Evaluate the withheld sample on top X cyclical
Joyce Hsiao
Last updated: 2018-07-04
Code version: 2498ce4
Load previously computed results
results_eval_top101 <- readRDS("../data/results/results_eval_top101.rds")
results_eval_top10 <- readRDS("../data/results/results_eval_top10.rds")Before removing moisy labels, on top 10 cyclical genes
source("../peco/R/utility.R")
source("../peco/R/fit.trendfilter.generic.R")
source("../peco/R/run_seurat.R")
mean(results_eval_top10$fit.supervised$diff_time)/2/pi[1] 0.1024371mean(results_eval_top10$fit.trend2.unsup$diff_time)/2/pi[1] 0.1027337mean(results_eval_top10$fit.seurat$diff_time)/2/pi[1] 0.1450893pve.wrapper <- function(results_list, methods_list) {
res <- lapply(1:length(results_list),
function(i) {
obj <- results_list[[i]]
out <- data.frame(
dapi=with(obj, get.pve(dapi[order(pred_time_shift)])$pve),
gfp=with(obj, get.pve(gfp[order(pred_time_shift)])$pve),
rfp=with(obj, get.pve(rfp[order(pred_time_shift)])$pve) )
})
names(res) <- methods_list
return(res)
}
results_list <- results_eval_top10
methods_list <- sapply(names(results_list), function(x) strsplit(x, split=".", fixed=TRUE)[[1]][2])
pve_eval_top10 <- do.call(rbind, pve.wrapper(results_list=results_eval_top10,
methods_list=methods_list))Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ... pve_eval_top10$genes_used <- "top10"
pve_eval_top10$methods <- methods_list
saveRDS(pve_eval_top10,
"../output/method-eval-withheld-explore.Rmd/pve_eval_top10.rds")pve_eval_top10 <- readRDS("../output/method-eval-withheld-explore.Rmd/pve_eval_top10.rds")
print(pve_eval_top10) dapi gfp rfp genes_used methods
supervised 0.125585308 0.0148850520 0.3092444 top10 supervised
trend2 0.113966791 0.0884220119 0.2200606 top10 trend2
bspline 0.101424270 0.1342596991 0.2563613 top10 bspline
loess 0.133801337 0.1079941543 0.2980371 top10 loess
seurat 0.007779095 0.0005450732 0.1126729 top10 seuratget.aov(yy=results_eval_top10$fit.seurat$dapi,
xx=results_eval_top10$fit.seurat$assignments)$pve
[1] 0.06984216
$pval
[1] 0.01161179get.aov(yy=results_eval_top10$fit.seurat$gfp,
xx=results_eval_top10$fit.seurat$assignments)$pve
[1] 0.0538643
$pval
[1] 0.03134803get.aov(yy=results_eval_top10$fit.seurat$rfp,
xx=results_eval_top10$fit.seurat$assignments)$pve
[1] 0.1285371
$pval
[1] 0.0003419435Seurat scores vs cell time predictions
seurat.S.sup <- with(results_eval_top10,
get.pve(fit.seurat$S[order(fit.supervised$pred_time_shift)]))
seurat.S.unsup <- with(results_eval_top10,
get.pve(fit.seurat$S[order(fit.trend2.unsup$pred_time_shift)]))
seurat.G2M.sup <- with(results_eval_top10,
get.pve(fit.seurat$G2M[order(fit.supervised$pred_time_shift)]))
seurat.G2M.unsup <- with(results_eval_top10,
get.pve(fit.seurat$G2M[order(fit.trend2.unsup$pred_time_shift)]))
seurat.S.owntime <- with(results_eval_top10,
get.pve(fit.seurat$S[order(fit.seurat$cell_times_est)]))
seurat.G2M.owntime <- with(results_eval_top10,
get.pve(fit.seurat$G2M[order(fit.seurat$cell_times_est)]))
save(seurat.S.sup, seurat.S.unsup,
seurat.G2M.sup, seurat.G2M.unsup,
seurat.S.owntime, seurat.G2M.owntime,
file = "../output/method-eval-withheld-explore.Rmd/seurat.time.top10.rda")load(file="../output/method-eval-withheld-explore.Rmd/seurat.time.top10.rda")
c(seurat.S.sup$pve, seurat.G2M.sup$pve)[1] 0.3896577 0.5287918c(seurat.S.unsup$pve, seurat.G2M.unsup$pve)[1] 0.2623815 0.5239003c(seurat.S.owntime$pve, seurat.G2M.owntime$pve)[1] 0.8272036 0.8565118with(results_eval_top10,
get.aov(yy=fit.seurat$G2M,xx=fit.seurat$assignments))$pve
[1] 0.6777578
$pval
[1] 1.471869e-15with(results_eval_top10,
get.aov(yy=fit.seurat$S,xx=fit.seurat$assignments))$pve
[1] 0.5747001
$pval
[1] 9.6592e-14Seurat cell class vs unsupervsied predicted time
cols <- c("orange", "red", "brown")
par(mfrow=c(1,3))
with(results_eval_top10,
hist(fit.trend2.unsup$pred_time_shift[fit.seurat$assignments=="G1"],
nclass=5, col=cols[1], main = "G1, 10 cells", xlim=c(0,2*pi), ylim=c(0,30),
xlab="Predicted cell time"))
with(results_eval_top10, hist(fit.trend2.unsup$pred_time_shift[fit.seurat$assignments=="S"],
nclass=10, col=cols[2],
main = "S, 66 cells", xlim=c(0,2*pi), ylim=c(0,30),
xlab="Predicted cell time"))
with(results_eval_top10, hist(fit.trend2.unsup$pred_time_shift[fit.seurat$assignments=="G2M"],
nclass=10, col=cols[3],
main = "G2M, 57 cells", xlim=c(0,2*pi), ylim=c(0,30),
xlab="Predicted cell time"))
title("Unsupervised cell time and Seurat classes", outer=TRUE, line=-1)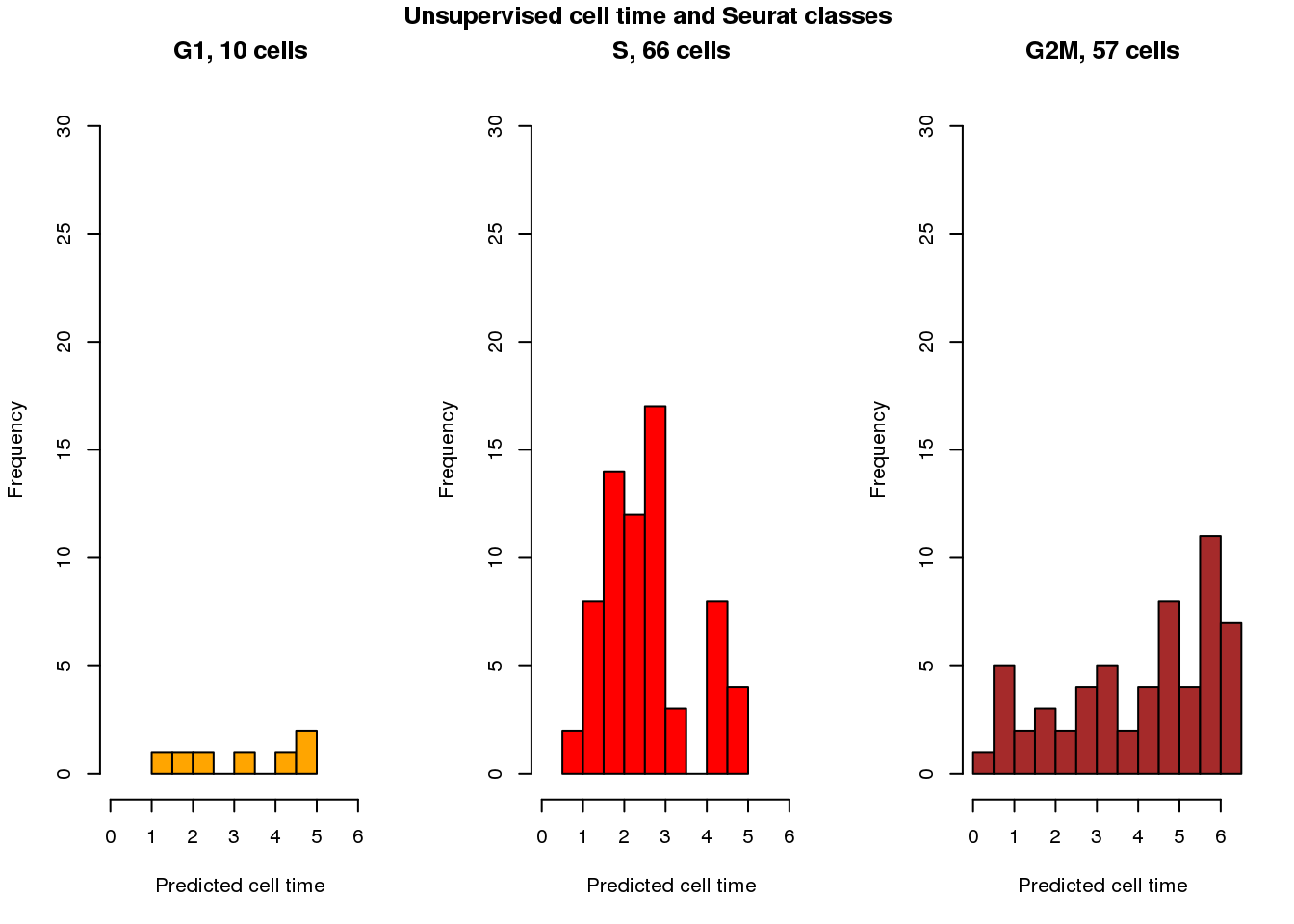
Seurat clases vs supervsied cell times
cols <- c("orange", "red", "brown")
par(mfrow=c(1,3))
with(results_eval_top10,
hist(fit.supervised$pred_time_shift[fit.seurat$assignments=="G1"],
nclass=5, col=cols[1], main = "G1, 10 cells", xlim=c(0,2*pi), ylim=c(0,30),
xlab="Predicted cell time"))
with(results_eval_top10, hist(fit.supervised$pred_time_shift[fit.seurat$assignments=="S"],
nclass=10, col=cols[2],
main = "S, 66 cells", xlim=c(0,2*pi), ylim=c(0,30),
xlab="Predicted cell time"))
with(results_eval_top10, hist(fit.supervised$pred_time_shift[fit.seurat$assignments=="G2M"],
nclass=10, col=cols[3],
main = "G2M, 57 cells", xlim=c(0,2*pi), ylim=c(0,30),
xlab="Predicted cell time"))
title("Supervised cell time and Seurat classes", outer=TRUE, line=-1)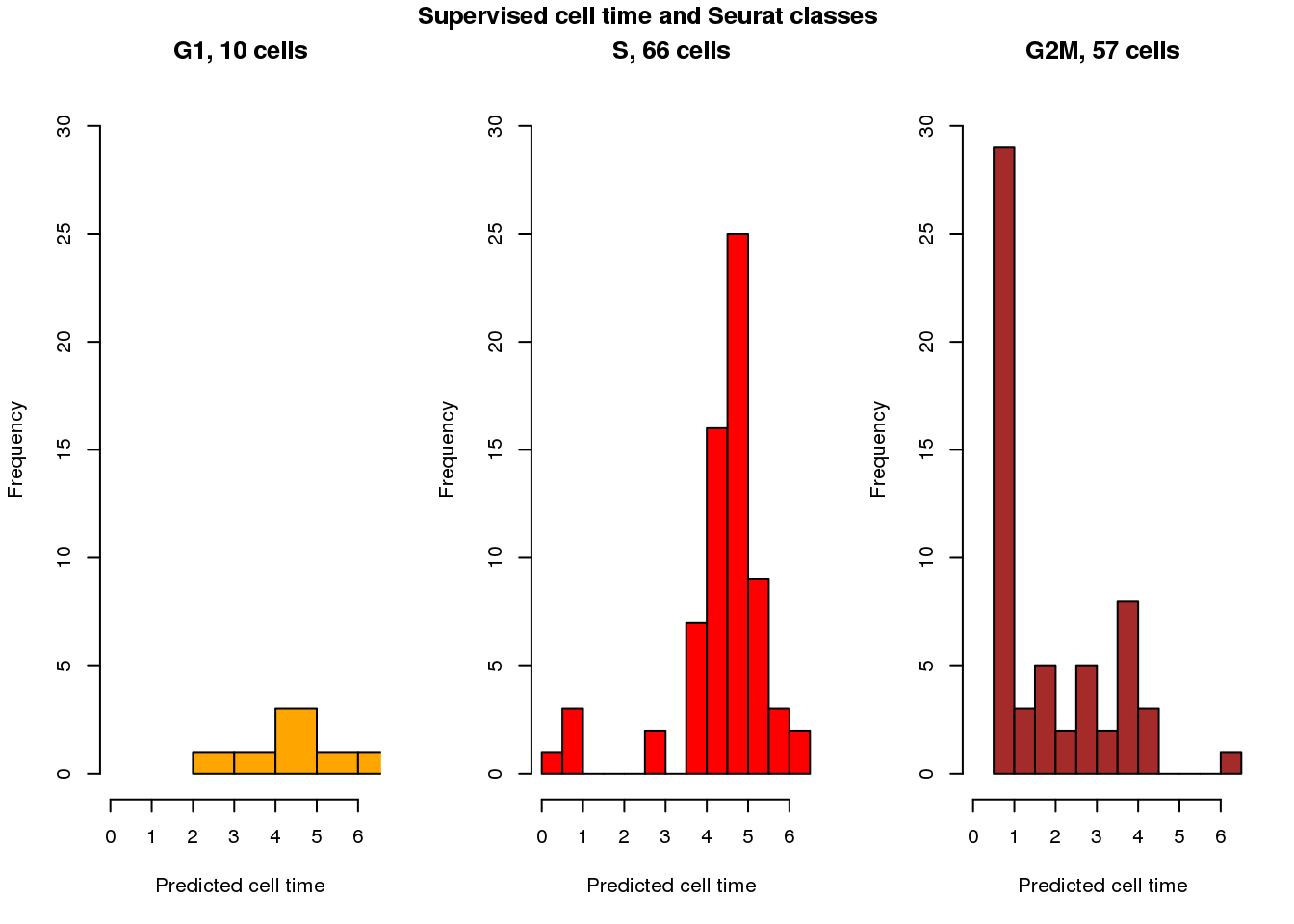
Before removing moisy labels, on top 101 cyclical genes
source("../peco/R/utility.R")
source("../peco/R/fit.trendfilter.generic.R")
source("../peco/R/run_seurat.R")
mean(results_eval_top101$fit.supervised$diff_time)/2/pi[1] 0.09062966mean(results_eval_top101$fit.trend2.unsup$diff_time)/2/pi[1] 0.1160692mean(results_eval_top101$fit.seurat$diff_time)/2/pi[1] 0.1450893pve.wrapper <- function(results_list, methods_list) {
res <- lapply(1:length(results_list),
function(i) {
obj <- results_list[[i]]
out <- data.frame(
dapi=with(obj, get.pve(dapi[order(pred_time_shift)])$pve),
gfp=with(obj, get.pve(gfp[order(pred_time_shift)])$pve),
rfp=with(obj, get.pve(rfp[order(pred_time_shift)])$pve) )
})
names(res) <- methods_list
return(res)
}
results_list <- results_eval_top101
methods_list <- sapply(names(results_list), function(x) strsplit(x, split=".", fixed=TRUE)[[1]][2])
pve_eval_top101 <- do.call(rbind, pve.wrapper(results_list=results_eval_top101,
methods_list=methods_list))Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ... pve_eval_top101$genes_used <- "top101"
pve_eval_top101$methods <- methods_list
saveRDS(pve_eval_top101,
"../output/method-eval-withheld-explore.Rmd/pve_eval_top101.rds")pve_eval_top101 <- readRDS("../output/method-eval-withheld-explore.Rmd/pve_eval_top101.rds")
print(pve_eval_top101) dapi gfp rfp genes_used methods
supervised 0.099929197 0.1208146060 0.2861203 top101 supervised
trend2 0.147145939 0.0948834158 0.1423536 top101 trend2
bspline 0.183418205 0.0020355877 0.3354409 top101 bspline
loess 0.004744912 0.0649387956 0.1642361 top101 loess
seurat 0.007779095 0.0005450732 0.1126729 top101 seuratget.aov(yy=results_eval_top101$fit.seurat$dapi,
xx=results_eval_top101$fit.seurat$assignments)$pve
[1] 0.06984216
$pval
[1] 0.01161179get.aov(yy=results_eval_top101$fit.seurat$gfp,
xx=results_eval_top101$fit.seurat$assignments)$pve
[1] 0.0538643
$pval
[1] 0.03134803get.aov(yy=results_eval_top101$fit.seurat$rfp,
xx=results_eval_top101$fit.seurat$assignments)$pve
[1] 0.1285371
$pval
[1] 0.0003419435Seurat scores vs cell time predictions
seurat.S.sup <- with(results_eval_top101,
get.pve(fit.seurat$S[order(fit.supervised$pred_time_shift)]))
seurat.S.unsup <- with(results_eval_top101,
get.pve(fit.seurat$S[order(fit.trend2.unsup$pred_time_shift)]))
seurat.G2M.sup <- with(results_eval_top101,
get.pve(fit.seurat$G2M[order(fit.supervised$pred_time_shift)]))
seurat.G2M.unsup <- with(results_eval_top101,
get.pve(fit.seurat$G2M[order(fit.trend2.unsup$pred_time_shift)]))
seurat.S.owntime <- with(results_eval_top101,
get.pve(fit.seurat$S[order(fit.seurat$cell_times_est)]))
seurat.G2M.owntime <- with(results_eval_top101,
get.pve(fit.seurat$G2M[order(fit.seurat$cell_times_est)]))
save(seurat.S.sup, seurat.S.unsup,
seurat.G2M.sup, seurat.G2M.unsup,
seurat.S.owntime, seurat.G2M.owntime,
file = "../output/method-eval-withheld-explore.Rmd/seurat.time.top101.rda")load(file="../output/method-eval-withheld-explore.Rmd/seurat.time.top101.rda")
c(seurat.S.sup$pve, seurat.G2M.sup$pve)[1] 0.4832478 0.6548333c(seurat.S.unsup$pve, seurat.G2M.unsup$pve)[1] 0.5051351 0.5050090c(seurat.S.owntime$pve, seurat.G2M.owntime$pve)[1] 0.8272036 0.8565118with(results_eval_top101,
get.aov(yy=fit.seurat$G2M,xx=fit.seurat$assignments))$pve
[1] 0.6777578
$pval
[1] 1.471869e-15with(results_eval_top101,
get.aov(yy=fit.seurat$S,xx=fit.seurat$assignments))$pve
[1] 0.5747001
$pval
[1] 9.6592e-14Seurat cell class vs unsupervsied predicted time
cols <- c("orange", "red", "brown")
par(mfrow=c(1,3))
with(results_eval_top101,
hist(fit.trend2.unsup$pred_time_shift[fit.seurat$assignments=="G1"],
nclass=5, col=cols[1], main = "G1, 10 cells", xlim=c(0,2*pi), ylim=c(0,30),
xlab="Predicted cell time"))
with(results_eval_top101, hist(fit.trend2.unsup$pred_time_shift[fit.seurat$assignments=="S"],
nclass=10, col=cols[2],
main = "S, 66 cells", xlim=c(0,2*pi), ylim=c(0,30),
xlab="Predicted cell time"))
with(results_eval_top101, hist(fit.trend2.unsup$pred_time_shift[fit.seurat$assignments=="G2M"],
nclass=10, col=cols[3],
main = "G2M, 57 cells", xlim=c(0,2*pi), ylim=c(0,30),
xlab="Predicted cell time"))
title("Unsupervised cell time and Seurat classes", outer=TRUE, line=-1)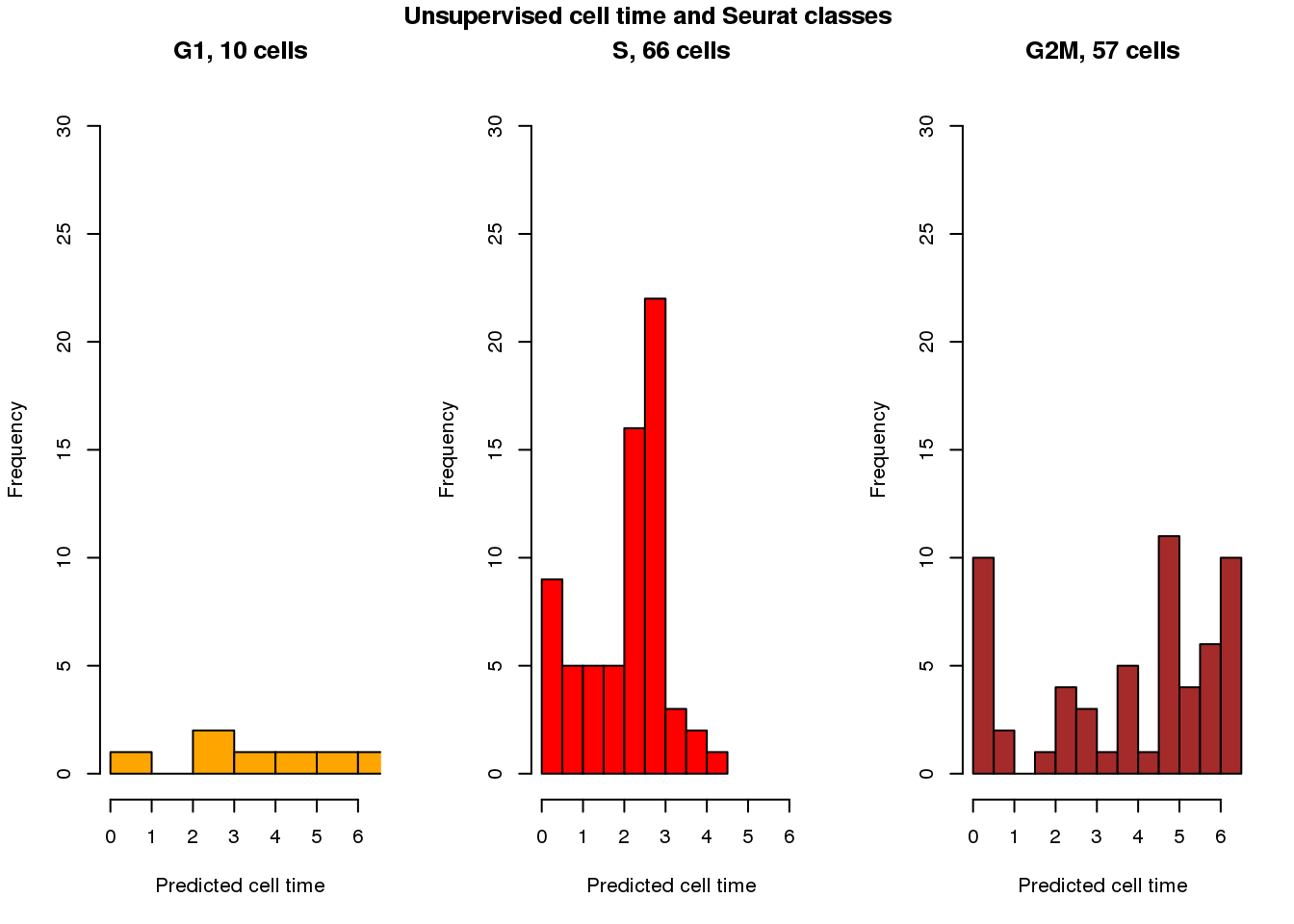
Seurat clases vs supervsied cell times
cols <- c("orange", "red", "brown")
par(mfrow=c(1,3))
with(results_eval_top101,
hist(fit.supervised$pred_time_shift[fit.seurat$assignments=="G1"],
nclass=5, col=cols[1], main = "G1, 10 cells", xlim=c(0,2*pi), ylim=c(0,30),
xlab="Predicted cell time"))
with(results_eval_top101, hist(fit.supervised$pred_time_shift[fit.seurat$assignments=="S"],
nclass=10, col=cols[2],
main = "S, 66 cells", xlim=c(0,2*pi), ylim=c(0,30),
xlab="Predicted cell time"))
with(results_eval_top101, hist(fit.supervised$pred_time_shift[fit.seurat$assignments=="G2M"],
nclass=10, col=cols[3],
main = "G2M, 57 cells", xlim=c(0,2*pi), ylim=c(0,30),
xlab="Predicted cell time"))
title("Supervised cell time and Seurat classes", outer=TRUE, line=-1)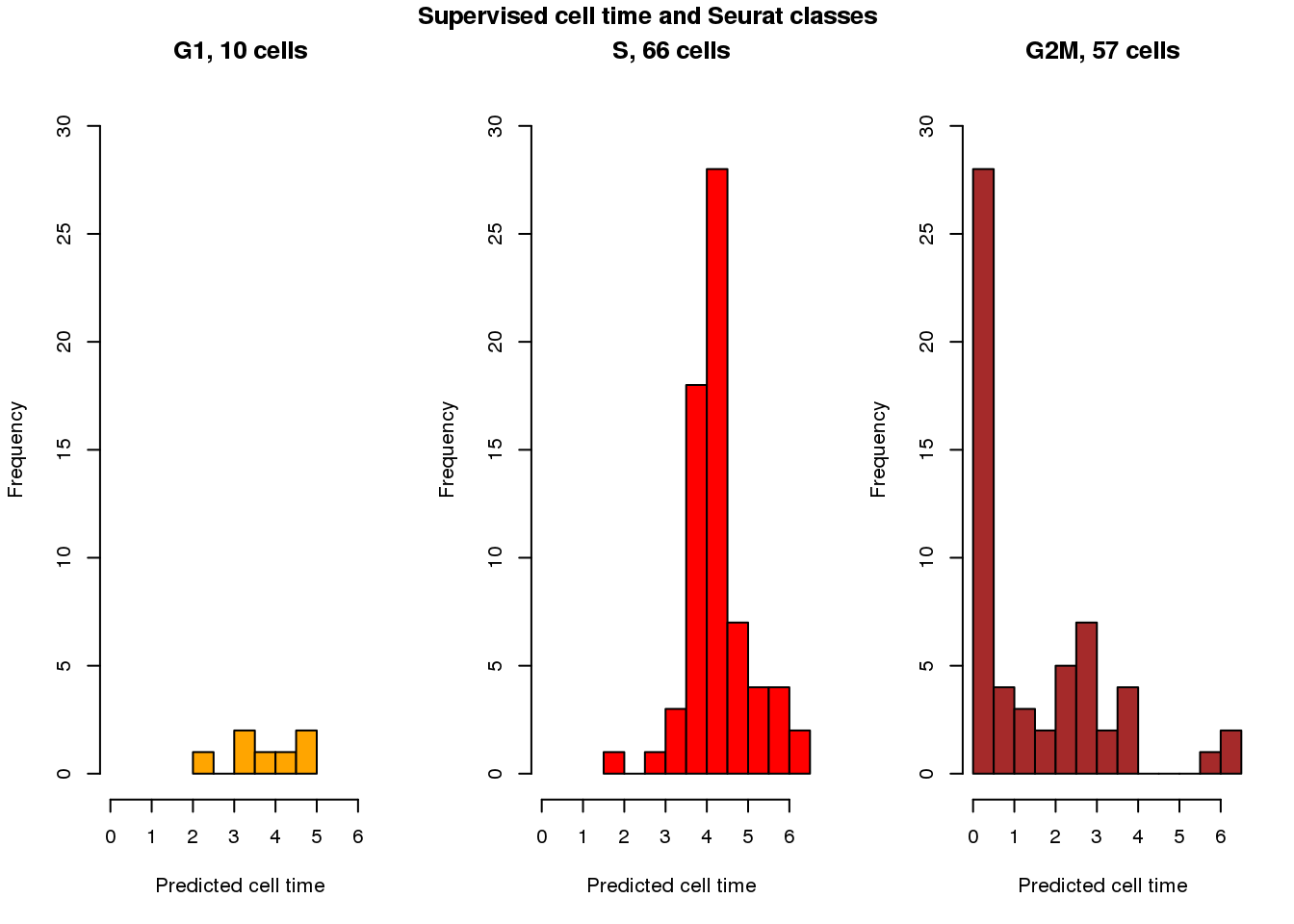
Session information
sessionInfo()R version 3.4.3 (2017-11-30)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] genlasso_1.3 igraph_1.2.1 Matrix_1.2-14 MASS_7.3-50
loaded via a namespace (and not attached):
[1] Rcpp_0.12.17 lattice_0.20-35 digest_0.6.15 rprojroot_1.3-2
[5] grid_3.4.3 backports_1.1.2 git2r_0.21.0 magrittr_1.5
[9] evaluate_0.10.1 stringi_1.2.3 rmarkdown_1.10 tools_3.4.3
[13] stringr_1.3.1 yaml_2.1.19 compiler_3.4.3 pkgconfig_2.0.1
[17] htmltools_0.3.6 knitr_1.20 This R Markdown site was created with workflowr