Predicting splicing effects with MTSplice
Last updated: 2022-08-10
Checks: 5 2
Knit directory: funcFinemapping/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210404) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| ~/projects/funcFinemapping/docs/assets/splicing/compare_MAFs_mtsplice.png | docs/assets/splicing/compare_MAFs_mtsplice.png |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 03e831f. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .ipynb_checkpoints/
Ignored: analysis/.Rhistory
Ignored: analysis/PTR_m6A.nb.html
Ignored: analysis/build_annotations_for_single_cell_data.nb.html
Ignored: analysis/lab4_prepare.nb.html
Ignored: analysis/ldsc_results.nb.html
Ignored: analysis/learn_archR.nb.html
Ignored: analysis/mtsplice_finemapping_results.nb.html
Ignored: analysis/results.nb.html
Ignored: analysis/snp_finemapping_results.nb.html
Ignored: analysis/splicing.nb.html
Ignored: analysis/susie_tutorial.nb.html
Untracked files:
Untracked: SCZ_pval_vs_MAF.png
Untracked: SNPs_categories,png
Untracked: SNPs_categories.png
Untracked: analysis/.ipynb_checkpoints/
Untracked: analysis/PTR_m6A.Rmd
Untracked: analysis/PTR_splicing_mtsplice.Rmd
Untracked: analysis/PTR_splicing_spliceAI.Rmd
Untracked: bmi_locus1410.pdf
Untracked: code/.ipynb_checkpoints/
Untracked: code/.snakemake/
Untracked: code/Euro_LD_Chunks.RData
Untracked: code/Snakefile
Untracked: code/config.yaml
Untracked: code/environment.yml
Untracked: code/ldsc.log
Untracked: code/ldsc.results
Untracked: code/ldsc_regression.sh
Untracked: code/make_joint_annotations.R
Untracked: code/make_plots.R
Untracked: code/out/
Untracked: code/run_ldsc.sh
Untracked: code/run_ldsc_with_bed.sh
Untracked: code/run_ldsc_with_bed_v2.sh
Untracked: code/run_susie.R
Untracked: code/run_torus.sh
Untracked: code/run_torus/
Untracked: code/sctype/
Untracked: code/split_vcf.sh
Untracked: code_backup/
Untracked: data/ScTypeDB_full.xlsx
Untracked: data/hg19_gtf_genomic_annots_ver2.gr.rds
Untracked: data/mmsplice_mtsplice_cutoffs.txt
Untracked: data/num_overlaps_finemapped_SNPs_and_ctcf.txt
Untracked: data/qqplot_SNPs_high_spliceAI.png
Untracked: data/qqplot_SNPs_high_spliceAI_scores_SCZ.png
Untracked: data/qqplot_SNPs_high_spliceAI_scores_aFib.png
Untracked: data/qqplot_SNPs_high_spliceAI_scores_allergy.png
Untracked: data/spliceAIandMAF.txt.gz
Untracked: data/torus_enrichment_novel_annot.est
Untracked: data/torus_joint_enrichment.est
Untracked: data/torus_joint_refined_enrichment.est
Untracked: enhancer_gene_feature.rmd
Untracked: fig1_panels.pdf
Untracked: fig2.pdf
Untracked: fig_panel2.pdf
Untracked: gene_mapping.pdf
Untracked: output/AAD/GMP_merge_stats.txt
Untracked: output/AAD/Wang2020_joint.results
Untracked: output/AAD/Wang2020_joint_T.results
Untracked: output/AAD/Wang2020_joint_tissueResT.results
Untracked: output/AAD/allergy/Ulirsch2019/GMP_merge_compare_old.est
Untracked: output/AAD/allergy/Ulirsch2019_disjoint_snps.sumstats
Untracked: output/AAD/allergy/Wang2020_T_subsets.est
Untracked: output/AAD/allergy/Wang2020_T_subsets_indiv.est
Untracked: output/AAD/allergy/Wang2020_T_tissueRes.est
Untracked: output/AAD/allergy/Wang2020_joint_T.results
Untracked: output/AAD/allergy/Wang2020_joint_tissueResT.results
Untracked: output/AAD/allergy/Wang2020_tissueResT.est
Untracked: output/AAD/allergy/torus_enrichment_CD4.est
Untracked: output/AAD/allergy/torus_enrichment_CD8.est
Untracked: output/AAD/allergy/torus_enrichment_non_tissueRes_T.est
Untracked: output/AAD/allergy/torus_enrichment_tissueMigraT.est
Untracked: output/AAD/allergy/torus_enrichment_tissueResT_C6.est
Untracked: output/AAD/allergy/torus_enrichment_tissueResT_C8.est
Untracked: output/AAD/allergy/torus_enrichment_tissueRes_T.est
Untracked: output/AAD/allergy/torus_enrichment_tissueResident_T_cells.est
Untracked: output/AAD/asthma_adult/Ulirsch2019/CD4_compare_old.est
Untracked: output/AAD/asthma_adult/Ulirsch2019/CD8_compare_old.est
Untracked: output/AAD/asthma_adult/Ulirsch2019/GMP_merge_compare_old.est
Untracked: output/AAD/asthma_adult/Wang2020_T_subsets.est
Untracked: output/AAD/asthma_adult/Wang2020_T_subsets_indiv.est
Untracked: output/AAD/asthma_adult/Wang2020_T_tissueRes.est
Untracked: output/AAD/asthma_adult/Wang2020_joint_T.results
Untracked: output/AAD/asthma_adult/Wang2020_joint_tissueResT.results
Untracked: output/AAD/asthma_adult/torus_enrichment_CD4.est
Untracked: output/AAD/asthma_adult/torus_enrichment_CD8.est
Untracked: output/AAD/asthma_adult/torus_enrichment_non_tissueRes_T.est
Untracked: output/AAD/asthma_adult/torus_enrichment_tissueMigraT.est
Untracked: output/AAD/asthma_adult/torus_enrichment_tissueResT_C6.est
Untracked: output/AAD/asthma_adult/torus_enrichment_tissueResT_C8.est
Untracked: output/AAD/asthma_adult/torus_enrichment_tissueRes_T.est
Untracked: output/AAD/asthma_adult/torus_enrichment_tissueResident_T_cells.est
Untracked: output/AAD/asthma_child/CD4_compare.est
Untracked: output/AAD/asthma_child/CD8_compare.est
Untracked: output/AAD/asthma_child/Ulirsch2019/GMP_merge_compare_old.est
Untracked: output/AAD/asthma_child/Ulirsch2019/torus_enrichment_CD4.est
Untracked: output/AAD/asthma_child/Ulirsch2019/torus_enrichment_CD8.est
Untracked: output/AAD/asthma_child/Wang2020_T_subsets.est
Untracked: output/AAD/asthma_child/Wang2020_T_subsets_indiv.est
Untracked: output/AAD/asthma_child/Wang2020_T_tissueRes.est
Untracked: output/AAD/asthma_child/Wang2020_joint_T.results
Untracked: output/AAD/asthma_child/Wang2020_joint_tissueResT.results
Untracked: output/AAD/asthma_child/torus_enrichment_CD4.est
Untracked: output/AAD/asthma_child/torus_enrichment_CD8.est
Untracked: output/AAD/asthma_child/torus_enrichment_non_tissueRes_T.est
Untracked: output/AAD/asthma_child/torus_enrichment_tissueMigraT.est
Untracked: output/AAD/asthma_child/torus_enrichment_tissueResT_C6.est
Untracked: output/AAD/asthma_child/torus_enrichment_tissueResT_C8.est
Untracked: output/AAD/asthma_child/torus_enrichment_tissueRes_T.est
Untracked: output/AAD/asthma_child/torus_enrichment_tissueResident_T_cells.est
Untracked: output/LDL_ukb_L10.gif
Untracked: output/LDL_ukb_L10.pdf
Untracked: output/background_SNPs_annotated_percent.txt
Untracked: output/ldsc
Untracked: output/locus_1452.gif
Untracked: output/locus_1452.pdf
Untracked: output/spliceAI_vs_MAF.png
Untracked: output/splicing/.ipynb_checkpoints/
Untracked: output/splicing/PTR_across_traits_annotations.results
Untracked: output/splicing/QQplot_mmsplice.png
Untracked: output/splicing/QQplot_mmsplice_top15.png
Untracked: output/splicing/QQplot_mmsplice_vs_mtsplice.png
Untracked: output/splicing/QQplot_mmsplice_vs_mtsplice_SCZ.png
Untracked: output/splicing/TSplice_scores_distribution.png
Untracked: output/splicing/header.txt
Untracked: output/splicing/mmsplice_vs_mtsplice_AA_heart.png
Untracked: output/splicing/mmsplice_vs_mtsplice_Hypothalamus_brain.png
Untracked: output/splicing/mmsplice_vs_mtsplice_LV_heart.png
Untracked: output/splicing/mmsplice_vs_mtsplice_across_tissues.png
Untracked: output/splicing/prior/
Untracked: output/splicing/scz_PTR_annotations.results
Untracked: output/splicing/scz_neuOCR_m6a_DMR.results
Untracked: output/splicing/scz_spliceAI0.03_hist.png
Untracked: output/splicing/scz_spliceAI0.03_scatterplot.png
Untracked: output/splicing/scz_spliceai_binary0.03.results
Untracked: output/splicing/torus_afib_spliceai.est
Untracked: output/splicing/torus_annotations_spliceai0.01.txt.gz
Untracked: output/splicing/torus_annotations_spliceai0.03.txt.gz
Untracked: output/splicing/torus_annotations_spliceai0.05.txt.gz
Untracked: output/splicing/torus_annotations_spliceai0.07.txt.gz
Untracked: output/splicing/torus_annotations_spliceai0.1.txt.gz
Untracked: output/splicing/torus_annotations_spliceai0.2.txt.gz
Untracked: output/splicing/torus_annotations_spliceai9.txt.gz
Untracked: output/splicing/torus_enrichment_joint_scz_mtsplice0.6_hypothalamus-brain.est
Untracked: output/splicing/torus_enrichment_joint_scz_spliceAI.est
Untracked: output/splicing/torus_spliceai0.01.enrichment
Untracked: output/splicing/torus_spliceai0.03.enrichment
Untracked: output/splicing/torus_spliceai0.05.enrichment
Untracked: output/splicing/torus_spliceai0.07.enrichment
Untracked: output/splicing/torus_spliceai0.1.enrichment
Untracked: output/splicing/torus_spliceai0.2.enrichment
Untracked: output/splicing/torus_zscores.txt.gz
Untracked: output/torus
Untracked: panel_figure2.pdf
Untracked: test.txt
Unstaged changes:
Deleted: .Rprofile
Modified: analysis/index.Rmd
Modified: analysis/lab4_prepare.Rmd
Modified: analysis/ldsc_PTR_results.Rmd
Deleted: output/AAD/Caldero2019_disjoint_snps.sumstats
Modified: output/AAD/allergy/Caldero2019_disjoint_snps.sumstats
Modified: output/AAD/allergy/Ulirsch2019/GMP_merge_compare.est
Modified: output/AAD/allergy/Wang2020_indiv.est
Modified: output/AAD/allergy/Wang2020_joint.results
Deleted: output/AAD/asthma/Caldero2019_diffDA_annot_percent.txt
Deleted: output/AAD/asthma/Caldero2019_stimuDA_annot_percent.txt
Deleted: output/AAD/asthma/celltype_specific_adult_lungs_torus.est
Deleted: output/AAD/asthma/diffe_adult_blood_torus.est
Deleted: output/AAD/asthma/joint_blood_immune_rest_vs_stimu.est
Deleted: output/AAD/asthma/joint_lung_vs_blood_immune_diff_torus.est
Deleted: output/AAD/asthma/joint_lung_vs_blood_immune_stimu_torus.est
Deleted: output/AAD/asthma/lung_clusters_dict.txt
Deleted: output/AAD/asthma/lung_clusters_info.txt
Deleted: output/AAD/asthma/stimu_adult_blood_torus.est
Deleted: output/AAD/asthma/torus_enrichment_all_rest.est
Deleted: output/AAD/asthma/torus_enrichment_all_stimulated.est
Deleted: output/AAD/asthma/zhang2021_annot_percent.txt
Deleted: output/AAD/asthma/zhang2021_cell_type_overlaps.txt
Deleted: output/AAD/asthma/zhang2021_peaks_per_celltype.txt
Modified: output/AAD/asthma_adult/Ulirsch2019/CD4_compare.est
Modified: output/AAD/asthma_adult/Ulirsch2019/CD8_compare.est
Deleted: output/AAD/asthma_adult/Ulirsch2019/GMP_merge_compare.est
Modified: output/AAD/asthma_adult/Wang2020_indiv.est
Modified: output/AAD/asthma_adult/Wang2020_joint.results
Modified: output/AAD/asthma_child/Ulirsch2019/GMP_merge_compare.est
Modified: output/AAD/asthma_child/Wang2020_indiv.est
Modified: output/AAD/asthma_child/Wang2020_joint.results
Deleted: output/asthma/Caldero2019_diffDA_annot_percent.txt
Deleted: output/asthma/Caldero2019_stimuDA_annot_percent.txt
Deleted: output/asthma/celltype_specific_adult_lungs_torus.est
Deleted: output/asthma/diffe_adult_blood_torus.est
Deleted: output/asthma/joint_lung_vs_blood_immune_diff_torus.est
Deleted: output/asthma/joint_lung_vs_blood_immune_stimu_torus.est
Deleted: output/asthma/lung_clusters_dict.txt
Deleted: output/asthma/lung_clusters_info.txt
Deleted: output/asthma/stimu_adult_blood_torus.est
Deleted: output/asthma/zhang2021_annot_percent.txt
Deleted: output/asthma/zhang2021_cell_type_overlaps.txt
Deleted: output/asthma/zhang2021_peaks_per_celltype.txt
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/PTR_splicing_mtsplice.Rmd) and HTML (docs/PTR_splicing_mtsplice.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | 03e831f | Jing Gu | 2022-08-02 | PTR_splicing |
| html | abda215 | Jing Gu | 2022-08-02 | PTR_mtsplice |
Backgrounds
Post-transcriptional regulatory (PTR) processes have been implicated in development and diseases, however, it is largely unknown how genetic variations are mediated through PTR processes. We propose to annotate GWAS variants using both experimental measurements and computational predictions. With this prior knowledge, we can further identify most likely causal variants through fine-mapping and then link them to genes.
Several post-transcriptonal features will be explored:
*** Alternative splicing** * RNA modification: m6A * RNA binding * Polyadenylation
Methods
MMSplice/MTSplice
A CNN method that aims to provide tissue-average or tissue-specific combined with the average predictions on how likely a given variant can alter splicing patterns based upon its sequence alone.
SNP effect predictions
The predictions of variant effects on post-transcriptional regulation were performed on 10 million SNPs after some QC criteria, from 1000 genome phase 3 project.
Enrichment analysis
We first tested annotations one at a time using both TORUS and LDSC.Then we jointly assessed a set of annotations.
Notably, we ran Torus without being limited to 1M hapmap SNPs. However, all analyses with LDSC were limited to 1M hapmap SNPs.
Results
Characteristics for MTSplice predictions
Out of 10M SNPs (MAF>=0.005), around 2000 SNPs have predicted score bigger than 2 or smaller than -2, which can be considered strong.
Compare MMSplice and MTSplice predictions 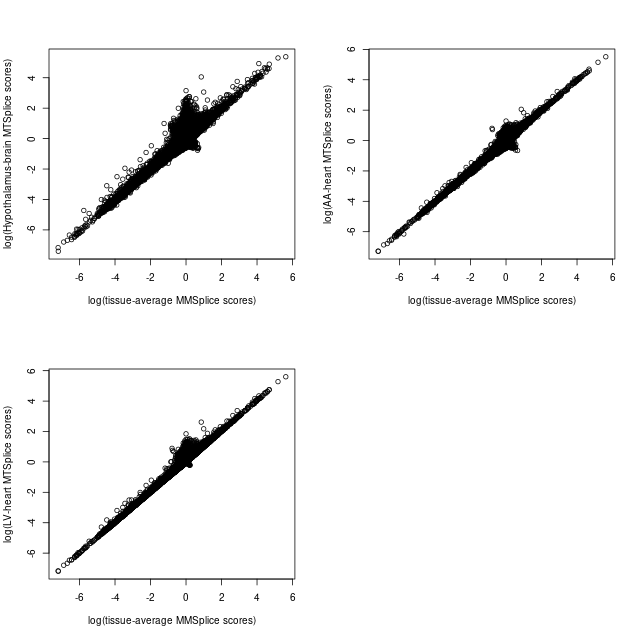
Table of prediction cutoffs
delta_logit_psi Hypothalamus...Brain Atrial.Appendage...Heart
85% 0.1231559 0.3461778 0.1902997
90% 0.1763513 0.4321360 0.2414605
95% 0.3011078 0.5868945 0.3530906
Left.Ventricle...Heart
85% 0.2160257
90% 0.2698549
95% 0.3789613Understand the distribution of predicted scores wrt. MAFs
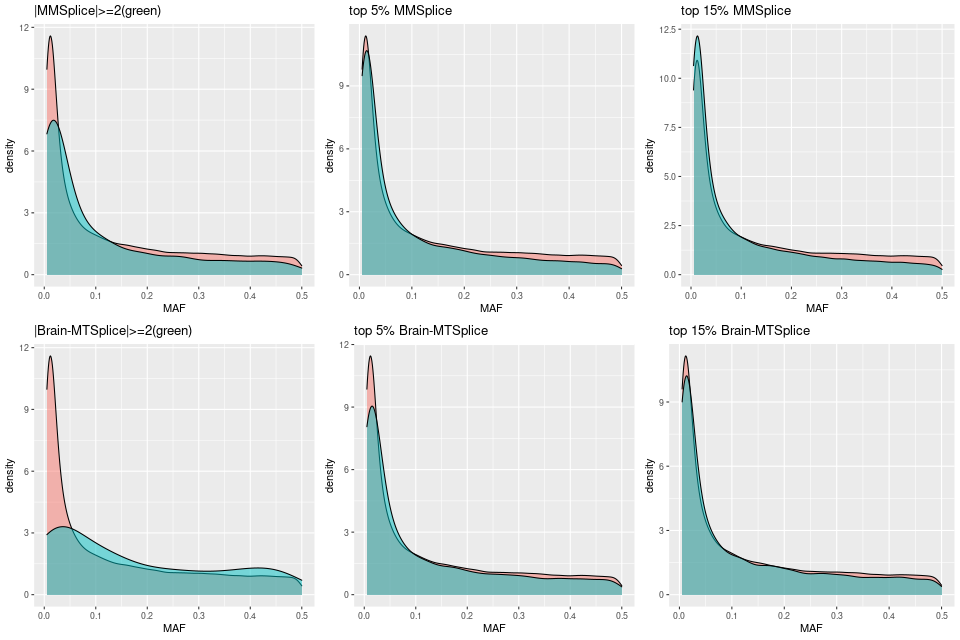
MAFs distribution
QQ Plots - comparing SNPs within annotations and the rest
Color scheme: * red - tissue specific * blue - mmsplice * black - background
Top-left: SCZ gwas
Top-right and Down-left: aFib gwas; AA - Atrial Appendage; LV -Left ventrical
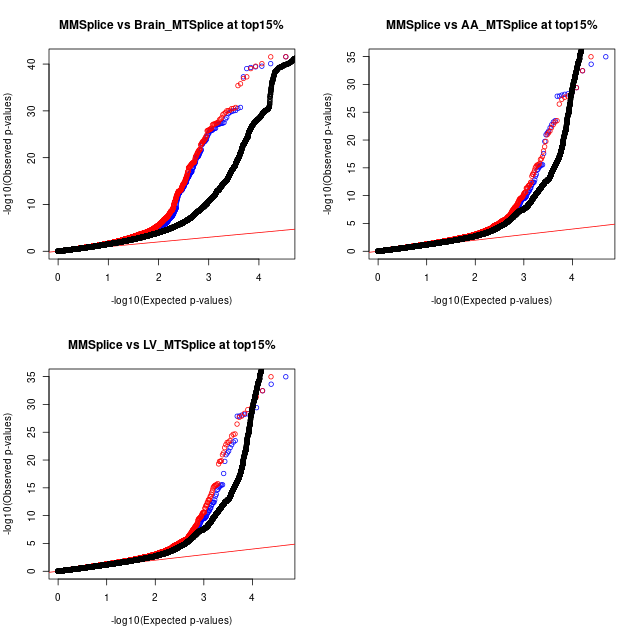
QQ plots for both SCZ and aFib traits
Enrichment analysis on SCZ GWAS
Legends for the plots:
- y-axis - annotations(% of SNPs within annotations)
- x-axis - fold of enrichment
- label on the plot - percent of SNP heritability or enrichment p-value
- dashed line - no enrichment
Evaluate features with LDSC
The defined set of baseline annotations for running LDSC are coding, promoter, 3’UTR, 5’UTR, each with a 500-bp extended region.
MMSplice vs. Brain-specific MTSplice
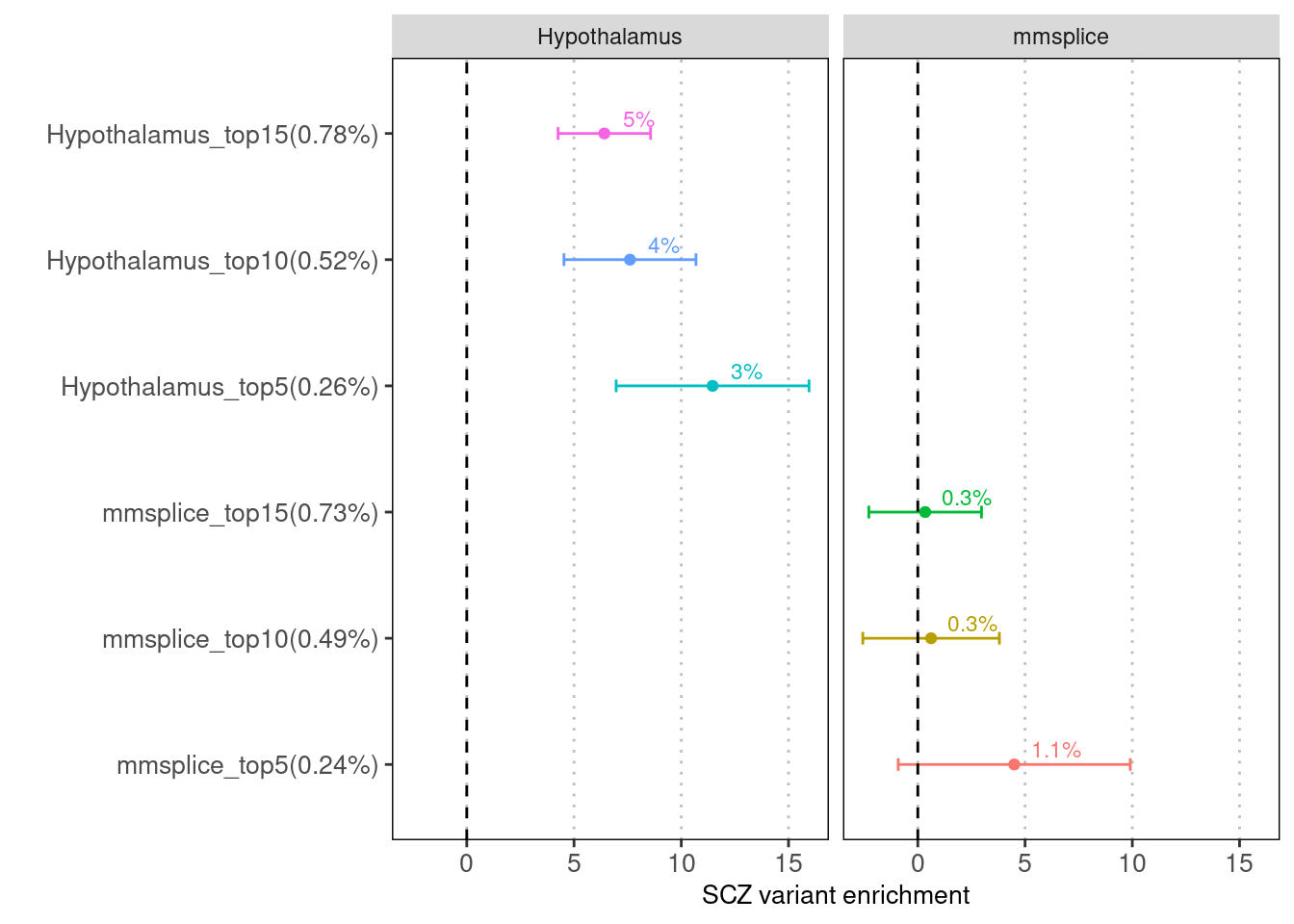
| Version | Author | Date |
|---|---|---|
| abda215 | Jing Gu | 2022-08-02 |
Fig 1.1 Enrichment of SCZ risk variants in tissue-average (mmsplice) versus Hypothalamus-specific predictions for altering splicing patterns (mtsplice) via LDSC.
The variants with predicted score within top 5%, 10% and 15% were compared for their enrichment of risk variants between the two methods.Examine baseline annotations
test annotation:top 10% Hypothalamus-specific MTSplice predictions
Warning: Removed 1 rows containing missing values (geom_text).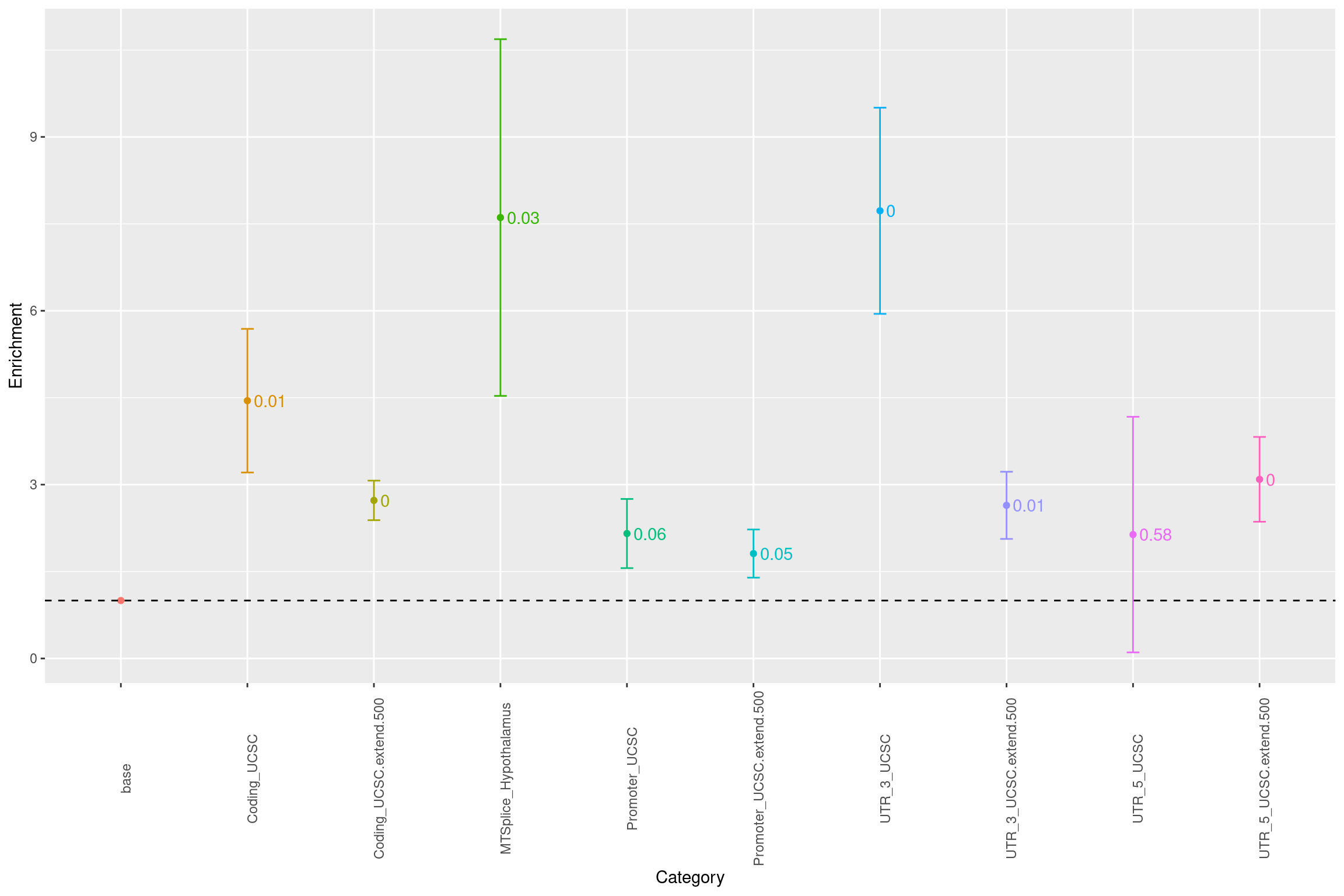
| Version | Author | Date |
|---|---|---|
| abda215 | Jing Gu | 2022-08-02 |
Fig 2.1 Baseline enrichment of SCZ risk variants from LDSC.
Conditional on coding, introns, promoters and UTR annotations, MTSplice predictions specific to Hypothalams shows significant enrichment with SCZ risk variants. The enrichment estimate of MTSplice prediction was not affected much by whether or not including introns.Evaluate features with Torus
Individual run at different cutoffs 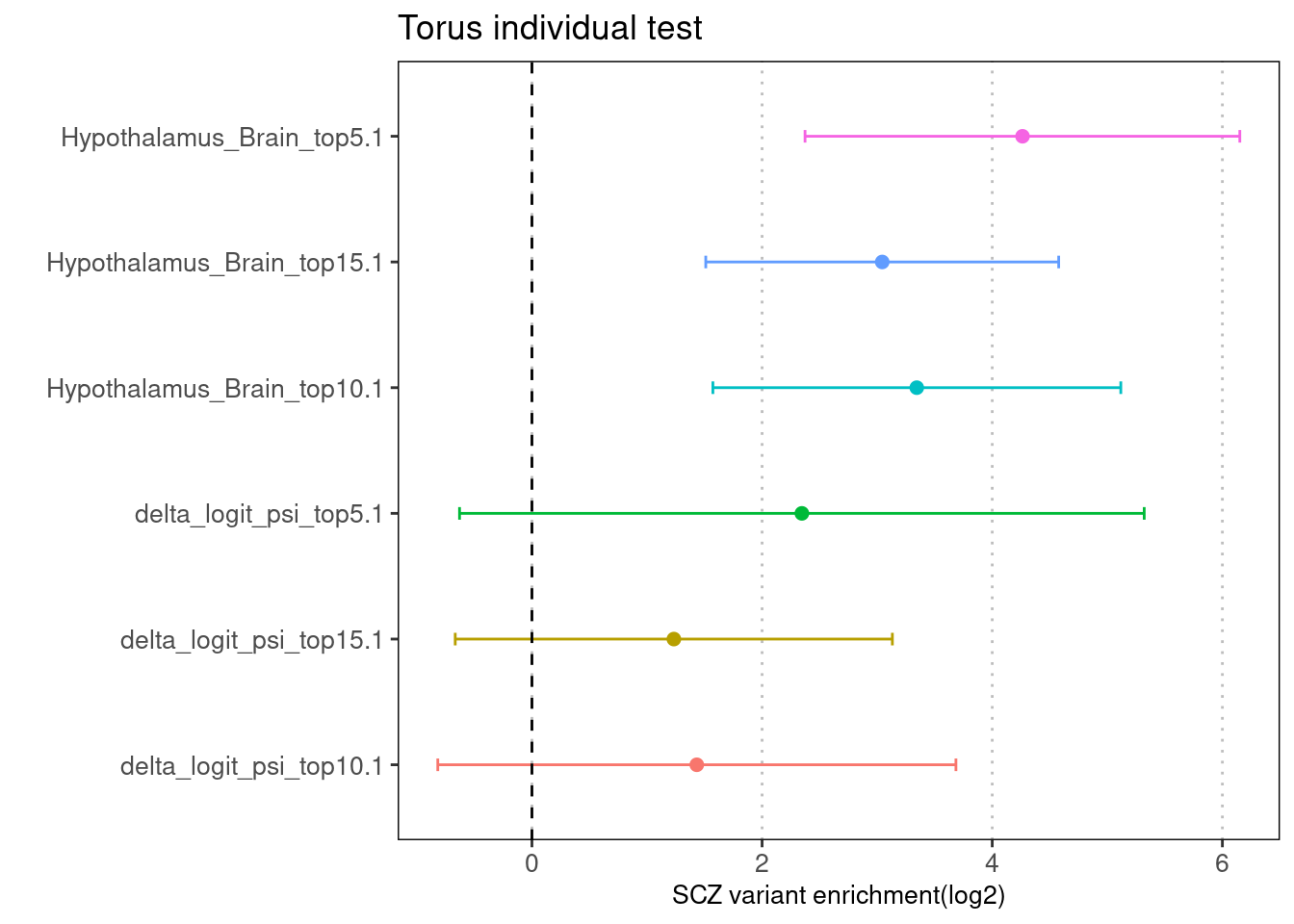
Fig 3.1 Enrichment of SCZ risk variants in individual annotation from Torus.
No baselines was provided for this run of analysis. For each of the two features, we ran torus at different cutoffs, from top 5% to top 15%.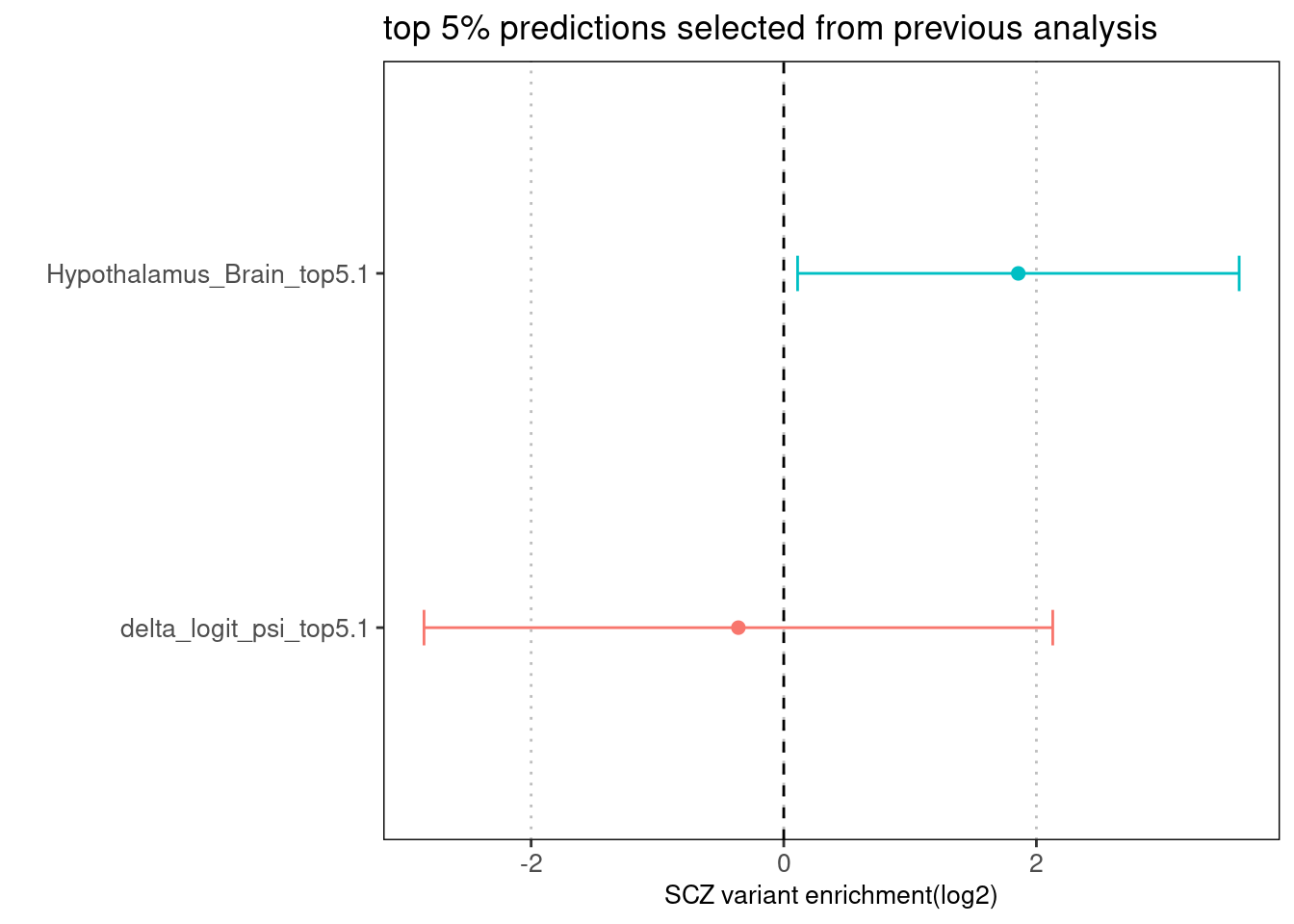
Fig 3.2 Enrichment of SCZ risk variants jointly run with baseline annotation via Torus.
The top 5% predictions were selected based on the best performance from the individual run. The enrichment for brain tissue-specific prediction remains conditional on baseline annotations.Joint run with other annotations
Sequentially adding annotation one at a time with the MTSplice prediction and the baseline set.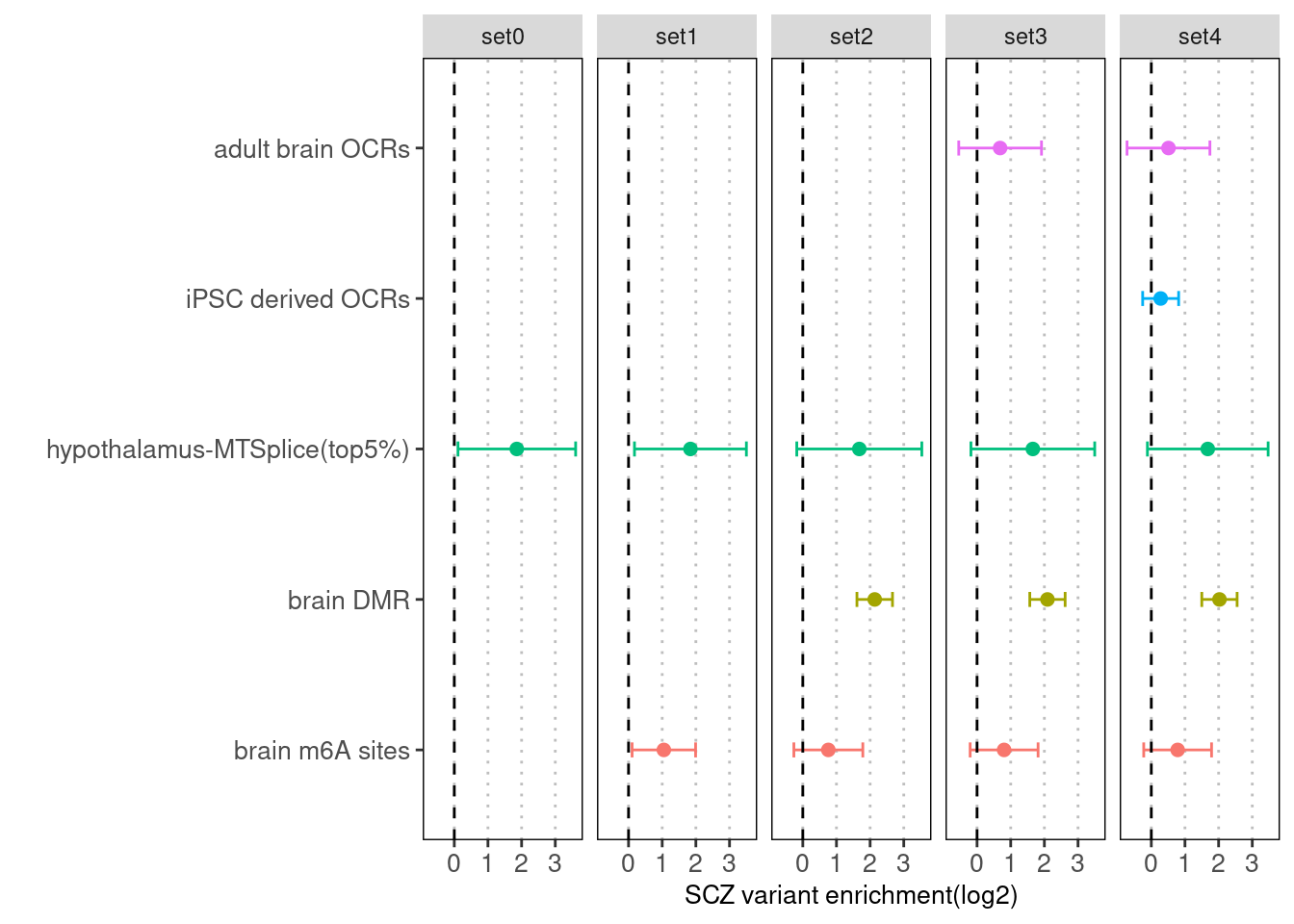
Fig 3.3 Enrichment of SCZ risk variants jointly run with other annotations and baselines via Torus.
After jointly run with adult brain OCRs, iPSC derived OCRs, differentially methylated sites across brain regions,and brain m6A sites, we observed slightly decrease in the enrichment of mtsplice predictions. The biggest decrease occurs when brain differentially methylated regions were added.Enrichment analysis on AFib GWAS
Evaluate features with LDSC
MMSplice vs. Heart-specific MTSplice
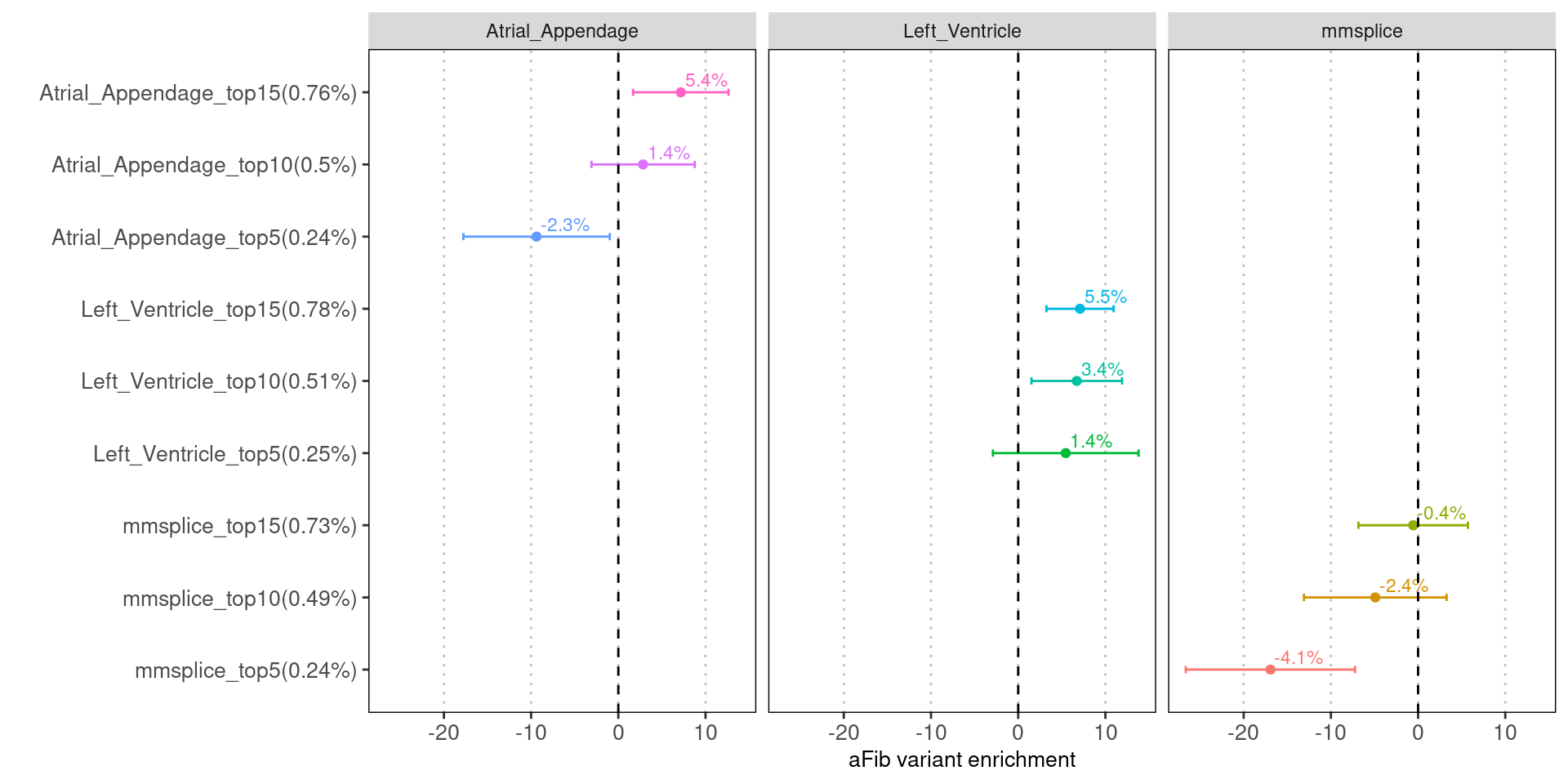
Fig 4.1 Enrichment of aFib risk variants in individual annotation from LDSC.
Tissue-specific predictions show much higher fold enrichment with risk variants than the tissue average ones.Evaluate features with Torus
No baselines
The enrichment results from LDSC were replicated in the torus run, probably due to different number of SNPs being tested. 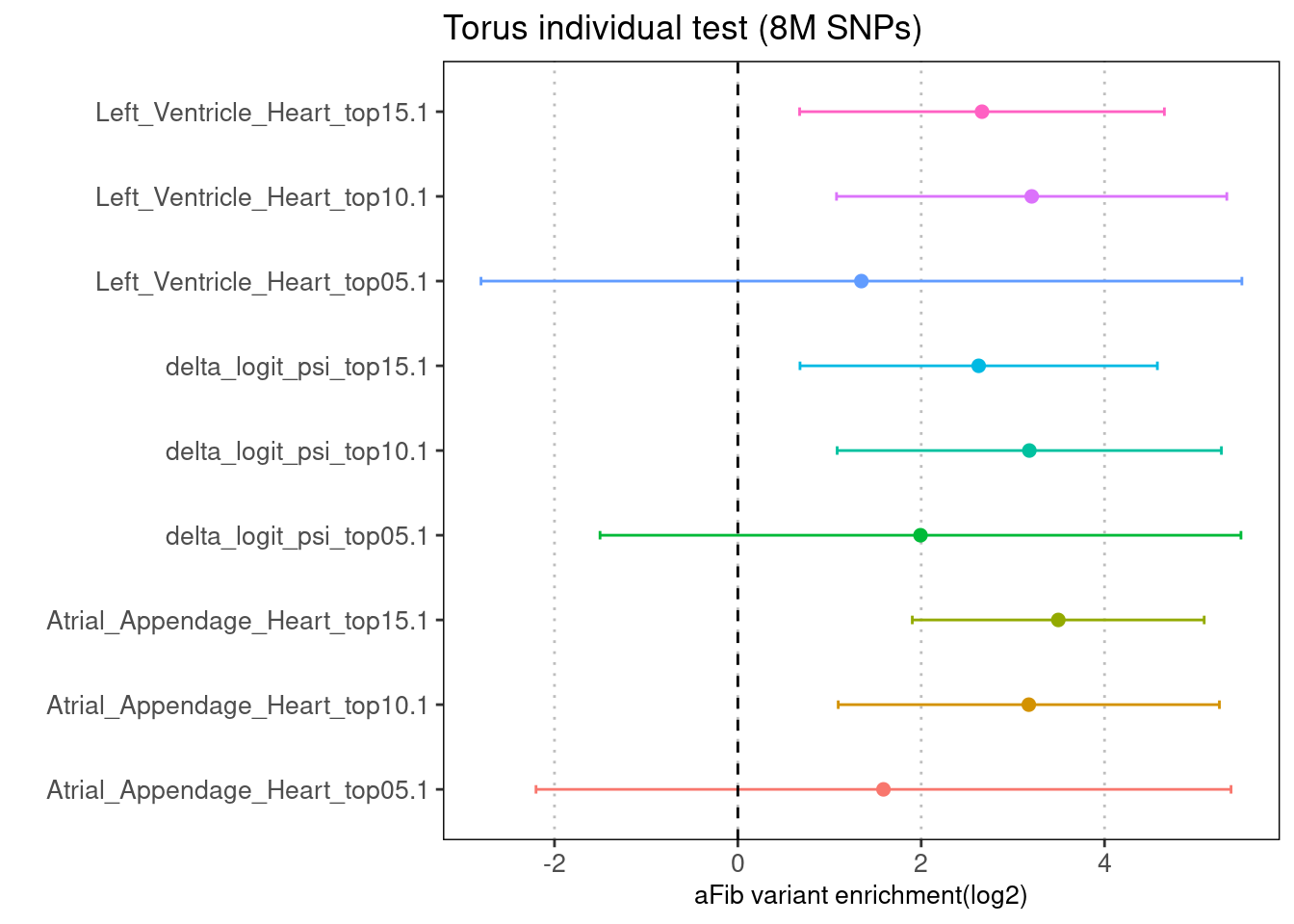
| Version | Author | Date |
|---|---|---|
| abda215 | Jing Gu | 2022-08-02 |
Fig 4.2 Torus individual test on 8M aFib GWAS SNPs
The Torus run across differnt types predictions shows similar results.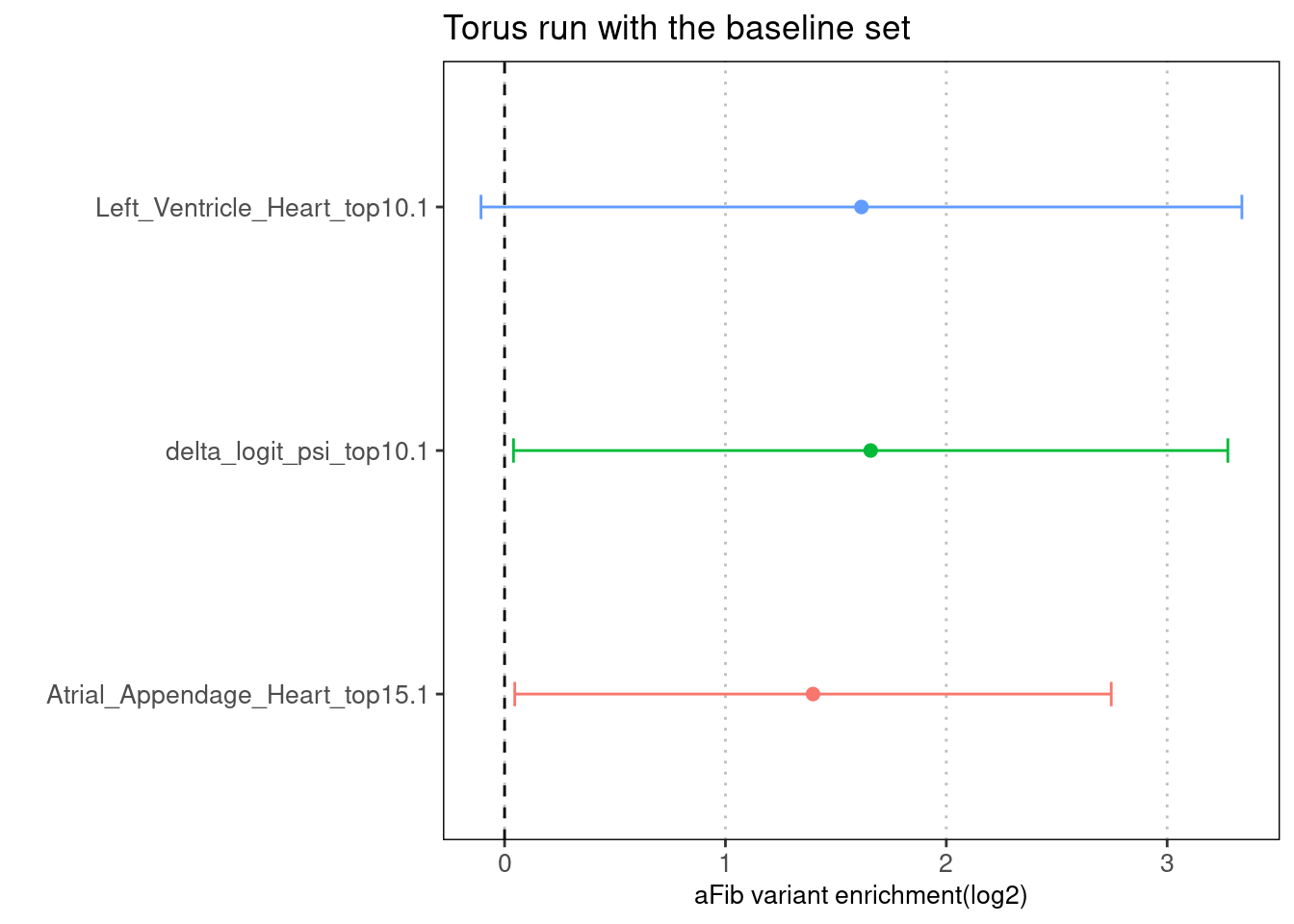
Fig 4.3 Torus individual run with the baseline set
For all three types of predictions, we see a decrease in enrichment estimates.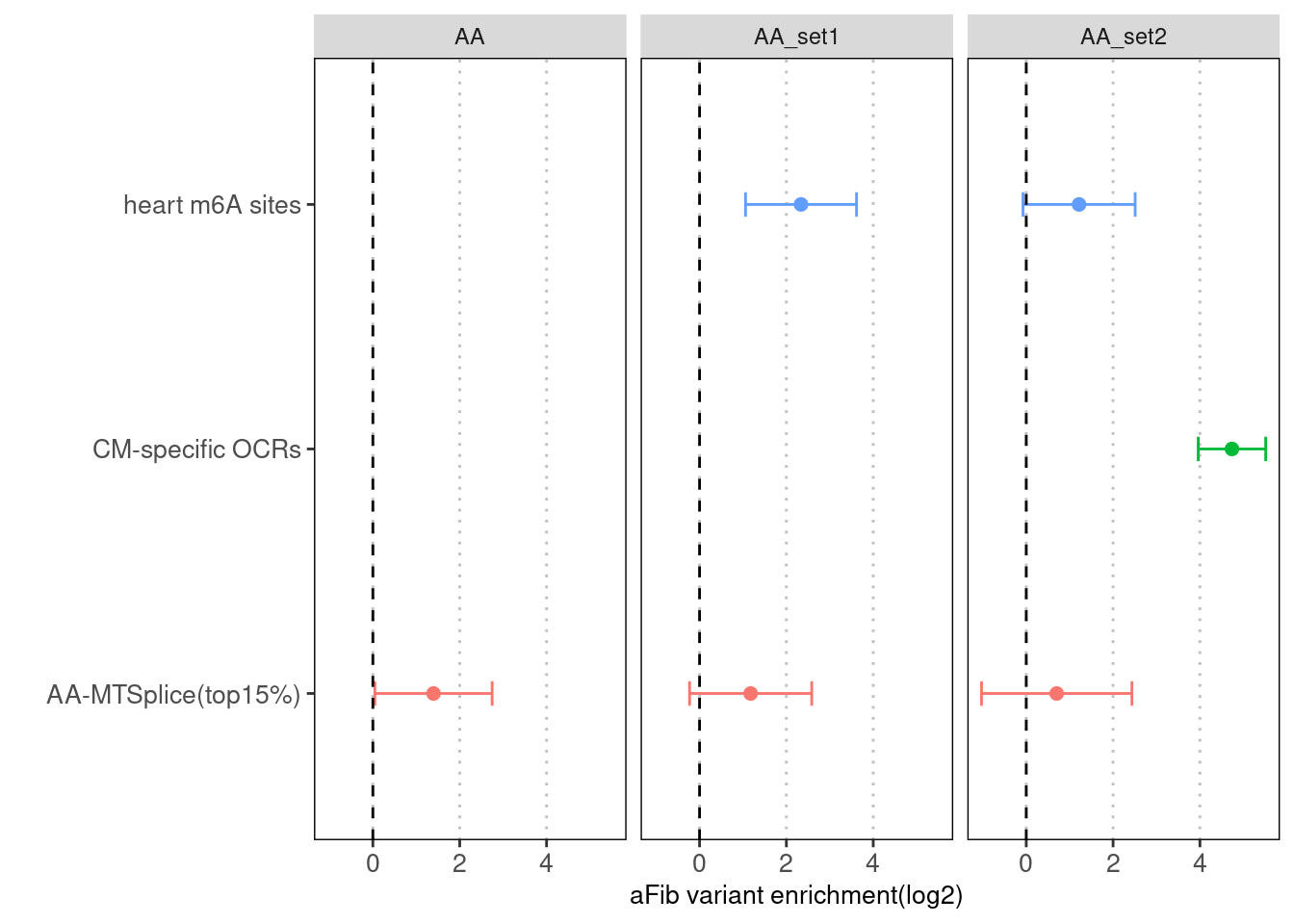
Fig 4.4 Torus joint run with other types of annotations for AA-specific MTSplice predictions
With CM-specific OCRs and m6A sites, AA-MTSplice predictions no longer shows significant enrichment.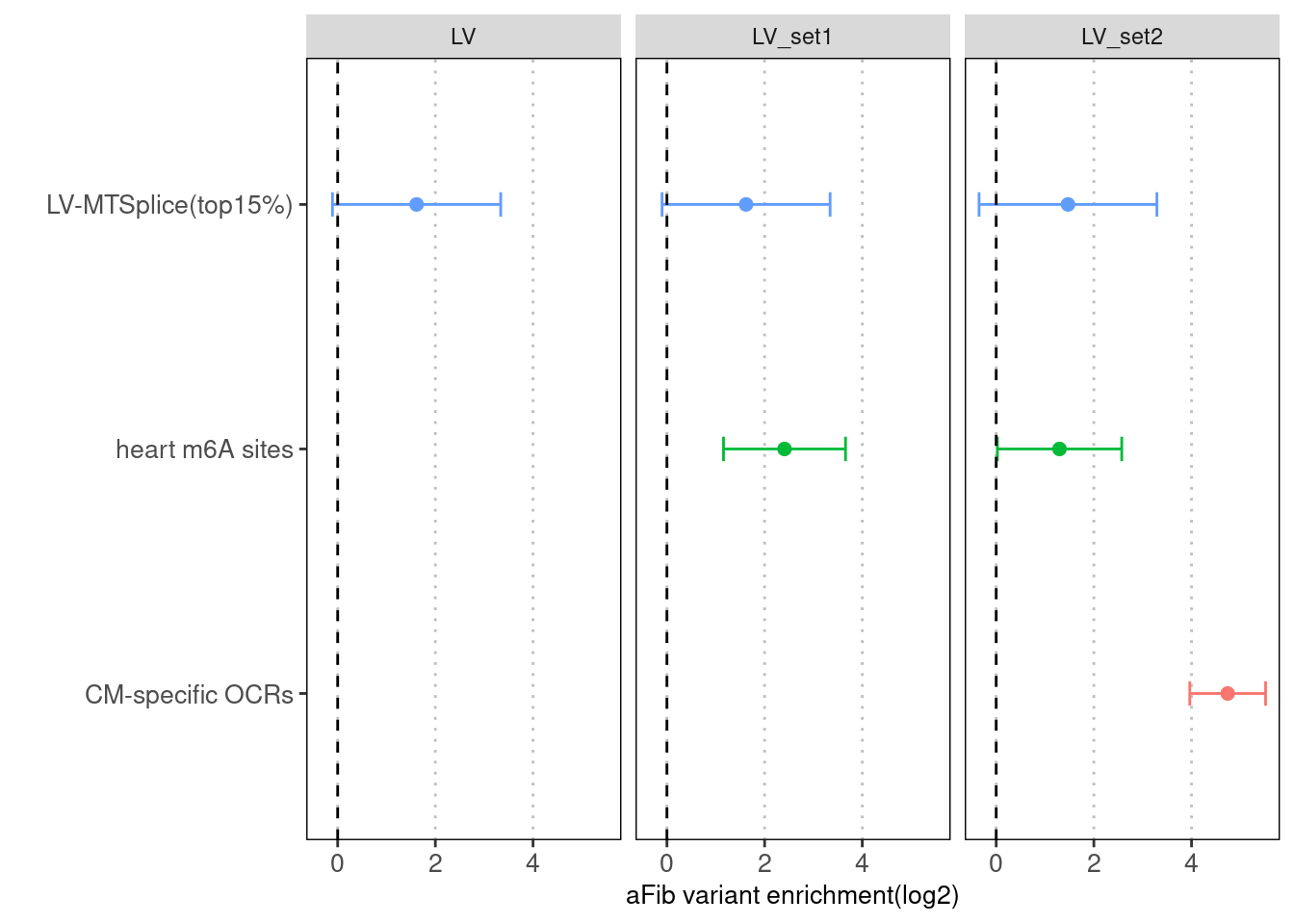
Fig 4.5 Torus joint run with other types of annotations for LV-specific MTSplice predictions
Similar trend as AA-specific MTSplice predictionsLDSC results across multiple traits
tissue-average prediction from MMSplice
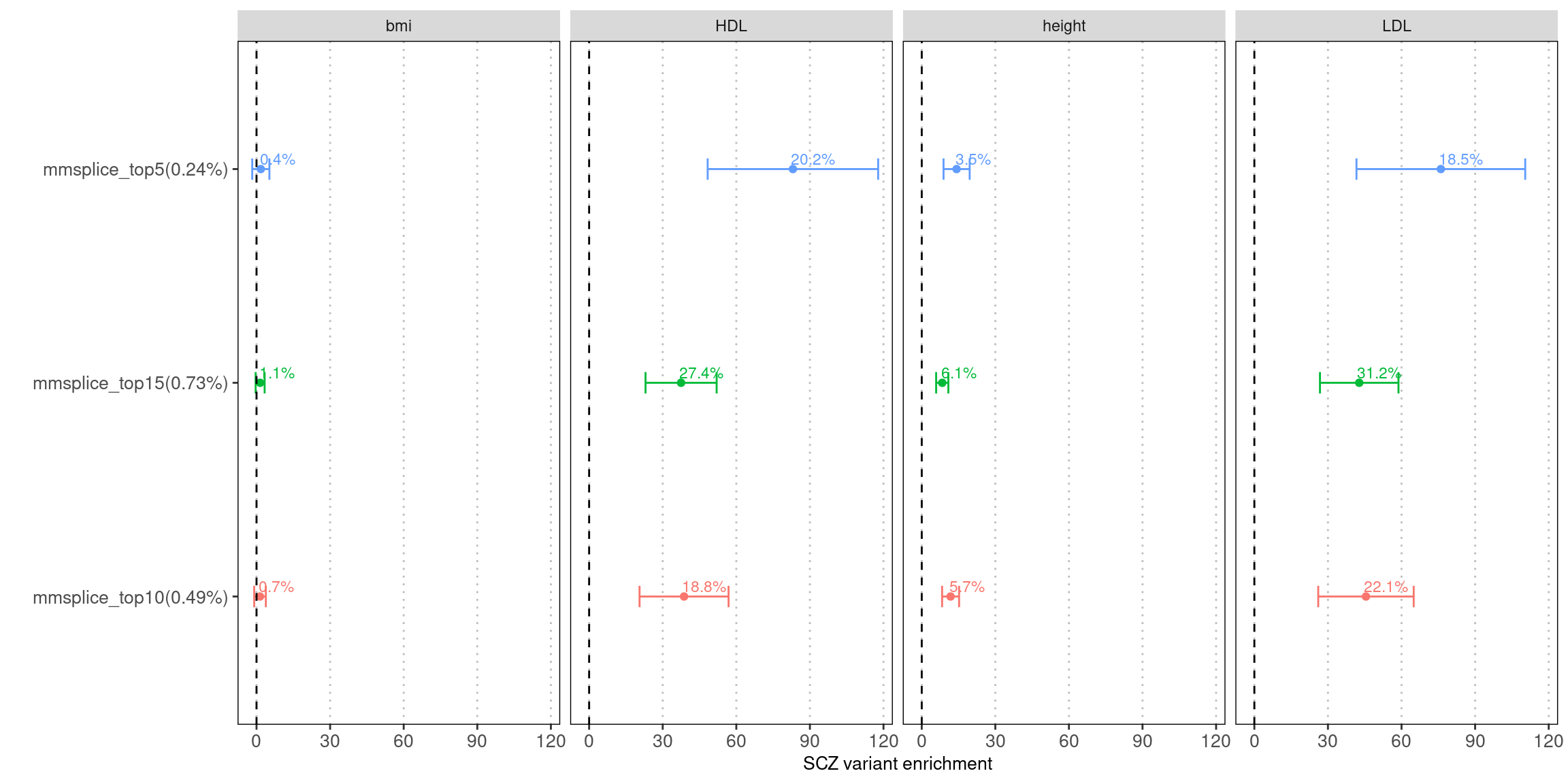
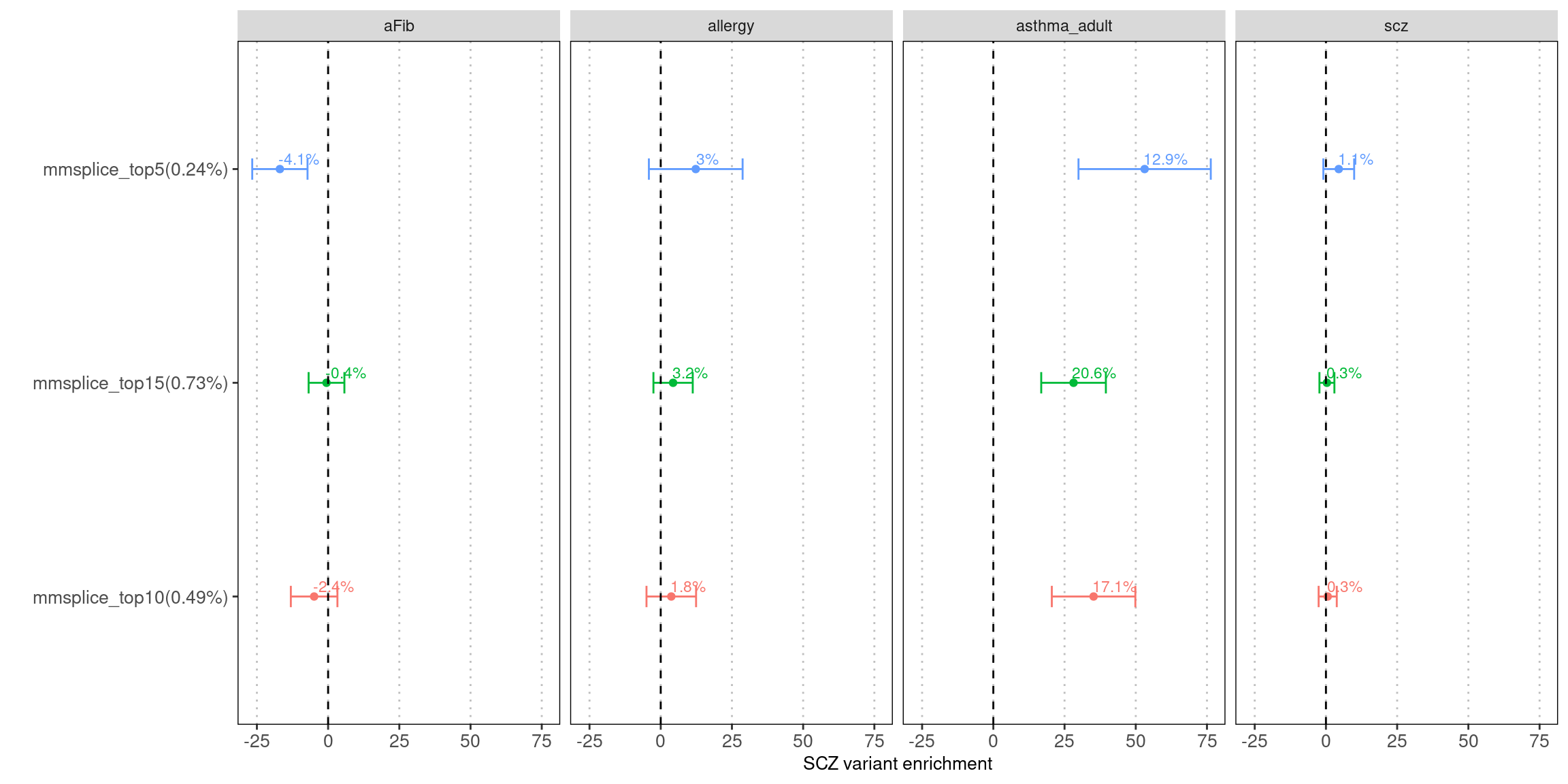
Fig 5.1 LDSC Enrichment results across traits
Enrichment estimates each with a 95% confidence interval for MMSplice/MTSplice predictions on splicing effects across various traits.
sessionInfo()R version 4.0.4 (2021-02-15)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.3.13-el7-x86_64/lib/libopenblas_haswellp-r0.3.13.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] gridExtra_2.3 data.table_1.14.2 ggplot2_3.3.3
loaded via a namespace (and not attached):
[1] Rcpp_1.0.8 pillar_1.5.0 compiler_4.0.4 bslib_0.2.4
[5] later_1.1.0.1 jquerylib_0.1.3 git2r_0.28.0 highr_0.8
[9] workflowr_1.6.2 tools_4.0.4 digest_0.6.27 gtable_0.3.0
[13] jsonlite_1.7.2 evaluate_0.14 lifecycle_1.0.0 tibble_3.0.6
[17] pkgconfig_2.0.3 rlang_1.0.1 DBI_1.1.1 cli_3.2.0
[21] rstudioapi_0.13 yaml_2.2.1 xfun_0.21 withr_2.4.3
[25] dplyr_1.0.4 stringr_1.4.0 knitr_1.31 generics_0.1.0
[29] fs_1.5.0 vctrs_0.3.8 sass_0.3.1 tidyselect_1.1.1
[33] rprojroot_2.0.2 grid_4.0.4 glue_1.6.1 R6_2.5.1
[37] fansi_1.0.2 rmarkdown_2.7 farver_2.1.0 purrr_0.3.4
[41] magrittr_2.0.1 whisker_0.4 scales_1.1.1 promises_1.2.0.1
[45] ellipsis_0.3.2 htmltools_0.5.1.1 assertthat_0.2.1 colorspace_2.0-2
[49] httpuv_1.5.5 labeling_0.4.2 utf8_1.2.2 stringi_1.5.3
[53] munsell_0.5.0 crayon_1.4.1