ABL_SM_CRISPR_Cut_Analyses
Haider Inam
2023-03-22
Last updated: 2023-08-30
Checks: 6 1
Knit directory: duplex_sequencing_screen/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of
the R Markdown file created these results, you’ll want to first commit
it to the Git repo. If you’re still working on the analysis, you can
ignore this warning. When you’re finished, you can run
wflow_publish to commit the R Markdown file and build the
HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200402) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version f9eea37. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/Consensus_Data/.Rhistory
Ignored: data/Consensus_Data/Novogene_lane11/sample1/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample1/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample1/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample1/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample1/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample1/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample2/archive/sscs_aligned_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample2/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample2/duplex/variant_caller_outputs/archive_ensembl/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample2/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample2/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample2/sscs/variant_caller_outputs/archive_ensembl/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample2/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample3/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample3/duplex/variant_caller_outputs/archive_ensembl/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample3/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample3/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample3/sscs/variant_caller_outputs/archive ensembl/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample3/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample4/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample4/duplex/variant_caller_outputs/archive ensembl/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample4/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample4/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample4/sscs/variant_caller_outputs/archive ensembl/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample4/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample5/variant_caller_outputs/sscs_L858R_aligned_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample5/variant_caller_outputs/sscs_L858R_aligned_filtered_sample5.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample6/archive/sscs_aligned_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample6/sscs_L858R_aligned_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample6/variant_caller_outputs/variants_ann_sample6.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample7/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample7/sscs/variant_caller_outputs/archive ensembl/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample7/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample1/low_sscscounts/sscs_aligned_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample1/low_sscscounts/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample1/sscs_aligned_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample1/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample3/sscs_combined_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample3/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample5/sscs_combined_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample5/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample7/sscs_combined_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample7/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample9/sscs_combined_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample9/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample1/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample1/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample1/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample1/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample10/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample10/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample10/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample10/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample10/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample10/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample11/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample11/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample11/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample11/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample11/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample11/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample12/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample12/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample12/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample12/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample12/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample12/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample2/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample2/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample3/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample3/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample4/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample4/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample5/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample5/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample6/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample6/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample7/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample7/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample7/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample7/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample7/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample7/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample8/
Ignored: data/Consensus_Data/Novogene_lane13/sample9/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample9/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample9/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample9/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample9/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample9/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample10_combined/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample10_combined/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample10_combined/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample10_combined/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample10_combined/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample10_combined/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample11/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample11/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample11/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample11/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample11/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample11/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample12/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample12/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample12/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample12/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample12/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample12/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample13/
Ignored: data/Consensus_Data/Novogene_lane14/sample14_combined/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample14_combined/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample14_combined/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample14_combined/sscs.filt_1.fa.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample14_combined/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample14_combined/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample14_combined/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample14b/
Ignored: data/Consensus_Data/Novogene_lane14/sample15/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample15/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample15/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample15/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample15/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample15/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample16/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample16/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample16/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample16/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample16/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample16/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample17/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample17/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample17/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample17/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample17/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample17/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample18/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample18/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample18/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample18/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample18/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample18/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample1_combined/
Ignored: data/Consensus_Data/Novogene_lane14/sample2_combined/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample2_combined/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample3/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample3/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample4/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample4/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample5/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample5/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample6/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample6/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample7/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample7/variant_caller_outputs/duplex/
Ignored: data/Consensus_Data/Novogene_lane14/sample7/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample8/
Ignored: data/Consensus_Data/Novogene_lane14/sample9/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample9/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample9/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample9/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample9/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample9/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane2/
Ignored: data/Consensus_Data/Novogene_lane3/
Ignored: data/Consensus_Data/Novogene_lane4/
Ignored: data/Consensus_Data/Novogene_lane5/
Ignored: data/Consensus_Data/Novogene_lane6/
Ignored: data/Consensus_Data/Novogene_lane7/
Ignored: data/Consensus_Data/Ranomics_Pooled/
Ignored: data/Consensus_Data/archive/
Ignored: data/Consensus_Data/novogene_lane15/sample_1/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_1/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_1/firstrun(lowsequencing)/
Ignored: data/Consensus_Data/novogene_lane15/sample_1/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_2/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_2/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_2/firstrun(lowsequencing)/sscs/
Ignored: data/Consensus_Data/novogene_lane15/sample_2/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_2/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/firstrun(lowsequencing)/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/firstrun(lowsequencing)/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/firstrun(lowsequencing)/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/firstrun(lowsequencing)/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/ngs/Sample3_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/ngs/sample3a(firsthalf)/Sample3_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/ngs/sample3a(firsthalf)/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/ngs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_4/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_4/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_4/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_4/firstrun(lowsequencing)/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_4/firstrun(lowsequencing)/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_4/firstrun(lowsequencing)/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_4/firstrun(lowsequencing)/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_4/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_4/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_4/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_5/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_5/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_5/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_5/firstrun(lowsequencing)/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_5/firstrun(lowsequencing)/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_5/firstrun(lowsequencing)/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_5/firstrun(lowsequencing)/sscs/variant_caller_outputs/.empty/
Ignored: data/Consensus_Data/novogene_lane15/sample_5/firstrun(lowsequencing)/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_5/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_5/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_5/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_6/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_6/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_6/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_6/firstrun(lowsequencing)/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_6/firstrun(lowsequencing)/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_6/firstrun(lowsequencing)/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_6/firstrun(lowsequencing)/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_6/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_6/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_6/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_7/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_7/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_7/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_7/firstrun(lowsequencing)/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_7/firstrun(lowsequencing)/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_7/firstrun(lowsequencing)/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_7/firstrun(lowsequencing)/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_7/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_7/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_7/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample10/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample10/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample10/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample10/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample11/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample11/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample11/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample11/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample12/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample12/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample12/sscs/
Ignored: data/Consensus_Data/novogene_lane16a/Sample13/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample13/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample13/sscs/
Ignored: data/Consensus_Data/novogene_lane16a/Sample14/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample14/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample14/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample14/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample1_combined/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample1_combined/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample1_combined/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample1_combined/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample2/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample2/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample2/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample3/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample3/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample3/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample3/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample4/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample4/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample4/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample4/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample5/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample5/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample5/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample5/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample6/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample6/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample6/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample6/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample7/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample7/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample7/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample7/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample8/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample8/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample8/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample8/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample9/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample9/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample9/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample9/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/duplex/
Ignored: data/Consensus_Data/novogene_lane16b/Sample10/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample10/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample10/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample10/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample11/
Ignored: data/Consensus_Data/novogene_lane16b/Sample15/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample15/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample15/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample15/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample1_combined/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample1_combined/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample1_combined/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample1_combined/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample2/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample2/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample2/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample2/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample3/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample3/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample3/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample3/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample4/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample4/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample4/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample4/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample5/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample5/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample5/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample5/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample6/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample6/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample6/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample6/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample7_combined/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample7_combined/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample7_combined/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample7_combined/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample8_combined/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample8_combined/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample8_combined/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample8_combined/sscs/variant_caller_outputs/archive/
Ignored: data/Consensus_Data/novogene_lane16b/Sample8_combined/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample9/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample9/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample9/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample9/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample10/duplex/
Ignored: data/Consensus_Data/novogene_lane17/sample10/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample10/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample11/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample11/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample11/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample11/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample1_combined/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample1_combined/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample1_combined/low_depth/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample1_combined/low_depth/duplex/low_depth/
Ignored: data/Consensus_Data/novogene_lane17/sample1_combined/low_depth/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample1_combined/low_depth/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample1_combined/low_depth/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample1_combined/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample1_combined/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample2/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample2/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample2/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample2/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample3/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample3/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample3/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample3/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample4/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample4/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample4/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample4/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample5/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample5/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample5/low_seq_depth/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample5/low_seq_depth/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample5/low_seq_depth/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample5/low_seq_depth/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample5/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample5/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample6/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample6/low_seq_depths/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample6/low_seq_depths/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample6/low_seq_depths/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample6/low_seq_depths/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample6/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample6/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample7/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample7/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample7/low_seq_depths/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample7/low_seq_depths/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample7/low_seq_depths/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample7/low_seq_depths/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample7/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample7/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample8/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample8/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample8/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample9/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample9/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample9/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample9/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17b/Sample1 copy 2/
Ignored: data/Consensus_Data/novogene_lane17b/Sample1 copy 3/
Ignored: data/Consensus_Data/novogene_lane17b/Sample1/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17b/Sample1/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17b/Sample1/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17b/Sample1/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17b/Sample2/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17b/Sample2/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17b/Sample2/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample1/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample1/l298l/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample1/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample1/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample1/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample1/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample1/nol298l/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample1/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample1/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample1/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample1/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample1/sscs/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample10/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample10/l298l/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample10/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample10/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample10/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample10/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample10/ngs/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample10/ngs/Sample_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample10/ngs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample10/nol298l/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample10/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample10/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample10/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample10/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample10/sscs/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample11/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample11/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample11/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample11/l298l/sscs/variant_caller_outputs/slow_variantcaller_outputs_forcomparison/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample11/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample11/ngs/Sample_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample11/ngs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample11/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample11/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample11/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample11/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample12/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample12/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample12/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample12/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample12/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample12/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample12/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample12/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample13/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample13/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample13/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample13/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample13/ngs/Sample_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample13/ngs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample13/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample13/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample13/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample13/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample14/duplex/
Ignored: data/Consensus_Data/novogene_lane18/sample14/l298l/duplex/
Ignored: data/Consensus_Data/novogene_lane18/sample14/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample14/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample14/ngs/Sample_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample14/ngs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample14/nol298l/duplex/
Ignored: data/Consensus_Data/novogene_lane18/sample14/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample14/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample15/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample15/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample15/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample15/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample15/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample15/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample15/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample15/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample16/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample16/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample16/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample16/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample16/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample16/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample16/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample16/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample17/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample17/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample17/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample17/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample17/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample17/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample17/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample17/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample18/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample18/l298l/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample18/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample18/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample18/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample18/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample18/nol298l/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample18/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample18/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample18/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample18/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample2/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample2/l298l/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample2/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample2/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample2/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample2/l298l/sscs/variant_caller_outputs/fastercaller/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample2/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample2/nol298l/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample2/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample2/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample2/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample2/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample2/sscs/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample3/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample3/l298l/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample3/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample3/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample3/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample3/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample3/nol298l/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample3/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample3/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample3/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample3/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample3/sscs/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample4/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample4/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample4/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample4/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample4/ngs/Sample_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample4/ngs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample4/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample4/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample4/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample4/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample5/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample5/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample5/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample5/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample5/ngs/Sample_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample5/ngs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample5/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample5/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample5/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample5/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample6/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample6/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample6/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample6/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample6/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample6/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample6/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample6/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample7/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample7/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample7/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample7/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample7/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample7/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample7/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample7/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample8/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample8/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample8/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample8/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample8/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample8/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample8/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample8/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample9/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample9/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample9/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample9/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample9/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample9/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample9/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample9/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample3/duplex/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample3/l298l/duplex/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample3/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample3/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample3/nol298l/duplex/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample3/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample3/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample3/sscs/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample5/duplex/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample5/l298l/duplex/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample5/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample5/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample5/nol298l/duplex/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample5/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample5/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample5/sscs/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample6/duplex/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample6/l298l/duplex/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample6/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample6/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample6/nol298l/duplex/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample6/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample6/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample6/sscs/
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample1/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample1/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample1/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample1/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample10/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample10/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample10/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample10/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample11/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample11/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample11/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample11/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample12/duplex/
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample12/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample12/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample2/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample2/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample2/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample2/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample3/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample3/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample3/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample3/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample4/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample4/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample4/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample4/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample5/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample5/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample5/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample5/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample6/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample6/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample6/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample6/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample7/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample7/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample7/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample7/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample8/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample8/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample8/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample8/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample9/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample9/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample9/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample9/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample1/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample1/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample1/sscs/
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample10/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample10/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample10/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample10/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample11/duplex/
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample11/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample11/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample12/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample12/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample12/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample12/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample13/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample13/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample13/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample13/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample14/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample14/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample14/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample14/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample15/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample15/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample15/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample15/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample16/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample16/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample16/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample16/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample2/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample2/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample2/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample2/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample3/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample3/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample3/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample3/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample4/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample4/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample4/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample4/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample5/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample5/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample5/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample5/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample6/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample6/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample6/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample6/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample7/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample7/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample7/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample7/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample8/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample8/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample8/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample8/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample9/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample9/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample9/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample9/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample10/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample10/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample10/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample10/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample11/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample11/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample11/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample11/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample2/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample2/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample2/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample2/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample3/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample3/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample3/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample3/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample4/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample4/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample4/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample4/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample5/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample5/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample5/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample5/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample6/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample6/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample6/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample6/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample7/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample7/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample7/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample7/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample8/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample8/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample8/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample8/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample9/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample9/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample9/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample9/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19b_Sample9/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19b_Sample9/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19b_Sample9/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19b_Sample9/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/.DS_Store
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample1/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample1/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample1/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample1/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample2/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample2/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample2/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample2/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample3/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample3/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample3/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample3/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample4/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample4/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample4/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample4/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample5/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample5/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample5/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample5/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample6/duplex/
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample6/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample6/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample7/
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample8/
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample13/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample13/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample13/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample13/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample14/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample14/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample14/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample14/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample15/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample15/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample15/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample15/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample16/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample16/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample16/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample16/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample17/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample17/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample17/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample17/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample18/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample18/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample18/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample18/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample19/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample19/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample19/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample19/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample20/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample20/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample20/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample20/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample21/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample21/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample21/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample21/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample22/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample22/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample22/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample22/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample23/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample23/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample23/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample23/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample24/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample24/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample24/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample24/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample25/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample25/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample25/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample25/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample26/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample26/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample26/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample26/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample27/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample27/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample27/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample27/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/sscs_dcs_comparisons/
Ignored: output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/NA/
Ignored: output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/baf3_IL3_low_rep1vsrep2/
Ignored: output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/baf3_Imat_Lowvsk562_Imat_Medium/
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/il3_indep_1.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/il3_indep_1.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/il3_indep_2.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/il3_indep_2.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/il3_indep_3.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/il3_indep_3.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/sorted_1.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/sorted_1.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/sorted_2.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/sorted_2.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/sorted_3.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/sorted_3.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/il3_indep_1.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/il3_indep_1.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/il3_indep_2.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/il3_indep_2.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/il3_indep_3.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/il3_indep_3.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/il3_indep_4.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/il3_indep_4.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/il3_indep_5.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/il3_indep_5.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_1.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_1.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_2.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_2.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_3.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_3.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_4.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_4.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_5.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_5.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_6.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_6.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_High_D2.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_High_D2.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_High_D4.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_High_D4.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_Low_D2.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_Low_D2.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_Low_D4.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_Low_D4.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_Medium_D2.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_Medium_D2.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_Medium_D4.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_Medium_D4.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_High_D2.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_High_D2.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_High_D4.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_High_D4.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_Low_D2.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_Low_D2.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_Low_D4.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_Low_D4.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_Medium_D2.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_Medium_D2.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_Medium_D4.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_Medium_D4.1.consensus.variant-calls.vcf.gz
Untracked files:
Untracked: analysis/ABL_sheared_libraries.Rmd
Untracked: analysis/figure/
Untracked: output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/baf3_IL3_rep1vsrep2_ft/screen_comparison_baf3_IL3_low_rep1vsrep2ft.csv
Unstaged changes:
Modified: .DS_Store
Modified: analysis/.DS_Store
Modified: analysis/ABL_SM_CRISPR_Cut_Analyses.Rmd
Modified: analysis/ABL_cosmic_analysis.Rmd
Modified: analysis/ABL_kinasefunction_data_parser.Rmd
Deleted: analysis/ABL_unevenness_analysis.Rmd
Modified: analysis/BCRABL_FunctionalKinaseAnalysis.Rmd
Modified: analysis/index.Rmd
Modified: code/compare_screens.R
Modified: data/.DS_Store
Modified: data/Consensus_Data/.DS_Store
Modified: data/Consensus_Data/novogene_lane18/tlane18a_sample6/.DS_Store
Modified: output/.DS_Store
Modified: output/ABLEnrichmentScreens/.DS_Store
Modified: output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/.DS_Store
Modified: output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/.DS_Store
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown
(analysis/ABL_SM_CRISPR_Cut_Analyses.Rmd) and HTML
(docs/ABL_SM_CRISPR_Cut_Analyses.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | f9eea37 | haiderinam | 2023-08-30 | 2023 Updates |
| html | f9eea37 | haiderinam | 2023-08-30 | 2023 Updates |
| Rmd | f04026b | haiderinam | 2023-04-18 | added further ROC analyses of BE screens |
| Rmd | 60b906b | haiderinam | 2023-04-10 | Added ROC analyses on BE-SM data |
| html | 60b906b | haiderinam | 2023-04-10 | Added ROC analyses on BE-SM data |
| Rmd | 88dabff | haiderinam | 2023-04-01 | Added lane 18 data of ABL Region 1 |
| Rmd | 6b51aa2 | haiderinam | 2023-03-25 | Added Lane 18 Data with ABL Region 1 SM Screen |
#Cleanup code for plotting
source("code/plotting/cleanup.R")
source("code/compare_screens.R")
source("code/plotting/heatmap_plotting_function.R")source("code/variantcaller/add_l298l.R")
for(i in c(1:18)){
sample=paste("sample",i,sep = "")
# sample="sample1"
input_df_nol298l=read.csv(paste("data/Consensus_Data/novogene_lane18/",sample,"/nol298l/duplex/variant_caller_outputs/variants_unique_ann.csv",sep=""))
input_df_l298l=read.csv(paste("data/Consensus_Data/novogene_lane18/",sample,"/l298l/duplex/variant_caller_outputs/variants_unique_ann.csv",sep=""))
output_df=add_l298l(input_df_nol298l,input_df_l298l)
write.csv(output_df,
paste("data/Consensus_Data/novogene_lane18/",sample,"/duplex/variant_caller_outputs/variants_unique_ann.csv",sep = ""))
}
input_df_nol298l=read.csv("data/Consensus_Data/novogene_lane18/sample9/nol298l/duplex/variant_caller_outputs/variants_unique_ann.csv")
input_df_l298l=read.csv("data/Consensus_Data/novogene_lane18/sample9/l298l/duplex/variant_caller_outputs/variants_unique_ann.csv")
output_df=add_l298l(input_df_nol298l,input_df_l298l)
write.csv(output_df,"data/Consensus_Data/novogene_lane18/sample9/duplex/variant_caller_outputs/variants_unique_ann.csv") # rm(list=ls())
comparisons=read.csv("data/Consensus_Data/novogene_lane18/TwistRegion1Screen_Comparisons_Todo.csv")
comparisons=comparisons%>%filter(Completed%in%"FALSE")
for(i in 1:nrow(comparisons)){
# i=10
dirname=comparisons$dirname[i]
pathname=paste("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/",dirname,sep = "")
# Create directory if it doesn't already exist
if (!file.exists(pathname)){
dir.create(pathname)
}
before_screen1_identifier=unlist(strsplit(comparisons$before_screen1_identifier[i],","))
after_screen1_identifier=unlist(strsplit(comparisons$after_screen1_identifier[i],","))
before_screen2_identifier=unlist(strsplit(comparisons$before_screen2_identifier[i],","))
after_screen2_identifier=unlist(strsplit(comparisons$after_screen2_identifier[i],","))
# length(after_screen1_identifier)
# screen_compare_means=compare_screens(comparisons$before_screen1_identifier[i],
# comparisons$after_screen1_identifier[i],
# comparisons$before_screen2_identifier[i],
# comparisons$after_screen2_identifier[i])
screen_compare_means=compare_screens(before_screen1_identifier,
after_screen1_identifier,
before_screen2_identifier,
after_screen2_identifier)
screen_compare_means_forexport=apply(screen_compare_means,2,as.character)
# write.csv(screen_compare_means_forexport,file = paste("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/",dirname,"/screen_comparison_",dirname,".csv",sep=""))
# Plot 1. What does the heatmap look like from the average of the net growth rate?
heatmap_plotting_function(screen_compare_means,242,321,fill_variable = "netgr_obs_mean",fill_name = "Net growth rate")
# ggsave(paste("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/",dirname,"/plot1_heatmap.pdf",sep=""),width=10,height=6,units="in",useDingbats=F)
# screen_compare_means2=screen_compare_means%>%filter(alt_codon%in%twist$Codon)
# Plot 2a: What do the correlations look like for net growth rate (show mutants in text)?
ggplot(screen_compare_means,aes(x=netgr_obs_screen1,y=netgr_obs_screen2,color=resmuts,label=species))+geom_text(size=2.5)+geom_abline()+cleanup+stat_cor(method = "pearson")+labs(color="Known\nResistant\nMutant")+scale_x_continuous("Net growth rate screen 1")+scale_y_continuous("Net growth rate screen 2")
# ggsave(paste("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/",dirname,"/plot2a_Netgrowthrate_correlations_text.pdf",sep=""),width=6,height=4,units="in",useDingbats=F)
# Plot 2b: What do the correlations look like for enrichment scores (show mutants in points)?
ggplot(screen_compare_means,aes(x=netgr_obs_screen1,y=netgr_obs_screen2,label=species))+geom_point(color="black",shape=21,size=2,aes(fill=resmuts))+geom_abline()+cleanup+stat_cor(method = "pearson")+labs(fill="Known\nResistant\nMutant")+scale_x_continuous("Net growth rate screen 1")+scale_y_continuous("Net growth rate screen 2")
# ggsave(paste("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/",dirname,"/plot2b_Netgrowthrate_correlations_points.pdf",sep=""),width=6,height=4,units="in",useDingbats=F)
# Plot 2c: What do the correlations look like for enrichment scores (show mutants in text)?
ggplot(screen_compare_means,aes(x=score_screen1,y=score_screen2,color=resmuts,label=species))+geom_text(size=2.5)+geom_abline()+cleanup+ stat_cor(method = "pearson")+labs(color="Known\nResistant\nMutant")+scale_x_continuous("Enrichment score screen 1")+scale_y_continuous("Enrichment score screen 2")
# ggsave(paste("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/",dirname,"/plot2c_Enrichmentscores_correlations_text.pdf",sep=""),width=6,height=4,units="in",useDingbats=F)
# Plot 3a: Plots: what are the overall net growth rate distributions?
ggplot(screen_compare_means,aes(x=netgr_obs_mean,fill=resmuts))+geom_density(alpha=0.7)+cleanup+labs(fill="Known\nResistant\nMutant")+scale_x_continuous("Mean net growth rate of screens")
# ggsave(paste("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/",dirname,"/plot3a_Netgrowthrate_distributions_resmuts.pdf",sep=""),width=6,height=4,units="in",useDingbats=F)
# Plot 3b: Plots: what are the net growth rate distributions?
library(reshape2)
screen_compare_melt=melt(screen_compare_means%>%dplyr::select(species,netgr_obs_screen1,netgr_obs_screen2),id.vars = "species",measure.vars =c("netgr_obs_screen1","netgr_obs_screen2"),variable.name = "Condition",value.name = "netgr_obs")
ggplot(screen_compare_melt,aes(x=netgr_obs,fill=Condition))+
geom_density(alpha=0.7)+
cleanup+
scale_x_continuous("Net growth rate observed")+
scale_fill_discrete(labels=c("Screen 1","Screen 2"))
# ggsave(paste("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/",dirname,"/plot3b_Netgrowthrate_distributions.pdf",sep=""),width=6,height=4,units="in",useDingbats=F)
}
# dirname="K562_Medium_rep1vs2"Doing some quick analyses on buried hydrophobic vs exposed residues
Quick analysis to do on the secondary structure: The buried hydrophobic core is probably going to be more susceptible to polar/hydrophilic substitutions, and not as much to hydrophobic substitutions… do you see that in the il3 independence data? Answer: yes, I see a slight singal but it’s not huge
dssp=read.csv("data/DSSP_SolventAccessibility_ABL/2hyy_dspp.csv",header = T)
dssp=dssp%>%mutate(RESIDUE=as.numeric(RESIDUE),
ACC=as.numeric(ACC),AA=gsub("<ca>","",AA),
SS=case_when(STRUCTURE%in%c("E","B")~"b-sheet",
STRUCTURE%in%c("H","G","I")~"a-helix",
STRUCTURE%in%"T"~"turn",
T~"undefined"))%>%dplyr::select(-"STRUCTURE")
dssp=dssp%>%rename(protein_start=RESIDUE)
il3=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/baf3_IL3_rep1vsrep2_ft/screen_comparison_baf3_IL3_low_rep1vsrep2_ft.csv")
il3=il3%>%dplyr::select(species,protein_start,netgr_obs_mean)
il3=il3%>%group_by(protein_start)%>%summarize(netgr_obs_mean=mean(netgr_obs_mean))
il3_dssp=merge(il3,dssp,by="protein_start")
il3_dssp=il3_dssp%>%mutate(exposed=case_when(ACC>=40~"Exposed",
T~"Buried"))
# il3_dssp=il3_dssp%>%filter(exposed%in%"Exposed")
ggplot(il3_dssp,aes(x=ACC))+geom_histogram()`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.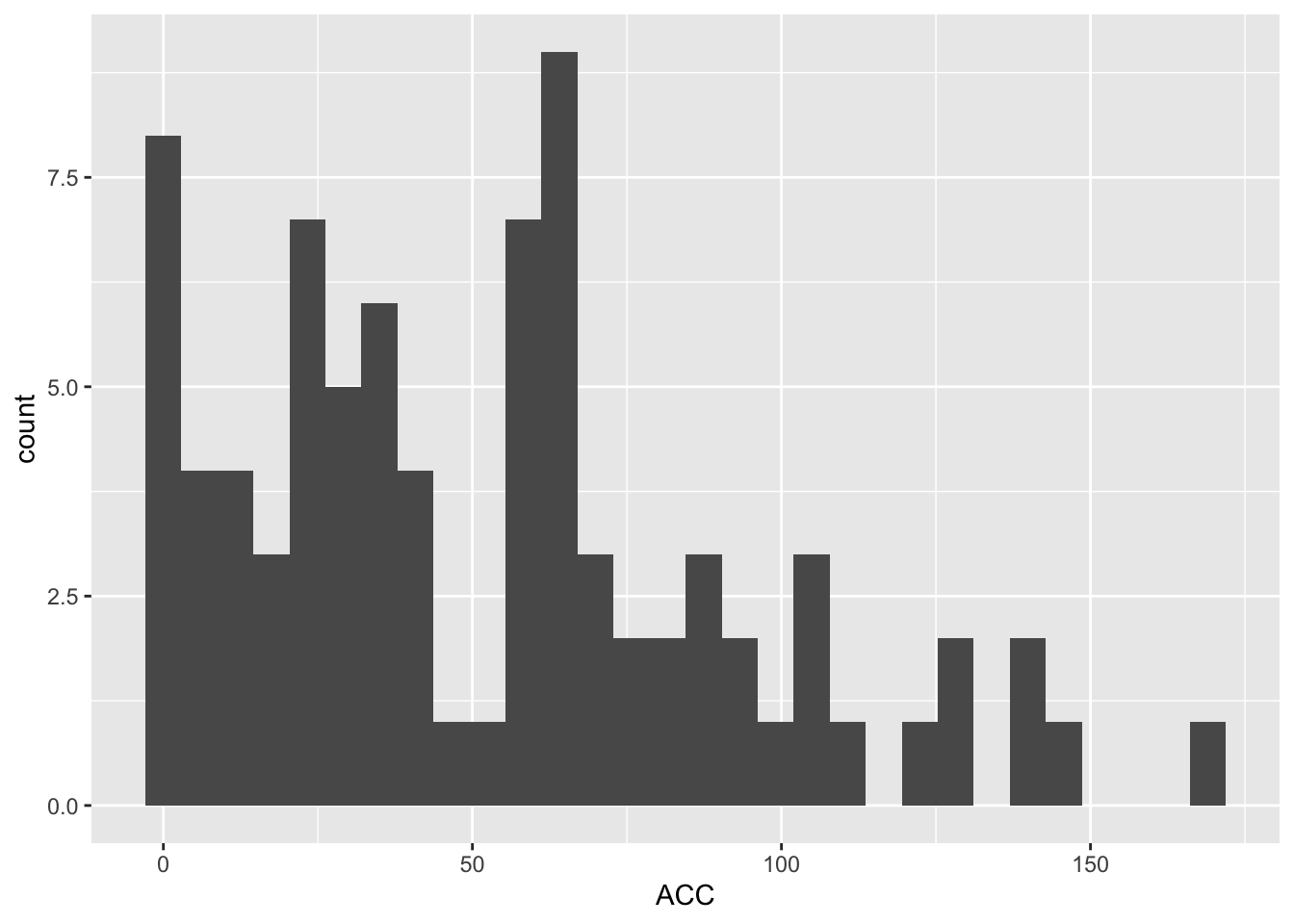
ggplot(il3_dssp,aes(x=protein_start,y=netgr_obs_mean,color=exposed))+
geom_point()+
facet_wrap(~exposed,nrow=2)+
scale_y_continuous("Mean net growth rate at residue")+
scale_x_continuous("Residue on the ABL Kinase")+
labs(color="Solvent \nAccessibility")+
cleanup+theme(legend.position = c(.9,.85))+
theme(
strip.background = element_blank(),
strip.text.x = element_blank()
)+theme(legend.background = element_rect(
size=0.5, linetype="solid",
colour ="black"))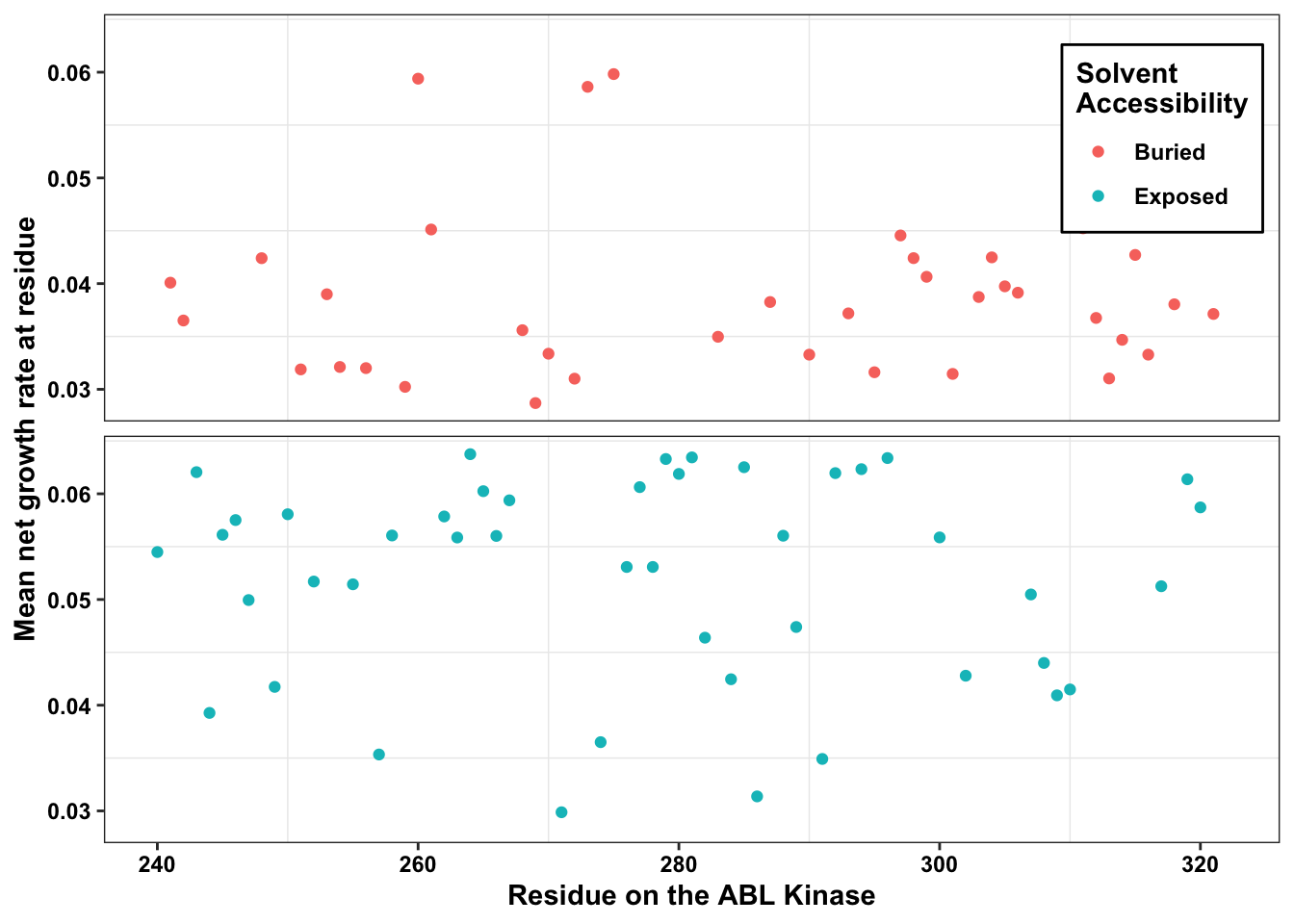
# ggsave("data/DSSP_SolventAccessibility_ABL/il3solvent_accessibility_v1.pdf",width=6,height = 4,units="in",useDingbats=F)
ggplot(il3_dssp,aes(x=protein_start,y=netgr_obs_mean,color=exposed))+
geom_point()+
# facet_wrap(~exposed,nrow=2)+
scale_y_continuous("Mean net growth rate at residue")+
scale_x_continuous("Residue on the ABL Kinase")+
labs(color="Solvent \nAccessibility")+cleanup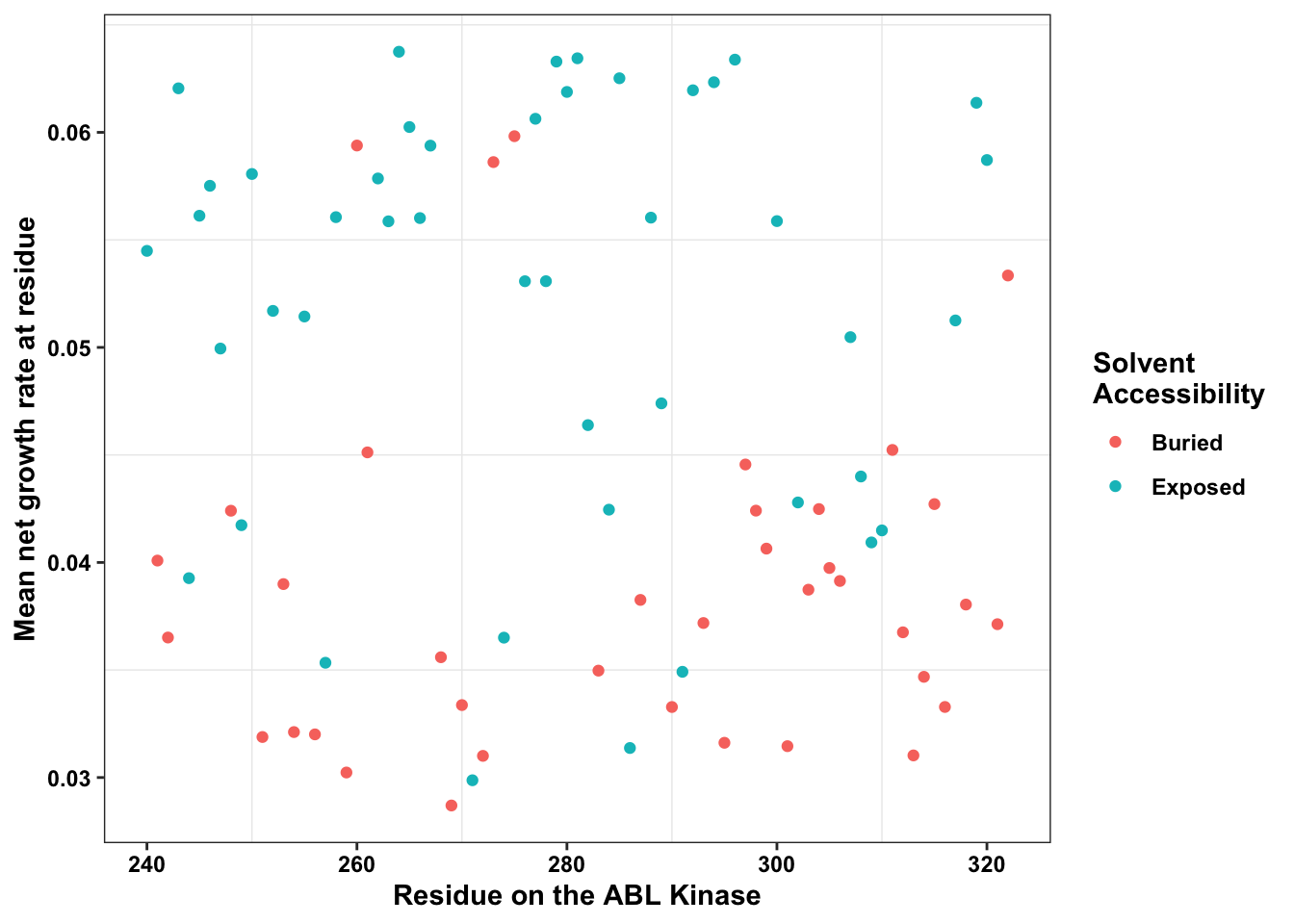
# ggsave("data/DSSP_SolventAccessibility_ABL/il3solvent_accessibility_v2.pdf",width=6,height = 4,units="in",useDingbats=F)
ggplot(il3_dssp,aes(x=ACC,y=netgr_obs_mean,color=exposed))+
geom_point()+
scale_y_continuous("Mean net growth rate at residue")+
scale_x_continuous("DSSP Solvent Accessibility at Residue")+
cleanup+
labs(color="Solvent \nAccessibility")+
theme(legend.position = c(.9,.25))+
theme(legend.background = element_rect(
size=0.5, linetype="solid",
colour ="black"))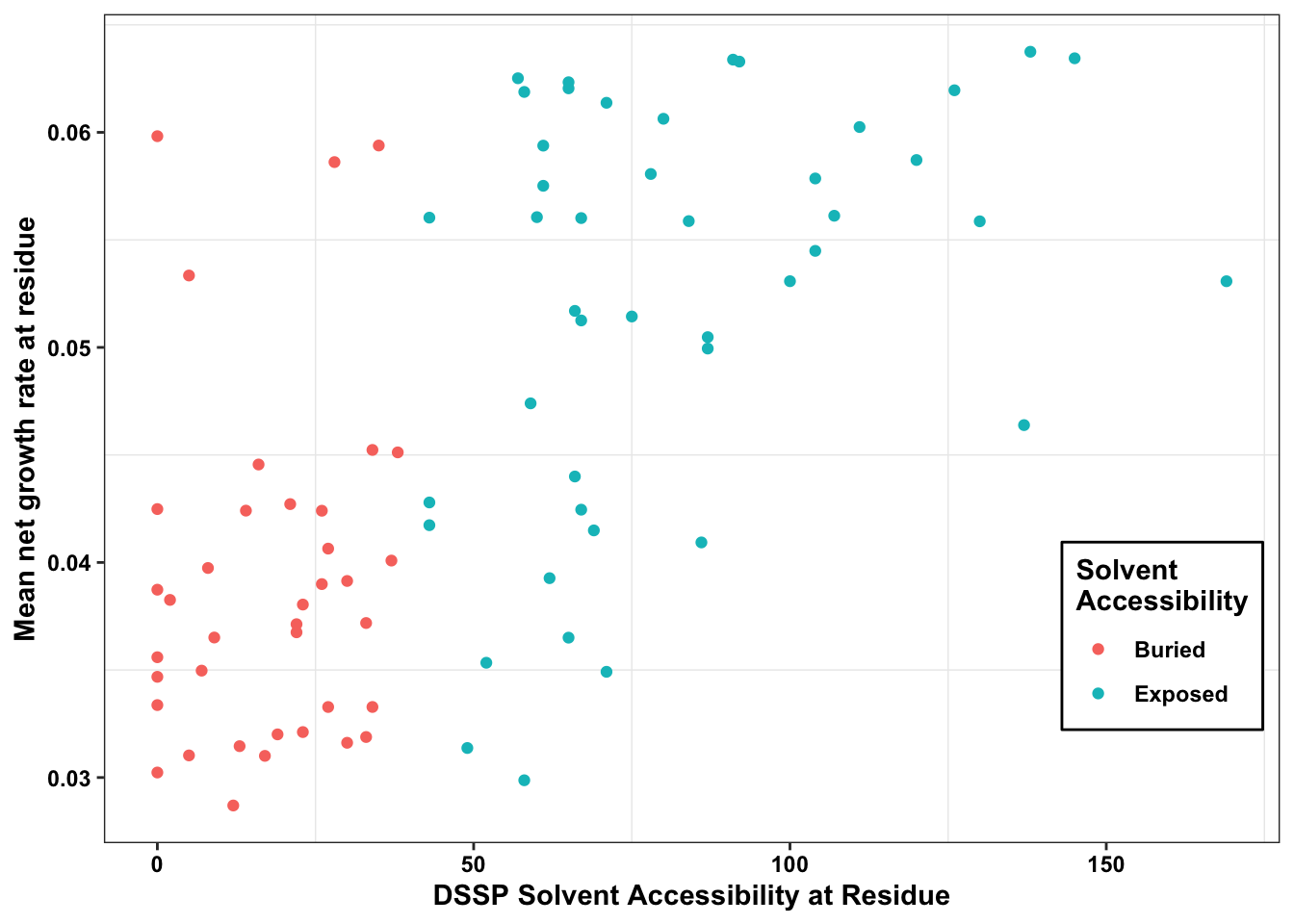
# ggsave("data/DSSP_SolventAccessibility_ABL/il3solvent_accessibility_v3.pdf",width=6,height = 4,units="in",useDingbats=F)
il3=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/baf3_IL3_rep1vsrep2_ft/screen_comparison_baf3_IL3_low_rep1vsrep2_ft.csv")
il3=il3%>%dplyr::select(species,ref_aa,protein_start,alt_aa,netgr_obs_mean)
il3_dssp=merge(il3,dssp,by.x=c("protein_start","ref_aa"),by.y=c("protein_start","AA"))
il3_dssp=il3_dssp%>%mutate(exposed=case_when(ACC>=40~"Exposed",
T~"Buried"))
# il3_dssp=il3_dssp%>%mutate(hydrophobic=case_when(alt_aa%in%c("A","V","I","M","L","F","Y","W")~"hydrophobic",
# T~"other"))
ggplot(il3_dssp,aes(x=alt_aa,y=netgr_obs_mean))+geom_boxplot()+facet_wrap(~exposed)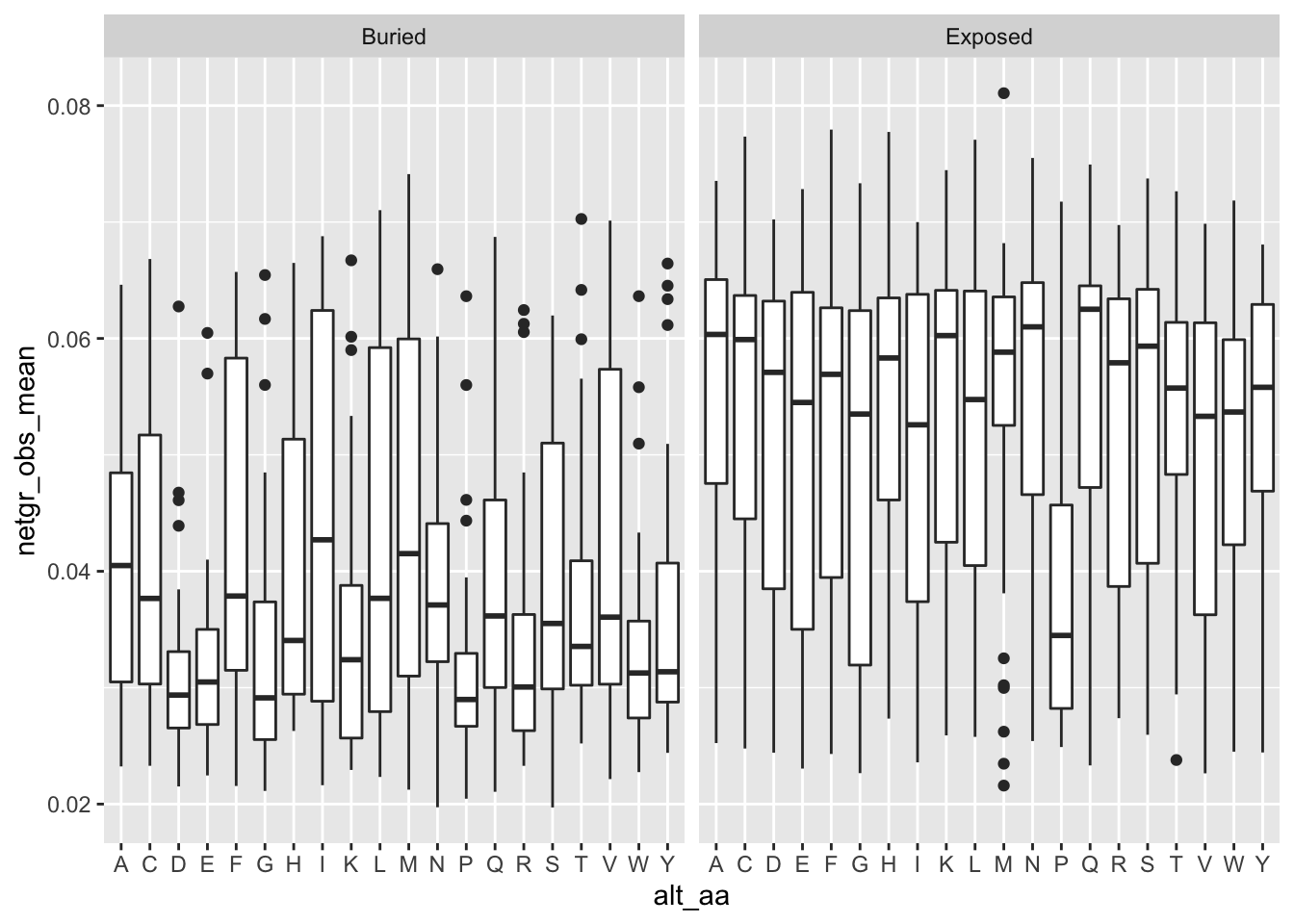
ggplot(il3_dssp,aes(x=exposed,y=netgr_obs_mean))+geom_boxplot()+facet_wrap(~exposed)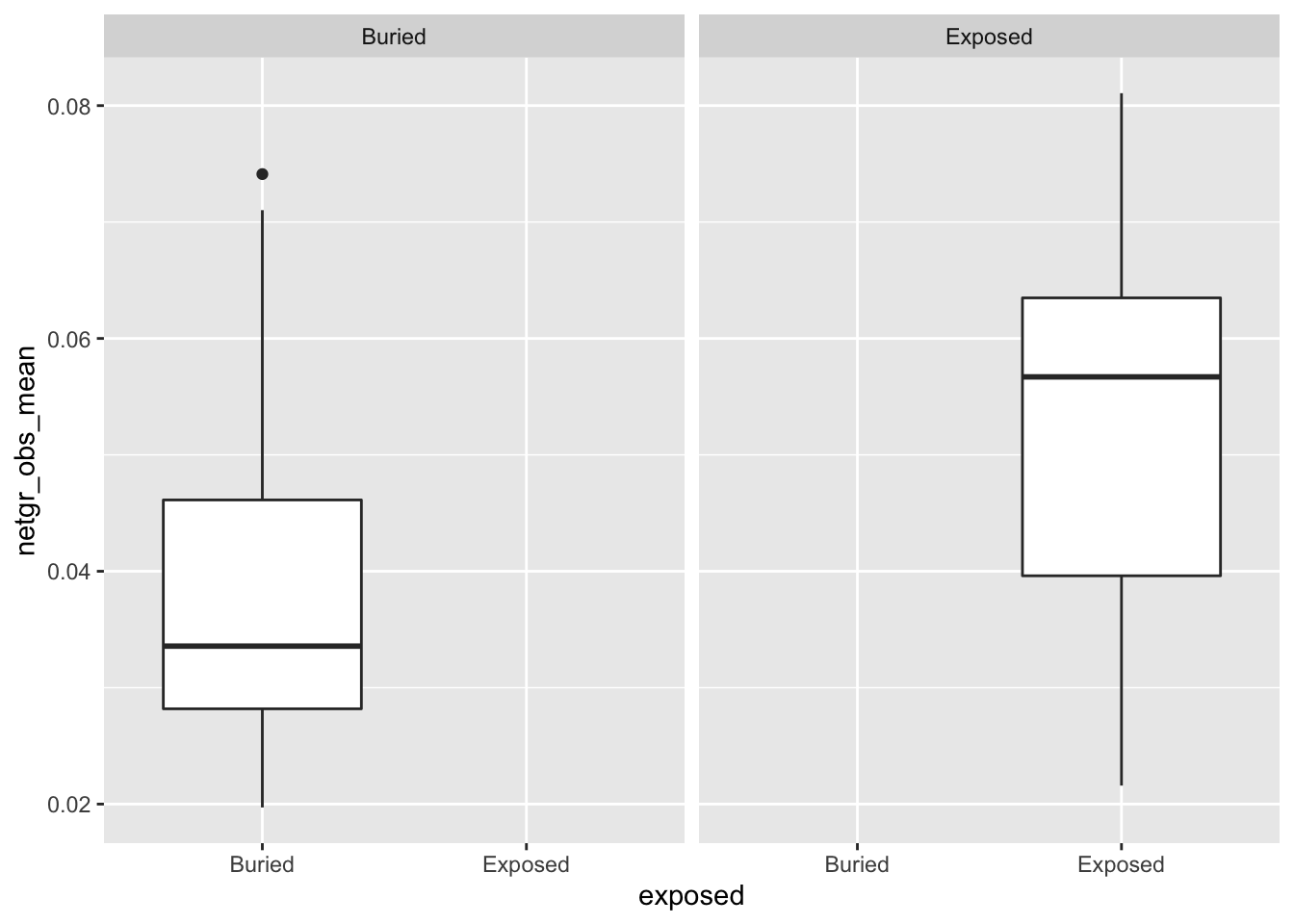
ggplot(il3_dssp,aes(x=factor(protein_start),y=netgr_obs_mean,fill=exposed))+
geom_boxplot()+
# facet_wrap(~exposed,nrow=2)+
scale_y_continuous("Mean net growth rate at residue")+
scale_x_discrete("Residue on the ABL Kinase")+
labs(fill="Solvent \nAccessibility")+cleanup+
theme(axis.text.x=element_text(angle=90, hjust=1))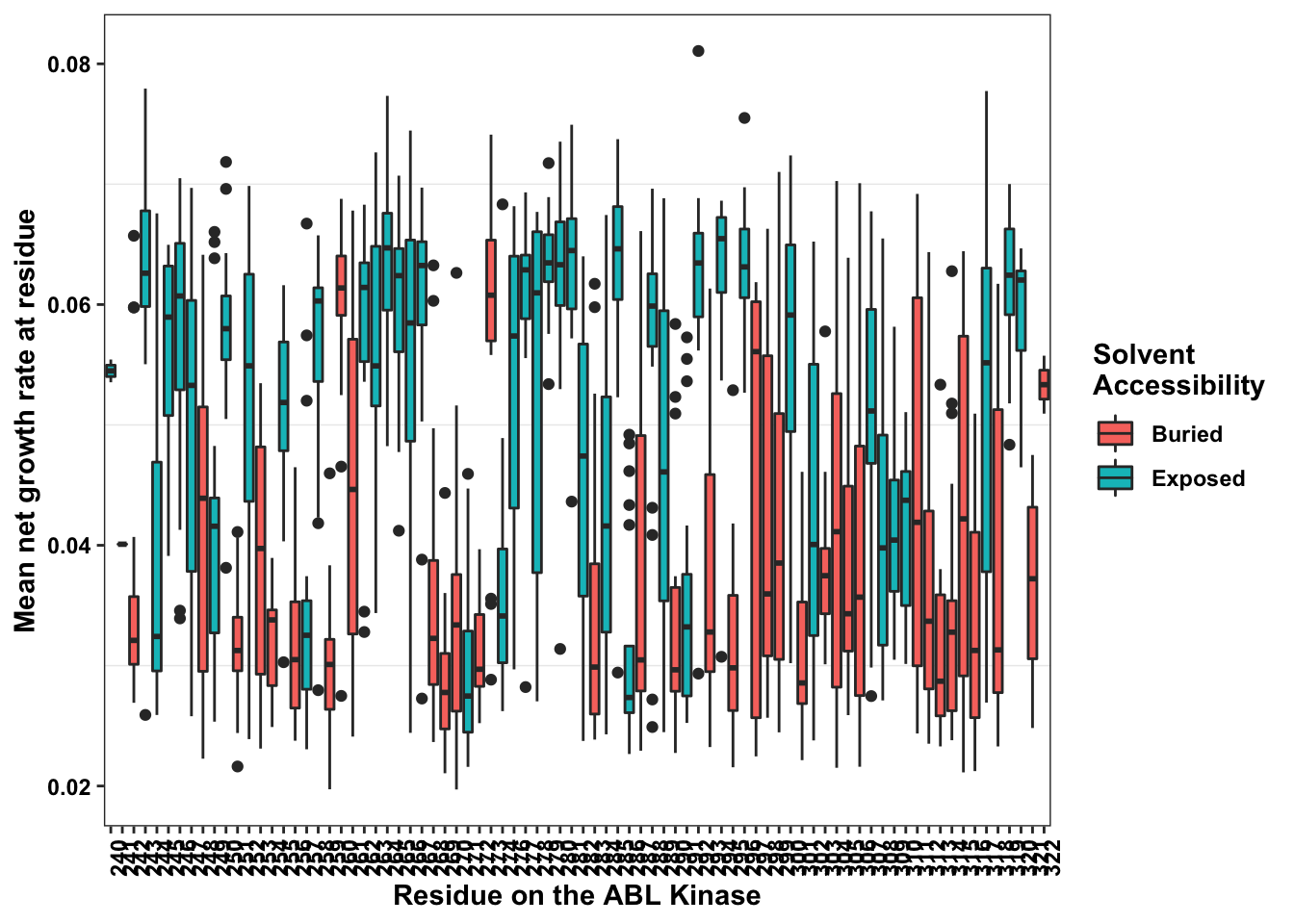
Doing some quick analyses on buried hydrophobic vs exposed residues
Doing some quick DDG analysis
ddg=read.table("data/DDG_ABL/2hyy_ddg.tsv")
colnames(ddg)=c("species","ddg","ddg_sd")
ddg=ddg%>%mutate(protein_start=234+as.numeric(gsub("[^0-9]","",species)),
ref_aa=substr(species,1,1),
alt_aa=sub(".+(.)$", "\\1", species),
species=paste(ref_aa,protein_start,alt_aa,sep = ""))
# class(ddg$protein_start)
ddg=ddg%>%filter(protein_start%in%c(242:322))
il3=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/baf3_IL3_rep1vsrep2_ft/screen_comparison_baf3_IL3_low_rep1vsrep2_ft.csv")
il3=il3%>%select(species,protein_start,netgr_obs_mean)
ddgil3=merge(ddg,il3,by=c("species","protein_start"))
ggplot(ddgil3,aes(x=ddg,y=netgr_obs_mean))+geom_point()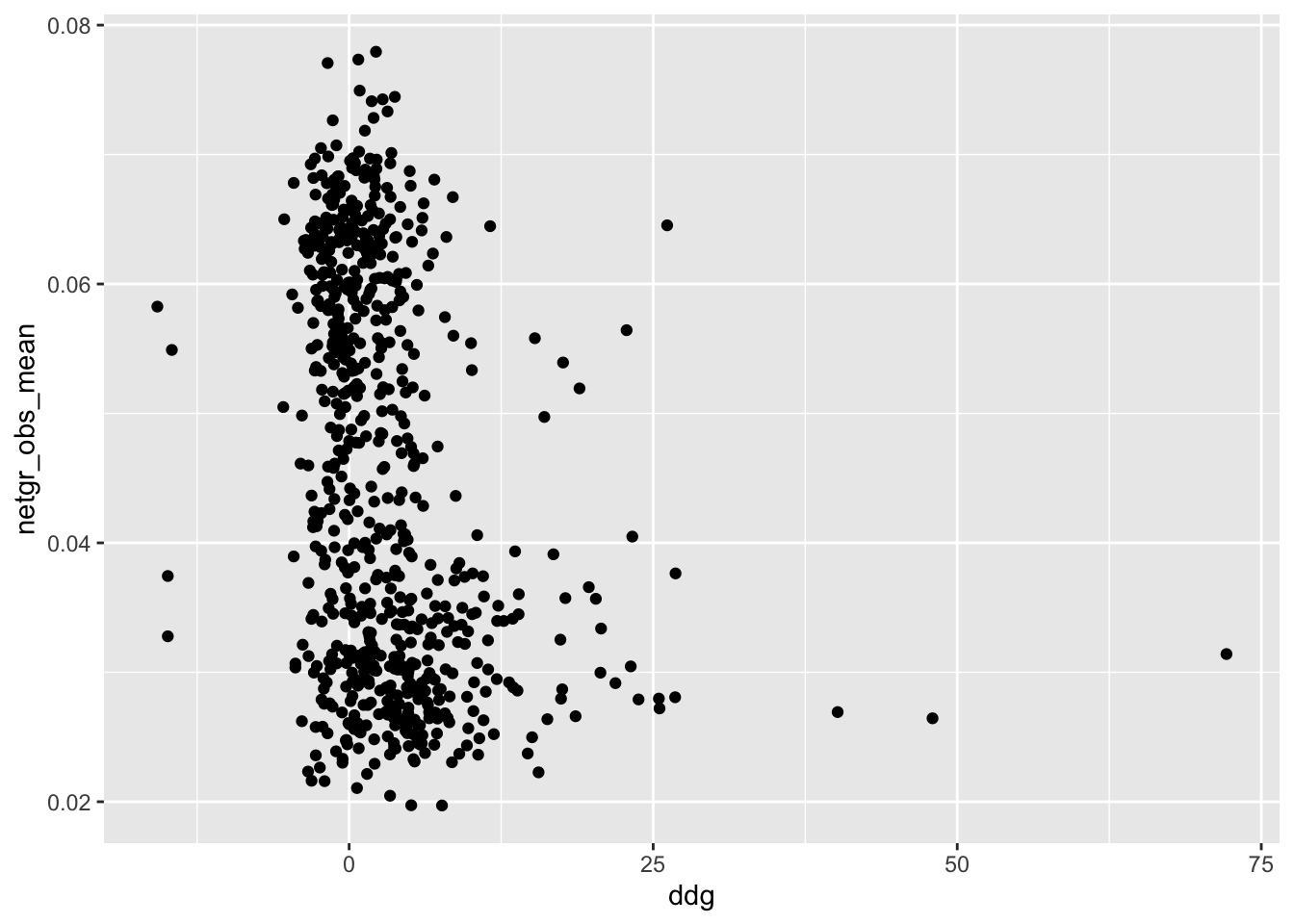
ggplot(ddgil3,aes(x=ddg,y=netgr_obs_mean,label=species))+geom_text()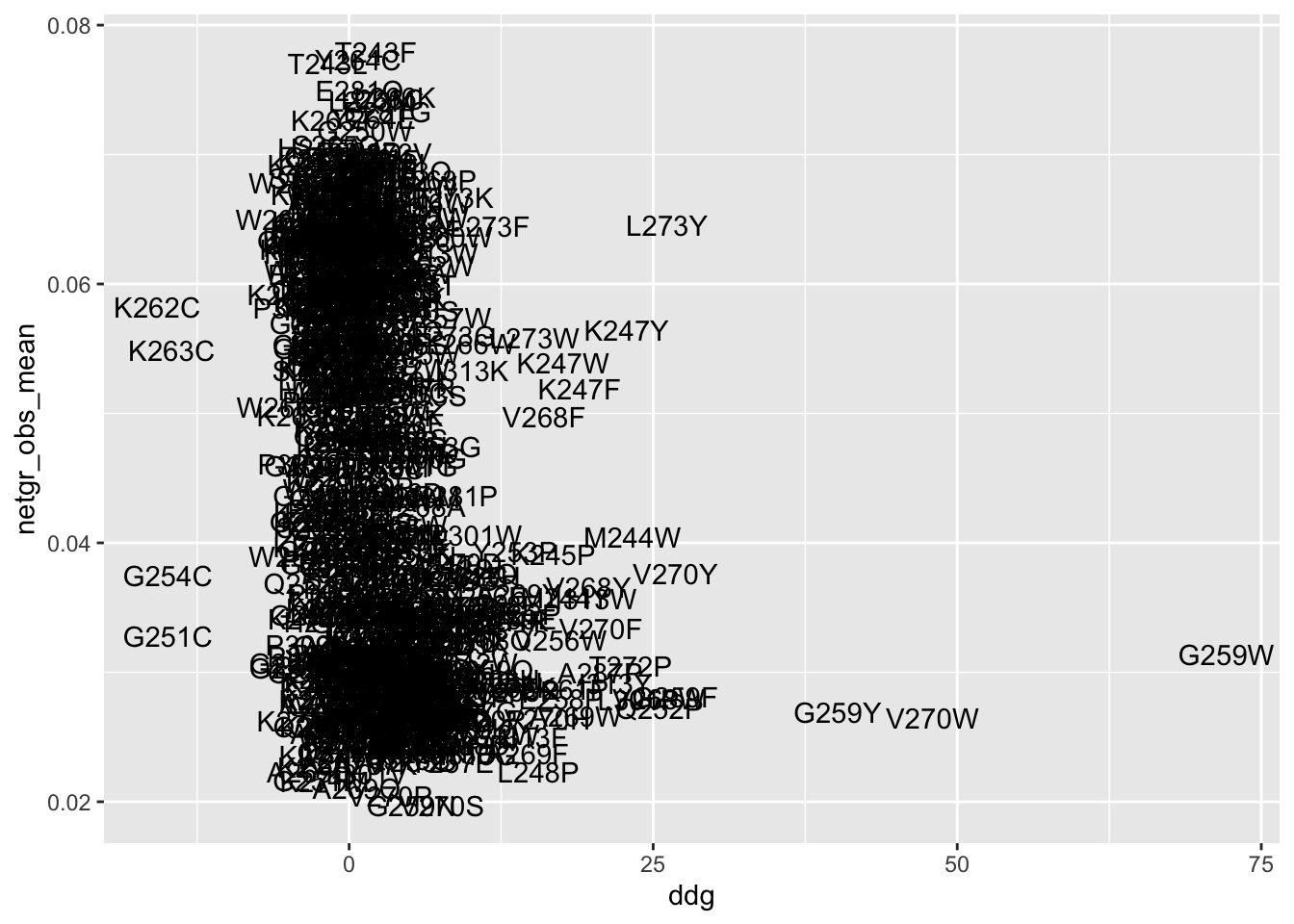
Comparing Fitness Scores with IC50 predictions
# ,include=F,eval=F
source("code/resmuts_adder.R")
ic50data_all_sum=read.csv("output/ic50data_all_confidence_intervals_raw_data.csv",row.names = 1)
imat=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/baf3_Imat_low_rep1vsrep2_ft/screen_comparison_baf3_Imat_low_rep1vsrep2_ft.csv")
# imat=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/k562_Imat_medium_rep1vsrep2/screen_comparison_k562_imat_medium_rep1vsrep2.csv")
imat=imat%>%select(species,ref_aa,protein_start,alt_aa,netgr_obs_mean)
# x=imat%>%filter(species%in%"V299L")
imat=resmuts_adder(imat)
resmuts_merged=merge(imat%>%filter(resmuts%in%"TRUE"),ic50data_all_sum%>%dplyr::select(species,netgr_pred_model,netgr_pred_mean),by="species")
ggplot(resmuts_merged,aes(x=netgr_pred_mean,y=netgr_obs_mean,label=species))+geom_text()+theme_bw()+geom_abline()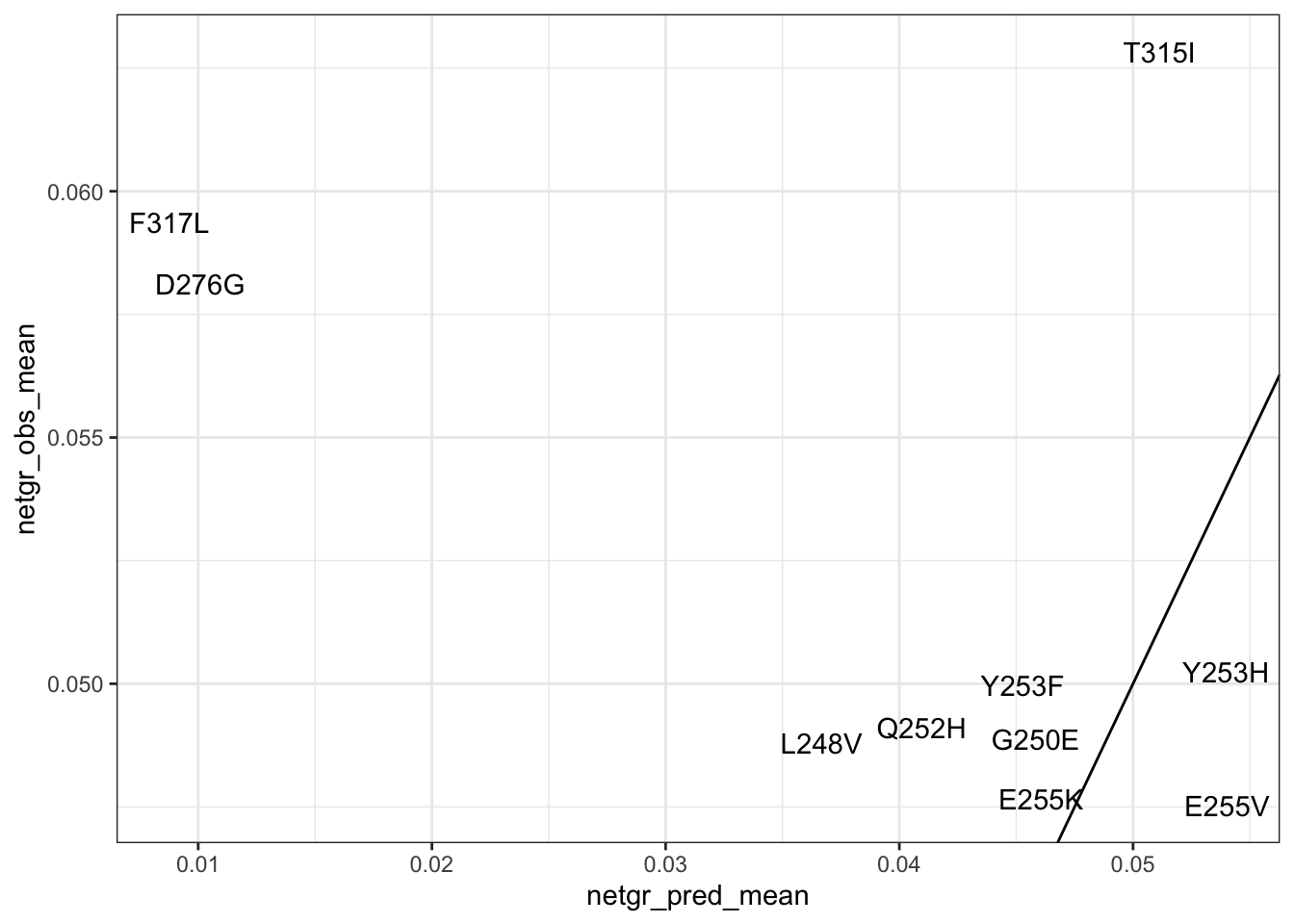
#######################Making Correlation Plots for Paper################
###########Cross Species Correlations###########
data=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-species/baf3_Imat_Lowvsk562_Imat_Medium/screen_comparison_baf3_Imat_Lowvsk562_Imat_Medium.csv",header = T,stringsAsFactors = F)
source("code/res_residues_adder.R")
source("code/cosmic_data_adder.R")
# data=read.csv("output/ABLEnrichmentScreens/Imat_Enrichment_bgmerged_2.22.23.csv",header = T,stringsAsFactors = F)
# class(data)
data=res_residues_adder(data)
data=cosmic_data_adder(data)
# data=data%>%rowwise()%>%mutate(netgr_mean=mean(netgr_obs.x,netgr_obs.y))
data=data%>%rowwise()%>%mutate(netgr_mean=mean(netgr_obs_screen1,netgr_obs_screen2))
data$resmut_cosmic="neither"
data[data$cosmic_present%in%TRUE,"resmut_cosmic"]="cosmic"
data[data$resmuts%in%TRUE,"resmut_cosmic"]="resmut"
data$resmut_cosmic=factor(data$resmut_cosmic,levels=c("neither","cosmic","resmut"))
ggplot(data%>%filter(protein_start>=242,protein_start<=322),aes(x=netgr_obs_screen1,y=netgr_obs_screen2))+
geom_point(color="black",shape=21,size=.75,aes(fill=resmuts))+
# geom_abline()+
scale_x_continuous(name="Net Growth Rate Baf3")+
scale_y_continuous(name="Net Growth Rate K562")+
labs(fill="Sanger\n Mutant")+
scale_fill_manual(values=c("gray90","red"))+
cleanup+theme(legend.position = "none")+
stat_cor(method="pearson")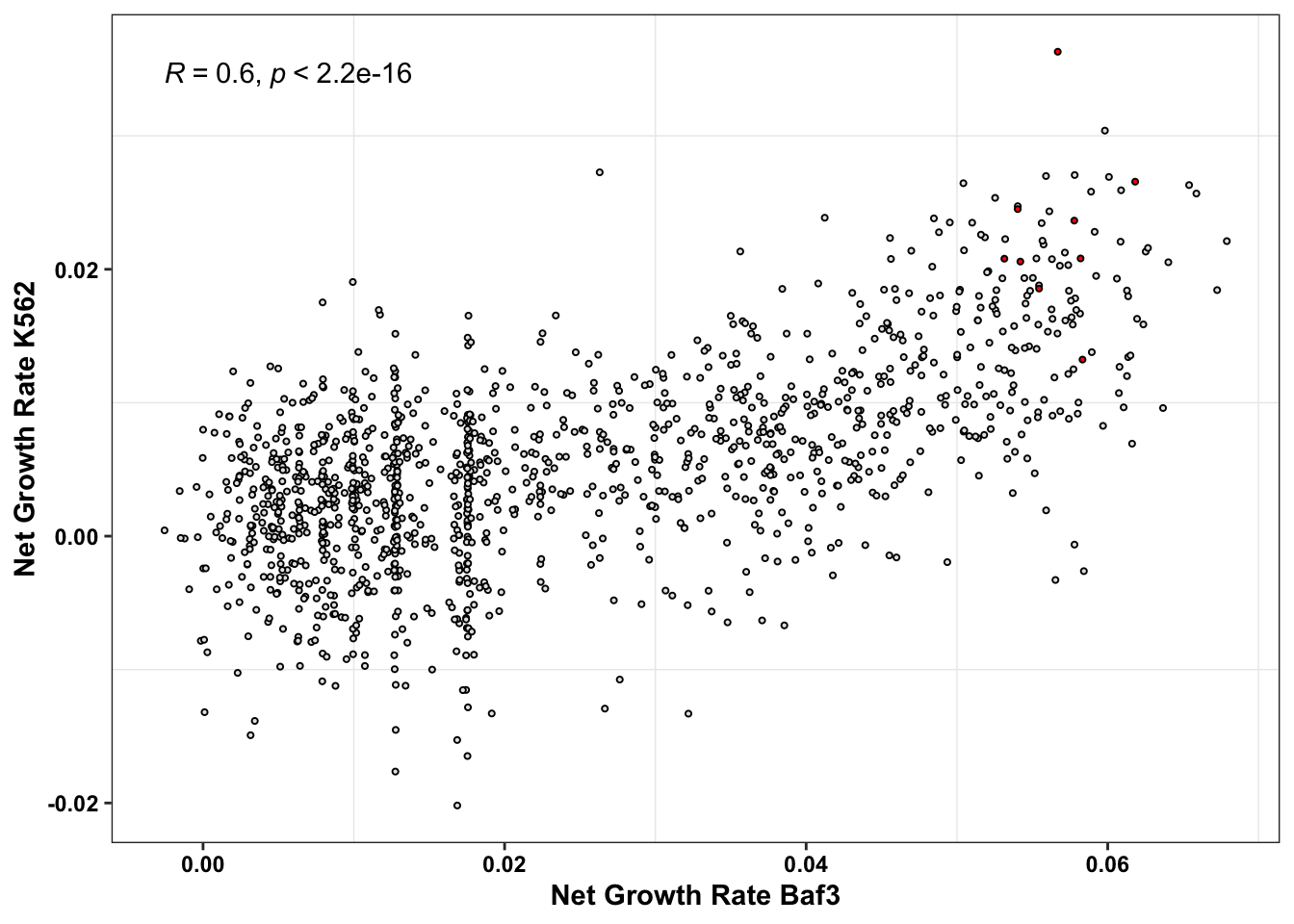
# ggsave("output/SM_Imatinib_Plots/Baf3vsK562_ImatinibEnrichment_Plot_Medium_4.13.23.pdf",width=3,height=3,units="in",useDingbats=F)
###########K562s###########
data=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/k562_imat_medium_rep1vsrep2/screen_comparison_k562_imat_medium_rep1vsrep2.csv",header = T,stringsAsFactors = F)
data=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/k562_imat_medium_rep1vsrep2/screen_comparison_k562_imat_medium_rep1vsrep2.csv",header = T,stringsAsFactors = F)
source("code/res_residues_adder.R")
source("code/cosmic_data_adder.R")
# data=read.csv("output/ABLEnrichmentScreens/Imat_Enrichment_bgmerged_2.22.23.csv",header = T,stringsAsFactors = F)
# class(data)
data=res_residues_adder(data)
data=cosmic_data_adder(data)
# data=data%>%rowwise()%>%mutate(netgr_mean=mean(netgr_obs.x,netgr_obs.y))
data=data%>%rowwise()%>%mutate(netgr_mean=mean(netgr_obs_screen1,netgr_obs_screen2))
data$resmut_cosmic="neither"
data[data$cosmic_present%in%TRUE,"resmut_cosmic"]="cosmic"
data[data$resmuts%in%TRUE,"resmut_cosmic"]="resmut"
data$resmut_cosmic=factor(data$resmut_cosmic,levels=c("neither","cosmic","resmut"))
# !ct_screen1_after%in%.5,!ct_screen2_after%in%.5,
ggplot(data%>%filter(protein_start>=242,protein_start<=322),aes(x=netgr_obs_screen1,y=netgr_obs_screen2))+
geom_point(color="black",shape=21,size=.75,aes(fill=resmuts))+
geom_abline()+
scale_x_continuous(name="Net Growth Rate Rep 1")+
scale_y_continuous(name="Net Growth Rate Rep 2")+
labs(fill="Sanger\n Mutant")+
scale_fill_manual(values=c("gray90","red"))+
cleanup+theme(legend.position = "none")+
stat_cor(method="pearson")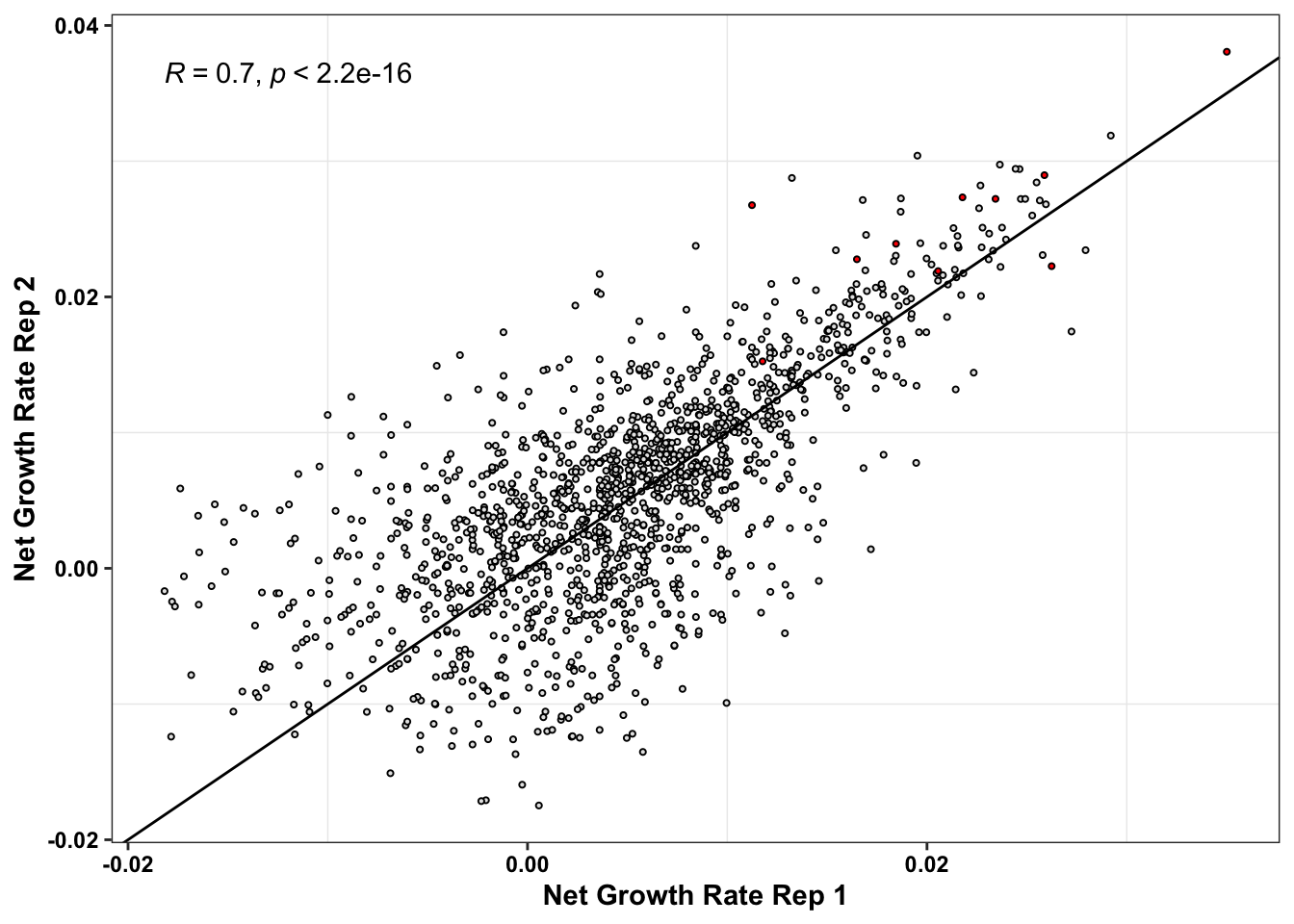
# ggsave("output/SM_Imatinib_Plots/K562_ImatinibEnrichment_Plot_Medium_4.13.23.pdf",width=3,height=3,units="in",useDingbats=F)
ggplot(data%>%filter(protein_start>=242,protein_start<=322),aes(x=netgr_obs_mean,fill=resmuts))+
geom_density(alpha=.7)+scale_x_continuous(bquote('Net Growth Rate '(Hours^-1)))+scale_y_continuous("Density")+scale_fill_manual(values=c("gray","red"))+cleanup+theme(legend.position = "none")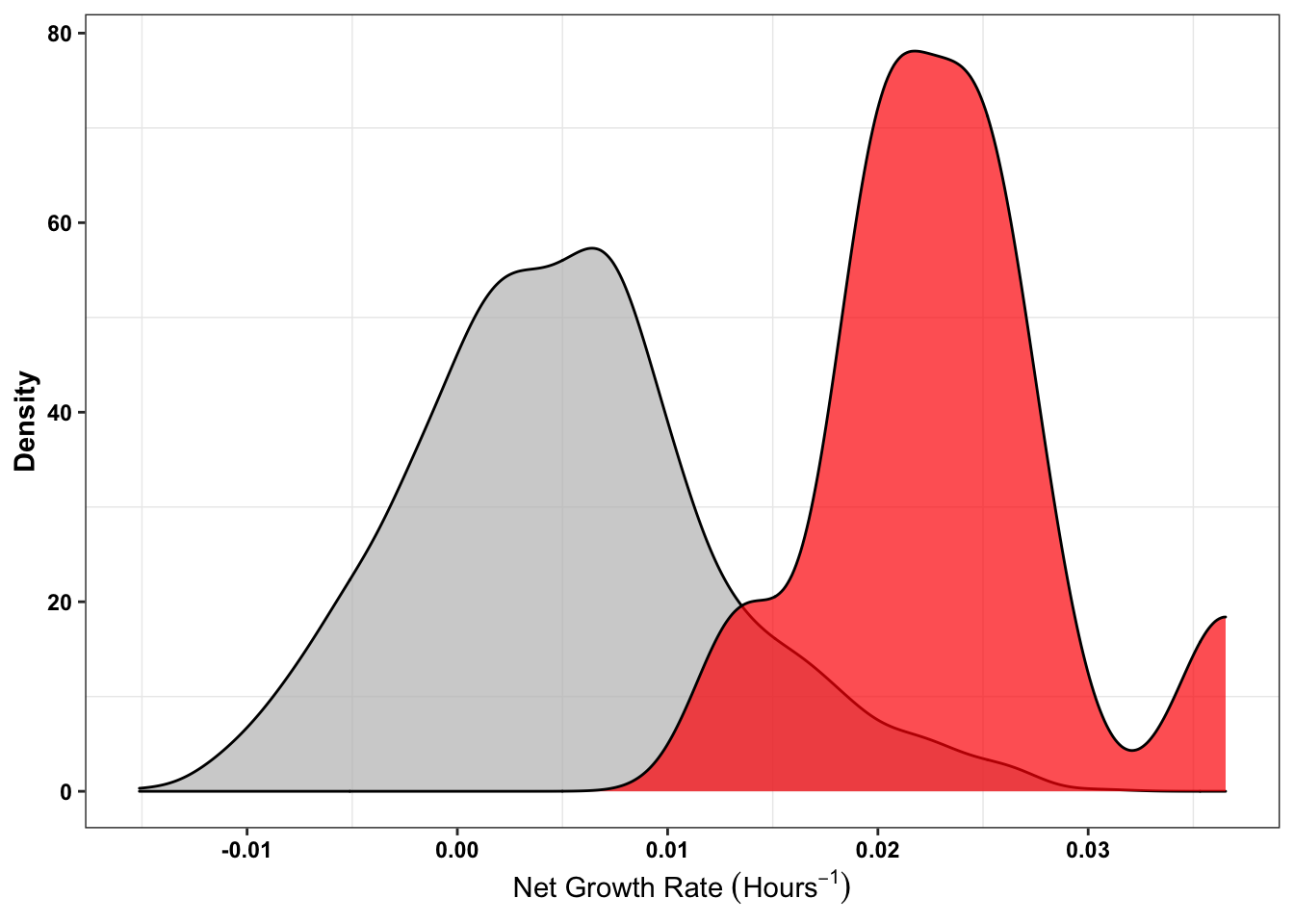
# ggsave("output/SM_Imatinib_Plots/K562_Density_Imat_netgr_4.19.23.pdf",width=4,height=3,units="in",useDingbats=F)
ggplot(data%>%filter(!species%in%"V299L",protein_start>=242,protein_start<=322,n_nuc_min%in%1),aes(x=resmut_cosmic,y=netgr_mean,fill=resmut_cosmic))+
# geom_violin(color="black")+
geom_boxplot(color="black")+
geom_jitter(color="black", size=1,width=.1, alpha=0.9)+
scale_fill_manual(values=c("gray90","orange","red"))+scale_y_continuous(name=bquote('Net Growth Rate '(Hours^-1)))+scale_x_discrete("Resistance Status",labels=c("Never\n Seen","Sanger","Known\nResistant"))+cleanup+theme(legend.position = "none")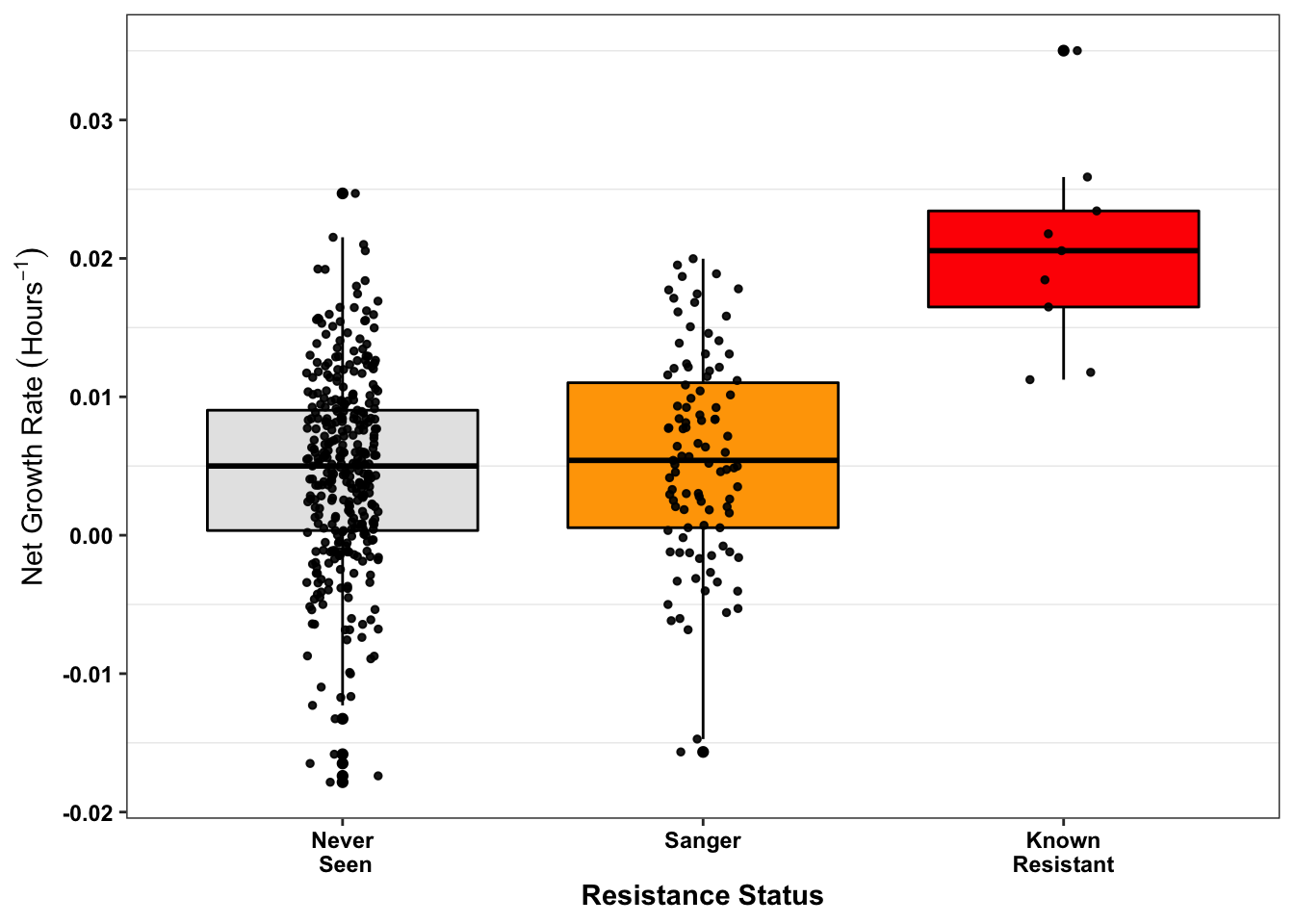
# ggsave("output/SM_Imatinib_Plots/ImatinibEnrichment_netgr_boxplot_k562s_4.21.23.pdf",width=3,height=3,units="in",useDingbats=F)
###########Baf3s###########
# data=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/baf3_Imat_low_rep1vsrep2_ft/screen_comparison_baf3_Imat_low_rep1vsrep2_ft.csv",header = T,stringsAsFactors = F)
data=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/baf3_Imat_high_rep1vsrep2_ft/screen_comparison_baf3_Imat_high_rep1vsrep2_ft.csv",header = T,stringsAsFactors = F)
# data=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/k562_imat_medium_rep1vsrep2/screen_comparison_k562_imat_medium_rep1vsrep2.csv",header = T,stringsAsFactors = F)
source("code/res_residues_adder.R")
source("code/cosmic_data_adder.R")
# data=read.csv("output/ABLEnrichmentScreens/Imat_Enrichment_bgmerged_2.22.23.csv",header = T,stringsAsFactors = F)
# class(data)
data=res_residues_adder(data)
data=cosmic_data_adder(data)
# data=data%>%rowwise()%>%mutate(netgr_mean=mean(netgr_obs.x,netgr_obs.y))
data=data%>%rowwise()%>%mutate(netgr_mean=mean(netgr_obs_screen1,netgr_obs_screen2))
data$resmut_cosmic="neither"
data[data$cosmic_present%in%TRUE,"resmut_cosmic"]="cosmic"
data[data$resmuts%in%TRUE,"resmut_cosmic"]="resmut"
data$resmut_cosmic=factor(data$resmut_cosmic,levels=c("neither","cosmic","resmut"))
# !ct_screen1_after%in%.5,!ct_screen2_after%in%.5,
ggplot(data%>%filter(protein_start>=242,protein_start<=322),aes(x=netgr_obs_screen1,y=netgr_obs_screen2))+
geom_point(color="black",shape=21,size=.75,aes(fill=resmuts))+
geom_abline()+
scale_x_continuous(name="Net Growth Rate Rep 1")+
scale_y_continuous(name="Net Growth Rate Rep 2")+
labs(fill="Sanger\n Mutant")+
scale_fill_manual(values=c("gray90","red"))+
cleanup+theme(legend.position = "none")+
stat_cor(method="pearson")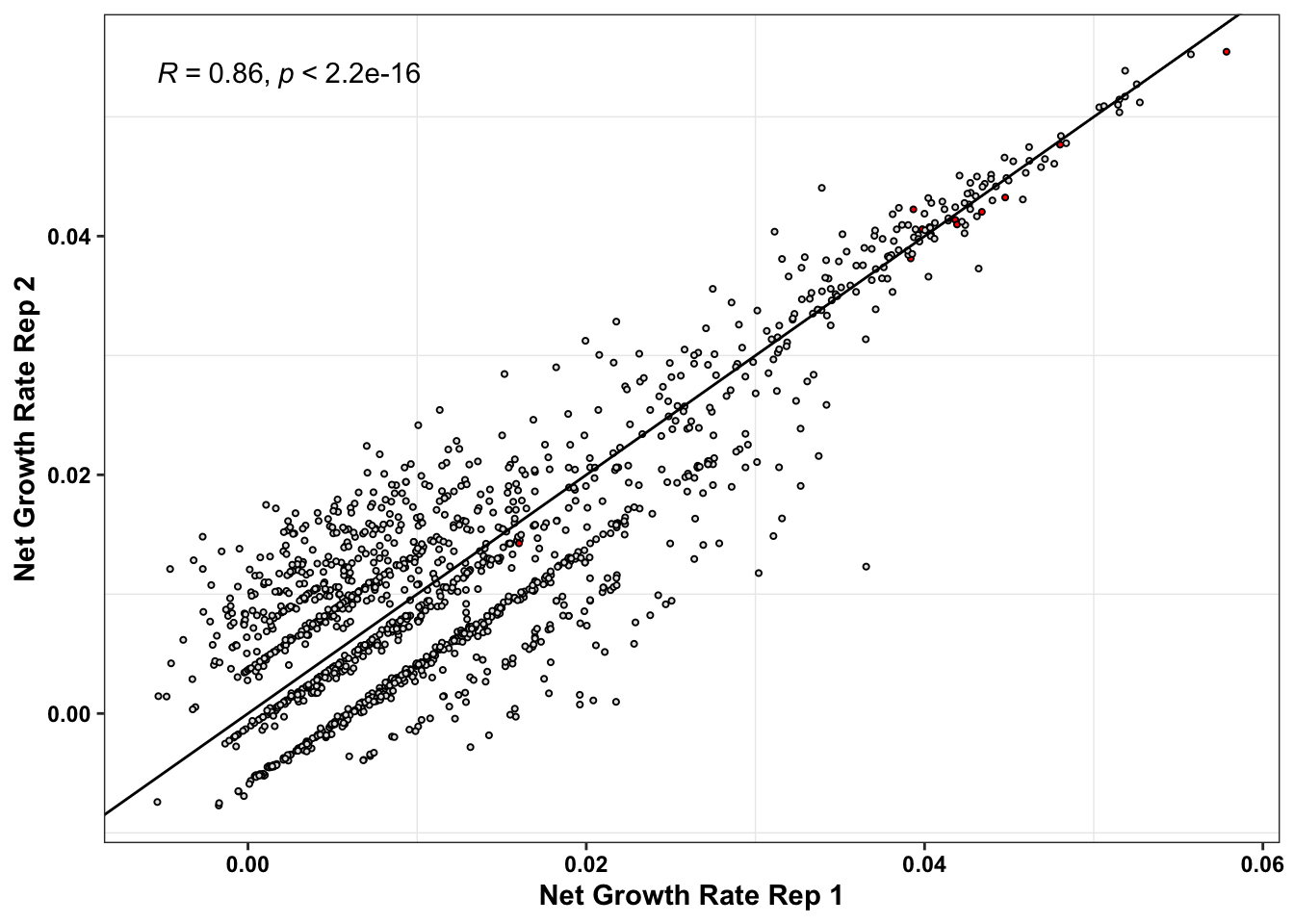
# ggsave("output/SM_Imatinib_Plots/BaF3_ImatinibEnrichment_Plot_Medium_4.13.23.pdf",width=3,height=3,units="in",useDingbats=F)
ggplot(data%>%filter(!species%in%"V299L",protein_start>=242,protein_start<=322),aes(x=netgr_obs_mean,fill=resmuts))+
geom_density(alpha=.7)+scale_x_continuous(bquote('Net Growth Rate '(Hours^-1)))+scale_y_continuous("Density")+scale_fill_manual(values=c("gray","red"))+cleanup+theme(legend.position = "none")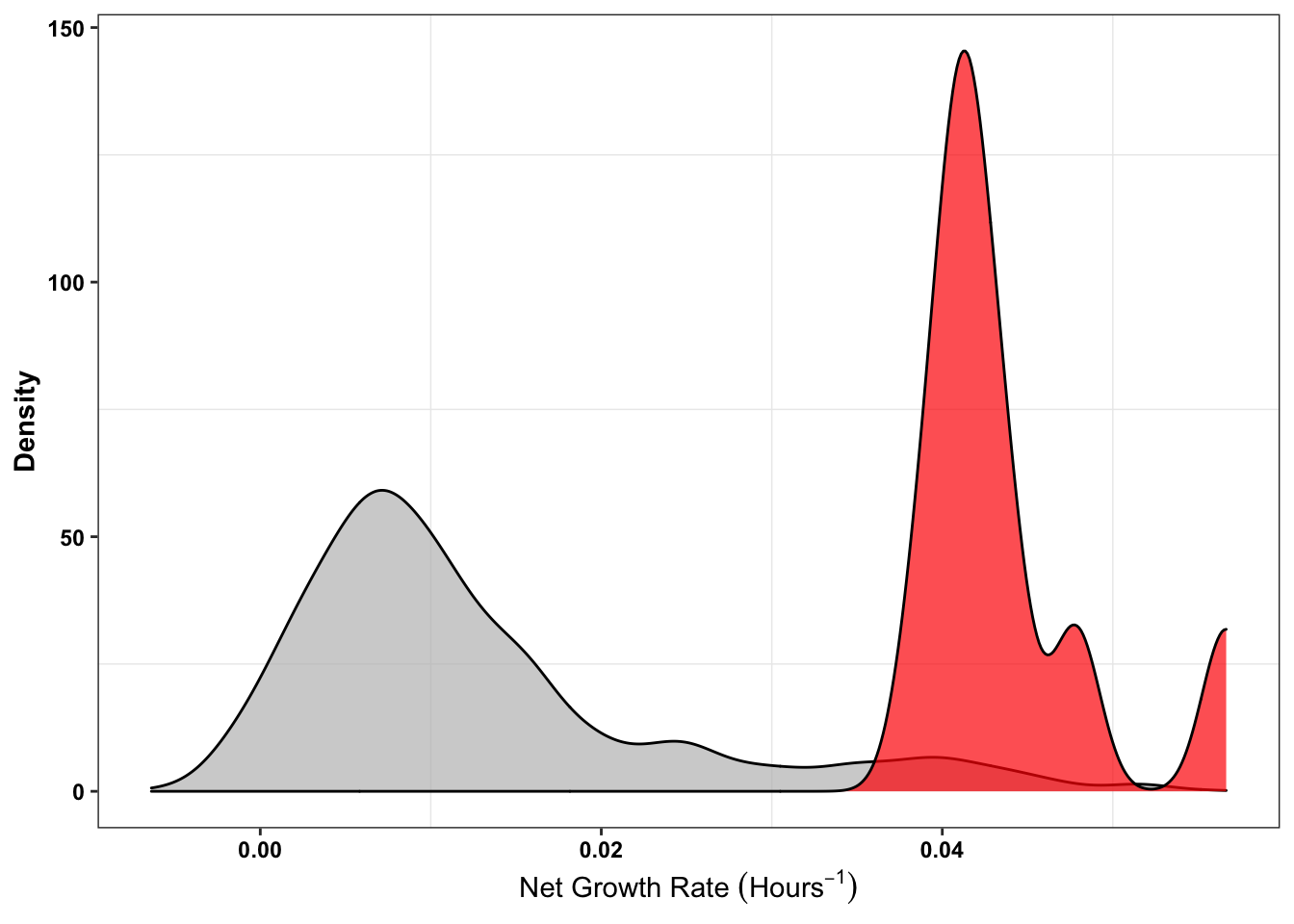
# ggsave("output/SM_Imatinib_Plots/Baf3_Density_Imat_netgr_4.19.23.pdf",width=4,height=3,units="in",useDingbats=F)
ggplot(data%>%filter(protein_start>=242,protein_start<=494,n_nuc_min%in%1),aes(x=reorder(species,-netgr_obs_mean),y=netgr_obs_mean,fill=resmut_cosmic))+geom_col()+theme(axis.text.x=element_text(angle=90, hjust=1))+scale_y_continuous(name="Enrichcment Score")+scale_x_discrete(name="Mutant")+scale_fill_manual(name="Resistance Status",labels=c("Never Seen","Sanger Mutant","Known Resistant Mutant"),values=c("gray","orange","red"))+cleanup+theme(legend.position = "none")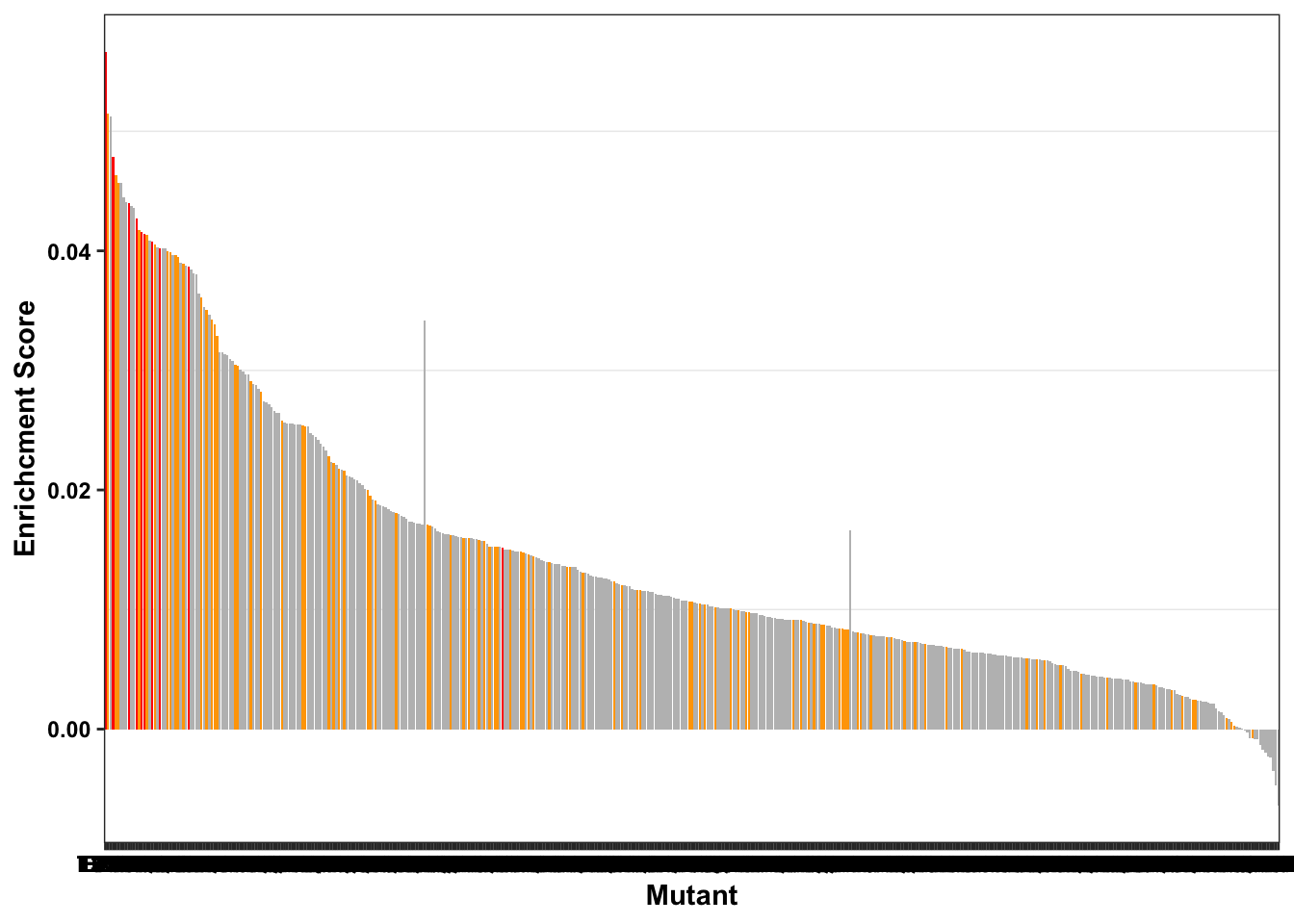
ggplot(data%>%group_by(species)%>%mutate(n_species=n())%>%filter(protein_start>=242,protein_start<=494,n_species%in%1),aes(x=reorder(species,-netgr_mean),y=netgr_mean,fill=resmut_cosmic))+geom_col()+theme(axis.text.x=element_text(angle=90, hjust=1))+scale_y_continuous(name=bquote('Net Growth Rate '(Hours^-1)))+scale_x_discrete(name="Mutant")+scale_fill_manual(name="Resistance Status",labels=c("Never Seen","Sanger Mutant","Known Resistant Mutant"),values=c("gray","orange","red"))+cleanup+theme(legend.position = "none")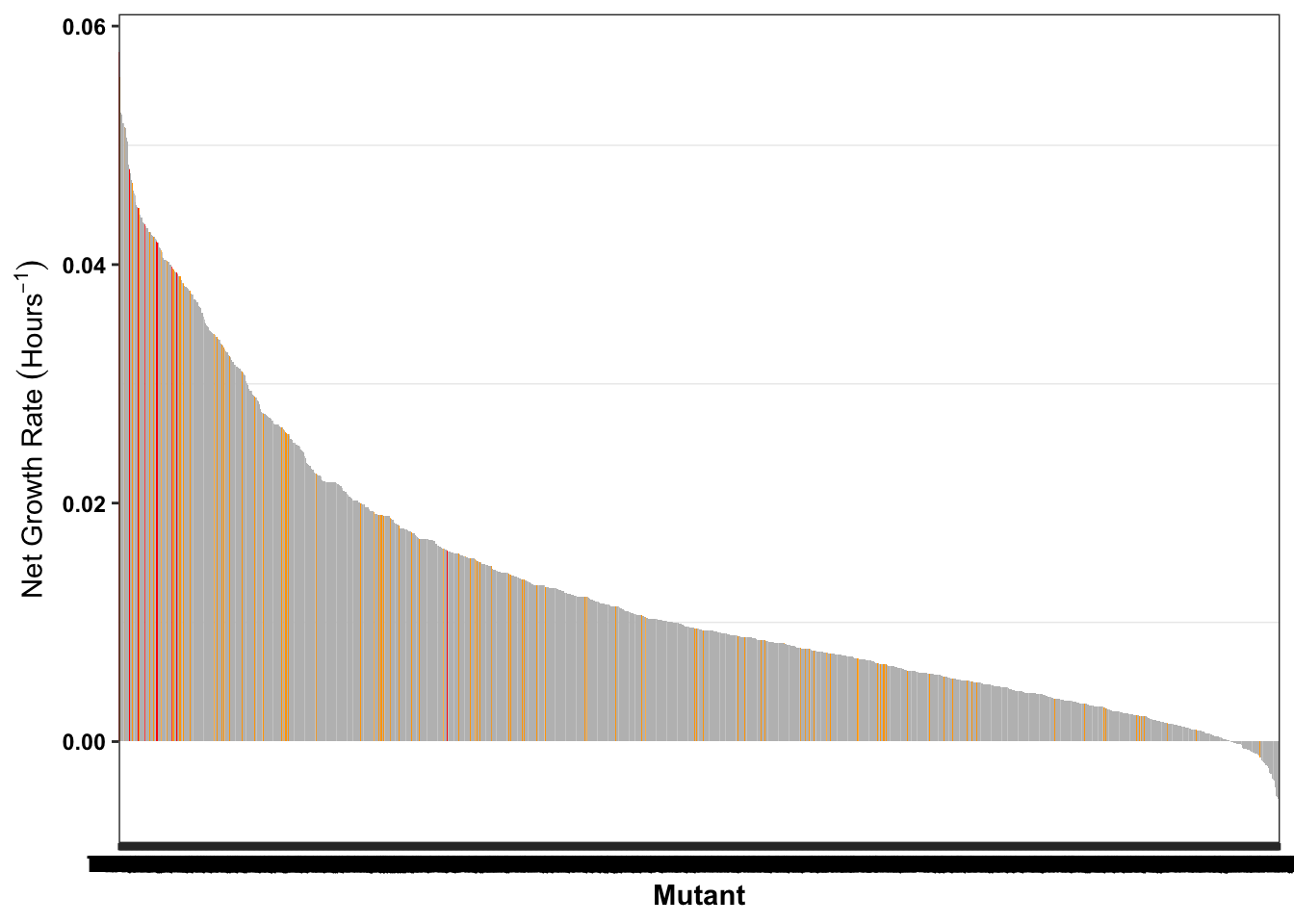
# ggsave("ImatinibEnrichment_Netgr_Distribution.pdf",width=8,height=4,units="in",useDingbats=F)
ggplot(data%>%filter(protein_start>=242,protein_start<=494,n_nuc_min%in%1,score_mean>=0),aes(x=reorder(species,-netgr_mean),y=netgr_mean,fill=resmut_cosmic))+geom_col()+
scale_y_continuous(name=bquote('Net Growth Rate '(Hours^-1)))+
scale_x_discrete(name="Mutant")+
scale_fill_manual(name="Resistance Status",labels=c("Never Seen","Sanger Mutant","Known Resistant Mutant"),values=c("gray","orange","red"))+
theme(axis.text.x=element_text(angle=90, hjust=1),
axis.text=element_text(size=6),
panel.grid.major = element_blank(),
panel.grid.major.y = element_blank(),
panel.background = element_blank())+
theme(legend.position = "none")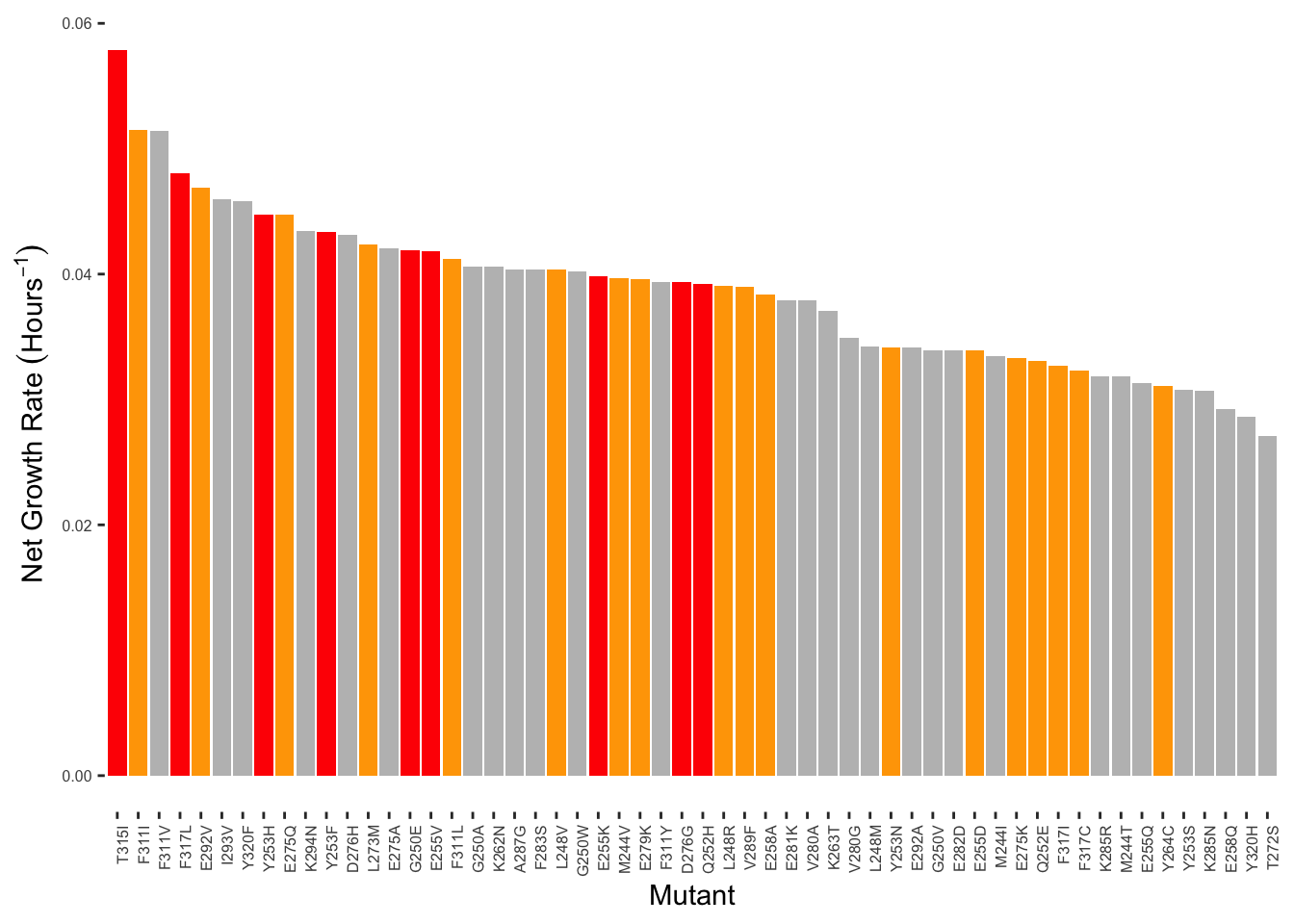
# ggsave("ImatinibEnrichment_Netgr_Distribution_zoom.pdf",width=6,height=4,units="in",useDingbats=F)
ggplot(data%>%filter(!species%in%"V299L",protein_start>=242,protein_start<=322,n_nuc_min%in%1),aes(x=resmut_cosmic,y=netgr_mean,fill=resmut_cosmic))+
# geom_violin(color="black")+
geom_boxplot(color="black")+
geom_jitter(color="black", size=1,width=.1, alpha=0.9)+
scale_fill_manual(values=c("gray90","orange","red"))+scale_y_continuous(name=bquote('Net Growth Rate '(Hours^-1)))+scale_x_discrete("Resistance Status",labels=c("Never\n Seen","Sanger","Known\nResistant"))+cleanup+theme(legend.position = "none")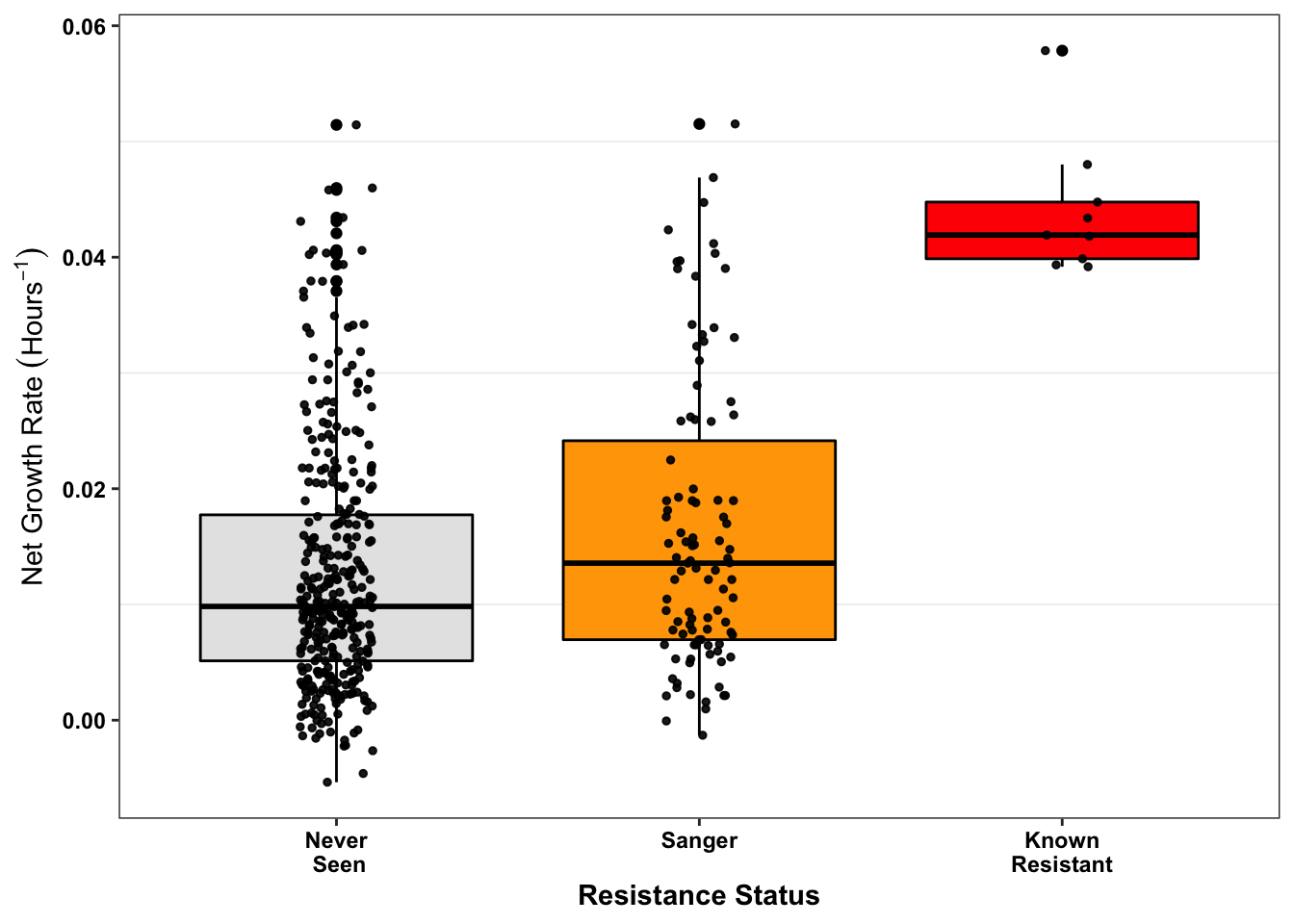
# ggsave("output/SM_Imatinib_Plots/ImatinibEnrichment_netgr_boxplot_baf3s_4.21.23.pdf",width=3,height=3,units="in",useDingbats=F)
plotly=ggplot(data%>%filter(protein_start>=242,protein_start<=494,n_nuc_min%in%1),aes(x=reorder(species,-netgr_mean),y=netgr_mean,fill=resmut_cosmic))+geom_col()+theme(axis.text.x=element_text(angle=90, hjust=1))+scale_y_continuous(name="Enrichcment Score")+scale_x_discrete(name="Mutant")+guides(fill = guide_legend(title = "Clinically\n Observed \n Resmut"))+scale_fill_manual(values=c("gray","orange","red"))
ggplotly(plotly)Looking at important sanger mutants across BaF3s and K562s
# K562
data=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/k562_imat_medium_rep1vsrep2/screen_comparison_k562_imat_medium_rep1vsrep2.csv",header = T,stringsAsFactors = F)
data=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/k562_imat_medium_rep1vsrep2/screen_comparison_k562_imat_medium_rep1vsrep2.csv",header = T,stringsAsFactors = F)
# BAf3
# data=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/baf3_Imat_high_rep1vsrep2_ft/screen_comparison_baf3_Imat_high_rep1vsrep2_ft.csv",header = T,stringsAsFactors = F)
source("code/res_residues_adder.R")
source("code/cosmic_data_adder.R")
# data=read.csv("output/ABLEnrichmentScreens/Imat_Enrichment_bgmerged_2.22.23.csv",header = T,stringsAsFactors = F)
# class(data)
data=res_residues_adder(data)
data=cosmic_data_adder(data)
# data=data%>%rowwise()%>%mutate(netgr_mean=mean(netgr_obs.x,netgr_obs.y))
data=data%>%rowwise()%>%mutate(netgr_mean=mean(netgr_obs_screen1,netgr_obs_screen2))
data$resmut_cosmic="neither"
data[data$cosmic_present%in%TRUE,"resmut_cosmic"]="cosmic"
data[data$resmuts%in%TRUE,"resmut_cosmic"]="resmut"
data$resmut_cosmic=factor(data$resmut_cosmic,levels=c("neither","cosmic","resmut"))
# x=data%>%filter(resmut_cosmic%in%"cosmic")
ggplot(data%>%filter(!species%in%"V299L",protein_start>=242,protein_start<=322,n_nuc_min%in%1),aes(x=resmut_cosmic,y=netgr_mean,fill=resmut_cosmic))+
# geom_violin(color="black")+
geom_boxplot(color="black")+
geom_jitter(color="black", size=1,width=.1, alpha=0.9)+
scale_fill_manual(values=c("gray90","orange","red"))+scale_y_continuous(name=bquote('Net Growth Rate '(Hours^-1)))+scale_x_discrete("Resistance Status",labels=c("Never\n Seen","Sanger","Known\nResistant"))+cleanup+theme(legend.position = "none")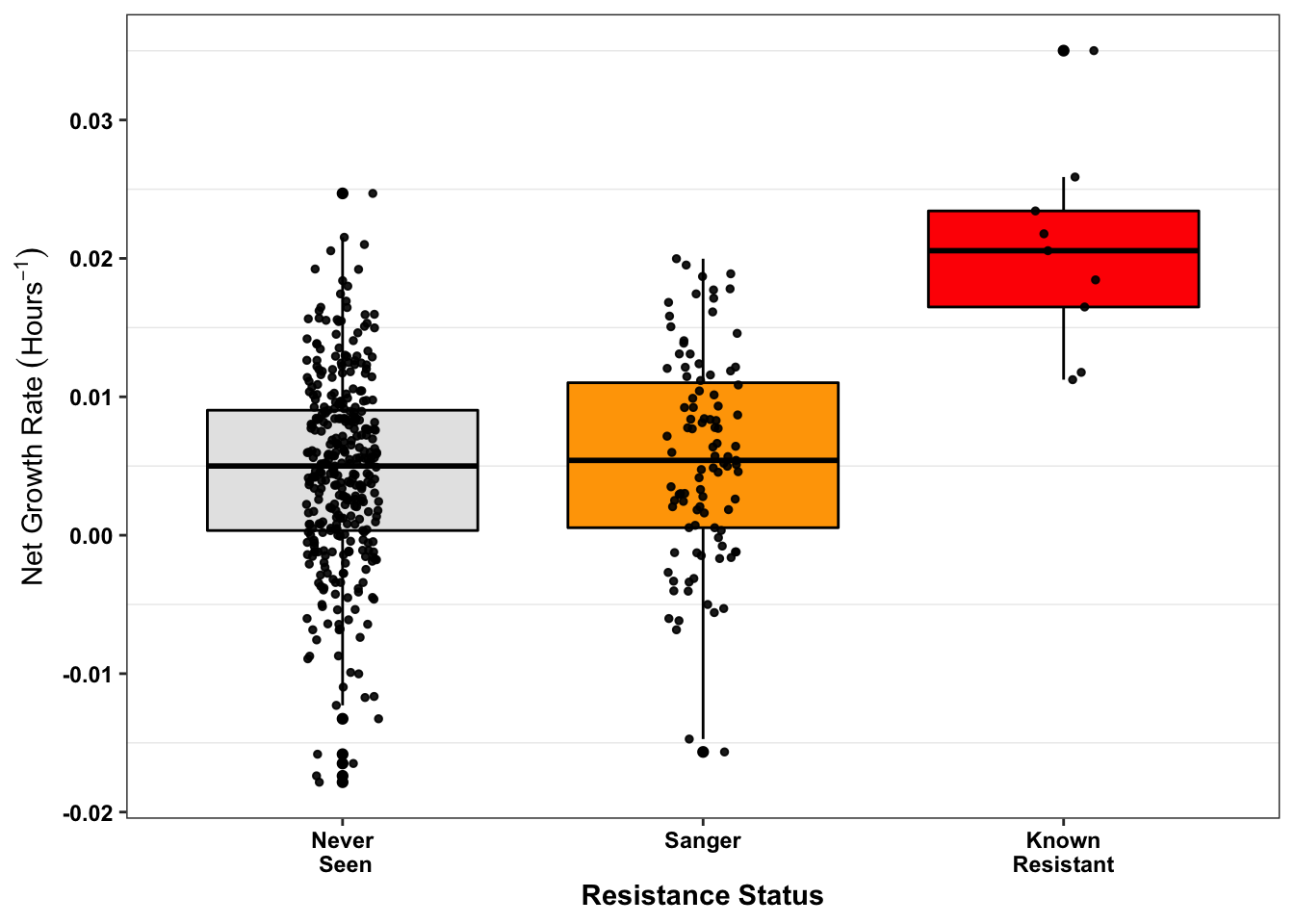
# ggsave("output/SM_Imatinib_Plots/ImatinibEnrichment_netgr_boxplot_baf3s_4.21.23.pdf",width=3,height=3,units="in",useDingbats=F)# Analyzing the Sanger data 4.22.2023
sanger=read.csv("data/Cosmic_ABL/ABL_Cosmic_Gene_mutations.csv",header = T)
sanger=sanger%>%dplyr::rename(protein_start=Position,species=AA.Mutation,sanger.count=Count)%>%filter(Type%in%"Substitution - Missense")%>%dplyr::select(protein_start,species,sanger.count)
sanger=sanger%>%mutate(species=gsub("p.","",species),ref_aa=substring(species,1,1),
alt_aa=substring(species, nchar(species), nchar(species)))
sanger=sanger%>%mutate(sanger.present=T)
sanger=sanger%>%filter(protein_start%in%242:520)
# sanger=resmuts_adder(sanger)
sum(sanger[sanger$resmuts%in%T,"sanger.count"])[1] 0sum(sanger[sanger$resmuts%in%F,"sanger.count"])[1] 0# There are a total of 1884 patients with mutations in the kianse. 1193 of these patients have known clinically resistant ImatR mutations. However, 691 of these patients fall into the other category
# K562
data=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/k562_imat_medium_rep1vsrep2/screen_comparison_k562_imat_medium_rep1vsrep2.csv",header = T,stringsAsFactors = F)
data=data%>%rowwise()%>%mutate(netgr_obs_mean.k562=mean(netgr_obs_screen1,netgr_obs_screen2),netgr_obs_sd.k562=sd(c(netgr_obs_screen1,netgr_obs_screen2)))
data.k562=data%>%dplyr::select(species,protein_start,ref_aa,alt_aa,n_nuc_min,netgr_obs_mean.k562,netgr_obs_sd.k562)
# Baf3
data=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/baf3_Imat_medium_rep1vsrep2_ft/screen_comparison_baf3_Imat_medium_rep1vsrep2_ft.csv",header = T,stringsAsFactors = F)
data=data%>%rowwise()%>%mutate(netgr_obs_mean.baf3=mean(netgr_obs_screen1,netgr_obs_screen2),netgr_obs_sd.baf3=sd(c(netgr_obs_screen1,netgr_obs_screen2)))
data.baf3=data%>%dplyr::select(species,protein_start,ref_aa,alt_aa,n_nuc_min,netgr_obs_mean.baf3,netgr_obs_sd.baf3)
data=merge(data.k562,data.baf3,by=c("species","protein_start","ref_aa","alt_aa","n_nuc_min"),all=T)
# data$sanger.present=F
# data$sanger.count=0
data=merge(data,sanger,by=c("species","protein_start","ref_aa","alt_aa"),all=T)
source("code/resmuts_adder.R")
data=resmuts_adder(data)
data=data%>%mutate(cosmic_present=sanger.present)
# data=data%>%mutate(alt_aa=substring(species, nchar(species), nchar(species)))
data$resmut_cosmic="neither"
data[data$cosmic_present%in%TRUE,"resmut_cosmic"]="cosmic"
data[data$resmuts%in%TRUE,"resmut_cosmic"]="resmut"
data$resmut_cosmic=factor(data$resmut_cosmic,levels=c("neither","cosmic","resmut"))
ggplot(data%>%filter(protein_start>=242,protein_start<=322),aes(x=resmut_cosmic,y=netgr_obs_mean.baf3,fill=resmut_cosmic))+
# geom_violin(color="black")+
geom_boxplot(color="black")+
geom_jitter(color="black", size=.5,width=.1, alpha=0.9)+
scale_fill_manual(values=c("gray90","orange","red"))+scale_y_continuous(name=bquote('Net Growth Rate '(Hours^-1)))+scale_x_discrete("Resistance Status",labels=c("Never\n Seen","Rare\n VUDR","Known\n Resistant\n Variant"))+cleanup+theme(legend.position = "none")Warning: Removed 33 rows containing non-finite values (stat_boxplot).Warning: Removed 33 rows containing missing values (geom_point).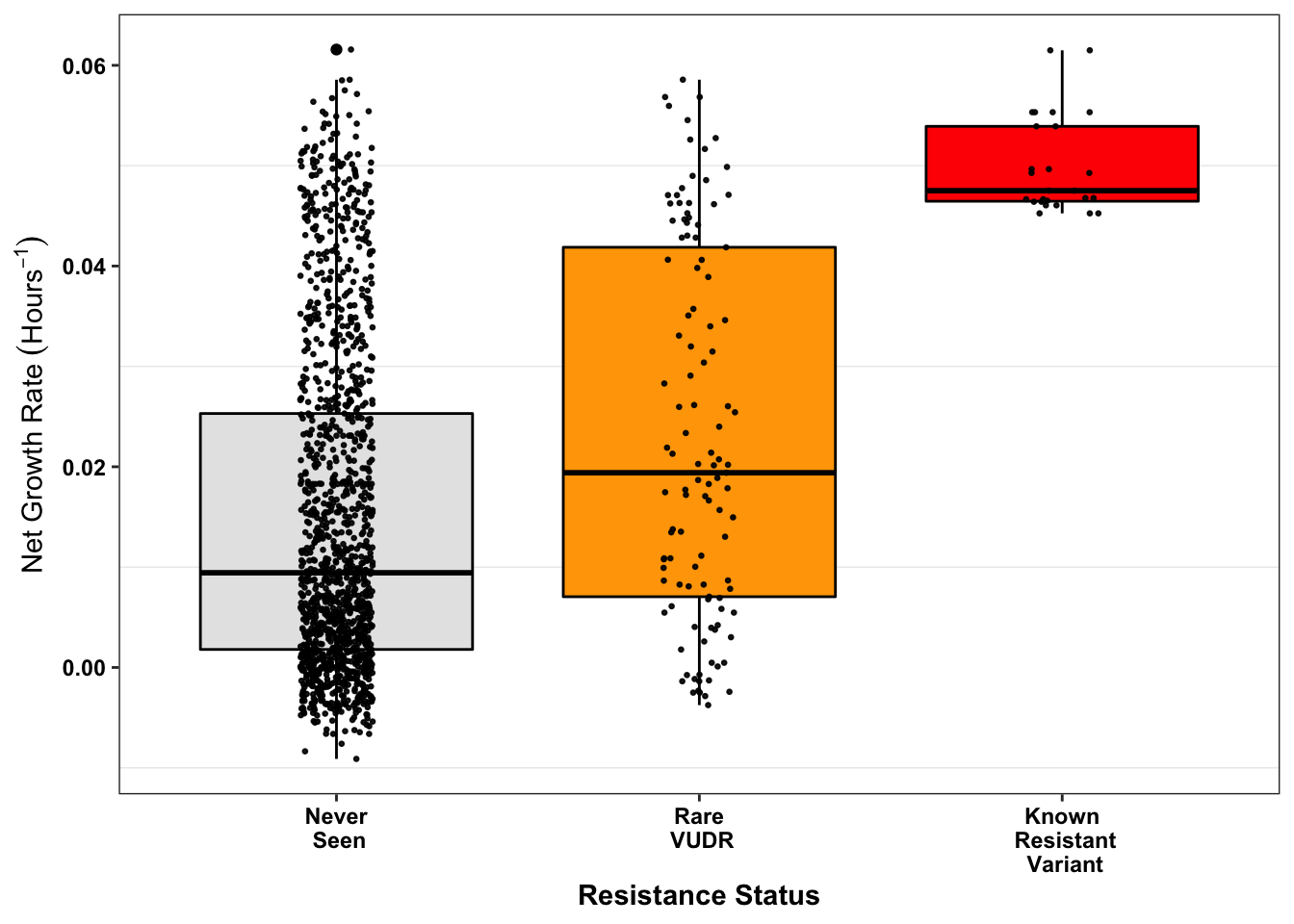
# ggsave("output/SM_Imatinib_Plots/ImatinibEnrichment_netgr_boxplot_baf3s_4.21.23.pdf",width=3,height=3.5,units="in",useDingbats=F)
ggplot(data%>%filter(protein_start>=242,protein_start<=322),aes(x=resmut_cosmic,y=netgr_obs_mean.k562,fill=resmut_cosmic))+
# geom_violin(color="black")+
geom_boxplot(color="black")+
geom_jitter(color="black", size=.5,width=.1, alpha=0.9)+
scale_fill_manual(values=c("gray90","orange","red"))+scale_y_continuous(name=bquote('Net Growth Rate '(Hours^-1)))+scale_x_discrete("Resistance Status",labels=c("Never\n Seen","Rare\n VUDR","Known\n Resistant\n Variant"))+cleanup+theme(legend.position = "none")Warning: Removed 26 rows containing non-finite values (stat_boxplot).Warning: Removed 26 rows containing missing values (geom_point).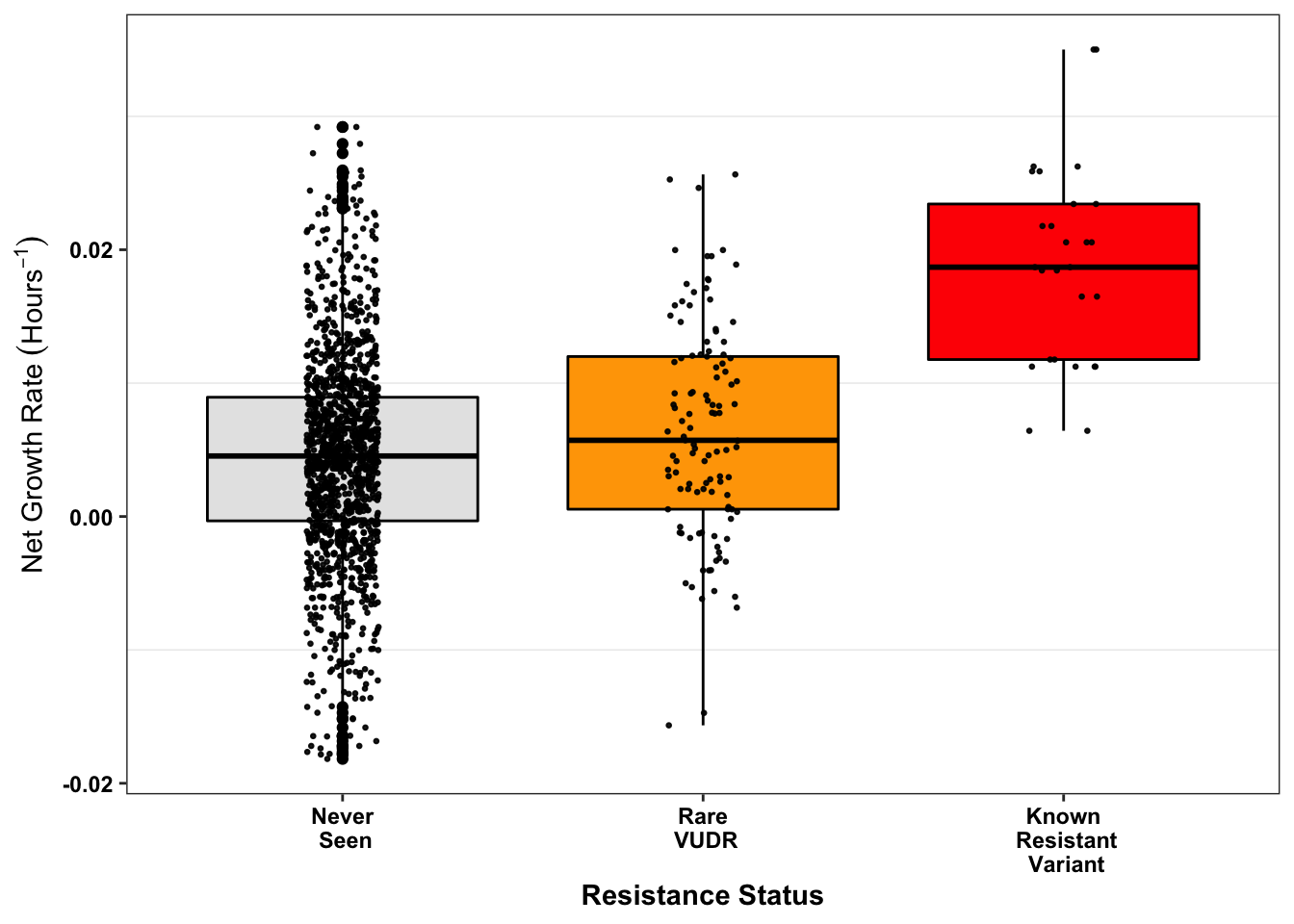
# ggsave("output/SM_Imatinib_Plots/ImatinibEnrichment_netgr_boxplot_k562s_4.21.23.pdf",width=3,height=3.5,units="in",useDingbats=F)
data.forttest=data%>%filter(protein_start>=242,protein_start<=322,n_nuc_min%in%1)
t.test(data.forttest[data.forttest$resmut_cosmic%in%"neither","netgr_obs_mean.baf3"],
data.forttest[data.forttest$resmut_cosmic%in%"cosmic","netgr_obs_mean.baf3"])
Welch Two Sample t-test
data: data.forttest[data.forttest$resmut_cosmic %in% "neither", "netgr_obs_mean.baf3"] and data.forttest[data.forttest$resmut_cosmic %in% "cosmic", "netgr_obs_mean.baf3"]
t = -2.1581, df = 158.9, p-value = 0.03242
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-0.0079228121 -0.0003510056
sample estimates:
mean of x mean of y
0.01752699 0.02166390 t.test(data.forttest[data.forttest$resmut_cosmic%in%"neither","netgr_obs_mean.baf3"],
data.forttest[data.forttest$resmut_cosmic%in%"resmut","netgr_obs_mean.baf3"])
Welch Two Sample t-test
data: data.forttest[data.forttest$resmut_cosmic %in% "neither", "netgr_obs_mean.baf3"] and data.forttest[data.forttest$resmut_cosmic %in% "resmut", "netgr_obs_mean.baf3"]
t = -25.642, df = 81.671, p-value < 2.2e-16
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-0.03481670 -0.02980322
sample estimates:
mean of x mean of y
0.01752699 0.04983696 t.test(data.forttest[data.forttest$resmut_cosmic%in%"neither","netgr_obs_mean.k562"],
data.forttest[data.forttest$resmut_cosmic%in%"cosmic","netgr_obs_mean.k562"])
Welch Two Sample t-test
data: data.forttest[data.forttest$resmut_cosmic %in% "neither", "netgr_obs_mean.k562"] and data.forttest[data.forttest$resmut_cosmic %in% "cosmic", "netgr_obs_mean.k562"]
t = -1.3254, df = 161.51, p-value = 0.1869
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-0.0027061696 0.0005325136
sample estimates:
mean of x mean of y
0.004696657 0.005783485 t.test(data.forttest[data.forttest$resmut_cosmic%in%"neither","netgr_obs_mean.k562"],
data.forttest[data.forttest$resmut_cosmic%in%"resmut","netgr_obs_mean.k562"])
Welch Two Sample t-test
data: data.forttest[data.forttest$resmut_cosmic %in% "neither", "netgr_obs_mean.k562"] and data.forttest[data.forttest$resmut_cosmic %in% "resmut", "netgr_obs_mean.k562"]
t = -9.6932, df = 29.511, p-value = 1.114e-10
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-0.01740694 -0.01134497
sample estimates:
mean of x mean of y
0.004696657 0.019072614 ggplot(data%>%filter(sanger.present%in%T,n_nuc_min%in%1),aes(x=netgr_obs_mean.baf3,y=netgr_obs_mean.k562))+
geom_point()+
stat_cor(method="pearson")Warning: Removed 2 rows containing non-finite values (stat_cor).Warning: Removed 2 rows containing missing values (geom_point).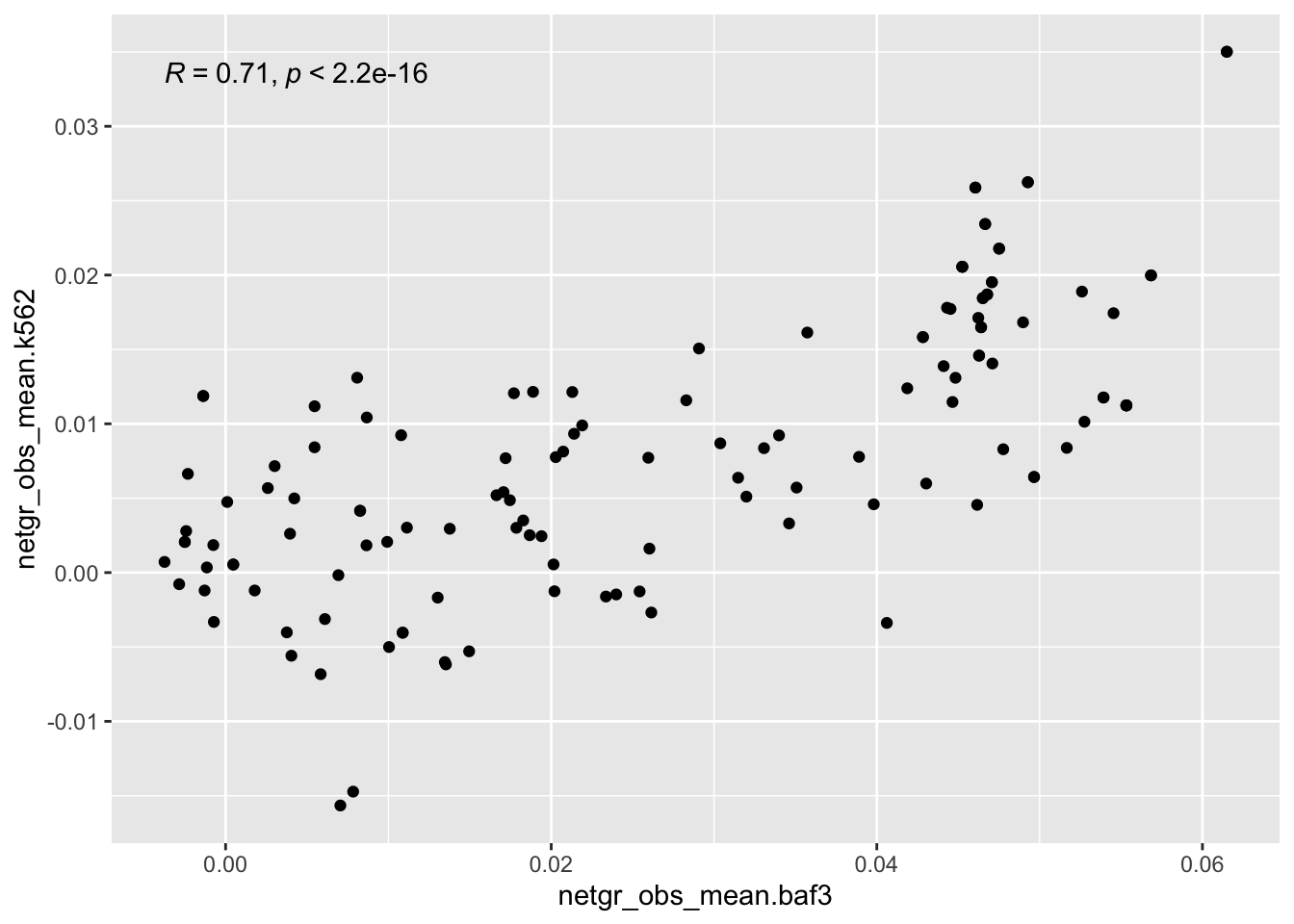
ggplot(data%>%filter(sanger.present%in%T,n_nuc_min%in%1),aes(x=netgr_obs_mean.baf3,y=netgr_obs_mean.k562,label=species,color=resmuts))+
geom_text(size=1.6)+
# stat_cor(method="pearson")+
cleanup+
scale_x_continuous("Net Growth Rate Baf3")+
scale_y_continuous("Net Growth Rate K562")+
scale_color_manual(values=c("orange","red"))+
theme(legend.position = "none")Warning: Removed 2 rows containing missing values (geom_text).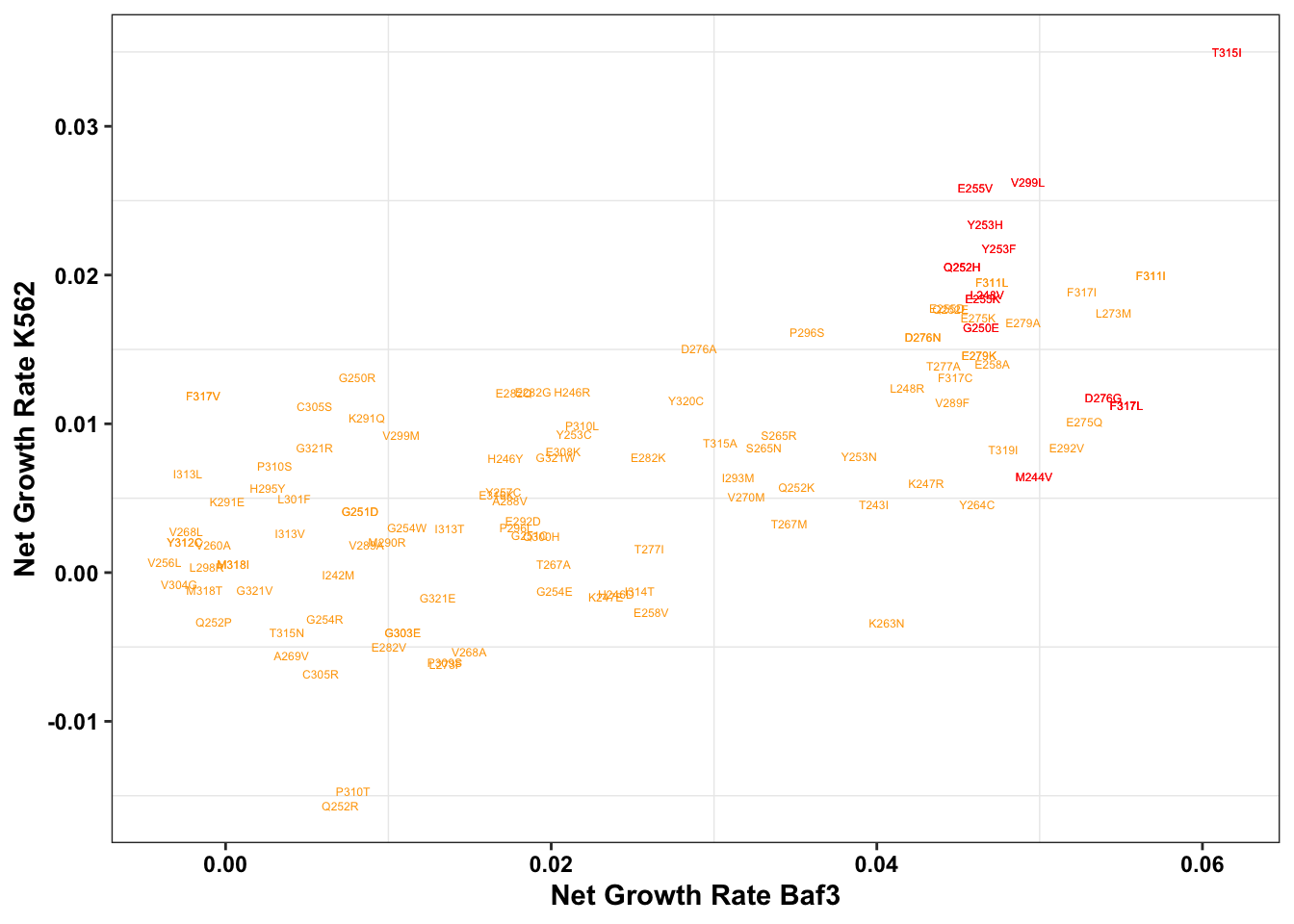
# ggsave("output/SM_Imatinib_Plots/baf3_k562_sanger_correlation.pdf",width=3,height=3,units = "in",useDingbats=F)
plotly=ggplot(data%>%filter(sanger.present%in%T,n_nuc_min%in%1),aes(x=netgr_obs_mean.baf3,y=netgr_obs_mean.k562,label=species,color=resmuts))+
geom_text()+
# stat_cor(method="pearson")+
cleanup+
scale_x_continuous("Net Growth Rate Baf3")+
scale_y_continuous("Net Growth Rate K562")+
scale_color_manual(values=c("orange","red"))+
theme(legend.position = "none")
ggplotly(plotly)x=data%>%filter(protein_start%in%c(242:322),n_nuc_min%in%1)%>%filter(resmut_cosmic%in%"neither")
x=data%>%filter(protein_start%in%c(242:322),n_nuc_min%in%1)%>%filter(resmut_cosmic%in%"cosmic")
x=data%>%filter(protein_start%in%c(242:322),n_nuc_min%in%1)%>%filter(resmut_cosmic%in%"cosmic")
# Making piechart that shows that VUDR take up 36%
sanger_forpiechart=resmuts_adder(sanger)
sanger_forpiechart$category="VUDR"
sanger_forpiechart=sanger_forpiechart%>%mutate(category=case_when(resmuts%in%T~species,
T~category))
sanger_forpiechart=sanger_forpiechart%>%group_by(category)%>%summarize(count=sum(sanger.count))
sanger_forpiechart=sanger_forpiechart%>%arrange(desc(count))
sanger_forpiechart$category=factor(sanger_forpiechart$category,levels = as.character(sanger_forpiechart$category[order((sanger_forpiechart$count),decreasing = T)]))
#Plotting the normalized dose response curves
getPalette = colorRampPalette(brewer.pal(22, "Spectral"))Warning in brewer.pal(22, "Spectral"): n too large, allowed maximum for palette Spectral is 11
Returning the palette you asked for with that many colorslibrary(RColorBrewer)
library("NatParksPalettes")
ggplot(sanger_forpiechart,aes(x="",y=count,fill=category))+
geom_bar(width=1,stat="identity")+
coord_polar("y", start=0)+
# theme_minimal()+
theme_void()+
# scale_fill_manual(values=natparks.pals("Yellowstone", 3))
scale_fill_manual(values = getPalette(length(unique(sanger_forpiechart$category))))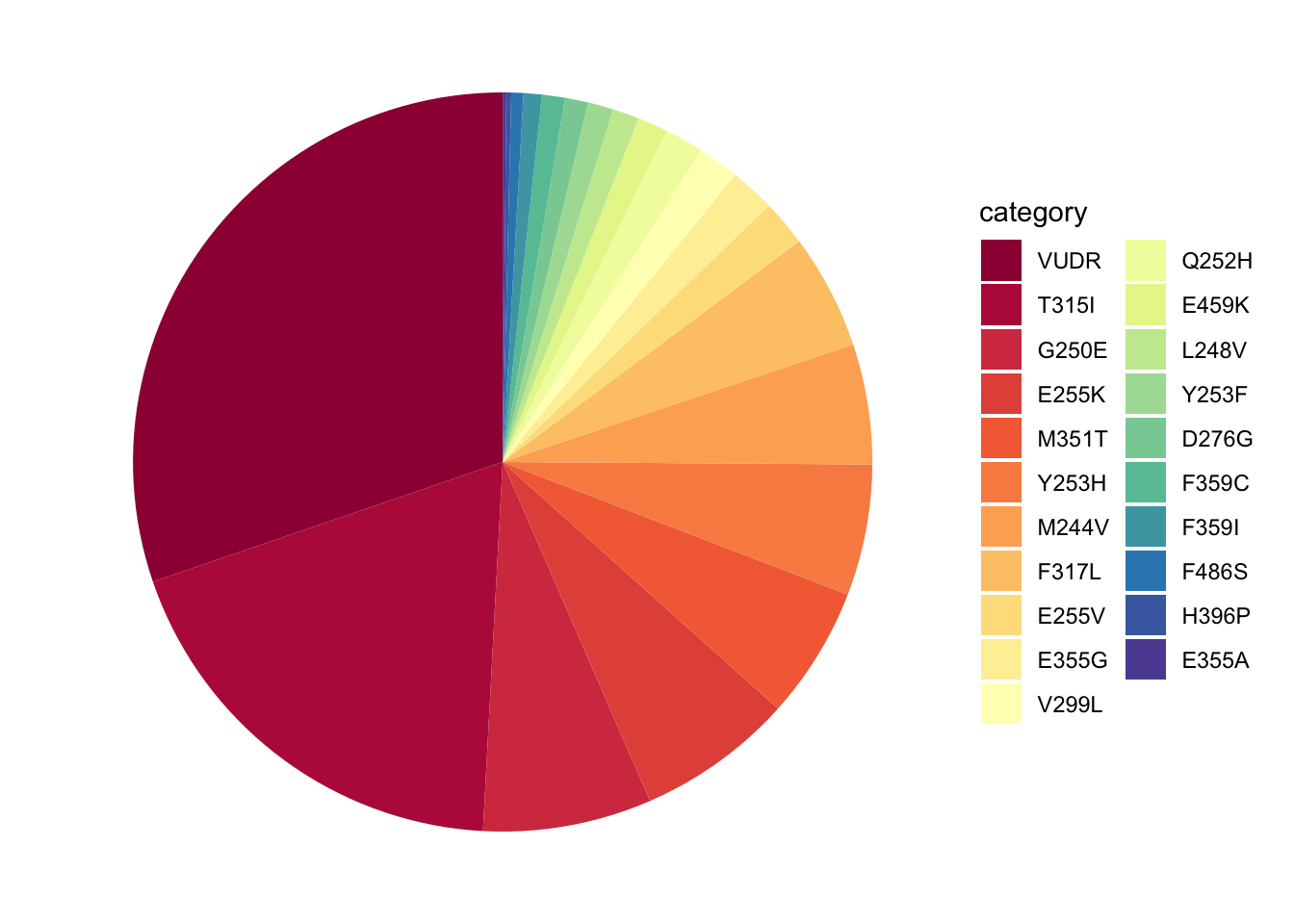
# ggsave("output/SM_Imatinib_Plots/sanger_piechart.pdf",width=4,height=3,units="in",useDingbats=F)
pie(sanger_forpiechart$count, labels = sanger_forpiechart$category)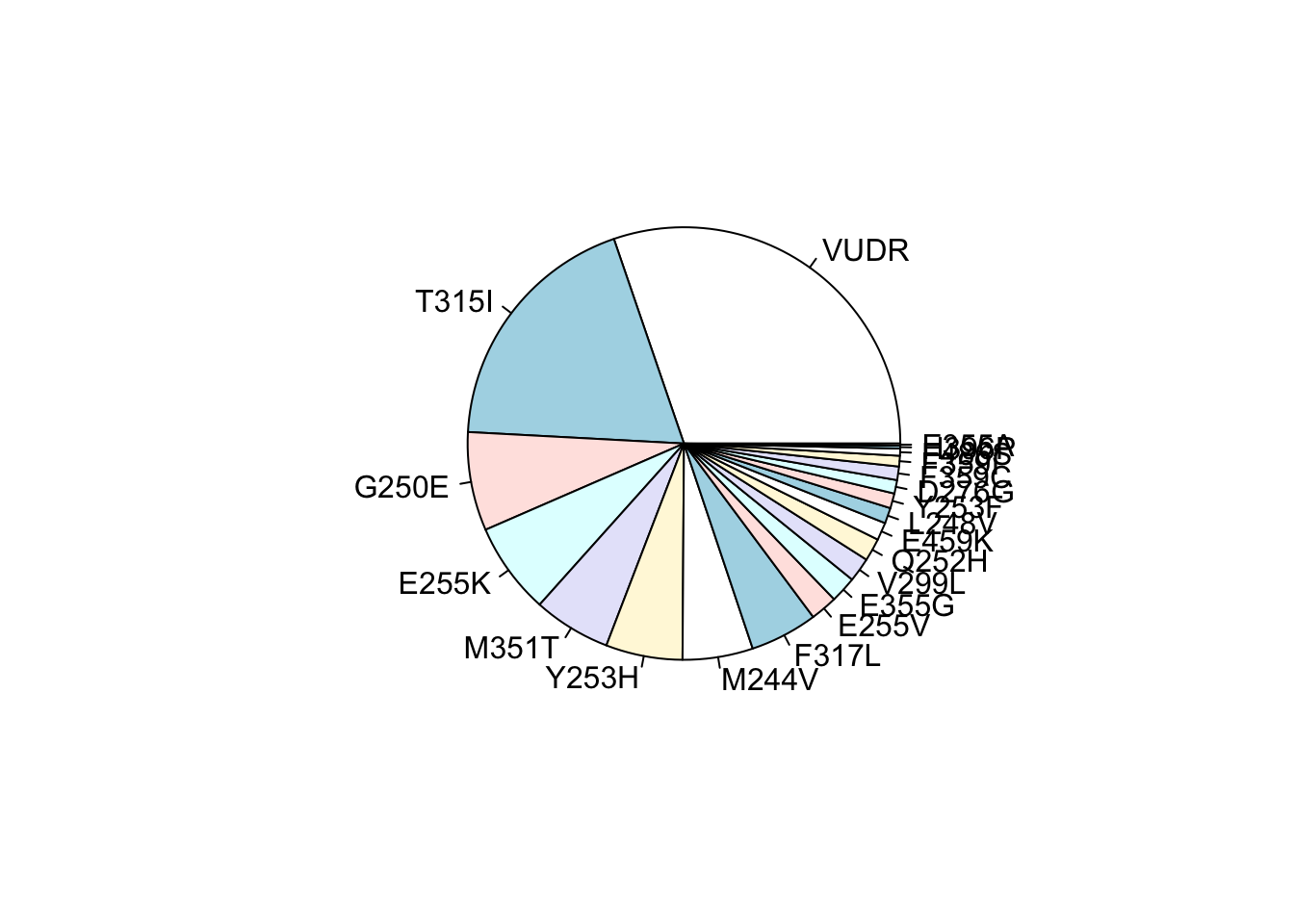
ggplot(data%>%filter(species%in%c("L273M","F311L","V260L","P296R","D276E", "K247W")),aes(x=netgr_obs_mean.baf3,y=netgr_obs_mean.k562,label=species,color=resmuts))+
geom_text(size=1.6)+
# stat_cor(method="pearson")+
cleanup+
scale_x_continuous("Net Growth Rate Baf3")+
scale_y_continuous("Net Growth Rate K562")+
scale_color_manual(values=c("orange","red"))+
theme(legend.position = "none")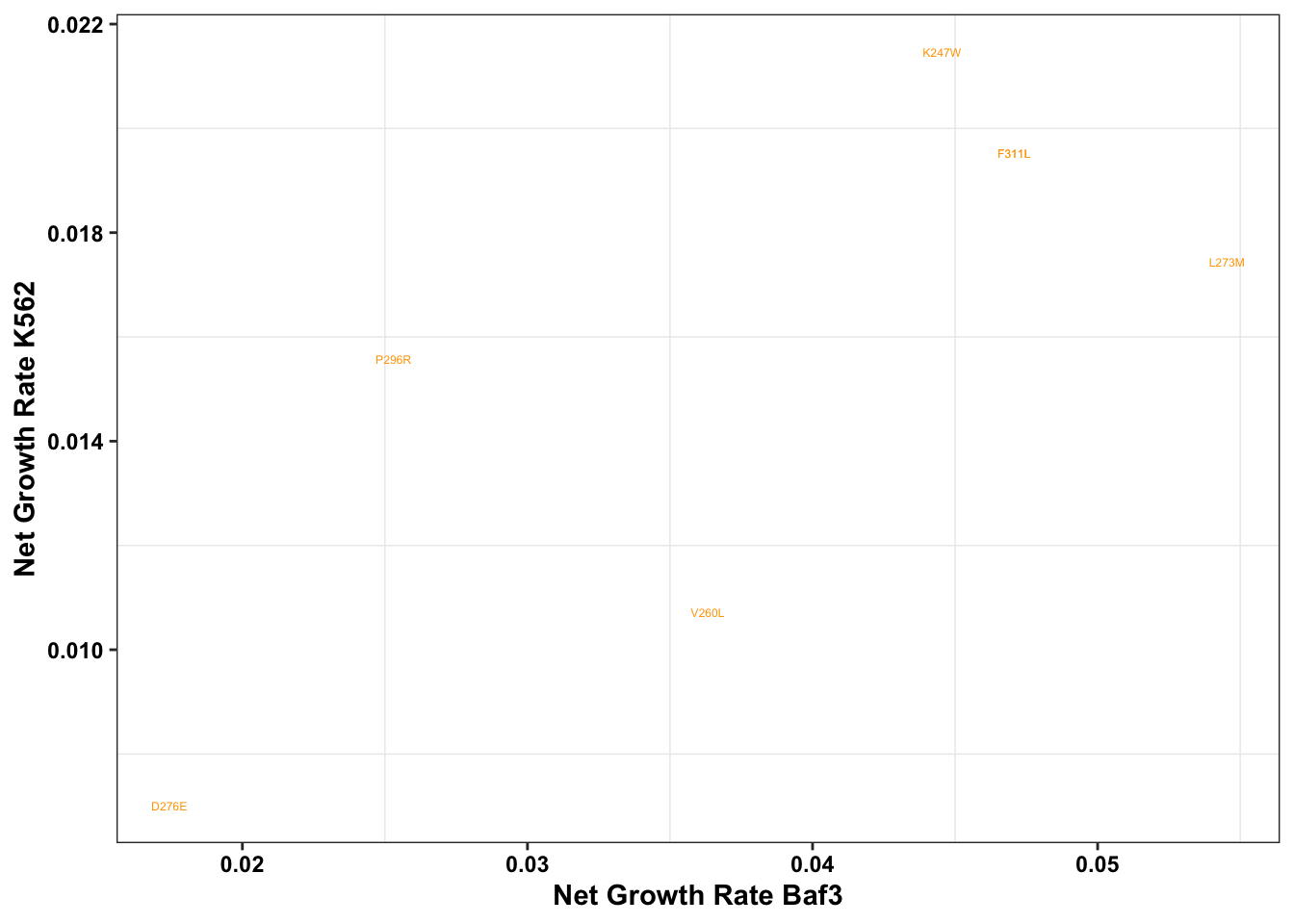 Making histogram of top clinically prevalent sanger mutants
Making histogram of top clinically prevalent sanger mutants
library(forcats)Warning: package 'forcats' was built under R version 4.0.2# Analyzing the Sanger data 4.22.2023
sanger=read.csv("data/Cosmic_ABL/ABL_Cosmic_Gene_mutations.csv",header = T)
sanger=sanger%>%dplyr::rename(protein_start=Position,species=AA.Mutation,sanger.count=Count)%>%filter(Type%in%"Substitution - Missense")%>%dplyr::select(protein_start,species,sanger.count)
sanger=sanger%>%mutate(species=gsub("p.","",species),ref_aa=substring(species,1,1),
alt_aa=substring(species, nchar(species), nchar(species)))
sanger=sanger%>%mutate(sanger.present=T)
sanger=sanger%>%group_by(species)%>%summarize(protein_start=mean(protein_start),sanger.count=sum(sanger.count))
sanger$resmuts=F
sanger[sanger$species%in%c("E355A","D276G","H396R","F317L","F359I","E459K","G250E","F359C","F359V","M351T","L248V","E355G","Q252H","Y253F","F486S","H396P","E255K","Y253H","T315I","E255V","M244V","V299L","L387M"),"resmuts"]=T
ggplot(sanger%>%filter(resmuts%in%T),aes(fct_reorder(species, -sanger.count),y=sanger.count,fill=resmuts))+
geom_col(color="black",alpha=0.8)+
scale_x_discrete("Mutant Name")+
scale_y_continuous("Clinical Abundance")+
scale_fill_manual(values=c("red"))+
cleanup+
theme(legend.position = "none",
axis.text.x=element_text(angle=90,hjust=.5,vjust=.5),
axis.title.x=element_blank())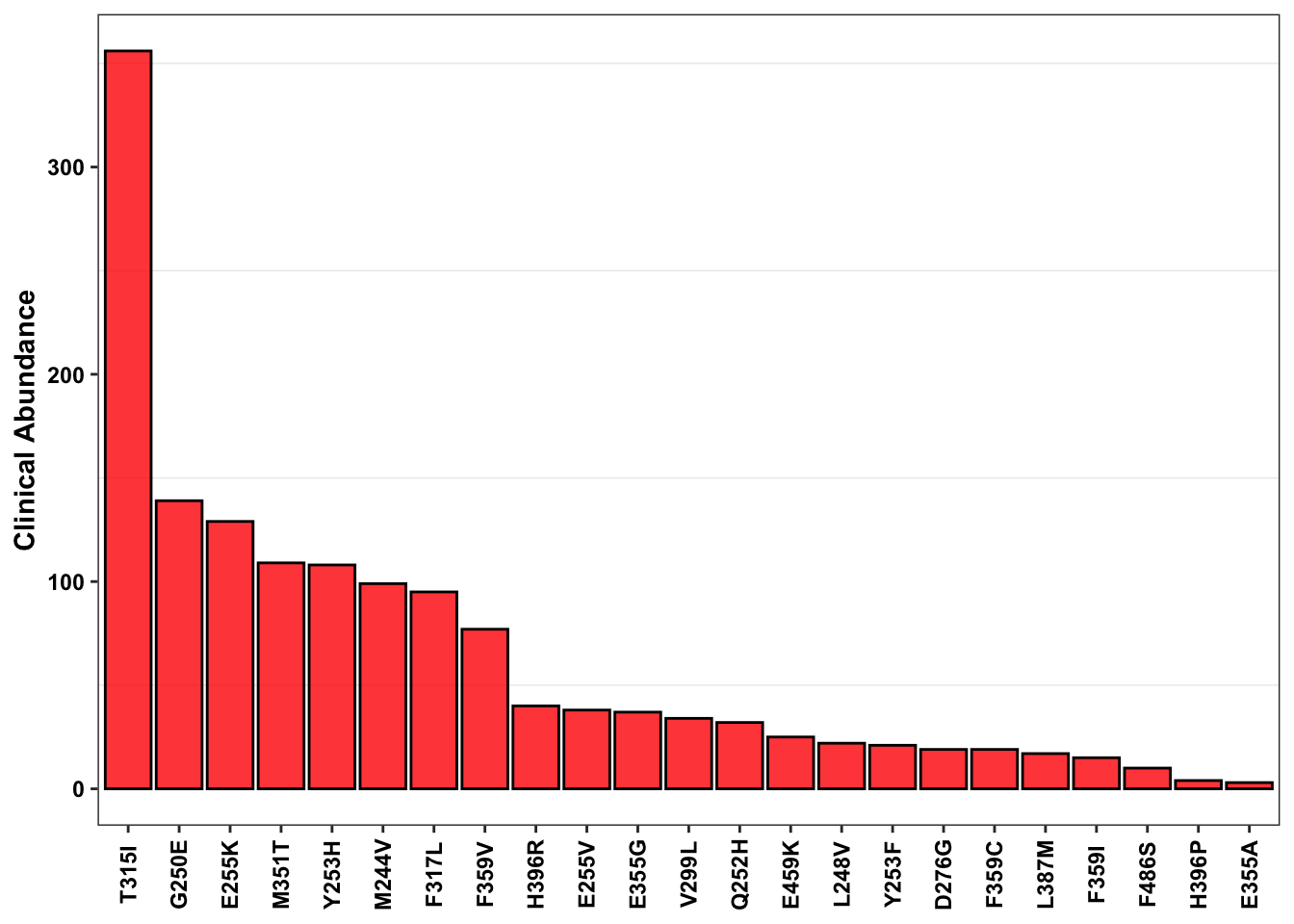
# ggsave("output/SM_Imatinib_Plots/clinical_abundance_zoom.pdf",width=5,height=3,units="in",useDingbats=F)
ggplot(sanger,aes(fct_reorder(species, -sanger.count),y=sanger.count,fill=resmuts))+
geom_col(alpha=0.8)+
scale_x_discrete("Mutant Name")+
scale_y_continuous("Clinical Abundance")+
scale_fill_manual(values=c("gray90","red"))+
# cleanup+
theme(legend.position = "none",
axis.text.x=element_text(size=2,angle=90,hjust=.5,vjust=.5),
axis.title.x=element_blank())
# ggsave("output/SM_Imatinib_Plots/clinical_abundance.pdf",width=14,height=3,units="in",useDingbats=F)
ggplot(sanger,aes(fct_reorder(species, -sanger.count),y=sanger.count,fill=resmuts))+
geom_col()+
scale_x_discrete("Mutant Name")+
scale_y_continuous("Clinical Abundance")+
scale_fill_manual(values=c("gray90","red"))+
theme(legend.position = "none",
axis.text.x=element_text(angle=90,hjust=.5,vjust=.5),
axis.title.x=element_blank())
Heatmaps
smdata=read.csv("output/ABLEnrichmentScreens/ABL_Region123_MergedwithOlddata/IL3_Enrichment_bgmerged_4.19.23.csv")
# Baf3s
smdata=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/baf3_Imat_low_rep1vsrep2_ft/screen_comparison_baf3_Imat_low_rep1vsrep2_ft.csv")
smdata=smdata%>%rowwise()%>%mutate(netgr_obs_mean=mean(netgr_obs_screen1,netgr_obs_screen2))
heatmap_plotting_function(smdata,290,297,fill_variable = "netgr_obs_mean",fill_name = "Net growth rate")+
scale_x_continuous(name="",expand=c(0,0),breaks=c(290:297),labels=c("290","291","292","293","294","295","296","297"))+
scale_fill_gradient(low ="darkblue", high ="red",name="Net growth rate")+theme(legend.position = "none",axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1))Scale for 'x' is already present. Adding another scale for 'x', which will
replace the existing scale.Scale for 'fill' is already present. Adding another scale for 'fill', which
will replace the existing scale.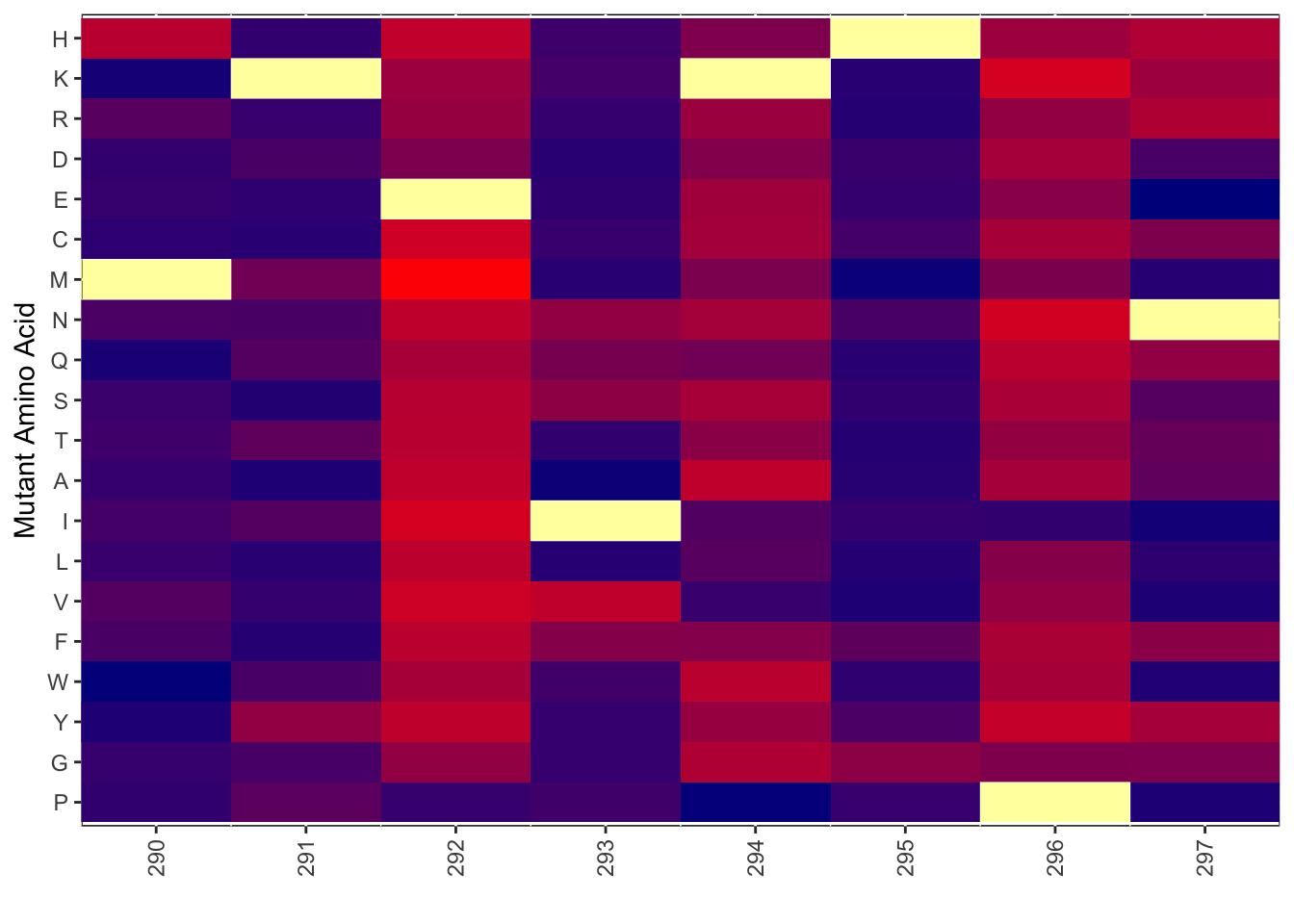
heatmap_plotting_function(smdata%>%mutate(netgr_obs_mean=case_when(netgr_obs_mean>=.06~.06,
T~netgr_obs_mean)),242,321,fill_variable = "netgr_obs_mean",fill_name = "Net growth rate")+
scale_x_continuous(name="Residue on the ABL Kinase",limits=c(242,321),expand=c(0,0))+
# scale_fill_gradient2(low ="darkblue",mid = "white",midpoint=0.04, high ="red",name="Net growth rate")+
# scale_fill_gradient(low ="lightblue", high ="red",name="Net growth rate")+
scale_fill_gradient2(low="blue",mid ="#9999FF",midpoint=.035, high ="red",name="Net growth rate")+theme(legend.position = "none")Scale for 'x' is already present. Adding another scale for 'x', which will
replace the existing scale.
Scale for 'fill' is already present. Adding another scale for 'fill', which
will replace the existing scale.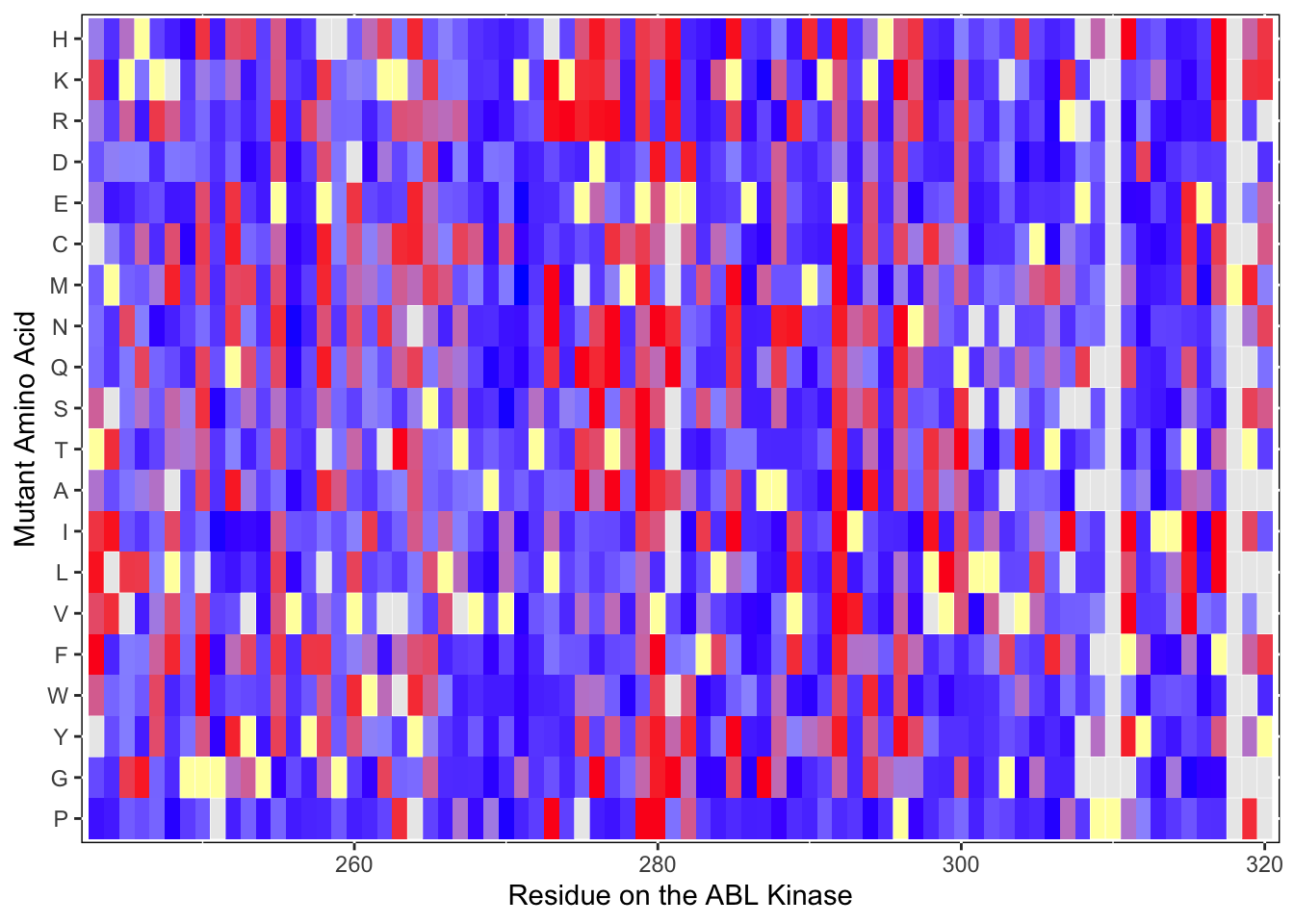
# ggsave("output/SM_Imatinib_Plots/heatmap_Imatinib_baf3.pdf",width=6,height = 4,units = "in",useDingbats=F)
####### K562 Low#######
smdata=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/k562_imat_low_rep1vsrep2/screen_comparison_k562_imat_low_rep1vsrep2.csv")
smdata=smdata%>%rowwise()%>%mutate(netgr_obs_mean=mean(netgr_obs_screen1,netgr_obs_screen2))
heatmap_plotting_function(smdata%>%mutate(netgr_obs_mean=case_when(netgr_obs_mean>=.045~.045,
T~netgr_obs_mean)),242,321,fill_variable = "netgr_obs_mean",fill_name = "Net growth rate")+
scale_x_continuous(name="Residue on the ABL Kinase",limits=c(242,321),expand=c(0,0))+
# scale_fill_gradient2(low ="darkblue",mid = "white",midpoint=0.04, high ="red",name="Net growth rate")+
# scale_fill_gradient(low ="lightblue", high ="red",name="Net growth rate")+
scale_fill_gradient2(low="blue",mid ="#9999FF",midpoint=.02, high ="red",name="Net growth rate")+theme(legend.position = "none")Scale for 'x' is already present. Adding another scale for 'x', which will
replace the existing scale.
Scale for 'fill' is already present. Adding another scale for 'fill', which
will replace the existing scale.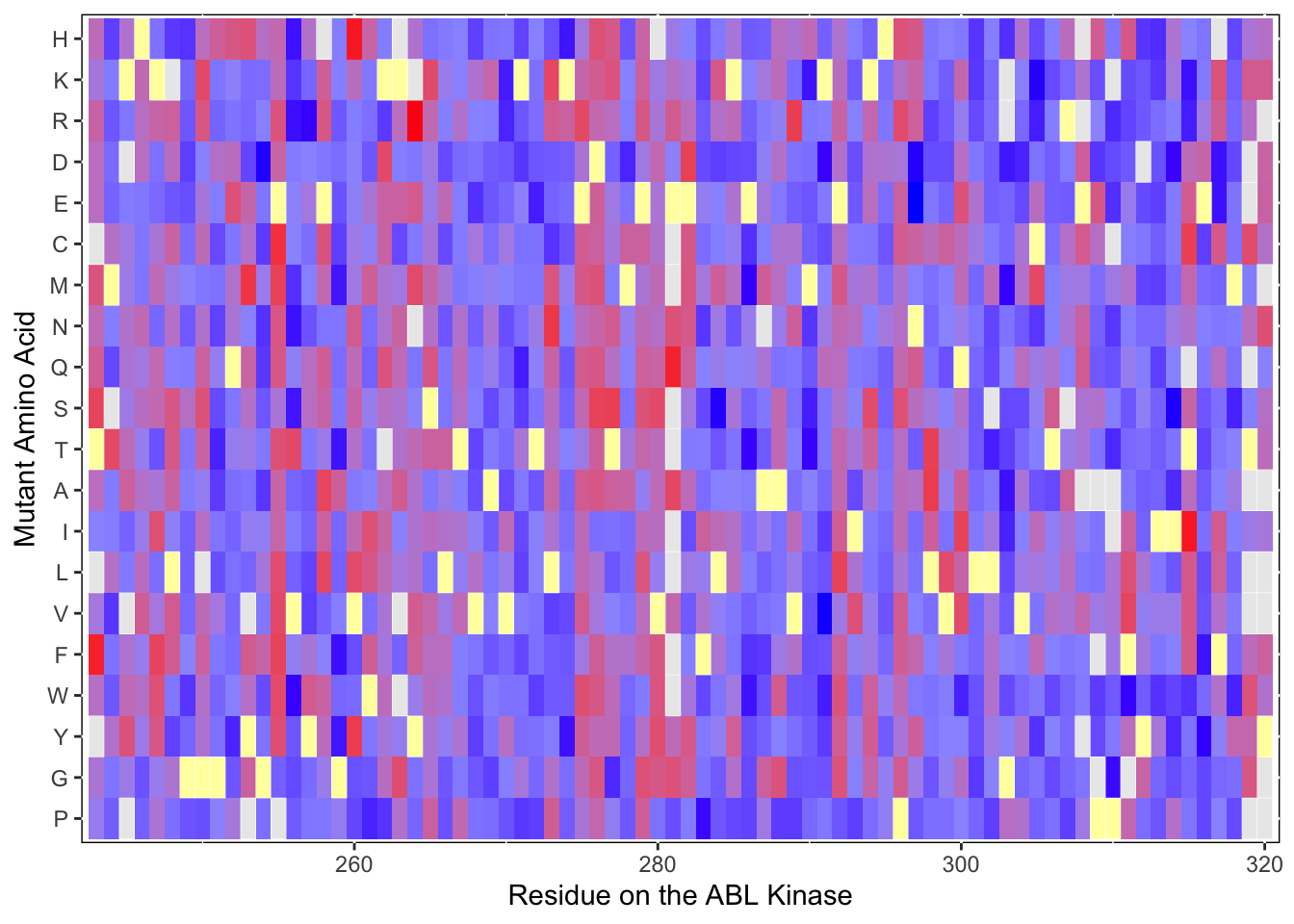
# ggsave("output/SM_Imatinib_Plots/heatmap_Imatinib_k562low.pdf",width=6,height = 4,units = "in",useDingbats=F)
####### K562 Medium#######
smdata=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/k562_imat_medium_rep1vsrep2/screen_comparison_k562_imat_medium_rep1vsrep2.csv")
smdata=smdata%>%rowwise()%>%mutate(netgr_obs_mean=mean(netgr_obs_screen1,netgr_obs_screen2))
heatmap_plotting_function(smdata,242,321,fill_variable = "netgr_obs_mean",fill_name = "Net growth rate")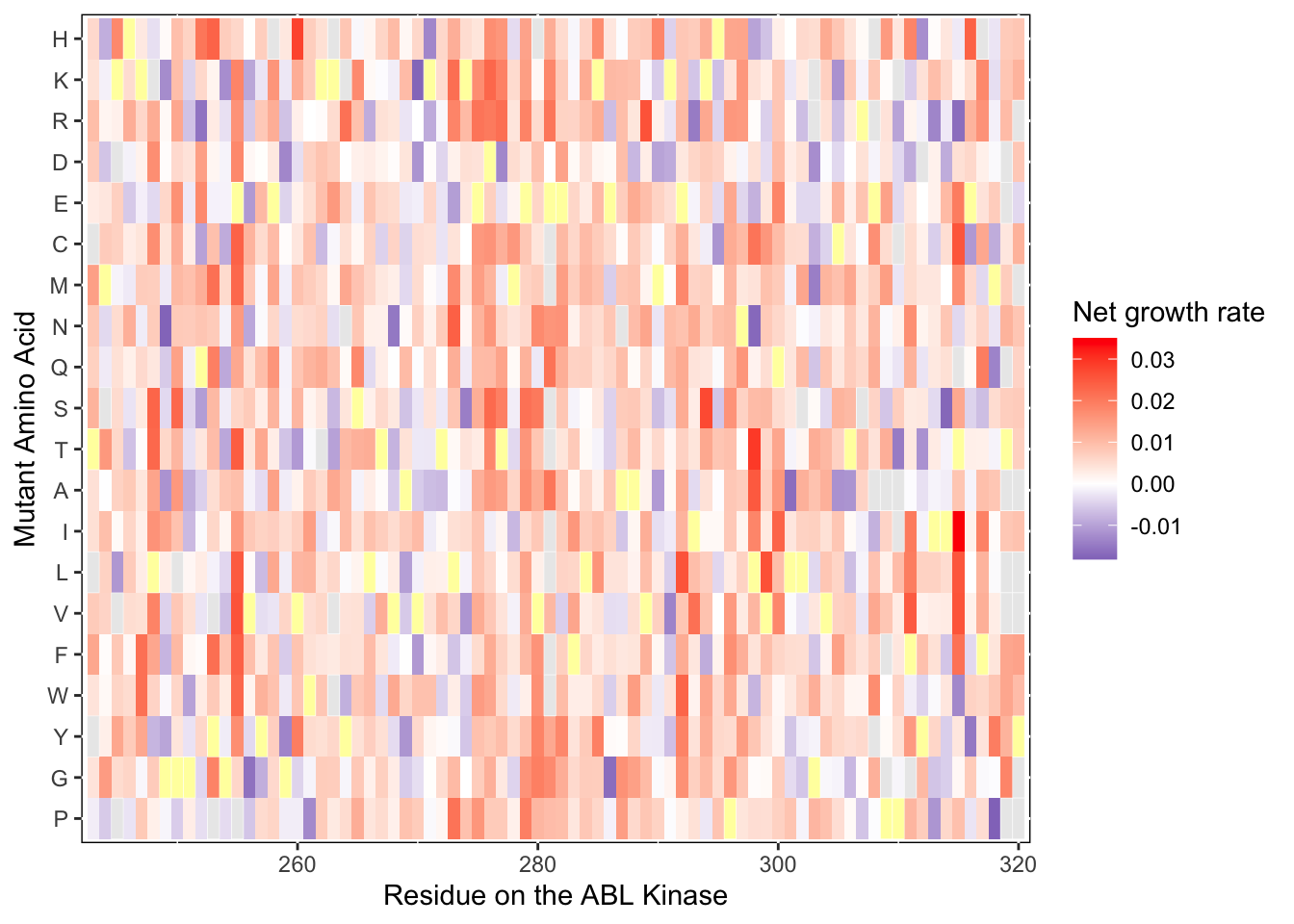
heatmap_plotting_function(smdata,242,321,fill_variable = "netgr_obs_mean",fill_name = "Net growth rate")+
scale_x_continuous(name="Residue on the ABL Kinase",limits=c(242,321),expand=c(0,0))+
# scale_fill_gradient2(low ="darkblue",mid = "white",midpoint=0.04, high ="red",name="Net growth rate")+
# scale_fill_gradient(low ="lightblue", high ="red",name="Net growth rate")+
scale_fill_gradient2(low="blue",mid ="#9999FF",midpoint=0, high ="red",name="Net growth rate")+theme(legend.position = "none")Scale for 'x' is already present. Adding another scale for 'x', which will
replace the existing scale.
Scale for 'fill' is already present. Adding another scale for 'fill', which
will replace the existing scale.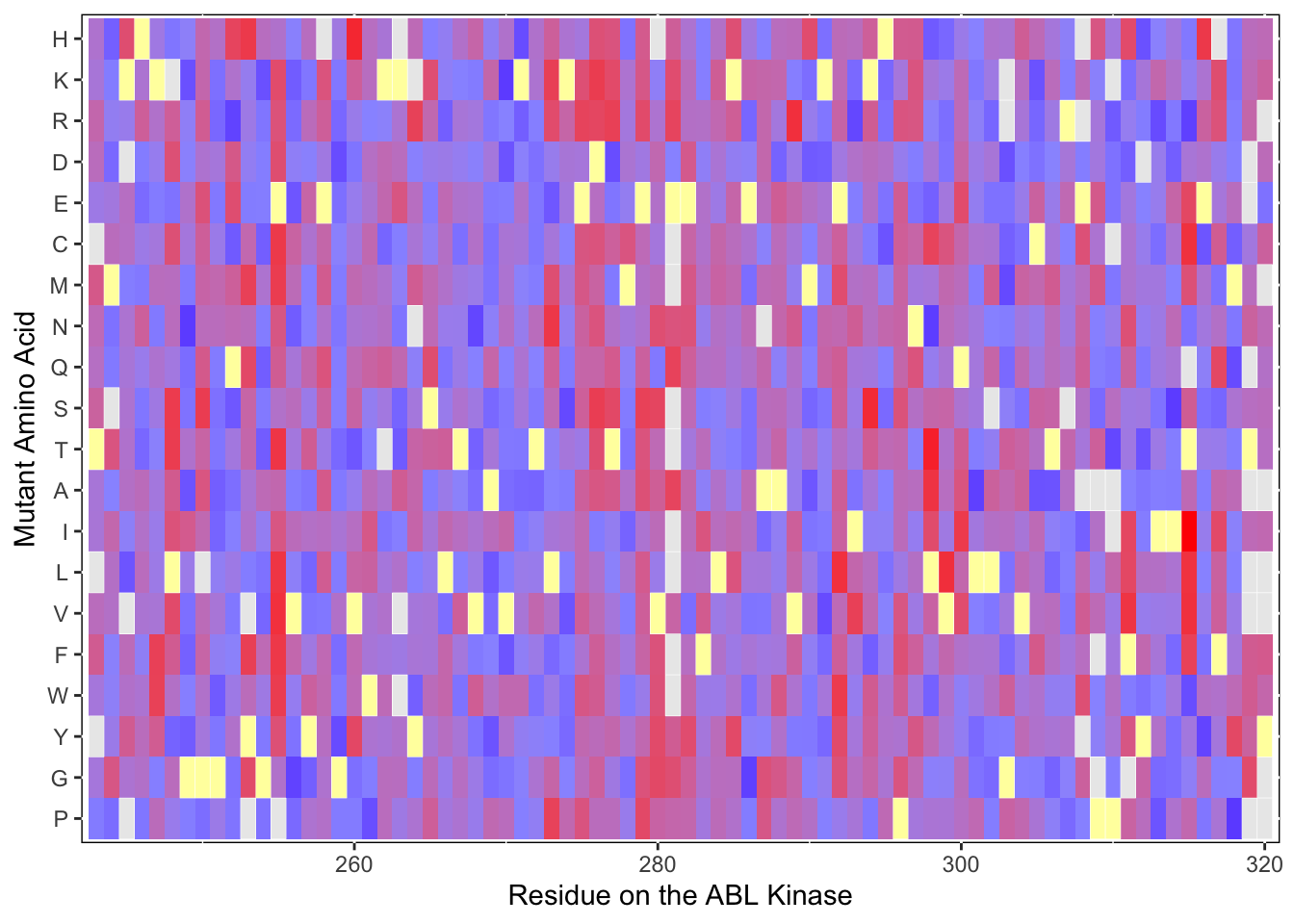
# ggsave("output/SM_Imatinib_Plots/heatmap_Imatinib_k562med.pdf",width=6,height = 4,units = "in",useDingbats=F)
####### K562 High#######
smdata=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/k562_imat_high_rep1vsrep2/screen_comparison_k562_imat_high_rep1vsrep2.csv")
smdata=smdata%>%rowwise()%>%mutate(netgr_obs_mean=mean(netgr_obs_screen1,netgr_obs_screen2))
heatmap_plotting_function(smdata,242,321,fill_variable = "netgr_obs_mean",fill_name = "Net growth rate")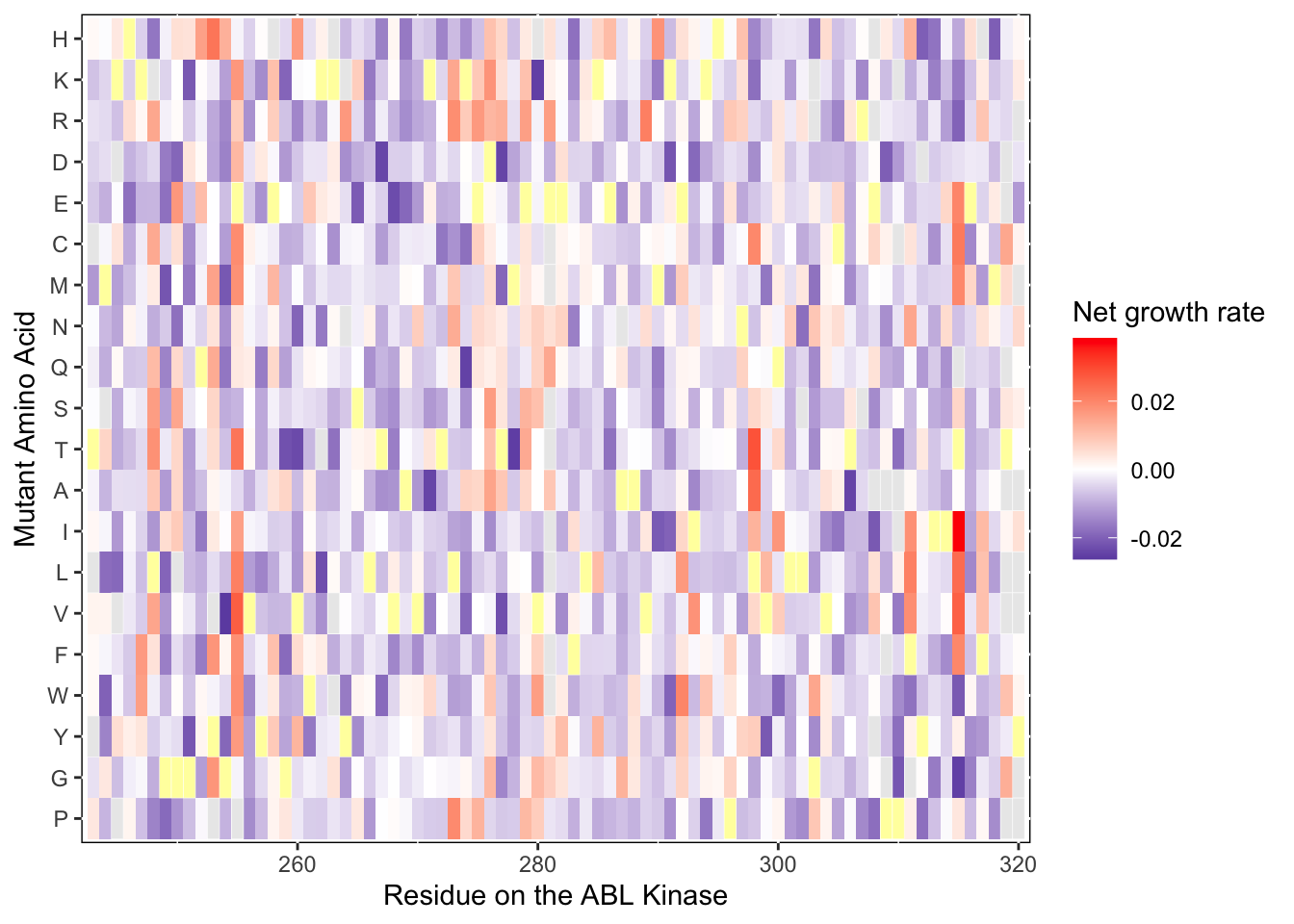
heatmap_plotting_function(smdata,242,321,fill_variable = "netgr_obs_mean",fill_name = "Net growth rate")+
scale_x_continuous(name="Residue on the ABL Kinase",limits=c(242,321),expand=c(0,0))+
# scale_fill_gradient2(low ="darkblue",mid = "white",midpoint=0.04, high ="red",name="Net growth rate")+
# scale_fill_gradient(low ="lightblue", high ="red",name="Net growth rate")+
scale_fill_gradient2(low="blue",mid ="#9999FF",midpoint=-0.005, high ="red",name="Net growth rate")+theme(legend.position = "none")Scale for 'x' is already present. Adding another scale for 'x', which will
replace the existing scale.
Scale for 'fill' is already present. Adding another scale for 'fill', which
will replace the existing scale.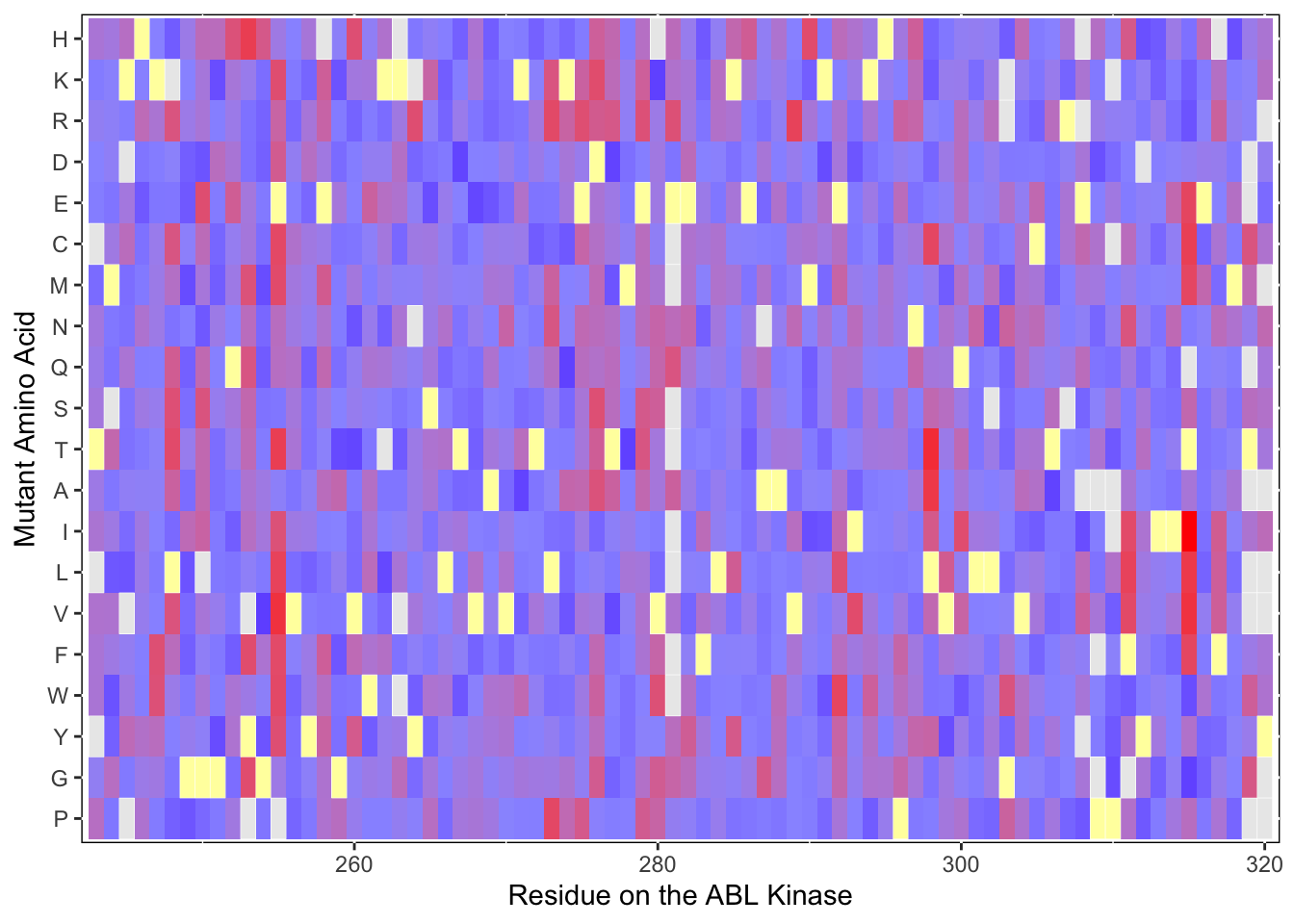
# ggsave("output/SM_Imatinib_Plots/heatmap_Imatinib_k562high.pdf",width=6,height = 4,units = "in",useDingbats=F)smdata.k562=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/k562_imat_high_rep1vsrep2/screen_comparison_k562_imat_high_rep1vsrep2.csv")
# smdata=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/k562_imat_low_rep1vsrep2/screen_comparison_k562_imat_low_rep1vsrep2.csv")
# smdata=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/k562_imat_medium_rep1vsrep2/screen_comparison_k562_imat_medium_rep1vsrep2.csv")
smdata=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/baf3_imat_low_rep1vsrep2_ft/screen_comparison_baf3_imat_low_rep1vsrep2_ft.csv")
# smdata=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/baf3_imat_medium_rep1vsrep2_ft/screen_comparison_baf3_imat_medium_rep1vsrep2_ft.csv")
smdata.baf3=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/baf3_imat_high_rep1vsrep2_ft/screen_comparison_baf3_imat_high_rep1vsrep2_ft.csv")
smdata.baf3=cosmic_data_adder(smdata.baf3)
smdata.k562=cosmic_data_adder(smdata.k562)
smdata.baf3=smdata%>%filter(!species%in%"V299L")
smdata.baf3=smdata.baf3%>%filter(n_nuc_min%in%1)
smdata.k562=smdata.k562%>%filter(n_nuc_min%in%1)
smdata.baf3=smdata.baf3%>%rowwise()%>%mutate(netgr_obs_mean=mean(netgr_obs_screen1,netgr_obs_screen2))
smdata.baf3=smdata.baf3%>%rowwise()%>%mutate(netgr_obs_mean=mean(netgr_obs_screen1,netgr_obs_screen2))
# Demonstrating that the SM data works better at predicting residue exposure than the BE data
# smdata_roc=smdata%
# glm.fit.sm=glm(as.numeric(smdata$cosmic_present)~smdata$netgr_obs_mean,family=binomial)
glm.fit.baf3=glm(as.numeric(smdata.baf3$resmuts)~smdata.baf3$netgr_obs_mean,family=binomial)
glm.fit.k562=glm(as.numeric(smdata.k562$resmuts)~smdata.k562$netgr_obs_mean,family=binomial)
smdata.baf3$glm_fits=glm.fit.baf3$fitted.values
smdata.k562$glm_fits=glm.fit.k562$fitted.values
# roc(as.numeric(smdata$cosmic_present),smdata$glm_fits,plot=T,legacy.axes=T,percent=T,xlab="False Positive Percentage",ylab="True
# Positive Percentage",print.auc=T)
roc(as.numeric(smdata.baf3$resmuts),smdata.baf3$glm_fits,plot=T,legacy.axes=T,percent=T,xlab="False Positive Percentage",ylab="True Positive Percentage",print.auc=T)Setting levels: control = 0, case = 1Setting direction: controls < cases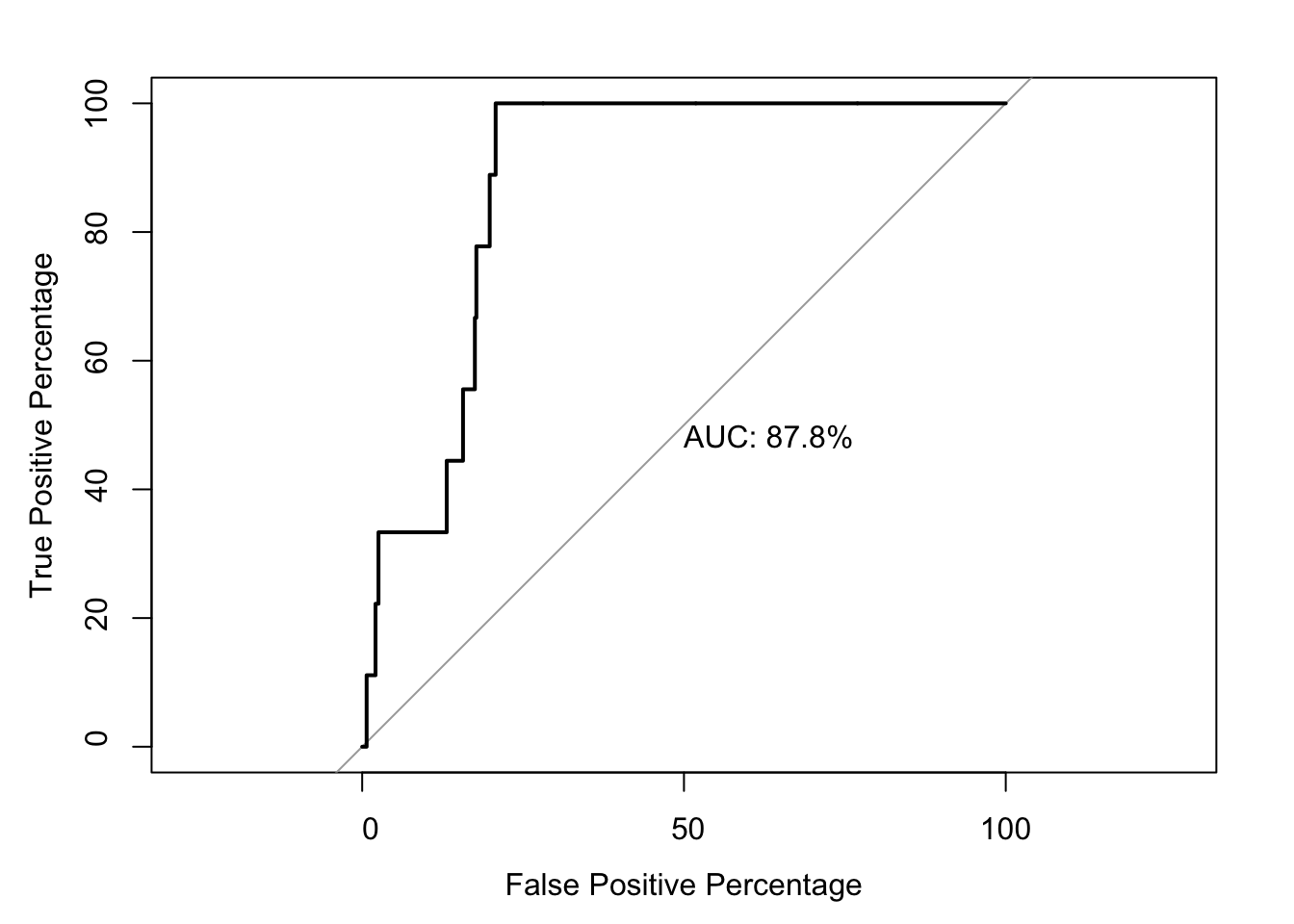
Call:
roc.default(response = as.numeric(smdata.baf3$resmuts), predictor = smdata.baf3$glm_fits, percent = T, plot = T, legacy.axes = T, xlab = "False Positive Percentage", ylab = "True Positive Percentage", print.auc = T)
Data: smdata.baf3$glm_fits in 434 controls (as.numeric(smdata.baf3$resmuts) 0) < 9 cases (as.numeric(smdata.baf3$resmuts) 1).
Area under the curve: 87.79%roc(as.numeric(smdata.k562$resmuts),smdata.k562$glm_fits,plot=T,legacy.axes=T,percent=T,xlab="False Positive Percentage",ylab="True Positive Percentage",print.auc=T)Setting levels: control = 0, case = 1
Setting direction: controls < cases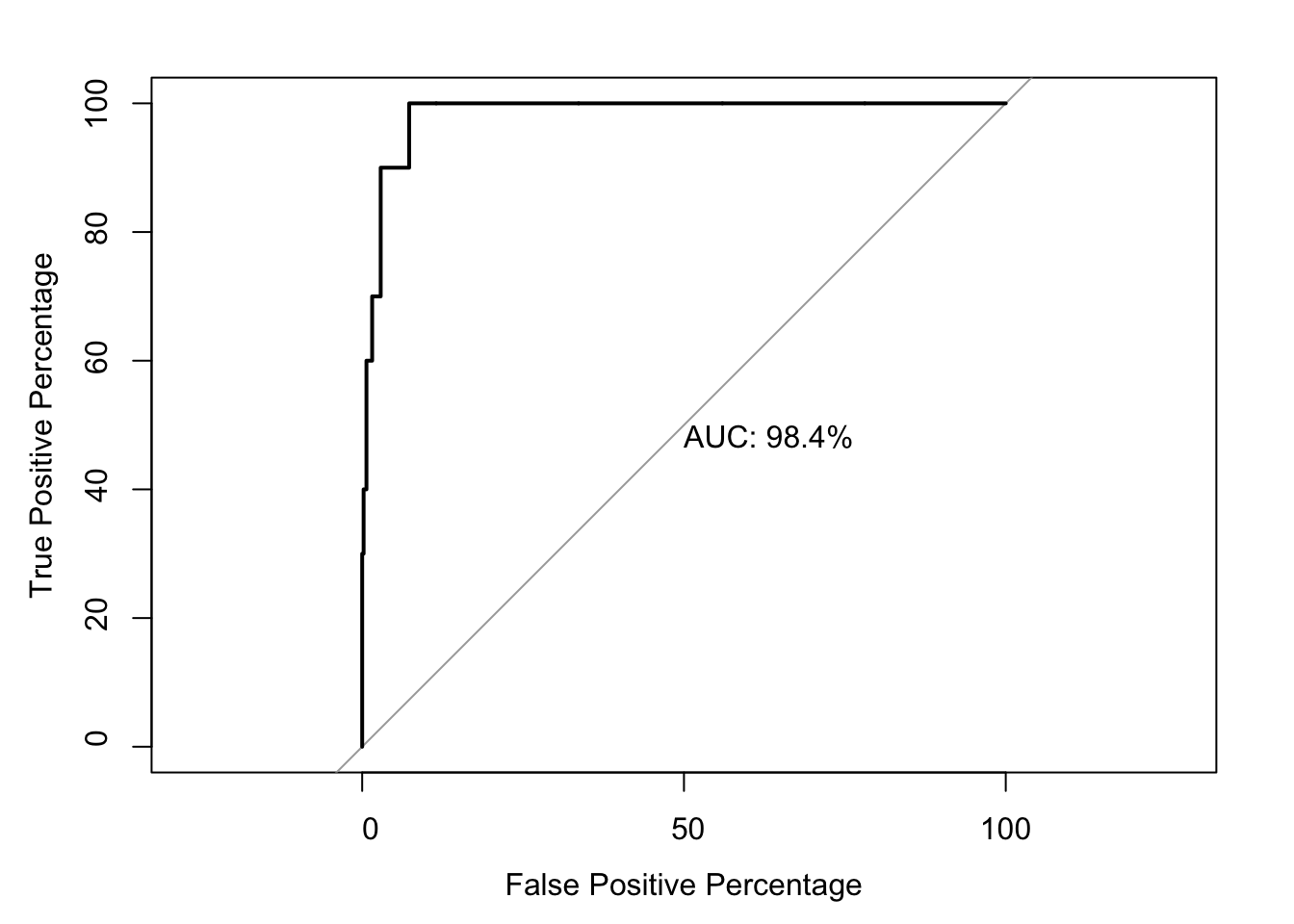
Call:
roc.default(response = as.numeric(smdata.k562$resmuts), predictor = smdata.k562$glm_fits, percent = T, plot = T, legacy.axes = T, xlab = "False Positive Percentage", ylab = "True Positive Percentage", print.auc = T)
Data: smdata.k562$glm_fits in 452 controls (as.numeric(smdata.k562$resmuts) 0) < 10 cases (as.numeric(smdata.k562$resmuts) 1).
Area under the curve: 98.38%# The GLM in this case looks a little weird, probably because there are so many negative datapoints
# ggplot(smdata.baf3,aes(x=netgr_obs_mean,y=as.numeric(resmuts)))+geom_point()+geom_line(aes(x=netgr_obs_mean,y=glm_fits))
# ggplot(smdata.k562,aes(x=netgr_obs_mean,y=as.numeric(resmuts)))+geom_point()+geom_line(aes(x=netgr_obs_mean,y=glm_fits))
par(pty="s")
roc(as.numeric(smdata.baf3$resmuts),smdata.baf3$glm_fits,plot=T,legacy.axes=T,percent=T,xlab="False Positive Percentage",ylab="True Positive Percentage",print.auc=T,col="#3937E3",lwd=4)Setting levels: control = 0, case = 1
Setting direction: controls < cases
Call:
roc.default(response = as.numeric(smdata.baf3$resmuts), predictor = smdata.baf3$glm_fits, percent = T, plot = T, legacy.axes = T, xlab = "False Positive Percentage", ylab = "True Positive Percentage", print.auc = T, col = "#3937E3", lwd = 4)
Data: smdata.baf3$glm_fits in 434 controls (as.numeric(smdata.baf3$resmuts) 0) < 9 cases (as.numeric(smdata.baf3$resmuts) 1).
Area under the curve: 87.79%# plot.roc(as.numeric(be_lfc.sm$DSSP.Buried),be_lfc.sm$glm_fits,legacy.axes=T,percent=T,xlab="False Positive Percentage",ylab="True Positive Percentage",print.auc=T,col="#378BE3",add=T,lwd=4,print.auc.y=45)
plot.roc(as.numeric(smdata.k562$resmuts),smdata.k562$glm_fits,legacy.axes=T,percent=T,xlab="False Positive Percentage",ylab="True Positive Percentage",print.auc=T,col="#378BE3",add=T,lwd=4,print.auc.y=45)Setting levels: control = 0, case = 1
Setting direction: controls < cases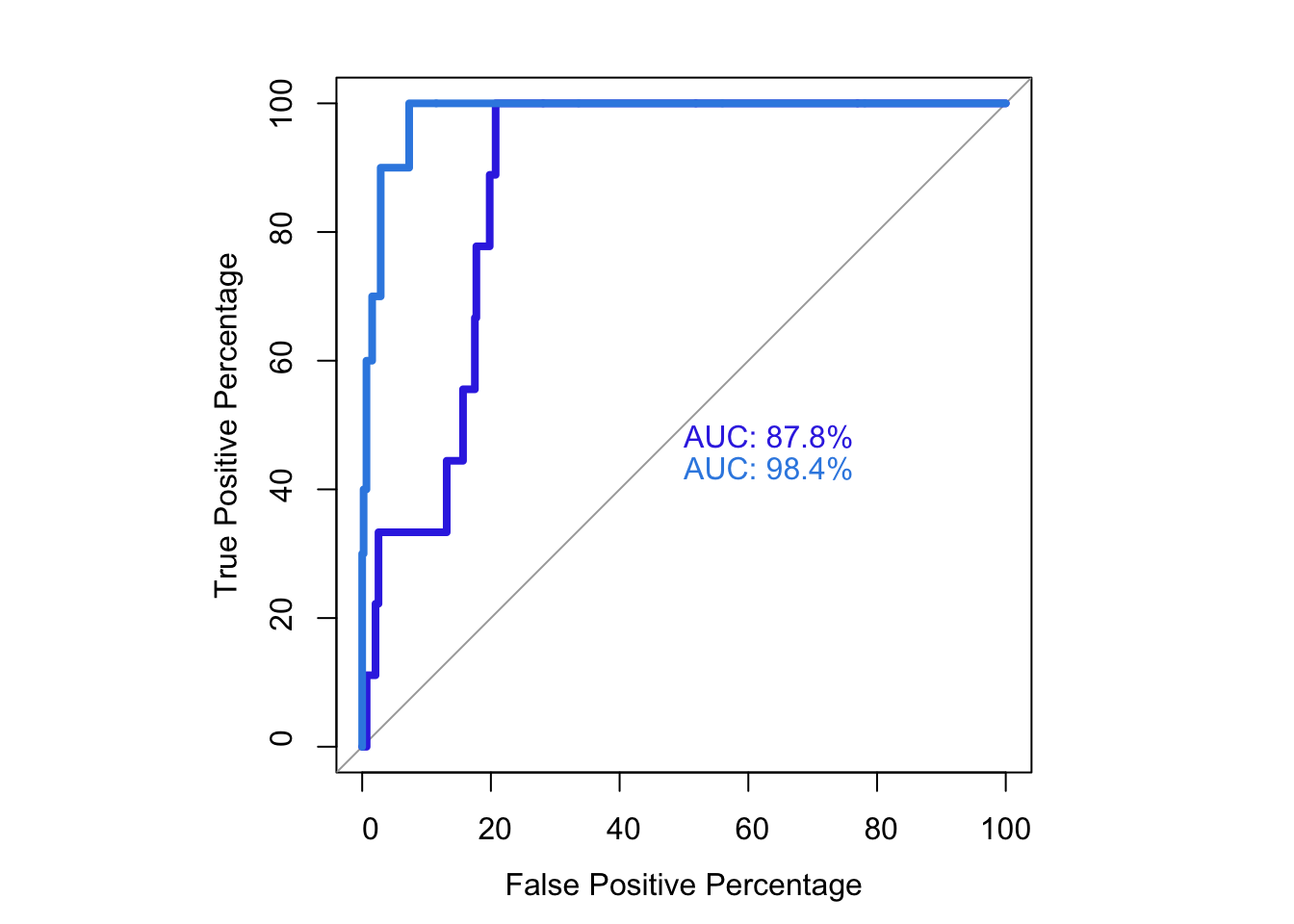
par(pty="m")Doing quick correlation of lane 18b sample 4 NGS vs SSCS
ngs=read.csv("data/Consensus_Data/novogene_lane18/sample4/ngs/variant_caller_outputs/variants_unique_ann.csv")
ngs=ngs%>%filter(protein_start%in%c(242:272))%>%mutate(maf=ct/depth)%>%dplyr::select(ref_aa,protein_start,alt_aa,alt_codon,maf)
sscs=read.csv("data/Consensus_Data/novogene_lane18/sample4/sscs/variant_caller_outputs/variants_unique_ann.csv")
sscs=sscs%>%filter(protein_start%in%c(242:272))%>%mutate(maf=ct/depth)%>%dplyr::select(ref_aa,protein_start,alt_aa,alt_codon,maf)
ngs_sscs=merge(ngs,sscs,by=c("ref_aa","protein_start","alt_aa","alt_codon"))
ngs_sscs=ngs_sscs%>%mutate(species=paste(ref_aa,protein_start,alt_aa,sep=""))
# ngs_sscs=ngs_sscs%>%filter(alt_codon%in%twist_alt_codon)
# plotly=ggplot(ngs_sscs,aes(x=maf.x,y=maf.y))+geom_point()+scale_x_continuous("NGS",trans="log10")+scale_y_continuous("SSCS",trans="log10")
# ggplotly(plotly)
plotly=ggplot(ngs_sscs,aes(x=maf.x,y=maf.y,label=species))+geom_text()+scale_x_continuous("NGS",trans="log10")+scale_y_continuous("SSCS",trans="log10")+geom_abline()
ggplotly(plotly)plotly=ggplot(ngs_sscs,aes(x=maf.x,y=maf.y,label=species))+geom_text()+scale_x_continuous("NGS")+scale_y_continuous("SSCS")+geom_abline()
ggplotly(plotly)x=ngs%>%filter(protein_start%in%255,alt_aa%in%"L",alt_codon%in%"CTG")
x=ngs_sscs%>%filter(species%in%"E255L")
codon_table=read.csv("data/codon_table.csv",header = T,stringsAsFactors = F)
twist_codons=codon_table%>%filter(Twist%in%T)
# twist_alt_codon=twist_codons[twist_codons$Letter%in%alt_aa,"Codon"]
twist_alt_codon=twist_codons[,"Codon"]sample10=read.csv("data/Consensus_Data/novogene_lane18/sample10/ngs/variant_caller_outputs/variants_unique_ann.csv")
# sample10=sample10%>%filter(protein_start%in%c(242:272))%>%mutate(maf=ct/depth)%>%dplyr::select(ref_aa,protein_start,alt_aa,alt_codon,maf.d0=maf)
sample10=sample10%>%
# filter(protein_start%in%c(242:272))%>%
filter(protein_start%in%c(242:322))%>%
mutate(maf=ct/depth,
species=paste(ref_aa,protein_start,alt_aa,sep=""))%>%
dplyr::select(type,ref_aa,protein_start,alt_aa,alt_codon,species,maf.d0=maf)
sample10=sample10%>%filter(alt_codon%in%twist_alt_codon)%>%
group_by(type,ref_aa,protein_start,alt_aa,alt_codon,species)%>%
summarize(maf.d0=max(maf.d0))`summarise()` has grouped output by 'type', 'ref_aa', 'protein_start', 'alt_aa', 'alt_codon'. You can override using the `.groups` argument.sample4=read.csv("data/Consensus_Data/novogene_lane18/sample4/ngs/variant_caller_outputs/variants_unique_ann.csv")
sample4=sample4%>%
# filter(protein_start%in%c(242:272))%>%
filter(protein_start%in%c(242:322))%>%
mutate(maf=ct/depth,
species=paste(ref_aa,protein_start,alt_aa,sep=""))%>%
dplyr::select(type,ref_aa,protein_start,alt_aa,alt_codon,species,maf.screen1=maf)
sample4=sample4%>%filter(alt_codon%in%twist_alt_codon)%>%
group_by(type,ref_aa,protein_start,alt_aa,alt_codon,species)%>%
summarize(maf.screen1=max(maf.screen1))`summarise()` has grouped output by 'type', 'ref_aa', 'protein_start', 'alt_aa', 'alt_codon'. You can override using the `.groups` argument.sample5=read.csv("data/Consensus_Data/novogene_lane18/sample5/ngs/variant_caller_outputs/variants_unique_ann.csv")
sample5=sample5%>%
# filter(protein_start%in%c(242:272))%>%
filter(protein_start%in%c(242:322))%>%
mutate(maf=ct/depth,
species=paste(ref_aa,protein_start,alt_aa,sep=""))%>%
dplyr::select(type,ref_aa,protein_start,alt_aa,alt_codon,species,maf.screen2=maf)
sample5=sample5%>%filter(alt_codon%in%twist_alt_codon)%>%
group_by(type,ref_aa,protein_start,alt_aa,alt_codon,species)%>%
summarize(maf.screen2=max(maf.screen2))`summarise()` has grouped output by 'type', 'ref_aa', 'protein_start', 'alt_aa', 'alt_codon'. You can override using the `.groups` argument.# sample5=sample5%>%filter(protein_start%in%c(242:272))%>%mutate(maf=ct/depth)%>%dplyr::select(ref_aa,protein_start,alt_aa,alt_codon,maf.screen2=maf)
baf3_low=merge(sample10,sample4,by=c("type","ref_aa","protein_start","alt_aa","alt_codon"))
baf3_low=merge(baf3_low,sample5,by=c("type","ref_aa","protein_start","alt_aa","alt_codon"))
# baf3_low=baf3_low%>%filter(alt_codon%in%twist_alt_codon)
baf3_low=baf3_low%>%mutate(netgr_screen1=log(maf.screen1*25546/(maf.d0*54))/144,
netgr_screen2=log(maf.screen2*22471/(maf.d0*54))/144)
plotly=ggplot(baf3_low,aes(x=netgr_screen1,y=netgr_screen2))+geom_point()+scale_x_continuous(trans="log10")+scale_y_continuous(trans="log10")+facet_wrap(~type)
ggplotly(plotly)Warning in self$trans$transform(x): NaNs producedWarning: Transformation introduced infinite values in continuous x-axisWarning in self$trans$transform(x): NaNs producedWarning: Transformation introduced infinite values in continuous y-axisWarning: `group_by_()` was deprecated in dplyr 0.7.0.
Please use `group_by()` instead.
See vignette('programming') for more helpsource("code/resmuts_adder.R")
baf3_low=resmuts_adder(baf3_low)
plotly=ggplot(baf3_low,aes(x=netgr_screen1,y=netgr_screen2,label=species,color=resmuts))+geom_text()+scale_x_continuous(trans="log10")+scale_y_continuous(trans="log10")
ggplotly(plotly)Warning in self$trans$transform(x): NaNs producedWarning: Transformation introduced infinite values in continuous x-axisWarning in self$trans$transform(x): NaNs producedWarning: Transformation introduced infinite values in continuous y-axiscor(baf3_low$netgr_screen1,baf3_low$netgr_screen2)[1] 0.9508483class(baf3_low$netgr_screen1)[1] "numeric"baf3_low=baf3_low%>%mutate(netgr_obs_mean=(netgr_screen1+netgr_screen2)/2)
glm.fit.baf3=glm(as.numeric(baf3_low$resmuts)~baf3_low$netgr_obs_mean,family=binomial)
baf3_low$glm_fits=glm.fit.baf3$fitted.values
roc(as.numeric(baf3_low$resmuts),baf3_low$glm_fits,plot=T,legacy.axes=T,percent=T,xlab="False Positive Percentage",ylab="True Positive Percentage",print.auc=T)Setting levels: control = 0, case = 1Setting direction: controls < cases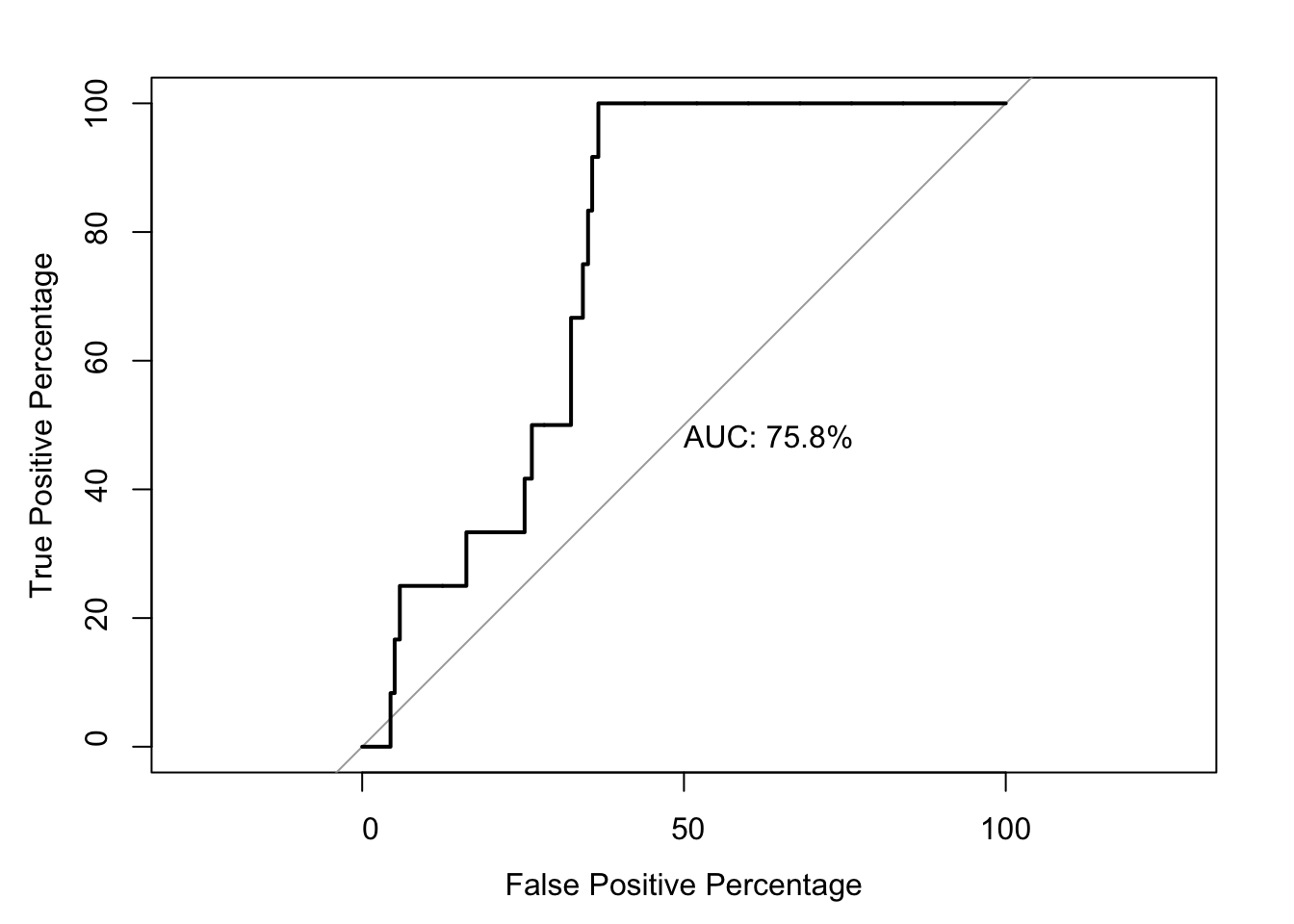
Call:
roc.default(response = as.numeric(baf3_low$resmuts), predictor = baf3_low$glm_fits, percent = T, plot = T, legacy.axes = T, xlab = "False Positive Percentage", ylab = "True Positive Percentage", print.auc = T)
Data: baf3_low$glm_fits in 1248 controls (as.numeric(baf3_low$resmuts) 0) < 12 cases (as.numeric(baf3_low$resmuts) 1).
Area under the curve: 75.85%baf3_low2=baf3_low%>%filter(!protein_start%in%c(289:306))%>%dplyr::select(ref_aa,protein_start,alt_aa,species,netgr_obs_mean)
# baf3_low=
smdata=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/baf3_imat_low_rep1vsrep2_ft/screen_comparison_baf3_imat_low_rep1vsrep2_ft.csv")
smdata=smdata%>%filter(!ct_screen1_after%in%.5,!ct_screen1_after%in%.5)
smdata=smdata%>%filter(!protein_start%in%c(289:306))%>%dplyr::select(ref_aa,protein_start,alt_aa,species,netgr_obs_mean)
baf3_low2=merge(baf3_low2,smdata,by=c("ref_aa","protein_start","alt_aa","species"),all = T)
baf3_low2=baf3_low2%>%filter(!ref_aa==alt_aa)
baf3_low2$ngsonly=F
baf3_low2[baf3_low2$netgr_obs_mean.y%in%NA,"ngsonly"]=T
baf3_low2[baf3_low2$netgr_obs_mean.y%in%NA,"netgr_obs_mean.y"]=.01
ggplot(baf3_low2,aes(x=netgr_obs_mean.x,y=netgr_obs_mean.y,label=species,color=ngsonly))+geom_text()+stat_cor(method = "pearson")Warning: Removed 3 rows containing non-finite values (stat_cor).Warning: Removed 3 rows containing missing values (geom_text).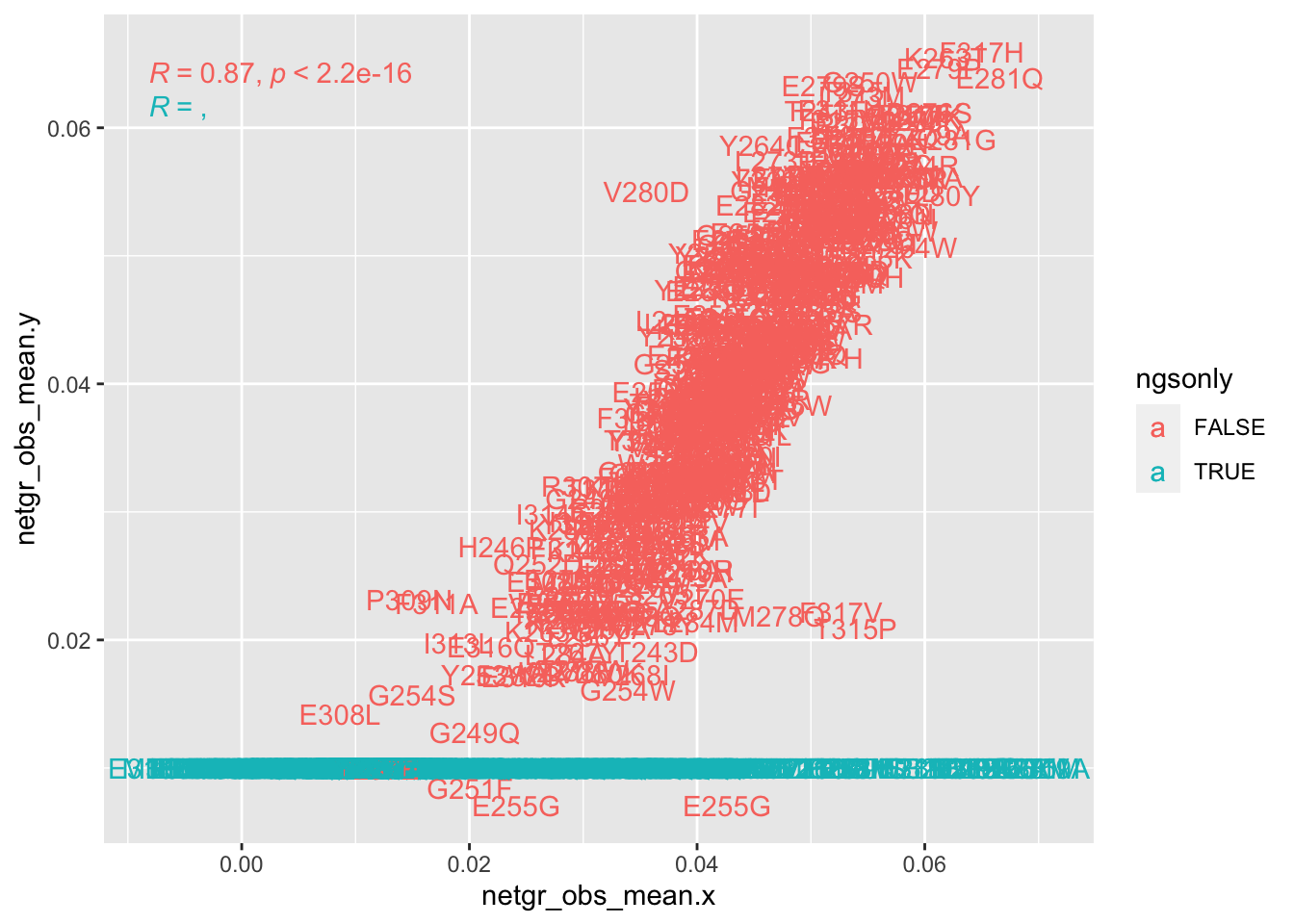
x=baf3_low2%>%filter(netgr_obs_mean.y%in%NA)
smdata=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/baf3_imat_low_rep1vsrep2_ft/screen_comparison_baf3_imat_low_rep1vsrep2_ft.csv")
ggplot(smdata,aes(x=ct_screen1_after,y=netgr_obs_mean))+geom_point()+scale_x_continuous(trans="log10")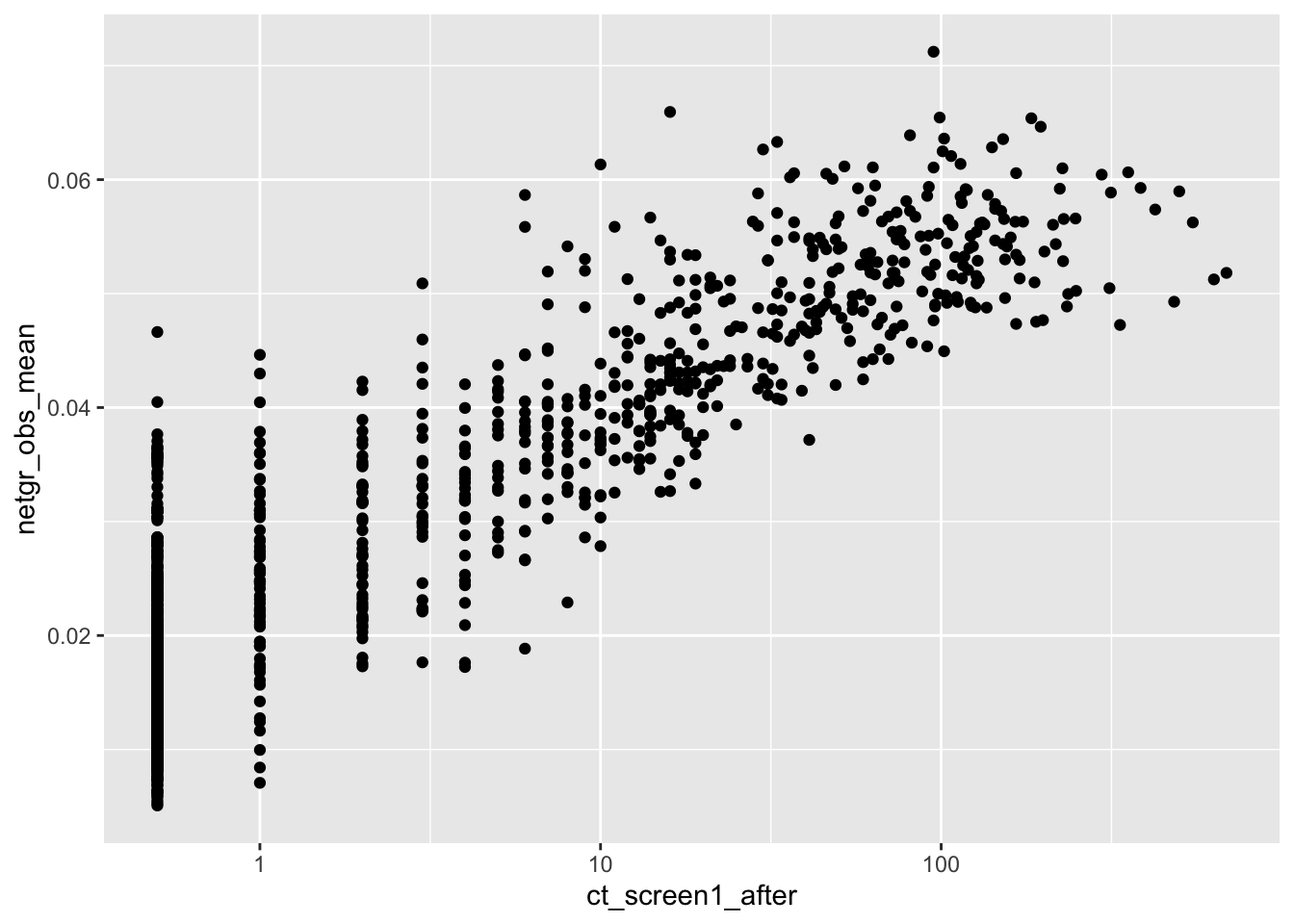
Merging ABL Lane 18 dataset
Plotting the short et al data for Twinstrand
#################################
short_mutants=read.csv("data/Short_et_al_fig1/short_et_al_3.12.23.csv",header = T)
short_mutants$preexisting_all=T
short_mutants=short_mutants%>%dplyr::select(-Index)%>%rename(species=Species)
data_merged=merge(data_merged,short_mutants,by="species",all.x = T)
data_merged[!data_merged$preexisting_all%in%T,"preexisting_all"]=F
nrow(data_merged%>%filter(preexisting_all%in%T))[1] 43# source("code/resmuts_adder.R")
data_merged=resmuts_adder(data_merged)
data_merged$resmut_cosmic="neither"
data_merged[data_merged$preexisting_all%in%TRUE,"resmut_cosmic"]="preexisting"
data_merged[data_merged$resmuts%in%TRUE,"resmut_cosmic"]="resmut"
data_merged$resmut_cosmic=factor(data_merged$resmut_cosmic,levels=c("neither","preexisting","resmut"))
ggplot(data_merged%>%filter(n_nuc_min%in%1),aes(x=resmut_cosmic,y=netgr_obs_mean,fill=resmut_cosmic))+
geom_violin(color="black")+
# geom_boxplot(color="black")+
geom_jitter(color="black", size=.5,width=.1, alpha=0.9)+
scale_fill_manual(values=c("gray90","gray50","red"))+scale_y_continuous(name=bquote('Net Growth Rate '(Hours^-1)))+scale_x_discrete("",labels=c("Undetected","Pre-existing\n ALL","Known\nResistant\n Variant"))+cleanup+theme(legend.position = "none")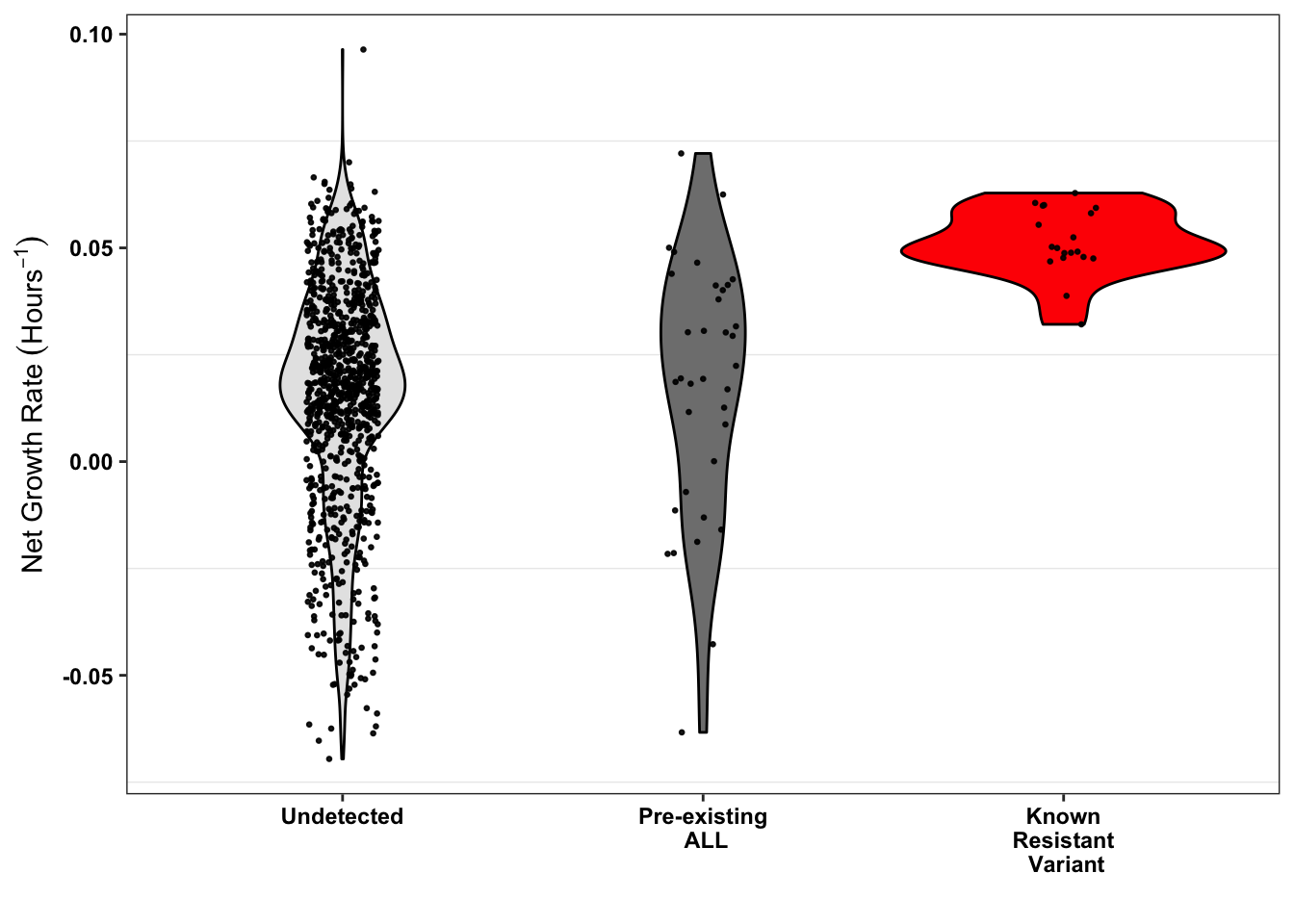
# ggsave("data/Short_et_al_fig1/shortetal_netgr_BoxPlot_4.14.23.pdf",width=4,height=4,units="in",useDingbats=F)
# ggsave("output/SM_Imatinib_Plots/shortetal_netgr_BoxPlot_4.14.23.pdf",width=3,height=3,units="in",useDingbats=F)
x=short_mutants%>%filter(!Classification%in%"Silent")
x=data_merged%>%filter(protein_start%in%c(242:322),preexisting_all%in%T)
x=data_merged%>%filter(protein_start>=242,protein_start<=322,n_nuc_min%in%1,preexisting_all%in%T,resmuts%in%F)
x=data_merged%>%filter(n_nuc_min%in%1,preexisting_all%in%T,resmuts%in%F)bi=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/baf3_IL3_rep1vsrep2_ft/screen_comparison_baf3_IL3_low_rep1vsrep2_ft.csv")
bi$dose=0
bi=bi%>%dplyr::select(ref_aa,protein_start,alt_aa,species,dose,netgr_obs_mean)
bl=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/baf3_Imat_low_rep1vsrep2/screen_comparison_baf3_Imat_low_rep1vsrep2.csv")
bl$dose=300
bl=bl%>%dplyr::select(ref_aa,protein_start,alt_aa,species,dose,netgr_obs_mean)
bm=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/baf3_Imat_medium_rep1vsrep2/screen_comparison_baf3_Imat_medium_rep1vsrep2.csv")
bm$dose=600
bm=bm%>%dplyr::select(ref_aa,protein_start,alt_aa,species,dose,netgr_obs_mean)
bh=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/baf3_Imat_high_rep1vsrep2/screen_comparison_baf3_Imat_high_rep1vsrep2.csv")
bh$dose=1200
bh=bh%>%dplyr::select(ref_aa,protein_start,alt_aa,species,dose,netgr_obs_mean)
b_merged=merge(bl,bm,by = c("ref_aa","protein_start","alt_aa","species"),suffixes = c(".low",".medium"))
b_merged=merge(b_merged,bh%>%rename(dose.high=dose,netgr_obs_mean.high=netgr_obs_mean),by = c("ref_aa","protein_start","alt_aa","species"))
b_merged=merge(b_merged,bi%>%rename(dose.il3=dose,netgr_obs_mean.il3=netgr_obs_mean),by = c("ref_aa","protein_start","alt_aa","species"))
source("code/cosmic_data_adder.R")
b_merged=cosmic_data_adder(b_merged)
b_merged=resmuts_adder(b_merged)
# write.csv(b_merged,"output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/baf3_smscreen_netgrowthrates_582023.csv")
# x=b_merged%>%filter(cosmic_present%in%T)
# kl=kl%>%mutate(netgr_obs_mean=netgr_obs_screen1)
# kl=cosmic_data_adder(kl)6.2.2022 I am adding replicate level growth rates for Scott to use
bi=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/baf3_IL3_rep1vsrep2_ft/screen_comparison_baf3_IL3_low_rep1vsrep2_ft.csv")
bi$dose=0
bi=bi%>%dplyr::select(ref_aa,protein_start,alt_aa,species,dose,netgr_obs_rep1=netgr_obs_screen1,netgr_obs_rep2=netgr_obs_screen2)
bl=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/baf3_Imat_low_rep1vsrep2/screen_comparison_baf3_Imat_low_rep1vsrep2.csv")
bl$dose=300
bl=bl%>%dplyr::select(ref_aa,protein_start,alt_aa,species,dose,netgr_obs_rep1=netgr_obs_screen1,netgr_obs_rep2=netgr_obs_screen2)
bm=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/baf3_Imat_medium_rep1vsrep2/screen_comparison_baf3_Imat_medium_rep1vsrep2.csv")
bm$dose=600
bm=bm%>%dplyr::select(ref_aa,protein_start,alt_aa,species,dose,netgr_obs_rep1=netgr_obs_screen1,netgr_obs_rep2=netgr_obs_screen2)
bh=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/baf3_Imat_high_rep1vsrep2/screen_comparison_baf3_Imat_high_rep1vsrep2.csv")
bh$dose=1200
bh=bh%>%dplyr::select(ref_aa,protein_start,alt_aa,species,dose,netgr_obs_rep1=netgr_obs_screen1,netgr_obs_rep2=netgr_obs_screen2)
b_merged=merge(bl,bm,by = c("ref_aa","protein_start","alt_aa","species"),suffixes = c(".low",".medium"))
b_merged=merge(b_merged,bh%>%rename(dose.high=dose,netgr_obs_rep1.high=netgr_obs_rep1,netgr_obs_rep2.high=netgr_obs_rep2),by = c("ref_aa","protein_start","alt_aa","species"))
b_merged=merge(b_merged,bi%>%rename(dose.il3=dose,netgr_obs_rep1.il3=netgr_obs_rep1,netgr_obs_rep2.il3=netgr_obs_rep2),by = c("ref_aa","protein_start","alt_aa","species"))
source("code/cosmic_data_adder.R")
source("code/resmuts_adder.R")
b_merged=cosmic_data_adder(b_merged)
b_merged=resmuts_adder(b_merged)
# write.csv(b_merged,"output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/baf3_smscreen_netgrowthrates_622023.csv")
# x=b_merged%>%filter(cosmic_present%in%T)
# kl=kl%>%mutate(netgr_obs_mean=netgr_obs_screen1)
# kl=cosmic_data_adder(kl)bi=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/baf3_IL3_rep1vsrep2_ft/screen_comparison_baf3_IL3_low_rep1vsrep2_ft.csv")
bi$dose=0
bi=bi%>%mutate(lfc_rep1=log2(maf_screen1_after/maf_screen1_before),lfc_rep2=log2(maf_screen2_after/maf_screen2_before))
bi=bi%>%dplyr::select(ref_aa,protein_start,alt_aa,species,n_nuc_min,dose,lfc_rep1,lfc_rep2)
bl=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/baf3_Imat_low_rep1vsrep2/screen_comparison_baf3_Imat_low_rep1vsrep2.csv")
bl$dose=300
bl=bl%>%mutate(lfc_rep1=log2(maf_screen1_after/maf_screen1_before),lfc_rep2=log2(maf_screen2_after/maf_screen2_before))
bl=bl%>%dplyr::select(ref_aa,protein_start,alt_aa,species,n_nuc_min,dose,lfc_rep1,lfc_rep2)
# bl=bl%>%dplyr::select(ref_aa,protein_start,alt_aa,species,dose,lfc_obs_rep1=lfc_obs_screen1,lfc_obs_rep2=lfc_obs_screen2)
bm=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/baf3_Imat_medium_rep1vsrep2/screen_comparison_baf3_Imat_medium_rep1vsrep2.csv")
bm$dose=600
bm=bm%>%mutate(lfc_rep1=log2(maf_screen1_after/maf_screen1_before),lfc_rep2=log2(maf_screen2_after/maf_screen2_before))
bm=bm%>%dplyr::select(ref_aa,protein_start,alt_aa,species,n_nuc_min,dose,lfc_rep1,lfc_rep2)
# bm=bm%>%dplyr::select(ref_aa,protein_start,alt_aa,species,dose,lfc_obs_rep1=lfc_obs_screen1,lfc_obs_rep2=lfc_obs_screen2)
bh=read.csv("output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/baf3_Imat_high_rep1vsrep2/screen_comparison_baf3_Imat_high_rep1vsrep2.csv")
bh$dose=1200
bh=bh%>%mutate(lfc_rep1=log2(maf_screen1_after/maf_screen1_before),lfc_rep2=log2(maf_screen2_after/maf_screen2_before))
bh=bh%>%dplyr::select(ref_aa,protein_start,alt_aa,species,n_nuc_min,dose,lfc_rep1,lfc_rep2)
# bh=bh%>%dplyr::select(ref_aa,protein_start,alt_aa,species,dose,lfc_obs_rep1=lfc_obs_screen1,lfc_obs_rep2=lfc_obs_screen2)
b_merged=merge(bl,bm,by = c("ref_aa","protein_start","alt_aa","species","n_nuc_min"),suffixes = c(".low",".medium"))
b_merged=merge(b_merged,bh%>%rename(dose.high=dose,lfc_rep1.high=lfc_rep1,lfc_rep2.high=lfc_rep2),by = c("ref_aa","protein_start","alt_aa","species","n_nuc_min"))
b_merged=merge(b_merged,bi%>%rename(dose.il3=dose,lfc_rep1.il3=lfc_rep1,lfc_rep2.il3=lfc_rep2),by = c("ref_aa","protein_start","alt_aa","species","n_nuc_min"))
source("code/cosmic_data_adder.R")
source("code/resmuts_adder.R")
b_merged=cosmic_data_adder(b_merged)
b_merged=resmuts_adder(b_merged)
# write.csv(b_merged,"output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/baf3_smscreen_lfc_682023.csv")
# x=b_merged%>%filter(cosmic_present%in%T)
# kl=kl%>%mutate(netgr_obs_mean=netgr_obs_screen1)
# kl=cosmic_data_adder(kl)
sessionInfo()R version 4.0.0 (2020-04-24)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS 10.16
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRblas.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] parallel stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] forcats_0.5.1 NatParksPalettes_0.2.0 pROC_1.16.2
[4] ggpubr_0.4.0 RColorBrewer_1.1-2 doParallel_1.0.15
[7] iterators_1.0.12 foreach_1.5.0 tictoc_1.0
[10] plotly_4.9.2.1 ggplot2_3.3.3 dplyr_1.0.6
[13] stringr_1.4.0
loaded via a namespace (and not attached):
[1] httr_1.4.2 sass_0.4.1 tidyr_1.1.3 jsonlite_1.7.2
[5] viridisLite_0.3.0 carData_3.0-3 bslib_0.3.1 assertthat_0.2.1
[9] cellranger_1.1.0 yaml_2.2.1 pillar_1.6.1 backports_1.1.7
[13] glue_1.4.1 digest_0.6.25 promises_1.1.0 ggsignif_0.6.0
[17] colorspace_1.4-1 htmltools_0.5.2 httpuv_1.5.2 plyr_1.8.6
[21] pkgconfig_2.0.3 broom_0.7.6 haven_2.4.1 purrr_0.3.4
[25] scales_1.1.1 whisker_0.4 openxlsx_4.1.5 later_1.0.0
[29] rio_0.5.16 git2r_0.27.1 tibble_3.1.2 generics_0.0.2
[33] farver_2.0.3 car_3.0-7 ellipsis_0.3.2 withr_2.4.2
[37] lazyeval_0.2.2 magrittr_2.0.1 crayon_1.4.1 readxl_1.3.1
[41] evaluate_0.14 fs_1.4.1 fansi_0.4.1 rstatix_0.6.0
[45] foreign_0.8-78 tools_4.0.0 data.table_1.12.8 hms_1.1.0
[49] lifecycle_1.0.0 munsell_0.5.0 zip_2.0.4 compiler_4.0.0
[53] jquerylib_0.1.4 rlang_0.4.11 grid_4.0.0 htmlwidgets_1.5.1
[57] crosstalk_1.1.0.1 labeling_0.3 rmarkdown_2.14 gtable_0.3.0
[61] codetools_0.2-16 abind_1.4-5 DBI_1.1.0 curl_4.3
[65] R6_2.4.1 knitr_1.28 fastmap_1.1.0 utf8_1.1.4
[69] workflowr_1.6.2 rprojroot_1.3-2 stringi_1.7.5 Rcpp_1.0.4.6
[73] vctrs_0.3.8 tidyselect_1.1.0 xfun_0.31