Explore Unexplained QTLs
Briana Mittleman
2/25/2019
Last updated: 2019-02-25
Checks: 6 0
Knit directory: threeprimeseq/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.2.0). The Report tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(12345) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/.DS_Store
Ignored: data/perm_QTL_trans_noMP_5percov/
Ignored: output/.DS_Store
Untracked files:
Untracked: KalistoAbundance18486.txt
Untracked: analysis/4suDataIGV.Rmd
Untracked: analysis/DirectionapaQTL.Rmd
Untracked: analysis/EvaleQTLs.Rmd
Untracked: analysis/YL_QTL_test.Rmd
Untracked: analysis/fixBWChromNames.Rmd
Untracked: analysis/groSeqAnalysis.Rmd
Untracked: analysis/ncbiRefSeq_sm.sort.mRNA.bed
Untracked: analysis/snake.config.notes.Rmd
Untracked: analysis/verifyBAM.Rmd
Untracked: analysis/verifybam_dubs.Rmd
Untracked: code/PeaksToCoverPerReads.py
Untracked: code/strober_pc_pve_heatmap_func.R
Untracked: data/18486.genecov.txt
Untracked: data/APApeaksYL.total.inbrain.bed
Untracked: data/AllPeak_counts/
Untracked: data/ApaQTLs/
Untracked: data/ApaQTLs_otherPhen/
Untracked: data/ChromHmmOverlap/
Untracked: data/DistTXN2Peak_genelocAnno/
Untracked: data/GM12878.chromHMM.bed
Untracked: data/GM12878.chromHMM.txt
Untracked: data/LianoglouLCL/
Untracked: data/LocusZoom/
Untracked: data/LocusZoom_proc/
Untracked: data/MatchedSnps/
Untracked: data/NuclearApaQTLs.txt
Untracked: data/PeakCounts/
Untracked: data/PeakCounts_noMP_5perc/
Untracked: data/PeakCounts_noMP_genelocanno/
Untracked: data/PeakUsage/
Untracked: data/PeakUsage_noMP/
Untracked: data/PeakUsage_noMP_GeneLocAnno/
Untracked: data/PeaksUsed/
Untracked: data/PeaksUsed_noMP_5percCov/
Untracked: data/QTL_overlap/
Untracked: data/RNAkalisto/
Untracked: data/RefSeq_annotations/
Untracked: data/Replicates_usage/
Untracked: data/TotalApaQTLs.txt
Untracked: data/Totalpeaks_filtered_clean.bed
Untracked: data/UnderstandPeaksQC/
Untracked: data/WASP_STAT/
Untracked: data/YL-SP-18486-T-combined-genecov.txt
Untracked: data/YL-SP-18486-T_S9_R1_001-genecov.txt
Untracked: data/YL_QTL_test/
Untracked: data/apaExamp/
Untracked: data/apaExamp_proc/
Untracked: data/apaQTL_examp_noMP/
Untracked: data/bedgraph_peaks/
Untracked: data/bin200.5.T.nuccov.bed
Untracked: data/bin200.Anuccov.bed
Untracked: data/bin200.nuccov.bed
Untracked: data/clean_peaks/
Untracked: data/comb_map_stats.csv
Untracked: data/comb_map_stats.xlsx
Untracked: data/comb_map_stats_39ind.csv
Untracked: data/combined_reads_mapped_three_prime_seq.csv
Untracked: data/diff_iso_GeneLocAnno/
Untracked: data/diff_iso_proc/
Untracked: data/diff_iso_trans/
Untracked: data/eQTLs_Lietal/
Untracked: data/ensemble_to_genename.txt
Untracked: data/example_gene_peakQuant/
Untracked: data/explainProtVar/
Untracked: data/filtPeakOppstrand_cov_noMP_GeneLocAnno_5perc/
Untracked: data/filtered_APApeaks_merged_allchrom_refseqTrans.closest2End.bed
Untracked: data/filtered_APApeaks_merged_allchrom_refseqTrans.closest2End.noties.bed
Untracked: data/first50lines_closest.txt
Untracked: data/gencov.test.csv
Untracked: data/gencov.test.txt
Untracked: data/gencov_zero.test.csv
Untracked: data/gencov_zero.test.txt
Untracked: data/gene_cov/
Untracked: data/joined
Untracked: data/leafcutter/
Untracked: data/merged_combined_YL-SP-threeprimeseq.bg
Untracked: data/molPheno_noMP/
Untracked: data/mol_overlap/
Untracked: data/mol_pheno/
Untracked: data/nom_QTL/
Untracked: data/nom_QTL_opp/
Untracked: data/nom_QTL_trans/
Untracked: data/nuc6up/
Untracked: data/nuc_10up/
Untracked: data/other_qtls/
Untracked: data/pQTL_otherphen/
Untracked: data/peakPerRefSeqGene/
Untracked: data/perm_QTL/
Untracked: data/perm_QTL_GeneLocAnno_noMP_5percov/
Untracked: data/perm_QTL_GeneLocAnno_noMP_5percov_3UTR/
Untracked: data/perm_QTL_diffWindow/
Untracked: data/perm_QTL_opp/
Untracked: data/perm_QTL_trans/
Untracked: data/perm_QTL_trans_filt/
Untracked: data/protAndAPAAndExplmRes.Rda
Untracked: data/protAndAPAlmRes.Rda
Untracked: data/protAndExpressionlmRes.Rda
Untracked: data/reads_mapped_three_prime_seq.csv
Untracked: data/smash.cov.results.bed
Untracked: data/smash.cov.results.csv
Untracked: data/smash.cov.results.txt
Untracked: data/smash_testregion/
Untracked: data/ssFC200.cov.bed
Untracked: data/temp.file1
Untracked: data/temp.file2
Untracked: data/temp.gencov.test.txt
Untracked: data/temp.gencov_zero.test.txt
Untracked: data/threePrimeSeqMetaData.csv
Untracked: data/threePrimeSeqMetaData55Ind.txt
Untracked: data/threePrimeSeqMetaData55Ind.xlsx
Untracked: data/threePrimeSeqMetaData55Ind_noDup.txt
Untracked: data/threePrimeSeqMetaData55Ind_noDup.xlsx
Untracked: data/threePrimeSeqMetaData55Ind_noDup_WASPMAP.txt
Untracked: data/threePrimeSeqMetaData55Ind_noDup_WASPMAP.xlsx
Untracked: data/~$threePrimeSeqMetaData55Ind_noDup_WASPMAP.xlsx
Untracked: output/deeptools_plots/
Untracked: output/picard/
Untracked: output/plots/
Untracked: output/qual.fig2.pdf
Unstaged changes:
Modified: analysis/28ind.peak.explore.Rmd
Modified: analysis/CompareLianoglouData.Rmd
Modified: analysis/NewPeakPostMP.Rmd
Modified: analysis/apaQTLoverlapGWAS.Rmd
Modified: analysis/cleanupdtseq.internalpriming.Rmd
Modified: analysis/coloc_apaQTLs_protQTLs.Rmd
Modified: analysis/dif.iso.usage.leafcutter.Rmd
Modified: analysis/diffIsoAnalysisNewMapping.Rmd
Modified: analysis/diff_iso_pipeline.Rmd
Modified: analysis/explainpQTLs.Rmd
Modified: analysis/explore.filters.Rmd
Modified: analysis/flash2mash.Rmd
Modified: analysis/mispriming_approach.Rmd
Modified: analysis/overlapMolQTL.Rmd
Modified: analysis/overlapMolQTL.opposite.Rmd
Modified: analysis/overlap_qtls.Rmd
Modified: analysis/peakOverlap_oppstrand.Rmd
Modified: analysis/peakQCPPlots.Rmd
Modified: analysis/pheno.leaf.comb.Rmd
Modified: analysis/pipeline_55Ind.Rmd
Modified: analysis/swarmPlots_QTLs.Rmd
Modified: analysis/test.max2.Rmd
Modified: analysis/test.smash.Rmd
Modified: analysis/understandPeaks.Rmd
Modified: code/Snakefile
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the R Markdown and HTML files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view them.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | d3dfe95 | Briana Mittleman | 2019-02-25 | look at pi1 |
| html | b74a969 | Briana Mittleman | 2019-02-25 | Build site. |
| Rmd | 2336f87 | Briana Mittleman | 2019-02-25 | add unexplained QTL analysis |
library(qvalue)
library(tidyverse)── Attaching packages ────────────────────────────────────────────────────────────────────────────────── tidyverse 1.2.1 ──✔ ggplot2 3.0.0 ✔ purrr 0.2.5
✔ tibble 1.4.2 ✔ dplyr 0.7.6
✔ tidyr 0.8.1 ✔ stringr 1.4.0
✔ readr 1.1.1 ✔ forcats 0.3.0Warning: package 'stringr' was built under R version 3.5.2── Conflicts ───────────────────────────────────────────────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()library(cowplot)
Attaching package: 'cowplot'The following object is masked from 'package:ggplot2':
ggsavelibrary(workflowr)This is workflowr version 1.2.0
Run ?workflowr for help getting startedOne original goal for this project was too see if APA qtls could explain a number of the unexplianed eQTLs Yang found in the integrated molQTL science paper. He has provided me a list of explained eQTLs (chromatin associatated) and unexplained eQTLs. As a first pass, I want to look at the loci/gene associations in my QTL data. If there is significant sharing I expect lower pvalues for the apa associatiations at these loci. I will start with all peaks in the e genes.
These data have 1163 explained loci and 801 unexplained loci.
I want to make a python script that can take either of these and the nominal results for my total or nuclear apaQTLs. It will extract any association for a peak in one of these genes.
First sort these. They are chr, pos, gene,
sort -k1,1 -k2,2n /project2/gilad/briana/threeprimeseq/data/eQTL_Lietal/explained_FDR10.txt > /project2/gilad/briana/threeprimeseq/data/eQTL_Lietal/explained_FDR10.sort.txt
sort -k1,1 -k2,2n /project2/gilad/briana/threeprimeseq/data/eQTL_Lietal/unexplained_FDR10.txt > /project2/gilad/briana/threeprimeseq/data/eQTL_Lietal/unexplained_FDR10.sort.txt
Look for sharing in associations
Take some of this code from this analysis
APApval4eQTL.py
def main(eQTL,apaQTL, outF):
fout=open(outF,"w")
geneNames=open("/project2/gilad/briana/genome_anotation_data/ensemble_to_genename.txt","r")
#gene name dictionary
geneDic={}
geneDicOpp={}
for i, ln in enumerate(geneNames):
if i >0:
ID=ln.split()[0]
gene=ln.split()[1]
if gene not in geneDic.keys():
geneDic[gene]=[ID]
else:
geneDic[gene].append(ID)
geneDicOpp[ID]=gene
qtl_dic={}
for ln in open(eQTL,"r"):
chrom=ln.split()[0][3:]
pos=ln.split()[1]
snp=chrom + ":" + pos
gene=ln.split()[2]
if gene not in geneDicOpp.keys():
continue
geneName=geneDicOpp[gene]
qtl_dic[snp]=geneName
for ln in open(apaQTL, "r"):
snp=ln.split()[1]
gene=ln.split()[0].split(":")[-1].split("_")[0]
peak=ln.split()[0].split(":")[-1].split("_")[-1]
pval=ln.split()[3]
if snp in qtl_dic.keys():
if qtl_dic[snp]==gene:
fout.write("%s\t%s\t%s\t%s\n"%(snp, gene, peak, pval))
fout.close()
if __name__ == "__main__":
import sys
fraction = sys.argv[1]
eqtl = sys.argv[2]
inQTL="/project2/gilad/briana/threeprimeseq/data/nominal_APAqtl_GeneLocAnno_noMP_5percUs/filtered_APApeaks_merged_allchrom_refseqGenes.GeneLocAnno_NoMP_sm_quant.%s.fixed.pheno_5perc.fc.gz.qqnorm_allNomRes.txt"%(fraction)
eQTLin="/project2/gilad/briana/threeprimeseq/data/eQTL_Lietal/%s_FDR10.sort.txt"%(eQTL)
outFile="/project2/gilad/briana/threeprimeseq/data/ExplaineQTLS/NomPval_%sApaQTLs_for%seQTLs.txt"%(fraction, eQTL)
main(eQTLin,inQTL,outFile)
Run this overall combinations:
runAPApval4eQTL.sh
#!/bin/bash
#SBATCH --job-name=runAPApval4eQTL
#SBATCH --account=pi-yangili1
#SBATCH --time=24:00:00
#SBATCH --output=runAPApval4eQTL.out
#SBATCH --error=runAPApval4eQTL.err
#SBATCH --partition=broadwl
#SBATCH --mem=10G
#SBATCH --mail-type=END
module load Anaconda3
source activate three-prime-env
python APApval4eQTL.py Total explained
python APApval4eQTL.py Total unexplained
python APApval4eQTL.py Nuclear explained
python APApval4eQTL.py Nuclear unexplained
Genes not in the switch gene name file:
geneNoName=read.table("../data/eQTLs_Lietal/genesNoName_uniq.txt", stringsAsFactors = F, col.names = c("GeneID"))Upload results:
resNames=c("SNP", "gene", "peak", "pval")
totUn=read.table("../data/eQTLs_Lietal/NomPval_TotalApaQTLs_forunexplainedeQTLs.txt", stringsAsFactors = F, col.names = resNames)
totEx=read.table("../data/eQTLs_Lietal/NomPval_TotalApaQTLs_forexplainedeQTLs.txt", stringsAsFactors = F, col.names = resNames)
nucUn=read.table("../data/eQTLs_Lietal/NomPval_NuclearApaQTLs_forunexplainedeQTLs.txt", stringsAsFactors = F, col.names = resNames)
nucEx=read.table("../data/eQTLs_Lietal/NomPval_NuclearApaQTLs_forexplainedeQTLs.txt", stringsAsFactors = F, col.names = resNames)ggplot(totUn, aes(x=pval)) + geom_density(fill="blue", alpha=.5) + geom_density(data=totEx,aes(x=pval), fill="red", alpha=.5 ) + labs(title="Total APA association pval for explained and unexplained eQTLs \n red=explained, blue=unexplained")
ggplot(nucUn, aes(x=pval)) + geom_density(fill="blue", alpha=.5) + geom_density(data=nucEx,aes(x=pval), fill="red", alpha=.5 ) + labs(title="Nuclear APA association pval for explained and unexplained eQTLs \n red=explained, blue=unexplained")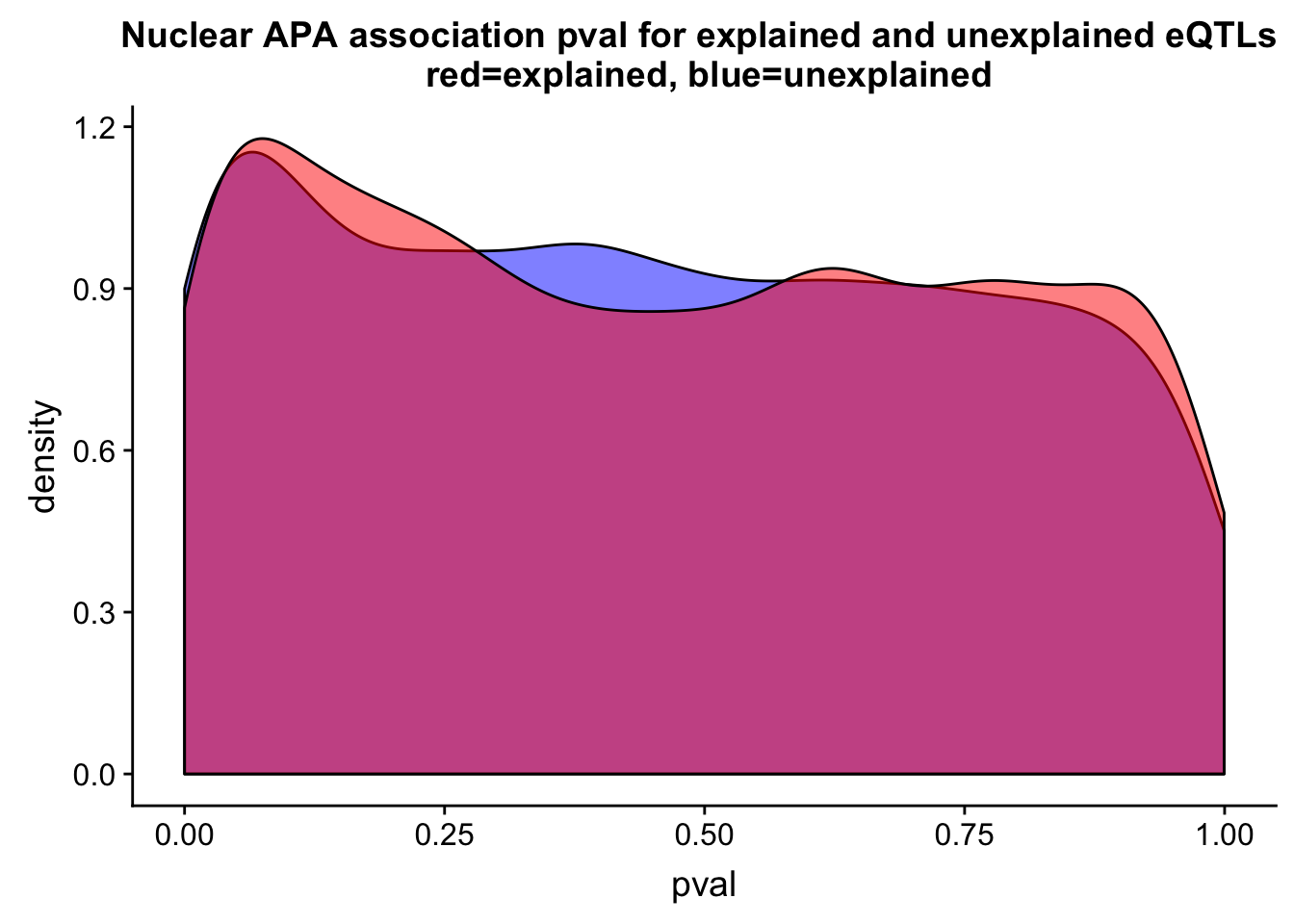
Pi1 values:
Tot Explained:
TotExPi=pi0est(totEx$pval, pi0.method = "bootstrap")
1-TotExPi$pi0[1] 0.1386882Tot unexplained:
TotUnPi=pi0est(totUn$pval, pi0.method = "bootstrap")
1-TotUnPi$pi0[1] 0.1043331Nuc Explained:
NucExPi=pi0est(nucEx$pval, pi0.method = "bootstrap")
1-NucExPi$pi0[1] 0.08966862Nuc unexplained:
NucUnPi=pi0est(nucUn$pval, pi0.method = "bootstrap")
1-NucUnPi$pi0[1] 0.1117647This is the naive version, I need to accont for the multiple peaks in the same gene.
totUn_fix=totUn %>% group_by(gene) %>% mutate(nPeaks=n()) %>% ungroup()
totEx_fix=totEx %>% group_by(gene) %>% mutate(nPeaks=n()) %>% ungroup()
nucUn_fix=nucUn %>% group_by(gene) %>% mutate(nPeaks=n()) %>% ungroup()
nucEx_fix=nucEx %>% group_by(gene) %>% mutate(nPeaks=n()) %>% ungroup()Direct overlap
I can use a similar LD anaylsis I used in the GWAS overlap. I will get all of the snps in LD with the eQTLs then look for overlap with my apaQTLs.
sessionInfo()R version 3.5.1 (2018-07-02)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS 10.14.1
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] workflowr_1.2.0 cowplot_0.9.3 forcats_0.3.0 stringr_1.4.0
[5] dplyr_0.7.6 purrr_0.2.5 readr_1.1.1 tidyr_0.8.1
[9] tibble_1.4.2 ggplot2_3.0.0 tidyverse_1.2.1 qvalue_2.12.0
loaded via a namespace (and not attached):
[1] tidyselect_0.2.4 reshape2_1.4.3 splines_3.5.1 haven_1.1.2
[5] lattice_0.20-35 colorspace_1.3-2 htmltools_0.3.6 yaml_2.2.0
[9] rlang_0.2.2 pillar_1.3.0 glue_1.3.0 withr_2.1.2
[13] modelr_0.1.2 readxl_1.1.0 bindrcpp_0.2.2 bindr_0.1.1
[17] plyr_1.8.4 munsell_0.5.0 gtable_0.2.0 cellranger_1.1.0
[21] rvest_0.3.2 evaluate_0.13 labeling_0.3 knitr_1.20
[25] broom_0.5.0 Rcpp_0.12.19 scales_1.0.0 backports_1.1.2
[29] jsonlite_1.6 fs_1.2.6 hms_0.4.2 digest_0.6.17
[33] stringi_1.2.4 grid_3.5.1 rprojroot_1.3-2 cli_1.0.1
[37] tools_3.5.1 magrittr_1.5 lazyeval_0.2.1 crayon_1.3.4
[41] whisker_0.3-2 pkgconfig_2.0.2 xml2_1.2.0 lubridate_1.7.4
[45] assertthat_0.2.0 rmarkdown_1.11 httr_1.3.1 rstudioapi_0.9.0
[49] R6_2.3.0 nlme_3.1-137 git2r_0.24.0 compiler_3.5.1