Update Meeting: 23-09-2021
Jenny Sjaarda
2021-09-20
Last updated: 2021-09-24
Checks: 7 0
Knit directory: proxyMR/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it's best to always run the code in an empty environment.
The command set.seed(20210602) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version fd45d13. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: _targets/
Ignored: analysis/bgenie_GWAS/
Ignored: analysis/data_setup/
Ignored: analysis/process_Neale.out
Ignored: analysis/traitMR/
Ignored: data/Neale_SGG_directory_12_07_2021.csv
Ignored: data/Neale_SGG_directory_15_07_2021.csv
Ignored: data/processed/
Ignored: data/raw/
Ignored: output/figures/
Ignored: output/tables/traitMR/
Ignored: proxyMR_comparison.RData
Ignored: proxyMR_figure_data.RData
Ignored: proxymr_100_clustermq.out
Ignored: proxymr_101_clustermq.out
Ignored: proxymr_102_clustermq.out
Ignored: proxymr_103_clustermq.out
Ignored: proxymr_104_clustermq.out
Ignored: proxymr_105_clustermq.out
Ignored: proxymr_106_clustermq.out
Ignored: proxymr_107_clustermq.out
Ignored: proxymr_108_clustermq.out
Ignored: proxymr_109_clustermq.out
Ignored: proxymr_10_clustermq.out
Ignored: proxymr_110_clustermq.out
Ignored: proxymr_111_clustermq.out
Ignored: proxymr_112_clustermq.out
Ignored: proxymr_113_clustermq.out
Ignored: proxymr_114_clustermq.out
Ignored: proxymr_115_clustermq.out
Ignored: proxymr_116_clustermq.out
Ignored: proxymr_117_clustermq.out
Ignored: proxymr_118_clustermq.out
Ignored: proxymr_119_clustermq.out
Ignored: proxymr_11_clustermq.out
Ignored: proxymr_120_clustermq.out
Ignored: proxymr_121_clustermq.out
Ignored: proxymr_122_clustermq.out
Ignored: proxymr_123_clustermq.out
Ignored: proxymr_124_clustermq.out
Ignored: proxymr_125_clustermq.out
Ignored: proxymr_126_clustermq.out
Ignored: proxymr_127_clustermq.out
Ignored: proxymr_128_clustermq.out
Ignored: proxymr_129_clustermq.out
Ignored: proxymr_12_clustermq.out
Ignored: proxymr_130_clustermq.out
Ignored: proxymr_131_clustermq.out
Ignored: proxymr_132_clustermq.out
Ignored: proxymr_133_clustermq.out
Ignored: proxymr_134_clustermq.out
Ignored: proxymr_135_clustermq.out
Ignored: proxymr_136_clustermq.out
Ignored: proxymr_137_clustermq.out
Ignored: proxymr_138_clustermq.out
Ignored: proxymr_139_clustermq.out
Ignored: proxymr_13_clustermq.out
Ignored: proxymr_140_clustermq.out
Ignored: proxymr_14_clustermq.out
Ignored: proxymr_15_clustermq.out
Ignored: proxymr_16_clustermq.out
Ignored: proxymr_17_clustermq.out
Ignored: proxymr_18_clustermq.out
Ignored: proxymr_19_clustermq.out
Ignored: proxymr_1_clustermq.out
Ignored: proxymr_20_clustermq.out
Ignored: proxymr_21_clustermq.out
Ignored: proxymr_22_clustermq.out
Ignored: proxymr_23_clustermq.out
Ignored: proxymr_24_clustermq.out
Ignored: proxymr_25_clustermq.out
Ignored: proxymr_26_clustermq.out
Ignored: proxymr_27_clustermq.out
Ignored: proxymr_28_clustermq.out
Ignored: proxymr_29_clustermq.out
Ignored: proxymr_2_clustermq.out
Ignored: proxymr_30_clustermq.out
Ignored: proxymr_31_clustermq.out
Ignored: proxymr_32_clustermq.out
Ignored: proxymr_33_clustermq.out
Ignored: proxymr_34_clustermq.out
Ignored: proxymr_35_clustermq.out
Ignored: proxymr_36_clustermq.out
Ignored: proxymr_37_clustermq.out
Ignored: proxymr_38_clustermq.out
Ignored: proxymr_39_clustermq.out
Ignored: proxymr_3_clustermq.out
Ignored: proxymr_40_clustermq.out
Ignored: proxymr_41_clustermq.out
Ignored: proxymr_42_clustermq.out
Ignored: proxymr_43_clustermq.out
Ignored: proxymr_44_clustermq.out
Ignored: proxymr_45_clustermq.out
Ignored: proxymr_46_clustermq.out
Ignored: proxymr_47_clustermq.out
Ignored: proxymr_48_clustermq.out
Ignored: proxymr_49_clustermq.out
Ignored: proxymr_4_clustermq.out
Ignored: proxymr_50_clustermq.out
Ignored: proxymr_51_clustermq.out
Ignored: proxymr_52_clustermq.out
Ignored: proxymr_53_clustermq.out
Ignored: proxymr_54_clustermq.out
Ignored: proxymr_55_clustermq.out
Ignored: proxymr_56_clustermq.out
Ignored: proxymr_57_clustermq.out
Ignored: proxymr_58_clustermq.out
Ignored: proxymr_59_clustermq.out
Ignored: proxymr_5_clustermq.out
Ignored: proxymr_60_clustermq.out
Ignored: proxymr_61_clustermq.out
Ignored: proxymr_62_clustermq.out
Ignored: proxymr_63_clustermq.out
Ignored: proxymr_64_clustermq.out
Ignored: proxymr_65_clustermq.out
Ignored: proxymr_66_clustermq.out
Ignored: proxymr_67_clustermq.out
Ignored: proxymr_68_clustermq.out
Ignored: proxymr_69_clustermq.out
Ignored: proxymr_6_clustermq.out
Ignored: proxymr_70_clustermq.out
Ignored: proxymr_71_clustermq.out
Ignored: proxymr_72_clustermq.out
Ignored: proxymr_73_clustermq.out
Ignored: proxymr_74_clustermq.out
Ignored: proxymr_75_clustermq.out
Ignored: proxymr_76_clustermq.out
Ignored: proxymr_77_clustermq.out
Ignored: proxymr_78_clustermq.out
Ignored: proxymr_79_clustermq.out
Ignored: proxymr_7_clustermq.out
Ignored: proxymr_80_clustermq.out
Ignored: proxymr_81_clustermq.out
Ignored: proxymr_82_clustermq.out
Ignored: proxymr_83_clustermq.out
Ignored: proxymr_84_clustermq.out
Ignored: proxymr_85_clustermq.out
Ignored: proxymr_86_clustermq.out
Ignored: proxymr_87_clustermq.out
Ignored: proxymr_88_clustermq.out
Ignored: proxymr_89_clustermq.out
Ignored: proxymr_8_clustermq.out
Ignored: proxymr_90_clustermq.out
Ignored: proxymr_91_clustermq.out
Ignored: proxymr_92_clustermq.out
Ignored: proxymr_93_clustermq.out
Ignored: proxymr_94_clustermq.out
Ignored: proxymr_95_clustermq.out
Ignored: proxymr_96_clustermq.out
Ignored: proxymr_97_clustermq.out
Ignored: proxymr_98_clustermq.out
Ignored: proxymr_99_clustermq.out
Ignored: proxymr_9_clustermq.out
Ignored: renv/library/
Ignored: renv/staging/
Untracked files:
Untracked: Rplots.pdf
Unstaged changes:
Modified: analysis/AM_MR_summary.Rmd
Modified: analysis/couple_selection.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/update_meeting_23_09_2021.Rmd) and HTML (docs/update_meeting_23_09_2021.html) files. If you've configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | fd45d13 | Jenny Sjaarda | 2021-09-24 | publish update meeting 24-09-21 |
| Rmd | 71c67df | jennysjaarda | 2021-09-24 | select the right columns for DT |
| Rmd | 529020f | jennysjaarda | 2021-09-24 | add update meeting 23-09-2021 |
| html | 529020f | jennysjaarda | 2021-09-24 | add update meeting 23-09-2021 |
1 Last meeting summary.
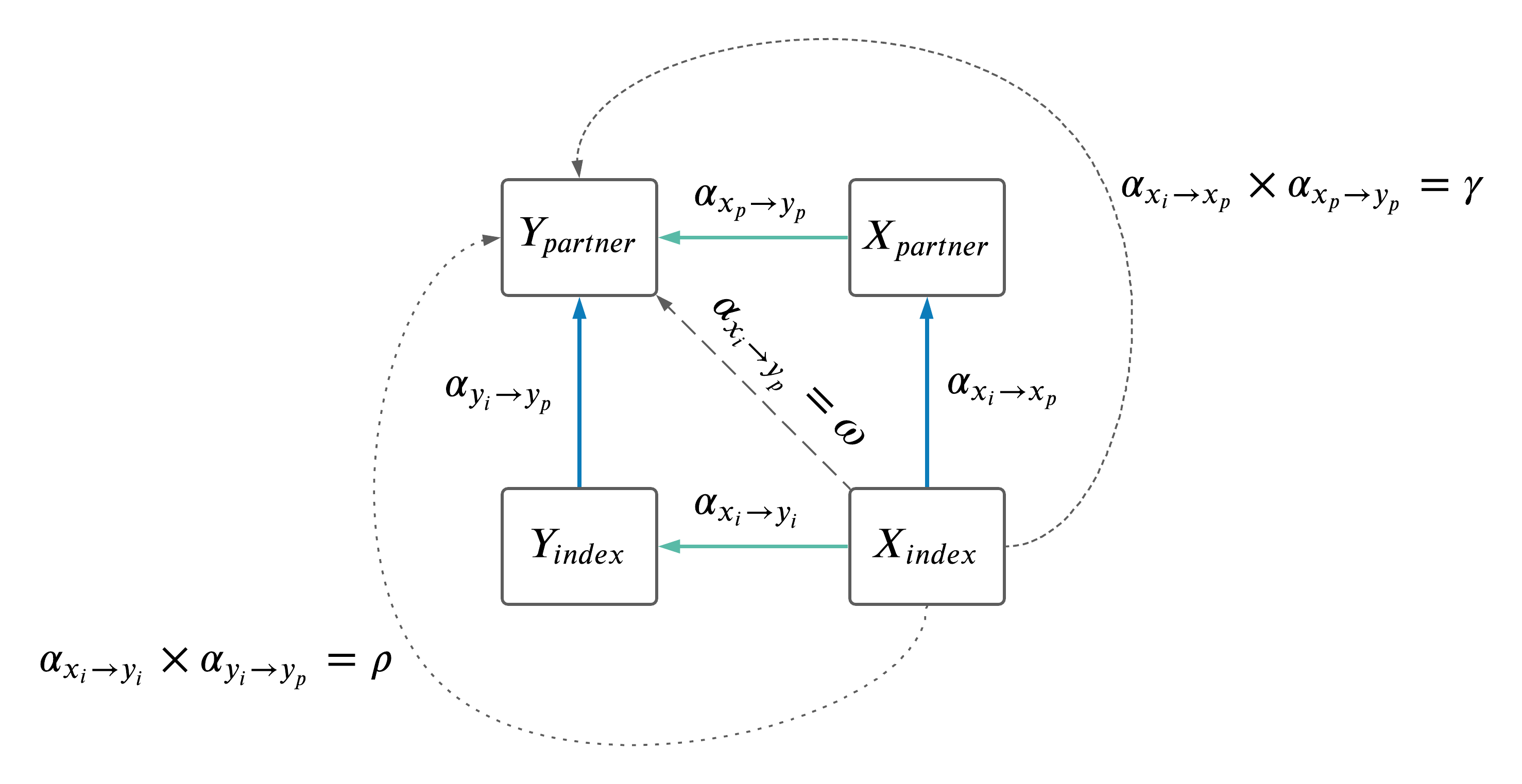
- Because the results comparing \(\omega\) to \(\rho + \gamma\) were slightly confusing (see last meeting summary here), decided we should explore the impact of \(X_i\) on the \(Y_i \rightarrow Y_p\) causal effects.
| Version | Author | Date |
|---|---|---|
| fda4843 | jennysjaarda | 2021-09-14 |
- Two things to explore:
- Identify overlap between \(X_i \rightarrow X_p\) IVs and \(Y_i \rightarrow Y_p\).
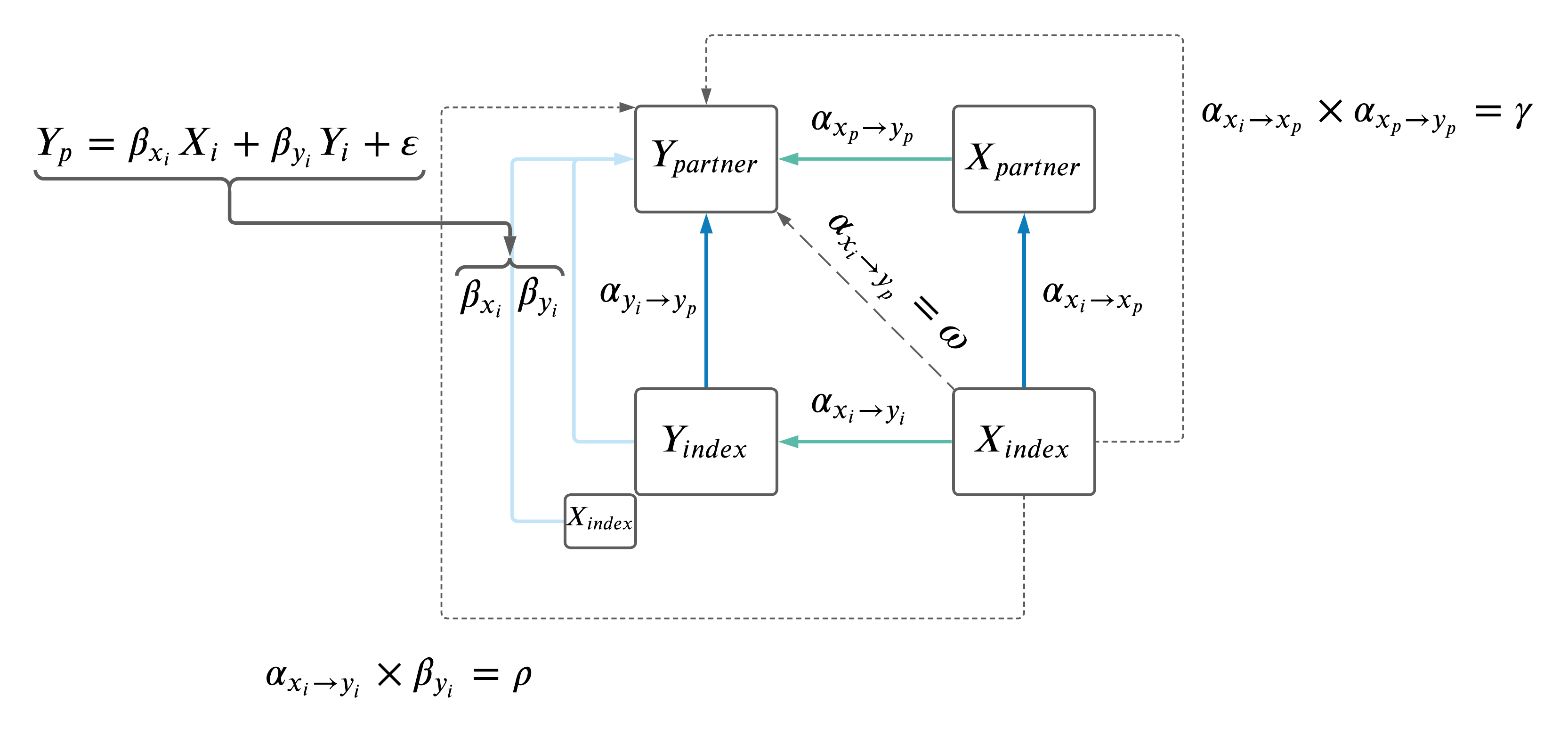
- Adjust the \(Y_i \rightarrow Y_p\) MR results for effects of \(X_i\) (as depicted below).
| Version | Author | Date |
|---|---|---|
| 529020f | jennysjaarda | 2021-09-24 |
2 Results.
2.1 Overlap between IVs for \(X\) and \(Y\).
Is simply calculating the SNP overlap good enough? Better to calculate signal overlap using LD at some \(r^2\) cutoff (what threshold would be useful?).
- For each IV used to estimate the causal effect \(Y_i \rightarrow Y_p\),
- Test if they are in LD with any IV used to estimate the causal effect \(X_i \rightarrow X_p\) at \(r^2\) >
0.9, - Repeat for all IVs for \(Y_i\) to calcualte the percentage overlap between the two sets of IVs.
- Define percentage overlap as: (# of \(Y_i\) IVs are in LD with at least one \(X_i\) IV at \(r^2\) >
0.9) / (total number of \(Y_i\) IVs used).
The results can be seen below. The YiXi_IV_exact_overlap column indicates the percentage of \(Y_i\) IVs that are an exact match with \(X_i\) IVs (same rs#). The YiXi_IV_sig_overlap column denotes the proportion overlap of signals, as defined above.
The results with the most overlap involve traits that are nearly identical.
2.2 Adjust \(Y_i \rightarrow Y_p\).
Performed MV MR \(Y_i \rightarrow Y_p\) relationships: \(Y_p \sim Y_i + X_i\). Where IVs were the same as the two trait MR (\(Y_p \sim Y_i\)) based on \(Y\), but the effect of the IVs on \(X_i\) were also included in the model.
We can compare the \(\alpha_{y_{i}\rightarrow y_{p}}\) before and after adjustment, and clearly see a band of estimates centered around \(y = 0\), which matches nearly perfectly the instances where \(Y\) IVs share more than 50% of \(X\) IVs. There are several cases where the adjusted results caused the \(abs(\alpha_{y_{i}\rightarrow y_{p}})\) to increase.
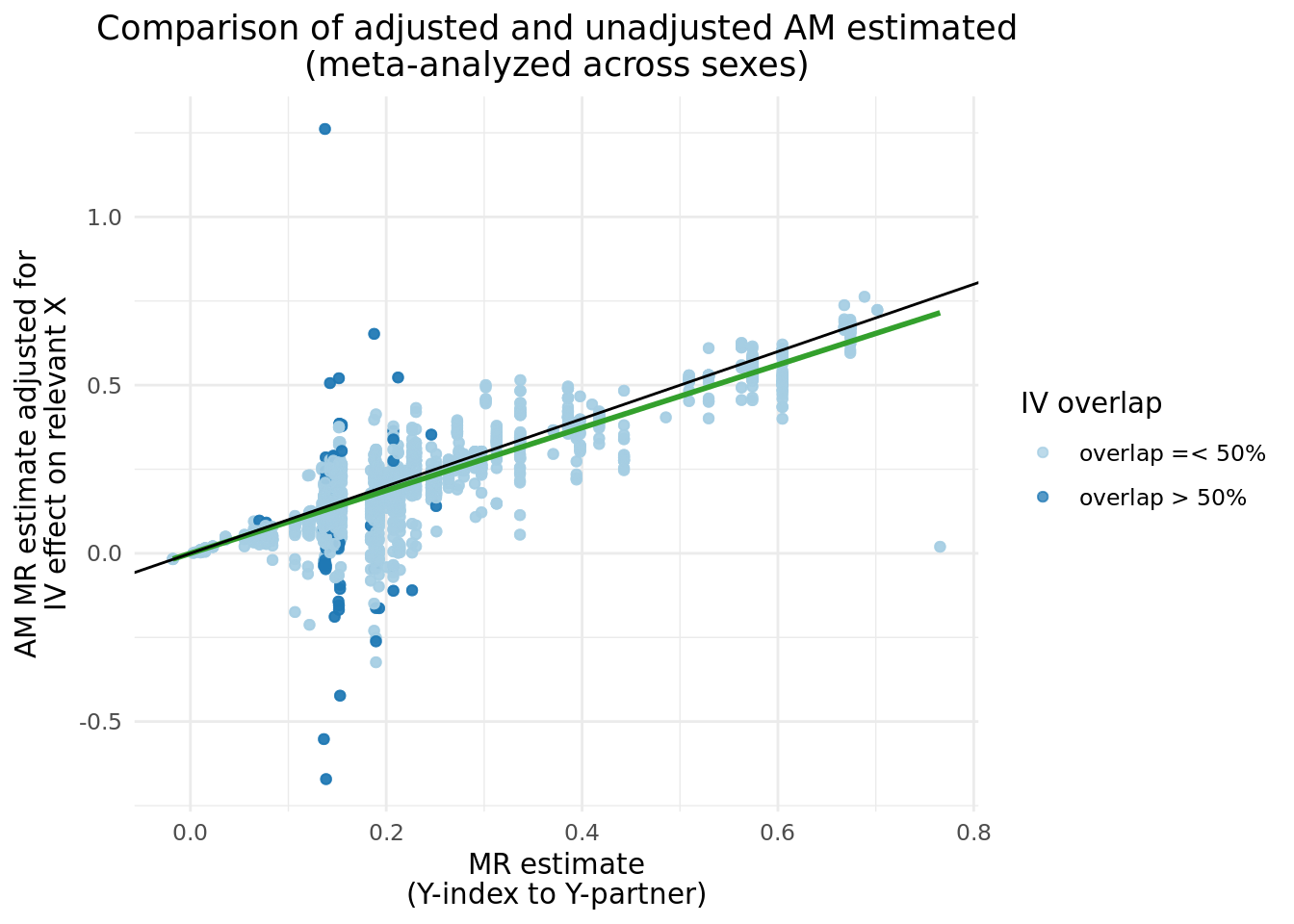
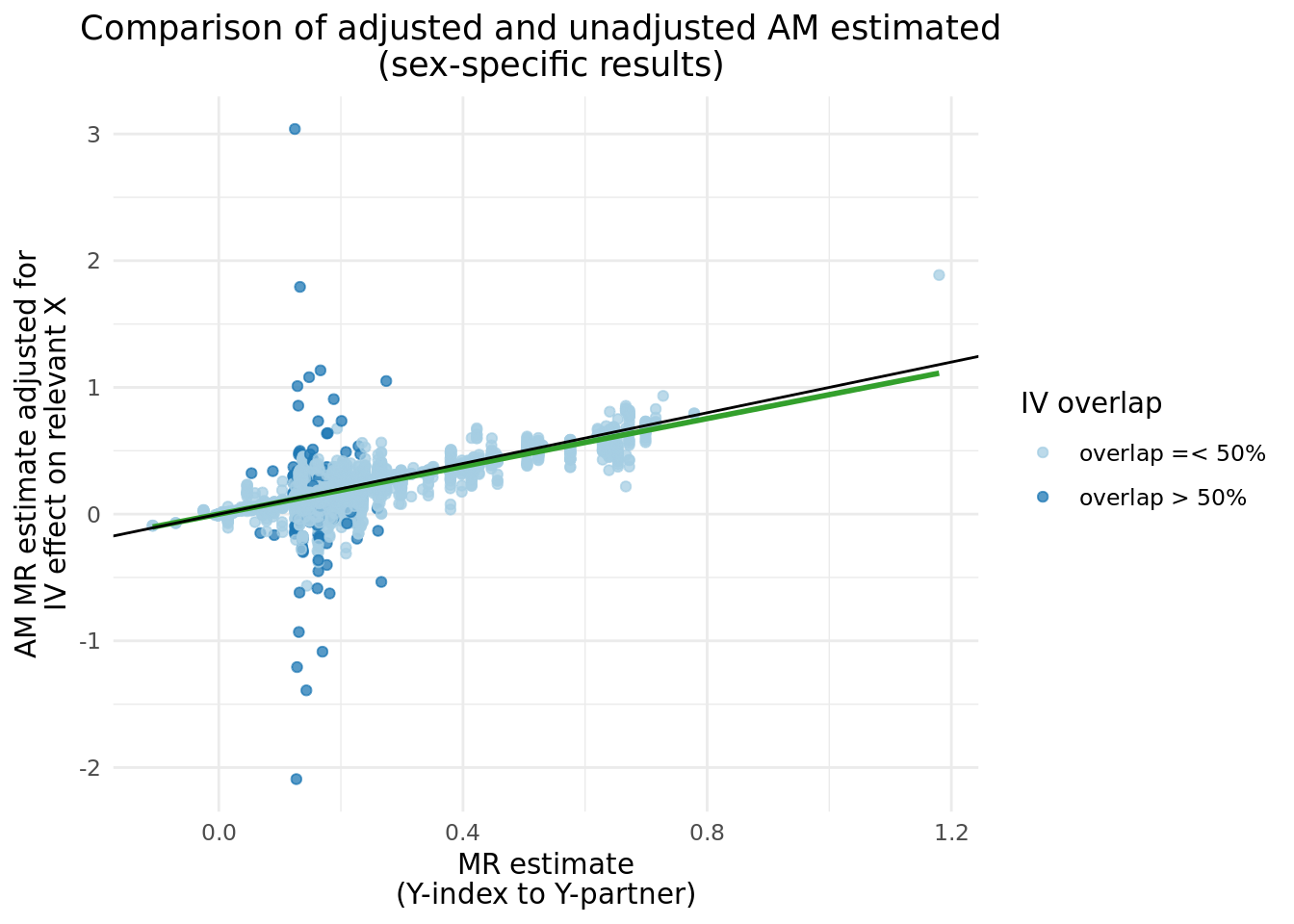
2.3 Updated two trait results with adjusted \(Y_i\).
The results below show the significant difference between \(\omega\) vs \(\rho\), \(\gamma\) and their sum. Concerned that most of the significant results involve cases where the adjusted \(abs(\alpha_{y_{i}\rightarrow y_{p}})\) is greater than the unadjusted case.
| omega_vs_gam_BF_sig_meta | Frq |
|---|---|
| FALSE | 1459 |
| TRUE | 438 |
| omega_vs_rho_BF_sig_meta | Frq |
|---|---|
| FALSE | 1366 |
| TRUE | 531 |
| omega_vs_gam_rho_BF_sig_meta | Frq |
|---|---|
| FALSE | 1477 |
| TRUE | 420 |
2.4 MVMR.
Have so far run / am running all MVMR models but need to sum up along all \(\beta\) from the MV model to create the \(\gamma_{mv}\). (Having some computing issues with pruning.)
Should we use the adjusted \(Y_i \rightarrow Y_p\) for calculating \(\rho\)?
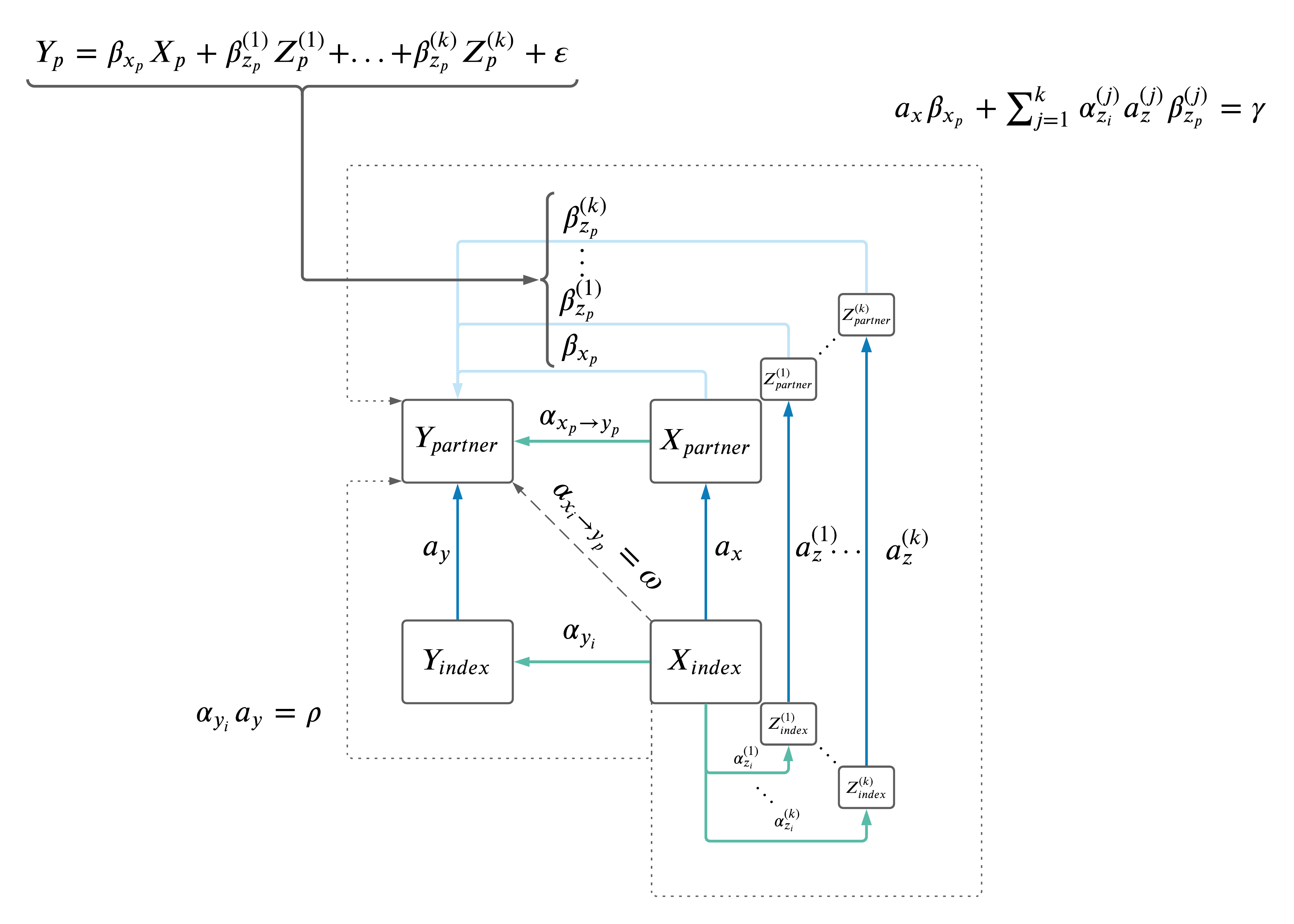
MV MR: \(X \rightarrow Z \rightarrow Y\).
- Drop Z's if not significant in MV MR (i.e. \(Y \sim X + Z_1 + Z_2 + Z_3...\))
- Do we drop from the model itself and recompute the beta-coefficients? Or simply not include them in the result sum?
| Version | Author | Date |
|---|---|---|
| 1afb033 | jennysjaarda | 2021-09-14 |
sessionInfo()R version 4.1.0 (2021-05-18)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS: /data/sgg2/jenny/bin/R-4.1.0/lib64/R/lib/libRblas.so
LAPACK: /data/sgg2/jenny/bin/R-4.1.0/lib64/R/lib/libRlapack.so
locale:
[1] en_CA.UTF-8
attached base packages:
[1] stats graphics grDevices datasets utils methods base
other attached packages:
[1] kableExtra_1.3.4 knitr_1.33 DT_0.18.1 forcats_0.5.1
[5] stringr_1.4.0 dplyr_1.0.7 purrr_0.3.4 readr_1.4.0
[9] tidyr_1.1.3 tibble_3.1.2 ggplot2_3.3.4 tidyverse_1.3.1
[13] targets_0.5.0.9001 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] httr_1.4.2 splines_4.1.0 jsonlite_1.7.2 viridisLite_0.4.0
[5] modelr_0.1.8 assertthat_0.2.1 highr_0.9 renv_0.13.2-62
[9] cellranger_1.1.0 yaml_2.2.1 lattice_0.20-44 pillar_1.6.1
[13] backports_1.2.1 glue_1.4.2 digest_0.6.27 promises_1.2.0.1
[17] rvest_1.0.0 colorspace_2.0-1 Matrix_1.3-3 htmltools_0.5.1.1
[21] httpuv_1.6.1 pkgconfig_2.0.3 broom_0.7.7 haven_2.4.1
[25] scales_1.1.1 webshot_0.5.2 processx_3.5.2 svglite_2.0.0
[29] whisker_0.4 later_1.2.0 git2r_0.28.0 mgcv_1.8-35
[33] farver_2.1.0 generics_0.1.0 ellipsis_0.3.2 withr_2.4.2
[37] cli_2.5.0 magrittr_2.0.1 crayon_1.4.1 readxl_1.3.1
[41] evaluate_0.14 ps_1.6.0 fs_1.5.0 fansi_0.5.0
[45] nlme_3.1-152 xml2_1.3.2 tools_4.1.0 data.table_1.14.0
[49] hms_1.1.0 lifecycle_1.0.0 munsell_0.5.0 reprex_2.0.0
[53] callr_3.7.0 compiler_4.1.0 systemfonts_1.0.2 rlang_0.4.11
[57] grid_4.1.0 rstudioapi_0.13 htmlwidgets_1.5.3 crosstalk_1.1.1
[61] igraph_1.2.6 labeling_0.4.2 rmarkdown_2.9 gtable_0.3.0
[65] codetools_0.2-18 DBI_1.1.1 R6_2.5.0 lubridate_1.7.10
[69] utf8_1.2.1 rprojroot_2.0.2 stringi_1.6.2 Rcpp_1.0.6
[73] vctrs_0.3.8 dbplyr_2.1.1 tidyselect_1.1.1 xfun_0.24