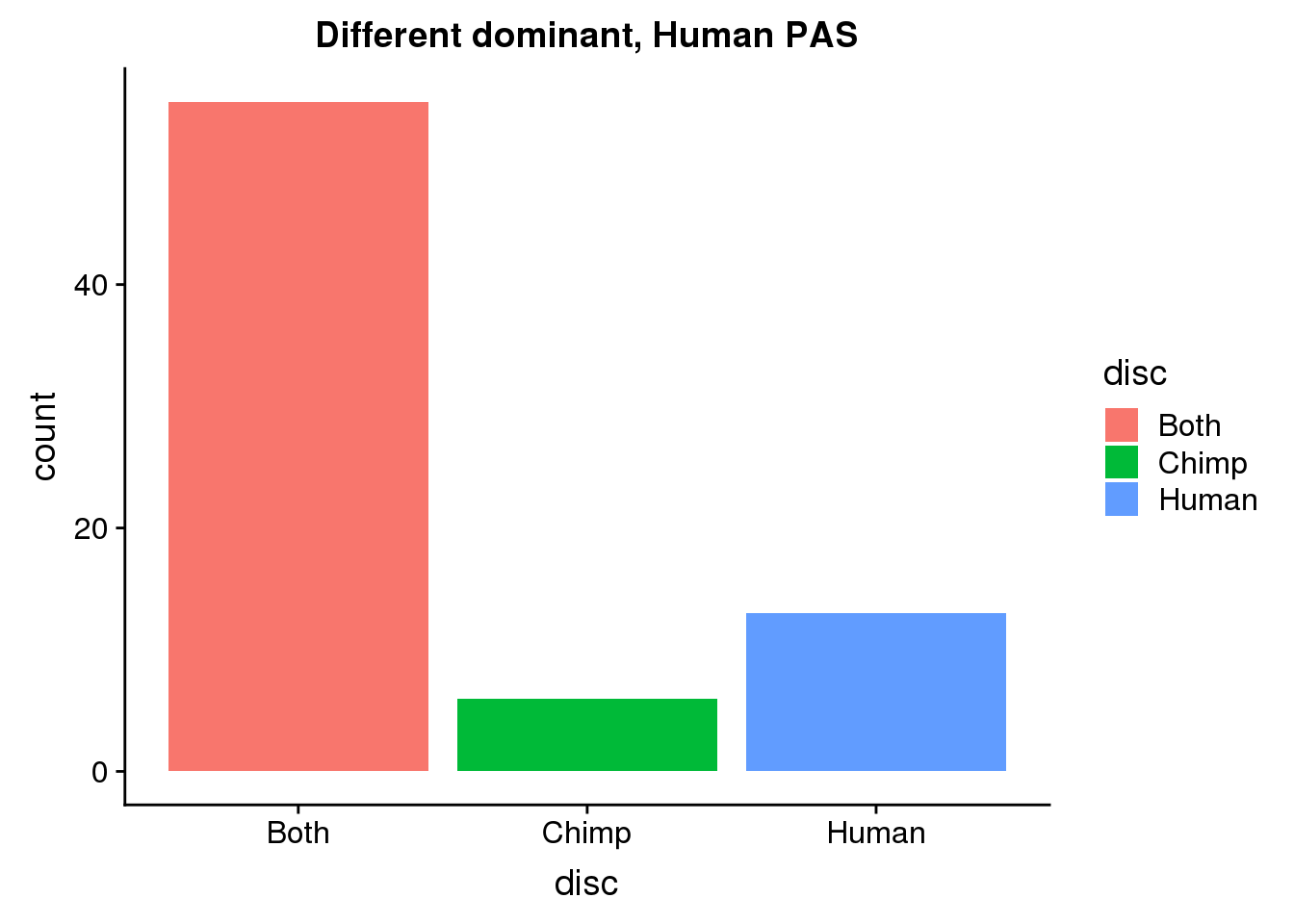
Problems:
- snorna (need to be removed)
- mispriming?
- why not disovered in the right species
sessionInfo()R version 3.5.1 (2018-07-02)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] cowplot_0.9.4 workflowr_1.6.0 forcats_0.3.0 stringr_1.3.1
[5] dplyr_0.8.0.1 purrr_0.3.2 readr_1.3.1 tidyr_0.8.3
[9] tibble_2.1.1 ggplot2_3.1.1 tidyverse_1.2.1
loaded via a namespace (and not attached):
[1] tidyselect_0.2.5 reshape2_1.4.3 haven_1.1.2
[4] lattice_0.20-38 colorspace_1.3-2 generics_0.0.2
[7] htmltools_0.3.6 yaml_2.2.0 rlang_0.4.0
[10] later_0.7.5 pillar_1.3.1 glue_1.3.0
[13] withr_2.1.2 RColorBrewer_1.1-2 modelr_0.1.2
[16] readxl_1.1.0 plyr_1.8.4 munsell_0.5.0
[19] gtable_0.2.0 cellranger_1.1.0 rvest_0.3.2
[22] evaluate_0.12 labeling_0.3 knitr_1.20
[25] httpuv_1.4.5 broom_0.5.1 Rcpp_1.0.2
[28] promises_1.0.1 scales_1.0.0 backports_1.1.2
[31] jsonlite_1.6 fs_1.3.1 hms_0.4.2
[34] digest_0.6.18 stringi_1.2.4 grid_3.5.1
[37] rprojroot_1.3-2 cli_1.1.0 tools_3.5.1
[40] magrittr_1.5 lazyeval_0.2.1 crayon_1.3.4
[43] whisker_0.3-2 pkgconfig_2.0.2 xml2_1.2.0
[46] lubridate_1.7.4 assertthat_0.2.0 rmarkdown_1.10
[49] httr_1.3.1 rstudioapi_0.10 R6_2.3.0
[52] nlme_3.1-137 git2r_0.26.1 compiler_3.5.1