Proportion eQTLs explained
Briana Mittleman
6/7/2019
Last updated: 2019-06-13
Checks: 6 0
Knit directory: apaQTL/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.3.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190411) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/.DS_Store
Ignored: output/.DS_Store
Untracked files:
Untracked: .Rprofile
Untracked: ._.DS_Store
Untracked: .gitignore
Untracked: _workflowr.yml
Untracked: analysis/._PASdescriptiveplots.Rmd
Untracked: analysis/._cuttoffPercUsage.Rmd
Untracked: analysis/QTLexampleplots.Rmd
Untracked: analysis/cuttoffPercUsage.Rmd
Untracked: analysis/eQTLoverlap.Rmd
Untracked: analysis/oldstuffNotNeeded.Rmd
Untracked: apaQTL.Rproj
Untracked: code/.NascentRNAdtPlotFirstintronicPAS.sh.swp
Untracked: code/._ApaQTL_nominalNonnorm.sh
Untracked: code/._BothFracDTPlotGeneRegions_normalized.sh
Untracked: code/._FC_NucintornUpandDown.sh
Untracked: code/._FC_UTR.sh
Untracked: code/._FC_intornUpandDownsteamPAS.sh
Untracked: code/._FC_newPeaks_olddata.sh
Untracked: code/._HMMpermuteTotal.py
Untracked: code/._HmmPermute.py
Untracked: code/._LC_samplegroups.py
Untracked: code/._NascentRNAdtPlot.sh
Untracked: code/._NascentRNAdtPlot3UTRPAS.sh
Untracked: code/._NascentRNAdtPlotExcludeFirstintronicPAS.sh
Untracked: code/._NascentRNAdtPlotNucPAS.sh
Untracked: code/._NascentRNAdtPlotTotPAS.sh
Untracked: code/._NascentRNAdtPlotintronicPAS.sh
Untracked: code/._NascnetRNAdtPlotPAS.sh
Untracked: code/._NetSeq_fourthintronDT.sh
Untracked: code/._QTL2bed.py
Untracked: code/._QTL2bed_withstrand.py
Untracked: code/._SnakefilePAS
Untracked: code/._SnakefilefiltPAS
Untracked: code/._TESplots100bp.sh
Untracked: code/._TESplots150bp.sh
Untracked: code/._TESplots200bp.sh
Untracked: code/._Untitled
Untracked: code/._ZipandTabPheno.sh
Untracked: code/._aAPAqtl_nominal39ind.sh
Untracked: code/._apaQTLCorrectPvalMakeQQ.R
Untracked: code/._apaQTL_Nominal.sh
Untracked: code/._apaQTL_permuted.sh
Untracked: code/._assignNucIntonpeak2intronlocs.sh
Untracked: code/._assignTotIntronpeak2intronlocs.sh
Untracked: code/._bam2BW_5primemost.sh
Untracked: code/._bed2saf.py
Untracked: code/._bothFracDTplot1stintron.sh
Untracked: code/._bothFracDTplot4thintron.sh
Untracked: code/._bothFrac_FC.sh
Untracked: code/._callPeaksYL.py
Untracked: code/._chooseAnno2SAF.py
Untracked: code/._chooseSignalSite
Untracked: code/._chooseSignalSite.py
Untracked: code/._cluster.json
Untracked: code/._clusterPAS.json
Untracked: code/._clusterfiltPAS.json
Untracked: code/._codingdms2bed.py
Untracked: code/._config.yaml
Untracked: code/._config2.yaml
Untracked: code/._configOLD.yaml
Untracked: code/._convertNominal2SNPLOC.py
Untracked: code/._convertNumeric.py
Untracked: code/._correctNomeqtl.R
Untracked: code/._dag.pdf
Untracked: code/._eQTLgenestestedapa.py
Untracked: code/._encodeRNADTplots.sh
Untracked: code/._extractGenotypes.py
Untracked: code/._extractseqfromqtlfastq.py
Untracked: code/._fc2leafphen.py
Untracked: code/._filter5perc.R
Untracked: code/._filter5percPheno.py
Untracked: code/._filterpeaks.py
Untracked: code/._finalPASbed2SAF.py
Untracked: code/._fix4su304corr.py
Untracked: code/._fix4su604corr.py
Untracked: code/._fix4sukalisto.py
Untracked: code/._fixExandUnexeQTL
Untracked: code/._fixExandUnexeQTL.py
Untracked: code/._fixFChead.py
Untracked: code/._fixFChead_bothfrac.py
Untracked: code/._fixH3k12ac.py
Untracked: code/._fixRNAhead4corr.py
Untracked: code/._fixRNAkalisto.py
Untracked: code/._fixgroupedtranscript.py
Untracked: code/._fixhead_netseqfc.py
Untracked: code/._getAPAfromanyeQTL.py
Untracked: code/._getApapval4eqtl.py
Untracked: code/._getApapval4eqtl_unexp.py
Untracked: code/._getDownstreamIntronNuclear.py
Untracked: code/._getIntronDownstreamPAS.py
Untracked: code/._getIntronUpstreamPAS.py
Untracked: code/._getQTLalleles.py
Untracked: code/._getQTLfastq.sh
Untracked: code/._getUpstreamIntronNuclear.py
Untracked: code/._grouptranscripts.py
Untracked: code/._keep5perMAF.py
Untracked: code/._keepSNP_vcf.sh
Untracked: code/._make5percPeakbed.py
Untracked: code/._makeFileID.py
Untracked: code/._makePheno.py
Untracked: code/._makeSAFbothfrac5perc.py
Untracked: code/._makeSNP2rsidfile.py
Untracked: code/._makeeQTLempirical_unexp.py
Untracked: code/._makeeQTLempiricaldist.py
Untracked: code/._makegencondeTSSfile.py
Untracked: code/._mergeAllBam.sh
Untracked: code/._mergeBW_norm.sh
Untracked: code/._mergeBamNascent.sh
Untracked: code/._mergeByFracBam.sh
Untracked: code/._mergePeaks.sh
Untracked: code/._mnase1stintron.sh
Untracked: code/._mnaseDT_fourthintron.sh
Untracked: code/._namePeaks.py
Untracked: code/._netseqDTplot1stIntron.sh
Untracked: code/._netseqFC.sh
Untracked: code/._peak2PAS.py
Untracked: code/._peakFC.sh
Untracked: code/._pheno2countonly.R
Untracked: code/._processYRIgen.py
Untracked: code/._qtlRegionseq.sh
Untracked: code/._qtlsPvalOppFrac.py
Untracked: code/._quantassign2parsedpeak.py
Untracked: code/._removeXfromHmm.py
Untracked: code/._removeloc_pheno.py
Untracked: code/._runCorrectNomEqtl.sh
Untracked: code/._runHMMpermuteAPAqtls.sh
Untracked: code/._runHMMpermuteeQTLS.sh
Untracked: code/._runMakeEmpiricaleQTL_unexp.sh
Untracked: code/._runMakeeQTLempirical.sh
Untracked: code/._run_getApaPval4eqtl.sh
Untracked: code/._run_getapafromeQTL.py
Untracked: code/._run_getapafromeQTL.sh
Untracked: code/._run_getapapval4eqtl_unexp.sh
Untracked: code/._run_leafcutterDiffIso.sh
Untracked: code/._run_sepUsagephen.sh
Untracked: code/._run_sepgenobychrom.sh
Untracked: code/._selectNominalPvalues.py
Untracked: code/._sepUsagePhen.py
Untracked: code/._sepgenobychrom.py
Untracked: code/._snakemakePAS.batch
Untracked: code/._snakemakefiltPAS.batch
Untracked: code/._submit-snakemakePAS.sh
Untracked: code/._submit-snakemakefiltPAS.sh
Untracked: code/._subsetApanoteGene.py
Untracked: code/._subsetUnexplainedeQTLs.py
Untracked: code/._subset_diffisopheno.py
Untracked: code/._subsetpermAPAwithGenelist.py
Untracked: code/._subtrachfiveprimeUTR.sh
Untracked: code/._subtractExons.sh
Untracked: code/._subtractfiveprimeUTR.sh
Untracked: code/._tabixSNPS.sh
Untracked: code/._utrdms2saf.py
Untracked: code/.snakemake/
Untracked: code/APAqtl_nominal.err
Untracked: code/APAqtl_nominal.out
Untracked: code/APAqtl_nominal_39.err
Untracked: code/APAqtl_nominal_39.out
Untracked: code/APAqtl_nominal_nonNorm.err
Untracked: code/APAqtl_nominal_nonNorm.out
Untracked: code/APAqtl_permuted.err
Untracked: code/APAqtl_permuted.out
Untracked: code/ApaQTL_nominalNonnorm.sh
Untracked: code/BothFracDTPlot1stintron.err
Untracked: code/BothFracDTPlot1stintron.out
Untracked: code/BothFracDTPlot4stintron.err
Untracked: code/BothFracDTPlot4stintron.out
Untracked: code/BothFracDTPlotGeneRegions.err
Untracked: code/BothFracDTPlotGeneRegions.out
Untracked: code/BothFracDTPlotGeneRegions_norm.err
Untracked: code/BothFracDTPlotGeneRegions_norm.out
Untracked: code/BothFracDTPlotGeneRegions_normalized.sh
Untracked: code/DistPAS2Sig.py
Untracked: code/EncodeRNADTPlotGeneRegions.err
Untracked: code/EncodeRNADTPlotGeneRegions.out
Untracked: code/FC_NucintornUpandDown.sh
Untracked: code/FC_NucintronPASupandDown.err
Untracked: code/FC_NucintronPASupandDown.out
Untracked: code/FC_UTR.err
Untracked: code/FC_UTR.out
Untracked: code/FC_UTR.sh
Untracked: code/FC_intornUpandDownsteamPAS.sh
Untracked: code/FC_intronPASupandDown.err
Untracked: code/FC_intronPASupandDown.out
Untracked: code/FC_newPAS_olddata.err
Untracked: code/FC_newPAS_olddata.out
Untracked: code/FC_newPeaks_olddata.sh
Untracked: code/HMMpermuteTotal.py
Untracked: code/HmmPermute.p
Untracked: code/HmmPermute.py
Untracked: code/LC_samplegroups.py
Untracked: code/NascentDTPlotGeneRegions.err
Untracked: code/NascentDTPlotGeneRegions.out
Untracked: code/NascentDTPlotPAS.err
Untracked: code/NascentDTPlotPAS.out
Untracked: code/NascentDTPlotPAS_3utr.err
Untracked: code/NascentDTPlotPAS_3utr.out
Untracked: code/NascentDTPlotPAS_firstintron.err
Untracked: code/NascentDTPlotPAS_firstintron.out
Untracked: code/NascentDTPlotPAS_intron.err
Untracked: code/NascentDTPlotPAS_intron.out
Untracked: code/NascentDTPlotPAS_nuc.err
Untracked: code/NascentDTPlotPAS_nuc.out
Untracked: code/NascentDTPlotPAS_tot.err
Untracked: code/NascentDTPlotPAS_tot.out
Untracked: code/NascentRNAdtPlot.sh
Untracked: code/NascentRNAdtPlot3UTRPAS.sh
Untracked: code/NascentRNAdtPlotExcludeFirstintronicPAS.sh
Untracked: code/NascentRNAdtPlotFirstintronicPAS.sh
Untracked: code/NascentRNAdtPlotNucPAS.sh
Untracked: code/NascentRNAdtPlotTotPAS.sh
Untracked: code/NascentRNAdtPlotintronicPAS.sh
Untracked: code/NascnetRNAdtPlotPAS.sh
Untracked: code/NetSeq_fourthintronDT.sh
Untracked: code/QTL2bed.py
Untracked: code/QTL2bed_withstrand.py
Untracked: code/README.md
Untracked: code/Rplots.pdf
Untracked: code/TESplots100bp.err
Untracked: code/TESplots100bp.out
Untracked: code/TESplots100bp.sh
Untracked: code/TESplots150bp.err
Untracked: code/TESplots150bp.out
Untracked: code/TESplots150bp.sh
Untracked: code/TESplots200bp.err
Untracked: code/TESplots200bp.out
Untracked: code/TESplots200bp.sh
Untracked: code/Untitled
Untracked: code/Upstream100Bases_general.py
Untracked: code/ZipandTabPheno.sh
Untracked: code/aAPAqtl_nominal39ind.sh
Untracked: code/apaQTLCorrectPvalMakeQQ_4pc.R
Untracked: code/apaQTL_Nominal_4pc.sh
Untracked: code/apaQTL_permuted.4pc.sh
Untracked: code/apafacetboxplots.R
Untracked: code/apaqtlfacetboxplots.R
Untracked: code/assignNucIntonpeak2intronlocs.sh
Untracked: code/assignPeak2Intronicregion.err
Untracked: code/assignPeak2Intronicregion.out
Untracked: code/assignTotIntronpeak2intronlocs.sh
Untracked: code/assigntotPeak2Intronicregion.err
Untracked: code/assigntotPeak2Intronicregion.out
Untracked: code/bam2BW_5primemost.sh
Untracked: code/bam2bw.err
Untracked: code/bam2bw.out
Untracked: code/bam2bw_5primemost.err
Untracked: code/bam2bw_5primemost.out
Untracked: code/bothFracDTplot1stintron.sh
Untracked: code/bothFracDTplot4thintron.sh
Untracked: code/bothFrac_FC.err
Untracked: code/bothFrac_FC.out
Untracked: code/bothFrac_FC.sh
Untracked: code/codingdms2bed.py
Untracked: code/convertNominal2SNPLOC.py
Untracked: code/correctNomeqtl.R
Untracked: code/dag.pdf
Untracked: code/dagPAS.pdf
Untracked: code/dagfiltPAS.pdf
Untracked: code/eQTLgenestestedapa.py
Untracked: code/encodeRNADTplots.sh
Untracked: code/extractGenotypes.py
Untracked: code/extractseqfromqtlfastq.py
Untracked: code/fc2leafphen.py
Untracked: code/finalPASbed2SAF.py
Untracked: code/findbuginpeaks.R
Untracked: code/fix4su304corr.py
Untracked: code/fix4su604corr.py
Untracked: code/fix4sukalisto.py
Untracked: code/fixExandUnexeQTL
Untracked: code/fixExandUnexeQTL.py
Untracked: code/fixFChead_bothfrac.py
Untracked: code/fixFChead_summary.py
Untracked: code/fixH3k12ac.py
Untracked: code/fixRNAhead4corr.py
Untracked: code/fixRNAkalisto.py
Untracked: code/fixgroupedtranscript.py
Untracked: code/fixhead_netseqfc.py
Untracked: code/genotypesYRI.gen.proc.keep.vcf.log
Untracked: code/genotypesYRI.gen.proc.keep.vcf.recode.vcf
Untracked: code/get100upPAS.py
Untracked: code/getAPAfromanyeQTL.py
Untracked: code/getApapval4eqtl.py
Untracked: code/getApapval4eqtl_unexp.py
Untracked: code/getDownstreamIntronNuclear.py
Untracked: code/getIntronDownstreamPAS.py
Untracked: code/getIntronUpstreamPAS.py
Untracked: code/getQTLalleles.py
Untracked: code/getQTLfastq.sh
Untracked: code/getSeq100up.sh
Untracked: code/getUpstreamIntronNuclear.py
Untracked: code/getseq100up.err
Untracked: code/getseq100up.out
Untracked: code/grouptranscripts.err
Untracked: code/grouptranscripts.out
Untracked: code/grouptranscripts.py
Untracked: code/keep5perMAF.py
Untracked: code/keepSNP_vcf.sh
Untracked: code/log/
Untracked: code/makeSAFbothfrac5perc.py
Untracked: code/makeSNP2rsidfile.py
Untracked: code/makeeQTLempirical_unexp.py
Untracked: code/makeeQTLempiricaldist.py
Untracked: code/makegencondeTSSfile.py
Untracked: code/mergeBW_norm.sh
Untracked: code/mergeBWnorm.err
Untracked: code/mergeBWnorm.out
Untracked: code/mergeBamNacent.err
Untracked: code/mergeBamNacent.out
Untracked: code/mergeBamNascent.sh
Untracked: code/mnase1stintron.sh
Untracked: code/mnaseDTPlot1stintron.err
Untracked: code/mnaseDTPlot1stintron.out
Untracked: code/mnaseDTPlot4thintron.err
Untracked: code/mnaseDTPlot4thintron.out
Untracked: code/mnaseDT_fourthintron.sh
Untracked: code/netDTPlot4thintron.out
Untracked: code/netseqDTplot1stIntron.sh
Untracked: code/netseqFC.err
Untracked: code/netseqFC.out
Untracked: code/netseqFC.sh
Untracked: code/neyDTPlot4thintron.err
Untracked: code/processYRIgen.py
Untracked: code/qtlFacetBoxplots.err
Untracked: code/qtlFacetBoxplots.out
Untracked: code/qtlRegionseq.sh
Untracked: code/qtlsPvalOppFrac.py
Untracked: code/removeXfromHmm.py
Untracked: code/removeloc_pheno.py
Untracked: code/runCorrectNomEqtl.sh
Untracked: code/runCorrectNomeqtl.err
Untracked: code/runCorrectNomeqtl.out
Untracked: code/runHMMpermute.err
Untracked: code/runHMMpermute.out
Untracked: code/runHMMpermuteAPAqtls.sh
Untracked: code/runHMMpermuteeQTLS.sh
Untracked: code/runHMMpermuteeQTLs.err
Untracked: code/runHMMpermuteeQTLs.out
Untracked: code/runMakeEmpiricaleQTL_unexp.sh
Untracked: code/runMakeEmpiricaleQTLs.err
Untracked: code/runMakeEmpiricaleQTLs.out
Untracked: code/runMakeEmpiricaleQTLsunex.err
Untracked: code/runMakeEmpiricaleQTLsunex.out
Untracked: code/runMakeeQTLempirical.sh
Untracked: code/run_DistPAS2Sig.err
Untracked: code/run_DistPAS2Sig.out
Untracked: code/run_distPAS2Sig.sh
Untracked: code/run_getAPAfromanyeQTL.err
Untracked: code/run_getAPAfromanyeQTL.out
Untracked: code/run_getApaPval4eQTLs.err
Untracked: code/run_getApaPval4eQTLs.out
Untracked: code/run_getApaPval4eQTLsunexplained.err
Untracked: code/run_getApaPval4eQTLsunexplained.out
Untracked: code/run_getApaPval4eqtl.sh
Untracked: code/run_getapafromeQTL.sh
Untracked: code/run_getapapval4eqtl_unexp.sh
Untracked: code/run_leafcutterDiffIso.sh
Untracked: code/run_leafcutter_ds.err
Untracked: code/run_leafcutter_ds.out
Untracked: code/run_qtlFacetBoxplots.sh
Untracked: code/run_sepUsagephen.sh
Untracked: code/run_sepgenobychrom.err
Untracked: code/run_sepgenobychrom.out
Untracked: code/run_sepgenobychrom.sh
Untracked: code/run_sepusage.err
Untracked: code/run_sepusage.out
Untracked: code/selectNominalPvalues.py
Untracked: code/sepUsagePhen.py
Untracked: code/sepgenobychrom.py
Untracked: code/seqQTLfastq.err
Untracked: code/seqQTLfastq.out
Untracked: code/seqQTLregion.err
Untracked: code/seqQTLregion.out
Untracked: code/snakePASlog.out
Untracked: code/snakefiltPASlog.out
Untracked: code/subsetApanoteGene.py
Untracked: code/subsetUnexplainedeQTLs.py
Untracked: code/subset_diffisopheno.py
Untracked: code/subsetpermAPAwithGenelist.py
Untracked: code/subtract5UTR.err
Untracked: code/subtract5UTR.out
Untracked: code/subtractExons.err
Untracked: code/subtractExons.out
Untracked: code/subtractExons.sh
Untracked: code/subtractfiveprimeUTR.sh
Untracked: code/tabixSNPS.sh
Untracked: code/tabixSNPs.err
Untracked: code/tabixSNPs.out
Untracked: code/transcriptdm2bed.py
Untracked: code/utrdms2saf.py
Untracked: code/vcf_keepsnps.err
Untracked: code/vcf_keepsnps.out
Untracked: code/zipandtabPhen.err
Untracked: code/zipandtabPhen.out
Untracked: data/._.DS_Store
Untracked: data/ApaByEgene/
Untracked: data/CompareOldandNew/
Untracked: data/DTmatrix/
Untracked: data/DiffIso/
Untracked: data/EncodeRNA/
Untracked: data/ExampleQTLPlots/
Untracked: data/GeuvadisRNA/
Untracked: data/HMMqtls/
Untracked: data/Li_eQTLs/
Untracked: data/NascentRNA/
Untracked: data/PAS/
Untracked: data/QTLGenotypes/
Untracked: data/QTLoverlap/
Untracked: data/QTLoverlap_nonNorm/
Untracked: data/README.md
Untracked: data/RNAseq/
Untracked: data/Reads2UTR/
Untracked: data/SignalSiteFiles/
Untracked: data/ThirtyNineIndQtl_nominal/
Untracked: data/apaQTLNominal/
Untracked: data/apaQTLNominal_4pc/
Untracked: data/apaQTLPermuted/
Untracked: data/apaQTLPermuted_4pc/
Untracked: data/apaQTLs/
Untracked: data/assignedPeaks/
Untracked: data/bam/
Untracked: data/bam_clean/
Untracked: data/bam_waspfilt/
Untracked: data/bed_10up/
Untracked: data/bed_clean/
Untracked: data/bed_clean_sort/
Untracked: data/bed_waspfilter/
Untracked: data/bedsort_waspfilter/
Untracked: data/bothFrac_FC/
Untracked: data/bw_norm/
Untracked: data/eQTLs/
Untracked: data/exampleQTLs/
Untracked: data/fastq/
Untracked: data/filterPeaks/
Untracked: data/fourSU/
Untracked: data/h3k27ac/
Untracked: data/highdiffsiggenes.txt
Untracked: data/inclusivePeaks/
Untracked: data/inclusivePeaks_FC/
Untracked: data/intronRNAratio/
Untracked: data/intron_analysis/
Untracked: data/mergedBG/
Untracked: data/mergedBW_byfrac/
Untracked: data/mergedBW_norm/
Untracked: data/mergedBam/
Untracked: data/mergedbyFracBam/
Untracked: data/motifdistrupt/
Untracked: data/netseq/
Untracked: data/nonNorm_pheno/
Untracked: data/nuc_10up/
Untracked: data/nuc_10upclean/
Untracked: data/overlapeQTL_try2/
Untracked: data/overlapeQTLs/
Untracked: data/peakCoverage/
Untracked: data/peaks_5perc/
Untracked: data/phenotype/
Untracked: data/phenotype_5perc/
Untracked: data/sigDiffGenes.txt
Untracked: data/sort/
Untracked: data/sort_clean/
Untracked: data/sort_waspfilter/
Untracked: nohup.out
Untracked: output/._.DS_Store
Untracked: output/._meanCorrelationPhenotypes.svg
Untracked: output/dtPlots/
Untracked: output/fastqc/
Untracked: output/meanCorrelationPhenotypes.svg
Unstaged changes:
Modified: analysis/Readdistagainstfeatures.Rmd
Modified: analysis/index.Rmd
Modified: analysis/nascenttranscription.Rmd
Modified: analysis/nucintronicanalysis.Rmd
Modified: analysis/overlapapaqtlsandeqtls.Rmd
Modified: analysis/rna_netseq_h3k12ac.Rmd
Modified: code/BothFracDTPlotGeneRegions.sh
Modified: code/Snakefile
Deleted: code/Upstream10Bases_general.py
Modified: code/apaQTLCorrectPvalMakeQQ.R
Modified: code/apaQTL_Nominal.sh
Modified: code/apaQTL_permuted.sh
Modified: code/apaQTLsnake.err
Modified: code/bam2bw.sh
Modified: code/bed2saf.py
Modified: code/cluster.json
Modified: code/clusterfiltPAS.json
Modified: code/config.yaml
Modified: code/environment.yaml
Modified: code/makePheno.py
Deleted: code/test.txt
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the R Markdown and HTML files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view them.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 2fd2b27 | brimittleman | 2019-06-13 | fix bug |
| html | b907ac1 | brimittleman | 2019-06-12 | Build site. |
| Rmd | 178c5dc | brimittleman | 2019-06-12 | new geno |
| html | 6b164c8 | brimittleman | 2019-06-07 | Build site. |
| Rmd | b39620d | brimittleman | 2019-06-07 | add bonfor results |
| html | 458e494 | brimittleman | 2019-06-07 | Build site. |
| Rmd | 32091ee | brimittleman | 2019-06-07 | more prop explained to new analysis |
library(tidyverse)── Attaching packages ─────────────────────────────────────────────────────────────── tidyverse 1.2.1 ──✔ ggplot2 3.1.1 ✔ purrr 0.3.2
✔ tibble 2.1.1 ✔ dplyr 0.8.0.1
✔ tidyr 0.8.3 ✔ stringr 1.3.1
✔ readr 1.3.1 ✔ forcats 0.3.0 ── Conflicts ────────────────────────────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()library(workflowr)This is workflowr version 1.3.0
Run ?workflowr for help getting startedlibrary(reshape2)
Attaching package: 'reshape2'The following object is masked from 'package:tidyr':
smithsI need to fix the explained_FDR10.sort.txt and unexplained_FDR10.sort.txt files because right now this file has multiple genes per snp.
python fixExandUnexeQTL.py ../data/Li_eQTLs/explained_FDR10.sort.txt ../data/Li_eQTLs/explained_FDR10.sort_FIXED.txt
python fixExandUnexeQTL.py ../data/Li_eQTLs/unexplained_FDR10.sort.txt ../data/Li_eQTLs/unexplained_FDR10.sort_FIXED.txtThere are 1195 explained and 814 unexplained eQTLs. I will next look at each of these in my apadata.
Convert nominal results to have snps rather than rsids:
python convertNominal2SNPLOC.py Total
python convertNominal2SNPLOC.py Nuclearmkdir ../data/overlapeQTL_try2
sbatch run_getapafromeQTL.sh
total
I can group the unexplained by gene and snp then I can ask if there is at least 1 significat peak for each of these.
I will use the bonforoni correction here and multiply the pvalue by the number of peaks in the gene:snp association.
nomnames=c("peakID", 'snp','dist', 'pval', 'slope')
totalapaUnexplained=read.table("../data/overlapeQTL_try2/apaTotal_unexplainedQTLs.txt", stringsAsFactors = F, col.names = nomnames)
totalapaUnexplained=totalapaUnexplained %>% separate(peakID, into=c("chr","start","end","geneID"), sep=":") %>% separate(geneID, into=c("gene", "loc", "strand", "PASnum"), sep="_") %>% group_by(gene, snp) %>% mutate(nPeaks=n(), adjPval=pval* nPeaks)%>% dplyr::slice(which.min(adjPval))
totalapaUnexplained_sig= totalapaUnexplained %>% filter(adjPval<.05)Look at distribution of these pvals:
ggplot(totalapaUnexplained, aes(x=adjPval)) + geom_histogram(bins=50)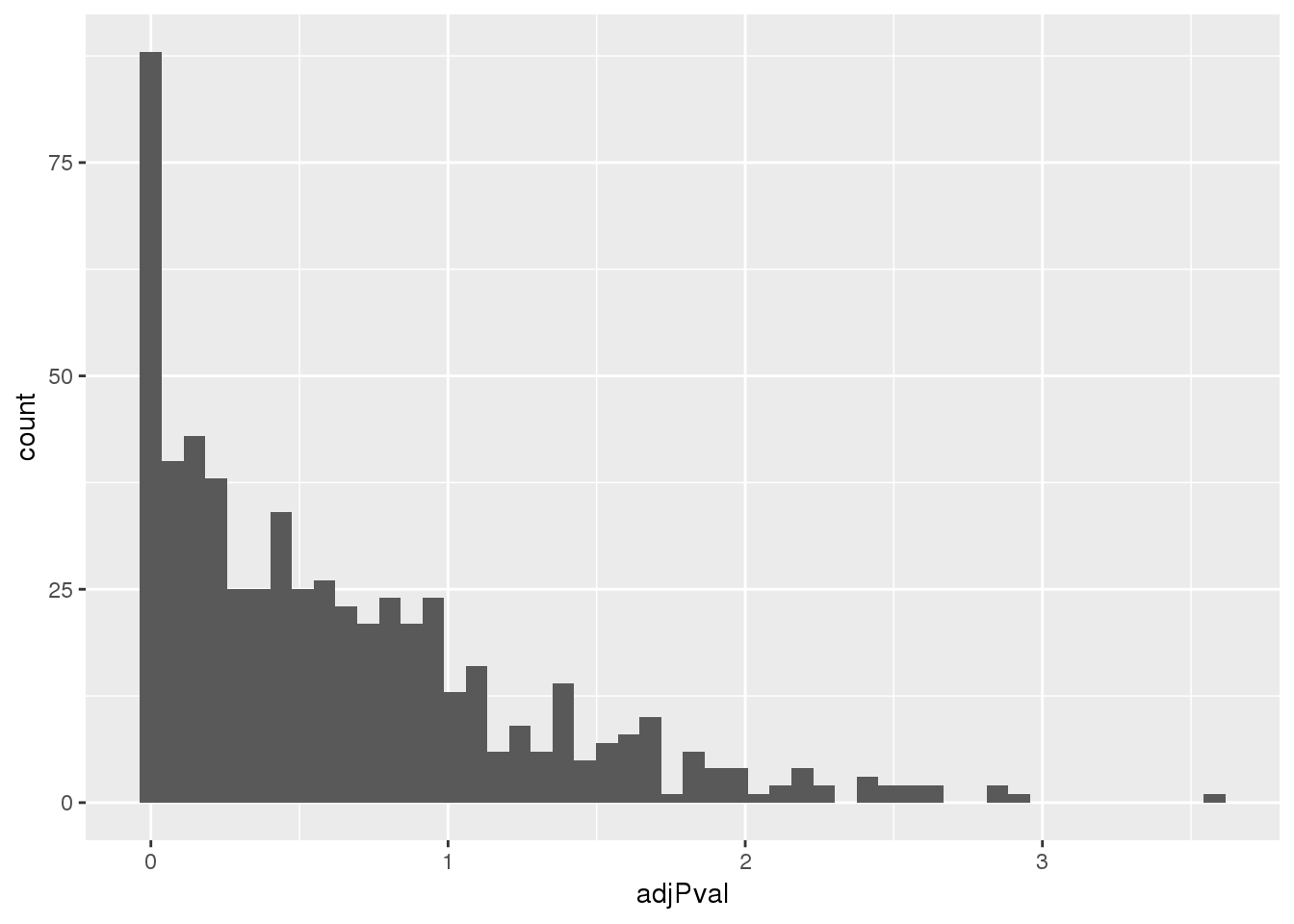
| Version | Author | Date |
|---|---|---|
| b907ac1 | brimittleman | 2019-06-12 |
Proportion explained:
nrow(totalapaUnexplained_sig)/nrow(totalapaUnexplained)[1] 0.1632653I tested 588 unexplained eQTLs in the total fraction and 96 have a bonforoni corrected significant peak.
Compare to explained eQTLS:
totalapaexplained=read.table("../data/overlapeQTL_try2/apaTotal_explainedQTLs.txt", stringsAsFactors = F, col.names = nomnames) %>% separate(peakID, into=c("chr","start","end","geneID"), sep=":") %>% separate(geneID, into=c("gene", "loc", "strand", "PASnum"), sep="_") %>% group_by(gene, snp) %>% mutate(nPeaks=n(), adjPval=pval* nPeaks) %>% dplyr::slice(which.min(adjPval))
totalapaexplained_sig= totalapaexplained %>% filter(adjPval<.05)
nrow(totalapaexplained_sig)/nrow(totalapaexplained)[1] 0.1304878I am testing 820 explained eQTLs and of those 107 have a bonforoni corrected significant peak.
difference of proportions:
prop.test(x=c(nrow(totalapaUnexplained_sig),nrow(totalapaexplained_sig)), n=c(nrow(totalapaUnexplained),nrow(totalapaexplained)))
2-sample test for equality of proportions with continuity
correction
data: c(nrow(totalapaUnexplained_sig), nrow(totalapaexplained_sig)) out of c(nrow(totalapaUnexplained), nrow(totalapaexplained))
X-squared = 2.722, df = 1, p-value = 0.09898
alternative hypothesis: two.sided
95 percent confidence interval:
-0.00641871 0.07197371
sample estimates:
prop 1 prop 2
0.1632653 0.1304878 ggplot(totalapaUnexplained_sig,aes(x=loc)) + geom_histogram(stat="count",aes(y=..count../sum(..count..))) + labs(y="Proportion", title = "Total apaQTLs explaining eQTLs")Warning: Ignoring unknown parameters: binwidth, bins, pad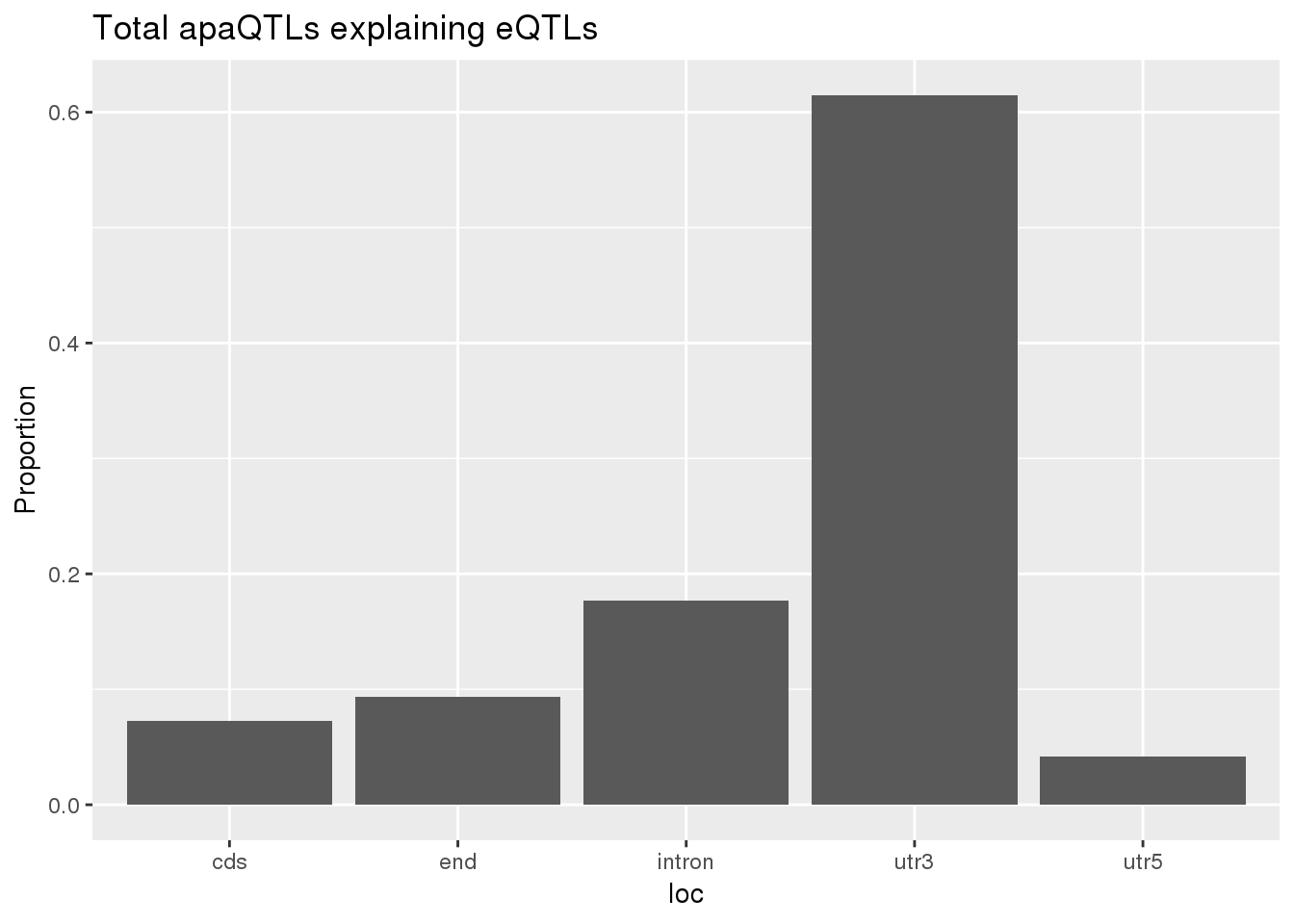
| Version | Author | Date |
|---|---|---|
| b907ac1 | brimittleman | 2019-06-12 |
totalapaUnexplained_sig_loc= totalapaUnexplained_sig %>% group_by(loc) %>% summarise(nLocTotalUn=n()) %>% mutate(propTotalUn=nLocTotalUn/nrow(totalapaUnexplained_sig))
totalapaexplained_sig_loc= totalapaexplained_sig %>% group_by(loc) %>% summarise(nLocTotalEx=n()) %>% mutate(propTotalEx=nLocTotalEx/nrow(totalapaexplained_sig))
BothTotalLoc=totalapaUnexplained_sig_loc %>% full_join(totalapaexplained_sig_loc,by="loc") %>% replace_na(list(propTotalUn = 0, nLocTotalUn = 0,propTotalEx=0,nLocTotalEx=0 ))
BothTotalLoc# A tibble: 5 x 5
loc nLocTotalUn propTotalUn nLocTotalEx propTotalEx
<chr> <dbl> <dbl> <dbl> <dbl>
1 cds 7 0.0729 8 0.0748
2 end 9 0.0938 7 0.0654
3 intron 17 0.177 20 0.187
4 utr3 59 0.615 70 0.654
5 utr5 4 0.0417 2 0.0187nuclear
nuclearapaUnexplained=read.table("../data/overlapeQTL_try2/apaNuclear_unexplainedQTLs.txt", stringsAsFactors = F, col.names = nomnames) %>% separate(peakID, into=c("chr","start","end","geneID"), sep=":") %>% separate(geneID, into=c("gene", "loc", "strand", "PASnum"), sep="_") %>% group_by(gene, snp) %>% mutate(nPeaks=n(), adjPval=pval* nPeaks) %>% dplyr::slice(which.min(adjPval))
nuclearapaUnexplained_sig= nuclearapaUnexplained %>% filter(adjPval<.05)
nrow(nuclearapaUnexplained_sig)/nrow(nuclearapaUnexplained)[1] 0.1649832I tested 594 unexplained eQTLs in the nuclear fraction and 98 have a bonforoni corrected significant peak.
nuclearapaexplained=read.table("../data/overlapeQTL_try2/apaNuclear_explainedQTLs.txt", stringsAsFactors = F, col.names = nomnames) %>% separate(peakID, into=c("chr","start","end","geneID"), sep=":") %>% separate(geneID, into=c("gene", "loc", "strand", "PASnum"), sep="_") %>% group_by(gene, snp) %>% mutate(nPeaks=n(), adjPval=pval* nPeaks) %>% dplyr::slice(which.min(adjPval))
nuclearapaexplained_sig= nuclearapaexplained %>% filter(adjPval<.05)
nrow(nuclearapaexplained_sig)/nrow(nuclearapaexplained)[1] 0.13269I tested 829 explained eQTLs in the nuclear fraction and 110 have a nominally significant peak. difference of proportions:
prop.test(x=c(nrow(nuclearapaUnexplained_sig),nrow(nuclearapaexplained_sig)), n=c(nrow(nuclearapaUnexplained),nrow(nuclearapaexplained)))
2-sample test for equality of proportions with continuity
correction
data: c(nrow(nuclearapaUnexplained_sig), nrow(nuclearapaexplained_sig)) out of c(nrow(nuclearapaUnexplained), nrow(nuclearapaexplained))
X-squared = 2.6386, df = 1, p-value = 0.1043
alternative hypothesis: two.sided
95 percent confidence interval:
-0.006890426 0.071476780
sample estimates:
prop 1 prop 2
0.1649832 0.1326900 ggplot(nuclearapaUnexplained_sig,aes(x=loc)) + geom_histogram(stat="count",aes(y=..count../sum(..count..))) + labs(title = "Nuclear apaQTLs explaining eQTLs", y="Proportion")Warning: Ignoring unknown parameters: binwidth, bins, pad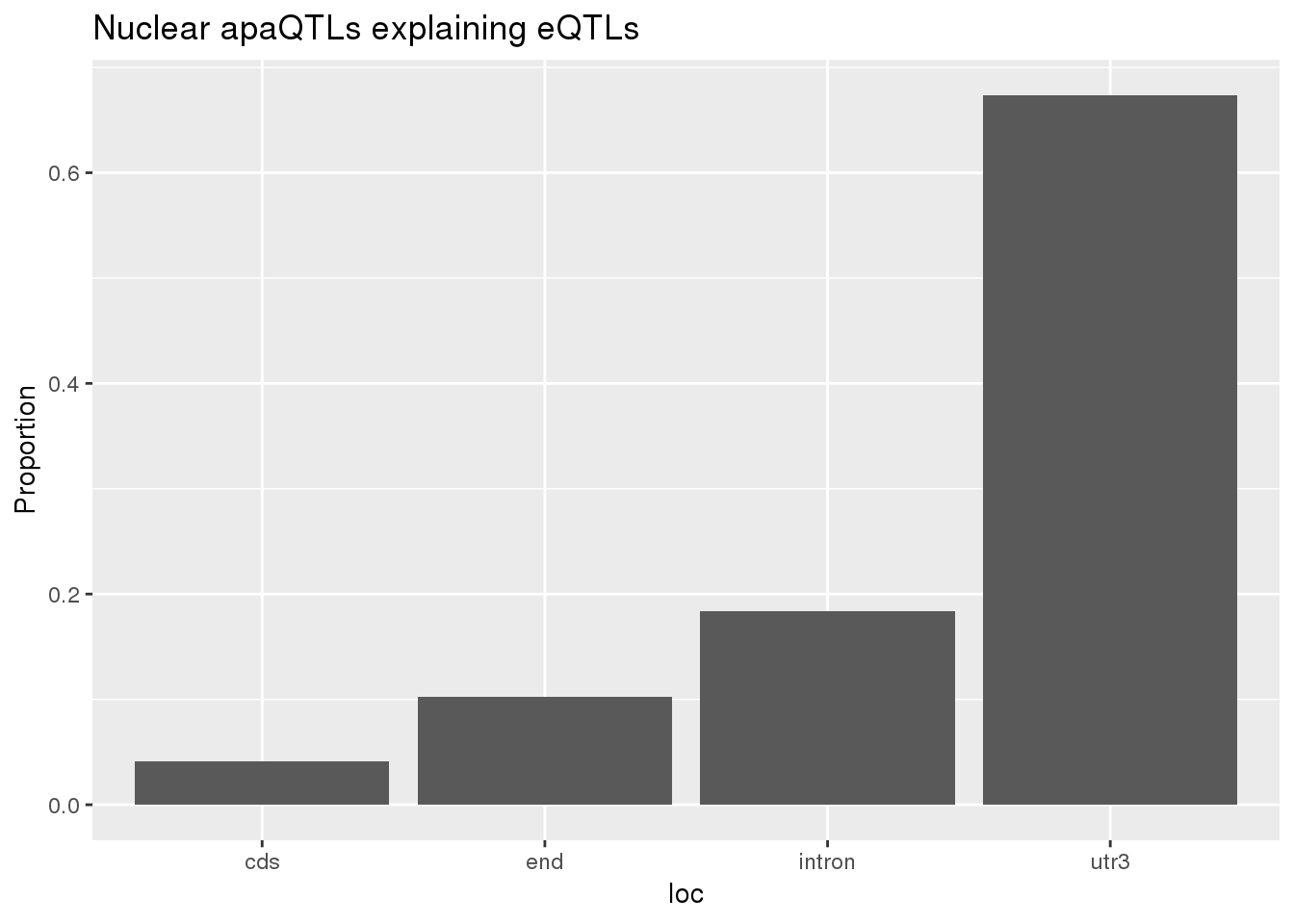
| Version | Author | Date |
|---|---|---|
| b907ac1 | brimittleman | 2019-06-12 |
nuclearapaUnexplained_sig_loc= nuclearapaUnexplained_sig %>% group_by(loc) %>% summarise(nLocnuclearUn=n()) %>% mutate(propnuclearUn=nLocnuclearUn/nrow(nuclearapaUnexplained_sig))
nuclearapaexplained_sig_loc= nuclearapaexplained_sig %>% group_by(loc) %>% summarise(nLocnuclearEx=n()) %>% mutate(propnuclearEx=nLocnuclearEx/nrow(nuclearapaexplained_sig))
BothnuclearLoc=nuclearapaUnexplained_sig_loc %>% full_join(nuclearapaexplained_sig_loc,by="loc") %>% replace_na(list(propnuclearUn = 0, nLocnuclearUn = 0,propnuclearEx=0,nLocnuclearEx=0 ))
BothnuclearLoc# A tibble: 5 x 5
loc nLocnuclearUn propnuclearUn nLocnuclearEx propnuclearEx
<chr> <dbl> <dbl> <dbl> <dbl>
1 cds 4 0.0408 3 0.0273
2 end 10 0.102 9 0.0818
3 intron 18 0.184 33 0.3
4 utr3 66 0.673 63 0.573
5 utr5 0 0 2 0.0182prop.test(x=c(18,33), n=c(nrow(nuclearapaUnexplained_sig),nrow(nuclearapaexplained_sig)))
2-sample test for equality of proportions with continuity
correction
data: c(18, 33) out of c(nrow(nuclearapaUnexplained_sig), nrow(nuclearapaexplained_sig))
X-squared = 3.1869, df = 1, p-value = 0.07423
alternative hypothesis: two.sided
95 percent confidence interval:
-0.240913267 0.008260206
sample estimates:
prop 1 prop 2
0.1836735 0.3000000 prop.test(x=c(66,63), n=c(nrow(nuclearapaUnexplained_sig),nrow(nuclearapaexplained_sig)))
2-sample test for equality of proportions with continuity
correction
data: c(66, 63) out of c(nrow(nuclearapaUnexplained_sig), nrow(nuclearapaexplained_sig))
X-squared = 1.8258, df = 1, p-value = 0.1766
alternative hypothesis: two.sided
95 percent confidence interval:
-0.03992433 0.24140856
sample estimates:
prop 1 prop 2
0.6734694 0.5727273 total v nuclear
prop.test(x=c(nrow(nuclearapaUnexplained_sig),nrow(totalapaUnexplained_sig)), n=c(nrow(nuclearapaUnexplained),nrow(totalapaUnexplained)))
2-sample test for equality of proportions with continuity
correction
data: c(nrow(nuclearapaUnexplained_sig), nrow(totalapaUnexplained_sig)) out of c(nrow(nuclearapaUnexplained), nrow(totalapaUnexplained))
X-squared = 1.4301e-06, df = 1, p-value = 0.999
alternative hypothesis: two.sided
95 percent confidence interval:
-0.04220475 0.04564046
sample estimates:
prop 1 prop 2
0.1649832 0.1632653 Differences in proportion by location
allLocProp=BothnuclearLoc %>% full_join(BothTotalLoc, by="loc") %>% select(loc,propnuclearUn,propnuclearEx,propTotalUn,propTotalEx )
allLocPropmelt= melt(allLocProp, id.vars = "loc") %>% mutate(Fraction=ifelse(grepl("Total", variable), "Total", "Nuclear"),eQTL=ifelse(grepl("Un", variable), "Unexplained", "Explained"))
ggplot(allLocPropmelt,aes(x=loc, fill=eQTL, y=value)) + geom_histogram(stat="identity", position = "dodge") + facet_grid(~Fraction)+ labs(y="Proportion of PAS", title="apaQTLs overlaping eQTLs by PAS location")Warning: Ignoring unknown parameters: binwidth, bins, pad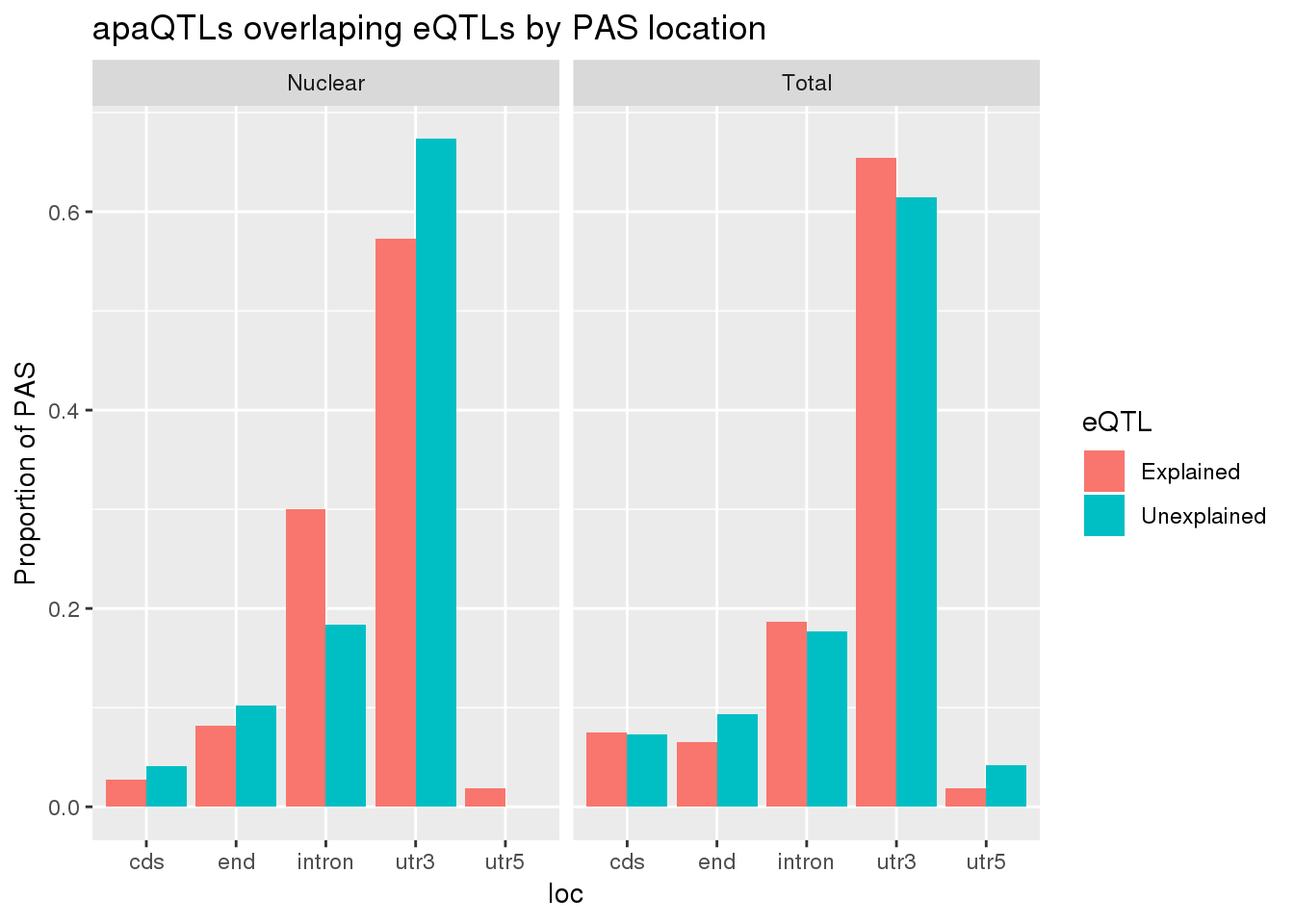
| Version | Author | Date |
|---|---|---|
| b907ac1 | brimittleman | 2019-06-12 |
This is a very stringent test. A less stringent way to get an upper bound would be to make an informed decision about which peak to use. This will make it so I am only testing one PAS per gene.
sessionInfo()R version 3.5.1 (2018-07-02)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] reshape2_1.4.3 workflowr_1.3.0 forcats_0.3.0 stringr_1.3.1
[5] dplyr_0.8.0.1 purrr_0.3.2 readr_1.3.1 tidyr_0.8.3
[9] tibble_2.1.1 ggplot2_3.1.1 tidyverse_1.2.1
loaded via a namespace (and not attached):
[1] Rcpp_1.0.0 cellranger_1.1.0 pillar_1.3.1 compiler_3.5.1
[5] git2r_0.25.2 plyr_1.8.4 tools_3.5.1 digest_0.6.18
[9] lubridate_1.7.4 jsonlite_1.6 evaluate_0.12 nlme_3.1-137
[13] gtable_0.2.0 lattice_0.20-38 pkgconfig_2.0.2 rlang_0.3.1
[17] cli_1.0.1 rstudioapi_0.10 yaml_2.2.0 haven_1.1.2
[21] withr_2.1.2 xml2_1.2.0 httr_1.3.1 knitr_1.20
[25] hms_0.4.2 generics_0.0.2 fs_1.2.6 rprojroot_1.3-2
[29] grid_3.5.1 tidyselect_0.2.5 glue_1.3.0 R6_2.3.0
[33] fansi_0.4.0 readxl_1.1.0 rmarkdown_1.10 modelr_0.1.2
[37] magrittr_1.5 whisker_0.3-2 backports_1.1.2 scales_1.0.0
[41] htmltools_0.3.6 rvest_0.3.2 assertthat_0.2.0 colorspace_1.3-2
[45] labeling_0.3 utf8_1.1.4 stringi_1.2.4 lazyeval_0.2.1
[49] munsell_0.5.0 broom_0.5.1 crayon_1.3.4