Asthma project
Last updated: 2021-09-23
Checks: 6 1
Knit directory: funcFinemapping/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it's best to always run the code in an empty environment.
The command set.seed(20210404) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| ~/projects/funcFinemapping/output/asthma/Caldero2019_stimuDA_annot_percent.txt | output/asthma/Caldero2019_stimuDA_annot_percent.txt |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version e5d6f32. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .ipynb_checkpoints/
Ignored: analysis/ldsc_results.nb.html
Ignored: analysis/results.nb.html
Ignored: analysis/snp_finemapping_results.nb.html
Ignored: analysis/splicing.nb.html
Untracked files:
Untracked: SNPs_categories,png
Untracked: SNPs_categories.png
Untracked: analysis/enhancer_gene_feature.Rmd
Untracked: analysis/feedback.Rmd
Untracked: analysis/gene_finemapping_results.Rmd
Untracked: analysis/learn_susie.Rmd
Untracked: analysis/notes.Rmd
Untracked: analysis/snp_finemapping_results.Rmd
Untracked: analysis/splicing.Rmd
Untracked: code/.ipynb_checkpoints/
Untracked: code/ldsc_regression.sh
Untracked: code/make_plots.R
Untracked: code/run_ldsc.sh
Untracked: code/run_ldsc_with_bed.sh
Untracked: code/run_torus.sh
Untracked: code/split_vcf.sh
Untracked: data/num_overlaps_finemapped_SNPs_and_ctcf.txt
Untracked: data/scz_2018
Untracked: data/torus_enrichment_novel_annot.est
Untracked: data/torus_joint_enrichment.est
Untracked: data/torus_joint_refined_enrichment.est
Untracked: enhancer_gene_feature.rmd
Untracked: fig1_panels.pdf
Untracked: fig2.pdf
Untracked: fig_panel2.pdf
Untracked: gene_mapping.pdf
Untracked: panel_figure2.pdf
Untracked: test.txt
Unstaged changes:
Modified: analysis/biology_bkg.Rmd
Modified: analysis/method_bkg.Rmd
Deleted: analysis/results.Rmd
Deleted: data/joint_torus_conservation_enrichment.est
Deleted: data/torus_enrichment.est
Deleted: data/torus_enrichment_DMR.est
Modified: data/torus_enrichment_ambigousSNPs.est
Deleted: data/torus_enrichment_m6A.est
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/asthma_results.Rmd) and HTML (docs/asthma_results.html) files. If you've configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | e5d6f32 | Jing Gu | 2021-09-23 | added percent of SNPs with annotations |
| html | 53a8774 | Jing Gu | 2021-09-22 | Build site. |
| html | 2a6fc91 | Jing Gu | 2021-09-22 | Build site. |
| Rmd | bbb2944 | Jing Gu | 2021-09-22 | analyzed Asthma GWAS with functional priors from single cell data |
| html | 41f53c8 | Jing Gu | 2021-09-22 | Build site. |
| Rmd | 33d4e5a | Jing Gu | 2021-09-22 | analyzed Asthma GWAS with functional priors from single cell data |
| html | b0b938b | Jing Gu | 2021-09-22 | Build site. |
| Rmd | 8a5656c | Jing Gu | 2021-09-22 | analyzed Asthma GWAS with functional priors from single cell data |
Overall Goal
Identify disease-relevant cell types and genes for Asthma
Enrichment estimates for individual annotation
Adult-onset asthma
- GWAS: Zhu et al. 2019 (case N=22296, control N=347481)
- cell-type specific Annotations: Zhang et al.2021 - single-cell ATAC-Seq data for adult lungs
- Entropy based method used to identify cell-type specific peaks
- For each cell type, peaks that pass 0.1% FDR cutoff were used to generate binary annotations
Enrichment results
| Version | Author | Date |
|---|---|---|
| b0b938b | Jing Gu | 2021-09-22 |
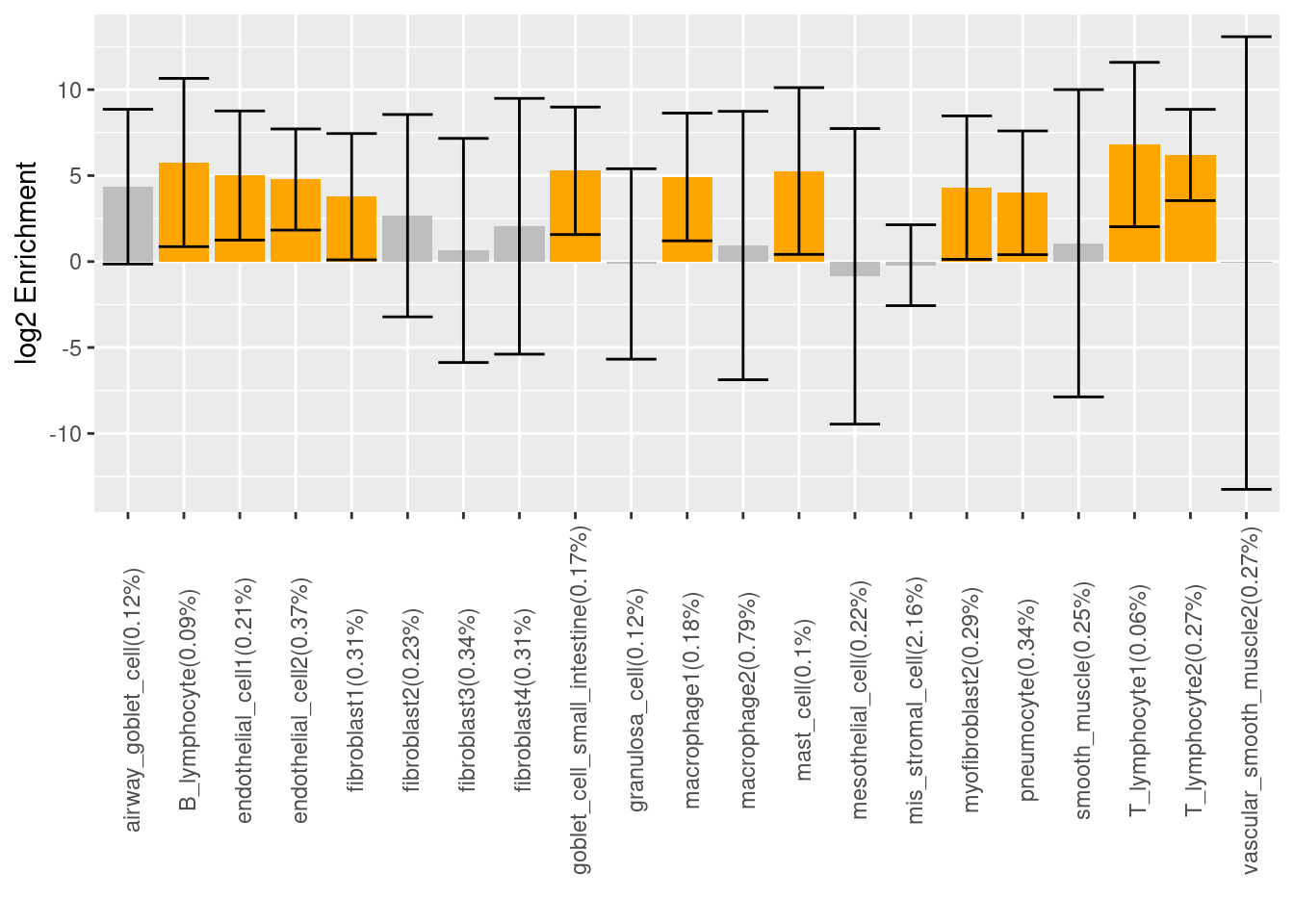
Log2 scaled enrichment estimates for each cell type, which was individually run using Torus. The highlighted bars show significant enrichment, while the bars in grey color do not. The x-axis tickmark labels consist of the annotation name and percent of all SNPs that have that annotation. Asthma-associated variants are significantly enriched in certain sub-clusters of macrophages and fibroblasts, but not in others. The cell types that show the highest enrichment in the chromatin accessibility peaks are immune-related cells.
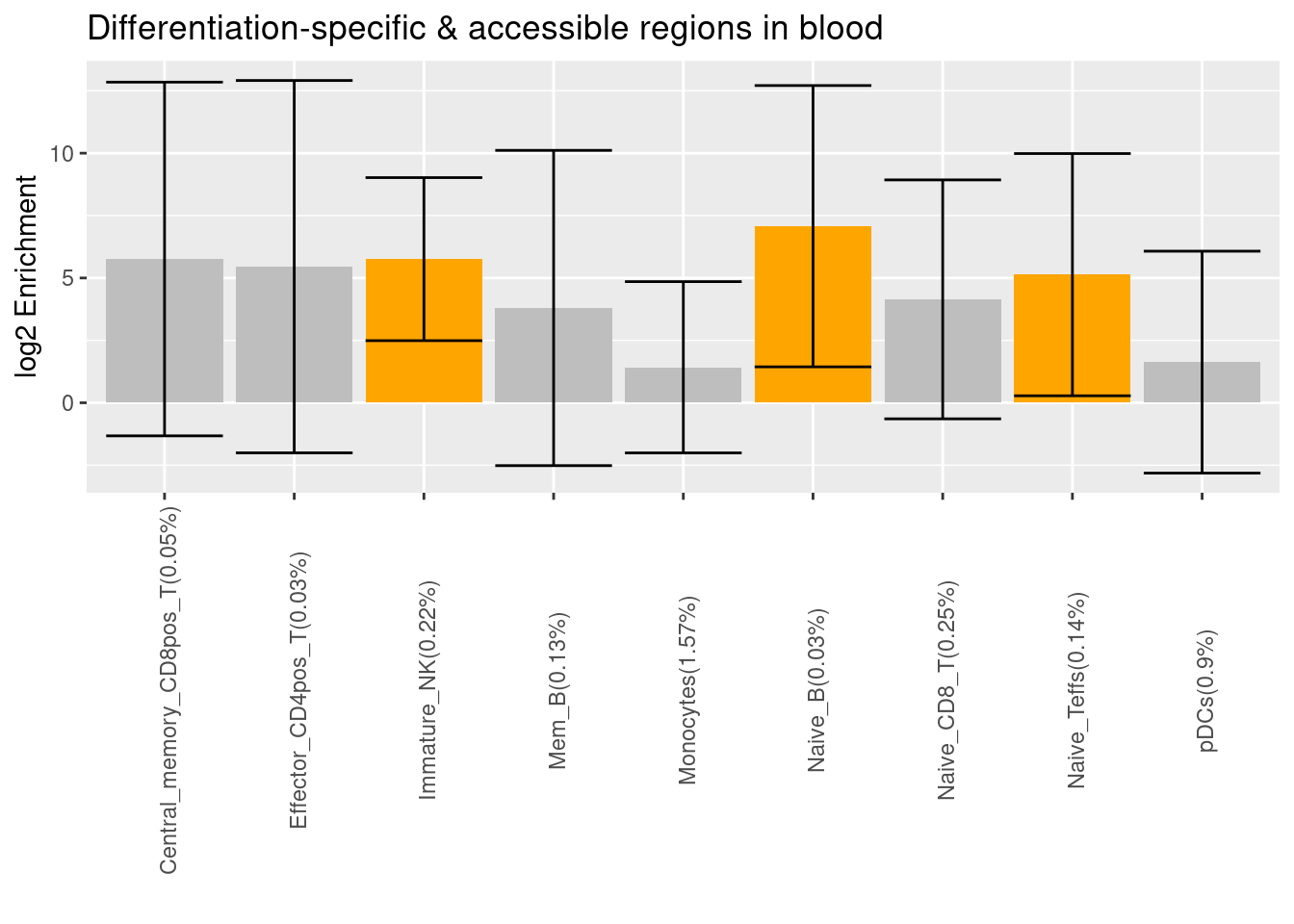
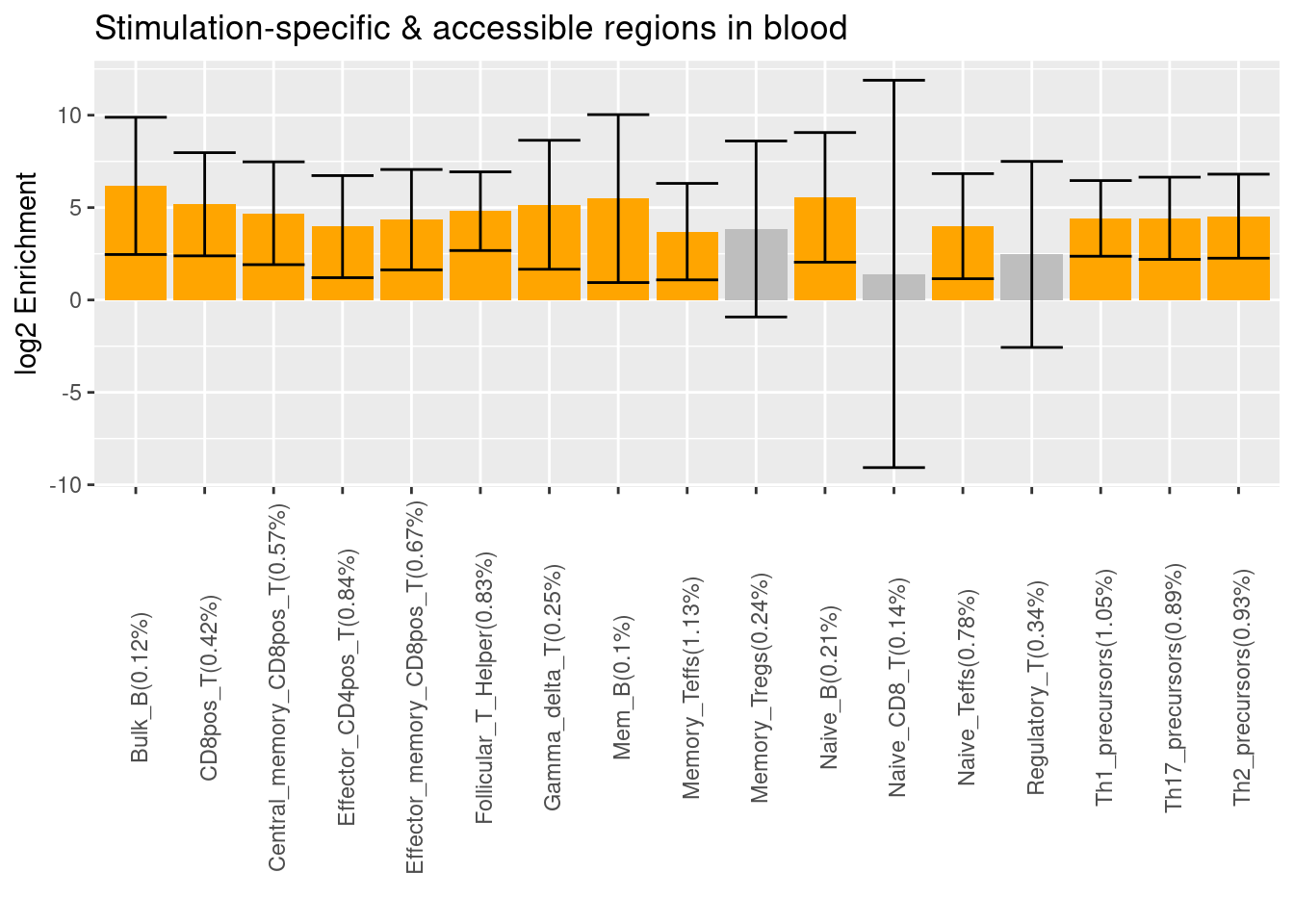
- Caldero et al. 2019 - ATAC-Seq data for FACS-sorted cells from whole blood
- Significant differentially accessible regions when compared to progenitor cells
- Significant differentially accessible regions under stimulation
| Version | Author | Date |
|---|---|---|
| b0b938b | Jing Gu | 2021-09-22 |
# bg_percent=c()
# for(i in df$term){
# fname<-strsplit(i, "_atac")[[1]][1]
# annot<-fread(sprintf("/home/jinggu/cluster/projects/torus_enrichment/asthma_adult/stimu_DA/torus_annotations_%s.txt.gz",fname))
# bg_percent<-rbind(bg_percent, c(fname, sum(annot[,2])))
# }
# bg_percent<-data.frame(bg_percent)
# bg_percent$X2<-as.numeric(bg_percent$X2)
# bg_percent["percent"]<-paste0(round(bg_percent$X2/dim(gwas)[1]*100,2), "%")
# write.table(bg_percent,"~/projects/funcFinemapping/output/asthma/Caldero2019_stimuDA_annot_percent.txt", quote = F,row.names=F, col.names = F, sep='\t')- Not sure why much fewer cell types show differentially accessiblity during differentiation.
| Version | Author | Date |
|---|---|---|
| b0b938b | Jing Gu | 2021-09-22 |
Upon stimulation, the enrichment signals prevail across a broader range of immune cells. There are more diverse stimulated cell types that show differential accessibility against the unstimulated ones.
Joint enrichment estimates across annotations
immune cells in lungs vs. immune cells in blood
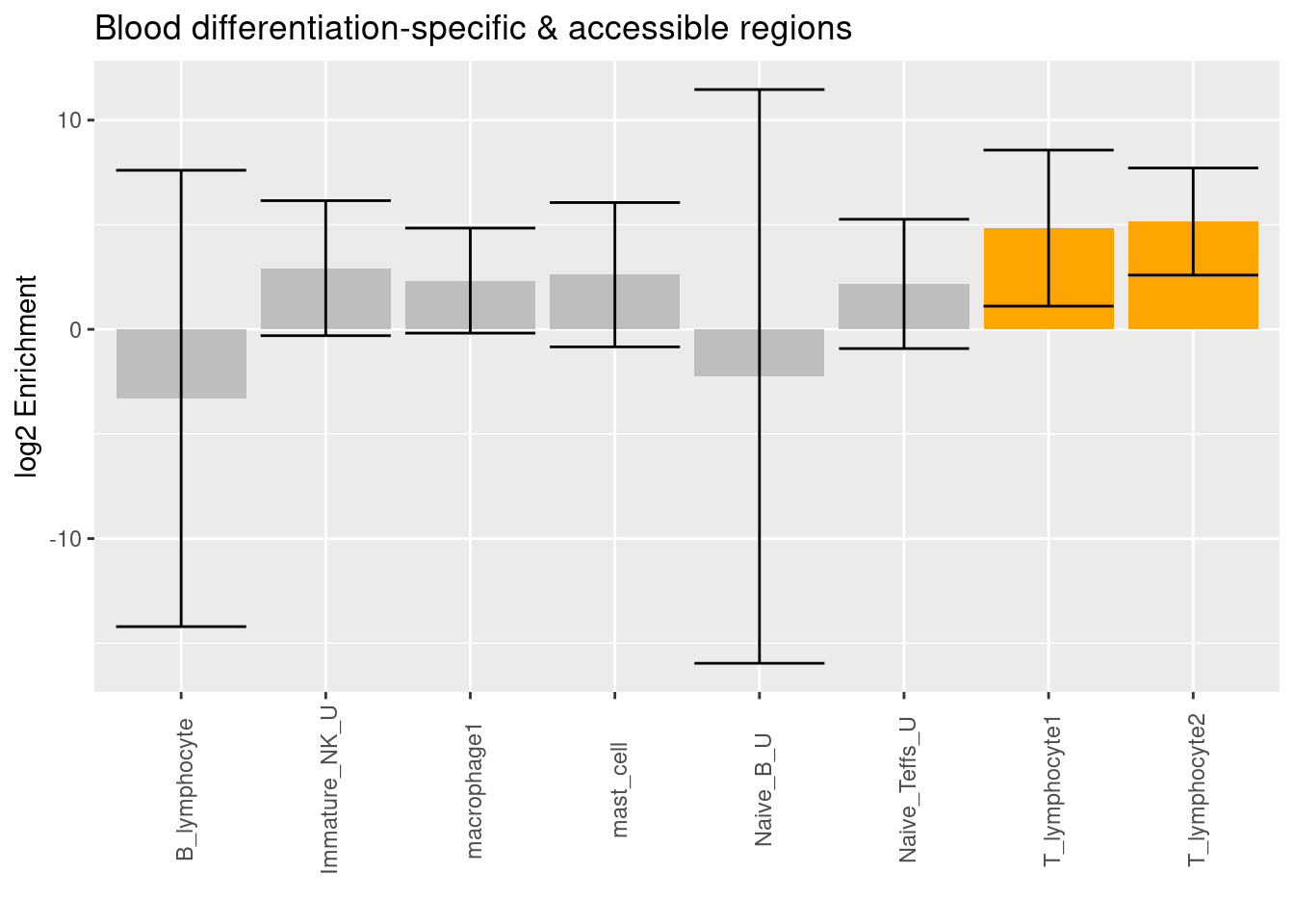
| Version | Author | Date |
|---|---|---|
| b0b938b | Jing Gu | 2021-09-22 |
Both clusters of cell-type specific accessible regions from T lymphocytes in lungs are significantly enriched with risk variants conditional on differentiation-specific regions identified from blood.
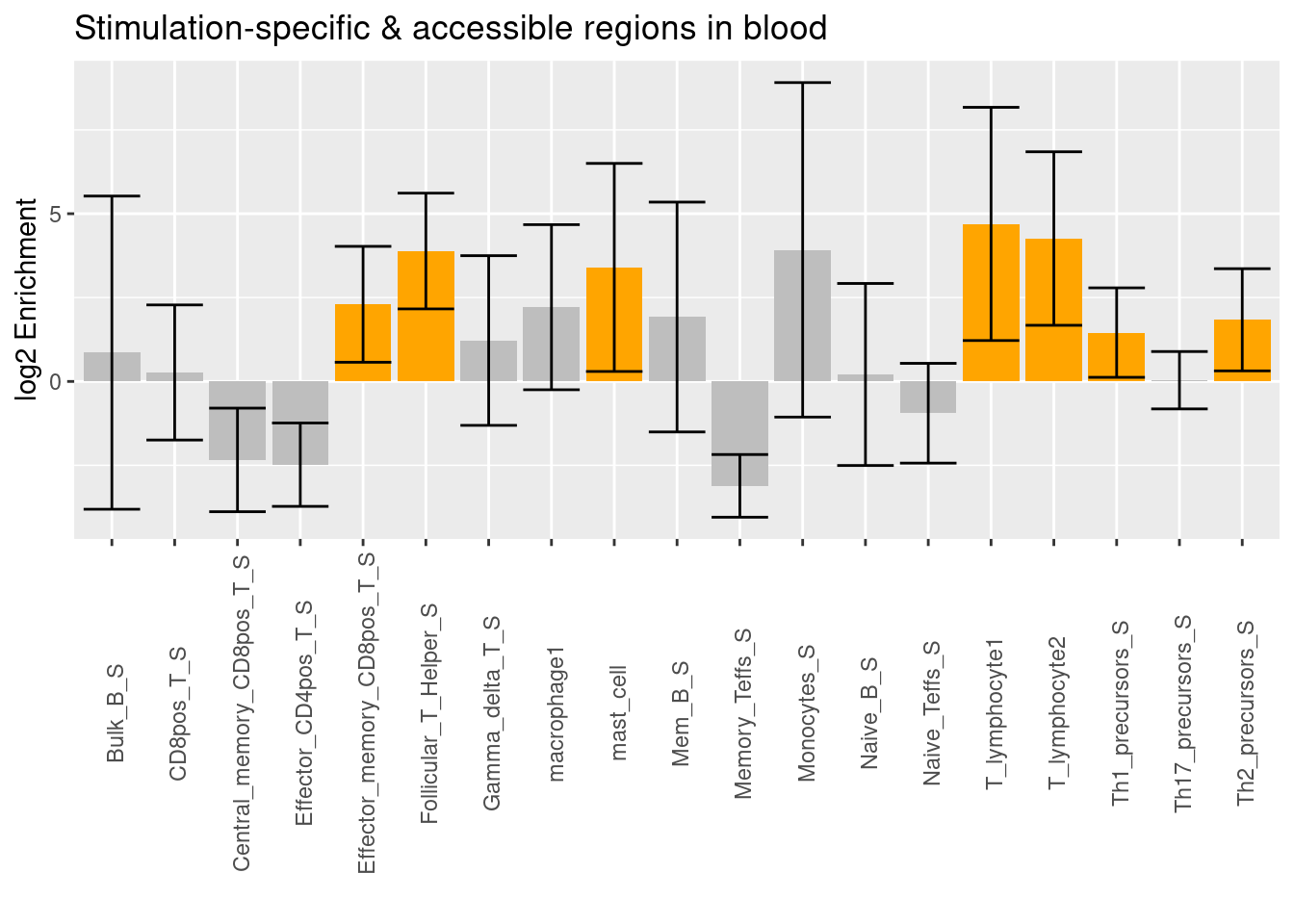 The enrichment signal of T lymphocytes remains after conditional on stimulated immune cell types in blood. The additional enrichment signals come from stimulated memoryCD8+ T-cells, T_helpler cells, mast cells, Th1 precursors, and Th2 precursors.
The enrichment signal of T lymphocytes remains after conditional on stimulated immune cell types in blood. The additional enrichment signals come from stimulated memoryCD8+ T-cells, T_helpler cells, mast cells, Th1 precursors, and Th2 precursors.
Children-onset Asthma
- GWAS: Zhu et al. 2019
- cell-type specific Annotations: Domcke et al.2020 - single-cell ATAC-Seq data for fetal lungs
sessionInfo()R version 4.0.4 (2021-02-15)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.3.13-el7-x86_64/lib/libopenblas_haswellp-r0.3.13.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] ggplot2_3.3.3 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] Rcpp_1.0.7 pillar_1.5.0 compiler_4.0.4 bslib_0.2.4
[5] later_1.1.0.1 jquerylib_0.1.3 git2r_0.28.0 highr_0.8
[9] tools_4.0.4 digest_0.6.27 jsonlite_1.7.2 evaluate_0.14
[13] lifecycle_1.0.0 tibble_3.0.6 gtable_0.3.0 pkgconfig_2.0.3
[17] rlang_0.4.11 DBI_1.1.1 yaml_2.2.1 xfun_0.21
[21] withr_2.4.2 dplyr_1.0.4 stringr_1.4.0 knitr_1.31
[25] generics_0.1.0 fs_1.5.0 vctrs_0.3.8 sass_0.3.1
[29] tidyselect_1.1.1 rprojroot_2.0.2 grid_4.0.4 glue_1.4.2
[33] R6_2.5.1 fansi_0.5.0 rmarkdown_2.7 farver_2.1.0
[37] purrr_0.3.4 magrittr_2.0.1 whisker_0.4 scales_1.1.1
[41] promises_1.2.0.1 ellipsis_0.3.2 htmltools_0.5.1.1 assertthat_0.2.1
[45] colorspace_2.0-2 httpuv_1.5.5 labeling_0.4.2 utf8_1.2.2
[49] stringi_1.5.3 munsell_0.5.0 crayon_1.4.1