cTWAS_analysis_for_white_blood_counts
Jing Gu
2023-08-11
Last updated: 2023-08-11
Checks: 6 1
Knit directory: m6A_in_disease_genetics/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20230331) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| ~/projects/m6A_in_disease_genetics/code/ctwas/ctwas_config_b37.R | code/ctwas/ctwas_config_b37.R |
| ~/projects/m6A_in_disease_genetics/code/ctwas/qiansheng/locus_plot.R | code/ctwas/qiansheng/locus_plot.R |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version d1d6b2a. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .ipynb_checkpoints/
Ignored: analysis/figure/
Ignored: analysis/m6A_switch_to_disease_h2g.nb.html
Ignored: data/plots/
Untracked files:
Untracked: analysis/.ipynb_checkpoints/
Untracked: analysis/IBD_E_S_m6A.Rmd
Untracked: analysis/IBD_E_S_m6A_output.Rmd
Untracked: analysis/LDL_E_S_m6A.Rmd
Untracked: analysis/LDL_m6A_output.Rmd
Untracked: analysis/RA_m6A_output.Rmd
Untracked: analysis/WhiteBlood_WholeBlood_E_M.Rmd
Untracked: analysis/lymph_m6A_output.Rmd
Untracked: analysis/lymph_m6A_output_hg19.Rmd
Untracked: analysis/pre_weights_m6AQTL.txt
Untracked: analysis/rbc_E_S_m6A_output.Rmd
Untracked: analysis/rbc_m6A_output.Rmd
Untracked: analysis/wbc_E_S_m6A_output.Rmd
Untracked: code/.ipynb_checkpoints/
Untracked: code/all_m6a_sites_with_paired_cisNATs_summary.csv
Untracked: code/check_double_strand.ipynb
Untracked: code/check_double_strand_v2.ipynb
Untracked: code/ctwas/
Untracked: code/figure/
Untracked: code/learn_gviz.Rmd
Untracked: code/learn_gviz.html
Untracked: code/learn_gviz.nb.html
Untracked: code/m6AQTL_finemapping.Rmd
Untracked: code/summary_TWAS_coloc_m6A_2023.Rmd
Untracked: code/test_gviz.ipynb
Untracked: code/twas_genes_PP4_0.3_immune_traits_trackplots.pdf
Untracked: data/.ipynb_checkpoints/
Untracked: data/ADCY7_gwas_input.tsv
Untracked: data/ADCY7_qtl_input.tsv
Untracked: data/Allergy_full_coloc.txt
Untracked: data/Asthma_full_coloc.txt
Untracked: data/CAD_full_coloc.txt
Untracked: data/Eosinophil_count_full_coloc.txt
Untracked: data/GSE125377_jointPeakReadCount.txt
Untracked: data/IBD_full_coloc.txt
Untracked: data/JointPeaks.bed
Untracked: data/Li2022_dsRNAs.xlsx
Untracked: data/Lupus_full_coloc.txt
Untracked: data/RA_full_coloc.txt
Untracked: data/TABLE1_hg19.txt
Untracked: data/TABLE1_hg19.txt.zip
Untracked: data/__MACOSX/
Untracked: data/coloc_blood_traits.csv
Untracked: data/crohns_disease_full_coloc.txt
Untracked: data/edit_sites_and_GE_neg_correlated.txt
Untracked: data/edit_sites_and_GE_pos_correlated.txt
Untracked: data/features
Untracked: data/human_EERs.csv
Untracked: data/human_EERs.txt
Untracked: data/lymph_full_coloc.txt
Untracked: data/m6A_TWAS_results.csv
Untracked: data/m6a_TWAS_genes.txt
Untracked: data/m6a_joint_calling_peaks.csv
Untracked: data/nat_sense_pairs.csv
Untracked: data/plt_full_coloc.txt
Untracked: data/rbc_full_coloc.txt
Untracked: data/rdw_full_coloc.txt
Untracked: data/reported_AS_targets_S1.txt
Untracked: data/reported_AS_wanowska.txt
Untracked: data/sig_coloc_results/
Untracked: data/test_locuscomparer.pdf
Untracked: data/ulcerative_colitis_full_coloc.txt
Untracked: data/wbc_full_coloc.txt
Untracked: output/.ipynb_checkpoints/
Untracked: output/all_m6a_sites_with_cisNATs.csv
Untracked: output/all_m6a_sites_with_paired_cisNATs_summary.csv
Untracked: output/all_m6a_sites_with_paired_cisNATs_summary_PP40.3.csv
Untracked: output/all_m6a_sites_with_paired_cisNATs_summary_PP40.5.csv
Untracked: output/all_m6a_sites_with_paired_cis_NATs.csv
Untracked: output/fine_mapped_m6AQTLs_TWAS_genes_highPP4.rds
Untracked: output/gene_summary.csv
Untracked: output/immune_related_m6A_targets.csv
Untracked: output/m6aQTL_dsRNAs_PPP2R3C_PRORP.pdf
Untracked: output/m6a_peaks_nearby_dsRNAs.csv
Untracked: output/m6a_sites_near_all_dsRNAs_twas.csv
Untracked: output/m6a_sites_near_dsRNAs_coloc.csv
Untracked: output/m6a_sites_near_dsRNAs_twas.csv
Untracked: output/m6a_sites_near_dsRNAs_twas_summary.csv
Untracked: output/m6a_sites_overlapping_NAT_twas.csv
Untracked: output/m6a_sites_overlapping_dsRNAs_coloc.csv
Untracked: output/m6a_sites_overlapping_dsRNAs_twas.csv
Untracked: output/m6a_sites_overlapping_dsRegions.csv
Untracked: output/m6a_sites_overlapping_dsRegions_coloc.csv
Untracked: output/negatively_correlated_genes.txt
Untracked: output/postively_correlated_genes.txt
Untracked: output/rs1806261_RABEP1-NUP88_focused_locusview.pdf
Untracked: output/rs1806261_RABEP1-NUP88_locusview.pdf
Untracked: output/rs3177647_MAPKAPK5-AS1-MAPKAPK5_locusview.pdf
Untracked: output/rs3204541_DDX55-EIF2B1_locusview.pdf
Untracked: output/rs7184802_ADCY7-BRD7_locusview.pdf
Untracked: output/rs7184802_ADCY7_locuscompare.pdf
Untracked: output/twas_genes_PP4_0.3_immune_traits_trackplots.pdf
Untracked: output/twas_genes_PP4_0.5_blood_traits_trackplots.pdf
Untracked: output/twas_m6a_sites_with_all_cisNATs.RDS
Untracked: output/twas_m6a_sites_with_cisNATs_range.RDS
Untracked: output/twas_m6a_sites_with_the_nearest_cisNAT.RDS
Untracked: twas_genes_PP4_0.3_immune_traits_trackplots.pdf
Unstaged changes:
Modified: analysis/m6A_switch_to_disease_h2g.Rmd
Modified: analysis/wbc_m6A_output.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/wbc_m6A_output_hg19.Rmd)
and HTML (docs/wbc_m6A_output_hg19.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | d1d6b2a | Jing Gu | 2023-08-11 | wflow_publish(c("analysis/wbc_m6A_output_hg19.Rmd", "analysis/index.Rmd", |
| Rmd | c94ee10 | Jing Gu | 2023-08-11 | wflow_publish(c("analysis/wbc_m6A_output_hg19.Rmd", "analysis/index.Rmd", |
Load ctwas results
# top 1 method
res <- impute_expr_z(z_snp, weight = weight, ld_R_dir = ld_R_dir,
method = NULL, outputdir = outputdir, outname = outname.e,
harmonize_z = T, harmonize_wgt = T, scale_by_ld_variance=F,
strand_ambig_action_z = "recover",
recover_strand_ambig_wgt = T
# lasso/elastic-net method
res <- impute_expr_z(z_snp, weight = weight, ld_R_dir = ld_R_dir,
method = NULL, outputdir = outputdir, outname = outname.e,
harmonize_z = T, harmonize_wgt = T, scale_by_ld_variance=F,
strand_ambig_action_z = "none",
recover_strand_ambig_wgt = FGWAS: UK Biobank GWAS summary statistics - European individuals
Weights: FUSION weights using top1, lasso, or elastic-net models were converted into PredictDB format and were not needed to do scaling when running ctwas.
Check convergence of parameters
cTWAS analysis on m6A alone
[1] "Check convergence for the top1 model:"
[1] "Table of group size:"
SNP gene
8713250 888
SNP gene
estimated_group_prior 2.481e-04 1.227e-02
estimated_group_prior_var 1.920e+01 2.631e+01
estimated_group_pve 1.184e-01 8.178e-04
attributable_group_pve 9.931e-01 6.858e-03
[1] "Check convergence for the lasso model:"
[1] "Table of group size:"
SNP gene
8713250 912
SNP gene
estimated_group_prior 2.414e-04 1.016e-02
estimated_group_prior_var 1.898e+01 3.699e+01
estimated_group_pve 1.139e-01 9.778e-04
attributable_group_pve 9.915e-01 8.513e-03$top1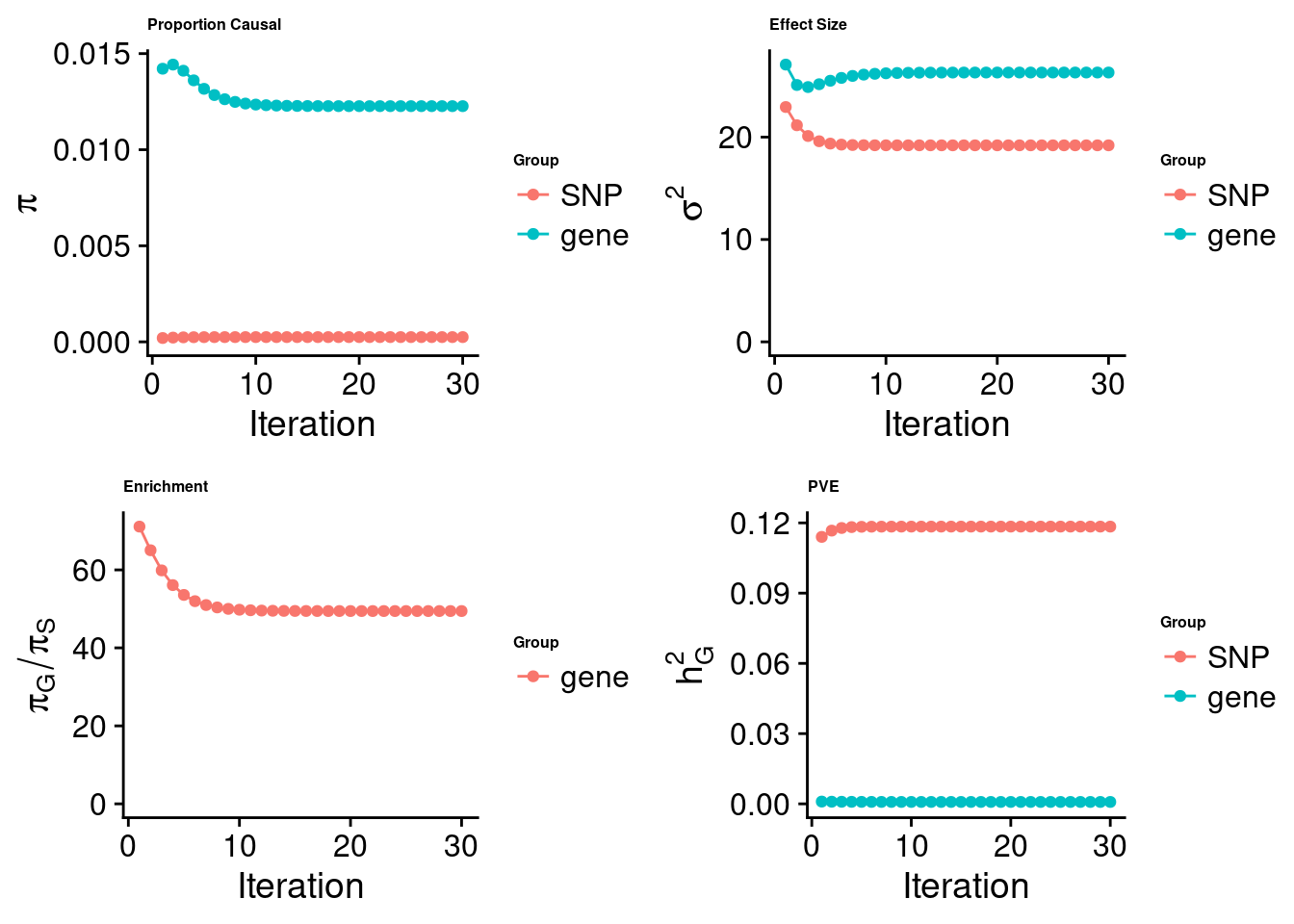
$lasso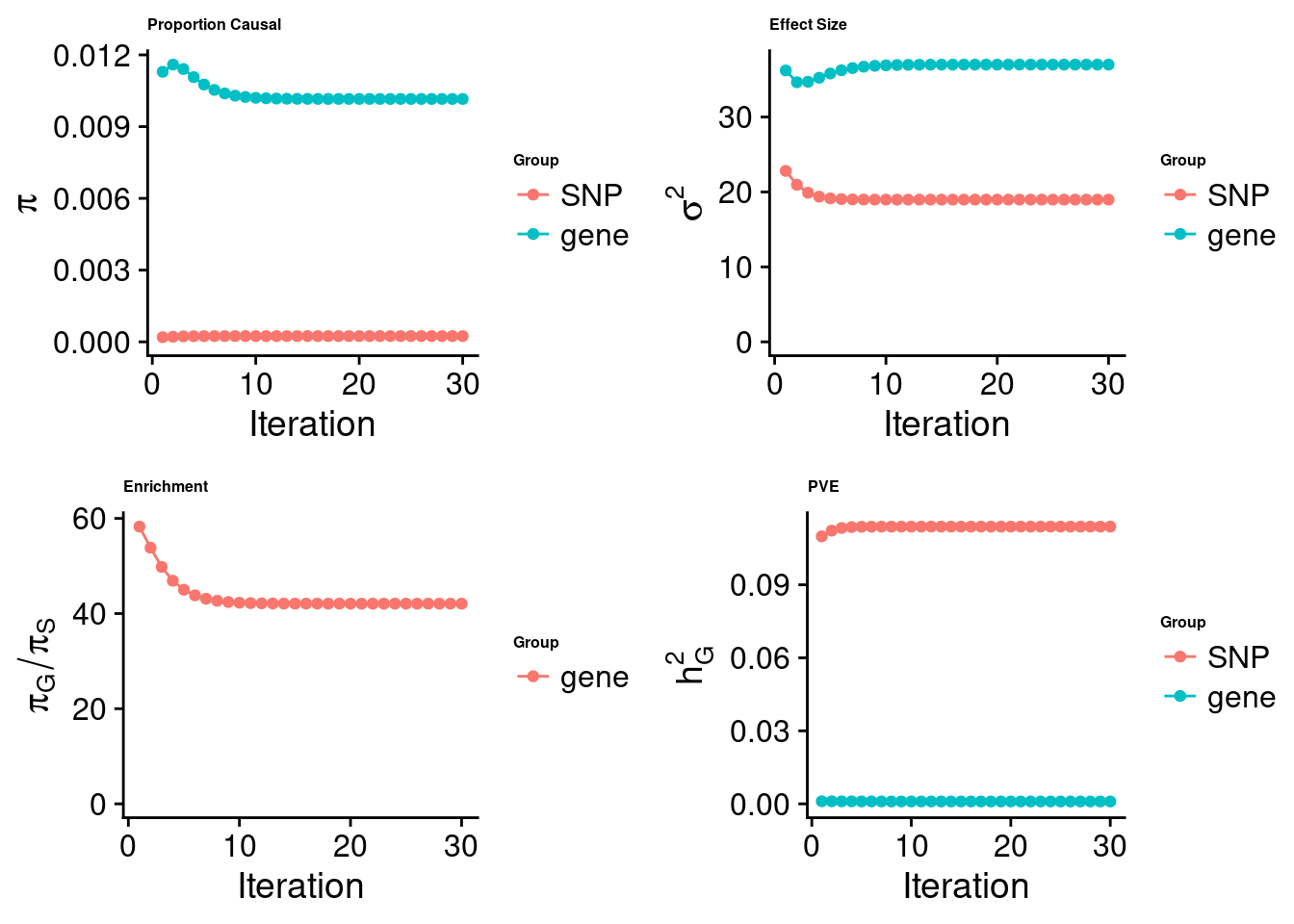
Joint analysis of expression, splicing and m6A
[1] "Check convergence for the top1 model when jointly analyzing expression, splicing and m6A:"
[1] "Table of group size before/after matching with UKBB SNPs:"
SNP eQTL sQTL m6AQTL
prior_group_size 9.324e+06 2005.0000 2191.000 918.0000
group_size 8.713e+06 1928.0000 2123.000 888.0000
percent_of_overlaps 9.345e-01 0.9616 0.969 0.9673
SNP eQTL sQTL m6AQTL
estimated_group_prior 2.406e-04 8.895e-03 0.012934 1.236e-02
estimated_group_prior_var 1.858e+01 1.683e+01 36.589120 2.554e+01
estimated_group_pve 1.112e-01 8.236e-04 0.002867 7.999e-04
attributable_group_pve 9.612e-01 7.120e-03 0.024783 6.916e-03$top1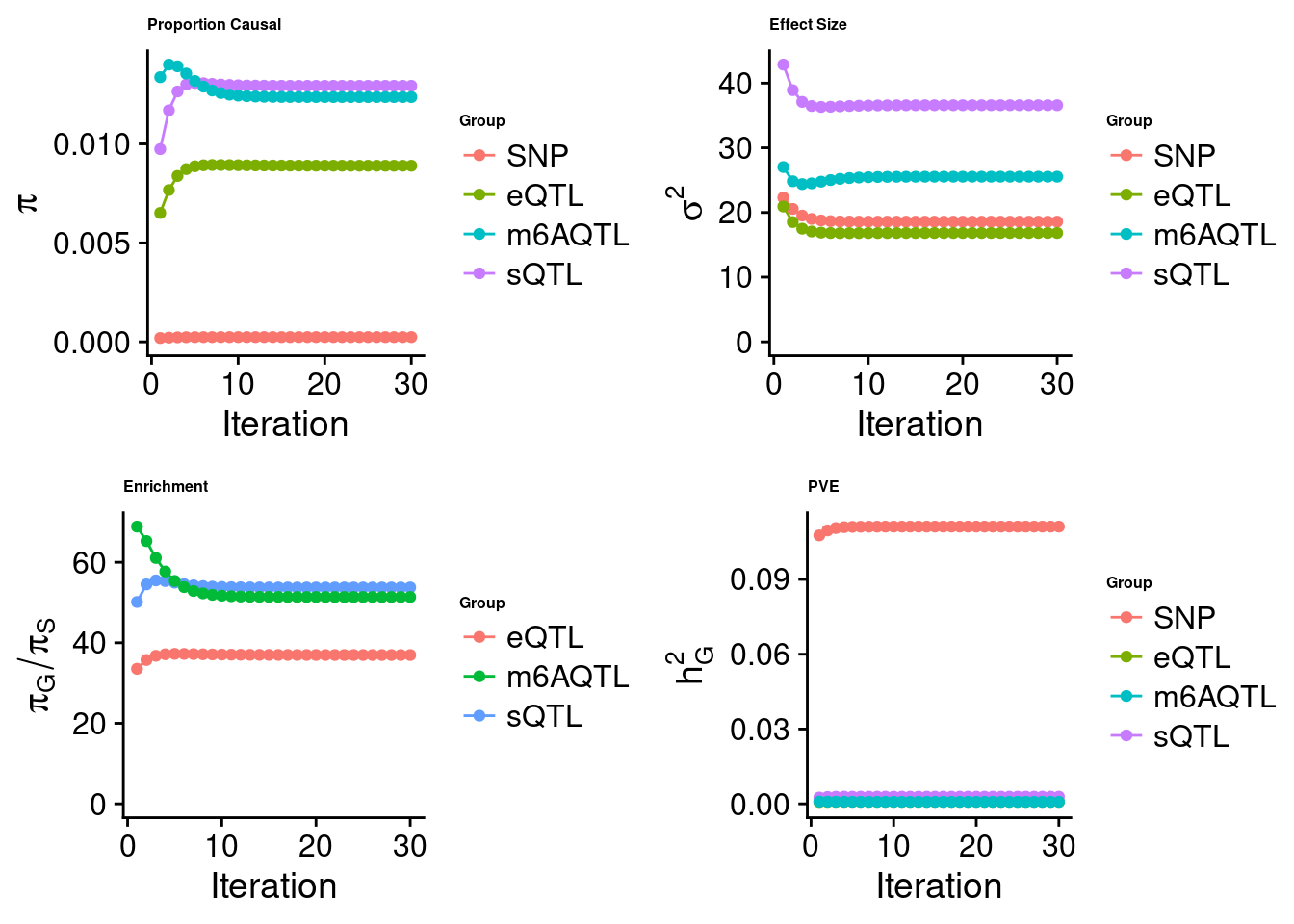
cTWAS results from individual analysis on m6A
Lasso model
Top m6A modification pip
genename region_tag susie_pip z
1 SLC9A3R1 17_42 0.9473 -7.630
2 ZKSCAN5 7_61 0.7976 7.112
3 ADCY7 16_27 0.7817 4.382
4 TRIT1 1_25 0.7516 5.554
5 THEMIS2 1_19 0.7034 6.243
6 BTN3A3 6_20 0.6855 -13.445
7 WAC-AS1 10_20 0.6102 11.178Summing up PIPs for m6A peaks located in the same gene
Top m6A PIPs by genes
# A tibble: 7 × 2
genename total_susie_pip
<chr> <dbl>
1 SLC9A3R1 0.947
2 ZKSCAN5 0.798
3 ADCY7 0.782
4 TRIT1 0.752
5 THEMIS2 0.703
6 BTN3A3 0.686
7 WAC-AS1 0.615Top expression/splicing/m6A units
For m6A or splicing QTLs, they are assigned to the nearest genes (m6A needs to be confirmed with Kevin).
Top SNPs or genes with PIP > 0.6
$eQTL
genename susie_pip group region_tag
1913 CSNK1G1 0.9587 eQTL 15_29
1916 RAPGEFL1 0.7362 eQTL 17_23
132 NDUFS2 0.6213 eQTL 1_81
$m6AQTL
genename susie_pip group region_tag
4938 SLC9A3R1 0.9539 m6AQTL 17_42
4922 ZKSCAN5 0.7863 m6AQTL 7_61
4067 THEMIS2 0.7816 m6AQTL 1_19
4312 TRAM2 0.7091 m6AQTL 6_39
$sQTL
genename susie_pip group region_tag
4000 RNF181 1.0000 sQTL 2_54
4011 MYO1G 0.9932 sQTL 7_33
2385 GSK3B 0.7962 sQTL 3_74
4033 PDLIM1 0.7387 sQTL 10_61
3788 ZNF428 0.6957 sQTL 19_30Top m6A modification pip
ZKSCAN5: RNA Polymerase II Cis-Regulatory Region Sequence-Specific DNA Binding (GO:0000978). THEMIS2 is involved in the biological process T Cell Receptor Signaling Pathway (GO:0050852). BANF: DNA binding factor|Regulation Of Innate Immune Response (GO:0045088). TRIT1 has the molecular function of Catalytic Activity, Acting On A tRNA (GO:0140101). TRIT1 is involved in the biological process RNA Modification (GO:0009451). S1PR2 is involved in the biological process Regulation Of Cell Population Proliferation (GO:0042127). WAC has the molecular function of RNA Polymerase II Complex Binding (GO:0000993). CD320 is involved in the biological process Regulation Of B Cell Proliferation (GO:0030888).
genename region_tag susie_pip z
1 SLC9A3R1 17_42 0.9539 -7.630
2 ZKSCAN5 7_61 0.7863 7.158
3 THEMIS2 1_19 0.7816 6.277
4 TRAM2 6_39 0.7091 5.233
5 BANF1 11_36 0.5786 6.174
6 TRIT1 1_25 0.5278 5.298
7 S1PR2 19_9 0.5220 9.939
8 WAC-AS1 10_20 0.4945 11.169
9 SQSTM1 5_108 0.4934 -4.857
10 CD320 19_8 0.3627 -4.062Summing up PIPs for m6A peaks located in the same gene
Top 10 m6A PIPs by genes
# A tibble: 800 × 2
genename total_susie_pip
<chr> <dbl>
1 SLC9A3R1 0.954
2 ZKSCAN5 0.786
3 THEMIS2 0.782
4 TRAM2 0.709
5 BANF1 0.579
6 TRIT1 0.528
7 S1PR2 0.522
8 WAC-AS1 0.513
9 SQSTM1 0.493
10 CD320 0.379
# ℹ 790 more rowsTop splicing PIPs
Some loci contain variants in the same credible set but having opposite z scores. For instance, the predicted splicing levels of two introns of CNN2 based on the same variant (position=1038445) have opposite associations with traits. Is this variant more likely to affect traits by altering the splicing levels of both transcripts, rather than one of them since they have equal PIP?
peak_id genename pos region_tag susie_pip z
1 chr2:85823772-85824227 RNF181 85818886 2_54 1.0000 5.009
2 chr7:45009474-45009639 MYO1G 45009341 7_33 0.9932 -11.719
3 chr3:119582452-119624602 GSK3B 119542297 3_74 0.7962 5.622
4 chr10:97007123-97023621 PDLIM1 97023552 10_61 0.7387 -7.331
5 chr19:44112259-44118381 ZNF428 44146930 19_30 0.6957 -4.929
6 chr5:122111457-122130961 SNX2 122088686 5_74 0.5628 -6.687
7 chr11:67120548-67124214 AP003419.1 67185596 11_37 0.5416 -4.432
8 chr6:29693820-29694660 HLA-F 29688501 6_23 0.5000 -16.046
9 chr6:29694781-29695734 HLA-F 29688501 6_23 0.5000 -16.046
10 chr11:47761655-47765505 FNBP4 47863119 11_29 0.4878 10.101
11 chr19:13885521-13886291 C19orf53 13942221 19_11 0.4797 6.500
12 chr19:13886427-13888866 C19orf53 13942221 19_11 0.4797 6.500
13 chr7:72986365-72987174 TBL2 72989390 7_47 0.4741 6.870
14 chr1:224544695-224548197 CNIH4 224630695 1_116 0.4698 8.830
15 chr19:1036561-1037624 CNN2 1038445 19_2 0.4643 6.170
16 chr19:1036999-1037624 CNN2 1038445 19_2 0.4643 -6.170
17 chr17:47288203-47295101 ABI3 47287067 17_28 0.4626 -4.041
18 chr19:49458856-49459455 BAX 49459104 19_34 0.4605 -4.118
19 chr7:5569315-5570155 ACTB 5556807 7_7 0.4288 -4.696
20 chr16:67690548-67690704 CARMIL2 67780829 16_36 0.3912 -3.955Summing up PIPs for spliced introns located in the same gene
Top 10 splicing PIPs by genes
# A tibble: 10 × 2
genename total_susie_pip
<chr> <dbl>
1 HLA-F 1.01
2 RNF181 1.00
3 MYO1G 0.993
4 C19orf53 0.959
5 CNN2 0.929
6 CD46 0.904
7 HNRNPK 0.811
8 GSK3B 0.796
9 ZNF428 0.748
10 PDLIM1 0.739Top genes by combined PIP
genename combined_pip expression_pip splicing_pip m6A_pip region_tag
1456 HLA-F 1.006 6.563e-06 1.0057 1.512e-05 6_23
2437 RNF181 1.000 0.000e+00 1.0000 0.000e+00 2_54
1891 MYO1G 0.993 0.000e+00 0.9932 0.000e+00 7_33
349 C19orf53 0.982 0.000e+00 0.9594 2.284e-02 19_11
605 CSNK1G1 0.969 9.587e-01 0.0000 1.015e-02 15_29
2656 SLC9A3R1 0.954 0.000e+00 0.0000 9.539e-01 17_42
1468 HNRNPK 0.950 0.000e+00 0.8109 1.388e-01 9_41
548 CNN2 0.929 0.000e+00 0.9287 0.000e+00 19_2
454 CD46 0.904 0.000e+00 0.9044 0.000e+00 1_107
1392 GSK3B 0.796 0.000e+00 0.7962 0.000e+00 3_74
3218 ZKSCAN5 0.786 0.000e+00 0.0000 7.863e-01 7_61
2868 THEMIS2 0.782 0.000e+00 0.0000 7.816e-01 1_19
2986 TRAM2 0.755 4.565e-02 0.0000 7.091e-01 6_39
3252 ZNF428 0.748 0.000e+00 0.7475 0.000e+00 19_30
2114 PDLIM1 0.739 0.000e+00 0.7387 0.000e+00 10_61
2360 RAPGEFL1 0.736 7.362e-01 0.0000 0.000e+00 17_23
285 BCL2A1 0.706 0.000e+00 0.7064 0.000e+00 15_37
547 CNIH4 0.690 0.000e+00 0.6904 0.000e+00 1_116
277 BAX 0.653 0.000e+00 0.6532 0.000e+00 19_34
1944 NDUFS2 0.621 6.213e-01 0.0000 0.000e+00 1_81Loading required package: gridWarning: replacing previous import 'utils::download.file' by
'restfulr::download.file' when loading 'rtracklayer'Locus plots for specific examples
genename combined_pip expression_pip splicing_pip m6A_pip region_tag
1944 NDUFS2 0.621 0.6213 0 0 1_81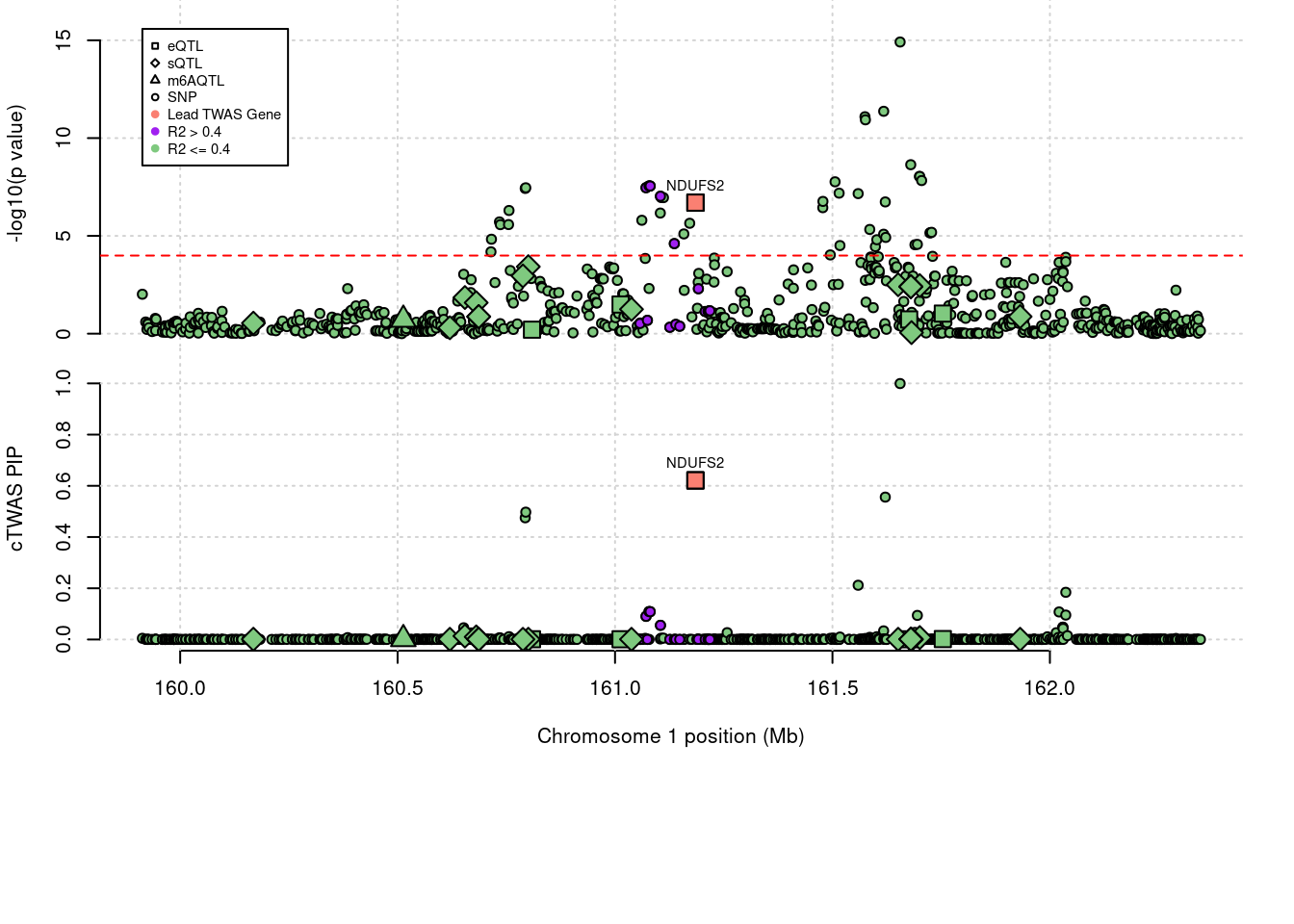
genename combined_pip expression_pip splicing_pip m6A_pip region_tag
605 CSNK1G1 0.969 0.9587 0 0.01015 15_29Warning in asMethod(object): sparse->dense coercion: allocating vector of size
1.1 GiB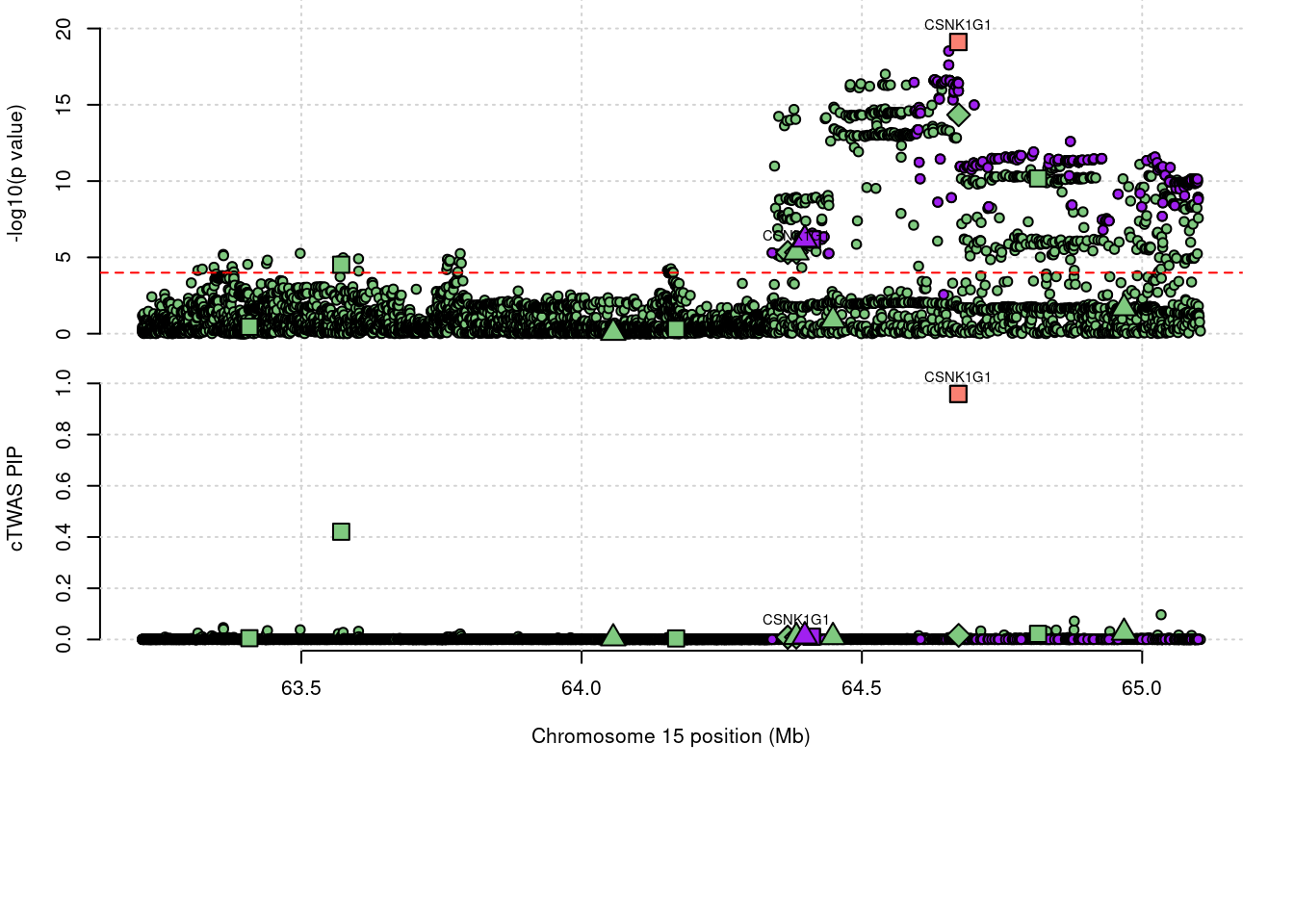
genename combined_pip expression_pip splicing_pip m6A_pip region_tag
2868 THEMIS2 0.782 0 0 0.7816 1_19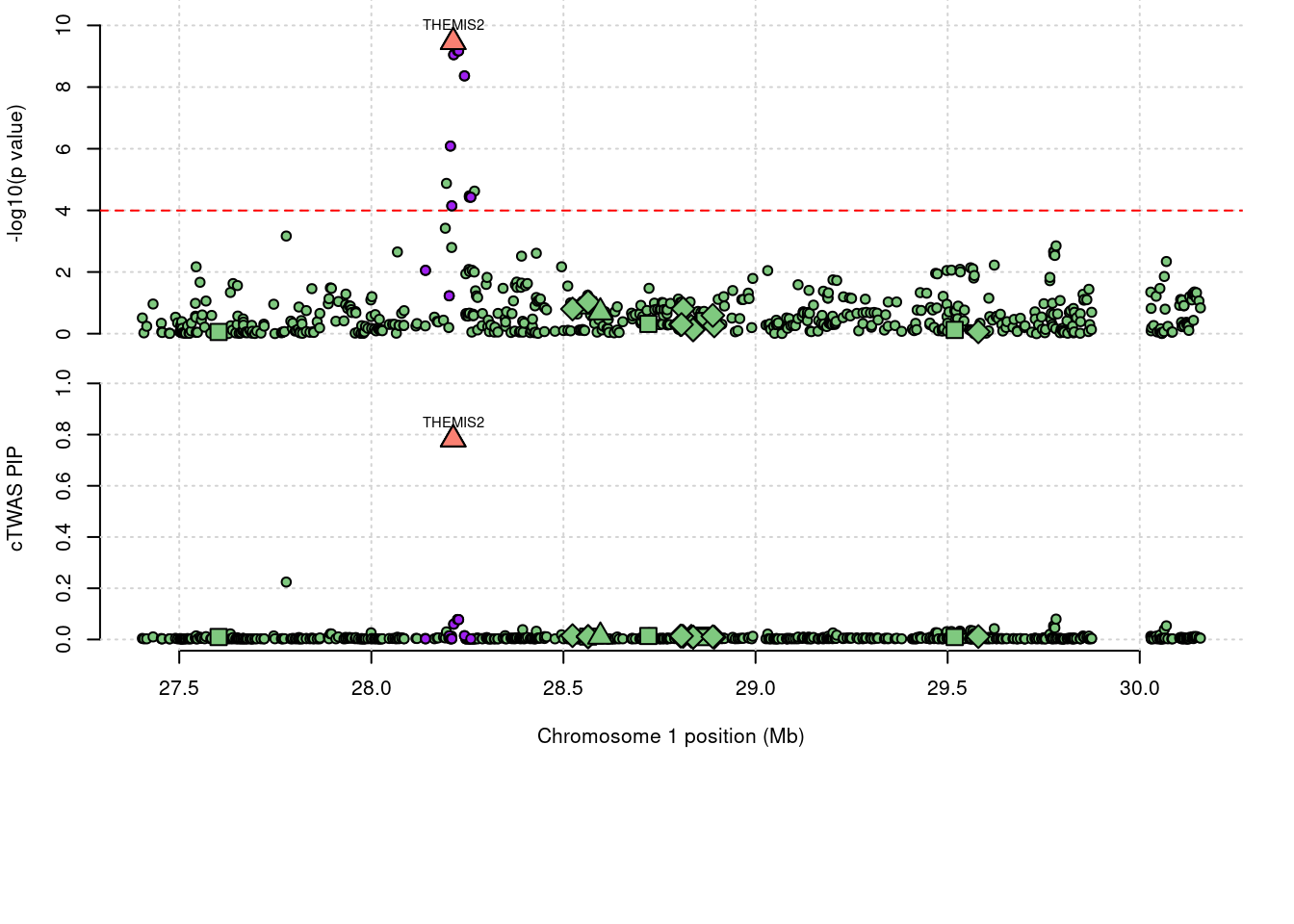
genename combined_pip expression_pip splicing_pip m6A_pip region_tag
3218 ZKSCAN5 0.786 0 0 0.7863 7_61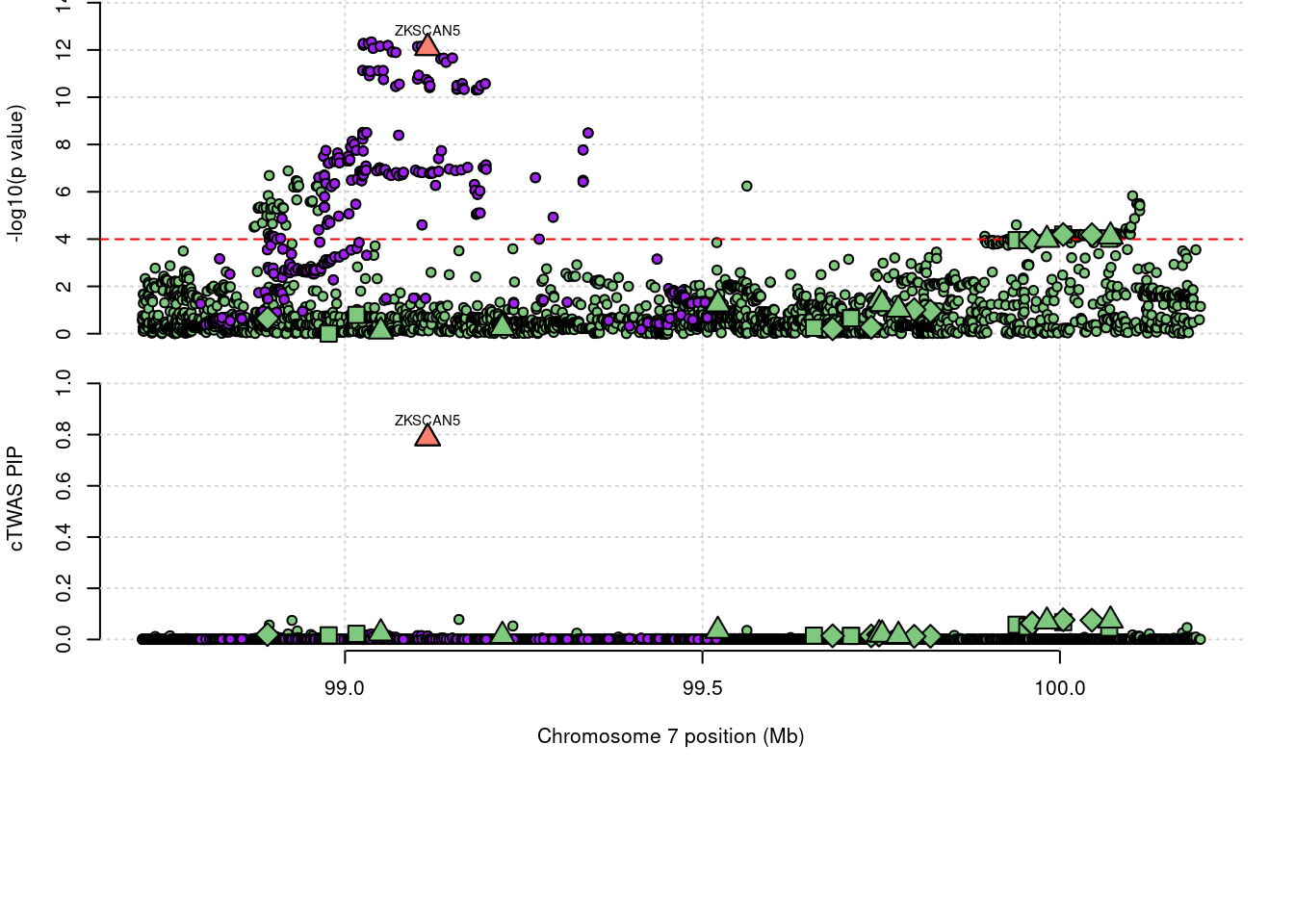
R version 4.2.0 (2022-04-22)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.3.13-el7-x86_64/lib/libopenblas_haswellp-r0.3.13.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=C
[4] LC_COLLATE=C LC_MONETARY=C LC_MESSAGES=C
[7] LC_PAPER=C LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=C LC_IDENTIFICATION=C
attached base packages:
[1] grid stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] Gviz_1.40.1 cowplot_1.1.1 ggplot2_3.4.2
[4] GenomicRanges_1.48.0 GenomeInfoDb_1.32.2 IRanges_2.30.1
[7] S4Vectors_0.34.0 BiocGenerics_0.42.0 ctwas_0.1.38
[10] dplyr_1.1.2 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] colorspace_2.1-0 deldir_1.0-6
[3] rjson_0.2.21 rprojroot_2.0.3
[5] biovizBase_1.44.0 htmlTable_2.4.0
[7] XVector_0.36.0 base64enc_0.1-3
[9] fs_1.6.3 dichromat_2.0-0.1
[11] rstudioapi_0.15.0 farver_2.1.1
[13] bit64_4.0.5 AnnotationDbi_1.58.0
[15] fansi_1.0.4 xml2_1.3.3
[17] codetools_0.2-18 logging_0.10-108
[19] cachem_1.0.8 knitr_1.39
[21] Formula_1.2-4 jsonlite_1.8.7
[23] Rsamtools_2.12.0 cluster_2.1.3
[25] dbplyr_2.3.3 png_0.1-7
[27] compiler_4.2.0 httr_1.4.6
[29] backports_1.4.1 lazyeval_0.2.2
[31] Matrix_1.6-0 fastmap_1.1.1
[33] cli_3.6.1 later_1.3.0
[35] htmltools_0.5.2 prettyunits_1.1.1
[37] tools_4.2.0 gtable_0.3.3
[39] glue_1.6.2 GenomeInfoDbData_1.2.8
[41] rappdirs_0.3.3 Rcpp_1.0.11
[43] Biobase_2.56.0 jquerylib_0.1.4
[45] vctrs_0.6.3 Biostrings_2.64.0
[47] rtracklayer_1.56.0 iterators_1.0.14
[49] xfun_0.30 stringr_1.5.0
[51] ps_1.7.0 lifecycle_1.0.3
[53] ensembldb_2.20.2 restfulr_0.0.14
[55] XML_3.99-0.14 getPass_0.2-2
[57] zlibbioc_1.42.0 scales_1.2.1
[59] BSgenome_1.64.0 VariantAnnotation_1.42.1
[61] ProtGenerics_1.28.0 hms_1.1.3
[63] promises_1.2.0.1 MatrixGenerics_1.8.0
[65] parallel_4.2.0 SummarizedExperiment_1.26.1
[67] AnnotationFilter_1.20.0 RColorBrewer_1.1-3
[69] yaml_2.3.5 curl_5.0.1
[71] memoise_2.0.1 gridExtra_2.3
[73] sass_0.4.1 biomaRt_2.52.0
[75] rpart_4.1.16 latticeExtra_0.6-30
[77] stringi_1.7.12 RSQLite_2.3.1
[79] highr_0.9 BiocIO_1.6.0
[81] foreach_1.5.2 checkmate_2.1.0
[83] GenomicFeatures_1.48.4 filelock_1.0.2
[85] BiocParallel_1.30.3 rlang_1.1.1
[87] pkgconfig_2.0.3 matrixStats_0.62.0
[89] bitops_1.0-7 evaluate_0.15
[91] lattice_0.20-45 htmlwidgets_1.5.4
[93] GenomicAlignments_1.32.0 labeling_0.4.2
[95] bit_4.0.5 processx_3.8.0
[97] tidyselect_1.2.0 magrittr_2.0.3
[99] R6_2.5.1 generics_0.1.3
[101] Hmisc_5.1-0 DelayedArray_0.22.0
[103] DBI_1.1.3 pgenlibr_0.3.6
[105] pillar_1.9.0 whisker_0.4
[107] foreign_0.8-82 withr_2.5.0
[109] KEGGREST_1.36.2 RCurl_1.98-1.7
[111] nnet_7.3-17 tibble_3.2.1
[113] crayon_1.5.2 interp_1.1-4
[115] utf8_1.2.3 BiocFileCache_2.4.0
[117] rmarkdown_2.14 jpeg_0.1-10
[119] progress_1.2.2 data.table_1.14.8
[121] blob_1.2.4 callr_3.7.3
[123] git2r_0.30.1 digest_0.6.33
[125] httpuv_1.6.5 munsell_0.5.0
[127] bslib_0.3.1