Ortho Exon evaluation as an annotation
Briana Mittleman
3/17/2020
Last updated: 2020-03-31
Checks: 7 0
Knit directory: Comparative_APA/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190902) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: code/chimp_log/
Ignored: code/human_log/
Ignored: data/.DS_Store
Ignored: data/TrialFiltersMeta.txt.sb-9845453e-R58Y0Q/
Ignored: data/mediation_prot/
Ignored: data/metadata_HCpanel.txt.sb-a5794dd2-i594qs/
Ignored: output/.DS_Store
Untracked files:
Untracked: ._.DS_Store
Untracked: Chimp/
Untracked: Human/
Untracked: analysis/CrossChimpThreePrime.Rmd
Untracked: analysis/DiffTransProtvsExpression.Rmd
Untracked: analysis/DiffUsedUTR.Rmd
Untracked: analysis/GvizPlots.Rmd
Untracked: analysis/HandC.TvN
Untracked: analysis/PhenotypeOverlap10.Rmd
Untracked: analysis/annotationBias.Rmd
Untracked: analysis/assessReadQual.Rmd
Untracked: analysis/diffExpressionPantro6.Rmd
Untracked: analysis/pol2.Rmd
Untracked: code/._BothFCMM.sh
Untracked: code/._BothFCMMPrim.sh
Untracked: code/._BothFCnewOInclusive.sh
Untracked: code/._ChimpStarMM2.sh
Untracked: code/._ClassifyLeafviz.sh
Untracked: code/._ClosestorthoEx.sh
Untracked: code/._Config_chimp.yaml
Untracked: code/._Config_chimp_full.yaml
Untracked: code/._Config_human.yaml
Untracked: code/._ConvertJunc2Bed.sh
Untracked: code/._CountNucleotides.py
Untracked: code/._CrossMapChimpRNA.sh
Untracked: code/._CrossMapThreeprime.sh
Untracked: code/._DiffSplice.sh
Untracked: code/._DiffSplicePlots.sh
Untracked: code/._DiffSplicePlots_gencode.sh
Untracked: code/._DiffSplice_gencode.sh
Untracked: code/._DiffSplice_removebad.sh
Untracked: code/._Filter255MM.sh
Untracked: code/._FilterPrimSec.sh
Untracked: code/._FindIntronForDomPAS.sh
Untracked: code/._FindIntronForDomPAS_DF.sh
Untracked: code/._GetMAPQscore.py
Untracked: code/._GetSecondaryMap.py
Untracked: code/._Lift5perPAS.sh
Untracked: code/._LiftFinalChimpJunc2Human.sh
Untracked: code/._LiftOrthoPAS2chimp.sh
Untracked: code/._MapBadSamples.sh
Untracked: code/._MismatchNumbers.sh
Untracked: code/._PAS_ATTAAA.sh
Untracked: code/._PAS_ATTAAA_df.sh
Untracked: code/._PAS_seqExpanded.sh
Untracked: code/._PASsequences.sh
Untracked: code/._PASsequences_DF.sh
Untracked: code/._PlotNuclearUsagebySpecies.R
Untracked: code/._PlotNuclearUsagebySpecies_DF.R
Untracked: code/._QuantMergedClusters.sh
Untracked: code/._RNATranscriptDTplot.sh
Untracked: code/._ReverseLiftFilter.R
Untracked: code/._RunFixLeafCluster.sh
Untracked: code/._RunNegMCMediation.sh
Untracked: code/._RunNegMCMediationDF.sh
Untracked: code/._RunPosMCMediationDF.err
Untracked: code/._RunPosMCMediationDF.sh
Untracked: code/._SAF2Bed.py
Untracked: code/._Snakefile
Untracked: code/._SnakefilePAS
Untracked: code/._SnakefilePASfilt
Untracked: code/._SortIndexBadSamples.sh
Untracked: code/._StarMM2.sh
Untracked: code/._TestFC.sh
Untracked: code/._assignPeak2Intronicregion
Untracked: code/._assignPeak2Intronicregion.sh
Untracked: code/._bed215upbed.py
Untracked: code/._bed2Bedbothstrand.py
Untracked: code/._bed2SAF_gen.py
Untracked: code/._buildIndecpantro5
Untracked: code/._buildIndecpantro5.sh
Untracked: code/._buildLeafviz.sh
Untracked: code/._buildLeafviz_leadAnno.sh
Untracked: code/._buildStarIndex.sh
Untracked: code/._chimpChromprder.sh
Untracked: code/._chimpMultiCov.sh
Untracked: code/._chimpMultiCov255.sh
Untracked: code/._chimpMultiCovInclusive.sh
Untracked: code/._chooseSignalSite.py
Untracked: code/._cleanbed2saf.py
Untracked: code/._cluster.json
Untracked: code/._cluster2bed.py
Untracked: code/._clusterLiftReverse.sh
Untracked: code/._clusterLiftReverse_removebad.sh
Untracked: code/._clusterLiftprimary.sh
Untracked: code/._clusterLiftprimary_removebad.sh
Untracked: code/._converBam2Junc.sh
Untracked: code/._converBam2Junc_removeBad.sh
Untracked: code/._extraSnakefiltpas
Untracked: code/._extractPhyloReg.py
Untracked: code/._extractPhyloRegGene.py
Untracked: code/._extractPhylopGeneral.ph
Untracked: code/._extractPhylopGeneral.py
Untracked: code/._extractPhylopReg200down.py
Untracked: code/._extractPhylopReg200up.py
Untracked: code/._filter5percPAS.py
Untracked: code/._filterNumChroms.py
Untracked: code/._filterPASforMP.py
Untracked: code/._filterPostLift.py
Untracked: code/._filterPrimaryread.py
Untracked: code/._filterSecondaryread.py
Untracked: code/._fixExonFC.py
Untracked: code/._fixFCheadforExp.py
Untracked: code/._fixLeafCluster.py
Untracked: code/._fixLiftedJunc.py
Untracked: code/._fixUTRexonanno.py
Untracked: code/._formathg38Anno.py
Untracked: code/._formatpantro6Anno.py
Untracked: code/._getRNAseqMapStats.sh
Untracked: code/._hg19MapStats.sh
Untracked: code/._humanChromorder.sh
Untracked: code/._humanMultiCov.sh
Untracked: code/._humanMultiCov255.sh
Untracked: code/._humanMultiCov_inclusive.sh
Untracked: code/._intersectLiftedPAS.sh
Untracked: code/._liftJunctionFiles.sh
Untracked: code/._liftPAS19to38.sh
Untracked: code/._liftedchimpJunc2human.sh
Untracked: code/._makeNuclearDapaplots.sh
Untracked: code/._makeNuclearDapaplots_DF.sh
Untracked: code/._makeSamplyGroupsHuman_TvN.py
Untracked: code/._mapRNAseqhg19.sh
Untracked: code/._mapRNAseqhg19_newPipeline.sh
Untracked: code/._maphg19.sh
Untracked: code/._maphg19_subjunc.sh
Untracked: code/._mediation_test.R
Untracked: code/._mergeChimp3prime_inhg38.sh
Untracked: code/._mergeandBWRNAseq.sh
Untracked: code/._mergedBam2BW.sh
Untracked: code/._nameClusters.py
Untracked: code/._negativeMediation_montecarlo.R
Untracked: code/._negativeMediation_montecarloDF.R
Untracked: code/._numMultimap.py
Untracked: code/._overlapMMandOrthoexon.sh
Untracked: code/._overlapPASandOrthoexon.sh
Untracked: code/._overlapapaQTLPAS.sh
Untracked: code/._parseHg38.py
Untracked: code/._postiveMediation_montecarlo_DF.R
Untracked: code/._prepareCleanLiftedFC_5perc4LC.py
Untracked: code/._prepareLeafvizAnno.sh
Untracked: code/._preparePAS4lift.py
Untracked: code/._primaryLift.sh
Untracked: code/._processhg38exons.py
Untracked: code/._quantJunc.sh
Untracked: code/._quantJunc_TEST.sh
Untracked: code/._quantJunc_removeBad.sh
Untracked: code/._quantLiftedPASPrimary.sh
Untracked: code/._quantMerged_seperatly.sh
Untracked: code/._recLiftchim2human.sh
Untracked: code/._revLiftPAShg38to19.sh
Untracked: code/._reverseLift.sh
Untracked: code/._runCheckReverseLift.sh
Untracked: code/._runChimpDiffIso.sh
Untracked: code/._runCountNucleotides.sh
Untracked: code/._runFilterNumChroms.sh
Untracked: code/._runHumanDiffIso.sh
Untracked: code/._runNuclearDiffIso_DF.sh
Untracked: code/._runNuclearDifffIso.sh
Untracked: code/._runTotalDiffIso.sh
Untracked: code/._run_chimpverifybam.sh
Untracked: code/._run_verifyBam.sh
Untracked: code/._snakemake.batch
Untracked: code/._snakemakePAS.batch
Untracked: code/._snakemakePASchimp.batch
Untracked: code/._snakemakePAShuman.batch
Untracked: code/._snakemake_chimp.batch
Untracked: code/._snakemake_human.batch
Untracked: code/._snakemakefiltPAS.batch
Untracked: code/._snakemakefiltPAS_chimp
Untracked: code/._snakemakefiltPAS_chimp.sh
Untracked: code/._snakemakefiltPAS_human.sh
Untracked: code/._spliceSite2Fasta.py
Untracked: code/._submit-snakemake-chimp.sh
Untracked: code/._submit-snakemake-human.sh
Untracked: code/._submit-snakemakePAS-chimp.sh
Untracked: code/._submit-snakemakePAS-human.sh
Untracked: code/._submit-snakemakefiltPAS-chimp.sh
Untracked: code/._submit-snakemakefiltPAS-human.sh
Untracked: code/._subset_diffisopheno_Nuclear_HvC.py
Untracked: code/._subset_diffisopheno_Nuclear_HvC_DF.py
Untracked: code/._subset_diffisopheno_Total_HvC.py
Untracked: code/._threeprimeOrthoFC.sh
Untracked: code/._transcriptDTplotsNuclear.sh
Untracked: code/._verifyBam4973.sh
Untracked: code/._verifyBam4973inHuman.sh
Untracked: code/._wrap_chimpverifybam.sh
Untracked: code/._wrap_verifyBam.sh
Untracked: code/._writeMergecode.py
Untracked: code/.snakemake/
Untracked: code/BothFCMM.err
Untracked: code/BothFCMM.out
Untracked: code/BothFCMM.sh
Untracked: code/BothFCMMPrim.err
Untracked: code/BothFCMMPrim.out
Untracked: code/BothFCMMPrim.sh
Untracked: code/BothFCnewOInclusive.sh
Untracked: code/BothFCnewOInclusive.sh.err
Untracked: code/BothFCnewOInclusive.sh.out
Untracked: code/ChimpStarMM2.err
Untracked: code/ChimpStarMM2.out
Untracked: code/ChimpStarMM2.sh
Untracked: code/ClassifyLeafviz.sh
Untracked: code/ClosestorthoEx.err
Untracked: code/ClosestorthoEx.out
Untracked: code/ClosestorthoEx.sh
Untracked: code/Config_chimp.yaml
Untracked: code/Config_chimp_full.yaml
Untracked: code/Config_human.yaml
Untracked: code/ConvertJunc2Bed.err
Untracked: code/ConvertJunc2Bed.out
Untracked: code/ConvertJunc2Bed.sh
Untracked: code/CountNucleotides.py
Untracked: code/CrossMapChimpRNA.sh
Untracked: code/CrossMapThreeprime.sh
Untracked: code/CrossmapChimp3prime.err
Untracked: code/CrossmapChimp3prime.out
Untracked: code/CrossmapChimpRNA.err
Untracked: code/CrossmapChimpRNA.out
Untracked: code/DiffSplice.err
Untracked: code/DiffSplice.out
Untracked: code/DiffSplice.sh
Untracked: code/DiffSplicePlots.err
Untracked: code/DiffSplicePlots.out
Untracked: code/DiffSplicePlots.sh
Untracked: code/DiffSplicePlots_gencode.sh
Untracked: code/DiffSplice_gencode.sh
Untracked: code/DiffSplice_removebad.err
Untracked: code/DiffSplice_removebad.out
Untracked: code/DiffSplice_removebad.sh
Untracked: code/Filter255.err
Untracked: code/Filter255.out
Untracked: code/Filter255MM.sh
Untracked: code/FilterPrimSec.err
Untracked: code/FilterPrimSec.out
Untracked: code/FilterPrimSec.sh
Untracked: code/FilterReverseLift.err
Untracked: code/FilterReverseLift.out
Untracked: code/FindIntronForDomPAS.err
Untracked: code/FindIntronForDomPAS.out
Untracked: code/FindIntronForDomPAS.sh
Untracked: code/FindIntronForDomPAS_DF.sh
Untracked: code/GencodeDiffSplice.err
Untracked: code/GencodeDiffSplice.out
Untracked: code/GetMAPQscore.py
Untracked: code/GetSecondaryMap.py
Untracked: code/HchromOrder.err
Untracked: code/HchromOrder.out
Untracked: code/IntersectMMandOrtho.err
Untracked: code/IntersectMMandOrtho.out
Untracked: code/IntersectPASandOrtho.err
Untracked: code/IntersectPASandOrtho.out
Untracked: code/JunctionLift.err
Untracked: code/JunctionLift.out
Untracked: code/JunctionLiftFinalChimp.err
Untracked: code/JunctionLiftFinalChimp.out
Untracked: code/Lift5perPAS.sh
Untracked: code/Lift5perPASbed.err
Untracked: code/Lift5perPASbed.out
Untracked: code/LiftClustersFirst.err
Untracked: code/LiftClustersFirst.out
Untracked: code/LiftClustersFirst_remove.err
Untracked: code/LiftClustersFirst_remove.out
Untracked: code/LiftClustersSecond.err
Untracked: code/LiftClustersSecond.out
Untracked: code/LiftClustersSecond_remove.err
Untracked: code/LiftClustersSecond_remove.out
Untracked: code/LiftFinalChimpJunc2Human.sh
Untracked: code/LiftOrthoPAS2chimp.sh
Untracked: code/LiftorthoPAS.err
Untracked: code/LiftorthoPASt.out
Untracked: code/Log.out
Untracked: code/MapBadSamples.err
Untracked: code/MapBadSamples.out
Untracked: code/MapBadSamples.sh
Untracked: code/MapStats.err
Untracked: code/MapStats.out
Untracked: code/MaxEntCode/
Untracked: code/MergeClusters.err
Untracked: code/MergeClusters.out
Untracked: code/MergeClusters.sh
Untracked: code/MismatchNumbers.err
Untracked: code/MismatchNumbers.out
Untracked: code/MismatchNumbers.sh
Untracked: code/PAS_ATTAAA.err
Untracked: code/PAS_ATTAAA.out
Untracked: code/PAS_ATTAAA.sh
Untracked: code/PAS_ATTAAADF.err
Untracked: code/PAS_ATTAAADF.out
Untracked: code/PAS_ATTAAA_df.sh
Untracked: code/PAS_seqExpanded.sh
Untracked: code/PAS_sequence.err
Untracked: code/PAS_sequence.out
Untracked: code/PAS_sequenceDF.err
Untracked: code/PAS_sequenceDF.out
Untracked: code/PASexpanded_sequenceDF.err
Untracked: code/PASexpanded_sequenceDF.out
Untracked: code/PASsequences.sh
Untracked: code/PASsequences_DF.sh
Untracked: code/PlotNuclearUsagebySpecies.R
Untracked: code/PlotNuclearUsagebySpecies_DF.R
Untracked: code/QuantMergeClusters
Untracked: code/QuantMergeClusters.err
Untracked: code/QuantMergeClusters.out
Untracked: code/QuantMergedClusters.sh
Untracked: code/RNATranscriptDTplot.err
Untracked: code/RNATranscriptDTplot.out
Untracked: code/RNATranscriptDTplot.sh
Untracked: code/Rev_liftoverPAShg19to38.err
Untracked: code/Rev_liftoverPAShg19to38.out
Untracked: code/ReverseLiftFilter.R
Untracked: code/RunFixCluster.err
Untracked: code/RunFixCluster.out
Untracked: code/RunFixLeafCluster.sh
Untracked: code/RunNegMCMediation.err
Untracked: code/RunNegMCMediation.sh
Untracked: code/RunNegMCMediationDF.err
Untracked: code/RunNegMCMediationDF.out
Untracked: code/RunNegMCMediationDF.sh
Untracked: code/RunNegMCMediationr.out
Untracked: code/RunPosMCMediation.err
Untracked: code/RunPosMCMediation.sh
Untracked: code/RunPosMCMediationDF.err
Untracked: code/RunPosMCMediationDF.out
Untracked: code/RunPosMCMediationDF.sh
Untracked: code/RunPosMCMediationr.out
Untracked: code/SAF215upbed_gen.py
Untracked: code/SAF2Bed.py
Untracked: code/Snakefile
Untracked: code/SnakefilePAS
Untracked: code/SnakefilePASfilt
Untracked: code/SortIndexBadSamples.err
Untracked: code/SortIndexBadSamples.out
Untracked: code/SortIndexBadSamples.sh
Untracked: code/StarMM2.err
Untracked: code/StarMM2.out
Untracked: code/StarMM2.sh
Untracked: code/TestFC.err
Untracked: code/TestFC.out
Untracked: code/TestFC.sh
Untracked: code/TotalTranscriptDTplot.err
Untracked: code/TotalTranscriptDTplot.out
Untracked: code/Upstream10Bases_general.py
Untracked: code/apaQTLsnake.err
Untracked: code/apaQTLsnake.out
Untracked: code/apaQTLsnakePAS.err
Untracked: code/apaQTLsnakePAS.out
Untracked: code/apaQTLsnakePAShuman.err
Untracked: code/assignPeak2Intronicregion.err
Untracked: code/assignPeak2Intronicregion.out
Untracked: code/assignPeak2Intronicregion.sh
Untracked: code/bam2junc.err
Untracked: code/bam2junc.out
Untracked: code/bam2junc_remove.err
Untracked: code/bam2junc_remove.out
Untracked: code/bed215upbed.py
Untracked: code/bed2Bedbothstrand.py
Untracked: code/bed2SAF_gen.py
Untracked: code/bed2saf.py
Untracked: code/bg_to_cov.py
Untracked: code/buildIndecpantro5
Untracked: code/buildIndecpantro5.sh
Untracked: code/buildLeafviz.err
Untracked: code/buildLeafviz.out
Untracked: code/buildLeafviz.sh
Untracked: code/buildLeafviz_leadAnno.sh
Untracked: code/buildLeafviz_leafanno.err
Untracked: code/buildLeafviz_leafanno.out
Untracked: code/buildStarIndex.sh
Untracked: code/callPeaksYL.py
Untracked: code/chimpChromprder.sh
Untracked: code/chimpMultiCov.err
Untracked: code/chimpMultiCov.out
Untracked: code/chimpMultiCov.sh
Untracked: code/chimpMultiCov255.sh
Untracked: code/chimpMultiCovInclusive.err
Untracked: code/chimpMultiCovInclusive.out
Untracked: code/chimpMultiCovInclusive.sh
Untracked: code/chooseAnno2Bed.py
Untracked: code/chooseAnno2SAF.py
Untracked: code/chooseSignalSite.py
Untracked: code/chromOrder.err
Untracked: code/chromOrder.out
Untracked: code/classifyLeafviz.err
Untracked: code/classifyLeafviz.out
Untracked: code/cleanbed2saf.py
Untracked: code/cluster.json
Untracked: code/cluster2bed.py
Untracked: code/clusterLiftReverse.sh
Untracked: code/clusterLiftReverse_removebad.sh
Untracked: code/clusterLiftprimary.sh
Untracked: code/clusterLiftprimary_removebad.sh
Untracked: code/clusterPAS.json
Untracked: code/clusterfiltPAS.json
Untracked: code/comands2Mege.sh
Untracked: code/converBam2Junc.sh
Untracked: code/converBam2Junc_removeBad.sh
Untracked: code/convertNumeric.py
Untracked: code/environment.yaml
Untracked: code/extraSnakefiltpas
Untracked: code/extractPhyloReg.py
Untracked: code/extractPhyloRegGene.py
Untracked: code/extractPhylopGeneral.py
Untracked: code/extractPhylopReg200down.py
Untracked: code/extractPhylopReg200up.py
Untracked: code/filter5perc.R
Untracked: code/filter5percPAS.py
Untracked: code/filter5percPheno.py
Untracked: code/filterBamforMP.pysam2_gen.py
Untracked: code/filterJuncChroms.err
Untracked: code/filterJuncChroms.out
Untracked: code/filterMissprimingInNuc10_gen.py
Untracked: code/filterNumChroms.py
Untracked: code/filterPASforMP.py
Untracked: code/filterPostLift.py
Untracked: code/filterPrimaryread.py
Untracked: code/filterSAFforMP_gen.py
Untracked: code/filterSecondaryread.py
Untracked: code/filterSortBedbyCleanedBed_gen.R
Untracked: code/filterpeaks.py
Untracked: code/fixExonFC.py
Untracked: code/fixFChead.py
Untracked: code/fixFChead_bothfrac.py
Untracked: code/fixFCheadforExp.py
Untracked: code/fixLeafCluster.py
Untracked: code/fixLiftedJunc.py
Untracked: code/fixUTRexonanno.py
Untracked: code/formathg38Anno.py
Untracked: code/generateStarIndex.err
Untracked: code/generateStarIndex.out
Untracked: code/generateStarIndexHuman.err
Untracked: code/generateStarIndexHuman.out
Untracked: code/getRNAseqMapStats.sh
Untracked: code/hg19MapStats.err
Untracked: code/hg19MapStats.out
Untracked: code/hg19MapStats.sh
Untracked: code/humanChromorder.sh
Untracked: code/humanFiles
Untracked: code/humanMultiCov.err
Untracked: code/humanMultiCov.out
Untracked: code/humanMultiCov.sh
Untracked: code/humanMultiCov255.err
Untracked: code/humanMultiCov255.out
Untracked: code/humanMultiCov255.sh
Untracked: code/humanMultiCovInclusive.err
Untracked: code/humanMultiCovInclusive.out
Untracked: code/humanMultiCov_inclusive.sh
Untracked: code/intersectAnno.err
Untracked: code/intersectAnno.out
Untracked: code/intersectAnnoExt.err
Untracked: code/intersectAnnoExt.out
Untracked: code/intersectLiftedPAS.sh
Untracked: code/leafcutter_merge_regtools_redo.py
Untracked: code/liftJunctionFiles.sh
Untracked: code/liftPAS19to38.sh
Untracked: code/liftoverPAShg19to38.err
Untracked: code/liftoverPAShg19to38.out
Untracked: code/log/
Untracked: code/make5percPeakbed.py
Untracked: code/makeFileID.py
Untracked: code/makeNuclearDapaplots.sh
Untracked: code/makeNuclearDapaplots_DF.sh
Untracked: code/makeNuclearPlots.err
Untracked: code/makeNuclearPlots.out
Untracked: code/makeNuclearPlotsDF.err
Untracked: code/makeNuclearPlotsDF.out
Untracked: code/makePheno.py
Untracked: code/makeSamplyGroupsChimp_TvN.py
Untracked: code/makeSamplyGroupsHuman_TvN.py
Untracked: code/mapRNAseqhg19.sh
Untracked: code/mapRNAseqhg19_newPipeline.sh
Untracked: code/maphg19.err
Untracked: code/maphg19.out
Untracked: code/maphg19.sh
Untracked: code/maphg19_new.err
Untracked: code/maphg19_new.out
Untracked: code/maphg19_sub.err
Untracked: code/maphg19_sub.out
Untracked: code/maphg19_subjunc.sh
Untracked: code/mediation_test.R
Untracked: code/merge.err
Untracked: code/mergeChimp3prime_inhg38.sh
Untracked: code/merge_leafcutter_clusters_redo.py
Untracked: code/mergeandBWRNAseq.sh
Untracked: code/mergeandsort_ChimpinHuman.err
Untracked: code/mergeandsort_ChimpinHuman.out
Untracked: code/mergedBam2BW.sh
Untracked: code/mergedbam2bw.err
Untracked: code/mergedbam2bw.out
Untracked: code/mergedbamRNAand2bw.err
Untracked: code/mergedbamRNAand2bw.out
Untracked: code/nameClusters.py
Untracked: code/namePeaks.py
Untracked: code/negativeMediation_montecarlo.R
Untracked: code/negativeMediation_montecarloDF.R
Untracked: code/nuclearTranscriptDTplot.err
Untracked: code/nuclearTranscriptDTplot.out
Untracked: code/numMultimap.py
Untracked: code/overlapMMandOrthoexon.sh
Untracked: code/overlapPAS.err
Untracked: code/overlapPAS.out
Untracked: code/overlapPASandOrthoexon.sh
Untracked: code/overlapapaQTLPAS.sh
Untracked: code/overlapapaQTLPAS_extended.sh
Untracked: code/overlapapaQTLPAS_samples.sh
Untracked: code/parseHg38.py
Untracked: code/peak2PAS.py
Untracked: code/pheno2countonly.R
Untracked: code/postiveMediation_montecarlo.R
Untracked: code/postiveMediation_montecarlo_DF.R
Untracked: code/prepareAnnoLeafviz.err
Untracked: code/prepareAnnoLeafviz.out
Untracked: code/prepareCleanLiftedFC_5perc4LC.py
Untracked: code/prepareLeafvizAnno.sh
Untracked: code/preparePAS4lift.py
Untracked: code/prepare_phenotype_table.py
Untracked: code/primaryLift.err
Untracked: code/primaryLift.out
Untracked: code/primaryLift.sh
Untracked: code/processhg38exons.py
Untracked: code/quantJunc.sh
Untracked: code/quantJunc_TEST.sh
Untracked: code/quantJunc_removeBad.sh
Untracked: code/quantLiftedPAS.err
Untracked: code/quantLiftedPAS.out
Untracked: code/quantLiftedPAS.sh
Untracked: code/quantLiftedPASPrimary.err
Untracked: code/quantLiftedPASPrimary.out
Untracked: code/quantLiftedPASPrimary.sh
Untracked: code/quatJunc.err
Untracked: code/quatJunc.out
Untracked: code/recChimpback2Human.err
Untracked: code/recChimpback2Human.out
Untracked: code/recLiftchim2human.sh
Untracked: code/revLift.err
Untracked: code/revLift.out
Untracked: code/revLiftPAShg38to19.sh
Untracked: code/reverseLift.sh
Untracked: code/runCheckReverseLift.sh
Untracked: code/runChimpDiffIso.sh
Untracked: code/runChimpDiffIsoDF.sh
Untracked: code/runCountNucleotides.err
Untracked: code/runCountNucleotides.out
Untracked: code/runCountNucleotides.sh
Untracked: code/runCountNucleotidesPantro6.err
Untracked: code/runCountNucleotidesPantro6.out
Untracked: code/runCountNucleotides_pantro6.sh
Untracked: code/runFilterNumChroms.sh
Untracked: code/runHumanDiffIso.sh
Untracked: code/runHumanDiffIsoDF.sh
Untracked: code/runNuclearDiffIso_DF.sh
Untracked: code/runNuclearDifffIso.sh
Untracked: code/runTotalDiffIso.sh
Untracked: code/run_Chimpleafcutter_ds.err
Untracked: code/run_Chimpleafcutter_ds.out
Untracked: code/run_Chimpverifybam.err
Untracked: code/run_Chimpverifybam.out
Untracked: code/run_Humanleafcutter_dF.err
Untracked: code/run_Humanleafcutter_dF.out
Untracked: code/run_Humanleafcutter_ds.err
Untracked: code/run_Humanleafcutter_ds.out
Untracked: code/run_Nuclearleafcutter_ds.err
Untracked: code/run_Nuclearleafcutter_ds.out
Untracked: code/run_Nuclearleafcutter_dsDF.err
Untracked: code/run_Nuclearleafcutter_dsDF.out
Untracked: code/run_Totalleafcutter_ds.err
Untracked: code/run_Totalleafcutter_ds.out
Untracked: code/run_chimpverifybam.sh
Untracked: code/run_verifyBam.sh
Untracked: code/run_verifybam.err
Untracked: code/run_verifybam.out
Untracked: code/slurm-62824013.out
Untracked: code/slurm-62825841.out
Untracked: code/slurm-62826116.out
Untracked: code/slurm-64108209.out
Untracked: code/slurm-64108521.out
Untracked: code/slurm-64108557.out
Untracked: code/snakePASChimp.err
Untracked: code/snakePASChimp.out
Untracked: code/snakePAShuman.out
Untracked: code/snakemake.batch
Untracked: code/snakemakeChimp.err
Untracked: code/snakemakeChimp.out
Untracked: code/snakemakeHuman.err
Untracked: code/snakemakeHuman.out
Untracked: code/snakemakePAS.batch
Untracked: code/snakemakePASFiltChimp.err
Untracked: code/snakemakePASFiltChimp.out
Untracked: code/snakemakePASFiltHuman.err
Untracked: code/snakemakePASFiltHuman.out
Untracked: code/snakemakePASchimp.batch
Untracked: code/snakemakePAShuman.batch
Untracked: code/snakemake_chimp.batch
Untracked: code/snakemake_human.batch
Untracked: code/snakemakefiltPAS.batch
Untracked: code/snakemakefiltPAS_chimp.sh
Untracked: code/snakemakefiltPAS_human.sh
Untracked: code/spliceSite2Fasta.py
Untracked: code/submit-snakemake-chimp.sh
Untracked: code/submit-snakemake-human.sh
Untracked: code/submit-snakemakePAS-chimp.sh
Untracked: code/submit-snakemakePAS-human.sh
Untracked: code/submit-snakemakefiltPAS-chimp.sh
Untracked: code/submit-snakemakefiltPAS-human.sh
Untracked: code/subset_diffisopheno.py
Untracked: code/subset_diffisopheno_Chimp_tvN.py
Untracked: code/subset_diffisopheno_Chimp_tvN_DF.py
Untracked: code/subset_diffisopheno_Huma_tvN.py
Untracked: code/subset_diffisopheno_Huma_tvN_DF.py
Untracked: code/subset_diffisopheno_Nuclear_HvC.py
Untracked: code/subset_diffisopheno_Nuclear_HvC_DF.py
Untracked: code/subset_diffisopheno_Total_HvC.py
Untracked: code/test
Untracked: code/test.txt
Untracked: code/threeprimeOrthoFC.out
Untracked: code/threeprimeOrthoFC.sh
Untracked: code/threeprimeOrthoFCcd.err
Untracked: code/transcriptDTplotsNuclear.sh
Untracked: code/transcriptDTplotsTotal.sh
Untracked: code/verifyBam4973.sh
Untracked: code/verifyBam4973inHuman.sh
Untracked: code/verifybam4973.err
Untracked: code/verifybam4973.out
Untracked: code/verifybam4973HumanMap.err
Untracked: code/verifybam4973HumanMap.out
Untracked: code/wrap_Chimpverifybam.err
Untracked: code/wrap_Chimpverifybam.out
Untracked: code/wrap_chimpverifybam.sh
Untracked: code/wrap_verifyBam.sh
Untracked: code/wrap_verifybam.err
Untracked: code/wrap_verifybam.out
Untracked: code/writeMergecode.py
Untracked: data/._.DS_Store
Untracked: data/._HC_filenames.txt
Untracked: data/._HC_filenames.txt.sb-4426323c-IKIs0S
Untracked: data/._HC_filenames.xlsx
Untracked: data/._MapPantro6_meta.txt
Untracked: data/._MapPantro6_meta.txt.sb-a5794dd2-Cskmlm
Untracked: data/._MapPantro6_meta.xlsx
Untracked: data/._OppositeSpeciesMap.txt
Untracked: data/._OppositeSpeciesMap.txt.sb-a5794dd2-mayWJf
Untracked: data/._OppositeSpeciesMap.xlsx
Untracked: data/._RNASEQ_metadata.txt
Untracked: data/._RNASEQ_metadata.txt.sb-4426323c-TE4ns3
Untracked: data/._RNASEQ_metadata.txt.sb-51f67ae1-HXp7Gq
Untracked: data/._RNASEQ_metadata_2Removed.txt
Untracked: data/._RNASEQ_metadata_2Removed.txt.sb-4426323c-a4lBwx
Untracked: data/._RNASEQ_metadata_2Removed.xlsx
Untracked: data/._RNASEQ_metadata_stranded.txt
Untracked: data/._RNASEQ_metadata_stranded.txt.sb-a5794dd2-D659m2
Untracked: data/._RNASEQ_metadata_stranded.txt.sb-a5794dd2-ImNMoY
Untracked: data/._RNASEQ_metadata_stranded.txt.sb-e4bf31f0-ZGnGgl
Untracked: data/._RNASEQ_metadata_stranded.xlsx
Untracked: data/._TrialFiltersMeta.txt
Untracked: data/._TrialFiltersMeta.txt.sb-9845453e-R58Y0Q
Untracked: data/._metadata_HCpanel.txt
Untracked: data/._metadata_HCpanel.txt.sb-a3d92a2d-b9cYoF
Untracked: data/._metadata_HCpanel.txt.sb-a5794dd2-i594qs
Untracked: data/._metadata_HCpanel.txt.sb-f4823d1e-qihGek
Untracked: data/._metadata_HCpanel.xlsx
Untracked: data/._metadata_HCpanel_frompantro5.xlsx
Untracked: data/._~$RNASEQ_metadata.xlsx
Untracked: data/._~$metadata_HCpanel.xlsx
Untracked: data/._.xlsx
Untracked: data/BaseComp/
Untracked: data/CleanLiftedPeaks_FC_primary/
Untracked: data/CompapaQTLpas/
Untracked: data/DNDS/
Untracked: data/DTmatrix/
Untracked: data/DiffExpression/
Untracked: data/DiffIso_Nuclear/
Untracked: data/DiffIso_Nuclear_DF/
Untracked: data/DiffIso_Total/
Untracked: data/DiffSplice/
Untracked: data/DiffSplice_liftedJunc/
Untracked: data/DiffSplice_removeBad/
Untracked: data/DominantPAS/
Untracked: data/DominantPAS_DF/
Untracked: data/EvalPantro5/
Untracked: data/HC_filenames.txt
Untracked: data/HC_filenames.xlsx
Untracked: data/Khan_prot/
Untracked: data/Li_eqtls/
Untracked: data/MapPantro6_meta.txt
Untracked: data/MapPantro6_meta.xlsx
Untracked: data/MapStats/
Untracked: data/NormalizedClusters/
Untracked: data/NuclearHvC/
Untracked: data/NuclearHvC_DF/
Untracked: data/OppositeSpeciesMap.txt
Untracked: data/OppositeSpeciesMap.xlsx
Untracked: data/OrthoExonBed/
Untracked: data/OverlapBenchmark/
Untracked: data/OverlappingPAS/
Untracked: data/PAS/
Untracked: data/PAS_SAF/
Untracked: data/PAS_doubleFilter/
Untracked: data/Peaks_5perc/
Untracked: data/Pheno_5perc/
Untracked: data/Pheno_5perc_DF_nuclear/
Untracked: data/Pheno_5perc_nuclear/
Untracked: data/Pheno_5perc_nuclear_old/
Untracked: data/Pheno_5perc_total/
Untracked: data/PhyloP/
Untracked: data/RNASEQ_metadata.txt
Untracked: data/RNASEQ_metadata_2Removed.txt
Untracked: data/RNASEQ_metadata_2Removed.xlsx
Untracked: data/RNASEQ_metadata_stranded.txt
Untracked: data/RNASEQ_metadata_stranded.txt.sb-e4bf31f0-ZGnGgl/
Untracked: data/RNASEQ_metadata_stranded.xlsx
Untracked: data/SignalSites/
Untracked: data/SignalSites_doublefilter/
Untracked: data/SpliceSite/
Untracked: data/TestAnnoBiasOE/
Untracked: data/TestMM2/
Untracked: data/TestMM2_PrimaryRead/
Untracked: data/TestMM2_SeondaryRead/
Untracked: data/TestMM2_mismatch/
Untracked: data/TestMM2_quality/
Untracked: data/Test_FC_methods/
Untracked: data/Threeprime2Ortho/
Untracked: data/TotalHvC/
Untracked: data/TrialFiltersMeta.txt
Untracked: data/TwoBadSampleAnalysis/
Untracked: data/Wang_ribo/
Untracked: data/apaQTLGenes/
Untracked: data/bioGRID/
Untracked: data/chainFiles/
Untracked: data/cleanPeaks_anno/
Untracked: data/cleanPeaks_byspecies/
Untracked: data/cleanPeaks_lifted/
Untracked: data/files4viz_nuclear/
Untracked: data/files4viz_nuclear_DF/
Untracked: data/gviz/
Untracked: data/leafviz/
Untracked: data/liftover_files/
Untracked: data/mediation/
Untracked: data/mediation_DF/
Untracked: data/metadata_HCpanel.txt
Untracked: data/metadata_HCpanel.xlsx
Untracked: data/metadata_HCpanel_frompantro5.txt
Untracked: data/metadata_HCpanel_frompantro5.xlsx
Untracked: data/multimap/
Untracked: data/primaryLift/
Untracked: data/reverseLift/
Untracked: data/testQuant/
Untracked: data/~$RNASEQ_metadata.xlsx
Untracked: data/~$metadata_HCpanel.xlsx
Untracked: data/.xlsx
Untracked: output/._.DS_Store
Untracked: output/dtPlots/
Untracked: projectNotes.Rmd
Untracked: proteinModelSet.Rmd
Unstaged changes:
Modified: analysis/DiffUsedIntronic.Rmd
Modified: analysis/ExploredAPA.Rmd
Modified: analysis/ExploredAPA_DF.Rmd
Modified: analysis/MMExpreiment.Rmd
Modified: analysis/OppositeMap.Rmd
Modified: analysis/SpliceSiteStrength.Rmd
Modified: analysis/TotalVNuclearBothSpecies.Rmd
Modified: analysis/annotationInfo.Rmd
Modified: analysis/changeMisprimcut.Rmd
Modified: analysis/comp2apaQTLPAS.Rmd
Modified: analysis/correlationPhenos.Rmd
Modified: analysis/dAPAandapaQTL_DF.Rmd
Modified: analysis/establishCutoffs.Rmd
Modified: analysis/investigatePantro5.Rmd
Modified: analysis/multiMap.Rmd
Modified: analysis/speciesSpecific.Rmd
Modified: analysis/upsetter_DF.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the R Markdown and HTML files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view them.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 721b56f | brimittleman | 2020-03-31 | add prop |
| html | 4d772b1 | brimittleman | 2020-03-31 | Build site. |
| Rmd | 8280da5 | brimittleman | 2020-03-31 | add distance plots |
| html | 1274a02 | brimittleman | 2020-03-30 | Build site. |
| html | 469010e | brimittleman | 2020-03-30 | Build site. |
| Rmd | ef52dfc | brimittleman | 2020-03-30 | add all overlap orthoexon |
| html | 3d4dd47 | brimittleman | 2020-03-27 | Build site. |
| Rmd | f92b89c | brimittleman | 2020-03-26 | add ortho exon and new mm |
library(tidyverse)── Attaching packages ────────────────────────────────────────────────────────────────────────── tidyverse 1.2.1 ──✔ ggplot2 3.1.1 ✔ purrr 0.3.2
✔ tibble 2.1.1 ✔ dplyr 0.8.0.1
✔ tidyr 0.8.3 ✔ stringr 1.3.1
✔ readr 1.3.1 ✔ forcats 0.3.0 ── Conflicts ───────────────────────────────────────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()I am still concerned about annotation. I want to see if using the ortho exon file as an annotation tool would be more appropriate. I want to look at it and see if there is a way to combine exons by gene and see if the spaces inbetween could be introns.
First look at the human one.
HumanOrtho=read.table("/project2/gilad/kenneth/OrthoExonPartialMapping/human.noM.gtf", sep="\t")Parse this into a bed file with a python script.
mkdir ../data/OrthoExonBed
python SAF2Bed.py /project2/gilad/kenneth/OrthoExonPartialMapping/human.noM.gtf ../data/OrthoExonBed/human.noM.bed
sort -k1,1 -k2,2n ../data/OrthoExonBed/human.noM.bed > ../data/OrthoExonBed/human.noM.sort.bed All PAS
sbatch overlapPASandOrthoexon.sh Results:
PASMeta=read.table("../data/PAS_doubleFilter/PAS_10perc_either_HumanCoord_BothUsage_meta_doubleFilter.txt",header = T,stringsAsFactors = F) %>% select(PAS, gene,loc)
OverlapPAS=read.table("../data/OrthoExonBed/allPASinOrtho.bed", col.names = c("chr", "start", "end", "PAS", "score", "strand"),stringsAsFactors = F)%>% group_by(PAS) %>% summarize(n=n())%>% mutate(OE="In") %>% inner_join(PASMeta,by="PAS")
nrow(OverlapPAS)[1] 25621NotOverlapPAS=read.table("../data/OrthoExonBed/allPAS_NOT_inOrtho.bed", col.names = c("chr", "start", "end", "PAS", "score", "strand"),stringsAsFactors = F)%>% group_by(PAS) %>% summarize(n=n())%>% mutate(OE="OUT") %>% inner_join(PASMeta,by="PAS")
nrow(NotOverlapPAS)[1] 16697ALLPAS_ortho=OverlapPAS %>% bind_rows(NotOverlapPAS)
ggplot(ALLPAS_ortho, aes(x=OE, by=loc,fill=loc)) +geom_bar(stat="count",position = "dodge") + labs(x="Is PAS overlapping ortho exon", title="All PAS in OrthoExon",y="Number of PAS")+ scale_fill_brewer(palette = "Dark2")
| Version | Author | Date |
|---|---|---|
| 469010e | brimittleman | 2020-03-30 |
Ok this is the expected distribution, We expect the coding and 3’ UTRs to be in the ortho exon file. What is more interesting is genes with and without exons in the ortho exon.
Take this to the gene level.
OrthoBed=read.table("../data/OrthoExonBed/human.noM.sort.bed", col.names = c("chr","start","end","gene", "nExon","strand"),stringsAsFactors = F) %>% group_by(gene) %>% summarise(nExon=n())Now I look to see which of the PAS are in genes not in the ortho exon.
PASMeta_GeneOE= PASMeta %>% mutate(OE=ifelse(gene %in% OrthoBed$gene, "Yes", "No"))
PASMeta_GeneOEgene= PASMeta_GeneOE %>% group_by(gene, OE) %>% summarise()
ggplot(PASMeta_GeneOEgene, aes(x=OE, fill=OE))+ geom_histogram(stat="count") + labs(x="Is gene in Ortho Exon", y="Genes") + scale_fill_brewer(palette = "Dark2")Warning: Ignoring unknown parameters: binwidth, bins, pad
| Version | Author | Date |
|---|---|---|
| 469010e | brimittleman | 2020-03-30 |
Look at the genes without:
PASMeta_GeneOE_NO= PASMeta_GeneOE %>% filter(OE=="No")
nrow(PASMeta_GeneOE_NO)[1] 5293ggplot(PASMeta_GeneOE_NO,aes(x=loc, fill=OE)) + geom_bar(stat="count")
Genesnotin=PASMeta_GeneOE_NO %>% group_by(gene) %>% summarise(n=n())1000 genes not in ortho exons.
#sno
Genesnotin %>% filter(grepl("SNO",gene)) %>% nrow()[1] 112#linc
Genesnotin %>% filter(grepl("LINC",gene)) %>% nrow()[1] 63#loc
Genesnotin %>% filter(grepl("LOC",gene)) %>% nrow()[1] 327Are these the genes with different dominant PAS.
allPAS= read.table("../data/PAS_doubleFilter/PAS_10perc_either_HumanCoord_BothUsage_meta_doubleFilter.txt", header = T)
ChimpPASwMean =allPAS %>% dplyr::select(-Human)
HumanPASwMean =allPAS %>% dplyr::select(-Chimp)
Chimp_Dom= ChimpPASwMean %>%
group_by(gene) %>%
top_n(1,Chimp) %>%
mutate(nPer=n()) %>%
filter(nPer==1) %>%
dplyr::select(gene,loc,PAS,Chimp) %>%
rename(ChimpLoc=loc, ChimpPAS=PAS)
Human_Dom= HumanPASwMean %>%
group_by(gene) %>%
top_n(1, Human) %>%
mutate(nPer=n()) %>%
filter(nPer==1) %>%
dplyr::select(gene,loc,PAS,Human) %>%
rename(HumanLoc=loc, HumanPAS=PAS)
#merge
BothDom= Chimp_Dom %>% inner_join(Human_Dom,by="gene")
DifDom= BothDom %>% filter(ChimpPAS!=HumanPAS)Plot this before I remove those genes.
DifDom_before=DifDom %>% select(gene, ChimpLoc, HumanLoc) %>% gather("species","loc",-gene)
ggplot(DifDom_before, aes(x=loc, by=species, fill=species))+geom_histogram(position = "dodge",stat = "count")+ labs(title="Location of PAS with different Dominant",y="PAS")+scale_fill_brewer(palette = "Dark2")Warning: Ignoring unknown parameters: binwidth, bins, pad
| Version | Author | Date |
|---|---|---|
| 469010e | brimittleman | 2020-03-30 |
Remove the not ortho genes:
DifDom_after=DifDom_before %>% anti_join(Genesnotin,by="gene")Warning: Column `gene` joining factor and character vector, coercing into
character vectorggplot(DifDom_after, aes(x=loc, by=species, fill=species))+geom_histogram(position = "dodge",stat = "count")+ labs(title="Location of PAS with different Dominant\n after removing genes not in orthoexon",y="PAS")+scale_fill_brewer(palette = "Dark2")Warning: Ignoring unknown parameters: binwidth, bins, pad
| Version | Author | Date |
|---|---|---|
| 469010e | brimittleman | 2020-03-30 |
That didnt help.
Check the matching ones (are these in ortho exon)
SameDom= BothDom %>% filter(ChimpPAS==HumanPAS)
nrow(SameDom)[1] 7638SameDom_after=SameDom %>% anti_join(Genesnotin,by="gene")Warning: Column `gene` joining factor and character vector, coercing into
character vectornrow(SameDom_after)[1] 6906Where did I lose genes.
6906/7638[1] 0.90416343432/4028[1] 0.8520357Lose more in the different dominant than in the same dominant. Percent lost=
(7638-6906)/7638[1] 0.09583661(4028-3432)/4028[1] 0.1479643MultiMap
I will check if some of the problem genes, I found in my pipeline are in this.
ChimpMM=read.table("../data/multimap/Chimp_Uniq_multimapPAS.txt", stringsAsFactors = F, header = T)
HumanMM=read.table("../data/multimap/Human_Uniq_multimapPAS.txt", stringsAsFactors = F, header = T)
BothMM=read.table("../data/multimap/Both_multimapPAS.txt",stringsAsFactors = F, header = T)
AllMM=ChimpMM %>% bind_rows(HumanMM) %>% bind_rows(BothMM)I will overlap this with the ortho exon file. I will look at those that overlap and those that do not. I need a bed file
AllMM_bed=AllMM %>% mutate(Name=paste(gene, PAS,loc, MultiMap, sep=":")) %>% select(chr, start,end, Name, Human, strandFix)
write.table(AllMM_bed,"../data/multimap/allMM.bed",col.names = F, quote = F, row.names = F, sep="\t")sort -k1,1 -k2,2n ../data/multimap/allMM.bed > ../data/multimap/allMM.sort.bedOverlap.
sbatch overlapMMandOrthoexon.sh Results:
InOrtho=read.table("../data/OrthoExonBed/allMMinOrtho.bed", col.names = c("chr", "start", "end", "name", "score", "strand")) %>% group_by(name) %>% summarize(n=n())%>% separate(name, into=c("gene", "PAS","loc", "MM"),sep=":") %>% mutate(OE="In")
NotInOrtho=read.table("../data/OrthoExonBed/allMM_NOT_inOrtho.bed", col.names = c("chr", "start", "end", "name", "score", "strand")) %>% group_by(name) %>% summarize(n=n()) %>% separate(name, into=c("gene", "PAS","loc", "MM"),sep=":") %>% mutate(OE="Out")
AllOrthores=InOrtho %>% bind_rows(NotInOrtho)
ggplot(AllOrthores, aes(x=OE, by=loc,fill=loc)) +geom_bar(stat="count",position = "dodge") + labs(x="Is PAS overlapping ortho exon", title="PAS impacted by multimapping and ortho exon",y="Number of PAS")+ scale_fill_brewer(palette = "Dark2")
| Version | Author | Date |
|---|---|---|
| 469010e | brimittleman | 2020-03-30 |
Proportion:
AllOrthores %>% group_by(OE) %>% summarise(n=n())# A tibble: 2 x 2
OE n
<chr> <int>
1 In 831
2 Out 578#look only at utr
AllOrthores %>% filter(loc=="utr3") %>% group_by(OE) %>% summarise(n=n())# A tibble: 2 x 2
OE n
<chr> <int>
1 In 492
2 Out 120Look at the UTR sequences that are in:
AllOrthores %>% filter(OE=="In", loc=="utr3")# A tibble: 492 x 6
gene PAS loc MM n OE
<chr> <chr> <chr> <chr> <int> <chr>
1 AARS2 human285010 utr3 Human 2 In
2 ABCB10 human30678 utr3 Both 2 In
3 ABHD17A human153108 utr3 Both 8 In
4 ABR chimp125493 utr3 Human 10 In
5 ACAD8 chimp63259 utr3 Chimp 6 In
6 ACAD8 human66039 utr3 Chimp 6 In
7 ACTB human299134 utr3 Both 7 In
8 ADAM9 chimp303728 utr3 Human 8 In
9 ADAM9 chimp303729 utr3 Human 8 In
10 ADH5 human247010 utr3 Chimp 2 In
# … with 482 more rowsdistance to next annotated ortho exon boundry
I can look at the intronic PAS in both species to see how far they are from an ortho exon.
I need to merge exons.
sort -k1,1 -k2,2n ../data/OrthoExonBed/chimp.noM.bed > ../data/OrthoExonBed/chimp.noM.sort.bed
bedtools merge -s -c 4,5,6 -o distinct,count,distinct -i ../data/OrthoExonBed/chimp.noM.sort.bed > ../data/OrthoExonBed/chimp.noM.sort.merged.bed
bedtools merge -s -c 4,5,6 -o distinct,count,distinct -i ../data/OrthoExonBed/human.noM.sort.bed > ../data/OrthoExonBed/human.noM.sort.merged.bed
mkdir ../data/TestAnnoBiasOENow I can map all of the intronic PAS
For intronic PAS in genes in the ortho exon, filter the bed files
PASMeta_GeneOE_intronYes= PASMeta_GeneOE %>% filter(loc=="intron", OE=="Yes")
HumanBed=read.table("../data/PAS_doubleFilter/PAS_doublefilter_either_HumanCoordHummanUsage.sort.bed", col.names = c("chr", 'start','end','PAS','usage','strand'),stringsAsFactors = F) %>% semi_join(PASMeta_GeneOE_intronYes, by="PAS")
write.table(HumanBed, "../data/TestAnnoBiasOE/HumanIntronicGeneinOE.bed",col.names = F, row.names = F, quote = F, sep="\t")
ChimpBed=read.table("../data/PAS_doubleFilter/PAS_doublefilter_either_ChimpCoordChimpUsage.sort.bed", col.names = c("chr", 'start','end','PAS','usage','strand'),stringsAsFactors = F) %>% semi_join(PASMeta_GeneOE_intronYes, by="PAS")
write.table(ChimpBed, "../data/TestAnnoBiasOE/ChimpIntronicGeneinOE.bed",col.names = F, row.names = F, quote = F, sep="\t")Find the closest:
same strand. look only upstream (-id) to say closest annotated 5’ splice site. then do only downstream to say closest annotated 3’ splice site.
sbatch ClosestorthoEx.shLook at upstream
If the gene is a different gene, make it 0
HumanUpstream=read.table("../data/TestAnnoBiasOE/HumanUpstream.intronic.txt",stringsAsFactors = F, col.names = c("chrPAS", "startPAS", "endPAS", "PAS", "HumanUsage", "strandPAS", "chrExon","startExon","endExon", "Orthogene", "n","strandIntron", "UpstreamdistancePAS2Exon")) %>% inner_join(PASMeta,by="PAS") %>% mutate(Fixed=ifelse(gene==Orthogene, UpstreamdistancePAS2Exon,0))
HumanDownstream=read.table("../data/TestAnnoBiasOE/HumanDownstream.intronic.txt",stringsAsFactors = F, col.names = c("chrPAS", "startPAS", "endPAS", "PAS", "HumanUsage", "strandPAS", "chrExon","startExon","endExon", "Orthogene", "n","strandIntron", "DownstreamdistancePAS2Exon")) %>% inner_join(PASMeta,by="PAS") %>% mutate(Fixed=ifelse(gene==Orthogene, DownstreamdistancePAS2Exon,0))
HumanBoth=as.data.frame(cbind(HumanUpstream[,1:6],gene=HumanUpstream[,14], "UpstreamHuman"=HumanUpstream[,16],"DownstreamHuman"=HumanDownstream[,16])) %>%
mutate(DominanceHuman=ifelse(PAS %in% BothDom$HumanPAS, "yes","no"))Chimp:
ChimpUpstream=read.table("../data/TestAnnoBiasOE/ChimpUpstream.intronic.txt",stringsAsFactors = F, col.names = c("chrPAS", "startPAS", "endPAS", "PAS", "ChimpUsage", "strandPAS", "chrExon","startExon","endExon", "Orthogene", "n","strandIntron", "UpstreamdistancePAS2Exon")) %>% inner_join(PASMeta,by="PAS") %>% mutate(Fixed=ifelse(gene==Orthogene, UpstreamdistancePAS2Exon,0))
ChimpDownstream=read.table("../data/TestAnnoBiasOE/ChimpDownstream.intronic.txt",stringsAsFactors = F, col.names = c("chrPAS", "startPAS", "endPAS", "PAS", "ChimpUsage", "strandPAS", "chrExon","startExon","endExon", "Orthogene", "n","strandIntron", "DownstreamdistancePAS2Exon")) %>% inner_join(PASMeta,by="PAS") %>% mutate(Fixed=ifelse(gene==Orthogene, DownstreamdistancePAS2Exon,0))
ChimpBoth=as.data.frame(cbind(PAS=ChimpUpstream[,4], 'UpstreamChimp'=ChimpUpstream[,16],'DownstreamChimp'=ChimpDownstream[,16])) %>%
mutate(DominanceChimp=ifelse(PAS %in% BothDom$ChimpPAS, "yes","no"))Join together:
BothSpeciesDistance= HumanBoth %>% inner_join(ChimpBoth, by="PAS")Warning: Column `PAS` joining character vector and factor, coercing into
character vectorBothSpeciesDistance$UpstreamChimp=as.numeric(as.character(BothSpeciesDistance$UpstreamChimp))
BothSpeciesDistance$DownstreamChimp=as.numeric(as.character(BothSpeciesDistance$DownstreamChimp))Look at distance based on human dominance then chimp dominance
ggplot(BothSpeciesDistance, aes(x=UpstreamHuman, y=UpstreamChimp, col=DominanceHuman)) + geom_point(alpha=.3)+ geom_abline(aes(slope=1,intercept=0))
| Version | Author | Date |
|---|---|---|
| 4d772b1 | brimittleman | 2020-03-31 |
ggplot(BothSpeciesDistance, aes(x=UpstreamHuman, y=UpstreamChimp, col=DominanceHuman)) + geom_point(alpha=.3) + ylim(c(-10000,0)) +xlim(c(-10000,0)) + geom_abline(aes(slope=1,intercept=0)) + scale_color_brewer(palette = "Dark2")Warning: Removed 1651 rows containing missing values (geom_point).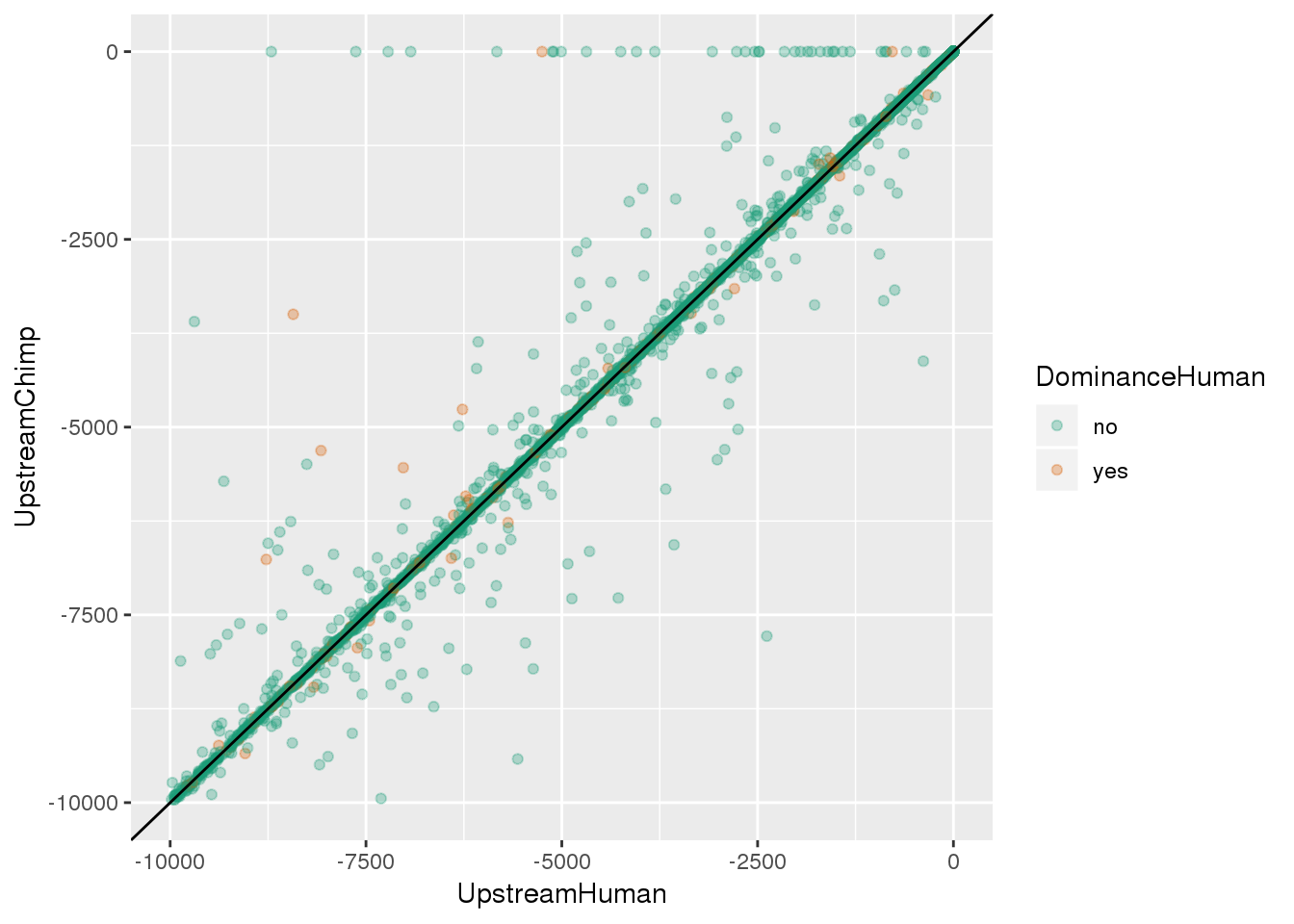
| Version | Author | Date |
|---|---|---|
| 4d772b1 | brimittleman | 2020-03-31 |
I want to color by dominant in human, chimp, both neither
BothSpeciesDistance_dom=BothSpeciesDistance %>% mutate(Dominance=ifelse(DominanceHuman=="yes", ifelse(DominanceChimp=="yes", "Both", "human"), ifelse(DominanceChimp=="yes", "chimp", "Neither")))Upstream Plots:
ggplot(BothSpeciesDistance_dom, aes(x=UpstreamHuman, y=UpstreamChimp, col=Dominance)) + geom_point(alpha=.3)+ geom_abline(aes(slope=1,intercept=0)) + geom_smooth(method="lm", alpha=.1)+scale_color_brewer(palette = "Dark2") 
| Version | Author | Date |
|---|---|---|
| 4d772b1 | brimittleman | 2020-03-31 |
ggplot(BothSpeciesDistance_dom, aes(x=UpstreamHuman, y=UpstreamChimp, col=Dominance)) + geom_point(alpha=.3)+ geom_abline(aes(slope=1,intercept=0))+ylim(c(-50000,0)) +xlim(c(-50000,0)) + scale_color_brewer(palette = "Dark2") + geom_smooth(method="lm", alpha=.1)Warning: Removed 122 rows containing non-finite values (stat_smooth).Warning: Removed 122 rows containing missing values (geom_point).Warning: Removed 1 rows containing missing values (geom_smooth).
| Version | Author | Date |
|---|---|---|
| 4d772b1 | brimittleman | 2020-03-31 |
Downstream
ggplot(BothSpeciesDistance_dom, aes(x=DownstreamHuman, y=DownstreamChimp, col=Dominance)) + geom_point(alpha=.3)+ geom_abline(aes(slope=1,intercept=0)) + scale_color_brewer(palette = "Dark2") + geom_smooth(method="lm", alpha=.1) + geom_smooth(method="lm", alpha=.1)
| Version | Author | Date |
|---|---|---|
| 4d772b1 | brimittleman | 2020-03-31 |
There are a lot of PAS that are far from an exon on the human side.
ggplot(BothSpeciesDistance_dom, aes(x=DownstreamHuman, y=DownstreamChimp, col=Dominance)) + geom_point(alpha=.3)+ geom_abline(aes(slope=1,intercept=0))+ylim(c(0,50000)) +xlim(c(0,50000)) + scale_color_brewer(palette = "Dark2") + geom_smooth(method="lm", alpha=.01)Warning: Removed 395 rows containing non-finite values (stat_smooth).Warning: Removed 395 rows containing missing values (geom_point).Warning: Removed 3 rows containing missing values (geom_smooth).
| Version | Author | Date |
|---|---|---|
| 4d772b1 | brimittleman | 2020-03-31 |
Look at the top examples:
BothSpeciesDistance_dom_TopDiffUp=BothSpeciesDistance_dom %>% mutate(Updiff=abs(UpstreamHuman-UpstreamChimp)) %>% select(PAS, gene, Updiff) %>% arrange(desc(Updiff))
head(BothSpeciesDistance_dom_TopDiffUp) PAS gene Updiff
1 human34001 SFMBT2 51262
2 chimp298751 ACTR3C 34423
3 human296634 TFB1M 33486
4 human175470 ANKRD36 31986
5 chimp298752 ACTR3C 31871
6 human175469 ANKRD36 31766Proportion of intron
I can look at where these are in the “intron”. Here I am calling the space between 2 orthologous exons in the same genes introns. I will remove PAS that have 0 distance on either side in either species first.
BothSpeciesDistance_dom_remove0= BothSpeciesDistance_dom %>% filter(UpstreamHuman!=0,DownstreamHuman!=0,UpstreamChimp!=0, DownstreamChimp!=0 )
nrow(BothSpeciesDistance_dom)-nrow(BothSpeciesDistance_dom_remove0)[1] 1123nrow(BothSpeciesDistance_dom_remove0)[1] 8229Lose 1123. Looking at 8229.
BothSpeciesDistance_dom_remove0_dist= BothSpeciesDistance_dom_remove0 %>% mutate(HumanLength=abs(UpstreamHuman)+ DownstreamHuman, HumanProp=abs(UpstreamHuman)/HumanLength, ChimpLength=abs(UpstreamChimp)+DownstreamChimp, ChimpProp=abs(UpstreamChimp)/ChimpLength)ggplot(BothSpeciesDistance_dom_remove0_dist,aes(x=HumanProp, y=ChimpProp,col=Dominance)) + geom_point(alpha=.3) + scale_color_brewer(palette = "Dark2") + geom_smooth(method="lm") + labs(x="Proportion of Human Intron",y= "Proportion of Chimp Intron", title="Intronic PAS between ortho exons")
BothSpeciesDistance_dom_remove0_dist_filt=BothSpeciesDistance_dom_remove0_dist %>% filter(Dominance !="Neither")
ggplot(BothSpeciesDistance_dom_remove0_dist_filt,aes(x=HumanProp, y=ChimpProp,col=Dominance)) + geom_point(alpha=.3) + scale_color_brewer(palette = "Dark2") + geom_smooth(method="lm") + labs(x="Proportion of Human Intron", y="Proportion of Chimp Intron", title="Intronic PAS between ortho exons") What about just length:
What about just length:
ggplot(BothSpeciesDistance_dom_remove0_dist,aes(x=HumanLength, y=ChimpLength,col=Dominance)) + geom_point(alpha=.3) + scale_color_brewer(palette = "Dark2") + geom_smooth(method="lm") + labs(x="Human Distance between ortho exon", "Chimp Distance between ortho exon", title="Intronic PAS between ortho exons")
sessionInfo()R version 3.5.1 (2018-07-02)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] forcats_0.3.0 stringr_1.3.1 dplyr_0.8.0.1 purrr_0.3.2
[5] readr_1.3.1 tidyr_0.8.3 tibble_2.1.1 ggplot2_3.1.1
[9] tidyverse_1.2.1
loaded via a namespace (and not attached):
[1] tidyselect_0.2.5 haven_1.1.2 lattice_0.20-38
[4] colorspace_1.3-2 generics_0.0.2 htmltools_0.3.6
[7] yaml_2.2.0 utf8_1.1.4 rlang_0.4.0
[10] later_0.7.5 pillar_1.3.1 glue_1.3.0
[13] withr_2.1.2 RColorBrewer_1.1-2 modelr_0.1.2
[16] readxl_1.1.0 plyr_1.8.4 munsell_0.5.0
[19] gtable_0.2.0 workflowr_1.6.0 cellranger_1.1.0
[22] rvest_0.3.2 evaluate_0.12 labeling_0.3
[25] knitr_1.20 httpuv_1.4.5 fansi_0.4.0
[28] broom_0.5.1 Rcpp_1.0.2 promises_1.0.1
[31] scales_1.0.0 backports_1.1.2 jsonlite_1.6
[34] fs_1.3.1 hms_0.4.2 digest_0.6.18
[37] stringi_1.2.4 grid_3.5.1 rprojroot_1.3-2
[40] cli_1.1.0 tools_3.5.1 magrittr_1.5
[43] lazyeval_0.2.1 crayon_1.3.4 whisker_0.3-2
[46] pkgconfig_2.0.2 xml2_1.2.0 lubridate_1.7.4
[49] assertthat_0.2.0 rmarkdown_1.10 httr_1.3.1
[52] rstudioapi_0.10 R6_2.3.0 nlme_3.1-137
[55] git2r_0.26.1 compiler_3.5.1