cross-tissue DE analysis with fastTopics
Jing Gu
2024-09-11
Last updated: 2024-09-11
Checks: 7 0
Knit directory: lung_lymph_scMultiomics/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20221229) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 163b422. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: analysis/.RData
Ignored: analysis/.Rhistory
Untracked files:
Untracked: analysis/.ipynb_checkpoints/
Untracked: analysis/test.pdf
Untracked: analysis/test_GO_enrichment.ipynb
Untracked: analysis/test_magma.Rmd
Untracked: analysis/u19_atac_fastTopics.Rmd
Untracked: analysis/u19_regulon_enrichment.Rmd
Untracked: code/run_magma/
Untracked: data/DA_peaks_Tsub_vs_others.RDS
Untracked: data/DA_peaks_by_cell_type.RDS
Untracked: data/TF_target_sizes_GRN.txt
Untracked: data/U19_T_cell_peaks_metadata.RDS
Untracked: data/Wang_2020_T_cell_peaks_metadata.RDS
Untracked: data/lung_GRN_CD4_T_edges.txt
Untracked: data/lung_GRN_CD8_T_edges.txt
Untracked: data/lung_GRN_Th17_edges.txt
Untracked: data/lung_GRN_Treg_edges.txt
Untracked: output/annotation_reference.txt
Untracked: output/fastTopics
Untracked: output/homer/
Untracked: output/ldsc_enrichment
Untracked: output/lung_immune_atac_peaks_high_ePIPs.RDS
Untracked: output/positions.bed
Untracked: output/topic1/
Untracked: output/topic10/
Untracked: output/topic11/
Untracked: output/topic12/
Untracked: output/topic2/
Untracked: output/topic3/
Untracked: output/topic4/
Untracked: output/topic5/
Untracked: output/topic6/
Untracked: output/topic7/
Untracked: output/topic8/
Untracked: output/topic9/
Untracked: test.pdf
Unstaged changes:
Modified: analysis/identify_regulatory_programs_u19_GRN.Rmd
Modified: analysis/rank_TFs_from_pairwise_comparison.ipynb
Modified: analysis/u19_h2g_enrichment.Rmd
Deleted: code/run_fastTopic.R
Deleted: lung_immune_fine_mapping.Rproj
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown
(analysis/cross_tissue_DE_u19_fastTopics.Rmd) and HTML
(docs/cross_tissue_DE_u19_fastTopics.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 163b422 | Jing Gu | 2024-09-11 | plotted heatmaps for enrichment results |
| html | 87c183b | Jing Gu | 2024-05-25 | Build site. |
| Rmd | b7a8317 | Jing Gu | 2024-05-25 | updated result page |
| html | 4e0028d | Jing Gu | 2024-05-23 | Build site. |
| Rmd | 36aceab | Jing Gu | 2024-05-23 | organized results |
| html | 744d4f0 | Jing Gu | 2024-05-22 | Build site. |
| Rmd | 5c6b22f | Jing Gu | 2024-05-22 | updated GSEA results |
| html | 3d73252 | Jing Gu | 2024-05-13 | Build site. |
| Rmd | f26e8ad | Jing Gu | 2024-05-13 | wflow_publish("analysis/cross_tissue_DE_u19_fastTopics.Rmd") |
| html | 95bfdff | Jing Gu | 2024-05-13 | Build site. |
| Rmd | 3e4f015 | Jing Gu | 2024-05-13 | wflow_publish("analysis/cross_tissue_DE_u19_fastTopics.Rmd") |
| html | 2e475c9 | Jing Gu | 2024-05-13 | Build site. |
| Rmd | 306ddb1 | Jing Gu | 2024-05-13 | wflow_publish("analysis/cross_tissue_DE_u19_fastTopics.Rmd") |
| html | f812a40 | Jing Gu | 2024-05-09 | Build site. |
| html | 68d9e18 | Jing Gu | 2024-05-09 | Build site. |
| Rmd | 7ca45f6 | Jing Gu | 2024-05-09 | cross-tissue comparison |
| Rmd | 7a45261 | Jing Gu | 2024-05-08 | cross-tissue comparison with topic modeling |
GoM DE analysis on u19 dataset
Model fitting
Parameters:
N_updates = 150 N_topics = 12 ## Model evaluation
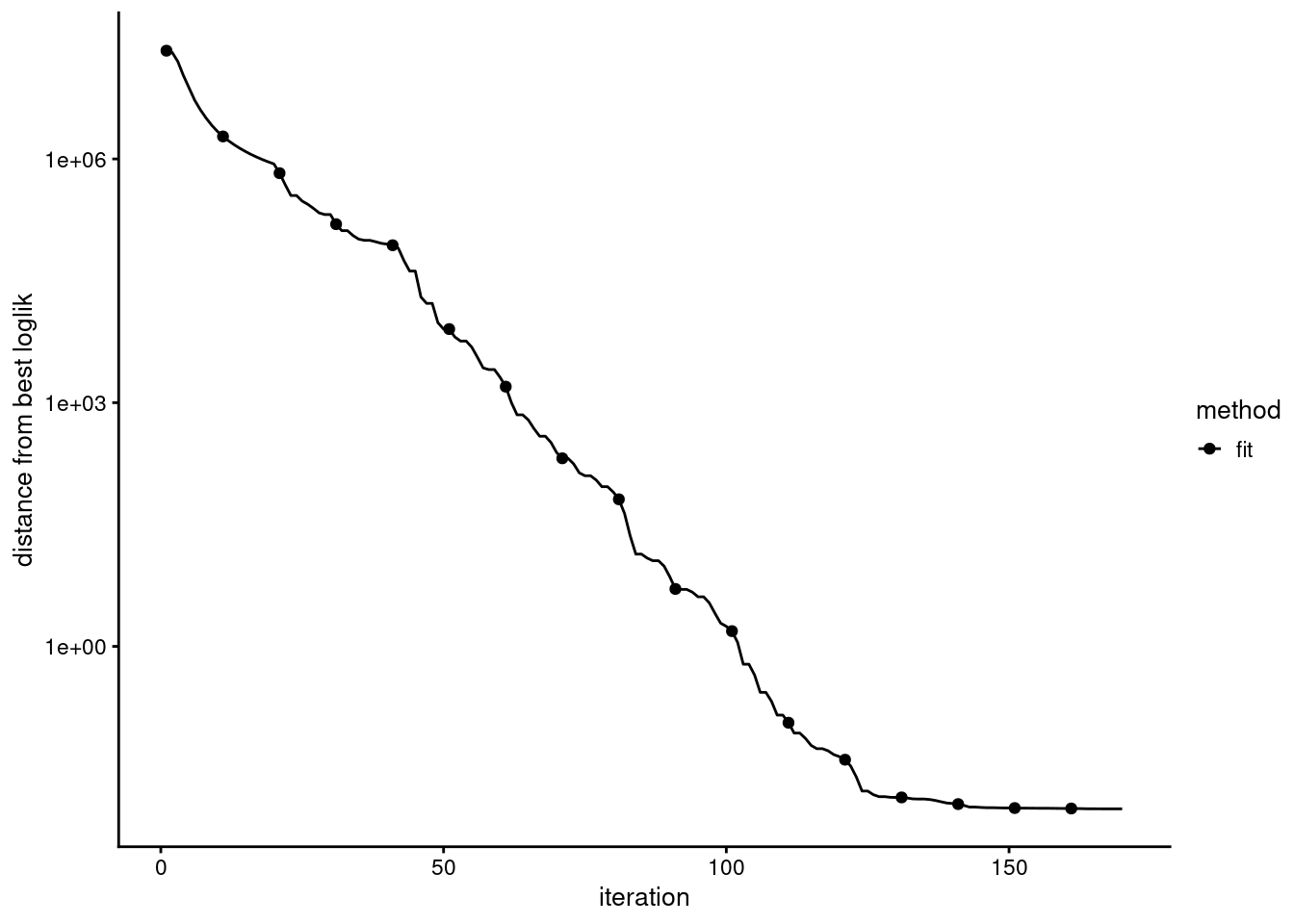
check the convergence
| Version | Author | Date |
|---|---|---|
| 68d9e18 | Jing Gu | 2024-05-09 |
Model overview:
Number of data rows, n: 53647
Number of data cols, m: 17420
Rank/number of topics, k: 12
Evaluation of model fit (170 updates performed):
Poisson NMF log-likelihood: -1.997995557900e+08
Multinomial topic model log-likelihood: -1.995509032083e+08
Poisson NMF deviance: +2.634951598369e+08
Max KKT residual: +1.430262e-02Set show.size.factors = TRUE, show.mixprops = TRUE and/or show.topic.reps = TRUE in print(...) for more information
| Version | Author | Date |
|---|---|---|
| 68d9e18 | Jing Gu | 2024-05-09 |
Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
ℹ Please use `linewidth` instead.
This warning is displayed once every 8 hours.
Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
generated.
| Version | Author | Date |
|---|---|---|
| 68d9e18 | Jing Gu | 2024-05-09 |
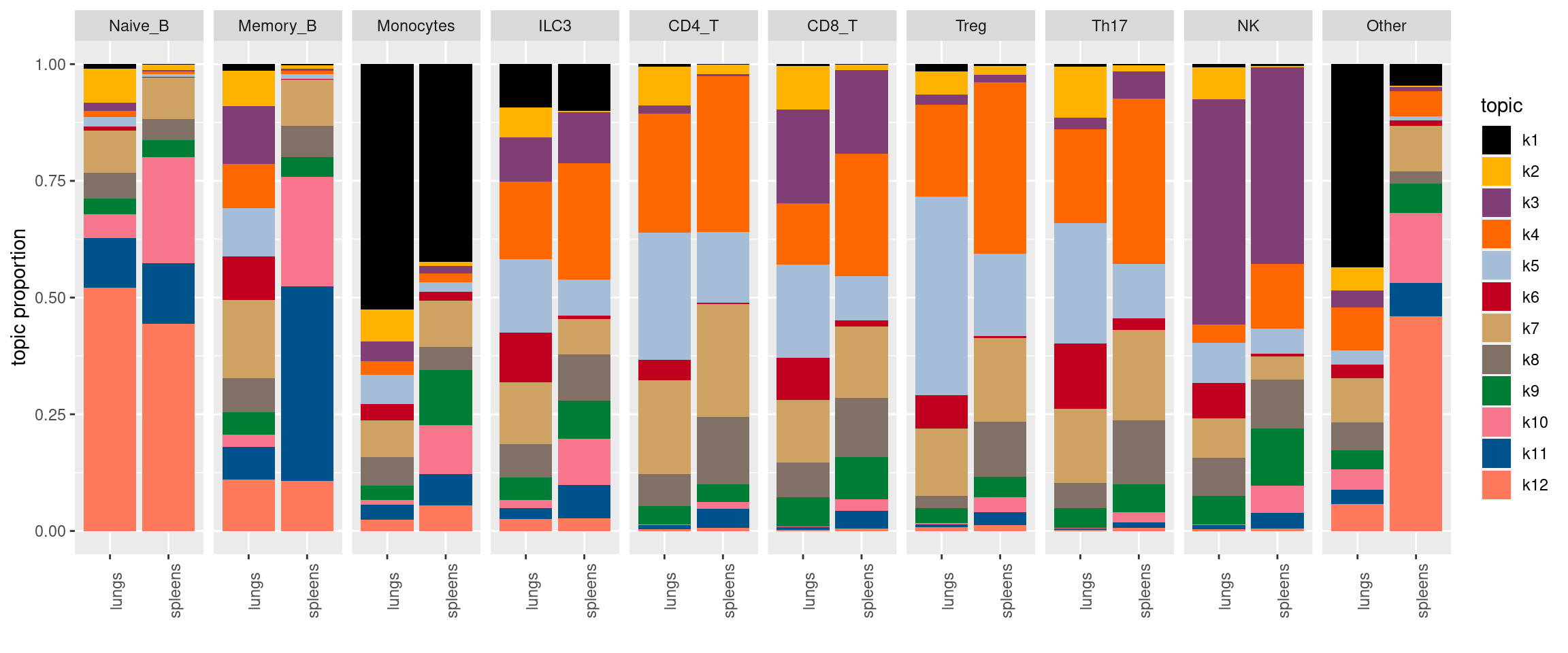
Visualize topics with structural plots
plot by tissue
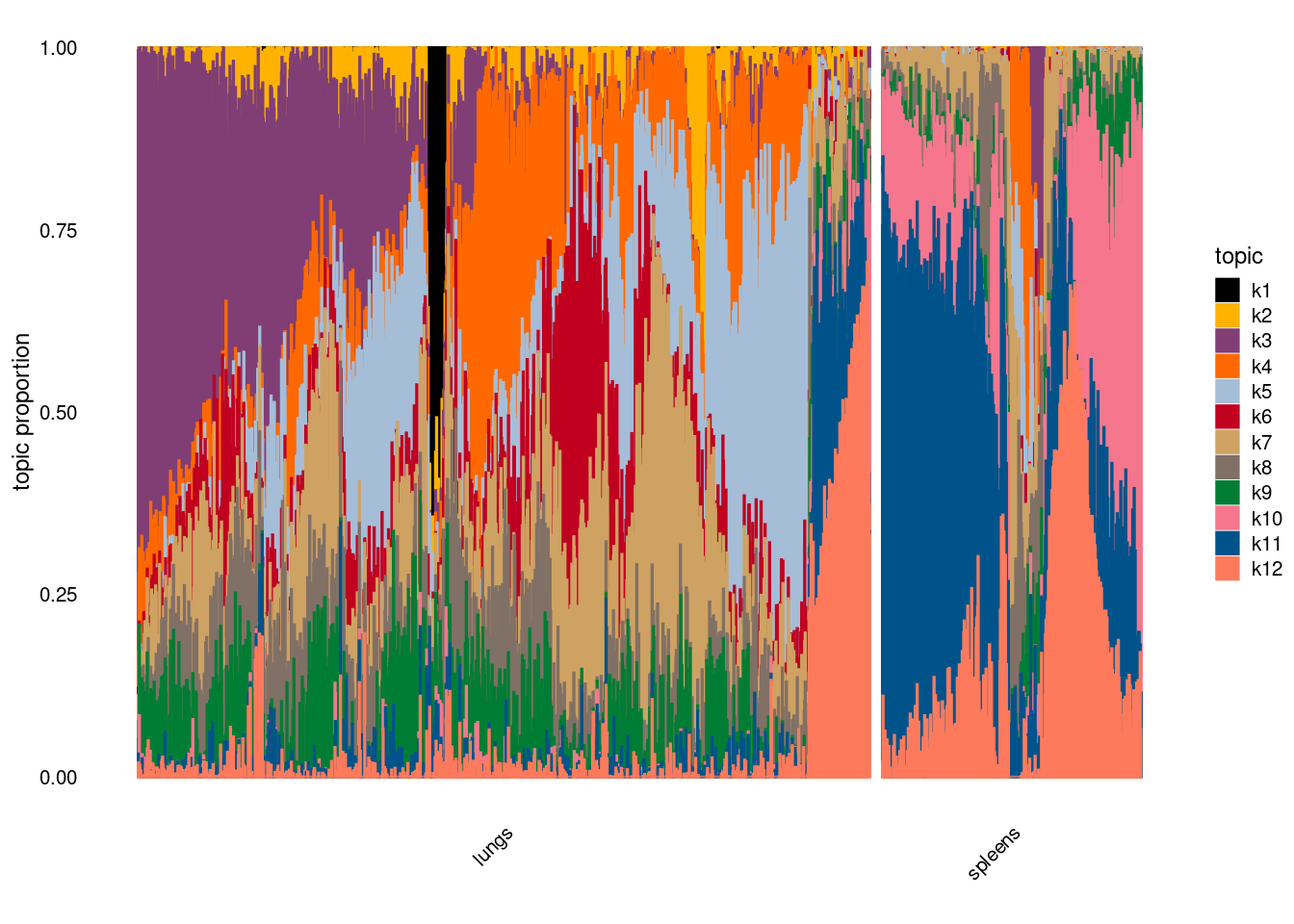
Version Author Date 68d9e18 Jing Gu 2024-05-09 plot by tissue and cell-type
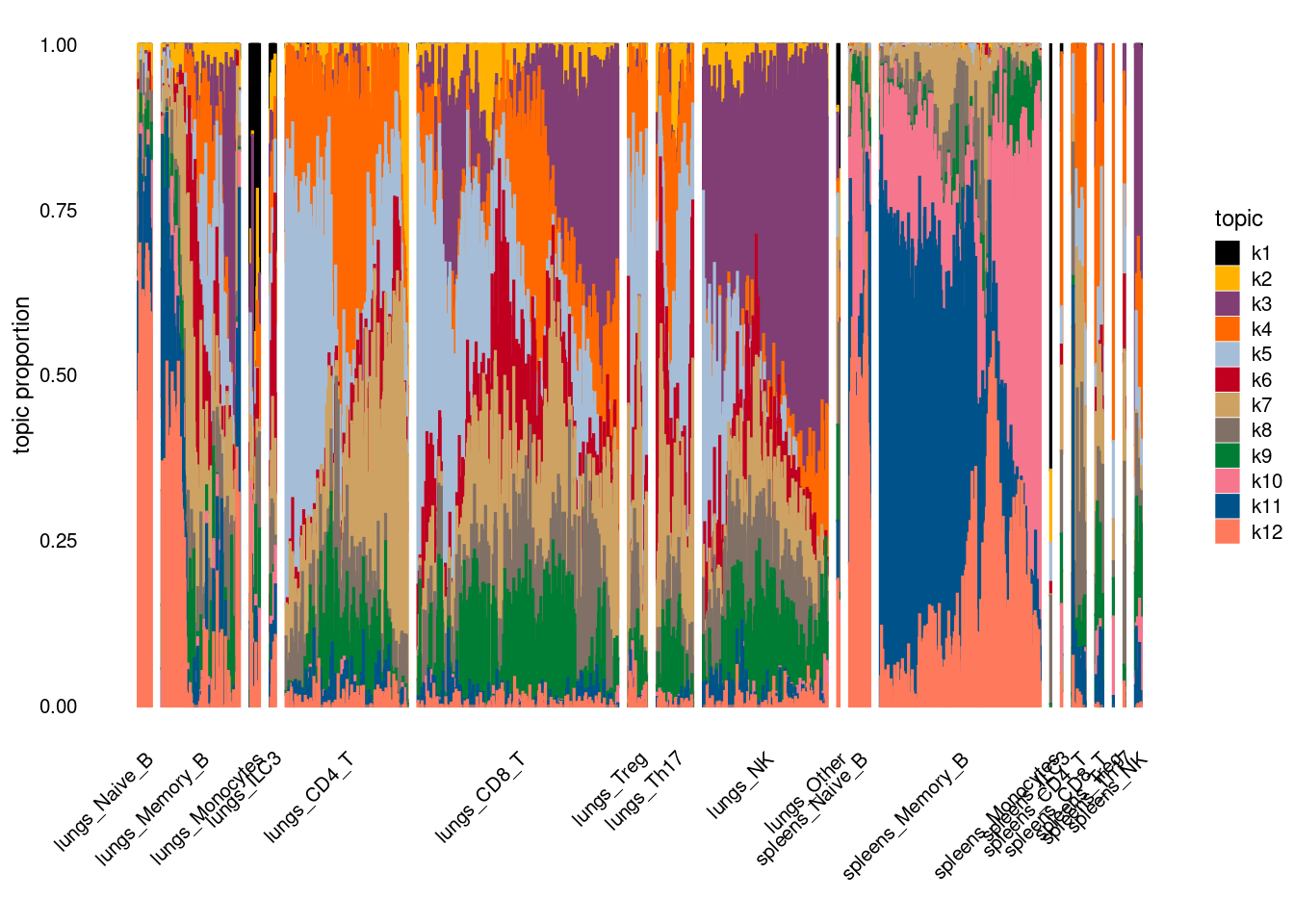
| Version | Author | Date |
|---|---|---|
| 68d9e18 | Jing Gu | 2024-05-09 |
Characterize topics with enrichment analysis
Two ways to perform enrichment test:
- hypergeometric test (ORA from WebGestaltR)
A test purely depends on occurrences, asking whether the genes related to a topic occur more frequently in GO term compared to the background genes.
- two-sample T test or regression (MAGMA)
It tests whether the genes in a set is more associated with phenotype than those outside of a set.
GO enrichment results (ORA method)
Top genes ranked by loadings
Input:
- Top 500 loading genes vs. all genes
- Database: Biological_Process, Cellular_Component, Molecular_Function
Top loading genes are enriched in large number of GO terms, which have broad functions.
Topic-specific genes through DE analysis
Input:
- Up-regulated genes specific to each topic (z > 0 and lfsr < 0.01) vs. all genes
- Database: Biological_Process, Cellular_Component, Molecular_Function
Volcano plots for GoM DE results
Axis: the z-scores for posterior mean log-fold change estimates vs. log-fold change
From volcano plots, we see several genes that encode different types of cytokines are present in topic 6. Chemokine ligands (CCL4, CCL20, etc.) are proteins that signal leukocyte migration, while cytokines (IL17A, IL22) are interleukins that signal immune cells to defend against pathogens.
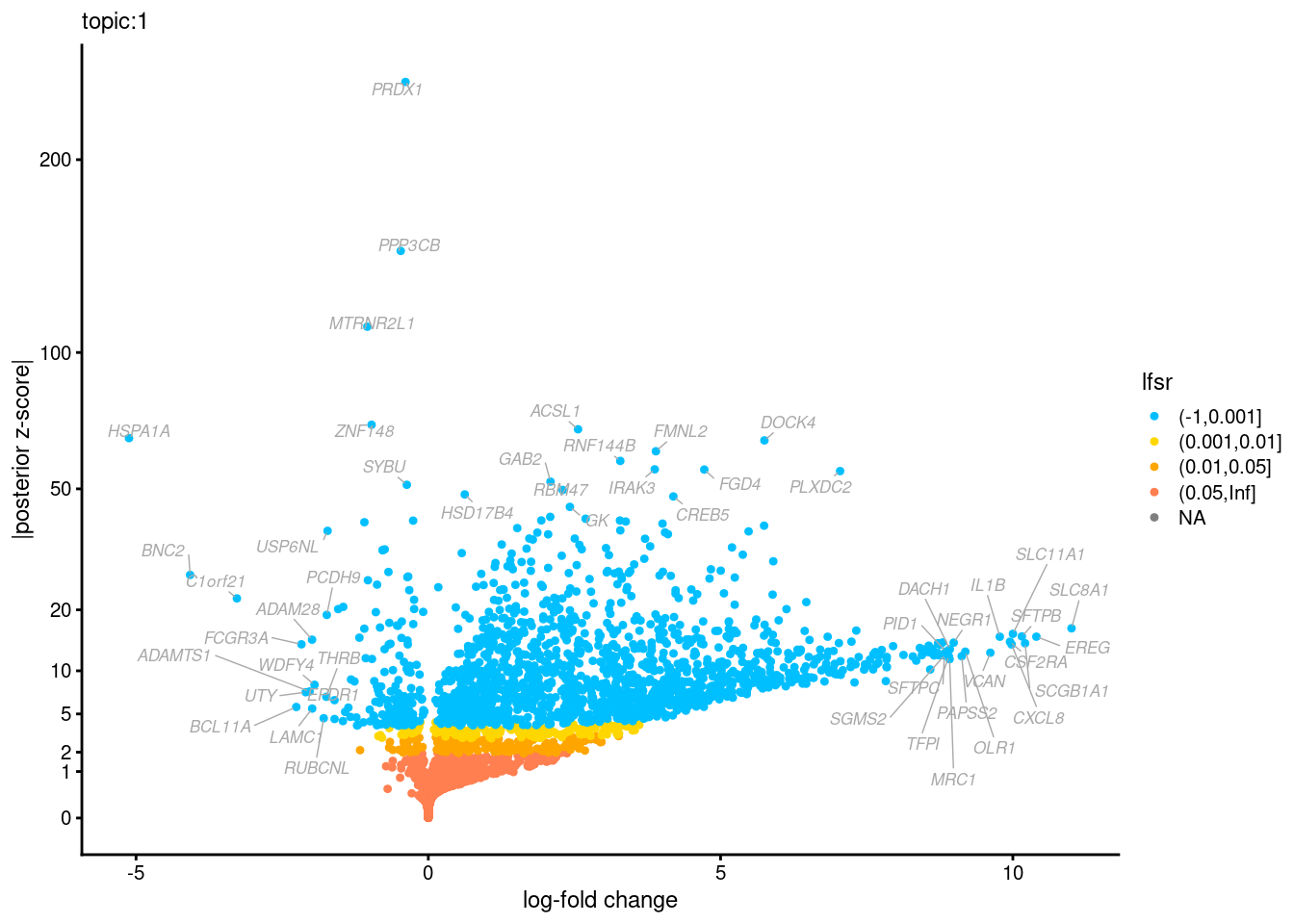
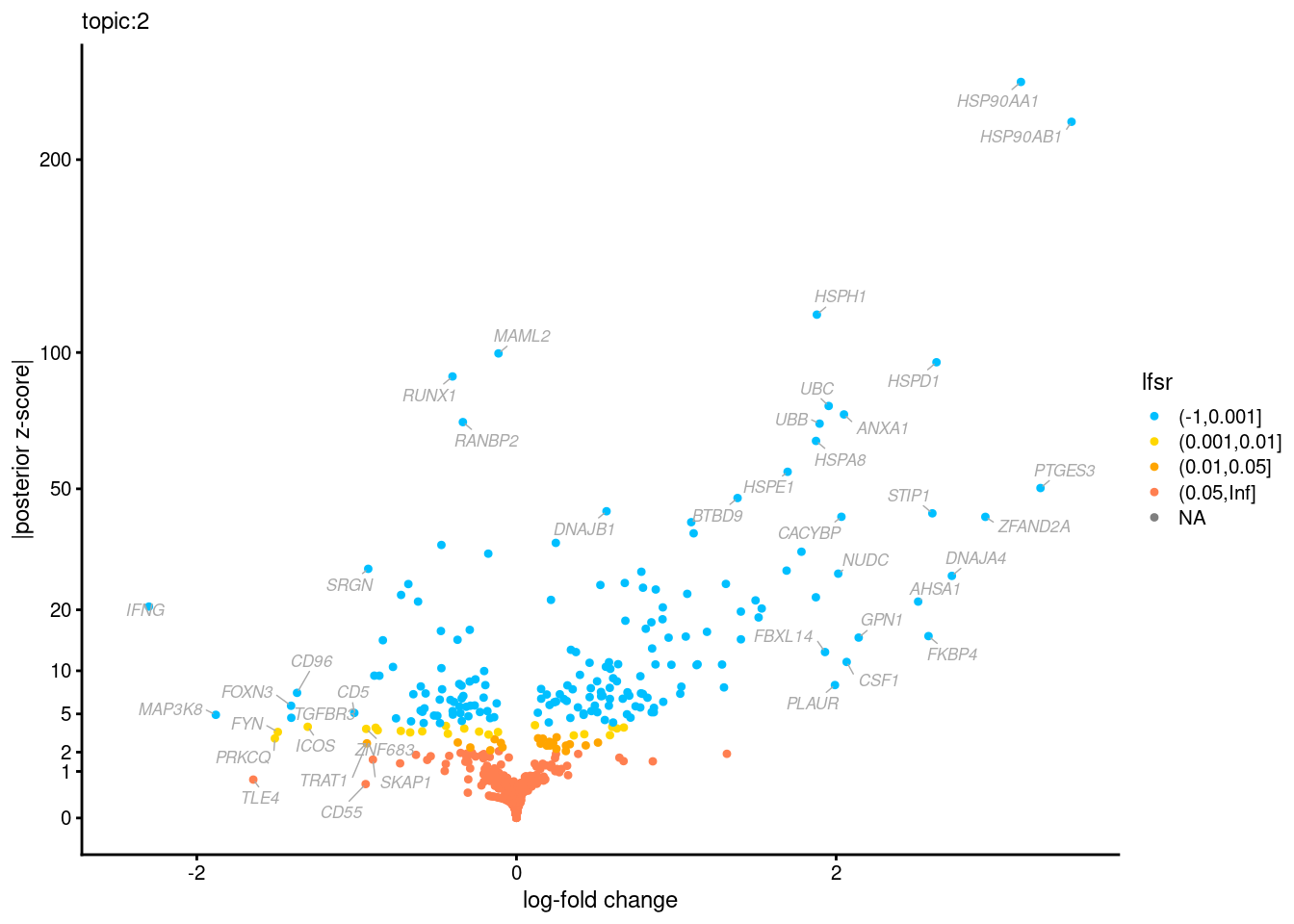
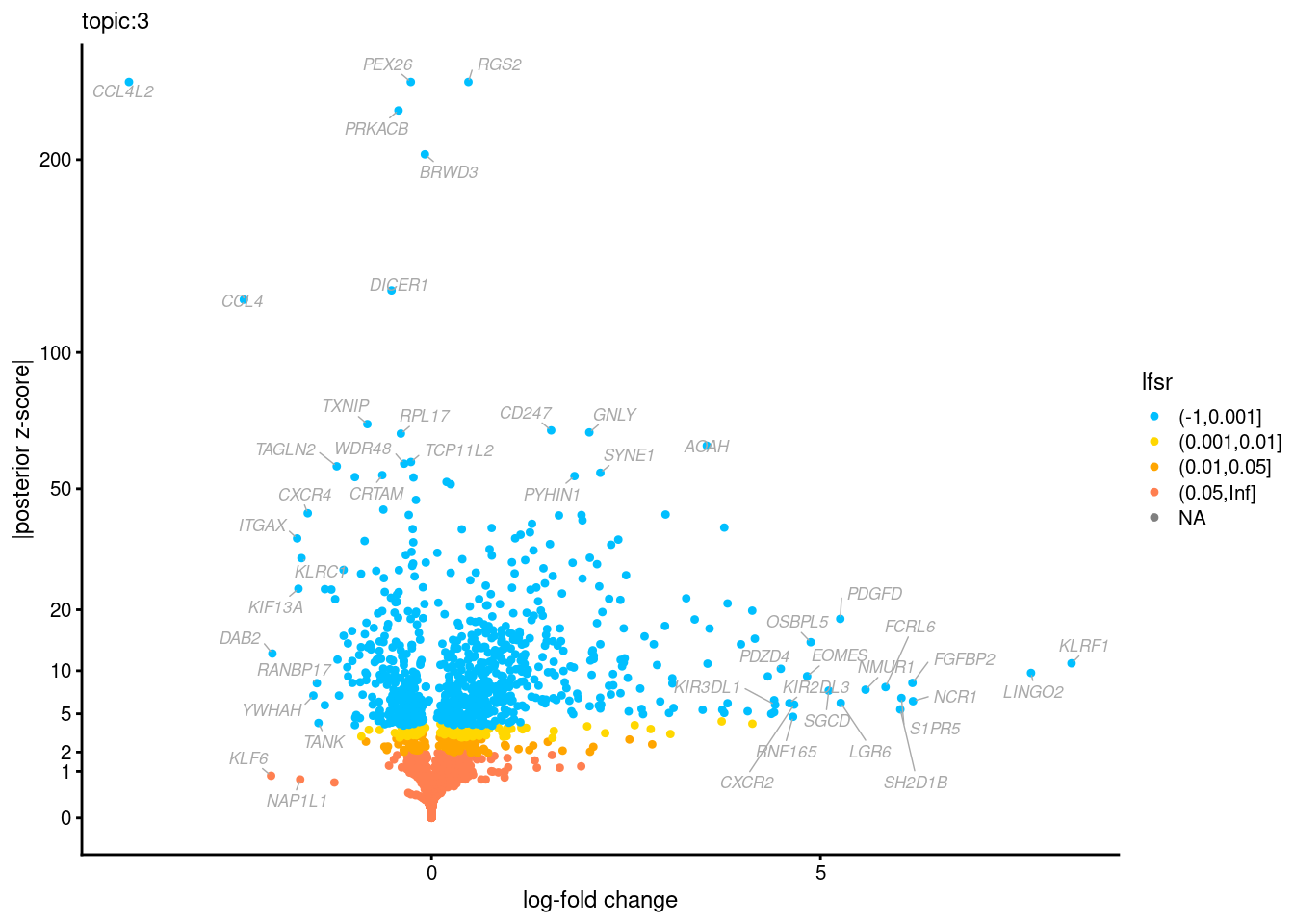
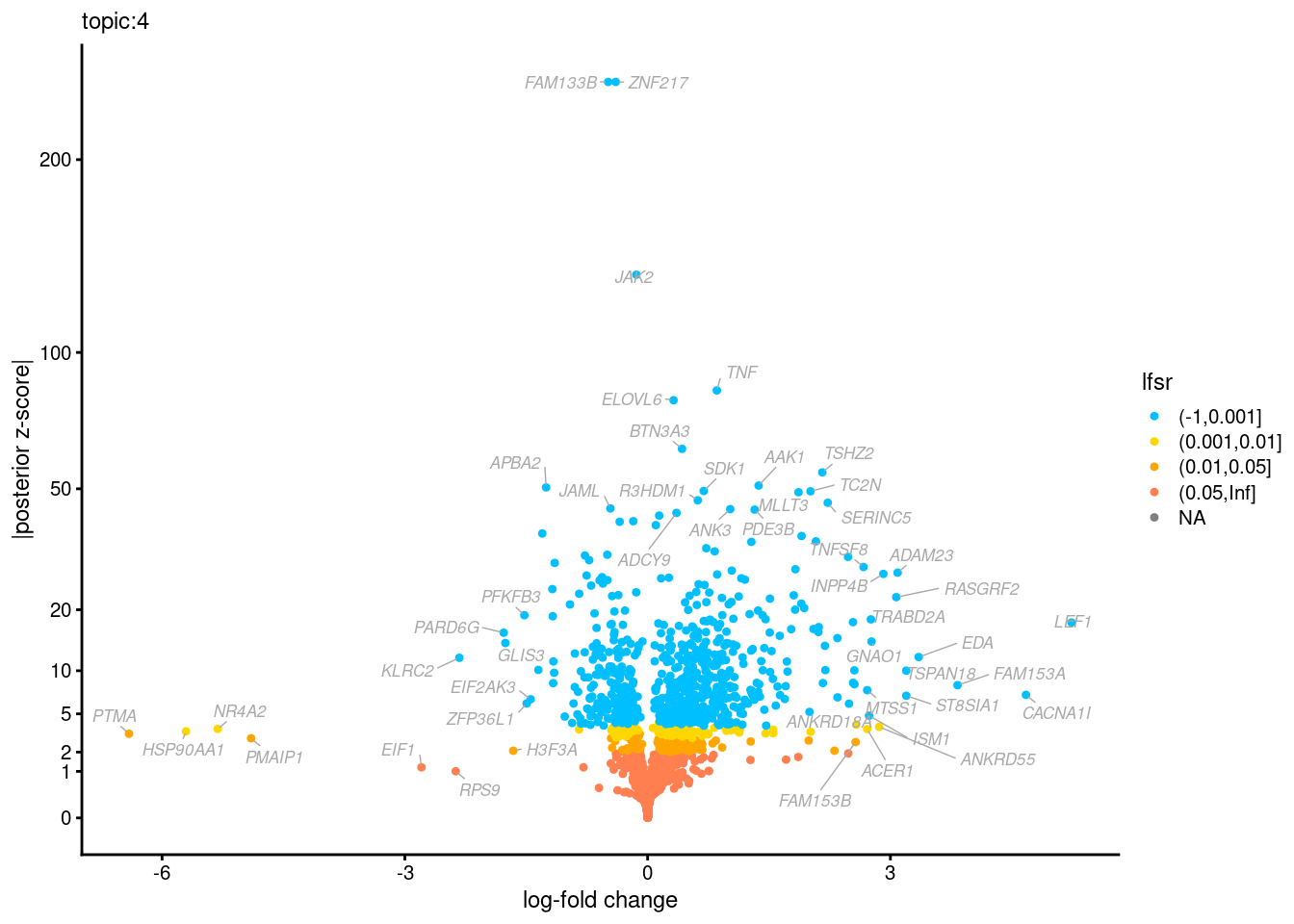
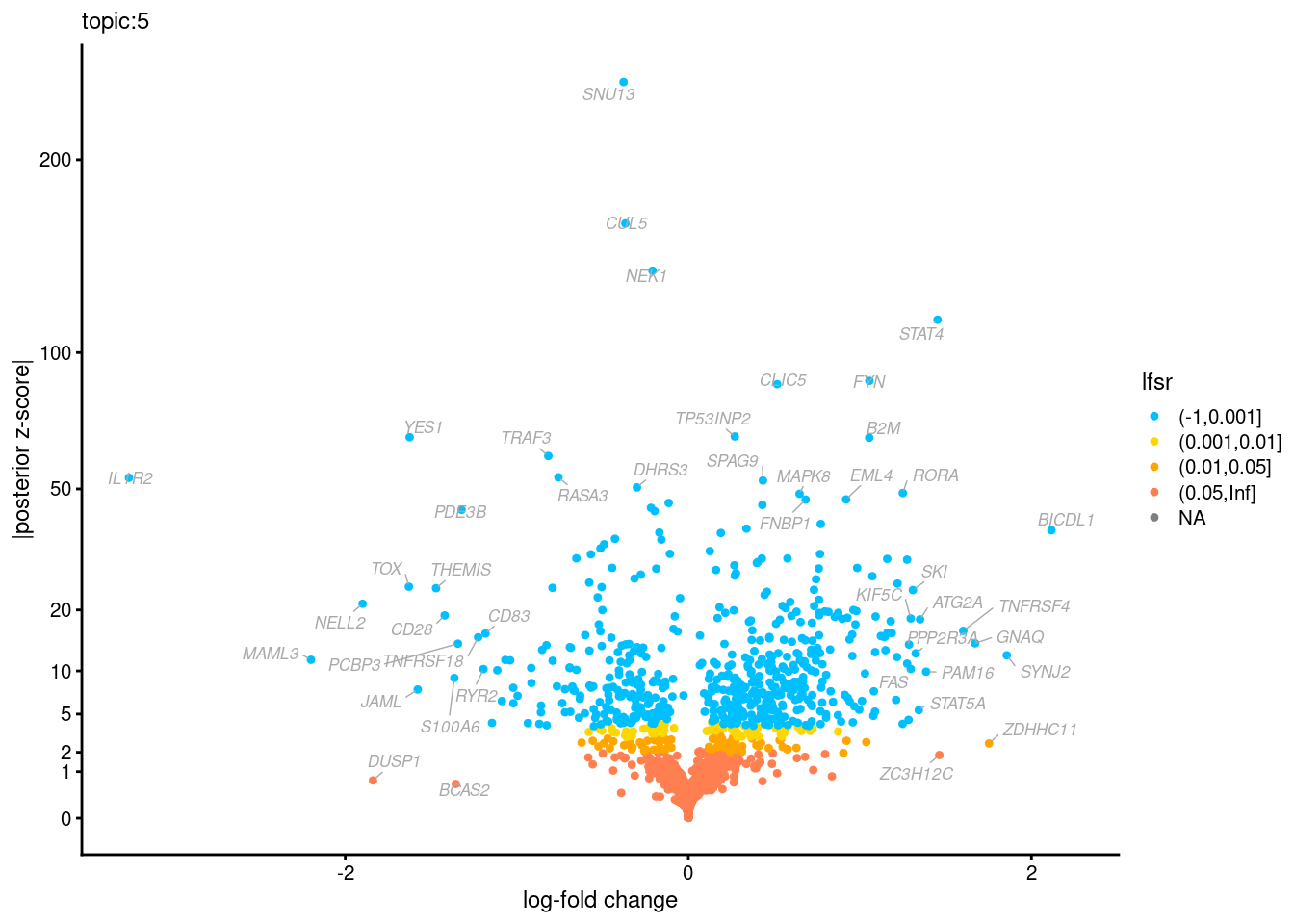
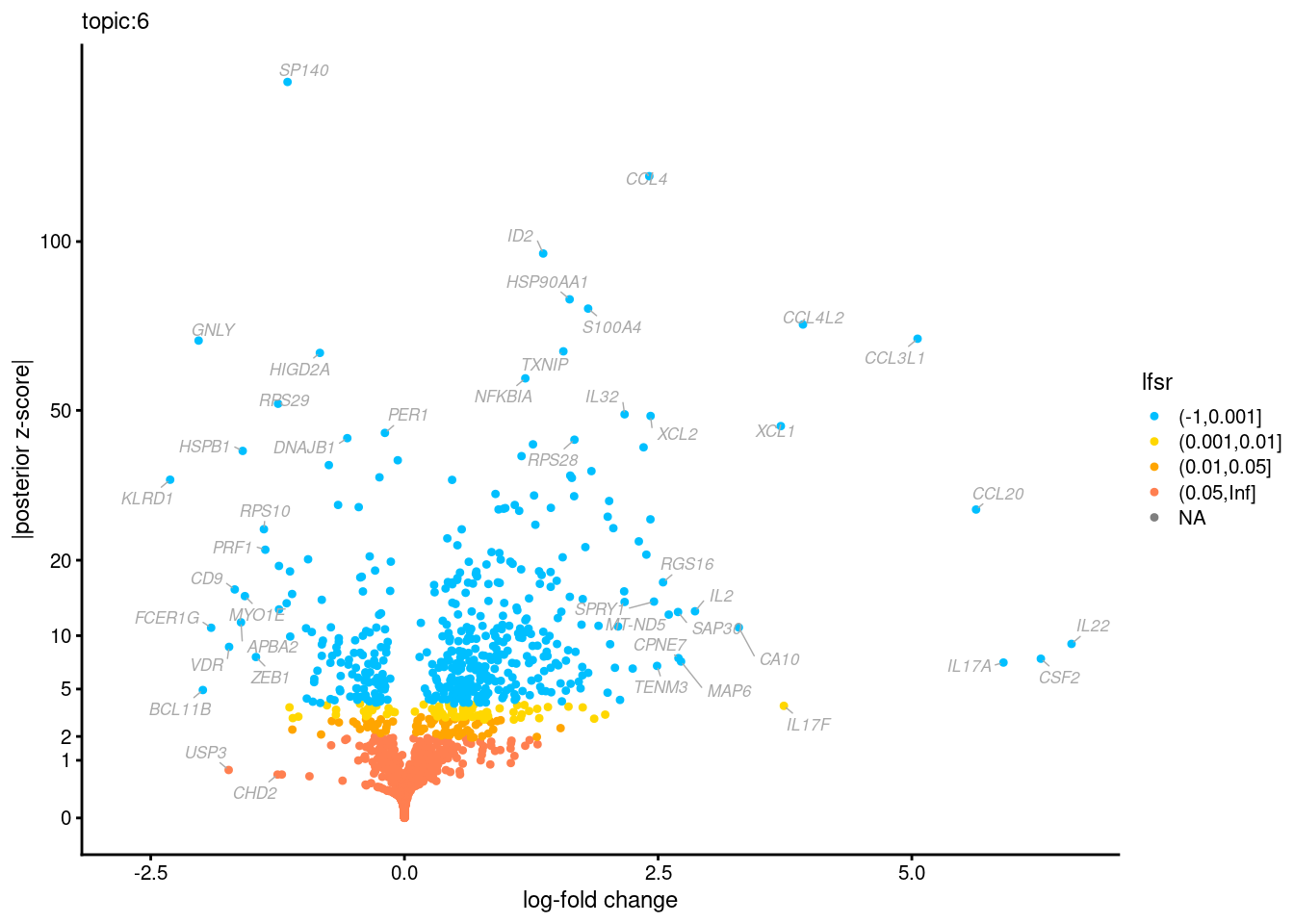
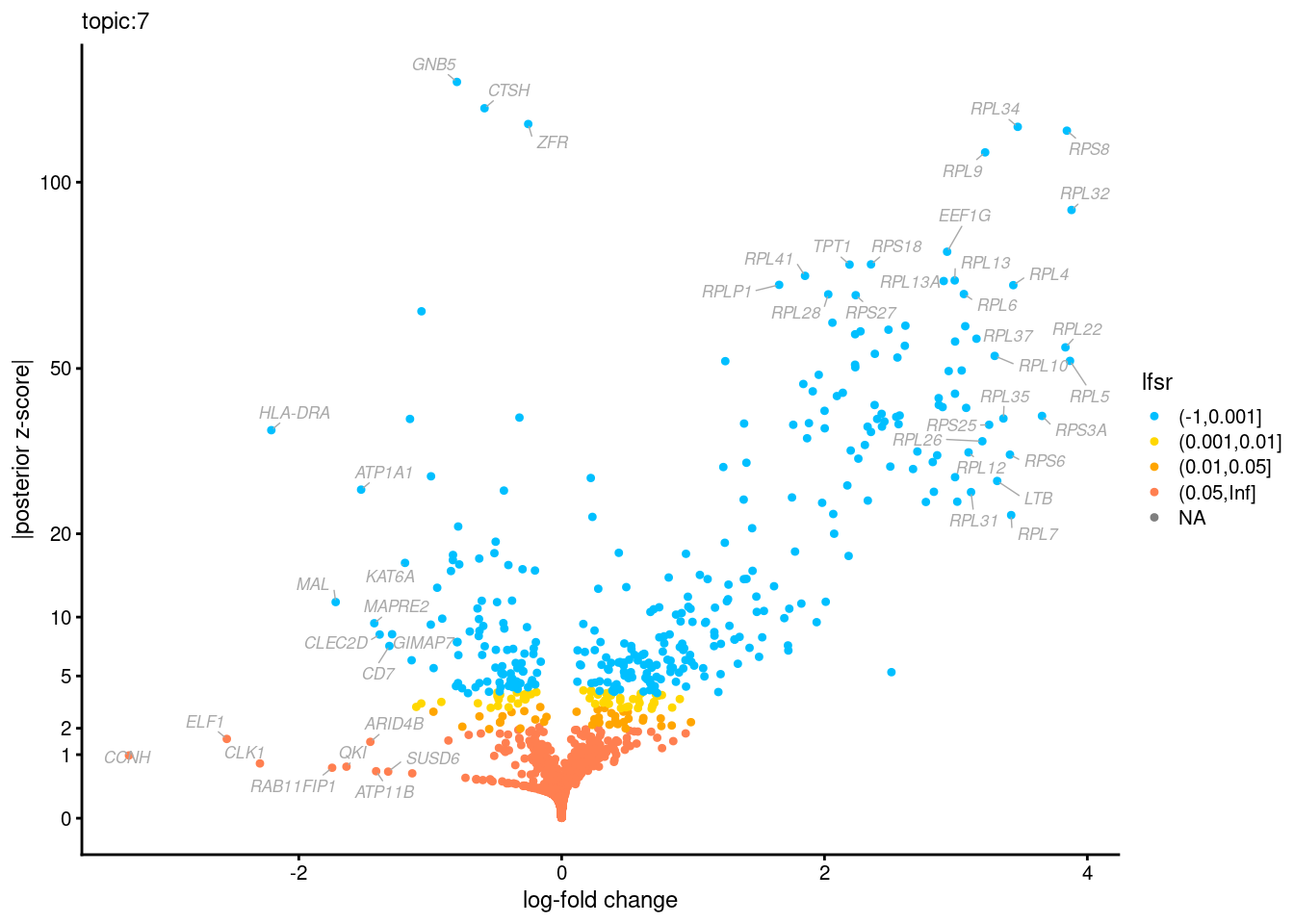
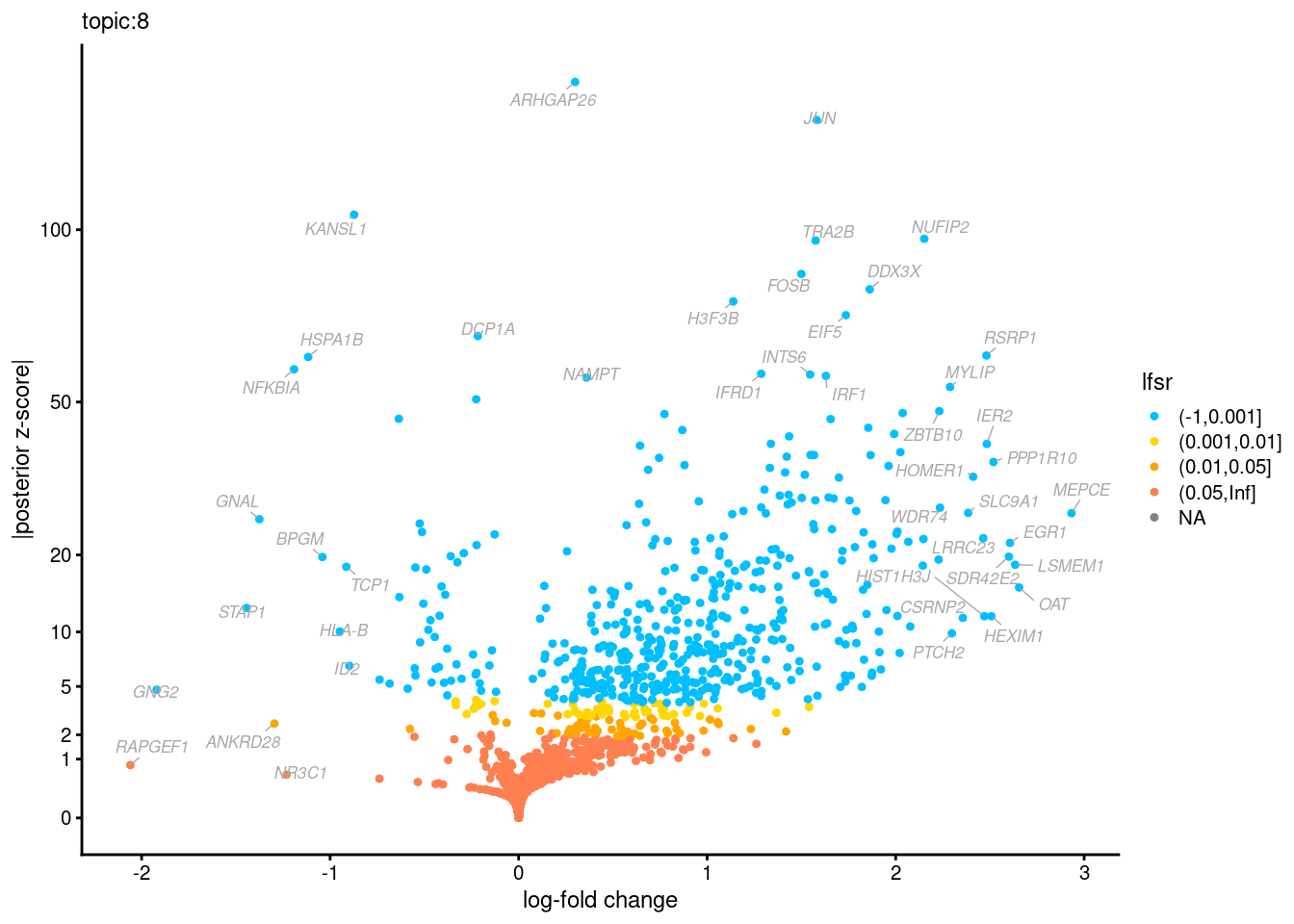
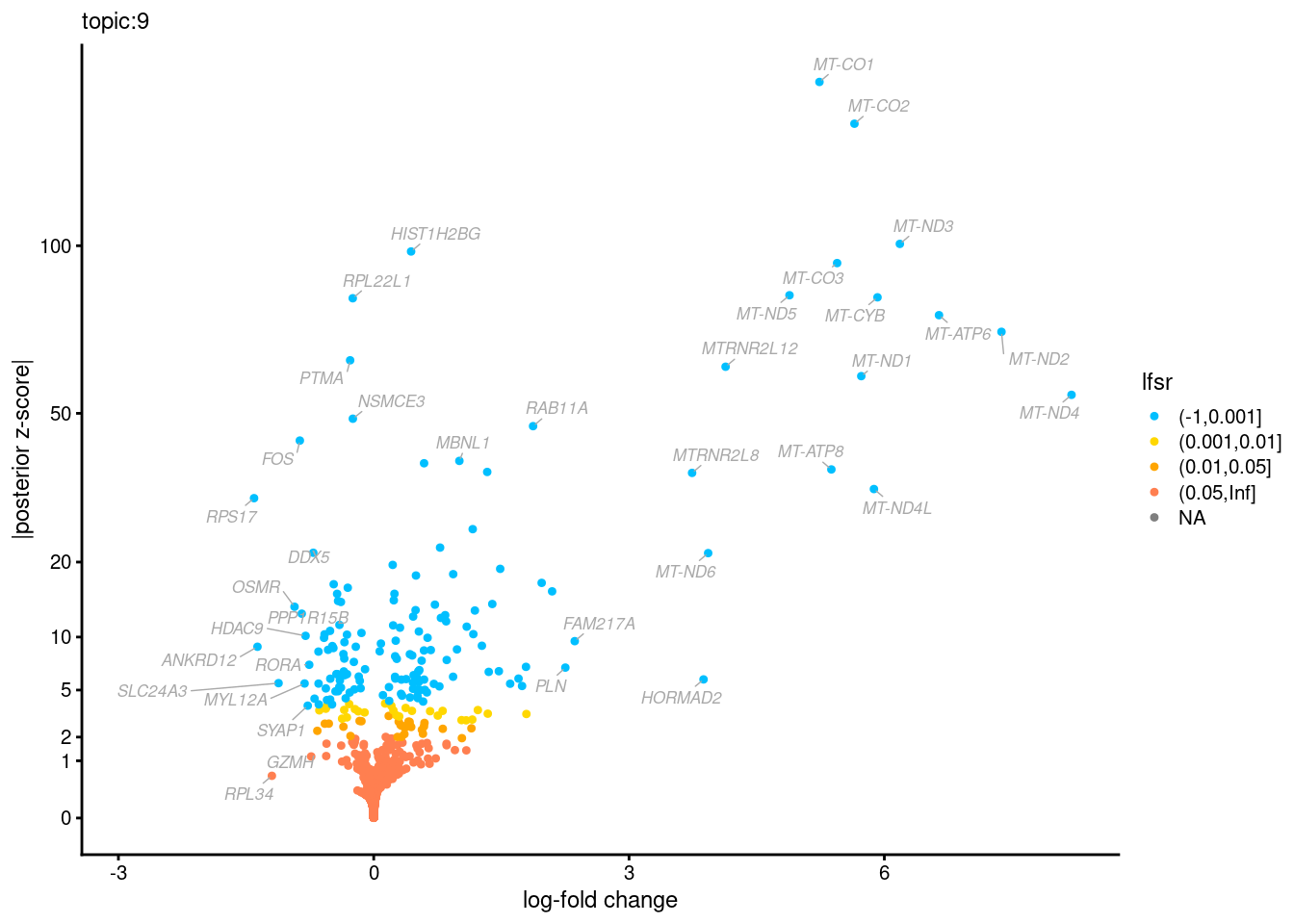
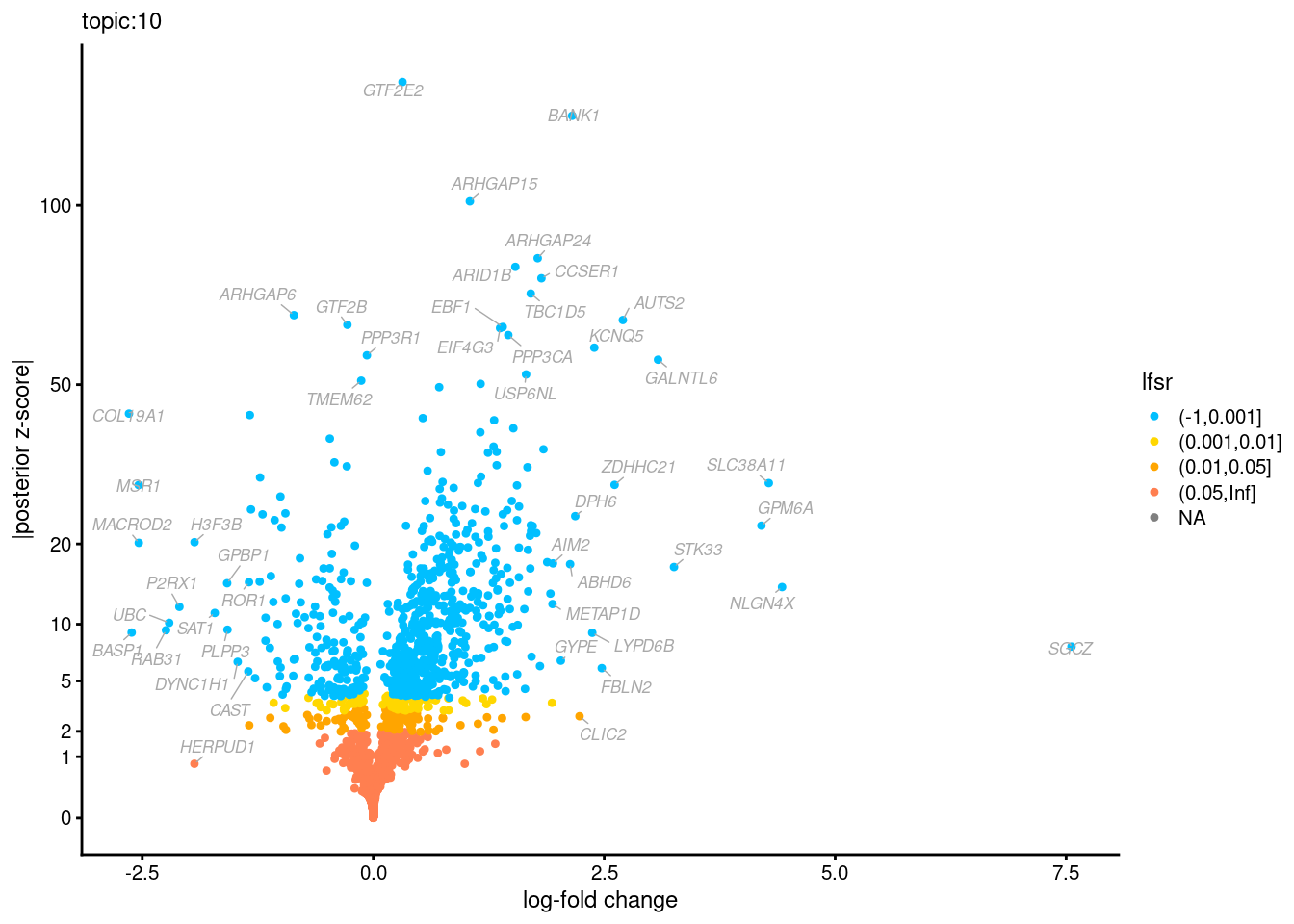
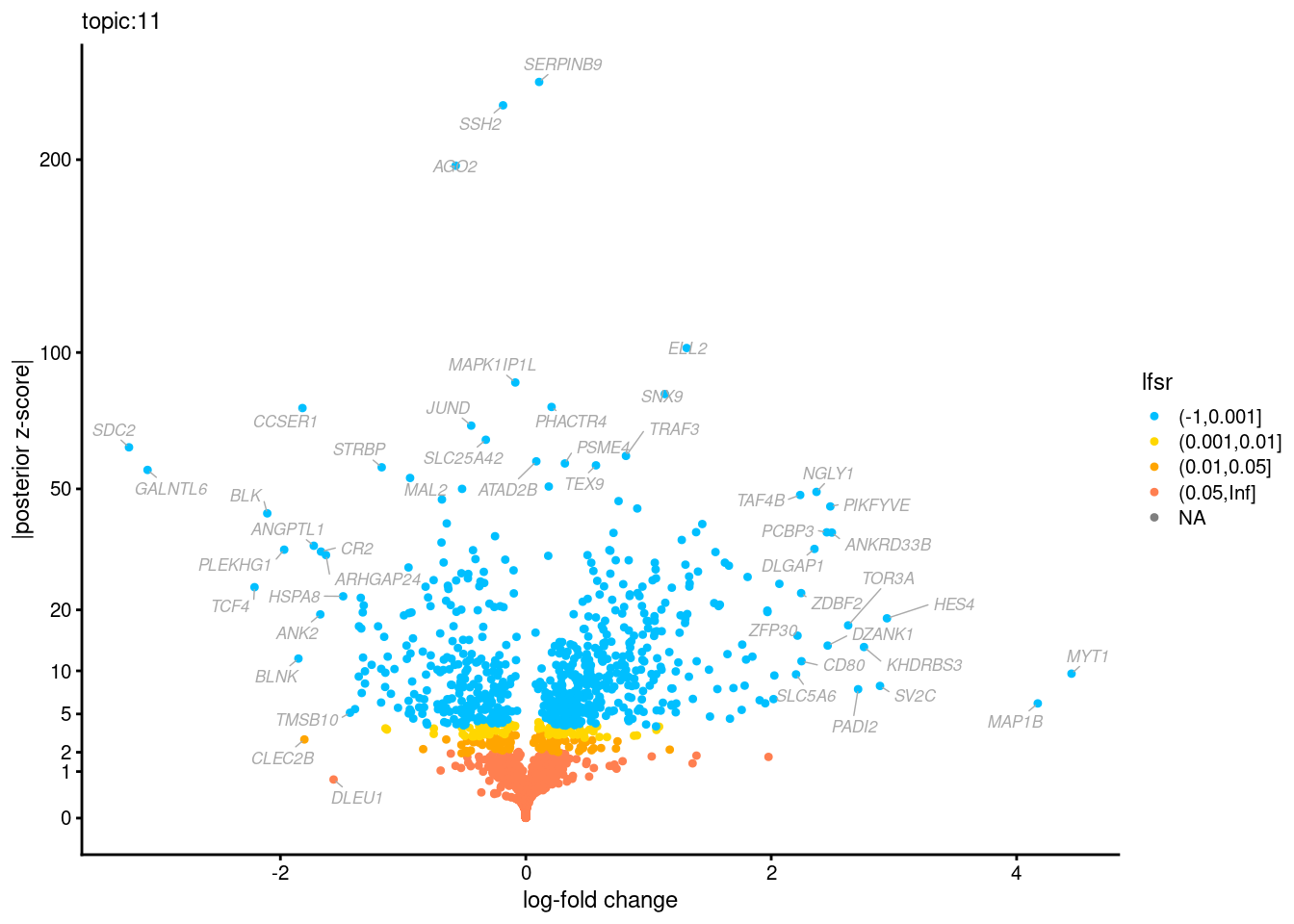
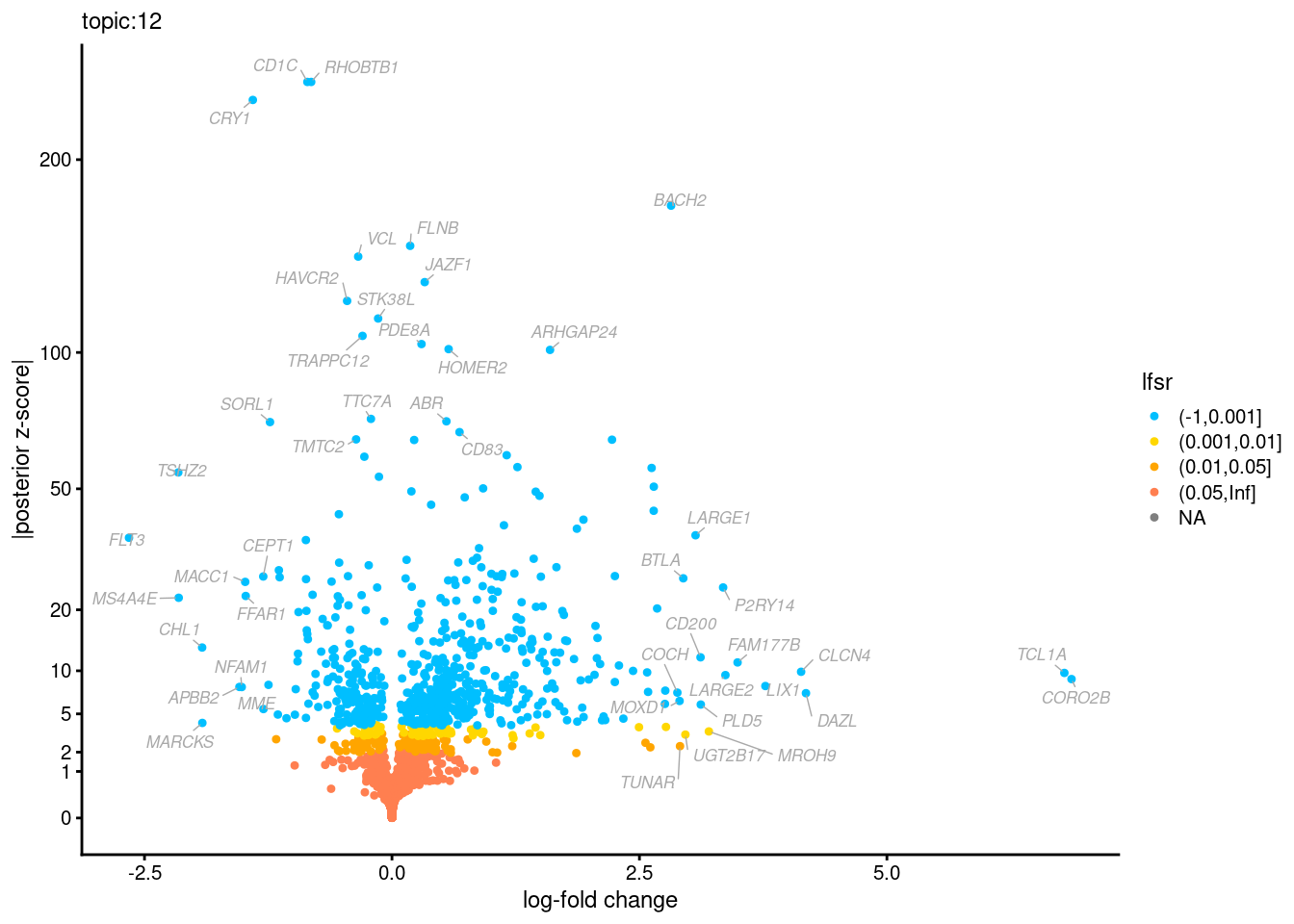
[1] "Number of up-regulated genes in each topic against the rest:" t1 t2 t3 t4 t5 t6 t7 t8 t9 t10 t11 t12
1602 122 594 562 391 381 267 505 111 626 465 489 GO Enrichment result table
All topics have fewer number of GO terms with enrichment, except for topic 1.
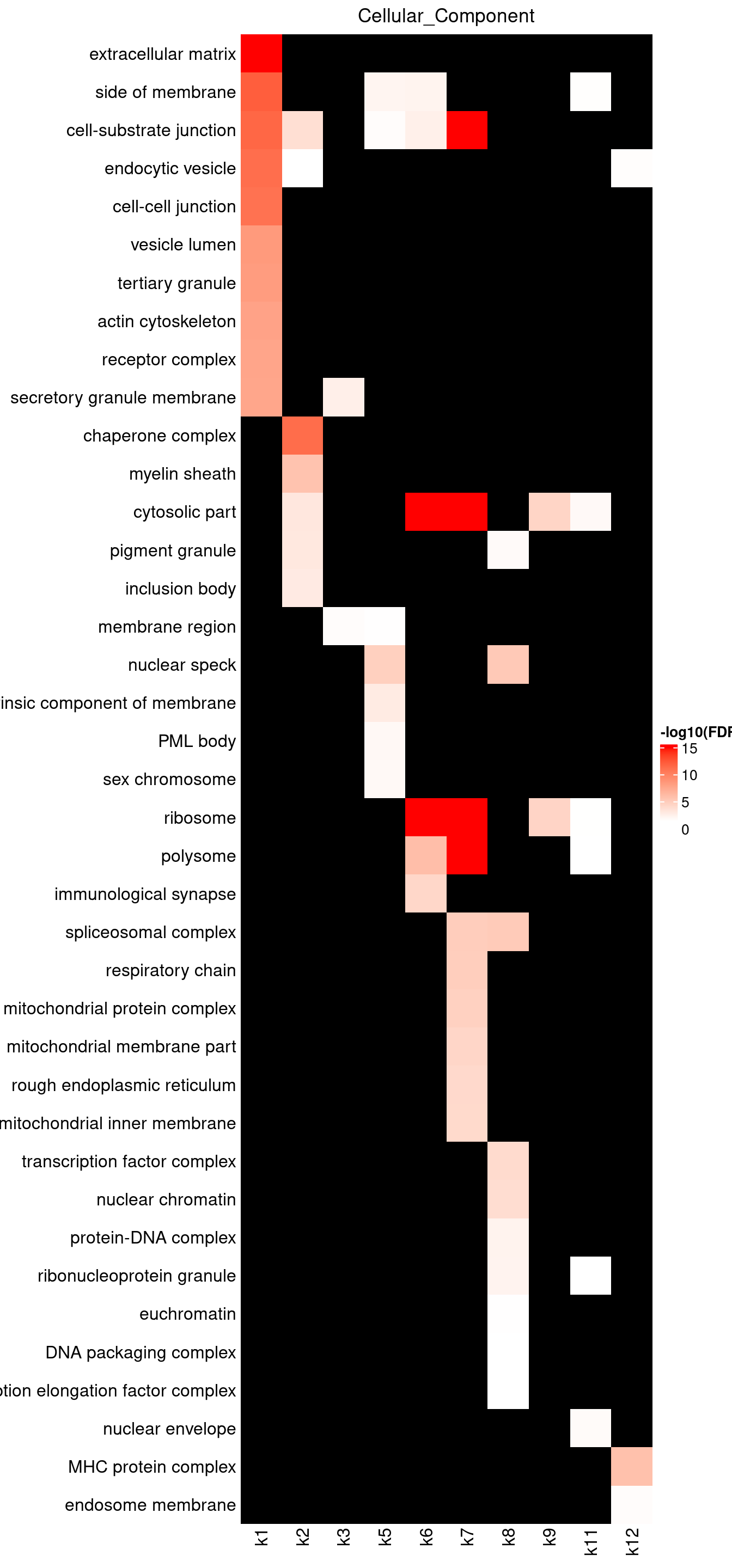
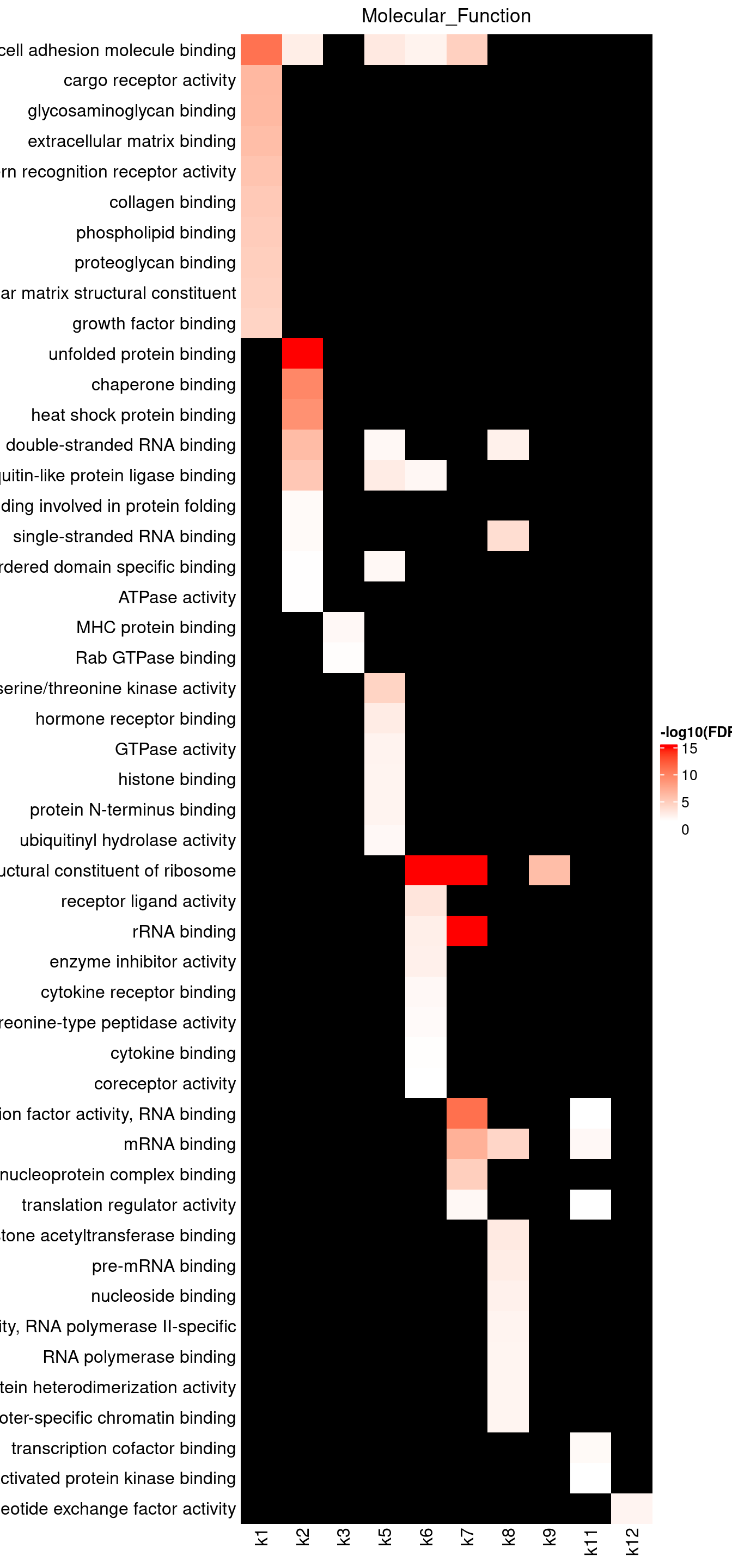
Visualizing GO enrichments with ComplexHeatmap
Parameters: - -log10(FDR) as value input - No clustering due to missing data - top 10 GO terms shown for each topic
Legend: - Color scale capped by FDR = \(10^{-15}\)
BP results show k1 enriched for granulocyte activation and neutrophil mediated immunity, as well as cell adhesion and motility. These pathways are highly relevant to Asthma. Due to high number of GO terms over-represented by k1 genes, we may repeat the topic modeling by increasing topic number. Several topics like k2, 6, 7, 9, 10 are strongly enriched with protein localization. Topics k3-6 have genes over-represented in T-cell activation, while k10, 12 in B-cell activation.
MF results show broad enrichment of molecular binding, with k7 highly enriched for cell adhesion molecular binding.
Selecting by FDR
Perform t-test while adjusting for confounders
Procedure
Test mean difference between tissue one donor at a time and then do meta-analysis with Fisher’s method
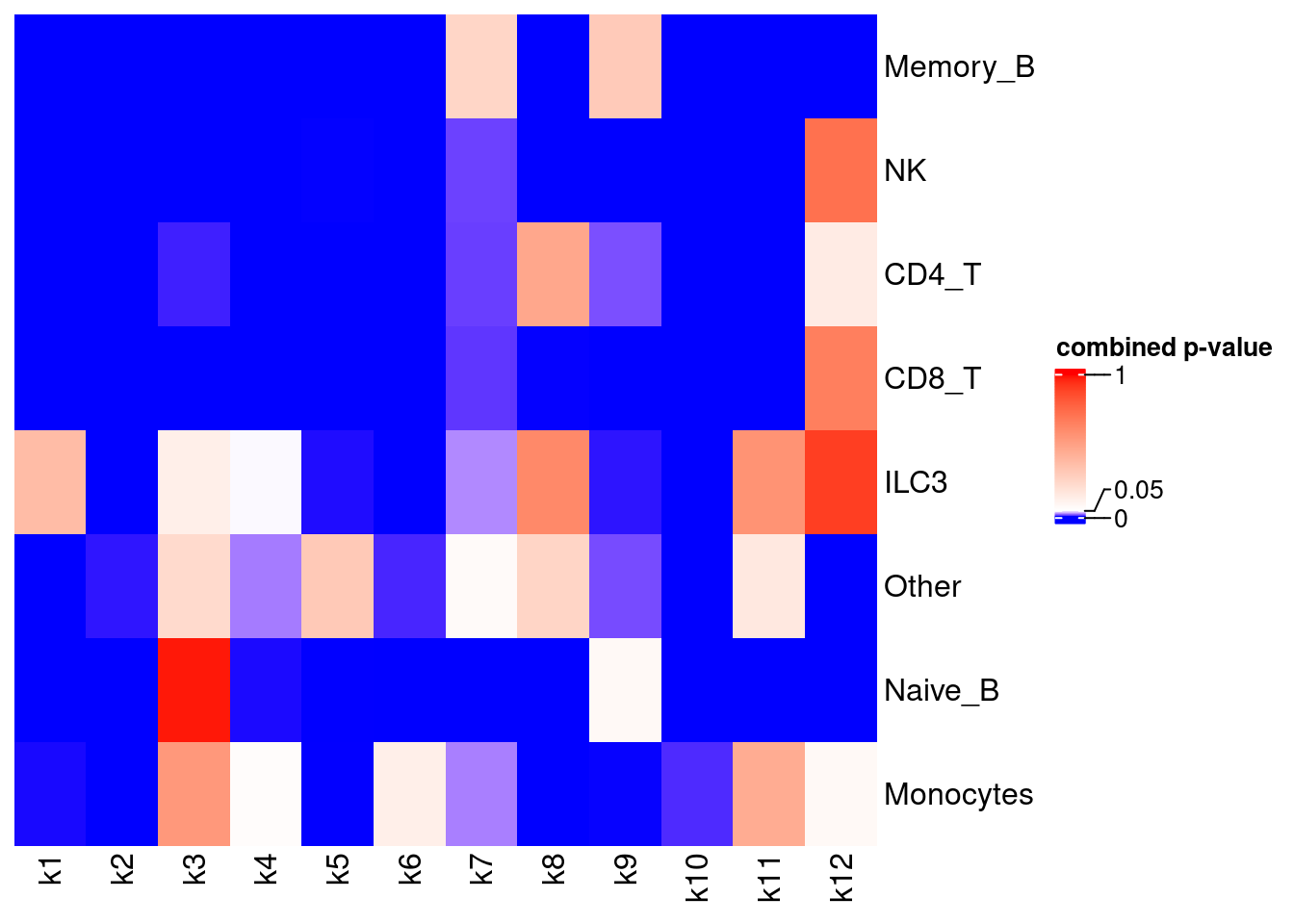
Results
X-axis denotes cell types and y-axis denotes the topics. For major cell types, we saw majority of topics have significant differences in proportions between tissue.
Find evidence for the function of TFs with motif enrichment in genes loaded on topic 5
Warning: The dot-dot notation (`..count..`) was deprecated in ggplot2 3.4.0.
ℹ Please use `after_stat(count)` instead.
This warning is displayed once every 8 hours.
Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
generated.
| Version | Author | Date |
|---|---|---|
| 744d4f0 | Jing Gu | 2024-05-22 |
R version 4.2.0 (2022-04-22)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.3.13-el7-x86_64/lib/libopenblas_haswellp-r0.3.13.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=C
[4] LC_COLLATE=C LC_MONETARY=C LC_MESSAGES=C
[7] LC_PAPER=C LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=C LC_IDENTIFICATION=C
attached base packages:
[1] grid stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] catecolors_0.1 ComplexHeatmap_2.14.0 colorRamp2_0.1.0
[4] tidyr_1.3.1 dplyr_1.1.4 poolr_1.1-1
[7] cowplot_1.1.3 ggplot2_3.4.0 fastTopics_0.6-175
[10] Matrix_1.6-5 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] matrixStats_1.2.0 fs_1.6.4 RColorBrewer_1.1-3
[4] doParallel_1.0.17 progress_1.2.3 httr_1.4.7
[7] rprojroot_2.0.4 tools_4.2.0 bslib_0.7.0
[10] DT_0.33 utf8_1.2.4 R6_2.5.1
[13] irlba_2.3.5.1 BiocGenerics_0.44.0 uwot_0.2.2
[16] lazyeval_0.2.2 colorspace_2.1-0 GetoptLong_1.0.5
[19] withr_3.0.0 tidyselect_1.2.1 prettyunits_1.2.0
[22] processx_3.8.3 compiler_4.2.0 git2r_0.33.0
[25] cli_3.6.2 Cairo_1.6-2 plotly_4.10.4
[28] labeling_0.4.3 sass_0.4.9 scales_1.3.0
[31] SQUAREM_2021.1 quadprog_1.5-8 callr_3.7.3
[34] pbapply_1.7-2 mixsqp_0.3-54 stringr_1.5.1
[37] digest_0.6.35 rmarkdown_2.26 RhpcBLASctl_0.23-42
[40] pkgconfig_2.0.3 htmltools_0.5.8.1 highr_0.10
[43] fastmap_1.1.1 invgamma_1.1 GlobalOptions_0.1.2
[46] htmlwidgets_1.6.4 rlang_1.1.3 rstudioapi_0.15.0
[49] farver_2.1.1 shape_1.4.6 jquerylib_0.1.4
[52] generics_0.1.3 jsonlite_1.8.8 crosstalk_1.2.1
[55] gtools_3.9.5 magrittr_2.0.3 S4Vectors_0.36.2
[58] Rcpp_1.0.12 munsell_0.5.1 fansi_1.0.6
[61] lifecycle_1.0.4 stringi_1.7.6 whisker_0.4.1
[64] yaml_2.3.8 mathjaxr_1.6-0 Rtsne_0.17
[67] parallel_4.2.0 promises_1.3.0 ggrepel_0.9.5
[70] crayon_1.5.2 lattice_0.22-5 circlize_0.4.15
[73] hms_1.1.3 knitr_1.46 ps_1.7.6
[76] pillar_1.9.0 rjson_0.2.21 stats4_4.2.0
[79] codetools_0.2-19 glue_1.7.0 evaluate_0.23
[82] getPass_0.2-2 data.table_1.15.4 RcppParallel_5.1.7
[85] vctrs_0.6.5 png_0.1-8 httpuv_1.6.14
[88] foreach_1.5.2 gtable_0.3.5 purrr_1.0.2
[91] clue_0.3-65 ashr_2.2-63 cachem_1.0.8
[94] xfun_0.43 later_1.3.2 viridisLite_0.4.2
[97] truncnorm_1.0-9 tibble_3.2.1 iterators_1.0.14
[100] IRanges_2.32.0 cluster_2.1.6