Differential gene expression analysis with DESeq2
2024-08-05
Last updated: 2024-08-05
Checks: 7 0
Knit directory: muse/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200712) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version d5e67bc. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: r_packages_4.3.3/
Ignored: r_packages_4.4.0/
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/deseq2.Rmd) and HTML
(docs/deseq2.html) files. If you’ve configured a remote Git
repository (see ?wflow_git_remote), click on the hyperlinks
in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | d5e67bc | Dave Tang | 2024-08-05 | Getting started with DESeq2 |
DESeq2 is used to:
Estimate variance-mean dependence in count data from high-throughput sequencing assays and test for differential expression based on a model using the negative binomial distribution.
Installation
Install using BiocManager::install().
if (!require("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("DESeq2")We will use data from the pasilla package so install it too.
BiocManager::install("pasilla")'getOption("repos")' replaces Bioconductor standard repositories, see
'help("repositories", package = "BiocManager")' for details.
Replacement repositories:
CRAN: https://p3m.dev/cran/__linux__/jammy/2024-06-13Bioconductor version 3.19 (BiocManager 1.30.23), R 4.4.0 (2024-04-24)Warning: package(s) not installed when version(s) same as or greater than current; use
`force = TRUE` to re-install: 'pasilla'Installation paths not writeable, unable to update packages
path: /usr/local/lib/R/library
packages:
KernSmooth, nlme, survivalExample data
Example dataset in the experiment data package {pasilla}.
fn <- system.file("extdata", "pasilla_gene_counts.tsv", package = "pasilla", mustWork = TRUE)
counts <- as.matrix(read.csv(fn, sep="\t", row.names = "gene_id"))
dim(counts)[1] 14599 7The matrix tallies the number of reads assigned for each gene in each sample.
tail(counts) untreated1 untreated2 untreated3 untreated4 treated1 treated2
FBgn0261570 3296 4910 2156 2060 5077 3069
FBgn0261571 0 0 0 0 1 0
FBgn0261572 4 13 4 11 7 3
FBgn0261573 2651 3653 1571 1612 3334 1848
FBgn0261574 6385 9318 3110 2819 10455 3508
FBgn0261575 6 53 1 3 42 3
treated3
FBgn0261570 3022
FBgn0261571 0
FBgn0261572 3
FBgn0261573 1908
FBgn0261574 3047
FBgn0261575 4Size factors
Estimate size factors.
DESeq2::estimateSizeFactorsForMatrix(counts)untreated1 untreated2 untreated3 untreated4 treated1 treated2 treated3
1.1382630 1.7930004 0.6495470 0.7516892 1.6355751 0.7612698 0.8326526 Mean-variance relationship
Variance versus mean for the (size factor adjusted)
counts data. The axes are logarithmic. Also shown are lines
through the origin with slopes 1 (green) and 2 (red).
sf <- DESeq2::estimateSizeFactorsForMatrix(counts)
ncounts <- t(t(counts) / sf)
# untreated samples
uncounts <- ncounts[, grep("^untreated", colnames(ncounts)), drop = FALSE]
ggplot(
tibble(
mean = rowMeans(uncounts),
var = rowVars(uncounts)
),
aes(x = log1p(mean), y = log1p(var))
) +
geom_hex() +
coord_fixed() +
theme_minimal() +
theme(legend.position = "none") +
geom_abline(slope = 1:2, color = c("forestgreen", "red"))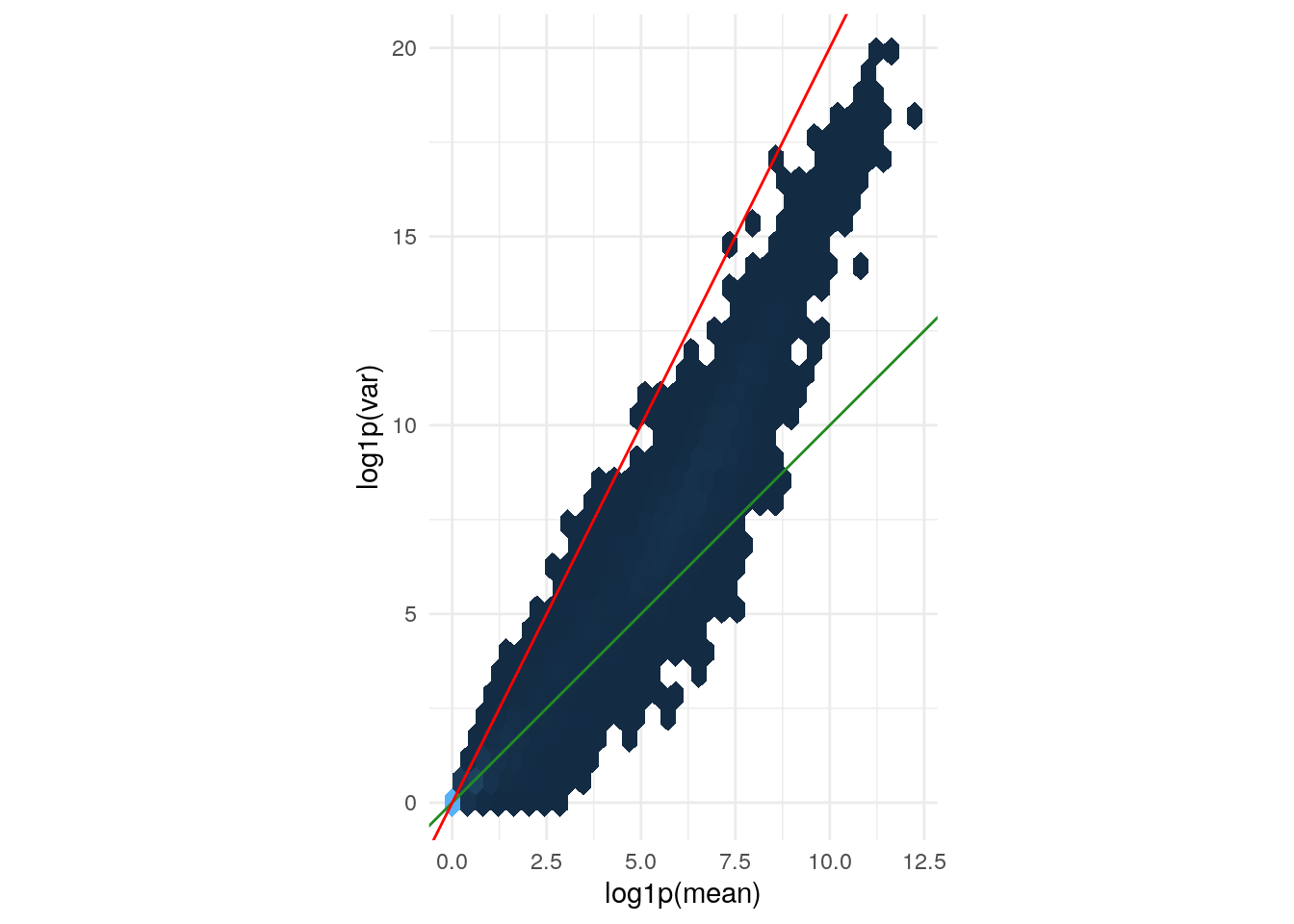
The green line is what we expect if the variance equals the mean, as is the case for a Poisson-distributed random variable. This approximately fits the data in the lower range. The red line corresponds to the quadratic mean-variance relationship \(v = m^2\). We can see that in the upper range of the data, the quadratic relationship approximately fits the data.
sessionInfo()R version 4.4.0 (2024-04-24)
Platform: x86_64-pc-linux-gnu
Running under: Ubuntu 22.04.4 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
time zone: Etc/UTC
tzcode source: system (glibc)
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] hexbin_1.28.3 pasilla_1.32.0
[3] DEXSeq_1.50.0 RColorBrewer_1.1-3
[5] AnnotationDbi_1.66.0 BiocParallel_1.38.0
[7] DESeq2_1.44.0 SummarizedExperiment_1.34.0
[9] Biobase_2.64.0 MatrixGenerics_1.16.0
[11] matrixStats_1.3.0 GenomicRanges_1.56.1
[13] GenomeInfoDb_1.40.1 IRanges_2.38.1
[15] S4Vectors_0.42.1 BiocGenerics_0.50.0
[17] lubridate_1.9.3 forcats_1.0.0
[19] stringr_1.5.1 dplyr_1.1.4
[21] purrr_1.0.2 readr_2.1.5
[23] tidyr_1.3.1 tibble_3.2.1
[25] ggplot2_3.5.1 tidyverse_2.0.0
[27] workflowr_1.7.1
loaded via a namespace (and not attached):
[1] DBI_1.2.3 bitops_1.0-7 httr2_1.0.1
[4] biomaRt_2.60.1 rlang_1.1.4 magrittr_2.0.3
[7] git2r_0.33.0 compiler_4.4.0 RSQLite_2.3.7
[10] getPass_0.2-4 png_0.1-8 callr_3.7.6
[13] vctrs_0.6.5 pkgconfig_2.0.3 crayon_1.5.2
[16] fastmap_1.2.0 dbplyr_2.5.0 XVector_0.44.0
[19] labeling_0.4.3 utf8_1.2.4 Rsamtools_2.20.0
[22] promises_1.3.0 rmarkdown_2.27 tzdb_0.4.0
[25] UCSC.utils_1.0.0 ps_1.7.6 bit_4.0.5
[28] xfun_0.44 zlibbioc_1.50.0 cachem_1.1.0
[31] jsonlite_1.8.8 progress_1.2.3 blob_1.2.4
[34] highr_0.11 later_1.3.2 DelayedArray_0.30.1
[37] parallel_4.4.0 prettyunits_1.2.0 R6_2.5.1
[40] bslib_0.7.0 stringi_1.8.4 genefilter_1.86.0
[43] jquerylib_0.1.4 Rcpp_1.0.12 knitr_1.47
[46] splines_4.4.0 httpuv_1.6.15 Matrix_1.7-0
[49] timechange_0.3.0 tidyselect_1.2.1 rstudioapi_0.16.0
[52] abind_1.4-5 yaml_2.3.8 codetools_0.2-20
[55] hwriter_1.3.2.1 curl_5.2.1 processx_3.8.4
[58] lattice_0.22-6 withr_3.0.0 KEGGREST_1.44.1
[61] evaluate_0.24.0 survival_3.5-8 BiocFileCache_2.12.0
[64] xml2_1.3.6 Biostrings_2.72.1 BiocManager_1.30.23
[67] filelock_1.0.3 pillar_1.9.0 whisker_0.4.1
[70] generics_0.1.3 rprojroot_2.0.4 hms_1.1.3
[73] munsell_0.5.1 scales_1.3.0 xtable_1.8-4
[76] glue_1.7.0 tools_4.4.0 annotate_1.82.0
[79] locfit_1.5-9.9 XML_3.99-0.16.1 fs_1.6.4
[82] grid_4.4.0 colorspace_2.1-0 GenomeInfoDbData_1.2.12
[85] cli_3.6.2 rappdirs_0.3.3 fansi_1.0.6
[88] S4Arrays_1.4.1 gtable_0.3.5 sass_0.4.9
[91] digest_0.6.35 SparseArray_1.4.8 farver_2.1.2
[94] geneplotter_1.82.0 memoise_2.0.1 htmltools_0.5.8.1
[97] lifecycle_1.0.4 httr_1.4.7 statmod_1.5.0
[100] bit64_4.0.5