Post-transcriptional regulation
Last updated: 2022-07-02
Checks: 5 2
Knit directory: funcFinemapping/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R Markdown file created these results, you'll want to first commit it to the Git repo. If you're still working on the analysis, you can ignore this warning. When you're finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it's best to always run the code in an empty environment.
The command set.seed(20210404) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| ~/projects/funcFinemapping/data/qqplot_SNPs_high_spliceAI_scores_allergy.png | data/qqplot_SNPs_high_spliceAI_scores_allergy.png |
| ~/projects/funcFinemapping/data/qqplot_SNPs_high_spliceAI.png | data/qqplot_SNPs_high_spliceAI.png |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version d96f416. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .ipynb_checkpoints/
Ignored: analysis/build_annotations_for_single_cell_data.nb.html
Ignored: analysis/lab4_prepare.nb.html
Ignored: analysis/ldsc_results.nb.html
Ignored: analysis/learn_archR.nb.html
Ignored: analysis/mtsplice_finemapping_results.nb.html
Ignored: analysis/results.nb.html
Ignored: analysis/snp_finemapping_results.nb.html
Ignored: analysis/splicing.nb.html
Ignored: analysis/susie_tutorial.nb.html
Untracked files:
Untracked: SNPs_categories,png
Untracked: SNPs_categories.png
Untracked: analysis/.ipynb_checkpoints/
Untracked: analysis/annotations_PTR.Rmd
Untracked: bmi_locus1410.pdf
Untracked: code/.ipynb_checkpoints/
Untracked: code/.snakemake/
Untracked: code/Euro_LD_Chunks.RData
Untracked: code/Snakefile
Untracked: code/config.yaml
Untracked: code/environment.yml
Untracked: code/ldsc.log
Untracked: code/ldsc.results
Untracked: code/ldsc_regression.sh
Untracked: code/make_plots.R
Untracked: code/run_ldsc.sh
Untracked: code/run_ldsc_with_bed.sh
Untracked: code/run_ldsc_with_bed_v2.sh
Untracked: code/run_susie.R
Untracked: code/run_torus.sh
Untracked: code/sctype/
Untracked: code/slurm-20815237.out
Untracked: code/slurm-20832976.out
Untracked: code/slurm-20832977.out
Untracked: code/slurm-20832978.out
Untracked: code/slurm-20832979.out
Untracked: code/slurm-20832980.out
Untracked: code/slurm-21213166.out
Untracked: code/slurm-21213167.out
Untracked: code/slurm-21213168.out
Untracked: code/slurm-21213169.out
Untracked: code/slurm-21213170.out
Untracked: code/slurm-21213181.out
Untracked: code/slurm-21213183.out
Untracked: code/slurm-21213184.out
Untracked: code/slurm-21213188.out
Untracked: code/slurm-21213195.out
Untracked: code/slurm-21213200.out
Untracked: code/slurm-21213204.out
Untracked: code/slurm-21213207.out
Untracked: code/slurm-21213212.out
Untracked: code/slurm-21213217.out
Untracked: code/slurm-21213224.out
Untracked: code/slurm-21213230.out
Untracked: code/slurm-21213231.out
Untracked: code/slurm-21213236.out
Untracked: code/slurm-21213240.out
Untracked: code/slurm-21213244.out
Untracked: code/slurm-21213246.out
Untracked: code/slurm-21213601.out
Untracked: code/slurm-21213602.out
Untracked: code/slurm-21213603.out
Untracked: code/slurm-21213604.out
Untracked: code/slurm-21214302.out
Untracked: code/slurm-21214303.out
Untracked: code/slurm-21214359.out
Untracked: code/slurm-21216841.out
Untracked: code/slurm-21216842.out
Untracked: code/slurm-21216843.out
Untracked: code/slurm-21216844.out
Untracked: code/slurm-21216845.out
Untracked: code/slurm-21217091.out
Untracked: code/slurm-21217092.out
Untracked: code/slurm-21217093.out
Untracked: code/slurm-21217094.out
Untracked: code/slurm-21217095.out
Untracked: code/slurm-21217137.out
Untracked: code/slurm-21217138.out
Untracked: code/slurm-21217139.out
Untracked: code/slurm-21217140.out
Untracked: code/slurm-21217141.out
Untracked: code/slurm-21217343.out
Untracked: code/slurm-21217344.out
Untracked: code/slurm-21217345.out
Untracked: code/slurm-21217346.out
Untracked: code/slurm-21217347.out
Untracked: code/slurm-21217382.out
Untracked: code/slurm-21217383.out
Untracked: code/slurm-21217384.out
Untracked: code/slurm-21217385.out
Untracked: code/slurm-21217386.out
Untracked: code/slurm-21217411.out
Untracked: code/slurm-21217412.out
Untracked: code/slurm-21217413.out
Untracked: code/slurm-21217414.out
Untracked: code/slurm-21217416.out
Untracked: code/slurm-21217455.out
Untracked: code/slurm-21217456.out
Untracked: code/slurm-21217457.out
Untracked: code/slurm-21217458.out
Untracked: code/slurm-21217459.out
Untracked: code/slurm-21217482.out
Untracked: code/slurm-21217489.out
Untracked: code/slurm-21217490.out
Untracked: code/slurm-21217491.out
Untracked: code/slurm-21217492.out
Untracked: code/slurm-21217531.out
Untracked: code/slurm-21217532.out
Untracked: code/slurm-21217533.out
Untracked: code/slurm-21217534.out
Untracked: code/slurm-21217536.out
Untracked: code/slurm-21217574.out
Untracked: code/slurm-21217575.out
Untracked: code/slurm-21217576.out
Untracked: code/slurm-21217577.out
Untracked: code/slurm-21217578.out
Untracked: code/slurm-21217601.out
Untracked: code/slurm-21217602.out
Untracked: code/slurm-21217603.out
Untracked: code/slurm-21217604.out
Untracked: code/slurm-21217607.out
Untracked: code/slurm-21217627.out
Untracked: code/slurm-21217628.out
Untracked: code/slurm-21217629.out
Untracked: code/slurm-21217630.out
Untracked: code/slurm-21217633.out
Untracked: code/slurm-21217669.out
Untracked: code/slurm-21217671.out
Untracked: code/slurm-21217672.out
Untracked: code/slurm-21217673.out
Untracked: code/slurm-21217674.out
Untracked: code/slurm-21217700.out
Untracked: code/slurm-21217701.out
Untracked: code/slurm-21217702.out
Untracked: code/slurm-21217703.out
Untracked: code/slurm-21217707.out
Untracked: code/slurm-21217747.out
Untracked: code/slurm-21217748.out
Untracked: code/slurm-21217749.out
Untracked: code/slurm-21217750.out
Untracked: code/slurm-21217751.out
Untracked: code/slurm-21217773.out
Untracked: code/slurm-21217783.out
Untracked: code/slurm-21217784.out
Untracked: code/slurm-21217785.out
Untracked: code/slurm-21217786.out
Untracked: code/slurm-21217975.out
Untracked: code/slurm-21217976.out
Untracked: code/slurm-21217977.out
Untracked: code/slurm-21217979.out
Untracked: code/slurm-21217980.out
Untracked: code/slurm-21218015.out
Untracked: code/slurm-21218016.out
Untracked: code/slurm-21218017.out
Untracked: code/slurm-21218018.out
Untracked: code/slurm-21218019.out
Untracked: code/slurm-21218038.out
Untracked: code/slurm-21218039.out
Untracked: code/slurm-21218040.out
Untracked: code/slurm-21218046.out
Untracked: code/slurm-21218047.out
Untracked: code/slurm-21218071.out
Untracked: code/slurm-21218072.out
Untracked: code/slurm-21218073.out
Untracked: code/slurm-21218076.out
Untracked: code/slurm-21218077.out
Untracked: code/slurm-21218081.out
Untracked: code/slurm-21218082.out
Untracked: code/slurm-21218083.out
Untracked: code/slurm-21218084.out
Untracked: code/slurm-21218085.out
Untracked: code/slurm-21218117.out
Untracked: code/slurm-21218118.out
Untracked: code/slurm-21218119.out
Untracked: code/slurm-21218120.out
Untracked: code/slurm-21218125.out
Untracked: code/slurm-21218129.out
Untracked: code/split_vcf.sh
Untracked: code_backup/
Untracked: data/ScTypeDB_full.xlsx
Untracked: data/hg19_gtf_genomic_annots_ver2.gr.rds
Untracked: data/num_overlaps_finemapped_SNPs_and_ctcf.txt
Untracked: data/qqplot_SNPs_high_spliceAI.png
Untracked: data/qqplot_SNPs_high_spliceAI_scores_SCZ.png
Untracked: data/qqplot_SNPs_high_spliceAI_scores_aFib.png
Untracked: data/qqplot_SNPs_high_spliceAI_scores_allergy.png
Untracked: data/torus_enrichment_novel_annot.est
Untracked: data/torus_joint_enrichment.est
Untracked: data/torus_joint_refined_enrichment.est
Untracked: enhancer_gene_feature.rmd
Untracked: fig1_panels.pdf
Untracked: fig2.pdf
Untracked: fig_panel2.pdf
Untracked: gene_mapping.pdf
Untracked: output/AAD/GMP_merge_stats.txt
Untracked: output/AAD/Wang2020_joint.results
Untracked: output/AAD/Wang2020_joint_T.results
Untracked: output/AAD/Wang2020_joint_tissueResT.results
Untracked: output/AAD/allergy/Ulirsch2019/GMP_merge_compare_old.est
Untracked: output/AAD/allergy/Ulirsch2019_disjoint_snps.sumstats
Untracked: output/AAD/allergy/Wang2020_T_subsets.est
Untracked: output/AAD/allergy/Wang2020_T_subsets_indiv.est
Untracked: output/AAD/allergy/Wang2020_T_tissueRes.est
Untracked: output/AAD/allergy/Wang2020_joint_T.results
Untracked: output/AAD/allergy/Wang2020_joint_tissueResT.results
Untracked: output/AAD/allergy/Wang2020_tissueResT.est
Untracked: output/AAD/allergy/torus_enrichment_CD4.est
Untracked: output/AAD/allergy/torus_enrichment_CD8.est
Untracked: output/AAD/allergy/torus_enrichment_non_tissueRes_T.est
Untracked: output/AAD/allergy/torus_enrichment_tissueMigraT.est
Untracked: output/AAD/allergy/torus_enrichment_tissueResT_C6.est
Untracked: output/AAD/allergy/torus_enrichment_tissueResT_C8.est
Untracked: output/AAD/allergy/torus_enrichment_tissueRes_T.est
Untracked: output/AAD/allergy/torus_enrichment_tissueResident_T_cells.est
Untracked: output/AAD/asthma_adult/Ulirsch2019/CD4_compare_old.est
Untracked: output/AAD/asthma_adult/Ulirsch2019/CD8_compare_old.est
Untracked: output/AAD/asthma_adult/Ulirsch2019/GMP_merge_compare_old.est
Untracked: output/AAD/asthma_adult/Wang2020_T_subsets.est
Untracked: output/AAD/asthma_adult/Wang2020_T_subsets_indiv.est
Untracked: output/AAD/asthma_adult/Wang2020_T_tissueRes.est
Untracked: output/AAD/asthma_adult/Wang2020_joint_T.results
Untracked: output/AAD/asthma_adult/Wang2020_joint_tissueResT.results
Untracked: output/AAD/asthma_adult/torus_enrichment_CD4.est
Untracked: output/AAD/asthma_adult/torus_enrichment_CD8.est
Untracked: output/AAD/asthma_adult/torus_enrichment_non_tissueRes_T.est
Untracked: output/AAD/asthma_adult/torus_enrichment_tissueMigraT.est
Untracked: output/AAD/asthma_adult/torus_enrichment_tissueResT_C6.est
Untracked: output/AAD/asthma_adult/torus_enrichment_tissueResT_C8.est
Untracked: output/AAD/asthma_adult/torus_enrichment_tissueRes_T.est
Untracked: output/AAD/asthma_adult/torus_enrichment_tissueResident_T_cells.est
Untracked: output/AAD/asthma_child/CD4_compare.est
Untracked: output/AAD/asthma_child/CD8_compare.est
Untracked: output/AAD/asthma_child/Ulirsch2019/GMP_merge_compare_old.est
Untracked: output/AAD/asthma_child/Ulirsch2019/torus_enrichment_CD4.est
Untracked: output/AAD/asthma_child/Ulirsch2019/torus_enrichment_CD8.est
Untracked: output/AAD/asthma_child/Wang2020_T_subsets.est
Untracked: output/AAD/asthma_child/Wang2020_T_subsets_indiv.est
Untracked: output/AAD/asthma_child/Wang2020_T_tissueRes.est
Untracked: output/AAD/asthma_child/Wang2020_joint_T.results
Untracked: output/AAD/asthma_child/Wang2020_joint_tissueResT.results
Untracked: output/AAD/asthma_child/torus_enrichment_CD4.est
Untracked: output/AAD/asthma_child/torus_enrichment_CD8.est
Untracked: output/AAD/asthma_child/torus_enrichment_non_tissueRes_T.est
Untracked: output/AAD/asthma_child/torus_enrichment_tissueMigraT.est
Untracked: output/AAD/asthma_child/torus_enrichment_tissueResT_C6.est
Untracked: output/AAD/asthma_child/torus_enrichment_tissueResT_C8.est
Untracked: output/AAD/asthma_child/torus_enrichment_tissueRes_T.est
Untracked: output/AAD/asthma_child/torus_enrichment_tissueResident_T_cells.est
Untracked: output/LDL_ukb_L10.gif
Untracked: output/LDL_ukb_L10.pdf
Untracked: output/background_SNPs_annotated_percent.txt
Untracked: output/ldsc
Untracked: output/locus_1452.gif
Untracked: output/locus_1452.pdf
Untracked: output/splicing/PTR_across_traits_annotations.results
Untracked: output/splicing/header.txt
Untracked: output/splicing/prior/
Untracked: output/splicing/scz_PTR_annotations.results
Untracked: output/splicing/scz_neuOCR_m6a_DMR.results
Untracked: output/splicing/scz_spliceAI0.03_hist.png
Untracked: output/splicing/scz_spliceAI0.03_scatterplot.png
Untracked: output/splicing/scz_spliceai_binary0.03.results
Untracked: output/splicing/torus_afib_spliceai.est
Untracked: output/splicing/torus_annotations_spliceai0.01.txt.gz
Untracked: output/splicing/torus_annotations_spliceai0.03.txt.gz
Untracked: output/splicing/torus_annotations_spliceai0.05.txt.gz
Untracked: output/splicing/torus_annotations_spliceai0.07.txt.gz
Untracked: output/splicing/torus_annotations_spliceai0.1.txt.gz
Untracked: output/splicing/torus_annotations_spliceai0.2.txt.gz
Untracked: output/splicing/torus_annotations_spliceai9.txt.gz
Untracked: output/splicing/torus_enrichment_joint_scz_mtsplice0.6_hypothalamus-brain.est
Untracked: output/splicing/torus_enrichment_joint_scz_spliceAI.est
Untracked: output/splicing/torus_spliceai0.01.enrichment
Untracked: output/splicing/torus_spliceai0.03.enrichment
Untracked: output/splicing/torus_spliceai0.05.enrichment
Untracked: output/splicing/torus_spliceai0.07.enrichment
Untracked: output/splicing/torus_spliceai0.1.enrichment
Untracked: output/splicing/torus_spliceai0.2.enrichment
Untracked: output/splicing/torus_zscores.txt.gz
Untracked: output/torus
Untracked: panel_figure2.pdf
Untracked: test.txt
Unstaged changes:
Deleted: .Rprofile
Modified: analysis/ldsc_PTR_results.Rmd
Deleted: output/AAD/Caldero2019_disjoint_snps.sumstats
Modified: output/AAD/allergy/Caldero2019_disjoint_snps.sumstats
Modified: output/AAD/allergy/Ulirsch2019/GMP_merge_compare.est
Modified: output/AAD/allergy/Wang2020_indiv.est
Modified: output/AAD/allergy/Wang2020_joint.results
Deleted: output/AAD/asthma/Caldero2019_diffDA_annot_percent.txt
Deleted: output/AAD/asthma/Caldero2019_stimuDA_annot_percent.txt
Deleted: output/AAD/asthma/celltype_specific_adult_lungs_torus.est
Deleted: output/AAD/asthma/diffe_adult_blood_torus.est
Deleted: output/AAD/asthma/joint_blood_immune_rest_vs_stimu.est
Deleted: output/AAD/asthma/joint_lung_vs_blood_immune_diff_torus.est
Deleted: output/AAD/asthma/joint_lung_vs_blood_immune_stimu_torus.est
Deleted: output/AAD/asthma/lung_clusters_dict.txt
Deleted: output/AAD/asthma/lung_clusters_info.txt
Deleted: output/AAD/asthma/stimu_adult_blood_torus.est
Deleted: output/AAD/asthma/torus_enrichment_all_rest.est
Deleted: output/AAD/asthma/torus_enrichment_all_stimulated.est
Deleted: output/AAD/asthma/zhang2021_annot_percent.txt
Deleted: output/AAD/asthma/zhang2021_cell_type_overlaps.txt
Deleted: output/AAD/asthma/zhang2021_peaks_per_celltype.txt
Modified: output/AAD/asthma_adult/Ulirsch2019/CD4_compare.est
Modified: output/AAD/asthma_adult/Ulirsch2019/CD8_compare.est
Deleted: output/AAD/asthma_adult/Ulirsch2019/GMP_merge_compare.est
Modified: output/AAD/asthma_adult/Wang2020_indiv.est
Modified: output/AAD/asthma_adult/Wang2020_joint.results
Modified: output/AAD/asthma_child/Ulirsch2019/GMP_merge_compare.est
Modified: output/AAD/asthma_child/Wang2020_indiv.est
Modified: output/AAD/asthma_child/Wang2020_joint.results
Deleted: output/asthma/Caldero2019_diffDA_annot_percent.txt
Deleted: output/asthma/Caldero2019_stimuDA_annot_percent.txt
Deleted: output/asthma/celltype_specific_adult_lungs_torus.est
Deleted: output/asthma/diffe_adult_blood_torus.est
Deleted: output/asthma/joint_lung_vs_blood_immune_diff_torus.est
Deleted: output/asthma/joint_lung_vs_blood_immune_stimu_torus.est
Deleted: output/asthma/lung_clusters_dict.txt
Deleted: output/asthma/lung_clusters_info.txt
Deleted: output/asthma/stimu_adult_blood_torus.est
Deleted: output/asthma/zhang2021_annot_percent.txt
Deleted: output/asthma/zhang2021_cell_type_overlaps.txt
Deleted: output/asthma/zhang2021_peaks_per_celltype.txt
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with wflow_publish() to start tracking its development.
Backgrounds
Post-transcriptional regulatory (PTR) processes have been implicated in development and diseases, however, it is largely unknown how genetic variations are mediated through PTR processes. We propose to annotate GWAS variants using both experimental measurements and computational predictions. With this prior knowledge, we can further identify most likely causal variants through fine-mapping and then link them to genes.
Several post-transcriptonal features will be explored:
- alternative splicing
- RNA modification: m6A
- RNA binding
- Polyadenylation
Methods
SNP effect predictions
The predictions of variant effects on post-transcriptional regulation were performed on 10 million SNPs after some QC criteria, from 1000 genome phase 3 project.
Enrichment analysis
We first tested annotations one at a time using both TORUS and S-LDSC.Then we jointly assessed a set of annotations.
For the early attempts, I ran torus and S-LDSC on different numbers of test SNPs. As an example, there are 6M SNPs for SCZ, but 8M SNPs for aFIb after the same filtering steps. However, for S-LDSC analysis, around 1M hapmap3 SNPs were tested across all traits.
Results
Enrichment analysis
To make it comparable, I run S-LDSC and Torus on the same set of test SNPs, which are around 1 million SNPs from hapmap3.
Legends for the plots:
- x-axis - annotations
- y-axis - fold of enrichment
- label on the plot - percent of narrow-sense heritability
- dashed line - no enrichment
S-LDSC
modified baselineLD model: genic features (introns excluded), LD, MAF, background selection, nucleotide diversity, promoter, recombination rate. annotations: spidex, spliceai, fetal brain m6a, neuronal OCR, differentially methylated regions in brains (DMR)
 Examine baseline annotations
Examine baseline annotations
- baseline annotations from m6A paper: baseline_gene_MAF_LD
- test annotation:spliceai score >=0.05
The intron region annotations explain ~50% of SNP heritability. To not interfere with our testing of splicing effects, I removed intron related annotations from baselines.
 Torus
Torus 
 With the same set of test SNPs, the enrichment estimates are similarly low between torus and LDSC. The enrichment signals seen in the previous runs for torus were gone, likely due to the background difference.
With the same set of test SNPs, the enrichment estimates are similarly low between torus and LDSC. The enrichment signals seen in the previous runs for torus were gone, likely due to the background difference.
Other traits - AFib
 For aFib, we see significant enrichment of variants with spliceAI scores >=0.05. Approximately, they contribute to 2.4% of SNP heritability based on taking a product of percent of variants within annotation and enrichment estimate. However, we did not see such enrichment in S-LDSC results. The possible reason might be the signal was drown in the baseline annotations.
For aFib, we see significant enrichment of variants with spliceAI scores >=0.05. Approximately, they contribute to 2.4% of SNP heritability based on taking a product of percent of variants within annotation and enrichment estimate. However, we did not see such enrichment in S-LDSC results. The possible reason might be the signal was drown in the baseline annotations.
QQ plots for comparing p-values
Comparing p-values of SNPs with high spliceAI scores against 1M genome-wide SNPs
black: 1M genome-wide SNPs with spliceAI scores >=0 blue: GWAS SNPs with spliceAI scores >=0.05
Schizophrenia
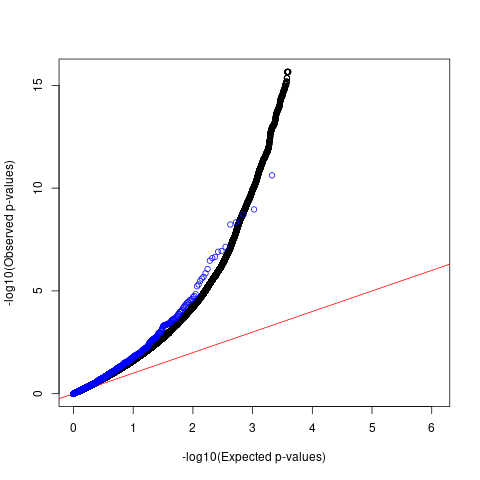
QQ plots
Afib
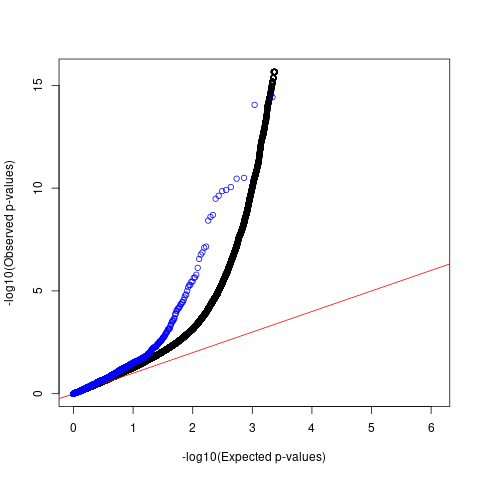
QQ plots
Allergy
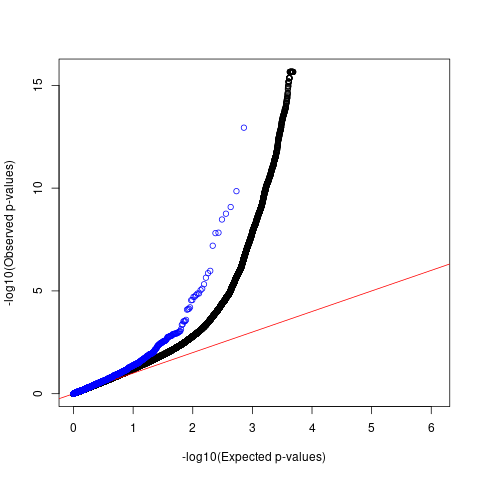
QQ plots
For SCZ, the p-values of GWAS SNPs with spliceiAI scores above the threshold deviate from the null in a similar way as compared to genome-wide SNPs with predicted spliceAI scores.
For AFib and allergy, we observed that p-values of SNPs with high spliceAI scores deviated more from the null compared to the genome-wide SNPs. However, this enrichemnt signal was gone after adjusting for baseline annotations by LDSC.
Across multiple traits


test annotations jointly 
sessionInfo()R version 4.0.4 (2021-02-15)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.3.13-el7-x86_64/lib/libopenblas_haswellp-r0.3.13.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] ggplot2_3.3.3
loaded via a namespace (and not attached):
[1] Rcpp_1.0.8 pillar_1.5.0 compiler_4.0.4 bslib_0.2.4
[5] later_1.1.0.1 jquerylib_0.1.3 git2r_0.28.0 highr_0.8
[9] workflowr_1.6.2 tools_4.0.4 digest_0.6.27 jsonlite_1.7.2
[13] evaluate_0.14 lifecycle_1.0.0 tibble_3.0.6 gtable_0.3.0
[17] pkgconfig_2.0.3 rlang_1.0.1 DBI_1.1.1 cli_3.2.0
[21] rstudioapi_0.13 yaml_2.2.1 xfun_0.21 withr_2.4.3
[25] dplyr_1.0.4 stringr_1.4.0 knitr_1.31 generics_0.1.0
[29] fs_1.5.0 vctrs_0.3.8 sass_0.3.1 tidyselect_1.1.1
[33] rprojroot_2.0.2 grid_4.0.4 glue_1.6.1 R6_2.5.1
[37] fansi_1.0.2 rmarkdown_2.7 farver_2.1.0 purrr_0.3.4
[41] magrittr_2.0.1 scales_1.1.1 promises_1.2.0.1 ellipsis_0.3.2
[45] htmltools_0.5.1.1 assertthat_0.2.1 colorspace_2.0-2 httpuv_1.5.5
[49] labeling_0.4.2 utf8_1.2.2 stringi_1.5.3 munsell_0.5.0
[53] crayon_1.4.1