ABL_sheared_libraries
Haider Inam
2023-01-31
Last updated: 2023-08-30
Checks: 6 1
Knit directory: duplex_sequencing_screen/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R
Markdown file created these results, you’ll want to first commit it to
the Git repo. If you’re still working on the analysis, you can ignore
this warning. When you’re finished, you can run
wflow_publish to commit the R Markdown file and build the
HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200402) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version f9eea37. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/Consensus_Data/.Rhistory
Ignored: data/Consensus_Data/Novogene_lane11/sample1/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample1/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample1/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample1/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample1/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample1/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample2/archive/sscs_aligned_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample2/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample2/duplex/variant_caller_outputs/archive_ensembl/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample2/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample2/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample2/sscs/variant_caller_outputs/archive_ensembl/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample2/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample3/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample3/duplex/variant_caller_outputs/archive_ensembl/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample3/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample3/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample3/sscs/variant_caller_outputs/archive ensembl/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample3/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample4/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample4/duplex/variant_caller_outputs/archive ensembl/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample4/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample4/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample4/sscs/variant_caller_outputs/archive ensembl/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample4/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample5/variant_caller_outputs/sscs_L858R_aligned_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample5/variant_caller_outputs/sscs_L858R_aligned_filtered_sample5.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample6/archive/sscs_aligned_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample6/sscs_L858R_aligned_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample6/variant_caller_outputs/variants_ann_sample6.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample7/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample7/sscs/variant_caller_outputs/archive ensembl/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane11/sample7/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample1/low_sscscounts/sscs_aligned_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample1/low_sscscounts/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample1/sscs_aligned_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample1/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample3/sscs_combined_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample3/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample5/sscs_combined_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample5/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample7/sscs_combined_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample7/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample9/sscs_combined_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane12/sample9/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample1/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample1/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample1/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample1/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample10/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample10/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample10/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample10/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample10/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample10/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample11/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample11/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample11/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample11/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample11/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample11/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample12/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample12/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample12/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample12/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample12/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample12/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample2/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample2/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample3/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample3/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample4/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample4/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample5/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample5/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample6/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample6/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample7/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample7/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample7/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample7/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample7/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample7/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample8/
Ignored: data/Consensus_Data/Novogene_lane13/sample9/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample9/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample9/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample9/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample9/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane13/sample9/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample10_combined/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample10_combined/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample10_combined/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample10_combined/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample10_combined/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample10_combined/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample11/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample11/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample11/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample11/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample11/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample11/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample12/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample12/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample12/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample12/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample12/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample12/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample13/
Ignored: data/Consensus_Data/Novogene_lane14/sample14_combined/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample14_combined/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample14_combined/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample14_combined/sscs.filt_1.fa.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample14_combined/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample14_combined/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample14_combined/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample14b/
Ignored: data/Consensus_Data/Novogene_lane14/sample15/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample15/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample15/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample15/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample15/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample15/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample16/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample16/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample16/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample16/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample16/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample16/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample17/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample17/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample17/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample17/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample17/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample17/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample18/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample18/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample18/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample18/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample18/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample18/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample1_combined/
Ignored: data/Consensus_Data/Novogene_lane14/sample2_combined/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample2_combined/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample3/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample3/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample4/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample4/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample5/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample5/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample6/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample6/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample7/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample7/variant_caller_outputs/duplex/
Ignored: data/Consensus_Data/Novogene_lane14/sample7/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample8/
Ignored: data/Consensus_Data/Novogene_lane14/sample9/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample9/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample9/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample9/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample9/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane14/sample9/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/Novogene_lane2/
Ignored: data/Consensus_Data/Novogene_lane3/
Ignored: data/Consensus_Data/Novogene_lane4/
Ignored: data/Consensus_Data/Novogene_lane5/
Ignored: data/Consensus_Data/Novogene_lane6/
Ignored: data/Consensus_Data/Novogene_lane7/
Ignored: data/Consensus_Data/Ranomics_Pooled/
Ignored: data/Consensus_Data/archive/
Ignored: data/Consensus_Data/novogene_lane15/sample_1/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_1/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_1/firstrun(lowsequencing)/
Ignored: data/Consensus_Data/novogene_lane15/sample_1/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_2/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_2/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_2/firstrun(lowsequencing)/sscs/
Ignored: data/Consensus_Data/novogene_lane15/sample_2/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_2/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/firstrun(lowsequencing)/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/firstrun(lowsequencing)/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/firstrun(lowsequencing)/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/firstrun(lowsequencing)/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/ngs/Sample3_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/ngs/sample3a(firsthalf)/Sample3_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/ngs/sample3a(firsthalf)/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/ngs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_3/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_4/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_4/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_4/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_4/firstrun(lowsequencing)/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_4/firstrun(lowsequencing)/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_4/firstrun(lowsequencing)/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_4/firstrun(lowsequencing)/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_4/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_4/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_4/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_5/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_5/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_5/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_5/firstrun(lowsequencing)/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_5/firstrun(lowsequencing)/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_5/firstrun(lowsequencing)/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_5/firstrun(lowsequencing)/sscs/variant_caller_outputs/.empty/
Ignored: data/Consensus_Data/novogene_lane15/sample_5/firstrun(lowsequencing)/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_5/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_5/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_5/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_6/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_6/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_6/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_6/firstrun(lowsequencing)/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_6/firstrun(lowsequencing)/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_6/firstrun(lowsequencing)/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_6/firstrun(lowsequencing)/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_6/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_6/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_6/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_7/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_7/duplex/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_7/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_7/firstrun(lowsequencing)/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_7/firstrun(lowsequencing)/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_7/firstrun(lowsequencing)/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_7/firstrun(lowsequencing)/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_7/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_7/sscs/variant_caller_outputs/archive/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane15/sample_7/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample10/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample10/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample10/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample10/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample11/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample11/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample11/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample11/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample12/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample12/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample12/sscs/
Ignored: data/Consensus_Data/novogene_lane16a/Sample13/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample13/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample13/sscs/
Ignored: data/Consensus_Data/novogene_lane16a/Sample14/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample14/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample14/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample14/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample1_combined/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample1_combined/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample1_combined/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample1_combined/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample2/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample2/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample2/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample3/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample3/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample3/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample3/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample4/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample4/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample4/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample4/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample5/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample5/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample5/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample5/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample6/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample6/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample6/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample6/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample7/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample7/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample7/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample7/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample8/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample8/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample8/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample8/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample9/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample9/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample9/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16a/Sample9/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16a/duplex/
Ignored: data/Consensus_Data/novogene_lane16b/Sample10/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample10/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample10/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample10/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample11/
Ignored: data/Consensus_Data/novogene_lane16b/Sample15/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample15/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample15/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample15/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample1_combined/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample1_combined/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample1_combined/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample1_combined/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample2/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample2/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample2/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample2/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample3/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample3/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample3/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample3/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample4/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample4/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample4/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample4/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample5/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample5/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample5/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample5/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample6/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample6/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample6/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample6/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample7_combined/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample7_combined/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample7_combined/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample7_combined/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample8_combined/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample8_combined/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample8_combined/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample8_combined/sscs/variant_caller_outputs/archive/
Ignored: data/Consensus_Data/novogene_lane16b/Sample8_combined/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample9/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample9/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample9/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane16b/Sample9/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample10/duplex/
Ignored: data/Consensus_Data/novogene_lane17/sample10/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample10/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample11/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample11/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample11/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample11/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample1_combined/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample1_combined/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample1_combined/low_depth/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample1_combined/low_depth/duplex/low_depth/
Ignored: data/Consensus_Data/novogene_lane17/sample1_combined/low_depth/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample1_combined/low_depth/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample1_combined/low_depth/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample1_combined/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample1_combined/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample2/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample2/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample2/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample2/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample3/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample3/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample3/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample3/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample4/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample4/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample4/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample4/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample5/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample5/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample5/low_seq_depth/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample5/low_seq_depth/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample5/low_seq_depth/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample5/low_seq_depth/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample5/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample5/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample6/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample6/low_seq_depths/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample6/low_seq_depths/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample6/low_seq_depths/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample6/low_seq_depths/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample6/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample6/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample7/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample7/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample7/low_seq_depths/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample7/low_seq_depths/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample7/low_seq_depths/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample7/low_seq_depths/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample7/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample7/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample8/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample8/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample8/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample9/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample9/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample9/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17/sample9/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17b/Sample1 copy 2/
Ignored: data/Consensus_Data/novogene_lane17b/Sample1 copy 3/
Ignored: data/Consensus_Data/novogene_lane17b/Sample1/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17b/Sample1/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17b/Sample1/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17b/Sample1/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17b/Sample2/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane17b/Sample2/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane17b/Sample2/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample1/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample1/l298l/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample1/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample1/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample1/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample1/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample1/nol298l/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample1/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample1/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample1/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample1/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample1/sscs/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample10/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample10/l298l/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample10/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample10/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample10/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample10/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample10/ngs/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample10/ngs/Sample_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample10/ngs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample10/nol298l/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample10/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample10/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample10/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample10/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample10/sscs/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample11/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample11/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample11/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample11/l298l/sscs/variant_caller_outputs/slow_variantcaller_outputs_forcomparison/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample11/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample11/ngs/Sample_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample11/ngs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample11/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample11/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample11/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample11/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample12/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample12/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample12/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample12/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample12/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample12/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample12/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample12/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample13/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample13/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample13/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample13/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample13/ngs/Sample_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample13/ngs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample13/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample13/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample13/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample13/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample14/duplex/
Ignored: data/Consensus_Data/novogene_lane18/sample14/l298l/duplex/
Ignored: data/Consensus_Data/novogene_lane18/sample14/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample14/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample14/ngs/Sample_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample14/ngs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample14/nol298l/duplex/
Ignored: data/Consensus_Data/novogene_lane18/sample14/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample14/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample15/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample15/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample15/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample15/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample15/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample15/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample15/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample15/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample16/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample16/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample16/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample16/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample16/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample16/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample16/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample16/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample17/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample17/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample17/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample17/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample17/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample17/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample17/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample17/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample18/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample18/l298l/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample18/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample18/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample18/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample18/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample18/nol298l/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample18/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample18/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample18/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample18/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample2/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample2/l298l/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample2/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample2/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample2/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample2/l298l/sscs/variant_caller_outputs/fastercaller/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample2/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample2/nol298l/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample2/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample2/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample2/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample2/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample2/sscs/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample3/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample3/l298l/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample3/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample3/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample3/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample3/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample3/nol298l/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample3/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample3/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample3/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample3/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample3/sscs/.DS_Store
Ignored: data/Consensus_Data/novogene_lane18/sample4/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample4/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample4/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample4/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample4/ngs/Sample_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample4/ngs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample4/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample4/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample4/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample4/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample5/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample5/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample5/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample5/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample5/ngs/Sample_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample5/ngs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample5/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample5/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample5/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample5/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample6/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample6/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample6/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample6/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample6/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample6/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample6/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample6/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample7/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample7/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample7/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample7/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample7/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample7/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample7/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample7/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample8/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample8/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample8/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample8/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample8/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample8/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample8/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample8/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample9/l298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample9/l298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample9/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample9/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample9/nol298l/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample9/nol298l/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample9/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/sample9/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample3/duplex/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample3/l298l/duplex/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample3/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample3/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample3/nol298l/duplex/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample3/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample3/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample3/sscs/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample5/duplex/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample5/l298l/duplex/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample5/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample5/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample5/nol298l/duplex/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample5/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample5/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample5/sscs/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample6/duplex/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample6/l298l/duplex/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample6/l298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample6/l298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample6/nol298l/duplex/
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample6/nol298l/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample6/nol298l/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane18/tlane18a_sample6/sscs/
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample1/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample1/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample1/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample1/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample10/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample10/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample10/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample10/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample11/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample11/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample11/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample11/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample12/duplex/
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample12/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample12/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample2/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample2/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample2/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample2/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample3/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample3/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample3/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample3/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample4/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample4/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample4/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample4/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample5/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample5/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample5/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample5/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample6/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample6/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample6/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample6/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample7/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample7/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample7/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample7/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample8/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample8/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample8/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample8/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample9/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample9/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample9/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19a_Sample9/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample1/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample1/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample1/sscs/
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample10/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample10/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample10/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample10/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample11/duplex/
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample11/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample11/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample12/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample12/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample12/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample12/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample13/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample13/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample13/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample13/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample14/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample14/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample14/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample14/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample15/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample15/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample15/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample15/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample16/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample16/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample16/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample16/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample2/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample2/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample2/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample2/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample3/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample3/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample3/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample3/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample4/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample4/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample4/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample4/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample5/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample5/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample5/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample5/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample6/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample6/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample6/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample6/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample7/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample7/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample7/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample7/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample8/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample8/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample8/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample8/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample9/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample9/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample9/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19/Ln19b_Sample9/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample10/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample10/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample10/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample10/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample11/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample11/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample11/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample11/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample2/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample2/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample2/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample2/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample3/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample3/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample3/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample3/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample4/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample4/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample4/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample4/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample5/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample5/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample5/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample5/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample6/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample6/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample6/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample6/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample7/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample7/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample7/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample7/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample8/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample8/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample8/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample8/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample9/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample9/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample9/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19a_Sample9/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19b_Sample9/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19b_Sample9/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19b_Sample9/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane19_20/Ln19b_Sample9/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/.DS_Store
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample1/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample1/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample1/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample1/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample2/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample2/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample2/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample2/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample3/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample3/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample3/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample3/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample4/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample4/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample4/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample4/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample5/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample5/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample5/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample5/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample6/duplex/
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample6/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample6/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample7/
Ignored: data/Consensus_Data/novogene_lane22/Lane22_Sample8/
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample13/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample13/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample13/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample13/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample14/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample14/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample14/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample14/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample15/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample15/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample15/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample15/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample16/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample16/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample16/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample16/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample17/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample17/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample17/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample17/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample18/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample18/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample18/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample18/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample19/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample19/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample19/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample19/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample20/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample20/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample20/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample20/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample21/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample21/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample21/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample21/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample22/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample22/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample22/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample22/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample23/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample23/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample23/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample23/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample24/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample24/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample24/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample24/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample25/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample25/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample25/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample25/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample26/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample26/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample26/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample26/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample27/duplex/duplex_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample27/duplex/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample27/sscs/sscs_sorted_filtered.tsv.gz
Ignored: data/Consensus_Data/novogene_lane23/Lane23_Sample27/sscs/variant_caller_outputs/variants_ann.csv.gz
Ignored: data/Consensus_Data/sscs_dcs_comparisons/
Ignored: output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/NA/
Ignored: output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/baf3_IL3_low_rep1vsrep2/
Ignored: output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/baf3_Imat_Lowvsk562_Imat_Medium/
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/il3_indep_1.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/il3_indep_1.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/il3_indep_2.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/il3_indep_2.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/il3_indep_3.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/il3_indep_3.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/sorted_1.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/sorted_1.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/sorted_2.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/sorted_2.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/sorted_3.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane3/sorted_3.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/il3_indep_1.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/il3_indep_1.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/il3_indep_2.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/il3_indep_2.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/il3_indep_3.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/il3_indep_3.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/il3_indep_4.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/il3_indep_4.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/il3_indep_5.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/il3_indep_5.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_1.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_1.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_2.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_2.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_3.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_3.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_4.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_4.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_5.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_5.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_6.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane4/sorted_6.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_High_D2.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_High_D2.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_High_D4.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_High_D4.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_Low_D2.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_Low_D2.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_Low_D4.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_Low_D4.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_Medium_D2.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_Medium_D2.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_Medium_D4.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP4_Im_Medium_D4.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_High_D2.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_High_D2.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_High_D4.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_High_D4.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_Low_D2.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_Low_D2.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_Low_D4.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_Low_D4.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_Medium_D2.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_Medium_D2.1.consensus.variant-calls.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_Medium_D4.1.consensus.variant-calls.genome.vcf.gz
Ignored: output/Twinstrand/ABL1AppOutput/Novogene_Lane5/RP5_Im_Medium_D4.1.consensus.variant-calls.vcf.gz
Untracked files:
Untracked: analysis/ABL_sheared_libraries.Rmd
Untracked: output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/baf3_IL3_rep1vsrep2_ft/screen_comparison_baf3_IL3_low_rep1vsrep2ft.csv
Unstaged changes:
Modified: .DS_Store
Modified: analysis/ABL_cosmic_analysis.Rmd
Modified: analysis/ABL_kinasefunction_data_parser.Rmd
Deleted: analysis/ABL_unevenness_analysis.Rmd
Modified: analysis/index.Rmd
Modified: code/compare_screens.R
Modified: data/.DS_Store
Modified: data/Consensus_Data/.DS_Store
Modified: data/Consensus_Data/novogene_lane18/tlane18a_sample6/.DS_Store
Modified: output/.DS_Store
Modified: output/ABLEnrichmentScreens/.DS_Store
Modified: output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/.DS_Store
Modified: output/ABLEnrichmentScreens/ABL_Region1_Lane18_Comparisons/cross-replicate/.DS_Store
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with
wflow_publish() to start tracking its development.
This code has some of the analysis done for libraries made using the enzymatic fragrmentation library prep approach (mostly data from lane 11-16)
source("code/merge_samples.R")
source("code/variants_parser.R")
source("code/shortest_codon_finder.R")
source("code/compare_samples.R")
source("code/depth_finder.R")
source("code/resmuts_adder.R")
source("code/res_residues_adder.R")
source("code/plotting/cleanup.R")For the density plots:
library(viridisLite)
library("MASS")Warning: package 'MASS' was built under R version 4.0.5
Attaching package: 'MASS'The following object is masked from 'package:plotly':
selectThe following object is masked from 'package:dplyr':
selecttheme_set(theme_bw(base_size = 16))
# Get density of points in 2 dimensions.
# @param x A numeric vector.
# @param y A numeric vector.
# @param n Create a square n by n grid to compute density.
# @return The density within each square.
get_density <- function(x, y, ...) {
dens <- MASS::kde2d(x, y, ...)
ix <- findInterval(x, dens$x)
iy <- findInterval(y, dens$y)
ii <- cbind(ix, iy)
return(dens$z[ii])
}
set.seed(1)
dat <- data.frame(
x = c(
rnorm(1e4, mean = 0, sd = 0.1),
rnorm(1e3, mean = 0, sd = 0.1)
),
y = c(
rnorm(1e4, mean = 0, sd = 0.1),
rnorm(1e3, mean = 0.1, sd = 0.2)
)
)Function to find the shortest possible ALT codon for a given amino acid substitution
TP3D6=read.csv("data/Consensus_Data/novogene_lane17/sample6/sscs/variant_caller_outputs/variants_unique_ann.csv",header = T,stringsAsFactors = F)
TP3D6=variants_parser(TP3D6,93000)
TP3D2=read.csv("data/Consensus_Data/novogene_lane17/sample7/sscs/variant_caller_outputs/variants_unique_ann.csv",header = T,stringsAsFactors = F)
TP3D2=variants_parser(TP3D2,839)
TP3D2.D6=compare_samples(TP3D2,TP3D6,.055,96)
# TP3D2.D6=TP3D2.D6%>%filter(ct.x>=3)
TP3D2.D6_clinrel=TP3D2.D6%>%filter(n_nuc_min==1)
TP3D2.D6_clinrel=resmuts_adder(TP3D2.D6_clinrel)
TP3D2.D6_clinrel=res_residues_adder(TP3D2.D6_clinrel)
resmuts=TP3D2.D6_clinrel%>%filter(resmuts%in%T)%>%mutate(species=paste(ref_aa,protein_start,alt_aa,sep = ""))
ic50data_all_sum=read.csv("output/ic50data_all_confidence_intervals_raw_data.csv",row.names = 1)
resmuts_merged=merge(resmuts,ic50data_all_sum,by="species")
ggplot(resmuts_merged,aes(x=netgr_pred_model,y=netgr_obs,label=species))+geom_text()+geom_abline()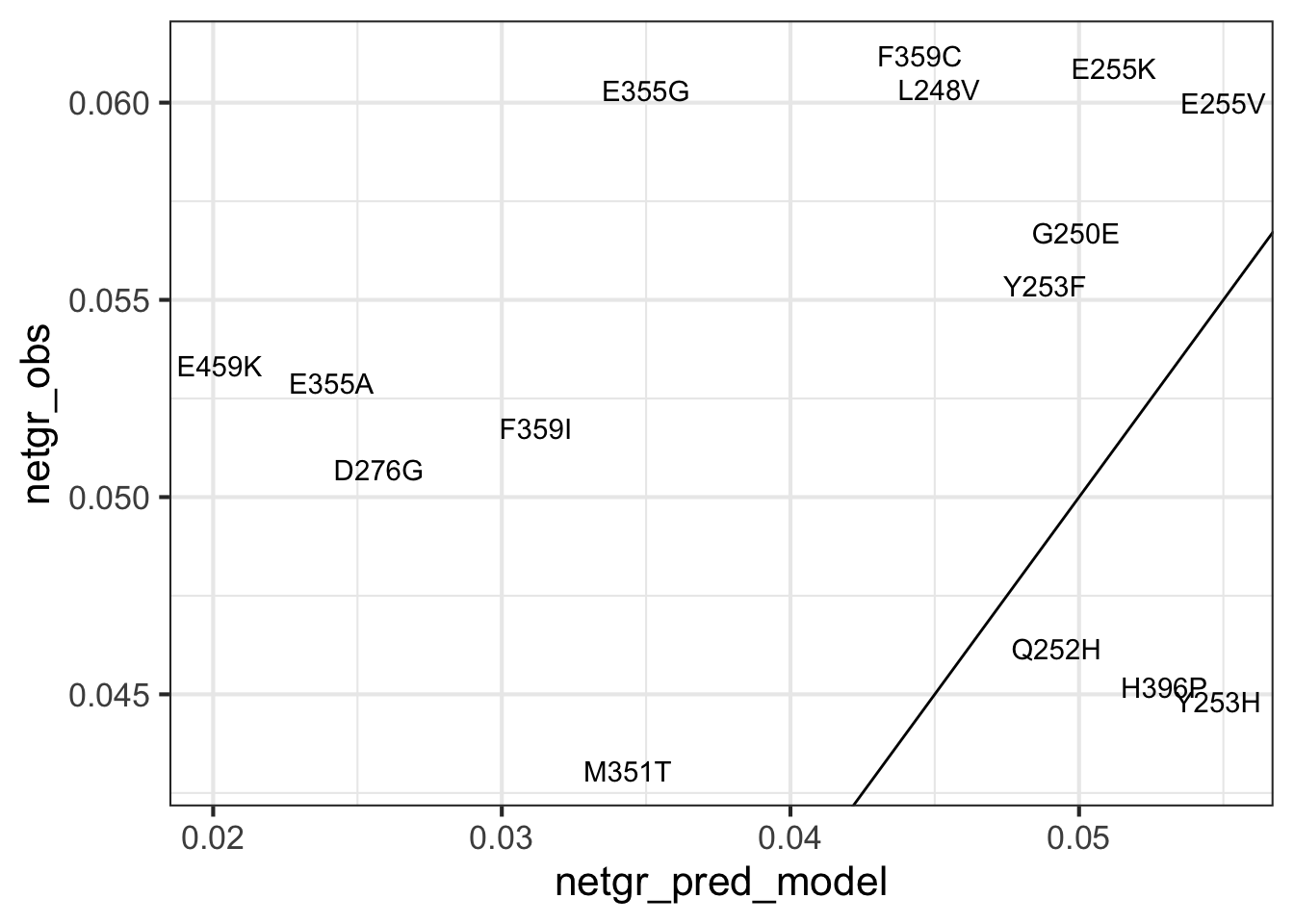
ggplot(resmuts_merged,aes(x=netgr_pred_model,y=score,label=species))+geom_text()+geom_abline()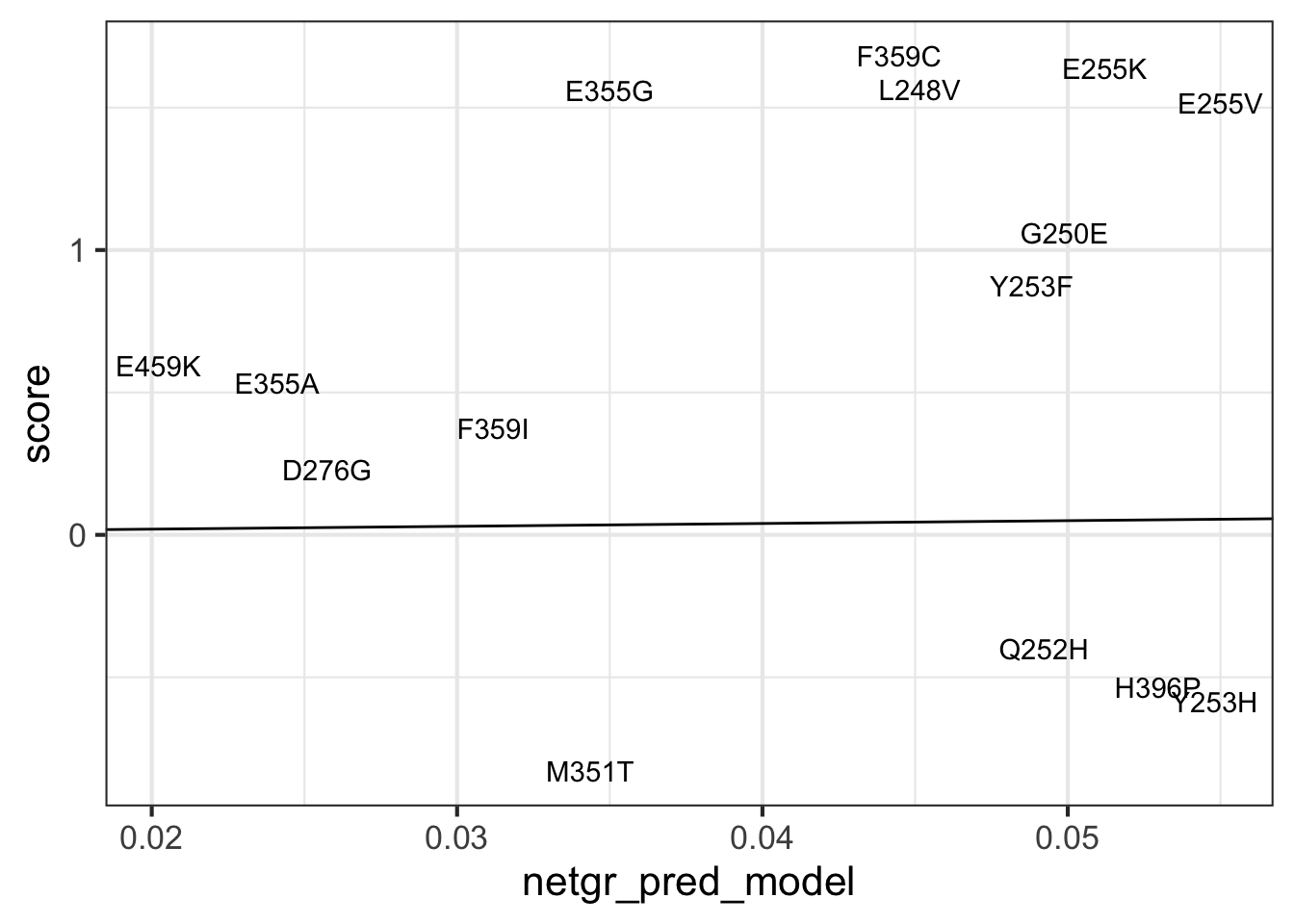
il3D0=merge_samples("Novogene_lane14/Sample10_combined/sscs","Novogene_lane13/sample7/sscs")
il3D0=merge_samples(il3D0,"Novogene_lane14/sample11/sscs")
il3D0=merge_samples(il3D0,"Novogene_lane15/sample_3/sscs")
# il3D0=merge_samples(il3D0,"Novogene_lane17/sample1_combined/sscs")
il3D0=variants_parser(il3D0,370)
imatD4=read.csv("data/Consensus_Data/novogene_lane15/sample_7/sscs/variant_caller_outputs/variants_unique_ann.csv",header = T,stringsAsFactors = F)
imatD4=variants_parser(imatD4,10000)
TSIID0.D4=compare_samples(il3D0,imatD4,.055,96)
# TSIID0.D4=TSIID0.D4%>%filter(ct.x>=3)
TSIID0.D4_clinrel=TSIID0.D4%>%filter(n_nuc_min==1)
TSIID0.D4_clinrel=resmuts_adder(TSIID0.D4_clinrel)
TSIID0.D4_clinrel=res_residues_adder(TSIID0.D4_clinrel)
resmuts=TSIID0.D4_clinrel%>%filter(resmuts%in%T)%>%mutate(species=paste(ref_aa,protein_start,alt_aa,sep = ""))
ic50data_all_sum=read.csv("output/ic50data_all_confidence_intervals_raw_data.csv",row.names = 1)
resmuts_merged=merge(resmuts,ic50data_all_sum,by="species")
ggplot(resmuts_merged,aes(x=netgr_pred_model,y=netgr_obs,label=species))+geom_text()+geom_abline()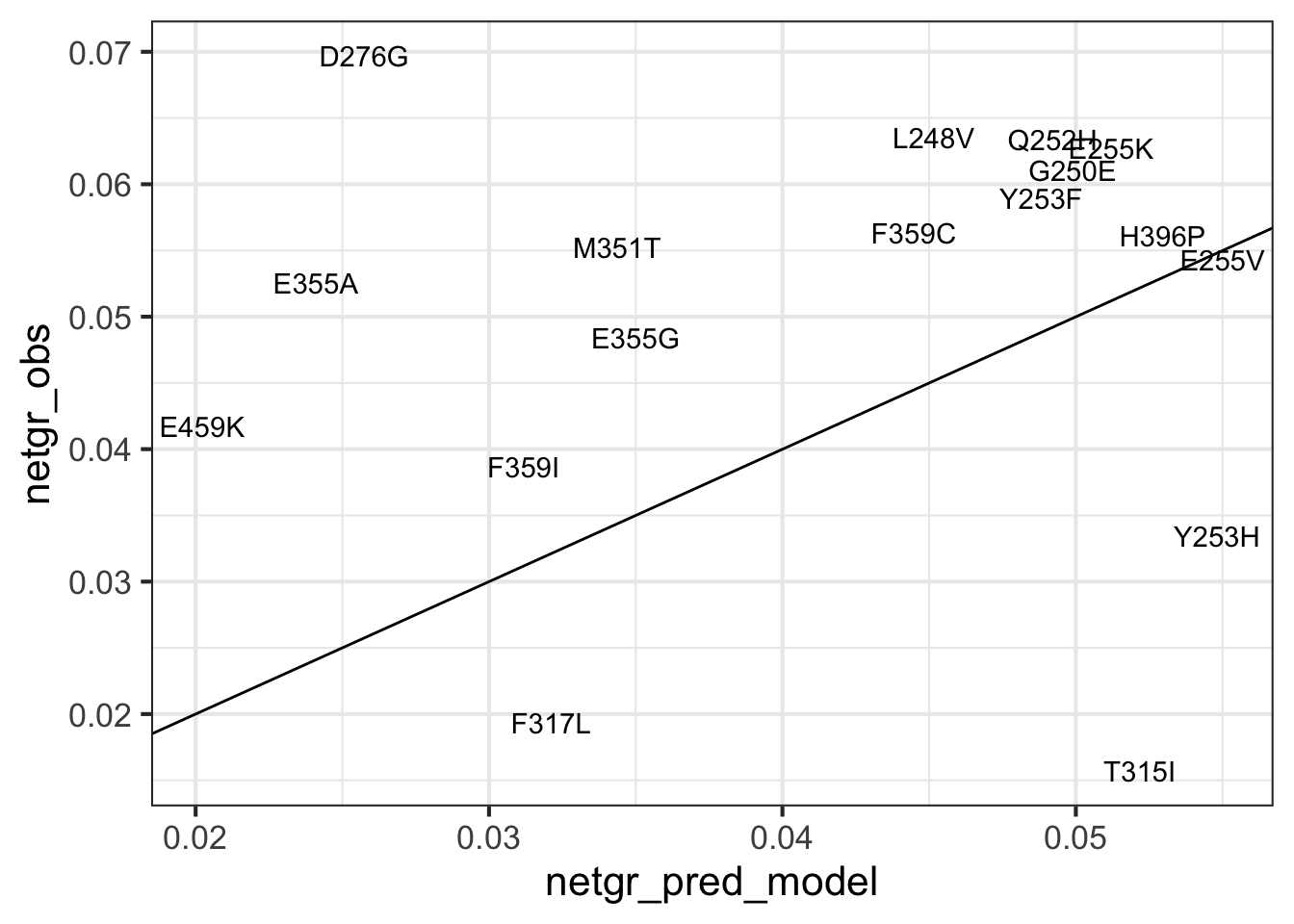
ggplot(resmuts_merged%>%filter(!species%in%(c("T315I","F317L"))),aes(x=netgr_pred_model,y=netgr_obs,label=species))+geom_text()+geom_abline()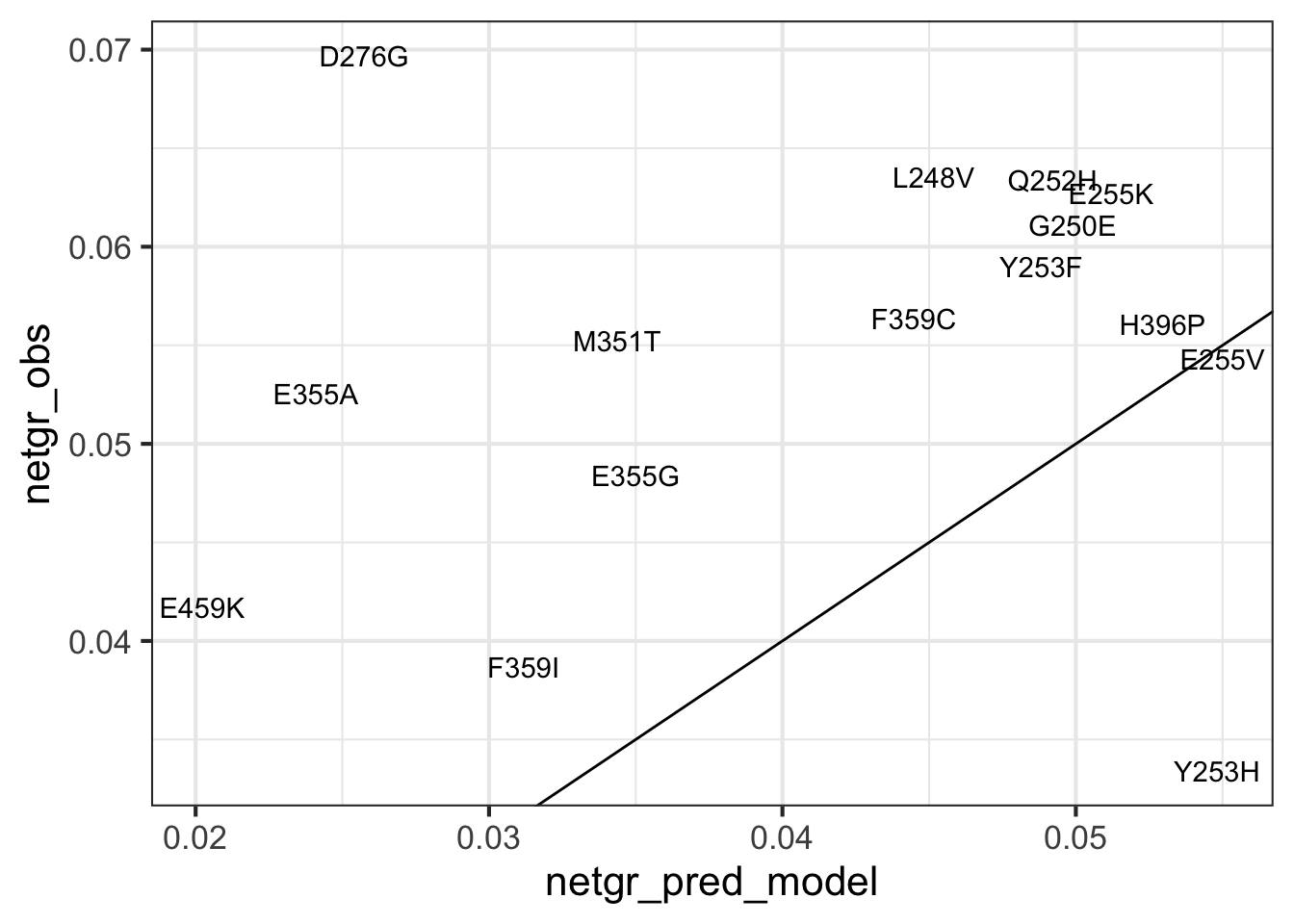
TSIID0.D4_clinrel_simple=TSIID0.D4_clinrel%>%dplyr::select(-c("ct.x","depth.x","maf.x","totalcells.x","totalmutant.x","ct.y","depth.y","maf.y","totalcells.y","totalmutant.y"))
TP3D2.D6_clinrel_simple=TP3D2.D6_clinrel%>%dplyr::select(-c("ct.x","depth.x","maf.x","totalcells.x","totalmutant.x","ct.y","depth.y","maf.y","totalcells.y","totalmutant.y"))
screen_compare=merge(TSIID0.D4_clinrel_simple,
TP3D2.D6_clinrel_simple,
by=c("ref_aa","protein_start","alt_aa","ref","alt","alt_start_pos","consequence_terms","ref_codon","alt_codon","alt_codon_shortest","n_nuc_min"),
all.x=T,
all.y=T)
screen_compare=resmuts_adder(screen_compare)
screen_compare=screen_compare%>%mutate(species=paste(ref_aa,protein_start,alt_aa,sep = ""))
plotly=ggplot(screen_compare,aes(x=score.x,y=score.y,label=species,color=resmuts))+geom_text()+geom_abline()
ggplotly(plotly)plotly=ggplot(screen_compare,aes(x=score.x,y=score.y,color=resmuts))+geom_point()+geom_abline()
ggplotly(plotly)# Side note: check how many of the top mutations are good by cosmic
# Check TP4 vs TP3 agreement. Check
# Next, I'm going to calculate the weighted mean of the scores across the concomitant vs in series screens
screen_compare_means=screen_compare%>%
filter(!score.x%in%NA,!score.y%in%NA,!score.x%in%NaN,!score.y%in%NaN)%>%
rowwise()%>%
mutate(score_mean=mean(c(score.x,score.y)))
a=screen_compare%>%
filter(!score.x%in%NA,!score.y%in%NA,!score.x%in%NaN,!score.y%in%NaN)TSID0.D2 TSID0.D4 TSID0.D6 Comparisons: D0.D2 vs D2 D4 vs D4 D6
Day 0 Day 2 vs Day 0 Day 4
###### TSI Day 0 vs Day 2 #########
delta_t=48 #hours
cells_before=107 #total cells at before time point
cells_after=1515 #total cells at after time point
netgr=.055
# before_timepoint=merge_samples("Novogene_lane14/Sample10_combined/sscs","Novogene_lane14/sample11/sscs")
before_timepoint=merge_samples("Novogene_lane14/Sample10_combined/sscs","Novogene_lane13/sample7/sscs")
before_timepoint=merge_samples(before_timepoint,"Novogene_lane14/sample11/sscs")
before_timepoint=merge_samples(before_timepoint,"Novogene_lane15/sample_3/sscs")
# before_timepoint=merge_samples("Novogene_lane15/sample_3/sscs","Novogene_lane13/sample7/sscs")
before_timepoint=variants_parser(before_timepoint,cells_before)
after_timepoint=merge_samples("Novogene_lane13/Sample9/sscs","Novogene_lane13/Sample10/sscs")
after_timepoint=merge_samples(after_timepoint,"Novogene_lane15/Sample_4/sscs")
after_timepoint=variants_parser(after_timepoint,cells_after)
before_after=compare_samples(before_timepoint,after_timepoint,netgr,delta_t)
before_after=before_after%>%filter(ct.x>=3)
before_after_screen1=before_after
###### TSI Day 0 vs Day 4 #########
delta_t=96 #hours
cells_before=107 #total cells at before time point
cells_after=17239 #total cells at after time point
netgr=.055
# before_timepoint=merge_samples("Novogene_lane15/sample_3/sscs","Novogene_lane13/sample7/sscs")
before_timepoint=merge_samples("Novogene_lane14/Sample10_combined/sscs","Novogene_lane13/sample7/sscs")
before_timepoint=merge_samples(before_timepoint,"Novogene_lane14/sample11/sscs")
before_timepoint=merge_samples(before_timepoint,"Novogene_lane15/sample_3/sscs")
before_timepoint=variants_parser(before_timepoint,cells_before)
after_timepoint=read.csv("data/Consensus_Data/novogene_lane15/sample_5/sscs/variant_caller_outputs/variants_unique_ann.csv")
after_timepoint=variants_parser(after_timepoint,cells_after)
before_after=compare_samples(before_timepoint,after_timepoint,netgr,delta_t)
before_after=before_after%>%filter(ct.x>=3)
before_after_screen2=before_after
before_after_screen1=before_after_screen1%>%
mutate(ct_screen1=ct.x)%>%
dplyr::select(-c("ct.x","depth.x","maf.x","totalcells.x","totalmutant.x","ct.y","depth.y","maf.y","totalcells.y","totalmutant.y"))
before_after_screen2=before_after_screen2%>%
mutate(ct_screen2=ct.x)%>%
dplyr::select(-c("ct.x","depth.x","maf.x","totalcells.x","totalmutant.x","ct.y","depth.y","maf.y","totalcells.y","totalmutant.y"))
screen_compare=merge(before_after_screen1,
before_after_screen2,
by=c("ref_aa","protein_start","alt_aa","ref","alt","alt_start_pos","consequence_terms","ref_codon","alt_codon","alt_codon_shortest","n_nuc_min"),
all.x=T,
all.y=T)
screen_compare=resmuts_adder(screen_compare)
screen_compare=res_residues_adder(screen_compare)
screen_compare=screen_compare%>%mutate(species=paste(ref_aa,protein_start,alt_aa,sep = ""))
############### Adding Count, Depth, and MAF columns for Ivan 2.20.23###########
# a=before_after_screen1
# b=before_after_screen2
# before_after_screen1=a
# before_after_screen2=b
# before_after_screen1=before_after_screen1%>%
# mutate(ct_screen1_before=ct.x,
# depth_screen1_before=depth.x,
# maf_screen1_before=maf.x,
# ct_screen1_after=ct.y,
# depth_screen1_after=depth.y,
# maf_screen1_after=maf.y)%>%
# dplyr::select(-c("ct.x","depth.x","maf.x","totalcells.x","totalmutant.x","ct.y","depth.y","maf.y","totalcells.y","totalmutant.y"))
#
# before_after_screen2=before_after_screen2%>%
# mutate(ct_screen2_before=ct.x,
# depth_screen2_before=depth.x,
# maf_screen2_before=maf.x,
# ct_screen2_after=ct.y,
# depth_screen2_after=depth.y,
# maf_screen2_after=maf.y)%>%
# dplyr::select(-c("ct.x","depth.x","maf.x","totalcells.x","totalmutant.x","ct.y","depth.y","maf.y","totalcells.y","totalmutant.y"))
#
# screen_compare=merge(before_after_screen1,
# before_after_screen2,
# by=c("ref_aa","protein_start","alt_aa","ref","alt","alt_start_pos","consequence_terms","ref_codon","alt_codon","alt_codon_shortest","n_nuc_min"),
# all.x=T,
# all.y=T,suffixes = c("_screen1","_screen2"))
#
# screen_compare=resmuts_adder(screen_compare)
# screen_compare=res_residues_adder(screen_compare)
# screen_compare=screen_compare%>%mutate(species=paste(ref_aa,protein_start,alt_aa,sep = ""))
#
# screen_compare_means=screen_compare%>%
# filter(!score_screen1%in%NA,!score_screen2%in%NA,!score_screen1%in%NaN,!score_screen2%in%NaN)%>%
# rowwise()%>%
# mutate(score_mean=mean(c(score_screen1,score_screen2)))
# write.csv(screen_compare_means,"IL3_Enrichment_bgmerged_2.20.23.csv")
##############
plotly=ggplot(screen_compare,aes(x=score.x,y=score.y,label=species,color=resmuts))+geom_text()+geom_abline()
ggplotly(plotly)plotly=ggplot(screen_compare,aes(x=score.x,y=score.y,color=resmuts))+geom_point()+geom_abline()
ggplotly(plotly)a=screen_compare%>%filter(!score.x%in%NA,!score.y%in%NA,!score.x%in%NaN,!score.y%in%NaN)
cor(a$score.x,a$score.y)[1] 0.8203084screen_compare_means=screen_compare%>%
filter(!score.x%in%NA,!score.y%in%NA,!score.x%in%NaN,!score.y%in%NaN)%>%
rowwise()%>%
mutate(score_mean=mean(c(score.x,score.y)),
score_weighteed_mean=weighted.mean(c(score.x,score.y),c(ct_screen1,ct_screen2)))
# write.csv(screen_compare_means,"IL3_Enrichment_2.12.23.csv")
ggplot(screen_compare%>%filter(protein_start>=242,protein_start<=494),aes(x=protein_start,y=alt_aa,fill=score.x))+geom_tile()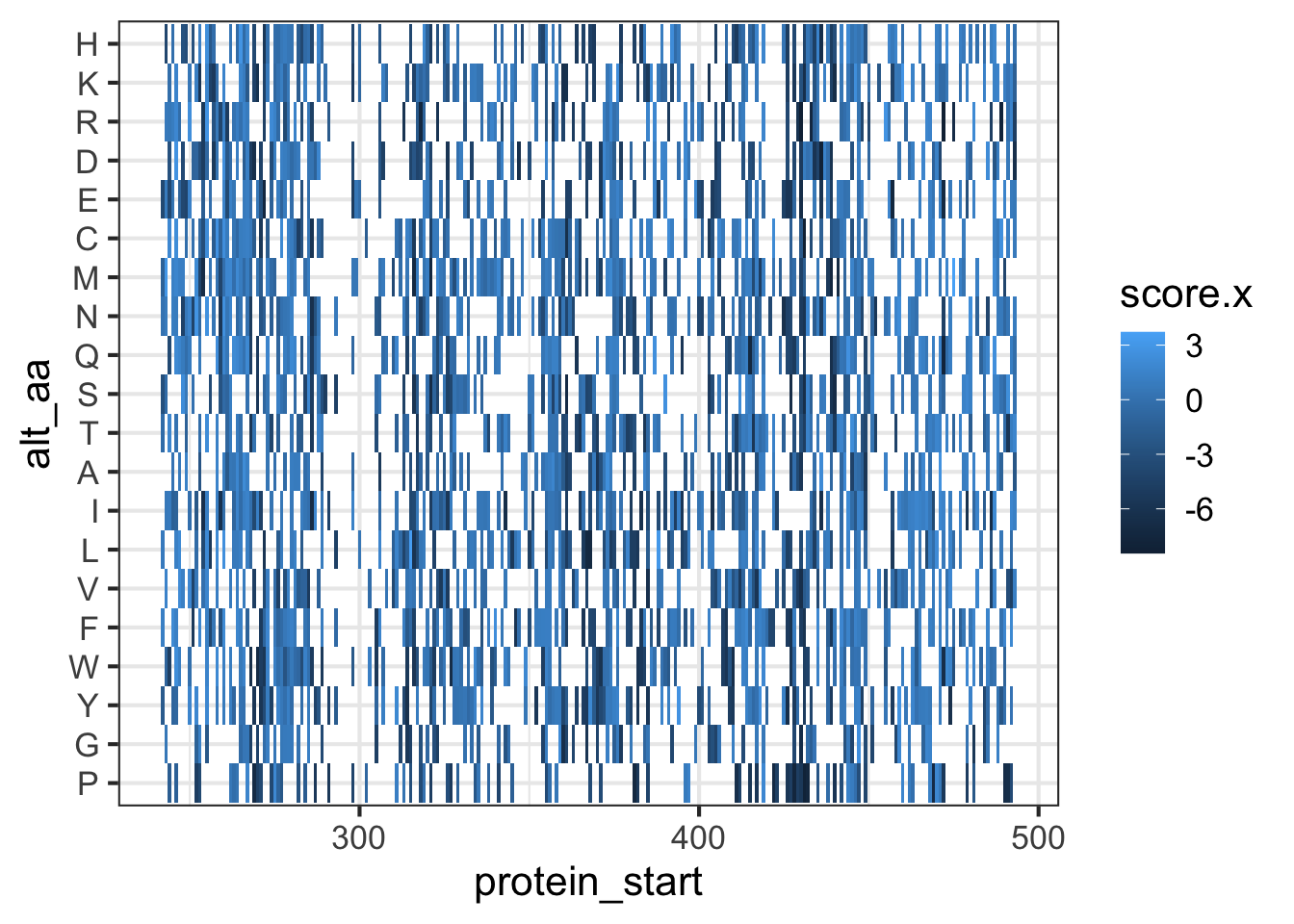
screen_compare=screen_compare%>%filter(!score.x%in%c(NA,NaN),!score.y%in%c(NA,NaN))
screen_compare$density=get_density(x = screen_compare$score.x,y=screen_compare$score.y,n=100)
ggplot(screen_compare,aes(x=score.x,y=score.y,color=density))+
geom_point()+
scale_color_viridis_c()+
# geom_jitter(position = position_jitter(width = 0.2, height = 0.2))+
# scale_x_continuous(name="Score Replicate 1",limits=c(-4,4))+
# scale_y_continuous(name="Score Replicate 2",limits=c(-4,4))+
geom_abline(linetype="dashed")+
theme(legend.position = "none")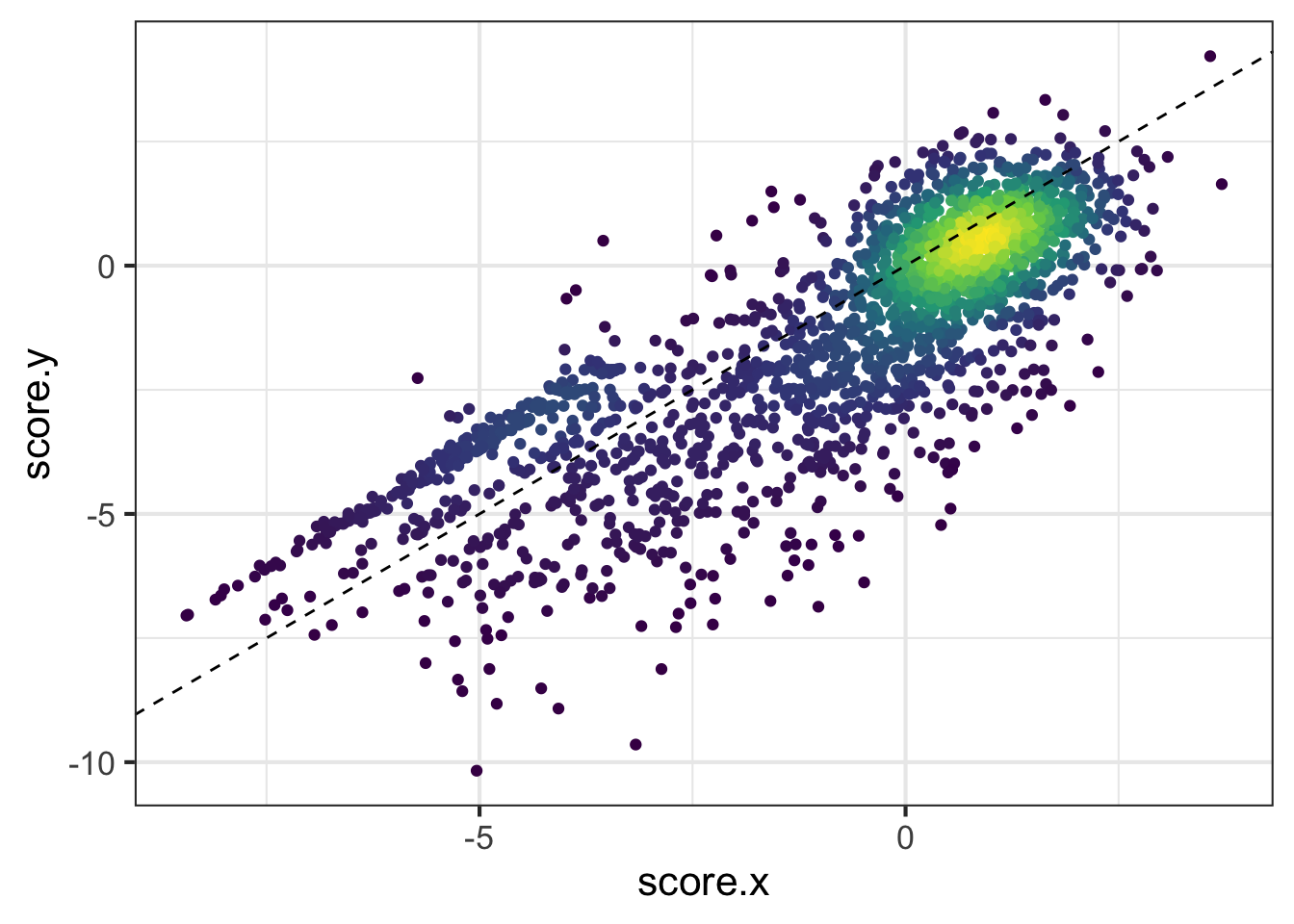
# ggsave("il3d0d2.d0d4_score_2.15.23.pdf",width=6,height=6,units="in",useDingbats=F)
a=screen_compare%>%filter(!score.x%in%c(NA,NaN),!score.y%in%c(NA,NaN))
cor(a$score.x,a$score.y)[1] 0.8203084# screen_compare$resmuts=factor(screen_compare$resmuts,levels=c("TRUE","FALSE"))
ggplot(screen_compare,aes(x=score.x,y=score.y))+
geom_point(color="black",shape=21,aes(fill=resmuts))+
geom_abline()+
scale_x_continuous(name="Score D0 D2")+
scale_y_continuous(name="Score D0 D4")+
labs(fill="Resistant\n Mutant")+
scale_fill_manual(values=c("gray90","red"))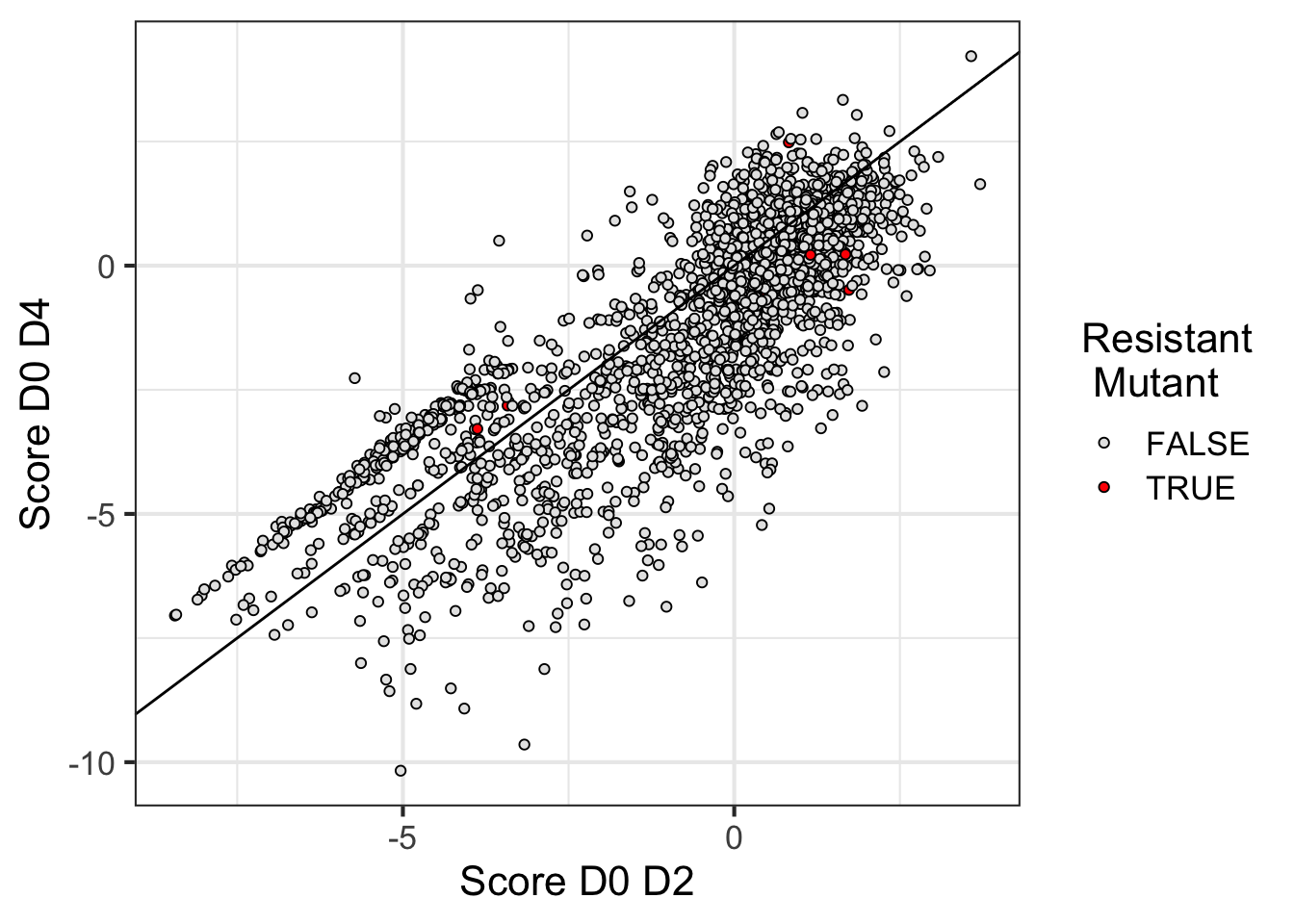
ggplot(screen_compare,aes(x=score.x,y=score.y))+
geom_point()+
geom_abline()+
scale_x_continuous(name="Score D0 D2")+
scale_y_continuous(name="Score D0 D4")+
labs(fill="Resistant\n Mutant")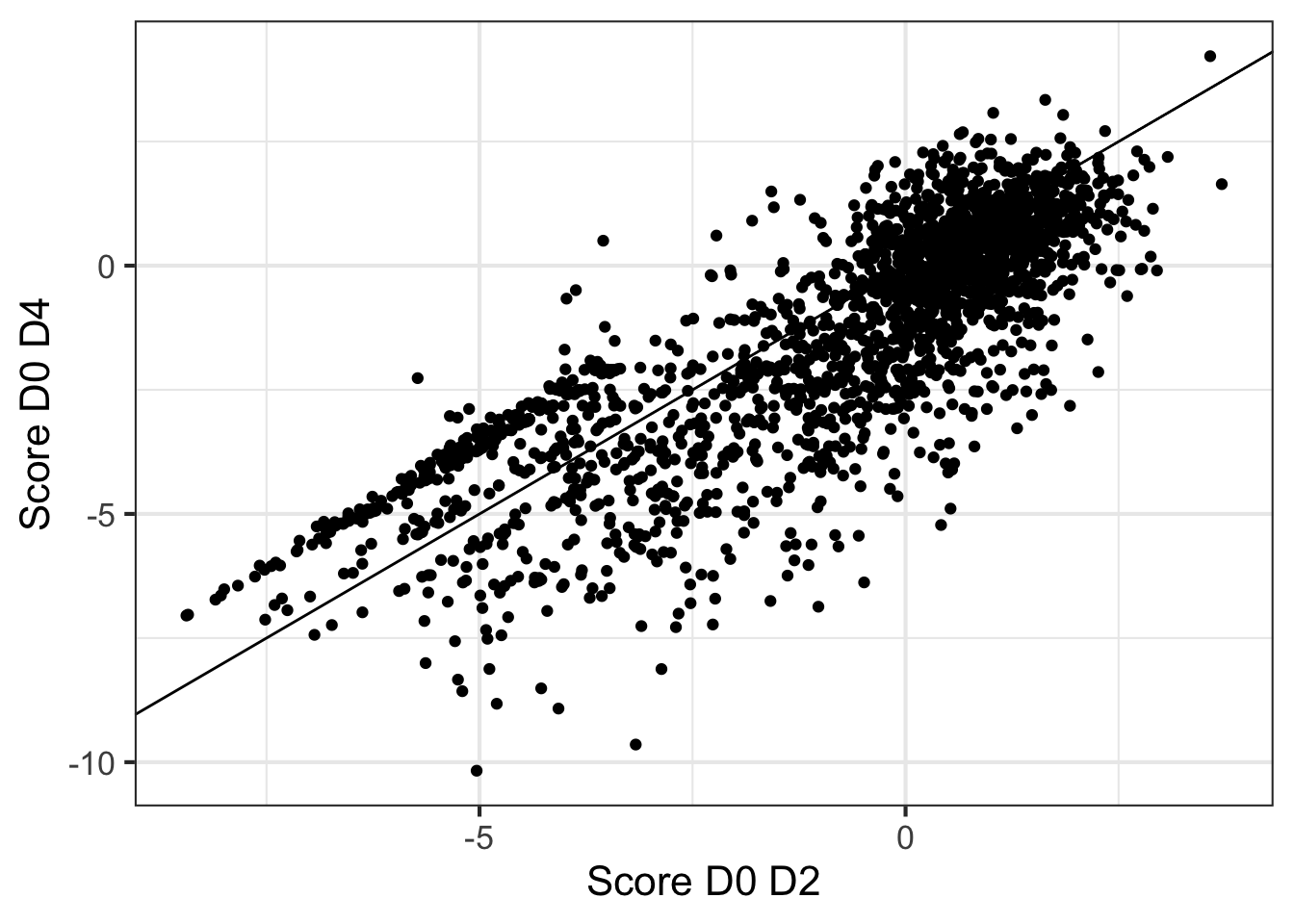
# ggsave("il3d0d2.d0d4_score_2.15.23.pdf",width=8,height=6,units="in",useDingbats=F)
data=read.csv("IL3_Enrichment_2.12.23.csv",header = T,stringsAsFactors = F)
data=data%>%filter(ct_screen1>=3,ct_screen2>=3)screen_compare[screen_compare$score.x%in%NA,"score.x"]=-6
#####Graying out unseen residues#####
df_grid = expand.grid(protein_start = c(242:493),alt_aa = unique(screen_compare$alt_aa))
###Trying to add in the score as WT for residues that are wt
reference_seq=read.table("data/Refs/ABL/abl_cds_translation.txt")
screen_compare=screen_compare%>%
rowwise()%>%
# group_by(protein_start)%>%
mutate(ref_aa=case_when(ref_aa%in%NA~substr(reference_seq,protein_start,protein_start),
T~ref_aa))%>%
mutate(wt=case_when(ref_aa==alt_aa~T,
T~F))
##########Plotting gray and yellow heatmap
# screen_compare=screen_compare%>%mutate(score.x)
screen_compare$alt_aa=factor(screen_compare$alt_aa,levels=c("P","G","Y","W","F","V","L","I","A","T","S","Q","N","M","C","E","D","R","K","H"))
screen_compare.filtered=screen_compare%>%filter(!protein_start%in%c(290:305))
# a=il3D0.D4.merge.filtered%>%filter(wt%in%T)
ggplot(screen_compare.filtered,aes(x=protein_start,y=alt_aa))+
geom_tile(data=subset(screen_compare.filtered,!is.na(score.x)),aes(fill=score.x))+
scale_fill_gradient2(low ="darkblue",midpoint=.03,mid="white", high ="red",name="Score")+
geom_tile(data=subset(screen_compare.filtered,is.na(score.x)&wt%in%F),aes(color="white"),linetype = "solid",color="white", fill = "gray90", alpha = 0.8)+
geom_tile(data=subset(screen_compare.filtered,is.na(score.x)&wt%in%T),aes(color="white"),linetype = "solid",color="white", fill = "yellow", alpha = 0.4)+
theme(panel.background=element_rect(fill="white", colour="black"))+scale_x_continuous(name="Residue on the ABL Kinase",limits=c(242,493),expand=c(0,0))+
ylab("Mutant Amino Acid")Warning: Removed 74 rows containing missing values (geom_tile).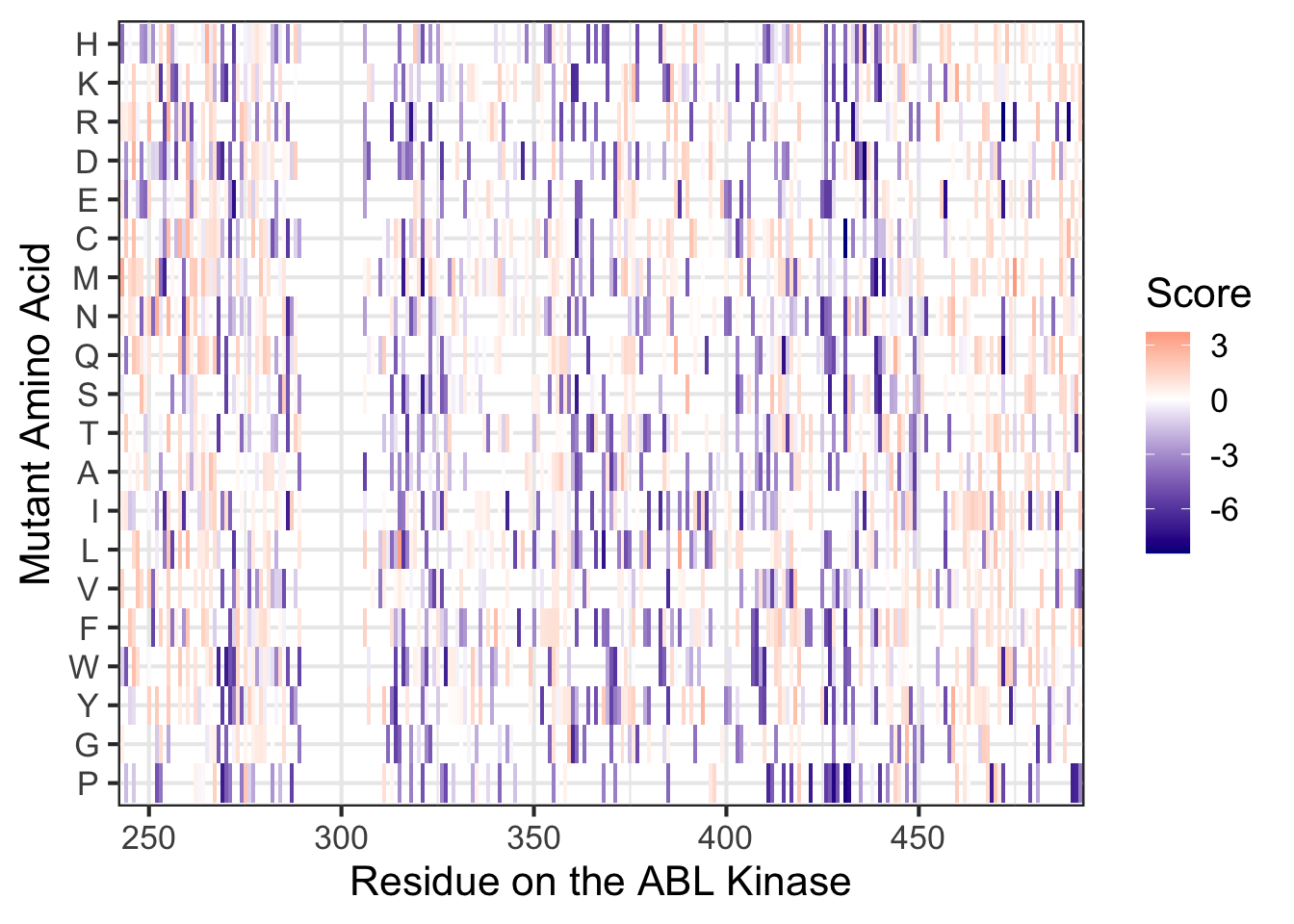
# ggsave("BCRABL_IL3_D0D4_2.12.23.pdf",height=6,width=24,units="in",useDingbats=F)
ggplot(screen_compare.filtered,aes(x=score.x,y=score.y))+geom_point()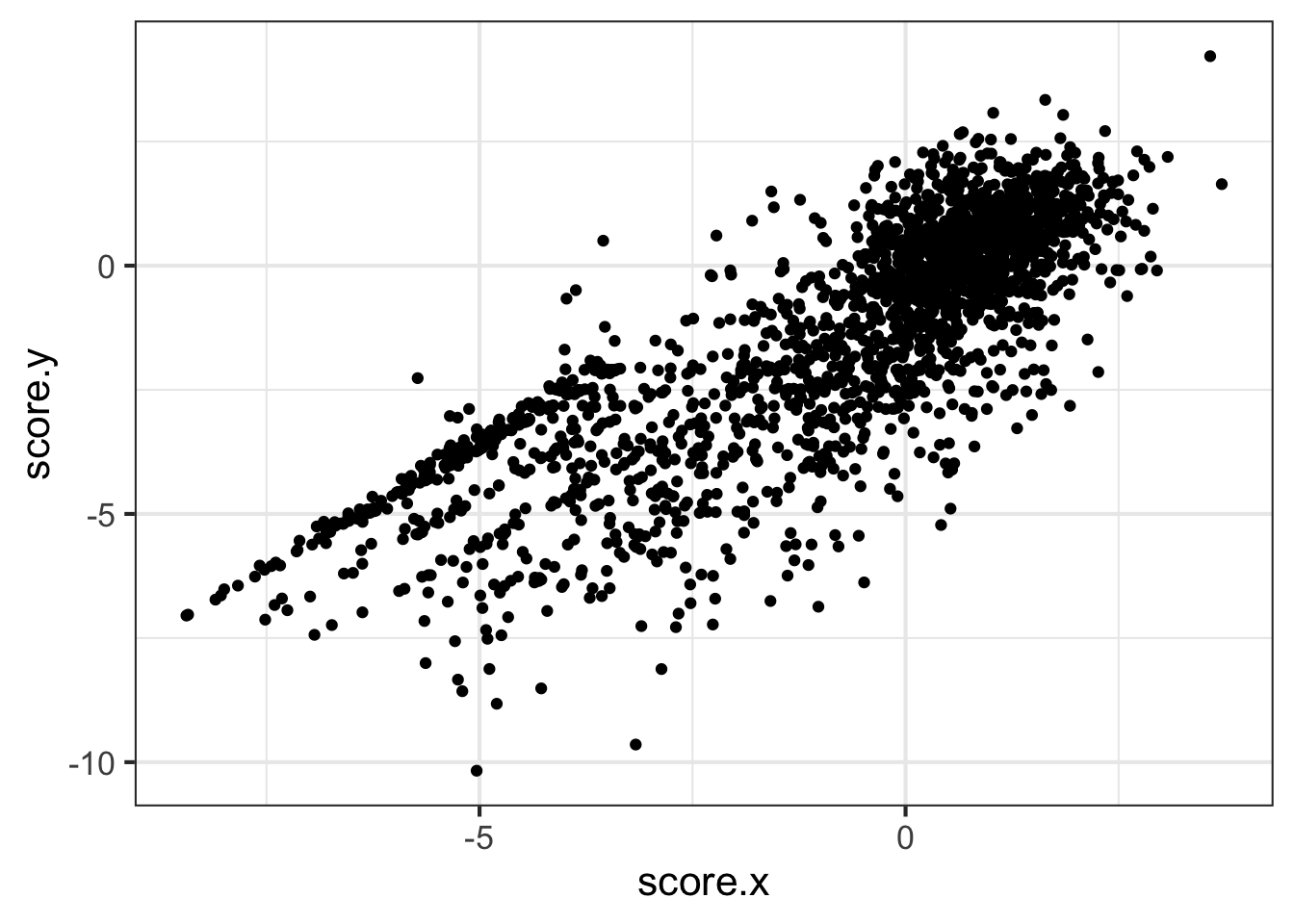
IL3 Day 0 Day 4 vs Day 2 Day 6
###### TSI Day 0 vs Day 4 #########
delta_t=96 #hours
cells_before=107 #total cells at before time point
cells_after=17239 #total cells at after time point
netgr=.055
before_timepoint=merge_samples("Novogene_lane14/Sample10_combined/sscs","Novogene_lane13/sample7/sscs")
before_timepoint=merge_samples(before_timepoint,"Novogene_lane14/sample11/sscs")
before_timepoint=merge_samples(before_timepoint,"Novogene_lane15/sample_3/sscs")
before_timepoint=variants_parser(before_timepoint,cells_before)
after_timepoint=read.csv("data/Consensus_Data/novogene_lane15/sample_5/sscs/variant_caller_outputs/variants_unique_ann.csv")
after_timepoint=variants_parser(after_timepoint,cells_after)
before_after=compare_samples(before_timepoint,after_timepoint,netgr,delta_t)
# before_after=before_after%>%filter(ct.x>=3)
before_after_screen1=before_after
###### TSI Day 2 vs Day 6 #########
delta_t=96 #hours
cells_before=1515 #total cells at before time point
cells_after=430713 #total cells at after time point
netgr=.055
before_timepoint=merge_samples("Novogene_lane13/Sample9/sscs","Novogene_lane13/Sample10/sscs")
before_timepoint=merge_samples(before_timepoint,"Novogene_lane15/Sample_4/sscs")
before_timepoint=variants_parser(before_timepoint,cells_before)
after_timepoint=read.csv("data/Consensus_Data/novogene_lane17/sample2/sscs/variant_caller_outputs/variants_unique_ann.csv")
after_timepoint=variants_parser(after_timepoint,cells_after)
before_after=compare_samples(before_timepoint,after_timepoint,netgr,delta_t)
# before_after=before_after%>%filter(ct.x>=3)
before_after_screen2=before_after
before_after_screen1=before_after_screen1%>%
mutate(ct_screen1=ct.x)%>%
dplyr::select(-c("ct.x","depth.x","maf.x","totalcells.x","totalmutant.x","ct.y","depth.y","maf.y","totalcells.y","totalmutant.y"))
before_after_screen2=before_after_screen2%>%
mutate(ct_screen2=ct.x)%>%
dplyr::select(-c("ct.x","depth.x","maf.x","totalcells.x","totalmutant.x","ct.y","depth.y","maf.y","totalcells.y","totalmutant.y"))
screen_compare=merge(before_after_screen1,
before_after_screen2,
by=c("ref_aa","protein_start","alt_aa","ref","alt","alt_start_pos","consequence_terms","ref_codon","alt_codon","alt_codon_shortest","n_nuc_min"),
all.x=T,
all.y=T)
screen_compare=resmuts_adder(screen_compare)
screen_compare=screen_compare%>%mutate(species=paste(ref_aa,protein_start,alt_aa,sep = ""))
plotly=ggplot(screen_compare,aes(x=score.x,y=score.y,label=species,color=resmuts))+geom_text()+geom_abline()
ggplotly(plotly)plotly=ggplot(screen_compare,aes(x=score.x,y=score.y,color=resmuts))+geom_point()+geom_abline()
ggplotly(plotly)screen_compare_means=screen_compare%>%
filter(!score.x%in%NA,!score.y%in%NA,!score.x%in%NaN,!score.y%in%NaN)%>%
rowwise()%>%
mutate(score_mean=mean(c(score.x,score.y)),
score_weighteed_mean=weighted.mean(c(score.x,score.y),c(ct_screen1,ct_screen2)))
# write.csv(screen_compare_means,"IL3_Enrichment_2.8.23.csv")
ggplot(screen_compare%>%filter(protein_start>=242,protein_start<=494),aes(x=protein_start,y=alt_aa,fill=score.x))+geom_tile()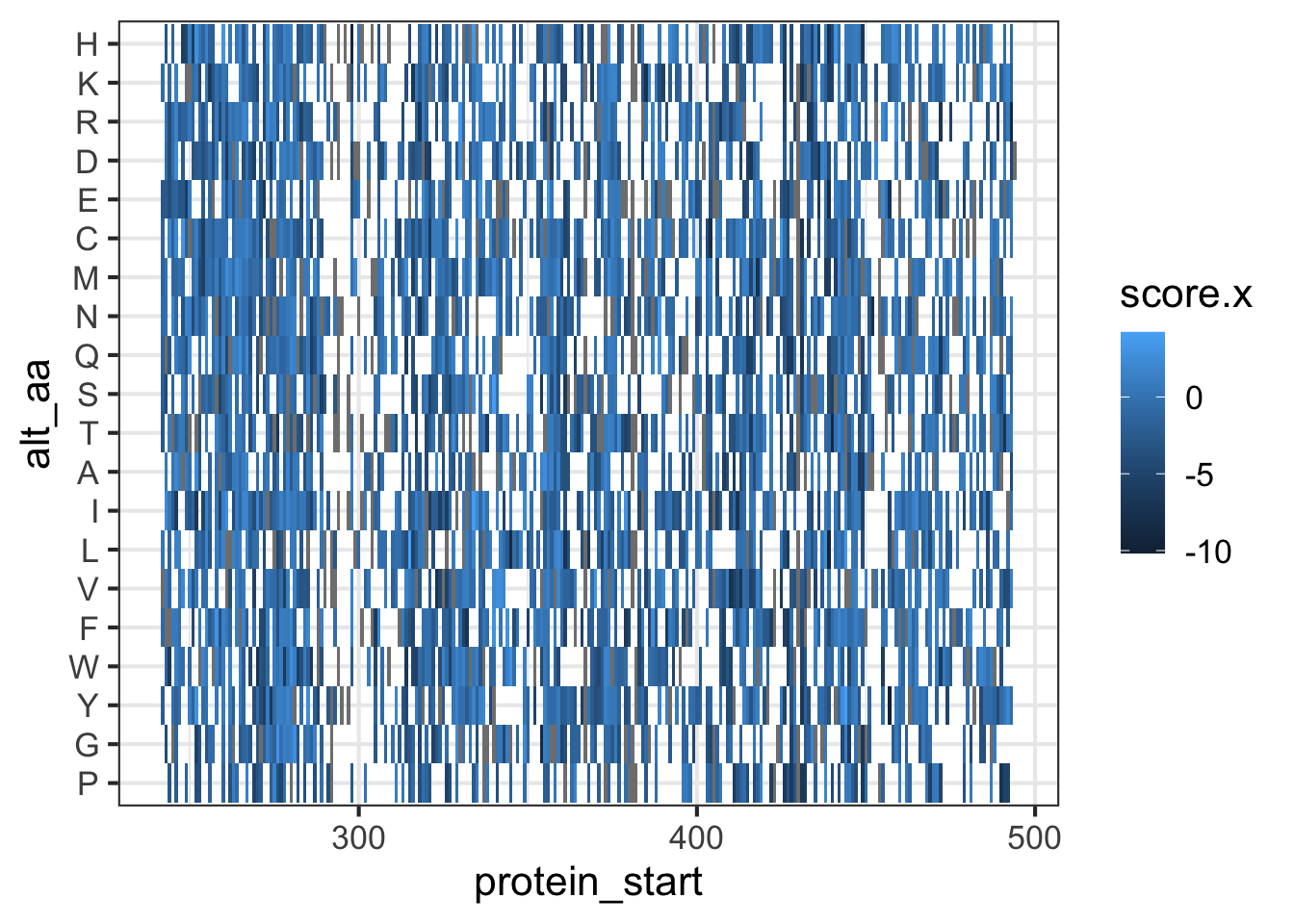
a=screen_compare%>%filter(!score.x%in%NA,!score.y%in%NA,!score.x%in%NaN,!score.y%in%NaN)
cor(a$score.x,a$score.y)[1] -0.2418853screen_compare[screen_compare$score.x%in%NA,"score.x"]=-6
#####Graying out unseen residues#####
df_grid = expand.grid(protein_start = c(242:493),alt_aa = unique(screen_compare$alt_aa))
###Trying to add in the score as WT for residues that are wt
reference_seq=read.table("data/Refs/ABL/abl_cds_translation.txt")
screen_compare=screen_compare%>%
rowwise()%>%
# group_by(protein_start)%>%
mutate(ref_aa=case_when(ref_aa%in%NA~substr(reference_seq,protein_start,protein_start),
T~ref_aa))%>%
mutate(wt=case_when(ref_aa==alt_aa~T,
T~F))
##########Plotting gray and yellow heatmap
# screen_compare=screen_compare%>%mutate(score.x)
screen_compare$alt_aa=factor(screen_compare$alt_aa,levels=c("P","G","Y","W","F","V","L","I","A","T","S","Q","N","M","C","E","D","R","K","H"))
screen_compare.filtered=screen_compare%>%filter(!protein_start%in%c(290:305))
# a=il3D0.D4.merge.filtered%>%filter(wt%in%T)
ggplot(screen_compare.filtered,aes(x=protein_start,y=alt_aa))+
geom_tile(data=subset(screen_compare.filtered,!is.na(score.x)),aes(fill=score.x))+
scale_fill_gradient2(low ="darkblue",midpoint=.03,mid="white", high ="red",name="Score")+
geom_tile(data=subset(screen_compare.filtered,is.na(score.x)&wt%in%F),aes(color="white"),linetype = "solid",color="white", fill = "gray90", alpha = 0.8)+
geom_tile(data=subset(screen_compare.filtered,is.na(score.x)&wt%in%T),aes(color="white"),linetype = "solid",color="white", fill = "yellow", alpha = 0.4)+
theme(panel.background=element_rect(fill="white", colour="black"))+scale_x_continuous(name="Residue on the ABL Kinase",limits=c(242,493),expand=c(0,0))+
ylab("Mutant Amino Acid")Warning: Removed 363 rows containing missing values (geom_tile).Warning: Removed 3 rows containing missing values (geom_tile).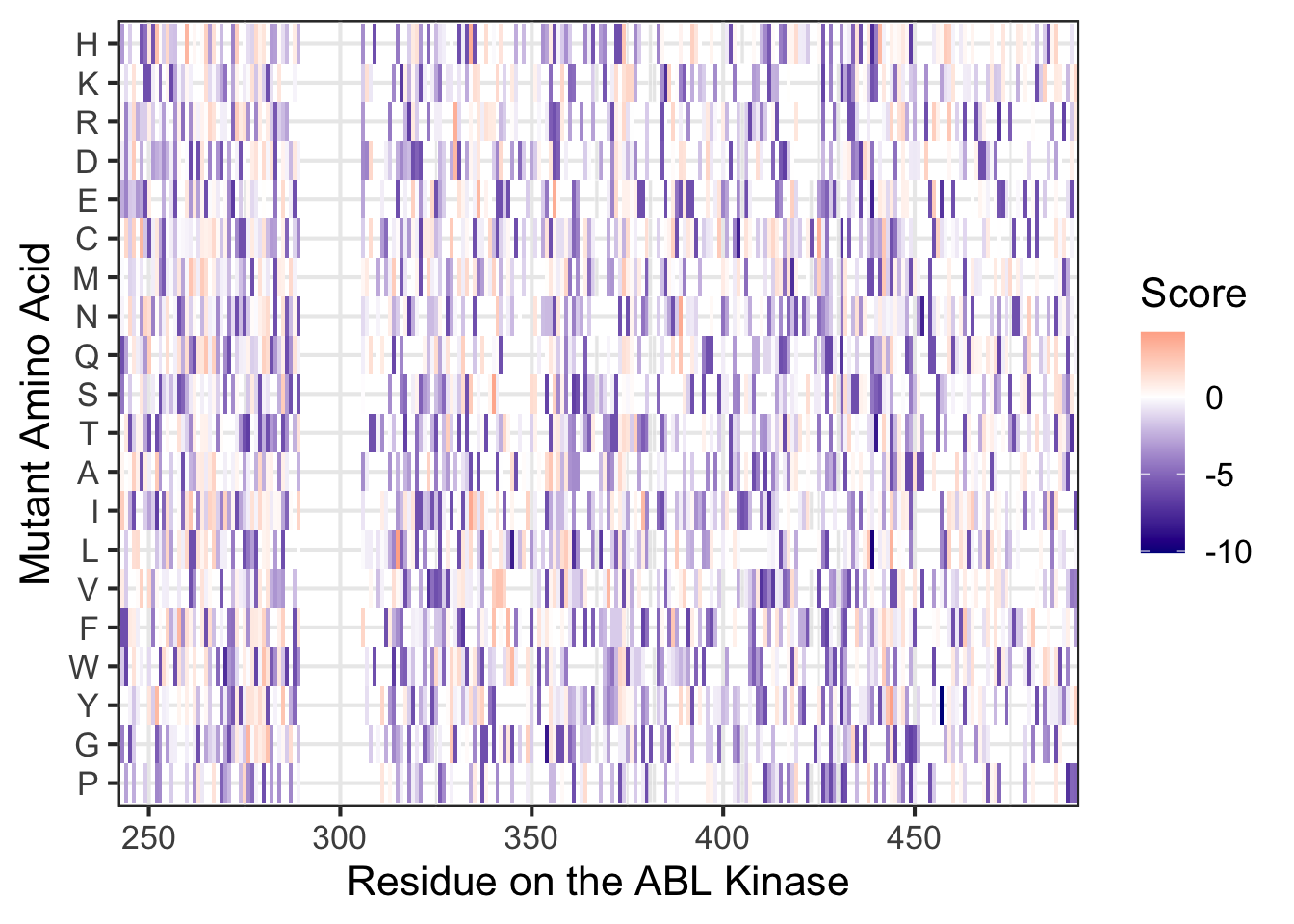
# ggsave("BCRABL_IL3_D0D4_2.8.23.pdf",height=6,width=24,units="in",useDingbats=F)
ggplot(screen_compare.filtered,aes(x=score.x,y=score.y))+geom_point()Warning: Removed 1215 rows containing missing values (geom_point).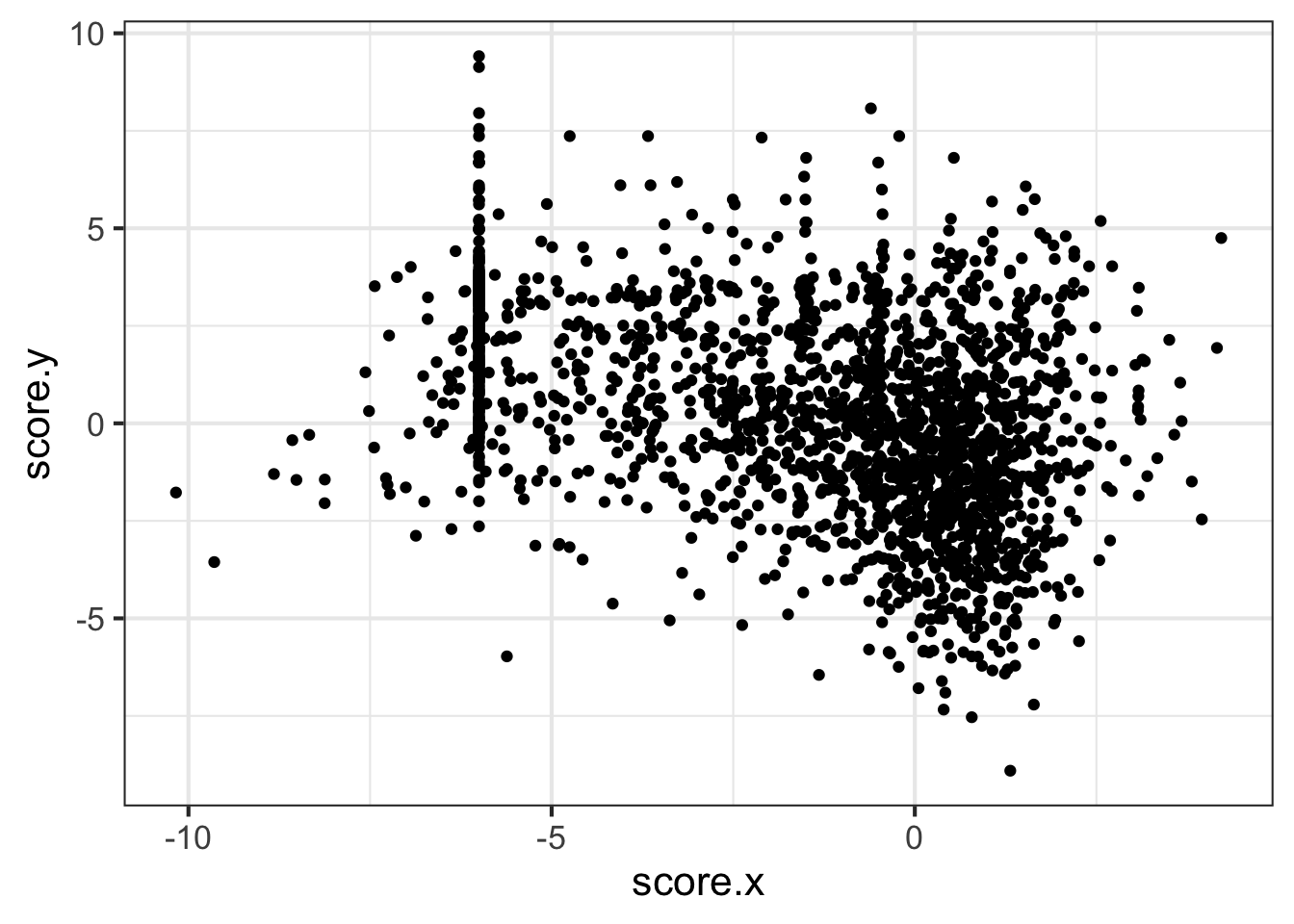
Imatinib Screens Day 0 Day 2 vs Day 0 Day 4
###### TSII Day 0 vs Day 2 #########
delta_t=48 #hours
cells_before=370 #total cells at before time point
cells_after=1192.25 #total cells at after time point
netgr=.055
# before_timepoint=merge_samples("Novogene_lane14/Sample10_combined/sscs","Novogene_lane14/sample11/sscs")
before_timepoint=merge_samples("Novogene_lane14/Sample10_combined/sscs","Novogene_lane13/sample7/sscs")
before_timepoint=merge_samples(before_timepoint,"Novogene_lane14/sample11/sscs")
before_timepoint=merge_samples(before_timepoint,"Novogene_lane15/sample_3/sscs")
# source()
before_timepoint=variants_parser(before_timepoint,cells_before)
after_timepoint=merge_samples("Novogene_lane15/sample_6/sscs","Novogene_lane14/sample12/sscs")
# after_timepoint=merge_samples(after_timepoint,"Novogene_lane15/Sample_4/sscs")
# after_timepoint=read.csv("data/Consensus_Data/novogene_lane15/sample_7/sscs/variant_caller_outputs/variants_unique_ann.csv")
after_timepoint=variants_parser(after_timepoint,cells_after)
before_after=compare_samples(before_timepoint,after_timepoint,netgr,delta_t)
before_after=before_after%>%filter(ct.x>=3)
before_after_screen1=before_after
###### TSII Day 0 vs Day 4 #########
delta_t=96 #hours
cells_before=370 #total cells at before time point
cells_after=10397 #total cells at after time point
netgr=.055
# before_timepoint=merge_samples("Novogene_lane15/sample_3/sscs","Novogene_lane13/sample7/sscs")
before_timepoint=merge_samples("Novogene_lane14/Sample10_combined/sscs","Novogene_lane13/sample7/sscs")
before_timepoint=merge_samples(before_timepoint,"Novogene_lane14/sample11/sscs")
before_timepoint=merge_samples(before_timepoint,"Novogene_lane15/sample_3/sscs")
before_timepoint=variants_parser(before_timepoint,cells_before)
after_timepoint=read.csv("data/Consensus_Data/novogene_lane15/sample_7/sscs/variant_caller_outputs/variants_unique_ann.csv")
# after_timepoint=read.csv("data/Consensus_Data/novogene_lane17/sample3/sscs/variant_caller_outputs/variants_unique_ann.csv")
after_timepoint=variants_parser(after_timepoint,cells_after)
before_after=compare_samples(before_timepoint,after_timepoint,netgr,delta_t)
# before_after=before_after%>%filter(ct.x>=3)
before_after_screen2=before_after
############### Adding Count, Depth, and MAF columns for Ivan 2.20.23###########
# a=before_after_screen1
# b=before_after_screen2
# before_after_screen1=a
# before_after_screen2=b
# before_after_screen1=before_after_screen1%>%
# mutate(ct_screen1_before=ct.x,
# depth_screen1_before=depth.x,
# maf_screen1_before=maf.x,
# ct_screen1_after=ct.y,
# depth_screen1_after=depth.y,
# maf_screen1_after=maf.y)%>%
# dplyr::select(-c("ct.x","depth.x","maf.x","totalcells.x","totalmutant.x","ct.y","depth.y","maf.y","totalcells.y","totalmutant.y"))
#
# before_after_screen2=before_after_screen2%>%
# mutate(ct_screen2_before=ct.x,
# depth_screen2_before=depth.x,
# maf_screen2_before=maf.x,
# ct_screen2_after=ct.y,
# depth_screen2_after=depth.y,
# maf_screen2_after=maf.y)%>%
# dplyr::select(-c("ct.x","depth.x","maf.x","totalcells.x","totalmutant.x","ct.y","depth.y","maf.y","totalcells.y","totalmutant.y"))
#
# screen_compare=merge(before_after_screen1,
# before_after_screen2,
# by=c("ref_aa","protein_start","alt_aa","ref","alt","alt_start_pos","consequence_terms","ref_codon","alt_codon","alt_codon_shortest","n_nuc_min"),
# all.x=T,
# all.y=T,suffixes = c("_screen1","_screen2"))
#
# screen_compare=resmuts_adder(screen_compare)
# screen_compare=res_residues_adder(screen_compare)
# screen_compare=screen_compare%>%mutate(species=paste(ref_aa,protein_start,alt_aa,sep = ""))
#
# screen_compare_means=screen_compare%>%
# filter(!score_screen1%in%NA,!score_screen2%in%NA,!score_screen1%in%NaN,!score_screen2%in%NaN)%>%
# rowwise()%>%
# mutate(score_mean=mean(c(score_screen1,score_screen2)))
# write.csv(screen_compare_means,"Imat_Enrichment_bgmerged_2.22.23.csv")
##############
before_after_screen1=before_after_screen1%>%
mutate(ct_screen1=ct.x)%>%
dplyr::select(-c("ct.x","depth.x","maf.x","totalcells.x","totalmutant.x","ct.y","depth.y","maf.y","totalcells.y","totalmutant.y"))
before_after_screen2=before_after_screen2%>%
mutate(ct_screen2=ct.x)%>%
dplyr::select(-c("ct.x","depth.x","maf.x","totalcells.x","totalmutant.x","ct.y","depth.y","maf.y","totalcells.y","totalmutant.y"))
screen_compare=merge(before_after_screen1,
before_after_screen2,
by=c("ref_aa","protein_start","alt_aa","ref","alt","alt_start_pos","consequence_terms","ref_codon","alt_codon","alt_codon_shortest","n_nuc_min"),
all.x=T,
all.y=T)
screen_compare=resmuts_adder(screen_compare)
screen_compare=screen_compare%>%mutate(species=paste(ref_aa,protein_start,alt_aa,sep = ""))
plotly=ggplot(screen_compare,aes(x=score.x,y=score.y,label=species,color=resmuts))+geom_text()+geom_abline()
ggplotly(plotly)a=screen_compare%>%filter(!score.x%in%NA,!score.y%in%NA,!score.x%in%NaN,!score.y%in%NaN)
cor(a$score.x,a$score.y)[1] 0.7306101screen_compare_means=screen_compare%>%
filter(!score.x%in%NA,!score.y%in%NA,!score.x%in%NaN,!score.y%in%NaN)%>%
rowwise()%>%
mutate(score_mean=mean(c(score.x,score.y)),
score_weighteed_mean=weighted.mean(c(score.x,score.y),c(ct_screen1,ct_screen2)))
# write.csv(screen_compare_means,"Imatinib_Enrichment_2.20.23_v2.csv")
ggplot(screen_compare%>%filter(protein_start>=242,protein_start<=494),aes(x=protein_start,y=alt_aa,fill=score.x))+geom_tile()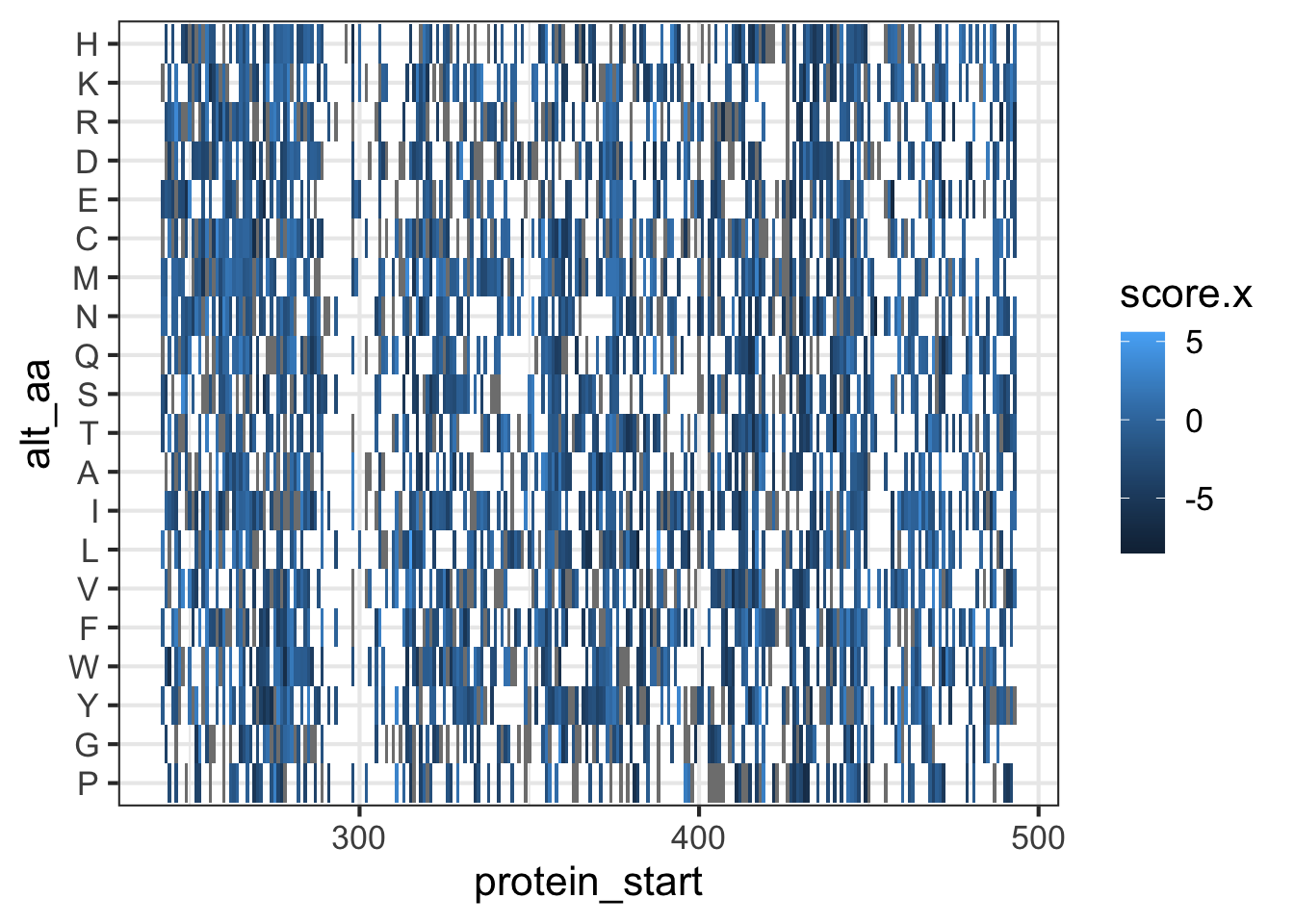
screen_compare=res_residues_adder(screen_compare)
plotly=ggplot(screen_compare,aes(x=score.x,y=score.y,label=species,color=resmuts))+geom_text()+geom_abline()
ggplotly(plotly)plotly=ggplot(screen_compare%>%filter(!species%in%"T315I"),aes(x=score.x,y=score.y,label=species,color=resmuts))+geom_text()+geom_abline()
ggplotly(plotly)ggplot(screen_compare%>%filter(!species%in%"T315I"),aes(x=score.x,y=score.y,label=species,color=resmuts))+geom_text()+geom_abline()+cleanupWarning: Removed 635 rows containing missing values (geom_text).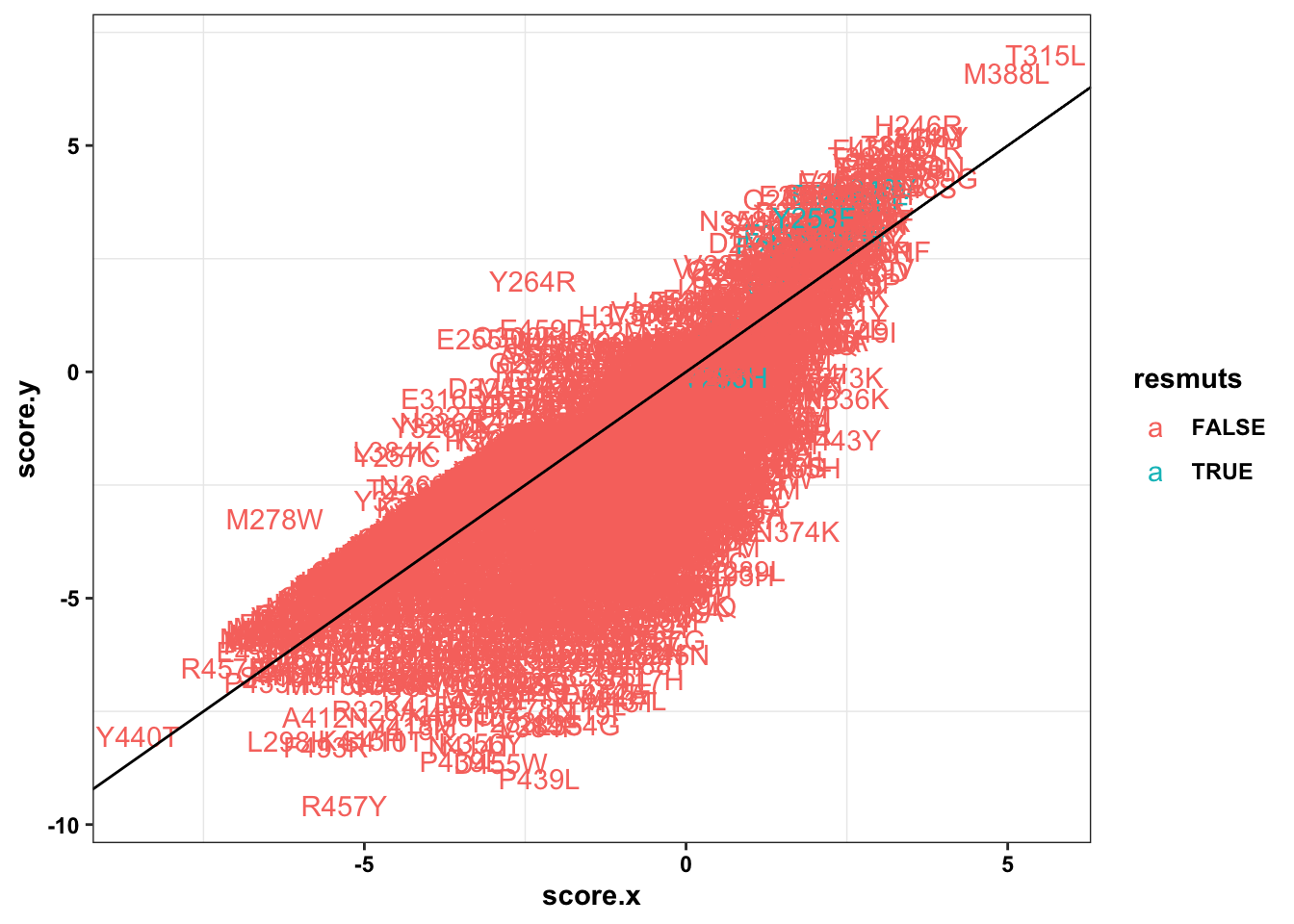
plotly=ggplot(screen_compare,aes(x=netgr_obs.x,y=netgr_obs.y,label=species,color=resmuts))+geom_text()+geom_abline()
ggplotly(plotly)screen_compare=screen_compare%>%filter(!score.x%in%c(NA,NaN),!score.y%in%c(NA,NaN))
screen_compare$density=get_density(x = screen_compare$score.x,y=screen_compare$score.y,n=50)
ggplot(screen_compare,aes(x=netgr_obs.x,y=netgr_obs.y,color=density))+
geom_point()+
scale_color_viridis_c()+
# geom_jitter(position = position_jitter(width = 0.1, height = 0.1))+
# scale_x_continuous(name="Score Replicate 1",limits=c(-5,5))+
# scale_y_continuous(name="Score Replicate 2",limits=c(-5,5))+
geom_abline(linetype="dashed")+
theme(legend.position = "none")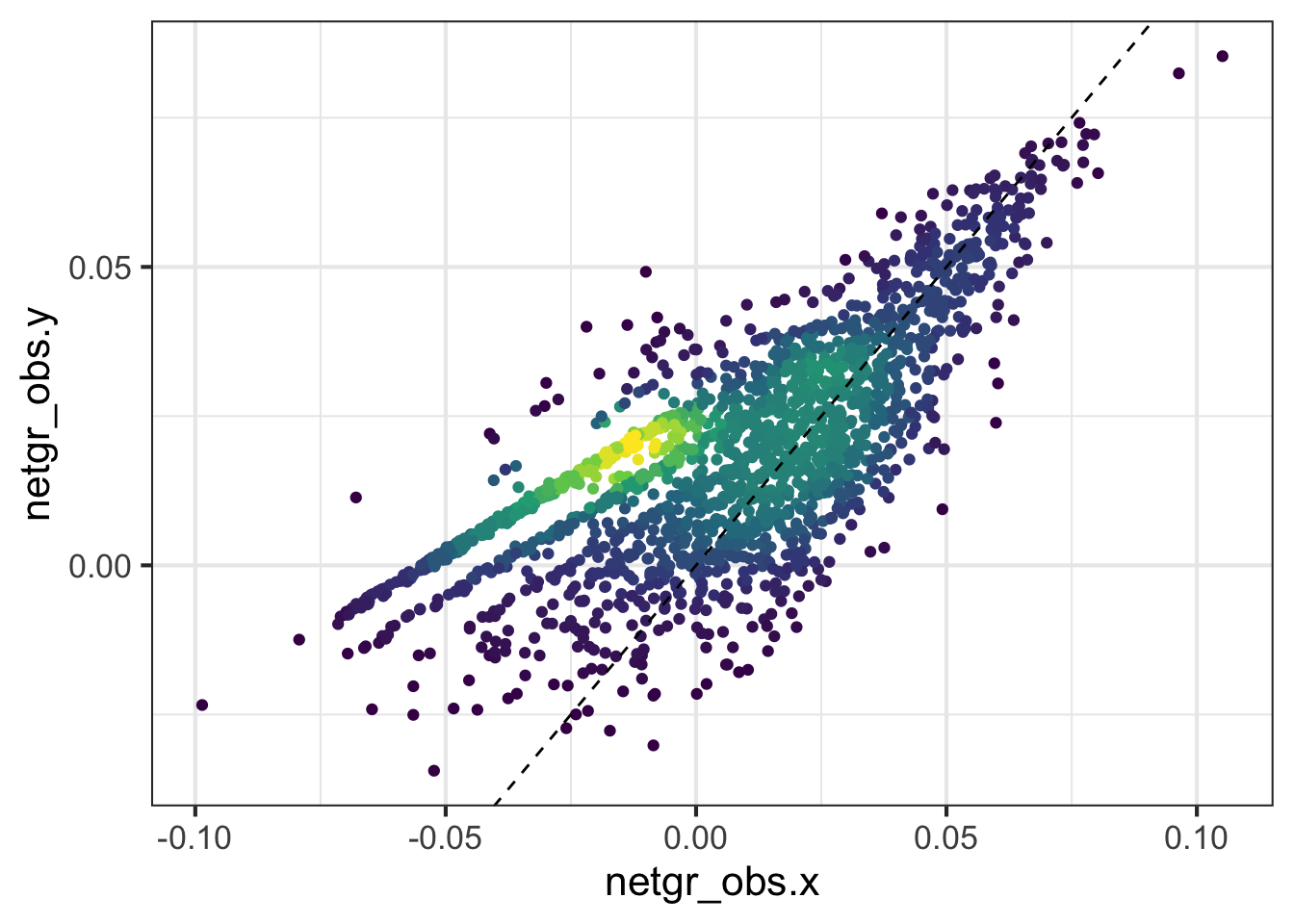
a=screen_compare%>%filter(!score.x%in%c(NA,NaN),!score.y%in%c(NA,NaN))
cor(a$score.x,a$score.y)[1] 0.7306101# cor(a$netgr_obs.x,a$netgr_obs.y)
plotly=ggplot(screen_compare,aes(x=score.x,y=score.y))+
geom_point(color="black",shape=21,aes(fill=resmuts))+geom_abline()
ggplotly(plotly)ggplot(screen_compare,aes(x=score.x,y=score.y))+
geom_point(color="black",shape=21,aes(fill=resmuts))+
geom_abline()+
scale_x_continuous(name="Score D0 D2")+
scale_y_continuous(name="Score D0 D4")+
labs(fill="Resistant\n Mutant")+
scale_fill_manual(values=c("gray90","red"))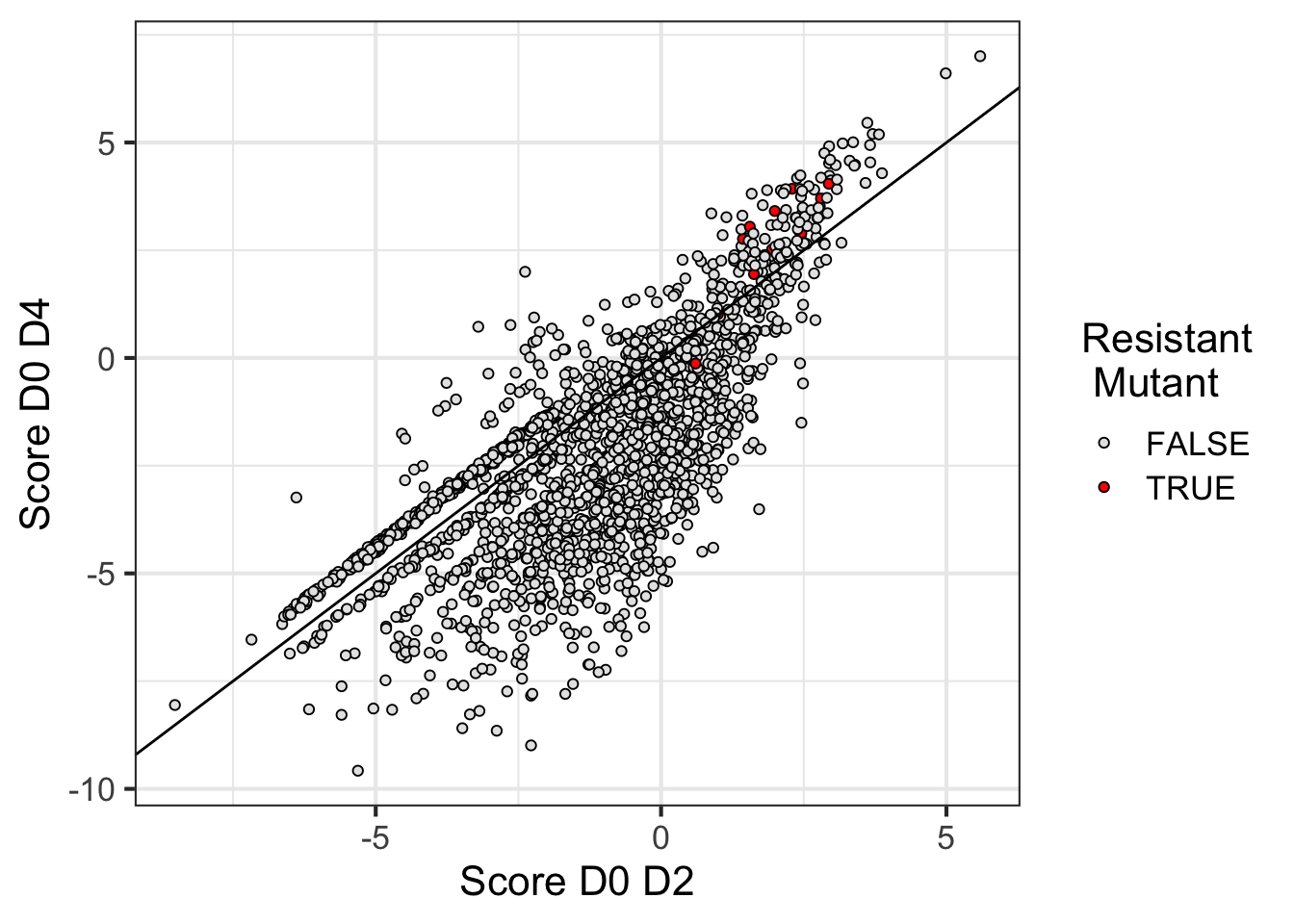
# ggsave("imatd0d2.d0d4_score_2.15.23.pdf",width=8,height=6,units="in",useDingbats=F)IL3 Screens for plotting Day 0 Day 2 vs Day 0 Day 4
###### TSII Day 0 vs Day 2 #########
delta_t=48 #hours
cells_before=107 #total cells at before time point
cells_after=1515 #total cells at after time point
netgr=.055
before_timepoint=merge_samples("Novogene_lane14/Sample10_combined/sscs","Novogene_lane13/sample7/sscs")
before_timepoint=merge_samples(before_timepoint,"Novogene_lane14/sample11/sscs")
before_timepoint=merge_samples(before_timepoint,"Novogene_lane15/sample_3/sscs")
before_timepoint=merge_samples(before_timepoint,"Novogene_lane16b/Sample9/sscs")
before_timepoint=merge_samples(before_timepoint,"Novogene_lane16b/Sample10/sscs")
before_timepoint=variants_parser(before_timepoint,cells_before)
##########TSI Imatinib D2###########
# after_timepoint=merge_samples("Novogene_lane15/sample_6/sscs","Novogene_lane14/sample12/sscs")
# after_timepoint=merge_samples(after_timepoint,"Novogene_lane15/Sample_7/sscs")
##########TSI D2###########
after_timepoint=merge_samples("Novogene_lane13/Sample9/sscs","Novogene_lane13/Sample10/sscs")
# after_timepoint=merge_samples(after_timepoint,"Novogene_lane15/Sample_4/sscs")
####################
after_timepoint=variants_parser(after_timepoint,cells_after)
before_after=compare_samples(before_timepoint,after_timepoint,netgr,delta_t)
# a=before_after%>%filter(protein_start%in%271)# a=before_after%>%filter(protein_start%in%271)
ggplot(before_after%>%filter(protein_start>=240,protein_start<=494),aes(x=protein_start,y=alt_aa,fill=score))+geom_tile()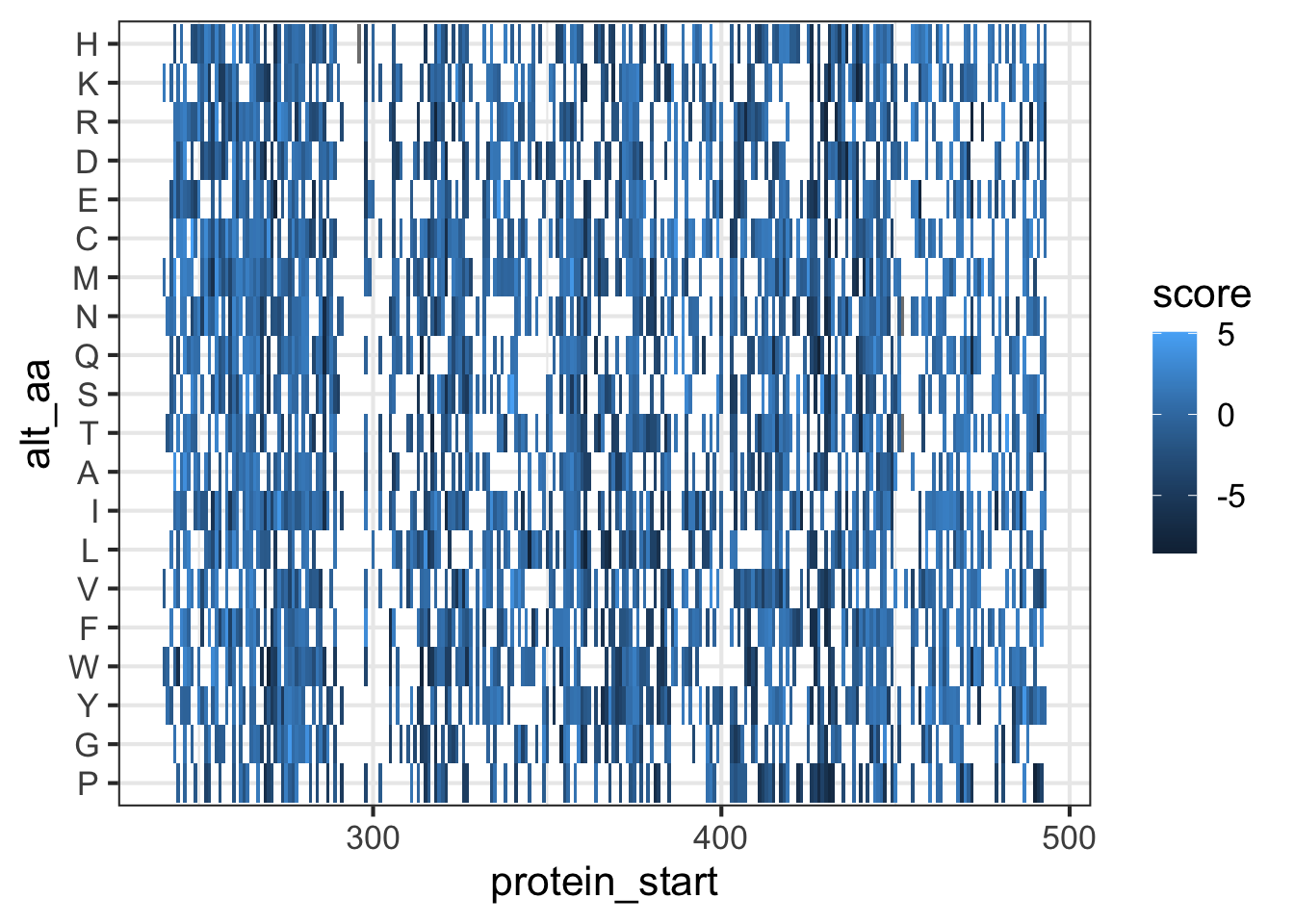
#####Focusing on conserved residues#####
ggplot(before_after%>%filter(protein_start%in%c(271,363,381:383)),aes(x=protein_start,y=alt_aa,fill=netgr_obs))+
geom_tile()+
theme(panel.background=element_rect(fill="white", colour="black"))+
scale_fill_gradient2(low ="blue",midpoint=0,mid="white", high ="red",name="Enrichment Score")+
scale_color_manual(values=c("black"))+scale_x_continuous(name="Residue on the ABL Kinase",limits=c(242,493),expand=c(0,0),breaks = c(248,250,256, 271,275,300,325,350,363,375,381,400,405,425,450,475))+
ylab("Mutant Amino Acid")+
theme(panel.grid.major.x = element_blank() ,
panel.grid.major.y = element_blank() )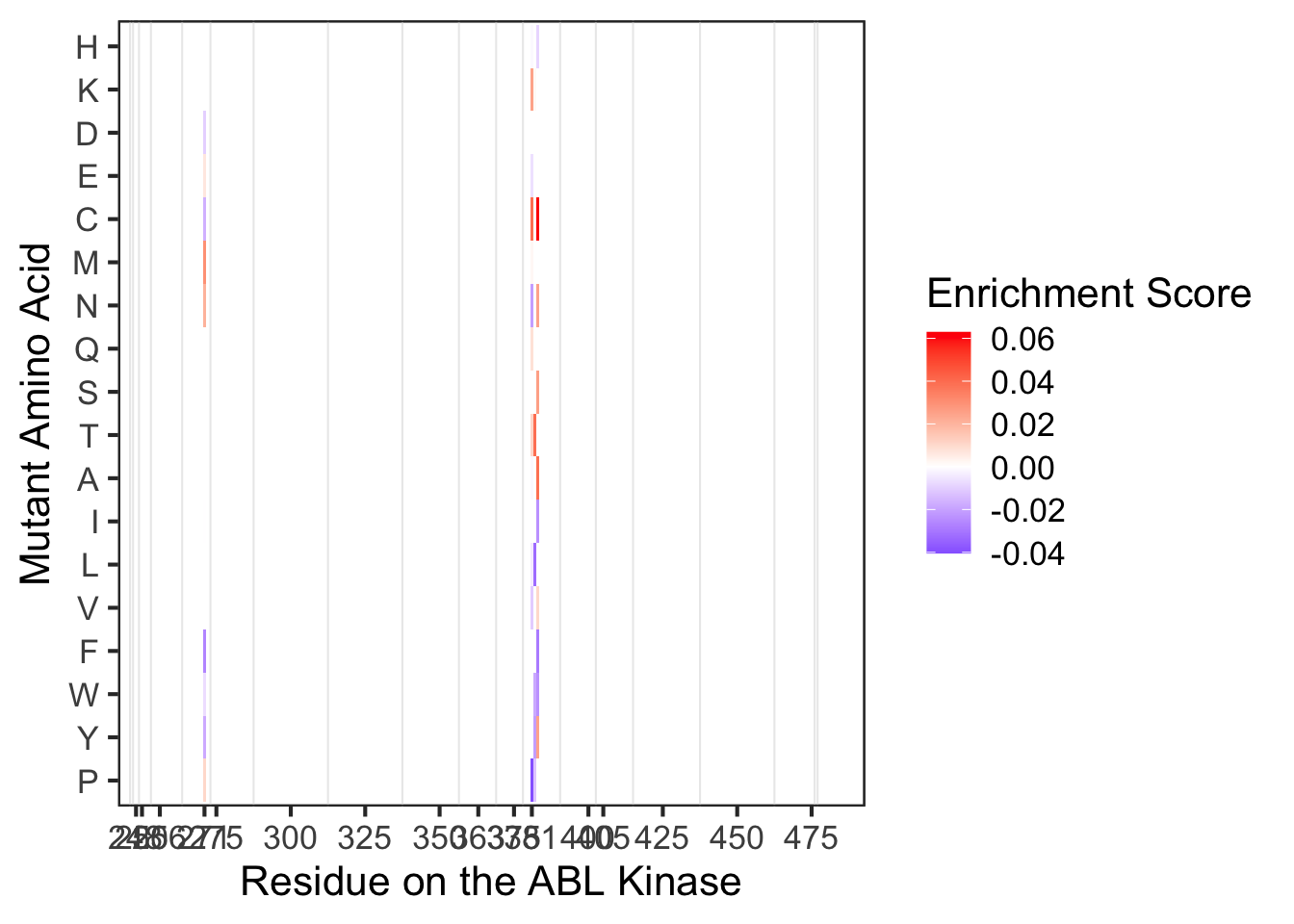
# ggsave("BCRABL_iL3Independence_D2_essential.pdf",height=6,width=24,units="in",useDingbats=F)
ggplot(before_after%>%filter(protein_start>=240,protein_start<=494),aes(x=protein_start,y=alt_aa,fill=score))+
geom_tile()+
theme(panel.background=element_rect(fill="white", colour="black"))+
scale_fill_gradient2(low ="blue",midpoint=0,mid="white", high ="red",name="Enrichment Score")+
scale_color_manual(values=c("black"))+scale_x_continuous(name="Residue on the ABL Kinase",limits=c(242,493),expand=c(0,0),breaks = c(248,250,256, 271,275,300,325,350,363,375,381,400,405,425,450,475))+
ylab("Mutant Amino Acid")+
theme(panel.grid.major.x = element_blank() ,
panel.grid.major.y = element_blank() )Warning: Removed 8 rows containing missing values (geom_tile).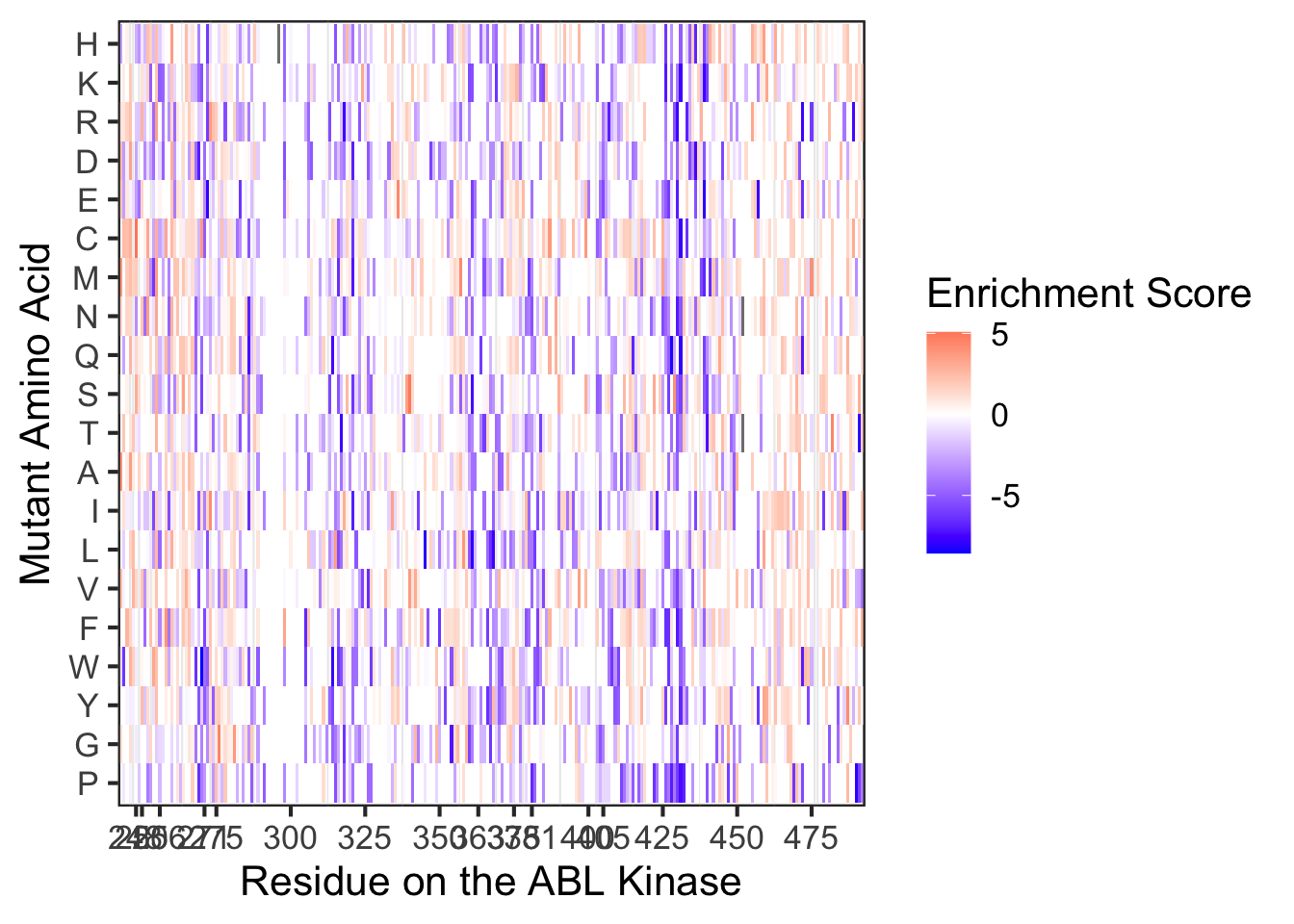
# ggsave("BCRABL_iL3Independence_D2.pdf",height=6,width=24,units="in",useDingbats=F)IMATINIB Screens for plotting Day 0 Day 2 vs Day 0 Day 4
###### TSII Day 0 vs Day 2 #########
source("code/merge_samples.R")
source("code/depth_finder.R")
delta_t=96 #hours
cells_before=370 #total cells at before time point
cells_after=10397 #total cells at after time point
netgr=.055
before_timepoint=merge_samples("Novogene_lane14/Sample10_combined/sscs","Novogene_lane13/sample7/sscs")
before_timepoint=merge_samples(before_timepoint,"Novogene_lane14/sample11/sscs")
before_timepoint=merge_samples(before_timepoint,"Novogene_lane15/sample_3/sscs")
before_timepoint=merge_samples(before_timepoint,"Novogene_lane16b/Sample9/sscs")
before_timepoint=merge_samples(before_timepoint,"Novogene_lane16b/Sample10/sscs")
before_timepoint=variants_parser(before_timepoint,cells_before)
##########TSI Imatinib D2###########
after_timepoint=merge_samples("Novogene_lane15/sample_6/sscs","Novogene_lane14/sample12/sscs")
after_timepoint=merge_samples(after_timepoint,"Novogene_lane15/Sample_7/sscs")
##########TSI D2###########
# after_timepoint=merge_samples("Novogene_lane13/Sample9/sscs","Novogene_lane13/Sample10/sscs")
# after_timepoint=merge_samples(after_timepoint,"Novogene_lane15/Sample_4/sscs")
####################
after_timepoint=variants_parser(after_timepoint,cells_after)
before_after=compare_samples(before_timepoint,after_timepoint,netgr,delta_t)
# a=before_after%>%filter(protein_start%in%271)
# a=before_after%>%filter(protein_start%in%426)# a=before_after%>%filter(resmuts%in%T)
before_after=resmuts_adder(before_after)
before_after=res_residues_adder(before_after)
ggplot(before_after%>%filter(protein_start>=240,protein_start<=494),aes(x=protein_start,y=alt_aa,fill=score))+geom_tile()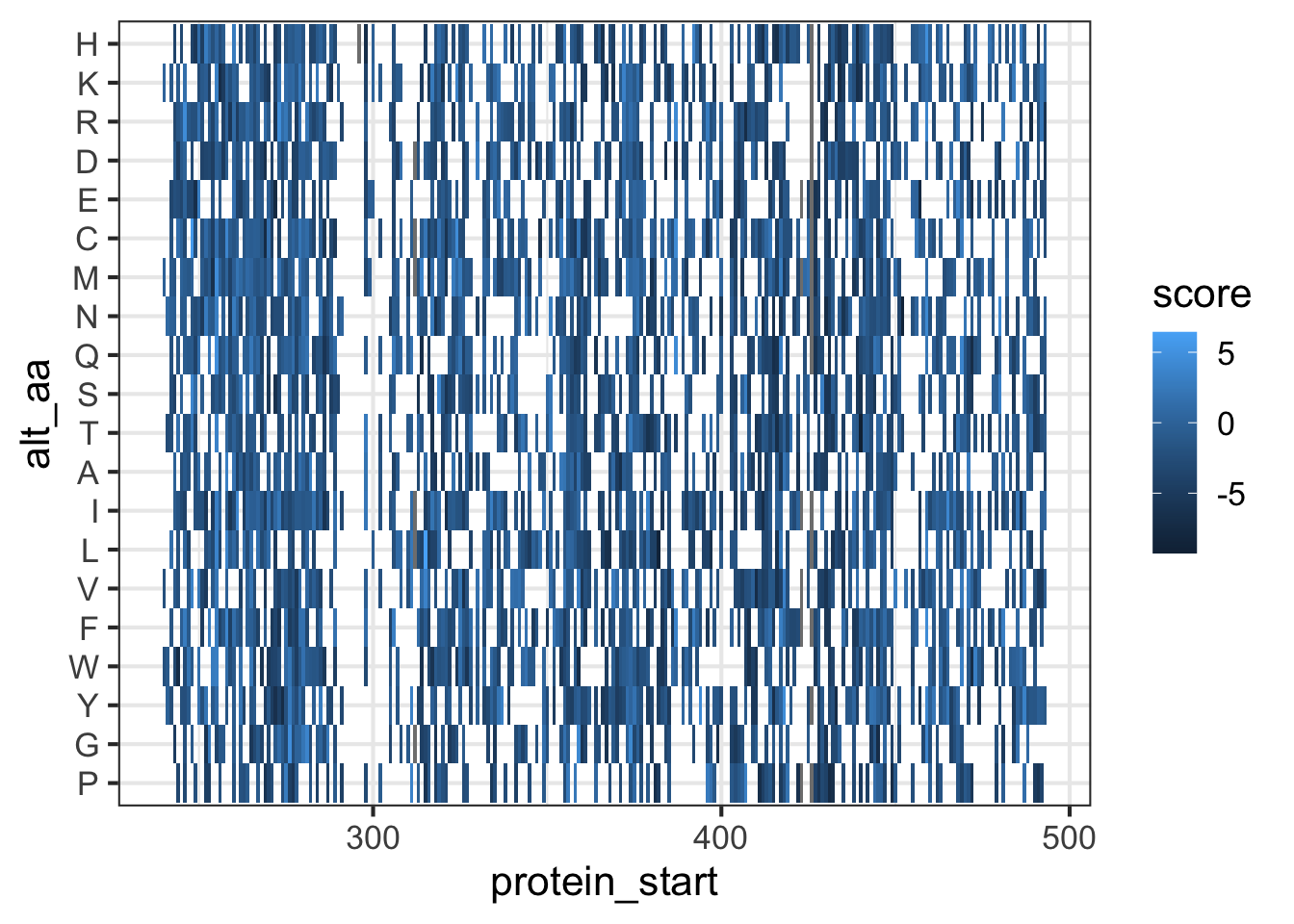
#####Focusing on RESISTANT residues#####
ggplot(before_after%>%filter(resmuts%in%T,!protein_start%in%"315"),aes(x=protein_start,y=alt_aa,fill=score))+
geom_tile()+
theme(panel.background=element_rect(fill="white", colour="black"))+
scale_fill_gradient2(low ="blue",midpoint=0,mid="white", high ="red",name="Enrichment Score")+
scale_color_manual(values=c("black"))+scale_x_continuous(name="Residue on the ABL Kinase",limits=c(242,493),expand=c(0,0),breaks=c(250,253,255,276,299,315,317,351,355,359,396,459,486))+
ylab("Mutant Amino Acid")+
theme(panel.grid.major.x = element_blank() ,
panel.grid.minor.x = element_blank() ,
panel.grid.major.y = element_blank() ,
panel.grid.minor.y = element_blank() )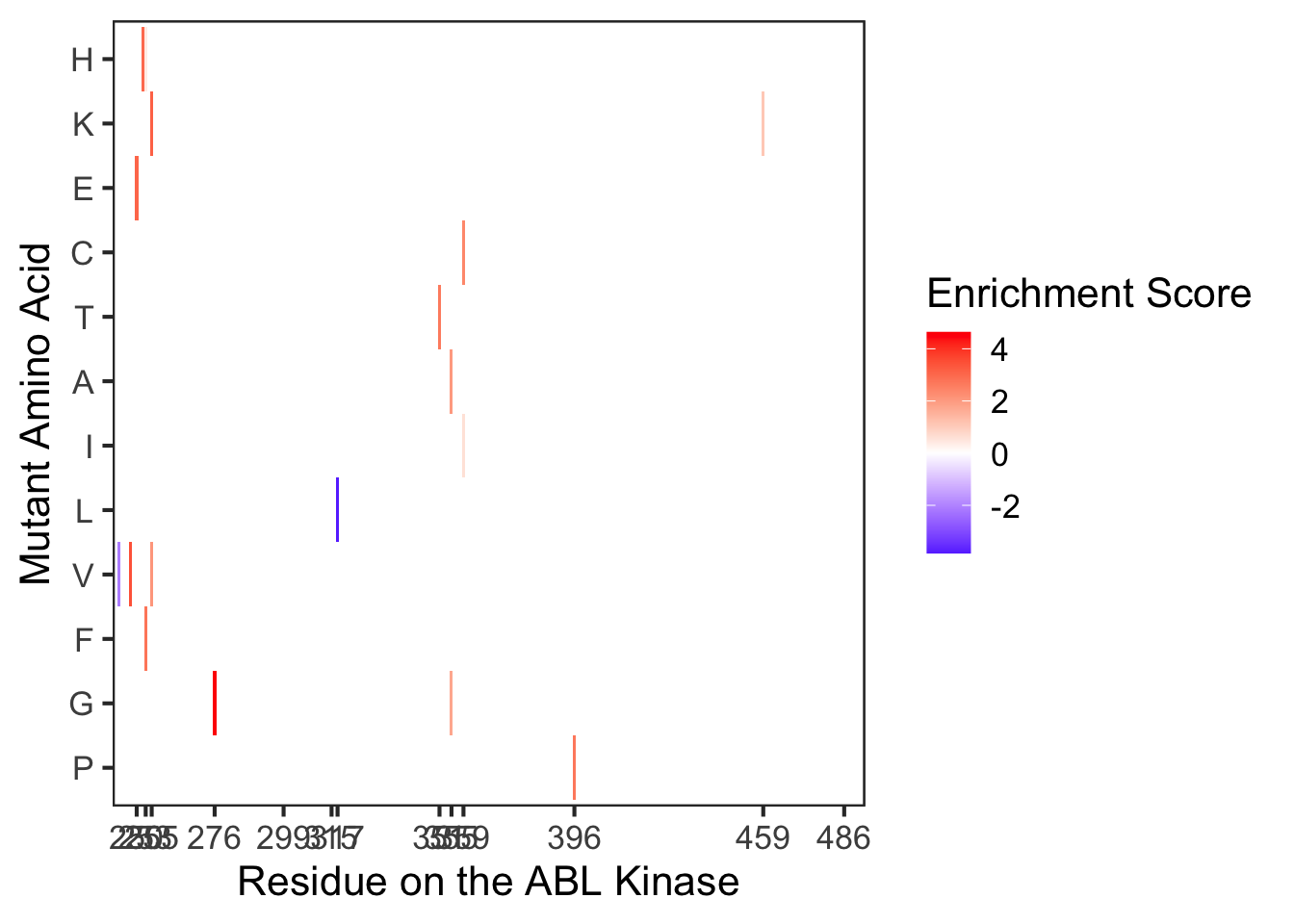
# ggsave("BCRABL_imatinib_D2_resistant.pdf",height=6,width=24,units="in",useDingbats=F)
ggplot(before_after%>%filter(protein_start>=240,protein_start<=494),aes(x=protein_start,y=alt_aa,fill=score))+
geom_tile()+
theme(panel.background=element_rect(fill="white", colour="black"))+
scale_fill_gradient2(low ="blue",midpoint=0,mid="white", high ="red",name="Enrichment Score")+
scale_color_manual(values=c("black"))+scale_x_continuous(name="Residue on the ABL Kinase",limits=c(242,493),expand=c(0,0),breaks = c(248,250,256, 271,275,300,325,350,363,375,381,400,405,425,450,475))+
ylab("Mutant Amino Acid")+
theme(panel.grid.major.x = element_blank() ,
panel.grid.minor.x = element_blank() ,
panel.grid.major.y = element_blank() ,
panel.grid.minor.y = element_blank() )Warning: Removed 8 rows containing missing values (geom_tile).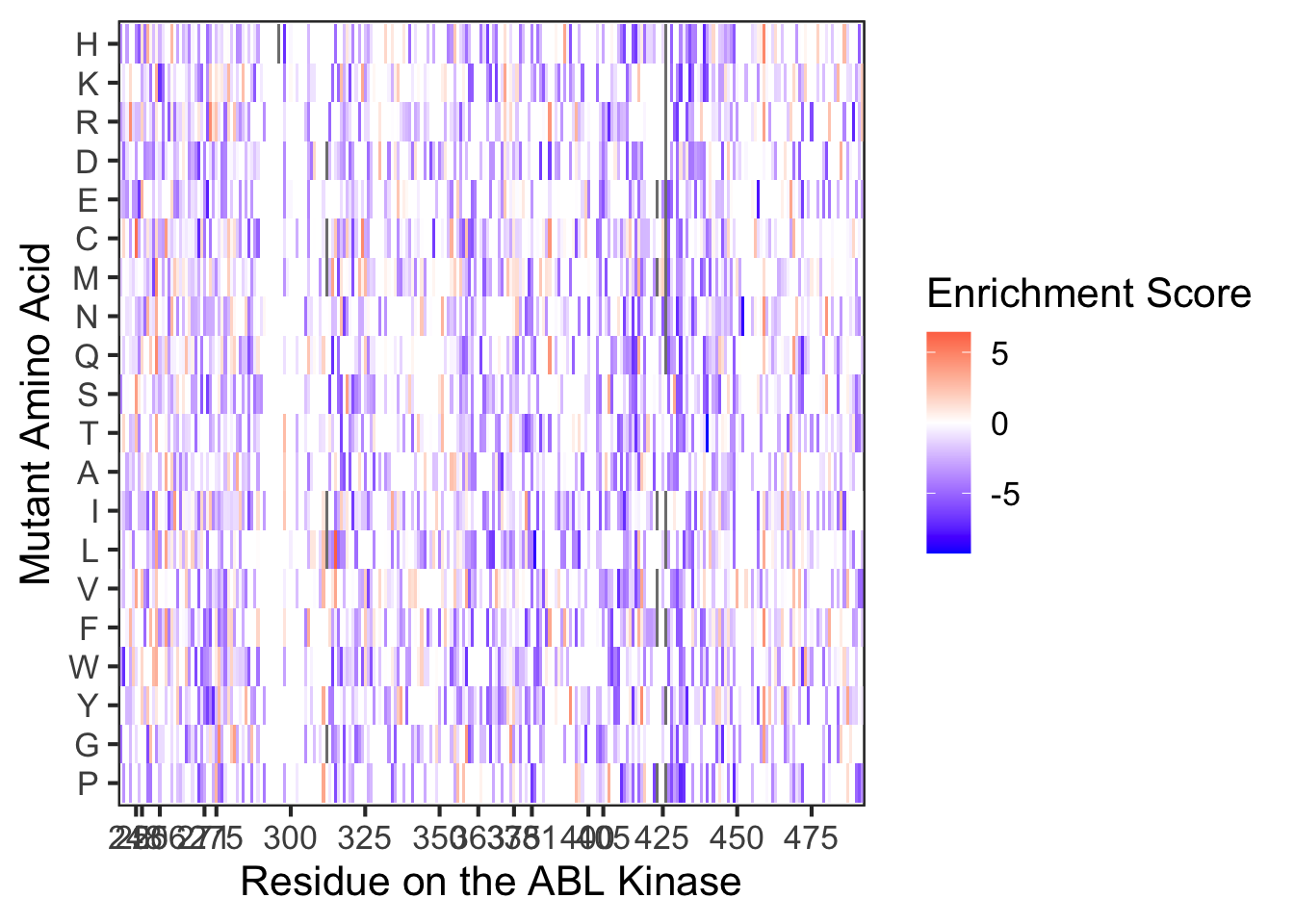
# ggsave("BCRABL_imat_D2.pdf",height=6,width=24,units="in",useDingbats=F)
ggplot(before_after%>%filter(protein_start>=240,protein_start<=494),aes(x=protein_start,y=alt_aa,fill=netgr_obs))+
geom_tile()+
theme(panel.background=element_rect(fill="white", colour="black"))+
scale_fill_gradient2(low ="blue",midpoint=0.035,mid="white", high ="red",name="Net Growth Rate")+
scale_color_manual(values=c("black"))+scale_x_continuous(name="Residue on the ABL Kinase",limits=c(242,493),expand=c(0,0),breaks = c(248,250,256, 271,275,300,325,350,363,375,381,400,405,425,450,475))+
ylab("Mutant Amino Acid")+
theme(panel.grid.major.x = element_blank() ,
panel.grid.minor.x = element_blank() ,
panel.grid.major.y = element_blank() ,
panel.grid.minor.y = element_blank())+
theme_dark()Warning: Removed 8 rows containing missing values (geom_tile).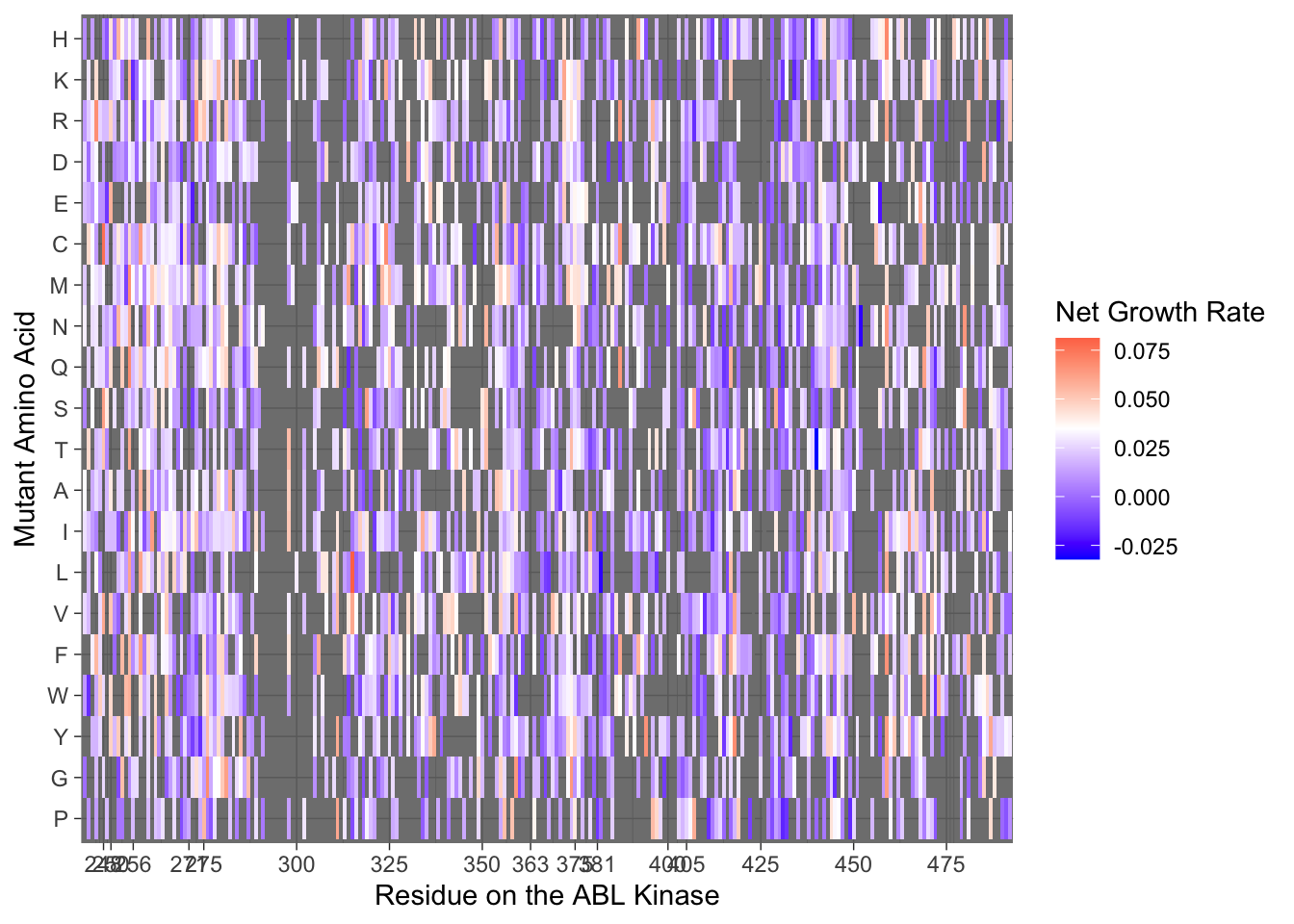
a=before_after%>%filter(protein_start>=240,protein_start<=494,!ct.x%in%.5,!ct.y%in%.5)
median(a$netgr_obs)[1] 0.02824059ggplot(before_after%>%filter(protein_start>=240,protein_start<=494),aes(x=netgr_obs))+geom_histogram()`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.Warning: Removed 27 rows containing non-finite values (stat_bin).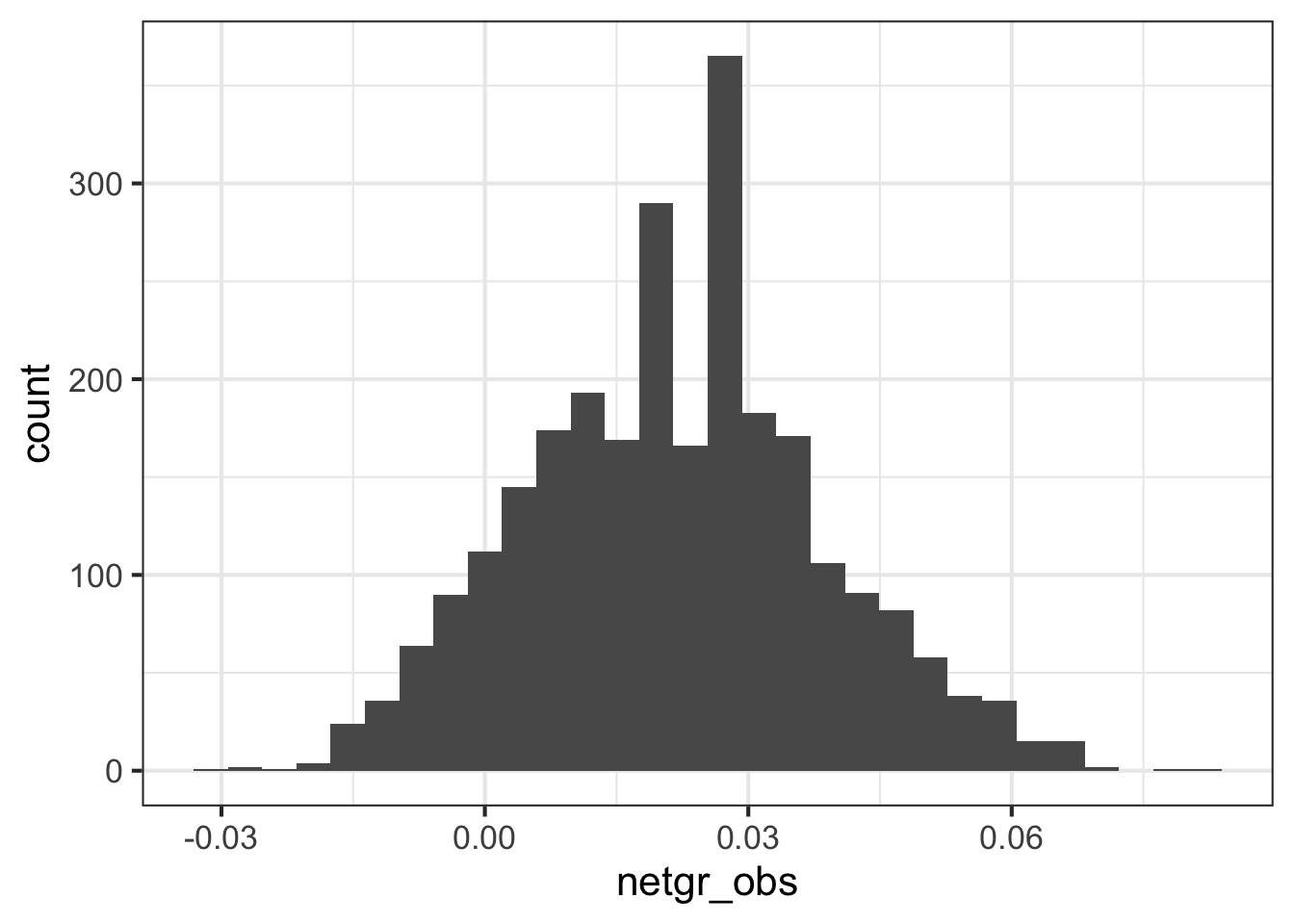
# ggsave("BCRABL_imat_D2_netgr.pdf",height=6,width=24,units="in",useDingbats=F)
sessionInfo()R version 4.0.0 (2020-04-24)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS 10.16
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRblas.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] parallel stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] MASS_7.3-55 viridisLite_0.3.0 RColorBrewer_1.1-2 doParallel_1.0.15
[5] iterators_1.0.12 foreach_1.5.0 tictoc_1.0 plotly_4.9.2.1
[9] ggplot2_3.3.3 dplyr_1.0.6 stringr_1.4.0
loaded via a namespace (and not attached):
[1] tidyselect_1.1.0 xfun_0.31 bslib_0.3.1 purrr_0.3.4
[5] colorspace_1.4-1 vctrs_0.3.8 generics_0.0.2 htmltools_0.5.2
[9] yaml_2.2.1 utf8_1.1.4 rlang_0.4.11 jquerylib_0.1.4
[13] later_1.0.0 pillar_1.6.1 glue_1.4.1 withr_2.4.2
[17] DBI_1.1.0 lifecycle_1.0.0 munsell_0.5.0 gtable_0.3.0
[21] workflowr_1.6.2 htmlwidgets_1.5.1 codetools_0.2-16 evaluate_0.14
[25] labeling_0.3 knitr_1.28 fastmap_1.1.0 crosstalk_1.1.0.1
[29] httpuv_1.5.2 fansi_0.4.1 Rcpp_1.0.4.6 promises_1.1.0
[33] backports_1.1.7 scales_1.1.1 jsonlite_1.7.2 farver_2.0.3
[37] fs_1.4.1 digest_0.6.25 stringi_1.7.5 rprojroot_1.3-2
[41] grid_4.0.0 tools_4.0.0 magrittr_2.0.1 sass_0.4.1
[45] lazyeval_0.2.2 tibble_3.1.2 crayon_1.4.1 tidyr_1.1.3
[49] pkgconfig_2.0.3 ellipsis_0.3.2 data.table_1.12.8 assertthat_0.2.1
[53] rmarkdown_2.14 httr_1.4.2 R6_2.4.1 git2r_0.27.1
[57] compiler_4.0.0