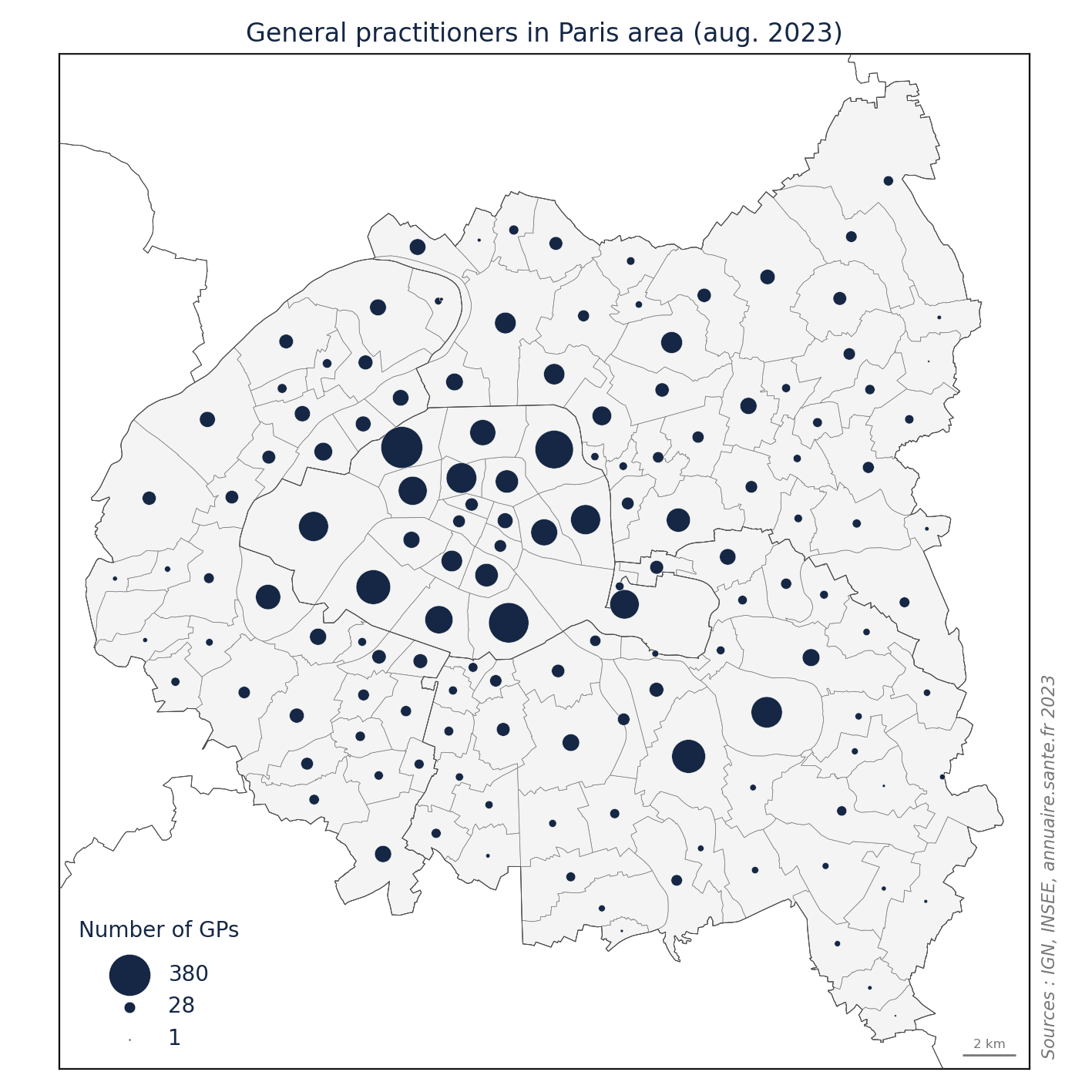
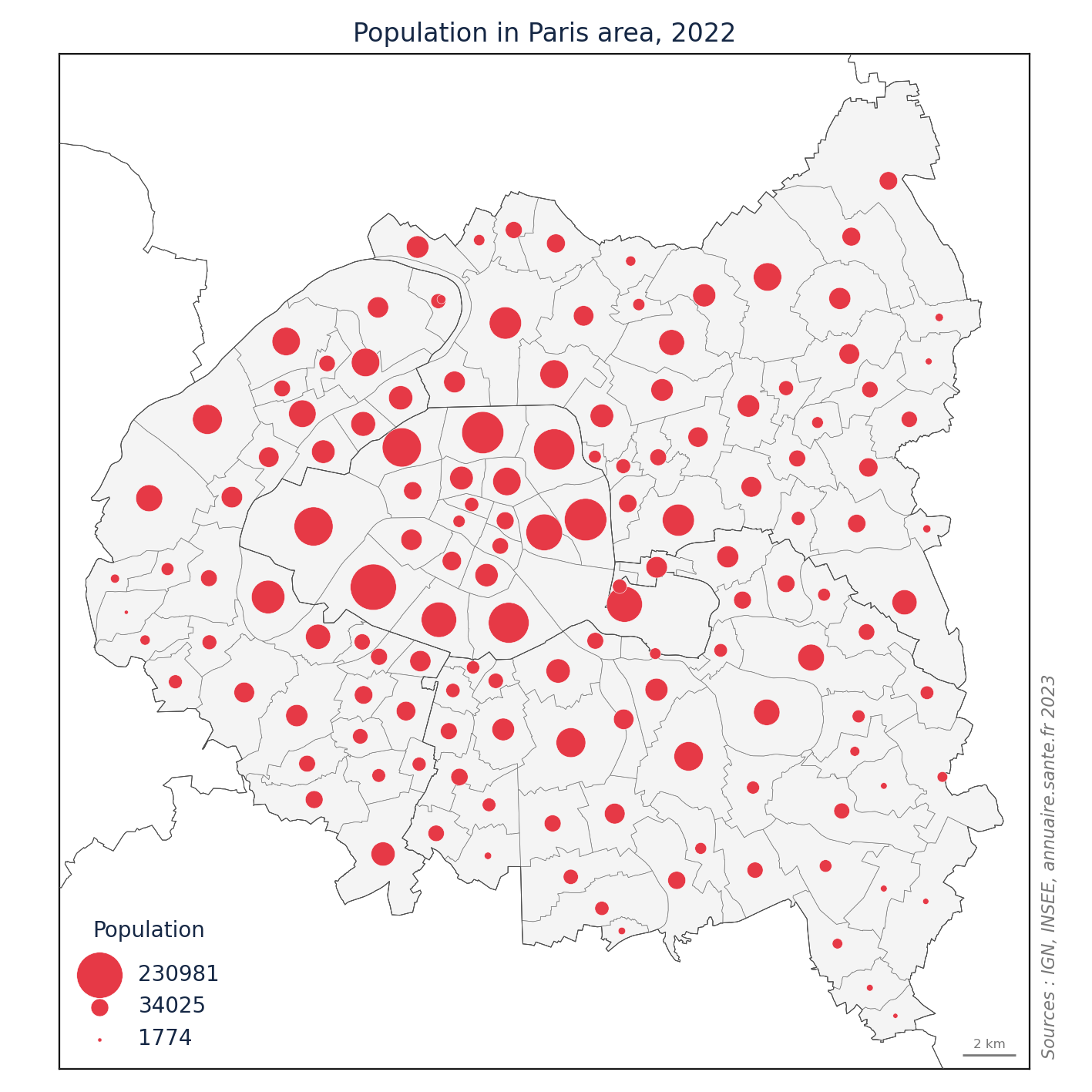
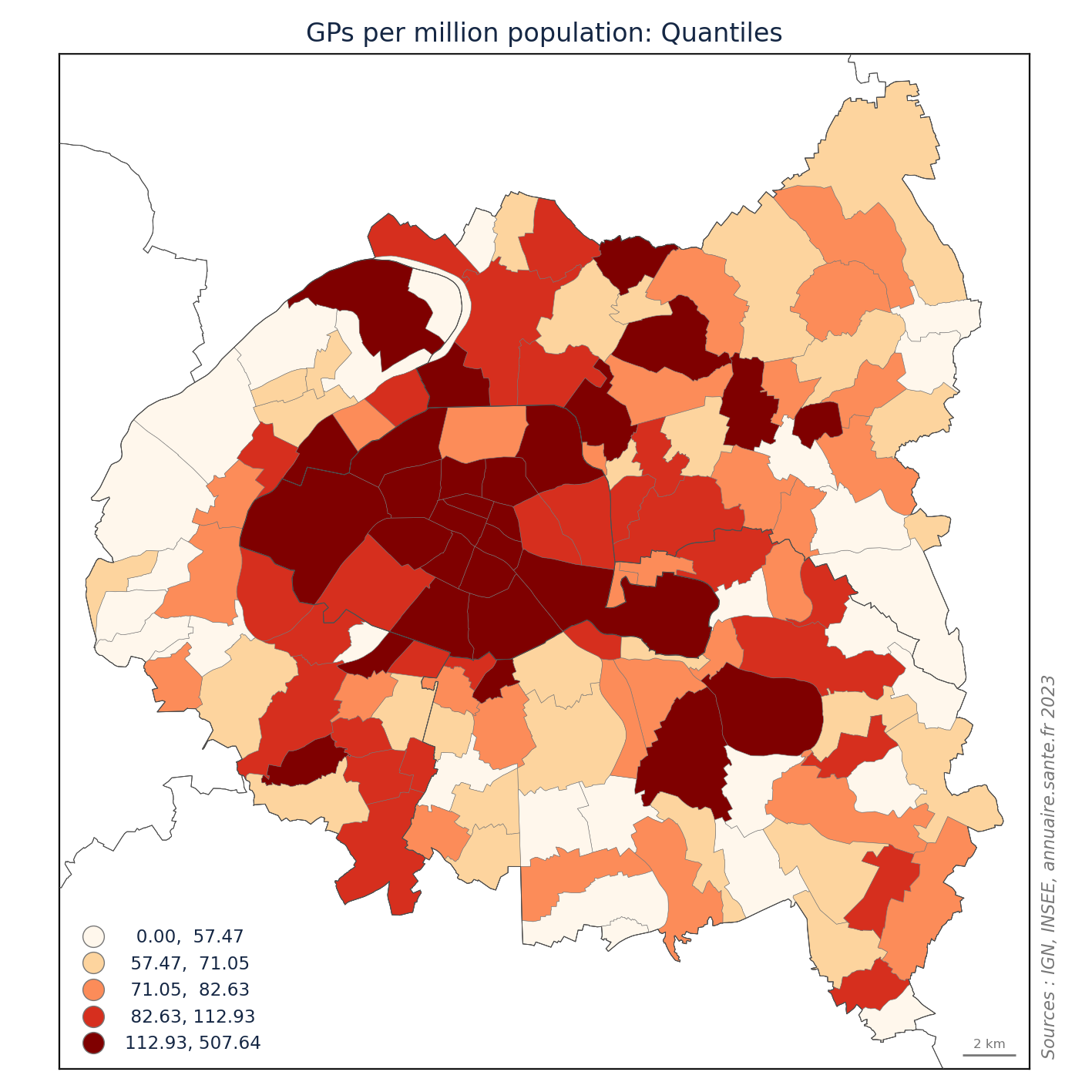
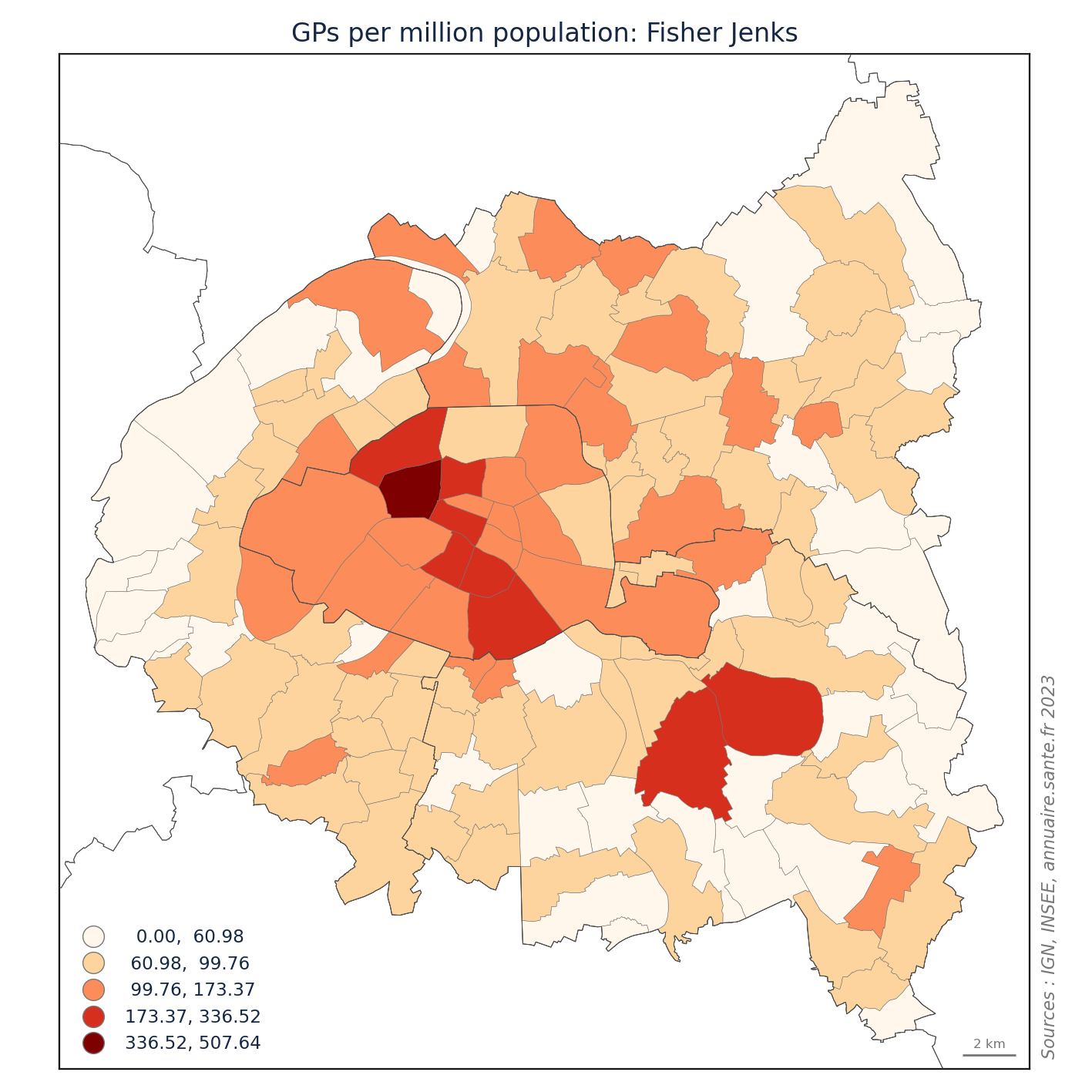
import matplotlib.pyplot as plt
def customize_axes(_ax, _fig):
for spine in ['top', 'right']:
_ax.spines[spine].set_visible(False)
for spine in _ax.spines.values():
spine.set_edgecolor(Colors.white)
_ax.tick_params(axis='x', colors=Colors.white, rotation=45)
_ax.tick_params(axis='y', colors=Colors.white)
_ax.set_facecolor('none')
_fig.patch.set_facecolor('none')
fig_box, ax_box = plt.subplots()
boxplot = geocoded.boxplot(column=['result_score'], vert=False, grid=False, ax=ax_box,
flierprops={'marker': 'o', 'markersize': 5, 'markerfacecolor': Colors.red, 'markeredgecolor': 'none'},
boxprops={'color': Colors.white}, medianprops={'color': Colors.red}, whiskerprops={'color': Colors.white})
ax_box.set_xlabel('')
ax_box.set_yticklabels([])
ax_box.set_xlim(0, 1)
ax_box.set_title('Geocoding results scores', color=Colors.white)
customize_axes(ax_box,fig_box)
fig_box.set_size_inches(fig_box.get_size_inches()[0]*1.5, fig_box.get_size_inches()[1] / 3)
plt.show()