Premature Termination QTL
Briana Mittleman
7/1/2019
Last updated: 2019-07-08
Checks: 7 0
Knit directory: apaQTL/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.4.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190411) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/.DS_Store
Ignored: output/.DS_Store
Untracked files:
Untracked: .Rprofile
Untracked: ._.DS_Store
Untracked: .gitignore
Untracked: _workflowr.yml
Untracked: analysis/._PASdescriptiveplots.Rmd
Untracked: analysis/._cuttoffPercUsage.Rmd
Untracked: analysis/QTLexampleplots.Rmd
Untracked: analysis/cuttoffPercUsage.Rmd
Untracked: analysis/eQTLoverlap.Rmd
Untracked: analysis/gwasOverlap.Rmd
Untracked: analysis/mergeRNA.Rmd
Untracked: analysis/oldstuffNotNeeded.Rmd
Untracked: apaQTL.Rproj
Untracked: code/.NascentRNAdtPlotFirstintronicPAS.sh.swp
Untracked: code/._ApaQTL_nominalNonnorm.sh
Untracked: code/._BothFracDTPlotGeneRegions_normalized.sh
Untracked: code/._EandPqtl_perm.sh
Untracked: code/._EandPqtls.sh
Untracked: code/._FC_NucintornUpandDown.sh
Untracked: code/._FC_UTR.sh
Untracked: code/._FC_intornUpandDownsteamPAS.sh
Untracked: code/._FC_nascentseq.sh
Untracked: code/._FC_newPeaks_olddata.sh
Untracked: code/._HMMpermuteTotal.py
Untracked: code/._HmmPermute.py
Untracked: code/._LC_samplegroups.py
Untracked: code/._LD_qtl.sh
Untracked: code/._NascentRNAdtPlot.sh
Untracked: code/._NascentRNAdtPlot3UTRPAS.sh
Untracked: code/._NascentRNAdtPlotExcludeFirstintronicPAS.sh
Untracked: code/._NascentRNAdtPlotNucPAS.sh
Untracked: code/._NascentRNAdtPlotTotPAS.sh
Untracked: code/._NascentRNAdtPlotintronicPAS.sh
Untracked: code/._NascnetRNAdtPlotPAS.sh
Untracked: code/._NetSeq_fourthintronDT.sh
Untracked: code/._NomResfromPASSNP.py
Untracked: code/._PrematureQTLNominal.sh
Untracked: code/._PrematureQTLPermuted.sh
Untracked: code/._QTL2bed.py
Untracked: code/._QTL2bed_withstrand.py
Untracked: code/._RNAbam2bw.sh
Untracked: code/._SnakefilePAS
Untracked: code/._SnakefilefiltPAS
Untracked: code/._TESplots100bp.sh
Untracked: code/._TESplots150bp.sh
Untracked: code/._TESplots200bp.sh
Untracked: code/._Untitled
Untracked: code/._ZipandTabPheno.sh
Untracked: code/._aAPAqtl_nominal39ind.sh
Untracked: code/._apaQTLCorrectPvalMakeQQ.R
Untracked: code/._apaQTL_Nominal.sh
Untracked: code/._apaQTL_permuted.sh
Untracked: code/._assignNucIntonpeak2intronlocs.sh
Untracked: code/._assignTotIntronpeak2intronlocs.sh
Untracked: code/._bam2BW_5primemost.sh
Untracked: code/._bed2saf.py
Untracked: code/._bothFracDTplot1stintron.sh
Untracked: code/._bothFracDTplot4thintron.sh
Untracked: code/._bothFrac_FC.sh
Untracked: code/._callPeaksYL.py
Untracked: code/._changenomQTLres2geneName.py
Untracked: code/._chooseAnno2SAF.py
Untracked: code/._chooseSignalSite
Untracked: code/._chooseSignalSite.py
Untracked: code/._cluster.json
Untracked: code/._clusterPAS.json
Untracked: code/._clusterfiltPAS.json
Untracked: code/._codingdms2bed.py
Untracked: code/._config.yaml
Untracked: code/._config2.yaml
Untracked: code/._configOLD.yaml
Untracked: code/._convertNominal2SNPLOC.py
Untracked: code/._convertNumeric.py
Untracked: code/._correctNomeqtl.R
Untracked: code/._dag.pdf
Untracked: code/._eQTL_switch2snploc.py
Untracked: code/._eQTLgenestestedapa.py
Untracked: code/._encodeRNADTplots.sh
Untracked: code/._extractGenotypes.py
Untracked: code/._extractseqfromqtlfastq.py
Untracked: code/._fc2leafphen.py
Untracked: code/._filter5perc.R
Untracked: code/._filter5percPheno.py
Untracked: code/._filterpeaks.py
Untracked: code/._finalPASbed2SAF.py
Untracked: code/._fix4su304corr.py
Untracked: code/._fix4su604corr.py
Untracked: code/._fix4sukalisto.py
Untracked: code/._fixExandUnexeQTL
Untracked: code/._fixExandUnexeQTL.py
Untracked: code/._fixFChead.py
Untracked: code/._fixFChead_bothfrac.py
Untracked: code/._fixH3k12ac.py
Untracked: code/._fixPASregionSNPs.py
Untracked: code/._fixRNAhead4corr.py
Untracked: code/._fixRNAkalisto.py
Untracked: code/._fixgroupedtranscript.py
Untracked: code/._fixhead_netseqfc.py
Untracked: code/._getAPAfromanyeQTL.py
Untracked: code/._getApapval4eqtl.py
Untracked: code/._getApapval4eqtl_unexp.py
Untracked: code/._getDownstreamIntronNuclear.py
Untracked: code/._getIntronDownstreamPAS.py
Untracked: code/._getIntronUpstreamPAS.py
Untracked: code/._getQTLalleles.py
Untracked: code/._getQTLfastq.sh
Untracked: code/._getUpstreamIntronNuclear.py
Untracked: code/._grouptranscripts.py
Untracked: code/._intersectVCFandupPAS.sh
Untracked: code/._keep5perMAF.py
Untracked: code/._keepSNP_vcf.sh
Untracked: code/._make5percPeakbed.py
Untracked: code/._makeFileID.py
Untracked: code/._makePheno.py
Untracked: code/._makeSAFbothfrac5perc.py
Untracked: code/._makeSNP2rsidfile.py
Untracked: code/._makeeQTLempirical_unexp.py
Untracked: code/._makeeQTLempiricaldist.py
Untracked: code/._makegencondeTSSfile.py
Untracked: code/._mapSSsnps2PAS.sh
Untracked: code/._mergRNABam.sh
Untracked: code/._mergeAllBam.sh
Untracked: code/._mergeBW_norm.sh
Untracked: code/._mergeBamNascent.sh
Untracked: code/._mergeByFracBam.sh
Untracked: code/._mergePeaks.sh
Untracked: code/._mnase1stintron.sh
Untracked: code/._mnaseDT_fourthintron.sh
Untracked: code/._namePeaks.py
Untracked: code/._netseqDTplot1stIntron.sh
Untracked: code/._netseqFC.sh
Untracked: code/._nucSpeceffectsize.py
Untracked: code/._peak2PAS.py
Untracked: code/._peakFC.sh
Untracked: code/._pheno2countonly.R
Untracked: code/._phenoQTLfromlist.py
Untracked: code/._processYRIgen.py
Untracked: code/._pttQTLsinapaQTL.py
Untracked: code/._qtlRegionseq.sh
Untracked: code/._qtlsPvalOppFrac.py
Untracked: code/._quantassign2parsedpeak.py
Untracked: code/._removeXfromHmm.py
Untracked: code/._removeloc_pheno.py
Untracked: code/._runCorrectNomEqtl.sh
Untracked: code/._runHMMpermuteAPAqtls.sh
Untracked: code/._runHMMpermuteeQTLS.sh
Untracked: code/._runMakeEmpiricaleQTL_unexp.sh
Untracked: code/._runMakeeQTLempirical.sh
Untracked: code/._run_bam2bw_all3prime.sh
Untracked: code/._run_bam2bw_extra3.sh
Untracked: code/._run_getApaPval4eqtl.sh
Untracked: code/._run_getapafromeQTL.py
Untracked: code/._run_getapafromeQTL.sh
Untracked: code/._run_getapapval4eqtl_unexp.sh
Untracked: code/._run_leafcutterDiffIso.sh
Untracked: code/._run_sepUsagephen.sh
Untracked: code/._run_sepgenobychrom.sh
Untracked: code/._selectNominalPvalues.py
Untracked: code/._sepUsagePhen.py
Untracked: code/._sepgenobychrom.py
Untracked: code/._snakemakePAS.batch
Untracked: code/._snakemakefiltPAS.batch
Untracked: code/._sortindexRNAbam.sh
Untracked: code/._submit-snakemakePAS.sh
Untracked: code/._submit-snakemakefiltPAS.sh
Untracked: code/._subsetAPAnotEorPgene.py
Untracked: code/._subsetApanoteGene.py
Untracked: code/._subsetUnexplainedeQTLs.py
Untracked: code/._subsetVCF_SS.sh
Untracked: code/._subsetVCF_noSSregions.sh
Untracked: code/._subsetVCF_upstreamPAS.sh
Untracked: code/._subset_diffisopheno.py
Untracked: code/._subsetpermAPAwithGenelist.py
Untracked: code/._subsetvcf_otherreg.sh
Untracked: code/._subsetvcf_permSS.sh
Untracked: code/._subtrachfiveprimeUTR.sh
Untracked: code/._subtractExons.sh
Untracked: code/._subtractfiveprimeUTR.sh
Untracked: code/._tabixSNPS.sh
Untracked: code/._totSeceffectsize.py
Untracked: code/._utrdms2saf.py
Untracked: code/._vcf2bed.py
Untracked: code/.snakemake/
Untracked: code/APAqtl_nominal.err
Untracked: code/APAqtl_nominal.out
Untracked: code/APAqtl_nominal_39.err
Untracked: code/APAqtl_nominal_39.out
Untracked: code/APAqtl_nominal_nonNorm.err
Untracked: code/APAqtl_nominal_nonNorm.out
Untracked: code/APAqtl_permuted.err
Untracked: code/APAqtl_permuted.out
Untracked: code/ApaQTL_nominalNonnorm.sh
Untracked: code/BothFracDTPlot1stintron.err
Untracked: code/BothFracDTPlot1stintron.out
Untracked: code/BothFracDTPlot4stintron.err
Untracked: code/BothFracDTPlot4stintron.out
Untracked: code/BothFracDTPlotGeneRegions.err
Untracked: code/BothFracDTPlotGeneRegions.out
Untracked: code/BothFracDTPlotGeneRegions_norm.err
Untracked: code/BothFracDTPlotGeneRegions_norm.out
Untracked: code/BothFracDTPlotGeneRegions_normalized.sh
Untracked: code/DistPAS2Sig.py
Untracked: code/EandPqtl.err
Untracked: code/EandPqtl.out
Untracked: code/EandPqtl_perm.sh
Untracked: code/EandPqtls.sh
Untracked: code/EncodeRNADTPlotGeneRegions.err
Untracked: code/EncodeRNADTPlotGeneRegions.out
Untracked: code/FC_NucintornUpandDown.sh
Untracked: code/FC_NucintronPASupandDown.err
Untracked: code/FC_NucintronPASupandDown.out
Untracked: code/FC_UTR.err
Untracked: code/FC_UTR.out
Untracked: code/FC_UTR.sh
Untracked: code/FC_intornUpandDownsteamPAS.sh
Untracked: code/FC_intronPASupandDown.err
Untracked: code/FC_intronPASupandDown.out
Untracked: code/FC_nascent.err
Untracked: code/FC_nascentout
Untracked: code/FC_nascentseq.sh
Untracked: code/FC_newPAS_olddata.err
Untracked: code/FC_newPAS_olddata.out
Untracked: code/FC_newPeaks_olddata.sh
Untracked: code/HMMpermuteTotal.py
Untracked: code/HmmPermute.p
Untracked: code/HmmPermute.py
Untracked: code/LC_samplegroups.py
Untracked: code/LD_qtl.sh
Untracked: code/LD_vcftools.hap.out
Untracked: code/NascentDTPlotGeneRegions.err
Untracked: code/NascentDTPlotGeneRegions.out
Untracked: code/NascentDTPlotPAS.err
Untracked: code/NascentDTPlotPAS.out
Untracked: code/NascentDTPlotPAS_3utr.err
Untracked: code/NascentDTPlotPAS_3utr.out
Untracked: code/NascentDTPlotPAS_firstintron.err
Untracked: code/NascentDTPlotPAS_firstintron.out
Untracked: code/NascentDTPlotPAS_intron.err
Untracked: code/NascentDTPlotPAS_intron.out
Untracked: code/NascentDTPlotPAS_nuc.err
Untracked: code/NascentDTPlotPAS_nuc.out
Untracked: code/NascentDTPlotPAS_tot.err
Untracked: code/NascentDTPlotPAS_tot.out
Untracked: code/NascentRNAdtPlot.sh
Untracked: code/NascentRNAdtPlot3UTRPAS.sh
Untracked: code/NascentRNAdtPlotExcludeFirstintronicPAS.sh
Untracked: code/NascentRNAdtPlotFirstintronicPAS.sh
Untracked: code/NascentRNAdtPlotNucPAS.sh
Untracked: code/NascentRNAdtPlotTotPAS.sh
Untracked: code/NascentRNAdtPlotintronicPAS.sh
Untracked: code/NascnetRNAdtPlotPAS.sh
Untracked: code/NetSeq_fourthintronDT.sh
Untracked: code/NomResfromPASSNP.py
Untracked: code/Nuclear_example.err
Untracked: code/Nuclear_example.out
Untracked: code/PrematureQTLNominal.sh
Untracked: code/PrematureQTLPermuted.sh
Untracked: code/Prematureqtl_nominal.err
Untracked: code/Prematureqtl_nominal.out
Untracked: code/Prematureqtl_permuted.err
Untracked: code/Prematureqtl_permuted.out
Untracked: code/QTL2bed.py
Untracked: code/QTL2bed_withstrand.py
Untracked: code/README.md
Untracked: code/RNABam2BW.err
Untracked: code/RNABam2BW.out
Untracked: code/RNAbam2bw.sh
Untracked: code/Rplots.pdf
Untracked: code/Script4NuclearQTLexamples.sh
Untracked: code/Script4TotalQTLexamples.sh
Untracked: code/TESplots100bp.err
Untracked: code/TESplots100bp.out
Untracked: code/TESplots100bp.sh
Untracked: code/TESplots150bp.err
Untracked: code/TESplots150bp.out
Untracked: code/TESplots150bp.sh
Untracked: code/TESplots200bp.err
Untracked: code/TESplots200bp.out
Untracked: code/TESplots200bp.sh
Untracked: code/Total_example.err
Untracked: code/Total_example.out
Untracked: code/Untitled
Untracked: code/Upstream100Bases_general.py
Untracked: code/ZipandTabPheno.sh
Untracked: code/aAPAqtl_nominal39ind.sh
Untracked: code/apaQTLCorrectPvalMakeQQ_4pc.R
Untracked: code/apaQTL_Nominal_4pc.sh
Untracked: code/apaQTL_permuted.4pc.sh
Untracked: code/apafacetboxplots.R
Untracked: code/apaqtlfacetboxplots.R
Untracked: code/assignNucIntonpeak2intronlocs.sh
Untracked: code/assignPeak2Intronicregion.err
Untracked: code/assignPeak2Intronicregion.out
Untracked: code/assignTotIntronpeak2intronlocs.sh
Untracked: code/assigntotPeak2Intronicregion.err
Untracked: code/assigntotPeak2Intronicregion.out
Untracked: code/bam2BW_5primemost.sh
Untracked: code/bam2bw.err
Untracked: code/bam2bw.out
Untracked: code/bam2bw_5primemost.err
Untracked: code/bam2bw_5primemost.out
Untracked: code/bothFracDTplot1stintron.sh
Untracked: code/bothFracDTplot4thintron.sh
Untracked: code/bothFrac_FC.err
Untracked: code/bothFrac_FC.out
Untracked: code/bothFrac_FC.sh
Untracked: code/changePermQTLres2geneName.py
Untracked: code/changenomQTLres2geneName.py
Untracked: code/codingdms2bed.py
Untracked: code/convertNominal2SNPLOC.py
Untracked: code/correctNomeqtl.R
Untracked: code/dag.pdf
Untracked: code/dagPAS.pdf
Untracked: code/dagfiltPAS.pdf
Untracked: code/eQTL_switch2snploc.py
Untracked: code/eQTLgenestestedapa.py
Untracked: code/encodeRNADTplots.sh
Untracked: code/extractGenotypes.py
Untracked: code/extractseqfromqtlfastq.py
Untracked: code/fc2leafphen.py
Untracked: code/finalPASbed2SAF.py
Untracked: code/findbuginpeaks.R
Untracked: code/fix4su304corr.py
Untracked: code/fix4su604corr.py
Untracked: code/fix4sukalisto.py
Untracked: code/fixExandUnexeQTL
Untracked: code/fixExandUnexeQTL.py
Untracked: code/fixFChead_bothfrac.py
Untracked: code/fixFChead_summary.py
Untracked: code/fixH3k12ac.py
Untracked: code/fixPASregionSNPs.py
Untracked: code/fixRNAhead4corr.py
Untracked: code/fixRNAkalisto.py
Untracked: code/fixgroupedtranscript.py
Untracked: code/fixhead_netseqfc.py
Untracked: code/genotypesYRI.gen.proc.keep.vcf.log
Untracked: code/genotypesYRI.gen.proc.keep.vcf.recode.vcf
Untracked: code/get100upPAS.py
Untracked: code/getAPAfromanyeQTL.py
Untracked: code/getApapval4eqtl.py
Untracked: code/getApapval4eqtl_unexp.py
Untracked: code/getDownstreamIntronNuclear.py
Untracked: code/getIntronDownstreamPAS.py
Untracked: code/getIntronUpstreamPAS.py
Untracked: code/getQTLalleles.py
Untracked: code/getQTLfastq.sh
Untracked: code/getSeq100up.sh
Untracked: code/getUpstreamIntronNuclear.py
Untracked: code/getseq100up.err
Untracked: code/getseq100up.out
Untracked: code/grouptranscripts.err
Untracked: code/grouptranscripts.out
Untracked: code/grouptranscripts.py
Untracked: code/intersectPAS_ssSNPS.err
Untracked: code/intersectPAS_ssSNPS.out
Untracked: code/intersectVCFPAS.err
Untracked: code/intersectVCFPAS.out
Untracked: code/intersectVCFandupPAS.sh
Untracked: code/keep5perMAF.py
Untracked: code/keepSNP_vcf.sh
Untracked: code/log/
Untracked: code/makeSAFbothfrac5perc.py
Untracked: code/makeSNP2rsidfile.py
Untracked: code/makeeQTLempirical_unexp.py
Untracked: code/makeeQTLempiricaldist.py
Untracked: code/makegencondeTSSfile.py
Untracked: code/mapSSsnps2PAS.sh
Untracked: code/mergRNABam.sh
Untracked: code/mergeBW_norm.sh
Untracked: code/mergeBWnorm.err
Untracked: code/mergeBWnorm.out
Untracked: code/mergeBamNacent.err
Untracked: code/mergeBamNacent.out
Untracked: code/mergeBamNascent.sh
Untracked: code/mergeRNAbam.err
Untracked: code/mergeRNAbam.out
Untracked: code/mnase1stintron.sh
Untracked: code/mnaseDTPlot1stintron.err
Untracked: code/mnaseDTPlot1stintron.out
Untracked: code/mnaseDTPlot4thintron.err
Untracked: code/mnaseDTPlot4thintron.out
Untracked: code/mnaseDT_fourthintron.sh
Untracked: code/netDTPlot4thintron.out
Untracked: code/netseqDTplot1stIntron.sh
Untracked: code/netseqFC.err
Untracked: code/netseqFC.out
Untracked: code/netseqFC.sh
Untracked: code/neyDTPlot4thintron.err
Untracked: code/nucSpeceffectsize.py
Untracked: code/phenoQTLfromlist.py
Untracked: code/processYRIgen.py
Untracked: code/pttQTLsinapaQTL.py
Untracked: code/pullTwoMechData.py
Untracked: code/qtlFacetBoxplots.err
Untracked: code/qtlFacetBoxplots.out
Untracked: code/qtlRegionseq.sh
Untracked: code/qtlsPvalOppFrac.py
Untracked: code/rLD_vcftools.hap.err
Untracked: code/removeXfromHmm.py
Untracked: code/removeloc_pheno.py
Untracked: code/runCorrectNomEqtl.sh
Untracked: code/runCorrectNomeqtl.err
Untracked: code/runCorrectNomeqtl.out
Untracked: code/runHMMpermute.err
Untracked: code/runHMMpermute.out
Untracked: code/runHMMpermuteAPAqtls.sh
Untracked: code/runHMMpermuteeQTLS.sh
Untracked: code/runHMMpermuteeQTLs.err
Untracked: code/runHMMpermuteeQTLs.out
Untracked: code/runMakeEmpiricaleQTL_unexp.sh
Untracked: code/runMakeEmpiricaleQTLs.err
Untracked: code/runMakeEmpiricaleQTLs.out
Untracked: code/runMakeEmpiricaleQTLsunex.err
Untracked: code/runMakeEmpiricaleQTLsunex.out
Untracked: code/runMakeeQTLempirical.sh
Untracked: code/run_DistPAS2Sig.err
Untracked: code/run_DistPAS2Sig.out
Untracked: code/run_bam2bw.err
Untracked: code/run_bam2bw.out
Untracked: code/run_bam2bw_all3prime.sh
Untracked: code/run_bam2bw_extra3.sh
Untracked: code/run_bam2bwexta.err
Untracked: code/run_bam2bwexta.out
Untracked: code/run_distPAS2Sig.sh
Untracked: code/run_getAPAfromanyeQTL.err
Untracked: code/run_getAPAfromanyeQTL.out
Untracked: code/run_getApaPval4eQTLs.err
Untracked: code/run_getApaPval4eQTLs.out
Untracked: code/run_getApaPval4eQTLsunexplained.err
Untracked: code/run_getApaPval4eQTLsunexplained.out
Untracked: code/run_getApaPval4eqtl.sh
Untracked: code/run_getapafromeQTL.sh
Untracked: code/run_getapapval4eqtl_unexp.sh
Untracked: code/run_leafcutterDiffIso.sh
Untracked: code/run_leafcutter_ds.err
Untracked: code/run_leafcutter_ds.out
Untracked: code/run_qtlFacetBoxplots.sh
Untracked: code/run_sepUsagephen.sh
Untracked: code/run_sepgenobychrom.err
Untracked: code/run_sepgenobychrom.out
Untracked: code/run_sepgenobychrom.sh
Untracked: code/run_sepusage.err
Untracked: code/run_sepusage.out
Untracked: code/selectNominalPvalues.py
Untracked: code/sepUsagePhen.py
Untracked: code/sepgenobychrom.py
Untracked: code/seqQTLfastq.err
Untracked: code/seqQTLfastq.out
Untracked: code/seqQTLregion.err
Untracked: code/seqQTLregion.out
Untracked: code/snakePASlog.out
Untracked: code/snakefiltPASlog.out
Untracked: code/sortindexRNABam.err
Untracked: code/sortindexRNABam.out
Untracked: code/sortindexRNAbam.sh
Untracked: code/subsetAPAnotEorPgene.py
Untracked: code/subsetApanoteGene.py
Untracked: code/subsetUnexplainedeQTLs.py
Untracked: code/subsetVCF_SS.sh
Untracked: code/subsetVCF_noSSregions.sh
Untracked: code/subsetVCF_upstreamPAS.sh
Untracked: code/subset_diffisopheno.py
Untracked: code/subsetpermAPAwithGenelist.py
Untracked: code/subsetvcf_SS.err
Untracked: code/subsetvcf_SS.out
Untracked: code/subsetvcf_noSS.err
Untracked: code/subsetvcf_noSS.out
Untracked: code/subsetvcf_otherreg.sh
Untracked: code/subsetvcf_pas.err
Untracked: code/subsetvcf_pas.out
Untracked: code/subsetvcf_perm.err
Untracked: code/subsetvcf_perm.out
Untracked: code/subsetvcf_permSS.sh
Untracked: code/subsetvcf_rand.err
Untracked: code/subsetvcf_rand.out
Untracked: code/subtract5UTR.err
Untracked: code/subtract5UTR.out
Untracked: code/subtractExons.err
Untracked: code/subtractExons.out
Untracked: code/subtractExons.sh
Untracked: code/subtractfiveprimeUTR.sh
Untracked: code/tabixSNPS.sh
Untracked: code/tabixSNPs.err
Untracked: code/tabixSNPs.out
Untracked: code/totSeceffectsize.py
Untracked: code/transcriptdm2bed.py
Untracked: code/utrdms2saf.py
Untracked: code/vcf2bed.py
Untracked: code/vcf_keepsnps.err
Untracked: code/vcf_keepsnps.out
Untracked: code/writeExampleQTLcode.py
Untracked: code/zipandtabPhen.err
Untracked: code/zipandtabPhen.out
Untracked: data/._.DS_Store
Untracked: data/ApaByEgene/
Untracked: data/ApaByPgene/
Untracked: data/Battle_pQTL/
Untracked: data/CompareOldandNew/
Untracked: data/DTmatrix/
Untracked: data/DiffIso/
Untracked: data/EncodeRNA/
Untracked: data/ExampleQTLPlots/
Untracked: data/GWAS_overlap/
Untracked: data/GeuvadisRNA/
Untracked: data/HMMqtls/
Untracked: data/Li_eQTLs/
Untracked: data/NascentRNA/
Untracked: data/NucSpeceQTLeffect/
Untracked: data/PAS/
Untracked: data/PolyA_DB/
Untracked: data/PreTerm_pheno/
Untracked: data/PrematureQTLNominal/
Untracked: data/PrematureQTLPermuted/
Untracked: data/QTLGenotypes/
Untracked: data/QTLoverlap/
Untracked: data/QTLoverlap_nonNorm/
Untracked: data/README.md
Untracked: data/RNAseq/
Untracked: data/Reads2UTR/
Untracked: data/SNPinSS/
Untracked: data/SignalSiteFiles/
Untracked: data/TF_motifdisruption/
Untracked: data/ThirtyNineIndQtl_nominal/
Untracked: data/apaQTLNominal/
Untracked: data/apaQTLNominal_4pc/
Untracked: data/apaQTLPermuted/
Untracked: data/apaQTLPermuted_4pc/
Untracked: data/apaQTLs/
Untracked: data/assignedPeaks/
Untracked: data/bam/
Untracked: data/bam_clean/
Untracked: data/bam_waspfilt/
Untracked: data/bed_10up/
Untracked: data/bed_clean/
Untracked: data/bed_clean_sort/
Untracked: data/bed_waspfilter/
Untracked: data/bedsort_waspfilter/
Untracked: data/bothFrac_FC/
Untracked: data/bw/
Untracked: data/bw_norm/
Untracked: data/eQTLs/
Untracked: data/exampleQTLs/
Untracked: data/fastq/
Untracked: data/filterPeaks/
Untracked: data/fourSU/
Untracked: data/h3k27ac/
Untracked: data/highdiffsiggenes.txt
Untracked: data/inclusivePeaks/
Untracked: data/inclusivePeaks_FC/
Untracked: data/intronRNAratio/
Untracked: data/intron_analysis/
Untracked: data/locusZoom/
Untracked: data/mergedBG/
Untracked: data/mergedBW_byfrac/
Untracked: data/mergedBW_norm/
Untracked: data/mergedBam/
Untracked: data/mergedbyFracBam/
Untracked: data/molPhenos/
Untracked: data/molQTLs/
Untracked: data/motifdistrupt/
Untracked: data/netseq/
Untracked: data/nonNorm_pheno/
Untracked: data/nuc_10up/
Untracked: data/nuc_10upclean/
Untracked: data/overlapeQTL_try2/
Untracked: data/overlapeQTLs/
Untracked: data/peakCoverage/
Untracked: data/peaks_5perc/
Untracked: data/phenotype/
Untracked: data/phenotype_5perc/
Untracked: data/pttQTL/
Untracked: data/sigDiffGenes.txt
Untracked: data/sort/
Untracked: data/sort_clean/
Untracked: data/sort_waspfilter/
Untracked: data/twoMech/
Untracked: docs/figure/pttgeneinAPA.Rmd/
Untracked: nohup.out
Untracked: output/._.DS_Store
Untracked: output/._meanCorrelationPhenotypes.svg
Untracked: output/dtPlots/
Untracked: output/fastqc/
Untracked: output/meanCorrelationPhenotypes.svg
Unstaged changes:
Modified: analysis/DiffIsoAnalysis.Rmd
Modified: analysis/NuclearSpecAPAqtl.Rmd
Modified: analysis/NuclearSpecIncludeNotTested.Rmd
Modified: analysis/QTLlocation.Rmd
Modified: analysis/Readdistagainstfeatures.Rmd
Modified: analysis/chromHHMQTL.Rmd
Modified: analysis/nonNormQTL.Rmd
Modified: analysis/nucSpecinEQTLs.Rmd
Modified: analysis/overlapapaqtlsandeqtls.Rmd
Modified: analysis/propeQTLs_explained.Rmd
Modified: code/BothFracDTPlotGeneRegions.sh
Modified: code/Snakefile
Deleted: code/Upstream10Bases_general.py
Modified: code/apaQTLCorrectPvalMakeQQ.R
Modified: code/apaQTL_Nominal.sh
Modified: code/apaQTL_permuted.sh
Modified: code/apaQTLsnake.err
Modified: code/bam2bw.sh
Modified: code/bed2saf.py
Modified: code/cluster.json
Modified: code/clusterfiltPAS.json
Modified: code/config.yaml
Modified: code/environment.yaml
Modified: code/makePheno.py
Deleted: code/test.txt
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the R Markdown and HTML files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view them.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | b1e6dd1 | brimittleman | 2019-07-08 | update ptt analysis |
| html | 429432a | brimittleman | 2019-07-02 | Build site. |
| Rmd | fe7b5dc | brimittleman | 2019-07-02 | add eQTL overlap |
| html | dad7bd8 | brimittleman | 2019-07-02 | Build site. |
| Rmd | fe41a93 | brimittleman | 2019-07-02 | add prop of tested genes |
| html | 2a63cde | brimittleman | 2019-07-01 | Build site. |
| Rmd | 8d36f9b | brimittleman | 2019-07-01 | add res |
| html | 5ba28ec | brimittleman | 2019-07-01 | Build site. |
| Rmd | 6db6003 | brimittleman | 2019-07-01 | add qtl code |
| html | a4a34bf | brimittleman | 2019-07-01 | Build site. |
| Rmd | 75b84f4 | brimittleman | 2019-07-01 | add code premature term |
library(reshape2)
library(workflowr)This is workflowr version 1.4.0
Run ?workflowr for help getting startedlibrary(tidyverse)── Attaching packages ───────────────────────────────────────────────────── tidyverse 1.2.1 ──✔ ggplot2 3.1.1 ✔ purrr 0.3.2
✔ tibble 2.1.1 ✔ dplyr 0.8.0.1
✔ tidyr 0.8.3 ✔ stringr 1.3.1
✔ readr 1.3.1 ✔ forcats 0.3.0 ── Conflicts ──────────────────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()Many papers have started to talk about premature termination. Premature terminated isoforms may be truncated protein or may be degraded. I am going to create a measure for this and test for genetic variation associated with it in my data. The measure will be sum of the reads in intronic PAS and the sum of the UTR reads. I will use leafcutter to put the ratios onto a normal distribution. I will then test for QTLs these ratios.
mkdir ../data/PreTerm_phenoPrepare phenotype
Total
gene start and end
genes=read.table("/project2/gilad/briana/genome_anotation_data/RefSeq_annotations/FullTranscriptByName.bed", col.names = c("chr", "Gene_start", "Gene_end", "gene", "score", "strand"),stringsAsFactors = F) %>% select(chr,Gene_start, Gene_end, gene)totalPAS=read.table("../data/phenotype_5perc/APApeak_Phenotype_GeneLocAnno.Total.5perc.fc.gz",stringsAsFactors = F,header = T)
totalPASPheno=totalPAS %>% melt(id.vars="chrom", variable.name="Ind", value.name = "ratio") %>% separate(ratio, into=c("count", "geneCount"), sep="/") %>% separate(chrom, into=c("chr", "start", "end", "geneID"), sep=":") %>% separate(geneID, into=c("gene", "loc","strand", "PAS"), sep="_") %>% filter(loc=="utr3" | loc=="intron") %>% inner_join(genes,by=c("chr", "gene"))%>% mutate(gene=paste(chr,Gene_start, Gene_end, gene,sep=":")) %>% group_by(Ind,gene,loc) %>% summarise(SumCount=sum(as.integer(count))) %>% ungroup() %>% group_by(Ind,gene) %>% mutate(nType=n()) %>% filter(nType==2) %>% spread(loc, SumCount) %>% mutate(total=intron+utr3,PreTermInt=paste(intron,total, sep="/"),PreTermUTR=paste(utr3,total, sep="/")) %>% select(-nType, -intron,-utr3,-total)
totalPASPheno_melt= totalPASPheno %>% melt(id.vars=c("Ind", "gene"), variable.name="Type", value.name = "Value") %>% mutate(chrom=paste(gene, Type, sep="_")) %>% spread(Ind, Value) %>% select(-gene, -Type)
#write.table(totalPASPheno_melt,"../data/PreTerm_pheno/Total_preterminationPheno.txt",quote=F, row.names=F,col.names=T, sep=" ")#python2
gzip ../data/PreTerm_pheno/Total_preterminationPheno.txt
python prepare_phenotype_table.py ../data/PreTerm_pheno/Total_preterminationPheno.txt.gz
#activate env
sh ../data/PreTerm_pheno/Total_preterminationPheno.txt.gz_prepare.sh
#top 2 pcs
head -n 3 ../data/PreTerm_pheno/Total_preterminationPheno.txt.gz.PCs > ../data/PreTerm_pheno/Total_preterminationPheno.txt.gz.2PCs Nuclear
nuclearPAS=read.table("../data/phenotype_5perc/APApeak_Phenotype_GeneLocAnno.Nuclear.5perc.fc.gz",stringsAsFactors = F,header = T)
nuclearPASPheno=nuclearPAS %>% melt(id.vars="chrom", variable.name="Ind", value.name = "ratio") %>% separate(ratio, into=c("count", "geneCount"), sep="/") %>% separate(chrom, into=c("chr", "start", "end", "geneID"), sep=":") %>% separate(geneID, into=c("gene", "loc","strand", "PAS"), sep="_") %>% filter(loc=="utr3" | loc=="intron") %>% inner_join(genes,by=c("chr", "gene"))%>% mutate(gene=paste(chr,Gene_start, Gene_end, gene,sep=":")) %>% group_by(Ind,gene,loc) %>% summarise(SumCount=sum(as.integer(count))) %>% ungroup() %>% group_by(Ind,gene) %>% mutate(nType=n()) %>% filter(nType==2) %>% spread(loc, SumCount) %>% mutate(total=intron+utr3,PreTermInt=paste(intron,total, sep="/"),PreTermUTR=paste(utr3,total, sep="/")) %>% select(-nType, -intron,-utr3,-total)
nuclearPASPheno_melt= nuclearPASPheno %>% melt(id.vars=c("Ind", "gene"), variable.name="Type", value.name = "Value") %>% mutate(chrom=paste(gene, Type, sep="_")) %>% spread(Ind, Value) %>% select(-gene, -Type)
#write.table(nuclearPASPheno_melt,"../data/PreTerm_pheno/Nuclear_preterminationPheno.txt",quote=F, row.names=F,col.names=T, sep=" ")#python2
gzip ../data/PreTerm_pheno/Nuclear_preterminationPheno.txt
python prepare_phenotype_table.py ../data/PreTerm_pheno/Nuclear_preterminationPheno.txt.gz
#env
sh ../data/PreTerm_pheno/Nuclear_preterminationPheno.txt.gz_prepare.sh
#top 2 pcs
head -n 3 ../data/PreTerm_pheno/Nuclear_preterminationPheno.txt.gz.PCs > ../data/PreTerm_pheno/Nuclear_preterminationPheno.txt.gz.2PCs Call QTLs
Sample list from previous work
mkdir ../data/PrematureQTLNominal
mkdir ../data/PrematureQTLPermutedsbatch PrematureQTLNominal.sh
sbatch PrematureQTLPermuted.shMay want to only test one number per gene but do this for now because I want to take advantage of the leafcutter normalization software.
cat ../data/PrematureQTLPermuted/Total_preterminationPheno.txt.gz.qqnorm_chr* > ../data/PrematureQTLPermuted/Total_preterminationPheno.txt.gz.qqnorm_AllChr.txt
cat ../data/PrematureQTLPermuted/Nuclear_preterminationPheno.txt.gz.qqnorm_chr* > ../data/PrematureQTLPermuted/Nuclear_preterminationPheno.txt.gz.qqnorm_AllChr.txtTot
totRes=read.table("../data/PrematureQTLPermuted/Total_preterminationPheno.txt.gz.qqnorm_AllChr.txt", stringsAsFactors = F,col.names = c("pid", "nvar", "shape1", "shape2", "dummy", "sid", "dist", "npval", "slope", "ppval", "bpval"))
totRes$bh=p.adjust(totRes$bpval, method="fdr")
totRes_sig=totRes %>% filter(-log10(bh)>1)
totRes_sig_genes=totRes_sig %>% separate(pid, into=c("chr","start","end", "geneID"), sep=":") %>% separate(geneID, into=c("gene", "Frac"),sep="_") %>% select(gene) %>% unique()
write.table(totRes, file = "../data/PrematureQTLPermuted/Total_preterminationPheno.txt.gz.qqnorm_AllChrBH.txt", col.names = T, row.names = F, quote = F)nrow(totRes_sig_genes)[1] 40Proportion of genes tested:
tottested_genes=totRes %>% separate(pid, into=c("chr","start","end", "geneID"), sep=":") %>% separate(geneID, into=c("gene", "Frac"),sep="_") %>% select(gene) %>% unique()
nrow(totRes_sig_genes)/nrow(tottested_genes)[1] 0.01162453qqplot:
qqplot(-log10(runif(nrow(totRes))), -log10(totRes$bpval),ylab="-log10 Total permuted pvalue", xlab="Uniform expectation", main="Total premature termination")
abline(0,1)
| Version | Author | Date |
|---|---|---|
| dad7bd8 | brimittleman | 2019-07-02 |
ggplot(totRes, aes(x=dist)) + geom_histogram(bins=100)Warning: Removed 340 rows containing non-finite values (stat_bin).
Nuclear:
nucRes=read.table("../data/PrematureQTLPermuted/Nuclear_preterminationPheno.txt.gz.qqnorm_AllChr.txt", stringsAsFactors = F,col.names = c("pid", "nvar", "shape1", "shape2", "dummy", "sid", "dist", "npval", "slope", "ppval", "bpval"))
nucRes$bh=p.adjust(nucRes$bpval, method="fdr")
nucRes_sig=nucRes %>% filter(-log10(bh)>1)
nucRes_sig_genes=nucRes_sig %>% separate(pid, into=c("chr","start","end", "geneID"), sep=":") %>% separate(geneID, into=c("gene", "Frac"),sep="_") %>% select(gene) %>% unique()
write.table(nucRes, file = "../data/PrematureQTLPermuted/Nuclear_preterminationPheno.txt.gz.qqnorm_AllChrBH.txt", col.names = T, row.names = F, quote = F)nrow(nucRes_sig_genes)[1] 103Proportion of genes tested:
nuctested_genes=nucRes %>% separate(pid, into=c("chr","start","end", "geneID"), sep=":") %>% separate(geneID, into=c("gene", "Frac"),sep="_") %>% select(gene) %>% unique()
nrow(nucRes_sig_genes)/nrow(nuctested_genes)[1] 0.02003501qqplot:
qqplot(-log10(runif(nrow(nucRes))), -log10(nucRes$bpval),ylab="-log10 Nuclear permuted pvalue", xlab="Uniform expectation", main="Nuclear premature termination")
abline(0,1)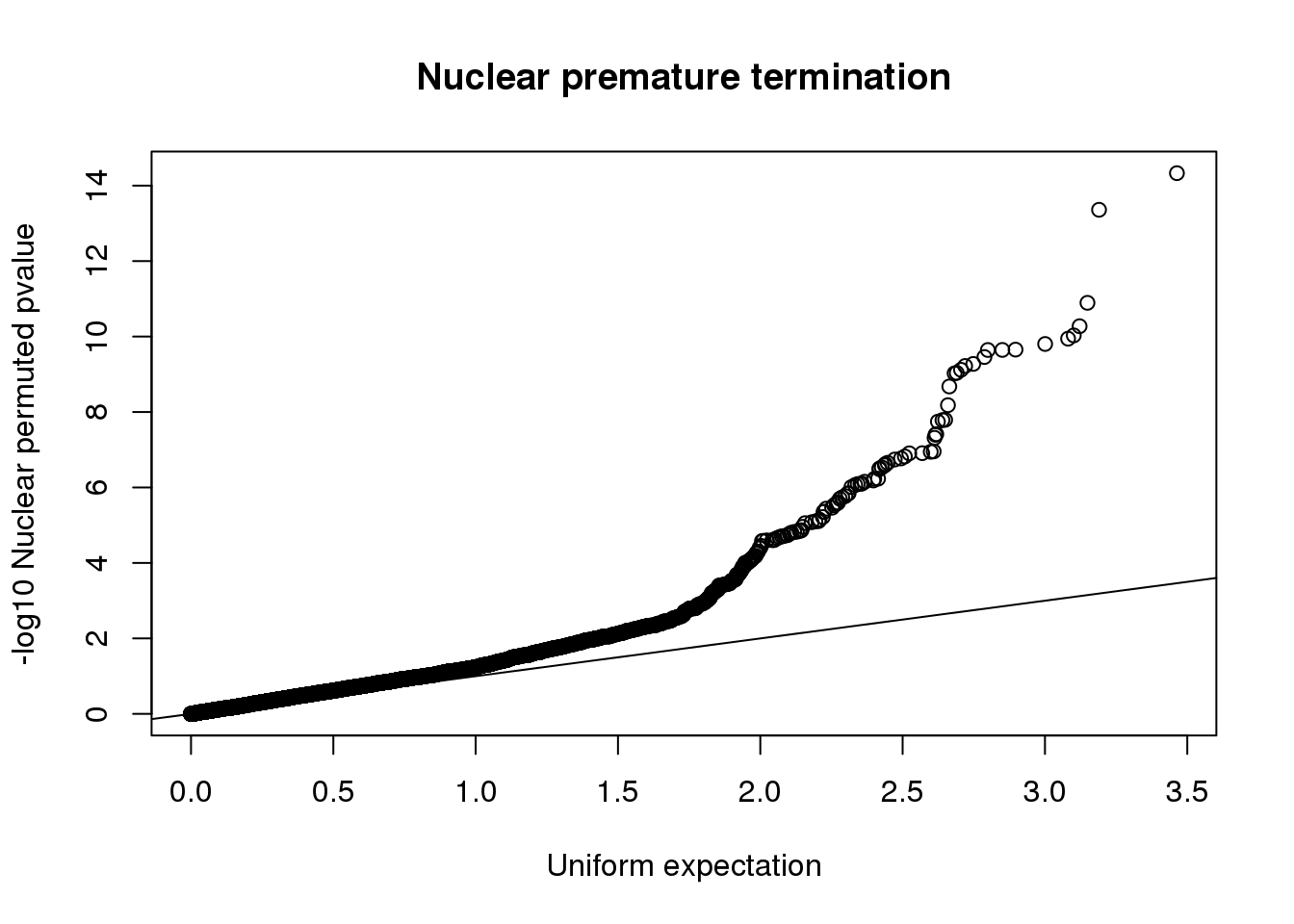 More likely in nuclear:
More likely in nuclear:
prop.test(x=c(nrow(nucRes_sig_genes),nrow(totRes_sig_genes)), n=c(nrow(nuctested_genes),nrow(tottested_genes)),alternative = "greater")
2-sample test for equality of proportions with continuity
correction
data: c(nrow(nucRes_sig_genes), nrow(totRes_sig_genes)) out of c(nrow(nuctested_genes), nrow(tottested_genes))
X-squared = 8.393, df = 1, p-value = 0.001883
alternative hypothesis: greater
95 percent confidence interval:
0.003767208 1.000000000
sample estimates:
prop 1 prop 2
0.02003501 0.01162453 overlap with eGenes
I next want to look at the proportion of eGenes.
explainedEgenes=read.table("../data/Li_eQTLs/explainedEgenes.txt", col.names = c("gene"), stringsAsFactors = F)
unexplainedEgenes=read.table("../data/Li_eQTLs/UnexplainedEgenes.txt", col.names = c("gene"), stringsAsFactors = F)
allEgenes=bind_rows(explainedEgenes, unexplainedEgenes)I want to test the proportion of overlap.
TotPre_uneGene=totRes_sig_genes %>% inner_join(unexplainedEgenes,by="gene")
NucPre_uneGene=nucRes_sig_genes %>% inner_join(unexplainedEgenes,by="gene")
TotPre_exeGene=totRes_sig_genes %>% inner_join(explainedEgenes,by="gene")
NucPre_exeGene=nucRes_sig_genes %>% inner_join(explainedEgenes,by="gene")
TotPre_alleGene=totRes_sig_genes %>% inner_join(allEgenes,by="gene")
NucPre_alleGene=nucRes_sig_genes %>% inner_join(allEgenes,by="gene")Proportion of eGenes explaiend by this:
#total
nrow(TotPre_uneGene)/nrow(unexplainedEgenes)[1] 0.007894737nrow(TotPre_exeGene)/nrow(explainedEgenes)[1] 0.006578947nrow(TotPre_alleGene)/nrow(allEgenes)[1] 0.007127193#nuclear
nrow(NucPre_uneGene)/nrow(unexplainedEgenes)[1] 0.01184211nrow(NucPre_exeGene)/nrow(explainedEgenes)[1] 0.01221805nrow(NucPre_alleGene)/nrow(allEgenes)[1] 0.0120614prop.test(x=c(nrow(NucPre_uneGene),nrow(TotPre_uneGene)), n=c(nrow(unexplainedEgenes),nrow(unexplainedEgenes)))
2-sample test for equality of proportions with continuity
correction
data: c(nrow(NucPre_uneGene), nrow(TotPre_uneGene)) out of c(nrow(unexplainedEgenes), nrow(unexplainedEgenes))
X-squared = 0.26932, df = 1, p-value = 0.6038
alternative hypothesis: two.sided
95 percent confidence interval:
-0.007305071 0.015199808
sample estimates:
prop 1 prop 2
0.011842105 0.007894737 prop.test(x=c(nrow(NucPre_exeGene),nrow(TotPre_exeGene)), n=c(nrow(explainedEgenes),nrow(explainedEgenes)))
2-sample test for equality of proportions with continuity
correction
data: c(nrow(NucPre_exeGene), nrow(TotPre_exeGene)) out of c(nrow(explainedEgenes), nrow(explainedEgenes))
X-squared = 1.2619, df = 1, p-value = 0.2613
alternative hypothesis: two.sided
95 percent confidence interval:
-0.003496441 0.014774636
sample estimates:
prop 1 prop 2
0.012218045 0.006578947 Conclusion:
Total- 13 overlaps with all eGenes, 7 ex, 6 unexplained Nuclear- 24 overlaps with all eGenes, 13 ex, 11 unexpained
All eGenes=1824 Unexplained=760 Explained=1064
Are the total in the nuclear:
totInuc=totRes_sig_genes %>% anti_join(nucRes_sig_genes,by="gene")
nrow(totRes_sig_genes)-nrow(totInuc)[1] 22#did we test all of the
totInucTESTEDnuc=totInuc %>% anti_join(nuctested_genes, by="gene")
nrow(totInucTESTEDnuc)[1] 2totInucTESTEDnuc gene
1 IPO5P1
2 ZNF718#all
totInuc %>% inner_join(allEgenes,by="gene") gene
1 ATF7IP2
2 MTHFSD
3 IPO5P1
4 ELMOD3
5 ANKRD44#explained
totInuc %>% inner_join(explainedEgenes,by="gene") gene
1 ATF7IP2
2 IPO5P1#unexplained
totInuc %>% inner_join(unexplainedEgenes,by="gene") gene
1 MTHFSD
2 ELMOD3
3 ANKRD44Direction of effect size:
nucRes_sig_dir= nucRes_sig %>% separate(pid, into=c("chr","start","end", "geneID"), sep=":") %>% separate(geneID, into=c("gene", "Frac"),sep="_")ggplot(nucRes_sig_dir, aes(x=Frac, y=slope)) + geom_boxplot() + geom_jitter(aes(col=Frac)) + labs(title="Nuclear premature APA QTL effect sizes", y="Effect size",x="") + scale_x_discrete(labels=c( "Intronic Usage","3' UTR Usage"))+ scale_color_discrete(name ="", labels=c( "Intronic Usage","3' UTR Usage")) 
totRes_sig_dir= totRes_sig %>% separate(pid, into=c("chr","start","end", "geneID"), sep=":") %>% separate(geneID, into=c("gene", "Frac"),sep="_")ggplot(totRes_sig_dir, aes(x=Frac, y=slope)) + geom_boxplot() + geom_jitter(aes(col=Frac))+ labs(title="Total premature APA QTL effect sizes", y="Effect size",x="") + scale_x_discrete(labels=c( "Intronic Usage","3' UTR Usage"))+ scale_color_discrete(name ="", labels=c( "Intronic Usage","3' UTR Usage"))  The difference may just be due to the numbers but most of the variants are associated with decreased utr usage and increase intronic usage.
The difference may just be due to the numbers but most of the variants are associated with decreased utr usage and increase intronic usage.
sessionInfo()R version 3.5.1 (2018-07-02)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] forcats_0.3.0 stringr_1.3.1 dplyr_0.8.0.1 purrr_0.3.2
[5] readr_1.3.1 tidyr_0.8.3 tibble_2.1.1 ggplot2_3.1.1
[9] tidyverse_1.2.1 workflowr_1.4.0 reshape2_1.4.3
loaded via a namespace (and not attached):
[1] Rcpp_1.0.0 cellranger_1.1.0 compiler_3.5.1 pillar_1.3.1
[5] git2r_0.25.2 plyr_1.8.4 tools_3.5.1 digest_0.6.18
[9] lubridate_1.7.4 jsonlite_1.6 evaluate_0.12 nlme_3.1-137
[13] gtable_0.2.0 lattice_0.20-38 pkgconfig_2.0.2 rlang_0.3.1
[17] cli_1.0.1 rstudioapi_0.10 yaml_2.2.0 haven_1.1.2
[21] withr_2.1.2 xml2_1.2.0 httr_1.3.1 knitr_1.20
[25] hms_0.4.2 generics_0.0.2 fs_1.2.6 rprojroot_1.3-2
[29] grid_3.5.1 tidyselect_0.2.5 glue_1.3.0 R6_2.3.0
[33] readxl_1.1.0 rmarkdown_1.10 modelr_0.1.2 magrittr_1.5
[37] whisker_0.3-2 backports_1.1.2 scales_1.0.0 htmltools_0.3.6
[41] rvest_0.3.2 assertthat_0.2.0 colorspace_1.3-2 labeling_0.3
[45] stringi_1.2.4 lazyeval_0.2.1 munsell_0.5.0 broom_0.5.1
[49] crayon_1.3.4