A testrun for MAGMA
Jing Gu
2024-09-19
Last updated: 2024-09-19
Checks: 7 0
Knit directory: lung_lymph_scMultiomics/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20221229) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 1e7e576. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: analysis/.RData
Ignored: analysis/.Rhistory
Ignored: analysis/figure/
Untracked files:
Untracked: analysis/.ipynb_checkpoints/
Untracked: analysis/test.pdf
Untracked: analysis/test_GO_enrichment.ipynb
Untracked: analysis/u19_atac_fastTopics.Rmd
Untracked: analysis/u19_regulon_enrichment.Rmd
Untracked: analysis/ukb-a-446.log
Untracked: code/run_magma/
Untracked: data/DA_peaks_Tsub_vs_others.RDS
Untracked: data/DA_peaks_by_cell_type.RDS
Untracked: data/TF_target_sizes_GRN.txt
Untracked: data/U19_T_cell_peaks_metadata.RDS
Untracked: data/Wang_2020_T_cell_peaks_metadata.RDS
Untracked: data/lung_GRN_CD4_T_edges.txt
Untracked: data/lung_GRN_CD8_T_edges.txt
Untracked: data/lung_GRN_Th17_edges.txt
Untracked: data/lung_GRN_Treg_edges.txt
Untracked: output/annotation_reference.txt
Untracked: output/fastTopics
Untracked: output/homer/
Untracked: output/ldsc_enrichment
Untracked: output/lung_immune_atac_peaks_high_ePIPs.RDS
Untracked: output/positions.bed
Untracked: output/topic1/
Untracked: output/topic10/
Untracked: output/topic11/
Untracked: output/topic12/
Untracked: output/topic2/
Untracked: output/topic3/
Untracked: output/topic4/
Untracked: output/topic5/
Untracked: output/topic6/
Untracked: output/topic7/
Untracked: output/topic8/
Untracked: output/topic9/
Untracked: test.pdf
Unstaged changes:
Modified: analysis/identify_regulatory_programs_u19_GRN.Rmd
Modified: analysis/rank_TFs_from_pairwise_comparison.ipynb
Modified: analysis/u19_h2g_enrichment.Rmd
Deleted: code/run_fastTopic.R
Deleted: lung_immune_fine_mapping.Rproj
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/test_magma.Rmd) and HTML
(docs/test_magma.html) files. If you’ve configured a remote
Git repository (see ?wflow_git_remote), click on the
hyperlinks in the table below to view the files as they were in that
past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 1e7e576 | Jing Gu | 2024-09-19 | look into k5 genes with high z scores |
| html | ab42a52 | Jing Gu | 2024-09-16 | Build site. |
| Rmd | a2b320f | Jing Gu | 2024-09-16 | test topic enrichment for genetic risks |
MAGMA
Gene analysis A linear principal component regression model that estimates whether there is genetic effect of gene g on the phenotype Y, conditional on all covariates. The model first projects genotype matrix for a gene g onto its PCs, pruning away PCs with very small eigenvalues. Then it performs F test in the regression of Y on SNP matrix and covariates to estimate genetic effect.
\[ Y = \alpha_{0g}\vec 1 + X_g^*\alpha_g + W\beta_g + \epsilon_g \] When inividual geneotype matrix not available, MAGMA performs gene test with mean \(X^2\) statistics and a gene p-value is then obtained by using a known approximation of the sampling distribution. Please refer to the following paper for details of approximation for the distribution of the weighted combination of p-values. This model requires summary statistics and reference LD panel.
Hou C (2005) A simple approximation for the distribution of the weighted combination of non-independent or independent probabilities. Stat Probabil Lett 73: 179–187.
Competitive gene-set analysis
One-sided Two-sample T test or linear regression in equivalence is applied to test whether the genes in a gene set are more strongly associated with Y or not.
Let Z denote the association z-score. Let \(\S_s\) be an indicator variable with element \(s_g = 1\) defined as for gene g in gene set s and 0 otherwise. The goal is to test whether \(\beta_s\) is greater than zero, which represents the difference in association between genes in the gene set and genes outside the gene set.
\[ Z = \beta_{0s}\vec 1 + S_s\beta_s + \epsilon \] This also be tested by unpaired two sample T-test, while two samples can have unequal variances and sample sizes.
Testrun
Procedure:
- annotate SNPs and genes
- gene-based analysis
- gene-set analysis
Results
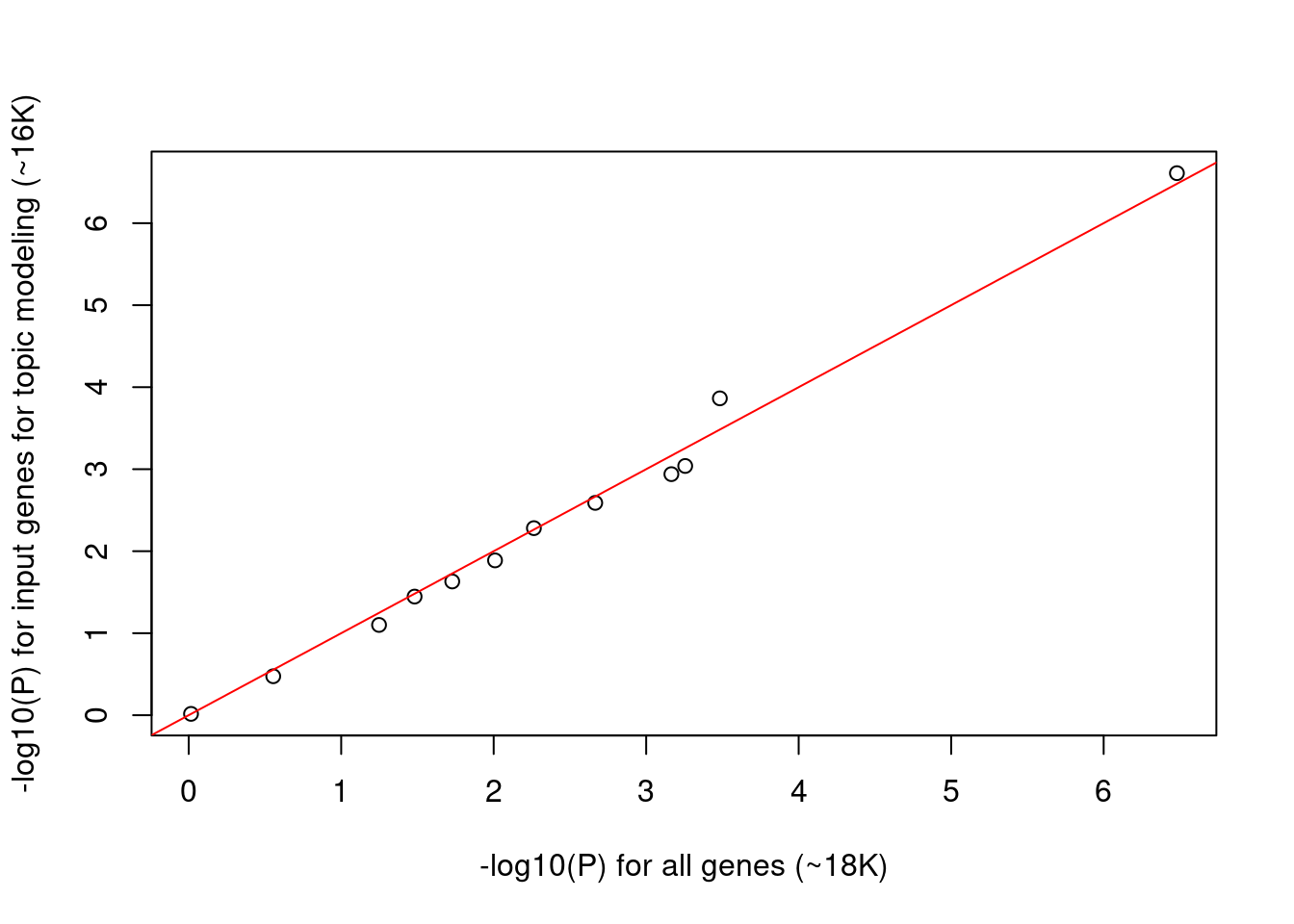
When corrected for multiple testing, tests will be significant if p-value lower than ~0.005. Around half of the tests show significant p-values, which makes us wonder if p-values are inflated. Then we try using the input genes for topic modeling rather than all genes as background so that they are more comparable.
- Total number of genes reduced from to 18K to ~16K
- P values for the reduced background genes are very similar to the full ones.
| Version | Author | Date |
|---|---|---|
| ab42a52 | Jing Gu | 2024-09-16 |
Test enrichment using top 100 genes from each topic
The supplementary table from MAGMA paper shows the mean type 1 error
rates are well controlled for a set of size 100. The MSigDB
canonical pathways contains 1320 gene sets from a number of
different databases. I can look into the average size of the gene sets.
Instead of all DE genes, I used top 100 up-regulated genes ranked by z scores to test the enrichment for each topic. Now we see only k3, k4, k5 and k12 (4/12) topics show significant enrichment after multiple testing correction.
Check the top genes contributing to topic 5 that have high MAGMA Z-scores
Genes with high Z-scores (p < 0.05) are found to show enrichment in the following gene sets:
- Regulation of signal transduction (adj.P = 0.01) from GO
- RORA, GATA3, CDC42SE2, SREBF2
- Interleukin-2 signaling pathway (adj.P = 0.003) from BioPlanet
- CD247, RORA, GATA3, TAB2, FOXO1,IL21R, YARS (overlapping genes)
- Th17 Cell Differentiation (adj.P = 0.0002) and IBD (adj.P = 0.001)
from KEGG
- CD247, RORA, GATA3,IL21R
Joining with `by = join_by(GENE)` X_SET4_ GENE CHR START STOP NSNPS NPARAM N ZSTAT
1 _SET4_ 60468 6 90636247 91006627 1026 117 336782 8.45050000
2 _SET4_ 919 1 167399877 167487847 355 73 336782 6.84930000
3 _SET4_ 2625 10 8087294 8117164 147 28 336782 6.12250000
4 _SET4_ 6095 15 60780483 61521502 2789 298 336782 5.60430000
5 _SET4_ 115426 9 6413151 6507054 357 33 336782 4.79490000
6 _SET4_ 340152 6 149768766 149806148 214 19 336782 4.01200000
7 _SET4_ 56990 5 130599702 130730383 280 35 336782 2.90970000
8 _SET4_ 23118 6 149639436 149732747 335 25 336782 2.78020000
9 _SET4_ 50615 16 27413483 27463363 229 57 336782 2.64820000
10 _SET4_ 10666 18 67530192 67624412 378 59 336782 2.36120000
11 _SET4_ 10318 5 150409504 150467221 256 43 336782 2.23780000
12 _SET4_ 6721 22 42229083 42303312 221 39 336782 2.18420000
13 _SET4_ 2308 13 41129801 41240734 323 29 336782 2.15450000
14 _SET4_ 1362 17 28705942 28796675 160 23 336782 1.73580000
15 _SET4_ 51230 20 34359923 34538292 585 24 336782 1.73200000
16 _SET4_ 23387 11 116714118 116969131 1087 35 336782 1.55350000
17 _SET4_ 8565 1 33240840 33283633 108 22 336782 1.55560000
18 _SET4_ 54331 14 52327022 52436518 577 66 336782 1.51080000
19 _SET4_ 59343 3 185300284 185348889 146 15 336782 1.33270000
20 _SET4_ 55833 9 33921691 34048947 497 32 336782 1.09100000
21 _SET4_ 2534 6 111981535 112194655 815 52 336782 1.04560000
22 _SET4_ 7879 3 128444975 128533641 327 29 336782 1.09670000
23 _SET4_ 678 2 43449541 43453745 13 7 336782 1.06920000
24 _SET4_ 23253 18 9136751 9285983 528 35 336782 1.05670000
25 _SET4_ 26136 7 115850547 115898840 180 17 336782 0.96789000
26 _SET4_ 51742 1 235330210 235491532 504 23 336782 0.93847000
27 _SET4_ 8556 1 100810598 100985833 444 38 336782 0.96840000
28 _SET4_ 9267 17 76670130 76778376 319 27 336782 0.94589000
29 _SET4_ 3685 2 187454058 187545629 201 39 336782 0.84031000
30 _SET4_ 23198 2 54091204 54197977 404 36 336782 0.85629000
31 _SET4_ 257415 7 92190072 92219706 86 18 336782 0.93003000
32 _SET4_ 57488 7 158523688 158622729 519 25 336782 0.85795000
33 _SET4_ 94120 6 159065931 159185908 546 66 336782 0.74631000
34 _SET4_ 5430 17 7387698 7417935 153 16 336782 0.78662000
35 _SET4_ 90268 5 14664783 14699842 89 21 336782 0.80570000
36 _SET4_ 8887 7 27778992 27869386 267 35 336782 0.74946000
37 _SET4_ 5906 1 112162405 112256807 420 36 336782 0.65849000
38 _SET4_ 4676 11 2965660 3013607 222 32 336782 0.56302000
39 _SET4_ 27436 2 42396490 42559688 630 31 336782 0.42312000
40 _SET4_ 51430 1 172501494 172580973 167 35 336782 0.33426000
41 _SET4_ 7409 19 6772679 6857377 332 55 336782 0.32158000
42 _SET4_ 3607 17 80477594 80562483 419 49 336782 0.25939000
43 _SET4_ 2288 12 2904108 2914589 11 4 336782 0.39082000
44 _SET4_ 56829 7 138728266 138794466 212 34 336782 0.19810000
45 _SET4_ 4603 8 67474410 67525484 113 18 336782 0.46371000
46 _SET4_ 9043 17 49039535 49198226 394 41 336782 0.24384000
47 _SET4_ 1106 15 93442286 93571237 485 46 336782 0.09673500
48 _SET4_ 923 11 60739113 60787849 230 49 336782 0.09332900
49 _SET4_ 80714 19 19672516 19729725 214 20 336782 0.20781000
50 _SET4_ 23074 12 100422233 100536642 395 37 336782 0.16546000
51 _SET4_ 10260 15 65952954 66084631 364 27 336782 0.09866700
52 _SET4_ 6775 2 191894302 192037404 407 56 336782 0.03275800
53 _SET4_ 284996 2 101887681 101925178 87 24 336782 -0.00084963
54 _SET4_ 10383 9 140135711 140138159 9 5 336782 0.27720000
55 _SET4_ 26051 20 37434348 37551667 411 72 336782 0.00594630
56 _SET4_ 6497 1 2160134 2241652 346 69 336782 -0.04317000
57 _SET4_ 58508 7 151832010 152133090 377 20 336782 0.13412000
58 _SET4_ 6732 6 35800811 35888957 295 23 336782 -0.04915800
59 _SET4_ 23130 11 64662004 64684722 62 11 336782 0.05085400
60 _SET4_ 4929 2 157180944 157189287 9 5 336782 -0.09094000
61 _SET4_ 29072 3 47057898 47205467 277 26 336782 -0.15736000
62 _SET4_ 8027 10 17686124 17758823 359 21 336782 -0.12647000
63 _SET4_ 60481 6 53132196 53213977 279 19 336782 -0.16222000
64 _SET4_ 9728 15 49280835 49338760 140 22 336782 -0.19923000
65 _SET4_ 80331 20 62526455 62567384 192 28 336782 -0.18511000
66 _SET4_ 51429 6 158137078 158366109 970 58 336782 -0.28369000
67 _SET4_ 54622 5 53180578 53606403 1813 122 336782 -0.27789000
68 _SET4_ 2776 9 80335189 80646219 863 57 336782 -0.29571000
69 _SET4_ 4928 11 3696240 3819022 430 33 336782 -0.28774000
70 _SET4_ 27161 8 141541264 141645646 467 68 336782 -0.42633000
71 _SET4_ 567 15 45003685 45010357 13 5 336782 -0.27618000
72 _SET4_ 63916 20 44994689 45035690 72 17 336782 -0.35065000
73 _SET4_ 253959 14 36007558 36278432 574 32 336782 -0.42536000
74 _SET4_ 960 11 35160417 35253949 413 73 336782 -0.50107000
75 _SET4_ 9585 10 91461264 91534700 315 19 336782 -0.49604000
76 _SET4_ 53405 6 45866188 46048085 739 90 336782 -0.54930000
77 _SET4_ 868 3 105374306 105589354 743 45 336782 -0.56987000
78 _SET4_ 7293 1 1146706 1149703 11 5 336782 -0.40596000
79 _SET4_ 79572 3 194123403 194188968 187 23 336782 -0.48753000
80 _SET4_ 5599 10 49514682 49647403 398 37 336782 -0.58362000
81 _SET4_ 26191 1 114356433 114414375 144 30 336782 -0.65354000
82 _SET4_ 55031 11 11862970 11980872 403 41 336782 -0.67810000
83 _SET4_ 125488 18 21572737 21715574 507 57 336782 -0.72006000
84 _SET4_ 5604 15 66679182 66783882 374 33 336782 -0.72970000
85 _SET4_ 1499 3 41236401 41281939 99 22 336782 -0.78735000
86 _SET4_ 829 1 113162075 113214241 121 29 336782 -0.85602000
87 _SET4_ 26524 13 21547175 21635722 325 32 336782 -0.87000000
88 _SET4_ 23347 18 2655886 2805015 678 33 336782 -0.95213000
89 _SET4_ 58476 20 33292147 33301240 19 10 336782 -1.04680000
90 _SET4_ 2782 1 1716725 1822552 217 39 336782 -1.08390000
91 _SET4_ 285513 4 90162427 90229161 237 44 336782 -1.14290000
92 _SET4_ 6935 10 31607825 31818742 408 30 336782 -0.97917000
93 _SET4_ 4363 16 16043434 16236931 811 67 336782 -1.28210000
94 _SET4_ 23048 9 132649458 132805473 654 46 336782 -1.20560000
95 _SET4_ 9147 14 50250528 50319763 128 41 336782 -1.24380000
96 _SET4_ 8428 13 99102455 99229396 485 46 336782 -1.91380000
P ZFITTED_BASE ZRESID_BASE SYMBOL
1 9.9039e-20 0.0000e+00 8.45050000 BACH2
2 1.0814e-13 0.0000e+00 6.84930000 CD247
3 1.5312e-11 0.0000e+00 6.12250000 GATA3
4 4.0321e-10 0.0000e+00 5.60430000 RORA
5 7.4517e-08 0.0000e+00 4.79490000 UHRF2
6 3.0972e-06 0.0000e+00 4.01200000 ZC3H12D
7 3.9436e-04 4.4409e-16 2.90970000 CDC42SE2
8 5.4278e-04 4.4409e-16 2.78020000 TAB2
9 7.6388e-04 4.4409e-16 2.64820000 IL21R
10 2.3719e-03 4.4409e-16 2.36120000 CD226
11 3.1237e-03 4.4409e-16 2.23780000 TNIP1
12 3.7568e-03 4.4409e-16 2.18420000 SREBF2
13 4.1285e-03 4.4409e-16 2.15450000 FOXO1
14 1.1793e-02 2.2204e-16 1.73580000 CPD
15 1.6984e-02 2.2204e-16 1.73200000 PHF20
16 2.0633e-02 2.2204e-16 1.55350000 SIK3
17 2.0987e-02 2.2204e-16 1.55560000 YARS
18 2.1930e-02 2.2204e-16 1.51080000 GNG2
19 3.5505e-02 2.2204e-16 1.33270000 SENP2
20 5.6123e-02 2.2204e-16 1.09100000 UBAP2
21 6.3251e-02 2.2204e-16 1.04560000 FYN
22 6.5841e-02 2.2204e-16 1.09670000 RAB7A
23 6.6380e-02 2.2204e-16 1.06920000 ZFP36L2
24 6.6452e-02 2.2204e-16 1.05670000 ANKRD12
25 7.2884e-02 2.2204e-16 0.96789000 TES
26 7.4910e-02 2.2204e-16 0.93847000 ARID4B
27 7.7287e-02 2.2204e-16 0.96840000 CDC14A
28 7.8260e-02 2.2204e-16 0.94589000 CYTH1
29 8.5962e-02 2.2204e-16 0.84031000 ITGAV
30 8.6831e-02 2.2204e-16 0.85629000 PSME4
31 9.0951e-02 2.2204e-16 0.93003000 FAM133B
32 9.7966e-02 2.2204e-16 0.85795000 ESYT2
33 1.0373e-01 2.2204e-16 0.74631000 SYTL3
34 1.0565e-01 2.2204e-16 0.78662000 POLR2A
35 1.0827e-01 2.2204e-16 0.80570000 OTULIN
36 1.0937e-01 2.2204e-16 0.74946000 TAX1BP1
37 1.2473e-01 2.2204e-16 0.65849000 RAP1A
38 1.4087e-01 2.2204e-16 0.56302000 NAP1L4
39 1.7093e-01 2.7756e-16 0.42312000 EML4
40 1.9850e-01 2.7756e-16 0.33426000 SUCO
41 2.0326e-01 2.7756e-16 0.32158000 VAV1
42 2.1586e-01 2.7756e-16 0.25939000 FOXK2
43 2.3176e-01 2.7756e-16 0.39082000 FKBP4
44 2.3304e-01 2.7756e-16 0.19810000 ZC3HAV1
45 2.3537e-01 2.7756e-16 0.46371000 MYBL1
46 2.3855e-01 2.7756e-16 0.24384000 SPAG9
47 2.6657e-01 2.6368e-16 0.09673500 CHD2
48 2.7075e-01 2.6368e-16 0.09332900 CD6
49 2.7184e-01 2.7756e-16 0.20781000 PBX4
50 2.8320e-01 2.7756e-16 0.16546000 UHRF1BP1L
51 2.8801e-01 2.6368e-16 0.09866700 DENND4A
52 2.9184e-01 2.6368e-16 0.03275800 STAT4
53 3.0147e-01 2.6476e-16 -0.00084963 RNF149
54 3.1265e-01 2.7756e-16 0.27720000 TUBB4B
55 3.1278e-01 2.6455e-16 0.00594630 PPP1R16B
56 3.2321e-01 2.6368e-16 -0.04317000 SKI
57 3.2898e-01 2.7756e-16 0.13412000 KMT2C
58 3.3295e-01 2.6368e-16 -0.04915800 SRPK1
59 3.4537e-01 2.6368e-16 0.05085400 ATG2A
60 3.6504e-01 2.6368e-16 -0.09094000 NR4A2
61 3.7147e-01 2.7756e-16 -0.15736000 SETD2
62 3.7562e-01 2.7756e-16 -0.12647000 STAM
63 3.7722e-01 2.7756e-16 -0.16222000 ELOVL5
64 3.8339e-01 2.7756e-16 -0.19923000 SECISBP2L
65 3.8868e-01 2.7756e-16 -0.18511000 DNAJC5
66 4.0245e-01 2.7756e-16 -0.28369000 SNX9
67 4.0338e-01 2.7756e-16 -0.27789000 ARL15
68 4.1351e-01 2.7756e-16 -0.29571000 GNAQ
69 4.1970e-01 2.7756e-16 -0.28774000 NUP98
70 4.6964e-01 2.7756e-16 -0.42633000 AGO2
71 4.7298e-01 2.7756e-16 -0.27618000 B2M
72 4.7943e-01 2.7756e-16 -0.35065000 ELMO2
73 5.0369e-01 2.7756e-16 -0.42536000 RALGAPA1
74 5.0731e-01 2.2204e-16 -0.50107000 CD44
75 5.1298e-01 2.7756e-16 -0.49604000 KIF20B
76 5.2310e-01 2.2204e-16 -0.54930000 CLIC5
77 5.2936e-01 2.2204e-16 -0.56987000 CBLB
78 5.3331e-01 2.7756e-16 -0.40596000 TNFRSF4
79 5.3378e-01 2.7756e-16 -0.48753000 ATP13A3
80 5.4428e-01 2.2204e-16 -0.58362000 MAPK8
81 5.5105e-01 2.2204e-16 -0.65354000 PTPN22
82 5.8017e-01 2.2204e-16 -0.67810000 USP47
83 5.9235e-01 2.2204e-16 -0.72006000 TTC39C
84 6.0026e-01 2.2204e-16 -0.72970000 MAP2K1
85 6.0696e-01 2.2204e-16 -0.78735000 CTNNB1
86 6.3320e-01 2.2204e-16 -0.85602000 CAPZA1
87 6.4814e-01 2.2204e-16 -0.87000000 LATS2
88 6.7139e-01 2.2204e-16 -0.95213000 SMCHD1
89 7.1615e-01 2.2204e-16 -1.04680000 TP53INP2
90 7.1974e-01 2.2204e-16 -1.08390000 GNB1
91 7.2113e-01 2.2204e-16 -1.14290000 GPRIN3
92 7.3190e-01 2.2204e-16 -0.97917000 ZEB1
93 7.7264e-01 2.2204e-16 -1.28210000 ABCC1
94 7.7937e-01 2.2204e-16 -1.20560000 FNBP1
95 7.8719e-01 2.2204e-16 -1.24380000 NEMF
96 9.1854e-01 2.2204e-16 -1.91380000 STK24
R version 4.2.0 (2022-04-22)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.3.13-el7-x86_64/lib/libopenblas_haswellp-r0.3.13.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=C
[4] LC_COLLATE=C LC_MONETARY=C LC_MESSAGES=C
[7] LC_PAPER=C LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=C LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] dplyr_1.1.4 data.table_1.15.4 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] Rcpp_1.0.12 highr_0.10 compiler_4.2.0 pillar_1.9.0
[5] bslib_0.7.0 later_1.3.2 git2r_0.33.0 jquerylib_0.1.4
[9] tools_4.2.0 getPass_0.2-2 digest_0.6.35 jsonlite_1.8.8
[13] evaluate_0.23 lifecycle_1.0.4 tibble_3.2.1 pkgconfig_2.0.3
[17] rlang_1.1.3 cli_3.6.2 rstudioapi_0.15.0 crosstalk_1.2.1
[21] yaml_2.3.8 xfun_0.43 fastmap_1.1.1 httr_1.4.7
[25] stringr_1.5.1 knitr_1.46 htmlwidgets_1.6.4 generics_0.1.3
[29] fs_1.6.4 vctrs_0.6.5 sass_0.4.9 DT_0.33
[33] tidyselect_1.2.1 rprojroot_2.0.4 glue_1.7.0 R6_2.5.1
[37] processx_3.8.3 fansi_1.0.6 rmarkdown_2.26 callr_3.7.3
[41] magrittr_2.0.3 whisker_0.4.1 ps_1.7.6 promises_1.3.0
[45] htmltools_0.5.8.1 httpuv_1.6.14 utf8_1.2.4 stringi_1.7.6
[49] cachem_1.0.8