mr.mash issue with larger number of conditions
Fabio Morgante
April 13, 2021
Last updated: 2021-04-13
Checks: 7 0
Knit directory: mr_mash_test/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200328) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 0091cf6. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .sos/
Ignored: analysis/.sos/
Ignored: code/.sos/
Ignored: code/assess_mrmash_speed.2835257.err
Ignored: code/assess_mrmash_speed.2835257.out
Ignored: code/assess_mrmash_speed.2914525.err
Ignored: code/assess_mrmash_speed.2914525.out
Ignored: code/fit_mr_mash.66662433.err
Ignored: code/fit_mr_mash.66662433.out
Ignored: data/cis_eqtl_analysis_ready
Ignored: data/gtex-v8-ids.txt
Ignored: data/gtex-v8-manifest-full-X-10tissues-missing-rate.txt
Ignored: data/gtex-v8-manifest-full-X.txt
Ignored: data/gtex-v8-manifest.txt
Ignored: dsc/.sos/
Ignored: dsc/dsc_mvreg_07_29_20.3339807.err
Ignored: dsc/dsc_mvreg_07_29_20.3339807.out
Ignored: dsc/dsc_mvreg_07_29_20.3340460.err
Ignored: dsc/dsc_mvreg_07_29_20.3340460.out
Ignored: dsc/dsc_mvreg_07_29_20.3341113.err
Ignored: dsc/dsc_mvreg_07_29_20.3341113.out
Ignored: dsc/dsc_mvreg_07_29_20.3352804.err
Ignored: dsc/dsc_mvreg_07_29_20.3352804.out
Ignored: dsc/functions/__pycache__/
Ignored: dsc/mvreg_test/rep1/
Ignored: dsc/mvreg_test/rep2/
Ignored: dsc/outfiles/
Ignored: output/dsc.html
Ignored: output/dsc/
Ignored: output/dsc_05_18_20.html
Ignored: output/dsc_05_18_20/
Ignored: output/dsc_07_29_20.html
Ignored: output/dsc_07_29_20/
Ignored: output/dsc_OLD.html
Ignored: output/dsc_OLD/
Ignored: output/dsc_test.html
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10.html
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_inter/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_lowPVE.html
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_lowPVE/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_lowPVE_inter/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10.html
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_inter/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE.html
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_inter/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_pipeline.html
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10.html
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_diffPVE.html
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_diffPVE/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_diffPVE_inter/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_inter/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_lowPVE.html
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_lowPVE/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_lowPVE_inter/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10.html
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_inter/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_lowPVE.html
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_lowPVE/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_lowPVE_inter/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10.html
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_inter/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_lowPVE.html
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_lowPVE/
Ignored: output/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_lowPVE_inter/
Ignored: output/mvreg_all_genes_prior_corrX_indepV_sharedB_2blocksr10.html
Ignored: output/mvreg_all_genes_prior_corrX_indepV_sharedB_2blocksr10/
Ignored: output/mvreg_all_genes_prior_corrX_indepV_sharedB_2blocksr10_inter/
Ignored: output/mvreg_all_genes_prior_highcorrX_indepV_sharedB_2blocksr10.html
Ignored: output/mvreg_all_genes_prior_highcorrX_indepV_sharedB_2blocksr10/
Ignored: output/mvreg_all_genes_prior_highcorrX_indepV_sharedB_2blocksr10_inter/
Ignored: output/mvreg_all_genes_prior_indepX_indepV_B_1causalrespr10_bigtestset.html
Ignored: output/mvreg_all_genes_prior_indepX_indepV_B_1causalrespr10_bigtestset/
Ignored: output/mvreg_all_genes_prior_indepX_indepV_B_1causalrespr10_bigtestset_inter/
Ignored: output/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10.html
Ignored: output/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10/
Ignored: output/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_bigtestset.html
Ignored: output/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_bigtestset/
Ignored: output/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_bigtestset_inter/
Ignored: output/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_inter/
Ignored: output/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_test.html
Ignored: output/mvreg_all_genes_prior_indepX_indepV_sharedB_3causalrespr10.html
Ignored: output/mvreg_all_genes_prior_indepX_indepV_sharedB_3causalrespr10/
Ignored: output/mvreg_all_genes_prior_indepX_indepV_sharedB_3causalrespr10_inter/
Ignored: output/test_dense_issue.rds
Ignored: output/test_sparse_issue.rds
Ignored: scripts/.sos/
Ignored: scripts/extract_gtex_ids.10523031.err
Ignored: scripts/extract_gtex_ids.10523031.out
Ignored: scripts/get_full_X_gtex_v8_manifest.10520041.err
Ignored: scripts/get_full_X_gtex_v8_manifest.10520041.out
Ignored: scripts/get_gtex_v8_manifest.10520039.err
Ignored: scripts/get_gtex_v8_manifest.10520039.out
Ignored: scripts/get_missing_Y_rate_10tissues_full_X_gtex_v8_manifest.6515875.err
Ignored: scripts/get_missing_Y_rate_10tissues_full_X_gtex_v8_manifest.6515875.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_lowPVE_pipeline.6297709.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_lowPVE_pipeline.6297709.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_lowPVE_pipeline.6297720.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_lowPVE_pipeline.6297720.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_lowPVE_pipeline.6300278.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_lowPVE_pipeline.6300278.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_lowPVE_pipeline.6300959.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_lowPVE_pipeline.6300959.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_lowPVE_pipeline.6312239.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_lowPVE_pipeline.6312239.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_pipeline.6030933.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_pipeline.6030933.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_pipeline.6030949.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_pipeline.6030949.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_pipeline.6034734.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_pipeline.6034734.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_pipeline.6035003.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_pipeline.6035003.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_pipeline.6190207.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_B_1causalrespr10_pipeline.6190207.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6318547.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6318547.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6318567.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6318567.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6322126.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6322126.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6322461.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6322461.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6401917.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6401917.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6433374.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6433374.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6449097.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6449097.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6450584.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_lowPVE_pipeline.6450584.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_pipeline.6192054.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_pipeline.6192054.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_pipeline.6192082.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_pipeline.6192082.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_pipeline.6193059.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_pipeline.6193059.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_pipeline.6194214.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_pipeline.6194214.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_pipeline.6219626.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_indepB_r10_pipeline.6219626.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_diffPVE_pipeline.6295843.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_diffPVE_pipeline.6295843.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_lowPVE_pipeline.6400836.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_lowPVE_pipeline.6400836.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_lowPVE_pipeline.6400839.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_lowPVE_pipeline.6400839.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_lowPVE_pipeline.6401592.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_lowPVE_pipeline.6401592.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_lowPVE_pipeline.6401599.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_lowPVE_pipeline.6401599.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_pipeline.5955005.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_pipeline.5955005.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_pipeline.5955011.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_pipeline.5955011.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_pipeline.5955473.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_pipeline.5955473.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_pipeline.5955517.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_2blocksr10_pipeline.5955517.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_lowPVE_pipeline.6342504.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_lowPVE_pipeline.6342504.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_lowPVE_pipeline.6342513.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_lowPVE_pipeline.6342513.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_lowPVE_pipeline.6350805.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_lowPVE_pipeline.6350805.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_lowPVE_pipeline.6350813.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_lowPVE_pipeline.6350813.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_lowPVE_pipeline.6355751.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_lowPVE_pipeline.6355751.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_pipeline.6017919.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_pipeline.6017919.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_pipeline.6017922.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_pipeline.6017922.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_pipeline.6019026.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_pipeline.6019026.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_pipeline.6019566.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_3causalrespr10_pipeline.6019566.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_lowPVE_pipeline.6372840.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_lowPVE_pipeline.6372840.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_lowPVE_pipeline.6372856.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_lowPVE_pipeline.6372856.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_lowPVE_pipeline.6373601.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_lowPVE_pipeline.6373601.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_lowPVE_pipeline.6373615.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_lowPVE_pipeline.6373615.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_lowPVE_pipeline.6382984.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_lowPVE_pipeline.6382984.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_lowPVE_pipeline.6400810.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_lowPVE_pipeline.6400810.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_pipeline.6225970.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_pipeline.6225970.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_pipeline.6226041.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_pipeline.6226041.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_pipeline.6228446.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_pipeline.6228446.out
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_pipeline.6228985.err
Ignored: scripts/mvreg_all_genes_prior_GTExrealX_indepV_sharedB_r10_pipeline.6228985.out
Ignored: scripts/mvreg_all_genes_prior_corrX_indepV_sharedB_2blocksr10_pipeline.5297178.err
Ignored: scripts/mvreg_all_genes_prior_corrX_indepV_sharedB_2blocksr10_pipeline.5297178.out
Ignored: scripts/mvreg_all_genes_prior_corrX_indepV_sharedB_2blocksr10_pipeline.5297184.err
Ignored: scripts/mvreg_all_genes_prior_corrX_indepV_sharedB_2blocksr10_pipeline.5297184.out
Ignored: scripts/mvreg_all_genes_prior_corrX_indepV_sharedB_2blocksr10_pipeline.5299552.err
Ignored: scripts/mvreg_all_genes_prior_corrX_indepV_sharedB_2blocksr10_pipeline.5299552.out
Ignored: scripts/mvreg_all_genes_prior_corrX_indepV_sharedB_2blocksr10_pipeline.5300596.err
Ignored: scripts/mvreg_all_genes_prior_corrX_indepV_sharedB_2blocksr10_pipeline.5300596.out
Ignored: scripts/mvreg_all_genes_prior_corrX_indepV_sharedB_2blocksr10_pipeline.5300796.err
Ignored: scripts/mvreg_all_genes_prior_corrX_indepV_sharedB_2blocksr10_pipeline.5300796.out
Ignored: scripts/mvreg_all_genes_prior_highcorrX_indepV_sharedB_2blocksr10_pipeline.5305725.err
Ignored: scripts/mvreg_all_genes_prior_highcorrX_indepV_sharedB_2blocksr10_pipeline.5305725.out
Ignored: scripts/mvreg_all_genes_prior_highcorrX_indepV_sharedB_2blocksr10_pipeline.5305727.err
Ignored: scripts/mvreg_all_genes_prior_highcorrX_indepV_sharedB_2blocksr10_pipeline.5305727.out
Ignored: scripts/mvreg_all_genes_prior_highcorrX_indepV_sharedB_2blocksr10_pipeline.5308354.err
Ignored: scripts/mvreg_all_genes_prior_highcorrX_indepV_sharedB_2blocksr10_pipeline.5308354.out
Ignored: scripts/mvreg_all_genes_prior_highcorrX_indepV_sharedB_2blocksr10_pipeline.5309181.err
Ignored: scripts/mvreg_all_genes_prior_highcorrX_indepV_sharedB_2blocksr10_pipeline.5309181.out
Ignored: scripts/mvreg_all_genes_prior_highcorrX_indepV_sharedB_2blocksr10_pipeline.5310380.err
Ignored: scripts/mvreg_all_genes_prior_highcorrX_indepV_sharedB_2blocksr10_pipeline.5310380.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_B_1causalrespr10_bigtestset_pipeline.5867284.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_B_1causalrespr10_bigtestset_pipeline.5867284.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_B_1causalrespr10_bigtestset_pipeline.5867300.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_B_1causalrespr10_bigtestset_pipeline.5867300.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_B_1causalrespr10_bigtestset_pipeline.5870492.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_B_1causalrespr10_bigtestset_pipeline.5870492.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_B_1causalrespr10_bigtestset_pipeline.5870684.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_B_1causalrespr10_bigtestset_pipeline.5870684.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_B_1causalrespr10_bigtestset_pipeline.5876670.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_B_1causalrespr10_bigtestset_pipeline.5876670.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_bigtestset_pipeline.5845435.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_bigtestset_pipeline.5845435.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_bigtestset_pipeline.5845459.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_bigtestset_pipeline.5845459.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_bigtestset_pipeline.5850880.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_bigtestset_pipeline.5850880.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_bigtestset_pipeline.5851060.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_bigtestset_pipeline.5851060.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_pipeline.5290364.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_pipeline.5290364.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_pipeline.5290365.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_pipeline.5290365.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_pipeline.5293048.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_pipeline.5293048.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_pipeline.5294079.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_pipeline.5294079.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_pipeline.5295637.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_pipeline.5295637.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_pipeline.5297100.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_2blocksr10_pipeline.5297100.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_3causalrespr10_pipeline.5315542.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_3causalrespr10_pipeline.5315542.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_3causalrespr10_pipeline.5315575.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_3causalrespr10_pipeline.5315575.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_3causalrespr10_pipeline.5318936.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_3causalrespr10_pipeline.5318936.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_3causalrespr10_pipeline.5374687.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_3causalrespr10_pipeline.5374687.out
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_3causalrespr10_pipeline.5374902.err
Ignored: scripts/mvreg_all_genes_prior_indepX_indepV_sharedB_3causalrespr10_pipeline.5374902.out
Ignored: scripts/pred_HHB_imputed.6496220.err
Ignored: scripts/pred_HHB_imputed.6496220.out
Ignored: scripts/pred_fullV_HHB_imputed.6503400.err
Ignored: scripts/pred_fullV_HHB_imputed.6503400.out
Untracked files:
Untracked: analysis/dense_issue_investigation2/
Untracked: code/20200502_Prepare_ED_prior.ipynb
Untracked: code/plot_test.R
Untracked: code/pred_fullV_LNP1.R
Untracked: dsc/mvreg_all_genes_prior_09_11_20_COPY.dsc
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/mrmash_larger_r_issue.Rmd) and HTML (docs/mrmash_larger_r_issue.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 0091cf6 | fmorgante | 2021-04-13 | Add more mash results |
| html | 446489f | fmorgante | 2021-04-12 | Build site. |
| Rmd | 0fe4f50 | fmorgante | 2021-04-12 | Add mash results |
| html | b2c3824 | fmorgante | 2021-03-25 | Build site. |
| Rmd | 5361380 | fmorgante | 2021-03-25 | Add a few things |
| html | cde23ac | fmorgante | 2021-03-24 | Build site. |
| Rmd | d5846a6 | fmorgante | 2021-03-24 | Add investigation on the issue with larger r |
library(mr.mash.alpha)
library(glmnet)
library(mashr)
options(stringsAsFactors = FALSE)
###Functions to compute g-lasso coefficients
compute_coefficients_glasso <- function(X, Y, standardize, nthreads, version=c("Rcpp", "R")){
version <- match.arg(version)
n <- nrow(X)
p <- ncol(X)
r <- ncol(Y)
Y_has_missing <- any(is.na(Y))
if(Y_has_missing){
###Extract per-individual Y missingness patterns
Y_miss_patterns <- mr.mash.alpha:::extract_missing_Y_pattern(Y)
###Initialize missing Ys
muy <- colMeans(Y, na.rm=TRUE)
for(l in 1:r){
Y[is.na(Y[, l]), l] <- muy[l]
}
###Compute V and its inverse
V <- mr.mash.alpha:::compute_V_init(X, Y, matrix(0, p, r), method="flash")
Vinv <- chol2inv(chol(V))
###Compute expected Y (assuming B=0)
mu <- matrix(rep(muy, each=n), n, r)
###Impute missing Ys
Y <- mr.mash.alpha:::impute_missing_Y(Y=Y, mu=mu, Vinv=Vinv, miss=Y_miss_patterns$miss, non_miss=Y_miss_patterns$non_miss,
version=version)$Y
}
##Fit group-lasso
if(nthreads>1){
doMC::registerDoMC(nthreads)
paral <- TRUE
} else {
paral <- FALSE
}
cvfit_glmnet <- glmnet::cv.glmnet(x=X, y=Y, family="mgaussian", alpha=1, standardize=standardize, parallel=paral)
coeff_glmnet <- coef(cvfit_glmnet, s="lambda.min")
##Build matrix of initial estimates for mr.mash
B <- matrix(as.numeric(NA), nrow=p, ncol=r)
for(i in 1:length(coeff_glmnet)){
B[, i] <- as.vector(coeff_glmnet[[i]])[-1]
}
return(list(B=B, Y=Y))
}In this website, we want to investigate an issue that happens with larger number of conditions than the 10 conditions we have used in the other investigations. The issue is that mr.mash fails to shrink to 0 coefficients that are actually 0. As we will see, some of them are pretty badly misestimated.
Let’s look at an example with n=150, p=400, r=25, PVE=0.2, p_causal=5, independent residuals, independent predictors, and equal effects across conditions from a single multivariate normal with variance 1.
set.seed(12)
n <- 200
ntest <- 50
p <- 400
p_causal <- 5
r <- 25
X_cor <- 0
pve <- 0.2
r_causal <- list(1:r)
B_cor <- 1
B_scale <- 1
w <- 1
###Simulate V, B, X and Y
out <- simulate_mr_mash_data(n, p, p_causal, r, r_causal, intercepts = rep(1, r),
pve=pve, B_cor=B_cor, B_scale=B_scale, w=w,
X_cor=X_cor, X_scale=1, V_cor=0)
colnames(out$Y) <- paste0("Y", seq(1, r))
rownames(out$Y) <- paste0("N", seq(1, n))
colnames(out$X) <- paste0("X", seq(1, p))
rownames(out$X) <- paste0("N", seq(1, n))
###Split the data in training and test sets
test_set <- sort(sample(x=c(1:n), size=ntest, replace=FALSE))
Ytrain <- out$Y[-test_set, ]
Xtrain <- out$X[-test_set, ]
Ytest <- out$Y[test_set, ]
Xtest <- out$X[test_set, ]mr.mash is run standardizing X and dropping mixture components with weight less than \(1e-8\). However, the results look the same without dropping any component. The mixture prior consists only of canonical matrices, scaled by a grid computed as in the mash paper. group-LASSO is used to initialize mr.mash and as a comparison.
standardize <- TRUE
nthreads <- 4
w0_threshold <- 1e-8
S0_can <- compute_canonical_covs(ncol(Ytrain), singletons=TRUE, hetgrid=c(0, 0.25, 0.5, 0.75, 1))
####Complete data####
###Compute grid of variances
univ_sumstats <- compute_univariate_sumstats(Xtrain, Ytrain, standardize=standardize,
standardize.response=FALSE, mc.cores=nthreads)
grid <- autoselect.mixsd(univ_sumstats, mult=sqrt(2))^2
###Compute prior with only canonical matrices
S0 <- expand_covs(S0_can, grid, zeromat=TRUE)
###Fit mr.mash
out_coeff <- compute_coefficients_glasso(Xtrain, Ytrain, standardize=standardize, nthreads=nthreads, version="Rcpp")
prop_nonzero_glmnet <- sum(out_coeff$B[, 1]!=0)/p
w0 <- c((1-prop_nonzero_glmnet), rep(prop_nonzero_glmnet/(length(S0)-1), (length(S0)-1)))
fit_mrmash <- mr.mash(Xtrain, Ytrain, S0, w0=w0, update_w0=TRUE, update_w0_method="EM", tol=1e-2,
convergence_criterion="ELBO", compute_ELBO=TRUE, standardize=standardize,
verbose=FALSE, update_V=TRUE, update_V_method="diagonal", e=1e-8,
w0_threshold=w0_threshold, nthreads=nthreads, mu1_init=out_coeff$B)
#####Coefficients plots######
layout(matrix(c(1, 1, 2, 2,
1, 1, 2, 2,
0, 3, 3, 0,
0, 3, 3, 0), 4, 4, byrow = TRUE))
###Plot estimated vs true coeffcients
##mr.mash
plot(out$B, fit_mrmash$mu1, main="mr.mash", xlab="True coefficients", ylab="Estimated coefficients",
cex=2, cex.lab=2, cex.main=2, cex.axis=2)
abline(0, 1)
##g-lasso
plot(out$B, out_coeff$B, main="g-lasso", xlab="True coefficients", ylab="Estimated coefficients",
cex=2, cex.lab=2, cex.main=2, cex.axis=2)
abline(0, 1)
###Plot mr.mash vs g-lasso estimated coeffcients
colorz <- matrix("black", nrow=p, ncol=r)
zeros <- apply(out$B, 2, function(x) x==0)
for(i in 1:ncol(colorz)){
colorz[zeros[, i], i] <- "red"
}
plot(fit_mrmash$mu1, out_coeff$B, main="mr.mash vs g-lasso",
xlab="mr.mash coefficients", ylab="g-lasso coefficients",
col=colorz, cex=2, cex.lab=2, cex.main=2, cex.axis=2)
abline(0, 1)
legend("topleft",
legend = c("Non-zero", "Zero"),
col = c("black", "red"),
pch = c(1, 1),
horiz = FALSE,
cex=2) 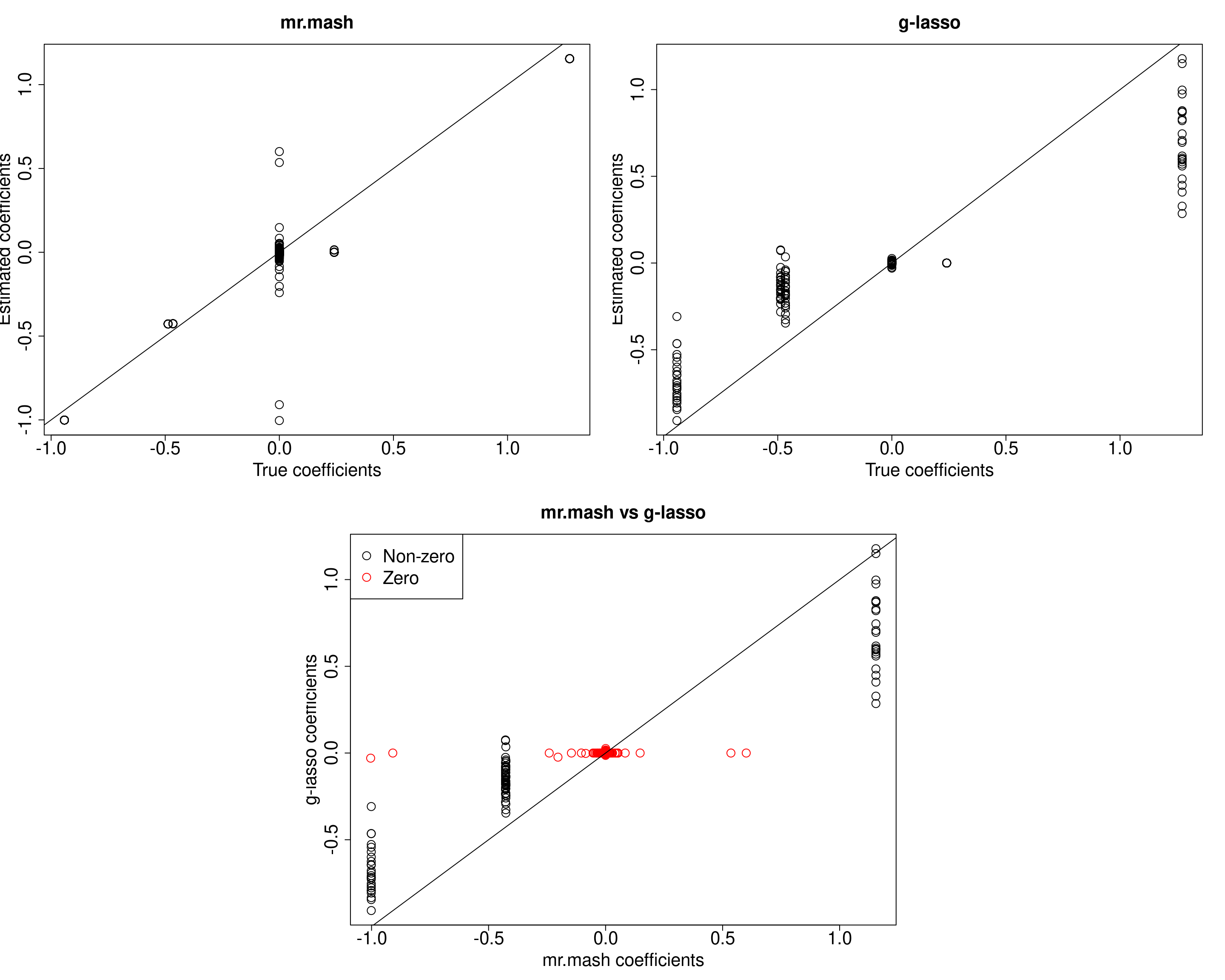
###Identify badly estimated variables
bad_est_1 <- which(out$B==0 & fit_mrmash$mu1 < -0.5, arr.ind = TRUE)
bad_est_2 <- which(out$B==0 & fit_mrmash$mu1 > 0.5, arr.ind = TRUE)
bad_est <- rbind(bad_est_1, bad_est_2)
cat("Bad variables:\n")Bad variables:bad_est row col
X274 274 10
X347 347 15
X183 183 10
X72 72 13mr.mash is much better than group-LASSO at correctly estimating the non-zero coefficients. However, it does much worse at shrinking the true null coefficients to 0. In particular, there are a few coeffcients that are pretty badly misestimated. We are going to look at those cases more closely.
###Estimated coefficients by mr.mash for the bad variables
cat("mr.mash estimated coefficients:\n")mr.mash estimated coefficients:fit_mrmash$mu1[bad_est[, 1], ] Y1 Y2 Y3 Y4 Y5
X274 -2.642841e-05 -2.663867e-05 -2.335354e-05 -2.653733e-05 -2.613332e-05
X347 6.491175e-07 2.916934e-06 1.739985e-06 2.150244e-06 2.321851e-06
X183 1.133417e-04 1.284499e-04 1.203046e-04 1.115160e-04 1.203380e-04
X72 9.529781e-06 -4.624202e-06 1.235475e-06 1.068643e-06 2.780234e-06
Y6 Y7 Y8 Y9 Y10
X274 -2.233829e-05 -2.511605e-05 -2.896884e-05 -2.870533e-05 -9.092047e-01
X347 1.451095e-06 6.299573e-06 3.501019e-06 6.391154e-07 6.646096e-05
X183 1.200349e-04 1.184511e-04 1.168999e-04 9.990242e-05 5.354676e-01
X72 1.060682e-05 1.326606e-05 7.372047e-06 -2.390740e-06 -6.613187e-04
Y11 Y12 Y13 Y14 Y15
X274 -2.773517e-05 -2.442013e-05 8.829146e-05 -3.148464e-05 6.064451e-05
X347 5.231139e-06 -3.038223e-05 -2.331779e-04 1.595482e-06 -1.003837e+00
X183 1.290409e-04 1.330501e-04 3.121302e-04 1.422738e-04 3.496837e-04
X72 9.218040e-06 -3.617465e-05 6.008830e-01 -6.688684e-06 1.265758e-03
Y16 Y17 Y18 Y19 Y20
X274 -2.554938e-05 -2.579409e-05 -2.871577e-05 -2.471882e-05 -2.455460e-05
X347 4.505555e-06 2.149654e-06 1.773809e-06 2.081464e-06 -8.069752e-08
X183 1.223263e-04 1.269347e-04 1.225920e-04 1.272094e-04 1.296489e-04
X72 5.131390e-06 3.996038e-06 2.568455e-06 4.814783e-06 9.623803e-06
Y21 Y22 Y23 Y24 Y25
X274 -2.592537e-05 4.360263e-04 -2.732129e-05 -1.867228e-05 -2.871367e-05
X347 9.000980e-06 -9.735566e-07 2.812928e-06 3.894336e-06 8.448670e-07
X183 1.170115e-04 -1.310602e-04 1.225248e-04 1.238062e-04 9.362710e-05
X72 -1.681922e-05 3.996730e-04 4.902352e-06 9.337151e-06 1.645090e-05We can see that the estimated coefficients for each one of those variables look like a singleton structure, where there is an effect in one condition and nowhere else. When we look at the prior weights at convergence, we can see that those singleton matrices get relatively large weight (see below).
###Estimated prior weights by mr.mash for the bad variables
cat("mr.mash estimated prior weights:\n")mr.mash estimated prior weights:head(sort(fit_mrmash$w0, decreasing=TRUE), 20) null shared1_grid11 singleton13_grid10
0.9346759513 0.0086745867 0.0069429060
singleton10_grid10 singleton22_grid10 singleton10_grid11
0.0065330516 0.0052718769 0.0051348432
singleton15_grid11 singleton15_grid10 shared1_grid10
0.0049436750 0.0047741792 0.0034326125
singleton22_grid9 singleton13_grid9 singleton13_grid11
0.0030192287 0.0018686026 0.0014915698
singleton15_grid9 singleton10_grid9 singleton22_grid8
0.0004987422 0.0004802332 0.0003878599
singleton13_grid8 singleton12_grid8 singleton24_grid8
0.0003738464 0.0002453590 0.0001796324
singleton12_grid7 singleton22_grid11
0.0001623117 0.0001445351 To see why that is happening, the next step will be to look at the data. While the best thing to do would be to look at the summary statistics from a simple multivariate regression with the same residual covariance matrix as in mr.mash, we are going to look at the univariate summary statistics that are readily available.
###Estimated coefficients and Z-scores by univariate regression for the bad variables
rownames(univ_sumstats$Bhat) <- colnames(out$X)
colnames(univ_sumstats$Bhat) <- colnames(out$Y)
cat("SLR estimated coefficients:\n")SLR estimated coefficients:univ_sumstats$Bhat[bad_est[, 1], ] Y1 Y2 Y3 Y4 Y5 Y6
X274 -0.1325482 -0.09893288 0.61508065 -0.1165136 -0.1069066 0.35151305
X347 -0.2427989 0.28123165 0.06309551 0.1649556 0.3030400 0.02980773
X183 -0.2428503 0.49439918 0.20907556 -0.2292677 0.2192696 0.17487806
X72 0.3468332 -0.38077183 -0.36862902 -0.2403712 -0.2716020 0.18512322
Y7 Y8 Y9 Y10 Y11 Y12
X274 0.1024990 -0.454970612 -0.25909065 -1.0881426 -0.2781424 0.0916439
X347 0.4888237 0.446450157 -0.08708036 0.2033015 0.6209967 -0.5022021
X183 0.1270435 0.003659657 -0.29134310 1.1836731 0.5654748 0.3028926
X72 0.2467956 0.150784203 -0.25086360 -0.1810409 0.2310031 -0.3425984
Y13 Y14 Y15 Y16 Y17 Y18
X274 0.2435822 -0.39061436 0.2271793 0.071133592 0.01441381 -0.58419877
X347 -0.3747620 0.04851022 -1.1350130 0.640749645 0.19264130 0.07550262
X183 0.3618534 0.62647779 0.4556610 0.337172801 0.62552105 0.39018457
X72 1.1717008 -0.33024120 0.5026009 0.009030604 -0.08879668 -0.23306137
Y19 Y20 Y21 Y22 Y23
X274 0.23015756 0.1516004 0.02275088 0.52279901 -0.226039596
X347 0.15321483 -0.1375850 0.51024287 0.07540335 0.296716744
X183 0.54839476 0.4148681 0.10061303 -0.04286237 0.308742629
X72 -0.01456058 0.1413060 -0.37970711 0.21641262 -0.006988736
Y24 Y25
X274 0.32427352 -0.3004417
X347 0.19882637 -0.0776774
X183 0.22366030 -0.4267544
X72 0.06677394 0.3970696###Estimated Z-scores by univariate regression for the bad variables
Zscores <- univ_sumstats$Bhat/univ_sumstats$Shat
cat("SLR estimated Z-scores:\n")SLR estimated Z-scores:Zscores[bad_est[, 1], ] Y1 Y2 Y3 Y4 Y5 Y6
X274 -0.4365736 -0.3252120 1.7394660 -0.3690672 -0.3517424 1.11406779
X347 -0.8009137 0.9267963 0.1766698 0.5227513 0.9999831 0.09408288
X183 -0.8010841 1.6391949 0.5860324 -0.7271807 0.7224003 0.55252082
X72 1.1466597 -1.2578544 -1.0357761 -0.7625325 -0.8956515 0.58496230
Y7 Y8 Y9 Y10 Y11 Y12
X274 0.3267747 -1.44776342 -0.8270740 -3.3784413 -0.8473222 0.2965010
X347 1.5706941 1.42027984 -0.2774153 0.6090926 1.9102079 -1.6389006
X183 0.4051023 0.01156436 -0.9305960 3.7011173 1.7357935 0.9828482
X72 0.7881596 0.47683468 -0.8006966 -0.5422600 0.7031937 -1.1126963
Y13 Y14 Y15 Y16 Y17 Y18
X274 0.732031 -1.1812598 0.696065 0.23658439 0.04131068 -1.9612099
X347 -1.129041 0.1460284 -3.621566 2.16389997 0.55268195 0.2503124
X183 1.089836 1.9086398 1.403033 1.12595782 1.81241787 1.3006204
X72 3.670433 -0.9973537 1.549786 0.03002948 -0.25454953 -0.7740532
Y19 Y20 Y21 Y22 Y23 Y24
X274 0.71527253 0.4970719 0.0678337 1.7837366 -0.74185722 1.0726073
X347 0.47569908 -0.4510520 1.5332637 0.2546203 0.97512125 0.6560840
X183 1.71812707 1.3676601 0.3000724 -0.1447153 1.01491063 0.7383136
X72 -0.04517348 0.4632679 -1.1370125 0.7319304 -0.02289468 0.2200577
Y25
X274 -0.932783
X347 -0.240511
X183 -1.328901
X72 1.235480plot(fit_mrmash$mu1[bad_est[, 1], ], univ_sumstats$Bhat[bad_est[, 1], ], main="mr.mash vs SLR -- bad variables",
xlab="mr.mash coefficients", ylab="SLR coefficients",
cex=2, cex.lab=2, cex.main=2, cex.axis=2)
abline(0, 1)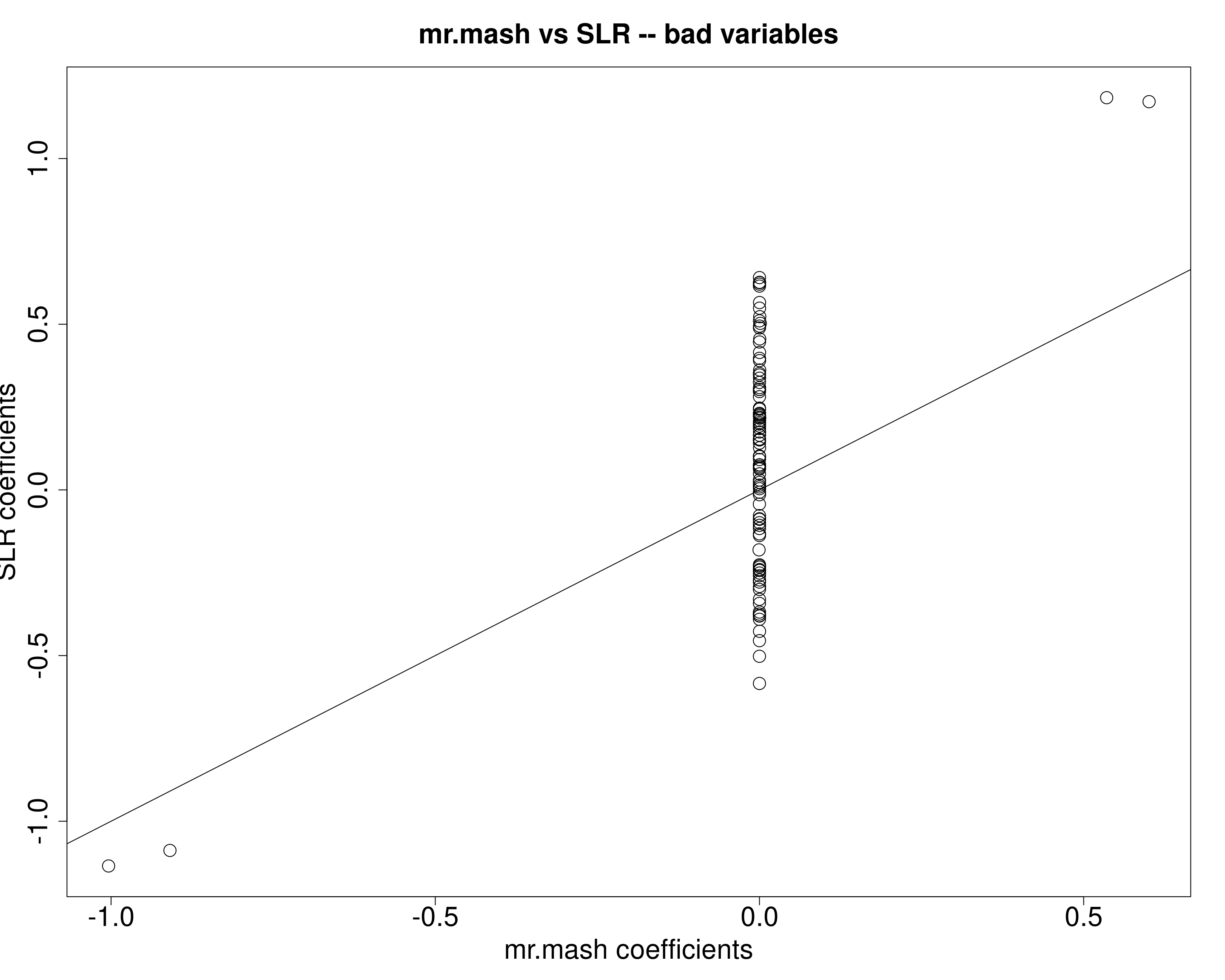
The data (wrongly) supports the singleton structure (i.e., large effect in one condition and smaller effects in the other conditions). mr.mash then estimates the coefficients accordingly, by shrinking to 0 the coefficients in the conditions with smaller effects and leaving the coefficent in the one condition almost unshrunken for two variables and a little shrunken for the other two variables.
If we do not include the singleton matrices in the prior, this behavior does not appear.
S0_can_nosing <- compute_canonical_covs(ncol(Ytrain), singletons=FALSE, hetgrid=c(0, 0.25, 0.5, 0.75, 1))
###Compute prior with only canonical matrices without singletons
S0_nosing <- expand_covs(S0_can_nosing, grid, zeromat=TRUE)
###Fit mr.mash
w0_nosing <- c((1-prop_nonzero_glmnet), rep(prop_nonzero_glmnet/(length(S0_nosing)-1), (length(S0_nosing)-1)))
fit_mrmash_nosing <- mr.mash(Xtrain, Ytrain, S0_nosing, w0=w0_nosing, update_w0=TRUE, update_w0_method="EM", tol=1e-2,
convergence_criterion="ELBO", compute_ELBO=TRUE, standardize=standardize,
verbose=FALSE, update_V=TRUE, update_V_method="diagonal", e=1e-8,
w0_threshold=w0_threshold, nthreads=nthreads, mu1_init=out_coeff$B)
#####Coefficients plots######
layout(matrix(c(1, 1, 2, 2,
1, 1, 2, 2,
0, 3, 3, 0,
0, 3, 3, 0), 4, 4, byrow = TRUE))
###Plot estimated vs true coeffcients
##mr.mash
plot(out$B, fit_mrmash_nosing$mu1, main="mr.mash", xlab="True coefficients", ylab="Estimated coefficients",
cex=2, cex.lab=2, cex.main=2, cex.axis=2)
abline(0, 1)
##g-lasso
plot(out$B, out_coeff$B, main="g-lasso", xlab="True coefficients", ylab="Estimated coefficients",
cex=2, cex.lab=2, cex.main=2, cex.axis=2)
abline(0, 1)
###Plot mr.mash vs g-lasso estimated coeffcients
colorz <- matrix("black", nrow=p, ncol=r)
zeros <- apply(out$B, 2, function(x) x==0)
for(i in 1:ncol(colorz)){
colorz[zeros[, i], i] <- "red"
}
plot(fit_mrmash_nosing$mu1, out_coeff$B, main="mr.mash vs g-lasso",
xlab="mr.mash coefficients", ylab="g-lasso coefficients",
col=colorz, cex=2, cex.lab=2, cex.main=2, cex.axis=2)
abline(0, 1)
legend("topleft",
legend = c("Non-zero", "Zero"),
col = c("black", "red"),
pch = c(1, 1),
horiz = FALSE,
cex=2)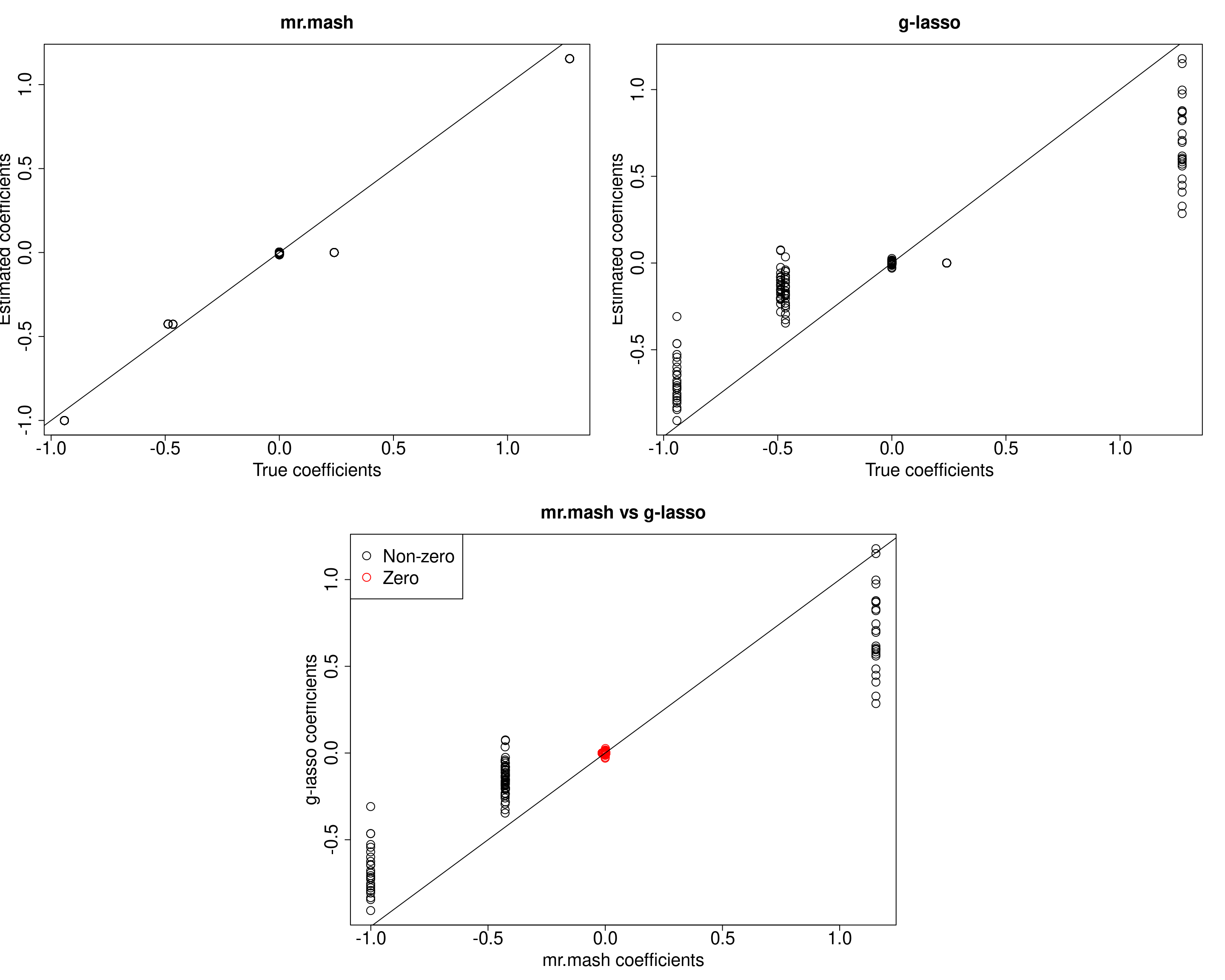
When comparing the ELBO of the two models, we can see that the ELBO for the model with the singletons is a little larger.
###ELBO with singletons
cat("mr.mash ELBO with singletons:\n")mr.mash ELBO with singletons:fit_mrmash$ELBO[1] -10083.11###ELBO with singletons
cat("mr.mash ELBO without singletons:\n")mr.mash ELBO without singletons:fit_mrmash_nosing$ELBO[1] -10085.82However, at least in this example, the variable/condition combinations displaying this problem are not so many. Interestingly, a few conditions (10, 13, 15, 22) are the only ones creating this issue.
badd <- which(abs(fit_mrmash$mu1 - out$B) > 0.05, arr.ind = TRUE)
unique_vars <- unique(badd[,1])
unique_vars <- unique_vars[-unique(which(out$B[unique_vars, ] !=0, arr.ind = TRUE)[, 1])]
unique_vars_bad_y <- badd[which(badd[, 1] %in% unique_vars), ]
unique_vars_bad_y row col
X141 141 10
X183 183 10
X274 274 10
X72 72 13
X342 342 13
X357 357 13
X142 142 15
X147 147 15
X332 332 15
X347 347 15
X187 187 22
X192 192 22
X194 194 22
X232 232 22
X327 327 22plot(out$B[unique_vars_bad_y], fit_mrmash$mu1[unique_vars_bad_y], xlab="True coefficents", ylab="Estimated coefficients", main="mr.mash vs truth\nmost bad variables/conditions")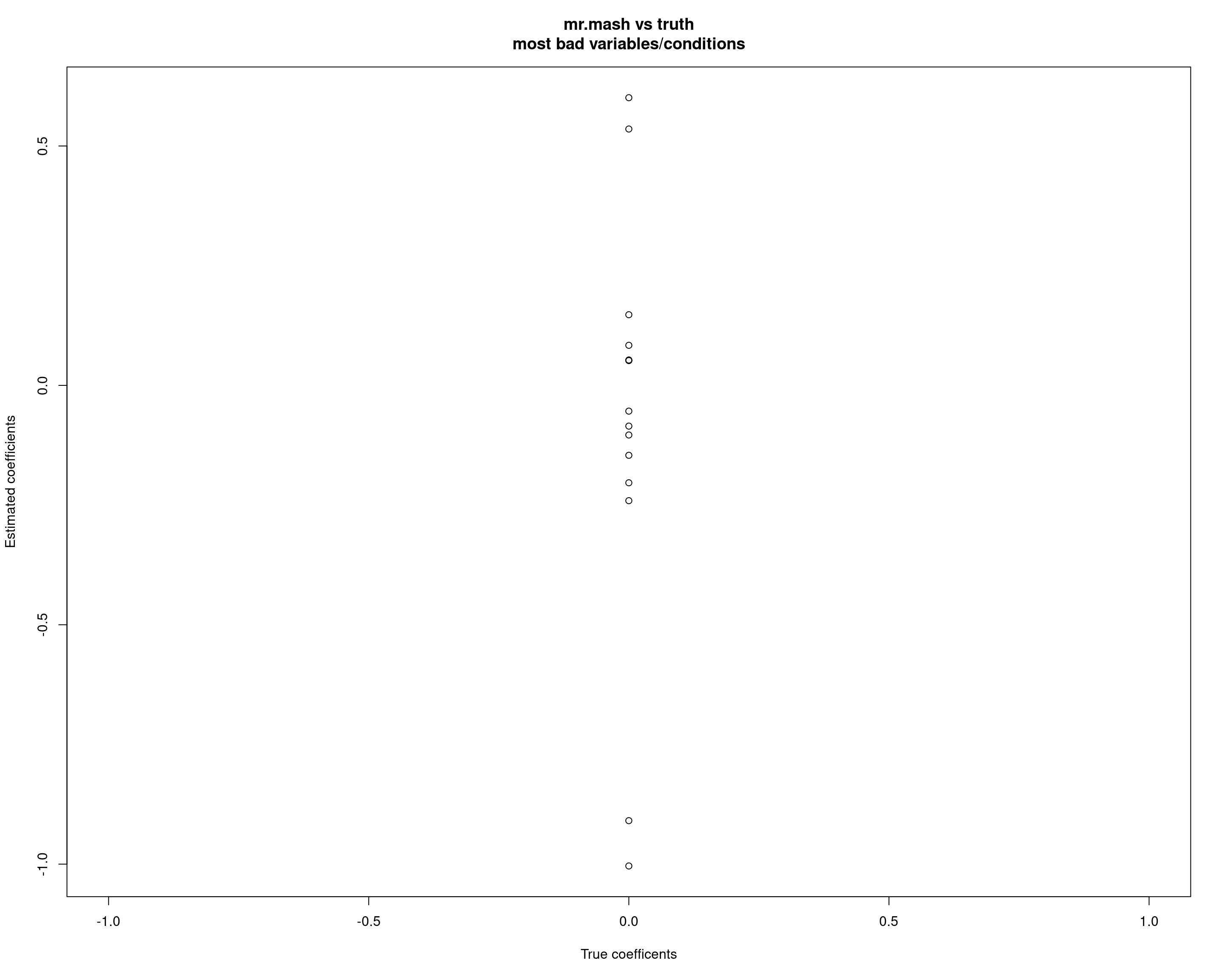
Let’s what happens if we run mash on the summary statistics. Since predictors are independent, so we should roughly get the same results as mr.mash.
dat_mash <- mash_set_data(univ_sumstats$Bhat, univ_sumstats$Shat)
mash_obj <- mash(dat_mash, S0_can) - Computing 400 x 451 likelihood matrix.
- Likelihood calculations took 1.52 seconds.
- Fitting model with 451 mixture components.
- Model fitting took 3.01 seconds.
- Computing posterior matrices.
- Computation allocated took 0.10 seconds.mash_pm <- get_pm(mash_obj)
cat("mash estimated coefficients:\n")mash estimated coefficients:mash_pm[bad_est[, 1], ] Y1 Y2 Y3 Y4 Y5
X274 -0.023650792 -0.023650792 -0.023650792 -0.023650792 -0.023650792
X347 0.057239661 0.057239661 0.057239661 0.057239661 0.057239661
X183 0.213057913 0.213057913 0.213057913 0.213057913 0.213057913
X72 0.006652372 0.006652372 0.006652372 0.006652372 0.006652372
Y6 Y7 Y8 Y9 Y10
X274 -0.023650792 -0.023650792 -0.023650792 -0.023650792 -0.023650792
X347 0.057239661 0.057239661 0.057239661 0.057239661 0.057239661
X183 0.213057913 0.213057913 0.213057913 0.213057913 0.213057913
X72 0.006652372 0.006652372 0.006652372 0.006652372 0.006652372
Y11 Y12 Y13 Y14 Y15
X274 -0.023650792 -0.023650792 -0.02250090 -0.023650792 -0.023650792
X347 0.057239661 0.057239661 0.05573754 0.057239661 0.057239661
X183 0.213057913 0.213057913 0.21306298 0.213057913 0.213057913
X72 0.006652372 0.006652372 0.45452574 0.006652372 0.006652372
Y16 Y17 Y18 Y19 Y20
X274 -0.023650792 -0.023650792 -0.023650792 -0.023650792 -0.023650792
X347 0.057239661 0.057239661 0.057239661 0.057239661 0.057239661
X183 0.213057913 0.213057913 0.213057913 0.213057913 0.213057913
X72 0.006652372 0.006652372 0.006652372 0.006652372 0.006652372
Y21 Y22 Y23 Y24 Y25
X274 -0.023642208 -0.023650792 -0.023650792 -0.023650792 -0.023650792
X347 0.057577792 0.057239661 0.057239661 0.057239661 0.057239661
X183 0.213058005 0.213057913 0.213057913 0.213057913 0.213057913
X72 0.006514678 0.006652372 0.006652372 0.006652372 0.006652372mash seems to have a different problem – it estimates that the effects of variables X274, X347, and X183 to be equal across conditions. However, similar behavior to mr.mash is shown for variable X72. Checking the prior weights, we can see that mash wrongly puts a lot of weight on equal effects components.
###Estimated prior weights by mr.mash for the bad variables
cat("mr.mash estimated prior weights:\n")mr.mash estimated prior weights:head(sort(get_estimated_pi(mash_obj, "all"), decreasing=TRUE), 20) shared1.6 null shared1.10 singleton13.10 shared1.11
0.7600049178 0.2209967909 0.0074711314 0.0061764252 0.0046510891
singleton21.11 singleton1.1 singleton2.1 singleton3.1 singleton4.1
0.0006996457 0.0000000000 0.0000000000 0.0000000000 0.0000000000
singleton5.1 singleton6.1 singleton7.1 singleton8.1 singleton9.1
0.0000000000 0.0000000000 0.0000000000 0.0000000000 0.0000000000
singleton10.1 singleton11.1 singleton12.1 singleton13.1 singleton14.1
0.0000000000 0.0000000000 0.0000000000 0.0000000000 0.0000000000 Let’s now look at the estimated effects from mash, and compare them to those from mr.mash.
#####Coefficients plots######
layout(matrix(c(1, 1, 2, 2,
1, 1, 2, 2), 2, 4, byrow = TRUE))
##mash
plot(out$B, mash_pm, main="mash", xlab="True coefficients", ylab="Estimated coefficients",
cex=2, cex.lab=2, cex.main=2, cex.axis=2)
abline(0, 1)
##mr.mash vs mash
plot(fit_mrmash$mu1, mash_pm, main="mr.mash vs mash -- all variables",
xlab="mr.mash coefficients", ylab="mash coefficients",
col=colorz, cex=2, cex.lab=2, cex.main=2, cex.axis=2)
abline(0, 1)
legend("topleft",
legend = c("Non-zero", "Zero"),
col = c("black", "red"),
pch = c(1, 1),
horiz = FALSE,
cex=2)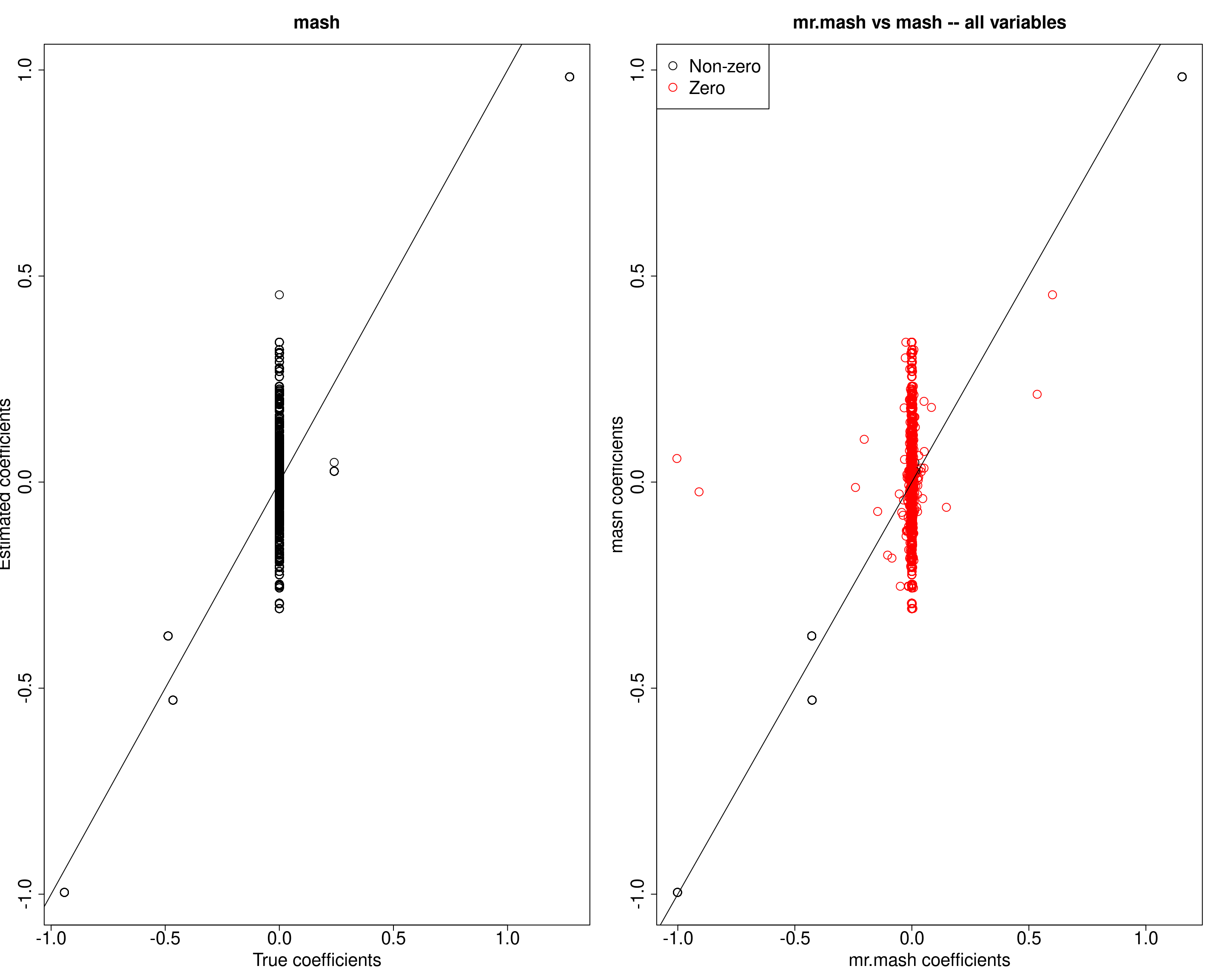
In general, mash seems to have more difficulty to shrink true 0 coefficients.
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] mashr_0.2.41 ashr_2.2-51 glmnet_3.0
[4] Matrix_1.2-18 mr.mash.alpha_0.2-6
loaded via a namespace (and not attached):
[1] foreach_1.4.4 RcppParallel_4.4.3 rmeta_3.0
[4] assertthat_0.2.1 horseshoe_0.2.0 mixsqp_0.3-43
[7] yaml_2.2.1 ebnm_0.1-35 pillar_1.4.2
[10] backports_1.1.4 lattice_0.20-38 quantreg_5.85
[13] glue_1.4.2 digest_0.6.20 promises_1.0.1
[16] colorspace_1.4-1 htmltools_0.3.6 httpuv_1.5.1
[19] plyr_1.8.6 conquer_1.0.2 pkgconfig_2.0.3
[22] invgamma_1.1 SparseM_1.81 purrr_0.3.4
[25] mvtnorm_1.1-1 scales_1.1.0 whisker_0.3-2
[28] later_0.8.0 MatrixModels_0.5-0 git2r_0.26.1
[31] tibble_2.1.3 ggplot2_3.2.1 lazyeval_0.2.2
[34] magrittr_1.5 crayon_1.4.1 mcmc_0.9-7
[37] evaluate_0.14 fs_1.3.1 MASS_7.3-51.4
[40] truncnorm_1.0-8 tools_3.6.1 doMC_1.3.7
[43] softImpute_1.4 lifecycle_1.0.0 matrixStats_0.58.0
[46] stringr_1.4.0 MCMCpack_1.5-0 GIGrvg_0.5
[49] trust_0.1-8 munsell_0.5.0 flashier_0.2.7
[52] MBSP_1.0 irlba_2.3.3 compiler_3.6.1
[55] rlang_0.4.10 grid_3.6.1 iterators_1.0.10
[58] rmarkdown_1.13 gtable_0.3.0 codetools_0.2-16
[61] abind_1.4-5 reshape2_1.4.3 flashr_0.6-7
[64] R6_2.5.0 knitr_1.23 dplyr_0.8.3
[67] workflowr_1.6.2 rprojroot_1.3-2 shape_1.4.4
[70] stringi_1.4.3 parallel_3.6.1 SQUAREM_2021.1
[73] Rcpp_1.0.6 tidyselect_1.1.0 xfun_0.8
[76] coda_0.19-4