Usage Effect Sizes
Briana Mittleman
5/24/2019
Last updated: 2019-05-28
Checks: 6 0
Knit directory: apaQTL/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.3.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190411) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: output/.DS_Store
Untracked files:
Untracked: .Rprofile
Untracked: ._.DS_Store
Untracked: .gitignore
Untracked: _workflowr.yml
Untracked: analysis/._PASdescriptiveplots.Rmd
Untracked: analysis/._cuttoffPercUsage.Rmd
Untracked: analysis/cuttoffPercUsage.Rmd
Untracked: analysis/nascentRNA.Rmd
Untracked: apaQTL.Rproj
Untracked: code/._ApaQTL_nominalNonnorm.sh
Untracked: code/._BothFracDTPlotGeneRegions_normalized.sh
Untracked: code/._FC_UTR.sh
Untracked: code/._FC_newPeaks_olddata.sh
Untracked: code/._LC_samplegroups.py
Untracked: code/._NetSeq_fourthintronDT.sh
Untracked: code/._SnakefilePAS
Untracked: code/._SnakefilefiltPAS
Untracked: code/._TESplots100bp.sh
Untracked: code/._TESplots150bp.sh
Untracked: code/._TESplots200bp.sh
Untracked: code/._Untitled
Untracked: code/._ZipandTabPheno.sh
Untracked: code/._aAPAqtl_nominal39ind.sh
Untracked: code/._apaQTLCorrectPvalMakeQQ.R
Untracked: code/._apaQTL_Nominal.sh
Untracked: code/._apaQTL_permuted.sh
Untracked: code/._assignNucIntonpeak2intronlocs.sh
Untracked: code/._assignTotIntronpeak2intronlocs.sh
Untracked: code/._bam2BW_5primemost.sh
Untracked: code/._bed2saf.py
Untracked: code/._bothFracDTplot1stintron.sh
Untracked: code/._bothFracDTplot4thintron.sh
Untracked: code/._bothFrac_FC.sh
Untracked: code/._callPeaksYL.py
Untracked: code/._chooseAnno2SAF.py
Untracked: code/._chooseSignalSite
Untracked: code/._chooseSignalSite.py
Untracked: code/._cluster.json
Untracked: code/._clusterPAS.json
Untracked: code/._clusterfiltPAS.json
Untracked: code/._codingdms2bed.py
Untracked: code/._config.yaml
Untracked: code/._config2.yaml
Untracked: code/._configOLD.yaml
Untracked: code/._convertNumeric.py
Untracked: code/._dag.pdf
Untracked: code/._encodeRNADTplots.sh
Untracked: code/._extractGenotypes.py
Untracked: code/._fc2leafphen.py
Untracked: code/._filter5perc.R
Untracked: code/._filter5percPheno.py
Untracked: code/._filterpeaks.py
Untracked: code/._finalPASbed2SAF.py
Untracked: code/._fix4su304corr.py
Untracked: code/._fix4su604corr.py
Untracked: code/._fix4sukalisto.py
Untracked: code/._fixFChead.py
Untracked: code/._fixFChead_bothfrac.py
Untracked: code/._fixH3k12ac.py
Untracked: code/._fixRNAhead4corr.py
Untracked: code/._fixRNAkalisto.py
Untracked: code/._fixgroupedtranscript.py
Untracked: code/._fixhead_netseqfc.py
Untracked: code/._grouptranscripts.py
Untracked: code/._make5percPeakbed.py
Untracked: code/._makeFileID.py
Untracked: code/._makePheno.py
Untracked: code/._makeSAFbothfrac5perc.py
Untracked: code/._makegencondeTSSfile.py
Untracked: code/._mergeAllBam.sh
Untracked: code/._mergeBW_norm.sh
Untracked: code/._mergeByFracBam.sh
Untracked: code/._mergePeaks.sh
Untracked: code/._mnase1stintron.sh
Untracked: code/._mnaseDT_fourthintron.sh
Untracked: code/._namePeaks.py
Untracked: code/._netseqDTplot1stIntron.sh
Untracked: code/._netseqFC.sh
Untracked: code/._peak2PAS.py
Untracked: code/._peakFC.sh
Untracked: code/._pheno2countonly.R
Untracked: code/._qtlsPvalOppFrac.py
Untracked: code/._quantassign2parsedpeak.py
Untracked: code/._removeloc_pheno.py
Untracked: code/._run_leafcutterDiffIso.sh
Untracked: code/._run_sepUsagephen.sh
Untracked: code/._selectNominalPvalues.py
Untracked: code/._sepUsagePhen.py
Untracked: code/._snakemakePAS.batch
Untracked: code/._snakemakefiltPAS.batch
Untracked: code/._submit-snakemakePAS.sh
Untracked: code/._submit-snakemakefiltPAS.sh
Untracked: code/._subset_diffisopheno.py
Untracked: code/._subtrachfiveprimeUTR.sh
Untracked: code/._subtractExons.sh
Untracked: code/._subtractfiveprimeUTR.sh
Untracked: code/._utrdms2saf.py
Untracked: code/.snakemake/
Untracked: code/APAqtl_nominal.err
Untracked: code/APAqtl_nominal.out
Untracked: code/APAqtl_nominal_39.err
Untracked: code/APAqtl_nominal_39.out
Untracked: code/APAqtl_nominal_nonNorm.err
Untracked: code/APAqtl_nominal_nonNorm.out
Untracked: code/APAqtl_permuted.err
Untracked: code/APAqtl_permuted.out
Untracked: code/ApaQTL_nominalNonnorm.sh
Untracked: code/BothFracDTPlot1stintron.err
Untracked: code/BothFracDTPlot1stintron.out
Untracked: code/BothFracDTPlot4stintron.err
Untracked: code/BothFracDTPlot4stintron.out
Untracked: code/BothFracDTPlotGeneRegions.err
Untracked: code/BothFracDTPlotGeneRegions.out
Untracked: code/BothFracDTPlotGeneRegions_norm.err
Untracked: code/BothFracDTPlotGeneRegions_norm.out
Untracked: code/BothFracDTPlotGeneRegions_normalized.sh
Untracked: code/DistPAS2Sig.py
Untracked: code/EncodeRNADTPlotGeneRegions.err
Untracked: code/EncodeRNADTPlotGeneRegions.out
Untracked: code/FC_UTR.err
Untracked: code/FC_UTR.out
Untracked: code/FC_UTR.sh
Untracked: code/FC_newPAS_olddata.err
Untracked: code/FC_newPAS_olddata.out
Untracked: code/FC_newPeaks_olddata.sh
Untracked: code/LC_samplegroups.py
Untracked: code/NetSeq_fourthintronDT.sh
Untracked: code/README.md
Untracked: code/Rplots.pdf
Untracked: code/TESplots100bp.err
Untracked: code/TESplots100bp.out
Untracked: code/TESplots100bp.sh
Untracked: code/TESplots150bp.err
Untracked: code/TESplots150bp.out
Untracked: code/TESplots150bp.sh
Untracked: code/TESplots200bp.err
Untracked: code/TESplots200bp.out
Untracked: code/TESplots200bp.sh
Untracked: code/Untitled
Untracked: code/Upstream100Bases_general.py
Untracked: code/ZipandTabPheno.sh
Untracked: code/aAPAqtl_nominal39ind.sh
Untracked: code/apaQTLCorrectPvalMakeQQ_4pc.R
Untracked: code/apaQTL_Nominal_4pc.sh
Untracked: code/apaQTL_permuted.4pc.sh
Untracked: code/apaqtlfacetboxplots.R
Untracked: code/assignNucIntonpeak2intronlocs.sh
Untracked: code/assignPeak2Intronicregion.err
Untracked: code/assignPeak2Intronicregion.out
Untracked: code/assignTotIntronpeak2intronlocs.sh
Untracked: code/assigntotPeak2Intronicregion.err
Untracked: code/assigntotPeak2Intronicregion.out
Untracked: code/bam2BW_5primemost.sh
Untracked: code/bam2bw.err
Untracked: code/bam2bw.out
Untracked: code/bam2bw_5primemost.err
Untracked: code/bam2bw_5primemost.out
Untracked: code/bothFracDTplot1stintron.sh
Untracked: code/bothFracDTplot4thintron.sh
Untracked: code/bothFrac_FC.err
Untracked: code/bothFrac_FC.out
Untracked: code/bothFrac_FC.sh
Untracked: code/codingdms2bed.py
Untracked: code/dag.pdf
Untracked: code/dagPAS.pdf
Untracked: code/dagfiltPAS.pdf
Untracked: code/encodeRNADTplots.sh
Untracked: code/extractGenotypes.py
Untracked: code/fc2leafphen.py
Untracked: code/finalPASbed2SAF.py
Untracked: code/findbuginpeaks.R
Untracked: code/fix4su304corr.py
Untracked: code/fix4su604corr.py
Untracked: code/fix4sukalisto.py
Untracked: code/fixFChead_bothfrac.py
Untracked: code/fixFChead_summary.py
Untracked: code/fixH3k12ac.py
Untracked: code/fixRNAhead4corr.py
Untracked: code/fixRNAkalisto.py
Untracked: code/fixgroupedtranscript.py
Untracked: code/fixhead_netseqfc.py
Untracked: code/get100upPAS.py
Untracked: code/getSeq100up.sh
Untracked: code/getseq100up.err
Untracked: code/getseq100up.out
Untracked: code/grouptranscripts.err
Untracked: code/grouptranscripts.out
Untracked: code/grouptranscripts.py
Untracked: code/log/
Untracked: code/makeSAFbothfrac5perc.py
Untracked: code/makegencondeTSSfile.py
Untracked: code/mergeBW_norm.sh
Untracked: code/mergeBWnorm.err
Untracked: code/mergeBWnorm.out
Untracked: code/mnase1stintron.sh
Untracked: code/mnaseDTPlot1stintron.err
Untracked: code/mnaseDTPlot1stintron.out
Untracked: code/mnaseDTPlot4thintron.err
Untracked: code/mnaseDTPlot4thintron.out
Untracked: code/mnaseDT_fourthintron.sh
Untracked: code/netDTPlot4thintron.out
Untracked: code/netseqDTplot1stIntron.sh
Untracked: code/netseqFC.err
Untracked: code/netseqFC.out
Untracked: code/netseqFC.sh
Untracked: code/neyDTPlot4thintron.err
Untracked: code/qtlFacetBoxplots.err
Untracked: code/qtlFacetBoxplots.out
Untracked: code/qtlsPvalOppFrac.py
Untracked: code/removeloc_pheno.py
Untracked: code/run_DistPAS2Sig.err
Untracked: code/run_DistPAS2Sig.out
Untracked: code/run_distPAS2Sig.sh
Untracked: code/run_leafcutterDiffIso.sh
Untracked: code/run_leafcutter_ds.err
Untracked: code/run_leafcutter_ds.out
Untracked: code/run_qtlFacetBoxplots.sh
Untracked: code/run_sepUsagephen.sh
Untracked: code/run_sepusage.err
Untracked: code/run_sepusage.out
Untracked: code/selectNominalPvalues.py
Untracked: code/sepUsagePhen.py
Untracked: code/snakePASlog.out
Untracked: code/snakefiltPASlog.out
Untracked: code/subset_diffisopheno.py
Untracked: code/subtract5UTR.err
Untracked: code/subtract5UTR.out
Untracked: code/subtractExons.err
Untracked: code/subtractExons.out
Untracked: code/subtractExons.sh
Untracked: code/subtractfiveprimeUTR.sh
Untracked: code/transcriptdm2bed.py
Untracked: code/utrdms2saf.py
Untracked: code/zipandtabPhen.err
Untracked: code/zipandtabPhen.out
Untracked: data/CompareOldandNew/
Untracked: data/DTmatrix/
Untracked: data/DiffIso/
Untracked: data/EncodeRNA/
Untracked: data/ExampleQTLPlots/
Untracked: data/GeuvadisRNA/
Untracked: data/PAS/
Untracked: data/QTLGenotypes/
Untracked: data/QTLoverlap/
Untracked: data/QTLoverlap_nonNorm/
Untracked: data/README.md
Untracked: data/RNAseq/
Untracked: data/Reads2UTR/
Untracked: data/SignalSiteFiles/
Untracked: data/ThirtyNineIndQtl_nominal/
Untracked: data/apaQTLNominal/
Untracked: data/apaQTLNominal_4pc/
Untracked: data/apaQTLPermuted/
Untracked: data/apaQTLPermuted_4pc/
Untracked: data/apaQTLs/
Untracked: data/assignedPeaks/
Untracked: data/bam/
Untracked: data/bam_clean/
Untracked: data/bam_waspfilt/
Untracked: data/bed_10up/
Untracked: data/bed_clean/
Untracked: data/bed_clean_sort/
Untracked: data/bed_waspfilter/
Untracked: data/bedsort_waspfilter/
Untracked: data/bothFrac_FC/
Untracked: data/bw_norm/
Untracked: data/exampleQTLs/
Untracked: data/fastq/
Untracked: data/filterPeaks/
Untracked: data/fourSU/
Untracked: data/h3k27ac/
Untracked: data/highdiffsiggenes.txt
Untracked: data/inclusivePeaks/
Untracked: data/inclusivePeaks_FC/
Untracked: data/intron_analysis/
Untracked: data/mergedBG/
Untracked: data/mergedBW_byfrac/
Untracked: data/mergedBW_norm/
Untracked: data/mergedBam/
Untracked: data/mergedbyFracBam/
Untracked: data/netseq/
Untracked: data/nonNorm_pheno/
Untracked: data/nuc_10up/
Untracked: data/nuc_10upclean/
Untracked: data/peakCoverage/
Untracked: data/peaks_5perc/
Untracked: data/phenotype/
Untracked: data/phenotype_5perc/
Untracked: data/sigDiffGenes.txt
Untracked: data/sort/
Untracked: data/sort_clean/
Untracked: data/sort_waspfilter/
Untracked: nohup.out
Untracked: output/._.DS_Store
Untracked: output/._meanCorrelationPhenotypes.svg
Untracked: output/dtPlots/
Untracked: output/fastqc/
Untracked: output/meanCorrelationPhenotypes.svg
Unstaged changes:
Modified: analysis/PASusageQC.Rmd
Modified: analysis/Readdistagainstfeatures.Rmd
Modified: analysis/choosePCs.Rmd
Modified: analysis/corrbetweenind.Rmd
Modified: analysis/nascenttranscription.Rmd
Modified: analysis/rerunQTL_changePC.Rmd
Modified: analysis/rna_netseq_h3k12ac.Rmd
Modified: code/BothFracDTPlotGeneRegions.sh
Modified: code/Snakefile
Deleted: code/Upstream10Bases_general.py
Modified: code/apaQTLCorrectPvalMakeQQ.R
Modified: code/apaQTL_permuted.sh
Modified: code/apaQTLsnake.err
Modified: code/bed2saf.py
Modified: code/cluster.json
Modified: code/config.yaml
Deleted: code/test.txt
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the R Markdown and HTML files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view them.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 6ece235 | brimittleman | 2019-05-28 | add example code |
| html | de2aa7e | brimittleman | 2019-05-28 | Build site. |
| Rmd | c04929f | brimittleman | 2019-05-28 | add example code |
| html | f4a2106 | brimittleman | 2019-05-28 | Build site. |
| Rmd | f10e64d | brimittleman | 2019-05-28 | add plot grid |
| html | 64a7d5d | brimittleman | 2019-05-28 | Build site. |
| Rmd | f5260bd | brimittleman | 2019-05-28 | add results |
| html | 28c8ca3 | brimittleman | 2019-05-24 | Build site. |
| Rmd | 3f63045 | brimittleman | 2019-05-24 | add code to prepare non norm qtl |
In order to compare effect sizes for the QTLs I have previously identified in an interpretable manner, I need to run the linear model with the non normalized usage. To do this I will separate the the usage (with annotation) files by chromosome and run fastqtl on these files.
library(workflowr)This is workflowr version 1.3.0
Run ?workflowr for help getting startedlibrary(tidyverse)── Attaching packages ───────────────────────────────────────────────── tidyverse 1.2.1 ──✔ ggplot2 3.1.1 ✔ purrr 0.3.2
✔ tibble 2.1.1 ✔ dplyr 0.8.0.1
✔ tidyr 0.8.3 ✔ stringr 1.3.1
✔ readr 1.3.1 ✔ forcats 0.3.0 ── Conflicts ──────────────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()library(cowplot)
Attaching package: 'cowplot'The following object is masked from 'package:ggplot2':
ggsavePrepare files
countsnum= APApeak_Phenotype_GeneLocAnno.Total.5perc.CountsNumeric, APApeak_Phenotype_GeneLocAnno.Nuclear.5perc.CountsNumeric
id file= APApeak_Phenotype_GeneLocAnno.Nuclear.5perc.fc.gz, APApeak_Phenotype_GeneLocAnno.Total.5perc.fc.gz
totAnno= read.table("../data/phenotype_5perc/APApeak_Phenotype_GeneLocAnno.Total.5perc.fc.gz", stringsAsFactors = F, header = T) %>% separate(chrom, into=c("Chrchrom", "Start", "End", "ID"),sep=":") %>% mutate(Chr=str_sub(Chrchrom, 4, str_length(Chrchrom)))
colnamesTot= colnames(totAnno)[5:58]
totUsage=read.table("../data/phenotype_5perc/APApeak_Phenotype_GeneLocAnno.Total.5perc.CountsNumeric", stringsAsFactors = F, header = F, col.names = colnamesTot)
totUsageAnno=as.data.frame(cbind(Chr=totAnno$Chr, start=totAnno$Start, end=totAnno$End, ID=totAnno$ID, totUsage ))
write.table(totUsageAnno,file="../data/nonNorm_pheno/TotalUsageAllChrom.txt", col.names = T, row.names = F, quote = F, sep="\t" )nucAnno= read.table("../data/phenotype_5perc/APApeak_Phenotype_GeneLocAnno.Nuclear.5perc.fc.gz", stringsAsFactors = F, header = T)%>% separate(chrom, into=c("Chrchrom", "Start", "End", "ID"),sep=":") %>% mutate(Chr=str_sub(Chrchrom, 4, str_length(Chrchrom)))
colnamesNuc= colnames(nucAnno)[5:58]
nucUsage=read.table("../data/phenotype_5perc/APApeak_Phenotype_GeneLocAnno.Nuclear.5perc.CountsNumeric", stringsAsFactors = F, header = F, col.names = colnamesNuc)
nucUsageAnno=as.data.frame(cbind(Chr=nucAnno$Chr, start=nucAnno$Start, end=nucAnno$End, ID=nucAnno$ID, nucUsage ))
write.table(nucUsageAnno,file="../data/nonNorm_pheno/NuclearUsageAllChrom.txt", col.names = T, row.names = F, quote = F, sep="\t" )Run QTL scripts
I will create a python script to seperate the file into each chromosome for running fastQTL.
sbatch run_sepUsagephen.sh
sbatch ZipandTabPheno.sh
sbatch ApaQTL_nominalNonnorm.shConcatinate files:
cat TotalUsageChrom*.nominal.out > TotalUsageChrom_Nominal_AllChrom.txt
cat NuclearUsageChrom*.nominal.out > NuclearUsageChrom_Nominal_AllChrom.txt
Pull out real total and nuc QLTs
python qtlsPvalOppFrac.py ../data/apaQTLs/Nuclear_apaQTLs4pc_5fdr.txt ../data/nonNorm_pheno/TotalUsageChrom_Nominal_AllChrom.txt ../data/QTLoverlap_nonNorm/NuclearQTLinTotalNominal_nonNorm.txt
python qtlsPvalOppFrac.py ../data/apaQTLs/Nuclear_apaQTLs4pc_5fdr.txt ../data/nonNorm_pheno/NuclearUsageChrom_Nominal_AllChrom.txt ../data/QTLoverlap_nonNorm/NuclearQTLinNuclearNominal_nonNorm.txt
python qtlsPvalOppFrac.py ../data/apaQTLs/Total_apaQTLs4pc_5fdr.txt ../data/nonNorm_pheno/TotalUsageChrom_Nominal_AllChrom.txt ../data/QTLoverlap_nonNorm/TotalQTLinTotalNominal_nonNorm.txt
python qtlsPvalOppFrac.py ../data/apaQTLs/Total_apaQTLs4pc_5fdr.txt ../data/nonNorm_pheno/NuclearUsageChrom_Nominal_AllChrom.txt ../data/QTLoverlap_nonNorm/TotalQTLinNuclearNominal_nonNorm.txt totAPAinNuc=read.table("../data/QTLoverlap_nonNorm/TotalQTLinNuclearNominal_nonNorm.txt", header = F, stringsAsFactors = F, col.names=c("peakID", "snp", "dist", "pval", "slope"))
nucAPAinTot=read.table("../data/QTLoverlap_nonNorm/NuclearQTLinTotalNominal_nonNorm.txt", header = F, stringsAsFactors = F, col.names=c("peakID", "snp", "dist", "pval", "slope"))
totAPAinTot=read.table("../data/QTLoverlap_nonNorm/TotalQTLinTotalNominal_nonNorm.txt", header = F, stringsAsFactors = F, col.names=c("peakID", "snp", "dist", "pval", "slope")) %>% dplyr::select(peakID, snp, slope) %>% dplyr::rename("Originalslope"=slope)
nucAPAinNuc=read.table("../data/QTLoverlap_nonNorm/NuclearQTLinNuclearNominal_nonNorm.txt", header = F, stringsAsFactors = F, col.names=c("peakID", "snp", "dist", "pval", "slope")) %>% dplyr::select(peakID, snp, slope)%>% dplyr::rename("Originalslope"=slope)Total
TotBoth= totAPAinNuc %>% inner_join(totAPAinTot,by=c("peakID", "snp"))
summary(lm(TotBoth$slope ~ TotBoth$Originalslope))
Call:
lm(formula = TotBoth$slope ~ TotBoth$Originalslope)
Residuals:
Min 1Q Median 3Q Max
-1.34029 -0.07935 -0.01337 0.06956 0.46743
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.019095 0.006562 2.91 0.00376 **
TotBoth$Originalslope 0.398243 0.013884 28.68 < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.1518 on 543 degrees of freedom
Multiple R-squared: 0.6024, Adjusted R-squared: 0.6017
F-statistic: 822.8 on 1 and 543 DF, p-value: < 2.2e-16totbothplot=ggplot(TotBoth, aes(x=Originalslope, y=slope))+geom_point() + geom_smooth(method="lm") + labs(title="Total apaQTL effect sizes", x="Effect size in Total",y="Effect size in Nucler") + geom_density_2d(col="red") + annotate("text", y=2, x=2, label="R2=.61, slope=0.4")Nuclear
NucBoth= nucAPAinTot %>% inner_join(nucAPAinNuc,by=c("peakID", "snp"))
summary(lm(NucBoth$slope ~ NucBoth$Originalslope))
Call:
lm(formula = NucBoth$slope ~ NucBoth$Originalslope)
Residuals:
Min 1Q Median 3Q Max
-0.74544 -0.03953 0.00153 0.04010 0.71657
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.001065 0.003189 -0.334 0.738
NucBoth$Originalslope 0.742666 0.011220 66.193 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.0998 on 1010 degrees of freedom
Multiple R-squared: 0.8127, Adjusted R-squared: 0.8125
F-statistic: 4381 on 1 and 1010 DF, p-value: < 2.2e-16Nucbothplot=ggplot(NucBoth, aes(x=Originalslope, y=slope))+geom_point() + geom_smooth(method="lm") + labs(title="Nuclear apaQTL effect sizes", x="Effect size in Nuclear",y="Effect size in Total") + geom_density_2d(col="red") + annotate("text", y=2, x=1, label="R2=.81, slope=0.74")plot_grid(totbothplot,Nucbothplot)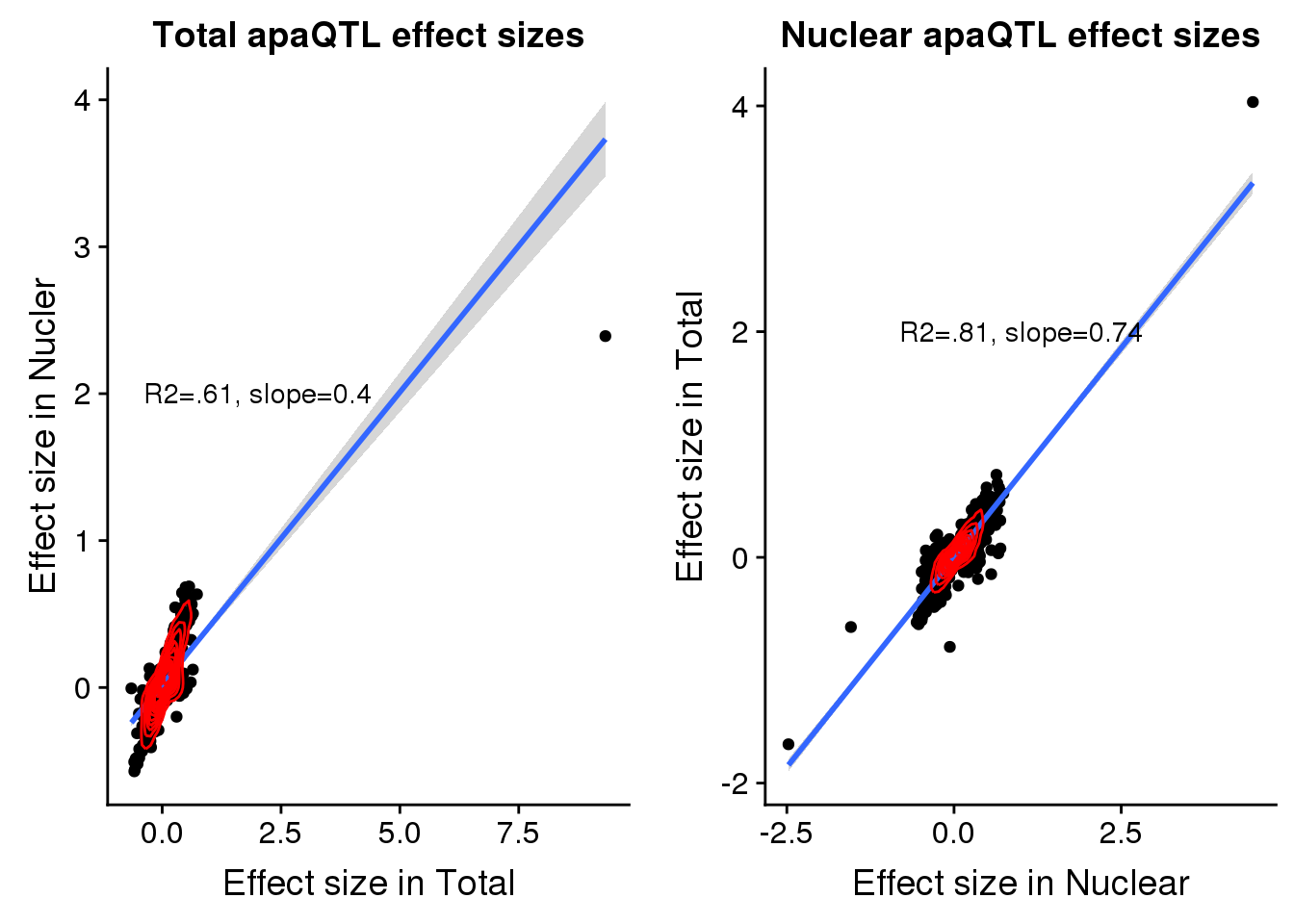
Remove Effect size > abs(1)
totAPAinNucFilt=read.table("../data/QTLoverlap_nonNorm/TotalQTLinNuclearNominal_nonNorm.txt", header = F, stringsAsFactors = F, col.names=c("peakID", "snp", "dist", "pval", "slope")) %>% filter(abs(slope)<= 1)
nucAPAinTotFilt=read.table("../data/QTLoverlap_nonNorm/NuclearQTLinTotalNominal_nonNorm.txt", header = F, stringsAsFactors = F, col.names=c("peakID", "snp", "dist", "pval", "slope")) %>% filter(abs(slope)<= 1)
totAPAinTotFilt=read.table("../data/QTLoverlap_nonNorm/TotalQTLinTotalNominal_nonNorm.txt", header = F, stringsAsFactors = F, col.names=c("peakID", "snp", "dist", "pval", "slope")) %>% dplyr::select(peakID, snp, slope) %>% filter(abs(slope)<= 1) %>% dplyr::rename("Originalslope"=slope)
nucAPAinNucFilt=read.table("../data/QTLoverlap_nonNorm/NuclearQTLinNuclearNominal_nonNorm.txt", header = F, stringsAsFactors = F, col.names=c("peakID", "snp", "dist", "pval", "slope")) %>% dplyr::select(peakID, snp, slope)%>% filter(abs(slope)<= 1) %>%dplyr::rename("Originalslope"=slope)TotBothFilt= totAPAinNucFilt %>% inner_join(totAPAinTotFilt,by=c("peakID", "snp"))
summary(lm(TotBothFilt$slope ~ TotBothFilt$Originalslope))
Call:
lm(formula = TotBothFilt$slope ~ TotBothFilt$Originalslope)
Residuals:
Min 1Q Median 3Q Max
-0.43593 -0.04769 -0.00197 0.05133 0.48722
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.004788 0.004634 1.033 0.302
TotBothFilt$Originalslope 0.767422 0.018322 41.885 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.1063 on 542 degrees of freedom
Multiple R-squared: 0.764, Adjusted R-squared: 0.7635
F-statistic: 1754 on 1 and 542 DF, p-value: < 2.2e-16totbothplotfilt=ggplot(TotBothFilt, aes(x=Originalslope, y=slope))+geom_point() + geom_smooth(method="lm") + labs(title="Total apaQTL effect sizes", y="Effect size in Nuclear",x="Effect size in Total") + geom_density_2d(col="red") + annotate("text", y=.75, x=.1, label="R2=.76, slope=0.77")+ geom_abline(slope=1,color="green")NucBothFilt= nucAPAinTotFilt %>% inner_join(nucAPAinNucFilt,by=c("peakID", "snp"))
summary(lm(NucBothFilt$slope ~ NucBothFilt$Originalslope))
Call:
lm(formula = NucBothFilt$slope ~ NucBothFilt$Originalslope)
Residuals:
Min 1Q Median 3Q Max
-0.74819 -0.03867 0.00179 0.04084 0.37901
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.0006196 0.0030789 -0.201 0.841
NucBothFilt$Originalslope 0.7059759 0.0133986 52.690 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.09539 on 1007 degrees of freedom
Multiple R-squared: 0.7338, Adjusted R-squared: 0.7336
F-statistic: 2776 on 1 and 1007 DF, p-value: < 2.2e-16Nucbothplotfilt=ggplot(NucBothFilt, aes(x=Originalslope, y=slope))+geom_point() + geom_smooth(method="lm") + labs(title="Nuclear apaQTL effect sizes", y="Effect size in Total",x="Effect size in Nuclear") + geom_density_2d(col="red") + annotate("text", y=.75, x=.1, label="R2=.73, slope=0.71")+ geom_abline(slope=1,color="green")plot_grid(totbothplotfilt,Nucbothplotfilt)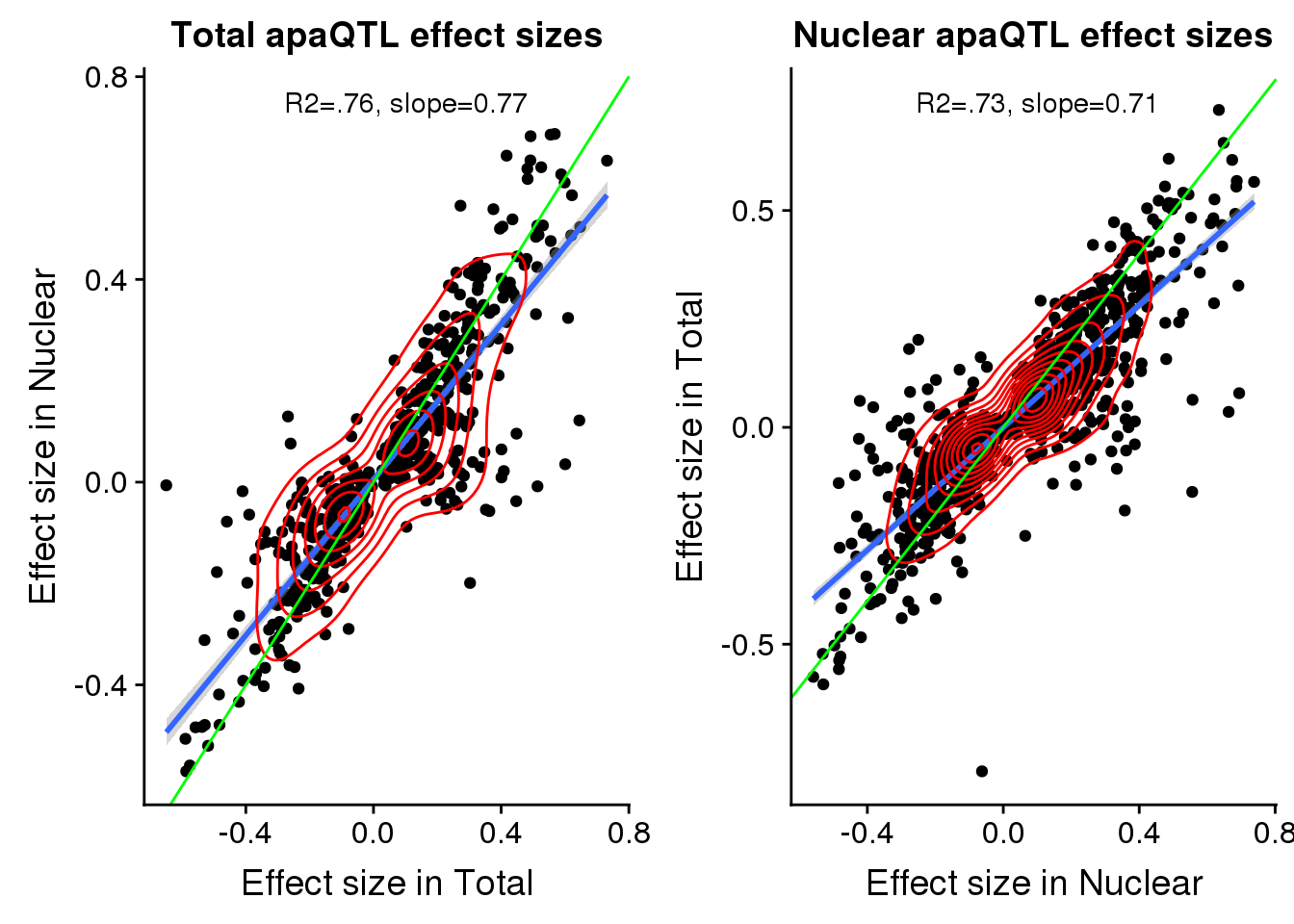
| Version | Author | Date |
|---|---|---|
| de2aa7e | brimittleman | 2019-05-28 |
Box plots to look at the outliers:
get headers:
less /project2/gilad/briana/YRI_geno_hg19/chr3.dose.filt.vcf.gz | head -n14 | tail -n1 > /project2/gilad/briana/apaQTL/data/ExampleQTLPlots/genotypeHeader.txt
less /project2/gilad/briana/apaQTL/data/phenotype_5perc/APApeak_Phenotype_GeneLocAnno.Total.5perc.fc.gz | head -n1 > /project2/gilad/briana/apaQTL/data/ExampleQTLPlots/phenotypeHeader.txtremove the hashtag in these
apaqtlfacetboxplots.R
library(tidyverse)
library(reshape2)
library(optparse)
library(cowplot)
option_list = list(
make_option(c("-P", "--pheno"), action="store", default=NA, type='character',
help="input pheno file"),
make_option(c("-G", "--geno"), action="store", default=NA, type='character',
help="input genotype"),
make_option(c("-g", "--gene"), action="store", default=NA, type='character',
help="gene"),
make_option(c("-p", "--peakID"), action="store", default=NA, type='character',
help="peakID"),
make_option(c("-o", "--output"), action="store", default=NA, type='character',
help="output file for plot")
)
opt_parser <- OptionParser(option_list=option_list)
opt <- parse_args(opt_parser)
opt_parser <- OptionParser(option_list=option_list)
opt <- parse_args(opt_parser)
phenohead=read.table("/project2/gilad/briana/apaQTL/data/ExampleQTLPlots/phenotypeHeader.txt", header = T,stringsAsFactors = F)
pheno=read.table(opt$pheno, col.names =colnames(phenohead),stringsAsFactors = F)
meltpheno=melt(pheno, id.vars = "chrom", value.name = "Ratio", variable.name = "Individual") %>% separate(Ratio, into=c("num", "denom"), sep="/") %>% separate(chrom, into=c("chrom", "start", "end", "peakID"),sep=":") %>% mutate(PeakLoc=paste(start, end, sep=":"))
meltpheno$Individual= as.character(meltpheno$Individual)
meltpheno$num= as.numeric(meltpheno$num)
meltpheno$denom=as.numeric(meltpheno$denom)
genoHead=read.table("/project2/gilad/briana/apaQTL/data/ExampleQTLPlots/genotypeHeader.txt", header = T,stringsAsFactors = F)
geno=read.table(opt$geno, col.names =colnames(genoHead),stringsAsFactors = F ) %>% select(ID,contains("NA"))
lettersGeno=read.table(opt$geno, col.names =colnames(genoHead),stringsAsFactors = F,colClasses = c("character")) %>% select(REF, ALT)
refAllele=lettersGeno$REF
altAllele=lettersGeno$ALT
genoMelt=melt(geno, id.vars = "ID", value.name = "FullGeno", variable.name = "Individual" ) %>% separate(FullGeno, into=c("geno","dose","extra1"), sep=":") %>% select(Individual, dose) %>% mutate(genotype=ifelse(round(as.integer(dose))==0, paste(refAllele, refAllele, sep=""), ifelse(round(as.integer(dose))==1, paste(refAllele,altAllele, sep=""), paste(altAllele,altAllele,sep=""))))
genoMelt$Individual= as.character(genoMelt$Individual)
pheno_qtlpeak=meltpheno %>% inner_join(genoMelt, by="Individual") %>% mutate(PAU=num/denom)
qtlplot=ggplot(pheno_qtlpeak, aes(x=genotype, y=PAU, fill=genotype)) + geom_boxplot(width=.5)+ geom_jitter(alpha=1) + facet_grid(~PeakLoc) +scale_fill_brewer(palette = "YlOrRd")
ggsave(plot=qtlplot, filename=opt$output, height=10, width=10)Code for boxplots:
run_qtlFacetBoxplots.sh
#!/bin/bash
#SBATCH --job-name=qtlFacetBoxplots
#SBATCH --account=pi-yangili1
#SBATCH --time=24:00:00
#SBATCH --output=qtlFacetBoxplots.out
#SBATCH --error=qtlFacetBoxplots.err
#SBATCH --partition=broadwl
#SBATCH --mem=18G
#SBATCH --mail-type=END
module load Anaconda3
source activate three-prime-env
Fraction=$1
gene=$2
chrom=$3
snp=$4
peakID=$5
less /project2/gilad/briana/apaQTL/data/phenotype_5perc/APApeak_Phenotype_GeneLocAnno.${Fraction}.5perc.fc.gz | grep ${gene}_ > /project2/gilad/briana/apaQTL/data/ExampleQTLPlots/${gene}_${Fraction}PeaksPheno.txt
less /project2/gilad/briana/YRI_geno_hg19/chr${chrom}.dose.filt.vcf.gz | grep ${snp} > /project2/gilad/briana/apaQTL/data/ExampleQTLPlots/${gene}_${Fraction}PeaksGenotype.txt
Rscript apaqtlfacetboxplots.R -P /project2/gilad/briana/apaQTL/data/ExampleQTLPlots/${gene}_${Fraction}PeaksPheno.txt -G /project2/gilad/briana/apaQTL/data/ExampleQTLPlots/${gene}_${Fraction}PeaksGenotype.txt --gene ${gene} -p ${peakID} -o /project2/gilad/briana/apaQTL/data/ExampleQTLPlots/${gene}_${Fraction}${SNP}${peakID}_boxplot.png
totAPAinTot %>% filter(abs(Originalslope)>1) peakID snp Originalslope
1 NBPF9_intron_-_peak7314 1:144701300 3.14235
2 HLA-DRB5_intron_+_peak113454 6:32486756 -207.01900
3 HLA-DRB6_end_+_peak113461 6:32538598 9.32331sbatch run_qtlFacetBoxplots.sh "Total" "NBPF9" "1" "1:144701300" "peak7314"
sbatch run_qtlFacetBoxplots.sh "Total" "HLA-DRB5" "6" "6:32486756" "peak113454"
sbatch run_qtlFacetBoxplots.sh "Total" "HLA-DRB6" "6" "6:32538598" "peak113461"
nucAPAinNuc %>% filter(abs(Originalslope)>1) peakID snp Originalslope
1 LINC00869_intron_-_peak7883 1:149598905 -2.47583
2 FRG1BP_end_-_peak80905 20:29641550 -1.54150
3 HLA-DRB5_intron_+_peak113456 6:32468906 4.46867sbatch run_qtlFacetBoxplots.sh "Nuclear" "LINC00869" "1" "1:149598905" "peak7883"
sbatch run_qtlFacetBoxplots.sh "Nuclear" "FRG1BP" "20" "20:29641550" "peak80905"
sbatch run_qtlFacetBoxplots.sh "Nuclear" "HLA-DRB5" "6" "6:32468906" "peak113456"
#test case
sbatch run_qtlFacetBoxplots.sh "Nuclear" "TAF3" "10" "10:7980931" "peak14035"
sessionInfo()R version 3.5.1 (2018-07-02)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] cowplot_0.9.4 forcats_0.3.0 stringr_1.3.1 dplyr_0.8.0.1
[5] purrr_0.3.2 readr_1.3.1 tidyr_0.8.3 tibble_2.1.1
[9] ggplot2_3.1.1 tidyverse_1.2.1 workflowr_1.3.0
loaded via a namespace (and not attached):
[1] Rcpp_1.0.0 cellranger_1.1.0 pillar_1.3.1 compiler_3.5.1
[5] git2r_0.23.0 plyr_1.8.4 tools_3.5.1 digest_0.6.18
[9] lubridate_1.7.4 jsonlite_1.6 evaluate_0.12 nlme_3.1-137
[13] gtable_0.2.0 lattice_0.20-38 pkgconfig_2.0.2 rlang_0.3.1
[17] cli_1.0.1 rstudioapi_0.10 yaml_2.2.0 haven_1.1.2
[21] withr_2.1.2 xml2_1.2.0 httr_1.3.1 knitr_1.20
[25] hms_0.4.2 generics_0.0.2 fs_1.2.6 rprojroot_1.3-2
[29] grid_3.5.1 tidyselect_0.2.5 glue_1.3.0 R6_2.3.0
[33] readxl_1.1.0 rmarkdown_1.10 modelr_0.1.2 magrittr_1.5
[37] whisker_0.3-2 MASS_7.3-51.1 backports_1.1.2 scales_1.0.0
[41] htmltools_0.3.6 rvest_0.3.2 assertthat_0.2.0 colorspace_1.3-2
[45] labeling_0.3 stringi_1.2.4 lazyeval_0.2.1 munsell_0.5.0
[49] broom_0.5.1 crayon_1.3.4