cTWAS_analysis_for_IBD
Jing Gu
2023-08-16
Last updated: 2023-08-16
Checks: 6 1
Knit directory: m6A_in_disease_genetics/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20230331) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| ~/projects/m6A_in_disease_genetics/code/ctwas/ctwas_config_b37.R | code/ctwas/ctwas_config_b37.R |
| ~/projects/m6A_in_disease_genetics/data/nasser_2021_ABC_IBD_genes.txt | data/nasser_2021_ABC_IBD_genes.txt |
| ~/projects/m6A_in_disease_genetics/data/m6A_TWAS_results.csv | data/m6A_TWAS_results.csv |
| ~/projects/m6A_in_disease_genetics/code/ctwas/qiansheng/locus_plot.R | code/ctwas/qiansheng/locus_plot.R |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 08eaf44. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .ipynb_checkpoints/
Ignored: analysis/m6A_switch_to_disease_h2g.nb.html
Ignored: data/plots/
Untracked files:
Untracked: HMGCR_locus_gene_tracks.pdf
Untracked: Rplots.pdf
Untracked: analysis/.ipynb_checkpoints/
Untracked: analysis/IBD_E_S_m6A.Rmd
Untracked: analysis/IBD_E_S_m6A_output.Rmd
Untracked: analysis/LDL_E_S_m6A.Rmd
Untracked: analysis/LDL_m6A_output.Rmd
Untracked: analysis/RA_m6A_output.Rmd
Untracked: analysis/WhiteBlood_WholeBlood_E_M.Rmd
Untracked: analysis/identify_m6A_mechanisms_with_finemapping.Rmd
Untracked: analysis/lymph_m6A_output.Rmd
Untracked: analysis/pre_weights_m6AQTL.txt
Untracked: analysis/rbc_E_S_m6A_output.Rmd
Untracked: analysis/rbc_m6A_output.Rmd
Untracked: analysis/rbc_m6A_output_hg19.Rmd
Untracked: analysis/wbc_E_S_m6A_output.Rmd
Untracked: code/.ipynb_checkpoints/
Untracked: code/all_m6a_sites_with_paired_cisNATs_summary.csv
Untracked: code/check_double_strand.ipynb
Untracked: code/check_double_strand_v2.ipynb
Untracked: code/ctwas/
Untracked: code/figure/
Untracked: code/learn_gviz.Rmd
Untracked: code/learn_gviz.html
Untracked: code/learn_gviz.nb.html
Untracked: code/m6AQTL_finemapping.Rmd
Untracked: code/summary_TWAS_coloc_m6A_2023.Rmd
Untracked: code/test_gviz.ipynb
Untracked: code/twas_genes_PP4_0.3_immune_traits_trackplots.pdf
Untracked: data/.ipynb_checkpoints/
Untracked: data/ADCY7_gwas_input.tsv
Untracked: data/ADCY7_qtl_input.tsv
Untracked: data/Allergy_full_coloc.txt
Untracked: data/Asthma_full_coloc.txt
Untracked: data/CAD_full_coloc.txt
Untracked: data/Eosinophil_count_full_coloc.txt
Untracked: data/GSE125377_jointPeakReadCount.txt
Untracked: data/HMGCR_ctwas_dat.Rd
Untracked: data/IBD_full_coloc.txt
Untracked: data/JointPeaks.bed
Untracked: data/Li2022_dsRNAs.xlsx
Untracked: data/Lupus_full_coloc.txt
Untracked: data/RA_full_coloc.txt
Untracked: data/TABLE1_hg19.txt
Untracked: data/TABLE1_hg19.txt.zip
Untracked: data/__MACOSX/
Untracked: data/coloc_blood_traits.csv
Untracked: data/crohns_disease_full_coloc.txt
Untracked: data/edit_sites_and_GE_neg_correlated.txt
Untracked: data/edit_sites_and_GE_pos_correlated.txt
Untracked: data/features
Untracked: data/human_EERs.csv
Untracked: data/human_EERs.txt
Untracked: data/lymph_full_coloc.txt
Untracked: data/m6A_TWAS_results.csv
Untracked: data/m6a_TWAS_genes.txt
Untracked: data/m6a_joint_calling_peaks.csv
Untracked: data/nasser_2021_ABC_IBD_genes.txt
Untracked: data/nat_sense_pairs.csv
Untracked: data/plt_full_coloc.txt
Untracked: data/rbc_full_coloc.txt
Untracked: data/rdw_full_coloc.txt
Untracked: data/reported_AS_targets_S1.txt
Untracked: data/reported_AS_wanowska.txt
Untracked: data/sig_coloc_results/
Untracked: data/test_locuscomparer.pdf
Untracked: data/ulcerative_colitis_full_coloc.txt
Untracked: data/wbc_full_coloc.txt
Untracked: data/zhao_silver_genes.csv
Untracked: output/.ipynb_checkpoints/
Untracked: output/HMGCR_gene_track_plot.pdf
Untracked: output/HMGCR_locus_plot.pdf
Untracked: output/all_m6a_sites_with_cisNATs.csv
Untracked: output/all_m6a_sites_with_paired_cisNATs_summary.csv
Untracked: output/all_m6a_sites_with_paired_cisNATs_summary_PP40.3.csv
Untracked: output/all_m6a_sites_with_paired_cisNATs_summary_PP40.5.csv
Untracked: output/all_m6a_sites_with_paired_cis_NATs.csv
Untracked: output/fine_mapped_m6AQTLs_TWAS_genes_highPP4.rds
Untracked: output/gene_summary.csv
Untracked: output/immune_related_m6A_targets.csv
Untracked: output/m6aQTL_dsRNAs_PPP2R3C_PRORP.pdf
Untracked: output/m6a_peaks_nearby_dsRNAs.csv
Untracked: output/m6a_sites_near_all_dsRNAs_twas.csv
Untracked: output/m6a_sites_near_dsRNAs_coloc.csv
Untracked: output/m6a_sites_near_dsRNAs_twas.csv
Untracked: output/m6a_sites_near_dsRNAs_twas_summary.csv
Untracked: output/m6a_sites_overlapping_NAT_twas.csv
Untracked: output/m6a_sites_overlapping_dsRNAs_coloc.csv
Untracked: output/m6a_sites_overlapping_dsRNAs_twas.csv
Untracked: output/m6a_sites_overlapping_dsRegions.csv
Untracked: output/m6a_sites_overlapping_dsRegions_coloc.csv
Untracked: output/negatively_correlated_genes.txt
Untracked: output/postively_correlated_genes.txt
Untracked: output/rs1806261_RABEP1-NUP88_focused_locusview.pdf
Untracked: output/rs1806261_RABEP1-NUP88_locusview.pdf
Untracked: output/rs3177647_MAPKAPK5-AS1-MAPKAPK5_locusview.pdf
Untracked: output/rs3204541_DDX55-EIF2B1_locusview.pdf
Untracked: output/rs7184802_ADCY7-BRD7_locusview.pdf
Untracked: output/rs7184802_ADCY7_locuscompare.pdf
Untracked: output/twas_genes_PP4_0.3_immune_traits_trackplots.pdf
Untracked: output/twas_genes_PP4_0.5_blood_traits_trackplots.pdf
Untracked: output/twas_m6a_sites_with_all_cisNATs.RDS
Untracked: output/twas_m6a_sites_with_cisNATs_range.RDS
Untracked: output/twas_m6a_sites_with_the_nearest_cisNAT.RDS
Untracked: twas_genes_PP4_0.3_immune_traits_trackplots.pdf
Unstaged changes:
Modified: analysis/m6A_switch_to_disease_h2g.Rmd
Modified: analysis/wbc_m6A_output.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/IBD_m6A_output_hg19.Rmd)
and HTML (docs/IBD_m6A_output_hg19.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 08eaf44 | Jing Gu | 2023-08-16 | run ctwas for multiple traits |
Load ctwas results
# top 1 method
res <- impute_expr_z(z_snp, weight = weight, ld_R_dir = ld_R_dir,
method = NULL, outputdir = outputdir, outname = outname.e,
harmonize_z = T, harmonize_wgt = T, scale_by_ld_variance=F,
strand_ambig_action_z = "recover",
recover_strand_ambig_wgt = T
# lasso/elastic-net method
res <- impute_expr_z(z_snp, weight = weight, ld_R_dir = ld_R_dir,
method = NULL, outputdir = outputdir, outname = outname.e,
harmonize_z = T, harmonize_wgt = T, scale_by_ld_variance=F,
strand_ambig_action_z = "none",
recover_strand_ambig_wgt = FGWAS: UK Biobank GWAS summary statistics - European individuals
Weights: FUSION weights using top1, lasso, or elastic-net models were converted into PredictDB format and were not needed to do scaling when running ctwas.
Check convergence of parameters
cTWAS analysis on m6A alone
[1] "Check convergence for the top1 model:"
[1] "Table of group size:"
SNP gene
7549540 843
SNP gene
estimated_group_prior 0.000248 0.0094812
estimated_group_prior_var 9.409679 7.2407510
estimated_group_pve 0.293787 0.0009652
attributable_group_pve 0.996725 0.0032748$top1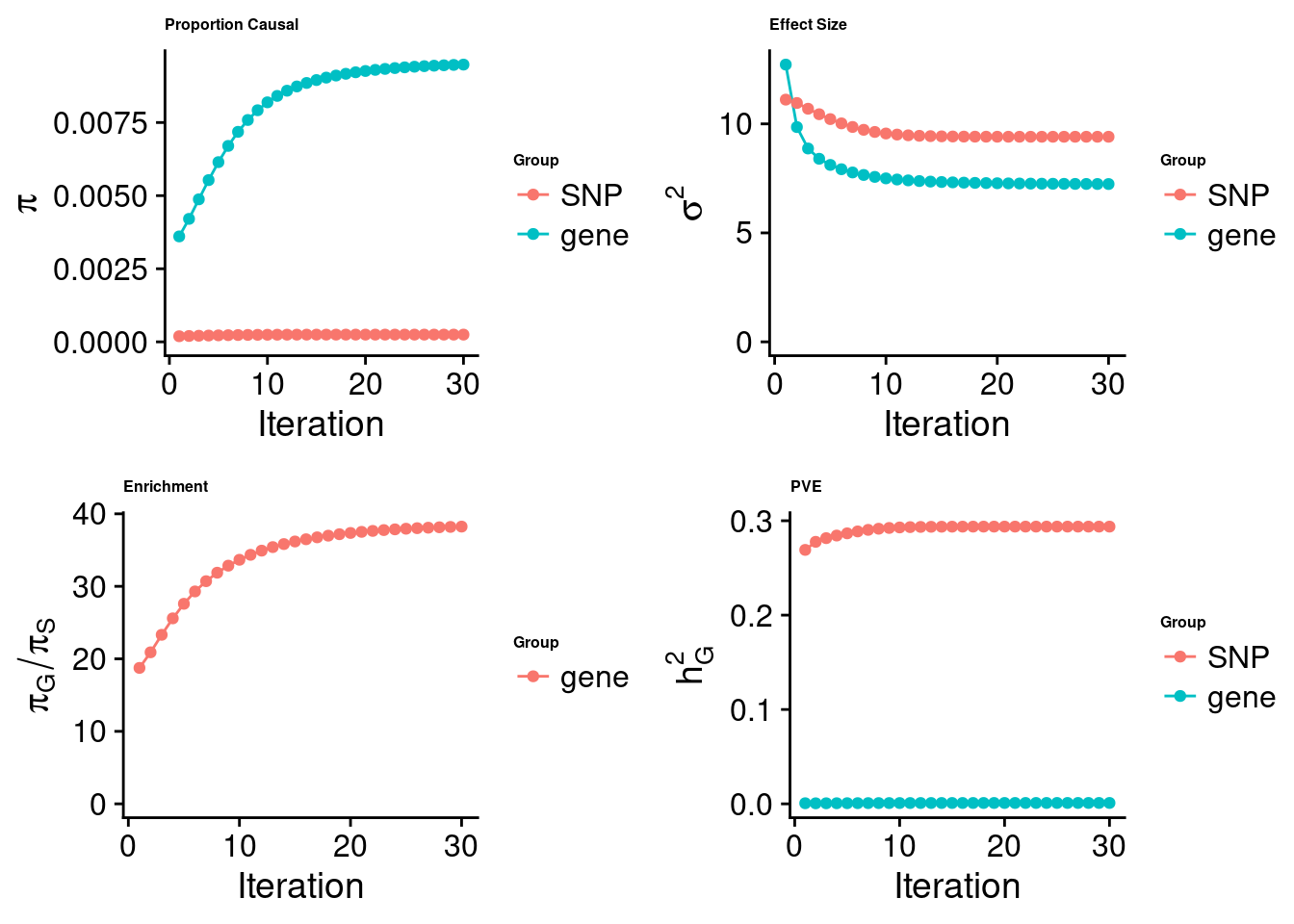
Joint analysis of expression, splicing and m6A
[1] "Check convergence for the top1 model when jointly analyzing expression, splicing and m6A:"
[1] "Table of group size before/after matching with UKBB SNPs:"
SNP eQTL sQTL m6AQTL
prior_group_size 9.324e+06 2005.0000 2191.0000 918.0000
group_size 7.550e+06 1834.0000 2039.0000 843.0000
percent_of_overlaps 8.097e-01 0.9147 0.9306 0.9183
SNP eQTL sQTL m6AQTL
estimated_group_prior 0.0002292 0.04629 0.001540 0.0061797
estimated_group_prior_var 9.2477617 7.45709 26.327399 7.4376303
estimated_group_pve 0.2669012 0.01056 0.001379 0.0006462
attributable_group_pve 0.9549740 0.03778 0.004933 0.0023122
[1] "Check convergence for the lasso model when jointly analyzing expression, splicing and m6A:"
[1] "Table of group size before/after matching with UKBB SNPs:"
SNP eQTL sQTL m6AQTL
prior_group_size 9.324e+06 2005.000 2191.0000 918.0000
group_size 7.550e+06 1993.000 2168.0000 906.0000
percent_of_overlaps 8.097e-01 0.994 0.9895 0.9869
SNP eQTL sQTL m6AQTL
estimated_group_prior 2.023e-04 0.031993 3.557e-05 0.019938
estimated_group_prior_var 1.037e+01 6.651734 3.954e+00 7.688136
estimated_group_pve 2.640e-01 0.007074 5.085e-06 0.002316
attributable_group_pve 9.656e-01 0.025873 1.860e-05 0.008472$top1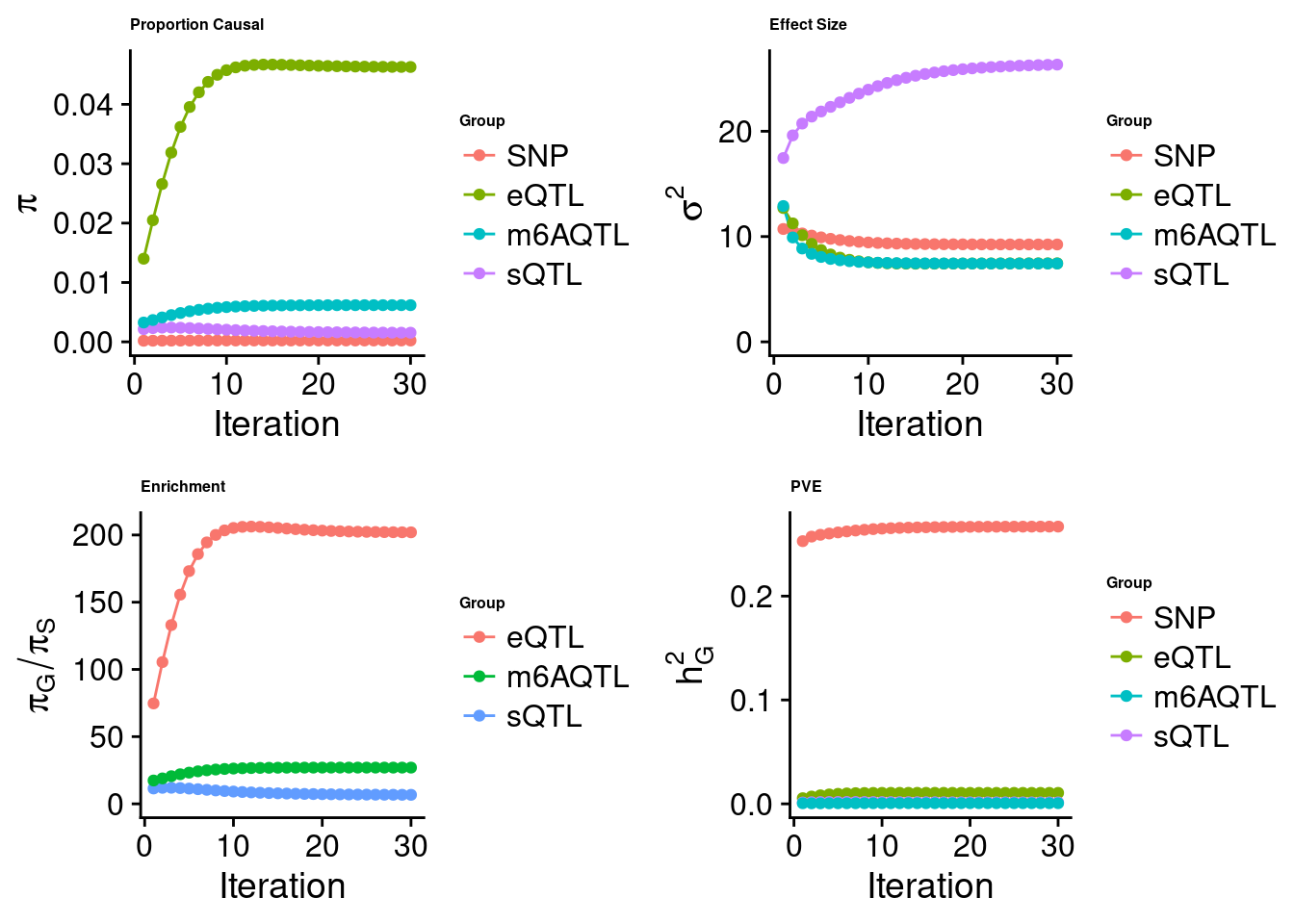
$lasso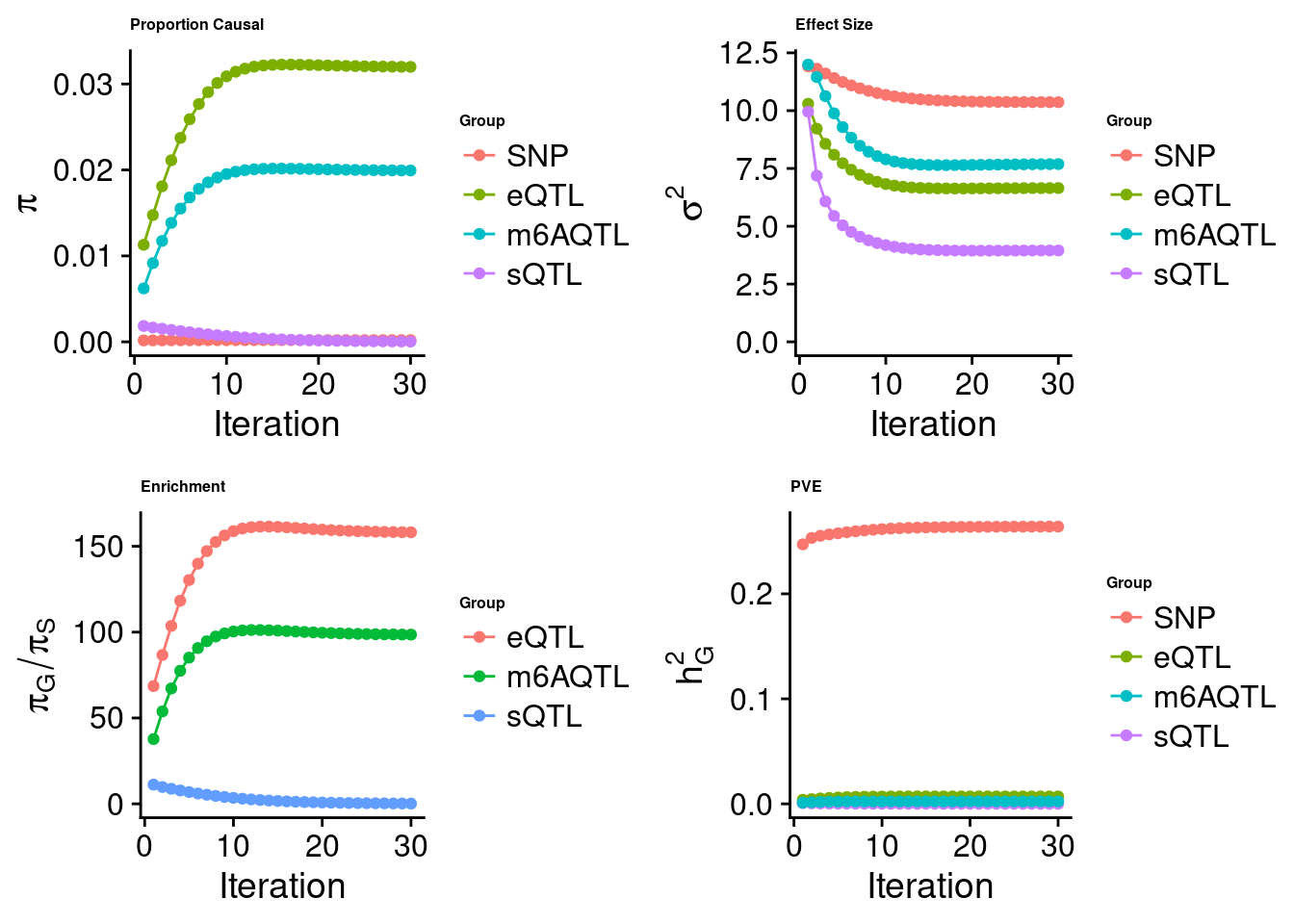
cTWAS results for individual analysis with m6A
top1 model
genename region_tag susie_pip z
1 IP6K2 3_34 0.8041 -4.767
2 ICOSLG 21_22 0.5716 9.960
3 CYTH1 17_44 0.3671 -4.097
4 MMP24-AS1 20_21 0.2748 -3.579
5 DHX38 16_38 0.2530 -3.560
6 RGS10 10_74 0.2472 -3.797
7 AKAP8 19_12 0.2463 -3.754
8 RNGTT 6_60 0.2267 2.851
9 TRIM8 10_66 0.2174 -3.259
10 CRTC3 15_42 0.2150 -3.841
11 ZFP36L2 2_27 0.2088 5.896
12 KIF21B 1_103 0.2059 -8.739
13 NEAT1 11_36 0.1911 -3.440
14 COG4 16_37 0.1902 2.841
15 TAF6L 11_35 0.1889 -3.091
16 STK24 13_50 0.1336 -2.750
17 C1orf43 1_77 0.1311 -2.570
18 CD320 19_8 0.1299 -2.217
19 DHX38 16_38 0.1269 -3.102
20 GSDMD 8_94 0.1252 2.543Summing up PIPs for m6A peaks located in the same gene
Top m6A PIPs by genes
# A tibble: 760 × 2
genename total_susie_pip
<chr> <dbl>
1 IP6K2 0.804
2 ICOSLG 0.615
3 DHX38 0.380
4 CYTH1 0.367
5 MMP24-AS1 0.275
6 GSDMD 0.250
7 RGS10 0.247
8 AKAP8 0.246
9 RNGTT 0.227
10 TRIM8 0.217
# ℹ 750 more rowscTWAS results for joint analysis using a lasso model
Top expression/splicing/m6A units
For m6A or splicing QTLs, they are assigned to the nearest genes (m6A needs to be confirmed with Kevin).
Top SNPs or genes with PIP > 0.6
$eQTL
genename susie_pip group region_tag
1960 GLRX 0.9484 eQTL 5_56
1974 PTGIR 0.9191 eQTL 19_34
1952 ENOPH1 0.9082 eQTL 4_56
1938 CD28 0.8974 eQTL 2_120
1936 ABL2 0.8072 eQTL 1_91
1964 KDELR2 0.7994 eQTL 7_9
1867 ENSG00000211659 0.7943 eQTL 22_5
1444 CBFB 0.7281 eQTL 16_37
29 TMCO4 0.6598 eQTL 1_13
9 TNFRSF14 0.6438 eQTL 1_2
1880 MTMR3 0.5969 eQTL 22_10
106 DR1 0.5717 eQTL 1_57
285 NDUFS1 0.5555 eQTL 2_122
795 SMIM19 0.5523 eQTL 8_37
476 DAP 0.5301 eQTL 5_9
419 TMEM128 0.5269 eQTL 4_5
1462 WWP2 0.5240 eQTL 16_37
1067 ENSG00000245532 0.5114 eQTL 11_36
$m6AQTL
genename susie_pip group region_tag
5047 IP6K2 0.9080 m6AQTL 3_35
5009 C21orf33 0.7936 m6AQTL 21_23
4946 AKAP8 0.7020 m6AQTL 19_12
4214 KIF21B 0.6662 m6AQTL 1_103
4890 CYTH1 0.5689 m6AQTL 17_44
4821 DHX38 0.5541 m6AQTL 16_38
$sQTL
[1] genename susie_pip group region_tag
<0 rows> (or 0-length row.names)Top m6A modification pip
genename region_tag susie_pip z
1 IP6K2 3_35 0.9080 -4.767
2 C21orf33 21_23 0.7936 -5.739
3 AKAP8 19_12 0.7020 -4.074
4 KIF21B 1_103 0.6662 -8.732
5 CYTH1 17_44 0.5689 -4.120
6 DHX38 16_38 0.5541 -3.918
7 CRTC3 15_42 0.4149 -4.466
8 DHX38 16_38 0.3841 -3.682
9 C19orf54 19_28 0.3077 -3.376
10 LHFPL2 5_46 0.3025 -2.940Summing up PIPs for m6A peaks located in the same gene
Top 10 m6A PIPs by genes
# A tibble: 814 × 2
genename total_susie_pip
<chr> <dbl>
1 DHX38 0.938
2 IP6K2 0.908
3 C21orf33 0.794
4 AKAP8 0.702
5 KIF21B 0.666
6 CYTH1 0.569
7 CRTC3 0.415
8 LHFPL2 0.382
9 CENPF 0.363
10 KIAA1147 0.329
# ℹ 804 more rowsTop splicing PIPs
Splicing PIPs are quite low.
peak_id genename pos region_tag susie_pip z
1 chr1:155226210-155226433 SCAMP3 155135335 1_79 0.022818 6.753
2 chr1:155226210-155226465 SCAMP3 155135335 1_79 0.013819 -6.456
3 chr2:231054571-231065601 SP110 231050715 2_135 0.012014 4.938
4 chr2:231110655-231112631 SP140 231099170 2_135 0.007161 -4.812
5 chr7:26233314-26235467 HNRNPA2B1 26151190 7_23 0.004200 4.279
6 chr11:1908544-1908704 LSP1 1841555 11_2 0.003660 4.304
7 chr2:231079833-231084589 SP140 230992212 2_135 0.003155 4.548
8 chr11:810039-810234 RPLP2 716765 11_1 0.002695 3.548
9 chr11:810357-811597 RPLP2 752059 11_1 0.002639 -3.523
10 chr10:114186117-114186976 ZDHHC6 114169664 10_70 0.002388 3.592Summing up PIPs for spliced introns located in the same gene
Top 10 splicing PIPs by genes
# A tibble: 10 × 2
genename total_susie_pip
<chr> <dbl>
1 SCAMP3 0.0369
2 SP140 0.0149
3 SP110 0.0120
4 RPLP2 0.00544
5 LSP1 0.00451
6 HNRNPA2B1 0.00420
7 EPSTI1 0.00348
8 ORAI2 0.00329
9 RPSA 0.00277
10 IMMP1L 0.00255Top genes by combined PIP
genename combined_pip expression_pip splicing_pip m6A_pip
1389 GLRX 1.044 0.9484 0.0000000 0.095273
725 DHX38 0.938 0.0000 0.0000000 0.938280
2370 PTGIR 0.919 0.9191 0.0000000 0.000000
847 ENOPH1 0.908 0.9082 0.0000000 0.000000
1586 IP6K2 0.908 0.0000 0.0000173 0.907997
462 CD28 0.897 0.8974 0.0000000 0.000000
21 ABL2 0.807 0.8072 0.0000000 0.000000
1623 KDELR2 0.799 0.7994 0.0000000 0.000000
368 C21orf33 0.794 0.0000 0.0000000 0.793555
905 ENSG00000211659 0.794 0.7943 0.0000000 0.000000
416 CBFB 0.728 0.7281 0.0000000 0.000000
111 AKAP8 0.702 0.0000 0.0000000 0.701968
1639 KIF21B 0.666 0.0000 0.0000000 0.666226
2977 TMCO4 0.660 0.6598 0.0000000 0.000000
3025 TNFRSF14 0.644 0.6438 0.0000000 0.000000
665 DAP 0.615 0.5301 0.0002289 0.084182
1934 MTMR3 0.597 0.5969 0.0000000 0.000000
2986 TMEM128 0.575 0.5269 0.0000000 0.048111
761 DR1 0.574 0.5717 0.0020552 0.000000
661 CYTH1 0.569 0.0000 0.0000000 0.568940
2005 NDUFS1 0.555 0.5555 0.0000000 0.000000
2748 SMIM19 0.552 0.5523 0.0000000 0.000000
3254 WWP2 0.532 0.5240 0.0000000 0.007832
1039 ENSG00000245532 0.511 0.5114 0.0000000 0.000000
1190 ENSG00000269958 0.496 0.4958 0.0000000 0.000000
2893 TAMM41 0.488 0.4883 0.0000000 0.000000
339 BTN3A3 0.482 0.4411 0.0000000 0.041386
2550 RPL9 0.481 0.4806 0.0000000 0.000000
2190 PEX7 0.474 0.4740 0.0000000 0.000000
985 ENSG00000233429 0.455 0.4545 0.0000000 0.000000
1659 KYAT3 0.450 0.4495 0.0000000 0.000000
2716 SLC37A2 0.449 0.1561 0.0000000 0.293129
2300 PPIH 0.445 0.4449 0.0000000 0.000000
3121 TTC32 0.443 0.4425 0.0000000 0.000000
2879 SYNGR1 0.432 0.4317 0.0000000 0.000000
1346 FRA10AC1 0.431 0.4309 0.0000000 0.000000
618 CRTC3 0.415 0.0000 0.0000000 0.414926
3299 ZFP90 0.415 0.4146 0.0000000 0.000000
220 ARPC5 0.413 0.2451 0.0002714 0.167322
360 C19orf54 0.410 0.1015 0.0009512 0.307729
region_tag
1389 5_56
725 16_38
2370 19_34
847 4_56
1586 3_35
462 2_120
21 1_91
1623 7_9
368 21_23
905 22_5
416 16_37
111 19_12
1639 1_103
2977 1_13
3025 1_2
665 5_9
1934 22_10
2986 4_5
761 1_57
661 17_44
2005 2_122
2748 8_37
3254 16_37
1039 11_36
1190 14_54
2893 3_9
339 6_20
2550 4_32
2190 6_90
985 7_23
1659 1_54
2716 11_77
2300 1_27
3121 2_12
2879 22_16
1346 10_60
618 15_42
3299 16_37
220 1_93
360 19_28Compared with the putative genes identified by ABC model
Only two genes annotated with ABC model are imputable with our current QTL datasets.
genename combined_pip expression_pip splicing_pip m6A_pip region_tag
1 NCF4 0.038 0.03762 2.803e-05 0 22_15
2 CD40 0.001 0.00000 8.489e-04 0 20_28Joining with `by = join_by(genename)`[1] "The causal genes identified by both methods:" genename m6A.PEAK.ID TWAS.P.Bonferroni m6A_pip
1 IP6K2 chr3:48731165-48731510_IP6K2_- 1.602e-03 0.9080
2 AKAP8 chr19:15483795-15484043_AKAP8_- 4.592e-02 0.7020
3 KIF21B chr1:200939513-200939662_KIF21B_- 2.367e-09 0.6662[1] "The causal genes specifically identified by ctwas:" genename combined_pip expression_pip splicing_pip m6A_pip region_tag
1 DHX38 0.938 0 0.00e+00 0.9383 16_38
2 IP6K2 0.908 0 1.73e-05 0.9080 3_35
3 C21orf33 0.794 0 0.00e+00 0.7936 21_23
4 AKAP8 0.702 0 0.00e+00 0.7020 19_12
5 KIF21B 0.666 0 0.00e+00 0.6662 1_103
6 CYTH1 0.569 0 0.00e+00 0.5689 17_44Locus plots for specific examples
Loading required package: gridWarning: replacing previous import 'utils::download.file' by
'restfulr::download.file' when loading 'rtracklayer' genename combined_pip expression_pip splicing_pip m6A_pip region_tag
1389 GLRX 1.044 0.9484 0 0.09527 5_56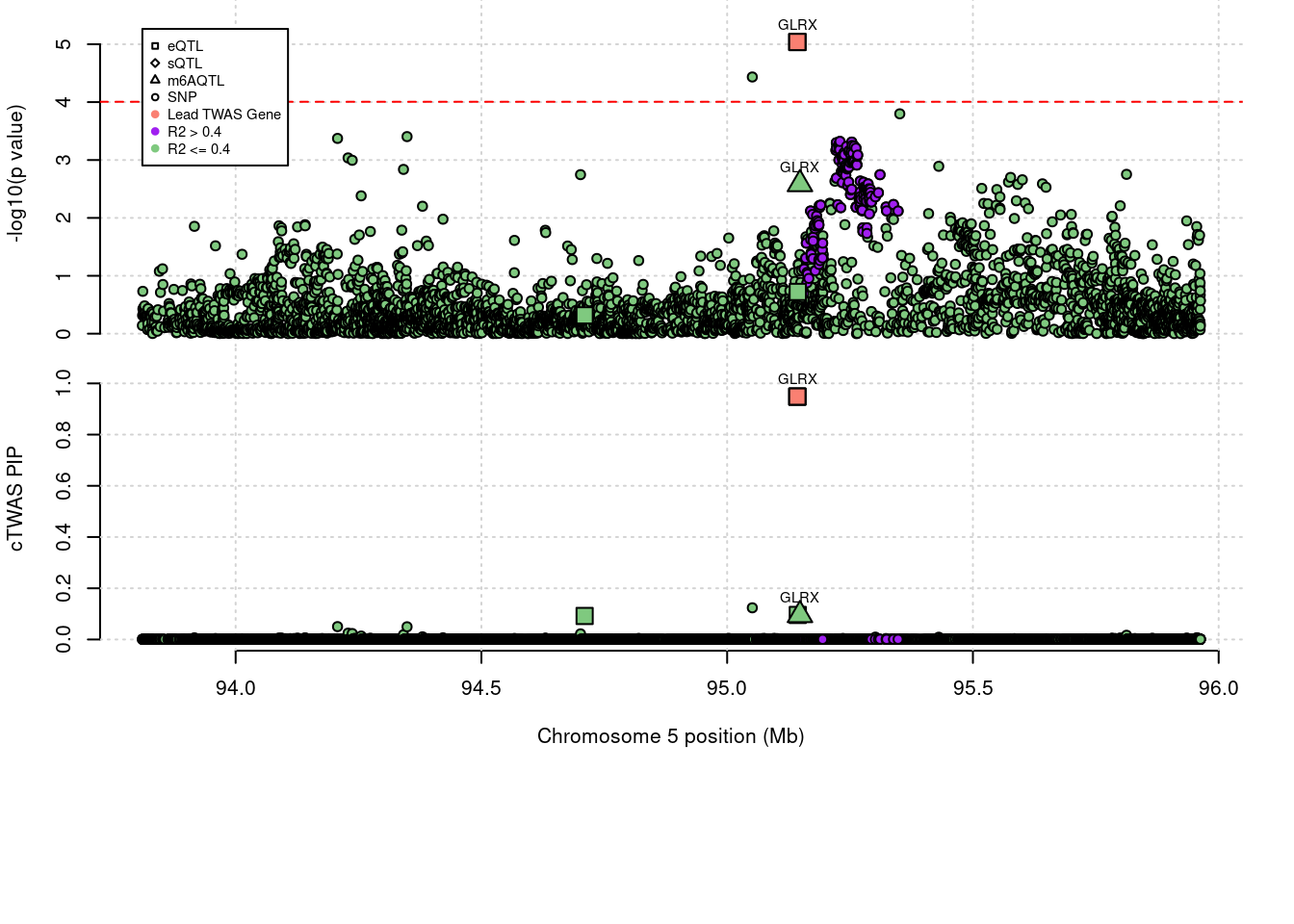
genename combined_pip expression_pip splicing_pip m6A_pip region_tag
725 DHX38 0.938 0 0 0.9383 16_38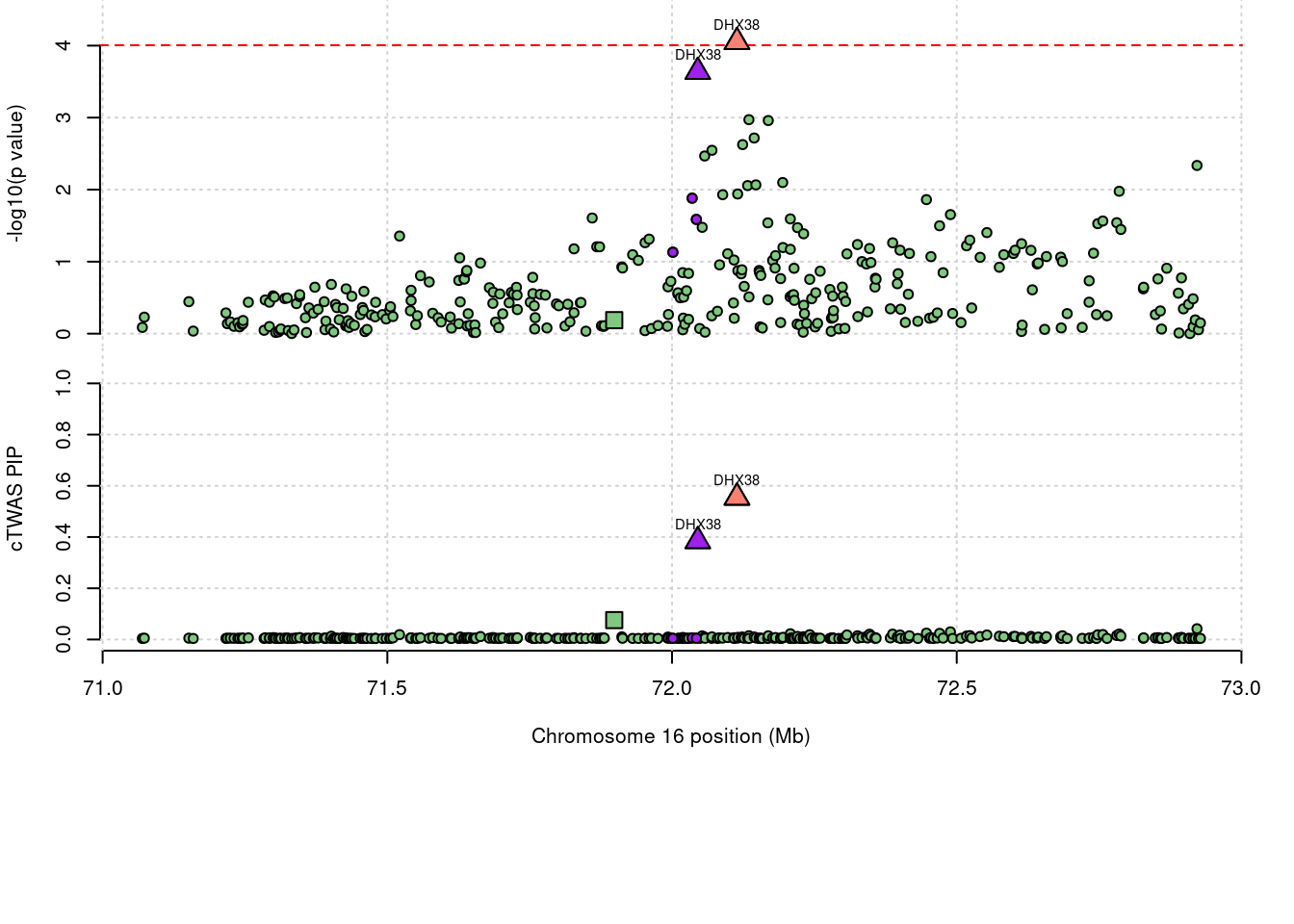
genename combined_pip expression_pip splicing_pip m6A_pip region_tag
1586 IP6K2 0.908 0 1.73e-05 0.908 3_35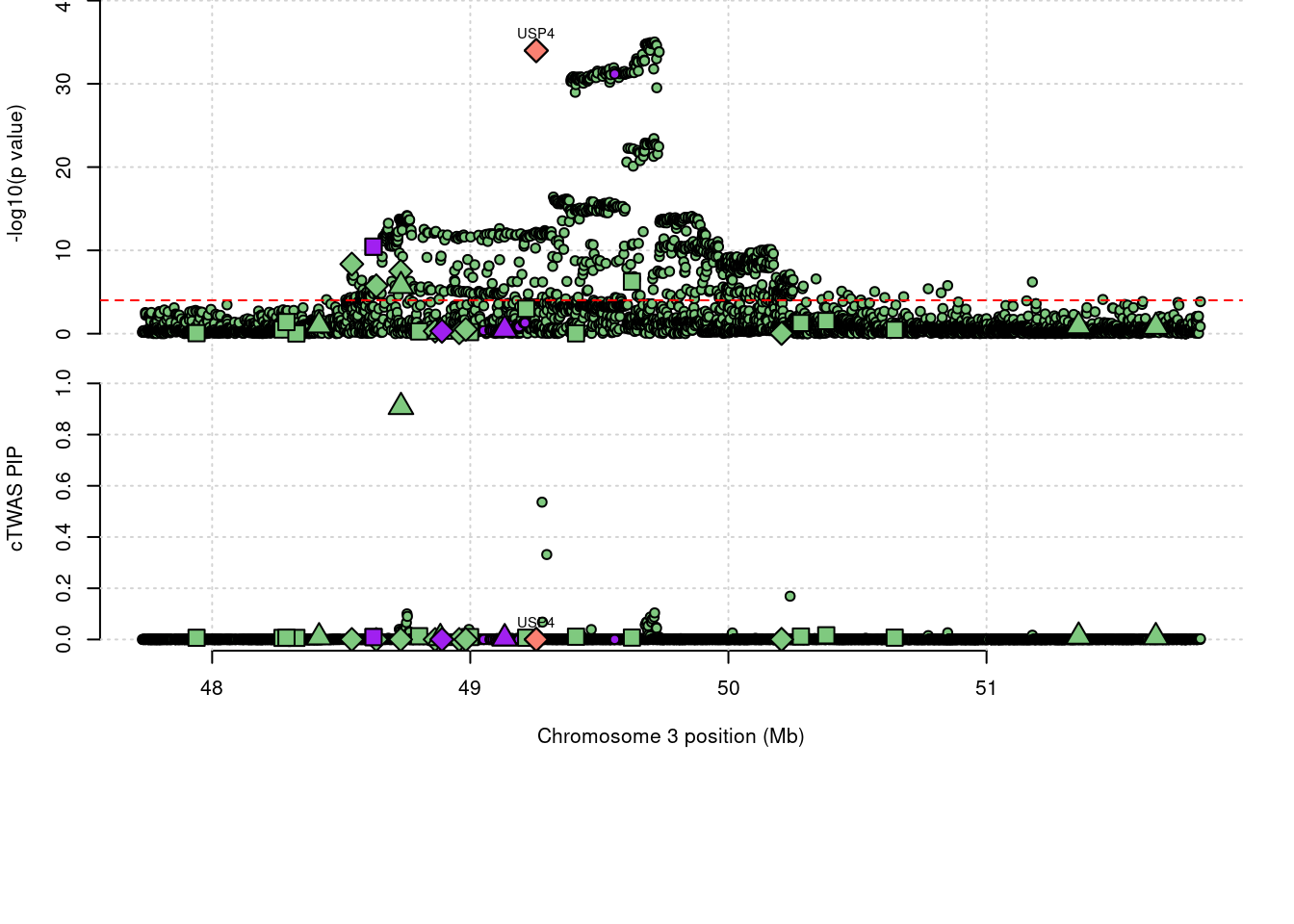
Locus plot for the prioritized TWAS genes with FUSION
genename combined_pip expression_pip splicing_pip m6A_pip region_tag
3065 TRAF3IP3 0.057 0 0.0002895 0.05718 1_108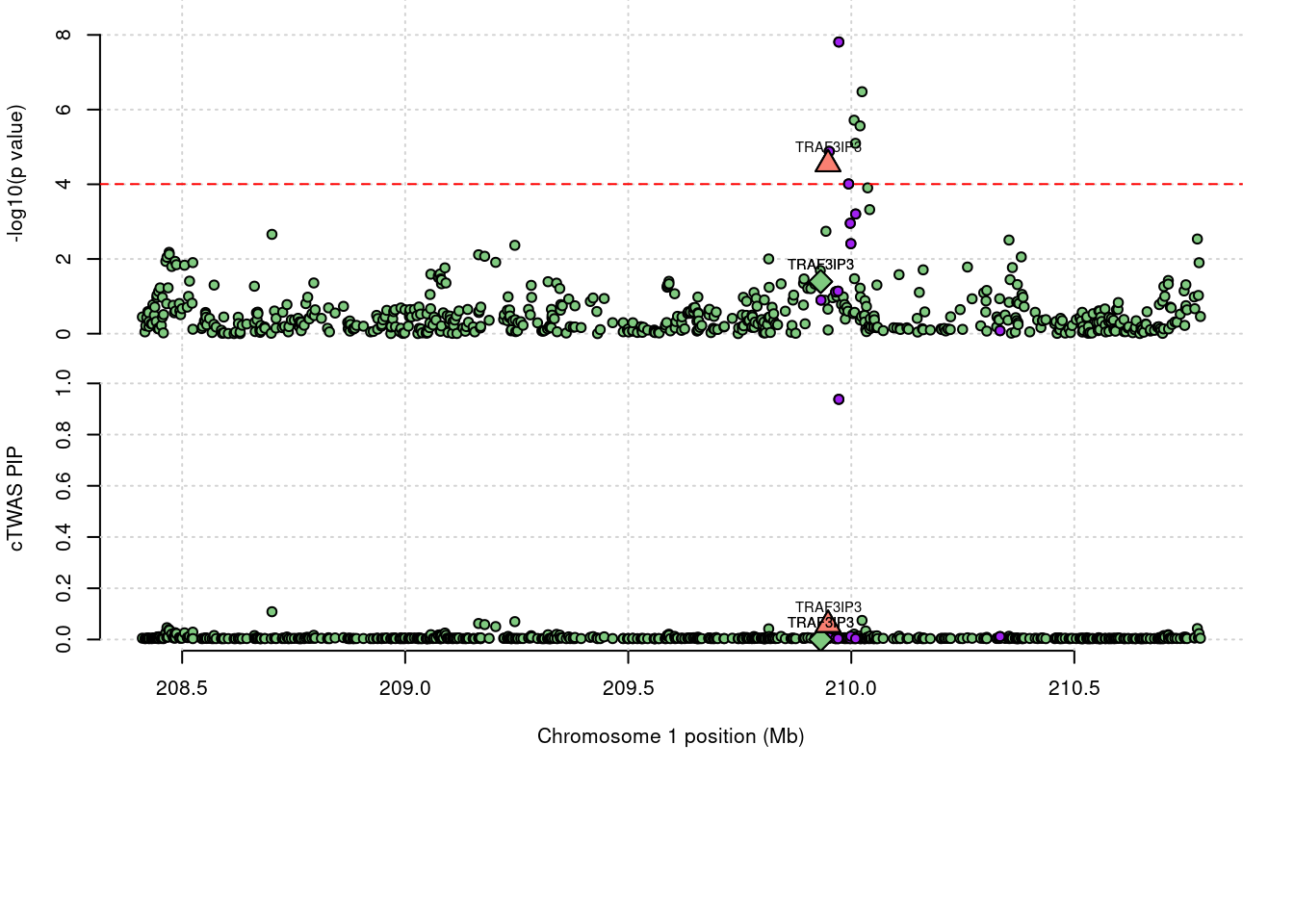
genename combined_pip expression_pip splicing_pip m6A_pip region_tag
86 ADCY7 0.098 0 0 0.09821 16_27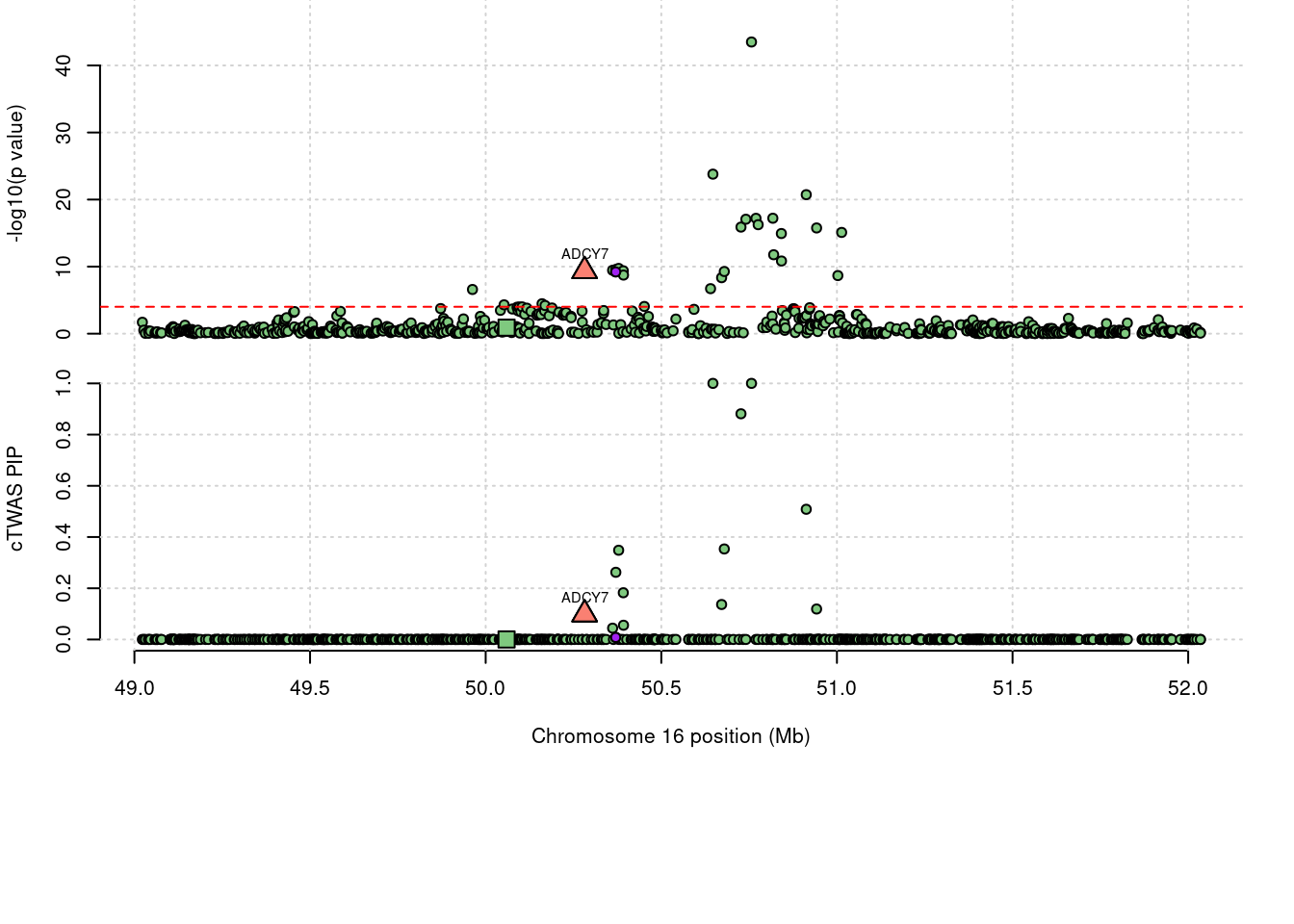
genename combined_pip expression_pip splicing_pip m6A_pip region_tag
1541 ICOSLG 0.052 0 0 0.05211 21_23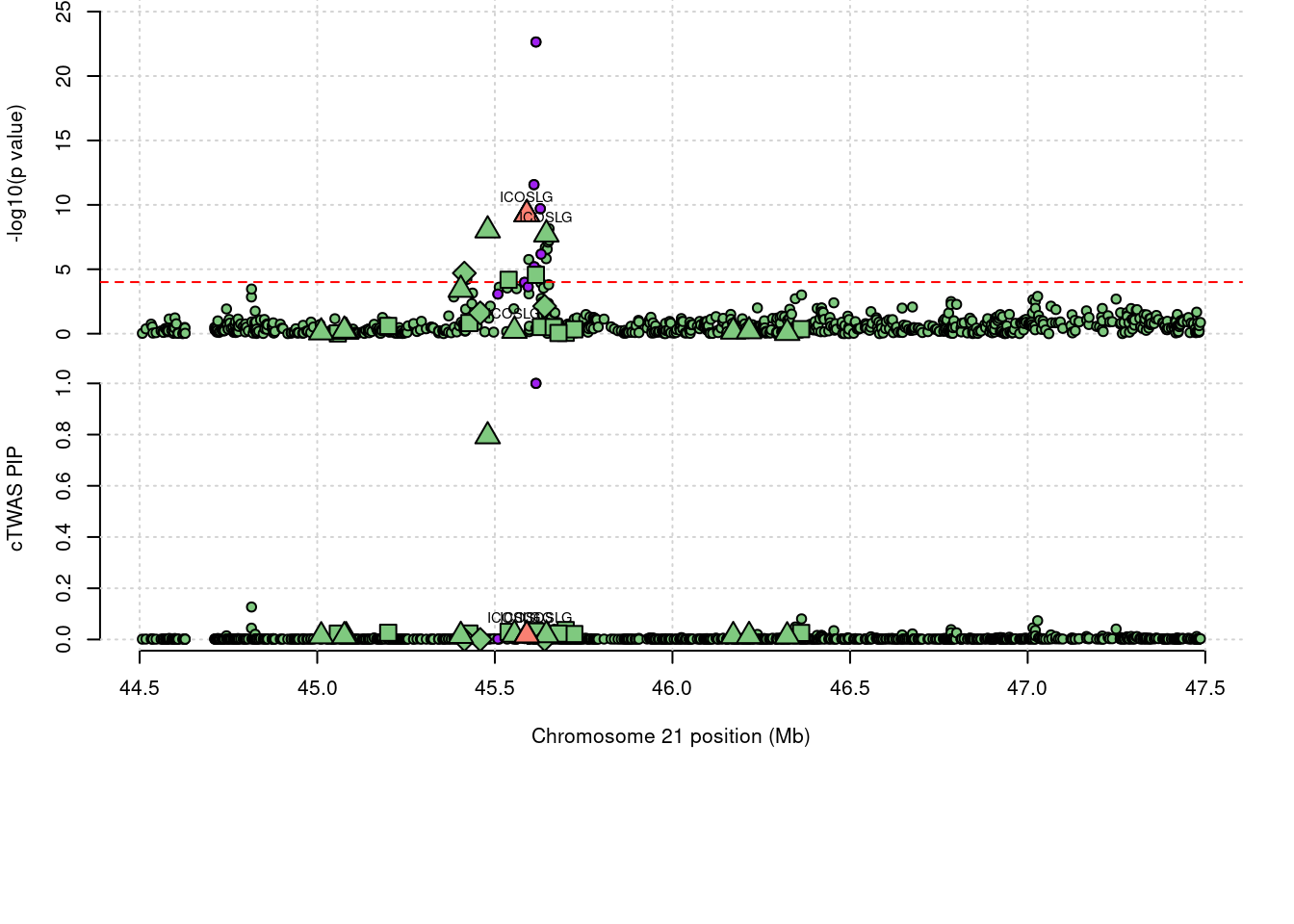
R version 4.2.0 (2022-04-22)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.3.13-el7-x86_64/lib/libopenblas_haswellp-r0.3.13.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=C
[4] LC_COLLATE=C LC_MONETARY=C LC_MESSAGES=C
[7] LC_PAPER=C LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=C LC_IDENTIFICATION=C
attached base packages:
[1] grid stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] biomaRt_2.52.0 Gviz_1.40.1 cowplot_1.1.1
[4] ggplot2_3.4.3 GenomicRanges_1.48.0 GenomeInfoDb_1.32.2
[7] IRanges_2.30.1 S4Vectors_0.34.0 BiocGenerics_0.42.0
[10] ctwas_0.1.38 dplyr_1.1.2 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] colorspace_2.1-0 deldir_1.0-6
[3] rjson_0.2.21 rprojroot_2.0.3
[5] biovizBase_1.44.0 htmlTable_2.4.0
[7] XVector_0.36.0 base64enc_0.1-3
[9] fs_1.6.3 dichromat_2.0-0.1
[11] rstudioapi_0.15.0 farver_2.1.1
[13] bit64_4.0.5 AnnotationDbi_1.58.0
[15] fansi_1.0.4 xml2_1.3.3
[17] codetools_0.2-18 logging_0.10-108
[19] cachem_1.0.8 knitr_1.39
[21] Formula_1.2-4 jsonlite_1.8.7
[23] Rsamtools_2.12.0 cluster_2.1.3
[25] dbplyr_2.3.3 png_0.1-7
[27] compiler_4.2.0 httr_1.4.6
[29] backports_1.4.1 lazyeval_0.2.2
[31] Matrix_1.6-1 fastmap_1.1.1
[33] cli_3.6.1 later_1.3.0
[35] htmltools_0.5.2 prettyunits_1.1.1
[37] tools_4.2.0 gtable_0.3.3
[39] glue_1.6.2 GenomeInfoDbData_1.2.8
[41] rappdirs_0.3.3 Rcpp_1.0.11
[43] Biobase_2.56.0 jquerylib_0.1.4
[45] vctrs_0.6.3 Biostrings_2.64.0
[47] rtracklayer_1.56.0 iterators_1.0.14
[49] xfun_0.30 stringr_1.5.0
[51] ps_1.7.0 lifecycle_1.0.3
[53] ensembldb_2.20.2 restfulr_0.0.14
[55] XML_3.99-0.14 getPass_0.2-2
[57] zlibbioc_1.42.0 scales_1.2.1
[59] BSgenome_1.64.0 VariantAnnotation_1.42.1
[61] ProtGenerics_1.28.0 hms_1.1.3
[63] promises_1.2.0.1 MatrixGenerics_1.8.0
[65] parallel_4.2.0 SummarizedExperiment_1.26.1
[67] AnnotationFilter_1.20.0 RColorBrewer_1.1-3
[69] yaml_2.3.5 curl_5.0.2
[71] memoise_2.0.1 gridExtra_2.3
[73] sass_0.4.1 rpart_4.1.16
[75] latticeExtra_0.6-30 stringi_1.7.12
[77] RSQLite_2.3.1 highr_0.9
[79] BiocIO_1.6.0 foreach_1.5.2
[81] checkmate_2.1.0 GenomicFeatures_1.48.4
[83] filelock_1.0.2 BiocParallel_1.30.3
[85] rlang_1.1.1 pkgconfig_2.0.3
[87] matrixStats_0.62.0 bitops_1.0-7
[89] evaluate_0.15 lattice_0.20-45
[91] htmlwidgets_1.5.4 GenomicAlignments_1.32.0
[93] labeling_0.4.2 bit_4.0.5
[95] processx_3.8.0 tidyselect_1.2.0
[97] magrittr_2.0.3 R6_2.5.1
[99] generics_0.1.3 Hmisc_5.1-0
[101] DelayedArray_0.22.0 DBI_1.1.3
[103] pgenlibr_0.3.6 pillar_1.9.0
[105] whisker_0.4 foreign_0.8-82
[107] withr_2.5.0 KEGGREST_1.36.2
[109] RCurl_1.98-1.7 nnet_7.3-17
[111] tibble_3.2.1 crayon_1.5.2
[113] interp_1.1-4 utf8_1.2.3
[115] BiocFileCache_2.4.0 rmarkdown_2.14
[117] jpeg_0.1-10 progress_1.2.2
[119] data.table_1.14.8 blob_1.2.4
[121] callr_3.7.3 git2r_0.30.1
[123] digest_0.6.33 httpuv_1.6.5
[125] munsell_0.5.0 bslib_0.3.1