Tidy data with tidyr
John Blischak
2019-10-15
Last updated: 2019-10-15
Checks: 7 0
Knit directory: wflow-r4ds/
This reproducible R Markdown analysis was created with workflowr (version 1.4.0.9001). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190925) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the R Markdown and HTML files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view them.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | d856862 | John Blischak | 2019-10-16 | Chapter 9 exercises on tidyr |
Setup
library(tidyverse)Tidy data
p. 151
- Using prose, describe how the variables and observations are organised in each of the sample tables.
table1- Each observation of a country in a given year is in its own row. The number of cases and the total population are columns.table2- This is a longer format. Each observation has two rows, one for the cases and one for the population.table3- Each observation of a country in a given year is in its own row. The number of cases and the total population are combined into a single column.table4a- Each country is a row. The number of cases in 1999 and 2000 are in separate columns.table4b- Each country is a row. The total population in 1999 and 2000 are in separate columns.
Compute the
ratefortable2, andtable4a+table4b. You will need to perform four operations:- Extract the number of TB cases per country per year.
- Extract the matching population per country per year.
- Divide cases by population, and multiply by 10000.
- Store back in the appropriate place.
Which representation is easiest to work with? Which is hardest? Why?
The example tables are included as data sets when the tidyr package is loaded (see ?table1).
tb <- table2 %>% filter(type == "cases") %>% select(country, year, tb = count)
pop <- table2 %>% filter(type == "population") %>% select(pop = count)
rate <- tb$tb / pop$pop * 10^4
cbind(tb, pop, rate) country year tb pop rate
1 Afghanistan 1999 745 19987071 0.372741
2 Afghanistan 2000 2666 20595360 1.294466
3 Brazil 1999 37737 172006362 2.193930
4 Brazil 2000 80488 174504898 4.612363
5 China 1999 212258 1272915272 1.667495
6 China 2000 213766 1280428583 1.669488tb <- c(table4a$`1999`, table4a$`2000`)
pop <- c(table4b$`1999`, table4b$`2000`)
rate <- tb / pop * 10^4
cbind(table4a$country, tb, pop, rate) tb pop rate
[1,] "Afghanistan" "745" "19987071" "0.372740958392553"
[2,] "Brazil" "37737" "172006362" "2.19393047799011"
[3,] "China" "212258" "1272915272" "1.66749511667419"
[4,] "Afghanistan" "2666" "20595360" "1.29446632639585"
[5,] "Brazil" "80488" "174504898" "4.61236337331918"
[6,] "China" "213766" "1280428583" "1.6694878795907" They were both kind of awkward.
- Recreate the plot showing change in cases over time using
table2instead oftable1. What do you need to do first?
Need to convert from long to wide. Kind of putting the cart before the horse. I guess maybe they want you to think about how painful this is to do manually.
I’ll reuse my code from above:
tb <- table2 %>% filter(type == "cases") %>% select(country, year, tb = count)
pop <- table2 %>% filter(type == "population") %>% select(pop = count)
rate <- tb$tb / pop$pop * 10^4
cbind(tb, pop, rate) %>%
ggplot(aes(x = year, y = tb)) +
geom_line(aes(group = country), color = "grey50") +
geom_point(aes(color = country))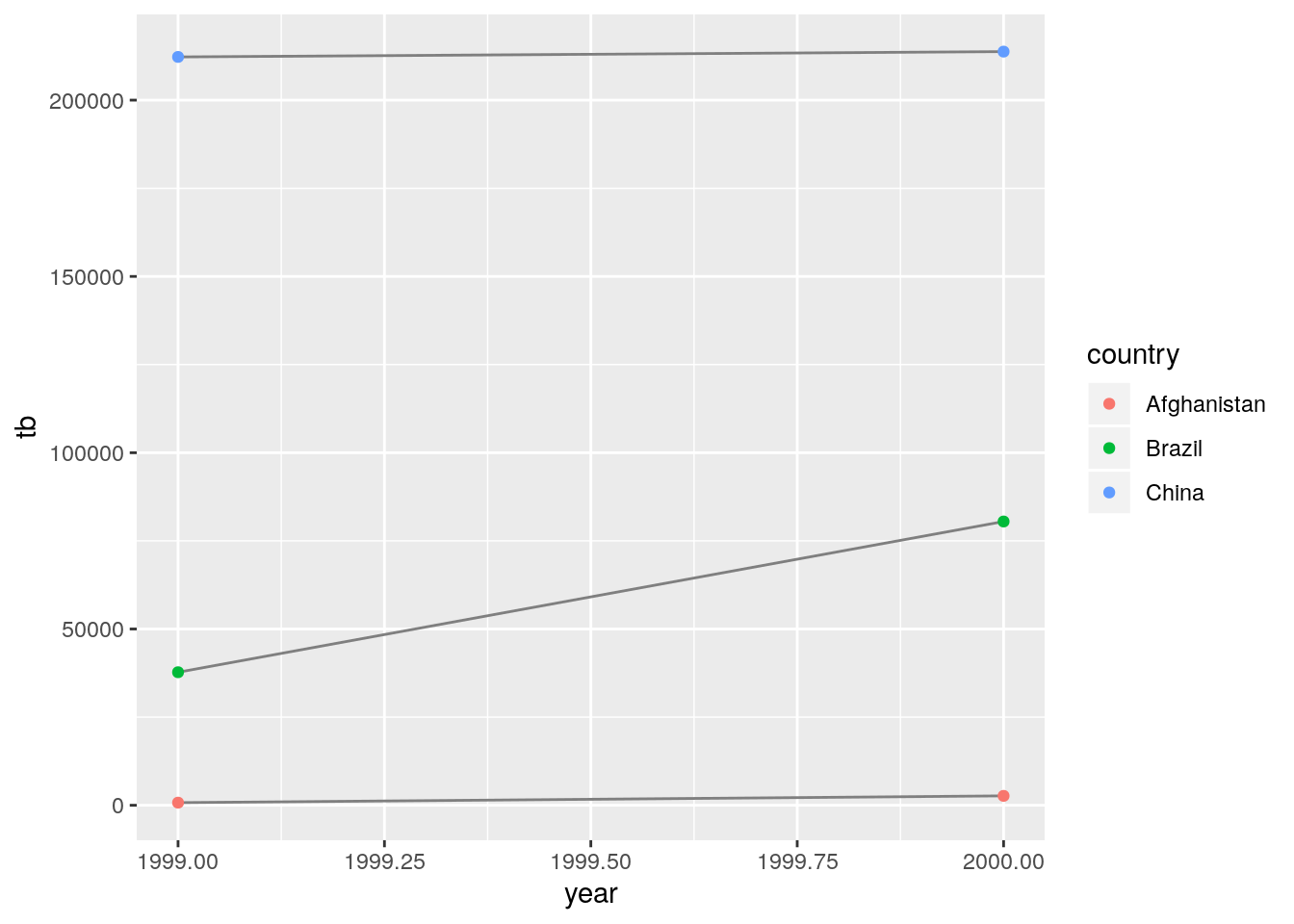
Spreading and gathering
p. 156
Why are
gather()andspread()not perfectly symmetrical?
Carefully consider the following example:stocks <- tibble( year = c(2015, 2015, 2016, 2016), half = c( 1, 2, 1, 2), return = c(1.88, 0.59, 0.92, 0.17) ) stocks %>% spread(year, return) %>% gather("year", "return", `2015`:`2016`)# A tibble: 4 x 3 half year return <dbl> <chr> <dbl> 1 1 2015 1.88 2 2 2015 0.59 3 1 2016 0.92 4 2 2016 0.17(Hint: look at the variable types and think about column names.)
Year is converted to a character vector after gather() moves them from column names to their own column.
Both `spread()` and `gather()` have a `convert` argument. What does it
do?If convert=TRUE (the default is FALSE), then utils::type.convert() will be run on the newly created columns. This results in year being converted to an integer, which is still different from its original status as a numeric vector.
stocks %>%
spread(year, return) %>%
gather("year", "return", `2015`:`2016`, convert = TRUE)# A tibble: 4 x 3
half year return
<dbl> <int> <dbl>
1 1 2015 1.88
2 2 2015 0.59
3 1 2016 0.92
4 2 2016 0.17Why does this code fail?
table4a %>% gather(1999, 2000, key = "year", value = "cases")Error in inds_combine(.vars, ind_list): Position must be between 0 and n
Because 1999 and 2000 are numbers, they are interpreted as indicating the columns to gather by their position. Using column position is an alternative way to specify the columns:
table4a %>%
gather(2, 3, key = "year", value = "cases")# A tibble: 6 x 3
country year cases
<chr> <chr> <int>
1 Afghanistan 1999 745
2 Brazil 1999 37737
3 China 1999 212258
4 Afghanistan 2000 2666
5 Brazil 2000 80488
6 China 2000 213766Why does spreading this tibble fail? How could you add a new column to fix the problem?
people <- tribble( ~name, ~key, ~value, #-----------------|--------|------ "Phillip Woods", "age", 45, "Phillip Woods", "height", 186, "Phillip Woods", "age", 50, "Jessica Cordero", "age", 37, "Jessica Cordero", "height", 156 )
It fails because there are two entries for name “Phillip Woods” and “age”. This could be fixed by adding a new column with a unique ID for each individual, so that people with the same name could be distinguished.
people %>%
spread(key, value)Each row of output must be identified by a unique combination of keys.
Keys are shared for 2 rows:
* 1, 3Tidy the simple tibble below. Do you need to spread or gather it? What are the variables?
preg <- tribble( ~pregnant, ~male, ~female, "yes", NA, 10, "no", 20, 12 )
The variables are pregnancy status (yes, no), gender (male, female), and count (the number of observations of each combination of pregnancy status and gender). It needs to be gathered so that the variable gender is its own column.
preg %>%
gather(key = "gender", value = "count", male, female)# A tibble: 4 x 3
pregnant gender count
<chr> <chr> <dbl>
1 yes male NA
2 no male 20
3 yes female 10
4 no female 12Separating and uniting
p. 160
Note in the print copy I have, the section is mistakenly named “Separating and pull”.
What do the
extraandfillarguments do inseparate()? Experiment with the various options for the following two toy datasets.tibble(x = c("a,b,c", "d,e,f,g", "h,i,j")) %>% separate(x, c("one", "two", "three")) tibble(x = c("a,b,c", "d,e", "f,g,i")) %>% separate(x, c("one", "two", "three"))
The argument extra “controls what happens when there are too many pieces”.
tibble(x = c("a,b,c", "d,e,f,g", "h,i,j"))# A tibble: 3 x 1
x
<chr>
1 a,b,c
2 d,e,f,g
3 h,i,j tibble(x = c("a,b,c", "d,e,f,g", "h,i,j")) %>%
separate(x, c("one", "two", "three"))Warning: Expected 3 pieces. Additional pieces discarded in 1 rows [2].# A tibble: 3 x 3
one two three
<chr> <chr> <chr>
1 a b c
2 d e f
3 h i j tibble(x = c("a,b,c", "d,e,f,g", "h,i,j")) %>%
separate(x, c("one", "two", "three"), extra = "drop")# A tibble: 3 x 3
one two three
<chr> <chr> <chr>
1 a b c
2 d e f
3 h i j tibble(x = c("a,b,c", "d,e,f,g", "h,i,j")) %>%
separate(x, c("one", "two", "three"), extra = "merge")# A tibble: 3 x 3
one two three
<chr> <chr> <chr>
1 a b c
2 d e f,g
3 h i j The argument fill “controls what happens when there are not enough pieces”.
tibble(x = c("a,b,c", "d,e", "f,g,i"))# A tibble: 3 x 1
x
<chr>
1 a,b,c
2 d,e
3 f,g,itibble(x = c("a,b,c", "d,e", "f,g,i")) %>%
separate(x, c("one", "two", "three"))Warning: Expected 3 pieces. Missing pieces filled with `NA` in 1 rows [2].# A tibble: 3 x 3
one two three
<chr> <chr> <chr>
1 a b c
2 d e <NA>
3 f g i tibble(x = c("a,b,c", "d,e", "f,g,i")) %>%
separate(x, c("one", "two", "three"), fill = "right")# A tibble: 3 x 3
one two three
<chr> <chr> <chr>
1 a b c
2 d e <NA>
3 f g i tibble(x = c("a,b,c", "d,e", "f,g,i")) %>%
separate(x, c("one", "two", "three"), fill = "left")# A tibble: 3 x 3
one two three
<chr> <chr> <chr>
1 a b c
2 <NA> d e
3 f g i - Both
unite()andseparate()have aremoveargument. What does it do? Why would you set it toFALSE?
The argument remove removes the original column(s) that was/were separated or united from the final output. You would set it to FALSE if you wanted to maintain these columns for your analysis. For example, it could be useful to have the 4-digit year, 2-digit year, and century in the same data frame.
- Compare and contrast
separate()andextract(). Why are there three variations of separation (by position, by separator, and with groups), but only one unite?
extract() uses capture groups from a regular expression. separate() uses a character or numeric position to split.
Because there are so many potential ways to separate a character string. On the other hand, uniting only requires the connecting character. And if you wanted to unite from subsets of the existing columns, you could first run separate() to create the new columns and then unite them.
Missing values
p.163
- Compare and contrast the
fillarguments tospread()andcomplete().
The argument fill to both functions provides the option of specifying the value to use for missing values (instead of NA). The difference is that spread() only accepts one argument; whereas, complete() expects a named list where the name corresponds to the column where the missing value should be replaced.
- What does the direction argument to
fill()do?
Determines whether to fill in missing values from top to bottom ("down", the default) or from bottom to top ("up"). There are also the options "downup" and "updown", but it is unclear to me how these work from the description, and the toy data set doesn’t reveal any difference with their shorter counterparts.
treatment <- tribble(
~ person, ~ treatment, ~response,
"Derrick Whitmore", 1, 7,
NA, 2, 10,
NA, 3, 9,
"Katherine Burke", 1, 4
)
treatment %>%
fill(person, .direction = "down")# A tibble: 4 x 3
person treatment response
<chr> <dbl> <dbl>
1 Derrick Whitmore 1 7
2 Derrick Whitmore 2 10
3 Derrick Whitmore 3 9
4 Katherine Burke 1 4treatment %>%
fill(person, .direction = "up")# A tibble: 4 x 3
person treatment response
<chr> <dbl> <dbl>
1 Derrick Whitmore 1 7
2 Katherine Burke 2 10
3 Katherine Burke 3 9
4 Katherine Burke 1 4treatment %>%
fill(person, .direction = "downup")# A tibble: 4 x 3
person treatment response
<chr> <dbl> <dbl>
1 Derrick Whitmore 1 7
2 Derrick Whitmore 2 10
3 Derrick Whitmore 3 9
4 Katherine Burke 1 4treatment %>%
fill(person, .direction = "updown")# A tibble: 4 x 3
person treatment response
<chr> <dbl> <dbl>
1 Derrick Whitmore 1 7
2 Katherine Burke 2 10
3 Katherine Burke 3 9
4 Katherine Burke 1 4Case study
p. 168
tidywho <- who %>%
gather(key, value, new_sp_m014:newrel_f65, na.rm = TRUE) %>%
mutate(key = stringr::str_replace(key, "newrel", "new_rel")) %>%
separate(key, c("new", "var", "sexage")) %>%
select(-new, -iso2, -iso3) %>%
separate(sexage, c("sex", "age"), sep = 1)- In this case study I set
na.rm = TRUEjust to make it easier to check that we had the correct values. Is this reasonable? Think about how missing values are represented in this dataset. Are there implicit missing values? What’s the difference between anNAand zero?
The data set contains both NA and zeros, e.g.
length(who$new_sp_f014)[1] 7240sum(is.na(who$new_sp_m014))[1] 4067sum(who$new_sp_m014 == 0, na.rm = TRUE)[1] 862tidywho_na <- who %>%
gather(key, value, new_sp_m014:newrel_f65, na.rm = FALSE) %>%
mutate(key = stringr::str_replace(key, "newrel", "new_rel")) %>%
separate(key, c("new", "var", "sexage")) %>%
select(-new, -iso2, -iso3) %>%
separate(sexage, c("sex", "age"), sep = 1)- What happens if you neglect the
mutate()step? (mutate(key = stringr::str_replace(key, "newrel", "new_rel")))
Because there aren’t enough pieces (2 instead of 3), all the variables with newrel have their sexage stored in var, which results in sex and age being missing for all the new cases of relapsed TB.
who %>%
gather(key, value, new_sp_m014:newrel_f65, na.rm = TRUE) %>%
separate(key, c("new", "var", "sexage")) %>%
tailWarning: Expected 3 pieces. Missing pieces filled with `NA` in 2580 rows
[73467, 73468, 73469, 73470, 73471, 73472, 73473, 73474, 73475, 73476,
73477, 73478, 73479, 73480, 73481, 73482, 73483, 73484, 73485, 73486, ...].# A tibble: 6 x 8
country iso2 iso3 year new var sexage value
<chr> <chr> <chr> <int> <chr> <chr> <chr> <int>
1 Venezuela (Bolivarian Republi… VE VEN 2013 newr… f65 <NA> 402
2 Viet Nam VN VNM 2013 newr… f65 <NA> 3110
3 Wallis and Futuna Islands WF WLF 2013 newr… f65 <NA> 2
4 Yemen YE YEM 2013 newr… f65 <NA> 360
5 Zambia ZM ZMB 2013 newr… f65 <NA> 669
6 Zimbabwe ZW ZWE 2013 newr… f65 <NA> 725- I claimed that
iso2andiso3were redundant withcountry. Confirm this claim.
who %>%
group_by(country) %>%
summarize(n_iso2 = n_distinct(iso2),
n_iso3 = n_distinct(iso3)) %>%
{stopifnot(.$n_iso2 == 1, .$n_iso3 == 1)}
# Note: need to surround last expression in {} because this prevents . from
# being passed as the first argument to stopifnot(), which obviously causes a
# failure since a tibble does not evaluate to TRUE
# https://stackoverflow.com/a/42386886/2483477- For each country, year, and sex compute the total number of cases of TB. Make an informative visualisation of the data.
tidywho %>%
group_by(country, year, sex) %>%
summarize(total_cases = sum(value)) %>%
ggplot(aes(x = year, y = total_cases)) +
geom_point(aes(color = country)) +
geom_smooth() +
facet_wrap(~sex) +
theme(legend.position = "none")`geom_smooth()` using method = 'gam' and formula 'y ~ s(x, bs = "cs")'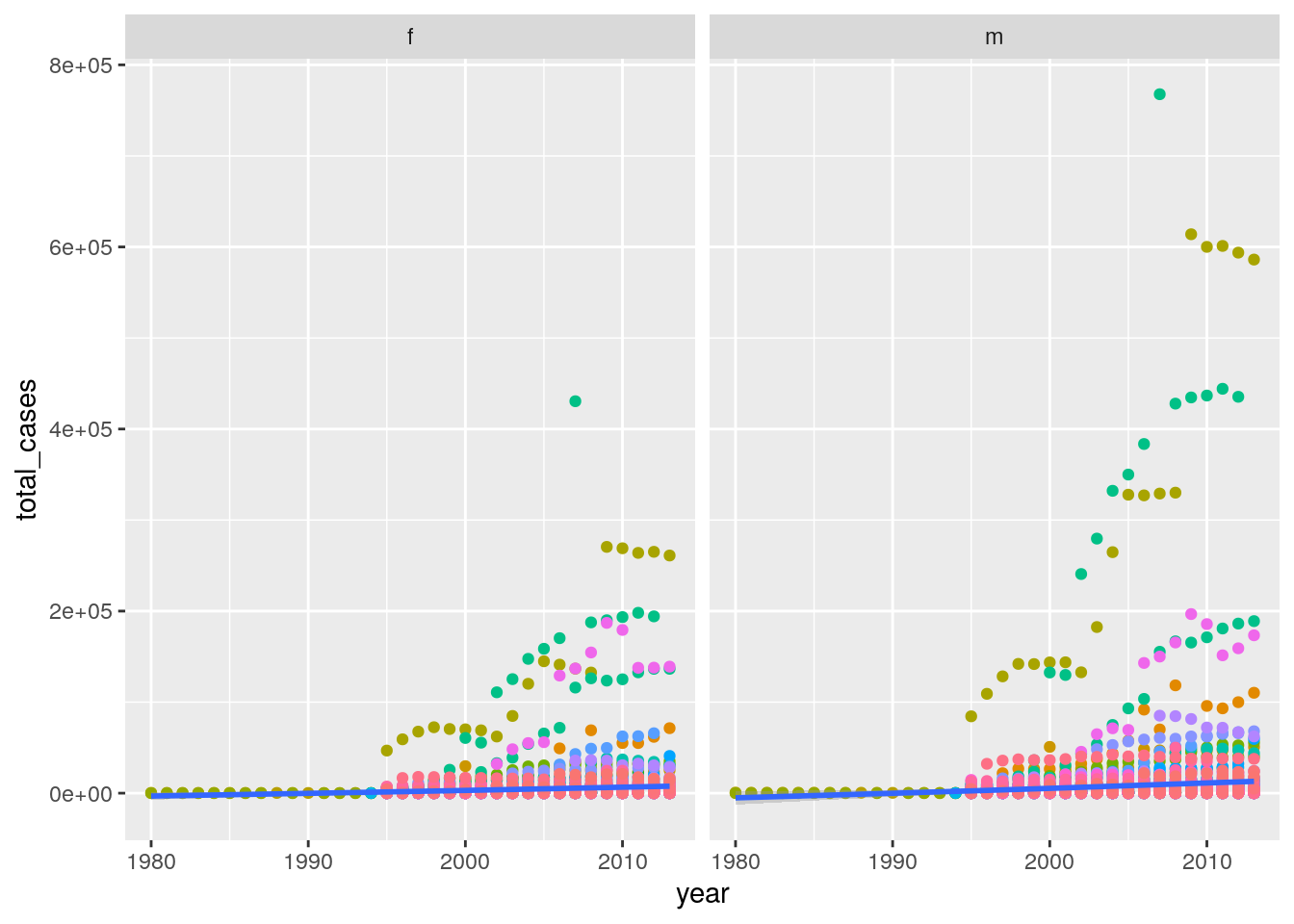
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 18.04.3 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/atlas/libblas.so.3.10.3
LAPACK: /usr/lib/x86_64-linux-gnu/atlas/liblapack.so.3.10.3
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] forcats_0.4.0 stringr_1.4.0 dplyr_0.8.3 purrr_0.3.2
[5] readr_1.3.1 tidyr_1.0.0 tibble_2.1.3 ggplot2_3.2.1
[9] tidyverse_1.2.1
loaded via a namespace (and not attached):
[1] tidyselect_0.2.5 xfun_0.10 splines_3.6.1
[4] haven_2.1.1 lattice_0.20-38 colorspace_1.4-1
[7] vctrs_0.2.0 generics_0.0.2 htmltools_0.4.0
[10] mgcv_1.8-29 yaml_2.2.0 utf8_1.1.4
[13] rlang_0.4.0 pillar_1.4.2 glue_1.3.1
[16] withr_2.1.2 modelr_0.1.5 readxl_1.3.1
[19] lifecycle_0.1.0 munsell_0.5.0 gtable_0.3.0
[22] workflowr_1.4.0.9001 cellranger_1.1.0 rvest_0.3.4
[25] evaluate_0.14 labeling_0.3 knitr_1.25
[28] fansi_0.4.0 broom_0.5.2 Rcpp_1.0.2
[31] scales_1.0.0 backports_1.1.5 jsonlite_1.6
[34] fs_1.3.1 hms_0.5.1 digest_0.6.21
[37] stringi_1.4.3 grid_3.6.1 rprojroot_1.2
[40] cli_1.1.0 tools_3.6.1 magrittr_1.5
[43] lazyeval_0.2.2 crayon_1.3.4 whisker_0.4
[46] pkgconfig_2.0.3 zeallot_0.1.0 Matrix_1.2-17
[49] ellipsis_0.3.0 xml2_1.2.2 lubridate_1.7.4
[52] assertthat_0.2.1 rmarkdown_1.15 httr_1.4.1
[55] rstudioapi_0.10 R6_2.4.0 nlme_3.1-141
[58] git2r_0.26.1.9000 compiler_3.6.1