QGT-Columbia-analysis-plan
Hae Kyung Im
2020-06-03
Last updated: 2020-06-08
Checks: 6 1
Knit directory: QGT-Columbia-lab/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200603) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version a2b9918. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: extras/.DS_Store
Unstaged changes:
Modified: analysis/analysis_plan.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/analysis_plan.Rmd) and HTML (docs/analysis_plan.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | a2b9918 | Hae Kyung Im | 2020-06-07 | testing, reducing size of genotype file |
| html | a2b9918 | Hae Kyung Im | 2020-06-07 | testing, reducing size of genotype file |
| Rmd | 2257a0f | Hae Kyung Im | 2020-06-06 | raw figure links |
| html | 2257a0f | Hae Kyung Im | 2020-06-06 | raw figure links |
| Rmd | efbfceb | Hae Kyung Im | 2020-06-06 | raw urls for figures |
| Rmd | 973fc2b | Hae Kyung Im | 2020-06-05 | prelim notes |
| Rmd | 409558f | Yanyu Liang | 2020-06-05 | updated some paths |
| html | 409558f | Yanyu Liang | 2020-06-05 | updated some paths |
| Rmd | 0deee3c | Hae Kyung Im | 2020-06-05 | added TWMR |
| html | 0deee3c | Hae Kyung Im | 2020-06-05 | added TWMR |
| Rmd | 064f6ee | Hae Kyung Im | 2020-06-05 | added figures and slides under extras |
| html | 064f6ee | Hae Kyung Im | 2020-06-05 | added figures and slides under extras |
| Rmd | 1b70eb7 | Hae Kyung Im | 2020-06-05 | updated prerequisites |
| html | 1b70eb7 | Hae Kyung Im | 2020-06-05 | updated prerequisites |
| Rmd | 339f40c | Hae Kyung Im | 2020-06-05 | added plan |
| html | 339f40c | Hae Kyung Im | 2020-06-05 | added plan |
| Rmd | a3aa03e | Hae Kyung Im | 2020-06-05 | prerequisites added |
| Rmd | 09f5dae | Hae Kyung Im | 2020-06-05 | fastenloc |
| Rmd | ccb1167 | Hae Kyung Im | 2020-06-05 | knit |
| html | ccb1167 | Hae Kyung Im | 2020-06-05 | knit |
| Rmd | d427aee | Hae Kyung Im | 2020-06-05 | minor comment sort1 |
| Rmd | 4ca65b9 | Hae Kyung Im | 2020-06-05 | edits 2 |
| Rmd | f59cd02 | Hae Kyung Im | 2020-06-04 | edits |
| html | f59cd02 | Hae Kyung Im | 2020-06-04 | edits |
| Rmd | d65c555 | Hae Kyung Im | 2020-06-04 | twmr |
| html | d65c555 | Hae Kyung Im | 2020-06-04 | twmr |
| html | 682b6e2 | Hae Kyung Im | 2020-06-04 | Build site. |
| Rmd | d3502e8 | Hae Kyung Im | 2020-06-04 | wflow_publish(“analysis/analysis_plan.Rmd”) |
| Rmd | 4420fc7 | Hae Kyung Im | 2020-06-04 | wflow_rename(“analysis/predixcan_analysis.Rmd”, “analysis/analysis_plan.Rmd”) |
| html | 4420fc7 | Hae Kyung Im | 2020-06-04 | wflow_rename(“analysis/predixcan_analysis.Rmd”, “analysis/analysis_plan.Rmd”) |
Set up
This information is also on the slides
- download data and software from Box. This will have copies of all the software repositories and the models
Linux is the operating system of choice to run bioinformatics software. Here are offering two options
- Option 1: full setup, recommended for the linux-savvy with full setup
- OPtion 2: pre-installed RStudio in Google cloud, recommended for people less familiar with linux
The latest version of the analysis plan markdown document that generated this page is on github here rendered here as an html page
Option 1
- install anaconda/miniconda
- define imlabtools conda environment how to here, which will install all the python modules needed for this analysis session
-
download software (copies of the repos are already included in the course folder QCT-Columbia-HKI/repos/)
- download metaxcan repo
- download torus repo
- download fastenloc repo
- download TMWR repo
- download prediction models from predictdb.org (a few models are included in the course folder QCT-Columbia-HKI/repos/)
- install R/RStudio/tidyverse package
- (optional) install workflowr package in R
- git clone https://github.com/hakyimlab/QGT-Columbia-HKI.git
- start Rstudio (if you installed workflowr, you can just open the QGT-Columbia-HKI.Rproj)
Option 2
- claim your Rstudio server IP address ()
- connect to the Rstudio server using the url you claimed (http://xxx.xxx.xxx.xxx:8787)
Both options
- update the analysis document
PRE="/home/student/"
cd $PRE/../lab/
git pull - activate the the imlabtools environment
conda activate imlabtools** Notice that the bash chunks need to be copy-pasted to the terminal, not performed within the chunk.
Summary of analysis plan
- predict whole blood expression
- check how well the prediction works with GEUVADIS expression data
- run association between predicted expression and a simulated phenotype
- calculate association between expression levels and coronary artery disease risk using s-predixcan
- fine-map the coronary artery disease gwas results using torus (need some preformatting)
- calculate colocalization probability using fastenloc
- run transcriptome-wide mendelian randomization in one locus of interest
library(tidyverse)── Attaching packages ──────────────────────────────────────────────── tidyverse 1.3.0 ──✓ ggplot2 3.3.0 ✓ purrr 0.3.4
✓ tibble 3.0.0 ✓ dplyr 0.8.5
✓ tidyr 1.0.2 ✓ stringr 1.4.0
✓ readr 1.3.1 ✓ forcats 0.5.0── Conflicts ─────────────────────────────────────────────────── tidyverse_conflicts() ──
x dplyr::filter() masks stats::filter()
x dplyr::lag() masks stats::lag()Transcriptome-wide association methods
print(getwd())[1] "/Users/haekyungim/Github/QGT-Columbia-lab"pre="~/Box/LargeFiles/QGT-Columbia-HKI"
#pre="/home/student/QGT-Columbia-HKI"
model.dir=glue::glue("{pre}/models")
metaxcan.dir=glue::glue("{pre}/repos/MetaXcan-master/software")
fastenloc.dir=glue::glue("{pre}/repos/fastenloc-master")
torus.dir=glue::glue("{pre}/repos/torus-master")
twmr.dir=glue::glue("{pre}/repos/TWMR-master")
results.dir=glue::glue("{pre}/results")
MODEL=glue::glue("{pre}/models")
DATA=glue::glue("{pre}/data")
RESULTS=glue::glue("{pre}/results")
METAXCAN=glue::glue("{pre}/repos/MetaXcan-master/software")
FASTENLOC=glue::glue("{pre}/repos/fastenloc-master")
TORUS=glue::glue("{pre}/repos/torus-master")
TWMR=glue::glue("{pre}/repos/TWMR-master")export PRE="/home/student/QGT-Columbia-HKI"
export DATA=$PRE/data
export MODEL=$PRE/models
export RESULTS=$PRE/results
export METAXCAN=$PRE/repos/MetaXcan-master/softwarepredict expression
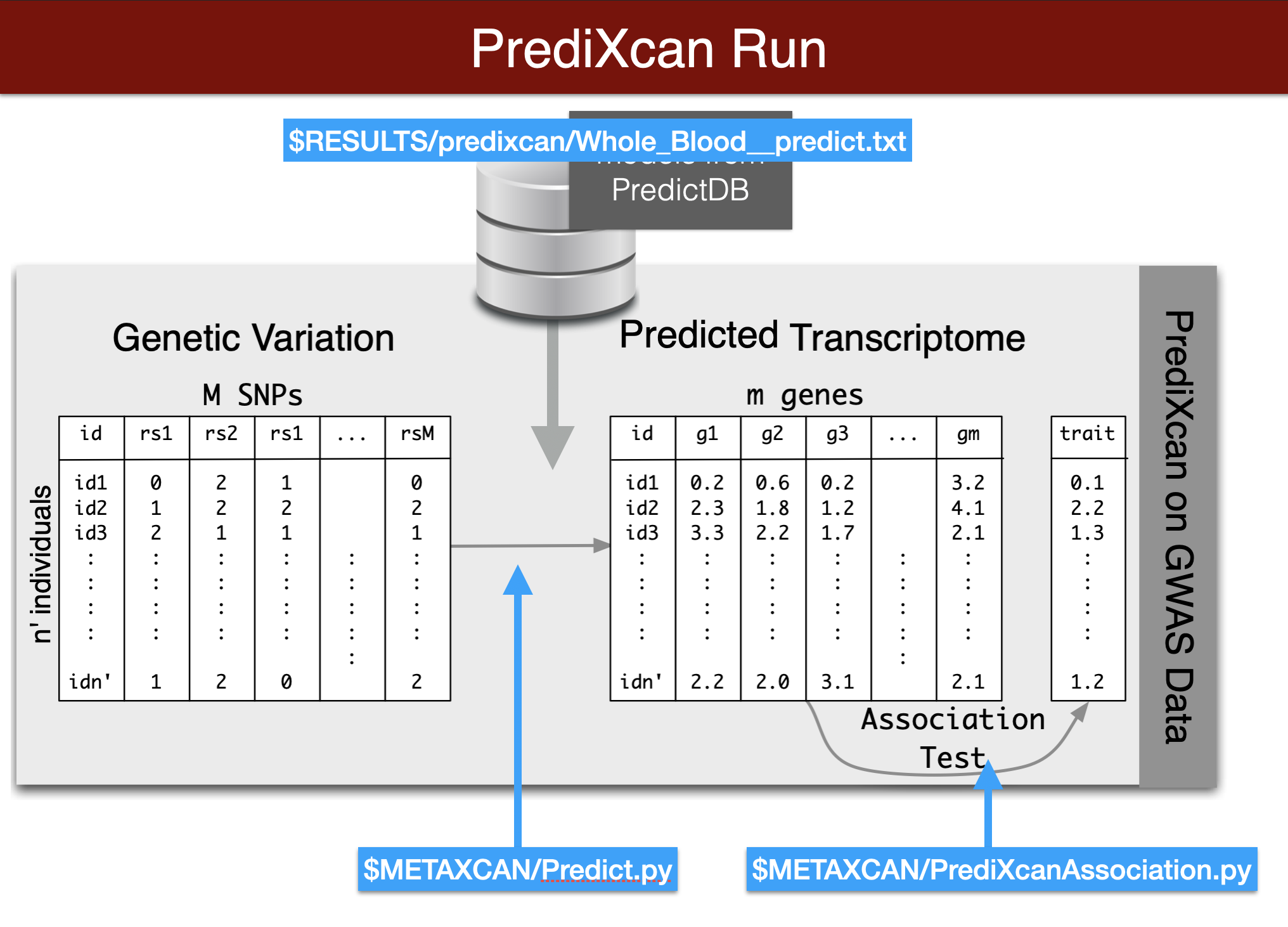
Visual summary of predixcan runs
Remember you need to copy and paste this code chunk into the terminal to run it. Also make sure you activated the imlabtools environment which has all the necessary python modules.
Make sure all the paths and file names are correct. This run should take about one minute.
<!-- PRE="/home/student/QGT-Columbia-HKI" -->
<!-- DATA=$PRE/data -->
<!-- MODEL=$PRE/models -->
<!-- METAXCAN=$PRE/repos/MetaXcan-master/software -->
<!-- RESULTS=$PRE/results -->
printf "Predict expression\n\n"
python3 $METAXCAN/Predict.py \
--model_db_path $PRE/models/gtex_v8_en/en_Whole_Blood.db \
--vcf_genotypes $DATA/predixcan/genotype/filtered.vcf.gz \
--vcf_mode genotyped \
--variant_mapping $DATA/predixcan/gtex_v8_eur_filtered_maf0.01_monoallelic_variants.txt.gz id rsid \
--on_the_fly_mapping METADATA "chr{}_{}_{}_{}_b38" \
--prediction_output $RESULTS/predixcan/Whole_Blood__predict.txt \
--prediction_summary_output $RESULTS/predixcan/Whole_Blood__summary.txt \
--verbosity 9 \
--throw
assess prediction performance (optional)
predicted_expression = read_tsv(glue::glue("{results.dir}/predixcan/Whole_Blood__predict.txt"))
dim(predicted_expression)
head(predicted_expression[,1:5])
prediction_summary = read_tsv(glue::glue("{results.dir}/predixcan/Whole_Blood__summary.txt"))
dim(prediction_summary)
head(prediction_summary)
## merge with GEUVADIS expression data
## calculate spearman correlation
## select a few genes and plot predicted vs observed expressionrun association with a phenotype
PHENO="sim.infinitesimal_pve0.1"
printf "association\n\n"
python3 $METAXCAN/PrediXcanAssociation.py \
--expression_file $RESULTS/predixcan/Whole_Blood__predict.txt \
--input_phenos_file $DATA/predixcan/phenotype/$PHENO.txt \
--input_phenos_column pheno \
--output $RESULTS/predixcan/$PHENO/Whole_Blood__association.txt \
--verbosity 9 \
--throw
PHENO="sim.spike_n_slab_0.01_pve0.1"
printf "association\n\n"
python3 $METAXCAN/PrediXcanAssociation.py \
--expression_file $RESULTS/predixcan/Whole_Blood__predict.txt \
--input_phenos_file $DATA/predixcan/phenotype/$PHENO.txt \
--input_phenos_column pheno \
--output $RESULTS/predixcan/$PHENO/Whole_Blood__association.txt \
--verbosity 9 \
--throw
PHENO="sim.spike_n_slab_0.1_pve0.05"
printf "association\n\n"
python3 $METAXCAN/PrediXcanAssociation.py \
--expression_file $RESULTS/predixcan/Whole_Blood__predict.txt \
--input_phenos_file $DATA/predixcan/phenotype/$PHENO.txt \
--input_phenos_column pheno \
--output $RESULTS/predixcan/$PHENO/Whole_Blood__association.txt \
--verbosity 9 \
--throw
read association results
## read association results
PHENO="sim.spike_n_slab_0.01_pve0.1"
predixcan_association = read_tsv(glue::glue("{results.dir}/predixcan/{PHENO}/Whole_Blood__association.txt"))
## take a look at the results
dim(predixcan_association)
predixcan_association %>% arrange(pvalue) %>% head
predixcan_association %>% arrange(pvalue) %>% ggplot(aes(pvalue)) + geom_histogram(bins=20)
## compare distribution against the null (uniform)
qq(predixcan_association$pvalue)
truebetas = read_tsv(glue::glue("{DATA}/predixcan/phenotype/gene-effects/{PHENO}.txt"))
predixcan_association %>% inner_join(truebetas,by=c("gene"="gene_id")) %>% ggplot(aes(effect,effect_size))+geom_point()+geom_abline()Exercise
-[ ] show top genes -[ ] compare with true effect sizes
library(qqman)For example usage please run: vignette('qqman')Citation appreciated but not required:Turner, S.D. qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. biorXiv DOI: 10.1101/005165 (2014).——-
Summary PrediXcan
Now we will use the summary results from a GWAS of coronary artery disease to calculate the association between the genetic component of the expression of genes and coronary artery disease risk. We will use the SPrediXcan.py.
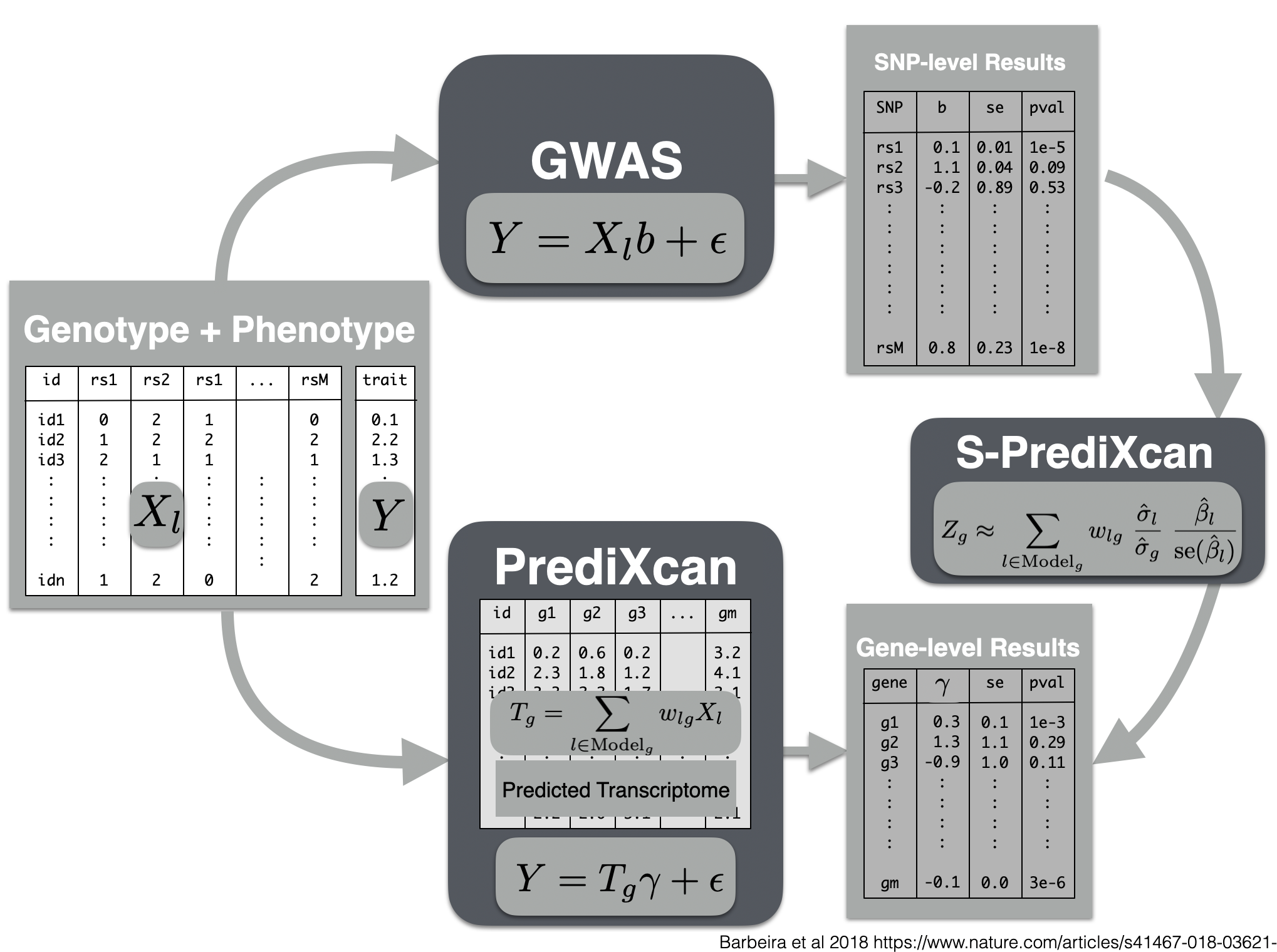
Visual summary of s-predixcan
## harmonized and imputed GWAS result for coronary artery disease is available in
# $PRE/s-predixcan/data/run s-predixcan
export PRE="/home/student/QGT-Columbia-HKI"
export DATA=$PRE/data/s-predixcan
export MODEL=$PRE/models
export METAXCAN=$PRE/repos/MetaXcan-master/software
export RESULTS=$PRE/results
python $METAXCAN/SPrediXcan.py \
--gwas_file $DATA/imputed_CARDIoGRAM_C4D_CAD_ADDITIVE.txt.gz \
--snp_column panel_variant_id --effect_allele_column effect_allele --non_effect_allele_column non_effect_allele --zscore_column zscore \
--model_db_path $MODEL/gtex_v8_mashr/mashr_Whole_Blood.db \
--covariance $MODEL/gtex_v8_mashr/mashr_Whole_Blood.txt.gz \
--keep_non_rsid --additional_output --model_db_snp_key varID \
--throw \
--output_file $RESULTS/spredixcan/eqtl/CARDIoGRAM_C4D_CAD_ADDITIVE__PM__Whole_Blood.csv
plot and interpret s-predixcan results
spredixcan_association = read_csv(glue::glue("{results.dir}/spredixcan/eqtl/CARDIoGRAM_C4D_CAD_ADDITIVE__PM__Whole_Blood.csv"))
dim(spredixcan_association)
spredixcan_association %>% arrange(pvalue) %>% head
spredixcan_association %>% arrange(pvalue) %>% ggplot(aes(pvalue)) + geom_histogram(bins=20)SORT1, considered to be a causal gene for LDL cholesterol and as a consequence of coronary artery disease, is not found here. Why? (tissue)
run multixcan (optional)
export MODEL=$PRE/models
export DATA=$PRE/data/s-predixcan
python $METAXCAN/SMulTiXcan.py \
--models_folder $MODEL/gtex_v8_mashr \
--models_name_pattern "mashr_(.*).db" \
--snp_covariance $MODEL/gtex_v8_expression_mashr_snp_covariance.txt.gz \
--metaxcan_folder $RESULTS/spredixcan/eqtl/ \
--metaxcan_filter "CARDIoGRAM_C4D_CAD_ADDITIVE__PM__(.*).csv" \
--metaxcan_file_name_parse_pattern "(.*)__PM__(.*).csv" \
--gwas_file $DATA/imputed_CARDIoGRAM_C4D_CAD_ADDITIVE.txt.gz \
--snp_column panel_variant_id --effect_allele_column effect_allele --non_effect_allele_column non_effect_allele --zscore_column zscore --keep_non_rsid --model_db_snp_key varID \
--cutoff_condition_number 30 \
--verbosity 7 \
--throw \
--output $RESULTS/smultixcan/eqtl/CARDIoGRAM_C4D_CAD_ADDITIVE_smultixcan.txt
Colocalization methods
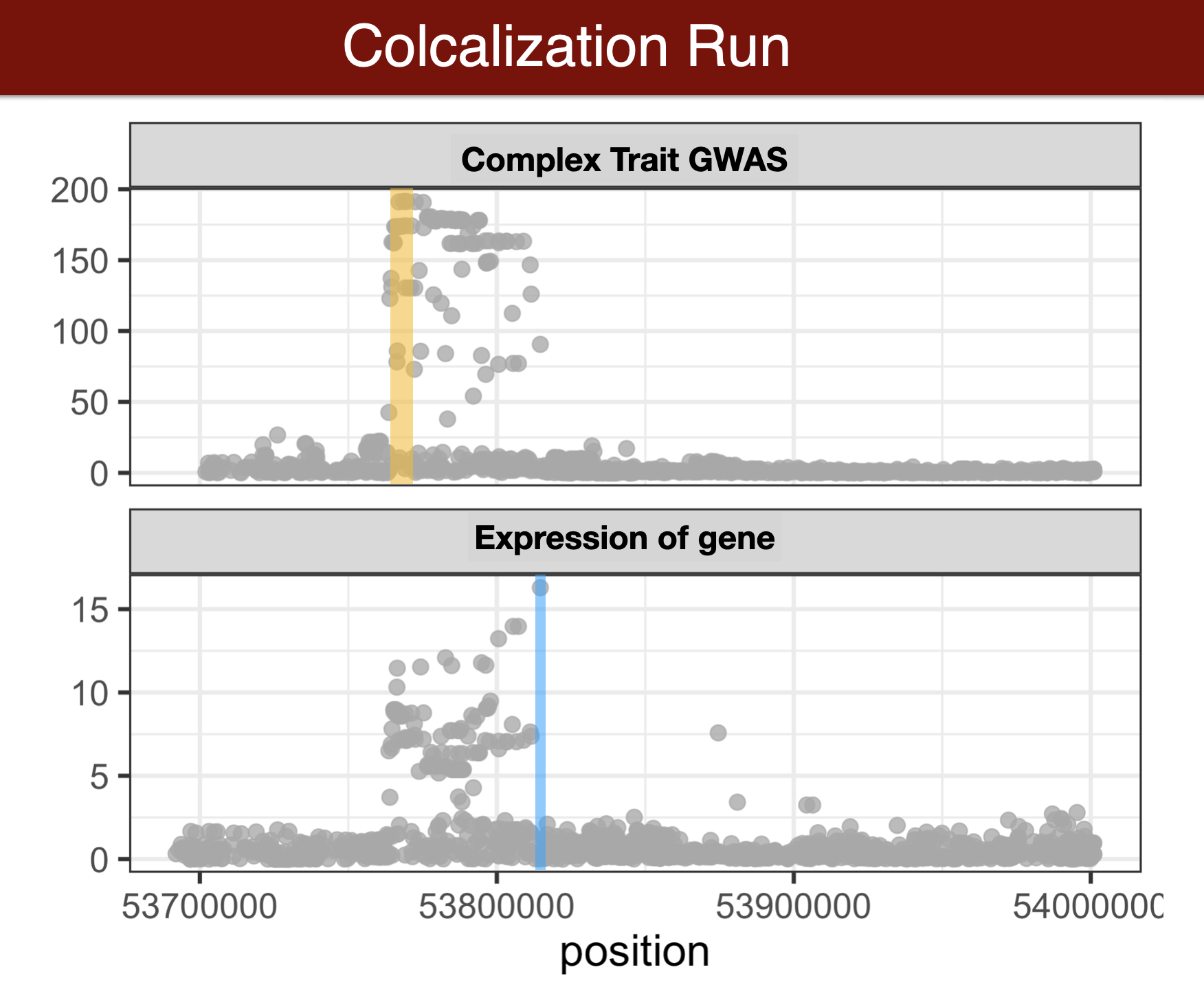
Visual summary of colocalization
fine-map GWAS results
We will run torus due to time limitation but ideally we would like to run a method that allows multiple causal variants per locus.
#torus -d Height.torus.zval.gz --load_zval -dump_pip Height.gwas.pip
#gzip Height.gwas.pip
TORUSOFT=torus
$TORUSOFT -d $PRE/data/fastenloc/Height.torus.zval.gz --load_zval -dump_pip $PRE/data/fastenloc/Height.gwas.pip
cd $PRE/data/fastenloc
gzip Height.gwas.pip
cd $PRE
calculate colocalization with fastENLOC
## check out tutorial https://github.com/xqwen/fastenloc/tree/master/tutorial
export eqtl_annotation_gzipped=$PRE/data/fastenloc/FASTENLOC-gtex_v8.eqtl_annot.vcf.gz
export gwas_data_gzipped=$PRE/data/fastenloc/Height.gwas.pip.gz
export TISSUE=Whole_Blood
export FASTENLOCSOFT=fastenloc
mkdir $RESULTS/fastenloc/
cd $RESULTS/fastenloc/
$FASTENLOCSOFT -eqtl $eqtl_annotation_gzipped -gwas $gwas_data_gzipped -t $TISSUE
#[-total_variants total_snp] [-thread n] [-prefix prefix_name] [-s shrinkage]
analyze results
## optional - compare with s-predixcan results-[] prepare
Mendelian randomization methods
run SMR (optional)
run TWMR (for a locus)
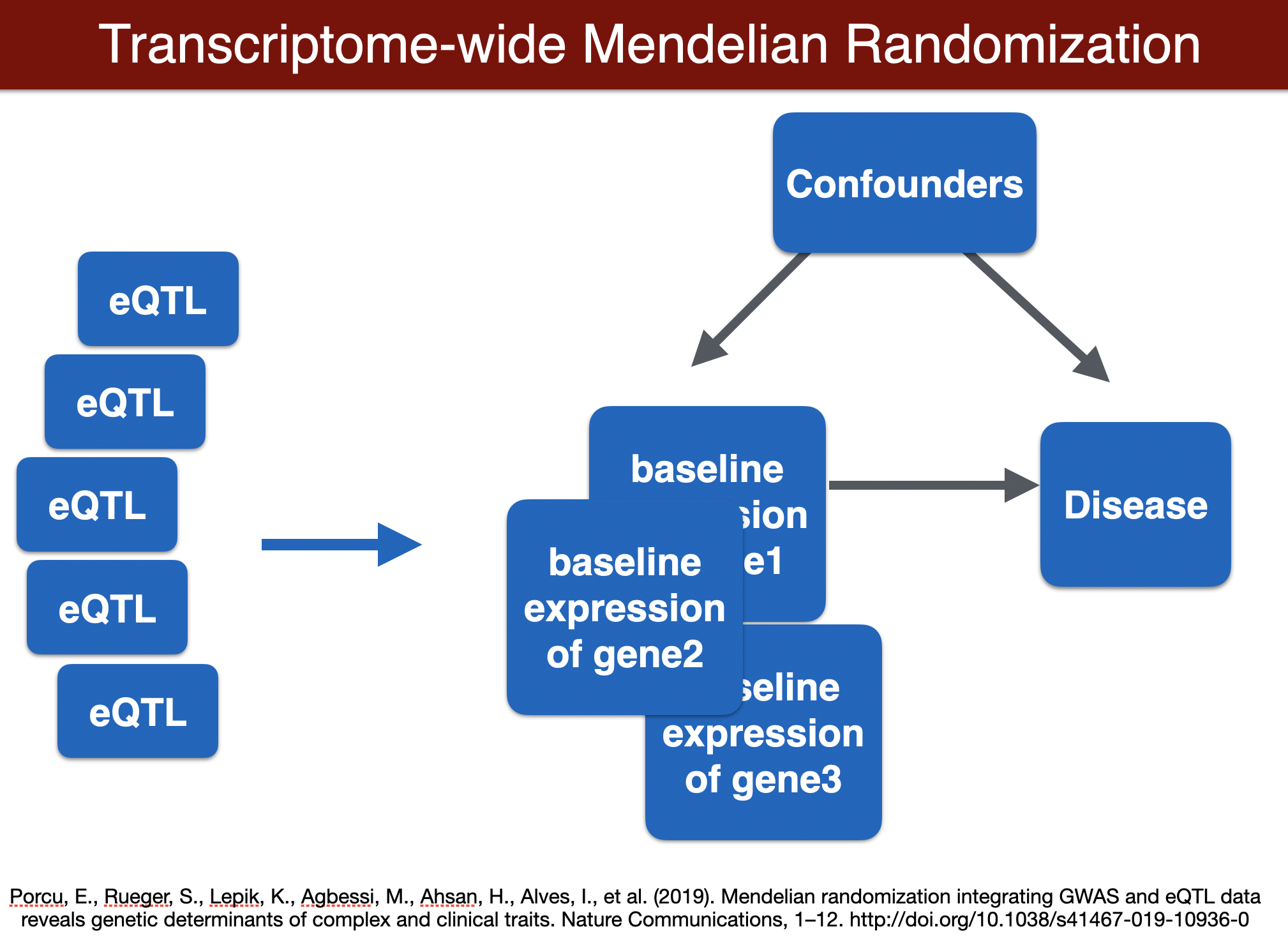
TWMR
export TWMR=$PRE/repos/TWMR-master
export OUTPUT=$PRE/results
GENE=ENSG00000002919
cd $TWMR
R < $TWMR/MR.R --no-save $GENE
cd $PRE
## output: /home/student/QGT-Columbia-HKI/repos/TWMR-master/ENSG00000002919.alpha
sessionInfo()R version 3.6.3 (2020-02-29)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS Catalina 10.15.2
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] qqman_0.1.4 forcats_0.5.0 stringr_1.4.0 dplyr_0.8.5
[5] purrr_0.3.4 readr_1.3.1 tidyr_1.0.2 tibble_3.0.0
[9] ggplot2_3.3.0 tidyverse_1.3.0
loaded via a namespace (and not attached):
[1] tidyselect_1.0.0 xfun_0.13 haven_2.2.0 lattice_0.20-41
[5] colorspace_1.4-1 vctrs_0.2.4 generics_0.0.2 htmltools_0.4.0
[9] yaml_2.2.1 rlang_0.4.5 later_1.0.0 pillar_1.4.3
[13] withr_2.1.2 glue_1.4.1 DBI_1.1.0 dbplyr_1.4.2
[17] calibrate_1.7.5 readxl_1.3.1 modelr_0.1.6 lifecycle_0.2.0
[21] cellranger_1.1.0 munsell_0.5.0 gtable_0.3.0 workflowr_1.6.2
[25] rvest_0.3.5 evaluate_0.14 knitr_1.28 httpuv_1.5.2
[29] fansi_0.4.1 broom_0.5.5 Rcpp_1.0.4.6 promises_1.1.0
[33] backports_1.1.6 scales_1.1.0 jsonlite_1.6.1 fs_1.4.1
[37] hms_0.5.3 digest_0.6.25 stringi_1.4.6 rprojroot_1.3-2
[41] grid_3.6.3 cli_2.0.2 tools_3.6.3 magrittr_1.5
[45] crayon_1.3.4 whisker_0.4 pkgconfig_2.0.3 MASS_7.3-51.5
[49] ellipsis_0.3.0 xml2_1.3.1 reprex_0.3.0 lubridate_1.7.8
[53] rstudioapi_0.11 assertthat_0.2.1 rmarkdown_2.1 httr_1.4.1
[57] R6_2.4.1 nlme_3.1-147 git2r_0.26.1 compiler_3.6.3