Evaluate training labels
Joyce Hsiao
- Results
- Extract data from the top 101 genes identified
- PVE between intensities in all samples
- PVE of cell times in all samples
- PC scores of samples versus prediction error
- Re-fitting after removing PC outliers, remove outliers from the same 5 folds
- PVE of cell times in all samples after removing PC outliers
- Re-fitting after removing PC outliers from the same 5 folds, top 50 genes
- Re-fitting after removing PC outliers from the same 5 folds, top 10 genes
- Re-fitting after removing PC outliers, odd number
- Re-fitting after removing PC outliers, even number
- Fitting using all samples, top 50 genes
- Fitting using all samples, top 10 genes
- Fitting using all samples, odd-numbered genes
- Fitting using all samples, even-numbered genes
- Session information
Last updated: 2018-06-24
Code version: 1f3cd60
Results
First we get a sense of the associations between the intensities. We do this by computing PVE of one intensities given another.
Then, we compute cell time labels based on GFP, RPF and based on DAPI, GFP, RFP. We then compute the PVE of intensities by these two sets of labels. Three together don’t do better in explaining RFP and GFP than two together.
- Then we consider in the training dataset, the associations between the PC score of the samples versus the predicted cell times. Are there more errors for cells at the center than the others?
- I looked at prediction error vs PC scores and dont’ see a clear association. To the point that there’s no need to statistical analysis.
- I then compute each cell’s point to the origin on the PC plot and see if prediction error is a function of distance of the origin. Again there doesn’t seem to be an obvious pattern for the relationship between prediction error and distance to the origin.
Notes: predicted time mostly doesn’t need rotations
Extract data from the top 101 genes identified
library(Biobase)
source("../peco/R/cycle.corr.R")
source("../peco/R/cycle.npreg.R")
df <- readRDS(file="../data/eset-final.rds")
pdata <- pData(df)
fdata <- fData(df)
# select endogeneous genes
counts <- exprs(df)[grep("ENSG", rownames(df)), ]
log2cpm.all <- t(log2(1+(10^6)*(t(counts)/pdata$molecules)))
# compare three cell times
theta_pca_triple <- prcomp(cbind(pdata$dapi.median.log10sum.adjust,
pdata$rfp.median.log10sum.adjust,
pdata$gfp.median.log10sum.adjust), scale=TRUE)
library(circular)
pdata$theta_triple <- coord2rad(cbind(theta_pca_triple$x[,1],
theta_pca_triple$x[,2]))
pdata$theta_triple <- as.numeric(pdata$theta_triple)
y1=pdata$theta
y2=pdata$theta_triple
y2shift <- rotation(y1, y2)$y2shift
par(mfrow=c(1,2))
plot(pdata$theta,
pdata$theta_triple, main="pre rotation",
xlab="based on GFP, RFP",
ylab="based on DAPI, GFP, RFP")
plot(y1,
y2shift, main = "post rotation",
xlab="based on GFP, RFP",
ylab="based on DAPI, GFP, RFP")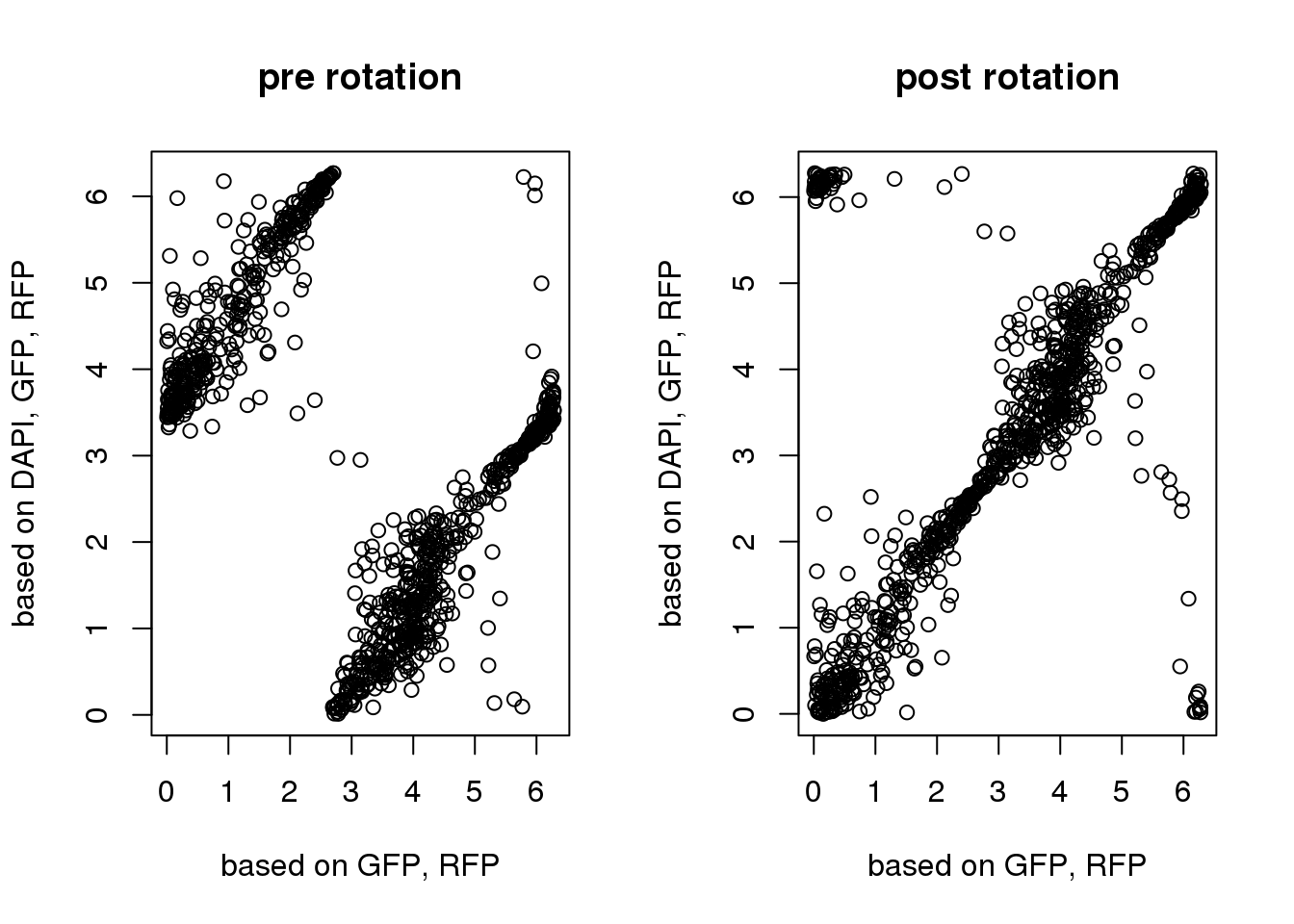
pdata$theta_triple <- y2shift
# rr <- rFLIndTestRand(pdata$theta, pdata$theta_triple, NR=10000)
# cor(y1, y2shift)
# select external validation samples
log2cpm.quant <- readRDS("../output/npreg-trendfilter-quantile.Rmd/log2cpm.quant.rds")
set.seed(99)
nvalid <- round(ncol(log2cpm.quant)*.15)
ii.valid <- sample(1:ncol(log2cpm.quant), nvalid, replace = F)
ii.nonvalid <- setdiff(1:ncol(log2cpm.quant), ii.valid)
log2cpm.quant.nonvalid <- log2cpm.quant[,ii.nonvalid]
log2cpm.quant.valid <- log2cpm.quant[,ii.valid]
sig.genes <- readRDS("../output/npreg-trendfilter-quantile.Rmd/out.stats.ordered.sig.101.rds")
expr.sig <- log2cpm.quant.nonvalid[rownames(log2cpm.quant.nonvalid) %in% rownames(sig.genes), ]
# get predicted times
# set training samples
source("../peco/R/primes.R")
source("../peco/R/partitionSamples.R")
parts <- partitionSamples(1:ncol(log2cpm.quant.nonvalid), runs=5,
nsize.each = rep(151,5))
part_indices <- parts$partitionsPVE between intensities in all samples
source("../peco/R/fit.trendfilter.generic.R")
source("../peco/R/utility.R")
with(pdata, summary(lm(dapi.median.log10sum.adjust~rfp.median.log10sum.adjust))$adj.r.squared)[1] 0.03743611with(pdata, summary(lm(dapi.median.log10sum.adjust~gfp.median.log10sum.adjust))$adj.r.squared)[1] 0.3433102with(pdata, summary(lm(rfp.median.log10sum.adjust~gfp.median.log10sum.adjust))$adj.r.squared)[1] -0.0004061214with(pdata, summary(lm(gfp.median.log10sum.adjust~rfp.median.log10sum.adjust))$adj.r.squared)[1] -0.0004061214with(pdata, summary(lm(rfp.median.log10sum.adjust~dapi.median.log10sum.adjust))$adj.r.squared)[1] 0.03743611with(pdata, summary(lm(gfp.median.log10sum.adjust~dapi.median.log10sum.adjust))$adj.r.squared)[1] 0.3433102plot(pdata$gfp.median.log10sum.adjust,
pdata$rfp.median.log10sum.adjust,
xlab="GFP log10 intensities",
ylab="RFP log10 intensities",
main = "RFP vs. GFP")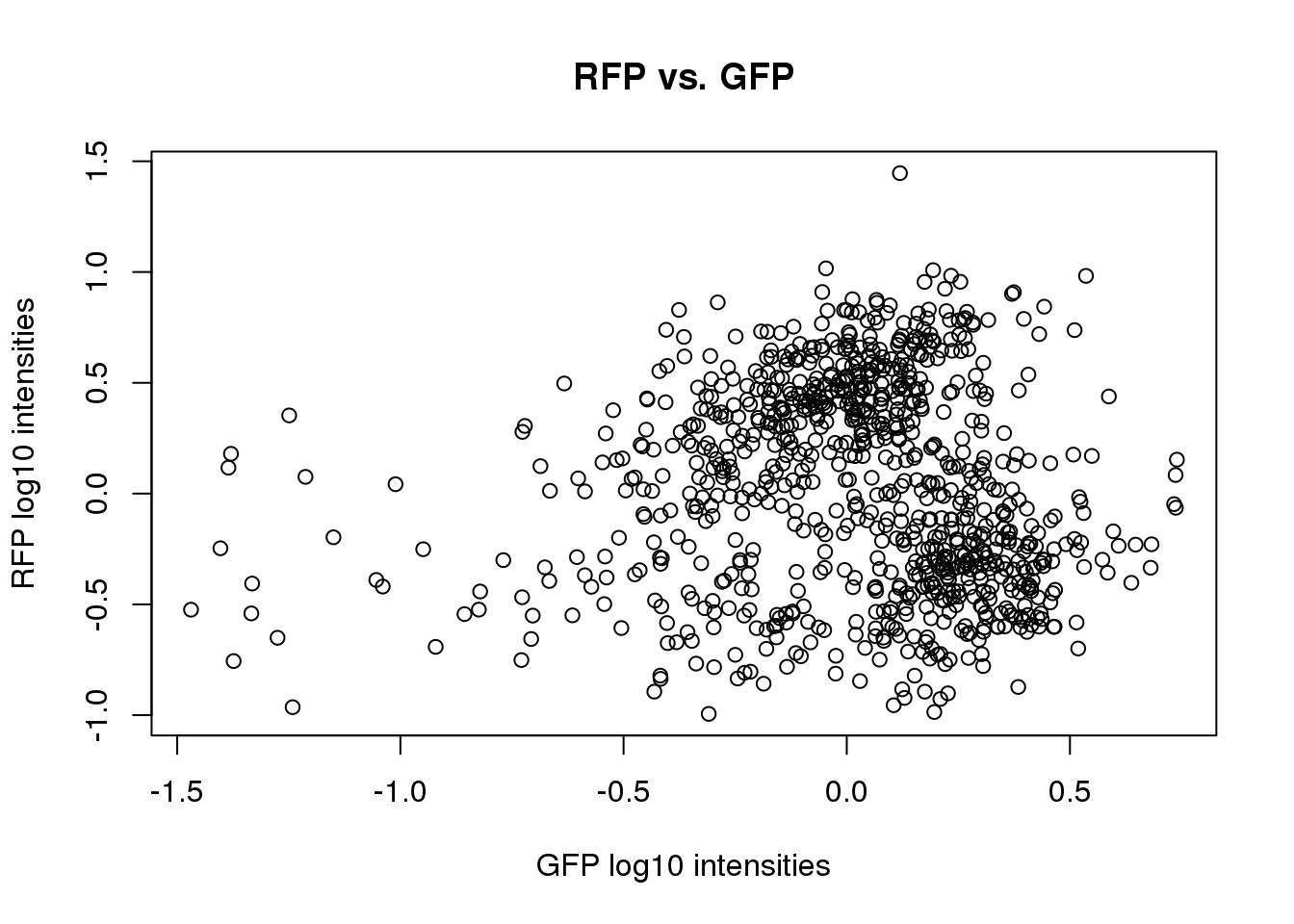
PVE of cell times in all samples
rfp_theta <- with(pdata, get.pve(pdata$rfp.median.log10sum.adjust[order(pdata$theta)]))
rfp_theta_triple <- with(pdata, get.pve(pdata$rfp.median.log10sum.adjust[order(pdata$theta_triple)]))
gfp_theta <- with(pdata, get.pve(pdata$gfp.median.log10sum.adjust[order(pdata$theta)]))
gfp_theta_triple <- with(pdata, get.pve(pdata$gfp.median.log10sum.adjust[order(pdata$theta_triple)]))
dapi_theta <- with(pdata, get.pve(pdata$dapi.median.log10sum.adjust[order(pdata$theta)]))
dapi_theta_triple <- with(pdata, get.pve(pdata$dapi.median.log10sum.adjust[order(pdata$theta_triple)]))
save(rfp_theta, rfp_theta_triple,
gfp_theta, gfp_theta_triple,
dapi_theta, dapi_theta_triple,
file = "../output/method-labels.Rmd/pve.rda")load(file = "../output/method-labels.Rmd/pve.rda")
c(rfp_theta, rfp_theta_triple,
gfp_theta, gfp_theta_triple,
dapi_theta, dapi_theta_triple)[1] 0.8652412 0.8585310 0.7651058 0.6466199 0.2696816 0.5925586PC scores of samples versus prediction error
Fitting using 101 genes
# first check the theta in pdata
pca <- prcomp(cbind(pdata$rfp.median.log10sum.adjust,
pdata$gfp.median.log10sum.adjust), scale=TRUE)
pca_df <- cbind(pca$x[,1],pca$x[,2])
rownames(pca_df) <- rownames(pdata)
theta_check <- as.numeric(coord2rad(pca_df))
theta_check <- 2*pi-theta_check
plot(theta_check, pdata$theta)
names(theta_check) <- rownames(pdata)fits <- vector("list", 5)
for (run in 1:5) {
print(run)
# fitting training data
Y_train <- expr.sig[,part_indices[[run]]$train]
theta_train <- theta_check[match(colnames(Y_train), rownames(pdata))]
names(theta_train) <- colnames(Y_train)
fit.train <- cycle.npreg.insample(Y = Y_train,
theta = theta_train,
ncores=10,
method.trend="npcirc.nw")
# fitting test data
Y_test <- expr.sig[,part_indices[[run]]$test]
theta_test <- theta_check[match(colnames(Y_test), rownames(pdata))]
names(theta_test) <- colnames(Y_test)
fit.test <- cycle.npreg.outsample(Y_test=Y_test,
sigma_est=fit.train$sigma_est,
funs_est=fit.train$funs_est,
method.grid = "uniform",
method.trend="npcirc.nw",
ncores=12)
fits[[run]] <- list(fit.train=fit.train,
fit.test=fit.test,
theta_test=theta_test)
}
for (i in 1:5) {
fits[[i]]$theta_est_shift <- rotation(fits[[i]]$theta_test, fits[[i]]$fit.test$cell_times_est)$y2shift
}
saveRDS(fits, file = "../output/method-labels.Rmd/fits.rds")fits <- readRDS(file = "../output/method-labels.Rmd/fits.rds")
source("../peco/R/utility.R")
diff_time <- lapply(1:length(fits), function(i) {
pmin(abs(fits[[i]]$theta_est_shift-fits[[i]]$theta_test),
abs(fits[[i]]$theta_est_shift-(2*pi-fits[[i]]$theta_test)))
})
mean(sapply(diff_time, mean)/2/pi)[1] 0.0962017pve <- lapply(1:length(fits), function(i) {
dap <- pdata$dapi.median.log10sum[match(names(fits[[i]]$theta_test),rownames(pdata))]
get.pve(dap[order(fits[[i]]$theta_est_shift)])
})Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ... mean(unlist(pve))[1] 0.1096017explore PCA outliers
pca_df_sub <- pca_df[match(names(fits[[2]]$theta_test),rownames(pca_df)),]
diff_bins <- cut(diff_time[[2]],
breaks=quantile(diff_time[[2]], prob=seq(0,1,.25)), include.lowest=TRUE)
summary(diff_bins)[0.000612,0.202] (0.202,0.436] (0.436,0.956] (0.956,2.44]
38 38 37 38 .43/2/pi[1] 0.06843663library(RColorBrewer)
library(scales)
cols <- brewer.pal(9, "YlGn")[c(3, 5, 7, 9)]
par(mfrow=c(1,2))
plot(fits[[2]]$theta_test,
diff_time[[2]]/2/pi, pch=16,
xlab="cell time labels", ylab="margin of error (% arc length)",
col=alpha(c("gray50", "royalblue")[(as.numeric(diff_bins)==4|as.numeric(diff_bins)==3)+1],.7),
main="Prediction error")
plot(pca_df_sub[,1], pca_df_sub[,2],
col=alpha(c("gray50", "royalblue")[(as.numeric(diff_bins)==4|as.numeric(diff_bins)==3)+1],.7), pch=16,
xlab="PC1", ylab="PC2",
main="PCs based on GFP, RFP")
abline(h=0,v=0, lty=3)
title(main="blue dots: diff time > 50th percentile", outer=TRUE, line=-1)
par(mfrow=c(1,2))
dist_to_origin <- sqrt(pca_df_sub[,1]^2+pca_df_sub[,2]^2)
#which(abs(scale(dist_to_origin))>2)
plot(dist_to_origin, diff_time[[2]]/2/pi,
xlab="Distance to the origin in the PC1 vs PC2 space",
ylab="Maring of error (% arc length)",
main = "Prediction error", pch=16,
col=alpha(c("gray50", "forestgreen"),.7)[(abs(scale(dist_to_origin))>1)+1])
plot(pca_df_sub[,1], pca_df_sub[,2],
col=alpha(c("gray50", "forestgreen"),.7)[(abs(scale(dist_to_origin))>1)+1],
pch=16,
xlab="PC1", ylab="PC2",
main="PC1 vs. PC2 based on GFP, RFP")
abline(h=0,v=0, lty=3)
title(main="Green points: 1 +/- SD of mean distance to the origin", outer=TRUE, line=-1)
Re-fitting after removing PC outliers, remove outliers from the same 5 folds
# first check the theta in pdata
pca <- prcomp(cbind(pdata$rfp.median.log10sum.adjust,
pdata$gfp.median.log10sum.adjust), scale=TRUE)
pca_df <- cbind(pca$x[,1],pca$x[,2])
rownames(pca_df) <- rownames(pdata)
theta_check <- as.numeric(coord2rad(pca_df))
theta_check <- 2*pi-theta_check
plot(theta_check, pdata$theta)
names(theta_check) <- rownames(pdata)
dist_to_origin <- sqrt(pca_df[,1]^2+pca_df[,2]^2)
which_out <- rownames(pdata)[which(abs(scale(dist_to_origin))>1)]
plot(pca_df[,1],
pca_df[,2],
col=c("gray50", "forestgreen")[(abs(scale(dist_to_origin))>1)+1], pch=16,
xlab="PC1",
ylab="PC2",
main="abs(Distance-to-Origin) > +/- 1")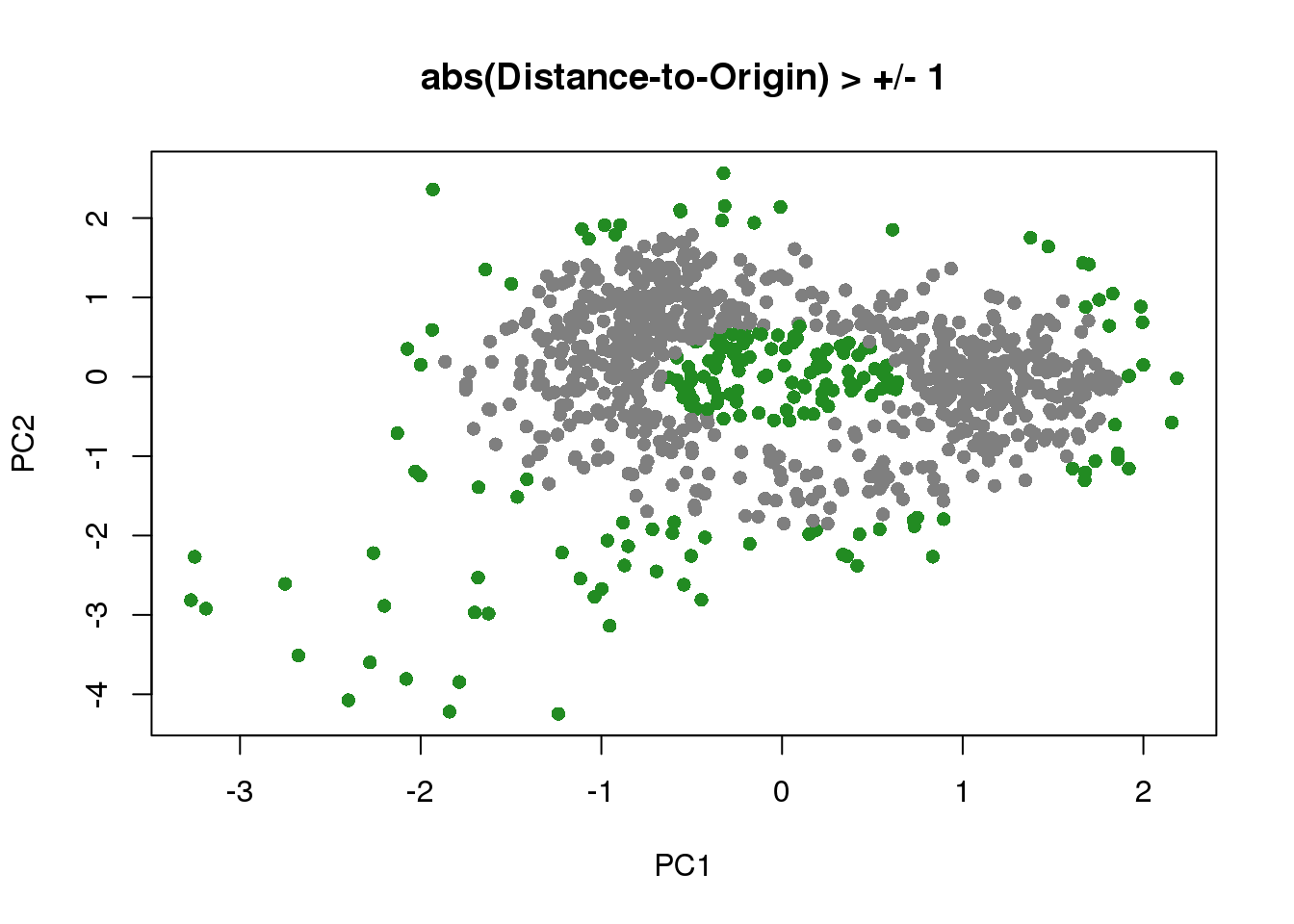
fits_sub <- vector("list", 5)
for (run in 1:5) {
print(run)
# fitting training data
Y_train <- expr.sig[,part_indices[[run]]$train]
theta_train <- theta_check[match(colnames(Y_train), rownames(pdata))]
names(theta_train) <- colnames(Y_train)
Y_train_sub <- Y_train[,which(!(colnames(Y_train) %in% which_out))]
theta_train_sub <- theta_train[which(!(names(theta_train) %in% which_out))]
fit.train <- cycle.npreg.insample(Y = Y_train_sub,
theta = theta_train_sub,
ncores=10,
method.trend="npcirc.nw")
# fitting test data
Y_test <- expr.sig[,part_indices[[run]]$test]
theta_test <- theta_check[match(colnames(Y_test), rownames(pdata))]
names(theta_test) <- colnames(Y_test)
Y_test_sub <- Y_test[,which(!(colnames(Y_test) %in% which_out))]
theta_test_sub <- theta_test[which(!(names(theta_test) %in% which_out))]
fit.test <- cycle.npreg.outsample(Y_test=Y_test_sub,
sigma_est=fit.train$sigma_est,
funs_est=fit.train$funs_est,
method.grid = "uniform",
method.trend="npcirc.nw",
ncores=12)
fits_sub[[run]] <- list(fit.train=fit.train,
fit.test=fit.test,
theta_test=theta_test_sub)
}
for (i in 1:5) {
fits_sub[[i]]$theta_est_shift <- rotation(fits_sub[[i]]$theta_test, fits_sub[[i]]$fit.test$cell_times_est)$y2shift
}
saveRDS(fits_sub, file = "../output/method-labels.Rmd/fits_sub.rds")fits_sub <- readRDS(file = "../output/method-labels.Rmd/fits_sub.rds")
diff_time <- lapply(1:5, function(i) {
pmin(abs(fits_sub[[i]]$theta_est_shift-fits_sub[[i]]$theta_test),
abs(fits_sub[[i]]$theta_est_shift-(2*pi-fits_sub[[i]]$theta_test)))
})
mean(sapply(diff_time, mean)/2/pi)[1] 0.08959582pve <- lapply(1:length(fits_sub), function(i) {
dap <- pdata$dapi.median.log10sum[match(names(fits_sub[[i]]$theta_test),rownames(pdata))]
get.pve(dap[order(fits_sub[[i]]$theta_est_shift)])
})Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ... mean(unlist(pve))[1] 0.1610684PVE of cell times in all samples after removing PC outliers
rfp_theta_sub <- with(pdata[which(!(rownames(pdata) %in% which_out)),],
get.pve(rfp.median.log10sum.adjust[order(theta)]))
gfp_theta_sub <- with(pdata[which(!(rownames(pdata) %in% which_out)),],
get.pve(gfp.median.log10sum.adjust[order(theta)]))
dapi_theta_sub <- with(pdata[which(!(rownames(pdata) %in% which_out)),],
get.pve(dapi.median.log10sum.adjust[order(theta)]))
save(rfp_theta_sub,
gfp_theta_sub,
dapi_theta_sub,
file = "../output/method-labels.Rmd/pve_no_pc_outlier.rda")load(file="../output/method-labels.Rmd/pve_no_pc_outlier.rda")
c(rfp_theta_sub,
gfp_theta_sub,
dapi_theta_sub) [1] 0.9343087 0.9233100 0.2767365Re-fitting after removing PC outliers from the same 5 folds, top 50 genes
expr_sub <- expr.sig[which(rownames(expr.sig) %in% rownames(sig.genes)[1:50]),]
fits_sub_top50 <- vector("list", 5)
for (run in 1:5) {
print(run)
# fitting training data
Y_train <- expr_sub[,part_indices[[run]]$train]
theta_train <- theta_check[match(colnames(Y_train), rownames(pdata))]
names(theta_train) <- colnames(Y_train)
Y_train_sub <- Y_train[,which(!(colnames(Y_train) %in% which_out))]
theta_train_sub <- theta_train[which(!(names(theta_train) %in% which_out))]
fit.train <- cycle.npreg.insample(Y = Y_train_sub,
theta = theta_train_sub,
ncores=10,
method.trend="npcirc.nw")
# fitting test data
Y_test <- expr_sub[,part_indices[[run]]$test]
theta_test <- theta_check[match(colnames(Y_test), rownames(pdata))]
names(theta_test) <- colnames(Y_test)
Y_test_sub <- Y_test[,which(!(colnames(Y_test) %in% which_out))]
theta_test_sub <- theta_test[which(!(names(theta_test) %in% which_out))]
fit.test <- cycle.npreg.outsample(Y_test=Y_test_sub,
sigma_est=fit.train$sigma_est,
funs_est=fit.train$funs_est,
method.grid = "uniform",
method.trend="npcirc.nw",
ncores=12)
fits_sub_top50[[run]] <- list(fit.train=fit.train,
fit.test=fit.test,
theta_test=theta_test_sub)
}
for (i in 1:5) {
fits_sub_top50[[i]]$theta_est_shift <- rotation(fits_sub_top50[[i]]$theta_test, fits_sub_top50[[i]]$fit.test$cell_times_est)$y2shift
}
saveRDS(fits_sub_top50, file = "../output/method-labels.Rmd/fits_sub_top50.rds")fits_sub_top50 <- readRDS(file = "../output/method-labels.Rmd/fits_sub_top50.rds")
diff_time <- lapply(1:5, function(i) {
pmin(abs(fits_sub_top50[[i]]$theta_est_shift-fits_sub_top50[[i]]$theta_test),
abs(fits_sub_top50[[i]]$theta_est_shift-(2*pi-fits_sub_top50[[i]]$theta_test)))
})
mean(sapply(diff_time, mean)/2/pi)[1] 0.08906205pve <- lapply(1:length(fits_sub_top50), function(i) {
dap <- pdata$dapi.median.log10sum[match(names(fits_sub_top50[[i]]$theta_test),
rownames(pdata))]
get.pve(dap[order(fits_sub_top50[[i]]$theta_est_shift)])
})Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ... mean(unlist(pve))[1] 0.1712479Re-fitting after removing PC outliers from the same 5 folds, top 10 genes
expr_sub <- expr.sig[which(rownames(expr.sig) %in% rownames(sig.genes)[1:10]),]
fits_sub_top10 <- vector("list", 5)
for (run in 1:5) {
print(run)
# fitting training data
Y_train <- expr_sub[,part_indices[[run]]$train]
theta_train <- theta_check[match(colnames(Y_train), rownames(pdata))]
names(theta_train) <- colnames(Y_train)
Y_train_sub <- Y_train[,which(!(colnames(Y_train) %in% which_out))]
theta_train_sub <- theta_train[which(!(names(theta_train) %in% which_out))]
fit.train <- cycle.npreg.insample(Y = Y_train_sub,
theta = theta_train_sub,
ncores=10,
method.trend="npcirc.nw")
# fitting test data
Y_test <- expr_sub[,part_indices[[run]]$test]
theta_test <- theta_check[match(colnames(Y_test), rownames(pdata))]
names(theta_test) <- colnames(Y_test)
Y_test_sub <- Y_test[,which(!(colnames(Y_test) %in% which_out))]
theta_test_sub <- theta_test[which(!(names(theta_test) %in% which_out))]
fit.test <- cycle.npreg.outsample(Y_test=Y_test_sub,
sigma_est=fit.train$sigma_est,
funs_est=fit.train$funs_est,
method.grid = "uniform",
method.trend="npcirc.nw",
ncores=12)
fits_sub_top10[[run]] <- list(fit.train=fit.train,
fit.test=fit.test,
theta_test=theta_test_sub)
}
for (i in 1:5) {
fits_sub_top10[[i]]$theta_est_shift <- rotation(fits_sub_top10[[i]]$theta_test, fits_sub_top10[[i]]$fit.test$cell_times_est)$y2shift
}
saveRDS(fits_sub_top10, file = "../output/method-labels.Rmd/fits_sub_top10.rds")fits_sub_top10 <- readRDS(file = "../output/method-labels.Rmd/fits_sub_top10.rds")
diff_time <- lapply(1:5, function(i) {
pmin(abs(fits_sub_top10[[i]]$theta_est_shift-fits_sub_top10[[i]]$theta_test),
abs(fits_sub_top10[[i]]$theta_est_shift-(2*pi-fits_sub_top10[[i]]$theta_test)))
})
mean(sapply(diff_time, mean)/2/pi)[1] 0.08471943pve <- lapply(1:length(fits_sub_top10), function(i) {
dap <- pdata$dapi.median.log10sum[match(names(fits_sub_top10[[i]]$theta_test),
rownames(pdata))]
get.pve(dap[order(fits_sub_top10[[i]]$theta_est_shift)])
})Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ... mean(unlist(pve))[1] 0.2258223Re-fitting after removing PC outliers, odd number
expr_sub <- expr.sig[which(rownames(expr.sig) %in% rownames(sig.genes)[seq(1,nrow(expr.sig),by=2)]),]
fits_sub_odd <- vector("list", 5)
for (run in 1:5) {
print(run)
# fitting training data
Y_train <- expr_sub[,part_indices[[run]]$train]
theta_train <- theta_check[match(colnames(Y_train), rownames(pdata))]
names(theta_train) <- colnames(Y_train)
Y_train_sub <- Y_train[,which(!(colnames(Y_train) %in% which_out))]
theta_train_sub <- theta_train[which(!(names(theta_train) %in% which_out))]
fit.train <- cycle.npreg.insample(Y = Y_train_sub,
theta = theta_train_sub,
ncores=10,
method.trend="npcirc.nw")
# fitting test data
Y_test <- expr_sub[,part_indices[[run]]$test]
theta_test <- theta_check[match(colnames(Y_test), rownames(pdata))]
names(theta_test) <- colnames(Y_test)
Y_test_sub <- Y_test[,which(!(colnames(Y_test) %in% which_out))]
theta_test_sub <- theta_test[which(!(names(theta_test) %in% which_out))]
fit.test <- cycle.npreg.outsample(Y_test=Y_test_sub,
sigma_est=fit.train$sigma_est,
funs_est=fit.train$funs_est,
method.grid = "uniform",
method.trend="npcirc.nw",
ncores=12)
fits_sub_odd[[run]] <- list(fit.train=fit.train,
fit.test=fit.test,
theta_test=theta_test_sub)
}
for (i in 1:5) {
fits_sub_odd[[i]]$theta_est_shift <- rotation(fits_sub_odd[[i]]$theta_test, fits_sub_odd[[i]]$fit.test$cell_times_est)$y2shift
}
saveRDS(fits_sub_odd, file = "../output/method-labels.Rmd/fits_sub_odd.rds")fits_sub_odd <- readRDS(file = "../output/method-labels.Rmd/fits_sub_odd.rds")
diff_time <- lapply(1:5, function(i) {
pmin(abs(fits_sub_odd[[i]]$theta_est_shift-fits_sub_odd[[i]]$theta_test),
abs(fits_sub_odd[[i]]$theta_est_shift-(2*pi-fits_sub_odd[[i]]$theta_test)))
})
mean(sapply(diff_time, mean)/2/pi)[1] 0.09553247pve <- lapply(1:length(fits_sub_odd), function(i) {
dap <- pdata$dapi.median.log10sum[match(names(fits_sub_odd[[i]]$theta_test),
rownames(pdata))]
get.pve(dap[order(fits_sub_odd[[i]]$theta_est_shift)])
})Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ... mean(unlist(pve))[1] 0.08201038Re-fitting after removing PC outliers, even number
expr_sub <- expr.sig[which(rownames(expr.sig) %in% rownames(sig.genes)[seq(2,nrow(expr.sig),by=2)]),]
fits_sub_even <- vector("list", 5)
for (run in 1:5) {
print(run)
# fitting training data
Y_train <- expr_sub[,part_indices[[run]]$train]
theta_train <- theta_check[match(colnames(Y_train), rownames(pdata))]
names(theta_train) <- colnames(Y_train)
Y_train_sub <- Y_train[,which(!(colnames(Y_train) %in% which_out))]
theta_train_sub <- theta_train[which(!(names(theta_train) %in% which_out))]
fit.train <- cycle.npreg.insample(Y = Y_train_sub,
theta = theta_train_sub,
ncores=10,
method.trend="npcirc.nw")
# fitting test data
Y_test <- expr_sub[,part_indices[[run]]$test]
theta_test <- theta_check[match(colnames(Y_test), rownames(pdata))]
names(theta_test) <- colnames(Y_test)
Y_test_sub <- Y_test[,which(!(colnames(Y_test) %in% which_out))]
theta_test_sub <- theta_test[which(!(names(theta_test) %in% which_out))]
fit.test <- cycle.npreg.outsample(Y_test=Y_test_sub,
sigma_est=fit.train$sigma_est,
funs_est=fit.train$funs_est,
method.grid = "uniform",
method.trend="npcirc.nw",
ncores=12)
fits_sub_even[[run]] <- list(fit.train=fit.train,
fit.test=fit.test,
theta_test=theta_test_sub)
}
for (i in 1:5) {
fits_sub_even[[i]]$theta_est_shift <- rotation(fits_sub_even[[i]]$theta_test, fits_sub_even[[i]]$fit.test$cell_times_est)$y2shift
}
saveRDS(fits_sub_even, file = "../output/method-labels.Rmd/fits_sub_even.rds")fits_sub_even <- readRDS(file = "../output/method-labels.Rmd/fits_sub_even.rds")
diff_time <- lapply(1:5, function(i) {
pmin(abs(fits_sub_even[[i]]$theta_est_shift-fits_sub_even[[i]]$theta_test),
abs(fits_sub_even[[i]]$theta_est_shift-(2*pi-fits_sub_even[[i]]$theta_test)))
})
mean(sapply(diff_time, mean)/2/pi)[1] 0.09370391pve <- lapply(1:length(fits_sub_even), function(i) {
dap <- pdata$dapi.median.log10sum[match(names(fits_sub_even[[i]]$theta_test),
rownames(pdata))]
get.pve(dap[order(fits_sub_even[[i]]$theta_est_shift)])
})Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ... mean(unlist(pve))[1] 0.22471Fitting using all samples, top 50 genes
expr_sub <- expr.sig[which(rownames(expr.sig) %in% rownames(sig.genes)[1:50]),]
fits_top50 <- vector("list", 5)
for (run in 1:5) {
print(run)
# fitting training data
Y_train <- expr_sub[,part_indices[[run]]$train]
theta_train <- theta_check[match(colnames(Y_train), rownames(pdata))]
names(theta_train) <- colnames(Y_train)
fit.train <- cycle.npreg.insample(Y = Y_train,
theta = theta_train,
ncores=10,
method.trend="npcirc.nw")
# fitting test data
Y_test <- expr_sub[,part_indices[[run]]$test]
theta_test <- theta_check[match(colnames(Y_test), rownames(pdata))]
names(theta_test) <- colnames(Y_test)
fit.test <- cycle.npreg.outsample(Y_test=Y_test,
sigma_est=fit.train$sigma_est,
funs_est=fit.train$funs_est,
method.grid = "uniform",
method.trend="npcirc.nw",
ncores=12)
fits_top50[[run]] <- list(fit.train=fit.train,
fit.test=fit.test,
theta_test=theta_test)
}
for (i in 1:5) {
fits_top50[[i]]$theta_est_shift <- rotation(fits_top50[[i]]$theta_test, fits_top50[[i]]$fit.test$cell_times_est)$y2shift
}
saveRDS(fits_top50, file = "../output/method-labels.Rmd/fits_top50.rds")fits_top50 <- readRDS(file = "../output/method-labels.Rmd/fits_top50.rds")
source("../peco/R/utility.R")
diff_time <- lapply(1:length(fits_top50), function(i) {
pmin(abs(fits_top50[[i]]$theta_est_shift-fits_top50[[i]]$theta_test),
abs(fits_top50[[i]]$theta_est_shift-(2*pi-fits_top50[[i]]$theta_test)))
})
mean(sapply(diff_time, mean)/2/pi)[1] 0.088139pve <- lapply(1:length(fits_top50), function(i) {
dap <- pdata$dapi.median.log10sum[match(names(fits_top50[[i]]$theta_test),
rownames(pdata))]
get.pve(dap[order(fits_top50[[i]]$theta_est_shift)])
})Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ... mean(unlist(pve))[1] 0.1356245Fitting using all samples, top 10 genes
expr_sub <- expr.sig[which(rownames(expr.sig) %in% rownames(sig.genes)[1:10]),]
fits_top10 <- vector("list", 5)
for (run in 1:5) {
print(run)
# fitting training data
Y_train <- expr_sub[,part_indices[[run]]$train]
theta_train <- theta_check[match(colnames(Y_train), rownames(pdata))]
names(theta_train) <- colnames(Y_train)
fit.train <- cycle.npreg.insample(Y = Y_train,
theta = theta_train,
ncores=10,
method.trend="npcirc.nw")
# fitting test data
Y_test <- expr_sub[,part_indices[[run]]$test]
theta_test <- theta_check[match(colnames(Y_test), rownames(pdata))]
names(theta_test) <- colnames(Y_test)
fit.test <- cycle.npreg.outsample(Y_test=Y_test,
sigma_est=fit.train$sigma_est,
funs_est=fit.train$funs_est,
method.grid = "uniform",
method.trend="npcirc.nw",
ncores=12)
fits_top10[[run]] <- list(fit.train=fit.train,
fit.test=fit.test,
theta_test=theta_test)
}
for (i in 1:5) {
fits_top10[[i]]$theta_est_shift <- rotation(fits_top10[[i]]$theta_test, fits_top10[[i]]$fit.test$cell_times_est)$y2shift
}
saveRDS(fits_top10, file = "../output/method-labels.Rmd/fits_top10.rds")fits_top10 <- readRDS(file = "../output/method-labels.Rmd/fits_top10.rds")
source("../peco/R/utility.R")
diff_time <- lapply(1:length(fits_top10), function(i) {
pmin(abs(fits_top10[[i]]$theta_est_shift-fits_top10[[i]]$theta_test),
abs(fits_top10[[i]]$theta_est_shift-(2*pi-fits_top10[[i]]$theta_test)))
})
mean(sapply(diff_time, mean)/2/pi)[1] 0.08583889pve <- lapply(1:length(fits_top10), function(i) {
dap <- pdata$dapi.median.log10sum[match(names(fits_top10[[i]]$theta_test),
rownames(pdata))]
get.pve(dap[order(fits_top10[[i]]$theta_est_shift)])
})Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ... mean(unlist(pve))[1] 0.1680728Fitting using all samples, odd-numbered genes
expr_sub <- expr.sig[which(rownames(expr.sig) %in% rownames(sig.genes)[seq(1,nrow(expr.sig),by=2)]),]
fits_top50_odd <- vector("list", 5)
for (run in 1:5) {
print(run)
# fitting training data
Y_train <- expr_sub[,part_indices[[run]]$train]
theta_train <- theta_check[match(colnames(Y_train), rownames(pdata))]
names(theta_train) <- colnames(Y_train)
fit.train <- cycle.npreg.insample(Y = Y_train,
theta = theta_train,
ncores=10,
method.trend="npcirc.nw")
# fitting test data
Y_test <- expr_sub[,part_indices[[run]]$test]
theta_test <- theta_check[match(colnames(Y_test), rownames(pdata))]
names(theta_test) <- colnames(Y_test)
fit.test <- cycle.npreg.outsample(Y_test=Y_test,
sigma_est=fit.train$sigma_est,
funs_est=fit.train$funs_est,
method.grid = "uniform",
method.trend="npcirc.nw",
ncores=12)
fits_top50_odd[[run]] <- list(fit.train=fit.train,
fit.test=fit.test,
theta_test=theta_test)
}
for (i in 1:5) {
fits_top50_odd[[i]]$theta_est_shift <- rotation(fits_top50_odd[[i]]$theta_test, fits_top50_odd[[i]]$fit.test$cell_times_est)$y2shift
}
saveRDS(fits_top50_odd, file = "../output/method-labels.Rmd/fits_top50_odd.rds")fits_top50_odd <- readRDS(file = "../output/method-labels.Rmd/fits_top50_odd.rds")
source("../peco/R/utility.R")
diff_time <- lapply(1:length(fits_top50_odd), function(i) {
pmin(abs(fits_top50_odd[[i]]$theta_est_shift-fits_top50_odd[[i]]$theta_test),
abs(fits_top50_odd[[i]]$theta_est_shift-(2*pi-fits_top50_odd[[i]]$theta_test)))
})
mean(sapply(diff_time, mean)/2/pi)[1] 0.09638676pve <- lapply(1:length(fits_top50_odd), function(i) {
dap <- pdata$dapi.median.log10sum[match(names(fits_top50_odd[[i]]$theta_test),
rownames(pdata))]
get.pve(dap[order(fits_top50_odd[[i]]$theta_est_shift)])
})Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ... mean(unlist(pve))[1] 0.09501819Fitting using all samples, even-numbered genes
expr_sub <- expr.sig[which(rownames(expr.sig) %in% rownames(sig.genes)[seq(2,nrow(expr.sig),by=2)]),]
fits_top50_even <- vector("list", 5)
for (run in 1:5) {
print(run)
# fitting training data
Y_train <- expr_sub[,part_indices[[run]]$train]
theta_train <- theta_check[match(colnames(Y_train), rownames(pdata))]
names(theta_train) <- colnames(Y_train)
fit.train <- cycle.npreg.insample(Y = Y_train,
theta = theta_train,
ncores=10,
method.trend="npcirc.nw")
# fitting test data
Y_test <- expr_sub[,part_indices[[run]]$test]
theta_test <- theta_check[match(colnames(Y_test), rownames(pdata))]
names(theta_test) <- colnames(Y_test)
fit.test <- cycle.npreg.outsample(Y_test=Y_test,
sigma_est=fit.train$sigma_est,
funs_est=fit.train$funs_est,
method.grid = "uniform",
method.trend="npcirc.nw",
ncores=12)
fits_top50_even[[run]] <- list(fit.train=fit.train,
fit.test=fit.test,
theta_test=theta_test)
}
for (i in 1:5) {
fits_top50_even[[i]]$theta_est_shift <- rotation(fits_top50_even[[i]]$theta_test, fits_top50_even[[i]]$fit.test$cell_times_est)$y2shift
}
saveRDS(fits_top50_even, file = "../output/method-labels.Rmd/fits_top50_even.rds")fits_top50_even <- readRDS(file = "../output/method-labels.Rmd/fits_top50_even.rds")
source("../peco/R/utility.R")
diff_time <- lapply(1:length(fits_top50_even), function(i) {
pmin(abs(fits_top50_even[[i]]$theta_est_shift-fits_top50_even[[i]]$theta_test),
abs(fits_top50_even[[i]]$theta_est_shift-(2*pi-fits_top50_even[[i]]$theta_test)))
})
mean(sapply(diff_time, mean)/2/pi)[1] 0.0941044pve <- lapply(1:length(fits_top50_even), function(i) {
dap <- pdata$dapi.median.log10sum[match(names(fits_top50_even[[i]]$theta_test),
rownames(pdata))]
get.pve(dap[order(fits_top50_even[[i]]$theta_est_shift)])
})Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ...
Fold 1 ... Fold 2 ... Fold 3 ... Fold 4 ... Fold 5 ... mean(unlist(pve))[1] 0.1702022Session information
sessionInfo()R version 3.4.3 (2017-11-30)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] parallel stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] scales_0.5.0 RColorBrewer_1.1-2 genlasso_1.3
[4] igraph_1.1.2 Matrix_1.2-14 MASS_7.3-50
[7] circular_0.4-93 Biobase_2.38.0 BiocGenerics_0.24.0
loaded via a namespace (and not attached):
[1] Rcpp_0.12.17 knitr_1.18 magrittr_1.5 munsell_0.4.3
[5] colorspace_1.3-2 lattice_0.20-35 plyr_1.8.4 stringr_1.2.0
[9] tools_3.4.3 grid_3.4.3 git2r_0.21.0 htmltools_0.3.6
[13] yaml_2.1.16 rprojroot_1.3-2 digest_0.6.15 evaluate_0.10.1
[17] rmarkdown_1.8 stringi_1.1.6 compiler_3.4.3 backports_1.1.2
[21] boot_1.3-20 mvtnorm_1.0-7 pkgconfig_2.0.1 This R Markdown site was created with workflowr